- 1State Key Laboratory of Chinese Medicine Modernization, Tianjin University of Traditional Chinese Medicine, Tianjin, China
- 2Key Laboratory of Cancer Prevention and Therapy, Tianjin’s Clinical Research Center for Cancer, National Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin, China
- 3Guangzhou Laboratory, Guangzhou, China
- 4Department of Pharmaceutical Engineering and Pharmaceutical Chemistry, College of Chemical Engineering, Qingdao University of Science and Technology, Qingdao, China
- 5Haihe Laboratory of Modern Chinese Medicine, Tianjin, China
Background: The unavoidable propagation of the coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has underscored the urgent requirement for efficacious therapeutic agents. The dried fruit of Terminalia chebula Retz., namely Chebulae Fructus, is widely used for treating bacterial and viral infectious diseases, which was witnessed to perform anti-SARS-CoV-2 activity in recommended Chinese patent medicine.
Aim: SARS-CoV-2 main protease (Mpro) and papain-like protease (PLpro) present essential effects on SARS-CoV-2 replication and transcription, considering as the attractive targets for therapeutic intervention. In this study, we focused on the dual-target to obtain broad-spectrum antiviral candidates from Chebulae Fructus.
Methods: The identified compounds from Chebulae Fructus were used to build a library in a previous study, which were evaluated by molecular docking to screen potential antiviral agents. The SARS-CoV-2 Mpro and PLpro were expressed in E. coli cells and purified. Fluorescence resonance energy transfer (FRET) and surface plasmon resonance (SPR) were utilized to verify the affinity with dual targets. SARS-CoV-2 wild-type, Omicron BA.5 and Omicron EG.5 variants were employed to validate their antiviral activities in vitro. Molecular dynamics simulation was conducted via Gromacs 2022 software in 500 ns to unveil the conformation stability.
Results: Targeting on Mpro and PLpro, eight compounds were screened as the potential dual-target inhibitors in molecular docking. In FRET and SPR assays, 1,2,3,4,6-penta-O-galloyl-β-D-glucose (PGG) and 1,2,3,6-tetra-O-galloyl-β-D-glucose (TGG) showed good inhibitory activities with IC50 values ranging from 1.33 to 27.37 μM, and affinity with KD values ranging from 0.442 to 0.776 μM. Satisfactorily, both PGG and TGG display antiviral activity in vitro with EC50 values ranging from 3.20 to 37.29 μM, suggesting as the promising candidates against SARS-CoV-2. In molecular dynamics simulation study, the complexes of Mpro-PGG, Mpro-TGG, PLpro-PGG, and PLpro-TGG exhibited stability over 500 ns period, unveiling the potential interactions.
Conclusion: PGG and TGG are the promising dual-target inhibitors of SARS-CoV-2, which may avoid drug resistance and have a good development prospect. The outcomes of this study provide an effective strategy to systematically explore the antiviral bioactive compounds from Chebulae Fructus.
1 Introduction
The quest for curbing the global proliferation of coronavirus disease 2019 (COVID-19), triggered by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), still represents a critical challenge. Since its initial outbreak in late 2019, it has led to over 775 million confirmed cases and 7.05 million deaths worldwide as of August 2024 (WHO, n.d.). The advent of vaccines was a pivotal milestone in preventing the spread of COVID-19. The emergence of SARS-CoV-2 variants has been accompanied by a notable immune escape, while the protective effects of vaccines have been found to be relatively weak (Jackson et al., 2020; Li et al., 2022; Yu et al., 2020). Moreover, the pace of antiviral drug development has not kept up with the rapid mutation rate of SARS-CoV-2 (Beigel et al., 2020). It underscores the importance of identifying more effective and biosafe broad-spectrum antiviral agents to treat COVID-19 effectively.
In the battlefield against SARS-CoV-2, the main protease (Mpro) (Liu et al., 2022) and the papain-like protease (PLpro) (Zang et al., 2023) devote essential part to cleaving the polyproteins pp1a and pp1ab to 16 non-structural proteins (Nsps), which is crucial for SARS-CoV-2 replication. Mpro comprises three domains (domains I, II, and III) and takes charge of 11 cleavage sites at least (Jin et al., 2020). Given high conservation in the active site across coronaviridae viruses and non-homology to mammalian proteins, Mpro is regarded as the most crucial target for the development of broad-spectrum antiviral candidates. In a previous study, the active unit of Mpro is a homodimer, whose substrate-binding site located within the C145-H41 catalytic dyad between domain I and II (Jin et al., 2020). The catalytic pocket is an important region for inhibitor binding. PLpro is another cleavage protease, belonging to a cysteine protease, and takes charge of other three cleavage sites to release Nsp1, Nsp2 and Nsp3. The PLpro contains two potential inhibitor binding pockets, including a classical catalytic triad (C111-H272-A286) and the BL2 domain. Unlike Mpro, the PLpro catalytic domain is normally off but opens when substrates close to it, making it harder for molecules to bind and giving low inhibitory activity. However, PLpro is capable of removing signaling proteins and ubiquitin-like interferon-stimulated gene 15 (ISG15) in the host, thereby suppressing innate immune responses (Shin et al., 2020). It gives the PLpro inhibitors with both antiviral and anti-inflammatory activity (Osipiuk et al., 2021), which is an important feature differing from Mpro inhibitors. Therefore, dual-target inhibitors of Mpro and PLpro offer a promising therapeutic strategy, akin to a combination therapy within a single medication, potentially circumventing drug resistance more effectively than single-target drugs in the fight against viral infections (Yang et al., 2005). Recent research has shown that simultaneous inhibition of Mpro and PLpro is markedly more efficacious in combating SARS-CoV-2 and its variants (Narayanan et al., 2022). Therefore, we focused on SARS-CoV-2 Mpro and PLpro to develop dual-target inhibitors. However, despite both enzymes being cysteine proteases, their substrate specificities and active site configurations differ, posing challenges for screening effective dual-target inhibitors (Rut et al., 2020).
Chinese herbal medicine is a great treasure, supplying numerous strategies to conquer diseases. In COVID-19 treatment, due to the wide application of traditional Chinese medicine, the mortality and severe disease incidence reduce significantly (Lyu et al., 2023). The dried fruit of Terminalia chebula Retz., namely Chebulae Fructus (CF), is referred to as the king of Tibetan medicine (Nigam et al., 2020), and is widely used in many countries for its antibacterial and antiviral effects, such as India, Bhutan, Maldives, Nepal and China (Bidikar et al., 2022). Notably, CF extract displays broad-spectrum antiviral activities, including herpes simplex virus-2 (Kesharwani et al., 2017), influenza A virus strains (Li et al., 2020), human immunodeficiency virus (Ahn et al., 2002), and SARS-CoV-2 (Upadhyay et al., 2020). Several effective Chinese patent medicines with CF as the main component were, respectively, recommended by the National Medical Products Administration (NMPA) and the Joint Prevention and Control Mechanism of the State Council, such as Tibetan medicine influenza pills, Cuitang granules, and Qingyan Dropping pills. With natural products’ diverse structures and complex compositions, they are invaluable for antiviral drug discovery. It is imperative to rapidly identify bioactive compounds from CF and clarify their antiviral mechanism, supplying the structural guidance for lead compound design, which is crucial for the development of antiviral drugs.
In this study, a total of 122 compounds derived from CF were subjected to molecular docking to assess their binding energy with both Mpro and PLpro. Of these, eight compounds exhibited strong binding affinity. Specifically, 1,2,3,4,6-penta-O-galloyl-β-D-glucose (PGG) and 1,2,3,6-tetra-O-galloyl-β-D-glucose (TGG) demonstrated high enzymatic activity against Mpro and PLpro by fluorescence resonance energy transfer (FRET). Furthermore, through surface plasmon resonance (SPR) assay, PGG and TGG possess good binding affinities for Mpro and PLpro. They exhibited potent antiviral activity against both SARS-CoV-2 wild-type strain, Omicron BA.5 and Omicron EG.5 variant in Vero E6 cells. In the end, molecular dynamics simulation was employed to unveil the potential binding mechanism, suggesting that PGG and TGG could serve as promising dual-target inhibitors of Mpro and PLpro against SARS-CoV-2.
2 Materials and methods
2.1 Chemicals and reagents
The tested compounds (PGG, TGG, punicalin, and 1,3,6-tri-O-galloylglucose) and three positive controls (PF-07321332, GRL0617, and S-217622) were acquired from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China) with purity of 98% by HPLC. CM5 sensor chips, amine-coupling kit, and running buffer were purchased from General Electric Company (Boston, MA, USA). Dulbecco’s modified Eagle’s medium and fetal bovine serum were purchased from Gibco Invitrogen Corp. (New York, USA).
