
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 24 February 2025
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1517772
This article is part of the Research TopicEmerging Antimicrobials: Sources, Mechanisms of Action, Spectrum of Activity, Combination Antimicrobial Therapy, and Resistance MechanismsView all 24 articles
 Arun Karnwal1*
Arun Karnwal1* Amar Yasser Jassim2
Amar Yasser Jassim2 Ameer Abbas Mohammed2
Ameer Abbas Mohammed2 Abdel Rahman Mohammad Said Al-Tawaha3
Abdel Rahman Mohammad Said Al-Tawaha3 Manickam Selvaraj4,5
Manickam Selvaraj4,5 Tabarak Malik6*
Tabarak Malik6*The COVID-19 pandemic underscored bacterial resistance as a critical global health issue, exacerbated by the increased use of antibiotics during the crisis. Notwithstanding the pandemic’s prevalence, initiatives to address bacterial medication resistance have been inadequate. Although an overall drop in worldwide antibiotic consumption, total usage remains substantial, requiring rigorous regulatory measures and preventive activities to mitigate the emergence of resistance. Although National Action Plans (NAPs) have been implemented worldwide, significant disparities persist, particularly in low- and middle-income countries (LMICs). Settings such as farms, hospitals, wastewater treatment facilities, and agricultural environments include a significant presence of Antibiotic Resistant Bacteria (ARB) and antibiotic-resistance genes (ARG), promoting the propagation of resistance. Dietary modifications and probiotic supplementation have shown potential in reshaping gut microbiota and reducing antibiotic resistance gene prevalence. Combining antibiotics with adjuvants or bacteriophages may enhance treatment efficacy and mitigate resistance development. Novel therapeutic approaches, such as tailored antibiotics, monoclonal antibodies, vaccines, and nanoparticles, offer alternate ways of addressing resistance. In spite of advancements in next-generation sequencing and analytics, gaps persist in comprehending the role of gut microbiota in regulating antibiotic resistance. Effectively tackling antibiotic resistance requires robust policy interventions and regulatory measures targeting root causes while minimizing public health risks. This review provides information for developing strategies and protocols to prevent bacterial colonization, enhance gut microbiome resilience, and mitigate the spread of antibiotic resistance.
In recent years, antibiotics have made a significant contribution to socioeconomic growth by promoting healthcare, avoiding deaths, and increasing animal productivity. However, the inadequate use of antibiotics has exacerbated the emergence of bacterial resistance, failed treatments, increased disease rates, and increased healthcare expenses (Bunduki et al., 2024; Gulumbe et al., 2023; Watkins and Bonomo, 2020). In 2018, the global use of veterinary antibiotics was approximately 76,704 tons (Ardakani et al., 2024; de Lagarde et al., 2022; Zeedan et al., 2023), while medical antibiotic consumption was 14.3 defined daily doses (DDDs) per thousand people per day (Zhang R. M. et al., 2023). Antibiotic-resistant bacteria (ARB) account for over 25% of nosocomial infections, posing a growing challenge to healthcare systems. Projections suggest that by 2050, there will be a shocking 10 million deaths caused by these bacteria (Asmare et al., 2024). Reducing the use of antibiotics is crucial to prevent the spread of resistance in various contexts, such as healthcare facilities, animals, the food chain, and the environment (Bunduki et al., 2024). It will help to minimize the dangers to public health.
The emergence of antimicrobial resistance in the environment, livestock, and humans increases the risk of human infection by resistant bacteria (Sachdeva et al., 2025). The human gut, rich in nutrients and maintaining an ideal temperature, fosters the spread of antibiotic resistance genes (ARGs) and antibiotic-resistant bacteria (ARB) due to its diverse microbiota (Das et al., 2022). ARGs and ARB within the human body pose a significant and growing health threat (Fang et al., 2023). This review analyzes global patterns in veterinary and clinical antibiotic use and presents National Action Plans (NAPs) (Figure 1) to combat antibiotic resistance. Additionally, it explores strategies for reversing resistance, reducing transmission post-colonization, and preventing the spread of resistant bacteria. Implementing these measures is crucial to mitigating antimicrobial resistance and its transmission to humans (Mendelson et al., 2024).
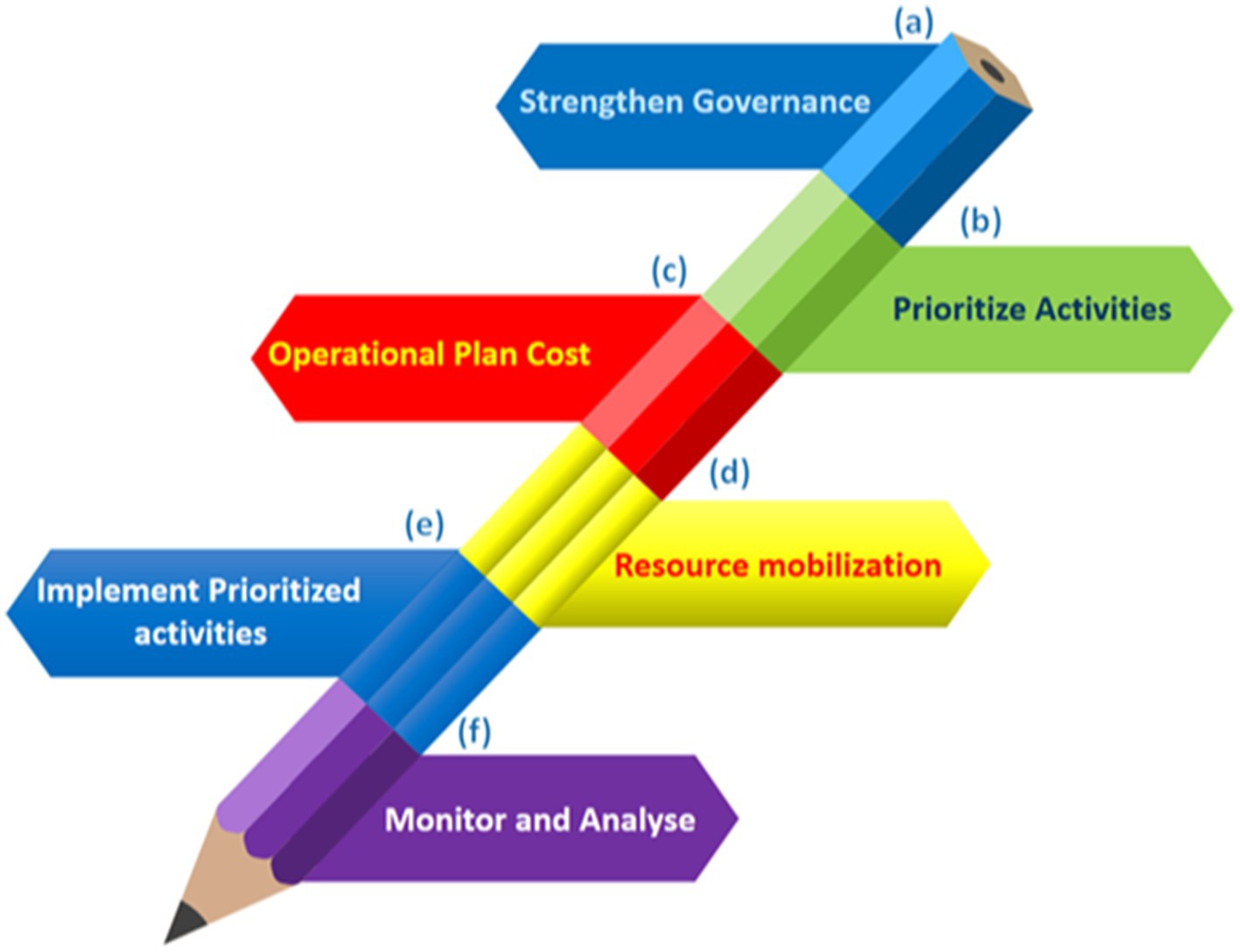
Figure 1. Six stages for the successful execution of NAPs on antimicrobial resistance in a sustainable manner. (A) Create an effective cross-sector coordination system and technical teams with defined roles, budget allocations, and accountability structures in place. (B) Conduct a consultative approach to select operations based on current conditions, resources, predicted impact, and feasibility. (C) Prioritize tasks, define responsibilities, dates, and locations, and incorporate existing financing streams into an operational plan. (D) Identify current and potential financiers, lobby to close the financing gap, and use indigenous funding through national initiatives and fiscal allocations when possible. (E) Work with internal and external stakeholders to sustainably implement prioritized initiatives. (F) Regularly evaluate and share progress and lessons from plan and endeavor execution.
Although a recent global decline in antibiotic use for veterinary purposes, excessive antibiotic consumption in clinical settings remains a major concern, particularly in low- and middle-income countries (Alabi et al., 2024; O’Leary et al., 2024; Liu L. et al., 2021; Liu Y. et al., 2021). While many countries have implemented National Action Plans (NAPs) to combat antibiotic resistance, approaches vary significantly (Villarreal et al., 2023) (Table 1). The World Health Organization (WHO), the Centers for Disease Control and Prevention (CDC), and the European Centre for Disease Prevention and Control (ECDC) have raised alarms over the growing threat of antimicrobial resistance (AMR) (World Health Organization, 2024; Cobar and Cobar, 2024). WHO advocates for stricter regulations on antimicrobial use in animals, particularly those critical to human medicine, while the CDC and ECDC emphasize a One Health approach that integrates human, animal, and environmental health (Mercy et al., 2024; Catteau et al., 2024). These organizations call for reduced antibiotic use in animals, enhanced surveillance, and improved stewardship practices to curb the spread of resistant pathogens (Vekemans et al., 2021).
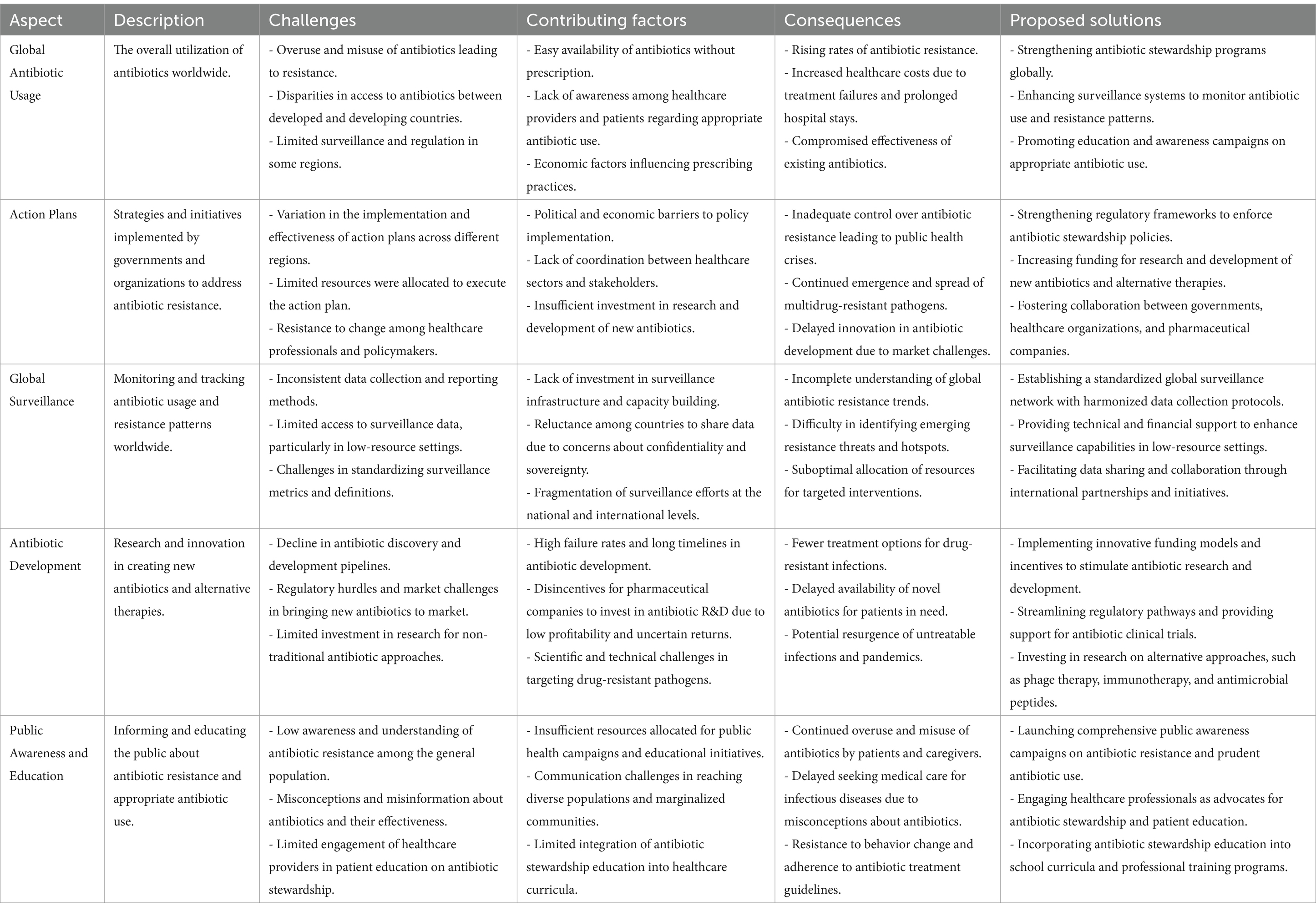
Table 1. Key challenges in global antibiotic usage and the limitations of current action plans in addressing antibiotic resistance (Cobar and Cobar, 2024; Berry, 2024; Bolbanabad et al., 2023; Khouja et al., 2022; Pham et al., 2024; Villarreal et al., 2023; Willemsen et al., 2022; Uddin et al., 2021).
In 2018, global veterinary antibiotic consumption was approximately 81,000 tons, with an estimated dosage of 75.16–82.56 mg per kilogram of animal weight (Sorbara et al., 2019; Sun et al., 2019). Ardakani et al. (2024) reported that in 2017, antimicrobial usage (AMU) in chickens, cattle, and pigs—comprising 93.75% of all food animals—amounted to 93,309 tonnes of active ingredients (Ardakani et al., 2024; Calayag et al., 2021). This figure is projected to rise by 11.5% to 104,079 tonnes by 2030, with pigs contributing the most to this increase (45%), followed by cattle (22%). In 2017, pigs consumed an average of 193 mg per population correction unit (PCU), while cattle had the lowest consumption at 42 mg/PCU (Rothrock et al., 2021).
Figure 2 illustrates trends in antibiotic usage across major countries over two-time points, highlighting shifts in consumption patterns and the rise in usage between the years under review. This comparison emphasizes the critical need for sustainable antibiotic use to combat antimicrobial resistance (AMR) (Ardakani et al., 2024; Al-Tawfiq et al., 2024). In 2017, chickens consumed an average of 68 mg/PCU, contributing to 33% of the global increase in AMU (Murray et al., 2021; Salam et al., 2023). Asia, the largest consumer of veterinary antibiotics in 2017, is expected to continue this trend, with its usage projected to grow by 10.3% by 2030, accounting for 68% of global usage by that time for all used antibiotics mentioned by Tiseo et al. (2020) and supported by Van Boeckel et al. (2015). Africa is forecast to experience the most significant increase, with an expected rise of 37% by 2030, although it will still account for just 6.1% of global consumption (Mercy et al., 2024). Meanwhile, Oceania, North America, and Europe are projected to see minimal growth in antimicrobial sales (Cheng et al., 2022; Iera et al., 2025). As the largest consumer in 2017, China is expected to maintain its position in 2030, with the top 10 consuming countries collectively accounting for 72% of global antimicrobial consumption by that year (Zuo et al., 2021) (see Tables 2,3).
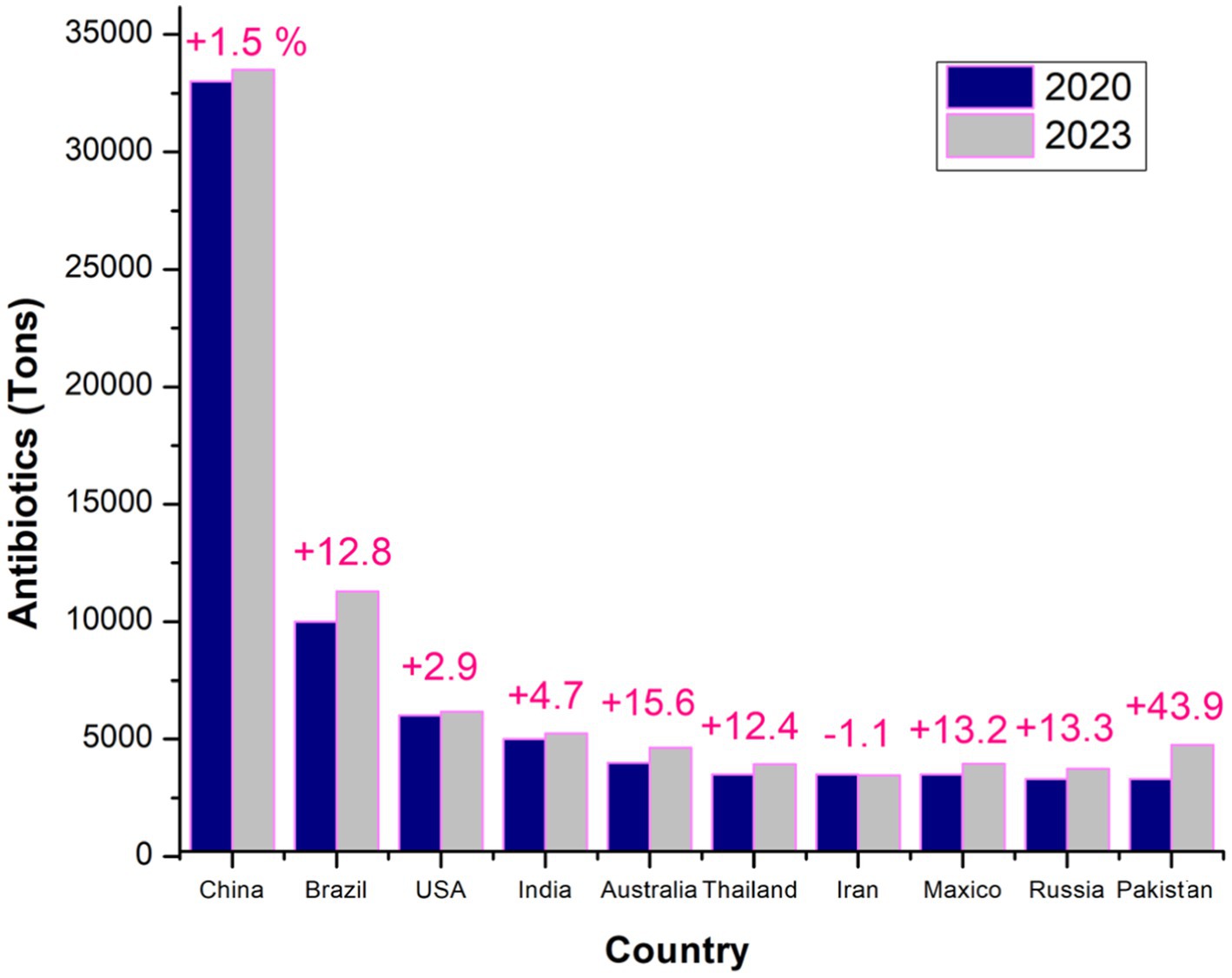
Figure 2. Global comparison of top antibiotic consumers in veterinary medicine: a detailed analysis of the leading countries in 2020 and 2023, highlighting trends in antibiotic usage and year variations (Ardakani et al., 2024).
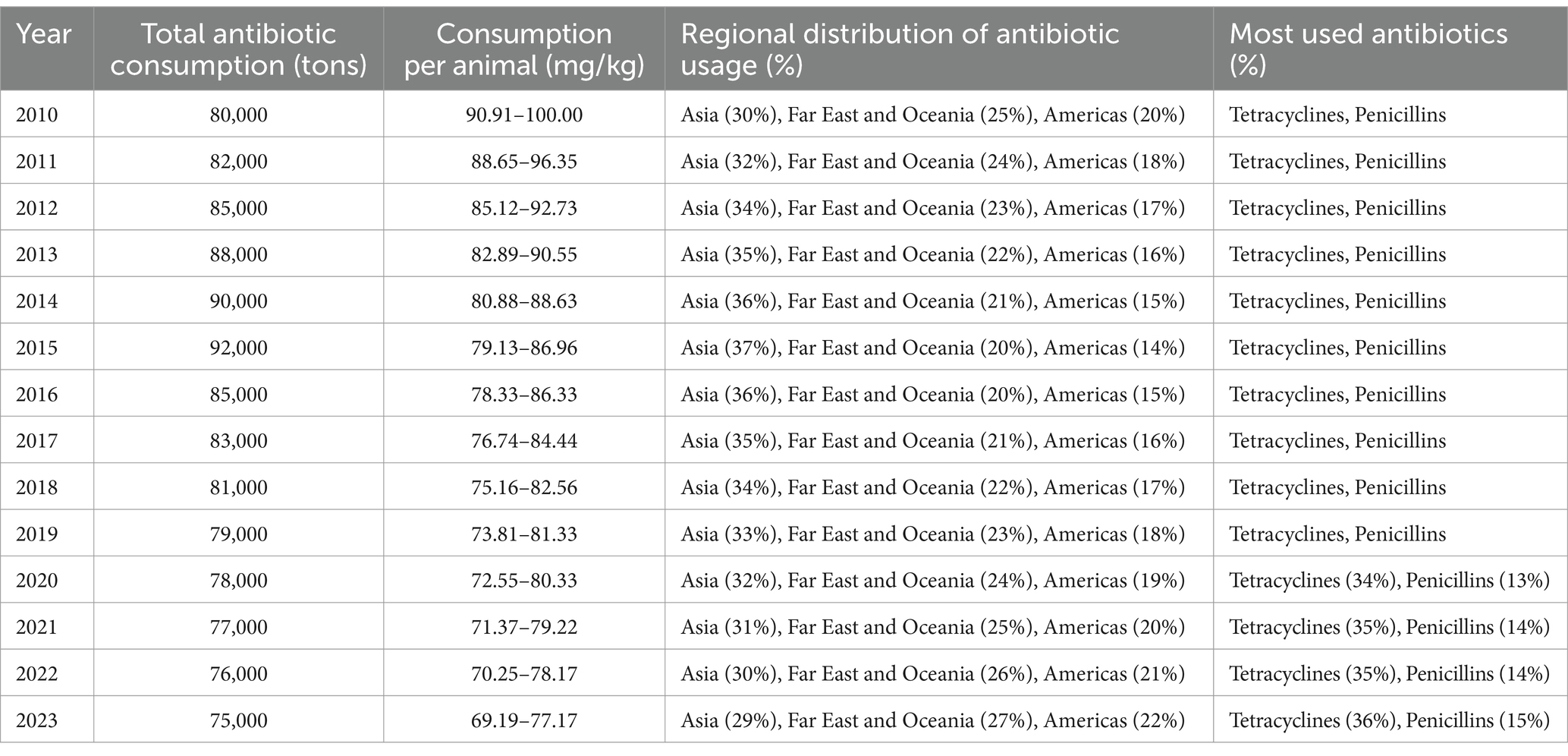
Table 2. Trends in global antibiotic consumption from 2010 to 2023: an analysis of usage patterns across different regions and antibiotic (Al-Taani et al., 2022; Al Meslamani, 2023; Ardakani et al., 2024; Cuevas et al., 2021; Kamere et al., 2022; Quaik et al., 2020).
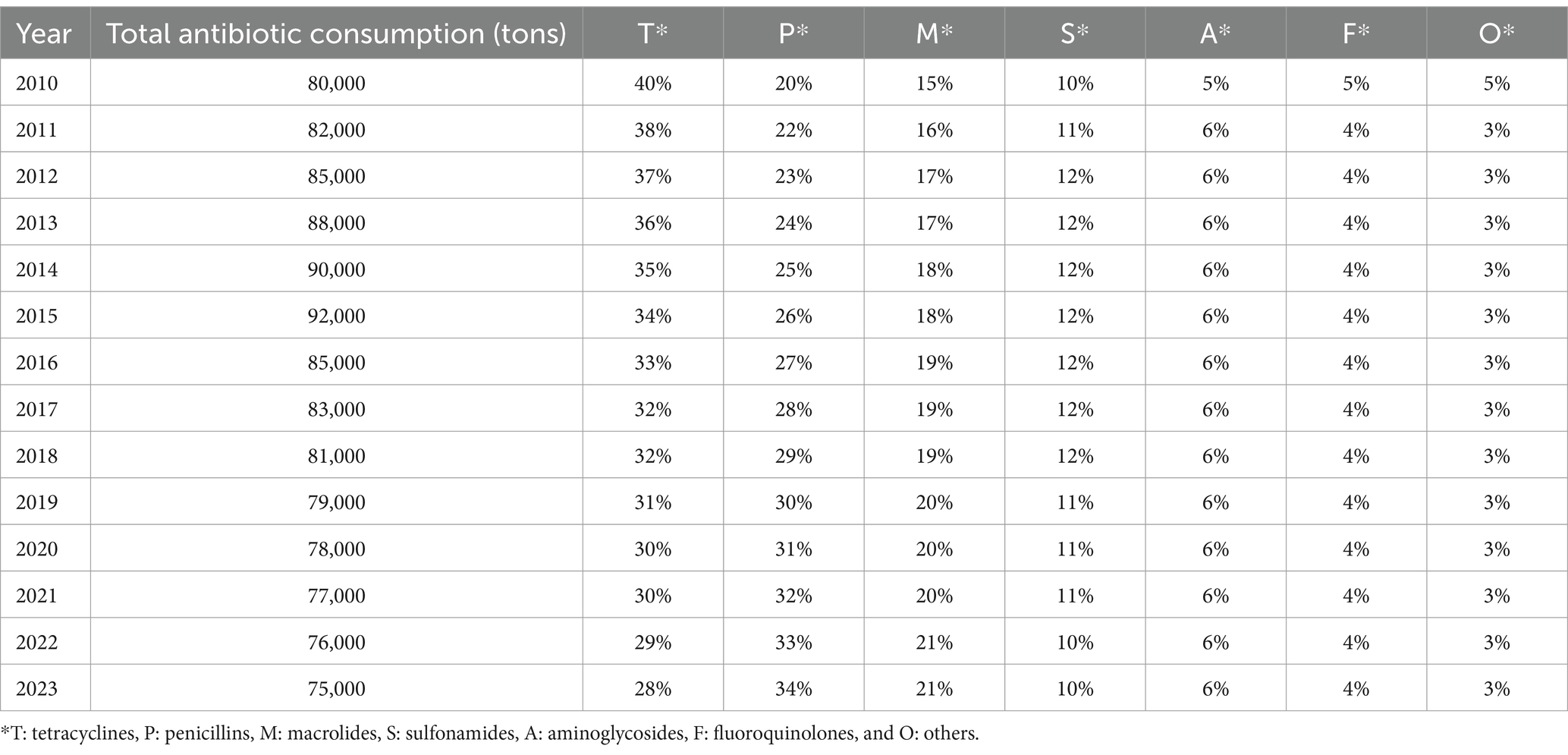
Table 3. Global antibiotic consumption trends by class from 2010 to 2023: a comparative analysis of usage patterns across different antibiotic classes (Al-Taani et al., 2022; Al Meslamani, 2023; Ardakani et al., 2024; Cuevas et al., 2021; Kamere et al., 2022; Quaik et al., 2020).
