
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 25 February 2025
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1549044
 Ayman Elbehiry1*
Ayman Elbehiry1* Eman Marzouk1
Eman Marzouk1 Adil Abalkhail1
Adil Abalkhail1 Moustafa H. Abdelsalam2
Moustafa H. Abdelsalam2 Mohamed E. A. Mostafa3
Mohamed E. A. Mostafa3 Mazen Alasiri4
Mazen Alasiri4 Mai Ibrahem5
Mai Ibrahem5 Abousree T. Ellethy6
Abousree T. Ellethy6 Abdulaziz Almuzaini7
Abdulaziz Almuzaini7 Sahar N. Aljarallah8
Sahar N. Aljarallah8 Akram Abu-Okail9
Akram Abu-Okail9 Naif Marzook10
Naif Marzook10 Satam Alhadyan11
Satam Alhadyan11 Husam M. Edrees2
Husam M. Edrees2Antimicrobial resistance (AMR) is recognized as one of the foremost global health challenges, complicating the treatment of infectious diseases and contributing to increased morbidity and mortality rates. Traditionally, microbiological culture and susceptibility testing methods, such as disk diffusion and minimum inhibitory concentration (MIC) assays, have been employed to identify AMR bacteria. However, these conventional techniques are often labor intensive and time consuming and lack the requisite sensitivity for the early detection of resistance. Recent advancements in molecular and genomic technologies—such as next-generation sequencing (NGS), matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), lateral flow immunoassays (LFIAs), PCR-based diagnostic methods, and CRISPR-based diagnostics—have revolutionized the diagnosis of AMR. These innovative approaches provide increased sensitivity, reduced turnaround times, and the ability to identify genetic resistance mechanisms. This review seeks to examine the advantages and disadvantages of both emerging technologies and traditional methods for detecting AMR, emphasizing the potential benefits and limitations inherent to each. By understanding the strengths and limitations of these technologies, stakeholders, including researchers, healthcare professionals, regulatory agencies, health authorities, financial managers, and patients, can make informed decisions aimed at preventing the emergence and dissemination of antibiotic-resistant strains, thereby ultimately increasing patient safety.
Since penicillin was discovered, antimicrobial drugs have greatly advanced medicine and community health (Breijyeh and Karaman, 2023; Gould, 2016; Shanmugakani et al., 2020; Zaffiri et al., 2012). They have saved millions of lives and controlled communicable diseases, which were leading causes of mortality before their introduction (Aminov, 2010; Aminov, 2017). The development of antimicrobials has been a key advancement in healthcare (Brown and Wright, 2016; Yamin et al., 2023). However, new antibiotic-resistant bacteria frequently emerge shortly after their introduction (Hu et al., 2020; Lobanovska and Pilla, 2017). Penicillin, recognized as the first beta-lactam antibiotic, fundamentally transformed the management of infectious diseases and facilitated the development of subsequent antibiotic classes, including sulfonamides and aminoglycosides, such as streptomycin and streptothricin. Since its initial discovery, antimicrobial research has progressed significantly, leading to the emergence of novel classes of antimicrobial agents (Ventola, 2015). Currently, a diverse array of antibiotics are commonly used, including modified beta-lactams, cephalosporins, tetracyclines, fluoroquinolones, and aminoglycosides. Nevertheless, the increasing incidence of antibiotic resistance has compelled researchers to investigate natural compounds as potential alternatives to address the therapeutic challenges that have arisen since the mid-20th century (Hunt and Kates, 2024). Although traditional antimicrobials are common in agriculture, new antimicrobial drugs are scarce (Miller et al., 2022). Misadministration of antimicrobials in humans and animals (Caneschi et al., 2023) contributes to an increase in the prevalence of antibiotic-resistant bacteria (Golding et al., 2019; Shanmugakani et al., 2020; Singer et al., 2016; Tangcharoensathien et al., 2018). Factors such as healthcare and environmental influences facilitate this spread (Ahmad et al., 2021; Bengtsson-Palme et al., 2018; Serweciñska, 2020), posing a significant threat to public health and the economy in both developed and developing countries (Naylor et al., 2018; Weldon and Hoffman, 2023).
Antimicrobial resistance is a major global health threat in the 21st century, requiring urgent action as per World Health Organization (WHO) recommendations (Vasala et al., 2020). This resistance has made common diseases, such as respiratory and cardiovascular illnesses, more prevalent and difficult to treat (Bassetti et al., 2017). In 2019, it was responsible for approximately 1.27 million deaths and contributed to an additional 4.95 million fatalities worldwide (Murray et al., 2022). Antibiotic-resistant microbes cause more than 33,000 deaths annually in Europe (Antoñanzas and Goossens, 2019; Peñalva-Moreno et al., 2022) and affect more than three million people in the Unites States., resulting in more than 35,000 deaths each year (Church and McKillip, 2021). In India, more than 50,000 infants die from sepsis due to resistant bacteria, with an infant dying every 9 min from such infections (Subramaniam and Girish, 2020).
In 2019, the number of deaths from methicillin-resistant Staphylococcus aureus (MRSA) surpassed 100,000 (Alghamdi et al., 2023). The WHO’s Global Antimicrobial Surveillance reported over 500,000 confirmed infections from bacteria with extensive AMR. ESKAPE pathogens—Enterococcus faecium, Staphylococcus aureus (S. aureus), Klebsiella pneumoniae (K. pneumoniae), Acinetobacter baumannii (A. baumannii), Pseudomonas aeruginosa (P. aeruginosa), and Enterobacter spp.—are the most commonly isolated resistant organisms in hospital environments (Miller and Arias, 2024). Healthcare-associated infections (HAIs) constitute a major health issue, affecting 5% to 10% of hospitalized patients and costing the United States healthcare system approximately USD 4 billion annually. A 2011 survey revealed that 4% of hospitalized patients had an HAI (Magill et al., 2018). AMR contributes to the increase in HAIs, complicating cost management (Salam et al., 2023b). Patients with drug-resistant pathogens face longer hospital stays and higher costs (Fongang et al., 2023). Strict infection prevention protocols, judicious antibiotic use, and monitoring for resistant bacteria are crucial for reducing costs and improving patient safety.
Antibiotic stewardship programs (ASPs) are essential for reducing global antibiotic resistance (Majumder et al., 2020). They promote appropriate antimicrobial use by identifying the most effective agents, durations, doses, and administration methods, thus minimizing side effects and costs (Ababneh et al., 2021). Successful ASPs require leadership, hospital management support, skilled infectious disease clinicians, ongoing education, and interdisciplinary collaboration (Baraka et al., 2019). The WHO is working with various organizations to develop strategies that raise awareness of AMR (World Health Organization, 2014). These strategies aim to reduce transmissible diseases through precautionary measures, improved antibiotic therapy, novel treatments, and enhanced antimicrobial drug efficacy (World Health Organization, 2014). Despite significant advancements in rapidly detecting antibiotic-resistant bacteria (Yamin et al., 2023), the effectiveness of antibiotics is declining (Chinemerem Nwobodo et al., 2022). Microbial infections are often hard to identify, causing treatment delays (Cansizoglu et al., 2019; Giordano et al., 2018). As a result, doctors may need to start antimicrobial treatment before a full evaluation, which can worsen the patient’s condition and contribute to AMR (van Belkum et al., 2019). There is an urgent need for rapid, accurate, and affordable tests to detect AMR (Salam et al., 2023b), enabling better-targeted medications and reducing the time for antimicrobial susceptibility testing (AST) (Cansizoglu et al., 2019). Screening is crucial in managing AMR (Khan et al., 2019).
Microbiological investigations often use phenotypic methods to assess infection sensitivity to antibiotics (Maugeri et al., 2019; van Belkum et al., 2020). Although these techniques are cost-effective and standardized, they can be labor intensive, causing treatment delays (Shin et al., 2019; Syal et al., 2017) that negatively impact patient management (Khan et al., 2019). Delayed antibiotic treatment is associated with higher mortality rates (Pak et al., 2023) and longer hospital stays (Nauclér et al., 2021). Comprehensive surveillance systems are vital for reducing AMR-related mortality and morbidity, guiding treatment decisions, and developing new antibacterial medications (Cantón et al., 2023). The Study for Monitoring AMR Trends (SMART) includes nearly 500,000 isolates from over 200 sites in more than 60 countries, addressing evolving medical needs (Cantón et al., 2023). SMART data have identified emerging resistance risks and informed clinical guidelines, making the database accessible to clinicians and researchers worldwide, particularly in resource-limited countries. The development of rapid tests for antibiotic susceptibility aims to improve patient treatment and manage AMR (Datar et al., 2022; Gulumbe et al., 2022; Zakhour et al., 2023) by quickly identifying pathogenic microorganisms and their susceptibility to antibiotics.
This review examines both traditional and contemporary methodologies for the detection of bacteria that have acquired resistance to antimicrobials. The objective of this review was to improve our understanding of the effectiveness of AST in assessing the susceptibility of bacteria to various antibiotics. We provide a thorough analysis of the advantages and limitations associated with current AST methods while also highlighting significant advancements in point-of-care testing. Additionally, we present forecasts regarding the most promising technologies expected to emerge in the future for ASTs. Figure 1 provides a summary of the conventional and modern technologies discussed in this review. To ensure the relevance of the included research, only articles published in English between 2002 and 2024 were considered.
Organizations such as the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on AST recommend phenotypic screening to identify antibiotic-resistant bacteria (Gajic et al., 2022; Maugeri et al., 2019). Traditional AST methods, including agar dilution, broth microdilution, and disk diffusion, involve exposing isolated bacteria to antimicrobial agents and observing their growth (Puttaswamy et al., 2018; Salam et al., 2023a). A key method for assessing resistance is determining the minimum inhibitory concentration (MIC) (Kowalska-Krochmal and Dudek-Wicher, 2021), which determines the lowest antibiotic concentration that inhibits bacterial growth via agar or broth. This technique mixes a standardized number of bacteria (typically 0.5 according to the McFarland standard) with antibiotic-diluted broth or agar (Wiegand et al., 2008). In brief, the MIC assay was conducted by standardizing a bacterial culture to 5 × 105 colony-forming units (CFU)/ml and exposing it to various antibiotic concentrations for 16–24 h at 37°C (Lewis and James, 2022). After the incubation period, bacterial growth was evaluated for each antibiotic concentration, and the MIC value was identified as the lowest concentration of the antimicrobial agent needed to inhibit visible growth of the bacterial strain being studied.
After incubation, bacteria may or may not grow, and the procedure is simple and inexpensive, requiring no specialized instruments. The first step in therapeutic treatment often involves measuring the MICs of antimicrobial agents against bacteria (Gajic et al., 2022). However, this technique has limitations in assessing resistance in non-cultivable yet viable microbial species, and its effectiveness depends on factors such as incubation duration, antimicrobial concentration, and initial microbial inoculum (Wiegand et al., 2008). The disk diffusion method involves placing an antimicrobial-infused disk on solid agar, creating a circular zone of inhibition that indicates bacterial growth suppression. This qualitative analysis classifies bacteria as susceptible, intermediate, or resistant. While effective for rapidly multiplying bacteria, the method has drawbacks, including low agar diffusion of some antimicrobial agents and difficulties in evaluating anaerobic and fastidious bacteria (Sejas et al., 2003). Clinical laboratories use a variety of antimicrobial susceptibility testing (AST) methods, depending on the specific equipment and range of laboratory tests they provide (Gajic et al., 2022).
The E-Test, the original gradient strip, a proprietary tool from bioMérieux, is widely used in clinical laboratories to guide antibiotic selection by indicating effective antimicrobial concentrations (Salam et al., 2023a). It combines elements of previous methods to generate MIC data via disk diffusion (Erfani et al., 2011). The E-test® features strips with an exponential gradient of antibiotic concentrations and a numeric scale. While it shares time-related limitations with earlier tests, it simplifies microbial susceptibility quantification (Yamin et al., 2023) and is particularly effective for hard-to-cultivate microorganisms such as Mycobacterium (obligate aerobe) and Haemophilus influenzae (facultatively anaerobic) (Hedberg, 2005). The rapid direct E test is an effective tool for obtaining preliminary AST data within a period of 5–6 h. This method is especially beneficial when it is employed alongside phenotypic or genotypic analyses to clarify essential mechanisms of resistance (Bianco et al., 2019b). The gradient strips are used for precise quantification of resistant strains in laboratory and clinical settings because of their stable concentration gradient (Khan et al., 2019). Di Bonaventura et al. (1998) reported that E-test MICs correlated well with agar dilution and disk diffusion methods for 248 P. aeruginosa isolates from bladder-catheterized patients, confirming their reliability. Using the EUCAST Rapid Antimicrobial Susceptibility Testing (RAST) technique, Bianco et al. (2022b) examined 676 positive blood cultures derived from gram-negative rods and gram-positive cocci. RAST was performed within 2 h of leaving the incubator, in accordance with EUCAST recommendations. Inhibition zones were evaluated at 4, 6, 8, and 16–20 h, and the results were interpreted according to EUCAST breakpoints (version 5.1). The results after 16–20 h suggest the potential for more effective antibacterial de-escalation therapy. Despite these limitations, traditional methods for detecting AMR are crucial for identifying treatment options for antibiotic resistance. Despite their drawbacks, traditional tests are inexpensive, easy to perform (requiring no special expertise), and still widely used in hospitals around the world (Kaprou et al., 2021; Yalew, 2020).
The sensitivity of optical equipment for ASTs can be enhanced with optoelectronic systems, fiber optics, microfluidics, and pH- or redox-sensitive indicator dyes (She and Bender, 2019; Trovato et al., 2022). FDA-approved programmable AST devices, such as the Sensititre™ ARIS™ 2X, Vitek 2 Compact System, Phoenix™ panel systems, and MicroScan WalkAway plus systems, significantly impact laboratory settings (Bhatnagar et al., 2023; Elbehiry et al., 2023; Fader et al., 2013; Leonard et al., 2018). These devices feature antibacterial panels for gram-positive and gram-negative infections and use redox indicators to identify bacteria and measure turbidity from multiwell plates. AliFax S.r.l. in Italy utilizes the ALFRED 60/AST system, which employs laser-light scattering technology to detect bacterial growth in liquid culture, providing susceptibility data within four to 6 h from positive blood cultures (López-Hernández et al., 2022). Broth dilution is used with AST cards containing varying antimicrobial concentrations and positive controls, allowing for analysis of MIC patterns in large microorganism clusters (Puttaswamy et al., 2018; Vasala et al., 2020).
The Microscan WalkAway system measures experimental progress via a photometer or fluorometer and can continuously monitor incubated samples (Shanmugakani et al., 2020). MicroScan panels use conventional 96-well microdilution plates for bacterial identification, typically within four hours, although slow-growing bacteria may take 6–42 h (Reller et al., 2009). Susceptibility results are available in 20 h, varying from 16.8 to 27.8 h on the basis of the bacteria type (Winstanley and Courvalin, 2011). In brief, a bacterial suspension corresponding to the 0.5 McFarland standard was prepared in saline via the direct colony suspension method. Subsequently, 5 μl of the diluted suspension was dispensed into each well. Before inoculation, the panels were checked for expiration dates, batch numbers, and packaging integrity. The inoculation was carried out via Prompt Inoculation System-D in conjunction with the RENOK system and Inoculators-D. The panels were then incubated at 35 ± 1°C for 16 h, after which readings were taken. The MIC was determined on the basis of standardized readings provided by the manufacturer. Successful antimicrobial susceptibility values were assessed and evaluated according to the European Committee on Antimicrobial Susceptibility Testing.
A pilot study in a tertiary care teaching hospital located in Rishikesh, Uttarakhand (Singh et al., 2019), in May and June 2019 used the MicroScan WalkAway 96 Plus ID/AST system and Mikrolatest MIC kit to assess colistin susceptibility in carbapenem-resistant gram-negative bacteria. The susceptibility rates were 71.4% for A. baumannii, 85.7% for P. aeruginosa, and 100% for Acinetobacter junii, Acinetobacter johnsonii, Escherichia coli (E. coli), and K. pneumoniae. Hernández-Durán et al. (2017) assessed bacterial susceptibility to antibiotics in hospitalized patients via the VITEK 2® Compact and MicroScan WalkAway® SI systems. The study included 20 gram-positive cocci, 34 g-negative rods, and 13 reference strains. Both techniques showed 90.2% concordance for gram-negative bacteria and 96.3% for gram-positive bacteria, identifying 89.5% of strains by species. VITEK 2 had a median result time of 6.5 h, whereas MicroScan took 12.5 h, indicating a significant delay.
The BD Phoenix Automated Microbiology System is a reliable tool for identifying clinical isolates in healthcare laboratories (Hirakata et al., 2005). It accurately tests most clinically relevant bacteria (Donay et al., 2004; Funke and Funke-Kissling, 2004) and detects ESBLs (Sanguinetti et al., 2003). The system analyzes 99 test panels, each with 84 wells of antibacterial agent dilutions (Yamin et al., 2023). Growth is monitored with a turbidimeter and calorimeter, enabling MIC estimation for various pathogens, including gram-negative and gram-positive bacteria, within 6–16 h (Shanmugakani et al., 2020). The Vitek system, developed in the late 1970s, automates AST and identification. After standardizing the main inoculum, all the necessary steps are performed, and the results are analyzed every 15 min for kinetic analysis (Funke et al., 1998; Ligozzi et al., 2002). The system records colorimetric, turbidity, and fluorescence signals via multichannel fluorimeters and photometers. It employs 64-well reagent cards containing diagnostic media and antimicrobial agents (Abalkhail and Elbehiry, 2022; Elbehiry et al., 2022b), allowing for 30–240 simultaneous assays for gram-negative and gram-positive bacteria within 4–10 h (Shanmugakani et al., 2020). The protocol for the Vitek system for the detection of AST is as follows: 2–3 fresh colonies of the tested bacteria were transferred to a 5 mL tube of sterile physiological sodium chloride solution. The 0.5 McFarland bacterial mixture was then diluted to 1.5 × 10^7 CFU/mL in 0.45% saline (0.50–0.63) via the DensiChek device (BioMérieux, Marcy l’Etoile, France). After filling, sealing, and inserting the AST cards into the VITEK 2 device (VITEK® 2, BioMerieux, France), they were incubated and read. The results were analyzed in accordance with the guidelines established by the CLSI.
