- 1College of Food Science and Technology, Yunnan Agricultural University, Kunming, China
- 2Sericulture and Apiculture Research Institute, Yunnan Academy of Agricultural Sciences, Mengzi, Yunnan, China
- 3Heqing County Inspection and Testing Institute, Heqing, Yunnan, China
Introduction: Exopolysaccharides (EPS) produced by Lactic acid bacteria have many health benefits and unique physicochemical properties. They are widely used in the food industry to improve viscosity, mouthfeel, and textural properties of foods. In our previous studies, Limosilactobacillus fermentum A51 (L. fermentum A51) isolated from yak yogurt exhibited high EPS production capacity and was applied to improve the texture of yogurt. In this study, whole genome sequencing analysis and corresponding in vitro assays were performed to investigate the probiotic potential and safety properties of L. fermentum A51.
Results: Scanning electron microscopy (SEM) observed that L. fermentum strain A51 adhered into clusters and its colony exhibited the obvious silk drawing phenomenon. Whole genome mapping revealed that L. fermentum A51 genome is 2,188,538 bp, and with an average guanine and cytosine (GC) content of 51.28%. PGAAP annotation identified 2,152 protein-encoding genes and 58 rRNAs, 15 tRNAs, and 5 5sRNAs. Hemolysis and antibiotic resistance tests, combined with the analysis of genes involved in antibiotic resistance, virulence factor, and hemolysins, suggested that L. fermentum A51 is safe. Fifty-one carbohydrate active enzyme genes in the whole genome sequence of L. fermentum A51 were annotated by carbohydrate active enzymes (CAZymes). Furthermore, L. fermentum A51 possesses adhesion, acid tolerance, bile salt tolerance, and heat tolerance genes (srtA, tuf, Bsh, nhaC, Ntn, cfa), antioxidant (nrfA, npr, nox2, tps), antibacterial genes (Idh and Dld) EPS synthesis-related genes (glf, epsG, gtf, Wzz, Wzx, Wzy), and signal molecule A1-2 synthesis-related genes (luxS, pfs). These probiotic genes were verified by quantitative real-time PCR. In vitro assays confirmed that L. fermentum A51 showed good tolerance to simulated gastrointestinal tract (8.49 log CFU/mL), 0.3% bile salt (39.06%), and possessed adhesion (86.92%), antioxidant (70.60–89.71%), and antimicrobial activities, as well as EPS and signaling molecule AI-2 synthesis capacities.
Conclusion: Collectively, our findings have confirmed that L. fermentum A51 is safe and exhibits good probiotic properties, thus recommending its potential application in the production of value-added fermented dairy products.
1 Introduction
LAB are generally recognized as safe (GRAS) microorganisms and are widely and safely used in the production of fermented dairy, fruit and vegetable products, and fermented meat products (Stefanovic et al., 2017; Saadat et al., 2019). Over the past decade, LAB have gained widespread attention for their health-promoting properties and are thus increasingly used as supplements in the food industry (Vasiee et al., 2018; Ma et al., 2019). The World Food Organisation (FAO) and the World Health Organisation (WHO) define probiotics as living microorganisms that produce health benefits for the host when consumed in sufficient quantities. Limosilactobacillus fermentum (L. fermentum) is one of the most versatile LAB strains. Studies have reported that L. fermentum exhibits probiotic activities such as antibacterial, antioxidant, anti-inflammatory, and immunomodulatory activities, in addition to the ability in prevention and treatment of hyperuricemia and gout, prevention of diseases and bile salt tolerance, among other functions (Zhao et al., 2019; Zhao et al., 2021a; Zhao et al., 2021b; Łepecka et al., 2023). The probiotic properties of L. fermentum are positively correlated with the functional metabolites that it produces. For instance, lactic acid and bacteriocin NQGPLGNAHR produced by L. fermentum show anti-adhesive and bactericidal activity against Staphylococcus aureus (Song et al., 2021). EPS produced by L. fermentum have many health benefits and unique physicochemical properties. They are widely used in the food industry to improve viscosity, mouthfeel, and textural properties of foods (Korcz and Varga, 2021). L. fermentum MWLf-4 and L. fermentum MWLp-12 have a high production of EPS, which can be used to increase the viscosity of fermented milk (Wang et al., 2023). A study reveals that L. fermentum U-21 possesses superior in-vivo and in-vitro antioxidant activity (Grishina et al., 2023). Notably, L. fermentum has been widely used in the food industry. In the recent past, studies have gradually extended from physiological and biochemical analyses to conducting further investigation of molecular mechanisms and genetic properties (Nordstrm et al., 2021).
LAB exhibits various probiotic activities; however, it is time-consuming and expensive to explore the health benefits of LAB through in-vitro and in-vivo experiments. Recently, the development of high-throughput genomic methods has provided the means for rapidly understanding the genetic and probiotic properties of LAB, including their genes, metabolic capabilities, and potential health benefits (Wang et al., 2023). For instance, Wang et al. (2023) identified tagE and glmU genes regulating EPS production in L. fermentum MWLf-4 and the cps gene cluster and galE gene in L. plantarum MWLp-12 using whole gene sequencing. Liu et al. (2022) identified potential genes involved in the probiotic functions of L. plantarum DMDL 9010, including stress responses, bile salt resistance, adhesion ability, EPS biosynthesis and plantaricin biosynthesis using complete genome sequencing. Gao et al. (2020) also employed whole genome sequencing to identify potential genes involved in the probiotic function of L. plantarum Y44, including stress-related genes (groES-groEL and CLIC), gastric and intestinal transit tolerance-related genes (Bsh and cfa), antioxidant activity-related genes (nox, nrdH, and trxB), and EPS synthesis-related genes (glf, epsD, gtf, Wzx, Wzy, and wzx). Whole genome sequencing results indicate that L. plantarum Y42 genome contains genes associated with LuxS/AI-2 quorum sensing (QS) system (Li et al., 2023). Meanwhile, whole genome sequencing has been used to rapidly analyze safety-related biological information of LAB, including antimicrobial resistance genes and virulence factor gene annotations (Li et al., 2023; Lu et al., 2023). Based on the above research, it is apparent that combining whole genome sequencing and phenotyping analyses can provide insights into the probiotic properties and safety of LAB (Gu et al., 2023).
The evaluation of the safety and probiotic properties of LABs is a prerequisite for their application in the food industry. In our previous study, L. fermentum A51 isolated from naturally fermented yak yogurt was proven to have good EPS production capacity (452.728 mg/L) and fermentation properties. Especially, L. fermentum A51 was successfully applied to improve the textural properties of yogurt (Wei et al., 2023). However, the safety and probiotic properties of L. fermentum A51 are still unknown. In this study, the whole genome of L. fermentum A51 was sequenced using the third-generation sequencing technique. The antibiotic sensitivity and hemolytic activity of L. fermentum A51 were determined. Furthermore, the probiotic activities and probiotic genes, including gastrointestinal digestive tolerance, adhesion, antioxidant, and antimicrobial activities, as well as EPS and signaling molecule AI-2 synthesis capacity were investigated. The results provide insights into the probiotic properties and safety of L. fermentum A51, and will guide its application in the production of value-added fermented dairy products with probiotic properties.
2 Materials and methods
2.1 Materials
2.1.1 Bacterial strains
L. fermentum A51 (Gene bank accession number: CP132542) used in the current study was isolated from naturally fermented yak yogurt in Yunnan province and identified by Gram stain reaction, morphological, and 16S rDNA sequence analysis. Subsequently, sequence similarity was assessed using BLAST tool, and a phylogenetic tree was constructed using the neighbor-joining method in Mega 7.0 software according to 16S rRNA gene sequences from 17 Lactobacillus strains. The strain has been deposited in the China Center for Type Culture Collection (CCTCC NO: M 2023861). The strain was stored at the College of Food Science and Technology, Yunnan Agricultural University, Kunming, China. The bacterial strains were preserved at −80°C in glycerol supplemented with 20% (v/v) nutrient broth. Lactobacillus casei Zhang was obtained from the Inner Mongolia Agricultural University. Vibrio harveyi BB170 (ATCC BAA-1117), which was used as an indicator bacteria, was purchased from Guangdong Microbial Culture Collection Center (GDMCC). Escherichia coli CICC 10003 and Listeria monocytogenes were purchased from the China Center of Industrial Culture Collection (Beijing). Staphylococcus aureus ATCC25923 was purchased from the American Type Culture Collection.
Columbia agar medium supplemented with 7% sterile defibrinated sheep blood and de Man-Rogosa-Sharpe (MRS) medium were purchased from Solarbio Science & Technology Co., Ltd., Beijing, China. Pepsin (from porcine gastric mucosa, ≥ 250 U/mg), trypsin (from bovine pancreas, ≥ 10,000 U/mg), and bile salts were purchased from Sigma-Aldrich® (St. Louis, MO, USA). In addition, 1,1-Diphenyl-2-picrylhydrazyl radical (DPPH) and 2,2′-diazo-bis (3-ethylbenzothiazolin-6-sulfonic acid) diammonium salt (ABTS) were purchased from Shanghai Jingchun Biochemical Technology Co., Ltd. (Shanghai, China).
2.2 Morphology of strains
One milliliter of third-generation L. fermentum A51 was gradient diluted to 10−6 folds, spread on MRS solid medium (Beijing Solarbio Science & Technology Co., Ltd. Beijing, China) and then incubated at 37°C for 48 h. Lengths of single colonies that were sticky and had obvious filament pulling with a sterilized gun were recorded. The colonies of L. fermentum A51 were Gram-stained and observed under a microscope (E200, Nikon, Japan).
The morphology of L. fermentum A51 was observed using biological scanning electron microscopy. Briefly, cells were obtained by centrifugation at 8,000 rcf, 4°C for 10 min, then washed twice with phosphate buffer (0.1 M, pH 7.2). The cells were fixed with 2.5% glutaraldehyde for 12 h and washed three times with phosphate buffer. Next, gradient dehydration with ethanol was performed for 10 min each time, and bacteria were precipitated by centrifugation at 8,000 rcf, 4°C for 10 min. Tert-butanol was used instead of ethanol for centrifugation. The bacteria were diluted with phosphate buffer, dropwise freeze-dried on slides, and sputter-coated with gold (Wu et al., 2015). Finally, cells were observed using a regulus 8,100 biological SEM (Hitachi Ltd., Tokyo, Japan).
2.3 Genome sequencing and annotation
L. fermentum A51 was sequenced using Illumina NovaSeq 6000 and PacBio Sequel II platforms by Biomarker Technologies Co, LTD. (Beijing, China). At least 1 μg of each genomic DNA sample was used for Illumina sequencing to construct a sequence library. DNA samples were sheared into 400–500 bp fragments using a Covaris M220 Focused Acoustic Shearer. Illumina sequencing libraries were prepared from the sheared fragments. The prepared libraries were then used for paired-end Illumina sequencing (2 × 150 bp) on an Illumina NovaSeq 6,000 machine. For PacBio sequencing, SMRTbell library inserts (20 kb) were sequenced, and subreads shorter than 500 bp were removed. The PacBio sequences were error-corrected, binned, and then assembled using the Canu assembler (version 2.2).1 Pilon software (version 1.242) was used for polishing assemblies from Illumina short reads to improve genome quality. The predicted gene sequences were translated and searched against the National Center for Biotechnology Information (NCBI) database.3 Infernal v1.1.3, tRNAscan-SE v2.0, CRT v1.2, and IslandPath-DIMOB v0.2 were used for the prediction of rRNA, tRNA, CRISPR sequence, and gene island, respectively. The non-redundant protein (Nr) database, the Gene Ontology (GO) database, the Kyoto encyclopedia of genes and genomes (KEGG) database, and the carbohydrate-active enzymes (CAZy) database were used for annotation. Putative virulence genes were identified by comparing the whole genome of L. fermentum A51 with Virulence Factors of Pathogenic Bacteria (VFDB) database.4 Antibiotic resistance genes of L. fermentum A51 were identified using Comprehensive Antibiotic Research (CARD) database5 (Liu et al., 2022).
2.4 Detection of antibiotic sensitivity
The antibiotic sensitivity of L. fermentum A51 was evaluated using the disk diffusion method (Lu et al., 2023). Briefly, an MRS agar plate was overlaid with L. fermentum A51 culture (200 μL) containing 108 CFU/mL and antibiotic discs containing erythromycin, chloromycetin, tetracycline, ciprofloxacin, clindamycin, ampicillin, gentamicin, kanamycin, vancomycin, and ofloxacin were placed on the plates under sterile conditions. After incubation at 37°C for 48 h, the diameter (mm) of the inhibition zones was measured and the level of antibiotic sensitivity of the strain was determined.