Tetracyclines and penicillins were the primary antibiotics used in animal health, accounting for 40.5 and 14.1% of total consumption, respectively (Ardakani et al., 2024). However, discrepancies in antimicrobial usage (AMU) data, particularly from China, highlight the need for further investigation into the accuracy of reported figures. Between 2015 and 2030, human AMU is projected to increase by 15%, paralleling the rising demand for food animals (Cherian et al., 2023). Interestingly, the expected surge in animal antimicrobial consumption is lower than previous estimates—Van Boeckel et al. (2019) had predicted a 53% rise by 2030. These discrepancies stem from variations in data sources, specifically in China, where recent reports from the Ministry of Agriculture indicate a significant decline in AMU, raising concerns about data reliability (Zhang K. et al., 2022). However, The fourth JIACRA report analyzed antimicrobial consumption (AMC) and antimicrobial resistance (AMR) trends across 2014–2021, based on data from EU surveillance networks. In 2021, the total AMC for humans was 125.0 mg/kg, while for food-producing animals, it was 92.6 mg/kg. Over this period, AMC in animals decreased by 44%, while human consumption remained stable (European Centre for Disease Prevention and Control (ECDC), 2024). Positive associations between AMC and AMR in both sectors were observed, indicating the influence of AMC on resistance patterns. The report also highlighted that reductions in AMC in both humans and animals were often associated with improved antimicrobial susceptibility in bacteria. These trends suggest that efforts to reduce AMC have been effective in many countries, although further actions are needed to maintain and strengthen these gains (European Centre for Disease Prevention and Control (ECDC), 2024). Measures such as vaccination and improved hygiene are crucial in reducing reliance on antimicrobials and promoting health.
In high-income countries (HICs), antimicrobial sales have declined due to stewardship programs promoting responsible antibiotic use (Ahmed et al., 2024). The United Kingdom and the United States, for instance, have successfully reduced veterinary antimicrobial use through targeted resistance strategies and stricter guidelines (Hawkins et al., 2022; Sachan et al., 2023; Ahmed et al., 2024). However, trends have been inconsistent across regions. Canada initially saw a reduction in antimicrobial sales but later experienced an increase. Globally, clinical antibiotic usage has risen, with daily defined doses (DDDs) per 1,000 people increasing from 9.8 in 2000 to 14.3 in 2018 (Browne et al., 2021). This trend raises serious concerns about the growing prevalence of antibiotic-resistant bacteria, which pose a significant public health threat.
A notable shift in global antibiotic consumption occurred during the COVID-19 pandemic. Between 2019 and 2020, antibiotic utilization dropped by 20.84%, from 2,928.33 units per 1,000 individuals to 2,317.94 units (Khouja et al., 2022). This decline coincided with pandemic-related control measures, suggesting a correlation between stricter healthcare protocols and reduced antibiotic use. Additionally, hospitalized patients exhibited a shift from using antibiotics in the “Access” category to more restrictive “Watch” and “Reserve” groups, reflecting changes in prescribing patterns (Hussein and Ali, 2021; Hussein et al., 2022).
Paradoxically, the early stages of the pandemic saw a surge in antimicrobial consumption, driven by concerns over bacterial co-infections and the precautionary use of antibiotics, despite COVID-19 being a viral disease (Kumar et al., 2023). Over time, stricter regulations led to a global decline in antibiotic consumption. However, the overuse and misuse of antibiotics during the pandemic exacerbated antimicrobial resistance (AMR), heightening the risk of treatment failure, prolonged hospital stays, and increased mortality (Hussein et al., 2022). The pandemic underscored the urgent need for stronger antibiotic stewardship, responsible prescribing practices, and enhanced global awareness to mitigate the threat of AMR. Without immediate intervention, AMR could undermine modern medicine, making once-treatable infections life-threatening (Gulumbe et al., 2023).
The Global Action Plan on Antimicrobial Resistance (AMR), endorsed by the 68th World Health Assembly in 2015, aimed to drive global efforts to combat antibiotic resistance (Gostin et al., 2020; Truché et al., 2020). Since then, numerous governments and regions have developed strategic initiatives to address AMR (Khouja et al., 2022; Bolbanabad et al., 2023). The AMR National Action Plan (NAP) Library was assessed on January 31, 2023 (Saleem et al., 2022), revealing that 134 out of 194 countries—comprising 69% of nations worldwide, including 40 non-English-speaking countries—have officially adopted NAPs. While most nations align with the Global Action Plan (GAP) framework, significant variations exist in the implementation of AMR programs (Charani et al., 2023; Chua et al., 2021). According to the World Organization for Animal Health (WOAH), approximately one-third of countries globally continued using antibiotics for livestock growth promotion in 2022 (Gehring et al., 2023). These disparities reflect inconsistencies in the design, execution, and monitoring of NAPs across different regions. The necessity of a well-structured, country-specific NAP tailored with targeted interventions to combat AMR is indisputable (Charani et al., 2023).
However, concerns persist regarding the effectiveness of current policies, particularly in low- and middle-income countries (LMICs), where AMR remains a pressing issue (Bolbanabad et al., 2023). Willemsen et al. (2022) emphasize that inadequate infrastructure, a lack of skilled professionals, and limited financial resources hinder the successful implementation of AMR management strategies. Furthermore, the increased interconnection between human populations, livestock, and agricultural ecosystems in LMICs exacerbates the risk of antibiotic resistance transmission, highlighting the urgent need to bridge these gaps (Checcucci et al., 2020) (see Table 4).
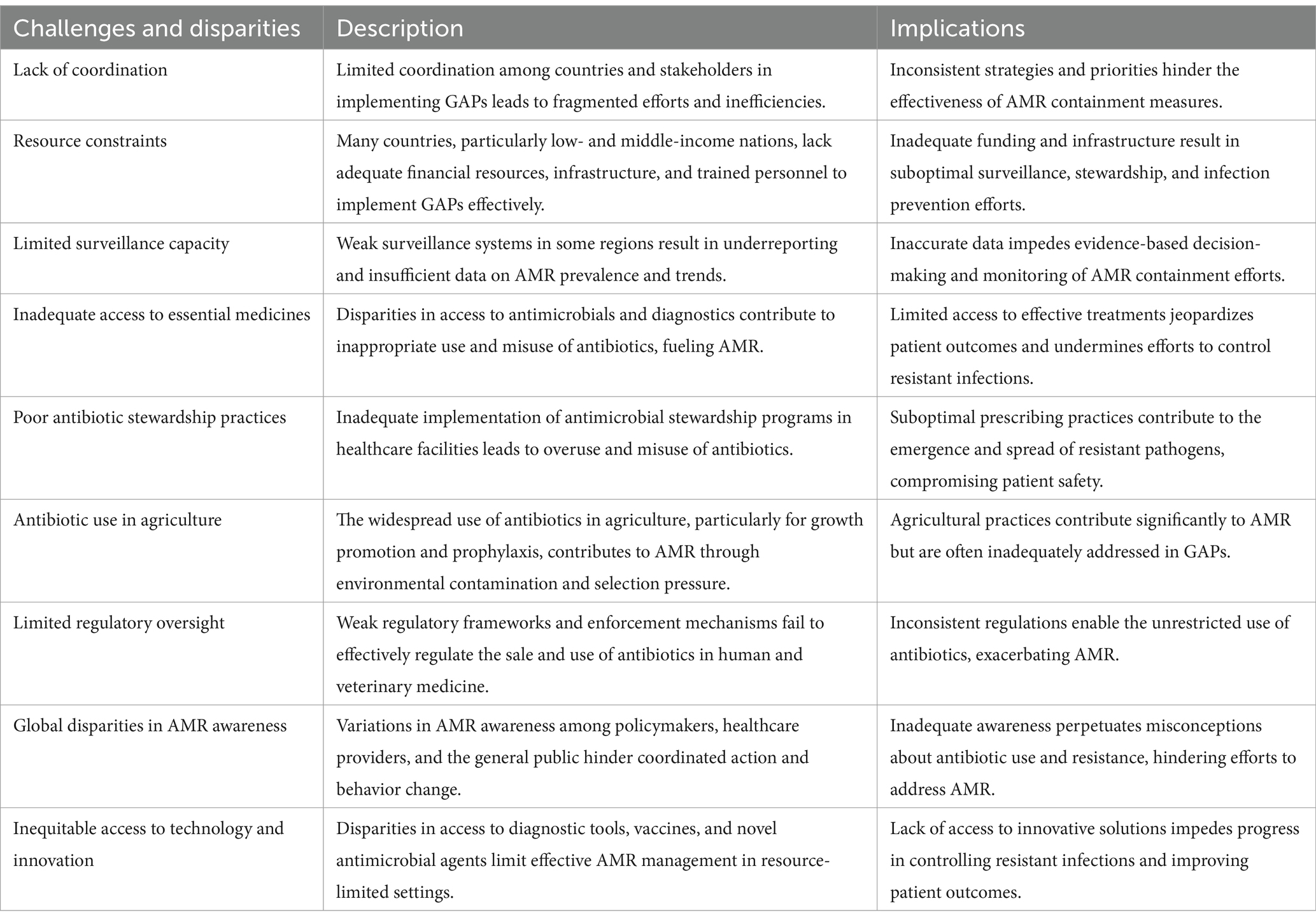
Table 4. A comprehensive overview of the key challenges, disparities, and barriers in the effective implementation of global action plans on antimicrobial resistance (AMR): A comparative analysis across regions and stakeholders (Anderson et al., 2019; Ashiru-Oredope et al., 2023; Bolbanabad et al., 2023; Willemsen et al., 2022; Chinemerem Nwobodo et al., 2022; Pham et al., 2024; Villarreal et al., 2023).
In 2019, agricultural employment accounted for an average of 23.51% of the global workforce, but this figure concealed stark contrasts between high- and low-income nations. In wealthier countries, where economies have shifted toward industry and services, agricultural employment was typically below 5%. In contrast, it exceeded 70% in many low- and middle-income nations, underscoring agriculture’s role as a primary livelihood source (Chen et al., 2023). The unregulated livestock trade exacerbates global challenges, particularly antibiotic resistance. In Colombia (2018), the widespread presence of cattle, along with the illegal sale of refrigerated chicken meat in Nigeria, illustrates how weak regulation enables the spread of resistant bacteria (Zapata-Cortés, 2020). Many countries attempt to mitigate environmental resistance through top-down strategies, such as Non-Aligned Party interventions. However, these measures often fall short in moderate- and low-income nations due to systemic barriers, including a shortage of skilled professionals, weak veterinary drug regulations, illegal livestock trade, and frequent human-animal interactions (Iera et al., 2025; Anderson et al., 2019). Effectively combating antibiotic resistance requires comprehensive national action plans that integrate both top-down policies and grassroots initiatives. While strong regulations and interventions are crucial for promoting responsible antibiotic use, community-driven strategies—such as public awareness campaigns and local management efforts—are equally vital, particularly in developing regions where agriculture remains central to the economy and infrastructure is limited (see Table 5).
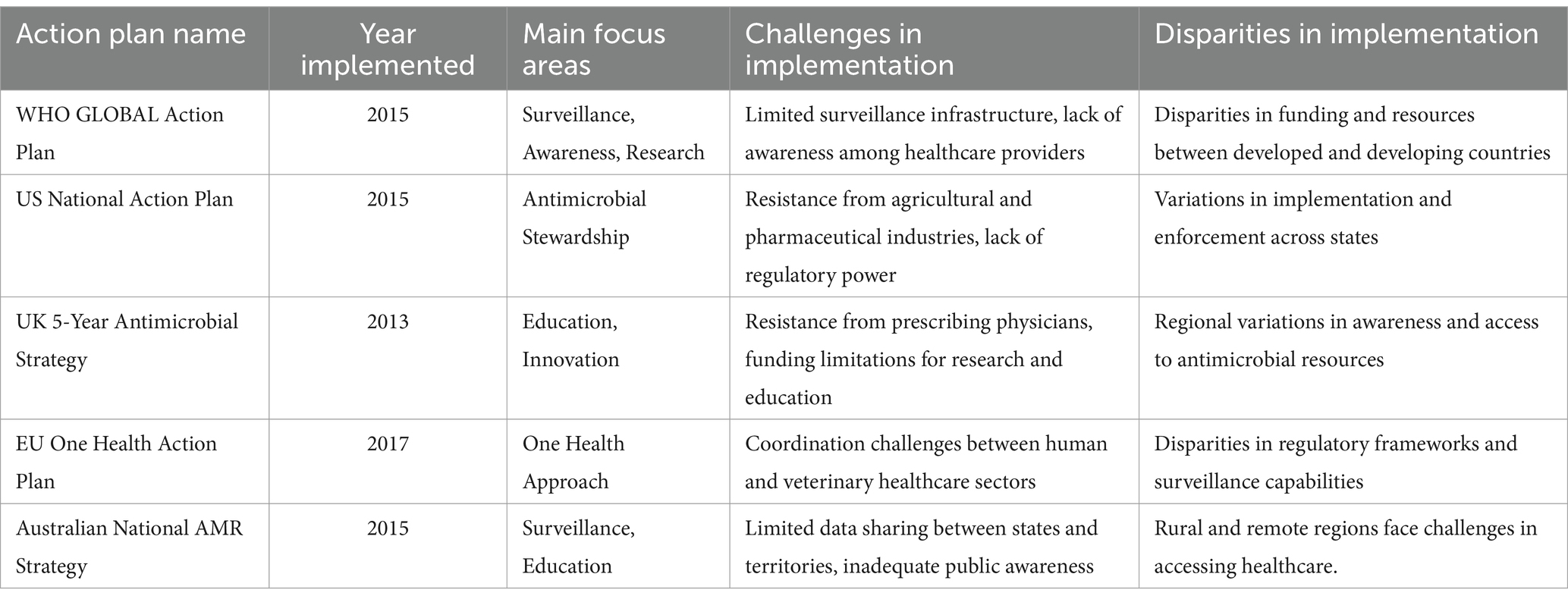
Table 5. A comprehensive overview of global efforts to combat antimicrobial resistance, highlighting common challenges and disparities across regions and countries (Catteau et al., 2024; Charani et al., 2023; Chua et al., 2021; Özçelik et al., 2022; Tanno and Demoly, 2020).
Previous studies (Checcucci et al., 2020; Hetman et al., 2022; Osman et al., 2021) have identified healthcare facilities, farmland, wastewater treatment plants (WWTPs), and agricultural areas as key sources of ARGs and ARB. These regions serve as significant reservoirs of resistant bacteria and genes, facilitating the transmission of bacterial resistance among animals, humans, and the environment (Bunduki et al., 2024; Mugerwa et al., 2021; Okpala et al., 2021). Consequently, they are crucial in antibiotic stewardship and resistance prevention efforts. Hospitals, in particular, are significant contributors to the spread of ARB and ARG (Haseeb et al., 2022). Table 6 provides a comprehensive overview of the sources and transmission pathways of antibiotic resistance in medical and livestock settings. It details various sources, including freshwater bodies, WWTPs, farms, hospitals, and their respective transmission mechanisms. Understanding these pathways is essential for developing effective strategies to curb the spread of antibiotic resistance.
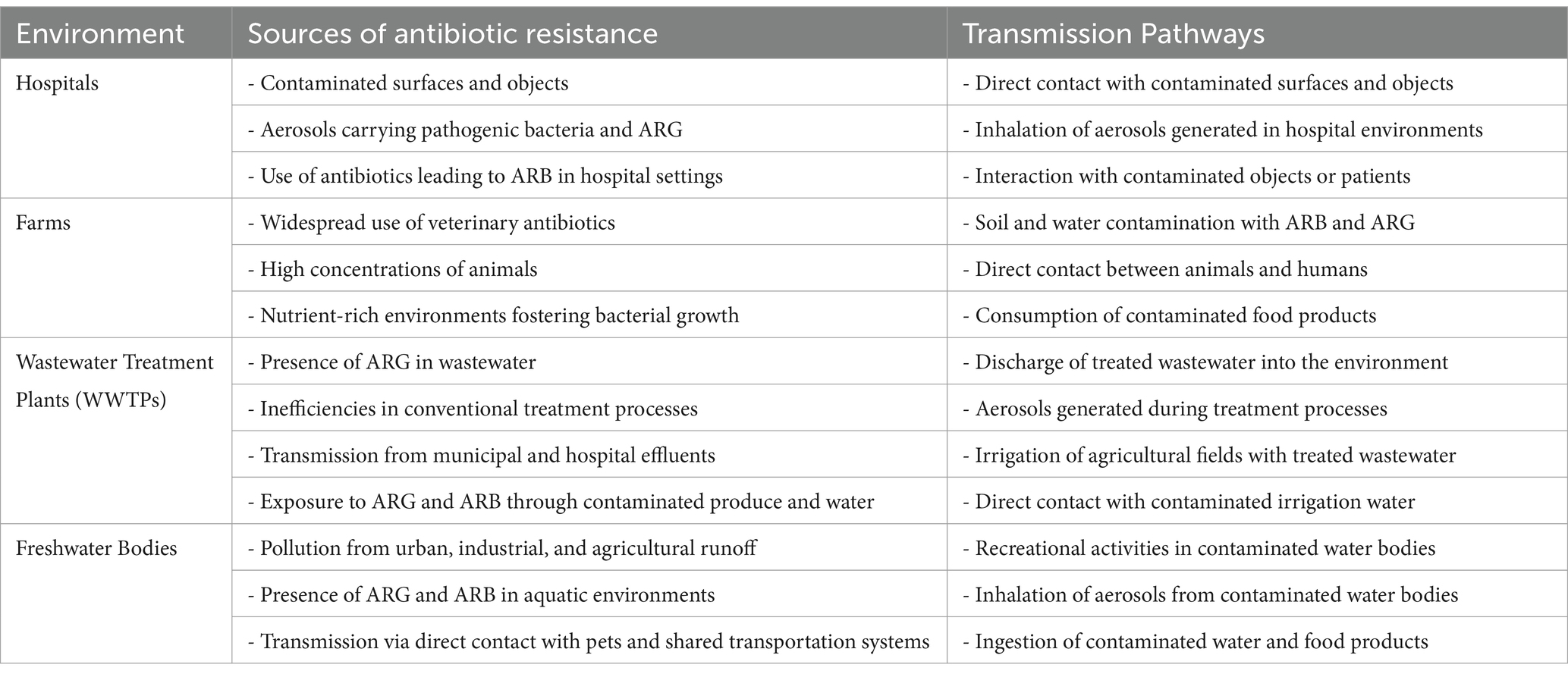
Table 6. Sources and transmission pathways of antibiotic resistance in various environments (Acolatse et al., 2022; Harun et al., 2024; Haseeb et al., 2022; Liu et al., 2018; Otieku et al., 2024).
Hospitals, due to their specialized functions, inevitably serve as reservoirs for infectious agents. Various healthcare environments, including water systems (sewers, taps, sinks), medical instruments (scissors, work tables, switches, infusion stands), and bedding (mattresses), are highly susceptible to microbial contamination and biofilm formation. The presence of antibiotic-resistant genes (ARGs) and antibiotic-resistant bacteria (ARBs) significantly increases the risk of pulmonary infections (Haseeb et al., 2022). Research by He et al. (2020) confirms that hospital bacteria commonly exhibit antibiotic resistance, reinforcing the role of healthcare facilities as hubs for ARG and ARB transmission. Additionally, exposure to pharmaceuticals, contaminated surfaces, and shared hospital wards facilitates ARB spread among healthcare workers and patients. Mitigating contamination requires strict antibiotic stewardship, rigorous sterilization protocols, and standardized medical waste management.
Similarly, agricultural environments—particularly farms with intensive veterinary antibiotic use, high animal densities, and nutrient-rich conditions—are hotspots for antibiotic resistance emergence and proliferation (Bai et al., 2022). Studies (Bai et al., 2022; Cui et al., 2024; Ding et al., 2022; Georgakakos et al., 2021) confirm the widespread presence of ARGs and ARBs in animal waste, wastewater, soil, and air. Resistance genes such as tet(X) (conferring tigecycline resistance) and mcr-1 (associated with colistin resistance) have been detected in humans, animals, and the environment (Khan et al., 2021). Additionally, resistant bacterial strains—including extended-spectrum beta-lactamase (ESBL)-producing E. coli and methicillin-resistant Staphylococcus aureus (MRSA)—have been identified in agricultural settings (Crespo-Piazuelo and Lawlor, 2021). Environmental factors such as antibiotics, pH, temperature, and oxygen levels facilitate ARG transfer between bacteria, contributing to cross-contamination risks for agricultural workers and surrounding communities (Carfora et al., 2016; Liu et al., 2016; Tong et al., 2022; Hounmanou et al., 2021).
Wastewater treatment plants (WWTPs), processing municipal and hospital effluents, are also major reservoirs of ARBs and ARGs (Bueno et al., 2020). Studies have detected ARGs such as sulI, qnrA, ermB, and blaCMY in wastewater (Herraiz-Carboné et al., 2022; Kayali and Icgen, 2020), highlighting the limitations of conventional treatment methods. Aerosols generated during wastewater processing can disperse ARGs into surrounding areas, while irrigation with contaminated water and agricultural use of animal waste further propagate their spread into food products (Liu and Song, 2019). Consequently, fresh produce and meat remain vulnerable to contamination throughout production and processing, posing potential health risks to consumers.
Freshwater bodies are similarly at risk of ARG and ARB contamination from urban and industrial discharges, as well as agricultural runoff. Human exposure occurs through direct contact during recreational activities, inhalation of contaminated aerosols, or ingestion of polluted water (Che et al., 2019). Additionally, close interactions between pet owners and animals, along with shared public spaces, may contribute to ARG and ARB dissemination. However, accurately assessing human exposure remains challenging due to the complexity and variability of these transmission pathways.
ARG and ARB in the human body pose substantial health hazards comparable to ticking time bombs. When someone is sick or has a weaker immune system, opportunistic infections can quickly multiply, which can cause antibiotics not to work effectively (Andrina et al., 2020). ARGs and ARB can arise within the human body through two primary mechanisms. The first mechanism involves de novo emergence via spontaneous mutation in the host microbiome or by introducing external ARBs and ARGs (Alame Emane et al., 2021). This can be colloquially described as “starting from scratch.” The second mechanism involves horizontal gene transfer (HGT) of ARGs within bacterial communities, a process denoted as “from less to more” (Zhang F. et al., 2022).
Moreover, it is essential to have practical approaches for restoring the sensitivity of ARB to prevent the emergence of additional resistance. Therefore, it is crucial to hinder the colonization of foreign ARG and ARB in humans, reduce the horizontal gene transfer (HGT) of the collection of resistance genes within the body, and reverse bacterial resistance (see Figure 3).
