- 1Department of Dermatology, The First Hospital of China Medical University, Shenyang, China
- 2NHC Key Laboratory of Immunodermatology, China Medical University, Shenyang, China
- 3Key Laboratory of Immunodermatology, China Medical University, Ministry of Education, Shenyang, China
- 4National and Local Joint Engineering Research Center of Immunodermatological Theranostics, Shenyang, China
With the increasing application of artificial intelligence (AI) in medical research, studies on the human immunodeficiency virus type 1(HIV-1) and acquired immunodeficiency syndrome (AIDS) have become more in-depth. Integrating AI with technologies like single-cell sequencing enables precise biomarker identification and improved therapeutic targeting. This review aims to explore the advancements in AI technologies and their applications across various facets of HIV research, including viral mechanisms, diagnostic innovations, therapeutic strategies, and prevention efforts. Despite challenges like data limitations and model interpretability, AI holds significant potential in advancing HIV-1 management and contributing to global health goals.
1 Introduction
The human immunodeficiency virus type 1 (HIV-1) causes acquired immunodeficiency syndrome (AIDS). Since the World Health Organization (WHO) published guidelines in 2002 for treating HIV, advancements in antiretroviral therapy (ART) have notably increased both the life expectancy and quality of life for people living with HIV (PLWH) (World Health Organization, 2015). However, the world is still not on track to achieve the Sustainable Development Goal of ending AIDS as a public health threat by 2030 (Godfrey-Faussett et al., 2022). This highlights the urgent need to deepen our understanding of HIV and develop more effective treatment strategies and vaccines to address challenges posed by the virus’s variability and drug resistance.
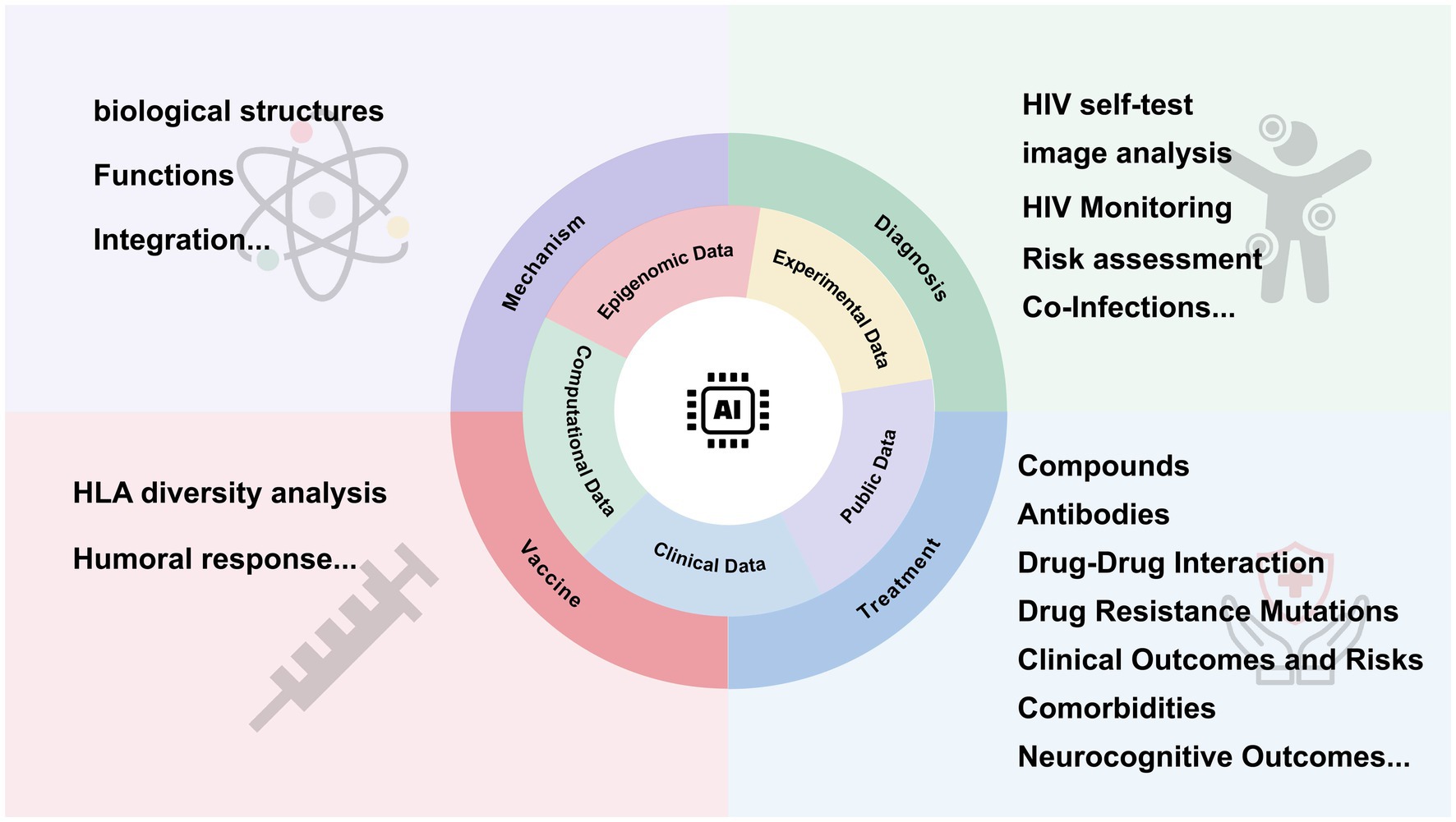
Recent advancements in AI have opened new avenues in biomedical research. AI, using machine learning (ML) algorithms, can analyze vast biological datasets to enhance early detection, personalized treatment, and vaccine development (Wang et al., 2024; Hu et al., 2024; Yu et al., 2022; Holzinger et al., 2019). Machine learning algorithms particularly excel at building complex nonlinear models to link features with disease-related risk factors in large datasets, demonstrating both efficiency and accuracy (Li et al., 2024). As a subfield of AI, ML contains a diverse range of algorithm classes, linking to different learning tasks. These include supervised learning, unsupervised learning and reinforcement learning. Among these, the family of artificial neural networks have flexible structure, which enables adaptation to various situations in all three types of machine learning (Janiesch et al., 2021). Deep learning (DL) involves neural networks with multiple layers of computational neurons, enabling it to handle unstructured data like images (Petersen et al., 2022). The emergence of single-cell technologies has increased the availability of full-length paired B cell receptor (BCR) sequences. Consequently, combining immune repertoire sequencing with AI holds significant potential for improving the diagnosis and treatment of immune-related and infectious diseases (Irvine and Reddy, 2024; Figure 1).
