- Jiangxi Provincial Key Laboratory of Agricultural Non-Point Source Pollution Control and Waste Comprehensive Utilization, Institute of Agricultural Applied Microbiology, Jiangxi Academy of Agricultural Sciences, Nanchang, China
Clubroot disease is an important disease of cruciferous crops worldwide caused by Plasmodiophora brassicae. The pathogen P. brassicae can infect almost all cruciferous crops, resulting in a reduction in yield and quality of the host plant. The first part of this review outlines the process of P. brassicae infestation, effectors, physiological pathotypes and identification systems. The latter part highlights and summarizes the various current control measures and research progress on clubroot. Finally, we propose a strategic concept for the sustainable management of clubroot. In conclusion, this paper will help to deepen the knowledge of P. brassicae and the understanding of integrated control measures for clubroot, and to lay a solid foundation for the sustainable management of clubroot.
1 Introduction
Clubroot disease caused by obligate biotrophic, parasitic protist Plasmodiophora brassicae Woronin is a major constraint in the current production of cruciferous crops (Ahmed et al., 2020; Hwang et al., 2012; Saharan et al., 2021). The host range of P. brassicae is wide, and all 330 genera and 3,700 species of the Brassicaceae family may be hosts of P. brassicae, including rape, Chinese cabbage, bok choy, kale, radish, cauliflower, mustard and other cultivated and wild species (Chai et al., 2014; Hwang et al., 2012). The infestation of P. brassicae in the host leads to the slow growth of tumors on the roots, which hinders the plant’s capacity to absorp water and nutrients. This may ultimately limit the host’s growth and, in severe situations, lead to death (Howard et al., 2010). It has been estimated that P. brassicae causes 10–15% yield loss in cruciferou crop production globally each year (Dixon, 2009a; Strehlow et al., 2014). In addition, the infection of P. brassicae also has an adverse effect on the quality of these crops (Howard et al., 2010). The spread of P. brassicae is extremely fast, and its geographical distribution is constantly expanding rapidly. It is reported that P. brassicae is widely distributed in more than 80 countries such as Europe, North America, Latin America, Oceania and Asia, causing economic losses of hundreds of millions every year (Javed et al., 2023).
The P. brassicae can infest both seedling and adult stages of the host plant causing damage. The absorption of water and nutrients in the infected plant is blocked, affecting the normal growth and development of the plant. In the early stage of infection, the aboveground part of the plant has no obvious symptoms, and some plants grow slowly. In the middle stage of infection, the plant is dwarfed and some leaves turn yellow and wither. In the late stage of infection, root galls of varying sizes, shapes and locations form on the roots (Figure 1), leading to plant death in severe cases (Greer et al., 2023; Zamani-Noor et al., 2022).
2 Process of infestation by Plasmodiophora brassicae
The lifecycle of P. brassicae consists of three phases, survival in soil, primary root hair infection, and secondary cortical infection (Feng et al., 2014; Ludwig-Müller et al., 2009). The resting spores are the main body of the soil survival stage and the initial stage of the life cycle of P. brassicae. Under appropriate conditions, resting spores of P. brassicae in the soil germinate and release primary zoospores with biflagellates, which subsequently move to the vicinity of the host plant roots to begin the process of infestation (Olszak et al., 2019; Schuller and Ludwig-Müller, 2016).
The process of infection of host plants by the P. brassicae has now been more clearly described (Ji et al., 2014; Kageyama and Asano, 2009). The infection of P. brassicae to host roots is generally divided into two stages. The first stage is the primary root hair infection, which lasts about 7 days. Around 1 day post infection (dpi), primary spores puncture the host cell wall and enter root hair cells or epidermal cells, producing uinucleate primary plasmodium. At 1–3 dpi, the uinucleate primary plasmodium develop into multinucleated zoosporangial plasmodium through mitosis. Then, at 3–4 dpi, large numbers of uninucleate secondary spores are produced with cytoplasmic cleavage. At 4–7 dpi, the uninucleate secondary zoospores were released into the living epidermal cells of the root hair cells, and finally two mononuclear secondary spores were combined in the epidermal cells of the roots to form a diploid uninucleate zygote. The second stage is secondary cortical infection, which takes about 17 days. At 8 dpi, uninucleate secondary plasmodium appeared in the cortical cells of the host, indicating the beginning of secondary infection. Next, at the 10 dpi, the uninucleate secondary plasmodium developed into binucleate secondary plasmodium. This then continues to develop into tetranucleate and multinucleate secondary plasmodium resulting in root expansion. The uninucleate resting spores are produced on day 24 and are released into the soil after maturation to complete a life cycle and enter the next infection cycle (Liu et al., 2020a; Liu et al., 2020b).
3 Effectors of Plasmodiophora brassicae
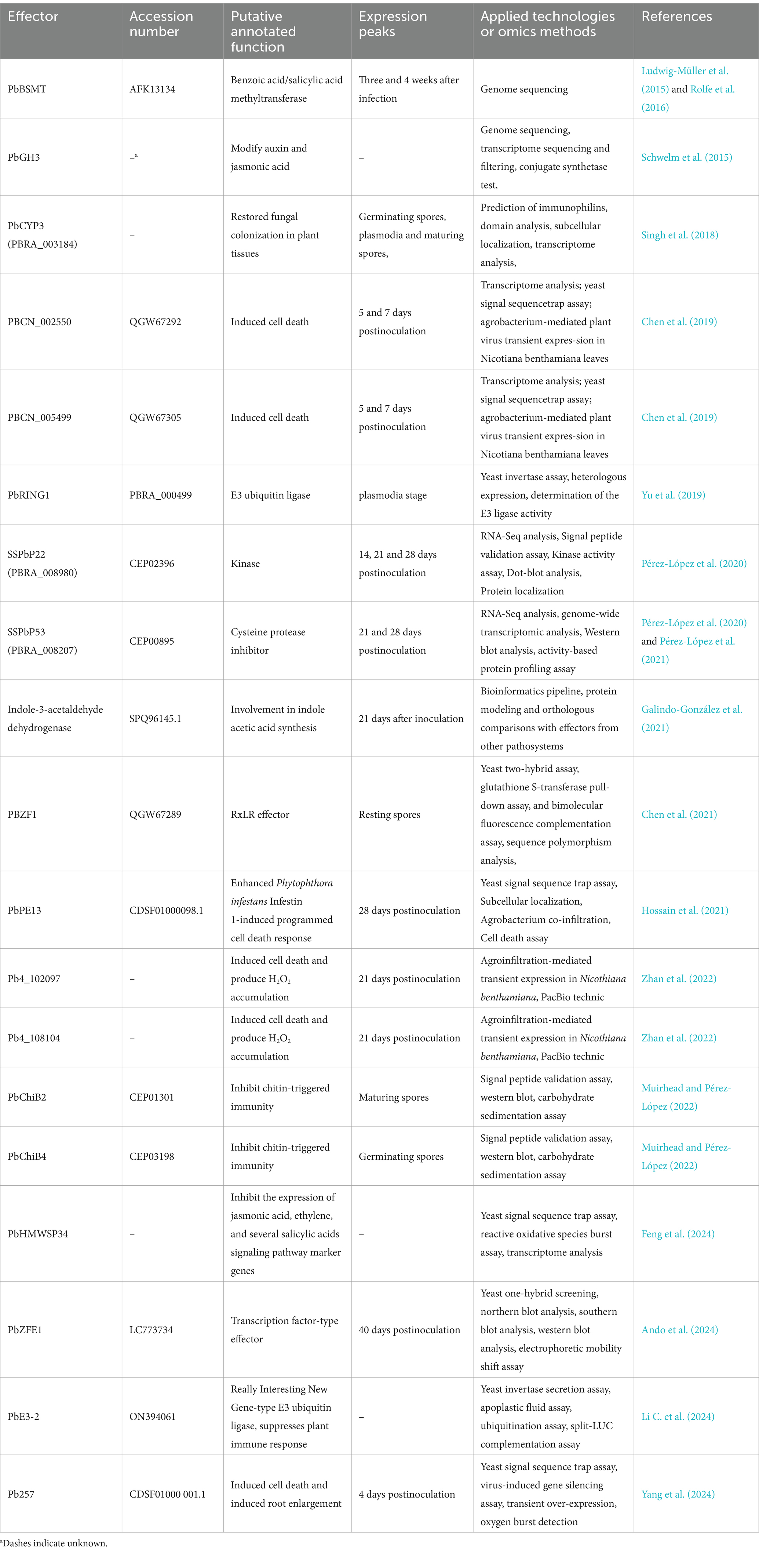
Plant defense against pathogens relies primarily on their own immune system, which recognizes the pathogens and initiates an effective defense response. Conversely, the key to successful infestation of a plant host by a pathogen lies in the manipulation of the host’s defense system through effectors (Cui et al., 2015; Dodds and Rathjen, 2010; Dodds et al., 2024). The identification of effectors is a prerequisite for understanding how pathogens manipulate immune responses and disease resistance genes in plants (Pérez-López et al., 2018). However, P. brassicae is an obligate biotroph cannot be cultured away from its host, representing a major challenge in the research of their effectors. With the rapid development of genomic, transcriptomic, proteomic and other biotechnologies, particularly the powerful predictive capabilities of bioinformatics tools, an increasing number of effectors from P. brassicae have been identified and predicted (Schwelm and Ludwig-Müller, 2021; Siemens et al., 2009). The identified and predicted putative effectors of P. brassicae are summarized in Table 1.
PbBSMT was reported to be the first effector of P. brassicae to be identified. It is a protein with homologies to plant SABATH-type methyltransferases that reduces or removes the host salicylic acid (SA) defense signaling pathway by methylating SA (Ludwig-Müller et al., 2015; Rolfe et al., 2016). Furthermore, transcriptome analyses revealed that among the genes, PbBSMT was one of the most highly expressed during P. brassicae infestation, suggesting that it plays an important role in the infestation process (Ciaghi et al., 2019). Chen et al. (2019) found that 28 secreted proteins expressed during the primary infection stage of P. brassicae infestation inhibited cell death to facilitate their propagation and colonization. The response mechanism of resistant and susceptible Brassica napus to P. brassicae infestation was assessed through transcriptomic analysis, which identified 21 gene sequences associated with secretory proteins that correlate with the virulence strength of P. brassicae. Among them, a gene similar to indole-3-aldehyde dehydrogenase, with ID SPQ96145.1, is involved in the synthesis of indole acetic acid and is a promising candidate effector. (Galindo-González et al., 2021). Yang et al. (2022a, 2022b) found that primary zoospores of P. brassicae released many effectors during infestation of B. rapa and their expression levels were significantly induced. In addition, 32 small secreted proteins have been reported to be highly expressed during the secondary infestation of P. brassica. Among these, SSPbP22 was localized in both the cytoplasm and nucleus and was validated as a kinase (Pérez-López et al., 2020), while SSPbP53 is a papain-like cysteine protease (PLCP) inhibitor that suppresses PLCP activity in six susceptible cruciferous hosts (Pérez-López et al., 2021). Zhan et al. (2022) screened 518 secretory proteins from the P. brassicae genome, among which 55 candidate effectors can inhibite cell death induced by Bcl-2-associated X protein, and 21 candidate effectors can inhibit immunity induced by the bacterial pathogen Pseudomonas syringae pv. tomato strain DC3000 of avrRps4 in Arabidopsis.
4 Physiological pathotypes of Plasmodiophora brassicae
Pathogens can rapidly adapt to changing environments, and the generation of pathogen types or races is often achieved through mutations in single effector genes. Horizontal gene transfer (HGT) is very common in protists (Sanders, 2006; Shi-Kunne et al., 2019), and HGT is also a means for pathogens to broaden their host range (Mehrabi et al., 2011; Ochman et al., 2000). Therefore, some scholars have suggested that P. brassicae is also a species that can proximally perform horizontal gene transfer (Zhang et al., 2015; Zheng et al., 2018).
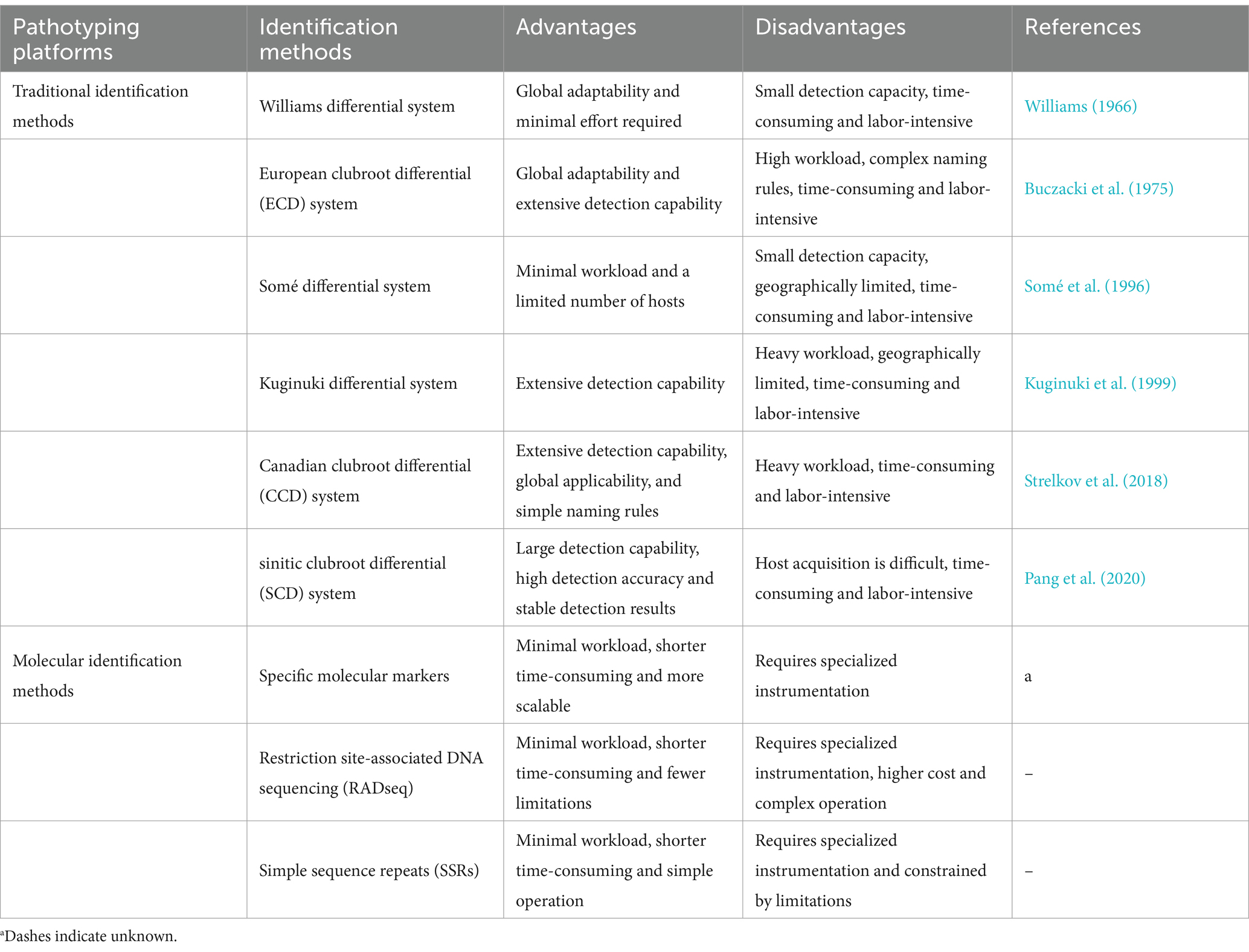
Pathogenic differentiation of physiological microspecies exists in P. brassicae. The P. brassicae isolated from different regional sources may have different genetic backgrounds, resulting in different disease phenotypic characteristics on specific host plants. The P. brassicae can be classified into different pathotypes (physiological races) based on their virulence patterns on different hosts plants (Liu et al., 2022). Studies have revealed that the population structure of P. brassicae is changing, conferring diversity and heterogeneity to its populations (Hasan et al., 2021). The genetic diversity of P. brassicae contributes to the mutation of its field populations into multiple pathotypes, and the large variation in pathogenicity of different pathotypes increase the difficulty of controlling these pathogens (Yang et al., 2020a). The identification of pathogen types is very important. This is due to the fact that without knowledge of the pathogen types and their virulence in the field, it is difficult to breed the appropriate resistance in host cultivars and for growers to choose the best variety or cultivar. Therefore, global efforts have been undertaken to develop accurate and reproducible identification systems or techniques for P. brassicae to ensure precise identification and control of different pathotypes (Hollman et al., 2023; Zamani-Noor and Jedryczka, 2024; Zeng et al., 2024).