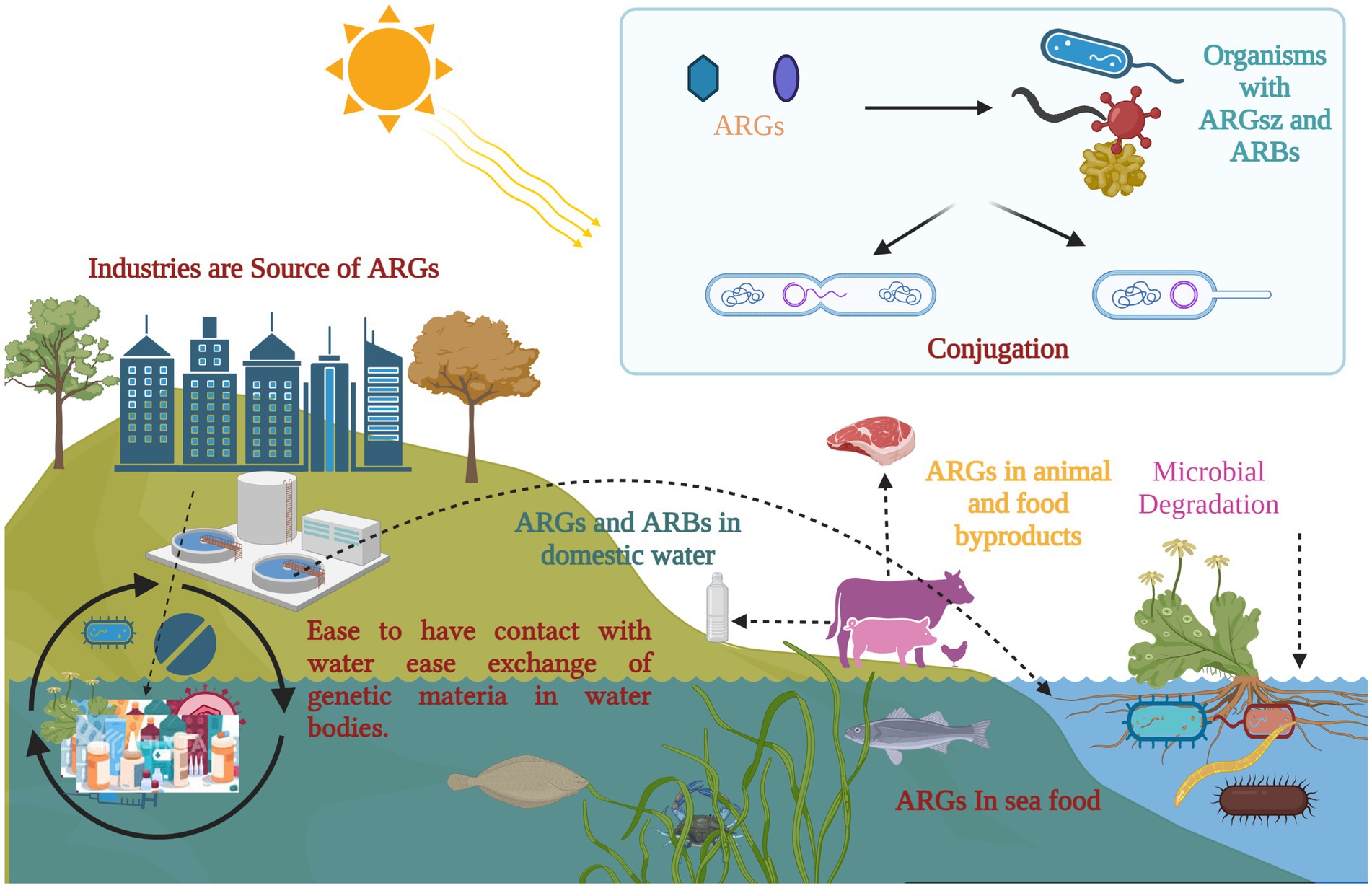
Figure 3. Antimicrobial resistance and its potential implications across organisms: exposure to selective pressure can promote the transmission of antibiotic resistance genes (ARGs) within microbial communities, including biofilms. Floating vegetation has the ability to absorb and harbor ARGs, which are introduced into aquatic environments from various sources. Once in these environments, ARGs can spread through microbial genetic exchange, food chains, and selective pressure. A potential pathway for ARGs includes transmission to humans and animals through contaminated water or infected fish. (Created with BioRender.com).
According to Salyers et al. (2004) and Lee et al. (2020), antibiotic-resistant bacteria (ARB) play a crucial role in transferring antibiotic resistance genes (ARGs) to other organisms, including commensal bacteria in the intestines. Pathogenic bacteria have developed various virulence factors to overcome colonization resistance, allowing them to establish themselves in the gut and cause infection or disease. These factors significantly impact the human microbiome.
Bacterial colonization is a complex process involving movement, response to chemical signals, attachment, penetration, and the use of a specialized secretion system known as the Type VI secretion system (T6SS) (Klassert et al., 2021) (Figure 4). Table 7 presents a detailed overview of strategies to reduce bacterial colonization capacity by targeting flagellar motility, disrupting bacterial adhesion, and leveraging the Type VI secretion system. Each strategy is supported by examples of methods or approaches for effective implementation (Borgeaud et al., 2015).
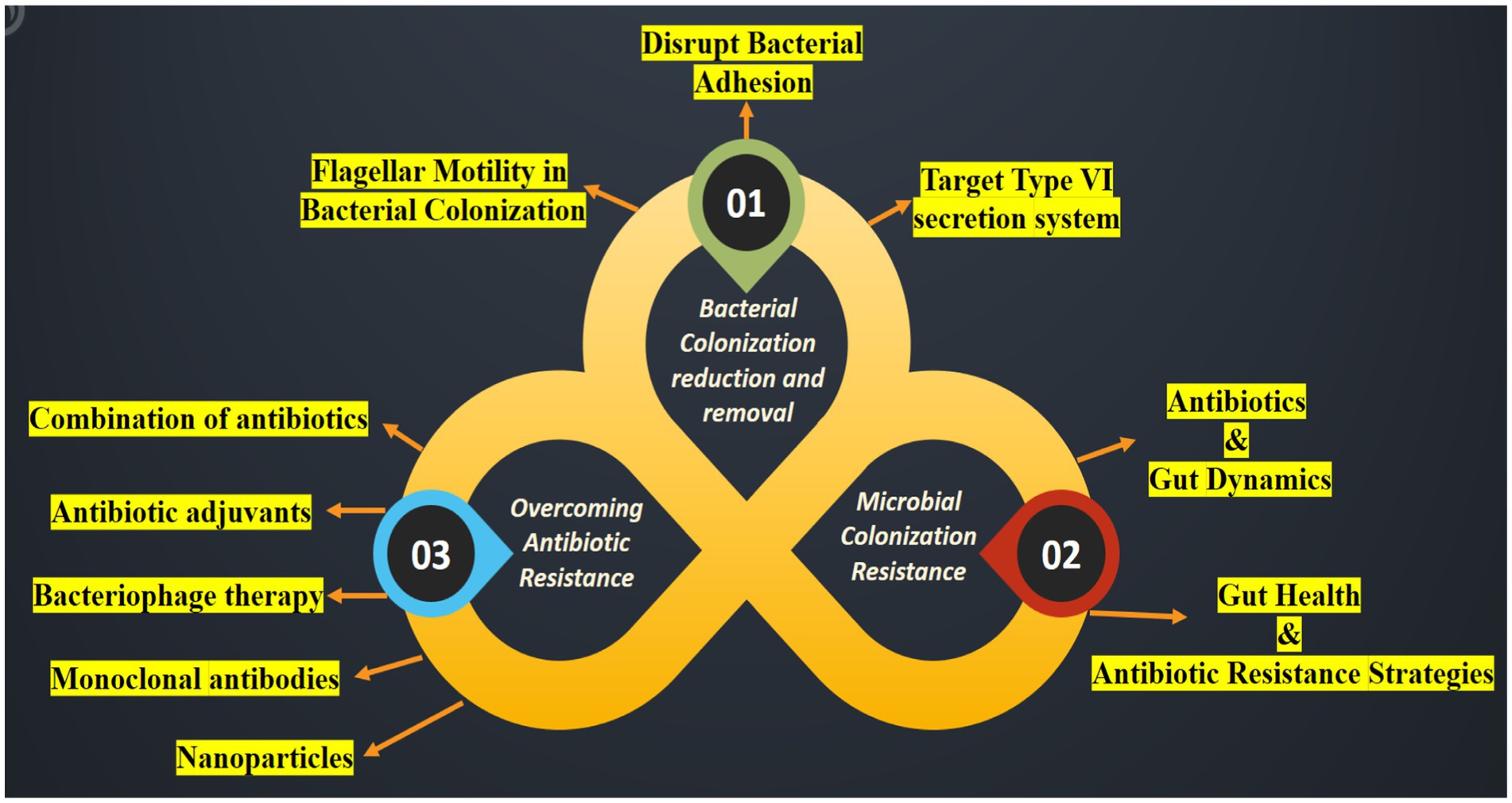
Figure 4. An illustration highlighting the suppression of colonization and multiplication by ARB and Antibiotic-Resistant Genes (ARG). Strategies to hinder the spread of the pathogen encompass the following: (01) reducing the ability of bacteria to colonize the microbiome; (02) strengthening resistance to colonization by the microbiome and preventing horizontal gene transfer; and (03) reversing bacterial resistance to antibiotics.
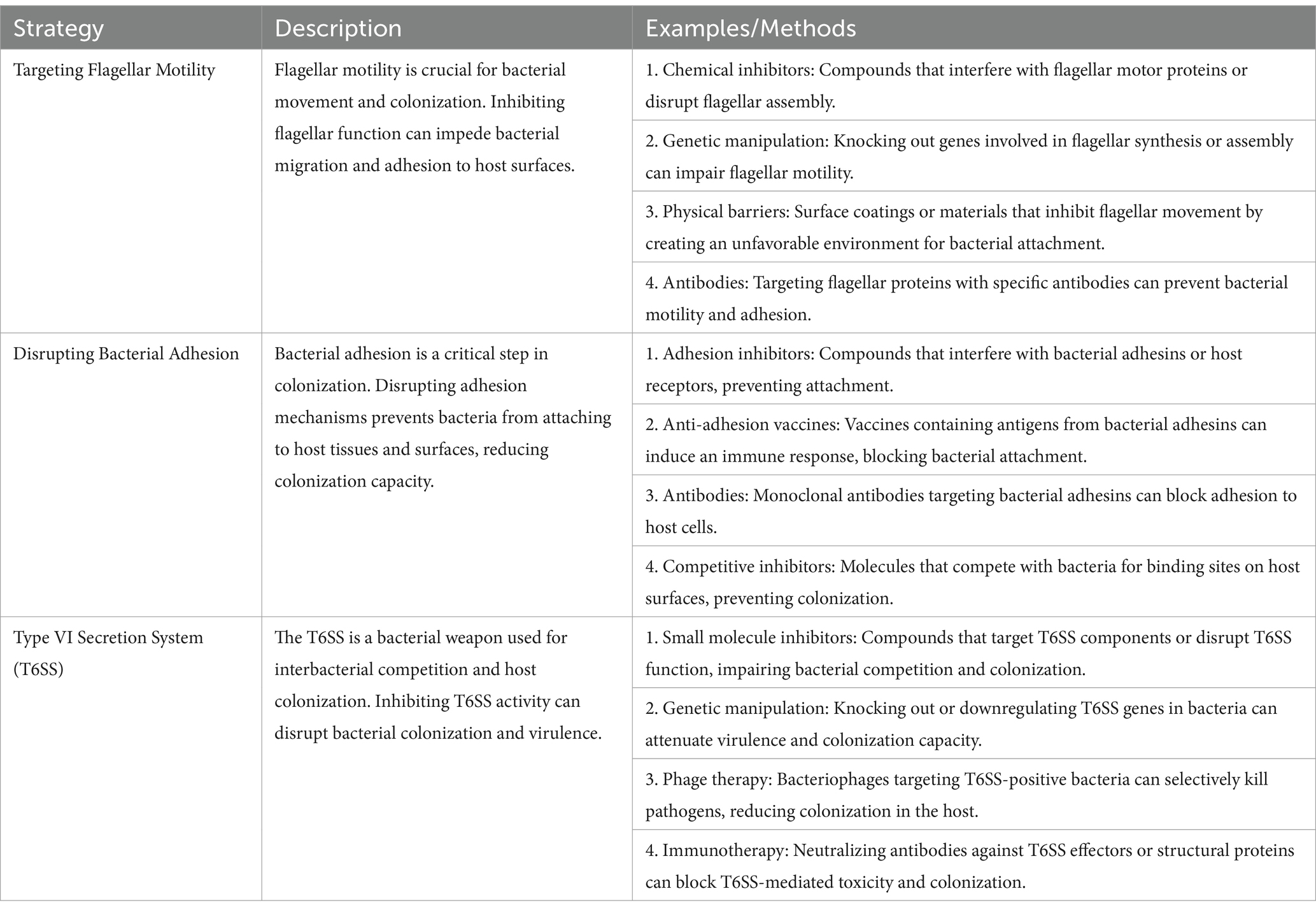
Table 7. Strategies to combat bacterial colonization: targeting motility, adhesion, and secretion mechanisms (Ducarmon et al., 2019; Hsieh et al., 2019; Klassert et al., 2021; Klassert et al., 2020).
Flagellar motility and chemotaxis play crucial roles in helping bacteria detect nutrient sources and thrive in favorable environmental conditions, enhancing their colonizing ability (Colin et al., 2021). The bacterial flagellum is a remarkable molecular machinery that facilitates bacterial or pathogenic movement. Through flagellar-mediated chemotaxis (Colin et al., 2021), bacteria can navigate toward specific regions within new hosts, thereby promoting colonization, invasion, and growth (Figure 5). Recent research by Kreuder et al. (2020) demonstrated that a short noncoding RNA, CjNC110, can influence the mobility of Campylobacter jejuni, enhancing its ability to establish in animal hosts. Consequently, modulating flagellar mobility may reduce bacterial adherence and motility.

Figure 5. Overview of the Bacterial Flagellum as an Adhesin (a) The flagellum facilitates bacterial attachment to eukaryotic cells indirectly by enabling motility, allowing bacteria to reach target sites. (b) It can also directly bind to epithelial cells on both the apical and basolateral surfaces. (c) The flagellum targets various receptors, including mucus, mucins, glycans on cell surfaces or in mucus, extracellular matrix (ECM) proteins, and the bacterial-secreted protein EtpA, which aids in adhesion to host cells. (d) Furthermore, the flagellum can mediate inter-bacterial adhesion, connecting different bacterial species. (Created with BioRender.com).
Additionally, studies by Šimunović et al. (2020) have shown that essential oils and ethanolic extracts can diminish Campylobacter jejuni invasion and motility on INT407 epithelial cells by targeting the LuxS system. Given the significance of flagellar motility and chemotaxis in bacterial colonization, interventions aimed at reducing bacterial or pathogen motility hold promise for colonization prevention. Bacteria with decreased motility are less likely to locate favorable environmental niches, limiting their ability to adhere to and penetrate host tissues (Colin et al., 2021).
To establish colonization and cause infection, Staphylococcus aureus must first adhere to and invade host tissues (Asadi et al., 2019). This process is facilitated by specific surface proteins known as adhesins, which play a crucial role in bacterial attachment. Among these, fibronectin-binding proteins (FnBPA and FnBPB) and laminin-binding proteins (Lmb) are particularly significant, aiding in persistent colonization of the nasal passages and intestines (Tewawong et al., 2020; Speziale and Pietrocola, 2020). Additionally, clumping factors A and B (ClfA and ClfB) enhance adhesion by binding to skin proteins, further promoting nasal colonization (Chen et al., 2020).
For successful infection, pathogens must firmly attach to the mucosal epithelium while overcoming host defenses such as secretions, movement, and peristalsis. Bacterial pili, encoded by multiple genes, are critical in this process, enhancing survival and biofilm formation. Type IV pili (T4P) are particularly essential for bacterial attachment, niche selection, and population expansion (Ligthart et al., 2020). Preventing bacterial adhesion can significantly reduce colonization and infection risk, strengthening the host’s ability to combat pathogens. Various strategies have been explored to disrupt bacterial attachment, including inhibiting pili formation, interfering with adhesion mechanisms, and utilizing vaccines or antibodies targeting adhesion factors (O'Brien et al., 2002).
One promising approach involves lactoferrin, which exhibits antibacterial properties by downregulating virulence genes associated with adhesion, such as flagellin, F18 fimbriae, and F4 fimbriae in intestinal epithelial cells (Dierick et al., 2020). Similarly, Ortiz et al. (2021) demonstrated that rifaximin, oregano extract, and carvacrol can modulate the expression of adhesion-related genes (ae, aggA, aap, pic, and aggR) in Escherichia coli, reducing its ability to adhere to HEp-2 cells. These findings offer promising insights into the molecular mechanisms governing bacterial colonization and highlight potential pathways for developing targeted anti-adhesion vaccines.
Further research by Von Mentzer et al. (2020) identified glycosphingolipids as potential receptors for enterotoxigenic E. coli (ETEC) colonization factor CS30. Additionally, studies by Akhtar et al. (2019), Leach et al. (2017), and Qadri et al. (2020) suggest that combining multiple antigens—such as colonization factors CS5, CS3, and CFA/I—with cross-reactive antibodies effectively inhibits bacterial attachment in humans. This multi-antigen strategy provides a comprehensive defense against bacterial colonization and infection.
Beyond pharmaceutical interventions, bioactive compounds found in food, such as proanthocyanidins and polyphenols, have demonstrated the ability to bind bacterial flagella and pili, preventing aggregation and adhesion. This targeted approach offers advantages over conventional antibiotics, as it selectively disrupts bacterial adhesins without inducing bacterial death, thereby reducing the risk of resistance development (Gowd et al., 2019). While sub-inhibitory antibiotic doses can also reduce bacterial adherence, they contribute to the spread of antibiotic resistance genes (ARGs) due to selective pressure. In contrast, natural bioactive compounds present a promising early-stage therapy alternative, as they do not promote antibiotic-resistant bacteria (Gurumallu and Javaraiah, 2021). Continued research into adhesion inhibitors and their mechanisms of action holds great potential for novel therapeutic interventions against bacterial infections.
The Type VI secretion system (T6SS) is a specialized mechanism employed by Gram-negative bacteria to inject toxic compounds into rival microbes, fostering microbial competition (Borgeaud et al., 2015). This system allows pathogens to bypass the natural defense mechanisms of the host’s commensal microbiota, granting them a competitive edge and enabling prolonged persistence within the host. Additionally, bacterial cell death triggered by T6SS releases DNA, serving as a substrate for horizontal gene transfer and facilitating the spread of antibiotic resistance genes (ARGs) (Hachani et al., 2016). By manipulating the microbiota in this manner, pathogenic bacteria can evade immune responses and outcompete beneficial microbes.
Outer membrane vesicles (OMVs) play a crucial role in bacterial competition, antibiotic resistance, and horizontal gene transfer. Recent findings by Li et al. (2022) highlight the importance of the lipopolysaccharide (LPS)-binding effector TeoL in T6SS-mediated recognition and utilization of OMVs. TeoL interacts with OMVs in the surrounding environment, enhancing bacterial adaptation. T6SS thus provides a substantial advantage for bacteria colonizing symbiotic microbiota niches (Figure 6), shaping microbial communities and influencing horizontal gene transfer. Exploring the interactions between T6SS, OMVs, and gene transfer mechanisms could provide deeper insights into the role of T6SS in the dissemination of ARGs.
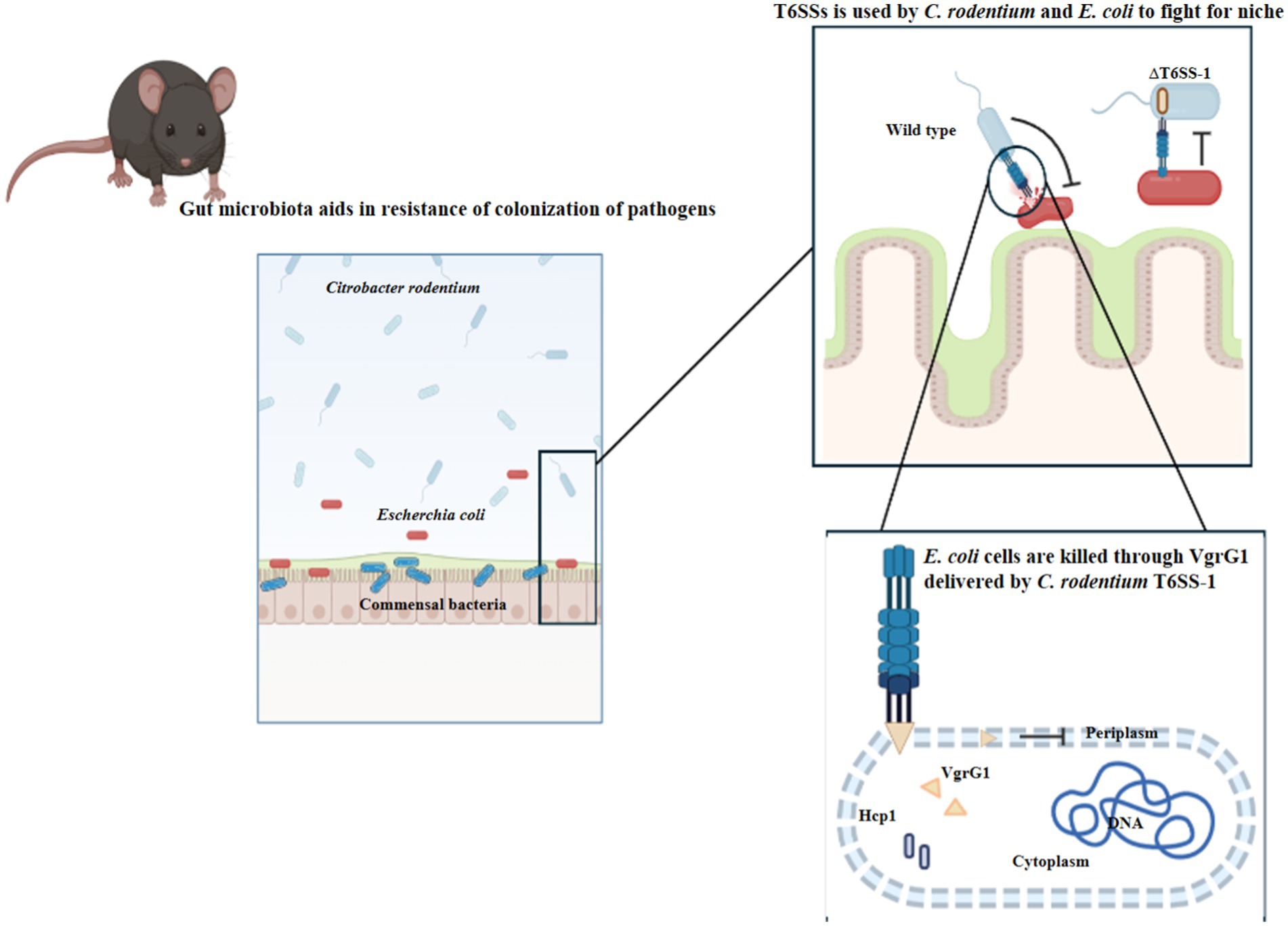
Figure 6. Pathogenic Escherichia coli (E. coli) and commensal Citrobacter rodentium (C. rodentium) employ Type VI secretion systems to facilitate their colonization and persistence within the gut environment. (Created with BioRender.com).
Efficient suppression of the Type VI Secretion System (T6SS) presents a promising strategy to control the infiltration of symbiotic bacteria into specific niches and regulate the invasion of foreign pathogens. Research by Sima et al. (2018) and Balta et al. (2021) has demonstrated that Auranta 3,001, a formulation containing organic acids and plant extracts, effectively reduces the infection potential and cecal colonization of Campylobacter coli and Campylobacter jejuni. This occurs through the downregulation of genes associated with T6SS. Similarly, paramyxoviruses can diminish their virulence when exposed to a combination of citrus extract, lactic acid, and citric acid (E330), which inhibits the T6SS-associated hcp1 and hcp2 genes. These findings suggest that reducing T6SS activity or minimizing direct interactions with unrelated bacteria can help protect bacterial populations while preserving microbial diversity and abundance. By counteracting T6SS-dependent mortality, this approach enhances the resilience of commensal bacteria, enabling them to better withstand colonization in the human host.
Colonization resistance (CR) is vital in host-microbial interactions. It actively hinders the establishment of foreign microbes in the body by many mechanisms, such as immune-mediated responses (Buffie and Pamer, 2013). The mechanisms involve the secretion of antimicrobial substances such as bacteriocins, bile acids, and short-chain fatty acids. They also include competing for necessary nutrients, maintaining the integrity of the intestinal barrier, and influencing the host’s immune response (Seekatz et al., 2022; Styczynski et al., 2023). Microbiota disruptions caused by variables, including antibiotic usage and non-antibiotic medications, such as antipsychotics and anti-diabetic drugs, might change the makeup of microbes, leading to a compromised ability to resist colonization (Seekatz et al., 2022).
The host’s microbiota typically demonstrates tolerance towards the majority of invading germs. Microbiomes with a higher diversity level show a more vital ability to withstand and recover from external bacterial assaults or infections. Hence, maintaining the composition and variety of indigenous microbiota improves its capacity to resist colonization by exogenous bacteria (Styczynski et al., 2023). ARGs can spread among bacterial species by horizontal gene transfer (HGT), particularly in diverse microbial communities like human and animal gut. Gaining a more profound comprehension of horizontal gene transfer (HGT) pathways in the gastrointestinal tract could expedite the advancement of medicines to diminish the dissemination of antibiotic-resistant genes (Ducarmon et al., 2019). Several studies (Buffie and Pamer, 2013; Khazaei et al., 2020; Le Guern et al., 2021) have confirmed a robust association between the distribution of ARGs and the evolutionary relationships of bacteria. In addition, the microbiome makeup is influenced by several factors that also impact the abundance of ARGs.
The human gut microbiome serves as a significant reservoir of antibiotic resistance genes (ARGs) and plays a crucial role in the emergence and spread of antibiotic-resistant bacteria (ARB) and ARGs (Zhang et al., 2024). Antibiotic use profoundly impacts the gut microbiome’s composition and resistance gene profile, often leading to the depletion of both beneficial and harmful bacteria. The extent of this impact depends on factors such as the specific antibiotic used, the mode of administration, and the patient’s preexisting gut microbiota (Ahuja et al., 2022). Broad-spectrum antibiotics, in particular, tend to reduce microbial diversity, facilitating the proliferation of antibiotic-specific ARGs. The influence of different antibiotics on the gut microbiome varies significantly. For instance, Willmann et al. (2019) demonstrated that cotrimoxazole administration led to a 148.1% increase in sulfonamide ARGs, whereas ciprofloxacin had little effect (Batra et al., 2017). Similarly, certain antibiotics, such as macrolides, glycopeptides, and β-lactams, can reduce populations of beneficial commensals like lactic acid bacteria and bifidobacteria, disrupting gut homeostasis (Li et al., 2023).
The distinction between bacteriostatic and bactericidal antibiotics further influences resistance development and dissemination. Bacteriostatic agents inhibit bacterial growth and replication without directly killing the bacteria, allowing the immune system to clear the infection. They primarily target processes like protein synthesis or metabolic pathways. In contrast, bactericidal agents directly kill bacteria by disrupting essential cellular structures, such as the cell wall, membrane, or DNA integrity (Hong P. Y. et al., 2020). The effectiveness of these agents depends on factors like bacterial species, antibiotic concentration, and environmental conditions. While bactericidal agents are often preferred for severe infections due to their rapid action, bacteriostatic agents are useful for controlling bacterial growth in less critical cases. Notably, bactericidal antibiotics tend to reduce mutation rates, thereby lowering the risk of resistance development.
