- 1Department of Pharmacy, Faculty of Mathematics and Natural Sciences, Universitas Islam Indonesia, Yogyakarta, Indonesia
- 2Department of Pharmacy, Sekolah Tinggi Ilmu Kesehatan Insan Sarjana Farmasi Indonesia (ISFI) Banjarmasin, Banjarmasin, Indonesia
- 3Department of Chemistry, Faculty of Mathematics and Natural Sciences, Lambung Mangkurat University, Banjarbaru, Indonesia
- 4Prodi Magister Kimia, Fakultas Matematika dan Ilmu Pengetahuan Alam (MIPA) Universitas Lambung Mangkurat, Banjarbaru, Indonesia
1 Introduction
Antimicrobial resistance (AMR) poses a critical global health threat, complicating infection management worldwide. Data on the prevalence of antibiotic resistance released by the World Health Organization (WHO) in 2019 has caused the deaths of 1.27 million people (Murray et al., 2022; WHO, 2023). Additionally, the World Bank estimates that the economic impact of AMR could reach a loss of up to US$ 1 trillion in healthcare costs by 2050 and a gross domestic product (GDP) loss of US$ 3.4 trillion by 2030 (Jonas et al., 2017).
The urgent need to discover new drugs to replace resistant antibiotics has become increasingly critical. One of the largest sources of new antibiotic producers comes from the soil, which harbors 99% of microbial species. Antimicrobial compounds are produced by microbes in soil that often remain unculturable in the laboratory due to the limitations of traditional cultivation techniques, which fail to replicate the microbes' natural habitats (Choi et al., 2015; Bhattacharjee, 2022). The type of soil that has great potential for obtaining new antibiotic agents is peat soil (Kujala et al., 2018; Liu et al., 2022; Atapattu et al., 2023). Peat soil contains organic deposits rich in nutrients that support microbial growth and diversity (Nawan and Wasito, 2020).
The abundant microbial content in peat soil needs to be effectively harnessed to develop new antibiotics. Current microbial cultivation techniques are generally limited to only a subset of microbes, restricting the isolation of secondary metabolites. Overcoming these limitations requires innovative approaches to cultivate antibiotic-producing microbes that remain unculturable under laboratory conditions. Uncultured Soil Technology (UST) or in situ incubation is one of the latest developments, which involves cultivation using natural growth factors present in the environment (Berdy et al., 2017; Chaudhary et al., 2019).
A prominent in situ incubation method is the isolation chip (iChip) technique, developed by D. Nichols in 2010, which led to his first discovery. This method enabled the discovery of Teixobactin, a groundbreaking antibiotic (Berdy et al., 2017; Zhao et al., 2023). This article reviews several relevant articles published between 2015 and 2024 to address the question, “How is the discovery of new antibiotics from peat soil using the iChip method?” This Opinion Article aims to provide a critical discussion, highlighting the opportunities and challenges in harnessing the potential application of iChip technology for the discovery of novel antibiotics from peat soil microbiomes. This article is intended to encourage original experimental studies focusing on peat soil, which holds significant potential.
2 Sample preparation
2.1 Sample collection
Peatlands, which encompass the most extensive total area in the world, are predominantly found on the Asian continent, accounting for 38.4% of the world's peatlands. The regions in Asia with the largest peatland areas are Asian Russia, Indonesia, and Malaysia. Following Asia, other regions that have large amounts of peatland after the Asian zone are North America (31.6%), Europe (12.5%), South America (11.5%), Africa (4.4%), and Australasia and Oceania (1.6%) (Xu et al., 2018).
Previous studies have discussed the optimal sampling depth for producing microbes. Sampling in research using the culture isolation method taken at 0 to 15 cm depth can isolate Actinomycetes microbes (Atapattu et al., 2023). Samples taken at a depth of 50 cm produced Enterobacteriaceae (46.4%), Bacillaceae (28.6%), Streptococcaceae (10.7%), Staphylococcaceae (10.7%), and Clostridiaceae (3.6%) microbes (Mahdiyah et al., 2020).