Despite its potential, AI in HIV research faces challenges such as limited data access due to privacy concerns (You et al., 2020), ethical considerations (Gillette et al., 2023; Garett and Young, 2021), high variability of HIV and its complex immune evasion mechanisms, which complicate training and validation (Poonia and Al-Alshaikh, 2024; Vieira et al., 2017).
This review seeks to deliver a thorough overview of the latest advancements in AI within HIV research. It will also critically examine current challenges and propose future research directions.
2 Structural insights and integration mechanisms of HIV
2.1 HIV protein structure analysis
As a ML technique, support vector machine (SVM) is capable of executing both linear and nonlinear classification tasks (Pouyanfar et al., 2018). Another common statistical model, logistic regression (LR), can be used for both classification and regression problems (Sarker, 2021). In the quest to better understand HIV proteins, Mei’s study utilized Chou’s pseudo amino acid composition and increment of diversity as features, employing SVM, LR, and multilayer perceptron models to predict HIV-1 and HIV-2 proteins (Mei and Zhao, 2018). All three models demonstrated superior performance. Concurrently, the tomoDRGN (Powell and Davis, 2024) captures structural heterogeneity in cryo-electron tomography (cryo-ET) datasets through low-dimensional continuous representations, enabling the reconstruction of data-driven heterogeneous structures. This approach demonstrates exceptional performance in analyzing diverse datasets, including elucidating the organization of HIV capsid complexes within virus-like particles and resolving structural heterogeneity among ribosomes in cellular imaging.
2.2 Integration and tropism mechanisms of HIV-1
The integration of the HIV-1 genome into the human genome is a critical step in the viral infection and replication cycle influencing the persistence of infected cells (Maldarelli et al., 2014). Understanding this mechanism is essential for strategies to control the infection and its long-term effects.
DeepHINT (Hu et al., 2019), an attention-based deep learning framework, improves the prediction of HIV integration sites compared to traditional models. Unlike conventional approaches, DeepHINT automatically learned genomic context from DNA sequences, either independently or in conjunction with epigenetic information, providing both high accuracy and mechanistic insights.
HIV can infect cells at various stages of their development, ranging from monocytes to macrophages. A ML model has been employed to differentiate viral genomes in monocytes and T cells based on envelope sequences. This model identified five key features in the C2V3C3 region that significantly distinguish proviruses in monocytes: positions 297, 326, 335, 355, and 395 (Peng and Zhu, 2023). Additionally, XGBpred offer valuable insights into disease progression, underscoring the importance of computational methods in HIV research (Chen et al., 2019).
3 HIV diagnosis and co-infection
3.1 Advances in HIV diagnosis and monitoring tools
HIV self-test (HIVST) is an underutilized innovation in differentiated service delivery for HIV, despite its high sensitivity and specificity. Concerns about false negatives could delay antiretroviral therapy initiation and/or lead to inappropriate use of HIV pre-exposure prophylaxis (PrEP), which in turn increases the risk of HIV drug resistance (Gibas et al., 2019).
Two studies demonstrated the potential of AI in improving HIVST sensitivity through image analysis. Roche’s research (Roche et al., 2024) found that AI identified four HIV infections that were missed by both pharmacy providers and customers, highlighting its higher sensitivity in detecting faint lines on HIVST results. Similarly, Valérian Turbé’s deep learning algorithm (Turbé et al., 2021), trained on 11,374 images of rapid HIV tests from rural South Africa, achieved high sensitivity (97.8%), outperforming human interpretation in a pilot field study using a mobile app. Both studies demonstrated that the sensitivity of the AI algorithms surpassed that of traditional visual interpretation.
3.2 HIV monitoring and risk assessment
In HIV monitoring, real-time electronic adherence monitoring (EAM) combined with machine learning significantly improved the accuracy of predicting HIV viral load (Benitez et al., 2020). Additionally, researchers proposed a selective testing strategy to reduce testing frequency while effectively identifying high-risk patients. Another study (Balzer et al., 2020) used HIV testing data from Kenya and Uganda to develop a machine learning-based HIV risk score, which improved sensitivity in identifying high-risk individuals compared to traditional methods. Moreover, Bao et al. (2021) developed a model using demographic and sexual behavior data to predict the risk of HIV and sexually transmitted infections (STIs) among men who had sex with other men, yielding promising results.
3.3 Advances in managing HIV co-infections
HIV co-infections are common but crucial to study because they can accelerate disease progression and complicate treatment for both HIV and the co-infecting pathogen. Common HIV co-infections include viral hepatitis (HBV, HCV) (Li et al., 2024; Laguno et al., 2004; Wei et al., 2019; Sun et al., 2014; Muriuki et al., 2013; Wang et al., 2023), tuberculosis (Shen, 2024), and oncogenic viruses like HPV and HHV-8. These infections can worsen the overall health outcomes for HIV-positive individuals and increase risks of severe complications, such as liver disease and certain cancers. While ART has significantly improved HIV management, it cannot completely mitigate the additional risks posed by co-infections (Brites et al., 2021).
Firstly, HIV-1 and HCV share transmission routes, leading to a high risk of co-infection (Laguno et al., 2004). Researchers combined Naïve Bayes (NB) and SVM algorithms with two molecular fingerprints (MACCS and ECFP6) to develop 60 classification models, predicting compounds effective against 11 HIV-1 and 4 HCV targets. Over 20 potential multi-target inhibitors were identified, including seven HIV-1 and four HCV-approved drugs (Wei et al., 2019).
Co-infection with HIV and HBV is also prevalent due to similar transmission routes. Patients with HIV/HBV co-infection often experience faster disease progression, significantly increasing their risk of developing chronic liver disease, cirrhosis, end-stage liver disease, and hepatocellular carcinoma (Sun et al., 2014; Muriuki et al., 2013). Researchers applied ML to identify six novel compounds targeting both HIV-1 and HBV, demonstrating the potential for virtual screening in co-infection treatment (Wang et al., 2023).
