Abstract
Introduction:
Fecal microbiota transplantation (FMT) has been described useful as an adjunct treatment for chronic enteropathy in dogs. Different protocols can be used to prepare and store FMT preparations, however, the effect of these methods on microbial viability is unknown. We aimed (1) to assess the viability of several core intestinal bacterial species by qPCR and (2) to assess Peptacetobacter (Clostridium) hiranonis viability through culture to further characterize bacterial viability in different protocols for FMT preparations.
Methods:
Bacterial abundances were assessed in feces from six healthy dogs by qPCR after propidium monoazide (PMA-qPCR) treatment for selective quantitation of viable bacteria. Conservation methods tested included lyophilization (stored at 4°C and at −20°C) and freezing with glycerol-saline solution (12.5%) and without any cryoprotectant (stored at −20°C). Additionally, the abundance of P. hiranonis was quantified using bacterial culture.
Results:
Using PMA-qPCR, the viability of Faecalibacterium, Escherichia coli, Streptococcus, Blautia, Fusobacterium, and P. hiranonis was reduced in lyophilized fecal samples kept at 4°C and −20°C up to 6 months (p < 0.05). In frozen feces without cryoprotectant, only Streptococcus and E. coli were not significantly reduced for up to 3 months (p > 0.05). Lastly, no differences were observed in the viability of those species in glycerol-preserved samples up to 6 months (p > 0.05). When using culture to evaluate the viability of P. hiranonis, we observed that P. hiranonis abundance was lower in lyophilized samples kept at 4°C than −20°C; and P. hiranonis abundance was higher in glycerol-preserved samples for up to 6 months than in samples preserved without glycerol for up to 3 months. Moreover, the highest abundance of P. hiranonis was observed in glycerol-preserved feces. After 3 months, P. hiranonis was undetectable by culture in 83% (5/6) of the frozen samples without glycerol.
Discussion:
While the lyophilization procedure initially reduced P. hiranonis abundance, P. hiranonis viability was stable thereafter for up to 6 months at −20°C. The higher bacterial viability detected in fecal samples preserved with glycerol confirms the use of this cryoprotectant as a reliable method to keep bacteria alive in the presence of fecal matrix for FMT purposes.
1 Introduction
Fecal microbiota transplantation (FMT) is defined as the transplantation of feces from a healthy donor to a recipient, aiming to modulate the recipient’s intestinal microbiota and potentially confer beneficial effects (Cammarota et al., 2017; Chaitman and Gaschen, 2021; Borody and Khoruts, 2011). In humans, FMT has been notably successful in treating Clostridioides difficile enteric infection and it has been pointed out as an efficient treatment for recurring C. difficile infections (Mullish et al., 2018; Costello et al., 2015; Monaghan et al., 2019; Bishop and Tiruvoipati, 2022). In veterinary medicine, FMT is an adjunct treatment for chronic and acute diarrhea, and it can be used in the recovery of companion animals with antibiotic-induced dysbiosis (Chaitman and Gaschen, 2021; Chaitman et al., 2020; Toresson et al., 2023; Collier et al., 2022). FMT has also been investigated as a tool to improve clinical signs in a myriad of diseases in humans and animal models, from obesity to cancer, with varying results (Bäckhed et al., 2004; Napolitano and Covasa, 2020; Ting et al., 2022).
The exact mechanism of how FMT works is currently unknown: live bacteria, bacterial products, metabolites, or a combination of those may be related to the success of FMT (Borody and Khoruts, 2011; Scaldaferri et al., 2016; Segal et al., 2020). Khoruts and Sadowsky postulated that the effectiveness of FMT in humans may be linked to or dependent on the ability of beneficial bacteria from the donor to establish themselves in the recipient’s intestine, a process known as colonization or engraftment (Khoruts and Sadowsky, 2016). For colonization to occur, the viability of those microorganisms linked to healthy status in donors must be conserved during the FMT procedure. In addition, fecal metabolites may impact the overall intestine health by signaling downstream pathways relevant to certain diseases’ pathogenesis (Suez and Elinav, 2017; Ott et al., 2017; Butzner et al., 1996). Modulation of certain metabolites present in the gut such as the provision of butyrate and modification of the intestinal bile acid pool, including high fecal concentration of secondary bile acid, have been correlated to gastrointestinal health improvement in animal models (Butzner et al., 1996; Buffie et al., 2015).
The quantitative evaluation of bacterial viability in FMT preparations can be assessed through classical bacteriology (culture) or molecular biology (qPCR and sequencing). When assessing viability through bacterial culture, identifying all different cultivable bacterial species within the fecal sample can be challenging (Oliver, 2005), especially for those species with unremarkable colony morphology; however, culture remains the gold standard for evaluating the viability of certain bacterial species (Costello et al., 2015). On the other hand, when assessing viability using molecular biology tools, important limitations arise in distinguishing between DNA from live bacteria, “free-floating” DNA, and DNA from viable but nonculturable bacteria within a sample. The use of propidium monoazide (PMA) has been described in human and veterinary literature as a potential methodology to assess bacterial viability (Papanicolas et al., 2019; Fouhy et al., 2015; Lee and Bae, 2018). Despite FMT being often used as a treatment in veterinary and human medicine, there is limited literature on the viability of bacteria in FMT preparations, regardless of the method used for bacterial viability assessment (Costello et al., 2015; Papanicolas et al., 2019; Fouhy et al., 2015; Costello et al., 2017).
Although many bacterial species are transplanted with FMT, only a few species have been consistently correlated with maintaining a healthy intestinal microbiome in dogs, such as Faecalibacterium, Fusobacterium, Turicibacter, Blautia—which are short-chain fatty acids (SCFA) producers (Vital et al., 2014; Liu et al., 2021; Lynch et al., 2023)—and Peptacetobacter (Clostridium) hiranonis, the main species responsible for the conversion of primary into secondary bile acid in dogs and cats through the 7α-dehydroxylation pathway (AlShawaqfeh et al., 2017; Hirano et al., 1981; Correa Lopes et al., 2024). The SCFAs, including acetate, propionate, and butyrate, play several roles in host metabolism, with butyrate being the primary energy source for colonocytes in the gut (Deleu et al., 2021). In addition, the metabolism of bile acids is an important pathway modulated by intestinal microbiota, regulating lipid and glucose metabolism, energy production, and inflammatory signaling (Chiang, 2013). Dysmetabolism of bile acids and reduction of P. hiranonis have been linked to severe dysbiosis (Sung et al., 2023; Pilla et al., 2020; Manchester et al., 2019) and intestinal colonization by pathogenic species such as C. difficile in dogs (Werner et al., 2023; Sulaiman et al., 2024).
In the context of bile acid metabolism, the gut microbiota plays a crucial role in two essential metabolic functions related to bile acids that escape enterohepatic reabsorption: deconjugation and conversion of primary to secondary fecal unconjugated bile acids (Hofmann, 2009; Vinithakumari et al., 2021). While many bacterial species deconjugate glycine- and taurine-conjugated bile acids through bile salt hydrolase (Song et al., 2019; Jones et al., 2008), only a few bacterial species have been identified as responsible for converting unconjugated primary bile acids—cholic acid and chenodeoxycholic acid—into unconjugated secondary bile acids—deoxycholic acid and lithocholic acid, respectively (Cai et al., 2022; Vital et al., 2019). Among these bacteria is P. hiranonis, which possesses the bile acid-inducible operon encoding enzymes responsible for converting bile acids through the 7α-dehydroxylation pathway (Hirano et al., 1981; Hirano and Masuda, 1982; Doerner et al., 1997; Wise and Cummings, 2022; Ridlon et al., 2020). C. hiranonis, recently renamed P. hiranonis (Chen et al., 2020), is an anaerobic, spore-forming, and gram-positive bacteria (Chen et al., 2020; Kitahara et al., 2001). In dogs, P. hiranonis is described as a biomarker for gastrointestinal functionality and is closely linked to maintaining a balanced gastrointestinal health (Félix et al., 2022; Suchodolski, 2022). P. hiranonis plays a pivotal role in the conversion of primary to secondary fecal unconjugated bile acids and is the main species with this ability within canine and feline gastrointestinal microbiomes (Correa Lopes et al., 2024; Pilla et al., 2020; Vinithakumari et al., 2021; Giaretta et al., 2018).