4.1 Identification methods
During the past decades of research on P. brassicae, researchers have developed several pathotype classification systems based on serial host sets of Brassica species that exhibit resistance or partial resistance to P. brassicae in different hosts. The Williams differential system was the earliest system developed and used four hosts (Badger shipper, Jersey Queen, Laurentian and Wilhelmsburger) for identification. Theoretically, this system can distinguish 16 pathotypes or physiological races (Williams, 1966). The host set for European clubroot differential (ECD) was identified by selecting five from each of B. rapa, B. napus and B. oleracea, and pathotypes of P. brassicae were coded using a binary notation system. This is currently the universal identification system for clubroot pathogens in Europe (Buczacki et al., 1975). Subsequently, Somé et al. (1996) selected three cultivars of B. napus for identification and classified the P. brassicae prototypes into five types. Kuginuki et al. (1999) developed a P. brassicae pathotype identification system that is more suitable for Japan by using 18 hosts of a Japanese clubroot-resistant (CR) F1 hybrid (F1) cultivars and Brassica rapa lines for identification. The Canadian Clubroot Differential (CCD) set selected 13 Brassica hosts for assessment of P. brassicae populations, with a total of 17 detectable pathotypes. The system uses a combination of letters and numbers to designate pathotype (Strelkov et al., 2018). Pang et al. (2020) used a set of eight differential inbred lines of Chinese cabbage that contain clubroot-resistant (CR) genes as hosts to develop a sinitic clubroot differential (SCD) system. This system was designed to identify hosts of P. brassicae in China and is a more suitable system for determining the pathotype of P. brassicae in the region.
Although the molecular basis for the classification of P. brassicae pathotype is unclear, one viewpoint that is generally shared by most researchers is that differences between pathotypes ultimately reflect differences in their genetic composition, that is, the presence, absence, or differential expression of certain pathogenicity-associated genes (Zhang et al., 2015). Therefore, classification of rhizobial pathotype at the molecular level is a relevant research trend (Kubo et al., 2017). The development of specific molecular markers related to specific pathotypes will serve as a crucial tool for identifying and monitoring P. brassicae populations. Previous studies have reported several molecular markers that can be used to identify different pathotypes. Manzanares-Dauleux et al. (2000) identified a specific random amplified polymorphic DNA (RAPD) marker, OPL141200, for pathotype 1 of P. brassicae based on the identification system established by Somé et al. (1996) that can be used to rapidly and reliably identify isolates of pathotype 1. The Cr811 gene can be used as a molecular marker to distinguish between pathotype 5 and other pathotypes based on the Williams differential set identification system (Zhang et al., 2015). Zhou et al. (2018) developed primers P5XF3/P5XR3 and TaqMan probe P5XP3 based on the 18S-ITS region of P. brassicae for rapid and reliable diagnosis and quantification of pathotype 5. Zheng et al. (2019) identified six genes unique to pathotype 4 identified by the Williams difference set, PBRA_003263 and PBRA_003268 were present in all P4 isolates, PBRA_000003 and Novel512 was present in P4-1, and Novel137 PBRA_005772 was present in P4-2. These genes can be used as molecular markers to distinguish pathotype 4 from other pathotypes and also to distinguish between different types of pathotype 4. Yang et al. (2020a) found three molecular marker genes that screened for pathotype 7. PBRA_000303 appeared only in pathotype 7 identified based on the Williams difference set; PBRA_006533 and PBRA_009559 appeared in pathotype 7 and one genotype of pathotype 4.
Restriction site-associated DNA sequencing (RADseq) employs high-throughput sequencing technology to identify polymorphic genetic markers within genomes. It not only facilitates the rapid and low-cost identification of a large number of polymorphisms within species, but also has fueled studies in ecological, evolutionary and conservation genomics (Davey and Blaxter, 2010; Andrews et al., 2016). Holtz et al. (2018) detected P. brassicae in Canada as two distinct and highly divergent populations using RADseq with the removal of the host’s DNA and the endophyte’s DNA. Simple sequence repeats (SSRs) are frequently used as markers for genetic analyses of many eukaryotic organisms (Guichoux et al., 2011). Kubo et al. (2017) developed SSR markers for P. brassicae to classify 24 isolates of P. brassicae from Japan, and found that there was a close genetic relationship between isolates of pathotype 2 and pathotype 4.
However, these identification method s have certain limitations (Table 2). Traditional identification systems typically require a considerable amount of time, usually around 2 months, making them both time-consuming and labor-intensive (Holtz et al., 2021; Manzanares-Dauleux et al., 2000). Moreover, the pathogenicity of P. brassicae spores is influenced not only by environmental factors, such as temperature and pH, but also by their maturity. This interplay can diminish the accuracy and consistency of pathotype identification (Sharma et al., 2011; Tso et al., 2022). Molecular identification methods differ in terms of sensitivity, scalability, accessibility, and operational costs. A major challenge is to identify polymorphisms that provide consistent genomic and phenotypic clustering of pathotypes. Moreover, it is difficult to distinguish individual pathotypes using unique sequences in one region of the genome and may be necessary to rely on multiple regions for identification. In addition, primers designed for molecular identification need to be specific enough to avoid the amplification of non-target genes (Tso et al., 2021).
4.2 Single-spore isolation technology
The P. brassicae in most studies originated from field populations, i.e., isolated from root galls or plants of field-incurred hosts. However, field populations typically consist of a mixture of different pathotypes, leading to the possibility that pathotypes with lower frequencies may be masked by others that occur at higher frequencies (Jones et al., 1982). This increases the difficulty of breeding resistant varieties and hinders research on resistance mechanisms of host against P. brassicae. In response to these challenges, isolation of a single spore from populations of P. brassicae is an effective method. Accurate identification of the pathotype of P. brassicae populations is essential for screening effective sources of genetic resistance. Additionally, the successful isolation of single spores is important for pathotype identification efforts (Klewer et al., 2001; Xue et al., 2008).
Methods for single spore isolation of P. brassicae are diverse and constantly being improved and optimized. The primary objective is to successfully obtain single spore isolates and enhance the success rate of single spore inoculation. A report in 1977 showed that single spores of P. brassicae were cultivated using liquid paraffin droplets as a medium and were successfully inoculated onto Brassica plants, although only 2 out of 250 inoculations were successful (Buczacki, 1977). Voorrips (1996) prepared a suspension of P. brassicae spores at a concentration of 500 spores/ml, but the success rate of inoculation was only 1.2%. Xue et al. (2008) prepared a spore suspension containing 5% glycerol to avoid spore aggregation and then inoculated the droplets onto the roots of cabbage seedlings that had germinated for 1–2 weeks. The seedlings were placed in petri dishes lined with filter paper and then incubated at 21°C and darkness for 2d before being transplanted into nutrient pots and incubated at 24°C in light and 18°C in darkness, and the success rate of inoculation was up to 16.95%. These studies were conducted by preparing highly diluted spore suspensions of P. brassicae and using droplet inoculation to ultimately obtain single spores. However, there are obstacles that cannot be ignored with the droplet approach to spore suspensions. For example, it is difficult to demonstrate that there is only one microscopic spore in a small droplet, and there may be problems with the absence of a single spore or multiple spores in a droplet. The surface tension of the droplet hinders the identification of stationary spores. In addition, small droplets evaporate easily, whereas large droplets are difficult to identify under high magnification (Askarian et al., 2021; Diederichsen et al., 2016).
Another major method for isolating of single spores of P. brassicae was performed using agar as a medium. Tinggal and Webster (1981) developed a technique to inoculate P. brassicae with agar in a petri dish, which had a higher success rate relative to the paraffin drop method. Follow-up studies have continually improved this method, resulting in the optimization of the technique for single spore isolation and continued improvement in the success of inoculation (Askarian et al., 2021; Jones et al., 1982). Follow-up studies have continually improved this method, resulting in the optimization of the technique for single spore isolation with success rates of up to 30% (Askarian et al., 2021; Jones et al., 1982). Somé et al. (1996) aspirated a diluted suspension of P. brassicae onto an agar block, and the agar block containing one spore was picked out with a needle by microscopic observation and then inoculated into the host root with a success rate of about 9.6%. Despite the high success rate of isolating single spore inoculations of P. brassicae using agar-based methods, there are still some limitations to this method. For example, special equipment and micromanipulators are necessary to cut and lift agar gel blocks that contain a single spore (Askarian et al., 2021). In addition, agar tends to solidify and agar blocks are prone to bubbling and can have problems with impurities, which can affect the efficiency and accuracy of single spore isolation.
In addition to these two isolation methods, researchers are continually exploring other techniques to isolate single spores of P. brassicae. Diederichsen et al. (2016) inoculated infected root hairs containing a single sporangiosorus of P. brassicae onto Brassica seedlings, and successfully obtained single pathotype isolates with a highest infection rate of 17.9%. Recently, a newly developed and efficient systematic protocol for isolation of single spores of P. brassicae could increase the rate of infection success up to 47.38%. The procedure is as follows: inoculation of 2-day-old seedlings in cryoboxes, addition of nutrient solution for culture, and microscopic observation of single spores prior to medium incubation (Lv et al., 2021).
Although the P. brassicae extracted from single spores is considered genetically homogeneous and it has been shown that single spores are genetically stable (Heo et al., 2009). However, the possibility of introducing heterogeneity in the reproduction process of P. brassicae still cannot be ruled out (Askarian et al., 2021). Graf et al. (2004) performed electrophoretic karyotyping of single spore isolates of P. brassicae using contour-clamped homogeneous electric field gel electrophoresis and found that they had chromosomally polymorphic. Therefore, the single spore isolation method was unable to obtain consistent and accurate DNA information. In addition, Sharp et al. (2018) sequenced the genomes of more than 200 replica lines of yeast (Saccharomyces cerevisiae) and analyzed the number, types, locations, and effects of thousands of mutations. They found that haploids were more prone to single-nucleotide mutations and mitochondrial mutations, while larger structural changes were more common in diploids (Diederichsen et al., 2016).
5 Management strategy of Plasmodiophora brassicae
As a soil-borne disease, P. brassicae is highly insidious and difficult to detected in the early stages of the disease. In addition, it is highly contagious and can spread rapidly with the movement of farm machinery operated in infested field, and can also be transmitted with wind-raised dust and seed translocation (Cao et al., 2009; Gossen et al., 2015; Rennie et al., 2015). The management of clubroot disease has always posed a significant challenge, and various measures have been used to control this disease. However, the implementation of these methods has demonstrated that none are sufficient to completely eradicate the pathogen (Struck et al., 2022).
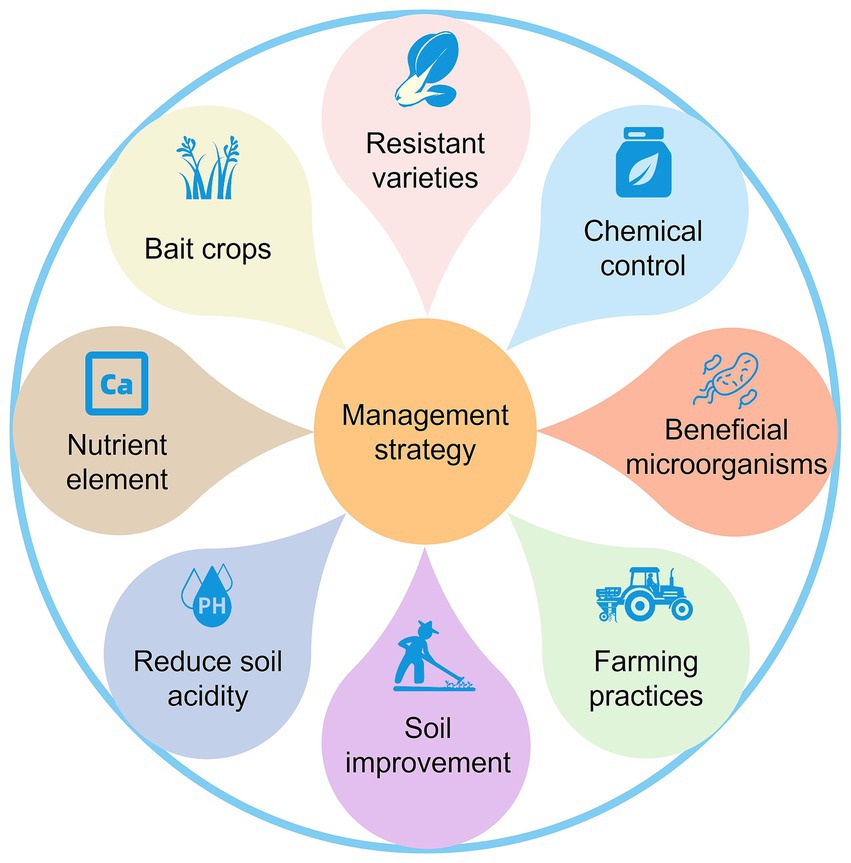
Due to the variations in pathotypes of P. brassicae populations, as well as differences in environmental conditions, farming patterns, and control policies across regions, it is challenging to establish a universally applicable and effective strategies for controlling clubroot. The development of locally adapted integrated strategies for controlling clubroot, tailored to the specific conditions of different regions, is a key focus for future efforts. We summarized various management strategies for P. brassicae based on previous studies (Figure 3).
5.1 Planting resistant varieties
Developing and cultivating resistant varieties is the most economical, effective and safe sustainable clubroot management strategy. The development of genetically resistant cultivars can mitigate the harmful effects of P. brassicae and reduce crop yield losses (Dolatabadian et al., 2022; Piao et al., 2009). Brassica plants consist mainly of three diploid basic species, including B. rapa (AA, 2n = 20), B. nigra (BB, 2n = 16), and B. oleracea (CC, 2n = 18) (Zhu et al., 2022). The genomes of all three of these basic species are likely to contain clubroot resistance genes. Hu et al. (2024) incorporated the clubroot-resistance gene Rcr1 into clubroot-susceptible elite canola lines using a modified CRISPR/Cas9-based intragenic vector system to develop selection-marker-free canola germplasms with a stable resistant phenotype in <2 years.
The development of technologies such as high-throughput sequencing, genomics, and molecular genetics has greatly promoted the rapid identification and application of clubroot resistance genes (Liégard et al., 2019). It has been reported that resistance in the A genome is usually controlled by a major dominant gene, whereas resistance in the C genome is usually regulated by multiple quantitative trait loci (Hasan et al., 2021). The A genome species contains numerous specific, single, dominant resistance genes and serves as the primary source of resistance genes for P. brassicae. Most resistance genes in the species with the C genome consist of a combination of major and minor genes, exhibiting a high degree of continuity (Dolatabadian et al., 2022; Neik et al., 2017). However, the number of resistance genes in the B genome is significantly lower than those in the A and C genomes. It was not until 2019 that the first single dominant gene for clubroot resistance, Rcr6, was identified. This gene is located on the B3_Canada chromosome and is homologous to chromosome A08 in the A genome (Chang et al., 2019). The identified clubroot resistant genes and their mapped chromosomes are summarized in Table 3.

Table 3. List of identified clubroot resistant genes to Plasmodiophora brassicae and their mapped chromosomes.
Most clubroot resistance genes have been identified in Brassica plants, although other cruciferous species also possess the potential genetic resources for resistance. Wang et al. (2023) reported a broad-spectrum clubroot resistance gene, WeiTsing (WTS), isolated from Arabidopsis, which is induced to up-regulate its expression in the pericycle upon P. brassicae infection to prevent the pathogen colonization in the stele. In addition, WTS encodes a small protein localized in the endoplasmic reticulum that oligomerizes to form a pentameric cation-selective ion channel permeable to Ca2+. The infestation of P. brassicae can activate this calcium-permeable channel and thereby triggering plant immunity.
At present, there are numerous detailing the mechanisms by which resistant varieties against P. brassicae. Most studies have revealed that P. brassicae can be recognized and triggers a complex series of immune responses when infesting resistant varieties (Mei et al., 2019; Ning et al., 2019; Ochoa et al., 2023). With the development of bioinformatics, multiple omics technologies have made certain progress in elucidating resistance mechanisms. This has not only allowed us to gain a deeper understanding of the dynamic changes that transpired during host-pathogen interactions, but also provided us with a wider perspective to explore the potential mechanisms of action (Ludwig-Müller, 2022; Shaw et al., 2022). A recent study revealed the resistance mechanism of the cabbage variety “Shangpin” with broad-spectrum immunity to P. brassicae. It may enhance disease resistance by recruiting beneficial microorganisms, such as Flavobacterium and Sphingomonas, to maintain the stability of the root microbial community structure while inhibiting the growth and reproduction of P. brassicae (Fang et al., 2024).
However, most sources of resistance are specific to the P. brassicae prototype and the main resistant varieties circulating on the market are single-gene, which makes resistance of these varieties not durable. In particular, the selection pressure exerted by P. brassicae populations on resistant varieties produces changes in virulence that may lead to loss of resistance (Botero-Ramírez et al., 2021). Faced with the pressure of resistance of from resistant varieties being overcome by P. brassicae, the primary coping strategies can be worked from the following ways. On the one hand, new resistance genes should be continuously explored and new resistant varieties should be developed. On the other hand, the polymerization of two or more resistance genes to develop superior resistant varieties. Before these resistance genes can be utilized in breeding efforts, their characteristics and relationships must be evaluated in detail. Li et al. (2022) used a marker-assisted selection method to polymerize two resistance genes, CRa and CRd, into Chinese cabbage to develop homozygous pyramided lines that showed strong resistance to six different pathotypes of P. brassicae.
