
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 February 2025
Sec. Food Microbiology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1519202
 Qinglei Meng1†
Qinglei Meng1† Lili Song2†
Lili Song2† Shanshan Chi1
Shanshan Chi1 Haifeng Wang3
Haifeng Wang3 Jie Li4
Jie Li4 Yunjiao Chen1
Yunjiao Chen1 Zhilin Liu1
Zhilin Liu1 Xin Zhang1
Xin Zhang1 Zelin Jia1
Zelin Jia1 Jiayu Cui1
Jiayu Cui1 Xueli Wang1*
Xueli Wang1*Objective: Bacillus cereus (B. cereus) can be used as a probiotic or produce a variety of toxins that are pathogenic to humans and animals. Environmental stressors can affect the growth process of B. cereus and the expression of its virulence genes. Due to the limitations of methods such as pharmacological disinfection methods (there are limits to the use of antibiotics) and chemical disinfection methods (chemical methods may produce residues), attempts can be made to remove and reduce B. cereus infections through environmental stress factors.
Methods: In this study, the expression of four virulence genes (nheA, hblD, cytK, and entFM) of bovine-origin lethal B. cereus was investigated by qPCR under the effect of different environmental stressors. The extent of pathological damage to various organs of mice by B. cereus was observed by pathological sections.
Results: The results showed that high temperature could inhibit the expression of B. cereus virulence genes. Expression of B. cereus virulence genes was affected under the influence of pH. Different salt concentrations could make the B. cereus virulence genes show low expression. Under a single environmental stressors, nheA, hblD, cytK, and entFM had the lowest expression at 40°C, pH 8.0, and were lowly expressed at all salt concentrations except the control group. The action of multiple environmental stressors affect the expression of virulence genes. Under multiple environmental stressors, nheA, hblD and cytK were least expressed at a temperature of 40°C, pH 6.0, and salt concentration of 3.0%, and entFM was least expressed at a temperature of 20°C, pH 8.0, and salt concentration of 1.5%. Animal pathogenicity tests have shown that environmental stressors affect the virulence of B. cereus.
Conclusion: The level of virulence gene expression in B. cereus can be reduced by environmental stress factors, thus further reducing the risk of B. cereus to human health. This study provides some reference for the prevention and control of B. cereus disease.
B. cereus is one of the more common conditionally pathogenic bacteria in the family Bacillaceae. Some B. cereus can be used as plant growth promoters and microbial additives for animal feeds (Elsayed et al., 2022; Gong et al., 2018). B. cereus produces vomiting enterotoxins and diarrhea enterotoxins that cause disease in humans and animals (Jia et al., 2022). Pathogenic B. cereus produces toxins that in turn cause disease in humans and animals, and in a few cases can be life-threatening. B. cereus is an increasing hazard to human health, and controlling the means of transmission of B. cereus can reduce B. cereus infections. The removal and reduction of B. cereus from the environment is important to ensure the safety of the food handling environment. B. cereus produces spores that are thermally stable, a property that allows B. cereus to survive in harsh environments (Bai et al., 2018). B. cereus is subject to acidity stress (including the combined effects of inorganic and organic acid stress) in environments with low pH. It has been shown that B. cereus has a lag period of 1, 2, and 5 h for experimental strains at pH 7.0, 5.3, and 4.9, respectively (Biesta-Peters et al., 2011). Other studies have shown that B. cereus can reproduce normally in higher salt concentrations (6.5–7.5%) (Jeon et al., 2016). In response to salt stress in B. cereus, the increase in Na+ and H+ components of the osmoprotectant and the activation of the oxidative stress response may create cross-protection against other stresses (Belaouni et al., 2022).
In this paper, four virulence genes (nheA, hblD, cytK, and entFM) of B. cereus were selected as targeted genes (it has been shown in the literature that these four virulence genes have high detection rates and high toxicity) (Chang et al., 2021; Berthold-Pluta et al., 2019; Rossi et al., 2018). The NHE complex contains three different proteins, NheA, NheB, and NheC, which are encoded by the nheA gene, the nheB gene and the nheC gene, respectively. Of these, NheA is considered to be the major diarrheal enterotoxin of B. cereus. The HblD is hemolytic and cytotoxic to intestinal and other cells. CytK is associated with foodborne diarrhea caused by B. cereus strains, it is cytotoxic and hemolytic, and the toxin may cause necrotizing enterocolitis in animals. EntFM is known as a cell wall peptidase and is involved in adhesion, motility, biofilm formation and cytotoxicity of B. cereus. In the current study, the relative expression of B. cereus virulence genes after 14 h of incubation under different stress conditions were determined by qPCR. A mouse model was used to evaluate the pathological damage caused by B. cereus to various organs of mice after 14 h of incubation under different stress conditions. The aim of this study was to identify environmental conditions from experimental data that are relatively unfavorable for the growth and expression of virulence genes in B. cereus. In this study, we controlled the expression levels of bacterial virulence genes through environmental stressors to reduce B. cereus infections in humans and animals. Prevention and control of B. cereus disease by controlling environmental stress factors is of clinical significance. This method can be used to prevent and control B. cereus disease by reducing the expression of B. cereus virulence genes. Common sterilization methods have limitations (for example, both pharmaceutical and chemical disinfection methods can produce residues; chemical disinfection methods have a higher odor, etc.). The method used in this study does not produce residues and is relatively safe to operate. This study is expected to provide some ideas for the prevention and control of B. cereus disease in the future.
