
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Microbiol. , 22 January 2025
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1516584
 Jia-Yi Lin1,2
Jia-Yi Lin1,2 Jia-Kai Lai1,3
Jia-Kai Lai1,3 Jian-Yi Chen1,2
Jian-Yi Chen1,2 Jia-Yu Cai1,4
Jia-Yu Cai1,4 Zhan-Dong Yang1,2
Zhan-Dong Yang1,2 Liu-Qingqing Yang1,5,6
Liu-Qingqing Yang1,5,6 Ze-Tao Zheng1,2
Ze-Tao Zheng1,2 Xu-Guang Guo1,4,7*
Xu-Guang Guo1,4,7*Background: Methicillin-resistant Staphylococcus aureus (MRSA) bloodstream infections (BSIs) pose a significant challenge to global public health, characterized by high morbidity and mortality rates, particularly in immunocompromised patients. Despite extensive research, the rapid development of MRSA antibiotic resistance has outpaced current treatment methods, increasing the difficulty of treatment. Therefore, reviewing research on MRSA BSIs is crucial.
Methods: This study conducted a bibliometric analysis, retrieving and analyzing 1,621 publications related to MRSA BSIs from 2006 to 2024. The literature was sourced from the Web of Science Core Collection (WoSCC), and data visualization and trend analysis were performed using VOSviewer, CiteSpace, and Bibliometrix software packages.
Results: The bibliometric analysis showed that research on MRSA BSIs was primarily concentrated in the United States, China, and Japan. The United States leads in research output and influence, with significant contributions from institutions such as the University of California system and the University of Texas system. The journal with the most publications is Antimicrobial Agents and Chemotherapy, while the most cited global publication is Vincent JL’s article “Sepsis in European Intensive Care Units: Results of the SOAP Study” published in Critical Care Medicine in 2006. Cosgrove SE’s article “Comparison of Mortality Associated with Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Bacteremia: A Meta-analysis” had the most co-citations. Key trends in the research include MRSA’s antibiotic resistance mechanisms, the application of new diagnostic technologies, and the impact of COVID-19 on MRSA studies. Additionally, artificial intelligence (AI) and machine learning are increasingly applied in MRSA diagnosis and treatment, and phage therapy and vaccine development have become future research hotspots.
Conclusion: Methicillin-resistant Staphylococcus aureus BSIs remain a major global public health challenge, especially with the increasing severity of antibiotic resistance. Although progress has been made in new treatments and diagnostic technologies, further validation is required. Future research will rely on integrating genomics, AI, and machine learning to drive personalized treatment. Strengthening global cooperation, particularly in resource-limited countries, will be key to effectively addressing MRSA BSIs.
Methicillin-resistant Staphylococcus aureus (MRSA) bloodstream infections (BSIs) are a major public health challenge worldwide (Brown et al., 2021; Mahmoud et al., 2020). As a pathogen resistant to ß-lactam antibiotics, MRSA has become a leading cause of hospital- and community-acquired infections (Kavanagh, 2019), and MRSA BSIs are often associated with high morbidity and mortality, particularly in immunocompromised patients (Hassoun et al., 2017). Despite ongoing research, the development of resistance to MRSA continues to outpace advances in available therapies, making it increasingly difficult to treat (Evans et al., 2021). The impact of MRSA infections on healthcare systems worldwide has prompted scientists to work toward understanding the mechanisms of resistance, the routes of transmission and the search for alternative therapeutic approaches (Brown et al., 2021).
Previous studies have highlighted the increasing incidence of MRSA BSIs and their impact on the healthcare environment (Popovich et al., 2021). In particular, the ability of MRSA to spread in high-risk areas such as intensive care units (ICUs) has been extensively studied (Kavanagh, 2019). Although antibiotics such as vancomycin and daptomycin have shown some efficacy in treatment, the resulting problem of drug resistance is increasing, leading to a gradual decline in therapeutic efficacy (Evans et al., 2021). In addition, delayed diagnosis is a key factor contributing to increased mortality, and early detection and timely treatment are critical to the survival of patients with MRSA BSIs (Brown et al., 2021). In recent years, new technologies such as whole-genome sequencing (WGS) and machine learning (ML) have opened new avenues for early detection and improved treatment (Evans et al., 2021).
However, there are still many gaps in current research into MRSA BSIs. In particular, there is a lack of in-depth understanding of the evolution of resistance patterns in different regions and the effectiveness of available treatments. In addition, although international collaborations have produced some results in mapping the global distribution of MRSA (Stefani et al., 2012; Tokajian, 2014), the role of multi-institutional collaborations and new diagnostic technologies (Gammel et al., 2021; Tang et al., 2022) in the management of MRSA has not been fully explored. A thorough understanding of these issues is essential for the development of effective global response strategies.
This study aims to comprehensively explore research trends in the field of MRSA BSIs through a bibliometric analysis of studies conducted between 2006 and 2024. Bibliometrics (Li et al., 2024) is a strategy used to investigate library and data science by conducting a quantitative analysis of bibliographic data. It helps researchers quickly understand their areas of interest and emerging trends, thereby laying a foundation for future research directions. Given the current lack of research and summaries on the future trends of MRSA BSIs, we conducted a bibliometric study based on published data in this field. We hope this will contribute to the development of MRSA BSI research and provide future researchers with guidance for broader and more effective applications in the future.
The Web of Science Core Collection (WoSCC) was searched for articles on MRSA and BSIs from 2006 to 2024. The literature search was completed in 1 day (24 August 2024) to eliminate citation oscillations caused by rapid publication updates. The search formula was set as ((TS = (MRSA)) OR TS = (Methicillin Resistant Staphylococcus aureus) AND (TS = (bloodstream infection) OR TS = (Bloodstream Infections) OR TS = (Infection, Bloodstream) OR TS = (Infection, Bloodstream) OR TS = (Septicemia) OR TS = (Septicemias) OR TS = (Blood Poisoning) OR TS = (Blood Poisonings) OR TS = (Poisonings, Blood) OR TS = (Severe Sepsis) OR TS = (Poisoning, Blood) OR TS = (Sepsis, Severe) OR TS = (Pyemia) OR TS = (Pyemias) OR TS = (Pyaemia) OR TS = (Pyaemias) OR TS = (Pyohemia) OR TS = (Pyohemias)). The only publication types used were Article and Review; in addition, the results of the study were documented in “plain text” format in the content of the “Full Record and Cited References.” The file contains: (i) author(s), (ii) abstract, (iii) address, (iv) ISSN/ISBN, (v) IDS number, (vi) funding information, (vii) PubMed ID, (viii) title, (ix) cited reference(s), (x) number of citations, (xi) count of cited references, (xii) language, (xiii) source, ((xiv) document type, (xv) keyword, (xvi) source abbreviation, (xvii) author identifier, (xviii) article information, (xix) publisher information, (xx) field of study, (xxi) usage count, and (xxii) high citation count. Finally, 1,621 original articles or reviews were included in the bibliometric analysis. Figure 1A depicts the detailed process.
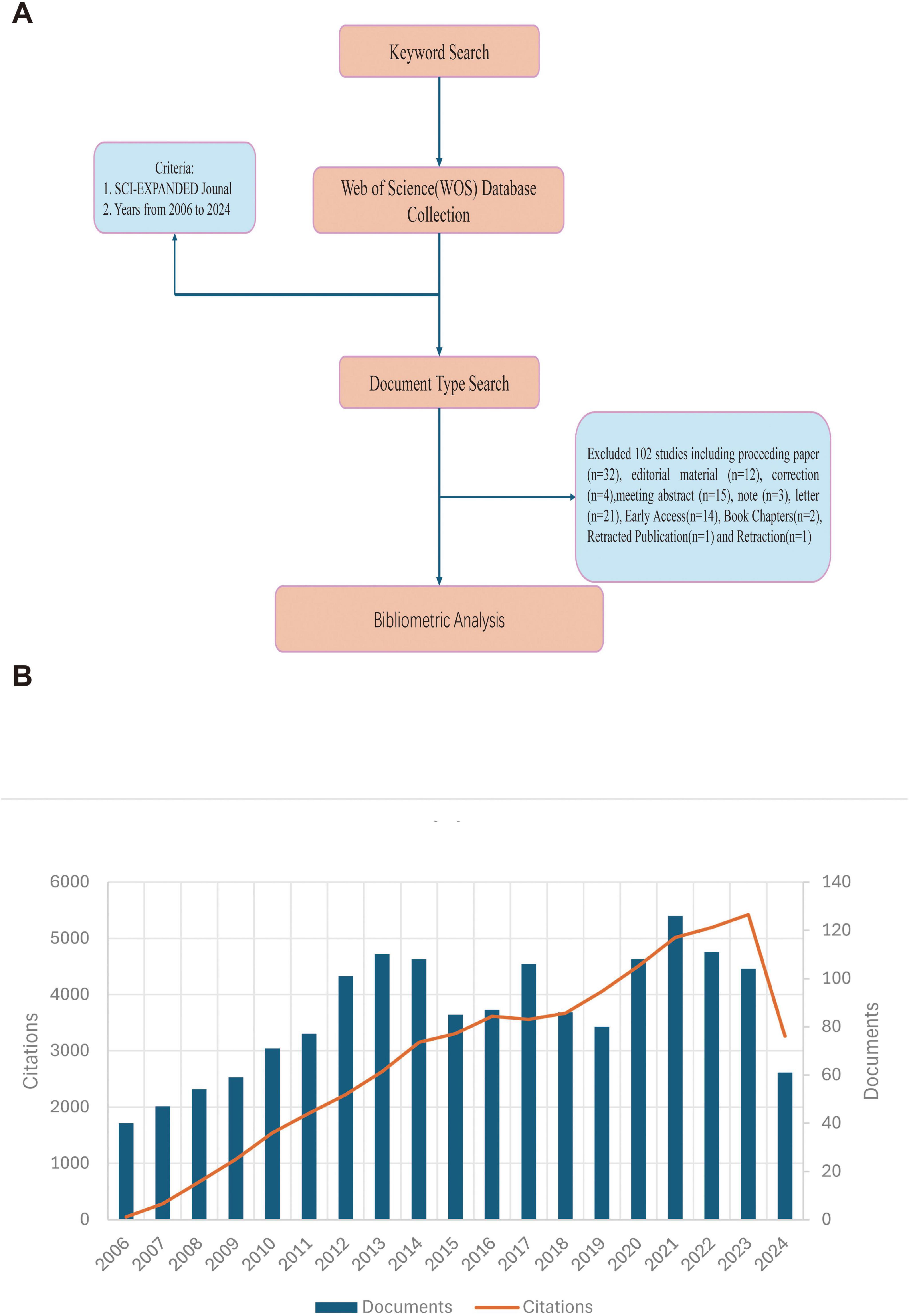
Figure 1. (A) A process flowchart. (B) The number of annual publications and citations of blood infection of MRSA research from 2006 to 2024 has been steadily increasing.
