
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 February 2025
Sec. Microbial Physiology and Metabolism
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1527727
Ferrichrome (Fc) acquisition in Schizosaccharomyces pombe is mediated by the cell-surface siderophore-iron transporter Str1. Here, we report that Str2, a protein homologous to Str1, localizes to the vacuolar membrane. Like Str1, Str2 expression is transcriptionally regulated in response to changes in iron concentrations. Both the str2+ and str1+ genes are induced under low-iron conditions and are repressed by the iron-responsive GATA-type transcription factor Fep1 when iron is abundant. Under high-iron conditions, chromatin immunoprecipitation (ChIP) assays reveal that TAP-Fep1 occupies the str2+ and str1+ promoters. Isolated vacuoles from str2Δ fep1Δ cells expressing GFP-tagged Str2 exhibit iron accumulation in vacuoles upon exposure to exogenous holo-Fc. sib1Δ sib2Δ cells deficient in Fc biosynthesis and lacking the str2+ gene (str2Δ) are unable to grow in the presence of exogenous Fc as a sole source of iron. Further analysis identified that conserved amino acids Tyr539 and Tyr553 in the last predicted loop of Str2 are required for supporting Fc-dependent growth of a sib1Δ sib2Δ mutant strain. Collectively, these findings indicate that the vacuolar Str2 protein plays a role in the consumption of Fc as an iron source, while also revealing the involvement of the vacuole in iron release from exogenous Fc after its assimilation.
The acquisition of the transition metal iron is essential for aerobic organisms. Due to its redox properties, iron serves as an indispensable cofactor in the catalytic centers of several enzymes, including those involved in the respiratory chain, energy production, amino acid biosynthesis, and the detoxification of reactive oxygen species (Puig et al., 2017; Galaris et al., 2019; Katsarou and Pantopoulos, 2020; Philpott et al., 2020). In aerobic environments, iron predominantly exists as ferric oxyhydroxides, which are poorly soluble and therefore not readily bioavailable (Aguiar et al., 2021). Given this limited iron availability, all organisms, including fungi, have evolved different mechanisms to mobilize and assimilate iron from their surroundings.
One strategy used by the fission yeast Schizosaccharomyces pombe is the synthesis, accumulation, and secretion of the hydroxamate-type siderophore ferrichrome (Fc) (Schrettl et al., 2004b; Haas et al., 2008; Mercier and Labbé, 2010; Plante and Labbé, 2019). The biosynthesis of Fc in S. pombe requires the Sib2, Sib3, and Sib1 proteins (Schrettl et al., 2004b; Brault et al., 2022). First, the ornithine-N5-oxygenase Sib2 catalyzes the N5 hydroxylation of the precursor ornithine. The resulting N5-hydroxyornithine is subsequently acylated by the N5-transacetylase Sib3, forming N5-acetyl-N5-hydroxyornithine. Finally, this compound, in combination with three glycine residues, is used to assemble Fc through the activity of the non-ribosomal peptide synthetase (NRPS) Sib1, a multimeric enzyme capable of generating peptide-based molecules (Schrettl et al., 2004a; Haas et al., 2008; Haas, 2014). Microscopic analyses of S. pombe cells expressing functional fluorescently tagged Sib1, Sib2, and Sib3 proteins have revealed that all three proteins share a common cytosolic subcellular localization under low-iron conditions (Brault et al., 2022). Furthermore, protein–protein interaction studies have shown that Sib2 and Sib3 are interacting partners when cells are cultured under iron-deficient conditions (Brault et al., 2022; Mbuya et al., 2024).
Once synthesized, a portion of Fc is excreted into the extracellular environment to capture ferric ions from various sources (Schrettl et al., 2004b). Despite the relatively low levels of extracellular Fc secreted by S. pombe (less than 5% of its total intracellular Fc content), this extracellular pool of Fc is sufficient to promote the growth of Saccharomyces cerevisiae fet3Δ arn1-4Δ cells expressing the Fc transporter Arn1, as well as Aspergillus nidulans sidAΔ cells, in cross-feeding co-culture assays (Schrettl et al., 2004b; Brault et al., 2022; Chiu et al., 2022). Fc-bound iron (holo-Fc) is subsequently retrieved by S. pombe cells via the cell-surface siderophore transporter Str1 (Pelletier et al., 2003; Plante and Labbé, 2019). Str1 is a member of the major facilitator superfamily (MFS) of transporters (Law et al., 2008). Studies have shown that heterologous expression of S. pombe Str1 complements the Fc assimilation deficiency of an S. cerevisiae mutant strain defective in holo-Fc uptake (Pelletier et al., 2003). Furthermore, experiments using an S. pombe sib1Δ sib2Δ strain, unable to synthesize Fc de novo, have shown that the Fc-dependent growth deficiency of this strain is rescued by Fc supplementation in the presence of a functional Str1 protein (Plante and Labbé, 2019).
The transcription of sib1+, sib2+, and str1+ genes is differentially regulated in response to changes in iron concentrations. Expression of these genes is induced under iron-starvation conditions and repressed when iron is abundant (Plante and Labbé, 2019; Brault et al., 2022). The iron-dependent repression of sib1+, sib2+, and str1+ is primarily governed by the iron-responsive transcription factor Fep1 (Pelletier et al., 2002; Pelletier et al., 2005; Plante and Labbé, 2019; Brault et al., 2022). Under iron-replete conditions, Fep1 interacts with its target genes by recognizing GATA-containing DNA sequences. In contrast, when cells undergo a transition from high to low iron concentrations, Fep1 loses its ability to bind to these GATA-binding elements in vivo, leading to the transcriptional induction of its target genes (Jbel et al., 2009).
The Genome Database for S. pombe, known as PomBase (Rutherford et al., 2024), indicates that Str1 has a paralog called Str2 (SPCC61.01c). Like Str1, Str2 is predicted to be a transmembrane protein, with an arrangement of transmembrane spans and hydrophilic loop regions that is predicted to form a tridimensional structure closely related to members of the major facilitator superfamily (MFS) of transporters (Law et al., 2008). The amino acid sequence identity between Str1 and Str2 is 29.0%, whereas their amino acid sequences show 48.8% similarity. Consistently, the predicted topological structures of these two transmembrane proteins exhibit a high degree of resemblance (Pelletier et al., 2003). However, the physiological role of Str2 in S. pombe remains unclear.
In this study, we determined that str2+ mRNA levels are increased in iron-starved cells, whereas under basal and iron-replete conditions, mRNA levels for this gene are down-regulated. Chromatin immunoprecipitation assays showed that Fep1 is recruited to the str2+ promoter in response to iron. Microscopic analysis revealed that a functional GFP-tagged Str2 localizes to the vacuole membrane in iron-deprived cells or in cells lacking Fep1. Purified vacuoles from str2Δ fep1Δ mutant cells exhibit reduced vacuolar iron content. Under low-iron conditions, str2Δ sib1Δ sib2Δ mutant cells deficient in Fc biosynthesis fail to grow when exogenous holo-Fc is used as the sole source of iron. Together, these results provide the first example of a resident vacuolar membrane-localized siderophore-iron transporter involved in Fc mobilization, which strengthens fission yeast cells against iron starvation.
The genotypes of S. pombe strains are listed in Table 1. Alleles were inactivated using a resistance gene cassette engineered for multiple use in fission yeast (Iwaki and Takegawa, 2004). This cassette contains the kanamycin/G418 resistance gene (kanMX) flanked by loxP sequences on either side. Each disruption cassette is flanked by short DNA segments homologous to the chromosomal sequences lying upstream and downstream of the gene to be deleted. After gene disruption, the cassette can be recycled by excision from the yeast genome using a Cre recombinase/loxP-mediated removal process (Gueldener et al., 2002). Yeast extract with supplements (YES) medium was used to grow yeast strains under standard conditions (Sabatinos and Forsburg, 2010). Edinburgh minimal medium (EMM) lacking specific nutrients was used to select transformed yeast strains carrying integrative or episomal plasmids.
For transcript and protein steady-state level assessments, yeast liquid cultures were seeded to an OD600 of 0.5. When the cultures reached an OD600 of 1.0, cells were either left untreated or treated with the iron chelator 2,2′-dipyridyl (Dip, 250 μM) or FeCl3 (100 μM) for 1.5 h or 3 h, unless otherwise stated. For vacuole purification, the indicated cultures were treated with Dip for 3 h, with holo-Fc (1 μM) added during the final hour of treatment. Growth assays on solid media were performed by growing cells in YES medium to an OD600 of 1.0, then spotting serial dilutions (6,000 cells/10 μL; 600 cells/10 μL; and, 60 cells/10 μL) onto media without Dip or Fc supplementation (control) or supplemented with Dip (140 μM) or a combination of Dip and Fc (0.1 or 1 μM).
To create the integrative plasmid pJK-1000str2+-GFP, a 2,791-bp EcoRI-BamHI PCR-amplified DNA fragment containing the str2+ allele and its promoter region (starting from 1,000 bp upstream the initiator codon to the penultimate codon) was cloned into the EcoRI and BamHI sites of pJK148 (Keeney and Boeke, 1994). This plasmid was named pJK-1000str2+nostop. The GFP coding sequence was then amplified by PCR using primers designed to introduce BamHI and NotI restriction sites at the 5′ and 3′ ends, respectively. The resulting BamHI-NotI PCR-amplified DNA fragment was inserted in-frame with str2+ at the corresponding sites in pJK-1000str2+nostop, generating the plasmid pJK-1000str2+-GFP. This plasmid was subsequently used to introduce mutations into the str2+ coding sequence. In the str2Y539A/Y553A mutant allele, the codons for tyrosine (Tyr) at positions 539 and 553 were replaced by nucleotide triplets encoding alanine residues. For the str2Y539A/R546A/Y553A mutant allele, the same substitutions as in str2Y539A/Y553A were made, with an additional mutation replacing the arginine (Arg) codon at position 546 with a codon for alanine. These site-specific substitutions were introduced using a PCR overlap extension method (Ho et al., 1989). The construction of the plasmids pJK-1478fep1+, pJK-1478TAPfep1+, and pSP-808abc3+-GFP has been described previously (Pelletier et al., 2005; Pouliot et al., 2010).
Total RNA was extracted from cell cultures using the hot phenol method, as previously described (Chen et al., 2003). To analyze the transcript levels of str2+ and str1+, real-time quantitative reverse transcription PCR (RT-qPCR) assays were performed, as previously described (Brault et al., 2022). The procedures for RT reactions, cDNA synthesis, and qPCR reactions were carried out as previously reported (Brault et al., 2022). Each target transcript was analyzed in experiments with a minimum of three biological replicates, with each sample reaction performed in triplicate. Results were considered valid if the target-specific fluorescent signal produced a Ct value ≤37 cycles, and all positive and negative control reactions yielded successful amplification and no amplification, respectively. Fold changes in str2+ or str1+ transcript levels between wild-type and fep1Δ mutant samples were calculated using the ΔΔCt method, normalized to the internal control act1+ (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008; Protacio et al., 2022). The following equation was used for calculations: ΔΔCt = [(Ct gene–Ct ref) in wild-type] versus [(Ct gene–Ct ref) in fep1Δ]; or, ΔΔCt = [(Ct gene–Ct ref) in sib1Δ sib2Δ] versus [(Ct gene–Ct ref) in sib1Δ sib2Δ fep1Δ], under the indicated experimental conditions related to iron availability. In the case of str2+, the primer pair allowed the detection of an amplicon corresponding to the coding region between positions +502 and + 601 down to the first nucleotide of the initiator codon. For str1+, the amplicon covered the coding region from positions +376 to +475, down to the A of the start codon. To detect act1+ expression, a primer pair was used to amplify the coding sequence between positions +173 and + 280 down to the first base of the ATG codon of act1+.