The mode of antibiotic administration also influences gut microbiota disruption and ARG selection. Studies by Fan et al. (2021), Guo et al. (2022), Kelly et al. (2021), and Zhang et al. (2013) indicate that injectable tetracycline and ampicillin have a lesser impact on intestinal ARGs compared to oral administration. Injections help maintain microbiota stability and reduce the depletion of beneficial bacteria. Additionally, the composition of the gut microbiome before antibiotic treatment plays a crucial role in determining treatment outcomes. Interestingly, non-antibiotic drugs can also contribute to ARG dissemination in clinical and environmental settings through plasmid-mediated transfer (Ding et al., 2022; Wang M. et al., 2022).
Given these complexities, a comprehensive assessment of antibiotic administration’s effects on the microbiome and resistome in real-world scenarios is essential. Translating laboratory and clinical findings into practical strategies is key to combating antibiotic resistance while minimizing harm to the host. Selecting antibiotics and delivery methods that exert minimal impact on the gut microbiota is crucial. Advances in precision medicine, such as supramolecular antibiotics, gold nanoparticles, bacteriophage therapy, antibody-antibiotic conjugates, and nanoparticle-based drug delivery, offer promising solutions for targeted antimicrobial therapy. Maier et al. (2021) identified compounds like benzbromarone, dicumarol, and tolfenamic acid, which protect Bacteroides from erythromycin without affecting other microbes. Such counteractive agents, along with optimized drug administration techniques and the use of specific antibiotics, can significantly reduce microbiota disruption and limit ARG proliferation. By integrating these approaches, we can mitigate the adverse effects of antibiotics while maintaining a balanced and resilient gut microbiome.
Maintaining a healthy gut microbiome is essential for defending against the invasion of harmful microorganisms. Diet plays a significant role in shaping the composition and resistance capabilities of the gut microbiota. Polyphenols, found in fruits, vegetables, cereals, tea, coffee, and wine, have garnered attention for their potential health benefits, including antioxidant, anti-inflammatory, and anticancer properties (de Llano et al., 2020; Gowd et al., 2019; Lal et al., 2021; Maisto et al., 2023). In vitro studies have shown that polyphenols can regulate the human gut microbiota by inhibiting harmful pathogens such as Helicobacter pylori and Staphylococcus sp., while promoting beneficial bacteria like Lactobacillus and Bifidobacteria (Gowd et al., 2019). Evidence from animal and clinical trials further supports that polyphenols influence gut microbial composition, diversity, and the Firmicutes to Bacteroidetes (F/B) ratio, largely due to their prebiotic-like effects (Man et al., 2020). Flavonoids such as anthocyanins, phenolic acids like epicatechins, p-coumaric acid, and o-coumaric acid, and other polyphenols, including quercetin, rutin, chlorogenic acid, and caffeic acid, have been shown to enhance beneficial gut bacteria like Bifidobacterium and Lactobacillus while reducing harmful bacterial colonization (Samarasinghe et al., 2019). Catechins, a subgroup of polyphenols, promote helpful bacteria such as Clostridium coccoides, Eubacterium rectale, and Bifidobacterium sp., while inhibiting harmful bacteria like Clostridium histolyticum (Kumar Singh et al., 2019).
However, antimicrobial treatments can disrupt the balance of gut bacteria, reducing beneficial species, particularly those that produce short-chain fatty acids (SCFAs). This imbalance can create a favorable environment for the growth of carbapenem-resistant Enterobacteriaceae (CRE) (Lagadinou et al., 2024; Korach-Rechtman et al., 2020; Sorbara et al., 2019). Additionally, diets high in sugar, fat, and protein have been linked to the promotion of skin bacteria, facilitating the spread of genes that confer antibiotic resistance. A study by Tan et al. (2022) found that dietary patterns could potentially enhance the expression of regulatory genes associated with the amplification and transfer of antibiotic resistance genes (ARG). Research has shown a strong correlation between high-fat diets and an increase in specific ARGs and mobile genetic elements (MGE), leading to dysbiosis and compromised immune function (Tan et al., 2022). Diets tailored to promote gut health, such as low-fat, high-fiber diets, can help maintain a healthy microbiome and prevent the development of antibiotic-resistant bacteria.
Further studies have shown that polyphenols, along with essential nutrients like vitamins A and D, and Omega-3 fatty acids, significantly impact gut microbiota and its barrier function (Cantorna et al., 2019; Duan et al., 2022; Gowd et al., 2019; Kawabata et al., 2013). Vitamin A, for instance, is suggested as an adjunctive treatment for infectious diseases and autism spectrum disorders (ASD), potentially by modulating the gut microbiota (Cantorna et al., 2019). Deficiency in vitamin A has been linked to reduced gut microbiota diversity and lower levels of butyrate-producing bacteria. On the other hand, supplementation with retinoic acid has been shown to inhibit murine Norovirus replication and lessen infection severity (Lee and Ko, 2016).
Higher calcium intake has been associated with a reduced prevalence of obesity, likely due to changes in gut flora linked to lean body composition (Surmeneva et al., 2019). Intervention trials have found that a daily calcium intake of 1,000 mg increases the presence of beneficial Clostridium XVIII in male fecal samples, indicating a higher abundance of butyrate-producing bacteria (Yang et al., 2020). In high-fat diet mouse studies, calcium supplementation at a dosage of 5.25 g/kg enhanced the diversity and abundance of Ruminococcaceae and Akkermansia bacteria in the fecal microbiome. Such interventions are essential to maintain the structural integrity of the intestinal lining and promote a healthy balance of beneficial bacteria, which is crucial for preventing the spread of harmful bacteria and antibiotic-resistant genes (Surmeneva et al., 2019). Non-nutritive sweeteners, as specific dietary additives, have antimicrobial properties and can mimic the effects of antibiotics on the gut microbiota (Yang et al., 2020). Therefore, it is critical to evaluate dietary choices and nutritional intake carefully to regulate the gut microbiome and its susceptibility to antibiotic resistance. More research is needed to determine how dietary interventions can mitigate health risks related to the load of ARGs and antibiotic resistance, as well as to identify which food components are most effective against different antibiotic-resistant bacteria (ARB).
Bacteria have developed a variety of mechanisms, both inherent and acquired, to mitigate the adverse effects of antibiotics (Table 8). Intrinsic resistance predominantly relies on three principal strategies: enzymatic inactivation or alteration of antibiotics, mutation of antibiotic targets, and the presence of cellular membrane barriers and efflux pumps that restrict antibiotic penetration (Zhang H. et al., 2023; Zhang R. M. et al., 2023). Conversely, acquired resistance primarily results from horizontal gene transfer (HGT) (Chen et al., 2021). This process encompasses mechanisms such as transformation (incorporation of exogenous DNA into the bacterial genome), transduction (transfer of DNA mediated by bacteriophages), conjugation (direct exchange of genetic elements between bacteria), and DNA transfer facilitated by membrane vesicle transport (Bai et al., 2022). Developing these mechanisms enables bacteria to acquire antibiotic resistance, leading to the emergence of highly resistant strains. Carbapenemase-producing colistin-resistant Klebsiella pneumoniae is an appropriate example of this phenomenon because it exhibits resistance to almost all of the currently available antibiotics (Weterings et al., 2015). The World Health Organization (WHO) has expressed serious concern regarding a critical shortage of effective antibiotics and a concern about the lack of new research and development (R&D) efforts in the field (World Health Organization, 2024; Bull et al., 2020; Bertagnolio et al., 2024). To combat the escalating threat of antibiotic resistance and ensure the continued efficacy of existing antibiotics, strategies beyond the sole development of novel drugs are crucial.
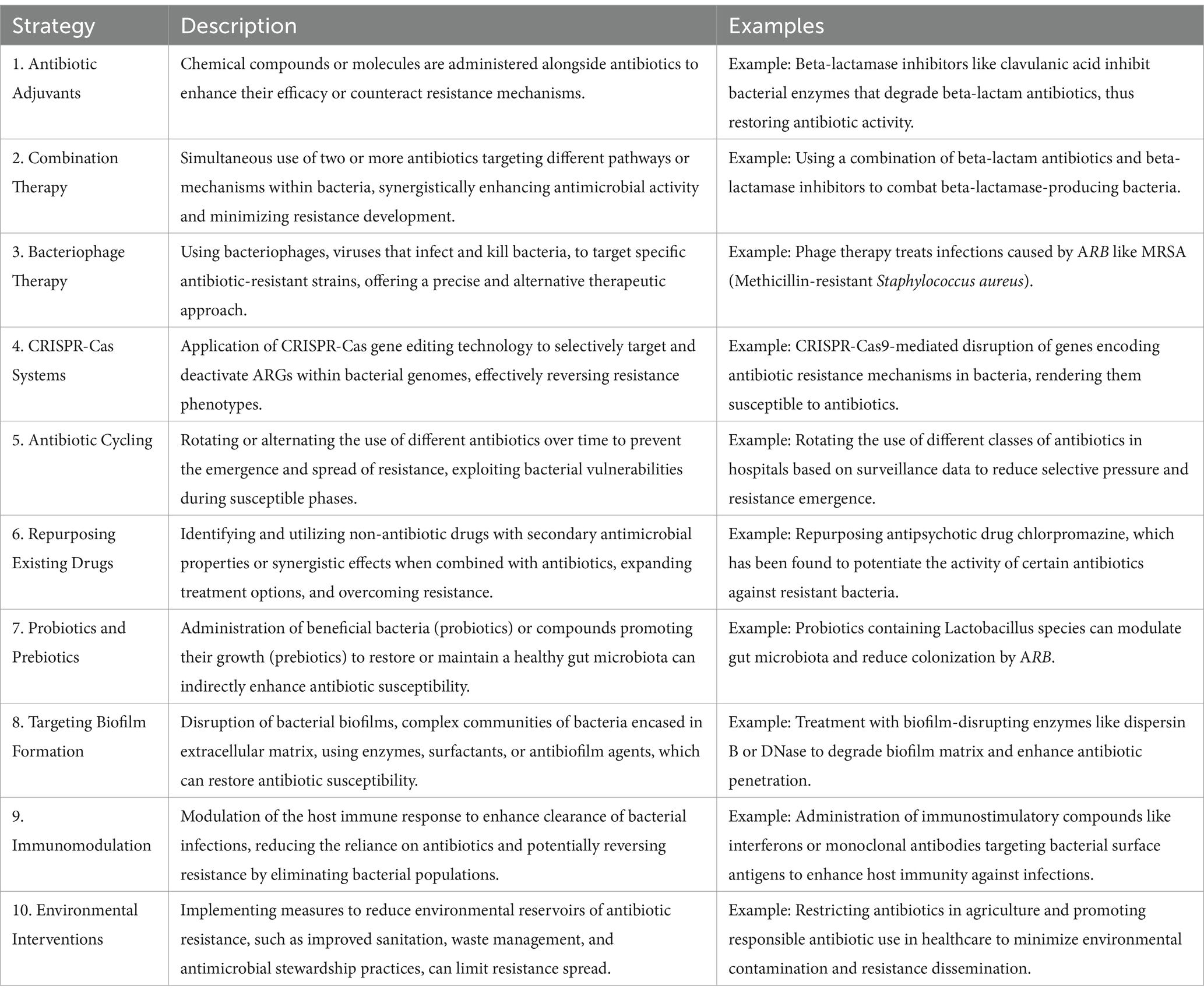
Table 8. Comprehensive strategies and mechanisms for combating antibiotic resistance: approaches, examples, and applications (Gholipour et al., 2024; Gitaka et al., 2020; Kasimanickam et al., 2021; Pattnaik et al., 2023).
Combination antibiotic therapy, commonly used in clinical practice, involves administering two or more antibiotics together to prevent bacterial resistance (Liu L. et al., 2021; Liu Y. et al., 2021). This approach offers advantages over monotherapy, including improved efficacy, broader bacterial coverage, and potentially fewer side effects (Eskenazi et al., 2022). Specifically, combinations of aminoglycoside antibiotics (AGAs), such as gentamicin, enhance treatment effectiveness, accelerate bacterial eradication, and combat antibiotic resistance through synergistic effects with β-lactam antibiotics. β-lactams facilitate the entry of AGAs into bacteria by causing non-lethal damage to the bacterial cell wall, amplifying their bactericidal activity (Wang N. et al., 2022). This strategy is particularly useful in treating severe hospital-acquired infections caused by multidrug-resistant organisms, including pneumonia and sepsis.
Azithromycin, a widely used macrolide with strong antibacterial properties and a long half-life, is effective when combined with AGAs like gentamicin, specifically in treating Pseudomonas aeruginosa infections. This combination allows for dose reductions of both antibiotics (Wang N. et al., 2022). Gentamicin enhances the bactericidal effect of azithromycin on both planktonic and biofilm cells, with notable success in treating genitourinary gonorrhoea. Additionally, antimicrobial peptides (AMPs), derived from plants and animals, play a crucial role as the body’s first line of defense against pathogens (Costa et al., 2021). With broad-spectrum resistance and rapid action, AMPs are emerging as promising antibacterial agents. Research shows that AMPs, like PMAP-36 or PRW4, when combined with gentamicin, produce synergistic effects against Escherichia coli and Staphylococcus aureus by disrupting the bacterial outer membrane, enhancing permeability, and facilitating gentamicin’s entry into the cytoplasmic membrane. AMPs also exhibit direct antibacterial activity through interactions with intracellular targets like DNA.
In treating sepsis and severe Pseudomonas infections caused by gram-negative bacteria, combination therapies involving beta-lactam antibiotics, aminoglycosides, and fluoroquinolones are common (Fish et al., 2002). Studies have also evaluated the combination of polymyxin B, doripenem, and rifampin against multidrug-resistant strains of A. baumannii and K. pneumoniae (Fish et al., 2002). However, clinical outcomes can vary, and the development of antibiotic resistance is influenced by factors such as drug interactions and the inadvertent selection of resistant bacterial subpopulations. Some studies suggest that combination therapy may contribute to the emergence of resistance, exacerbating the spread of resistant strains (Liu et al., 2020; Bassetti et al., 2022). The increasing prevalence of antibiotic resistance, coupled with the slow pace of new antibiotic discovery, highlights the need for innovative therapeutic strategies, focusing on next-generation antibiotics and alternative treatments. While combination therapy can enhance antimicrobial treatment efficacy, it may also lead to unintended consequences (Liu L. et al., 2021; Liu Y. et al., 2021). In some cases, antibiotics may interact antagonistically, reducing each other’s effectiveness. This is particularly concerning when drugs target the same bacterial pathways or have overlapping mechanisms of action (Thu et al., 2021). For instance, combining beta-lactams with tetracyclines can reduce bacterial cell wall synthesis, as beta-lactams inhibit cell wall formation, while tetracyclines prevent protein synthesis (Hamadamin et al., 2024). This may impair the bacteria’s ability to respond to the cell wall damage caused by beta-lactams, reducing treatment efficacy.
Combination therapy, commonly used to treat bacterial infections, offers benefits such as a broader activity spectrum and reduced resistance risk. However, it can pose challenges like reduced bioavailability and increased toxicity (Stielow et al., 2023). When multiple drugs are combined, one may interfere with the absorption or distribution of another, decreasing its effectiveness. For instance, rifampin can reduce the bioavailability of protease inhibitors by inducing liver enzymes that accelerate their metabolism, lowering plasma concentrations (Zhuang et al., 2022). Additionally, combining drugs with overlapping toxicities, such as aminoglycosides and vancomycin, can increase nephrotoxicity and ototoxicity risks. Hence, carefully evaluating the risks and benefits of combination therapy is crucial to avoid reducing therapeutic efficacy or causing harmful side effects. While combination therapies are widely used to treat bacterial infections, they can lead to adverse effects, particularily in patients with impaired organ function or those on multiple medications (Thu et al., 2021). Antibiotics with different mechanisms of action may increase resistance, as seen in the use of ampicillin and ceftriaxone for Enterococcus faecalis infections, which has led to vancomycin-resistant enterococci colonization (Cusumano et al., 2022). Research by Okoliegbe et al. (2021) showed that combination therapy can enhance bacterial fitness and resistance, particularly through mexR gene inactivation in P. aeruginosa, which reduces antibiotic susceptibility. Mathematical models by Berríos-Caro et al. (2021) also demonstrated how resource scarcity and bacterial competition could drive resistance expression, resulting in double resistance under certain conditions.
The synergy in antimicrobial resistance (AMR) can have contradictory effects, accelerating pathogen elimination while promoting single-drug resistance. Carolus et al. (2024) found that collateral sensitivity could alter AMR development, potentially reducing resistance evolution. Anusha et al. (2023) highlighted that genetic evolution and cross-resistance are key when considering combination therapies. The development of resistance depends on factors like drug interactions and both host and pathogen characteristics. Angst et al. (2021) suggested that combination therapies are more effective than monotherapies if double resistance does not emerge, as resistance mutations usually occur independently, and fitness costs of resistance-associated genes are influenced by microbial competition. Future research should focus on understanding the mechanisms of resistance in combination therapies and exploring new combinations that minimize resistance development (Wang et al., 2022). Identifying synergistic effects and aligning treatments with the pathogen’s behavior can optimize therapeutic outcomes. Future studies should assess the impact of these strategies on clinical endpoints, such as infection resolution, patient morbidity, and long-term resistance patterns (Liu L. et al., 2021; Liu Y. et al., 2021; Hamadamin et al., 2024).
The notion of “adjuvant therapy” holds promise in antimicrobial therapies, yet its exploration remains constrained (Douafer et al., 2019). Thus, it is imperative to reassess the reliance solely on antibiotics for treating bacterial infections and instead explore non-antibiotic substances that could mitigate the emergence of antibiotic resistance (Douafer et al., 2019). Antibiotic adjuvants are compounds designed to bolster the effectiveness of antibiotics by either reducing or directly inhibiting mechanisms that confer resistance to them. The concept of antibiotic adjuvants draws from the successful use of synergistic combinations of two or more antibiotics in clinical settings (Hu Y. et al., 2023). These approaches, informed by empirical evidence, aim to harness the combined effects of multiple agents, broaden the spectrum of efficacy, and overcome resistance mechanisms. Unlike conventional antibiotic combinations, antibiotic adjuvants exhibit minimal or no antibacterial activity.
Antibiotic adjuvants can be classified into three primary types based on their target profiles: direct, indirect, and host-modulating resistance breakers (Douafer et al., 2019; Song et al., 2020). Direct antibiotic adjuvants target various active and passive resistance mechanisms in bacteria. These mechanisms fall into three main categories: outer membrane permeabilizers, efflux pump inhibitors, and β-lactamase inhibitors (Figure 7). β-lactamase inhibitors, such as diazabicyclooctanones (DBOs) and boronate-based compounds, have garnered attention for enhancing the efficacy of β-lactam antibiotics (Farhat and Khan, 2022). However, the emergence of reduced Susceptibility to DBOs, mainly due to KPC mutations, underscores the need for ongoing exploration of novel combinations. Efflux pump inhibitors (EPIs) that target specific pumps, such as NorA, AcrAB-TolC, and MexAB-OprM, have shown promise in combating multidrug resistance, as evidenced by studies conducted by Brawley et al. (2022), Tsutsumi et al. (2019), and Sulavik et al. (2001). Notwithstanding extensive investigation, none of these experimental pharmaceutical interventions (EPIs) have progressed to clinical utilization.
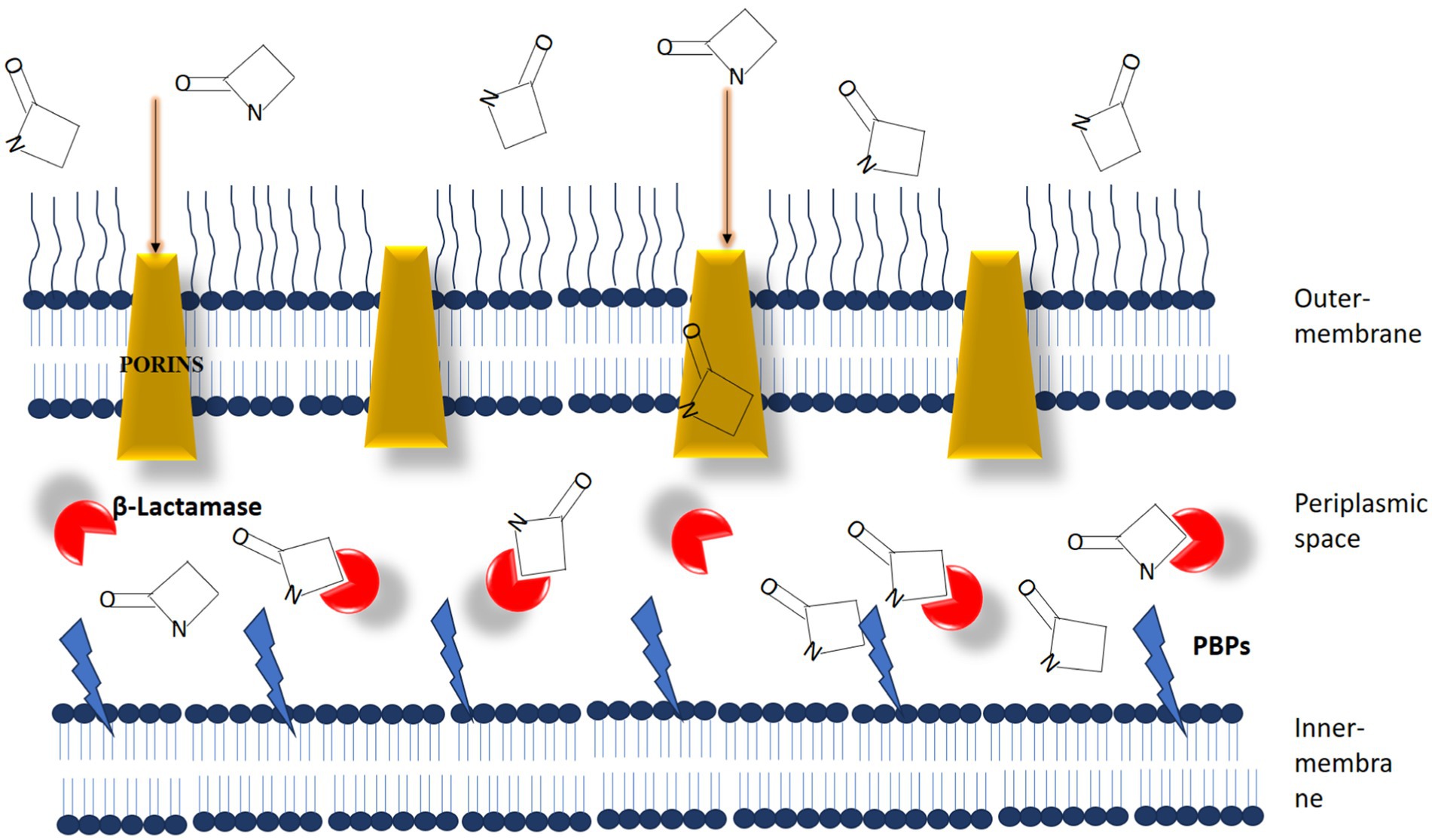
Figure 7. The dynamic interaction between β-lactam antibiotics and β-lactam interactive proteins in gram-negative bacteria plays a crucial role. β-lactamases function as interceptors, impeding the efficient binding of antibiotic molecules to penicillin-binding proteins (PBPs). [Source: Bush K. and Bradford P.A./Clinical Microbiology Reviews, 2020].
Polymyxins, surfactants, antimicrobial peptides, and other outer membrane permeabilizers make it easier for antibiotics to enter bacteria by making their membranes more permeable. Studies by researchers (Song et al., 2020; Song et al., 2022; Song et al., 2023) have validated the efficacy of SLAP-S25 and nordihydroguaiaretic acid (NDGA) as broad-spectrum antibiotic adjuvants. Combining adjuvants with antibiotics represents a proactive strategy against multidrug-resistant (MDR) pathogens. These combinations augment the effectiveness of existing antibiotics and help forestall the development of novel antibiotic resistance mechanisms.
Highly specific for their bacterial targets, bacteriophages (phages) have emerged as a promising alternative therapeutic strategy, particularly for combating multidrug-resistant (MDR) bacteria (Moghadam et al., 2020) (Figure 8). While introducing antibiotics initially overshadowed Felix d’Herelle’s pioneering discovery of phages in 1910 (Vandamme and Mortelmans, 2019), the alarming rise of antibiotic resistance has reignited interest in phage therapy. Unlike broad-spectrum antibiotics, phages exhibit remarkable selectivity, lysing only targeted bacterial strains, including MDR pathogens, while preserving the commensal microbiota. This unique characteristic has spurred exploring various phage-based therapeutic approaches, including phage cocktails, combination therapies with antibiotics, and genetically modified phages (Moghadam et al., 2020; Yacoby et al., 2007). Phage cocktails, offering a broader antimicrobial spectrum, have demonstrated superior efficacy in eradicating bacterial populations compared to single-phage therapies, potentially reducing the emergence of resistance.
