- 1The Fourth School of Clinical Medicine, Zhejiang Chinese Medical University (Hangzhou First People’s Hospital), Hangzhou, China
- 2Affiliated Hangzhou First People’s Hospital, Westlake University School of Medicine, Hangzhou, China
The demand for early disease detection, treatment monitoring, and personalized medicine is increasing, making it more imperative than ever to create effective, accurate, portable, intelligent, multifunctional diagnostic equipment. Bacteria possess a remarkable perception of their surroundings and have the capacity to adapt by altering the expression of specific genes. Bacteria interact with target substances and produce detectable signals in response to their presence or concentration. This unique property has been harnessed in the development of bacterial biosensors. Due to groundbreaking advancements in synthetic biology, genetic engineering now enables the creation of bacteria tailored with exceptional detecting traits. In addition to meeting a wide range of application needs, this allows quick and precise detection in intricate settings and offers a strong technological basis for early disease diagnosis and treatment monitoring. This article reviews the applications and recent advancements of bacterial biosensors in the medical field and discusses the challenges and obstacles that remain in their research and application.
1 Introduction
With the rapid advancement of biotechnology and growing demands for affordable healthcare, particularly in resource limited settings, traditional diagnostic methods reliant on complex instrumentation such as PCR and mass spectrometry face challenges in achieving rapid, sensitive, and portable testing solutions (Law et al., 2015). Bacterial biosensors, which employ engineered bacteria as programmable sensing elements, offer a cost effective and scalable alternative. These devices detect target analytes including pathogens, metabolites and biomolecules, through synthetic genetic circuits, converting biological signals into quantifiable outputs such as electrical currents or fluorescence via electrochemical or optical interfaces (Nakamura et al., 2008). Recent advances in synthetic biology, such as CRISPR-based gene circuits and synthetic circuit, have enabled precise tuning of bacterial sensing pathways. Such innovations expand medical applications from early disease diagnosis to personalized therapeutics (Riglar and Silver, 2018). However, critical barriers such as biosafety concerns and functional stability hinder clinical translation. This article broadly examines recent advances in bacterial biosensor design, highlights their emerging roles in precision medicine, and critically discusses unresolved technical and regulatory hurdles.
2 Principles and mechanisms of bacterial biosensors
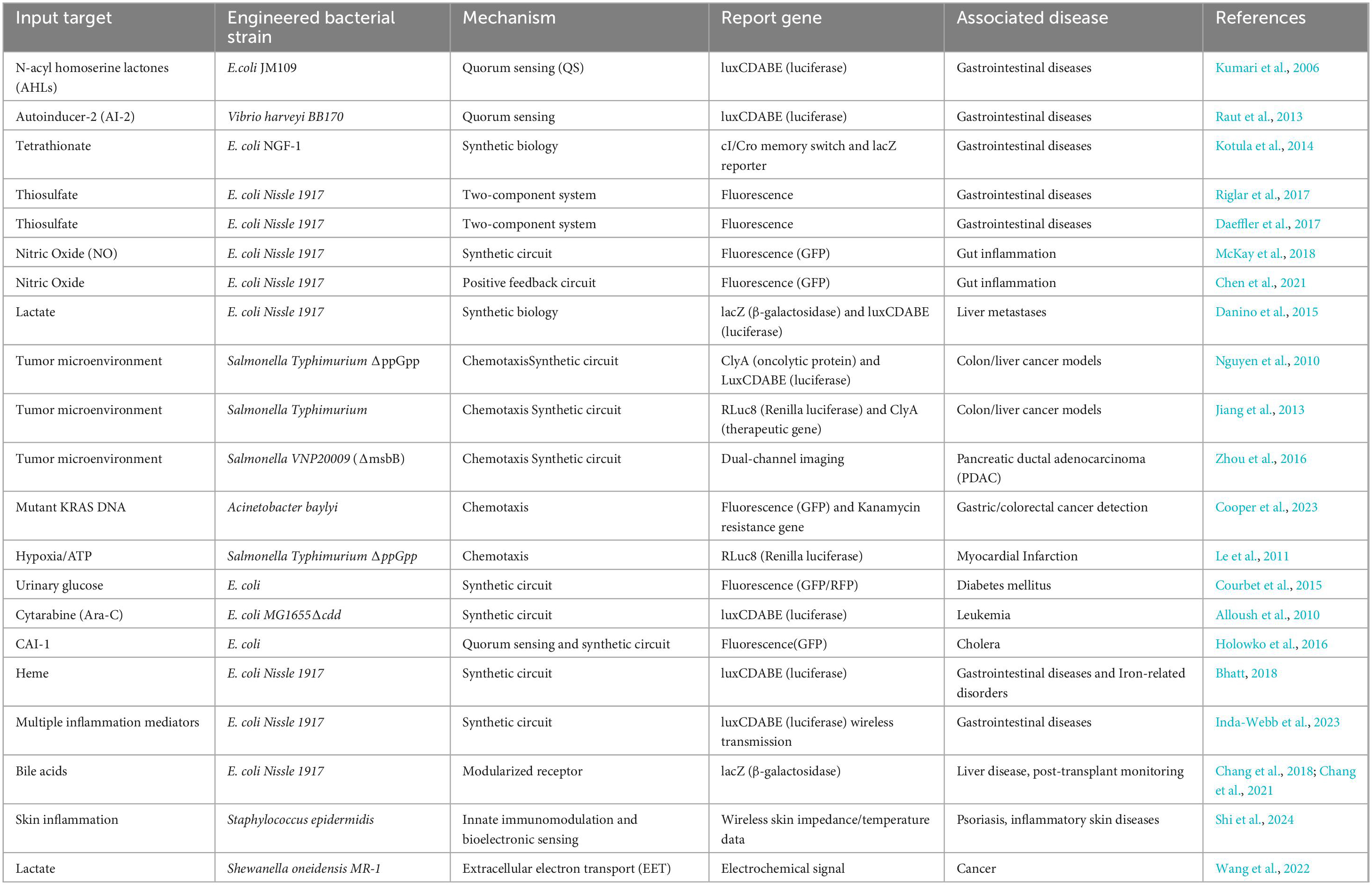
Bacteria possess remarkable environmental adaptability, allowing them to detect and react to environmental alterations, such as variations in chemical concentrations, temperature, and pH levels (Andrianantoandro et al., 2006; Park et al., 2013). This unique trait makes bacteria one of the most suitable biological recognition components for biosensors (Soltani Zarrin et al., 2018). Bacterial biosensors achieve specific detection of target molecules or environmental factors by converting biological responses into quantifiable signals. Their functionality relies on the coordinated operation of three core components: the input module (sensing unit), signal transduction module (processing unit), and output module (response unit) (Chen et al., 2023). The input module of bacterial biosensors functions as the sensing element responsible for the specific recognition of and response to target signals, relying on selective molecular interactions between biomolecules (Saltepe and Kehribar et al., 2018). This module employs both naturally occurring components such as transcription factors and membrane receptors and engineered constructs, including aptamers or nucleic acid switches, to directly capture external stimuli (e.g., chemical signals, physical cues, or biomarkers) and convert them into intracellular signals amenable to processing (Ding et al., 2021). Upon target recognition, the sensing elements activate the signal transduction module through distinct triggering mechanisms: conformational changes, induced dimerization, conditional stabilization, or enzymatic reactions (Miller et al., 2022). For example, conformational changes occur when transcription factors or membrane receptors bind target molecules, inducing structural rearrangements that initiate downstream responses (Serganov and Nudler, 2013; Zhong et al., 2016); induced dimerization involves the binding of two monomeric molecules under specific conditions to form a functional dimer, thereby activating signaling pathways (Capra and Laub, 2012; Lazar and Tabor, 2021); conditional stabilization refers to molecular stability regulated by environmental parameters such as temperature (Schuster and Greenberg, 2008); and enzymatic amplification leverages enzyme-catalyzed substrate to product conversions to enhance detection sensitivity (Miller et al., 2022). Acting as the central hub, the signal transduction module bridges the input and output modules by transforming initial detection signals into processable intracellular signals while enabling amplification, integration, or logical operations. Natural bacterial systems predominantly utilize pathways such as two-component systems (TCS), in which histidine kinases (HK) recognize extracellular signals via their sensor domains, undergo autophosphorylation at histidine residues, and transfer phosphate groups to aspartate residues on response regulators (RR), thereby activating RR’s DNA-binding or enzymatic functions to regulate gene expression (Lazar and Tabor, 2021; Salis et al., 2009; Tanna et al., 2021); quorum sensing (QS), a density-dependent communication mechanism mediated by autoinducers (AIs) that accumulate to threshold concentrations to trigger QS-regulated behaviors such as bioluminescence, virulence factor production, or biofilm formation (Kumari et al., 2006; Lee et al., 2013); and chemotaxis systems, which direct bacterial motility toward nutrient-rich environments or away from harmful substances (Karmakar, 2021). The output module translates processed intracellular signals into detectable and quantifiable physical, chemical, or biological responses (Lopreside et al., 2019). Optical outputs such as fluorescence (green fluorescent protein, GFP) or bioluminescence (luciferase) rely on promoter-driven reporter gene expression, where fluorescence intensity correlates linearly with target concentration to achieve high-sensitivity detection (Belkin, 2003; Roda et al., 2011; van der Meer and Belkin, 2010). Chromogenic outputs exploit enzymatic cleavage of substrates, including X-gal hydrolysis by lacZ-encoded β-galactosidase to produce a blue chromogen (Mascher et al., 2004), enabling semi-quantitative visual or spectrophotometric analysis without specialized equipment, a feature particularly advantageous for point-of-care or resource-limited settings. Electrochemical outputs detect target-induced changes in redox reactions, ion concentrations, or charge distributions at electrode surfaces, with signals quantified through current, voltage, or impedance measurements (Johnson et al., 2017; Zhu et al., 2023). Collectively, these modular frameworks enable bacterial biosensors to address diverse biomedical challenges by balancing sensitivity, specificity, and practicality for applications spanning diagnostics, environmental monitoring, and therapeutic evaluation.
Synthetic biology holds significant potential to advance the development of bacterial biosensors. A cornerstone of this advancement lies in the strategic deployment of gene-editing tools, particularly CRISPR-Cas9 technology (Doudna and Charpentier, 2017). By enabling targeted knockout of genes responsible for non-specific responses or background interference, CRISPR-Cas9 enhances sensor specificity through noise reduction (de la Fuente-Núñez and Lu, 2017). Complementarily, gene knock-in techniques integrate functional genetic elements to amplify both sensitivity and specificity, permitting reliable detection even at ultralow target concentrations. The redesign of endogenous signaling circuits constitutes another critical strategy (Jung et al., 2018). Native bacterial signal transduction pathways, composed of receptor proteins, transcription factors, and effector proteins, are systematically engineered to maintain their intrinsic efficiency while enhancing analytical versatility (Jung et al., 2018; Raut et al., 2013; Skjoedt et al., 2016). For example, modifying receptor binding sites to accommodate structurally analogous targets expands the detectable analyte spectrum, thereby addressing diverse diagnostic needs. Synthetic genetic circuit construction further introduces novel functionalities (Sedlmayer et al., 2018; Wang et al., 2013). Modular components, including AND, OR, and NOR logic gates, enable coordinated multi-signal processing, allowing biosensors to function with high precision in complex matrices (Bonnet et al., 2013; Tang et al., 2021; Wang et al., 2011). Memory modules such as transcription factor-based toggle switches and recombinase-mediated memory circuits confer bacteria with programmable “memory storage,” recording prior exposure to specific analytes (Riglar et al., 2017). This capability facilitates accelerated, context-dependent responses upon re-exposure, which is essential for monitoring dynamic fluctuations in environmental conditions or disease biomarkers (Archer et al., 2012; Park et al., 2013). To optimize performance, circuit design integrates signal amplification coupled with feedback control mechanisms. Positive feedback loops serve as biological amplifiers (Wan et al., 2019), heightening sensitivity to low-abundance signals, whereas negative feedback mechanisms act as stabilizers, preventing signal oversaturation and preserving cellular homeostasis (Jia et al., 2019). This dual regulatory framework ensures sustained operational stability across prolonged and variable detection scenarios (Tiwari et al., 2013). Collectively, these innovations position synthetic biology-driven bacterial biosensors as robust tools for applications spanning environmental monitoring to clinical diagnostics Figure 1.