5.2 Chemical control
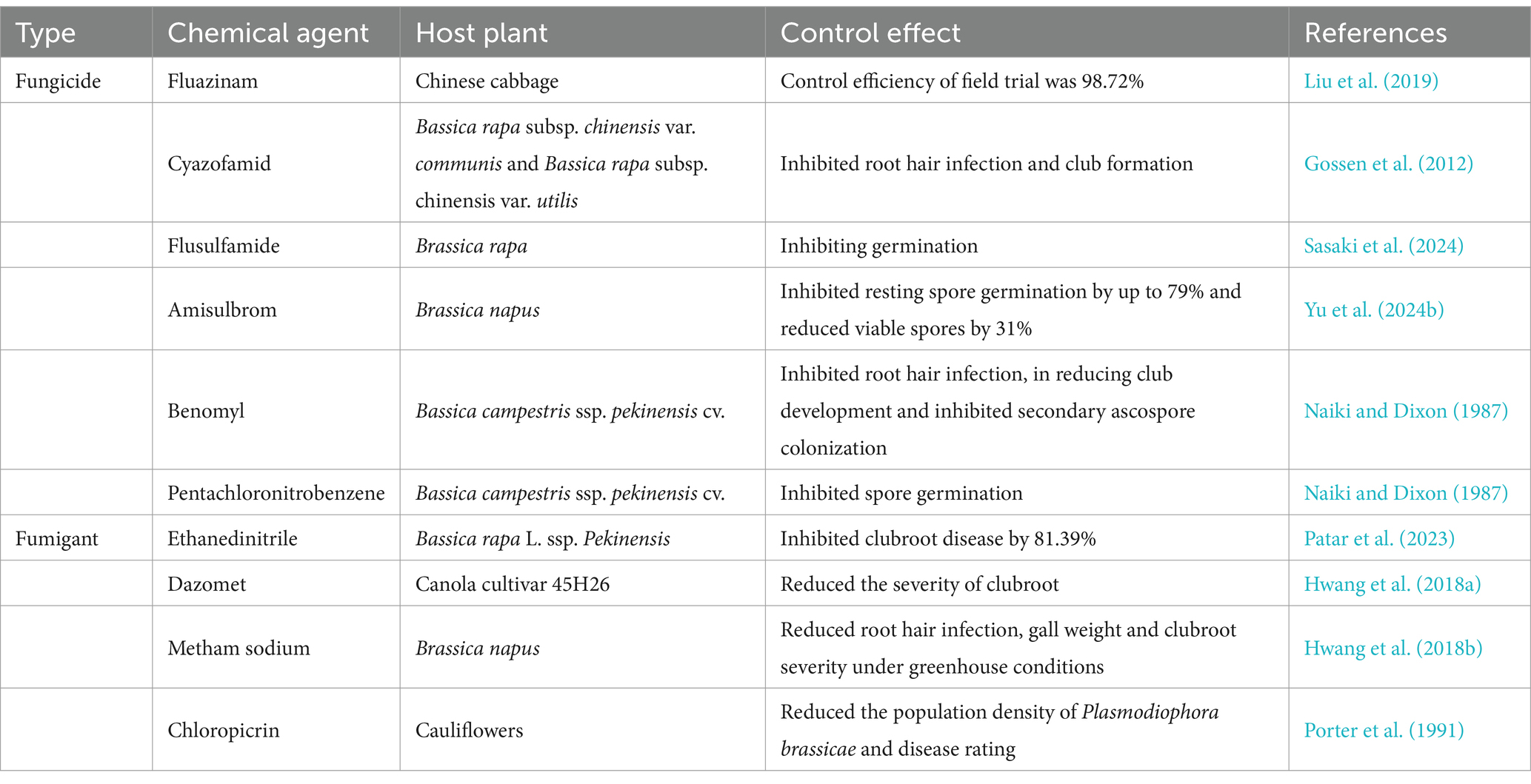
Chemical control is a crucial and effective method for managing clubroot disease in the field and is the primary choice for growers aiming to combat this disease. There are two main types of chemical agents used to control P. brassicae: namely fungicides and fumigants. To date, efficacy assessments have been conducted for a wide range of chemicals, including fungicides and fumigants. Detailed information on the effectiveness of different chemicals against P. brassicae is shown in Table 4.
Fungicides are an attractive option for clubroot management. A range of fungicides have been identified as effective against P. brassicae, but only a few have been registered. It has been reported that benomyl and pentachloronitrobenzene can reduce root-hair infection and clubroot development (Naiki and Dixon, 1987). Flusulfamide has been registered as a fungicide against clubroot in several countries. It adsorbs to the cell walls of resting spores to inhibit their germination. The mechanism of action has been reported to involve the inhibition of germination by inducing the overexpression of the immunophilin gene PbCyp3. This overexpression leads to an unusual accumulation of the PbCYP3 protein, which causes aberrant folding of the proteins involved in primary zoospores (Sasaki et al., 2024; Tanaka et al., 1999). Fluazinam is a broad-spectrum fungicide developed by Ishihara Sangyo Kaisha Ltd. of Japan, which belongs to chemical group of the 2,6-dinitroanilines. Liu et al. (2019) found that fluazinam reduced the disease index of clubroot to 0.95% and provided 98.72% control efficiency of P. brassicae. Liao et al. (2022) reported a control efficacy of 59.81% against clubroot disease using fluazinam, along with a 21.29% reduction in the abundance of P. brassicae in the soil after 3 weeks of treatment. Cyazofamid effectively inhibits root hair infection and club formation by directly inhibiting the germination of resting spores (Mitani et al., 2003). In the presence of high inoculum pressure, the drench application of cyazofamid was shown to substantially reduce the incidence and severity of clubroot in Shanghai pak choy and Chinese flowering cabbage (Gossen et al., 2012). Amisulbrom is a quinone inside inhibitor. In vitro experiments have shown that it inhibits resting spore germination by up to 79% and reduces spore viability by 31%. In addition, application of amisulbrom under both greenhouse and field conditions significantly reduced the clubroot severity (Yu et al., 2024b).
Soil fumigation prior to crop planting is an effective and reliable method for controlling soil-borne diseases and alleviating the obstacles to continuous cropping. Methyl bromide, an earlier fumigant used for clubroot control, has been restricted due to its deleterious effects on ozone concentrations (Ruzo, 2006; White and Buczacki, 1977). Previous studies have reported that chloropicrin is effective in reducing clubroot, however, it has been banned in several countries due to potential risks to non-target organisms and human health (Pesonen and Vähäkangas, 2020; Porter et al., 1991). Dazomet, a solid fumigant, reduces the severity of clubroot under both greenhouse and field conditions, but excessive concentrations (e.g., 0.4–0.8 t a.i. ha−1) can lead to a reduction in seedling emergence and yield (Hwang et al., 2018a). Metham sodium is a broad-spectrum dithiocarbamate fumigant that rapidly decomposes to produce biologically active volatile methyl isothiocyanate when applied to soil. The application of 0.4 to 1.6 mL of metham sodium per liter of soil under greenhouse conditions has been shown to effectively reduce root hair infections, gall weight, and clubroot severity, while also improving the health of canola crops (Hwang et al., 2018b). It is important to note that metham sodium has limited mobility in the soil and requires adequate fumigation time, as well as appropriate soil moisture and temperature. Ethanedinitrile, a cyano-fumigant, was shown in a recent study to inhibit clubroot disease by 81.39%, with complete control of the disease when treated for 12 h at rates of 42 and 50 g/m3 or as well as at a rate of 35 g/m3 for 48 h (Patar et al., 2023).
Despite the fact that chemicals have certain advantages in terms of disease control effectiveness, there are also limitations that cannot be ignored. The most notable limitations are the adverse effects of exposure to the ecosystem and the potential risks to human health (Zhang J. H. et al., 2024). Moreover, chemicals do not offer advantages in terms of production costs, and their lower cost-effectiveness is a major limiting factor. Chemicals are suitable for the control of clubroot disease in high-yield facility vegetable crops. However, for lower-value field crops such as oilseed rape, neither chemical fungicides nor chemical fumigants are typically the preferred options for growers (Zuzak et al., 2023). In addition, the control effect of chemicals is susceptible to receive the influence of many factors such as spore number, physiological pathotypes, application method and soil temperature, moisture, pH, and weather conditions (Hwang et al., 2014a; Zuzak et al., 2023).
5.3 Beneficial microorganisms
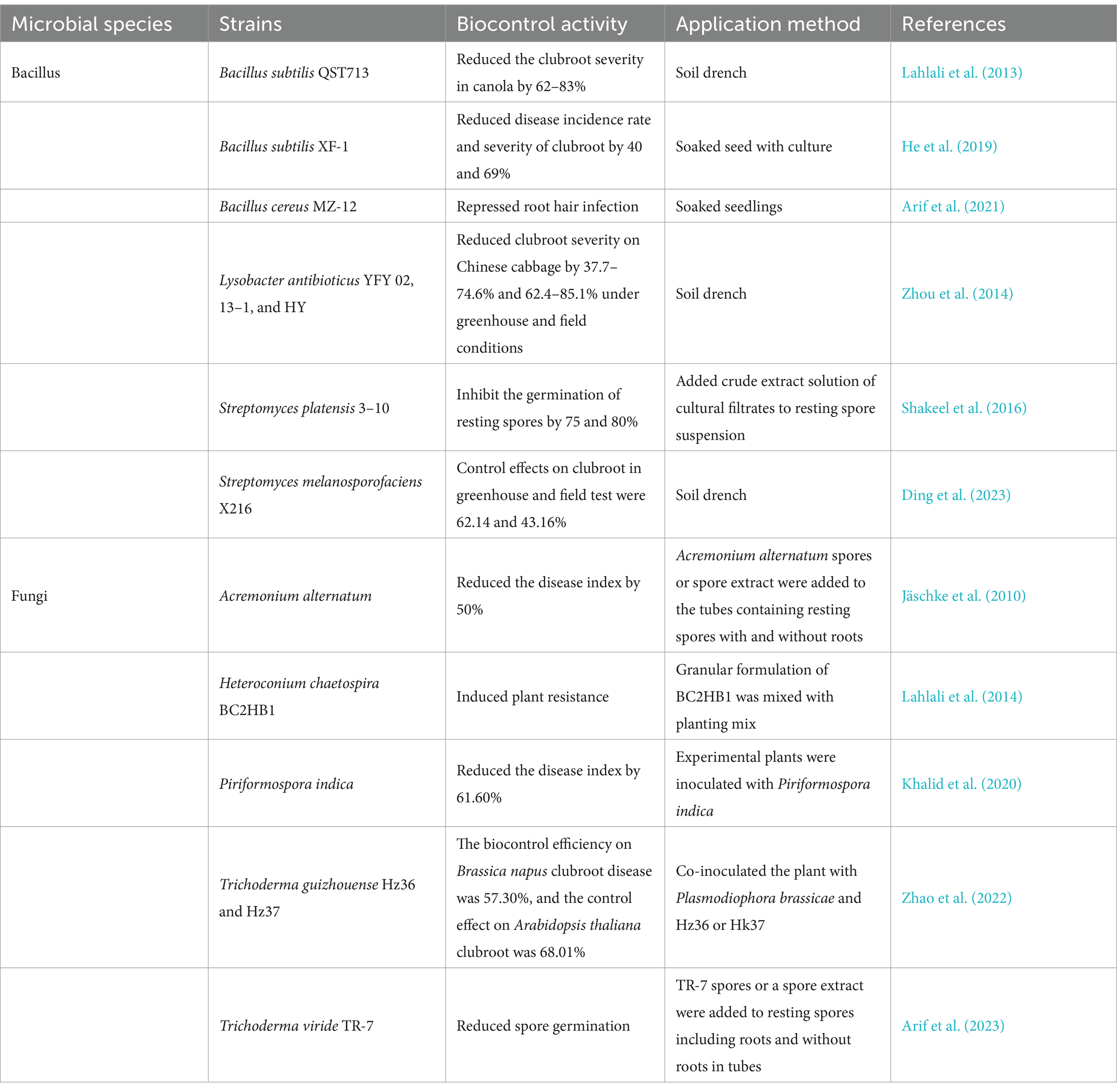
The use of microbial resources for clubroot control is an economical, ecologically friendly, and sustainable management strategy that is considered an alternative to chemical control. In recent years, there has been an increasing interest in utilizing beneficial microorganisms as biological control agents to control plant diseases. The great potential of antagonistic microorganisms in reducing clubroot severity and inhibiting P. brassicae infestation has been demonstrated in numerous studies. In addition, the ability of microorganisms to colonize the soil or within the plant may provide durable protection against P. brassicae damage (Dombrowski et al., 2017). Microorganisms control pathogens through various mechanisms, mainly including antibiosis, parasitism, production of active substances, induction of plant resistance and or competition for resources (Xu et al., 2011). Detailed information on the efficacy of different microbial strains against P. brassicae is shown in Table 5.
Currently, a large number of investigations have revealed that bacterial biocontrol strains have a controlling effect on P. brassicae. Serenade, a biofungicide consisting of Bacillus subtilis QST713, when applied as a soil drench, has been reported to reduce the clubroot severity in canola by 62–83% (Lahlali et al., 2013). B. subtilis XF-1 has strong inhibitory effects on both the survival and germination of dormant spores of P. brassicae. Seed soaking with a culture of XF-1 reduced disease incidence rate and severity of clubroot by 40 and 69%, respectively (He et al., 2019). Arif et al. (2021) isolated a strain of the endophytic bacterium B. cereus MZ-12 from rhizosphere soil of pak choi that was able to colonize cabbage roots to inhibit P. brassicae infestation, and also inhibited infestation through direct inhibition of zoospores. Zhou et al. (2014) screened three strains of Lysobacter antibioticus, YFY 02, 13–1, and HY, from vegetable rhizosphere soils. The cell-free culture filtrates of these strains reduced clubroot severity on Chinese cabbage by 37.7–74.6% under greenhouse conditions and by 62.4–85.1% under field conditions. There are also a number of streptomycetes that have been tapped for their potential to control clubroot. The culture conditions of Streptomyces platensis 3–10 were optimized to inhibit the germination of resting spores by 75 and 80% for its culture filtrates and crude extracts, respectively (Shakeel et al., 2016). Ding et al. (2023) isolated a strain of S. melanosporofaciens X216 from fields of oilseed rape, which showed a lethality of 56.59% against resting spores, and was effective in controlling clubroot disease of oilseed rape by 62.14% in greenhouse pots and 43.16% and under field conditions, respectively.
Fungi are another group of microorganisms that have been extensively investigated for P. brassica control. Jäschke et al. (2010) found that inoculation of A. thaliana with both P. brassicae and the endophytic fungus Acremonium alternatum at the same time induced a 50% reduction in the disease index compared to inoculation with P. brassicae only. The endophytic fungus Heteroconium chaetospira has been reported to penetrate canola roots and colonize the cortical tissues, promoting the up-regulated expression of genes involved in the biosynthesis of jasmonic acid, ethylene, and auxin, thereby inducing resistance to P. brassicae in canola (Lahlali et al., 2014). Piriformospora indica was able to colonize B. campestris roots, and co-inoculation with P. indica and P. brassicae resulted in a 61.6% reduction in the disease index of the plant compared to inoculated with P. brassicae only (Khalid et al., 2020). Zhao et al. (2022) screened two Trichoderma strains, T. guizhouense (Hz36) and T. koningiopsis (Hz37), which significantly inhibited the germination of resting spores. The biocontrol efficiency of 44.29 and 57.30% against clubroot in oilseed rape, respectively, and for the biocontrol efficiency of 52.18 and 68.01% in A. thaliana, respectively. A strain of T. viride TR-7 was isolated from tomato rhizosphere soil, which could reduce the germination rate of resting spores to 25.3%. Co-inoculation with TR-7 and P. brassicae resulted in a 56.7% reduction in gall growth compared to pak choi treated with P. brassicae alone (Arif et al., 2023).
