
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Microbiol., 11 March 2025
Sec. Infectious Agents and Disease
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1472124
This article is part of the Research TopicMicroRNA: the swift development in infectious diseasesView all 6 articles
Background: Sepsis is a life-threatening condition caused by a dysregulated immune response to infection and remains a major cause of mortality in intensive care units (ICUs). Recent studies have identified microRNAs (miRNAs), a class of small RNA molecules, as potential biomarkers for diagnosing and predicting outcomes in sepsis patients. However, the results of these studies have been inconsistent. This meta-analysis aims to comprehensively evaluate the diagnostic and prognostic value of miRNAs in predicting sepsis-related mortality.
Methods: A comprehensive literature search was performed across major databases, including PubMed, Cochrane Library, EMBASE, and CNKI, up to April 7, 2024. Data extraction and meta-analysis were conducted using Meta-disk 1.4 and STATA 15.1, employing both fixed- and random-effects models to ensure robust statistical analysis.
Results: A total of 55 studies met the inclusion criteria and were analyzed. The pooled sensitivity, specificity, and area under the summary receiver operating characteristic (SROC) curve for miRNA detection were calculated. The overall performance of total miRNA detection demonstrated a sensitivity of 0.76 (95% confidence interval [CI]: 0.74–0.77), a specificity of 0.72 (95% CI: 0.71–0.73), and an SROC value of 0.83. Subgroup analyses revealed that miR-133a-3p exhibited the highest diagnostic accuracy, with a pooled sensitivity of 0.83 (95% CI: 0.70–0.92), specificity of 0.79 (95% CI: 0.71–0.86), and an SROC value of 0.90. Additionally, other miRNAs, including miR-146a, miR-21, miR-210, miR-223-3p, miR-155, miR-25, miR-122, miR-125a, miR-125b, and miR-150, also demonstrated high SROC values (0.84 to 0.76).
Conclusion: This meta-analysis underscores the potential of several microRNAs (miRNAs) as reliable biomarkers for predicting sepsis mortality. Specifically, miR-133a-3p, miR-146a, miR-21, miR-210, miR-223-3p, miR-155, miR-25, miR-122, miR-125b, and miR-150 emerge as promising candidates for clinical applications in sepsis prognosis.
Sepsis is a leading cause of mortality in Intensive Care Units (ICUs) (Venkatesh et al., 2018). It is characterized by a dysregulated immune response to infection that often results in organ failure and high mortality (Singer et al., 2016). Early identification of patients at high risk for sepsis is therefore vital for reducing mortality and improving outcomes (Yang et al., 2022). Current diagnostic and management strategies rely on clinical assessments, monitoring of vital signs, and laboratory parameters, supported by scoring systems such as the Quick Sequential Organ Failure Assessment Score (qSOFA) and the National Early Warning Score (NEWS) (Zhang et al., 2021). Despite their utility, these systems have inherent limitations, necessitating continuous clinical monitoring and highlighting the urgent need for novel predictive indicators of sepsis mortality.
MicroRNAs (miRNAs), a class of small non-coding RNA molecules, have gained attention for their significant role in the regulation of gene expression and immune responses in sepsis (Salim et al., 2020; Sankar et al., 2022; Wang and Han, 2021; Wang et al., 2015). A recent meta-analysis validated the diagnostic utility of miRNAs and identified miR-223-3p as a potential biomarker for sepsis (Shen et al., 2020). However, the predictive value of miRNAs in sepsis mortality remains controversial due to inconsistent findings across studies. To address this gap, this meta-analysis was conducted to systematically evaluate the potential of miRNAs as predictors of sepsis mortality.
This meta-analysis followed a predefined protocol, as recommended by Deeks (2001). The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement was followed for data collection and analysis (Supplementary Table S1) (Moher et al., 2009). The protocol was prospectively registered in PROSPERO (CRD42024530167).
A systematic search was conducted in the following databases: PubMed, EMBASE, CNKI, and the Cochrane Central Register of Controlled Trials. The search was conducted up to April 7, 2024. The search terms used were (“sepsis” OR “septicemia” OR “pyemia”) AND (“MicroRNAs” OR “MicroRNA” OR “miRNAs” OR “miRNA”). For the PubMed database, the search query was: (Sepsis[MeSH Terms] OR septicemia OR pyemia) AND (MicroRNAs[MeSH Terms] OR MicroRNA OR miRNAs OR miRNA). Detailed search strategies for EMBASE, Cochrane, and CNKI are provided in the Supplementary material. Only studies published in English or Chinese were considered.
The initial step involved the screening of titles and abstracts. Full texts of potentially relevant studies were then retrieved to assess inclusion/exclusion criteria.
The inclusion criteria were as follows: (1) all sepsis patients were confirmed by diagnosis criteria; (2) trials evaluating the expression of microRNAs were included; (3) the data of receiver operating characteristic (ROC) curve and the essential sample size were contained; (4) all reports had specific numbers of sepsis patients who died and survived; and (5) the full text was published in English or Chinese.
The following criteria were used to exclude studies: (1) reviews, letters, conference articles, or case reports; (2) inadequate data for analysis; and (3) duplicated studies.
Two independent investigators (Yuxi Jin and Yue Zhang) screened titles and abstracts, with discrepancies resolved by a third reviewer (Yifei Li). Study quality was assessed using the QUADAS-2 checklist (Whiting et al., 2011). Any discrepancies in quality assessments were discussed with the third reviewer. Subsequently, data extraction for sensitivity, specificity, and the number of true positives (TP), false positives (FP), false negatives (FN), and true negatives (TN) was performed by Xiaolan Zheng and Yuxi Jin.
Sensitivity, specificity, diagnostic odds ratio (DOR), and area under the summary ROC (SROC) curve were calculated. The SROC curve was constructed using sensitivity and specificity, with the area under the curve (AUC) serving as a measure of global performance (Moses et al., 1993).
Publication bias was quantitatively evaluated using STATA version 15.1, based on funnel plots and Deeks’ test (StataCorp, Texas, United States). In the event that publication bias was detected (p < 0.05) (Deeks et al., 2005), the trim-and-fill method was employed to assess its influence. Consistency of results before and after the trim-and-fill adjustment was interpreted as evidence of stability and reliability (Lin and Chu, 2018).
Heterogeneity was assessed using the Chi-squared (χ2) test for pooled sensitivity and specificity, and the Cochran Q test for pooled DOR. Statistical heterogeneity was defined as p < 0.05. The I2 test was also employed to measure the proportion of variability across studies, with values of 25, 50, and 75% representing low, moderate, and high heterogeneity, respectively (Higgins and Thompson, 2002). When heterogeneity was detected, meta-regression analyses were performed using STATA 15.1 to explore potential sources, such as sample type and population differences. A p value < 0.05 was considered significant.
Sensitivity analysis was performed using STATA 15.1 to evaluate the influence of individual studies on overall results. Subgroup analyses were performed using Meta-Disc 1.4.
Meta-Disc 1.4 was used for data processing and for the analysis of threshold effects. Publication bias was assessed using STATA 15.1. For homogeneous results, a fixed-effects model was employed, while for heterogeneous results (I2 > 50%), a random-effects model was applied. The generation of forest plots was instrumental in the visualization of the results.
The initial search yielded 3,912 papers, of which 249 articles were deemed suitable for full-text review following an initial assessment of their titles and abstracts. However, further evaluation revealed that 22 articles were excluded due to inappropriate article types, and 119 studies lacked data on TP, FP, FN, and TN cases. Additionally, 53 articles did not include a comparison between survival and non-survival sepsis patients. The study selection process is detailed in Figure 1. Ultimately, 55 studies were included in the meta-analysis (Yang et al., 2022; Salim et al., 2020; Sankar et al., 2022; Wang et al., 2012; Yang et al., 2015; Yao et al., 2015; Yang et al., 2016; Huo et al., 2017; Lin, 2017; Rahmel et al., 2018; Zhang, 2018; Zhang et al., 2018; Lin et al., 2019; Sun et al., 2019; Xue et al., 2019; Chen L. et al., 2020; Chen W. et al., 2020; Dou et al., 2020; Lin et al., 2020; Liu et al., 2020; Na et al., 2020; Qiu et al., 2020; Wang H. et al., 2020; Wang J. et al., 2020; Wang L. et al., 2020; Wang Q. et al., 2020; Xu et al., 2020; Yang J. et al., 2020; Yang Y. et al., 2020; Zhao et al., 2020; Zhu, 2020; Deng et al., 2021; Guo et al., 2021; Ma et al., 2021; Pan et al., 2021; Wang H. et al., 2021; Xu et al., 2021; Yang et al., 2021; Yao et al., 2021; Deng et al., 2022; Li et al., 2022; Pan et al., 2022; Yu et al., 2022; Zhang et al., 2022; Zhao et al., 2022; Ali et al., 2023; Bao and Zhang, 2023; Bian and Pang, 2023; Li N. et al., 2023; Liu and Yang, 2023; Yao et al., 2023; Zhang et al., 2023; Behroozizad et al., 2024; Hao et al., 2024; Zhang et al., 2024), encompassing a total of 6,443 sepsis patients, of whom 2,047 were non-survivors and 4,396 were survivors. These studies analyzed the roles of 41 different microRNAs (miRNAs), 11 of which (miR-133a-3p, miR-146a, miR-21, miR-210, miR-223-3p, miR-155, miR-25, miR-122, miR-125a, miR-125b, and miR-150) were examined in more than two investigations. Full-text articles published in both English and Chinese were included. Of the 55 studies analyzed, 27 (Yang et al., 2015; Yang et al., 2016; Huo et al., 2017; Zhang, 2018; Zhang et al., 2018; Sun et al., 2019; Xue et al., 2019; Qiu et al., 2020; Wang H. et al., 2020; Wang L. et al., 2020; Xu et al., 2020; Yang J. et al., 2020; Pan et al., 2021; Yang et al., 2021; Deng et al., 2022; Li et al., 2022; Pan et al., 2022; Yu et al., 2022; Zhang et al., 2022; Zhao et al., 2022; Bao and Zhang, 2023; Bian and Pang, 2023; Li N. et al., 2023; Liu and Yang, 2023; Yao et al., 2023; Zhang et al., 2023; Zhang et al., 2024) were published in Chinese, ensuring a comprehensive review of data from both English and Chinese sources. The patient population spanned a range of ages, with four studies focusing on neonates (less than 28 days old), five on children older than one month, and 46 on adults. Sample types included plasma (26 studies), serum (25 studies), peripheral blood mononuclear cells (PBMCs) (2 studies), and whole blood (2 studies). The geographical distribution of the studies was as follows: 52 studies were conducted in Asia (50 in China, 1 in India, and 1 in Iran), 2 in Africa (Egypt), and 1 in Europe (Germany). Among the 55 studies, 30 had a sample size of ≥100, while 25 included fewer than 100 participants. Diagnostic criteria for sepsis varied significantly, with two studies adhering to Sepsis-1.0, 13 studies using Sepsis-2.0, and 38 studies adopting Sepsis-3.0 criteria. Two studies did not specify the criteria utilized. All included studies employed quantitative reverse transcription PCR (qRT-PCR) for miRNA detection, ensuring high sensitivity and specificity in the quantification of these molecules across different sample types. Regarding reference genes for qRT-PCR, 41 studies used U6, 8 employed non-U6 genes, and 6 did not specify the reference gene. Table 1 summarizes the characteristics of the included studies.
