
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 24 February 2025
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1529157
This article is part of the Research TopicHorizontal Transfer of Antibiotic Resistance Genes in the Environment: Dynamic, Contributing Factors, and ControlView all 8 articles
 Bin Han1,2,3,4
Bin Han1,2,3,4 Chunlin Feng1,2,3
Chunlin Feng1,2,3 Yuan Jiang1,2,3
Yuan Jiang1,2,3 Caihong Ye1,2,3
Caihong Ye1,2,3 Yueshuai Wei1,2,3
Yueshuai Wei1,2,3 Jinbo Liu1,2,3,4*
Jinbo Liu1,2,3,4* Zhangrui Zeng1,2,3*
Zhangrui Zeng1,2,3*Klebsiella pneumoniae is an opportunistic pathogen primarily associated with nosocomial infections, characterized by a propensity for multi-drug resistance and the potential evolution into hypervirulent strains. Based on its phenotypic and genotypic characteristics, K. pneumoniae can be classified into two types: classical K. pneumoniae (cKP) and hypervirulent K. pneumoniae (hvKP). The spread of mobile genetic elements (MGEs) in K. pneumoniae has led to the emergence of carbapenem-resistant K. pneumoniae (CRKP) and carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP). The emergence of CR-hvKP is particularly concerning due to its multidrug resistance, high pathogenicity, and increased transmissibility. This review summarizes the types of MGEs present in K. pneumoniae, the mechanisms of horizontal gene transfer (HGT) mediated by these mobile elements, their roles in the dissemination of antibiotic resistance genes (ARGs) and virulence genes, and the relationships among MGEs that resemble Russian dolls or exhibit hybrid characteristics. Additionally, the clinical treatment and epidemiological characteristics of CR-hvKP are discussed. Given the high variability and transmissibility of MGEs, continuous monitoring and control of the variation and transmission of such genetic material in K. pneumoniae should be prioritized.
K. pneumoniae is a well-known opportunistic and hospital-acquired pathogen capable of causing both pulmonary and non-pulmonary infections. These infections can be invasive, including liver abscesses, endophthalmitis, and meningitis (Chew et al., 2017). K. pneumoniae strains can generally be classified into cKPs and hvKPs based on their disease profiles and genetic characteristics (Zou and Li, 2021). The pathogenicity of K. pneumoniae is closely linked to various virulence factors, including the capsule, which resists neutrophil phagocytosis and serum complement-mediated sterilization (Cortés et al., 2002; Ares et al., 2019); Lipopolysaccharide (LPS), a type of endotoxin that resists immune phagocytosis and induces fever (Papo and Shai, 2005); type I and III fimbriae, which enable adherence to host cells and facilitate infection (Schroll et al., 2010; Wu et al., 2012); and siderophores or iron acquisition systems (Martin and Bachman, 2018). HvKP is responsible for community-acquired infections that predominantly affect young and adult hosts, such as pyogenic liver abscesses, endophthalmitis, and meningitis, but it has historically been susceptible to antibiotics. CRKP is commonly associated with hospital-acquired urinary tract infections, pneumonia, sepsis, and soft tissue infections. The outbreaks and rapid spread of CRKP in hospitals have become a major public health challenge due to the lack of effective antimicrobial treatments. HvKP and CRKP first emerged as distinct lineages during the early stages of K. pneumoniae evolution. However, with the widespread dissemination of MGEs in K. pneumoniae, the current boundary that separates these two pathotypes is diminishing. CR-hvKP can arise when hvKP or CRKP acquires plasmids carrying carbapenem resistance genes or virulence genes, or when cKP acquires hybrid plasmids containing both types of genes. CR-hvKP has been widely reported in Asia, particularly in China, due to its multidrug resistance, high virulence, and contagious nature, which poses a significant threat to clinical treatment.
MGEs play a critical role in both antibiotic resistance and pathogenicity in CR-hvKP. Virulence and resistance genes are typically located within MGEs and are horizontally transferred among bacteria. MGEs are widely distributed throughout the bacterial genome and facilitate the intra-or inter-bacterial transfer of virulence and resistance genes. The primary mechanisms of HGT are transformation, conjugation, and transduction in prokaryotes (Li P. et al., 2022). MGEs are essential for maintaining bacterial genome stability, enhancing environmental adaptation, and increasing gene diversity through the functional genes they carry (Frost et al., 2005).
Plasmids are primary MGEs that facilitate the dissemination of ARGs, virulence genes, and other functional genes in K. pneumoniae. Plasmids are categorized as conjugative or mobilizable, depending on their ability to self-transfer. However, this does not imply that all plasmids are capable of conjugation. Furthermore, recent studies have underscored the importance of extracellular vehicles (EVs) in mediating the transfer of these functional genes (Mathieu et al., 2019).
Research on resistance genes, virulence genes, and associated MGEs is crucial for mitigating the global health threat posed by K. pneumoniae. This review is organized around the concept of MGEs and is divided into four sections. The first section provides an overview of the primary types of MGEs found in K. pneumoniae and their roles in the transmission of resistance and virulence genes. The second section examines the mechanisms of HGT involving MGEs, including transformation, transduction, conjugation, and vesiduction. The third section explores the interrelationships between MGEs, drawing analogies to Russian nesting dolls or intersections. Finally, the fourth section discusses the impact of MGEs on the clinical management of K. pneumoniae.
The mobile genome of bacteria encompasses all MGEs within a bacterial genome, including gene cassettes, integrons, plasmids, transposable elements, insertion sequences (IS), transposons (Tn), prophages, and integrative and conjugative elements (ICE), among others (Figure 1).
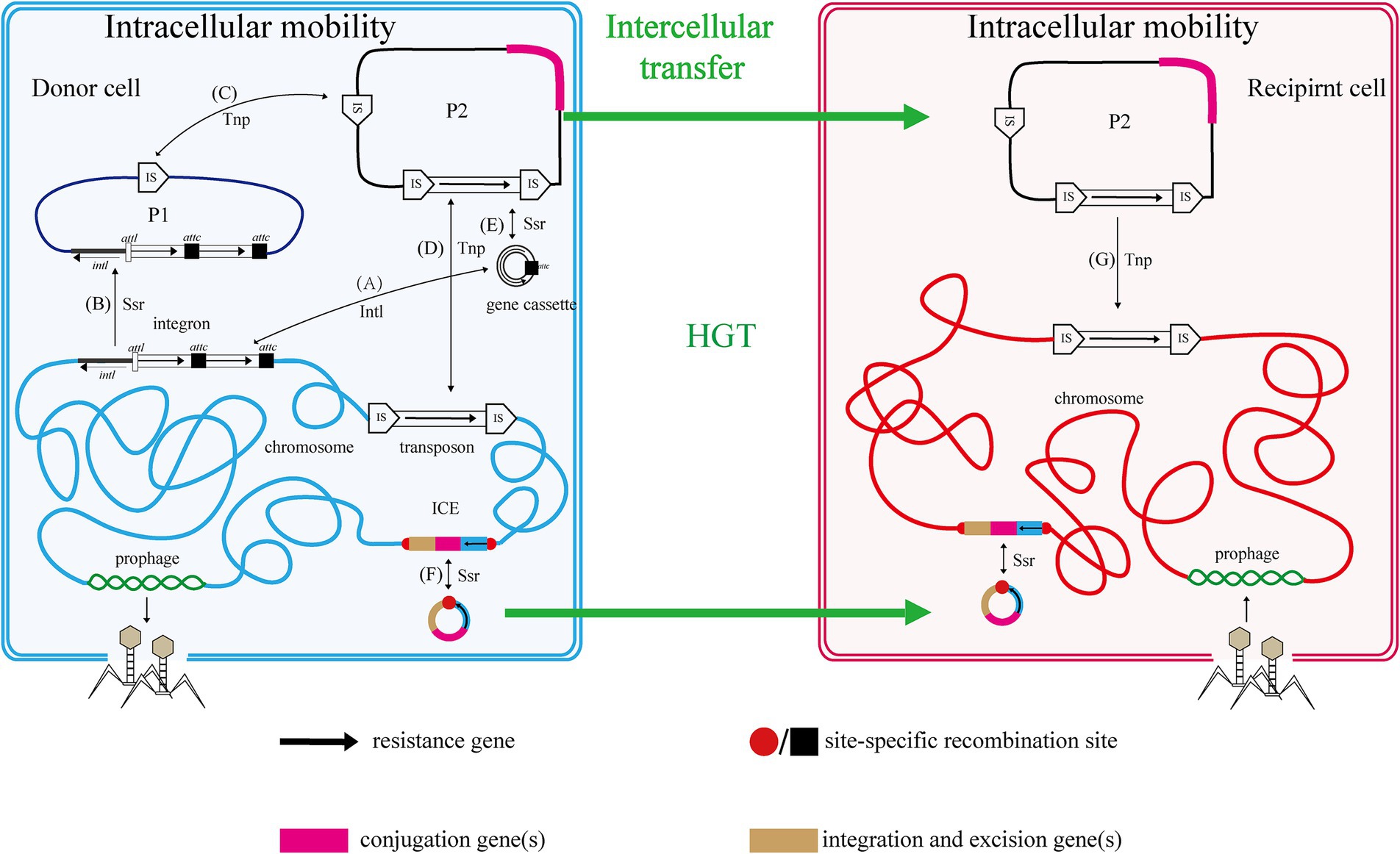
Figure 1. Relationship and mobility of MGEs. Two cells, one donor (blue envelope and chromosome with plasmids P1 and P2) and one recipient (red), are shown with various mobile genetic elements (MGEs). The functions of genes and resistance genes they carry are indicated by color coding and fat black arrows, respectively. Thin black arrows indicate intracellular processes, with those mediated by an integrase protein labeled Intl, a transposase protein labeled Tnp and those mediated by a site-specific recombinase protein labeled Ssr. Thick green arrows represent HGT. Capital letters in parentheses represent the process of intracellular MGEs transfer in cells. Refer to the text for more details.
A gene cassette, a small MGE, consists of multiple genes and recombination sites and can be captured by integrons (Partridge et al., 2009; Xu et al., 2018). Currently, the genes within gene cassettes are primarily associated with drug resistance traits. Integrons serve as assembly platforms that capture gene cassettes and incorporate exogenous genes through site-specific recombination, allowing for the internalization and expression of these foreign genes (Hall and Collis, 1995; Mazel, 2006; Escudero et al., 2015). Although integrases cannot excise it from the chromosome, rendering the integron itself immobile, they can depend on recombinases or transposases for genome-wide migration.
Transposon elements, including insertion sequences (ISs) and transposons (Tns), are MGEs that mediate intracellular gene transfer (Partridge et al., 2018). ISs and Tns are discrete, mobile DNA segments that can relocate within the genome through a series of processes, including excision and reintegration.
Insertion sequences, which are essentially short DNA sequences, represent the smallest and most abundant autonomous transposable elements (Siguier et al., 2015). They play a crucial role in the evolution of the host genome, contributing to gene sequestration, transmission, mutation, and activation, as well as facilitating plasmid and chromosome rearrangements. In recent years, several transposable elements (TEs) closely related to known insertion sequences (IS) have been identified, referred to as transporter IS (tIS). However, these tIS carry passenger genes that are not directly involved in transposition (Siguier et al., 2009). Passenger genes include transcription regulators (e.g., ISNha5, a member of the IS1595 family), methyltransferases (e.g., IS220, IS1380 family), and antibiotic resistance genes (e.g., ISCgl1, IS481 family). IS that mediate the movement of drug-resistant genes have been documented in K. pneumoniae (Cienfuegos-Gallet et al., 2017; Yang Y. et al., 2021; Pu et al., 2023b; Li et al., 2024). The addition or deletion of insertion sequences in plasmids may serve as a potential mechanism for mediating their evolution or rearrangement. For instance, plasmid Eco-N-1-p results from the deletion of the blaOXA-carrying IS26 from plasmid GX34p4_OXA-181 and the addition of IS26 from the chromosome that carries blaSHV, along with an IS5-like sequence from the chromosome that “hijacks” the blaNDM located in other plasmids (Zhang et al., 2023). These plasmids can be transferred into K. pneumoniae. The carbapenem-resistant plasmid pK186_KP is derived from the cointegration of the IncN and IncFII plasmids, an event that is entirely dependent on the transposition of IS26 (Chen Q. et al., 2022). Additionally, IS-mediated transposition in the mgrB gene may explain K. pneumoniae’s resistance to colistin (Aris et al., 2020; Yan et al., 2021; Sánchez-León et al., 2023).
Tns and ISs function similarly in bacterial genomes by facilitating genetic mobility. Composite transposons carry specific genes, such as ARGs (Mansour et al., 2017; Zhou et al., 2017; Abril et al., 2021; Tian et al., 2022; Shi et al., 2024), flanked by identical or highly homologous ISs. In contrast, unit transposons are larger elements than ISs, consisting of a transposase gene and an internal “passenger” gene, which may encode antibiotic resistance. They are flanked by inverted repeats (IRs) rather than a pair of ISs. The beta-lactamase gene blaKPC-2 in a K. pneumoniae isolate from the United States was found on a novel transposon called Tn4401 (Naas et al., 2008). This transposon was 10 kb long and contained two 39-bp imperfect inverted repeat sequences. In addition to the blaKPC-2 gene, Tn4401 also carried a transposase gene (Garbari et al., 2015; Yoon et al., 2018), a resolvase gene, and two novel insertion sequences, ISKpn6 and ISKpn7. In Switzerland, a Tn3-like transposon harboring blaVIM-1 was identified in a new plasmid (pOW16C2) from a K. pneumoniae strain isolated from river water (Zurfluh et al., 2015). The blaOXA-48 gene in plasmid pOXA-48a, isolated from K. pneumoniae, had been integrated through the acquisition of the Tn1999 composite transposon (Poirel et al., 2012; Li W. et al., 2022). The dissemination of the Tn125 transposon facilitated the spread of the blaNDM gene in Enterobacteriaceae, Acetobacteriaceae, and Pseudomonadaceae (Bonnin et al., 2014; Li et al., 2020).
Phages can be classified into two types, virulent and temperate phages, based on their distinct life cycles within the host (Casjens, 2003). Virulent phages follow a classical cycle consisting of five stages: adsorption, invasion, proliferation, assembly, and lysis, ultimately leading to the release of progeny phages. In contrast, temperate phages integrate their genetic material into the host genome and can adopt a lysogenic state. Under specific inducing conditions, temperate phages can enter the lytic phase, resulting in the lysis of the host bacterium. Prophages are the nucleic acids of temperate phages that integrate into the host genome at specific sites, allowing them to replicate alongside the host chromosomes (Canchaya et al., 2003). HGT occurs through the imprecise excision of the prophage, which can carry host genes, leading to cell lysis under induced conditions and subsequent infection of a new host. Prophage-mediated HGT is facilitated by transduction, which includes specific transduction, generalized transduction, and lateral transduction (as described in the section on transduction below). Multiple prophages have been identified on the chromosomes and plasmids of K. pneumoniae, exhibiting diversity and widespread dissemination (Gabashvili et al., 2020; Shen et al., 2020; Kang et al., 2023). Notably, a prophage containing the blaKPC gene has been discovered in the chromosomes of KPC-producing K. pneumoniae strains (Chen et al., 2015).
K. pneumoniae, which mediates the transfer of resistance genes, is primarily associated with various classes of antibiotics to which it develops resistance. These include tetracyclines (tet), sulfonamides (sul), β-lactams (bla), aminoglycosides (AAC), mucormycetes (MCR), quinolones (QNR), and multidrug-resistant bacteria (MDR). Information regarding these plasmids, as collected from the literature, is presented in Table 1. This table was utilized to create plasmid maps that illustrate the relevant resistance gene information in K. pneumoniae. Furthermore, plasmids also facilitate the transfer of genes that encode functions enabling recipient cells to better adapt and survive in their environments (Bellanger et al., 2014). To better understand the contribution of plasmids in HGT, it is important to distinguish between transmissible and non-transmissible plasmids. Transmissible plasmids include conjugative plasmids and mobilizable plasmids (Fang and Zhou, 2020). Specifically, conjugative plasmids carry all the essential genes for self-transmission through conjugation, including a Type IV secretion system (T4SS), a T4SS coupling protein (T4CP), relaxasome accessory factors (RAFs), and a relaxase gene (Ramsay and Firth, 2017). Essential genes are shown on the K. pneumoniae plasmid map in the following description.
The dissemination of antimicrobial resistance in K. pneumoniae is primarily associated with the exchange of DNA both inter-and intra-specifically, particularly through the horizontal transfer of plasmid-located resistance genes. These plasmids can confer resistance to a wide range of antimicrobial classes, including β-lactams, aminoglycosides, chloramphenicol, macrolides, sulfonamides, trimethoprim, tetracyclines, and quinolones (Carattoli, 2009). Based on plasmid incompatibility, drug-resistant plasmids in K. pneumoniae can be categorized into several groups, including IncF, IncN, IncX, IncA/C, IncL/M, IncR, IncP, IncH, IncI, and IncW. Plasmids within the same incompatibility (Inc) group are characterized by shared elements of their replication or partition systems, which prevent their stable coexistence within the same cell (Novick, 1987). The major plasmid incompatibility groups associated with drug resistance genes in K. pneumoniae include IncF, IncN, IncA/C, IncL/M, and IncX. (Plasmid group maps should be inserted separately into the appropriate sections of the article; however, IncR, IncQ, IncP, and IncH will be included in the other incompatibility groups).
The predominant incompatibility group among Enterobacteriaceae is the IncF plasmids, which have been described globally (Carattoli, 2009; Mathers et al., 2015). The prevalent resistance genes found on IncF plasmids include extended-spectrum β-lactamases (ESBLs) (Shen et al., 2008), carbapenemase-encoding genes, aminoglycoside-modifying enzyme genes, and plasmid-mediated quinolone resistance (PMQR) genes. However, IncF plasmids are typically low-copy-number plasmids, >100 kb in size, and often carry more than one replicon that promotes the initiation of replication, relying on both self-encoded and host-encoded factors for replication (Villa et al., 2010; Ogbolu et al., 2013). IncF plasmids are primarily associated with ESBLs, particularly blaCTX-M and blaTEM. Additionally, IncF plasmids have been implicated in the transmission of carbapenemase genes, specifically blaKPC and blaNDM, within the Enterobacteriaceae family (e.g., K. pneumoniae and Escherichia coli). In many countries, the IncF group dominates the spread of KPC-2 and KPC-3. IncF plasmids facilitate the dissemination of resistance genes by encoding regions essential for conjugative transfer, replication, and segregational stability in K. pneumoniae and other species. This group has been reported in numerous studies focusing on KPC-, NDM-, and OXA-producing K. pneumoniae across different countries. By searching for the keyword and limiting the scope to K. pneumoniae, we collected information on 20 specific reported plasmids (Figure 2; Supplementary Figure S1).