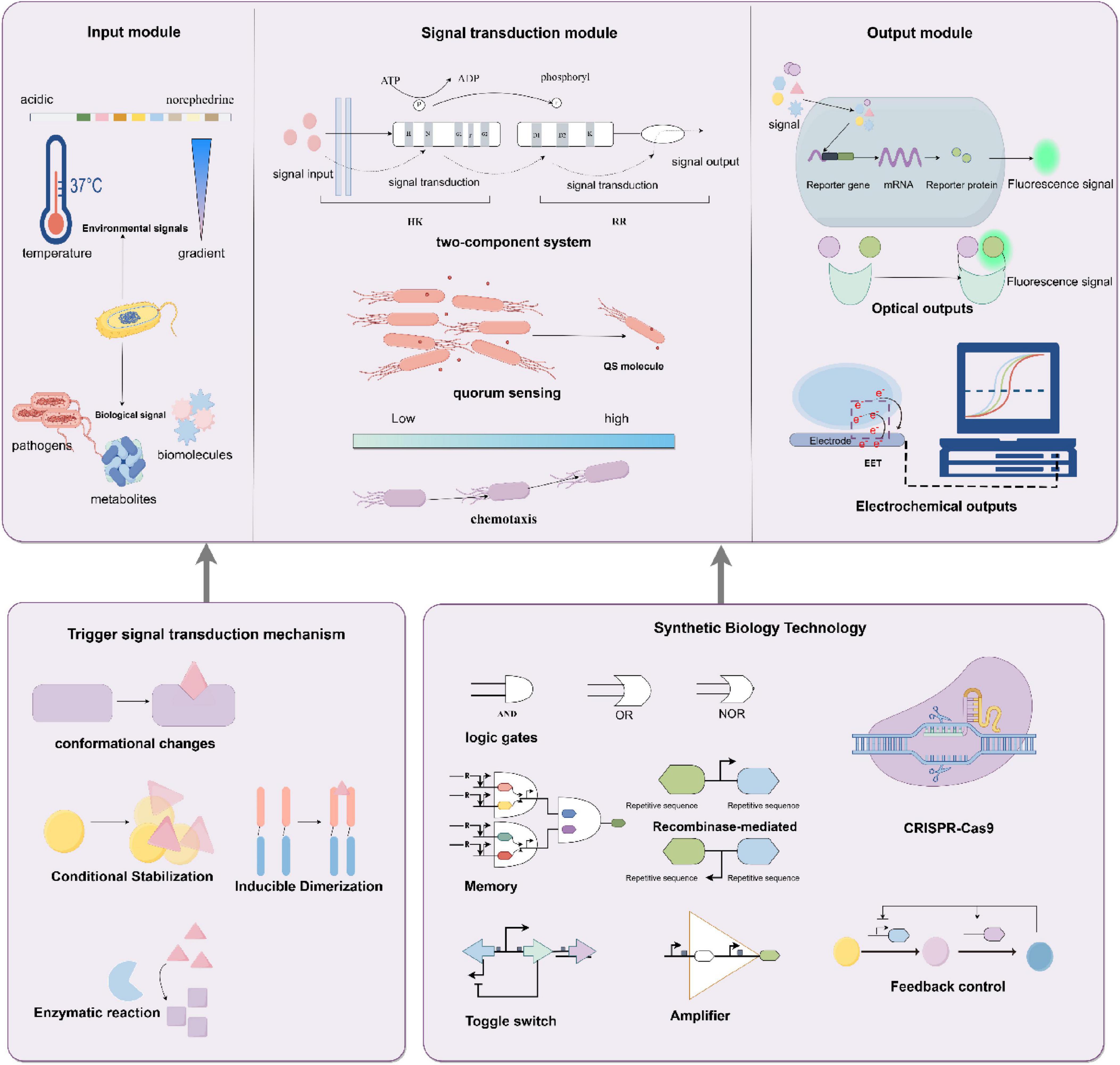
Figure 1. Principles and mechanisms of bacterial biosensors. The input module receives environmental signals (e.g., temperature gradients, acidic environments) and biological signals (e.g., pathogens, metabolites), triggering signal transduction through mechanisms such as conformational changes, conditional stabilization, or enzymatic reactions. The signal transduction module, which comprises two-component systems, quorum sensing, and chemotaxis, processes these signals, while the output module converts them into optical (e.g., fluorescence) or electrochemical responses. Synthetic biology techniques (e.g., logic gates, memory switches, feedback regulation, and CRISPR-Cas9 gene editing) further optimize the sensor’s detection sensitivity and dynamic response range. By Figdraw.
3 Applications of bacterial biosensors in disease detection and treatment monitoring
Bacterial biosensors are an innovative biotechnological tool with significant potential across various fields (Plotnikova et al., 2016). In environmental monitoring, they are used to detect organic pollutants, heavy metal ions, and other harmful substances in soil and water (Plotnikova et al., 2016). These sensors employ engineered bacteria to generate specific biological reactions to pollutants, which are then converted into measurable signals (Coelho et al., 2015). For instance, bacterial biosensors can detect organic pollutants like phenolic compounds and polycyclic aromatic hydrocarbons (Plotnikova et al., 2016), as well as heavy metals such as mercury (Chen et al., 2020), arsenic (Hui et al., 2023), and chromium (Francisco et al., 2019). Their high sensitivity and selectivity enable rapid detection and early warning of environmental pollutants. In food safety, bacterial biosensors offer a faster, more cost-effective alternative to traditional methods (Gao et al., 2023), detecting harmful substances like antibiotics, hydrogen peroxide, pesticide residues, and pathogenic bacteria (Poikulainen et al., 2020). These sensors allow for quick screening and early warning, ensuring food safety and reliability (Liao et al., 2006). Additionally, bacterial biosensors are gaining attention in the medical field for their role in diagnosing gastrointestinal diseases, cancer, and other applications Table 1. We will now explore their specific uses and potential value in these areas.
3.1 Applications of bacterial biosensors in gastrointestinal diseases
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder driven by genetic susceptibility, immune dysregulation, and gut microbiota dysbiosis (Bourgonje et al., 2020; Reese et al., 2018; York, 2023). Quorum sensing (QS), a bacterial communication mechanism mediated by signaling molecules such as AI-2, AHLs, and AIP, plays a dual role in gut homeostasis (Ge et al., 2020; Raut et al., 2013). Early studies established the clinical relevance of QS molecules in gastrointestinal diseases. For instance, Kumari et al. engineered E. coli JM109 as a biosensor by introducing Pseudomonas aeruginosa QS regulatory systems (LasR/RhlR) through plasmids pSB406 and pSB1075, coupled with the luxCDABE bioluminescent reporter. This system enabled sensitive detection of N-acyl homoserine lactones (AHLs) in human saliva and stool samples, achieving a detection limit of 1 × 10–9 M without extensive sample preparation. Their work demonstrated that AHLs are present in both healthy individuals and Crohn’s disease patients, with levels correlating to microbial dysbiosis (Kumari et al., 2006, 2008). The use of E. coli as a chassis organism highlighted its adaptability in heterologous QS circuit engineering, though the role of AHLs in IBD-associated inflammation remained to be fully elucidated. While QS coordinates beneficial microbial interactions, its dysregulation may promote pathogenic behaviors and amplify inflammation in IBD (Chang, 2015; Zhang J. et al., 2023). To leverage QS for IBD monitoring, Nilesh Raut’s team developed a biosensor using Vibrio harveyi BB170. In this system, AI-2 binds to the cytoplasmic receptor LuxP, triggering a phosphorylation cascade (LuxQ→ LuxU→ LuxO) that activates the luxCDABE promoter, producing bioluminescence proportional to AI-2 concentration (Kumari et al., 2006; Raut et al., 2013; Figure 2). Feces provide limited spatiotemporal resolution for monitoring intestinal inflammation, despite reflecting gut microbiota dynamics (Chang, 2015; Naydich et al., 2019). To overcome this, synthetic biology strategies deploy engineered bacteria as living biosensors capable of in situ signal recording (Chang, 2015; Naydich et al., 2019). Kotula et al. (2014) designed and constructed a two-part system that comprised a “trigger element” and a “memory element.” Kotula et al.’s (2014) team pioneered a two-component memory system in E. coli, comprising a trigger element (tetracycline-responsive promoter) and a λ phage-derived cI/Cro memory switch. In tetracycline-treated mice, the trigger activated Cro expression, irreversibly switching the bacterial state from cI (silent) to Cro (active), which persisted for ≥ 5 days without inducer (Burrill et al., 2012; Kotula et al., 2014; Moon et al., 2012). This system laid the foundation for chronic gut monitoring. Thiosulfate and tetrathionate is a transient product of reactive oxygen species (ROS), which are produced during inflammation (Winter and Bäumler, 2014; Winter et al., 2013). Expanding on this, Riglar et al. (2017) integrated the Salmonella Typhimurium PttrBCA promoter (responsive to tetrathionate) with the λ memory module and lacZ reporter in E. coli NGF-1. Upon detecting tetrathionate, β-galactosidase hydrolyzed X-gal to generate blue fecal colonies. Remarkably, this sensor functioned for > 6 months in mice, demonstrating sustained in vivo operation (Riglar et al., 2017). In parallel, Daeffler et al. (2017) engineered an E. coli Nissle 1917 biosensor using a Shewanella oneidensis-derived two-component system (TsrA/TsrR) to detect thiosulfate, a biomarker of gut inflammation. The sensor activates a fluorescent reporter via phosphorylation cascades, achieving a detection limit of 50 μM within 2 h. In murine colitis models, thiosulfate levels correlated strongly with histopathological scores (r = 0.68, p < 0.01) (Daeffler et al., 2017). These advances demonstrate the potential of synthetic biology in developing multiplexed, long-term gut surveillance systems capable of non-invasively tracking dynamic inflammatory processes.