Peat soil characteristics, such as acidic pH (3.5 to 4.1) (Nawan and Wasito, 2020; Goh et al., 2022; Atapattu et al., 2023), temperature (tropical climate with an average temperature of 28°C), nutrient availability, and substrate composition, influence microbial abundance and diversity, are located in lowlands, and are inundated due to excessive rainfall (average rainfall 200 cm3 per year) (Nawan and Wasito, 2020; Paul et al., 2021; Goh et al., 2022). In addition, peat soil has waterlogged conditions and low nutrient availability, which influence the characteristics of the secondary metabolites produced (Weeraphan et al., 2023).
2.2 Sample preparation
Peat soil samples were collected from a depth of 0–50 cm. Sampling was conducted beneath trees, with an emphasis on root systems to enhance microbial diversity. When feasible, the in situ incubation process was initiated directly at the sampling site. If sample transportation to another location was required, the samples were preserved at 4°C in darkness until the in situ incubation process could proceed. This cooling step is essential for maintaining sample integrity by preserving their chemical and biological properties and preventing alterations caused by external factors such as light, elevated temperatures, or desiccation. This methodology aims to create a controlled environment that replicates the microbes' natural habitat, thereby promoting their survival and metabolic activity during incubation (Preston and Basiliko, 2016; Ong et al., 2020; Tate, 2021; Goh et al., 2022; Polrot et al., 2022).
3 Isolation chip (iChip) technology for isolation peat soil
3.1 Environmental condition: collection and preparation
The sample underwent serial dilutions before being intervened in an agar medium. The agar medium that can be used is Tryptic Soy Agar (TSA). Agar plates were then incubated for 3–5 days at room temperature. After incubation, the number of colonies was counted to determine the optimal dilution amount before inoculating a single bacterial cell into the diffusion chamber (Polrot et al., 2022). The minimum value of CFU/g peat soil in previous studies was log 5.7, and the maximum value was 8.0, so it can be used as a reference in carrying out serial dilutions (Ramata-Stunda et al., 2015; Glushakova et al., 2021). This step is carried out in the laboratory to determine the abundance of bacteria outside their natural environmental conditions.
The iChip design comprises several parts developed by Berdy et al. (2017). The parts of the iChip include (1) a middle plate containing through-holes, (2) semipermeable membranes affixed to both the front and back of the middle plate to separate the plate from the environment, and (3) a membrane seal designed as a panel board, to reinforce the structure (Berdy et al., 2017).
All iChip components must be sterilized prior to assembly and use in the incubation process. Sterilization is performed by soaking the components in 70% ethanol for 15 min, followed by air drying at room temperature until completely dry. Additionally, a sediment bucket filled with peat soil is prepared for the incubation of the iChip device (Polrot et al., 2022).
3.2 In situ incubation: iChip
The iChip technology consists of hundreds of diffusion chambers containing an agarose medium or other media, such as molten SMS medium (0.125 g casein, 0.1 g potato starch, 1 g casamino acids, 20 g bacto-agar, dissolved in 1 liter of water). This medium is designed to support bacterial growth during the incubation process for Grasses and sediment samples, which can be applied to peat soil samples, but further experiments are needed (Ling et al., 2015; Polrot et al., 2022). Another medium that can be used and has been proven in previous studies for peat soil samples is TSA, with a concentration of 10% (according to the acidic conditions of peat soil) (Liu H. et al., 2021; Goh et al., 2022).
The initial step of in situ incubation involves immersing the iChip device in the prepared medium, which has been supplemented with samples at the optimal concentration, as determined in Section 3.1. This procedure ensures the sample's distribution into the through-holes on the iChip plate. After immersion, the agarose medium containing the sample is allowed to solidify.
Subsequently, the diffusion chamber is sealed by placing a plate directly over the through-holes and applying petroleum jelly around the edges to secure the sample's position within the chambers during the incubation period. Once prepared, the iChip device is wrapped with a parafilm protector and placed in a sediment bucket containing peat soil to begin the incubation process. During incubation, the diffusion chambers are positioned as close as possible to their natural environmental conditions. This setup is achieved by separating the sample from the external environment using a semipermeable membrane, which allows for nutrient and waste exchange while maintaining microbial viability. The incubation process lasts for 7 days (Polrot et al., 2022).
