- Department of Gastroenterology, The Second Affiliated Hospital of Guangxi Medical University, Nanning, China
Non-Alcoholic Fatty Liver Disease (NAFLD) affects approximately 32.4% of the global population and poses a significant health concern. Emerging evidence underscores the pivotal role of the gut microbiota—including bacteria, viruses, fungi, and parasites—in the development and progression of NAFLD. Dysbiosis among gut bacteria alters key biological pathways that contribute to liver fat accumulation and inflammation. The gut virome, comprising bacteriophages and eukaryotic viruses, significantly shapes microbial community dynamics and impacts host metabolism through complex interactions. Similarly, gut fungi maintain a symbiotic relationship with bacteria; the relationship between gut fungi and bacteria is crucial for overall host health, with certain fungal species such as Candida in NAFLD patients showing detrimental associations with metabolic markers and liver function. Additionally, the “hygiene hypothesis” suggests that reduced exposure to gut parasites may affect immune regulation and metabolic processes, potentially influencing conditions like obesity and insulin resistance. This review synthesizes current knowledge on the intricate interactions within the gut microbiota and their associations with NAFLD. We highlight the therapeutic potential of targeting these microbial communities through interventions such as probiotics, prebiotics, and fecal microbiota transplantation. Addressing the complexities of NAFLD requires comprehensive strategies that consider the multifaceted roles of gut microorganisms in disease pathology.
Highlights
• Lytic and lysogenic bacteriophages disrupt the gut microbiota by causing bacterial lysis, which leads to the release of LPS and subsequent inflammation.
• Candida albicans and Mucor ambiguus activate NF-κB-mediated inflammation affecting insulin resistance and lipid metabolism.
• Saccharomyces cerevisiae and Schizosaccharomyces pombe improve insulin resistance by activating anti-inflammatory pathway through β-glucans.
• Blastocystis ST7 interacts with gut bacteria, disrupting tight junctions and increasing pro-inflammatory cytokines, thereby influencing NAFLD development.
Introduction
Non-Alcoholic Fatty Liver Disease (NAFLD) is a common liver disorder affecting approximately 32.4% of the global population (Riazi et al., 2022). It is characterized by excessive fat accumulation in the liver, which leads to various health complications. Although the precise causes of NAFLD remain unclear, several key risk factors have been identified, including obesity, type 2 diabetes, high cholesterol, insulin resistance, and poor dietary habits (Kneeman et al., 2012; Basaranoglu et al., 2015; Mirmiran et al., 2017; Abenavoli et al., 2022). Additionally, gut microbiota dysbiosis are recognized as significant contributors to the onset and progression of NAFLD (Hrncir et al., 2021).
The human gut is a complex ecosystem that hosts a vast array of microorganisms, collectively known as the gut microbiome. This microbiome includes bacteria, viruses, fungi, and parasites, all of which coexist in a delicate equilibrium (Fay et al., 2017). Notably, the microorganisms in the gut outnumber human cells by a substantial margin, with estimates ranging between 1013 and 1014 cells—approximately 10 times more than the number of human cells (Bäckhed et al., 2005; Neish, 2009). These microorganisms also carry a genetic material that is roughly 100 times greater than the human genome. In addition to their abundance, these microorganisms produce metabolites that have significant effects on human health. The complex interactions among gut microbiota are closely linked with various metabolic disorders, and the metabolites they produce have far-reaching impacts on health (Boulangé et al., 2016; Dabke et al., 2019; Breuninger et al., 2021).
Within the gut ecosystem, the virome plays a key role. The gut virome consists of prokaryotic viruses, primarily bacteriophages, which infect bacteria, as well as eukaryotic viruses that target human gut cells, fungi, and protozoa (Cao et al., 2022). Viruses infecting prokaryotes dominate the gut virome, making up 97.8% of its composition, including bacteriophages at 97.7% and archaeal viruses at 0.1%, while eukaryotic viruses account for the remaining 2.1% (Spencer et al., 2022). The interactions between bacteriophages and bacteria regulate the gut microenvironment and have a profound influence on human health.
Gut fungi also play a crucial role in this ecosystem. They help regulate the immune system, produce metabolites that impact metabolic diseases, and influence both the composition and function of the gut microbiota (van Tilburg Bernardes et al., 2020; Pérez, 2021; Zhang F. et al., 2022). Protozoa, which can act as both beneficial commensals and potential disease-causing agents, add another layer of complexity to this finely balanced system (Chabé et al., 2017; Dubik et al., 2022). Indeed, all microorganisms in the gut—including bacteria, fungi, viruses, and protozoa—have dual roles, contributing to host health while also possessing the potential to cause disease under certain conditions.
Emerging evidence suggests that probiotics may have a beneficial effect on NAFLD by modulating the gut microbiota and reducing inflammation. Studies indicate that the administration of probiotics could help restore the balance of gut microbiota, reduce hepatic inflammation, and improve liver health by preventing gut dysbiosis (Wang L. et al., 2023). Probiotics, particularly those that target gut bacteria and improve bile acid metabolism, have been shown to alleviate hepatic steatosis and support metabolic health in patients with NAFLD (Xie and Halegoua-DeMarzio, 2019; Mims et al., 2021). The modulation of the gut-liver axis through probiotics offers a promising approach for the prevention and treatment of NAFLD (Abenavoli et al., 2022).
Additionally, noninvasive biomarkers and surrogate scores are becoming increasingly important for diagnosing and assessing the severity of NAFLD, particularly in the context of liver fibrosis. Tools such as the NAFLD Fibrosis Score (NFS), Fibrosis-4 (FIB-4), and NAFLD Fibrosis Protein Panel (NFPP) provide valuable information regarding the extent of liver damage and help guide clinical decisions (Elsayed et al., 2022). Identifying patients at risk for developing severe liver complications through these noninvasive measures could allow for early interventions that target inflammation and fibrosis, ultimately preventing the progression of NAFLD.