All valid data were imported into Microsoft Office Excel 2019, the R-based package Bibliometrix 4.3.3, VOSviewer (version 1.6.20) and CiteSpace (version 6.3.R1) for visualization and analysis.
VOSviewer is another bibliometric analysis software developed by Nees Jan van Eck and Ludo Waltman for web-based data construction and viewing of bibliometric maps, from which key information can be extracted from a wide range of publications (10) (van Eck and Waltman, 2010). It allows the creation of visual representations of bibliometric networks, including collaboration between countries and institutions. The different clusters are indicated by the color of the nodes, the number of publications by the size of the nodes, and the strength of the relationships by the thickness of the lines.
CiteSpace, developed by Professor Chaomei Chen, is a general-purpose JAVA-based program for bibliometric and visual analysis (11) (Chen, 2004). CiteSpace is used to perform cluster analysis, create bursts of citations and keywords, and timeline views. By categorizing the keywords and references, cluster analysis of keywords can reveal the key areas of MRSA blood infection. Silhouette >0.5 and modularity Q > 0.3 indicate that the clustering results are adequate and convincing. The clustering of keywords and references can be used to discover new trends in MRSA blood infection research.
Bibliometrix packages is a bibliometric analysis tool based on the R programming language for mathematical and statistical calculations of publication frequency, percentage and number of citations for each journal, country, etc.
According to our research strategy, a total of 1,621 publications related to blood infection of MRSA from 2006 to 2024 were obtained from the WoSCC database. Figure 1B illustrates the annual number of publications and citations for blood infection of MRSA research from 2006 to 2024. Thus, in 2006, the field of research was already well established. The steady increase in publications from 2006 to 2013 reflects the tremendous attention and strong interest in this area of research, but after 2014, there is an overall steady trend in the number of publications, suggesting that research on blood infection of MRSA continues to be hot but not explosive. The trend of increasing citations also suggests that research in this area is not only numerous but also far-reaching, continuing to attract scholars to conduct research related to blood infection of MRSA, and that more prospective research is needed in this area in the future, highlighting its global relevance.
A total of 111 countries and 2,726 institutions were involved in research on MRSA BSIs, and the 10 most prolific countries in terms of NP- and TC-based publications on MRSA BSIs were ranked (Table 1), with the United States leading the way in research on MRSA BSIs, with a total of 511 publications, followed by China (181) and Japan (74). The United States also had the highest total number of citations, highlighting its influence in this area of research. Multinational publications (MCP) describes the proportion of publications in a specific field that involve contributions from multiple countries, and it is used to evaluate the level of international collaboration of a country in a particular research domain. It is worth noting that Germany and the Netherlands have a high percentage of MCP (Figure 2), suggesting a significant contribution to international collaboration. Although the UK has fewer publications, it has very close research exchanges with other countries (Table 1).
A minimum number of articles of 13 was set to filter out the 30 countries that met the set parameters, and the close cooperation between the countries is shown in Figures 3A, B, which shows the extensive network of cooperation between the countries, with the United States, China, and the European countries forming the key hubs in the global research network. Figure 3B further confirms the prominence of these countries in terms of the number of collaborations and publications between them in this research area.
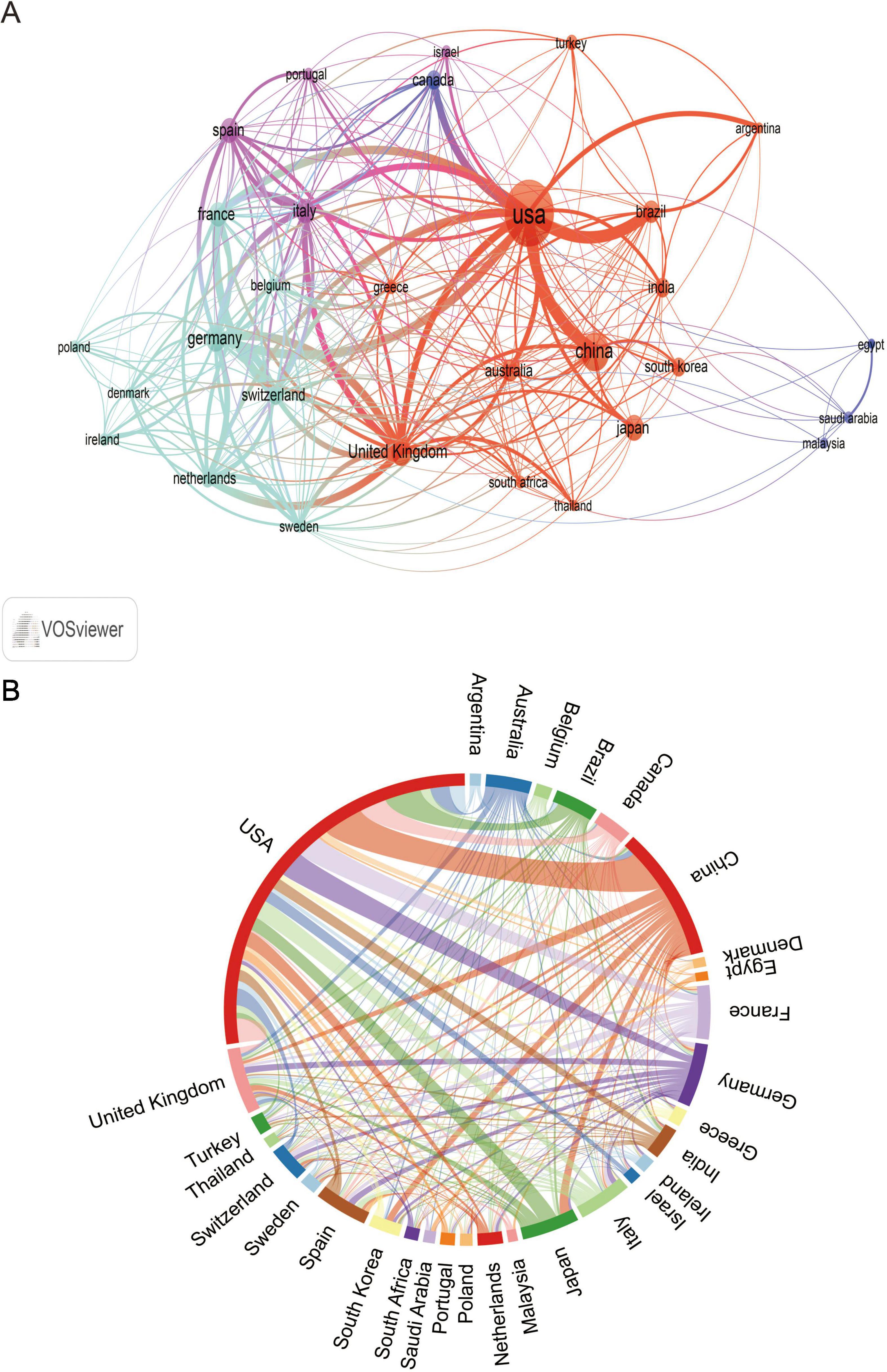
Figure 3. The analysis of countries related to blood infection of MRSA. (A) Co-authors–countries collaboration visualization in blood infection of MRSA. (B) The network map of cooperation between countries.
Among the top 10 institutions in terms of number of publications, the University of California system is the most productive institution with 116 publications and 3,217 total citations, followed by the University of Texas system (NP: 71) and Harvard University (NP: 66) (Table 2). Not only do these institutions contribute significantly in terms of publication volume, they are equally significant in terms of impact. Among them, although ASSISTANCE PUBLIQUE HOPITAUX PARIS (APHP) is only ranked fourth in terms of the number of publications (NC: 61), it is ranked second in terms of the total number of citations (TC: 2,782), which indicates that the quality and impact of its publications are very high.
Figure 4A illustrates the top 16 institutions with citation outbreaks, with Zhejiang University and New York University experiencing citation outbreaks in the recent past, suggesting that they have a lot of potential in the field of MRSA BSIs today. Co-occurrence graph (Figure 4B) Node size indicates co-occurrence frequency, while links indicate co-occurrence relationships. Nodes with purple rounded corners indicate high mediator centrality (≥0.1). The critical role of institutions with high mediator centrality (e.g., the University of California system) in connecting diverse research communities.
