- 1School of Biosciences, University of Nottingham, Nottingham, United Kingdom
- 2School of Veterinary Medicine and Science, University of Nottingham, Nottingham, United Kingdom
Introduction: Canine otitis externa (OE) is a frequently-diagnosed condition in veterinary practices worldwide. Pseudomonas aeruginosa is commonly associated with chronic and recalcitrant canine OE, but studies with detailed genomic and phenotypic characterisation of clinical isolates are lacking.
Methods: Pseudomonas aeruginosa canine OE isolates (n = 253) were collected from different geographical locations in Europe and characterised with respect to antimicrobial resistance and biofilm formation. A subset (n = 35) were genome sequenced then characterised with respect to diversity, and complement of virulence, antimicrobial resistance, and biofilm-associated genes.
Results: Genome-sequenced P. aeruginosa strains were distributed among phylogroups, showing no obvious clonality. However, two isolates belonged to ST111 and ST244 respectively,—MLST sequence types associated with AMR nosocomial infections in humans. Resistance to fluoroquinolones was detected in 25% of isolates, and multidrug resistance detected in 1.6%, though this did not always correlate with the presence of antimicrobial resistance genes. Additionally, 82% of isolates were characterised as forming strong biofilms.
Discussion: For the first time, this study has characterised a large multinational collection of P. aeruginosa isolates from canine otitis with a combination of whole genome sequencing, phenotypic screening and bioinformatic analysis. These strains did not cluster together based on genomic diversity or virulence gene complement, supporting their likely environmental origin. However, the identification of ST111 and ST244, important ‘high-risk’ sequence types, could suggest potential spread between humans and dogs. Furthermore, we found that most strains were formed strong biofilms, and exhibited a significant level of resistance towards critically important antimicrobials. These findings could assist in the selection of appropriate treatments for canine OE as well as possibly identifying one health risks of these infections for cohabiting pets and humans.
Introduction
Canine otitis externa (OE) is recognised as inflammation of the ear canal and is one of the most common diagnoses affecting between 7.3–10.2% of dogs attending UK primary care practices (O’Neill et al., 2014; O’Neill et al., 2021). A similar prevalence ranging from 6.3–13%, has been reported worldwide (Lund et al., 1999; Kim et al., 2018). OE is thought to result from secondary infection following a multifactorial primary inflammation (Paterson and Matyskiewicz, 2018). Pseudomonas aeruginosa is the most common pathogen isolated from chronic and recalcitrant canine OE; associated with up to 35% of cases (Nuttall and Cole, 2007). Pseudomonas aeruginosa is a Gram-negative, opportunistic pathogen of humans and animals. In humans, it is closely associated with cystic fibrosis patients, where in the UK 13.1% of adults suffer from chronic P. aeruginosa infections and 18.8% from intermittent infections (Naito et al., 2023). A similar prevalence can be seen globally in the US, Canada and Germany (Cystic Fibrosis Canada, 2023; Cystic Fibrosis Foundation, 2023; Nährlich et al., 2023).
Treatment of P. aeruginosa canine OE usually requires the use of antimicrobials as part of combined steroid, antimicrobial, antifungal products. Three classes of antimicrobials are often used; fluoroquinolones such as marbofloxacin; aminoglycosides (particularly gentamicin); and polymyxin B (Secker et al., 2023). Treatment is challenging because of tissue swelling, hyperplasia and antimicrobial resistance (AMR). Pseudomonas aeruginosa has intrinsic AMR mechanisms such as limited outer membrane permeability, efflux pumps and a chromosomally encoded β-lactamase (Cox and Wright, 2013). It can also rapidly accrue mutations conferring resistance to most antimicrobials used in clinical therapy for OE (López-Causapé et al., 2018) or acquire antimicrobial resistance genes (ARG) through horizontal gene transfer (Pang et al., 2019).
The majority (>90%) of P. aeruginosa isolates from clinical cases of OE in both humans and dogs have been shown to produce biofilms (Fusconi et al., 2011; Robinson et al., 2019). This reduces treatment success as P. aeruginosa cells within a biofilm can be many times more resistant to antimicrobials than planktonic cells (Thöming and Häussler, 2022). Consequently, this can result in incurable chronic infections which—in extreme cases—require surgery (Pye, 2018).
Whole genome sequencing has not been used to evaluate P. aeruginosa isolates from clinical cases of canine OE to our knowledge. As such, little is known about the full genetic diversity of these isolates. Likewise, few studies have investigated the presence and role of virulence genes in P. aeruginosa isolates from canine OE. One study found a high prevalence of five virulence genes from canine P. aeruginosa isolates, though these were not exclusively from OE infections (Hattab et al., 2021). Multilocus Sequence Typing (MLST) studies have found a range of sequence types (ST) linked to P. aeruginosa from canine otitis and pyoderma which hinted at the underlying genetic diversity amongst these isolates (Hyun et al., 2018; Elfadadny et al., 2023).
In the present study, antimicrobial resistance and biofilm formation was assessed for 253 European isolates of P. aeruginosa from clinical cases of canine OE. Subsequently, a subset of 35 isolates representing different phenotypes were selected for whole genome sequencing to determine their complement of virulence and antimicrobial resistance genes (ARG) and to gain a fuller understanding of their genomic diversity.
Materials and methods
Bacterial strain isolation and laboratory maintenance
A collection (n = 253) of geographically diverse Pseudomonas aeruginosa isolates from dogs with otitis externa were collected from the UK Royal Veterinary College (RVC; n = 48), CAPL Nationwide Laboratories, UK (n = 99), and an EU collection from the University of Copenhagen (n = 106). Confirmation of P. aeruginosa identity was performed as described by the UK Health Security Agency standards for microbiology investigations (UKHSA, 2024) using cetrimide agar and the oxidase test, followed by 16S PCR. Pseudomonas aeruginosa PAO1 was used as a positive control for all assays in this study.
Prior to each experiment, agar plates were prepared [Lysogeny Broth (LB), Miller, 25 g/L, Sigma-Aldrich; supplemented with agar, 15 g/L, Fisher Scientific], and were inoculated with the required P. aeruginosa strain and streaked to obtain single colonies. Plates were inverted and incubated statically at 37° C for 16 h. Following incubation, 10 mL of LB was inoculated with a single colony using a sterile inoculating loop and incubated at 37° C with shaking at 200 rpm for 16 h.
Antimicrobial sensitivity testing
Antimicrobial sensitivity testing (AST) was performed as described by the Clinical & Laboratory Standards Institute (CLSI) using the disk diffusion method (CLSI, 2012). A suspension of bacterial cells was prepared by inoculating 8 mL of Mueller-Hinton broth (MH; Sigma-Aldrich) with bacterial colonies selected from an LB plate until a turbidity equivalent to a 0.5 McFarland standard had been reached, as determined by visual comparison using a Wickerham card. A swab was then used to inoculate the surface of a MH agar 2 plate (Sigma-Aldrich) ensuring the entire plate is covered evenly by rotating the plate 90° between swabbing the first time with a final diagonal swab. The plates were then incubated at 37°C for 16 h, and the results recorded by measuring the inhibition zone diameter using a digital calliper.
AST was performed using an antimicrobial panel comprising six antimicrobial classes (Supplementary Table 1). All antimicrobials were tested against all strains except for Ticarcillin + Clavulanic acid which was discontinued part way through this study, and were acquired from two suppliers: ProLabs and Oxoid. E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were the control strains used in this assay. Antimicrobial sensitivity was determined using breakpoints outlined by the CLSI, where available canine veterinary breakpoints were used (CLSI, 2018b) otherwise human breakpoints were used (CLSI, 2018a).
Submerged biofilm assay
The ability of clinical P. aeruginosa isolates to form biofilm was tested in vitro as described previously with some modifications (Coffey and Anderson, 2014). Briefly, overnight cultures were diluted 1 in 100 into LB and 100 μL aliquots were separately transferred to wells of the 96-well microtiter plates in triplicate in the same microtiter plate; two microtiter plates were used for each technical repeat. A negative control, comprising three wells containing sterile LB, was included on each plate. The cultures were then grown statically at 37° C for 24 h. Following incubation, planktonic cells were removed, and the wells were washed using 125 μL of Ca/HEPES buffer; the wells were then stained using 0.1% (v/v) crystal violet solution. Excess crystal violet was removed, and the wells were then de-stained using ethanol (100%), and absorbance measured at 595 nm using a Tecan GENios Pro. The assay was repeated to achieve three biological repeats. Biofilm production was scored as either as either strong, moderate, weak, or non-biofilm producing as previously described (Stepanović et al., 2000).
Swarming motility assay
Swarming motility assays were modified according to (Ugurlu et al., 2016). Briefly, 1 μL of bacterial culture was spotted into the centre of swarming motility agar (8 g/L nutrient broth, Oxoid; 5 g/L agar select, Sigma-Aldrich; 0.5% v/v glucose, Fisher Scientific), and then incubated at 37° C for 16 h. The distance travelled from the point of inoculation was measured using a digital calliper, and swarming motility assessed by phenotype on the plates in combination with the distance travelled from the point of inoculation.
Pseudomonas aeruginosa genome sequencing
Whole genome sequencing of 35 isolates was performed using both short (Illumina MiSeq) and long (Oxford Nanopore) read technologies by MicrobesNG (Birmingham, United Kingdom). Isolates were selected for sequencing to ensure isolates from the CALP nationwide laboratories (n = 13), Denmark (n = 7) and the RVC (n = 9) were represented (PRJNA1180571) and included some sequences from previous work (n = 6; Gigante et al., 2024; PRJNA107813).