In another study, Onywera et al. (2020) used LEfSe analysis to find that men co-infected with HIV and HPV showed a more diverse penile microbiota, although no significant association between HIV status and microbiota diversity was found.
Machine learning was also applied to predict early cervical cancer (CC) in women living with HIV, identifying key predictive factors, such as the duration of ART, WHO clinical stage, TPT status, viral load status, and family planning history. A Random Forest (RF) model suggested that ML can improve early detection rates for CC, reduce healthcare costs, and enhance preventive strategies (Namalinzi et al., 2024).
4 Innovations in treatment
4.1 Advancements in discovering HIV compounds and antibodies
Protease inhibitors remain an important component of ART for the treatment of HIV-1 infection (Mulato et al., 2024). Researchers have utilized AI techniques to screen datasets for potential HIV-1 protease ligands. These preliminary findings, validated through docking and molecular dynamics simulations, led to the discovery of a novel ligand outside known HIV-1 protease inhibitor classes (Arrigoni et al., 2023). Another study developed a gradient boosting model using a large dataset of HIV-1 protease inhibitors to predict ligand binding affinity. Enhanced by structural and potency data, it showed high accuracy. Shapley value analysis revealed that van der Waals interactions with key protein residues were critical to ligand potency (Leidner et al., 2019). Similarly, another study (Bitencourt-Ferreira et al., 2021) incorporated affinity data to further optimize small molecule drugs.
An approach integrated Long Short-Term Memory (LSTM) networks and variational autoencoders to accelerate HIV drug discovery to help identify candidate compounds and validate their interactions with HIV, offering a cost-effective drug development pathway (Kutsal et al., 2024).
Drug–drug interaction (DDI) prediction offers crucial insights for managing complex treatments effectively. Researchers have developed Deep-ARV, a tool designed to predict four categories of DDIs. To address the imbalance in DDI severity distribution, the model incorporates undersampling and ensemble learning techniques. This approach can help identify high-risk drug pairings, enhance the drug screening process, and support clinical drug development targeting DDI risks (Pham et al., 2024).
During the COVID-19 pandemic, researchers focused on identifying potential drugs targeting SARS-CoV-2, specifically the 3-chymotrypsin-like protease (3CLpro), an essential enzyme that plays a critical role in viral replication. Through computational screening methods, two compounds emerged as promising candidates against 3CLpro. These compounds are considered strong candidates for further development, offering a new direction for COVID-19 therapeutic strategies (Nand et al., 2020).
In antibody research, broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope (Env) glycoprotein have shown promise for prevention and treatment. Using Rapid Automated Identification of bNAbs (RAIN) (Foglierini et al., 2024), researchers identified several bNAbs targeting the CD4 binding site of the HIV-1 Env glycoproteins. Structural analysis of bNAb4251, using cryo-electron microscopy, revealed that unconventional mutations are key to HIV-1 bNAbs’ function, offering new insights into HIV-1 immune evasion mechanisms.
4.2 Innovative predictive modeling and mutation analysis for addressing HIV drug resistance
To tackle HIV drug resistance, researchers have developed ML models to predict novel compounds that may inhibit HIV (Zorn et al., 2019). Due to HIV-1’s high sequence diversity and mutation rate, isolates often develop resistance to bNAbs. Traditionally, identifying resistant strains requires time-consuming in vitro assays. Reda Rawi used ML to predict HIV-1 resistance to 33 bNAbs, identifying key epitope features using gradient boosting machines. This in silico tool could streamline decision-making regarding antibody use and enable sequence-based monitoring of viral escape (Rawi et al., 2019). Steiner et al. (2020) evaluated three machine learning architectures—MLP, bidirectional recurrent neural network (Bi-RNN), and CNN—to predict resistance across 18 antiretroviral drugs. By combining sequence data with biological analysis, they enhanced the interpretation of drug resistance predictions.
Additionally, RF and SVM were employed to analyze resistance linked to 21 mutated residues in HIV target proteins. By using different kernel functions [linear, polynomial, and radial basis function (RBF)], these models offer insights into the impact of novel mutations on treatment efficacy (Cai et al., 2021).
Most antiretroviral drugs targeting HIV focus on reverse transcriptase (RT), yet HIV can develop drug resistance mutations (DRMs) under treatment pressure, limiting treatment options on a population level (Branda et al., 2024; Gupta et al., 2018). Traditionally, researchers have identified resistance mutations by comparing viral sequences from treated and untreated individuals (Willim et al., 2022), but this method tests mutations individually and fails to reveal interactions between them. In Luc Blassel’s study, ML methods were applied to analyze approximately 55,000 RT sequences from the UK. The analysis revealed six new mutations associated with resistance, providing candidates for further validation. They indicated that mutation interactions are primarily confined to the traditional paradigm, where primary DRMs confer resistance, while associated mutations modify resistance levels and/or offset the fitness costs linked to DRMs (Blassel et al., 2021).
4.3 Innovations in predictive modeling to address clinical outcomes and risks
Because of the challenges associated with gathering large cohort samples and comprehensive genetic data in clinical environments, data imbalance remains one of the significant challenges in applying ML to HIV research. To address this, researchers have combined ML with undersampling techniques, such as MAREV-1 and MAREV-2, aimed at identifying associations between Vif protein motifs and HIV clinical outcomes. These methods effectively identify genetic variants linked to HIV prognosis and mutations in accessory protein-coding regions, guiding new therapeutic strategies (Altamirano-Flores et al., 2023).
A meta-analysis of 24 studies involving 401,389 people living with HIV (PWH) evaluated machine learning models like random survival forests and SVM for predicting mortality risk. Machine learning showed strong potential for assessing long-term mortality risk, enhancing clinical decision-making (Li et al., 2024).
The persistent viral reservoir, particularly in CD4 T cells, remains a major obstacle to a cure. The complex interaction between the HIV reservoir and the host immune system made its characterization essential. A study (Semenova et al., 2024) identified significant correlations between immune cell populations and HIV reservoir size, including CD127 expression in CD4 T cells. Both intact and total proviral DNA levels showed a positive correlation with T cell activation and exhaustion, whereas ART duration and HIV-specific CD4 T cell responses were negatively correlated with intact provirus levels.