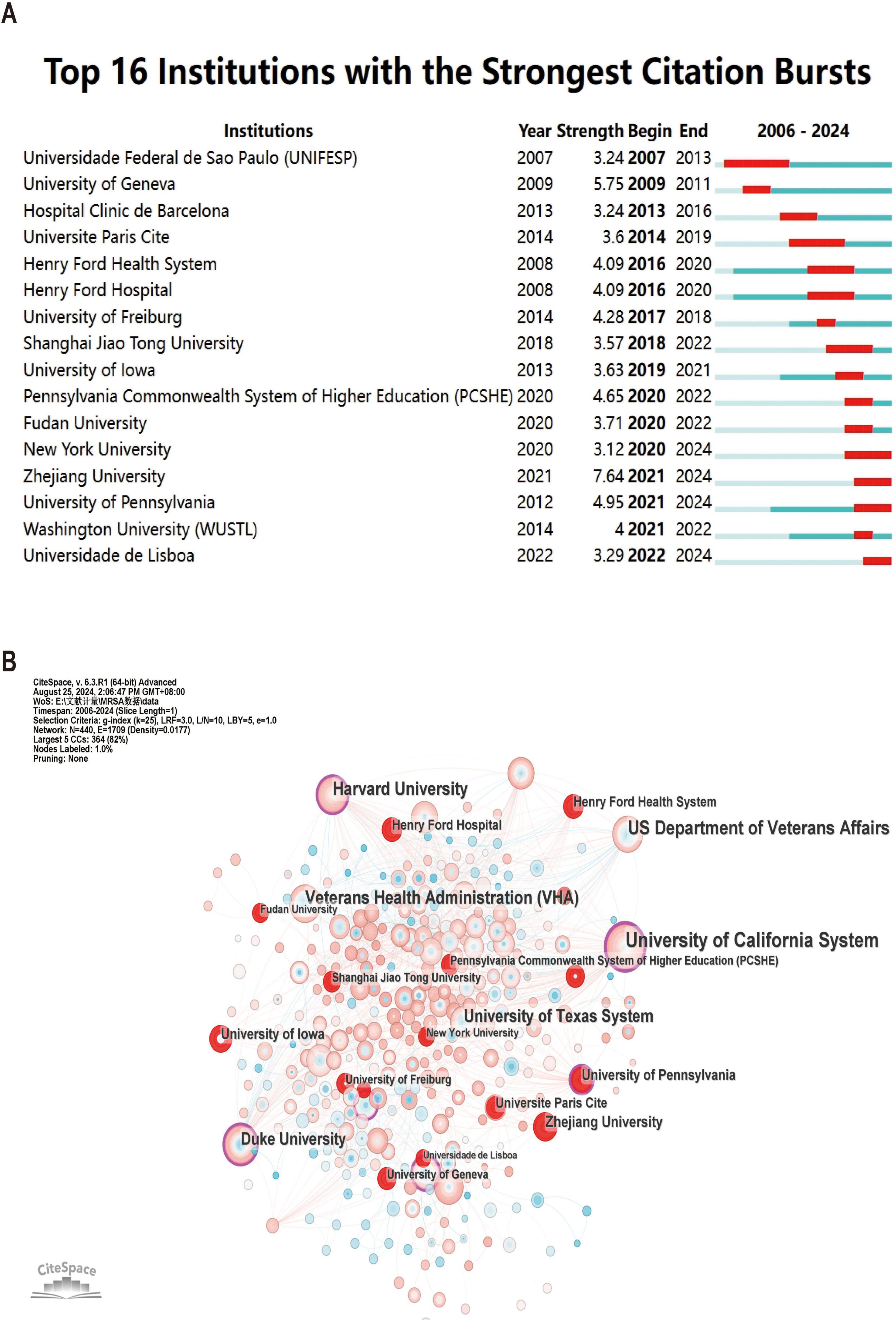
Figure 4. The analysis of research institutions related to blood infection of MRSA. (A) Co-authors–institute collaboration visualization in blood infection of MRSA. (B) The co-occurrence map of research institutions. The node size means the co-occurrence frequencies, while the linkages mean the co-occurrence relationships. Nodes with purple round mean high betweenness centrality (≥0.1).
To show active and influential journals on MRSA BSIs, a visual analysis of published journals was conducted. We found 1,621 publications related to MRSA BSIs published in 494 academic journals. Table 3 and Figure 5 show that the journal with the most publications was ANTIMICROBIAL AGENTS AND CHEMOTHERAPY (NC: 76), followed by INFECTION CONTROL AND HOSPITAL EPIDEMIOLOGY (NC: 55), and CLINICAL INFECTIOUS DISEASES (NC: 53). In addition, CLINICAL MICROBIOLOGY AND INFECTION has the highest impact factor (10.9) among the top 10 journals, but his impact in the field of MRSA BSIs is not exceptional.
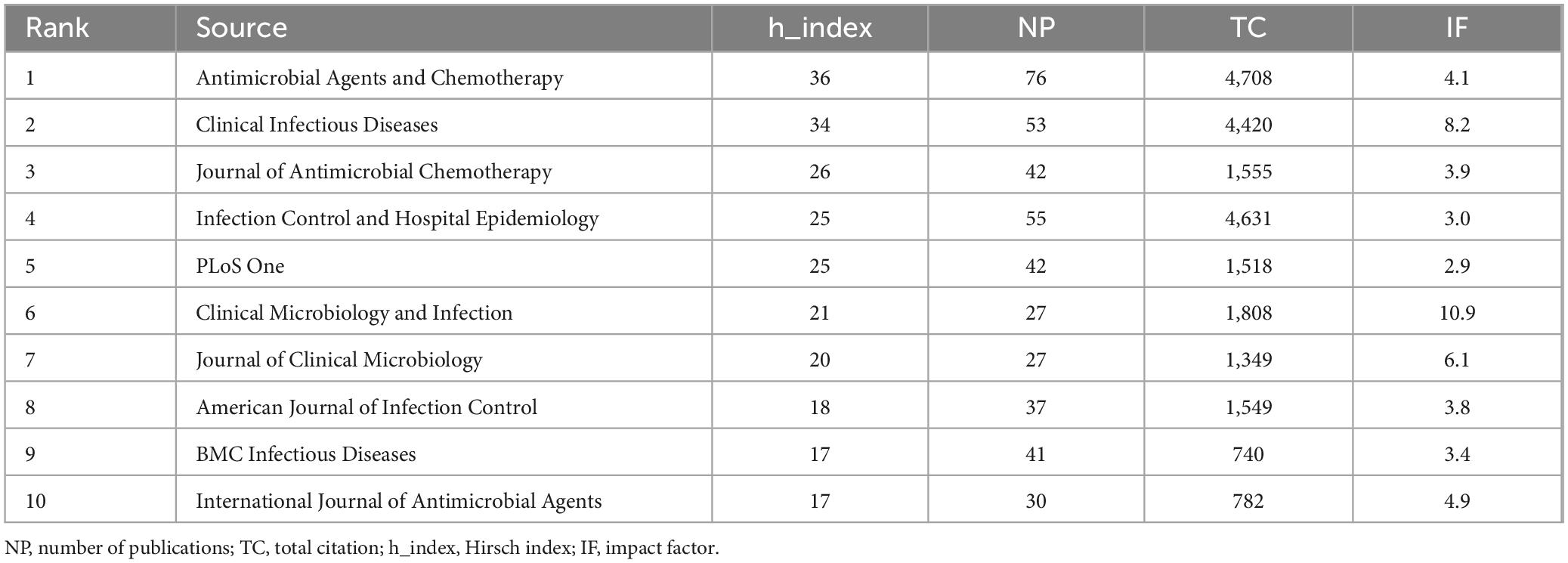
Table 3. The top 10 influential academic journals with publications concerning blood infection of MRSA.
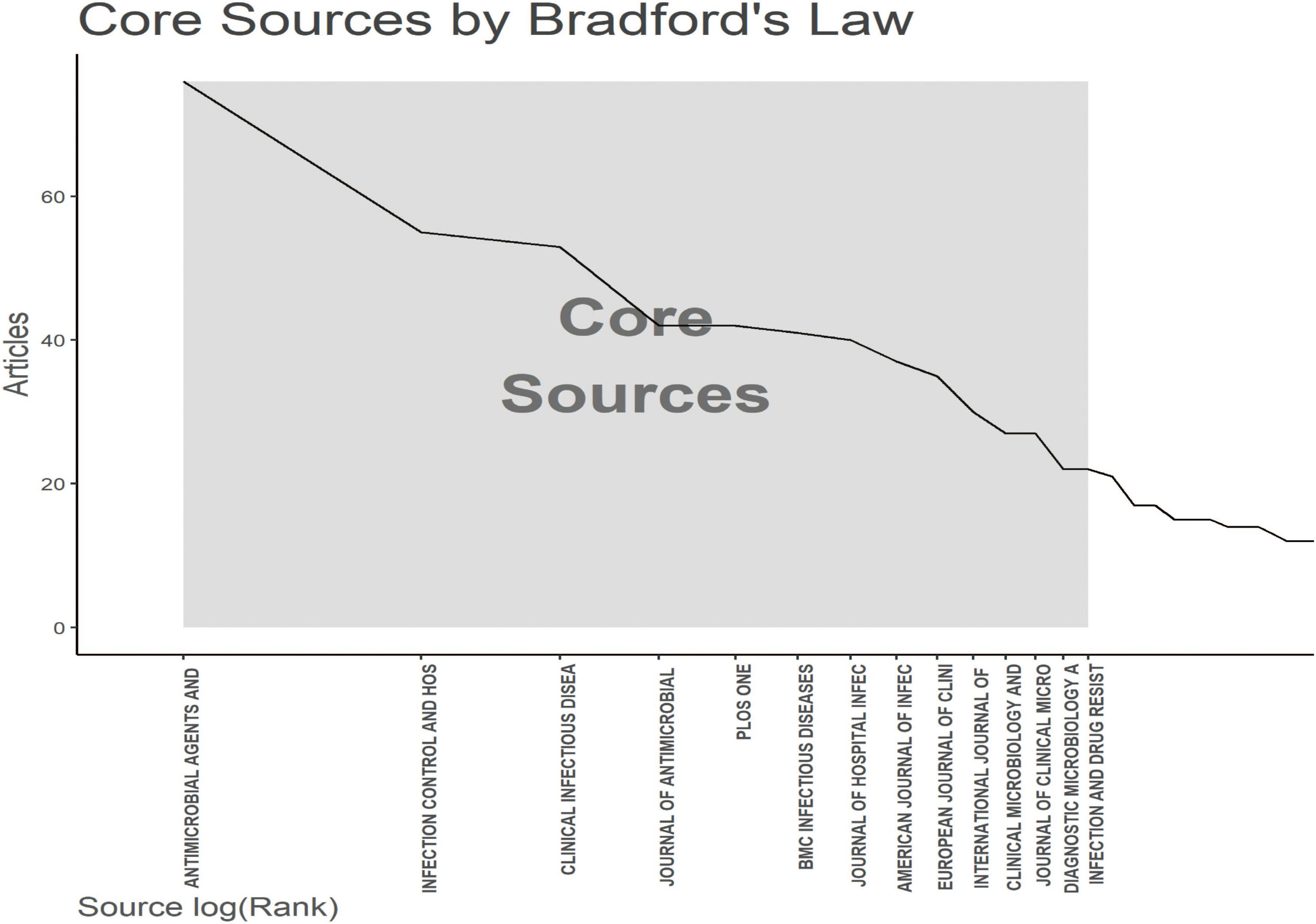
Figure 5. The analysis of academic journals related to blood infection of MRSA. Bradford’s law according to the academic journals.
Figure 5 applies Bradford’s law to show the core journals in MRSA BSIs research.
A total of 9,425 authors were involved in the study of MRSA BSIs. Table 4 shows the top 10 most relevant authors in MRSA BSI research. We can see that Fowler VG leads with 16 articles and 867 citations, followed by Rybak MJ with 17 articles and 507 citations. It is worth noting that Lodise TP has a small number of articles but the largest number of citations, which shows that his articles are very influential in the field. The interconnectedness of these authors is further illustrated by the co-occurrence diagram in Figure 6, showing the collaborative network between authors that provides expert information for finding research partners, with the 21 colors representing the 21 clusters in Figure 6. Fowler, Vance G. and Rybak, Michael J. are at the heart of the collaborative network. Authors actively collaborate on MRSA BSIs, especially between authors in the same cluster, e.g. Kaye, Keith S. and Rybak, Michael J. Close co-operation between authors from different clusters can also be observed, e.g., Fowler, Vance G. and Kern, Winfried V.