2.2 Molecular docking
The crystal structures of SARS-CoV-2 Mpro (PDB ID: 6 LU7), SARS-CoV-2 PLpro (PDB ID: 7CJM), MERS-CoV Mpro (PDB ID: 9BOO), and HCoV 229E Mpro (PDB ID: 7YRZ) were obtained from the RCSB PDB.1 The 122 compounds in CF, which were identified in a previous study (Wang et al., 2023), were employed as ligands. The structural details of the ligands were obtained from PubChem2 database and ChemDraw in SDF or mol2 formats, respectively.
Ligands and proteins were prepared by utilizing Discovery Studio 2020 software. The protein preparation contains the removal of extraneous water molecules, the addition of hydrogen atoms, and the repair of missing residues. The binding site was identified based on the location of the native ligand, which was subsequently excised from the protein model. Subsequently, docking simulations were conducted using the cdocker mode. The docking results were visualized by PyMOL version 2.6. N3, the native ligand of Mpro, and GRL0617, the native ligand of PLpro, served as positive controls (Fu et al., 2021; Jin et al., 2020). Each ligand was docked into its respective protein.
2.3 Cloning, protein expression, and purification of SARS-CoV-2 Mpro and PLpro
The plasmids encoding the full-length genes of SARS-CoV-2 Mpro and PLpro were successfully constructed and then individually transformed into Escherichia coli BL21 (DE3) cells. A 100 μL aliquot of competent cells was thawed on ice and mixed with either the Mpro or PLpro plasmid solution. The mixture was incubated on ice for 30 min before undergoing a heat shock at 42°C for 90 s. After the heat shock, 900 μL of fresh LB medium was added to the cells to facilitate recovery at 37°C 220 rpm for 1 h. The supernatant was then cleanly and quickly pipetted off after centrifugation. The recovered cultures were spread onto LB agar plates containing the appropriate antibiotic at 37°C for 12–16 h. As s shown in Supplementary Figure S1, these transformed cells were then cultured in Luria broth medium supplemented with 100 μg/mL ampicillin for Mpro expression and 50 μg/mL kanamycin for PLpro expression. Cultivation was carried out at 37°C for 6–8 h. Subsequently, 500 μM isopropyl-beta-D-thiogalactopyranoside (IPTG) was added to induce protein expression at 16°C for 16–20 h. Following induction, the cells were harvested by centrifugation at 1,500 × g for 20 min at 4°C. The resultant cell pellet was lysed using a high-pressure homogenizer and centrifugation at 16,000 × g for 1.5 h at 4°C, and the cell supernatant was collected. The proteins were eluted by running buffer containing 20 mM Tris–HCl (pH 8.0), 150 mM NaCl, 5% glycerol, and 300 mM imidazole through the Ni-NTA affinity chromatography column (5 mL, QIAGEN, Shenzhen, China) at a flow rate of 5 mL/min. The C-terminal 6 × His tag of Mpro was excised by human rhinovirus 3C protease, while the small ubiquitin-like modifier moiety attached to PLpro was cleaved with small ubiquitin-like modifier enzyme. For the ion exchange chromatography, a HiTrap™ Q HP column (5 mL, GE Healthcare, Little Chalfont, UK) at a flow rate of 1 mL/min was utilized with a buffer system comprising 20 mM Tris–HCl (pH 8.0) and 1 M NaCl. The elution was performed using a gradient of NaCl concentration from 50 mM to 1 M to effectively separate the proteins based on their different charges. The size-exclusion chromatography was conducted on a Superdex 200 Increase 10/300 GL column (GE Healthcare, Little Chalfont, Buckinghamshire, UK) at a flow rate of 0.5 mL/min with a buffer system containing 20 mM Tris–HCl and 150 mM NaCl at pH 8.0. Using SDS-PAGE, the purity of Mpro was determined to be approximately 88.3%, and PLpro was about 88.5%.
2.4 Determination of enzymatic inhibition activities on SARS-CoV-2 Mpro and PLpro
Via a continuous kinetic assay, the enzymatic activity of SARS-CoV-2 Mpro was quantified by employing the specific substrate MCA-AVLQSGFR-Lys (Dnp)-Lys-NH2, which is synthesized by GL Biochem Co., Ltd. (Shanghai, China). This excitation and emission wavelengths were 340 nm and 460 nm, respectively. The enzymatic reaction system of Mpro includes 20 mM Tris–HCl, 150 mM NaCl, 2 mM DTT, 0.2 μM Mpro and variable substrate concentration ranging from 4 to 36 μM at a pH of 8.0. For SARS-CoV-2 PLpro, the substrate Dabcyl-FTLKGGYAPTKVTE-Edans was also provided by GL Biochem Co., Ltd. The excitation and emission wavelengths were 336 nm and 490 nm, respectively. The enzymatic reaction system of PLpro contains 20 mM Tris–HCl, 150 mM NaCl, 10 mM DTT, 1.0 μM PLpro, and variable substrate concentrations ranging from 10 to 100 μM at a pH of 8.0. Fluorescence intensity was meticulously monitored using a Spark multimode microplate reader from Tecan (Männedorf, ZH, Switzerland).
For enzymatic inhibitor screening, the sample solutions of the tested compounds (40 μM) were preliminarily used to evaluate their inhibitory activities, and the compounds with inhibitory activity over 60% were employed for determining IC50 values (Yi et al., 2022). The enzymatic system comprised 0.2 μM Mpro or 1.0 μM PLpro, 20 μM substrate for Mpro or 10 μM substrate for PLpro, and various concentrations of the tested compounds. All experimental results were rigorously analyzed by employing GraphPad Prism 8, with triple replicates of each experiment to ensure data reliability.
2.5 SPR assay
The binding affinity was assessed via SPR assay, conducted on Biacore T200. Mpro and PLpro were, respectively, immobilized onto the surface of an activated CM5 sensor chip through an amine coupling reaction. Initially, the compounds were prepared in DMSO at a concentration of 2.5 mM, serving as the standard stock solution, which was subsequently diluted 20-fold with 1.05 × PBS-P to achieve an initial concentration of 125 μM. Then, they were stepwise diluted by running buffer (1.05 × PBS-P containing 5% DMSO), yielding a range of concentrations from 0.030 μM to 125 μM. The assay conditions were optimized with a flow rate of 30 μL/min, while the association and dissociation phases were set at 120 s and 180 s, respectively. Data analysis was carried out via BIAevaluation software version 2.0 in the kinetics analysis mode to elucidate the binding properties.
2.6 Antiviral activity assay
The antiviral activity of the tested compounds was assessed through the inhibition of cytopathic effect (CPE) in Vero E6 cells, which was conducted in the Biosafety Level 3 (BSL-3) laboratory in Guangzhou Customs Inspection and Quarantine Technology Center. Vero E6 cells were inoculated in a 96-well plate at a density of 1 × 105 cells per well and maintained in DMEM culture solution containing 10% fetal bovine serum, 100 μg/mL streptomycin, and 100 U/mL penicillin at 37°C with an atmosphere comprising 5% CO2 and 95% air. Firstly, the cytotoxicity was evaluated by the CCK8 assay to obtain the non-cytotoxicity dose, which was calculated according to the half-cytotoxic concentration (CC50) value. Subsequently, at the safe dose, cells were mixed with SARS-CoV-2 wild-type, Omicron BA.5 or Omicron EG.5 strain (MOI = 0.01) and the tested compounds at varying concentrations (0.0031 to 50.00 μM) and incubated for 48 h. The scores quantifying CPE were obtained by the Celigo Image Cytometer. The 50% tissue culture infectious dose (TCID50) was calculated by employing the Reed-Muench formula, serving as a reference to facilitate the determination of the half-maximal effective concentration (EC50) of the tested compounds.
2.7 Molecular dynamics simulation
After the evaluation of the molecular docking and experimental verification results, a molecular dynamics simulation was performed to analyze the stability of protein-ligand complexes and unveil the interaction mechanism. The CHARMM-GUI was used to construct four simulation systems, including SARS-CoV-2 Mpro-PGG, SARS-CoV-2 Mpro-TGG, SARS-CoV-2 PLpro-PGG, and SARS-CoV-2 PLpro-TGG. The dimensions of the simulation boxes were 12 × 12 × 12 nm3, with the total number of atoms varying from approximately 100,000 to 150,000. CHARMM36m force field was applied, and the simulation parameters were the same as in a previous study (Cao et al., 2023; Huang et al., 2017). A physiological environment was mimicked by incorporating 0.15 M NaCl into the simulation systems. During the simulation phase, initial energy minimization was performed using the steepest descent algorithm for 20,000 steps, then followed by a 300 ns pre-equilibration six times to optimize the system. NVT (constant volume and pressure) and NPT (constant pressure and temperature) simulations were employed by 100 ns. Finally, molecular dynamics simulation was performed for each complex at 500 ns. Electrostatic interactions were described via the Particle Mesh Ewald (PME) method, with a cutoff radius of 1.2–1.4 nm. The LINear Constraint Solver (LINCS) algorithm was employed to constrain chemical bonds. Pressure was maintained at 1 bar in the x, y, and z directions using the C-rescale pressure coupling, and temperature was kept constantly at the 310 K using the V-rescale thermostat. All simulations were conducted using the GROMACS2022 software package (Valdés-Tresanco et al., 2021). The root means square deviation (RMSD), root mean square fluctuation (RMSF), and the radius of gyration (Rg) were analyzed.