3.3 Isolation of secondary metabolite screening for activity
The iChip device that has completed the incubation process is continued to the microbial isolation stage. The iChip is rinsed with sterile distilled water and subsequently incubated at 20°C for several weeks under room temperature conditions and in the dark (Polrot et al., 2022). The grown isolates were cultured in seed broth containing 15 g glucose, 10 g malt extract, 10 g soluble starch, 2.5 g yeast extract, 5 g casamino acids, and 0.2 g CaCl2•2H2O per 1 liter of deionized H2O with pH adjustment of 7.0. The cultures were diluted 1:20 into four different types of fermentation media (Ling et al., 2015; Quigley et al., 2020).
The fermentation process is carried out over 11 days at 29°C with continuous stirring. Following fermentation, the culture was dried and dissolved in 100% dimethyl sulfoxide (DMSO). The resulting extract was tested for antimicrobial activity against Staphylococcus aureus using Mueller-Hinton Agar (MHA) plates, which are incubated for 20 h at 37°C. Antimicrobial activity was assessed by observing the presence of a clear inhibition zone around the test area (Ling et al., 2015).
The fermentation and purification processes are based on the methodology for producing the secondary metabolite teixobactin, as outlined by Ling et al. (2015). Large-scale production employs the Sartorius Biostat Cultibag STR 50/200 bioreactor. Subsequently, extraction and purification are conducted using specific solvents and methods tailored to the metabolite being isolated (Ling et al., 2015).
4 Secondary metabolite
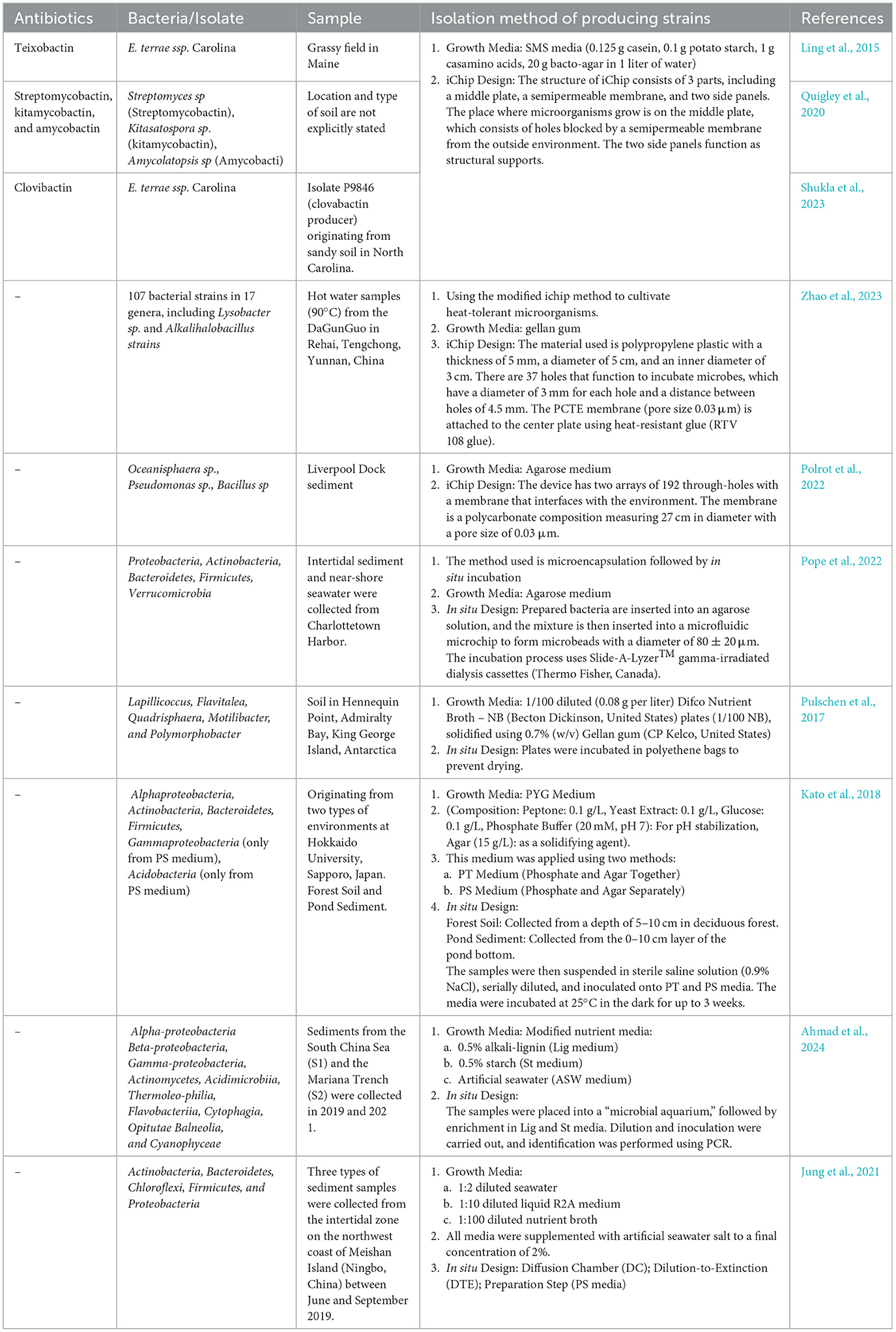