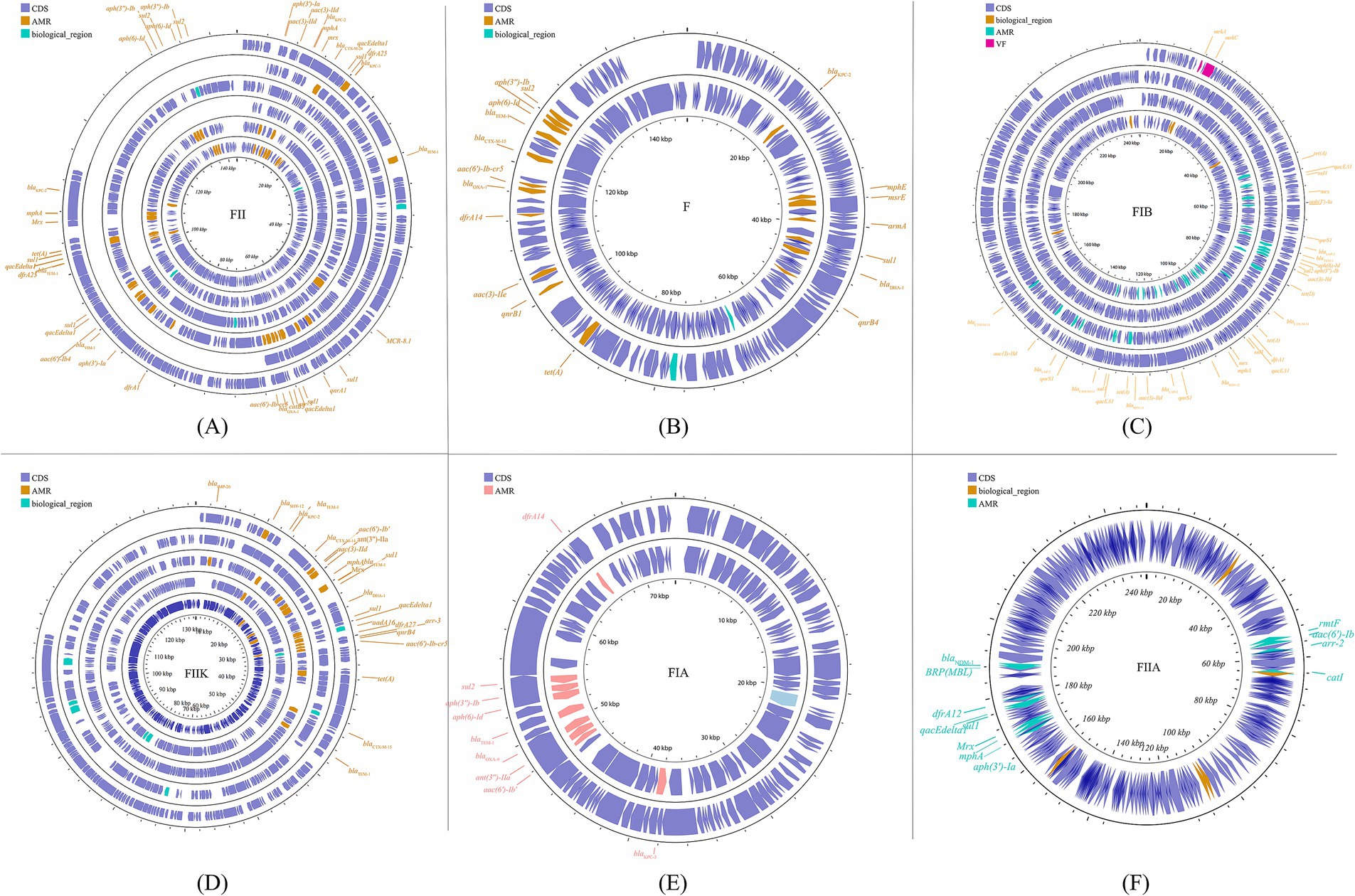
Figure 2. The IncF plasmid group contains multiple subfamilies, which in this paper include incFIA, incFIB, incFII, incFIIA, and incFIIK. again, for the specific plasmid names in the different subtypes are listed behind: FII (A): p0716-KPC, p12181-KPC, pKP91, pKpn-431cz, pKpn1693-CTXM, pKpQIL; F (B): pKP048, pKPX-2; FIB (C): p1642-tetA, p911021-tetA, pA1705-qnrS, pKAM260_1; FIIK (D): p1220-CTXM, pBK32179, pIMP1572, pKPN3, pKPN4; FIA (E): pBK30661, pBK30683; FIIA (F): pKPX-1. Plasmids of the same plasmid group were placed in the same plasmid profile, and we marked their resistance genes and virulence genes in the plasmid profile. The sequence of plasmids in the plasmid profile from inside to outside is (A): p0716-kpc, p12181-kpc, pkp91, pkpn-431cz, pkpn1693-ctxm, pkpqil; (B): pKP048, pKPX-2; (C): p1642-tetA, p911021-tetA, pA1705-qnrS, pKAM260_1; (D): p1220-CTXM, pBK32179, pIMP1572, pKPN3, pKPN4; (E): pBK30661, pBK30683; (F): pKPX-1. CDS, coding DNA sequence; AMR, antimicrobial resistance; VF, virulence factor.
PKpQIL was the first documented occurrence of an IncF plasmid harboring blaKPC in K. pneumoniae ST258 (Leavitt et al., 2007). Plasmid pKpQIL is a multi-replicon, self-transferable plasmid with a size of 113,637 bp and belongs to the IncFII group. It contains numerous drug resistance and mercury resistance genes, including blaKPC-2/-3, blaNDM-1, blaSHV-11, blaTEM-1, ΔblaOXA-9, ΔaadA1, merA, merC, merD, and merE (Leavitt et al., 2010; Hammad et al., 2023). Subsequently, new variants of pKpQIL have emerged globally, including pKpQIL isolated in Egypt showing its role in the transmission of PMQRs and blaNDM (Hammad et al., 2023); pKpQIL-IT, isolated in Italy, which contains blaKPC-3, blaTEM-1, and blaSHV-11 genes and exhibits resistance to kanamycin (García-Fernández et al., 2012); and several isolates from hospitals in New Jersey and New York City, including pKpQIL, -03, -04, -10, -234, -Ec, and -Ea, all of which exhibited comparable profiles of antimicrobial and mercury resistance genes, such as blaKPC-2/-3, blaTEM-1, blaOXA-9, aadA1, merA, merC, merD, and merE (Chen et al., 2014). Examples of IncF plasmids in K. pneumoniae carrying the blaNDM gene include pKPX-1 (250,444 bp) and pKPX-2 (250,444 bp), which were isolated from a Taiwanese patient in New Delhi (Huang et al., 2013). The former is equipped with β-lactam resistance (blaNDM-1) and aminoglycoside resistance genes (rmt, aac (69)-Ib, aph(39)-I, and aadA2), while the latter exhibits β-lactam resistance (blaCTX-M-15, blaNDM-1, blaTEM-1, and blaOXA-1), aminoglycoside resistance (aac(69)-Ib-cr, aac(3)-II, and strB), and quinolone resistance determinants (aminoglycoside acetyltransferase aac(69)-Ib-cr and qnrB). Based on sequence annotation, both pKPX-1 and pKPX-2 harbor genes related to conjugation functions. However, experimental findings indicate that pKPX-2 is indeed a conjugative plasmid, whereas pKPX-1 appears to lack the ability to conjugate.
With the spread of resistant plasmids, there has been an emergence of increasingly resistant strains of K. pneumoniae, which exhibit resistance to a broader range of antibiotics as these plasmids disseminate. Resistance genes such as blaKPC, blaNDM, blaOXA, blaTME, blaSHV, and blaCTX-M are located within the plasmids of K. pneumoniae and mediate the transmission of these genes. This phenomenon contributes to the widespread prevalence of bacterial resistance, undermines the effectiveness of antibiotics, and complicates the diagnosis and treatment of affected patients.
IncN plasmids are typically broad host ranges and self-conjugative plasmids with a size ranging from 30 to 70 kb (García-Fernández et al., 2011). These plasmids have been associated with genes conferring resistance to ESBLs, sulfonamides, quinolones, aminoglycosides, tetracyclines and streptomycin. These groups have been described as three subgroups: IncN1, IncN2 and IncN3. The same collection method as above (Figure 3; Supplementary Figure S2) was used.
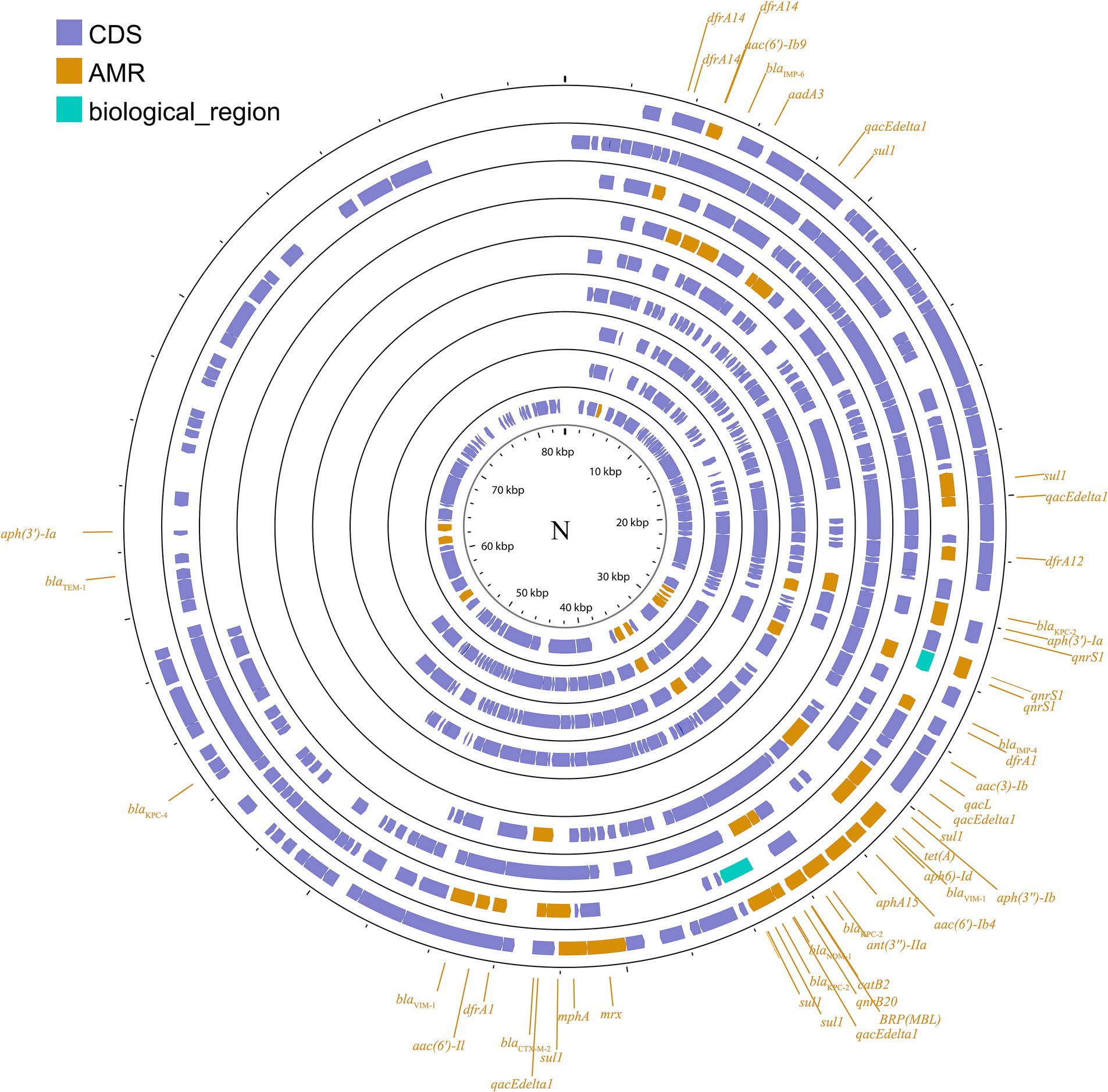
Figure 3. Plasmid profile of IncN. Including pBK31551, pFCF3SP, pFCF1305, pIMP-HZ1, pK186_KPC, pKPI-6, pNDM-BTR, pNL194, pOW16C2. Plasmids of the same plasmid group were placed in the same plasmid profile, and we marked their resistance genes and virulence genes in the plasmid profile. The sequence of plasmids in the plasmid profile from inside to outside is pBK31551, pFCF3SP, pFCF1305, pIMP-HZ1, pK186_KPC, pKPI-6, pNDM-BTR, pNL194, pOW16C2. CDS, coding DNA sequence; AMR, antimicrobial resistance.
Imipenemase (IMP), an Ambler class B metallo-β-lactamase, is an important carbapenemase that confers resistance to almost all β-lactams. pIMP-4-BKP19 is a blaIMP-4-harboring IncN plasmid from a K. pneumoniae ST1873 strain discovered in China (Xu et al., 2020). The results of the conjugation experiment indicated that pIMP4-BKP19 does not possess conjugative abilities. However, it appears that the IS26-associated class 1 integron plays a role in disseminating the blaIMP-4 gene to various plasmids. Another blaIMP-4-carrying IncN plasmid reported in China is pIMP-HZ1 (Lo et al., 2013). It is a conjugative plasmid equipped with qnrS1. Additionally, the West China Hospital discovered the presence of a self-transmissible IncN plasmid from K. pneumoniae strain WCHKP020034 harboring resistance genes: blaNDM-1, dfrA14, and qnrS (Liu et al., 2018). For K. pneumoniae, pNDM-BTR is also a blaNDM-1-encoding plasmid with a size of ~59.6 kb and belongs to IncN, which was found in Taiwan, China (Lai et al., 2018). IncN plasmid has been found in many countries. In Brazil, two blaKPC-2-carrying IncN plasmids (pFCF1305 and pFCF3SP) were isolated from K. pneumoniae strain ST442 (Pérez-Chaparro et al., 2014) and the blaKPC-containing conjugative IncN plasmids pBHKPC93_3 and pBHKPC104_3 from K. pneumoniae strains BHKPC93 and BHKPC104 were found to be transferable to Escherichia coli (Boralli et al., 2023). Similarly, IncN plasmids from K. pneumoniae have been reported to mediate the transfer of blaKPC among Enterobacteria in several Colombian hospitals (Rada et al., 2020). Additionally, the complete nucleotide sequencing of the self-transmissible plasmid pKPI-6, which encodes the blaIMP-6 and blaCTX-M-2 genes, was performed in Japan and revealed a classical IncN plasmid backbone (Kayama et al., 2015). The IncN plasmid pBK31551, originating from New Jersey, has a size of 84 kb and carries several resistance genes, including blaKPC-4, blaTEM-1, qnrB2, aac(3)-Ib, aph (3′)-I, qacF, sul1, and dfrA14. These genes confer resistance to various antibiotics such as β-lactams, quinolones, aminoglycosides, quaternary ammonium compounds, and co-trimoxazole (Chen et al., 2013). Furthermore, IncN plasmids have also been associated with blaVIM gene, as seen in a blaVIM-carrying IncN plasmid found at a university hospital in North Norway (Naseer et al., 2012). Similarly, pNL194, which encodes VIM-1 and was isolated from K. pneumoniae NL194, was found to be a self-transferable IncN plasmid (Miriagou et al., 2010). Overall, in different countries, IncN plasmids have been shown to mediate the dissemination of various antibiotic resistance genes in K. pneumoniae, enhancing its environmental adaptation and posing more challenges for clinical treatment.
The IncX plasmid is a narrow host range plasmid of the Enterobacteriaceae family that encodes type IV fimbriae, enabling self-conjugative transfer and providing auxiliary functions to host bacteria, such as resistance to antimicrobials and the ability to form biofilms (Johnson et al., 2012). The IncX group is usually associated with the transmission of K. pneumoniae carbapenemase genes, especially blaKPC, blaNDM, and blaOXA-181 (Ho et al., 2013; Campos et al., 2015; Yang et al., 2015; Fortini et al., 2016; Kim et al., 2017). Several subgroups have been successively identified: IncX1, IncX2 (Jones et al., 1993), IncX3, IncX4 (Johnson et al., 2012), and IncX5 (Chen et al., 2013), among others. IncX3 is the major subgroup carrying blaKPC and blaNDM (Guo et al., 2022). The IncX plasmid pKpS90 is a transferable plasmid that carries blaKPC-2 and blaSHV-12 in K. pneumoniae, and its complete nucleotide sequence has been reported (Kassis-Chikhani et al., 2013). The plasmid pBK31567, which belongs to a novel IncX subgroup (IncX5), is 47 kb in length and harbors blaKPC-5, dfrA5, qacEΔ1, and sul1 (Chen et al., 2013). The IncX group has not been valued in previous studies, but genetic analysis of it has revealed several surprising traits that were directly recruited from the chromosome of K. pneumoniae (Burmølle et al., 2012). The plasmids described above are shown in Figure 4; Supplementary Figure S3.
