- 1Department of Biological Sciences, Northern Illinois University, DeKalb, IL, United States
- 2Food and Feed Safety Research Unit, USDA/ARS, Southern Regional Research Center, New Orleans, LA, United States
- 3Department of Biological Sciences and Evolutionary Studies Initiative, Vanderbilt University, Nashville, TN, United States
Introduction: Aspergillus flavus is an opportunistic pathogenic fungus that infects oilseed crops worldwide. When colonizing plants, it produces mycotoxins, including carcinogenic compounds such as aflatoxins. Mycotoxin contamination results in an important economic and health impact. The design of new strategies to control A. flavus colonization and mycotoxin contamination is paramount.
Methods: The biocontrol potential of a promising new isolate of Pseudomonas spp., 20EI1 against A. flavus was assessed using bioassays and microscopy. To further elucidate the nature of this bacterial-fungal interaction, we also performed chemical and transcriptomics analyses.
Results: In the present study, Pseudomonas spp., 20EI1 was able to reduce the growth of A. flavus. Furthermore, we determined that this growth inhibition is iron-dependent. In addition, Pseudomonas 20EI1 reduced or blocked the production of aflatoxin, as well as cyclopiazonic acid and kojic acid. Expression of iron-related genes was altered in the presence of the bacteria and genes involved in the production of aflatoxin were down-regulated. Iron supplementation partially reestablished their expression. Expression of other secondary metabolite (SM) genes was also reduced by the bacteria, including genes of clusters involved in cyclopiazonic acid, kojic acid and imizoquin biosynthesis, while genes of the cluster corresponding to aspergillicin, a siderophore, were upregulated. Interestingly, the global SM regulatory gene mtfA was significantly upregulated by 20EI1, which could have contributed to the observed alterations in SM.
Discussion: Our results suggest that Pseudomonas 20EI1 is a promising biocontrol against A. flavus, and provide further insight into this iron-dependent bacterial-fungal interaction affecting the expression of numerous genes, among them those involved in SM.
1 Introduction
Aspergillus flavus is an opportunistic phytopathogen found colonizing oil seed crops such as peanut, corn, sorghum, tree nuts and cotton (Robens and Cardwell, 2003). This fungus efficiently disseminates by forming air-borne conidia present on specialized structures denominated conidiophores. The fungus also forms resistant structures termed sclerotia, which allows survival under harsh conditions for several years (Horn et al., 2014). Sclerotia germinate to form additional hyphae or conidia, further facilitating further spreading in the field (Hedayati et al., 2007; Amaike and Keller, 2011).
Upon colonization of the plant substrate, A. flavus synthesizes several mycotoxins, including aflatoxins (AFs) (Cary et al., 2015; Bhatnagar et al., 2018). Exposure to mycotoxins can have lasting health impacts. AF is classified as a Group-1 carcinogen by the International Agency for Research on Cancer (Kale et al., 2008). AFB1 has been linked to liver, lung, and kidney cancers, immunosuppression, aflatoxicosis and growth retardation in children (Williams et al., 2004; Amaike and Keller, 2011; Hyde et al., 2018; Kinyungu et al., 2019). AF contamination is a worldwide problem (Mamo et al., 2022). Numerous countries enforce strict AF limits in crops to reduce health impacts, leading to major economic losses, as contaminated crops are often destroyed or reduced in value. Annually contaminated crops account for $1 billion US in economic losses (Robens and Cardwell, 2003; Wu et al., 2008; Cary et al., 2019); AF contamination caused the loss of approximately 25% of the world’s food crops yearly (WHO, 2018). Some developing countries do not have legislation to control AF limits, and 4.5 billion people are at risk of ingesting AF (Drott et al., 2021). A. flavus also produces other toxic secondary metabolites (SMs), such as cyclopiazonic acid (CPA), which can also result in health impacts. CPA inhibits mammalian Ca2+-ATPases and disrupts normal intracellular calcium flux (Chang and Ehrlich, 2011). Normal calcium flux is vital to cellular life and its disruption leads to cell death. Usually, toxicity can occur through ingestion and exposure to contaminated food and feed products (Norred et al., 1988; Dorner et al., 1994; Oliveira et al., 2008; Chang and Ehrlich, 2011).
Current control methods for reducing mycotoxin crop contamination are still limited. Azole fungicides are applied abundantly to crops, leading to the acquisition of antifungal resistance through environmental routes (Kleinkauf et al., 2013) that could reduce their efficacy. Furthermore, azoles are one of the main forms of antifungal drugs used in the medical field, and therefore their extensive use in agriculture could reduce their efficacy when treating fungal infections in humans and animals (Snelders et al., 2009; Chowdhary et al., 2014; Ahangarkani et al., 2020; Barber et al., 2020). Thus, it is important to develop new methodologies, including biocontrols, to reduce fungal growth and dissemination, as well as mycotoxin contamination of crops. Recent studies have examined the impact of non-aflatoxigenic strains being introduced to outcompete toxigenic species (Amaike and Keller, 2011; Kinyungu et al., 2019; Moore, 2022), and although applying an non-aflatoxigenic biocontrol to corn post-harvest shows a decrease in A. flavus contamination (Kinyungu et al., 2019), many regions remain infected by A. flavus as non-aflatoxigenic strains are not adapted to all environmental conditions.
Thus, finding new strategies to protect crops from fungal infections and mycotoxin contamination is key to prevent their negative impact. Previous studies showed that compounds of plant origin present inhibitory activity against pathogens of plants and animals, including humans (Radha et al., 2024). Also, some nanomaterials with antifungal effect, such as copper nanoparticles (Qin et al., 2023), have been biologically synthesized in fungi (Gaba et al., 2022). These mycogenic copper oxide nanoparticles also showed antifungal activity. Additionally, bacteria and their bioactive metabolites have the potential to be utilized as biocontrol agents against fungal plant pathogens (Veliz et al., 2017; Chalivendra and Ham, 2019; Legein et al., 2020). The use of bacterial biocontrol agents presents certain advantages over other methods. For instance, bacteria can adapt to many environments and often outcompete pathogens for resources and nutrients due to being better acclimatized (Elnahal et al., 2022), and even improve plant health by increasing the plant’s ability to endure abiotic and biotic stresses (Elnahal et al., 2022). Previous reports have indicated the potential of bacteria and bacterial compounds against numerous fungal species. For example, antimicrobials from Bacillus species have been shown to reduce Fusarium spp. infection (Gong et al., 2015; Alberts et al., 2016; Palazzini et al., 2016). Bacillus spp. (including the recategorized Lysinibacillus spp.) have also been used against A. flavus and A. parasiticus, negatively affecting fungal growth as well as aflatoxin production (Raksha Rao et al., 2017; Xia et al., 2017; Chalivendra et al., 2018; Shu et al., 2018). Additionally, Sporosarcina sp., Pantoea sp., Lysinibacillus fusiformis and Staphylococcus warneri have been utilized to degrade aflatoxins using in vitro assays (Adebo et al., 2016; Xie et al., 2019). A. flavus growth and expression of aflatoxin biosynthetic genes were reduced in Priestia megaterium and Wickerhamomyces anomalus co-cultures (Kong et al., 2014; Hua et al., 2019; Gupta et al., 2020). Strains of B. subtilis and Vibrio gazogenes were also shown to inhibit expression of the AF genes aflD and aflR (Al-Saad et al., 2016; Kandel et al., 2022). However, the factors causing this inhibition remains largely unknown.
Various strains of Pseudomonas fluorescens have demonstrated its potential as biocontrol (Kerr, 1999; Nagarajkumar et al., 2004; Barahona et al., 2011). For example, P. fluorescens causes fungal growth inhibition in Fusarium oxysporum, Macrophomina phaseolina, Magnaporthe grisea, Cercosporidium personatum, and Rhizoctonia solani (Ganeshan and Manoj Kumar, 2005; Gull and Hafeez, 2012). F. verticillioides had reduced incidence when maize was treated with P. fluorescens (Chandra Nayaka et al., 2009). P. fluorescens reduces cell wall degradation in radish and carnation by F. oxysporum (Ganeshan and Manoj Kumar, 2005). Also, P. fluorescens-treated rice seeds showed a reduction in Xanthomonas oryzae colonization (Ganeshan and Manoj Kumar, 2005). In addition, rice blast incidence from M. grisea was reduced by P. fluorescens. Strains of P. fluorescens have shown to inhibit mycelial growth of sheath blight fungus R. solani (Ganeshan and Manoj Kumar, 2005; Gull and Hafeez, 2012). Several factors could be involved in the inhibition of fungal growth caused by P. fluorescens, such as the production of antifungal metabolites (Chin-A-Woeng et al., 2000; Dutta et al., 2020). For example, 2,4-diacetylphloroglucinol, phenazines, pyoluteorin, pyoverdin, pyrrolnitrin, siderophores, lipopeptides and hydrogen cyanide are all SMs produced by P. fluorescens that have antimicrobial and antifungal properties. Many of these metabolites have been found to inhibit hyphal formation, hydrolyze the fungal cell wall, interfere with cell membrane permeability and nutrient acquisition in fungal phytopathogens (Ganeshan and Manoj Kumar, 2005; Gull and Hafeez, 2012; Veliz et al., 2017; Legein et al., 2020; Kandel et al., 2022).
Other Pseudomonas species have also been evaluated as possible biocontrols, including P. aeruginosa, P. corrugata, P. chlororaphis, and P. protegens. P. aeruginosa is a well-studied biocontrol against fungi. It produces a variety of antifungal SMs including phenazine, hydrogen cyanide, siderophores, pyoverdine, pyochelin and rhamnolipid-type biosurfactants (Höfte, 2021). Pseudomonas corrugata is another biocontrol that inhibits growth of filamentous fungi (Höfte, 2021). For instance, P. corrugata inhibited fungal growth of Gaeumannomycis graminis var. tritici in wheat (Harrison et al., 1993), and reduces disease on strawberry and tomato by F. oxysporum and Phytophthora cactorum (Barahona et al., 2011). Pseudomonas chlororaphis is yet another biocontrol against filamentous fungi. P. chlororaphis has been shown to inhibit mycelial growth in F. graminearum, Colletotrichum gloeosporioides, and Botrytis cinerea (Huang et al., 2018). P. chlororaphis also induces resistance in cucumber toward Corynespora cassicola (Kim et al., 2004). P. protegens represses growth of R. solani and P. ultimum on cotton (Howell, 1979; Howell and Stipanovic, 1980). This organism also inhibits growth of F. oxysporum in vitro (Takeuchi et al., 2014). Additionally, P. protegens reduced incidence of P. ultimum in cucumber (Takeuchi and Someya, 2019).
As mentioned above, Pseudomonas species are important biocontrols antagonistic to fungal phytopathogens. The present study provides further insight into the mechanism of action of a new isolate of Pseudomonas, 20EI1, against A. flavus, and its possible dependency on iron acquisition, using both chemical and transcriptomics approaches. An in-depth understanding of the bacterial-fungal interaction is essential for the development of a successful control strategy to decrease A. flavus phytopathogenicity and mycotoxin contamination.
2 Materials and methods
2.1 Isolation and identification of Pseudomonas 20EI1
The environmental isolate 20EI1 was isolated from a lagoon in DeKalb, Illinois, United States, at a depth of 0.5 m. The 20EI1 genomic DNA was extracted and used as template for PCR. The amplification was carried out using primers 515F-Y (GTGYCAGCMGCCGCGGTAA) (Parada et al., 2016) and DG74 (AGGAGGTGATCCAACCGCA) (Greisen et al., 1994). These primers amplified 16S rRNA. The PCR product was sequenced and a Blastn analysis revealed a match with the genus Pseudomonas (Altschul et al., 1990; Camacho et al., 2009).