Changes in gastrointestinal microbiota composition and abundance of P. hiranonis induced by antibiotic use can disrupt bile acid metabolism (Chaitman et al., 2020; Manchester et al., 2019; Blake et al., 2019). Dysbiosis and dysmetabolism of bile acids have been linked to chronic enteropathy in dogs (Guard et al., 2019; Vecchiato et al., 2023) and other chronic inflammatory diseases in humans (Prawitt et al., 2011; Wang et al., 2017). While the link between intestinal healthy microbiota and P. hiranonis, Faecalibacterium, Fusobacterium, Turicibacter, and Blautia in dogs has been outlined (Pilla et al., 2020; Suchodolski, 2022; Guard et al., 2019), our understanding of how their viability is affected by the FMT manufacturing process remains not completely elucidated. This study aims: (1) to evaluate the effect of lyophilization, freezing, and storage in different temperatures on bacterial survival by assessing the viability of core bacteria Faecalibacterium, Fusobacterium, Turicibacter, Blautia, P. hiranonis, Streptococcus, and Escherichia coli as well as total bacteria in fecal samples of potential FMT donors using PMA-qPCR; and (2) to evaluate the effect of the previously mentioned methods of FMT conservation on the viability of P. hiranonis using bacterial culture.
2 Materials and methods
2.1 Sampling, conservation methods, and time points
Fecal samples were collected from six clinically healthy dogs undergoing screening as potential FMT donors (Supplementary Table S1). Fecal samples were collected immediately post-defecation and transported to the laboratory in an insulated cooler using ice packs. All samples were processed within 24 h of defecation. Samples were divided into multiple vials for lyophilization, freezing with glycerol, and freezing without any cryoprotectants. The lyophilization process began by freezing the samples for 1 h at −80°C, followed by overnight drying. After lyophilization, the aliquoted samples were stored at 4°C and − 20°C for further analysis. For glycerol-preserved samples, we followed the protocol described by Chen et al. (2021), which involved adding 10 g of saline with glycerol (final concentration of glycerol: 12.5%) to 1 g of stool. These samples were kept frozen at −20°C until further analysis. Finally, aliquots without cryoprotectants were stored at −20°C. Throughout the study, samples preserved at both 4°C and − 20°C were evaluated by culture and PMA-qPCR at specific time points: fresh feces, and after 1 week, 1, 3, and 6 months (Table 1). Unfortunately, there were not enough feces for the evaluation of the last time point for fecal samples conserved without cryoprotectants.
Table 1
| Methods of FMT preparation | Temperature of storage | Time points |
|---|---|---|
| Lyophilization | 4°C and −20°C | F, 1W, 1M, 3M, and 6M |
| Freezing with glycerol | –20°C | F, 1W, 1M, 3M, and 6M |
| Freezing without cryoprotectant | –20°C | F, 1M, and 3M |
Methods of conservation and temperature of storage to preserve bacterial viability in fecal samples from healthy dogs evaluated at different time points by culture and PMA-qPCR.
F, fresh fecal sample; 1W, 1 week; 1M, 1 month; 3M, 3 months; 6M, 6 months.
2.2 Propidium monoazide treatment of fecal samples for viability qPCR
Fecal samples were 10-fold diluted in phosphate-buffered saline (0.008 M sodium phosphate, 0.002 M potassium phosphate, 0.14 M sodium chloride, 0.01 M potassium chloride, pH 7.4, 500 mL; Pierce™, IL), vortexed for 5 min, and incubated on an ice rack for 15 min. After incubation, 500 μL from the supernatant was added to a new tube and 5 μL of propidium monoazide (2.5 mM) was added. Samples were mixed for 10 min (60 rpm) and exposed to light to cross-link PMAxx to the DNA for 15 min. After centrifugation at 5000 × g for 10 min, pelleted cells were re-suspended and used for DNA extraction (Nye et al., 2023).
2.3 DNA extraction and qPCR assays
The DNA from fecal samples were isolated using MoBio Power Soil Kit (MoBio Laboratories), following the manufacturer’s instructions. The qPCR assay was performed using SsoFast EvaGreen® Supermix (Bio-Rad Laboratories) as described by AlShawaqfeh et al. (2017) and Sung et al. (2022). Briefly, the concentration of the extracted DNA was assessed using a spectrophotometer (NanoDrop 1,000; Thermo Scientific) and normalized to 5 ng/μl. For the PCR reaction, 2 μL normalized DNA (5 ng/μl) was added to 5 μL SsoFast EvaGreen Supermix (Bio-Rad Laboratories), along with 0.4 μL each of forward and reverse primers (400 nM) (Supplementary Table S2), and 2.2 μL DNA-free water. The Bio-Rad CFX384 Touch Thermal Cycler (Bio-Rad Laboratories) was used in this study. The cycling protocol is described as follows: initial denaturation at 98°C for 2 min; 35 cycles with denaturation at 98°C for 3 s; and annealing for 3 s. All samples were analyzed in duplicate (CFX MaestroTM 1.1 Software) and the average of the two results was used for the analysis. The quantification of the log amount of DNA (number of copies) per 10 ng of total isolated DNA was based on a standard curve. In summary, a 10-fold dilution series of purified plasmids containing the sequences for each bacterial target was used to construct the standard curve for DNA quantification. The amplicon length, melting peak temperature, efficiency of the qPCR assay, and the coefficient of determination (R2) of the calibration curve were previously described (AlShawaqfeh et al., 2017; Sung et al., 2022).
2.4 Peptacetobacter hiranonis culture, identification and quantification
The viability of P. hiranonis was assessed by bacterial culture. Fecal samples were serially 10-fold diluted in phosphate-buffered saline (0.008 M sodium phosphate, 0.002 M potassium phosphate, 0.14 M sodium chloride, 0.01 M potassium chloride, pH 7.4, 500 mL; Pierce™, IL), and plated onto Brucella blood agar plates (Anaerobic Systems, CA). For the quantification of P. hiranonis, all colonies morphologically compatible with P. hiranonis (Supplementary Figure S1) were collected for further identification using molecular methods. DNA extraction from each colony was performed as described by Dashti et al. (2009), and confirmation of P. hiranonis by qPCR was conducted following the protocol described by AlShawaqfeh et al. (2017).
2.5 Statistical analysis
Shapiro–Wilk’s test was employed for the normality assessment of the data. Comparison between time points (fresh fecal sample, 1 week, 1, 3, and 6 months) and different FMT preparation methods (lyophilization stored at 4°C and −20°C, feces preserved with and without glycerol stored at −20°C) were evaluated using Repeated Measures One-way ANOVA and Dunnett’s multiple comparison test. Spearman’s test was employed to assess the correlation between the abundance of P. hiranonis assessed by PMA-qPCR, expressed in log DNA, and viability of P. hiranonis assessed by bacterial culture, expressed in log CFU/g of feces, through the time points and different methods of FMT preparations. The statistical analysis was performed in GraphPad Prism (Version 9.4.1). A p-value of less than 0.05 was considered statistically significant.
3 Results
3.1 Bacterial viability assessed by PMA-qPCR
Using PMA-qPCR to assess bacterial viability, abundances of Faecalibacterium, E. coli, Streptococcus, Blautia, Fusobacterium, and P. hiranonis reduced significantly in lyophilized samples stored at both 4°C and −20°C after 6 months (p < 0.05). In frozen feces without glycerol, only Streptococcus and E. coli were not significantly reduced after 3 months (p > 0.05). No statistically significant differences were observed in glycerol-preserved samples for all assessed bacteria taxa over 6 months of storage (p > 0.05) (Table 2 and Figure 1). Detailed results for all tested bacterial taxa in each one of the conditions over time can be found in Supplementary Figure S2.
Table 2
| Fresh fecal sample | LYO 4°C 6M | LYO –20°C 6M | GLY –20°C 6M | w/o GLY –20°C 3M | |
|---|---|---|---|---|---|
| Median (Range) | Median (Range) Adjusted p-value |
Median (Range) Adjusted p-value |
Median (Range) Adjusted p-value |
Median (Range) Adjusted p-value |
|
| Universal | 10.14 (9.87–10.53) | 8.13 (7.64–9.12) 0.0010 |
9.25 (8.04–9.65) 0.0260 |
10.51 (10.09–10.66) 0.3285 |
9.58 (8.09–10.28) 0.1704 |
| Faecalibacterium | 6.37 (5.97–7.04) | 2.72 (2.48–4.77) 0.0003 |
4.57 (3.16–5.04) 0.0037 |
6.87 (6.60–6.97) 0.1701 |
4.28 (3.09–5.83) 0.0021 |
| Turicibacter | 6.33 (5.02–7.56) | 6.04 (4.37–7.10) 0.5354 |
6.47 (5.34–7.64) 0.8720 |
7.27 (5.87–7.65) 0.1884 |
7.34 (5.81–8.42) 0.0040 |
| Streptococcus | 2.59 (2.09–3.39) | 1.27 (0.33–1.48) 0.0171 |
1.55 (1.10–1.93) 0.0135 |
2.85 (1.94–3.76) 0.7315 |
2.06 (1.05–2.75) 0.1051 |
| E. coli | 3.61 (2.51–7.06) | 1.67 (0.88–4.83) 0.0185 |
2.60 (1.32–4.68) 0.0313 |
4.07 (3.26–6.70) 0.8319 |
3.20 (1.74–6.59) 0.2908 |
| Blautia | 9.82 (9.30–10.17) | 6.91 (6.53–7.94) <0.0001 |
8.09 (6.85–8.45) 0.0054 |
9.82 (9.53–10.25) 0.9976 |
8.62 (7.37–9.24) 0.0017 |
| Fusobacterium | 8.59 (7.65–9.31) | 5.67 (5.27–7.37) 0.0012 |
6.28 (5.48–8.30) 0.0083 |
8.41 (7.89–8.97) 0.8871 |
6.22 (6.13–7.67) 0.0028 |
| P. hiranonis | 5.47 (4.96–5.94) | 0.47 (0.10–3.08) 0.0015 |
3.48 (1.13–3.58) 0.0121 |
5.78 (5.08–6.37) 0.1425 |
3.48 (1.89–4.61) 0.0140 |
Bacterial abundances, expressed in log DNA, assessed in fecal samples by PMA-qPCR at the last time point for each conservation method: lyophilized fecal samples stored at 4°C and −20°C, and freezing with glycerol and without cryoprotectants stored at −20°C.