This review aims to thoroughly explore the multifaceted roles of the gut microbiota in relation to NAFLD. It focuses on understanding how the gut environment influences the development and progression of the disease. Maintaining a stable gut microenvironment is critical to understanding the pathogenesis of NAFLD. A detailed examination of the relationship between the gut environment and NAFLD reveals the interconnectedness of various factors contributing to the disease. By understanding the key roles played by gut microbiota—including bacteria, viruses, fungi, and parasites—researchers and healthcare professionals can gain valuable insights into the complex mechanisms driving the onset and progression of NAFLD (Mims et al., 2021; Wang L. et al., 2023). Furthermore, the identification and use of noninvasive biomarkers can enhance the early detection of disease and provide better strategies for managing NAFLD (Elsayed et al., 2022). The review emphasizes the need for further research into these intricate relationships in order to develop more effective strategies for both the prevention and treatment of NAFLD.
Unraveling the role of gut bacteria in NAFLD
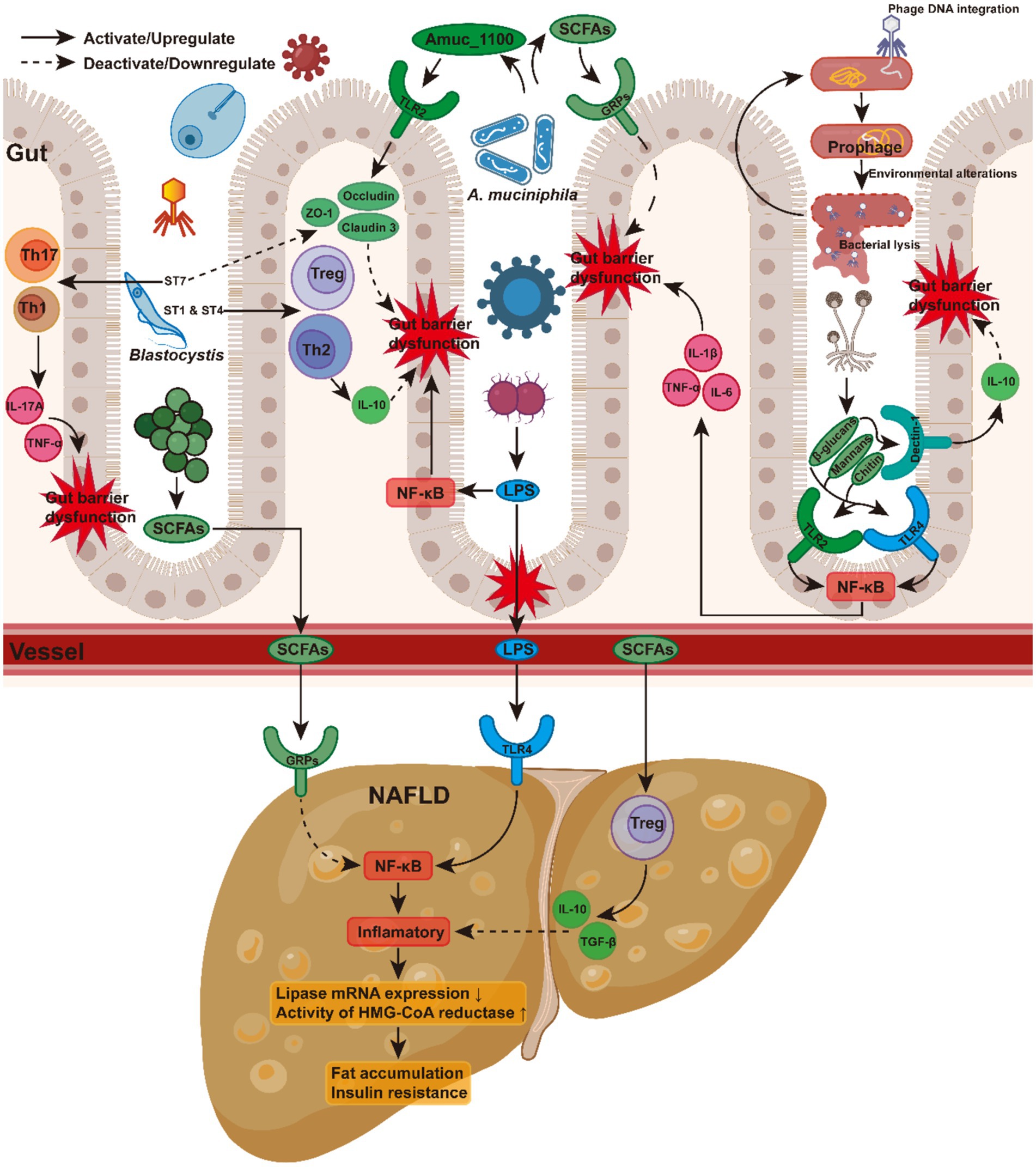
In patients with NAFLD, compelling evidence suggests significant dysbiosis in the gut bacteria, accompanied by a marked reduction in alpha diversity (Khan et al., 2021; Fang et al., 2022). NAFLD patients frequently exhibit a gut microbiota predominantly dominated by Firmicutes, resulting in an elevated Firmicutes-to-Bacteroidetes ratio compared to healthy controls (Hrncir et al., 2021; Aron-Wisnewsky et al., 2020; Tokuhara, 2021; Jasirwan et al., 2021). The increased abundance of Bacteroides vulgatus, Escherichia coli, and Klebsiella pneumoniae in NAFLD is associated with metabolic alterations, particularly in obesity and insulin resistance. B. vulgatus and E. coli promote hepatic fat accumulation and exacerbate insulin resistance by producing lipopolysaccharides (LPS), which activates the Toll-like receptor 4 (TLR4) pathway in the liver and nuclear factor kappa B (NF-κB) signaling pathways (Aron-Wisnewsky et al., 2020; Shi et al., 2006; Cai et al., 2005). Under inflammatory conditions, there is a decrease in hepatic lipase mRNA expression, along with an upregulation of the expression and activity of HMG-CoA reductase, the rate-limiting enzyme in cholesterol synthesis. As a result, lipid hydrolysis in the liver is reduced, whereas cholesterol production is increased (Feingold et al., 1993, 1999). K. pneumoniae increases oxidative stress and pro-inflammatory cytokine release, directly damaging hepatocytes (Aron-Wisnewsky et al., 2020). Ethanol also activates the NF-κB and apoptosis-related pathways, accelerating the progression of NAFLD to non-alcoholic steatohepatitis (NASH) (Mandrekar and Szabo, 2009). Conversely, beneficial bacteria such as Akkermansia muciniphila, Faecalibacterium prausnitzii, and Bifidobacterium are depleted in NAFLD patients (Aron-Wisnewsky et al., 2020). A. muciniphila improves intestinal barrier function by downregulating the NF-κB pathway, regulating the mucus layer, and modulating tight junction proteins, leading to reduced LPS absorption (Everard et al., 2013). Furthermore, Amuc_1100, a specific protein isolated from the outer membrane of A. muciniphila, can interact with Toll-like receptor 2 (TLR2), regulate various tight-junction proteins including occludin and claudin 3, contributing to the improvement of the intestinal barrier (Plovier et al., 2017). In addition, A. muciniphila produces short-chain fatty acids (SCFAs), which activate G protein-coupled receptors (GRPs), thereby