Luria-Bertani (LB) agar medium (tryptone 10 g/L, yeast extract 5 g/L, sodium chloride 10 g/L, agar powder 15 g/L), LB broth medium (tryptone 10 g/L, yeast extract 5 g/L, sodium chloride 10 g/L). Sodium chloride, nutrient agar, glucose, and agar powder were purchased from Beijing Chemical Factory; yeast extract, tryptone were purchased from Zhengzhou Dening Biotechnology Company; multifunctional constant temperature shaker, constant temperature incubator were purchased from Henan Wolin Instrument Company; RNAprep pure Bacteria Kit, Bacterial genomic DNA extraction kit, TB Green® Premix Ex Taq™ II (Tli RNaseH Plus), DL2000 DNA marker, PCR mastermix, nucleic acid dye, agarose, 50 × Tris Acetate-EDTA buffer were purchased from TaKaRa Company (Bai et al., 2018; Li, 2017). Automatic gel imaging analysis system ZF-258 was purchased from TwisDxTM Company; StepOnePlus™ Real-Time Fluorescence PCR System and Evolution™ Pro UV–Vis spectrophotometer were purchased from Thermo Fisher Company. B. cereus strain was kindly provided by Laboratory of Veterinary Pathology, College of Animal Science and Technology, Inner Mongolia Minzu University, and the strain (we named the strain lycx) was isolated from the spleens of the sudden death cattle. A total of 200 Kunming mice (6–8 weeks old, 25–30 g weight) were purchased from Liaoning Changsheng Biotechnology Company. McFarland turbidimeter (Bacterial turbidimeter), McFarland turbidimeter tube were purchased from Shanghai Kunquan Biotechnology Company. The bacterial genomic DNA of experimental strains were uploaded in the sequence of the experimental strain to GenBank (Serial numbers are CP129005.1 and CP129006.1). Isolates were deposited at China General Microbiological Culture Collection Center (CGMCC No.17626).
B. cereus was incubated with LB broth medium for 14 h with shaking (37°C, 160 rpm), and the OD value of the bacterial culture was measured every 2 h and the growth curve of the bacteria was plotted (Use of GraphPad Prism version 5.01). Cultures of B. cereus were carried out in three replicates.
DNA of the strain was extracted using a Bacterial genomic DNA extraction kit. Primers were synthesized with reference to the relevant literature (as shown in Table 1) and detected using agarose gel electrophoresis after PCR amplification (Hansen and Hendriksen, 2001; Ngamwongsatit et al., 2008; Agata et al., 1995; Asano et al., 1997; Kim et al., 2013; Wang and Liu, 2018).
B. cereus was cultured (37°C, 14 h) using LB broth medium. Temperature group medium: LB broth medium was incubated at 20, 30, and 40°C, and the experimental groups were named as “group 20,” “group 30,” and “group 40.” pH group medium: The pH of LB broth medium was adjusted to 6.0, 7.0, and 8.0, and the experimental groups were named as “group 6,” “group 7,” and “group 8.” Salt concentration group medium: The salt concentration of LB broth medium was adjusted to 0, 1.5, and 3.0%, and the experimental groups were named as “group 0,” “group 1.5,” and “group 3.” Total RNA was extracted from the strains using an RNA extraction kit, and the RNA concentration was determined and then the RNA was reverse transcribed to cDNA. Synthetic primers were designed according to the relevant literature (as shown in part b of Table 1; Xu, 2023; Jing et al., 2018). The cDNA was used as a template, and PCR amplification of the gatB_Yqey gene was verified by agarose gel electrophoresis (Reiter et al., 2011). The expression of four virulence genes, nheA, hblD, entFM, and cytK, was determined using gat B_Yqey as an internal reference gene. Reactions were performed using qPCR kits (3 replicates of qPCR reactions were performed for each test group) and experimental data were saved for subsequent analysis (Use of GraphPad Prism version 5.01).