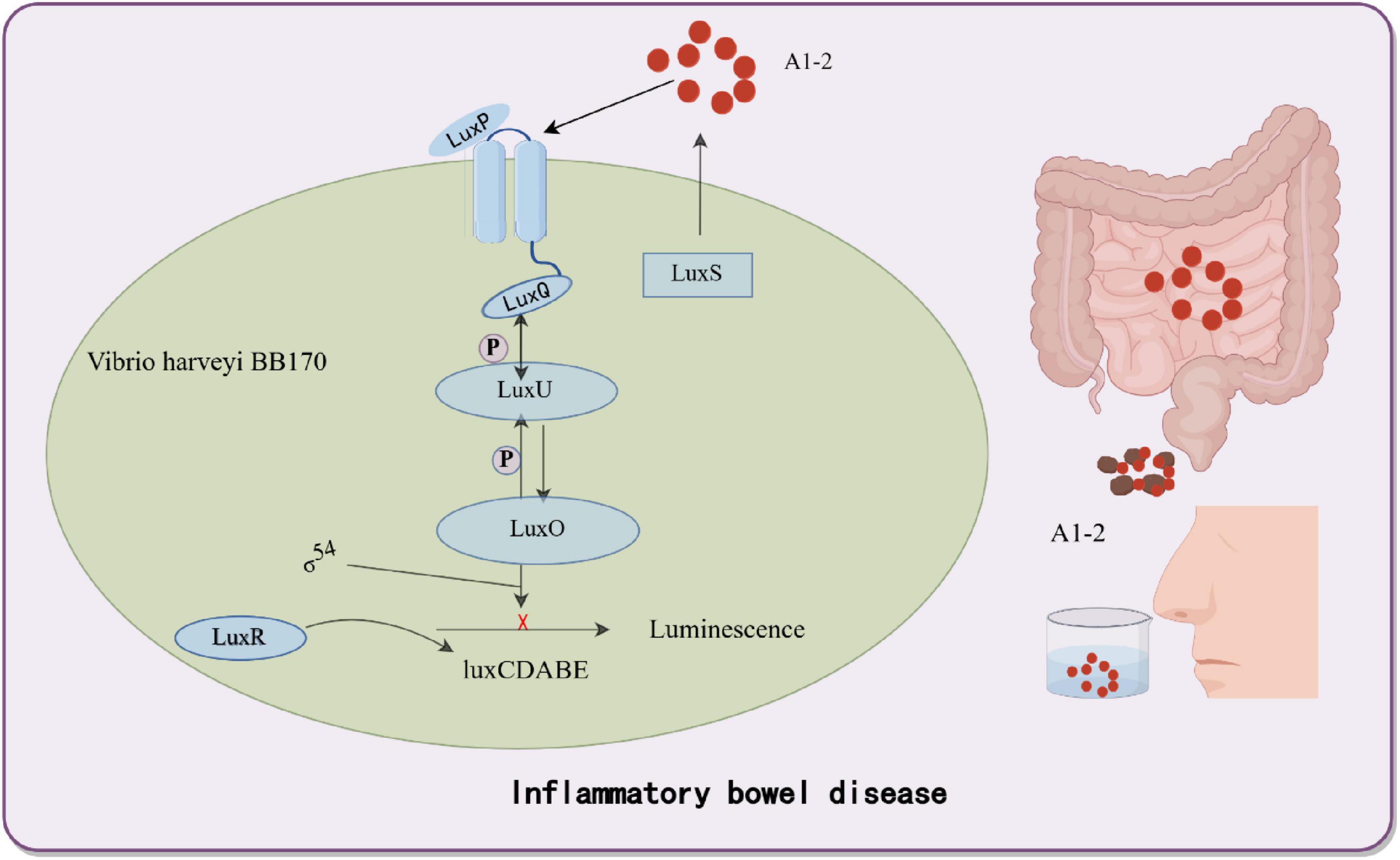
Figure 2. Bacterial biosensor based on a quorum sensing system for inflammatory bowel disease detection. Utilizing the quorum sensing system of Vibrio harveyi BB170, this biosensor detects the inflammatory bowel disease (IBD)-associated autoinducer AI-2, triggering the expression of bioluminescent genes to generate quantitatively detectable fluorescent signals. By Figdraw.
The interactions between the gut microbiota and the human host, as well as changes in its metabolic products, have been linked to a variety of pathological conditions, such as metabolic disorders, immunological diseases, cancer, neurological diseases, and behavioral disorders (Pesce et al., 2022). Bacterial biosensors represent non-invasive instruments pivotal in intestinal health monitoring, capable of detecting intestinal metabolites and translating these signals into readily observable outcomes, encompassing colorimetric changes, fluorescence emission, and electrical signals, thereby facilitating accurate assessment of gut health status (Pesce et al., 2022; Winter et al., 2010). Transient molecules in the gastrointestinal system, such as nitric oxide (NO) and hydrogen sulfide (H2S), are critical but elusive inflammation markers due to their short half-life and high reactivity (Riglar et al., 2017). At the start, To address NO detection challenges, McKay et al. (2018) engineered a dual-plasmid system in E. coli Nissle 1917. The first plasmid contains a NO-sensitive promoter (PnorV) driving T7 RNA polymerase (T7Pol) expression, while the second plasmid utilizes a T7/lac hybrid promoter to control green fluorescent protein (GFP) expression. This cascade amplification enables visualization of gut NO levels with a detection limit of 10 nM within 1 h, validated in murine models (McKay et al., 2018). Subsequently, Chen et al.’s (2021) team enhanced sensitivity by designing a positive feedback circuit, with the NO-responsive transcription factor NorR activates its own expression alongside superfolder GFP (sfGFP) under the PnorV promoter. Elevated NO concentrations trigger NorR self-amplification, reducing the detection threshold to 2 nM and accelerating response time by 3-fold (Chen et al., 2021). These ingestible biosensors exemplify the potential of synthetic biology in real-time gut monitoring.
3.2 Applications of bacterial biosensors in cancer
Cancer is a leading cause of global mortality, with liver metastases occurring in > 50% of patients with gastrointestinal malignancies and associated with a 5 years survival rate < 15% due to delayed detection (Groen, 1999; Liang et al., 2022). Current interventions (surgical resection, radiotherapy, etc.) target macroscopic lesions (> 1 cm) but fail to eliminate micrometastases (< 1 mm), which evade detection by conventional imaging (CT/MRI) and seed recurrent tumors (Chien et al., 2021; Schroeder et al., 2011). To address this, bacterial biosensors engineered as “living diagnostics” have emerged. In engineered E. coli Nissle 1917, synthetic genetic circuits employ lactate-responsive promoters to drive tumor-specific expression of β-galactosidase (lacZ) and bioluminescent reporters (luxCDABE) (Meighen, 1991). The oral delivery strategy leverages the gut-liver axis, through which EcN crosses the intestinal barrier via bile acid transporters and colonizes hepatic metastases within 24 h, thereby avoiding systemic toxicity (Chien et al., 2021). In preclinical models, the engineered EcN produced bioluminescence, enabling longitudinal imaging of liver metastases and improving visualization of metastatic lesions. Additionally, in mice fed with these probiotics, the bacteria colonized liver tumors specifically within 24 h while being cleared from healthy organs. Upon confirming colonization, LuGal (D-luciferin-O-β-galactoside) was administered. The β-galactosidase produced by EcN hydrolyzes LuGal into luciferin, which is subsequently excreted in urine. A complementary urinary detection system achieved rapid diagnosis using only 1 μL of urine, with detectable signals within 24 h post-administration. Remarkably, no adverse effects on mouse health were observed during 12 months of monitoring (Danino et al., 2015; Schultz et al., 2005; Figure 3). Despite promising sensitivity, clinical translation requires optimization of bacterial containment strategies to prevent horizontal gene transfer and validation in human trials. Genetically engineered Salmonella enterica serovar Typhimurium ΔppGpp exhibits tumor-specific colonization by exploiting the nutrient-rich tumor microenvironment (TME), making it a promising platform for theranostic applications (Nguyen and Min, 2017). Nguyen et al. (2010) engineered this strain to co-express cytolysin A (ClyA) and bacterial luciferase (LuxCDABE) under the control of an L-arabinose-inducible PBAD promoter. In murine colon and liver cancer models, intravenous administration led to selective tumor colonization, with oral L-arabinose triggering ClyA-mediated tumor lysis (62% volume reduction, p < 0.01) and intraperitoneal D-luciferin enabling real-time tracking imaging (R2 = 0.85 vs. tumor burden) (Nguyen et al., 2010). To enhance spatiotemporal control, Jiang’s et al. (2013) team integrated a Tet-On system, where tetracycline dose-dependently activates RLuc8 (reporter) and ClyA (therapeutic gene) through Ptet promoters. This dual-function design allowed simultaneous imaging-guided therapy: RLuc8 bioluminescence correlated with ClyA efficacy (30% apoptosis increase per 1 μg/mL tetracycline), enabling personalized dosing (Jiang et al., 2013).
Figure 3. Bacterial biosensors for non-invasive detection of liver cancer. Genetically engineered bacteria are designed to colonize the liver cancer microenvironment and induce the expression of fluorescent proteins upon sensing specific biomarkers (e.g., LuGal). The fluorescent molecules are excreted via the bloodstream into urine, thereby enabling non-invasive detection of liver cancer biomarkers. By Figdraw.
Pancreatic ductal adenocarcinoma (PDAC), characterized by its aggressive nature and poor prognosis (5 years survival < 10%), remains a major clinical challenge despite advancements in multimodal therapies (Principe et al., 2017). To address this, Zhou et al. (2016) engineered an attenuated Salmonella strain (VNP20009) through deletion of endotoxin-producing genes (msbB), enhancing its safety profile while preserving tumor-targeting capability (Low et al., 2004). This modified strain was designed as a theranostic agent, combining therapeutic potential with real-time diagnostic imaging. In their study, human PDAC cells (CFPAC-1) stably expressing the far-red fluorescent protein mKate2 (λem ≈ 635 nm) were implanted in mice to establish tumors (Piatkevich et al., 2010). Following intratumoral injection, the engineered Salmonella expressing bacterial luciferase (luxCDABE) colonized the tumor microenvironment, enabling dual-channel tracking via the IVIS Spectral Imaging System (Cheng et al., 2014; Toso et al., 2002). The distinct emission spectra of mKate2 (minimizing tissue autofluorescence) and luciferase (λem ≈ 490 nm) allowed simultaneous quantification of tumor burden and bacterial localization. Through longitudinal imaging, researchers mapped tumor growth kinetics and evaluated the spatiotemporal distribution of co-administered chemotherapeutics, demonstrating a correlation between bacterial density and drug efficacy (Hiddemann and Büchner, 2001; Zhou et al., 2016). This integrated approach highlights the potential of bacterial-based theranostics to refine PDAC management, though clinical translation requires further validation of safety and targeting specificity. Horizontal gene transfer (HGT) is the process by which organisms transfer genetic material to other cells rather than their offspring, in contrast to vertical transmission. HGT happens often in microorganisms and between prokaryotes and eukaryotes (Mell and Redfield, 2014; Soucy et al., 2015). However, the broad field of bacteria utilizing HGT mechanisms to detect mammals and respond to their DNA remains largely unexplored. Cooper’s et al. (2023) research team proposed the Cellular Assay for Targeted CRISPR-discriminated Horizontal Gene Transfer (CATCH), which is based on this principle. This technique integrates target DNA into the genome of engineered Acinetobacter baylyi, which functions as a biosensor to identify certain extracellular DNA. The KRAS gene is a typical oncogene. It controls cell growth normally, but when it is mutated, it causes uncontrolled cell proliferation and dysregulation (Priestley et al., 2019; Vogelstein et al., 1988). To eliminate false positive results produced by natural KRAS binding, the CRISPR technique efficiently addresses this issue. The CRISPR-Cas system was designed as a bacterial defensive mechanism for cutting foreign DNA, but it can be purposefully altered to modify its cutting positions and targets. The CATCH technique uses a specially designed sgRNA as a guide to accurately target the DNA sequence through complementary pairing. The CRISPR-Cas system is specifically designed to cut just the mutant KRAS gene that lacks a specific PAM sequence, with no effect on the normal KRAS gene (Cooper and Hasty, 2020). By integrating the CRISPR-Cas system and reporter genes (GFP or kanamycin resistance genes) into bacteria, the novel bacterial biosensor may co-culture with tumor cells in a variety of complex situations. The detection of the target DNA is demonstrated by the monitoring of GFP expression and bacterial growth in the kanamycin-selective media (Cooper et al., 2023). In the future, such engineered bacteria will have limitless potential for detecting cancer and precancerous lesions, and they are expected to help in the prevention of stomach and colorectal cancer. It is also possible to further modify these bacterial biosensors to create novel detectors with a variety of detecting goals in mind.