Illumina paired-end reads were quality-assessed and trimmed using FastQC v0.11.8 (Andrews, 2010) and FastP v0.12.4 (Chen et al., 2018) respectively. Subsequently, contigs were assembled de novo using Flye v2.9.2-b1786 (Kolmogorov et al., 2019), followed by post-processing with Circlator v.1.5.5 (Hunt et al., 2015) and Bandage v0.8.1 (Wick et al., 2015). This was followed by one round of long read polishing and two rounds of short read polishing using Medaka v1.11.1 (ONT, 2023), Polypolish v0.5.0 (Wick and Holt, 2022) and POLCA from the MaSuRCA toolkit v4.0.9 (Zimin and Salzberg, 2020) respectively. Finally, genomes were reoriented to begin with dnaA using dnaapler v0.4.0 (Bouras et al., 2024) and annotated using Bakta v1.8.2 (Schwengers et al., 2021).
Bioinformatic analysis
To aid in the comparison of isolates from the present study to a wider population of P. aeruginosa isolates, PanACoTA v1.4.0 (Perrin and Rocha, 2021) was used to download and filter P. aeruginosa genomes from the RefSeq database as previously described (Botelho et al., 2023). Briefly, P. aeruginosa sequences were selected using taxid (−T 287) and filtered so that genomes with more than 100 contigs were removed (--nbcont 100). Subsequently, redundant and misclassified genomes were removed using Mash distances (--min_dist 0.0001; --max_dist 0.05). Metadata for the RefSeq genomes was downloaded using NCBI Datasets v16.40.1 (O’Leary et al., 2024).
Whole genome average nucleotide identity (ANI) was calculated using FastANI v1.34 (Jain et al., 2018). This was used in combination with hierarchical clustering using Euclidean distance and complete linkage to assign isolates to a phylogroup (Botelho et al., 2023; Supplementary Figure 1). A neighbour joining phylogenetic tree was constructed using Mashtree v1.4.6 (Katz et al., 2019) using bootstrapping with 100 replicates and mindepth 0. Trees were then visualised using iTOL (Letunic and Bork, 2024).
Multilocus sequence typing (MLST) was performed in silico using the PubMLST database (Jolley et al., 2018). Where an unknown ST was identified, genome sequences were uploaded to the PubMLST database, and a new ST was assigned.
Screening of virulence genes was performed using ABRicate (Seemann, 2015) using the VFDB (Liu et al., 2021). Acquired antimicrobial resistance genes were identified using AMRFinderPlus v4.0.3 using assembled nucleotide sequences (−n), this included searching for point mutations (−O; Feldgarden et al., 2021).
Biofilm associated genes were identified using a custom database, created as described by (Seemann, 2015). Genes located within the psl, pel and alginate (alg) operons (Franklin et al., 2011) in addition to other associated biofilm genes (Liu et al., 2021; Kiel et al., 2022) were downloaded from NCBI using P. aeruginosa PAO1 (NC_002516.2). A complete list of the genes is shown in Supplementary Table 2.
Results
Genomic diversity of Pseudomonas aeruginosa from canine otitis infections
A total of 10,300 genomes were downloaded from the RefSeq database and filtered for quality and genetic distance. The resulting set of 3,721 genomes was then randomly subsampled to yield a subset of 1,000 genomes. This included P. aeruginosa PAO1 (GCF_000006765.1) and PA14 (GCF_000014625.1) but PA7 (GCF_000017205.1) was removed based on Mash genetic distance which is in line with previous reports proposing that this strain be reclassified to a novel species Pseudomonas paraeruginosa (Rudra et al., 2022). These genomes in addition to 35 from the present study were compared using Mashtree. The distances calculated were then used to construct a phylogenetic tree (Figure 1). The strains were ascribed to three phylogroups; A (n = 765), containing P. aeruginosa PAO1; B (n = 244), containing P. aeruginosa PA14, and a third minor group, C (n = 26). The classification of P. aeruginosa into three phylogroups has been described previously (Stewart et al., 2014; Ozer et al., 2019); with one study suggesting that phylogroup C should be split into three groups (Freschi et al., 2019). However, with the exclusion of PA7, the results in the present study are in line with a recent study of P. aeruginosa phylogeny (Botelho et al., 2023).
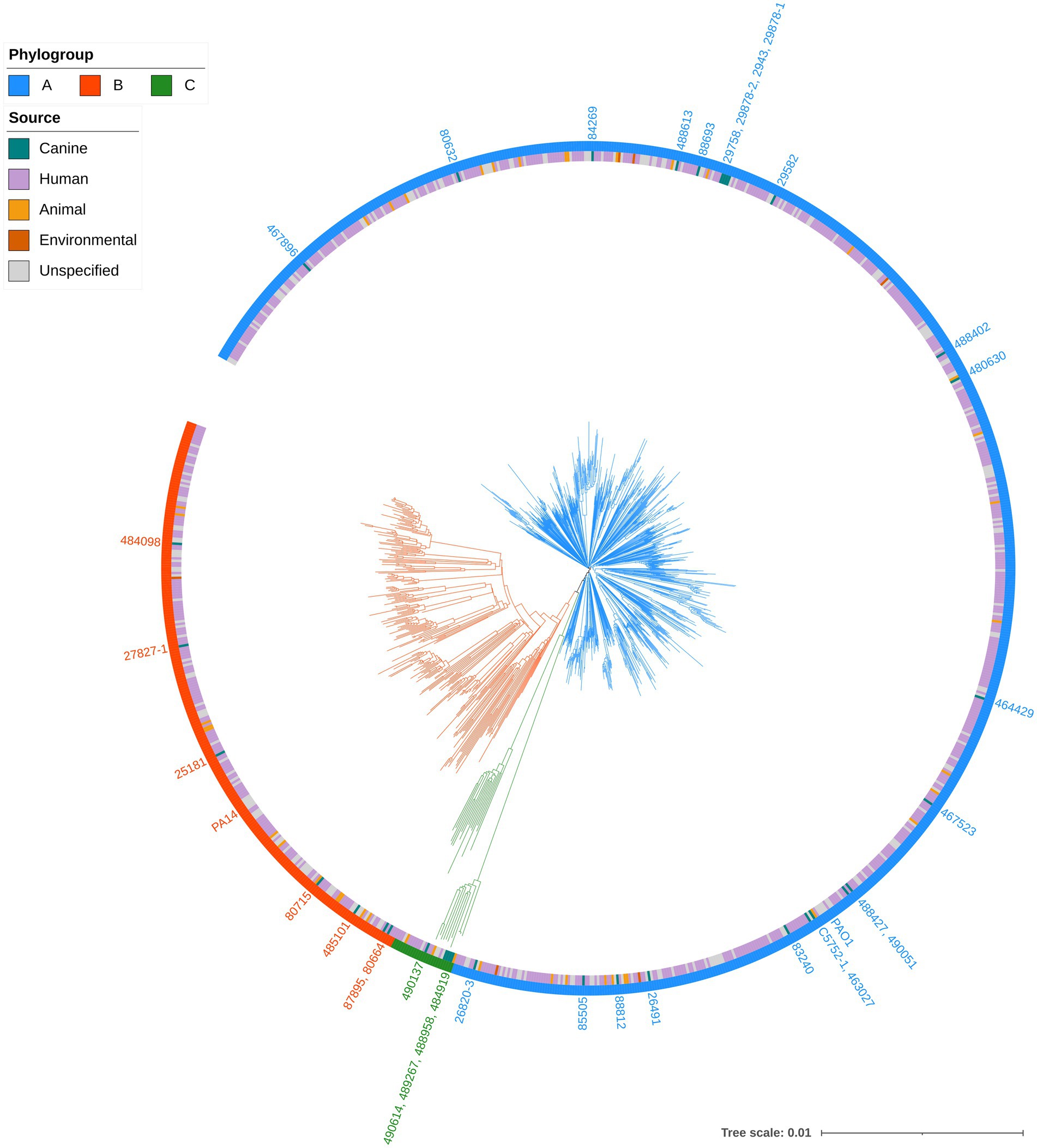
Figure 1. Phylogenetic tree of Pseudomonas aeruginosa strains. Mashtree with bootstrapping (100 replicates) was used to calculate the phylogenetic distances of the P. aeruginosa isolates from this study in addition to 1,000 isolates from other sources to create a neighbour joining tree which was visualised using iTOL. The scale represents 1 base difference per 100 bases. The outer ring represents the phylogroups while the inner ring shows the source of the isolates (where available). Strains from the present study, in addition to PAO1 and PA14, are labelled on the tree, coloured according to their phylogroup.
Canine strains from the present study were distributed across all three phylogroups indicating that they were not closely related and did not cluster according to host source or geographical origin. This was supported by MLST analysis which identified 30 different STs among the clinical isolates. One isolate, 488613, was assigned a novel ST (5180). The most common ST was 788 which was identified in 9% (3/35) of clinical isolates.
Antimicrobial sensitivity testing revealed resistance to up to six antimicrobials
Antimicrobial resistance (AMR) in P. aeruginosa from cases of canine otitis externa was determined using the disk diffusion method with 14 antimicrobials from 6 antimicrobial classes (Figure 2). Isolate 29130 failed to grow under the tested conditions and was excluded from this assay. The highest level of resistance was recorded for enrofloxacin (25.4%; 64/252) while tobramycin showed the highest sensitivity (99.6%; 251/252). Intermediate resistance was mainly seen against ticarcillin + clavulanic acid (60.7%; 127/252). Finally, 8.3% (21/252) and 0.79% (2/252) of isolates were resistant to imipenem and meropenem, respectively.