Using an orthogonal test, a 3-factor (temperature, pH, salt concentration), 3-level manipulation was designed (Wang and Liu, 2018). The factors and levels are shown in part “a” of Table 2, and the operational protocol for the orthogonal test is shown in part “b” of Table 2. Experimental data were analyzed using GraphPad Prism version 5.01. It has been shown that the optimum growth temperature of B. cereus is 28–35°C (Reiter et al., 2011). We chose three temperatures, 20, 30, and 40°C, to conduct the experiment in this study. B. cereus grows more slowly when temperatures are too high or too low, and it is less harmful to humans and animals. Have a neutral pH, so gradients of 6.0, 7.0, and 8.0 were chosen for the test. Bacteria grown under mild salt stress conditions showed a slight decrease in growth rate compared to normal growth conditions. Severe salt stress may lead to a lag period in bacterial growth followed by a resumption of growth, so the salt concentration used should not be too high (Li, 2017).
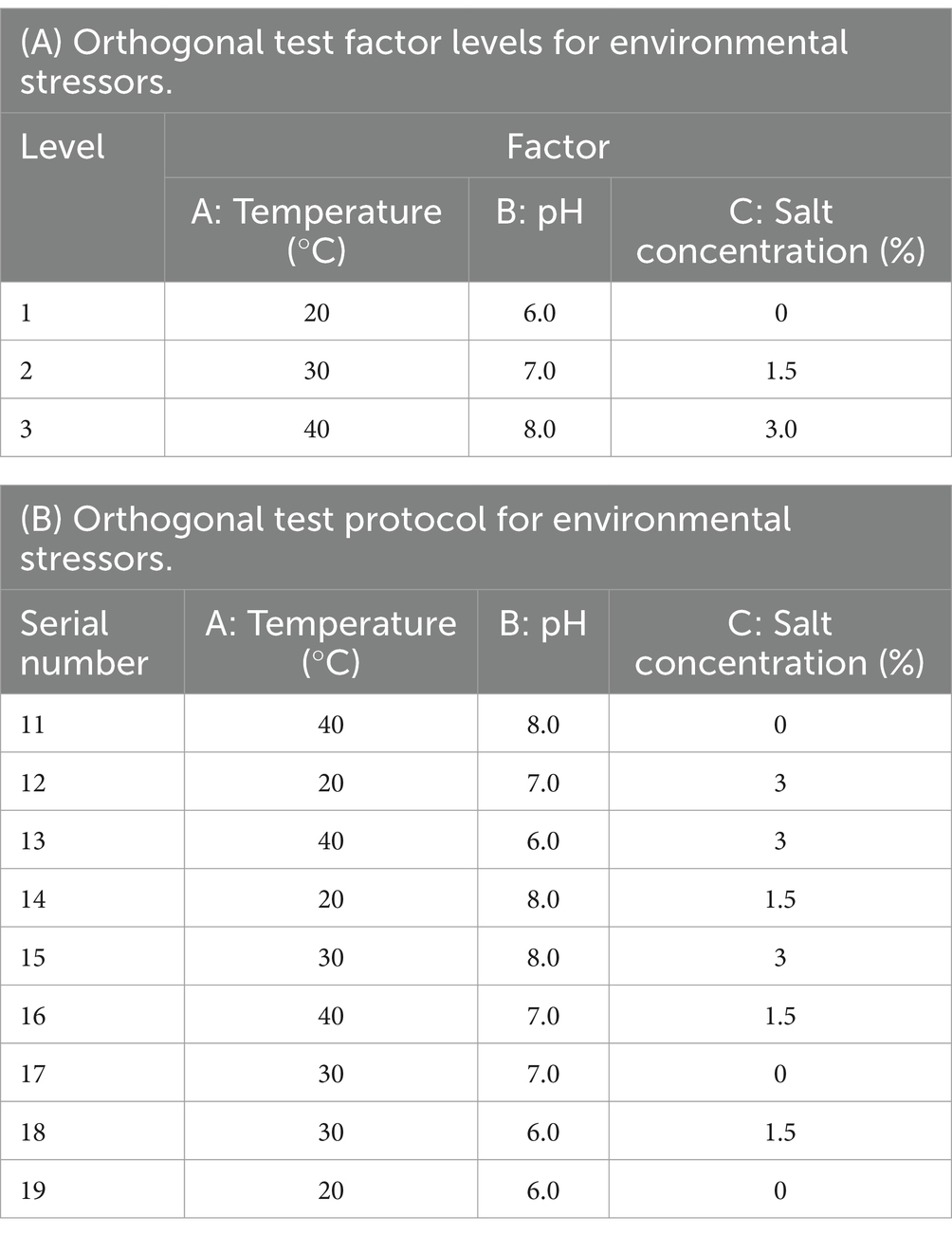
Table 2. Factor levels and experimental program for orthogonal testing of environmental stress factors.
Total bacterial RNA was extracted and reverse transcribed into cDNA using B. cereus cultured under the above conditions. The quality of cDNA was assessed through endpoint PCR targeting the housekeeping gene gatB_Yqey, followed by analysis using agarose gel electrophoresis. The validated cDNA was then utilized for qPCR reactions.
Effect of unifactorial environmental stressors on the pathogenicity of B. cereus. Mice were given intraperitoneal injections of B. cereus bacterial culture (Each mouse was injected intraperitoneally with 0.3 mL of bacterial culture at a concentration of 109 CFU/mL) in 9 groups (groups 0, 1.5, 3, 6, 7, 8, 20, 30, 40). There were 10 mice in each group. Twenty more mice were also selected as a control group (10 injected with LB broth medium; 10 injections of bacterial culture cultured using the conditions of 1.2.1, with a salt concentration of 1%, a pH of 7 and an incubation temperature of 37°C). The salt concentrations were 0, 1.5, and 3.0% in groups 0, 1.5, and 3, respectively; in the remaining groups the salt concentration was 1%. Groups 6, 7, 8 corresponded to the following culture media pH: 6, 7, and 8, respectively; the rest of the groups had pH 7.2. Groups 20, 30, and 40 were incubated at 20, 30, and 40°C, respectively; the remaining groups were incubated at 37°C. The time of death was recorded for each group of mice. Mice were dissected immediately after death. Mice that did not die after 3 d (d: day) were anesthetized with 0.3% pentobarbital (5 mg/100 g, intraperitoneal injection), whereas the mice were then executed using the cervical dislocation method and immediately dissected after execution (National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals, 2011). The organs of dissected mice were immersed and fixed for 3 days in 10% formaldehyde solution. Pathologic tissue sections were made using mouse organs (Hematoxylin–Eosin staining).
Effect of multifactorial environmental stressors on the pathogenicity of B. cereus. Mice were given intraperitoneal injections of B. cereus bacterial culture (groups 11, 12, 13, 14, 15, 16, 17, 18, 19), 10 in each group. The salt concentrations of groups 11, 12, 13, 14, 15, 16, 17, 18, and 19 were 0, 3.0, 3.0, 1.5, 3.0, 1.5%, 0, 1.5%, and 0, respectively. The pH of each group was 8.0, 7.0, 6.0, 8.0, 8.0, 7.0, 7.0, 6.0, and 6.0, respectively. The incubation temperatures were 40, 20, 40, 20, 30, 40, 30, 30, and 20°C, respectively. Pathologic tissue sections were made using mouse organs (Hematoxylin–Eosin staining). Mice were executed in the same way as in the previous paragraph. The organs of the mice were handled in the same way as in the previous paragraph.
The genome sequence number of the test strain was CP129005.1 and the genome size was 5,484,835 bp (GC content 35.39%). The plasmid sequence of the test strain was CP129006.1 with a plasmid size of 609,501 bp (GC content of 32.16%).
Growth curves of B. cereus over 24 h were plotted (as shown in Figure 1A). Incubation time was taken as the horizontal coordinate and OD600 as the vertical coordinate. The results showed that B. cereus had slow cell division in 0–2 h (lag phase), rapid cell division in 2–12 h (logarithmic phase), cell division reached the peak of reproduction at 12 h (stationary phase), followed by a slowdown of cell division (decline phase), and gradual cell death after stabilization in 20–24 h.