2.5 Hemolytic activity
The hemolytic activity of L. fermentum A51 was measured as described by Li et al. (2023). Briefly, L. fermentum A51 was inoculated on Columbia agar medium supplemented with 7% sheep blood and cultured anaerobically at 37°C for 48 h. Lactobacillus casei Zhang was used as the control strain. Hemolytic rings were observed to confirm the hemolytic activity of the strains. A transparent area meant β-hemolysis and has hemolysis, while the absence of a transparent zone (γ-hemolysis) or a green zone (α-hemolysis) meant non-hemolysis strain.
2.6 Probiotic properties evaluation of Limosilactobacillus fermentum A51
2.6.1 Bile salt tolerance test
The bile salt tolerance of L. fermentum A51 was determined according to the method reported by Yan et al. (2023). The bacterial solution (1.0 mL, 109 CFU/mL) was added to 9 mL MRS medium (with 0.1, 0.2, 0.3 and 0.4% w/v bile salt) and incubated at 37°C for 4 h. After the incubation, 100 μL of the fermentation broth was taken separately, and the number of viable bacteria was counted after appropriate dilution on MRS agar plates. The medium without added bile salts was used as a control and the survival rate was calculated.
2.6.2 Gastrointestinal fluid tolerance test
The tolerance of L. fermentum A51 in in vitro simulated gastrointestinal was investigated according to the method reported by Fei et al. (2018) with slight modifications. The gastric juice (3 mg/mL pepsin) was prepared in phosphate buffered saline (PBS, pH 3.0). The intestinal fluid (0.1 mg/mL trypsin, 0.15% (w/v) bile salts) was prepared in PBS (pH 6.8). Activated bacterial solution was added to the artificial gastric juice and incubated at 37°C for 2 h before plate colony counting. After incubating in the simulated gastric fluid for 2 h at 37°C, 1 mL of activated bacterial solution was added to 9 mL of simulated artificial intestinal fluid (pH 8.0). After incubating at 37°C for 2 h, the intestinal fluid tolerance was determined by counting total viable cells. Distilled water was used as a blank control. Lactobacillus casei Zhang was used as the control strain.
2.6.3 Detection of adhesion capacity
The adhesion capacity of L. fermentum A51 was assessed by hydrophobicity and self-aggregation tests (Peng et al., 2023). Lactobacillus casei Zhang was used as the control strain. Briefly, the washed bacterial sludge was taken, and the concentration of bacterial suspension was adjusted to 106–107 CFU/mL with saline; the absorbance at 600 nm was measured and counted as A0. Then 1 mL each of xylene, chloroform, and ethyl acetate were added to 3 mL of bacterial suspension and mixed well. The aqueous phase was taken after 20 min of standing and its absorbance at 600 nm was measured and counted as A1. The hydrophobicity of the strain to the solvent was calculated according to the following formula:
A total of 5 mL of the bacterial suspension was pipetted into an EP tube, vortexed for 30 s, and then left to stand for 5 h. The supernatant was aspirated at 1 h intervals and the absorbance value at 600 nm was determined. The auto aggregation capability of the strain was calculated according to the following formula:
where At represents the absorbance value of the bacterial suspension at 8, 12, and 24 h and A0 represents the absorbance value at 0 h.
2.6.4 Assays for antioxidant activities
The radical scavenging activity of L. fermentum A51 against DPPH radical, ABTS radical, and reducing capability were determined following previous methods (Sun et al., 2022; Liu et al., 2021). Lactobacillus casei Zhang was used as the control strain.
2.6.5 Evaluation of antibacterial activity
The antimicrobial activity of L. fermentum A51 against Escherichia coli (E. coli), Staphylococcus aureus (S. aureus) and Listeria monocytogenes (L. monocytogenes) was determined by oxford cup assay (Delgado et al., 2007). Briefly, the E. coli, S. aureus and L. monocytogenes cultured to a mid-log phase were mixed with LB agar broth at 37°C and then poured into Petri dishes. Then, 150 μL of L. fermentum A51 cell-free supernatant (CFS) was added to an 8-mm LB agar plate. The plates were then incubated at 37°C and then checked for inhibition after 12 h. Lactobacillus casei Zhang was used as the control strain.
2.6.6 Determination of EPS production
The EPS synthesis ability of L. fermentum A51 was measured according to Li M. et al. (2022). Briefly, L. fermentum A51 was inoculated at 5% (V/V) in fresh MRS medium and incubated at 37°C for 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, and 26 h. The fermentation broth was centrifuged at 6250 rcf for 15 min at 4°C to obtain CFS. Then, trichloroacetic acid (TCA) at a final concentration of 4% was added to the CFS to remove proteins. The solution was centrifuged at 8,000 rcf for 15 min at 4°C to obtain supernatant. The supernatant was concentrated to one-third of its original volume; and after 99.95% chilled ethanol at three times its volume was added, it was stored overnight at 4°C. The precipitate was collected and redissolved in deionized water. After dialysis and lyophilization, EPS produced by L. fermentum A51 was obtained, and the EPS content was determined by phenol-sulfuric acid method.
2.6.7 Determination of activity of signaling molecule AI-2
The ability of L. fermentum A51 to secrete the signaling molecule AI-2 was determined based on the method of Ma et al. (2022). Vibrio harveyi (V. harveyi) BB170 was cultured overnight in AB medium until the OD600 reached 0.9–1.1. The V. harveyi BB170 solution was diluted with fresh autoinducer bioassay (AB) medium at a volume ratio of 1:100. Sterile supernatants (obtained by filtering through a 0.22 μm microbial filter) of L. fermentum A51 incubated at 37°C for 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, and 26 h, negative control and medium control were mixed with the diluted V. harveyi BB170 solution at a volume ratio of 1:50 and incubated at 100 rpm and 30°C in a shaker (Shanghai Zhichu Instrument Co., Ltd. Shanghai, China). The AI-2 content of each test group was calculated when the fluorescence value of the negative control is the lowes. The content of AI-2 is the ratio of the fluorescence value of the experimental group to the fluorescence value of the medium group.
2.7 Determination of expression levels of probiotic genes
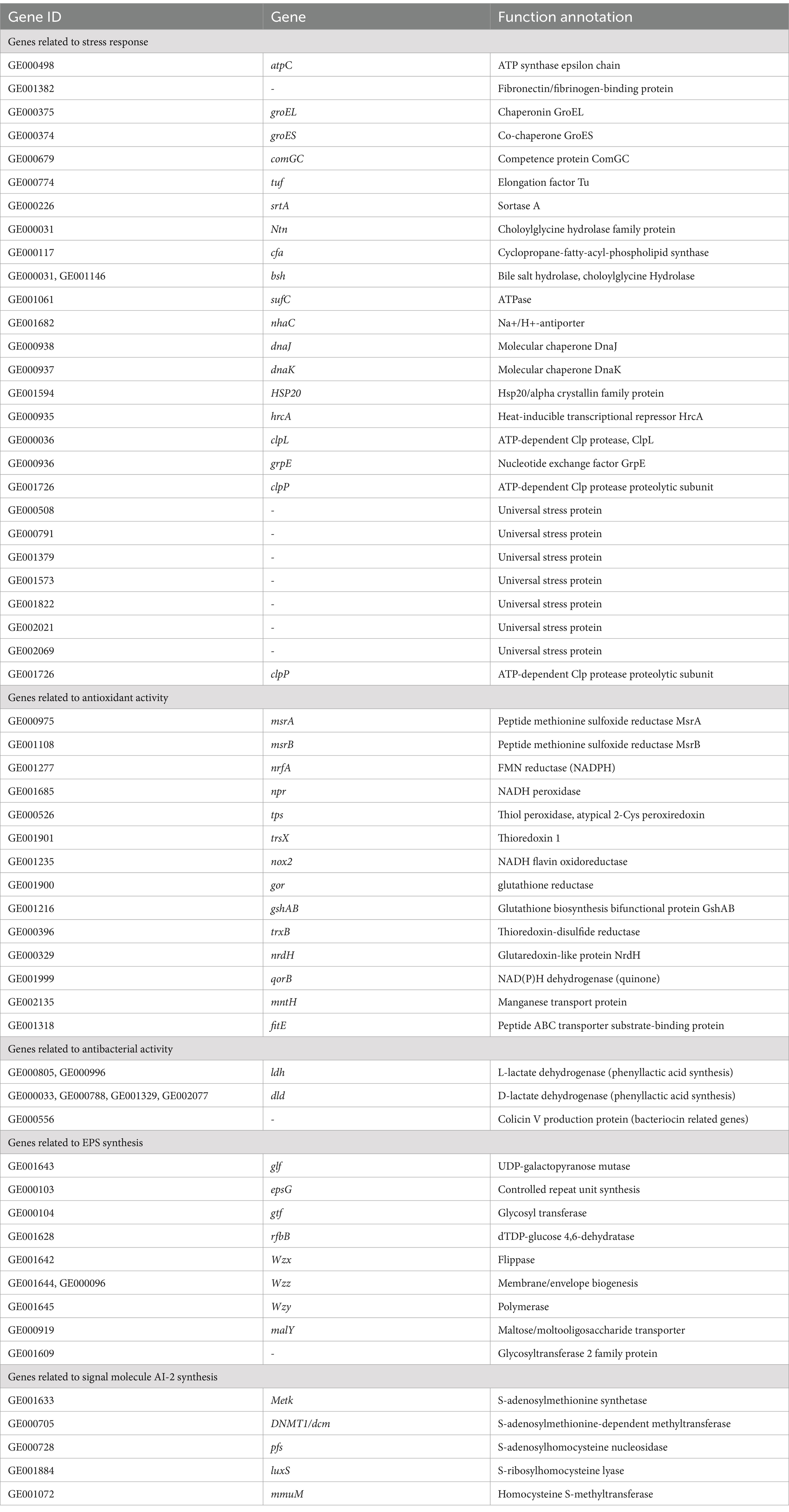
Total RNA was isolated from L. fermentum A51 (incubated for 20 h) using RNA Extraction Kit (Beijing Tsingke Biotech Co., Ltd.). The A260/280 ratios of the extracted RNA were about 2.1. The total RNA was reverse-transcribed into cDNA using Goldenstar RT6 cDNA Synthesis Kit Ver 2 (Qingke Biotechnology [Beijing] Co., Ltd., China). Real-time PCR analysis of candidate genes, including luxS, glf, epsG, gtf, Cfa, Wzz, Wzx, bsh, nhaC, npr, nox2, Idh, and Dld, was performed using ABI, QuantStudioTM 1 Plus real-time fluorescence quantitative PCR instrument (Thermo Fisher, USA). Gene-specific primers used in the experiment are listed in Table 1. Cfa gene was used as an internal reference gene. Probiotic genes were analyzed using the 2-ΔΔCt method.
2.8 Data analysis
Experimental data are presented as mean ± SD of triplicate measurements. Data analysis was performed using SPSS 23.0 (SPSS Inc., USA). Statistical significance differences (p < 0.05) between treatments were determined by one-way analysis of variance (ANOVA). All the figures were generated using the Origin Pro software (Origin-Lab, version 2021, USA).
3 Results and discussion
3.1 Morphology of Limosilactobacillus fermentum A51
The colony morphology of high EPS-producing LAB grown on MRS solid media is known to be mainly filamentous, mucilaginous, and annular (Xu, 2019). As can be seen in Figures 1A,B, the colony of L. fermentum A51 was rounded and greyish-white with neat, elevated, mucilaginous annular morphologies, and had better drawing properties, which was similar to the colony morphology of EPS-producing LAB reported by Ruas-Madiedo and de Los Reyes-Gavilán (2005) and Trabelsi et al. (2015). As shown in Figure 1C, L. fermentum A51 could be observed as short purple rods, which is an indication that the strain is Gram-positive bacteria. L. fermentum A51 was observed as a slender rod structure, typical morphology of the species under SEM. Interestingly, the colonies distribution was dense, which can be attributed to the secreted EPS of L. fermentum, promoting the adhesion of the colonies into clusters (Figures 1D–F; Zhang et al., 2023).