The quality of the included studies was assessed using the QUADAS-2 tool, which indicated a high risk of bias for the index test (Supplementary Figure S1).
The overall predictive performance of total mixed microRNAs (T miR) for identifying sepsis is shown in Figure 2. The pooled sensitivity was 0.76 (95% CI: 0.74–0.77), accompanied by significant heterogeneity (p < 0.0001, χ2 = 357.42, I2 = 75.4%, Figure 2A). The pooled specificity was 0.72 (95% CI: 0.71–0.73), also exhibiting notable heterogeneity (p < 0.0001, χ2 = 901.34, I2 = 90.2%, Figure 2B). The pooled diagnostic odds ratio (DOR) was 10.21 (95% CI: 8.60–12.14), with substantial heterogeneity (p < 0.0001, Cochran Q = 218.42, I2 = 59.7%, Figure 2C). The area under the curve (AUC) value was 0.83 (Figure 2D). The absence of a curvilinear form in the SROC curve indicates that no threshold effect was detected.
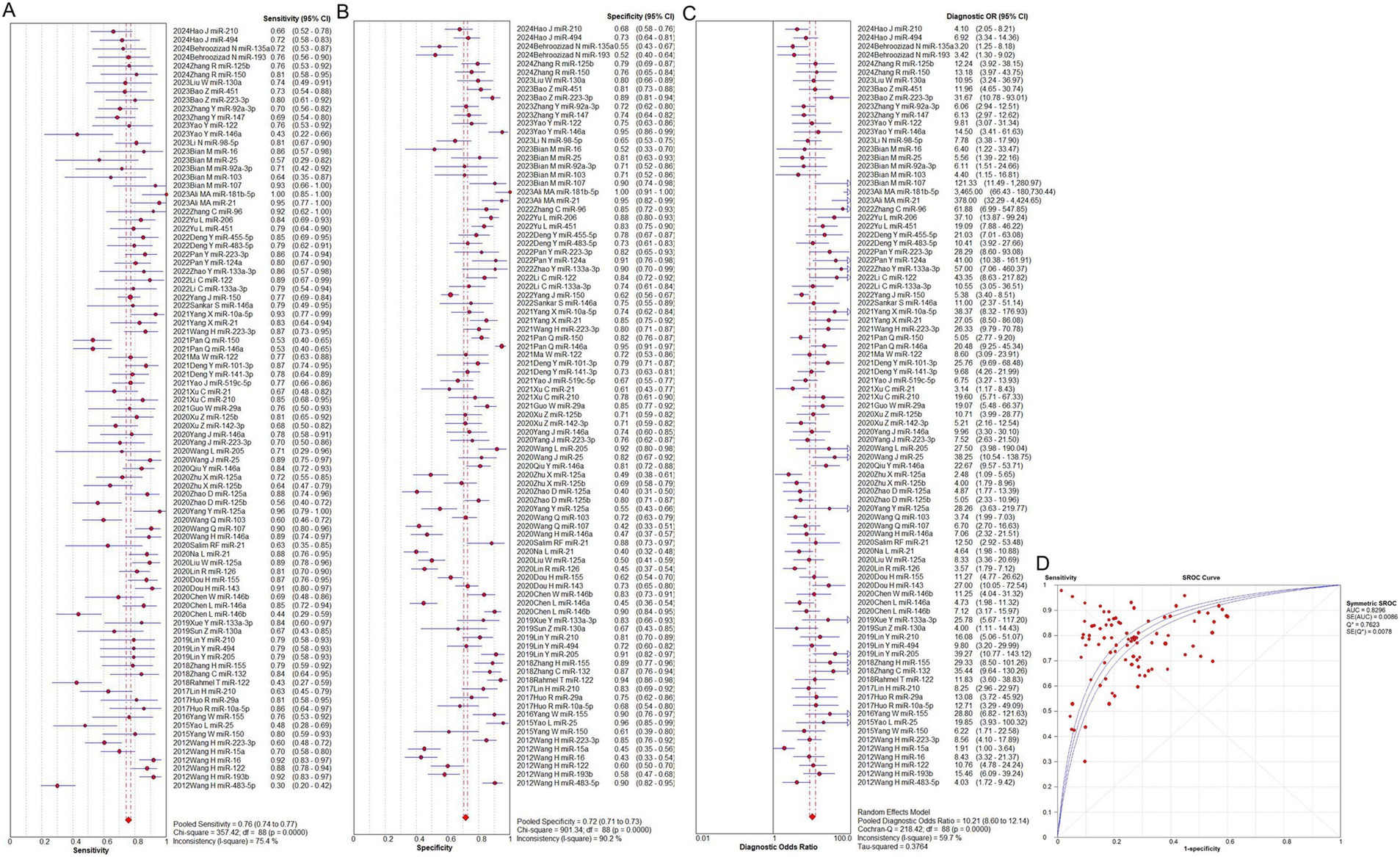
Figure 2. Performance of total miRNAs detection for sepsis diagnosis. (A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets. CI, confidence interval; DOR, predicting odds ratio; miR, mircoRNA; OR, odds ratio; SROC, summary receiver operating characteristic curves value.
To identify potential sources of heterogeneity, a meta-regression analysis was conducted, evaluating factors such as sample type, geographical location, sepsis diagnostic criteria, reference genes for qRT-PCR, patient age, follow-up duration, miRNA expression levels (upregulated or downregulated), and total sample size. Population (p = 0.013, t = −2.53, 95% CI: 0.39–0.89, Figure 3A) and total sample size (p = 0.003, t = −3.01, 95% CI: 0.40–0.83, Figure 3B) were identified as significant contributors to heterogeneity, whereas the remaining factors were not significant (p > 0.05, Figures 3C–H).
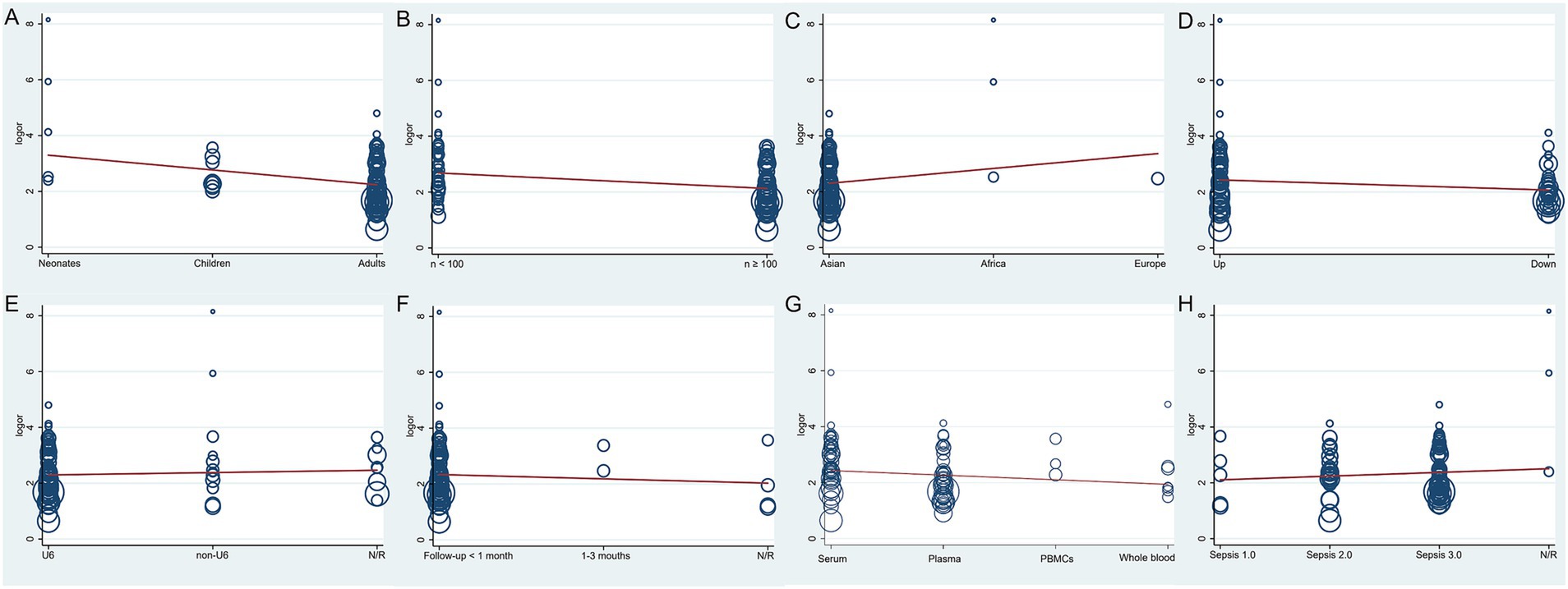
Figure 3. The meta-regression of the enrolled studies. (A) For the population, the meta-regression found a significant impact on the homogeneity of the included studies, p = 0.013, t = −2.53, 95%CI 0.39–0.89. (B) For the total sample size, the meta-regression found it was a dramatic impact on the homogeneity of the enrolled studies, p = 0.003, t = −3.01, 95%CI 0.40–0.83. (C) For the region, the meta-regression did not detect it had a dramatic impact on the homogeneity of the enrolled studies, p = 0.174, t = 1.37, 95%CI 0.79–3.68. (D) For the miRNA expression level, the meta-regression did not detect it was a dramatic impact on the homogeneity of the enrolled studies, p = 0.06, t = −1.90, 95%CI 0.48–1.02. (E) For the qRT-PCR reference genes, the meta-regression did not find it was a dramatic impact on the homogeneity of the enrolled studies, p = 0.565, t = 0.58, 95%CI 0.81–1.46. (F) For the follow-up time, the meta-regression did not detect it was a dramatic impact on the homogeneity of the included studies, p = 0.462, t = −0.74, 95%CI 0.57–1.30. (G) For the type of samples, the meta-regression did not detect it was a dramatic impact on the homogeneity of the enrolled studies, p = 0.154, t = −1.44, 95%CI 0.68–1.06. (H) For the sepsis diagnostic criteria, the meta-regression did not detect it was a dramatic impact on the homogeneity of the enrolled studies, p = 0.372, t = 0.90, 95%CI 0.85–1.53.