3 Results
3.1 High-throughput screening of compounds with potential inhibitory activity against Mpro and PLpro by molecular docking
The binding energies between the tested compounds and Mpro or PLpro were determined by Discovery Studio 2020 based on SARS-CoV-2 Mpro (PDB ID: 6 LU7) and PLpro (PDB ID: 7CJM) crystal structures (Figure 1A), respectively. The molecular docking results showed that 16 out of the 122 compounds exhibited the lower binding affinity to Mpro than the positive control (N3), which were classified into three groups based on their chemical structures: gallotannins, chebulic ellagitannins, and ellagitannins (Figure 1B). Meanwhile, 27 out of the 122 compounds performed a lower binding affinity to PLpro than the positive control (GRL0617), which were classified into five categories: gallotannins, ellagitannins, phenolcarboxylic acids, flavonoids, and chebulic ellagitannins (Figure 1C). To our surprise, eight compounds showed the potential dual-target inhibitory activities (Figure 1D), which need to be verified experimentally.
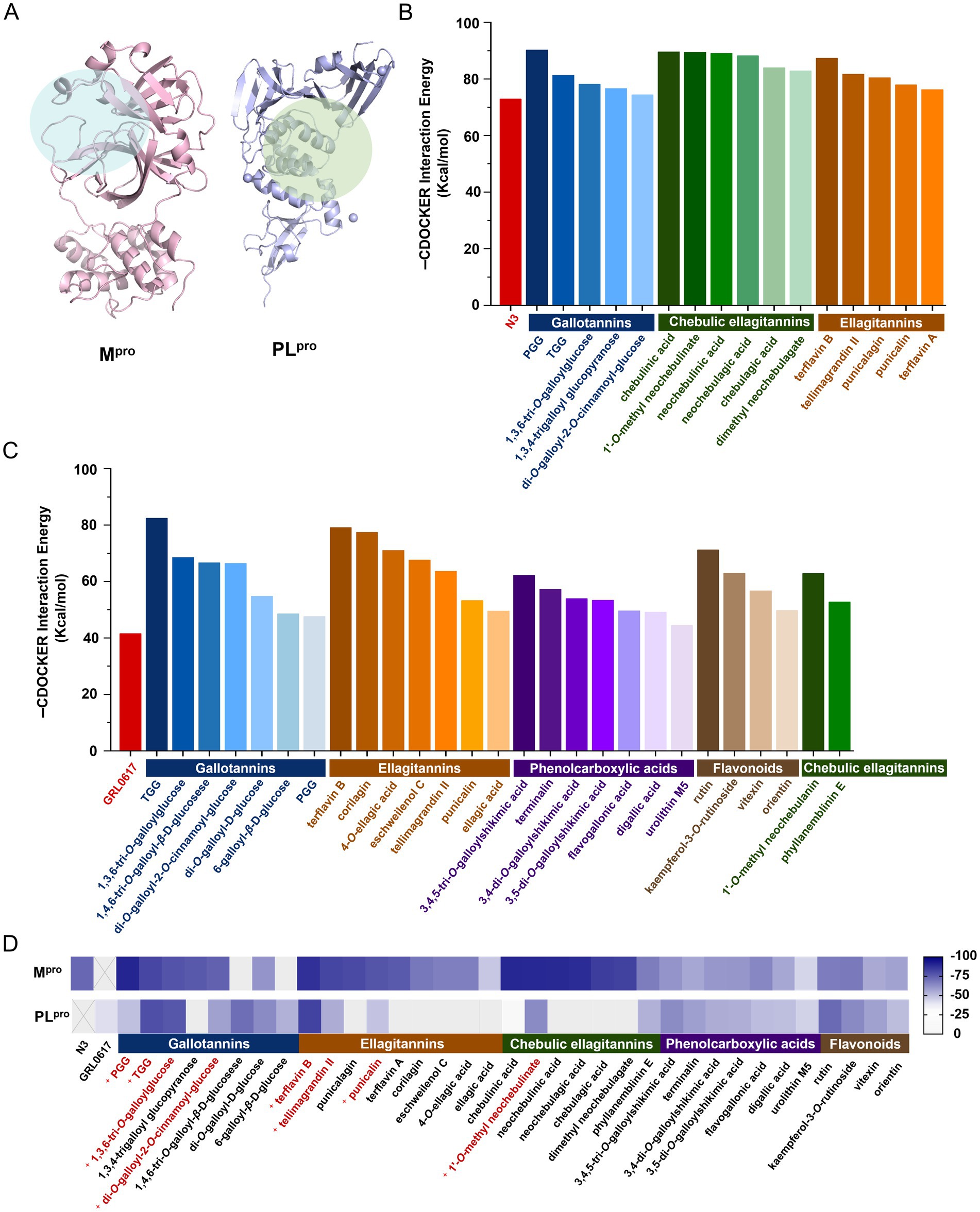
Figure 1. Binding affinity of the tested compounds with SARS-CoV-2 Mpro or PLpro by molecular docking. (A) Mpro and PLpro and their active pockets. The cdocker interaction energy of the tested compounds bound to Mpro (B) or PLpro (C). (D) The heatmap analysis of cdocker interaction energy to screen the potential dual-target inhibitors.
3.2 Evaluation of binding affinity with SARS-CoV-2 Mpro and PLpro by FRET and SPR assays
Recombinant SARS-CoV-2 Mpro and PLpro expressed in E. coli cells were purified (Supplementary Figures S1A,B,D,E). The enzymatic catalytic efficiency (kcat/Km) for substrates of SARS-CoV-2 Mpro and PLpro was 12,516 M−1·S−1 and 1,845 M−1·S−1, respectively (Supplementary Figures 1C,F).
In the FRET assay, there are four compounds (PGG, TGG, 1,3,6-tri-O-galloylglucose, and punicalagin) available, which can be employed to determine the inhibitory activities. In 40 μM, PGG and TGG exhibited inhibition effects over 60% on both Mpro and PLpro (Figure 2A). PF-07321332 is reported as the inhibitor of Mpro, which can be employed as the positive control (Owen et al., 2021). GRL0617, used as the positive control, effectively inhibited PLpro activity (Shin et al., 2020). Interestingly, these two compounds shared the similar structures. Compared with TGG, PGG possesses an additional galloyl unit at the 4th substitution base (Figure 2B). The results showed that PF-07321332, PGG, and TGG inhibited Mpro with IC50 values of 0.04 μM, 1.89 μM, and 1.33 μM, respectively (Figures 2C–E). GRL0617, PGG, and TGG inhibited PLpro with IC50 values of 2.78 μM, 14.09 μM, and 27.37 μM, respectively (Figures 2F–H).