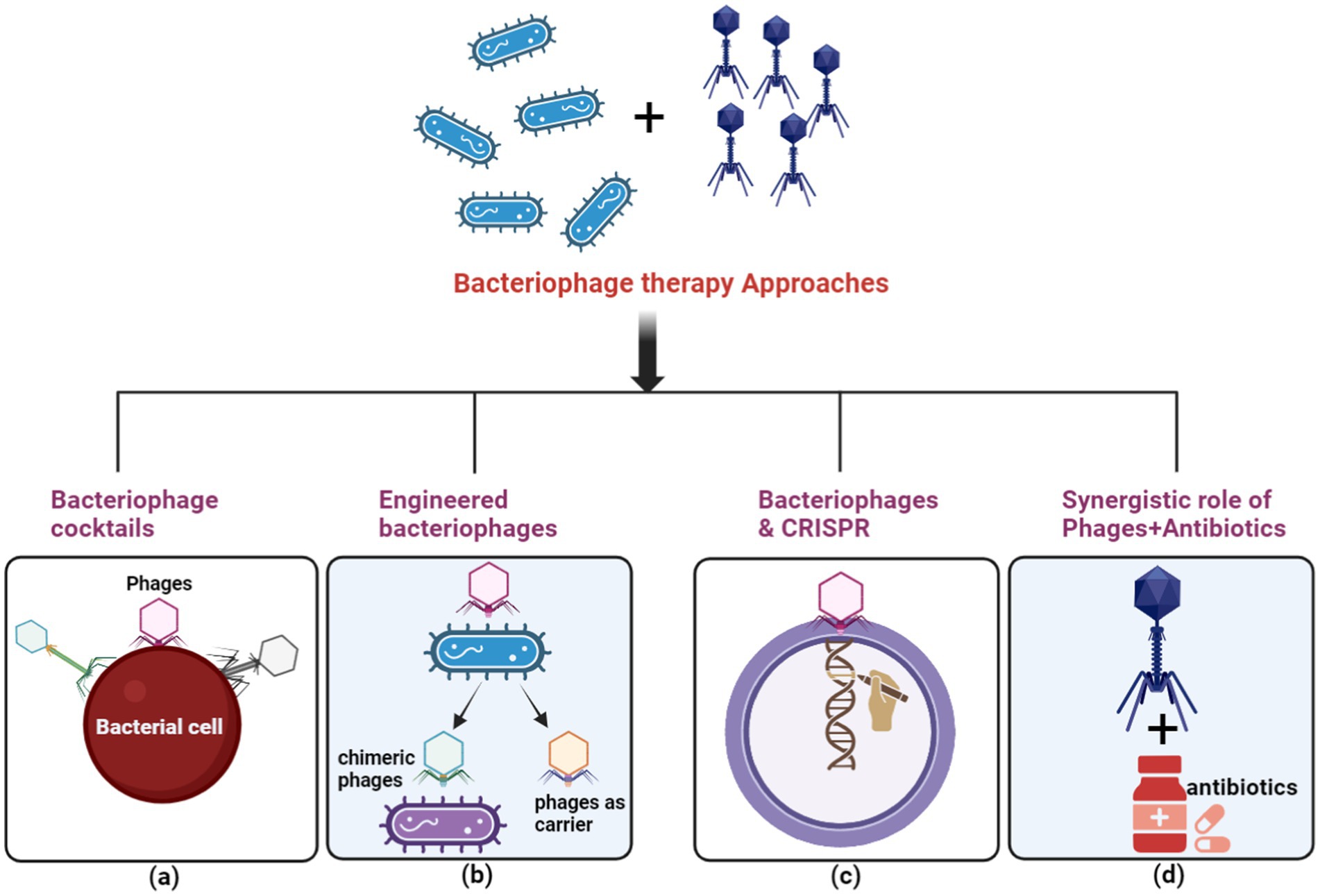
Figure 8. The principal methods for executing phage therapy include. (A) Utilizing a variety of bacteriophages in a phage cocktail to selectively target ARB. (B) Employing bacteriophage engineering to augment the efficacy of the response against ARB. (C) Employing bacteriophages to deliver the clustered regularly interspaced short palindromic repeats-associated (CRISPR-Cas) system for host cell elimination. (D) Depicting the synergistic effect between bacteriophages and antibiotics with a green circle symbol. (Created with BioRender.com).
Clinical successes have been documented in the treatment of Mycobacterium abscessus infections in cystic fibrosis patients and Pseudomonas aeruginosa infections in burn victims (Dedrick et al., 2023). Despite these achievements, significant challenges remain in the use of phage cocktails for antimicrobial therapy (Kering et al., 2019). Key areas of ongoing research focus on identifying optimal multi-component mixtures that target specific pathogens and expanding the host range of phages. Additionally, the co-evolution of bacteria and phages requires continual efforts to address the emergence of phage resistance within bacterial populations.
Phage-antibiotic synergy (PAS) presents a promising approach to enhance antibacterial efficacy while mitigating resistance development (Comeau et al., 2007). PAS exploits the phenomenon whereby sub-inhibitory antibiotic concentrations can promote phage replication, leading to a synergistic reduction in bacterial populations. Combining phage therapy with antibiotics has shown promise in treating infections caused by multi-drug resistant (MDR) bacteria, including MRSA and pan-drug-resistant Klebsiella pneumoniae (Eskenazi et al., 2022; Zalewska-Piątek and Piątek, 2020). However, it is crucial to carefully assess each phage-antibiotic combination, as not all are synergistic; some antibiotics may inadvertently hinder phage replication and reduce their effectiveness.
Bioengineered phages offer exciting possibilities for enhancing antibiotic efficacy and specifically targeting MDR bacterial infections (Cui et al., 2023). These phages can be engineered in various ways, such as increasing host diversity, altering host specificity, delivering foreign genes, and modifying capsids. For instance, modifying phages to deliver antibiotics directly into bacterial cells has led to substantial therapeutic improvements (Kumar et al., 2023). Additionally, the use of CRISPR-Cas systems to target virulence factors and antibiotic resistance genes (ARGs) in bacteria shows promise for restoring antibiotic susceptibility in resistant strains (Wu et al., 2021). By introducing the long tail fibre gene from bacteriophage IP008 into T2 bacteriophage, the host range of E. coli can be broadened (Dunne et al., 2021). Pires et al. (2016) developed a hybrid bacteriophage that combines IP008’s broad host range with T2’s potent cell destruction capability. Similarly, Tabib-Salazar et al. (2017) modified E. coli bacteriophage T7 to produce enzyme protein B, which disrupts biofilms and enhances host cell invasion, reducing cellular biofilms by over 100-fold.
Moreover, bioengineered bacteriophages can improve antibiotic selectivity and address the challenge of multidrug-resistant infections. A pediatric cystic fibrosis patient with a life-threatening infection from antibiotic-resistant Mycobacterium abscessus was successfully treated using a combination of three genetically modified bacteriophages after conventional treatments had failed (Adesanya et al., 2020). This novel treatment led to a favorable prognosis without significant adverse effects. Bacteriophages are increasingly being used as theranostic platforms, with advancements in synthetic biology and drug delivery strategies. For instance, Yacoby et al. (2007) developed a delivery method that attaches chloramphenicol molecules to lysed bacteriophages, resulting in a 2000-fold increase in treatment efficacy while reducing side effects. Furthermore, phages can be employed to transport photosensitizers for more efficient photodynamic inactivation of harmful bacteria, minimizing damage to the normal microbial community (Sheng et al., 2022). In vivo studies have shown the effectiveness of this method in treating infections caused by antimicrobial-resistant bacteria and Candida albicans.
Despite these advancements, some bacteria have developed resistance to bacteriophages through mechanisms such as CRISPR-Cas immunity, restriction-modification systems, and receptor mutations that prevent phage attachment (Hasan and Ahn, 2022). For example, Escherichia coli can acquire CRISPR-Cas spacers to recognize and degrade phage DNA, conferring resistance to certain bacteriophages (Mitić et al., 2023). To overcome these challenges, researchers are developing genetically engineered bacteriophages that bypass bacterial defense mechanisms, enhance phage lysis of resistant bacteria, and deliver antimicrobial genes. Ongoing research, including the engineering of Mycobacteriophage D29 to target antibiotic-resistant Mycobacterium tuberculosis, suggests that these phages could provide an alternative to traditional antibiotics in combating antimicrobial resistance (Pal et al., 2024; Biswas et al., 2024).
Bacteriophage-based technology also has significant potential in the field of cancer treatment. By displaying antibodies, peptides, or proteins on the surfaces of bacteriophages, researchers can utilize them to study the molecular characteristics of tumor cells and the tumor microenvironment. Phages serve as effective delivery vehicles for imaging agents and therapeutic treatments, making them valuable tools in tumor immunology. Additionally, phages have been engineered to deliver suicide genes into cancer cells, significantly improving the efficacy of gene therapy for anti-tumor treatments. Bioengineered bacteriophages hold great promise across a variety of biomedical applications. However, further research is needed to fully exploit their therapeutic potential and to overcome the challenges posed by bacterial resistance mechanisms.
Due to their targeted action, monoclonal antibodies (mAbs) exhibit promise in treating severe bacterial infections (Imran et al., 2021). Clinical investigations are underway for humanized mAb CMTX-001, which targets the DNABII protein crucial for biofilm formation, thereby enhancing antibiotic efficacy and bacterial eradication by disrupting biofilms (Ding et al., 2023).
Moreover, mAbs can target bacterial virulence factors. For instance, panobacumab neutralizes the outer membrane and lipopolysaccharide (LPS) of Pseudomonas aeruginosa (Figure 9), while the KB001 fusion antibody shows potential in targeting bacterial virulence factors (Nagy et al., 2017). In spite of their therapeutic potential, the development and clinical utilization of mAbs are challenging due to high production costs and the need for intravenous administration.
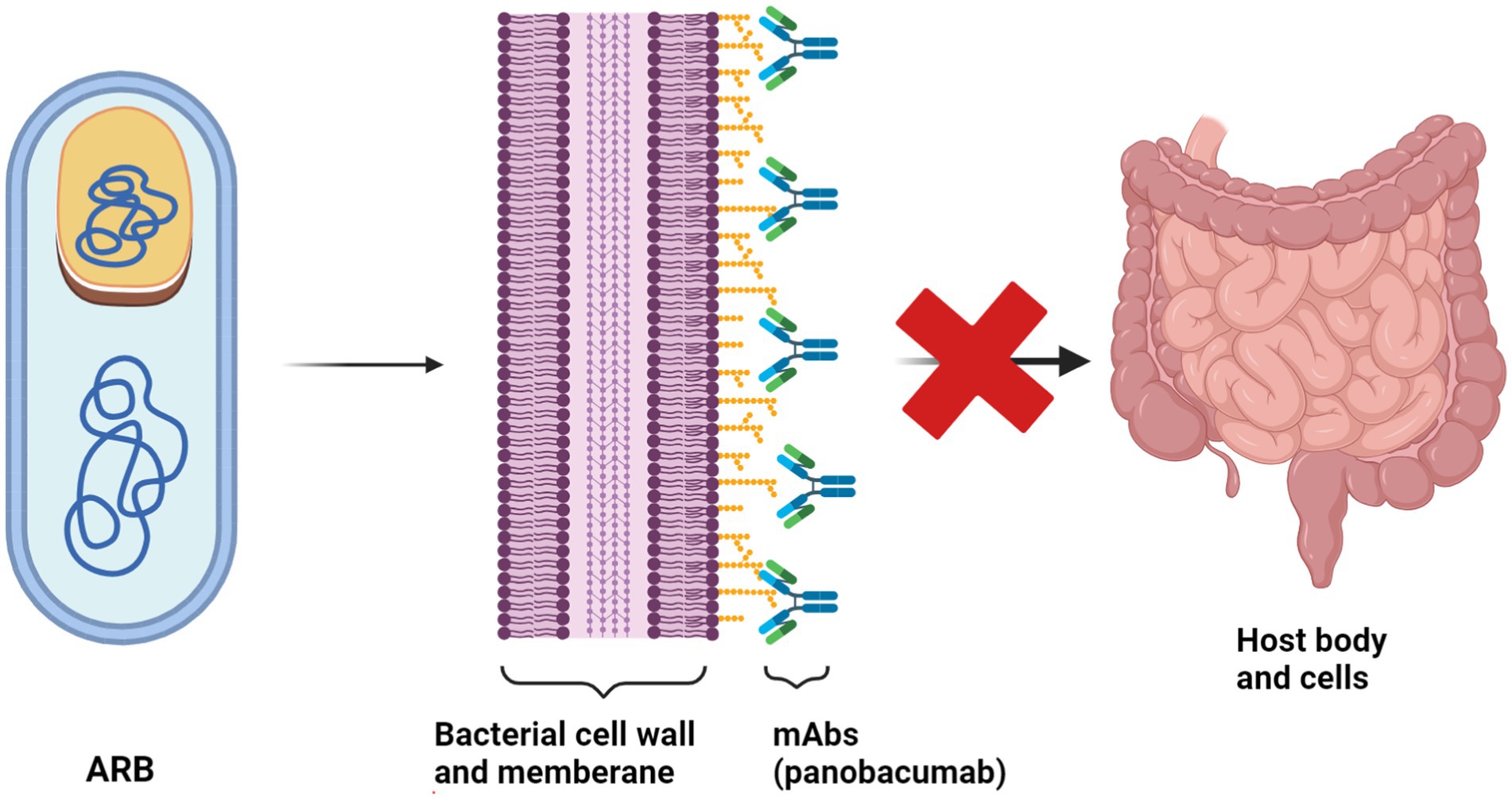
Figure 9. The mechanism of action of Panobacumab monoclonal antibody against Pseudomonas aeruginosa Panobacumab operates through a unique mechanism to combat infections caused by P. aeruginosa, a notorious pathogen known for its resistance to multiple antibiotics. This monoclonal antibody targets a specific antigen on the surface of P. aeruginosa bacteria, namely, the PcrV protein, a critical component of the type III secretion system (T3SS). (Created with BioRender.com).
In addition to combating bacterial infections, vaccines play a pivotal role in reducing antibiotic usage and resistance. The success of Streptococcus pneumoniae vaccination has reduced antibiotic-resistant pneumococcal strains and antibiotic consumption by bolstering herd immunity (Saeed et al., 2023). Similarly, the introduction of conjugated Haemophilus influenzae type b (Hib) vaccinations has substantially decreased Hib-related illnesses and beta-lactamase-producing strains, thus mitigating antibiotic resistance (Storz et al., 2016). These achievements underscore the importance of ongoing vaccine research in combating bacterial diseases. While both vaccines and mAbs offer the potential to fight bacterial infections, mAbs face challenges related to cost and administration (Imran et al., 2021). Conversely, preventive vaccines can potentially significantly decrease antibiotic use and resistance. A comprehensive and practical approach to bacterial pathogens necessitates continued research and development in vaccine and mAb domains.
Regardless of promising findings in treating multidrug-resistant (MDR) bacterial infections, mAb-based immunotherapy encounters significant development hurdles (Seixas et al., 2022). Challenges include the potential for humanized mAbs to elicit a human anti-chimeric antibody (HACA) response, rendering therapeutic efficacy uncertain (Almagro et al., 2018). Moreover, mAb targets are often specific to bacterial antigens, emphasizing the importance of rapid pathogen detection. Additionally, mAbs may be less effective if the target antigen is expressed only in certain circulating strains, specific organ infections, or disease phases. Exopolysaccharide architectures, particularly serotype-specific variations, pose challenges for mAb efficacy against diverse bacterial strains. For instance, Streptococcus pneumoniae serotypes exhibit differences in exopolysaccharide capsule content and structure, complicating mAb efficacy across all serotypes (Wang-Lin and Balthasar, 2018).
Furthermore, PNAG (β-1-6-linked poly-N-acetyl-d-glucosamine), a conserved exopolysaccharide in numerous pathogens, influences microbial survival and biofilm formation (Champion et al., 2019). Although mAb F598 effectively minimized microbial issues across various models and microorganisms, further clinical studies were not pursued. Addressing commercial hurdles and aligning development efforts with disease market size is imperative for advancing broad-spectrum mAb development. Future research endeavours are warranted to address these challenges comprehensively.
The combination of antibiotics with nanomaterials, particularly silver nanoparticles (AgNPs), has a combinatorial antibacterial effect (Figure 10) (Scandorieiro et al., 2022). Antibiotics commonly promote bacterial cell death by stimulating the generation of reactive oxygen species (ROS) (Hong Y. et al., 2020). Studies show that gentamicin significantly enhances the production of reactive oxygen species (ROS) by silver nanoparticles (AgNPs) (Mazur et al., 2020). Luminol chem-iluminescence (CL) indicates that reactive oxygen species (ROS) are generated when Tween-stabilized AgNPs have an antibacterial impact (Wang N. et al., 2022). The concurrent use of gentamicin and Tween-stabilized AgNPs exhibits a synergistic effect in combating gentamicin-resistant Staphylococcus epidermidis, as observed in the study by Mazur et al. (2020). The combination of AgNPs plus antibiotics can efficiently eradicate microbes through numerous pathways, enhancing their antibacterial effectiveness (Mohammed and Abdel Aziz, 2019; Scandorieiro et al., 2022). Silver nanoparticles (AgNPs) have been reported to infiltrate the bacteria’s cell wall, disrupt the cellular membrane, and trigger bacterial death. The study by Katva et al. (2017) revealed a noteworthy synergistic antibacterial impact when combining various antibiotics and AgNPs, specifically gentamicin and chloramphenicol, with AgNPs in the context of E. faecalis (Ef) infection (Katva et al., 2017).
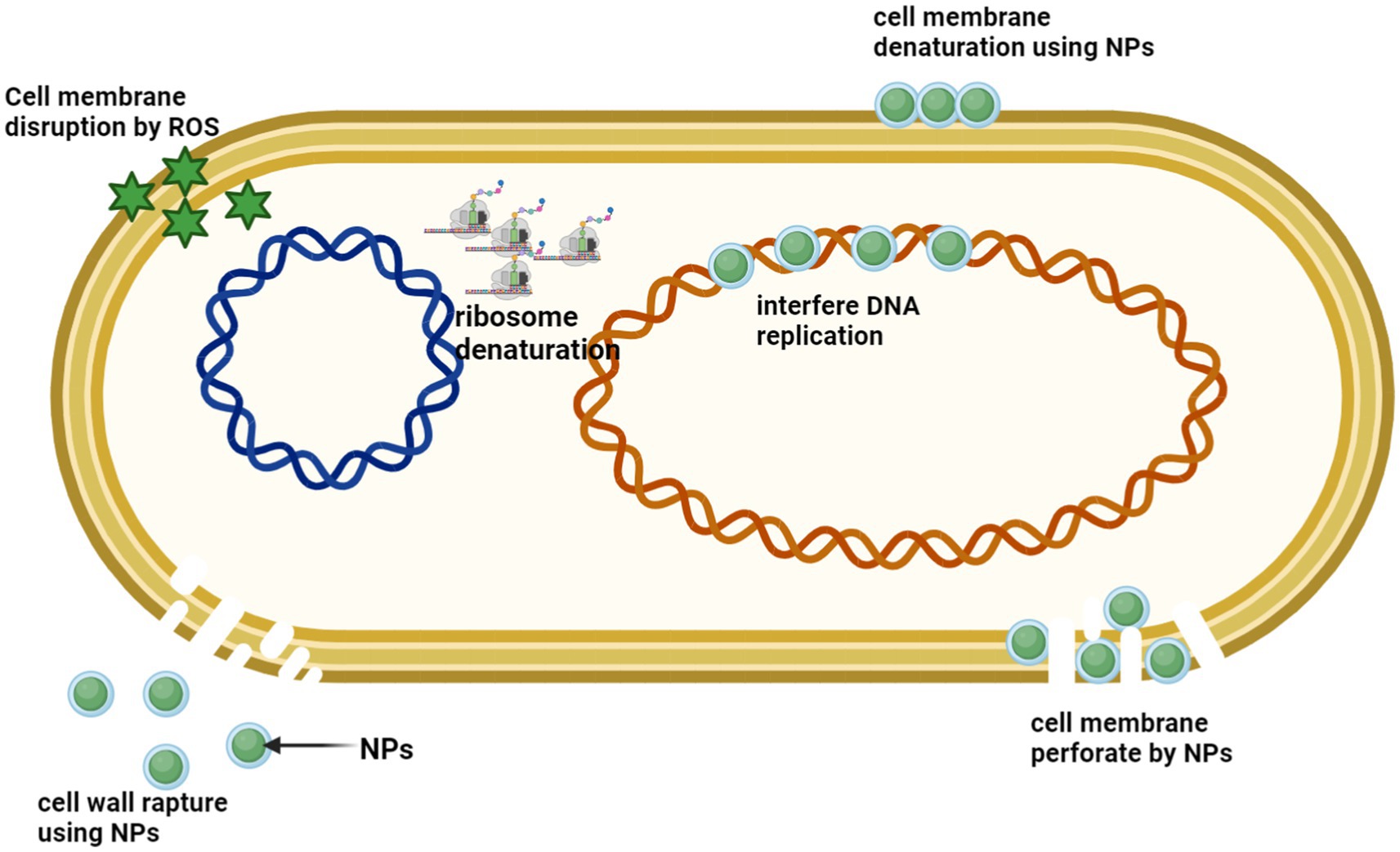
Figure 10. Silver nanoparticles (AgNPs) exhibit a range of antimicrobial properties through multiple mechanisms, including disruption of the cell wall and cytoplasmic membrane, denaturation of ribosomes, generation of reactive oxygen species (ROS) leading to membrane disruption, interference with DNA replication, and membrane denaturation and perforation. (Created with BioRender.com).
Graphene is a well-known substance that is compatible with living organisms and has several uses in the fields of antibacterial activity, biosensing, cancer therapy, and biocarriers (Hu X. et al., 2023; Huang et al., 2022; Saha, Visconti, Desipio, & Saha et al., 2020; Zhang H. et al., 2023). The study (Wang N. et al., 2022) The study investigated the antibacterial properties of TOB-GO-Ag, a water-soluble hybrid composed of silver nanoparticles (AgNPs), graphene oxide (GO), and tobramycin,. The investigation focused on its effectiveness against multidrug-resistant gram-negative E. coli bacteria. The results revealed that TOB-GO-Ag demonstrated the highest antibacterial efficacy compared to GO, AgNPs, and tobramycin, which were used separately. Scientists (Wang N. et al., 2022) revealed how the TOB-GO-Ag composite fights bacteria. The combination disrupts the bacterial cell wall, allowing silver ions (Ag+) and graphene oxide (GO) to enter the cells. This one-two punch triggers oxidative stress within the bacteria, ultimately leading to their death. Additionally, the tobramycin component further hinders bacterial growth by blocking protein-synthesis (Ullah et al., 2018). Some compounds containing bismuth (Bi) have synergistic antibacterial properties when combined with antibiotics (Keogan and Griffith, 2014). Studies (Ma et al., 2017) have demonstrated that Bi2S3 nanoparticles do not possess antibacterial properties when tested against Staphylococcus aureus and methicillin-resistant S. aureus (MRSA), with minimum inhibitory concentration (MIC) values exceeding 1,024 μg/mL. Nevertheless, combining gentamicin with Bi2S3 nanoparticles demonstrates a synergistic antibacterial impact on MRSA. The reaction between Bi2S3 and gentamicin is the key strategy for this interaction. It leads to bacterial cell membrane rupturing, increased gentamicin accumulation, and the production of reactive oxygen species (ROS) (Ma et al., 2017).
CCNPs, or microscopic calcium carbonate particles, possess a consistent structure consisting of crystal grains measuring around 62.5 nanometers in diameter. Research has demonstrated that these CCNPs can function as carriers for the antibiotic gentamicin, effectively prolonging its release duration by up to 24 h. In addition, CCNPs significantly enhance the bactericidal efficacy of gentamicin (Pan et al., 2018). According to Pan et al. (2018), examining zeta potential and microscopic observations have shown that when CCNPs are absorbed into bacterial surfaces, they cause more harm to the bacterial cell wall and make the membrane more permeable. This ultimately results in more bacterial death.
Fecal microbiota transplantation (FMT) has emerged as a promising intervention for addressing a range of gastrointestinal disorders, with potential applications in reducing the impact of antimicrobial resistance (AMR) (Lou et al., 2023; Singh et al., 2022). AMR, where bacteria evolve to resist the effects of antibiotics, poses a significant threat to global health by rendering many existing antibiotics ineffective. The rise of resistant infections is exacerbated by the overuse and misuse of antibiotics in human and veterinary medicine. FMT, a procedure that involves transplanting fecal bacteria from a healthy donor into the gastrointestinal tract of a recipient, has gained attention as a potential tool in the fight against AMR (Wang et al., 2021). The rationale behind FMT’s possible role in reducing AMR lies in its ability to restore a healthy, diverse microbiome, which can be crucial in preventing infections and reducing the need for antibiotics. Antibiotic resistance worsens outcomes in cirrhosis, and fecal microbiota transplant (FMT) may reduce antibiotic resistance gene (ARG) burden. The study bt Bajaj et al. (2021) analyzed ARG abundance in cirrhotic patients undergoing capsule or enema FMT across two trials. Capsule FMT reduced beta-lactamase and rifamycin ARGs, while enema FMT initially increased ARGs post-antibiotics but decreased them by day 15. Overall, FMT lowered ARG abundance compared to baseline and non-FMT groups, showing its potential in decompensated cirrhosis. In another study Rashidi et al. (2024) conducted a randomized placebo-controlled trial analyzed 226 stool samples from 100 patients undergoing intensive cancer therapy to assess the short- and long-term effects of fecal microbiota transplantation (FMT) on antibiotic resistance genes (ARGs). Initially, low-level transfer of ARGs from donor microbiota occurred, followed by long-term resistance to new ARGs as stable microbial communities formed. This suggests FMT may aid in reducing multidrug-resistant organism colonization in high-risk patients. Further research is needed to determine its clinical implications, particularly in preventing infections during intensive therapy. Woodworth et al. (2023) reported that Faecal microbiota transplantation (FMT) is a promising strategy for decolonising multidrug-resistant organisms (MDROs) like vancomycin-resistant enterococci (VRE) and carbapenemase-producing Enterobacteriaceae (CPE). This study assessed the genetic response of MDROs to FMT in 29 patients, showing a significant decrease in resistance gene expression, particularly VanA and blaNDM. Both culture-dependent and independent methods confirmed gene downregulation over time. FMT was well tolerated, with no adverse events, highlighting its potential for reducing MDRO carriage in infected patients.