At the Prince of Wales Hospital in Hong Kong, China, Ling et al. (2003) conducted a study on bacterial identification and susceptibility to various antibiotics from July 21 to December 15, 2002. Using the VITEK 2 system, they performed direct identification and susceptibility testing on an aerobic bottle from a positive blood culture set. The results of the susceptibility testing were available 3.3 h postincubation, while identification reports were also provided 3.3 h after incubation. A unique method for assessing antibiotic resistance involves detecting volatile organic molecules in the headspace of bacterial cultures. Therefore, Bianco et al. (2024) evaluated the VITEK® REVEAL system on 128 positive blood cultures, including 95 Enterobacterales, 21 P. aeruginosa, and 12 A. baumannii complex samples. The study compared 22 antimicrobials with reference techniques across 2,220 strain-antibiotic combinations, revealing 1,091 resistant pairings (48.7%). The categorical agreement and essential agreement rates were 97.6 and 97.7%, respectively. In the ESBL phenotype screening test, positive, indeterminate, and negative results were found for 13.7, 32.6, and 27.4% of the Enterobacterales isolates, respectively, with 100% concordance with the reference technique. This system effectively assesses antimicrobial susceptibility in major gram-negative species from positive blood cultures, delivering results in under 8 h. Charnot-Katsikas et al. (2018) evaluated the Accelerate Pheno system at the University of Chicago Medicine for identifying bacterial and yeast species and conducting AST. When the blood culture results were compared with those of routine care within 0–8 h of growth detection, the system showed 95.6% sensitivity and 99.5% specificity for identification, with 95.1% agreement in essential susceptibility tests and 95.5% agreement in categorical susceptibility tests. The accelerated Pheno system reduces the susceptibility testing time by an average of 41.86 h, enabling quicker delivery of clinically relevant information.
Traditional phenotyping techniques are slow because of the multiple stages of plating and cultivation, making them unsuitable for rapid results (Elbehiry et al., 2017; Elbehiry et al., 2022a). Researchers have developed genetic methods to detect AMR to address this issue (Anjum et al., 2018; Sahoo et al., 2023). The term “molecular diagnostics” was introduced by Pauling et al. (1949). Genetic methods, such as multiplex targeting, offer advantages over phenotypic testing in identifying antibiotic resistance genes (Yamin et al., 2023). These methods are useful when sensitivity breakpoints are unknown and can use non-purified samples, allowing for faster responses to new resistance components (Vasala et al., 2020). Genetic approaches have limitations, including low sensitivity and narrow scope, and often fail to overlook many AMR genes (Gajic et al., 2022). Additionally, molecular diagnostics can be costly (Singh et al., 2021). However, advancements in techniques such as nucleic acid amplification and hybridization are improving detection (Bai et al., 2023). Overall, molecular methods are effective and sensitive for identifying AMR genes (Kaprou et al., 2021; Rentschler et al., 2021).
Genetic evaluations, such as virulence genotyping and multilocus sequence typing, are more suitable for outbreak investigations than are ASTs (Vasala et al., 2020). These methods require large amounts of pure nucleic acids, making them impractical for rapid diagnosis. In contrast, molecular beacon systems and hybridization-based strategies are effectively integrated into NAATs (Yamin et al., 2023). The CDC states that commercial NAATs are used mainly in hospitals, health agencies, and private labs (Ling et al., 2008). Many low-income countries still rely on in-house PCR tests, which cost $15 each (Roos et al., 1998). These nations often have the highest case numbers, limiting their access to expensive technologies. In the last two decades, NAATs have become essential for detecting microorganisms, greatly assisting in diagnostic testing and research on infectious diseases (Apfalter et al., 2005). Global migration, climate change, urbanization, and the ongoing pandemic emphasize the significance of NAATs. Although NAATs are known for their high accuracy and sensitivity, their centralized structure and dependence on qualified personnel can impede effective pathogen management during emergencies (Narasimhan et al., 2023).
Nucleic acid amplification tests are highly effective for identifying microbial species, especially when combined with a strong molecular test (Patel, 2022). Recent diagnostic panels developed by companies such as BioMérieux, Qiagen, and Becton Dickinson can identify genes related to antibiotic resistance (Trotter et al., 2019; Vasala et al., 2020) and provide clinically relevant data even when antimicrobial treatment is not needed (Vasala et al., 2020). Among drug-resistant pathogens, only a few, such as Neisseria gonorrhea, Chlamydia trachomatis, and Mycoplasma, have shown a certain degree of resistance (Unemo and Jensen, 2017). Identifying genes linked to antibiotic-resistant bacteria is promising but does not confirm resistance (Salyers and Amabile-Cuevas, 1997). A key limitation of NAATs is their inability to specify MICs or recommend antibiotics (Yu et al., 2023). Nonetheless, NAATs can be easily adapted to address resistance factors and emerging infections (Bang et al., 2023). Techniques such as isothermal amplification, polymerase chain reaction (PCR), multiplex PCR, reverse transcriptase PCR (RT-PCR), and quantitative PCR (qPCR) can be used to identify antibiotic resistance genes.
The isothermal DNA amplification technique is a breakthrough in molecular biology that eliminates the need for thermal cycling in conventional PCR (Lee et al., 2019; Zhu et al., 2020). Techniques such as transcription-mediated amplification (TMA) and loop-mediated isothermal amplification (LAMP) have been developed (Glökler et al., 2021; Srivastava and Prasad, 2023), leading to next-generation genetic diagnostics that better serve patients (Lee et al., 2019). Isothermal methods bypass thermocycling, reducing analysis times and energy consumption (Oliveira et al., 2021), and alternative heat control methods have rendered thermocyclers obsolete (Zou et al., 2020). Additionally, isothermal amplification offers higher rates of amplification and specificity than PCR does (Zanoli and Spoto, 2012). A recent study investigated the efficacy of the Amplex eazyplex® LAMP assay for rapidly identifying A. baumannii and its predominant acquired carbapenemases directly from blood culture bottles in less than 30 min (Comini et al., 2021). The findings of this study indicate that the Amplex eazyplex® LAMP assay is proficient at detecting A. baumannii and carbapenemases associated with carbapenem resistance. In numerous international airports, hospitals, and testing facilities globally, LAMP has emerged as a viable alternative to qPCR (Subsoontorn et al., 2020). However, these methods face challenges, including less effective multiplexing due to experimental complexity (Dhama et al., 2014) and the need for more primers in complex reactions.
Polymerase chain reaction is an in vitro technique used to amplify DNA and RNA sequences exponentially (Bej et al., 1991). False positives may arise from infections or cross-reactivity with similar microorganisms (Hahn et al., 2020). Therefore, quality control measures for PCR assays, particularly in diagnostics, start with established laboratory practices that reduce contamination and increase reproducibility (Kitchin and Bootman, 1993). A significant advantage of PCR over traditional culture methods is its ability to amplify genes from uncultivable or dying organisms (Adzitey et al., 2013). Overall, PCR is effective for tracking AMR infections, identifying known resistance genes, such as mecA (methicillin resistance) in S. aureus (Abalkhail and Elbehiry, 2022), blaSHV (beta-lactam resistance) in Enterobacteriaceae (Zorbozan and Kimiran, 2022), tetM (tetracycline resistance) (Domínguez-Pérez et al., 2018), and vanA (vancomycin resistance) in Enterococcus (Wagner et al., 2023; Wardal et al., 2023). An earlier investigation assessed the effectiveness of ELITe MGB assays in detecting the main carbapenemase and ESBL genes and mec genes associated with S. aureus in blood cultures within a timeframe of less than three hours (Bianco et al., 2019a). The findings of the study indicated a concordance between genotypic and conventional phenotypic outcomes. Boattini et al. (2021) conducted a study on molecular tests to enhance antibiotic treatment for critically ill patients with carbapenemase- and/or CTX-M-producing pneumonia. They examined 197 bronchoalveolar lavage (BAL) samples for carbapenem-resistant Enterobacteriaceae (CRE) and ESBL using ELITe MGB® assays and compared them to standard culture methods. Twenty (10.2%) strains tested positive for blaKPC–like genes, and twelve (6.1%) strains tested positive for blaCTX–M–like genes. The CRE ELITe MGB Kit demonstrated a positive predictive value (PPV) of 85% (95% CI: 64.9–94.6) and a negative predictive value (NPV) of 100%. The ESBL ELITe MGB Kit had a PPV of 75% (95% CI: 49.4–90.2) and an NPV of 100%.
Multiplex PCR may outperform conventional and quantitative PCR in increasing the possibility of cross-contamination during multiplexing (Adler et al., 2008). The use of different primers in a solution blend allows the identification of various bacteria in a single test run, reducing costs and time (Pishnian et al., 2019). Multiplex PCR can effectively identify specific mutations in AMR genes (Banerjee and Patel, 2023; Chahorm and Prakitchaiwattana, 2018) if one primer interacts with the mutation points. It has been developed to quickly and cost-effectively identify AMR genes in infections and multiple organisms in patient samples (Sigmund et al., 2019). However, information on its effectiveness for detecting antibiotic resistance indicators in healthcare settings is limited (Sigmund et al., 2020).
Reverse transcriptase polymerase chain reaction converts RNA into complementary DNA (cDNA) and amplifies it via PCR. This method is efficient, responsive, and reliable (Adzitey et al., 2013; Rodrigues et al., 2018). cDNA produced from RNA is free of impurities such as proteins that can skew analyses, allowing for precise identification with primers. RT-PCR also detects viable microbes, including antibiotic-resistant organisms (Rajapaksha et al., 2019), and is highly specific for replicating cells. A study was conducted at the Hospital of Tropical Diseases in Ho Chi Minh City by Dung et al. (2024). They collected tracheal aspirates and sputum from patients with lower respiratory tract infections to identify and detect AMR genes for various bacteria. The Microbiology Department at the hospital used RT-PCR to identify six bacterial pathogens and their AMR genes. The results revealed that this technique is rapid, straightforward, and highly specific. Treating infections caused by antibiotic-resistant microorganisms, especially those with high AMR gene concentrations, is challenging. Real-time PCR and reverse transcription-PCR are currently used together to evaluate gene expression variations.
The NGS method has been developed for sequencing DNA and RNA and detecting mutations and variants (Qin, 2019; Satam et al., 2023). NGS can sequence thousands of genes or entire genomes in a short time (Goodwin et al., 2016). Integrating DNA and RNA next-generation sequencing (NGS) is the most effective strategy for understanding and adapting to the evolving resistance mechanisms to targeted therapies (Reita et al., 2021). NGS is particularly recognized for evaluating organisms with slow progression and unusual resistance patterns, such as highly resistant Mycobacterium tuberculosis isolates (Iketleng et al., 2018). Since the 1995 sequencing of Haemophilus influenzae type B (Fleischmann et al., 1995), advances in genomic technology have transformed infection prevention, control, and healthcare delivery (Purushothaman et al., 2022). Key methodologies include whole-genome sequencing (WGS), which analyzes the complete genome of a single bacterial colony, and metagenomics, which examines microbial communities in a sample without considering the cultural background (Chiu and Miller, 2019; Eyre, 2022). The cost of nucleotide sequencing has decreased significantly since the early 21st century (Brown et al., 2021; Dymond et al., 2020) owing to advancements in sequencing capabilities, affordable technologies, improved laboratory automation, and standardized procedures (Perez-Sepulveda et al., 2021). WGS is now used as a diagnostic tool in various laboratories for medical microbiology diagnosis and monitoring (Bonsall et al., 2020; Smetana and Brož, 2022; Van Belkum and Rochas, 2018). Its ability to quickly identify resistance factors has influenced treatment strategies (Iketleng et al., 2018).
However, recent challenges have limited the widespread use of NGS in resistance screening (Ávila-Ríos et al., 2020). NGS faces challenges such as high costs and limited user-friendly bioinformatics systems (Sherry et al., 2013). Nevertheless, high-quality sequencing can be achieved within days to a week (Sherry et al., 2013). NGS is anticipated to replace standard cultivation methods in the medium or long term (Femerling et al., 2023) and serve as a practical alternative for routine microbiological testing to identify bacterial infections and predict antibiotic susceptibility (Dunne et al., 2012). Additionally, NGS has the potential to provide both typing results and the ability to detect resistance genes in a single experiment (Veenemans et al., 2014). Many innovative NGS applications have recently transitioned from research to clinical use (Di Resta and Ferrari, 2018), including cell-free DNA analysis in prenatal testing, circulating tumor DNA testing, HLA typing, microbial analysis (Yohe and Thyagarajan, 2017), RNA sequencing, and methylation analysis. Despite ongoing challenges, these technologies are increasingly employed for therapeutic purposes.
Whole-genome sequencing antibiotic susceptibility testing is a rapid and accurate method for detecting antibiotic resistance (MacLean et al., 2020). The correlation between genotypes and clinical phenotypes is not always accurate, for example, mechanisms involving inducible resistance, gene expression and regulation, posttranslational modifications or combinations thereof. Consequently, rapid phenotypic testing and growth-based susceptibility testing are expected to be required initially to confirm an NGS result (Ellington et al., 2017). Most resistance strategies involve multiple genes and complex cellular networks, which are not well understood. WGS provides a novel approach to studying these systems by examining the entire genomes of microorganisms in clinical settings. A systematic review by Yusof et al. (2022) revealed that WGS is commonly used to detect variants in colistin resistance genes among K. pneumoniae isolates. WGS significantly advances AMR research by analyzing all contributing factors and identifying relationships between resistance genes and their host elements. WGS enables genome-wide studies of multiple genes and alterations (De Been et al., 2015; Kohl et al., 2014; Lin et al., 2022). Katiyar et al. (2020) used WGS to explore resistance genes and their symptom correlations, aiding in targeted treatment selection. These findings revealed that fluoroquinolone-resistant isolates presented changes in the gyrA, gyrB, parC, and parE genes. By predicting resistance polymorphisms and their phenotypic relationships, WGS enhances understanding and facilitates efficient identification of AMR correlates, allowing for antibiotic use on the basis of genetic factors (Wan Makhtar et al., 2021).
Peptide fingerprinting analytical technique (PFAT) can be used to identify proteins in microorganisms, including bacteria and fungi (Elbehiry et al., 2022a; Elbehiry et al., 2022c; Elbehiry et al., 2023). It breaks down target proteins into smaller peptides and measures the mass-charge ratio (m/z), such as matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). This method is valuable for assessing AMR (Figure 2), bridging the gap between species identification and resistance evaluation (Florio et al., 2020; Vrioni et al., 2018). Since 2010, healthcare settings have utilized MALDI-TOF MS, which is easier, faster, more accurate, and less expensive than traditional biochemical methods (Elbehiry et al., 2022a). The San Lazaro Hospital-Nagasaki Collaborative Research Laboratory analyzed approximately 13,000 bacterial and fungal isolates via MALDI-TOF MS over a span of 5 years (Osa et al., 2021). This system is able to detect resistance mechanisms and identify biomarkers, making it vital for monitoring resistant infections and facilitating rapid diagnosis (Rodríguez-Sánchez et al., 2019). Compared with traditional methods, MALDI-TOF accelerates resistance identification (Vrioni et al., 2018). The MBT-ASTRA evaluates bacterial growth by comparing the area under the curve (AUC) of bacterial peaks in antibiotic-treated (AUCBAC+ATB) versus untreated (AUCBAC) spectra. The ratio (AUCBAC+ATB)/(AUCBAC) is known as relative growth (RG). An RG close to zero indicates susceptibility, whereas an RG near one suggests resistance (Lange et al., 2014; Sparbier et al., 2016).

Figure 2. A workflow for the detection of antimicrobial resistance (AMR) using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS).
Matrix-assisted laser desorption ionization-time of flight mass spectrometry has shown potential for detecting antibiotic resistance in clinical samples, but its use is limited by low sensitivity and a focus on relatively small resistance pathways (Yamin et al., 2023). Recent machine learning advancements have improved the ability of these methods to recognize drug-resistant bacteria (Griffin et al., 2012). MBT-ASTRA quickly analyzes spectra, providing AST results within 2–3 h (Vrioni et al., 2018). A study conducted by Rhoads et al. (2016) evaluated the utility of the 2,415 m/z peak in clinical isolates of S. aureus and Staphylococcus epidermidis as a predictor of mecA gene carriage. Although numerous molecular assays are available for the rapid detection of mecA, these tests are predominantly performed on blood cultures. Nevertheless, in certain staphylococcal species, MALDI-TOF MS may serve as an effective and expedient method for the detection of mecA carriage. Timely diagnosis of mecA is crucial for facilitating appropriate medical intervention and enhancing antibiotic stewardship. By analyzing data obtained from routine MALDI-TOF MS testing, it is feasible to identify methicillin resistance in a subset of clinically isolated staphylococci. A recent study conducted by Bianco et al. (2022a) in a tertiary teaching hospital in northwestern Italy over a 3 years period (November 2019–October 2022) revealed that the MALDI Biotyper® platform can accurately and effectively identify K. pneumoniae carbapenemase (KPC) producers without the need for additional procedures or extra costs. This MALDI-based method is suitable for high-throughput laboratories. Depending on the local prevalence of non-pKpQIL-encoded KPC and other carbapenemases, molecular or immunochromatographic tests should be utilized.
The advantages of MALDI-TOF MS include precision, reliability, affordability in terms of cost/sample run, and simplicity (Vrioni et al., 2018). However, spectrum analysis software usually requires programming skills that many microbiology laboratory specialists do not possess (Welker and Van Belkum, 2019). The technique also requires purification and sample preparation, making it unsuitable for mixed materials, and relies on additional chemicals such as those in the matrix (Wang et al., 2018). Databases should differentiate between susceptible and resistant strains (Dubourg and Raoult, 2016). Mass spectrometry can identify key resistance genes, such as vanA, blaKPC, and mecA (Neil et al., 2021). Although Bruker Daltonics has created a prototype software package, it is not yet available (Wilhelm et al., 2023). Therefore, there is considerable interest in developing a modified version of this test for hospitals in need of faster AST results.
The fluorescence in situ hybridization (FISH) technique is effective for quantifying target microorganisms (Swidsinski et al., 2022; Vasala et al., 2020). PNA-FISH, which uses peptide nucleic acid (PNA) probes, offers rapid and specific binding advantages over DNA or RNA probes (Almeida et al., 2009; Vasala et al., 2020). Its electrically neutral backbone minimizes non-specific attachment, making it reliable for routine diagnostics, although PNA probes are often more expensive. Enroth et al. (2019) highlighted the use of PNA-FISH in QuickFish technology (OpGen, United States), which targets 16S rRNA for identification. FISH-based identification of resistance determinants is a fast, simple, and affordable method. Many AMR genes, such as blaCTX–M (Khan et al., 2020), mecA (Zhu and Li, 1994), and vanA (Conwell et al., 2021), can be detected via FISH. Salimnia et al. (2014) revealed that XpressFISH can identify the mecA gene in MRSA alongside QuickFish, allowing the diagnosis of methicillin resistance within 2 h of a positive blood culture. FISH can be used to assess resistance directly from raw materials such as tissues, making it suitable for resource-limited settings. Two conditions must be satisfied for FISH to be effective in identifying resistance determinants. First, the high abundance of ribosomal RNA copies in living cells results in ribosomally mediated resistance, which influences the efficacy of antibiotics such as macrolides and linezolid and is particularly suitable for FISH analysis. Second, FISH can be effectively employed when resistance is conferred by only one or a few variable bases, thereby eliminating the necessity for a large array of probes (Frickmann et al., 2014).