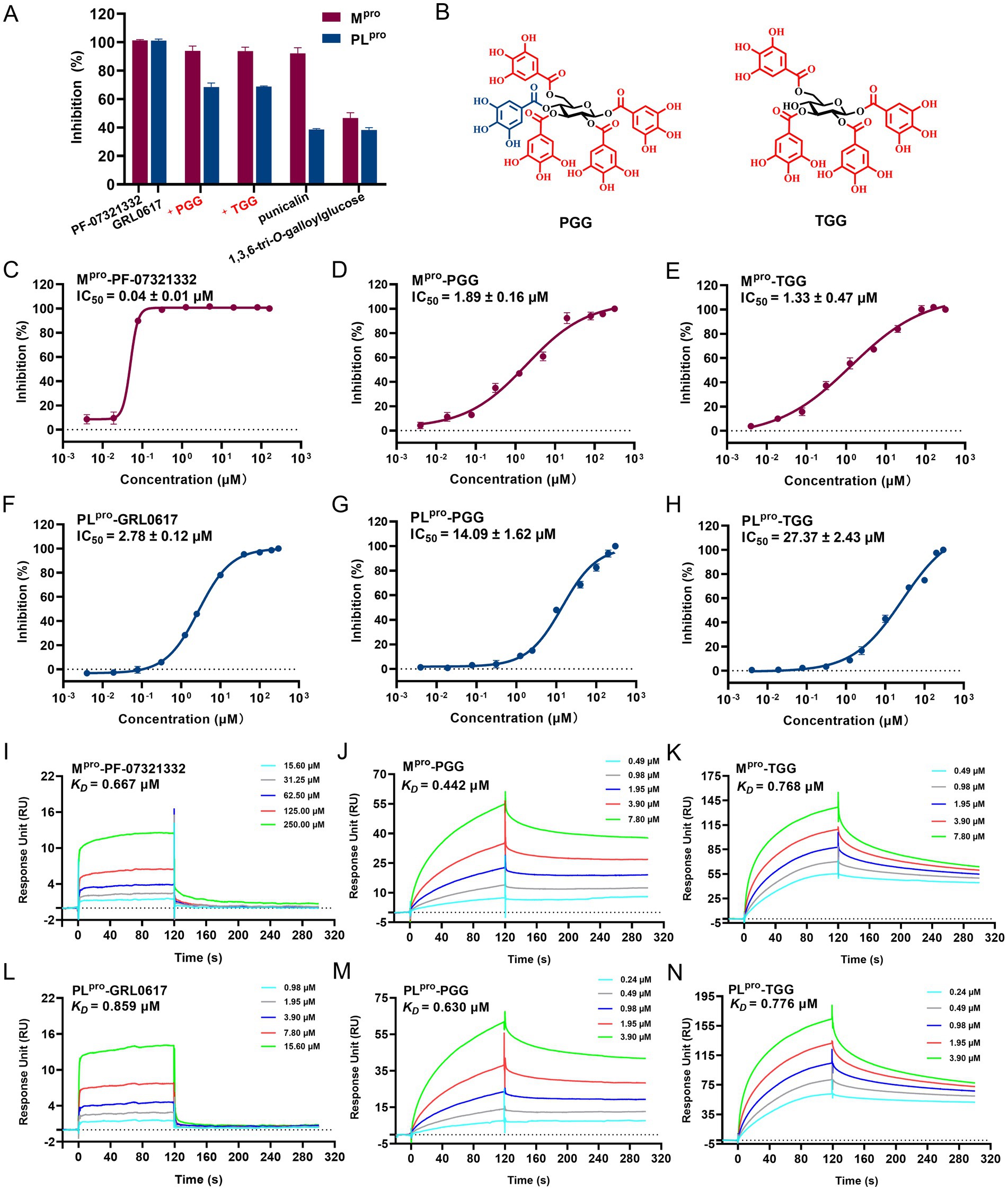
Figure 2. The inhibitory activities of the tested compounds against SARS-CoV-2 Mpro and PLpro. (A) The inhibitory activities of the tested compounds (40 μM) against Mpro and PLpro. (B) Chemical structures of 1,2,3,4,6-penta-O-galloyl-β-D-glucose (PGG) and 1,2,3,6-tetra-O-galloyl-β-D-glucose (TGG). The IC50 curves of PF-07321332 (C), PGG (D), and TGG (E) against Mpro. The IC50 curves of GRL0617 (F), PGG (G), and TGG (H) against PLpro. SPR assay for the binding affinity of the tested PF-07321332 (I), PGG (J), and TGG (K) with Mpro. SPR assay for binding affinity of the tested GRL0617 (L), PGG (M), and TGG (N) with PLpro. (mean ± SD, n = 3).
The binding affinities of various compounds to the Mpro or PLpro were quantitatively assessed via SPR, a pivotal method for monitoring ligand-enzyme interactions in real time. The lower dissociation constant (KD) signifies a stronger affinity of compounds binding to the target protein, highlighting its promise as a therapeutic agent. In this study, the KD values for PF-07321332, PGG, and TGG with Mpro were determined to be 0.667 μM, 0.442 μM, and 0.768 μM, respectively (Figures 2I–K). The KD values for GRL0617, PGG, and TGG with PLpro were evaluated to be 0.859 μM, 0.630 μM, and 0.776 μM, respectively (Figures 2L–N). In summary, the integration of FRET and SPR underscores the potential activities of PGG and TGG against SARS-CoV-2, which may hold promise in the development of novel therapeutic strategies.
3.3 Evaluation of antiviral activity on SARS-CoV-2 wild type, omicron BA.5 and omicron EG.5 in vitro
As the potential dual-target inhibitors, PGG and TGG showed excellent safety on Vero E6 cells with CC50 > 100 μM (Figure 3A). In antiviral activity assay, S-217622, a nonpeptidic inhibitor targeting Mpro, was used as a positive control drug (Unoh et al., 2022). As illustrated in Figure 3B, the EC50 values for S-217622, PGG, and TGG against the SARS-CoV-2 wild-type strain are presented as 0.05 μM, 8.22 μM, and 13.00 μM, respectively. Against the Omicron BA.5 variant, these compounds display EC50 values of 0.09 μM for S-217622, 3.20 μM for PGG, and 25.72 μM for TGG. Furthermore, when the compounds were assessed against the Omicron EG.5 variant, the EC50 values are 0.06 μM for S-217622, 15.44 μM for PGG, and 37.29 μM for TGG. It suggested that PGG and TGG performed cellular protection by antiviral activity.
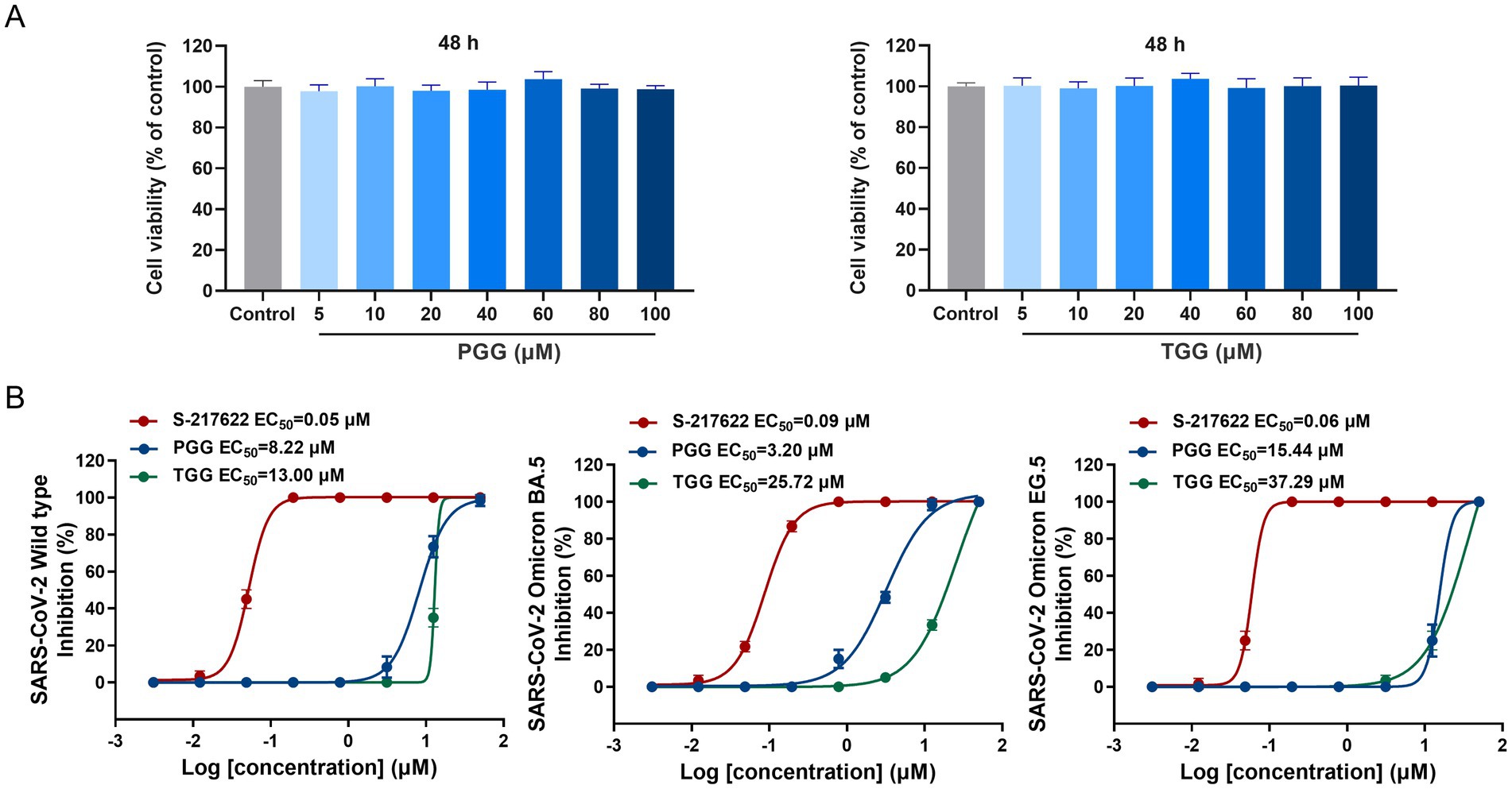
Figure 3. Cytotoxic effect and antiviral activity of the tested compounds. (A) Effect of 1,2,3,4,6-penta-O-galloyl-β-D-glucose (PGG) and 1,2,3,6-tetra-O-galloyl-β-D-glucose (TGG) on the viability of Vero E6 cells by CCK8. (B) The antiviral activities of S-217622, PGG, and TGG against SARS-CoV-2 wild-type, Omicron BA.5 and Omicron EG.5 in vitro. (mean ± SD, n = 3).