2.2 Microbial strains, culture conditions and assessment of fungal and bacterial growth
The new isolate of Pseudomonas 20EI1, and A. flavus AF70 wild-type strain were used in this study. Bacteria were inoculated and grown in overnight cultures. Bacterial cells and fungal spores were co-cultured on solid PDA or modified Czapek-Dox (CZ, 0.5% glucose, 0.1% yeast extract, pH 7.3 ± 0.3), conducive to the growth of both organisms as indicated. Top agarose (0.7%) was inoculated with 0.3 mL overnight bacterial culture in 4.7 mL top agarose for a total of 5 mL and poured onto 20 mL solid medium plates. Then, 2 μL of fungal spore suspension (in concentration of 106 spores/mL) was point-inoculated at the center of the plates. In a separate experiment, plates were top-agarose inoculated with 106 spores/mL of A. flavus. Then 2 μL of bacterial suspension was point-inoculated on these plates. Plates were incubated at 30°C in the dark. The experiments were carried out in triplicate.
Both organisms were also co-cultured in liquid PDB medium (adjusted pH to 7.3 ± 0.3). In this case, in order to test whether antifungal activity was iron-dependent, liquid cultures were set with and without iron supplementation (200 μg/mL FeCl3). Controls of fungal cultures without the bacterial treatment and bacterial cultures without the fungus were included. Inoculation was carried out with 24 h bacterial and fungal seed cultures growing in PDB medium at 30°C and 150 rpm shaking conditions. Three mL of bacterial culture and about 0.12 g of mycelia were added to fresh PDB. Filter sterilized FeCl3 solution was then added to the cultures. Cultures were then grown for 48 h in a shaker incubator at 150 rpm and 30°C. The experiments included three replicates. Separation of mycelia from the bacteria was carried out by filtration using miracloth. Mycelia was collected to measure dry weight. Bacterial OD600 readings were acquired to assess bacterial growth using a Denovix DS-11 Spectrophotometer (Denovix, CT).
2.3 Scanning electron microscopy
To obtain scanning electron microscopy (SEM) micrographs, liquid co-cultures and monoculture controls were grown as previously described. Liquid cultures were aliquoted into 1.5 mL Eppendorf tubes. Samples underwent fixation as described by Simon et al. (2015). Briefly, samples were fixed for 1 h with aldehyde fixative, followed by 1 h in osmium fixative. Samples were sequentially dehydrated using ethanol at 30, 50 and 70% concentrations, for 20 min each and then changed to 100% for 10 min, three times. A critical point dryer (Tousimis CPD) was used to replace ethanol with CO2. Samples were then sputter coated with gold using Denton Desk IV Sputter Coater. Images were taken using a Hitachi S3400N SEM (NUANCE, Evanston, IL) at the NUANCE facility at Northwestern University.
2.4 Chemical analysis
Liquid cultures were set as described above. Five mL of supernatant was collected into a 50 mL falcon tube. Five mL of chloroform was added and extracted overnight. The bottom organic layer was collected and put into a clean beaker and allowed to evaporate. Once evaporated the extract was collected with chloroform and spotted on a TLC plate to detect AF. The plate was run in a solvent system of 85 chloroform:15 acetone (v/v) for 15 min. After 15 min the plate was sprayed with 12.5% AlCl3 and baked at 80°C for 10 min. The TLC plate was then visualized and imaged with a UV light.
In a separate experiment, broth cultures (50 mL) were filtered, the filtrates lyophilized (Freezemobile, SP Scientific), and extracted with 20 mL 80% acetonitrile: 19.5% water: 0.5% formic acid on a shaker (200 rpm) for 2 h. Portions of the filtered mycelia (0.15–0.2 g) were also lyophilized then extracted with 1 mL methanol on a shaker (200 rpm) overnight. The extracts were centrifuged to pellet the particulates, and the supernatant was analyzed using a Waters ACQUITY UPLC system (1 μL injections, 40% methanol in water isocratic solvent system on a BEH C18 1.7 μm, 2.1 mm × 50 mm column) using fluorescence detection (Ex = 365 nm, Em = 440 nm). Samples were diluted if the aflatoxin signal saturated the detector. Analytical standards (Sigma-Aldrich, St. Louis, MO, United States) were used to identify and quantify aflatoxins: aflatoxin B1 (AFB1); aflatoxin B2 (AFB2). Aflatoxin content was expressed in ppb (ng AF/mL broth or ng AF/g mycelia).
CPA and other metabolites in the extracts were analyzed on a Waters Acquity UPLC system coupled to a Waters Xevo G2 XS QTOF mass spectrometer. Extract injections (1 μL) were separated on a Waters BEH C18 1.7 μm, 2.1 × 50 mm column with the following gradient solvent system: (0.5 mL/min, solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile): 5% B (0–1.25 min), gradient to 25% B (1.25–1.5 min), gradient to 100% B (1.5–5.0 min), 100% B (5.0–7.5 min), then column equilibration to 5% B (7.6–10.1 min). The Z-spray ionization source was run in ESI+ mode using MassLynx 4.2 software with the following settings: source temperature: 100°C, desolvation temperature: 250°C, desolvation gas flow: 600 L/h, cone gas flow: 50 L/h, capillary voltage: 3.0 kV, sampling cone voltage: 40 V. Analyses were performed in sensitivity and continuum mode, with a mass range of m/z 50–1,200 and a scan time of 0.1 s. A data-independent acquisition method with elevated collision energy (MSE) was used with 6 eV low energy and a high energy ramp from 15–45 eV. Data were analyzed on Waters UNIFI 1.9.4 software and quantified using “Quantify Assay Tof 2D” analysis method with lock mass corrected by UNIFI. CPA was purchased from Sigma-Aldrich (St. Louis, MO, United States) and used for quantification. Aflatrems and aflavinine standards were kind gifts from James Gloer (University of Iowa). Toxin content was expressed in ppb (ng toxin/mL broth or ng toxin/g mycelia).
Kojic acid (KA) analyses were conducted on the Waters Acquity UPLC system coupled to a Waters Xevo G2 XS QTOF system with the same mass spectrometer settings that was used for CPA analysis. The filtered broth culture extracts were diluted 10-fold with LC/MS grade water. Injections (1 μL) were separated on a Waters BEH C18 1.7 μm, 2.1 × 50 mm column with the following solvent system: 0.5 mL/min, solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile; pump settings: isocratic at 100% A for 2.5 min (0–2.5 min), gradient to 95% A over 0.5 min (2.5–3.0 min), gradient to 80% A over 1.0 min (3.0–4.0 min), isocratic at 80% A for 1 min (4.0–5.0 min) then column equilibration to 100% A for 2.5 min (5.0–7.5 min). Mass data were collected for the first 2 min of the 7.5 min run, imported into Waters UNIFI 1.9.4 software, and quantified using “Quantify Assay Tof 2D” analysis method with lock mass corrected by UNIFI. Kojic acid was purchased from Sigma-Aldrich (St. Louis, MO, United States) and used for quantification. KA content was expressed in ppb (ng KA/mL broth).
2.5 Transcriptome analysis
2.5.1 RNA preparation and sequencing
Liquid co-cultures of P. fluorescens and A. flavus and their respective monoculture controls were carried out as described in the section above. The amount of iron used was also 200 μg/mL FeCl3. Control cultures without iron supplementation were also included. Fungal mycelia were collected from these cultures for RNA extraction. Bacterial and fungal biomass from co-cultures were separated before RNA extraction. To separate bacteria from mycelia, the samples were filtered using miracloth. After that, samples were frozen in liquid nitrogen and stored at −80°C. RNA extraction was carried out as follows: For extraction of RNA from fungal samples, a Maxwell RSC Plant RNA Kit was used following the manufacturer’s directions. Briefly, 20–50 mg of mycelium was placed in a bead tube. 1-T/Homogenization solution was added to the tube and bullet-blended. This step was repeated and then centrifuged at maximum speed for 2 min. The homogenate was then transferred to a microcentrifuge tube and lysis buffer added. Vortexed for 15 s and incubated at room temperature for 10 min. Samples were centrifuged and supernatant was transferred to wells. DNase was added to a separate well. Nuclease free water was added to the bottom of each elution tube.
RNA quality and quantity were determined using Qubit fluorometer (Invitrogen) and analyzed for integrity using Agilent 4200 TapeStation. RNA libraries were prepared using CORALL Total RNA-seq Library Prep Kit with Unique Dual Indices with RiboCop HMRv2 rRNA Depletion Kit (Lexogen). After cDNA synthesis, the 3′ ends of first-strand cDNA fragments were ligated with a linker containing Illumina-compatible P5 sequences and Unique Molecular Identifiers (UMIs). During the following steps of second strand cDNA synthesis and ds cDNA amplification, i7 and i5 indices as well as complete adapter sequences required for cluster generation were added. The number of PCR amplification cycles was 14 as determined by qPCR using a small pre-amplification library aliquot for each individual sample.
Final amplified libraries were purified and quantified, and average fragment sizes were examed by gel electrophoresis using 4200 TapeStation and D5000 Screen Tape (Agilent). The concentration of the final library pool was confirmed by qPCR and the pool was subjected to test sequencing on MiniSeq instrument (Illumina) in order to check sequencing efficiencies and adjust accordingly proportions of individual libraries. Sequencing was carried out on NovaSeq X 10B (Illumina), 2/150 bp reads.
2.5.2 RNAseq read alignment and differential expression calculation
RNAseq reads were quality checked before and after filtering with Trimmomatic (v0.39) (Bolger et al., 2014) using FastQC (v0.12.1, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Each sample was aligned to the NRRL3357 reference (GCA_014117465.1) using STAR (v2.711) (Dobin et al., 2013). Read counts per strain were calculated using HTseq (v2.0.5) (Anders et al., 2014). For all software, default settings were used unless otherwise specified.
Using DESEQ2 (v1.42.0) (Love et al., 2014), differential expression was calculated using a multifactored approach. Each treatment group (fungus only, fungus + iron, fungus + bacteria, and fungus +bacteria + iron) had three replicates used to calculate differential expression. The effects of iron and bacteria were determined using the respective single factor treatment groups (“~iron” and “~bacteria”). Using the formula “~bacteria + iron + bacteria:iron” the combined effects of bacteria and iron were determined using all of the treatment groups. Differential expression results were shrunk using the “ashr” shrinkage estimator. Downstream visualizations and analyses were performed using R (v4.4.1, http://www.r-project.org/).
2.5.3 Functional annotation and referencing
GO terms and protein function were determined by running Interproscan (v5.61-93.0) (Jones et al., 2014) on the NCBI reference GCA_014117465.1 using default parameters. For comparison with prior datasets and previously determined genes in biosynthetic gene clusters (BGC), reciprocal DIAMOND (v2.1.6) (Buchfink et al., 2021) BLAST was used between the current NCBI reference (GCA_014117465.1) and an older version (GCF_000006275.2). Hits with the highest bitscore were retained resulting in 11,278 matches. Any genes of interest identified were evaluated on the percent identity (>70%) for reporting.
2.6 Statistical analysis
ANOVA tests were used followed by a Tukey’s honest significant difference test, unless otherwise indicated. This post hoc test allowed for multiple comparisons to confirm where differences occurred between treatments. GraphPad Prism (version 10.3.1 (509)) was the software used to carry out the statistical analyses.