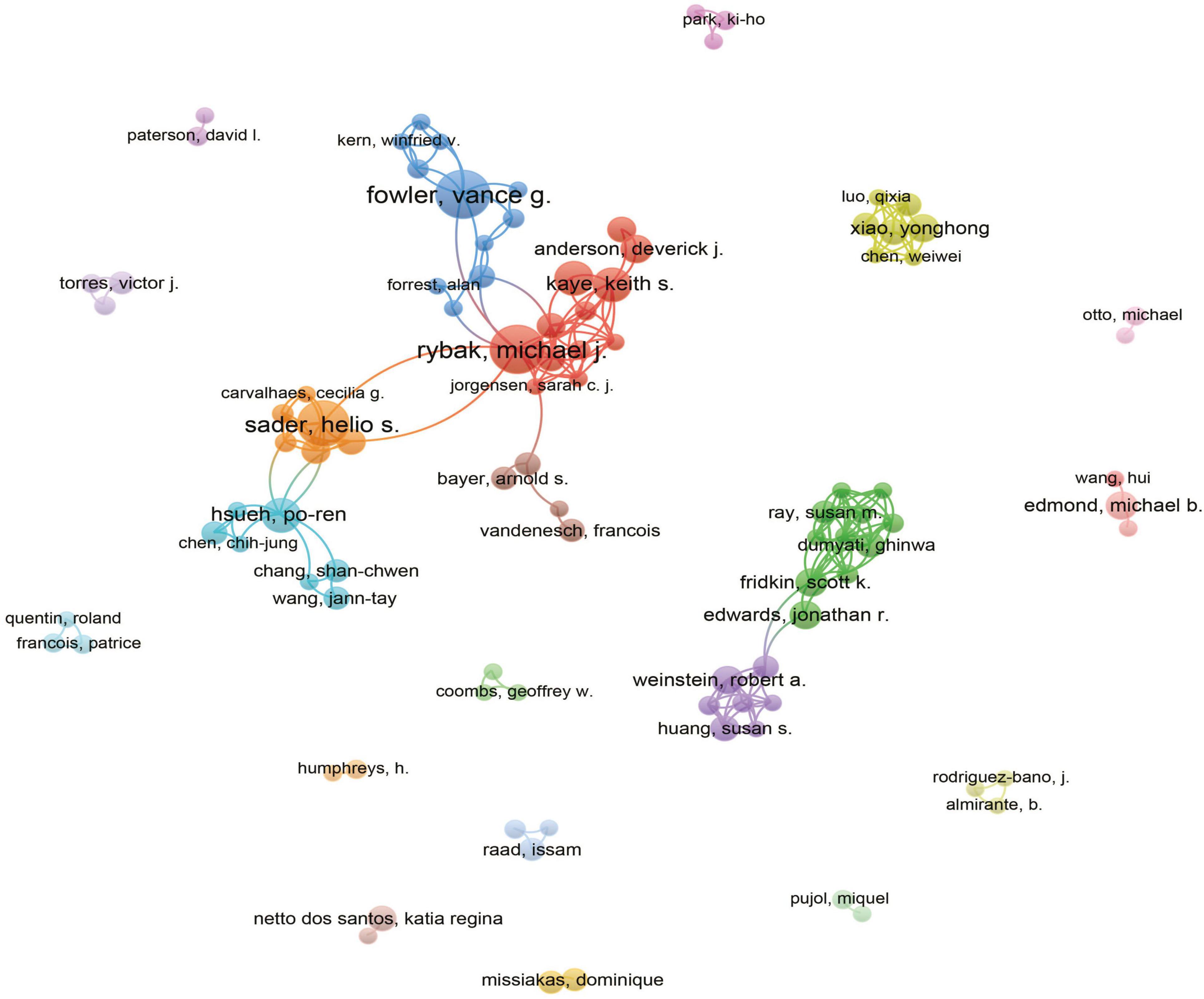
Figure 6. The analysis of authors related to blood infection of MRSA. The co-occurrence authors’ map of blood infection of MRSA research. The varied colored nodes 766 reflect the authors in various clusters. The node size means the co-occurrence frequencies, while the 767 linkages mean the co-occurrence relationships between authors.
Table 5 shows the top 10 most cited articles on MRSA BSIs, with the top 4 articles having more than 1,000 citations. The most cited article globally was Vincent JL’s article “Sepsis in European intensive care units: results of the SOAP study” published in Critical Care Medicine in 2006 with 1,954 citations (Vincent et al., 2006). In this article, MRSA was firstly introduced as one of the important pathogens leading to sepsis, which laid the foundation for subsequent studies on MRSA BSIs.
The co-cited references (Table 6) highlight the foundational studies by authors such as Cosgrove et al. (2003) and Liu et al. (2011), which are often cited together, demonstrating their important role in the literature. Table 6 shows the top 10 co-cited papers related to MRSA BSIs and shows the number of times each paper was cited. Of these, four articles were cited more than 100 times. The most cited article was COSGROVE SE’s “Comparison of Mortality Associated with Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Bacteremia: a Meta-analysis,” published in the journal Clinical Infectious Diseases in 2003 (Cosgrove et al., 2003). This article introduced MRSA BSIs and laid the foundation for subsequent research on bacteremia. Liu et al. (2011) published in the journal Clinical Infectious Diseases “Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections in Adults and Children,” was cited 125 times.
Figure 7A illustrates the 25 core literatures with a significant increase in citation frequency, reflecting the significant impact and cutting edge of these literatures on the relevant research field in a given time period. Earlier literature, such as Cosgrove et al. (2003) and Cardo (2004), experienced a strong citation explosion between 2006 and 2010, suggesting that they featured prominently in the topics of nosocomial infection control, antimicrobial drug use, and so on, during that period. These literatures laid the foundation for research on antimicrobial resistance and infection prevention, providing a theoretical framework and empirical support for subsequent studies.
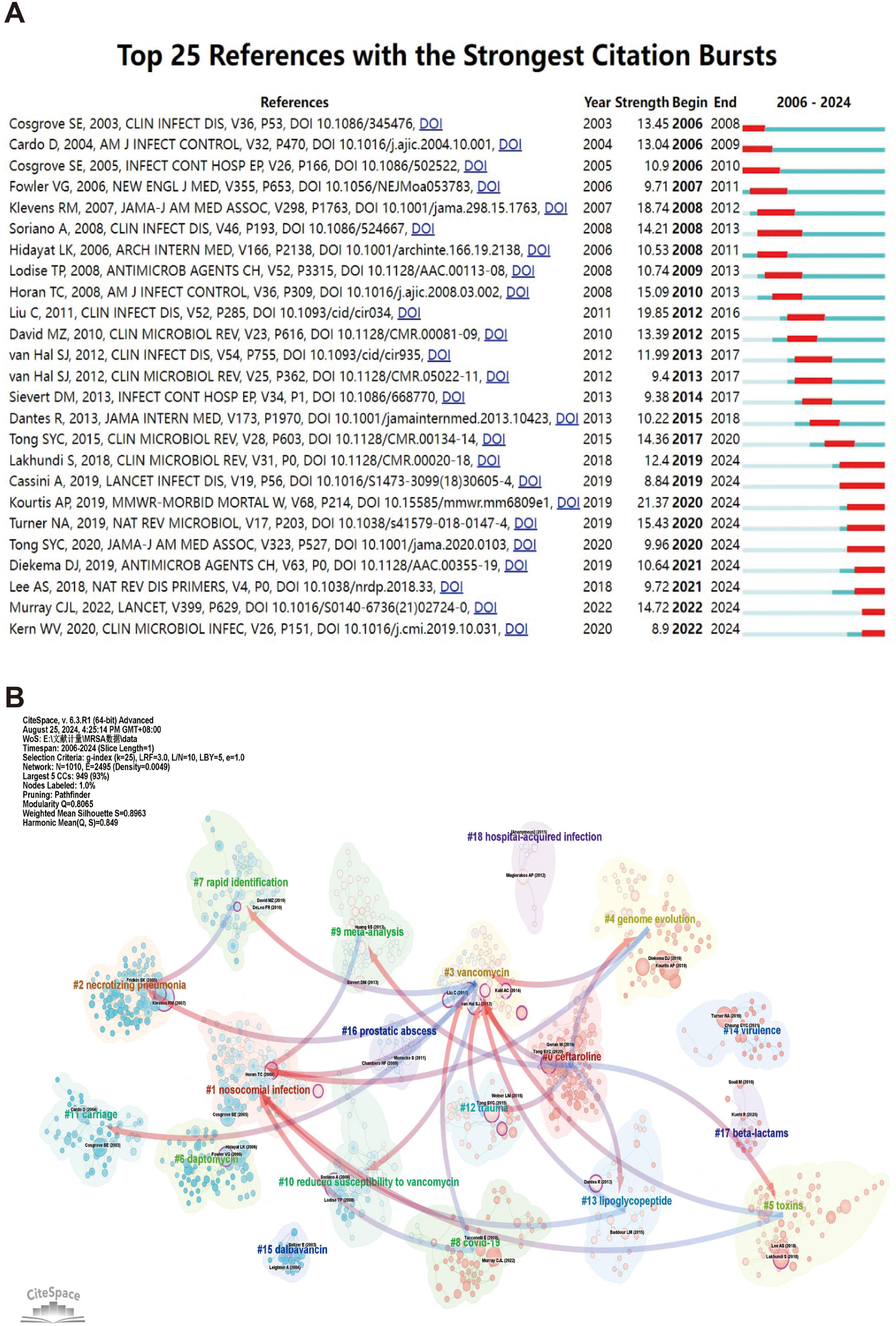
Figure 7. The analysis of references related to blood infection of MRSA. (A) The top 25 references with strong citation bursts. A red bar means high citations in the corresponding year. (B) Clustering of references based on the similarity between references, including #0 ceftaroline, #1 nosocomial infection, #2 necrotizing pneumonia, #3 vancomycin, #4 genome evolution, #5 toxins, #6 daptomycin, #7 rapid identification, #8 COVID-19, and so on. While linkages refer to the connections that develop between different clusters, and the blue groups on the line evolve from the red ones.