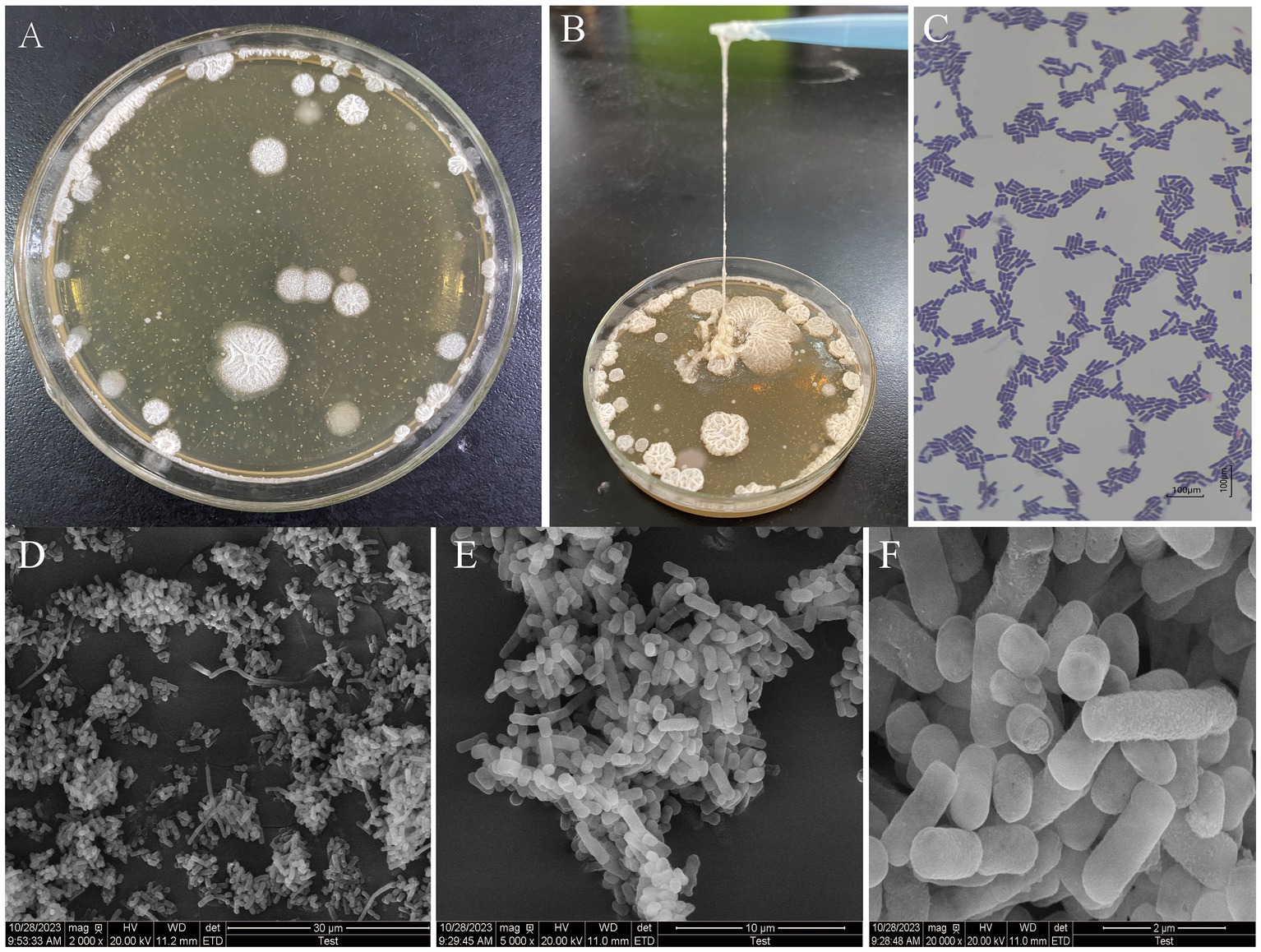
Figure 1. Morphology of L. fermentum A51: (A) Colony morphology observed on MRS solid plates. (B) Colony thread-drawing observed on MRS solid plates. (C) Gram-staining; Scanning electron microscope image. (D) 2,000 × magnification. (E) 5,000 × magnification. (F) 20,000 × magnification.
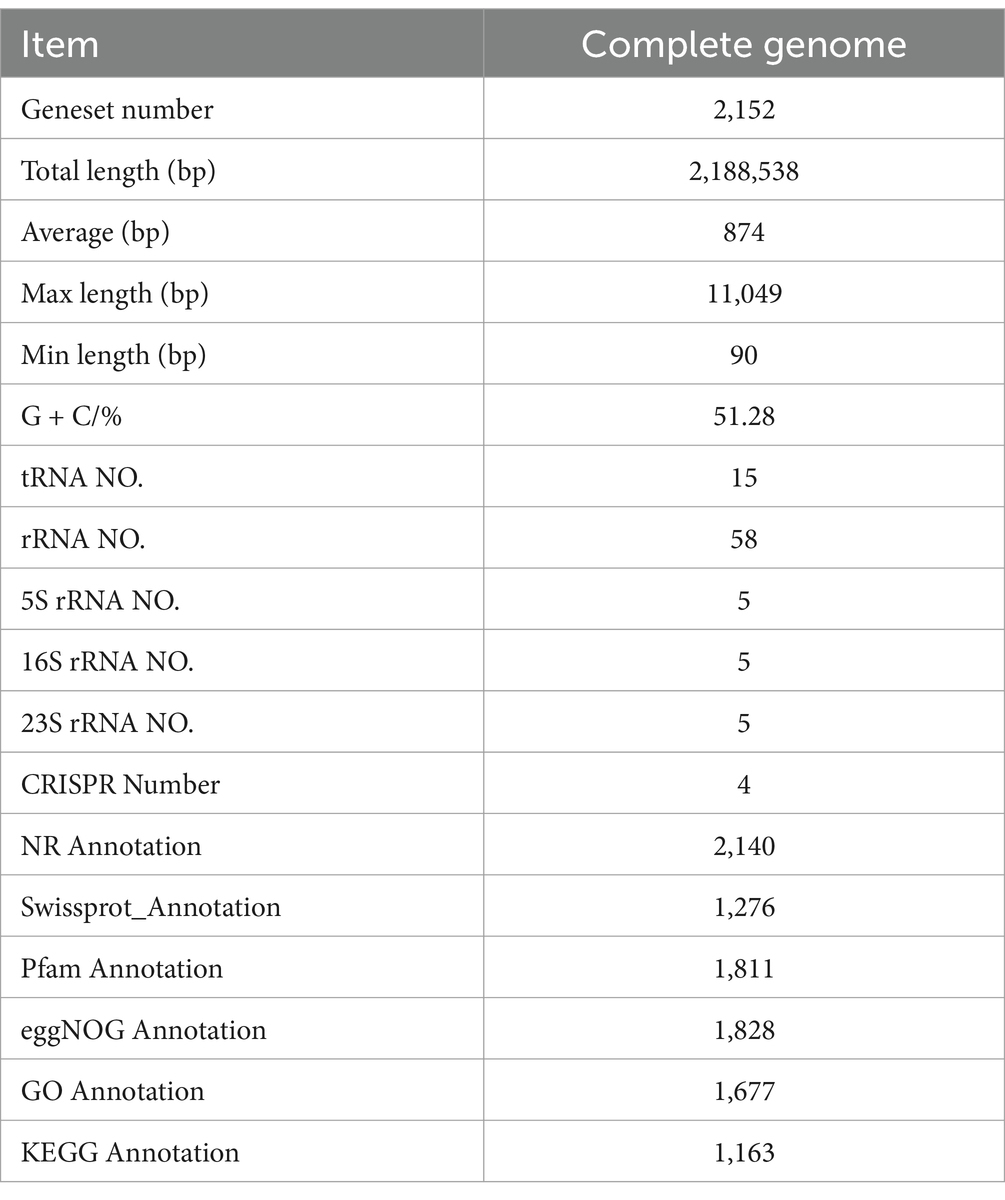
3.2 Genome features of Limosilactobacillus fermentum A51
3.2.1 General genome features
The phylogenetic tree based on 16S rRNA sequences is shown in Figure 2A. The phylogenetic analysis indicated that A51 is closely related to the strain L. fermentum CIP 102980. Consequently, A51 was identified as L. fermentum and named Limosilactobacillus fermentum A51.
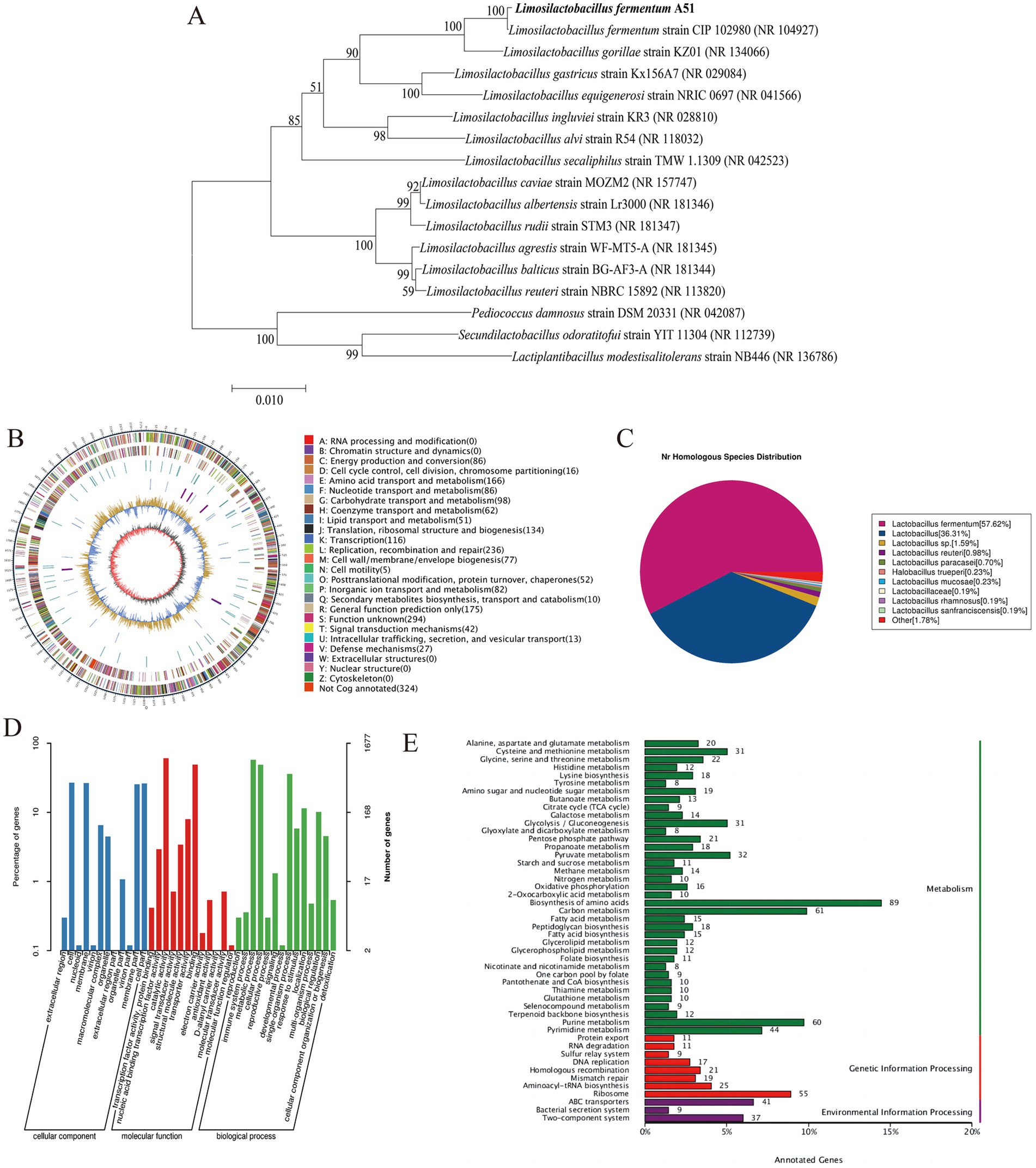
Figure 2. (A) Phylogenetic tree was carried out using 1,000 replicates and the number on the nodes represents the support proportion of each branch. (B) Circular genome maps. (C) NR database-annotated statistics chart. (D) GO annotation classification statistics chart, and (E) KEGG database annotation of L. fermentum A51.
In order to obtain a comprehensive understanding of the genome features of L. fermentum A51, the whole genome mapping of L. fermentum A51 was conducted using PacBio triple sequencing technology, and the sequencing results are shown in Figure 2B and Table 2. The complete genome of L. fermentum A51 consisted of a circular chromosome of 2,188,538 bp and had an average gene length of 874 bp and an average GC content of 51.28%. A total of 2,152 genes were predicted by PGAAP and found to include 2,079 protein-coding sequences, 58 rRNAs, 15 tRNAs, and 5 5sRNAs genes. Three incomplete pre-phage sequences with lengths of 85,965 bp, 26,620 bp, and 46,719 bp were predicted in L. fermentum A51. The complete genome of L. fermentum A51 has been deposited in the NCBI GenBank database (Accession No: CP132542). In addition, four plausible CRISPR regions in L. fermentum A51 were predicted. In addition, the NR database annotation results showed that the top 11 species in the NR database with genes matched genes encoding L. fermentum A51 were: Lactobacillus fermentum, Lactobacillus (57.62%), Lactobacillus sp. (35.67%), Lactobacillus reuteri (1.55%), Lactobacillus paracasei (1.01%), Lactobacillus mucosae (0.69%), Halobacillus trueperi (0.27%), Lactobacillus rhamnosus (0.23%), Lactobacillus sanfranciscensis (0.18%), Lactobacillaceae (0.18%), and other (1.74%) (Figure 2C). Therefore, the strain A51 was identified as Limosilactobacillus fermentum, which is consistent with the results of the phylogenetic tree analysis.
