- 1Department of Animal Production, School of Veterinary Medicine and Sciences, University of Ngaoundere, Ngaoundere, Cameroon
- 2Department of Comparative Biomedicine and Food Science, University of Padua, Legnaro, Italy
- 3National Veterinary Laboratory, Garoua, Cameroon
Introduction: Poultry production accounts for 42% of Cameroonian meat production. However, infectious diseases represent the main hindrance in this sector, resulting in overuse and misuse of antimicrobials that can contribute to the emergence and dissemination of antimicrobial resistance (AMR). This study aimed to evaluate the prevalence of antimicrobial resistance genes (ARGs) conferring resistance to carbapenems (blaVIM-2 and blaNDM), (fluoro) quinolones (qnrS, qnrA, and qnrB), polymyxins (mcr1 to mcr5), and macrolides (ermA and ermB) in the poultry farm environment. Additionally, the study examined the relationship between these ARGs and biosecurity implementation, as well as farmers’ knowledge, attitudes, and practices toward antimicrobial use (AMU) and AMR, including their perception of AMR risk.
Materials and methods: Fecal, drinking water, and biofilm samples from drinking water pipelines were collected from 15 poultry farms and subsequently analyzed by real-time PCR and 16S rRNA NGS.
Results: All samples tested positive for genes conferring resistance to (fluoro) quinolones, 97.8% to macrolides, 64.4% to polymyxins, and 11.1% to carbapenems. Of concern, more than half of the samples (64.4%) showed a multi-drug resistance (MDR) pattern (i.e., resistance to ≥3 antimicrobial classes). Drinking water and biofilm microbial communities significantly differed from the one of the fecal samples, both in term of diversity (α-diversity) and composition (β-diversity). Furthermore, opportunistic pathogens (i.e., Comamonadaceae and Sphingomonadaceae) were among the most abundant bacteria in drinking water and biofilm. The level of biosecurity implementation was intermediate, while the knowledge and attitude of poultry farmers toward AMU were insufficient and unsuitable, respectively. Good practices toward AMU were found to be correlated with a reduction in polymyxins and MDR.
Discussion: This study provides valuable information on resistance to medically important antimicrobials in poultry production in Cameroon and highlights their potential impact on human and environmental health.
1 Introduction
The poultry industry represents a vital sector in many African countries (Ochieng et al., 2021). In Cameroon, the second highest poultry producer in West and Central Africa (Monamele et al., 2019) the sector accounts for 4% of Gross Domestic Product (GDP). Eggs and poultry represent 14% of the population protein intake (MINEPIA, 2019) and 42% of the total meat production [GIZ (Deutsche Gesellschaft fur Internationale Zusammenarbeit GmbH), 2018], respectively. However, similarly to other livestock sectors, infectious diseases represent the main hindrance in Cameroonian poultry production, leading to increased antimicrobial use (AMU) for disease prevention and control (Moffo et al., 2020). The poultry sector is the main user of antimicrobials in Cameroon (Mouiche et al., 2020). Factors such as population growth (Demography - Cameroon Statista, 2024) and rising consumer demand for poultry products, along with the transition from small-to large-scale intensive production systems (Klein et al., 2018), are driving the increased use of antimicrobials.
In poultry farming, drinking water represents the most common route for administering antimicrobial drugs (AMDs). However, drinking water and its distribution systems can be contaminated with microbiological constituents (e.g., biofilm), affecting the stability and availability of AMDs. Biofilms can capture antimicrobials, contributing to treatment failures and the spread of antimicrobial resistance (AMR) (Sparks, 2009). To complicate the scenario, the overuse and misuse of antimicrobials, including medically important antimicrobials for human medicine, in Cameroonian poultry farms, can contribute to the emergence and dissemination of AMR (Moffo et al., 2022; Vougat Ngom et al., 2024a).
AMR poses a serious public health concern all over the world (FDA, 2020), with a higher impact recorded in Africa (Sartorius et al., 2024). Resistant genes and bacteria from livestock can reach humans through direct contact (e.g., farmers), the food chain, or drainage into water basins and water supplies (Berendonk et al., 2015; Hruby et al., 2016; Alban et al., 2017; Collineau et al., 2020). Biosecurity and farmers’ education are key strategies to reduce AMU, by mitigating the risk of introduction and spread of infectious diseases and by increasing their awareness on AMDs use and AMR, respectively (Postma et al., 2017; Rodrigues Da Costa et al., 2019). Improved biosecurity can also enhance the technical performances of reared flocks (Rodrigues Da Costa et al., 2019). Many phenotypic studies in Cameroon (Djuikoue et al., 2023; Leinyuy et al., 2023) have demonstrated widespread AMR in poultry farms. However, a recent review reported a lack of molecular studies to identify resistance genes (ARGs) to medically important antimicrobials from livestock (Vougat Ngom et al., 2024a). Furthermore, only one previous study investigated the knowledge the risk perception on AMR of Cameroonian poultry farmers (Moffo et al., 2020). This gap may be due to insufficient infrastructure for molecular analyses, yet this approach is highly recommended to improve the understanding of AMR dissemination in primary production.
In this context, the present study aimed to evaluate for the first time the presence of resistance determinants to medically important antimicrobials for human medicine in feces, drinking water, and biofilm from Cameroonian poultry farms. Additionally, it investigated the relationship between the ARGs and biosecurity implementation, as well as farmers’ knowledge, attitudes, and practices regarding AMU and their knowledge and perception of AMR risk.
2 Materials and methods
2.1 Study area
This study was conducted in the Center region of Cameroon (3° 52′- 6°14’ LN and 11°31′- 12°93′ LE) in August 2023 (Figure 1). The Center region was selected due to its significant poultry production, accounting for 15.8% of the national poultry population, following the West (23.2%) and North West (18.7%) regions. It also has the highest rate of chicken meat and egg consumption in the country (MINEPIA, 2019). The study was carried out in 6 of the 10 divisions in this region: Méfou-et-Afamba, Méfou-et-Akono, Lekié, Mfoundi, Nyong-et-Mfoumou and Mbam-et-Kim. Authorization for the study was obtained from the regional delegation in charge of livestock (N 00120/L/MINEPIA.SG/DREPIA-CE).
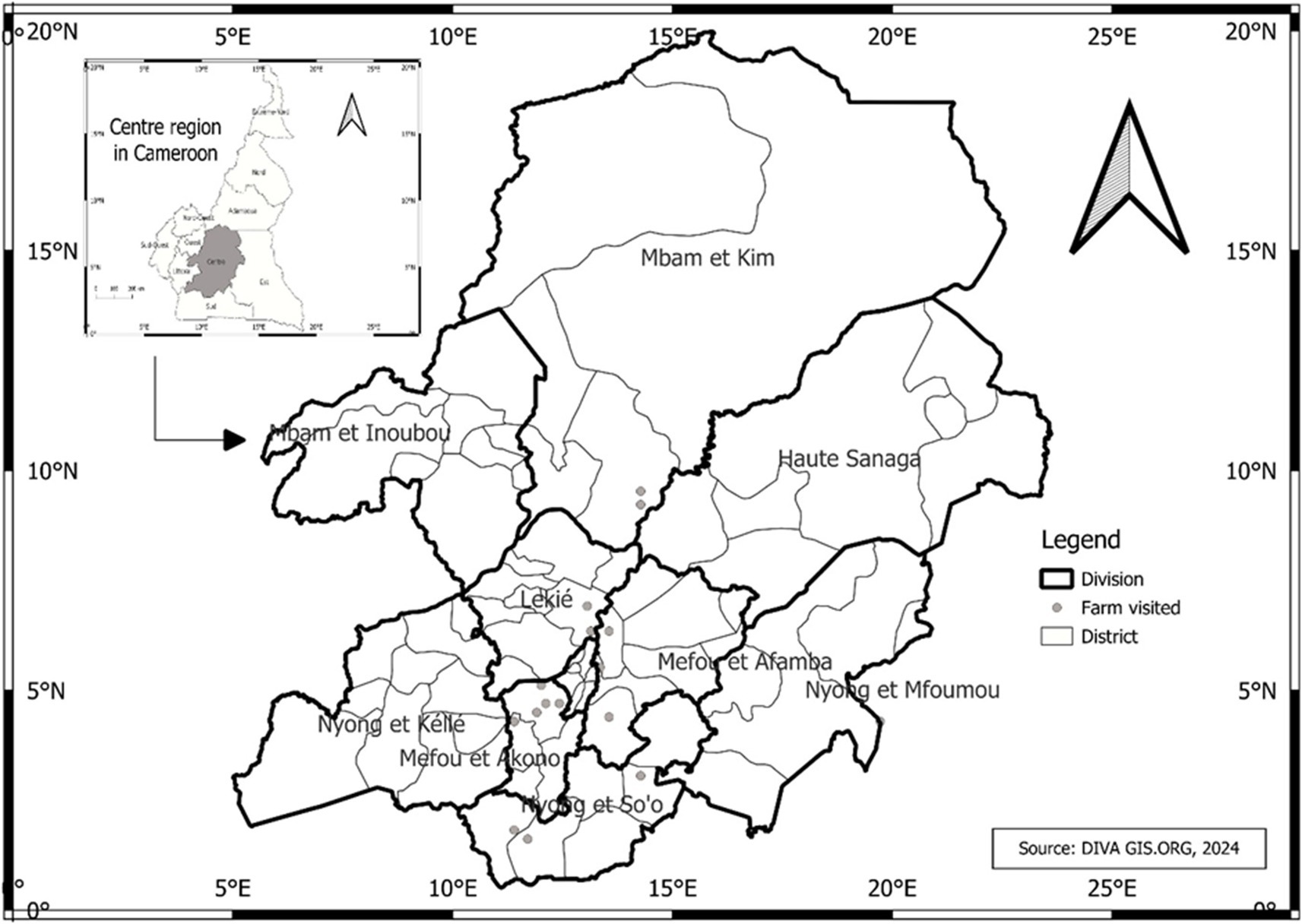
Figure 1. Map reporting the locations of farms in the Center Region of Cameroon. The poultry farms included in this study were in 6 (Méfou-et-Afamba, Méfou-et-Akono, Lekié, Mfoundi, Nyong-et-Mfoumou and Mbam-et-Kim) out of 10 divisions of the Center Region of Cameroon.