The p-values were calculated by comparing the last time point for each conservation method with bacterial abundances assessed in fresh fecal samples (first column). Adjusted p-values from Dunnett’s multiple comparison tests were reported. LYO, lyophilized feces; 3M, 3 months; 6M, 6 months; GLY, feces preserved with glycerol; w/o GLY, feces conserved without cryoprotectants.
Figure 1
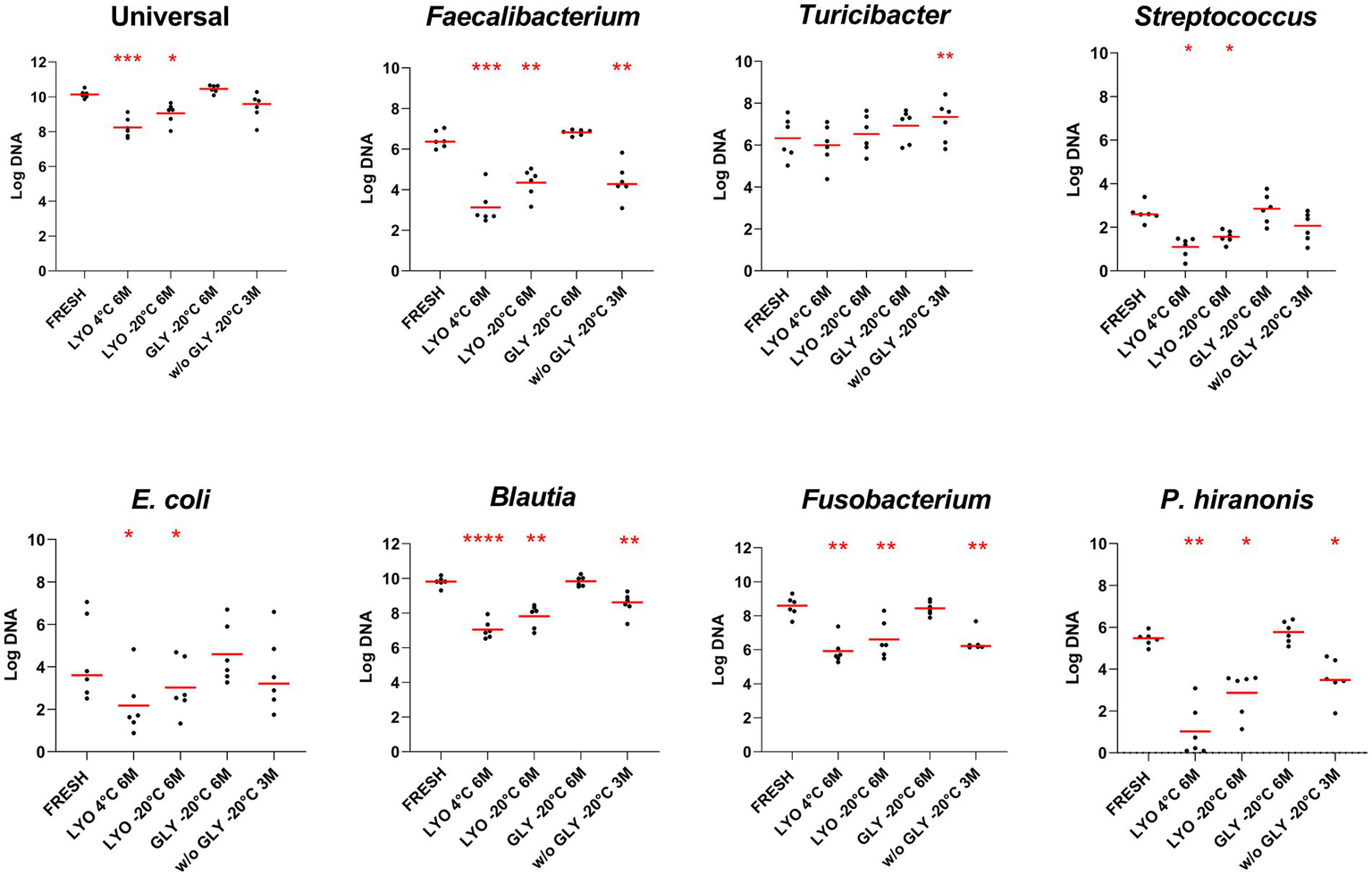
Bacterial abundances assessed in fecal samples by PMA-qPCR at the last time point for each conservation method: lyophilization stored at 4°C (LYO 4°C 6 M) and −20°C (LYO –20°C 6 M), and freezing with glycerol (GLY –20°C 6 M) and without cryoprotectant stored at −20°C (w/o GLY –20°C 3 M). The red line indicates the median value for each evaluated condition. The p-values are denoted as follows: * for p ≤ 0.05, ** for p ≤ 0.01, *** for p ≤ 0.001, and **** for p ≤ 0.0001.
3.2 Quantification of Peptacetobacter hiranonis by bacterial culture
A decrease in the abundance of P. hiranonis was observed by culture across all tested conditions (p < 0.0001). After 6 months of storage, the abundance of P. hiranonis was lower in lyophilized samples stored at both 4°C [median (range): 0 (0–0) log colony-forming unit (CFU)/g] compared to samples stored at −20°C [2.69 (0–3.66)]. Additionally, P. hiranonis abundance was higher in glycerol-preserved samples [5.94 (0–6.70)] after 6 months, as compared to frozen samples without cryoprotectants [0 (0–3.30)] after 3 months (Figure 2).
Figure 2
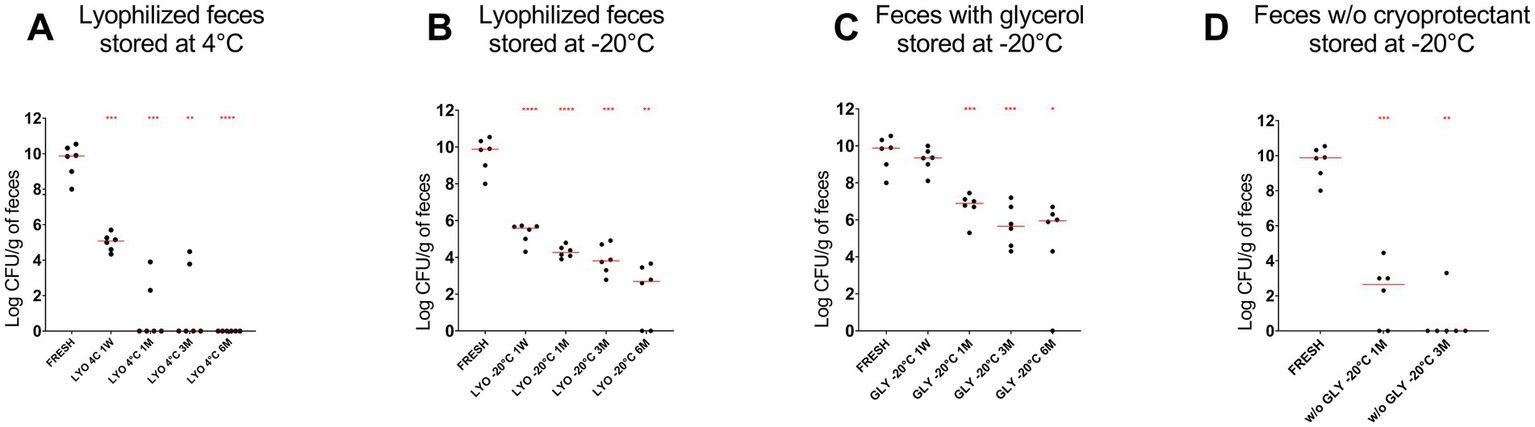
The abundance of P. hiranonis in feces assessed by culture at different time points: fresh, after 1 week of storage, and after 1, 3, and 6 months of storage. The abundance of P. hiranonis is expressed in log CFU/g of feces. The evaluated conservation methods were lyophilization stored at 4°C (A) and −20°C (B), and freezing with glycerol (C) and without cryoprotectant (D) stored at −20°C. LYO, lyophilized feces; 1W, 1 week; 1M, 1 month; 3M, 3 months; 6M, 6 months; GLY: feces conserved with glycerol; w/o GLY: feces conserved without cryoprotectant. The red lines indicate the median value for each evaluated condition. The p-values are denoted as follows: * for p ≤ 0.05, ** for p ≤ 0.01, *** for p ≤ 0.001, and **** for p ≤ 0.0001.