3.2.2 Annotation of functional genes of Limosilactobacillus fermentum A51
To further understand the gene function of L. fermentum A51, we annotated the identified genes using the GO and KEGG databases. In total, 1,828 genes were annotated by the protein database eggNOG, 1,677 genes were annotated by the GO database, and 1,163 functional genes were annotated by the KEGG database (Table 2). The GO database has an inverted root structure for biological functions, with the three top-level functional nodes being: (1) cellular component (CC), (2) molecular function (MF), and (3) biological process (BP). Among the whole genome sequence of L. fermentum A51, 1,677 genes were annotated by the GO database (Figure 2D). These included 1,975 genes related to cellular component CC (427 membrane component, 440 cellular component, 109 macromolecular complex, 75 organelle, and 18 organelle component, etc.), 2,127 genes related to molecular function MF (1,016 catalytic activity, 825 binding, 134 transporter activity, 57 structural molecular activity, 12 signal transducer activity, 9 antioxidant activity, etc.), and 2,976 genes related to biological processes BP (965 metabolism, 816 cellular processes, 602 single organism processes, 170 biomodulation, 98 stress response, 22 signaling, etc.). Although these genes are involved in every part of cellular metabolism, they have different intensities, indicating their involvement of different processes in the cell is different.
There were 1,163 genes in the whole genome sequence of L. fermentum A51 annotated by KEGG, of which metabolism-related genes were the most numerous genes, including 60 for purine metabolism, 44 for pyrimidine metabolism, 32 for pyruvate metabolism, amino acid metabolism (pyruvate metabolism 31, glycine, serine and threonine metabolism 22, alanine, aspartate and glutamate metabolism 20, histidine metabolism 12), and gluconeogenesis (glycolysis/gluconeogenesis 31, pentose phosphate pathway 21, galactose metabolism 14 and phosphotransferase system (PTS) 7). These numbers show that the bacterium has a strong protein and sugar catabolism and metabolism function (Liu et al., 2022). In addition, biosynthesis-related genes were also annotated by KEGG: 89 genes for amino acid biosynthesis, 18 for lysine biosynthesis, 18 for peptidoglycan biosynthesis, 15 for fatty acid biosynthesis, 12 for arginine biosynthesis and 11 for folate biosynthesis (Figure 2E).
3.3 Safety assessment of Limosilactobacillus fermentum A51
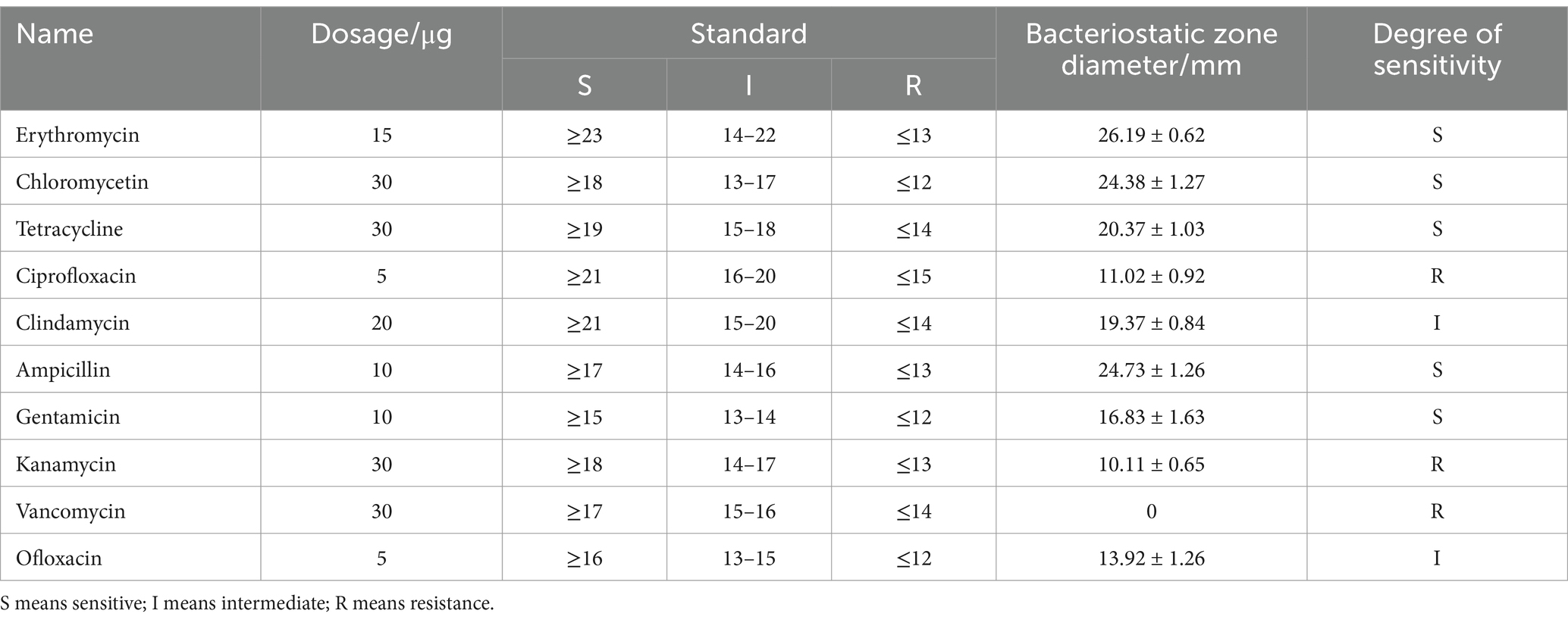
3.3.1 Antibiotic sensitivity
As potential probiotics, LAB should not exhibit transferable antibiotic resistance. In the current study, the susceptibility of L. fermentum A51 to ten common antibiotics was determined (Table 3). The results show that L. fermentum A51 is sensitive to erythromycin, chloromycetin, tetracycline, ampicillin, and gentamicin, intermediate to clindamycin and ofloxacin, resistant against ciprofloxacin, kanamycin, and vancomycin. Lactobacilli are resistant to vancomycin because their peptidoglycan contains D-Ala-D-lactic acid rather than D-Ala-D-Ala dipeptide (Oh et al., 2022). Notably, some studies have confirmed that many of the resistance attributes are inherent and cannot be transferred (Salminen et al., 1998). Thus, L. fermentum A51 is considered safe.
3.3.2 Hemolytic activity
The hemolytic activity of probiotics is one of the most important criteria for their safety assessment. The hemolysis assay showed that a transparent ring was observed around the bacteria colonies of S. aureus CICC 10384 (positive control), which indicates β-hemolysis (Supplementary Figure S1C). A grass-green haemolytic ring found around the bacteria colonies of E. coli, indicating α-hemolysis (Supplementary Figure S1D). While, no hemolytic ring found around the bacteria colonies of L. fermentum A51 (Supplementary Figure S1A), which was consistent with the control strain Lactobacillus casei Zhang (Supplementary Figure S1B). Therefore, we considered the L. fermentum A51 to be γ-hemolytic, which is consistent with the results of Wang et al. (2023), who reported that Limosilactobacillus fermentum MWLf-4 from human milk was γ-hemolytic. This finding suggests that L. fermentum A51 is safe for human applications based on its hemolytic activity.
3.3.3 Antibiotic resistance genes of Limosilactobacillus fermentum A51
The CARD database search showed that L. fermentum A51 has only one potential antibiotic resistance gene (poxtA) that is resistant to tetracycline, oxazolidinone, and phenicol. However, the results of the antibiotic resistance test showed that L. fermentum A51 is sensitive to tetracycline, indicating that the tetracycline-encoding gene may not be expressed. In addition to the phenotypic results of the antibiotic resistance test, these findings suggest that L. fermentum A51 is not resistant to common antibiotics. This may be related to the origin of the strain. L. fermentum A51 is derived from fermented yak milk, which is made from fresh yak milk, sterilized, fermented, and post-cooked without the addition of preservatives during production.
3.3.4 Virulence factor genes of Limosilactobacillus fermentum A51
The virulence factor gene in L. fermentum A51 was annotated through the VFDB database. A total of 290 potential virulence factor genes were annotated in L. fermentum A5. However, most of the putative virulence factor genes exhibited less than 50% similarity to those in the VFDB database, and only five genes were annotated with similarity greater than 60% (but all≤75%) (Supplementary data 1). In addition, no genes encoding hemolysins (cylA, cylB, cylM, Hbl, Nhe, and cereulide-Ces) were identified in the L. fermentum A51 genome, which is consistent with the absence of hemolytic activity. Based on the annotation of the KEGG and COG functional databases, most of these putative virulence genes were annotated as genes involved in carbohydrate transport, ABC transport systems, two-component systems, bacterial adhesion, stress response, and quorum sensing systems. ABC transport system plays a key role in normal substance transportation and release of exogenous toxic compounds and waste metabolites in vivo (Zhu et al., 2019). The two-component system is involved in the regulation of various physiological and biochemical functions of lactic acid bacteria, which is an important regulatory system for their metabolic activities. In addition, most of these putative virulence genes were associated with carbohydrate transport, indicating that L. fermentum A51 has a strong sugar metabolism ability (Choi et al., 2008). The annotated adhesion genes allow L. fermentum A51 to better adhere to and colonize the host cells (Altermann et al., 2005; Senan et al., 2015). Meanwhile, some virulence factor genes are associated with probiotic functions in LAB. For example, the luxS gene is associated with the LuxS/AI-2 type quorum sensing system in LAB, which regulates tolerance and adhesion of LAB (Meng et al., 2022). The gene bsh is positively associated with bile salt tolerance in LAB (Choi and Chang, 2015). In pathogenic bacteria, genes associated with biofilm formation may be identified as pathogens. However, for LAB, these genes are associated with adhesion and resistance (Meng et al., 2022). Therefore, based on the results of hemolysis and antibiotic resistance tests, as well as the analysis of genes involved in antibiotic resistance, virulence factor, and hemolytic activity assay, we can postulate that L. fermentum A51 is safe.
3.4 Probiotic gene features of Limosilactobacillus fermentum A51
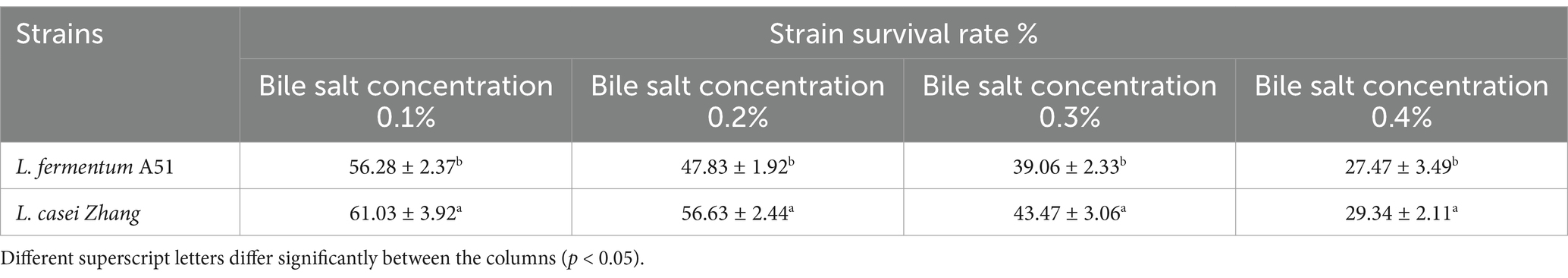
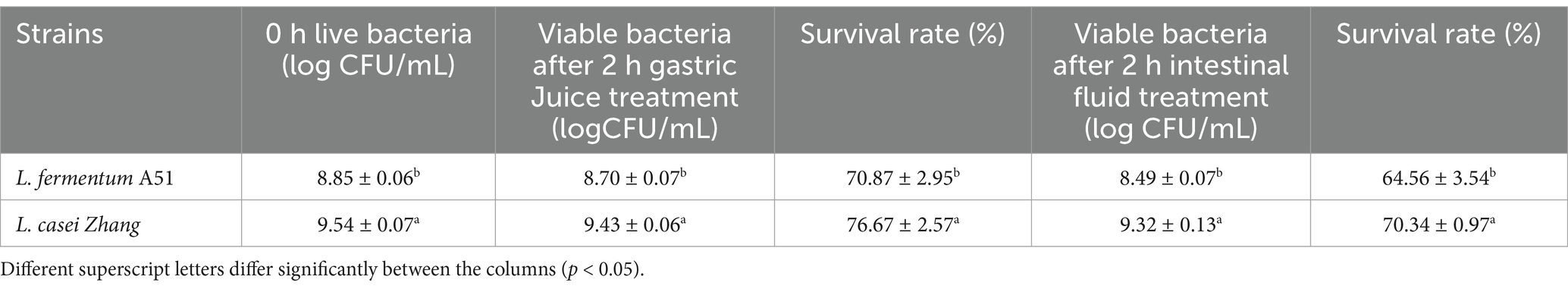
3.4.1 Genes related to stress response of Limosilactobacillus fermentum A51
A prerequisite that allows probiotics to exhibit its phenotypic characteristics is their ability to survive the passage through the human gastrointestinal tract. Therefore, LAB need to adapt to their acidic fermentation environment and also tolerate the acidic conditions and bile salt of the gastrointestinal tract. The survival rate of L. fermentum A51 in bile salt and in-vitro simulated gastrointestinal digestion is shown in Tables 4, 5. After treated in 0.3% bile salt for 4 h, the survival rate of L. fermentum A51 was 39.06%, which was lower than that of L. casei Zhang (43. 47%) but higher than that of L. plantarum ZFM4 (35.5%) (Yan et al., 2023), suggesting that L. fermentum A51 has better bile salt tolerance (Table 4). After digestion with simulated gastric juice for 2 h, the viable counts of L. fermentum A51 and L. casei Zhang (control) were 70.87% (viable counts was 8.70 log CFU/mL) and 76.67% (viable counts was 9.43 log CFU/mL), respectively, which were not significantly different (p < 0.05; Table 5). After being exposed to intestinal fluid for 2 h, the survival rates of L. fermentum A51 and L. casei Zhang decreased to 64.56% (viable counts was 8.49 log CFU/mL) and 70.34% (viable counts was 9.32 log CFU/mL) respectively. After gastric juice and intestinal fluid treatment, the viable counts of strains remained above 8 log CFU/mL, indicating that both strains were well tolerated. However, the tolerability to the gastrointestinal environment of L. casei Zhang is slightly better than that of L. fermentum A51. Previous studies have shown that EPS-producing LAB are more resistant and tolerant to bile salts than free LAB (Gu et al., 2020). L. casei Zhang is a highly EPS-producing strain (546.3 ± 31.5 mg/L; Ba et al., 2021). The gastrointestinal digestive tolerance of L. fermentum A51 and L. casei Zhang is related to their EPS production capacity. EPS produced by these two strains wrapped around the strains, forming a protective layer that enhances their environmental tolerance. In addition, bile salt hydrolase (Bsh) and choloylglycine hydrolase family protein (Ntn) produced in some LAB are important for the survival of the bacteria in the intestinal tract (Choi and Chang, 2015). Cyclopropane-fatty-acyl-phospholipid synthase (CFA), cystathionine-β-lyase, and cystathionine-γ-synthase protect LAB from adverse conditions such as acidity (Ma et al., 2019; Charnchai et al., 2017). Na+/H+-antiporter acts as a regulator of intracellular pH, enabling the bacteria to survive in an acidic environment (Iyer et al., 2002). Genes encoding bile salt hydrolase (Bsh), Na+/H+-antiporter (nhaC), choloylglycine hydrolase family protein (Ntn), cyclopropane-fatty-acyl-phospholipid synthase (cfa), and ATPase (sufC) were found in the L. fermentum A51 genome. The presence of these genes suggests that L. fermentum A51 can tolerate bile salts and acids (Table 6).
