- 1Key Laboratory of Protection and Utilization of Biological Resources in Tarim Basin of Xinjiang Production & Construction Corps, Xinjiang, China
- 2College of Life Sciences and Technology, Tarim University, Xinjiang, China
Introduction: The microbial community in desert ecosystems is a vital and highly active component. Streptomyces, one of the dominant genera within this community, exhibits significant capabilities in metabolic degradation and the synthesis of secondary metabolites.
Methods: To investigate the metabolic potential of Streptomyces desertis, two strains of Streptomyces were isolated from rhizosphere soil samples of Populus euphratica in the Taklimakan Desert during the initial phase of this study, TRM 70351T and TRM 70361T.
Results: The identification of these strains as belonging to the genus Streptomyces was confirmed through 16S rRNA sequencing. All calculated Average Nucleotide Identity (ANI) values were below the 95% cut-off recommended for distinguishing distinct species, and the estimated digital DNA-DNA hybridization (dDDH) values were all below the suggested threshold of 70% for species delineation. Results from phylogenetic, genomic, phenotypic, and chemotaxonomic analyses confirmed that TRM 70351T and TRM 70361T represent two new species within the genus Streptomyces, designated as Streptomyces solitudinis sp. nov. and Streptomyces rubellus sp. nov. The type strain for Streptomyces solitudinis sp. nov. is TRM 70351T ( = CCTCC AA 2020049T = LMG 32404T), while the type strain for Streptomyces rubellus sp. nov. is TRM 70361T ( = CCTCC AA 2020043T = JCM 35793T). Notably, Among the identified gene clusters of TRM 70351T, cluster 12.1 was predicted to be the biosynthetic gene cluster responsible for producing the aminoglycoside compound streptomycin, exhibiting a similarity of 55%. In this study, HSQC-TOCSY was employed to detect the presence of aminoglycosides in fermentation medium No. 1, while LC-MS/MS was utilized to analyze the molecular fragments of neomycin and streptomycin in the alkaline aqueous phase sample of the fermentation product. The mixture was eluted using methanol and ammonia water in a 3:1 ratio, leading to the further separation of the compounds daidzein and Tridec-1-ene.
Discussion: This study has enhanced the species resources of Streptomyces deserticum and the diversity of aminoglycoside compound-producing bacteria. TRM 70351T exhibited unique metabolic potential, indicating that further studies could be conducted in the future.
Introduction
Few plants and animals can survive in deserts, particularly in the desert hinterland, making the microbial community the most active and crucial component of this ecosystem. Actinomycetes, characterized by their strong spore-producing capabilities, metabolic degradation abilities, synthesis of secondary metabolites, and various ultraviolet repair mechanisms, are among the dominant phyla in desert environments, with Streptomyces being one of the predominant genera (Ensign, 1978; Gao and Garcia-Pichel, 2011; Makhalanyane et al., 2013; McCarthy and Williams, 1992; Neilson et al., 2012). Consequently, Streptomyces has become a popular germplasm resource studied by experts and scholars worldwide. Between 2000 and 2021, a total of 129 new species of actinomycetes were discovered and documented across 35 desert environments globally. These actinomycetes have produced over 50 new compounds with potential applications in medicine, agriculture, and industry (Xie and Pathom-Aree, 2021). It has been found that Streptomyces asenjonii sp. nov. can produce new compounds Asenjonamides A-C (Abdelkader et al., 2018). This underscores the untapped potential of desert microorganisms and their secondary metabolites. As the predominant species of desert actinomycetes and a primary source of aminoglycosides, Streptomyces merits further exploration and utilization.
Natural aminoglycosides (AGs) are classified based on the primary sources of their strains. These include antibiotics derived from Streptomyces cultures, such as streptomycin and kanamycin, as well as those derived from Microsporum cultures, such as gentamicin and cissomycin (Becker and Cooper, 2013). Aminoglycosides offer significant advantages over other antibiotics; they are bactericidal rather than bacteriostatic for Gram-negative bacteria and can be effectively combined with other drugs, such as ampicillin, to enhance bacterial eradication. Furthermore, aminoglycosides are hypoallergenic and suitable for use in treating acute infections (Xie et al., 2011). Despite their critical role in clinical settings, the widespread use of these drugs has contributed to the emergence of drug resistance.
To discover new aminoglycosides, genome-guided metabolic potential mining serves as a crucial approach. For instance, crexazone 2 identified by genome mining of Streptomyces sp. CS057 (Prado-Alonso et al., 2024). Due to the highly polar and basic nature of aminoglycosides, these compounds lack chromophores and consequently exhibit no UV or fluorescence absorption, complicating their isolation and detection. To overcome these challenges, cation exchange resins are commonly employed for separation and purification. For example, Li et al. utilized a D4 cation exchange resin to isolate and purify kanamycin b, increasing its purity from 2.5 to 6% in the fermentation broth (Li et al., 2002). In terms of detecting kanamycin b, Li et al. applied a combination of one- and two-dimensional nuclear magnetic resonance (NMR) techniques to comparatively analyze the structural features of three aminoglycosides: etilmicin, 1,3-di-N-ethylgentamicin, and 1,6-di-N-ethylgentamicin, thereby establishing a method for the rapid and precise structural analysis of these antibiotics. The high sensitivity and selectivity of HPLC-MS/MS, along with the weakly basic characteristics of these compounds, contribute to a robust positive ion response during mass spectrometric analysis (Li et al., 2017; Yang et al., 2006). Additionally, an ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) method has been developed for the simultaneous determination of nine aminoglycosides, including streptomycin, dihydrostreptomycin, neomycin, and gentamicin (Ma et al., 2020).
In this study, we conducted a comprehensive characterization and phylogenetic analysis on two novel strains, namely TRM 70351T and TRM 70361T, which were isolated from poplar rhizomes collected in the Taklamakan Desert, Xinjiang, China. We further explored the metabolic potential of TRM 70351T by detecting aminoglycoside signals using LC-MS, and then isolated and purified compounds daidzein and tridec-1-ene.
Materials and methods
Isolation and preserve
Soil samples were collected from poplar rhizomes in the Taklamakan Desert (37.813°N, 80.430°E), Xinjiang, China. The samples were then air-dried for 7 days. TRM 70351T and TRM 70361T were isolated on Gao’s agar containing sea salt (sea salt 33.3g, K2HPO4 0.5g, MgSO47H2O 0.5g, CaCO3 1g, Agar 17g, H2O 1L of water Add a trace of salt 1ml before pouring the plate). The strains were preserved in tubes containing 20% glycerol or 20% milk, respectively.
16S rRNA gene phylogeny
The 16S rRNA gene was amplified from strains TRM 70351T and TRM 70361T using universal primers 27F (5′-AGAGTTTGATCCTGGCTC-3′) and 1492R (5′-CGGCTACCTTGTTACGACTT-3′) (Qingke Biotechnology, China) following the extraction of genomic DNA with a Bacterial DNA Isolation KitSequencing was performed by sanger sequencing using the 3730XL sequencing platform. The sequence was assembled using SeqMan software (available from DNA Star and accessible at https://www.dnastar.com/software/lasergene/seqman-ultra/). The complete 16S rRNA sequence was compared to the 16S rRNA gene sequences of other species of Streptomyces in the EZbiocloud database, and the 16S rRNA sequence of the strain was stored in the GenBank/EMBL/DDBJ database (PQ758399, PQ758398). Phylogenetic trees were generated using three methodologies, namely neighbor-joining (NJ) (Saitou and Nei, 1987), maximum-likelihood (ML) (Felsenstein, 1981), and maximum-parsimony (MP) (Mount, 2008), which were facilitated by MEGA X software (Kumar et al., 2018). A multilocus sequence analysis (MLSA) tree was constructed by utilizing five housekeeping genes, namely atpD, gyrB, recA, rpoB, and trpB1 (Komaki, 2022). The genetic distance was conducted utilizing Kimura’s two-parameter model (Kimura, 1980). Bootstrap values were obtained in a test of tree topology stability with 1,000 replicates (Berner, 2009). Strain TRM 70351T and TRM 70361T genome was sequenced using the Illumina HiSeq 2000 platform (Hu et al., 2015). The genomic of the strains was assembled by using Abyss 2.0 (Jackman et al., 2017). The genome was carried out with the rapid annotations tool, facilitated by Prokka (Seemann, 2014). The digital DNA-DNA hybridization (dDDH) values were calculated using formula 2 on the GGDC website.2 The average nucleotide identity (ANI) was determined using OrthoANI, with default parameters (Yoon et al., 2017). Cross-checks such as ANIb, ANIm were also used in this study to better determine the new species status of the strains3 (Riesco and Trujillo, 2024). In this study, we evaluated the potential of TRM 70351T to synthesize aminoglycosides using a comparative genomics approach to elucidate the biosynthetic potential of the strain (Blin et al., 2021).