Also, it is hypothesized that PGG and TGG played antiviral activity mainly through inhibiting the activities of Mpro and PLpro to prevent the viral replication. Moreover, PGG and TGG exhibit favorable solubility and stability, rendering them as promising candidates for the development of antiviral inhibitors.
3.4 Unveiling of the antiviral mechanism for PGG and TGG by in silico approach
To elucidate the binding modes involved in the interaction of the focused compounds and targets, molecular docking was conducted. For SARS-CoV-2 Mpro, the active site is defined at the C145-H41 catalytic dyad, which is originally occupied by N3. As depicted in Figures 4A,B, PGG and TGG both align in the binding pocket in preferred conformation, and the cdocker interaction energy is −90.21 and −81.24 kcal/mol, which is originated by the intermolecular hydrogen bonds. These bonds were formed with the residues H413.8, S462.7, C1453.2, G1663.1, and A1882.9 in the SARS-CoV-2 Mpro-PGG. Similarly, in the SARS-CoV-2 Mpro-TGG complex, hydrogen bonds were formed with the residues H413.2, L1413.4, S1442.6, C1453.6, and G1663.0.
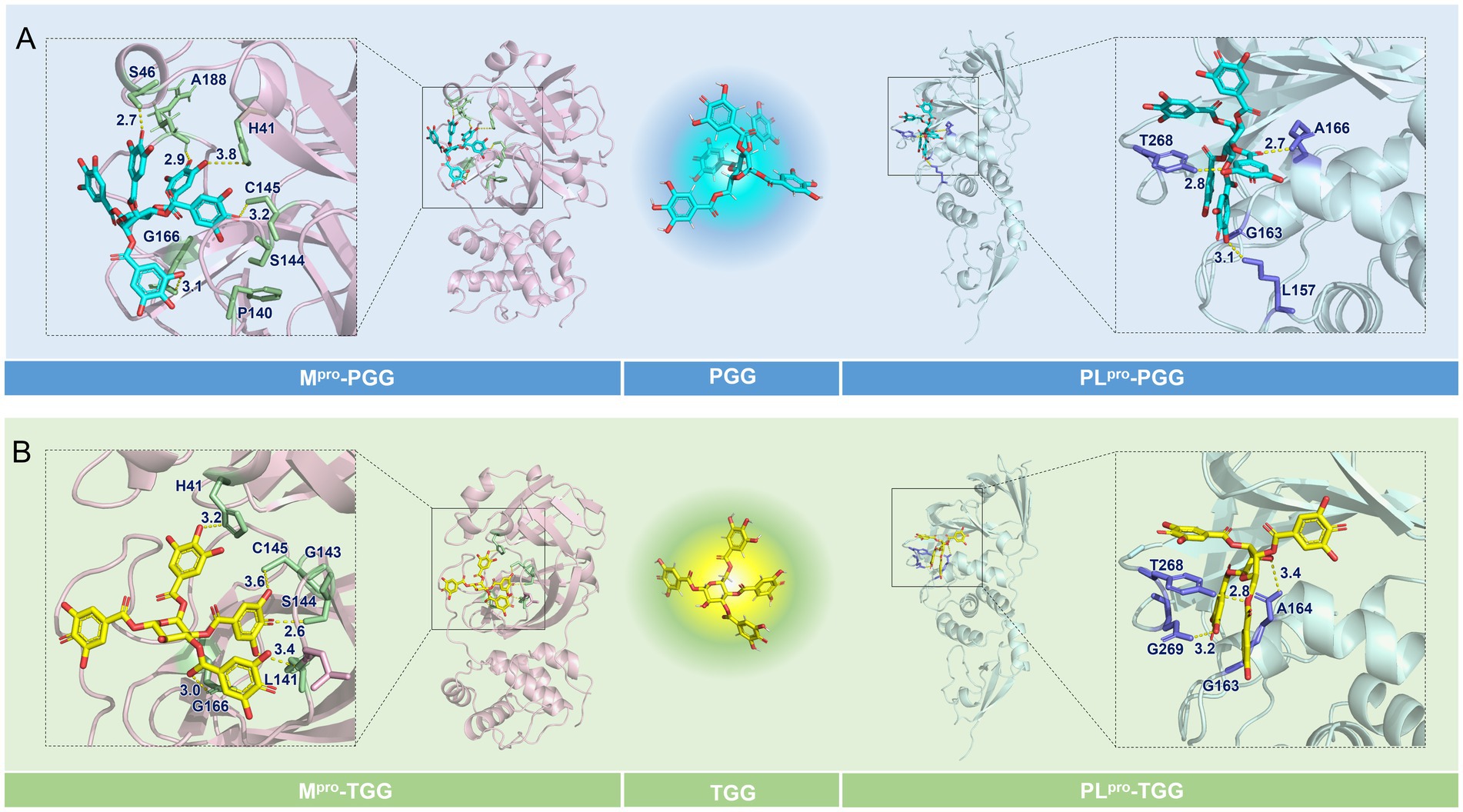
Figure 4. Binding modes of the focused compounds and the interesting targets. (A) The predicted binding modes of the Mpro-1,2,3,4,6-penta-O-galloyl-β-D-glucose (PGG) and PLpro-PGG complexes. (B) The predicted binding modes of the Mpro-1,2,3,6-tetra-O-galloyl-β-D-glucose (TGG) and PLpro-TGG complexes.
The structure of SARS-CoV-2 PLpro is split into four sub-domains: the N-terminal ubiquitin-like domain (Ubl, β1-3), the α-helical thumb domain (α2-7), the β-stranded finger domain (β4-7), and the palm domain (β8-13). As shown in Figures 4A,B, PGG and TGG fit into the cleft formed between the BL2 loop and the α3-α4 loop, which is almost identical to GRL0617 (Gao et al., 2021), and the cdocker interaction energy is −49.56 and −79.11 kcal/mol, respectively. Hydrogen bonds are also the dominant interactions. In the SARS-CoV-2 PLpro-PGG complex, PGG formed multiple hydrogen bonds with the residues L1573.1, A1662.7, and T2682.8. Similarly, in the SARS-CoV-2 PLpro-TGG complex, TGG interacted with the residues A1643.4, T2682.8, and T2693.2 to form hydrogen bonds, contributing to execution of inhibition activities.
Additionally, as shown in Supplementary Figures S3A,B, docking studies of PGG and TGG with MERS-CoV Mpro (PDB ID: 9BOO) yielded interaction energies of −83.38 and −93.61 kcal/mol, respectively. Key interactions involve hydrogen bonds with residues S147, C148, G167, G169, and H194 for PGG, and P143, G146, G169, H175, L191, and G192 for TGG. Docking results for the HCoV-229E Mpro (PDB ID: 7YRZ) showed interaction energies of −83.09 and −94.67 kcal/mol for PGG and TGG, respectively, with critical hydrogen bonds involving residues H41, G142, A143, C144, G163, A186, and G191 for PGG, and A141, G142, A143, C144, G165, and G191 for TGG (Supplementary Figures 3C,D). Hence, the findings suggest that PGG and TGG may perform the potential inhibitory effects on MERS-CoV and HCoV-229E.
