- 1Department of Bacterial Molecular Genetics, Faculty of Biology, University of Gdańsk, Gdańsk, Poland
- 2Department of Neuroscience, Uppsala University, Uppsala, Sweden
Introduction: The stringent response is one of the fundamental mechanisms that control and modulate bacterial adaptation to stress conditions, such as nutrient limitation. The accumulation of stringent response effectors, (p)ppGpp, causes differential expression of approximately 500 genes, including genes of bacterial endogenous toxin-antitoxin (TA) systems. However, the exact link between (p)ppGpp and toxin-antitoxin systems’ activation, as well as toxin-antitoxin role in stress adaptation remains disputed.
Methods: In this study, we performed a complex analysis of changes (RNA-Seq) in the toxin-antitoxin operons’ transcription in response to nitrogen, fatty acid, or carbon starvation, in bacteria with different abilities of (p)ppGpp accumulation.
Results and discussion: Although we observed that in some cases (p)ppGpp accumulation appears to be crucial for transcriptional activation of TA genes (e.g., ghoST, ryeA), our data indicates that the general pattern of chromosomally encoded TA gene expression in E. coli differs depending on the nutrient distribution in the environment, regardless of the alarmone accumulation.
Introduction
The availability of nutrients is a key element regulating the cell’s entire metabolism; thus, the deficiency of nutrients activates specific adaptation mechanisms. The stringent response is one of the fundamental mechanisms that control and modulate adaptation to stress conditions in bacteria. The stringent response effectors are specific nucleotides, guanosine tetraphosphate and guanosine pentaphosphate, collectively abbreviated as (p)ppGpp. These nucleotides, in E. coli, are synthesized by RelA and SpoT proteins using two different pathways, where RelA produces (p)ppGpp in response to the presence of uncharged tRNA in the ribosomal A-site, during amino acid starvation, or in response to pyruvate depletion during fatty acid starvation (Kushwaha et al., 2019; Sinha et al., 2019). On the other hand, SpoT is responsible for accumulation of (p)ppGpp in response to glucose or fatty acid starvation and several other stress conditions (Potrykus and Cashel, 2008). Moreover, SpoT also acts as a (p)ppGpp hydrolase (Potrykus and Cashel, 2008).
The (p)ppGpp nucleotides, acting as cellular alarmones, regulate various crucial cellular processes such as: transcription, translation, and initiation of DNA replication, directly and indirectly. In E. coli (p)ppGpp exerts its effect mainly by binding to the core subunits of the RNA polymerase to modulate its transcriptional properties, affecting in this way numerous processes (Irving et al., 2021). Accumulation of (p)ppGpp causes differential expression of approximately 500 genes (Costanzo and Ades, 2006; Traxler et al., 2008, 2011), including toxin-antitoxin (TA) encoding genes (Christensen-Dalsgaard et al., 2010; Shan et al., 2017; LeRoux et al., 2020). Beside RelA/SpoT homologs (RSH), many bacteria encode additional enzymes engaged in alarmone biosynthesis and hydrolysis, namely small alarmone synthetases (SAS) and small alarmone hydrolases (SAH), which makes the model of (p)ppGpp-mediated regulation of transcription even more complicated (Potrykus and Cashel, 2018). Surprisingly, in 2020 it has even been proposed that some of the small alarmone synthetases (SAS) from Cellulomonas marina can itself act as a toxic part of TA systems (Jimmy et al., 2020).
Since the mid-80s TA systems were considered to be linked, along with the (p)ppGpp alarmone, to bacterial persistent phenotype (Pacios et al., 2020; Brown, 2019; Niu, 2024). Persister cells are a subpopulation of bacteria that are nongrowing, dormant bacteria exhibiting transient high levels of tolerance to antibiotics (Pacios et al., 2020). However, a few years ago the hypothesis about TA systems’ role in persister cell formation was challenged (Van Melderen and Wood, 2017; Pacios et al., 2020; Brown, 2019; Niu, 2024). Additionally, although numerous studies have reported TA operons’ transcriptional activation in response to diverse stress conditions (Christensen-Dalsgaard et al., 2010; Muthuramalingam et al., 2016; Shan et al., 2017; Ronneau and Helaine, 2019; LeRoux et al., 2020), this was not necessarily accompanied by detection of a given toxin activity (LeRoux et al., 2020). Therefore, the link between the stringent response and TA systems’ activity remains elusive (Fraikin et al., 2019; LeRoux et al., 2020; Sanchez-Torres and Wood, 2024).
To shed some light on the linkage between (p)ppGpp and transcriptional regulation of toxin-antitoxin systems, we performed a complex RNA-seq analysis of E. coli bearing relA and spoT mutations, cultured under different limited nutritional conditions. We noted expression of 28 TA operons from 35 experimentally validated and one bioinformatically predicted in E. coli K-12 MG1655 strain (Guan et al., 2024). We find that, among them the expression of 20 TA operons is not significantly changed (−2 > log2FD > 2), or statistically insignificant, whereas several TA genes are differentially expressed in response to carbon or fatty acid starvation in the tested strains. According to our data (p)ppGpp appears to affect expression of several TA systems (ghoST, relBE, ryeA, yefM-yoeB, chpBS, topAi-yjhQ), while the expression of other TA genes (tisB, hokB, mqsR) seems to be independent of the alarmone accumulation. Therefore, we propose that alterations in intracellular (p)ppGpp levels cannot be considered as the main element of transcriptional regulation of all TA systems.
