- 1Engineering Research Center of Southwest Animal Disease Prevention and Control Technology, Ministry of Education of the People's Republic of China, Chengdu, China
- 2Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
- 3International Joint Research Center for Animal Disease Prevention and Control of Sichuan Province, Chengdu, China
- 4Institute of Veterinary Medicine and Immunology, Sichuan Agricultural University, Chengdu, China
- 5Research Center of Avian Disease, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
During viral infection, the transport of various proteins between the nucleus and cytoplasm plays an important role in the viral lifecycle. Shuttling proteins are key factors in the transmission of nucleocytoplasmic information within cells and usually contain nuclear localization signals and nuclear export signals to mediate correct positioning for themselves and other proteins. The nucleocytoplasmic transport process is carried out through the nuclear pore complex on the nuclear envelope and is mediated by specific protein carriers. The viral proteins that function through nucleocytoplasmic shuttling in herpesviruses have gradually been identified as research advances. This article provides an overview of how shuttling proteins utilize nucleocytoplasmic shuttling signals and nuclear transport receptors for nucleocytoplasmic transport, as well as discusses how herpesvirus shuttling proteins enhance the effective infection of viruses by affecting their lifecycle and participating in innate immunity, this review provides a reference for understanding the pathogenesis of herpesvirus infection and determining new antiviral strategies.
1 Introduction
Herpesviruses belong to the Herpesviridae family and are enveloped viruses. They have a large double-stranded nuclear genome. Herpesviruses can infect humans and other vertebrates, and employ a biphasic replication cycle consisting of latent and lytic phases, allowing the virus to infect the host and persist within it (Zhong et al., 1996; Davis et al., 2001; Fakhari and Dittmer, 2002; Klass et al., 2005; Fu and Pan, 2024). The herpesvirus family can be classified based on different physicochemical characteristics, such as cell tropism, pathogenicity, and latent sites, and can be divided into the α, β, and γ herpesvirus subfamilies (McGeoch et al., 1995; Boyne and Whitehouse, 2006a; Ilouze et al., 2006; Santos, 2016; Rathbun and Szpara, 2021). The α herpesvirus subfamily include herpes simplex virus type 1 and type 2 (HSV-1 and HSV-2), varicella zoster virus (VZV), which infect humans, as well as pseudorabies virus (PRV), Marek's disease virus (MDV), and bovine herpesvirus-1 (BHV-1), which infect animals. These viruses typically establish latency in sensory neurons (Biswas et al., 2013; Couteaudier and Denesvre, 2014; Depledge et al., 2018b; Suzich and Cliffe, 2018; Tognarelli et al., 2019; Zheng et al., 2022). The β herpesvirus subfamily, which include mainly human cytomegalovirus (HCMV), human herpesvirus 6 (HHV-6), and human herpesvirus 7 (HHV-7), which typically establish latency in mononuclear (Hahn et al., 1998; Agut et al., 2016; Elder and Sinclair, 2019; Hamada et al., 2023; Ijezie et al., 2023). The γ herpesvirus subfamily establishes a latent period in lymphocytes, including Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV), which infect humans; and alcelaphine herpesvirus 1 (AlHV-1) and herpesvirus saimiri (HVS), which infect animals (Russo et al., 1996; Dupin et al., 1999; Boshoff and Weiss, 2001; Bechtel et al., 2003; Thorley-Lawson et al., 2013; Myster et al., 2020; Damania et al., 2022). The herpesvirus particles are composed of a double-stranded DNA genome, capsid, tegument, and envelope from inside to outside, the herpesvirus genome contains multiple open reading frames (ORFs) that encode various viral proteins. The herpesvirus genome is covalently linked by a unique long region (UL) and a unique short region (US), with repetitive sequences at both ends of each unique region, including terminal repeat (TR) sequences, internal repeat (IR) sequences, and direct repeat (DR) sequences. Due to the variations in the location and quantity of repetitive sequences among different herpesviruses, the genome structures of the herpesviruses discussed in this review are briefly distinguished (see Figure 1; McGeoch et al., 1988, 1991; Russo et al., 1996; Rigoutsos et al., 2003). These viral proteins work separately and coordinate with each other to promote the proliferation and transmission of herpesvirus. The production of mature viral particles and the normal function of viral proteins are closely related to their correct subcellular location, viruses need to overcome multiple barriers, such as the plasma membrane and nuclear membrane, in host cells to enter the site of viral replication or assembly. Some viral proteins regulate the transcription and translation of host cells, innate immunity, and other biological functions by shuttling back and forth in the cytoplasm and nucleus and mediate viral mRNA transport, viral particle assembly, and interactions among various viral proteins to participate in the life cycle of the virus, thereby promoting the proliferation of the virus itself (Sandri-Goldin, 2001; Cullen, 2003; Lake and Hutt-Fletcher, 2004; Batisse et al., 2005; da Silva et al., 2012).
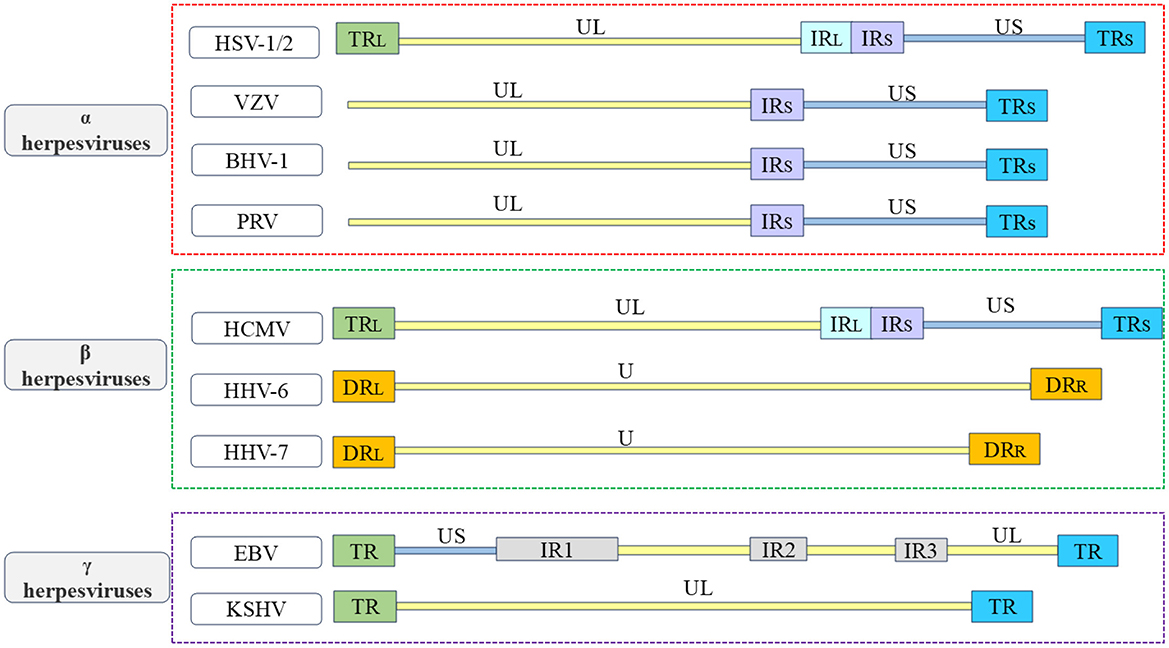
Figure 1. Overview of the structure of herpesvirus genome. In order to better understand the various types of herpesviruses and their associated viral proteins, the figure provides a brief overview of the genome structures of common α herpesviruses, β herpesviruses, and γ herpesviruses. The α herpesviruses include HSV-1, HSV-2, VZV, PRV, and BHV-1 (Davison and Scott, 1986; McGeoch et al., 1988, 1991; Klupp et al., 2005; Xu et al., 2015; Liu et al., 2022; Shitrit et al., 2023); the β herpesviruses include HCMV, HHV-6, and HHV-7 (Gompels et al., 1995; Dominguez et al., 1999; Dunn et al., 2003; Sijmons et al., 2014; Verbeek et al., 2024); the γ herpesviruses include EBV and KSHV (Russo et al., 1996; Kwok and Chiang, 2016; Majerciak et al., 2019). The repetitive sequences in the US are referred to as IRS sequences and TRS sequences. The repetitive sequences in the UL are referred to as IRL sequences and TRL sequences. The unique regions of the HHV-6 and HHV-7 genomes are collectively known as U.
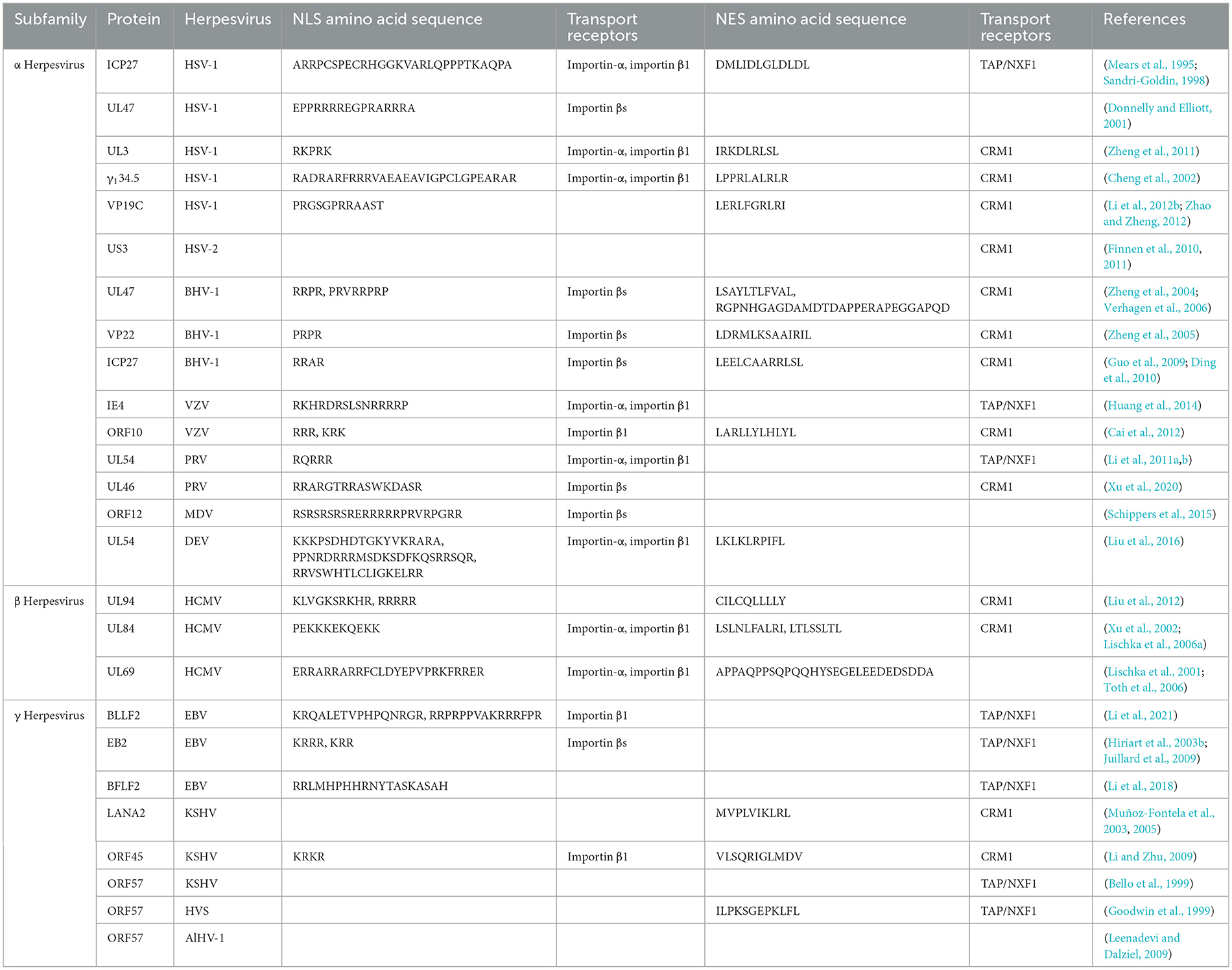
Throughout the lifecycle of herpesviruses, multiple viral proteins must function directly or indirectly at various locations. For example, the α herpesvirus HSV-1 UL47 protein and BHV-1 UL47 protein not only participate in the replication process as nuclear proteins but also play a role in primary envelopment during the maturation of viral particles (Donnelly et al., 2007; Liu et al., 2014b; Zhang et al., 2015, 2016). These viral proteins, which use specific sequences to dynamically change their localization and perform different functions, are called nucleocytoplasmic shuttling proteins, and their different localizations at different stages strongly impact their functions throughout the viral lifecycle. The specific shift sequences of shuttling proteins typically contain nuclear localization signals (NLSs) and nuclear export signals (NESs; Michael, 2000). In addition, a variety of receptors and adapter proteins are involved in the process of nucleocytoplasmic transport. Numerous herpesvirus shuttling proteins have been documented, classified according to α, β, and γ herpesviruses (Table 1). Among them, the ICP27 protein family is conserved in α, β, and γ herpesviruses, this family functions as a shuttling protein that regulates mRNA transport and influences the viral lifecycle (Patel et al., 2015; Tunnicliffe et al., 2015). A common feature of all herpesviruses is that most of the viral genes expressed in the lytic stage lack introns (Zheng, 2003). Since the herpesvirus gene is transcribed into mRNA in the nucleus of the host cell, it is necessary to export the intron-free viral mRNA to the cytoplasm for translation (Reed and Hurt, 2002; Le Hir et al., 2003). The ICP27 protein family translates viral mRNAs in the cytoplasm by hijacking the components of the host mRNA processing and export mechanism to ensure the expression and stability of herpesvirus intronless genes (Malik et al., 2004; Sandri-Goldin, 2008; Toth and Stamminger, 2008; Majerciak et al., 2014). In addition, the HSV-1 VP19C protein (Person and Desai, 1998; Adamson et al., 2006), HCMV UL94 protein, and KSHV ORF45 protein also affect the assembly of viral particles (Li and Zhu, 2009; Liu et al., 2009, 2012; Wang et al., 2015). Herpesviruses cause lifelong infection by evading the host immune system and establishing latent infection. Multiple viral proteins, including the PRV UL46 protein (Xu et al., 2020), HSV-1 γ134.5 protein (Cheng et al., 2002; Verpooten et al., 2009; Pan et al., 2018), HCMV UL94 protein, and KSHV LANA2 protein, regulate the localization of specific signal molecules to inhibit or block signal transduction pathways (Seo et al., 2004; Zou et al., 2020). This strategy is crucial for viruses to evade immune responses. Based on this, shuttling proteins are important factors in dynamically coordinating nucleocytoplasmic life activities, and herpesvirus shuttling proteins balance NLSs and NESs in space and time to optimize the time and amplitude of viral gene expression. The NLS and NES of herpesvirus shuttling proteins and their transport mechanisms must be determined to analyze the process of viral proliferation of herpesvirus. This review summarizes the transport mechanism of shuttling proteins and discusses the roles of various shuttling proteins in the various stages of the virus lifecycle in herpesvirus.
2 The nucleocytoplasmic shuttling mechanism of shuttling proteins
Herpesvirus shuttling proteins have nucleocytoplasmic shuttling mechanisms similar to that of most other shuttling proteins. Each step is accompanied by many interactions between proteins and is subject to strict and complex regulation, as will be detailed below (Mattaj and Englmeier, 1998).
2.1 Channels for nucleocytoplasmic shuttling
The cytoplasm and nucleus are separated by nuclear envelope (NE), providing a physical barrier for the diffusion of large molecules between the cytoplasm and nucleus (Dey and Baum, 2021). NE is penetrated by multiple supramolecular structures, which are called nuclear pore complex (NPC), NPCs are complex structures composed of nucleoporins (Nups) that penetrate and bridge the inner and outer nuclear membranes, where they mediate nucleocytoplasmic transport (Strambio-De-Castillia et al., 2010; Knockenhauer and Schwartz, 2016; Fontana et al., 2022).
2.2 Transport receptors of nucleocytoplasmic shuttling
The nucleocytoplasmic shuttling of proteins around NPCs is mediated by specific protein carriers, collectively known as nuclear transport receptors (NTRs), such as karyopherins, which are responsible for transporting nucleocytoplasmic cargo (Bednenko et al., 2003; Mosammaparast and Pemberton, 2004; Conti et al., 2006). The functions of karyopherins can be divided into two different categories: importins and exportins, importins are heterodimers, which are divided into importin-αs and importin βs (Goldfarb et al., 2004; Xu et al., 2010). The importin-αs protein recognizes the NLS in the cargo protein for transport (Goldfarb et al., 2004). It has three key domains: its N-terminal domain is an IBB domain that binds importin β1, its middle domain is an Armadillo (ARM) repeat sequence that binds NLS-cargo, and its C-terminal region interacts with nuclear export factor CAS and nucleoporin 50 (Nup50) to mediate the nucleocytoplasmic transport of the protein (Görlich et al., 1996; Weis et al., 1996; Kutay et al., 1997; Herold et al., 1998).
Importin βs, another family of proteins involved in the nucleocytoplasmic transport of proteins and RNA. Almost all importin βs contain two conserved domains: the central HEAT domain and the importin β N-terminal domain (IBN), HEAT is named by the initials of the four proteins that initially found this repetitive motif (Figure 2; Xu et al., 2010). Importin βs include nuclear import receptors (importins), export receptors (exportins), and bidirectional receptors. For example, the human genome encodes 20 importin βs, of which 10 (importin β1, transportin 1, transportin 2, importin 4, importin 5, importin 7, importin 8, importin 9, importin 11, and importin 12) are importins, 7 (exportin 1/CRM1, exportin 2/CAS, exportin 5, exportin 6, exportin 7, exportin t, and RanBP17) are exportins, 2 (importin 13 and exportin 4) are bidirectional receptors, and 1 (RanBP6) has not been characterized (Nehrbass and Blobel, 1996; Kimura and Imamoto, 2014).
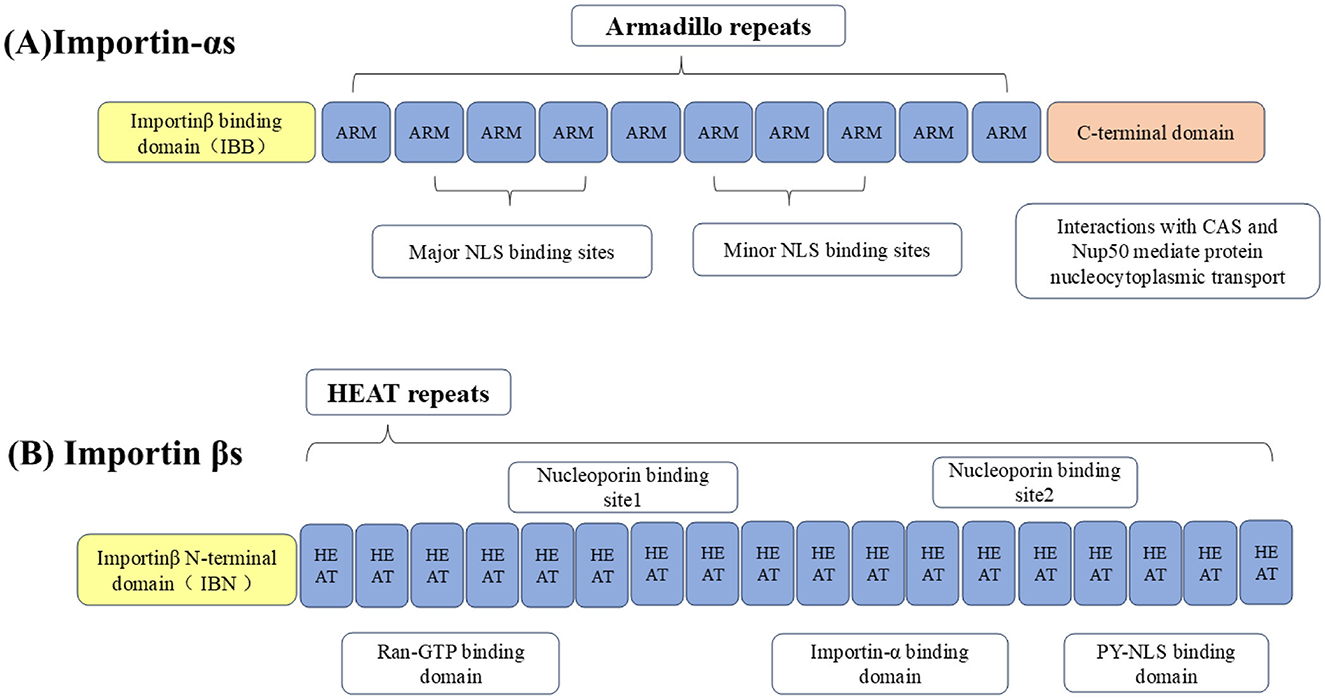
Figure 2. Importins are divided into (A) importin-αs and (B) importin βs. The N-terminus of importin-αs binds to the IBB domain of importin β1, the middle part is the Arm repeat sequence that bind to NLS-containing cargo, and the C-terminal region interacts with CAS and Nup50 to mediate nucleocytoplasmic transport of the protein. Importin βs contain two conserved domains, including the central HEAT domain and the importin β N-terminal domain (IBN).