However, a major challenge in the use of microorganisms to control soil-borne diseases is their limited stability. The biological activity of antagonistic microorganisms may be diminished or rendered ineffective by various of environmental factors, such as temperature, humidity, pH, and light (Nicholson, 2002). To solve this dilemma, plant pathologists are constantly searching and experimenting with methods to protect and enhance the biological activity of microorganisms. Encapsulation or embedding of microorganisms using biocompatible materials is an effective strategy. Pelletization of cabbage seeds with microencapsulated Paenibacillus polymyxa ZF129 has been reported to enhance stability and improve bioactivity against clubroot disease under greenhouse conditions, achieving a control efficacy of 71.23% (Abdukerim et al., 2023). Kang et al. (2024) prepared macrobeads embedded with P. polymyxa ZF129 (chitosan: carrageenan, 1:1) using ionotropic gelation. This method enhanced stability compared to free bacteria and showed higher control efficacy in controlling clubroot disease than the ZF129 culture both in greenhouse and field conditions. Another promising solution is to combine different microbial strains into a microbial consortium. Microbial consortia consisting of two or more beneficial microbial strains are better adapted to the pressures of the rhizosphere environment and are more effective in suppressing soil-borne pathogens when the compatibility and modes of action of the microbial strains are carefully evaluated (Niu et al., 2020). A field trial revealed that the microbial consortia of B. cereus BT-23, L. antibioticus 13–6, and L. capsici ZST1-2 reduced the incidence of clubroot in Chinese cabbage by decreasing soil acidity and reshaping the diversity and structure of the rhizobial community, achieving a biocontrol effect of 65.78% (Zhang et al., 2022).
5.4 Farming practices
Agricultural producers recognized early on that some farming practices, such as the timely removal of diseased plants, improved drainage, and deep plowing and tilling, could reduce the infestation rate of P. brassicae or the severity of clubroot disease (Botero et al., 2019). It should be noted that infected seeds are an important pathway leading to the long-distance dissemination of P. brassicae, making seed cleaning particularly important (Greer et al., 2023). However, they often ignore the fact that the resting spores can easily spread in the field with the soil adhering to agricultural machinery and their own boots (Gossen et al., 2013a).
Crop rotation is a time-honored farming practice that helps maintain soil structure and organic matter, reduces soil erosion, and also reduces plant diseases caused by soil-borne pathogens (Janvier et al., 2007). Selection of appropriate crop rotations must begin with an understanding of the host range of the pathogen. To reduce the risk of plant diseases, developing effective rotation patterns necessitates the selection of non-host crops, the design of rational rotation sequences, and the establishment of appropriate intervals (Krupinsky et al., 2002). Compared to Chinese cabbage monoculture, rotating of cabbage with potato onion reduced the disease incidence and disease index of clubroot by 34.3 and 37.5%, respectively (Chen et al., 2018). Yang et al. (2020b) selected leguminous crops (soybean, clover), gramineous crops (rice, maize) and cruciferous crops (oilseed rape, Chinese cabbage) as preceding crops to rotate with oilseed rape. They found that the soybean-oilseed rape rotation pattern was the most effective for controlling clubroot, resulting in the lowest density of resting spores in the soil (< 2.0 × 106 spores per gram of soil). This rotation led to a 50% reduction in the incidence rate and a 40% decrease in the disease index.
Weather conditions, particularly temperature and rainfall, vary considerably depending on the seeding dates. It is well known that soil temperature and moisture play a crucial role in the development of clubroot. Therefore, different seeding dates are bound to have different effects on rhizoctonia. Gossen et al. (2012) showed that seeding date had a significant effect on clubroot incidence and severity on both Shanghai pak choy and Chinese flowering cabbage. Therefore, the risk of clubroot can be minimized by choosing an appropriate seeding date to avoid the optimum temperature conditions required by P. brassicae. A field experiment conducted in Ontario, Canada, showed that temperature and rainfall influence the germination and development of P. brassicae. Additionally, seeding brassica crops as early as possible in the region can reduce the severity of clubroot (Gossen et al., 2017).
The use of solarization combined with mulching is also a management method to control clubroot in agricultural practices. Hong et al. (2023) evaluated the effect of plastic mulching on clubroot under greenhouse and field conditions. The results showed that mulching increased the soil temperature in the 0–20 cm layer and reduced the P. brassicae population in the soil, which effectively reduced the incidence and severity of clubroot.
Cutting off transmission route of pathogens is one of the basic strategies for controlling clubroot. Although the possibility of P. brassicae entering clean fields has been reduced by cultural controls, such as maintaining farm and nursery sanitation and seed disinfection. However, the reality is that the rate of spread speed and extent of infestation of P. brassicae are still increasing, and it is difficult to achieve complete interruption of the spread of P. brassicae. In addition, conventional farming practices have limitations in controlling clubroot disease, especially when confronted with excessive inoculum pressure, and these measures are often unsatisfactory (Peng et al., 2014).
5.5 Soil improvement
Appropriate amelioration of infested soils not only improves soil properties but also benefits the control of soil-borne diseases, making it a promising strategy for improving agricultural sustainability. The benefits of using soil amendments to control soilborne diseases are gradually apparent, usually slower in onset but longer in duration, and the effects can be cumulative (Bailey and Lazarovits, 2003). In addition, soil amendments can directly or indirectly influence the balance between beneficial and detrimental microbial populations in the soil. They achieve this by enhancing the biomass, activity, diversity, and structure of soil microorganisms, which in turn improves the general suppression of pathogens in the soil (Chen et al., 2020). Zhang et al. (2019) found that soil amendments, including quicklime and organic fertilizers, altered soil pH and available nutrients. These amendments also enhanced the relative abundance of bacteria with biocontrol potential, such as Xanthomonadales, Pseudomonas, and Bacillus, as well as increased the enzyme activities of urease and polyphenol oxidase in the topsoil of Chinese cabbage fields.
There is a wide range of soil amendments commonly used in agricultural production, mainly including various organic amendments (e.g., chemical fertilizers, animal manure, solid waste and composts) and crop residues. Calcium cyanamide (CaCN2) is a time-honored and slow-release nitrogen fertilizer that not only provides a source of nitrogen and carbon for crops but also produces degradation products that increase soil pH (Dixon, 2017). Therefore, calcium cyanamide is widely used as a soil amendment to manage clubroot disease. A study conducted in Australia assessed the efficacy of calcium cyanamide against clubroot disease under field and greenhouse conditions. The results indicated that broadcasting 1,000 kg/ha of small particles (98% w/w < 300 μm) of calcium cyanamide effectively controlled clubroot disease (Donald et al., 2004). Niwa et al. (2007) conducted a seven-year continuous field trial and highly reproducible bioassay experiments, demonstrating that the application of farmyard manure (compost mixed with cattle feces and rice straw) and food factory sludge compost (compost mixed with dehydrated activated sludge and corn gluten feed discharged from a cornstarch factory) effectively suppressed clubroot. The main mechanism of action was identified as the calcium enrichment of organic fertilizers, which led to an increase in soil pH.
The utilization of crop residues is an important measure of residue management in sustainable agricultural development and serves as one of the main sources of organic matter in agricultural soils. Straw is a natural soil amendment material commonly used in agricultural production and is a severely underestimated source of organic carbon. It not only plays an important role in regulating the soil environment but also contributes positively to the control soil-borne diseases (Hu et al., 2023). A recent study showed that applying rice straw enhanced the level of available nutrients, pH, and electrical conductivity in the soil of Chinese cabbage field. However, it also decreased the alpha diversity of the bacterial community and altered its community composition, which helped to suppress the incidence of clubroot disease (Han et al., 2021). Similarly, a two-year field trial showed that maize, rice, and wheat straw all promoted the growth of Chinese cabbage plants, improved the rhizosphere microbiome, and reduced the abundance of P. brassicae (Di et al., 2023).
The purpose of soil amendment is to maintain soil health, with the suppression of pathogens serving as a crucial indicator of soil health. Soil amendments have a significant impact on soil health, including plant health, by altering soil physicochemical properties and influencing soil microbial communities. However, the inhibitory activity of soil is a complex process influenced by many biotic and abiotic factors, and the mechanism underlying the inhibitory properties are not yet fully understood. In addition, the application of soil amendments has been shown to have varying effects on soil-borne diseases. Although reports on the inhibitory properties of soil amendments against pathogens are generally encouraging, issues such as reduced or failing inhibitory activity, as well as inconsistency and unpredictability, continue to pose significant challenges to the application of soil amendments (De Corato, 2023). Previous studies have demonstrated the variable control efficacy of soil amendment applications in different regions and under different levels of disease pressure, highlighting the limitations of relying solely on this control measure (McGrann et al., 2016).
5.6 Reduce soil acidity
P. brassicae usually grows in acidic soils environments, which promotes the germination of resting spores when the soil pH ranges from 5 to 7 (Rashid et al., 2013). Previous studies have revealed that both pH and calcium content significantly impact the longevity and germination of resting spores. Additionally, these factors strongly influence on the primary infestation stage of P. brassicae and colonization of the root hairs (Dixon, 2014). Bhering et al. (2017) found a direct relationship between soil acidity on reduction of the healthy root volumes of Brazilian cauliflower and increase of the root volumes with clubroot. For this reason, reducing soil acidity is beneficial in inhibiting the development of P. brassicae. Controlling clubroot by lowering soil acidity is one of the oldest and most widely used management strategies. The application of lime-based substances has been the most commonly used measure to achieve this (Dobson et al., 1983).
Previously, there have been numerous reports on the successful application of lime-based products to control the development of clubroot. For example, it has been reported that applying hydrated lime or calcitic limestone dust to raise the pH value of field soil above 7.0 prior to planting cauliflower can effectively reduce the disease indices (Tremblay et al., 2005). Hennig et al. (2022) found that applying hydrated lime to raise soil pH from an initial 5.2–5.5 to 7.2 reduced the severity of clubroot disease in clubroot-susceptible canola cultivar by 34–36%. Additionally, it decreased the density of resting spores by 48–80% and increased yield by 70–98%. Fox et al. (2022) evaluated the control effect of lime products against P. brassicae under both greenhouse and field conditions. In greenhouse experiments, the disease index of clubroot in susceptible and resistant oilseed canola cultivars was 0% following the application of hydrated lime [Ca(OH)₂] at doses of 4.7, 8.1, 11.4, and 14.8 tonnes/ha. In contrast, limestone (CaCO₃) only reduced the disease index at inoculum concentrations of <1 × 104 spores g−1. In field trials, hydrated lime reduced the clubroot severity. However, control was affected by rainfall and sowing time. Previous studies have indicated that lime can be effective in mitigating clubroot; however, the degree of control afforded may be influenced by the type of lime used, the timing of application, and many environmental and soil factors (Murakami et al., 2002).
However, merely increasing soil pH is not sufficient to control clubroot disease and infestation. This is due to the influence of various of environmental factors, such as nutrient availability, moisture levels, and soil temperature, which all affect the extent of P. brassicae infestation and proliferation (McDonald and Westerveld, 2008). Gossen et al. (2013b) evaluated the effects of a wide range of temperature and pH conditions on P. brassicae infestation in canola under controlled conditions. They found that applying lime to raise soil pH prevented clubroot disease to some extent; however, moderate to severe clubroot still occurred even at a pH of 8.0, provided that the temperatures were suitable (20–25°C) and soil moisture was adequate.
5.7 Nutrient element
Nutrients are necessary for the normal growth and development of plants. They mainly include the massive elements of nitrogen, phosphorus, and potassium; the medium elements of calcium, magnesium; and sulfur; the trace elements of iron, molybdenum, zinc, copper, boron, and manganese. In addition to providing nutritive value to the host, nutrients in the soil environment such as calcium, boron, magnesium, and silicon play an important role in the relationship between the host and clubroot (Dixon, 2009b).
There is a broad consensus on the various roles that calcium plays in inhibiting P. brassicae. Earlier studies have demonstrated that calcium inhibits sporangial dehiscence at high inoculum levels of P. brassicae spores and also inhibits sporangial development at low inoculum levels (Webster and Dixon, 1991a). A study by Niwa et al. (2008) provided direct evidence that high levels of calcium under neutral conditions can inhibit the resting spore germination of P. brassicae. In addition, calcium is essential for the structural strength of cell walls, the normal structure of membranes, and the transport and retention of ions. The stability of cell walls and cell membranes is likely to inhibit the invasion of P. brassicae (Donald and Porter, 2009).
Boron is an essential micronutrient for plant growth, and Brassica crops, especially oilseed rape, have a high demand for this element. However, excessive application of Boron can result in toxicity symptoms (Brown and Shelp, 1997; Kaur et al., 2006). Numerous studies have revealed that boron plays a significant role in crop inhibition of clubroot (Ruaro et al., 2009; Webster and Dixon, 1991b). Deora et al. (2011) found that under controlled conditions, increased boron application reduced root hair infections as well as primary and secondary infections. However, when applied at rates higher than 2 kg/ha, canola seedlings exhibited symptoms of toxicity. In addition, boron reduced both the incidence and severity of clubroot in the field, with an application rate of 4 kg/ha proving to be the most effective without causing any phytotoxicity symptoms.
Silicon plays a positive role in plant-pathogen interactions by mediating plant resistance to pathogens. It does this by serving as a physical barrier, activating the activity of defense-related enzymes, and regulating the expression of genes involved in the defense response (Wang et al., 2017). A recent study showed that silicon s reduced the disease index of clubroot in spring canola and improved shoot height and root length in spring rape under greenhouse conditions (Sarkar et al., 2023).
In addition to the above several nutrient elements, magnesium ion concentration also has a certain effect on inhibiting clubroot. Myers and Campbell (1985) showed that the infestation of broccoli by P. brassicae and the development of clubbing were inhibited when calcium or magnesium concentrations were increased from 0.5 mM to 2.5 mM (Myers and Campbell, 1985).
The interrelationships between nutrient elements in the soil system are very complex. Together with the host and P. brassicae, they form a complex nutrient delivery system that plays a crucial role in maintaining material balance and ecological stability within the soil microenvironmental. Although many nutrient elements have been reported to inhibit P. brassicae, the mechanisms of action for each element against P. brassicae and their interactions with one another remain unclear. This is a huge challenge for future research on nutrient control of clubroot.
5.8 Bait crops
The surface of resting spores of P. brassicae is covered with a thick shell layer that helps resist most environmental stresses. However, resting spores germinate into primary zoospores, which are short-lived and sensitive to environmental conditions. They can perish quickly if they fail to successfully infect a host. Germination of resting spores is necessary for root infestation by P. brassicae. It is generally believed that the root exudates of plants are key factors in inducing the germination of resting spores (Rashid et al., 2013). However, it is worth noting that these root exudates do not exhibit host specificity in stimulating the germination of resting spores; root exudates from non-host plants can also induce spore germination. Friberg et al. (2005) investigated the stimulation of resting spores by the root exudates of the host plant Brassica rapa and four non-host plants, finding that the root exudates of Lolium perenne exhibited the strongest stimulatory capacity. Some plants can induce germination of resting spores without infection or with only root hair infectionand are considered bait crops (Ren et al., 2016). Typically, bait crops are primarily non-host crops of P. brassicae. However, host plants can also be used as bait crops in cases where they are killed prior to resting spore infestation or the formation of clubs (Hwang et al., 2014b).
In the absence of a host, stimulation the germination of resting spore helps accelerate the reduction of spore numbers, as pathogens cannot complete their life cycle without living hosts. Therefore, planting bait crops to stimulate the germination of resting spores without causing infestation is one potential strategy for managing clubroot in large-scale fields. Murakami et al. (2000) found that potted Raphanus sativus can reduce the number of resting spores by 71%, and can reduce the number of resting spores in the field by 94%. A recent study demonstrated that, compared to the bare soil control, Bromus inermis, B. riparius, and L. perenne were able to reduce spore concentration by more than 50% under pot conditions. Additionally, Triticum aestivum, Hordeum vulgare, and Pisum sativum decreased spore concentrations by 61, 43, and 39%, respectively (Drury et al., 2022).
However, the actual efficacy and potential of growing bait crops for clubroot control remain inconclusive. Studies have indicated that the impact of bait crops on clubroot severity is minimal and inconsistent (Friberg et al., 2006). Ahmed et al. (2011) evaluated the impact of bait crops, including non-cruciferous host crops (Trifolium pratense, L. perenne, Dactylis glomerata, Agrotis palustris) and on-host crops (H. vulgare, T. aestivum) on the resting spore populations and clubroot severity under both greenhouse and field conditions, and reached similar conclusions. That is, bait crops may be beneficial at moderate or low levels of resting spore inoculum; however, they had no effect on clubroot severity when field spore levels were high (1 × 106 spores per gram of soil).