Secondary metabolites produced by the genus Streptomyces obtained from peat soil of the Nong Jum Rung peat swamp forest (Rayong Province, Thailand) include geldanamycin, 17-O-demethylgeldanamycin, reblastatin, 17-demethoxyreblastatin, nocardamine, and dehydroxynocardamine (Weeraphan et al., 2023). Secondary metabolites produced using in situ incubation methods and similar methods can be seen in Table 1.
5 Comparative analysis in microbial isolation
The application of iChip technology in studies involving marine water columns and soil microorganisms has demonstrated superior results compared to traditional cultivation methods using standard Petri dishes. Comparison of tests resulted in several findings, including (1) Increased microbial recovery: the recovery of colonies was five times higher with the iChip method, with 40%−50% of cells incubated in iChips forming microcolonies or 5 to 300 times more compared to traditional methods (Berdy et al., 2017), (2) access to novel microorganisms and reduced cultivation bias: the iChip method yielded a much higher level of phylogenetic uniqueness, increasing the richness and uniformity of isolates that are representative microorganisms of the microbial community is biased (Berdy et al., 2017; Lodhi et al., 2018; Liu X. et al., 2021; Liu H. et al., 2021), and (3) the iChip has been shown to produce new antibiotics such as Teixobactin and Clovabactin, which is produced by a previously unknown soil bacteria (tentatively named Eleftheria terrae), and N-Acyltyrosine from Alteromonas sp. (Ling et al., 2015; Sherpa et al., 2015; MacIntyre et al., 2019; Shukla et al., 2023). The flow of the in situ incubation method using iChip on peat soil can be seen in Figure 1.