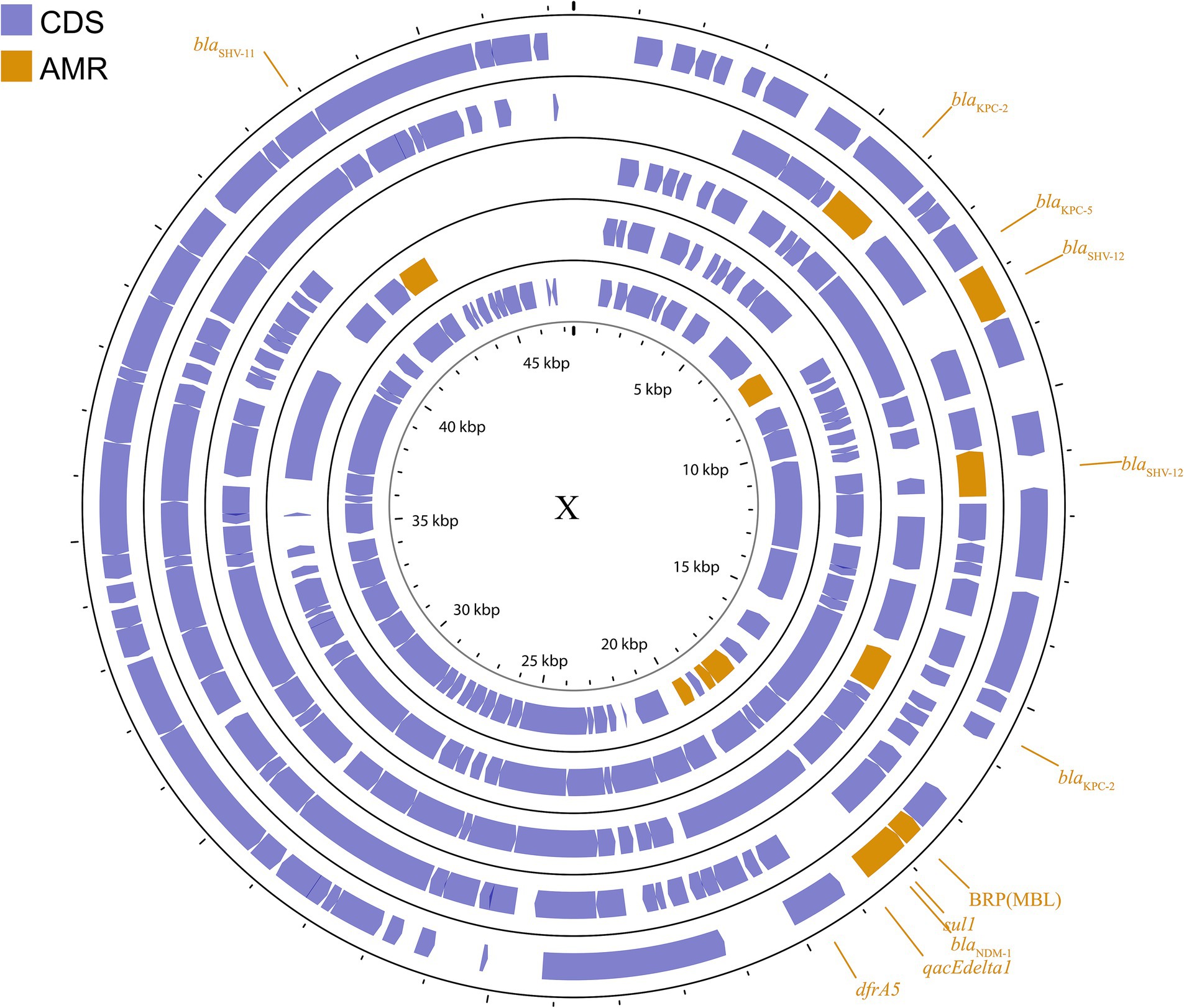
Figure 4. Plasmid profile of IncX. Including pBK31567, pIncX-SHV, pKPC-NY79, pKpS90, pNDM-HN380. Plasmids of the same plasmid group were placed in the same plasmid profile, and we marked their resistance genes and virulence genes in the plasmid profile. The sequence of plasmids in the plasmid profile from inside to outside is pBK31567, pIncX-SHV, pKPC-NY79, pKpS90, pNDM-HN380. CDS, coding DNA sequence; AMR, antimicrobial resistance.
IncA/C is a group of broad-host-range, low-copy-number, self-transferable plasmids in the size range of 40–230 kb, including two subgroups: A/C1 and A/C2 (Carraro et al., 2014). Although the coexistence of IncA and IncC has been demonstrated, they are categorized in the same incompatible plasmid group (Datta and Hedges, 1973; Hedges, 1974). IncA/C group plasmids have a significant role in the transmission of ARGs in K. pneumoniae. The IncA/C1 plasmid pIncAC_KP4898, carrying the extended spectrum β-lactamase blaSHV-12 and carbapenem-hydrolyzing metallo-β-lactamase blaVIM-1 genes, contributed to the transmission of ARGs in K. pneumoniae at the Naples Hospital (Esposito et al., 2017). Additionally, the discovery of pKP-Gr642 (162,787 bp) and pKP-Gr8143 (154,395 bp), containing blaCMY-2 and blaVIM-19, respectively, belonging to the IncA/C2 subgroup in K. pneumoniae strain ST384 from Greece, has been reported (Papagiannitsis et al., 2016a). Likewise, the IncA/C2 subgroup plasmid carrying blaIMP-4 gene, pIMP-PH114, was reported in the Philippines (Ho et al., 2014). In Taiwan, the IncA/C plasmid is the main plasmid type responsible for the transmission of ESBL genes in K. pneumoniae, along with the presence of blaOXA-48 (Ma et al., 2015; Chang et al., 2017). Furthermore, the IncA/C plasmid containing blaNDM-1 has been found in K. pneumoniae collected from outpatients in Thailand (Assawatheptawee et al., 2023). The conjugative plasmids of the IncA/C group pose a substantial threat due to their wide host range, the extensive spectrum of antimicrobial resistance they impart, their prevalence in enteric bacteria, and their highly efficient spread through conjugation (Carraro et al., 2014). These plasmids are visible in Figure 5 and Supplementary Figure S4.
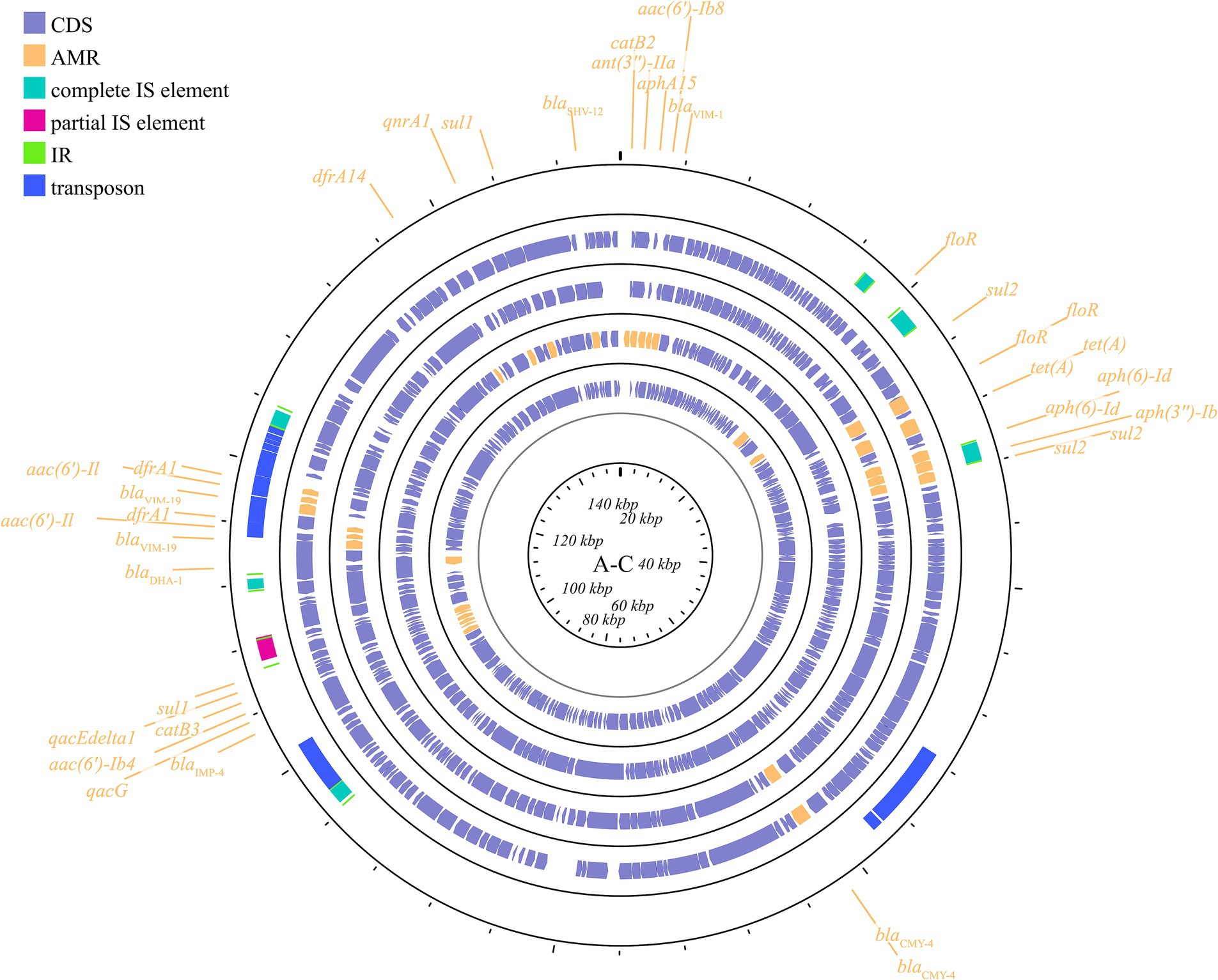
Figure 5. Plasmid profile of IncA-C. Including pIMP-PH114, pIncAC-KP4898, pKP-Gr642, pKP-Gr8143. Plasmids of the same plasmid group were placed in the same plasmid profile, and we marked their resistance genes and virulence genes in the plasmid profile. The sequence of plasmids in the plasmid profile from inside to outside is pIMP-PH114, pIncAC-KP4898, pKP-Gr642, pKP-Gr8143. CDS, coding DNA sequence; AMR, antimicrobial resistance.
Another important broad-host-range incompatibility type is the L/M group, which consists of low-copy-number and transferable plasmids. IncL/M plasmids play an important role in mediating the transfer of blaOXA-48 in K. pneumoniae, as reported in Germany, China, France, Italy, Croatia, Spain, South America, the Arabian Peninsula, and Ireland (Berger et al., 2013; Liu et al., 2015; Guo L. et al., 2016; Loucif et al., 2016; Pérez-Vázquez et al., 2016; Gaibani et al., 2017; Al-Baloushi et al., 2018; Jelić et al., 2018; Schwanbeck et al., 2021; Salazar et al., 2022). In addition to carrying blaOXA-48, IncL/M plasmids have been reported to carry other ARGs in K. pneumoniae, such as blaKPC-2 in a Chinese teaching hospital, blaIMP-68 in Japan, blaCTX-M-3 in Bulgaria, and blaCTX-M-14 in Egypt (Markovska et al., 2014; Liu et al., 2015; Kubota et al., 2019; Edward et al., 2022). These plasmids are shown in Figure 6; Supplementary Figure S5. This plasmid group is relatively small compared to the IncF, but it plays a significant role in driving resistance in K. pneumoniae, limiting the range of available antibiotics.
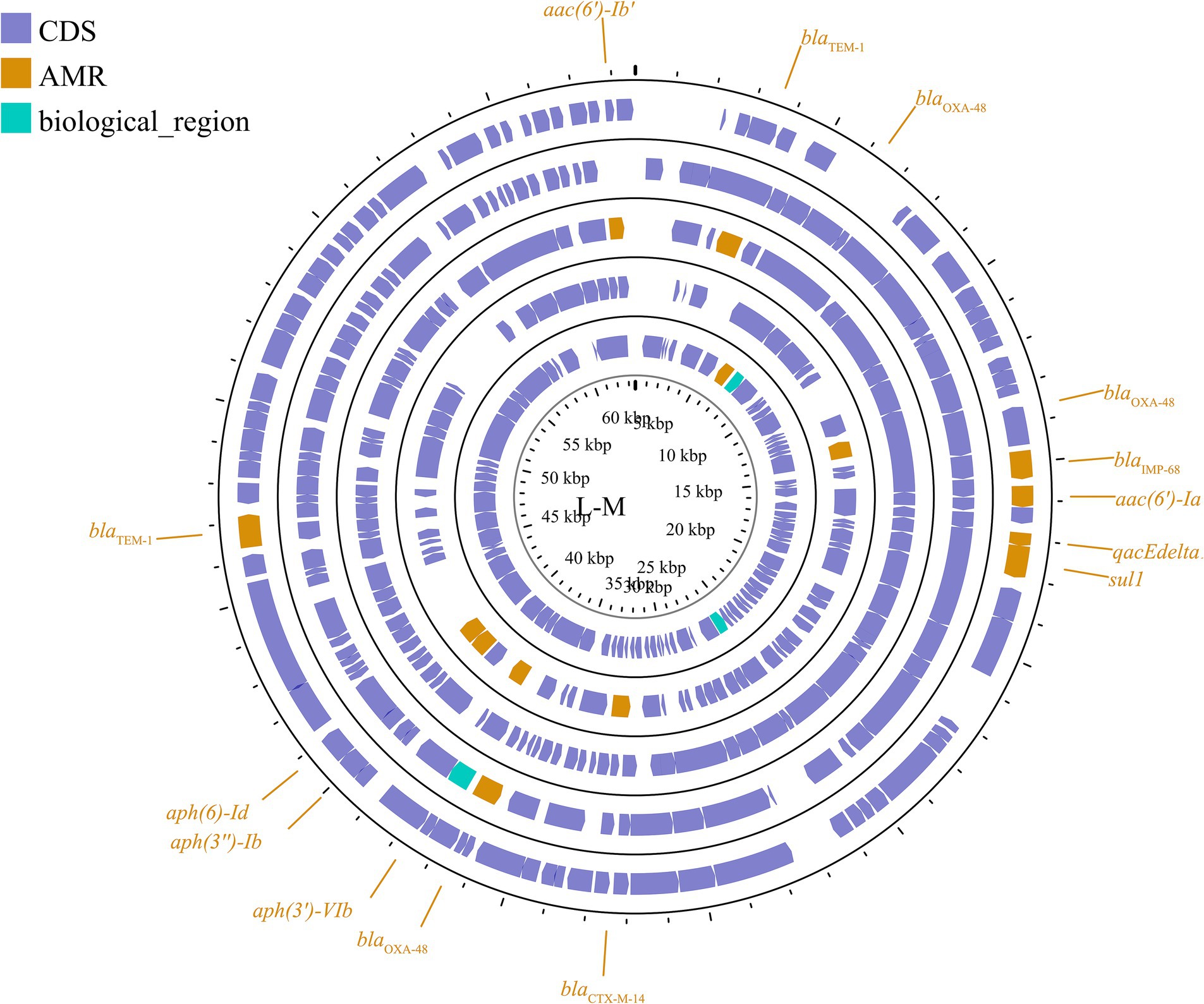
Figure 6. Plasmid profile of IncL/M. Including pE71T, pEGY22_CTX-M-14, pFOX-7a, pKPoxa-48N1, pTMTA63632. Plasmids of the same plasmid group were placed in the same plasmid profile, and we marked their resistance genes and virulence genes in the plasmid profile. The sequence of plasmids in the plasmid profile from inside to outside is pE71T, pEGY22_CTX-M-14, pFOX-7a, pKPoxa-48N1, pTMTA63632. CDS, coding DNA sequence; AMR, antimicrobial resistance.
Other plasmid incompatibility groups also play a significant role in K. pneumoniae by mediating the spread of drug-resistant genes. Broad-host-range and mobilizable IncR plasmids are responsible for transferring multiple drug-resistant genes (Chen et al., 2006; Bielak et al., 2011; Compain et al., 2014; Guo Q. et al., 2016). Additionally, IncP, H, Y, and W plasmids have been reported to contribute to the transmission of drug-resistant genes (Almeida et al., 2012; Villa et al., 2012; Zhao et al., 2017; Ahmad et al., 2019) (Figure 7; Supplementary Figure S6).
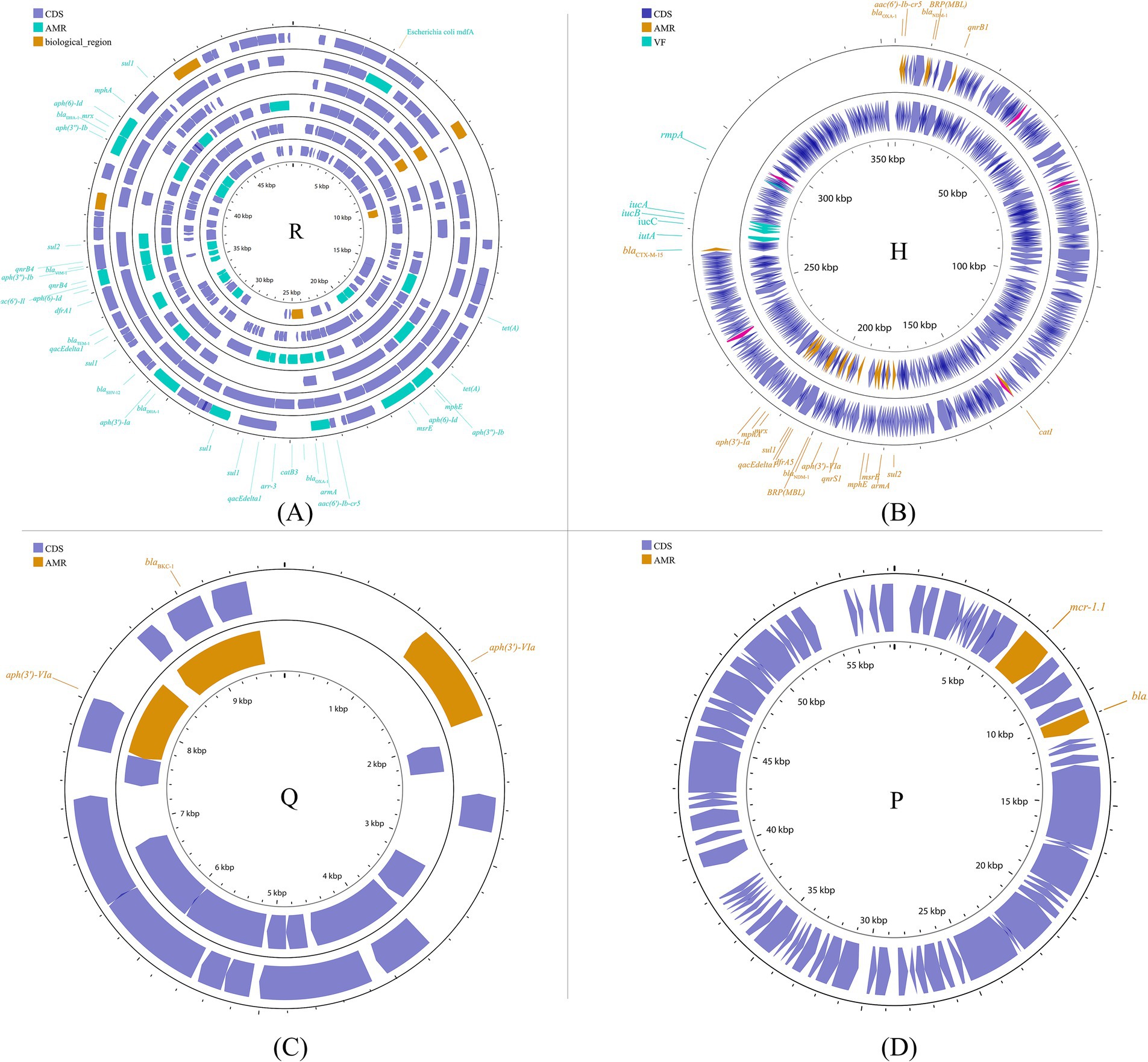
Figure 7. Plasmid profile of the other plasmid groups. Including IncR (A): pKP1780, pKPC-LK30, pKPS30, pKPS77, pR50-74, pYDC676; IncH (B): p51015_NDM-1, pNDM-MAR; IncQ (C): p60136, pKPN535a; IncP (D): pMCR_KP1511. Plasmids of the same plasmid group were placed in the same plasmid profile, and we marked their resistance genes and virulence genes in the plasmid profile. The sequence of plasmids in the plasmid profile from inside to outside is (A): pKP1780, pKPC-LK30, pKPS30, pKPS77, pR50-74, pYDC676; (B): p51015_NDM-1, pNDM-MAR; (C): p60136, pKPN535a; (D): pMCR_KP1511. CDS, coding DNA sequence; AMR, antimicrobial resistance; VF, virulence factor.