3.4.2 Adhesion capacity-related genes
Hydrophobicity and autoaggregation capability are the evaluation indexes of non-specific adhesion of LAB to the intestine (Peng et al., 2023). The hydrophobicity and autoaggregation capability of L. fermentum A51 was measured (Figures 3A,B). As shown in Figure 3A, the hydrophobicity of L. fermentum A51 in chloroform, xylene, and ethyl acetate were all significantly higher than that of L. casei Zhang (p < 0.05). In particular, L. fermentum A51 had the highest hydrophobicity of 83.03 ± 0.08% in chloroform. The autoaggregation rate of the strains all showed an increasing trend with the increase in adhesion time (Figure 3B). However, the autoaggregation capability of L. fermentum A51 remained higher than that of L. casei Zhang. At 24 h, the autoaggregation capability of L. fermentum A51 was 86.92 ± 0.34% (>50%), indicating it had good autoaggregation capability (Montoro et al., 2016). Whole-genome sequencing analysis showed that L. fermentum A51 contained adhesion-related genes, including genes encoding fibronectin/fibrinogen-binding protein, sortase A (srtA), competence protein ComGC (ComGC), chaperonin GroEL (GroEL) and elongation factor Tu (tuf), which allows L. fermentum A51 to better adhere to and colonize in the host cells (Altermann et al., 2005; Senan et al., 2015; Table 6).
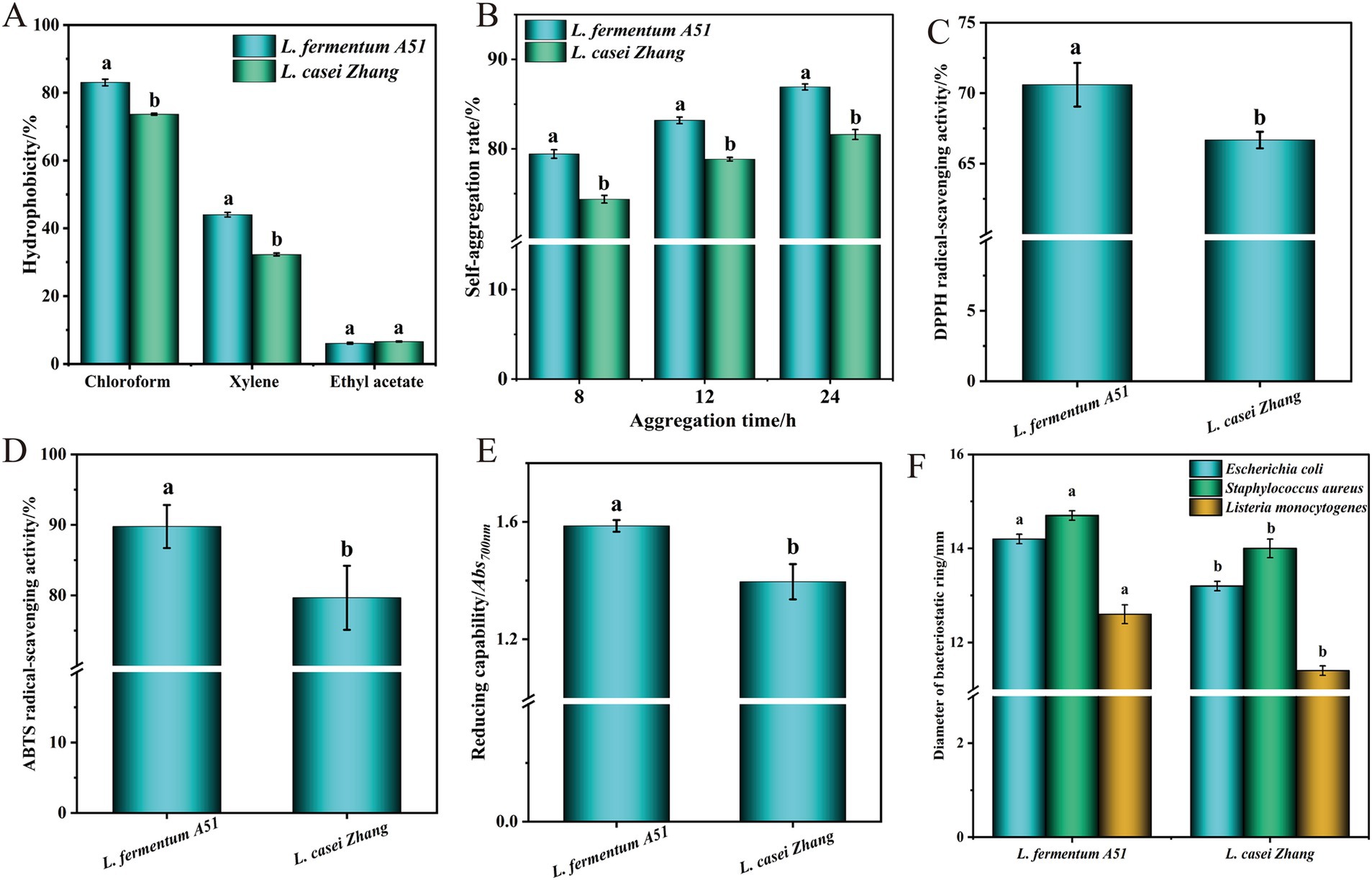
Figure 3. Probiotic properties of L. fermentum A51. (A) Hydrophobic capability. (B) Self aggregation capability. (C) DPPH free radical scavenging activity. (D) ABTS free radical scavenging activity. (E) Reducing capability. (F) Inhibitory activity against E. coli and S. aureus. Different lower case letters represent significant differences with p < 0.05 between L. fermentum A51 and L. casei Zhang.
3.4.3 Antioxidant activity-related genes
Many studies have shown that LAB has antioxidant capacity. In the current study, the in vitro antioxidant activity of L. fermentum A51 was measured, and the results are shown in Figures 3C–E. As shown in Figure 3C, the CFS of L. fermentum A51 showed that it had a DPPH radical scavenging activity of 70.60%, which is significantly higher (p < 0.05) than that of the control (66.67% for L. casei Zhang and 28.02% for Lactiplantibacillus plantarumisolated D444 from Chinese traditional fermented foods) (Zhang et al., 2010; Sun et al., 2022). In addition, the ABTS radical scavenging activity and reducing capability of L. fermentum A51 were significantly stronger than that of the control (p < 0.05) (Figures 3D–E), indicating that L. fermentum A51 has a strong antioxidant capacity. The antioxidant activity of L. fermentum A51 may be due to its ability to produce superoxide dismutase (SOD), reduced glutathione (GSH), ferulic acid esterase (FAE), ferulic acid (FA), NADH oxidase and NADH peroxidase (Cheng et al., 2020). Whole genome mapping analysis showed that L. fermentum A51 carried genes encoding thioredoxin reductase (NADPH), glutathione reductase, peptide methionine sulfoxide reductase msrA/msrB (msrA, msrB), thiol peroxidase (tps) and thioredoxin 1 (trsX) (Table 6), implying that L. fermentum A51 possesses antioxidant activity (Papadimitriou et al., 2016). NADH oxidase is directly involved in detoxifying hydrogen peroxide and ROS. The antioxidant enzyme glutathione reductase is a crucial antioxidant enzyme responsible for the maintenance of glutathione, one of the main antioxidant metabolites (Liang et al., 2020). The expression of thioredoxin genes can increase the activity of antioxidant enzymes, such as superoxide dismutase (SOD) and catalase (CAT). In addition, under certain conditions, thioredoxin can promote the conversion of oxidized glutathione to reduced glutathione (Shi et al., 2010). For non-enzymatic antioxidants, probiotics secreting EPS and antioxidant peptides (e.g., glutathione) can reduce oxidative damage and thus can prevent aging and various chronic diseases (Zhang et al., 2022; Xue et al., 2023). All the above findings indicate that L. fermentum A51 is a potential probiotic with antioxidant ability.
3.4.4 Antibacterial activity-related genes
The antibacterial activity of L. fermentum A51 against E. coli, S. aureus and L. monocytogenes was evaluated, and the results are shown in Figure 3F. The diameters of the inhibitory zones of L. fermentum A51 against E. coli, S. aureus and L. monocytogene were 14.2 ± 0.1 mm, 14.7 ± 0.1 mm and 12.6 ± 0.2 mm, respectively, which were higher (p < 0.05) than those of control (L. casei Zhang, 13.2 ± 0.1 mm, 14.0 ± 0.2 mm and 11.4 ± 0.1 mm). The antibacterial capacity of L. fermentum A51 is possibly related to the protein and non-protein antibacterial components, such as bacteriocins, organic acids, hydrogen peroxide and EPS, generated by the strain. Organic acid antibacterial substances, including lactic acid, phenyl lactic acid, citric acid, and acetic acid, are metabolized by LAB during fermentation. Organic acids inhibit the growth of pathogenic bacteria by increasing their outer membrane permeability, altering intracellular osmotic pressure, and inhibiting DNA synthesis. Preliminary experiments demonstrated that the antimicrobial substances in L. fermentum A51 were not bacteriocins, hydrogen peroxide, and EPS, but organic acids. Moreover, L. fermentum A51 contained genes involved in phenyl lactate synthesis (two ldh genes and four dld genes) (Table 6). Therefore, it can be assumed that the inhibitory effect of L. fermentum A51 against E. coli and S. aureus is also due to organic acids. Overall, these findings indicate that ldh and dld genes identified in the genome of L. fermentum A51 may help in its antimicrobial activity (Zhou et al., 2020).