3.3 Comparison between Peptacetobacter hiranonis quantification by PMA-qPCR and bacterial culture
Using PMA-qPCR to assess bacterial viability, abundance of P. hiranonis was reduced in lyophilized samples kept at 4°C and −20°C after 6 months and in frozen samples without glycerol after 3 months (p < 0.05). Moreover, decreased P. hiranonis abundance was observed for all tested conditions by culture (p < 0.05) (Figure 3 and Table 3). Correlation between PMA-qPCR and P. hiranonis viability assessed by cultured demonstrate a positive correlation Spearman r = 0.6732 (p < 0.0001), being affected by several samples (17/84, 20%) which P. hiranonis was not recovered by culture but detected by PMA-qPCR.
Figure 3
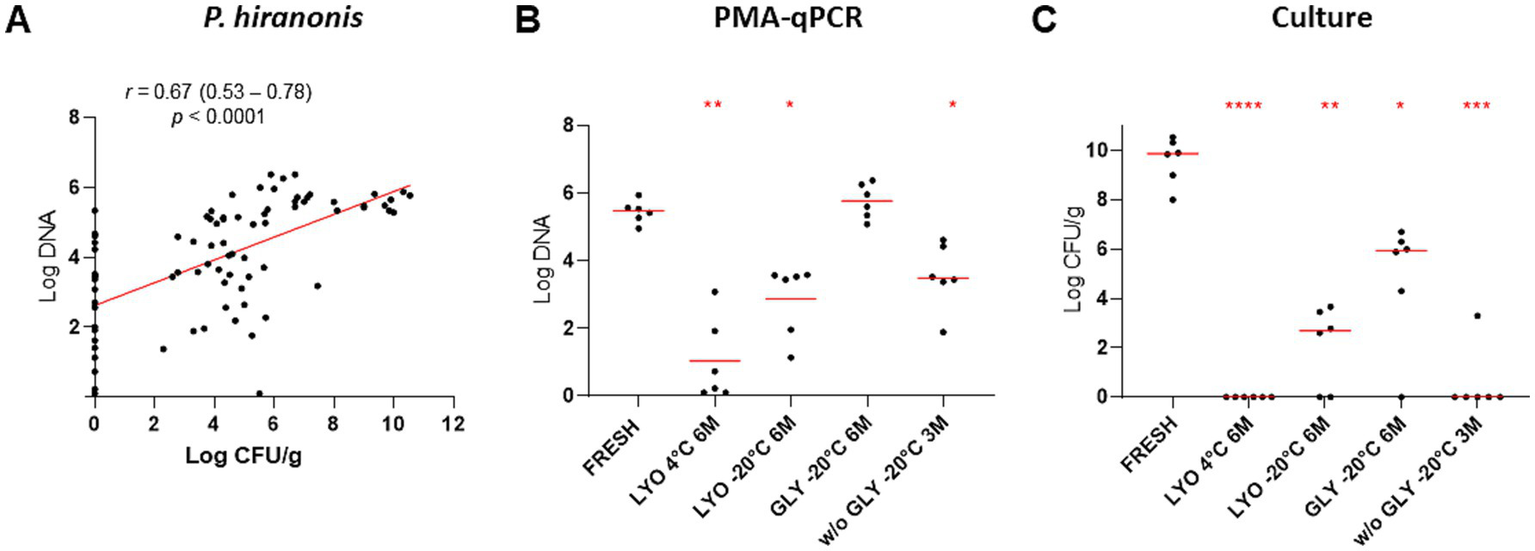
Comparison between the quantification of P. hiranonis by PMA-qPCR, expressed in log DNA, and bacterial culture, expressed in log CFU/g of feces. (A) Spearman’s correlation between the abundance of P. hiranonis quantified by PMA-qPCR and culture. Spearman’s correlation coefficient (r), 95% confidence interval, and p-value were provided. (B) Abundance of P. hiranonis in lyophilized fecal samples stored at 4°C and −20°C, glycerol-preserved fecal samples, and fecal samples preserved without cryoprotectant stored at −20°C assessed by PMA-qPCR at the last time point for each conservation method. (C) Abundance of P. hiranonis in lyophilized samples stored at 4°C and −20°C, glycerol-preserved fecal samples, and fecal samples preserved without cryoprotectant stored at −20°C assessed by culture at the last time point for each conservation method. LYO, lyophilized feces; GLY, feces conserved with glycerol; w/o GLY, feces conserved without cryoprotectant; 3M, 3 months; 6M, 6 months. The p-values are denoted as follows: * for p ≤ 0.05, ** for p ≤ 0.01, *** for p ≤ 0.001, and **** for p ≤ 0.0001.
Table 3
| Conservation methods |
P. hiranonis
(log CFU/g of feces) |
|
|---|---|---|
| Median (range) | Adjusted p-value | |
| Fresh fecal sample | 9.87 (8–10.54) | - |
| Lyophilized fecal samples kept at 4°C 6M | 0 (0–0) | <0.0001 |
| Lyophilized fecal samples kept at −20°C 6M | 2.69 (0–3.66) | 0.0016 |
| Glycerol-preserved fecal samples kept at −20°C 6M | 5.94 (0–6.70) | 0.0192 |
| Fecal samples kept at −20°C 3M | 0 (0–3.30) | 0.0002 |
The abundance of P. hiranonis, expressed in log CFU/g of feces, assessed in fecal samples by culture at the last time point for each conservation method: lyophilized feces stored at 4°C and −20°C, and freezing with glycerol and without cryoprotectants stored at −20°C.
The p-values were calculated by comparing the last time point for each conservation method to bacterial abundances assessed in fresh fecal samples (first row). Adjusted p-values from Dunnett’s multiple comparison tests were reported. 3M, 3 months; 6M, 6 months.
4 Discussion
In this study, we assessed the overall reduction in viability of several bacteria in healthy canine feces under different conservation methods. The bacterial viability was evaluated in feces stored at −20°C with and without the addition of glycerol and in lyophilized feces stored at 4°C and −20°C for up to 6 months. Storage at −20°C seems more adequate for preservation of bacterial viability compared to storage at 4°C. The bacteria quantified included E. coli, Faecalibacterium, Blautia, P. hiranonis, Fusobacterium, Turicibacter, and Streptococcus, which were evaluated through PMA-qPCR. In addition to viability assessment by PMA-qPCR, P. hiranonis viability was also quantified through bacterial culture, with both methods indicating a reduction in the viability of P. hiranonis across time points.
In veterinary medicine, there is a lack of information regarding bacterial viability in fecal samples intended for FMT use (Chaitman and Gaschen, 2021; Collier et al., 2022; Papanicolas et al., 2019). Guidelines for FMT in veterinary medicine have been recently published, however, the screening criteria and analysis of the donor microbiota are not consistent across veterinary clinics (Chaitman and Gaschen, 2021; Winston et al., 2024). Clinical response is the ultimate outcome to evaluate the efficacy of FMT preparations, however, testing each small protocol variation in clinical trials is not feasible. While it is currently unknown whether live bacteria are required for FMT to result in clinical response and to restore gut microbiota functionality, which confer gut health benefits (Khoruts and Sadowsky, 2016), it is reasonable to ensure that specific keystone bacteria remain viable for their beneficial effects. In particular, re-colonization with P. hiranonis, which plays an essential role in bile acid metabolism (Ridlon et al., 2020; Kitahara et al., 2001) and is commonly missing in severe dysbiosis, is a desired outcome of FMT (Chaitman et al., 2020; Toresson et al., 2023). Additionally, the bacterial taxa Faecalibacterium, Fusobacterium, Turicibacter, and Blautia display a key role in the production of SCFAs and have been correlated to the maintenance of a healthy intestinal microbiome (Vital et al., 2014; Liu et al., 2021; Lynch et al., 2023; Miquel et al., 2013).