2.3 The energy sources for nucleocytoplasmic transport
The transport process of shuttling proteins to nuclear transport receptors depends on the participation of an active energy transport mechanism, which is generally driven by the RanGTP-RanGDP gradient during nuclear envelopment. Ran is the most abundant member of the Ras-GTPase superfamily and is a key regulator of the import protein importin β, which is crucial for protein and RNA transport between the nucleus and cytoplasm (Izaurralde et al., 1997; Görlich and Kutay, 1999). To sum up, NPC performs complex biological functions through the spatial and temporal interactions of cargo proteins, Ran GTPases, and NTR.
2.4 Nucleocytoplasmic shuttling of proteins into the nucleus
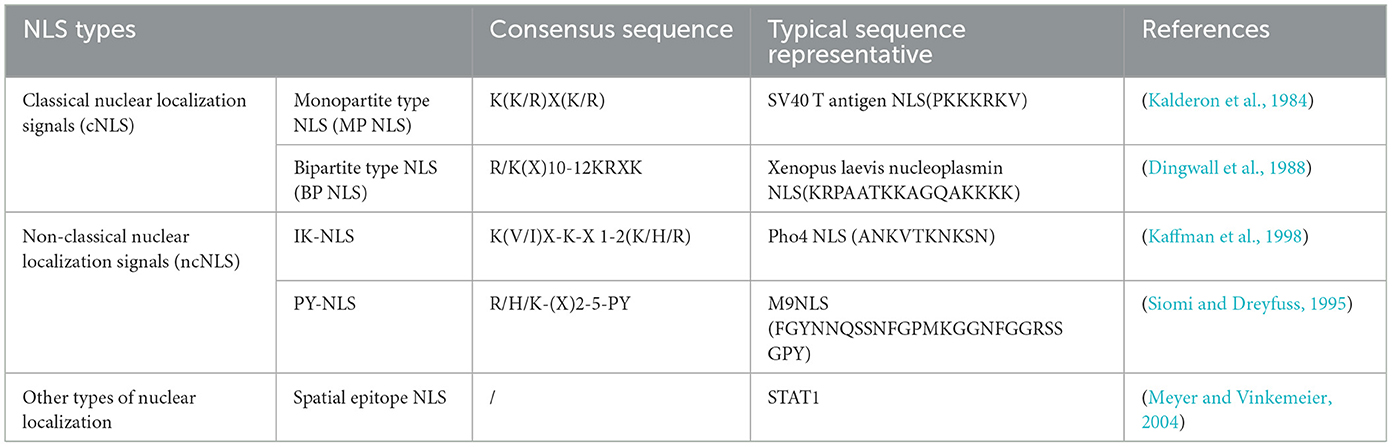
The NLS is a short peptide containing a special amino acid sequence that acts as a signal fragment that mediates the transport of shuttling proteins from the cytoplasm to the nucleus through the NPC, with “localization” and “orientation.” The NLS is divided into classical nuclear localization signals (cNLSs) and non-classical nuclear localization signals (ncNLSs; Table 2; Bradley et al., 2007). Unlike cNLSs (Robbins et al., 1991; Arregi et al., 2011). Many proteins contain NLSs with irregular characteristic structures, known as ncNLSs. There are many types of ncNLSs. These can generally be divided into NLSs rich in arginine (IK-NLSs), NLSs containing proline and tyrosine (PY-NLSs), and spatial epitope NLSs (Fagerlund et al., 2002; Meyer and Vinkemeier, 2004). The IK-NLS is recognized by importin5 (Kobayashi and Matsuura, 2013), while the PY-NLS is more complex than the cNLS and is recognized by transportin 1 (Lee et al., 2006; Wang et al., 2012; Mallet and Bachand, 2013; Soniat and Chook, 2016).
In the classical nuclear import process, cNLS-cargo in the cytoplasm first binds to importin-α and then binds to importin β1 through the IBB domain form the cNLS-Cargo-importin-α-importin β1 trimer. Importin β1 then facilitates the transport of the trimer into the nucleus. The energy consumption of the trimer passing through the NPC is provided by the hydrolysis of GTP by Ran GTPase, and the combination of Ran GTP and importin β1 results in conformational changes in importin-α and cNLS-cargo, respectively, which are released from the trimer. cNLS-cargo is transported along the internal skeleton solid phase and remains in the nucleus (Lange et al., 2007; Stewart, 2007; Fontana et al., 2022). Non-classical nuclear entry is generally mediated by importin βs binding to the ncNLS, of which transportin 1 may be second only to importin-α1. The protein containing the PY-NLS appears to be imported specifically by transportin 1 (Siomi et al., 1997; Twyffels et al., 2014; Hwang et al., 2022). In summary, the process of NLS-mediated protein entry into the nucleus is a necessary factor for the proper functioning of the protein.
2.5 Nucleocytoplasmic shuttling of proteins out of the nucleus
After the shuttling protein has completed its function in the nucleus, the process of gradual transport from the nucleus to the cytoplasm requires the NES to interact with exportins. The NES is a short, leucine-rich, or hydrophobic amino acid motif (Fischer et al., 1995; Wen et al., 1995). Among the seven types of exportins, CRM1, also known as export 1 or Xpo1, is the best characterized. The nuclear export process of shuttling proteins largely relies on CRM1 (Fornerod et al., 1997; Fukuda et al., 1997). First, the NES on the shuttling protein binds to CRM1 to form the CRM1-NES cargo RanGTP ternary export complex. The ternary complex then binds to various nuclear pore proteins and crosses the NPC to the cytoplasm. Finally, CRM1 returns to the nucleus through the NPC for the next export cycle (Kehlenbach et al., 1999; Seewald et al., 2002; Bernad et al., 2004; Hutten and Kehlenbach, 2006). In summary, the NES also plays an important role in shuttling protein transport, and the dynamic processes mediated by the NLS and NES are the basis for shuttling proteins to perform different functions (Figure 3).
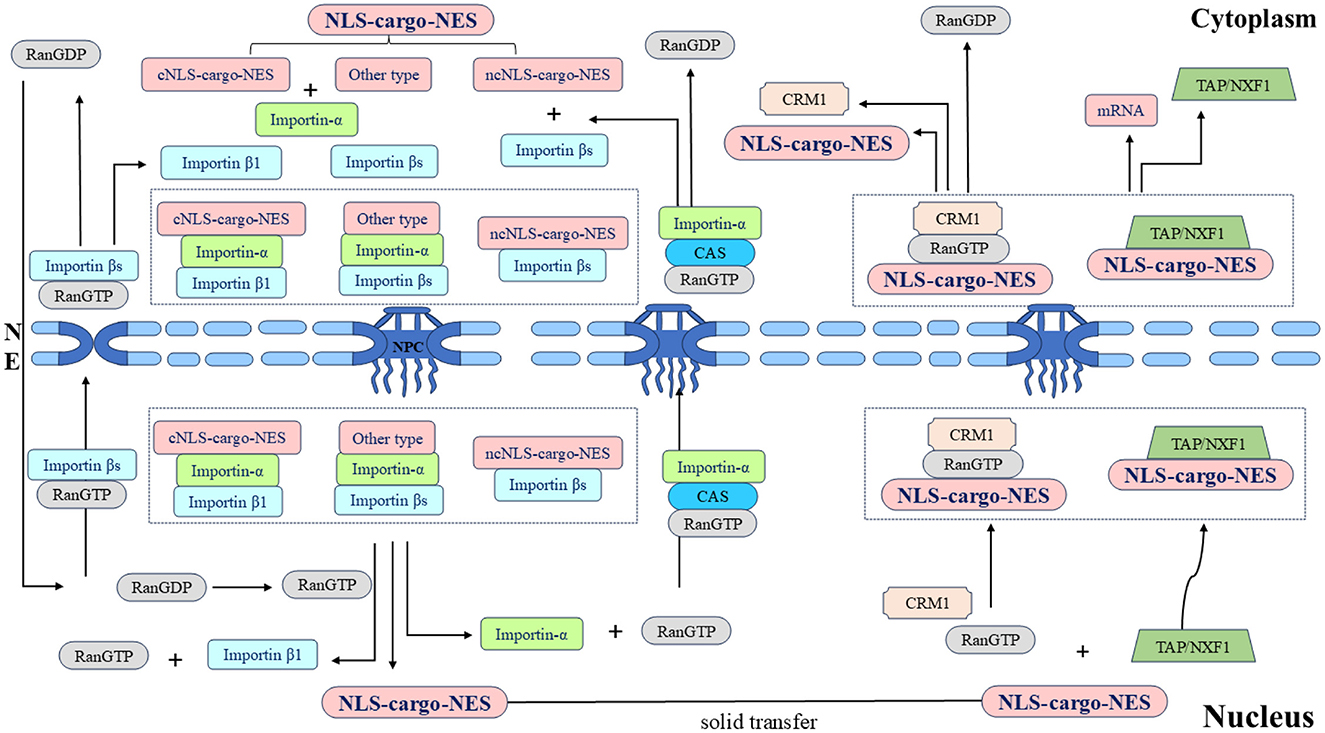
Figure 3. Schematic diagram of the nucleocytoplasmic transport of the shuttling protein NLS-cargo-NES protein complex and its associated protein molecular circulation. Different types of NLSs on shuttling proteins are recognized by different importins, and transport complexes are formed under the action of multiple factors. Importins directs the complex to the NPC and transfers it to the nucleus. Shuttling proteins undergo solid-phase transport within the nucleus, and different types of NESs on shuttling proteins are recognized by different exportins. The transport complex is formed under the coordination of various proteins, this complex is transported through the NPC to the cytoplasm, and the whole shuttling process is thus completed.
3 The lifecycle of herpesviruses
3.1 Herpesvirus latency
When the virus is in the latency, the viral DNA is stably maintained in the nucleus of the cell as multiple copies of circular episomes, with the exception being the HHV-6, which has the ability to integrate into telomeric regions of host chromosomes (Arbuckle et al., 2010). In α herpesvirus, the latency-associated transcript (LAT) is the only viral gene expressed by HSV-1 (Whitley et al., 1998; Roizman et al., 2011; Wang et al., 2023). Infected cell protein 0 (ICP0) is an immediate-early regulatory protein of HSV-1 that promotes the lytic infection and reactivation of viral genomes (Leib et al., 1989; Halford and Schaffer, 2001; Thompson and Sawtell, 2006; Roizman et al., 2011). The expression of latent viral gene in VZV is also limited to VZV latency-associated transcript (VLT) and VLT-ORF63 RNA (VLT63) fusion transcripts (Depledge et al., 2018a; Ouwendijk et al., 2020; Braspenning et al., 2021). The tegument protein VZV IE62, encoded by the VZV gene, is delivered to the newly infected cell nucleus, where it initiates VZV lytic replication by transactivating viral immediate early, early, and late genes (Kinchington et al., 1992, 2001).
In β and γ herpesviruses, In HCMV, one of the mechanisms controlling the balance between latency and reactivation or lytic replication is the IE1 and IE2 proteins encoded by the HCMV UL123 and UL122, respectively. IE1 is essential for the establishment of lytic infection and the reactivation of viral latency (Tarrant-Elorza et al., 2014; Arend et al., 2016), IE2 initiates the subsequent cascade of viral gene expression (Malone et al., 1990; Dooley and O'Connor, 2020). Epstein-Barr nuclear antigen 1 (EBNA1) was the first reported EBV latency protein. The EBNA1 protein tethers the latent viral episomes to the host chromosome. According to the expression of viral genes, the latent infection of EBV is more complex and variable than those of other herpesviruses. The latent form of EBV infection is further categorized into five patterns: Latency 0, I, IIa, IIb, and III (McKenzie and El-Guindy, 2015; Thorley-Lawson, 2015). The lytic cycle activation of EBV is controlled by two viral transcription factors Zta and Rta, which are immediate early proteins encoded by EBV genes BRLF1 and BZLF1, respectively (Murata et al., 2021; Ali et al., 2022). The latency of KSHV also expresses only a limited number of viral genes, latency-associated nuclear antigen 1 (LANA1), encoded by the KSHV gene, is the main regulator of latency (Katano, 2018). The key viral protein that regulates the transition from latency to lysis replication is the transcriptional activator regulator (RTA), which is encoded by the KSHV UL50 gene (Miller et al., 2007; Srivastava et al., 2023).
To sum up, there are differences in the latency of herpesviruses. The α herpesviruses (such as HSV-1 and VZV) lurk in non-dividing neuronal cells. It is not necessary to express viral proteins to tether the viral genome to chromosomes. The β and γ herpesviruses (such as HCMV, EBV, and KSHV) that maintain latency in dividing cells express viral proteins, tethering the viral genome to chromosomes so that episomes are partitioned to daughter cells. Therefore, the viral proteins required for maintaining latency and facilitating reactivation are also distinct. Despite these differences, herpesviruses of the three subfamilies also have similar lifecycles. The differences and similarities among various herpesviruses during lytic replication are outlined below to provide a clear understanding of the effects of herpesvirus shuttling proteins on the viral lifecycle.
3.2 Lysis replication of herpesviruses
3.2.1 Entry of herpesviruses
The entry of α, β, and γ herpesviruses into cells requires the coordinated interaction of multiple glycoproteins on the surface of viral particles, these glycoproteins were named with letters and prefix g (glycosylation). First, herpesviruses attach to host cells through a variety of viral glycoproteins and multiple binding receptors but do not trigger entry. Then, herpesviruses directly fuse with the host cell membrane through the viral envelope (or enter specific cells through endocytosis; Eisenberg et al., 2012; Sathiyamoorthy et al., 2017; Weed and Nicola, 2017). Although herpesviruses of different types (such as HSV-1, HCMV, and EBV) infect a variety of host cells, they share a common entry mechanism: glycoprotein gB and the heterodimer gH-gL. But different herpesvirus subfamilies use distinct viral glycoprotein combinations (gD, gp42, and gH-gL) to bind to various cellular receptors (Compton et al., 1992; Miller and Hutt-Fletcher, 1992; Gerna et al., 2004; Ryckman et al., 2006; Hutt-Fletcher, 2007).
3.2.2 Dissociation of tegument proteins and transport of capsids
After the herpesviruses enter the host cell, the viral capsid, and some tegument proteins enter the cytoplasm, and some tegument proteins are quickly dissociated from the capsid. Some tegument proteins are still attached to the capsid after entering the cell and interact with microtubules to transport the capsid to the nucleus. An opening is formed at a single vertex of the capsid. Some virus-encoded capsid proteins bind to the nuclear pore proteins Nup214 and Nup358 on the surface of cytoplasmic NPC to ensure that the capsid opening is docked with the nuclear pore and stimulates DNA release. Because the tightly packed negatively charged genome inside the capsid causes internal pressure, the high internal pressure, dozens of times the atmospheric pressures, promotes ejection of the genome from the capsid, overcoming the NPC permeability barrier and releasing the viral DNA into the nucleus (Jovasevic et al., 2008; Copeland et al., 2009; Pasdeloup et al., 2009; Huffman et al., 2017; Brandariz-Nuñez et al., 2019; Dünn-Kittenplon et al., 2021). The α herpesviruses (such as HSV-1), some tegument proteins need to be dissociated from the viral particles to function. The outer tegument proteins are first dissociated, and then the inner tegument proteins are dissociated. The first dissociated outer tegument protein is VP16 (Wysocka and Herr, 2003; Bohannon et al., 2013). The second dissociated tegument protein is VP13/14, an outer tegument protein encoded by the UL47 gene, which is also a shuttling protein that will be highlighted below. Followed by VP22 protein (encoded by UL49 gene; Morrison et al., 1998), the tegument proteins dissociated in sequence are involved in viral replication, among which UL47 and UL49 genes are unique to α herpesvirus. However, the dissociation patterns of tegument proteins in VZV, the β herpesviruses, and γ herpesviruses have not been extensively studied.
3.2.3 Transcription, replication, and capsid assembly of viral genomes
After the viral DNA is released into the nucleus, the linear viral DNA begins to cyclize and replicate through the rolling circle process to form concatemer (McVoy and Adler, 1994; Colletti et al., 2004; Gualtiero et al., 2013; Packard and Dembowski, 2021). When the viral genome completes transcription, translation, and DNA replication, the capsid proteins enter the nucleus to form the basic assembly unit of the capsid and assemble into the nucleocapsid in the nucleus (Heming et al., 2017). The viral genome is transcribed in a cascade manner in the nucleus. The viral genes are divided into the immediate early (IE), early (E), and late (L) genes according to the order of transcriptional regulation. The IE gene is first transcribed and expressed under the action of a transcription activator, and its transcription occurs before viral DNA replication. First, the post-transcriptional IE viral mRNA is exported from the nucleus and synthesizes the IE protein in the cytoplasm. Then, the IE protein re-enters the nucleus to activate the transcription of the E gene. Finally, the E protein initiates and guides the transcription and expression of the L gene (Honess and Roizman, 1974; Rixon et al., 1996; Gruffat et al., 2016; Dembowski and DeLuca, 2018). In general, the IE protein initiates and guides the transcription and expression of E gene and L gene through nuclear export and nuclear import pathways (Hiriart et al., 2003b; Donnelly et al., 2007; Liu et al., 2009, 2014b). The ICP27 protein family plays a crucial role in the transport of viral mRNA during the viral lifecycle, which will be discussed in detail below.
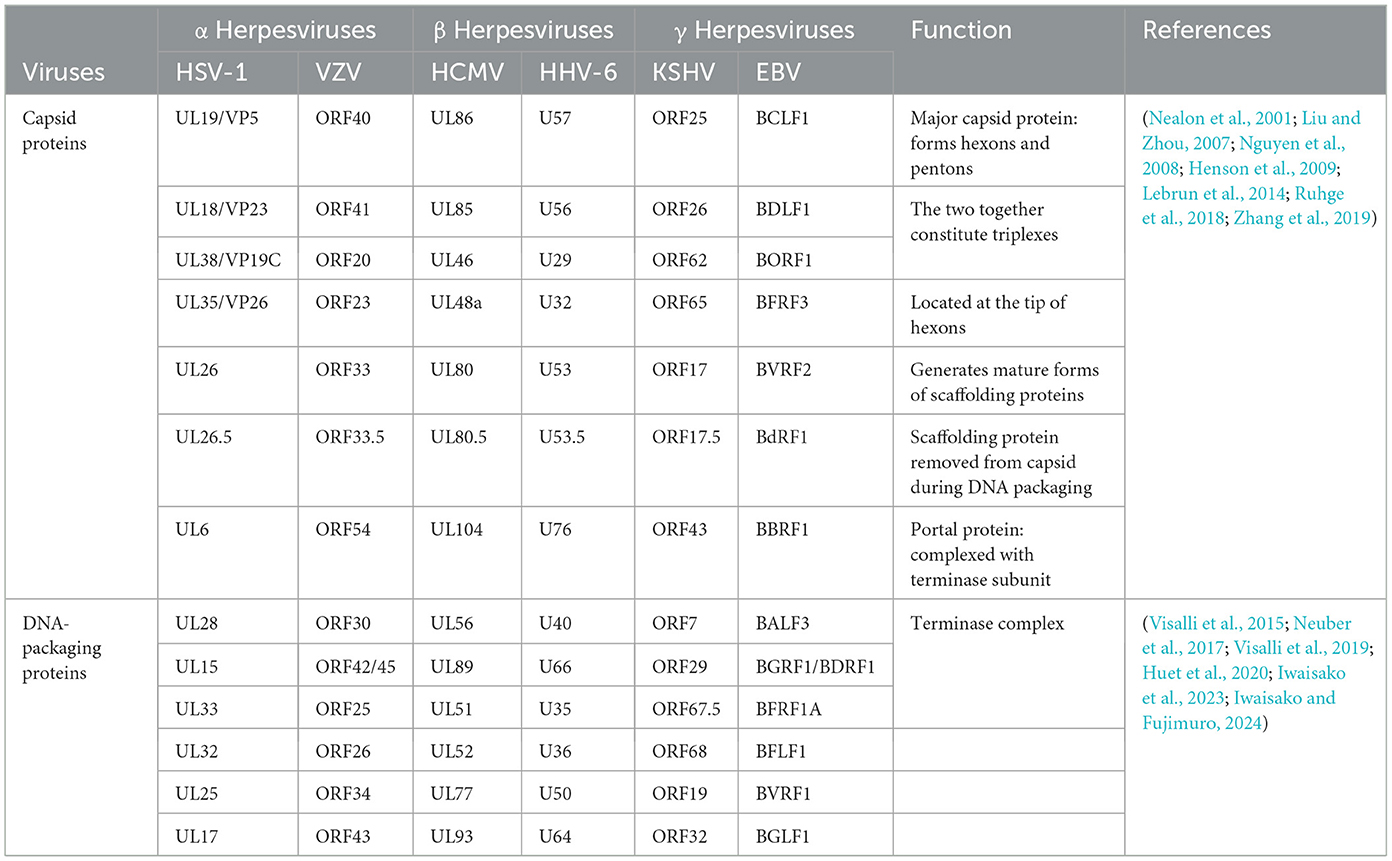
The capsid assembly mechanism is considered highly similar in herpesviruses (Lye et al., 2017). The structure and function of the main capsid proteins in the Herpesviridae are also highly conserved (see Table 3), the following is an example of HSV-1. The major capsid proteins encoded by HSV-1 include the VP5, VP23, VP19C, VP26, UL26, UL26.5, and UL6 proteins (Brown and Newcomb, 2011; Döhner et al., 2021; Villanueva-Valencia et al., 2021). After the capsid protein is encoded in the cytoplasm, it forms a subcomplex of VP5-UL26.5 and VP23-VP19C. The NLS dependent on the VP19C and VP5 proteins is coenters the nucleus, and the capsid is assembled in the nucleus (Newcomb et al., 1993; Booy et al., 1994; Newcomb et al., 2001; Beard et al., 2002). The terminase complex (UL15, UL28, and UL33 proteins) recognizes the cis-regulatory “pac” motif in viral DNA. The viral DNA is cut twice and cut into a unit-length genome (Beard et al., 2002). The viral DNA is released into the capsid by interacting with UL6 protein (Varmuza and Smiley, 1985; Beard et al., 2002). Among these, the HSV-1 VP19C protein is the shuttling protein that will be highlighted next.