3 Results
3.1 Co-culture assays
Fungal growth inhibition was observed in solid and liquid cultures of A. flavus AF70 incubated in the presence of the 20EI1 bacteria (Supplementary Figures S1, S2; Figure 1), the most striking results were detected in solid cultures, where 20EI1 inhibited the formation of A. flavus colonies (Supplementary Figure S1B). Additionally, in co-culture studies with a fungal lawn, a growth inhibition halo was observed around 20EI1 (Supplementary Figure S2), whereas this effect was not detected when the E. coli control was used or in the absence of bacteria (Supplementary Figure S2C). In liquid cultures, measurement of mycelial dry weight indicated reduction of fungal growth in the co-culture compared to that of the fungus-only control (Figure 1B). Interestingly, fungal growth inhibition was partially lost in liquid co-cultures supplemented with iron (Figure 1A); A. flavus growth was reduced 84.6% in co-culture without iron, and 45.9% when in co-culture with iron. In addition, OD600 readings indicated that bacterial growth was reduced in the co-cultures with iron (Figure 1C). Addition of iron did not affect the growth of A. flavus or Pseudomonas 20EI1 when cultured individually.
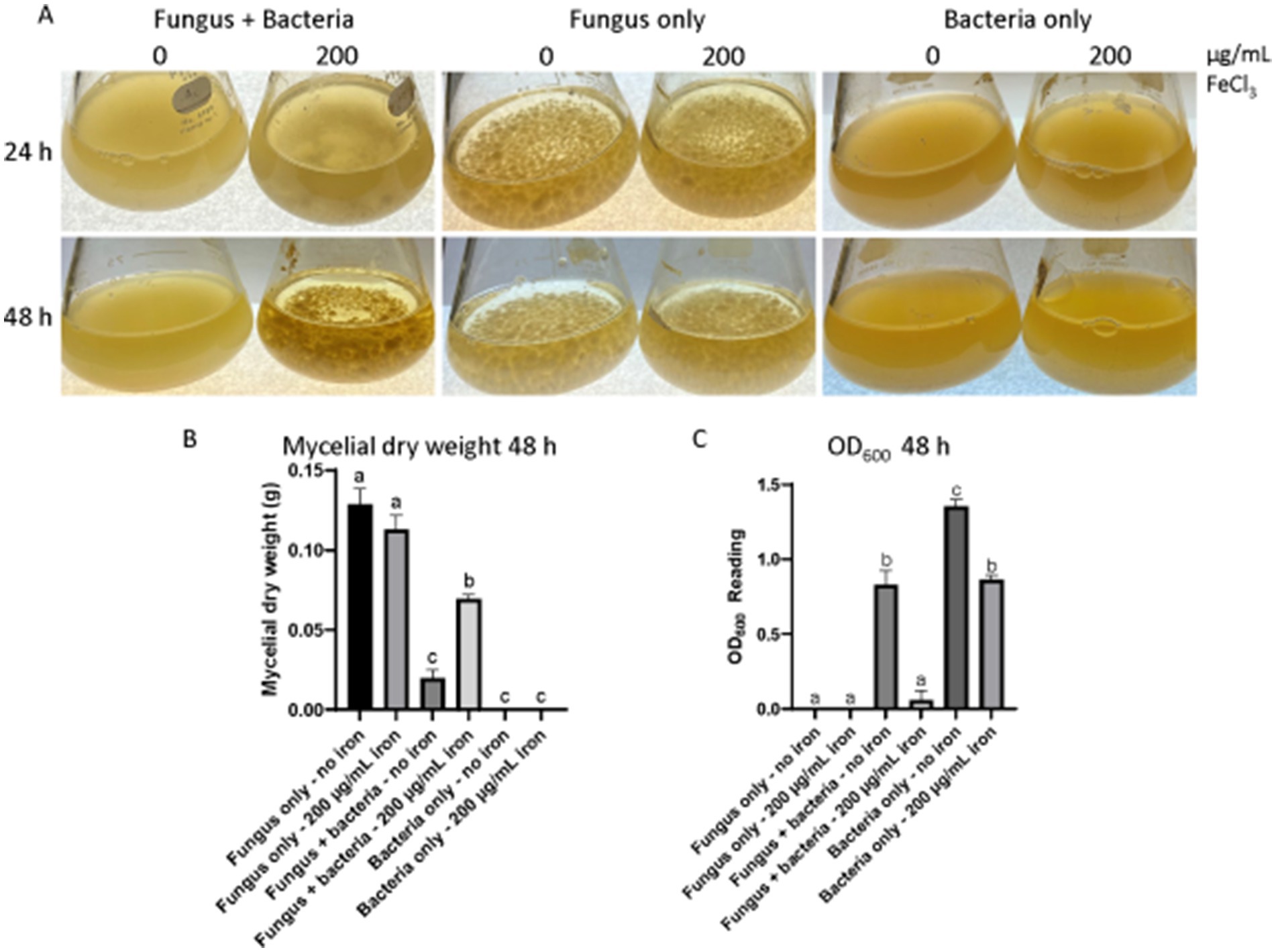
Figure 1. Liquid co-culture assay of Pseudomonas 20EI1 and Aspergillus flavus. (A) A. flavus with and without bacterial and iron treatment grown at 30°C for 48 h at 150 rpm. Both organisms were co-cultured in PDB. Controls of fungal cultures without the bacterial treatment and bacterial cultures without the fungus were included. Inoculation was carried out with 24 h bacterial and fungal seed cultures growing in PDB at 30°C and 150 rpm shaking conditions. Filter sterile liquid FeCl3 was then added to the corresponding cultures. The experiments included three replicates. (B) Dry weight of mycelia collected from cultures in (A). (C) Bacterial OD600 readings acquired to assess bacterial growth.
3.2 Scanning electron microscopy of Pseudomonas 20EI1 attachment to Aspergillus flavus hyphae
Some Pseudomonas species, such as P. fluorescens, has been found to damage the hyphae of A. flavus in previous studies (Akocak et al., 2015), we investigated whether 20EI1 had a similar effect. Pseudomonas 20EI1 presence in the co-cultures did not result in hyphal leakage, however, a strong attachment of the bacteria to the hyphae was observed (Figure 2). This is not detected in the co-cultures of fungus and bacteria with supplemented iron.
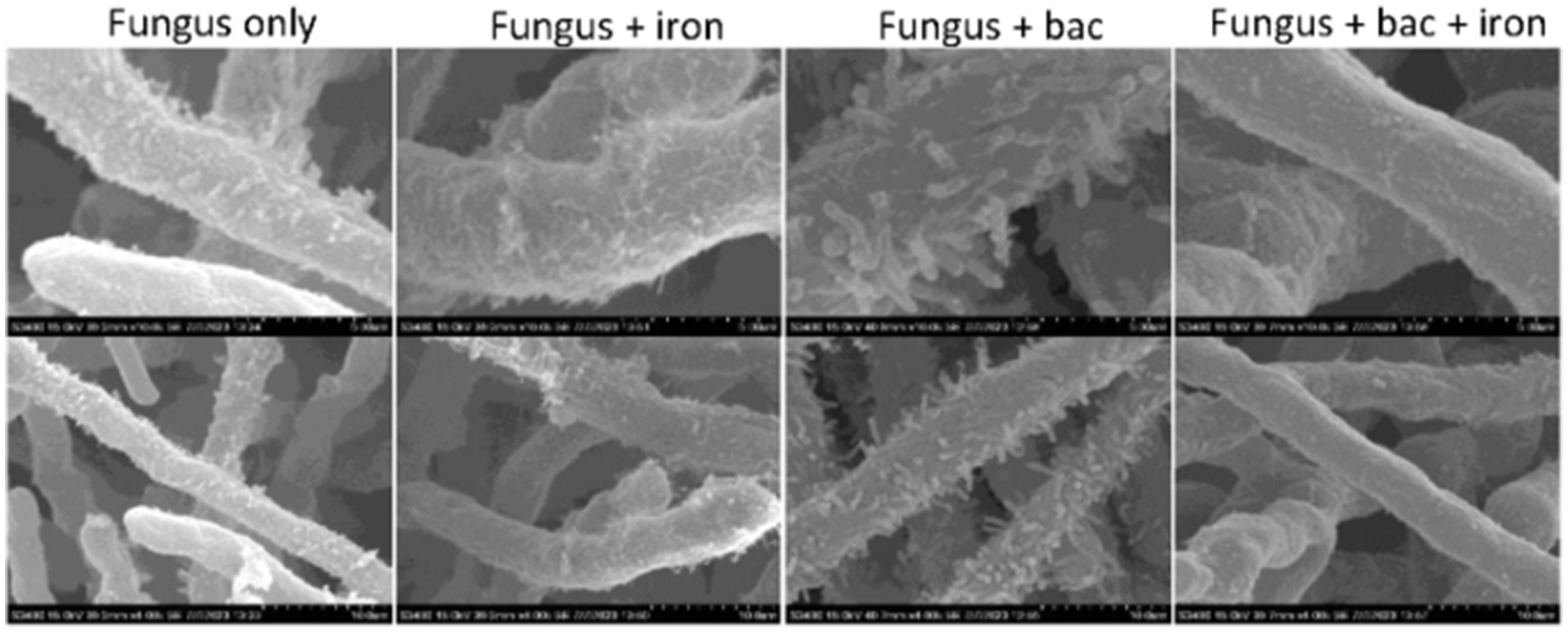
Figure 2. SEM micrographs of Aspergillus flavus and Pseudomonas 20EI1. Mycelia collected from liquid cultures was fixed using aldehyde and osmium. Samples were sequentially dehydrated using ethanol, critical point dried to replace ethanol with CO2 and sputter coated with gold. Micrographs in the top row were taken with ×10k magnification, and those in the bottom row were taken with ×4k magnification.
3.3 Aspergillus flavus secondary metabolism is altered by Pseudomonas 20EI1 and iron
Our chemical analysis indicated that Pseudomonas 20EI1 and iron impacted production of SMs in A. flavus. TLC analysis of culture supernatant extracts revealed that AF production was reduced in fungal cultures with added iron. Furthermore, the presence of Pseudomonas 20EI1 in the fungal culture nearly blocked aflatoxin biosynthesis in this fungus in both supplemented and non-supplemented iron cultures (Figure 3A). UPLC results confirmed these findings, showing a similar pattern of AF production as that shown in the TLC analysis (Figures 3A,B), with a 99.8% reduction in the fungus and bacteria culture. Addition of iron to the co-culture did not rescue AF production, still showing a 99.7% decrease. In addition, iron supplementation resulted in an 82.2% reduction in AF content in the fungal monoculture (Figure 3B).