6 Conclusions and prospects
The increasing harm caused by P. brassicae to cruciferous crops has raised major concerns. By examining and condensing earlier reports, there is an enhanced comprehension of the biological characteristics and infestation mechanisms of P. brassicae. An increasing number of effectors associated with P. brassicae infestation have been identified, enhancing our understanding of the process by which P. brassicae infest their host. A variety of management strategies including planting resistant varieties, chemical control, biological control, farming practices and soil amendments have been shown to play an important role in controlling P. brassicae damage. Nonetheless, numerous issues remain to be addressed in the management of clubroot. First, the prevention of clubroot disease is essential. Accurate quantification of active spores is essential for predicting the severity of clubroot disease incidence. This places high demands on the accuracy and efficiency of quantification techniques. Establishing an accurate early prediction techniques system to assess disease risk is the focus and challenge of prevention work for clubroot. Second, the management of clubroot disease. There are very few registered chemicals available, and there is an urgent need to develop new, safe, efficient and economical agents. Biologists have consistently maintained great interest and enthusiasm in developing biocontrol agents. However, most biocontrol agents that have shown promising results in laboratory tests have yielded disappointing outcomes in practice practical applications. Nonetheless, researchers remain extremely passionate about trying to improve the stability of biocontrol agents. Most breeders believe that identifying resistance genes for use in breeding resistant varieties is the most reliable measure against P. brassicae infestation. However, resistant varieties have a risk of resistance break-down in the face of constantly evolving populations of P. brassicae. For this reason, scientists are constantly screening for new and effective resistance genes or polymerizing multiple resistance genes to develop resistant varieties to avoid risks.
The rapid development of bioinformatics and the emergence of new biotechnologies have accelerated the level of understanding of P. brassicae. On one hand, these technologies have deepened our understanding of the biological characteristics, pathogenic mechanisms, and catastrophic patterns of P. brassicae in a more comprehensive and scientific manner. On the other hand, they accelerate the analysis of the interaction between P. brassicae and host plants, as well as the interrelationships within the soil ecosystems in which they are located. Due to variations in soil acidity, cropping patterns, and field management across different regions, as well as potential differences in the physiological pathotypes of P. brassicae, developing a uniform management strategy to control this pathogen is challenging. Moreover, individual control measures are often inadequate for managing clubroot. With careful consideration to cost-effectiveness, the integrated application of multiple control measures, along with the development of region-specific clubroot management strategies, can significantly minimize the losses caused by P. brassicae. Specifically, it is the comprehensive use of ecological management, healthy cultivation, biological control, physical control, and the use of safe, efficient, and economical pesticides, and other measures to manage clubroot effectively. This approach aims to maximize social, economic, and ecological benefits (Figure 2). It is important to note that the actual effectiveness of such a management strategy that integrates multiple measures needs to be evaluated in detail and accurately across a large number of trials. In conclusion, it is essential to actively seek improvements in the current integrated control system for clubroot disease while minimizing reliance on chemical agents. This approach aims to prevent catastrophic outbreaks in the cruciferous plant production system caused by the P. brassicae and ultimately achieve sustainable management of clubroot disease.

Figure 3. Integrated management of clubroot disease. (A) resting spores; (B) primary zoospores; (C) uinucleate primary plasmodium; (D) multinucleated zoosporangial plasmodium; (E) diploid uninucleate zygote; (F) secondary zoospores; (G) uinucleate secondary plasmodium; (H) multinucleated secondary plasmodium; (I) resting sporangial plasmodium; (J) resting spore formation.
Author contributions
XX: Conceptualization, Data curation, Formal analysis, Writing – original draft, Writing – review & editing. CW: Software, Supervision, Writing – review & editing. FZ: Supervision, Writing – review & editing. JY: Supervision, Writing – review & editing. LF: Software, Writing – review & editing. ZL: Software, Writing – review & editing. YY: Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Jiangxi Rapeseed Industry Technology System (JXARS-08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdukerim, R., Xiang, S., Shi, Y. X., Xie, X. W., Li, L., Chai, A., et al. (2023). Seed pelleting with gum arabic-encapsulated biocontrol bacteria for effective control of clubroot disease in Pak choi. Plan. Theory 12:3702. doi: 10.3390/plants12213702
Ahmed, H. U., Hwang, S. F., Strelkov, S. E., Gossen, B. D., Peng, G., Howard, R. J., et al. (2011). Assessment of bait crops to reduce inoculum of clubroot (Plasmodiophora brassicae) of canola. Can. J. Plant Sci. 91, 545–551. doi: 10.4141/cjps10200
Ahmed, A., Munir, S., He, P. F., Li, Y. M., He, P. B., Wu, Y. X., et al. (2020). Biocontrol arsenals of bacterial endophyte: an imminent triumph against clubroot disease. Microbiol. Res. 241:126565. doi: 10.1016/j.micres.2020.126565
Ando, S., Otawara, S., Tabei, Y., and Tsushima, S. (2024). Plasmodiophora brassicae affects host gene expression by secreting the transcription factor-type effector PbZFE1. J. Exp. Bot. 75, 454–467. doi: 10.1093/jxb/erad377
Andrews, K. R., Good, J. M., Miller, M. R., Luikart, G., and Hohenlohe, P. A. (2016). Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 17, 81–92. doi: 10.1038/nrg.2015.28
Arif, S., Liaquat, F., Yang, S. L., Shah, I. H., and Zhao, L. N. (2021). Exogenous inoculation of endophytic bacterium Bacillus cereus suppresses clubroot (Plasmodiophora brassicae) occurrence in pak choi (Brassica campestris sp. chinensis L.). Planta 253:25. doi: 10.1007/s00425-020-03546-4
Arif, S., Munis, M. F. H., Liaquat, F., Gulzar, S., Haroon, U., Zhao, L. A., et al. (2023). Trichoderma viride establishes biodefense against clubroot (Plasmodiophora brassicae) and fosters plant growth via colonizing root hairs in pak choi (Brassica campestris spp. chinesnsis). Biol. Control 183:105265. doi: 10.1016/j.biocontrol.2023.105265
Askarian, H., Akhavan, A., Manolii, V. P., Cao, T., Hwang, S. F., and Strelkov, S. E. (2021). Virulence spectrum of single-spore and field isolates of Plasmodiophora brassicae able to overcome resistance in canola (Brassica napus). Plant Dis. 105, 43–52. doi: 10.1094/PDIS-03-20-0471-RE
Bailey, K. L., and Lazarovits, G. (2003). Suppressing soil-borne diseases with residue management and organic amendments. Soil Tillage Res. 72, 169–180. doi: 10.1016/S0167-1987(03)00086-2
Bhering, A. D., do Carmo, M. G. F., Matos, T. D., Lima, E. S. A., and Sobrinho, N. M. B. D. (2017). Soil factors related to the severity of clubroot in Rio de Janeiro, Brazil. Plant Dis. 101, 1345–1353. doi: 10.1094/PDIS-07-16-1024-SR
Botero, A., García, C., Gossen, B. D., Strelkov, S. E., Todd, C. D., Bonham-Smith, P. C., et al. (2019). Clubroot disease in Latin America: distribution and management strategies. Plant Pathol. 68, 827–833. doi: 10.1111/ppa.13013
Botero-Ramírez, A., Laperche, A., Guichard, S., Jubault, M., Gravot, A., Strelkov, S. E., et al. (2021). Clubroot symptoms and resting spore production in a doubled haploid population of oilseed rape (Brassica napus) are controlled by four main QTLs. Front. Plant Sci. 11:604527. doi: 10.3389/fpls.2020.604527
Brown, P. H., and Shelp, B. J. (1997). Boron mobility in plants. Plant Soil 193, 85–101. doi: 10.1023/A:1004211925160
Buczacki, S. T. (1977). Root infections from single resting spores of Plasmodiophora brassicae. Trans. Br. Mycol. Soc. 69, 328–329. doi: 10.1016/S0007-1536(77)80059-4
Buczacki, S. T., Toxopeus, H., Mattusch, P., Johnston, T. D., Dixon, G. R., and Hobolth, L. A. (1975). Study of physiologic specialization in Plasmodiophora brassicae: proposals for attempted rationalization through an international approach. Trans. Br. Mycol. Soc. 65, 295–303. doi: 10.1016/S0007-1536(75)80013-1
Cao, T., Manolii, V. P., Hwang, S. F., Howard, R. J., and Strelkov, S. E. (2009). Virulence and spread of Plasmodiophora brassicae [clubroot] in Alberta, Canada. Can. J. Plant Pathol. 31, 321–329. doi: 10.1080/07060660909507606
Ce, F. Q., Mei, J. Q., He, H. Y., Zhao, Y., Hu, W. H., Yu, F. Q., et al. (2021). Identification of candidate genes for clubroot-resistance in Brassica oleracea using quantitative trait loci-sequencing. Front. Plant Sci. 12:703520. doi: 10.3389/fpls.2021.703520
Chai, A. L., Xie, X. W., Shi, Y. X., and Li, B. J. (2014). Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. 36, 142–153. doi: 10.1080/07060661.2013.868829
Chang, A., Lamara, M., Wei, Y. D., Hu, H., Parkin, I. A. P., Gossen, B. D., et al. (2019). Clubroot resistance gene Rcr6 in Brassica nigra resides in a genomic region homologous to chromosome A08 in B. rapa. BMC Plant Biol. 19:224. doi: 10.1186/s12870-019-1844-5
Chen, J. J., Jing, J., Zhan, Z. X., Zhang, T., Zhang, C. Y., and Piao, Z. Y. (2013). Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS One 8:e85307. doi: 10.1371/journal.pone.0085307
Chen, W., Li, Y., Yan, R. B., Ren, L., Liu, F., Zeng, L. Y., et al. (2021). SnRK1.1-mediated resistance of Arabidopsis thaliana to clubroot disease is inhibited by the novel Plasmodiophora brassicae effector PBZF1. Mol. Plant Pathol. 22, 1057–1069. doi: 10.1111/mpp.13095
Chen, W., Li, Y., Yan, R. B., Xu, L., Ren, L., Liu, F., et al. (2019). Identification and characterization of Plasmodiophora brassicae primary infection effector candidates that suppress or induce cell death in host and nonhost plants. Phytopathology 109, 1689–1697. doi: 10.1094/PHYTO-02-19-0039-R
Chen, D. L., Wang, X. X., Zhang, W., Zhou, Z. G., Ding, C. F., Liao, Y. W. K., et al. (2020). Persistent organic fertilization reinforces soil-borne disease suppressiveness of rhizosphere bacterial community. Plant Soil 452, 313–328. doi: 10.1007/s11104-020-04576-3
Chen, S. C., Zhou, X. G., Yu, H. J., and Wu, F. Z. (2018). Root exudates of potato onion are involved in the suppression of clubroot in a Chinese cabbage-potato onion-Chinese cabbage crop rotation. Eur. J. Plant Pathol. 150, 765–777. doi: 10.1007/s10658-017-1307-5
Choi, S. R., Oh, S. H., Chhapekar, S. S., Dhandapani, V., Lee, C. Y., Rameneni, J. J., et al. (2020). Quantitative trait locus mapping of clubroot resistance and Plasmodiophora brassicae pathotype banglim-specific marker development in Brassica rapa. Int. J. Mol. Sci. 21:4157. doi: 10.3390/ijms21114157
Chu, M. G., Song, T., Falk, K. C., Zhang, X. G., Liu, X. J., Chang, A., et al. (2014). Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genomics 15:1166. doi: 10.1186/1471-2164-15-1166
Ciaghi, S., Schwelm, A., and Neuhauser, S. (2019). Transcriptomic response in symptomless roots of clubroot infected kohlrabi (Brassica oleracea var. gongylodes) mirrors resistant plants. BMC Plant Biol. 19:288. doi: 10.1186/s12870-019-1902-z
Cui, H. T., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Dakouri, A., Zhang, X. G., Peng, G., Falk, K. C., Gossen, B. D., Strelkov, S. E., et al. (2018). Analysis of genome-wide variants through bulked segregant RNA sequencing reveals a major gene for resistance to Plasmodiophora brassicae in Brassica oleracea. Sci. Rep. 8:17657. doi: 10.1038/s41598-018-36187-5
Davey, J. W., and Blaxter, M. L. (2010). RADSeq: next-generation population genetics. Brief. Funct. Genomics 10, 108–423. doi: 10.1093/bfgp/elr007
De Corato, U. (2023). Governance of soil amendment to enhance suppression to soil-borne plant pathogens from a long-term perspective. Appl. Soil Ecol. 182:104721. doi: 10.1016/j.apsoil.2022.104721
Deora, A., Gossen, B. D., Walley, F., and McDonald, M. R. (2011). Boron reduces development of clubroot in canola. Can. J. Plant Sci. 33, 475–484. doi: 10.1080/07060661.2011.620630
Di, C. Q., Han, Z., Chai, C., Sun, J., Wu, F. Z., and Pan, K. (2023). Improvement of straw changed soil microbial flora composition and suppressed Chinese cabbage (Brassica rapa L ssp. pekinensis) clubroot disease. Agronomy 13:1688. doi: 10.3390/agronomy13071688
Diederichsen, E., Wagenblatt, B., and Schallehn, V. (2016). Production of pure genotype isolates of Plasmodiophora brassicae Wor. - comparison of inoculations with root hairs containing single sporangiosori or with single resting spores. Eur. J. Plant Pathol. 145, 621–627. doi: 10.1007/s10658-016-0876-z
Ding, L., Zhou, H., Liang, H. D., Tan, L., Zhao, H., Chen, X. J., et al. (2023). Efficacy of Streptomyces melanosporofaciens strain X216 at controlling clubroot disease on oilseed rape. Front. Microbiol. 14:1249813. doi: 10.3389/fmicb.2023.1249813
Dixon, G. R. (2009a). Plasmodiophora brassicae in its environment. J. Plant Growth Regul. 28, 212–228. doi: 10.1007/s00344-009-9098-3
Dixon, G. R. (2009b). The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 28, 194–202. doi: 10.1007/s00344-009-9090-y
Dixon, G. R. (2014). Clubroot (Plasmodiophora brassicae Woronin) – an agricultural and biological challenge worldwide. Can. J. Plant Pathol. 36, 5–18. doi: 10.1080/07060661.2013.875487
Dixon, G. R. (2017). Managing clubroot disease (caused by Plasmodiophora brassicae Wor.) by exploiting the interactions between calcium cyanamide fertilizer and soil microorganisms. J. Agric. Sci. 155, 527–543. doi: 10.1017/S0021859616000800
Dobson, R. L., Gabrielson, R. L., Baker, A. S., and Bennett, L. (1983). Effects of lime particle size and distribution and fertilizer formulation on clubroot disease caused by Plasmodiophora brassicae. Plant Dis. 67, 50–52. doi: 10.1094/PD-67-50