Phenotypic characterization
After 7 days of incubation in the International Streptomyces Program medium (Waksman, 1967), Gao’s medium (Kelly, 1964), PDA, NA, and Czapek-Dox Agar, the colony morphology of TRM 70351T and TRM 70361T was observed at 37°C. The morphological characteristics of the colonies were examined using optical microscopy (BX41, Olympus) and scanning electron microscopy (JSM-6610LV, JEOL) for 7 days. The colors of the colonies were established based on established color standards and nomenclature. The experiments using carbon sources were conducted according to the methodology outlined by Pridham and Gottlieb (1948) and Shirling and Gottlieb (1966). Streptomyces has a variety of physiological and biochemical characteristics, as previously describied (McCarthy and Williams, 1992; Pridham and Gottlieb, 1948). The growth capabilities of strains TRM 70351T and TRM 70361T were assessed across a temperature range of 10–55°C (specifically at 10, 12, 15, 20, 25, 28, 30, 37, 40, 45, 50, and 55°C) and a pH range of 4–12 (including pH values of 4, 5, 6, 7, 8, 9, 10, 11, and 12). Additionally, their tolerance to NaCl concentrations ranging from 0 to 10% (0, 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10%, w/v) was evaluated using Gao’s broth as the basal medium. The production of peroxidase, urease, esterase, and catalase was tested utilizing the method described by Gerhardt et al. (1994). Determination of optimum conditions by observation and mycelial centrifugal weighing.
Phenotypic and chemotaxonomic characterization
To study cell biomass, cells are grown in flasks with liquid Gause’s medium for 7 days at 37°C. Then, the cells are spun around at 12,000 rpm for 10 minutes (Centrifuge: cence-H1750R, Hunan) and then washed with distilled water twice. The Whole-cell sugars were analyzed using procedures developed by Kim (Freeze-dryer: BMC FD-1B-50+Beijing) (Kim and Goodfellow, 1999). Menaquinones were extracted from freeze-dried biomass and purified following the protocol established by Collins et al. (1977). Cellular fatty acid analyses were conducted in accordance with the protocol of the Sherlock Microbial ID System.4 Standard methods were used to determine the types of amino acids and total sugars in the cell wall hydrolysates (Li et al., 2019). Polar lipids were prepared according to Minnikin’s method, extracted (Spin Vapour Ankeyq N-1100), detected by two-dimensional thin-layer chromatography (Display laminate: TLC Silica gel 60), and analyzed with 10% molybdate ethanol (Zhang et al., 2018).
TRM 70351 fermentation crude extract activity and HSQC-TOCSY detection
The uncontaminated seed liquid was inoculated into a pre-configured fermentation medium. One liter of each of the four mediums—namely millet medium, P4 medium, Streptomycin fermentation medium No. 1 (Han and Guirong, 2008) and Streptomycin fermentation medium No. 2 (Li et al., 2013) was selected, with an inoculum volume of 1%. This selection was made in conjunction with the literature on streptomycin fermentation and the growth state of the medium. The fermentation was conducted for 7 days at a temperature of 35°C. Another bottle of medium (200 mL) was used as a blank control. The samples were dried by freezing. After freezeing, the samples were soaked in 60% methanol, ultrasonicated, concentrated, and analyzed for activity using the Oxford cup method, while 5 mg of frozen samples were sent for HSQC-TOCSY to detect aminoglycosides (Bruker Ascend 500 MHz). Based on the activity and HSQC-TOCSY findings, Streptomycin fermentation medium No.1 of 70 L was selected for batch fermentation (fermenter:BIOTECH-100JS-7000, Shanghai) (Li et al., 2017).
Isolation and identification of compounds
After fermentation, the crude product obtained after fermentation was redissolved in water and subsequently filtered through four layers of gauze. The resulting filtrate was adsorbent with D101 macroporous resin and D4 cation exchange resin. The D101 column was mixed with 80% methanol and the extract was concentrated in the methanol phase at low pressure. For the cation exchange resin column, elution was performed using a methanol:ammonia (3:1) elution and the extract was concentrated under reduced pressure in the alkaline aqueous. A total of 5 mg of the sample, along with standards of neomycin sulfate and streptomycin sulfate, were prepared by employing D2O as the solvent. The sample was diluted to a concentration of 5 μg/mL, and detection was performed using LC-MS/MS in an ESI scanning mode with positive ion detection (Ma et al., 2020; Yang et al., 2006).
In order to obtain additional compounds, the spreading agent was selected as dichloromethane:methanol, using the traditional separation method TLC. The spreading ratio for the spreading layer is determined by 1:1, 3:1, 5:1, 10:1, 20:1, 30:1, 40:1, and 50:1. Select sulfuric acid as the color development agent. The mass ratio of crude extracts to silica gel was 1:1, and according to the retention factor (Rf), dichloromethane:methanol 30:1 was selected as the eluent for the elution. A total of eight groups of samples were collected. The yield of compound 1 was obtained by concentrating sample 7. The concentrated samples 1–5 were utilized for subsequent elution, with a ratio of 5:1 between dichloromenthane and ethyl acetate. One group of 500 mL was selected, and the color developer was chosen to be sulfuric acid, Compound 2 was obtained by concentrating sample 4. The compounds were identified using mass spectrometry and nuclear magnetic resonance (NMR) techniques (Li et al., 2017). A comparative analysis was conducted to investigate the structures of the monomer compounds in conjunction with existing literature.
Results
Polyphasic classification and genetic characterization of strains TRM 70351T and TRM 70361T
The 16S rRNA gene sequence of strain TRM 70351T showed that the strain was closely related to Streptomyces chumphonensis K1-2T ( = JCM 18522T, 98.34%). Analysis of the phylogenetic tree and genome and 16S rRNA gene sequences revealed that strain TRM 70351T shared the same node with S. chumphonensis K1-2T (Supplementary Figure S1). The 16S rRNA gene sequence of strain TRM 70361T showed a close relationship with Streptomyces barkulensis RC 1831T( = JCM 18754T, 98.15%). The physiological tree of MLSA and genome and 16S rRNA gene sequences analysis indicated that strain TRM 70361T shared the same node with S.carminius TRM SA0054T. Both TRM 70351T and TRM 70361T are clustered with similar strains, suggesting that they are potential new species.
The analysis of the MLSA tree revealed that strain TRM 70351T was situated in close proximity to Streptomyces chumphonensis K1-2T (Figure 1). Based on the highest 16S rRNA gene similarity, 16S rRNA gene tree, MLSA results, and phylogenetic tree, the type strains of S.chumphonensis K1-2T and S.durbertensis NEAU-S1GS20T were selected for further comparisons with strain TRM 70351T. The results of the MLSA tree analysis indicated that strain TRM 70361T was situated at the same node as S. carminius TRM SA0054T. Based on the highest 16S rRNA gene similarity, 16S rRNA gene tree, MLSA results, and phylogenetic tree, the type strains of S.barkulensis RC 1831T and S.carminius TRM SA0054T were chosen for further comparison with strain TRM 70361T (Supplementary Figure S2).