3.5 Molecular dynamics simulation
Molecular dynamics simulation is pivotal for post-docking analysis, providing insights into the structural stability of the ligand-receptor complex. Here, we employed the CHARMM36m force field, which is commonly used in ligand-receptor molecular dynamics simulation study. A series of molecular dynamics simulations were performed for the SARS-CoV-2 Mpro-PGG, SARS-CoV-2 Mpro-TGG, SARS-CoV-2 PLpro-PGG, and SARS-CoV-2 PLpro-TGG complexes spanning a period of 500 ns. As shown in Figure 5A, the RMSD values of Mpro range between 1.2 and 3.0 Å in the Mpro-PGG complex and 1.4 and 2.5 Å in the Mpro-TGG complex, meanwhile the RMSD values of PLpro range from 1.5 to 2.8 Å for the PLpro-PGG complex and 1.5 to 2.5 Å for the PLpro-TGG complex, which were witnessed to display structural stability. Moreover, we examined the RMSF for each amino acid involved in these receptor-ligand interactions. As shown in Figure 5B, it revealed that the maximum RMSF values is about 4 Å in the Mpro-TGG, PLpro-PGG, and PLpro-TGG complexes. Nevertheless, the Mpro-PGG complex RMSF value shows slight fluctuations. Additionally, the stability of the complex was further assessed using the Rg value, which reflects the structural compactness. The measured Rg displayed a narrow fluctuation, suggesting the good compactness of the tested compounds (Figure 5C). Generally, the simulation results indicated that the ligands, PGG and TGG, were bound to the receptor stability.
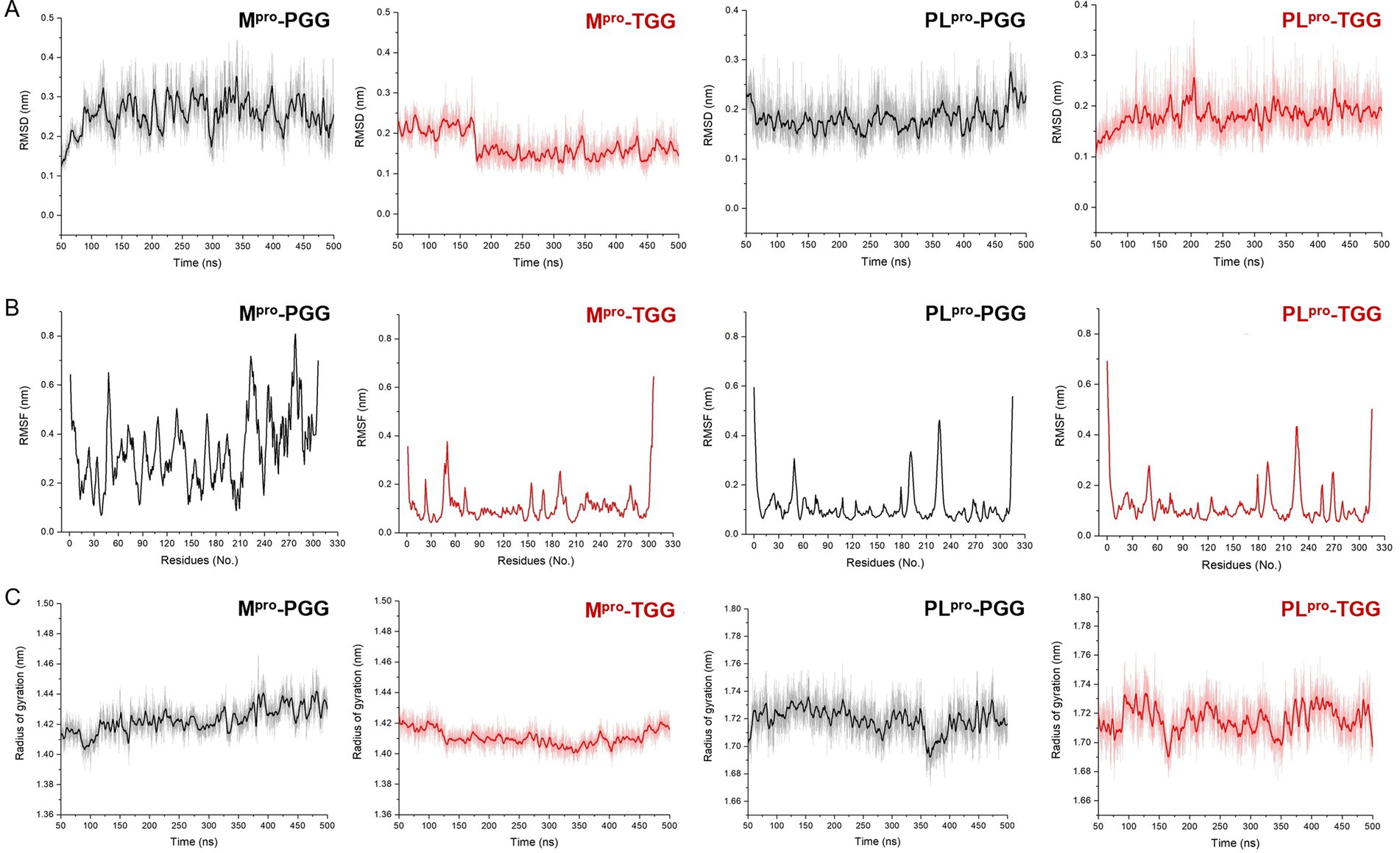
Figure 5. The molecular dynamics simulation for 1,2,3,4,6-penta-O-galloyl-β-D-glucose (PGG) and 1,2,3,6-tetra-O-galloyl-β-D-glucose (TGG) inside the active site of Mpro and PLpro, respectively. The analysis of root mean square deviation (RMSD) (A), root mean square fluctuation (RMSF) (B), and radius of gyration (Rg) (C) for Mpro-PGG, Mpro-TGG, PLpro-PGG, and PLpro-TGG complexes. The molecular dynamics simulation for each system was performed for 500 ns.
The binding sites and pocket of Mpro are shown in Figure 6A. Initially, PGG and TGG exhibit obvious interactions with Mpro and are well-encased by H41, S46, L141, C145, G166, and A191, which highlights the binding stability of the Mpro-PGG (Figure 6B) and Mpro-TGG complexes (Figure 6C). Similarly, the active sites and pocket of PLpro are depicted in Figure 6D. Here, PGG and TGG also show evident interactions with PLpro and are effectively surrounded by L157, G163, A164, A166, T268, and G269. This result demonstrates the stability of the PLpro-PGG (Figure 6E) and PLpro-TGG complexes (Figure 6F) during molecular dynamics simulation. The simulation trajectories showed that PGG and TGG could be bound stably inside the active pockets, which is consistent with the experimental results.
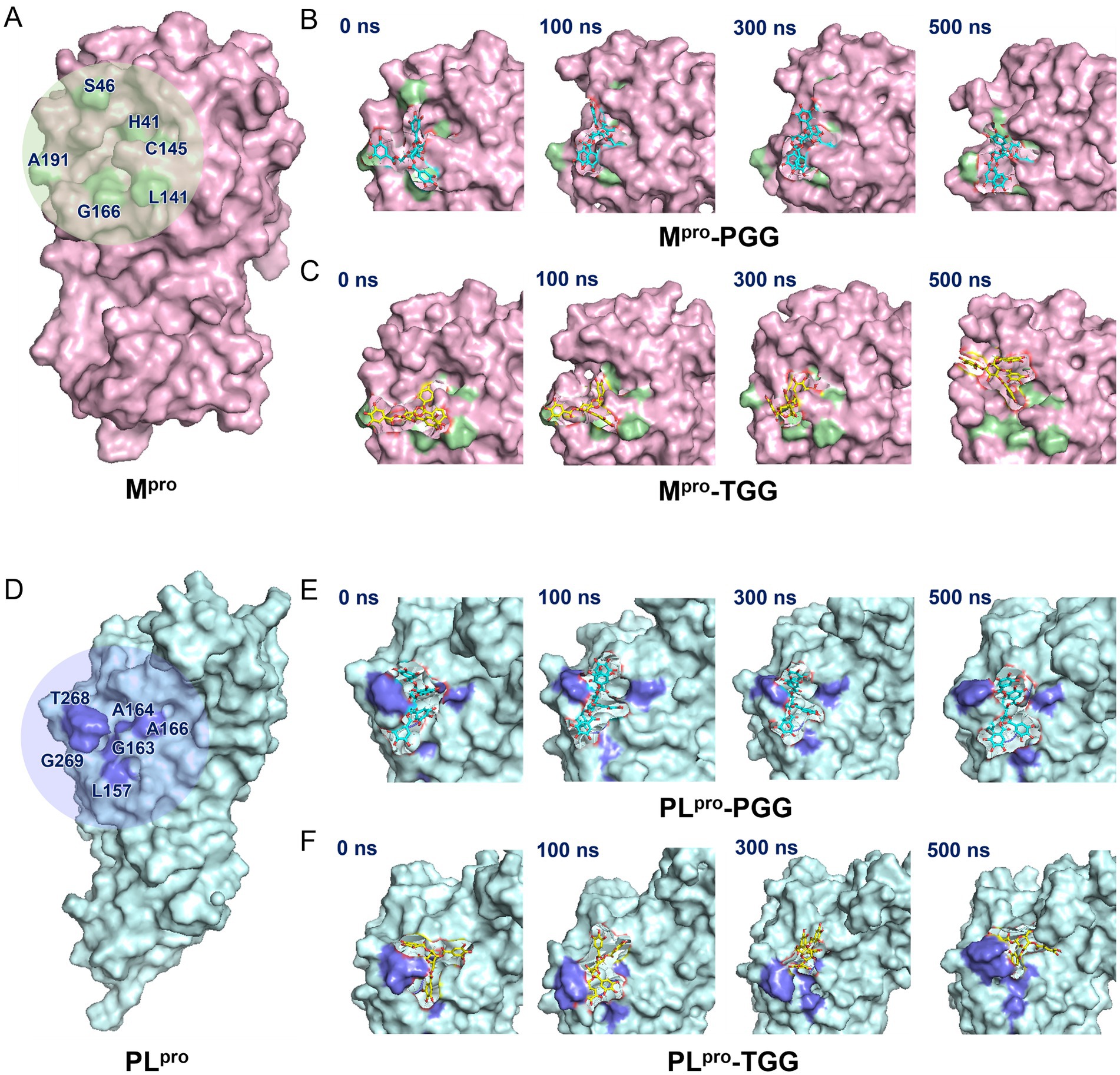
Figure 6. Illustration on 1,2,3,4,6-penta-O-galloyl-β-D-glucose (PGG) and 1,2,3,6-tetra-O-galloyl-β-D-glucose (TGG)‘s conformations inside the substrate-binding sites of Mpro and PLpro. (A) Mpro active site. (B) The surface view of Mpro-PGG complex in molecular dynamics simulation. (C) The surface view of Mpro-TGG complex in molecular dynamics simulation. (D) PLpro active site. (E) The surface view of PLpro-PGG complex in molecular dynamics simulation. (F) The surface view of PLpro-TGG complex in molecular dynamics simulation. The snapshot was taken at 0, 100, 300, and 500 ns, respectively.