modulating energy metabolism and providing protection against NAFLD (Kimura et al., 2014). F. prausnitzii produces anti-inflammatory SCFAs, particularly butyrate, which inhibits the NF-κB signaling pathway and reduces the release of pro-inflammatory cytokines (Miquel et al., 2013). Through SCFA production, Bifidobacterium enhances gut barrier function, reducing endotoxin translocation from entering the bloodstream. The reduction in hepatic inflammation is further supported by Bifidobacterium’s ability to enhance regulatory T cell (Treg) activity and increase the production of IL-10 and TGF-β, thereby helping to suppress inflammatory responses (Round and Mazmanian, 2009; Kwon et al., 2010).
These microbial shifts indicate complex interactions in metabolic pathways, which exert their influence on NAFLD via several mechanisms. These include modulation of SCFA levels (Rau et al., 2018; Parada Venegas et al., 2019; Dai et al., 2020; Liu et al., 2022; Portincasa et al., 2022; Wang et al., 2022; Xiong et al., 2022), regulation of intestinal barrier integrity (Chang et al., 2014; Deleu et al., 2021; Forlano et al., 2022; Quesada-Vázquez et al., 2022; Liu et al., 2023; Fagundes et al., 2024), influence of host immune responses (Parada Venegas et al., 2019; Kim, 2021; Visekruna and Luu, 2021; Yao et al., 2022; Zhang Z. et al., 2022; Sarkar et al., 2023), and alteration of bile acid metabolism (Mouzaki et al., 2016; Appleby et al., 2019; Gottlieb and Canbay, 2019; Collins et al., 2023; Figure 1). However, as these complex biological pathways have been comprehensively reviewed elsewhere (Aron-Wisnewsky et al., 2020), we will not discuss them in detail here.
These findings highlight the critical role of gut bacteria in shaping NAFLD pathogenesis, influencing a range of biological processes. The bacterial composition within the gut has substantial effects on various aspects of NAFLD, including the regulation of metabolic pathways, immune modulation, and gut barrier integrity (Aron-Wisnewsky et al., 2020).
Nonetheless, our understanding of the role of bacteria in NAFLD remains incomplete. In addition to bacteria, other microbial components—including viruses, fungi, and parasites—contribute to this complex narrative (Iliev and Cadwell, 2021). The roles of these additional microbial elements in the development of NAFLD introduce further complexity. Investigating their specific contributions presents a promising opportunity to advance our understanding of NAFLD progression. This review seeks to elucidate these understudied aspects, potentially identifying novel therapeutic targets for the managing and preventing NAFLD through comprehensive modulation of the microbiota. Continued exploration of these multifactorial interactions holds the potential to uncover innovative interventions, benefiting those at risk of or currently suffering from NAFLD.
Decoding the gut virome: bacteriophages, eukaryotic viruses, and the unexplored interactions in NAFLD pathogenesis
The gut virome—which comprises a diverse range of viruses, including eukaryotic viruses, bacteriophages and archaeal viruses acquired from diet and environmental factors—is a fundamental component of the gut microbiome. Its presence plays a pivotal role in regulating the balance of the human gut microbiota. The virome significantly influences the structure and functionality of the microbiome, affecting health and disease states. Research highlights the virome’s critical role in modulating the dynamics and functions of the gut microbiota, with implications for overall human health (Liang and Bushman, 2021). A summary of the key points is provided in Table 1.