Early logarithmic fep1Δ php4Δ cells expressing untagged or TAP-tagged fep1+ alleles were grown in the presence of FeCl3 (Fe, 75 μM). When cultures reached an OD600 of 0.5, they were washed and incubated with either Dip (250 μM) or FeCl3 (Fe, 100 μM) for 3 h. Following the treatments, in vivo chemical cross-linking of proteins was performed by incubating the cell cultures in the presence of 1% formaldehyde for 20 min. The crosslinking reaction was neutralized by adding glycine (0.45 M), and cell lysates were prepared by glass bead disruption, as previously described (Larochelle et al., 2012; Brault et al., 2016). The samples were subsequently sonicated to shear the chromatin DNA into fragments of 500 to 1,000 bp. Immunoprecipitation of TAP-tagged Fep1 bound to chromatin was conducted using immunoglobulin G (IgG)-Sepharose beads, following the procedures as previously described (Jacques et al., 2014). Bead manipulation, which includes washings, elution, reversal cross-linking, and DNA precipitation, was performed in accordance with the protocols previously described (Adam et al., 2001; Jbel et al., 2009). Quantification of the immunoprecipitated DNA was conducted by quantitative real-time PCR (qPCR) using different sets of primers targeting the promoter regions of str2+ and str1+. The occupancy of TAP-Fep1 at the str2+ or str1+ loci was determined by calculating the enrichment of the specific str2+ and str1+ promoter regions relative to a GATA-free 18S ribosomal DNA coding region, which served as an internal background control. The primer pairs used for amplifying the str2+ and str1+ promoter regions were: str2-105 (5’-CCAACTTCATTAAACATCTCGGTTAG-3′)/str2-2 (5’-CAGAGTGTATGGTAAATGGCAGTA-3′), and str1-921 (5’-GACAGTCCCGTACAAGGAAGAA-3′)/str1-812 (5’-AAGATGGAGGTGAAGGCAACTT-3′), respectively. The primers used for amplifying the 18S ribosomal DNA coding region were described previously (Brault et al., 2016). Each qPCR reaction was performed in triplicate using the Perfecta SYBR Green Fast mix (Quanta) on a CFX96 Touch Real-Time PCR instrument (BioRad). All ChIP experiments were repeated at least three times with independent chromatin preparations.
For all growth conditions, phenylmethylsulfonylfluoride (PMSF) (1 mM) was added directly to the cell cultures 15 min before harvesting to protect proteins from proteolysis. Whole cell extracts were prepared with glass beads using a FastPrep-24 instrument (MP Biomedicals, Solon, OH). Cells were lysed in HEGN100 buffer containing 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.9, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 10% glycerol, 0.1 mM Na3VO4, 1 mM PMSF, 1 mM dithiothreitol (DTT), and a complete protease inhibitor mixture (Sigma-Aldrich; P8340). Cell lysates were incubated in the presence of Triton X-100 (1%) for 30 min at 4°C. Equal concentrations of each sample were resuspended in loading buffer (100 mM Tris–HCl, pH 7.5, 1.4 M β-mercaptoethanol, 140 mM sodium dodecyl sulfate (SDS), 5 mM EDTA, 4 M urea, 1 M thiourea, and 0.72 mM bromophenol blue), and proteins were resolved by electrophoresis on 6% SDS-polyacrylamide gels. Proteins were then transferred to nitrocellulose membranes, and the following antibodies were used for immunodetection of Str2-GFP, Abc3-GFP, and α-tubulin: monoclonal anti-GFP antibody B-2 (Santa Cruz Biotechnology) and monoclonal anti-α-tubulin antibody B-5-1-2. After incubation, the membranes were washed and incubated with appropriate horseradish peroxidase-conjugated secondary antibodies, developed with enhanced chemiluminescence (ECL) reagents (Amersham Biosciences), and visualized using an ImageQuant LAS 4000 instrument (GE Healthcare).
Cells were subjected to microscopic analysis using 1,000× magnification using the following filters: 340–380 nm (bimane-GS), 465–495 nm (GFP-tagged proteins), and 510–560 nm (FM4-64). Both fluorescence and differential interference contrast (DIC) images (Nomarski) were captured using a Nikon Eclipse E800 epifluorescence microscope equipped with a Hamamatsu ORCA-ER digital cooled camera. The representative fields shown in the images were obtained from a minimum of three independent experiments. Furthermore, the cell fields shown represent protein localization in 200 cells tested per condition.
The indicated strains were subjected to cell wall digestion, cell-surface membrane disruption, and then cell fractionation using a differential centrifugation method as previously described (Sooksa-Nguan et al., 2009). After the second Percoll step gradient (50%, v/v), the integrity of the isolated vacuoles was assessed by monitoring their ability to actively retain the fluorescent compound bimane-GS. This fluorescent compound is derived from the nonfluorescent, membrane-permeant monochlorobimane. Upon entering the cells, monochlorobimane is glutathionylated by cellular glutathione S-transferases, resulting in the production of bimane-GS, which is actively transported into the vacuoles, where it becomes fluorescent (Sarry et al., 2007).
Vacuole preparations were used either to quantify iron content or to detect the presence of GFP-tagged Str2 and Abc3 proteins. For protein detection, purified vacuoles were disrupted using glass beads in Thorner buffer (40 mM Tris–HCl, pH 6.8, 0.1 mM EDTA, 5% SDS and 8 M urea) supplemented with 1% Triton X-100. Vacuole lysates were incubated in a thermomixer at 37°C for 15 min with periodic rotation at 600 rpm, followed by an additional round of glass bead disruption using a MP-24 FastPrep instrument. The extracted vacuolar proteins were then subjected to immunoblot assays to detect Str2-GFP and Abc3-GFP.
For measuring vacuole iron content, the vacuole preparations were washed and lysed in a Tris–HCl solution (50 mM, pH 7.5) supplemented with Triton X-100 (1%), using glass beads disruption. The protein concentrations of the vacuolar lysates were determined using the Bradford assay, and equal amounts of these extracts were treated with citric acid (100 mM, pH 2.0) and incubated at 60°C for 4 h, as previously described (Rad et al., 2007). After centrifugation, the supernatant was mixed with an equivalent volume of citric acid and transferred to a fresh tube. BPS (5 mM) and freshly prepared ascorbic acid (100 mM) were then added. After 45 min of incubation at 25°C in the dark, the absorbance of the samples containing iron was measured, and the total iron content was calculated using a separate calibration curve, as previously described (Rad et al., 2007).
Although previous studies have identified target genes of Fep1 (Pelletier et al., 2003; Rustici et al., 2007), several remain poorly characterized in terms of their roles in iron homeostasis, including the str2+-encoded putative siderophore transporter. To validate that str2+ expression is repressed by iron repletion, we monitored str2+ transcript levels in wild-type or sib1Δ sib2Δ strains either left untreated or treated with the iron chelator 2,2′-dipyridyl (Dip, 250 μM) or FeCl3 (Fe, 100 μM) for 90 min. As a control, we concurrently analyzed str1+ transcripts, which are known to be down-regulated under iron-replete conditions (Pelletier et al., 2003; Plante and Labbé, 2019). For both strains, results showed that str2+ and str1+ transcript levels were highly expressed in the presence of Dip (Figures 1A–D). In contrast, their expression markedly decreased under basal and iron-replete conditions. In the wild-type strain, str2+ and str1+ mRNA levels were repressed 2.4- and 37.0-fold, respectively, in response to iron compared to levels under iron-starved conditions (Figures 1A,B). Similarly, in the sib1Δ sib2Δ mutant strain, str2+ and str1+ transcript levels were repressed 2.0- and 27.0-fold, respectively, by iron repletion compared to levels under low-iron conditions (Figures 1C,D). To examine the role of Fep1 in str2+ gene regulation, similar experiments were performed using isogenic fep1Δ and sib1Δ sib2Δ fep1Δ strains. In both strains lacking Fep1, str2+ and str1+ transcripts exhibited high and constitutive expression levels that were unresponsive to iron for repression (Figures 1A–D). However, when the wild-type fep1+ allele or a functional TAP-tagged fep1+ allele was reintroduced by integration into fep1Δ and sib1Δ sib2Δ fep1Δ strains, the ability to repress str2+ and str1+ gene expression under basal and iron-replete conditions was restored (Figures 1C,D). Collectively, these results demonstrated that str2+ is an iron-regulated gene, with its expression moderately repressed by Fep1 in response to iron.
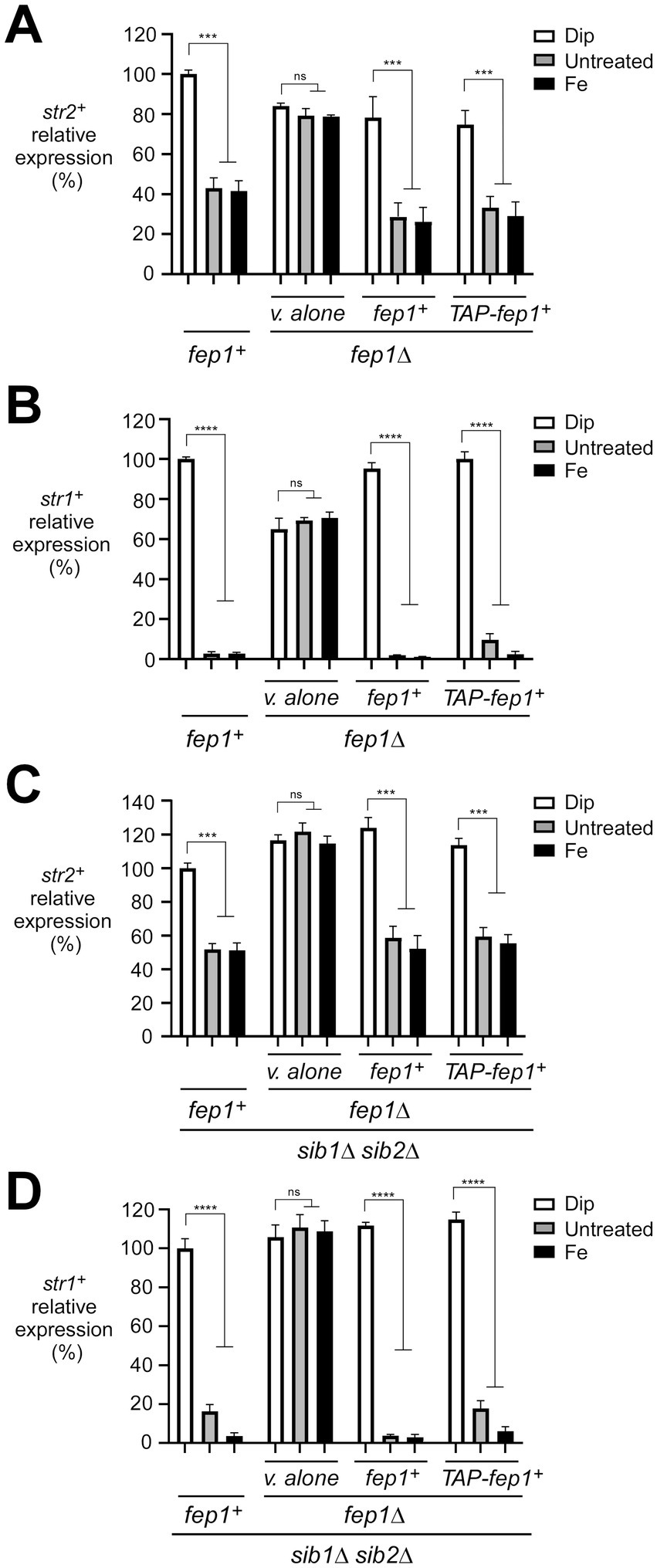
Figure 1. Assessment of the str2+ and str1+ transcript levels in response to iron availability. (A–D) Representative expression profiles of the str2+ and str1+ mRNAs in wild-type (fep1+), fep1Δ, sib1Δ sib2Δ, and fep1Δ sib1Δ sib2Δ strains. The cells were grown in YES medium to an OD600 of 1.0, followed by treatment with either Dip (250 μM) or FeCl3 (Fe, 100 μM) for 90 min, or left untreated. For the fep1Δ or fep1Δ sib1Δ sib2Δ strains, cells were transformed with either an empty integrative plasmid (v. alone) or a plasmid containing an untagged fep1+ or TAP-tagged fep1+ allele. After RNA isolation, the steady-state mRNA levels of str2+ and str1+ were analyzed by RT-qPCR assays. The graphs represent the quantification from three independent RT-qPCR experiments, with error bars indicating standard deviation (± SD; error bars). Statistical significance is represented by asterisks: p < 0.001 (***) and p < 0.0001 (****) (two-way ANOVA with Tukey’s multiple comparisons test, comparing the indicated strains grown under low-iron conditions), whereas “ns” denotes no significant difference. str1+ was analyzed as a control gene, known to be repressed by iron.