Dodds, P. N., Chen, J., and Outram, M. A. (2024). Pathogen perception and signaling in plant immunity. Plant Cell 36, 1465–1481. doi: 10.1093/plcell/koae020
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dolatabadian, A., Cornelsen, J., Huang, S. L., Zou, Z. W., and Fernando, W. G. D. (2022). Sustainability on the farm: breeding for resistance and management of major canola diseases in Canada contributing towards an IPM approach. Can. J. Plant Pathol. 44, 157–190. doi: 10.1080/07060661.2021.1991480
Dombrowski, N., Schlaeppi, K., Agler, M. T., Hacquard, S., Kemen, E., Garrido-Oter, R., et al. (2017). Root microbiota dynamics of perennial Arabis alpina are dependent on soil residence time but independent of flowering time. ISME J. 11, 43–55. doi: 10.1038/ismej.2016.109
Donald, E. C., Lawrence, J. M., and Porter, I. J. (2004). Influence of particle size and application method on the efficacy of calcium cyanamide for control of clubroot of vegetable brassicas. Crop Prot. 23, 297–303. doi: 10.1016/j.cropro.2003.08.016
Donald, C., and Porter, I. (2009). Integrated control of clubroot. J. Plant Growth Regul. 28, 289–303. doi: 10.1007/s00344-009-9094-7
Drury, S. C., Sedaghatkish, A., Gossen, B. D., and McDonald, M. R. (2022). Grasses and field crops reduce the concentration of resting spores of Plasmodiophora brassicae in soil under controlled conditions. Plant Pathol. 71, 1793–1800. doi: 10.1111/ppa.13601
Fang, T. Y., Han, X. Y., and Yue, Y. L. (2024). Disease-resistant varieties of Chinese cabbage (Brassica rapa L. ssp. pekinensis) inhibit Plasmodiophora brassicae infestation by stabilising root flora structure. Front. Plant Sci. 15:1328845. doi: 10.3389/fpls.2024.1328845
Feng, J., Cao, T., Rennie, D. C., Strelkov, S. E., and Hwang, S. F. (2014). Host-parasite interactions in clubroot of crucifers. Can. J. Plant Pathol. 36, 113–121. doi: 10.1080/07060661.2013.860192
Feng, Y. Q., Yang, X. Y., Cai, G. L., Wang, S. T., Liu, P. G., Li, Y., et al. (2024). Identification and characterization of high-molecular-weight proteins secreted by Plasmodiophora brassicae that suppress plant immunity. J. Fungi 10:462. doi: 10.3390/jof10070462
Fox, N. M., Hwang, S. F., Manolii, V. P., Turnbull, G., and Strelkov, S. E. (2022). Evaluation of lime products for clubroot (Plasmodiophora brassicae) management in canola (Brassica napus) cropping systems. Can. J. Plant Pathol. 44, 21–38. doi: 10.1080/07060661.2021.1940590
Fredua-Agyeman, R., Jiang, J. Y., Hwang, S. F., and Strelkov, S. E. (2020). QTL mapping and inheritance of clubroot resistance genes derived from Brassica rapa subsp. rapifera (ECD 02) reveals resistance loci and distorted segregation ratios in two F2 populations of different crosses. Front. Plant Sci. 11:899. doi: 10.3389/fpls.2020.00899
Friberg, H., Lagerlöf, J., and Rämert, B. (2005). Germination of Plasmodiophora brassicae resting spores stimulated by a non-host plant. Eur. J. Plant Pathol. 113, 275–281. doi: 10.1007/s10658-005-2797-0
Friberg, H., Lagerlöf, J., and Rämert, B. (2006). Usefulness of nonhost plants in managing Plasmodiophora brassicae. Plant Pathol. 55, 690–695. doi: 10.1111/j.1365-3059.2006.01408.x
Galindo-González, L., Hwang, S. F., and Strelkov, S. E. (2021). Candidate effectors of Plasmodiophora brassicae pathotype 5X during infection of two Brassica napus genotypes. Front. Microbiol. 12:742268. doi: 10.3389/fmicb.2021.742268
Gossen, B. D., Adhikari, K. K. C., and Mcdonald, M. R. (2012). Effect of seeding date on development of clubroot in vegetable Brassica crops. Can. J. Plant Pathol. 34, 516–523. doi: 10.1080/07060661.2012.722129
Gossen, B. D., Cranmer, T. J., Gludovacz, T. V., and Mcdonald, M. R. (2017). Weather thresholds for clubroot development on canola and brassica vegetables. Can. J. Plant Pathol. 39, 475–485. doi: 10.1080/07060661.2017.1369459
Gossen, B. D., Kasinathan, H., Cao, T., Manolii, V. P., Strelkov, S. E., Hwang, S. F., et al. (2013b). Interaction of pH and temperature affect infection and symptom development of Plasmodiophora brassicae in canola. Can. J. Plant Pathol. 35, 294–303. doi: 10.1080/07060661.2013.804882
Gossen, B. D., Mcdonald, M. R., Hwang, S. F., Strelkov, S. E., and Peng, G. (2013a). A comparison of clubroot development and management on canola and Brassica vegetables. Can. J. Plant Pathol. 35, 175–191. doi: 10.1080/07060661.2013.763293
Gossen, B. D., Strelkov, S. E., Manolii, V. P., Rennie, D. C., Cao, T., Hwang, S. F., et al. (2015). Spread of Plasmodiophora brassicae on canola in Canada, 2003-2014: old pathogen, new home. Can. J. Plant Pathol. 37, 403–413. doi: 10.1080/07060661.2015.1105871
Graf, H., Fähling, M., and Siemens, J. (2004). Chromosome polymorphism of the obligate biotrophic parasite Plasmodiophora brassicae. J. Phytopathol. 152, 86–91. doi: 10.1046/j.1439-0434.2003.00805.x
Gravot, A., Liégard, B., Quadrana, L., Veillet, F., Aigu, Y., Bargain, T., et al. (2024). Two adjacent NLR genes conferring quantitative resistance to clubroot disease in Arabidopsis are regulated by a stably inherited epiallelic variation. Plant Commun. 5:100824. doi: 10.1016/j.xplc.2024.100824
Greer, S. F., Surendran, A., Grant, M., and Lillywhite, R. (2023). The current status, challenges, and future perspectives for managing diseases of brassicas. Front. Microbiol. 14:1209258. doi: 10.3389/fmicb.2023.1209258
Guichoux, E., Lagache, L., Wagner, S., Chaumeil, P., Léger, P., Lepais, O., et al. (2011). Current trends in microsatellite genotyping. Mol. Ecol. Resour. 11, 591–611. doi: 10.1111/j.1755-0998.2011.03014.x
Han, Z., Di, C. Q., Rahman, M. K. U., Gao, D. M., Wu, F. Z., and Pan, K. (2021). Repeated application of rice straw stabilizes soil bacterial community composition and inhibits clubroot disease. Agriculture 11:108. doi: 10.3390/agriculture11020108
Hasan, J., Megha, S., and Rahman, H. (2021). Clubroot in Brassica: recent advances in genomics, breeding, and disease management. Genome 64, 735–760. doi: 10.1139/gen-2020-0089
He, P. J., Cui, W. Y., Munir, S., He, P. F., Li, X. Y., Wu, Y. X., et al. (2019). Plasmodiophora brassicae root hair interaction and control by Bacillus subtilis XF-1 in Chinese cabbage. Biol. Control 128, 56–63. doi: 10.1016/j.biocontrol.2018.09.020
Hennig, B. C., Hwang, S. F., Manolii, V. P., Turnbull, G., Robinson, S. V. J., and Strelkov, S. E. (2022). Evaluation of host resistance, hydrated lime, and weed control to manage clubroot in canola. Horticulturae 8:215. doi: 10.3390/horticulturae8030215
Heo, S. H., Jang, S. J., Choi, J. S., Jang, C. S., Song, J. Y., and Kim, H. G. (2009). Chinese cabbage clubroot pathogen, Plasmodiophora brassicae, is genetically stable. Mycobiology 37, 225–229. doi: 10.4489/MYCO.2009.37.3.225
Hirai, M., Harada, T., Kubo, N., Tsukada, M., Suwabe, K., and Matsumoto, S. (2004). A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor. Appl. Genet. 108, 639–643. doi: 10.1007/s00122-003-1475-x
Hirani, A. H., Gao, F., Liu, J., Fu, G. H., Wu, C. R., McVetty, P. B. E., et al. (2018). Combinations of independent dominant loci conferring clubroot resistance in all four turnip accessions (Brassica raps) from the European clubroot differential set. Front. Plant Sci. 9:1628. doi: 10.3389/fpls.2018.01628
Hollman, K. B., Manolii, V. P., Aigu, Y., Harding, M.W., Hwang, S.F., and Strelkov, S.E. (2023). Characterization of Plasmodiophora brassicae pathotypes from western Canada in 2019-2020. Can. J. Plant Pathol. 45, 475–484. doi: 10.1080/07060661.2023.2212639
Holtz, M. D., Hwang, S. F., Manolii, V. P., Strelkov, I. S., and Strelkov, S. E. (2021). Development of molecular markers to identify distinct populations of Plasmodiophora brassicae. Eur. J. Plant Pathol. 159, 637–654. doi: 10.1007/s10658-020-02194-4
Holtz, M. D., Hwang, S. F., and Strelkov, S. E. (2018). Genotyping of Plasmodiophora brassicae reveals the presence of distinct populations. BMC Genomics 19:254. doi: 10.1186/s12864-018-4658-1
Hong, Y. Z., Wang, S. Q., Feng, J., and Liang, Y. (2023). Effect of mulching on soil temperatures and its impact on Plasmodiophora brassicae and clubroot. Plant Dis. 107, 3731–3736. doi: 10.1094/PDIS-05-23-0960-SC
Hossain, M. M., Pérez-López, E., Todd, C. D., Wei, Y. D., and Bonham-Smith, P. C. (2021). Endomembrane-targeting Plasmodiophora brassicae effectors modulate PAMP triggered immune responses in plants. Front. Microbiol. 12:651279. doi: 10.3389/fmicb.2021.651279
Howard, R. J., Strelkov, S. E., and Harding, M. W. (2010). Clubroot of cruciferous crops - new perspectives on an old disease. Can. J. Plant Pathol. 32, 43–57. doi: 10.1080/07060661003621761
Hu, J., Sun, Y. F., Liu, M. J., Zhao, Y. M., Lv, H. F., Wang, Y. F., et al. (2023). Drip fertigation with straw incorporation promotes soil microbial network complexity and potentially reduces pathogen abundance in greenhouse vegetable production systems. Agric. Ecosyst. Environ. 351:108501. doi: 10.1016/j.agee.2023.108501
Hu, H., Zhang, Y., and Yu, F. Q. (2024). A CRISPR/Cas9-based vector system enables the fast breeding of selection-marker-free canola with Rcr1-rendered clubroot resistance. J. Exp. Bot. 75, 1347–1363. doi: 10.1093/jxb/erad471
Huang, Z., Peng, G., Gossen, B. D., and Yu, F. (2019). Fine mapping of a clubroot resistance gene from turnip using SNP markers identified from bulked segregant RNA-Seq. Mol. Breeding 39:131. doi: 10.1007/s11032-019-1038-8
Huang, Z., Peng, G., Liu, X. J., Deora, A., Falk, K. C., Gossen, B. D., et al. (2017). Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA sequencing. Front. Plant Sci. 8:1448. doi: 10.3389/fpls.2017.01448
Hwang, S. F., Ahmed, H. U., Strelkov, S. E., Zhou, Q., Gossen, B. D., McDonald, M. R., et al. (2018a). Suppression of clubroot by dazomet fumigant. Can. J. Plant Sci. 98, 172–182. doi: 10.1139/cjps-2017-0099
Hwang, S. F., Ahmed, H. U., Strelkov, S. E., Zhou, Q., Gossen, B. D., Peng, G., et al. (2018b). Effects of rate and application method on the efficacy of metam sodium to reduce clubroot (Plasmodiophora brassicae) of canola. Eur. J. Plant Pathol. 150, 341–349. doi: 10.1007/s10658-017-1281-y
Hwang, S. F., Ahmed, H. U., Zhou, Q., Strelkov, S. E., Gossen, B. D., Peng, G., et al. (2014a). Efficacy of Vapam fumigant against clubroot (Plasmodiophora brassicae) of canola. Plant Pathol. 63, 1374–1383. doi: 10.1111/ppa.12207
Hwang, S. F., Howard, R. J., Strelkov, S. E., Gossen, B. D., and Peng, G. (2014b). Management of clubroot (Plasmodiophora brassicae) on canola (Brassica napus) in western Canada. Can. J. Plant Pathol. 36, 49–65. doi: 10.1080/07060661.2013.863806
Hwang, S. F., Strelkov, S. E., Feng, J., Gossen, B. D., and Howard, R. J. (2012). Plasmodiophora brassicae: a review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Pathol. 13, 105–113. doi: 10.1111/j.1364-3703.2011.00729.x
Janvier, C., Villeneuve, F., Alabouvette, C., Edel-Hermann, V., Mateille, T., and Steinberg, C. (2007). Soil health through soil disease suppression: which strategy from descriptors to indicators? Soil Biol. Biochemist. 39, 1–23. doi: 10.1016/j.soilbio.2006.07.001
Jäschke, D., Dugassa-Gobena, D., Karlovsky, P., Vidal, S., and Ludwig-Müller, J. (2010). Suppression of clubroot (Plasmodiophora brassicae) development in Arabidopsis thaliana by the endophytic fungus Acremonium alternatum. Plant Pathol. 59, 100–111. doi: 10.1111/j.1365-3059.2009.02199.x
Javed, M. A., Schwelm, A., Zamani-Noor, N., Salih, R., Vañó, M. S., Wu, J. X., et al. (2023). The clubroot pathogen Plasmodiophora brassicae: A profile update. Mol. Plant Pathol. 24, 89–106. doi: 10.1111/mpp.13283
Ji, R., Zhao, L., Xing, M., Shen, X., Bi, Q., Peng, S., et al. (2014). Infection of Plasmodiophora brassicae in Chinese cabbage. Genet. Mol. Res. 13, 10976–10982. doi: 10.4238/2014.December.19.20
Jones, D. R., Ingram, D. S., and Dixon, G. R. (1982). Characterization of isolates derived from single resting spores of Plasmodiophora brassicae and studies of their interaction. Plant Pathol. 31, 239–246. doi: 10.1111/j.1365-3059.1982.tb01274.x
Kageyama, K., and Asano, T. (2009). Life cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 28, 203–211. doi: 10.1007/s00344-009-9101-z
Kamei, A., Tsuro, M., Kubo, N., Hayashi, T., Wang, N., Fujimura, T., et al. (2010). QTL mapping of clubroot resistance in radish (Raphanus sativus L.). Theor. Appl. Genet. 120, 1021–1027. doi: 10.1007/s00122-009-1230-z
Kang, H. J., Fan, T. F., Lin, Z. H., Shi, Y. X., Xie, X. W., Li, L., et al. (2024). Development of chitosan/carrageenan macrobeads for encapsulation of Paenibacillus polymyxa and its biocontrol efficiency against clubroot disease in Brassica crops. Int. J. Biol. Macromol. 264:130323. doi: 10.1016/j.ijbiomac.2024.130323
Karim, M. M., Dakouri, A., Zhang, Y., Chen, Q. L., Peng, G., Strelkov, S. E., et al. (2020). Two clubroot-resistance genes, Rcr3 and Rcr9wa, mapped in Brassica rapa using bulk segregant RNA sequencing. Int. J. Mol. Sci. 21:5033. doi: 10.3390/ijms21145033
Karim, M. M., and Yu, F. Q. (2023). Identification of QTLs for resistance to 10 pathotypes of Plasmodiophora brassicae in Brassica oleracea cultivar ECD11 through genotyping-by-sequencing. Theor. Appl. Genet. 136:249. doi: 10.1007/s00122-023-04483-y
Kaur, S., Nicolas, M. E., Ford, R., Norton, R. M., and Taylor, P. W. J. (2006). Selection of Brassica rapa genotypes for tolerance to boron toxicity. Plant Soil 285, 115–123. doi: 10.1007/s11104-006-0063-7
Khalid, M., Hui, N., Saeed-ur-Rahman, H., and Huang, D. F. (2020). Suppression of clubroot (Plasmodiophora brassicae) development in Brassica campestris sp. chinensis L. via exogenous inoculation of Piriformospora indica. J. Radiat. Res. Appl. Sci. 13, 180–190. doi: 10.1080/16878507.2020.1719337
Klewer, A., Luerben, H., Graf, H., and Siemens, J. (2001). Restriction fragment length polymorphism markers to characterize Plasmodiophora brassicae single-spore isolates with different virulence patterns. J. Phytopathol. 149, 121–127. doi: 10.1046/j.1439-0434.2001.00595.x
Krupinsky, J. M., Bailey, K. L., Mc Mullen, M. P., Gossen, B. D., and Turkington, K. T. (2002). Managing plant disease risk in diversified cropping systems. Agron. J. 94, 198–209. doi: 10.2134/agronj2002.1980
Kubo, N., Onnazaka, K., Ono, U., and Tsuji, G. (2017). Development of simple sequence repeat markers for the classification of the clubroot pathogen Plasmodiophora brassicae. Eur. J. Plant Pathol. 149, 733–738. doi: 10.1007/s10658-017-1203-z