3.3 Applications of bacterial biosensors in myocardial infarction
Myocardial infarction (MI), characterized by coronary artery occlusion leading to ischemic necrosis of cardiac tissue, remains a leading cause of global mortality (Tsao et al., 2023). The auxotrophic Salmonella Typhimurium ΔppGpp strain exhibits selective tropism to infarcted myocardium, driven by hypoxia-responsive chemotaxis and necrotic cell-derived ATP gradients (Yi et al., 2020). To enable real-time imaging, this strain was engineered with a RLuc8 construct, where the E. coli L-arabinose-inducible promoter strictly controls expression of Renilla luciferase variant RLuc8. Upon systemic L-arabinose administration, RLuc8 is activated exclusively in bacteria colonizing ischemic regions, generating localized bioluminescence signals detectable within 2 h post-injection (Loening et al., 2006). In murine MI models, this approach achieved a spatial resolution of 0.5 mm, identifying subendocardial infarcts comprising as little as 3% of left ventricular mass—a significant improvement over SPECT’s 10% threshold (Le et al., 2011). Importantly, intravenous delivery of ΔppGpp Salmonella induced minimal systemic inflammation and no histopathological evidence of myocardial damage, underscoring its biosafety (Chang et al., 2021).
3.4 Applications of bacterial biosensors in diabetes mellitus
Diabetes mellitus, characterized by chronic hyperglycemia, requires rigorous glucose monitoring to prevent complications (Aloraynan et al., 2022). While fingerstick tests and continuous glucose monitors (CGMs) remain clinical standards, their invasiveness and cost drive demand for alternative methods (Liu et al., 2020). Urinary glucose (glycosuria) serves as a non-invasive proxy for hyperglycemia, though its utility is limited by a 1–2 h lag behind blood glucose levels and inter-individual renal threshold variations (de Sousa Vieira et al., 2022). To address this, Courbet et al. (2015) engineered E. coli to detect urinary glucose via a synthetic cpxP promoter—a stress-responsive element repurposed to activate GFP/RFP expression upon glucose uptake. The bacteria were encapsulated in alginate-PVA hydrogel beads, maintaining 90% fluorescence stability over 72 h in urine while preventing bacterial leakage (Harpaz et al., 2023). A genetic AND gate circuit further enhanced specificity: simultaneous glucose detection and hypoxia (mimicking bladder conditions) triggered GFP expression, achieving a detection limit of 0.1 mM glucose (equivalent to blood glucose ∼180 mg/dL) with 88.9% sensitivity and 96.3% specificity in diabetic urine samples (n = 150) (Courbet et al., 2015). Despite these advances, urinary glucose monitoring cannot replace real-time blood measurements due to physiological lag.
3.5 Applications of bacterial biosensors in monitoring cytarabine in leukemia
Bacterial biosensors have emerged as transformative tools for rapid drug sensitivity testing, particularly in predicting leukemia patients’ responses to cytarabine (Ara-C) (Hiddemann and Büchner, 2001). A key innovation is the engineering of an E. coli MG1655 cytidine deaminase-deficient mutant (Δcdd), which cannot metabolize Ara-C to its inactive form (Ara-U). This strain was integrated with the luxCDABE operon to generate a bioluminescent reporter system responsive to intracellular Ara-CTP levels—the active metabolite of Ara-C that inhibits DNA polymerase α and induces leukemic cell death (Wang et al., 1998; Yamauchi et al., 2009). In co-culture assays with patient-derived leukemic cells, the biosensor quantifies bioluminescence intensity, which correlates with Ara-CTP accumulation and drug efficacy. Compared to traditional MTT assays requiring 3–5 days, this system delivers results within 8 h, achieving 85% sensitivity and 92% specificity in identifying Ara-C-resistant patients (n = 50) (Yamauchi et al., 2009). The Δcdd mutation ensures bacterial viability by preventing Ara-C detoxification, enabling continuous signal generation without interference from host cell metabolites (Alloush et al., 2010). By enabling rapid, low-cost drug sensitivity testing, this biosensor platform exemplifies the potential of synthetic biology to bridge precision medicine and global health accessibility, particularly in resource-limited settings.
3.6 Applications of bacterial biosensors in cholera
Cholera, caused by toxigenic Vibrio cholerae, demands rapid diagnostics to curb its high transmission risk. Conventional methods like culture enrichment require > 24 h and lack sensitivity (Rafique et al., 2016). Synthetic biology offers innovative solutions: Holowko et al. (2016) engineered non-pathogenic E. coli to detect V. cholerae-specific CAI-1 (10 nM detection limit) by integrating its QS system (CqsS sensor kinase and response regulators) with a CRISPRi-based genetic inverter. In this system, dCas9 represses GFP expression in the absence of CAI-1, while CAI-1 binding relieves repression, enabling fluorescence readout within 2 h—100-fold faster than ELISA (Duan and March, 2010; Holowko et al., 2016; Ng et al., 2011; Yamasaki et al., 2017). Parallelly, Mao et al. (2018) developed a probiotic Lactococcus lactis biosensor using a TetR-regulated mCherry reporter. CAI-1 inactivates TetR via allosteric displacement, inducing a 60-fold mCherry increase. For field applications, they replaced fluorescence with β-lactamase secretion: hydrolysis of nitrocefin triggers a yellow-to-red color shift in 15 min, achieving 95% concordance with PCR in clinical stool samples (Mao et al., 2018). Beyond detection, Jayaraman et al. (2017) engineered a “sense-and-kill” E. coli that secretes Art-085 lysin via the YebF pathway upon CAI-1 detection. In murine models, this system reduced intestinal V. cholerae loads by 3 logs within 6 h, outperforming oral rehydration alone (Jayaraman et al., 2017).
4 Applications of bacterial biosensors in personalized medicine
Personalized medicine aims to revolutionize healthcare by tailoring diagnostic and therapeutic strategies to individual patients through real-time, dynamic monitoring of biomarkers. For instance, IBD patients require frequent monitoring of intestinal inflammation to optimize anti-inflammatory therapies, yet current techniques cannot provide continuous, non-invasive insights into biomarker dynamics. Bacterial biosensors, engineered to detect specific molecules in situ, offer a transformative solution (Neurath, 2017). Bhatt (2018) developed ingestible microbial electronic devices (IMBED) for personalized gut monitoring. They engineered E. coli Nissle 1917 to express heme-responsive genetic circuits (Phas promoter) and the luxCDABE operon, enabling bioluminescence upon heme detection. The IMBED encapsulates bacteria in a chamber with a nanoporous membrane (pore size < 50 nm), allowing metabolite influx while preventing bacterial escape (Liu et al., 2021). A silicon photodetector converts bioluminescence into wireless signals transmitted to smartphones, enabling real-time tracking of gut biomarkers (Mimee et al., 2018). In murine models, IMBED achieved a heme detection limit of 1 μM within 30 min, demonstrating potential for personalized management of iron-related disorders (Bhatt, 2018). However, IMBED requires enteric coating to neutralize gastric acid, and its long-term reliability may be compromised by intestinal peristalsis or biofilm formation. Future iterations could integrate pH-resistant circuits and anti-fouling membranes to enhance clinical viability. Recent advances in bacterial biosensors have enabled real-time tracking of transient gastrointestinal molecules for personalized medicine. Inda-Webb et al. (2023) engineered an ingestible electronic capsule (< 1.4 cm3) integrating E. coli Nissle 1917 biosensors with silicon photodiode arrays. The bacteria were modified to express engineered sensing proteins and recombinase-based memory circuits, allowing continuous recording of oxidative stress markers. A low-power module wirelessly transmits bioluminescent signals (triggered by luxCDABE promoter) to smartphones, achieving real-time monitoring of multiple inflammation mediators, including thiosulfate, tetrathionate, hydrogen peroxide (H2O2), and nitric oxide (NO). For instance, the system detects H2O2 with a limit of 10 nM and responds within < 5 min, enabling dynamic tracking of redox imbalance during disease progression. The capsule, validated in porcine colitis models, demonstrated a direct correlation between H2O2 concentration spikes and disease flare severity, enabling data-driven adjustments to antioxidant therapies (Inda-Webb et al., 2023; Liu et al., 2023). This platform represents a closed-loop theranostic system that integrates biomarker detection, therapeutic decision-making, and treatment response monitoring, thereby advancing non-invasive and individualized approaches to gastrointestinal care. MeRALD (Engineered Modularized Receptors Activated by Ligand-induced Dimerization) platform to address critical gaps in liver disease management (Chang et al., 2018). By engineering E. coli Nissle 1917 with programmable receptors sensitive to bile acids, their system detects pathological concentrations of these hepatic biomarkers through ligand-induced dimerization mechanisms (Chang et al., 2021). When bile acids bind to the modularized TcpP18 receptor, conformational changes trigger β-galactosidase expression via a LacZ reporter, generating quantifiable colorimetric signals in fecal samples (Abdelbasset et al., 2023). This innovation demonstrated clinical utility in liver transplant monitoring, while enabling smartphone-based color analysis for home testing. Notably, the platform’s modular design allows rapid adaptation to other biomarkers through receptor reprogramming, as evidenced by its parallel success in detecting gut inflammation markers like thiosulfate in IBD patients (Sicard et al., 2014). By overcoming traditional limitations of centralized laboratory diagnostics, this work exemplifies how synthetic biology can bridge precision medicine with global health accessibility, though challenges persist in ensuring sensor stability across diverse microbiota environments.