To further investigate whether Fep1 physically occupies the str2+ and str1+ promoters in an iron-dependent manner, we used a fep1Δ php4Δ mutant strain expressing either an untagged or TAP-tagged fep1+ allele, which had been re-integrated. Since this mutant strain lacks Php4, it ensures that the transcription of the re-integrated fep1+ or TAP-fep1+ alleles occurs independently of Php4, allowing their constitutive expression regardless of cellular iron levels (Mercier et al., 2008; Jbel et al., 2009; Mercier and Labbé, 2009). Thus, this biological system enabled us to separate the iron-dependent DNA binding activity of Fep1 and TAP-Fep1 from potential changes in their gene expression. Under this setup context, we used a ChIP approach to test whether the presence of TAP-Fep1 could be detected at the str2+ and str1+ promoters in vivo (Figure 2A). We probed for promoter occupancy using primers specific to the str2+ and str1+ promoter regions, which are known to contain GATA elements that are bound by Fep1 in vitro (Pelletier et al., 2003). ChIP analysis revealed that TAP-Fep1 occupied the str2+ and str1+ promoters at maximum levels when cells were treated with FeCl3 (100 μM). Under these conditions, TAP-Fep1 occupancy at the str2+ and str1+ promoters increased 7.9- and 10.7-fold, respectively, compared to a control region encoding the 18S ribosomal RNA (Figure 2B). In contrast, when fep1Δ php4Δ cells expressing TAP-Fep1 were treated with Dip (250 μM), only low levels of str2+ and str1+ promoter fragments were immunoprecipitated (Figure 2B). In iron-replete cells, TAP-Fep1 exhibited 8.8- and 13.4-fold higher binding to the str2+ and str1+ promoter regions, respectively, compared to iron-starved cells expressing TAP-Fep1 (Figure 2B). As negative controls, untagged Fep1 immunoprecipitated only background levels of the str2+ and str1+ promoter regions (Figure 2B). Collectively, these results showed that TAP-Fep1 is recruited to str2+ and str1+ promoters under conditions of high levels of iron.
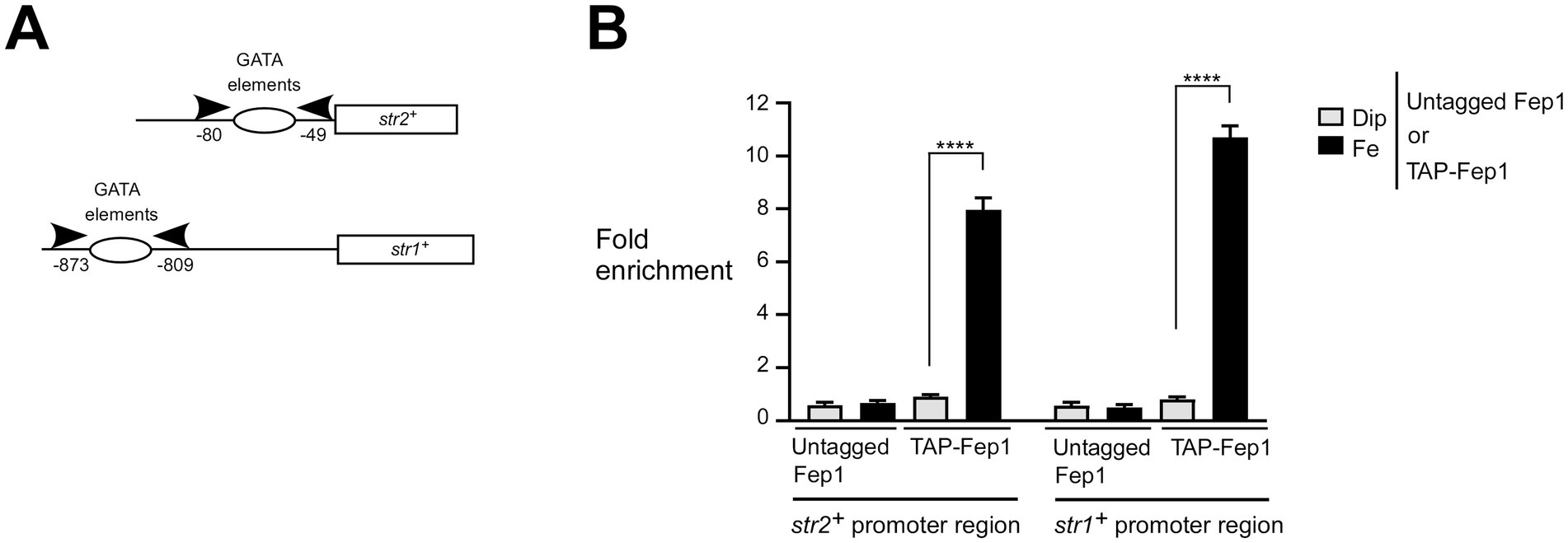
Figure 2. Fep1 binds to str2+ and str1+ promoters under iron-replete conditions. (A) Schematic representation of the str2+ and str1+ promoter regions. Arrowheads show primer positions for qPCR analysis. Nucleotide numbers correspond to the positions of primer binding relative to the A of the initiator codon of the str2+ or str1+ gene. Empty ovals depict promoter regions containing GATA elements, which are known to serve as binding sites for Fep1. (B) Logarithmic phase fep1Δ php4Δ cells expressing untagged or TAP-tagged fep1+ alleles were incubated in the presence of Dip (250 μM) or FeCl3 (Fe, 100 μM) for 3 h. Following chromatin preparation and immunoprecipitation using Sepharose-bound anti-mouse IgG antibodies, specific regions of the str2+ and str1+ promoters were analyzed by qPCR to assess TAP-Fep1 occupancy. TAP-Fep1 binding to the str2+ (positions −80 to −49) and str1+ (positions −873 to −809) promoter regions was calculated by measuring the enrichment of specific amplified str2+ and str1+ promoter fragments relative to an 18S ribosomal DNA coding region. ChIP data were calculated as values of the largest amount of chromatin measured (fold enrichment). Results are shown as averages ± SD from three independent experiments, each performed in biological triplicate. Asterisks indicate statistical significance (****p < 0.0001, one-way ANOVA with Dunnett’s multiple comparisons test, comparing iron-replete cells expressing TAP-Fep1).
The iron- and Fep1-dependent regulation of str2+ expression led us to investigate whether the steady-state protein levels of Str2 mirrored the changes in str2+ transcript levels as a function of iron availability. A functional str2+-GFP allele, expressed under the control of its own promoter, was integrated into the genomes of str2Δ, str2Δ fep1Δ, str2Δ sib1Δ sib2Δ, and str2Δ fep1Δ sib1Δ sib2Δ mutant strains. The first two strains were grown to an OD600 of 1.0 and either left untreated or treated with Dip (250 μM) or FeCl3 (100 μM) for 3 h. The last two strains, which are deficient in Fc biosynthesis, were grown under same conditions as the first two. However, when they reached an OD600 of 1.0, Dip-treated cells were either incubated without Fc supplementation or supplemented with holo-Fc (1 μM) during the final hour of treatment. Whole cell extracts were then prepared and analyzed by immunoblotting. The results showed that Str2-GFP steady-state levels correlated with str2+ transcript levels, increasing in the presence of Dip but remaining low in untreated or iron-treated str2Δ and str2Δ sib1Δ sib2Δ cells harboring a str2+-GFP allele (Figures 3A,B). In contrast, in str2Δ fep1Δ and str2Δ fep1Δ sib1Δ sib2Δ cells expressing str2+-GFP, the steady-state levels of Str2-GFP remained higher in both untreated and iron-treated conditions compared to strains with a functional fep1+ allele (Figures 3A,B). Furthermore, steady-state levels of Str2-GFP were detected when str2Δ sib1Δ sib2Δ and str2Δ fep1Δ sib1Δ sib2Δ strains harboring an integrated str2+-GFP allele were incubated in the presence of Dip with Fc supplementation (1 μM) (Figure 3B).