Metabolites, such as bile acids, amino acids, short and long-chain fatty acids, vitamins, and polysaccharides, are transplanted in association with bacteria in FMT, however different methods of FMT preparations (e.g., freezing, freeze-drying, and not frequently used, dehydration) might affect metabolite concentration in the fecal content (Reygner et al., 2020; Martinez-Gili et al., 2020). In this study, we have not evaluated the effect of FMT preparation methods on the fecal metabolites, although, previous reports have shown limited or no effects on SCFA (Reygner et al., 2020) or bile acid concentration (unpublished data). Fecal filtrated transplant (FFT), lacking viable bacteria but containing bacterial debris and metabolites, has been described as an alternative to FMT, since FMT may represent a risk for immunocompromised patients when screening methods for FMT donors fail to detect potential risks, i.e., pathogens. In addition, the presence of bacteriophages has been linked with beneficial effects observed after FFT in humans and animal models (Ott et al., 2017; Brunse et al., 2021). In summary, the FMT mechanisms are complex, and clinical effectiveness may be associated with different factors from live bacteria to metabolites.
While few studies describe bacterial viability (Papanicolas et al., 2019; Barko et al., 2024), most relied on PMA-qPCR for assessment rather than traditional bacterial cultivation methods. Although PMA-qPCR is a well-established method to assess bacterial viability, it is important to consider limitations related to false-positive results, since PMA will allow the detection of viable-non-culturable bacteria by qPCR (van Frankenhuyzen et al., 2011; Wolffs et al., 2005). In our study, we evaluated the viability of P. hiranonis through bacterial culture, and in a substantial fraction of the samples (17/84, 20%), P. hiranonis was detected by PMA-qPCR, but not by culture. Parallel to limitations associated with PMA, P. hiranonis is considered a fastidious bacterium and false-negative results may be observed. However, it is important to note that live P. hiranonis is needed for colonization and further conversion of bile acids. We focused solely on P. hiranonis, a strictly anaerobic bacterium highly sensitive to oxygen exposure (Chen et al., 2020; Kitahara et al., 2001). However, assessing P. hiranonis viability through culture may serve as a proxy for other anaerobic bacteria, considering their shared characteristic of sensitivity to oxygen exposure (see Supplementary Figure S3).
Sequencing methods are commonly applied to evaluate microbial composition in fecal samples. The quantification of bacteria in different taxonomic levels is expressed in relative abundance, and the generated data are treated as compositional (Gloor et al., 2017). In our study, a target qPCR method was used for absolute quantification of core bacterial genera and species present in the intestinal microbiome of dogs. In the article published by Barko et al. (2024), long-read 16S rRNA amplicon sequencing was used, and relative abundances of Turicibacter sanguinis, Streptococcus pasteuri, and Blautia glucerasea were found to increase over time in samples cryopreserved in glycerol and stored at −20°C. In contrast, our results indicated no significant difference in the absolute abundance of Turicibacter spp., Streptococcus spp., or Blautia spp. in glycerol-preserved samples stored at −20°C.
Targeted methods, such as qPCR, are more reliable for the quantification of bacterial absolute abundance compared to sequencing methods (Wang et al., 2021), which are known for presenting variable results affected by sample collection method, storage, DNA extraction, and sequencing method, and further by the bioinformatics tools applied (Tourlousse et al., 2021). In addition, considering the limitations of the PMA-qPCR methodology previously mentioned, more studies are needed to define whether the observed increase or decrease in the abundances of Turicibacter and Blautia reflects the abundance of live bacteria in FMT preparations over time and across different storage temperatures. Turicibacter and Blautia are Gram-positive strictly anaerobic bacteria capable of growth between 30 and 45°C. An observed increase in abundance over time may indicate PMA saturation, a scenario where the amount of PMA used is insufficient to bind all DNA from non-viable or membrane-damaged cells (Liu et al., 2021; Maki and Looft, 2022).
A decreased relative abundance of Fusobacterium spp. over time was reported in samples kept at −20°C with glycerol using long-read 16S rRNA amplicon sequencing (Barko et al., 2024). However, our target qPCR did not detect a reduction in Fusobacterium preserved with glycerol at −20°C, only in lyophilized feces stored at 4°C and −20°C and feces preserved without cryoprotectant stored at −20°C. Finally, while Barko et al. (2024) reported an increased relative abundance of P. hiranonis over time in glycerol-preserved samples, our study found the absolute abundance of P. hiranonis unchanged by PMA-qPCR or decreased by culture in fecal samples after 6 months of storage (see Figures 1, 2). Differences between data reported by Barko and this current publication may be explained by the different methodologies applied: qPCR and sequencing. It is important to mention that sequencing methods have inherent limitations in terms of quantification, repeatability, and reproducibility; therefore, results need to be evaluated carefully and, when possible, confirmed by targeted assays.
The processing methods employed in this study require a homogenization step, exposing the fecal sample to ambient air and consequently to high levels of oxygen. Homogenization and filtration are required steps in several consensus guidelines for FMT preparations in humans (Cammarota et al., 2017; Mullish et al., 2018) and in veterinary medicine (Chaitman and Gaschen, 2021). While the deleterious effects of oxygen exposure can be mitigated when processing the samples in an anaerobic environment (i.e., anaerobic chamber), it is important to note that ensuring an oxygen-free environment requires specialized and trained personnel and specific equipment, potentially increasing operational costs for the FMT preparation and rendering it inaccessible to clinicians outside of referral centers. Though more studies are needed to clarify if the exposure to oxygen during preparation could be detrimental to FMT effectivity, our goal was to analyze viability in preparation methods that were commonly used and easily applicable in clinical practice without the need for specialized equipment, i.e., preparation in the presence of oxygen, use of glycerol as a cryopreservative, and storage at −20°C.
Besides the effect of oxygen exposure, different storage temperatures may affect bacterial viability in long-term storage. The temperatures used in this study reflect the temperature available in most veterinary practices (refrigeration at 4°C and freezing with household-grade freezers at –20°C), because −80°C freezers are usually not available outside of laboratory facilities. Costello et al. reported that long-term storage of FMT preparation, more than two months, preserved with glycerol at –80°C presented satisfactory results when used to prevent C. difficile recurrence (Costello et al., 2015). In another study published by Lee et al., fresh and frozen stored at –20°C feces were compared leading to similar outcomes – recovery from C. difficile infection (Lee et al., 2016). Although there is a lack of similar studies in the veterinary field, research in humans points out that similar outcomes are accomplished when using fresh and frozen feces for FMT. Therefore, in this study we chose to test storage at -20°C to reflect the current use of FMT preparation methodologies in veterinary clinics, making our results translatable to clinical practice.
Faecalibacterium, Blautia, Turicibacter, and Fusobacterium are involved in SCFA production, a metabolite that is crucial for nourishing colonocytes (Vital et al., 2014; Liu et al., 2021; Lynch et al., 2023; Miquel et al., 2013; Hosomi et al., 2022). These bacterial genera are strict anaerobes and are among the most abundant bacteria taxa in the gut microbiota of dogs and cats (AlShawaqfeh et al., 2017; Sung et al., 2023; Sung et al., 2022). Dysbiosis often leads to a reduction in the abundance of strictly anaerobic bacteria, accompanied by an increase in facultative anaerobes such as E. coli and Streptococcus (AlShawaqfeh et al., 2017; Pilla et al., 2020; Guard et al., 2019; Sung et al., 2022; Rivera-Chavez et al., 2017). Although the precise mechanism by which the resident microbiota limits pathogen expansion remains unclear (Deleu et al., 2021), SCFA appears to play an important role in maintaining gut homeostasis, intestinal barrier integrity, as well as overall metabolism and energy balance (Portincasa et al., 2022; Ma et al., 2022). Therefore, the survival of these SCFA-producing taxa in FMT preparations may directly impact FMT outcomes.
While frozen feces preserved with glycerol presented better results in conserving P. hiranonis viability, lyophilization is an alternative methodology for FMT pill manufacture, given that most capsules dissolve upon contact with water or humidity. The absence of water in lyophilized feces during the manufacture of FMT capsules ensures the preservation of the capsules’ structure. Lyophilization can be performed with or without cryoprotectants (Zain et al., 2022). In our study, we have chosen to perform the lyophilization without any cryoprotectants, as this aligns with a more easily replicable method in veterinary practice facilities. However, further research is necessary to elucidate the effect of cryoprotectants on bacterial viability in lyophilized feces.
Similar to a previous study, while there was a reduction of potentially beneficial bacteria, facultative anaerobic bacteria, i.e., E. coli and Streptococcus, assessed in this study did not increase over time (Papanicolas et al., 2019). Increased Enterobacteriaceae abundance in the intestine is associated with an inflammatory response and it has been linked to inflammatory bowel disease, type 2 diabetes, and colorectal cancer in humans (Baldelli et al., 2021), as well as chronic enteropathy and antibiotic-induced dysbiosis in dogs and cats (AlShawaqfeh et al., 2017; Giaretta et al., 2018; Blake et al., 2019; Guard et al., 2019; Sung et al., 2022). In addition, elevated Streptococcus abundance has been correlated to antibiotic-induced dysbiosis and chronic enteropathy in dogs and cats (Chaitman et al., 2020; Pilla et al., 2020; Sung et al., 2022), highlighting the role of increased oxygen levels in the intestinal lumen as a potent cause and trigger for inflammation and severe dysbiosis (Rivera-Chavez et al., 2017). Moreover, gastrointestinal dysmotility and malabsorption have been associated with the overgrowth of facultative anaerobic bacterial species, including increased E. coli and Streptococcus (Bala et al., 2006), where impaired fat absorption and motility might contribute to intestinal inflammation (Singh et al., 2021).