Kuginuki, Y., Yoshikawa, H., and Hirai, M. (1999). Variation in virulence of Plasmodiophora brassicae in Japan tested with clubroot resistant cultivars of Chinese cabbage (Brassica rapa var. pekinensis). Eur. J. Plant Pathol. 105, 327–332. doi: 10.1023/A:1008705413127
Lahlali, R., McGregor, L., Song, T., Gossen, B. D., Narisawa, K., and Peng, G. (2014). Heteroconium chaetospira induces resistance to clubroot via upregulation of host genes involved in jasmonic acid, ethylene, and auxin biosynthesis. PLoS One 9:e94144. doi: 10.1371/journal.pone.0094144
Lahlali, R., Peng, G., Gossen, B. D., McGregor, L., Yu, F. Q., Hynes, R. K., et al. (2013). Evidence that the biofungicide serenade (Bacillus subtilis) suppresses clubroot on canola via antibiosis and induced host resistance. Phytopathology 103, 245–254. doi: 10.1094/PHYTO-06-12-0123-R
Laila, R., Park, J. I., Robin, A. H. K., Natarajan, S., Vijayakumar, H., Shirasawa, K., et al. (2019). Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage (Brassica rapa L.). BMC Plant Biol. 19:13. doi: 10.1186/s12870-018-1615-8
Landry, B. S., Hubert, N., Crete, R., Chang, M. S., Lincoln, S. E., and Etoh, T. (1992). A genetic map for Brassica oleracea based on RFLP markers detected with expressed DNA sequences and mapping of resistance genes to race 2 of Plasmodiophora brassicae (Woronin). Genome 35, 409–420. doi: 10.1139/g92-061
Li, C., Luo, S., Feng, L., Wang, Q., Cheng, J., Xie, J., et al. (2024). Protist ubiquitin ligase effector PbE3-2 targets cysteine protease RD21A to impede plant immunity. Plant Physiol. 194, 1764–1778. doi: 10.1093/plphys/kiad603
Li, K. Q., Wang, K., Shi, Y. J., Liang, F. H., Li, X. R., Bao, S. J., et al. (2024). BjuA03.BNT1 plays a positive role in resistance to clubroot disease in resynthesized Brassica juncea L. Plant Sci. 349:112268. doi: 10.1016/j.plantsci.2024.112268
Li, X. A., Wei, Y. X., Ma, Y. M., Cao, G. Z., Ma, S. W., Zhang, T. Y., et al. (2022). Marker-assisted pyramiding of CRa and CRd genes to improve the clubroot resistance of Brassica rapa. Genes 13:2414. doi: 10.3390/genes13122414
Liao, J. J., Luo, L. Y., Zhang, L., Wang, L. Z., Shi, X. D., Yang, H., et al. (2022). Comparison of the effects of three fungicides on clubroot disease of tumorous stem mustard and soil bacterial community. J. Soils Sediments 22, 256–271. doi: 10.1007/s11368-021-03073-z
Liégard, B., Baillet, V., Etcheverry, M., Joseph, E., Lariagon, C., Lemoine, J., et al. (2019). Quantitative resistance to clubroot infection mediated by transgenerational epigenetic variation in Arabidopsis. New Phytol. 222, 468–479. doi: 10.1111/nph.15579
Liu, L. J., Qin, L., Cheng, X. H., Zhang, Y., Xu, L., Liu, F., et al. (2020a). Comparing the infection biology of Plasmodiophora brassicae in clubroot susceptible and resistant hosts and non-hosts. Front. Microbiol. 11:507036. doi: 10.3389/fmicb.2020.507036
Liu, L. J., Qin, L., Zhou, Z. Q., Hendriks, W. G. H. M., Liu, S. Y., and Wei, Y. D. (2020b). Refining the life cycle of Plasmodiophora brassicae. Phytopathology 110, 1704–1712. doi: 10.1094/PHYTO-02-20-0029-R
Liu, X. T., Strelkov, S. E., Sun, R. F., Hwang, S. F., Fredua-Agyeman, R., Li, F., et al. (2022). Histopathology of the Plasmodiophora brassicae-Chinese cabbage interaction in hosts carrying different sources of resistance. Front. Plant Sci. 12:783550. doi: 10.3389/fpls.2021.783550
Liu, C. M., Yang, Z. F., He, P. F., Munir, S., He, P. B., Wu, Y. X., et al. (2019). Fluazinam positively affected the microbial communities in clubroot cabbage rhizosphere. Sci. Hortic. 256:108519. doi: 10.1016/j.scienta.2019.05.046
Ludwig-Müller, J. (2022). What can we learn from -omics approaches to understand clubroot disease? Int. J. Mol. Sci. 23:6293. doi: 10.3390/ijms23116293
Ludwig-Müller, J., Jülke, S., Geiss, K., Richter, F., Mithöfer, A., Sola, I., et al. (2015). A novel methyltransferase from the intracellular pathogen Plasmodiophora brassicae methylates salicylic acid. Mol. Plant Pathol. 16, 349–364. doi: 10.1111/mpp.12185
Ludwig-Müller, J., Prinsen, E., Rolfe, S., and Scholes, J. (2009). Metabolism and plant hormone action during the clubroot disease. J. Plant Growth Regul. 28, 229–244. doi: 10.1007/s00344-009-9089-4
Lv, M. C., Liu, Y. F., Wu, Y., Zhang, J., Liu, X. Y., Ji, R. Q., et al. (2021). An improved technique for isolation and characterization of single-spore isolates of Plasmodiophora brassicae. Plant Dis. 105, 3932–3938. doi: 10.1094/PDIS-03-21-0480-RE
Manzanares-Dauleux, M. J., Barret, P., and Thomas, G. (2000). Development of a pathotype specific SCAR marker in Plasmodiophora brassicae. Eur. J. Plant Pathol. 106, 781–787. doi: 10.1023/A:1026586803761
Matsumoto, E., Yasui, C., Ohi, M., and Tsukada, M. (1998). Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis). Euphytica 104, 79–86. doi: 10.1023/A:1018370418201
McDonald, M. R., and Westerveld, S. M. (2008). Temperature prior to harvest influences the incidence and severity of clubroot on two Asian Brassica vegetables. HortScience 43, 1509–1513. doi: 10.21273/HORTSCI.43.5.1509
McGrann, G. R. D., Gladders, P., Smith, J. A., and Burnett, F. (2016). Control of clubroot (Plasmodiophora brassicae) in oilseed rape using varietal resistance and soil amendments. Field Crop Res. 186, 146–156. doi: 10.1016/j.fcr.2015.11.013
Mehrabi, R., Bahkali, A. H., Abd-Elsalam, K. A., Moslem, M., Ben, M.' B., et al. (2011). Horizontal gene and chromosome transfer in plant pathogenic fungi affecting host range. FEMS Microbiol. Rev. 35, 542–554. doi: 10.1111/j.1574-6976.2010.00263.x
Mei, J. Q., Guo, Z., Wang, J. H., Feng, Y. X., Ma, G. H., Zhang, C. Y., et al. (2019). Understanding the resistance mechanism in Brassica napus to clubroot caused by Plasmodiophora brassicae. Phytopathology 109, 810–818. doi: 10.1094/PHYTO-06-18-0213-R
Mitani, S., Sugimoto, K., Hayashi, H., Takii, Y., Ohshima, T., and Matsuo, N. (2003). Effects of cyazofamid against Plasmodiophora brassicae Woronin on Chinese cabbage. Pest Manag. Sci. 59, 287–293. doi: 10.1002/ps.627
Muirhead, K., and Pérez-López, E. (2022). Plasmodiophora brassicae CBM18 proteins bind chitin and suppress chitin-triggered immunity. PhytoFrontiers 2, 21–29. doi: 10.1094/PHYTOFR-04-21-0032-R
Murakami, H., Tsushima, S., Akimoto, T., Murakami, K., Goto, I., and Shishido, Y. (2000). Effects of growing leafy daikon (Raphanus sativus) on populations of Plasmodiophora brassicae (clubroot). Plant Pathol. 49, 584–589. doi: 10.1046/j.1365-3059.2000.00495.x
Murakami, H., Tsushima, S., Kuroyanagi, Y., and Shishido, Y. (2002). Reduction of resting spore density of Plasmodiophora brassicae and clubroot disease severity by liming. Soil Sci. Plant Nutr. 48, 685–691. doi: 10.1080/00380768.2002.10409258
Myers, D. F., and Campbell, R. N. (1985). Lime and the control of clubroot of crucifers: effects of pH, calcium, magnesium, and their interactions. Phytopathology 75, 670–673. doi: 10.1094/Phyto-75-670
Naiki, T., and Dixon, G. R. (1987). 1999). The effects of chemicals on developmental stages of Plasmodiophora brassicae (clubroot. Plant Pathol. 36, 316–327. doi: 10.1111/j.1365-3059.1987.tb02238.x
Neik, T. X., Barbetti, M. J., and Batley, J. (2017). Current status and challenges in identifying disease resistance genes in Brassica napus. Front. Plant Sci. 8:1788. doi: 10.3389/fpls.2017.01788
Nicholson, W. L. (2002). Roles of Bacillus endospores in the environment. Cell. Mol. Life Sci. 59, 410–416. doi: 10.1007/s00018-002-8433-7
Ning, Y., Wang, Y., Fang, Z. Y., Zhuang, M., Zhang, Y. Y., Lv, H. H., et al. (2019). Comparative transcriptome analysis of cabbage (Brassica oleracea var. capitata) infected by Plasmodiophora brassicae reveals drastic defense response at secondary infection stage. Plant Soil 443, 167–183. doi: 10.1007/s11104-019-04196-6
Niu, B., Wang, W. X., Yuan, Z. B., Sederoff, R. R., Sederoff, H., Chiang, V. L., et al. (2020). Microbial interactions within multiple-strain biological control agents impact soil-borne plant disease. Front. Microbiol. 11:585404. doi: 10.3389/fmicb.2020.585404
Niwa, R., Kumei, T., Nomura, Y., Yoshida, S., Osaki, M., and Ezawa, T. (2007). Increase in soil pH due to ca-rich organic matter application causes suppression of the clubroot disease of crucifers. Soil Biol. Biochem. 39, 778–785. doi: 10.1016/j.soilbio.2006.09.027
Niwa, R., Nomura, Y., Osaki, M., and Ezawa, T. (2008). Suppression of clubroot disease under neutral pH caused by inhibition of spore germination of Plasmodiophora brassicae in the rhizosphere. Plant Pathol. 57, 445–452. doi: 10.1111/j.1365-3059.2007.01817.x
Ochman, H., Lawrence, J. G., and Groisman, E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304. doi: 10.1038/35012500
Ochoa, J. C., Mukhopadhyay, S., Bieluszewski, T., Jedryczka, M., Malinowski, R., and Truman, W. (2023). Natural variation in Arabidopsis responses to Plasmodiophora brassicae reveals an essential role for resistance to Plasmodiophora brasssicae 1 (RPB1). Plant J. 116, 1421–1440. doi: 10.1111/tpj.16438
Olszak, M., Truman, W., Stefanowicz, K., Sliwinska, E., Ito, M., Walerowski, P., et al. (2019). Transcriptional profiling identifies critical steps of cell cycle reprogramming necessary for Plasmodiophora brassicae-driven gall formation in Arabidopsis. Plant J. 97, 715–729. doi: 10.1111/tpj.14156
Pang, W. X., Fu, P. Y., Li, X. N., Zhan, Z. X., Yu, S., and Piao, Z. Y. (2018). Identification and mapping of the clubroot resistance gene CRd in Chinese cabbage (Brassica rapa ssp. pekinensis). Front. Plant Sci. 9:653. doi: 10.3389/fpls.2018.00653
Pang, W. X., Liang, Y., Zhan, Z. X., Li, X. N., and Piao, Z. Y. (2020). Development of a sinitic clubroot differential set for the pathotype classification of Plasmodiophora brassicae. Front. Plant Sci. 11:568771. doi: 10.3389/fpls.2020.568771
Pang, W. X., Zhang, X., Ma, Y. B., Wang, Y. J., Zhan, Z. X., and Piao, Z. Y. (2022). Fine mapping and candidate gene analysis of CRA3.7 conferring clubroot resistance in Brassica rapa. Theor. Appl. Genet. 135, 4541–4548. doi: 10.1007/s00122-022-04237-2
Patar, U. R., Manasova, M., Hnatek, J., Wenzlova, J., and Zouhar, M. (2023). Evaluation of ethanedinitrile as a potential fumigant against Plasmodiophora brassicae, the clubroot pathogen. Agronomy 13:1177. doi: 10.3390/agronomy13041177
Peng, G., Lahlali, R., Hwang, S. F., Pageau, D., Hynes, R. K., Mcdonald, M. R., et al. (2014). Crop rotation, cultivar resistance, and fungicides/biofungicides for managing clubroot (Plasmodiophora brassicae) on canola. Can. J. Plant Pathol. 36, 99–112. doi: 10.1080/07060661.2013.860398
Pérez-López, E., Hossain, M. M., Tu, J. Y., Waldner, M., Todd, C. D., Kusalik, A. J., et al. (2020). Transcriptome analysis identifies Plasmodiophora brassicae secondary infection effector candidates. J. Eukaryot. Microbiol. 67, 337–351. doi: 10.1111/jeu.12784
Pérez-López, E., Hossain, M. M., Wei, Y. D., Todd, C. D., and Bonham-Smith, P. C. (2021). A clubroot pathogen effector targets cruciferous cysteine proteases to suppress plant immunity. Virulence 12, 2327–2340. doi: 10.1080/21505594.2021.1968684
Pérez-López, E., Waldner, M., Hossain, M., Kusalik, A. J., Wei, Y., Bonham-Smith, P. C., et al. (2018). Identification of Plasmodiophora brassicae effectors - A challenging goal. Virulence 9, 1344–1353. doi: 10.1080/21505594.2018.1504560
Pesonen, M., and Vähäkangas, K. (2020). Chloropicrin-induced toxicity in the respiratory system. Toxicol. Lett. 323, 10–18. doi: 10.1016/j.toxlet.2020.01.022
Piao, Z. Y., Deng, Y. Q., Choi, S. R., Park, Y. J., and Lim, Y. P. (2004). SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 108, 1458–1465. doi: 10.1007/s00122-003-1577-5
Piao, Z., Ramchiary, N., and Lim, Y. P. (2009). Genetics of clubroot resistance in Brassica species. J. Plant Growth Regul. 28, 252–264. doi: 10.1007/s00344-009-9093-8
Porter, I. J., Merriman, P. R., and Keane, P. J. (1991). Soil solarisation combined with low rates of soil fumigants controls clubroot of cauliflowers, caused by Plasmodiophora brassicae Woron. Aust. J. Exp. Agric. 31, 843–851. doi: 10.1071/EA9910843
Rahaman, M., Strelkov, S. E., Hu, H., Gossen, B. D., and Yu, F. Q. (2022). Identification of a genomic region containing genes involved in resistance to four pathotypes of Plasmodiophora brassicae in Brassica rapa turnip ECD02. Plant Genome 15:e20245. doi: 10.1002/tpg2.20245
Rashid, A., Ahmed, H. U., Xiao, Q., Hwang, S. F., and Strelkov, S. E. (2013). Effects of root exudates and pH on Plasmodiophora brassicae resting spore germination and infection of canola (Brassica napus L.) root hairs. Crop Prot. 48, 16–23. doi: 10.1016/j.cropro.2012.11.025
Ren, L., Xu, L., Liu, F., Chen, K. R., Sun, C. C., Li, J., et al. (2016). Host range of Plasmodiophora brassicae on cruciferous crops and weeds in China. Plant Dis. 100, 933–939. doi: 10.1094/PDIS-09-15-1082-RE
Rennie, D. C., Holtz, M. D., Turkington, T. K., Leboldus, J. M., Hwang, S. F., Howard, R. J., et al. (2015). Movement of Plasmodiophora brassicae resting spores in windblown dust. Can. J. Plant Pathol. 37, 188–196. doi: 10.1080/07060661.2015.1036362
Rocherieux, J., Glory, P., Giboulot, A., Boury, S., Barbeyron, G., Thomas, G., et al. (2004). Isolate-specific and broad-spectrum QTLs are involved in the control of clubroot in Brassica oleracea. Theor. Appl. Genet. 108, 1555–1563. doi: 10.1007/s00122-003-1580-x
Rolfe, S. A., Strelkov, S. E., Links, M. G., Clarke, W. E., Robinson, S. J., Djavaheri, M., et al. (2016). The compact genome of the plant pathogen Plasmodiophora brassicae is adapted to intracellular interactions with host brassica spp. BMC Genomics 17:272. doi: 10.1186/s12864-016-2597-2
Ruaro, L., Lima, V. D., and Ribeiro, P. J. (2009). Influence of boron, nitrogen sources and soil pH on the control of club root of crucifers caused by Plasmodiophora brassicae. Trop. Plant Pathol. 34, 231–238. doi: 10.1590/S1982-56762009000400005
Ruzo, L. O. (2006). Physical, chemical and environmental properties of selected chemical alternatives for the pre-plant use of methyl bromide as soil fumigant. Pest Manag. Sci. 62, 99–113. doi: 10.1002/ps.1135
Saharan, G. S., Mehta, N., and Meena, P. D. (2021). Club root disease of crucifers: Biology, ecology and disease management. Singapore: Springer Verlag.