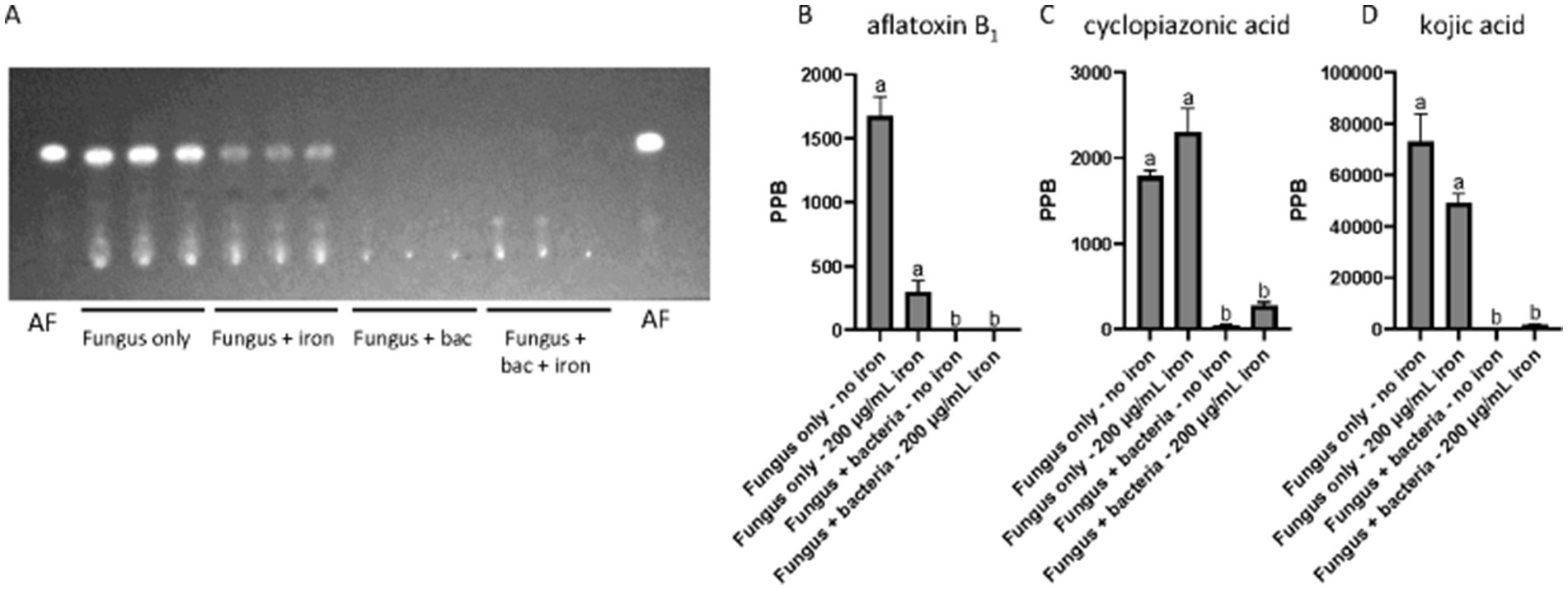
Figure 3. Chemical analyses of Pseudomonas 20EI1 and A. flavus cultures. (A) TLC evaluating production of aflatoxin B1 (AFB1—indicated as AF in this panel) in liquid cultures. Supernatant was collected and extracted with an equal amount of chloroform. Extracts were spotted on a TLC silica plate and separate in an 85 chloroform:15 acetone (v/v) solvent system. (B–D) UPLC quantification of aflatoxin B1 (B), cyclopiazonic acid (C) and kojic acid (D). Conditions for UPLC analysis were as described in the materials and methods section.
The presence of the bacteria also altered production of other SMs. Our analysis revealed that CPA production was dramatically reduced in A. flavus cultures challenged with Pseudomonas 20EI1, independently of iron supplementation, with a 97.6% reduction in the fungus and bacteria co-culture, and an 84.8% reduction in the fungus, bacteria and iron culture (Figure 3C). Similarly, production of KA was also reduced 99.9% in the A. flavus cultures in the presence of the bacteria without iron and 97.9% in the co-culture supplemented with iron (Figure 3D). Addition of iron also reduced KA production in the A. flavus culture by 32.9% compared to the control (Figure 3D).
3.4 Gene expression is influenced by Pseudomonas 20EI1 and iron in Aspergillus flavus
To understand the effect of Pseudomonas 20EI1, iron, and combination of both on A. flavus transcriptome we performed RNA-sequencing (Supplementary Table S1). Iron presented the smallest effect on overall gene expression, with 454 genes differentially expressed (1 ≥ Log2FoldChange ≤ −1; padj ≤ 0.05), 176 of which were unique to iron treatment (Figures 4A,D). Due to the relatively small number of differentially expressed genes between the control and iron treatment, there was no enrichment of genes from any functional categories using either gene set enrichment (GSEA) or GO enrichment. Co-culture with Pseudomonas 20EI1 had the greatest effect on differential expression, with 1,836 genes differentially expressed, 907 unique to Pseudomonas treatment (Figures 4B,E). Based on GSEA, genes involved in copper ion binding and transporter activity were over-represented for genes that are positively expressed, and molybdenum ion binding, heme binding, and iron ion binding were over-represented for genes with reduced expression. The combined effect of Pseudomonas and iron resulted in 1,238 genes differentially expressed, with 308 genes unique to the combined effects of Pseudomonas and iron (Figures 4C,F). GSEA analysis revealed an overrepresentation of genes annotated in the suppression of branch chained amino acid synthesis that was recapitulated with subsequent GO enrichment along with reduced expression in genes involved in transmembrane transporter activity.
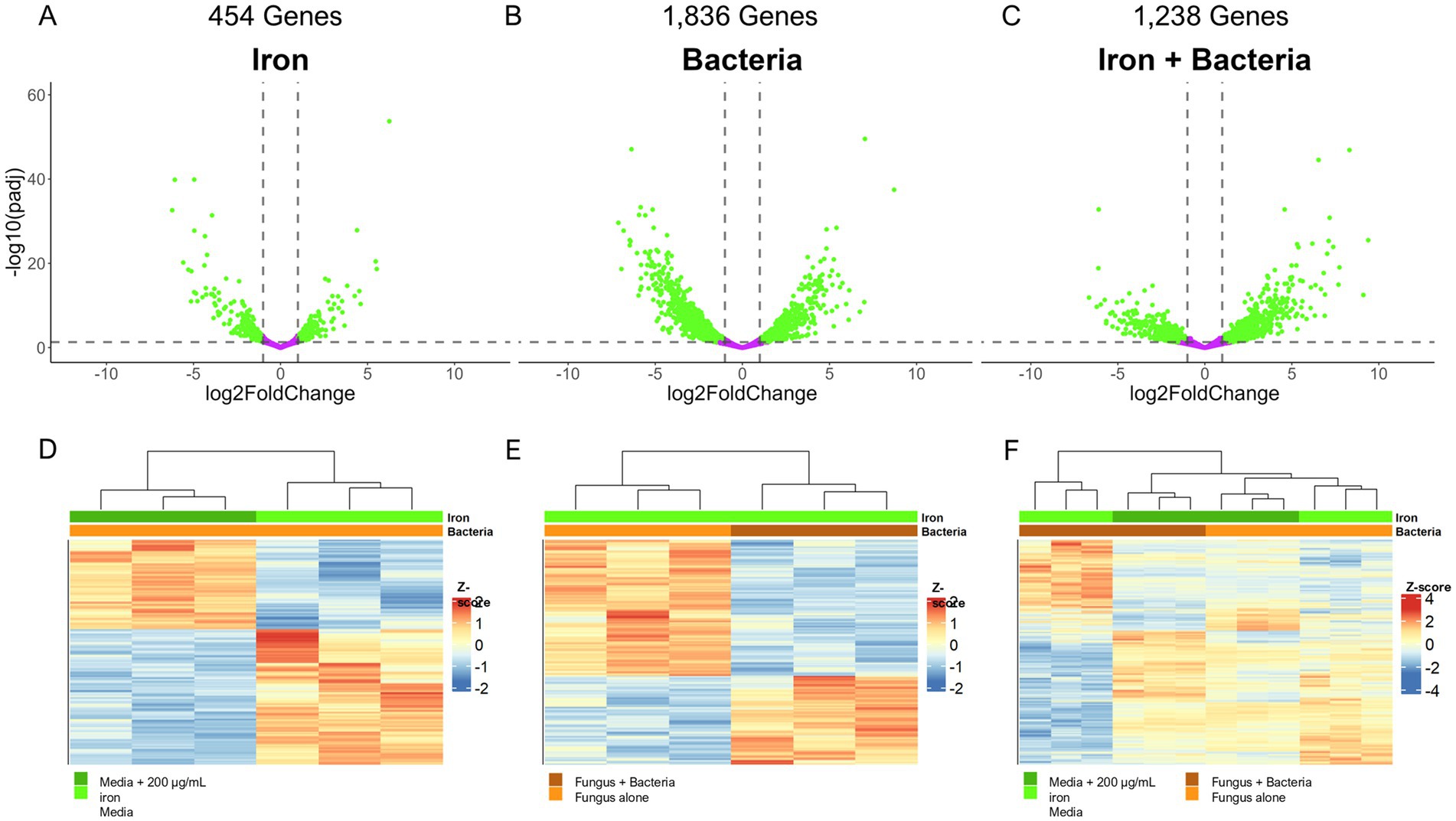
Figure 4. (A–C) Volcano plots depicting the significance and fold change of calculated differential expression across all three effect groups: iron (A), bacteria (B), and iron + bacteria (C). Genes significantly, differentially expressed (1 ≥ Log2FoldChange ≤ −1, padj ≤ 0.05) are colored green while genes not differentially expressed are colored purple. Dotted lines indicate the significance cut off points. The number of genes differentially expressed are noted above the plots. (D–F) Heatmaps depicting the z-score of each differentially expressed gene for the effect groups: iron (D), bacteria (E), and iron + bacteria (F). Dendrograms above the plots indicate the sample clustering based on hierarchical clustering of the calculated z-scores, and the colored bars beneath the dendrogram indicate the treatment of each sample (green = iron with dark green indicating 200 μg/mL of iron and light green as the control; orange = Pseudomonas treatment with dark orange indicating co-culture conditions and light orange as the control).
Our analysis revealed that some A. flavus putative homologs to genes with verified roles in iron homeostasis in other fungal species (Misslinger et al., 2021) were differentially expressed in the presence of the bacteria. Specifically, we found that Pseudomonas 20EI1 increases the expression (>3-fold) of the siderophore transporters mirB, mirD, and sit1, as well as atrH (Figure 5A). Concomitantly, the genes responsible for the biosynthesis of the siderophore triacetylfusarinine C (TAFC), sidI, sidH, sidF, and sidD, are all upregulated (>3-fold) in the presence of 20EI1 (Figure 5B). For both, these siderophore transporters and biosynthetic genes, differential expression is reversed when iron is added to the co-culture (Figure 5).
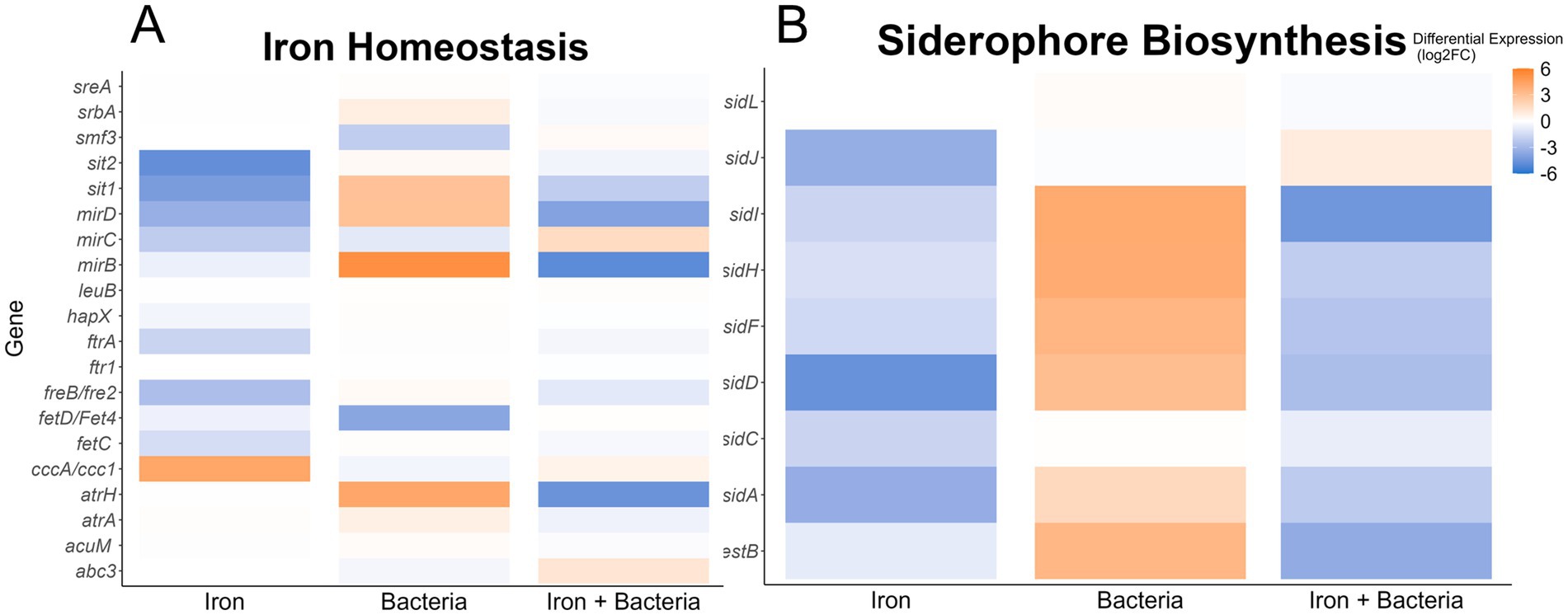
Figure 5. Heatmaps showing differential expression of genes homologous to Aspergillus fumigatus iron homeostasis (A) or siderophore biosynthetic genes (B). The conditions used to calculate differential expression are indicated along the x-axis. Color of the heatmap corresponds to the Log2FoldChange of the differential expression.