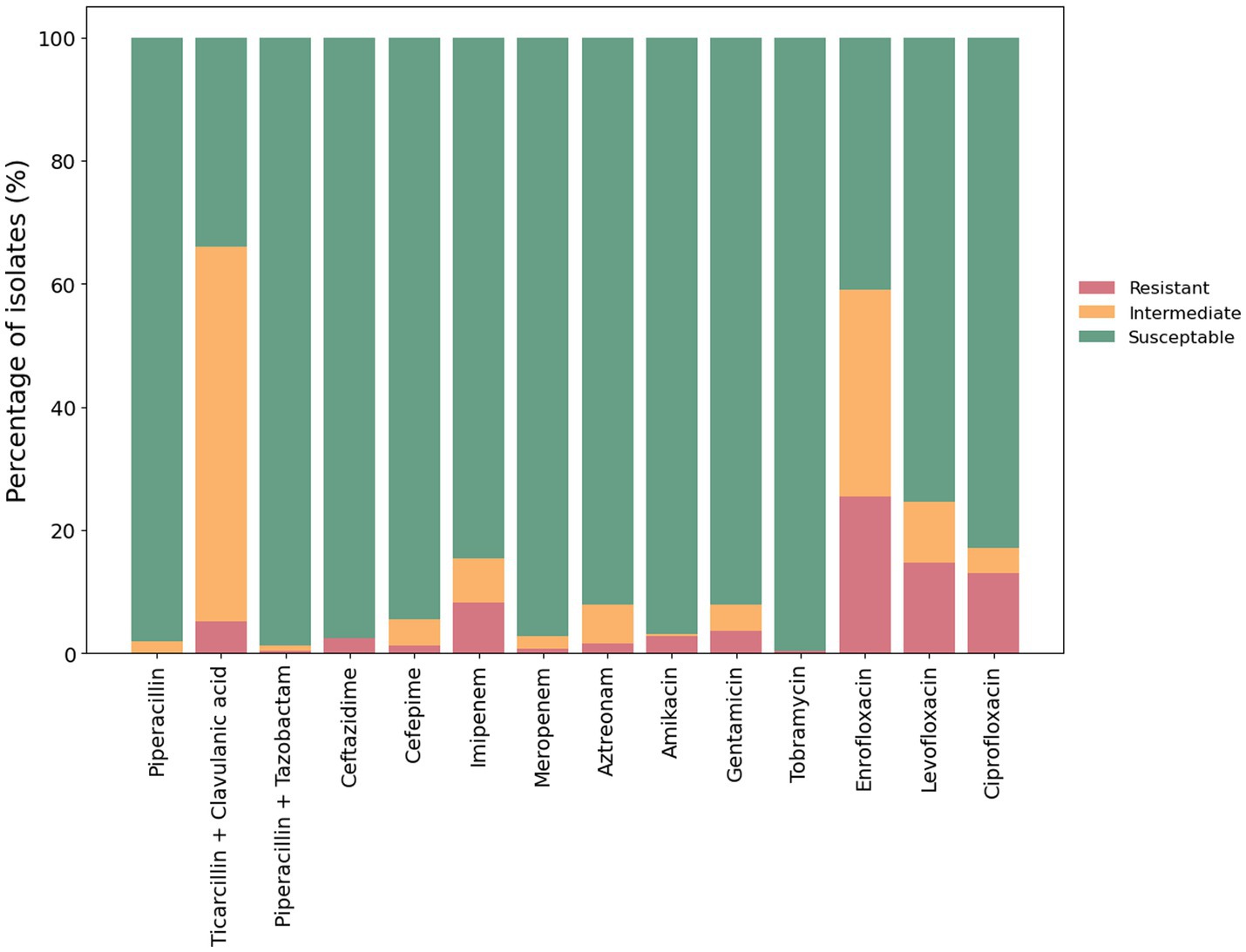
Figure 2. Antimicrobial susceptibility of P. aeruginosa isolates. The antimicrobial susceptibility of 252 P. aeruginosa isolates from clinical cases of canine otitis externa was determined using the disk diffusion method (CLSI, 2012). The percentage of sensitive, intermediate and resistant isolates are shown with red, amber and green bars, respectively.
Of the 252 isolates tested, 65% (164/252) were susceptible to all of the tested antimicrobials. The remaining 35% (88/252) were resistant to at least one antimicrobial. Moreover, 1.6% (4/252) were classified as multidrug resistant—defined as resistant to three or more different classes of antimicrobials (Sweeney et al., 2018; Figure 3).
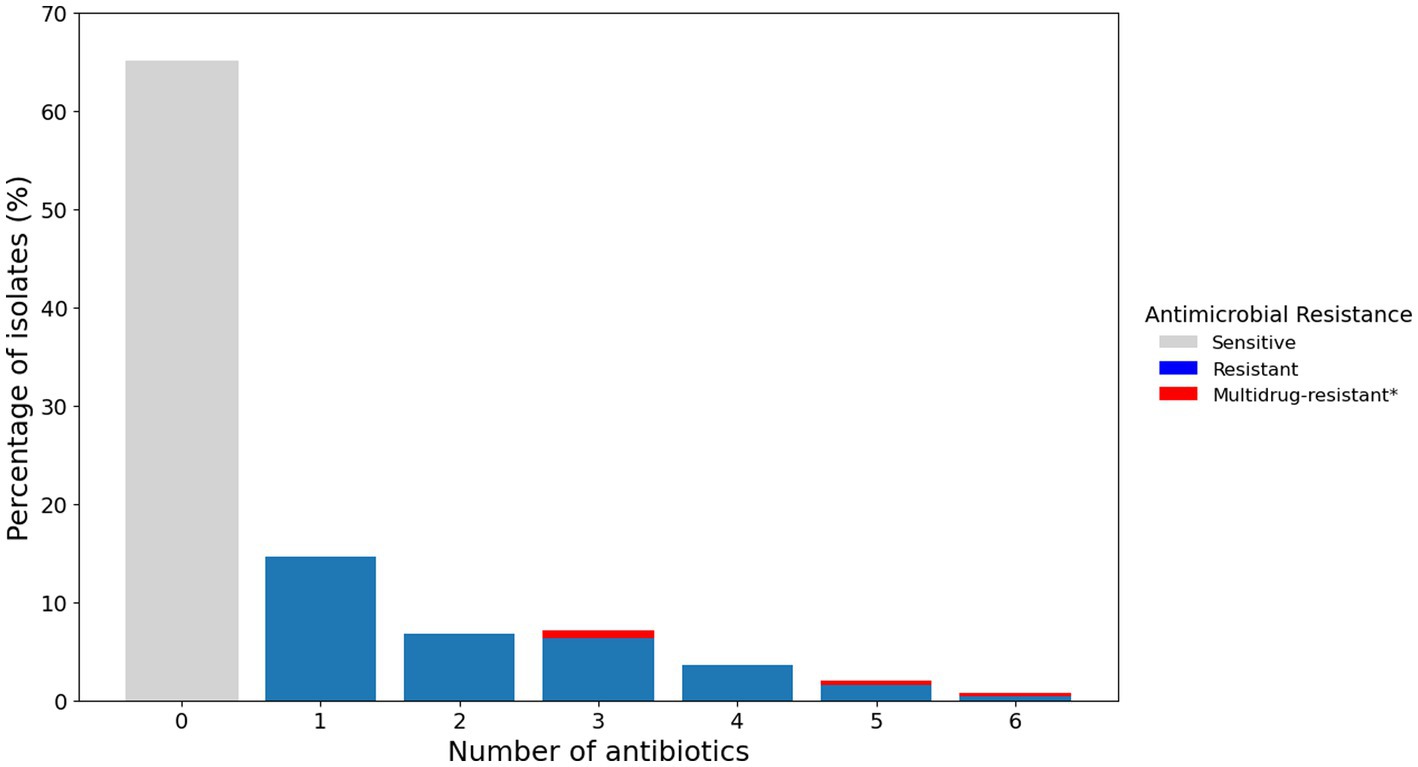
Figure 3. Percentage of P. aeruginosa isolates from canine otitis externa resistant to multiple antimicrobials. Bar chart representing the percentage of isolates resistant to a different number of antimicrobials as determined by the disk diffusion method. *Multidrug resistance is reported for resistance to three or more classes of antimicrobials.
Comparison of genotypic and phenotypic antimicrobial resistance
The 35 genome sequences of clinical P. aeruginosa isolates from canine OE were screened for the presence of ARG (Figure 4). Despite the variation in phenotypic susceptibility to antimicrobials between the strains, their complement of antimicrobial resistance genes remained remarkably consistent. Five genes, aph(3′)-IIb, fosA, catB7, blaOXA and blaPDC were detected in almost all of the isolates. Some notable exceptions are the presence of sul1, aadA7 and qacEΔ1 in 25181 and the absence of a blaOXA gene in 84269. The genes aadA7 and sul1 are linked to integrons and encode for a aminoglycoside nucleotidyltransferase that confers resistance to streptomycin and spectinomycin (Ahmed et al., 2004) and a sulfonamide resistant dihydropteroate synthase, respectively, (Sköld, 2000).