Among the limitations of our study, we were only able to analyze frozen feces without cryoprotectants for a period of up to 3 months. Unfortunately, there were not enough sample aliquots available for further evaluation. Additionally, other cryoprotectants described in the literature were not assessed in this study (Hubalek, 2003), as well as additional storage temperatures, such as −80°C or controlled room temperature (20–25°C). However, lyophilized feces and frozen feces with and without glycerol for preservation are the most common methods used for FMT preparation in veterinary medicine and are considered more accessible for veterinarians (Toresson et al., 2023). Finally, another important limitation is associated with the use of the PMA-qPCR methodology, which may yield false-positive results (Miquel et al., 2013; van Frankenhuyzen et al., 2011). False positive results using PMA-qPCR are linked to incomplete inhibition of amplification of free-floating DNA due to insufficient PMA in a sample rich in free-floating DNA. In addition, dead bacterial cells with intact cell membranes cannot be differentiated from viable cells by PMA-qPCR. Finally, the small sample size evaluated in this study reduces the statistical power, and further studies containing larger sample sizes are needed.
In summary, a reduction of Faecalibacterium, E. coli, Streptococcus, Blautia, Fusobacterium, and P. hiranonis abundances was observed by culture and or PMA-qPCR across the time points evaluated. Better conservation of bacterial viability can be achieved by freezing fecal samples with glycerol. Despite decreases in viability, lyophilized feces stored for up to 6 months at −20°C may represent a valid alternative for manufacturing FMT capsules. Finally, clinical studies are needed to elucidate how these differences in bacterial viability affect FMT outcomes.
Statements
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
BCL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. JT: Data curation, Investigation, Visualization, Writing – review & editing. MKT: Conceptualization, Methodology, Visualization, Writing – review & editing. PRG: Visualization, Writing – review & editing. JSS: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Visualization, Writing – review & editing. RP: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The microbiome research is supported in part by the Purina PetCare Research Excellence Fund.
Conflict of interest
BCL, JT, MKT, PRG, JS, and RP are employed by the Gastrointestinal Laboratory at Texas A&M University, which provides assays for intestinal function and microbiota analysis on a fee-for-service basis.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1502452/full#supplementary-material
References
1
AlShawaqfeh M. K. Wajid B. Minamoto Y. Markel M. Lidbury J. A. Steiner J. M. et al . (2017). A dysbiosis index to assess microbial changes in fecal samples of dogs with chronic inflammatory enteropathy. FEMS Microbiol. Ecol.93, 1–8. doi: 10.1093/femsec/fix136
2
Bäckhed F. Ding H. Wang T. Hooper L. V. Koh G. Y. Nagy A. et al . (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci.101, 15718–15723. doi: 10.1073/pnas.0407076101
3
Bala L. Ghoshal U. C. Ghoshal U. Tripathi P. Misra A. Gowda G. A. N. et al . (2006). Malabsorption syndrome with and without small intestinal bacterial overgrowth: a study on upper-gut aspirate using 1H NMR spectroscopy. Magnetic Resonance Med.56, 738–744. doi: 10.1002/mrm.21041
4
Baldelli V. Scaldaferri F. Putignani L. del Chierico F. (2021). The role of Enterobacteriaceae in gut microbiota Dysbiosis in inflammatory bowel diseases. Microorganisms9, 1–15. doi: 10.3390/microorganisms9040697
5
Barko P. Nguyen-Edquilang J. Williams D. A. Gal A. (2024). Fecal microbiome composition and diversity of cryopreserved canine stool at different duration and storage conditions. PLoS One19:e0294730. doi: 10.1371/journal.pone.0294730
6
Bishop E. J. Tiruvoipati R. (2022). Management of Clostridioides difficile infection in adults and challenges in clinical practice: review and comparison of current IDSA/SHEA, ESCMID and ASID guidelines. J. Antimicrob. Chemother.78, 21–30. doi: 10.1093/jac/dkac404
7
Blake A. B. Guard B. C. Honneffer J. B. Lidbury J. A. Steiner J. M. Suchodolski J. S. (2019). Altered microbiota, fecal lactate, and fecal bile acids in dogs with gastrointestinal disease. PLoS One14:e0224454. doi: 10.1371/journal.pone.0224454
8
Borody T. J. Khoruts A. (2011). Fecal microbiota transplantation and emerging applications. Nat. Rev. Gastroenterol. Hepatol.9, 88–96. doi: 10.1038/nrgastro.2011.244
9
Brunse A. Deng L. Pan X. Hui Y. Castro-Mejía J. L. Kot W. et al . (2021). Fecal filtrate transplantation protects against necrotizing enterocolitis. ISME J.16, 686–694. doi: 10.1038/s41396-021-01107-5
10
Buffie C. G. Bucci V. Stein R. R. McKenney P. T. Ling L. Gobourne A. et al . (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature517, 205–208. doi: 10.1038/nature13828
11
Butzner J. D. Parmar R. Bell C. J. Dalal V. (1996). Butyrate enema therapy stimulates mucosal repair in experimental colitis in the rat. Gut38, 568–573. doi: 10.1136/gut.38.4.568
12
Cai J. Rimal B. Jiang C. Chiang J. Y. L. Patterson A. D. (2022). Bile acid metabolism and signaling, the microbiota, and metabolic disease. Pharmacol. Ther.237:108238. doi: 10.1016/j.pharmthera.2022.108238
13
Cammarota G. Ianiro G. Tilg H. Rajilić-Stojanović M. Kump P. Satokari R. et al . (2017). European consensus conference on faecal microbiota transplantation in clinical practice. Gut66, 569–580. doi: 10.1136/gutjnl-2016-313017
14
Chaitman J. Gaschen F. (2021). Fecal microbiota transplantation in dogs. Vet. Clin. North Am. Small Anim. Pract.51, 219–233. doi: 10.1016/j.cvsm.2020.09.012
15
Chaitman J. Ziese A. L. Pilla R. Minamoto Y. Blake A. B. Guard B. C. et al . (2020). Fecal microbial and metabolic profiles in dogs with acute diarrhea receiving either fecal microbiota transplantation or Oral metronidazole. Front. Vet. Sci.7:192. doi: 10.3389/fvets.2020.00192
16
Chen X. J. Wang Z. Q. Zhou Z. Y. Zeng N. Y. Huang Q. F. Wang Z. W. et al . (2020). Characterization of Peptacetobacter hominis gen. Nov., sp. nov., isolated from human faeces, and proposal for the reclassification of Clostridium hiranonis within the genus Peptacetobacter. Int. J. Syst. Evol. Microbiol.70, 2988–2997. doi: 10.1099/ijsem.0.003925
17
Chen J. Zaman A. Ramakrishna B. Olesen S. W. (2021). Stool banking for fecal microbiota transplantation: methods and operations at a large stool bank. Front. Cell. Infect. Microbiol.11:622949. doi: 10.3389/fcimb.2021.622949
18
Chiang J. Y. (2013). Bile acid metabolism and signaling. Compr. Physiol.3, 1191–1212. doi: 10.1002/cphy.c120023
19
Collier A. J. Gomez D. E. Monteith G. Plattner B. L. Verbrugghe A. Webb J. et al . (2022). Investigating fecal microbial transplant as a novel therapy in dogs with inflammatory bowel disease: a preliminary study. PLoS One17:e0276295. doi: 10.1371/journal.pone.0276295
20
Correa Lopes B. Chen C. C. Sung C. H. Ishii P. E. Medina L. F. C. Gaschen F. P. et al . (2024). Correlation between Peptacetobacter hiranonis, the baiCD gene, and secondary bile acids in dogs. Animals14:216. doi: 10.3390/ani14020216
21
Costello S. P. Conlon M. A. Vuaran M. S. Roberts-Thomson I. C. Andrews J. M. (2015). Faecal microbiota transplant for recurrent Clostridium difficile infection using long-term frozen stool is effective: clinical efficacy and bacterial viability data. Aliment. Pharmacol. Ther.42, 1011–1018. doi: 10.1111/apt.13366
22
Costello S. P. Soo W. Bryant R. V. Jairath V. Hart A. L. Andrews J. M. (2017). Systematic review with meta-analysis: faecal microbiota transplantation for the induction of remission for active ulcerative colitis. Aliment. Pharmacol. Ther.46, 213–224. doi: 10.1111/apt.14173
23
Dashti A. A. Jadaon M. M. Abdulsamad A. M. Dashti H. M. (2009). Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med. J.41, 117–122.