4.4 Machine learning insights into comorbidities and neurocognitive outcomes in HIV
Research involving PWH must consider comorbidities like cardiovascular disease, coinfections, and neurocognitive disorders, as these conditions often confound the primary effects of HIV (Long et al., 2016; Meyer-Rath et al., 2013; Calon et al., 2020).
Chronic HIV infection can lead to HIV-associated neurocognitive disorder (HAND). In a study conducted by Ogishi and Yotsuyanagi (2018), ML models identified key genetic features in the HIV env gene that predict HAND status. Specifically, three amino acid positions within the gp120 glycoprotein were predictive of HAND, with positions 291 and 315 being predictive of HIV-associated dementia (Holman and Gabuzda, 2012). These findings could guide the development of combination antiretroviral therapy (cART) regimens for HAND-associated quasispecies.
Peripheral nerve disorders (PNP) have been acknowledged for a long time as a characteristic of HIV-1 infection, becoming most evident as the disease progresses to AIDS (Kaku and Simpson, 2018). A study assessed PNP symptoms and signs through demographic, laboratory, and clinical variables. Using univariate and multivariate logistic regression combined with ML techniques, it identified key predictive factors for PNP. The most significant predictors included the duration of HIV-1 infection, peak plasma viral load, age, and low CD4+ T cell levels. These models improved classification performance and uncovered both known and novel factors, such as the duration of exposure to stavudine (Tu et al., 2021).
A subset of children with perinatal HIV (pHIV) faces persistent neurocognitive challenges (Ezeamama et al., 2016; Garvie et al., 2017; Bunupuradah et al., 2012). Recent studies have applied ML for neurocognitive development in pHIV children, demonstrating the feasibility of identifying those at risk for suboptimal outcomes. They also highlighted the interaction between HIV infection and mental health issues as early indicators of later neurocognitive challenges in pHIV children (Paul et al., 2020).
White matter changes in HIV-infected individuals can be quantified using brain-age gap (BAG) (Cole et al., 2017; Cole et al., 2018). Kalen J. Petersen used a Gaussian process regression model, trained on diffusion magnetic resonance imaging data from publicly available normative controls to assess BAG. Results indicated a significant interactive effect between BAG and detectable viral load (VL): PWH who had detectable VL exhibited an accumulation of +1.5 years in BAG per decade compared to HIV-negative controls (Petersen et al., 2022).
Studies have shown that the effects of smoking on DNA methylation in white blood cells (WBCs) can be detected using epigenome-wide association studies (EWAS) (You et al., 2020). Researchers used a DNA methylation-based ML approach to identify smoking-associated methylation sites predicting HIV prognosis and mortality. The chosen features successfully distinguished between favorable and poor HIV-related clinical outcomes in an independent sample. Additionally, the DNA methylation index derived from these CpGs was linked to mortality in the HIV-infected population. Notably, this study is the first to describe smoking-related DNA methylation associations in the HIV-infected population, demonstrating the utility of methylation-based machine learning in linking molecular information to clinical outcomes (Zhang et al., 2018).
5 Advancements in vaccine design
Over the past 15 years, numerous HIV-1 vaccine trials have failed to produce significant evidence of efficacy. Despite extensive efforts, the challenge of eliciting bNAbs has limited the success of these vaccine trials (Klasse and Moore, 2022).
Ed McGowan proposed a novel approach by analyzing HLA diversity and CD8 T cell immune responses, to identify key antigens (McGowan et al., 2021). Additionally, predicting viral strain sensitivity to monoclonal antibodies (mAbs) is critical for HIV vaccine and therapy development. Using amino acid sequence data and deep learning, researchers (Dănăilă and Buiu, 2022) have advanced the identification of potent antibody combinations by predicting HIV sensitivity to mAbs.
In a related effort, researchers developed a non-linear RF model using clinical and demographic data to predict the humoral response of PLWH to SARS-CoV-2 mRNA vaccines. This model suggests that additional booster doses may be necessary for PLWH, offering a tailored approach to vaccine strategies in clinical settings (Montesi et al., 2024).
6 Discussion
While AI is widely used and shows promising potential in HIV research, several challenges must be addressed.
AI models rely heavily on large, high-quality datasets to train algorithms effectively (Zou et al., 2023). Insufficient data can lead to incomplete training of AI models, which in turn affects the accuracy and reliability. Due to concerns over patient privacy, ethical issues, and the fragmentation of healthcare data, access to comprehensive HIV datasets remains limited especially across different populations and regions. In Emma Gillette’s study, participants identified accidental disclosure, stigma, and discrimination as significant risks of participating in research (Gillette et al., 2023). If participants lack confidence in researchers’ ability to protect data privacy and confidentiality, they may be discouraged from engaging in studies (Garett and Young, 2021). To ensure data security, one study implemented secure data governance rules and data encryption (Olatosi et al., 2019). Additionally, Zucchi proposed that, in some areas, adolescents need parental consent to participate in HIV research. Since it involved sensitive topics like sexual behavior, this requirement might violate their privacy and lead to embarrassment or even worse situations. Researchers suggested eliminating parental consent in ethical reviews to better respect, protect, and assist the participants (Zucchi et al., 2023).
Furthermore, HIV’s high genetic diversity and ability to evade the immune system present unique challenges (Dingens et al., 2017). HIV’s rapid mutation rate demands more sophisticated models to accurately predict viral behavior and other mechanisms, such as supporting long-term treatment and prevention strategies.
More importantly, the stigma surrounding AIDS (Guarnieri et al., 2024; Britton et al., 2024) places specific demands on AI technology. To be specific, false negatives may delay the initiation of ART (Gibas et al., 2019; Roche et al., 2024). Also, specificity is needed to avoid false positives that may cause psychological distress. Another important point is that AI models need to be accessible, facilitating testing in community and home settings to increase testing uptake and reduce stigma. In this way, the method can be considered highly practical by making testing more convenient and inclusive. It is advisable to conduct multicenter datasets and integrate a variety of ML algorithms for training. The goal is to create a more profound and powerful system to achieve a higher accuracy in analysis.
Collaboration among AI researchers, clinicians, and policymakers is of great importance. As mentioned above, removing parental consent can help not only protect adolescents but also enhance the size of datasets. Establishing ethical frameworks for data sharing, improving model transparency, and protecting data privacy are key to unlocking AI’s full potential in HIV research. With the advancement of AI models, personalized medicine and vaccines development could become a reality.