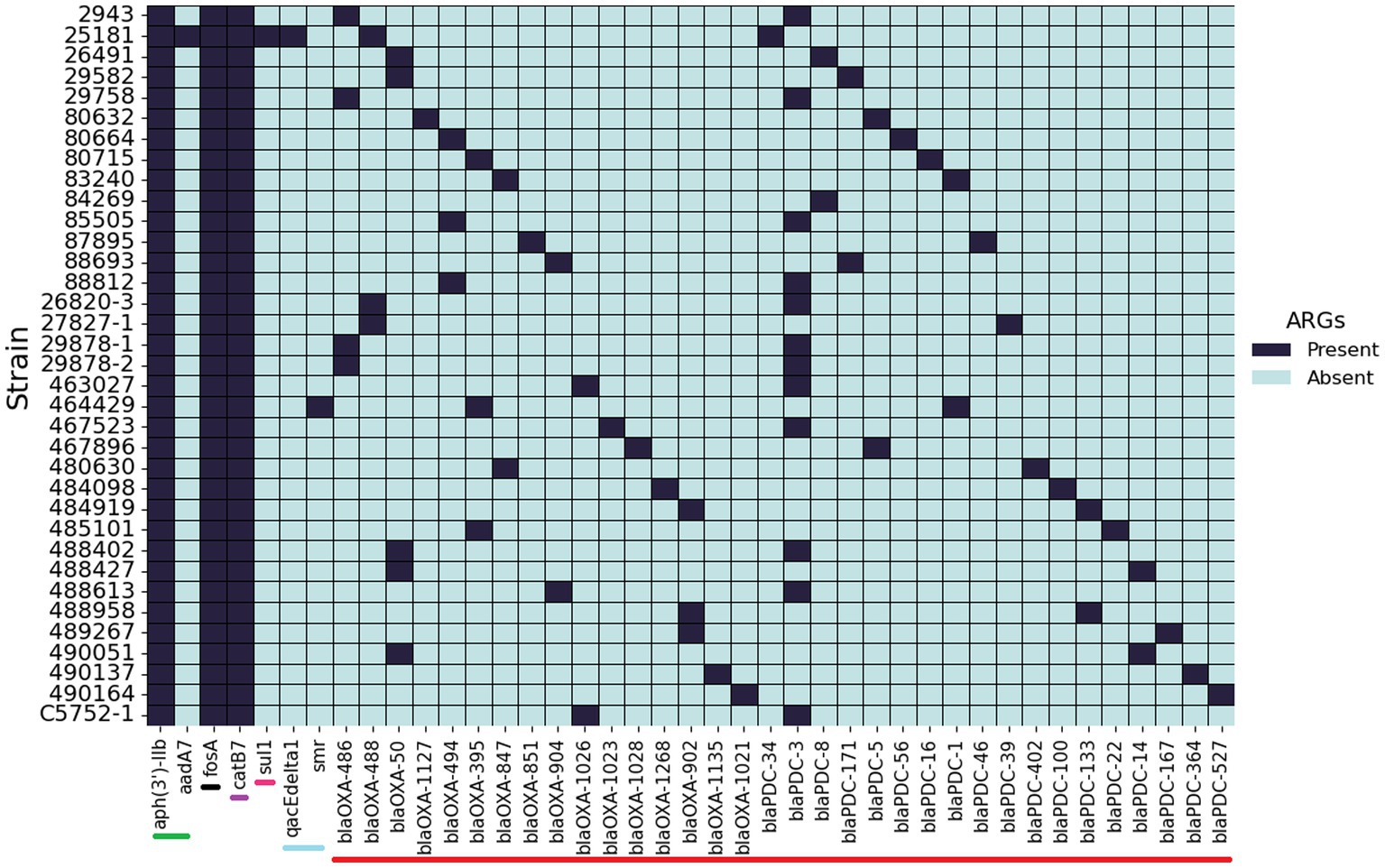
Figure 4. Presence and absence of antimicrobial resistance genes. Screening of 35 P. aeruginosa isolates from canine otitis infections for the presence of antimicrobial resistance genes. Bars at the base of the figure indicate the antibiotic class: Green—aminoglycoside, Black—Fosfomycin, Purple—chloramphenicol, Pink—sulfonamide, Light blue—Quaternary ammonium, Red—β-lactam.
Development of resistance through the acquisition of mutations is one of the main drivers of resistance in P. aeruginosa (López-Causapé et al., 2018). Twelve different mutations were identified in the present study (Supplementary Table 3); the most common occurring in oprD. Other common mutations included those in the quinolone resistance-determining region (QRDR), which are the primary way in which P. aeruginosa can become resistant to fluoroquinolones (Piddock, 1999). In E. coli these mutations occur between amino acid residues 67–107 for GyrA, and 63–102 for ParC (Egorov et al., 2018). Studies with P. aeruginosa have reported the QRDR regions in GyrB as 429–585 and ParE as 357–503 (Bruchmann et al., 2013).
QRDR mutations were identified in almost all of the isolates that were resistant to at least one fluoroquinolone antimicrobial—with the exception of 84269 that was resistant to enrofloxacin but did not contain a QRDR mutation. When examining ARG individually, gyrA mutations were the most common, occurring in 20% (7/35) of isolates. The mutation T83I in GyrA was the most common among strains resistant to at least one fluoroquinolone. S466F, S87L/S331T and D533E were the most common mutations in GyrB, ParC and ParE respectively. These are within the QRDR regions, and have been previously associated with resistance (Akasaka et al., 2001; Bruchmann et al., 2013). Furthermore, some of the isolates contained more than one QRDR mutation which is known to further increase the MIC of ciprofloxacin (Bruchmann et al., 2013).
Two of the sequenced strains were resistant to gentamycin. Resistance to aminoglycosides can be due to inactivation by enzymes but also though reduced permeability or increased efflux (Poole, 2005). More recently, mutations to fusA1 in P. aeruginosa have been shown to confer resistance to aminoglycosides (Bolard et al., 2018). One chromosomal aminoglycoside resistance gene, 3′-phosphotransferase (aph(3′)-IIb), was identified which conferred resistance to kanamycin A and B, neomycin B and C, butirosin and seldomycin F5 in all the sequenced isolates (Zeng and Jin, 2003). However, aph(3′)-IIb does not provide resistance to the clinically relevant aminoglycosides tested in the present study (Atassi et al., 2023).
Only a small number of sequenced isolates (2/35) harboured a mutation in fusA1. Strain 488958, which was resistant to all of the tested aminoglycosides, contained T671A. Mutations in domains II, IV and V have been reported to increase the MIC of aminoglycosides up to 16-fold. T671A has been shown to significantly decrease the susceptibility of P. aeruginosa to aminoglycosides including gentamicin, amikacin and tobramycin (Bolard et al., 2018). Conversely, strain 87895 contained a Q678L mutation in the fusA1 gene which is known to result in a four-fold increase in the minimum inhibitory concentration (MIC) of tobramycin, however this strain was sensitive to tobramycin, and other tested aminoglycosides, in the present study (Scribner et al., 2020).
Other mutations identified in the present study include the introduction of a premature stop codon in mexZ in 8.6% (3/35) of the isolates. Inactivation of mexZ results in the overproduction of the MexXY-OprM efflux pump which can result in decreased susceptibility to fluoroquinolones, aminoglycosides and cefepime (López-Causapé et al., 2018). Finally, 14% (5/35) of the isolates harboured a V15I mutation in the pmrB gene, previously identified in clinical strains with an increased MIC of colistin (Lee and Ko, 2014) although this antibiotic was not included in the present study.
Biofilm-forming ability of Pseudomonas aeruginosa isolates
Clinical isolates of P. aeruginosa were tested for their ability to form a biofilm in vitro (Figure 5; Supplementary Figure 2). In total, 82% (207/253) of strains produced strong levels of biofilm, 9% (23/253) produced moderate biofilm, 5% (13/253) produced weak biofilm and 4% (10/253) made no quantifiable biofilm under the tested conditions.
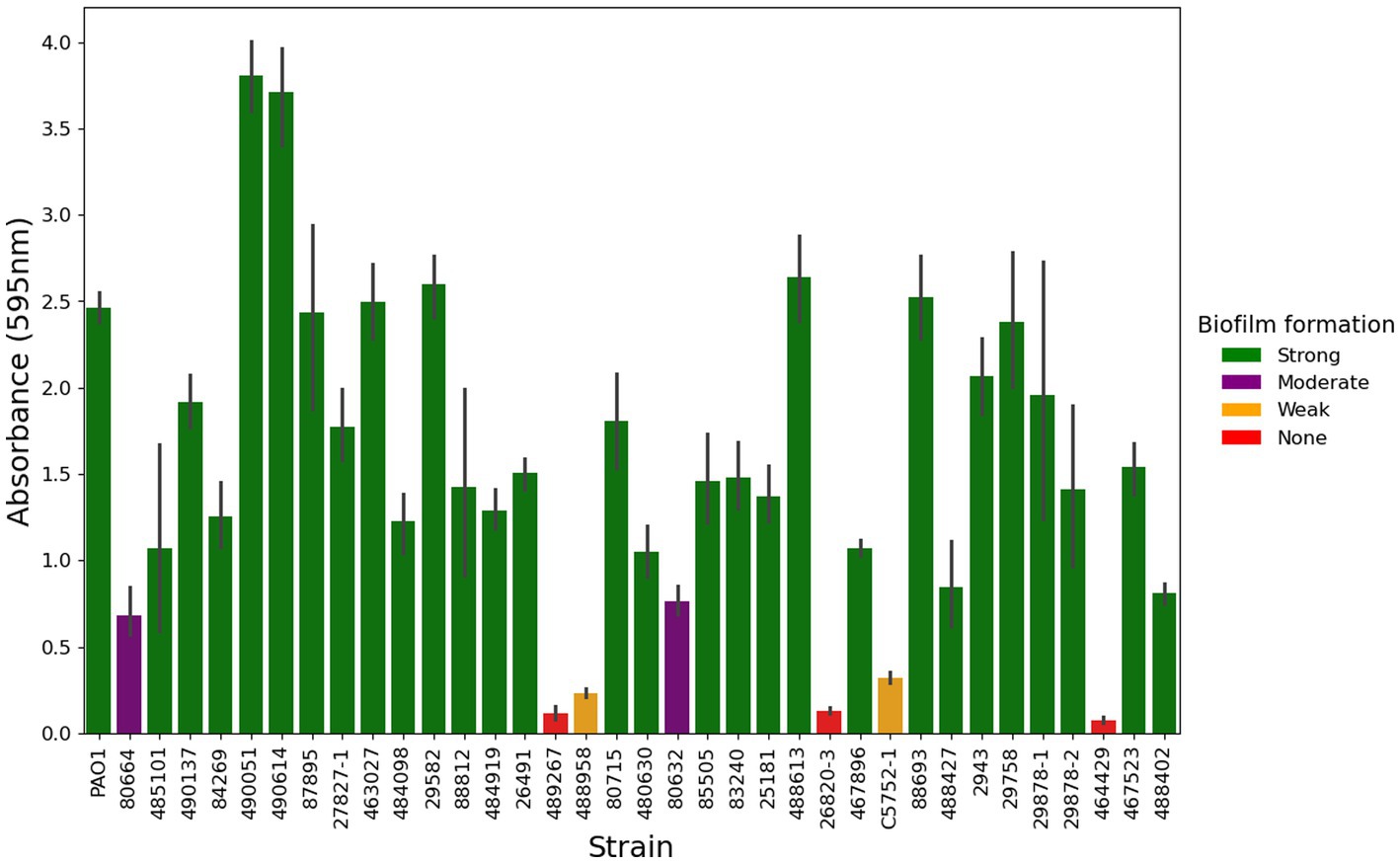
Figure 5. Biofilm formation of P. aeruginosa from canine otitis externa. The biofilm forming ability of 253 P. aeruginosa strains was tested in a crystal violet, 96 well plate assay, a representative subset of 35 strains is shown here. Green—Strong biofilm producing, Purple—Moderate biofilm producing, Amber—Weak biofilm producing, Red—No quantifiable biofilm. The values here are from three biological and six technical repeats represented by the mean with 95% confidence interval.