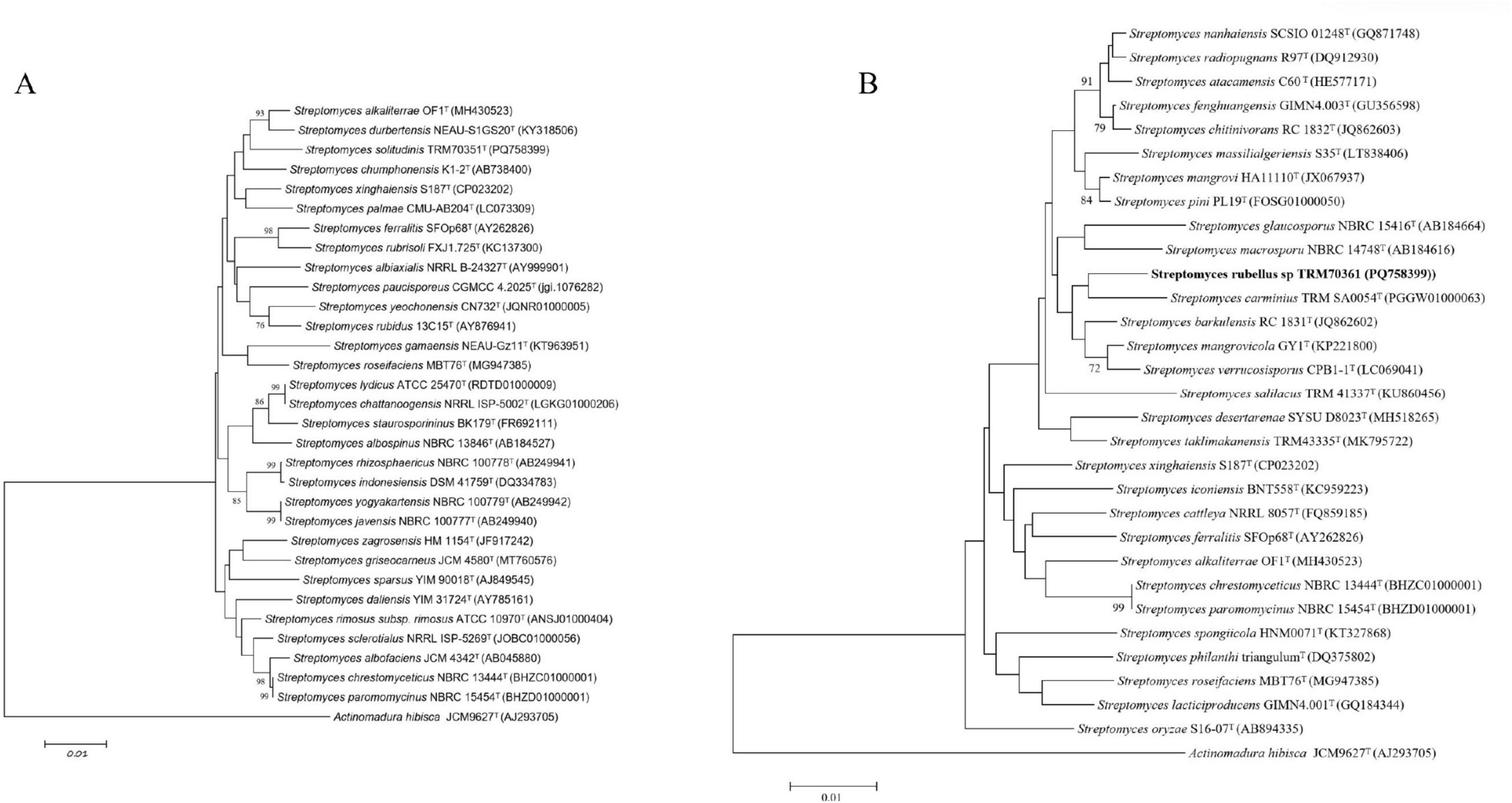
Figure 1. Neighbor-joining phylogenetic tree based on nearly complete 16S rRNA gene sequences. (A) The relationships between strain TRM 70351. (B) TRM 70361, and the type strains of closely related Streptomyces species were analyzed. Actinomadura hibisca JCM 9627T was used as the outgroup.
All of the calculated ANI values and ANIb and ANIm were below the 95% threshold for determining distinct species (Supplementary Table S2). Moreover, all the estimated dDDH values were below the threshold for species differentiation (70%) (Chun et al., 2018),. The summary of these findings has been presented in Table 1. Based on the ANI and dDDH values, it appears that the strains TRM 70351T and TRM 70361T are novel Streptomyces species.
Physiology, morphological characteristics
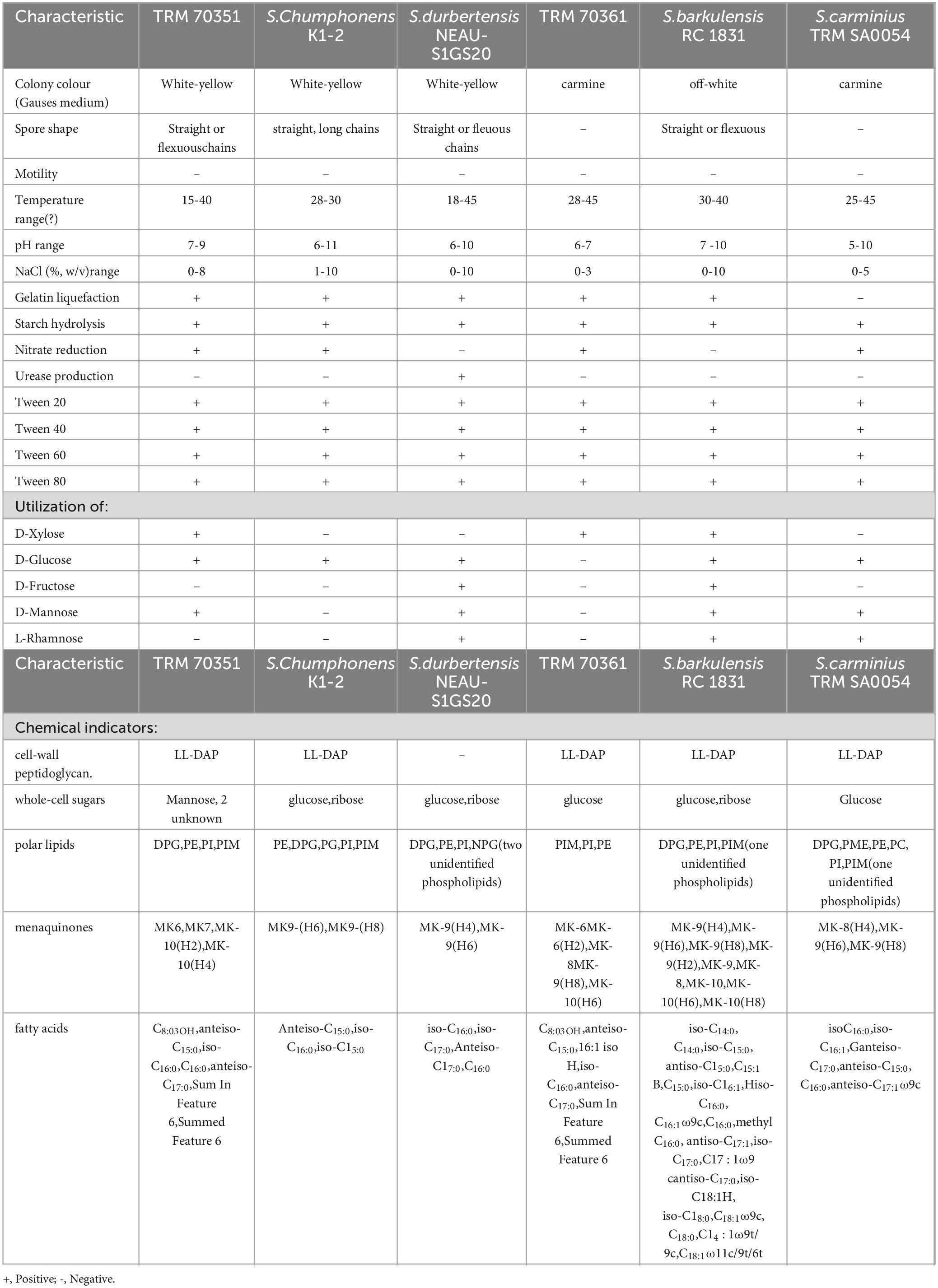
TRM 70351T produced straight, lengthy chains of non-motile spores (Rectiflexibiles) with rough surfaces (Figures 2A,B), whereas TRM 70361T has sporangia, each of which is rosebud-shaped, with hairs on the surface of the mycelium (Figures 2C,D). TRM 70351T demonstrated excellent growth on ISP 1, 2, 4, Czapek’s and Gao’s medium (Table 2). The aerial mycelium and substrate mycelium of TRM 70351T were developed without fragmentation, and the aerial mycelium exhibited a white to off-white appearance on all medium, while the substrate mycelium exhibited a white to yellow-white appearance. This strain has an optimal growth at pH 7–8. The optimal growth temperature ranged from 28to 40?. The growth occurred in the presence of 0–8% NaCl (w/v) as indicated in Table 2. TRM 70361T demonstrated significant growth on both ISP 2, 4, and Gao’s medium. TRM 70361T was developed without fragmentation of the aerial mycelium and substrate mycelium. The aerial mycelium appeared to be white to carmine on all media, and the substrate mycelium appeared to be white to yellow-white. The optimal pH ranges from 6 to 7. The optimal growth temperature was 30?. The growth was observed in the presence of 0-3% NaCl (w/v). The detailed physiological and biochemical properties are shown in Table 2 and in the species description.