3.4.5 Genes involved EPS synthesis
Fifty-one carbohydrate active enzyme genes in the whole genome sequence of L. fermentum A51 were annotated by CAZy, including auxiliary oxidoreductases (AAs, 3 genes), carbohydrate esterases (CEs, 9 genes), functional domains of carbohydrate-binding modules (CBMs, 9 genes), glycoside hydrolases (GHs, 17 genes) and glycosyltransferases (GTs, 13 genes) (Figures 4A,B). However, the annotation did not indicate polysaccharide lyase (PLs) family genes in the L. fermentum A51 genome. GHs have the potential to hydrolyze complex carbohydrates and are considered to be key enzymes responsible for carbohydrate metabolism. The 17 GH genes in L. fermentum A51 were predicted to belong to nine different families, according to the CAZy database, which suggests that L. fermentum A51 can utilize a wide range of carbohydrates. Alpha-galactosidase [EC 3.2.1.22] and beta-galactosidase [EC 3.2.1.23], members of the GH2 and GH36 family, are responsible for the metabolism of D-galactose and lactose (Su et al., 2010; Graebin et al., 2016). In this study, a variety of genes encoding GTs were found in L. fermentum A51, with GT2 and GT4 being the most abundant genes. GT2 and GT4 enzymes are associated with the synthesis of disaccharides such as sucrose, as well as the synthesis of lipopolysaccharides, cellulose, and chitosan. The high content of GT2 and GT4 (galactosyltransferase, glucosyltransferase, rhamnosyltransferase, and N-acetylglucosaminyltransferase [EC 2.4.1]) indicates that L. fermentum A51 has a high capacity in sugar and EPS synthesis. In summary, alpha/beta-glucosidases hydrolyze the glucosidic bonds to produce monosaccharides, providing precursor materials for EPS biosynthesis, whereas glycosyltransferases are responsible for transferring EPS from the intracellular space to the extracellular matrix. The KEGG annotation also identified L. fermentum A51 as rich in carbohydrate transport-and metabolism-related genes, including glycolysis/gluconeogenesis genes (31), phosphotransferase system genes (21) and galactose metabolism genes (14) (Supplementary Table S1). In particular, L. fermentum A51 possesses a complete galactose and glucose metabolic pathway. It can be implied that L. fermentum A51 can synthesize EPS using galactose and glucose and thus could potentially be used in value-added fermented dairy products (Figure 4C).
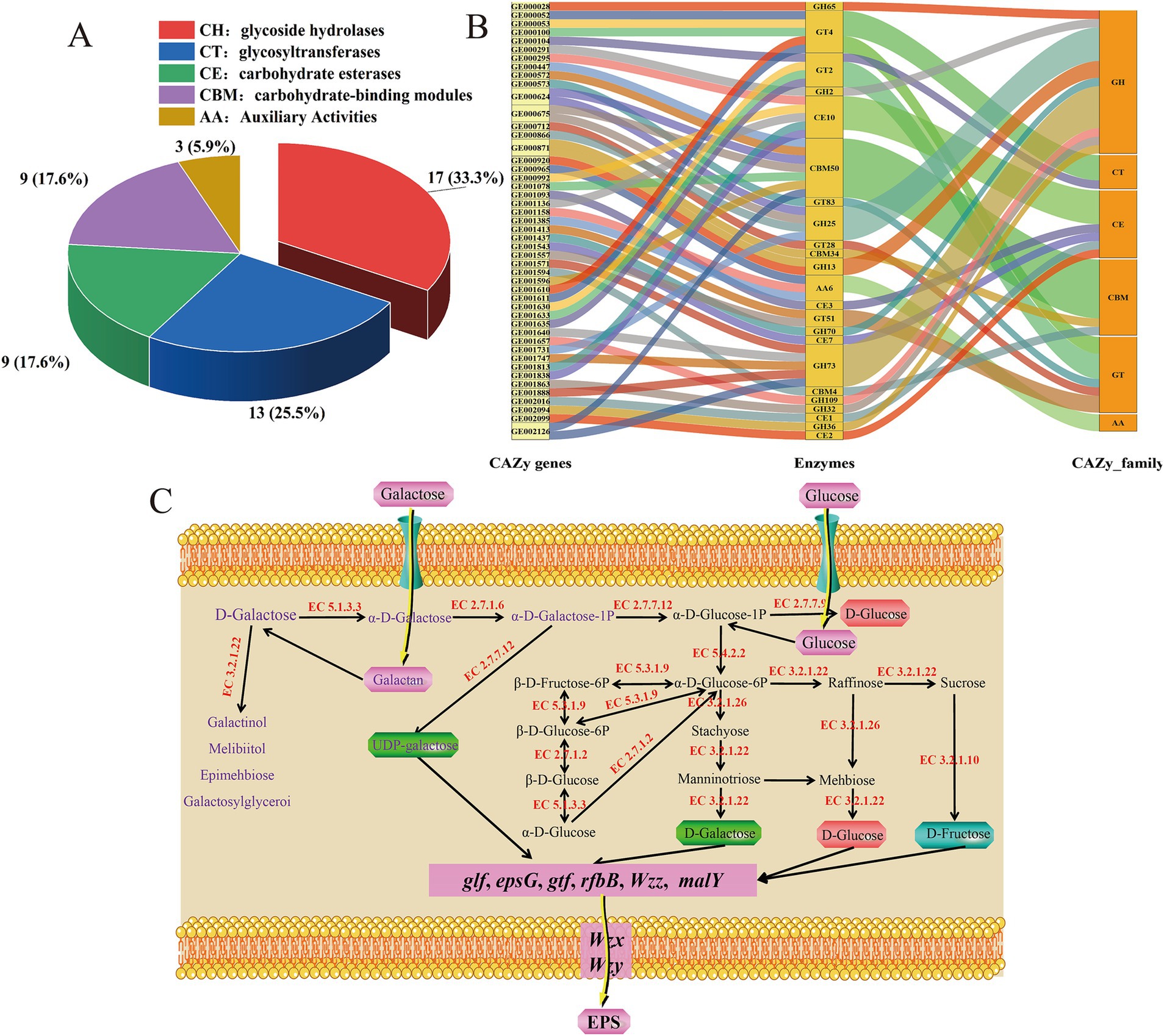
Figure 4. Prediction of genes encoding carbohydrate-active enzymes prediction. (A) Carbohydrate-active enzymes database (CAZy)-annotated statistics chart. (B) Classification of predicted CAZy from L. fermentum A51. (C) Carbohydrate metabolic systems in L. fermentum A51 genome. “EC + number” indicates the annotation of enzymes in L. fermentum A51. EPS, exopolysaccharides.
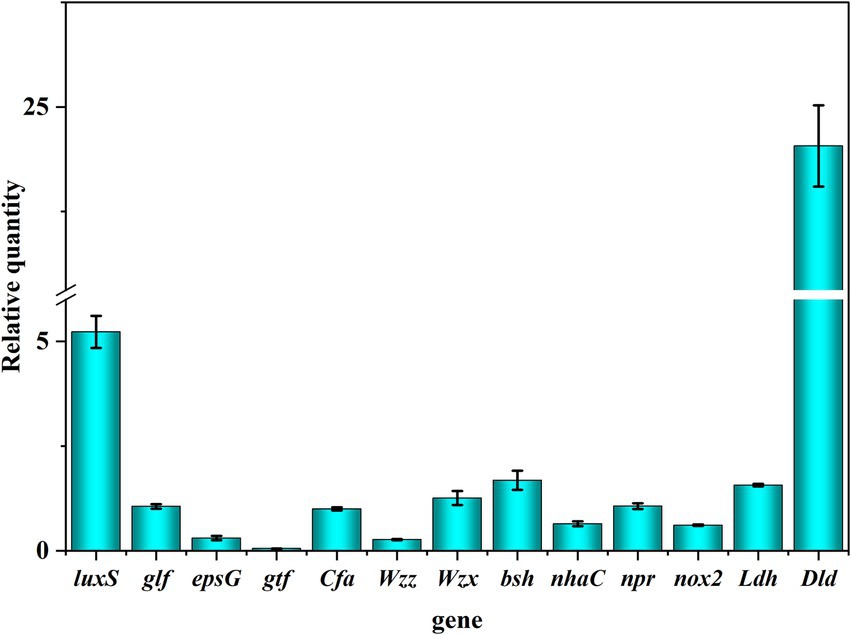
Whole-genome sequencing analysis showed that L. fermentum A51 contained EPS synthesis-regulating genes, including glf (GE001643), epsG (GE000103), Wzx (GE001642), Wzz (GE001644), Wzy (GE001645), pgk (GE000425) and glf (GE001643), suggesting that EPS synthesis by L. fermentum A51 relies on the Wzx/wzy-dependent pathway (Wu et al., 2022; Figure 4C; Table 6). The steps for the synthesis of EPS by L. fermentum A51 were as follows: phosphotransferases transfer extracellular glucose and galactose into the cell for phosphorylation. The pgk and glf genes are involved in the production of sugar nucleotides. The epsG genes encode glycosyltransferases, which are responsible for the translocation of monosaccharides to lipid carriers and the synthesis of repeating units. In the polymerization of repeats, the chain length-determining protein, Wzz, controls the length of the polysaccharide chain by controlling the number of repeats that are added. The repeating units are transferred by flippase to the periplasmic space or outer membranes. The repeating units are polymerized into long chains by Wzy polymerase. Eventually, the long-chain EPS is released into the extracellular space by Wzx flippase. Genotypic analysis showed that L. fermentum A51 was rich in carbohydrate transport and metabolism-related genes, as well as key EPS synthesis regulating genes. These genes allow L. fermentum A51 to synthesize EPS.
The EPS synthesis capability of L. fermentum A51 in MRS liquid medium was determined, and the results are shown in Figure 5A. The production of EPS increased significantly during the first 14 h and reached a maximum value of 482.728 ± 12.304 mg/L at 20 h, but then decreased as fermentation progressed further (Figure 5A). The decrease of EPS production in late fermentation may be caused by the degradation of glycohydrolases presented in the culture for EPS synthesis. Our results agree with the findings of Li M. et al. (2022), who found that EPS production of Lactococcus lactis subsp. lactis IMAU11823 in MRS liquid medium increased at the fastest rate during the first 12 h and gradually increased to a maximum value of 201.44 mg/L at 24 h.
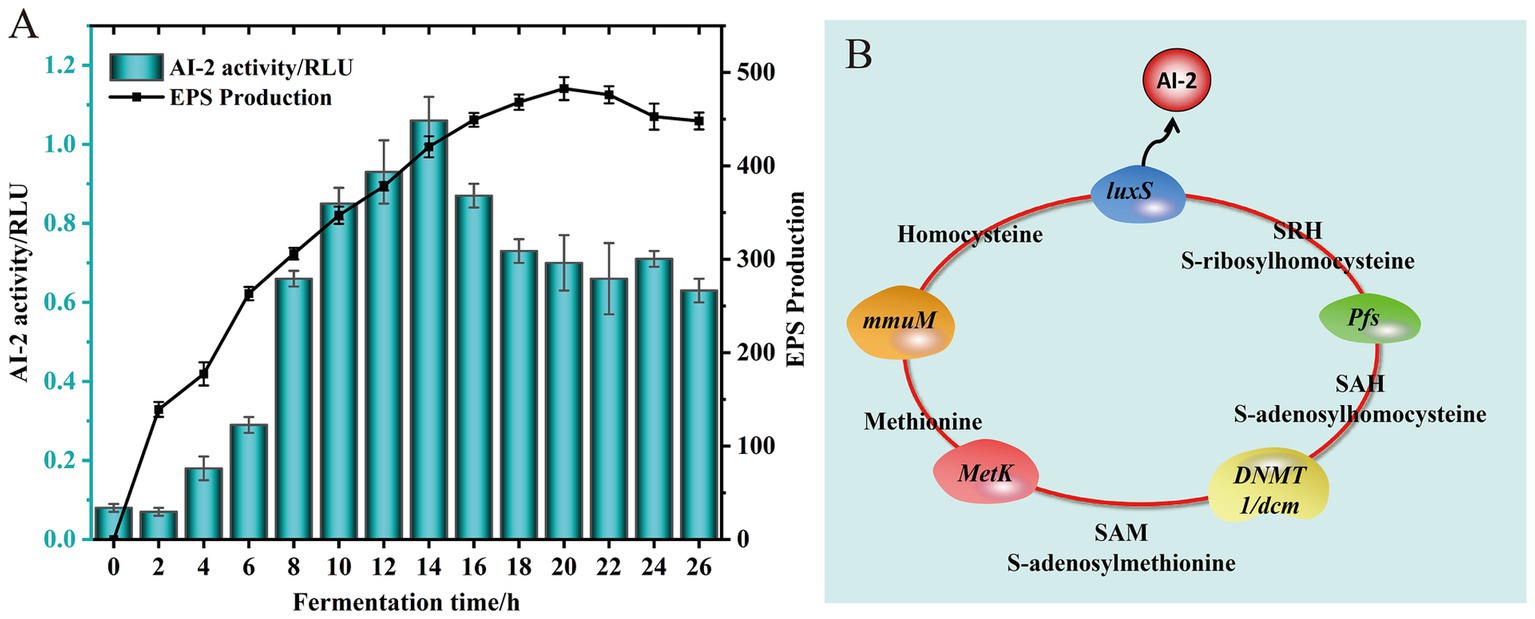
Figure 5. EPS production and AI-2 activity of L. fermentum A51 at different culture time (A). Signaling molecule AI-2 synthesis pathway of L. fermentum A51 (B).