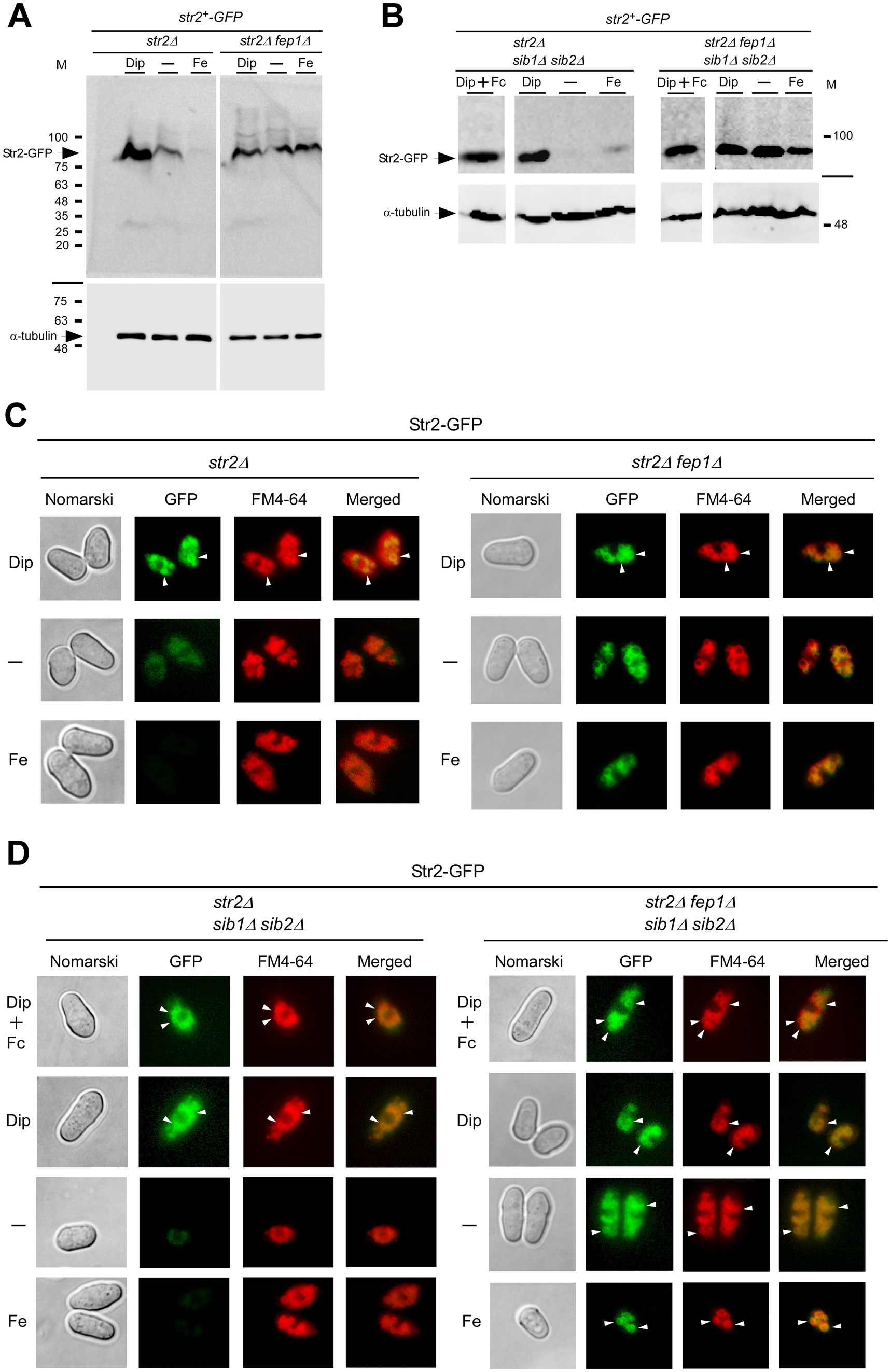
Figure 3. Effect of fep1Δ deletion on Str2 protein expression and localization. (A,B) str2Δ, str2Δ fep1Δ, str2Δ sib1Δ sib2Δ, and str2Δ fep1Δ sib1Δ sib2Δ strains expressing Str2-GFP were grown to an OD600 of 1.0. The cultures were then either left untreated (−) or treated with Dip (250 μM) or FeCl3 (Fe, 100 μM) for 3 h. In the case of str2Δ sib1Δ sib2Δ and str2Δ fep1Δ sib1Δ sib2Δ strains, Dip-treated cells were either left without further supplementation or supplemented with holo-Fc (1 μM) during the final hour of treatment. Whole cell extracts were analyzed by immunoblot assays with anti-GFP and anti-α-tubulin antibodies. The positions of molecular weight markers (in kDa) are indicated on the right side. (C,D) Fluorescence microscopy was performed on cells incubated from each group of cultures described in panels A and B to visualize the localization of Str2-GFP (center left). Cell morphology was examined using Nomarski optics (far left). White arrowheads point to examples of vacuole membranes. FM4-64 staining (center right), a marker of vacuolar membranes, was also visualized by fluorescence microscopy. Merged images of Str2-GFP and FM4-64 are shown in the far-right panels. The microscopy results are representative of three independent experiments, each performed in biological triplicate.
Next, we aimed to determine the subcellular localization of Str2-GFP when expressed in untreated, iron-replete, and iron-starved str2Δ, str2Δ fep1Δ, str2Δ sib1Δ sib2Δ, and str2Δ fep1Δ sib1Δ sib2Δ strains. Moreover, we examined the Str2-GFP fluorescent signal in str2Δ sib1Δ sib2Δ and str2Δ fep1Δ sib1Δ sib2Δ strains that had been incubated with exogenous Fc (1 μM) under low-iron conditions. Fluorescence microscopy analysis of iron-starved str2Δ and str2Δ sib1Δ sib2Δ cells expressing Str2-GFP revealed that Str2-GFP-mediated fluorescence was localized to the vacuole membranes, regardless of the presence of exogenous Fc in the case of str2Δ sib1Δ sib2Δ cells (Figures 3C,D). The Str2-GFP signal colocalized with the vacuole-staining dye FM4-64, which served as a marker for the vacuolar membrane (Figures 3C,D). Consistent with iron-dependent repression of str2+ expression, Str2-GFP fluorescence levels were markedly reduced in Str2-GFP-expressing str2Δ and str2Δ sib1Δ sib2Δ cells grown under basal or high-iron conditions (Figures 3C,D). In contrast, the fluorescence signal at the vacuole membrane persisted when GFP-tagged str2+ was expressed in str2Δ fep1Δ and str2Δ fep1Δ sib1Δ sib2Δ strains under all tested conditions (Figures 3C,D). Taken together, these results led us to conclude that Str2 functions at the vacuole membrane in iron-starved cells, regardless of whether they are Fc prototrophic or auxotrophic. Furthermore, the vacuolar localization of Str2 remains unchanged in the presence of exogenous Fc.
When the fep1+ gene is disrupted, fep1 null strains exhibit substantially increased expression of several genes encoding proteins involved in cellular iron homeostasis, such as iron transporters, iron–sulfur proteins, and iron-consuming proteins (Rustici et al., 2007; Brault et al., 2015). Based on these previous findings, we used str2Δ fep1Δ and abc3Δ fep1Δ mutant strains expressing re-integrated str2+-GFP and abc3+-GFP alleles, respectively. Moreover, for the str2Δ fep1Δ strain, an integrative empty vector was transformed as a negative control. Cells were grown to an OD600 of 1.0 and then incubated with Dip (250 μM) for 3 h. During the final hour of treatment, holo-Fc (1 μM) was added, and aliquots of the cells were subsequently visualized using fluorescence microscopy. The results showed that Str2-GFP-associated fluorescence was detected at the vacuolar membrane (Figure 4A). To further validate the vacuolar localization of Str2-GFP, the vacuolar-sequestered fluorescent compound bimane-GS was used as a marker (Sarry et al., 2007). Merged images showed that Str2-GFP and bimane-GS shared a similar subcellular localization pattern within the cells (Figure 4A). As an additional control, we examined the vacuolar localization of Abc3-GFP, a known vacuolar membrane transporter (Pouliot et al., 2010). Under identical growth conditions, Abc3-GFP fluorescence was observed at the vacuoles and co-localized with the bimane-GS marker (Figure 4A). In contrast, no green fluorescence signal was detected in str2Δ fep1Δ cells containing an empty vector (Figure 4A). To distinguish protein localization at the vacuole from other organelles, we used cox4Δ fep1Δ cells expressing Cox4-Cherry, a protein known to be a mitochondrial resident marker. Results showed that the fluorescence associated with Cox4-Cherry exhibited a distinct localization pattern compared to the subcellular localization of Str2-GFP and showed no overlap with the bimane-GS signal (Figure 4A).
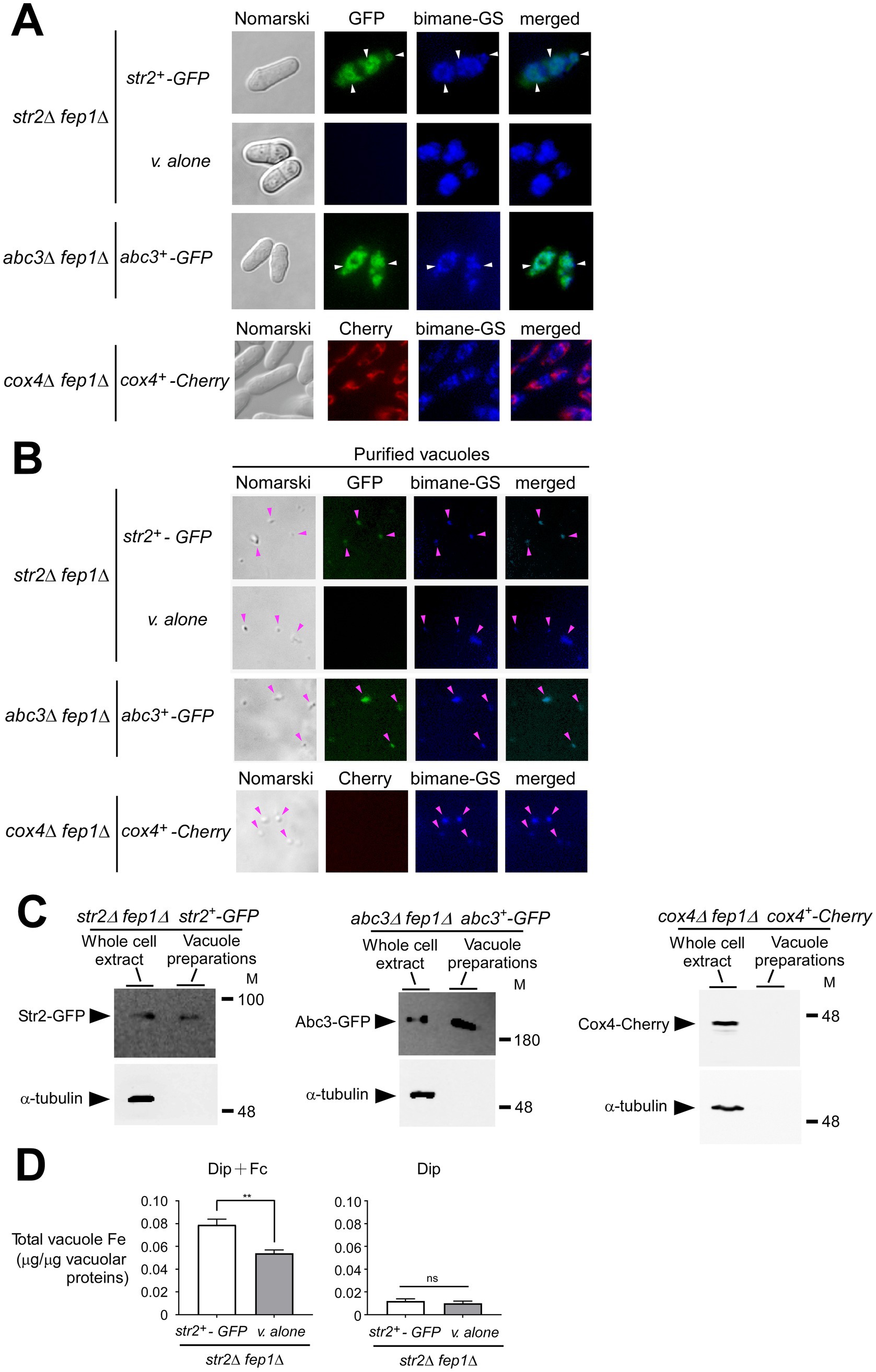
Figure 4. Str2 co-purifies with yeast vacuoles that exhibit iron accumulation. (A) str2Δ fep1Δ cells expressing either an empty plasmid (v. alone) or the str2+-GFP allele were grown in YES medium to an OD600 of 1.0. The cultures were then incubated with Dip (250 μM) for 3 h. In the final hour of this treatment, holo-Fc (1 μM) was added, followed by vacuole isolation from each group of cultures. For the abc3Δ fep1Δ and cox4Δ fep1Δ mutant strains, the abc3+-GFP and cox4+-Cherry alleles were reintroduced, and these cells were cultured under the same conditions as the str2Δ fep1Δ cells expressing GFP-tagged Str2. The above-mentioned cultures were examined by fluorescence microscopy to visualize the cellular localization of Str2-GFP, Abc3-GFP, and Cox4-Cherry (center left), along with the accumulation of fluorescent bimane-GS (center right). Merged images of GFP or Cherry and bimane-GS fluorescent signals are shown in the far-right panels. Cell morphology was examined using Nomarski optics (far left). White arrowheads indicate examples of vacuole membranes. (B) Vacuoles were purified from each group of cultures described in panel A and visualized by fluorescence microscopy to observe Str2-GFP, Abc3-GFP, Cox4-Cherry, and bimane-GS fluorescent signals. Merged images of GFP and bimane-GS fluorescent signals are displayed in the far-right panels. Pink arrowheads point to examples of purified vacuoles. (C) Aliquots of total cell extracts and vacuole preparations from each group of cultures were analyzed by immunoblotting using anti-GFP, anti-Cherry, and anti-α-tubulin antibodies. Abc3-GFP was used as a known vacuolar membrane marker, whereas the absence of the Cox4-Cherry and α-tubulin signals confirmed the specificity of the vacuole preparations. (D) Purified vacuoles from str2Δ fep1Δ cells expressing an empty plasmid (v. alone) or the str2+-GFP allele were analyzed using a BPS-based spectrophotometric method to quantitatively measure iron levels. Cells were incubated with Dip (250 μM) for 3 h. In the final hour of this treatment, holo-Fc (1 μM) was added or omitted. Results are representative of three independent experiments. Data are presented as mean ± SD. Statistical significance is indicated by asterisks, with **p < 0.01 (determined by one-way ANOVA with Dunnett’s multiple comparisons test, comparing against cells expressing Str2-GFP).