Bacteriophages
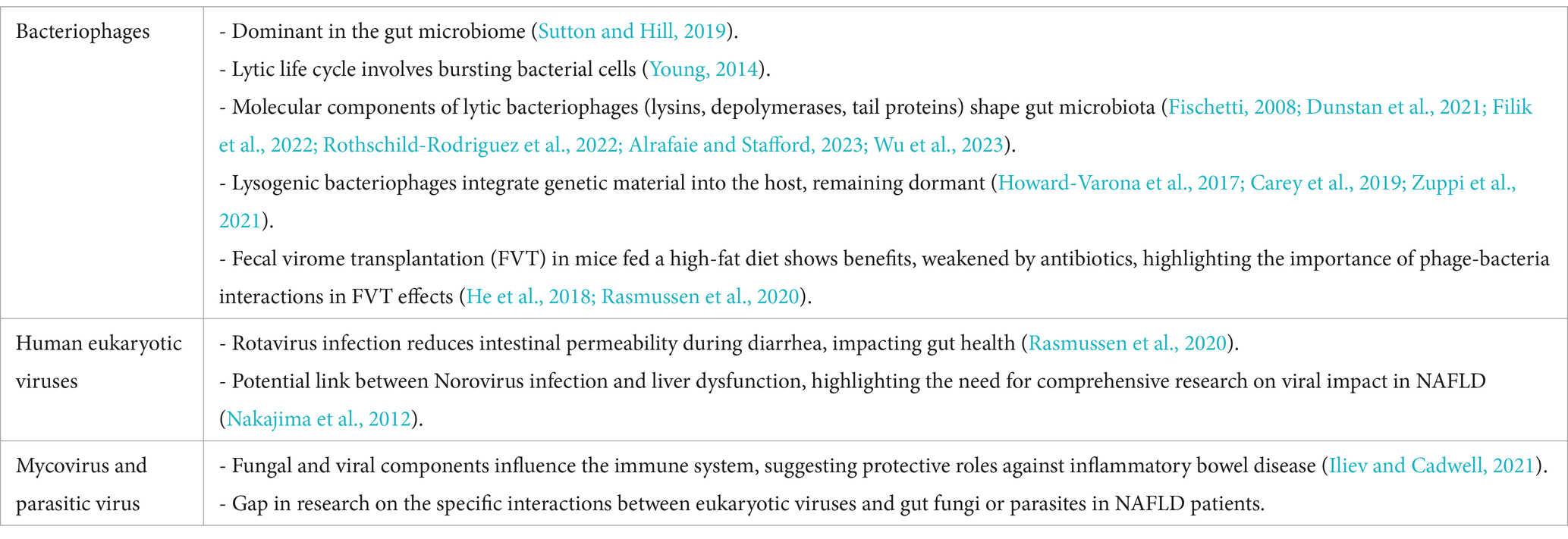
Bacteriophages—viruses that infect and replicate within bacteria—dominate the viral component of the gut microbiome (Sutton and Hill, 2019). However, due to the difficulty in identifying specific bacteriophage hosts, our understanding of their impact on NAFLD remains limited.
Bacteriophages follow two primary life cycles: lytic and lysogenic. In the lytic cycle, bacteriophages infect bacterial cells, causing the cells to burst and release newly formed phages (Young, 2014). Lytic bacteriophages use a “kill the winner” strategy, targeting dominant bacterial species while sparing less abundant ones. This strategy helps maintain microbial diversity and stability within the gut (Silveira and Rohwer, 2016; Sutcliffe et al., 2023). The effectiveness of this process is facilitated by molecular components within lytic bacteriophages, including lysins, depolymerases, and tail proteins (Filik et al., 2022; Rothschild-Rodriguez et al., 2022; Alrafaie and Stafford, 2023). Lysins degrade the bacterial cell wall, compromising its structural integrity (Fischetti, 2008). Depolymerases break down bacterial capsules, which shield certain bacteria, allowing phages to infect their hosts (Dunstan et al., 2021; Wu et al., 2023). Tail proteins enable phages to attach to the bacterial cell surface and inject viral genetic material (Taslem Mourosi et al., 2022). These mechanisms are central to understanding how lytic bacteriophages shape the gut microbiota and influence its composition, with broad implications for human health.
Lysogenic bacteriophages, by contrast, integrate their genetic material into the host genome using specialized enzymes called integrases (Howard-Varona et al., 2017; Carey et al., 2019). This process, known as lysogeny, results in the formation of a dormant prophage within the host cell, which can remain inactive for extended periods (Zuppi et al., 2021). Environmental factors such as unhealthy diet, chemical exposure, or stress can trigger prophage activation, initiating a switch to the lytic cycle (Pasechnek et al., 2020). This activation leads to bacterial cell lysis and the release of inflammatory compounds such as LPS, which potentially contributing to NAFLD development (Lin and Lin, 2019; Hsu et al., 2021).
In various ecosystems, bacteriophages display distinct reproductive strategies. Lytic phages, found in favorable environments, exhibit high virulence and cause rapid bacterial lysis. In contrast, temperate phages, prevalent in nutrient-poor environments, have lower virulence and smaller burst sizes, preferring to infect slow-growing bacterial populations. These phages often enter a lysogenic state, contributing to the co-evolution and stability of microbial communities. The ratio of lytic to temperate bacteriophages may serve as an indicator of gut microbiota health (Howard-Varona et al., 2017; Manrique et al., 2017).
NAFLD patients often exhibit unbalanced diets, characterized by high intake of saturated fats and low consumption of micronutrients (Yasutake et al., 2014; Lian et al., 2020). These dietary habits are also prevalent in urban populations, where they are associated with higher Firmicutes-to-Bacteroidetes ratios compared to rural counterparts (De Filippo et al., 2010). In both animal models and individuals with non-alcoholic steatohepatitis (NASH), high-fat diets have been linked to changes in the gut virome, including increased lytic phage activity, which further exacerbates gut dysbiosis (Zhu et al., 2013; He et al., 2018).
Fecal virome transplantation (FVT) is gaining recognition as a potential therapeutic strategy for correcting gut microbiota imbalances, particularly in metabolic disorders like NAFLD. Preclinical studies highlight FVT’s efficacy in improving metabolic parameters such as enhanced glucose tolerance and promoting weight reduction, especially in high-fat diet models. However, several challenges limit the clinical translation of FVT. Donor variability remains a major concern, as differences in virome composition can lead to inconsistent therapeutic outcomes. In addition, safety concerns regarding the possible transfer of pathogenic eukaryotic viruses necessitate rigorous donor screening, increasing both complexity and cost (Raeisi et al., 2023; Rasmussen et al., 2024; Zuppi et al., 2024). In one study, FVT in high-fat diet-fed mice significantly improved metabolic outcomes; however, the benefits were reduced when antibiotics were administered prior to the procedure, underscoring the critical role of phage-bacteria interactions in the therapeutic efficacy of FVT. Addressing these hurdles is vital for the reliable implementation of FVT as a standard treatment for NAFLD (Rasmussen et al., 2020).