Shi et al. (2024) developed a bacterial biosensor leveraging the skin commensal bacterium Staphylococcus epidermidis to diagnose and treat inflammatory diseases such as psoriasis (Cau et al., 2021; Severn and Horswill, 2022). The biosensor integrates S. epidermidis within a dual-network hydrogel matrix composed of gelatin and tapioca starch, mimicking natural biofilm structures to sustain bacterial viability for over 4 days (Xu et al., 2021). Bioinspired double network hydrogels: from covalent double network hydrogels via hybrid double network hydrogels to physical double network hydrogels. This living hydrogel not only adheres conformally to skin but also modulates the immune microenvironment by downregulating pro-inflammatory cytokines (e.g., IL-17, TNF-α) and reducing T-cell infiltration, addressing the root cause of psoriasis. The bacteria’s innate ability to regulate skin homeostasis was enhanced through electrostatic interactions with the conductive polymer, which lowered charge transfer resistance, optimizing electron transfer for real-time monitoring of skin impedance, temperature, and humidity (Zhang P. et al., 2023). A wireless bioelectronic interface enabled on-demand electrical stimulation to control bacterial activity, ensuring biosafety by disinfecting pathogens (e.g., S. aureus) via reactive oxygen species generation. The biosensor demonstrated therapeutic efficacy in a psoriasis mouse model, reducing epidermal hyperplasia by 60% and restoring skin microbiota diversity without genetic modification. By synergizing bacterial immunomodulation with bioelectronic sensing, this platform exemplifies the potential of living bacterial systems in precision medicine, offering a drug-free approach to inflammation management while minimizing biohazard risks (Shi et al., 2024). Wang et al. (2022) developed a biosensor based on the electroactive bacterium Shewanella oneidensis MR-1, which demonstrates significant potential in medical diagnostics. This system utilizes the bacterium’s intrinsic lactate oxidation capability, where electrons generated from metabolic activity are transferred via outer-membrane cytochrome complexes (e.g., MtrCAB) and riboflavin-mediated extracellular electron transport (EET), enabling label-free lactate detection without genetic engineering (Gurdeep et al., 2021). Electrostatic integration of the conductive polymer poly PMNT enhanced biofilm formation and reduced charge transfer resistance from 226 to 12 Ω, achieving a lactate detection limit of 78 μM in physiological fluids (sweat, urine, plasma). By exploiting the Warburg effect—a hallmark of cancer metabolism characterized by excessive lactate secretion—the biosensor indirectly quantified HeLa, MCF-7, and A549 cancer cells with a sensitivity of 2.9 × 104 cells and an error rate < 10% (Wang et al., 2022). Integration with a flexible wearable platform and wireless signal transmission highlights its clinical applicability for real-time monitoring. Future engineering of S. oneidensis to recognize diverse biomarkers (e.g., inflammatory cytokines or pathogens) could expand the utility of living bacterial sensors in precision medicine (Li et al., 2021).
5 Challenges and future prospects
Bacterial biosensors exhibit distinct performance advantages over conventional diagnostic technologies, enabled by synthetic biology-driven detection of ultralow-concentration biomarkers in complex biological matrices (Hicks et al., 2020). These systems achieve real-time/near-real-time monitoring of dynamic analyte fluctuations and support multiplexed detection through engineered genetic circuits (Hicks et al., 2020; Table 2). Notably, advanced prototypes integrate diagnostic and therapeutic capabilities, forming closed-loop theranostic systems with feedback-controlled intervention (Jin et al., 2025). However, their medical application faces three primary challenges: functional stability, biosafety, and clinical translation (Riglar and Silver, 2018). Engineered bacteria withstand extreme host environments that involve immune attacks, resource competition with commensal microbiota, and fluctuating physiological conditions, all of which collectively destabilize sensor functionality (Dana et al., 2012). While synthetic genetic circuits provide novel sensing capabilities, they disrupt native metabolic balance, triggering compensatory mutations such as fluorescence reporter inactivation in engineered E. coli after serial passages and increasing susceptibility to endogenous signal interference (Sleight and Sauro, 2013; Sleight et al., 2010). Biosafety concerns remain unresolved as conventional suicide switches show limited effectiveness in complex human microenvironments, particularly in intestinal hypoxia zones where occasional bacterial escape may occur (Piraner et al., 2017). Physical encapsulation strategies, though effective in restricting microbial spread, reduce detection sensitivity due to molecular permeability barriers caused by suboptimal material interfaces (Hirota et al., 2017). Clinical translation is further challenged by the lack of standardized validation protocols, rigorous requirements for classifying these systems as “live medical devices” necessitating extensive virulence testing, and difficulties in assessing unpredictable long-term ecological risks. Emerging nanotechnology integration addresses existing limitations through bacterial-nanomaterial co-immobilization strategies. Nanostructure fixation enhances sensitivity by extending cellular electron transfer distances while biomimetic nanocapsules with dynamically tunable pores enable immune evasion and selective molecular permeation (Liu et al., 2024). Concurrently, artificial intelligence-driven platforms revolutionize biosensor development as deep learning models deciphering gene sequence-function correlations enable precise prediction of genetic editing outcomes (Angenent-Mari et al., 2020; Kim et al., 2020). These advancements are combined with microfluidic high-throughput screening to establish Design-Build-Test-Learn (DBTL) closed-loop optimization systems (Berlanda et al., 2021; Orsi et al., 2021). Dynamic evolution systems that simulate in vivo pressures accelerate directed bacterial adaptation in microfluidic chips, significantly reducing development timelines. Enhanced biosafety protocols incorporate dual-lock containment mechanisms to reduce escape probability alongside chassis genome minimization strategies for metabolic stability. Regulatory frameworks are evolving through multicenter standardized testing platforms and refined classifications of “live medical device” based on colonization capacity. Researchers are integrating bacterial biosensors with mobile health platforms like smartphones and cloud systems, enabling continuous physiological tracking and remote care delivery via real-time wireless networks (Chang et al., 2017; Raut et al., 2012). While most systems remain experimental, retrospective studies confirm their diagnostic potential in chronic disease management and cancer biomarker detection. Transitioning to clinical practice requires multicenter trials validating sensor stability and specificity in diverse patient populations. Successful validation could translate prototypes into standardized diagnostic modules for healthcare integration. This technological shift promises to enhance detection of complex biomarkers while supporting adaptive treatment protocols through persistent health data streams, ultimately advancing intelligent closed-loop diagnostic-therapeutic systems.
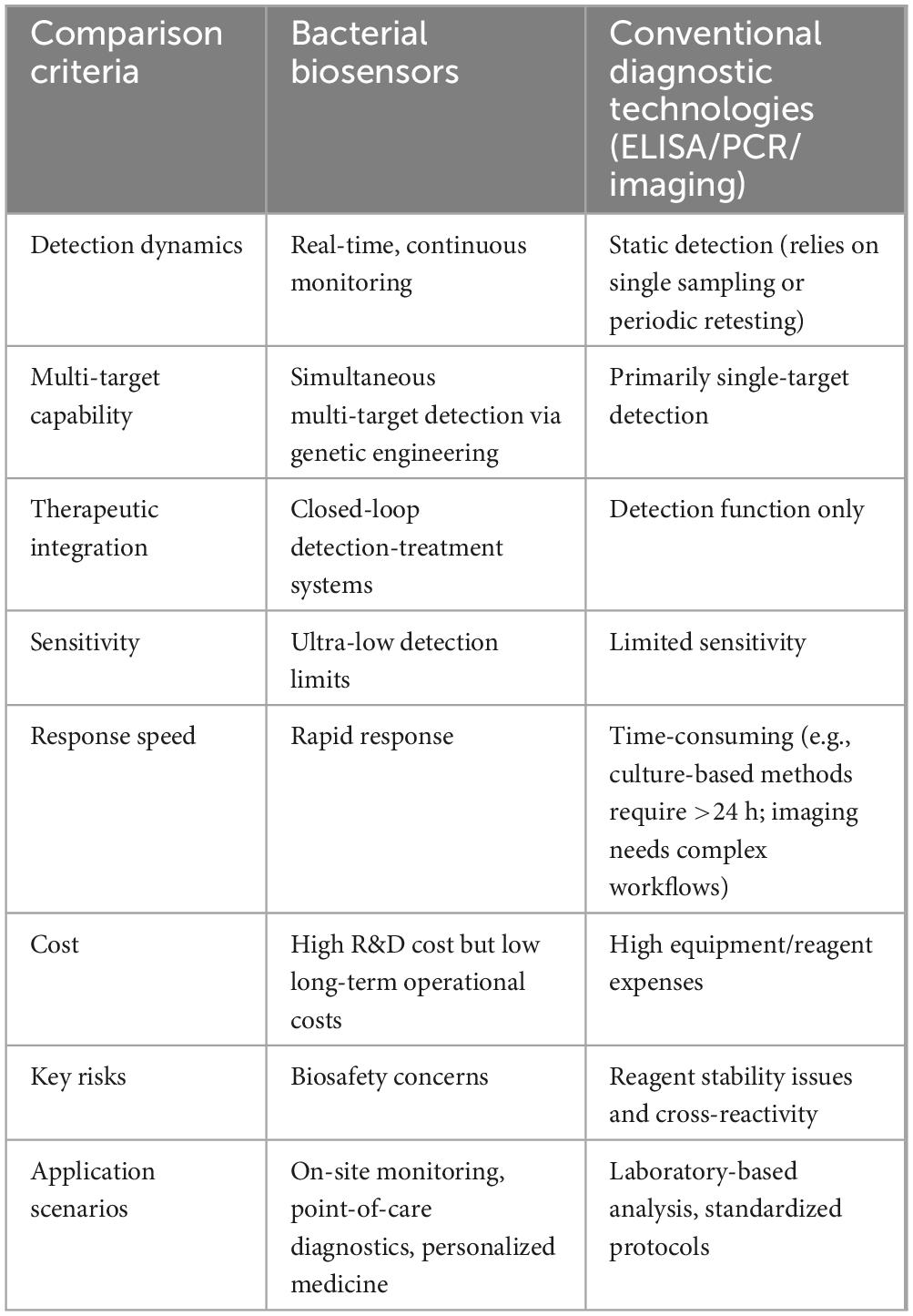
Table 2. Performance comparison between bacterial biosensors and conventional diagnostic technologies.