Vacuoles were purified from the above-mentioned cultures, and sample aliquots were examined by fluorescence microscopy. The results indicated that the vacuoles maintained their integrity throughout the purification process, as the bimane-GS-associated blue fluorescence was retained in most of the isolated vacuoles (Figure 4B). Furthermore, some of these isolated vacuoles exhibited a green fluorescence signal when purified from str2Δ fep1Δ and abc3Δ fep1Δ cells expressing str2+-GFP and abc3+-GFP, respectively (Figure 4B). In contrast, vacuoles isolated from str2Δ fep1Δ cells containing an empty vector or cox4Δ fep1Δ cells expressing Cox4-Cherry lacked any green and red fluorescence signal, respectively.
Proteins were extracted from vacuole preparations and analyzed by immunoblot assays. Str2-GFP was detected as vacuolar protein when yeast vacuoles were isolated from str2Δ fep1Δ cells expressing str2+-GFP (Figure 4C). Similarly, Abc3-GFP, a known vacuolar membrane protein, was detected when vacuolar proteins were analyzed by immunoblotting from Abc3GFP-expressing abc3Δ fep1Δ cells (Figure 4C). In contrast, when proteins were extracted from vacuole preparations, immunoblot experiments consistently showed no signal corresponding to Cox4-Cherry.
Vacuoles isolated from str2Δ fep1Δ cells expressing str2+-GFP or containing an empty plasmid were analyzed for their iron content by a BPS-based spectrophotometric method (Rad et al., 2007; Pouliot et al., 2010). In str2Δ fep1Δ cells treated with Fc and expressing str2+-GFP, purified vacuoles exhibited a total iron concentration of 0.079 μg per μg of vacuolar proteins (Figure 4D). In contrast, vacuoles from str2Δ fep1Δ cells harboring an empty plasmid showed 31.7% less iron (0.054 μg iron/μg of vacuolar proteins) compared to those isolated from Str2GFP-expressing str2Δ fep1Δ cells (Figure 4D). As a control, vacuoles isolated from str2Δ fep1Δ cells expressing str2+-GFP or carrying an empty plasmid, and incubated with Dip without Fc supplementation, displayed very low total iron concentrations (0.012 and 0.010 μg iron/μg of vacuolar proteins, respectively) (Figure 4D). Interestingly, these results showed that adding Fc in the absence of Str2-GFP led to an increase in vacuolar iron concentration, although not to the same extent as when Str2-GFP was expressed. This observation suggests the possible existence of an alternative transport system that facilitates iron transport to the vacuole, independent of Str2. Nonetheless, these findings strongly suggested that Str2 plays a role in mobilizing iron within the vacuole when cells are grown with holo-Fc as the sole source of iron.
The Fc biosynthetic pathway is essential for S. pombe survival under iron-limiting conditions (Mercier and Labbé, 2010; Brault et al., 2022). The sib1+ and sib2+ genes are necessary for Fc production in S. pombe (Schrettl et al., 2004b; Mercier and Labbé, 2010; Brault et al., 2022). Yeast strains with deletions of these two genes (sib1Δ sib2Δ) are unable to grow on iron-poor media supplemented with Dip (Figure 5A; Mercier and Labbé, 2010; Brault et al., 2022). Notably, this growth defect due to iron deficiency was rescued by adding exogenous Fc (0.1 and 1 μM) to the medium (Figure 5A). To determine whether Str2 is required for the utilization of exogenous Fc, the str2+ gene was disrupted in the sib1Δ sib2Δ strain and tested on Dip-supplemented media containing 0.1 and 1 μM Fc. As shown in Figure 5A, sib1Δ sib2Δ str2Δ mutant cells exhibited a severe growth defect on this Fc-supplemented medium compared to wild-type and sib1Δ sib2Δ cells expressing str2+ (Figure 5A). Given the established role of Str1 in the uptake of exogenous Fc (Plante and Labbé, 2019; Brault et al., 2022), we validated that its inactivation (str1Δ) in the sib1Δ sib2Δ strain led to an inability to grow on Dip-supplemented media containing 0.1 and 1 μM Fc (Figure 5A). Moreover, the results consistently showed that the sib1Δ sib2Δ str1Δ str2Δ quadruple mutant strain was unable to grow in the presence of exogenous Fc under low-iron conditions (Figure 5A).
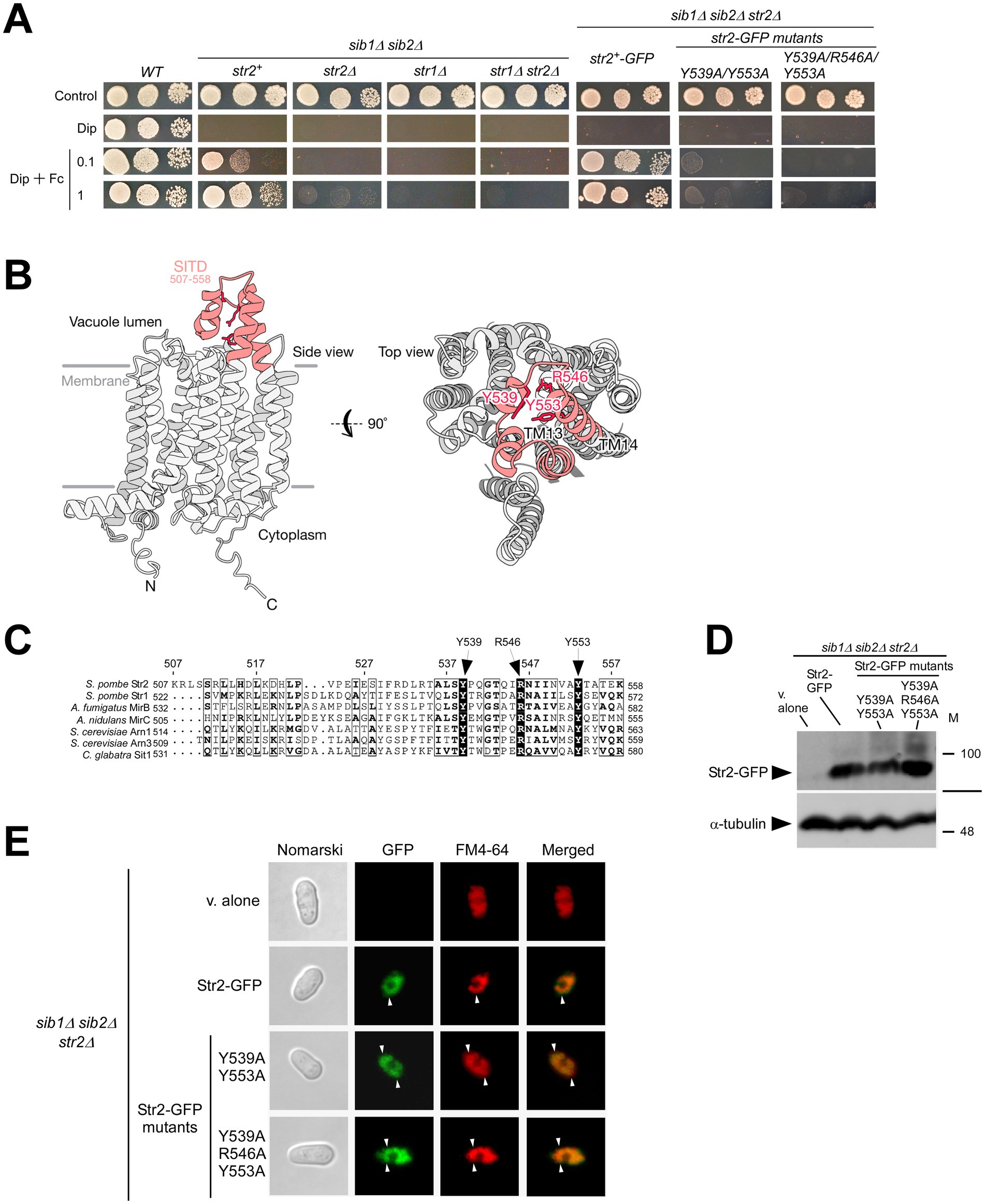
Figure 5. sib1Δ sib2Δ mutant cells require Str2 for Fc-dependent growth. (A) Wild-type (WT), sib1Δ sib2Δ, sib1Δ sib2Δ str2Δ, sib1Δ sib2Δ str1Δ, and sib1Δ sib2Δ str1Δ str2Δ cells, as well as sib1Δ sib2Δ str2Δ cells expressing str2+-GFP, str2-Y539A/Y553A-GFP or str2-Y539A/R546A/Y553A-GFP alleles were grown in YES medium to an OD600 of 1.0, and then spotted in serial dilutions (6,000 cells/10 μL; 600 cells/10 μL; and, 60 cells/10 μL) onto medium without Dip or Fc supplementation (control) or supplemented with Dip (140 μM) or a combination of Dip and Fc (0.1 or 1 μM). (B) A predicted three-dimensional structure of Str2 is shown, with potential transmembrane-spanning domains indicated in gray and the STID highlighted in red. (C) Amino acid alignment of the predicted carboxyl-terminal final loop of S. pombe Str2 with other predicted final loops found in S. pombe Str1, A. fumigatus MirB, A. nidulans MirC, C. glabrata Sit1, and S. cerevisiae Arn1 and Arn3. Arrows indicate three highly conserved Tyr (Y) and Arg (R) residues. Amino acid sequence numbers refer to their position relative to the first amino acid of each protein. (D) Whole extracts from aliquots of iron-starved sib1Δ sib2Δ str2Δ cells expressing an empty plasmid (v. alone), str2+-GFP, str2-Y539A/Y553A-GFP or str2-Y539A/R546A/Y553A-GFP alleles were analyzed by immunoblotting using anti-GFP and anti-α-tubulin antibodies. The positions of molecular weight markers (in kDa) are indicated on the right side. (E) sib1Δ sib2Δ str2Δ cells expressing an empty plasmid (v. alone), str2+-GFP, str2-Y539A/Y553A-GFP or str2-Y539A/R546A/Y553A-GFP alleles treated with Dip were analyzed by fluorescence microscopy to detect GFP fluorescence (center left), along with FM4-64 staining (center right). Merged images of GFP and FM4-64 signals are shown in the far-right panels. Cell morphology was examined using Nomarski optics (far left). White arrowheads indicate examples of vacuole membranes.