Materials and methods
Bacterial growth and RNA-Seq
All experiments were conducted using the Escherichia coli K-12 MG1655 strain bearing a single relA deletion (ΔrelA), double deletion of the relA and spoT genes (ΔrelA ΔspoT), or ΔrelA spoT203 mutations (Xiao et al., 1991; Jishage et al., 2002; Potrykus et al., 2011). Bacteria were grown to the early logarithmic phase in M9 medium supplemented with glucose and casamino acids as carbon sources. Upon reaching OD600 = 0.2, carbon, nitrogen, and fatty acid starvation were induced. To induce carbon and nitrogen stress, cells were centrifuged at 4,000 × g for 10 min and then resuspended in fresh medium lacking carbon sources (glucose and casamino acids). For nitrogen starvation, following centrifugation, bacteria were cultured in a nitrogen-limited minimal medium without (NH4)2SO4 as the nitrogen source. Fatty acids’ stress was induced by adding triclosan (2 μg/mL). RNA-seq samples were collected after 1.5 cell generations. A volume of 2.5 mL from each culture was collected and mixed with 5% AquaPhenol in 96% ethanol. The samples were then rapidly frozen in liquid nitrogen and stored at −80°C. For RNA extraction, the frozen samples were thawed on ice and centrifuged at 4,000 × g for 5 min. The cell pellet was resuspended in 100 μL of Tris-EDTA (TE) buffer (10 mM Tris-Cl, 1 mM EDTA, pH 8.0) containing lysozyme (1 mg/mL) and incubated at room temperature for 10 min. Subsequent purification was performed using the RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer’s instructions. To ensure complete removal of DNA, a DNAse I (Qiagen) treatment was applied during the column-based purification process. RNA was eluted in 50 μL of RNase-free water, and the purified RNA samples were stored at −80°C until analysis. Quality was checked by employing Bioanalyzer 2100. The sequencing runs were conducted on the Illumina NovaSeq6000 platform. Thirty million pair-end reads per sample were assessed with 101 pb read length.
RNA-Seq processing and analysis
Raw fastq files were processed with fastp software using default parameters (Chen et al., 2018). Reads were aligned to the Escherichia coli reference genome NC_000913.3 using Rsubread v2.6.4 package (Liao et al., 2019). In total, 99.5% of reads were uniquely mapped to the reference genome. Resulting BAM files were summarized into counts with Rsubread v2.6.4. Differential gene expression (DE) analysis was performed with DESeq2 package v1.44.0 (Love et al., 2014). Adjusted p value for multiple testing was calculated with Benjamini-Hochberg correction. If not stated otherwise, significant DE genes were selected with |log2FC| ≥ 1 and adjusted p value < 0.05 thresholds. Analysis using median average deviation (MAD; Chung et al., 2008) used median ± 3*MAD. GO terms and KEGG pathways were analyzed with clusterProfiler package v 4.12.6 (Yu et al., 2012). Heat maps were generated with pheatmap package v1.0.12 and volcano plots were generated with EnhancedVolcano package v1.22.0. In all analyses R version 4.4.1 was used.
Results and discussion
No significant changes in TA transcription were observed during nitrogen starvation regardless of the (p)ppGpp intracellular level
To test the effect of different intracellular (p)ppGpp level on TA operons’ transcriptional regulation, we assessed the whole transcriptome of three E. coli strains. Bacteria bearing double ΔrelA ΔspoT mutations (so called ppGpp0 or null strain) are unable to synthesize (p)ppGpp under any conditions; the ΔrelA mutant, which bears an active SpoT and a deletion of the relA gene, allows for the complete elimination of the effects caused by amino acid starvation; and ΔrelA spoT203 is a strain with physiologically approximately sevenfold higher basal (p)ppGpp level when compared to the wild type strain (Sarubbi et al., 1988). Surprisingly, we find that there are no significant differences (−2 < log2FD < 2) in TA transcription in all tested strains cultured under nitrogen limiting conditions irrespectively of the bacterial ability of (p)ppGpp accumulation (Figure 1).
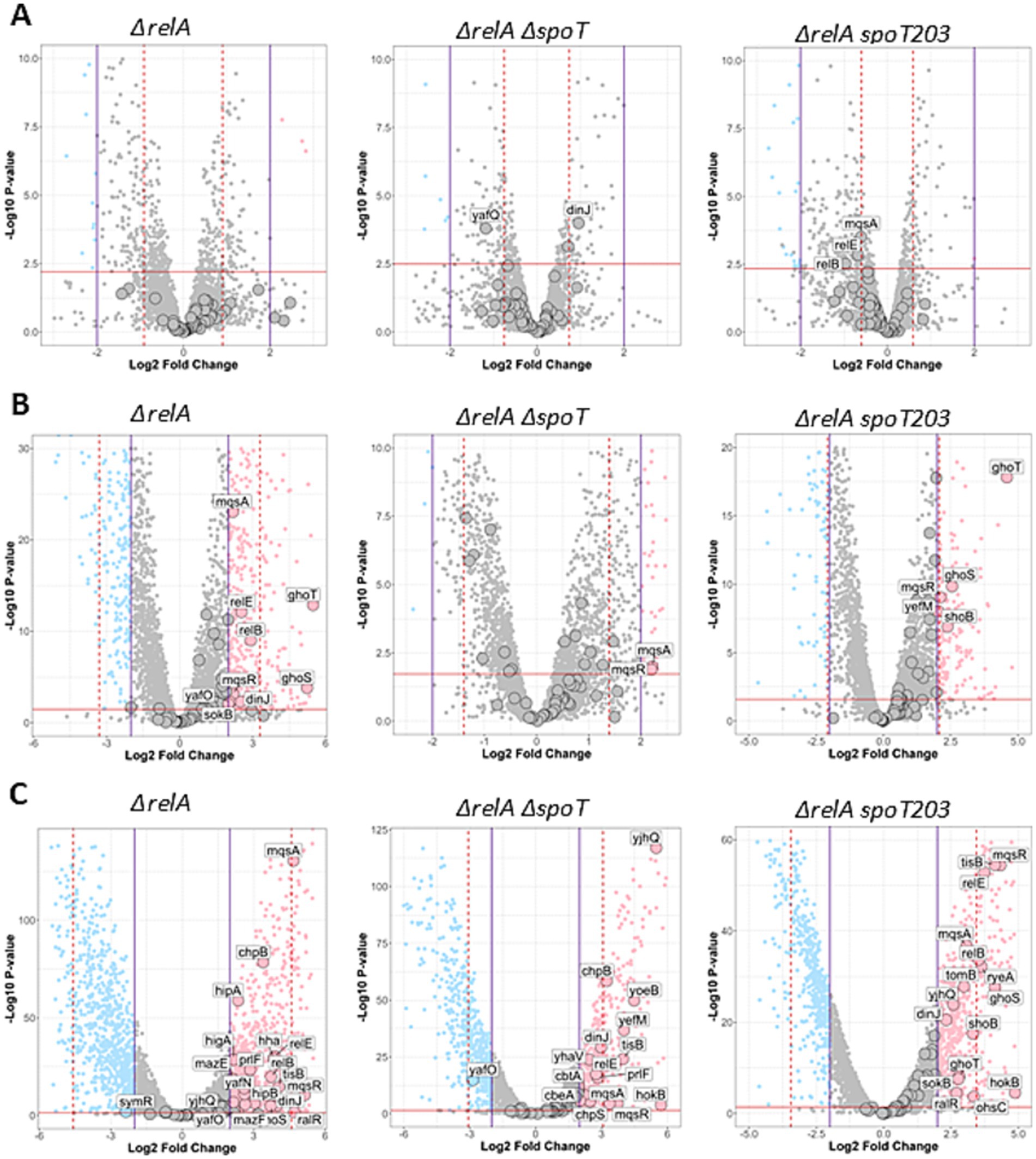
Figure 1. Overview of gene expression changes in ΔrelA, ΔrelA ΔspoT, and ΔrelA spoT203 mutants upon nutritional stress. Stress conditions: nitrogen (A), fatty acid deprivation (B), and carbohydrate deprivation (NH4+) deprivation (C) compared to no stress (normal). Volcano plots illustrate log2 fold change (x-axis) and statistical significance of changes (y-axis) as calculated by DESeq2 package. Horizontal dashed red line indicates p value corresponding to adjusted p value = 0.05. Purple vertical lines indicate casual log2FC = 2 threshold and red dashed vertical lines indicate MAD thresholds calculated for each comparison. Genes which are significantly up or downregulated (|log2FC| >2 and adj. p < 0.05) are colored in pink and blue respectively, whereas genes colored in gray are not significant. TA genes are indicated by large points. Labels are shown only for significantly altered TA genes.