Multiple virulence genes are present in the genome of K. pneumoniae, affecting bacterial adhesion, colonization, invasion, and growth on the surfaces of host organs or tissues. These genes establish mechanisms for immune evasion, allowing the bacteria to avoid destruction by the host’s immune system. Currently, the most extensively studied virulence factors include capsules, lipopolysaccharides, fimbriae, and iron uptake systems. All known capsule variants of K. pneumoniae are synthesized by the Wzx/Wzy polysaccharide polymerization machinery, which is encoded by a single capsule polysaccharide (CPS) biosynthesis locus (Pan et al., 2015). The regulator of mucoid phenotype A (rmpA), which encodes regulatory proteins for CPS synthesis, was first identified in K. pneumoniae as a determinant controlling CPS biosynthesis (Nassif and Sansonetti, 1986). The capsule polysaccharides of K. pneumoniae resist phagocytosis by obstructing phagocytic action and lack a recognition site (the 2,3-mannose structure) for macrophage surface lectins (Huang et al., 2022). Lipopolysaccharide is an endotoxin composed of lipid A, O-antigen, and an oligosaccharide core, encoded by the lpx, wbb, and waa gene clusters, respectively (Merino et al., 2000; Frirdich and Whitfield, 2005). The lipopolysaccharide O side chain of K. pneumoniae prevents complement C1q or C3b from binding to the bacterial cell membrane, thereby protecting the bacteria from complement-mediated membrane damage and cell death (Merino et al., 1992; Albertí et al., 1996; Zhang et al., 2017). In K. pneumoniae, type 1 and 3 fimbriae, encoded by the fim and mrk genes respectively, are the major adhesive structures that have been characterized as pathogenicity factors (Di Martino et al., 1996; Tarkkanen et al., 1998; Struve et al., 2008). Several siderophores are expressed in K. pneumoniae, including enterobactin (encoded by the entABCDEF genome), yersiniabactin (irp gene cluster), salmochelin (iro gene cluster), and aerobactin (iucABCD gene cluster) (Bach et al., 2000; Hsieh et al., 2008; Müller et al., 2009). They promote bacterial metabolism and are prerequisites for bacterial infection of the host.
Studies have shown that plasmids can mediate the dissemination of virulence genes, resulting in the emergence of HVKP, or even hypervirulent and drug-resistant K. pneumoniae (Jin et al., 2021). Generally, these virulence plasmids carry many virulence-encoding genes, including capsular polysaccharide (CPS) regulator genes (rmpA and rmpA2) and several siderophore gene clusters (iucABCD-iutA, iroBCDN, ybtAEPQTUX, and entABCDEFS clusters) (Struve et al., 2015). However, the classical virulence plasmids pLVPK and pK2044 are non-conjugative due to the absence of the tra gene, which is responsible for the conjugation transfer function (Yang X. et al., 2021). The specific mechanism by which pLVPK-like virulence plasmids are delivered remains unclear. Nevertheless, some studies have suggested that virulence plasmids can be transferred with the assistance of helper plasmids that contain all the necessary genes for self-transmission via conjugation (Yang et al., 2022). The transfer mechanisms include transfer alone, co-transfer with a conjugative plasmid, and hybrid plasmid formation resulting from two rounds of single-strand exchanges at specific 28-bp fusion sites or homologous recombination (Xu et al., 2021). This process is analogous to the mobilization process described earlier. Furthermore, the integration of a virulence-associated gene fragment from a virulence plasmid into a conjugative plasmid results in the formation of a new conjugative plasmid that may carry both resistance and virulence genes. This new plasmid plays a crucial role in the development of hypervirulent and drug-resistant K. pneumoniae.
The best-known virulence plasmids are the 219 kb plasmid pLVPK from K. pneumoniae CG43 (Chen et al., 2004) and the 224 kb plasmid pK2044 (Wu et al., 2009) from K. pneumoniae NTUH-K2044, with 96% sequence coverage and 99.39% homology, respectively (Hu, 2021). Virulence-associated genes identified in pLVPK include the CPS synthesis regulator gene rmpA and its homolog rmpA2, as well as multiple iron-acquisition system genes, including iucABCD, iutA, iroBCDN, fepBC, and fecIRA. Additionally, several gene clusters homologous to copper, silver, lead, and tellurite resistance genes found in other bacteria were also identified. Furthermore, the presence of high concentrations of heavy metals within cells can lead to the formation of nonspecific complex compounds, resulting in toxic effects.
Although the classical virulence plasmid itself is incapable of mediating the dissemination of virulence-related genes via conjugation, some reports show that fragments of the virulence plasmid can be integrated into other plasmids to generate a conjugative virulence plasmid that can be transferred. For example, the virulence plasmid p15WZ-82_Vir is a conjugative plasmid formed by the integration of a fragment of the pLVPK virulence plasmid into a conjugative IncFIB-type plasmid (Yang et al., 2019). It can be transferred in K. pneumoniae and is even able to be horizontally transferred into CRKP, leading to the emergence of hypervirulent drug-resistant strains. In addition, the fusion plasmid (p17ZR-91-Vir-KPC) is a conjugative plasmid encoding virulence and resistance formed by a non-conjugative pLVPK-like virulence plasmid and a conjugation-resistant plasmid, allowing the evolution of an ordinary strain into an HVKP (Xie et al., 2021). However, recombination between gene fragments and insertion elements serves as a mechanism for the formation of hybrid plasmids (Han et al., 2022). (Potential mechanisms can be found in Figure 8). The high expression of the adhesion-associated factor rmpA probably restricts the transfer of the virulence plasmid (Xu et al., 2021). Additionally, the virulence plasmid can be transferred with the support of outer membrane vesicles (Wang et al., 2022).
Integrative and conjugative elements (ICEs) are prevalent mobile units carrying modules responsible for excision, maintenance, conjugative transfer, and integration into the genome of a new host (Johnson and Grossman, 2015). ICEs are plasmid-like MGEs that intercept chromosomal genes from the host, which are transferred to recipient cells via conjugation, and subsequently integrate into the recipient chromosome (Thomas and Nielsen, 2005). A key distinction between ICEs and plasmids is that ICEs typically do not replicate independently; rather, they integrate into the chromosome and replicate in conjunction with it. ICE gene clusters are classified into four classical modules based on their functions: integration and excision (int-xis), mating-pair formation (mpf), mobilization and processing (mob), and regulatory (reg). Additionally, ICEs contain hot spots (HS) and variable regions (VR) that may include exogenous genes. The processes of ICE segregation and integration are associated with the genes for dissociase (xis) and integrase (int), respectively (Hirano et al., 2011). The widespread presence of the yersiniabactin gene cluster and its associated integrative conjugative elements (ICEKp) has been documented among clinical isolates of K. pneumoniae (Lam et al., 2018; Octavia et al., 2019; Jati et al., 2023). ICEKp1, an integrative and conjugative element identified in K. pneumoniae, plays a significant role in the transmission of the high-pathogenicity island (HPI) and contributes to the genomic heterogeneity associated with K. pneumoniae pyogenic liver abscess (PLA) infections (Lin et al., 2008). ICEKp1 can be divided into three distinct regions: one region resembles the high-pathogenicity island of Yersinia species and encodes the siderophore yersiniabactin; the second region resembles a portion of the large virulence plasmid pLVPK and encodes the glycosidophile salmochelin (iroBCDN) and the capsular polysaccharide regulator RmpA; the third region encodes the type IV secretion system (T4SS) (Shen et al., 2019). Due to their critical role in the transfer of carbapenemase genes (Botelho et al., 2020), ICEs pose a significant threat to human health by potentially facilitating the emergence of hypervirulent, drug-resistant K. pneumoniae.
Integrative and mobilizable elements (IMEs) exhibit structural similarities to integrative conjugative elements (ICEs), paralleling the relationship between conjugative plasmids and mobilizable plasmids. Both IMEs and ICEs are classified as mobile genetic elements (MGEs) that possess their own excision and integration modules. IMEs can utilize the mating pair formation (Mpf) machinery of conjugative elements, whether plasmids or ICEs, to facilitate their transfer between bacterial cells (Guédon et al., 2017). Importantly, the IME should not be regarded as a disabled component of the ICE; rather, it functions as a hitchhiking passenger.
HGT is an important way for the expansion of drug resistance, and its main process is shown in Figure 9.
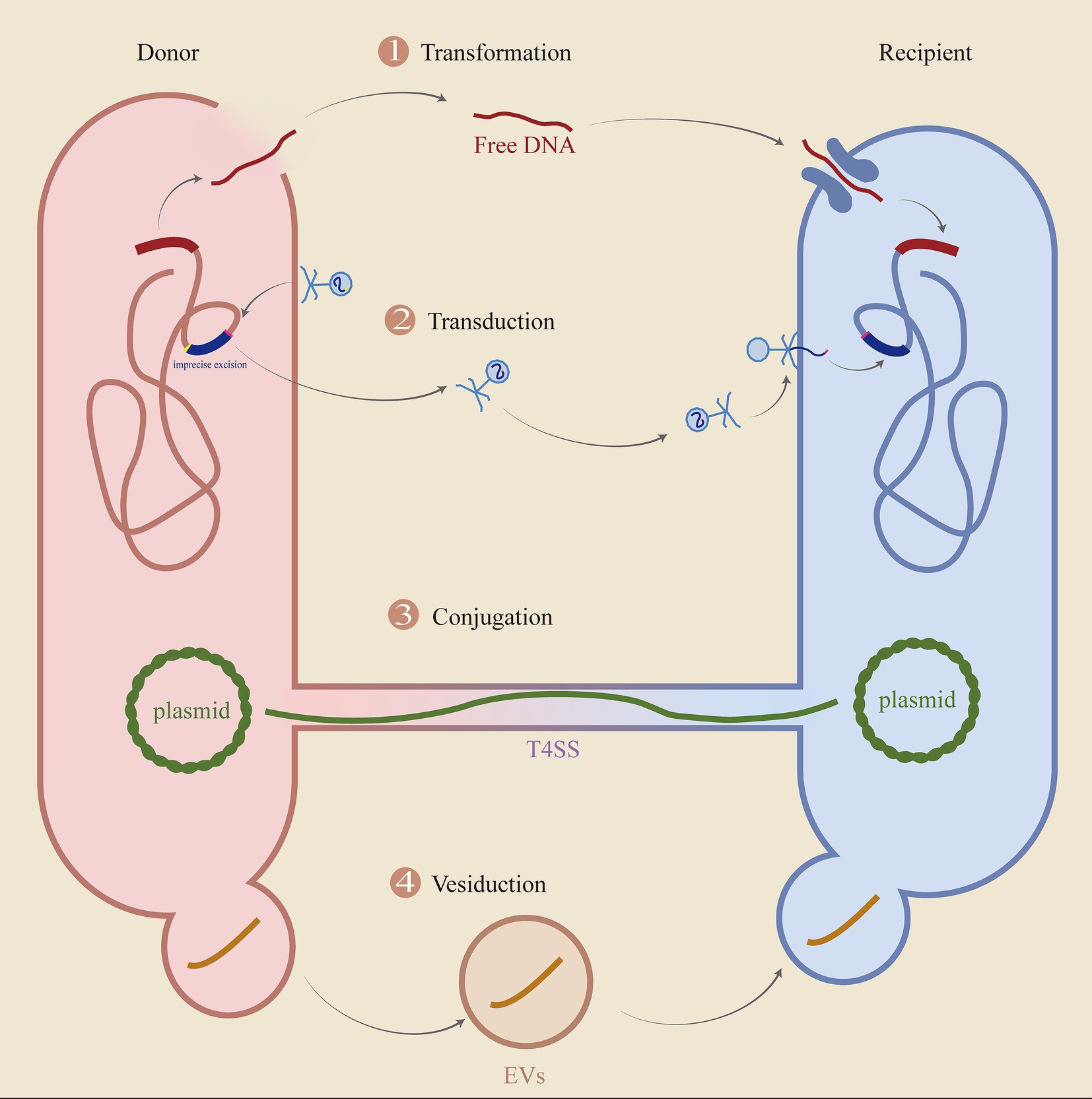
Figure 9. The key processes of HGT. The diagram delineates the fundamental processes involved in horizontal gene transfer (HGT), encompassing transformation, transduction, conjugation, and vesiduction. (1) Transformation: Cells are lysed, allowing DNA to be released and captured by recipient cells. (2) Transduction: The imprecise excision of the original bacteriophage takes genes from the donor cell, discards some of its genes, and then enters the recipient. (3) Conjugation: Transfer of plasmids or ICE via T4SS. (4) Vesiduction: Gene transfer mediated by vesicles, which is more conducive to protecting genetic material.
Natural bacterial transformation refers to the process by which a recipient bacterium directly assimilates a specific DNA fragment containing genes from a donor bacterium present in the environment. The blaIMP-68 gene, identified in K. pneumoniae strain TA6363 isolated in Tokyo, was inserted into the pHSG398 vector plasmid and subsequently used to transform E. coli DH5α cells. The minimum inhibitory concentration (MIC) of carbapenems was observed to increase in the recipient strain (Kubota et al., 2019). This finding suggests that when K. pneumoniae undergoes fragmentation, plasmids harboring resistance genes can escape and be transformed under specific conditions, facilitating the transfer of resistance genes.
This process can be delineated into four distinct sequential stages: competence induction, DNA uptake, translocation of DNA across the inner membrane (IM) and outer membrane (OM), and the integration or recombination of the incoming DNA into the genome or plasmid (Lorenz and Wackernagel, 1994; Averhoff et al., 2021). Research has been conducted to construct a comprehensive transformation mechanism by analyzing specific transformation processes in various bacterial species (Johnston et al., 2014). In a study by Averhoff et al. (2021), preliminary transformation models were developed for the systems of Acinetobacter and Thermus thermophilus. The transformation process can be summarized as follows: a bacterial pseudopilus penetrates a designated channel in the outer membrane and carries a DNA receptor at its tip, which enables recognition and binding of DNA from the external environment. Subsequently, the pseudopilus translocates the DNA through the secretin channel into the periplasm, where it is transported to the inner membrane by binding to a periplasmic protein. At this juncture, the DNA associates with an inner membrane protein that functions as a channel protein, facilitating the transfer of DNA into the cytoplasm. It is noteworthy that there are variations in the functional proteins involved in transformation systems across different species. In the extensively studied Acinetobacter, the functional proteins implicated in the transformation process include the fimbrillin proteins ComP, ComB, ComE, ComF, PilV, PilX, and FimT. The pili tip proteins are represented by ComC, while the secretin channel proteins consist of the multimeric secretin subunit known as PilQ. Furthermore, Acinetobacter employs the periplasmic binding protein ComEA and the endosomal protein ComA.
Transduction is a biological process whereby a bacteriophage transfers genetic material from a host bacterium to another bacterium during the infection cycle. K. pneumoniae exhibits a high degree of lysogenicity and harbors prophages, which are likely associated with phage transduction (Wang et al., 2019; Kang et al., 2023). This genetic transfer involves the integration of the acquired genetic material into the genome of the recipient bacterium. The incorporation of the phage genome into the bacterial chromosome is referred to as lysogeny. Prior research has elucidated the mechanisms by which bacteriophages facilitate HGT (Borodovich et al., 2022). This overview aims to summarize key aspects of phage propagation, which are essential for understanding the transduction process. During replication, the phage genome undergoes replication and translation to produce structural and functional proteins, leading to the formation of concatemers. These concatemers are subsequently introduced into an empty viral capsid, where the terminase complex excises a copy for packaging. The filled capsid then assembles with the tail to form a progeny phage, while the terminase complex binds to the next empty capsid to continue the packaging process (Rao and Feiss, 2015). Transduction can be categorized into three distinct types based on the mechanisms involved: (i) specialized transduction, which occurs in temperate phages when the prophage integrated into the bacterial chromosome undergoes imprecise excision, resulting in the replication and packaging of adjacent host chromosomal genes into the phage. This process is restricted to genes located near the prophage and may lead to the loss of phage genes; (ii) generalized transduction, primarily observed in pac-type phages, involves the terminase complex recognizing a homologous region at the pac site within the host genome, excising a gene fragment of phage genome size, and packaging it into the phage; and (iii) lateral transduction, where the phage genome is replicated within the host chromosome without excision. In this case, the terminase complex acts on the pac site located in the middle of the phage genome, packaging the replicated DNA into daughter phages that contain both host and phage genes, resulting in multiple phages that incorporate segments of the host genome.
Conjugation represents a significant mechanism of HGT that necessitates direct contact between bacterial cells. As previously noted, in K. pneumoniae, MGEs such as conjugative plasmids and conjugation elements (ICEs) can be exchanged between bacterial cells through a cell-to-cell conjugation system (Navon-Venezia et al., 2017). This process is facilitated by various modules encoded by the plasmid, which include genes responsible for the type IV secretion system (T4SS), the T4SS coupling protein (T4CP), relaxosome accessory factors (RAF), and relaxase genes. Further details regarding the F-plasmid conjugation process and the roles of the associated molecules have been comprehensively documented by Virolle et al. (2020).
The initial phase of conjugation involves the expression of the tra gene, which is situated within the plasmid transfer region. This gene is responsible for the synthesis of all proteins associated with the conjugative pilus and the T4SS required for the establishment of the mating pair. Additionally, the tra gene produces components of relaxase that facilitate the processing of the plasmid before its transfer. In the subsequent phase, conjugative pilus is generated, which senses the extracellular environment and identifies recipient cells, thereby facilitating their aggregation with donor cells to form the mating pair (Curtiss, 1969; Folkhard et al., 1979; Willetts and Skurray, 1980; Clarke et al., 2008). Notably, it has been observed that the pilus may also serve as a conduit for the direct transfer of single-stranded DNA (Brinton, 1965). Following the establishment of the mating pair, the relaxosome—a complex comprising relaxases and auxiliary proteins—initiates the processing of the plasmid, which includes relaxosome cleavage and the isolation of T-DNA (Lanka and Wilkins, 1995; Dostál and Schildbach, 2010). The relaxosome binds to and cleaves a specific site known as the nik site located in the origin region of the plasmid, resulting in the relaxation of the double-stranded DNA. The processing of the plasmid entails the cleavage by the relaxosome and the subsequent isolation of the T-DNA (Cohen et al., 1968; Willetts and Skurray, 1980). The T-DNA subsequently forms a covalent bond with the relaxase, which, in conjunction with the Type IV coupling protein (T4CP) anchored to the cell membrane, facilitates its recruitment to the conjugative pore (Curtiss, 1969; de la Cruz et al., 2010). Thereafter, the T-DNA and relaxase are transferred to the recipient cell through the conjugation pore, with the leading sequence of the plasmid being the first to be transferred to the recipient (Frost et al., 1994). This concludes the description of plasmid transfer.