As the study continued, subsequent literature, such as Liu et al. (2011) and David and Daum (2010), saw a significant increase in citation frequency between 2012 and 2016. This period coincided with a surge in global interest in drug-resistant microbial infections and antimicrobial stewardship, and Liu et al.’s (2011) study has had a profound impact on the clinical infection field, providing valuable insights into the rational use of antibiotics and mechanisms of resistance, while David and Daum’s (2010) study has further explored the link between community-acquired and hospital-acquired infections, advancing the field of microbial drug resistance. David and Daum’s (2010) study further explored the link between community-acquired infections and hospital-acquired infections and contributed to the in-depth research in the field of microbial resistance.
In recent years, some of the more recent literature has begun to spark a sustained citation explosion post-2020 as the problem of emerging infectious diseases and antibiotic resistance intensifies. For example, literature such as Kourtis et al. (2019) and Turner et al. (2019) experienced significant citation growth between 2020 and 2024, demonstrating its importance in the context of the COVID-19 pandemic. Kourtis et al.’s (2019) research on emerging infectious diseases provided key insights into outbreak response strategies, while Turner et al.’s (2019) work on provide insights into the mechanisms of transmission of drug-resistant bacteria. In addition, the study by Murray et al. (2022) reflects changes in the global health burden during an outbreak, and its findings are widely used in current public health decision-making, making it a cutting-edge literature in the field.
The use of co-citation cluster analysis provides an objective indication of the knowledge structure of the research area. To further describe the co-cited reference groups, we created a network diagram. The degree of association between articles is divided into 18 categories, which are the basis of the cluster classification. In the diagram, the co-citation cluster analysis clearly reveals the knowledge structure of the research area. In order to fully describe the co-cited literature groups, we constructed a complex network diagram (shown in Figure 7B). Research topics were classified into 18 categories based on co-citation relationships, forming a clear cluster structure. One nosocomial infection is the largest cluster in the diagram, indicating that nosocomial infection-related research occupies a central position in the field and has broad academic influence. This cluster is closely linked to three vancomycin by multiple connecting lines, suggesting that studies of vancomycin treatment and nosocomial infections co-occur frequently in the literature, demonstrating a strong correlation between the two.
Evolution between the study clusters over time is also evident. For example, eight the COVID-19 cohort evolved gradually from studies of the one nosocomial infection and three vancomycin cohorts, and nosocomial infection and antibiotic resistance studies have expanded further in the neo-crowning context, especially with the global epidemic of COVID-19. This demonstrates the intersection and correlation between emerging infectious diseases and traditional antibiotic resistance research. In addition, 10 reduced susceptibility to vancomycin groups also evolved from red groups (e.g., three vancomycin), and this evolution reflects the further refinement of antimicrobial drug resistance research, which has gradually shifted from the use of vancomycin to an in-depth exploration of its resistance mechanisms.
In addition, four genome evolution groups have gradually diverged from those associated with antimicrobial drug use (e.g., three vancomycin), evolving into the research field of pathogen genome evolution. This trend clearly indicates that with advances in genomics technology, researchers have begun to pay more attention to the mechanisms of genetic evolution of pathogens and have attempted to better understand changes in drug resistance and infectivity through genomic analyses.
In addition to these major evolutionary trends, a number of smaller groups reflect the continued development of specific research directions. For example, the 13 lipoglycopeptide group has evolved to target studies of resistance mechanisms to specific antimicrobial drugs through its close association with three vancomycin. This suggests that research is not only focusing on widely used antibiotics, but is also progressively expanding toward refined antibiotic class resistance.
Usually, keywords reflect the research topic and content of an article. By analyzing keywords, we can quickly understand the research trend in a certain field. We listed four common keywords for MRSA: infections, blood stream infections, methicillin resistance, and Staphylococcus aureus. These four main keywords were focused on the mechanism of MRSA resistance, the transmission route of the infection, and the prevention and treatment of related diseases.
Subsequently, we performed keyword cluster analysis and obtained eight clusters (Figure 8). These eight clusters are as follows: #0 strains, #1 nosocomial infections, #2 nasal carriage, #3 daptomycin, #4 mortality, #5 dalbavancin, #6 antimicrobial resistance, #7 septic shock. #0 strains is the largest cluster, and the main keywords in the field are strains and infections.
The heatmap analysis in Figure 9A details the popularity of specific keywords across years, revealing the evolution of research topics over time. The color shades in the heatmap reflect the research intensity of the keyword, with red areas indicating high research intensity for the keyword in a given year and purple areas showing relatively low research activity. Keywords such as “antibiotic resistance,” “antimicrobial stewardship,” and “blood culture” are shown. “Blood culture” have come to the forefront of research in recent years. This suggests that as clinical needs change, research is expanding from the issue of antibiotic resistance to how to effectively respond to infections through antimicrobial stewardship and new diagnostic approaches.
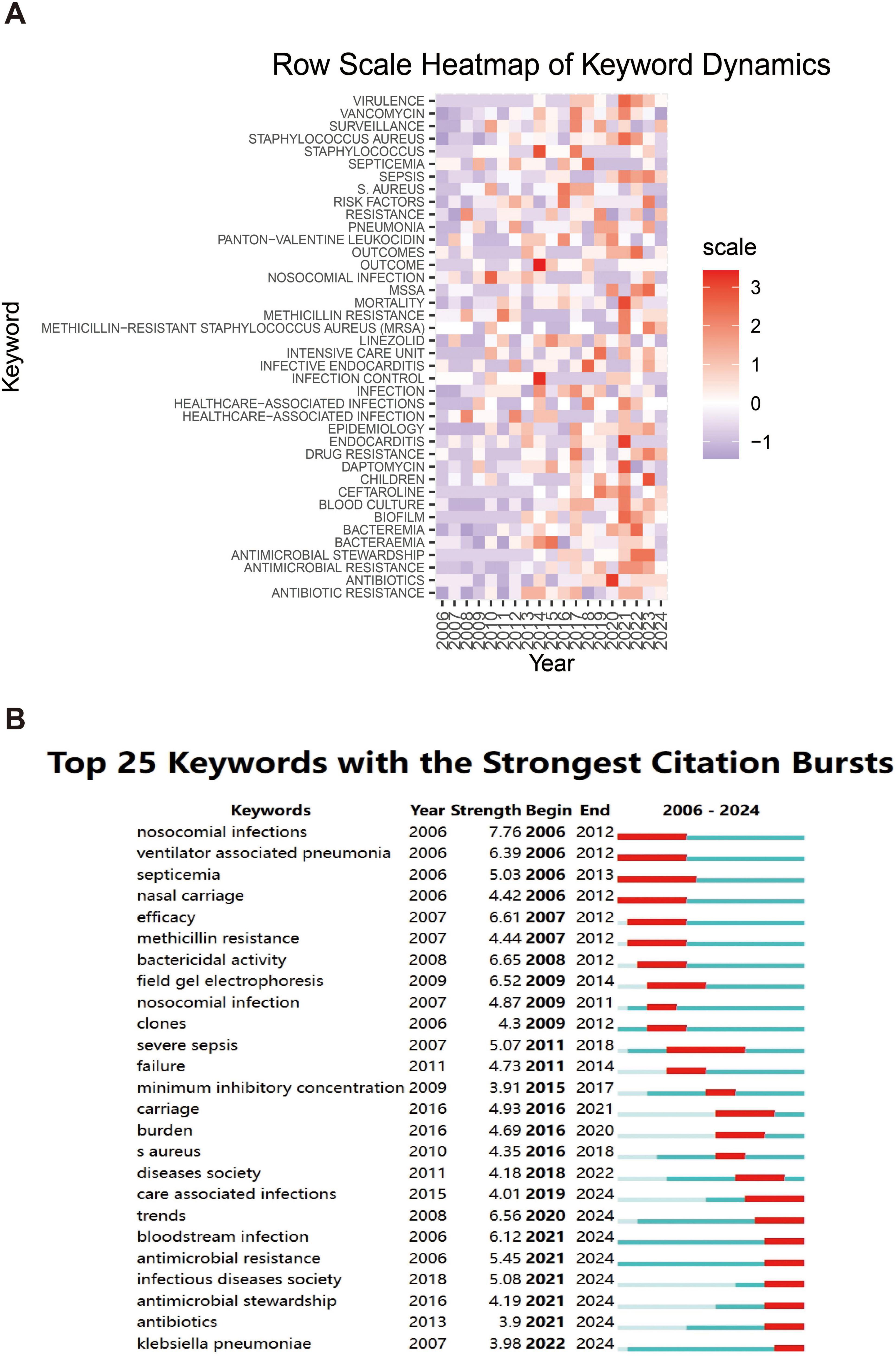
Figure 9. Heatmap analysis of keywords. (A) The annual heatmap related to blood infection of MRSA research. The annual popularity of the keyword is measured as the standardized frequency of a keyword’s occurrence in a given year. (B) The top 25 keywords with the strongest citation bursts.
Figure 9B further demonstrates the dynamics of research hotspots through the citation bursts of keywords. Keywords with strong citation bursts indicate an explosive growth of research in these areas over a specific time period. The figure shows that keywords such as, “antibiotics,” “antimicrobial resistance,” and “care associated infections” (care-associated infections) show a strong citation burst between 2021 and 2024, with research related to COVID-19 in particular further driving the intensity of research on these keywords. In addition, keywords related to “MRSA” BSIs, such as “methicillin resistance” and “clones” (clones), showed a high citation intensity between 2006 and 2012, indicating that the genome evolution study of MRSA attracted widespread attention at that time.
An overall analysis of the two graphs shows that the focus of infectious disease research is gradually expanding from the traditional pathogen resistance to more complex clinical management and diagnostic techniques, particularly driven by the recent outbreaks, where researchers have shown a great deal of interest in how to control nosocomial infections and the spread of drug-resistant strains.
From 2006 to 2024, research on MRSA BSIs has grown significantly. This growth reflects the global academic community’s high level of concern over MRSA bacteremia as a serious public health issue (Hassoun et al., 2017). MRSA bacteremia, defined as BSIs caused by MRSA, has extremely high morbidity and mortality rates (Jaganath et al., 2018), particularly in immunocompromised patients. Globally, the United States, China, and European countries are the primary contributors to this field, showcasing their research strengths in addressing the global threat posed by MRSA bacteremia. The United States undoubtedly leads in terms of research output in this area, with 511 related publications, demonstrating not only its vast research scale but also its global influence in MRSA BSI research. At the same time, China and Japan contributed 181 and 74 papers, respectively, indicating the growing importance of Asian countries in this field, particularly in their rapid rise in studying MRSA bacteremia.