2.2 Study design and data collection
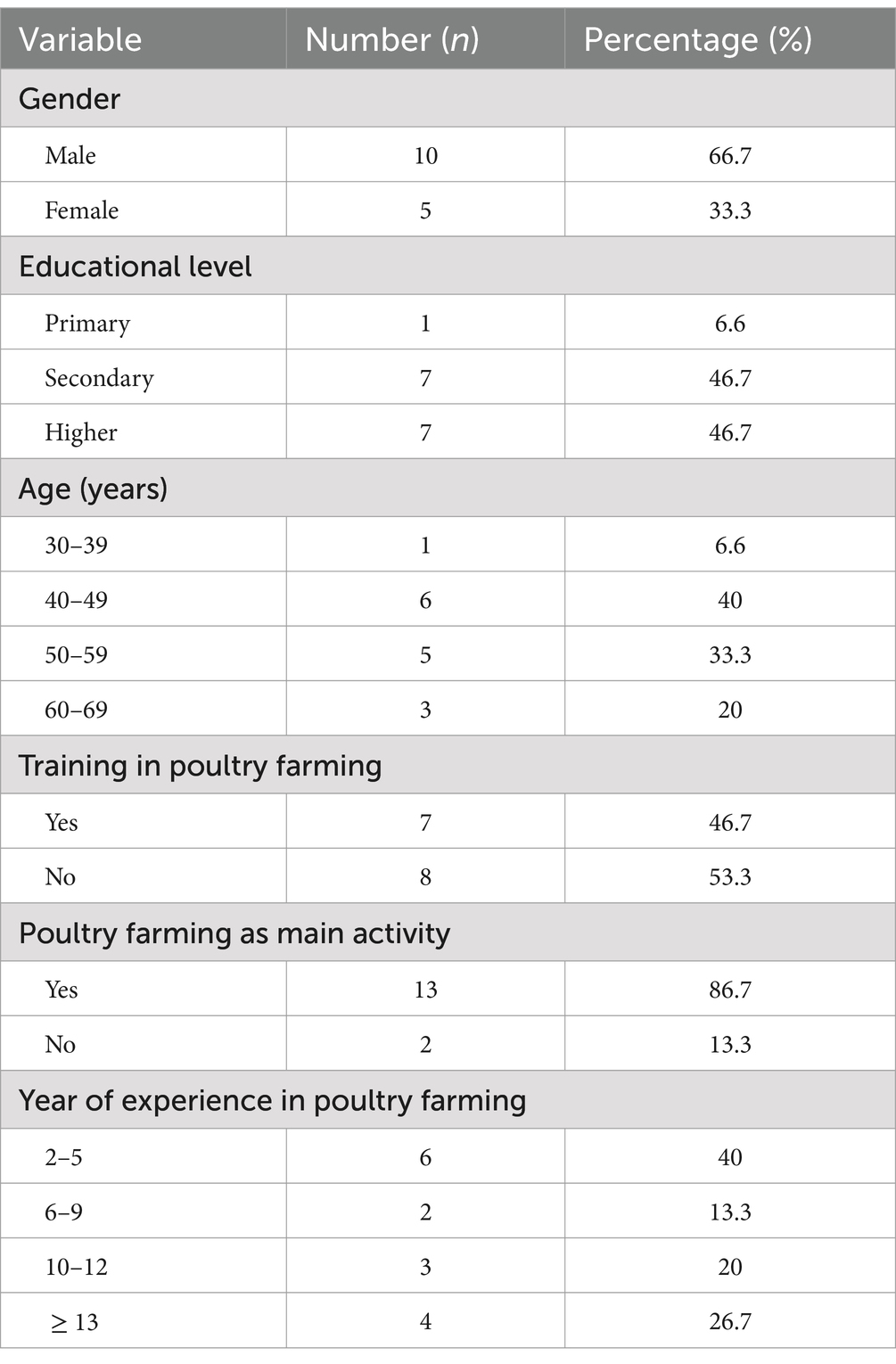
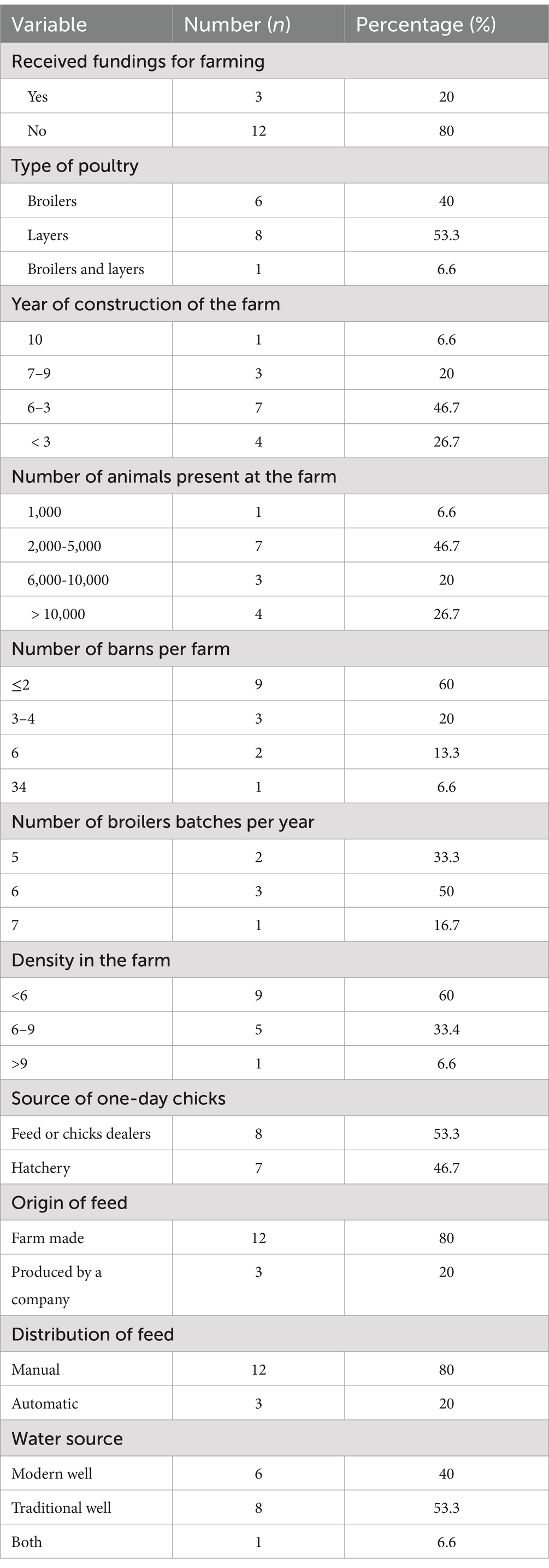
A cross-sectional survey was conducted in poultry farms using drinking water distribution systems (i.e., nipple drinkers, automatic bell water drinker, and pendular drinking system). Based on lists of poultry farmers from the division Delegations of the Ministry of Livestock, Fisheries and Animal Industries (MINEPIA), 18 poultry farmers were identified, and 15 agreed to participate. The purpose of the study was explained to the farmers before data collection. The demographics of participants are summarized in Table 1, indicating that the majority were male (66.7%), over 39 years of age (53.4%), had at least a secondary education level (66.7%), and had more than 5 years of experience in poultry farming (60.0%). Most farms were constructed before 2020 (73.3%) using the farmers’ own funds (80.0%) (Table 2). The number of chickens reared varied from 1,000 to 20,000 per cycle, with most farms (46.7%) housing between 2,000 and 5,000 birds, and having at least three barns (60.0%). Eight farms reared layers, six broilers and one both. Broiler farms reared at least five batches per year. Day-old chicks were purchased from feed (antibiotic-free) or chick sellers (53.3%) or directly from hatcheries (46.7%). Feeds were generally farm made and distributed manually. Farms sourced water from traditional (53.3%) and modern (40.0%) wells, or both (6.6%).
A questionnaire (Supplementary material 1) adapted from Moffo et al. (2020, 2022) and Chowdhury et al. (2023) was used for data collection. The questionnaire, pre-tested in five farms (not included in the paper), contained different sections related to farm characteristics, biosecurity practices, farmers’ knowledge, attitudes and practices on AMU, and knowledge and risk perception on AMR. Biosecurity (i.e., internal and external) implementation was assessed using a risk-based scoring tool (Moffo et al., 2020). Data collection involved face-to-face interviews with the farm owners or managers using a hard copy of the questionnaire, complemented by direct observations. Interviews targeted workers involved in the decision process of antimicrobial administration and were conducted by two trained final-year veterinary students under the supervision of a full researcher. Each farm visit lasted about 2 hours. Questionnaires are available upon request to the first author.
2.3 Sampling procedure
Fecal, drinking water, and biofilm samples were collected in each farm. Samples were taken from all barns if one or two were present, and from two randomly selected barns if more than two were present. At least 25 mg of fecal droppings were collected from 10 locations using sterile spatulas. Biofilm samples were collected by swabbing the inside of 10 randomly selected water lines using sterile cotton swabs, which were placed in a separate bottle after removing the wooden shaft. Swabs collected from the same farm were pooled together before DNA extraction. Two liters of drinking water were collected at the end of all water lines using sterile bottles. Samples were labeled, stored on ice, and either processed immediately (<12 h) or stored at −80°C until analyzed. To minimize disease transmission risk, a maximum of two farms per day were sampled, with strict biosecurity measures observed before (e.g., take an appointment, no visit to other farms on the same day), during (e.g., cleaning and disinfection, parking away from the farms, wear farm-specific clothing and shoes,) and after visiting each farm (e.g., cleaning and disinfection, no visit to other farms on the same day).
2.4 DNA extraction
Water samples were filtered using a vacuum pump and 0.45 μm filter membranes (Sartorius Stedim Biotech GmbH, Germany). Filters were cut into small pieces and placed in ZR BashingBeadTM Lysis Tubes with 750 μL of ZymoBIOMICS lysis solution. For fecal samples, 200 mg were directly placed in lysis tubes. Swabs were placed in ZR BashingBead™ Lysis Tube with 750 μL of ZymoBIOMICS™ lysis solution and vortexed for 30 min before removing the swabs. DNA was extracted using the DNA Miniprep Kit (ZymoBIOMICS™) according to the manufacturer’s instructions at the National Veterinary Laboratory (LANAVET) annex of Yaoundé, Cameroon. DNA extracts were shipped at controlled temperature to the Department of Comparative Biomedicine and Food Science of the University of Padua (Italy) for further analyses. DNA quantity was assessed using the Qubit dsDNA High Sensitivity kit (Thermo fisher Scientific) (Supplementary material 2). On a subset of samples (n = 10) representative of all sample-type and range of DNA concentration (i.e., low, medium, high), DNA quality was assessed using the Agilent 2,100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
2.5 16S rRNA gene amplification, sequencing, and data analysis
Microbial communities were investigated by amplifying the V3-V4 regions of the 16S rRNA gene and sequencing using next-generation sequencing (NGS). Libraries were prepared as described by Laconi et al. (2022a) and sequenced using the Illumina MiSeq sequencing platform (San Diego, California, USA) with a 2 × 300 bp paired-end approach. Data analysis and taxonomic assignment of microbial communities were performed using the DADA2 package (Callahan et al., 2016; Bolyen et al., 2019) and the SILVA-Naive Bayes sklearn trained database (Yilmaz et al., 2014), respectively, within the Quantitative Insights into Microbial Ecology 2 (QIIME2 version 2023.5). After data filtering, total sum normalization (TSS), and SquareRoot transformation, microbial communities were assessed using MicrobiomeAnalyst1. In detail, the microbial diversity within each group (α-diversity) was assessed using Shannon’s and Simpson’s indexes, while the Permutational Multivariable Analysis of Variance (PERMANOVA) based on the Bray-Curtis dissimilar measure was used to assess differences among groups (β-diversity). β-diversity was visualized using Principal Coordinate Analysis (PCoA) and Non-metric Multidimensional Scaling (NMDS), while microbial communities’ composition was investigated using heatmaps (pheatmap v1.0.12 package within Rstudio v2024.04.2). Linear discriminant analysis (LDA) effect size (LEfSe) was used to identify taxa associated with different sample matrices. Raw sequence reads are deposited in the NCBI Short Read Archive under the accession number PRJNA1123609. After the quality-filter step, removal of chimeric fragments and reads merging, a total of 396,859 reads were obtained, with 12,501 different features and an average of 8,819 sequences per individual sample.