Unlike PCR, FISH can pinpoint specific resistance mechanisms in bacteria, but its applications are limited, and the results may be inconsistent (Frickmann et al., 2014). Resistance testing with FISH lacks standardization, posing challenges. Additionally, tissue autofluorescence must be considered, requiring expert interpretation of the results (Vasala et al., 2020). Expertise is required to interpret outcomes, especially when dealing with tissue autofluorescence. Counterstaining with a paneubacterial FISH probe and non-specific DNA stains is crucial for confirming nucleic acids associated with detected illnesses (Swidsinski, 2006). Owing to its limitations, FISH may serve as a temporary solution until more user-friendly and cost-effective amplification-based alternatives become available (Frickmann et al., 2014). Future research that investigates the mechanisms underlying drug resistance and the failure of eradication efforts in developing countries, particularly regarding clarithromycin and its related medications, may benefit from the application of the FISH approach (Frickmann et al., 2017).
Microarray analysis is a valuable method for examining genomic AMR in microorganisms (Waskito et al., 2022). It allows for the purification and amplification of specific RNA molecules, facilitating gene expression analysis and gene function determination (Galhano et al., 2021; Hung and Weng, 2017). Microarrays are effective in gene transcriptome studies and can identify AMR genes via multiple hybridization procedures on the same substrate. DNA samples can be extracted without culturing bacterial cells (Nsofor, 2014). However, microarrays are increasingly being replaced by next-generation sequencing (Galhano et al., 2021). A key limitation is the requirement for prior knowledge of the genetic region being studied, which may result in overlooked data (Kanzi et al., 2020). Additionally, analyzing target genes can be complicated because of hybridization of similar sequences (Jaksik et al., 2015).
Baumgartner et al. (2014) used the microarray method to identify genes linked to resistance in foodborne pathogens and reported that mecA, vanB, msr, aadD, and cat confer resistance to methicillin, vancomycin, macrolides, tobramycin, and chloramphenicol, respectively. Vogt et al. (2014) reported strains of E. coli resistant to third-generation cephalosporins in various meats and identified genes associated with resistance to trimethoprim (dfrA), sulfonamides (sul), tetracycline (tet), and chloramphenicol (cmlA1-like). Conventional resistance tests for Mycobacterium tuberculosis are time-consuming, delaying treatment (Asmare and Erkihun, 2023). Microarray analysis can quickly identify AMR by detecting mutations in the rpoB and katG genes linked to rifampin and isoniazid resistance (Herrera-Rodriguez et al., 2013). It can also identify ESBL- and carbapenemase-producing Enterobacteriaceae (Dortet et al., 2014). Microarrays are more effective and faster at detecting AMR genes in both gram-positive and gram-negative microbes (Fink et al., 2019). Recently, microarrays have emerged as widely utilized diagnostic techniques in biological research and clinical laboratories around the world (Rabiee et al., 2021). Song et al. (2019) analyzed 416 clinical samples from the Chinese PLA General Hospital via microarray technology, which directly identified eight carbapenemase genes across diverse samples. This method is recognized for its user-friendly design, adaptability for high-throughput detection, and clinical application potential. In 2019, Fink et al. (2019) evaluated a DNA microarray for the detection of AMR genes in 240 g-positive and gram-negative bacterial isolates. These genes included plasmid-encoded extended-spectrum β-lactamases and carbapenemases, as well as mecA, vanA, and vanB. The isolates were sourced from the Microbiology Services of two hospitals in Valencia, Spain. The assay demonstrated 100% sensitivity and specificity for all target genes. Although the manufacturing of DNA microarrays has advanced significantly and has been commercialized in recent decades, the constraints associated with high-resolution scanning, along with the time-intensive and expensive processes involved in microarray production, continue to restrict their accessibility to many laboratories (Xu et al., 2024).
Flow cytometry, introduced in the 1960s, is an effective single-cell analytical method that has significantly impacted fields such as immunology, molecular biology, chemotherapy, healthcare microbiology, ecological microbiology, and the food and beverage industries (Marutescu, 2023). Advances in cytometry have transformed life sciences and biomedicine by enabling rapid evaluation of bacterial cells without cultivation (Robinson et al., 2023). Researchers are developing strategies and tests to combat AMR, using flow cytometry to identify antibiotic-resistant microorganisms in healthcare and environmental samples (Godeux et al., 2018; Zhang et al., 2023). Flow cytometry analysis in healthcare can assess a microorganism’s susceptibility, resistance, or intermediate status to antibiotics (Martins-Oliveira et al., 2020; Silva-Dias et al., 2021). It quickly identifies AMR patterns in hospital samples (Filbrun et al., 2022; Inglis et al., 2020; Mulroney et al., 2022; Velican et al., 2020) and uses fluorescent dyes to evaluate bacterial cell viability after antibiotic treatment.
In accordance with the recommendations of Thermo Fisher Scientific, the flow cytometry-AST workflow comprises several critical steps. Initially, bacteria are collected and suspended in an appropriate solution to minimize background noise for the Attune (NxT) flow cytometer. The bacteria are subsequently exposed to various antibiotics, and a vital dye is applied to facilitate detection. The preinstalled software then analyzes the gated populations, monitoring alterations in size, shape, and color intensity induced by the antibiotics. The Attune (NxT) flow cytometer effectively illustrates the impact of varying antibiotic concentrations on these parameters, rendering this method more efficient than traditional culture-based MIC determination. However, limitations exist, such as non-specific dye binding, variable microorganism–antimicrobial interactions, and inadequate computational power for diverse populations (Waagsbø et al., 2022). Recently, commercial flow cytometry-based ASTs have been introduced, providing results in under 2 h, whereas traditional methods require 24–48 h (Martins-Oliveira et al., 2020; Silva et al., 2019; Silva-Dias et al., 2021). Unlike conventional culture-based methods, flow cytometric tests can determine microorganism fatality in medical samples treated rapidly with pharmaceuticals in less than 24 h (Marutescu, 2023).
Flow cytometric assays assess bacterial susceptibility, resistance, or intermediate responses to antibiotics in clinical microbiology. In 2020, Velican et al. (2020) evaluated the antibiotic susceptibility of 29 E. coli strains from urine samples of hospitalized patients in Bucharest, Romania. The technique effectively quantified susceptibility to nitrofurantoin, trimethoprim-sulfamethoxazole, ciprofloxacin, and ceftriaxone. In 2021, Kállai and his team at Budapest (Kállai et al., 2021) developed AST protocols using flow cytometry on six bacterial strains: E. coli, K. pneumoniae, P. aeruginosa, S. aureus, Streptococcus pyogenes, and Enterococcus faecalis. A flow cytometer was used to monitor bacterial growth to determine the optimal AST timing. The bacteria were tested against 12 antibiotics, with MIC values compared with microdilution values as a reference. Using EUCAST clinical breakpoints, susceptibility profile-matched microdilution results in more than 92% of the cases, indicating that flow cytometry-AST is an efficient method for generating susceptibility profiles with a low failure rate. Costa-de-Oliveira et al. (2017) studied the effectiveness of FASTinov® testing for managing gram-negative bacteremia at Centro Hospitalar S. João in Porto, Portugal, in 2015. An analysis of 102 positive blood cultures via routine methods, the Vitek2 test, the FASTinov® kit, and the gold standard microdilution method revealed that the FASTinov® kit yielded significantly faster results than did the Vitek2 test or the standard method. Although molecular ASTs are faster and can predict phenotypic resistance, varying sensitivity levels necessitate phenotypic screening to identify emerging resistance pathways (Marutescu, 2023).
Microbiology laboratories and healthcare settings have greatly benefited from advancements in optical techniques (Kaprou et al., 2021), particularly infrared (IR) spectroscopy and microscopy, which enhance the collection of molecular-level biological data on microbes (Beć et al., 2020). Fourier transform infrared spectroscopy (FTIR) is an innovative tool for biochemical evaluation that provides detailed information on a substance’s chemical composition (Chirman and Pleshko, 2021; Theakstone et al., 2021). By measuring infrared light absorption by proteins, carbohydrates, lipids, and genetic material, FTIR generates a spectrum that accurately reflects the sample’s composition (Candoğan et al., 2021; Vogt et al., 2019). Its application is increasingly used to identify chemical changes linked to AMR in prokaryotic organisms (Novais et al., 2019; Salman et al., 2017).
In 2017, Sharaha et al. (2017) used FTIR to assess antibiotic susceptibility in bacteria through IR spectroscopy. After 24 h of culturing bacterial colonies from patient samples, they employed a computer classification method and an IR microscope to evaluate E. coli sensitivity to gentamicin, ceftazidime, nitrofurantoin, nalidixic acid, and ofloxacin. Approximately 85% of bacteria are classified as susceptible or resistant, highlighting the technique’s potential for rapid testing. Abu-Aqil et al. (2024) employed FTIR spectroscopy in conjunction with machine learning techniques to identify 636 strains of K. pneumoniae from urine samples collected from patients with urinary tract infections and to assess their antibiotic susceptibility. An analysis of a total of 27,966 spectra via an XGBoost classifier revealed that the identification accuracy exceeded 95%, with the sensitivity for antibiotic susceptibility ranging from 74 to 81%. These results suggest that the proposed system has the potential to reduce the risks associated with bacterial resistance; however, further research is necessary to validate these findings. Research by Kochan and colleagues indicated that S. aureus has evolved chemically, leading to resistance against daptomycin and vancomycin. They utilized atomic force microscopy with infrared spectroscopy, a novel single-cell nanoscale technique (dos Santos et al., 2023; Li, 2023), which enhances the precision and imaging of atomic-scale structures within cells.
In 2022, Wang-Wang et al. (2022) assessed FTIR as a primary tool for identifying extended-spectrum β-lactamase-producing K. pneumoniae outbreaks in hospitals. Compared with traditional methods, FTIR identified more true clustering relationships (38 out of 42 vs. 24 out of 42, p = 0.001), highlighting its potential as a rapid and cost-effective detection tool, with WGS for confirmation. This integration can enhance infection control in clinical microbiology laboratories. FTIR provides advantages in AMR research, such as speed, cost-effectiveness, reliability, and environmental friendliness (Kaprou et al., 2021; Mashhadikhan et al., 2022; Yamin et al., 2023). However, its application in resource-limited settings as a point-of-care tool for testing AMR is hindered by the instrument’s size and cost (Kaprou et al., 2021). Accurate results require prioritizing sample purification, culturing, and processing, along with accessible datasets that distinguish between sensitive and resistant isolates.
Microfluidics-based lab-on-a-chip technologies benefit various fields, including environmental surveillance (Fernandez-Gavela et al., 2019), food security (Tsougeni et al., 2019), and healthcare diagnostics (Papadakis et al., 2019), and have recently enabled the identification of antibiotic-resistant microorganisms (Li et al., 2020). MIC values are essential for analyzing bacterial phenotypic resistance, assessing new antimicrobial agents, and monitoring global drug resistance (Zhang et al., 2020). This technique determines if an antibiotic can inhibit pathogen growth, even if slowly. However, this delay prolongs the selection of effective treatments, leading to worse clinical outcomes and higher patient mortality rates (Li et al., 2017). To address this issue, developing technologies for the early detection of antibiotic resistance during therapy is crucial. In 2018, Zhang et al. (2018) developed a microfluidic chip platform to identify bacteria and detect antibiotic resistance genes in 108 cerebrospinal fluid culture broths from Beijing Tiantan Hospital and Capital Medical University, China. The platform achieved a 94.44% concordance rate with traditional methods and demonstrated over 90% sensitivity and specificity for carbapenemase and ESBL resistance genes. The study concluded that the platform is rapid, accurate, and user friendly, with strong potential for treating postneurosurgical meningitis. Ardila et al. (2023) conducted a systematic review examining the potential clinical applications of microfluidics-based lab-on-a-chip technology for the identification and assessment of antibiotic susceptibility in Enterococcus faecalis associated with endodontic infections. Although these platforms demonstrate significant potential for enhancing the diagnosis and treatment of Enterococcus faecalis, their effective implementation in clinical practice requires comprehensive research, development, and validation to ascertain their efficacy and reliability. Figure 3 shows recent microfluidic techniques for quick bacterial ASTs, including genotypic methods (such as droplet digital technology for resistance detection) and phenotypic methods (such as microfluidic single bacterial culture).
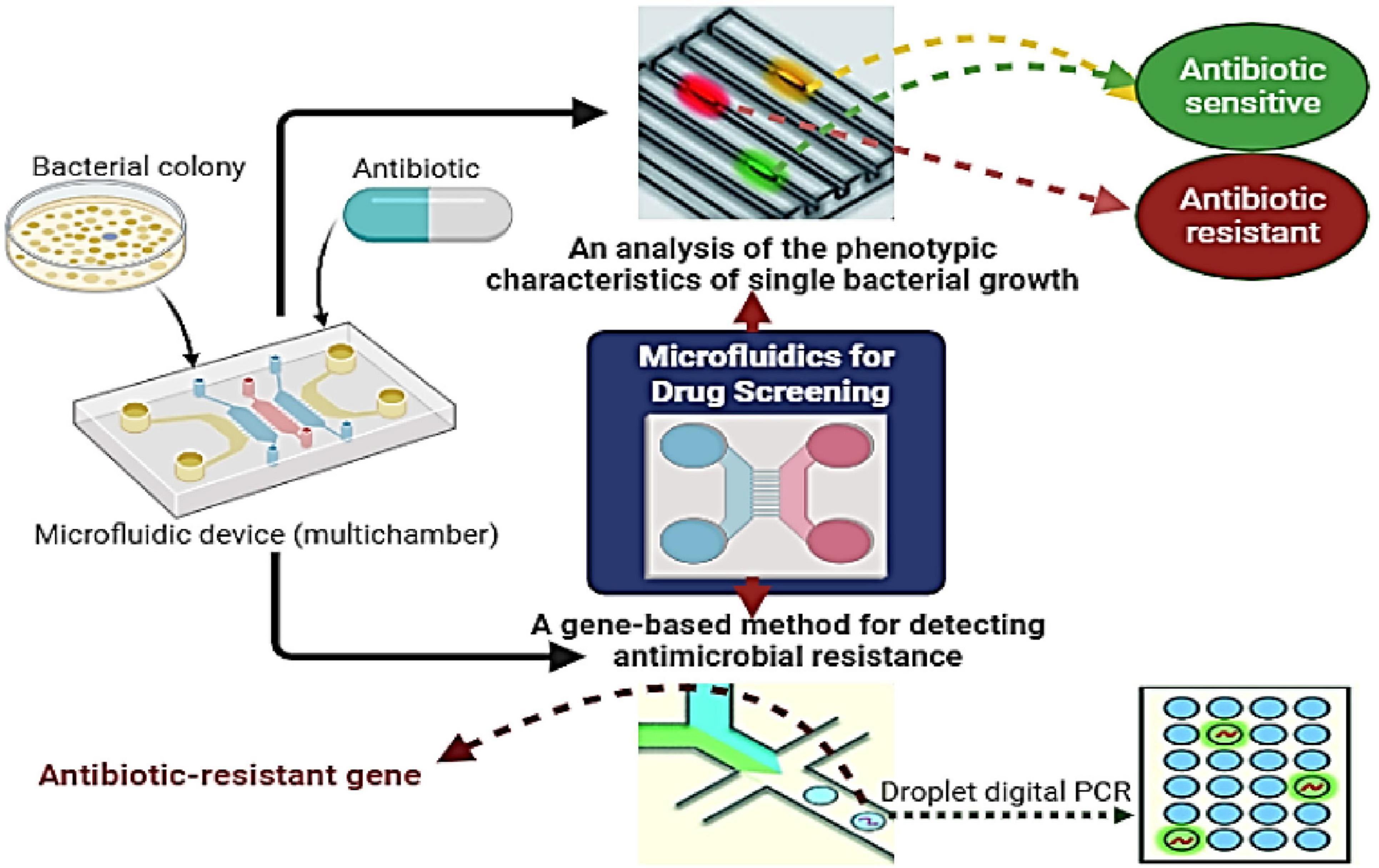
Figure 3. Procedures for the rapid screening of individual bacteria utilizing microfluidics encompass gene-based detection of antimicrobial resistance (AMR) through droplet digital assessment, as well as phenotypic evaluation via microfluidic-based single bacterial culture.
Lab-on-a-chip technologies offer advantages over macroscale methods, including rapid output evaluation, precise fluid control, cost-effectiveness, the utilization of minimal amounts of reagents/samples/solvents (few μL), automation, mobility, and process integration (Fernandez-Gavela et al., 2019). Microfluidic identification techniques fall into two categories: phenotypic and molecular analyses. Loop-mediated isothermal amplification enables rapid results by analyzing genetic markers such as AMR genes (Zhang et al., 2018). Integrating nucleic acid detection assays into microfluidic devices provides a cost-effective solution for diagnostics in the clinic, food safety, and environmental monitoring. These devices, which are designed for resource-limited settings and point-of-care use, require amplification of target nucleic acids for sensitive detection. Traditional PCR methods are limited by temperature cycling. Therefore, novel isothermal amplification techniques will be developed to integrate sample preparation and target identification using minimally invasive samples such as saliva, blood, or urine (Giuffrida and Spoto, 2017; Zhang et al., 2019).
Phenotypic microfluidic methods reliably track microorganism growth for AST in the presence of antimicrobial agents (Campbell et al., 2016). These methods utilize small amounts of microorganisms in droplets, channels, or chambers (Needs et al., 2020) and can encapsulate them in agarose-filled capsules or immobilize them using antibodies on magnetized substrates (Dong and Zhao, 2015). Hydrodynamic capture is one technique for immobilization, offering dense trap arrays and easy integration, but it has low effectiveness (Kaprou et al., 2021). Antibodies are costly and may not be accessible for all bacteria, whereas droplet-based methods often require complex reading techniques. Although agarose-based methods can use standard multiwell plates, they complicate automatic identification and information processing (Kaprou et al., 2021). Further research is needed before these devices can be commercially viable.