Sakamoto, K., Saito, A., Hayashida, N., Taguchi, G., and Matsumoto, E. (2008). Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theor. Appl. Genet. 117, 759–767. doi: 10.1007/s00122-008-0817-0
Sanders, I. R. (2006). Rapid disease emergence through horizontal gene transfer between eukaryotes. Trends Ecol. Evol. 21, 656–658. doi: 10.1016/j.tree.2006.10.006
Sarkar, A., Kisiala, A., Adhikary, D., Basu, U., Emery, R. J. N., Rahman, H., et al. (2023). Silicon ameliorates clubroot responses in canola (Brassica napus): A “multi-omics”-based investigation into possible mechanisms. Physiol. Plantar. 175:e13900. doi: 10.1111/ppl.13900
Sasaki, T., Naito, R., Ohara, T., Sakane, K., Tanaka, S., Sasaki, K., et al. (2024). Effects of flusulfamide on spore germination of Plasmodiophora brassicae. J. Pestic. Sci. 49, 52–57. doi: 10.1584/jpestics.D23-031
Schuller, A., and Ludwig-Müller, J. (2016). Histological methods to detect the clubroot pathogen Plasmodiophora brassicae during its complex life cycle. Plant Pathol. 65, 1223–1237. doi: 10.1111/ppa.12520
Schwelm, A., Fogelqvist, J., Knaust, A., Jülke, S., Lilja, T., Bonilla-Rosso, G., et al. (2015). The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci. Rep. 5:11153. doi: 10.1038/srep11153
Schwelm, A., and Ludwig-Müller, J. (2021). Molecular pathotyping of Plasmodiophora brassicae—genomes, marker genes, and obstacles. Pathogens 10:259. doi: 10.3390/pathogens10030259
Shakeel, Q., Lyu, A., Zhang, J., Wu, M. D., Chen, S. W., Chen, W. D., et al. (2016). Optimization of the cultural medium and conditions for production of antifungal substances by Streptomyces platensis 3-10 and evaluation of its efficacy in suppression of clubroot disease (Plasmodiophora brassicae) of oilseed rape. Biol. Control 101, 59–68. doi: 10.1016/j.biocontrol.2016.06.007
Sharma, K., Gossen, B. D., and McDonald, M. R. (2011). Effect of temperature on cortical infection by Plasmodiophora brassicae and clubroot severity. Phytopathology 101, 1424–1432. doi: 10.1094/PHYTO-04-11-0124
Sharp, N. P., Sandell, L., James, C. G., and Otto, S. P. (2018). The genome-wide rate and spectrum of spontaneous mutations differ between haploid and diploid yeast. Proc. Natl. Acad. Sci. USA 115, E5046–E5055. doi: 10.1073/pnas.1801040115
Shaw, R. K., Shen, Y. S., Yu, H. F., Sheng, X. G., Wang, J. S., and Gu, H. H. (2022). Multi-omics approaches to improve clubroot resistance in Brassica with a special focus on Brassica oleracea L. Int. J. Mol. Sci. 23:9280. doi: 10.3390/ijms23169280
Shi-Kunne, X., van Kooten, M., Depotter, J. R. L., Thomma, B. P. H. J., and Seidl, M. F. (2019). The genome of the fungal pathogen Verticillium dahliae reveals extensive bacterial to fungal gene transfer. Genome Biol. Evol. 11, 855–868. doi: 10.1093/gbe/evz040
Siemens, J., Bulman, S., Rehn, F., and Sundelin, T. (2009). Molecular biology of Plasmodiophora brassicae. J. Plant Growth Regul. 28, 245–251. doi: 10.1007/s00344-009-9091-x
Singh, K., Tzelepis, G., Zouhar, M., Ryšánek, P., and Dixelius, C. (2018). The immunophilin repertoire of Plasmodiophora brassicae and functional analysis of PbCYP3 cyclophilin. Mol. Genet. Genomics 293, 381–390. doi: 10.1007/s00438-017-1395-0
Somé, A., Manzanares, M. J., Laurens, F., Baron, F., Thomas, G., and Rouxel, F. (1996). Variation for virulence on Brassica napus L. amongst Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathol. 45, 432–439. doi: 10.1046/j.1365-3059.1996.d01-155.x
Strehlow, B., de Mol, F., and Struck, C. (2014). Risk potential of clubroot disease on winter oilseed rape. Plant Dis. 99, 667–675. doi: 10.1094/PDIS-05-14-0482-RE
Strelkov, S. E., Hwang, S. F., Manolii, V. P., Cao, T. S., Fredua-Agyeman, R., Harding, M. W., et al. (2018). Virulence and pathotype classification of Plasmodiophora brassicae populations collected from clubroot resistant canola (Brassica napus) in Canada. Can. J. Plant Pathol. 40, 284–298. doi: 10.1080/07060661.2018.1459851
Struck, C., Rüsch, S., and Strehlow, B. (2022). Control strategies of clubroot disease caused by Plasmodiophora brassicae. Microorganisms 10:620. doi: 10.3390/microorganisms10030620
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Fujimura, M., Nunome, T., et al. (2003). Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 107, 997–1002. doi: 10.1007/s00122-003-1309-x
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., Kondo, M., Fujimura, M., et al. (2006). Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics 173, 309–319. doi: 10.1534/genetics.104.038968
Tanaka, S., Kochi, S., Kunita, H., Ito, S., and Kameya-Iwaki, M. (1999). Biological mode of action of the fungicide, flusulfamide, against Plasmodiophora brassicae (clubroot). Eur. J. Plant Pathol. 105, 577–584. doi: 10.1023/A:1008722005570
Tinggal, S. H., and Webster, J. (1981). Technique for single spore infection by Plasmodiophora brassicae. Tran. Br. Mycol. Soc. 76, 187–190. doi: 10.1016/S0007-1536(81)80137-4
Tremblay, N., Bélec, C., Coulombe, J., and Godin, C. (2005). Evaluation of calcium cyanamide and liming for control of clubroot disease in cauliflower. Crop Prot. 24, 798–803. doi: 10.1016/j.cropro.2004.12.013
Tso, H. H., Galindo-González, L., Locke, T., and Strelkov, S. E. (2022). Protocol: rhPCR and SNaPshot assays to distinguish Plasmodiophora brassicae pathotype clusters. Plant Methods 18:91. doi: 10.1186/s13007-022-00923-w
Tso, H. H., Galindo-González, L., and Strelkov, S. E. (2021). Current and future pathotyping platforms for Plasmodiophora brassicae in Canada. Plan. Theory 10:1446. doi: 10.3390/plants10071446
Voorrips, R. E. (1996). Production, characterization and interaction of single-spore isolates of Plasmodiophora brassicae. Eur. J. Plant Pathol. 102, 377–383. doi: 10.1007/BF01878132
Voorrips, R. E., Jongerius, M. C., and Kanne, H. J. (1997). Mapping of two genes for resistance to clubroot (Plasmodiophora brassicae) in a population of doubled haploid lines of Brassica oleracea by means of RFLP and AFLP markers. Theor. Appl. Genet. 94, 75–82. doi: 10.1007/s001220050384
Wang, M., Gao, L. M., Dong, S. Y., Sun, Y. M., Shen, Q. R., and Guo, S. W. (2017). Role of silicon on plant-pathogen interactions. Front. Plant Sci. 8:701. doi: 10.3389/fpls.2017.00701
Wang, W., Qin, L., Zhang, W. J., Tang, L. H., Zhang, C., Dong, X. J., et al. (2023). WeiTsing, a pericycle-expressed ion channel, safeguards the stele to confer clubroot resistance. Cell 186, 2656–2671. doi: 10.1016/j.cell.2023.05.023
Wang, Y. Y., Xiang, X. Y., Huang, F., Yu, W. L., Zhou, X. Q., Li, B. J., et al. (2022). Fine mapping of clubroot resistance loci CRA8.1 and candidate gene analysis in Chinese cabbage (Brassica rapa L.). Front. Plant Sci. 13:898108. doi: 10.3389/fpls.2022.898108
Webster, M. A., and Dixon, G. R. (1991a). Calcium, pH and inoculum concentration influencing colonization by Plasmodiophora brassicae. Mycol. Res. 95, 64–73. doi: 10.1016/S0953-7562(09)81362-2
Webster, M. A., and Dixon, G. R. (1991b). Boron, pH and inoculum concentration influencing colonization by Plasmodiophora brassicae. Mycol. Res. 95, 74–79. doi: 10.1016/S0953-7562(09)81363-4
Wei, X. C., Li, J. D., Zhang, X. W., Zhao, Y. Y., Nath, U. K., Mao, L. X., et al. (2022). Fine mapping and functional analysis of major QTL, CRq for clubroot resistance in Chinese cabbage (Brassica rapa ssp. pekinensis). Agronomy 12:1172. doi: 10.3390/agronomy12051172
White, J. G., and Buczacki, S. T. (1977). The control of clubroot by soil partial sterilization: A review. Ann. Appl. Biol. 85, 287–300. doi: 10.1111/j.1744-7348.1977.tb01802.x
Williams, P. H. (1966). A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 56, 624–626.
Xu, X. M., Jeffries, P., Pautasso, M., and Jeger, M. J. (2011). A numerical study of combined use of two biocontrol agents with different biocontrol mechanisms in controlling foliar pathogens. Phytopathology 101, 1032–1044. doi: 10.1094/PHYTO-10-10-0267
Xue, S., Cao, T., Howard, R. J., Hwang, S. F., and Strelkov, S. E. (2008). Isolation and variation in virulence of single-spore isolates of Plasmodiophora brassicae from Canada. Plant Dis. 92, 456–462. doi: 10.1094/PDIS-92-3-0456
Yang, X. X., Huang, X. Q., Wu, W. X., Xiang, Y. J., Du, L., Zhang, L., et al. (2020b). Effects of different rotation patterns on the occurrence of clubroot disease and diversity of rhizosphere microbes. J. Integr. Agric. 19, 2265–2273. doi: 10.1016/S2095-3119(20)63186-0
Yang, Z. Q., Jiang, Y. F., Gong, J. F., Li, Q., Dun, B. C., Liu, D. X., et al. (2022a). R gene triplication confers European fodder turnip with improved clubroot resistance. Plant Biotechnol. J. 20, 1502–1517. doi: 10.1111/pbi.13827
Yang, H., Liao, R., Jin, C., Feng, X. Y., Zhang, Y., Wang, X. Y., et al. (2024). The conserved GOLD domain in the Plasmodiophora brassicae effector Pb257 is required for triggering cell death and root swelling. Phytopathol. Res. 6:18. doi: 10.1186/s42483-024-00236-x
Yang, H., Sun, Q. Y., Zhang, Y. H., Zhang, Y., Zhao, Y. S., Wang, X. Y., et al. (2022b). Comparing the infection biology and gene expression differences of Plasmodiophora brassicae primary and secondary zoospores. Front. Microbiol. 13:1002976. doi: 10.3389/fmicb.2022.1002976
Yang, H., Zheng, J., Fu, Y. D., Zhang, Y. H., Yi, C. L., Jin, C., et al. (2020a). Specific genes and sequence variation in pathotype 7 of the clubroot pathogen Plasmodiophora brassicae. Eur. J. Plant Pathol. 157, 17–28. doi: 10.1007/s10658-020-01968-0
Yu, Z. Y., Fredua-Agyeman, R., Strelkov, S. E., and Hwang, S. F. (2024a). RNA-Seq bulked segregant analysis of an exotic B. napus ssp. napobrassica (rutabaga) F2 population reveals novel QTLs for breeding clubroot-resistant canola. Int. J. Mol. Sci. 25:4596. doi: 10.3390/ijms25094596
Yu, Z. Y., Strelkov, S. E., and Hwang, S. F. (2024b). Evaluation of amisulbrom products for the management of clubroot of canola (Brassica napus). Plan. Theory 13:28. doi: 10.3390/plants13010028
Yu, F. W., Wang, S. Y., Zhang, W., Tang, J., Wang, H., Yu, L., et al. (2019). Genome-wide identification of genes encoding putative secreted E3 ubiquitin ligases and functional characterization of PbRING1 in the biotrophic protist Plasmodiophora brassicae. Curr. Genet. 65, 1355–1365. doi: 10.1007/s00294-019-00989-5
Yu, F. Q., Zhang, X. G., Peng, G., Falk, K. C., Strelkov, S. E., and Gossen, B. D. (2017). Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 7:4516. doi: 10.1038/s41598-017-04903-2
Yu, F. Q., Zhang, Y., Wang, J. H., Chen, Q. L., Karim, M. M., Gossen, B. D., et al. (2022). Identification of two major QTLs in Brassica napus lines with introgressed clubroot resistance from turnip cultivar ECD01. Front. Plant Sci. 12:785989. doi: 10.3389/fpls.2021.785989
Zamani-Noor, N., Brand, S., and Söchting, H. P. (2022). Effect of pathogen virulence on pathogenicity, host range, and reproduction of Plasmodiophora brassicae, the causal agent of clubroot disease. Plant Dis. 106, 57–64. doi: 10.1094/PDIS-02-21-0410-RE
Zamani-Noor, N., and Jedryczka, M. (2024). Pathotyping systems and pathotypes of Plasmodiophora brassicae-navigating toward the optimal classification. Pathogens 13:313. doi: 10.3390/pathogens13040313
Zeng, L. Y., Zhang, Y., Wu, Y. P., Zhang, X., Zhao, C. J., Ren, L., et al. (2024). Pathotype characterization of Plasmodiophora brassicae by European clubroot differential and Williams sets in China. Plant Dis. 108, 847–851. doi: 10.1094/PDIS-08-23-1571-SC
Zhan, Z. X., Liu, H. S., Yang, Y., Liu, S., Li, X. A., and Piao, Z. Y. (2022). Identification and characterization of putative effectors from Plasmodiophora brassicae that suppress or induce cell death in Nicotiana benthamiana. Front. Plant Sci. 13:881992. doi: 10.3389/fpls.2022.881992
Zhang, J. H., Ahmed, W., Dai, Z. L., Zhou, X. H., He, Z. L., Wei, L. F., et al. (2022). Microbial consortia: an engineering tool to suppress clubroot of Chinese cabbage by changing the rhizosphere bacterial community composition. Biology 11:918. doi: 10.3390/biology11060918
Zhang, H., Feng, J., Manolii, V. P., Strelkov, S. E., and Hwang, S. F. (2015). Characterization of a gene identified in pathotype 5 of the clubroot pathogen Plasmodiophora brassicae. Phytopathology 105, 764–770. doi: 10.1094/PHYTO-10-14-0270-R
Zhang, X. L., Han, F. Q., Li, Z. S., Wen, Z. H., Cheng, W. J., Shan, X. Z., et al. (2024). Map-based cloning and functional analysis of a major quantitative trait locus, BolC.Pb9.1, controlling clubroot resistance in a wild Brassica relative (Brassica macrocarpa). Theor. Appl. Genet. 137:41. doi: 10.1007/s00122-024-04543-x
Zhang, D., Jiang, B. W., and Lv, S. (2019). The importance of soil conditioners controlling bacterial biodiversity of vegetable fields under continuous cropping. Appl. Ecol. Environ. Res. 17, 4655–4676. doi: 10.15666/aeer/1702_46554676
Zhang, H., Liu, X. T., Zhou, J. Y., Strelkov, S. E., Fredua-Agyeman, R., Zhang, S. F., et al. (2024). Identification of clubroot (Plasmodiophora brassicae) resistance loci in Chinese cabbage (Brassica rapa ssppekinensis) with recessive character. Genes 15:274. doi: 10.3390/genes15030274
Zhang, J. H., Zhou, X. H., Zhang, Y., Dai, Z. L., He, Z. L., Qiu, Y., et al. (2024). Pre-soil fumigation with ammonium bicarbonate and lime modulates the rhizosphere microbiome to mitigate clubroot disease in Chinese cabbage. Front. Microbiol. 15:1376579. doi: 10.3389/fmicb.2024.1376579
Zhao, Y. L., Chen, X. F., Cheng, J. S., Xie, J. T., Lin, Y., Jiang, D. H., et al. (2022). Application of Trichoderma Hz36 and Hk37 as biocontrol agents against clubroot caused by Plasmodiophora brassicae. J. Fungi 8:777. doi: 10.3390/jof8080777
Zheng, J., Wang, X. L., Li, Q., Yuan, S., Wei, S. Q., Tian, X. Y., et al. (2018). Characterization of five molecular markers for pathotype identification of the clubroot pathogen Plasmodiophora brassicae. Phytopathology 108, 1486–1492. doi: 10.1094/PHYTO-11-17-0362-R
Zheng, J., Wang, X. L., Xiao, Y., Wei, S. Q., Wang, D., Huang, Y., et al. (2019). Specific genes identified in pathotype 4 of the clubroot pathogen Plasmodiophora brassicae. Plant Dis. 103, 495–503. doi: 10.1094/PDIS-05-18-0912-RE
Zhou, Q., Hwang, S. F., Strelkov, S. E., Fredua-Agyeman, R., and Manolii, V. P. (2018). A molecular marker for the specific detection of new pathotype 5-like strains of Plasmodiophora brassicae in canola. Plant Pathol. 67, 1582–1588. doi: 10.1111/ppa.12868
Zhou, L. H., Zhang, L. H., He, Y. Q., Liu, F., Li, M., Wang, Z. S., et al. (2014). Isolation and characterization of bacterial isolates for biological control of clubroot on Chinese cabbage. Eur. J. Plant Pathol. 140, 159–168. doi: 10.1007/s10658-014-0451-4
Zhu, M. Z., Yang, L. M., Zhang, Y. Y., Zhuang, M., Ji, J. L., Hou, X. L., et al. (2022). Introgression of clubroot resistant gene into Brassica oleracea L. from Brassica rapa based on homoeologous exchange. Hortic. Res. 9:uhac195. doi: 10.1093/hr/uhac195
Keywords: clubroot disease, Plasmodiophora brassicae , infection process, effectors, physiological pathotypes, management strategies
Citation: Xu X, Wu C, Zhang F, Yao J, Fan L, Liu Z and Yao Y (2025) Comprehensive review of Plasmodiophora brassicae: pathogenesis, pathotype diversity, and integrated control methods. Front. Microbiol. 16:1531393. doi: 10.3389/fmicb.2025.1531393
Edited by:
Md. Motaher Hossain, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Xiefeng Yao, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaRunmao Lin, Hainan University, China
Prabhu Dayal Meena, Indian Council of Agricultural Research (ICAR), India
Copyright © 2025 Xu, Wu, Zhang, Yao, Fan, Liu and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingjuan Yao, eWFveWluZ2p1YW4yMDA4QDE2My5jb20=
 Xueliang Xu
Xueliang Xu Caiyun Wu
Caiyun Wu Yingjuan Yao
Yingjuan Yao