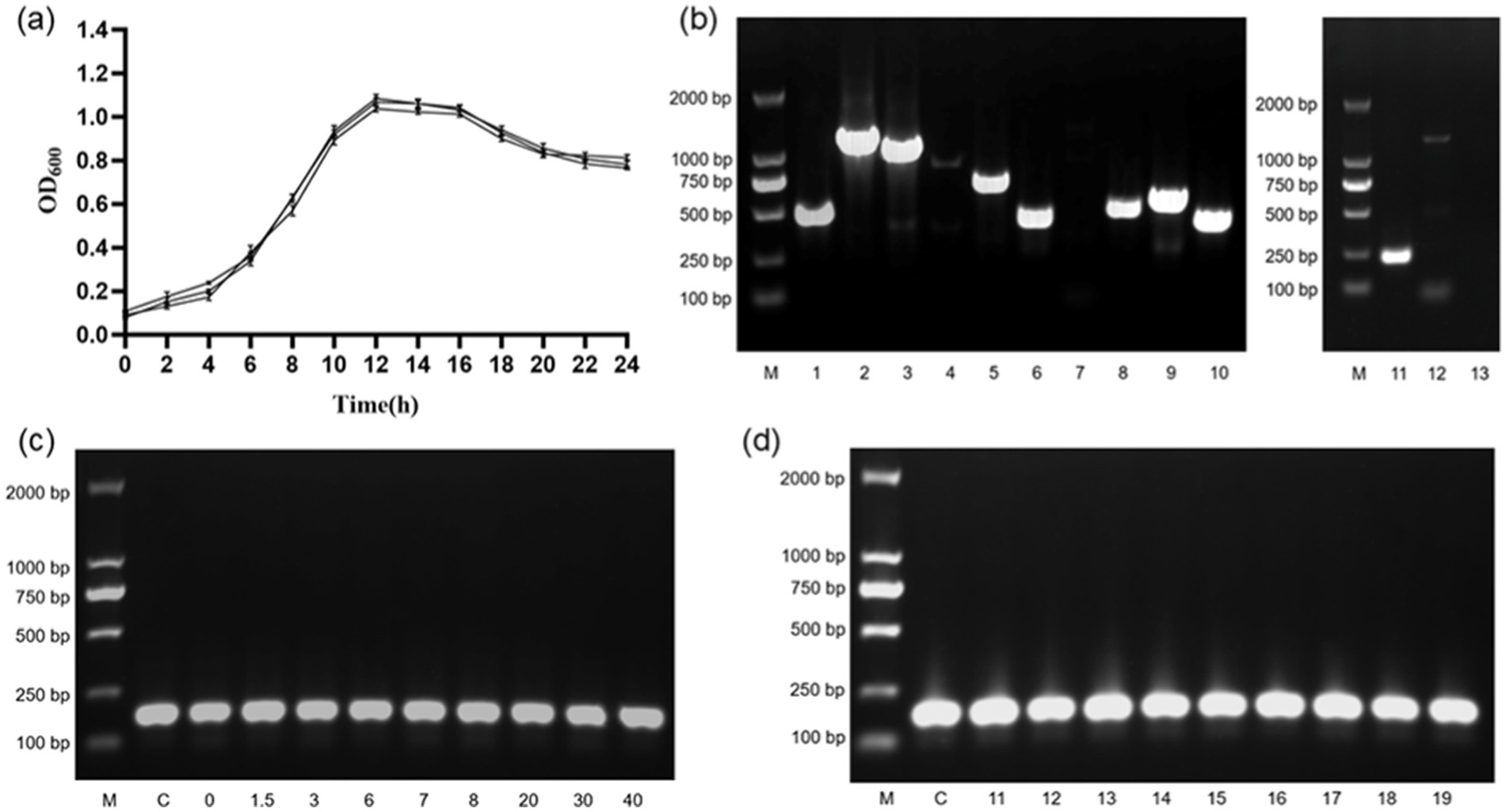
Figure 1. B. cereus growth curves and genetic validation results. (A) 1–24 h growth curve of B. cereus. (B) Results of the B. cereus virulence gene assay. M, DL2000 DNAMarker; 1, nheA gene; 2, nheB gene; 3, nheC gene; 4, hblA gene; 5, hblC gene; 6, hblD gene; 7, ces gene; 8, plcR gene; 9, cytK gene; 10, bceT gene; 11, groEL gene; 12, entFM gene; 13, negative control. (C) RNA validation of single factor stressors. M, DL2000 DNAMarker; C, Control group; 0, group 0; 1.5, group 1.5; 3, group 3; 6, group 6; 7, group 7; 8, group 8; 20, group 20; 30, group 30; 40, group 40. (D) RNA validation of multifactorial stressors. M, DL2000 DNAMarker; C, Control group; 11, group 11; 12, group 12; 13, group 13; 14, group 14; 15, group 15; 16, group 16; 17, group 17; 18, group 18; 19, group 19.
The results (as shown in Figure 1B) showed that the house-keeping gene groEL, the hemolytic enterotoxin genes hblA, hblC and hblD, the nonhemolytic enterotoxin genes nheA, nheB and nheC, the enterotoxin genes bceT and entFM, the cytotoxin gene cytK, and the pleiotropic regulator plcR were presence in our B. cereus strain. The vomitoxin gene ces was not presence in our B. cereus strain.
The cDNA obtained from the reverse transcription reaction was used as a template for PCR amplification of the target gene (gat B_Yqey). The target bands (approximately 175 bp) were visible in all groups (as shown in Figures 1C,D). This indicates that gat B_Yqey gene expression is stable and suitable for use as an internal reference.
The results of the analysis of the test data are shown in Figures 2A–D. The expression of both nheA gene and cytK gene were higher than that of the control group when incubated at 20°C, and the difference were significant (p < 0.01) compared with that of the control group (37°C). The expression of hblD gene and entFM gene were lower than that of the control group, and none of them were statistically different from the control group (p > 0.05). The expression of the four virulence genes, nheA, cytK, hblD and entFM, were higher than that of the control group when incubated at 30°C, and the difference between each group and the control group (37°C) were highly significant (p < 0.001). The relative expression of both cytK and hblD genes were lower than that of the control when cultured at 40°C, and the differences were both highly significant (p < 0.001) compared with the control (37°C). The expression of nheA gene was lower than that of the control group, and the difference was significant (p < 0.01) compared with the control group (37°C). The expression of entFM gene was lower than that of the control group and was statistically different (p < 0.05) from that of the control group (37°C).