3.4.6 LuxS/AI-2 quorum sensing system-related genes
Recently, several studies have reported that the probiotic properties of LAB, which include resistance to harsh environmental conditions, biofilm formation, phenyl lactic acid synthesis, conjugated linoleic acid production, adhesion and colonization, and bacteriocin synthesis are closely related to the regulation of the LuxS/AI-2 quorum sensing (QS) system (Ma et al., 2022; Meng et al., 2022; Chai et al., 2023). The LuxS/AI-2-QS system is a density-dependent regulatory system that relies on a cascade reaction to regulate the expression of target genes. This cascade response is triggered by an extracellular signaling molecule, AI-2, at a certain threshold concentration (Moreno-Gámez et al., 2017). The activity of signaling molecule AI-2 during fermentation of L. fermentum A51 was determined (Figure 5A). During the first 12 h of fermentation, the fluorescence intensity of the signal molecule AI-2 produced by L. fermentum A51 gradually increased. Starting from 12 h, the signal molecule AI-2 was higher than the positive relative fluorescence intensity, demonstrating that L. fermentum A51 could produce the signal molecule AI-2. The activity of the signal molecule AI-2 was highest at 14 h of incubation and then decreased with increasing incubation time. The reduced activity of the signal molecule AI-2 at a later stage may be related to the unstable and easily decomposed character of the signal molecule AI-2 (Chen et al., 2002).
Genomic results showed that L. fermentum A51 contained complete genes involved in the synthesis of signaling molecule AI-2, including luxS (GE001943), pfs (GE000728), DNMT1/dcm (GE000705), MetK (GE001633), and mmuM (GE001072) genes (Figure 5B; Table 6). This finding suggests that L. fermentum A5 has the ability to secrete the signaling molecule AI-2 (Duanis-Assaf et al., 2016). The secreted signaling molecule AI-2 can then activate the LuxS/AI-2-QS system. Whole genome sequencing analysis and phenotypic experiments confirmed that L. fermentum A51 showed good tolerance to simulated gastrointestinal tract and possessed antioxidant and antimicrobial activities, as well as EPS synthesis capacities. Previous studies have indicated that the LuxS/AI-2-QS system positively regulates the expression of the nhaC gene (Na+/H+ reverse transporter protein) and enhances the survival of Lactobacillus acidophilus CICC 6074 in intestinal juice (Li X. et al., 2022). Another study discovered that the LuxS/AI-2-QS system mediates the synthesis of phenyl lactic acid in L. plantarum L3 by regulating the expression of key proteins, including S-ribosomal homocysteine lyase (LuxS), aminotransferase (araT) and lactate dehydrogenase (ldh) (Chai et al., 2023). Wang (2019) demonstrated that the LuxS/AI-2-QS system mediates the expression of S-layer proteins of Lactobacillus acidophilus CICC6074 and its adhesion to the intestine. In this study, the LuxS/AI-2-QS system was positively associated with the probiotic properties of LAB. However, the regulatory relationship between the LuxS/AI-2-QS system and the probiotic properties of L. fermentum A51 needs to be further investigated.
3.5 Validation of probiotic genes using RT-qPCR
In the current study, the expression of probiotic genes was determined, including EPS synthesis-related genes (glf, epsG, gtf, Wzz, and Wzx) and quorum sensing system-related genes (luxS), tolerance-related genes (bsh, cfa, and nhaC), antioxidant genes (npr and nox2) and antibacterial genes (Idh and Dld). As shown in Figure 6, genes related to probiotic properties were expressed, including EPS synthesis-related genes (glf), quorum sensing system genes (luxS), tolerance-related genes (bsh, cfa, and nhaC) and antimicrobial genes (Idh and Dld). These results suggest that the expression of probiotic genes contributes to the probiotic properties of L. fermentum A51 (Figure 7).
4 Conclusion
In summary, we investigated the safety and probiotic properties of L. fermentum A51 by whole genome mapping and phenotypic analyses, which proved that L. fermentum A51 is a safe strain with no antibiotic resistance genes and virulence factor genes in its genome. In addition, L. fermentum A51 was found to possess genes implicated in the synthesis of EPS and signaling molecule AI-2, stress response, antioxidant, and antibacterial activities, all of which led to enhanced probiotic activities, including good tolerance to simulated gastrointestinal tract, strong antioxidant and antimicrobial activities. Collectively, our findings have confirmed that L. fermentum A5 is safe and exhibits good probiotic properties and high exopolysaccharide production, thus recommending its potential application in the production of value-added fermented dairy products.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: L. fermentum A51 (Gene bank accession number: CP132542).
Author contributions
GuW: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft. DW: Conceptualization, Investigation, Methodology, Validation, Writing – original draft. TW: Conceptualization, Data curation, Writing – original draft. GaW: Formal analysis, Methodology, Writing – original draft. YC: Conceptualization, Data curation, Writing – original draft. YL: Conceptualization, Methodology, Writing – review & editing. MM: Conceptualization, Methodology, Writing – original draft. HW: Investigation, Methodology, Writing – original draft. AH: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the National Natural Science Foundation of China (No. 32260578), the Major Science and Technology Project in Yunnan Province of China (202102AE090027-1), the Yun-Ling Industrial Technology Leading Talent program (Grant No. 2014, 1782).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1498352/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Evaluation of hemolysis of L. fermentum A51 (A), Lactobacillus casei Zhang (B), Escherichia coli (C), and Staphylococcus aureus (D). Positive control (Escherichia coli and Staphylococcus aureus); Negative control (Lactobacillus casei Zhang).
SUPPLEMENTARY TABLE S1 | Gene prediction and annotation of carbohydrate transport and metabolism related genes in L. fermentum A51.
SUPPLEMENTARY DATA 1 | The virulence factor genes of L. fermentum A51.
Abbreviations
LAB, Lactic acid bacteria; GRAS, Generally recognized as safe; L. fermentum, Limosilactobacillus fermentum; EPS, Exopolysaccharides; L. plantarum, Lactobacillus plantarum; E. coli, Escherichia coli; S. aureus, Staphylococcus aureus; GO, Gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; NR, Non-redundant protein sequence database; VFDB, Virulence Factors of Pathogenic Bacteria; CARD, Comprehensive Antibiotic Research; CFS, Cell-free supernatant; TCA, Trichloroacetic acid; DPPH, 2,2-Biphenyl-1-picrylhydrazino; ABTS, 2,2′-diazo-bis (3-ethylbenzothiazolin-6-sulfonic acid) diammonium salt; GHs, Glycoside hydrolases; GTs, Glycosyltransferases; PLs, Polysaccharide lyase; CEs, Carbohydrate esterases; AAs, Auxiliary activities; CBMs, Carbohydrate-binding modules; qRT-PCR, Quantitative real time polymerase chain reaction; QS, Quorum sensing
Footnotes
1. ^https://github.com/marbl/canu
2. ^https://github.com/broadinstitute/pilon
References
Altermann, E., Russell, W. M., Peril, M. A., Barrangou, R., Buck, B. L., Mcauliffe, O., et al. (2005). Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proceedings of the National Academy of Sciences of the United States of America. USA 102, 3906–3912. doi: 10.1073/pnas.0409188102
Ba, M., Huang, T., Guo, S., Wang, Y. J., Han, Z. H., Li, M., et al. (2021). Effects of extracellular polysaccharide in yogurt produced by the probiotic bacteria, lactobacillus casei zhang and bifidobacterium animalis subsp. lactis v9 on rheological properties, texture and stability. J. Chin. Inst. Food Sci. Technol. 21, 193–202. doi: 10.16429/j.1009-7848.2021.04.023
Chai, Y. M., Ma, Q. W., Nong, X., Mu, X. Y., and Huang, A. X. (2023). Dissecting LuxS/AI-2 quorum sensing system-mediated phenyllactic acid production mechanisms of Lactiplantibacillus plantarum L3. Food Res. Int. 166:112582. doi: 10.1016/j.foodres.2023.112582
Charnchai, P., Jantama, S. S., and Jantama, K. (2017). Genome analysis of food-processing stressful-resistant probiotic Bifidobacterium animalis subsp. lactis BF052, and itspotential application in fermented soymilk. FEMS Microbiol. Lett. 364. doi: 10.1093/femsle/fnx180
Chen, X., Schauder, S., Potier, N., Dorsselaer, A. V., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. doi: 10.1038/415545a
Cheng, Y., Wu, T., Chu, X., Tang, S., Cao, W., Liang, F., et al. (2020). Fermented blueberry pomace with antioxidant properties improves fecal microbiota community structure and short chain fatty acids production in an in vitro mode. LWT 125:109260. doi: 10.1016/j.lwt.2020.109260
Choi, E. A., and Chang, H. C. (2015). Cholesterol-lowering effects of a putative probiotic strain lactobacillus plantarum em isolated from kimchi. LWT 62, 210–217. doi: 10.1016/j.lwt.2015.01.019
Choi, K. S., Veeraragouda, Y., Cho, K. M., Lee, S. O., and Lee, K. (2008). Effect of gacs and gaca mutations on colony architecture, surface motility, biofilm formation and chemical toxicity in Pseudomonas sp. kl28. J. Microbiol. 45, 492–498. doi: 10.1016/j.mimet.2007.09.016
Delgado, S., O’Sullivan, E., Fitzgerald, G., and Mayo, B. (2007). Subtractive screening for probiotic properties of lactobacillus species from the human gastrointestinal tract in the search for new probiotics. J. Food Sci. 72, 310–315. doi: 10.1111/j.1750-3841.2007.00479.x
Duanis-Assaf, D., Steinberg, D., Chai, Y., and Shemesh, M. (2016). The luxs based quorum sensing governs lactose induced biofilm formation by Bacillus subtilis. Front. Microbiol. 6, 6:1517. doi: 10.3389/fmicb.2015.01517
Fei, Y., Li, L., Zheng, Y., Liu, D., Zhou, Q., and Fu, L. (2018). Characterization of Lactobacillus amylolyticus L6 as potential probiotics based on genome sequence and corresponding phenotypes. LWT 90, 460–468. doi: 10.1016/j.lwt.2017.12.028
Gao, Y., Liu, Y., Sun, M., Zhang, H., and Tuo, Y. (2020). Physiological function analysis of Lactobacillus plantarum y44 based on genotypic and phenotypic characteristics. J. Dairy Sci. 103, 5916–5930. doi: 10.3168/jds.2019-18047
Graebin, N., Schöffer, J., Diandra, A., Plinho, H., Marco, A., and Rafael, R. (2016). Immobilization of glycoside hydrolase families GH1, GH13, and GH70: state of the art and perspectives. Molecules 21, 21:1074. doi: 10.3390/molecules21081074
Grishina, Y. V., Vatlin, A. A., Mavletova, D. A., Odorskaya, M. V., Senkovenko, A. M., Ilyasov, R. A., et al. (2023). Metabolites potentially determine the high antioxidant properties of limosilactobacillus fermentum U-21. Biotech 12:39. doi: 10.3390/BIOTECH12020039
Gu, Y., Tian, J., Zhang, Y., Wu, R., and He, Y. (2020). Dissecting signal molecule AI-2 mediated biofilm formation and environmental tolerance in lactobacillus plantarum. J. Biosci. Bioeng. 131, 153–160. doi: 10.1016/j.jbiosc.2020.09.015
Gu, Y., Zhang, B., Tian, J., Li, L., and He, Y. (2023). Physiology, quorum sensing, and proteomics of lactic acid bacteria were affected by Saccharomyces cerevisiae YE4. Food Res. Int. 166:112612. doi: 10.1016/j.foodres.2023.112612
Iyer, R., Iverson, T. M., Accardi, A., and Miller, C. (2002). A biological role for prokaryotic clc chloride channels. Nature 419, 715–718. doi: 10.1038/nature01000
Korcz, E., and Varga, L. (2021). Exopolysaccharides from lactic acid bacteria: techno-functional application in the food industry. Trends Food Sci. Tech. 110, 375–384. doi: 10.1016/j.tifs.2021.02.014
Łepecka, A., Szymański, P., Okoń, A., and Zielińska, D. (2023). Antioxidant activity of environmental lactic acid bacteria strains isolated from organic raw fermented meat products. LWT 174:114440. doi: 10.1016/j.lwt.2023.114440
Li, X., Fan, X., Shi, Z., Xu, J., Cao, Y., Zhang, T., et al. (2022). AI-2E family transporter protein in Lactobacillus acidophilus exhibits AI-2 exporter activity and relate with intestinal juice resistance of the strain. Front. Microbiol. 13:908145. doi: 10.3389/fmicb.2022.908145
Li, M., Li, W., Li, D., Tian, J., Xiao, L., Kwok, L. Y., et al. (2022). Structure characterization, antioxidant capacity, rheological characteristics and expression of biosynthetic genes of exopolysaccharides produced by Lactococcus lactis subsp. lactis IMAU11823. Food Chem. 384:132566. doi: 10.1016/J.FOODCHEM.2022.132566
Li, J. Y., Zhang, L. J., Mu, G. Q., and Tuo, Y. F. (2023). Interpretation of safety and potential probiotic properties of Lactiplantibacillus plantarum Y42 based on genome-wide sequencing. Food Biosci. 56:103249. doi: 10.1016/j.fbio.2023.103249
Liang, M.-H., Jiang, J.-G., Wang, L., and Zhu, J. (2020). Transcriptomic insights into the heat stress response of Dunaliella bardawil. Enzym. Microb. Technol. 132:109436. doi: 10.1016/j.enzmictec.2019.109436
Liu, D. M., Huang, Y. Y., and Liang, M. H. (2022). Analysis of the probiotic characteristics and adaptability of lactiplantibacillus plantarum dmdl 9010 to gastrointestinal environment by complete genome sequencing and corresponding phenotypes. LWT 158:113129. doi: 10.1016/j.lwt.2022.113129
Liu, X. Y., Xie, D., Ma, L. Z., and Xu, J. (2021). Compatative study on active xomponets and antioxidant capacity of Rosa roxburghii tratt.Fruit residue before and after fermentation. Food Sci. Technol. 46, 16–24.