The rise of antimicrobial-resistant bacteria has prompted the development of lateral flow immunoassays (LFIAs) that target enzyme-mediated resistance in key pathogens (Boutal et al., 2022). Despite advancements, identifying pathogenic bacteria and detecting antibiotic resistance remain complex (Shanmugakani et al., 2020). In the last decade, LFIAs have improved the diagnosis of antimicrobial resistance, proving essential for detecting resistance mechanisms in gram-negative bacteria, especially beta-lactamases (Bernabeu et al., 2020; Boutal et al., 2018; Han et al., 2021). One LFIA identified the AAC (6’)-Iae enzyme from P. aeruginosa with a sensitivity of 10^5 CFU per test (Kitao et al., 2010). Another LFIA targets the ArmA 16S rRNA methylase, which is prevalent in resistant gram-negative bacteria such as A. baumannii and E. coli (Oshiro et al., 2015). Early identification of methicillin resistance relies on detecting penicillin-binding protein 2a (PBP2a), which affects all beta-lactams. The affinity of S. aureus protein A for mammalian immunoglobulins complicates antibody-based detection. Nevertheless, Yamada et al. (2013) used an LFIA, whereas Amini et al. (2020) utilized IgY anti-PBP2a antibodies. For VanA vancomycin-resistant Enterococcus (VRE) isolates, LFIA is 100% sensitive and specific, with detection limits of 6.3 × 10^6 or 4.9 × 10^5 cfu per test on either MH or ChromID® VRE plates, respectively (Oueslati et al., 2021).
Lateral flow immunoassays have been developed for the detection of KPC and OXA-48-like enzymes (Coris Bioconcept, Gembloux, Belgium) (Wareham et al., 2016), as well as IMP, which is recognized as the most prevalent metallo-β-lactamase in Japan (Notake et al., 2013), and New Delhi metallo-beta-lactamase 1 (NDM) (Boutal et al., 2017; Tada et al., 2019). These assays demonstrate 100% sensitivity and specificity when utilized with isolated colonies obtained from agar plates. Comini et al. (2022) evaluated a rapid diagnostic method using MALDI-TOF MS, LFIAs, and molecular testing to identify gram-negative bacteria and key β-lactamases from positive blood cultures. The NG-Test® CARBA 5 and NG-Test® CTX-M MULTI LFIAs had sensitivities of 92.2 and 91.6%, respectively, whereas the Easyplex® SuperBug CRE showed 100% sensitivity for blaKPC mutations linked to ceftazidime/avibactam resistance. No false positives were found, making this method a cost-effective solution for quickly identifying gram-negative organisms and resistance indicators. In a French clinical laboratory, the NG-Test CTX-M MULTI identified 98% of ESBL producers from colonies and positive blood cultures (Bernabeu et al., 2020). While CTX-M enzymes are primarily responsible for extended-spectrum cephalosporin (ESC) resistance, some uncommon enzymes, such as plasmid-encoded AmpC and certain carbapenemases, may not be detected. The LFIA-CTX test showed 99.1% sensitivity and 100% specificity for detecting ESC hydrolytic activity in colonies (Moguet et al., 2022).
A recent evaluation of ESC hydrolysis compared three assays: the ESBL NDP test, the ZG-Lacta™ test, and the LFIA-CTX test with the NG-Test CTX-M-Multi. LFIA-CTX demonstrated increased sensitivity and specificity, particularly for Enterobacterales. LFIA-CTX offers the advantage of detecting more than just ESBLs, unlike the NDP test. Its integration with NG-CTX-M-Multi effectively enables the hydrolysis of extended-spectrum cephalosporins (ESCs) and ESBLs. Moguet et al. (2022) introduced a novel LFIA strip that combines the LFIA-CTX and NG-CTX-M-Multi assays. This strip allows clinical microbiology laboratories to detect extended-spectrum cephalosporin (ESC) hydrolysis directly from colonies grown on culture media used for CTX-M-like enzymes, which represent 98% of ESBLs. This improvement in antimicrobial stewardship allows for early treatment with new medications. NG Biotech’s LFIA can detect MCR-1 within 15 min via the use of streptavidin-labeled anti-MCR-1 mouse monoclonal antibodies. A multicentric validation study revealed that this test successfully identified 109 true-positive MCR-1 cases with no false-negative results. Additionally, the assay detected three E. coli isolates that produce MCR-2. In contrast, isolates producing MCR-3, MCR-4, or MCR-5 tested negative (Smelikova et al., 2022; Volland et al., 2019).
A novel strategy is urgently needed to address the rise of antibiotic resistance and infectious diseases caused by bacteria. The CRISPR/Cas system, an RNA-guided adaptive immune mechanism in prokaryotes, identifies and neutralizes invasive genetic elements such as plasmids and phages (Tao et al., 2022). This technology is being developed to prevent and control antimicrobial resistance, effectively targeting DNA sequences that carry resistance genes (Aslam et al., 2020; Duan et al., 2021; Wu et al., 2021). Over the past few decades, CRISPR/Cas has become increasingly popular for gene modification. Some research suggests that it could be more effective in addressing antibiotic resistance genes in certain cell types and experimental platforms (Manghwar et al., 2019). Antibiotic-resistant bacteria pose a threat to human health and hinder the progress of modern medicine (Okaiyeto et al., 2024). By swiftly and accurately identifying resistance, healthcare practitioners can administer appropriate treatment faster, optimize the use of current antibiotics, and avoid resorting to “last resort” medications (Raro et al., 2024). Therefore, the development of technology capable of rapidly and accurately detecting drug resistance genes is crucial for the advancement of modern medicine. Since its discovery in 2012 in Streptococcus pyogenes, the Cas9 protein in the CRISPR system has led to the development of numerous novel applications (Jinek et al., 2012). The detection of antibiotic resistance genes and bacterial infections has also become increasingly prevalent in recent years.
Müller et al. (2016) developed an optical DNA mapping technique utilizing a single plasmid within a nanofluidic tube. This method leverages the cleavage capabilities of Cas9 to convert a circular plasmid into a linear form, thereby facilitating the detection of resistance genes. The CRISPR/Cas9 system incorporates a gene-specific RNA (gRNA) that assists in the identification of target genes. The optical DNA mapping technique can identify this gRNA by linearizing it at a predetermined location on the circular plasmid. Through their investigations, researchers have examined the potential for enhancing this detection technology for future clinical applications. They integrate multiple gRNAs that target various genes associated with antibiotic resistance. In a study conducted by Nyblom et al., strains of E. coli and K. pneumoniae were directly identified in patient samples via this approach (Nyblom et al., 2023). By targeting antibiotic resistance genes with the Cas9 protein, researchers have successfully identified specific strains or subtypes, as well as plasmids associated with these strains. This optical DNA profiling technique enables the rapid acquisition of comprehensive diagnostic information, thereby optimizing antibiotic treatment regimens and paving the way for the future of precision medicine management.
In 2023, (Qin et al. (2023). developed a method using CRISPR/Cas9-induced isothermal exponential amplification reactions (IEXPARs) to discover antibiotic resistance genes. They achieved this by cleaving the genes into two short fragments with free 3’-OH ends. The cleaved DNA templates trigger exponential amplification of IEXPAR under isothermal conditions, enabling the direct detection of antibiotic resistance genes. This rapid and precise method relies on the CRISPR/Cas9 system, which cleaves DNA only in the presence of antibiotic resistance genes. This ensures high specificity, as antibiotic-sensitive bacteria lack these genes. After approximately half an hour of amplification, the method has a detection limit of 81 fM, allowing for the identification of antibiotic resistance genes at concentrations as low as 100 fM. This method is effective in detecting both antibiotic-resistant and antibiotic-sensitive bacteria in real biological samples under isothermal conditions.
A study conducted in 2023 by Gao et al. (2023) introduced a CRISPR/Cas12a-based colorimetric paper sensor. This sensor utilizes the trans-cleavage activity of Cas12a, which is enhanced through rolling circle replication, to generate a 3D DNAzyme that strongly adheres to the paper. This resulted in a sensor with a high concentration of functional DNAzymes, making it highly bioactive. The assay was designed to swiftly detect the antibiotic resistance gene NDM with exceptional sensitivity. In the absence of the gene, the 3D DNAzyme produced a blue color. Conversely, the presence of the gene triggered collateral Cas12a cleavage activity, causing the circular template to be cleaved and preventing the formation of the 3D DNAzyme, leading to no colorimetric signal. This paper introduces a sensor capable of rapidly and affordably detecting antibiotic resistance genes in pathogenic microorganisms with femtomolar sensitivity. The results are visible to the naked eye, and the analysis can be completed in under one and a half hours. The programmable CRISPR probe design shown in this study has potential for swift responses to emerging global epidemics. At present, CRISPR-based antimicrobials are not widely utilized, so there have been no reports of resistance to this type of antimicrobial in clinically significant bacteria (Mayorga-Ramos et al., 2023). Research indicates that CRISPR-Cas systems could serve as next-generation antimicrobials, although their clinical application remains largely unexplored. Concerns regarding the safety and environmental implications of CRISPR-Cas technology highlight the need for risk assessments and strict regulatory measures for therapeutic purposes (Caplan et al., 2015). It is crucial to engage and educate the medical community and the public about this emerging technology to ensure its responsible and safe utilization (Javed et al., 2018; Pursey et al., 2018).
Collaboration among scientists, risk administrators, government agencies, and businesses is essential to address the AMR crisis and improve ASTs. Current instruments have limitations, such as the need for specimens before therapy and a lack of integration and mobility. Extensive biological methods remain the only viable options for infection detection. Innovative screening technologies are essential for expediting approval and commercialization. They should provide improved accuracy, faster turnaround times, lower costs, enhanced accessibility, and scalability in various healthcare settings. Faster and more reliable detection will benefit both healthcare practitioners and patients. Investing in the improvement of existing procedures and tools is necessary. A MarketsandMarkets Research Pvt. An Ltd. report predicts that the AST market will reach USD 4.2 billion by 2025 (Kaprou et al., 2021). However, the high costs of programmed AST systems limit their use, especially among budget-constrained organizations, despite their effectiveness in reducing incubation and identification periods (MarketsandMarkets™, 2021). In 2019, manual AST devices were popular because of their lower costs, with the disk diffusion method holding the largest market share due to the variety and affordability of disks. Healthcare facilities and testing laboratories account for the largest segment of the AST market (MarketResearch.com, 2021). AST has been proven to reduce hospital stays and improve patient outcomes (Cassini et al., 2019). However, the cost of molecular testing, which ranges from $100–$250 per test (Li et al., 2017), presents a significant barrier to its widespread implementation. Additionally, NAATs can result in annual reagent costs exceeding $500,000 for a 500-bed community hospital (She and Bender, 2019). In contrast, AST using mass spectrometry costs approximately €79 per patient, with reagent costs of approximately $1 (Patel et al., 2017). The costs associated with Microscan and Vitek susceptibility testing typically range between €30 and €50 (European Union, 2013). The variability in pricing for different AST methodologies further complicates the estimation of overall expenses (Vasala et al., 2020).
While the methods and technologies mentioned here show promise in addressing antimicrobial resistance, several questions remain. What percentage are consistently effective? Do they meet industry standards? When will new techniques be widely available? Although many claims to detect resistance within hours, they often neglect time-consuming steps such as culture isolation and enrichment before treatment. The traditional AST takes 18 to 36 h and provides MICs (Gajic et al., 2022); however, it is unsuitable for non-culturable microorganisms. The automated systems currently available do not accommodate these organisms and have response times of 2–24 h. Some methods can measure MICs (Kasas et al., 2021), while MALDI-TOF MS can yield results in 2–4 h, although it shares limitations with traditional methods and lacks processing software (Elbehiry et al., 2022a). NAATs can be used with ASTs when culture is not possible (Johnson et al., 2002; Theel and Schuetz, 2022), and new AMR genes can be detected. WGS is a promising tool for rapid AST but requires extensive datasets, making bioinformatics a major challenge (Bogaerts et al., 2019). An overview of the benefits and drawbacks of these techniques is provided in Table 1.
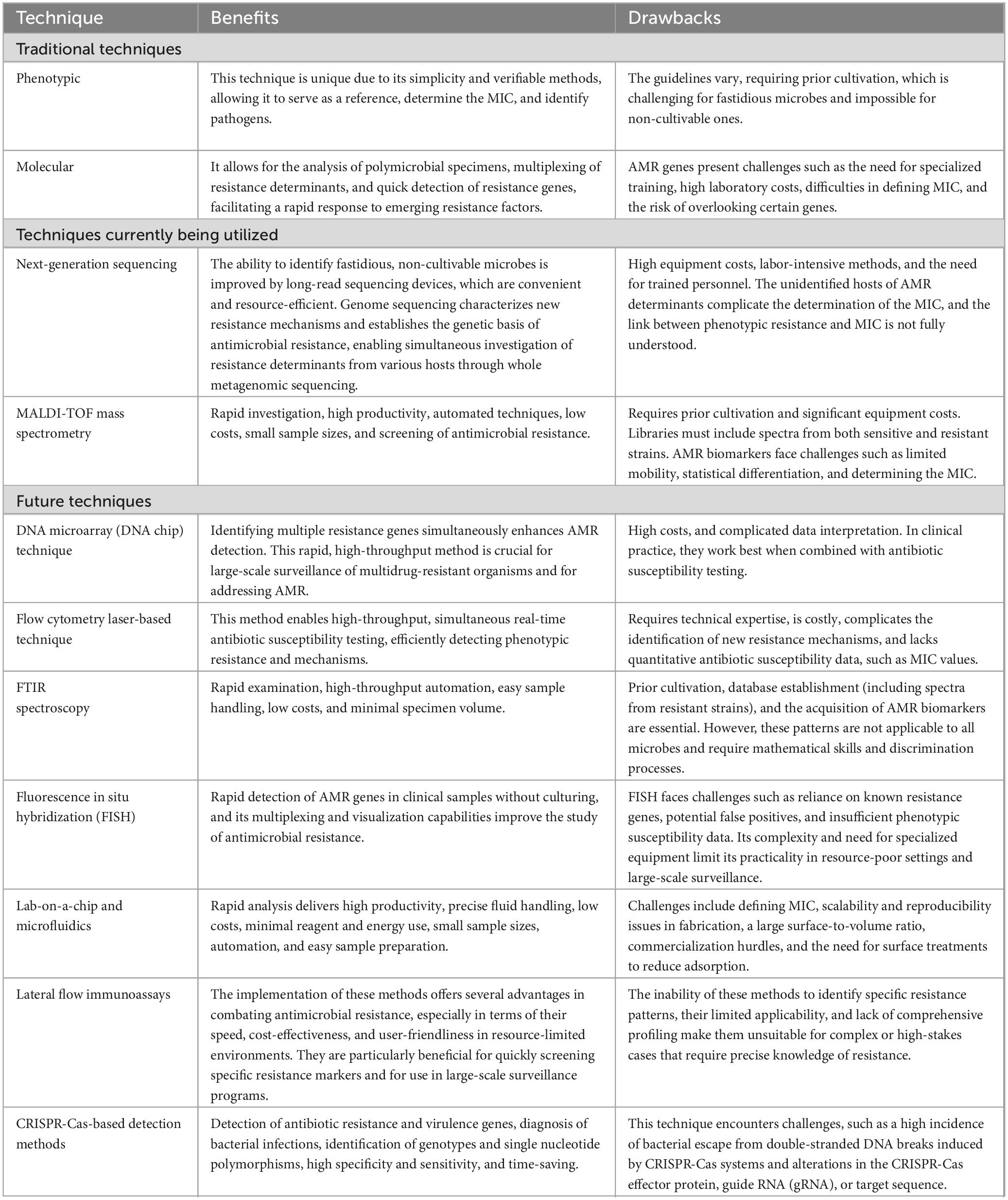
Table 1. An analysis of the benefits and drawbacks of conventional and non-conventional methods used to detect antimicrobial resistance.
Meeting the demands for rapid ASTs is challenging, and no existing methods are ideal. However, some may significantly influence the quick AST market. This market includes point-of-need services offered by well-equipped labs in healthcare and research facilities, which can integrate methods such as whole-genome sequencing, PCR, and automated AST systems. Alternatively, portable microfluidic AST systems may be better suited for smaller labs and medical professionals, offering cost-effectiveness, mobility, and quick turnaround times.
Identifying AMR is crucial in combating resistant infections. Advances in diagnostic technology have revolutionized AMR identification and treatment. While traditional methods such as disk diffusion and MIC testing are accessible and cost-effective, they often lack the speed and sensitivity required to detect new resistance mechanisms. Modern technologies, such as CRISPR-based systems, PCR diagnostics, MALDI-TOF mass spectrometry, and next-generation sequencing (NGS), have significantly improved AMR diagnosis by enabling comprehensive genomic analyses and providing real-time results. However, challenges such as cost, complexity, and the need for specialized training persist. The future of AMR detection will likely involve a combination of traditional and modern methods, allowing for quicker and more accurate diagnostics. This integrated approach can improve treatment decisions, enhance surveillance, and ultimately lead to better patient outcomes in the global fight against antimicrobial resistance. Continued research, investment in infrastructure, and policy support are essential for the effective clinical application of these advancements.
AyE: Data curation, Formal Analysis, Methodology, Validation, Visualization, Writing – original draft, Writing – review and editing. EM: Data curation, Formal Analysis, Methodology, Validation, Writing – original draft, Writing – review and editing. AAb: Writing – original draft, Writing – review and editing. MHA: Writing – original draft, Writing – review and editing. MEAM: Writing – original draft, Writing – review and editing. MA: Writing – original draft, Writing – review and editing. MI: Writing – original draft, Writing – review and editing. AbTE: Writing – original draft, Writing – review and editing. AAl: Writing – original draft, Writing – review and editing. SNA: Writing – original draft, Writing – review and editing. AA-O: Writing – original draft, Writing – review and editing. HE: Writing – original draft, Writing – review and editing. NM: Writing – review & editing, Validation, Funding acquisition. SAl: Writing – review & editing, Validation, Funding acquisition.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We would like to thank the Deanship of Graduate Studies and Scientific Research at Qassim University for financial support (QU-APC-2025).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ababneh, M., Nasser, S., and Rababa’h, A. M. (2021). A systematic review of antimicrobial stewardship program implementation in middle Eastern countries. Int. J. Infect. Dis. 105, 746–752. doi: 10.1016/j.ijid.2021.03.035
Abalkhail, A., and Elbehiry, A. (2022). Methicillin-resistant Staphylococcus aureus in diabetic foot infections: Protein profiling, virulence determinants, and antimicrobial resistance. Appl. Sci. 12:10803.
Abu-Aqil, G., Suleiman, M., Lapidot, I., Huleihel, M., and Salman, A. (2024). Infrared spectroscopy-based machine learning algorithms for rapid detection of Klebsiella pneumoniae isolated directly from patients’ urine and determining its susceptibility to antibiotics. Spectrochimica Acta A Mol. Biomol. Spectroscopy 314:124141. doi: 10.1016/j.saa.2024.124141
Adler, M., Wacker, R., and Niemeyer, C. (2008). Sensitivity by combination: Immuno-PCR and related technologies. Analyst 133, 702–718.