The mqsRA and ghoST operons are differentially expressed in bacteria with varying SpoT activity during fatty acid starvation
Induction of fatty acid starvation significantly affected the transcription of several TA modules in tested bacteria, including mqsRA, ghoST, hokB-sokB, relBE, dinJ-yafQ, tomB-hha in bacteria bearing ΔrelA mutation, mqsRA in ppGpp0 strain, and mqsRA, ghoST, yefM-yoeB and shoB in ΔrelA ΔspoT203 double mutant (Figures 1, 2). Among those, the expression pattern of two TA modules was particularly interesting.
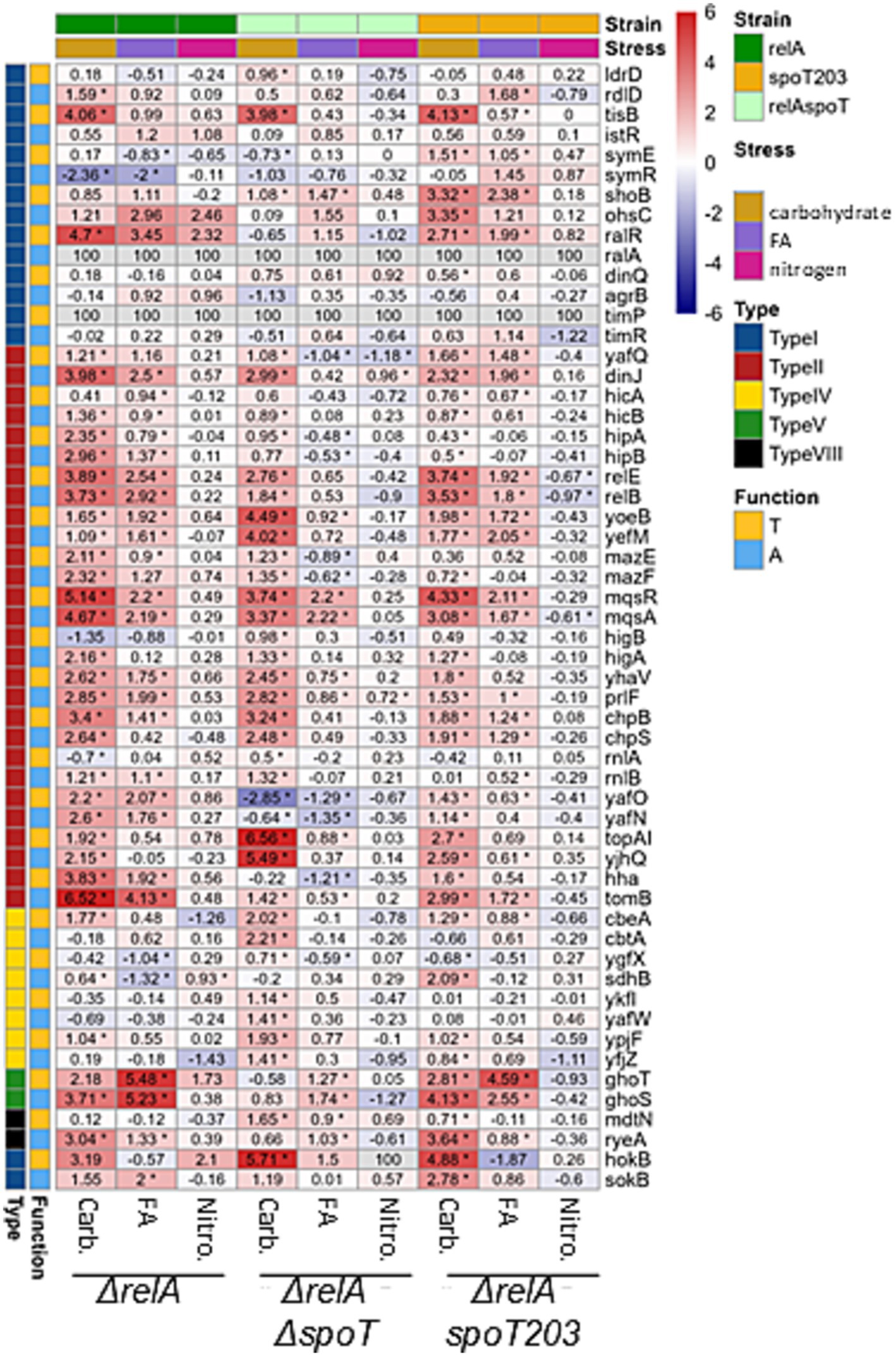
Figure 2. Changes in Toxin and Antitoxin Transcript Levels Due to Nutritional Stress. The heatmap illustrates the log2 fold change (log2FC) in transcript levels of toxin-antitoxin (TA) systems in cells subjected to various nutritional stresses for 1.5 generations, as assessed by Illumina NGS RNA-seq. Stress conditions: carbohydrate deprivation (Carb.), fatty acid deprivation (FA), and nitrogen (NH4+) deprivation (Nitro.) compared to no stress (normal). Mutant Strains: ΔrelA; ΔrelA, ΔspoT (ppGpp null); ΔrelA, spoT203 (ppGpp high). Top Sidebar: Indicates the stress condition and mutant strain and left Sidebar: Indicates TA type and gene function (antitoxin or toxin). Color scale represents the exact log2FC values for the comparison of stress vs. normal conditions. Log2FC values for genes, mutants and conditions are provided in cells. Asterisk indicates statistical significance evaluated by DESeq2, adjusted p < 0.05. Small RNAs ralA and timA were not annotated in the genome version used in this study thus data for them were not available (n.a.).
