
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Microbiol. , 17 February 2025
Sec. Food Microbiology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1545266
 Mengrong Chen
Mengrong Chen Qiling Chen*
Qiling Chen*Oenococcus oeni is an important engineering microbe in winemaking. Detailed knowledge of its growth and metabolism in harsh wine environments could contribute to breeding elite O. oeni varieties. However, further studies on this topic do not appear to be sustained due to the lack of stable and reproducible technology to perform gene manipulation on O. oeni. Therefore, this research was designed to study gene function by exploring a newly applicable transformation technique that could perform stably and reproducibly on O. oeni. By using gene gun technology with detonation nanodiamonds as a plasmid DNA carrier, we achieved stable and reproducible plasmid DNA transformation in O. oeni. In addition, the plasmid with the chloramphenicol resistance gene allowed O. oeni SX-1b to thrive in chloramphenicol medium.
Oenococcus oeni is an important engineering microbe in winemaking due to its excellent fermentative behavior (Bech-Terkilsen et al., 2020; Cappello et al., 2014; Guzzon et al., 2009; Liu et al., 2024). Selected strains of this species have been traditionally chosen as microorganisms for use in wine starter cultures (Grandvalet, 2017; St'Ana and Lemos, 2024; Zhang et al., 2024). In-depth investigations of the functional genes involved in growth and metabolism in harsh wine environments of this species could contribute to breeding more elite O. oeni varieties. However, despite multiple techniques available for the transformation of bacteria, several species, including O. oeni, are still resistant to the introduction of foreign DNA (Beltramo et al., 2004). However, the plasmid pSR7Rep, which was constructed using the pRS7 obtained from O. oeni itself, could not achieve the transformation (Rodriguez et al., 2015). Therefore, current research is primarily focused on “big data omics” that encompass genomics, metabolomics, transcriptomics and proteomics (Chen et al., 2022; Liang et al., 2020; Liu et al., 2017; Margalef-Catala et al., 2016; Sternes et al., 2017) or gene function validation by heterologous expression (Morel et al., 2001; Qi et al., 2020; Yuan et al., 2019; Zhao et al., 2019a,b; Zheng et al., 2024). Thus, stable and reproducible transformation of foreign DNA is a prerequisite to achieving the genetic manipulation of O. oeni.
Gene gun technology was developed to enhance DNA delivery, and gene gun-mediated gene transfer is widely used in plants, yeast, gene vaccines and gene therapy (Ghogare et al., 2021; Imai, 2021; Jafari et al., 2012; Slon-Campos et al., 2020). Moreover, O. oeni has a diameter of <500 nm (Wang et al., 2019), and nanoscale material is needed as a vehicle (Osipov et al., 2020; Stehlik et al., 2016). Therefore, this research was designed to tentatively study the gene functions of O. oeni based on the precondition that gene gun combined with detonation nanodiamonds (DNDs) could achieve a stable and reproducible transformation into O. oeni.
O. oeni is an important engineering bacterium in wine making (Bech-Terkilsen et al., 2020; Guzzon et al., 2009). Thus, intensive study on this useful industrial microorganism is necessary. However, in-depth research is sluggish and proceeds gradually due to the lack of efficient gene-transfer techniques (Beltramo et al., 2004; Rodriguez et al., 2015). Therefore, stable and reproducible transformation of foreign DNA is an important prerequisite to enable genetic manipulation of O. oeni.
Figure 1 provides a flowchart of the particle bombardment on O. oeni by the gene gun. The detailed procedure is presented in the materials and methods section. To ensure the successful transformation of the exogenous plasmid coated with DNDs by gene gun on O. oeni, a recombination plasmid pLCNICK-citP with a size of 14 kbp and carrying an exogenous gene that is specific for Lactiplantibacillus plantarum WCFS1 and therefore must be absent in O. oeni, was transformed into O. oeni SX-1b, SX-1a and SD-2a cells collected at various growth stages. After bombardment, the cells were transferred to 2.8% agar ATB with erythromycin resistance and incubated in at 26°C.