Extending our study, we analyzed the expression changes in genes with functional annotations that involve iron interaction (Supplementary Table S2). Of the 333 genes annotated to interact with iron, 29 were differentially expressed in the presence of iron alone. Of these 29 differentially expressed genes, 13/24 of the predicted iron transporters were differentially expressed (Figure 5; Supplementary Figure S2). The presence of Pseudomonas 20EI1 had a greater impact on the 333 iron related genes (Supplementary Table S2), as 70 of them were found differentially expressed in the co-culture, with a similar ratio of downregulated genes to upregulated genes as the iron treated group (44/70 downregulated). Iron transporters were least impacted as only 6 were differentially expressed (4 negatively expressed 2 positively expressed) (Figure 5; Supplementary Figure S2). Interestingly, 39 iron-binding, cytochrome P450 genes were differentially expressed, with the majority (29 genes) displaying reduced expression in the presence of Pseudomonas. The combined effect of Pseudomonas and added iron resulted in 45/333 iron related genes differentially expressed, with a nearly even split between up and downregulated genes (22 downregulated 23 upregulated). This trend of even expression changes is shared by the annotated iron transporters, with 5 differentially expressed transporters downregulated when both iron and Pseudomonas 20EI1 are present (Figure 5; Supplementary Figure S2). Similarly, there is a marked reduction in differentially expressed (only 16 in this treatment) cytochrome P450 genes, and the majority (11/16) of those genes are positively expressed.
Based on the observed impact of Pseudomonas and iron on the production of secondary metabolites, such as AF, CPA and kojic acid (Figure 3), we analyzed the expression of A. flavus biosynthetic gene clusters (BGCs). Our study revealed that of the more than 60 previously identified BGCs (Georgianna et al., 2010; Cary et al., 2018; Uka et al., 2020), 14 have more than 50% of their genes differentially expressed in one of the three experimental groups (4 for iron, 13 for Pseudomonas, and 8 for Pseudomonas + iron) (Supplementary Table S3). Notably, Pseudomonas 20EI1 overwhelmingly reduces the expression of A. flavus BGCs that are differentially expressed. In contrast, the addition of iron during co-culture increases the expression of many of the BGCs with reduced expression in co-culture alone. Specific analysis of BGCs with known products revealed heavily reduced expression of the AF, CPA, kojic acid and imizoquin BGCs (Figures 6A,B, 7B,C). Notably, Pseudomonas co-culture increases the expression of genes in the aspergillicin BGC in A. flavus (Figure 7A). Adding iron had the opposite effect on all of the BGCs mentioned, other than AF which was only greatly affected by Pseudomonas alone (Figure 6A). Iron only has a significant effect (defined by the number of genes differentially expressed) on the aspergillicin BGC, resulting in reduced expression of many genes in this cluster (Figure 6A).
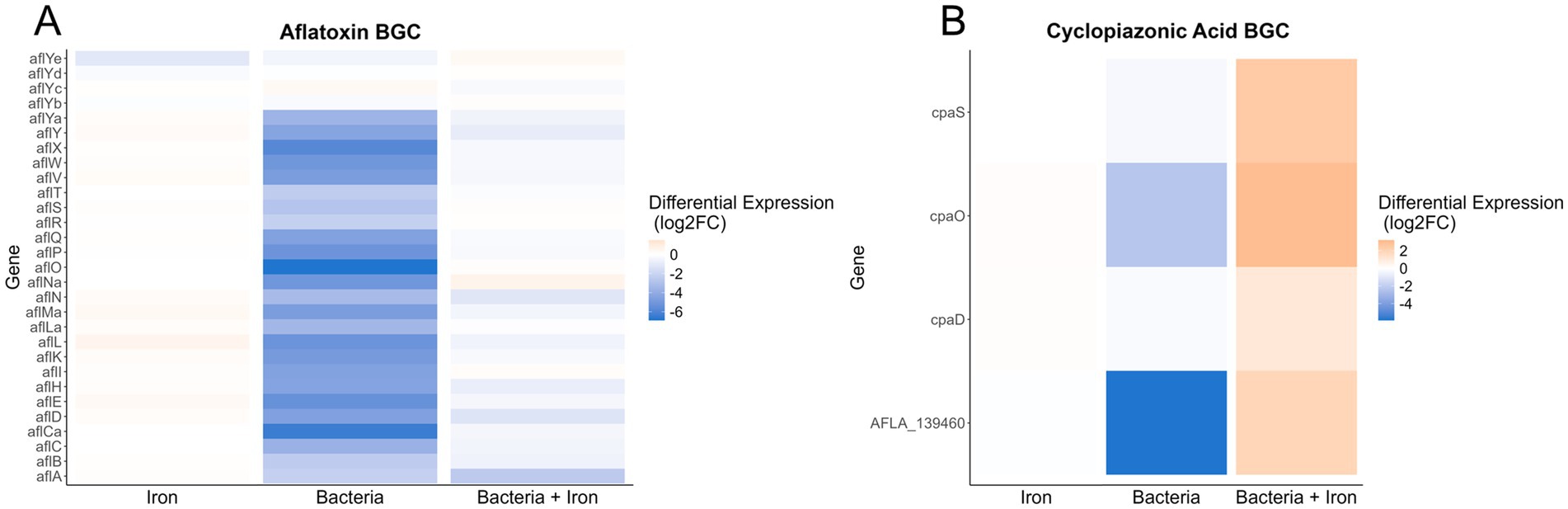
Figure 6. Heatmaps indicating differential expression of all annotated genes within the aflatoxin (A) and cyclopiazonic acid (B) BGCs. The conditions used to calculate differential expression are indicated along the x-axis. Color of the heatmap corresponds to the Log2FoldChange of the differential expression.

Figure 7. Heatmaps depicting differential expression of all annotated genes within the aspergillicin (A), imizoquin (B), and kojic acid (C) BGCs. The conditions used to calculate differential expression are indicated along the x-axis. Color of the heatmap corresponds to the Log2FoldChange of the differential expression.
In this study we also examined the expression of a group of global regulatory genes (Supplementary Table S4), including velvet genes (Eom et al., 2018), as well as laeA (Amaike and Keller, 2009), hbx1 (Cary et al., 2019), rtfA (Lohmar et al., 2019), and mtfA (Zhuang et al., 2016). Interestingly, our result revealed that expression of the A. flavus putative C2H2 transcription factor gene mtfA was significantly upregulated in the presence of Pseudomonas 20EI1.
4 Discussion
Aspergillus flavus is a plant pathogen that impacts oil seed crops, such as corn, peanuts, tree nuts and sorghum. Current treatments to reduce A. flavus colonization and AF contamination of crops are still limited, and resistance to antifungal chemicals is emerging (Kleinkauf et al., 2013). Biocontrol methods have reached certain success, for example the use of non-aflatoxigenic A. flavus strains in the field (Cotty, 1990). In addition, treatments with bacteria and bacterial metabolites present promising potential as controls against fungal growth and to decrease toxin biosynthesis (Peles et al., 2021). Examples of these bacterial biocontrol treatments are Pseudomonas spp., such as P. fluorescens, as studies indicated their potential against several fungi (Ganeshan and Manoj Kumar, 2005; Gull and Hafeez, 2012; Höfte, 2021). In our study, a new isolate that we identified to belong to the genus Pseudomonas, 20EI1, demonstrated a substantial effect on A. flavus, reducing or blocking its growth when both organisms were incubated together.
Competition between A. flavus and Pseudomonas 20EI1 for nutrients, including iron, might occur in the co-cultures. Both organisms could be producing siderophores to capture a limited amount of this element to survive, and eliminating the competing organism by starvation (Haas, 2012). In our study Pseudomonas 20EI1 eliminates or reduces the presence of A. flavus from the environment (Figure 1; Supplementary Figures S1, S2); perhaps a more efficient iron acquisition by the bacteria might starve A. flavus from this element, leading to a drastic decrease in its growth. On the other hand, during iron sufficiency, in iron-supplemented cultures, the fungus survives, while the bacterial presence decreases over time. In this case, where iron is not limited, other factors might be at play where the fungus is able to kill the bacteria, possibly by starvation of other limited nutrients or by involving antibiotic production. Chemical warfare between fungi and bacteria has been previously described, for instance, in a study by Spraker et al. (2018), a SM produced by Fusarium fujikuroi accumulates near the site of infection by the ralsolamycin-producing R. solanacearum GMI1000. Fungal growth is reduced by the bacteria, however the fungus produces several metabolites with antibacterial activity. Additionally, it is worth noting that when iron is in abundance in the bacterial monoculture, the bacterial biomass decreases (Figure 1C). It is possible that above certain levels, iron could become toxic for the bacteria (Andrews et al., 2003). The mechanism behind this has been investigated in E. coli; when an increase in intracellular free iron occurs, this causes an increase in the Fenton reaction, producing hydroxyl radicals and ultimately causing DNA damage (Touati, 2000).
In a recent study, A. flavus was co-cultured with a novel Pseudomonas spp. isolated from soil and sugarcane juice in Brazil (Rabiço et al., 2024). This study showed reduced growth of A. flavus due to volatile organic compounds (VOCs) by an estimated 31%. This decrease was less drastic compared to what we observed in our cultures using 20EI1. In another similar study, VOCs released by Pseudomonas stutzeri YM6 inhibited A. flavus growth (Gong et al., 2022). However, in that study only VOC in air were used, instead of co-cultures (Gong et al., 2022). Although we did not examine Pseudomonas 20EI1 VOCs here, it is possible that VOCs could contribute to some of the observed A. flavus growth inhibition. Future studies could provide additional insight into this possible mechanism.