Figure 2. Relative mRNA expression of virulence genes under unifactorial environmental stresses (A–D). The expression of virulence genes (nheA, cytK, hblD and entFM) of B. cereus in different environments (20, 30, and 40°C), respectively (A–D). The expression of virulence genes (nheA, cytK, hblD and entFM) of B. cereus at different pH (6.0, 7.0, and 8.0), respectively (E–H). The expression of virulence genes (nheA, cytK, hblD, and entFM) of B. cereus at different salt concentrations (0, 1.5, and 3.0%), respectively (I–L). The data obtained above are mean ± standard deviation, where *P < 0.05; **p < 0.01; ***p < 0.001 indicates statistically significant differences.
The results of the analysis of the test data are shown in Figures 2E–H. The nheA gene, cytK gene and entFM gene showed low expression compared to the control group. The expression of hblD gene was higher than the control at pH 6.0 and lower than the control at both pH 7.0 and 8.0. The difference between the groups and the control group was highly significant (p < 0.001).
The results of the analysis of the test data are shown in Figures 2I–L. The expression of nheA, cytK, hblD, and entFM genes were lower than that of the control group (salt concentration of 1%). The difference between the groups and the control group were highly significant (p < 0.001).
The results show (as shown in Figure 3A). The expression of nheA gene was higher in “group 17” than in the control group, and lower in the other groups than in the control group. In “group 11,” the expression of nheA gene was significantly different from that of the control group (p < 0.01). In “group 12,” “group 13,” “group 14,” “group 15,” “group 16,” “group 17,” “group 18,” and “group 19,” the expression of nheA gene were all highly significantly different from the control group (p < 0.001). The results of Type III variance (ANOVA) in orthogonal tests are shown in Table 3A. The effect of the three factors on nheA expression was NaCl > pH > temperature (the magnitude of the effect was determined by the F-value). There was no significant difference between the three factors (p > 0.05).
Figure 3. Relative mRNA expression of virulence genes under multifactorial environmental stresses. (A) The relative mRNA expression of the nheA gene under multifactorial environmental stress. (B) The relative mRNA expression of the hblD gene under multifactorial environmental stress. (C) The relative mRNA expression of the cytK gene under multifactorial environmental stress. (D) The relative mRNA expression of the entFM gene under multifactorial environmental stress. The data obtained above are mean ± standard deviation, where *p < 0.05; **p < 0.01; ***p < 0.001 indicates statistically significant differences.
The results show (as shown in Figure 3B). The expression of hblD gene was higher in “group 15” than in the control group, while the expression in other groups was lower than in the control group. There was no statistically significant difference between “group 15” and “group 18” and the control group (p > 0.05). “Group 11,” “group 12,” “group 13,” “group 14,” “group 16,” “group 17,” and “group 19” showed highly significant differences compared to the control group (p < 0.001). The results of Type III variance (ANOVA) in orthogonal tests are shown in Table 3B. The effect of the three factors on hblD expression was temperature > pH > NaCl (the magnitude of the effect was determined by the F-value). There was no significant difference between the three factors (p > 0.05).
The results show (as shown in Figure 3C). The expression of cytK gene was higher than that of the control group in “group 15” and “group 17,” and lower than that of the control group in the other groups. “Group 13,” “group 16,” “group 17,” “group 18,” and “group 19” showed highly significant differences compared to the control group (p < 0.001). “Group 14” and “group 15” were statistically different from the control group (p < 0.05). There was no statistically significant difference between “group 11” and “group 12” and the control group (p > 0.05). The results of Type III variance (ANOVA) in orthogonal tests are shown in Table 3C. The effect of the three factors on cytK expression was NaCl > pH > temperature (the magnitude of the effect was determined by the F value). There was no significant difference between the three factors (p > 0.05).
The results show (as shown in Figure 3D). The expression of entFM gene was lower than that of the control group in all groups. There was a statistically significant difference between “group 13” and the control group (p < 0.05). “Group 11,” “group 12,” “group 14,” and “group 15,” “group 16,” “group 17,” “group 18,” and “group 19” showed highly significant differences compared to the control group (p < 0.001). The results of Type III variance (ANOVA) in orthogonal tests are shown in Table 3D. The effect of the three factors on entFM expression was temperature > NaCl > pH (the magnitude of the effect was determined by the F-value). There was no significant difference between the three factors (p > 0.05).
Some mice in groups 20 and 30 in the unifactorial group died within 3–6 h after inoculation with the bacterial culture. Mice in groups 6 and 7 partially died about 6 h after inoculation with the bacterial culture. No mice died in the LB medium control group and 2 mice died in the bacterial culture control group. Three mice died in group 20 and two mice died in group 30. Two mice died in group 6 and three mice died in group 7. One mouse died in groups 40, 8, and 1.5, and no mice died in groups 0 and 3. Some of the mice in groups 11, 15, 17, and 18 of the multifactorial group appeared to die within 3–6 h after inoculation with the bacterial culture, and the numbers of dead mice were 2, 3, 2, and 3, respectively. The number of dead mice in “group 12,” “group 13,” “group 14,” “group 16,” and “group 19” were 1, 0, 0, 1, and 0, respectively.
The results of the pathological sections (as shown in Figures 4, 5) showed a small differences in the pathological changes in the organs of the mice in each group. We repeated the histopathological profiles shown in 10 mice included in each group of studies. Mice in the bacterial culture injection group showed more severe pathological damage than mice in the normal control group. It can be seen that the myocardial fibers of the mice in the bacterial injection group were swollen and broken, with inflammatory cell infiltration between the myocardial fibers and disappearance of myocardial cell nuclei. Widening of the hepatic blood sinusoids, narrowing of the central vein, necrosis of individual hepatocytes, loss of nucleolysis. Necrosis and disappearance of lymphocytes in some areas of the red medulla of the spleen. A few alveolar lumens appear dilated, with dilated capillaries in the alveolar walls and massive leukocytosis. Proliferation of glomerular mesangial cells, narrowing of the lumen of dense plaques, detachment and necrosis of tubular epithelial cells. The mucosal epithelium of the small intestine was detached and individual intestinal glandular cells were necrotic and disappeared.