Figure 1. Technical processes involved in transforming detonation nanodiamond (DND)-coated plasmids into Oenococcus oeni by gene gun.
Transformed colonies appeared after 7 days of incubation. As shown in Figures 2A,B, all plates grew dense colonies (>400), regardless of the different O. oeni strains (SX-1a, SX-1b and SD-2a), gradient concentrations of plasmid (2 μg/μl, 1 μg/μl and 0.5 μg/μl), or different collection times (20 and 45 h). Except for plate bombardment with 0.5 μg/μl plasmid, SX-1b cells collected at 45 h grew to fewer than 10 single colonies. Then, single colonies were randomly selected and identified through Sanger sequencing with pairs of primers (citP-test) (Figures 2C,D). Overall, the transformation efficiency was approximately 40% by colony PCR at either low or high concentrations of plasmid. Thus, gene gun combined with DND technology may be employed as an efficient transformation method for O. oeni.
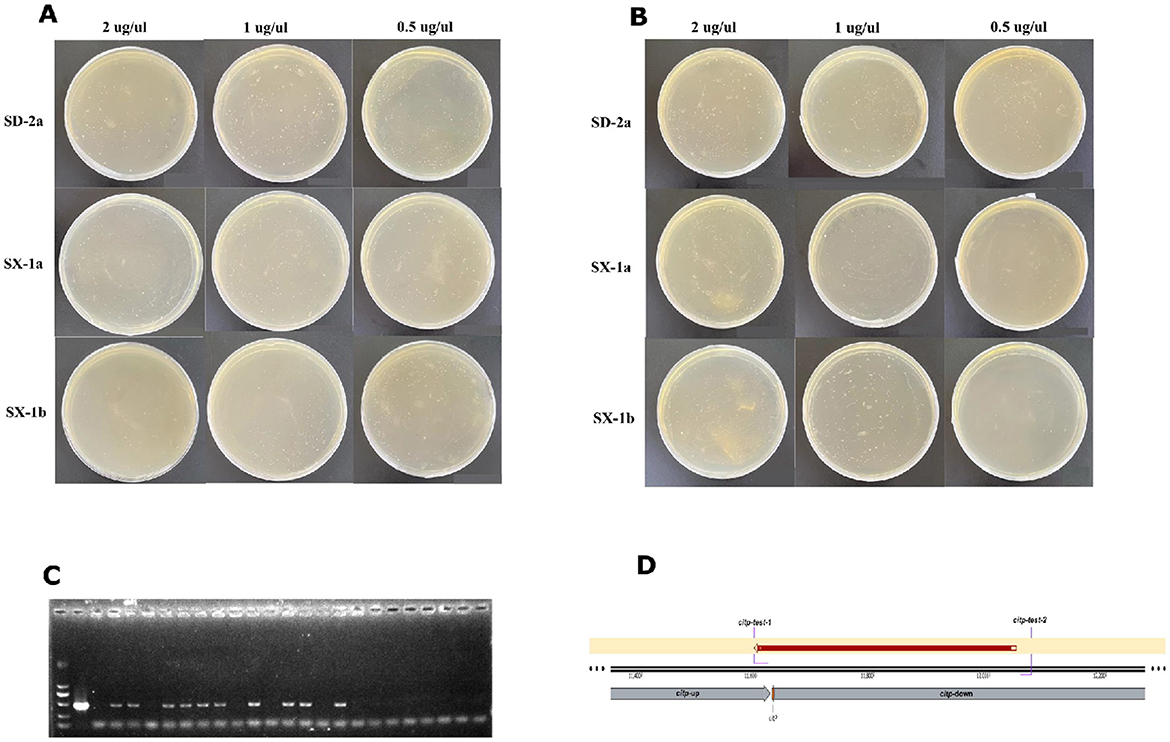
Figure 2. Results of the bombardment experiment of O. oeni by gene gun. Transformed colonies appeared on the selection plates after 7 days of incubation. (A) Transformed cells collected at 20 h; and (B) transformed cells collected at 45 h. (C) Randomly selected transformants were verified by colony PCR, and the 477 bp products were checked via electrophoresis to confirm the correct construct. (D) Single colony clones were validated by DNA sequencing, with the sequence perfectly matching the criteria.