Metagenomic analysis of the bacteriophage community in NAFLD patients has revealed notable findings (Lang et al., 2020). In patients with advanced liver fibrosis, phages infecting Lactococcus and Leuconostoc were markedly reduced, correlating with an increase in their bacterial hosts. Conversely, phages infecting Streptococcus, Escherichia, Enterobacteria, and Lactobacillus increased, while the abundance of these bacteria decreased. Lactococcus and Leuconostoc, commonly found in foods, can mitigate NAFLD through mechanisms such as modulating the gut microbiota, reducing LPS levels, producing SCFAs, regulating lipid metabolism, and alleviating inflammatory responses, ultimately aiding in weight reduction, decreasing liver fat content, and improving insulin resistance (Castro-Rodríguez et al., 2020; Naudin et al., 2020; Sun et al., 2020; Mendes et al., 2021; Lee et al., 2024). In contrast, Streptococcus, Escherichia, and Enterobacteria are associated with fatty liver disease (Munukka et al., 2014; Naka et al., 2014; Boursier et al., 2016; Aron-Wisnewsky et al., 2020). These shifts in bacteriophage abundance were inversely correlated with their bacterial hosts, highlighting the complexity of bacteriophage-bacteria interactions. This complexity necessitates further study, including the exploration of “viral dark matter”—unexplored viral components that may provide crucial insights into the role of bacteriophages in NAFLD pathogenesis.
Human eukaryotic viruses
Eukaryotic viruses, part of the gut virome, also significantly impact human health. Highly contagious viruses such as Norovirus and Rotavirus can cause gastrointestinal inflammation, leading to vomiting, diarrhea, nausea, abdominal pain, fever, and dehydration. These symptoms are particularly severe in young children and vulnerable adults. While bacterial diarrhea often increase intestinal permeability, Rotavirus has been shown to decrease permeability via 5-HT and neurotrophic factors (Hagbom et al., 2020).
Additionally, case reports suggest a potential link between Norovirus and liver dysfunction, though more research is needed to elucidate this relationship (Nakajima et al., 2012). Rotavirus infections are commonly associated with elevated liver transaminases, with about 20% of affected individuals showing increased ALT and AST levels (Nakajima et al., 2012). Both Norovirus (Pearson et al., 2019) and Rotavirus (Pane et al., 2014a,b; Pearson et al., 2019) have also been implicated in type 1 diabetes through their effects on the immune system. Type 1 diabetes primarily involves autoimmune destruction of insulin-secreting cells, leading to insulin deficiency. However, some individuals with type 1 diabetes may develop insulin resistance due to obesity or long-term insulin therapy, a condition sometimes referred to as “double diabetes,” which may increase the risk of developing NAFLD (Memaj and Jornayvaz, 2022).
This interconnection between enteric viral infections, liver dysfunction, and metabolic disorders underscores the need for comprehensive research into the interactions between gut viruses and systemic health. Understanding these mechanisms is crucial for developing preventive and therapeutic strategies.
Mycovirus and parasitic virus
The interaction between eukaryotic viruses, gut fungi, and parasites in the pathogenesis of NAFLD is an underexplored yet promising area of research. While fungal and viral components of the gut microbiome are known to influence immune modulation and metabolic processes, their specific roles in NAFLD remain largely undefined.
Fungal viruses can reduce the pathogenicity of their host fungi by inducing hypovirulence. These viruses spread horizontally through hyphal anastomosis and vertically via spore transmission, which can be either asexual or sexual. Some viruses can influence the metabolic pathways of fungal cells; for example, Penicillium chrysogenum virus (PcV) affects the expression of the host’s metabolic genes, leading to changes in glycolysis pathways and lipid metabolism. These metabolic alterations may impact the fungi’s responses to environmental stresses and the production of secondary metabolites, such as antibiotics (Ghabrial et al., 2015).
Specific fungi like Candida and Saccharomyces play a role in maintaining gut homeostasis and potentially protecting against inflammatory disorders (Iliev and Cadwell, 2021). However, these fungi may also contribute to dysbiosis in NAFLD patients, though the precise mechanisms remain unclear (Wu et al., 2021).
Eukaryotic viruses, which include those that infect fungi and parasites, could further complicate these interactions. Some studies suggest that parasitic protozoa, such as Blastocystis and Giardia, may alter the composition of the gut microbiota and exacerbate immune responses. This dysregulation could lead to compromised gut barrier function and increased susceptibility to metabolic diseases, including NAFLD (Wu et al., 2021).
Despite these insights, specific research on how eukaryotic viruses influence the gut’s fungal and parasitic communities in NAFLD patients remains minimal. The intricate relationships among viruses, fungi, and parasites in the gut microbiome are not well understood, particularly regarding their collective impact on NAFLD pathogenesis.
Therefore, expanding research in this area is crucial. Investigating these interactions could uncover critical insights into the mechanisms of NAFLD, offering new therapeutic targets through the modulation of these eukaryotic agents within the gut microbiome. Understanding this complex triad could help develop more comprehensive strategies for managing gut health and metabolic disorders like NAFLD.