In the donor cell, the single-stranded cyclic plasmid is restored to its original structure through the process of rolling circle replication (RCR) (Waters and Guiney, 1993). In contrast, upon transfer to the recipient cell, the T-DNA initially forms a cyclized single-stranded DNA via the ligation of both ends of the oriT, a process facilitated by relaxases (Draper et al., 2005; Chandler et al., 2013). Subsequently, this cyclized single-stranded DNA is converted into double-stranded DNA through the coordinated action of the host’s DNA and RNA polymerases (Baharoglu and Mazel, 2014). In response to the incoming T-DNA, the host activates various defense mechanisms, perceiving it as exogenous DNA. These defense mechanisms encompass restriction modifications, nucleic acid exonucleases, recombinant systems, and adaptive immunity (Casjens, 2003). Importantly, the host’s single-stranded binding protein (SSB) plays a crucial role in protecting the T-DNA from nuclease degradation by enveloping it (Virolle et al., 2020). Additionally, the plasmid may encode PsiB proteins that inhibit SOS induction, thereby providing further protection for the plasmid (Bailone et al., 1988).
Extracellular vesicles (EVs), which are generated through cellular metabolic activities, are spherical nanoparticles that encapsulate nucleic acids, proteins, lipids, and various other biomolecules (Kalluri and LeBleu, 2020). These vesicles play a crucial role in evading the immune response, transmitting virulence factors, facilitating intercellular communication, mediating HGT, facilitating nutrient and electron transfer, and promoting biofilm formation. Bacteria secrete a diverse array of extracellular vesicles, including outer membrane vesicles (OMVs), outer-inner membrane vesicles (OIMVs), and explosive outer membrane vesicles (EOMVs) (Toyofuku et al., 2019).
OMVs are nanoscale proteoliposomes characterized by a single membrane and are secreted by bacteria. These vesicles predominantly consist of components derived from the bacterial outer membrane and the periplasmic space (Amano et al., 2010; Ellis and Kuehn, 2010; Toyofuku et al., 2023). OMVs facilitate the differential intracellular release of various toxins and virulence factors, including adhesins, invasins, outer membrane proteins, lipopolysaccharides (LPS), flagellin, and proteases. Additionally, they may encapsulate cytoplasmic proteins and DNA (Kulp and Kuehn, 2010). Recently, a novel class of double-membrane bilayer EVs, known as outer-inner membrane vesicles (OIMVs), has been identified in Gram-negative bacteria (Pérez-Cruz et al., 2015). OIMVs not only contain the traditional components found in OMVs but also incorporate cytoplasmic elements, including DNA and plasmids. Consequently, OIMVs have been proposed as a significant type of EV involved in DNA (Toyofuku et al., 2019). The mechanisms underlying the production of EVs and the packaging of their contents remain inadequately understood; however, several hypotheses have been proposed regarding the presence of genetic material within these vesicles (Pérez-Cruz et al., 2013). The first hypothesis suggests that extracellular free DNA is internalized into the vesicles via a mechanism similar to bacterial transformation (Renelli et al., 2004). The second hypothesis posits that DNA is transported across the inner membrane and cell wall into the extraplasmic space, where it is subsequently encapsulated within the OMV (Kadurugamuwa and Beveridge, 1995). The third hypothesis, which has gained widespread acceptance, proposes that the formation of OIMVs involves the rolling of DNA from the cytoplasm of the bacterial cell (Pérez-Cruz et al., 2015). Thus, EVs, encompassing both OMVs and OIMVs, have the potential to facilitate horizontal gene transfer.
EVs have emerged as a novel mechanism of HGT in various bacterial species, functioning as carriers for the transfer of genetic material (Dorward et al., 1989; Yaron et al., 2000; Rumbo et al., 2011). These vesicles can adhere to the outer membrane of recipient cells, thereby facilitating the transmission of genetic elements between different bacterial species, which may result in the acquisition of new pathogenic traits or drug resistance. The uptake pathways for extracellular vesicles into host cells may involve several mechanisms, including macropinocytosis, clathrin-mediated endocytosis, caveolin-mediated endocytosis, or non-caveolin, non-clathrin-mediated endocytosis (O’Donoghue and Krachler, 2016). Research on K. pneumoniae has demonstrated that outer membrane vesicles (OMVs) play a critical role in mediating the transfer of drug-resistant or virulence plasmids both within and between bacterial species. Dell’Annunziata et al. (2021) was the first to report the transfer of plasmids containing resistance genes via OMVs derived from K. pneumoniae. Furthermore, research conducted by Wang et al. (2022) indicated that OMVs facilitate the transfer of the virulence plasmid phvK2115, which harbors various virulence genes, including rmpAp, rmpA2p, iucA, iroB, and peg344, within and among HVKP species. Additionally, it has been demonstrated that OMVs can disseminate two CRK3022 resistance plasmids, which contain genes such as blaKPC-2 and blaCTX-M-1, among K. pneumoniae and Escherichia coli strains within the Enterobacteriaceae family (Tang et al., 2023). Notably, all of the aforementioned plasmids possess conjugation transfer regions, yet it remains unclear whether these regions play a corresponding role in the fusion of the outer vesicle with the recipient cell. OMVs isolated from K. pneumoniae NUHL30457 (K2, ST86) have also been shown to transfer both resistance and virulence genes, resulting in a phenotype characterized by increased drug resistance and virulence (Li P. et al., 2022). Moreover, the plasmid IncFIBpKPHS1, which carries blaNDM-1 within OMVs, is also transmitted among K. pneumoniae strains (Tang et al., 2023). It is important to note that the aforementioned studies must exclude the possibility of free plasmids entering K. pneumoniae through transformation during the experimental procedures. Furthermore, the specific type of extracellular vesicles involved has not been delineated; it remains uncertain whether they are single-membrane OMVs or double-membrane outer-inner membrane vesicles (OIMVs). Therefore, future investigations should aim to differentiate between these vesicle types to enhance our understanding of the mechanisms by which genetic material enters outer membrane vesicles and to ascertain which type of outer membrane vesicles predominantly mediates gene transfer.
EVs can mediate the transmission of drug-resistance genes and virulence genes between K. pneumoniae, presenting additional challenges for clinical treatment. However, EVs offer several advantages over traditional plasmid-mediated gene transfer (Bonnington and Kuehn, 2014). Firstly, EVs do not require direct cell-to-cell contact for long-distance transport, significantly enhancing transfer efficiency. Secondly, they protect the molecular biological components contained within them from external environmental factors, thereby improving the stability of these components. For instance, EVs safeguard DNA from degradation by extracellular nucleases (Pérez-Cruz et al., 2015).
The nested structure of these MGEs is analogous to the Russian doll model and exhibits cross-cutting relationships (Figure 1). In this context, MGEs are categorized into intracellular and intercellular types. Intracellular MGEs encompass gene cassettes, integrons, and transposon elements, which exhibit an inclusion relationship; specifically, gene cassettes can be integrated into integrons, and integrons may also form part of the transposon structure (Di Pilato et al., 2015). Conversely, intercellular MGEs include prophages, ICE/IME, and plasmids. These elements facilitate the horizontal transfer of genetic material between cells through mechanisms such as transduction and conjugation, thereby expressing the associated traits. As previously mentioned, intracellular MGEs possess the ability to mobilize across the host genome, allowing for their presence within intercellular MGEs. Notably, integrons and transposable elements can be found within prophages, ICE/IME, and plasmids during their transfer. Furthermore, outer vesicles, which have recently been identified as vehicles for the dissemination of genetic material, may also encapsulate a variety of the aforementioned MGEs.
On May 17, 2024, the WHO published an updated list of drug-resistant bacteria that are most threatening to human health. Carbapenem-resistant K. pneumoniae was ranked first in the critical priority group. MGEs are the culprits for the emergence of carbapenem-resistant K. pneumoniae, as they help spread resistance genes and virulence factors widely among bacteria (Naim et al., 2018).
Although CRKP poses a major health threat worldwide, there are differences in drug resistance patterns and treatment options in different countries and regions. In China, the main drug resistance mechanism of CRKP is plasmid-mediated antibiotic resistance genes, and the main resistance gene is blaKPC-2. Therefore, most CRKP strains are susceptible to tigecycline, polymyxin B and ceftazidime-avibactam (Hu et al., 2024). In the United States, the main resistance mechanism is also plasmid-mediated resistance gene transfer, mainly associated with pColKP3-type and Inc-type plasmids (Cerón et al., 2023). Initially, polymyxin B and colistin were recommended as treatment options (Cerón et al., 2023); however, the nephrotoxic effects of polymyxin B and colistin have led to a shift in treatment options to the use of ceftazidime-avibactam in combination with aztreonam or cefdirox for infections with NDM enzyme-producing K. pneumoniae; ceftazidime-avibactam alone for urinary tract infections that produce OXA-48 enzymes; while meropenem-vaborbactam, ceftazidime-avibactam, and imipenem-cilastatin-relebactam are the preferred treatment options for infections with Enterobacteriaceae that produce KPC enzymes (Tamma et al., 2024). In Europe, the emergence of CRKP is mainly associated with the blaKPC gene, followed by blaOXA-48 and blaNDM-1 (Budia-Silva et al., 2024) and the spread of these resistance genes is associated with IncF type and ColRNAI plasmids (Loconsole et al., 2020). However, the resistance genes in some European countries also show different manifestations: Romania and Turkey mainly show blaNDM-1, Spain mainly shows blaKPC genes, Serbia mainly shows blaOXA-48, while Greece and Italy show a large number of blaKPC and other resistance genes, including blaVIM, blaNDM and blaOXA-48, Colistin is the last resort antibiotic for the treatment of Enterobacteriaceae infections (Loconsole et al., 2020; Afolayan et al., 2023). In Russia, the main resistance genes include blaOXA-48, followed by blaNDM-1 and blaKPC-3, and colistin is the main treatment option (Shamina et al., 2020). In India, blaOXA-48-like genes are the most common, followed by blaNDM-1 and blaNDM-5 (Nagaraj et al., 2021). More detailed information on drug resistance patterns, current treatment options in different regions, and the transmission patterns of drug resistance genes and virulence genes. They are shown in Table 2.
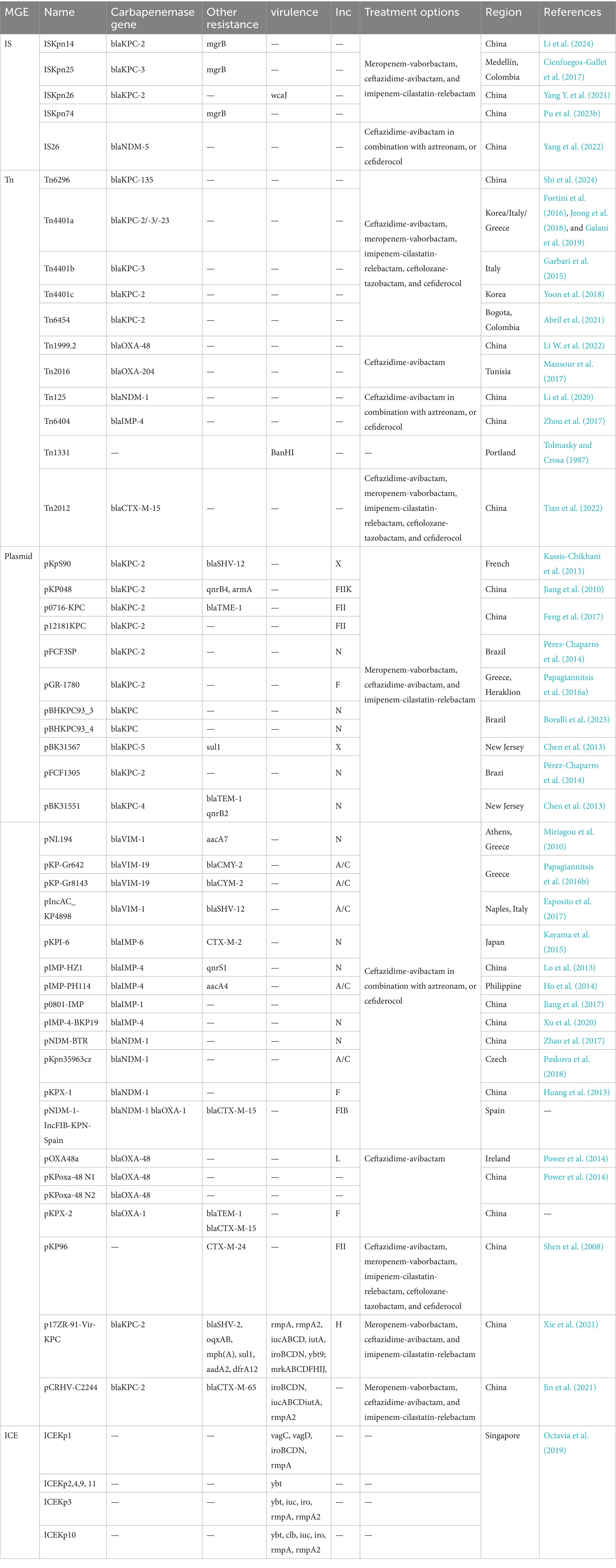
Table 2. Summary of MGEs and their associated ARGs, virulence genes, regional distribution, and treatment options.
The emergence of hypervirulent drug-resistant K. pneumoniae poses a significant public health threat, driven by three primary pathways: (i) the acquisition of virulence plasmids by drug-resistant strains, (ii) the transfer of drug-resistant plasmids to highly virulent strains, and (iii) the simultaneous acquisition of both drug-resistant and virulence plasmids by cKP strains. The rise of the “superbug” CR-hvKP is particularly concerning due to the limited therapeutic options available (Pu et al., 2023a). Potential treatment strategies for CR-hvKP infections include novel combinations of β-lactam antibiotics with β-lactamase inhibitors, as well as the combination of polymyxin B with minocycline or rifampicin, and the use of zidovudine in conjunction with rifampicin (Lei et al., 2024). However, prolonged use of these antibiotics may lead to emergence of new resistant strains, such as polymyxin-resistant variants (Chen X. et al., 2022). Therefore, there is an urgent need for research and development of novel antibiotics, alongside a focus on rational and standardized antibiotic stewardship.
Given the ongoing emergence of “superbug” strains and the escalating threat to antimicrobial defenses, it is imperative to prioritize the development and implementation of infection prevention and control policies for CR-hvKP, alongside robust antibiotic stewardship programs. Effective antimicrobial stewardship involves the optimal selection, dosage, and duration of an antimicrobial agent that yields the best clinical outcomes for the treatment or prevention of infections, while minimizing toxicity to the patient and reducing the potential for subsequent resistance (Dyar et al., 2017). In the absence of antibiotic pressure, it is plausible that we could effectively prevent the emergence of drug-resistant strains at their source. The implementation of infection prevention strategies is particularly critical in the context of drug-resistant strains (Okeah et al., 2021). The study implemented several strategies to manage patients colonized or infected with CRKP: (1) isolating of affected patients in individual rooms; (2) cohorting CRKP patients with specialized nursing staff, along with screening patients in proximity to newly identified CRKP carriers; (3) executing of weekly active surveillance for patients in the intensive care unit; and (4) selectively surveilling of patients admitted to the emergency department. To evaluate the impact of these interventions on the CRKP epidemic, interrupted regression analysis and change-point analysis were employed (Cohen et al., 2011).
With the continuous deepening of research on mobile genetic elements (MGEs), their transmission mechanism has become clearer, but their research is also facing more and more challenges. With the frequent and intensified transmission and combination between MGEs, the diversity and structural complexity of MGEs make detection more and more complicated. For example, metagenomic assembly is considered to be the main bottleneck of MGE identification. The existing MGE database is not comprehensive and cannot cover all types of MGEs, resulting in many MGEs being unable to be accurately annotated. It is not yet fully clear whether the interactive transmission of mobile genetic elements in different hosts is affected by the host’s immune system and physiological state, as well as the long-term ecological and evolutionary impact on the host. The uncertainty of cross-species transmission increases the complexity of drug resistance transmission and the difficulty of prevention and control. In addition, some potential mechanisms of MGE transmission have not been fully elucidated, including the process of extracellular vesicles encapsulating genetic material and its interaction mechanism with target cells, which need to be further clarified. These issues pose major challenges to the evolutionary study of MGEs and the prevention and control monitoring of antibiotic resistance. Therefore, exploring unknown types of MGEs, revealing their structure and function; studying the interaction between MGEs and host genomes, revealing their impact on host adaptability and evolution; and evaluating the impact of MGEs spread in different ecosystems on global health and ecological balance will become the direction of our future research. Through technology optimization and interdisciplinary cooperation, the development of resistance transmission mechanisms and prevention and control strategies, MGEs research is expected to provide new ideas and methods for solving resistance problems and promoting the development of genome engineering technology.
The widespread spread of mobile genetic elements (MGEs) among K. pneumoniae has led to the emergence of multi-drug resistant K. pneumoniae (MDRKP) and carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP), which has seriously aggravated the global difficulties in the treatment and prevention of Klebsiella pneumoniae. Therefore, the monitoring and transmission mechanism of mobile genetic elements in K. pneumoniae should be strengthened, the barriers of different disciplines should be broken, the cooperation of researchers in different fields should be promoted, and exploration should be made in all aspects, such as detection technology, gene transmission, host influence, and MGEs evolution, to provide a new scheme for solving the problems of clinical diagnosis and treatment, prevention and control, and promoting the development of genome engineering technology of mobile genetic elements.