The plasmid pMG36ck11 (modified from plasmid pMG36e, and have a chloramphenicol resistance gene) was successfully introduced into O. oeni SX-1b, with colonies emerging after over 12 days on chloramphenicol selection plates. This delay in colony formation is likely due to the inhibitory effects of chloramphenicol on protein synthesis, which can slow down bacterial growth. Growth curve analyses (Figure 3) further supported these findings, showing that the mutant strain O. oeni SX-1b-pMG36ck11 was able to thrive in chloramphenicol, unlike the wild strain. This difference in growth behavior between the wild and transformed strains provides strong evidence of successful plasmid transfer and expression of the chloramphenicol resistance gene.
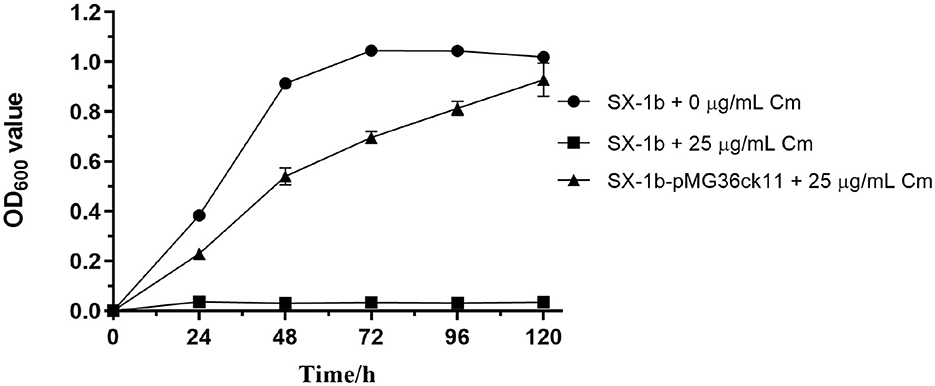
Figure 3. Proliferative capacities of O. oeni mutants and wild-type strains cultivated in chloramphenicol medium. The absorbance values (OD600 nm) were calculated as the tested samples' OD reading minus the negative control's OD reading. Error bars represent the SD of three biological repeats.
Contrary to expectations, the qPCR results were disappointing and discouraging because the Ct values were very high for the antibiotic resistance gene [mean Ct value was 34.67 for the chloramphenicol gene, whereas the mean Ct value was 17.09 for the reference gene ddl (Costantini et al., 2011)], although significant differences were found for the survival ratio between mutants and wild O. oeni SX-1b (Figure 3). This also indicates from another perspective that there are mechanisms within O. oeni cells that can recognize and rapidly degrade foreign genes. Such mechanisms may protect the cells from potential toxic effects by limiting the expression of foreign genes (Leinyuy et al., 2023).
The knockout plasmid pLCNICK-1070 was created for O. oeni SX-1b and successfully introduced into the bacteria. Transformants were cultured in ATB medium with erythromycin, but gene knockout was not achieved. This may also due to the strain's weak acceptance of foreign DNA.
In summary, the results appear to indicate that the lower expression of genes located on the plasmid and transferred into O. oeni is not due to the vector itself or the source of the DNA. In addition, previous research in our laboratory showed that O. oeni transferred with a GFP fluorescence plasmid did not present GFP fluorescence. It seems that O. oeni may have a restriction system that could interfere with exogenous genes, as speculated by previous researchers (Beltramo et al., 2004; Rodriguez et al., 2015), and this possibility deserves further investigation. In addition, we speculated that although this restriction system might strongly inhibit exogenous DNA replication, exogenous DNAs could continue to be functional, which was also supported by the findings reported by RNA silencing (Darsonval et al., 2016).