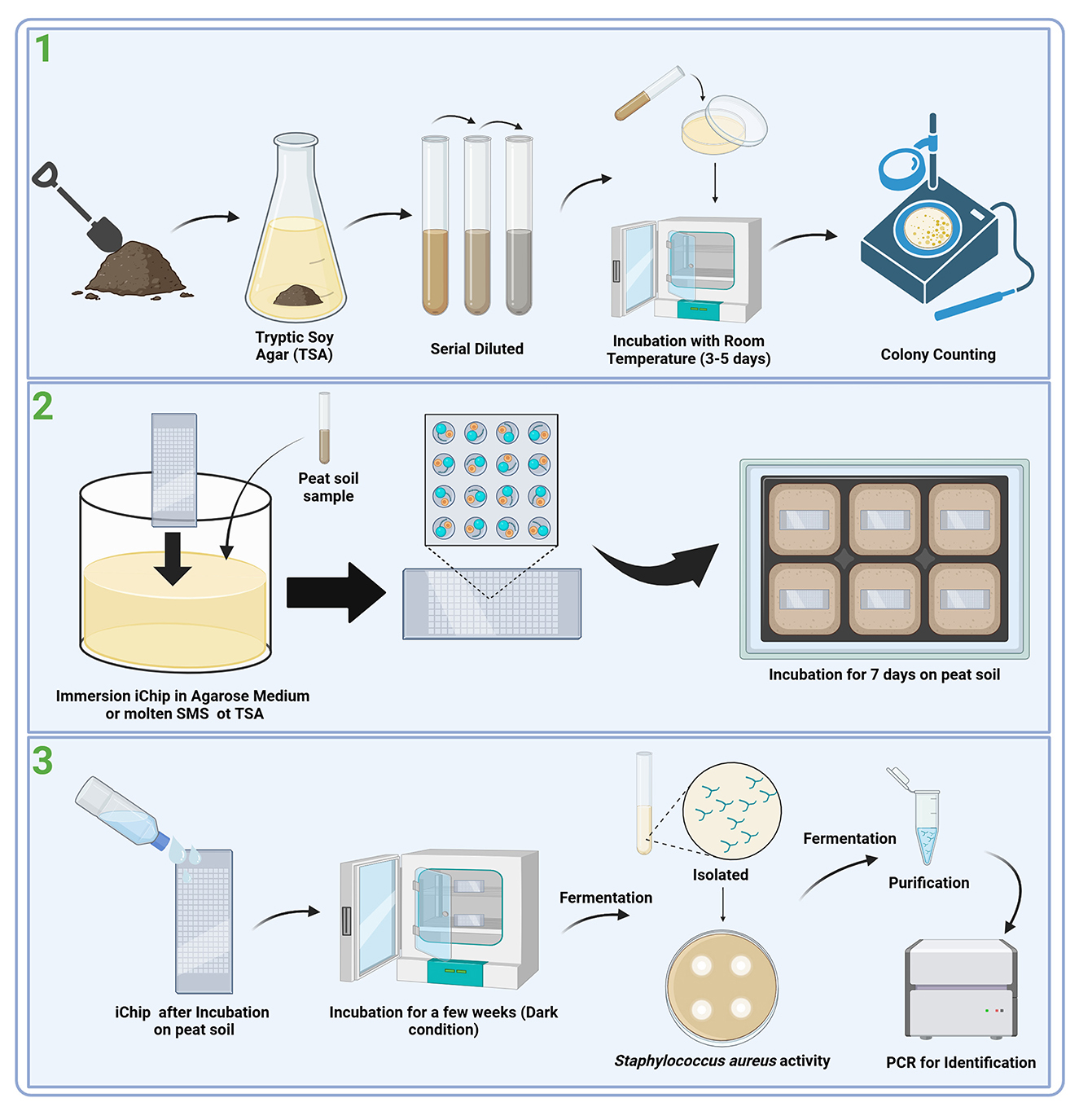
Figure 1. Workflow for the discovery of novel antibiotic agents from peat soil (Modolon et al., 2023; Zhang and Zhang, 2023; Ahmad et al., 2024). (1) Environmental condition: Collection and preparation, (2) In situ incubation: iChip, and (3) Isolation of secondary metabolite screening for activity.
6 Conclusion
The in situ incubation method represents an innovative approach to discovering new antibiotic agents to address the growing issue of drug resistance. One notable application of this method is the iChip technology, which has been employed across various soil types. Among these, peat soil stands out as a highly promising source of secondary metabolites due to the rich microbial diversity it harbors, yet it remains largely unexplored.