Adzitey, F., Huda, N., and Ali, G. (2013). Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. Biotechnol. J. 3, 97–107. doi: 10.1007/s13205-012-0074-4
Ahmad, I., Malak, H., and Abulreesh, H. (2021). Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resistance 27, 101–111.
Alghamdi, B., Al-Johani, I., Al-Shamrani, J., Alshamrani, H., Al-Otaibi, B., Almazmomi, K., et al. (2023). Antimicrobial resistance in methicillin-resistant staphylococcus aureus. Saudi J. Biol. Sci. 30:103604.
Almeida, C., Azevedo, N., Iversen, C., Fanning, S., Keevil, C., and Vieira, M. (2009). Development and application of a novel peptide nucleic acid probe for the specific detection of Cronobacter genomospecies (Enterobacter sakazakii) in powdered infant formula. Appl. Environ. Microbiol. 75, 2925–2930. doi: 10.1128/AEM.02470-08
Amini, M., Pourmand, M., Faridi-Majidi, R., Heiat, M., Mohammad Nezhady, M., Safari, M., et al. (2020). Optimizing effective parameters to improve performance quality in lateral flow immunoassay for detection of PBP2a in methicillin-resistant Staphylococcus aureus (MRSA). J. Exp. Nanosci. 15, 266–279.
Aminov, R. (2017). History of antimicrobial drug discovery: Major classes and health impact. Biochem. Pharmacol. 133, 4–19.
Aminov, R. I. (2010). A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 1:134. doi: 10.3389/fmicb.2010.00134
Anjum, M., Zankari, E., and Hasman, H. (2018). “Molecular methods for detection of antimicrobial resistance,” in Antimicrobial Resistance in Bacteria from Livestock and Companion Animals, eds S. Schwarz, L. M. Cavaco, and J. Shen (Hoboken, NJ: John Wiley & Sons), 33–50.
Antoñanzas, F., and Goossens, H. (2019). The economics of antibiotic resistance: A claim for personalized treatments. Eur. J. Health Econ. 20, 483–485.
Apfalter, P., Reischl, U., and Hammerschlag, M. (2005). In-house nucleic acid amplification assays in research: How much quality control is needed before one can rely upon the results? J. Clin. Microbiol. 43, 5835–5841. doi: 10.1128/JCM.43.12.5835-5841.2005
Ardila, C., Jiménez-Arbeláez, G., and Vivares-Builes, A. (2023). The potential clinical applications of a microfluidic lab-on-a-chip for the identification and antibiotic susceptibility testing of Enterococcus faecalis-associated endodontic infections: A systematic review. Dentistry J. 12:5. doi: 10.3390/dj12010005
Aslam, B., Rasool, M., Idris, A., Muzammil, S., Alvi, R., Khurshid, M., et al. (2020). CRISPR-Cas system: A potential alternative tool to cope antibiotic resistance. Antimicrob. Resist. Infect. Control 9, 1–3. doi: 10.1186/s13756-020-00795-6
Asmare, Z., and Erkihun, M. (2023). Recent application of DNA microarray techniques to diagnose infectious disease. Pathol. Lab. Med. Int. 15, 77–82.
Ávila-Ríos, S., Parkin, N., Swanstrom, R., Paredes, R., Shafer, R., Ji, H., et al. (2020). Next-generation sequencing for HIV drug resistance testing: Laboratory, clinical, and implementation considerations. Viruses 12:617.
Bai, H., Wang, Y., Li, X., and Guo, J. (2023). Electrochemical nucleic acid sensors: Competent pathways for mobile molecular diagnostics. Biosensors Bioelectronics 237:115407. doi: 10.1016/j.bios.2023.115407
Banerjee, R., and Patel, R. (2023). Molecular diagnostics for genotypic detection of antibiotic resistance: Current landscape and future directions. JAC Antimicrob. Resist. 5:dlad018. doi: 10.1093/jacamr/dlad018
Bang, E., Oh, S., Cho, H., Park, D. H., Chang, H. E., Park, J. S., et al. (2023). Development of diagnostic tests for pathogen identification and detection of antimicrobial resistance on WHO global priority pathogens using modular real-time nucleic acid amplification test. Int. Microbiol. 26, 563–577. doi: 10.1007/s10123-023-00321-9
Baraka, M., Alsultan, H., Alsalman, T., Alaithan, H., Islam, M., and Alasseri, A. (2019). Health care providers’ perceptions regarding antimicrobial stewardship programs (AMS) implementation—facilitators and challenges: A cross-sectional study in the Eastern province of Saudi Arabia. Ann. Clin. Microbiol. Antimicrob. 18, 1–10. doi: 10.1186/s12941-019-0325-x
Bassetti, M., Poulakou, G., Ruppe, E., Bouza, E., Van Hal, S., and Brink, A. (2017). Antimicrobial resistance in the next 30 years, humankind, bugs and drugs: A visionary approach. Intensive Care Med. 43, 1464–1475. doi: 10.1007/s00134-017-4878-x
Baumgartner, A., Niederhauser, I., and Johler, S. (2014). Virulence and resistance gene profiles of Staphylococcus aureus strains isolated from ready-to-eat foods. J. Food Protect. 77, 1232–1236. doi: 10.4315/0362-028X.JFP-14-027
Beć, K., Grabska, J., and Huck, C. (2020). Biomolecular and bioanalytical applications of infrared spectroscopy–A review. Anal. Chim. Acta 1133, 150–177. doi: 10.1016/j.aca.2020.04.015
Bej, A., Mahbubani, M., and Atlas, R. (1991). Amplification of nucleic acids by polymerase chain reaction (PCR) and other methods and their applications. Crit. Rev. Biochem. Mol. Biol. 26, 301–334.
Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42:fux053.
Bernabeu, S., Ratnam, K., Boutal, H., Gonzalez, C., Vogel, A., Devilliers, K., et al. (2020). A lateral flow immunoassay for the rapid identification of CTX-M-producing Enterobacterales from culture plates and positive blood cultures. Diagnostics 10:764. doi: 10.3390/diagnostics10100764
Bhatnagar, A., Machado, M., Patterson, L., Anderson, K., Abelman, R., Bateman, A., et al. (2023). Antimicrobial resistance laboratory network’s multisite evaluation of the ThermoFisher Sensititre GN7F broth microdilution panel for antimicrobial susceptibility testing. J. Clin. Microbiol. 61:e0799-23. doi: 10.1128/jcm.00799-23
Bianco, G., Boattini, M., Comini, S., Bondi, A., Curtoni, A., Piccinini, G., et al. (2024). Detection of volatile organic compounds as new paradigm to accelerate antimicrobial susceptibility testing: Performance evaluation of VITEK® REVEAL™. J. Antimicrob. Chemother. 79, 2237–2245. doi: 10.1093/jac/dkae219
Bianco, G., Boattini, M., Iannaccone, M., Sidoti, F., Cavallo, R., and Costa, C. (2019a). Detection of antibiotic resistance genes from blood cultures: Performance assessment and potential impact on antibiotic therapy management. J. Hosp. Infect. 102, 465–469. doi: 10.1016/j.jhin.2019.03.007
Bianco, G., Comini, S., Boattini, M., Ricciardelli, G., Guarrasi, L., Cavallo, R., et al. (2022a). Based approaches for direct identification of gram-negative bacteria and Bla KPC-carrying plasmid detection from blood cultures: A three-year single-centre study and proposal of a diagnostic algorithm. Microorganisms 11:91. doi: 10.3390/microorganisms11010091
Bianco, G., Iannaccone, M., Boattini, M., Cavallo, R., and Costa, C. (2019b). Assessment of rapid direct E-test on positive blood culture for same-day antimicrobial susceptibility. Braz. J. Microbiol. 50, 953–959. doi: 10.1007/s42770-019-00139-6
Bianco, G., Lombardo, D., Ricciardelli, G., Boattini, M., Comini, S., Cavallo, R., et al. (2022b). Multicenter evaluation of the EUCAST rapid antimicrobial susceptibility testing (RAST) extending analysis to 16–20 hours reading time. Antibiotics 11:1404. doi: 10.3390/antibiotics11101404
Boattini, M., Bianco, G., Iannaccone, M., Zanotto, E., Sidoti, F., Almeida, A., et al. (2021). Detection of carbapenemase and CTX-M encoding genes directly from bronchoalveolar lavage using the CRE and ESBL ELITe MGB assays: Toward early and optimal antibiotic therapy management of critically ill patients with pneumonia. Microbial Drug Resist. 27, 241–246. doi: 10.1089/mdr.2020.0199
Bogaerts, B., Winand, R., Fu, Q., Van Braekel, J., Ceyssens, P., Mattheus, W., et al. (2019). Validation of a bioinformatics workflow for routine analysis of whole-genome sequencing data and related challenges for pathogen typing in a European National Reference Center: Neisseria meningitidis as a proof-of-concept. Front. Microbiol. 10:362. doi: 10.3389/fmicb.2019.00362
Bonsall, D., Golubchik, T., de Cesare, M., Limbada, M., Kosloff, B., MacIntyre-Cockett, G., et al. (2020). A comprehensive genomics solution for HIV surveillance and clinical monitoring in low-income settings. J. Clin. Microbiol. 58:e00382-20.
Boutal, H., Moguet, C., Pommiès, L., Simon, S., Naas, T., and Volland, H. (2022). The revolution of lateral flow assay in the field of AMR detection. Diagnostics 12:1744. doi: 10.3390/diagnostics12071744
Boutal, H., Naas, T., Devilliers, K., Oueslati, S., Dortet, L., Bernabeu, S., et al. (2017). Development and validation of a lateral flow immunoassay for rapid detection of NDM-producing Enterobacteriaceae. J. Clin. Microbiol. 55, 2018–2029. doi: 10.1128/JCM.00248-17
Boutal, H., Vogel, A., Bernabeu, S., Devilliers, K., Creton, E., Cotellon, G., et al. (2018). A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP-and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 73, 909–915. doi: 10.1093/jac/dkx521
Breijyeh, Z., and Karaman, R. (2023). Design and synthesis of novel antimicrobial agents. Antibiotics 12:628.
Brown and Wright, G. (2016). Antibacterial drug discovery in the resistance era. Nature 529, 336–343.
Brown, B., Allard, M., Bazaco, M., Blankenship, J., and Minor, T. (2021). An economic evaluation of the whole-genome sequencing source tracking program in the US. PLoS One 16:e0258262. doi: 10.1371/journal.pone.0258262
Campbell, J., McBeth, C., Kalashnikov, M., Boardman, A., Sharon, A., and Sauer-Budge, A. (2016). Microfluidic advances in phenotypic antibiotic susceptibility testing. Biomed. Microdevices 18:103.
Candoğan, K., Altuntas, E., and İğci, N. (2021). Authentication and quality assessment of meat products by Fourier transform infrared (FTIR) spectroscopy. Food Eng. Rev. 13, 66–91.
Caneschi, A., Bardhi, A., Barbarossa, A., and Zaghini, A. (2023). The use of antibiotics and antimicrobial resistance in veterinary medicine, a complex phenomenon: A narrative review. Antibiotics 12:487. doi: 10.3390/antibiotics12030487
Cansizoglu, M., Tamer, Y., Farid, M., Koh, A., and Toprak, E. (2019). Rapid ultrasensitive detection platform for antimicrobial susceptibility testing. PLoS Biol. 17:e3000291. doi: 10.1371/journal.pbio.3000291
Cantón, R., Gottlieb, T., Coombs, G., Woo, P., Korman, T., Garcia-Castillo, M., et al. (2023). Antimicrobial surveillance: A 20-year history of the SMART approach to addressing global antimicrobial resistance into the future. Int. J. Antimicrob. Agents 62:107014. doi: 10.1016/j.ijantimicag.2023.107014
Caplan, A., Parent, B., Shen, M., and Plunkett, C. (2015). No time to waste—the ethical challenges created by CRISPR: CRISPR/Cas, being an efficient, simple, and cheap technology to edit the genome of any organism, raises many ethical and regulatory issues beyond the use to manipulate human germ line cells. EMBO Rep. 16, 1421–1426. doi: 10.15252/embr.201541337
Cassini, A., Högberg, L., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G., et al. (2019). Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modeling analysis. Lancet Infect. Dis. 19, 56–66. doi: 10.1016/S1473-3099(18)30605-4
Chahorm, K., and Prakitchaiwattana, C. (2018). Application of reverse transcriptase-PCR-DGGE as a rapid method for routine determination of Vibrio spp. in foods. Int. J. Food Microbiol. 264, 46–52. doi: 10.1016/j.ijfoodmicro.2017.10.014
Charnot-Katsikas, A., Tesic, V., Love, N., Hill, B., Bethel, C., Boonlayangoor, S., et al. (2018). Use of the accelerate pheno system for identification and antimicrobial susceptibility testing of pathogens in positive blood cultures and impact on time to results and workflow. J. Clin. Microbiol. 56, e1166–e1117.
Chinemerem Nwobodo, D., Ugwu, M., Oliseloke Anie, C., Al-Ouqaili, M. T., Chinedu Ikem, J., Victor Chigozie, U., et al. (2022). Antibiotic resistance: The challenges and some emerging strategies for tackling a global menace. J. Clin. Lab. Anal. 36:e24655.
Chirman, D., and Pleshko, N. (2021). Characterization of bacterial biofilm infections with Fourier transform infrared spectroscopy: A review. Appl. Spectroscopy Rev. 56, 673–701.
Church, N., and McKillip, J. (2021). Antibiotic resistance crisis: Challenges and imperatives. Biologia 76, 1535–1550.
Comini, S., Bianco, G., Boattini, M., Banche, G., Ricciardelli, G., Allizond, V., et al. (2022). Evaluation of a diagnostic algorithm for rapid identification of gram-negative species and detection of extended-spectrum β-lactamase and carbapenemase directly from blood cultures. J. Antimicrob. Chemother. 77, 2632–2641.
Comini, S., Bianco, G., Boattini, M., Iannaccone, M., Casale, R., Banche, G., et al. (2021). Evaluation of the Amplex eazyplex SuperBug acineto test for direct detection of multidrug-resistant Acinetobacter baumannii bloodstream infections in high endemicity settings. J. Hosp. Infect. 117, 179–181. doi: 10.1016/j.jhin.2021.09.015
Conwell, M., Dooley, J. S., and Naughton, P. J. (2021). A novel biofilm model system to visualise conjugal transfer of vancomycin resistance by environmental enterococci. Microorganisms. 9:789.
Costa-de-Oliveira, S., Teixeira-Santos, R., Silva, A., Pinho, E., Mergulhão, P., Silva-Dias, A., et al. (2017). Potential impact of flow cytometry antimicrobial susceptibility testing on the clinical management of gram-negative bacteremia using the FASTinov® kit. Front. Microbiol. 8:2455. doi: 10.3389/fmicb.2017.02455
Datar, R., Orenga, S., Pogorelcnik, R., Rochas, O., Simner, P., and van Belkum, A. (2022). Recent advances in rapid antimicrobial susceptibility testing. Clin. Chem. 68, 91–98.
De Been, M., Pinholt, M., Top, J., Bletz, S., Mellmann, A., Van Schaik, W., et al. (2015). Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J. Clin. Microbiol. 53, 3788–3797. doi: 10.1128/JCM.01946-15
Dhama, K., Karthik, K., Chakraborty, S., Tiwari, R., Kapoor, S., Kumar, A., et al. (2014). Loop-mediated isothermal amplification of DNA (LAMP): A new diagnostic tool lights the world of diagnosis of animal and human pathogens: A review. Pak. J. Biol. Sci. 17, 151–166. doi: 10.3923/pjbs.2014.151.166
Di Bonaventura, G., Ricci, E., Della Loggia, N., Catamo, G., and Piccolomini, R. (1998). Evaluation of the E test for antimicrobial susceptibility testing of Pseudomonas aeruginosa isolates from patients with long-term bladder catheterization. J. Clin. Microbiol. 36, 824–826. doi: 10.1128/JCM.36.3.824-826.1998
Di Resta, C., and Ferrari, M. (2018). Next generation sequencing: From research area to clinical practice. EJIFCC 29, 215–220.
Domínguez-Pérez, R., De la Torre-Luna, R., Ahumada-Cantillano, M., Vázquez-Garcidueñas, M. S., Pérez-Serrano, R. M., Martínez-Martínez, R. E., et al. (2018). Detection of the antimicrobial resistance genes blaTEM-1, cfxA, tetQ, tetM, tetW and ermC in endodontic infections of a Mexican population. J. Glob. Antimicrob. Resist. 15, 20–24. doi: 10.1016/j.jgar.2018.05.011
Donay, J., Mathieu, D., Fernandes, P., Pregermain, C., Bruel, P., Wargnier, A., et al. (2004). Evaluation of the automated Phoenix system for potential routine use in the clinical microbiology laboratory. J. Clin. Microbiol. 42, 1542–1546.
Dong, T., and Zhao, X. (2015). Rapid identification and susceptibility testing of uropathogenic microbes via immunosorbent ATP-bioluminescence assay on a microfluidic simulator for antibiotic therapy. Anal. Chem. 87, 2410–2418. doi: 10.1021/ac504428t
Dortet, L., Bréchard, L., Cuzon, G., Poirel, L., and Nordmann, P. (2014). Strategy for rapid detection of carbapenemase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 58, 2441–2445. doi: 10.1128/AAC.01239-13
dos Santos, A. C., Hondl, N., Ramos-Garcia, V., Kuligowski, J., Lendl, B., and Ramer, G. (2023). AFM-IR for nanoscale chemical characterization in life sciences: Recent developments and future directions. Measurement Sci. Au 3, 301–314. doi: 10.1021/acsmeasuresciau.3c00010
Duan, C., Cao, H., Zhang, L., and Xu, Z. (2021). Harnessing the CRISPR-Cas systems to combat antimicrobial resistance. Front. Microbiol. 12:716064. doi: 10.3389/fmicb.2021.716064
Dubourg, G., and Raoult, D. (2016). Emerging methodologies for pathogen identification in positive blood culture testing. Exp. Rev. Mol. Diag. 16, 97–111.
Dung, T., Phat, V., Vinh, C., Lan, N., Phuong, N., Ngan, L., et al. (2024). Development and validation of multiplex real-time PCR for simultaneous detection of six bacterial pathogens causing lower respiratory tract infections and antimicrobial resistance genes. BMC Infect. Dis. 24:164. doi: 10.1186/s12879-024-09028-2
Dunne, W., Westblade, L., and Ford, B. (2012). Next-generation and whole-genome sequencing in the diagnostic clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1719–1726.