Subgroup analysis was performed based on population and sample size to investigate the sources of heterogeneity. Furthermore, a comprehensive analysis of 11 specific microRNAs (namely, miR-133a-3p, miR-146a, miR-21, miR-210, miR-223-3p, miR-155, miR-25, miR-122, miR-125a, miR-125b, and miR-150) was conducted in studies that have examined them in more than two investigations. The results of these investigations are presented in Table 2 and Supplementary Figures S2–S14.
The population was segmented into three distinct categories: neonates, children, and adults. In the neonate population (see Supplementary Figure S2), the pooled sensitivity was 0.87 (95%CI 0.78–0.93), indicating moderate heterogeneity (p = 0.0046, χ2 = 15.03, I2 = 73.4%, see Supplementary Figure S2A). Similarly, the pooled specificity was 0.89 (95%CI 0.84–0.93), also demonstrating moderate heterogeneity (p = 0.0037, χ2 = 15.56, I2 = 74.3%, see Supplementary Figure S2B). The pooled DOR was 60.89 (95%CI 10.99–337.27), with moderate heterogeneity (p = 0.0094, Cochran Q = 13.42, I2 = 70.2%, Supplementary Figure S2C). The area under the curve (AUC) value was 0.96 ± 0.04 (Supplementary Figure S2D).
For the pediatric population, the pooled sensitivity was 0.80 (95%CI 0.75–0.85), exhibiting no heterogeneity (p = 0.7472, χ2 = 4.28, I2 = 0%, Supplementary Figure S2E). The pooled specificity was 0.77 (95%CI 0.73–0.80), with no heterogeneity (p = 0.4442, χ2 = 6.85, I2 = 0%, Supplementary Figure S2F). The pooled DOR was 13.02 (95%CI 9.28–18.27), with no heterogeneity (p = 0.3805, Cochran Q = 7.48, I2 = 6.4%, Supplementary Figure S2G). The AUC value was 0.84 ± 0.02 (Supplementary Figure S2H).
In adults, the pooled sensitivity was 0.75 (95%CI 0.73–0.76), with high heterogeneity (p < 0.0001, χ2 = 326.86, I2 = 77.1%, Supplementary Figure S2I). The pooled specificity was 0.71 (95%CI 0.70–0.72), also with notable heterogeneity (p < 0.0001, χ2 = 834.66, I2 = 91.0%, Supplementary Figure S2J). The pooled DOR was 9.49 (95%CI 7.93–11.36), with moderate heterogeneity (p < 0.0001, Cochran Q = 183.54, I2 = 59.1%, Supplementary Figure S2K). The AUC value was 0.82 ± 0.01 (Supplementary Figure S2L).
The predictive performance of studies with small sample sizes is shown in Supplementary Figures S3A–D. The pooled sensitivity was 0.78 (95% CI: 0.75–0.80), with low heterogeneity (p = 0.0008, χ2 = 70.4, I2 = 47.5%, Supplementary Figure S3A). The pooled specificity was 0.81 (95% CI: 0.79–0.83), with moderate heterogeneity (p < 0.0001, χ2 = 115.15, I2 = 67.9%, Supplementary Figure S3B). The pooled DOR was 14.18 (95% CI: 11.61–17.30), with low heterogeneity (p = 0.0048, Cochran Q = 63.08, I2 = 41.3%, Supplementary Figure S3C). The AUC value was 0.86 ± 0.01 (Supplementary Figure S3D).
In studies with large sample sizes, the pooled sensitivity was 0.75 (95% CI: 0.73–0.77), with significant heterogeneity (p < 0.0001, χ2 = 284.60, I2 = 82.4%, Supplementary Figure S3E). The pooled specificity was 0.69 (95% CI: 0.68–0.70), with substantial heterogeneity (p < 0.0001, χ2 = 686.24, I2 = 92.7%, Supplementary Figure S3F). The pooled DOR was 8.42 (95% CI: 6.90–10.28), with moderate heterogeneity (p < 0.0001, Cochran Q = 133.42, I2 = 62.5%, Supplementary Figure S3G). The AUC value was 0.81 ± 0.01 (Supplementary Figure S3H).
Three reports (Xue et al., 2019; Li et al., 2022; Zhao et al., 2022) assessed miR-133a-3p. The pooled sensitivity was 0.83 (95% CI: 0.70–0.92), with no heterogeneity (p = 0.8599, χ2 = 0.30, I2 = 0%, Supplementary Figure S4A). The pooled specificity was 0.79 (95% CI: 0.71–0.86), with low heterogeneity (p = 0.1964, χ2 = 3.26, I2 = 38.6%, Supplementary Figure S4B). The pooled DOR was 18.87 (95% CI: 8.19–43.48), with no heterogeneity (p = 0.3531, Cochran Q = 2.08, I2 = 3.9%, Supplementary Figure S4C). The AUC was 0.90 ± 0.03 (Supplementary Figure S4D).
Seven studies (Sankar et al., 2022; Chen L. et al., 2020; Qiu et al., 2020; Wang H. et al., 2020; Yang J. et al., 2020; Pan et al., 2021; Yao et al., 2023) analyzed miR-146a. The pooled sensitivity was 0.73 (95% CI: 0.67–0.78), and specificity was 0.73 (95% CI: 0.70–0.77), both showing significant heterogeneity (sensitivity: p < 0.0001, χ2 = 35.20, I2 = 83.0%; specificity: p < 0.0001, χ2 = 162.24, I2 = 96.3%, Supplementary Figures S5A,B). The pooled DOR was 11.14 (95% CI: 7.63–16.28), with low heterogeneity (p = 0.1539, Cochran Q = 9.37, I2 = 36.0%, Supplementary Figure S5C). The AUC was 0.84 ± 0.02 (Supplementary Figure S5D).
Five reports (Salim et al., 2020; Na et al., 2020; Xu et al., 2021; Yang et al., 2021; Ali et al., 2023) examined miR-21. The pooled sensitivity was 0.81 (95% CI: 0.74–0.87), with significant heterogeneity (p = 0.0136, χ2 = 12.56, I2 = 68.2%, Supplementary Figure S6A). The specificity was 0.63 (95% CI: 0.57–0.68), also showing high heterogeneity (p < 0.0001, χ2 = 85.61, I2 = 95.3%, Supplementary Figure S6B). The pooled DOR was 13.13 (95% CI: 3.94–43.80), with high heterogeneity (p = 0.0007, Cochran Q = 19.11, I2 = 79.1%, Supplementary Figure S6C). The AUC was 0.87 ± 0.07 (Supplementary Figure S6D).
Four reports (Lin, 2017; Lin et al., 2019; Xu et al., 2021; Hao et al., 2024) evaluated miR-210. The pooled sensitivity was 0.72 (95% CI: 0.64–0.79), with low heterogeneity (p = 0.1132, χ2 = 5.97, I2 = 49.7%, Supplementary Figure S7A). The specificity was 0.75 (95% CI: 0.70–0.80), with moderate heterogeneity (p = 0.1074, χ2 = 6.09, I2 = 50.7%, Supplementary Figure S7B). The pooled DOR was 9.09 (95% CI: 4.24–19.47), with moderate heterogeneity (p = 0.0739, Cochran Q = 6.94, I2 = 56.8%, Supplementary Figure S7C). The AUC was 0.83 ± 0.05 (Supplementary Figure S7D).
Five studies (Wang et al., 2012; Yang J. et al., 2020; Wang H. et al., 2021; Pan et al., 2022; Bao and Zhang, 2023) assessed miR-223-3p. The pooled sensitivity was 0.75 (95% CI: 0.69–0.81), with significant heterogeneity (p = 0.0030, χ2 = 16.00, I2 = 75.0%, Supplementary Figure S8A). The pooled specificity was 0.83 (95% CI: 0.79–0.87), showing no heterogeneity (p = 0.2676, χ2 = 5.20, I2 = 23.0%, Supplementary Figure S8B). The pooled DOR was 16.24 (95% CI: 8.65–30.49), with moderate heterogeneity (p = 0.0892, Cochran Q = 8.06, I2 = 50.4%, Supplementary Figure S8C). The AUC was 0.89 ± 0.02 (Supplementary Figure S8D).
Three studies (Yang et al., 2016; Zhang et al., 2018; Dou et al., 2020) assessed the predictive performance of microRNA-155 (Supplementary Figure S9). The pooled sensitivity was 0.83 (95%CI 0.74–0.89), with no heterogeneity (p = 0.4159, χ2 = 1.75, I2 = 0%, Supplementary Figure S9A). The pooled specificity was 0.73 (95%CI 0.67–0.78), with high heterogeneity (p < 0.0001, χ2 = 23.59, I2 = 91.5%, Supplementary Figure S9B). The pooled DOR was 16.34 (95%CI 8.76–30.46), with no heterogeneity (p = 0.3379, Cochran-Q = 2.17, I2 = 7.8%, Supplementary Figure S9C). The AUC value was 0.89 ± 0.02 (Supplementary Figure S9D).
The overall predicting performance of miR-25 was analyzed using three studies (Yao et al., 2015; Wang J. et al., 2020; Bian and Pang, 2023) (Supplementary Figure S10). The pooled sensitivity was 0.70 (95%CI 0.59–0.80), and the specificity was 0.87 (95%CI 0.79–0.92). The presence of significant heterogeneity was identified (sensitivity: p = 0.0007, χ2 = 14.59, I2 = 86.3%; specificity: p = 0.0581, χ2 = 5.69, I2 = 64.9%, Supplementary Figures S10A,B). The pooled DOR was 16.31 (95%CI 5.05–52.68), with moderate heterogeneity (p = 0.1307, Cochran-Q = 4.07, I2 = 50.8%, Supplementary Figure S10C). The AUC value was 0.89 ± 0.07 (Supplementary Figure S10D).