Author contributions
LC: Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft. TR: Conceptualization, Methodology, Software, Writing – original draft, Writing – review & editing. MF: Visualization, Writing – review & editing. EK: Formal analysis, Writing – review & editing. DL: Validation, Writing – review & editing. SA: Resources, Writing – review & editing. NN: Data curation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding provided by the Hibah Penelitian Unggulan, Directorate of Research and Community Service, Universitas Islam Indonesia, Grant No. 001/Dir/DPPM/70/Pen.Unggulan/III/2022, Department of Pharmacy, Faculty of Mathematics and Natural Sciences, Universitas Islam Indonesia, Yogyakarta, Indonesia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Gen AI was used in the creation of this manuscript. The author(s) verify and take full responsibility for the use of generative AI in the preparation of this manuscript. Generative AI was used to enhance the grammatical accuracy and improve the academic language of the manuscript using Grammarly. No content, data, or scientific interpretation was generated or altered by the AI tools, and the author(s) take full responsibility for the integrity and originality of the work presented.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, T., Ishaq, S. E., Liang, L., Xie, R., Wang, Y., and Wang, F. (2024). A diffusion-based integrative approach for culturing previously uncultured bacteria from marine sediments. Mar. Life Sci. Technol. doi: 10.1007/s42995-024-00240-2
Atapattu, G., Obeng, S. A., Battersby, T., Giltrap, M., and Tian, F. (2023). Effect of ‘peatland-use' type on culturable microbial groups in irish peatlands in the midlands. Land 12:1614. doi: 10.3390/land12081614
Berdy, B., Spoering, A. L., Ling, L. L., and Epstein, S. S. (2017). In situ cultivation of previously uncultivable microorganisms using the ichip. Nat. Protoc. 12, 2232–2242. doi: 10.1038/nprot.2017.074
Bhattacharjee, M. K. (2022). Chemistry of Antibiotics and Related Drugs. New York: Springer International Publishing. doi: 10.1007/978-3-031-07582-7
Chaudhary, D. K., Khulan, A., and Kim, J. (2019). Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 9:6666. doi: 10.1038/s41598-019-43182-x
Choi, E. J., Nam, S. J., Paul, L., Beatty, D., Kauffman, C. A., Jensen, P. R., et al. (2015). Previously uncultured marine bacteria linked to novel alkaloid production. Chem. Biol. 22, 1270–1279. doi: 10.1016/j.chembiol.2015.07.014
Glushakova, A. M., Lysak, L. V., Kachalkin, A. V., Ivanova, A. E., Umarova, A. B., Abramyan, I. A., et al. (2021). Transformation of microbial complexes in components of soil constructions of different origin (soil, peat, sand) during freezing-thawing processes. Microbiology 90, 176–186. doi: 10.1134/S002626172102003X
Goh, C. B. S., Goh, C. H. P., Wong, L. W., Cheng, W. T., Yule, C. M., Ong, K. S., et al. (2022). A three-dimensional (3D) printing approach to fabricate an isolation chip for high throughput in situ cultivation of environmental microbes. Lab. Chip. 22, 387–402. doi: 10.1039/D1LC00723H
Jonas, O. B., Irwin, A., Berthe, F. C. J., Le Gall, F. G., and Marquez, P. V. (2017). Drug-Resistant Infections : A Threat to Our Economic Future (Vol. 2): Final Report (English). Washington, D.C. Available at: http://documents.worldbank.org/curated/en/323311493396993758/final-report (accessed November 4, 2024).