Detection of biofilm associated genes
To further understand the differences between levels of biofilm formation, the genomes of 35 P. aeruginosa isolates were screened for the presence of 53 genes known to be associated with biofilm formation (Supplementary Figure 3). All of the genes were highly conserved when compared to PAO1, with the exception of algP—a histone-like regulatory protein whose function in alginate synthesis is disputed and may be strain-dependent (Cross et al., 2020). All but one of the isolates contained all 53 genes, with the exception of 464429 which did not contain pslABCD. These genes encode the psl polysaccharide, which contributes to submerged biofilm (Friedman and Kolter, 2004), which may explain the poor biofilm forming ability of this strain.
Swarming motility
Most strains (71%, 179/253) tested positive for swarming motility (Figure 6). The positive control strain (PAO1) consistently migrated to the perimeter of the agar plate within 16 h of incubation, and 16% (41/253) of the clinical isolates showed a similar swarming ability. Of the 205 isolates that produced strong levels of biofilm, 72% (147) also tested positive for swarming. Interestingly, strain 29051—the strongest biofilm producer—tested negative for swarming. When the relationship between biofilm formation and swarming motility was investigated no correlation was identified (r2 = 0.002; Supplementary Figure 4).
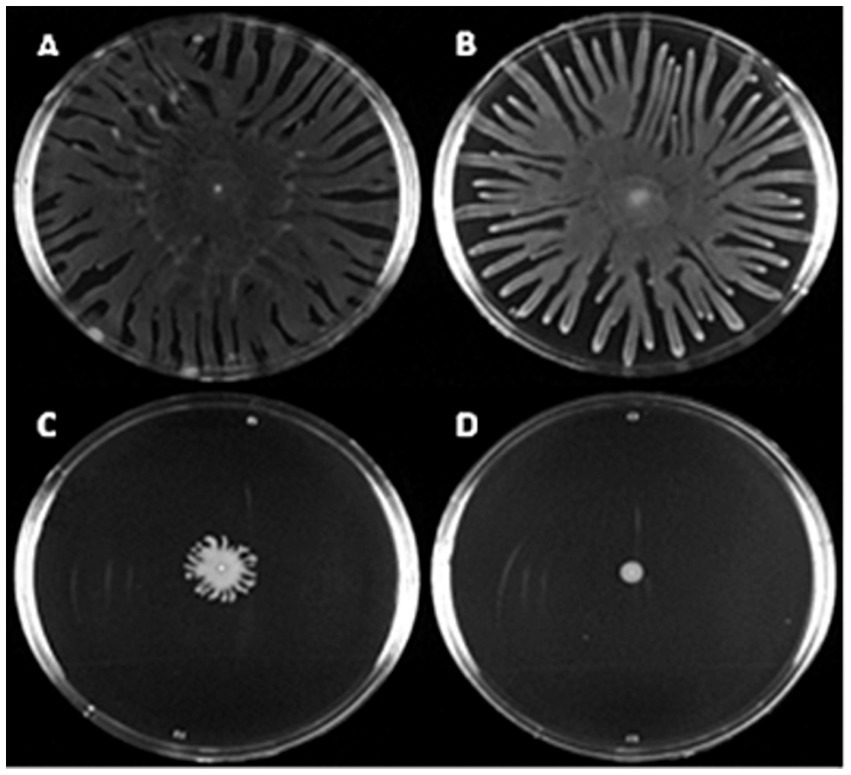
Figure 6. Canine OE isolates exhibit a range of swarming motility levels. Overnight cultures grown in LB were spotted onto swarming motility agar and incubated at 37° C for 16 h before imaging. Above shows selection of P. aeruginosa clinical canine otitis externa isolates with varying levels of swarming motility. (A–C) Shows strains PAO1, 83240 and C3524. (D) Shows strain 29775 which is swarming negative under the tested conditions. Images shown are representative of three biological repeats.
Presence of virulence genes in clinical isolates
The presence and absence of virulence genes was investigated using ABRicate with VFDB. Comparison of the canine OE isolates from this study to other P. aeruginosa isolates from human, animal and environmental sources using hierarchical clustering highlighted that there is no specific virulence profile associated with canine OE infection (Figure 7). Additionally, this highlighted that the strains did not cluster within phylogroups based on virulence factors. However, one notable exception is a cluster of 12 strains– including some from canines—which lacked numerous virulence factors, namely a series of genes involved in type III secretion systems. The majority (92%; 11/12), of these isolates belonged to phylogroup C, specifically, the section of the tree identified as group 5 by Freschi et al., 2019. These results are in line with the aforementioned study as they also identified that isolates in this group were missing 36 genes that encode for a Type III secretion system. The remaining isolate belonged to phylogroup A and was missing 32/36 of the type three secretion genes.
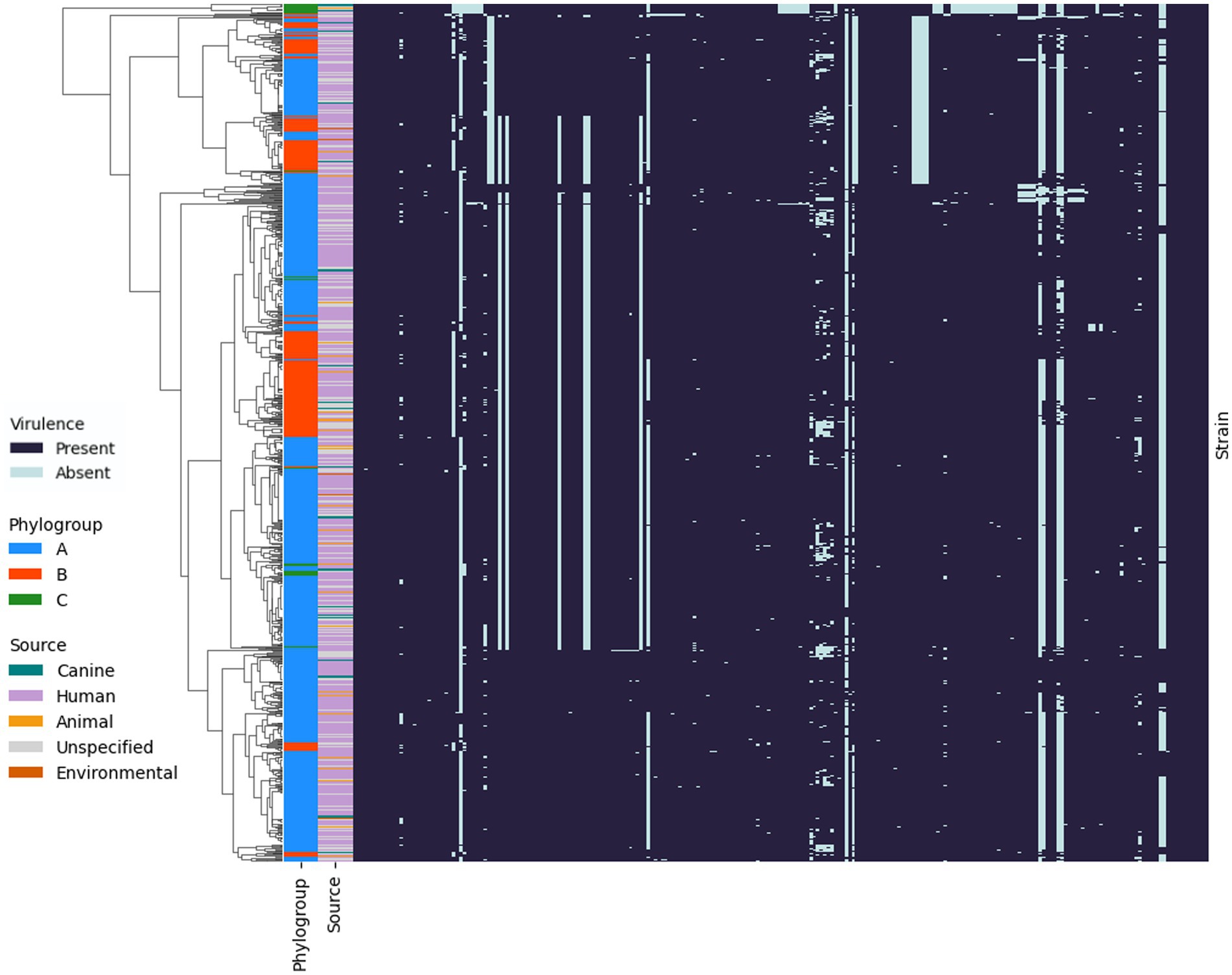
Figure 7. Presence and absence of virulence genes in the genomes of P. aeruginosa isolates. Hierarchical clustering using Jaccard distance and average linkage is shown on the left. Phylogroups A (blue), B (orange) and C (green) for each strain are shown in the outer band, with the source of the strains indicated in the inner band.
Discussion
Canine otitis caused by P. aeruginosa is a very common infection seen in veterinary practices worldwide which is frequently resistant to antimicrobial treatment. To our knowledge, this is the first study to use a combination of whole genome sequencing, phenotypic screening and bioinformatic analysis to characterise a large collection of clinical OE isolates of P. aeruginosa from diverse geographical origins. This revealed extensive genomic diversity across strains, as well as high levels of resistance to antimicrobials—particularly fluoroquinolones which are often used in the treatment of canine otitis. Furthermore, the MLST sequence type of two isolates (ST111, ST244) were identical to STs known to be associated with AMR nosocomial infections in people (Oliver et al., 2024), which may pose a risk of zoonotic spread, particularly to immunocompromised individuals. Interestingly, ST111 has been shown to have a low affinity to cystic fibrosis cases while ST244 has an intermediate affinity (Weimann et al., 2024).