Author contributions
ML: Conceptualization, Writing – original draft, Writing – review and editing. WY: Writing – review and editing. WZ: Writing – review and editing. DY: Conceptualization, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Natural Science Foundation of Zhejiang Province (grant number LZ22H190002) and the Health & Medical Sci-Tech Project of Health Commission of Hangzhou Municipal (grant numbers B20231745, B20231819, A20230151).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelbasset, W., Savina, S., Mavaluru, D., Shichiyakh, R., Bokov, D., and Mustafa, Y. (2023). Smartphone based aptasensors as intelligent biodevice for food contamination detection in food and soil samples: Recent advances. Talanta 252:123769. doi: 10.1016/j.talanta.2022.123769
Alloush, H., Anderson, E., Martin, A., Ruddock, M., Angell, J., Hill, P., et al. (2010). A bioluminescent microbial biosensor for in vitro pretreatment assessment of cytarabine efficacy in leukemia. Clin. Chem. 56, 1862–1870. doi: 10.1373/clinchem.2010.145581
Aloraynan, A., Rassel, S., Xu, C., and Ban, D. A. (2022). Single wavelength mid-infrared photoacoustic spectroscopy for noninvasive glucose detection using machine learning. Biosensors (Basel) 12:166. doi: 10.3390/bios12030166
Andrianantoandro, E., Basu, S., Karig, D., and Weiss, R. (2006). Synthetic biology: New engineering rules for an emerging discipline. Mol. Syst. Biol. 2:2006.0028. doi: 10.1038/msb4100073
Angenent-Mari, N., Garruss, A., Soenksen, L., Church, G., and Collins, J. J. A. (2020). deep learning approach to programmable RNA switches. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-020-18677-1
Archer, E., Robinson, A., and Süel, G. (2012). Engineered E. coli that detect and respond to gut inflammation through nitric oxide sensing. ACS Synth Biol. 1, 451–457. doi: 10.1021/sb3000595
Belkin, S. (2003). Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 6, 206–212. doi: 10.1016/s1369-5274(03)00059-6
Berlanda, S., Breitfeld, M., Dietsche, C., and Dittrich, P. (2021). Recent advances in microfluidic technology for bioanalysis and diagnostics. Anal. Chem. 93, 311–331. doi: 10.1021/acs.analchem.0c04366
Bhatt, A. (2018). Digesting new developments in biosensors. N. Engl. J. Med. 379, 686–688. doi: 10.1056/NEJMcibr1806952
Bonnet, J., Yin, P., Ortiz, M., Subsoontorn, P., and Endy, D. (2013). Amplifying genetic logic gates. Science 340, 599–603. doi: 10.1126/science.1232758
Bourgonje, A., Feelisch, M., Faber, K., Pasch, A., Dijkstra, G., and van Goor, H. (2020). Oxidative stress and redox-modulating therapeutics in inflammatory bowel disease. Trends Mol. Med. 26, 1034–1046. doi: 10.1016/j.molmed.2020.06.006
Burrill, D., Inniss, M., Boyle, P., and Silver, P. (2012). Synthetic memory circuits for tracking human cell fate. Genes Dev. 26, 1486–1497. doi: 10.1101/gad.189035.112
Capra, E. J., and Laub, M. T. (2012). Evolution of two-component signal transduction systems. Annu Rev Microbiol. 66, 325–347. doi: 10.1146/annurev-micro-092611-150039
Cau, L., Williams, M., Butcher, A., Nakatsuji, T., Kavanaugh, J., Cheng, J., et al. (2021). Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J. Allergy Clin. Immunol. 147, 955–966.e16. doi: 10.1016/j.jaci.2020.06.024
Chang, H., Mayonove, P., Zavala, A., De Visch, A., Minard, P., Cohen-Gonsaud, M., et al. (2018). A modular receptor platform to expand the sensing repertoire of bacteria. ACS Synth. Biol. 7, 166–175. doi: 10.1021/acssynbio.7b00266
Chang, H., Voyvodic, P., Zúñiga, A., and Bonnet, J. (2017). Microbially derived biosensors for diagnosis, monitoring and epidemiology. Microb. Biotechnol. 10, 1031–1035. doi: 10.1111/1751-7915.12791
Chang, H., Zúñiga, A., Conejero, I., Voyvodic, P., Gracy, J., Fajardo-Ruiz, E., et al. (2021). Programmable receptors enable bacterial biosensors to detect pathological biomarkers in clinical samples. Nat. Commun. 12:5216. doi: 10.1038/s41467-021-25538-y
Chang, S. (2015). Disease monitoring in inflammatory bowel disease. World J. Gastroenterol. 21:11246. doi: 10.3748/wjg.v21.i40.11246
Chen, F., Warnock, R., Van der Meer, J., and Wegner, S. (2020). Bioluminescence-triggered photoswitchable bacterial adhesions enable higher sensitivity and dual-readout bacterial biosensors for mercury. ACS Sens. 5, 2205–2210. doi: 10.1021/acssensors.0c00855
Chen, S., Chen, X., Su, H., Guo, M., and Liu, H. (2023). Advances in synthetic-biology-based whole-cell biosensors: Principles, genetic modules, and applications in food safety. Int. J. Mol. Sci. 24:7989. doi: 10.3390/ijms24097989
Chen, X., Wang, B., Thompson, I., and Huang, W. (2021). Rational design and characterization of nitric oxide biosensors in E. coli Nissle 1917 and mini simcells. ACS Synth. Biol. 10:2566–2578. doi: 10.1021/acssynbio.1c00223
Cheng, X., Zhang, X., Zhou, Y., Zhang, C., and Hua, Z. C. A. (2014). Salmonella Typhimurium mutant strain capable of RNAi delivery: Higher tumor-targeting and lower toxicity. Cancer Biol. Ther. 15, 1068–1076. doi: 10.4161/cbt.29185
Chien, T., Harimoto, T., Kepecs, B., Gray, K., Coker, C., Hou, N., et al. (2021). Enhancing the tropism of bacteria via genetically programmed biosensors. Nat. Biomed. Eng. 6, 94–104. doi: 10.1038/s41551-021-00772-3
Coelho, C., Branco, R., Natal-da-Luz, T., Sousa, J., and Morais, P. (2015). Evaluation of bacterial biosensors to determine chromate bioavailability and to assess ecotoxicity of soils. Chemosphere 128, 62–69. doi: 10.1016/j.chemosphere.2014.12.026
Cooper, R., and Hasty, J. (2020). One-day construction of multiplex arrays to harness natural CRISPR-Cas systems. ACS Synth. Biol. 9, 1129–1137. doi: 10.1021/acssynbio.9b00489
Cooper, R., Wright, J., Ng, J., Goyne, J., Suzuki, N., Lee, Y., et al. (2023). Engineered bacteria detect tumor DNA. Science 381, 682–686. doi: 10.1126/science.adf3974
Courbet, A., Endy, D., Renard, E., Molina, F., and Bonnet, J. (2015). Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci. Transl. Med. 7:289ra83. doi: 10.1126/scitranslmed.aaa3601
Daeffler, K., Galley, J., Sheth, R., Ortiz-Velez, L., Bibb, C., Shroyer, N., et al. (2017). Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol. 13:923. doi: 10.15252/msb.20167416
Dana, G., Kuiken, T., Rejeski, D., and Snow, A. (2012). Synthetic biology: Four steps to avoid a synthetic-biology disaster. Nature 483, 29–29. doi: 10.1038/483029a
Danino, T., Prindle, A., Kwong, G., Skalak, M., Li, H., Allen, K., et al. (2015). Programmable probiotics for detection of cancer in urine. Sci. Transl. Med. 7:289ra84. doi: 10.1126/scitranslmed.aaa3519
de la Fuente-Núñez, C., and Lu, T. K. (2017). CRISPR-Cas9 technology: Applications in genome engineering, development of sequence-specific antimicrobials, and future prospects. Integr. Biol. 9, 109–122. doi: 10.1039/c6ib00140h
de Sousa Vieira, E., Silveira, L., Carvalho, H., Bispo, J., Fernandes, F., and Fernandes, A. (2022). Biochemical analysis of urine samples from diabetic and hypertensive patients without renal dysfunction using spectrophotometry and raman spectroscopy techniques aiming classification and diagnosis. Bioengineering (Basel) 9:500. doi: 10.3390/bioengineering9100500
Ding, N., Zhou, S., and Deng, Y. (2021). [Progress in transcription factor-based metabolite biosensors]. Sheng Wu Gong Cheng Xue Bao 37, 911–922. doi: 10.13345/j.cjb.200641
Doudna, J., and Charpentier, E. (2017). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
Duan, F., and March, J. (2010). Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc. Natl. Acad. Sci. U S A. 107, 11260–11264. doi: 10.1073/pnas.1001294107
Francisco, R., Branco, R., Schwab, S., Baldani, I., and Morais, P. (2019). Two plant-hosted whole-cell bacterial biosensors for detection of bioavailable Cr(VI). World J. Microbiol. Biotechnol. 35:129. doi: 10.1007/s11274-019-2703-0
Gao, Y. Z., Wang, Y., and Ji, M. (2023). A whole-cell hydrogen peroxide biosensor and its application in visual food analysis. Innov. Life 1:100011. doi: 10.59717/j.xinn-life.2023.100011
Ge, C., Sheng, H., Chen, X., Shen, X., Sun, X., Yan, Y., et al. (2020). Quorum sensing system used as a tool in metabolic engineering. Biotechnol. J. 15:e1900360. doi: 10.1002/biot.201900360
Groen, K. (1999). Primary and metastatic liver cancer. Semin. Oncol. Nurs. 15, 48–57. doi: 10.1016/s0749-2081(99)80039-4
Gurdeep, R., Nishtha, K., and Kumar, M. V. (2021). Lactate detection sensors for food, clinical and biological applications: A review. Environ. Chem. Lett. 19, 1135–1152.