24
Deleu S. Machiels K. Raes J. Verbeke K. Vermeire S. (2021). Short chain fatty acids and its producing organisms: an overlooked therapy for IBD?EBioMedicine66:103293. doi: 10.1016/j.ebiom.2021.103293
25
Doerner K. C. Takamine F. LaVoie C. P. Mallonee D. H. Hylemon P. B. (1997). Assessment of fecal bacteria with bile acid 7 alpha-dehydroxylating activity for the presence of bai-like genes. Appl. Environ. Microbiol.63, 1185–1188. doi: 10.1128/aem.63.3.1185-1188.1997
26
Félix A. P. Souza C. M. M. de Oliveira S. G. (2022). Biomarkers of gastrointestinal functionality in dogs: a systematic review and meta-analysis. Anim. Feed Sci. Technol.283:115183. doi: 10.1016/j.anifeedsci.2021.115183
27
Fouhy F. Deane J. Rea M. C. O’Sullivan Ó. Ross R. P. O’Callaghan G. et al . (2015). The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One10:e0119355. doi: 10.1371/journal.pone.0119355
28
Giaretta P. R. Rech R. R. Guard B. C. Blake A. B. Blick A. K. Steiner J. M. et al . (2018). Comparison of intestinal expression of the apical sodium-dependent bile acid transporter between dogs with and without chronic inflammatory enteropathy. J. Vet. Intern. Med.32, 1918–1926. doi: 10.1111/jvim.15332
29
Gloor G. B. Macklaim J. M. Pawlowsky-Glahn V. Egozcue J. J. (2017). Microbiome datasets are compositional: and this is not optional. Front. Microbiol.8:2224. doi: 10.3389/fmicb.2017.02224
30
Guard B. C. Honneffer J. B. Jergens A. E. Jonika M. M. Toresson L. Lawrence Y. A. et al . (2019). Longitudinal assessment of microbial dysbiosis, fecal unconjugated bile acid concentrations, and disease activity in dogs with steroid-responsive chronic inflammatory enteropathy. J. Vet. Intern. Med.33, 1295–1305. doi: 10.1111/jvim.15493
31
Hirano S. Masuda N. (1982). Enhancement of the 7 alpha-dehydroxylase activity of a gram-positive intestinal anaerobe by Bacteroides and its significance in the 7-dehydroxylation of ursodeoxycholic acid. J. Lipid Res.23, 1152–1158. doi: 10.1016/S0022-2275(20)38052-4
32
Hirano S. Nakama R. Tamaki M. Masuda N. Oda H. (1981). Isolation and characterization of thirteen intestinal microorganisms capable of 7 alpha-dehydroxylating bile acids. Appl. Environ. Microbiol.41, 737–745. doi: 10.1128/aem.41.3.737-745.1981
33
Hofmann A. F. (2009). The enterohepatic circulation of bile acids in mammals: form and functions. Front. Biosci.14, 2584–2598. doi: 10.2741/3399
34
Hosomi K. Saito M. Park J. Murakami H. Shibata N. Ando M. et al . (2022). Oral administration of Blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat. Commun.13:4477. doi: 10.1038/s41467-022-32015-7
35
Hubalek Z. (2003). Protectants used in the cryopreservation of microorganisms. Cryobiology46, 205–229. doi: 10.1016/S0011-2240(03)00046-4
36
Jones B. V. Begley M. Hill C. Gahan C. G. M. Marchesi J. R. (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA105, 13580–13585. doi: 10.1073/pnas.0804437105
37
Khoruts A. Sadowsky M. J. (2016). Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol.13, 508–516. doi: 10.1038/nrgastro.2016.98
38
Kitahara M. Takamine F. Imamura T. Benno Y. (2001). Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7alpha-dehydroxylating activity. Int. J. Syst. Evol. Microbiol.51, 39–44. doi: 10.1099/00207713-51-1-39
39
Lee S. Bae S. (2018). Molecular viability testing of viable but non-culturable bacteria induced by antibiotic exposure. Microb. Biotechnol.11, 1008–1016. doi: 10.1111/1751-7915.13039
40
Lee C. H. Steiner T. Petrof E. O. Smieja M. Roscoe D. Nematallah A. et al . (2016). Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: a randomized clinical trial. JAMA315, 142–149. doi: 10.1001/jama.2015.18098
41
Liu X. Mao B. Gu J. Wu J. Cui S. Wang G. et al . (2021). Blautia—a new functional genus with potential probiotic properties?Gut Microbes13, 1–21. doi: 10.1080/19490976.2021.1875796
42
Lynch J. B. Gonzalez E. L. Choy K. Faull K. F. Jewell T. Arellano A. et al . (2023). Gut microbiota Turicibacter strains differentially modify bile acids and host lipids. Nat. Commun.14:3669. doi: 10.1038/s41467-023-39403-7
43
Ma J. Piao X. Mahfuz S. Long S. Wang J. (2022). The interaction among gut microbes, the intestinal barrier and short chain fatty acids. Anim. Nutr.9, 159–174. doi: 10.1016/j.aninu.2021.09.012
44
Maki J. J. Looft T. (2022). Turicibacter bilis sp. nov., a novel bacterium isolated from the chicken eggshell and swine ileum. Int. J. Syst. Evol. Microbiol.72, 1–9. doi: 10.1099/ijsem.0.005153
45
Manchester A. C. Webb C. B. Blake A. B. Sarwar F. Lidbury J. A. Steiner J. M. et al . (2019). Long-term impact of tylosin on fecal microbiota and fecal bile acids of healthy dogs. J. Vet. Intern. Med.33, 2605–2617. doi: 10.1111/jvim.15635
46
Martinez-Gili L. McDonald J. K. Liu Z. Kao D. Allegretti J. R. Monaghan T. M. et al . (2020). Understanding the mechanisms of efficacy of fecal microbiota transplant in treating recurrent Clostridioides difficile infection and beyond: the contribution of gut microbial-derived metabolites. Gut Microbes12:1810531. doi: 10.1080/19490976.2020.1810531
47
Miquel S. Martín R. Rossi O. Bermúdez-Humarán L. G. Chatel J. M. Sokol H. et al . (2013). Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol.16, 255–261. doi: 10.1016/j.mib.2013.06.003
48
Monaghan T. Mullish B. H. Patterson J. Wong G. K. S. Marchesi J. R. Xu H. et al . (2019). Effective fecal microbiota transplantation for recurrent Clostridioides difficile infection in humans is associated with increased signalling in the bile acid-farnesoid X receptor-fibroblast growth factor pathway. Gut Microbes10, 142–148. doi: 10.1080/19490976.2018.1506667
49
Mullish B. H. Quraishi M. N. Segal J. P. McCune V. L. Baxter M. Marsden G. L. et al . (2018). The use of faecal microbiota transplant as treatment for recurrent or refractory Clostridium difficile infection and other potential indications: joint British Society of Gastroenterology (BSG) and Healthcare Infection Society (HIS) guidelines. Gut67, 1920–1941. doi: 10.1136/gutjnl-2018-316818
50
Napolitano M. Covasa M. (2020). Microbiota transplant in the treatment of obesity and diabetes: current and future perspectives. Front. Microbiol.11:590370. doi: 10.3389/fmicb.2020.590370
51
Nye A. K. Suchodolski J. Hong M. P. Park S. Y. Thieman Mankin K. M. (2023). Pilot clinical trial: propidium monoazide PCR quantifies reduction of the viable bacterial load after antiseptic preparation of canine oral mucosa. Am. J. Vet. Res.84, 1–7. doi: 10.2460/ajvr.23.02.0029
52
Oliver J. D. (2005). The viable but nonculturable state in bacteria. J. Microbiol.43, 93–100
53
Ott S. J. Waetzig G. H. Rehman A. Moltzau-Anderson J. Bharti R. Grasis J. A. et al . (2017). Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology152, 799–811.e7. doi: 10.1053/j.gastro.2016.11.010
54
Papanicolas L. E. Choo J. M. Wang Y. Leong L. E. X. Costello S. P. Gordon D. L. et al . (2019). Bacterial viability in faecal transplants: which bacteria survive?EBioMedicine41, 509–516. doi: 10.1016/j.ebiom.2019.02.023
55
Pilla R. Gaschen F. P. Barr J. W. Olson E. Honneffer J. Guard B. C. et al . (2020). Effects of metronidazole on the fecal microbiome and metabolome in healthy dogs. J. Vet. Intern. Med.34, 1853–1866. doi: 10.1111/jvim.15871
56
Portincasa P. Bonfrate L. Vacca M. de Angelis M. Farella I. Lanza E. et al . (2022). Gut microbiota and short chain fatty acids: implications in glucose homeostasis. Int. J. Mol. Sci.23. doi: 10.3390/ijms23031105
57