Firstly, the expression of toxin-encoding gene mqsR, which was significantly upregulated (log2FC > 2) in all tested strains growing under fatty acid limiting conditions (Figures 1, 2), suggesting that its transcriptional regulation can be independent of (p)ppGpp accumulation. Since toxin and antitoxin transcription is usually coupled, often the upregulation of toxin is accompanied by increased antitoxin expression. However, in the strain with elevated intracellular alarmone level (ΔrelA spoT203), we noted significant mqsR gene upregulation (log2FC = 2.11) and only minor changes in mqsA transcription (log2FC = 1.67) (Figures 1, 2). This could suggest MqsR toxin activation via toxin mRNA enrichment and, as a result, unbalanced T:A intracellular ratio. Still, not without controversies, MqsA has been shown to possess a role in repression of the general stress response and curli/biofilm formation, while MqsR was shown to play a role in bile-acid stress reduction, biofilm formation, phage defense and regulation of type V ghoST TA system (Sanchez-Torres and Wood, 2024). In the proposed model, in the presence of MqsR, the ghoS-encoding 5′ region of the ghoST transcript is degraded, thereby enriching for the ghoT-encoding 3′ region (Wang et al., 2013). Interestingly, besides changes in the mqsRA genes expression, we also observed upregulation of ghoST system in the ΔrelA and ΔrelA spoT203 strains. Curiously, we noted a distinctive pattern of ghoST expression in bacteria with (p)ppGpp excess (ΔrelA spoT203): the GhoT toxin transcription level was almost twice as high as GhoS antitoxin (log2FC 4.59 vs. 2.55) (Figures 1, 2). This indirectly confirms the MqsR-mediated mechanism of post-transcriptional regulation of GhoT activity under stress conditions.
To sum up, we noted upregulation of the whole mqsRA system in ΔrelA and ppGpp0 strains, and imbalance in MqsR toxin and MqsA antitoxin expression in ΔrelA spoT203, during fatty acid starvation. Elevated mqsR transcription was accompanied by unbalanced ghoS and ghoT transcription only in ΔrelA spoT203 strain. Therefore, we suppose that although the upregulation of mqsR gene appears to be independent of (p)ppGpp accumulation, the activation of MqsR toxin and further regulation of ghoST expression require high intracellular (p)ppGpp level.
The mqsRA operon is negatively autoregulated by an antitoxin binding directly to its promoter region, and toxin and antitoxin encoding genes are co-transcribed (Brown et al., 2013). Thus, the unbalanced toxin and antitoxin mRNA level noted in this study appears to be enigmatic and requires further investigation, since there is a possibility of additional post-transcriptional regulation of mqsRA, e.g., by RNase-mediated degradation of mqsA-encoding mRNA during fatty acid starvation. In the recently proposed model, stress induces conformational changes in nascent and zinc-free MqsA, resulting in exposure of the ClpX recognition motif for ClpXP-mediated degradation of the antitoxin, leading to MqsR activation (Vos et al., 2022). We believe that the MqsR toxin activation may require both elevated (p)ppGpp level and other concomitant factors, such as MqsA degradation by protease and/or depletion of mqsA encoding mRNA.
SpoT activity affects transcriptional regulation of some but not all TA systems under carbon starvation
After induction of carbon starvation, we observed significant changes (log2FC >3) in expression of several TA modules (Figures 1, 2), some of which were previously indicated as upregulated in response to stress conditions (mqsRA, relBE, yefM-yoeB, hokB, tisB) (Christensen et al., 2001; Wang et al., 2013; Janssen et al., 2015; Verstraeten et al., 2015; Su et al., 2022; Vos et al., 2022). In all tested strains, irrespective of SpoT activity, we noted tisB, hokB and mqsRA upregulation (Figures 1, 2), suggesting that the transcriptional activation of these genes is (p)ppGpp independent, at least under tested conditions.
One interesting observation was that the expression of hokB was negligible in all but carbohydrate stress conditions (Figures 1, 2). HokB is a toxin of a type I TA system, in which antitoxin is represented by a small antisense RNA (SokB) that binds the toxin mRNA and inhibits its translation. In the case of type I TA systems, transcriptional activation of only the toxin-encoding gene probably means that there are not enough antitoxin molecules to inhibit toxin translation, leading to its activation. Some data suggest that HokB activation may lead to persister cell formation (Yokoyama et al., 2023; Verstraeten et al., 2015). One of the proposed mechanisms of HokB-mediated persistence was based on the activity of the Obg protein that interacts with SpoT and stimulates (p)ppGpp synthetase activity of SpoT. According to this model, an elevated alarmone level leads to enhanced expression of the pore-forming HokB and, as a result, to a collapse of the membrane potential causing ATP leakage associated with persistence (Verstraeten et al., 2015). In fact, some data suggest that (p)ppGpp induction during carbon starvation affects bacterial persistence formation in the presence of ciprofloxacin (Svenningsen et al., 2021). However, we observed hokB upregulation in all tested bacterial strains in response to carbon starvation, including the ppGpp0 strain (log2FC = 5.71) (Figures 1, 2), which suggests that hokB transcriptional activation can be independent of (p)ppGpp intracellular level. Moreover, some studies claim that it is highly unlikely that this single gene would be the central element in the mechanism of persistent phenotype manifestation (Song and Wood, 2020). Especially since the hok-sok loci occasionally degenerate and appear to be non-functional in some bacterial strains (Fozo et al., 2010). Therefore, we suppose that the upregulation of hokB gene and possible toxin activation during growth in carbon limited environment is rather a secondary effect of more complexed bacterial response to nutritional limitation, than a conserved part of a specific pattern of bacterial response governed by (p)ppGpp under these environmental conditions.