2.6 Antimicrobial resistance genes (ARGs) detection by real-time PCR
All samples were tested for 12 ARGs, conferring resistance to carbapenems (blaVIM-2 and blaNDM), macrolides (ermA and ermB), (fluoro) quinolones (oqxA, oqxB, and qnrS), and polymyxins (mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5), using real-time PCR with PowerUp™ SYBR® Green Master Mix (Thermo Fisher Scientific) in a LightCycler®480 Roche (Roche, Basel, Switzerland) as previously described (Laconi et al., 2022b). The assays, primers pairs, melting temperatures, and positive controls are available in Supplementary material 3.
2.7 Statistical analysis
Binomial analysis assessed differences in ARGs occurrence (binary outcome variable), class-level resistance (i.e., at least one ARG per antimicrobial class) and MDR (i.e., resistance to at least three antimicrobial classes) across sample types, farms, water source types, water treatments, and AMU using Fisher’s exact test. To assess the association between the relative abundance of microbial taxa at family level and the ARGs detected at a minimum prevalence level of 10% in all samples, multivariate regression analysis was used to jointly regress the dependent variables (i.e., log-transformed relative abundances taxa) on the same independent variables (i.e., presence/absence of ARGs). Multiple logistic regression analysis was used to test for significance differences in ARGs prevalence, class-level resistance, and MDR across all the aforementioned explanatory variables, along with biosecurity scores and farmers’ knowledge, practices, AMU and AMR awareness, and risk perception regarding AMR. Descriptive statistics summarized data concerning farmers’ knowledge and risk perception on AMU and AMR. Knowledge, attitude and practices data were analyzed as described by Moffo et al. (2020). Briefly, answers of the interviewees were coded into binary outcomes, with 1 representing sufficient knowledge, desirable attitude, appropriate practice toward AMU/AMR and risk perception of AMR, and 0 represented insufficient knowledge, unsuitable attitude, inappropriate practice, and risk perception. The sum of responses recorded as binary outcome for each farmer for each individual category was divided by the total number of items in each category to obtain the average score for each (Caudell et al., 2019). Biosecurity scores were categorized using the following 3-level scoring system; “low” for scores ≤40%; “intermediate” 40% < scores <80%; and “high” scores ≥80%. Statistical analysis and data visualization were carried out in GraphPad Prism (version 10.2.3)2.
3 Results
3.1 Bacterial communities’ composition and diversity
The microbial communities’ composition of fecal, drinking water and biofilm samples was explored using 16S rRNA gene sequencing. The α-diversity, assessed at operational taxonomic unit (OTU) level and expressed using Shannon’s and Simpson’s indexes, was significantly higher in feces compared to drinking water (p = 0.0005) and biofilm (p = 0.0024), (Figures 2A,B). PERMANOVA analysis indicated significant differences in microbial communities among the three matrices (p = 0.0010). However, NMDS and PCoA graphs (Figures 2C,D) showed that biofilm and drinking water microbiota were less distant from each other (p = 0.0170) compared to fecal samples (p = 0.0010). No significant differences in α-diversity and β-diversity were observed within the sample type between farms that administered antimicrobials to birds in the 2 weeks preceding the sampling (AMU_Farms) and those that did not (No_AMU_Farms) (Supplementary material 4). The heatmap at the family level (Figure 3) clustered samples primarily by sample type rather than antimicrobials used. Indeed, two main clusters could be observed, one including the majority of fecal samples and one including mainly biofilm and drinking water samples, interspersed with one another. Lactobacillaceae and Entorobacteriaceae dominated fecal samples, while Burkholderiaceae, Xanthobacteraceae, and Comamondaceae were dominant in drinking water and biofilm. Indeed, among the six taxa significantly more abundant in fecal samples, LEfSe analysis identified Enterobacteriaceae (linear discriminant analysis (LDA) = 6.01) and Lactobacillaceae (LDA = 5.96) (Supplementary material 5). Other notable taxa associated with the fecal microbiota were Oscillospiraceae (LDA = 5.79), Ruminococcaceae (LDA = 5.60), Spiromaceae (LDA = 5.45), and Peptostreptococcaceae (LDA = 5.39). Xanthobacteraceae (LDA = 6.05), Reyranellaceae (LDA = 5.95), Comamonadaceae (LDA = 5.83), and Sphinomonadaceae (LDA = 5.64) were more abundant in drinking water, while Burkholderiaceae (LDA = 5.93), Chromobacteriaceae (LDA = 5.69), Weeksellaceae (LDA = 5.63), Lachnospiraceae (LDA = 5.45), and Oxalobacteraceae (LDA = 5.37) in biofilm.
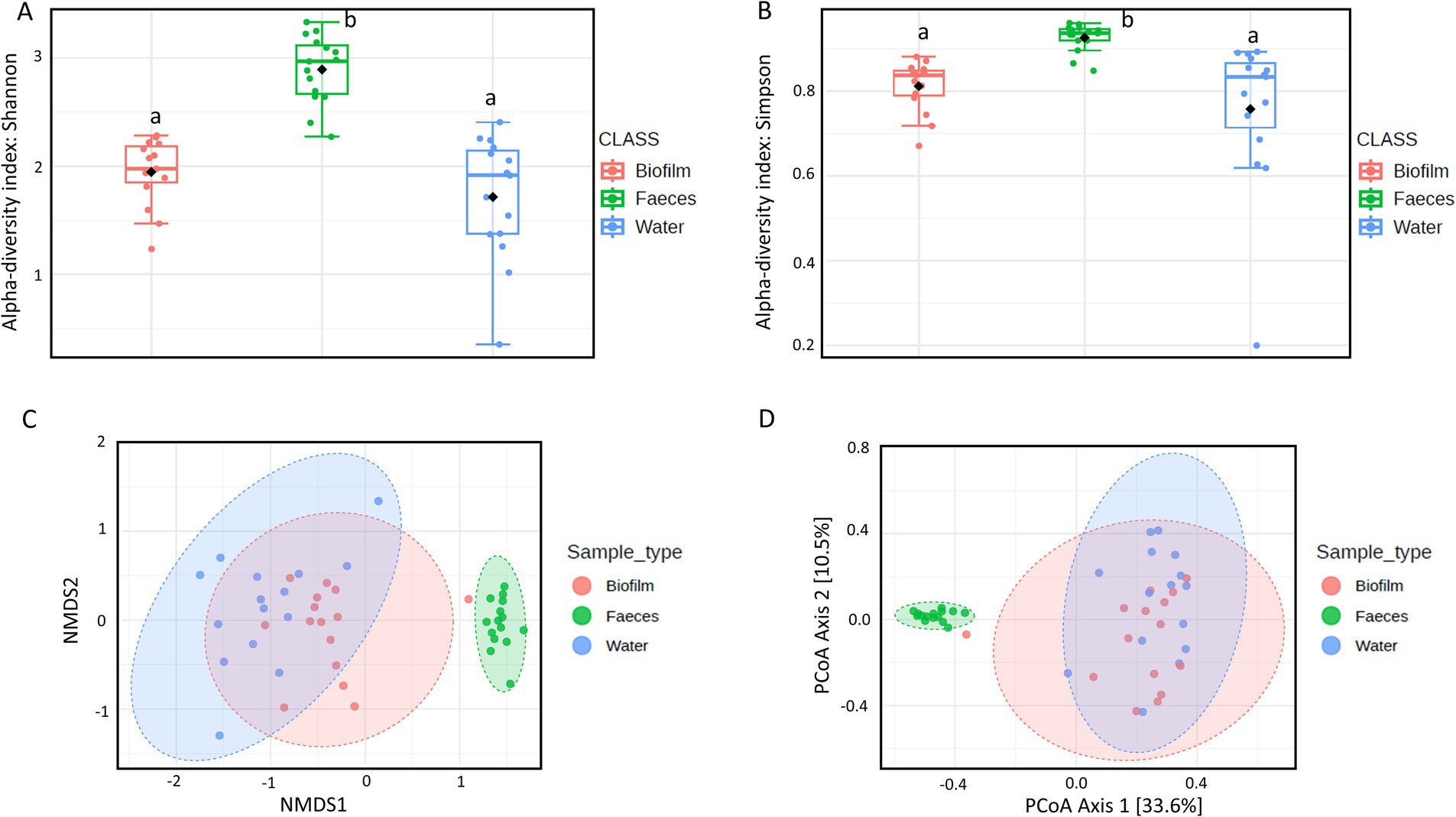
Figure 2. α-Diversity within each sample using (A) Shannon’s and (B) Simpson’s indexes. Boxplots represent 25th to 75th percentiles; different letters indicate significant differences within the α-diversity indexes (p < 0.05). β-Diversity between sample types according to Bray-Curtis distances using (C) Non-metric Multidimensional Scaling (NMDS) and (D) Principal Coordinate Analysis (PCoA).