Fungal players in NAFLD: unraveling the complex interactions and potential impact of Candida albicans, Saccharomyces cerevisiae, and Schizosaccharomyces pombe
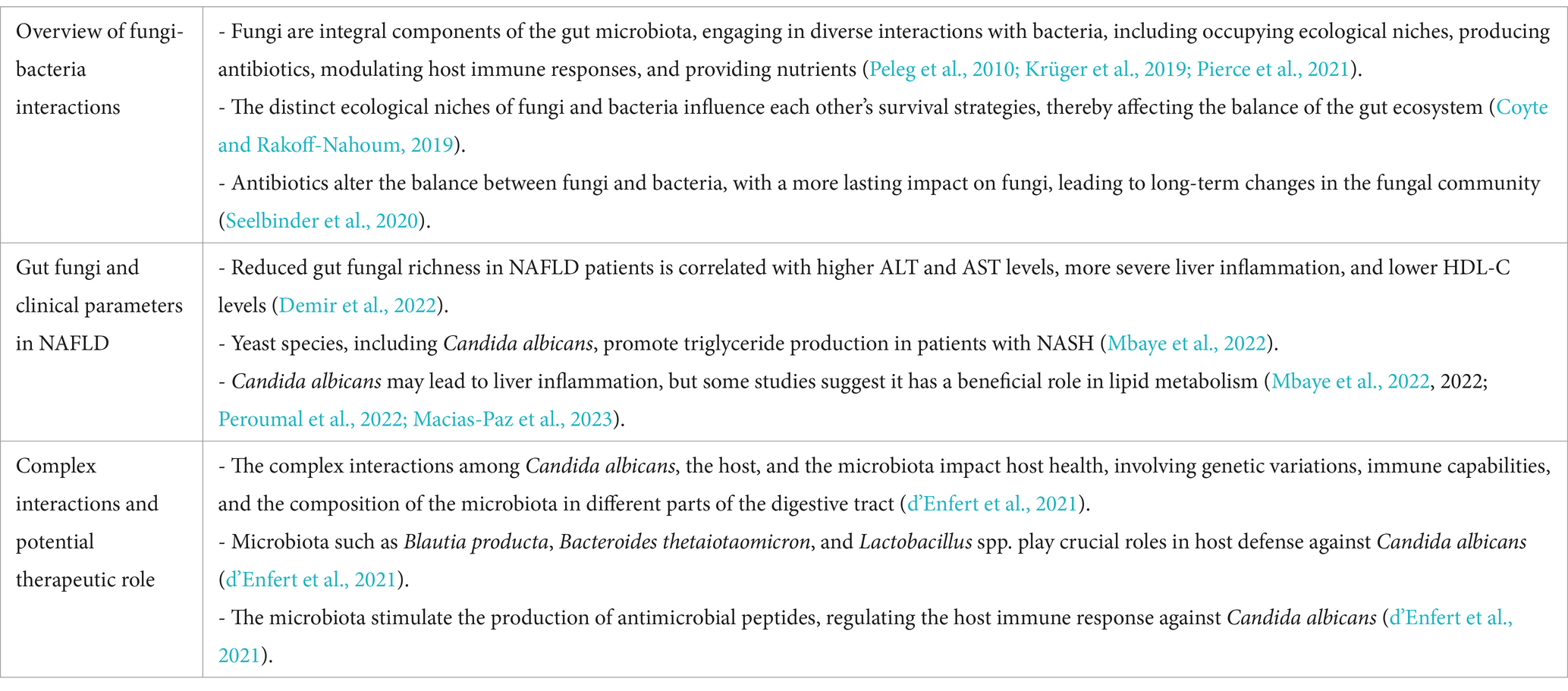
Fungi play a crucial role in the gut microbiota by interacting with bacteria through competition for niches, nutrient acquisition, and immune modulation (Peleg et al., 2010; Krüger et al., 2019; Pierce et al., 2021). These interactions significantly influence gut health, affecting immune responses and overall microbial balance. In NAFLD, fungal balance is often disrupted, with specific species influencing metabolic factors like insulin resistance and liver inflammation (Coyte and Rakoff-Nahoum, 2019). External factors, such as medications, diet, and environmental changes, can disturb this balance, leading to long-term shifts in the fungal community (Coyte and Rakoff-Nahoum, 2019). Fungal dysbiosis has been linked to NAFLD (Coyte and Rakoff-Nahoum, 2019; Demir et al., 2022), with some species associated with key metabolic dysfunctions (Szóstak et al., 2023).
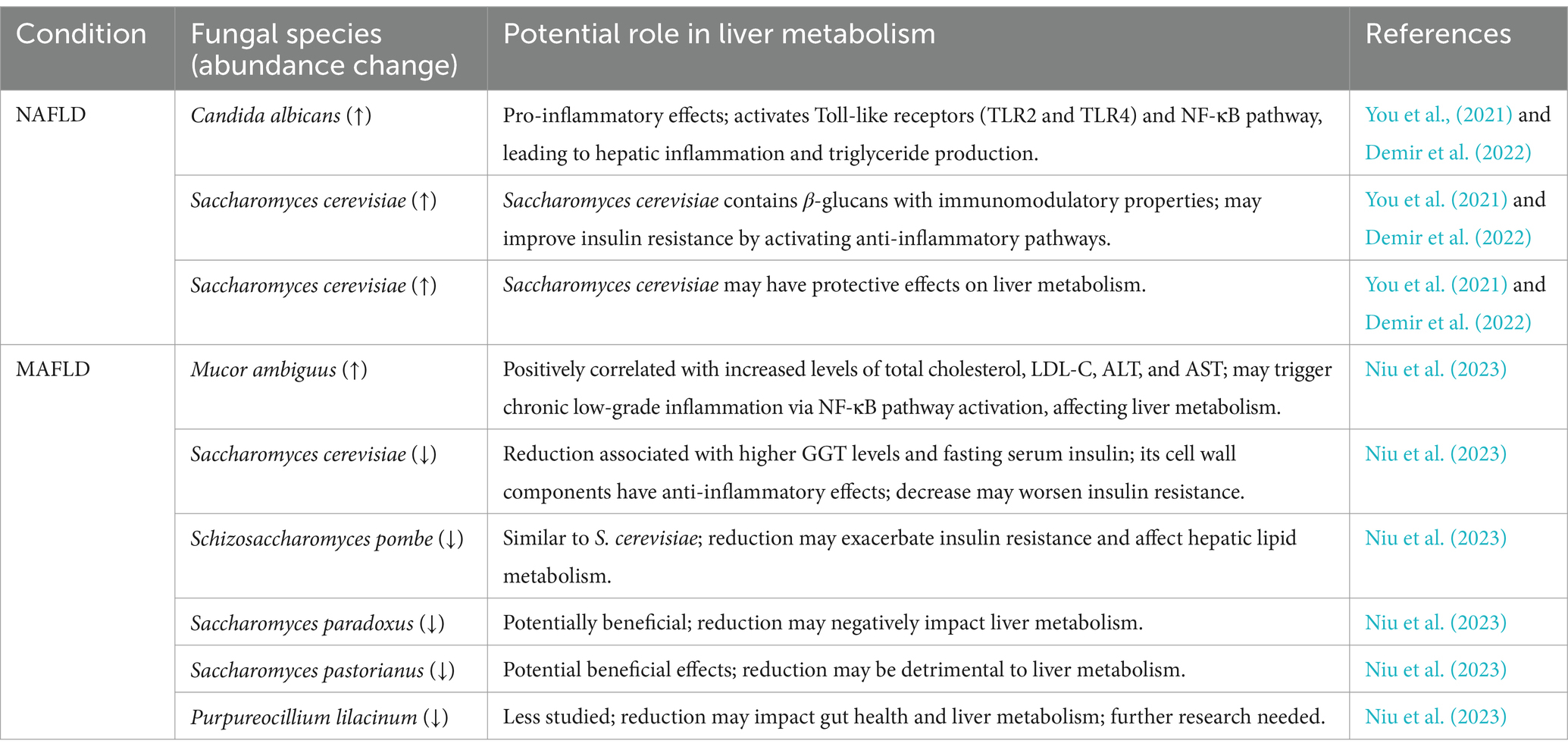
Research on gut fungi in NAFLD is limited. Given the limited research on gut fungi in NAFLD, exploring related conditions like metabolic dysfunction-associated fatty liver disease (MAFLD) becomes important. The diagnostic criteria for NAFLD and MAFLD highlight distinct approaches to defining fatty liver disease, as requested by the reviewer. NAFLD is diagnosed by exclusion, with fatty liver defined as fat accumulation in more than 5% of hepatocytes in the absence of significant alcohol intake, viral hepatitis, or other specific liver diseases. In contrast, MAFLD is a newer concept that emphasizes metabolic dysfunction as the central factor in fatty liver disease. MAFLD’s diagnosis is based on the presence of metabolic dysfunction, regardless of other potential causes, and requires meeting at least one of the following criteria: overweight or obesity, type 2 diabetes, or metabolic dysregulation (e.g., abnormal blood pressure, lipid profile, or insulin resistance). Unlike NAFLD, MAFLD does not rely on ruling out other liver diseases or alcohol intake. Due to the substantial overlap in pathogenesis and clinical characteristics between NAFLD and MAFLD, researchers often integrate findings from both conditions to gain a comprehensive understanding of the pathophysiological processes involved in fatty liver disease (Eslam et al., 2020; Shiha et al., 2021). Specific fungal species associated with NAFLD, such as Candida albicans and Mucor ambiguus have been found to be enriched in patients with NAFLD, while Saccharomyces cerevisiae and Schizosaccharomyces pombe show lower abundance in the gut of NAFLD patients (Table 2).