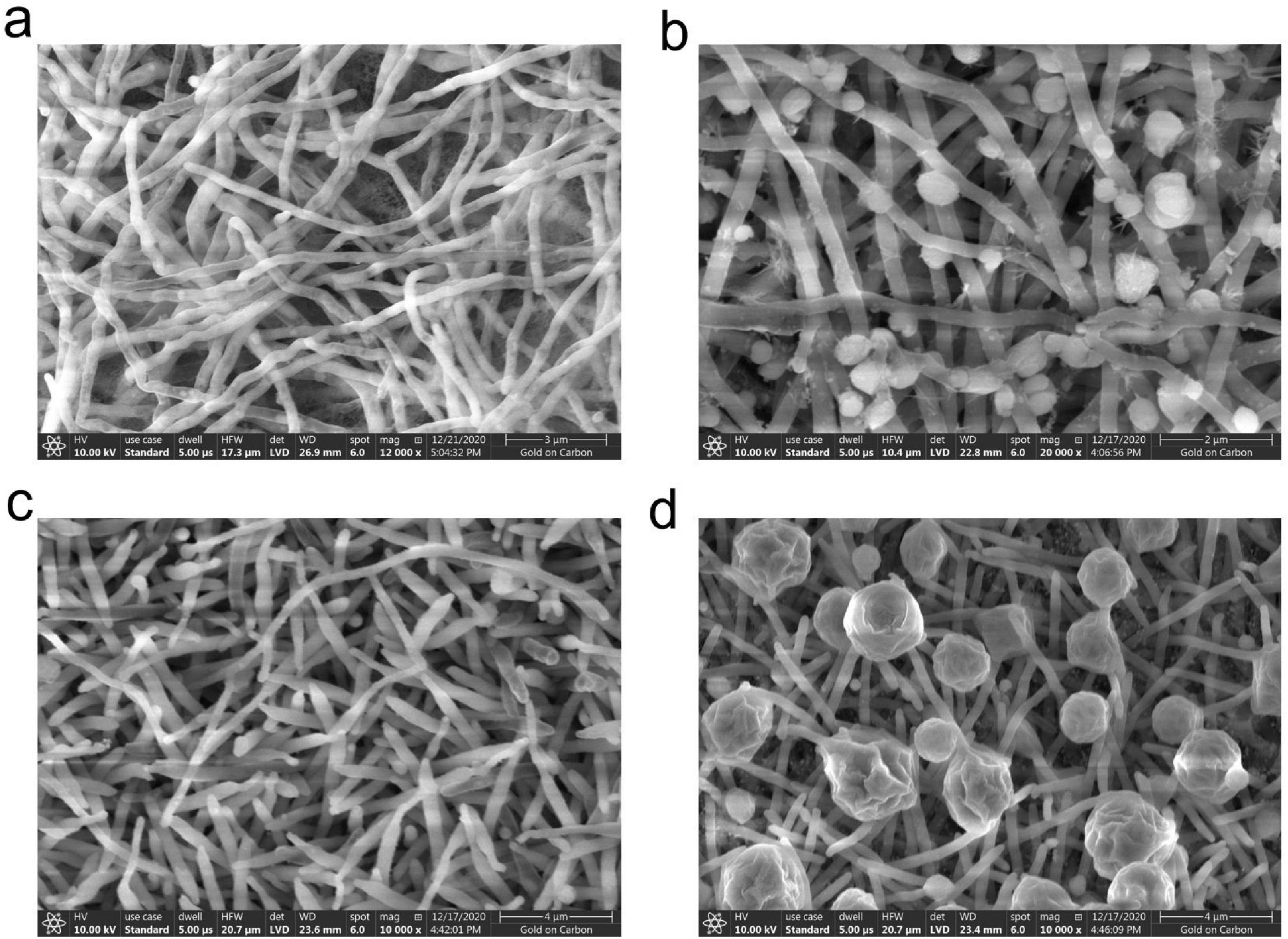
Figure 2. Scanning electron microscopy image of strain TRM 70351T grown on Gao’s agar at 37°C for 7 days. (A) Mycelium of strain TRM 70351T. (B) Spores of strain TRM 70351T. (C) Mycelium of strain TRM 70361T. (D) Spores of strain TRM 70361T.
Chemical composition characteristics
TRM 70351T contained LL-diaminopimelic acid in its cell-wall peptidoglycan and mainly contained mannitol as whole-cell hydrolysates, along with two unidentified hydrolysates. As illustrated in Supplementary Figure S3, the major polar lipids of TRM 70351T included phosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, and phosphatidylinositol mannosides. The dominant menaquinones in TRM 70351T were MK6, MK7, MK-10(H2), and MK-10(H4). The principal cellular fatty acids were identified as anteiso-C15:0, iso-C16:0, C16:0, and anteiso-C17:0, along with summed feature 6 (Table 2; Supplementary Table S3). TRM 70361T contained LL-diaminopimelic acid in the cell-wall peptidoglycan and primarily contained glucose as whole-cell hydrolysates. The major polar lipids of TRM 70361T comprised phosphatidylethanolamine, phosphatidylinositol, and phosphatidylinositol mannosides, as depicted in Supplementary Figure S3. The predominant menaquinones present in TRM 70361T were identified as MK6, MK6(H2), MK-8, MK-9(H8), and MK-10(H6). The major cellular fatty acids identified in Table 2. It is C8:0 3OH, anteiso-C15:0, 16:1 iso H, iso-C16:0, and anteiso-C17:0 (Table 2; Supplementary Table S3).
Analysis of the genomic metabolic capacity and biosynthetic gene cluster of TRM 70351T is conducted
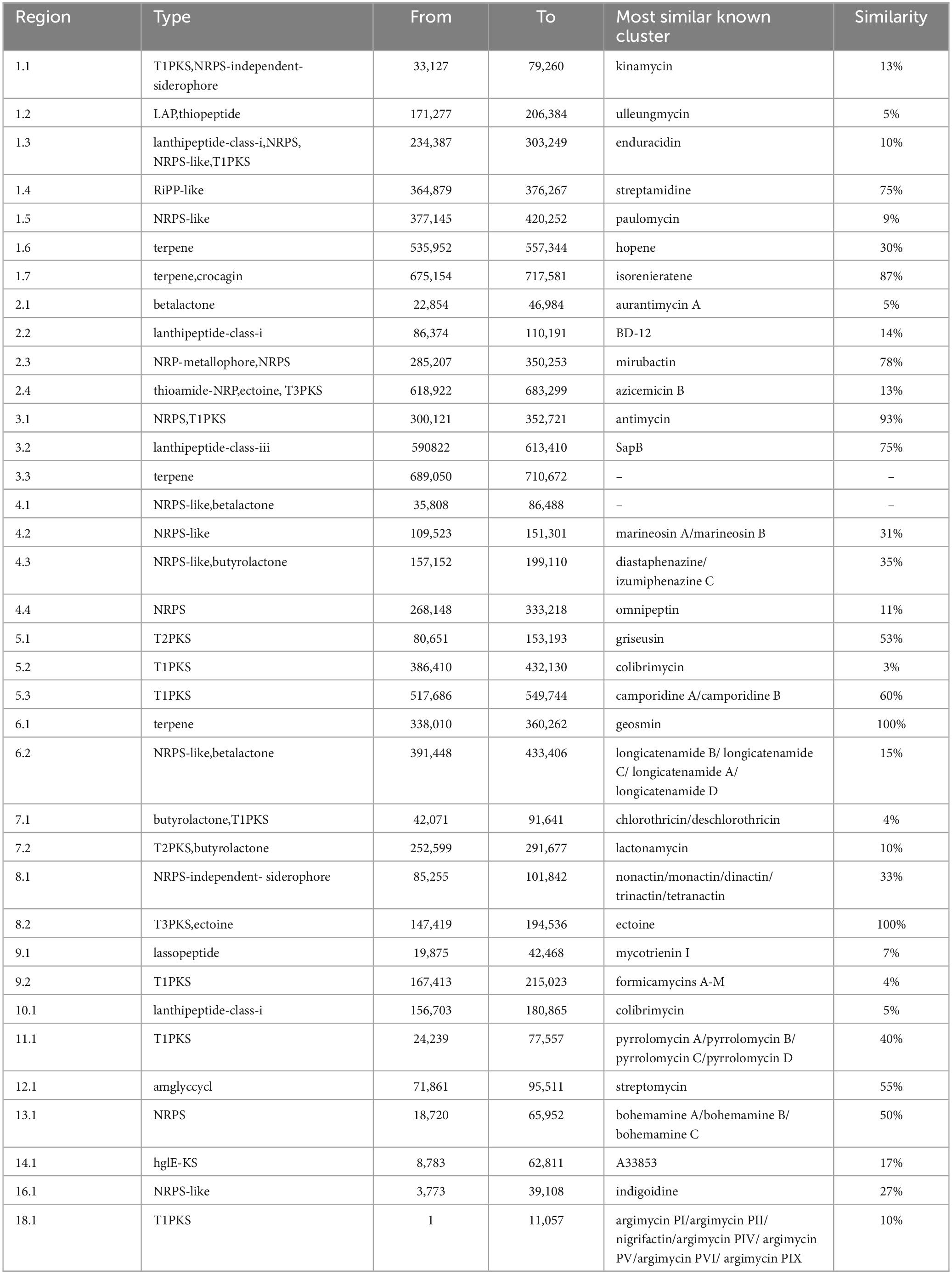
TRM 70351T has a genome size of 5,940,221 bp, a coding sequence count of 5,223, and a G+C content of 73.69 mol%. A total of 36 biosynthetic gene clusters were predicted within the TRM 70351T genome (Table 3). These include one amglyccycl, betalactone, hglE-KS, lanthipeptide-class-iii, lassopeptide, NRPS-independent-siderophore, and T2PKS. Additionally, there are two NRPS biosynthetic gene cluster, two lanthipeptide-class-i biosynthetic gene cluster, three NRPS-like biosynthetic gene cluster, three terpene types, five T1PKS types, and thirteen other heterozygous types in total. Among these, gene clusters 6.1 and 8.2 show 100% similarity to known biosynthetic gene clusters, indicating that the strain is capable of producing the corresponding secondary metabolites. However, antiSMASH did not predict gene clusters 3.3 and 4.1 because they were not similar to known biosynthetic gene clusters groups. It is proposed that these gene clusters may indeed produce natural products that differ in both structure and synthesis mechanisms from those of known secondary metabolites. Clusters 1.4, 1.7, 2.3, 3.1, 3.2, 5.1, 5.3, 12.1, and 13.1 exhibit a high degree of similarity (>50%) to known gene clusters, indicating a greater likelihood of producing corresponding compounds. It is notable that gene cluster 1.4 is predicted to produce ribosome-synthesized and post-translationally modified peptides (RiPPs), such as streptamidine, with a similarity of 75%. Streptamidine is recognized as a structurally diverse natural product with a wide range of biological activities, and is recognized as a structurally diverse natural product. Gene cluster 3.1 corresponds to antimycins (93%) belonging to the NRPS class, which possess antifungal, insecticidal, and antibiotic properties. Gene cluster 5.3 is predicted to correspond to a type 1 polyketide synthase compound, camporidine, which exhibits 60% similarity. Camporidine is a polyketide alkaloid with anti-inflammatory properties. Gene cluster 13.1 is predicted to yield a non-ribosomal peptide synthase (NRPS) compound, bohemamine, with a 50% similarity to NRPS. Bohemamine is classified within the alkaloid group and exhibits a diverse- range of biological activities, including antibacterial properties against gram-negative bacteria. Furthermore, there are a substantial number of antibiotic-like gene clusters with less than 50% similarity, including kinamycin, azicemicin B, and formicamycins A-M, among various others. Additionally, TRM 70351T holds significant potential for the discovery of secondary metabolites.