Dymond, A., Davies, H., Mealing, S., Pollit, V., Coll, F., Brown, N., et al. (2020). Genomic surveillance of methicillin-resistant Staphylococcus aureus: A mathematical early modeling study of cost-effectiveness. Clin. Infect. Dis. 70, 1613–1619. doi: 10.1093/cid/ciz480
Elbehiry, A., Aldubaib, M., Abalkhail, A., Marzouk, E., ALbeloushi, A., Moussa, I., et al. (2022a). How MALDI-TOF mass spectrometry technology contributes to microbial infection control in healthcare settings. Vaccines 10:1881. doi: 10.3390/vaccines10111881
Elbehiry, A., Aldubaib, M., Al Rugaie, O., Marzouk, E., Abaalkhail, M., Moussa, I., et al. (2022b). Proteomics-based screening and antibiotic resistance assessment of clinical and subclinical Brucella species: An evolution of brucellosis infection control. PLos One 17:e0262551. doi: 10.1371/journal.pone.0262551
Elbehiry, A., Marzouk, E., Aldubaib, M., Moussa, I., Abalkhail, A., Ibrahem, M., et al. (2022c). Pseudomonas species prevalence, protein analysis, and antibiotic resistance: An evolving public health challenge. AMB Express 12, 1–14. doi: 10.1186/s13568-022-01390-1
Elbehiry, A., Marzouk, E., Hamada, M., Al-Dubaib, M., Alyamani, E., Moussa, I., et al. (2017). Application of MALDI-TOF MS fingerprinting as a quick tool for identification and clustering of foodborne pathogens isolated from food products. New Microbiol. 40, 269–278.
Elbehiry, A., Marzouk, E., Moussa, I., Anagrayyah, S., AlGhamdi, A., Alqarni, A., et al. (2023). Using protein fingerprinting for identifying and discriminating methicillin resistant Staphylococcus aureus isolates from inpatient and outpatient clinics. Diagnostics 13:2825. doi: 10.3390/diagnostics13172825
Ellington, M., Ekelund, O., Aarestrup, F., Canton, R., Doumith, M., Giske, C., et al. (2017). The role of whole-genome sequencing in antimicrobial susceptibility testing of bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 23, 2–22. doi: 10.1016/j.cmi.2016.11.012
Enroth, H., Retz, K., Andersson, S., Andersson, C., Svensson, K., Ljungström, L., et al. (2019). Evaluation of QuickFISH and maldi Sepsityper for identification of bacteria in bloodstream infection. Infect. Dis. 51, 249–258.
Erfani, Y., Rasti, A., Mirsalehian, A., Mirafshar, S., and Ownegh, V. E. - (2011). test versus disk diffusion method in determining multidrug resistant strains of Escherichia coli in urinary tract infection. Afr. J. Microbiol. Res. 5, 608–611.
European Union (2013). Commission Implementing Decision 2013/653 of 12 November 2013 as Regards a Union Financial aid Toward a Coordinated Control Plan for Antimicrobial Resistance Monitoring in Zoonotic Agents in 2014 (notified under Document C (2013) 7289). Brussels: European Union.
Eyre, D. (2022). Infection prevention and control insights from a decade of pathogen whole-genome sequencing. J. Hosp. Infect. 122, 180–186.
Fader, R., Weaver, E., Fossett, R., Toyras, M., Vanderlaan, J., Gibbs, D., et al. (2013). Multilaboratory study of the biomic automated well-reading instrument versus MicroScan WalkAway for reading MicroScan antimicrobial susceptibility and identification panels. J. Clin. Microbiol. 51, 1548–1554. doi: 10.1128/JCM.03088-12
Femerling, G., Van Oosterhout, C., Feng, S., Bristol, R., Zhang, G., Groombridge, J., et al. (2023). Genetic load and adaptive potential of a recovered avian species that narrowly avoided extinction. Mol. Biol. Evol. 40:msad256. doi: 10.1093/molbev/msad256
Fernandez-Gavela, A., Herranz, S., Chocarro, B., Falke, F., Schreuder, E., Leeuwis, H., et al. (2019). Full integration of photonic nanoimmunosensors in portable platforms for on-line monitoring of ocean pollutants. Sensors Actuators B Chem. 297:126758.
Filbrun, A., Richardson, J., Khanal, P., Tzeng, Y., and Dickson, R. (2022). Rapid, label-free antibiotic susceptibility determined directly from positive blood culture. Cytometry Part A 101, 564–576.
Fink, I., Palop, N., Salvador, R., Gómez, J., Cardona, C., and Ortega, D. (2019). Evaluation of the DNA microarray “AMR direct flow chip kit” for detection of antimicrobial resistance genes from gram-positive and gram-negative bacterial isolated colonies. Enfermedades Infecc. Microbiol. Clin. 37, 454–457. doi: 10.1016/j.eimc.2018.12.015
Fleischmann, R., Adams, M., White, O., Clayton, R., Kirkness, E., Kerlavage, A., et al. (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512. doi: 10.1126/science.7542800
Florio, W., Baldeschi, L., Rizzato, C., Tavanti, A., Ghelardi, E., and Lupetti, A. (2020). Detection of antibiotic-resistance by MALDI-TOF mass spectrometry: An expanding area. Front. Cell. Infect. Microbiol. 10:572909. doi: 10.3389/fcimb.2020.572909
Fongang, H., Mbaveng, A., and Kuete, V. (2023). Global burden of bacterial infections and drug resistance. Adv. Bot. Res. 106, 1–20.
Frickmann, H., Masanta, W., and Zautner, A. (2014). Emerging rapid resistance testing methods for clinical microbiology laboratories and their potential impact on patient management. BioMed Res. Int. 2014:375681.
Frickmann, H., Zautner, A., Moter, A., Kikhney, J., Hagen, R., Stender, H., et al. (2017). Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: A review. Crit. Rev. Microbiol. 43, 263–293.
Funke, G., and Funke-Kissling, P. (2004). Use of the BD PHOENIX automated microbiology system for direct identification and susceptibility testing of gram-negative rods from positive blood cultures in a three-phase trial. J. Clin. Microbiol. 42, 1466–1470. doi: 10.1128/JCM.42.4.1466-1470.2004
Funke, G., Monnet, D., deBernardis, C., von Graevenitz, A., and Freney, J. (1998). Evaluation of the VITEK 2 system for rapid identification of medically relevant gram-negative rods. J. Clin. Microbiol. 36, 1948–1952.
Gajic, I., Kabic, J., Kekic, D., Jovicevic, M., Milenkovic, M., Mitic Culafic, D., et al. (2022). Antimicrobial susceptibility testing: A comprehensive review of currently used methods. Antibiotics 11:427.
Galhano, B., Ferrari, R., Panzenhagen, P., de Jesus, A., and Conte-Junior, C. (2021). Antimicrobial resistance gene detection methods for bacteria in animal-based foods: A brief review of highlights and advantages. Microorganisms 9:923. doi: 10.3390/microorganisms9050923
Gao, H., Li, Y., Li, Y., Qu, K., Zhang, K., and Li, J. (2023). Detection of antibiotic-resistance genes in bacterial pathogens using a Cas12a/3D DNAzyme colorimetric paper sensor. Fundamental Res. 14:31.
Giordano, C., Piccoli, E., Brucculeri, V., and Barnini, S. (2018). A prospective evaluation of two rapid phenotypical antimicrobial susceptibility technologies for the diagnostic stewardship of sepsis. BioMed Res. Int. 2018:6976923. doi: 10.1155/2018/6976923
Giuffrida, M., and Spoto, G. (2017). Integration of isothermal amplification methods in microfluidic devices: Recent advances. Biosensors Bioelectronics. 90, 174–186.
Glökler, J., Lim, T., Ida, J., and Frohme, M. (2021). Isothermal amplifications–A comprehensive review on current methods. Crit. Rev. Biochem. Mol. Biol. 56, 543–586. doi: 10.1080/10409238.2021.1937927
Godeux, A., Lupo, A., Haenni, M., Guette-Marquet, S., Wilharm, G., Laaberki, M., et al. (2018). Fluorescence-based detection of natural transformation in drug-resistant Acinetobacter baumannii. J. Bacteriol. 200: e00181-18. doi: 10.1128/jb.
Golding, S., Ogden, J., and Higgins, H. (2019). Shared goals, different barriers: A qualitative study of UK veterinarians’ and farmers’ beliefs about antimicrobial resistance and stewardship. Front. Vet. Sci. 6:132. doi: 10.3389/fvets.2019.00132
Goodwin, S., McPherson, J., and McCombie, W. (2016). Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351. doi: 10.1038/nrg.2016.49
Gould, K. (2016). Antibiotics: From prehistory to the present day. J. Antimicrob. Chemother. 71, 572–575. doi: 10.1093/jac/dkv484
Griffin, P., Price, G., Schooneveldt, J., Schlebusch, S., Tilse, M., Urbanski, T., et al. (2012). Use of matrix-assisted laser desorption ionization–time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J. Clin. Microbiol. 50, 2918–2931.
Gulumbe, B., Haruna, U., Almazan, J., Ibrahim, I., Faggo, A., and Bazata, A. (2022). Combating the menace of antimicrobial resistance in Africa: A review on stewardship, surveillance and diagnostic strategies. Biol. Procedures Online 24, 1–13. doi: 10.1186/s12575-022-00182-y
Hahn, A., Podbielski, A., Meyer, T., Zautner, A., Loderstädt, U., Schwarz, N., et al. (2020). On detection thresholds–a review on diagnostic approaches in the infectious disease laboratory and the interpretation of their results. Acta Trop. 205:105377. doi: 10.1016/j.actatropica.2020.105377
Han, R., Guo, Y., Peng, M., Shi, Q., Wu, S., Yang, Y., et al. (2021). Evaluation of the immunochromatographic NG-Test Carba 5, RESIST-5 OOKNV, and IMP K-SeT for rapid detection of KPC-, NDM-, IMP-, VIM-type, and OXA-48-like carbapenemase among Enterobacterales. Front. Microbiol. 11:609856. doi: 10.3389/fmicb.2020.609856
Hedberg, M. (2005). Antibiotic sensitivity testing of anaerobic bacteria–Workshop arranged by ESGARAB. Clin. Microbiol. Infect. 11:2.
Hernández-Durán, M., López-Jácome, L., Colín-Castro, C., Cerón-González, G., Ortega-Peña, S., Vanegas-Rodríguez, E., et al. (2017). Comparison of the MicroScan WalkAway and VITEK 2 compact systems for the identification and susceptibility of clinical gram-positive and gram-negative bacteria. Invest. Discapacidad. 6, 105–114.
Herrera-Rodriguez, S., Elizondo-Quiroga, D., and Alvarez-Maya, I. (2013). Infectious diseases detection by microarray: An overview of clinical relevant infections. J. Biomed. Sci. Eng. 6, 1006–1013.
Hirakata, Y., Matsuda, J., Nakano, M., Hayashi, T., Tozaka, S., Takezawa, T., et al. (2005). Evaluation of the BD phoenix automated microbiology system SMIC/ID panel for identification and antimicrobial susceptibility testing of Streptococcus spp. Diagnostic Microbiol. Infect. Dis. 53, 169–173. doi: 10.1016/j.diagmicrobio.2005.06.003
Hu, X., Logue, M., and Robinson, N. (2020). Antimicrobial resistance is a global problem–a UK perspective. Eur. J. Integr. Med. 36:101136. doi: 10.1016/j.eujim.2020.101136
Hung, J., and Weng, Z. (2017). Analysis of microarray and RNA-seq expression profiling data. Cold Spring Harbor Protoc. 2017.
Hunt, D., and Kates, O. S. A. (2024). Brief history of antimicrobial resistance. AMA J. Ethics 26, 408–417.
Iketleng, T., Lessells, R., Dlamini, M., Mogashoa, T., Mupfumi, L., Moyo, S., et al. (2018). Mycobacterium tuberculosis next-generation whole-genome sequencing: Opportunities and challenges. Tuberculosis Res. Treat. 2018:1298542. doi: 10.1155/2018/1298542
Inglis, T., Paton, T., Kopczyk, M., Mulroney, K., and Carson, C. (2020). Same-day antimicrobial susceptibility test using acoustic-enhanced flow cytometry visualized with supervised machine learning. J. Med. Microbiol. 69, 657–669. doi: 10.1099/jmm.0.001092
Jaksik, R., Iwanaszko, M., Rzeszowska-Wolny, J., and Kimmel, M. (2015). Microarray experiments and factors which affect their reliability. Biol. Dir. 10, 1–14.
Javed, M., Sadaf, M., Ahmed, T., Jamil, A., Nawaz, M., Abbas, H., et al. (2018). CRISPR-Cas system: History and prospects as a genome editing tool in microorganisms. Curr. Microbiol. 75, 1675–1683. doi: 10.1007/s00284-018-1547-4
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Johnson, R., Newhall, W., Papp, J., Knapp, J., Black, C., Gift, T., et al. (2002). Screening tests to detect Chlamydia trachomatis and Neisseria gonorrheae infections—2002. MMWR Recomm. Rep. 51, 1–38.
Kállai, A., Kelemen, M., Molnár, N., Tropotei, A., Hauser, B., Iványi, Z., et al. (2021). MICy: A novel flow cytometric method for rapid determination of minimal inhibitory concentration. Microbiol. Spectr. 9, e901–e921. doi: 10.1128/spectrum.00901-21
Kanzi, A., San, J., Chimukangara, B., Wilkinson, E., Fish, M., Ramsuran, V., et al. (2020). Next generation sequencing and bioinformatics analysis of family genetic inheritance. Front. Genet. 11:544162. doi: 10.3389/fgene.2020.544162
Kaprou, G., Bergšpica, I., Alexa, E., Alvarez-Ordóñez, A., and Prieto, M. (2021). Rapid methods for antimicrobial resistance diagnostics. Antibiotics 10:209.
Kasas, S., Malovichko, A., Villalba, M., Vela, M., Yantorno, O., and Willaert, R. (2021). Nanomotion detection-based rapid antibiotic susceptibility testing. Antibiotics 10:287. doi: 10.3390/antibiotics10030287
Katiyar, A., Sharma, P., Dahiya, S., Singh, H., Kapil, A., and Kaur, P. (2020). Genomic profiling of antimicrobial resistance genes in clinical isolates of Salmonella Typhi from patients infected with Typhoid fever in India. Sci. Rep. 10:8299. doi: 10.1038/s41598-020-64934-0
Khan, M., Thurgood, N., Faheem, S., Rais, N., Ansari, M., Kaleem, S., et al. (2020). Occurrence of extended spectrum beta-lactamase gram-negative bacteria from nonclinical sources in Dubai, United Arab Emirates. Water 12:2562.
Khan, Z., Siddiqui, M., and Park, S. (2019). Current and emerging methods of antibiotic susceptibility testing. Diagnostics 9:49.
Kitao, T., Miyoshi-Akiyama, T., Shimada, K., Tanaka, M., Narahara, K., Saito, N., et al. (2010). Development of an immunochromatographic assay for the rapid detection of AAC (6’)-Iae-producing multidrug-resistant Pseudomonas aeruginosa. J. Antimicrob. Chemother. 65, 1382–1386. doi: 10.1093/jac/dkq148
Kitchin, P., and Bootman, J. (1993). Quality control of the polymerase chain reaction. Rev. Med. Microbiol. 3, 107–114.
Kohl, T., Diel, R., Harmsen, D., Rothgänger, J., Walter, K., Merker, M., et al. (2014). Whole-genome-based Mycobacterium tuberculosis surveillance: A standardized, portable, and expandable approach. J. Clin. Microbiol. 52, 2479–2486. doi: 10.1128/JCM.00567-14
Kowalska-Krochmal, B., and Dudek-Wicher, R. (2021). The minimum inhibitory concentration of antibiotics: Methods, interpretation, clinical relevance. Pathogens 10:165.
Lange, C., Schubert, S., Jung, J., Kostrzewa, M., and Sparbier, K. (2014). Quantitative matrix-assisted laser desorption ionization–time of flight mass spectrometry for rapid resistance detection. J. Clin. Microbiol. 52, 4155–4162.
Lee, S., Park, S. M., Kim, B. N., Kwon, O. S., Rho, W.-Y., and Jun, B.-H. (2019). Emerging ultrafast nucleic acid amplification technologies for next-generation molecular diagnostics. Biosensors Bioelectronics 141:111448. doi: 10.1016/j.bios.2019.111448
Leonard, H., Colodner, R., Halachmi, S., and Segal, E. (2018). Recent advances in the race to design a rapid diagnostic test for antimicrobial resistance. ACS Sensors 3, 2202–2217. doi: 10.1021/acssensors.8b00900
Lewis, I., and James, S. (2022). Performance Standards for Antimicrobial Susceptibility Testing, 35th Edn. Berwyn, PA: CLSI, 428.
Li, L., Wang, C., Nie, Y., Yao, B., and Hu, H. (2020). Nanofabrication enabled lab-on-a-chip technology for the manipulation and detection of bacteria. TrAC Trends Anal. Chem. 127:115905.
Li, M. (2023). Combining atomic force microscopy with complementary techniques for multidimensional single-cell analysis. J. Microscopy 290, 69–96. doi: 10.1111/jmi.13183
Li, Y., Yang, X., and Zhao, W. (2017). Emerging microtechnologies and automated systems for rapid bacterial identification and antibiotic susceptibility testing. SLAS Technol. Transl. Life Sci. Innov. 22, 585–608. doi: 10.1177/2472630317727519
Ligozzi, M., Bernini, C., Bonora, M., De Fatima, M., Zuliani, J., and Fontana, R. (2002). Evaluation of the VITEK 2 system for identification and antimicrobial susceptibility testing of medically relevant gram-positive cocci. J. Clin. Microbiol. 40, 1681–1686. doi: 10.1128/JCM.40.5.1681-1686.2002
Lin, X., Yang, Y., Melton, P., Singh, V., Simpson-Yap, S., Burdon, K., et al. (2022). Integrating genetic structural variations and whole-genome sequencing into clinical neurology. Neurol. Genet. 8:e200005.
Ling, D., Flores, L., Riley, L., and Pai, M. (2008). Commercial nucleic-acid amplification tests for diagnosis of pulmonary tuberculosis in respiratory specimens: Meta-analysis and meta-regression. PLoS One 3:e1536. doi: 10.1371/journal.pone.0001536
Ling, T., Liu, Z., and Cheng, A. (2003). Evaluation of the VITEK 2 system for rapid direct identification and susceptibility testing of gram-negative bacilli from positive blood cultures. J. Clin. Microbiol. 41, 4705–4707.