MLST has been used previously to show that canine P. aeruginosa isolates are diverse (Hyun et al., 2018; Elfadadny et al., 2023). This is consistent with research on Pseudomonas infections in people, which also revealed a predominantly non-clonal population structure interspersed with highly successful epidemic clones and clonal complexes (Maatallah et al., 2011). These ‘high-risk’ clones include STs such as ST235, ST111, ST233 and ST244 (Oliver et al., 2024). Transmission of Pseudomonas between dogs and people has been reported in both directions (Fernandes et al., 2018; Santaniello et al., 2020). The identification of high-risk clones in dogs is particularly concerning considering the level of AMR detected, many of which are important in human medicine. Should this occur, the associated risks may increase over time given the ageing population in many countries, and higher dog ownership levels following the Covid-19 pandemic.
Antimicrobial resistance of isolates in this study was highest for fluoroquinolones. The levels of resistance reported here are similar to previous studies (Arais et al., 2016; Petrov et al., 2019; KuKanich et al., 2022). Fluoroquinolones are defined by the WHO as Critically Important Antimicrobials for human medicine, to be used only when alternative antimicrobials were ineffective (EMA, 2019). All strains resistant to ciprofloxacin were also resistant to levofloxacin and enrofloxacin suggesting that it is the most effective fluoroquinolone against Pseudomonas in vitro. This is most likely due to its lower affinity to efflux pumps compared to other fluoroquinolones (Teresa Tejedor et al., 2003). This is supported by strain 26491, which was resistant to enrofloxacin and levofloxacin but not ciprofloxacin and contained a mutation that is known to result in the overexpression of the MexXY-OprM efflux pump. Similarly, continuous exposure studies using E. coli from canine infections shows that QRDR mutations occur after treatment with marbofloxacin that can provide protection against other fluoroquinolones (Gebru et al., 2011). This is important because marbofloxacin is a component of antimicrobial containing otic treatments used in the United Kingdom.
Antimicrobial resistance gene (ARG) profiles amongst the isolates, as determined using AMRFinderPlus, were very similar. However, two integron associated genes, aadA7 and sul1, were identified. Although these genes did not account for resistance to the antimicrobials tested in the present study, it does highlight that P. aeruginosa isolates in dogs can carry mobile genetic elements carrying antimicrobial resistance genes. This led to the investigation of mutations which identified five point mutations in the QRDR region that had previously been linked to a resistant phenotype. The presence of QRDR mutations has been studied in P. aeruginosa isolates from dog infections in Korea and Brazil where, similar to the present study, GyrA T83I was identified as a prevalent mutation (Arais et al., 2016; Park et al., 2020). This suggests that the high levels of fluoroquinolone resistance seen in the present study is likely due to QRDR mutations. Ultimately, this suggests that the use of antimicrobials in veterinary medicine can select for the development of mutations to clinically important human antimicrobials.
QRDR mutations do not account for all of the resistance seen in this study. Pseudomonas aeruginosa 84269 and 26491 were resistant to enrofloxacin/enrofloxacin and levofloxacin, respectively, but did not contain any mutations in the genes investigated. However, other well-characterised fluoroquinolone resistance mechanisms such as efflux pump overexpression and decreased outer membrane permeability could be responsible for the resistance observed as a mutation that introduced a premature stop codon into mexZ, which is known to result in efflux pump overexpression, was identified in 26491, suggesting that increased efflux might be the cause of this resistance (Hooper and Jacoby, 2016; López-Causapé et al., 2018). Moreover, 88812, which also had a mexZ mutation, was resistant to all the tested fluoroquinolones, in addition to amikacin and gentamicin. Although this isolate did have mutations in the QRDR, no resistance mechanisms for aminoglycosides were identified, again highlighting efflux as a potential resistance mechanism. However, it is thought that up to 200 genes could be involved in resistance to ciprofloxacin so it is possible that there are unknown mechanisms contributing to fluoroquinolone resistance (Bruchmann et al., 2013).
Worryingly, resistance to two carbapenem antimicrobials, meropenem and imipenem, was identified, with the latter present in 8.33% of isolates. This is significant as the World Health Organisation (WHO) recognises a critical need for new antimicrobials for the treatment of carbapenem resistant P. aeruginosa (WHO, 2017). Of the sequenced isolates in the present study, only one showed resistance to imipenem although no carbapenemase genes were identified. Carbapenemases have been found previously in a P. aeruginosa isolate from a case of canine otitis externa in Korea (Hyun et al., 2018).
Biofilm formation has been found in 40–100% of clinical P. aeruginosa from canine OE (Pye et al., 2013; Chan et al., 2019; Robinson et al., 2019). In our study, 96% of strains produced biofilm in vitro, with 82% classified as strong biofilm formers. Of the 4% of strains producing no detectable biofilm, only one (464429) was missing any of the biofilm genes that were screened for (specifically pslABCD).
Deletion of pslD can eliminate the ability of P. aeruginosa PAO1 to attach to a microtiter dish (Colvin et al., 2012). Some P. aeruginosa isolates have been shown to be deficient in psl production; most notably PA14, which is also missing pslABCD (Friedman and Kolter, 2004). As a result, PA14 uses pel as its primary biofilm matrix polysaccharide. Despite this, P. aeruginosa PA14 is still able to adhere to a microtiter dish and form a biofilm after 24 h (Colvin et al., 2012). This could mean that the strains that produce no quantifiable biofilm in the present study have an attachment deficiency. The presence of non-biofilm producing clinical strains is not yet fully understood, particularly given most of these have a full complement of biofilm-related genes. This might be explained by divergent expression in different environments, or deficiencies in transcriptional regulators such as LasR (Lima et al., 2018; Kamali et al., 2020).
Little is known about virulence factors specific to the development of canine OE. Investigation of five virulence genes in P. aeruginosa from canine sources, including the ear canal, found that three genes, lasB, aprA, and plcH, were present in all of the tested isolates, while exoS and toxA were present in 87.5 and 91.7%, respectively (Hattab et al., 2021). Similarly, in the present study lasB, aprA, and plcH were identified in all of the isolates and toxA in 91%.
Pseudomonas aeruginosa isolates usually harbour either exoU or exoS, although the reason for this is unclear. The distinction is important because exoU+ strains are associated with more severe infections, and chronic OM infections in people (Park et al., 2017; Ozer et al., 2019). In the present study, exoS+ strains constituted 69% (24/35) of the population with 14% (5/35) exoU+. Interesting, one isolate 3% (1/35) carried both genes and 14% (5/35) had neither. All the exoU+ isolates in addition to the isolate containing both genes were located on the same branch of the phylogenetic tree as PA14 as previously described (Ozer et al., 2019). Although, other groups have reported isolates with both genes in phylogroup A (Botelho et al., 2023). None of the virulence genes identified in the canine isolates could be implicated in specifically having a role in this disease, as similar genes were also identified in environmental and human disease isolates.
In conclusion, our study has applied genome sequencing, phenotypic and bioinformatic analysis to a large collection of P. aeruginosa isolates from canine otitis externa. Although two of the isolates shared an MLST sequence type with high-risk STs from nosocomial human infections, there was little overlap of STs with previous studies. This genetic diversity was further supported by phylogenetic analysis which revealed no specific clustering of canine isolates; supporting the hypothesis that infections are acquired from environmental sources. Antimicrobial resistance was common, particularly towards the fluoroquinolones, which has implications for veterinary and human therapeutic failure. Although strains in this study did harbour known virulence and biofilm-associated genes, none of these were specifically associated with canine OE. These findings may help stimulate further research into the fundamental mechanisms of canine OE, and the optimising of therapy.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
BS: Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. SS: Funding acquisition, Supervision, Writing – review & editing. LH: Funding acquisition, Investigation, Methodology, Supervision, Project administration, Resources, Writing – review & editing. RA: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/M008770/1].
Acknowledgments
We are grateful to Steven Steen (CALP NationWide Laboratories), Arshnee Moodley (University of Copenhagen) and Anette Loeffler (Royal Veterinary College) for providing the clinical isolates of Pseudomonas aeruginosa used in this study. We are grateful to Adam Blanchard (University of Nottingham) for providing bioinformatics support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1526843/full#supplementary-material
References
Ahmed, A. M., Nakagawa, T., Arakawa, E., Ramamurthy, T., Shinoda, S., and Shimamoto, T. (2004). New aminoglycoside acetyltransferase gene, aac(3)-id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J. Antimicrob. Chemother. 53, 947–951. doi: 10.1093/jac/dkh221
Akasaka, T., Tanaka, M., Yamaguchi, A., and Sato, K. (2001). Type II topoisomerase mutations in fluoroquinolone-resistant clinical strains of Pseudomonas aeruginosa isolated in 1998 and 1999: role of target enzyme in mechanism of fluoroquinolone resistance. Antimicrob. Agents Chemother. 45, 2263–2268. doi: 10.1128/aac.45.8.2263-2268.2001
Andrews, S. (2010). Babraham bioinformatics - FastQC a quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed December 21, 2023).