3.2.4 Nuclear egress, secondary envelopment and release
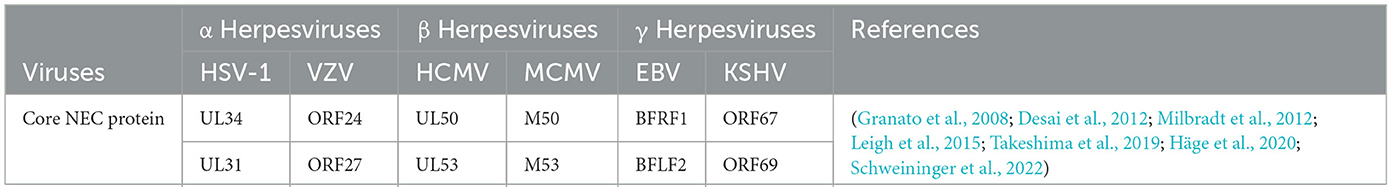
The assembled nucleocapsid interacts with the nuclear envelope, budding at the inner nuclear membrane (INM) to complete primary envelopment (Mettenleiter et al., 2013), and then forms a nucleocapsid virion structure wrapped by the primary envelope in the perinuclear space, called perinuclear virions (PEVSs), followed by the de-envelopment of virions at the outer nuclear membrane (ONM; Bigalke and Heldwein, 2016). Eventually, naked capsids are released into the cytosol (Sonntag et al., 2017; Lv et al., 2019). The nucleocapsids released into the cytoplasm by all herpesvirus subfamilies will bind to tegument proteins in an orderly manner, obtain the secondary envelope by budding into the trans-Golgi network, and form mature virus particles (Hogue, 2021; Roller and Johnson, 2021). The secondary envelopment of different herpesviruses generally occurs in the Golgi apparatus, early endosomes, or autophagosomes. The Golgi apparatus is widely considered to be the site of secondary envelope formation (Gershon et al., 1994; Zhu et al., 1995; Granzow et al., 1997; Hambleton et al., 2004; Wisner and Johnson, 2004; Sugimoto et al., 2008; Hogue et al., 2014). Subsequently, the mature virus particles are transported to the cell membrane by cytoplasmic vesicles, and the virus particles are released to the outside of the cell through exocytosis, and finally the transmission of the virus particles between the cells is completed. Herpesviruses express two conserved and essential nuclear egress regulatory proteins (see Table 4), The homologous BFLF2 protein of the UL31 protein in EBV is a shuttling protein that will be discussed later.
4 The role of herpesvirus shuttling proteins in the viral lifecycle
The mechanisms by which herpesviruses of different subfamilies infect host cells are slightly different. Herpesviruses have evolved different strategies to regulate the nucleocytoplasmic transport of NPC and proteins, creating an environment conducive to virus proliferation. Virus proteins target transport receptors through the NLS, NES, and other key functional domains, hijacking nucleocytoplasmic pathways to promote virus proliferation. Virus lifecycles depend on the transcription and replication in the nucleus of the host cell (Knipe, 1989; Boehmer and Lehman, 1997), and newly formed virus particles assemble into capsids in the nucleus (Nii et al., 1998). Herpesviruses must target transcription factors, scaffold proteins, and capsid proteins to the nucleus to participate in viral transcription, genome replication, capsid assembly (Malik et al., 1996; Rixon et al., 1996). The newly assembled viral particles from the capsid must subsequently leave the nucleus and continue to mature in the cytoplasm using tegument proteins and glycoproteins, the NES plays an important role in guiding the export of viral proteins into the cytoplasm. In summary, the nucleocytoplasmic shuttling proteins of herpesvirus ensure the correct cellular compartmentalization of herpesvirus proteins through interactions between the NLS and NES and transport receptors, which play important roles in the herpesvirus lifecycle. Mutations in NLS and NES affect the efficiency of nucleocytoplasmic transport to varying degrees. A detailed discussion of how herpesvirus shuttling proteins participate in processes throughout the lifecycle and what functions they play will be presented later (Figure 4).
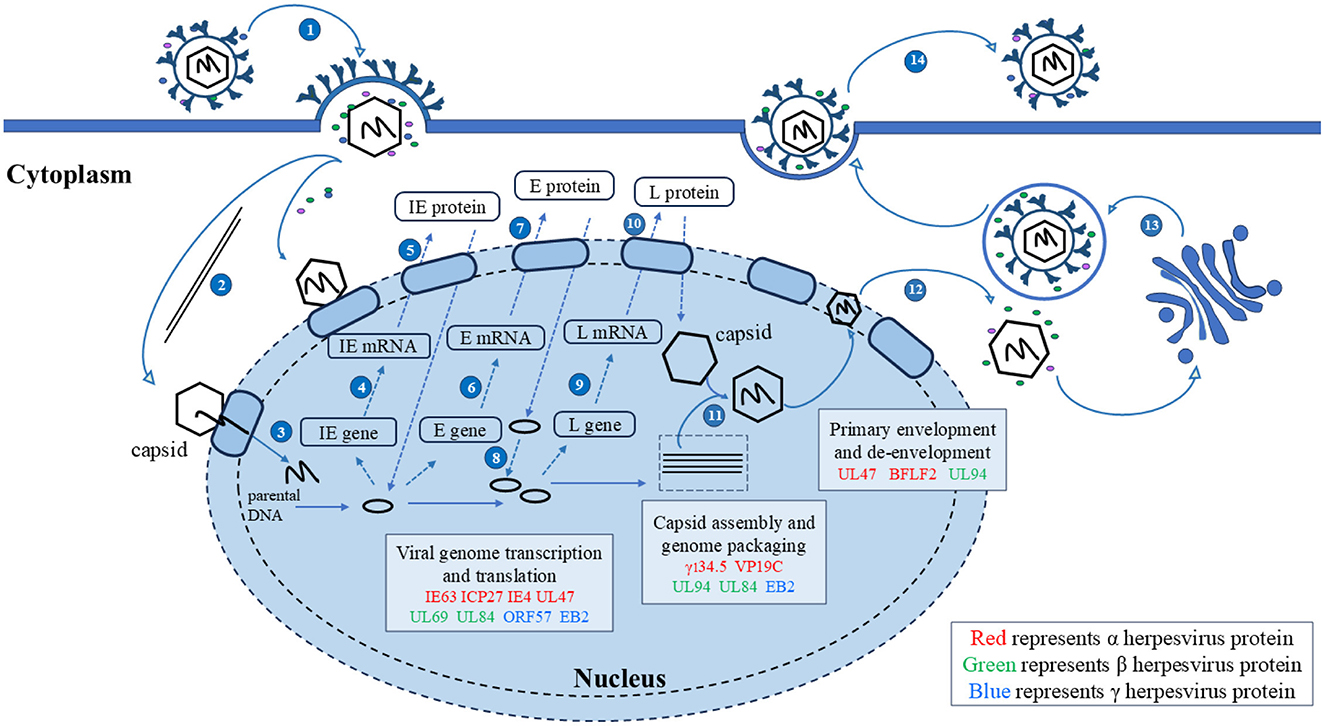
Figure 4. Herpesvirus shuttling proteins involved in the lifecycle of herpesviruses. 1. Entry; 2. The nucleocapsid enters the nucleus through microtubules; 3. Linear DNA is released into the nucleus; 4, 5. Transcription and translation of viral IE genes; 6, 7. Transcription and translation of viral E genes; 8. The viral genome replicates within the nucleus; 9, 10. Transcription and translation of viral IE genes; 11. Nucleocapsid formation and viral DNA packaging; 12. Primary envelopment and de-envelopment; 13. Secondary envelopment; 14. Release. Adapted from the literature (Sucharita et al., 2023; Li et al., 2024).
4.1 Herpesvirus shuttling proteins affect viral replication
4.1.1 Herpesvirus shuttling proteins regulate mRNA transport
mRNA transport is closely related to transcription, splicing, post-transcriptional modification, and translation, these processes are not isolated from each other but are highly continuous, dynamic, and complex. Most herpesvirus genes have no introns, and their replication depends on the selective nuclear export of intronless viral mRNAs. Since intronless virus mRNA cannot recruit mRNA export factors through a splicing-dependent mechanism, regulating nuclear mRNA export is crucial for the replication and pathogenesis of the herpesvirus (Gales et al., 2020). Research has shown that herpesvirus uses Aly/REF, TAP/NXF1, CRM1, and other export pathways to alter cellular and viral mRNA export and utilizes some regulatory proteins to promote viral mRNA nuclear export (Fornerod et al., 1997; Stade et al., 1997; Grüter et al., 1998), such as the ICP27 protein family encoded by herpesvirus, which includes the HSV-1 ICP27 protein (Johnson et al., 2009; Johnson and Sandri-Goldin, 2009; Koffa et al., 2023), VZV IE4 protein (Ote et al., 2009), BHV-1 ICP27 (Guo et al., 2009; Ding et al., 2010), HCMV UL69 protein (Zielke et al., 2011), EBV EB2 protein (Hiriart et al., 2003a), HVS ORF57 protein (Williams et al., 2005), and KSHV ORF57 protein (Boyne et al., 2008). They act as viral mRNA export factors, mediating the nucleocytoplasmic transport of viral transcripts (Figure 5).
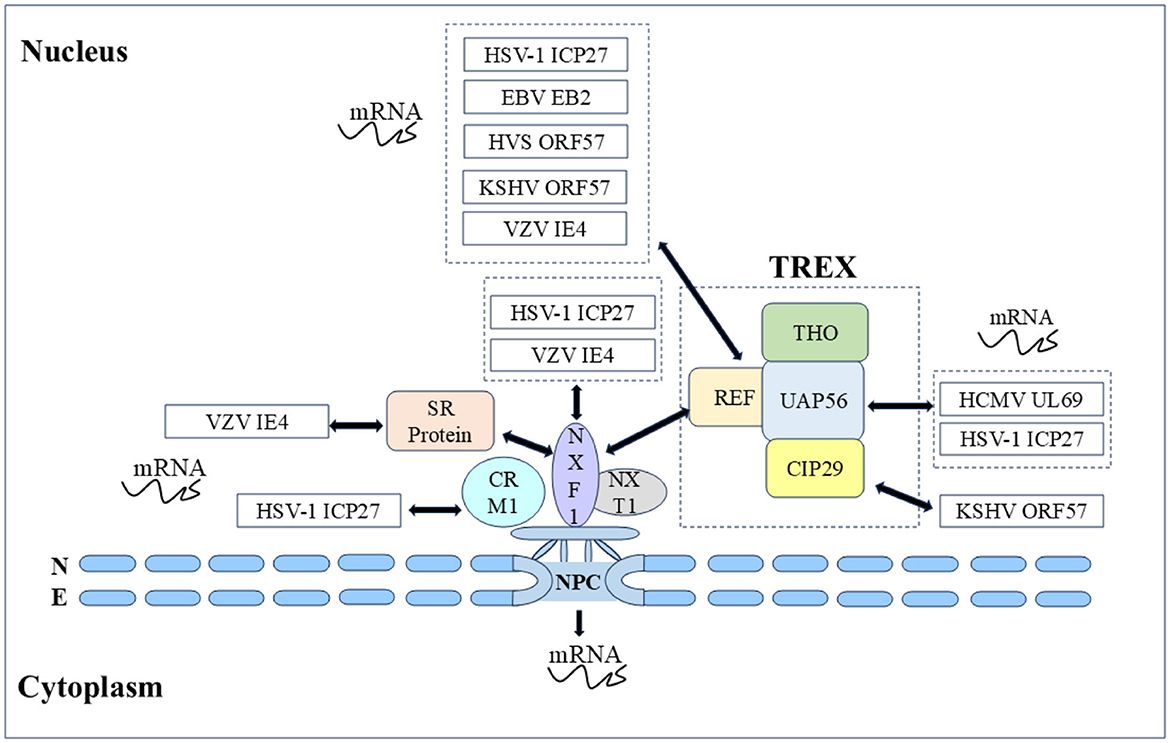
Figure 5. Herpesvirus shuttling proteins export mRNA by hijacking host mRNA export mechanisms (NXF1-mediated and CRM1-mediated). Double-headed arrow represents interaction.
In α herpesvirus, the HSV-1 ICP27 protein shuttle is coupled with viral mRNA export to the cytoplasm, and the nucleocytoplasmic shuttle activity depends on the NLS and NES as well as the other regions. Many studies have shown that the ICP27 protein recruits export factors such as Aly/REF, UAP56, and TAP to viral mRNA and promotes mRNA export through the TAP/NXF1 pathway. The ICP27 protein has been shown to affect the nucleocytoplasmic transport of UL15, UL17, and UL48 mRNAs, as well as the cytoplasmic levels of UL30, UL29, UL42, and UL5 mRNAs (Phelan et al., 1996; Uprichard and Knipe, 1996; Soliman et al., 1997). Initially, to screen the functional regions of the ICP27 protein, some scholars constructed 16 HSV-1 ICP27 mutants and finally reported that the mutations M11, M15, and M16 in these three regions completely inactivated the basic functions of the ICP27 protein (M11, M15, and M16 mutations led to amino acid substitutions at positions 340/341, 465/466, and 488, respectively). The shuttle ability of five HSV-1 ICP27 mutant proteins was subsequently studied, it was found that the M15 mutation affects the interaction between ICP27 and the core nucleoporin Nup62, causing M15 to be unable to re-export and shuttle continuously. After the M15 mutation completely blocks the shuttling activity of ICP27, it is not able to induce some viral late mRNAs, and seems owing to the loss of this key function, M15 mutant viruses are replication defective. In addition, the ICP27 NES mutation (Dleu) was found to weaken the shuttle, and Dleu mutant viruses were also replication defective (Rice and Lam, 1994; Mears and Rice, 1998; Lengyel et al., 2002; Malik et al., 2012). Subsequently, it was further found that the M15 mutant significantly reduced the interaction between ICP27 and mRNA, which explained the close relationship between the nucleocytoplasmic shuttling of ICP27 protein and mRNA (Sokolowski et al., 2003; Srinivas et al., 2021). Notably, the nucleocytoplasmic shuttling activity of the HSV-1 ICP27 protein is related to the activation of the late gene of the virus. How the shuttle of ICP27 facilitates the transcriptional activation of the late gene of the virus and whether ICP27 stimulates the nuclear export of the viral gene transcript by binding to the viral gene transcript in the nucleus and escorting it to the cytoplasm still need to be explored (Mears and Rice, 1998). The VZV IE4 protein possesses transactivating properties, and the IE4 protein can recruit the TAP/NXF1 receptor into viral mRNA to mediate mRNA export by interacting with the cellular export adapter protein SR protein. Some scholars have demonstrated that three domains of the IE4 protein, Ra, Rb, and Rc, mediate the interaction between the IE4 protein and its mRNA, Rb, and Rc not only participate in the transactivating properties of the IE4 protein but also mediate interactions with transcription factors such as p50 and TFIIB. Since the IE4 NLS is located in the Rb domain, the IE4 protein loses the ability to migrate to the nucleus when binding to mRNA and is exported to the cytoplasm, it is reasonable to speculate that only the IE4 NLS can localize to the nucleus to promote interaction with transcription factors (Baudoux et al., 2000; Ote et al., 2009, 2010; Huang et al., 2014). In addition, in the α herpesvirus UL47 protein family, the nucleocytoplasmic shuttling behaviors of the HSV-1 UL47 protein and BHV-1 UL47 protein are similar and are mediated by importin β for nuclear import (McLean et al., 1990; Donnelly and Elliott, 2001; Zheng et al., 2004), HSV-1 UL47 serves as an effective RNA-binding protein and its NLS and RNA binding sequence are NLS determined by the same motif. After binding to mRNA in vivo, the HSV-1 UL47 protein targets the main nuclear domain where viral mRNA transcription occurs (Donnelly et al., 2007).
In β herpesvirus, HCMV UL69 protein, as a post-transcriptional transactivator, promotes the nuclear export of mRNA through nucleocytoplasmic shuttling and the ability to recruit components of the cell mRNA export mechanism. UAP56 and URH49 are closely related RNA helicases that function in selective mRNA processing and export pathways, subsequent testing of the mRNA export activity of the HCMV UL69 protein revealed that the NLS and RNA binding motifs partially overlap with the UAP56/URH49 interaction motif, which is crucial for mRNA export activity, and that UAP56 binding-deficient viruses exhibit strong replication defects. In addition, the UL69 NES mutant, whose nucleocytoplasmic shuttling ability was lost, also lost the ability to activate gene expression and export mRNA (Lischka et al., 2001, 2006b; Stamminger, 2008; Zielke et al., 2011). Other UL69 homologous proteins (chimpanzee cytomegalovirus C69 protein, rhesus cytomegalovirus Rh69 protein, human herpesvirus 6 U42 protein, and elephant endotheliotropic herpesvirus U42 protein) in β herpesviruses were subsequently studied. Nucleocytoplasmic shuttling and homodimerization are conserved features of the HCMV UL69 homologous protein family of herpesviruses, whereas heterodimerization and recruitment of the cell mRNA export factors UAP56 and URH49 are limited to members of the UL69 protein (C69 protein and Rh69 protein) of the cytomegalovirus genus. All of these proteins function as viral mRNA export factors. In addition, HCMV UL84 protein is also a shuttling protein in HCMV that affects mRNA export, HCMV UL84 protein enhances the accumulation of viral transcripts encoding replication proteins in the cytoplasm. The UL84 protein enters the nucleus through interactions between unconventional nuclear-targeting domains and importin-α and participates in virus replication compartments by interacting with HCMV UL44 protein (polymerase accessory protein), which plays a direct role in DNA replication (Xu et al., 2002; Lischka et al., 2006a; Gao et al., 2010; Strang et al., 2012). UL84 is also an RNA binding protein, the construction of a non-shuttle mutant of UL84 NES revealed that the accumulation of HCMV IRS1 mRNA in the cytoplasm was reduced. The IRS1 protein is an immediate early protein encoded by the IRS1 ORF in the HCMV inverted repeat sequence. The IRS1 protein is involved in blocking the α group phosphorylation of eukaryotic initiation factor 2 (eIF2α) and the shutdown of cellular protein synthesis, and eIF2 is a key translation regulator. These results indicate that UL84 protein nucleocytoplasmic shuttling mediates IRS1 mRNA export and plays an important role in viral replication (Hakki et al., 2006; Gao et al., 2010). Members of the DExD box protein family are involved in various aspects of RNA metabolism, with many DExD/H proteins shuttling RNA from the nucleus to the cytoplasm. UL84 is a putative member of the DExD/H box protein family, further enhancing our understanding of UL84-mediated mRNA processes (Colletti et al., 2005).