This review is a comprehensive study of AI applications in HIV research. It focuses on the integration of ML with other technologies, which previous literature has not explored enough. It aims to fill the gap in understanding how AI can enhance HIV researches and offer a new framework for future research. Also, it highlights the potential of AI to address current challenges in HIV management and contribute to global health goals.
Author contributions
RJ: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. LZ: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by National Key R&D Program of China (2023YFC2508200); Funding sources: National Natural Science Foundation of China (82273538) and the Public Health Research and Development Program of the Shenyang Science and Technology Bureau (22–321–33-12).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Altamirano-Flores, J. S., Alvarado-Hernández, L. Á., Cuevas-Tello, J. C., Tino, P., Guerra-Palomares, S. E., and Garcia-Sepulveda, C. A. (2023). Identification of clinically relevant HIV VIF protein motif mutations through machine learning and undersampling. Cells 12:772. doi: 10.3390/cells12050772
Arrigoni, R., Santacroce, L., Ballini, A., and Palese, L. L. (2023). AI-aided search for new HIV-1 protease ligands. Biomol. Ther. 13:858. doi: 10.3390/biom13050858
Balzer, L. B., Havlir, D. V., Kamya, M. R., Chamie, G., Charlebois, E. D., Clark, T. D., et al. (2020). Machine learning to identify persons at high-risk of human immunodeficiency virus acquisition in Rural Kenya and Uganda. Clin. Infect. Dis. 71, 2326–2333. doi: 10.1093/cid/ciz1096
Bao, Y., Medland, N. A., Fairley, C. K., Wu, J., Shang, X., Chow, E. P. F., et al. (2021). Predicting the diagnosis of HIV and sexually transmitted infections among men who have sex with men using machine learning approaches. J. Infect. 82, 48–59. doi: 10.1016/j.jinf.2020.11.007
Benitez, A. E., Musinguzi, N., Bangsberg, D. R., Bwana, M. B., Muzoora, C., Hunt, P. W., et al. (2020). Super learner analysis of real-time electronically monitored adherence to antiretroviral therapy under constrained optimization and comparison to non-differentiated care approaches for persons living with HIV in rural Uganda. J. Int. AIDS Soc. 23:e25467. doi: 10.1002/jia2.25467
Bitencourt-Ferreira, G., Rizzotto, C., and de Azevedo Junior, W. F. (2021). Machine learning-based scoring functions, development and applications with SAnDReS. Curr. Med. Chem. 28, 1746–1756. doi: 10.2174/0929867327666200515101820.32410551
Blassel, L., Tostevin, A., Villabona-Arenas, C. J., Peeters, M., Hué, S., Gascuel, O., et al. (2021). Using machine learning and big data to explore the drug resistance landscape in HIV. PLoS Comput. Biol. 17:e1008873. doi: 10.1371/journal.pcbi.1008873
Branda, F., Giovanetti, M., Sernicola, L., Farcomeni, S., Ciccozzi, M., and Borsetti, A. (2024). Comprehensive analysis of HIV-1 integrase resistance-related mutations in African countries. Pathogens 13:102. doi: 10.3390/pathogens13020102
Brites, C., Borges, Á. H., Sprinz, E., and Page, K. (2021). Editorial: HIV and viral co-infections. Front. Microbiol. 12:731337. doi: 10.3389/fmicb.2021.731337
Britton, M. K., Lembo, M., Li, Y., Porges, E. C., Cook, R. L., Cohen, R. A., et al. (2024). HIV stigma is associated with two-year decline in cognitive performance among people with HIV. AIDS Behav. 29, 90–100. doi: 10.1007/s10461-024-04508-7.39397137
Bunupuradah, T., Puthanakit, T., Kosalaraksa, P., Kerr, S. J., Kariminia, A., Hansudewechakul, R., et al. (2012). Poor quality of life among untreated Thai and Cambodian children without severe HIV symptoms. AIDS Care 24, 30–38. doi: 10.1080/09540121.2011.592815
Cai, Q., Yuan, R., He, J., Li, M., and Guo, Y. (2021). Predicting HIV drug resistance using weighted machine learning method at target protein sequence-level. Mol. Divers. 25, 1541–1551. doi: 10.1007/s11030-021-10262-y
Calon, M., Menon, K., Carr, A., Henry, R. G., Rae, C. D., Brew, B. J., et al. (2020). Additive and synergistic cardiovascular disease risk factors and HIV disease Markers' effects on white matter microstructure in virally suppressed HIV. J. Acquir. Immune Defic. Syndr. 84, 543–551. doi: 10.1097/QAI.0000000000002390
Chen, X., Wang, Z. X., and Pan, X. M. (2019). HIV-1 tropism prediction by the XGboost and HMM methods. Sci. Rep. 9:9997. doi: 10.1038/s41598-019-46420-4
Cole, J. H., Poudel, R. P. K., Tsagkrasoulis, D., Caan, M. W. A., Steves, C., Spector, T. D., et al. (2017). Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. Neuroimage 163, 115–124. doi: 10.1016/j.neuroimage.2017.07.059
Cole, J. H., Ritchie, S. J., Bastin, M. E., Valdés Hernández, M. C., Muñoz Maniega, S., Royle, N., et al. (2018). Brain age predicts mortality. Mol. Psychiatry 23, 1385–1392. doi: 10.1038/mp.2017.62
Dănăilă, V. R., and Buiu, C. (2022). Prediction of HIV sensitivity to monoclonal antibodies using aminoacid sequences and deep learning. Bioinformatics 38, 4278–4285. doi: 10.1093/bioinformatics/btac530
Dingens, A. S., Haddox, H. K., Overbaugh, J., and Bloom, J. D. (2017). Comprehensive mapping of HIV-1 escape from a broadly neutralizing antibody. Cell Host Microbe 21, 777–787.e4. doi: 10.1016/j.chom.2017.05.003.28579254