Arais, L. R., Barbosa, A. V., Carvalho, C. A., and Cerqueira, A. M. F. (2016). Antimicrobial resistance, integron carriage, and gyrA and gyrB mutations in Pseudomonas aeruginosa isolated from dogs with otitis externa and pyoderma in Brazil. Vet. Dermatol. 27, 113–e31. doi: 10.1111/vde.12290
Atassi, G., Medernach, R., Scheetz, M., Nozick, S., Rhodes, N. J., Murphy-Belcaster, M., et al. (2023). Genomics of aminoglycoside resistance in Pseudomonas aeruginosa bloodstream infections at a United States academic hospital. Microbiology Spectrum 11:e0508722. doi: 10.1128/spectrum.05087-22
Bolard, A., Plésiat, P., and Jeannot, K. (2018). Mutations in gene fusA1 as a novel mechanism of aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62, e01835–e01817. doi: 10.1128/AAC.01835-17
Botelho, J., Tüffers, L., Fuss, J., Buchholz, F., Utpatel, C., Klockgether, J., et al. (2023). Phylogroup-specific variation shapes the clustering of antimicrobial resistance genes and defence systems across regions of genome plasticity in Pseudomonas aeruginosa. EBioMedicine 90:104532. doi: 10.1016/j.ebiom.2023.104532
Bouras, G., Grigson, S. R., Papudeshi, B., Mallawaarachchi, V., and Roach, M. J. (2024). Dnaapler: a tool to reorient circular microbial genomes. J. Open Source Software 9:5968. doi: 10.21105/joss.05968
Bruchmann, S., Dötsch, A., Nouri, B., Chaberny, I. F., and Häussler, S. (2013). Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob. Agents Chemother. 57, 1361–1368. doi: 10.1128/aac.01581-12
Chan, W. Y., Hickey, E. E., Page, S. W., Trott, D. J., and Hill, P. B. (2019). Biofilm production by pathogens associated with canine otitis externa, and the antibiofilm activity of ionophores and antimicrobial adjuvants. Vet Pharm & Therapeutics 42, 682–692. doi: 10.1111/jvp.12811
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
CLSI (2012). M02-A11: Performance standards for antimicrobial disk susceptibility tests; approved standard—Eleventh edition. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute.
CLSI (2018a). CLSI M100: Performance standards for antimicrobial susceptibility testing. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute.
CLSI (2018b). VLSI VET01s: Performance standards for antimicrobial disk and dilution susceptibility tests for Bacteria isolated from animals. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute.
Coffey, B. M., and Anderson, G. G. (2014). “Biofilm formation in the 96-well microtiter plate” in Pseudomonas methods and protocols. eds. A. Filloux and J.-L. Ramos (New York, NY: Springer), 631–641.
Colvin, K. M., Irie, Y., Tart, C. S., Urbano, R., Whitney, J. C., Ryder, C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x
Cox, G., and Wright, G. D. (2013). Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 303, 287–292. doi: 10.1016/j.ijmm.2013.02.009
Cross, A. R., Csatary, E. E., Raghuram, V., Diggle, F. L., Whiteley, M., Wuest, W. M., et al. (2020). The histone-like protein AlgP regulon is distinct in mucoid and nonmucoid Pseudomonas aeruginosa and does not include alginate biosynthesis genes. Microbiology 166, 861–866. doi: 10.1099/mic.0.000923
Cystic Fibrosis Canada (2023). The Canadian cystic fibrosis registry 2022 annual data report. Toronto, Canada: Cystric Fibrosis Canada.
Cystic Fibrosis Foundation (2023). Cystic Fibrosis Foundation patient registry 2023 annual data report. Bethesda, Maryland: Cystic Fibrosis Foundation.
Egorov, A. M., Ulyashova, M. M., and Rubtsova, M. Y. (2018). Bacterial enzymes and antibiotic resistance. Acta Nat. 10, 33–48. doi: 10.32607/20758251-2018-10-4-33-48
Elfadadny, A., Uchiyama, J., Goto, K., Imanishi, I., Ragab, R. F., Nageeb, W. M., et al. (2023). Antimicrobial resistance and genotyping of Pseudomonas aeruginosa isolated from the ear canals of dogs in Japan. Front Vet Sci 10:1074127. doi: 10.3389/fvets.2023.1074127
EMA (2019). Categorisation of antibiotics in the European Union. Available at: https://www.ema.europa.eu/en/documents/report/categorisation-antibiotics-european-union-answer-request-european-commission-updating-scientific-advice-impact-public-health-and-animal-health-use-antibiotics-animals_en.pdf (Accessed October 31, 2024).
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11:12728. doi: 10.1038/s41598-021-91456-0
Fernandes, M. R., Sellera, F. P., Moura, Q., Carvalho, M. P. N., Rosato, P. N., Cerdeira, L., et al. (2018). Zooanthroponotic transmission of drug-resistant Pseudomonas aeruginosa, Brazil. Emerg. Infect. Dis. 24, 1160–1162. doi: 10.3201/eid2406.180335
Franklin, M., Nivens, D., Weadge, J., and Howell, P. (2011). Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2:167. doi: 10.3389/fmicb.2011.00167
Freschi, L., Vincent, A. T., Jeukens, J., Emond-Rheault, J.-G., Kukavica-Ibrulj, I., Dupont, M.-J., et al. (2019). The Pseudomonas aeruginosa Pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol. Evol. 11, 109–120. doi: 10.1093/gbe/evy259
Friedman, L., and Kolter, R. (2004). Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186, 4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004
Fusconi, M., Petrozza, V., Taddei, A. R., Vinciguerra, V., de, A., Chiarini, F., et al. (2011). Is biofilm the cause of chronic otitis externa? Laryngoscope 121, 2626–2633. doi: 10.1002/lary.22348
Gebru, E., Choi, M.-J., Lee, S.-J., Damte, D., and Park, S. C. (2011). Mutant-prevention concentration and mechanism of resistance in clinical isolates and enrofloxacin/marbofloxacin-selected mutants of Escherichia coli of canine origin. J. Med. Microbiol. 60, 1512–1522. doi: 10.1099/jmm.0.028654-0
Gigante, A. M., Hadis, M. A., Secker, B., Shaw, S. C., Cooper, P. R., Palin, W. M., et al. (2024). Exposure to blue light reduces antimicrobial resistant Pseudomonas aeruginosa isolated from dog ear infections. Front. Microbiol. 15:1414412. doi: 10.3389/fmicb.2024.1414412
Hattab, J., Mosca, F., di, C., Aste, G., Marruchella, G., Guardiani, P., et al. (2021). Occurrence, antimicrobial susceptibility, and pathogenic factors of Pseudomonas aeruginosa in canine clinical samples. Vet World 14, 978–985. doi: 10.14202/vetworld.2021.978-985
Hooper, D. C., and Jacoby, G. A. (2016). Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 6:a025320. doi: 10.1101/cshperspect.a025320
Hunt, M., Silva, N. D., Otto, T. D., Parkhill, J., Keane, J. A., and Harris, S. R. (2015). Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16:294. doi: 10.1186/s13059-015-0849-0
Hyun, J.-E., Chung, T.-H., and Hwang, C.-Y. (2018). Identification of VIM-2 metallo-β-lactamase-producing Pseudomonas aeruginosa isolated from dogs with pyoderma and otitis in Korea. Vet. Dermatol. 29, 186–e68. doi: 10.1111/vde.12534
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1
Kamali, E., Jamali, A., Ardebili, A., Ezadi, F., and Mohebbi, A. (2020). Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC. Res. Notes 13:27. doi: 10.1186/s13104-020-4890-z
Katz, L. S., Griswold, T., Morrison, S. S., Caravas, J. A., and Zhang, S. (2019). Mashtree: a rapid comparison of whole genome sequence files. J. Open Source Software 4:1762. doi: 10.21105/joss.01762
Kiel, A., Creutz, I., Rückert, C., Kaltschmidt, B. P., Hütten, A., Niehaus, K., et al. (2022). Genome-based analysis of virulence factors and biofilm formation in Novel P. aeruginosa strains isolated from household appliances. Microorganisms 10:2508. doi: 10.3390/microorganisms10122508
Kim, E., Choe, C., Yoo, J. G., Oh, S.-I., Jung, Y., Cho, A., et al. (2018). Major medical causes by breed and life stage for dogs presented at veterinary clinics in the Republic of Korea: a survey of electronic medical records. PeerJ 6:e5161. doi: 10.7717/peerj.5161
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
KuKanich, K. S., Bagladi-Swanson, M., and KuKanich, B. (2022). Pseudomonas aeruginosa susceptibility, antibiogram and clinical interpretation, and antimicrobial prescribing behaviors for dogs with otitis in the Midwestern United States. J. Vet. Pharmacol. Ther. 45, 440–449. doi: 10.1111/jvp.13077
Lee, J.-Y., and Ko, K. S. (2014). Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn. Microbiol. Infect. Dis. 78, 271–276. doi: 10.1016/j.diagmicrobio.2013.11.027
Letunic, I., and Bork, P. (2024). Interactive tree of life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82. doi: 10.1093/nar/gkae268
Lima, J. L. D. C., Alves, L. R., Jacomé, P. R. L. D. A., Bezerra Neto, J. P., and Maciel, M. A. V. (2018). Biofilm production by clinical isolates of Pseudomonas aeruginosa and structural changes in LasR protein of isolates non biofilm-producing. Braz. J. Infect. Dis. 22, 129–136. doi: 10.1016/j.bjid.2018.03.003
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2021). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
López-Causapé, C., Cabot, G., del Barrio-Tofiño, E., and Oliver, A. (2018). The versatile mutational Resistome of Pseudomonas aeruginosa. Front. Microbiol. 9:685. doi: 10.3389/fmicb.2018.00685
Lund, E. M., Armstrong, P. J., Kirk, C. A., Kolar, L. M., and Klausner, J. S. (1999). Health status and population characteristics of dogs and cats examined at private veterinary practices in the United States. J. Am. Vet. Med. Assoc. 214, 1336–1341. doi: 10.2460/javma.1999.214.09.1336
Maatallah, M., Cheriaa, J., Backhrouf, A., Iversen, A., Grundmann, H., do, T., et al. (2011). Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS One 6:e25617. doi: 10.1371/journal.pone.0025617
Nährlich, L., Burkhart, M., and Wosniok, J. (2023). German cystic fibrosis registry 2022 annual report. In den Dauen, Bonn, Germany: Mukoviszidose e.V.
Naito, Y., Adams, F., Charman, S., Duckers, J., Davies, G., and Clarke, S. (2023). UK cystic fibrosis registry 2023 annual data report. London: Cystric Fibrosis Trust.