Compared to past research on antimicrobial resistance, the global distribution trend in MRSA BSI research is similar to that of other BSIs caused by antibiotic-resistant pathogens. North America and Europe have historically been the major drivers of antibiotic resistance research, with their research institutions and academic journals playing crucial roles in global scientific progress. Key institutions and journals, such as the University of California system and Antimicrobial Agents and Chemotherapy, have made significant contributions to advancing MRSA bacteremia research. Their abundance of research resources and investment in public health (Fan et al., 2017) have enabled them to maintain a leading position in combating MRSA bacteremia and other antibiotic-resistant BSIs.
From bibliometric analysis, European countries like Germany and the Netherlands stood out in international collaboration, particularly with their high percentage of multi-country publications. The United Kingdom, although producing fewer publications, maintained a close research network with other countries, underscoring its importance in advancing global MRSA bacteremia research. These international collaborations not only help in the dissemination and sharing of knowledge but also strengthen the synergy among countries in effectively addressing the global challenge of MRSA BSIs.
Regarding research institutions, top global institutions have been particularly prominent in both output and influence. For example, the University of California system’s significant contribution to MRSA bacteremia research is reflected in its 116 publications and 3,217 total citations, ranking first in both metrics. The University of Texas system and Harvard University also performed exceptionally in publication volume and citation impact, indicating their pivotal role in advancing research. However, in recent years, as Asian countries have risen in research capabilities, particularly China’s Zhejiang University, which has seen rapid growth in MRSA bacteremia research, these countries have begun to take a more dominant role in the field. Although there is strong cooperation among research institutions within the same country, particularly among U.S. institutions, cross-national collaboration remains relatively limited. This lack of collaboration might hinder the overall progress in MRSA BSI research. Therefore, we strongly recommend strengthening international collaborations between research institutions from different countries to accelerate global research progress on MRSA BSIs.
Compared to other research on antimicrobial infections, studies on MRSA bacteremia are not only concentrated in developed countries but also show an increasing trend of global collaboration. With advancements in technology and resource sharing, more research outputs from low- and middle-income countries are expected to emerge in the future. These countries often face a higher risk of MRSA BSIs (Mohamad Farook et al., 2022), yet research resources are relatively scarce. Strengthening global scientific cooperation and resource sharing can promote MRSA BSI research in these regions and provide a more diverse perspective and data support for the global fight against MRSA bacteremia.
In terms of journals, Antimicrobial Agents and Chemotherapy has published the most studies on MRSA BSIs, far exceeding other academic journals. Additionally, Infection Control and Hospital Epidemiology also leads in both publication and citation counts in this field, demonstrating its significant influence in MRSA BSI research. We also found that most of the top 10 journals are focused on infection and antimicrobial fields, such as the Journal of Antimicrobial Chemotherapy and Infection Control and Hospital Epidemiology. This indicates that current research is largely centered on clinical treatment and infection control (Köck et al., 2010; Kullar et al., 2016), which holds substantial practical significance for improving the diagnosis and treatment of MRSA bacteremia.
From the perspective of authors, Fowler VG from the United States has published the most articles on MRSA BSIs. Rybak MJ, also from the United States, ranks first among co-authors, highlighting his prominent position in this research field. Lodise TP, another American author, has the most citations in this field, exploring various aspects of MRSA’s multidrug resistance and treatment strategies. He and his team have also summarized research related to the epidemiology (Tong et al., 2020; Turner et al., 2019), transmission (David et al., 2014), genetic diversity (Kanafani and Fowler, 2006), as well as monitoring and treatment (Holland et al., 2014) of MRSA BSIs. They have emphasized that although the incidence of MRSA has recently declined in some regions, MRSA still poses a formidable clinical threat (Rasmussen et al., 2011), with morbidity and mortality rates remaining persistently high.
One of the key roles of bibliometrics is to process and analyze large amounts of data, providing researchers with information about research trends (Xiao et al., 2017). Analyzing frequently occurring keywords can reveal changing trends and prominent themes, which are important for understanding the evolution of this academic field. MRSA BSIs are a major challenge for global health systems. They are typically associated with high mortality, high hospitalization rates, and complex treatment plans (Alhurayri et al., 2021; Datta and Huang, 2008). As a result, current research hotspots focus on several key areas, including antibiotic resistance mechanisms, new diagnostic tools, hospital infection control strategies, and the impact of COVID-19 on MRSA bacteremia. In-depth analysis of these research hotspots helps better understand the progress in MRSA BSI research and predicts future developments.
Antibiotic resistance is a central challenge in the treatment of MRSA bacteremia. With the increasing resistance of MRSA to methicillin and other commonly used antibiotics (such as vancomycin), treating MRSA BSIs has become increasingly complex (Hassoun et al., 2017). Current research focuses on understanding the resistance mechanisms in MRSA bacteremia and developing new treatment options (Holubar et al., 2020). In recent years, with advances in genomics, researchers have utilized WGS to analyze genetic variations and resistance genes in MRSA strains. These studies provide valuable references for personalized treatment of MRSA bacteremia.
Studies have shown that MRSA strains have developed increasing resistance to vancomycin, prompting researchers to explore alternative therapies and combination therapies for treating MRSA BSIs (Li et al., 2023; Rose et al., 2021). For example, new antibiotics such as linezolid (Dodds and Hawke, 2009) and daptomycin (Adamu et al., 2024) have shown high efficacy in treating MRSA bacteremia. However, as these drugs are increasingly used, resistance is gradually emerging. Future research will likely focus on exploring more effective combinations of antibiotics or other alternative therapies to maximize treatment outcomes and slow the development of resistance.
In addition to hospital-acquired MRSA (HA-MRSA), the rapid spread of community-acquired MRSA (CA-MRSA) strains has exacerbated the public health challenge of MRSA bacteremia (Del Giudice, 2020). These new MRSA strains pose infection risks even to healthy individuals, further complicating the global issue of antibiotic resistance. In response, researchers are actively developing new treatment strategies to address these emerging strains and reduce the overuse of antibiotics.
Early diagnosis of MRSA bacteremia is crucial for effective treatment. Traditional bacterial culture methods are time-consuming and typically take several days, which can lead to treatment delays and increased mortality risk. Therefore, researchers are developing faster and more accurate diagnostic tools that can identify MRSA bacteremia early and guide treatment.
Molecular diagnostic tools based on PCR technology have been widely used in recent years, capable of detecting MRSA strains (Motoshima et al., 2010) within hours. However, advancements in next-generation sequencing (NGS) and genotyping have significantly improved diagnostic efficiency and accuracy (Miao et al., 2017). These new diagnostic tools can quickly identify MRSA BSIs and determine their resistance genotypes, helping physicians formulate more precise treatment plans.
Artificial intelligence is also being introduced into the diagnosis of MRSA BSIs (Gammel et al., 2021). These technologies analyze vast amounts of genomic and clinical data, enabling rapid detection of patterns associated with MRSA bacteremia and improving the speed and accuracy of diagnosis. The application of AI has great potential in enhancing infection control and personalized treatment, and future AI-driven diagnostic technologies are expected to significantly improve the early detection and treatment of MRSA bacteremia.
Hospital-acquired MRSA bacteremia remains a significant challenge in global hospital systems, especially in high-risk areas such as ICUs. To reduce the spread of MRSA in hospital environments, researchers have proposed a series of control strategies, including universal screening, patient isolation, hand hygiene management, and environmental disinfection.
Decolonization treatments (e.g., using mupirocin) have been shown to be effective (Poyraz et al., 2022) in reducing the nasal carriage of MRSA strains in high-risk patients. However, long-term use of these drugs may lead to resistance, so future research needs to balance the effectiveness of decolonization treatments with the risk of resistance. Additionally, hospital management policies play a critical role in reducing the transmission of MRSA bacteremia. Studies have shown that implementing strict infection control policies can effectively lower MRSA infection rates (Bertrand et al., 2012) in hospitals.
The COVID-19 pandemic has exacerbated the risk of bacterial infections in hospital environments, particularly the high incidence of MRSA bacteremia in severe COVID-19 patients. This has drawn researchers’ attention to the management (Tabah and Laupland, 2022) of bacterial and viral co-infections. COVID-19 patients, due to their compromised immune systems (Jeon et al., 2022), are more susceptible to bacterial co-infections, with MRSA bacteremia being a common complication, further complicating treatment.
Research on co-infections of COVID-19 and MRSA has become one of the current hotspots. Studies show that the widespread use of antibiotics during the pandemic has exacerbated the problem of resistance (Jeon et al., 2022), raising the bar for future hospital infection control measures. By deeply studying co-infections of COVID-19 and MRSA, scientists will be better prepared to manage similar co-infection issues in future pandemics and improve current infection control strategies.
In the future, the application of genomics technology (Dyzenhaus et al., 2023) will continue to drive the progress of MRSA bacteremia research. WGS provides researchers with the opportunity to deeply understand the evolution and resistance development of MRSA strains. By analyzing the genomic data of MRSA strains, researchers can identify key gene mutations (Lindsay, 2013) that lead to resistance and predict the emergence of resistant strains. This information is crucial for developing new treatment strategies, especially for personalized treatment of MRSA bacteremia patients.
In personalized medicine, future research will focus on using genomic data to develop more precise antibiotic treatment plans. By conducting genomic analysis, physicians can select the most appropriate combination of antibiotics based on the specific resistance genes present in the MRSA strain infecting the patient, avoiding the overuse of broad-spectrum antibiotics (Hassoun et al., 2017). Genomics-driven precision treatment not only improves therapeutic efficacy but also helps slow the development of resistance, which is critical for patients with severe infections (Rose et al., 2021).
Moreover, as the cost of genomic sequencing technology (van Belkum and Rochas, 2018) continues to decline, these personalized medical techniques will become increasingly integrated into clinical practice. Physicians will be able to quickly obtain the genomic information of MRSA strains infecting patients and use this data to develop individualized treatment plans. This precision medicine will play a core role in the future management of MRSA bacteremia, helping reduce patient mortality and complications.
Artificial intelligence and ML (Nigo et al., 2024) will play an increasingly important role in the research and clinical management of MRSA bacteremia. These technologies process vast amounts of genomic and clinical information (Lu et al., 2021), accelerating the early diagnosis of MRSA infections and the prediction of antibiotic resistance. For example, AI algorithms can analyze a patient’s genomic information and medical history to predict their risk of developing MRSA bacteremia and provide personalized treatment suggestions based on the predictions.