The gene cluster 12.1 is predicted to be an aminoglycoside biosynthetic gene cluster for streptomycin, exhibiting 55% similarity (see Figure 3). Aminoglycosides have a broad range of antimicrobial and bacteriostatic properties. Upon further examination of this gene cluster, two core genes, nine supplementary genes, and various other types of coding genes were identified within the biosynthetic gene cluster. Using BLAST to compare the gene functions of core genes and additional genes (Figure 3), it was found that the core genes have the same function as the genes in the streptomycin biosynthetic gene cluster of Streptomyces griseus, and they are both responsible for encoding dTDP-dihydrostreptose-streptidine-6-phosphate and alkaline phosphatase, the similarities are 98.18 and 97.84%. In addition, there are 9 additional genes, among which inosamine-phosphate amidinotransferase 1 and beta-ketoacyl-ACP reductase are unique to strain TRM 70351T, with similarities of 100 and 96.15%. The remaining genes have the same functions as those in the streptomycin biosynthetic gene cluster of Streptomyces griseus, including dTDP-4-dehydrorhamnose 3,5-epimerase, dTDP-4-dehydrorhamnose reductase, glucose-1-phosphate thymidylyltransferase, etc., with a similarity of 98.50, 97.04, and 98.59%. TRM 70351T were transporter-related genes: ABC transporter ATP-binding protein and ABC transporter ATP-binding protein/permease. Given that TRM 70351T has the core genes for streptomycin synthesis, as well as the synthesis genes for related aminotransferases and other key enzymes, it is speculated that this strain is extremely Streptomycin or structurally similar compounds may be synthesized.
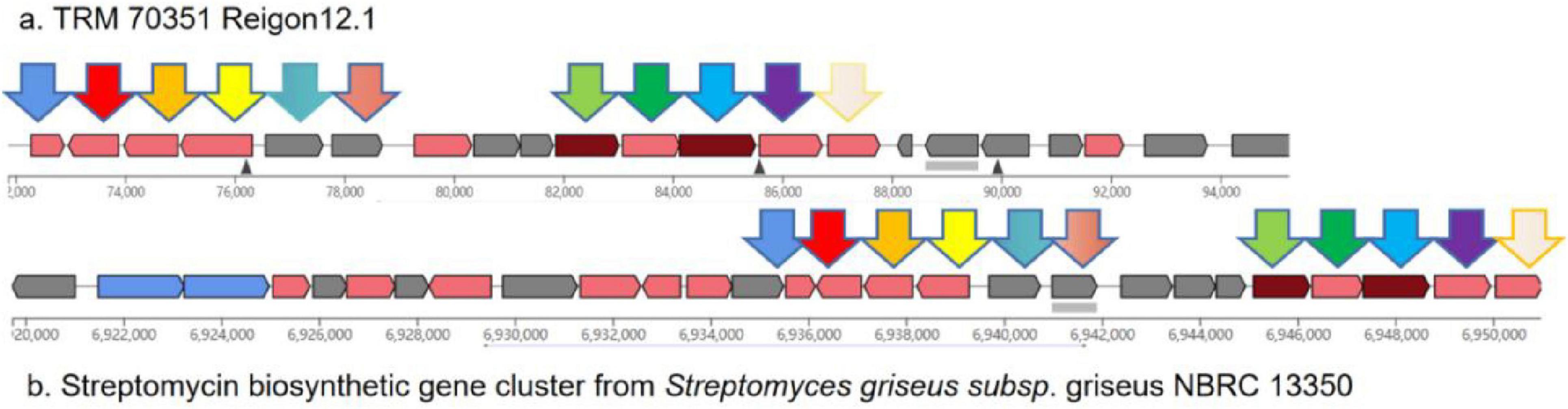
Figure 3. Comparison of gene cluster 12.1 predicted genes for TRM 70351T strain. (A) TRM 70351 Reigon12.1. (B) Streptomycin biosynthetic gene cluster from Streptomyces griseus subsp. Griseus NBRC 13350.
Selection of aminoglycoside fermentation media based on activity and spectral signals
Based on the antiSMASH prediction indicating the presence of the streptomycin biosynthetic gene cluster in the strain, four fermentation media were selected for HSQC-TOCSY analysis. Additional media were chosen based on activity screening results. The streptomycin fermentation samples showed significant activity against Staphylococcus aureus, Klebsiella pneumoniae, Salmonella, Acinetobacter baumannii, Pseudomonas amylovora, and Pseudomonas aeruginosa. In contrast, millet fermentation samples exhibited activity against Enterococcus faecalis, Escherichia coli, and Klebsiella pneumoniae. The ISP4 fermentation medium showed efficacy against Erwinia amylovora, while streptomycin fermentation medium No.2 (Li et al., 2013) demonstrated activity against Staphylococcus aureus, as illustrated in Supplementary Table S1 and Supplementary Figure S4. ISP4, known for its low-nutrient composition, did not exhibit aminoglycoside region signals post-fermentation, unlike the other three fermentation media samples. Considering both effectiveness and spectral signals, streptomycin fermentation medium No.1 (Han and Guirong, 2008) was chosen for the batch fermentation of TRM 70351T.