Five studies (Wang et al., 2012; Rahmel et al., 2018; Ma et al., 2021; Li et al., 2022; Yao et al., 2023) assessed the predictive performance of miR-122 (see Supplementary Figure S11). The pooled sensitivity was 0.75 (95% CI 0.69–0.81), and the specificity was 0.76 (95% CI 0.71–0.81). Notably, significant heterogeneity was observed in the sensitivity analysis (sensitivity: p < 0.0001, χ2 = 28.93, I2 = 86.2%; specificity: p < 0.0001, χ2 = 29.85, I2 = 86.6%, Supplementary Figures S11A,B). The pooled DOR was 11.81 (95%CI 7.44–18.73), with no heterogeneity (p = 0.5561, Cochran-Q = 3.01, I2 = 0%, Supplementary Figure S11C). The AUC value was 0.84 ± 0.02 (Supplementary Figure S11D).
Four studies (Liu et al., 2020; Yang Y. et al., 2020; Zhao et al., 2020; Zhu, 2020) were included to analyze the overall predicting performance of miR-125a (Supplementary Figure S12). The pooled sensitivity was 0.86 (95%CI 0.79–0.91), with moderate heterogeneity (p = 0.0369, χ2 = 8.49, I2 = 64.7%, Supplementary Figure S12A). The pooled specificity was 0.48 (95%CI 0.43–0.53), with low heterogeneity (p = 0.2191, χ2 = 4.43, I2 = 32.2%, Supplementary Figure S12B). The pooled DOR was 5.69 (95%CI 2.50–12.95), with moderate heterogeneity (p = 0.0725, Cochran-Q = 6.98, I2 = 57.0%, Supplementary Figure S12C). The AUC value was 0.51 ± 0.12 (Supplementary Figure S12D).
Four studies (Xu et al., 2020; Zhao et al., 2020; Zhu, 2020; Zhang et al., 2024) assessed the predictive performance of microRNA-125b (Supplementary Figure S13). The pooled sensitivity was 0.68 (95%CI 0.60–0.76), with moderate heterogeneity (p = 0.0828, χ2 = 6.68, I2 = 55.1%, Supplementary Figure S13A). The pooled specificity was 0.76 (95%CI 0.71–0.80), with low heterogeneity (p = 0.2584, χ2 = 4.03, I2 = 25.5%, Supplementary Figure S13B). The pooled DOR was 6.36 (95%CI 4.18–9.68), with no heterogeneity (p = 0.2667, Cochran-Q = 3.95, I2 = 24.1%, Supplementary Figure S13C). The AUC value was 0.80 ± 0.03 (Supplementary Figure S13D).
Four studies (Yang et al., 2022; Yang et al., 2015; Pan et al., 2021; Zhang et al., 2024) were included in order to analyze the overall predictive performance of microRNA-150 (see Supplementary Figure S14). The pooled sensitivity was 0.71 (95% CI 0.65–0.77), and the specificity was 0.70 (95% CI 0.66–0.74). Notably, significant heterogeneity was observed in the sensitivity analysis (sensitivity: p = 0.0024, χ2 = 14.37, I2 = 79.0%; specificity: p < 0.0001, χ2 = 25.07, I2 = 88.0%, Supplementary Figures S14A,B). The pooled DOR was 5.75 (95%CI 4.21–7.87), with no heterogeneity (p = 0.5495, Cochran-Q = 2.11, I2 = 0%, Supplementary Figure S14C). The AUC value was 0.76 ± 0.02 (Supplementary Figure S14D).
None of the individual studies significantly affected the overall results, as confirmed by STATA 15.1 (Figure 4). The Egger’s regression test yielded a p-value of less than 0.001, a t-value of 3.88, and a 95% confidence interval ranging from 7.18 to 22.25 (Figure 5A). This finding suggests the presence of publication bias. However, the implementation of the trim-and-fill method to address publication bias revealed no substantial alteration in the findings when compared to the results obtained after the inclusion of 28 studies (p < 0.001, Z = 20.08, 95%CI 1.74–2.12), suggesting that the meta-analysis outcomes remained resilient to potential bias (Figure 5B).

Figure 4. Sensitivity analysis for the results TmiRs. DOR, predicting odds ratio; ESS, effective sample size; miR, mircoRNA; TmiRs, total mixed miRNAs.
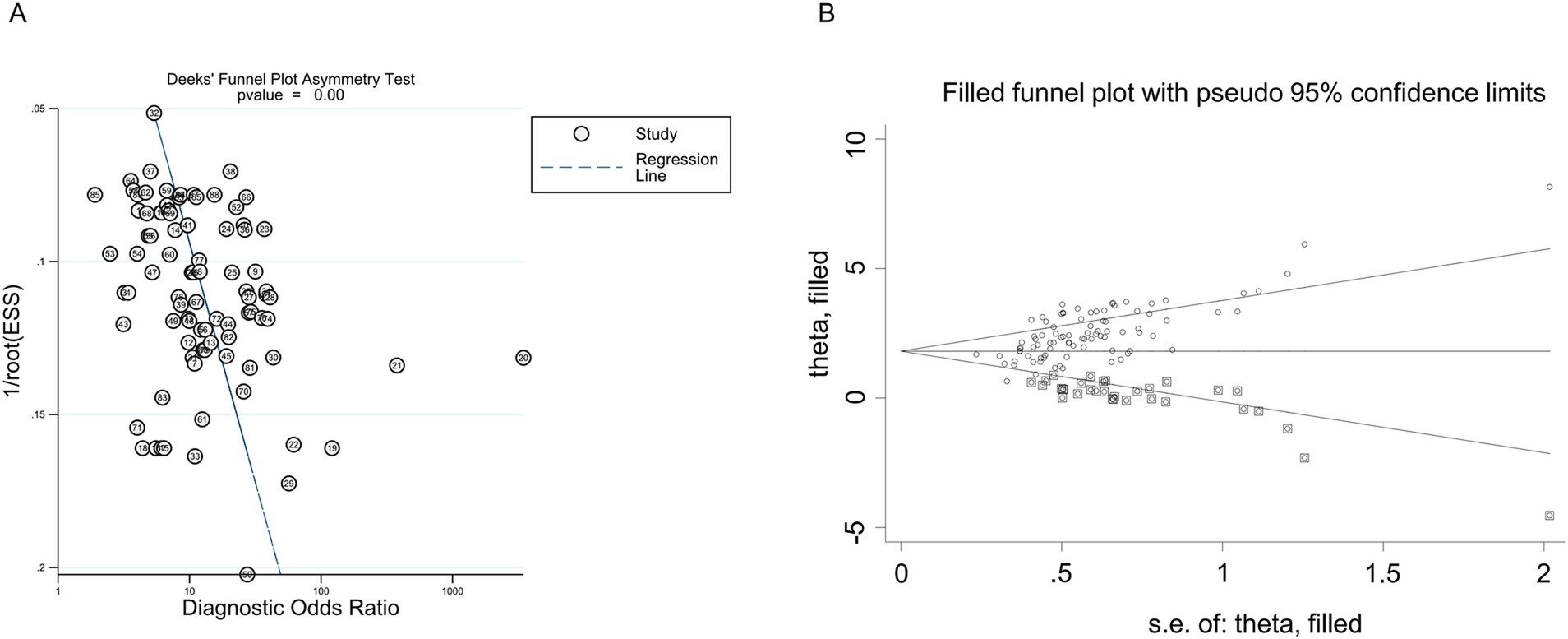
Figure 5. Publication bias of the individual trials on the results TmiRs. (A) Egger’s publication bias plots for the assessment of potential publication bias. Asymmetry of the dot distribution between regression lines showed potential publication bias, p < 0.001, t = 3.88, 95% CI 7.18–22.25. (B) The funnel plot of publication bias by the trim-and-fill method. After filled 28 potentially missing studies, the funnel plots were symmetrical. CI, confidence interval; DOR, predicting odds ratio; ESS, effective sample size; miR, mircoRNA; TmiRs, total mixed miRNAs.
Since their discovery approximately 10 years ago, numerous studies have investigated the potential of miRNAs as biomarkers for predicting sepsis mortality. In this meta-analysis, we have included 55 studies involving 2,047 non-survival and 4,396 survival sepsis patients, covering 41 different miRNAs. Our analysis revealed that TmiRs had a combined AUC of 0.83, with 76% sensitivity and 72% specificity, indicating that miRNAs exhibit moderate predictive accuracy as biomarkers for sepsis mortality. Furthermore, we conducted an assessment of individual miRNAs and identified that miR-133a-3p, miR-146a, miR-21, miR-210, miR-223-3p, miR-155, miR-25, miR-122, miR-125a, miR-125b, and miR-150 have been the most frequently examined in recent studies. Among these, miR-133a-3p exhibited the highest AUC in the SROC analysis, with a pooled sensitivity of 0.83 (95%CI 0.70–0.92), a pooled specificity of 0.79 (95%CI 0.71–0.86), and an SROC value of 0.90. Other microRNAs, including miR-146a, miR-21, miR-210, miR-223-3p, miR-155, miR-25, miR-122, miR-125a, miR-125b, and miR-150, also exhibited SROC values of 0.84, 0.87, 0.83, 0.89, 0.89, 0.89, 0.84, 0.51, 0.80, and 0.76, respectively. The findings of this study suggest that microRNAs (miRNAs), particularly miR-133a-3p, miR-146a, miR-21, miR-210, miR-223-3p, miR-155, miR-25, miR-122, miR-125b, and miR-150, have the potential to serve as useful biomarkers for predicting sepsis mortality. This meta-analysis is, to the best of our knowledge, the first to evaluate the accuracy of miRNAs in predicting sepsis mortality.