One the other hand, recently novel biological functions were proposed for another TA system upregulated in our experiment. Namely, it appears that tisB-istR participates in blocking the import of host and/or microbial-produced genotoxic compounds, allowing for increased survival in the mammalian gut (Su et al., 2022). Moreover, it has been demonstrated that TisB toxin is involved in bacterial response to high doses of ofloxacin, even though it is not required for ofloxacin inhibitory effect (Cayron et al., 2024). Therefore, the subject of TA system influence on bacterial cell physiology remains elusive and should be addressed carefully.
Besides tisB, hokB, and mqsRA, only three TA systems were highly upregulated (log2FC > 3) after induction of carbon starvation in the ppGpp0 strain: topAI-yjhQ, yefM-yoeB, and chpBS. The toxin of the topAI-yjhQ system inhibits topoisomerase I activity (Yamaguchi and Inouye, 2015), while YoeB and ChpB toxins are endoribonucleases that probably affect ribosome biosynthesis (Culviner et al., 2020). However, it has been claimed that the upregulation of TA operons is not necessarily accompanied by the corresponding toxin activity (LeRoux et al., 2020), thus further studies are needed to make any definitive conclusions here. The relBE operon as well as ghoS and ryeA antitoxins were upregulated both in the ΔrelA and ΔrelA spoT203 bacterial strains, but not in the ppGpp0 strain. Therefore, we conclude that transcriptional activation of relBE, ghoST, and ryeA under carbon starvation conditions requires (p)ppGpp synthesis. RyeA is a novel type VIII antitoxin (Choi et al., 2018) and has been shown to be most strongly expressed in minimal medium during exponential growth (Gupta et al., 2019) and upon acid exposure (Gupta et al., 2020). Recently, it has been demonstrated that during acid stress ryeA-sdsR system expression is controlled by a third factor—the GcvB small RNA (Ibrahim et al., 2024). It cannot be excluded that GcvB stabilizes the RyeA antitoxin also during carbon starvation and, since we detected no ryeA upregulation in (p)ppGpp deficient bacteria, that this protective effect is dependent on (p)ppGpp.
Since relBE system is encoded within DNA of the Qin cryptic prophage, its genetic localization suggests that this system’s activity could be connected to Qin DNA stabilization (Wang and Wood, 2016). Interestingly, we observed upregulation of some genes associated with the Qin prophage, including the ydfD gene (log2FC = 4.01 in ΔrelA and log2FC = 2.01 in ppGpp0 strain growing under fatty acids starvation, data not shown), encoding phage lysis protein (Lu et al., 2022). However, this observation seems to be devoid of correlation with the relBE expression, since no changes in relBE expression were observed in these conditions (Figures 1, 2).
To summarize, we observed that transcriptional activation of some TA genes during nutrient limitation seems to be independent of (p)ppGpp availability (tisB, hokB, mqsR), while the upregulation of others requires (p)ppGpp induction (e.g., ghoST, relBE, ryeA). In 2020 it has been claimed that although (p)ppGpp is sufficient to induce TA transcription, it was not essential for this induction (LeRoux et al., 2020). Moreover, it has been stated that the transcriptional activation of TA operons is not necessarily accompanied by a corresponding toxin activity (LeRoux et al., 2020). Our data agrees with these observations. We believe that nutritional stress or (p)ppGpp accumulation cannot be considered as universal factors inducing gene transcription of all TA systems resident within bacterial genome. Furthermore, we are convinced that transcriptional activation of a TA operon does not automatically lead to toxin activation. However, we suppose that in some cases (p)ppGpp induction can indirectly lead to alterations in the toxin and antitoxin ratio. The activity of a toxin can be the effect of imbalanced equilibrium of toxin and antitoxin molecules. This could be the result of an intensified toxin biosynthesis (e.g., due to enrichment in toxin mRNA), antitoxin preferential degradation by cellular proteases or both factors (reviewed in Boss and Kędzierska, 2023). In plasmid-encoded TA systems the post-segregational killing phenomenon (PSK) has been proven to exist (Fraikin and Van Melderen, 2024) as a result of uneven plasmid distribution into bacterial daughter cells (Figure 3A). In the case of chromosomal type II TA modules, it has been assumed frequently that since the TAs’ promoters are repressed by free antitoxin or TA complexes, if a toxin is activated, there should be less antitoxin and higher transcriptional activity of the whole operon (Christensen-Dalsgaard et al., 2010; Shan et al., 2017; LeRoux et al., 2020). In our opinion, that would be the moment when the toxin and antitoxin molecules distribution returns to the equilibrium state. Therefore, we think that to elucidate the mechanism of toxin activation, it is worth searching for conditions that trigger TA modules’ downregulation, since this would be the situation analogous to plasmid loss (Figure 3B). Nonetheless, we did not observe significant downregulation of any TA genes in tested bacteria during nutrient limitation (log2FC < −3, Figure 2). Therefore, this subject requires further research.