Patients with MAFLD show a decrease in fungal alpha diversity compared to healthy individuals, indicating a reduction in the richness and abundance of fungal species in the gut. Specifically, MAFLD patients exhibit increased levels of Mucor ambiguus within their gut, which is positively linked with total cholesterol, low-density lipoprotein cholesterol (LDL-C), alanine aminotransferase (ALT), and aspartate aminotransferase (AST) (Niu et al., 2023). The proliferation of Mucor ambiguus may trigger a host immune response, leading to chronic low-grade inflammation. Chronic inflammation can influence cholesterol synthesis through activating the NF-κB pathway, which increases the production of pro-inflammatory cytokines including TNF-α, IL-1, and IL-6 (Brown and Gordon, 2001; Netea et al., 2006). Interestingly, a high-fiber diet resulted in a reduction in the presence of the pathogenic fungus Mucor ambiguus (Wang T. et al., 2023).
Conversely, Saccharomyces cerevisiae and Schizosaccharomyces pombe display lower quantities in the feces of MAFLD patients. The reduced presence of these fungi was associated with higher gamma-glutamyl transferase (GGT) levels and fasting serum insulin (Niu et al., 2023). The cell wall components of Saccharomyces cerevisiae primarily include β-glucans, mannans, and chitin, which possess immunomodulatory properties (Brown and Gordon, 2001). In particular, β-glucans can combine with Dectin-1 on intestinal immune cells, promoting the secretion of anti-inflammatory cytokines such as interleukin-10 (IL-10), thereby suppressing chronic inflammation (Brown and Gordon, 2001; Gantner et al., 2005; Dillon et al., 2006). These findings suggest a potential relationship between these fungi and the improvement of insulin resistance, as evaluated through the Homeostatic Model Assessment of Insulin Resistance (HOMA-IR), calculated using the formula:
Another study also revealed reduced gut fungal richness in NAFLD patients, although this reduction did not reach statistical significance. The log-ratio of Candida albicans to Saccharomyces cerevisiae significantly correlated with higher ALT and AST levels, more severe liver inflammation, and lower levels of high-density lipoprotein cholesterol (HDL-C) (Dillon et al., 2006). Furthermore, yeast species such as Candida albicans, Pichia kudriavzevii, and Candida glabrata in the gut contributed to triglyceride production in patients with NASH (Mbaye et al., 2022). This may be attributed to the cell wall components of Candida albicans, such as mannans and β-glucans, which activate host Toll-like receptors, particularly TLR2 and TLR4. This activation triggers the NF-κB pathway, ultimately resulting in the release of pro-inflammatory cytokines (Netea et al., 2002, 2006). In an animal study, the inclusion of Candida albicans in a high-fat diet led to an enrichment of Akkermansia muciniphila—a probiotic known to improve insulin resistance and typically reduced in patients with NAFLD (Tokuhara, 2021; Peroumal et al., 2022). An explanation for this phenomenon is that Akkermansia muciniphila is an obligate mucin-degrading bacterium that utilizes mucin in the intestinal mucus as a carbon and energy source (Belzer and de Vos, 2012). When the intestinal barrier is compromised, the metabolic environment of the mucus layer changes, providing more mucin resources for Akkermansia muciniphila growth. Consequently, when Candida albicans disrupts the intestinal barrier and thickens the mucus layer, the abundance of Akkermansia muciniphila may increase accordingly.