Considering the substantial heterogeneity observed in the analysis, a meta-regression was conducted to explore potential sources of variability. The analysis encompassed a comprehensive set of factors, including specimen type (serum, plasma, PBMCs), patient characteristics (neonates, children, adults), measurement techniques for microRNA (e.g., variations in qRT-PCR reference genes and extraction methods), geographical region, and total sample size. The analysis revealed that population differences and sample size contributed significantly to the heterogeneity, likely due to variations in immune responses and disease progression across different age groups and populations. Inconsistent sepsis diagnostic criteria (e.g., Sepsis 1.0, 2.0, 3.0) and methodological differences in RNA extraction and quantification further compounded the findings. Despite conducting subgroup analyses based on population and sample type, heterogeneity persisted, potentially due to the uneven distribution of studies. To address this challenge, a random-effects model was employed to mitigate the impact of heterogeneity. These findings underscore the necessity for standardized study designs in future research endeavors. The harmonization of diagnostic criteria, the standardization of measurement techniques, and the assurance of adequate sample sizes are imperative to mitigate bias and enhance the reliability of miRNAs as prognostic biomarkers in sepsis.
In a recent meta-analysis of 26 studies, Wang C. et al. (2022) evaluated the prognostic accuracy of the qSOFA, NEWS, and SIRS criteria for sepsis. Their findings indicated that qSOFA exhibited superior overall prognostic accuracy compared to SIRS and NEWS. The pooled sensitivity of qSOFA was 0.46 (95%CI 0.39–0.53), specificity was 0.82 (95%CI 0.76–0.86), and the AUC value was 0.69, which is lower than the results obtained in this study. Furthermore, several studies (Yang et al., 2022; Sun et al., 2019; Qiu et al., 2020; Wang L. et al., 2020; Ma et al., 2021; Wang H. et al., 2021; Xu et al., 2021) incorporated within the present meta-analysis assessed the collective predictive capacity of miRNAs, IL-6, PCT, CRP, SOFA score, and additional markers for sepsis mortality. Despite the unfeasibility of a comprehensive combined analysis of microRNAs (miRNAs) and other indicators, the integrated predictive approach employed in this study demonstrates considerable promise. Concurrent with this, previous studies (Yang et al., 2022; Wang H. et al., 2021; Xu et al., 2021; Li et al., 2022) underscored the significance of combined prediction in sepsis management, though further clinical research is necessary to elucidate its benefits.
Recent studies have underscored the critical roles of miRNAs and other non-coding RNAs (ncRNAs) in regulating immune responses and driving the progression of sepsis. MiRNAs such as miR-150, miR-146a, and miR-223 have been consistently associated with sepsis pathophysiology, with several studies confirming their potential as biomarkers for sepsis mortality (Formosa et al., 2022; Antonakos et al., 2022). Concurrently, the potential of miR-155, miR-21, miR-223-3p, miR-146a, and miR-125a as sepsis indicators has been previously demonstrated in our study (Zheng et al., 2023). The present study further corroborates the predictive value of miR-155, miR-21, miR-223-3p, and miR-146a, in addition to miR-125a, suggesting that these microRNAs may play pivotal roles in sepsis pathogenesis. Among these, miR-146a has been the subject of particular study, as evidenced by seven publications in our meta-analysis that affirm its predictive value for sepsis mortality. However, further research is needed to clarify the molecular mechanisms underlying its function. Future studies should prioritize exploring these microRNAs at the molecular level to enhance our understanding of their roles in sepsis. MiR-146a has been demonstrated to modulate immune responses during sepsis by targeting pivotal components of the NF-κB and TNF signaling pathways. It has been demonstrated that MiR-146a suppresses excessive inflammation by inhibiting the expression of interleukin-1 receptor-associated kinase (IRAK1) and TNF receptor-associated factor 6 (TRAF6), both of which are critical mediators in these pathways (Gilyazova et al., 2023). This regulatory function helps prevent an overactive immune response, which could otherwise lead to tissue damage and organ failure. Furthermore, it has been demonstrated that polymorphisms in miR-146a are associated with altered inflammatory responses, thereby reinforcing the complex interplay between genetic and environmental factors in the pathogenesis of these conditions.
In a similar manner, microRNA-133a-3p has been shown to offer protection against sepsis-induced acute respiratory distress syndrome (ARDS) by modulating SIRT1, a pivotal regulator of inflammation and oxidative stress. The downregulation of miR-133a-3p in septic patients has been associated with increased lung injury, while its upregulation has been linked to reduced inflammatory damage and improved lung function (Hui et al., 2024). These findings underscore the pivotal role of miR-133a-3p in moderating the inflammatory cascade in sepsis, thereby substantiating its potential as a therapeutic target.
MiR-150 has also been shown to modulate immune responses in septic patients (Formosa et al., 2022). Notably, the function of miR-150 in cancer is dual, serving as either a tumor suppressor or an oncogene, depending on the specific cancer type and the cellular context. For instance, in liver, ovarian, and colorectal cancers, miR-150 acts as a tumor suppressor, whereas in breast cancer it promotes tumor progression by regulating processes like epithelial-mesenchymal transition through matrix metalloproteinases and cell adhesion molecules (Ameri et al., 2023; Forterre et al., 2020). This dual role underscores the complexity of miR-150’s function across different diseases, including sepsis, where its dysregulation may significantly impact immune and inflammatory pathways (Mazziotta et al., 2023). Other microRNAs identified in our analysis, such as miR-125b and miR-193, have also been implicated in cancer and various other diseases, thereby reinforcing their broader regulatory roles (Forterre et al., 2020; Mazziotta et al., 2023). A comprehensive understanding of the roles of these miRNAs in both cancer and sepsis could offer valuable insights into their potential as biomarkers and therapeutic targets.
Furthermore, miR-223-3p has been shown to play a pivotal role in modulating immune responses during sepsis through multiple mechanisms. Specifically, it has been observed that miR-223-3p exerts its regulatory influence over autophagy in CD4+ T lymphocytes, a process that is achieved by directly targeting Forkhead box O1 (FOXO1). Overexpression of miR-223-3p has been shown to suppress FOXO1 expression, thereby reducing autophagic activity and preventing immune cell dysfunction in sepsis (Xiang et al., 2024). Moreover, the influence of miR-223-3p on the polarization of macrophages is characterized by its promotion of an anti-inflammatory M2 phenotype, thereby contributing to the alleviation of sepsis severity in experimental models (Dang and Leelahavanichkul, 2020). Another critical role of miR-223-3p is its suppression of the NLRP3 inflammasome, a key mediator of inflammatory responses, which reduces pro-inflammatory cytokine release and attenuates hyperinflammation (Shi et al., 2023). These findings underscore the central role of miR-223-3p in orchestrating both innate and adaptive immune responses during sepsis.
MiR-210 has been identified as a pivotal regulator of sepsis pathophysiology, particularly through its involvement in hypoxia-related pathways and immune responses. As a hypoxia-inducible microRNA, miR-210 is significantly upregulated in septic conditions, reflecting the tissue hypoxia characteristic of the disease. Elevated circulating levels of miR-210 have been demonstrated to be strongly associated with disease severity and mortality, underscoring its potential as a prognostic biomarker (Powell et al., 2022). Mechanistically, the disruption of mitochondrial function by miR-210 is attributed to its targeting of ISCU, a scaffold protein that is essential for iron–sulfur cluster assembly. This disruption leads to mitochondrial dysfunction, oxidative stress, and cardiomyocyte apoptosis, exacerbating myocardial injury (Chen et al., 2021). Furthermore, miR-210 has been shown to induce glycolytic reprogramming in macrophages, thereby enhancing pro-inflammatory cytokine production. This metabolic shift, while supporting acute inflammation, also contributes to systemic damage and multi-organ dysfunction (Virga et al., 2021). Collectively, these findings underscore the dual role of miR-210 in mediating inflammation and organ damage in sepsis.
Additionally, miR-122 plays a central role in sepsis by modulating immune responses, inflammation, and coagulation pathways, as well as protecting against organ damage. A recent study demonstrated that miR-122 mitigates sepsis-induced liver injury by targeting the BCL2A1 signaling pathway, thereby reducing macrophage apoptosis and alleviating inflammatory responses (Liu et al., 2024). Furthermore, the role of miR-122 extends to the regulation of pyroptosis, a form of programmed cell death associated with inflammation. By targeting NLRP1, miR-122-3p suppresses LPS-induced pyroptosis in macrophages, limiting the release of pro-inflammatory cytokines and reducing systemic inflammation (Li M. et al., 2023). Additionally, miR-122 influences coagulation and inflammatory pathways during sepsis. Through its interactions with MASP1 and HO-1 genes, miR-122-5p modulates coagulation abnormalities and systemic inflammation, demonstrating its dual role in regulating immune and hemostatic responses (Wang H. et al., 2022). Collectively, these findings emphasize the multifaceted functions of miR-122 in mitigating organ damage, controlling systemic inflammation, and regulating coagulation in sepsis, thereby establishing its potential as both a biomarker and a therapeutic target.
In addition to microRNAs (miRNAs), non-coding RNAs (ncRNAs) such as long non-coding RNAs (lncRNAs) have been found to regulate sepsis-induced organ dysfunction. Among these, MALAT1 and NEAT1 have garnered significant attention due to their distinct roles in modulating inflammatory pathways (Wang C. et al., 2021; Li et al., 2021). MALAT1 is associated with promoting inflammatory responses by facilitating NF-κB nuclear translocation, exacerbating lung injury during sepsis (Cui et al., 2021). Conversely, NEAT1 exhibits dual regulatory effects, either promoting or suppressing inflammation depending on its molecular interactions. For instance, NEAT1 acts as a molecular sponge for miR-124-3p, inhibiting STAT3-mediated pro-inflammatory signaling (Ghafouri-Fard et al., 2021). These findings highlight the intricate regulatory mechanisms of lncRNAs in sepsis, offering potential therapeutic targets warranting further investigation.
To enhance diagnostic accuracy, recent studies have investigated the potential of integrating miRNAs with traditional biomarkers such as IL-6, PCT, and CRP. The integration of multi-biomarker panels, encompassing miRNAs, has been shown to yield substantial improvements in diagnostic precision (Teggert et al., 2020). The integration of these multi-biomarker panels with miRNAs has been shown to yield substantial improvements in diagnostic accuracy (Teggert et al., 2020; Bradley and Bhalla, 2023). Future research should prioritize validating these multi-biomarker panels in clinical settings to maximize their utility.