Figure 4. Pathological sections of the control group and the single-factor environmental stress group (400×).
B. cereus produces a variety of virulence factors to participate in the pathogenic process of bacteria (Mazzantini et al., 2020). B. cereus also forms endospores and biofilms to help B. cereus adapt to harsh environments (Bressuire-Isoard et al., 2018; Caro-Astorga et al., 2020). The bovine lethal B. cereus used in this experiment harbors multiple virulence genes (e.g., hblA, hblC, and hblD). In this study, a preliminary study of four virulence genes (nheA, hblD, cytK, and entFM) of B. cereus was conducted. More in-depth studies are needed in the future to determine the interaction patterns of proteins associated with B. cereus virulence genes, with a view to determining, at the molecular level, the reasons why environmental stressors alter the expression of B. cereus virulence genes. More virulence genes need to be studied to further understand the pathogenic mechanism of B. cereus for better prevention and control of B. cereus disease.
The influence of different factors on bacterial growth and pathogenicity is very complex and therefore appropriate strategies need to be developed to prevent and control bacterial infections (Hosseini et al., 2024; Ehling-Schulz et al., 2015). Changes in temperature, pH and salt concentration all affect the survival of B. cereus and the expression of its virulence genes. When temperature was used as a univariate variable, the results showed that high temperature inhibited the expression of B. cereus virulence genes. However, the control of B. cereus bacterial disease has limitations if it is done by temperature alone (High-temperature sterilization methods cannot be used in some cases, e.g., temperature-sensitive plastics). The use of 20°C and 30°C conditions was not effective in controlling B. cereus disease, while the use of 40°C conditions to control B. cereus disease may result in the development of heat resistance in the bacteria. In summary, B. cereus disease cannot be controlled by temperature alone, but temperature can be used as an aid to accomplish some of the disinfection. When pH was used as a variable it was evident that the bacterium had very low expression at pH 8.0, with the most prominent changes in nheA, cytK and entFM. High pH can be used to control B. cereus when pH is used as a one-way variable. When salt concentration was used as a variable (at salt concentrations of 0, 1.5, and 3%), all virulence genes showed degrees of low expression compared to the control (the salt concentration in the control group was 1%). Control of B. cereus can be achieved to a certain extent by a single stressor, but prolonged use of a single stressor may lead to the development of tolerance in B. cereus. Under multiple environmental stressors, nheA, hblD and cytK were least expressed at a temperature of 40°C, pH 6.0, and a salt concentration of 3.0%, and entFM was least expressed at a temperature of 20°C, pH 8.0, and a salt concentration of 1.5%. It is evident that different stress conditions inhibit the expression of B. cereus virulence genes (nheA, cytK, hblD, entFM). The time of death of mice indicated that bacteria cultured at pH 6.0 and 7.0 were relatively more virulent, and those cultured at 20 and 30°C were more virulent; among the multifactorial groups, B. cereus cultured in groups 11 (high temperature and high pH), 15 (medium temperature and high pH and high salt), 17 (medium temperature and neutral pH), and 18 (medium temperature and low pH and medium salt concentration) were relatively more virulent. In summary, it can be seen that different environmental stressors coercion may result in the expression of virulence genes and pathogenicity of B. cereus being affected. A single virulence gene showing low expression may not attenuate the pathogenicity of B. cereus, or even low expression of some virulence genes may enhance the pathogenicity of B. cereus. This may be due to the relatively complex pathogenic mechanisms of B. cereus (Multiple genes collectively affect the virulence of B. cereus; weak expression of a single gene may not affect the virulence of B. cereus). The specific reasons for these phenomena need to be further studied. In the future, there is a need to find suitable stress conditions to suppress the expression of virulence genes and the pathogenicity of strains of B. cereus.
The results of Wang et al. were significantly different from those of this paper (different expression of virulence genes) (Wang and Liu, 2018). It is possible that the isolation environments of the two strains were different or the biological characteristics of the strains were somewhat different, which led to different expression of virulence genes in the two bacterial strains. A study by and others showed that pu-erh tea broth significantly reduced the expression levels of cytK, nheA and hblD genes and attenuated the damage caused by B. cereus to the liver cells of mice (Sha, 2018). This result is consistent with the findings of the present study, suggesting that inhibition of virulence gene expression is one of the effective ways to reduce the pathogenicity of B. cereus. To prevent the development of resistance in B. cereus, the use of antibiotics is severely restricted (Zheng et al., 2024). Therefore, it is clinically relevant to explore the control of environmental stressors to accomplish food-handling-related appliances and farm environmental decontamination. The method used in this study reduces the probability of human and animal infection with B. cereus disease by decreasing B. cereus virulence gene expression. The method used in this study is non-toxic and is expected to be used in the future for the disinfection of utensils related to food handling (kitchen utensils, slaughterhouse equipment) and the environment of farms. Both pharmaceutical and chemical disinfection methods can produce residues and chemical disinfection has a high odor; this makes it unsuitable for sterilizing some utensils and environments (kitchenware, slaughterhouse equipment and farm environments) (Wildenthal et al., 2023; Doyle et al., 2021). Bacteriophages have been effective in the control of B. cereus disease (Huang et al., 2024). However, bacteriophages may affect the growth of beneficial bacteria when used, thereby disrupting the flora balance of the environment. It is also possible that bacteriophages could lead to some resistance to B. cereus, which would increase the cost of B. cereus disease prevention and control (bacteriophage enhancement or replacement with new phages is needed to achieve control of B. cereus disease). The potential safety risks of bacteriophages in use need to be further evaluated. The method used in this study does not produce residues and is relatively safe to operate. It is believed that with further research, environmental stressors can be gradually used in the future to replace antibiotics to reduce B. cereus in the environment. New acidic disinfectants can be developed to accomplish disinfection, and the salt concentration in the disinfectant can be increased appropriately to enhance its effectiveness. This study provides a reference for the development of new disinfectants and the prevention and control of bacterial diseases of B. cereus.