FMT works by reintroducing a balanced microbiome into a recipient’s gut, often after it has been disrupted by factors such as antibiotic use, disease, or other environmental factors. The gut microbiome is a complex ecosystem of bacteria, viruses, fungi, and other microorganisms that play a vital role in digestion, immune function, and protection against pathogenic microbes (Bajaj et al., 2021). When this microbiome is disrupted, often by antibiotics, the balance of microorganisms in the gut is disturbed, allowing pathogenic bacteria, including antibiotic-resistant strains, to proliferate. By restoring this balance, FMT can help re-establish microbial communities that outcompete harmful pathogens, including antibiotic-resistant ones, and may reduce the need for further antibiotic treatment (Minkoff et al., 2023). One of the most well-established uses of FMT is in treating Clostridium difficile infection (CDI), a common and serious bacterial infection often triggered by antibiotic use (Gupta et al., 2022; Nooij et al., 2021). Antibiotic treatments for CDI can disrupt the gut microbiome, leading to recurrent infections. FMT is highly effective in treating recurrent CDI, with up to 90% success rates. This success is largely attributed to the ability of FMT to restore a healthy microbiome that can prevent the overgrowth of C. difficile (Gupta et al., 2022). This microbiome restoration helps control CDI and reduces reliance on antibiotics, which is crucial in the fight against AMR.
FMT also promises to reduce the colonization and transmission of multidrug-resistant organisms (MDROs). Studies (Nooij et al., 2021; Chiu et al., 2021) have shown that restoring a healthy gut microbiota through FMT can reduce the colonization of resistant bacteria, such as extended-spectrum beta-lactamase (ESBL)-producing E. coli and vancomycin-resistant Enterococcus (VRE), in patients who are carriers of these pathogens. The process works by rebalancing the microbiota and restoring microbial diversity, which can help limit the dominance of resistant bacteria, potentially reducing their spread within healthcare settings.
Furthermore, FMT may contribute to reducing AMR by promoting the natural resilience of the microbiome to pathogenic and resistant bacteria. A healthy microbiome is a barrier to pathogenic organisms by outcompeting them for resources and producing substances that inhibit their growth. This resilience could help prevent infections from developing or becoming severe, thus reducing the need for antibiotics and limiting the opportunity for the emergence of resistance (Bajaj et al., 2021). While FMT shows significant promise in addressing AMR, its widespread use still has challenges and limitations. Standardizing FMT protocols, ensuring the safety and quality of donor samples, and understanding the long-term effects of microbiome restoration are all areas that require further research.
Additionally, FMT may not be appropriate for all patients, particularly those with compromised immune systems or severe underlying health conditions (Lou et al., 2023). Fecal microbiota transplantation represents a novel and promising approach to reducing the impact of AMR. By restoring a healthy microbiome, FMT can lessen the prevalence of antibiotic-resistant infections, lower the need for antibiotics, and potentially prevent the spread of resistant pathogens (Wang et al., 2021). As more research is conducted, FMT may become an integral part of strategies to combat AMR, providing a valuable alternative to traditional antibiotic treatments and preserving antibiotic efficacy for future generations.
In conclusion, the COVID-19 pandemic has highlighted the urgent need to tackle bacterial resistance as a global health threat. Despite a decrease in global antibiotic consumption, the overall volume remains high, underscoring the need for stricter regulations and proactive prevention strategies. Although National Action Plans (NAPs) have been implemented worldwide, significant disparities persist, specifically in developing countries where limited resources and healthcare infrastructure hinder effective action. Key reservoirs for antibiotic-resistant bacteria (ARB) and resistance genes (ARGs) include farms, hospitals, wastewater treatment plants, and agricultural settings, necessitating targeted mitigation strategies.
While promising approaches such as dietary modifications, probiotic supplementation, and combination therapies with adjuvants or phages offer potential, challenges remain in their large-scale implementation, regulatory approval, and long-term effectiveness. Dietary modifications may be ineffective if they do not specifically target pathogenic resistance mechanisms or modify the microbiome in beneficial ways. Probiotics may struggle to outcompete resistant bacteria, specifically in severe cases of antimicrobial resistance (ABR). Combination therapies could also face resistance if pathogens evolve against phages or adjuvants, or if biofilm formation or immune system interference hinders phage efficacy.
Emerging treatments like customized antibiotics, monoclonal antibodies, vaccines, and nanoparticles offer alternatives but face challenges in cost, technological complexity, and accessibility. Despite advances in next-generation sequencing and bioinformatics, there are still gaps in understanding the role of gut microbiota in resistance dynamics, indicating a need for further research. Effectively addressing antibiotic resistance requires comprehensive policies and regulatory frameworks that balance public health protection with sustainable antibiotic use. However, global coordination remains challenging due to differences in governance, economics, and healthcare priorities. Strengthening surveillance, raising public awareness, and fostering interdisciplinary collaboration will be key to overcoming these barriers. Through evidence-based strategies and global cooperation, we can work toward managing antibiotic resistance and ensuring the continued efficacy of antibiotics for future generations.
AK: Conceptualization, Formal analysis, Investigation, Resources, Supervision, Writing – original draft. AJ: Formal analysis, Methodology, Resources, Validation, Writing – review & editing. AM: Data curation, Methodology, Validation, Visualization, Writing – review & editing. AA-T: Data curation, Formal analysis, Investigation, Resources, Visualization, Writing – review & editing. MS: Formal analysis, Resources, Data curation, Writing – review & editing. TM: Data curation, Formal analysis, Investigation, Resources, Validation, Writing – original draft.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
I am obliged to Graphic Era (Deemed to be University), Dehradun, Jimma University, Ethiopia to help me conduct my research. The authors extend their appreciation to the Deanship of Scientific Research and Graduate Studies at King Khalid University for funding this work through the Large Project number R.G.P. 2/66/46 and the authors acknowledge the Center of Bee Research And its Products (CBRP) at King Khalid University, Saudi Arabia for their valuable technical support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acolatse, J. E. E., Portal, E. A. R., Boostrom, I., Akafity, G., Dakroah, M. P., Chalker, V. J., et al. (2022). Environmental surveillance of ESBL and carbapenemase-producing gram-negative bacteria in a Ghanaian tertiary hospital. Antimicrob. Resist. Infect. Control 11:49. doi: 10.1186/s13756-022-01090-2
Adesanya, O., Oduselu, T., Akin-Ajani, O., Adewumi, O. M., and Ademowo, O. G. J. A. (2020). An exegesis of bacteriophage therapy: An emerging player in the fight against anti-microbial resistance. AIMS Microbiol. 6, 204–230. doi: 10.3934/microbiol.2020014
Ahmed, S. K., Hussein, S., Qurbani, K., Ibrahim, R. H., Fareeq, A., Mahmood, K. A., et al. (2024). Antimicrobial resistance: impacts, challenges, and future prospects. J. Med. Surg. Public Health 2:100081. doi: 10.1016/j.glmedi.2024.100081
Ahuja, A., Saraswathy, M. P., Nandakumar, S., Arul Prakash, F., Gurpreet, K. N., and Dhanalekshmi, U. M. (2022). Role of the gut microbiome in diabetes and cardiovascular diseases including restoration and targeting approaches-a review. Drug Metab. Bioanal. Lett. 15, 133–149. doi: 10.2174/2949681015666220615120300
Akhtar, M., Chowdhury, M. I., Bhuiyan, T. R., Kaim, J., Ahmed, T., Rafique, T. A., et al. (2019). Evaluation of the safety and immunogenicity of the oral inactivated multivalent enterotoxigenic Escherichia coli vaccine ETVAX in Bangladeshi adults in a double-blind, randomized, placebo-controlled phase I trial using electrochemiluminescence and ELISA assays for immunogenicity analyses. Vaccine 37, 5645–5656. doi: 10.1016/j.vaccine.2018.11.040
Al Meslamani, A. Z. (2023). Antibiotic resistance in low- and middle-income countries: current practices and its global implications. Expert Rev. Anti Infect. Ther. 21, 1281–1286. doi: 10.1080/14787210.2023.2268835
Alabi, O. J., Makinde, O. J., Egena, S. S. A., Mbajiorgu, F. E., and Adewara, O. A. (2024). Antibiotics in broilers chicken production: a review of impacts, challenges and potential alternatives. Vet. Integr. Sci. 22, 559–578. doi: 10.12982/VIS.2024.039
Alame Emane, A. K., Guo, X., Takiff, H. E., and Liu, S. (2021). Drug resistance, fitness and compensatory mutations in Mycobacterium tuberculosis. Tuberculosis 129:102091. doi: 10.1016/j.tube.2021.102091
Almagro, J. C., Daniels-Wells, T. R., and Perez-Tapia, S. M. J. F. (2018). Progress and challenges in the design and clinical development of antibodies for Cancer therapy. Front. Immunol. 8:309477. doi: 10.3389/fimmu.2017.01751
Al-Taani, G. M., Al-Azzam, S., Karasneh, R. A., Ababneh, M., Al-Batayneh, O. B., Khader, Y. S., et al. (2022). Antibiotic use and resistance: information sources and application by dentists in Jordan. J. Infect. Dev. Ctries. 16, 1607–1613. doi: 10.3855/jidc.16540
Al-Tawfiq, J. A., Ebrahim, S. H., and Memish, Z. A. (2024). Preventing antimicrobial resistance together: reflections on AMR week 2023. J. Epidemiol. Global Health 14, 249–251. doi: 10.1007/s44197-023-00178-1
Anderson, M., Schulze, K., Cassini, A., Plachouras, D., and Mossialos, E. (2019). A governance framework for development and assessment of national action plans on antimicrobial resistance. Lancet Infect. Dis. 19, e371–e384. doi: 10.1016/S1473-3099(19)30415-3
Andrina, N., Lionel, C., von Ulrich, B., Ulrich, M., Andrea, T., Federica, A., et al. (2020). Characterisation of clinical manifestations and treatment strategies for invasive beta-haemolytic streptococcal infections in a Swiss tertiary hospital. Swiss Med. Wkly. 150:20378. doi: 10.4414/smw.2020.20378
Angst, D. C., Tepekule, B., Sun, L., Bogos, B., and Bonhoeffer, S. (2021). Comparing treatment strategies to reduce antibiotic resistance in an in vitro epidemiological setting. Proc. Natl. Acad. Sci. 118:e2023467118. doi: 10.1073/pnas.2023467118
Anusha, M., Tejaswini, V., Kumar, S. U., Prashantha, C. N., Vasudevan, K., and Doss, C. G. P. (2023). Gene network interaction analysis to elucidate the antimicrobial resistance mechanisms in the Clostridium difficile. Microb. Pathog. 178:106083. doi: 10.1016/j.micpath.2023.106083
Ardakani, Z., Aragrande, M., and Canali, M. J. (2024). Global antimicrobial use in livestock farming: an estimate for cattle, chickens, and pigs. Animal 18:101060. doi: 10.1016/j.animal.2023.101060
Asadi, A., Razavi, S., Talebi, M., and Gholami, M. (2019). A review on anti-adhesion therapies of bacterial diseases. Infection 47, 13–23. doi: 10.1007/s15010-018-1222-5
Ashiru-Oredope, D., Cunningham, N., Casale, E., Muller-Pebody, B., Hope, R., Brown, C. S., et al. (2023). Reporting England's progress towards the ambitions in the UK action plan for antimicrobial resistance: the English surveillance programme for antimicrobial utilisation and resistance (ESPAUR). J. Antimicrob. Chemother. 78, 2387–2391. doi: 10.1093/jac/dkad248
Asmare, Z., Awoke, T., Genet, C., Admas, A., Melese, A., and Mulu, W. (2024). Incidence of catheter-associated urinary tract infections by gram-negative bacilli and their ESBL and carbapenemase production in specialized hospitals of Bahir Dar, Northwest Ethiopia. Antimicrob. Resist. Infect. Control 13:10. doi: 10.1186/s13756-024-01368-7
Bai, H., He, L. Y., Wu, D. L., Gao, F. Z., Zhang, M., Zou, H. Y., et al. (2022). Spread of airborne antibiotic resistance from animal farms to the environment: dispersal pattern and exposure risk. Environ. Int. 158:106927. doi: 10.1016/j.envint.2021.106927
Bajaj, J. S., Shamsaddini, A., Fagan, A., Sterling, R. K., Gavis, E., Khoruts, A., et al. (2021). Fecal microbiota transplant in cirrhosis reduces gut microbial antibiotic resistance genes: analysis of two trials. Hepatol. Commun. 5, 258–271. doi: 10.1002/hep4.1639
Balta, I., Linton, M., Pinkerton, L., Kelly, C., Ward, P., Stef, L., et al. (2021). The effect of natural antimicrobials on the Campylobacter coli T6SS+/− during in vitro infection assays and on their ability to adhere to chicken skin and carcasses. Int. J. Food Microbiol. 338:108998. doi: 10.1016/j.ijfoodmicro.2020.108998
Bassetti, S., Tschudin-Sutter, S., Egli, A., and Osthoff, M. (2022). Optimizing antibiotic therapies to reduce the risk of bacterial resistance. Eur. J. Intern. Med. 99, 7–12. doi: 10.1016/j.ejim.2022.01.029
Batra, P., Deo, V., Mathur, P., and Gupta, A. K. (2017). Cotrimoxazole, a wonder drug in the era of multiresistance. Case Rep. Rev. Lit. 9, 210–213. doi: 10.4103/0974-2727.208261
Berríos-Caro, E., Gifford, D. R., and Galla, T. (2021). Competition delays multi-drug resistance evolution during combination therapy. J. Theor. Biol. 509:110524. doi: 10.1016/j.jtbi.2020.110524
Berry, I. (2024). Selection of antibiotics as prophylaxis for close contacts of patients with meningococcal disease in areas with ciprofloxacin resistance—United States, 2024. Morbidity and Mortality Weekly Report: MMWR.
Bertagnolio, S., Dobreva, Z., Centner, C. M., Olaru, I. D., Donà, D., Burzo, S., et al. (2024). WHO global research priorities for antimicrobial resistance in human health. The lancet. Microbe 5:100902. doi: 10.1016/S2666-5247(24)00134-4
Biswas, S. K., Sumon, M. M. H., Ahmed, S., Ruma, R. A., Parvin, A., Paul, D. K., et al. (2024). Beyond antibiotics: exploring the potential of bacteriophages and phage therapy. PHAGE 5, 186–202. doi: 10.1089/phage.2024.0005
Bolbanabad, A. M., Moradi, G., Abbasi, M., Heydari, M., Shokri, A., Moradpour, F., et al. (2023). An overview of the action plans of the Islamic Republic of Iran toward elimination of viral hepatitis B and C. J. Mazandaran Univ. Med. Sci. 33, 214–226.
Borgeaud, S., Metzger, L. C., Scrignari, T., and Blokesch, M. (2015). The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67. doi: 10.1126/science.1260064
Brawley, D. N., Sauer, D. B., Li, J., Zheng, X., Koide, A., Jedhe, G. S., et al. (2022). Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus. Nat. Chem. Biol. 18, 706–712. doi: 10.1038/s41589-022-00994-9
Browne, A. J., Chipeta, M. G., Haines-Woodhouse, G., Kumaran, E. P. A., Hamadani, B. H. K., Zaraa, S., et al. (2021). Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study. Lancet Planet. Health 5, e893–e904. doi: 10.1016/S2542-5196(21)00280-1
Bueno, I., Verdugo, C., Jimenez-Lopez, O., Alvarez, P. P., Gonzalez-Rocha, G., Lima, C. A., et al. (2020). Role of wastewater treatment plants on environmental abundance of antimicrobial resistance genes in Chilean rivers. Int. J. Hyg. Environ. Health 223, 56–64. doi: 10.1016/j.ijheh.2019.10.006
Buffie, C. G., and Pamer, E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801. doi: 10.1038/nri3535
Bull, F. C., Al-Ansari, S. S., Biddle, S., Borodulin, K., Buman, M. P., Cardon, G., et al. (2020). World Health Organization 2020 guidelines on physical activity and sedentary behaviour. Br. J. Sports Med. 54, 1451–1462. doi: 10.1136/bjsports-2020-102955
Bunduki, G. K., Masoamphambe, E., Fox, T., Musaya, J., Musicha, P., and Feasey, N. (2024). Prevalence, risk factors, and antimicrobial resistance of endemic healthcare-associated infections in Africa: a systematic review and meta-analysis. BMC Infect. Dis. 24:158. doi: 10.1186/s12879-024-09038-0
Calayag, A. M. B., Widmer, K. W., and Rivera, W. L. (2021). Antimicrobial susceptibility and frequency of Bla and qnr genes in Salmonella enterica isolated from slaughtered pigs. Antibiotics 10:442. doi: 10.3390/antibiotics10121442
Cantorna, M. T., Snyder, L., and Arora, J. (2019). Vitamin a and vitamin D regulate the microbial complexity, barrier function, and the mucosal immune responses to ensure intestinal homeostasis. Crit. Rev. Biochem. Mol. Biol. 54, 184–192. doi: 10.1080/10409238.2019.1611734
Carfora, V., Giacinti, G., Sagrafoli, D., Marri, N., Giangolini, G., Alba, P., et al. (2016). Methicillin-resistant and methicillin-susceptible Staphylococcus aureus in dairy sheep and in-contact humans: An intra-farm study. J. Dairy Sci. 99, 4251–4258. doi: 10.3168/jds.2016-10912
Carolus, H., Sofras, D., Boccarella, G., Jacobs, S., Biriukov, V., Goossens, L., et al. (2024). Collateral sensitivity counteracts the evolution of antifungal drug resistance in Candida auris. Nat. Microbiol. 9, 2954–2969. doi: 10.1038/s41564-024-01811-w
Catteau, L., Latour, K., Pearcy, M., and Catry, B. (2024). Antimicrobial use in Belgian acute care hospitals: results of the 2022 ECDC point prevalence survey. Antimicrob. Stewardship Healthc. Epidemiol. 4, s150–s151. doi: 10.1017/ash.2024.328
Champion, A. E., Catanzaro, K. C. F., Bandara, A. B., and Inzana, T. J. J. S. R. (2019). Formation of the Francisella tularensis biofilm is affected by cell surface glycosylation, growth medium, and a glucan exopolysaccharide. Sci. Rep. 9:12252. doi: 10.1038/s41598-019-48697-x
Charani, E., Mendelson, M., Pallett, S. J. C., Ahmad, R., Mpundu, M., Mbamalu, O., et al. (2023). An analysis of existing national action plans for antimicrobial resistance—gaps and opportunities in strategies optimising antibiotic use in human populations. Lancet Glob. Health 11, e466–e474. doi: 10.1016/S2214-109X(23)00019-0
Che, Y., Xia, Y., Liu, L., Li, A. D., Yang, Y., and Zhang, T. (2019). Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing. Microbiome 7:44. doi: 10.1186/s40168-019-0663-0
Checcucci, A., Trevisi, P., Luise, D., Modesto, M., Blasioli, S., Braschi, I., et al. (2020). Exploring the animal waste Resistome: the spread of antimicrobial resistance genes through the use of livestock manure. Front. Microbiol. 11:1416. doi: 10.3389/fmicb.2020.01416
Chen, M. L., An, X. L., Yang, K., and Zhu, Y. G. (2021). Soil phage and their mediation on the horizontal transfer of antibiotic resistance genes: a review. Chin. J. Appl. Ecol. 32, 2267–2274. doi: 10.13287/j.1001-9332.202106.031
Chen, M., Huang, X., Cheng, J., Tang, Z., and Huang, G. J. C. (2023). Urbanization and vulnerable employment: empirical evidence from 163 countries in 1991–2019. Cities 135:104208. doi: 10.1016/j.cities.2023.104208
Chen, L., Tang, Z. Y., Cui, S. Y., Ma, Z. B., Deng, H., and Kong, W. L., Zeng, Z. L. (2020). Biofilm production ability, virulence and antimicrobial resistance genes in Staphylococcus aureus from various veterinary hospitals. Pathogens 9:264. doi: 10.3390/pathogens9040264
Cheng, Z. K., Yang, J. S., Lü, M., and Luo, X. S. (2022). Antibiotics used in livestock and poultry breeding and its environmental and health risks in China: a review. J. Agric. Resourc. Environ. 39, 1253–1262. doi: 10.13254/j.jare.2021.0567
Cherian, T., Ragavendran, C., Vijayan, S., Kurien, S., and Peijnenburg, W. J. G. M. (2023). A review on the fate, human health and environmental impacts, as well as regulation of antibiotics used in aquaculture. Environ. Adv. 13:100411. doi: 10.1016/j.envadv.2023.100411
Chinemerem Nwobodo, D., Ugwu, M. C., Oliseloke Anie, C., Al-Ouqaili, M. T., Chinedu Ikem, J., Victor Chigozie, U., et al. (2022). Antibiotic resistance: the challenges and some emerging strategies for tackling a global menace. J. Clin. Lab. Anal. 36:e24655. doi: 10.1002/jcla.24655
Chiu, C. W., Tsai, P. J., Lee, C. C., Ko, W. C., and Hung, Y. P. (2021). Application of microbiome management in therapy for Clostridioides difficile infections: from fecal microbiota transplantation to probiotics to microbiota-preserving antimicrobial agents. Pathogens 10:649. doi: 10.3390/pathogens10060649
Chua, A. Q., Verma, M., Hsu, L. Y., and Legido-Quigley, H. (2021). An analysis of national action plans on antimicrobial resistance in Southeast Asia using a governance framework approach. Lancet Regnl. Health Western Pac. 7:100084. doi: 10.1016/j.lanwpc.2020.100084
Cobar, O., and Cobar, S. (2024). Omicron variants world prevalence, WHO COVID-19 dashboard, ECDC communicable disease threat report, and CDC COVID data tracker review. Available at: https://www.researchgate.net/publication/381638924_Omicron_Variants_World_Prevalence_168_WHO_COVID-19_Dashboard_ECDC_Communicable_Disease_Threat_Report_and_CDC_COVID_Data_Tracker_Review
Colin, R., Ni, B., Laganenka, L., and Sourjik, V. (2021). Multiple functions of flagellar motility and chemotaxis in bacterial physiology. FEMS Microbiol. Rev. 45:fuab038. doi: 10.1093/femsre/fuab038
Comeau, A. M., Tétart, F., Trojet, S. N., Prère, M. F., and Krisch, H. M. (2007). Phage-antibiotic synergy (PAS): β-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS One 2:e799. doi: 10.1371/journal.pone.0000799
Costa, B., Martínez-De-tejada, G., Gomes, P. A. C., Martins, M. C. L., and Costa, F. (2021). Antimicrobial peptides in the battle against orthopedic implant-related infections: a review. Pharmaceutics 13:918. doi: 10.3390/pharmaceutics13111918
Crespo-Piazuelo, D., and Lawlor, P. G. (2021). Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir. Vet. J. 74:21. doi: 10.1186/s13620-021-00200-7
Cuevas, C., Batura, N., Wulandari, L. P. L., Khan, M., and Wiseman, V. (2021). Improving antibiotic use through behaviour change: a systematic review of interventions evaluated in low- and middle-income countries. Health Policy Plan. 36, 754–773. doi: 10.1093/heapol/czab021
Cui, Y., Song, K., Liu, X., Xu, H., Wang, X., Cheng, G., et al. (2024). Research on bacterial diversity and antibiotic resistance in the dairy farm environment in a part of Shandong Province. Animals 14:160. doi: 10.3390/ani14010160
Cui, L., Veeranarayanan, S., Thitiananpakorn, K., and Wannigama, D. L. J. P. (2023). Bacteriophage bioengineering: A transformative approach for targeted drug discovery and beyond. Pathogens 12:1179. doi: 10.3390/pathogens12091179
Cusumano, J. A., Daffinee, K. E., Piehl, E. C., García-Solache, M., Desbonnet, C., Rice, L. B., et al. (2022). Meropenem plus ceftaroline is active against Enterococcus faecalis in an in vitro pharmacodynamic model using humanized dosing simulations. Antimicrob. Agents Chemother. 66, e00426–e00422. doi: 10.1128/aac.00426-22
Das, D., Bordoloi, A., Achary, M. P., Caldwell, D. J., and Suri, R. P. S. (2022). Degradation and inactivation of chromosomal and plasmid encoded resistance genes/ARBs and the impact of different matrices on UV and UV/H2O2 based advanced oxidation process. Sci. Total Environ. 833:155205. doi: 10.1016/j.scitotenv.2022.155205
de Lagarde, M., Fairbrother, J. M., Archambault, M., Dufour, S., Francoz, D., Massé, J., et al. (2022). Impact of a regulation restricting critical antimicrobial usage on prevalence of antimicrobial resistance in Escherichia coli isolates from fecal and manure pit samples on dairy farms in Québec, Canada. Front. Vet. Sci. 9:838498. doi: 10.3389/fvets.2022.838498
de Llano, D. G., Moreno-Arribas, M. V., and Bartolomé, B. (2020). Cranberry polyphenols and prevention against urinary tract infections: relevant considerations. Molecules 25:523. doi: 10.3390/molecules25153523
Dedrick, R. M., Smith, B. E., Cristinziano, M., Freeman, K. G., Jacobs-Sera, D., Belessis, Y., et al. (2023). Phage therapy of MycobacteriumInfections: compassionate use of phages in 20 patients with drug-resistant mycobacterial disease. Clin. Infect. Dis. 76, 103–112. doi: 10.1093/cid/ciac453
Dierick, M., Weken, H. V. D., Rybarczyk, J., Vanrompay, D., Devriendt, B., and Cox, E. (2020). Porcine and bovine forms of Lactoferrin inhibit growth of porcine Enterotoxigenic Escherichia coli and degrade its virulence factors. Appl. Environ. Microbiol. 86, 1–15. doi: 10.1128/AEM.0524-20
Ding, D., Wang, B., Zhang, X., Zhang, J., Zhang, H., Liu, X., et al. (2023). The spread of antibiotic resistance to humans and potential protection strategies. Ecotoxicol. Environ. Saf. 254:114734. doi: 10.1016/j.ecoenv.2023.114734
Ding, D., Zhu, J., Gao, Y., Yang, F., Ma, Y., Cheng, X., et al. (2022). Effect of cattle farm exposure on oropharyngeal and gut microbial communities and antibiotic resistance genes in workers. Sci. Total Environ. 806:150685. doi: 10.1016/j.scitotenv.2021.150685
Douafer, H., Andrieu, V., Phanstiel, O., and Brunel, J. M. (2019). Antibiotic adjuvants: make antibiotics great again! J. Med. Chem. 62, 8665–8681. doi: 10.1021/acs.jmedchem.8b01781
Duan, H., Yu, L., Tian, F., Zhai, Q., Fan, L., and Chen, W. (2022). Antibiotic-induced gut dysbiosis and barrier disruption and the potential protective strategies. Crit. Rev. Food Sci. Nutr. 62, 1427–1452. doi: 10.1080/10408398.2020.1843396
Ducarmon, Q. R., Zwittink, R. D., Hornung, B. V. H., Van Schaik, W., Young, V. B., and Kuijper, E. J. (2019). Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol. Mol. Biol. Rev. 83, e00007–e00019. doi: 10.1128/MMBR.00007-19
Dunne, M., Prokhorov, N. S., Loessner, M. J., and Leiman, P. G. (2021). Reprogramming bacteriophage host range: design principles and strategies for engineering receptor binding proteins. Curr. Opin. Biotechnol. 68, 272–281. doi: 10.1016/j.copbio.2021.02.006
Eskenazi, A., Lood, C., Wubbolts, J., Hites, M., Balarjishvili, N., Leshkasheli, L., et al. (2022). Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Commun. 13:302. doi: 10.1038/s41467-021-27656-z
European Centre for Disease Prevention and Control (ECDC) (2024). Antimicrobial consumption and resistance in bacteria from humans and food-producing animals: fourth joint inter-agency report on integrated analysis of antimicrobial agent consumption and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals in the EU/EEA JIACRA IV–2019− 2021. EFSA J. 22:e8589. doi: 10.2903/j.efsa.2024.p220201
Fan, H., Wu, S., Dong, W., Li, X., Dong, Y., Wang, S., et al. (2021). Characterization of tetracycline-resistant microbiome in soil-plant systems by combination of H218O-based DNA-stable isotope probing and metagenomics. J. Hazard. Mater. 420:126440. doi: 10.1016/j.jhazmat.2021.126440
Fang, H., Tian, L., Ye, N., and Zhang, S. (2023). Alizarin enhancement of the abundance of ARGs and impacts on the microbial community in water. Water Sci. Technol. 87, 2250–2264. doi: 10.2166/wst.2023.138
Farhat, N., and Khan, A. U. (2022). “β-Lactamase inhibitor combinations targeting antibiotic resistance in gram-negative Bacteria” in Beta-lactam resistance in gram-negative Bacteria: Threats and challenges. eds. M. Shahid, A. Singh, and H. Sami (Cham: Springer), 269–286.