In γ herpesvirus, the EBV EB2 protein has a similar ability to export mRNA, and the EB2 DN region binds to the export factor REF, which is critical for mRNA export (Hiriart et al., 2003b; Juillard et al., 2009). The nucleolus is the center of ribosome biogenesis and is involved in viral replication, and nucleolar localization sequence (NoLS) can target viral proteins to the nucleolus. The HVS ORF57 protein binds to viral mRNA for nucleocytoplasmic shuttling, and the nucleolar localization of ORF57 is crucial for nuclear mRNA export and gene activation. It has been reported that the ORF57 NLS mutants cannot be transport to the nucleolus reportedly, resulting in the loss of viral mRNA nuclear export, explaining the direct functional role of the nucleolar transport of viral proteins in viral mRNA nuclear export (Boyne and Whitehouse, 2006b). Notably, BHV-1 ICP27 NLS or NoLS single deletion mutants did not eliminate the transactivation activity of the glycoprotein gC promoter, whereas NLS and NoLS double deletion mutants lost this function. These results suggest that the nuclear and nucleolar localization of BHV-1 ICP27 may be involved in the regulation of viral RNA transport (Guo et al., 2009; Ding et al., 2010). The KSHV ORF57 protein interacts with the REF to recruit the transcription-export (TREX) complex to intron-free viral mRNA, enabling efficient mRNA export. During mRNA export, the ORF57 protein shuttles through the nucleolus, and the TREX complex is located in the nucleolus via the ORF57 protein. However, the exact cause of nucleolar localization during shuttling remains to be elucidated (Boyne et al., 2008; Boyne and Whitehouse, 2009; Jackson et al., 2011; Li et al., 2012a). Given the above, the ICP27 family, as posttranscriptional transactivators, promotes the nuclear export of mRNAs through nucleocytoplasmic shuttling and the ability to recruit components of the cellular mRNA export mechanism, strongly demonstrating the importance of nucleocytoplasmic shuttling in mediating the function of the ICP27 protein family.
4.1.2 Herpesvirus shuttling proteins regulate mRNA stability
mRNA is easily destroyed by physical, chemical, and enzymatic factors, and its stability in cells is called mRNA stability, which is one of the basic mechanisms regulating gene expression and determines the final mRNA concentration at the post-transcriptional level (Liu et al., 2014a; Radhakrishnan and Green, 2016; Li et al., 2022; Watson and Thoreen, 2022). VHS is an endoribonuclease that is important for viral replication. VHS degrades cells and viral mRNA, the degradation of cellular mRNA to increase the availability of cellular translation mechanisms to promote the synthesis of viral proteins, viral mRNA degradation helps regulate the sequence expression of different viral genes (Taddeo and Roizman, 2006; Taddeo et al., 2006). In α herpesvirus, the HSV-1 UL47 protein interacts with VHS, assisting in the translocation of some VHS to the nucleus. In transfected cells, the VHS NES mutant can degrade stable mRNA, but it does not degrade in infected cells, suggesting that UL47 protein greatly weakens the degradation of viral mRNA (Shu et al., 2013a,b). In addition, KSHV ORF59 is a viral DNA polymerase processivity factor, and the KSHV ORF57 protein has been reported that interacts with the RNA export cofactors RBM15 and OTT3 through the NLS2 and NLS3 region to offset the nuclear accumulation of KSHV ORF59 mRNA, thereby promoting the expression of intronless ORF59 genes. RNA decay analyses after actinomycin D-mediated suppression of polymerase II transcription revealed that the ORF57 protein enhances the stability of ORF59 mRNA by increasing its half-life in cells (Nekorchuk et al., 2007; Majerciak et al., 2011). Overall, the ICP27 protein family plays a role in post-transcription, promoting mRNA nuclear export into the cytoplasm and acting as a posttranscriptional transactivator. However, the main function of the KSHV ORF57 protein is to stabilize mRNA. The HSV-1 UL47 protein regulates the cascade activation mechanism of viral genes and maintains the stability of viral mRNA by mediating the localization of intracellular VHS.
4.2 Herpesvirus shuttling protein affects virus particle assembly
The maturation of virions requires capsid assembly, primary envelopment, secondary envelopment, and other processes. Owing to the large nucleocapsid of herpesvirus, it passes through the INM and ONM through the nuclear pore. Therefore, herpesviruses have evolved a unique nuclear export mechanism. After successful assembly of the nucleocapsid, the offspring virions enter the perinuclear space between the INM and ONM for primary envelopment through budding, after which the nucleocapsid of the envelope fuses with the ONM, releasing the de-enveloped nucleocapsid into the cytosol. The unique nuclear export mechanism of viral particles allows tegument proteins to attach directly or indirectly to the nucleocapsid and exert their functions, facilitating the expulsion and maturation of viral particles from the nucleus (Mettenleiter et al., 2013; Owen et al., 2015).
In α herpesvirus, the HSV-1 UL47 protein and BHV-1 UL47 protein are the main components of the viral particle tegument and not only affect replication but also participate in virus assembly. Phosphorylated BHV-1 UL47 protein affects its cellular localization plays a role in viral DNA encapsidation and secondary virion incorporation (Zhang et al., 2015, 2016). The HSV-1 UL47 ineffective virus affects the nuclear export of virus particles, resulting in a decrease in the proportion of primary enveloped virus particles in the perinuclear space. According to reports, the accumulation of nuclear capsids and the lack of primary enveloped virions in the perinuclear space in the absence of the UL47 protein likely reflect an imbalance between the rate of virion delivery into the perinuclear space and the rate of egress from this region. The UL47 protein appears to be required for efficient primary envelopment of nucleocapsids in HSV-1 nuclear export, the cytoplasmic shuttle characteristic of the UL47 protein may be the cause of this phenomenon (Liu et al., 2014b). In addition, the HSV-1 UL47 protein interacts with the UL31/UL34 complex and US3, regulating their functions to promote the primary envelope of viral particles (Liu et al., 2014b). The HSV-1 γ134.5 protein facilitates nuclear egress, and the absence of the γ134.5 protein leads to the accumulation of nucleocapsids, further research revealed that deleting the amino-terminal nuclear egress domain (including nucleolar localization signals) of this protein increases the accumulation of capsid in the nucleus, indicating the importance of this domain for the function of the γ134.5 protein, it is possible that this cis-element is required to direct virus egress from the nucleus to the cytoplasm (Brown et al., 1994; Cheng et al., 2002; Mao and Rosenthal, 2002; Jing et al., 2004). The HSV-1 VP19C protein is a structural protein of HSV-1 viral particles and is crucial for the assembly of the capsid. The VP19C protein utilizes NLS to bind to importin βs to cause nuclear import, and NES binds to CRM1 to cause export, becoming the first herpesvirus capsid protein with nucleocytoplasmic shuttling properties. The VP19C protein mediates its involvement in the assembly of the capsid structure, which can non-specifically bind to viral DNA and may play a role in anchoring DNA to the capsid in the nucleus (Braun et al., 1984; Tatman et al., 1994; Person and Desai, 1998). Another important function of the VP19C protein is to correctly transport component proteins to the capsid assembly site, where they serve as carriers to transport cytoplasmic capsid proteins (such as the VP23 and VP5 proteins) to the nucleus and promote capsid assembly. The nucleocytoplasmic shuttling ability of the VP19C protein is the direct condition for its function, and its NLS and NES mutations in recombinant viruses have demonstrated that the nuclear import of the VP19C protein is necessary for efficient production of HSV-1 (Rixon et al., 1996; Adamson et al., 2006; Okoye et al., 2006).
In β herpesvirus, the pp28 protein is a tegument protein encoded by HCMV UL99, HCMV pp28 mediates the capsid envelopment of HCMV and is an essential tegument protein required for HCMV assembly (Jones and Lee, 2004; Seo and Britt, 2006). The HCMV UL94 protein has been shown to interact with pp28 in the virus assembly compartment (AC), and a mutation at 343 aa has been found to alter the cytoplasmic distribution of the UL94 protein and disrupt its nucleocytoplasmic shuttling. The 343 aa site of the UL94 protein is the nuclear import site and a key site for interaction with pp28, which can serve as a bridge connecting the capsid and envelope. The UL94 protein partially guides pp28 to the assembly complex and participates in the secondary envelope of viral particles, demonstrating the importance of UL94's nucleocytoplasmic shuttling properties in mediating pp28's participation in the assembly process of the virus (Liu et al., 2009, 2012).
The shuttling proteins affecting viral assembly in γ herpesviruses include the KSHV ORF45 protein, the EBV EB2 protein, and the EBV BFLF2 protein. The KSHV ORF45 protein is both a tegument protein and an immediate early protein that is necessary for the entry and release of the targeted viral capsid. The viral particle protein content was significantly reduced in the BAC-45rc mutant in which the ORF45 NLS was disrupted (Li and Zhu, 2009). The lysine in the NLS often serves as a ubiquitin acceptor site (Chan et al., 2006), Lys297 in the ORF45 NLS is critical for ORF45 targeting of lipid rafts (LRs) and subsequently guides viral particles to the Golgi and endosome membrane for budding (Wang et al., 2015). In BCBL-1 cells, a portion of the ORF45 protein was found to be colocalized with the viral replication compartment, suggesting that the binding of the ORF45 protein to the capsid may participate in the maturation processes of the virus. However, further studies are needed to clarify the role of the ORF45 protein in the late stages of viral replication (Kuang et al., 2008; Li and Zhu, 2009). EB2 is a nuclear protein that mediates mRNA nuclear export. Studies have shown that in the absence of EB2, the newly replicated viral DNA is not properly encapsidated and is eventually completely digested by DNase I, suggesting that the correct balance of protein expression involved in intranuclear capsid assembly and maturation can be obtained only in the presence of EB2. In addition, as EB2 is crucial for the nuclear export of most late mRNAs and these genes are converted into proteins involved in nuclear capsid assembly and maturation, EB2 is essential for the correct assembly of nuclear capsids (Batisse et al., 2005). The EBV BFLF2 protein belongs to the herpesvirus UL31 protein family, and the EBV BFRF1 protein belongs to the herpesvirus UL34 protein family. The UL31 protein and the UL34 protein family are conserved among all herpesviruses and are the main viral components involved in the primary envelope of the virus, the UL31 protein and the UL34 protein family form a nuclear export complex to mediate the nuclear export of herpesvirus. BFLF2 is associated with the nuclear export of TAP and interacts with importin-α7, importin β1, and transportin 1, promoting the entry of the EBV nucleocapsid from the nucleus into the cytoplasm for the subsequent maturation of viral particles. Transfection of BFLF2 and BFRF1 interacting region deletion mutants, as well as NLS mutants, resulted in a significant decrease in the level of secreted viral particles. Both the nuclear targeting of BFLF2 and the BFRF1-interacting domains of BFLF2 are necessary for EBV maturation and secretion (Gonnella et al., 2005; Li et al., 2018; Dai et al., 2020). To sum up, nucleocytoplasmic shuttling proteins in herpesviruses affect the assembly of viral particles by participating in nucleocapsid assembly and influencing the primary envelopment of the virus.
4.3 Interactions of herpesvirus shuttling proteins with other proteins affect viral replication
Herpesvirus shuttling proteins interact with other proteins and regulate orderly in different compartments. In α herpesvirus, the UL47 protein forms a stable complex with the US3 protein in HSV-1 infected cells, and the UL47 protein is required for effective nuclear localization of the US3 protein. HSV-1 US3 plays an important role in the viral lifecycle by phosphorylating a variety of viral and host proteins, such as regulating cell morphology or microtubule networks and promoting the nuclear egress of progeny nucleocapsids. A study showed that phosphorylation of UL47 Ser77 (adjacent to the UL47 NLS) by US3 regulates the nuclear localization of the UL47 protein, and mutations in this site affect the effective viral replication of HSV-1 in mice (Kato et al., 2011). Further study demonstrated that the UL47 protein, UL34/UL31 complex and US3 protein colocalize to the nuclear envelope and promote viral nuclear egress (Liu et al., 2014b). The HSV-1 proteins ICP27 and UL47 are both nucleocytoplasmic shuttling proteins and RNA binding proteins, and their binding with the poly(A)-binding protein PABP1 ultimately leads to Paip2 translocation and the nuclear accumulation of PABPC1. PAPBC1 plays an important role in effective translation initiation. Under normal circumstances, it binds to the poly(A) tail of mRNA in the cytoplasm and enhances the stability and translation efficiency of mRNA. Abnormal nuclear localization of PAPBC may affect the processing or export steps of viral or host mRNAs (Dobrikova et al., 2010). The lytic replication of herpesvirus is activated by the viral transcriptional activator VP16, which activates the immediate-early genes of the virus and initiates a complex cascade of gene expression (Johnson et al., 1999). Studies have shown that the PRV UL46 protein interacts with VP16 through the NLS to regulate the function of VP16 (Xu et al., 2020).
In β herpesvirus, the HCMV UL94 protein interacts with the HCMV pp28 protein through nuclear import sites to affect viral particle assembly (Liu et al., 2009). In addition, the HCMV IE2 protein is the main transactivator and is encoded by the HCMV gene. The HCMV UL84 protein interacts with the IE2 protein to interfere with IE2-mediated transactivation of early gene promoters. In HCMV UL84 NES non-shuttle mutant transfected cells, the IE2 protein presented an abnormal localization pattern in the nucleus and failed to be correctly assigned to the replication chamber. The nucleocytoplasmic shuttling of the UL84 protein may indirectly regulate IE2 translation and localization (Colletti et al., 2004). In γ herpesvirus, the expression of the EBV BFLF2 protein alone is limited to the nucleus, while the expression of the EBV BFRF1 protein alone is located in the cytoplasm and perinucleus, the two proteins are co-localized at the nuclear rim, and their interaction plays a key role in the viral envelopment (Lake and Hutt-Fletcher, 2004). In general, the interaction network among viral proteins is complex and extensive. The interactions between these proteins can influence their localization, expression, and function. Some shuttling proteins play a regulatory role by affecting both their own localization and that of other proteins.
5 Other functions
5.1 Herpesvirus shuttling proteins regulate apoptosis
Apoptosis is a type of programmed cell death associated with characteristic morphological and biochemical changes in cells. It plays a role in preventing virus transmission and spread in the early stages of viral infection while promoting virus replication and export in the later stages of viral infection (Zhou et al., 2017). In α herpesvirus, BHV-1 UL47 protein does not inhibit the phosphorylation of SMC1 in the cytoplasm, but it inhibits SMC1 phosphorylation in the nucleus, leading to disruption of the ATM/NBS1/SMC1 pathway and inhibition of DNA repair. SMC1 is a component of the DNA damage network. Studies have shown that blocking SMC1 phosphorylation reduces cell survival after DNA damage. In BHV-1 UL47 protein-transfected cells, DNA damage-induced apoptosis increased, so the BHV-1 UL47 protein inhibits SMC1 phosphorylation and DNA repair, demonstrating a potential role in regulating apoptosis (Kitagawa et al., 2004; Vasilenko et al., 2012; Afroz et al., 2018). BHV-1 VP22 is a tegument protein encoded by the UL49 gene. The mitochondria-targeting sequence at the C terminus of the VP22 protein is found within the VP22 NES (Zheng et al., 2005; Zhu et al., 2005). VP22 has been reported to induce apoptosis (Qiu et al., 2005), and mitochondria playing a central role in this process (Jeong and Seol, 2008). Therefore, it is reasonable to speculate that the VP22 protein is exported to the cytoplasm via the NES and subsequently enters the mitochondria to influence apoptosis. Furthermore, in γ herpesvirus, a portion of the KSHV genome specifically mediates the maintenance of the virus in the B-cell environment, LANA2 is a nuclear latent protein detected only in B cells infected with KSHV, which inhibits p53 tumor suppressor gene protein-dependent transcriptional transactivation and apoptosis, as well as PKR-dependent apoptosis (Chao et al., 2000; Rivas et al., 2001; Chipuk and Green, 2003). LANA2 NLS mutants resulting in increased cytoplasmic localization are unable to inhibit the apoptosis induced by p53 activation. Subsequently, it was found that LANA2 phosphorylation inhibits the function of NES and affects the ability to inhibit p53-dependent apoptosis. Indicating that the nuclear localization of the LANA2 protein is crucial for the inhibition of p53-induced apoptosis and LANA2's nucleocytoplasmic shuttling mediates its ectopic location to the nucleus to inhibit apoptosis and promote the proliferation of virions (Muñoz-Fontela et al., 2003, 2005). In general, some herpesvirus shuttling proteins affect node proteins in apoptosis-related pathways through nucleocytoplasmic shuttling to mediate apoptosis.
5.2 Herpesvirus shuttling proteins participate in immune escape
Herpesvirus infection is lifelong and highly host-specific, to resist virus invasion, hosts use pattern recognition receptors (PRRs), such as retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), to recruit a series of signal transduction molecules, such as stimulator of interferon genes (STING) and mitochondrial antiviral signaling protein (MAVS). These proteins transfer signals to downstream molecules in different signaling pathways, ultimately leading to the activation and translocation of multiple transcription factors, including NF-κB, IFN regulatory factor 3 (IRF3), and IFN regulatory factor 7 (IRF7), into the nucleus, inducing the expression of type I interferons (IFN-I) and proinflammatory cytokines to activate downstream JAK-STAT signaling pathways, which results in the expression of multiple interferon-stimulated genes (ISGs), ultimately resulting in antiviral immune response (Pestka et al., 2004; Stark and Darnell, 2012). Therefore, during latent infection, the herpesvirus gradually evolves various host immune evasion strategies to ensure its survival to evade the host immune response. Some herpesvirus shuttling proteins participate in the virus immune evasion process by inhibiting the production of IFN-I, blocking downstream IFN signaling, regulating specific ISGs, antagonizing the host antiviral innate immune response and achieving effective virus transmission and pathogenicity.
In α herpesvirus, the nuclear localization of the BHV-1 ICP27 protein is necessary to inhibit IFN-β promoter activity in transfected cells, and the BHV-1 ICP27 Δ N (Δ NLS+NoLS) mutation interferes with the ability to inhibit IFN-β1 and IFN-β3 promoter activity (da Silva et al., 2012). On the other hand, the BHV-1 ICP27 protein regulates 3′ mRNA processing, whether ICP27 affects immunity due to its potential interference with the 3′ processing of cellular factors necessary for IFN-β-dependent transcription remains to be investigated (Singh et al., 1996). Daxx is an important component of intrinsic cellular immunity, herpesviruses achieve replication and immune escape in host cells by regulating Daxx, and the HSV-1 ICP27 protein has been reported to ectopic Daxx from the nucleus to the cytoplasm, increasing the interaction between p65 NF-κB and Daxx, thereby inhibiting NF-κB activity and regulating innate immune processes (Schreiner and Wodrich, 2013; Kim et al., 2017). Transcription activator 1 (STAT1) is a potential cytoplasmic transcription factor that directly participates in the IFN-I-mediated signaling pathway. Studies have shown that NLS deficiency in the BHV-1 UL47 protein inhibits the nuclear accumulation of STAT1 after IFN-β stimulation and ultimately downregulates IFN-I signaling (Afroz et al., 2016). In addition, the HSV-1 ICP27 protein can also inhibit IFN-I signaling by inhibiting STAT1 phosphorylation and nuclear accumulation (Johnson et al., 2008). Further investigated how the cellular localization of ICP27 affects the inhibition of IFN expression by using ICP27 ΔNES or ICP27 ΔNLS mutant viruses and reported that full inhibition of the IFN response depends on the cytosolic position of ICP27 after shuttling (Christensen et al., 2016). PRV UL46 protein helps viruses evade host innate immunity by regulating STING function in the cytoplasm (Xu et al., 2020). The HSV-1 γ134.5 protein interrupts the translocation of RIG-I from the cytoplasm to mitochondria and disrupts the translocation of STING from the endoplasmic reticulum to the Golgi apparatus, the γ134.5 protein also inhibits IRF3 phosphorylation and nuclear translocation, resulting in downregulation of the IFN response (Verpooten et al., 2009; Pan et al., 2018; Liu et al., 2021).
In β herpesvirus, the HCMV UL94 protein inhibits the antiviral innate immune response by targeting STING. STING recruits TBK1 and IRF3, in which IRF3 is phosphorylated and activated by TBK1, resulting in the subsequent induction of IFN-I. The UL94 protein selectively impairs the recruitment of TBK1 to STING signalosomes, resulting in the inhibition of downstream signal transduction (Zou et al., 2020). In γ herpesvirus, the KSHV LANA2 protein mediates the nuclear translocation of NF-κB, affecting NF-κB activity (Seo et al., 2004). The KSHV ORF45 protein inhibits virus-induced IFN-I production by blocking its phosphorylation and nuclear translocation through interaction with IRF7 (Zhu et al., 2002). In summary, proteins shuttled by herpesvirus participate in immune escape by regulating the localization of signal transduction molecules and transcription factors in the cytoplasm or nucleus and ultimately affect IFN activity, demonstrating the clever use of the cellular regulatory mechanisms of viral proteins to antagonize innate immune responses.