Siderophores also present an important antifungal activity, for example in the case of P. aeruginosa against A. niger, A. flavus, A. oryzae, F. oxysporum, and Sclerotium rolfsii (Manwar et al., 2004). Interestingly, our A. flavus transcriptome analysis indicated alteration of iron-related genes by the bacteria. Specifically, 70 of 333 annotated genes were found differentially expressed in the co-culture, including fungal siderophore transporters and biosynthetic genes that were highly upregulated by the bacteria, consistent with Pseudomonas 20EI1 possibly reducing the available iron in the environment and showing the attempt of A. flavus to capture this metal. Specifically, expression of transporter genes, such as mirB, mirD, sit1, and atrH, as well as the biosynthesic genes sidI, sidH, sidF, and sidD of the TAFC siderophore were highly upregulated in the presence of 20EI1, in agreement with the increased need for iron capture. The addition of iron to the co-cultures reduced the expression of the TAFC-related genes in an equal but opposite direction. Very little is known about the effect of iron on A. flavus. Our study also showed that iron alone also influences the expression of iron-related genes in this fungus. These findings closely resemble those from the model yeast Saccharomyces cerevisiae growing in an environment with high iron concentration, where the fungus would not need to use these iron-related gene products to acquire additional iron and thus those genes are downregulated (Philpott, 2006). It is also likely that the mechanism in A. flavus is similar to that of A. fumigatus (Misslinger et al., 2021), where the siderophore-mediated iron acquisition (SIA) pathways is downregulated in the presence of high levels of extracellular iron.
In addition, during co-culture bacteria are strongly attached to the fungal hyphae when iron is not supplemented, as shown by our SEM images. Akocak et al. (2015) reported that P. fluorescens and Bacillus spp. present chitinolytic activity that causes hyphal leakage of A. flavus, however, in the case of Pseudomonas 20EI1 no signs of leakage where observed. When iron was supplemented, the bacterial population became reduced in the co-cultures, and attachment to fungal hyphae was not observed.
Importantly, our transcriptome analysis also revealed that Pseudomonas 20EI1 affects SM in A. flavus. More than 50% of the fungal genes in 13 BGCs are differentially expressed in the presence of Pseudomonas 20EI1, causing downregulation in most cases, for example, a reduced expression of AF genes, including the endogenous cluster regulators aflR and aflS (previous called aflJ) (Chang, 2003; Wang et al., 2022). Our results were concomitant with a drastic decrease in AFB1 production. Addition of iron to the co-culture presented a modulating opposite effect on gene expression and although AF production in these co-cultures with iron was still almost completely inhibited, growth of A. flavus was not affected, pointing to inhibition mechanisms unrelated to growth. Decrease of AF production has also been observed in A. flavus exposed to P. fluorescens across media (PDB and peanut medium) and on peanuts, where a 97.8, 99.4 and 55.8% reduction rate was observed, respectively (Yang et al., 2017). Interestingly, the levels of AFB1 were also reduced in the fungal culture alone with added iron. This is a novel role of iron in relationship with AF production in A. flavus, as current literature mostly focuses on the influence of dietary iron enhancing the effect of AFB1 in causing liver cancer (Asare et al., 2007). Wiemann et al. (2014) showed that the synthesis of specific SMs was largely influenced by the availability of iron in A. fumigatus, and its effect is mediated by the iron-dependent transcription factors SreA and HapX (Haas, 2012; Wiemann et al., 2014). The corresponding homologs in A. flavus, AFLA_132440 and AFLA_006720, have not been investigated in A. flavus. These genes were slightly upregulated in the presence of the bacteria. Whether they play a role in SM in A. flavus has not been determined. Although supplemented iron might also have contributed to the decrease in AF in the fungal-bacterial co-culture, it is likely that the bacteria also caused AF inhibition in the co-culture, as the reduction in AF was more dramatic in these cultures compared to those of the fungal culture plus iron in the absence of bacteria.
In addition to the effect of Pseudomonas 20EI1 on the expression of the AF BGC, the bacteria also downregulated the expression of genes in the CPA, KA and imizoquin BGCs. Downregulation of A. flavus BGCs by bacteria has been previously observed, for example by P. megaterium. In co-culture, this bacterium downregulated 19 of these clusters (Kong et al., 2014; Gupta et al., 2020), impacting AFB1, and also CPA production. CPA and KA are both metal-binding metabolites. The most common CPA toxin, α-CPA, is an inhibitor of mammalian Ca2+-ATPases disrupting intracellular calcium flux (Chang and Ehrlich, 2011), while a closely related analog also produced by A. flavus, ß-CPA, has been shown to bind iron and affect the growth of Pseudomonas aeruginosa (Guo et al., 2023). KA, usually utilized in food industry, also presents antibacterial activity (Mahmoud et al., 2024) and binds iron (Sudhir et al., 2005). Imizoquin has been shown to present a protective role against oxidative stress in A. flavus (Khalid et al., 2018). Of these three compounds, CPA and KA were detected in our chemical analysis, and coinciding with the downregulation of their respective BGCs, production of these compounds was dramatically decreased in A. flavus by the bacteria in an iron-independent manner—although iron caused an increase in the expression of genes in those BGCs in co-culture, it was not sufficient to establish CPA and KA production suggesting another bacterial mode of action preventing their production.
A different expression pattern was observed in co-culture with Pseudomonas 20EI1, where the expression of genes in the aspergillicin BGC increased, while addition of iron decreased their expression. Aspergillicins are siderophores produced by A. flavus (Greco et al., 2019). It is likely that under iron starvation caused by the bacteria in the culture, A. flavus increased expression of these genes in an attempt to capture iron, and in the cases of cultures with abundant iron those genes were found repressed to prevent further iron acquisition.
Interestingly, while examining the effect of Pseudomonas 20EI1 on global regulatory genes in A. flavus, our transcriptome data indicated that the putative C2H2 transcription factor gene, mtfA, was significantly upregulated in the presence of the bacteria. The mtfA gene was first characterized in the model fungus A. nidulans where it was demonstrated to control development and secondary metabolism (Ramamoorthy et al., 2013), including the expression of genes involved in the synthesis of sterigmatocystin, penicillin and terrequinone. In A. flavus, mtfA overexpression results in a drastic reduction or elimination of several secondary metabolites, including AFB₁ (Zhuang et al., 2016). This decrease in AF was also accompanied by a reduction in aflR expression. It is possible that the effect of Pseudomonas 20EI1 on A. flavus SM could be in part mediated by MtfA, as a response to the presence of the bacteria in the environment, resulting in alterations of the A. flavus SM profile.
5 Conclusion
In conclusion, we have established that Pseudomonas 20EI1 is a strong biocontrol that affects A. flavus growth and the expression of BGCs in this agriculturally important fungus, resulting in alterations in the production of SM, that includes a drastic reduction of AF, CPA and KA levels. Interestingly, the effect of 20EI1 on A. flavus occurs in an iron-dependent manner, affecting the expression of numerous fungal iron-related genes, including those encoding iron transporters and siderophore biosynthetic genes. It is likely that in the warfare of iron-related proteins, a more efficient iron sequestration by the bacteria starves A. flavus, a condition that is remediated by increasing the abundance of iron in the environment. Here we also show for the first time that iron itself reduces AF production in this fungus. Additionally, the presence of Pseudomonas 20EI1 induced the upregulation of the global regulatory gene mtfA in A. flavus, which could contribute to the observed decrease of SM production, including AF. Future studies will focus on the application of this promising biocontrol to crops in laboratory and agricultural settings with the goal of reducing the detrimental effects of A. flavus and possibly of other mycotoxigenic fungi.
Data availability statement
The dataset generated and analyzed during this study are available in the Sequence Read Archive (SRA), accessible at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1207321, Accession: PRJNA1207321.
Author contributions
EW: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. SG: Investigation, Writing – original draft, Writing – review & editing, Methodology. MG: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. ML: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. JL: Conceptualization, Data curation, Validation, Writing – original draft, Writing – review & editing. JC: Conceptualization, Writing – original draft, Writing – review & editing. TS: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. AR: Writing – original draft, Writing – review & editing, Methodology, Supervision, Validation. AC: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the United States Department of Agriculture (USDA Grant 58-6054-4-040) and Northern Illinois University.
Acknowledgments
The authors thank Farzana Hossain for technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1514950/full#supplementary-material
References
Adebo, O. A., Njobeh, P. B., and Mavumengwana, V. (2016). Degradation and detoxification of AFB1 by Staphylocococcus warneri, Sporosarcina sp. and Lysinibacillus fusiformis. Food Control 68, 92–96. doi: 10.1016/j.foodcont.2016.03.021
Ahangarkani, F., Puts, Y., Nabili, M., Khodavaisy, S., Moazeni, M., Salehi, Z., et al. (2020). First azole-resistant Aspergillus fumigatus isolates with the environmental TR(46)/Y121F/T289A mutation in Iran. Mycoses 63, 430–436. doi: 10.1111/myc.13064
Akocak, P. B., Churey, J. J., and Worobo, R. W. (2015). Antagonistic effect of chitinolytic Pseudomonas and Bacillus on growth of fungal hyphae and spores of aflatoxigenic Aspergillus flavus. Food Biosci. 10, 48–58. doi: 10.1016/j.fbio.2015.01.005
Alberts, J. F., van Zyl, W. H., and Gelderblom, W. C. (2016). Biologically based methods for control of fumonisin-producing Fusarium species and reduction of the fumonisins. Front. Microbiol. 7:548. doi: 10.3389/fmicb.2016.00548
Al-Saad, L. A., Al-Badran, A. I., Al-Jumayli, S. A., Magan, N., and Rodríguez, A. (2016). Impact of bacterial biocontrol agents on aflatoxin biosynthetic genes, aflD and aflR expression, and phenotypic aflatoxin B1 production by Aspergillus flavus under different environmental and nutritional regimes. Int. J. Food Microbiol. 217, 123–129. doi: 10.1016/j.ijfoodmicro.2015.10.016
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/s0022-2836(05)80360-2
Amaike, S., and Keller, N. P. (2009). Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell 8, 1051–1060. doi: 10.1128/ec.00088-09
Amaike, S., and Keller, N. P. (2011). Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133. doi: 10.1146/annurev-phyto-072910-095221
Anders, S., Pyl, P. T., and Huber, W. (2014). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/s0168-6445(03)00055-x
Asare, G. A., Bronz, M., Naidoo, V., and Kew, M. C. (2007). Interactions between aflatoxin B1 and dietary iron overload in hepatic mutagenesis. Toxicology 234, 157–166. doi: 10.1016/j.tox.2007.02.009
Barahona, E., Navazo, A., Martínez-Granero, F., Zea-Bonilla, T., Pérez-Jiménez, R. M., Martín, M., et al. (2011). Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl. Environ. Microbiol. 77, 5412–5419. doi: 10.1128/AEM.00320-11
Barber, A. E., Riedel, J., Sae-Ong, T., Kang, K., Brabetz, W., Panagiotou, G., et al. (2020). Azole use in agriculture reduces Aspergillus fumigatus abundance but does not alter its population structure. bioRxiv. Available at: https://doi.org/10.1101/2020.05.26.116616. [Epub ahead of preprint].