In our meta-analysis, we comprehensively examined studies that utilized diverse sample types, including plasma, PBMCs, and whole blood, as outlined in Table 1. This diversity enables a comprehensive evaluation of the predictive utility of miRNAs across various biological sources. However, due to the limited number of studies focusing on PBMCs, the findings predominantly reflect circulating miRNAs found in plasma and serum. Circulating miRNAs are primarily transported in extracellular vesicles (EVs), where they are shielded from degradation and can function as systemic signaling molecules, mirroring systemic inflammatory and immune responses in sepsis (Real et al., 2018). For instance, studies have demonstrated that plasma-derived EVs in septic patients carry miRNAs implicated in inflammation and cell cycle regulation, underscoring their significance in sepsis pathophysiology (Real et al., 2018). Conversely, PBMC-derived miRNAs provide insights into cell-specific regulatory processes, such as immune cell activation. While underrepresented in this study, PBMC-derived miRNAs warrant further exploration to uncover their potential role in sepsis pathophysiology (Antonakos et al., 2022). Integrating data from both circulating and PBMC-derived miRNAs could advance diagnostic and therapeutic approaches (Xu et al., 2018).
The present meta-analysis has several notable strengths. Firstly, it is the inaugural meta-analysis to systematically evaluate the prognostic accuracy of miRNAs in predicting sepsis mortality. Secondly, it encompasses a substantial number of studies, with 55 studies included, which is more than most previous meta-analyses on sepsis mortality prediction. Thirdly, we evaluated the accuracy of nine miRNAs—namely, miR-133a-3p, miR-146a, miR-21, miR-210, miR-223-3p, miR-155, miR-25, miR-122, and miR-125b—that have the potential to serve as sepsis biomarkers.
While the present study underscores the potential of miRNAs as biomarkers for sepsis, several challenges exist that limit their clinical applicability. Variability in the expression of miRNAs across diverse populations, influenced by factors such as age, underlying health conditions, and disease progression, may compromise diagnostic accuracy and reduce generalizability. Furthermore, the evidence supporting each individual miRNA indicator is derived from a limited number of studies, ranging from three to seven, which introduces potential bias. The lack of standardized methodologies for detecting miRNAs, including inconsistencies in sample collection, RNA extraction, and qRT-PCR reference genes, adds another layer of complexity. Although meta-regression has been employed to address some of these issues, significant heterogeneity remains due to differences in the indicators of miRNAs and the study methodologies. Moreover, the predominance of studies conducted in Asian populations may limit the generalizability of our findings, considering the well-documented disparities in genetics and healthcare practices across different regions. To enhance the reproducibility and global relevance of these biomarkers, future studies should aim to incorporate more diverse populations and adopt standardized protocols.
In summary, our meta-analysis demonstrated that microRNAs (miRNAs), particularly miR-133a-3p, miR-155-5p, miR-146a, miR-21, miR-210, miR-223-3p, and miR-155, could serve as useful biomarkers for predicting sepsis mortality. To improve reliability, future research should focus on standardizing protocols, conducting longitudinal studies, and developing subgroup-specific miRNA panels for neonates, children, and adults. These advancements are crucial for transforming miRNAs into robust and universally applicable biomarkers in sepsis management.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
YJ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft. YZ: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft. YL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft. XZ: Conceptualization, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1472124/full#supplementary-material
Ali, M. A., Khamis Hussein, S., Ali Mohamed, E., Ezzat, M. A., Abdelmoktader, A., Habib, M. A., et al. (2023). Diagnostic and prognostic values of miR181b-5p and miR21-5p for neonatal sepsis risk and their link to SNAP II score and disease mortality. Non-coding RNA Res. 8, 115–125. doi: 10.1016/j.ncrna.2022.11.001
Ameri, A., Ahmed, H. M., Pecho, R. D. C., Arabnozari, H., Sarabadani, H., Esbati, R., et al. (2023). Diverse activity of miR-150 in tumor development: shedding light on the potential mechanisms. Cancer Cell Int. 23:261. doi: 10.1186/s12935-023-03105-3
Antonakos, N., Gilbert, C., Théroude, C., Schrijver, I. T., and Roger, T. (2022). Modes of action and diagnostic value of miRNAs in sepsis. Front. Immunol. 13:951798. doi: 10.3389/fimmu.2022.951798
Bao, Z., and Zhang, Y. (2023). Changes of miR-451 and miR-223 in patients with sepsis and their prognostic value. Chin. J. Lab. Diagn. :27(8). doi: 10.3969/j.issn.1007-4287.2023.08.026
Behroozizad, N., Mahmoodpoor, A., Shadvar, K., Ardebil, R., Pahnvar, A., Sohrabifar, N., et al. (2024). Evaluation of circulating levels of miR-135a and miR-193 in patients with sepsis. Mol. Biol. Rep. 51:282. doi: 10.1007/s11033-024-09225-x
Bian, M., and Pang, L. (2023). Value of peripheral blood microRNAs in the diagnosis and prognosis evaluation of pneumonia complicated with sepsis. J. Clin. Pulmonary Med. 78-82:116. doi: 10.3969/j.issn.1009-6663.2023.01.017
Bradley, Z., and Bhalla, N. (2023). Point-of-care diagnostics for sepsis using clinical biomarkers and microfluidic technology. Biosens. Bioelectron. 227:115181. doi: 10.1016/j.bios.2023.115181
Chen, D., Hou, Y., and Cai, X. (2021). MiR-210-3p enhances Cardiomyocyte apoptosis and mitochondrial dysfunction by targeting the NDUFA4 gene in Sepsis-induced myocardial dysfunction. Int. Heart J. 62, 636–646. doi: 10.1536/ihj.20-512
Chen, W., Liu, L., Yang, J., and Wang, Y. (2020). MicroRNA-146b correlates with decreased acute respiratory distress syndrome risk, reduced disease severity, and lower 28-day mortality in sepsis patients. J. Clin. Lab. Anal. 34:e23510. doi: 10.1002/jcla.23510
Chen, L., Yu, L., Zhang, R., Zhu, L., and Shen, W. (2020). Correlation of microRNA-146a/b with disease risk, biochemical indices, inflammatory cytokines, overall disease severity, and prognosis of sepsis. Medicine 99:e19754. doi: 10.1097/MD.0000000000019754
Cui, N., Liang, Y., Wang, J., Liu, B., Wei, B., and Zhao, Y. (2021). Minocycline attenuates oxidative and inflammatory injury in a intestinal perforation induced septic lung injury model via down-regulating lncRNA MALAT1 expression. Int. Immunopharmacol. 100:108115. doi: 10.1016/j.intimp.2021.108115
Dang, C. P., and Leelahavanichkul, A. (2020). Over-expression of miR-223 induces M2 macrophage through glycolysis alteration and attenuates LPS-induced sepsis mouse model, the cell-based therapy in sepsis. PLoS One 15:e0236038. doi: 10.1371/journal.pone.0236038
Deeks, J. J. (2001). Systematic reviews in health care: systematic reviews of evaluations of diagnostic and screening tests. BMJ (Clinical research ed). 323, 157–162. doi: 10.1136/bmj.323.7305.157
Deeks, J. J., Macaskill, P., and Irwig, L. (2005). The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J. Clin. Epidemiol. 58, 882–893. doi: 10.1016/j.jclinepi.2005.01.016
Deng, Y., Qiu, C., Wu, R., Kuang, X., and Lu, L. (2021). Significances of the plasma expression of microRNA-101-3p and microRNA-141-3p in children with sepsis. Chinese J. App. Clin. Pediatr. 36, 1383–1388. doi: 10.3760/cma.j.cn101070-20200429-00753
Deng, Y., Qiu, C., Wu, R., Kuang, X., and Lu, L. (2022). Clinical application of plasma miR-455-5p and miR-483-5p in children with Sepsis. J. Mod. Lab. Med. 37, 79–82. doi: 10.3969/j.issn.1671-7414.2022.03.016
Dou, H., Hu, F., Wang, W., Ling, L., Wang, D., and Liu, F. (2020). Serum MiR-155 and MiR-143 can be used as prognostic markers for severe sepsis/septic shock in the elderly. Int. J. Clin. Exp. Med. 13, 3771–3780.
Formosa, A., Turgeon, P., and Dos Santos, C. C. (2022). Role of miRNA dysregulation in sepsis. Mol. Med. 28:99. doi: 10.1186/s10020-022-00527-z
Forterre, A., Komuro, H., Aminova, S., and Harada, M. (2020). A comprehensive review of Cancer MicroRNA therapeutic delivery strategies. Cancers 12:1852. doi: 10.3390/cancers12071852
Ghafouri-Fard, S., Khoshbakht, T., Hussen, B. M., Taheri, M., and Arefian, N. (2021). Regulatory role of non-coding RNAs on immune responses during Sepsis. Front. Immunol. 12:798713. doi: 10.3389/fimmu.2021.798713
Gilyazova, I., Asadullina, D., Kagirova, E., Sikka, R., Mustafin, A., Ivanova, E., et al. (2023). MiRNA-146a-a key player in immunity and diseases. Int. J. Mol. Sci. 24:12767. doi: 10.3390/ijms241612767
Guo, W., Li, Y., and Li, Q. (2021). Relationship between miR-29a levels in the peripheral blood and sepsis-related encephalopathy. Am. J. Transl. Res. 13, 7715–7722
Hao, J., Liang, L., Ma, Y., Xu, M., and Li, Q. (2024). Identification and analysis of genes associated with the severity and prognosis of sepsis. Technol. Health Care 32, 989–996. doi: 10.3233/THC-230363
Higgins, J. P., and Thompson, S. G. (2002). Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558. doi: 10.1002/sim.1186
Hui, Q., Zhang, Q., Li, X., Wang, K., Zhang, J., and Zhou, Z. (2024). Down-regulation of miR-133a-3p protects lung tissue against sepsis-induced acute respiratory distress syndrome by up-regulating SIRT1. Arch. Med. Sci. 20, 289–301. doi: 10.5114/aoms.2020.94410
Huo, R., Dai, M., Fan, Y., Zhou, J., Li, L., and Zu, J. (2017). Predictive value of miRNA-29a and miRNA-10a-5p for 28-day mortality in patients with sepsis-induced acute kidney injury. J. Southern Med. Univ. 37, 645–651. doi: 10.3969/j.issn.1673-4254.2017.05.13
Li, M., Hu, L., Ke, Q., Ruan, C., and Liu, X. (2023). miR-122-3p alleviates LPS-induced Pyroptosis of macrophages via targeting NLRP1. Ann. Clin. Lab. Sci. 53, 578–586.