5.3 Cytoskeleton rearrangement induced by herpesvirus shuttling proteins
The cytoskeleton is involved in maintaining cell integrity and structure and remodeling surface structure and movement, including actin filaments, microtubules, and intermediate filaments. Herpesvirus remodels actin during the process of host cell entry, assembly and release, and transmits between host cells by destroying the cytoskeleton (Miranda-Saksena et al., 2018). The US3 protein is a multifunctional protein that participates in the modification of the cytoskeleton and nuclear egress of the herpesvirus capsid during the viral replication cycle, directly interacting with actin regulatory mechanisms. In α herpesvirus, HSV-2 US3 is the determining factor involved in the reorganization of the actin cytoskeleton into filamentous processes (FPs), and the formation of FPs is related to an increase in the cell-to-cell spread of the virus infection. HSV-2 US3 inhibits the formation of FPs if it cannot be exported to the cytoplasm. Furthermore, NES mutations in HSV-2 US3 contribute to its kinase activity, which is required for HSV-2 US3-induced FP formation and disassembly of actin stress fibers, thus coupling the nucleocytoplasmic shuttling properties of US3 with functional play. In addition, the loss of the HSV-2 US3 protein disrupts nuclear egress (Finnen et al., 2010, 2011). In summary, the HSV-2 US3 protein triggers remodeling of the host cytoskeleton during the life cycle through nucleocytoplasmic shuttling to promote the effective entry and transmission of the virus into the host cell.
6 Conclusion and prospects
The replication process of herpesviruses includes adsorption, entry, uncoating, biosynthesis, virion assembly, maturation, and release. Herpesvirus shuttling proteins play important roles in regulating viral mRNA transport, new capsid assembly and primary envelopment. These viral proteins are transported to different compartments through nucleocytoplasmic shuttling and play a role in different stages of viral maturation, which is conducive to maximizing self-regulatory functions. Although there are few relevant reports, it is reasonable to speculate that nucleocytoplasmic shuttling is essential for some viral proteins to perform multiple functions. Some herpesvirus nucleocytoplasmic shuttling proteins hijack the host pathway to export viral mRNA, inhibit host mRNA export, promote protein expression necessary for further replication and assembly of new viral particles, and block the expression of host-encoded cellular defense protein mRNAs.
At present, many available information about the function of shuttling proteins in the lifecycle of herpesviruses is based on α herpesviruses, especially HSV-1. Although some of this information is about HCMV and KSHV, if more shuttling proteins from β and γ herpesviruses can be identified in future research, a greater understanding of how shuttling proteins coordinate the viral lifecycle and whether they have similar functions can be achieved. Most of the characterized herpesvirus shuttling proteins interact with host transport receptors through the NLS and NES, utilizing intracellular transport mechanisms to shuttle between the nucleus and cytoplasm, promote virus proliferation, and evade host antiviral responses. First, NLS mediate transport to the nucleus, where they participate in virus replication, gene expression regulation, and assembly; in the later stage, NLS rely mainly on the export of NES to the cytoplasm for further maturation. Dysfunction of the NLS or NES can block or damage the production of infectious viral particles, demonstrating the roles of the NLS and NES in the viral lifecycle. Nucleocytoplasmic transport involving NLS and NES (including the transport of viral capsids, viral genes, viral polymerases, and some transcriptional regulators) is an important factor in viral transmission and regulates host-virus interactions. In summary, as the shuttling proteins of herpesvirus play multiple functions throughout the virus lifecycle, further elucidation of their mechanisms provide important references for identifying therapeutic targets. Targeting the binding region between the shuttling proteins and their nuclear transport receptors to inhibit the expression of nucleocytoplasmic function has broad importance for the development of antiviral drugs.
Author contributions
HC: Methodology, Validation, Writing – original draft, Writing – review & editing. MW: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. AC: Software, Writing – original draft, Writing – review & editing. BT: Investigation, Supervision, Writing – original draft, Writing – review & editing. QY: Validation, Writing – original draft, Writing – review & editing. XO: Funding acquisition, Writing – original draft, Writing – review & editing. DS: Visualization, Writing – original draft, Writing – review & editing. YH: Methodology, Writing – original draft, Writing – review & editing. ZW: Visualization, Writing – original draft, Writing – review & editing. XZ: Software, Writing – original draft, Writing – review & editing. YW: Data curation, Writing – original draft, Writing – review & editing. SZ: Conceptualization, Writing – original draft, Writing – review & editing. JH: Investigation, Writing – original draft, Writing – review & editing. YY: Software, Writing – original draft, Writing – review & editing. LZ: Investigation, Writing – original draft, Writing – review & editing. SC: Software, Writing – original draft, Writing – review & editing. ML: Software, Writing – original draft, Writing – review & editing. DZ: Resources, Writing – original draft, Writing – review & editing. RJ: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from China Agriculture Research System of MOF and MARA (CARS-42-17) and the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adamson, W. E., McNab, D., Preston, V. G., and Rixon, F. J. (2006). Mutational analysis of the herpes simplex virus triplex protein VP19C. J. Virol. 80, 1537–1548. doi: 10.1128/JVI.80.3.1537-1548.2006
Afroz, S., Brownlie, R., Fodje, M., and van Drunen Littel-van den Hurk, S. (2016). VP8, the major tegument protein of bovine herpesvirus 1, interacts with cellular STAT1 and inhibits interferon beta signaling. J. Virol. 90, 4889–4904. doi: 10.1128/JVI.00017-16
Afroz, S., Garg, R., Fodje, M., and van Drunen Littel-van den Hurk, S. (2018). The major tegument protein of bovine herpesvirus 1, VP8, interacts with DNA damage response proteins and induces apoptosis. J. Virol. 92:e00773-18. doi: 10.1128/JVI.00773-18
Agut, H., Bonnafous, P., and Gautheret-Dejean, A. (2016). Human herpesviruses 6A, 6B, and 7. Microbiol. Spectr. 4, 157–176. doi: 10.1128/microbiolspec.DMIH2-0007-2015
Ali, A., Ohashi, M., Casco, A., Djavadian, R., Eichelberg, M., Kenney, S. C., et al. (2022). Rta is the principal activator of Epstein-Barr virus epithelial lytic transcription. PLoS Pathog. 18:e1010886. doi: 10.1371/journal.ppat.1010886
Arbuckle, J. H., Medveczky, M. M., Luka, J., Hadley, S. H., Luegmayr, A., Ablashi, D., et al. (2010). The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 107, 5563–5568. doi: 10.1073/pnas.0913586107
Arend, K. C., Ziehr, B., Vincent, H. A., and Moorman, N. J. (2016). Multiple transcripts encode full-length human cytomegalovirus IE1 and IE2 proteins during lytic infection. J. Virol. 90, 8855–8865. doi: 10.1128/JVI.00741-16
Arregi, I., Falces, J., Bañuelos, S., Urbaneja, M. A., and Taneva, S. G. (2011). The nuclear transport machinery recognizes nucleoplasmin-histone complexes. Biochemistry 50, 7104–7110. doi: 10.1021/bi2008867
Batisse, J., Manet, E., Middeldorp, J., Sergeant, A., and Gruffat, H. (2005). Epstein-Barr virus mRNA export factor EB2 is essential for intranuclear capsid assembly and production of gp350. J. Virol. 79, 14102–14111. doi: 10.1128/JVI.79.22.14102-14111.2005
Baudoux, L., Defechereux, P., Rentier, B., and Piette, J. (2000). Gene activation by Varicella-zoster virus IE4 protein requires its dimerization and involves both the arginine-rich sequence, the central part, and the carboxyl-terminal cysteine-rich region. J. Biol. Chem. 275, 32822–32831. doi: 10.1074/jbc.M001444200
Beard, P. M., Taus, N. S., and Baines, J. D. (2002). DNA cleavage and packaging proteins encoded by genes U(L)28, U(L)15, and U(L)33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76, 4785–4791. doi: 10.1128/JVI.76.10.4785-4791.2002
Bechtel, J. T., Liang, Y., Hvidding, J., and Ganem, D. (2003). Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77, 6474–6481. doi: 10.1128/JVI.77.11.6474-6481.2003
Bednenko, J., Cingolani, G., and Gerace, L. (2003). Nucleocytoplasmic transport: navigating the channel. Traffic 4, 127–135. doi: 10.1034/j.1600-0854.2003.00109.x
Bello, L. J., Davison, A. J., Glenn, M. A., Whitehouse, A., Rethmeier, N., Schulz, T. F., et al. (1999). The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80(Pt. 12), 3207–3215. doi: 10.1099/0022-1317-80-12-3207
Bernad, R., van der Velde, H., Fornerod, M., and Pickersgill, H. (2004). Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 24, 2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004
Bigalke, J. M., and Heldwein, E. E. (2016). Nuclear exodus: herpesviruses lead the way. Annu. Rev. Virol. 3, 387–409. doi: 10.1146/annurev-virology-110615-042215
Biswas, S., Bandyopadhyay, S., Dimri, U., and Patra, P. H. (2013). Bovine herpesvirus-1 (BHV-1) - a re-emerging concern in livestock: a revisit to its biology, epidemiology, diagnosis, and prophylaxis. Vet. Q. 33, 68–81. doi: 10.1080/01652176.2013.799301
Boehmer, P. E., and Lehman, I. R. (1997). Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66, 347–384. doi: 10.1146/annurev.biochem.66.1.347
Bohannon, K. P., Jun, Y., Gross, S. P., and Smith, G. A. (2013). Differential protein partitioning within the herpesvirus tegument and envelope underlies a complex and variable virion architecture. Proc. Natl. Acad. Sci. USA. 110, E1613–E1620. doi: 10.1073/pnas.1221896110
Booy, F. P., Trus, B. L., Newcomb, W. W., Brown, J. C., Conway, J. F., and Steven, A. C. (1994). Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus). Proc. Natl. Acad. Sci. USA. 91, 5652–5656. doi: 10.1073/pnas.91.12.5652
Boshoff, C., and Weiss, R. A. (2001). Epidemiology and pathogenesis of Kaposi's sarcoma-associated herpesvirus. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 356, 517–534. doi: 10.1098/rstb.2000.0778
Boyne, J. R., Colgan, K. J., and Whitehouse, A. (2008). Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4:e1000194. doi: 10.1371/journal.ppat.1000194
Boyne, J. R., and Whitehouse, A. (2006a). gamma-2 herpes virus post-transcriptional gene regulation. Clin. Microbiol. Infect. 12, 110–117. doi: 10.1111/j.1469-0691.2005.01317.x
Boyne, J. R., and Whitehouse, A. (2006b). Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA. 103, 15190–15195. doi: 10.1073/pnas.0604890103
Boyne, J. R., and Whitehouse, A. (2009). Nucleolar disruption impairs Kaposi's sarcoma-associated herpesvirus ORF57-mediated nuclear export of intronless viral mRNAs. FEBS Lett. 583, 3549–3556. doi: 10.1016/j.febslet.2009.10.040
Bradley, K. J., Bowl, M. R., Williams, S. E., Ahmad, B. N., Partridge, C. J., Patmanidi, A. L., et al. (2007). Parafibromin is a nuclear protein with a functional monopartite nuclear localization signal. Oncogene 26, 1213–1221. doi: 10.1038/sj.onc.1209893
Brandariz-Nuñez, A., Liu, T., Du, T., and Evilevitch, A. (2019). Pressure-driven release of viral genome into a host nucleus is a mechanism leading to herpes infection. Elife 8:e47212. doi: 10.7554/eLife.47212.016
Braspenning, S. E., Lebbink, R. J., Depledge, D. P., Schapendonk, C. M. E., Anderson, L. A., Verjans, G., et al. (2021). Mutagenesis of the varicella-zoster virus genome demonstrates that VLT and VLT-ORF63 proteins are dispensable for lytic infection. Viruses 13:2289. doi: 10.3390/v13112289
Braun, D. K., Batterson, W., and Roizman, B. (1984). Identification and genetic mapping of a herpes simplex virus capsid protein that binds DNA. J. Virol. 50, 645–648. doi: 10.1128/jvi.50.2.645-648.1984
Brown, J. C., and Newcomb, W. W. (2011). Herpesvirus capsid assembly: insights from structural analysis. Curr. Opin. Virol. 1, 142–149. doi: 10.1016/j.coviro.2011.06.003
Brown, S. M., MacLean, A. R., Aitken, J. D., and Harland, J. (1994). ICP34.5 influences herpes simplex virus type 1 maturation and egress from infected cells in vitro. J. Gen. Virol. 75(Pt. 12), 3679–3686. doi: 10.1099/0022-1317-75-12-3679
Cai, M., Wang, S., Long, J., and Zheng, C. (2012). Probing of the nuclear import and export signals and subcellular transport mechanism of varicella-zoster virus tegument protein open reading frame 10. Med. Microbiol. Immunol. 201, 103–111. doi: 10.1007/s00430-011-0211-4
Chan, W. M., Mak, M. C., Fung, T. K., Lau, A., Siu, W. Y., and Poon, R. Y. (2006). Ubiquitination of p53 at multiple sites in the DNA-binding domain. Mol. Cancer Res. 4, 15–25. doi: 10.1158/1541-7786.MCR-05-0097
Chao, C., Saito, S., Kang, J., Anderson, C. W., Appella, E., and Xu, Y. (2000). p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 19, 4967–4975. doi: 10.1093/emboj/19.18.4967
Cheng, G., Brett, M. E., and He, B. (2002). Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the gamma(1)34.5 protein of herpes simplex virus type 1. J. Virol. 76, 9434–9445. doi: 10.1128/JVI.76.18.9434-9445.2002
Chipuk, J. E., and Green, D. R. (2003). p53's believe it or not: lessons on transcription-independent death. J. Clin. Immunol. 23, 355–361. doi: 10.1023/A:1025365432325
Christensen, M. H., Jensen, S. B., Miettinen, J. J., Luecke, S., Prabakaran, T., Reinert, L. S., et al. (2016). HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J. 35, 1385–1399. doi: 10.15252/embj.201593458
Colletti, K. S., Xu, Y., Cei, S. A., Tarrant, M., and Pari, G. S. (2004). Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication. J. Virol. 78, 9203–9214. doi: 10.1128/JVI.78.17.9203-9214.2004
Colletti, K. S., Xu, Y., Yamboliev, I., and Pari, G. S. (2005). Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J. Biol. Chem. 280, 11955–11960. doi: 10.1074/jbc.C400603200
Compton, T., Nepomuceno, R. R., and Nowlin, D. M. (1992). Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 191, 387–395. doi: 10.1016/0042-6822(92)90200-9
Conti, E., Müller, C. W., and Stewart, M. (2006). Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 16, 237–244. doi: 10.1016/j.sbi.2006.03.010
Copeland, A. M., Newcomb, W. W., and Brown, J. C. (2009). Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83, 1660–1668. doi: 10.1128/JVI.01139-08
Couteaudier, M., and Denesvre, C. (2014). Marek's disease virus and skin interactions. Vet. Res. 45:36. doi: 10.1186/1297-9716-45-36
Cullen, B. R. (2003). Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28, 419–424. doi: 10.1016/S0968-0004(03)00142-7
da Silva, L. F., Sinani, D., and Jones, C. (2012). ICP27 protein encoded by bovine herpesvirus type 1 (bICP27) interferes with promoter activity of the bovine genes encoding beta interferon 1 (IFN-β1) and IFN-β3. Virus Res. 169, 162–168. doi: 10.1016/j.virusres.2012.07.023
Dai, Y. C., Liao, Y. T., Juan, Y. T., Cheng, Y. Y., Su, M. T., Su, Y. Z., et al. (2020). The novel nuclear targeting and BFRF1-interacting domains of BFLF2 are essential for efficient Epstein-Barr virus virion release. J. Virol. 94:e01498-19. doi: 10.1128/JVI.01498-19
Damania, B., Kenney, S. C., and Raab-Traub, N. (2022). Epstein-Barr virus: biology and clinical disease. Cell 185, 3652–3670. doi: 10.1016/j.cell.2022.08.026
Davis, D. A., Rinderknecht, A. S., Zoeteweij, J. P., Aoki, Y., Read-Connole, E. L., Tosato, G., et al. (2001). Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97, 3244–3250. doi: 10.1182/blood.V97.10.3244
Davison, A. J., and Scott, J. E. (1986). The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67(Pt. 9), 1759–1816. doi: 10.1099/0022-1317-67-9-1759
Dembowski, J. A., and DeLuca, N. A. (2018). Temporal viral genome-protein interactions define distinct stages of productive herpesviral infection. mBio 9:e01182-18. doi: 10.1128/mBio.01182-18
Depledge, D. P., Ouwendijk, W. J. D., Sadaoka, T., Braspenning, S. E., Mori, Y., Cohrs, R. J., et al. (2018a). A spliced latency-associated VZV transcript maps antisense to the viral transactivator gene 61. Nat. Commun. 9:1167. doi: 10.1038/s41467-018-03569-2
Depledge, D. P., Sadaoka, T., and Ouwendijk, W. J. D. (2018b). Molecular aspects of varicella-zoster virus latency. Viruses 10:349. doi: 10.20944/preprints201806.0036.v1
Desai, P. J., Pryce, E. N., Henson, B. W., Luitweiler, E. M., and Cothran, J. (2012). Reconstitution of the Kaposi's sarcoma-associated herpesvirus nuclear egress complex and formation of nuclear membrane vesicles by coexpression of ORF67 and ORF69 gene products. J. Virol. 86, 594–598. doi: 10.1128/JVI.05988-11
Dey, G., and Baum, B. (2021). Nuclear envelope remodelling during mitosis. Curr. Opin. Cell Biol. 70, 67–74. doi: 10.1016/j.ceb.2020.12.004
Ding, Q., Guo, H., Lin, F., Pan, W., Ye, B., and Zheng, A. C. (2010). Characterization of the nuclear import and export mechanisms of bovine herpesvirus-1 infected cell protein 27. Virus Res. 149, 95–103. doi: 10.1016/j.virusres.2010.01.009
Dingwall, C., Robbins, J., Dilworth, S. M., Roberts, B., and Richardson, W. D. (1988). The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 107, 841–849. doi: 10.1083/jcb.107.3.841
Dobrikova, E., Shveygert, M., Walters, R., and Gromeier, M. (2010). Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its subcellular distribution. J. Virol. 84, 270–279. doi: 10.1128/JVI.01740-09
Döhner, K., Cornelius, A., Serrero, M. C., and Sodeik, B. (2021). The journey of herpesvirus capsids and genomes to the host cell nucleus. Curr. Opin. Virol. 50, 147–158. doi: 10.1016/j.coviro.2021.08.005
Dominguez, G., Dambaugh, T. R., Stamey, F. R., Dewhurst, S., Inoue, N., and Pellett, P. E. (1999). Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73, 8040–8052. doi: 10.1128/JVI.73.10.8040-8052.1999
Donnelly, M., and Elliott, G. (2001). Nuclear localization and shuttling of herpes simplex virus tegument protein VP13/14. J. Virol. 75, 2566–2574. doi: 10.1128/JVI.75.6.2566-2574.2001
Donnelly, M., Verhagen, J., and Elliott, G. (2007). RNA binding by the herpes simplex virus type 1 nucleocytoplasmic shuttling protein UL47 is mediated by an N-terminal arginine-rich domain that also functions as its nuclear localization signal. J. Virol. 81, 2283–2296. doi: 10.1128/JVI.01677-06
Dooley, A. L., and O'Connor, C. M. (2020). Regulation of the MIE locus during HCMV latency and reactivation. Pathogens 9:869. doi: 10.3390/pathogens9110869
Dunn, W., Chou, C., Li, H., Hai, R., Patterson, D., Stolc, V., et al. (2003). Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA. 100, 14223–14228. doi: 10.1073/pnas.2334032100
Dünn-Kittenplon, D., Ashkenazy-Titelman, A., Kalt, I., Lellouche, J. P., Shav-Tal, Y., and Sarid, R. (2021). The portal vertex of KSHV promotes docking of capsids at the nuclear pores. Viruses 13:597. doi: 10.3390/v13040597
Dupin, N., Fisher, C., Kellam, P., Ariad, S., Tulliez, M., Franck, N., et al. (1999). Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA. 96, 4546–4551. doi: 10.1073/pnas.96.8.4546
Eisenberg, R. J., Atanasiu, D., Cairns, T. M., Gallagher, J. R., Krummenacher, C., and Cohen, G. H. (2012). Herpes virus fusion and entry: a story with many characters. Viruses 4, 800–832. doi: 10.3390/v4050800
Elder, E., and Sinclair, J. (2019). HCMV latency: what regulates the regulators? Med. Microbiol. Immunol. 208, 431–438. doi: 10.1007/s00430-019-00581-1
Fagerlund, R., Mélen, K., Kinnunen, L., and Julkunen, I. (2002). Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J. Biol. Chem. 277, 30072–30078. doi: 10.1074/jbc.M202943200
Fakhari, F. D., and Dittmer, D. P. (2002). Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 76, 6213–6223. doi: 10.1128/JVI.76.12.6213-6223.2002
Finnen, R. L., Johnston, S. M., Neron, C. E., and Banfield, B. W. (2011). Nucleocytoplasmic shuttling of the HSV-2 serine/threonine kinase Us3. Virology 417, 229–237. doi: 10.1016/j.virol.2011.06.011