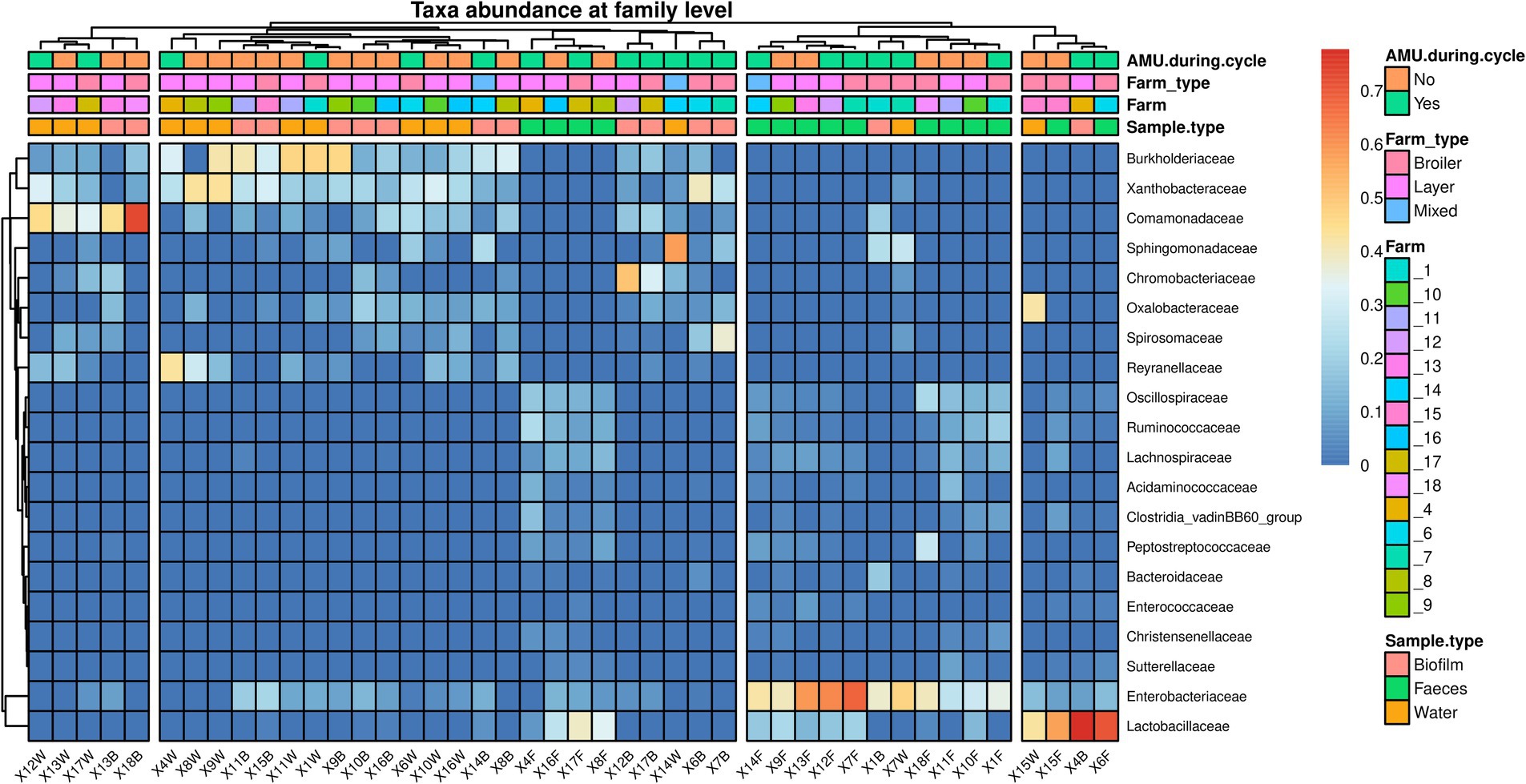
Figure 3. Heatmap representing the microbial community composition of fecal, water, and biofilm samples at family level. Each row represents a different taxon, while each column represents an individual sample. Samples are clustered according to the abundance of taxa. Taxa are displayed in a color gradient from red to blue, representing high and low abundance, respectively.
3.2 ARGs prevalence
The presence of ARGs in fecal, drinking water and biofilm samples was assessed by real-time PCR. 11 out of 12 ARGs were detected in at least one sample, with blaVIM2, encoding resistance to carbapenems, being the only one undetected. All samples tested positive for qnrS, while ermB (97.8, 95% confidence interval (CI) 93.3–100%) was the second most prevalent ARG, followed by oqxA (68.9, 95% CI 54.8–82.9%), mcr-1 (60.0, 95%CI 45.1–74.9%), ermA (20.0, 95% CI 6.0–40.4%), mcr-3 (17.8, 95% CI 6.2–29.4%), oqxB (13.3, 95% CI 3.0–23.7%), mcr-5 and blaNDM (11.1%; 95% CI 1.6–20.7%), and mcr-2 and mcr-4 (2.2, 95% CI 0–6.7%) (Figure 4A). When considering antimicrobial classes, all samples tested positive for at least one gene conferring resistance to (fluoro) quinolones, 97.8% (95% CI 93.3–100%) to macrolides, 64.4% (95% CI 49.9–79.0%) to polymyxins, and 11.1% (95% CI 1.6–20.7%) to carbapenems. Additionally, 64.4% (95% CI 49.9–79.0%) of the samples showed resistance to at least three antimicrobial classes (MDR).
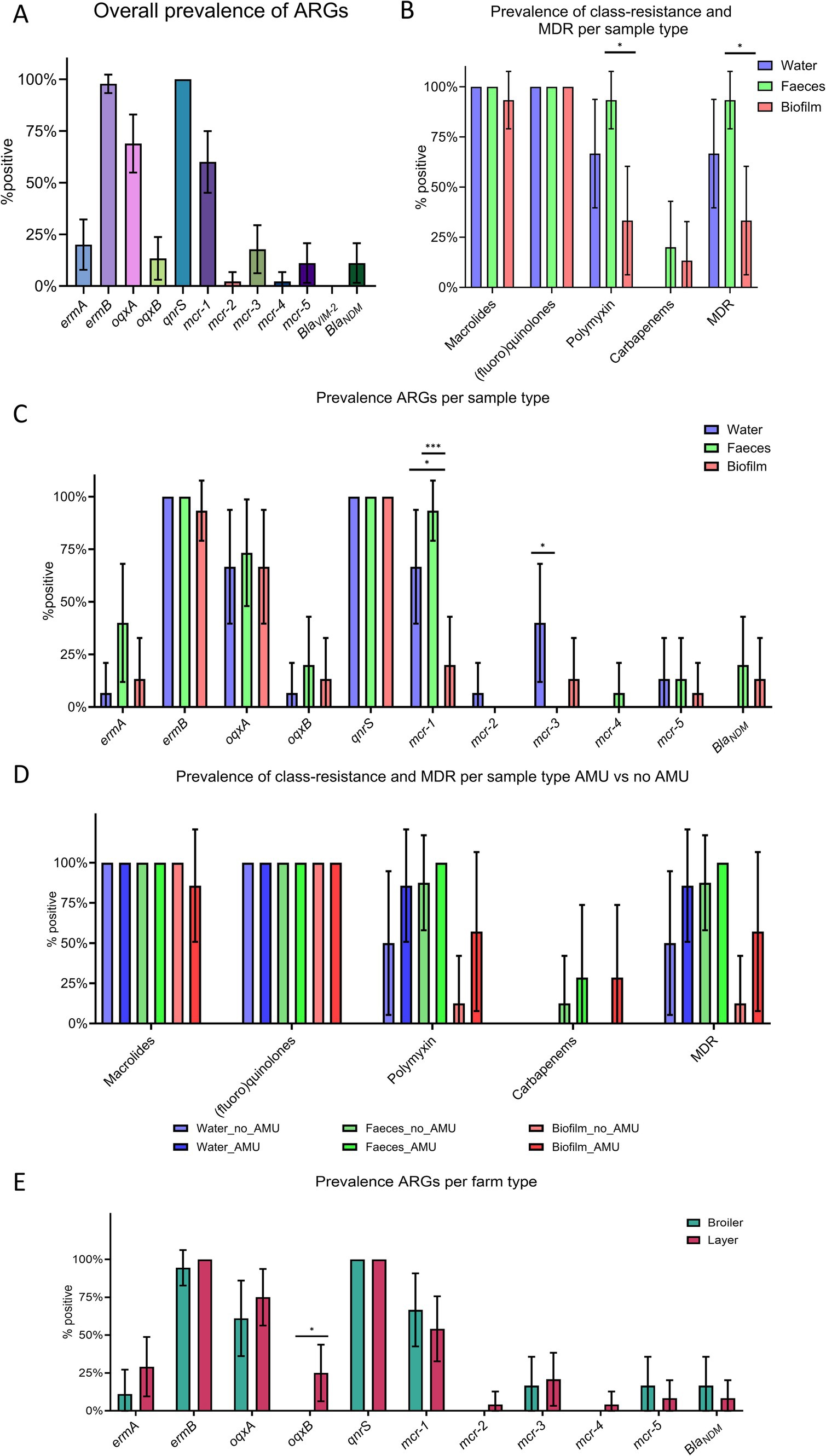
Figure 4. Prevalence of target genes in fecal, water, and biofilm samples. (A) Overall prevalence of antimicrobial resistance genes (ARGs). (B) Prevalence of class-resistance and multi-drug resistance (MDR) according to sample type. (C) Prevalence of ARGs according to sample type. (D) Prevalence of class-resistance and MDR according to sample type and antimicrobial use. (E) Prevalence of ARGs according to farm type (i.e., broilers and layers). p < 0.05 shown as * and p < 0.001 as ***. p-values referred to the binomial analysis. Whiskers represent 95% CI.
Although ARGs, class-level resistance, and MDR showed a similar distribution in the different matrices, some differences were observed (Figures 4B,C). Binomial analysis indicated that mcr-1 (p = 0.0001), polymyxins resistance genes (i.e., mcr-1 to mcr-5) (p = 0.0017), and MDR (p = 0.0017) were more prevalent in fecal samples compared to biofilm. Logistic regression analysis confirmed that biofilm samples were less likely to harbor mcr-1 (p = 0.0011, odd ratio (OR) = 0.0012, 95% CI 0.0001–0.0552), genes conferring resistance to polymyxins (p = 0.0025, OR = 0.0021, 95% CI 0.0003–0.0552), and MDR (p = 0.0024, OR = 0.0021, 95% CI 0.0002–0.05519) compared to feces. Binomial analysis showed that mcr-1 was also significantly less prevalent in drinking water (66.7, 95% CI 39.6–93.7%, p = 0.0253) compared to fecal samples. However, multiple logistic regression did not confirm this difference (OR = 0.0558, 95% CI 0.0139–0.7018, p = 0.0537). Furthermore, mcr-3 was detected only in drinking water and biofilm, blaNDM only in feces and biofilm, mcr-2 only in drinking water, and mcr-4 only in fecal samples.
Differences between farm type (i.e., broiler and layer farms) was also investigated and the binomial analysis identified a positive association between layer farms and the oqxB gene, encoding resistance to (fluoro) quinolones (Figure 4E). The analysis investigating the effect of antimicrobial treatments 14 days prior to sampling did not show any statistical difference in the prevalence of ARGs, class-level resistance, nor MDR between AMU_Farms and No_AMU_Farms (Figure 4D and Supplementary material 6). However, within each matrix, a trend of increasing prevalence of polymyxins and MDR was observed in farms where antimicrobials were recently used, and blaNDM was detected in biofilms from the AMU_Farms group only. Farms using different water sources (i.e., drilling or well water) and/or water treatments showed similar distributions of ARGs, class-level, and MDR (p > 0.05).
3.3 Association between microbial communities and ARGs
Multivariate regression analysis explored possible associations between the most abundant taxa at family level and the presence/absence of ARGs. Among the nine ARGs included (i.e., prevalence >10% of samples), only mcr-1 was positively associated with Xanthobacteraceae (p = 0.0141, O.D. = 4.467, 95% CI 1.62–19.46).
3.4 Farmers’ knowledge, practices and attitude toward AMU
The average knowledge and attitude scores on AMU of poultry farmers were insufficient (0.41 ± 0.14) and unsuitable (0.43 ± 0.11), respectively, while the average score on AMU practices was appropriate (0.74 ± 0.07). Sixty percent of participants had heard about antimicrobials, but only 33.3% could provide a proper definition (Figure 5A). Only 26.7% of respondents agreed that antimicrobial misuse could lead to residues in poultry meat. Most farms (93.3%) had farm workers administering antimicrobials, and 80% of farmers used antibiotics for preventive purposes. More than half (53.3%) administered antimicrobials on the selling day and consumed dead chicken shortly after treatment. Almost half of the layer farmers (44.4%) sold eggs while birds were under antimicrobial treatment (Figure 5A).