Ezeamama, A. E., Kizza, F. N., Zalwango, S. K., Nkwata, A. K., Zhang, M., Rivera, M. L., et al. (2016). Perinatal HIV status and executive function during school-age and adolescence: a comparative study of Long-term cognitive capacity among children from a high HIV prevalence setting. Medicine 95:e3438. doi: 10.1097/MD.0000000000003438
Foglierini, M., Nortier, P., Schelling, R., Winiger, R. R., Jacquet, P., O’Dell, S., et al. (2024). RAIN: machine learning-based identification for HIV-1 bNAbs. Nat. Commun. 15:5339. doi: 10.1038/s41467-024-49676-1
Garett, R., and Young, S. D. (2021). Geolocation, ethics, and HIV research. Health Technol. 11, 1305–1309. doi: 10.1007/s12553-021-00611-0.34722103
Garvie, P. A., Brummel, S. S., Allison, S. M., Malee, K. M., Mellins, C. A., Wilkins, M. L., et al. (2017). Roles of medication responsibility, executive and adaptive functioning in adherence for children and adolescents with perinatally acquired HIV. Pediatr. Infect. Dis. J. 36, 751–757. doi: 10.1097/INF.0000000000001573
Gibas, K. M., van den Berg, P., Powell, V. E., and Krakower, D. S. (2019). Drug resistance during HIV pre-exposure prophylaxis. Drugs 79, 609–619. doi: 10.1007/s40265-019-01108-x
Gillette, E., Naanyu, V., Nyandiko, W., Chory, A., Scanlon, M., Aluoch, J., et al. (2023). HIV-related stigma shapes research participation for youth living with HIV in Kenya. J. Int. Assoc. Provid. AIDS Care 22:23259582231170732. doi: 10.1177/23259582231170732.37101381
Godfrey-Faussett, P., Frescura, L., Abdool Karim, Q., Clayton, M., and Ghys, P. D.on Behalf of the 2025 Prevention Targets Working Group (2022). HIV prevention for the next decade: appropriate, person-centred, prioritised, effective, combination prevention. PLoS Med. 19:e1004102. doi: 10.1371/journal.pmed.1004102
Guarnieri, R., Botelho, F. C., Silva, L. A. V. D., and Zucchi, E. M. (2024). Social representations of HIV and healthcare among recently diagnosed youth. Rev. Saude Publica 58:6s. doi: 10.11606/s1518-8787.2024058005594
Gupta, R. K., Gregson, J., Parkin, N., Haile-Selassie, H., Tanuri, A., Andrade Forero, L., et al. (2018). HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression. 2018;18(2):139. doi: 10.1016/S1473-3099(17)30749-1. Lancet Infect. Dis. 18, 346–355. doi: 10.1016/S1473-3099(17)30702-8
Holman, A. G., and Gabuzda, D. (2012). A machine learning approach for identifying amino acid signatures in the HIV env gene predictive of dementia. PLoS One 7:e49538. doi: 10.1371/journal.pone.0049538
Holzinger, A., Langs, G., Denk, H., Zatloukal, K., and Müller, H. (2019). Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 9:e1312. doi: 10.1002/widm.1312
Hu, W., Joseph, S., Li, R., Woods, E., Sun, J., Shen, M., et al. (2024). Population impact and cost-effectiveness of artificial intelligence-based diabetic retinopathy screening in people living with diabetes in Australia: a cost effectiveness analysis. EClinicalMedicine 67:102387. doi: 10.1016/j.eclinm.2023.102387
Hu, H., Xiao, A., Zhang, S., Li, Y., Shi, X., Jiang, T., et al. (2019). DeepHINT: understanding HIV-1 integration via deep learning with attention. Bioinformatics 35, 1660–1667. doi: 10.1093/bioinformatics/bty842
Irvine, E. B., and Reddy, S. T. (2024). Advancing antibody engineering through synthetic evolution and machine learning. J. Immunol. 212, 235–243. doi: 10.4049/jimmunol.2300492
Janiesch, C., Zschech, P., and Heinrich, K. (2021). Machine learning and deep learning. Electron. Mark. 31, 685–695. doi: 10.1007/s12525-021-00475-2
Kaku, M., and Simpson, D. M. (2018). Neuromuscular complications of HIV infection. Handb. Clin. Neurol. 152, 201–212. doi: 10.1016/B978-0-444-63849-6.00016-5
Klasse, P. J., and Moore, J. P. (2022). Reappraising the value of HIV-1 vaccine correlates of protection analyses. J. Virol. 96:e0003422. doi: 10.1128/jvi.00034-22
Kutsal, M., Ucar, F., and Kati, N. (2024). Computational drug discovery on human immunodeficiency virus with a customized long short-term memory variational autoencoder deep-learning architecture. CPT Pharmacometrics Syst. Pharmacol. 13, 308–316. doi: 10.1002/psp4.13085
Laguno, M., Murillas, J., Blanco, J. L., Martínez, E., Miquel, R., Sánchez-Tapias, J. M., et al. (2004). Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for treatment of HIV/HCV co-infected patients. AIDS 18, 27–F36. doi: 10.1097/00002030-200409030-00003
Leidner, F., Kurt Yilmaz, N., and Schiffer, C. A. (2019). Target-specific prediction of ligand affinity with structure-based interaction fingerprints. J. Chem. Inf. Model. 59, 3679–3691. doi: 10.1021/acs.jcim.9b00457
Li, Y., Feng, Y., He, Q., Ni, Z., Hu, X., Feng, X., et al. (2024). The predictive accuracy of machine learning for the risk of death in HIV patients: a systematic review and meta-analysis. BMC Infect. Dis. 24:474. doi: 10.1186/s12879-024-09368-z
Li, J., Hao, Y., Liu, Y., Wu, L., Liang, H., Ni, L., et al. (2024). Supervised machine learning algorithms to predict the duration and risk of long-term hospitalization in HIV-infected individuals: a retrospective study. Front Public Health 11:1282324. doi: 10.3389/fpubh.2023.1282324
Li, X., Xu, L., Lu, L., Liu, X., Yang, Y., Wu, Y., et al. (2024). Differential T-cell profiles determined by hepatitis B surface antigen decrease among people with human immunodeficiency virus /hepatitis B virus coinfection on treatment. J. Transl. Med. 22:901. doi: 10.1186/s12967-024-05681-y