Li, C., Li, N., Lv, H., Wei, G., and Xia, H. (2022). Correlation of microrNA-122a and microrNA-133A-3p with severity and prognosis of sepsis patients. Chinese J. Clin. 50, 586–588. doi: 10.3969/j.issn.2095-8552.2022.05.025
Li, J., Zhang, Y., Zhang, D., and Li, Y. (2021). The role of Long non-coding RNAs in Sepsis-induced cardiac dysfunction. Front. Cardiovasc. Med. 8:684348. doi: 10.3389/fcvm.2021.684348
Li, N., Zhong, C., Zhang, J., Liu, Y., and Long, B. (2023). Serum miR-98-5p, ESM-1, sTM and VE-cad in sepsis patients with ARDS and their relationship with prognosis. Chin. J. Nosocom. 33, 2241–2245. doi: 10.11816/cn.ni.2023-230059
Lin, H. (2017). Assessment on miR-15b, miR120 and miR-486 as elderly sepsis diagnosis and prognosis biomarkers : Guangzhou University of Chinese Medicine.
Lin, L., and Chu, H. (2018). Quantifying publication bias in meta-analysis. Biometrics 74, 785–794. doi: 10.1111/biom.12817
Lin, Y., Ding, Y., Song, S., Li, M., Wang, T., and Guo, F. (2019). Expression patterns and prognostic value of miR-210, miR-494, and miR-205 in middle-aged and old patients with sepsis-induced acute kidney injury. Bosn. J. Basic Med. Sci. 19, 249–256. doi: 10.17305/bjbms.2019.4131
Lin, R., Hu, H., Li, L., Chen, G., Luo, L., and Rao, P. (2020). The potential of microRNA-126 in predicting disease risk, mortality of sepsis, and its correlation with inflammation and sepsis severity. J. Clin. Lab. Anal. 34:e23408. doi: 10.1002/jcla.23408
Liu, W., Geng, F., and Yu, L. (2020). Long non-coding RNA MALAT1/microRNA 125a axis presents excellent value in discriminating sepsis patients and exhibits positive association with general disease severity, organ injury, inflammation level, and mortality in sepsis patients. J. Clin. Lab. Anal. 34:e23222. doi: 10.1002/jcla.23222
Liu, S., Xie, J., Duan, C., Zhao, X., Feng, Z., Dai, Z., et al. (2024). ADAR1 inhibits macrophage apoptosis and alleviates Sepsis-induced liver injury through miR-122/BCL2A1 signaling. J. Clin. Transl. Hepatol. 12, 134–150. doi: 10.14218/JCTH.2023.00171
Liu, W., and Yang, Z. (2023). Relationship between the expression of serum lncRNA XIST and miRNA-130a and the severity and prognosis of patients with sepsis. Lab. Med. Clin. 20, 2537–2542. doi: 10.3969/j.issn.1672-9455.2023.17.018
Ma, W., Yang, W., Lin, Y., Zhang, L., and Wei, F. (2021). Predictive efficiency of miR-122a expression and HMGB1 level in serum on prognosis of children with SALI. Shandong Med. J. 61, 20–24. doi: 10.3969/j.issn.1002-266X.2021.18.005
Mazziotta, C., Cervellera, C. F., Lanzillotti, C., Touzé, A., Gaboriaud, P., Tognon, M., et al. (2023). MicroRNA dysregulations in Merkel cell carcinoma: molecular mechanisms and clinical applications. J. Med. Virol. 95:e28375. doi: 10.1002/jmv.28375
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6:e1000097. doi: 10.1371/journal.pmed.1000097
Moses, L. E., Shapiro, D., and Littenberg, B. (1993). Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Stat. Med. 12, 1293–1316. doi: 10.1002/sim.4780121403
Na, L., Ding, H., Xing, E., Zhang, Y., Gao, J., Liu, B., et al. (2020). The predictive value of microRNA-21 for sepsis risk and its correlation with disease severity, systemic inflammation, and 28-day mortality in sepsis patients. J. Clin. Lab. Anal. 34:e23103. doi: 10.1002/jcla.23103
Pan, Q., Guo, H., Zhao, Y., Ruan, L., and Ke, J. (2021). Influecing factors of 28d progrosis of patients with acute peritonitis and sepsis and the evaluation value of miR-150 and miR-146a on the condition of disease. Chin. J. Nosocomiol. 31, 3065–3068. doi: 10.11816/cn.ni.2021-210065
Pan, Y., Wu, X., Tang, X., and Chen, S. (2022). Predictive values of peripheral blood miR-223 and miR-124a in 28-day prognosis of septic shock. Chi. J. Emerg. Resusc. Disast. Med. 17, 1357–1360. doi: 10.3969/j.issn.1673-6966.2022.10.024
Powell, R. E., Tai, Y. Y., Kennedy, J. N., Seymour, C. W., and Chan, S. Y. (2022). Circulating hypoxia-dependent miR-210 is increased in clinical sepsis subtypes: a cohort study. J. Transl. Med. 20:448. doi: 10.1186/s12967-022-03655-6
Qiu, Y., Sun, Y., Gong, B., Lin, S., and Guan, W. (2020). Value of miR-146a and APACHE II score in predicting 28-day mortality in sepsis patients with acute kidney injury. Chinese J. Crit. Care Med. 40, 942–946. doi: 10.3969/j.issn.1002-1949.2020.10.007
Rahmel, T., Schäfer, S. T., Frey, U. H., Adamzik, M., and Peters, J. (2018). Increased circulating microrna-122 is a biomarker for discrimination and risk stratification in patients defined by sepsis-3 criteria. PLoS One 13:e0197637. doi: 10.1371/journal.pone.0197637
Real, J. M., Ferreira, L. R. P., Esteves, G. H., Koyama, F. C., Dias, M. V. S., Bezerra-Neto, J. E., et al. (2018). Exosomes from patients with septic shock convey miRNAs related to inflammation and cell cycle regulation: new signaling pathways in sepsis? Crit. Care 22:68. doi: 10.1186/s13054-018-2003-3
Salim, R. F., Sobeih, A. A., and Abd El Kareem, H. M. (2020). Evaluation of the clinical value of circulating miR-101, miR-187 and miR-21 in neonatal sepsis diagnosis and prognosis. Egypt. J. Med. Hum. Genet. 21:12. doi: 10.1186/s43042-020-00052-w
Sankar, S., Maruthai, K., Bobby, Z., and Adhisivam, B. (2022). MicroRNA expression in neonates with late-onset Sepsis–a cross-sectional comparative study. Immunol. Investig. 51, 1647–1659. doi: 10.1080/08820139.2021.2020282
Shen, X., Zhang, J., Huang, Y., Tong, J., Zhang, L., Zhang, Z., et al. (2020). Accuracy of circulating microRNAs in diagnosis of sepsis: a systematic review and meta-analysis. J. Intensive Care 8:84. doi: 10.1186/s40560-020-00497-6
Shi, M., Lu, Q., Zhao, Y., Ding, Z., Yu, S., Li, J., et al. (2023). miR-223: a key regulator of pulmonary inflammation. Front. Med. 10:1187557. doi: 10.3389/fmed.2023.1187557
Singer, M., Deutschman, C. S., Seymour, C. W., Shankar-Hari, M., Annane, D., Bauer, M., et al. (2016). The third international consensus definitions for Sepsis and septic shock (Sepsis-3). JAMA 315, 801–810. doi: 10.1001/jama.2016.0287
Sun, Z., Liu, D., Wang, X., and Pan, X. (2019). Prognostic value of serum microRNA-130a combined with APACHE II score for predicting death in septic patients with thrombocytopenia. Chinese J. Hyg. Rescue (Electronic Ed.) 5, 315–329. doi: 10.3877/cma.j.issn.2095-9133.2019.06.002
Teggert, A., Datta, H., and Ali, Z. (2020). Biomarkers for point-of-care diagnosis of sepsis. Micromachines 11:286. doi: 10.3390/mi11030286
Venkatesh, B., Finfer, S., Cohen, J., Rajbhandari, D., Arabi, Y., Bellomo, R., et al. (2018). Adjunctive glucocorticoid therapy in patients with septic shock. N. Engl. J. Med. 378, 797–808. doi: 10.1056/NEJMoa1705835
Virga, F., Cappellesso, F., Stijlemans, B., Henze, A. T., Trotta, R., Van Audenaerde, J., et al. (2021). Macrophage miR-210 induction and metabolic reprogramming in response to pathogen interaction boost life-threatening inflammation. Sci. Adv. 7:eabf0466. doi: 10.1126/sciadv.abf0466
Wang, Q., Feng, Q., Zhang, Y., Zhou, S., and Chen, H. (2020). Decreased microRNA 103 and microRNA 107 predict increased risks of acute respiratory distress syndrome and 28-day mortality in sepsis patients. Medicine 99:e20729. doi: 10.1097/MD.0000000000020729
Wang, D., and Han, L. (2021). Downregulation of miR-1184 serves as a diagnostic biomarker in neonatal sepsis and regulates LPS-induced inflammatory response by inhibiting IL-16 in monocytes. Exp. Ther. Med. 21:350. doi: 10.3892/etm.2021.9781
Wang, C., Liang, G., Shen, J., Kong, H., Wu, D., Huang, J., et al. (2021). Long non-coding RNAs as biomarkers and therapeutic targets in Sepsis. Front. Immunol. 12:722004. doi: 10.3389/fimmu.2021.722004
Wang, L., Tian, X., Fu, G., Xiao, F., and Li, L. (2020). Relationship between expressions of miR-205 and HMGB1 in serum of patients with sepsis and its significance. J. Clin. Emerg. (China) 21, 287–191. doi: 10.16781/j.0258-879x.2020.02.0135
Wang, H., Wang, L., and Guo, D. F. (2020). Correlation analysis of plasma microRNA-146a with risk, inflammatory cytokine levels and prognosis in sepsis patients. Acad. J. Second Mil. Univ. 41, 135–140.
Wang, X., Wang, X., Liu, X., Wang, X., Xu, J., Hou, S., et al. (2015). miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. Int. J. Clin. Exp. Med. 8, 5683–5690
Wang, H., Xiao, H., and Chen, X. (2021). Value of combined detection of plasma miR-223-3p, PCT, IL-6 and CRP levels in experimental diagnosis and prognosis of sepsis. J. Mod. Lab. Med. 36, 51–54+61. doi: 10.1590/1678-4685-gmb-2020-0009
Wang, C., Xu, R., Zeng, Y., Zhao, Y., and Hu, X. (2022). A comparison of qSOFA, SIRS and NEWS in predicting the accuracy of mortality in patients with suspected sepsis: a meta-analysis. PLoS One 17:e0266755. doi: 10.1371/journal.pone.0266755
Wang, J., Yan, X., Wang, H., and Xu, T. (2020). Evaluation vaule of serum microRNA-25 on severity and prognosis of sepsis patients. J. Clin. Emerg. (China) 21, 493–498.