Lobanovska, M., and Pilla, G. (2017). Focus: Drug development: Penicillin’s discovery and antibiotic resistance: Lessons for the future? Yale J. Biol. Med. 90, 135–145.
López-Hernández, I., López-Cerero, L., Fernández-Cuenca, F., and Pascual, Á (2022). The role of the microbiology laboratory in the diagnosis of multidrug-resistant gram-negative bacilli infections. The importance of the determination of resistance mechanisms. Med. Intensiva 46, 455–464. doi: 10.1016/j.medine.2022.05.003
MacLean, E., Kohli, M., Weber, S., Suresh, A., Schumacher, S., Denkinger, C., et al. (2020). Advances in molecular diagnosis of tuberculosis. J. Clin. Microbiol. 58:e01582-19.
Magill, S., O’Leary, E., Janelle, S., Thompson, D., Dumyati, G., Nadle, J., et al. (2018). Changes in prevalence of health care–associated infections in US hospitals. New Engl. J. Med. 379, 1732–1744.
Majumder, M., Rahman, S., Cohall, D., Bharatha, A., Singh, K., Haque, M., et al. (2020). Antimicrobial stewardship: Fighting antimicrobial resistance and protecting global public health. Infect. Drug Resist. 13, 4713–4738.
Manghwar, H., Lindsey, K., Zhang, X., and Jin, S. (2019). CRISPR/Cas system: Recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125.
MarketResearch.com. (2021). Antimicrobial Susceptibility Testing Market by Product (Manual Testing, Automated AST, Consumable), Method (Disk Diffusion, Dilution), Application (Clinical Diagnosis, Epidemiology), End User (Diagnostic Laboratories, Hospital)—Forecast to 2027. Available online at: https://www.marketresearch.com/MarketsandMarkets-v3719/Antimicrobial-Susceptibility-Testing-Product-Manual-33730279/ (accessed January 15, 2021).
MarketsandMarkets™. (2021). Antimicrobial Susceptibility Testing Market by Product (Manual, Automated Susceptibility Testing System), Type (Antibacterial, Antifungal), Application (Clinical Diagnostics), Method (ETEST, Disk Diffusion), End-User—Global Forecasts to 2025. Available online at: https://www.marketresearch.com/Business-Research-Company-v4006/Antimicrobial-Susceptibility-Testing-Global-39316175/ (accessed January 15, 2021).
Martins-Oliveira, I., Pérez-Viso, B., Quintas, S., Silva-Dias, A., Gomes, R., Rodrigues, A., et al. (2020). Evaluation of ultrarapid susceptibility testing of ceftolozane-tazobactam by a flow cytometry assay directly from positive blood cultures. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1907–1914. doi: 10.1007/s10096-020-03926-4
Marutescu, L. (2023). Current and future flow cytometry applications contributing to antimicrobial resistance control. Microorganisms 11:1300. doi: 10.3390/microorganisms11051300
Mashhadikhan, S., Amooghin, A., Sanaeepur, H., and Shirazi, M. M. A. (2022). A critical review on cadmium recovery from wastewater toward environmental sustainability. Desalination 535:115815.
Maugeri, G., Lychko, I., Sobral, R., and Roque, A. (2019). Identification and antibiotic-susceptibility profiling of infectious bacterial agents: A review of current and future trends. Biotechnol. J. 14:1700750. doi: 10.1002/biot.201700750
Mayorga-Ramos, A., Zúñiga-Miranda, J., Carrera-Pacheco, S., Barba-Ostria, C., and Guamán, L. P. (2023). CRISPR-Cas-based antimicrobials: Design, challenges, and bacterial mechanisms of resistance. ACS Infect. Dis. 9, 1283–1302. doi: 10.1021/acsinfecdis.2c00649
Miller, S., Ferreira, J., and LeJeune, J. (2022). Antimicrobial use and resistance in plant agriculture: A one health perspective. Agriculture 12:289.
Miller, W., and Arias, C. A. (2024). ESKAPE pathogens: Antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat. Rev. Microbiol. 22, 598–616.
Moguet, C., Gonzalez, C., Sallustrau, A., Gelhaye, S., Naas, T., Simon, S., et al. (2022). Detection of expanded-spectrum cephalosporin hydrolysis by lateral flow immunoassay. Microb. Biotechnol. 15, 603–612.
Müller, V., Rajer, F., Frykholm, K., Nyberg, L., Quaderi, S., Fritzsche, J., et al. (2016). Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci. Rep. 6:37938. doi: 10.1038/srep37938
Mulroney, K., Kopczyk, M., Carson, C., Paton, T., Inglis, T., and Chakera, A. (2022). Same-day confirmation of infection and antimicrobial susceptibility profiling using flow cytometry. EBioMedicine 82:104145. doi: 10.1016/j.ebiom.2022.104145
Murray, C., Ikuta, K., Sharara, F., Swetschinski, L., Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629–655.
Narasimhan, V., Kim, H., Lee, S., Kang, H., Siddique, R., Park, H., et al. (2023). Nucleic acid amplification-based technologies (NAAT)—toward accessible, autonomous, and mobile diagnostics. Adv. Mater. Technol. 8:2300230.
Nauclér, P., Huttner, A., Van Werkhoven, C., Singer, M., Tattevin, P., Einav, S., et al. (2021). Impact of time to antibiotic therapy on clinical outcome in patients with bacterial infections in the emergency department: Implications for antimicrobial stewardship. Clin. Microbiol. Infect. 27, 175–181. doi: 10.1016/j.cmi.2020.02.032
Naylor, N., Atun, R., Zhu, N., Kulasabanathan, K., Silva, S., Chatterjee, A., et al. (2018). Estimating the burden of antimicrobial resistance: A systematic literature review. Antimicrob. Resistance Infect. Control. 7, 1–17.
Needs, S., Donmez, S., Bull, S., McQuaid, C., Osborn, H., and Edwards, A. (2020). Challenges in microfluidic and point-of-care phenotypic antimicrobial resistance tests. Front. Mech. Eng. 6:73. doi: 10.3389/fmech.2020.00073
Neil, J., Verma, A., Kronewitter, S., McGee, W., Mullen, C., Viirtola, M., et al. (2021). Rapid MRSA detection via tandem mass spectrometry of the intact 80 kDa PBP2a resistance protein. Sci. Rep. 11:18309. doi: 10.1038/s41598-021-97844-w
Notake, S., Matsuda, M., Tamai, K., Yanagisawa, H., Hiramatsu, K., and Kikuchi, K. (2013). Detection of IMP metallo-β-lactamase in carbapenem-nonsusceptible Enterobacteriaceae and non-glucose-fermenting gram-negative rods by immunochromatography assay. J. Clin. Microbiol. 51, 1762–1768.
Novais, Â, Freitas, A. R., Rodrigues, C., and Peixe, L. (2019). Fourier transform infrared spectroscopy: Unlocking fundamentals and prospects for bacterial strain typing. Eur. J. Clin. Microbiol. Infect. Dis. 38, 427–448. doi: 10.1007/s10096-018-3431-3
Nsofor, C. A. (2014). DNA microarrays and their applications in medical microbiology. Biotechnol. Mol. Biol. Rev. 9, 1–11.
Nyblom, M., Johnning, A., Frykholm, K., Wrande, M., Müller, V., Goyal, G., et al. (2023). Strain-level bacterial typing directly from patient samples using optical DNA mapping. Commun. Med. 3:31. doi: 10.1038/s43856-023-00259-z
Okaiyeto, S., Sutar, P., Chen, C., Ni, J., Wang, J., Mujumdar, A., et al. (2024). Antibiotic resistant bacteria in food systems: Current status, resistance mechanisms, and mitigation strategies. Agric. Commun. 2:100027.
Oliveira, B., Veigas, B., and Baptista, P. (2021). Isothermal amplification of nucleic acids: The race for the next “gold standard”. Front. Sens. 2:752600. doi: 10.3389/fsens.2021.752600
Osa, M., Belo, M., Dela Merced, Z., Villanueva, A., Mauhay, J., Celis, A., et al. (2021). Performance of MALDI–TOF mass spectrometry in the Philippines. Trop. Med. Infect. Dis. 6:112.
Oshiro, S., Tada, T., Kameoka, Y., Suzuki, K., Ohmagari, N., Miyoshi-Akiyama, T., et al. (2015). Development and evaluation of immunochromatography to detect gram-negative bacteria producing ArmA 16S rRNA methylase responsible for aminoglycoside resistance. J. Microbiol. Methods 118, 159–163. doi: 10.1016/j.mimet.2015.09.005
Oueslati, S., Volland, H., Cattoir, V., Bernabeu, S., Girlich, D., Dulac, D., et al. (2021). Development and validation of a lateral flow immunoassay for rapid detection of VanA-producing enterococci. J. Antimicrob. Chemother. 76, 146–151. doi: 10.1093/jac/dkaa413
Pak, T., Young, J., McKenna, C., Agan, A., DelloStritto, L., Filbin, M., et al. (2023). Risk of misleading conclusions in observational studies of time-to-antibiotics and mortality in suspected sepsis. Clin. Infect. Dis. 77, 1534–1543. doi: 10.1093/cid/ciad450
Papadakis, G., Pantazis, A., Ntogka, M., Parasyris, K., Theodosi, G., Kaprou, G., et al. (2019). 3D-printed point-of-care platform for genetic testing of infectious diseases directly in human samples using acoustic sensors and a smartphone. ACS Sensors 4, 1329–1336. doi: 10.1021/acssensors.9b00264
Patel, R. (2022). Advances in testing for infectious diseases—looking back and projecting forward. Clin. Chem. 68, 10–15. doi: 10.1093/clinchem/hvab110
Patel, T., Kaakeh, R., Nagel, J., Newton, D., and Stevenson, J. (2017). Cost analysis of implementing matrix-assisted laser desorption ionization–time of flight mass spectrometry plus real-time antimicrobial stewardship intervention for bloodstream infections. J. Clin. Microbiol. 55, 60–67. doi: 10.1128/JCM.01452-16
Pauling, L., Itano, H., Singer, S., and Wells, I. (1949). Sickle cell anemia, a molecular disease. Science 110, 543–548.
Peñalva-Moreno, G., Crespo-Robledo, P., Molvik, M., López-Navas, A., Kacelnik, O., and Cisneros, J. M. (2022). A step forward in antibiotic use and resistance monitoring: A quarterly surveillance system pilot in 11 European Union/European Economic Area countries, September 2017 to May 2020. Euro Surveill. 27:2200082. doi: 10.2807/1560-7917.ES.2022.27.46.2200082
Perez-Sepulveda, B., Heavens, D., Pulford, C., Predeus, A., Low, R., Webster, H., et al. (2021). An accessible, efficient and global approach for the large-scale sequencing of bacterial genomes. Genome Biol. 22, 1–18.
Pishnian, Z., Haeili, M., and Feizi, A. (2019). Prevalence and molecular determinants of colistin resistance among commensal Enterobacteriaceae isolated from poultry in northwest of Iran. Gut Pathog. 11, 1–8. doi: 10.1186/s13099-019-0282-0
Pursey, E., Sünderhauf, D., Gaze, W., Westra, E., and van Houte, S. (2018). CRISPR-Cas antimicrobials: Challenges and future prospects. PLoS Pathogens 14:e1006990. doi: 10.1371/journal.ppat.1006990
Purushothaman, S., Meola, M., and Egli, A. (2022). Combination of whole-genome sequencing and metagenomics for microbiological diagnostics. Int. J. Mol. Sci. 23:9834.
Puttaswamy, S., Gupta, S., Regunath, H., Smith, L., and Sengupta, S. (2018). A comprehensive review of the present and future antibiotic susceptibility testing (AST) systems. Arch. Clin. Microbiol. 9:83.
Qin, D. (2019). Next-generation sequencing and its clinical application. Cancer Biol. Med. 16, 4–10.
Qin, K., Zhang, P., and Li, Z. (2023). Specific detection of antibiotic-resistant bacteria using CRISPR/Cas9 induced isothermal exponential amplification reaction (IEXPAR). Talanta 253:124045.
Rabiee, M., Rostami, A., Rabiee, N., and Bagherzadeh, M. (2021). Microarray Technologies. Biomedical Applications of Microfluidic Devices. Amsterdam: Elsevier, 77–98.
Rajapaksha, P., Elbourne, A., Gangadoo, S., Brown, R., Cozzolino, D., and Chapman, J. (2019). A review of methods for the detection of pathogenic microorganisms. Analyst 144, 396–411.
Raro, O., Bouvier, M., Kerbol, A., Poirel, L., and Nordmann, P. (2024). MultiRapid ATB NP test for detecting concomitant susceptibility and resistance of last-resort novel antibiotics available to treat multidrug-resistant Enterobacterales infections. Int. J. Antimicrob. Agents 64:107206. doi: 10.1016/j.ijantimicag.2024.107206
Reita, D., Pabst, L., Pencreach, E., Guérin, E., Dano, L., Rimelen, V., et al. (2021). Molecular mechanism of EGFR-TKI resistance in EGFR-mutated non-small cell lung cancer: Application to biological diagnostic and monitoring. Cancers 13:4926. doi: 10.3390/cancers13194926
Reller, L., Weinstein, M., Jorgensen, J., and Ferraro, M. (2009). Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 49, 1749–1755.
Rentschler, S., Kaiser, L., and Deigner, H. (2021). Emerging options for the diagnosis of bacterial infections and the characterization of antimicrobial resistance. Int. J. Mol. Sci. 22:456.
Rhoads, D., Wang, H., Karichu, J., and Richter, S. (2016). The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diag. Microbiol. Infect. Dis. 86, 257–261. doi: 10.1016/j.diagmicrobio.2016.08.001
Robinson, J., Ostafe, R., Iyengar, S., Rajwa, B., and Fischer, R. (2023). Flow cytometry: The next revolution. Cells 12:1875.
Rodrigues, P., Ferrari, R., and Conte-Junior, C. (2018). Application of molecular tools to elucidate the microbiota of seafood. J. Appl. Microbiol. 124, 1347–1365. doi: 10.1111/jam.13701
Rodríguez-Sánchez, B., Cercenado, E., Coste, A., and Greub, G. (2019). Review of the impact of MALDI-TOF MS in public health and hospital hygiene, 2018. Eurosurveillance 24:1800193. doi: 10.2807/1560-7917.ES.2019.24.4.1800193
Roos, B., Van Cleeff, M., Githui, W., Kivihya-Ndugga, L., Odhiambo, J., Kibuga, D., et al. (1998). Cost-effectiveness of the polymerase chain reaction versus smear examination for the diagnosis of tuberculosis in Kenya: A theoretical model. Int. J. Tuberculosis Lung Dis. 2, 235–241.
Sahoo, R., Jadhav, S., and Nema, V. (2023). Journey of technological advancements in the detection of antimicrobial resistance. J. Formosan Med. Assoc. 123, 430–441. doi: 10.1016/j.jfma.2023.08.008
Salam, M., Al-Amin, M., Pawar, J., Akhter, N., and Lucy, I. (2023a). Conventional methods and future trends in antimicrobial susceptibility testing. Saudi J. Biol. Sci. 30:103582.
Salam, M., Al-Amin, M., Salam, M., Pawar, J., Akhter, N., Rabaan, A., et al. (2023b). Antimicrobial resistance: A growing serious threat for global public. Healthcare 11:1946.
Salimnia, H., Fairfax, M., Lephart, P., Morgan, M., Gilbreath, J., Butler-Wu, S., et al. (2014). An international, prospective, multicenter evaluation of the combination of AdvanDx Staphylococcus Quick FISH BC with mecA Xpress FISH for detection of methicillin-resistant Staphylococcus aureus isolates from positive blood cultures. J. Clin. Microbiol. 52, 3928–3932. doi: 10.1128/JCM.01811-14
Salman, A., Sharaha, U., Rodriguez-Diaz, E., Shufan, E., Riesenberg, K., Bigio, I., et al. (2017). Detection of antibiotic resistant Escherichia Coli bacteria using infrared microscopy and advanced multivariate analysis. Analyst 142, 2136–2144. doi: 10.1039/c7an00192d
Salyers, A., and Amabile-Cuevas, C. (1997). Why are antibiotic resistance genes so resistant to elimination? Antimicrob. Agents Chemother. 41, 2321–2325.
Sanguinetti, M., Posteraro, B., Spanu, T., Ciccaglione, D., Romano, L., Fiori, B., et al. (2003). Characterization of clinical isolates of Enterobacteriaceae from Italy by the BD Phoenix extended-spectrum β-lactamase detection method. J. Clin. Microbiol. 41, 1463–1468.
Satam, H., Joshi, K., Mangrolia, U., Waghoo, S., Zaidi, G., Rawool, S., et al. (2023). Next-generation sequencing technology: Current trends and advancements. Biology 12:997.
Sejas, L., Silbert, S., Reis, A., and Sader, H. (2003). Avaliação da qualidade dos discos com antimicrobianos para testes de disco-difusão disponíveis comercialmente no Brasil. J. Bras. Patol. Med. Lab. 39, 27–35.
Serweciñska, L. (2020). Antimicrobials and antibiotic-resistant bacteria: A risk to the environment and to public health. Water 12:3313.
Shanmugakani, R., Srinivasan, B., Glesby, M., Westblade, L., Cárdenas, W., Raj, T., et al. (2020). Current state of the art in rapid diagnostics for antimicrobial resistance. Lab. Chip. 20, 2607–2625.
Sharaha, U., Rodriguez-Diaz, E., Riesenberg, K., Bigio, I., Huleihel, M., and Salman, A. (2017). Using infrared spectroscopy and multivariate analysis to detect antibiotics’ resistant Escherichia coli bacteria. Anal. Chem. 89, 8782–8790. doi: 10.1021/acs.analchem.7b01025
She, R., and Bender, J. (2019). Advances in rapid molecular blood culture diagnostics: Healthcare impact, laboratory implications, and multiplex technologies. J. Appl. Lab. Med. 3, 617–630. doi: 10.1373/jalm.2018.027409
Sherry, N., Porter, J., Seemann, T., Watkins, A., Stinear, T., and Howden, B. (2013). Outbreak investigation using high-throughput genome sequencing within a diagnostic microbiology laboratory. J. Clin. Microbiol. 51, 1396–1401. doi: 10.1128/JCM.03332-12
Shin, D., Andini, N., Hsieh, K., Yang, S., and Wang, T. (2019). Emerging analytical techniques for rapid pathogen identification and susceptibility testing. Annu. Rev. Anal. Chem. 12, 41–67.