Amino acid sequence analysis of Str2 suggests that the protein belongs to the MFS-type transporter family (Rutherford et al., 2024). Topological models of Str2 predict the presence of 14 transmembrane spans connected by hydrophilic loops (Figure 5B). The final loop is predicted to contain a putative siderophore transporter domain (SITD) with highly conserved amino acid residues (Nevitt and Thiele, 2011). Among these conserved residues, Tyr539, Arg546, and Tyr553 in Str2 are found within the SITD of other predicted or known hydroxamate-type siderophore transporters, including Str1 (S. pombe), MirB (Aspergillus fumigatus), MirC (Aspergillus nidulans), Sit1 (Candida glabrata), Arn1, and Arn3 (S. cerevisiae) (Figure 5C; Yun et al., 2000; Kim et al., 2002; Kim et al., 2005; Philpott, 2006; Haas et al., 2008; Nevitt and Thiele, 2011; Raymond-Bouchard et al., 2012; Plante and Labbé, 2019). Based on previous studies that had demonstrated the functional importance of conserved Tyr residues in the SITD domain of S. pombe Str1 (Tyr553 and Tyr567) and C. glabrata Sit1 (Tyr575) (Nevitt and Thiele, 2011; Plante and Labbé, 2019), we generated two mutant derivatives of Str2. In the first mutant, Tyr539 and Tyr553 were substituted with Ala residues, whereas in the second mutant, Tyr539, Arg546, and Tyr553 were replaced by Ala residues. To assess the role of Str2 in Fc-dependent growth under iron-deficient conditions, spot assays were performed using sib1Δ sib2Δ str2Δ cells expressing either str2+-GFP, str2-Y539A/Y553A-GFP, or str2-Y539A/R546A/Y553A-GFP allele. As shown in Figure 5A, sib1Δ sib2Δ str2Δ cells expressing str2+-GFP exhibited growth on Dip-supplemented media containing 0.1 and 1 μM Fc. In contrast, sib1Δ sib2Δ str2Δ cells expressing str2-Y539A/Y553A-GFP or str2-Y539A/R546A/Y553A-GFP allele displayed a severe growth defect when spotted on iron-depleted medium supplemented with Fc (0.1 and 1 μM) compared to those expressing str2+-GFP (Figure 5A).
To confirm that all GFP-tagged str2 alleles were expressed in sib1Δ sib2Δ str2Δ cells, steady-state protein levels of Str2-GFP and its mutant derivatives were analyzed by immunoblot assays. The results showed that all these proteins were expressed in the sib1Δ sib2Δ str2Δ strain (Figure 5D). As a negative control, whole cell extracts from sib1Δ sib2Δ str2Δ cells transformed with an empty plasmid were also analyzed by immunoblotting.
To ensure that the mutated forms, Str2-Y539A/Y553A-GFP and Str2-Y539A/R546A/Y553A-GFP, exhibited the same subcellular localization as the wild-type Str2-GFP protein, microscopic analyses were performed on the two GFP-tagged mutants alongside Str2-GFP. The results showed that Str2-GFP and its mutant derivatives displayed similar fluorescence patterns, localizing to the vacuoles and colocalizing with the vacuole-staining dye FM4-64 (Figure 5E). Taken together, the results showed that Str2 is required for sustaining cell growth in the presence of exogenous holo-Fc under iron-starvation conditions. Furthermore, the conserved amino acid residues Tyr539, Arg546, and Tyr553, located within the predicted final loop of Str2, are critical for its Fc-related function.
We next assessed the impact of the Str2-Y539A/Y553A-GFP and Str2-Y539A/R546A/Y553A-GFP mutants on the ability of str2Δ fep1Δ cells to mobilize iron within vacuoles. Logarithmic phase str2Δ fep1Δ cells expressing the str2+-GFP, str2-Y539A/Y553A-GFP, or str2-Y539A/R546A/Y553A-GFP alleles were treated with Dip (250 μM) for 3 h. During the final hour of treatment, holo-Fc (1 μM) was added, followed by vacuole isolation from each culture. Purified vacuoles were analyzed using a BPS-based spectrophotometric method to quantify iron concentrations. In str2Δ fep1Δ cells expressing the str2-Y539A/Y553A-GFP and str2-Y539A/R546A/Y553A-GFP alleles, purified vacuoles contained 0.062 and 0.054 μg iron/μg of vacuolar proteins, respectively (Figure 6). These values represent 40.4 and 48.1% less iron compared to str2Δ fep1Δ cells expressing the str2+-GFP allele, which had 0.104 μg iron/μg of vacuolar proteins (Figure 6). Taken together, these results indicated that substituting the Tyr539, Arg546, and Tyr553 residues with alanines in Str2 lead to a reduction in vacuolar iron levels when iron-starved cells are grown with holo-Fc as the sole source of iron.
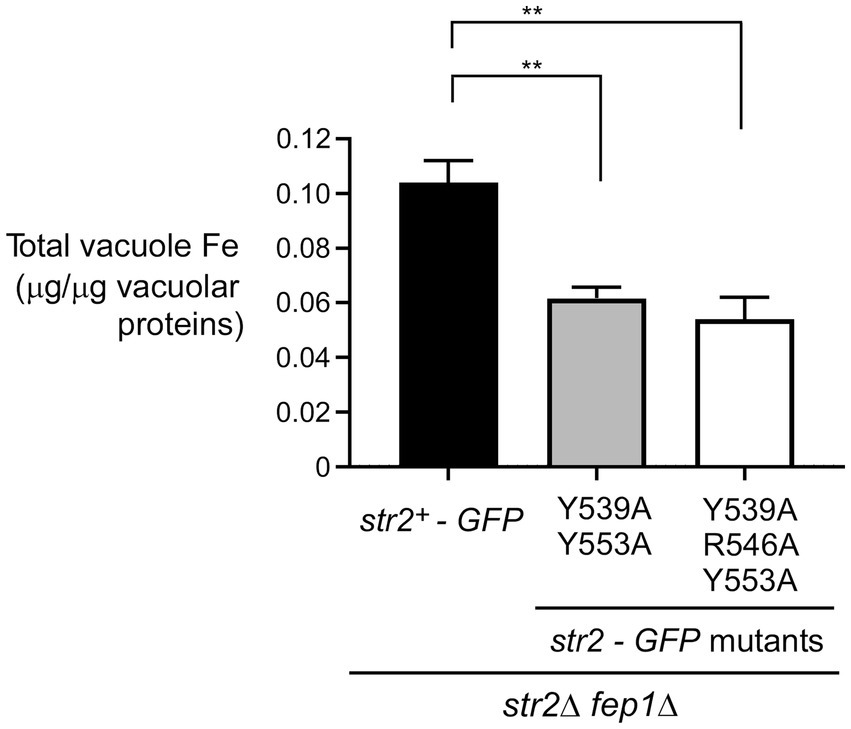
Figure 6. The Tyr539, Arg546, and Tyr553 residues of Str2 play an important role for maximal iron accumulation in vacuoles. str2Δ fep1Δ cells expressing str2+-GFP, str2-Y539A/Y553A-GFP or str2-Y539A/R546A/Y553A-GFP alleles were grown in YES medium to an OD600 of 1.0. The cultures were then incubated with Dip (250 μM) for 3 h. In the final hour of this treatment, holo-Fc (1 μM) was added, followed by vacuole isolation from each group of cultures. Purified vacuoles from each culture were analyzed using a BPS-based spectrophotometric method to quantitatively measure iron levels. Results are representative of three independent experiments. Data are presented as mean ± SD. Statistical significance is indicated by asterisks, with **p < 0.01 (determined by one-way ANOVA with Dunnett’s multiple comparisons test, comparing against cells expressing Str2-GFP).
Unlike Saccharomyces cerevisiae, S. pombe synthesizes and secretes Fc (Schrettl et al., 2004b). Consistently, S. pombe possesses a cell-surface transporter, Str1, that can take up Fc from the extracellular environment (Plante and Labbé, 2019). Although S. cerevisiae lacks the proteins necessary to produce siderophores, it can assimilate various types of siderophores, including Fc, ferrioxamine B, triacetylfusarinine C, and enterobactin secreted by other microbes (Philpott, 2006). S. cerevisiae expresses four siderophore-specific transporters (Arn1, Arn2/Taf1, Arn3/Sit1, and Arn4/Enb1) that mediate the uptake of siderophore-bound iron from the environment (Lesuisse et al., 1998; Heymann et al., 1999, 2000a, 2000b; Yun et al., 2000; Philpott, 2006). Among them, Arn1 transports Fc and other hydroxamate-type siderophores (Heymann et al., 2000b; Yun et al., 2000). Recent studies have shown that Fc produced by S. pombe promotes the growth of Arn1-expressing S. cerevisiae cells when Fc is used as the sole iron source (Brault et al., 2022). Due to fundamental differences between the two yeasts, their pathways involved in Fc metabolism differ in some aspects.