Lu, J., Mao, Y. R., Ma, T., Liu, X. L., Cheng, X. Y., et al. (2023). Screening and genome analysis of lactic acid bacteria with high exopolysaccharide production and good probiotic properties. Food Biosci. 56:103211. doi: 10.1016/J.FBIO.2023.103211
Ma, Q. W., Chai, Y. M., Yang, Z. B., and Huang, A. X. (2022). Deciphering the mechanisms of limosilactobacillus fermentum L1 involved in conjugated linoleic acid regulated by LuxS/AI-2 quorum sensing-sciencedirect. LWT 154:112736. doi: 10.1016/j.lwt.2021.112736
Ma, Y., Pan, C., and Wang, Q. (2019). Crystal structure of bacterial cyclopropane-fatty-acyl-phospholipid synthase with phospholipid. J. Biochem. 166, 139–147. doi: 10.1093/jb/mvz018
Meng, F., Zhao, M., and Lu, Z. (2022). The LuxS/AI-2 system regulates the probiotic activities of lactic acid bacteria. Trends Food Sci. Tech. 127, 272–279. doi: 10.1016/j.tifs.2022.05.014
Montoro, B. P., Benomar, N., Lerma, L. L., Gutiérrez, S. C., Gálvez, A., and Hikmate, A. (2016). Fermented aloreña table olives as a source of potential probiotic lactobacillus pentosus strains. Front. Microbiol. 7, 7:1583. doi: 10.3389/fmicb.2016.01583
Moreno-Gámez, S., Sorg, R. A., Domenech, A., Kjos, M., Weissing, F. J., van Doorn, G. S., et al. (2017). Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat. Commun. 8, 854–812. doi: 10.1038/S41467-017-00903-Y
Nordstrm, E. A., Teixeira, C., Montelius, C., Jeppsson, B., and Larsson, N. (2021). Lactiplantibacillus plantarum 299v 608(LP299V): three decades of research. Benef. Microbes. 12, 1–26. doi: 10.3920/BM2020.0191
Oh, Y. J., Kim, S. A., Yang, S. H., Kim, D. H., Cheng, Y. Y., Kang, J., et al. (2022). Integrated genome-based assessment of safety and probiotic characteristics of Lactiplantibacillus plantarum PMO 08 isolated from kimchi. PLoS One 17:e0273986. doi: 10.1371/JOURNAL.PONE.0273986
Papadimitriou, K., Alegría, A., Bron, P. A., De Angelis, M., Gobbetti, M., Kleerebezem, M., et al. (2016). Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. R. 80, 837–890. doi: 10.1128/MMBR.00076-15
Peng, Y. Y., Zhong, S. Y., Xu, X. L., and Liu, D. M. (2023). Analysis of the safety and probiotic properties of Bifidobacterium longum B2-01 by complete genome sequencing combined with corresponding phenotypes. LWT. 189:115445. doi: 10.1016/J.LWT.2023.115445
Ruas-Madiedo, P., and de Los Reyes-Gavilán, C. G. (2005). Invited review: methods for the screening, isolation, and characterization of exopolysaccharides produced by lactic acid bacteria. J. Dairy Sci. 88, 843–856. doi: 10.3168/jds.S0022-0302(05)72750-8
Saadat, Y. R., Khosroushahi, A. Y., and Gargari, B. P. (2019). A comprehensive review of anticancer, immunomodulatory and health beneficial effects of the lactic acid bacteria exopolysaccharides. Carbohyd. Polym. 217, 79–89. doi: 10.1016/j.carbpol.2019.04.025
Salminen, S., Von Wright, A., and Morelli, L. (1998). Demonstration of safety of probiotics - a review. Inte. J. Food Microbiol. 44, 93–106. doi: 10.1016/S0168-1605(98)00128-7
Senan, S., Prajapati, J. B., and Joshi, C. G. (2015). Whole-genome based validation of the adaptive properties of indian origin probiotic lactobacillus helveticus mtcc 5463. J. Sci. Food Agr. 95, 321–328. doi: 10.1002/jsfa.6721
Shi, X. T., Greetham, D., Raeth, S., and Perrone, G. (2010). The thioredoxin-thioredoxin reductase system canfunction in vivo as an alternative system to reduce oxidized glutathione in Saccharomyces cerevisiae. J. Biol. Chem. 285, 6118–6126. doi: 10.1074/jbc.M109.062844
Song, J., Peng, S., Yang, J., Zhou, F., and Suo, H. (2021). Isolation and identification of novel antibacterial peptides produced by Lactobacillus fermentum shy10 in chinese pickles. Food Chem. 348:Article 129097. doi: 10.1016/j.foodchem.2021.129097
Stefanovic, E., Fitzgerald, G., and McAuliffe, O. (2017). Advances in the genomics and metabolomics of dairy lactobacilli: a review. Food Microbiol. 61, 33–49. doi: 10.1016/10.1016/j.fm.2016.08.009
Su, E., Xia, T., Gao, L., Dai, Q., and Zhang, Z. (2010). Immobilization of β-glucosidase and its aroma-increasing effect on tea beverage. Food Bioprod. Process. 88, 83–89. doi: 10.1016/j.fbp.2009.04.001
Sun, Y., Zhang, S., Li, H., Zhu, J., Liu, Z., Hu, X., et al. (2022). Assessments of probiotic potentials of Lactiplantibacillus plantarum strains isolated from chinese traditional fermented food: phenotypic and genomic analysis. Front. Microbiol. 13:895132. doi: 10.3389/fmicb.2022.895132
Trabelsi, I., Slima, S. B., Chaabane, H., and Salah, R. B. (2015). Purification and characterization of a novel exopolysaccharides produced by lactobacillus sp. ca6. Int. J. Biol. Macromol. 74, 541–546. doi: 10.1016/j.ijbiomac.2014.12.045
Vasiee, A., Behbahani, B. A., Yazdi, F. T., Mortazavi, S. A., and Noorbakhsh, H. (2018). Diversity and probiotic potential of lactic acid bacteria isolated from horreh, a traditional iranian fermented food. Springer 10, 258–268. doi: 10.1007/s12602-017-9282-x
Wang, T. (2019). Effect of AI-2/LuxS quorum-sensing on S-layer protein expression and adhesion of L. acidophilus. Nanjing: Nanjing Normal University.
Wang, Y. J., Yang, H. Y., Mu, G. Q., and Wu, X. M. (2023). Safety evaluation and complete genome analysis emphasis on extracellular polysaccharide of two strains of Limosilactobacillus fermentum MWLf-4 and Lactipiantibacillus plantarum MWLp-12 from human milk. Food Biosci. 51:102356. doi: 10.1016/J.FBIO.2023.102356
Wei, G. Q., Dai, X. Y., Zhao, B., Li, Z. Y., Tao, J. F., Wang, T., et al. (2023). Structure-activity relationship of exopolysaccharides produced by limosilactobacillus fermentum A51 and the mechanism contributing to the textural properties of yogurt. Food Hydrocoll. 144:108993. doi: 10.1016/j.foodhyd.2023.108993
Wu, J., Han, X., Ye, M., Li, Y., Wang, X., and Zhong, Q. (2022). Exopolysaccharides synthesized by lactic acid bacteria: biosynthesis pathway, structure-function relationship, structural modification and applicability. Crit. Rev. Food Sci. Nutr. 63, 7043–7064. doi: 10.1080/10408398.2022.2043822
Wu, C. C., Lin, C. T., Wu, C. Y., Peng, W. S., Lee, M. J., and Tsai, Y. C. (2015). Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation. Mol Oral Microbiol 30, 16–26. doi: 10.1111/omi.12063
Xu, X. Q. (2019). Sourdough fermented by exopolysaccharide forming strain: Application and mechanism studies in frozen dough[D]. Wuxi: Jiangnan University.
Xue, Z. P., Cu, X., Xu, K., Peng, J. H., Liu, H. R., Zhao, R. T., et al. (2023). The effect of glutathione biosynthesis of Streptococcus thermophilus ST-1 on cocultured Lactobacillus delbrueckii ssp. bulgaricus ATCC11842. J. Dairy Sci. 106, 884–896. doi: 10.3168/JDS.2022-22123
Yan, J., Huang, Y., Gao, Z., Zhang, Z., Gu, Q., and Li, P. (2023). Probiotic potential of lactiplantibacillus plantarum zfm4 isolated from pickles and its effects on human intestinal microecology. LWT 184:114954. doi: 10.1016/j.lwt.2023.114954
Zhang, Y., Du, R. T., Wang, L. F., and Zhang, H. P. (2010). The antioxidative effects of probiotic Lactobacillus casei zhang on the hyperlipidemic rats. Eur. Food Res. Technol. 231, 151–158. doi: 10.1007/s00217-010-1255-1
Zhang, T., Guo, Y. X., Fan, X. K., Liu, M. Z., Xu, J., Zeng, X. Q., et al. (2023). Protection mechanism of metal ion pre-stress on Lactobacillus acidophilus CICC 6074 under acid tolerance. J. Agri. Food Chem 71, 13304–13315. doi: 10.1021/ACS.JAFC.3C01970
Zhang, M., Zeng, S., Hao, L., Yao, S., Wang, D., Yang, H., et al. (2022). Structural characterization and bioactivity of novel exopolysaccharides produced by Tetragenococcus halophilus. Food Res. Int. 155:111083. doi: 10.1016/j.foodres.2022.111083
Zhao, Y., Hong, K., Zhao, J., Zhang, H., Zhai, Q., and Chen, W. (2019). Lactobacillus fermentum and its potential immunomodulatory properties. J. Funct. Foods 56, 21–32. doi: 10.1016/j.jff.2019.02.044
Zhao, Y., Yu, L., Tian, F., Zhao, J., Zhang, H., Chen, W., et al. (2021a). An optimized culture medium to isolate Lactobacillus fermentum strains from the human intestinal tract. J. Funct. Foods 12, 6740–6754. doi: 10.1039/D1FO00209K
Zhao, Y., Zhang, C. C., Yu, L. L., Tian, F. W., Zhao, J. X., Zhang, H., et al. (2021b). Phylogenetic and comparative genomic analysis of Lactobacillus fermentum strains and the key genes related to their intestinal anti-inflammatory effects. Engineering 17, 170–182. doi: 10.1016/j.eng.2020.09.016
Zhou, Q., Gu, R., Li, P., Lu, Y., and Gu, Q. (2020). Anti-salmonella mode of action of natural l-phenyl lactic acid purified from lactobacillus plantarum ZJ316. Appl. Microbiol. Biot. 104, 5283–5292.
Keywords: Limosilactobacillusfermentum A51, whole genome sequencing, probiotic properties, phenotype analysis, safety
Citation: Wei G, Wang D, Wang T, Wang G, Chai Y, Li Y, Mei M, Wang H and Huang A (2025) Probiotic potential and safety properties of Limosilactobacillus fermentum A51 with high exopolysaccharide production. Front. Microbiol. 16:1498352. doi: 10.3389/fmicb.2025.1498352
Edited by:
Gerard M. Moloney, University College Cork, IrelandReviewed by:
E. Emma Tymczyszyn, National University of Quilmes, ArgentinaDigambar Kavitake, National Institute of Nutrition (ICMR), India
Copyright © 2025 Wei, Wang, Wang, Wang, Chai, Li, Mei, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aixiang Huang, YWl4aWFuZ2h1YW5nQDEyNi5jb20=
†These authors have contributed equally to this work
 Guangqiang Wei
Guangqiang Wei Daodian Wang1,2†
Daodian Wang1,2† Aixiang Huang
Aixiang Huang