Sigmund, I., Renz, N., Feihl, S., Morgenstern, C., Cabric, S., and Trampuz, A. (2020). Value of multiplex PCR for detection of antimicrobial resistance in samples retrieved from patients with orthopedic infections. BMC Microbiol. 20:88. doi: 10.1186/s12866-020-01741-7
Sigmund, I., Windhager, R., Sevelda, F., Staats, K., Puchner, S., Stenicka, S., et al. (2019). Multiplex PCR Unyvero i60 ITI application improves detection of low-virulent microorganisms in periprosthetic joint infections. Int. Orthopedics 43, 1891–1898. doi: 10.1007/s00264-018-4136-z
Silva, D. F., Silva-Dias, A., Gomes, R., Martins-Oliveira, I., Ramos, M., Rodrigues, A., et al. (2019). Evaluation of rapid colistin susceptibility directly from positive blood cultures using a flow cytometry assay. Int. J. Antimicrob. Agents 54, 820–823. doi: 10.1016/j.ijantimicag.2019.08.016
Silva-Dias, A., Pérez-Viso, B., Martins-Oliveira, I., Gomes, R., Rodrigues, A., Cantón, R., et al. (2021). Evaluation of FASTinov ultrarapid flow cytometry antimicrobial susceptibility testing directly from positive blood cultures. J. Clin. Microbiol. 59:e0054421. 00544-21 doi: 10.1128/jcm
Singer, A., Shaw, H., Rhodes, V., and Hart, A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7:1728. doi: 10.3389/fmicb.2016.01728
Singh, R., Bhatia, M., Anusha, K., Singh, V., Omar, B., and Gupta, P. (2019). Comparative evaluation of microscan walkaway 96 plus ID/AST system and mikrolatest broth microdilution kit in assessing In vitro colistin susceptibility of carbapenem-resistant clinical gram-negative bacterial isolates: Experience from a tertiary care teaching hospital in Rishikesh. Uttarakhand. Ind. J. Med. Microbiol. 37, 502–508. doi: 10.4103/ijmm.IJMM_19_437
Singh, S., Numan, A., and Cinti, S. (2021). Point-of-care for evaluating antimicrobial resistance through the adoption of functional materials. Anal. Chem. 94, 26–40. doi: 10.1021/acs.analchem.1c03856
Smelikova, E., Tkadlec, J., and Krutova, M. (2022). How to: Screening for mcr-mediated resistance to colistin. Clin. Microbiol. Infect. 28, 43–50.
Smetana, J., and Brož, P. (2022). National genome initiatives in Europe and the United Kingdom in the era of whole-genome sequencing: A comprehensive review. Genes 13:556. doi: 10.3390/genes13030556
Song, Y., Dou, F., He, S., Zhou, Y., and Liu, Q. (2019). Laboratory and clinical evaluation of DNA microarray for the detection of carbapenemase genes in gram-negative bacteria from hospitalized patients. BioMed Res. Int. 2019:8219748.
Sparbier, K., Schubert, S., and Kostrzewa, M. (2016). MBT-ASTRA A suitable tool for fast antibiotic susceptibility testing? Methods 104, 48–54. doi: 10.1016/j.ymeth.2016.01.008
Srivastava, P., and Prasad, D. (2023). Isothermal nucleic acid amplification and its uses in modern diagnostic technologies. 3 Biotech 13:200. doi: 10.1007/s13205-023-03628-6
Subramaniam, G., and Girish, M. (2020). Antibiotic resistance—A cause for reemergence of infections. Indian J. Pediatrics 87, 937–944. doi: 10.1007/s12098-019-03180-3
Subsoontorn, P., Lohitnavy, M., and Kongkaew, C. (2020). The diagnostic accuracy of isothermal nucleic acid point-of-care tests for human coronaviruses: A systematic review and meta-analysis. Sci. Rep. 10:22349. doi: 10.1038/s41598-020-79237-7
Swidsinski, A. (2006). Standards for bacterial identification by fluorescence in situ hybridization within eukaryotic tissue using ribosomal rRNA-based probes. Inflammatory Bowel Dis. 12, 824–826. doi: 10.1097/00054725-200608000-00018
Swidsinski, A., Guschin, A., Corsini, L., Loening-Baucke, V., Tisakova, L., Swidsinski, S., et al. (2022). Antimicrobial susceptibility of microbiota in bacterial vaginosis using fluorescence in situ hybridization. Pathogens 11:456. doi: 10.3390/pathogens11040456
Syal, K., Mo, M., Yu, H., Iriya, R., Jing, W., Guodong, S., et al. (2017). Current and emerging techniques for antibiotic susceptibility tests. Theranostics 7:1795.
Tada, T., Sekiguchi, J., Watanabe, S., Kuwahara-Arai, K., Mizutani, N., Yanagisawa, I., et al. (2019). Assessment of a newly developed immunochromatographic assay for NDM-type metallo-β-lactamase producing gram-negative pathogens in Myanmar. BMC Infect. Dis. 19:565. doi: 10.1186/s12879-019-4147-4.
Tangcharoensathien, V., Chanvatik, S., and Sommanustweechai, A. (2018). Complex determinants of inappropriate use of antibiotics. Bull. World Health Organ. 96, 141–144.
Tao, S., Chen, H., Li, N., and Liang, W. (2022). The application of the CRISPR-Cas system in antibiotic resistance. Infect. Drug Resist. 15, 4155–4168.
Theakstone, A., Rinaldi, C., Butler, H., Cameron, J., Confield, L., Rutherford, S., et al. (2021). Fourier-transform infrared spectroscopy of biofluids: A practical approach. Transl. Biophotonics 3:e202000025. doi: 10.1002/jbio.201400018
Theel, E., and Schuetz, A. (2022). Interacting with the Clinical Microbiology Laboratory. A Rational Approach to Clinical Infectious Diseases. Amsterdam: Elsevier, 12–33.
Trotter, A., Aydin, A., Strinden, M., and O’grady, J. (2019). Recent and emerging technologies for the rapid diagnosis of infection and antimicrobial resistance. Curr. Opin. Microbiol. 51, 39–45.
Trovato, V., Sfameni, S., Rando, G., Rosace, G., Libertino, S., Ferri, A., et al. (2022). A review of stimuli-responsive smart materials for wearable technology in healthcare: Retrospective, perspective, and prospective. Molecules 27:5709. doi: 10.3390/molecules27175709
Tsougeni, K., Kastania, A., Kaprou, G., Eck, M., Jobst, G., Petrou, P., et al. (2019). A modular integrated lab-on-a-chip platform for fast and highly efficient sample preparation for foodborne pathogen screening. Sensors Actuators B Chem. 288, 171–179.
Unemo, M., and Jensen, J. (2017). Antimicrobial-resistant sexually transmitted infections: Gonorrhea and Mycoplasma genitalium. Nat. Rev. Urol. 14, 139–152. doi: 10.1038/nrurol.2016.268
Van Belkum, A., and Rochas, O. (2018). Laboratory-based and point-of-care testing for MSSA/MRSA detection in the age of whole-genome sequencing. Front. Microbiol. 9:1437. doi: 10.3389/fmicb.2018.01437
van Belkum, A., Bachmann, T., Lüdke, G., Lisby, J., Kahlmeter, G., Mohess, A., et al. (2019). Developmental roadmap for antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 17, 51–62. doi: 10.1038/s41579-018-0098-9
van Belkum, A., Burnham, C., Rossen, J., Mallard, F., Rochas, O., and Dunne, W. M. (2020). Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 18, 299–311.
Vasala, A., Hytönen, V., and Laitinen, O. (2020). Modern tools for rapid diagnostics of antimicrobial resistance. Front. Cell. Infect. Microbiol. 10:308. doi: 10.3389/fcimb.2020.00308
Veenemans, J., Overdevest, I., Snelders, E., Willemsen, I., Hendriks, Y., Adesokan, A., et al. (2014). Next-generation sequencing for typing and detection of resistance genes: Performance of a new commercial method during an outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli. J. Clin. Microbiol. 52, 2454–2460. doi: 10.1128/JCM.00313-14
Velican, A., Mãruţescu, L., Kamerzan, C., Cristea, V., Banu, O., Borcan, E., et al. (2020). Rapid detection and antibiotic susceptibility of uropathogenic Escherichia coli by flow cytometry. Microorganisms 8:1233. doi: 10.3390/microorganisms8081233
Ventola, C. (2015). The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Therapeutics 40, 277–283.
Vogt, D., Overesch, G., Endimiani, A., Collaud, A., Thomann, A., and Perreten, V. (2014). Occurrence and genetic characteristics of third-generation cephalosporin-resistant Escherichia coli in Swiss retail meat. Microb. Drug Resist. 20, 485–494. doi: 10.1089/mdr.2013.0210
Vogt, S., Löffler, K., Dinkelacker, A., Bader, B., Autenrieth, I., Peter, S., et al. (2019). Fourier transform infrared (FTIR) spectroscopy for typing of clinical Enterobacter cloacae complex isolates. Front. Microbiol. 10:2582. doi: 10.3389/fmicb.2019.02582
Volland, H., Dortet, L., Bernabeu, S., Boutal, H., Haenni, M., Madec, J., et al. (2019). Development and multicentric validation of a lateral flow immunoassay for rapid detection of MCR-1-producing Enterobacteriaceae. J. Clin. Microbiol. 57:e01454-18.
Vrioni, G., Tsiamis, C., Oikonomidis, G., Theodoridou, K., Kapsimali, V., and Tsakris, A. (2018). MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: Current achievements and future perspectives. Ann. Transl. Med. 6:240. doi: 10.21037/atm.2018.06.28
Waagsbø, B., Stuve, N., Afset, J., Klepstad, P., Mo, S., Heggelund, L., et al. (2022). High levels of discordant antimicrobial therapy in hospital-acquired bloodstream infections is associated with increased mortality in an intensive care, low antimicrobial resistance setting. Infect. Dis. 54, 738–747. doi: 10.1080/23744235.2022.2083672
Wagner, T., Janice, J., Schulz, M., Ballard, S., da Silva, A., Coombs, G., et al. (2023). Reversible vancomycin susceptibility within emerging ST1421 Enterococcus faecium strains is associated with rearranged vanA-gene clusters and increased vanA plasmid copy number. Int. J. Antimicrob. Agents 62:106849. doi: 10.1016/j.ijantimicag.2023.106849
Wan Makhtar, W., Bharudin, I., Samsulrizal, N., and Yusof, N. (2021). Whole genome sequencing analysis of Salmonella enterica Serovar Typhi: History and current approaches. Microorganisms 9:2155. doi: 10.3390/microorganisms9102155
Wang, K., Li, S., Petersen, M., Wang, S., and Lu, X. (2018). Detection and characterization of antibiotic-resistant bacteria using surface-enhanced Raman spectroscopy. Nanomaterials 8:762.
Wang-Wang, J., Bordoy, A., Martró, E., Quesada, M., Pérez-Vázquez, M., Guerrero-Murillo, M., et al. (2022). Evaluation of Fourier transform infrared spectroscopy as a first-line typing tool for the identification of extended-Spectrum β-lactamase-producing Klebsiella pneumoniae outbreaks in the hospital setting. Front. Microbiol. 13:897161. doi: 10.3389/fmicb.2022.897161
Wardal, E., Żabicka, D., Skalski, T., Kubiak-Pulkowska, J., Hryniewicz, W., and Sadowy, E. (2023). Characterization of a tigecycline-, linezolid-and vancomycin-resistant clinical enteroccoccus faecium isolate, carrying vanA and vanB genes. Infect. Dis. Therapy 12, 2545–2565. doi: 10.1007/s40121-023-00881-3
Wareham, D., Shah, R., Betts, J., Phee, L., and Momin, M. (2016). Evaluation of an immunochromatographic lateral flow assay (OXA-48 K-SeT) for rapid detection of OXA-48-like carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 54, 471–473. doi: 10.1128/JCM.02900-15
Waskito, L., Rezkitha, Y., Vilaichone, R. K., Wibawa, I. D. N., Mustika, S., Sugihartono, T., et al. (2022). Antimicrobial resistance profile by metagenomic and Metatranscriptomic approach in clinical practice: Opportunity and challenge. Antibiotics 11:654. doi: 10.3390/antibiotics11050654
Weldon, I., and Hoffman, S. (2023). “Antimicrobial resistance,” in Global Health Law Policy: Ensuring Justice for a Healthier World, eds L. Gostin and B. M. Meier (Oxford: Oxford University Press), 395.
Welker, M., and Van Belkum, A. (2019). One system for all: Is mass spectrometry a future alternative for conventional antibiotic susceptibility testing? Front. Microbiol. 10:2711. doi: 10.3389/fmicb.2019.02711
Wiegand, I., Hilpert, K., and Hancock, R. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175.
Wilhelm, C., Carneiro, M. D. S., Inamine, E., and Barth, A. L. (2023). A rapid and easy method of MALDI Biotyper antibiotic susceptibility test rapid assay to provide early meropenem susceptibility profile in Enterobacterales. Microbiol. Spectr. 11:e0437522. doi: 10.1128/spectrum.04375-22
Winstanley, T., and Courvalin, P. (2011). Expert systems in clinical microbiology. Clin. Microbiol. Rev. 24, 515–556.
World Health Organization (2014). Antimicrobial Resistance: Global Report on Surveillance. Geneva: World Health Organization.
Wu, Y., Battalapalli, D., Hakeem, M., Selamneni, V., Zhang, P., Draz, M., et al. (2021). Engineered CRISPR-Cas systems for the detection and control of antibiotic-resistant infections. J. Nanobiotechnol. 19, 1–26.
Xu, J., Chun, H., Wang, L., Mei, H., Chen, S., and Huang, X. (2024). DNA microarray chips: Fabrication and cutting-edge applications. Chem. Eng. J. 499:155937.
Yalew, S. (2020). Review on antibiotic resistance: Resistance mechanisms, methods of detection and its controlling strategies. Biomed. J. Sci. Technical Res. 24, 18651–18657.
Yamada, K., Wanchun, J., Ohkura, T., Murai, A., Hayakawa, R., Kinoshita, K., et al. (2013). Detection of methicillin-resistant Staphylococcus aureus using a specific anti-PBP2a chicken IgY antibody. Jpn. J. Infect. Dis. 66, 103–108. doi: 10.7883/yoken.66.103
Yamin, D., Uskoković, V., Wakil, A., Goni, M., Shamsuddin, S., Mustafa, F., et al. (2023). Current and future technologies for the detection of antibiotic-resistant bacteria. Diagnostics 13:3246.
Yohe, S., and Thyagarajan, B. (2017). Review of clinical next-generation sequencing. Arch. Pathol. Lab. Med. 141, 1544–1557.
Yu, M., Shi, H., Shen, H., Chen, X., Zhang, L., Zhu, J., et al. (2023). Simple and rapid discrimination of methicillin-resistant Staphylococcus aureus based on gram staining and machine vision. Microbiol. Spectrum 11:e0528222. doi: 10.1128/spectrum.05282-22
Yusof, N., Norazzman, N., Hakim, S. N., Azlan, M. M., Anthony, A. A., Mustafa, F. H., et al. (2022). Prevalence of mutated colistin-resistant Klebsiella pneumoniae: A systematic review and meta-analysis. Trop. Med. Infect. Dis. 7:414. doi: 10.3390/tropicalmed7120414
Zaffiri, L., Gardner, J., and Toledo-Pereyra, L. (2012). History of antibiotics. From salvarsan to cephalosporins. J. Invest. Surg. 25, 67–77.
Zakhour, J., Haddad, S., Kerbage, A., Wertheim, H., Tattevin, P., Voss, A., et al. (2023). Diagnostic stewardship in infectious diseases: A continuum of antimicrobial stewardship in the fight against antimicrobial resistance. Int. J. Antimicrob. Agents 62:106816. doi: 10.1016/j.ijantimicag.2023.106816
Zanoli, L., and Spoto, G. (2012). Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 3, 18–43.
Zhang, G., Zheng, G., Zhang, Y., Ma, R., and Kang, X. (2018). Evaluation of a micro/nanofluidic chip platform for the high-throughput detection of bacteria and their antibiotic resistance genes in postneurosurgical meningitis. Int. J. Infect. Dis. 70, 115–120. doi: 10.1016/j.ijid.2018.03.012
Zhang, H., Song, J., Zheng, Z., Li, T., Shi, N., Han, Y., et al. (2023). Fungicide exposure accelerated horizontal transfer of antibiotic resistance genes via plasmid-mediated conjugation. Water Res. 233, 119789. doi: 10.1016/j.watres.2023.119789
Zhang, H., Xu, Y., Fohlerova, Z., Chang, H., Iliescu, C., and Neuzil, P. (2019). LAMP-on-a-chip: Revising microfluidic platforms for loop-mediated DNA amplification. TrAC Trends Anal. Chem. 113, 44–53. doi: 10.1016/j.trac.2019.01.015
Zhang, K., Qin, S., Wu, S., Liang, Y., and Li, J. (2020). Microfluidic systems for rapid antibiotic susceptibility tests (ASTs) at the single-cell level. Chem. Sci. 11, 6352–6361. doi: 10.1039/d0sc01353f
Zhu, H., Zhang, H., Xu, Y., Laššáková, S., Korabeèná, M., and Neužil, P. (2020). PCR past, present and future. Biotechniques 69, 317–325.
Zhu, Y., and Li, X. (1994). Detection of meticillin-resistant Staphylococcus aureus by fluorescence in situ hybridization and flow cytometry. Chin. J. Nosocomiol. 24:1994.
Zorbozan, H., and Kimiran, A. (2022). Detection of beta-lactam antibiotic resistance in aquatic Enterobacteriaceae isolates. Water Supply 22, 8557–8571.
Keywords: antibiotic resistance, detection, contemporary technologies, genomics, microbial community
Citation: Elbehiry A, Marzouk E, Abalkhail A, Abdelsalam MH, Mostafa MEA, Alasiri M, Ibrahem M, Ellethy AT, Almuzaini A, Aljarallah SN, Abu-Okail A, Marzook N, Alhadyan S and Edrees HM (2025) Detection of antimicrobial resistance via state-of-the-art technologies versus conventional methods. Front. Microbiol. 16:1549044. doi: 10.3389/fmicb.2025.1549044
Received: 30 December 2024; Accepted: 11 February 2025;
Published: 25 February 2025.
Edited by:
Kristina Kadlec, Independent researcher, Wunstorf, GermanyReviewed by:
Valerie J. Carabetta, Cooper Medical School of Rowan University, United StatesCopyright © 2025 Elbehiry, Marzouk, Abalkhail, Abdelsalam, Mostafa, Alasiri, Ibrahem, Ellethy, Almuzaini, Aljarallah, Abu-Okail, Marzook, Alhadyan and Edrees. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ayman Elbehiry, YXIuZWxiZWhpcnlAcXUuZWR1LnNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.