In the case of S. pombe, to dissociate its capacity to acquire exogenous Fc from its ability to synthesize endogenous Fc, we used a strain with deletions in the sib1+ and sib2+ genes (sib1Δ sib2Δ), which blocks de novo Fc biosynthesis by eliminating these enzymes from the pathway. Therefore, sib1Δ sib2Δ cells rely solely on their cell surface Fc transporter Str1 to acquire exogenous Fc. In the present study, microscopic analyses from str2Δ and str2Δ sib1Δ sib2Δ cells expressing str2+-GFP showed that Str2-GFP fluorescent signal is primarily observed to the vacuolar membrane under low-iron conditions. Furthermore, in the case of str2Δ sib1Δ sib2Δ cells, the vacuolar localization of Str2-GFP remains unchanged when the cells are exposed to exogenous Fc. These findings are different from those found in the case of siderophore transporters in S. cerevisiae (Philpott, 2006). None of the four siderophore transporters (Arn1 to Arn4) is a permanent resident vacuolar protein in S. cerevisiae (Philpott, 2006). The Arn1 and Arn3 transporters are localized to the trans-Golgi network where they are sorted to endosomal secretory vesicles (Kim et al., 2002; Moore et al., 2003; Kim et al., 2005). A model posits that exogenous Fc initially enters the cell through fluid-phase endocytosis, where it encounters Arn1 in the early endosome. Binding of Fc to Arn1 triggers its relocalization from the endosome to the plasma membrane. Once at the plasma membrane, extracellular Fc can bind Arn1, promoting its ubiquitination, internalization, and cycling between the plasma membrane and endosomes, while mediating the transport of Fc into the cell (Philpott, 2006). In contrast, when siderophores are no longer available outside the cell, the Arn1, Arn2, and Arn3 transporters located in the late Golgi network are sorted to the vacuole for degradation. In the case of Arn4, however, it exhibits a distinct trafficking pattern, being directed to the cell surface even in the absence of its siderophore, enterobactin (Philpott and Protchenko, 2008).
Our results showed that S. pombe cells deficient in Fc biosynthesis (sib1Δ sib2Δ) require str2+ for Fc-dependent growth under low-iron conditions. Since Str2 localizes to the vacuolar membrane, this suggests that, after Fc uptake through the plasma membrane by Str1, it must be delivered to Str2 at the vacuole. In the case of S. pombe Str1, it is unclear whether this siderophore transporter undergoes intracellular trafficking upon Fc binding. In S. cerevisiae, Fc binding to Arn1 triggers its internalization. In the endosome, Fc bound to Arn1 is thought to be translocated into the cytosol, where iron is released from the siderophore likely through degradation of Fc (Philpott, 2006). One possibility is that a similar Fc-mediated internalization of Str1 occurs in S. pombe. However, this would require that Fc maintains its integrity after its translocation into the cytosol, allowing it to subsequently bind to Str2 and be transported into the vacuole, where iron would then be dissociated from the siderophore.
Studies in budding and fission yeasts have shown that the vacuole serves as a storage compartment for metal ions, either to detoxify the cell or to act as a reservoir, enabling cell growth under conditions of metal ion deficiency (Ramsay and Gadd, 1997; Bellemare et al., 2002; Rees et al., 2004; Simm et al., 2007; Singh et al., 2007). In S. cerevisiae, different proteins are involved in the mobilization of vacuolar iron stores. Under iron-replete conditions, the vacuolar iron transporter Ccc1 transfers iron from the cytosol to the vacuole (Li et al., 2001). Conversely, in cells undergoing a transition from high to low iron levels, the vacuolar iron transporter Smf3 mobilizes stored iron from the vacuole to the cytosol (Portnoy et al., 2000). Copper-and iron-deficient cells also activate the expression of Fre6, a cupric/ferric reductase found on the vacuolar membrane that reduces vacuolar Cu2+ and Fe3+ ions (Rees and Thiele, 2007; Singh et al., 2007). The reduced iron (Fe2+) is then transported out of the vacuole by the Fet5/Fth1 oxidase/permease heteromeric complex (Urbanowski and Piper, 1999). In S. pombe, the mechanism of vacuolar iron mobilization in response to iron deficiency is not well understood and may differ due to the absence of orthologs for Smf3, Fre6, Fth1, and Fet5. S. pombe has a single Ccc1-like protein, Pcl1, which is thought to mediate vacuolar iron storage. Deletion of the pcl1+ gene (pcl1Δ) results in a mutant strain with reduced cellular iron content compared to the wild-type strain (Pouliot et al., 2010). However, the definitive role of Pcl1 in vacuolar iron storage has yet to be confirmed.
In this study, vacuolar iron concentration is lower in str2Δ fep1Δ cells lacking Str2 compared to control str2Δ fep1Δ cells expressing a functional str2+-GFP allele. The iron appears to be in an inorganic form, dissociated from Fc within the vacuole, as we were unable to detect holo-Fc in intact chelated form. MFS-type transporters contain two bundles of six or seven membrane-spanning alpha-helices. These two bundles come together to form a central pore, and the transporters operate via an alternating-access mechanism. This mechanism involves a rocker-switch-like movement, described as alternating between outward-open and inward-open conformations, triggered by substrate binding. Notably, the last two transmembrane domains and the final hydrophilic loop are unique to MFS-type fungal siderophore-iron transporters and are not shared by other related MFS transporters (Kim et al., 2005). Considering this, it is plausible that the SITD region of Str2, located in the final predicted loop on the luminal side of the vacuole, plays a critical role in facilitating the conformational change to the inward-open state. Fc may sequentially enter through the rocker-switch-like movement and subsequently bind to the C-terminal Tyr-Arg-Tyr residues of the Str2 SITD domain. In this way, the SITD region likely attracts Fc, forming a sink for Fc to bind to, before iron is extracted from Fc by an unknown mechanism and stored in the vacuole.
Iron accumulation in vacuoles when holo-Fc is available to Str2-GFP-expressing str2Δ fep1Δ cells is specific to Str2-GFP, as isogenic cells expressing str2-Y539A/Y553A-GFP and str2-Y539A/R546A/Y553A-GFP mutant alleles exhibited a decrease in vacuolar iron content. Based on a primary sequence alignment of Str2 with other fungal MFS-type siderophore-iron transporters, Tyr539, Arg546, and Tyr553 residues were mutated, as they were predicted to be located in a conserved loop that encompasses a putative siderophore transporter domain (SITD). For example, the SITD in the Fc importer Sit1 in C. glabrata contains a Tyr575 residue corresponding to Tyr553 in S. pombe Str2. Substituting Tyr575 with Ala in Sit1 significantly reduces the ability of C. glabrata to use exogenous Fc as an iron source for growth (Nevitt and Thiele, 2011). Similarly, in A. fumigatus (strain ATCC 13073), the siderophore transporter MirB contains a conserved Tyr577 residue corresponding to Tyr553 in S. pombe Str2. Substitution of Tyr577 with Ala in MirB resulted in a dramatic loss of siderophore transport activity (Raymond-Bouchard et al., 2012). The last extracellular loop of the S. cerevisiae Fc transporter Arn1 has undergone comprehensive mutagenesis (Kim et al., 2005). Alanine substitutions of Phe540 and Tyr544 (where Tyr544 corresponds to Tyr539 in Str2) resulted in a complete loss of low-affinity Fc binding, with a significant reduction in Fc uptake activity in cells expressing the mutant F540A/Y544A allele (Kim et al., 2005). Moreover, when Gln550, Arg551, Tyr558, and Arg563 were replaced with alanines (where Arg551 and Tyr558 in S. cerevisiae Arn1 correspond to Arg546 and Tyr553 in S. pombe Str2), there was a dramatic defect in Fc binding along with a complete loss of Fc transport and Fc-dependent growth in cells expressing this mutant allele (Kim et al., 2005). Our previous studies on the S. pombe MFS-type transporter Str1 revealed the critical importance of two conserved Tyr residues in the last predicted loop of this Fc importer (Plante and Labbé, 2019). Conserved Tyr553 and Tyr567 in Str1 correspond to Tyr539 and Tyr553 in Str2. Fungal spores expressing a mutant version of Str1, in which residues Tyr553 and Tyr567 were replaced with alanines, were unable to complete the outgrowth phase in a timely manner compared to control spores expressing wild-type Str1 in the presence of exogenous Fc (Plante and Labbé, 2019). Together, findings from previous studies and the present work underscore the necessity of conserved Tyr residues within the last loop of MFS-type fungal siderophore-iron transporters for transporting environmental Fc into cells or other subcellular targets. Based on the extended amino acid sequence similarity between Str2 and Str1, particularly within the regions encompassing the predicted transmembrane domains, it would be interesting to identify which motif of the intracellular Fc transporter Str2 is required for its sorting to the vacuolar membrane, in contrast to Str1, which is sorted to the plasma membrane to transport Fc into the cell.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
BM: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. SP: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. TV: Conceptualization, Formal analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – review & editing. AB: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. SL: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC, grant #RGPIN-2020/2025-04802) to SL.