Detection and characterization of metabolites
The analysis of alkaline aqueous-phase extracts from the fermentation medium of Streptomycin No. 1 using the HSQC-TOCSY assay identified aminosugar signals in the region of δH 3.0-4.0 ppm andδC 45.0-55.0 ppm, as depicted in Figure 4. The HSQC-TOCSY region summary plots for the reference compounds, streptomycin sulfate and neomycin sulfate, are detailed in Figure 4. LC-MS/MS analysis was performed on standard and TRM 70351T samples to determine the molecular weights of meomycin (615.0) and streptomycin (582.0) (see Figure 5). The targeted detection of TRM 70351T revealed signals for neomycin (615.0, 160.9, and 293.0 m/z) and streptomycin (582.0, 263.0, and 246.0 m/z), confirming the presence of these aminoglycosides. In conclusion, it was estabilished that strain TRM 70351T harbors neomycin and streptomycin.
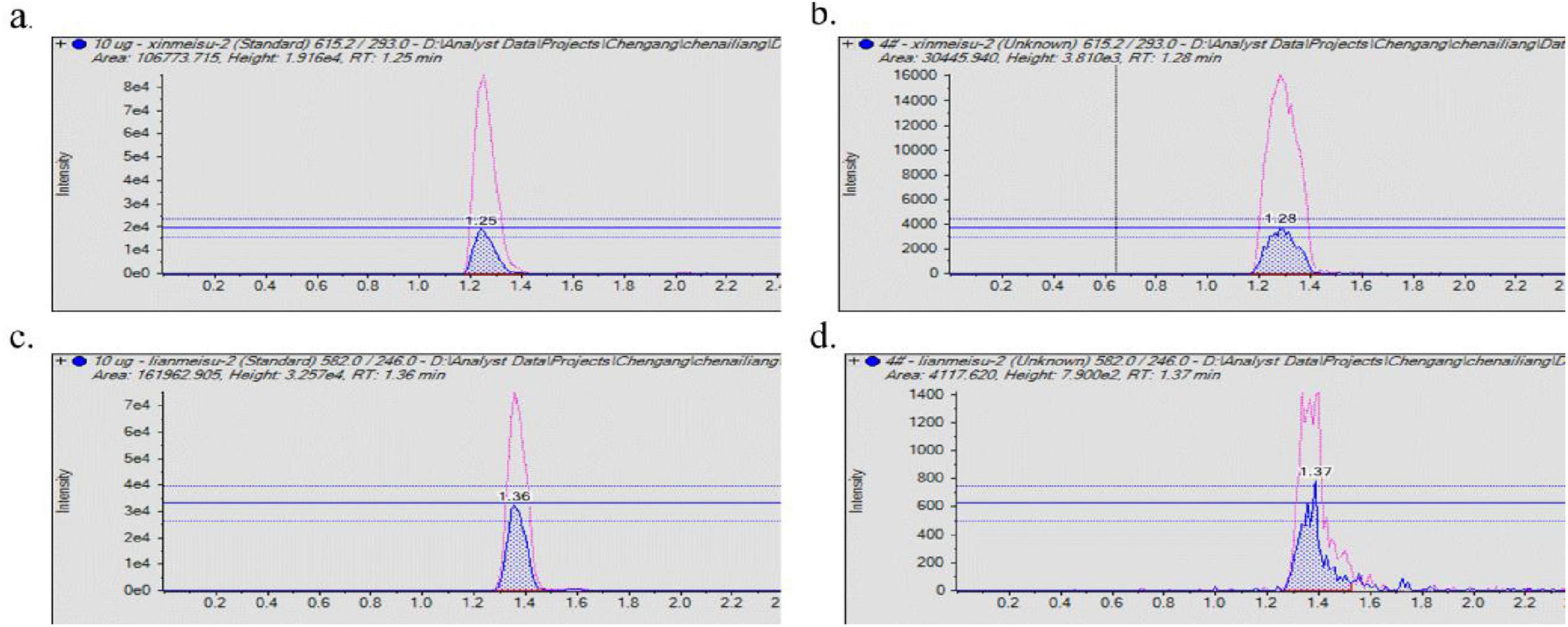
Figure 4. LC-MS/MS of alkaline aqueous extracts and standards. (A) Neomycin sulfate LC-MS/MS results. (B) TRM 70351 LC-MS/MS results. (C) Streptomycin sulfate LC-MS/MS results. (D) TRM 70351 LC-MS/MS results. The red peaks in the graph represent the response of quantitative ions and the blue peaks represent the response of qualitative ions.
Compounds 1 and 2 were isolated through conventional silica gel columns chromatography (Supplementary Figure S5), In this study, we mainly explored the large polar substances, so we chose methanol for the pre-extraction. Their structures were elucidated by NMR analysis and literature comparison. The pertinent data are presented belows. Compound 1, depicted by a hydrocarbon spectrum in Supplementary Figure S4, consists of light yellow crystals, molecular formula: C15H10O4. soluble in DMSO.1H NMR (500 MHz, DMSO-d6) δ 10.77 (s, 1H), 9.55 (s, 1H), 8.28 (s, 1H), 7.97 (d, J = 8.74 Hz, 1H), 7.39 (d, J = 8.43 Hz, 2H), 6.94 (dd, J = 8.76, 2.23 Hz, 1H), 6.86 (d, J = 2.23 Hz, 1H). = 8.43 Hz, 2H), 6.94 (dd, J = 8.76, 2.23 Hz, 1H), 6.86 (d, J = 2.23 Hz, 1H), 6.82 (d, J = 8.47 Hz, 2H). After conducting a literature review, the spectral data of this compound aligned perfectly with it, leading to the identification of compound as daidzein (Soidinsalo and Wähälä, 2004) (a soybean glycoside) with the structural formula depicted in Supplementary Figure S6.
Compound 2 is a colorless oil that exhibits solubility in petroleum ether, chloroform, and other similar solvents. Molecular formula C13H26. 1H-NMR (500 MHz, Chloroform-d)δ5.03-4.90 (m, 1H), 2.04 (q, J = 7.31, 7.17, 7.17 Hz, 1H), 1.37 (t, J = 7.32, 7.32 Hz, 1H), 1.30 (t, J = 12.28, 12.28 Hz, 1H), 1.28 (s, 4H), 1.26 (s, 9H), 1.21 (s, 1H), 1.18 (s, 3H), 0.91-0.78 (m, 4H). After literature review, the spectral data of the compound was in complete agreement with it, so the compound was identified as tridec-1-ene (Neipihoi et al., 2021) with the structural formula shown in Supplementary Figure S7.
Description of Streptomyces solitudinis sp. nov
Streptomyces solitudinis (solitudin’is N.L. masc. adj, the strain is a new species from the Taklamakan Desert in China, where desert means solitude).
Streptomyces solitudinis, a Gram-positive actinomycete, is an aerobic, mesophilic microorganism that forms straight or flexuous chains of spores with rough surfaces and lacks motility. It displays vigorous growth on ISP media 1, 2, 4, as well as Czapek’s and Gao’s medium. The species thrives optimally at temperatures ranging from 15 to 40°C and pH levels between 7 and 9. Growth is supported in the presence of 0 to 8% NaCl (w/v). It metabolizes D-Xylose, D-Glucose, and D-Mannose, while showing no utilization of D-Fructose or L-Rhamnose. Nitrate undergoes conversion to nitrite with gelatin liquefaction showing variability but stability starch hydrolysis demonstrating positivity, and urease production exhibiting negativity. The cell wall’s peptidoglycancomprises LL-diaminopimelic acid, while mannitol and two unidentified hydrolysates constitute the whole-cell sugars. The primary phospholipids consist of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, and phosphatidylinositol mannosides. The prevalent menaquinones include MK6, MK7, MK-10(H2), and MK-10(H4). Major fatty acids detected are C8:0 3OH, anteiso-C15:0, iso-C16:0, C16:0, and anteiso-C17:0.
The type strain for Streptomyces solitudinis sp. nov. is TRM 70351T ( = CCTCC AA 2020049T = LMG 32404T), was recovered from China.
Description of Streptomyces rubellus sp. nov
Streptomyces rubellus (rubellus N.L. masc. adj, the strain was red in color and resembled rubella in the electron micrographs, and was named Streptomyces rubellus based on the color morphology).