Gene gun technology with detonation nanodiamonds successfully and consistently transformed DNA into O. oeni cells. Overexpressing the pMG36ck11 plasmid (which contains a chloramphenicol resistance gene) enabled O. oeni SX-1b to survive in chloramphenicol medium. The reduced expression of plasmid genes may be linked to a restriction system in O. oeni, warranting further study. This research establishes a basis for gene manipulation in O. oeni species.
O. oeni was grown in ATB medium (pH 4.8, 26°C) and subcultured twice before being bombarded by a gene gun. One milliliter of cells in logarithmic phase (20 h for mid-logarithmic phase and 45 h for late logarithmic phase, with the OD600 adjusted at 1.0 by dilution or concentration with fresh ATB medium) was harvested by centrifugation (8000 rpm, 10 min) and washed with distilled water. Subsequently, the cells were collected by centrifugation again and then suspended in 0.02% sodium hyaluronate solution. Plasmids for the bombardment experiment were concentrated in plasmid DNA (2, 1, and 0.5 μg/mL) by freeze-drying. The collected cells were transiently transformed by particle bombardment using a gene gun (Bio–Rad, Biolistic PDS-1000/He). The experimental protocol for bombardment is based on the work of Grichko et al. (2006), with minor modifications.
The DNDs were purchased from Sigma-Aldrich (America, Cat No. 900180, 5 nm average part. size (DLS), 10 mg/mL in H2O, carboxylated). To prepare DND carriers for plasmid DNA delivery by particle bombardment, 0.1 mL DND hydrosol was diluted by adding 0.9 mL of distilled water. Then, the DNDs were repeatedly washed in distilled water through cycles of centrifugation and sterilized by autoclaving to generate 1 mL of approximately 10% hydrosol, which was stored at 4°C until use.
On the day of the experiment, 5 μL of different plasmid concentrations in Tris buffer (pH 8), 50 μL of 2.5 M CaCl2, and 20 μL of freshly prepared 0.1 M spermidine were added to 50 μL of DND hydrosol while continuously vortexing. The suspension was vortexed at low speed for 5 min and kept on ice for 10 min, and plasmid-coated DNDs were collected by centrifugation, washed in absolute ethanol, and resuspended in 50 μL of absolute ethanol.
Cells were bombarded with a Biolistic PDS-1000/He gene gun system (Bio–Rad) according to the manufacturer's instructions. The suspensions were settled in a sample disc and then placed into the target chamber of the gene gun chamber. Ten microliters of the suspension was placed on the center of a Bio–Rad macrocarrier disk and air-dried. A Bio–Rad PDS-1000/He instrument was operated according to the manufacturer's instructions using 1100 psi He pressure and approximately 28 in. Hg vacuum. Penetration of DNDs along with plasmids into O. oeni was confirmed by PCR with gene-specific primers. When the vacuum in the target chamber was below 28 MPa, the strains in the sample tray were bombarded with DND particles (5 nm average size) with plasmid. Then, the cooling (approximately 40°C) solid medium was poured onto the plastic dished with the bombarded strains. The plates were then incubated at 26°C.
The O. oeni strains used in this research were collected and preserved in our laboratory. Plasmids used in this research: pLCNICK (encodes erythromycin resistance and kanamycin double resistance) and pMG36ck11 (modified from plasmid pMG36e by adding a kanamycin resistance gene, replacing the P32 promoter with P11, and replacing the erythromycin gene with the chloramphenicol gene). The primers used in this study are shown in Table 1.