Bhatnagar, D., Rajasekaran, K., Gilbert, M., Cary, J. W., and Magan, N. (2018). Advances in molecular and genomic research to safeguard food and feed supply from aflatoxin contamination. World Mycotoxin J. 11, 47–72. doi: 10.3920/WMJ2017.2283
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Buchfink, B., Reuter, K., and Drost, H.-G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Cary, J. W., Entwistle, S., Satterlee, T., Mack, B. M., Gilbert, M. K., Chang, P. K., et al. (2019). The transcriptional regulator Hbx1 affects the expression of thousands of genes in the aflatoxin-producing fungus Aspergillus flavus. G3 9, 167–178. doi: 10.1534/g3.118.200870
Cary, J. W., Gilbert, M. K., Lebar, M. D., Majumdar, R., and Calvo, A. M. (2018). Aspergillus flavus secondary metabolites: more than just aflatoxins. Food Saf. 6, 7–32. doi: 10.14252/foodsafetyfscj.2017024
Cary, J. W., Han, Z., Yin, Y., Lohmar, J. M., Shantappa, S., Harris-Coward, P. Y., et al. (2015). Transcriptome analysis of Aspergillus flavus reveals veA-dependent regulation of secondary metabolite gene clusters, including the novel aflavarin cluster. Eukaryot. Cell 14, 983–997. doi: 10.1128/ec.00092-15
Chalivendra, S., DeRobertis, C., Reyes Pineda, J., Ham, J. H., and Damann, K. (2018). Rice phyllosphere Bacillus species and their secreted metabolites suppress Aspergillus flavus growth and aflatoxin production in vitro and in maize seeds. Toxins 10:159. doi: 10.3390/toxins10040159
Chalivendra, S., and Ham, J. H. (2019). “Bacilli in the biocontrol of mycotoxins” in Bacilli and agrobiotechnology: phytostimulation and biocontrol. eds. M. T. Islam, M. M. Rahman, P. Pandey, M. H. Boehme, and G. Haesaert (Cham: Springer), 49–62.
Chandra Nayaka, S., Udaya Shankar, A. C., Reddy, M. S., Niranjana, S. R., Prakash, H. S., Shetty, H. S., et al. (2009). Control of Fusarium verticillioides, cause of ear rot of maize, by Pseudomonas fluorescens. Pest Manag. Sci. 65, 769–775. doi: 10.1002/ps.1751
Chang, P. K. (2003). The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol. Gen. Genomics 268, 711–719. doi: 10.1007/s00438-003-0809-3
Chang, P. K., and Ehrlich, K. C. (2011). Cyclopiazonic acid biosynthesis by Aspergillus flavus. Toxin Rev. 30, 79–89. doi: 10.3109/15569543.2011.576795
Chin-A-Woeng, T. F., Bloemberg, G. V., Mulders, I. H., Dekkers, L. C., and Lugtenberg, B. J. (2000). Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant Microbe Interact. 13, 1340–1345. doi: 10.1094/MPMI.2000.13.12.1340
Chowdhary, A., Sharma, C., Kathuria, S., Hagen, F., and Meis, J. F. (2014). Azole-resistant Aspergillus fumigatus with the environmental TR46/Y121F/T289A mutation in India. J. Antimicrob. Chemother. 69, 555–557. doi: 10.1093/jac/dkt397
Cotty, P. (1990). Effect of atoxigenic strains of Aspergillus flavus on aflatoxin contamination of developing cottonseed. Plant Dis. 74, 233–235. doi: 10.1094/PD-74-0233
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dorner, J. W., Cole, R. J., Erlington, D. J., Suksupath, S., McDowell, G. H., and Bryden, W. L. (1994). Cyclopiazonic acid residues in milk and eggs. J. Agric. Food Chem. 42, 1516–1518. doi: 10.1021/jf00043a023
Drott, M. T., Rush, T. A., Satterlee, T. R., Giannone, R. J., Abraham, P. E., Greco, C., et al. (2021). Microevolution in the pansecondary metabolome of Aspergillus flavus and its potential macroevolutionary implications for filamentous fungi. Proc. Natl. Acad. Sci. U.S.A. 118:e2021683118. doi: 10.1073/pnas.2021683118
Dutta, S., Yu, S. M., Jeong, S. C., and Lee, Y. H. (2020). High-throughput analysis of genes involved in biocontrol performance of Pseudomonas fluorescens NBC275 against gray mold. J. Appl. Microbiol. 128, 265–279. doi: 10.1111/jam.14475
Elnahal, A. S. M., El-Saadony, M. T., Saad, A. M., Desoky, E.-S. M., El-Tahan, A. M., Rady, M. M., et al. (2022). The use of microbial inoculants for biological control, plant growth promotion, and sustainable agriculture: a review. Eur. J. Plant Pathol. 162, 759–792. doi: 10.1007/s10658-021-02393-7
Eom, T.-J., Moon, H., Yu, J.-H., and Park, H.-S. (2018). Characterization of the velvet regulators in Aspergillus flavus. J. Microbiol. 56, 893–901. doi: 10.1007/s12275-018-8417-4
Gaba, S., Rai, A. K., Varma, A., Prasad, R., and Goel, A. (2022). Biocontrol potential of mycogenic copper oxide nanoparticles against Alternaria brassicae. Front. Chem. 10:966396. doi: 10.3389/fchem.2022.966396
Ganeshan, G., and Manoj Kumar, A. (2005). Pseudomonas fluorescens, a potential bacterial antagonist to control plant diseases. J. Plant Interact. 1, 123–134. doi: 10.1080/17429140600907043
Georgianna, D. R., Fedorova, N. D., Burroughs, J. L., Dolezal, A. L., Bok, J. W., Horowitz-Brown, S., et al. (2010). Beyond aflatoxin: four distinct expression patterns and functional roles associated with Aspergillus flavus secondary metabolism gene clusters. Mol. Plant Pathol. 11, 213–226. doi: 10.1111/j.1364-3703.2009.00594.x
Gong, A. D., Lei, Y. Y., He, W. J., Liao, Y. C., Ma, L., Zhang, T. T., et al. (2022). The inhibitory effect of Pseudomonas stutzeri YM6 on Aspergillus flavus growth and aflatoxins production by the production of volatile dimethyl trisulfide. Toxins 14:788. doi: 10.3390/toxins14110788
Gong, A.-D., Li, H.-P., Yuan, Q.-S., Song, X.-S., Yao, W., He, W.-J., et al. (2015). Antagonistic mechanism of iturin A and plipastatin A from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS One 10:e0116871. doi: 10.1371/journal.pone.0116871
Greco, C., Pfannenstiel, B. T., Liu, J. C., and Keller, N. P. (2019). Depsipeptide aspergillicins revealed by chromatin reader protein deletion. ACS Chem. Biol. 14, 1121–1128. doi: 10.1021/acschembio.9b00161
Greisen, K., Loeffelholz, M., Purohit, A., and Leong, D. (1994). PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32, 335–351. doi: 10.1128/jcm.32.2.335-351.1994
Gull, M., and Hafeez, F. Y. (2012). Characterization of siderophore producing bacterial strain Pseudomonas fluorescens Mst 8.2 as plant growth promoting and biocontrol agent in wheat. Afr. J. Microbiol. Res. 6, 6308–6318. doi: 10.5897/AJMR12.1285
Guo, Y., Ying, Y., Wu, Q., Wei, B., Chen, J., and Wang, H. (2023). β-cyclopiazonic acid binds iron demonstrating siderophore-like activity and promotes growth in Pseudomonas aeruginosa. J. Oceanol. Limnol. 41, 1159–1167. doi: 10.1007/s00343-022-2007-3
Gupta, R. S., Patel, S., Saini, N., and Chen, S. (2020). Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the subtilis and cereus clades of species. Int. J. Syst. Evol. Microbiol. 70, 5753–5798. doi: 10.1099/ijsem.0.004475
Haas, H. (2012). Iron—a key nexus in the virulence of Aspergillus fumigatus. Front. Microbiol. 3:28. doi: 10.3389/fmicb.2012.00028
Harrison, L. A., Letendre, L., Kovacevich, P., Pierson, E., and Weller, D. (1993). Purification of an antibiotic effective against Gaeumannomyces graminis var. tritici produced by a biocontrol agent, Pseudomonas aureofaciens. Soil Biol. Biochem. 25, 215–221. doi: 10.1016/0038-0717(93)90029-B
Hedayati, M. T., Pasqualotto, A. C., Warn, P. A., Bowyer, P., and Denning, D. W. (2007). Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153, 1677–1692. doi: 10.1099/mic.0.2007/007641-0
Höfte, M. (2021). “The use of Pseudomonas spp. as bacterial biocontrol agents to control plant disease” in Microbial bioprotectants for plant disease management (Cambridge: Burleigh Dodds Science Publishing).
Horn, B. W., Sorensen, R. B., Lamb, M. C., Sobolev, V. S., Olarte, R. A., Worthington, C. J., et al. (2014). Sexual reproduction in Aspergillus flavus sclerotia naturally produced in corn. Phytopathology 104, 75–85. doi: 10.1094/PHYTO-05-13-0129-R
Howell, C. R. (1979). Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology 69, 480–482. doi: 10.1094/Phyto-69-480
Howell, C. R., and Stipanovic, R. D. (1980). Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 70:712. doi: 10.1094/Phyto-70-712
Hua, S. S. T., Sarreal, S. B. L., Chang, P.-K., and Yu, J. (2019). Transcriptional regulation of aflatoxin biosynthesis and conidiation in Aspergillus flavus by Wickerhamomyces anomalus WRL-076 for reduction of aflatoxin contamination. Toxins 11:81. doi: 10.3390/toxins11020081
Huang, R., Feng, Z., Chi, X., Sun, X., Lu, Y., Zhang, B., et al. (2018). Pyrrolnitrin is more essential than phenazines for Pseudomonas chlororaphis G05 in its suppression of Fusarium graminearum. Microbiol. Res. 215, 55–64. doi: 10.1016/j.micres.2018.06.008
Hyde, K. D., Al-Hatmi, A. M. S., Andersen, B., Boekhout, T., Buzina, W., Dawson, T. L., et al. (2018). The world’s ten most feared fungi. Fungal Divers. 93, 161–194. doi: 10.1007/s13225-018-0413-9
Jones, P., Binns, D., Chang, H. -Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics. 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kale, S. P., Milde, L., Trapp, M. K., Frisvad, J. C., Keller, N. P., and Bok, J. W. (2008). Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 45, 1422–1429. doi: 10.1016/j.fgb.2008.06.009
Kandel, S. L., Jesmin, R., Mack, B. M., Majumdar, R., Gilbert, M. K., Cary, J. W., et al. (2022). Vibrio gazogenes inhibits aflatoxin production through downregulation of aflatoxin biosynthetic genes in Aspergillus flavus. PhytoFrontiers 2, 218–229. doi: 10.1094/PHYTOFR-09-21-0067-R
Kerr, J. R. (1999). Bacterial inhibition of fungal growth and pathogenicity. Microb. Ecol. Health Dis. 11, 129–142. doi: 10.1080/089106099435709
Khalid, S., Baccile, J. A., Spraker, J. E., Tannous, J., Imran, M., Schroeder, F. C., et al. (2018). NRPS-derived isoquinolines and lipopetides mediate antagonism between plant pathogenic fungi and bacteria. ACS Chem. Biol. 13, 171–179. doi: 10.1021/acschembio.7b00731
Kim, M. S., Kim, Y. C., and Cho, B. H. (2004). Gene expression analysis in cucumber leaves primed by root colonization with Pseudomonas chlororaphis O6 upon challenge-inoculation with Corynespora cassiicola. Plant Biol. 6, 105–108. doi: 10.1055/s-2004-817803
Kinyungu, S., Isakeit, T., Ojiambo, P. S., and Woloshuk, C. P. (2019). Spread of Aspergillus flavus and aflatoxin accumulation in postharvested maize treated with biocontrol products. J. Stored Prod. Res. 84:101519. doi: 10.1016/j.jspr.2019.101519
Kleinkauf, N., Verweij, P. E., Arendrup, M. C., Donnelly, P. J., Cuenca-Estrella, M., Fraaije, B. A., et al. (2013). Risk assessment on the impact of environmental usage of triazoles on the development and spread of resistance to medical triazoles in Aspergillus species (ECDC technical report). Stockholm: ECDC.