Jung, D., Liu, B., He, X., Owen, J. S., Liu, L., Yuan, Y., et al. (2021). Accessing previously uncultured marine microbial resources by a combination of alternative cultivation methods. Microb. Biotechnol. 14, 1148–1158. doi: 10.1111/1751-7915.13782
Kato, S., Yamagishi, A., Daimon, S., Kawasaki, K., Tamaki, H., Kitagawa, W., et al. (2018). Isolation of previously uncultured slow-growing bacteria by using a simple modification in the preparation of agar media. Appl. Environ. Microbiol. 84:e00807–18. doi: 10.1128/AEM.00807-18
Kujala, K., Mikkonen, A., Saravesi, K., Ronkanen, A. K., and Tiirola, M. (2018). Microbial diversity along a gradient in peatlands treating mining-affected waters. FEMS Microbiol. Ecol. 94:fiy145. doi: 10.1093/femsec/fiy145
Ling, L. L., Schneider, T., Peoples, A. J., Spoering, A. L., Engels, I., Conlon, B. P., et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. doi: 10.1038/nature14098
Liu, H., Xue, R., Wang, Y., Stirling, E., Ye, S., Xu, J., et al. (2021). FACS-iChip: a high-efficiency iChip system for microbial ‘dark matter' mining. Mar. Life Sci. Technol. 3, 162–168. doi: 10.1007/s42995-020-00067-7
Liu, L., Wang, Z., Ma, D., Zhang, M., and Fu, L. (2022). Diversity and distribution characteristics of soil microbes across forest–peatland ecotones in the permafrost regions. Int. J. Environ. Res. Public Health 19:14782. doi: 10.3390/ijerph192214782
Liu, X., Wang, M., Nie, Y., and Wu, X. (2021). Isolation chip increases culturable bacterial diversity and reduces cultivation bias. Curr. Microbiol. 78, 2025–2032. doi: 10.1007/s00284-021-02474-0
Lodhi, A. F., Zhang, Y., Adil, M., and Deng, Y. (2018). Antibiotic discovery: combining isolation chip (iChip) technology and co-culture technique. Appl. Microbiol. Biotechnol. 102, 7333–7341. doi: 10.1007/s00253-018-9193-0
MacIntyre, L. W., Charles, M. J., Haltli, B. A., Marchbank, D. H., and Kerr, R. G. (2019). An iChip-domesticated sponge bacterium produces an N-acyltyrosine bearing an α-methyl substituent. Org. Lett. 21, 7768–7771. doi: 10.1021/acs.orglett.9b02710
Mahdiyah, D., Farida, H., Riwanto, I., Mustofa, M., Wahjono, H., Laksana Nugroho, T., et al. (2020). Screening of Indonesian peat soil bacteria producing antimicrobial compounds. Saudi J. Biol. Sci. 27, 2604–2611. doi: 10.1016/j.sjbs.2020.05.033
Modolon, F., Schultz, J., Duarte, G., Vilela, C. L. S., Thomas, T., and Peixoto, R. S. (2023). In situ devices can culture the microbial dark matter of corals. iScience 26:108374. doi: 10.1016/j.isci.2023.108374
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Nawan, I., and Wasito, E. B. (2020). Antimicrobial activity of streptomyces sp. Isolated from acidic peatlands against extended spectrum beta lactamase (ESBL) producing escherichia coli. Res. J. Pharm. Technol. 13, 1121–1126. doi: 10.5958/0974-360X.2020.00206.1
Ong, K. S., Letchumanan, V., Law, J. W. F., Yule, C. M., and Lee, S. M. (2020). Microbes from peat swamp forest — the hidden reservoir for secondary metabolites? Progr. Micr. Molec. Biol. 3:77. doi: 10.36877/pmmb.a0000077
Paul, A., Hussain, M., and Ramu, B. (2021). The physicochemical properties and microstructural characteristics of peat and their correlations: reappraisal. Int. J. Geotechn. Eng. 15, 692–703. doi: 10.1080/19386362.2018.1483099
Polrot, A., Kirby, J. R., Olorunniji, F. J., Birkett, J. W., and Sharples, G. P. (2022). iChip increases the success of cultivation of TBT-resistant and TBT-degrading bacteria from estuarine sediment. World J. Microbiol. Biotechnol. 38:180. doi: 10.1007/s11274-022-03297-2
Pope, E., Cartmell, C., Haltli, B., Ahmadi, A., and Kerr, R. G. (2022). Microencapsulation and in situ incubation methodology for the cultivation of marine bacteria. Front. Microbiol. 13:958660. doi: 10.3389/fmicb.2022.958660