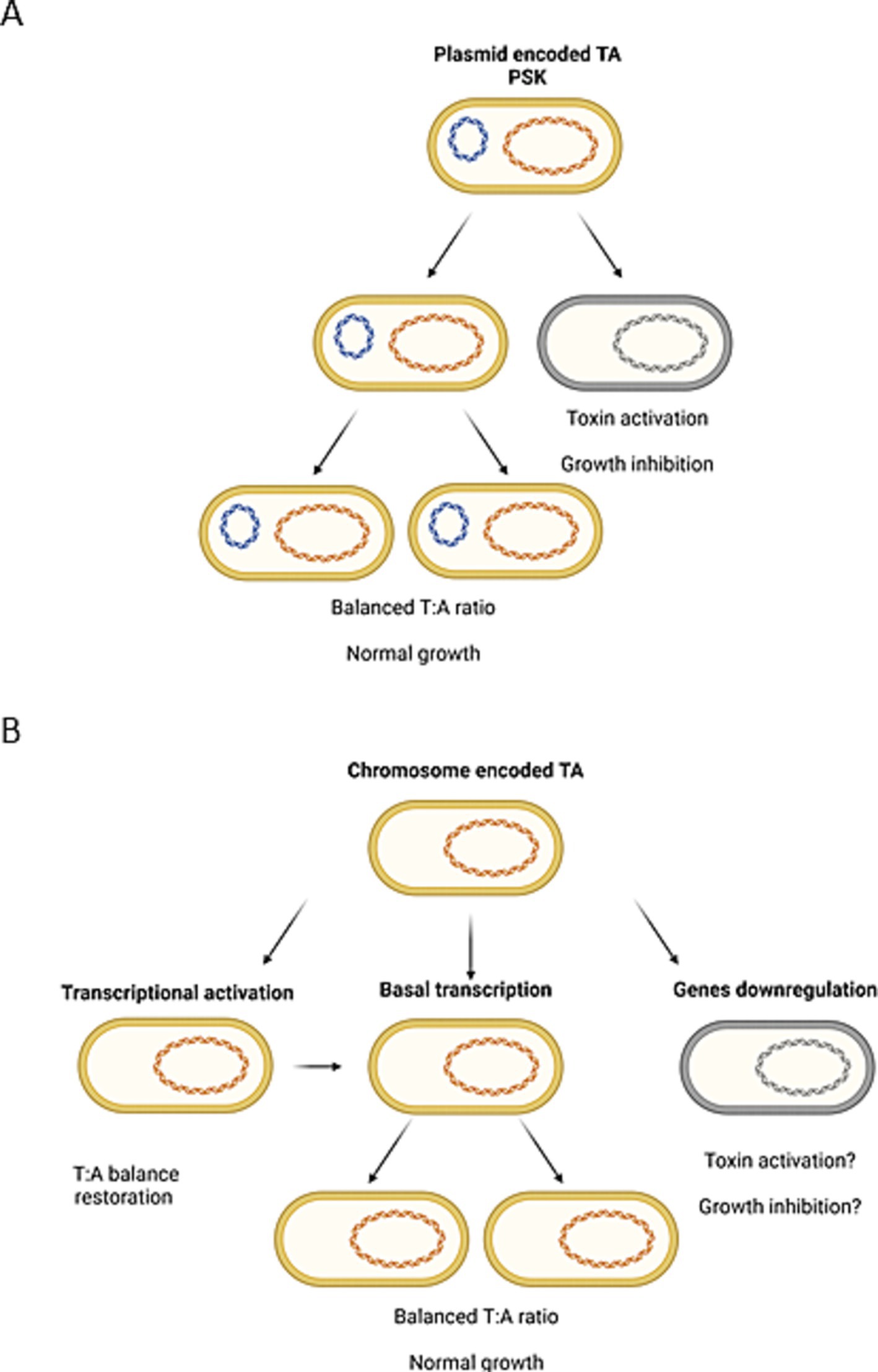
Figure 3. Illustration of the possible analogy between (A) the generally accepted mechanism of bacteria killing by plasmid-encoded toxin activation due to post-segregational killing (PSK) mechanism and (B) differential expression of chromosomally encoded TA systems. Created with Biorender.com.
Overall, our data indicates that the general pattern of E. coli chromosomally encoded TA genes expression differs depending on the nutrients’ distribution in the environment, regardless of (p)ppGpp presence. The greatest number of TA genes is considerably changed (upregulated) in all tested strains after induction of carbon starvation, while the effect of fatty acid or nitrogen starvation is much less spectacular. It is worth keeping in mind that the regulation of gene expression covers both transcription and translation control. In 2018 it has been demonstrated that bacterial cells regulate these processes differently depending on which nutrient is being limited. For example, it has been shown that carbon starvation mostly affects the rate of transcription that is coupled to translation and thus slows down both processes. On the other hand, nitrogen starvation slows down translation rate independently of transcription, and translation seems to be uncoupled from transcription. Interestingly, it appears that during growth under nitrogen limited conditions, bacteria regulate translation rate by an unknown (p)ppGpp-independent mechanism (Iyer et al., 2018; Li et al., 2018; Potrykus and Cashel, 2018). Both situations described above seem to be reflected in our data. By contrast, generally TA systems are rather upregulated than downregulated when bacteria are cultured under nutrient limited conditions. However, it appears that modulation of TA gene expression in response to carbon starvation relates to modification of transcription level, while only a few TA modules were upregulated fatty acid limitation and no significant changes in TA transcription were observed after induction of nitrogen starvation. Furthermore, the role of (p)ppGpp in regulation of TA gene expression during nitrogen starvation was rather minor. Therefore, we hypothesize that under these conditions most of TA genes are either unaffected or regulated at the translation level, as proposed by Li et al. for other genes (Li et al., 2018).
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
MM-D: Conceptualization, Formal analysis, Project administration, Resources, Supervision, Validation, Writing – review & editing, Data curation, Funding acquisition, Investigation, Methodology. PO: Formal analysis, Methodology, Writing – review & editing, Visualization. MK: Formal analysis, Methodology, Visualization, Writing – review & editing. LB: Conceptualization, Formal analysis, Project administration, Resources, Supervision, Validation, Writing – review & editing, Visualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Science Center, Poland (grant Sonata UMO-2016/23/D/NZ1/02601 to MM-D). The APC was funded by University of Gdańsk.