Finnen, R. L., Roy, B. B., Zhang, H., and Banfield, B. W. (2010). Analysis of filamentous process induction and nuclear localization properties of the HSV-2 serine/threonine kinase Us3. Virology 397, 23–33. doi: 10.1016/j.virol.2009.11.012
Fischer, U., Huber, J., Boelens, W. C., Mattaj, I. W., and Lührmann, R. (1995). The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82, 475–483. doi: 10.1016/0092-8674(95)90436-0
Fontana, P., Dong, Y., Pi, X., Tong, A. B., Hecksel, C. W., Wang, L., et al. (2022). Structure of cytoplasmic ring of nuclear pore complex by integrative cryo-EM and AlphaFold. Science 376:eabm9326. doi: 10.1126/science.abm9326
Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I. W. (1997). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051–1060. doi: 10.1016/S0092-8674(00)80371-2
Fu, H., and Pan, D. (2024). Mechanisms of HSV gene regulation during latency and reactivation. Virology 602:110324. doi: 10.1016/j.virol.2024.110324
Fukuda, M., Asano, S., Nakamura, T., Adachi, M., Yoshida, M., Yanagida, M., et al. (1997). CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308–311. doi: 10.1038/36894
Gales, J. P., Kubina, J., Geldreich, A., and Dimitrova, M. (2020). Strength in diversity: nuclear export of viral RNAs. Viruses 12:1014. doi: 10.3390/v12091014
Gao, Y., Kagele, D., Smallenberg, K., and Pari, G. S. (2010). Nucleocytoplasmic shuttling of human cytomegalovirus UL84 is essential for virus growth. J. Virol. 84, 8484–8494. doi: 10.1128/JVI.00738-10
Gerna, G., Baldanti, F., and Revello, M. G. (2004). Pathogenesis of human cytomegalovirus infection and cellular targets. Hum. Immunol. 65, 381–386. doi: 10.1016/j.humimm.2004.02.009
Gershon, A. A., Sherman, D. L., Zhu, Z., Gabel, C. A., Ambron, R. T., and Gershon, M. D. (1994). Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68, 6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994
Goldfarb, D. S., Corbett, A. H., Mason, D. A., Harreman, M. T., and Adam, S. A. (2004). Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514. doi: 10.1016/j.tcb.2004.07.016
Gompels, U. A., Nicholas, J., Lawrence, G., Jones, M., Thomson, B. J., Martin, M. E. D., et al. (1995). The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209, 29–51. doi: 10.1006/viro.1995.1228
Gonnella, R., Farina, A., Santarelli, R., Raffa, S., Feederle, R., Bei, R., et al. (2005). Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 79, 3713–3727. doi: 10.1128/JVI.79.6.3713-3727.2005
Goodwin, D. J., Hall, K. T., Stevenson, A. J., Markham, A. F., and Whitehouse, A. (1999). The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J. Virol. 73, 10519–10524. doi: 10.1128/JVI.73.12.10519-10524.1999
Görlich, D., Henklein, P., Laskey, R. A., and Hartmann, E. (1996). A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15, 1810–1817. doi: 10.1002/j.1460-2075.1996.tb00530.x
Görlich, D., and Kutay, U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660. doi: 10.1146/annurev.cellbio.15.1.607
Granato, M., Feederle, R., Farina, A., Gonnella, R., Santarelli, R., Hub, B., et al. (2008). Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 82, 4042–4051. doi: 10.1128/JVI.02436-07
Granzow, H., Weiland, F., Jöns, A., Klupp, B. G., Karger, A., and Mettenleiter, T. C. (1997). Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71, 2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997
Gruffat, H., Marchione, R., and Manet, E. (2016). Herpesvirus late gene expression: a viral-specific pre-initiation complex is key. Front. Microbiol. 7:869. doi: 10.3389/fmicb.2016.00869
Grüter, P., Tabernero, C., von Kobbe, C., Schmitt, C., Saavedra, C., Bachi, A., et al. (1998). TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1, 649–659. doi: 10.1016/S1097-2765(00)80065-9
Gualtiero, A., Jans, D. A., Camozzi, D., Avanzi, S., Loregian, A., Ripalti, A., et al. (2013). Regulated transport into the nucleus of herpesviridae DNA replication core proteins. Viruses 5, 2210–2234. doi: 10.3390/v5092210
Guo, H., Ding, Q., Lin, F., Pan, W., Lin, J., and Zheng, A. C. (2009). Characterization of the nuclear and nucleolar localization signals of bovine herpesvirus-1 infected cell protein 27. Virus Res. 145, 312–320. doi: 10.1016/j.virusres.2009.07.024
Häge, S., Sonntag, E., Borst, E. M., Tannig, P., Seyler, L., Bäuerle, T., et al. (2020). Patterns of autologous and nonautologous interactions between core nuclear egress complex (NEC) proteins of α-, β- and γ-herpesviruses. Viruses 12:303. doi: 10.3390/v12030303
Hahn, G., Jores, R., and Mocarski, E. S. (1998). Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA. 95, 3937–3942. doi: 10.1073/pnas.95.7.3937
Hakki, M., Marshall, E. E., De Niro, K. L., and Geballe, A. P. (2006). Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J. Virol. 80, 11817–11826. doi: 10.1128/JVI.00957-06
Halford, W. P., and Schaffer, P. A. (2001). ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75, 3240–3249. doi: 10.1128/JVI.75.7.3240-3249.2001
Hamada, N., Shigeishi, H., Oka, I., Sasaki, M., Kitasaki, H., Nakamura, M., et al. (2023). Associations between oral human herpesvirus-6 and−7 and periodontal conditions in older adults. Life 13:324. doi: 10.3390/life13020324
Hambleton, S., Gershon, M. D., and Gershon, A. A. (2004). The role of the trans-Golgi network in varicella zoster virus biology. Cell. Mol. Life Sci. 61, 3047–3056. doi: 10.1007/s00018-004-4269-7
Heming, J. D., Conway, J. F., and Homa, F. L. (2017). Herpesvirus capsid assembly and DNA packaging. Adv. Anat. Embryol. Cell Biol. 223, 119–142. doi: 10.1007/978-3-319-53168-7_6
Henson, B. W., Perkins, E. M., Cothran, J. E., and Desai, P. (2009). Self-assembly of Epstein-Barr virus capsids. J. Virol. 83, 3877–3890. doi: 10.1128/JVI.01733-08
Herold, A., Truant, R., Wiegand, H., and Cullen, B. R. (1998). Determination of the functional domain organization of the importin alpha nuclear import factor. J. Cell Biol. 143, 309–318. doi: 10.1083/jcb.143.2.309
Hiriart, E., Bardouillet, L., Manet, E., Gruffat, H., Penin, F., Montserret, R., et al. (2003a). A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J. Biol. Chem. 278, 37790–37798. doi: 10.1074/jbc.M305925200
Hiriart, E., Farjot, G., Gruffat, H., Nguyen, M. V., Sergeant, A., and Manet, E. (2003b). A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278, 335–342. doi: 10.1074/jbc.M208656200
Hogue, I. B. (2021). Tegument assembly, secondary envelopment and exocytosis. Curr. Issues Mol. Biol. 42, 551–604. doi: 10.21775/cimb.042.551
Hogue, I. B., Bosse, J. B., Hu, J. R., Thiberge, S. Y., and Enquist, L. W. (2014). Cellular mechanisms of alpha herpesvirus egress: live cell fluorescence microscopy of pseudorabies virus exocytosis. PLoS Pathog. 10:e1004535. doi: 10.1371/journal.ppat.1004535
Honess, R. W., and Roizman, B. (1974). Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14, 8–19. doi: 10.1128/jvi.14.1.8-19.1974
Huang, Y., Zhang, J., Halawa, M. A., and Yao, S. (2014). Nuclear localization signals of varicella zoster virus ORF4. Virus Genes 48, 243–251. doi: 10.1007/s11262-013-1006-z
Huet, A., Huffman, J. B., Conway, J. F., and Homa, F. L. (2020). Role of the herpes simplex virus CVSC proteins at the capsid portal vertex. J. Virol. 94:e01534-20. doi: 10.1128/JVI.01534-20
Huffman, J. B., Daniel, G. R., Falck-Pedersen, E., Huet, A., Smith, G. A., Conway, J. F., et al. (2017). The C terminus of the herpes simplex virus UL25 protein is required for release of viral genomes from capsids bound to nuclear pores. J. Virol. 91:e00641-17. doi: 10.1128/JVI.00641-17
Hutten, S., and Kehlenbach, R. H. (2006). Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell. Biol. 26, 6772–6785. doi: 10.1128/MCB.00342-06
Hutt-Fletcher, L. M. (2007). Epstein-Barr virus entry. J. Virol. 81, 7825–7832. doi: 10.1128/JVI.00445-07
Hwang, W. Y., Kostiuk, V., González, D. P., Lusk, C. P., and Khokha, M. K. (2022). Kap-β2/Transportin mediates β-catenin nuclear transport in Wnt signaling. Elife 11:e70495. doi: 10.7554/eLife.70495.sa2
Ijezie, E. C., O'Dowd, J. M., Kuan, M. I., Faeth, A. R., and Fortunato, E. A. (2023). HCMV infection reduces nidogen-1 expression, contributing to impaired neural rosette development in brain organoids. J. Virol. 97:e0171822. doi: 10.1128/jvi.01718-22
Ilouze, M., Dishon, A., Kahan, T., and Kotler, M. (2006). Cyprinid herpes virus-3 (CyHV-3) bears genes of genetically distant large DNA viruses. FEBS Lett. 580, 4473–4478. doi: 10.1016/j.febslet.2006.07.013
Iwaisako, Y., and Fujimuro, M. (2024). The terminase complex of each human herpesvirus. Biol. Pharm. Bull. 47, 912–916. doi: 10.1248/bpb.b23-00717
Iwaisako, Y., Watanabe, T., Suzuki, Y., Nakano, T., and Fujimuro, M. (2023). Kaposi's sarcoma-associated herpesvirus ORF67.5 functions as a component of the terminase complex. J. Virol. 97:e0047523. doi: 10.1128/jvi.00475-23
Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I. W., and Görlich, D. (1997). The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16, 6535–6547. doi: 10.1093/emboj/16.21.6535
Jackson, B. R., Boyne, J. R., Noerenberg, M., Taylor, A., Hautbergue, G. M., Walsh, M. J., et al. (2011). An interaction between KSHV ORF57 and UIF provides mRNA-adaptor redundancy in herpesvirus intronless mRNA export. PLoS Pathog. 7:e1002138. doi: 10.1371/journal.ppat.1002138
Jeong, S. Y., and Seol, D. W. (2008). The role of mitochondria in apoptosis. BMB Rep. 41, 11–22. doi: 10.5483/BMBRep.2008.41.1.011
Jing, X., Cerveny, M., Yang, K., and He, B. (2004). Replication of herpes simplex virus 1 depends on the gamma 134.5 functions that facilitate virus response to interferon and egress in the different stages of productive infection. J. Virol. 78, 7653–7666. doi: 10.1128/JVI.78.14.7653-7666.2004
Johnson, K. E., Song, B., and Knipe, D. M. (2008). Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374, 487–494. doi: 10.1016/j.virol.2008.01.001
Johnson, K. M., Mahajan, S. S., and Wilson, A. C. (1999). Herpes simplex virus transactivator VP16 discriminates between HCF-1 and a novel family member, HCF-2. J. Virol. 73, 3930–3940. doi: 10.1128/JVI.73.5.3930-3940.1999
Johnson, L. A., Li, L., and Sandri-Goldin, R. M. (2009). The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J. Virol. 83, 6335–6346. doi: 10.1128/JVI.00375-09
Johnson, L. A., and Sandri-Goldin, R. M. (2009). Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. J. Virol. 83, 1184–1192. doi: 10.1128/JVI.02010-08
Jones, T. R., and Lee, S. W. (2004). An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J. Virol. 78, 1488–1502. doi: 10.1128/JVI.78.3.1488-1502.2004
Jovasevic, V., Liang, L., and Roizman, B. (2008). Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82, 3311–3319. doi: 10.1128/JVI.01919-07
Juillard, F., Hiriart, E., Sergeant, N., Vingtdeux-Didier, V., Drobecq, H., Sergeant, A., et al. (2009). Epstein-Barr virus protein EB2 contains an N-terminal transferable nuclear export signal that promotes nucleocytoplasmic export by directly binding TAP/NXF1. J. Virol. 83, 12759–12768. doi: 10.1128/JVI.01276-09
Kaffman, A., Rank, N. M., and O'Shea, E. K. (1998). Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12, 2673–2683. doi: 10.1101/gad.12.17.2673
Kalderon, D., Roberts, B. L., Richardson, W. D., and Smith, A. E. (1984). A short amino acid sequence able to specify nuclear location. Cell 39(Pt. 2), 499–509. doi: 10.1016/0092-8674(84)90457-4
Katano, H. (2018). Pathological features of kaposi's sarcoma-associated herpesvirus infection. Adv. Exp. Med. Biol. 1045, 357–376. doi: 10.1007/978-981-10-7230-7_16
Kato, A., Liu, Z., Minowa, A., Imai, T., Tanaka, M., Sugimoto, K., et al. (2011). Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells. J. Virol. 85, 9599–9613. doi: 10.1128/JVI.00845-11
Kehlenbach, R. H., Dickmanns, A., Kehlenbach, A., Guan, T., and Gerace, L. (1999). A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145, 645–657. doi: 10.1083/jcb.145.4.645
Kim, J. A., Choi, M. S., Min, J. S., Kang, I., Oh, J., Kim, J. C., et al. (2017). HSV-1 ICP27 represses NF-κB activity by regulating Daxx sumoylation. BMB Rep. 50, 275–280. doi: 10.5483/BMBRep.2017.50.5.010
Kimura, M., and Imamoto, N. (2014). Biological significance of the importin-β family-dependent nucleocytoplasmic transport pathways. Traffic 15, 727–748. doi: 10.1111/tra.12174
Kinchington, P. R., Fite, K., Seman, A., and Turse, S. E. (2001). Virion association of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, requires expression of the VZV open reading frame 66 protein kinase. J. Virol. 75, 9106–9113. doi: 10.1128/JVI.75.19.9106-9113.2001
Kinchington, P. R., Hougland, J. K., Arvin, A. M., Ruyechan, W. T., and Hay, J. (1992). The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J. Virol. 66, 359–366. doi: 10.1128/jvi.66.1.359-366.1992
Kitagawa, R., Bakkenist, C. J., McKinnon, P. J., and Kastan, M. B. (2004). Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 18, 1423–1438. doi: 10.1101/gad.1200304
Klass, C. M., Krug, L. T., Pozharskaya, V. P., and Offermann, M. K. (2005). The targeting of primary effusion lymphoma cells for apoptosis by inducing lytic replication of human herpesvirus 8 while blocking virus production. Blood 105, 4028–4034. doi: 10.1182/blood-2004-09-3569
Klupp, B. G., Granzow, H., Klopfleisch, R., Fuchs, W., Kopp, M., Lenk, M., et al. (2005). Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79, 3831–3840. doi: 10.1128/JVI.79.6.3831-3840.2005
Knipe, D. M. (1989). The role of viral and cellular nuclear proteins in herpes simplex virus replication. Adv. Virus Res. 37, 85–123. doi: 10.1016/S0065-3527(08)60833-7
Knockenhauer, K. E., and Schwartz, T. U. (2016). The nuclear pore complex as a flexible and dynamic gate. Cell 164, 1162–1171. doi: 10.1016/j.cell.2016.01.034
Kobayashi, J., and Matsuura, Y. (2013). Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p. J. Mol. Biol. 425, 1852–1868. doi: 10.1016/j.jmb.2013.02.035
Koffa, M. D., Clements, J. B., Izaurralde, E., Wadd, S., Wilson, S. A., Mattaj, I. W., et al. (2023). Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 42:e114021. doi: 10.15252/embj.2023114021
Kuang, E., Tang, Q., Maul, G. G., and Zhu, F. (2008). Activation of p90 ribosomal S6 kinase by ORF45 of Kaposi's sarcoma-associated herpesvirus and its role in viral lytic replication. J. Virol. 82, 1838–1850. doi: 10.1128/JVI.02119-07
Kutay, U., Bischoff, F. R., Kostka, S., Kraft, R., and Görlich, D. (1997). Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90, 1061–1071. doi: 10.1016/S0092-8674(00)80372-4
Kwok, H., and Chiang, A. K. (2016). From conventional to next generation sequencing of Epstein-Barr virus genomes. Viruses 8:60. doi: 10.3390/v8030060
Lake, C. M., and Hutt-Fletcher, L. M. (2004). The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320, 99–106. doi: 10.1016/j.virol.2003.11.018
Lange, A., Mills, R. E., Lange, C. J., Stewart, M., Devine, S. E., and Corbett, A. H. (2007). Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282, 5101–5105. doi: 10.1074/jbc.R600026200
Le Hir, H., Nott, A., and Moore, M. J. (2003). How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28, 215–220. doi: 10.1016/S0968-0004(03)00052-5
Lebrun, M., Thelen, N., Thiry, M., Riva, L., Ote, I., Condé, C., et al. (2014). Varicella-zoster virus induces the formation of dynamic nuclear capsid aggregates. Virology 454–455, 311–327. doi: 10.1016/j.virol.2014.02.023
Lee, B. J., Cansizoglu, A. E., Süel, K. E., Louis, T. H., Zhang, Z., and Chook, Y. M. (2006). Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543–558. doi: 10.1016/j.cell.2006.05.049
Leenadevi, T., and Dalziel, R. G. (2009). The alcelaphine herpesvirus-1 ORF 57 encodes a nuclear shuttling protein. Vet. Res. Commun. 33, 409–419. doi: 10.1007/s11259-008-9187-y
Leib, D. A., Coen, D. M., Bogard, C. L., Hicks, K. A., Yager, D. R., Knipe, D. M., et al. (1989). Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63, 759–768. doi: 10.1128/jvi.63.2.759-768.1989
Leigh, K. E., Sharma, M., Mansueto, M. S., Boeszoermenyi, A., Filman, D. J., Hogle, J. M., et al. (2015). Structure of a herpesvirus nuclear egress complex subunit reveals an interaction groove that is essential for viral replication. Proc. Natl. Acad. Sci. USA. 112, 9010–9015. doi: 10.1073/pnas.1511140112
Lengyel, J., Guy, C., Leong, V., Borge, S., and Rice, S. A. (2002). Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: importance of the leucine-rich nuclear export signal and RGG Box RNA-binding domain. J. Virol. 76, 11866–11879. doi: 10.1128/JVI.76.23.11866-11879.2002
Li, D. J., Verma, D., and Swaminathan, S. (2012a). Binding of cellular export factor REF/Aly by Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 protein is not required for efficient KSHV lytic replication. J. Virol. 86, 9866–9874. doi: 10.1128/JVI.01190-12
Li, J., Guo, Y., Deng, Y., Hu, L., Li, B., Deng, S., et al. (2021). Subcellular localization of Epstein-Barr virus BLLF2 and its underlying mechanisms. Front. Microbiol. 12:672192. doi: 10.3389/fmicb.2021.672192
Li, M., Chen, T., Zou, X., Xu, Z., Wang, Y., Wang, P., et al. (2018). Characterization of the nucleocytoplasmic transport mechanisms of Epstein-Barr virus BFLF2. Cell. Physiol. Biochem. 51, 1500–1517. doi: 10.1159/000495641
Li, M., Wang, S., Cai, M., Guo, H., and Zheng, C. (2011a). Characterization of molecular determinants for nucleocytoplasmic shuttling of PRV UL54. Virology 417, 385–393. doi: 10.1016/j.virol.2011.06.004
Li, M., Wang, S., Cai, M., and Zheng, C. (2011b). Identification of nuclear and nucleolar localization signals of pseudorabies virus (PRV) early protein UL54 reveals that its nuclear targeting is required for efficient production of PRV. J. Virol. 85, 10239–10251. doi: 10.1128/JVI.05223-11
Li, S., Xie, Y., Yu, C., Zheng, C., and Xu, Z. (2024). The battle between host antiviral innate immunity and immune evasion by cytomegalovirus. Cell. Mol. Life Sci. 81:341. doi: 10.1007/s00018-024-05369-y
Li, W., Deng, X., and Chen, J. (2022). RNA-binding proteins in regulating mRNA stability and translation: roles and mechanisms in cancer. Semin. Cancer Biol. 86(Pt. 2), 664–677. doi: 10.1016/j.semcancer.2022.03.025
Li, X., and Zhu, F. (2009). Identification of the nuclear export and adjacent nuclear localization signals for ORF45 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 83, 2531–2539. doi: 10.1128/JVI.02209-08
Li, Y., Zhao, L., Wang, S., Xing, J., and Zheng, C. (2012b). Identification of a novel NLS of herpes simplex virus type 1 (HSV-1) VP19C and its nuclear localization is required for efficient production of HSV-1. J. Gen. Virol. 93(Pt. 9), 1869–1875. doi: 10.1099/vir.0.042697-0
Lischka, P., Rauh, C., Mueller, R., and Stamminger, T. (2006a). Human cytomegalovirus UL84 protein contains two nuclear export signals and shuttles between the nucleus and the cytoplasm. J. Virol. 80, 10274–10280. doi: 10.1128/JVI.00995-06
Lischka, P., Rosorius, O., Trommer, E., and Stamminger, T. (2001). A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20, 7271–7283. doi: 10.1093/emboj/20.24.7271
Lischka, P., Toth, Z., Thomas, M., Mueller, R., and Stamminger, T. (2006b). The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 26, 1631–1643. doi: 10.1128/MCB.26.5.1631-1643.2006
Liu, C., Cheng, A., Wang, M., Chen, S., Jia, R., Zhu, D., et al. (2016). Characterization of nucleocytoplasmic shuttling and intracellular localization signals in Duck Enteritis virus UL54. Biochimie 127, 86–94. doi: 10.1016/j.biochi.2016.05.003
Liu, C. Y., Jin, M., Guo, H., Zhao, H. Z., Hou, L. N., Yang, Y., et al. (2022). Concurrent gene insertion, deletion, and inversion during the construction of a novel attenuated BoHV-1 using CRISPR/Cas9 genome editing. Vet. Sci. 9:166. doi: 10.3390/vetsci9040166
Liu, F., and Zhou, Z. H. (2007). “Comparative virion structures of human herpesviruses,” in Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, eds. A. Arvin, G. Campadelli-Fiume, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley, et al. (Cambridge: Cambridge University Press Copyright © Cambridge University Press 2007), 3.