Harpaz, D., Zoabi, K., and Eltzov, E. (2023). Characterization and optimization of bioluminescent bacterial cells immobilization process in calcium alginate hydrogel tablets. J. Appl. Microbiol. 134:lxad070. doi: 10.1093/jambio/lxad070
Hicks, M., Bachmann, T. T., and Wang, B. (2020). Synthetic biology enables programmable cell-based biosensors. ChemPhysChem 21, 132–144. doi: 10.1002/cphc.201900739
Hiddemann, W., and Büchner, T. (2001). Current status and perspectives of therapy for acute myeloid leukemia. Semin. Hematol. 38, 3–9. doi: 10.1016/s0037-1963(01)90150-7
Hirota, R., Abe, K., Katsuura, Z., Noguchi, R., Moribe, S., Motomura, K., et al. (2017). A novel biocontainment strategy makes bacterial growth and survival dependent on phosphite. Sci. Rep. 7: 44748. doi: 10.1038/srep44748
Holowko, M., Wang, H., Jayaraman, P., and Poh, C. (2016). Biosensing Vibrio cholerae with Genetically Engineered Escherichia coli. ACS Synth. Biol. 5, 1275–1283. doi: 10.1021/acssynbio.6b00079
Hui, C., Hu, S., Yang, X., and Guo, Y. (2023). A panel of visual bacterial biosensors for the rapid detection of genotoxic and oxidative damage: A proof of concept study. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 888:503639. doi: 10.1016/j.mrgentox.2023.503639
Inda-Webb, M., Jimenez, M., Liu, Q., Phan, N., Ahn, J., Steiger, C., et al. (2023). Sub-1.4 cm3 capsule for detecting labile inflammatory biomarkers in situ. Nature 620, 386–392. doi: 10.1038/s41586-023-06369-x
Jayaraman, P., Holowko, M., Yeoh, J., Lim, S., and Poh, C. (2017). Repurposing a two-component system-based biosensor for the killing of Vibrio cholerae. ACS Synth. Biol. 6, 1403–1415. doi: 10.1021/acssynbio.7b00058
Jia, X., Bu, R., Zhao, T., and Wu, K. (2019). Sensitive and specific whole-cell biosensor for arsenic detection. Appl. Environ. Microbiol. 85:e00694-19. doi: 10.1128/AEM.00694-19
Jiang, S., Park, S., Lee, H., Zheng, J., Kim, H., Bom, H., et al. (2013). Engineering of bacteria for the visualization of targeted delivery of a cytolytic anticancer agent. Mol. Ther. 21, 1985–1995. doi: 10.1038/mt.2013.183
Jin, Z., Zhang, Y., Hu, H., Li, Q., Zhang, L., Zhao, K., et al. (2025). Closed-loop theranostic microgels for immune microenvironment modulation and microbiota remodeling in ulcerative colitis. Biomaterials 314:122834. doi: 10.1016/j.biomaterials.2024.122834
Johnson, A., Gonzalez-Villanueva, M., Wong, L., Steinbüchel, A., Tee, K., and Xu, P. (2017). Design and application of genetically-encoded malonyl-CoA biosensors for metabolic engineering of microbial cell factories. Metab. Eng. 44, 253–264. doi: 10.1016/j.ymben.2017.10.011
Jung, K., Fabiani, F., Hoyer, E., and Lassak, J. (2018). Bacterial transmembrane signalling systems and their engineering for biosensing. Open Biol. 8:180023. doi: 10.1098/rsob.180023
Karmakar, R. (2021). State of the art of bacterial chemotaxis. J. Basic Microbiol. 61, 366–379. doi: 10.1002/jobm.202000661
Kim, H., Seong, W., Rha, E., Lee, H., Kim, S., Kwon, K., et al. (2020). Machine learning linked evolutionary biosensor array for highly sensitive and specific molecular identification. Biosens. Bioelectron. 170:112670. doi: 10.1016/j.bios.2020.112670
Kotula, J., Kerns, S., Shaket, L., Siraj, L., Collins, J., Way, J., et al. (2014). Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. U S A. 111, 4838–4843. doi: 10.1073/pnas.1321321111
Kumari, A., Pasini, P., and Daunert, S. (2008). Detection of bacterial quorum sensing N-acyl homoserine lactones in clinical samples. Anal. Bioanal. Chem. 391, 1619–1627. doi: 10.1007/s00216-008-2002-3
Kumari, A., Pasini, P., Deo, S., Flomenhoft, D., Shashidhar, H., and Daunert, S. (2006). Biosensing systems for the detection of bacterial quorum signaling molecules. Anal. Chem. 78, 7603–7609. doi: 10.1021/ac061421n
Law, J., Ab Mutalib, N., Chan, K., and Lee, L. (2015). Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi: 10.3389/fmicb.2014.00770
Lazar, J., and Tabor, J. (2021). Bacterial two-component systems as sensors for synthetic biology applications. Curr. Opin. Syst. Biol. 28:100398. doi: 10.1016/j.coisb.2021.100398
Le, U., Kim, H., Kwon, J., Kim, M., Nguyen, V., Jiang, S., et al. (2011). Engineering and visualization of bacteria for targeting infarcted myocardium. Mol. Ther. 19, 951–959. doi: 10.1038/mt.2011.25
Lee, J., Wu, J., Deng, Y., Wang, J., Wang, C., Wang, J., et al. (2013). A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 9, 339–343. doi: 10.1038/nchembio.1225
Li, Q., Zhang, Y., Fan, H., Gong, Y., Xu, Y., Lv, Q., et al. (2021). In vitro and in vivo detection of lactate with nanohybrid-functionalized Pt microelectrode facilitating assessment of tumor development. Biosens. Bioelectron. 191:113474. doi: 10.1016/j.bios.2021.113474
Liang, S., Wang, C., Shao, Y., Wang, Y., Xing, D., and Geng, Z. (2022). Recent advances in bacteria-mediated cancer therapy. Front. Bioeng. Biotechnol. 10:1026248. doi: 10.3389/fbioe.2022.1026248
Liao, V., Chien, M., Tseng, Y., and Ou, K. (2006). Assessment of heavy metal bioavailability in contaminated sediments and soils using green fluorescent protein-based bacterial biosensors. Environ. Pollut. 142, 17–23. doi: 10.1016/j.envpol.2005.09.021
Liu, Q., Jimenez, M., and Eugenia Inda, M. (2023). A threshold-based bioluminescence detector with a CMOS-integrated photodiode array in 65 nm for a multi-diagnostic ingestible capsule. IEEE J. Solid State Circuits 58, 838–851. doi: 10.1109/JSSC.2022.3197465
Liu, X., Yang, Y., Inda, M., Lin, S., Wu, J., Kim, Y., et al. (2021). Magnetic living hydrogels for intestinal localization, retention, and diagnosis. Adv. Funct. Mater. 31:2010918. doi: 10.1002/adfm.202010918
Liu, Y., Huang, H., Gao, R., and Liu, Y. (2020). Dynamic phenotypes and molecular mechanisms to understand the pathogenesis of diabetic nephropathy in two widely used animal models of type 2 diabetes mellitus. Front. Cell. Dev. Biol. 8:172. doi: 10.3389/fcell.2020.00172
Liu, Y., Lin, Z., Wang, Y., Chen, L., Wang, Y., and Luo, C. (2024). Nanotechnology in inflammation: Cutting-edge advances in diagnostics, therapeutics and theranostics. Theranostics 14, 2490–2525. doi: 10.7150/thno.91394
Loening, A., Fenn, T., Wu, A., and Gambhir, S. (2006). Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng. Des. Sel. 19, 391–400. doi: 10.1093/protein/gzl023
Lopreside, A., Wan, X., Michelini, E., Roda, A., and Wang, B. (2019). Comprehensive profiling of diverse genetic reporters with application to whole-cell and cell-free biosensors. Anal. Chem. 91, 15284–15292. doi: 10.1021/acs.analchem.9b04444
Low, K., Ittensohn, M., Luo, X., Zheng, L., King, I., Pawelek, J., et al. (2004). Construction of VNP20009: A novel, genetically stable antibiotic-sensitive strain of tumor-targeting Salmonella for parenteral administration in humans. Methods Mol. Med. 90, 47–60. doi: 10.1385/1-59259-429-8:47
Mao, N., Cubillos-Ruiz, A., Cameron, D., and Collins, J. (2018). Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med. 10:eaao2586. doi: 10.1126/scitranslmed.aao2586
Mascher, T., Zimmer, S., Smith, T., and Helmann, J. (2004). Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48, 2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004
McKay, R., Hauk, P., Quan, D., and Bentley, W. (2018). Development of cell-based sentinels for nitric oxide: Ensuring marker expression and unimodality. ACS Synth. Biol. 7, 1694–1701. doi: 10.1021/acssynbio.8b00146
Meighen, E. (1991). Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55, 123–142. doi: 10.1128/mr.55.1.123-142.1991
Mell, J., and Redfield, R. (2014). Natural competence and the evolution of DNA uptake specificity. J. Bacteriol. 196, 1471–1483. doi: 10.1128/JB.01293-13
Miller, C., Ho, J., and Bennett, M. (2022). Strategies for improving small-molecule biosensors in bacteria. Biosensors (Basel) 12:64. doi: 10.3390/bios12020064
Mimee, M., Nadeau, P., Hayward, A., Carim, S., Flanagan, S., Jerger, L., et al. (2018). An ingestible bacterial-electronic system to monitor gastrointestinal health. Science 360, 915–918. doi: 10.1126/science.aas9315
Moon, T., Lou, C., Tamsir, A., Stanton, B., and Voigt, C. (2012). Genetic programs constructed from layered logic gates in single cells. Nature 491, 249–253. doi: 10.1038/nature11516
Nakamura, H., Shimomura-Shimizu, M., and Karube, I. (2008). Development of microbial sensors and their application. Adv. Biochem. Eng. Biotechnol. 109, 351–394. doi: 10.1007/10_2007_085
Naydich, A., Nangle, S., Bues, J., Trivedi, D., Nissar, N., Inniss, M., et al. (2019). Synthetic gene circuits enable systems-level biosensor trigger discovery at the host-microbe interface. mSystems 4:e00125-19. doi: 10.1128/mSystems.00125-19
Neurath, M. F. (2017). Current and emerging therapeutic targets for IBD. Nat. Rev. Gastroenterol. Hepatol. 14, 269–278. doi: 10.1038/nrgastro.2016.208
Ng, W., Perez, L., Wei, Y., Kraml, C., Semmelhack, M., and Bassler, B. (2011). Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 79, 1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x
Nguyen, V., and Min, J. (2017). Salmonella-mediated cancer therapy: Roles and potential. Nucl. Med. Mol. Imaging 51, 118–126. doi: 10.1007/s13139-016-0415-z
Nguyen, V., Kim, H., Ha, J., Hong, Y., Choy, H., and Min, J. (2010). Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res. 70, 18–23. doi: 10.1158/0008-5472.CAN-09-3453
Orsi, E., Claassens, N., Nikel, P., and Lindner, S. (2021). Growth-coupled selection of synthetic modules to accelerate cell factory development. Nat. Commun. 12:5295. doi: 10.1038/s41467-021-25665-6
Park, M., Tsai, S., and Chen, W. (2013). Microbial biosensors: Engineered microorganisms as the sensing machinery. Sensors (Basel) 13, 5777–5795. doi: 10.3390/s130505777
Pesce, M., Seguella, L., Del Re, A., Lu, J., Palenca, I., Corpetti, C., et al. (2022). Next-generation probiotics for inflammatory bowel disease. Int. J. Mol. Sci. 23:5466. doi: 10.3390/ijms23105466
Piatkevich, K., Hulit, J., Subach, O., Wu, B., Abdulla, A., Segall, J., et al. (2010). Monomeric red fluorescent proteins with a large stokes shift. Proc. Natl. Acad. Sci. U S A. 107, 5369–5374. doi: 10.1073/pnas.0914365107
Piraner, D., Abedi, M., Moser, B., Lee-Gosselin, A., and Shapiro, M. (2017). Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat. Chem. Biol. 13, 75–80. doi: 10.1038/nchembio.2233
Plotnikova, E., Shumkova, E., and Shumkov, M. (2016). [Whole-cell bacterial biosensors for the detection of aromatic hydrocarbons and their chlorinated derivatives]. Prikl Biokhim Mikrobiol. 52, 353–364. doi: 10.1134/S0003683816040128
Poikulainen, E., Tienaho, J., Sarjala, T., and Santala, V. A. (2020). panel of bioluminescent whole-cell bacterial biosensors for the screening for new antibacterial substances from natural extracts. J. Microbiol. Methods 178:106083. doi: 10.1016/j.mimet.2020.106083
Priestley, P., Baber, J., Lolkema, M., Steeghs, N., de Bruijn, E., Shale, C., et al. (2019). Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575, 210–216. doi: 10.1038/s41586-019-1689-y
Principe, M., Borgoni, S., Cascione, M., Chattaragada, M., Ferri-Borgogno, S., Capello, M., et al. (2017). Alpha-enolase (ENO1) controls alpha v/beta 3 integrin expression and regulates pancreatic cancer adhesion, invasion, and metastasis. J. Hematol. Oncol. 10:16. doi: 10.1186/s13045-016-0385-8
Rafique, R., Rashid, M., Monira, S., Rahman, Z., Mahmud, M., Mustafiz, M., et al. (2016). Transmission of Infectious Vibrio cholerae through drinking water among the household contacts of cholera patients (CHoBI7 Trial). Front. Microbiol. 7:1635. doi: 10.3389/fmicb.2016.01635
Raut, N., O’Connor, G., Pasini, P., and Daunert, S. (2012). Engineered cells as biosensing systems in biomedical analysis. Anal. Bioanal. Chem. 402, 3147–3159. doi: 10.1007/s00216-012-5756-6
Raut, N., Pasini, P., and Daunert, S. (2013). Deciphering bacterial universal language by detecting the quorum sensing signal, autoinducer-2, with a whole-cell sensing system. Anal. Chem. 85, 9604–9609. doi: 10.1021/ac401776k
Reese, A., Cho, E., Klitzman, B., Nichols, S., Wisniewski, N., Villa, M., et al. (2018). Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut. Elife 7:e35987. doi: 10.7554/eLife.35987
Riglar, D. T., and Silver, P. A. (2018). Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 16, 214–225.