Acknowledgments
We are grateful to Joanna Morcinek-Orłowska for advice on figures and to Katarzyna Potrykus for critical reading of the manuscript and helpful advice.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Boss, L., and Kędzierska, B. (2023). Bacterial toxin-antitoxin systems’ cross-interactions—implications for practical use in medicine and biotechnology | enhanced reader. Toxins (Basel) 15:380. doi: 10.3390/toxins15060380
Brown, D. R. (2019). Nitrogen starvation induces persister cell formation in Escherichia coli. J. Bacteriol. 201:e00622-18. doi: 10.1128/JB.00622-18
Brown, B. L., Lord, D. M., Grigoriu, S., Peti, W., and Page, R. (2013). The Escherichia coli toxin MqsR destabilizes the transcriptional repression complex formed between the antitoxin MqsA and the mqsRA operon promoter. J. Biol. Chem. 288, 1286–1294. doi: 10.1074/jbc.M112.421008
Cayron, J., Oms, T., Schlechtweg, T., Zedek, S., and Van Melderen, L. (2024). TisB protein is the single molecular determinant underlying multiple downstream effects of ofloxacin in Escherichia coli. Sci. Adv. 10:eadk1577. doi: 10.1126/SCIADV.ADK1577
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Choi, J. S., Kim, W., Suk, S., Park, H., Bak, G., Yoon, J., et al. (2018). The small RNA, SdsR, acts as a novel type of toxin in Escherichia coli. RNA Biol. 15, 1319–1335. doi: 10.1080/15476286.2018.1532252
Christensen, S. K., Mikkelsen, M., Pedersen, K., and Gerdes, K. (2001). RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98, 14328–14333. doi: 10.1073/PNAS.251327898
Christensen-Dalsgaard, M., Jørgensen, M. G., and Gerdes, K. (2010). Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75, 333–348. doi: 10.1111/j.1365-2958.2009.06969.x
Chung, N., Zhang, X. D., Kreamer, A., Locco, L., Kuan, P.-F., Bartz, S., et al. (2008). Median absolute deviation to improve hit selection for genome-scale RNAi screens. J. Biomol. Screen. 13, 149–158. doi: 10.1177/1087057107312035
Costanzo, A., and Ades, S. E. (2006). Growth phase-dependent regulation of the extracytoplasmic stress factor, σE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 188, 4627–4634. doi: 10.1128/JB.01981-05
Culviner, P. H., Guegler, C. K., and Laub, M. T. (2020). A Simple, Cost-Effective, and Robust Method for rRNA Depletion in RNA-Sequencing Studies. mBio 11, e00010–20. doi: 10.1128/mBio.00010-20
Fozo, E. M., Makarova, K. S., Shabalina, S. A., Yutin, N., Koonin, E. V., and Storz, G. (2010). Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 38, 3743–3759. doi: 10.1093/nar/gkq054
Fraikin, N., Rousseau, C. J., Goeders, N., and Van Melderen, L. (2019). Reassessing the role of the type II MqsRA toxin-antitoxin system in stress response and biofilm formation: MqsA is transcriptionally uncoupled from mqsR. MBio 10, 1–13. doi: 10.1128/mBio.02678-19
Fraikin, N., and Van Melderen, L. (2024). Single-cell evidence for plasmid addiction mediated by toxin-antitoxin systems. Nucleic Acids Res. 52, 1847–1859. doi: 10.1093/nar/gkae018
Guan, J., Chen, Y., Goh, Y. X., Wang, M., Tai, C., Deng, Z., et al. (2024). TADB 3.0: an updated database of bacterial toxin–antitoxin loci and associated mobile genetic elements. Nucleic Acids Res. 52, D784–D790. doi: 10.1093/NAR/GKAD962
Gupta, A. K., Siddiqui, N., and Dutta, T. (2020). A novel mechanism of RyeA/SraC induction under acid stress. Biochem. Biophys. Res. Commun. 525, 298–302. doi: 10.1016/j.bbrc.2020.02.085
Gupta, A. K., Siddiqui, N., Yadav, D., Arora, L., and Dutta, T. (2019). Regulation of RyeA/SraC expression in Escherichia coli. Biochem. Biophys. Res. Commun. 516, 661–665. doi: 10.1016/j.bbrc.2019.06.110
Ibrahim, A., Begum, A., and Dutta, T. (2024). Regulation of an RNA toxin-antitoxin system, SdsR-RyeA, by a small RNA GcvB. Biochem. Biophys. Res. Commun. 733:150688. doi: 10.1016/j.bbrc.2024.150688
Irving, S. E., Choudhury, N. R., and Corrigan, R. M. (2021). The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 19, 256–271. doi: 10.1038/s41579-020-00470-y
Iyer, S., Le, D., Park, B. R., and Kim, M. (2018). Distinct mechanisms coordinate transcription and translation under carbon and nitrogen starvation in Escherichia coli. Nat. Microbiol. 3, 741–748. doi: 10.1038/s41564-018-0161-3
Janssen, B. D., Garza-Sánchez, F., and Hayes, C. S. (2015). YoeB toxin is activated during thermal stress. Microbiology 4, 682–697. doi: 10.1002/MBO3.272
Jimmy, S., Saha, C. K., Kurata, T., Stavropoulos, C., Oliveira, S. R. A., Koh, A., et al. (2020). A widespread toxin−antitoxin system exploiting growth control via alarmone signaling. Proc. Natl. Acad. Sci. USA 117, 10500–10510. doi: 10.1073/pnas.1916617117
Jishage, M., Kvint, K., Shingler, V., and Nyström, T. (2002). Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16, 1260–1270. doi: 10.1101/gad.227902
Kushwaha, G. S., Bange, G., and Bhavesh, N. S. (2019). Interaction studies on bacterial stringent response protein RelA with uncharged tRNA provide evidence for its prerequisite complex for ribosome binding. Curr. Genet. 65, 1173–1184. doi: 10.1007/s00294-019-00966-y
LeRoux, M., Culviner, P. H., Liu, Y. J., Littlehale, M. L., and Laub, M. T. (2020). Stress can induce transcription of toxin-antitoxin systems without activating toxin. Mol. Cell 79, 280–292.e8. doi: 10.1016/j.molcel.2020.05.028
Li, S. H.-J., Li, Z., Park, J. O., King, C. G., Rabinowitz, J. D., Wingreen, N. S., et al. (2018). Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat. Microbiol. 3, 939–947. doi: 10.1038/s41564-018-0199-2
Liao, Y., Smyth, G. K., and Shi, W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47:e47. doi: 10.1093/nar/gkz114