Wang, H., Zhang, P., Chen, W., Feng, D., Jia, Y., and Xie, L. (2012). Serum microRNA signatures identified by Solexa sequencing predict sepsis patients' mortality: a prospective observational study. PLoS One 7:e38885. doi: 10.1371/journal.pone.0038885
Wang, H., Zhang, C., Zhang, C., Wang, Y., Zhai, K., and Tong, Z. (2022). MicroRNA-122-5p regulates coagulation and inflammation through MASP1 and HO-1 genes. Infect. Genet. Evol. 100:105268. doi: 10.1016/j.meegid.2022.105268
Whiting, P. F., Rutjes, A. W., Westwood, M. E., Mallett, S., Deeks, J. J., Reitsma, J. B., et al. (2011). QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 155, 529–536. doi: 10.7326/0003-4819-155-8-201110180-00009
Xiang, G., Li, Q., Lian, D., Su, C., Li, X., Deng, S., et al. (2024). FOXO1-mediated autophagy regulation by miR-223 in sepsis-induced immunosuppression. Front. Pharmacol. 15:1469286. doi: 10.3389/fphar.2024.1469286
Xu, J., Feng, Y., Jeyaram, A., Jay, S. M., Zou, L., and Chao, W. (2018). Circulating plasma extracellular vesicles from septic mice induce inflammation via MicroRNA- and TLR7-dependent mechanisms. J. Immunol. (Baltimore, Md: 1950) 201, 3392–3400. doi: 10.4049/jimmunol.1801008
Xu, Z., Xie, X., Zhu, Y., Zhou, Z., and Lei, Z. (2020). Relationship between the expression of serum miR-125b and miR-142-3p and the prognosis of sepsis and its predictive value. Chinese J. Difficult Complicated Cases 19, 714–718. doi: 10.3969/j.issn.1671-6450.2020.07.016
Xu, C., Zhou, G., Wang, X., Zhang, B., Zhao, T., and Wu, L. (2021). Correlation analysis of serum miR-21 and miR-210 with hs-CRP, TNF-alpha, IL-6, and ICAM-1 in patients with sepsis after burns. Burns 48, 633–638. doi: 10.1016/j.burns.2021.05.026
Xue, Y., Xue, X., and He, B. (2019). Diagnostic and predictive value of micro RNA-133a and micro RNA-499a-5p in patients with sepsis induced cardiomyopathy. Int. J. Anesthesiol. Resuscitat. 40, 756–764. doi: 10.3760/cma.j.issn.1673-4378.2019.08.011
Yang, J., Liao, Y., Dai, Y., Hu, L., and Cai, Y. (2022). Prediction of prognosis in sepsis patients by the SOFA score combined with miR-150. Adv. Clin. Exp. Med. 31, 9–15. doi: 10.17219/acem/142536
Yang, X., Wu, Z., Liu, J., Zhou, Y., Liu, J., and Chen, Z. (2021). Combination of miR-10a and miR-21 to predict mortality of elderly patients with severe sepsis. Chinese J. Immunol. 37, 850–855. doi: 10.3969/j.issn.1000-484X.2021.07.015
Yang, W., Wu, H., Zhang, H., Liu, H., Wei, Y., and Shi, B. (2015). Prognostic value of Picco monitoring combined with plasma microRNA-150 detection in septic shock patients. J. Zhejiang Univ. (Med. Sci.) 44, 659–664. doi: 10.3785/j.issn.1008-9292.2015.11.10
Yang, J., Xu, Y., and Xie, X. (2020). Changes and significance of miR-146a and miR-223 in peripheral blood of children with sepsis. Shandong Med. J. 60, 57–60. doi: 10.1002/jcla.23509
Yang, Y., Yang, L., Liu, Z., Wang, Y., and Yang, J. (2020). Long noncoding RNA NEAT 1 and its target microRNA-125a in sepsis: correlation with acute respiratory distress syndrome risk, biochemical indexes, disease severity, and 28-day mortality. J. Clin. Lab. Anal. 34:e23509. doi: 10.1002/jcla.23509
Yang, W., Zhang, H., Liu, H., Liu, C., Ye, D., Shi, B., et al. (2016). Change rule and significance of platelet microparticles and microRNA-155 level in patients with sepsis. Chongqing Med. 45:622. doi: 10.3969/j.issn.1671-8348.2016.05.014
Yao, L., Liu, Z., Zhu, J., Li, B., Chai, C., and Tian, Y. (2015). Clinical evaluation of circulating microRNA-25 level change in sepsis and its potential relationship with oxidative stress. Int. J. Clin. Exp. Pathol. 8, 7675–7684
Yao, J., Lui, K. Y., Hu, X., Liu, E., Zhang, T., Tong, L., et al. (2021). Circulating microRNAs as novel diagnostic biomarkers and prognostic predictors for septic patients. Infect. Genet. Evol. 95:105082. doi: 10.1016/j.meegid.2021.105082
Yao, Y., Yao, S., Zou, H., Yu, L., and Yao, F. (2023). Relationship between levels o fmiR-122 and miR-146a in peripheral blood and coagulation functionin sepsis patients and their predictive value for prognosis. Chin. J. Nosocom. 33, 2246–2250. doi: 10.11816/cn.ni.2023-230002
Yu, L., Zhang, L., Zhang, X., and Li, L. (2022). Correlation of serum miR-206 and miR-451 levels with disease severity and prognosis in elderly patients with Sepsis. J. Mod. Lab. Med. 37, 96–99. doi: 10.3969/j.issn.1671-7414.2022.03.020
Zhang, C. (2018). Expression levels of microRNA-132 and high-mobility group box-1 protein in peripheral blood mononuclear cells of children with sepsis. Lab. Med. 33, 815–818. doi: 10.3969/j.issn.1673-8640.2018.09.010
Zhang, C., Li, X., Liu, N., and Zhang, C. (2022). Relationship between microRNA-96 and inflammatory response in neonatal sepsis. Int. J. Lab. Med. 43, 2131–2440. doi: 10.3969/j.issn.1673-4130.2022.17.017
Zhang, R., Li, Q., and Su, L. (2024). Expression and study of miR-125b and miR-150 in peripheral blood of patients with sepsis. Chin. J. Emerg. Resusc. Disast. Med. 19, 194–198.
Zhang, H., Wang, J., and Sun, X. (2018). Value of serum levels of microRNA-155 and IL-35 in the assessment of condition and prognosis of patients with sepsis. Shandong Med. J. 58, 12–16. doi: 10.3969/j.issn.1002-266X.2018.29.004
Zhang, Y., Wang, Q., Yang, Z., Liu, H., and Wang, X. (2023). The relationship between serum miR-92a-3p, miR-147 and severity of illness, inflammatory response in patients with sepsis and their value for prognosis. Int. J. Lab. Med. 44, 1541–1546. doi: 10.3969/j.issn.1673-4130.2023.13.002
Zhang, K., Zhang, X., Ding, W., Xuan, N., Tian, B., Huang, T., et al. (2021). National Early Warning Score Does not Accurately Predict Mortality for patients with infection outside the intensive care unit: a systematic review and Meta-analysis. Front. Med. 8:704358. doi: 10.3389/fmed.2021.704358
Zhao, D., Li, S., Cui, J., Wang, L., Ma, X., and Li, Y. (2020). Plasma miR-125a and miR-125b in sepsis: correlation with disease risk, inflammation, severity, and prognosis. J. Clin. Lab. Anal. 34:e23036. doi: 10.1002/jcla.23036
Zhao, Y., Xu, Z., Wu, M., and Gu, W. (2022). Expression of serum miR-133a in sepsis patients with acute respiratory distress syndrome and its relationship with prognosis. Chin. J. Lung Dis. (Electronic Ed.) 15, 38–41. doi: 10.3877/cma.j.issn.1674-6902.2022.01.009
Zheng, X., Zhang, Y., Lin, S., Li, Y., Hua, Y., and Zhou, K. (2023). Diagnostic significance of microRNAs in sepsis. PLoS One 18:e0279726. doi: 10.1371/journal.pone.0279726
Zhu, X. (2020). MiR-125b but not miR-125a is upregulated and exhibits a trend to correlate with enhanced disease severity, inflammation, and increased mortality in sepsis patients. J. Clin. Lab. Anal. 34:e23094. doi: 10.1002/jcla.23094
CRP – C-reactive protein
PCT – Procalcitonin
miRNAs – microRNAs
CI – Confidence interval
SROC – Summary receiver operating characteristic curves value
AUC – Area under the curve
qSOFA – Quick sequential organ failure assessment score
NEWS – National early warning score
PRISMA – Preferred reporting items for systematic reviews and meta-analyses
CNKI – China national knowledge infrastructure
CENTRAL – Cochrane central register of controlled trials
ROC – Receiver operating characteristic
TP – True positive false positive
FP – False positive
FN – False negative
TN – True negative
QUADAS – Quality assessment of diagnostic accuracy studies
DOR – Diagnostic odds ratio
I2 – Inconsistency index
TSA – Trial sequential analysis
RIS – Required information size
PBMCs – Peripheral blood mononuclear cells
SIRS – Systemic inflammatory response syndrome
TmiRs – Total mixed miRNAs
OR – Odds ratio
ESS – Effective sample size
HC – Healthy control
N/R – Not report
SE – Standard error
Keywords: sepsis, miRNA, mortality, prognosis, meta-analysis
Citation: Jin Y, Zhang Y, Li Y and Zheng X (2025) Significances of miRNAs for predicting sepsis mortality: a meta-analysis. Front. Microbiol. 16:1472124. doi: 10.3389/fmicb.2025.1472124
Received: 28 July 2024; Accepted: 19 February 2025;
Published: 11 March 2025.
Edited by:
Sabreena Safuan, Universiti Sains Malaysia Health Campus, MalaysiaReviewed by:
Hamid Reza Jahantigh, Emory University, United StatesCopyright © 2025 Jin, Zhang, Li and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolan Zheng, NDE4ODI3MjAwQHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.