Nuttall, T., and Cole, L. K. (2007). Evidence-based veterinary dermatology: a systematic review of interventions for treatment of Pseudomonas otitis in dogs. Vet. Dermatol. 18, 69–77. doi: 10.1111/j.1365-3164.2007.00575.x
O’Leary, N. A., Cox, E., Holmes, J. B., Anderson, W. R., Falk, R., Hem, V., et al. (2024). Exploring and retrieving sequence and metadata for species across the tree of life with NCBI datasets. Sci Data 11:732. doi: 10.1038/s41597-024-03571-y
O’Neill, D. G., Church, D. B., McGreevy, P. D., Thomson, P. C., and Brodbelt, D. C. (2014). Prevalence of disorders recorded in dogs attending primary-care veterinary practices in England. PLoS One 9:e90501. doi: 10.1371/journal.pone.0090501
O’Neill, D. G., James, H., Brodbelt, D. C., Church, D. B., and Pegram, C. (2021). Prevalence of commonly diagnosed disorders in UK dogs under primary veterinary care: results and applications. BMC Vet. Res. 17:69. doi: 10.1186/s12917-021-02775-3
Oliver, A., Rojo-Molinero, E., Arca-Suarez, J., Beşli, Y., Bogaerts, P., Cantón, R., et al. (2024). Pseudomonas aeruginosa antimicrobial susceptibility profiles, resistance mechanisms and international clonal lineages: update from ESGARS-ESCMID/ISARPAE group. Clin. Microbiol. Infect. 30, 469–480. doi: 10.1016/j.cmi.2023.12.026
ONT (2023). Medaka. Available at: https://github.com/nanoporetech/medaka (Accessed December 21, 2023).
Ozer, E. A., Nnah, E., Didelot, X., Whitaker, R. J., and Hauser, A. R. (2019). The population structure of Pseudomonas aeruginosa is characterized by genetic isolation of exoU+ and exoS+ lineages. Genome Biol. Evol. 11, 1780–1796. doi: 10.1093/gbe/evz119
Pang, Z., Raudonis, R., Glick, B. R., Lin, T.-J., and Cheng, Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013
Park, M.-H., Kim, S. Y., Roh, E. Y., and Lee, H. S. (2017). Difference of type 3 secretion system (T3SS) effector gene genotypes (exoU and exoS) and its implication to antibiotics resistances in isolates of Pseudomonas aeruginosa from chronic otitis media. Auris Nasus Larynx 44, 258–265. doi: 10.1016/j.anl.2016.07.005
Park, Y., Oh, J., Park, S., Sum, S., Song, W., Chae, J., et al. (2020). Antimicrobial resistance and novel mutations detected in the gyrA and parC genes of Pseudomonas aeruginosa strains isolated from companion dogs. BMC Vet. Res. 16:111. doi: 10.1186/s12917-020-02328-0
Paterson, S., and Matyskiewicz, W. (2018). A study to evaluate the primary causes associated with Pseudomonas otitis in 60 dogs. J. Small Anim. Pract. 59, 238–242. doi: 10.1111/jsap.12813
Perrin, A., and Rocha, E. P. C. (2021). PanACoTA: a modular tool for massive microbial comparative genomics. NAR Genomics and Bioinformatics 3:lqaa106. doi: 10.1093/nargab/lqaa106
Petrov, V., Zhelev, G., Marutsov, P., Koev, K., Georgieva, S., Toneva, I., et al. (2019). Microbiological and antibacterial resistance profile in canine otitis externa – a comparative analysis. BJVM 22, 447–456. doi: 10.15547/bjvm.2151
Piddock, L. J. V. (1999). Mechanisms of fluoroquinolone resistance: an update 1994–1998. Drugs 58, 11–18. doi: 10.2165/00003495-199958002-00003
Poole, K. (2005). Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49, 479–487. doi: 10.1128/AAC.49.2.479-487.2005
Pye, C. C., Yu, A. A., and Weese, J. S. (2013). Evaluation of biofilm production by Pseudomonas aeruginosa from canine ears and the impact of biofilm on antimicrobial susceptibility in vitro. Vet. Dermatol. 24, 446–e99. doi: 10.1111/vde.12040
Robinson, V. H., Paterson, S., Bennett, C., and Steen, S. I. (2019). Biofilm production of Pseudomonas spp. isolates from canine otitis in three different enrichment broths. Vet. Dermatol. 30, 218–e67. doi: 10.1111/vde.12738
Rudra, B., Duncan, L., Shah, A. J., Shah, H. N., and Gupta, R. S. (2022). Phylogenomic and comparative genomic studies robustly demarcate two distinct clades of Pseudomonas aeruginosa strains: proposal to transfer the strains from an outlier clade to a novel species Pseudomonas paraeruginosa sp. nov. Int. J. Syst. Evol. Microbiol. 72:005542. doi: 10.1099/ijsem.0.005542
Santaniello, A., Sansone, M., Fioretti, A., and Menna, L. F. (2020). Systematic review and Meta-analysis of the occurrence of ESKAPE Bacteria Group in Dogs, and the related zoonotic risk in animal-assisted therapy, and in animal-assisted activity in the health context. Int. J. Environ. Res. Public Health 17:3278. doi: 10.3390/ijerph17093278
Schwengers, O., Jelonek, L., Dieckmann, M. A., Beyvers, S., Blom, J., and Goesmann, A. (2021). Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microbial Genomics 7:000685. doi: 10.1099/mgen.0.000685
Scribner, M. R., Santos-Lopez, A., Marshall, C. W., Deitrick, C., and Cooper, V. S. (2020). Parallel evolution of tobramycin resistance across species and environments. MBio 11, e00932–e00920. doi: 10.1128/mBio.00932-20
Secker, B., Shaw, S., and Atterbury, R. J. (2023). Pseudomonas spp. in canine otitis externa. Microorganisms 11:2650. doi: 10.3390/microorganisms11112650
Seemann, T. (2015). ABRicate. Available at: https://github.com/tseemann/abricate (Accessed December 21, 2023).
Sköld, O. (2000). Sulfonamide resistance: mechanisms and trends. Drug Resist. Updat. 3, 155–160. doi: 10.1054/drup.2000.0146
Stepanović, S., Vuković, D., Dakić, I., Savić, B., and Švabić-Vlahović, M. (2000). A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40, 175–179. doi: 10.1016/S0167-7012(00)00122-6
Stewart, L., Ford, A., Sangal, V., Jeukens, J., Boyle, B., Kukavica-Ibrulj, I., et al. (2014). Draft genomes of 12 host-adapted and environmental isolates of Pseudomonas aeruginosa and their positions in the core genome phylogeny. Pathogens and Disease 71, 20–25. doi: 10.1111/2049-632X.12107
Sweeney, M. T., Lubbers, B. V., Schwarz, S., and Watts, J. L. (2018). Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 73, 1460–1463. doi: 10.1093/jac/dky043
Teresa Tejedor, M., Martín, J. L., Navia, M., Freixes, J., and Vila, J. (2003). Mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from canine infections. Vet. Microbiol. 94, 295–301. doi: 10.1016/s0378-1135(03)00129-9
Thöming, J. G., and Häussler, S. (2022). Pseudomonas aeruginosa is more tolerant under biofilm than under planktonic growth conditions: a multi-isolate survey. Front. Cell. Infect. Microbiol. 12:851784. doi: 10.3389/fcimb.2022.851784
Ugurlu, A., Karahasan Yagci, A., Ulusoy, S., Aksu, B., and Bosgelmez-Tinaz, G. (2016). Phenolic compounds affect production of pyocyanin, swarming motility and biofilm formation of Pseudomonas aeruginosa. Asian Pac. J. Trop. Biomed. 6, 698–701. doi: 10.1016/j.apjtb.2016.06.008
UKHSA (2024). UK standards for microbiology investigations - identification of Pseudomonas species and other nonglucose fermenters. Available at: https://www.rcpath.org/static/bfd560d2-eb3a-4260-bf1f63feee21325a/UK-SMI-ID-17i4-Identification-of-Pseudomonas-species-and-other-non-glucose-fermenters-September-2024.pdf (Accessed November 8, 2024).
Weimann, A., Dinan, A. M., Ruis, C., Bernut, A., Pont, S., Brown, K., et al. (2024). Evolution and host-specific adaptation of Pseudomonas aeruginosa. Science 385:eadi0908. doi: 10.1126/science.adi0908
WHO Publishes list of bacteria for which new antibiotics are urgently needed (2017). Available at: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (Accessed March 20, 2024).
Wick, R. R., and Holt, K. E. (2022). Polypolish: short-read polishing of long-read bacterial genome assemblies. PLoS Comput. Biol. 18:e1009802. doi: 10.1371/journal.pcbi.1009802
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Zeng, L., and Jin, S. (2003). Aph(3′)-IIb, a gene encoding an aminoglycoside-modifying enzyme, is under the positive control of surrogate regulator HpaA. Antimicrob. Agents Chemother. 47, 3867–3876. doi: 10.1128/aac.47.12.3867-3876.2003
Keywords: pseudomonas, canine, otitis externa, genome, AMR, biofilm, one health, nosocomial
Citation: Secker B, Shaw S, Hobley L and Atterbury RJ (2025) Genomic and phenotypic characterisation of Pseudomonas aeruginosa isolates from canine otitis externa reveals high-risk sequence types identical to those found in human nosocomial infections. Front. Microbiol. 16:1526843. doi: 10.3389/fmicb.2025.1526843
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Costas C. Papagiannitsis, University of Thessaly, GreeceBurkhard Tümmler, Hannover Medical School, Germany
Copyright © 2025 Secker, Shaw, Hobley and Atterbury. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert J. Atterbury, cm9iZXJ0LmF0dGVyYnVyeUBub3R0aW5naGFtLmFjLnVr; Laura Hobley, bGF1cmEuaG9ibGV5QG5vdHRpbmdoYW0uYWMudWs=
 Bailey Secker
Bailey Secker Stephen Shaw
Stephen Shaw Laura Hobley
Laura Hobley Robert J. Atterbury
Robert J. Atterbury