Kong, Q., Chi, C., Yu, J., Shan, S., Li, Q., Li, Q., et al. (2014). The inhibitory effect of Bacillus megaterium on aflatoxin and cyclopiazonic acid biosynthetic pathway gene expression in Aspergillus flavus. Appl. Microbiol. Biotechnol. 98, 5161–5172. doi: 10.1007/s00253-014-5632-8
Legein, M., Smets, W., Vandenheuvel, D., Eilers, T., Muyshondt, B., Prinsen, E., et al. (2020). Modes of action of microbial biocontrol in the phyllosphere. Front. Microbiol. 11:1619. doi: 10.3389/fmicb.2020.01619
Lohmar, J. M., Puel, O., Cary, J. W., and Calvo, A. M. (2019). The Aspergillus flavus rtfA gene regulates plant and animal pathogenesis and secondary metabolism. Appl. Environ. Microbiol. 85:e02446. doi: 10.1128/aem.02446-18
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mahmoud, G. A., Abdel Shakor, A. B., Kamal-Eldin, N. A., and Zohri, A. A. (2024). Production of kojic acid by Aspergillus flavus OL314748 using box-Behnken statistical design and its antibacterial and anticancer applications using molecular docking technique. BMC Microbiol. 24:140. doi: 10.1186/s12866-024-03289-2
Mamo, F. T., Shang, B., Selvaraj, J. N., Zheng, Y., and Liu, Y. (2022). Biocontrol efficacy of atoxigenic Aspergillus flavus strains against aflatoxin contamination in peanut field in Guangdong province, South China. Mycology 13, 143–152. doi: 10.1080/21501203.2021.1978573
Manwar, A. V., Khandelwal, S. R., Chaudhari, B. L., Meyer, J. M., and Chincholkar, S. B. (2004). Siderophore production by a marine Pseudomonas aeruginosa and its antagonistic action against phytopathogenic fungi. Appl. Biochem. Biotechnol. 118, 243–252. doi: 10.1385/ABAB:118:1-3:243
Misslinger, M., Hortschansky, P., Brakhage, A. A., and Haas, H. (2021). Fungal iron homeostasis with a focus on Aspergillus fumigatus. Biochim. Biophys. Acta 1868:118885. doi: 10.1016/j.bbamcr.2020.118885
Moore, G. G. (2022). Practical considerations will ensure the continued success of pre-harvest biocontrol using non-aflatoxigenic Aspergillus flavus strains. Crit. Rev. Food Sci. Nutr. 62, 4208–4225. doi: 10.1080/10408398.2021.1873731
Nagarajkumar, M., Bhaskaran, R., and Velazhahan, R. (2004). Involvement of secondary metabolites and extracellular lytic enzymes produced by Pseudomonas fluorescens in inhibition of Rhizoctonia solani, the rice sheath blight pathogen. Microbiol. Res. 159, 73–81. doi: 10.1016/j.micres.2004.01.005
Norred, W. P., Porter, J. K., Dorner, J. W., and Cole, R. J. (1988). Occurrence of the mycotoxin cyclopiazonic acid in meat after oral administration to chickens. J. Agric. Food Chem. 36, 113–116. doi: 10.1021/jf00079a028
Oliveira, C. A. F., Sebastião, L. S., Fagundes, H., Rosim, R. E., and Fernandes, A. M. (2008). Aflatoxins and cyclopiazonic acid in feed and milk from dairy farms in São Paulo, Brazil. Food Addit. Contam. B 1, 147–152. doi: 10.1080/02652030802382865
Palazzini, J. M., Dunlap, C. A., Bowman, M. J., and Chulze, S. N. (2016). Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: genome sequencing and secondary metabolite cluster profiles. Microbiol. Res. 192, 30–36. doi: 10.1016/j.micres.2016.06.002
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Peles, F., Sipos, P., Kovács, S., Győri, Z., Pócsi, I., and Pusztahelyi, T. (2021). Biological control and mitigation of aflatoxin contamination in commodities. Toxins 13:104. doi: 10.3390/toxins13020104
Philpott, C. C. (2006). Iron uptake in fungi: a system for every source. Biochim. Biophys. Acta 1763, 636–645. doi: 10.1016/j.bbamcr.2006.05.008
Qin, Z., Peng, Y., Pu, Y., Liu, T., Qian, K., and Tang, H. (2023). Study on nanomaterials with inhibitory effects on the growth of Aspergillus niger. Polymers 15:3820. doi: 10.3390/polym15183820
Rabiço, F., Borelli, T. C., Alnoch, R. C., MLTM, P., da Silva, R. R., Silva-Rocha, R., et al. (2024). Novel Pseudomonas species prevent the growth of the phytopathogenic fungus Aspergillus flavus. BioTech 13:8. doi: 10.3390/biotech13020008
Radha,, Kumari, N., Prakash, S., Sharma, N., Puri, S., Thakur, M., et al. (2024). Medicinal and aromatic plants as potential sources of bioactives along with health-promoting activities. Curr. Food Sci. Tech. Rep. 2, 359–376. doi: 10.1007/s43555-024-00042-8
Raksha Rao, K., Vipin, A. V., Hariprasad, P., Anu Appaiah, K. A., and Venkateswaran, G. (2017). Biological detoxification of aflatoxin B1 by Bacillus licheniformis CFR1. Food Control 71, 234–241. doi: 10.1016/j.foodcont.2016.06.040
Ramamoorthy, V., Dhingra, S., Kincaid, A., Shantappa, S., Feng, X., and Calvo, A. M. (2013). The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS One 8:e74122. doi: 10.1371/journal.pone.0074122
Robens, J., and Cardwell, K. (2003). The costs of mycotoxin management to the USA: management of aflatoxins in the United States. J. Toxicol. Toxin Rev. 22, 139–152. doi: 10.1081/TXR-120024089
Shu, X., Wang, Y., Zhou, Q., Li, M., Hu, H., Ma, Y., et al. (2018). Biological degradation of aflatoxin B1 by cell-free extracts of Bacillus velezensis DY3108 with broad PH stability and excellent thermostability. Toxins 10:330. doi: 10.3390/toxins10080330
Simon, A., Bindschedler, S., Job, D., Wick, L. Y., Filippidou, S., Kooli, W. M., et al. (2015). Exploiting the fungal highway: development of a novel tool for the in situ isolation of bacteria migrating along fungal mycelium. FEMS Microbiol. Ecol. 91:fiv116. doi: 10.1093/femsec/fiv116
Snelders, E., Huis In’t Veld, R. A., Rijs, A. J., Kema, G. H., Melchers, W. J., and Verweij, P. E. (2009). Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl. Environ. Microbiol. 75, 4053–4057. doi: 10.1128/aem.00231-09
Spraker, J. E., Wiemann, P., Baccile, J. A., Venkatesh, N., Schumacher, J., Schroeder, F. C., et al. (2018). Conserved responses in a war of small molecules between a plant-pathogenic bacterium and fungi. mBio 9:e00820. doi: 10.1128/mBio.00820-18
Sudhir, P.-R., Wu, H.-F., and Zhou, Z.-C. (2005). Probing the interaction of kojic acid antibiotics with iron(III) chloride by using electrospray tandem mass spectrometry. Rapid Commun. Mass Spectrom. 19, 209–212. doi: 10.1002/rcm.1773
Takeuchi, K., Noda, N., and Someya, N. (2014). Complete genome sequence of the biocontrol strain Pseudomonas protegens Cab57 discovered in Japan reveals strain-specific diversity of this species. PLoS One 9:e93683. doi: 10.1371/journal.pone.0093683
Takeuchi, K., and Someya, N. (2019). An overview of recent genomic research on biocontrol pseudomonad strains isolated from the field in Japan. Jpn. Agric. Res. Q. 53, 87–91. doi: 10.6090/jarq.53.87
Touati, D. (2000). Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373, 1–6. doi: 10.1006/abbi.1999.1518
Uka, V., Cary, J. W., Lebar, M. D., Puel, O., De Saeger, S., and Diana Di Mavungu, J. (2020). Chemical repertoire and biosynthetic machinery of the Aspergillus flavus secondary metabolome: a review. Compr. Rev. Food Sci. Food Saf. 19, 2797–2842. doi: 10.1111/1541-4337.12638
Veliz, E. A., Martínez-Hidalgo, P., and Hirsch, A. M. (2017). Chitinase-producing bacteria and their role in biocontrol. AIMS Microbiol. 3, 689–705. doi: 10.3934/microbiol.2017.3.689
Wang, P., Xu, J., Chang, P.-K., Liu, Z., and Kong, Q. (2022). New insights of transcriptional regulator AflR in Aspergillus flavus physiology. Microbiol. Spectr. 10:e0079121. doi: 10.1128/spectrum.00791-21
WHO (2018). New food safety series launched in February 2018. Available at: https://web.archive.org/web/20180918200124/https://www.who.int/foodsafety/foodsafetydigest/en/
Wiemann, P., Lechner, B. E., Baccile, J. A., Velk, T. A., Yin, W. B., Bok, J. W., et al. (2014). Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Aspergillus fumigatus. Front. Microbiol. 5:530. doi: 10.3389/fmicb.2014.00530
Williams, J. H., Phillips, T. D., Jolly, P. E., Stiles, J. K., Jolly, C. M., and Aggarwal, D. (2004). Human aflatoxicosis in developing countries: a review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr. 80, 1106–1122. doi: 10.1093/ajcn/80.5.1106
Wu, F., Liu, Y., and Bhatnagar, D. (2008). Cost-effectiveness of aflatoxin control methods: economic incentives. Toxin Rev. 27, 203–225. doi: 10.1080/15569540802393690
Xia, X., Zhang, Y., Li, M., Garba, B., Zhang, Q., Wang, Y., et al. (2017). Isolation and characterization of a Bacillus subtilis strain with aflatoxin B1 biodegradation capability. Food Control 75, 92–98. doi: 10.1016/j.foodcont.2016.12.036
Xie, Y., Wang, W., and Zhang, S. (2019). Purification and identification of an aflatoxin B1 degradation enzyme from Pantoea sp. T6. Toxicon 157, 35–42. doi: 10.1016/j.toxicon.2018.11.290
Yang, X., Zhang, Q., Chen, Z.-Y., Liu, H., and Li, P. (2017). Investigation of Pseudomonas fluorescens strain 3JW1 on preventing and reducing aflatoxin contaminations in peanuts. PLoS One 12:e0178810. doi: 10.1371/journal.pone.0178810
Keywords: Aspergillus flavus, Pseudomonas, biocontrol, transcriptome, secondary metabolism, aflatoxin, iron, MtFA
Citation: Wyman EM, Grayburn WS, Gilbert MK, Lebar MD, Lohmar JM, Cary JW, Sauters TJC, Rokas A and Calvo AM (2025) An environmental isolate of Pseudomonas, 20EI1, reduces Aspergillus flavus growth in an iron-dependent manner and alters secondary metabolism. Front. Microbiol. 15:1514950. doi: 10.3389/fmicb.2024.1514950
Edited by:
Bin Ji, Wuhan University of Science and Technology, ChinaReviewed by:
Sanjay Kumar Singh Patel, Hemwati Nandan Bahuguna Garhwal University, IndiaPhilipp Wiemann, Solugen, Inc., United States
Copyright © 2025 Wyman, Grayburn, Gilbert, Lebar, Lohmar, Cary, Sauters, Rokas and Calvo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana M. Calvo, YW1jYWx2b0BuaXUuZWR1
 Elizabeth M. Wyman1
Elizabeth M. Wyman1 W. Scott Grayburn
W. Scott Grayburn Matthew D. Lebar
Matthew D. Lebar Jessica M. Lohmar
Jessica M. Lohmar Antonis Rokas
Antonis Rokas Ana M. Calvo
Ana M. Calvo