Riglar, D., Giessen, T., Baym, M., Kerns, S., Niederhuber, M., Bronson, R., et al. (2017). Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 35, 653–658. doi: 10.1038/nbt.3879
Roda, A., Roda, B., Cevenini, L., Michelini, E., Mezzanotte, L., Reschiglian, P., et al. (2011). Analytical strategies for improving the robustness and reproducibility of bioluminescent microbial bioreporters. Anal. Bioanal. Chem. 401, 201–211. doi: 10.1007/s00216-011-5091-3
Salis, H., Tamsir, A., and Voigt, C. (2009). Engineering bacterial signals and sensors. Contrib. Microbiol. 16, 194–225. doi: 10.1159/000219381
Saltepe, B., and Kehribar, E. Ş., Su Yirmibeşoğlu S. S., and Şafak Şeker U. Ö. (2018). Cellular biosensors with engineered genetic circuits. ACS Sens. 3, 13–26. doi: 10.1021/acssensors.7b00728
Schroeder, A., Heller, D., Winslow, M., Dahlman, J., Pratt, G., and Langer, R. (2011). Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer 12, 39–50. doi: 10.1038/nrc3180
Schultz, M., Watzl, S., Oelschlaeger, T., Rath, H., Göttl, C., Lehn, N., et al. (2005). Green fluorescent protein for detection of the probiotic microorganism Escherichia coli strain Nissle 1917 (EcN) in vivo. J. Microbiol. Methods 61, 389–398. doi: 10.1016/j.mimet.2005.01.007
Schuster, M., and Greenberg, E. P. (2008). “LuxR-type proteins in Pseudomonas aeruginosa quorum sensing: Distinct mechanisms with global implications,” in Chemical communication among bacteria, eds S. C. Winans and B. L. Bassler. doi: 10.1128/9781555815578.ch9
Sedlmayer, F., Aubel, D., and Fussenegger, M. (2018). Synthetic gene circuits for the detection, elimination and prevention of disease. Nat. Biomed. Eng. 2, 399–415. doi: 10.1038/s41551-018-0215-0
Serganov, A., and Nudler, E. A. (2013). decade of riboswitches. Cell 152, 17–24. doi: 10.1016/j.cell.2012.12.024
Severn, M., and Horswill, A. (2022). Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nat. Rev. Microbiol. 21, 97–111. doi: 10.1038/s41579-022-00780-3
Shi, J., Kim, S., Li, P., Dong, F., Yang, C., Nam, B., et al. (2024). Active biointegrated living electronics for managing inflammation. Science 384, 1023–1030. doi: 10.1126/science.adl1102
Sicard, C., Shek, N., White, D., Bowers, R., Brown, R., and Brennan, J. D. (2014). A rapid and sensitive fluorimetric β-galactosidase assay for coliform detection using chlorophenol red-β-D-galactopyranoside. Anal. Bioanal. Chem. 406, 5395–5403. doi: 10.1007/s00216-014-7935-0
Skjoedt, M., Snoek, T., Kildegaard, K., Arsovska, D., Eichenberger, M., Goedecke, T., et al. (2016). Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat. Chem. Biol. 12, 951–958. doi: 10.1038/nchembio.2177
Sleight, S., and Sauro, H. (2013). Visualization of evolutionary stability dynamics and competitive fitness of Escherichia coli engineered with randomized multigene circuits. ACS Synth. Biol. 2, 519–528. doi: 10.1021/sb400055h
Sleight, S., Bartley, B., Lieviant, J., and Sauro, H. (2010). Designing and engineering evolutionary robust genetic circuits. J. Biol. Eng. 4:12. doi: 10.1186/1754-1611-4-12
Soltani Zarrin, P., Jamal, F., Guha, S., Wessel, J., Kissinger, D., and Wenger, C. (2018). Design and fabrication of a BiCMOS dielectric sensor for viscosity measurements: A possible solution for early detection of COPD. Biosensors (Basel) 8:78. doi: 10.3390/bios8030078
Soucy, S., Huang, J., and Gogarten, J. (2015). Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 16, 472–482. doi: 10.1038/nrg3962
Tang, T. C., An, B., Huang, Y., Vasikaran, S., Wang, Y., Jiang, X., et al. (2021). Materials design by synthetic biology. Nat. Rev. Mater. 6, 332–350. doi: 10.1038/s41578-020-00265-w
Tanna, T., Ramachanderan, R., and Platt, R. (2021). Engineered bacteria to report gut function: Technologies and implementation. Curr. Opin. Microbiol. 59, 24–33. doi: 10.1016/j.mib.2020.07.014
Tiwari, A., Balázsi, G., Gennaro, M., and Igoshin, O. (2013). The interplay of multiple feedback loops with post-translational kinetics results in bistability of mycobacterial stress response. Phys. Biol. 7:036005. doi: 10.1088/1478-3975/7/3/036005
Toso, J., Gill, V., Hwu, P., Marincola, F., Restifo, N., Schwartzentruber, D., et al. (2002). Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J. Clin. Oncol. 20, 142–152. doi: 10.1200/JCO.2002.20.1.142
Tsao, C., Aday, A., Almarzooq, Z., Anderson, C., Arora, P., Avery, C., et al. (2023). Heart disease and stroke statistics-2023 update: A report from the american heart association. Circulation 147:e93–e621. doi: 10.1161/CIR.0000000000001123
van der Meer, J., and Belkin, S. (2010). Where microbiology meets microengineering: Design and applications of reporter bacteria. Nat. Rev. Microbiol. 8, 511–522. doi: 10.1038/nrmicro2392
Vogelstein, B., Fearon, E., Hamilton, S., Kern, S., Preisinger, A., Leppert, M., et al. (1988). Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319, 525–532. doi: 10.1056/NEJM198809013190901
Wan, X., Ho, Y., and Wang, B. (2019). “Engineering prokaryote synthetic biology biosensors,” in Handbook of Cell Biosensors, ed. G. Thouand (Berlin: Springer), doi: 10.1007/978-3-319-47405-2_131-1
Wang, B., Barahona, M., and Buck, M. A. (2013). modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosens. Bioelectron. 40, 368–376. doi: 10.1016/j.bios.2012.08.011
Wang, B., Kitney, R., Joly, N., and Buck, M. (2011). Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology. Nat. Commun. 2:508. doi: 10.1038/ncomms1516
Wang, J., Neuhard, J., and Eriksson, S. (1998). An Escherichia coli system expressing human deoxyribonucleoside salvage enzymes for evaluation of potential antiproliferative nucleoside analogs. Antimicrob. Agents Chemother. 42, 2620–2625. doi: 10.1128/AAC.42.10.2620
Wang, Z., Bai, H., Yu, W., Gao, Z., Chen, W., Yang, Z., et al. (2022). Flexible bioelectronic device fabricated by conductive polymer-based living material. Sci. Adv. 8:eabo1458. doi: 10.1126/sciadv.abo1458
Winter, S., and Bäumler, A. (2014). Dysbiosis in the inflamed intestine: Chance favors the prepared microbe. Gut Microbes 5, 71–73. doi: 10.4161/gmic.27129
Winter, S., Thiennimitr, P., Winter, M., Butler, B., Huseby, D., Crawford, R., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Winter, S., Winter, M., Xavier, M., Thiennimitr, P., Poon, V., Keestra, A., et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711. doi: 10.1126/science.1232467
Xu, X., Jerca, V., and Hoogenboom, R. (2021). Bioinspired double network hydrogels: From covalent double network hydrogels via hybrid double network hydrogels to physical double network hydrogels. Mater. Horiz. 8, 1173–1188. doi: 10.1039/d0mh01514h
Yamasaki, E., Sakamoto, R., Matsumoto, T., Maiti, B., Okumura, K., Morimatsu, F., et al. (2017). Detection of cholera toxin by an immunochromatographic test strip. Methods Mol. Biol. 1600, 1–7. doi: 10.1007/978-1-4939-6958-6_1
Yamauchi, T., Negoro, E., Kishi, S., Takagi, K., Yoshida, A., Urasaki, Y., et al. (2009). Intracellular cytarabine triphosphate production correlates to deoxycytidine kinase/cytosolic 5’-nucleotidase II expression ratio in primary acute myeloid leukemia cells. Biochem. Pharmacol. 77, 1780–1786. doi: 10.1016/j.bcp.2009.03.011
Yi, X., Zhou, H., Chao, Y., Xiong, S., Zhong, J., Chai, Z., et al. (2020). Bacteria-triggered tumor-specific thrombosis to enable potent photothermal immunotherapy of cancer. Sci. Adv. 6:eaba3546. doi: 10.1126/sciadv.aba3546
York, A. (2023). Intelligent bacteria for IBD. Nat. Rev. Microbiol. 21:129. doi: 10.1038/s41579-023-00854-w
Zhang, J., Guo, C., Xuan, C., Gu, J., Cui, Z., Zhang, J., et al. (2023). High-throughput, quantitative screening of quorum-sensing inhibitors based on a bacterial biosensor. ACS Chem. Biol. 18, 2544–2554. doi: 10.1021/acschembio.3c00537
Zhang, P., Zhou, X., Wang, X., and Li, Z. (2023). Enhanced bidirectional extracellular electron transfer based on biointerface interaction of conjugated polymers-bacteria biohybrid system. Colloids Surf. B Biointerfaces 228:113383. doi: 10.1016/j.colsurfb.2023.113383
Zhong, G., Wang, H., Bailey, C., Gao, G., and Farzan, M. (2016). Rational design of aptazyme riboswitches for efficient control of gene expression in mammalian cells. Elife 5:e18858. doi: 10.7554/eLife.18858
Zhou, S., Zhao, Z., Lin, Y., Gong, S., Li, F., Pan, J., et al. (2016). Suppression of pancreatic ductal adenocarcinoma growth by intratumoral delivery of attenuated Salmonella typhimurium using a dual fluorescent live tracking system. Cancer Biol. Ther. 17, 732–740. doi: 10.1080/15384047.2016.1177683
Keywords: bacteria, biosensors, disease diagnosis, personalized medicine, real-time monitoring
Citation: Liu M, Yang W, Zhu W and Yu D (2025) Innovative applications and research advances of bacterial biosensors in medicine. Front. Microbiol. 16:1507491. doi: 10.3389/fmicb.2025.1507491
Received: 07 October 2024; Accepted: 24 March 2025;
Published: 23 April 2025.
Edited by:
Thomas Maskow, Helmholtz Association of German Research Centres (HZ), GermanyReviewed by:
Huan Chen, Zhejiang Chinese Medical University, ChinaArun Karnwal, Graphic Era University, India
Copyright © 2025 Liu, Yang, Zhu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daojun Yu, eXVkYW9qdW45OEAxNjMuY29t
 Mengting Liu1,2
Mengting Liu1,2 Daojun Yu
Daojun Yu