BH: Writing – original draft, Methodology. CF: Data curation, Software, Writing – review & editing. YJ: Software, Writing – review & editing. CY: Data curation, Formal analysis, Writing – review & editing. YW: Validation, Writing – review & editing. JL: Conceptualization, Writing – review & editing. ZZ: Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Scientific Research Project of Southwest Medical University (202310632024), Luzhou Science and Technology Program (No. 2023SYF135), the Strategic Cooperation Science and Technology Project between Luzhou City and Southwest Medical University (2023LZXNYDJ041), Southwest Medical University Industry-University Training Program (No. 22001), and Sichuan Provincial Medical Research Project Plan (S22092).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1529157/full#supplementary-material
Abril, D., Vergara, E., Palacios, D., Leal, A. L., Marquez-Ortiz, R. A., Madroñero, J., et al. (2021). Within patient genetic diversity of bla(KPC) harboring Klebsiella pneumoniae in a Colombian hospital and identification of a new NTE(KPC) platform. Sci. Rep. 11:21409. doi: 10.1038/s41598-021-00887-2
Afolayan, A. O., Rigatou, A., Grundmann, H., Pantazatou, A., Daikos, G., and Reuter, S. (2023). Three Klebsiella pneumoniae lineages causing bloodstream infections variably dominated within a Greek hospital over a 15 year period. Microb. Genom. 9:mgen001082. doi: 10.1099/mgen.0.001082
Ahmad, N., Ali, S. M., and Khan, A. U. (2019). Molecular characterization of novel sequence type of carbapenem-resistant New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in the neonatal intensive care unit of an Indian hospital. Int. J. Antimicrob. Agents 53, 525–529. doi: 10.1016/j.ijantimicag.2018.12.005
Al-Baloushi, A. E., Pál, T., Ghazawi, A., and Sonnevend, A. (2018). Genetic support of carbapenemases in double carbapenemase producer Klebsiella pneumoniae isolated in the Arabian Peninsula. Acta Microbiol. Immunol. Hung. 65, 135–150. doi: 10.1556/030.65.2018.005
Albertí, S., Alvarez, D., Merino, S., Casado, M. T., Vivanco, F., Tomás, J. M., et al. (1996). Analysis of complement C3 deposition and degradation on Klebsiella pneumoniae. Infect. Immun. 64, 4726–4732. doi: 10.1128/iai.64.11.4726-4732.1996
Almeida, A. C., de Sá Cavalcanti, F. L., Vilela, M. A., Gales, A. C., de Morais, M. A. Jr., and de Morais, M. M. C. (2012). Escherichia coli ST502 and Klebsiella pneumoniae ST11 sharing an IncW plasmid harbouring the bla(KPC-2) gene in an intensive care unit patient. Int. J. Antimicrob. Agents 40, 374–376. doi: 10.1016/j.ijantimicag.2012.05.022
Amano, A., Takeuchi, H., and Furuta, N. (2010). Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes Infect. 12, 791–798. doi: 10.1016/j.micinf.2010.05.008
Ares, M. A., Sansabas, A., Rodríguez-Valverde, D., Siqueiros-Cendón, T., Rascón-Cruz, Q., Rosales-Reyes, R., et al. (2019). The interaction of Klebsiella pneumoniae with lipid rafts-associated cholesterol increases macrophage-mediated phagocytosis due to down regulation of the capsule polysaccharide. Front. Cell. Infect. Microbiol. 9:255. doi: 10.3389/fcimb.2019.00255
Aris, P., Robatjazi, S., Nikkhahi, F., and Amin Marashi, S. M. (2020). Molecular mechanisms and prevalence of colistin resistance of Klebsiella pneumoniae in the Middle East region: a review over the last 5 years. J. Glob. Antimicrob. Resist. 22, 625–630. doi: 10.1016/j.jgar.2020.06.009
Assawatheptawee, K., Sowanna, N., Treebupachatsakul, P., Na-Udom, A., Luangtongkum, T., and Niumsup, P. R. (2023). Presence and characterization of bla(NDM-1)-positive carbapenemase-producing Klebsiella pneumoniae from outpatients in Thailand. J. Microbiol. Immunol. Infect. 56, 612–623. doi: 10.1016/j.jmii.2023.01.018
Averhoff, B., Kirchner, L., Pfefferle, K., and Yaman, D. (2021). Natural transformation in gram-negative bacteria thriving in extreme environments: from genes and genomes to proteins, structures and regulation. Extremophiles 25, 425–436. doi: 10.1007/s00792-021-01242-z
Bach, S., de Almeida, A., and Carniel, E. (2000). The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183, 289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x
Baharoglu, Z., and Mazel, D. (2014). SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol. Rev. 38, 1126–1145. doi: 10.1111/1574-6976.12077
Bailone, A., Bäckman, A., Sommer, S., Célérier, J., Bagdasarian, M. M., Bagdasarian, M., et al. (1988). PsiB polypeptide prevents activation of RecA protein in Escherichia coli. Mol. Gen. Genet. 214, 389–395. doi: 10.1007/bf00330471
Bellanger, X., Payot, S., Leblond-Bourget, N., and Guédon, G. (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol. Rev. 38, 720–760. doi: 10.1111/1574-6976.12058
Berger, S., Alauzet, C., Aissa, N., Hénard, S., Rabaud, C., Bonnet, R., et al. (2013). Characterization of a new blaOXA-48-carrying plasmid in Enterobacteriaceae. Antimicrob. Agents Chemother. 57, 4064–4067. doi: 10.1128/aac.02550-12
Bielak, E., Bergenholtz, R. D., Jørgensen, M. S., Sørensen, S. J., Hansen, L. H., and Hasman, H. (2011). Investigation of diversity of plasmids carrying the blaTEM-52 gene. J. Antimicrob. Chemother. 66, 2465–2474. doi: 10.1093/jac/dkr331
Bonnin, R. A., Poirel, L., and Nordmann, P. (2014). New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol. 9, 33–41. doi: 10.2217/fmb.13.69
Bonnington, K. E., and Kuehn, M. J. (2014). Protein selection and export via outer membrane vesicles. Biochim. Biophys. Acta 1843, 1612–1619. doi: 10.1016/j.bbamcr.2013.12.011
Boralli, C., Paganini, J. A., Meneses, R. S., Mata, C., Leite, E. M. M., Schürch, A. C., et al. (2023). Characterization of bla(KPC-2) and bla(NDM-1) plasmids of a K. pneumoniae ST11 outbreak clone. Antibiotics 12:926. doi: 10.3390/antibiotics12050926
Borodovich, T., Shkoporov, A. N., Ross, R. P., and Hill, C. (2022). Phage-mediated horizontal gene transfer and its implications for the human gut microbiome. Gastroenterol. Rep. 10:goac012. doi: 10.1093/gastro/goac012
Botelho, J., Mourão, J., Roberts, A. P., and Peixe, L. (2020). Comprehensive genome data analysis establishes a triple whammy of carbapenemases, ICEs and multiple clinically relevant bacteria. Microb. Genom. 6:mgen000424. doi: 10.1099/mgen.0.000424
Brinton, C. C. Jr. (1965). The structure, function, synthesis and genetic control of bacterial pili and a molecular model for DNA and RNA transport in gram negative bacteria. Trans. N. Y. Acad. Sci. 27, 1003–1054. doi: 10.1111/j.2164-0947.1965.tb02342.x
Budia-Silva, M., Kostyanev, T., Ayala-Montaño, S., Bravo-Ferrer Acosta, J., Garcia-Castillo, M., Cantón, R., et al. (2024). International and regional spread of carbapenem-resistant Klebsiella pneumoniae in Europe. Nat. Commun. 15:5092. doi: 10.1038/s41467-024-49349-z
Burmølle, M., Norman, A., Sørensen, S. J., and Hansen, L. H. (2012). Sequencing of IncX-plasmids suggests ubiquity of mobile forms of a biofilm-promoting gene cassette recruited from Klebsiella pneumoniae. PLoS One 7:e41259. doi: 10.1371/journal.pone.0041259
Campos, J. C., da Silva, M. J., dos Santos, P. R., Barros, E. M., Pereira Mde, O., Seco, B. M., et al. (2015). Characterization of Tn3000, a transposon responsible for blaNDM-1 dissemination among Enterobacteriaceae in Brazil, Nepal, Morocco, and India. Antimicrob. Agents Chemother. 59, 7387–7395. doi: 10.1128/aac.01458-15
Canchaya, C., Proux, C., Fournous, G., Bruttin, A., and Brüssow, H. (2003). Prophage genomics. Microbiol. Mol. Biol. Rev. 67, 238–276. doi: 10.1128/mmbr.67.2.238-276.2003
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/aac.01707-08
Carraro, N., Matteau, D., Luo, P., Rodrigue, S., and Burrus, V. (2014). The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 10:e1004714. doi: 10.1371/journal.pgen.1004714
Casjens, S. (2003). Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49, 277–300. doi: 10.1046/j.1365-2958.2003.03580.x
Cerón, S., Salem-Bango, Z., Contreras, D. A., Ranson, E. L., and Yang, S. (2023). Clinical and genomic characterization of carbapenem-resistant Klebsiella pneumoniae with concurrent production of NDM and OXA-48-like Carbapenemases in Southern California, 2016–2022. Microorganisms 11:1717. doi: 10.3390/microorganisms11071717
Chandler, M., de la Cruz, F., Dyda, F., Hickman, A. B., Moncalian, G., and Ton-Hoang, B. (2013). Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat. Rev. Microbiol. 11, 525–538. doi: 10.1038/nrmicro3067
Chang, C. Y., Lin, H. J., Chang, L. L., Ma, L., Siu, L. K., Tung, Y. C., et al. (2017). Characterization of extended-spectrum β-lactamase-carrying plasmids in clinical isolates of Klebsiella pneumoniae from Taiwan. Microb. Drug Resist. 23, 98–106. doi: 10.1089/mdr.2015.0212
Chen, Y. T., Chang, H. Y., Lai, Y. C., Pan, C. C., Tsai, S. F., and Peng, H. L. (2004). Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 337, 189–198. doi: 10.1016/j.gene.2004.05.008
Chen, L., Chavda, K. D., DeLeo, F. R., Bryant, K. A., Jacobs, M. R., Bonomo, R. A., et al. (2015). Genome sequence of a Klebsiella pneumoniae sequence type 258 isolate with prophage-Encoded K. pneumoniae carbapenemase. Genome Announc. 3:e00659. doi: 10.1128/genomeA.00659-15
Chen, L., Chavda, K. D., Fraimow, H. S., Mediavilla, J. R., Melano, R. G., Jacobs, M. R., et al. (2013). Complete nucleotide sequences of blaKPC-4-and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob. Agents Chemother. 57, 269–276. doi: 10.1128/aac.01648-12
Chen, L., Chavda, K. D., Melano, R. G., Jacobs, M. R., Koll, B., Hong, T., et al. (2014). Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 58, 2871–2877. doi: 10.1128/aac.00120-14
Chen, X., Li, P., Sun, Z., Xu, X. J., and Su, J. (2022). Insertion sequence mediating mrgB disruption is the major mechanism of polymyxin resistance in carbapenem-resistant Klebsiella pneumoniae isolates from China. J. Glob. Antimicrob. Resist. 30, 357–362. doi: 10.1016/j.jgar.2022.07.002
Chen, Q., Liu, L., Hu, X., Jia, X., Gong, X., Feng, Y., et al. (2022). A small KPC-2-producing plasmid in Klebsiella pneumoniae: implications for diversified vehicles of carbapenem resistance. Microbiol. Spectr. 10:e0268821. doi: 10.1128/spectrum.02688-21
Chen, Y. T., Shu, H. Y., Li, L. H., Liao, T. L., Wu, K. M., Shiau, Y. R., et al. (2006). Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-beta-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob. Agents Chemother. 50, 3861–3866. doi: 10.1128/aac.00456-06
Chew, K. L., Lin, R. T. P., and Teo, J. W. P. (2017). Klebsiella pneumoniae in Singapore: hypervirulent infections and the carbapenemase threat. Front. Cell. Infect. Microbiol. 7:515. doi: 10.3389/fcimb.2017.00515
Cienfuegos-Gallet, A. V., Chen, L., Kreiswirth, B. N., and Jiménez, J. N. (2017). Colistin resistance in carbapenem-resistant Klebsiella pneumoniae mediated by chromosomal integration of plasmid DNA. Antimicrob. Agents Chemother. 61:e00404. doi: 10.1128/aac.00404-17
Clarke, M., Maddera, L., Harris, R. L., and Silverman, P. M. (2008). F-pili dynamics by live-cell imaging. Proc. Natl. Acad. Sci. USA 105, 17978–17981. doi: 10.1073/pnas.0806786105
Cohen, M. J., Block, C., Levin, P. D., Schwartz, C., Gross, I., Weiss, Y., et al. (2011). Institutional control measures to curtail the epidemic spread of carbapenem-resistant Klebsiella pneumoniae: a 4-year perspective. Infect. Control Hosp. Epidemiol. 32, 673–678. doi: 10.1086/660358
Cohen, A., Fisher, W. D., Curtiss, R. 3rd, and Adler, H. I. (1968). DNA isolated from Escherichia coli minicells mated with F+ cells. Proc. Natl. Acad. Sci. U.S.A. 61, 61–68. doi: 10.1073/pnas.61.1.61
Compain, F., Frangeul, L., Drieux, L., Verdet, C., Brisse, S., Arlet, G., et al. (2014). Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 58, 4207–4210. doi: 10.1128/aac.02773-13
Cortés, G., Borrell, N., de Astorza, B., Gómez, C., Sauleda, J., and Albertí, S. (2002). Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect. Immun. 70, 2583–2590. doi: 10.1128/iai.70.5.2583-2590.2002
Curtiss, R. 3rd (1969). Bacterial conjugation. Ann. Rev. Microbiol. 23, 69–136. doi: 10.1146/annurev.mi.23.100169.000441
Datta, N., and Hedges, R. W. (1973). R factors of compatibility group a. J. Gen. Microbiol. 74, 335–336. doi: 10.1099/00221287-74-2-335
de la Cruz, F., Frost, L. S., Meyer, R. J., and Zechner, E. L. (2010). Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol. Rev. 34, 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
Dell’Annunziata, F., Dell’Aversana, C., Doti, N., Donadio, G., Dal Piaz, F., Izzo, V., et al. (2021). Outer membrane vesicles derived from Klebsiella pneumoniae are a driving force for horizontal gene transfer. Int. J. Mol. Sci. 22:8732. doi: 10.3390/ijms22168732
Di Martino, P., Livrelli, V., Sirot, D., Joly, B., and Darfeuille-Michaud, A. (1996). A new fimbrial antigen harbored by CAZ-5/SHV-4-producing Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 64, 2266–2273. doi: 10.1128/iai.64.6.2266-2273.1996
Di Pilato, V., Pollini, S., and Rossolini, G. M. (2015). Tn 6249, a new Tn6162 transposon derivative carrying a double-integron platform and involved with acquisition of the blaVIM-1 metallo-β-lactamase gene in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 59, 1583–1587. doi: 10.1128/aac.04047-14
Dorward, D. W., Garon, C. F., and Judd, R. C. (1989). Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J. Bacteriol. 171, 2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989
Dostál, L., and Schildbach, J. F. (2010). Single-stranded DNA binding by F TraI relaxase and helicase domains is coordinately regulated. J. Bacteriol. 192, 3620–3628. doi: 10.1128/jb.00154-10
Draper, O., César, C. E., Machón, C., de la Cruz, F., and Llosa, M. (2005). Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl. Acad. Sci. U.S.A. 102, 16385–16390. doi: 10.1073/pnas.0506081102
Dyar, O. J., Huttner, B., Schouten, J., and Pulcini, C. (2017). What is antimicrobial stewardship? Clin. Microbiol. Infect. 23, 793–798. doi: 10.1016/j.cmi.2017.08.026
Edward, E. A., Mohamed, N. M., and Zakaria, A. S. (2022). Whole genome characterization of the high-risk clone ST383 Klebsiella pneumoniae with a simultaneous carriage of bla(CTX-M-14) on IncL/M plasmid and bla(CTX-M-15) on convergent IncHI1B/IncFIB plasmid from Egypt. Microorganisms 10:1097. doi: 10.3390/microorganisms10061097
Ellis, T. N., and Kuehn, M. J. (2010). Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74, 81–94. doi: 10.1128/mmbr.00031-09
Escudero, J. A., Loot, C., Nivina, A., and Mazel, D. (2015). The Integron: adaptation on demand. Microbiol. Spectr. 3:MDNA3-0019-2014. doi: 10.1128/microbiolspec.MDNA3-0019-2014
Esposito, E. P., Gaiarsa, S., Del Franco, M., Crivaro, V., Bernardo, M., Cuccurullo, S., et al. (2017). A novel IncA/C1 group conjugative plasmid, encoding VIM-1 Metallo-Beta-lactamase, mediates the Acquisition of Carbapenem Resistance in ST104 Klebsiella pneumoniae isolates from neonates in the intensive care unit of V Monaldi Hospital in Naples. Front. Microbiol. 8:2135. doi: 10.3389/fmicb.2017.02135
Fang, Z., and Zhou, H. (2020). Identification of the conjugative and mobilizable plasmid fragments in the plasmidome using sequence signatures. Microb. Genom. 6:mgen000459. doi: 10.1099/mgen.0.000459
Feng, J., Yin, Z., Zhao, Q., Zhao, Y., Zhang, D., Jiang, X., et al. (2017). Genomic characterization of novel IncFII-type multidrug resistant plasmids p0716-KPC and p12181-KPC from Klebsiella pneumoniae. Sci. Rep. 7:5830. doi: 10.1038/s41598-017-06283-z
Folkhard, W., Leonard, K. R., Malsey, S., Marvin, D. A., Dubochet, J., Engel, A., et al. (1979). X-ray diffraction and electron microscope studies on the structure of bacterial F pili. J. Mol. Biol. 130, 145–160. doi: 10.1016/0022-2836(79)90423-6
Fortini, D., Villa, L., Feudi, C., Pires, J., Bonura, C., Mammina, C., et al. (2016). Double copies of bla(KPC-3)::Tn4401a on an IncX3 plasmid in Klebsiella pneumoniae successful clone ST512 from Italy. Antimicrob. Agents Chemother. 60, 646–649. doi: 10.1128/aac.01886-15
Frirdich, E., and Whitfield, C. (2005). Lipopolysaccharide inner core oligosaccharide structure and outer membrane stability in human pathogens belonging to the Enterobacteriaceae. J. Endotoxin Res. 11, 133–144. doi: 10.1179/096805105x46592
Frost, L. S., Ippen-Ihler, K., and Skurray, R. A. (1994). Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58, 162–210. doi: 10.1128/mr.58.2.162-210.1994
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Gabashvili, E., Osepashvili, M., Koulouris, S., Ujmajuridze, L., Tskhitishvili, Z., and Kotetishvili, M. (2020). Phage transduction is involved in the intergeneric spread of antibiotic resistance-associated bla(CTX-M), mel, and tetM loci in natural populations of some human and animal bacterial pathogens. Curr. Microbiol. 77, 185–193. doi: 10.1007/s00284-019-01817-2
Gaibani, P., Scaltriti, E., Benni, C., Pongolini, S., Ambretti, S., Landini, M. P., et al. (2017). Characterization of an IncL/M plasmid carrying blaOXA-48 in a Klebsiella pneumoniae strain from Italy. New Microbiol. 40, 284–285
Galani, I., Antoniadou, A., Karaiskos, I., Kontopoulou, K., Giamarellou, H., and Souli, M. (2019). Genomic characterization of a KPC-23-producing Klebsiella pneumoniae ST258 clinical isolate resistant to ceftazidime-avibactam. Clin. Microbiol Infect. 25, 763.e765–763.e768. doi: 10.1016/j.cmi.2019.03.011
Garbari, L., Busetti, M., Dolzani, L., Petix, V., Knezevich, A., Bressan, R., et al. (2015). pKBuS13, a KPC-2-encoding plasmid from Klebsiella pneumoniae sequence type 833, carrying Tn4401b inserted into an Xer site-specific recombination locus. Antimicrob. Agents Chemother. 59, 5226–5231. doi: 10.1128/aac.04543-14
García-Fernández, A., Villa, L., Carta, C., Venditti, C., Giordano, A., Venditti, M., et al. (2012). Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56, 2143–2145. doi: 10.1128/aac.05308-11
García-Fernández, A., Villa, L., Moodley, A., Hasman, H., Miriagou, V., Guardabassi, L., et al. (2011). Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 66, 1987–1991. doi: 10.1093/jac/dkr225
Guédon, G., Libante, V., Coluzzi, C., Payot, S., and Leblond-Bourget, N. (2017). The obscure world of integrative and mobilizable elements, highly widespread elements that pirate bacterial conjugative systems. Genes 8:337. doi: 10.3390/genes8110337
Guo, L., An, J., Ma, Y., Ye, L., Luo, Y., Tao, C., et al. (2016). Nosocomial outbreak of OXA-48-producing Klebsiella pneumoniae in a Chinese hospital: clonal transmission of ST147 and ST383. PLoS One 11:e0160754. doi: 10.1371/journal.pone.0160754
Guo, X., Chen, R., Wang, Q., Li, C., Ge, H., Qiao, J., et al. (2022). Global prevalence, characteristics, and future prospects of IncX3 plasmids: a review. Front. Microbiol. 13:979558. doi: 10.3389/fmicb.2022.979558
Guo, Q., Spychala, C. N., McElheny, C. L., and Doi, Y. (2016). Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. J. Antimicrob. Chemother. 71, 882–886. doi: 10.1093/jac/dkv444
Hall, R. M., and Collis, C. M. (1995). Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15, 593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x
Hammad, H. A., Mohamed, I. S., El-Badawy, O., Zakaria, A. M., Shabaan, L., and Aly, S. A. (2023). pKpQIL-like plasmid contributes to the dissemination of bla(NDM-1) and plasmid mediated quinolone resistance determinants among multi drug resistant Klebsiella pneumoniae in Assiut university hospital, Egypy. Iran. J. Microbiol. 15, 208–218. doi: 10.18502/ijm.v15i2.12471
Han, Y. L., Wen, X. H., Zhao, W., Cao, X. S., Wen, J. X., Wang, J. R., et al. (2022). Epidemiological characteristics and molecular evolution mechanisms of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 13:1003783. doi: 10.3389/fmicb.2022.1003783
Hedges, R. W. (1974). R factors from Providence. J. Gen. Microbiol. 81, 171–181. doi: 10.1099/00221287-81-1-171
Hirano, N., Muroi, T., Takahashi, H., and Haruki, M. (2011). Site-specific recombinases as tools for heterologous gene integration. Appl. Microbiol. Biotechnol. 92, 227–239. doi: 10.1007/s00253-011-3519-5
Ho, P. L., Cheung, Y. Y., Lo, W. U., Li, Z., Chow, K. H., Lin, C. H., et al. (2013). Molecular characterization of an atypical IncX3 plasmid pKPC-NY79 carrying bla KPC-2 in a Klebsiella pneumoniae. Curr. Microbiol. 67, 493–498. doi: 10.1007/s00284-013-0398-2
Ho, P. L., Lo, W. U., Chan, J., Cheung, Y. Y., Chow, K. H., Yam, W. C., et al. (2014). pIMP-PH114 carrying bla IMP-4 in a Klebsiella pneumoniae strain is closely related to other multidrug-resistant IncA/C2 plasmids. Curr. Microbiol. 68, 227–232. doi: 10.1007/s00284-013-0471-x
Hsieh, P. F., Lin, T. L., Lee, C. Z., Tsai, S. F., and Wang, J. T. (2008). Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 197, 1717–1727. doi: 10.1086/588383
Hu, Y. (2021). “Molecular mechanism of pLVPK-like virulence plasmid mutation and evolution in carbapenem-resistant Klebsiella pneumoniae (CRKP)” in Master of degree (Zhengzhou: Zhengzhou University).