PrimerSTAR® Max DNA Polymerase (Takara) was used to amplify clones of genes from O. oeni fragments, which were ligated into vectors by a NovoRec® Plus PCR one-step cloning kit (NR005, Novoprotein). The application and ligation system and program followed the manufacturer's instructions. DNA was recovered using a DNA purification kit (Tiangen Biotech, Beijing, China) for purification of PCR DNA fragments. The plasmids were extracted using a Plasmid Extraction kit (Tiangen Biotech, Beijing, China).
Cells were grown to an OD600 of 1.0 prior to induction, then the cells were harvested by centrifugation and resuspended in distilled water twice. Mutants and wild strain were cultured in the pH 4.8 ATB medium contained 25 μg/mL chloramphenicol with a 4% inoculation amount, besides wild SX-1b was cultured in pH 4.8 ATB medium without any resistance.
Culture samples (5 mL) were collected at the mid-logarithmic phase of growth after introduction to ATB medium (containing 25 μg/mL chloramphenicol). RNA was extracted using AG RNAex Pro Reagent, and total RNA was reverse transcribed using an Evo M-MLV RT Kit with gDNA Clean for qPCR II. Fluorescence quantitative PCR and fluorescence quantitative PCR were performed according to the procedure of the SYBR® Green Premix Pro Taq HS qPCR Kit. All kits were purchased from Accurate Biotechnology (Hunan) Co., Ltd.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
MC: Writing – original draft. QC: Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We would like to thank the members of Shuwen Liu laboratory of College of Enology, Northwest A&F University for their help and support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bech-Terkilsen, S., Westman, J. O., Swiegers, J. H., and Siegumfeldt, H. (2020). Oenococcus oeni, a species born and moulded in wine: a critical review of the stress impacts of wine and the physiological responses. Aust. J. Grape Wine R. 26, 188–206. doi: 10.1111/ajgw.12436
Beltramo, C., Oraby, M., Bourel, G., Garmyn, D., and Guzzo, J. (2004). A new vector, pGID052, for genetic transfer in Oenococcus oeni. Fems. Microbiol. Lett. 236, 53–60. doi: 10.1111/j.1574-6968.2004.tb09626.x
Cappello, M. S., De Domenico, S., Logrieco, A., and Zapparoli, G. (2014). Bio-molecular characterisation of indigenous Oenococcus oeni strains from Negroamaro wine, Food Microbiol. 42, 142–148. doi: 10.1016/j.fm.2014.02.004
Chen, Q. L., Yang, X. K., Meng, Q., Zhao, L. L., Yuan, Y. X., Chi, W., He, L., Shi, K., and Liu, S. W. (2022). Integrative multiomics analysis of the acid stress response of Oenococcus oeni mutants at different growth stages. Food Microbiol. 102:103905. doi: 10.1016/j.fm.2021.103905
Costantini, A., Vaudano, E., Rantsiou, K., Cocolin, L., and Garcia-Moruno, E. (2011). Quantitative expression analysis of mleP gene and two genes involved in the ABC transport system in Oenococcus oeni during rehydration. Appl. Microbiol. Biotechnol. 91, 1601–1609. doi: 10.1007/s00253-011-3498-6
Darsonval, M., Msadek, T., Alexandre, H., and Grandvalet, C. (2016). The antisense RNA approach: a new application for in vivo investigation of the stress response of Oenococcus oeni, a wine-associated lactic acid bacterium. Appl. Environ. Microbiol. 82, 18–26. doi: 10.1128/AEM.02495-15