Preston, M. D., and Basiliko, N. (2016). Carbon mineralization in peatlands: does the soil microbial community composition matter? Geomicrobiol. J. 33, 151–162. doi: 10.1080/01490451.2014.999293
Pulschen, A. A., Bendia, A. G., Fricker, A. D., Pellizari, V. H., Galante, D., and Rodrigues, F. (2017). Isolation of uncultured bacteria from antarctica using long incubation periods and low nutritional media. Front. Microbiol. 8:1346. doi: 10.3389/fmicb.2017.01346
Quigley, J., Peoples, A., Sarybaeva, A., Hughes, D., Ghiglieri, M., Achorn, C., et al. (2020). Novel antimicrobials from uncultured bacteria acting against mycobacterium tuberculosis. MBio 11, 1–13. doi: 10.1128/mBio.01516-20
Ramata-Stunda, A., Petrina, Z., Mekss, P., Kizane, G., Silamikele, B., Muiznieks, I., et al. (2015). Microbiological characterization and sterilization-induced changes in the profile of the hydrophobic organic substances in Latvian balneological peat. Int. J. Environ. Sci. Technol. 12, 2371–2380. doi: 10.1007/s13762-014-0638-4
Sherpa, R. T., Reese, C. J., and Aliabadi, H. M. (2015). Application of iChip to grow “uncultivable” microorganisms and its impact on antibiotic discovery. J. Pharm. Pharm. Sci. 18, 303–15. doi: 10.18433/J30894
Shukla, R., Peoples, A. J., Ludwig, K. C., Maity, S., Derks, M. G. N., De Benedetti, S., et al. (2023). An antibiotic from an uncultured bacterium binds to an immutable target. Cell 186, 4059–4073.e27. doi: 10.1016/j.cell.2023.07.038
Weeraphan, T., Supong, K., Sripreechasak, P., Jutakanoke, R., Kowinthanaphat, S., Tanasupawat, S., et al. (2023). Streptomyces rugosispiralis sp. nov., a novel actinobacterium isolated from peat swamp forest soil that produces ansamycin derivatives and nocardamines. Antibiotics 12:1467. doi: 10.3390/antibiotics12091467
WHO (2023). Antimicrobial resistance. Available at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed November 4, 2024).
Xu, J., Morris, P. J., Liu, J., and Holden, J. (2018). PEATMAP: Refining estimates of global peatland distribution based on a meta-analysis. Catena 160, 134–140. doi: 10.1016/j.catena.2017.09.010
Zhang, Y., and Zhang, T. (2023). Culturing the uncultured microbial majority in activated sludge: a critical review. Crit. Rev. Environ. Sci. Technol. 53, 601–624. doi: 10.1080/10643389.2022.2077063
Keywords: antimicrobial resistance, microorganisms, peatlands, microbial cultivation techniques, co-culture, in situ culturing, uncultured microorganisms, environmental microbes
Citation: Chabib L, Rustandi T, Fawwazi MHAF, Kumalasari E, Lestari DA, Amalia SP and Normilawati N (2025) Harnessing iChip technology for novel antibiotic discovery from peat soil microbiomes to combat antimicrobial resistance. Front. Microbiol. 16:1530273. doi: 10.3389/fmicb.2025.1530273
Received: 18 November 2024; Accepted: 05 February 2025;
Published: 21 February 2025.
Edited by:
Piotr Majewski, Medical University of Bialystok, PolandReviewed by:
Yuana Nurulita, Riau University, IndonesiaCopyright © 2025 Chabib, Rustandi, Fawwazi, Kumalasari, Lestari, Amalia and Normilawati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tedi Rustandi, dGVkaXJ1c3RhbmRpMjZAZ21haWwuY29t
†These authors have contributed equally to this work and share first authorship
‡ORCID: Lutfi Chabib orcid.org/0000-0002-1250-1406
Tedi Rustandi orcid.org/0000-0002-5322-4476
Muhammad Hafizh Abiyyu Fathin Fawwazi orcid.org/0000-0003-3387-3433
Eka Kumalasari orcid.org/0009-0003-8711-6697
Desy Ayu Lestari orcid.org/0009-0007-9955-9769
Senya Puteri Amalia orcid.org/0009-0000-9023-4991
Normilawati Normilawati orcid.org/0009-0001-8475-3610
 Lutfi Chabib
Lutfi Chabib Tedi Rustandi
Tedi Rustandi Muhammad Hafizh Abiyyu Fathin Fawwazi
Muhammad Hafizh Abiyyu Fathin Fawwazi Eka Kumalasari
Eka Kumalasari Desy Ayu Lestari
Desy Ayu Lestari Senya Puteri Amalia
Senya Puteri Amalia Normilawati Normilawati
Normilawati Normilawati