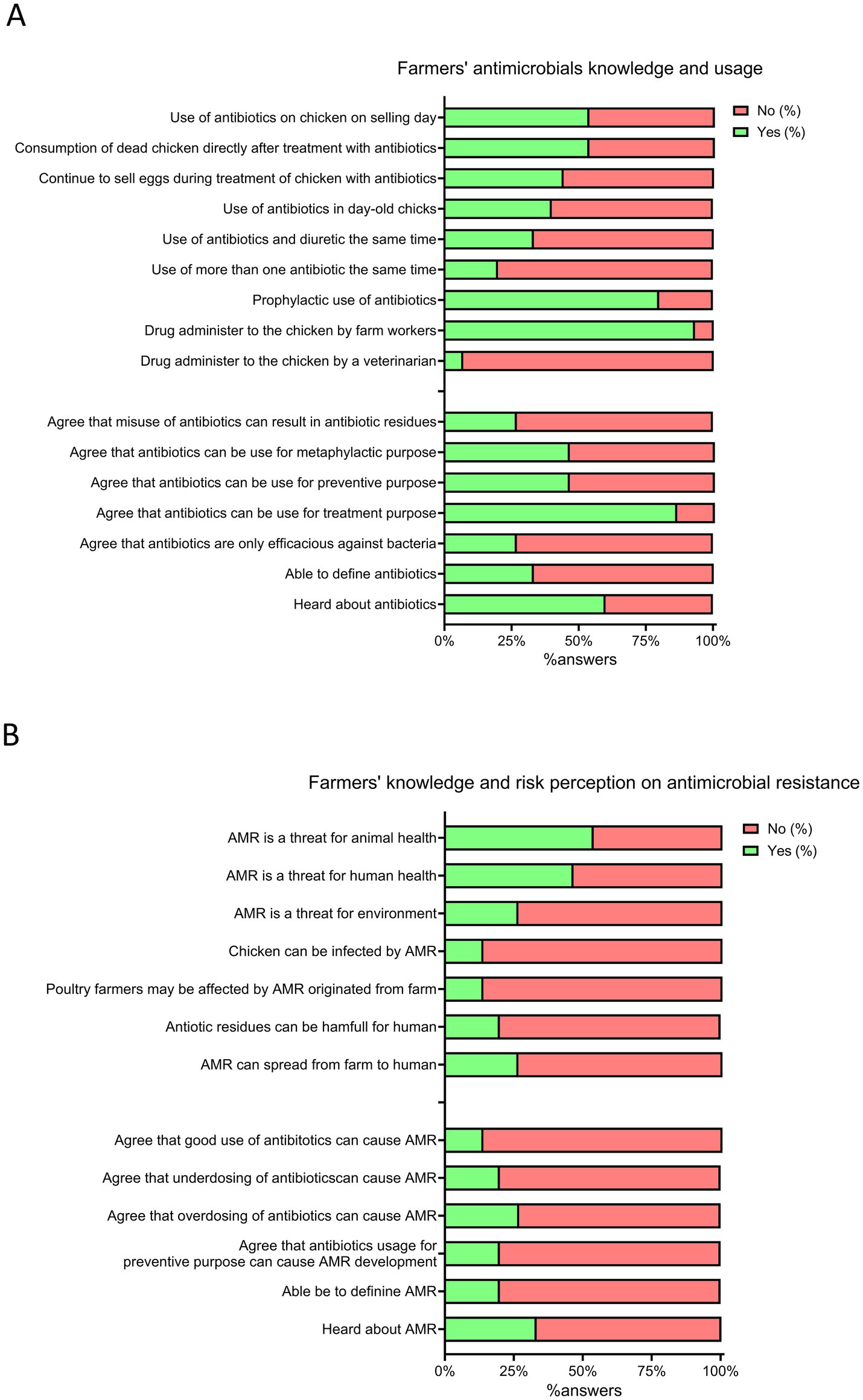
Figure 5. Farmer’s knowledge, practices and perception about antimicrobial use (AMU) and antimicrobial resistance (AMR). (A) Farmer’s antimicrobial knowledge and usage. (B) Farmer’s knowledge and risk perception toward antimicrobial resistance. Positive answers (%) are reported in green, while negative answers (%) are reported in red.
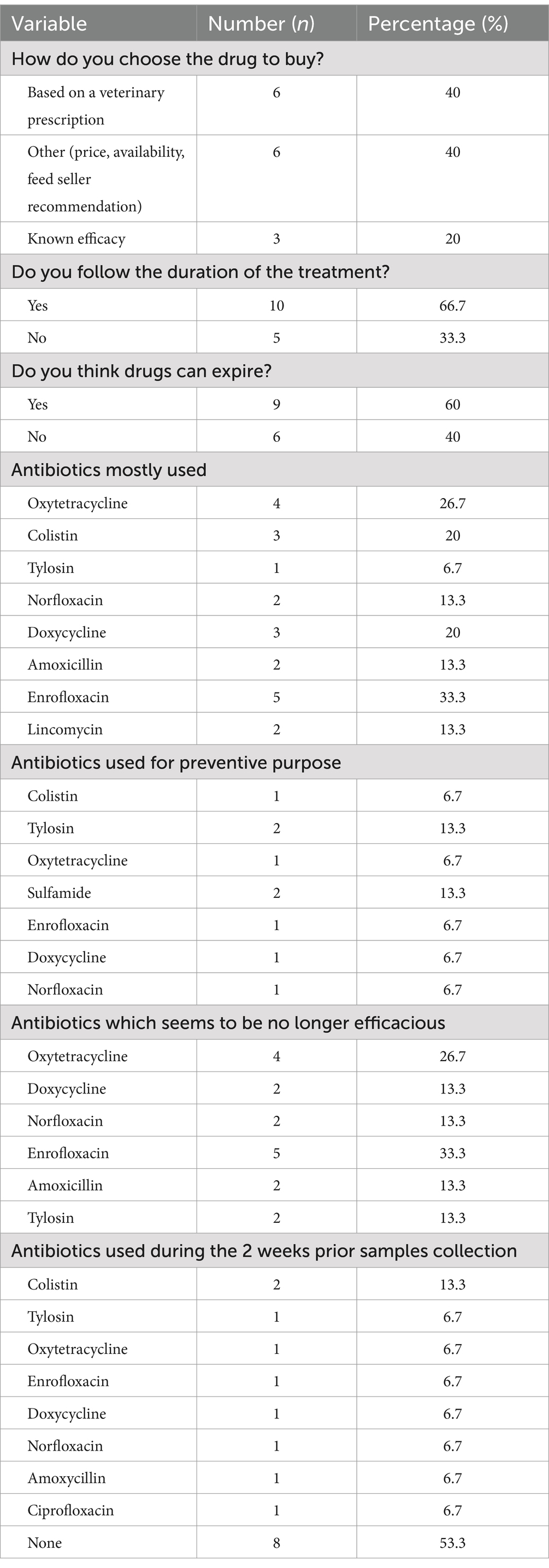
Table 3 reports the AMDs used in the visited farms. Less than half of farmers (40.0%) bought AMDs based on a veterinarian prescription. Notably, among the most commonly used antimicrobials, 50% are listed as medically important antimicrobials and are also used for prophylaxis treatments. Enrofloxacin and colistin, used by 33.3 and 20.0% of farmers respectively, are listed as highest priority critically important antimicrobials (HPCIA). The respondents declared that, except for colistin, none of these antimicrobials seemed to be effective for the treatment of bacterial infections.
3.5 Farmer’s knowledge and risk perception of AMR
The mean AMR knowledge and risk perception scores of poultry farmers were insufficient (0.22 ± 0.20) and inappropriate (0.34 ± 0.12), respectively. Only 33% of farmers were aware of AMR; indeed, less than a quarter of respondents (13.3%) recognized that antimicrobial misuse could lead to AMR development and that AMR could be harmful to themselves and their birds (Figure 5B). A minority acknowledged AMR as a threat to humans (46.7%) and the environment (26.7%), and only 26.7% mentioned AMR could spread from the farm environment to humans.
3.6 Level of implementation of biosecurity measures in poultry farms
Overall, the level of biosecurity implementation was intermediate (55.2 ± 10.5%), with higher internal (60.3 ± 9.1%) than external (50.1 ± 9.7%) biosecurity. Layer farms (61.9 ± 8.1%) were more compliant than broiler farms (45.2 ± 8.5%). Microbiological and physico-chemical controls of drinking water (e.g., pH, alkalinity, aerobic mesophilic counts) were routinely performed only in 13.3% of visited farms.
3.7 Association between biosecurity, AMU and AMR
Associations between ARGs prevalence, class-level resistance, and MDR and several correlates were investigated using multiple logistic regression analysis. No association between the scores of internal and external biosecurity and any of the independent variables considered (i.e., ARGs, class-level resistance, and MDR) were observed. Similarly, farmers’ knowledge on AMR and risk perception did not correlate with ARGs, class-level resistance, or MDR. However, when considering farmers’ attitude, knowledge, and practices on AMU, the latter was found to be associated with decreased resistance to polymyxin (p = 0.0401, OR = 0.8084, 95% CI 0.6223–0.9606) and MDR (p = 0.0374 and OR = 0.7576, 95% CI 0.5519–0.9148).
4 Discussion
4.1 Microbial communities differ among feces, drinking water and biofilm
The microbial community composition of poultry fecal samples aligns with that described in previous studies (Laconi et al., 2022a; Jensen et al., 2023; Meng et al., 2023), where Lactobacillaceae and Enterobacteriaceae were dominant. Similarly, the dominant taxa in drinking water and biofilm samples are consistent with those detected in drinking water distribution systems for human and livestock consumption (Kelly et al., 2014; Van Assche et al., 2019; Zhu et al., 2021). Notably, bacteria belonging to the most abundant families in drinking water and biofilm (i.e., Comamonadaceae and Sphingomonadaceae) can act as opportunistic pathogens in poultry carrying multiple resistance determinants (Willems, 2014; Ma et al., 2017). This finding suggests that more effort should be put into proper application of cleaning and disinfection procedures of the water pipelines, such as performing chlorine treatments between cycle, to reduce the abundance of these bacteria (Meng et al., 2023). While microbial communities of drinking water and biofilm seem to partially overlap, significant differences in richness and composition between these communities and the fecal microbiota were observed. In accordance with previous studies (Piccirillo et al., 2024), the latter showed higher richness and diversity (α-diversity) compared to the former. These differences in microbiota composition and diversity might be due to the unfavorable environment (e.g., lower temperature) of water and its lack of nutrients, which can reduce the ability of some bacteria commonly present in the chicken gut to survive and multiply (McAllister and Topp, 2012). Interestingly, AMU seemed not to have a significant effect on the microbial communities of any of the investigated samples. Indeed, within each same sample type, neither richness nor diversity were affected by AMU. Several studies investigating the effects of AMU on the chicken gut microbiota reported both reduced and increased diversity, as well as no changes in the microbial composition and structure (Schokker et al., 2017; Le Roy et al., 2019; Laconi et al., 2022a). Increased or comparable diversity in the chicken gut after treatments has been previously attributed to the proliferation of resistant bacteria compensating for the loss of non-resistant species (Laconi et al., 2022a). However, in this study, specific taxa neither disappeared nor bloomed in the AMU_group, suggesting a widespread dissemination of resistant bacteria in Cameroonian poultry farms, as supported by the high prevalence of ARGs observed.