In diagnostics, AI can automate the processing of large amounts of laboratory data, quickly identifying MRSA bacteremia patients and providing accurate diagnostic results. AI can also analyze historical case data to predict the evolutionary trends of MRSA strains and the possible future emergence of resistant strains. This predictive ability can help healthcare institutions better manage the use of antibiotics and prevent the spread of resistant strains (Lin et al., 2023; Lu et al., 2021).
Artificial intelligence will also play a role in drug development. By analyzing mutation patterns in MRSA strains, AI can help researchers identify potential new drug targets and accelerate the development of novel antibiotics. This data-driven research approach will significantly improve the efficiency of diagnosing and treating MRSA bacteremia, especially as antibiotic effectiveness declines. The integration of AI will provide strong support for combating resistance (Bueschbell et al., 2022; Liu et al., 2024).
Phage therapy (Strathdee et al., 2023) is an emerging research direction for combating MRSA bacteremia. Bacteriophages are viruses that specifically infect and kill bacteria. They have a high specificity for MRSA strains, making them a promising alternative to antibiotics, particularly in the context of increasing antibiotic resistance. Phage therapy has shown great potential in the treatment of MRSA bacteremia (Plumet et al., 2022), as it can selectively kill MRSA strains without disrupting the body’s normal microbiota.
Although phage therapy has demonstrated good results in laboratory studies, it still faces some challenges in clinical applications. Future research will focus on optimizing the safety and efficacy of phage therapy and exploring its potential combination with traditional antibiotics. For example, by combining phages with existing antibiotics, researchers hope to enhance the treatment effectiveness for MRSA bacteremia while reducing antibiotic use, thus delaying the development of resistance.
Additionally, phage therapy has potential in preventing MRSA infections. Future research may develop phage sprays or coatings to reduce the risk of MRSA transmission in hospital environments. By combining phage therapy with existing infection control measures, future strategies for the prevention and treatment of MRSA bacteremia will become more diverse and effective.
The development of MRSA vaccines (Chand et al., 2023) is a key area in the future fight against MRSA bacteremia. Although there is currently no widely available MRSA vaccine, the successful application of mRNA vaccine technology has made the development prospects for MRSA vaccines more optimistic. Researchers are working to develop vaccines that can elicit long-lasting immune responses to prevent the occurrence of MRSA bacteremia, particularly in immunocompromised patients.
A successful MRSA vaccine would help reduce the incidence of MRSA BSIs and lower the reliance on antibiotics, thereby curbing the spread of antibiotic resistance globally (Lindsay, 2007). The use of MRSA vaccines would be especially beneficial for long-term hospitalized patients and high-risk populations, significantly reducing their risk of developing MRSA bacteremia. Future research will focus on optimizing vaccine efficacy and ensuring that it can combat the diversity and rapid evolution of MRSA strains.
With continuous advancements in vaccine technology, MRSA vaccines are expected to enter clinical trials in the coming years and eventually gain approval (Miller et al., 2020). This would be a significant breakthrough in the global fight against MRSA bacteremia, not only significantly reducing infection rates but also effectively controlling antibiotic misuse and delaying the development of resistance.
While this study provides valuable insights into the research trends and future directions of MRSA BSIs, it has several limitations. First, the data used in this bibliometric analysis were sourced solely from the WoSCC, potentially omitting relevant publications indexed in other databases such as Scopus or PubMed. This may lead to an incomplete representation of the global research landscape on MRSA BSIs.
Methicillin-resistant Staphylococcus aureus BSIs remain a significant global public health challenge, particularly in the context of increasing antibiotic resistance. Despite notable advances in antimicrobial therapy, rapid diagnostic techniques, and emerging therapies such as phage therapy and vaccine development, further clinical validation and optimization are still needed. Future research directions will increasingly rely on the integration of genomics, AI, and ML to enhance the precision and effectiveness of personalized treatments.
Global collaboration will be a key driver of MRSA BSI research, especially in low- and middle-income countries, where the risk of infection is higher but research resources are relatively limited. By strengthening global cooperation and promoting the sharing of scientific resources, there is hope for effectively curbing the spread of MRSA BSIs worldwide and improving the ability to tackle antibiotic resistance.
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding author.
J-YL: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. J-KL: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. J-YChen: Formal analysis, Methodology, Writing – original draft, Writing – review & editing. J-YCai: Formal analysis, Methodology, Writing – original draft. Z-DY: Investigation, Writing – review & editing. L-QY: Investigation, Writing – review & editing. Z-TZ: Investigation, Writing – review & editing. X-GG: Project administration, Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors acknowledge all the colleagues who helped me in this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1516584/full#supplementary-material
Adamu, Y., Puig-Asensio, M., Dabo, B., and Schweizer, M. L. (2024). Comparative effectiveness of daptomycin versus vancomycin among patients with methicillin-resistant Staphylococcus aureus (MRSA) bloodstream infections: a systematic literature review and meta-analysis. PLoS One 19:e0293423. doi: 10.1371/journal.pone.0293423
Alhurayri, F., Porter, E., Douglas-Louis, R., Minejima, E., Wardenburg, J. B., and Wong-Beringer, A. (2021). Increased risk of thrombocytopenia and death in patients with bacteremia caused by high alpha toxin-producing methicillin-resistant Staphylococcus aureus. Toxins (Basel) 13:726. doi: 10.3390/toxins13100726
Bertrand, X., Lopez-Lozano, J. M., Slekovec, C., Thouverez, M., Hocquet, D., and Talon, D. (2012). Temporal effects of infection control practices and the use of antibiotics on the incidence of MRSA. J. Hosp. Infect. 82, 164–169. doi: 10.1016/j.jhin.2012.07.013
Brown, N. M., Goodman, A. L., Horner, C., Jenkins, A., and Brown, E. M. (2021). Treatment of methicillin-resistant Staphylococcus aureus (MRSA): updated guidelines from the UK. JAC Antimicrob. Resist. 3:dlaa114. doi: 10.1093/jacamr/dlaa114
Bueschbell, B., Caniceiro, A. B., Suzano, P. M. S., Machuqueiro, M., Rosário-Ferreira, N., and Moreira, I. S. (2022). Network biology and artificial intelligence drive the understanding of the multidrug resistance phenotype in cancer. Drug Resist. Updat. 60:100811. doi: 10.1016/j.drup.2022.100811
Cardo, D. (2004). National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32, 470–485. doi: 10.1016/j.ajic.2004.10.001
Chand, U., Priyambada, P., and Kushawaha, P. K. (2023). Staphylococcus aureus vaccine strategy: promise and challenges. Microbiol. Res. 271:127362. doi: 10.1016/j.micres.2023.127362
Chen, C. (2004). Searching for intellectual turning points: progressive knowledge domain visualization. Proc. Natl. Acad. Sci. U.S.A. 101 Suppl 1(Suppl. 1), 5303–5310. doi: 10.1073/pnas.0307513100
Cosgrove, S. E., Sakoulas, G., Perencevich, E. N., Schwaber, M. J., Karchmer, A. W., and Carmeli, Y. (2003). Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36, 53–59. doi: 10.1086/345476
Datta, R., and Huang, S. S. (2008). Risk of infection and death due to methicillin-resistant Staphylococcus aureus in long-term carriers. Clin. Infect. Dis. 47, 176–181. doi: 10.1086/589241
David, M. Z., and Daum, R. S. (2010). Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23, 616–687. doi: 10.1128/CMR.00081-09
David, M. Z., Daum, R. S., Bayer, A. S., Chambers, H. F., Fowler, V. G. Jr., Miller, L. G., et al. (2014). Staphylococcus aureus bacteremia at 5 US academic medical centers, 2008-2011: significant geographic variation in community-onset infections. Clin. Infect. Dis. 59, 798–807. doi: 10.1093/cid/ciu410
Del Giudice, P. (2020). Skin infections caused by Staphylococcus aureus. Acta Derm. Venereol. 100:adv00110. doi: 10.2340/00015555-3466
Dodds, T. J., and Hawke, C. I. (2009). Linezolid versus vancomycin for MRSA skin and soft tissue infections (systematic review and meta-analysis). ANZ J. Surg. 79, 629–635. doi: 10.1111/j.1445-2197.2009.05018.x
Dyzenhaus, S., Sullivan, M. J., Alburquerque, B., Boff, D., van de Guchte, A., Chung, M., et al. (2023). MRSA lineage USA300 isolated from bloodstream infections exhibit altered virulence regulation. Cell Host Microbe 31, 228–242.e8. doi: 10.1016/j.chom.2022.12.003
Evans, L., Rhodes, A., Alhazzani, W., Antonelli, M., Coopersmith, C. M., French, C., et al. (2021). Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Intens. Care Med. 47, 1181–1247. doi: 10.1007/s00134-021-06506-y
Fan, G., Han, R., Zhang, H., He, S., and Chen, Z. (2017). Worldwide research productivity in the field of minimally invasive spine surgery: a 20-year survey of publication activities. Spine (Phila Pa 1976) 42, 1717–1722. doi: 10.1097/brs.0000000000001393
Gammel, N., Ross, T. L., Lewis, S., Olson, M., Henciak, S., Harris, R., et al. (2021). Comparison of an Automated Plate Assessment System (APAS Independence) and Artificial Intelligence (AI) to Manual Plate Reading of Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus CHROMagar Surveillance Cultures. J. Clin. Microbiol. 59:e0097121. doi: 10.1128/jcm.00971-21