4 Discussion
With the global spread of COVID-19, it is urgent to develop specific broad-spectrum antiviral drug against SARS-CoV-2. In Chinese medicine, it gives a great opportunity to mine antiviral candidates from the structurally diverse natural products by the established method, which not only develops a strategy for rapid identification of antiviral candidate but also offers a compound library for responding to other viral outbreaks.
Among the verified therapeutic targets, Mpro and PLpro were recognized as the most essential targets for their highly conservation in the active site from SARS-CoV-2 wide-type strain to Omicron variants (Zang et al., 2023; Rosales et al., 2024). Upon comparing Mpro and PLpro sequences including SARS-CoV-2 wild-type, Omicron BA.5, and Omicron EG.5 with those of the currently circulating strain SARS-CoV-2 Omicron JN.1, high sequence similarity was observed (Supplementary Figures S4, S5). Given that PGG and TGG exhibit inhibitory effects on SARS-CoV-2 wild-type, Omicron BA.5, and Omicron EG.5, we hypothesize that PGG and TGG may also serve as potential inhibitors against Omicron JN.1. A number of compounds were reported to specifically inhibit Mpro or PLpro, resulting in prevention of viral replication. Importantly, the dual-target inhibitors present a promising therapeutic approach, potentially outmaneuvering drug resistance more effectively than single-target alternatives. There are a few natural compounds reported with dual-target inhibitory activity, such as schaftoside (Yi et al., 2022), chrysin 7-O-β-D-glucuronide (Yi et al., 2024), ginkgolic acid, anacardic acid (Chen et al., 2021), and (−)–epigallocatechin gallate (Wang et al., 2022) unit now. Fortunately, we found another potential dual-target compounds, PGG and TGG, as tannins. PGG possesses five galloyl substitutions, and TGG has four. PGG has been reported to have antiviral activities on herpes simplex virus type 1, influenza A virus, human respiratory syncytial virus, human rhinoviruses, and so on (Torres et al., 2017). In our study, the increased number of hydroxyl groups in the tested compounds might enhance their interaction with the protease via intermolecular hydrogen bonds. However, interactions between ligands and receptors are complex and may also arise from hydrogen bonds, π-π conjugation, and so on. Through systematic research, we hope to correlate the structure of tannins with their activity, which could facilitate the discovery of antiviral compounds.
Accordingly, PGG and TGG are regarded as noncovalent inhibitors. Compared with covalent inhibitors, noncovalent inhibitors often exhibit better membrane permeability and high affinity for target proteins (Zhang et al., 2015). Additionally, the reversible binding reduces the risk of undesirable toxic effects associated with interactions with host proteins and nucleic acids (El-Baba et al., 2020). PGG and TGG act as competitive inhibitors on Mpro and PLpro of SARS-CoV-2, which are abundant in CF, conducing to the illumination of CF efficacy against SARS-CoV-2.
In order to improve the screening efficiency and reliability, we built a high-throughput screening system of virtual screening, molecular dynamics simulation combining with experimental verification, which has been demonstrated to be a successful strategy (Supplementary Figure S6). In general, PGG and TGG are considered as the promising antiviral compounds derived from CF. The structure of PGG and TGG also provides guidance for the design of antiviral compounds.
5 Conclusion
In summary, the dual-target inhibitors, PGG and TGG, were identified to act effectively against SARS-CoV-2 via targeting SARS-CoV-2 Mpro and PLpro, which satisfactorily inhibit the SARS-CoV-2 wild-type strain, Omicron BA.5 and Omicron EG.5 variant on Vero E6 cells, suggesting as the promising antiviral inhibitors. The comprehensive approach, encompassing molecular docking, FRET assay, SPR assay, antiviral activity validations, and molecular dynamics simulation, provides an effective strategy and allows us to systematically explore the antiviral bioactive compounds from CF, conducting to the discovery of antiviral compounds from Chinese medicine.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
CW: Writing – original draft. YC: Writing – original draft. QY: Writing – original draft. XW: Writing – original draft. ZY: Writing – original draft. JY: Writing – review & editing. XL: Writing – original draft. BL: Writing – original draft. YW: Writing – review & editing. MZ: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (Grant Nos. 82104372 and 31900894), the Science and Technology Project of Haihe Laboratory of Modern Chinese Medicine (22HHZYJC00007), Shandong Provincial Natural Science Foundation (ZR2021LZY032), Tianjin Graduate Research Innovation Project (2022BKY182), TUTCM Graduate Research Innovation Project (YJSKC-20221011).