Environmental stressors (temperature, salt concentration, acidity and alkalinity) have an effect on the expression of four virulence genes (nheA, cytK, hblD, entFM) in B. cereus. Under a single environmental stressors, nheA, hblD, cytK and entFM had the lowest expression at 40°C, pH 8.0, and were lowly expressed at all salt concentrations except the control group. Under multiple environmental stressors, nheA, hblD and cytK were least expressed at a temperature of 40°C, pH 6.0, and salt concentration of 3.0%, and entFM was least expressed at a temperature of 20°C, pH 8.0, and salt concentration of 1.5%. New disinfectants can be developed to control the spread of the bacteria based on the above results (focusing on the development of acidic disinfectants, while the salt concentration in the disinfectant can be increased appropriately).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, CP129005; https://www.ncbi.nlm.nih.gov/genbank/, CP129006.
The animal study was approved by Institutional Animal Care and Ethical Committee of Inner Mongolia Minzu University. The study was conducted in accordance with the local legislation and institutional requirements.
QM: Data curation, Investigation, Writing – original draft. LS: Data curation, Investigation, Writing – original draft. SC: Data curation, Investigation, Writing – original draft. HW: Writing – review & editing. JL: Writing – review & editing. YC: Writing – review & editing. ZL: Writing – review & editing. XZ: Writing – review & editing. ZJ: Writing – review & editing. JC: Writing – review & editing. XW: Conceptualization, Funding acquisition, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Funding was provided by the Inner Mongolia Autonomous Region Higher School Science and Technology Research Key Project (NJZZ19147), the Inner Mongolia Natural Science Foundation (2021LHMS03008), the Multidisciplinary Cross-Research Project of Basic Scientific Research Business Fees of Universities Directly under Inner Mongolia (GXKY22003). Funds were utilized to acquire materials for experimentation and analyze resultant data.
We would like to express our gratitude to the College of Animal Science and Technology at Inner Mongolia Minzu University for their assistance to the authors. We thank the Department of Grassland Ecology and Animal Husbandry and Veterinary, Xilingol Vocational College for their assistance to the authors. We thank the Tongliao Institute of Agriculture and Animal Husbandry Sciences for their assistance to the authors. We thank the Hure Banner Animal Disease Prevention and Control Center for their assistance to the authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agata, N., Ohta, M., Arakawa, Y., and Mori, M. (1995). The bceT gene of Bacillus cereus encodes an enterotoxic protein. Microbiology 141, 983–988. doi: 10.1099/13500872-141-4-983
Asano, S. I., Nukumizu, Y., Bando, H., Iizuka, T., and Yamamoto, T. (1997). Cloning of novel enterotoxin genes from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 63, 1054–1057. doi: 10.1128/aem.63.3.1054-1057.1997
Bai, F. L., Chen, S., Luo, M. Y., Zeng, B. F., and Tang, J. N. (2018). Identification and virulence genes detection of Bacillus cereus food isolates. Mod Food Sci. Technol. 34:247-252+204. doi: 10.13982/j.mfst.1673-9078.2018.10.033
Belaouni, H. A., Compant, S., Antonielli, L., Nikolic, B., Zitouni, A., and Sessitsch, A. (2022). In-depth genome analysis of Bacillus sp. BH32, a salt stress-tolerant endophyte obtained from a halophyte in a semiarid region. Appl. Microbiol. Biotechnol. 106, 3113–3137. doi: 10.1007/s00253-022-11907-0
Berthold-Pluta, A., Pluta, A., Garbowska, M., and Stefańska, I. (2019). Prevalence and toxicity characterization of Bacillus cereus in food products from Poland. Food Secur. 8:269. doi: 10.3390/foods8070269
Biesta-Peters, E. G., Mols, M., Reij, M. W., and Abee, T. (2011). Physiological parameters of Bacillus cereus marking the end of acid-induced lag phases. Int. J. Food Microbiol. 148, 42–47. doi: 10.1016/j.ijfoodmicro.2011.04.024
Bressuire-Isoard, C., Broussolle, V., and Carlin, F. (2018). Sporulation environment influences spore properties in Bacillus: evidence and insights on underlying molecular and physiological mechanisms. FEMS Microbiol. Rev. 42, 614–626. doi: 10.1093/femsre/fuy021
Caro-Astorga, J., Álvarez-Mena, A., Hierrezuelo, J., Guadix, J. A., Heredia-Ponce, Z., Arboleda-Estudillo, Y., et al. (2020). Two genomic regions encoding exopolysaccharide production systems have complementary functions in B. cereus multicellularity and host interaction. Sci. Rep. 10:1000. doi: 10.1038/s41598-020-57970-3
Chang, Y., Xie, Q., Yang, J., Ma, L., and Feng, H. (2021). The prevalence and characterization of Bacillus cereus isolated from raw and pasteurized buffalo milk in southwestern China. J. Dairy Sci. 104, 3980–3989. doi: 10.3168/jds.2020-19432
Doyle, S., Meade, E., Gao, J., O'Hagan, B., Callan, J. F., Garvey, M., et al. (2021). A rapid antimicrobial photodynamic water treatment strategy utilizing a xanthene dye with subsequent removal by goethite nanoparticles. Chemosphere 280:130764. doi: 10.1016/j.chemosphere.2021.130764
Ehling-Schulz, M., Frenzel, E., and Gohar, M. (2015). Food-bacteria interplay: pathometabolism of emetic Bacillus cereus. Front. Microbiol. 6:704. doi: 10.3389/fmicb.2015.00704