Streptomyces rubellus, a Gram-stain-positive actinomycete is an aerobic and mesophilic organism that is non-motile and lacks spores. It thrives on various media including ISP1, ISP2, ISP4, NA, and Gao’s medium. The species metabolizes D-Xylose exclusively, while showing no utilization D-Glucose, D-Mannose, D-Fructose, or L-Rhamnose. Nitrate is enzymatically reduced to nitrite. Optimal growth transpires within a temperature ranges of 28 to 45°C, and within a pH range of 6–7. Growth is sustained in the presence of NaCl concentrations ranging from 0 to 3% (w/v). The liquefaction of gelatin exhibits variablility but remains stable, whereas starch hydrolysis shows a positive response, however, urease production is negative. LL-diaminopimelic acid is detected in the cell wall, with glucose identified as the predominant whole-cell sugar. The primary phospholipids include phosphatidyl ethanolamine, phosphatidylinosito, and phosphotidylinositol mannosides. The main menaquinones present are MK6, MK6(H2), MK-8, MK-9(H8) and MK-10(H6). Predominant fatty acids consist of C8:0 3OH, anteiso-C15:0, 16:1 iso H, iso-C16:0 and anteiso-C17:0.
The type strain for Streptomyces rubellus sp. nov. is TRM 70361T( = CCTCC AA 2020043T = JCM 35793T), was recovered from China.
Discussion
In this investigation, two novel strains of Streptomyces species, namely TRM 70351T and TRM 70361T, were discovered in rhizosphere soil samples collected from Populus euphratica in the Taklimakan Desert, Xinjiang, China. Through contemporary taxonomic techniques, their genotypic, phenotypic, and chemical profiles were delineated, leading to their designation as Streptomyces solitudinis and Streptomyces rubella, correspondingly. Notably, the strain TRM 70351T exhibited the production of aminoglycoside secondary metabolites during fermentation.
The DNA G+C content of TRM 70351 is 73.69 mol%, whereas TRM 70361T exhibits a G+C content of 73.42%. Strains closely related to TRM 70351 include Streptomyces chumphonensis K1-2T (Yu et al., 2018), Streptomyces alkaliterrae OF1T (Świecimska et al., 2020), and Streptomyces durbertensis NEAU- S1GS20T (Yu et al., 2018). There strains were studied: K1-2T, collected from marine environments in Thu Phuong Province, Thailand; OF1T, isolated from alkaline soil near a meteoric alkaline soda lake in India; and NEAU- S1GS20T, obtained from saline soil in Heilongjiang Province, northeastern China. Related strains included Streptomyces barkulensis RC 1831T (Ray et al., 2014) and Streptomyces carminius TRM SA0054T (Wang et al., 2018). RC 1831T was isolated from sediments at a fish dumpsite near the brackish Chilika Lake in Balkur village, Kulda district, Orissa, India; while TRM SA0054T was sourced from various locations, all of which were bitter ginseng roots in Alar, Xinjiang. Despite their disparate origins, both strains were sourced from alkaline environments, indicating shared survival conditions that underpin their similarities (Smithburn, 1936).
Preliminary statistics indicate the discovery of 155 new natural products from streptomycetes isolated in extreme environments like the deep sea, deserts, volcanoes, and polar regions during the period from 2009 to 2020 (Duan et al., 2022). Streptomycetes are also known as the primary sources of aminoglycosides, exemplified by streptomycin from S. griseus (Rao, 1948) and chloramphenicol from S. hygroscopicus (Tabakov et al., 1994). The identification of new streptomycetes from desert environments holds significant research value. This study utilized Mass spectrometry to analyze strain TRM 70351T, confirming its production of aminoglycosides such as neomycin and streptomycin. Furthermore, besides the isolation tridec-1-ene and daidzein, both known for their beneficial effects in related research, the study also explored the mining of other compounds. Tridec-1-ene shows promise as a chemopreventive agent by potentially activating Nrf2/ARE-mediated phase II enzymes (Bhat et al., 2022). Additionally, daidzein demonstrates notable antimicrobial properties (Saeed Kotb et al., 2024).
Conclusion
In this investigation, two novel species of Streptomyces, namely strains TRM 70351T and TRM 70361T originating from the Taklamakan Desert, were characterized. The metabolic capacity for aminoglycoside compounds in strain TRM 70351 was explored, leading to the isolation of the isolation of the bioactive compounds tridec-1-ene and Daidzein using contemporary extraction methods. This discovery has significantly augmented the repository of strains for aminoglycoside exploration.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/, accession numbers PQ758399 and PQ758398; genome accession numbers PRJNA1068773 and PRJNA1068755.
Author contributions
J-xZ: Methodology, Resources, Validation, Writing – review & editing. Y-hC: Data curation, Writing – original draft, Funding acquisition. XL: Investigation, Resources, Writing – original draft. L-lZ: Writing – review & editing, Methodology, Supervision. X-xL: Data curation, Formal analysis, Writing – review & editing, Funding acquisition, Project administration, Supervision.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Tarim University President gene project “Preparation and application of phosphonic acid producing biocontrol agents” (TDZKZD202202), supported by Study on the biosynthesis mechanism and combination biosynthesis of indole oxazole antibiotics in actinomycetes (32360009), and Diversity of typical soil actinomycetes in different habitats of the Tarim River and screening of phosphonic acid producing strains (2022-XBQNXZ-019), and Tarim University Innovation Program (BTYJXM-2024-K30).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1530786/full#supplementary-material
References
Abdelkader, M. S. A., Philippon, T., Asenjo, J. A., Bull, A. T., Goodfellow, M., Ebel, R., et al. (2018). Asenjonamides A-C, antibacterial metabolites isolated from Streptomyces asenjonii strain KNN 42.f from an extreme-hyper arid Atacama desert soil. J. Antibiot. 71, 425–431. doi: 10.1038/s41429-017-0012-0
Becker, B., and Cooper, M. A. (2013). Aminoglycoside antibiotics in the 21st century. ACS Chem. Biol. 8, 105–115. doi: 10.1021/cb3005116
Berner, D. (2009). Correction of a bootstrap approach to testing for evolution along lines of least resistance. J. Evol. Biol. 22, 2563–2565. doi: 10.1111/j.1420-9101.2009.01869.x
Bhat, B. A., Mir, W. R., Sheikh, B. A., Rather, M. A., Dar, T. U. H., and Mir, M. A. (2022). In vitro and in silico evaluation of antimicrobial properties of Delphinium cashmerianum L., a medicinal herb growing in Kashmir. India. J. Ethnopharmacol. 291:115046. doi: 10.1016/j.jep.2022.115046
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). AntiSmash 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Collins, M., Goodfellow, M., and Minnikin, D. (1977). Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230. doi: 10.1099/00221287-100-2-221
Duan, Z., Liao, L., and Chen, B. (2022). Complete genome analysis reveals secondary metabolite biosynthetic capabilities of Streptomyces sp. R527F isolated from the Arctic Ocean. Mar. Genomics 63:100949. doi: 10.1016/j.margen.2022.100949
Ensign, J. C. (1978). Formation, properties, and germination of actinomycete spores. Annu. Rev. Microbiol. 32, 185–219. doi: 10.1146/annurev.mi.32.100178.001153
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Gao, Q., and Garcia-Pichel, F. (2011). Microbial ultraviolet sunscreens. Nat. Rev. Microbiol. 9, 791–802. doi: 10.1038/nrmicro2649
Gerhardt, P., Murray, R., Wood, W., Smibert, R., and Krieg, N. (1994). Methods for General and Molecular Bacteriology. Washington, DC: American Society for Microbiology.
Han, Y., and Guirong, J. (2008). Optimization of streptomycin-producing medium for Streptomyces griseus. Chinese Brewing 18, 32–34.
Hu, Z., Cheng, L., and Wang, H. (2015). The Illumina-solexa sequencing protocol for bacterial genomes. Methods Mol. Biol. 1231, 91–97. doi: 10.1007/978-1-4939-1720-4_6
Jackman, S. D., Vandervalk, B. P., Mohamadi, H., Chu, J., Yeo, S., Hammond, S. A., et al. (2017). ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27, 768–777. doi: 10.1101/gr.214346.116
Kelly, K. (1964). Inter-Society Color Council - National Bureau of Standards Color Name Charts Illustrated with Centroid Colors. Washington, DC: US Government Printing Office.