Acknowledgments
We are grateful to the staff at Research Centre of Modern Analytical Technology, Tianjin University of Science and Technology to support surface plasmon resonance assay.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the preparation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1510665/full#supplementary-material
Footnotes
References
Ahn, M. J., Kim, C. Y., Lee, J. S., Kim, T. G., Kim, S. H., Lee, C. K., et al. (2002). Inhibition of HIV-1 integrase by galloyl glucoses from Terminalia chebula and flavonol glycoside gallates from Euphorbia pekinensis. Planta Med. 68, 457–459. doi: 10.1055/s-2002-32070
Beigel, J. H., Tomashek, K. M., Dodd, L. E., Mehta, A. K., Zingman, B. S., Kalil, A. C., et al. (2020). Remdesivir for the treatment of COVID-19-final report. N. Engl. J. Med. 383, 1813–1826. doi: 10.1056/NEJMoa2007764
Bidikar, C. M., Hurkadale, P. J., Nandanwadkar, S. M., and Hegde, H. V. (2022). A validated spectro densitometric regulatory compliant USP-HP-TLC protocol for quantification of polyphenols and antioxidants from polyherbal formulations containing Terminalia species. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1207:123379. doi: 10.1016/j.jchromb.2022.123379
Cao, Y., Wei, H., Jiang, S., Lu, T., Nie, P., Yang, C., et al. (2023). Effect of AQP4 and its palmitoylation on the permeability of exogenous reactive oxygen species: insights from computational study. Int. J. Biol. Macromol. 253:127568. doi: 10.1016/j.ijbiomac.2023.127568
Chen, Z., Cui, Q., Cooper, L., Zhang, P., Lee, H., Chen, Z., et al. (2021). Ginkgolic acid and anacardic acid are specific covalent inhibitors of SARS-CoV-2 cysteine proteases. Cell Biosci. 11:45. doi: 10.1186/s13578-021-00564-x
El-Baba, T. J., Lutomski, C. A., Kantsadi, A. L., Malla, T. R., John, T., Mikhailov, V., et al. (2020). Allosteric inhibition of the SARS-CoV-2 main protease: insights from mass spectrometry based assays. Angew. Chem. Int. Ed. 59, 23544–23548. doi: 10.1002/anie.202010316
Fu, Z., Huang, B., Tang, J., Liu, S., Liu, M., Ye, Y., et al. (2021). The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 12:488. doi: 10.1038/s41467-020-20718-8
Gao, X., Qin, B., Chen, P., Zhu, K., Hou, P., Wojdyla, J. A., et al. (2021). Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B 11, 237–245. doi: 10.1016/j.apsb.2020.08.014
Huang, J., Rauscher, S., Nawrocki, G., Ran, T., Feig, M., de Groot, B. L., et al. (2017). CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73. doi: 10.1038/nmeth.4067
Jackson, L. A., Anderson, E. J., Rouphael, N. G., Roberts, P. C., Makhene, M., Coler, R. N., et al. (2020). An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 383, 1920–1931. doi: 10.1056/NEJMoa2022483
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020). Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293. doi: 10.1038/s41586-020-2223-y
Kesharwani, A., Polachira, S. K., Nair, R., Agarwal, A., Mishra, N. N., and Gupta, S. K. (2017). Anti-HSV-2 activity of Terminalia chebula Retz extract and its constituents, chebulagic and chebulinic acids. BMC Compl. Altern. Med. 17:110. doi: 10.1186/s12906-017-1620-8
Li, P., Du, R., Wang, Y., Hou, X., Wang, L., Zhao, X., et al. (2020). Identification of chebulinic acid and chebulagic acid as novel influenza viral neuraminidase inhibitors. Front. Microbiol. 11:182. doi: 10.3389/fmicb.2020.00182
Li, X., Wichai, N., Wang, J., Liu, X., Yan, H., Wang, Y., et al. (2022). Regulation of innate and adaptive immunity using herbal medicine: benefits for the COVID-19 vaccination. Acupunct. Herb. Med. 2, 196–206. doi: 10.1097/HM9.0000000000000046
Liu, H., Iketani, S., Zask, A., Khanizeman, N., Bednarova, E., Forouhar, F., et al. (2022). Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19. Nat. Commun. 13:1891. doi: 10.1038/s41467-022-29413-2
Lyu, M., Xiao, G., Wang, S., Wang, R., Tan, L., Ma, S., et al. (2023). “Three medicines and three formulas” in COVID-19: from bench to bedside. Acupunct. Herb. Med. 3, 309–322. doi: 10.1097/HM9.0000000000000082
Narayanan, A., Narwal, M., Majowicz, S. A., Varricchio, C., Toner, S. A., Ballatore, C., et al. (2022). Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in-cell-protease assay. Commun. Biol. 5:169. doi: 10.1038/s42003-022-03090-9
Nigam, M., Mishra, A. P., Adhikari-Devkota, A., Dirar, A. I., Hassan, M. M., Adhikari, A., et al. (2020). Fruits of Terminalia chebula Retz.: a review on traditional uses, bioactive chemical constituents and pharmacological activities. Phytother. Res. 34, 2518–2533. doi: 10.1002/ptr.6702
Osipiuk, J., Azizi, S. A., Dvorkin, S., Endres, M., Jedrzejczak, R., Jones, K. A., et al. (2021). Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 12:743. doi: 10.1038/s41467-021-21060-3
Owen, D. R., Allerton, C. M. N., Anderson, A. S., Aschenbrenner, L., Avery, M., Berritt, S., et al. (2021). An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 374, 1586–1593. doi: 10.1126/science.abl4784
Rosales, R., McGovern, B. L., Rodriguez, M. L., Leiva-Rebollo, R., Diaz-Tapia, R., Benjamin, J., et al. (2024). Nirmatrelvir and molnupiravir maintain potent in vitro and in vivo antiviral activity against circulating SARS-CoV-2 omicron subvariants. Antivir. Res. 230:105970. doi: 10.1016/j.antiviral.2024.105970
Rut, W., Lv, Z., Zmudzinski, M., Patchett, S., Nayak, D., Snipas, S. J., et al. (2020). Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti-COVID-19 drug design. Sci. Adv. 6:eabd4596. doi: 10.1126/sciadv.abd4596
Shin, D., Mukherjee, R., Grewe, D., Bojkova, D., Baek, K., Bhattacharya, A., et al. (2020). Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587, 657–662. doi: 10.1038/s41586-020-2601-5
Torres, C., Ventura, J., Serna, L., Ascacio, J. A., Contreras, J., and Aguilar, C. N. (2017). Pentagalloylglucose (PGG): a valuable phenolic compound with functional properties. J. Funct. Foods 37, 176–189. doi: 10.1016/j.jff.2017.07.045
Unoh, Y., Uehara, S., Nakahara, K., Nobori, H., Yamatsu, Y., Yamamoto, S., et al. (2022). Discovery of S-217622, a noncovalent Oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J. Med. Chem. 65, 6499–6512. doi: 10.1021/acs.jmedchem.2c00117
Upadhyay, S., Tripathi, P. K., Singh, M., Raghavendhar, S., Bhardwaj, M., and Patel, A. K. (2020). Evaluation of medicinal herbs as a potential therapeutic option against SARS-CoV-2 targeting its main protease. Phytother. Res. 34, 3411–3419. doi: 10.1002/ptr.6802
Valdés-Tresanco, M. S., Valdés-Tresanco, M. E., Valiente, P. A., and Moreno, E. (2021). gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 17, 6281–6291. doi: 10.1021/acs.jctc.1c00645
Wang, X., Xu, J., Zhang, L. H., Yang, W., Yu, H., Zhang, M., et al. (2023). Global profiling of the antioxidant constituents in chebulae fructus based on an integrative strategy of UHPLC/IM-QTOF-MS, MS/MS molecular networking, and spectrum-effect correlation. Antioxidants 12:2093. doi: 10.3390/antiox12122093
Wang, C., Yang, Z., Chai, X., Wang, Y., Wang, W., and Zhang, M. (2022). Tea as a natural gift for discovering antiviral candidates. Acupunct. Herb. Med. 2, 211–220. doi: 10.1097/HM9.0000000000000048
WHO WHO coronavirus disease (COVID-19) pandemic, (n.d.) Available at https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (Accessed August 28, 2024).
Yang, H., Xie, W., Xue, X., Yang, K., Ma, J., Liang, W., et al. (2005). Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 3:e324. doi: 10.1371/journal.pbio.0030324
Yi, Y., Yu, R., Xue, H., Jin, Z., Zhang, M., Bao, Y. O., et al. (2024). Chrysin 7-O-β-D-glucuronide, a dual inhibitor of SARS-CoV-2 3CL(pro) and PL(pro), for the prevention and treatment of COVID-19. Int. J. Antimicrob. Agents 63:107039. doi: 10.1016/j.ijantimicag.2023.107039
Yi, Y., Zhang, M., Xue, H., Yu, R., Bao, Y. O., Kuang, Y., et al. (2022). Schaftoside inhibits 3CL(pro) and PL(pro) of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19. Acta Pharm. Sin. B 12, 4154–4164. doi: 10.1016/j.apsb.2022.07.017
Yu, J., Tostanoski, L. H., Peter, L., Mercado, N. B., McMahan, K., Mahrokhian, S. H., et al. (2020). DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811. doi: 10.1126/science.abc6284
Zang, Y., Su, M., Wang, Q., Cheng, X., Zhang, W., Zhao, Y., et al. (2023). High-throughput screening of SARS-CoV-2 main and papain-like protease inhibitors. Protein Cell 14, 17–27. doi: 10.1093/procel/pwac016
Keywords: SARS-CoV-2, main protease, papain-like protease, 1,2,3,4,6-penta-O-galloyl-β-D-glucose (PGG), 1,2,3,6-tetra-O-galloyl-β-D-glucose (TGG)
Citation: Wang C, Cao Y, Yang Q, Wang X, Yang Z, Yang J, Li X, Li B, Wang Y and Zhang M (2024) High-throughput screening of dual-target inhibitors for SARS-CoV-2 main protease and papain-like protease from Chebulae Fructus: in silico prediction and experimental verification. Front. Microbiol. 15:1510665. doi: 10.3389/fmicb.2024.1510665
Edited by:
Premkumar Lakshmanane, University of North Carolina at Chapel Hill, United StatesReviewed by:
Kei Miyakawa, National Institute of Infectious Diseases (NIID), JapanShamkant B. Badgujar, Independent Researcher, Jalgaon, India
Copyright © 2024 Wang, Cao, Yang, Wang, Yang, Yang, Li, Li, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuefei Wang, d2FuZ3lmMDYyMkB0anV0Y20uZWR1LmNu; Min Zhang, emhhbmdtMDM2QHRqdXRjbS5lZHUuY24=
†These authors have contributed equally to this work
 Changjian Wang
Changjian Wang Yipeng Cao
Yipeng Cao Qi Yang3
Qi Yang3 Yuefei Wang
Yuefei Wang Min Zhang
Min Zhang