TV is recipient of a studentship from the Fonds de Recherche du Québec - Santé (FRQ-S).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adam, M., Robert, F., Larochelle, M., and Gaudreau, L. (2001). H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21, 6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001
Aguiar, M., Orasch, T., Misslinger, M., Dietl, A. M., Gsaller, F., and Haas, H. (2021). The siderophore transporters Sit1 and Sit2 are essential for utilization of ferrichrome-, ferrioxamine-and coprogen-type siderophores in Aspergillus fumigatus. J. Fungi 7:768. doi: 10.3390/jof7090768
Bellemare, D. R., Shaner, L., Morano, K. A., Beaudoin, J., Langlois, R., and Labbé, S. (2002). Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 277, 46676–46686. doi: 10.1074/jbc.M206444200
Brault, A., Mbuya, B., and Labbé, S. (2022). Sib1, Sib2, and Sib3 proteins are required for ferrichrome-mediated cross-feeding interaction between Schizosaccharomyces pombe and Saccharomyces cerevisiae. Front. Microbiol. 13:962853. doi: 10.3389/fmicb.2022.962853
Brault, A., Mourer, T., and Labbé, S. (2015). Molecular basis of the regulation of iron homeostasis in fission and filamentous yeasts. IUBMB Life 67, 801–815. doi: 10.1002/iub.1441
Brault, A., Rallis, C., Normant, V., Garant, J. M., Bahler, J., and Labbé, S. (2016). Php4 is a key player for iron economy in meiotic and sporulating cells. G3 6, 3077–3095. doi: 10.1534/g3.116.031898
Chen, D., Toone, W. M., Mata, J., Lyne, R., Burns, G., Kivinen, K., et al. (2003). Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214–229. doi: 10.1091/mbc.E02-08-0499
Chiu, P.-C., Nakamura, Y., Nishimura, S., Tabuchi, T., Yashiroda, Y., Hirai, G., et al. (2022). Ferrichrome, a fungal-type siderophore, confers high ammonium tolerance to fission yeast. Sci. Rep. 12:17411. doi: 10.1038/s41598-022-22108-0
Galaris, D., Barbouti, A., and Pantopoulos, K. (2019). Iron homeostasis and oxidative stress: an intimate relationship. Biochim. Biophys. Acta Mol. Cell Res. 1866:118535. doi: 10.1016/j.bbamcr.2019.118535
Gueldener, U., Heinisch, J., Koehler, G. J., Voss, D., and Hegemann, J. H. (2002). A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23, 23e–223e. doi: 10.1093/nar/30.6.e23
Haas, H. (2014). Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 31, 1266–1276. doi: 10.1039/c4np00071d
Haas, H., Eisendle, M., and Turgeon, B. G. (2008). Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46, 149–187. doi: 10.1146/annurev.phyto.45.062806.094338
Heymann, P., Ernst, J. F., and Winkelmann, G. (1999). Identification of a fungal triacetylfusarinine C siderophore transport gene (TAF1) in Saccharomyces cerevisiae as a member of the major facilitator superfamily. Biometals 12, 301–306. doi: 10.1023/a:1009252118050
Heymann, P., Ernst, J. F., and Winkelmann, G. (2000a). A gene of the major facilitator superfamily encodes a transporter for enterobactin (Enb1p) in Saccharomyces cerevisiae. Biometals 13, 65–72. doi: 10.1023/a:1009250017785
Heymann, P., Ernst, J. F., and Winkelmann, G. (2000b). Identification and substrate specificity of a ferrichrome-type siderophore transporter (Arn1p) in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 186, 221–227. doi: 10.1111/j.1574-6968.2000.tb09108.x
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. doi: 10.1016/0378-1119(89)90358-2
Iwaki, T., and Takegawa, K. (2004). A set of loxP marker cassettes for Cre-mediated multiple gene disruption in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 68, 545–550. doi: 10.1271/bbb.68.545
Jacques, J. F., Mercier, A., Brault, A., Mourer, T., and Labbé, S. (2014). Fra2 is a co-regulator of Fep1 inhibition in response to iron starvation. PLoS One 9:e98959. doi: 10.1371/journal.pone.0098959
Jbel, M., Mercier, A., Pelletier, B., Beaudoin, J., and Labbé, S. (2009). Iron activates in vivo DNA binding of Schizosaccharomyces pombe transcription factor Fep1 through its amino-terminal region. Eukaryot. Cell 8, 649–664. doi: 10.1128/EC.00001-09
Katsarou, A., and Pantopoulos, K. (2020). Basics and principles of cellular and systemic iron homeostasis. Mol. Asp. Med. 75:100866. doi: 10.1016/j.mam.2020.100866
Keeney, J. B., and Boeke, J. D. (1994). Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136, 849–856. doi: 10.1093/genetics/136.3.849
Kim, Y., Lampert, S. M., and Philpott, C. C. (2005). A receptor domain controls the intracellular sorting of the ferrichrome transporter, ARN1. EMBO J. 24, 952–962. doi: 10.1038/sj.emboj.7600579
Kim, Y., Yun, C. W., and Philpott, C. C. (2002). Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J. 21, 3632–3642. doi: 10.1093/emboj/cdf382
Larochelle, M., Lemay, J. F., and Bachand, F. (2012). The THO complex cooperates with the nuclear RNA surveillance machinery to control small nucleolar RNA expression. Nucleic Acids Res. 40, 10240–10253. doi: 10.1093/nar/gks838
Law, C. J., Maloney, P. C., and Wang, D. N. (2008). Ins and outs of major facilitator superfamily antiporters. Ann. Rev. Microbiol. 62, 289–305. doi: 10.1146/annurev.micro.61.080706.093329
Lesuisse, E., Simon-Casteras, M., and Labbé, P. (1998). Siderophore-mediated iron uptake in Saccharomyces cerevisiae: the SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144, 3455–3462. doi: 10.1099/00221287-144-12-3455
Li, L., Chen, O. S., McVey Ward, D., and Kaplan, J. (2001). CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 276, 29515–29519. doi: 10.1074/jbc.M103944200
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mbuya, B., Plante, S., Ammar, F., Brault, A., and Labbé, S. (2024). The Schizosaccharomyces pombe ornithine-N(5)-oxygenase Sib2 interacts with the N(5)-transacetylase Sib3 in the ferrichrome biosynthetic pathway. Front. Microbiol. 15:1467397. doi: 10.3389/fmicb.2024.1467397
Mercier, A., and Labbé, S. (2009). Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J. Biol. Chem. 284, 20249–20262. doi: 10.1074/jbc.M109.009563
Mercier, A., and Labbé, S. (2010). Iron-dependent remodeling of fungal metabolic pathways associated with ferrichrome biosynthesis. Appl. Environ. Microbiol. 76, 3806–3817. doi: 10.1128/AEM.00659-10
Mercier, A., Watt, S., Bahler, J., and Labbé, S. (2008). Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell 7, 493–508. doi: 10.1128/EC.00446-07
Moore, R. E., Kim, Y., and Philpott, C. C. (2003). The mechanism of ferrichrome transport through Arn1p and its metabolism in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100, 5664–5669. doi: 10.1073/pnas.1030323100
Nevitt, T., and Thiele, D. J. (2011). Host iron withholding demands siderophore utilization for Candida glabrata to survive macrophage killing. PLoS Pathog. 7:e1001322. doi: 10.1371/journal.ppat.1001322
Pelletier, B., Beaudoin, J., Mukai, Y., and Labbé, S. (2002). Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J. Biol. Chem. 277, 22950–22958. doi: 10.1074/jbc.M202682200
Pelletier, B., Beaudoin, J., Philpott, C. C., and Labbé, S. (2003). Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res. 31, 4332–4344. doi: 10.1093/nar/gkg647
Pelletier, B., Trott, A., Morano, K. A., and Labbe, S. (2005). Functional characterization of the iron-regulatory transcription factor Fep1 from Schizosaccharomyces pombe. J. Biol. Chem. 280, 25146–25161. doi: 10.1074/jbc.M502947200
Philpott, C. C. (2006). Iron uptake in fungi: a system for every source. Biochim. Biophys. Acta 1763, 636–645. doi: 10.1016/j.bbamcr.2006.05.008
Philpott, C. C., Patel, S. J., and Protchenko, O. (2020). Management versus miscues in the cytosolic labile iron pool: the varied functions of iron chaperones. Biochim. Biophys. Acta, Mol. Cell Res. 1867:118830. doi: 10.1016/j.bbamcr.2020.118830
Philpott, C. C., and Protchenko, O. (2008). Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7, 20–27. doi: 10.1128/EC.00354-07
Plante, S., and Labbé, S. (2019). Spore germination requires ferrichrome biosynthesis and the siderophore transporter Str1 in Schizosaccharomyces pombe. Genetics 211, 893–911. doi: 10.1534/genetics.118.301843
Portnoy, M. E., Liu, X. F., and Culotta, V. C. (2000). Saccharomyces cerevisiae expresses three functionally distinct homologues of the Nramp family of metal transporters. Mol. Cell. Biol. 20, 7893–7902. doi: 10.1128/.20.21.7893-7902.2000
Pouliot, B., Jbel, M., Mercier, A., and Labbé, S. (2010). abc3+ encodes an iron-regulated vacuolar ABC-type transporter in Schizosaccharomyces pombe. Eukaryot. Cell 9, 59–73. doi: 10.1128/EC.00262-09
Protacio, R. U., Mukiza, T. O., Davidson, M. K., and Wahls, W. P. (2022). Molecular mechanisms for environmentally induced and evolutionarily rapid redistribution (plasticity) of meiotic recombination. Genetics 220:312371. doi: 10.1093/genetics/iyab212
Puig, S., Ramos-Alonso, L., Romero, A. M., and Martínez-Pastor, M. T. (2017). The elemental role of iron in DNA synthesis and repair. Metallomics 9, 1483–1500. doi: 10.1039/c7mt00116a
Rad, A. M., Janic, B., Iskander, A. S., Soltanian-Zadeh, H., and Arbab, A. S. (2007). Measurement of quantity of iron in magnetically labeled cells: comparison among different UV/VIS spectrometric methods. BioTechniques 43, 627–628, 630, 632. doi: 10.2144/000112599
Ramsay, L. M., and Gadd, G. M. (1997). Mutants of Saccharomyces cerevisiae defective in vacuolar function confirm a role for the vacuole in toxic metal ion detoxification. FEMS Microbiol. Lett. 152, 293–298. doi: 10.1111/j.1574-6968.1997.tb10442.x
Raymond-Bouchard, I., Carroll, C. S., Nesbitt, J. R., Henry, K. A., Pinto, L. J., Moinzadeh, M., et al. (2012). Structural requirements for the activity of the MirB ferrisiderophore transporter of Aspergillus fumigatus. Eukaryot. Cell 11, 1333–1344. doi: 10.1128/ec.00159-12
Rees, E. M., Lee, J., and Thiele, D. J. (2004). Mobilization of intracellular copper stores by the Ctr2 vacuolar copper transporter. J. Biol. Chem. 279, 54221–54229. doi: 10.1074/jbc.M411669200
Rees, E. M., and Thiele, D. J. (2007). Identification of a vacuole-associated metalloreductase and its role in Ctr2-mediated intracellular copper mobilization. J. Biol. Chem. 282, 21629–21638. doi: 10.1074/jbc.M703397200
Rustici, G., van Bakel, H., Lackner, D. H., Holstege, F. C., Wijmenga, C., Bahler, J., et al. (2007). Global transcriptional responses of fission and budding yeast to changes in copper and iron levels: a comparative study. Genome Biol. 8:R73. doi: 10.1186/gb-2007-8-5-r73
Rutherford, K. M., Lera-Ramírez, M., and Wood, V. (2024). PomBase: a global core biodata resource-growth, collaboration, and sustainability. Genetics 227:iyae007. doi: 10.1093/genetics/iyae007
Sabatinos, S. A., and Forsburg, S. L. (2010). Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 470, 759–795. doi: 10.1016/S0076-6879(10)70032-X
Sarry, J. E., Chen, S., Collum, R. P., Liang, S., Peng, M., Lang, A., et al. (2007). Analysis of the vacuolar luminal proteome of Saccharomyces cerevisiae. FEBS J. 274, 4287–4305. doi: 10.1111/j.1742-4658.2007.05959.x
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schrettl, M., Bignell, E., Kragl, C., Joechl, C., Rogers, T., Arst, H. N. Jr., et al. (2004a). Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200, 1213–1219. doi: 10.1084/jem.20041242
Schrettl, M., Winkelmann, G., and Haas, H. (2004b). Ferrichrome in Schizosaccharomyces pombe-an iron transport and iron storage compound. Biometals 17, 647–654. doi: 10.1007/s10534-004-1230-z
Simm, C., Lahner, B., Salt, D., LeFurgey, A., Ingram, P., Yandell, B., et al. (2007). Saccharomyces cerevisiae vacuole in zinc storage and intracellular zinc distribution. Eukaryot. Cell 6, 1166–1177. doi: 10.1128/EC.00077-07
Singh, A., Kaur, N., and Kosman, D. J. (2007). The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J. Biol. Chem. 282, 28619–28626. doi: 10.1074/jbc.M703398200
Sooksa-Nguan, T., Yakubov, B., Kozlovskyy, V. I., Barkume, C. M., Howe, K. J., Thannhauser, T. W., et al. (2009). Drosophila ABC transporter, DmHMT-1, confers tolerance to cadmium. DmHMT-1 and its yeast homolog, SpHMT-1, are not essential for vacuolar phytochelatin sequestration. J. Biol. Chem. 284, 354–362. doi: 10.1074/jbc.M806501200
Urbanowski, J. L., and Piper, R. C. (1999). The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 274, 38061–38070. doi: 10.1074/jbc.274.53.38061
Keywords: fission yeast, ferrichrome, iron, iron-regulatory GATA-type transcription factor, siderophore transporter
Citation: Mbuya B, Plante S, Vahsen T, Brault A and Labbé S (2025) Fission yeast cells deficient in siderophore biosynthesis require Str2 for ferrichrome-dependent growth. Front. Microbiol. 16:1527727. doi: 10.3389/fmicb.2025.1527727
Received: 13 November 2024; Accepted: 27 January 2025;
Published: 06 February 2025.
Edited by:
Marie-Joelle Virolle, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Vineet Kumar, The University of Texas at Austin, United StatesCopyright © 2025 Mbuya, Plante, Vahsen, Brault and Labbé. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simon Labbé, U2ltb24uTGFiYmVAVVNoZXJicm9va2UuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.