Prawitt J. Caron S. Staels B. (2011). Bile acid metabolism and the pathogenesis of type 2 diabetes. Curr. Diab. Rep.11, 160–166. doi: 10.1007/s11892-011-0187-x
58
Reygner J. Charrueau C. Delannoy J. Mayeur C. Robert V. Cuinat C. et al . (2020). Freeze-dried fecal samples are biologically active after long-lasting storage and suited to fecal microbiota transplantation in a preclinical murine model of Clostridioides difficile infection. Gut Microbes11, 1405–1422. doi: 10.1080/19490976.2020.1759489
59
Ridlon J. M. Devendran S. Alves J. M. Doden H. Wolf P. G. Pereira G. V. et al . (2020). The 'in vivo lifestyle' of bile acid 7alpha-dehydroxylating bacteria: comparative genomics, metatranscriptomic, and bile acid metabolomics analysis of a defined microbial community in gnotobiotic mice. Gut Microbes11, 381–404. doi: 10.1080/19490976.2019.1618173
60
Rivera-Chavez F. Lopez C. A. Baumler A. J. (2017). Oxygen as a driver of gut dysbiosis. Free Radic. Biol. Med.105, 93–101. doi: 10.1016/j.freeradbiomed.2016.09.022
61
Scaldaferri F. Pecere S. Petito V. Zambrano D. Fiore L. Lopetuso L. R. et al . (2016). Efficacy and mechanisms of action of fecal microbiota transplantation in ulcerative colitis: pitfalls and promises from a first Meta-analysis. Transplant. Proc.48, 402–407. doi: 10.1016/j.transproceed.2015.12.040
62
Segal J. P. Mullish B. H. Quraishi M. N. Iqbal T. Marchesi J. R. Sokol H. (2020). Mechanisms underpinning the efficacy of faecal microbiota transplantation in treating gastrointestinal disease. Ther. Adv. Gastroenterol.13:1756284820946904. doi: 10.1177/1756284820946904
63
Singh R. Zogg H. Wei L. Bartlett A. Ghoshal U. C. Rajender S. et al . (2021). Gut microbial Dysbiosis in the pathogenesis of gastrointestinal Dysmotility and metabolic disorders. J. Neurogastroenterol. Motil.27, 19–34. doi: 10.5056/jnm20149
64
Song Z. Cai Y. Lao X. Wang X. Lin X. Cui Y. et al . (2019). Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome7:9. doi: 10.1186/s40168-019-0628-3
65
Suchodolski J. S. (2022). Analysis of the gut microbiome in dogs and cats. Vet. Clin. Pathol.50, 6–17. doi: 10.1111/vcp.13031
66
Suez J. Elinav E. (2017). The path towards microbiome-based metabolite treatment. Nat. Microbiol.2:17075. doi: 10.1038/nmicrobiol.2017.75
67
Sulaiman J. E. Thompson J. Qian Y. Vivas E. I. Diener C. Gibbons S. M. et al . (2024). Elucidating human gut microbiota interactions that robustly inhibit diverse Clostridioides difficile strains across different nutrient landscapes. Nat. Commun.15:7416. doi: 10.1038/s41467-024-51062-w
68
Sung C. H. Marsilio S. Chow B. Zornow K. A. Slovak J. E. Pilla R. et al . (2022). Dysbiosis index to evaluate the fecal microbiota in healthy cats and cats with chronic enteropathies. J. Feline Med. Surg.24, e1–e12. doi: 10.1177/1098612X221077876
69
Sung C. H. Pilla R. Chen C. C. Ishii P. E. Toresson L. Allenspach-Jorn K. et al . (2023). Correlation between targeted qPCR assays and untargeted DNA shotgun metagenomic sequencing for assessing the fecal microbiota in dogs. Animals (Basel)13. doi: 10.3390/ani13162597
70
Ting N. L. Lau H. C. Yu J. (2022). Cancer pharmacomicrobiomics: targeting microbiota to optimise cancer therapy outcomes. Gut71, 1412–1425. doi: 10.1136/gutjnl-2021-326264
71
Toresson L. Spillmann T. Pilla R. Ludvigsson U. Hellgren J. Olmedal G. et al . (2023). Clinical effects of Faecal microbiota transplantation as adjunctive therapy in dogs with chronic enteropathies-a retrospective case series of 41 dogs. Vet. Sci.10, 1–18. doi: 10.3390/vetsci10040271
72
Tourlousse D. M. Narita K. Miura T. Sakamoto M. Ohashi A. Shiina K. et al . (2021). Validation and standardization of DNA extraction and library construction methods for metagenomics-based human fecal microbiome measurements. Microbiome9:95. doi: 10.1186/s40168-021-01048-3
73
van Frankenhuyzen J. K. Trevors J. T. Lee H. Flemming C. A. Habash M. B. (2011). Molecular pathogen detection in biosolids with a focus on quantitative PCR using propidium monoazide for viable cell enumeration. J. Microbiol. Methods87, 263–272. doi: 10.1016/j.mimet.2011.09.007
74
Vecchiato C. G. Pinna C. Sung C. H. Borrelli de Andreis F. Suchodolski J. S. Pilla R. et al . (2023). Fecal microbiota, bile acids, sterols, and fatty acids in dogs with chronic enteropathy fed a home-cooked diet supplemented with coconut oil. Animals13:502. doi: 10.3390/ani13030502
75
Vinithakumari A.A. Hernandez B. G. Ghimire S. Adams S. Stokes C. Jepsen I. et al . (2021). A model screening pipeline for bile acid converting anti-Clostridioides difficile bacteria reveals unique biotherapeutic potential of Peptoacetobacter hiranonis. BioRxiv. 30. doi: 10.1101/2021.09.29.462466
76
Vital M. Howe A. C. Tiedje J. M. (2014). Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio5:e00889. doi: 10.1128/mBio.00889-14
77
Vital M. Rud T. Rath S. Pieper D. H. Schlüter D. (2019). Diversity of Bacteria exhibiting bile acid-inducible 7alpha-dehydroxylation genes in the human gut. Comput. Struct. Biotechnol. J.17, 1016–1019. doi: 10.1016/j.csbj.2019.07.012
78
Wang X. Howe S. Deng F. Zhao J. (2021). Current applications of absolute bacterial quantification in microbiome studies and decision-making regarding different biological questions. Microorganisms9:1797. doi: 10.3390/microorganisms9091797
79
Wang B. Yao M. Lv L. Ling Z. Li L. (2017). The human microbiota in health and disease. Engineering3, 71–82. doi: 10.1016/J.ENG.2017.01.008
80
Werner M. Ishii P. E. Pilla R. Lidbury J. A. Steiner J. M. Busch-Hahn K. et al . (2023). Prevalence of Clostridioides difficile in canine feces and its association with intestinal Dysbiosis. Animals (Basel)13. doi: 10.3390/ani13152441
81
Winston J. A. Suchodolski J. S. Gaschen F. Busch K. Marsilio S. Costa M. C. et al . (2024). Clinical guidelines for fecal microbiota transplantation in companion animals. Advances Small Animal Care5, 79–107. doi: 10.1016/j.yasa.2024.06.006
82
Wise J. L. Cummings B. P. (2022). The 7-alpha-dehydroxylation pathway: an integral component of gut bacterial bile acid metabolism and potential therapeutic target. Front. Microbiol.13:1093420.
83
Wolffs P. Norling B. Radstrom P. (2005). Risk assessment of false-positive quantitative real-time PCR results in food, due to detection of DNA originating from dead cells. J. Microbiol. Methods60, 315–323. doi: 10.1016/j.mimet.2004.10.003
84
Zain N. M. M. ter Linden D. Lilley A. K. Royall P. G. Tsoka S. Bruce K. D. et al . (2022). Design and manufacture of a lyophilised faecal microbiota capsule formulation to GMP standards. J. Control. Release350, 324–331. doi: 10.1016/j.jconrel.2022.08.012
Summary
Keywords
Clostridium hiranonis , dysbiosis, canine, cryoprotectant, PMA, bacterial culture, lyophilization, bile acid metabolism
Citation
Correa Lopes B, Turck J, Tolbert MK, Giaretta PR, Suchodolski JS and Pilla R (2025) Prolonged storage reduces viability of Peptacetobacter (Clostridium) hiranonis and core intestinal bacteria in fecal microbiota transplantation preparations for dogs. Front. Microbiol. 15:1502452. doi: 10.3389/fmicb.2024.1502452
Received
26 September 2024
Accepted
17 December 2024
Published
07 January 2025
Volume
15 - 2024
Edited by
Lorenzo Nissen, University of Bologna, Italy
Reviewed by
Jing Liu, Agricultural Research Service (USDA), United States
Jia Xu, Jinhua Polytechnic, China
Updates
Copyright
© 2025 Correa Lopes, Turck, Tolbert, Giaretta, Suchodolski and Pilla.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bruna Correa Lopes, brunalopes@tamu.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.