Fish, D. N., Choi, M. K., and Jung, R. (2002). Synergic activity of cephalosporins plus fluoroquinolones against Pseudomonas aeruginosa with resistance to one or both drugs. J. Antimicrob. Chemother. 50, 1045–1049. doi: 10.1093/jac/dkf211
Gehring, R., Mochel, J. P., and Schmerold, I. J. F. (2023). Understanding the background and clinical significance of the WHO, WOAH, and EMA classifications of antimicrobials to mitigate antimicrobial resistance. Front. Vet. Sci. 10:1153048. doi: 10.3389/fvets.2023.1153048
Georgakakos, C. B., Hicks, B. J., and Walter, M. T. (2021). Farmer perceptions of dairy farm antibiotic use and transport pathways as determinants of contaminant loads to the environment. J. Environ. Manag. 281:111880. doi: 10.1016/j.jenvman.2020.111880
Gholipour, S., Shamsizadeh, Z., Halabowski, D., Gwenzi, W., and Nikaeen, M. (2024). Combating antibiotic resistance using wastewater surveillance: significance, applications, challenges, and future directions. Sci. Total Environ. 908:168056. doi: 10.1016/j.scitotenv.2023.168056
Gitaka, J., Kamita, M., Mureithi, D., Ndegwa, D., Masika, M., Omuse, G., et al. (2020). Combating antibiotic resistance using guidelines and enhanced stewardship in Kenya: a protocol for an implementation science approach. BMJ Open 10:e030823. doi: 10.1136/bmjopen-2019-030823
Gostin, L. O., Hodge, J. G., Bloom, B. R., El-Mohandes, A., Fielding, J., Hotez, P., et al. (2020). The public health crisis of underimmunisation: a global plan of action. Lancet Infect. Dis. 20, e11–e16. doi: 10.1016/S1473-3099(19)30558-4
Gowd, V., Karim, N., Shishir, M. R. I., Xie, L., and Chen, W. (2019). Dietary polyphenols to combat the metabolic diseases via altering gut microbiota. Trends Food Sci. Technol. 93, 81–93. doi: 10.1016/j.tifs.2019.09.005
Gulumbe, B. H., Sahal, M. R., Abdulrahim, A., Faggo, A. A., Yusuf, Z. M., Sambo, K. H., et al. (2023). Antibiotic resistance and the COVID-19 pandemic: a dual crisis with complex challenges in LMICs. Health. Sci. Rep. 6:1566. doi: 10.1002/hsr2.1566
Guo, T., Li, Z., Shao, Y., Fu, Y., Zhang, W., Shao, Y., et al. (2022). Effects of Oxytetracycline/Lead pollution alone and in the combined form on antibiotic resistance genes, Mobile genetic elements, and microbial communities in the soil. Int. J. Environ. Res. Public Health 19:619. doi: 10.3390/ijerph192315619
Gupta, K., Tappiti, M., Nazir, A. M., Koganti, B., Memon, M. S., Zahid, M. B. A., et al. (2022). Fecal microbiota transplant in recurrent clostridium difficile infections: a systematic review. Cureus 14:24754. doi: 10.7759/cureus.24754
Gurumallu, S. C., and Javaraiah, R. (2021). Bioactives in disease prevention and health promotion: exploiting combinatorial effects. Curr. Bioact. Comp. 17, 299–317. doi: 10.2174/1573407216999200612103526
Hachani, A., Wood, T. E., and Filloux, A. (2016). Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29, 81–93. doi: 10.1016/j.mib.2015.11.006
Hamadamin, H. Z., Shallal, A. F., and Qader, I. N. (2024). Synergistic role of extended-spectrum beta-lactamases (ESBL) and bacterial structure on antibacterial drugs. Jabirian J. Biointerface Res. Pharm. Appl. Chem. 1, 26–36. doi: 10.55559/jjbrpac.v1i3.293
Harun, M. G. D., Sumon, S. A., Hasan, I., Akther, F. M., Islam, M. S., and Anwar, M. M. U. (2024). Barriers, facilitators, perceptions and impact of interventions in implementing antimicrobial stewardship programs in hospitals of low-middle and middle countries: a scoping review. Antimicrob. Resist. Infect. Control 13:8. doi: 10.1186/s13756-024-01369-6
Hasan, M., and Ahn, J. (2022). Evolutionary dynamics between phages and bacteria as a possible approach for designing effective phage therapies against antibiotic-resistant bacteria. Antibiotics 11:915. doi: 10.3390/antibiotics11070915
Haseeb, A., Faidah, H. S., Algethamy, M., Alghamdi, S., Alhazmi, G. A., Alshomrani, A. O., et al. (2022). Antimicrobial usage and resistance in Makkah region hospitals: a regional point prevalence survey of public hospitals. Int. J. Environ. Res. Public Health 19:254. doi: 10.3390/ijerph19010254
Hawkins, O., Scott, A. M., Montgomery, A., Nicholas, B., Mullan, J., van Oijen, A., et al. (2022). Comparing public attitudes, knowledge, beliefs and behaviours towards antibiotics and antimicrobial resistance in Australia, United Kingdom, and Sweden (2010-2021): a systematic review, meta-analysis, and comparative policy analysis. PLoS One 17:e0261917. doi: 10.1371/journal.pone.0261917
He, P., Wu, Y., Huang, W., Wu, X., Lv, J., Liu, P., et al. (2020). Characteristics of and variation in airborne ARGs among urban hospitals and adjacent urban and suburban communities: a metagenomic approach. Environ. Int. 139:105625. doi: 10.1016/j.envint.2020.105625
Herraiz-Carboné, M., Cotillas, S., Lacasa, E., Cañizares, P., Rodrigo, M. A., and Sáez, C. (2022). Depletion of ARGs in antibiotic-resistance Klebsiella, Pseudomonas and Staphylococcus in hospital urines by electro and photo-electro disinfection. J. Water Process Eng. 49:103035. doi: 10.1016/j.jwpe.2022.103035
Hetman, B. M., Pearl, D. L., Barker, D. O. R., Robertson, J., Nash, J. H. E., Reid-Smith, R., et al. (2022). Combining analytical epidemiology and genomic surveillance to identify risk factors associated with the spread of antimicrobial resistance in Salmonella enterica subsp. enterica serovar Heidelberg. Microbial. Genomics 8:891. doi: 10.1099/mgen.0.000891
Hong, Y., Li, Q., Gao, Q., Xie, J., Huang, H., Drlica, K., et al. (2020). Reactive oxygen species play a dominant role in all pathways of rapid quinolone-mediated killing. J. Antimicrob. Chemother. 75, 576–585. doi: 10.1093/jac/dkz485
Hong, P. Y., Wang, C., and Mantilla-Calderon, D. (2020). Mitigating antimicrobial resistance risks when using reclaimed municipal wastewater for agriculture. Handb. Environ. Chem. 91, 245–265. doi: 10.1007/698_2020_473
Hounmanou, Y. M. G., Bortolaia, V., Dang, S. T. T., Truong, D., Olsen, J. E., and Dalsgaard, A. (2021). ESBL and AmpC β-lactamase encoding genes in E. coli from pig and pig farm Workers in Vietnam and Their Association with Mobile Genetic Elements. Front. Microbiol. 12:629139. doi: 10.3389/fmicb.2021.629139
Hsieh, P. F., Lu, Y. R., Lin, T. L., Lai, L. Y., and Wang, J. T. (2019). Klebsiella pneumoniae type VI secretion system contributes to bacterial competition, cell invasion, Type-1 fimbriae expression, and in vivo colonization. J. Infect. Dis. 219, 637–647. doi: 10.1093/infdis/jiy534
Hu, X., Xu, Y., Liu, S., Gudda, F. O., Ling, W., Qin, C., et al. (2023). Graphene quantum dots nonmonotonically influence the horizontal transfer of extracellular antibiotic resistance genes via bacterial transformation. Small 19:e2301177. doi: 10.1002/smll.202301177
Hu, Y., Zhang, X., Deng, S., Yue, C., Jia, X., and Lyu, Y. (2023). Non-antibiotic prevention and treatment against Acinetobacter baumannii infection: are vaccines and adjuvants effective strategies? Front. Microbiol. 14:1049917. doi: 10.3389/fmicb.2023.1049917
Huang, S., Zhong, Y., Fu, Y., Zheng, X., Feng, Z., and Mo, A. (2022). Graphene and its derivatives: “one stone, three birds” strategy for orthopedic implant-associated infections. Biomater. Sci. 11, 380–399. doi: 10.1039/d2bm01507b
Hussein, A. F. A., and Ali, H. E. (2021). Estimating awareness of health care workers and degree of implementation of antimicrobial resistance policies; a cross-sectional study from Egypt. Egypt. J. Med. Microbiol. 30, 15–19. doi: 10.51429/EJMM30203
Hussein, R. R. S., Rabie, A. S. I., Shaman, M. B., Shaaban, A. H., Fahmy, A. M., Sofy, M. R., et al. (2022). Antibiotic consumption in hospitals during COVID-19 pandemic: a comparative study. J. Infect. Dev. Ctries. 16, 1679–1686. doi: 10.3855/jidc.17148
Iera, J., Isonne, C., Seghieri, C., Tavoschi, L., Ceparano, M., Sciurti, A., et al. (2025). Availability and key characteristics of National Early Warning Systems for emerging profiles of antimicrobial resistance in high-income countries: systematic review. JMIR Public Health Surveill. 11:e57457. doi: 10.2196/57457
Imran, M., Dasgupta, A., and Chopra, S. (2021). Monoclonal antibody targeting S. aureus α-toxin prevention of S. aureus nosocomial pneumonia. Drugs Future 46, 885–889. doi: 10.1358/dof.2021.46.11.3294555
Kamere, N., Garwe, S. T., Akinwotu, O. O., Tuck, C., Krockow, E. M., Yadav, S., et al. (2022). Scoping review of National Antimicrobial Stewardship Activities in eight African countries and adaptable recommendations. Antibiotics 11:149. doi: 10.3390/antibiotics11091149
Kasimanickam, V., Kasimanickam, M., and Kasimanickam, R. (2021). Antibiotics use in food animal production: escalation of antimicrobial resistance: where are we now in combating AMR? Med. Sci. 9:14. doi: 10.3390/medsci9010014
Katva, S., Das, S., Moti, H. S., Jyoti, A., and Kaushik, S. J. P. (2017). Antibacterial synergy of silver nanoparticles with gentamicin and chloramphenicol against Enterococcus faecalis. Pharmacogn. Mag. 13:S828. doi: 10.4103/pm.pm_120_17
Kawabata, K., Sugiyama, Y., Sakano, T., and Ohigashi, H. (2013). Flavonols enhanced production of anti-inflammatory substance(s) by bifidobacterium adolescentis: prebiotic actions of galangin, quercetin, and fisetin. Biofactors 39, 422–429. doi: 10.1002/biof.1081
Kayali, O., and Icgen, B. (2020). Untreated HWWs emerged as hotpots for ARGs. Bull. Environ. Contam. Toxicol. 104, 386–392. doi: 10.1007/s00128-020-02792-2
Kelly, S. A., Nzakizwanayo, J., Rodgers, A. M., Zhao, L., Weiser, R., Tekko, I. A., et al. (2021). Antibiotic therapy and the gut microbiome: investigating the effect of delivery route on gut pathogens. ACS Infect. Dis. 7, 1283–1296. doi: 10.1021/acsinfecdis.1c00081
Keogan, D. M., and Griffith, D. M. J. M. (2014). Current and potential applications of bismuth-based drugs. Molecules 19, 15258–15297. doi: 10.3390/molecules190915258
Kering, K. K., Kibii, B. J., and Wei, H. (2019). Biocontrol of phytobacteria with bacteriophage cocktails. Pest Manag. Sci. 75, 1775–1781. doi: 10.1002/ps.5324
Khan, H., Liu, M., Kayani, M. U. R., Ahmad, S., Liang, J., and Bai, X. (2021). DNA phosphorothioate modification facilitates the dissemination of mcr-1 and blaNDM-1 in drinking water supply systems. Environ. Pollut. 268:115799. doi: 10.1016/j.envpol.2020.115799
Khazaei, Z., Ghorbani, P., Namaei, M. H., Rezaei, Y., and Yousefi, M. (2020). Prevalence of Escherichia coli K1 rectovaginal colonization among pregnant women in Iran: virulence factors and antibiotic resistance properties. Microb. Drug Resist. 26, 1201–1207. doi: 10.1089/mdr.2020.0006
Khouja, T., Mitsantisuk, K., Tadrous, M., and Suda, K. J. (2022). Global consumption of antimicrobials: impact of the WHO global action plan on antimicrobial resistance and 2019 coronavirus pandemic (COVID-19). J. Antimicrob. Chemother. 77, 1491–1499. doi: 10.1093/jac/dkac028
Klassert, T. E., Leistner, R., Zubiria-Barrera, C., Stock, M., López, M., Neubert, R., et al. (2021). Bacterial colonization dynamics and antibiotic resistance gene dissemination in the hospital environment after first patient occupancy: a longitudinal metagenetic study. Microbiome 9:169. doi: 10.1186/s40168-021-01109-7
Klassert, T. E., Zubiria-Barrera, C., Kankel, S., Stock, M., Neubert, R., Lorenzo-Diaz, F., et al. (2020). Early bacterial colonization and antibiotic resistance Gene Acquisition in newborns. Front. Cell. Infect. Microbiol. 10:332. doi: 10.3389/fcimb.2020.00332
Korach-Rechtman, H., Hreish, M., Fried, C., Gerassy-Vainberg, S., Azzam, Z. S., Kashi, Y., et al. (2020). Intestinal dysbiosis in carriers of carbapenem-resistant enterobacteriaceae. mSphere 5, e00173–e00120. doi: 10.1128/MSPHERE.00173-20
Kreuder, A. J., Ruddell, B., Mou, K., Hassall, A., Zhang, Q., and Plummera, P. J. (2020). Small noncoding RNA cjnc110 influences motility, autoagglutination, ai-2 localization, hydrogen peroxide sensitivity, and chicken colonization in campylobacter jejuni. Infect. Immun. 88, e00245–e00220. doi: 10.1128/IAI.00245-20
Kumar Singh, A., Cabral, C., Kumar, R., Ganguly, R., Kumar Rana, H., Gupta, A., et al. (2019). Beneficial effects of dietary polyphenols on gut microbiota and strategies to improve delivery efficiency. Nutrients 11:2216. doi: 10.3390/nu11092216
Kumar, V., Yasmeen, N., Chaudhary, A. A., Alawam, A. S., Al-Zharani, M., Suliman Basher, N., et al. (2023). Specialized pro-resolving lipid mediators regulate inflammatory macrophages: a paradigm shift from antibiotics to immunotherapy for mitigating COVID-19 pandemic. Front. Mol. Biosci. 10:1104577. doi: 10.3389/fmolb.2023.1104577
Lagadinou, M., Amerali, M., Michailides, C., Chondroleou, A., Skintzi, K., Spiliopoulou, A., et al. (2024). Antibiotic resistance trends in Carbapenem-resistant gram-negative pathogens and eight-year surveillance of XDR bloodstream infections in a Western Greece tertiary hospital. Pathogens 13:1136. doi: 10.3390/pathogens13121136
Lal, A., Singh, S., Franco, F., and Bhatia, S. (2021). Potential of polyphenols in curbing quorum sensing and biofilm formation in gram-negative pathogens. Asian Pac. J. Trop. Biomed. 11, 231–243. doi: 10.4103/2221-1691.314044
Le Guern, R., Stabler, S., Gosset, P., Pichavant, M., Grandjean, T., Faure, E., et al. (2021). Colonization resistance against multi-drug-resistant bacteria: a narrative review. J. Hosp. Infect. 118, 48–58. doi: 10.1016/j.jhin.2021.09.001
Leach, S., Lundgren, A., Carlin, N., Löfstrand, M., and Svennerholm, A. M. (2017). Cross-reactivity and avidity of antibody responses induced in humans by the oral inactivated multivalent enterotoxigenic Escherichia coli (ETEC) vaccine ETVAX. Vaccine 35, 3966–3973. doi: 10.1016/j.vaccine.2017.06.006
Lee, K., Kim, D. W., Lee, D. H., Kim, Y. S., Bu, J. H., Cha, J. H., et al. (2020). Mobile resistome of human gut and pathogen drives anthropogenic bloom of antibiotic resistance. Microbiome 8:2. doi: 10.1186/s40168-019-0774-7
Lee, H., and Ko, G. J. S. (2016). Antiviral effect of vitamin a on norovirus infection via modulation of the gut microbiome. Sci. Rep. 6:25835. doi: 10.1038/srep25835
Li, Z., Li, T., Tang, J., Huang, L., Ding, Y., Zeng, Z., et al. (2023). Antibacterial activity of Surfactin and synergistic effect with conventional antibiotics against methicillin-resistant Staphylococcus aureus isolated from patients with diabetic foot ulcers. Diabetes Metab. Syndr. Obesity 16, 3727–3737. doi: 10.2147/DMSO.S435062
Li, C., Zhu, L., Wang, D., Wei, Z., Hao, X., Wang, Z., et al. (2022). T6SS secretes an LPS-binding effector to recruit OMVs for exploitative competition and horizontal gene transfer. ISME J. 16, 500–510. doi: 10.1038/s41396-021-01093-8
Ligthart, K., Belzer, C., de Vos, W. M., and Tytgat, H. L. P. (2020). Bridging Bacteria and the gut: functional aspects of type IV pili. Trends Microbiol. 28, 340–348. doi: 10.1016/j.tim.2020.02.003
Liu, L., Li, J., Xin, Y., Huang, X., and Liu, C. (2021). Evaluation of wetland substrates for veterinary antibiotics pollution control in lab-scale systems. Environ. Pollut. 269:116152. doi: 10.1016/j.envpol.2020.116152
Liu, B. T., and Song, F. J. (2019). Emergence of two Escherichia coli strains co-harboring mcr-1 and blaNDM in fresh vegetables from China. Infect. Drug Resist. 12, 2627–2635. doi: 10.2147/IDR.S211746
Liu, Y., Tong, Z., Shi, J., Li, R., Upton, M., and Wang, Z. (2021). Drug repurposing for next-generation combination therapies against multidrug-resistant bacteria. Theranostics 11, 4910–4928. doi: 10.7150/thno.56205
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Liu, S., Wang, M., Zheng, L., and Guan, W. (2018). Antimicrobial resistance profiles of nosocomial pathogens in regional China: a brief report from two tertiary hospitals in China. Med. Sci. Monit. 24, 8602–8607. doi: 10.12659/MSM.911229
Liu, D., Zeng, L., Yan, Z., Jia, J., Gao, J., and Wei, Y. (2020). The mechanisms and safety of probiotics against toxigenic clostridium difficile. Expert Rev. Anti-Infect. Ther. 18, 967–975. doi: 10.1080/14787210.2020.1778464
Lou, X., Xue, J., Shao, R., Yang, Y., Ning, D., Mo, C., et al. (2023). Fecal microbiota transplantation and short-chain fatty acids reduce sepsis mortality by remodeling antibiotic-induced gut microbiota disturbances. Front. Immunol. 13:1063543. doi: 10.3389/fimmu.2022.1063543
Ma, L., Wu, J., Wang, S., Yang, H., Liang, D., Lu, Z. J. J., et al. (2017). Synergistic antibacterial effect of Bi2S3 nanospheres combined with ineffective antibiotic gentamicin against methicillin-resistant Staphylococcus aureus. J. Inorg. Biochem. 168, 38–45. doi: 10.1016/j.jinorgbio.2016.12.005
Maier, L., Goemans, C. V., Wirbel, J., Kuhn, M., Eberl, C., Pruteanu, M., et al. (2021). Unravelling the collateral damage of antibiotics on gut bacteria. Nature 599, 120–124. doi: 10.1038/s41586-021-03986-2
Maisto, M., Iannuzzo, F., Novellino, E., Schiano, E., Piccolo, V., and Tenore, G. C. (2023). Natural polyphenols for prevention and treatment of urinary tract infections. Int. J. Mol. Sci. 24:277. doi: 10.3390/ijms24043277
Man, A. W. C., Zhou, Y., Xia, N., and Li, H. (2020). Involvement of gut microbiota, microbial metabolites and interaction with polyphenol in host immunometabolism. Nutrients 12, 1–29. doi: 10.3390/nu12103054
Mazur, P., Skiba-Kurek, I., Mrowiec, P., Karczewska, E., and Drożdż, R. J. I. (2020). Synergistic ROS-associated antimicrobial activity of silver nanoparticles and gentamicin against Staphylococcus epidermidis. Int. J. Nanomed. 15, 3551–3562. doi: 10.2147/IJN.S246484
Mendelson, M., Lewnard, J. A., Sharland, M., Cook, A., Pouwels, K. B., Alimi, Y., et al. (2024). Ensuring progress on sustainable access to effective antibiotics at the 2024 UN general assembly: a target-based approach. Lancet 403, 2551–2564. doi: 10.1016/S0140-6736(24)01019-5
Mercy, K., Salyer, S. J., Mankga, C., Hedberg, C., Zondo, P., and Kebede, Y. (2024). Establishing an early warning event management system at Africa CDC. PLoS Digital Health 3:e0000546. doi: 10.1371/journal.pdig.0000546
Minkoff, N. Z., Aslam, S., Medina, M., Tanner-Smith, E. E., Zackular, J. P., Acra, S., et al. (2023). Fecal microbiota transplantation for the treatment of recurrent Clostridioides difficile (Clostridium difficile). Cochrane Database Syst. Rev. 4:CD013871. doi: 10.1002/14651858.CD013871.pub2
Mitić, D., Bolt, E. L., and Ivančić-Baće, I. (2023). CRISPR-Cas adaptation in Escherichia coli. Biosci. Rep. 43:BSR20221198. doi: 10.1042/BSR20221198
Moghadam, M. T., Khoshbayan, A., Chegini, Z., Farahani, I., and Shariati, A. (2020). Bacteriophages, a new therapeutic solution for inhibiting multidrug-resistant bacteria causing wound infection: lesson from animal models and clinical trials. Drug Des. Devel. Ther. 14, 1867–1883. doi: 10.2147/DDDT.S251171
Mohammed, A. N., and Abdel Aziz, S. A. A. (2019). Novel approach for controlling resistant Listeria monocytogenes to antimicrobials using different disinfectants types loaded on silver nanoparticles (AgNPs). Environ. Sci. Pollut. Res. 26, 1954–1961. doi: 10.1007/s11356-018-3773-5
Mugerwa, I., Nabadda, S. N., Midega, J., Guma, C., Kalyesubula, S., and Muwonge, A. (2021). Antimicrobial resistance situational analysis 2019–2020: design and performance for human health surveillance in Uganda. Trop. Med. Infect. Dis. 6:178. doi: 10.3390/tropicalmed6040178
Murray, M., Salvatierra, G., Dávila-Barclay, A., Ayzanoa, B., Castillo-Vilcahuaman, C., Huang, M., et al. (2021). Market chickens as a source of antibiotic-resistant Escherichia coli in a Peri-Urban Community in Lima, Peru. Front. Microbiol. 12:635871. doi: 10.3389/fmicb.2021.635871
Nagy, E., Nagy, G., Power, C. A., and Badarau, A. (2017). Anti-bacterial monoclonal antibodies. Adv. Exp. Med. Biol. 1053, 119–153. doi: 10.1007/978-3-319-72077-7_7
Nooij, S., Ducarmon, Q. R., Laros, J. F., Zwittink, R. D., Norman, J. M., Smits, W. K., et al. (2021). Fecal microbiota transplantation influences procarcinogenic Escherichia coli in recipient recurrent Clostridioides difficile patients. Gastroenterology 161, 1218–1228.e5. doi: 10.1053/j.gastro.2021.06.009
O’Leary, E. N., Neuhauser, M. M., McLees, A., Paek, M., Tappe, J., and Srinivasan, A. (2024). An update from the National Healthcare Safety Network on hospital antibiotic stewardship programs in the United States, 2014–2021. Open Forum Infect. Dis. 11:ofad684. doi: 10.1093/ofid/ofad684
O'Brien, L. M., Walsh, E. J., Massey, R. C., Peacock, S. J., and Foster, T. J. (2002). Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell. Microbiol. 4, 759–770. doi: 10.1046/j.1462-5822.2002.00231.x
Okoliegbe, I. N., Hijazi, K., Cooper, K., Ironside, C., and Gould, I. M. (2021). Trends of antimicrobial resistance and combination susceptibility testing of multidrug-resistant Pseudomonas aeruginosa isolates from cystic fibrosis patients: a 10-year update. Antimicrob. Agents Chemother. 65, 10–1128. doi: 10.1128/AAC.02483-20
Okpala, C. O. R., Anyanwu, M. U., Łukanko, S., Nwobi, O. C., Korzeniowska, M., and Ezeonu, I. M. (2021). Animal-food-human antimicrobial resistance fundamentals, prevention mechanisms and global surveillance trends: a terse review. Appl. Food Biotechnol. 8, 89–102. doi: 10.22037/afb.v8i2.32206
Ortiz, Y., García-Heredia, A., Merino-Mascorro, A., García, S., Solís-Soto, L., and Heredia, N. (2021). Natural and synthetic antimicrobials reduce adherence of enteroaggregative and enterohemorrhagic Escherichia coli to epithelial cells. PLoS One 16:e0251096. doi: 10.1371/journal.pone.0251096
Osman, E. A., El-Amin, N. I., Al-Hassan, L. L., and Mukhtar, M. (2021). Multiclonal spread of Klebsiella pneumoniae across hospitals in Khartoum, Sudan. J. Glob. Antimicrob. Resist. 24, 241–245. doi: 10.1016/j.jgar.2020.12.004
Otieku, E., Kurtzhals, J. A. L., Fenny, A. P., Ofori, A. O., Labi, A. K., and Enemark, U. (2024). Healthcare provider cost of antimicrobial resistance in two teaching hospitals in Ghana. Health Policy Plan. 39, 178–187. doi: 10.1093/heapol/czad114
Özçelik, E. A., Doucet, C., Kang, H., Levy, N., Feldhaus, I., Hashiguchi, T. C. O., et al. (2022). A comparative assessment of action plans on antimicrobial resistance from OECD and G20 countries using natural language processing. Health Policy 126, 522–533. doi: 10.1016/j.healthpol.2022.03.011
Pal, N., Sharma, P., Kumawat, M., Singh, S., Verma, V., Tiwari, R. R., et al. (2024). Phage therapy: An alternative treatment modality for MDR bacterial infections. Infect. Dis. 56, 785–817. doi: 10.1080/23744235.2024.2379492
Pan, X., Chen, S., Li, D., Rao, W., Zheng, Y., Yang, Z., et al. (2018). The synergistic antibacterial mechanism of gentamicin-loaded CaCO 3 nanoparticles. Front. Chem. 5:130. doi: 10.3389/fchem.2017.00130
Pattnaik, S., Mishra, M., and Naik, P. K. (2023). Alternative strategies for combating antibiotic resistance in microorganisms. In S. Busi and R. Prasad Antimicrobial Photodynamic Therapy: Concepts and Applications (pp. 65–109). London: Routledge
Pham, G. N., Dang, T. T. H., Nguyen, T. A., Zawahir, S., Le, H. T. T., Negin, J., et al. (2024). Health system barriers to the implementation of the national action plan to combat antimicrobial resistance in Vietnam: a scoping review. Antimicrob. Resist. Infect. Control 13:12. doi: 10.1186/s13756-024-01364-x
Pires, D. P., Cleto, S., Sillankorva, S., Azeredo, J., Lu, T. K. J. M., and Reviews, M. B. (2016). Genetically engineered phages: a review of advances over the last decade. Microbiol. Mol. Biol. Rev. 80, 523–543. doi: 10.1128/MMBR.00069-15
Qadri, F., Akhtar, M., Bhuiyan, T. R., Chowdhury, M. I., Ahmed, T., Rafique, T. A., et al. (2020). Safety and immunogenicity of the oral, inactivated, enterotoxigenic Escherichia coli vaccine ETVAX in Bangladeshi children and infants: a double-blind, randomised, placebo-controlled phase 1/2 trial. Lancet Infect. Dis. 20, 208–219. doi: 10.1016/S1473-3099(19)30571-7
Quaik, S., Embrandiri, A., Ravindran, B., Hossain, K., Al-Dhabi, N. A., Arasu, M. V., et al. (2020). Veterinary antibiotics in animal manure and manure laden soil: scenario and challenges in Asian countries. J. King Saud Univ. Sci. 32, 1300–1305. doi: 10.1016/j.jksus.2019.11.015
Rashidi, A., Ebadi, M., Rehman, T. U., Elhusseini, H., Kazadi, D., Halaweish, H., et al. (2024). Long-and short-term effects of fecal microbiota transplantation on antibiotic resistance genes: results from a randomized placebo-controlled trial. Gut Microbes 16:2327442. doi: 10.1080/19490976.2024.2327442
Rothrock, M. J., Min, B. R., Castleberry, L., Waldrip, H., Parker, D., Brauer, D., et al. (2021). Antibiotic resistance, antimicrobial residues, and bacterial community diversity in pasture-raised poultry, swine, and beef cattle manures. J. Anim. Sci. 99:skab144. doi: 10.1093/jas/skab144
Sachan, R. S. K., Bala, R., Al-Tawaha, A. R. M., Khanum, S., and Karnwal, A. (2023). “Antimicrobial drugs obtained from marine algae” in Current trends in the identification and development of antimicrobial agents. eds. Amin-ul Mannan, M., and Kumar, G. (Sharjah, U.A.E: Bentham Science Publishers), 213–247.