Kim, S. B., and Goodfellow, M. (1999). Reclassification of Amycolatopsis rugosa Lechevalier et al. 1986 as Prauserella rugosa gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49(Pt 2), 507–512. doi: 10.1099/00207713-49-2-507
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Komaki, H. (2022). Resolution of housekeeping gene sequences used in MLSA for the genus Streptomyces and reclassification of Streptomyces anthocyanicus and Streptomyces tricolor as heterotypic synonyms of Streptomyces violaceoruber. Int. J. Syst. Evol. Microbiol. 72:5370. doi: 10.1099/ijsem.0.005370
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, H. Y., Liu, L., and Wang, S. Y. (2013). Optimization of streptomycin-producing fermentation medium of Streptomyces griseus. Sci. Technol. Eng. 8202–8205.
Li, J., Li, P. F., Wang, X. W., Yin, P., Yen, D. D., Shao, L., et al. (2017). Application of nuclear magnetic resonance technique in the structural study of aminoglycoside antibiotics. J. Changzhou Univer. 1, 50–55.
Li, L. Y., Yang, Z. W., Asem, M. D., Fang, B. Z., Salam, N., Alkhalifah, D. H. M., et al. (2019). Streptomyces desertarenae sp. nov., a novel actinobacterium isolated from a desert sample. Antonie van Leeuwenhoek 112, 367–374. doi: 10.1007/s10482-018-1163-0
Li, Z., Guo, Y., Lin, H., Meng, C., and Shi, X. (2002). A new process for isolation and extraction of kanamycin from fermentation broth. Chin. J. Antibiotics 6, 328–331.
Ma, F. Y., Cai, Y. Q., Huang, X. W., Wu, C. B., Yang, K., and Li, L. B. (2020). Determination of nine aminoglycoside residues in honey by ultra performance liquid chromatography-tandem mass spectrometry. Food Fermentation Industry 18, 203–208. doi: 10.13995/j.cnki.11-1802/ts.023958
Makhalanyane, T. P., Valverde, A., Lacap, D. C., Pointing, S. B., Tuffin, M. I., and Cowan, D. A. (2013). Evidence of species recruitment and development of hot desert hypolithic communities. Environ. Microbiol. Rep. 5, 219–224. doi: 10.1111/1758-2229.12003
McCarthy, A. J., and Williams, S. T. (1992). Actinomycetes as agents of biodegradation in the environment–A review. Gene 115, 189–192. doi: 10.1016/0378-1119(92)90558-7
Mount, D. W. (2008). Maximum parsimony method for phylogenetic prediction. CSH Protoc. doi: 10.1101/pdb.top32
Neilson, J. W., Quade, J., Ortiz, M., Nelson, W. M., Legatzki, A., Tian, F., et al. (2012). Life at the hyperarid margin: Novel bacterial diversity in arid soils of the Atacama desert. Chile. Extremophiles 16, 553–566. doi: 10.1007/s00792-012-0454-z
Neipihoi, Y., Narzary, B., Saikia, S., Saikia, S., Tamuli, K. J., Sahoo, R. K., et al. (2021). Anticancer and antimicrobial compounds from Croton caudatus Gieseler and Eurya acuminata DC: Two edible plants used in the traditional medicine of the Kuki tribes. Nat. Prod. Res. 35, 6025–6029. doi: 10.1080/14786419.2020.1815737
Prado-Alonso, L., Ye, S., Pérez-Victoria, I., Montero, I., Riesco, P., Ortiz-López, F. J., et al. (2024). Genome mining for Diazo-synthesis-related genes in Streptomyces sp. CS057 unveiled the cryptic biosynthetic gene Cluster crx for the novel 3,4-AHBA-derived compound Crexazone 2. Biomolecules 14:1084. doi: 10.3390/biom14091084
Pridham, T., and Gottlieb, D. (1948). The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J. Bacteriol. 56, 107–114. doi: 10.1128/jb.56.1.107-114.1948
Rao, R. R. (1948). Utilization of wheat bran for streptomycin production by S. griseus. Nature 162:820. doi: 10.1038/162820a0
Ray, L., Mishra, S. R., Panda, A. N., Rastogi, G., Pattanaik, A. K., Adhya, T. K., et al. (2014). Streptomyces barkulensis sp. nov., isolated from an estuarine lake. Int. J. Syst. Evol. Microbiol. 64(Pt 4), 1365–1372. doi: 10.1099/ijs.0.056614-0
Riesco, R., and Trujillo, M. E. (2024). Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 74:006300. doi: 10.1099/ijsem.0.006300
Saeed Kotb, S., Ayoub, I. M., El-Moghazy, S. A., and Singab, A. N. B. (2024). Phytochemical analysis of Pithecellobium dulce (Roxb) Benth Bark via UPLC-ESI-MS/MS and evaluation of its biological activity. Nat. Prod. Res. 38, 1424–1429. doi: 10.1080/14786419.2022.2140153
Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shirling, E., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340.
Smithburn, K. C. (1936). The colony morphology of tubercle bacilli : V. influence of the Ph of the culture medium on colony form. J. Exp. Mede 63, 95–108. doi: 10.1084/jem.63.1.95
Soidinsalo, O., and Wähälä, K. (2004). Synthesis of phytoestrogenic isoflavonoid disulfates. Steroids 69, 613–616. doi: 10.1016/j.steroids.2004.03.015
Świecimska, M., Golińska, P., Nouioui, I., Wypij, M., Rai, M., Sangal, V., et al. (2020). Streptomyces alkaliterrae sp. nov., isolated from an alkaline soil, and emended descriptions of Streptomyces alkaliphilus. Streptomyces calidiresistens and Streptomyces durbertensis. Syst. Appl. Microbiol. 43: 126153. doi: 10.1016/j.syapm.2020.126153
Tabakov, V. I., Voeikova, T. A., Tokmakova, I. L., Bolotin, A. P., Vavilova, E. I., and Lomovskaia, D. (1994). Mezhrodovaia kon”iugatsiia Escherichia coli-Streptomyces kak sposob peredachi kon”iugativnykh plazmid v produtsenty antibiotikov khlortetratsiklina i bialafosa [Intergeneric Escherichia coli-Streptomyces conjugation as a means for the transfer of conjugative plasmids into producers of antibiotics chlortetracycline and bialaphos]. Genetika 30, 57–61.
Wang, Y., Xia, Z., Liu, Z., Wan, C., Luo, X., and Zhang, L. (2018). Streptomyces carminius sp. nov., a novel actinomycete isolated from Sophora alopecuroides in Xinjiang. China. Antonie van Leeuwenhoek. 111, 1807–1814. doi: 10.1007/s10482-018-1069-x
Xie, F., and Pathom-Aree, W. (2021). Actinobacteria from desert: Diversity and biotechnological applications. Front. Microbiol. 12:765531. doi: 10.3389/fmicb.2021.765531
Xie, J., Talaska, A. E., and Schacht, J. (2011). New developments in aminoglycoside therapy and ototoxicity. Hear. Res. 281, 28–37. doi: 10.1016/j.heares.2011.05.008
Yang, L. H., Hu, C. Q., and Liu, W. Y. (2006). Mass spectrometry analysis of 14 aminoglycoside antibiotics. J. Pharm. Anal. 8, 1050–1057.
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Yu, Y., Fu, Y., Guo, X., Yan, R., Wang, H., Zhao, J., et al. (2018). Streptomyces durbertensis sp. nov., isolated from saline-alkali soil. Int. J. Syst. Evol. Microbiol. 68, 3635–3640. doi: 10.1099/ijsem.0.003047
Keywords: Taklimakan Desert, Streptomyces solitudinis sp. nov, genome analysis, Streptomyces rubellus sp. nov, aminoglycosides
Citation: Zhang J-x, Chen Y-h, Luo X, Zhang L-l and Luo X-x (2025) Isolated from Populus euphratica rhizosphere soil, and mining their metabolites. Front. Microbiol. 16:1530786. doi: 10.3389/fmicb.2025.1530786
Received: 19 November 2024; Accepted: 09 January 2025;
Published: 19 February 2025.
Edited by:
Sandra Gabriela Gomez De Saravia, National University of La Plata, ArgentinaReviewed by:
D. Ipek Kurtböke, University of the Sunshine Coast, AustraliaCristina Galisteo, University of León, Spain
Copyright © 2025 Zhang, Chen, Luo, Zhang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxia Luo, eHhsdW80MTVAMTYzLmNvbQ==; Li-li Zhang, NDUxMDE2ODM3QHFxLmNvbQ==
†These authors share first authorship
 Jia-xin Zhang1,2†
Jia-xin Zhang1,2† Yi-huang Chen
Yi-huang Chen