Hu, F., Pan, Y., Li, H., Han, R., Liu, X., Ma, R., et al. (2024). Carbapenem-resistant Klebsiella pneumoniae capsular types, antibiotic resistance and virulence factors in China: a longitudinal, multi-Centre study. Nat. Microbiol. 9, 814–829. doi: 10.1038/s41564-024-01612-1
Huang, T. W., Chen, T. L., Chen, Y. T., Lauderdale, T. L., Liao, T. L., Lee, Y. T., et al. (2013). Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS One 8:e62774. doi: 10.1371/journal.pone.0062774
Huang, X., Li, X., An, H., Wang, J., Ding, M., Wang, L., et al. (2022). Capsule type defines the capability of Klebsiella pneumoniae in evading Kupffer cell capture in the liver. PLoS Pathog. 18:e1010693. doi: 10.1371/journal.ppat.1010693
Jati, A. P., Sola-Campoy, P. J., Bosch, T., Schouls, L. M., Hendrickx, A. P. A., Bautista, V., et al. (2023). Widespread detection of yersiniabactin gene cluster and its encoding integrative conjugative elements (ICEKp) among nonoutbreak OXA-48-producing Klebsiella pneumoniae clinical isolates from Spain and the Netherlands. Microbiol. Spectr. 11:e0471622. doi: 10.1128/spectrum.04716-22
Jelić, M., Škrlin, J., Bejuk, D., Košćak, I., Butić, I., Gužvinec, M., et al. (2018). Characterization of isolates associated with emergence of OXA-48-producing Klebsiella pneumoniae in Croatia. Microb. Drug Resist. 24, 973–979. doi: 10.1089/mdr.2017.0168
Jeong, S., Kim, J. O., Yoon, E. J., Bae, I. K., Lee, W., Lee, H., et al. (2018). Extensively Drug-Resistant Escherichia coli Sequence Type 1642 Carrying an IncX3 Plasmid Containing the blaKPC-2 Gene Associated with Transposon Tn4401a. Ann. Lab. Med. 38, 17–22. doi: 10.3343/alm.2018.38.1.17
Jiang, X., Yin, Z., Yin, X., Fang, H., Sun, Q., Tong, Y., et al. (2017). Sequencing of bla(IMP)-Carrying IncN2 Plasmids, and Comparative Genomics of IncN2 Plasmids Harboring Class 1 Integrons. Front. Cell Infect. Microbiol. 7:102. doi: 10.3389/fcimb.2017.00102
Jiang, Y., Yu, D., Wei, Z., Shen, P., Zhou, Z., and Yu, Y. (2010). Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob Agents Chemother 54, 3967–3969. doi: 10.1128/aac.00137-10
Jin, L., Wang, R., Gao, H., Wang, Q., and Wang, H. (2021). Identification of a novel hybrid plasmid encoding KPC-2 and virulence factors in Klebsiella pneumoniae sequence type 11. Antimicrob. Agents Chemother. 65:e02435. doi: 10.1128/aac.02435-20
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Johnson, C. M., and Grossman, A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49, 577–601. doi: 10.1146/annurev-genet-112414-055018
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J. P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196. doi: 10.1038/nrmicro3199
Jones, C. S., Osborne, D. J., and Stanley, J. (1993). Molecular comparison of the IncX plasmids allows division into IncX1 and IncX2 subgroups. J. Gen. Microbiol. 139, 735–741. doi: 10.1099/00221287-139-4-735
Kadurugamuwa, J. L., and Beveridge, T. J. (1995). Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177, 3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367:eaau6977. doi: 10.1126/science.aau6977
Kang, F., Chai, Z., Li, B., Hu, M., Yang, Z., Wang, X., et al. (2023). Characterization and diversity of Klebsiella pneumoniae prophages. Int. J. Mol. Sci. 24:9116. doi: 10.3390/ijms24119116
Kassis-Chikhani, N., Frangeul, L., Drieux, L., Sengelin, C., Jarlier, V., Brisse, S., et al. (2013). Complete nucleotide sequence of the first KPC-2-and SHV-12-encoding IncX plasmid, pKpS90, from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57, 618–620. doi: 10.1128/aac.01712-12
Kayama, S., Shigemoto, N., Kuwahara, R., Oshima, K., Hirakawa, H., Hisatsune, J., et al. (2015). Complete nucleotide sequence of the IncN plasmid encoding IMP-6 and CTX-M-2 from emerging carbapenem-resistant Enterobacteriaceae in Japan. Antimicrob. Agents Chemother. 59, 1356–1359. doi: 10.1128/aac.04759-14
Kim, J. O., Song, S. A., Yoon, E. J., Shin, J. H., Lee, H., Jeong, S. H., et al. (2017). Outbreak of KPC-2-producing Enterobacteriaceae caused by clonal dissemination of Klebsiella pneumoniae ST307 carrying an IncX3-type plasmid harboring a truncated Tn 4401a. Diagn. Microbiol. Infect. Dis. 87, 343–348. doi: 10.1016/j.diagmicrobio.2016.12.012
Kubota, H., Suzuki, Y., Okuno, R., Uchitani, Y., Ariyoshi, T., Takemura, N., et al. (2019). IMP-68, a novel IMP-type metallo-β-lactamase in imipenem-susceptible Klebsiella pneumoniae. mSphere 4:e00736. doi: 10.1128/mSphere.00736-19
Kulp, A., and Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Ann. Rev. Microbiol. 64, 163–184. doi: 10.1146/annurev.micro.091208.073413
Lai, C. C., Chen, C. C., Lu, Y. C., Chen, H. J., Su, B. A., Weng, T. C., et al. (2018). Simultaneous three Enterobacteriaceae with different bla (NDM-1)-encoding plasmids in a patient transferred from mainland China to Taiwan. Infect. Drug Resist. 11, 2555–2560. doi: 10.2147/idr.S179024
Lam, M. M. C., Wyres, K. L., Duchêne, S., Wick, R. R., Judd, L. M., Gan, Y. H., et al. (2018). Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 9:2703. doi: 10.1038/s41467-018-05114-7
Lanka, E., and Wilkins, B. M. (1995). DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64, 141–169. doi: 10.1146/annurev.bi.64.070195.001041
Leavitt, A., Chmelnitsky, I., Carmeli, Y., and Navon-Venezia, S. (2010). Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob. Agents Chemother. 54, 4493–4496. doi: 10.1128/aac.00175-10
Leavitt, A., Navon-Venezia, S., Chmelnitsky, I., Schwaber, M. J., and Carmeli, Y. (2007). Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51, 3026–3029. doi: 10.1128/aac.00299-07
Lei, T. Y., Liao, B. B., Yang, L. R., Wang, Y., and Chen, X. B. (2024). Hypervirulent and carbapenem-resistant Klebsiella pneumoniae: a global public health threat. Microbiol. Res. 288:127839. doi: 10.1016/j.micres.2024.127839
Li, W., Guo, H., Gao, Y., Yang, X., Li, R., Li, S., et al. (2022). Comparative genomic analysis of plasmids harboring bla (OXA-48)-like genes in Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 12:1082813. doi: 10.3389/fcimb.2022.1082813
Li, J., Hu, X., Yang, L., Lin, Y., Liu, Y., Li, P., et al. (2020). New Delhi metallo-β-lactamase 1-producing Klebsiella pneumoniae ST719 isolated from a neonate in China. Microb. Drug Resist. 26, 492–496. doi: 10.1089/mdr.2019.0058
Li, Z., Lei, Z., Liu, X., Zhang, F., Yang, X., Wu, Y., et al. (2024). Disruption of mgrB gene by ISkpn14 sourced from a bla(KPC-2) carrying plasmid mediating polymyxin resistance against carbapenem-resistant Klebsiella pneumoniae during treatment: study on the underlying mechanisms. BMC Microbiol. 24:422. doi: 10.1186/s12866-024-03572-2
Li, P., Luo, W., Xiang, T. X., Jiang, Y., Liu, P., Wei, D. D., et al. (2022). Horizontal gene transfer via OMVs co-carrying virulence and antimicrobial-resistant genes is a novel way for the dissemination of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 13:945972. doi: 10.3389/fmicb.2022.945972
Lin, T. L., Lee, C. Z., Hsieh, P. F., Tsai, S. F., and Wang, J. T. (2008). Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J. Bacteriol. 190, 515–526. doi: 10.1128/jb.01219-07
Liu, L., Feng, Y., Long, H., McNally, A., and Zong, Z. (2018). Sequence type 273 Carbapenem-resistant Klebsiella pneumoniae carrying bla(NDM-1) and bla(IMP-4). Antimicrob. Agents Chemother. 62:e00160. doi: 10.1128/aac.00160-18
Liu, Y., Wan, L. G., Deng, Q., Cao, X. W., Yu, Y., and Xu, Q. F. (2015). First description of NDM-1-, KPC-2-, VIM-2-and IMP-4-producing Klebsiella pneumoniae strains in a single Chinese teaching hospital. Epidemiol. Infect. 143, 376–384. doi: 10.1017/s0950268814000995
Lo, W. U., Cheung, Y. Y., Lai, E., Lung, D., Que, T. L., and Ho, P. L. (2013). Complete sequence of an IncN plasmid, pIMP-HZ1, carrying blaIMP-4 in a Klebsiella pneumoniae strain associated with medical travel to China. Antimicrob. Agents Chemother. 57, 1561–1562. doi: 10.1128/aac.02298-12
Loconsole, D., Accogli, M., De Robertis, A. L., Capozzi, L., Bianco, A., Morea, A., et al. (2020). Emerging high-risk ST101 and ST307 carbapenem-resistant Klebsiella pneumoniae clones from bloodstream infections in southern Italy. Ann. Clin. Microbiol. Antimicrob. 19:24. doi: 10.1186/s12941-020-00366-y
Lorenz, M. G., and Wackernagel, W. (1994). Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58, 563–602. doi: 10.1128/mr.58.3.563-602.1994
Loucif, L., Kassah-Laouar, A., Saidi, M., Messala, A., Chelaghma, W., and Rolain, J. M. (2016). Outbreak of OXA-48-producing Klebsiella pneumoniae involving a sequence type 101 clone in Batna university hospital, Algeria. Antimicrob. Agents Chemother. 60, 7494–7497. doi: 10.1128/aac.00525-16
Ma, L., Wang, J. T., Wu, T. L., Siu, L. K., Chuang, Y. C., Lin, J. C., et al. (2015). Emergence of OXA-48-producing Klebsiella pneumoniae in Taiwan. PLoS One 10:e0139152. doi: 10.1371/journal.pone.0139152
Mansour, W., Haenni, M., Saras, E., Grami, R., Mani, Y., Ben Haj Khalifa, A., et al. (2017). Outbreak of colistin-resistant carbapenemase-producing Klebsiella pneumoniae in Tunisia. J. Glob. Antimicrob. Resist. 10, 88–94. doi: 10.1016/j.jgar.2017.03.017
Markovska, R., Schneider, I., Ivanova, D., Mitov, I., and Bauernfeind, A. (2014). Predominance of IncL/M and IncF plasmid types among CTX-M-ESBL-producing Escherichia coli and Klebsiella pneumoniae in Bulgarian hospitals. APMIS 122, 608–615. doi: 10.1111/apm.12204
Martin, R. M., and Bachman, M. A. (2018). Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 8:4. doi: 10.3389/fcimb.2018.00004
Mathers, A. J., Peirano, G., and Pitout, J. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/cmr.00116-14
Mathieu, M., Martin-Jaular, L., Lavieu, G., and Théry, C. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 21, 9–17. doi: 10.1038/s41556-018-0250-9
Mazel, D. (2006). Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4, 608–620. doi: 10.1038/nrmicro1462
Merino, S., Altarriba, M., Izquierdo, L., Nogueras, M. M., Regué, M., and Tomás, J. M. (2000). Cloning and sequencing of the Klebsiella pneumoniae O5 wb gene cluster and its role in pathogenesis. Infect. Immun. 68, 2435–2440. doi: 10.1128/iai.68.5.2435-2440.2000
Merino, S., Camprubí, S., Albertí, S., Benedí, V. J., and Tomás, J. M. (1992). Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect. Immun. 60, 2529–2535. doi: 10.1128/iai.60.6.2529-2535.1992
Miriagou, V., Papagiannitsis, C. C., Kotsakis, S. D., Loli, A., Tzelepi, E., Legakis, N. J., et al. (2010). Sequence of pNL194, a 79.3-kilobase IncN plasmid carrying the blaVIM-1 metallo-beta-lactamase gene in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 54, 4497–4502. doi: 10.1128/aac.00665-10
Müller, S. I., Valdebenito, M., and Hantke, K. (2009). Salmochelin, the long-overlooked catecholate siderophore of Salmonella. Biometals 22, 691–695. doi: 10.1007/s10534-009-9217-4
Naas, T., Cuzon, G., Villegas, M. V., Lartigue, M. F., Quinn, J. P., and Nordmann, P. (2008). Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob. Agents Chemother. 52, 1257–1263. doi: 10.1128/aac.01451-07
Nagaraj, G., Shamanna, V., Govindan, V., Rose, S., Sravani, D., Akshata, K. P., et al. (2021). High-resolution genomic profiling of carbapenem-resistant Klebsiella pneumoniae isolates: a multicentric retrospective Indian study. Clin. Infect. Dis. 73, S300–s307. doi: 10.1093/cid/ciab767
Naim, H., Rizvi, M., Gupta, R., Azam, M., Taneja, N., Shukla, I., et al. (2018). Drug resistance and molecular epidemiology of carbapenem resistant gram-negative bacilli isolates. J. Glob. Infect. Dis. 10, 133–139. doi: 10.4103/jgid.jgid_74_17
Naseer, U., Eriksen, B. O., Sundsfjord, A., and Samuelsen, Ø. (2012). Fecal colonization of VIM-1-producing Klebsiella pneumoniae and in vivo transfer of multidrug-resistant IncN plasmid in a renal transplant patient. Diagn. Microbiol. Infect. Dis. 72, 363–366. doi: 10.1016/j.diagmicrobio.2011.12.010
Nassif, X., and Sansonetti, P. J. (1986). Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54, 603–608. doi: 10.1128/iai.54.3.603-608.1986
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Novick, R. P. (1987). Plasmid incompatibility. Microbiol. Rev. 51, 381–395. doi: 10.1128/mr.51.4.381-395.1987
O’Donoghue, E. J., and Krachler, A. M. (2016). Mechanisms of outer membrane vesicle entry into host cells. Cell. Microbiol. 18, 1508–1517. doi: 10.1111/cmi.12655
Octavia, S., Kalisvar, M., Venkatachalam, I., Ng, O. T., Xu, W., Sridatta, P. S. R., et al. (2019). Klebsiella pneumoniae and Klebsiella quasipneumoniae define the population structure of blaKPC-2Klebsiella: a 5 year retrospective genomic study in Singapore. J. Antimicrob. Chemother. 74, 3205–3210. doi: 10.1093/jac/dkz332
Ogbolu, D. O., Daini, O. A., Ogunledun, A., Terry Alli, O. A., and Webber, M. A. (2013). Dissemination of IncF plasmids carrying beta-lactamase genes in gram-negative bacteria from Nigerian hospitals. J. Infect. Dev. Ctries. 7, 382–390. doi: 10.3855/jidc.2613
Okeah, B. O., Morrison, V., and Huws, J. C. (2021). Antimicrobial stewardship and infection prevention interventions targeting healthcare-associated Clostridioides difficile and carbapenem-resistant Klebsiella pneumoniae infections: a scoping review. BMJ Open 11:e051983. doi: 10.1136/bmjopen-2021-051983
Pan, Y. J., Lin, T. L., Chen, C. T., Chen, Y. Y., Hsieh, P. F., Hsu, C. R., et al. (2015). Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 5:15573. doi: 10.1038/srep15573
Papagiannitsis, C. C., Di Pilato, V., Giani, T., Giakkoupi, P., Riccobono, E., Landini, G., et al. (2016a). Characterization of KPC-encoding plasmids from two endemic settings. Greece and Italy. J. Antimicrob. Chemother 71, 2824–2830. doi: 10.1093/jac/dkw227
Papagiannitsis, C. C., Dolejska, M., Izdebski, R., Giakkoupi, P., Skálová, A., Chudějová, K., et al. (2016b). Characterisation of IncA/C2 plasmids carrying an In416-like integron with the blaVIM-19 gene from Klebsiella pneumoniae ST383 of Greek origin. Int. J. Antimicrob. Agents 47, 158–162. doi: 10.1016/j.ijantimicag.2015.12.001
Papo, N., and Shai, Y. (2005). A molecular mechanism for lipopolysaccharide protection of gram-negative bacteria from antimicrobial peptides. J. Biol. Chem. 280, 10378–10387. doi: 10.1074/jbc.M412865200
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088. doi: 10.1128/cmr.00088-17
Partridge, S. R., Tsafnat, G., Coiera, E., and Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33, 757–784. doi: 10.1111/j.1574-6976.2009.00175.x