Long, L. C., Fox, M. P., Sauls, C., Evans, D., Sanne, I., and Rosen, S. B. (2016). The high cost of HIV-positive inpatient Care at an Urban Hospital in Johannesburg, South Africa. PLoS One 11:e0148546. doi: 10.1371/journal.pone.0148546
Maldarelli, F., Wu, X., Su, L., Simonetti, F. R., Shao, W., Hill, S., et al. (2014). HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183. doi: 10.1126/science.1254194
McGowan, E., Rosenthal, R., Fiore-Gartland, A., Macharia, G., Balinda, S., Kapaata, A., et al. (2021). Utilizing computational machine learning tools to understand immunogenic breadth in the context of a CD8 T-cell mediated HIV response. Front. Immunol. 12:609884. doi: 10.3389/fimmu.2021.609884
Mei, J., and Zhao, J. (2018). Prediction of HIV-1 and HIV-2 proteins by using Chou's pseudo amino acid compositions and different classifiers. Sci. Rep. 8:2359. doi: 10.1038/s41598-018-20819-x
Meyer-Rath, G., Brennan, A. T., Fox, M. P., Modisenyane, T., Tshabangu, N., Mohapi, L., et al. (2013). Rates and cost of hospitalization before and after initiation of antiretroviral therapy in urban and rural settings in South Africa. J. Acquir. Immune Defic. Syndr. 62, 322–328. doi: 10.1097/QAI.0b013e31827e8785
Montesi, G., Augello, M., Polvere, J., Marchetti, G., Medaglini, D., and Ciabattini, A. (2024). Predicting humoral responses to primary and booster SARS-CoV-2 mRNA vaccination in people living with HIV: a machine learning approach. J. Transl. Med. 22:432. doi: 10.1186/s12967-024-05147-1
Mulato, A., Lansdon, E., Aoyama, R., Voigt, J., Lee, M., Liclican, A., et al. (2024). Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability. Antimicrob. Agents Chemother. 68:e0137323. doi: 10.1128/aac.01373-23
Muriuki, B. M., Gicheru, M. M., Wachira, D., Nyamache, A. K., and Khamadi, S. A. (2013). Prevalence of hepatitis B and C viral co-infections among HIV-1 infected individuals in Nairobi, Kenya. BMC Res. Notes 6:363. doi: 10.1186/1756-0500-6-363
Namalinzi, F., Galadima, K. R., Nalwanga, R., Sekitoleko, I., and Uwimbabazi, L. F. R. (2024). Prediction of precancerous cervical cancer lesions among women living with HIV on antiretroviral therapy in Uganda: a comparison of supervised machine learning algorithms. BMC Womens Health 24:393. doi: 10.1186/s12905-024-03232-7
Nand, M., Maiti, P., Joshi, T., Chandra, S., Pande, V., Kuniyal, J. C., et al. (2020). Virtual screening of anti-HIV1 compounds against SARS-CoV-2: machine learning modeling, chemoinformatics and molecular dynamics simulation based analysis. Sci. Rep. 10:20397. doi: 10.1038/s41598-020-77524-x
Ogishi, M., and Yotsuyanagi, H. (2018). Prediction of HIV-associated neurocognitive disorder (HAND) from three genetic features of envelope gp120 glycoprotein. Retrovirology 15:12. doi: 10.1186/s12977-018-0401-x
Olatosi, B., Zhang, J., Weissman, S., Hu, J., Haider, M. R., and Li, X. (2019). Using big data analytics to improve HIV medical care utilisation in South Carolina: a study protocol. BMJ Open 9:e027688. doi: 10.1136/bmjopen-2018-027688.31326931
Onywera, H., Williamson, A. L., Cozzuto, L., Bonnin, S., Mbulawa, Z. Z. A., Coetzee, D., et al. (2020). The penile microbiota of black south African men: relationship with human papillomavirus and HIV infection. BMC Microbiol. 20:78. doi: 10.1186/s12866-020-01759-x
Paul, R. H., Cho, K. S., Belden, A. C., Mellins, C. A., Malee, K. M., Robbins, R. N., et al. (2020). Machine-learning classification of neurocognitive performance in children with perinatal HIV initiating de novo antiretroviral therapy. AIDS 34, 737–748. doi: 10.1097/QAD.0000000000002471
Peng, X., and Zhu, B. (2023). Machine learning identified genetic features associated with HIV sequences in the monocytes. Chin. Med. J. 136, 3002–3004. doi: 10.1097/CM9.0000000000002932.38018159
Petersen, K. J., Strain, J., Cooley, S., Vaida, F., and Ances, B. M. (2022). Machine learning quantifies accelerated white-matter aging in persons with HIV. J. Infect. Dis. 226, 49–58. doi: 10.1093/infdis/jiac156
Pham, T., Ghafoor, M., Grañana-Castillo, S., Marzolini, C., Gibbons, S., Khoo, S., et al. (2024). NPJ Syst. Biol. Appl. 10:48. doi: 10.1038/s41540-024-00374-0
Poonia, R. C., and Al-Alshaikh, H. A. (2024). Ensemble approach of transfer learning and vision transformer leveraging explainable AI for disease diagnosis: an advancement towards smart healthcare 5.0. Comput. Biol. Med. 179:108874. doi: 10.1016/j.compbiomed.2024.108874
Pouyanfar, S., Sadiq, S., Yan, Y., Tian, H., Tao, Y., Reyes, M. P., et al. (2018). A survey on deep learning: algorithms, techniques, and applications. ACM Comput. Surv. 51, 1–36. doi: 10.1145/3234150
Powell, B. M., and Davis, J. H. (2024). Learning structural heterogeneity from cryo-electron sub-tomograms with tomoDRGN. Nat. Methods 21, 1525–1536. doi: 10.1038/s41592-024-02210-z
Rawi, R., Mall, R., Shen, C. H., Farney, S., Shiakolas, A., Zhou, J., et al. (2019). Accurate prediction for antibody resistance of clinical HIV-1 isolates. Sci. Rep. 9:14696. doi: 10.1038/s41598-019-50635-w
Roche, S. D., Ekwunife, O. I., Mendonca, R., Kwach, B., Omollo, V., Zhang, S., et al. (2024). Measuring the performance of computer vision artificial intelligence to interpret images of HIV self-testing results. Front. Public Health 12:1334881. doi: 10.3389/fpubh.2024.1334881
Sarker, I. H. (2021). Machine learning: algorithms, real-world applications and research directions. SN Comput. Sci. 2:160. doi: 10.1007/s42979-021-00592-x