Sachdeva, A., Tomar, T., Malik, T., Bains, A., and Karnwal, A. (2025). Exploring probiotics as a sustainable alternative to antimicrobial growth promoters: mechanisms and benefits in animal health. Front. Sustain. Food Syst. 8:1523678. doi: 10.3389/fsufs.2024.1523678
Saeed, U., Insaf, R. A., and Piracha, Z. Z. J. F. (2023). Crisis averted: a world united against the menace of multiple drug-resistant superbugs-pioneering anti-AMR vaccines RNA interference, nanomedicine, CRISPR-based antimicrobials, bacteriophage therapies, and clinical artificial intelligence strategies to safeguard global antimicrobial arsenal. Front. Microbiol. 14:1270018. doi: 10.3389/fmicb.2023.1270018
Saha, D., Visconti, M. C., Desipio, M. M., and Thorpe, R. (2020). Inactivation of antibiotic resistance gene by ternary nanocomposites of carbon nitride, reduced graphene oxide and iron oxide under visible light. Chem. Eng. J. 382:122857. doi: 10.1016/j.cej.2019.122857
Salam, M. A., Al-Amin, M. Y., Salam, M. T., Pawar, J. S., Akhter, N., Rabaan, A. A., et al. (2023). Antimicrobial resistance: a growing serious threat for global public health. Healthcare 11:1946. doi: 10.3390/healthcare11131946
Saleem, Z., Godman, B., Azhar, F., Kalungia, A. C., Fadare, J., and Opanga, S., Hassali, M. A. (2022). Progress on the national action plan of Pakistan on antimicrobial resistance (AMR): a narrative review and the implications. Expert Rev. Anti-Infect. Ther. 20:71–93. doi: 10.1080/14787210.2021.1935238
Salyers, A. A., Gupta, A., and Wang, Y. (2004). Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12, 412–416. doi: 10.1016/j.tim.2004.07.004
Samarasinghe, S., Reid, R., and Al-Bayati, M. (2019). The anti-virulence effect of cranberry active compound proanthocyanins (PACs) on expression of genes in the third-generation cephalosporin-resistant Escherichia coli CTX-M-15 associated with urinary tract infection. Antimicrob. Resist. Infect. Control 8:181. doi: 10.1186/s13756-019-0637-9
Scandorieiro, S., Rodrigues, B. C. D., Nishio, E. K., Panagio, L. A., de Oliveira, A. G., Durán, N., et al. (2022). Biogenic silver nanoparticles strategically combined with Origanum vulgare derivatives: antibacterial mechanism of action and effect on multidrug-resistant strains. Front. Microbiol. 13:842600. doi: 10.3389/fmicb.2022.842600
Seekatz, A. M., Safdar, N., and Khanna, S. (2022). The role of the gut microbiome in colonization resistance and recurrent Clostridioides difficile infection. Ther. Adv. Gastroenterol. 15:396. doi: 10.1177/17562848221134396
Seixas, A. M., Sousa, S. A., and Leitão, J. H. J. V. (2022). Antibody-based immunotherapies as a tool for tackling multidrug-resistant bacterial infections. Vaccines 10:1789. doi: 10.3390/vaccines10111789
Sheng, L., Li, X., and Wang, L. (2022). Photodynamic inactivation in food systems: a review of its application, mechanisms, and future perspective. Trends Food Sci. Technol. 124, 167–181. doi: 10.1016/j.tifs.2022.04.001
Sima, F., Stratakos, A. C., Ward, P., Linton, M., Kelly, C., Pinkerton, L., et al. (2018). A novel natural antimicrobial can reduce the in vitro and in vivo pathogenicity of T6SS positive campylobacter jejuni and campylobacter coli chicken isolates. Front. Microbiol. 9:2139. doi: 10.3389/fmicb.2018.02139
Šimunović, K., Ramić, D., Xu, C., and Možina, S. S. (2020). Modulation of Campylobacter jejuni motility, adhesion to polystyrene surfaces, and invasion of int407 cells by quorum-sensing inhibition. Microorganisms 8:104. doi: 10.3390/microorganisms8010104
Singh, P., Alm, E. J., Kelley, J. M., Cheng, V., Smith, M., Kassam, Z., et al. (2022). Effect of antibiotic pretreatment on bacterial engraftment after fecal microbiota transplant (FMT) in IBS-D. Gut Microbes 14:2020067. doi: 10.1080/19490976.2021.2020067
Song, S., Han, M., Wang, X., Wang, S., Qin, W., Zhang, Y., et al. (2023). Fate of antibiotic resistance genes in cultivation substrate and its association with bacterial communities throughout commercial production of Agaricus bisporus. Ecotoxicol. Environ. Saf. 249:114360. doi: 10.1016/j.ecoenv.2022.114360
Song, M., Liu, Y., Huang, X., Ding, S., Wang, Y., Shen, J., et al. (2020). A broad-spectrum antibiotic adjuvant reverses multidrug-resistant gram-negative pathogens. Nat. Microbiol. 5, 1040–1050. doi: 10.1038/s41564-020-0723-z
Song, Q., Wang, B., Han, Y., and Zhou, Z. (2022). Metagenomics reveals the diversity and taxonomy of carbohydrate-active enzymes and antibiotic resistance genes in Suancai bacterial communities. Genes 13:773. doi: 10.3390/genes13050773
Sorbara, M. T., Dubin, K., Littmann, E. R., Moody, T. U., Fontana, E., Seok, R., et al. (2019). Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J. Exp. Med. 216, 84–98. doi: 10.1084/jem.20181639
Speziale, P., and Pietrocola, G. J. F. (2020). The multivalent role of fibronectin-binding proteins a and B (FnBPA and FnBPB) of Staphylococcus aureus in host infections. Front. Microbiol. 11:572022. doi: 10.3389/fmicb.2020.02054
Stielow, M., Witczyńska, A., Kubryń, N., Fijałkowski, Ł., Nowaczyk, J., and Nowaczyk, A. (2023). The bioavailability of drugs—the current state of knowledge. Molecules 28:8038. doi: 10.3390/molecules28248038
Storz, C., Schutz, C., Tluway, A., Matuja, W., Schmutzhard, E., and Winkler, A. S. (2016). Clinical findings and management of patients with meningitis with an emphasis on Haemophilus influenzae meningitis in rural Tanzania 366, 52–58. doi: 10.1016/j.jns.2016.04.044
Styczynski, A., Herzig, C., Luvsansharav, U. O., McDonald, L. C., and Smith, R. M. (2023). Using colonization to understand the burden of antimicrobial resistance across low- and middle-income countries. Clin. Infect. Dis. 77, S70–S74. doi: 10.1093/cid/ciad224
Sulavik, M. C., Houseweart, C., Cramer, C., Jiwani, N., Murgolo, N., Greene, J., et al. (2001). Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45, 1126–1136. doi: 10.1128/AAC.45.4.1126-1136.2001
Sun, L., Liu, S., Wang, J., and Wang, L. (2019). Analysis of risk factors for multiantibiotic-resistant infections among surgical patients at a children's hospital. Microb. Drug Resist. 25, 297–303. doi: 10.1089/mdr.2018.0279
Surmeneva, M., Lapanje, A., Chudinova, E., Ivanova, A., Koptyug, A., Loza, K., et al. (2019). Decreased bacterial colonization of additively manufactured Ti6Al4V metallic scaffolds with immobilized silver and calcium phosphate nanoparticles. Appl. Surf. Sci. 480, 822–829. doi: 10.1016/j.apsusc.2019.03.003
Tabib-Salazar, A., Liu, B., Shadrin, A., Burchell, L., Wang, Z., Wang, Z., et al. (2017). Full shut-off of Escherichia coli RNA-polymerase by T7 phage requires a small phage-encoded DNA-binding protein. Nucleic Acids Res. 45, 7697–7707. doi: 10.1093/nar/gkx370
Tan, R., Jin, M., Shao, Y., Yin, J., Li, H., Chen, T., et al. (2022). High-sugar, high-fat, and high-protein diets promote antibiotic resistance gene spreading in the mouse intestinal microbiota. Gut Microbes 14:442. doi: 10.1080/19490976.2021.2022442
Tanno, L. K., and Demoly, P. (2020). Action plan to ensure global availability of adrenaline autoinjectors. J. Invest. Allergol. Clin. Immunol. 30, 77–85. doi: 10.18176/jiaci.0346
Tewawong, N., Kowaboot, S., Pimainog, Y., Watanagul, N., Thongmee, T., and Poovorawan, Y. (2020). Distribution of phylogenetic groups, adhesin genes, biofilm formation, and antimicrobial resistance of uropathogenic Escherichia coli isolated from hospitalized patients in Thailand. PeerJ 8:e10453. doi: 10.7717/peerj.10453
Thu, P. N. T., Huong, M. N. T., Thi, N. T., Thanh, H. N., and Minh, K. P. (2021). Combination antibiotic therapy versus monotherapy in the treatment of acute exacerbations of chronic obstructive pulmonary disease: an open-label randomized trial. BMC Infect. Dis. 21, 1–8. doi: 10.1186/s12879-021-06687-3
Tiseo, K., Huber, L., Gilbert, M., Robinson, T. P., and Van Boeckel, T. P. (2020). Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics 9:918. doi: 10.3390/antibiotics9120918
Tong, C., Xiao, D., Xie, L., Yang, J., Zhao, R., Hao, J., et al. (2022). Swine manure facilitates the spread of antibiotic resistome including tigecycline-resistant tet(X) variants to farm workers and receiving environment. Sci. Total Environ. 808:152157. doi: 10.1016/j.scitotenv.2021.152157
Truché, P., Shoman, H., Reddy, C. L., Jumbam, D. T., Ashby, J., Mazhiqi, A., et al. (2020). Globalization of national surgical, obstetric and anesthesia plans: the critical link between health policy and action in global surgery. Glob. Health 16:1. doi: 10.1186/s12992-019-0531-5
Tsutsumi, K., Yonehara, R., Ishizaka-Ikeda, E., Miyazaki, N., Maeda, S., Iwasaki, K., et al. (2019). Structures of the wild-type MexAB–OprM tripartite pump reveal its complex formation and drug efflux mechanism. Nat. Commun. 10:1520. doi: 10.1038/s41467-019-09463-9
Uddin, T. M., Chakraborty, A. J., Khusro, A., Zidan, B. R. M., Mitra, S., Emran, T. B., et al. (2021). Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 14, 1750–1766. doi: 10.1016/j.jiph.2021.10.020
Ullah, S., Ahmad, A., Subhan, F., Jan, A., Raza, M., Khan, A. U., et al. (2018). Tobramycin mediated silver nanospheres/graphene oxide composite for synergistic therapy of bacterial infection. J. Photochem. Photobiol. B 183, 342–348. doi: 10.1016/j.jphotobiol.2018.05.009
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. 112, 5649–5654. doi: 10.1073/pnas.1503141112
Van Boeckel, T. P., Pires, J., Silvester, R., Zhao, C., Song, J., Criscuolo, N. G., et al. (2019). Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 365:aaw1944. doi: 10.1126/science.aaw1944
Vandamme, E. J., and Mortelmans, K. (2019). A century of bacteriophage research and applications: impacts on biotechnology, health, ecology and the economy! J. Chem. Technol. Biotechnol. 94, 323–342. doi: 10.1002/jctb.5810
Vekemans, J., Hasso-Agopsowicz, M., Kang, G., Hausdorff, W. P., Fiore, A., Tayler, E., et al. (2021). Leveraging vaccines to reduce antibiotic use and prevent antimicrobial resistance: a World Health Organization action framework. Clin. Infect. Dis. 73, e1011–e1017. doi: 10.1093/cid/ciab062
Villarreal, C. L., Price, M. A., Moreno, A. N., Zenteno, A., Saenz, C., Toppo, A., et al. (2023). Regulatory challenges in conducting human subjects research in emergency settings: the National Trauma Research Action Plan (NTRAP) scoping review. Trauma Surg. Acute Care Open 8:e001044. doi: 10.1136/tsaco-2022-001044
Von Mentzer, A., Zalem, D., Chrienova, Z., and Teneberg, S. (2020). Colonization factor CS30 from enterotoxigenic Escherichia coli binds to sulfatide in human and porcine small intestine. Virulence 11, 381–390. doi: 10.1080/21505594.2020.1749497
Wang, M., Cao, J., Gong, C., Amakye, W. K., Yao, M., and Ren, J. (2021). Exploring the microbiota-Alzheimer’s disease linkage using short-term antibiotic treatment followed by fecal microbiota transplantation. Brain Behav. Immun. 96, 227–238. doi: 10.1016/j.bbi.2021.06.003
Wang, N., Luo, J., Deng, F., Huang, Y., and Zhou, H. (2022). Antibiotic combination therapy: a strategy to overcome bacterial resistance to aminoglycoside antibiotics. Front. Pharmacol. 13:839808. doi: 10.3389/fphar.2022.839808
Wang, M., Yao, M., and Zhu, Y. G. (2022). Antibiotic resistance genes and antibiotic sensitivity in bacterial aerosols and their comparisons with known respiratory pathogens. J. Aerosol Sci. 161:105931. doi: 10.1016/j.jaerosci.2021.105931
Wang-Lin, S. X., and Balthasar, J. P. J. A. (2018). Pharmacokinetic and Pharmacodynamic considerations for the use of monoclonal antibodies in the treatment of bacterial infections. Antibodies 7:5. doi: 10.3390/antib7010005
Watkins, R. R., and Bonomo, R. A. (2020). Overview: the ongoing threat of antimicrobial resistance. Infect. Dis. Clin. N. Am. 34, 649–658. doi: 10.1016/j.idc.2020.04.002
Weterings, V., Zhou, K., Rossen, J. W., van Stenis, D., Thewessen, E., Kluytmans, J., et al. (2015). An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1647–1655. doi: 10.1007/s10096-015-2401-2
Willemsen, A., Reid, S., and Assefa, Y. (2022). A review of national action plans on antimicrobial resistance: strengths and weaknesses. Antimicrob. Resist. Infect. Control 11:90. doi: 10.1186/s13756-022-01130-x
Willmann, M., Vehreschild, M. J. G. T., Biehl, L. M., Vogel, W., Dörfel, D., Hamprecht, A., et al. (2019). Distinct impact of antibiotics on the gut microbiome and resistome: a longitudinal multicenter cohort study. BMC Biol. 17:76. doi: 10.1186/s12915-019-0692-y
Woodworth, M. H., Conrad, R. E., Haldopoulos, M., Pouch, S. M., Babiker, A., Mehta, A. K., et al. (2023). Fecal microbiota transplantation promotes reduction of antimicrobial resistance by strain replacement. Sci. Transl. Med. 15:eabo2750. doi: 10.1126/scitranslmed.abo2750
World Health Organization. (2024). Global research agenda for antimicrobial resistance in human health. Available at: https://www.who.int/publications/m/item/global-research-agenda-for-antimicrobial-resistance-in-human-health (Accessed May 20, 2024).
Wu, Y., Battalapalli, D., Hakeem, M. J., Selamneni, V., Zhang, P., Draz, M. S., et al. (2021). Engineered CRISPR-Cas systems for the detection and control of antibiotic-resistant infections. J. Nanobiotechnol. 19:401. doi: 10.1186/s12951-021-01132-8
Yacoby, I., Bar, H., and Benhar, I. (2007). Targeted drug-carrying bacteriophages as antibacterial nanomedicines. Antimicrob. Agents Chemother. 51, 2156–2163. doi: 10.1128/AAC.00163-07
Yang, Q., Liang, Q., Balakrishnan, B., Belobrajdic, D. P., Feng, Q.-J., and Zhang, W. J. N. (2020). Role of dietary nutrients in the modulation of gut microbiota: a narrative review. Nutrients 12:381. doi: 10.3390/nu12020381
Zalewska-Piątek, B., and Piątek, R. (2020). Phage therapy as a novel strategy in the treatment of urinary tract infections caused by E Coli. Antibiotics 9, 1–20. doi: 10.3390/antibiotics9060304
Zapata-Cortés, O. L. (2020). Reflection on the development plans in Colombia. Bitacora Urbano Territorial 30, 233–246. doi: 10.15446/BITACORA.V30N3.86811
Zeedan, G. S. G., Abdalhamed, A. M., and Ghazy, A. A. (2023). Strategies for prevention and control of multidrug-resistant Bacteria in ruminants. World's Vet. J. 13, 45–56. doi: 10.54203/scil.2023.wvj5
Zhang, L., Huang, Y., Zhou, Y., Buckley, T., and Wang, H. H. (2013). Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob. Agents Chemother. 57, 3659–3666. doi: 10.1128/AAC.00670-13
Zhang, K., Li, K., Xin, R., Han, Y., Guo, Z., Zou, W., et al. (2022). Antibiotic resistomes in water supply reservoirs sediments of Central China: main biotic drivers and distribution pattern. Environ. Sci. Pollut. Res. 29, 37712–37721. doi: 10.1007/s11356-021-18095-w
Zhang, H., Li, L., Xing, Q., Chen, J., Fan, Y., Dan, L., et al. (2023). nZnO-based graphene/graphene oxide compounds inhibit methane metabolic pathways and lower the development of antibiotic resistance genes and virulence factors. J. Clean. Prod. 428:139413. doi: 10.1016/j.jclepro.2023.139413
Zhang, R. M., Lian, X. L., Shi, L. W., Jiang, L., Chen, S. S., Haung, W. Q., et al. (2023). Dynamic human exposure to airborne bacteria-associated antibiotic resistomes revealed by longitudinal personal monitoring data. Sci. Total Environ. 904:166799. doi: 10.1016/j.scitotenv.2023.166799
Zhang, F., Wu, S., Lei, T., Wu, Q., Zhang, J., Huang, J., et al. (2022). Presence and characterization of methicillin-resistant Staphylococcus aureus co-carrying the multidrug resistance genes cfr and lsa(E) in retail food in China. Int. J. Food Microbiol. 363:109512. doi: 10.1016/j.ijfoodmicro.2021.109512
Zhang, H., Zha, X., Zhang, B., Zheng, Y., Elsabagh, M., Wang, H., et al. (2024). Gut microbiota contributes to bisphenol A-induced maternal intestinal and placental apoptosis, oxidative stress, and fetal growth restriction in pregnant ewe model by regulating gut-placental axis. Microbiome 12:28. doi: 10.1186/s40168-024-01749-5
Zhuang, X., Li, L., Liu, T., Zhang, R., Yang, P., Wang, X., et al. (2022). Mechanisms of isoniazid and rifampicin-induced liver injury and the effects of natural medicinal ingredients: a review. Front. Pharmacol. 13:1037814. doi: 10.3389/fphar.2022.1037814
Keywords: antibiotic consumption, National Action Plans (NAPs), antibiotic resistance Bacteria (ARB), antibiotic-resistance genes (ARG), combination therapies, next-generation sequencing, resistance transmission, human health impacts
Citation: Karnwal A, Jassim AY, Mohammed AA, Al-Tawaha ARMS, Selvaraj M and Malik T (2025) Addressing the global challenge of bacterial drug resistance: insights, strategies, and future directions. Front. Microbiol. 16:1517772. doi: 10.3389/fmicb.2025.1517772
Received: 27 October 2024; Accepted: 10 February 2025;
Published: 24 February 2025.
Edited by:
John Osei Sekyere, University of Pretoria, South AfricaReviewed by:
Shakil Ahmed Polash, RMIT University, AustraliaCopyright © 2025 Karnwal, Jassim, Mohammed, Al-Tawaha, Selvaraj and Malik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arun Karnwal, YXJ1bmthcm53YWxAZ21haWwuY29t; Tabarak Malik, bWFsaWtpdHJjQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.