Elsayed, A., Abdelsattar, A. M., Heikal, Y. M., and El-Esawi, M. A. (2022). Synergistic effects of Azospirillum brasilense and Bacillus cereus on plant growth, biochemical attributes and molecular genetic regulation of steviol glycosides biosynthetic genes in Stevia rebaudiana. Plant Physiol. Biochem. 189, 24–34. doi: 10.1016/j.plaphy.2022.08.016
Gong, L., Wang, B., Mei, X., Xu, H., Qin, Y., Li, W., et al. (2018). Effects of three probiotic Bacillus on growth performance, digestive enzyme activities, antioxidative capacity, serum immunity, and biochemical parameters in broilers. Anim. Sci. J. 89, 1561–1571. doi: 10.1111/asj.13089
Hansen, B. M., and Hendriksen, N. B. (2001). Detection of enterotoxic Bacillus cereus and Bacillus thuringiensis strains by PCR analysis. Appl. Environ. Microbiol. 67, 185–189. doi: 10.1128/AEM.67.1.185-189.2001
Hosseini, H., Mahmoudi, R., Pakbin, B., Manafi, L., Hosseini, S., Pilevar, Z., et al. (2024). Effects of intrinsic and extrinsic growth factors on virulence gene expression of foodborne pathogens in vitro and in food model systems; a review. Food Sci. Nutr. 12, 6093–6107. doi: 10.1002/fsn3.4281
Huang, Z., Yuan, X., Zhu, Z., Feng, Y., Li, N., Yu, S., et al. (2024). Isolation and characterization of Bacillus cereus bacteriophage DZ1 and its application in foods. Food Chem. 431:137128. doi: 10.1016/j.foodchem.2023.137128
Jeon, H. H., Jung, J. Y., Chun, B. H., Kim, M. D., Baek, S. Y., Moon, J. Y., et al. (2016). Screening and characterization of potential Bacillus starter cultures for fermenting low-salt soybean paste (Doenjang). J. Microbiol. Biotechnol. 26, 666–674. doi: 10.4014/jmb.1512.12014
Jia, W. J., Song, L. L., Zhang, L. Y., and Wang, X. L. (2022). Research progress of the toxins from Bacillus cereus. Chin. J. Antibiot. 47, 534–542. doi: 10.13461/j.cnki.cja.007350
Jing, D. W., Ma, H. L., Liu, F. C., Du, Z. Y., Jia, H. H., Ma, B. Y., et al. (2018). Effects of inoculating plant growth-promoting rhizobacteria on the micro-environmental characteristics of the rhizosphere soil of Suaeda glauce Bge under salt stress. Soil Fertil. Sci. China 4, 34–39.
Kim, G. H., Forghani, F., and Oh, D. H. (2013). Rapid detection of emetic toxin producing Bacillus cereus strains using triple-primer polymerase chain reaction (PCR) assay. Afr. J. Microbiol. Res. 58, 1286–1294. doi: 10.1007/s13197-020-04637-6
Li, F. (2017). Research on virulence genes of foodborne Bacillus cereus and thenaked eye detection method based on aPCR [dissertation/master's thesis]. Nanchang: Nanchang University, 2017.
Mazzantini, D., Fonnesu, R., Celandroni, F., Calvigioni, M., Vecchione, A., Mrusek, D., et al. (2020). GTP-dependent FlhF homodimer supports secretion of a hemolysin in Bacillus cereus. Front. Microbiol. 11:879. doi: 10.3389/fmicb.2020.00879
National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals (2011). Guide for the care and use of laboratory animals. 8th Edn. Washington (DC): National Academies Press.
Ngamwongsatit, P., Buasri, W., Pianariyanon, P., Pulsrikarn, C., Ohba, M., Assavanig, A., et al. (2008). Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK, and entFM) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int. J. Food Microbiol. 121, 352–356. doi: 10.1016/j.ijfoodmicro.2007.11.013
Reiter, L., Kolstø, A. B., and Piehler, A. P. (2011). Reference genes for quantitative, reverse-transcription PCR in Bacillus cereus group strains throughout the bacterial life cycle. J. Microbiol. Methods 86, 210–217. doi: 10.1016/j.mimet.2011.05.006
Rossi, G. A. M., Silva, H. O., Aguilar, C. E. G., Rochetti, A. L., Pascoe, B., Méric, G., et al. (2018). Comparative genomic survey of Bacillus cereus sensu stricto isolates from the dairy production chain in Brazil. FEMS Microbiol. Lett. 365. doi: 10.1093/femsle/fnx283
Sha, P. P. (2018). Effect of pu-erh tea decoction on Bacillus enterotoxin gene expression and prevention of colitis in mice [dissertation/master's thesis]. Nanchang: Nanchang University, 2018.
Wang, Y., and Liu, J. L. (2018). Effects of environmental factors on virulence genes expression and pathogenicity of Bacillus cereus. Jiangsu Agric. Sci. 46, 38–43. doi: 10.15889/j.issn.1002-1302.2018.12.008
Wildenthal, J. A., Schwartz, D. J., Nolan, N. S., Zhao, L., Robinson, J. I., Jones, E., et al. (2023). Everything but the kitchen sink: an analysis of bacterial and chemical contaminants found in syringe residue from people who inject drugs. Open Forum Infect. Dis. 11:ofad628. doi: 10.1093/ofid/ofad628
Xu, H. Q. (2023). Analysis of verification results by detection method for Bacillus cereus in food. J. Anhui Agric. Sci. 51, 184–189. doi: 10.3969/j.issn.0517-6611.2023.09.044
Keywords: environmental stressors, lethal B. cereus, fluorescence quantitative PCR, expression of virulence genes, pathological sections
Citation: Meng Q, Song L, Chi S, Wang H, Li J, Chen Y, Liu Z, Zhang X, Jia Z, Cui J and Wang X (2025) Effect of environmental stress factors on the expression of virulence genes and pathogenicity of lethal Bacillus cereus of bovine origin. Front. Microbiol. 16:1519202. doi: 10.3389/fmicb.2025.1519202
Received: 29 October 2024; Accepted: 30 January 2025;
Published: 13 February 2025.
Edited by:
Abd El-Latif Hesham, Beni-Suef University, EgyptReviewed by:
Angelica Bianco, Experimental Zooprophylactic Institute of Puglia and Basilicata (IZSPB), ItalyCopyright © 2025 Meng, Song, Chi, Wang, Li, Chen, Liu, Zhang, Jia, Cui and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueli Wang, d2FuZ3hsOTU3N0BhbGl5dW4uY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.