4.2 High prevalence and different profiles of ARGs among sample types
Aiming to understand the level of resistance and the distribution of ARGs in Cameroonian poultry farms, the prevalence of selected genes conferring resistance to medically important antimicrobials was assessed in drinking water and biofilm collected from drinking water distribution systems and fecal samples by using a culture-independent approach (i.e., real-time PCR). On one hand, the paucity of studies investigating AMR in poultry production in Central Africa (Vougat Ngom et al., 2024a) using culture-independent methods highlights the novelty and potential impact of the present study. On the other hand, it hampers a straightforward comparison between the prevalence of ARGs observed here and previous studies conducted in the same geographical area. However, some considerations can be drawn. In this study, for instance, a higher prevalence of qnrS (100%) was detected compared to previous observations, since this gene has been identified in less than 50% of bacteria isolated from chickens, fecal droppings and/or farm environment (Ayandiran et al., 2018; Nchawa et al., 2019b; Kimera et al., 2021; Leinyuy et al., 2023). Moreover, even when adopting a culture-independent strategy (i.e., metagenomics), the level of resistance to (fluoro) quinolones in Ghanaian poultry farms was lower compared to our findings (Jensen et al., 2023). Previous studies failed to identify resistance genes to polymyxins other than mcr-1 (Nchawa et al., 2019a; Vounba et al., 2019b; Kimera et al., 2021); however, four additional mcr genes were detected across the different samples in this study, representing a significant concern for public health. Of further concern is the detection of the blaNDM gene in feces and biofilm. While genes encoding for carbapenemases (e.g., blaOXA) have been previously detected in bacteria isolated from poultry farms in African countries (Nchawa et al., 2019b; Vounba et al., 2019a; Aworh et al., 2021), this represents the first ever detection of blaNDM. Moreover, most samples (64, 95% CI 49.9–79.0%) showed a MDR profile, being resistant to at least three different antimicrobial classes. Since one of main drivers of the emergence of AMR is the use of AMDs (Holmes et al., 2016), the observed high level of resistance could be reasonably attributed to the misuse and overuse of antimicrobials in the farms under investigation. Indeed, while only 7 out of 15 farmers reported using at least one AMD (e.g., norfloxacin, colistin, and/or tylosin), belonging to one of the antimicrobial classes investigated in the 2 weeks prior sampling, antimicrobials were routinely administered for metaphylaxis and prophylaxis in all sampled farms. Supporting the key role played by the selective pressure exerted by antimicrobials on AMR, farms where AMDs were more lately used showed a trend of increased resistance to polymyxins, carbapenems, and multi-drugs. Furthermore, farms scoring high on good practices on AMU were associated (p < 0.05) with decreased resistance to polymyxins and MDR. This finding suggests that education of farmers on AMU and the risks related to AMDs overuse and misuse may contribute to the reduction of AMR in poultry production, as previously pointed out by Caekebeke et al. (2021). However, the emergence and persistence of antimicrobial resistant bacteria and genes in the farm environment is a complex phenomenon and factors other than AMU can contribute to it. For instance, since all farms used traditional and/or modern wells as their water source, the high prevalence of ARGs detected in this study may also reflect AMR environmental contamination. Notably, some of these resistance genes (e.g., ermA, ermB, oqxA, and qnrS) are capable of persisting and accumulating in fertilized soil (Laconi et al., 2020). ARGs in amended soil can reach waterways and water sources through drainage (Hruby et al., 2016), potentially perpetuating a self-sustained cycle of AMR maintenance in poultry farms and their surrounding environment. For instance, since mcr-1 was more prevalent (p < 0.05) in feces and drinking water compared to biofilm, drinking water may represent a possible source of this ARG. On the other hand, although showing prevalence as high as 40 and 15% in drinking water and biofilm samples, respectively, mcr-3 was not found in feces. Meanwhile, blaNDM was detected only in feces and biofilm, with similar prevalence in both sample types, suggesting that biofilm may play a role in its persistence and spread within poultry farms. Overall, these findings seem to further confirm that the dissemination of antimicrobial determinants in the farm environment is a complex phenomenon. While drinking water and biofilm in drinking water distribution systems may represent an important reservoir of resistance genes and bacteria, other sources (e.g., feed, pests, wildlife, and farmers) can contribute to their transmission and proliferation in the chicken gut (Daehre et al., 2017; Apostolakos et al., 2019).
4.3 Farmers’ knowledge and behavior toward AMU and perception on AMR
This study revealed that all poultry farmers used AMDs on their farms and the farmers’ knowledge on AMU and AMR was assessed as insufficient. This finding is worrying given that the majority of farmers, besides having an acceptable level of education (most of them were at least at a secondary school level), had more than 6 years of experience in poultry farming. This result can be associated with the low contact of the farmers with animal health specialists due to their limited number and the high fees of their services (Caudell et al., 2019; Tasmim et al., 2023). Indeed, only 40% of farmers obtained veterinary drugs based on a veterinarian prescription, aligning with those of Moffo et al. (2020) and Sawadogo et al. (2023) in the Center Region of Cameroon and Ouagadougou in Burkina Faso, respectively. Furthermore, the lack of awareness of the risk posed by AMR among poultry farmers detected in this study is similar to that detected across the African continent (Al Sattar et al., 2023). Such widespread unawareness may contribute to increasing AMR in African livestock, with a potential impact also on human and environmental health. Surprisingly, the mean score on AMU practices was found appropriate. Moffo et al. (2020) reported similar results in the same study area, whereas an inappropriate AMU was recorded by Chah et al. (2022) in Nigeria. In our study, 80% of farmers used antibiotics for prevention and 50% of the most commonly used antibiotics were medically important. The use of these drugs is of great concern, since they are commonly used to treat severe and life-threatening infections in humans. Misuse of AMDs can heavily contribute to the high level of resistance detected in this study, as also highlighted in a recent review considering different livestock species in Africa (Vougat Ngom et al., 2024a).
Farmers’ risk perception on AMR was inappropriate with only 20% of farmers perceiving that misuse of AMDs can contribute to the emergence of AMR. In addition, less than a quarter was aware that AMR could be harmful for both themselves and their animals. These findings corroborate the results of a previous study on risk perceptions of poultry farmers in Kwara state in Nigeria (Al-Mustapha et al., 2023). Interventions for mitigating AMR in Cameroon should focus on raising awareness on AMR in poultry farmers. This is crucial considering the increasing number of farmers in this sector due the increased consumer demand of chicken meat and eggs. In addition, improving knowledge and awareness of AMU and AMR through targeted and contextualized education, as well as improving legislation and regulations of veterinary drug purchase and use can also contribute to improving the current status.
4.4 Biosecurity implementation
The overall level of biosecurity implementation in the studied farms was intermediate. This is probably associated with the lack of strict legislation on farm biosecurity in Cameroon. Farmers’ knowledge and attitude toward biosecurity implementation may also contribute to this result (Klein et al., 2023). Moffo et al. (2022) reported a low biosecurity level in broiler farms in the same study area. The conflict among these results may be attributed to the presence of layer farms in our study. Since layer farms require more investment, farmers might be more aware of the risk and the cost associated with disease occurrence (Islam et al., 2023). Low biosecurity levels in poultry farms have also been reported in other African countries (Waktole et al., 2023). Poor biosecurity in poultry farms has been associated with an increase of animal diseases (Vougat Ngom et al., 2024b) and AMU in Africa (Moffo et al., 2022). A higher level of implementation of internal rather than external biosecurity was recorded in this study. This suggests that farmers put more effort to control than to prevent the occurrence of infectious diseases in their farms. This finding suggests a need for effective training, advising and communication on biosecurity for farmers. Similar findings have also been found in poultry farms in Africa (Moffo et al., 2022; Waktole et al., 2024), Europe (Laconi et al., 2023; Souillard et al., 2024), and Asia (Tanquilut et al., 2020; Mirzaie et al., 2023). The higher implementation of internal biosecurity may be associated with the farmers’ knowledge and perception on biosecurity and a stronger link between internal biosecurity and disease occurrence. This result highlights the need for improving biosecurity legislation and farmer’s awareness on biosecurity in Cameroon.
4.5 Limitations of the study
In the present study, the antimicrobial resistance, the level of biosecurity implementation, the farmers’ knowledge, attitude, and practices toward AMU, and their knowledge and perception of AMR risk were investigated in poultry farms in the Center region of Cameroon. While the results of this study cannot be generalized to the entire country, due to the limited sample size, they provide valuable insights for future research. Furthermore, while this study represents the first attempt to investigate AMR and microbial community composition in Cameroonian farms using a culture-independent approach and advanced molecular biology techniques (i.e., real-time PCR and next-generation sequencing), limitations in DNA quantity and quality may have hindered the quantification of ARGs and the characterization of microbiota at taxonomic levels higher than family.
5 Conclusion
Considering the complexity of AMR in livestock production and its impact on environmental and human health, adopting culture-independent and one health approaches are required to understand the prevalence and diversity of ARGs in poultry farms. However, due to financial constraints, inadequate laboratory facilities, and/or lack of trained personnel, implementing these methods may be challenging in developing countries. Efforts should be made to support cooperation and knowledge transfer between high-and low-income countries. In the present study, more than half of the samples showed MDR and carried ARGs for last resort antimicrobials (i.e., mcr and blaNDM genes), posing thus a great concern, not only for animals, but also for public and environmental health. Indeed, most farmers declared that only polymyxins seem to be still effective for treating bacterial infections in their farms. The use of AMDs, including highest priority critically important antimicrobials, for metaphylactic and prophylactic treatments, may have contributed to the high prevalence of ARGs in Cameroonian poultry farms. Notably, good AMU practices appear to be associated with a reduction in polymyxins resistance and MDR. Farmers’ knowledge and attitude toward AMU, and their knowledge and perception of the risks posed by AMR, have been found to be scarce. This, coupled with an overall modest level of biosecurity compliance, needs improvements to reduce the AMDs use and mitigate the AMR risk effectively. Education programs for farmers, along with the development of more stringent and enforced regulations on AMU and biosecurity at national level, are strongly advocated. Overall, the study underscores the need for improved practices and regulations to tackle AMR in the poultry sector in Cameroon. By addressing these issues through education, better biosecurity practices, and international cooperation, significant progress can be made in controlling AMR and ensuring sustainable poultry production.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1123609.