Liu, H., Luo, M., and Wen, J. K. (2014a). mRNA stability in the nucleus. J. Zhejiang Univ. Sci. B 15, 444–454. doi: 10.1631/jzus.B1400088
Liu, X., Ma, Y., Voss, K., van Gent, M., Chan, Y. K., Gack, M. U., et al. (2021). The herpesvirus accessory protein γ134.5 facilitates viral replication by disabling mitochondrial translocation of RIG-I. PLoS Pathog. 17:e1009446. doi: 10.1371/journal.ppat.1009446
Liu, Y., Cui, Z., Zhang, Z., Wei, H., Zhou, Y., Wang, M., et al. (2009). The tegument protein UL94 of human cytomegalovirus as a binding partner for tegument protein pp28 identified by intracellular imaging. Virology 388, 68–77. doi: 10.1016/j.virol.2009.03.007
Liu, Y., Zhang, Z., Zhao, X., Wei, H., Deng, J., Cui, Z., et al. (2012). Human cytomegalovirus UL94 is a nucleocytoplasmic shuttling protein containing two NLSs and one NES. Virus Res. 166, 31–42. doi: 10.1016/j.virusres.2012.02.023
Liu, Z., Kato, A., Shindo, K., Noda, T., Sagara, H., Kawaoka, Y., et al. (2014b). Herpes simplex virus 1 UL47 interacts with viral nuclear egress factors UL31, UL34, and Us3 and regulates viral nuclear egress. J. Virol. 88, 4657–4667. doi: 10.1128/JVI.00137-14
Lv, Y., Zhou, S., Gao, S., and Deng, H. (2019). Remodeling of host membranes during herpesvirus assembly and egress. Protein Cell 10, 315–326. doi: 10.1007/s13238-018-0577-9
Lye, M. F., Wilkie, A. R., Filman, D. J., Hogle, J. M., and Coen, D. M. (2017). Getting to and through the inner nuclear membrane during herpesvirus nuclear egress. Curr. Opin. Cell Biol. 46, 9–16. doi: 10.1016/j.ceb.2016.12.007
Majerciak, V., Lu, M., Li, X., and Zheng, Z. M. (2014). Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA 20, 1747–1758. doi: 10.1261/rna.045500.114
Majerciak, V., Uranishi, H., Kruhlak, M., Pilkington, G. R., Massimelli, M. J., Bear, J., et al. (2011). Kaposi's sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J. Virol. 85, 1528–1540. doi: 10.1128/JVI.01709-10
Majerciak, V., Yang, W., Zheng, J., Zhu, J., and Zheng, Z. M. (2019). A genome-wide Epstein-Barr virus polyadenylation map and its antisense RNA to EBNA. J. Virol. 93:e01593-18. doi: 10.1128/JVI.01593-18
Malik, A. K., Shao, L., Shanley, J. D., and Weller, S. K. (1996). Intracellular localization of the herpes simplex virus type-1 origin binding protein, UL9. Virology 224, 380–389. doi: 10.1006/viro.1996.0545
Malik, P., Blackbourn, D. J., and Clements, J. B. (2004). The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279, 33001–33011. doi: 10.1074/jbc.M313008200
Malik, P., Tabarraei, A., Kehlenbach, R. H., Korfali, N., Iwasawa, R., Graham, S. V., et al. (2012). Herpes simplex virus ICP27 protein directly interacts with the nuclear pore complex through Nup62, inhibiting host nucleocytoplasmic transport pathways. J. Biol. Chem. 287, 12277–12292. doi: 10.1074/jbc.M111.331777
Mallet, P. L., and Bachand, F. (2013). A proline-tyrosine nuclear localization signal (PY-NLS) is required for the nuclear import of fission yeast PAB2, but not of human PABPN1. Traffic 14, 282–294. doi: 10.1111/tra.12036
Malone, C. L., Vesole, D. H., and Stinski, M. F. (1990). Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64, 1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990
Mao, H., and Rosenthal, K. S. (2002). An N-terminal arginine-rich cluster and a proline-alanine-threonine repeat region determine the cellular localization of the herpes simplex virus type 1 ICP34.5 protein and its ligand, protein phosphatase 1. J. Biol. Chem. 277, 11423–11431. doi: 10.1074/jbc.M111553200
Mattaj, I. W., and Englmeier, L. (1998). Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67, 265–306. doi: 10.1146/annurev.biochem.67.1.265
McGeoch, D. J., Cook, S., Dolan, A., Jamieson, F. E., and Telford, E. A. (1995). Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247, 443–458. doi: 10.1006/jmbi.1995.0152
McGeoch, D. J., Cunningham, C., McIntyre, G., and Dolan, A. (1991). Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J. Gen. Virol. 72(Pt. 12), 3057–3075. doi: 10.1099/0022-1317-72-12-3057
McGeoch, D. J., Dalrymple, M. A., Davison, A. J., Dolan, A., Frame, M. C., McNab, D., et al. (1988). The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69(Pt. 7), 1531–1574. doi: 10.1099/0022-1317-69-7-1531
McKenzie, J., and El-Guindy, A. (2015). Epstein-Barr virus lytic cycle reactivation. Curr. Top. Microbiol. Immunol. 391, 237–261. doi: 10.1007/978-3-319-22834-1_8
McLean, G., Rixon, F., Langeland, N., Haarr, L., and Marsden, H. (1990). Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J. Gen. Virol. 71(Pt. 12), 2953–2960. doi: 10.1099/0022-1317-71-12-2953
McVoy, M. A., and Adler, S. P. (1994). Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 68, 1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994
Mears, W. E., Lam, V., and Rice, S. A. (1995). Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 69, 935–947. doi: 10.1128/jvi.69.2.935-947.1995
Mears, W. E., and Rice, S. A. (1998). The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242, 128–137. doi: 10.1006/viro.1997.9006
Mettenleiter, T. C., Müller, F., Granzow, H., and Klupp, B. G. (2013). The way out: what we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 15, 170–178. doi: 10.1111/cmi.12044
Meyer, T., and Vinkemeier, U. (2004). Nucleocytoplasmic shuttling of STAT transcription factors. Eur. J. Biochem. 271, 4606–4612. doi: 10.1111/j.1432-1033.2004.04423.x
Michael, W. M. (2000). Nucleocytoplasmic shuttling signals: two for the price of one. Trends Cell Biol. 10, 46–50. doi: 10.1016/S0962-8924(99)01695-5
Milbradt, J., Auerochs, S., Sevvana, M., Muller, Y. A., Sticht, H., and Marschall, M. (2012). Specific residues of a conserved domain in the N terminus of the human cytomegalovirus pUL50 protein determine its intranuclear interaction with pUL53. J. Biol. Chem. 287, 24004–24016. doi: 10.1074/jbc.M111.331207
Miller, G., El-Guindy, A., Countryman, J., Ye, J., and Gradoville, L. (2007). Lytic cycle switches of oncogenic human gammaherpesviruses. Adv. Cancer Res. 97, 81–109. doi: 10.1016/S0065-230X(06)97004-3
Miller, N., and Hutt-Fletcher, L. M. (1992). Epstein-Barr virus enters B cells and epithelial cells by different routes. J. Virol. 66, 3409–3414. doi: 10.1128/jvi.66.6.3409-3414.1992
Miranda-Saksena, M., Denes, C. E., Diefenbach, R. J., and Cunningham, A. L. (2018). Infection and transport of herpes simplex virus type 1 in neurons: role of the cytoskeleton. Viruses 10:92. doi: 10.3390/v10020092
Morrison, E. E., Wang, Y. F., and Meredith, D. M. (1998). Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72, 7108–7114. doi: 10.1128/JVI.72.9.7108-7114.1998
Mosammaparast, N., and Pemberton, L. F. (2004). Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14, 547–556. doi: 10.1016/j.tcb.2004.09.004
Muñoz-Fontela, C., Collado, M., Rodriguez, E., García, M. A., Alvarez-Barrientos, A., Arroyo, J., et al. (2005). Identification of a nuclear export signal in the KSHV latent protein LANA2 mediating its export from the nucleus. Exp. Cell Res. 311, 96–105. doi: 10.1016/j.yexcr.2005.08.022
Muñoz-Fontela, C., Rodríguez, E., Nombela, C., Arroyo, J., and Rivas, C. (2003). Characterization of the bipartite nuclear localization signal of protein LANA2 from Kaposi's sarcoma-associated herpesvirus. Biochem. J. 374(Pt. 2), 545–550. doi: 10.1042/bj20021890
Murata, T., Sugimoto, A., Inagaki, T., Yanagi, Y., Watanabe, T., Sato, Y., et al. (2021). Molecular basis of Epstein-Barr virus latency establishment and lytic reactivation. Viruses 13:2344. doi: 10.3390/v13122344
Myster, F., Gong, M. J., Javaux, J., Suárez, N. M., Wilkie, G. S., Connelley, T., et al. (2020). Alcelaphine herpesvirus 1 genes A7 and A8 regulate viral spread and are essential for malignant catarrhal fever. PLoS Pathog. 16:e1008405. doi: 10.1371/journal.ppat.1008405
Nealon, K., Newcomb, W. W., Pray, T. R., Craik, C. S., Brown, J. C., and Kedes, D. H. (2001). Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J. Virol. 75, 2866–2878. doi: 10.1128/JVI.75.6.2866-2878.2001
Nehrbass, U., and Blobel, G. (1996). Role of the nuclear transport factor p10 in nuclear import. Science 272, 120–122. doi: 10.1126/science.272.5258.120
Nekorchuk, M., Han, Z., Hsieh, T. T., and Swaminathan, S. (2007). Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 81, 9990–9998. doi: 10.1128/JVI.00896-07
Neuber, S., Wagner, K., Goldner, T., Lischka, P., Steinbrueck, L., Messerle, M., et al. (2017). Mutual interplay between the human cytomegalovirus terminase subunits pUL51, pUL56, and pUL89 promotes terminase complex formation. J. Virol. 91:e02384-16. doi: 10.1128/JVI.02384-16
Newcomb, W. W., Juhas, R. M., Thomsen, D. R., Homa, F. L., Burch, A. D., Weller, S. K., et al. (2001). The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75, 10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001
Newcomb, W. W., Trus, B. L., Booy, F. P., Steven, A. C., Wall, J. S., and Brown, J. C. (1993). Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232, 499–511. doi: 10.1006/jmbi.1993.1406
Nguyen, N. L., Loveland, A. N., and Gibson, W. (2008). Nuclear localization sequences in cytomegalovirus capsid assembly proteins (UL80 proteins) are required for virus production: inactivating NLS1, NLS2, or both affects replication to strikingly different extents. J. Virol. 82, 5381–5389. doi: 10.1128/JVI.02697-07
Nii, S., Uno, F., Yoshida, M., and Akatsuka, K. (1998). [Structure and assembly of human beta herpesviruses]. Nippon. Rinsho 56, 22–28.
Okoye, M. E., Sexton, G. L., Huang, E., McCaffery, J. M., and Desai, P. (2006). Functional analysis of the triplex proteins (VP19C and VP23) of herpes simplex virus type 1. J. Virol. 80, 929–940. doi: 10.1128/JVI.80.2.929-940.2006
Ote, I., Lebrun, M., Vandevenne, P., Bontems, S., Medina-Palazon, C., Manet, E., et al. (2009). Varicella-zoster virus IE4 protein interacts with SR proteins and exports mRNAs through the TAP/NXF1 pathway. PLoS ONE 4:e7882. doi: 10.1371/journal.pone.0007882
Ote, I., Piette, J., and Sadzot-Delvaux, C. (2010). The Varicella-Zoster virus IE4 protein: a conserved member of the herpesviral mRNA export factors family and a potential alternative target in antiherpetic therapies. Biochem. Pharmacol. 80, 1973–1980. doi: 10.1016/j.bcp.2010.07.011
Ouwendijk, W. J. D., Depledge, D. P., Rajbhandari, L., Lenac Rovis, T., Jonjic, S., Breuer, J., et al. (2020). Varicella-zoster virus VLT-ORF63 fusion transcript induces broad viral gene expression during reactivation from neuronal latency. Nat. Commun. 11:6324. doi: 10.1101/2020.09.11.294280
Owen, D. J., Crump, C. M., and Graham, S. C. (2015). Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses 7, 5084–5114. doi: 10.3390/v7092861
Packard, J. E., and Dembowski, J. A. (2021). HSV-1 DNA replication-coordinated regulation by viral and cellular factors. Viruses 13:2015. doi: 10.3390/v13102015
Pan, S., Liu, X., Ma, Y., Cao, Y., and He, B. (2018). Herpes simplex virus 1 γ(1)34.5 protein inhibits STING activation that restricts viral replication. J. Virol. 92:e01015-18. doi: 10.1128/JVI.01015-18
Pasdeloup, D., Blondel, D., Isidro, A. L., and Rixon, F. J. (2009). Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83, 6610–6623. doi: 10.1128/JVI.02655-08
Patel, V., Dahlroth, S. L., Rajakannan, V., Ho, H. T., Cornvik, T., and Nordlund, P. (2015). Structure of the C-terminal domain of the multifunctional ICP27 protein from herpes simplex virus 1. J. Virol. 89, 8828–8839. doi: 10.1128/JVI.00441-15
Person, S., and Desai, P. (1998). Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frames of herpes simplex virus type 1 but are not formed in a null mutant of UL38 (VP19C). Virology 242, 193–203. doi: 10.1006/viro.1997.9005
Pestka, S., Krause, C. D., and Walter, M. R. (2004). Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202, 8–32. doi: 10.1111/j.0105-2896.2004.00204.x
Phelan, A., Dunlop, J., and Clements, J. B. (1996). Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J. Virol. 70, 5255–5265. doi: 10.1128/jvi.70.8.5255-5265.1996
Qiu, Z., Zhu, J., Harms, J. S., Friedrichsen, J., and Splitter, G. A. (2005). Bovine herpesvirus VP22 induces apoptosis in neuroblastoma cells by upregulating the expression ratio of Bax to Bcl-2. Hum. Gene Ther. 16, 101–108. doi: 10.1089/hum.2005.16.101
Radhakrishnan, A., and Green, R. (2016). Connections underlying translation and mRNA stability. J. Mol. Biol. 428, 3558–3564. doi: 10.1016/j.jmb.2016.05.025
Rathbun, M. M., and Szpara, M. L. (2021). A holistic perspective on herpes simplex virus (HSV) ecology and evolution. Adv. Virus Res. 110, 27–57. doi: 10.1016/bs.aivir.2021.05.001
Reed, R., and Hurt, E. (2002). A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108, 523–531. doi: 10.1016/S0092-8674(02)00627-X
Rice, S. A., and Lam, V. (1994). Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J. Virol. 68, 823–833. doi: 10.1128/jvi.68.2.823-833.1994
Rigoutsos, I., Novotny, J., Huynh, T., Chin-Bow, S. T., Parida, L., Platt, D., et al. (2003). In silico pattern-based analysis of the human cytomegalovirus genome. J. Virol. 77, 4326–4344. doi: 10.1128/JVI.77.7.4326-4344.2003
Rivas, C., Thlick, A. E., Parravicini, C., Moore, P. S., and Chang, Y. (2001). Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75, 429–438. doi: 10.1128/JVI.75.1.429-438.2001
Rixon, F. J., Addison, C., McGregor, A., Macnab, S. J., Nicholson, P., Preston, V. G., et al. (1996). Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Neurovirol. J. Gen. Virol. 77(Pt. 9), 2251–2260. doi: 10.1099/0022-1317-77-9-2251
Robbins, J., Dilworth, S. M., Laskey, R. A., and Dingwall, C. (1991). Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64, 615–623. doi: 10.1016/0092-8674(91)90245-T
Roizman, B., Zhou, G., and Du, T. (2011). Checkpoints in productive and latent infections with herpes simplex virus 1: conceptualization of the issues. J. Neurovirol. 17, 512–517. doi: 10.1007/s13365-011-0058-x
Roller, R. J., and Johnson, D. C. (2021). Herpesvirus nuclear egress across the outer nuclear membrane. Viruses 13:2356. doi: 10.3390/v13122356
Ruhge, L. L., Huet, A. G. E., Conway, J. F., and Smith, G. A. (2018). The apical region of the herpes simplex virus major capsid protein promotes capsid maturation. J. Virol. 92:e00821-18. doi: 10.1128/JVI.00821-18
Russo, J. J., Bohenzky, R. A., Chien, M. C., Chen, J., Yan, M., Maddalena, D., et al. (1996). Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA. 93, 14862–14867. doi: 10.1073/pnas.93.25.14862
Ryckman, B. J., Jarvis, M. A., Drummond, D. D., Nelson, J. A., and Johnson, D. C. (2006). Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 80, 710–722. doi: 10.1128/JVI.80.2.710-722.2006
Sandri-Goldin, R. M. (1998). ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12, 868–879. doi: 10.1101/gad.12.6.868
Sandri-Goldin, R. M. (2001). Nuclear export of herpes virus RNA. Curr. Top. Microbiol. Immunol. 259, 2–23. doi: 10.1007/978-3-642-56597-7_1
Sandri-Goldin, R. M. (2008). The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13, 5241–5256. doi: 10.2741/3078
Santos, C. A. (2016). Cytomegalovirus and other β-herpesviruses. Semin. Nephrol. 36, 351–361. doi: 10.1016/j.semnephrol.2016.05.012
Sathiyamoorthy, K., Chen, J., Longnecker, R., and Jardetzky, T. S. (2017). The complexity in herpesvirus entry. Curr. Opin. Virol. 24, 97–104. doi: 10.1016/j.coviro.2017.04.006
Schippers, T., Jarosinski, K., and Osterrieder, N. (2015). The ORF012 gene of Marek's disease virus type 1 produces a spliced transcript and encodes a novel nuclear phosphoprotein essential for virus growth. J. Virol. 89, 1348–1363. doi: 10.1128/JVI.02687-14
Schreiner, S., and Wodrich, H. (2013). Virion factors that target Daxx to overcome intrinsic immunity. J. Virol. 87, 10412–10422. doi: 10.1128/JVI.00425-13
Schweininger, J., Kriegel, M., Häge, S., Conrad, M., Alkhashrom, S., Lösing, J., et al. (2022). The crystal structure of the varicella-zoster Orf24-Orf27 nuclear egress complex spotlights multiple determinants of herpesvirus subfamily specificity. J. Biol. Chem. 298:101625. doi: 10.1016/j.jbc.2022.101625
Seewald, M. J., Körner, C., Wittinghofer, A., and Vetter, I. R. (2002). RanGAP mediates GTP hydrolysis without an arginine finger. Nature 415, 662–666. doi: 10.1038/415662a
Seo, J. Y., and Britt, W. J. (2006). Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 80, 5611–5626. doi: 10.1128/JVI.02630-05
Seo, T., Park, J., Lim, C., and Choe, J. (2004). Inhibition of nuclear factor kappaB activity by viral interferon regulatory factor 3 of Kaposi's sarcoma-associated herpesvirus. Oncogene 23, 6146–6155. doi: 10.1038/sj.onc.1207807