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, Z., Wang, B., Qiu, Z., Zhang, R., Zheng, J., and Jia, Z. (2022). YdfD, a lysis protein of the Qin prophage, is a specific inhibitor of the IspG-catalyzed step in the MEP pathway of Escherichia coli. Int. J. Mol. Sci. 23:1560. doi: 10.3390/ijms23031560
Muthuramalingam, M., White, J. C., and Bourne, C. R. (2016). Toxin-antitoxin modules are pliable switches activated by multiple protease pathways. Toxins (Basel) 8, 1–16. doi: 10.3390/toxins8070214
Niu, H., Gu, J., and Zhang, Y. (2024). Bacterial persisters: molecular mechanisms and therapeutic development. Signal Transduct. Target. Ther. 9:174. doi: 10.1038/s41392-024-01866-5
Pacios, O., Blasco, L., Bleriot, I., Fernandez-Garcia, L., Ambroa, A., López, M., et al. (2020). (p)ppGpp and its role in bacterial persistence: new challenges. Antimicrob. Agents Chemother. 64, 1–14. doi: 10.1128/AAC.01283-20
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Ann. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Potrykus, K., and Cashel, M. (2018). Growth at best and worst of times. Nat. Microbiol. 3, 862–863. doi: 10.1038/s41564-018-0207-6
Potrykus, K., Murphy, H., Philippe, N., and Cashel, M. (2011). ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 13, 563–575. doi: 10.1111/j.1462-2920.2010.02357.x
Ronneau, S., and Helaine, S. (2019). Clarifying the link between toxin–antitoxin modules and bacterial persistence. J. Mol. Biol. 431, 3462–3471. doi: 10.1016/j.jmb.2019.03.019
Sanchez-Torres, V., and Wood, T. K. (2024). Diverse physiological roles of the MqsR / MqsA toxin / antitoxin system. Sustain. Microbiol. 1:qvae006. doi: 10.1093/sumbio/qvae006
Shan, Y., Brown Gandt, A., Rowe, S. E., Deisinger, J. P., Conlon, B. P., Lewis, K., et al. (2017). ATPdependent persister formation in Escherichia coli. MBio 8, 1–14. doi: 10.1128/mBio.02267-16
Sarubbi, E., Rudd, KE., and Cashel, M. (1988). Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet 213, 214–22. doi: 10.1007/BF00339584
Sinha, A. K., Winther, K. S., Roghanian, M., and Gerdes, K. (2019). Fatty acid starvation activates RelA by depleting lysine precursor pyruvate. Mol. Microbiol. 112, 1339–1349. doi: 10.1111/mmi.14366
Song, S., and Wood, T. K. (2020). Toxin/antitoxin system paradigms: toxins bound to antitoxins are not likely activated by preferential antitoxin degradation. Adv. Biosyst. 4, e1900290–e1900295. doi: 10.1002/adbi.201900290
Su, W. L., Bredèche, M. F., Dion, S., Dauverd, J., Condamine, B., Gutierrez, A., et al. (2022). TisB protein protects Escherichia coli cells suffering massive DNA damage from environmental toxic compounds. MBio 13:e0038522. doi: 10.1128/MBIO.00385-22
Svenningsen, M. S., Svenningsen, S. L., Sørensen, M. A., and Mitarai, N. (2021). Existence of log-phase Escherichia coli persisters and lasting memory of a starvation pulse. Life Sci. Alliance 5:e202101076. doi: 10.26508/LSA.202101076
Traxler, M. F., Summers, S. M., Nguyen, H.-T., Zacharia, V. M., Hightower, G. A., Smith, J. T., et al. (2008). The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x
Traxler, M. F., Zacharia, V. M., Marquardt, S., Summers, S. M., Nguyen, H.-T., Stark, S. E., et al. (2011). Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the “feast to famine” gradient in Escherichia coli. Mol. Microbiol. 79, 830–845. doi: 10.1111/j.1365-2958.2010.07498.x
Van Melderen, L., and Wood, T. K. (2017). Commentary: what is the link between stringent response, endoribonuclease encoding type II toxin-antitoxin systems and persistence? Front. Microbiol. 8:191. doi: 10.3389/fmicb.2017.00191
Verstraeten, N., Knapen, J., Kint, I., Jansen, M., Fauvart, M., and Correspondence, J. M. (2015). Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 59, 9–21. doi: 10.1016/j.molcel.2015.05.011
Vos, M. R., Piraino, B., Labreck, C. J., Rahmani, N., Trebino, C. E., Schoenle, M., et al. (2022). Degradation of the E. coli antitoxin MqsA by the proteolytic complex ClpXP is regulated by zinc occupancy and oxidation. J. Biol. Chem. 298:101557. doi: 10.1016/j.jbc.2021.101557
Wang, X., Lord, D. M., Hong, S. H., Peti, W., Benedik, M. J., Page, R., et al. (2013). Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 15, 1734–1744. doi: 10.1111/1462-2920.12063
Wang, X., and Wood, T. K. (2016). Cryptic prophages as targets for drug development. Drug Resist. Updat. 27, 30–38. doi: 10.1016/j.drup.2016.06.001
Xiao, H., Kalman, M., Ikehara, K., Zemel, S., Glaser, G., and Cashel, M. (1991). Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990. doi: 10.1016/s0021-9258(19)67694-5
Yamaguchi, Y., and Inouye, M. (2015). An endogenous protein inhibitor, YjhX (TopAI), for topoisomerase I from Escherichia coli. Nucleic Acids Res. 43, gkv1197–gkv10396. doi: 10.1093/NAR/GKV1197
Yokoyama, T., Yamagata, Y., Honna, S., Mizuno, S., Katagiri, S., Oi, R., et al. (2023). S2P intramembrane protease RseP degrades small membrane proteins and suppresses the cytotoxicity of intrinsic toxin HokB. mBio 14:e0108623. doi: 10.1128/mbio.01086-23
Keywords: toxin-antitoxin, relA, spoT203, ppGpp, RNA-seq, hokB, mqsRA, ghoST
Citation: Maciąg-Dorszyńska M, Olszewski P, Karczewska M and Boss L (2025) Toxin-antitoxin genes are differentially expressed in Escherichia coli relA and spoT mutans cultured under nitrogen, fatty acid, or carbon starvation conditions. Front. Microbiol. 15:1528825. doi: 10.3389/fmicb.2024.1528825
Edited by:
Ulrike Mäder, University Medicine Greifswald, GermanyReviewed by:
Michael Benedik, Texas A&M University, United StatesAbdul Basit, University of Jhang, Pakistan
Copyright © 2025 Maciąg-Dorszyńska, Olszewski, Karczewska and Boss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lidia Boss, bGlkaWEuYm9zc0B1Zy5lZHUucGw=
 Monika Maciąg-Dorszyńska
Monika Maciąg-Dorszyńska Paweł Olszewski
Paweł Olszewski Monika Karczewska1
Monika Karczewska1 Lidia Boss
Lidia Boss