Ghogare, R., Ludwig, Y., Bueno, G. M., Slamet-Loedin, I. H., and Dhingra, A. (2021). Genome editing reagent delivery in plants. Transgenic Res. 30, 321–335. doi: 10.1007/s11248-021-00239-w
Grandvalet, C. (2017). Oenococcus oeni: queen of the cellar, nightmare of geneticists. Microbiology 163, 297–299. doi: 10.1099/mic.0.000456
Grichko, V., Grishko, V. I., and Shenderova, O. (2006). Nanodiamond bullets and their biological targets. Nanobiotechnology 2, 37–42. doi: 10.1007/s12030-006-0005-8
Guzzon, R., Poznanski, E., Conterno, L., Vagnoli, P., Krieger-Weber, S., and Cavazza, A. (2009). Selection of a new highly resistant strain for malolactic fermentation under difficult conditions. S. Afr. J. Enol. Vitic. 30, 133–141. doi: 10.21548/30-2-1433
Imai, T. (2021). Single amino acid deletion at N-terminus of the target antigen in DNA vaccine induces altered CD8(+) T cell responses against tumor antigen. Vaccines 9:540. doi: 10.3390/vaccines9060540
Jafari, M., Soltani, M., Naahidi, S., Karunaratne, D. N., and Chen, P. (2012). Nonviral approach for targeted nucleic acid delivery. Curr. Med. Chem. 19, 197–208. doi: 10.2174/092986712803414141
Leinyuy, J. F., Ali, I. M., Ousenu, K., and Tume, C. B. (2023). Molecular characterization of antimicrobial resistance related genes in E. coli, Salmonella and Klebsiella isolates from broilers in the West Region of Cameroon. PLoS ONE 18:e0280150. doi: 10.1371/journal.pone.0280150
Liang, H. Y., Su, N., Guo, K., Wang, Y., and Yang, D. Y. (2020). Effects of Saccharomyces cerevisiae strains on chemical profiles of Cabernet Sauvignon wines: based on the combined results of H-1 NMR, HS-SPME/GC-MS and HPLC-DAD-ESI-MS/MS. Curr. Top. Nutraceut. R. 18, 115–131. doi: 10.37290/ctnr2641-452X.18:115-131
Liu, L. X., Zhao, H. Y., Peng, S., Wang, T., Su, J., Liang, Y. Y., et al. (2017). Transcriptomic analysis of Oenococcus oeni SD-2a response to acid shock by RNA-Seq. Front. Microbiol. 8:1586. doi: 10.3389/fmicb.2017.01586
Liu, X. W., Fu, J. W., Ma, W., and Jin, G. (2024). Screening and evaluation of high stress tolerance, high esterase activity and safety of Oenococcus oeni strains adapt to challenging conditions in Northwest China wine. LWT Food Sci. Technol. 213:116975. doi: 10.1016/j.lwt.2024.116975
Margalef-Catala, M., Araque, I., Bordons, A., Reguant, C., and Bautista-Gallego, J. (2016). Transcriptomic and proteomic analysis of Oenococcus oeni adaptation to wine stress conditions. Front. Microbiol. 7:1554. doi: 10.3389/fmicb.2016.01554
Morel, F., Delmas, F., Jobin, M. P., Divies, C., and Guzzo, J. (2001). Improved acid tolerance of a recombinant strain of Escherichia coli expressing genes from the acidophilic bacterium Oenococcus oeni. Lett Appl Microbiol. 33, 126–130. doi: 10.1046/j.1472-765x.2001.00960.x
Osipov, V. Y., Shakhov, F. M., Bogdanov, K. V., Takai, K., Hayashi, T., Treussart, F., et al. (2020). High-quality green-emitting nanodiamonds fabricated by HPHT sintering of polycrystalline shockwave diamonds. Nanoscale Res. Lett. 15:209. doi: 10.1186/s11671-020-03433-7
Qi, Y. M., Liu, D., Yu, H. P., Zhang, G. Q., and Fan, M. T. (2020). Identification and characterization of the small heat shock protein Hsp20 from Oenococcus oeni SD-2a. Curr. Microbiol. 77, 3595–3602. doi: 10.1007/s00284-020-02168-z