Hassoun, A., Linden, P. K., and Friedman, B. (2017). Incidence, prevalence, and management of MRSA bacteremia across patient populations-a review of recent developments in MRSA management and treatment. Crit. Care 21:211. doi: 10.1186/s13054-017-1801-3
Holland, T. L., Arnold, C., and Fowler, V. G. Jr. (2014). Clinical management of Staphylococcus aureus bacteremia: a review. JAMA 312, 1330–1341. doi: 10.1001/jama.2014.9743
Holubar, M., Meng, L., Alegria, W., and Deresinski, S. (2020). Bacteremia due to methicillin-resistant Staphylococcus aureus: an update on new therapeutic approaches. Infect. Dis. Clin. North Am. 34, 849–861. doi: 10.1016/j.idc.2020.04.003
Jaganath, D., Jorakate, P., Makprasert, S., Sangwichian, O., Akarachotpong, T., Thamthitiwat, S., et al. (2018). Staphylococcus aureus bacteremia incidence and methicillin resistance in rural Thailand, 2006-2014. Am. J. Trop. Med. Hyg. 99, 155–163. doi: 10.4269/ajtmh.17-0631
Jeon, K., Jeong, S., Lee, N., Park, M. J., Song, W., Kim, H. S., et al. (2022). Impact of COVID-19 on antimicrobial consumption and spread of multidrug-resistance in bacterial infections. Antibiotics (Basel) 11:535. doi: 10.3390/antibiotics11040535
Kanafani, Z. A., and Fowler, V. G. Jr. (2006). [Staphylococcus aureus infections: new challenges from an old pathogen]. Enferm. Infecc. Microbiol. Clin. 24, 182–193. doi: 10.1157/13086552
Kavanagh, K. T. (2019). Control of MSSA and MRSA in the United States: protocols, policies, risk adjustment and excuses. Antimicrob. Resist. Infect. Control 8:103. doi: 10.1186/s13756-019-0550-2
Köck, R., Becker, K., Cookson, B., van Gemert-Pijnen, J. E., Harbarth, S., Kluytmans, J., et al. (2010). Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill 15:19688. doi: 10.2807/ese.15.41.19688-en
Kourtis, A. P., Hatfield, K., Baggs, J., Mu, Y., See, I., Epson, E., et al. (2019). Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections — United States. MMWR Morb. Mortal. Wkly. Rep. 68, 214–219. doi: 10.15585/mmwr.mm6809e1
Kullar, R., Sakoulas, G., Deresinski, S., and van Hal, S. J. (2016). When sepsis persists: a review of MRSA bacteraemia salvage therapy. J. Antimicrob. Chemother. 71, 576–586. doi: 10.1093/jac/dkv368
Li, J. G., Chen, X. F., Lu, T. Y., Zhang, J., Dai, S. H., Sun, J., et al. (2023). Increased activity of β-lactam antibiotics in combination with carvacrol against MRSA bacteremia and catheter-associated biofilm infections. ACS Infect. Dis. 9, 2482–2493. doi: 10.1021/acsinfecdis.3c00338
Li, X., Ning, X., and Li, Z. (2024). Global research trends of uranium-containing wastewater treatment based on bibliometric review. J. Environ. Manage. 354:120310. doi: 10.1016/j.jenvman.2024.120310
Lin, X., Geng, R., Menke, K., Edelson, M., Yan, F., Leong, T., et al. (2023). Machine learning to predict risk for community-onset Staphylococcus aureus infections in children living in southeastern United States. PLoS One 18:e0290375. doi: 10.1371/journal.pone.0290375
Lindsay, J. A. (2007). Prospects for a MRSA vaccine. Fut. Microbiol. 2, 1–3. doi: 10.2217/17460913.2.1.1
Lindsay, J. A. (2013). Hospital-associated MRSA and antibiotic resistance-what have we learned from genomics? Int. J. Med. Microbiol. 303, 318–323. doi: 10.1016/j.ijmm.2013.02.005
Liu, C., Bayer, A., Cosgrove, S. E., Daum, R. S., Fridkin, S. K., Gorwitz, R. J., et al. (2011). Clinical practice guidelines by the infectious diseases society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 52, e18–e55. doi: 10.1093/cid/ciq146
Liu, G. Y., Yu, D., Fan, M. M., Zhang, X., Jin, Z. Y., Tang, C., et al. (2024). Antimicrobial resistance crisis: could artificial intelligence be the solution? Mil. Med. Res. 11:7. doi: 10.1186/s40779-024-00510-1
Lu, M., Parel, J. M., and Miller, D. (2021). Interactions between staphylococcal enterotoxins A and D and superantigen-like proteins 1 and 5 for predicting methicillin and multidrug resistance profiles among Staphylococcus aureus ocular isolates. PLoS One 16:e0254519. doi: 10.1371/journal.pone.0254519
Mahmoud, E., Al Mansour, S., Bosaeed, M., Alharbi, A., Alsaedy, A., Aljohani, S., et al. (2020). Ceftobiprole for treatment of MRSA blood stream infection: a case series. Infect. Drug Resist. 13, 2667–2672. doi: 10.2147/idr.S254395
Miao, J., Chen, L., Wang, J., Wang, W., Chen, D., Li, L., et al. (2017). Current methodologies on genotyping for nosocomial pathogen methicillin-resistant Staphylococcus aureus (MRSA). Microb. Pathog. 107, 17–28. doi: 10.1016/j.micpath.2017.03.010
Miller, L. S., Fowler, V. G., Shukla, S. K., Rose, W. E., and Proctor, R. A. (2020). Development of a vaccine against Staphylococcus aureus invasive infections: evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol. Rev. 44, 123–153. doi: 10.1093/femsre/fuz030
Mohamad Farook, N. A., Argimón, S., Abdul Samat, M. N., Salleh, S. A., Sulaiman, S., Tan, T. L., et al. (2022). Diversity and dissemination of Methicillin-Resistant Staphylococcus aureus (MRSA) genotypes in Southeast Asia. Trop. Med. Infect. Dis. 7:438. doi: 10.3390/tropicalmed7120438
Motoshima, M., Yanagihara, K., Morinaga, Y., Matsuda, J., Sugahara, K., Yamada, Y., et al. (2010). Genetic diagnosis of community-acquired MRSA: a multiplex real-time PCR method for Staphylococcal cassette chromosome mec typing and detecting toxin genes. Tohoku J. Exp. Med. 220, 165–170. doi: 10.1620/tjem.220.165
Murray, C. J. L., Ikuta, K. S., and Sharara, F. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Nigo, M., Rasmy, L., Mao, B., Kannadath, B. S., Xie, Z., and Zhi, D. (2024). Deep learning model for personalized prediction of positive MRSA culture using time-series electronic health records. Nat. Commun. 15:2036. doi: 10.1038/s41467-024-46211-0
Plumet, L., Ahmad-Mansour, N., Dunyach-Remy, C., Kissa, K., Sotto, A., Lavigne, J. P., et al. (2022). Bacteriophage therapy for Staphylococcus aureus infections: a review of animal models, treatments, and clinical trials. Front. Cell Infect. Microbiol. 12:907314. doi: 10.3389/fcimb.2022.907314
Popovich, K. J., Green, S. J., Okamoto, K., Rhee, Y., Hayden, M. K., Schoeny, M., et al. (2021). MRSA transmission in intensive care units: genomic analysis of patients, their environments, and healthcare workers. Clin. Infect. Dis. 72, 1879–1887. doi: 10.1093/cid/ciaa731
Poyraz, O., Sater, M. R. A., Miller, L. G., McKinnell, J. A., Huang, S. S., Grad, Y. H., et al. (2022). Modelling methicillin-resistant Staphylococcus aureus decolonization: interactions between body sites and the impact of site-specific clearance. J. R. Soc. Interface 19:20210916. doi: 10.1098/rsif.2021.0916
Rasmussen, R. V., Fowler, V. G. Jr., Skov, R., and Bruun, N. E. (2011). Future challenges and treatment of Staphylococcus aureus bacteremia with emphasis on MRSA. Fut. Microbiol. 6, 43–56. doi: 10.2217/fmb.10.155
Rose, W., Fantl, M., Geriak, M., Nizet, V., and Sakoulas, G. (2021). Current paradigms of combination therapy in Methicillin-Resistant Staphylococcus aureus (MRSA) bacteremia: does it work, which combination, and for which patients? Clin. Infect. Dis. 73, 2353–2360. doi: 10.1093/cid/ciab452
Stefani, S., Chung, D. R., Lindsay, J. A., Friedrich, A. W., Kearns, A. M., Westh, H., et al. (2012). Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents 39, 273–282. doi: 10.1016/j.ijantimicag.2011.09.030
Strathdee, S. A., Hatfull, G. F., Mutalik, V. K., and Schooley, R. T. (2023). Phage therapy: from biological mechanisms to future directions. Cell 186, 17–31. doi: 10.1016/j.cell.2022.11.017
Tabah, A., and Laupland, K. B. (2022). Update on Staphylococcus aureus bacteraemia. Curr. Opin. Crit. Care 28, 495–504. doi: 10.1097/mcc.0000000000000974
Tang, R., Luo, R., Tang, S., Song, H., and Chen, X. (2022). Machine learning in predicting antimicrobial resistance: a systematic review and meta-analysis. Int. J. Antimicrob. Agents 60:106684. doi: 10.1016/j.ijantimicag.2022.106684
Tokajian, S. (2014). New epidemiology of Staphylococcus aureus infections in the Middle East. Clin. Microbiol. Infect. 20, 624–628. doi: 10.1111/1469-0691.12691
Tong, S. Y. C., Lye, D. C., Yahav, D., Sud, A., Robinson, J. O., Nelson, J., et al. (2020). Effect of vancomycin or daptomycin with vs without an antistaphylococcal β-lactam on mortality, bacteremia, relapse, or treatment failure in patients with MRSA bacteremia: a randomized clinical trial. JAMA 323, 527–537.
Turner, N. A., Sharma-Kuinkel, B. K., Maskarinec, S. A., Eichenberger, E. M., Shah, P. P., Carugati, M., et al. (2019). Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol. 17, 203–218. doi: 10.1038/s41579-018-0147-4
van Belkum, A., and Rochas, O. (2018). Laboratory-based and point-of-care testing for MSSA/MRSA detection in the age of whole genome sequencing. Front. Microbiol. 9:1437. doi: 10.3389/fmicb.2018.01437
van Eck, N. J., and Waltman, L. (2010). Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 84, 523–538. doi: 10.1007/s11192-009-0146-3
Vincent, J. L., Sakr, Y., Sprung, C. L., Ranieri, V. M., Reinhart, K., Gerlach, H., et al. (2006). Sepsis in European intensive care units: results of the SOAP study. Crit. Care Med. 34, 344–353. doi: 10.1097/01.ccm.0000194725.48928.3a
Keywords: MRSA, bloodstream infection, antibiotic resistance, artificial intelligence in diagnosis, personalized medicine
Citation: Lin J-Y, Lai J-K, Chen J-Y, Cai J-Y, Yang Z-D, Yang L-Q, Zheng Z-T and Guo X-G (2025) Global insights into MRSA bacteremia: a bibliometric analysis and future outlook. Front. Microbiol. 15:1516584. doi: 10.3389/fmicb.2024.1516584
Received: 29 October 2024; Accepted: 23 December 2024;
Published: 22 January 2025.
Edited by:
Takashi Azuma, Osaka Medical College, JapanReviewed by:
Ursula Waack, United States Food and Drug Administration, United StatesCopyright © 2025 Lin, Lai, Chen, Cai, Yang, Yang, Zheng and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu-Guang Guo, Z3lzeWd4Z0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.