Author contributions
RV: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Resources, Visualization, Writing – original draft, Writing – review & editing. AL: Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. RT: Data curation, Investigation, Writing – review & editing. AA: Investigation, Visualization, Writing – review & editing. SZ: Investigation, Writing – review & editing. VK: Investigation, Writing – review & editing. AP: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was performed under the Coimbra Group Scholarship Programme for young professors and researchers from Africa (2023–2024) and the Visiting Scientist Fellowship funded by the Department of Comparative Biomedicine and Food Science - University of Padua, within the Excellence Project SENTINEL, both granted to the first author. The post-doctoral position of the third author was funded by Fondazione Cassa di Risparmio di Padova e Rovigo, grant “PHD@UNIPD” for the project “Drinking water distribution systems and biofilms as reservoirs and spreaders of antimicrobial resistance - DARES.”
Acknowledgments
The authors extend their gratitude to all the farmers who participated in the study, and to the Ministry of Higher Education of Cameroon through the University of Ngaoundéré for the authorization granted to the first author during these fellowships. We also acknowledge the services of the Ministry of Livestock, Fisheries and Animal Industries for the research permission and assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1517159/full#supplementary-material
SUPPLEMENTARY MATERIAL 1 | Copy of the questionnaire used to collect information regarding farms’ characteristics, biosecurity practices, farmers’ knowledge, attitudes and practices on antimicrobial use (AMU), and knowledge and risk perception on antimicrobial resistance (AMR).
SUPPLEMENTARY MATERIAL 2 | DNA concentration (ng/μl) of the analyzed samples.
SUPPLEMENTARY MATERIAL 3 | The list of assays, primers pairs, melting temperatures, and positive controls used in the study.
SUPPLEMENTARY MATERIAL 4 | α-and β-Diversity according to sample type and antimicrobial use.
SUPPLEMENTARY MATERIAL 5 | Linear discriminant analysis (LDA) scores of LDA effect size (LEfSe) comparison analysis between fecal, drinking water, and biofilm samples. Red shading depicts bacterial taxa significantly higher in a given sample type.
SUPPLEMENTARY MATERIAL 6 | Prevalence (%) of ARGs according to sample type and antimicrobial use. Whiskers represent 95% CI.
Footnotes
References
Al Sattar, A., Chisty, N. N., Irin, N., Uddin, M. H., Hasib, F. M. Y., and Hoque, M. A. (2023). Knowledge and practice of antimicrobial usage and resistance among poultry farmers: A systematic review, meta-analysis, and meta-regression. Vet. Res. Commun. 47, 1047–1066. doi: 10.1007/s11259-023-10082-5
Alban, L., Ellis-Iversen, J., Andreasen, M., Dahl, J., and Sönksen, U. W. (2017). Assessment of the risk to public health due to use of antimicrobials in pigs-an example of pleuromutilins in Denmark. Front. Vet. Sci. 4:74. doi: 10.3389/fvets.2017.00074
Al-Mustapha, A. I., Raufu, I. A., Ogundijo, O. A., Odetokun, I. A., Tiwari, A., Brouwer, M. S. M., et al. (2023). Antibiotic resistance genes, mobile elements, virulence genes, and phages in cultivated ESBL-producing Escherichia coli of poultry origin in Kwara state, north Central Nigeria. Int. J. Food Microbiol. 389:110086. doi: 10.1016/j.ijfoodmicro.2023.110086
Apostolakos, I., Mughini-Gras, L., Fasolato, L., and Piccirillo, A. (2019). Assessing the occurrence and transfer dynamics of ESBL/pAmpC-producing Escherichia coli across the broiler production pyramid. PLoS One 14, e0217174–e0217113. doi: 10.1371/journal.pone.0217174
Aworh, M. K., Kwaga, J. K. P., Hendriksen, R. S., Okolocha, E. C., and Thakur, S. (2021). Genetic relatedness of multidrug resistant Escherichia coli isolated from humans, chickens and poultry environments. Antimicrob. Resist. Infect. Control 10, 58–13. doi: 10.1186/s13756-021-00930-x
Ayandiran, T. O., Falgenhauer, L., Schmiede, J., Chakraborty, T., and Ayeni, F. A. (2018). High resistance to tetracycline and ciprofloxacin in bacteria isolated from poultry farms in Ibadan, Nigeria. J. Infect. Dev. Ctries. 12, 462–470. doi: 10.3855/jidc.9862
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317. doi: 10.1038/nrmicro3439
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Caekebeke, N., Ringenier, M., Jonquiere, F. J., Tobias, T. J., Postma, M., van den Hoogen, A., et al. (2021). Coaching belgian and dutch broiler farmers aimed at antimicrobial stewardship and disease prevention. Antibiotics 10:590. doi: 10.3390/antibiotics10050590
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caudell, M. A., Charoonsophonsak, P. V., Miller, A., Lyimo, B., Subbiah, M., Buza, J., et al. (2019). Narrative risk messages increase uptake and sharing of health interventions in a hard-to-reach population: A pilot study to promote milk safety among Maasai pastoralists in Tanzania. Pastoralism 9:7. doi: 10.1186/s13570-019-0142-z
Chah, J. M., Nwankwo, S. C., Uddin, I. O., and Chah, K. F. (2022). Knowledge and practices regarding antibiotic use among small-scale poultry farmers in Enugu state, Nigeria. Heliyon 8:e09342. doi: 10.1016/j.heliyon.2022.e09342
Chowdhury, T., Ahmed, J., Hossain, M. T., Roy, M. C., Ashik-Uz-Zaman, M., Uddin, M. N., et al. (2023). Knowledge, attitudes and biosecurity practices among the small-scale dairy farmers in Sylhet district, Bangladesh. Vet. Med. Sci. 9, 2221–2229. doi: 10.1002/vms3.1199
Collineau, L., Chapman, B., Bao, X., Sivapathasundaram, B., Carson, C. A., Fazil, A., et al. (2020). A farm-to-fork quantitative risk assessment model for Salmonella Heidelberg resistant to third-generation cephalosporins in broiler chickens in Canada. Int. J. Food Microbiol. 330:108559. doi: 10.1016/j.ijfoodmicro.2020.108559
Daehre, K., Projahn, M., Semmler, T., Roesler, U., and Friese, A. (2017). Extended-Spectrum Beta-lactamase-/AmpC Beta-lactamase-producing Enterobacteriaceae in broiler farms: transmission dynamics at farm level. Microb. Drug Resist. 24, 511–518. doi: 10.1089/mdr.2017.0150
Demography - Cameroon Statista. (2024). https://fr.statista.com/outlook/co/socioeconomic-indicators/demography/cameroon (last visited février 26, 2024) Food and Drug Administration FDA Task Force on Antimicrobial Resistance: Key Recommendations and Report. Available online: http://fda.gov/downloads/ForConsumers/ConsumerUpdates/UCM143458.pdf (Accessed November 30, 2020).
Djuikoue, C. I., Nana, C. D. S., Nzenya, J., Tomi, C., Chounna, N., Pomte, O., et al. (2023). Intestinal carriage of extended Spectrum Beta-lactamase-producing Salmonella enterica from chickens and poultry farmers in Dschang, in the Western region of Cameroon. Bacteria 2, 37–47. doi: 10.3390/bacteria2010003
GIZ (Deutsche Gesellschaft fur Internationale Zusammenarbeit GmbH). (2018). Poultry production in Cameroon: How the import restriction affects the cameroonian poultry sector.p12 gizbmz-eu-agrarpolitik-huehner_250518_en.pdf (https://www.snrd-africa.net/). (Accessed May 2021).
Holmes, A. H., Moore, L. S. P., Sundsfjord, A., Steinbakk, M., Regmi, S., Karkey, A., et al. (2016). Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187. doi: 10.1016/S0140-6736(15)00473-0
Hruby, C. E., Soupir, M. L., Moorman, T. B., Shelley, M., and Kanwar, R. S. (2016). Effects of tillage and poultry manure application rates on Salmonella and fecal indicator bacteria concentrations in tiles draining Des Moines lobe soils. J. Environ. Manag. 171, 60–69. doi: 10.1016/j.jenvman.2016.01.040
Islam, Z., Hsan, K., Ripon, R. K., Madhu, J., Hossain, S., Masud, A., et al. (2023). Assessment of biosecurity measures in commercial poultry farms of Rajshahi district in Bangladesh. Prev. Vet. Med. 219:106027. doi: 10.1016/j.prevetmed.2023.106027
Jensen, E. E. B., Sedor, V., Eshun, E., Njage, P., Otani, S., and Aarestrup, F. M. (2023). The resistomes of rural and urban pigs and poultry in Ghana. mSystems 8, 1–14. doi: 10.1128/msystems.00629-23
Kelly, J. J., Minalt, N., Culotti, A., Pryor, M., and Packman, A. (2014). Temporal variations in the abundance and composition of biofilm communities colonizing drinking water distribution pipes. PLoS One 9:e98542. doi: 10.1371/journal.pone.0098542
Kimera, Z. I., Mgaya, F. X., Misinzo, G., Mshana, S. E., Moremi, N., and Matee, M. I. N. (2021). Multidrug-resistant, including extended-Spectrum Beta Lactamase-Producing and Quinolone-Resistant, Escherichia coli isolated from poultry and domestic pigs in Dar es Salaam, Tanzania. Antibiotics 10:406. doi: 10.3390/antibiotics10040406
Klein, L., Hessling-Zeinen, S., Adler, F., Gerdes, U., Blome, S., Beilage, E., et al. (2023). Exploring pig farmers’ decision-making concerning biosecurity measures against African swine fever. Prev. Vet. Med. 217:105949. doi: 10.1016/j.prevetmed.2023.105949
Klein, E. Y., Van Boeckel, T. P., Martinez, E. M., Pant, S., Gandra, S., Levin, S. A., et al. (2018). Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 115, E3463–E3470. doi: 10.1073/pnas.1717295115
Laconi, A., Mughini-Gras, L., Tolosi, R., Grilli, G., Trocino, A., Carraro, L., et al. (2020). Microbial community composition and antimicrobial resistance in agricultural soils fertilized with livestock manure from conventional farming in northern Italy. Sci. Total Environ. 760:3404. doi: 10.1016/j.scitotenv.2020.143404
Laconi, A., Tilli, G., Galuppo, F., Grilli, G., Souillard, R., and Piccirillo, A. (2023). Stakeholders’ perceptions of biosecurity implementation in Italian poultry farms. Animals 13:3246. doi: 10.3390/ani13203246
Laconi, A., Tolosi, R., Mughini-Gras, L., Cuccato, M., Cannizzo, F. T., and Piccirillo, A. (2022a). Amoxicillin and thiamphenicol treatments may influence the co-selection of resistance genes in the chicken gut microbiota. Sci. Rep. 12, 20413–20414. doi: 10.1038/s41598-022-24927-7
Laconi, A., Tolosi, R., Mughini-Gras, L., Mazzucato, M., Ferrè, N., Carraro, L., et al. (2022b). Beehive products as bioindicators of antimicrobial resistance contamination in the environment. Sci. Total Environ. 823:151131. doi: 10.1016/j.scitotenv.2021.151131
Le Roy, C. I., Woodward, M. J., Ellis, R. J., La Ragione, R. M., and Claus, S. P. (2019). Antibiotic treatment triggers gut dysbiosis and modulates metabolism in a chicken model of gastro-intestinal infection. BMC Vet. Res. 15, 37–13. doi: 10.1186/s12917-018-1761-0
Leinyuy, J. F., Ali, I. M., Ousenu, K., and Tume, C. B. (2023). Molecular characterization of antimicrobial resistance related genes in E. coli, Salmonella and Klebsiella isolates from broilers in the west region of Cameroon. PLoS One 18, e0280150–e0280114. doi: 10.1371/journal.pone.0280150
Ma, L., Li, B., Jiang, X. T., Wang, Y. L., Xia, Y., Li, A. D., et al. (2017). Catalogue of antibiotic resistome and host-tracking in drinking water deciphered by a large scale survey. Microbiome 5:154. doi: 10.1186/s40168-017-0369-0
McAllister, T. A., and Topp, E. (2012). Role of livestock in microbiological contamination of water: commonly the blame, but not always the source. Anim. Front. 2, 17–27. doi: 10.2527/af.2012-0039
Meng, W. S., Sui, X., Xiao, Y., Zou, Q., Cui, Y., Wang, T., et al. (2023). Regulating effects of chlorinated drinking water on cecal microbiota of broiler chicks. Poult. Sci. 102:103140. doi: 10.1016/j.psj.2023.103140
MINEPIA. (2019). unpublished. Statistical yearbook of the Ministry of Livestock, Fisheries and Animal Industries of Cameroon. 2018.