Paskova, V., Medvecky, M., Skalova, A., Chudejova, K., Bitar, I., Jakubu, V., et al. (2018). Characterization of NDM-Encoding Plasmids From Enterobacteriaceae Recovered From Czech Hospitals. Front Microbiol. 9:1549. doi: 10.3389/fmicb.2018.01549
Pérez-Chaparro, P. J., Cerdeira, L. T., Queiroz, M. G., de Lima, C. P., Levy, C. E., Pavez, M., et al. (2014). Complete nucleotide sequences of two blaKPC-2-bearing IncN plasmids isolated from sequence type 442 Klebsiella pneumoniae clinical strains four years apart. Antimicrob. Agents Chemother. 58, 2958–2960. doi: 10.1128/aac.02341-13
Pérez-Cruz, C., Carrión, O., Delgado, L., Martinez, G., López-Iglesias, C., and Mercade, E. (2013). New type of outer membrane vesicle produced by the gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl. Environ. Microbiol. 79, 1874–1881. doi: 10.1128/aem.03657-12
Pérez-Cruz, C., Delgado, L., López-Iglesias, C., and Mercade, E. (2015). Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS One 10:e0116896. doi: 10.1371/journal.pone.0116896
Pérez-Vázquez, M., Oteo, J., García-Cobos, S., Aracil, B., Harris, S. R., Ortega, A., et al. (2016). Phylogeny, resistome and mobile genetic elements of emergent OXA-48 and OXA-245 Klebsiella pneumoniae clones circulating in Spain. J. Antimicrob. Chemother. 71, 887–896. doi: 10.1093/jac/dkv458
Poirel, L., Bonnin, R. A., and Nordmann, P. (2012). Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56, 559–562. doi: 10.1128/aac.05289-11
Power, K., Wang, J., Karczmarczyk, M., Crowley, B., Cotter, M., Haughton, P., et al. (2014). Molecular analysis of OXA-48-carrying conjugative IncL/M-like plasmids in clinical isolates of Klebsiella pneumoniae in Ireland. Microb. Drug Resist. 20, 270–274. doi: 10.1089/mdr.2013.0022
Pu, D., Zhao, J., Chang, K., Zhuo, X., and Cao, B. (2023a). “Superbugs” with hypervirulence and carbapenem resistance in Klebsiella pneumoniae: the rise of such emerging nosocomial pathogens in China. Sci. Bull. 68, 2658–2670. doi: 10.1016/j.scib.2023.09.040
Pu, D., Zhao, J., Lu, B., Zhang, Y., Wu, Y., Li, Z., et al. (2023b). Within-host resistance evolution of a fatal ST11 hypervirulent carbapenem-resistant Klebsiella pneumoniae. Int. J. Antimicrob. Agents 61:106747. doi: 10.1016/j.ijantimicag.2023.106747
Rada, A. M., De La Cadena, E., Agudelo, C., Capataz, C., Orozco, N., Pallares, C., et al. (2020). Dynamics of bla(KPC-2) dissemination from non-CG258 Klebsiella pneumoniae to other Enterobacterales via IncN plasmids in an area of high endemicity. Antimicrob. Agents Chemother. 64:e01743. doi: 10.1128/aac.01743-20
Ramsay, J. P., and Firth, N. (2017). Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr. Opin. Microbiol. 38, 1–9. doi: 10.1016/j.mib.2017.03.003
Rao, V. B., and Feiss, M. (2015). Mechanisms of DNA packaging by large double-stranded DNA viruses. Annu. Rev. Virol. 2, 351–378. doi: 10.1146/annurev-virology-100114-055212
Renelli, M., Matias, V., Lo, R. Y., and Beveridge, T. J. (2004). DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150, 2161–2169. doi: 10.1099/mic.0.26841-0
Rumbo, C., Fernández-Moreira, E., Merino, M., Poza, M., Mendez, J. A., Soares, N. C., et al. (2011). Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 3084–3090. doi: 10.1128/aac.00929-10
Salazar, C., Antelo, V., Vieytes, M., Dávila, C., Grill, F., Galiana, A., et al. (2022). First detection and origin of multi-drug resistant Klebsiella pneumoniae ST15 harboring OXA-48 in South America. J. Glob. Antimicrob. Resist. 30, 480–484. doi: 10.1016/j.jgar.2022.08.005
Sánchez-León, I., García-Martínez, T., Diene, S. M., Pérez-Nadales, E., Martínez-Martínez, L., and Rolain, J. M. (2023). Heteroresistance to Colistin in clinical isolates of Klebsiella pneumoniae producing OXA-48. Antibiotics 12:1111. doi: 10.3390/antibiotics12071111
Schroll, C., Barken, K. B., Krogfelt, K. A., and Struve, C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. doi: 10.1186/1471-2180-10-179
Schwanbeck, J., Bohne, W., Hasdemir, U., Groß, U., Pfeifer, Y., Bunk, B., et al. (2021). Detection of a new resistance-mediating plasmid chimera in a bla(OXA-48)-positive Klebsiella pneumoniae strain at a German university hospital. Microorganisms 9:720. doi: 10.3390/microorganisms9040720
Shamina, O. V., Kryzhanovskaya, O. A., Lazareva, A. V., Alyabieva, N. M., Polikarpova, S. V., Karaseva, O. V., et al. (2020). Emergence of a ST307 clone carrying a novel insertion element MITEKpn1 in the mgrB gene among carbapenem-resistant Klebsiella pneumoniae from Moscow, Russia. Int. J. Antimicrob. Agents 55:105850. doi: 10.1016/j.ijantimicag.2019.11.007
Shen, Z., Gao, Q., Qin, J., Liu, Y., and Li, M. (2019). Emergence of an NDM-5-producing hypervirulent Klebsiella pneumoniae sequence type 35 strain with chromosomal integration of an integrative and conjugative element, ICEKp1. Antimicrob. Agents Chemother. 64:e01675. doi: 10.1128/aac.01675-19
Shen, P., Jiang, Y., Zhou, Z., Zhang, J., Yu, Y., and Li, L. (2008). Complete nucleotide sequence of pKP96, a 67 850 bp multiresistance plasmid encoding qnrA1, aac(6′)-Ib-cr and blaCTX-M-24 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 62, 1252–1256. doi: 10.1093/jac/dkn397
Shen, J., Zhou, J., Xu, Y., and Xiu, Z. (2020). Prophages contribute to genome plasticity of Klebsiella pneumoniae and may involve the chromosomal integration of ARGs in CG258. Genomics 112, 998–1010. doi: 10.1016/j.ygeno.2019.06.016
Shi, Q., Shen, S., Tang, C., Ding, L., Guo, Y., Yang, Y., et al. (2024). Molecular mechanisms responsible KPC-135-mediated resistance to ceftazidime-avibactam in ST11-K47 hypervirulent Klebsiella pneumoniae. Emerg. Microbes Infect. 13:2361007. doi: 10.1080/22221751.2024.2361007
Siguier, P., Gagnevin, L., and Chandler, M. (2009). The new IS1595 family, its relation to IS1 and the frontier between insertion sequences and transposons. Res. Microbiol. 160, 232–241. doi: 10.1016/j.resmic.2009.02.003
Siguier, P., Gourbeyre, E., Varani, A., Ton-Hoang, B., and Chandler, M. (2015). Everyman’s guide to bacterial insertion sequences. Microbiol. Spectr. 3:MDNA3-0030-2014. doi: 10.1128/microbiolspec.MDNA3-0030-2014
Struve, C., Bojer, M., and Krogfelt, K. A. (2008). Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 76, 4055–4065. doi: 10.1128/iai.00494-08
Struve, C., Roe, C. C., Stegger, M., Stahlhut, S. G., Hansen, D. S., Engelthaler, D. M., et al. (2015). Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6:e00630. doi: 10.1128/mBio.00630-15
Tamma, P. D., Heil, E. L., Justo, J. A., Mathers, A. J., Satlin, M. J., and Bonomo, R. A. (2024). Infectious Diseases Society of America 2024 guidance on the treatment of antimicrobial-resistant gram-negative infections. Clin. Infect. Dis. 2024:ciae403. doi: 10.1093/cid/ciae403
Tang, B., Yang, A., Liu, P., Wang, Z., Jian, Z., Chen, X., et al. (2023). Outer membrane vesicles transmitting bla(NDM-1) mediate the emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Antimicrob. Agents Chemother. 67:e0144422. doi: 10.1128/aac.01444-22
Tarkkanen, A. M., Westerlund-Wikström, B., Erkkilä, L., and Korhonen, T. K. (1998). Immunohistological localization of the MrkD adhesin in the type 3 fimbriae of Klebsiella pneumoniae. Infect. Immun. 66, 2356–2361. doi: 10.1128/iai.66.5.2356-2361.1998
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721. doi: 10.1038/nrmicro1234
Tian, C., Xing, M., Zhao, Y., Fan, X., Bai, Y., Fu, L., et al. (2022). Whole genome sequencing of OXA-232-producing wzi93-KL112-O1 carbapenem-resistant Klebsiella pneumoniae in human bloodstream infection co-harboring chromosomal ISEcp1-based bla (CTX-M-15) and one rmpA2-associated virulence plasmid. Front. Cell. Infect. Microbiol. 12:984479. doi: 10.3389/fcimb.2022.984479
Tolmasky, M. E., and Crosa, J. H. (1987). Tn1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob Agents Chemother 31, 1955–1960. doi: 10.1128/aac.31.12.1955
Toyofuku, M., Nomura, N., and Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17, 13–24. doi: 10.1038/s41579-018-0112-2
Toyofuku, M., Schild, S., Kaparakis-Liaskos, M., and Eberl, L. (2023). Composition and functions of bacterial membrane vesicles. Nat. Rev. Microbiol. 21, 415–430. doi: 10.1038/s41579-023-00875-5
Villa, L., García-Fernández, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Villa, L., Poirel, L., Nordmann, P., Carta, C., and Carattoli, A. (2012). Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J. Antimicrob. Chemother. 67, 1645–1650. doi: 10.1093/jac/dks114
Virolle, C., Goldlust, K., Djermoun, S., Bigot, S., and Lesterlin, C. (2020). Plasmid transfer by conjugation in gram-negative bacteria: from the cellular to the community level. Genes 11:1239. doi: 10.3390/genes11111239
Wang, F., Wang, D., Hou, W., Jin, Q., Feng, J., and Zhou, D. (2019). Evolutionary diversity of prophage DNA in Klebsiella pneumoniae chromosomes. Front. Microbiol. 10:2840. doi: 10.3389/fmicb.2019.02840
Wang, Z., Wen, Z., Jiang, M., Xia, F., Wang, M., Zhuge, X., et al. (2022). Dissemination of virulence and resistance genes among Klebsiella pneumoniae via outer membrane vesicle: an important plasmid transfer mechanism to promote the emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Transbound. Emerg. Dis. 69, e2661–e2676. doi: 10.1111/tbed.14615
Waters, V. L., and Guiney, D. G. (1993). Processes at the nick region link conjugation, T-DNA transfer and rolling circle replication. Mol. Microbiol. 9, 1123–1130. doi: 10.1111/j.1365-2958.1993.tb01242.x
Willetts, N., and Skurray, R. (1980). The conjugation system of F-like plasmids. Annu. Rev. Genet. 14, 41–76. doi: 10.1146/annurev.ge.14.120180.000353
Wu, K. M., Li, L. H., Yan, J. J., Tsao, N., Liao, T. L., Tsai, H. C., et al. (2009). Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 191, 4492–4501. doi: 10.1128/jb.00315-09
Wu, C. C., Lin, C. T., Cheng, W. Y., Huang, C. J., Wang, Z. C., and Peng, H. L. (2012). Fur-dependent MrkHI regulation of type 3 fimbriae in Klebsiella pneumoniae CG43. Microbiology 158, 1045–1056. doi: 10.1099/mic.0.053801-0
Xie, M., Yang, X., Xu, Q., Ye, L., Chen, K., Zheng, Z., et al. (2021). Clinical evolution of ST11 carbapenem resistant and hypervirulent Klebsiella pneumoniae. Commun. Biol. 4:650. doi: 10.1038/s42003-021-02148-4
Xu, J., Lin, W., Chen, Y., and He, F. (2020). Characterization of an IMP-4-producing Klebsiella pneumoniae ST1873 strain recovered from an infant with a bloodstream infection in China. Infect. Drug Resist. 13, 773–779. doi: 10.2147/idr.S247341
Xu, T., Wang, J., Ying, J., Zhu, T., Liu, Y., Xu, L., et al. (2018). Characterisation of a class 1 integron associated with the formation of quadruple bla(GES-5) cassettes from an IncP-1β group plasmid in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 52, 485–491. doi: 10.1016/j.ijantimicag.2018.07.002
Xu, Y., Zhang, J., Wang, M., Liu, M., Liu, G., Qu, H., et al. (2021). Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 13:119. doi: 10.1186/s13073-021-00936-5
Yan, W., Zhang, Q., Zhu, Y., Jing, N., Yuan, Y., Zhang, Y., et al. (2021). Molecular mechanism of polymyxin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli isolates from Henan province, China: a multicenter study. Infect Drug Resist 14, 2657–2666. doi: 10.2147/idr.S314490
Yang, X., Dong, N., Chan, E. W., Zhang, R., and Chen, S. (2021). Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 29, 65–83. doi: 10.1016/j.tim.2020.04.012
Yang, Q., Fang, L., Fu, Y., Du, X., Shen, Y., and Yu, Y. (2015). Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One 10:e0129454. doi: 10.1371/journal.pone.0129454
Yang, X., Wai-Chi Chan, E., Zhang, R., and Chen, S. (2019). A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat. Microbiol. 4, 2039–2043. doi: 10.1038/s41564-019-0566-7
Yang, X., Xie, M., Xu, Q., Ye, L., Yang, C., Dong, N., et al. (2022). Transmission of pLVPK-like virulence plasmid in Klebsiella pneumoniae mediated by an Incl1 conjugative helper plasmid. iScience 25:104428. doi: 10.1016/j.isci.2022.104428
Yang, Y., Yang, Y., Chen, G., Lin, M., Chen, Y., He, R., et al. (2021). Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg. Microbes Infect. 10, 700–709. doi: 10.1080/22221751.2021.1906163
Yaron, S., Kolling, G. L., Simon, L., and Matthews, K. R. (2000). Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66, 4414–4420. doi: 10.1128/aem.66.10.4414-4420.2000
Yoon, E. J., Kim, J. O., Kim, D., Lee, H., Yang, J. W., Lee, K. J., et al. (2018). Klebsiella pneumoniae Carbapenemase producers in South Korea between 2013 and 2015. Front. Microbiol. 9:56. doi: 10.3389/fmicb.2018.00056
Zhang, F., Li, Z., Liu, X., Luo, G., Wu, Y., Li, C., et al. (2023). Molecular characteristics of an NDM-4 and OXA-181 co-producing K51-ST16 Carbapenem-resistant Klebsiella pneumoniae: study of its potential dissemination mediated by conjugative plasmids and insertion sequences. Antimicrob. Agents Chemother. 67:e0135422. doi: 10.1128/aac.01354-22
Zhang, R., Wu, L., Eckert, T., Burg-Roderfeld, M., Rojas-Macias, M. A., Lütteke, T., et al. (2017). Lysozyme’s lectin-like characteristics facilitates its immune defense function. Q. Rev. Biophys. 50:e9. doi: 10.1017/s0033583517000075
Zhao, F., Feng, Y., Lü, X., McNally, A., and Zong, Z. (2017). IncP plasmid carrying colistin resistance gene mcr-1 in Klebsiella pneumoniae from hospital sewage. Antimicrob. Agents Chemother. 61:e02229. doi: 10.1128/aac.02229-16
Zhou, K., Yu, W., Shen, P., Lu, H., Wang, B., Rossen, J. W. A., et al. (2017). A novel Tn1696-like composite transposon (Tn6404) harboring bla (IMP-4) in a Klebsiella pneumoniae isolate carrying a rare ESBL gene bla (SFO-1). Sci. Rep. 7:17321. doi: 10.1038/s41598-017-17641-2
Zou, Q., and Li, Y. (2021). Hypervirulent Klebsiella pneumoniae. N. Engl. J. Med. 385:833. doi: 10.1056/NEJMicm2101602
Keywords: mobile genetic elements, antibiotic resistance genes, virulence genes, extracellular vesicles, K. pneumoniae
Citation: Han B, Feng C, Jiang Y, Ye C, Wei Y, Liu J and Zeng Z (2025) Mobile genetic elements encoding antibiotic resistance genes and virulence genes in Klebsiella pneumoniae: important pathways for the acquisition of virulence and resistance. Front. Microbiol. 16:1529157. doi: 10.3389/fmicb.2025.1529157
Received: 16 November 2024; Accepted: 06 February 2025;
Published: 24 February 2025.
Edited by:
Xiaoqin Zhou, University of Science and Technology Beijing, ChinaReviewed by:
Huaiqi Jing, Chinese Center for Disease Control and Prevention, ChinaCopyright © 2025 Han, Feng, Jiang, Ye, Wei, Liu and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinbo Liu, bGl1amI3MjAzQHN3bXUuZWR1LmNu; Zhangrui Zeng, emVuZ3poYW5ncnVpQHN3bXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.