Semenova, L., Wang, Y., Falcinelli, S., Archin, N., Cooper-Volkheimer, A. D., Margolis, D. M., et al. (2024). Machine learning approaches identify immunologic signatures of total and intact HIV DNA during long-term antiretroviral therapy. eLife 13:RP94899. doi: 10.7554/eLife.94899
Shen, Y. (2024). Mycobacterium tuberculosis and HIV co-infection: a public health problem that requires ongoing attention. Viruses 16:1375. doi: 10.3390/v16091375
Steiner, M. C., Gibson, K. M., and Crandall, K. A. (2020). Drug resistance prediction using deep learning techniques on HIV-1 sequence data. Viruses 12:560. doi: 10.3390/v12050560
Sun, H. Y., Sheng, W. H., Tsai, M. S., Lee, K. Y., Chang, S. Y., and Hung, C. C. (2014). Hepatitis B virus coinfection in human immunodeficiency virus-infected patients: a review. World J. Gastroenterol. 20, 14598–14614. doi: 10.3748/wjg.v20.i40.14598
Tu, W., Johnson, E., Fujiwara, E., Gill, M. J., Kong, L., and Power, C. (2021). Predictive variables for peripheral neuropathy in treated HIV type 1 infection revealed by machine learning. AIDS 35, 1785–1793. doi: 10.1097/QAD.0000000000002955
Turbé, V., Herbst, C., Mngomezulu, T., Meshkinfamfard, S., Dlamini, N., Mhlongo, T., et al. (2021). Deep learning of HIV field-based rapid tests. Nat. Med. 27, 1165–1170. doi: 10.1038/s41591-021-01384-9
Vieira, S., Pinaya, W. H., and Mechelli, A. (2017). Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: methods and applications. Neurosci. Biobehav. Rev. 74, 58–75. doi: 10.1016/j.neubiorev.2017.01.002
Wang, Y., Li, Y., Chen, X., and Zhao, L. (2023). HIV-1/HBV coinfection accurate multitarget prediction using a graph neural network-based ensemble predicting model. Int. J. Mol. Sci. 24:7139. doi: 10.3390/ijms24087139
Wang, Y., Liu, C., Hu, W., Luo, L., Shi, D., Zhang, J., et al. (2024). Economic evaluation for medical artificial intelligence: accuracy vs. cost-effectiveness in a diabetic retinopathy screening case. NPJ Digit Med 7:43. doi: 10.1038/s41746-024-01032-9
Wei, Y., Li, W., Du, T., Hong, Z., and Lin, J. (2019). Targeting HIV/HCV coinfection using a machine learning-based multiple quantitative structure-activity relationships (multiple QSAR) method. Int. J. Mol. Sci. 20:3572. doi: 10.3390/ijms20143572
Willim, R., Shadabi, E., Sampathkumar, R., Li, L., Balshaw, R., Kimani, J., et al. (2022). High level of pre-treatment HIV-1 drug resistance and its association with HLA class I-mediated restriction in the Pumwani sex worker cohort. Viruses 14:273. doi: 10.3390/v14020273
World Health Organization (2015). Guideline on when to start antiretroviral therapy and on pre-exposure prophylaxis for HIV. Geneva: World Health Organization.
You, W. X., Comins, C. A., Jarrett, B. A., Young, K., Guddera, V., Phetlhu, D. R., et al. (2020). Facilitators and barriers to incorporating digital technologies into HIV care among cisgender female sex workers living with HIV in South Africa. Mhealth 6:15. doi: 10.21037/mhealth.2019.12.07.32270007
You, C., Wu, S., Zheng, S. C., Zhu, T., Jing, H., Flagg, K., et al. (2020). A cell-type deconvolution meta-analysis of whole blood EWAS reveals lineage-specific smoking-associated DNA methylation changes. Nat. Commun. 11:4779. doi: 10.1038/s41467-020-18618-y
Yu, Z., Nguyen, J., Nguyen, T. D., Kelly, J., Mclean, C., Bonnington, P., et al. (2022). Early melanoma diagnosis with sequential Dermoscopic images. IEEE Trans. Med. Imaging 41, 633–646. doi: 10.1109/TMI.2021.3120091
Zhang, X., Hu, Y., Aouizerat, B. E., Peng, G., Marconi, V., Corley, M., et al. (2018). Machine learning selected smoking-associated DNA methylation signatures that predict HIV prognosis and mortality. Clin. Epigenetics 10:155. doi: 10.1186/s13148-018-0591-z
Zorn, K. M., Lane, T. R., Russo, D. P., Clark, A. M., Makarov, V., and Ekins, S. (2019). Multiple machine learning comparisons of HIV cell-based and reverse transcriptase data sets. Mol. Pharm. 16, 1620–1632. doi: 10.1021/acs.molpharmaceut.8b01297
Zou, H., Lu, Z., Weng, W., Yang, L., Yang, L., Leng, X., et al. (2023). Diagnosis of neurosyphilis in HIV-negative patients with syphilis: development, validation, and clinical utility of a suite of machine learning models. EClinicalMedicine 62:102080. doi: 10.1016/j.eclinm.2023.102080
Zucchi, E. M., Ferguson, L., Magno, L., Dourado, I., Greco, D., Ferraz, D., et al. (2023). When ethics and the law collide: a multicenter demonstration cohort study of pre-exposure prophylaxis provision to adolescent men who have sex with men and transgender women in Brazil. J. Adolesc. Health 73, S11–S18. doi: 10.1016/j.jadohealth.2023.08.002
Keywords: HIV-human immunodeficiency virus, acquired immuno deficiency syndrome (AIDS), artificial intelligence - AI, machine learning, virology, deep learning
Citation: Jin R and Zhang L (2025) AI applications in HIV research: advances and future directions. Front. Microbiol. 16:1541942. doi: 10.3389/fmicb.2025.1541942
Edited by:
Anna Luganini, University of Turin, ItalyReviewed by:
Yuko Morikawa, Kitasato University, JapanTexca Tatevari Méndez López, Laboratorio Estatal de Salud Publica de Michoacan Vigilancia Epidemiologica, Mexico
Rebecca Batorsky, Tufts University, United States
Copyright © 2025 Jin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Zhang, bGl6aGFuZ18xMDAxQDEyNi5jb20=
 Ruyi Jin
Ruyi Jin Li Zhang
Li Zhang