Shitrit, A., Nisnevich, V., Rozenshtein, N., Kobo, H., Phan, H. V., Tay, S., et al. (2023). Shared sequence characteristics identified in non-canonical rearrangements of HSV-1 genomes. J. Virol. 97:e0095523. doi: 10.1128/jvi.00955-23
Shu, M., Taddeo, B., and Roizman, B. (2013a). The nuclear-cytoplasmic shuttling of virion host shutoff RNase is enabled by pUL47 and an embedded nuclear export signal and defines the sites of degradation of AU-rich and stable cellular mRNAs. J. Virol. 87, 13569–13578. doi: 10.1128/JVI.02603-13
Shu, M., Taddeo, B., Zhang, W., and Roizman, B. (2013b). Selective degradation of mRNAs by the HSV host shutoff RNase is regulated by the UL47 tegument protein. Proc. Natl. Acad. Sci. USA. 110, E1669–E1675. doi: 10.1073/pnas.1305475110
Sijmons, S., Van Ranst, M., and Maes, P. (2014). Genomic and functional characteristics of human cytomegalovirus revealed by next-generation sequencing. Viruses 6, 1049–1072. doi: 10.3390/v6031049
Singh, M., Fraefel, C., Bello, L. J., Lawrence, W. C., and Schwyzer, M. (1996). Identification and characterization of BICP27, an early protein of bovine herpesvirus 1 which may stimulate mRNA 3' processing. J. Gen. Virol. 77(Pt. 4), 615–625. doi: 10.1099/0022-1317-77-4-615
Siomi, H., and Dreyfuss, G. (1995). A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129, 551–560. doi: 10.1083/jcb.129.3.551
Siomi, M. C., Eder, P. S., Kataoka, N., Wan, L., Liu, Q., and Dreyfuss, G. (1997). Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 138, 1181–1192. doi: 10.1083/jcb.138.6.1181
Sokolowski, M., Scott, J. E., Heaney, R. P., Patel, A. H., and Clements, J. B. (2003). Identification of herpes simplex virus RNAs that interact specifically with regulatory protein ICP27 in vivo. J. Biol. Chem. 278, 33540–33549. doi: 10.1074/jbc.M302063200
Soliman, T. M., Sandri-Goldin, R. M., and Silverstein, S. J. (1997). Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71, 9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997
Soniat, M., and Chook, Y. M. (2016). Karyopherin-β2 recognition of a PY-NLS variant that lacks the proline-tyrosine motif. Structure 24, 1802–1809. doi: 10.1016/j.str.2016.07.018
Sonntag, E., Milbradt, J., Svrlanska, A., Strojan, H., Häge, S., Kraut, A., et al. (2017). Protein kinases responsible for the phosphorylation of the nuclear egress core complex of human cytomegalovirus. J. Gen. Virol. 98, 2569–2581. doi: 10.1099/jgv.0.000931
Srinivas, K. P., Depledge, D. P., Abebe, J. S., Rice, S. A., Mohr, I., and Wilson, A. C. (2021). Widespread remodeling of the m(6)A RNA-modification landscape by a viral regulator of RNA processing and export. Proc. Natl. Acad. Sci. USA. 118:e2104805118. doi: 10.1073/pnas.2104805118
Srivastava, A., Srivastava, A., and Singh, R. K. (2023). Insight into the epigenetics of Kaposi's sarcoma-associated herpesvirus. Int. J. Mol. Sci. 24:14955. doi: 10.3390/ijms241914955
Stade, K., Ford, C. S., Guthrie, C., and Weis, K. (1997). Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90, 1041–1050. doi: 10.1016/S0092-8674(00)80370-0
Stamminger, T. (2008). Interactions of human cytomegalovirus proteins with the nuclear transport machinery. Curr. Top. Microbiol. Immunol. 325, 167–185. doi: 10.1007/978-3-540-77349-8_10
Stark, G. R., and Darnell, J. E. Jr. (2012). The JAK-STAT pathway at twenty. Immunity 36, 503–514. doi: 10.1016/j.immuni.2012.03.013
Stewart, M. (2007). Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208. doi: 10.1038/nrm2114
Strambio-De-Castillia, C., Niepel, M., and Rout, M. P. (2010). The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11, 490–501. doi: 10.1038/nrm2928
Strang, B. L., Bender, B. J., Sharma, M., Pesola, J. M., Sanders, R. L., Spector, D. H., et al. (2012). A mutation deleting sequences encoding the amino terminus of human cytomegalovirus UL84 impairs interaction with UL44 and capsid localization. J. Virol. 86, 11066–11077. doi: 10.1128/JVI.01379-12
Sucharita, S., Krishnagopal, A., and van Drunen Littel-van den Hurk, S. (2023). Comprehensive analysis of the tegument proteins involved in capsid transport and virion morphogenesis of alpha, beta and gamma herpesviruses. Viruses 15:2058. doi: 10.3390/v15102058
Sugimoto, K., Uema, M., Sagara, H., Tanaka, M., Sata, T., Hashimoto, Y., et al. (2008). Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82, 5198–5211. doi: 10.1128/JVI.02681-07
Suzich, J. B., and Cliffe, A. R. (2018). Strength in diversity: understanding the pathways to herpes simplex virus reactivation. Virology 522, 81–91. doi: 10.1016/j.virol.2018.07.011
Taddeo, B., and Roizman, B. (2006). The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J. Virol. 80, 9341–9345. doi: 10.1128/JVI.01008-06
Taddeo, B., Zhang, W., and Roizman, B. (2006). The U(L)41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. USA. 103, 2827–2832. doi: 10.1073/pnas.0510712103
Takeshima, K., Arii, J., Maruzuru, Y., Koyanagi, N., Kato, A., and Kawaguchi, Y. (2019). Identification of the capsid binding site in the herpes simplex virus 1 nuclear egress complex and its role in viral primary envelopment and replication. J. Virol. 93:e01290-19. doi: 10.1128/JVI.01290-19
Tarrant-Elorza, M., Rossetto, C. C., and Pari, G. S. (2014). Maintenance and replication of the human cytomegalovirus genome during latency. Cell Host Microbe 16, 43–54. doi: 10.1016/j.chom.2014.06.006
Tatman, J. D., Preston, V. G., Nicholson, P., Elliott, R. M., and Rixon, F. J. (1994). Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J. Gen. Virol. 75(Pt. 5), 1101–1113. doi: 10.1099/0022-1317-75-5-1101
Thompson, R. L., and Sawtell, N. M. (2006). Evidence that the herpes simplex virus type 1 ICP0 protein does not initiate reactivation from latency in vivo. J. Virol. 80, 10919–10930. doi: 10.1128/JVI.01253-06
Thorley-Lawson, D. A. (2015). EBV persistence–introducing the virus. Curr. Top. Microbiol. Immunol. 390(Pt. 1), 151–209. doi: 10.1007/978-3-319-22822-8_8
Thorley-Lawson, D. A., Hawkins, J. B., Tracy, S. I., and Shapiro, M. (2013). The pathogenesis of Epstein-Barr virus persistent infection. Curr. Opin. Virol. 3, 227–232. doi: 10.1016/j.coviro.2013.04.005
Tognarelli, E. I., Palomino, T. F., Corrales, N., Bueno, S. M., Kalergis, A. M., and González, P. A. (2019). Herpes simplex virus evasion of early host antiviral responses. Front. Cell. Infect. Microbiol. 9:127. doi: 10.3389/fcimb.2019.00127
Toth, Z., Lischka, P., and Stamminger, T. (2006). RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 34, 1237–1249. doi: 10.1093/nar/gkl007
Toth, Z., and Stamminger, T. (2008). The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front Biosci 13, 2939–2949. doi: 10.2741/2899
Tunnicliffe, R. B., Schacht, M., Levy, C., Jowitt, T. A., Sandri-Goldin, R. M., and Golovanov, A. P. (2015). The structure of the folded domain from the signature multifunctional protein ICP27 from herpes simplex virus-1 reveals an intertwined dimer. Sci. Rep. 5:11234. doi: 10.1038/srep11234
Twyffels, L., Gueydan, C., and Kruys, V. (2014). Transportin-1 and transportin-2: protein nuclear import and beyond. FEBS Lett. 588, 1857–1868. doi: 10.1016/j.febslet.2014.04.023
Uprichard, S. L., and Knipe, D. M. (1996). Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J. Virol. 70, 1969–1980. doi: 10.1128/jvi.70.3.1969-1980.1996
Varmuza, S. L., and Smiley, J. R. (1985). Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell 41, 793–802. doi: 10.1016/S0092-8674(85)80060-X
Vasilenko, N. L., Snider, M., Labiuk, S. L., Lobanov, V. A., Babiuk, L. A., and van Drunen Littel-van den Hurk, S. (2012). Bovine herpesvirus-1 VP8 interacts with DNA damage binding protein-1 (DDB1) and is monoubiquitinated during infection. Virus Res. 167, 56–66. doi: 10.1016/j.virusres.2012.04.005
Verbeek, R., Vandekerckhove, L., and Van Cleemput, J. (2024). Update on human herpesvirus 7 pathogenesis and clinical aspects as a roadmap for future research. J. Virol. 98:e0043724. doi: 10.1128/jvi.00437-24
Verhagen, J., Donnelly, M., and Elliott, G. (2006). Characterization of a novel transferable CRM-1-independent nuclear export signal in a herpesvirus tegument protein that shuttles between the nucleus and cytoplasm. J. Virol. 80, 10021–10035. doi: 10.1128/JVI.01322-06
Verpooten, D., Ma, Y., Hou, S., Yan, Z., and He, B. (2009). Control of TANK-binding kinase 1-mediated signaling by the gamma(1)34.5 protein of herpes simplex virus 1. J. Biol. Chem. 284, 1097–1105. doi: 10.1074/jbc.M805905200
Villanueva-Valencia, J. R., Tsimtsirakis, E., and Evilevitch, A. (2021). Role of HSV-1 capsid vertex-specific component (CVSC) and viral terminal DNA in capsid docking at the nuclear pore. Viruses 13:2515. doi: 10.3390/v13122515
Visalli, M. A., House, B. L., Lahrman, F. J., and Visalli, R. J. (2015). Intermolecular complementation between two varicella-zoster virus pORF30 terminase domains essential for DNA encapsidation. J. Virol. 89, 10010–10022. doi: 10.1128/JVI.01313-15
Visalli, R. J., Schwartz, A. M., Patel, S., and Visalli, M. A. (2019). Identification of the Epstein Barr virus portal. Virology 529, 152–159. doi: 10.1016/j.virol.2019.01.003
Wang, L., Li, M., Cai, M., Xing, J., Wang, S., and Zheng, C. (2012). A PY-nuclear localization signal is required for nuclear accumulation of HCMV UL79 protein. Med. Microbiol. Immunol. 201, 381–387. doi: 10.1007/s00430-012-0243-4
Wang, S., Song, X., Rajewski, A., Santiskulvong, C., and Ghiasi, H. (2023). Stacking the odds: multiple sites for HSV-1 latency. Sci. Adv. 9:eadf4904. doi: 10.1126/sciadv.adf4904
Wang, X., Zhu, N., Li, W., Zhu, F., Wang, Y., and Yuan, Y. (2015). Mono-ubiquitylated ORF45 mediates association of KSHV particles with internal lipid rafts for viral assembly and egress. PLoS Pathog. 11:e1005332. doi: 10.1371/journal.ppat.1005332
Watson, M. J., and Thoreen, C. C. (2022). Measuring mRNA decay with roadblock-qPCR. Curr. Protoc. 2:e344. doi: 10.1002/cpz1.344
Weed, D. J., and Nicola, A. V. (2017). Herpes simplex virus membrane fusion. Adv. Anat. Embryol. Cell Biol. 223, 29–47. doi: 10.1007/978-3-319-53168-7_2
Weis, K., Ryder, U., and Lamond, A. I. (1996). The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 15, 1818–1825. doi: 10.1002/j.1460-2075.1996.tb00531.x
Wen, W., Meinkoth, J. L., Tsien, R. Y., and Taylor, S. S. (1995). Identification of a signal for rapid export of proteins from the nucleus. Cell 82, 463–473. doi: 10.1016/0092-8674(95)90435-2
Whitley, R. J., Kimberlin, D. W., and Roizman, B. (1998). Herpes simplex viruses. Clin. Infect. Dis. 26, 541–553; quiz 554-545. doi: 10.1086/514600
Williams, B. J., Boyne, J. R., Goodwin, D. J., Roaden, L., Hautbergue, G. M., Wilson, S. A., et al. (2005). The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 387(Pt. 2), 295–308. doi: 10.1042/BJ20041223
Wisner, T. W., and Johnson, D. C. (2004). Redistribution of cellular and herpes simplex virus proteins from the trans-golgi network to cell junctions without enveloped capsids. J. Virol. 78, 11519–11535. doi: 10.1128/JVI.78.21.11519-11535.2004
Wysocka, J., and Herr, W. (2003). The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28, 294–304. doi: 10.1016/S0968-0004(03)00088-4
Xu, A., Qin, C., Lang, Y., Wang, M., Lin, M., Li, C., et al. (2015). A simple and rapid approach to manipulate pseudorabies virus genome by CRISPR/Cas9 system. Biotechnol. Lett. 37, 1265–1272. doi: 10.1007/s10529-015-1796-2
Xu, D., Farmer, A., and Chook, Y. M. (2010). Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr. Opin. Struct. Biol. 20, 782–790. doi: 10.1016/j.sbi.2010.09.008
Xu, J. J., Gao, F., Wu, J. Q., Zheng, H., Tong, W., Cheng, X. F., et al. (2020). Characterization of nucleocytoplasmic shuttling of pseudorabies virus protein UL46. Front. Vet. Sci. 7:484. doi: 10.3389/fvets.2020.00484
Xu, Y., Colletti, K. S., and Pari, G. S. (2002). Human cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments. J. Virol. 76, 8931–8938. doi: 10.1128/JVI.76.17.8931-8938.2002
Zhang, K., Afroz, S., Brownlie, R., Snider, M., and van Drunen Littel-van den Hurk, S. (2015). Regulation and function of phosphorylation on VP8, the major tegument protein of bovine herpesvirus 1. J. Virol. 89, 4598–4611. doi: 10.1128/JVI.03180-14
Zhang, K., Brownlie, R., Snider, M., and van Drunen Littel-van den Hurk, S. (2016). Phosphorylation of bovine herpesvirus 1 VP8 plays a role in viral DNA encapsidation and is essential for its cytoplasmic localization and optimal virion incorporation. J. Virol. 90, 4427–4440. doi: 10.1128/JVI.00219-16
Zhang, Y., Liu, W., Li, Z., Kumar, V., Alvarez-Cabrera, A. L., Leibovitch, E. C., et al. (2019). Atomic structure of the human herpesvirus 6B capsid and capsid-associated tegument complexes. Nat. Commun. 10:5346. doi: 10.1038/s41467-019-13064-x
Zhao, L., and Zheng, C. (2012). The first identified nucleocytoplasmic shuttling herpesviral capsid protein: herpes simplex virus type 1 VP19C. PLoS ONE 7:e41825. doi: 10.1371/journal.pone.0041825
Zheng, C., Brownlie, R., Babiuk, L. A., and van Drunen Littel-van den Hurk, S. (2004). Characterization of nuclear localization and export signals of the major tegument protein VP8 of bovine herpesvirus-1. Virology 324, 327–339. doi: 10.1016/j.virol.2004.03.042
Zheng, C., Brownlie, R., Babiuk, L. A., and van Drunen Littel-van den Hurk, S. (2005). Characterization of the nuclear localization and nuclear export signals of bovine herpesvirus 1 VP22. J. Virol. 79, 11864–11872. doi: 10.1128/JVI.79.18.11864-11872.2005
Zheng, C., Lin, F., Wang, S., and Xing, J. (2011). A novel virus-encoded nucleocytoplasmic shuttling protein: the UL3 protein of herpes simplex virus type 1. J. Virol. Methods 177, 206–210. doi: 10.1016/j.jviromet.2011.08.004
Zheng, H. H., Fu, P. F., Chen, H. Y., and Wang, Z. Y. (2022). Pseudorabies virus: from pathogenesis to prevention strategies. Viruses 14:1638. doi: 10.3390/v14081638
Zheng, Z. M. (2003). Split genes and their expression in Kaposi's sarcoma-associated herpesvirus. Rev. Med. Virol. 13, 173–184. doi: 10.1002/rmv.387
Zhong, W., Wang, H., Herndier, B., and Ganem, D. (1996). Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA. 93, 6641–6646. doi: 10.1073/pnas.93.13.6641
Zhou, X., Jiang, W., Liu, Z., Liu, S., and Liang, X. (2017). Virus infection and death receptor-mediated apoptosis. Viruses 9:316. doi: 10.3390/v9110316
Zhu, F. X., King, S. M., Smith, E. J., Levy, D. E., and Yuan, Y. (2002). A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. USA. 99, 5573–5578. doi: 10.1073/pnas.082420599
Zhu, J., Qiu, Z., Wiese, C., Ishii, Y., Friedrichsen, J., Rajashekara, G., et al. (2005). Nuclear and mitochondrial localization signals overlap within bovine herpesvirus 1 tegument protein VP22. J. Biol. Chem. 280, 16038–16044. doi: 10.1074/jbc.M500054200
Zhu, Z., Gershon, M. D., Hao, Y., Ambron, R. T., Gabel, C. A., and Gershon, A. A. (1995). Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J. Virol. 69, 7951–7959. doi: 10.1128/jvi.69.12.7951-7959.1995
Zielke, B., Thomas, M., Giede-Jeppe, A., Müller, R., and Stamminger, T. (2011). Characterization of the betaherpesviral pUL69 protein family reveals binding of the cellular mRNA export factor UAP56 as a prerequisite for stimulation of nuclear mRNA export and for efficient viral replication. J. Virol. 85, 1804–1819. doi: 10.1128/JVI.01347-10
Keywords: herpesvirus, shuttling proteins, nuclear localization signal, nuclear export signal, life cycle, innate immune escape, apoptosis
Citation: Cao H, Wang M, Cheng A, Tian B, Yang Q, Ou X, Sun D, He Y, Wu Z, Zhao X, Wu Y, Zhang S, Huang J, Yu Y, Zhang L, Chen S, Liu M, Zhu D and Jia R (2025) The functions of herpesvirus shuttling proteins in the virus lifecycle. Front. Microbiol. 16:1515241. doi: 10.3389/fmicb.2025.1515241
Received: 23 October 2024; Accepted: 10 January 2025;
Published: 05 February 2025.
Edited by:
Hye-Ra Lee, Korea University, Republic of KoreaReviewed by:
A. Alwin Prem Anand, University of Tübingen, GermanyMadavaraju Krishnaraju, Northwestern University, United States
Ji-Seung Yoo, Kyungpook National University, Republic of Korea
Copyright © 2025 Cao, Wang, Cheng, Tian, Yang, Ou, Sun, He, Wu, Zhao, Wu, Zhang, Huang, Yu, Zhang, Chen, Liu, Zhu and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingshu Wang, bXNod2FuZ0AxNjMuY29t
 Huijun Cao
Huijun Cao Mingshu Wang
Mingshu Wang Anchun Cheng
Anchun Cheng Bin Tian
Bin Tian Qiao Yang
Qiao Yang Xumin Ou
Xumin Ou Di Sun
Di Sun Yu He1,2,3,4,5
Yu He1,2,3,4,5 Shaqiu Zhang
Shaqiu Zhang Juan Huang
Juan Huang Shun Chen
Shun Chen Mafeng Liu
Mafeng Liu Dekang Zhu
Dekang Zhu