Mirzaie, K., Rabiee, M. H., Bashashati, M., Ghalyanchi, A., Shoushtari, A., Parsai, A., et al. (2023). Biosecurity practices on commercial layer farms in Abyek county, Qazvin, Iran: a cross-sectional study. One Health Bull. 3, 1–6. doi: 10.4103/2773-0344.380552
Moffo, F., Mouiche, M. M. M., Djomgang, H. K., Tombe, P., Wade, A., Kochivi, F. L., et al. (2022). Associations between antimicrobial use and antimicrobial resistance of Escherichia coli isolated from poultry litter under field conditions in Cameroon. Prev. Vet. Med. 204:105668. doi: 10.1016/j.prevetmed.2022.105668
Moffo, F., Mouliom Mouiche, M. M., Kochivi, F. L., Dongmo, J. B., Djomgang, H. K., Tombe, P., et al. (2020). Knowledge, attitudes, practices and risk perception of rural poultry farmers in Cameroon to antimicrobial use and resistance. Prev. Vet. Med. 182:105087. doi: 10.1016/j.prevetmed.2020.105087
Monamele, G. C., Phalla, Y., Albert, E., Vernet, M., and Wade, A. (2019). Evidence of exposure and human seroconversion during an outbreak of avian influenza A (H5N1) among poultry in Cameroon. Emerg. Microbes Infect. 8, 186–196. doi: 10.1080/22221751.2018.1564631
Mouiche, M. M. M., Moffo, F., Betsama, J. D. B., Mapiefou, N. P., Mbah, C. K., Mpouam, S. E., et al. (2020). Challenges of antimicrobial consumption surveillance in food-producing animals in sub-Saharan African countries: patterns of antimicrobials imported in Cameroon from 2014 to 2019. J. Glob. Antimicrob. Resist. 22, 771–778. doi: 10.1016/j.jgar.2020.06.021
Nchawa, Y. Y., Bassey, E. B., Inyang-etoh, P., Useh, M. F., Asuquo, A., and Angela, O.-E. (2019a). Antimicrobial resistance pattern in Salmonella enterica from clinical and poultry sources in Calabar, Nigeria. J. Microbiol. Antimicrob. 11, 5–10. doi: 10.5897/jma2019.0413
Nchawa, Y. Y., Bassey, E. B., Inyang-etoh, P., Useh, M. F., Asuquo, A., and Angela, O.-E. (2019b). Extended spectrum beta-lactamase production and plasmid mediated quinolone resistance among lactose non-fermenting nterobacteriaceae isolated from poultry sources in Calabar, Nigeria. Afr. J. Microbiol. Res. 13, 400–406. doi: 10.5897/ajmr2019.9137
Ochieng, P. E., Scippo, M. L., Kemboi, D. C., Croubels, S., Okoth, S., Kang’ethe, E. K., et al. (2021). Mycotoxins in poultry feed and feed ingredients from sub-saharan africa and their impact on the production of broiler and layer chickens: A review. Toxins 13:633. doi: 10.3390/toxins13090633
Piccirillo, A., Tolosi, R., Mughini-Gras, L., Kers, J. G., and Laconi, A. (2024). Drinking water and biofilm as sources of antimicrobial resistance in free-range organic broiler farms. Antibiotics 13:808. doi: 10.3390/antibiotics13090808
Postma, M., Vanderhaeghen, W., Sarrazin, S., Maes, D., and Dewulf, J. (2017). Reducing antimicrobial usage in pig production without jeopardizing production parameters. Zoonoses Public Health 64, 63–74. doi: 10.1111/zph.12283
Rodrigues Da Costa, M., Gasa, J., Calderón Díaz, J. A., Postma, M., Dewulf, J., McCutcheon, G., et al. (2019). Using the biocheck. UGent™ scoring tool in Irish farrow-to-finish pig farms: assessing biosecurity and its relation to productive performance. Porcine Health Manag. 5, 4–9. doi: 10.1186/s40813-018-0113-6
Sartorius, B., Gray, A. P., Davis Weaver, N., Robles Aguilar, G., Swetschinski, L. R., Ikuta, K. S., et al. (2024). The burden of bacterial antimicrobial resistance in the WHO African region in 2019: a cross-country systematic analysis. Lancet Glob. Health 12, e201–e216. doi: 10.1016/S2214-109X(23)00539-9
Sawadogo, A., Kagambèga, A., Moodley, A., Ouedraogo, A. A., Barro, N., and Dione, M. (2023). Knowledge, attitudes, and practices related to antibiotic use and antibiotic resistance among poultry farmers in urban and Peri-urban areas of Ouagadougou, Burkina Faso. Antibiotics 12:133. doi: 10.3390/antibiotics12010133
Schokker, D., Jansman, A. J. M., Veninga, G., de Bruin, N., Vastenhouw, S. A., de Bree, F. M., et al. (2017). Perturbation of microbiota in one-day old broiler chickens with antibiotic for 24 hours negatively affects intestinal immune development. BMC Genomics 18, 241–214. doi: 10.1186/s12864-017-3625-6
Souillard, R., Allain, V., Dufay-Lefort, A. C., Rousset, N., Amalraj, A., Spaans, A., et al. (2024). Biosecurity implementation on large-scale poultry farms in Europe: A qualitative interview study with farmers. Prev. Vet. Med. 224:106119. doi: 10.1016/j.prevetmed.2024.106119
Sparks, N. H. C. (2009). The role of the water supply system in the infection and control of Campylobacter in chicken. Worlds Poult. Sci. J. 65, 459–474. doi: 10.1017/S0043933909000324
Tanquilut, N. C., Espaldon, M. V. O., Eslava, D. F., Ancog, R. C., Medina, C. D. R., Paraso, M. G. V., et al. (2020). Quantitative assessment of biosecurity in broiler farms using biocheck. UGent in Central Luzon, Philippines. Poult. Sci. 99, 3047–3059. doi: 10.1016/j.psj.2020.02.004
Tasmim, S. T., Hasan, M. M., Talukder, S., Mandal, A. K., Parvin, M. S., Ali, M. Y., et al. (2023). Sociodemographic determinants of use and misuse of antibiotics in commercial poultry farms in Bangladesh. IJID Reg. 7, 146–158. doi: 10.1016/j.ijregi.2023.01.001
Van Assche, A., Crauwels, S., De Brabanter, J., Willems, K. A., and Lievens, B. (2019). Characterization of the bacterial community composition in water of drinking water production and distribution systems in Flanders, Belgium. Microbiology 8, e00726–e00711. doi: 10.1002/mbo3.726
Vougat Ngom, R., Ayissi, G. J., Akoussa, A. M. M., Laconi, A., Jajere, S. M., Zangue, H. A., et al. (2024b). A systematic review and Meta-analysis of the efficacy of biosecurity in disease prevention and control in livestock farms in Africa. Transbound. Emerg. Dis. 2024:8683715. doi: 10.1155/2024/8683715
Vougat Ngom, R., Jajere, S. M., Ayissi, G. J., Tanyienow, A., Moffo, F., Watsop, H. M., et al. (2024a). Unveiling the landscape of resistance against high priority critically important antimicrobials in food-producing animals across Africa: A scoping review. Prev. Vet. Med. 226:106173. doi: 10.1016/j.prevetmed.2024.106173
Vounba, P., Arsenault, J., Bada-Alambédji, R., and Fairbrother, J. M. (2019a). Prevalence of antimicrobial resistance and potential pathogenicity, and possible spread of third generation cephalosporin resistance, in Escherichia coli. PLoS One 14:e0214304. doi: 10.1371/journal.pone.0214304
Vounba, P., Rhouma, M., Arsenault, J., Bada Alambédji, R., Fravalo, P., and Fairbrother, J. M. (2019b). Prevalence of colistin resistance and mcr-1/mcr-2 genes in extended-spectrum β-lactamase/AmpC-producing Escherichia coli isolated from chickens in Canada, Senegal and Vietnam. J. Glob. Antimicrob. Resist. 19, 222–227. doi: 10.1016/j.jgar.2019.05.002
Waktole, H., Ayele, Y., Ayalkibet, Y., Teshome, T., Muluneh, T., Ayane, S., et al. (2024). Prevalence, molecular detection, and antimicrobial resistance of Salmonella isolates from poultry farms across Central Ethiopia: a cross-sectional study in urban and Peri-urban areas. Microorganisms 12:767. doi: 10.3390/microorganisms12040767
Waktole, H., Muluneh, T., Miressa, Y., Ayane, S., Berhane, G., Kabeta, T., et al. (2023). Quantitative assessment of major biosecurity challenges of poultry production in Central Ethiopia. Animals 13:3719. doi: 10.3390/ani13233719
Willems, A. (2014). “The family Comamonadaceae” in The prokaryotes: Alphaproteobacteria and Betaproteobacteria. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer), 777–851.
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: antimicrobial resistance, antimicrobial use, resistance genes, poultry, Africa
Citation: Vougat Ngom R, Laconi A, Tolosi R, Akoussa AMM, Ziebe SD, Kouyabe VM and Piccirillo A (2025) Resistance to medically important antimicrobials in broiler and layer farms in Cameroon and its relation with biosecurity and antimicrobial use. Front. Microbiol. 15:1517159. doi: 10.3389/fmicb.2024.1517159
Edited by:
Catherine M. Logue, University of Georgia, United StatesReviewed by:
Grazieli Maboni, University of Georgia, United StatesJohn J. Maurer, Virginia Tech, United States
Copyright © 2025 Vougat Ngom, Laconi, Tolosi, Akoussa, Ziebe, Kouyabe and Piccirillo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Laconi, YW5kcmVhLmxhY29uaUB1bmlwZC5pdA==
 Ronald Vougat Ngom
Ronald Vougat Ngom Andrea Laconi
Andrea Laconi Roberta Tolosi2
Roberta Tolosi2 Alessandra Piccirillo
Alessandra Piccirillo