Rodriguez, M. C., Alegre, M. T., Martin, M. C., and Mesas, J. M. (2015). The use of the replication region of plasmid pRS7 from Oenococcus oeni as a putative tool to generate cloning vectors for lactic acid bacteria. Plasmid 77, 28–31. doi: 10.1016/j.plasmid.2014.11.004
Slon-Campos, J. L., Poggianella, M., Zentilin, L., and Burrone, O. R. (2020). Use of adeno-associated viral vectors to improve delivery of a DNA vaccine against dengue virus. J. Gen. Virol. 101, 73–78. doi: 10.1099/jgv.0.001351
St'Ana, A. S., and Lemos, W. J. R. (2024). Microbial synergies and their impact on economic and quality innovation in sustainable winemaking: yeast and lactic acid bacteria interconnections. Food Biosci. 62:105238. doi: 10.1016/j.fbio.2024.105238
Stehlik, S., Varga, M., Ledinsky, M., Miliaieva, D., Kozak, H., Skakalova, V., et al. (2016). High-yield fabrication and properties of 1.4 nm nanodiamonds with narrow size distribution. Sci. Rep. 6:38419. doi: 10.1038/srep38419
Sternes, P. R., Costello, P. J., Chambers, P. J., Bartowsky, E. J., and Borneman, A. R. (2017). Whole transcriptome RNAseq analysis of Oenococcus oeni reveals distinct intra-specific expression patterns during malolactic fermentation, including genes involved in diacetyl metabolism. Int. J. Food Microbiol. 257, 216–224. doi: 10.1016/j.ijfoodmicro.2017.06.024
Wang, J. F., He, L., An, W., Yu, D. L., Liu, S. W., and Shi, K. (2019). Lyoprotective effect of soluble extracellular polymeric substances from Oenococcus oeni during its freeze-drying process. Process Biochem. 84, 205–212. doi: 10.1016/j.procbio.2019.05.026
Yuan, L., Zhao, H. Y., Liu, L. X., Peng, S., Li, H., and Wang, H. (2019). Heterologous expression of the puuE from Oenococcus oeni SD-2a in Lactobacillus plantarum WCFS1 improves ethanol tolerance. J. Basic Microb. 59, 1134–1142. doi: 10.1002/jobm.201900339
Zhang, B. Y., Liu, D. D., Liu, H., Shen, J. X., Zhang, J. X., He, L., et al. (2024). Impact of indigenous Oenococcus oeni and Lactiplantibacillus plantarum species co-culture on Cabernet Sauvignon wine malolactic fermentation: kinetic parameters, color and aroma. Food Chem. X 22:101369. doi: 10.1016/j.fochx.2024.101369
Zhao, H. Y., Liu, L. X., Peng, S., Yuan, L., Li, H., and Wang, H. (2019a). Heterologous expression of argininosuccinate synthase from Oenococcus oeni enhances the acid resistance of Lactobacillus plantarum. Front. Microbiol. 10:1393. doi: 10.3389/fmicb.2019.01393
Zhao, H. Y., Yuan, L., Hu, K., Liu, L. X., Peng, S., Li, H., and Wang, H. (2019b). Heterologous expression of ctsR from Oenococcus oeni enhances the acid-ethanol resistance of Lactobacillus plantarum. Fems Microbiol Lett. 366:fnz192. doi: 10.1093/femsle/fnz192
Keywords: Oenococcus oeni, gene gun, detonation nanodiamonds, stable genetic transfer, wine
Citation: Chen M and Chen Q (2025) Gene manipulation in Oenococcus oeni based on a newly applicable gene gun technology. Front. Microbiol. 16:1545266. doi: 10.3389/fmicb.2025.1545266
Received: 14 December 2024; Accepted: 17 January 2025;
Published: 17 February 2025.
Edited by:
Laurent Dufossé, Université de la Réunion, FranceReviewed by:
Wenchao Cai, Shihezi University, ChinaCopyright © 2025 Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiling Chen, Y2hlbnFpbGluZ0Bud2FmdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.