However, a study suggest a beneficial role of Candida albicans. It is involved in lipid metabolism due to the secretion of lipase, which can hydrolyze triglycerides (Macias-Paz et al., 2023).
An existing review delves into the intricate interactions among Candida albicans, the host, and the microbiota and how they impact the host’s health status (d’Enfert et al., 2021). These relationships are influenced by genetic and phenotypic variations among diverse Candida albicans isolates, the host’s immune capabilities, and the varying composition of the microbiota across different sections of the digestive tract.
The microbiota plays a pivotal role in supporting the host’s defense mechanisms against Candida albicans by producing butyrate. Butyrate stimulates the generation of antimicrobial peptide IL-37 and nitric oxide, both crucial in combating Candida albicans. Furthermore, specific members of the gut microbiota, such as Blautia producta and Bacteroides thetaiotaomicron, contribute significantly to host resilience against Candida albicans by promoting the production of IL-37. Lactobacillus stimulates the production of IL-22, instrumental in conferring resistance against the colonization of Candida albicans. IL-37, an anti-inflammatory cytokine, modulates immune responses by interacting with the IL-18 receptor and IL-1R8 (Su and Tao, 2021). This signaling cascade reduces excessive inflammation while enhancing the gut epithelial barrier function, which indirectly hinders C. albicans colonization by strengthening the physical and immunological defenses of the gut. IL-37 signaling also influences the activity of macrophages and dendritic cells, leading to increased production of antimicrobial peptides and enhancing the resilience of the host against fungal pathogens like C. albicans (Wang et al., 2024). In parallel, IL-22 plays a key role in gut epithelial defense by binding to the IL-22 receptor complex on intestinal epithelial cells (Zheng et al., 2008; Pickert et al., 2009). This stimulates the production of antimicrobial peptides, such as RegIIIβ and RegIIIγ, which directly target microbial pathogens (Zheng et al., 2008). IL-22 signaling also promotes epithelial cell proliferation and tight junction integrity, creating a robust barrier that resists fungal colonization (Pickert et al., 2009). These antimicrobial peptides not only exert direct fungistatic effects but also create an inhospitable environment for C. albicans, reducing its ability to establish a foothold in the gut. Lacticaseibacillus rhamnosus ATCC 53103 modulates the host’s pro-inflammatory response to Candida albicans by reducing the production of inflammatory substances such as IL-1α and granulocyte-macrophage colony-stimulating factor (GM-CSF), thus regulating the immune reaction within the host (d’Enfert et al., 2021).
Overall, these findings underscore the complex interactions between gut fungi and the development and progression of NAFLD. Further research is needed to comprehensively understand the relationship between fungi and NAFLD (Table 3).
Parasites in the gut: exploring the potential impact of protozoans Blastocystis, Giardia lamblia, and Entamoeba histolytica on NAFLD and metabolic disorders
Parasites are organisms that live on or within a host, including various types such as protozoans and helminths. They engage in complex interactions with their hosts, potentially causing diseases but also modulating the host’s immune system functionality to a certain extent. The “hygiene hypothesis” posits that increased hygiene practices lead to reduced early-life exposure to parasites, including protozoans and helminths. This reduction may heighten susceptibility to allergies, asthma, and autoimmune diseases (Bach, 2018). Gut parasites are considered may play a role in human health because they activate the immune system to recognize and combat pathogens, thereby regulating both innate and adaptive immune pathways (Ianiro et al., 2022). A comprehensive review has illuminated the interaction between helminths and the gut microbiota, elucidating their impact on human health (Loke and Harris, 2023).
Protozoans are single-celled organisms that inhabit the human gut and can infect individuals at various life stages. Their prevalence varies globally, with higher infection rates observed in developing countries (Baron, 1996). For example, studies have reported that 46.8% of schoolchildren in the Merhabete District of Central Ethiopia and 42.3% in Sanandaj City, Iran, are infected with intestinal protozoans (Bahmani et al., 2017; Dagne and Alelign, 2021). In the Laghouat province of Southern Algeria, prevalence ranges from 13.3 to 17.3% (Sebaa et al., 2021). These high prevalence rates underscore the potential impact of protozoans on human health in these regions. The high prevalence of protozoan infections in regions like Laghouat Province may significantly impact human health by altering the gut microbiota, potentially increasing the risk of metabolic diseases. Protozoan infections can disrupt the balance of gut microbiota, leading to dysbiosis, which has been linked to metabolic disorders, including NAFLD. In regions with already limited healthcare resources, these disruptions may exacerbate health outcomes by affecting the gut-liver axis, contributing to liver inflammation, and impairing metabolic health. Thus, protozoan infections in these specific regions could have far-reaching implications not only for general health but also for metabolic diseases such as NAFLD.
Although protozoans significantly affect human health by modulating gut microbiota and host immunity, there is a scarcity of research exploring their role in diseases such as NAFLD and metabolic disorders. Given that protozoans can influence gut barrier function and metabolic processes (Ianiro et al., 2022), it is important to investigate their potential impact on metabolic health and liver diseases.
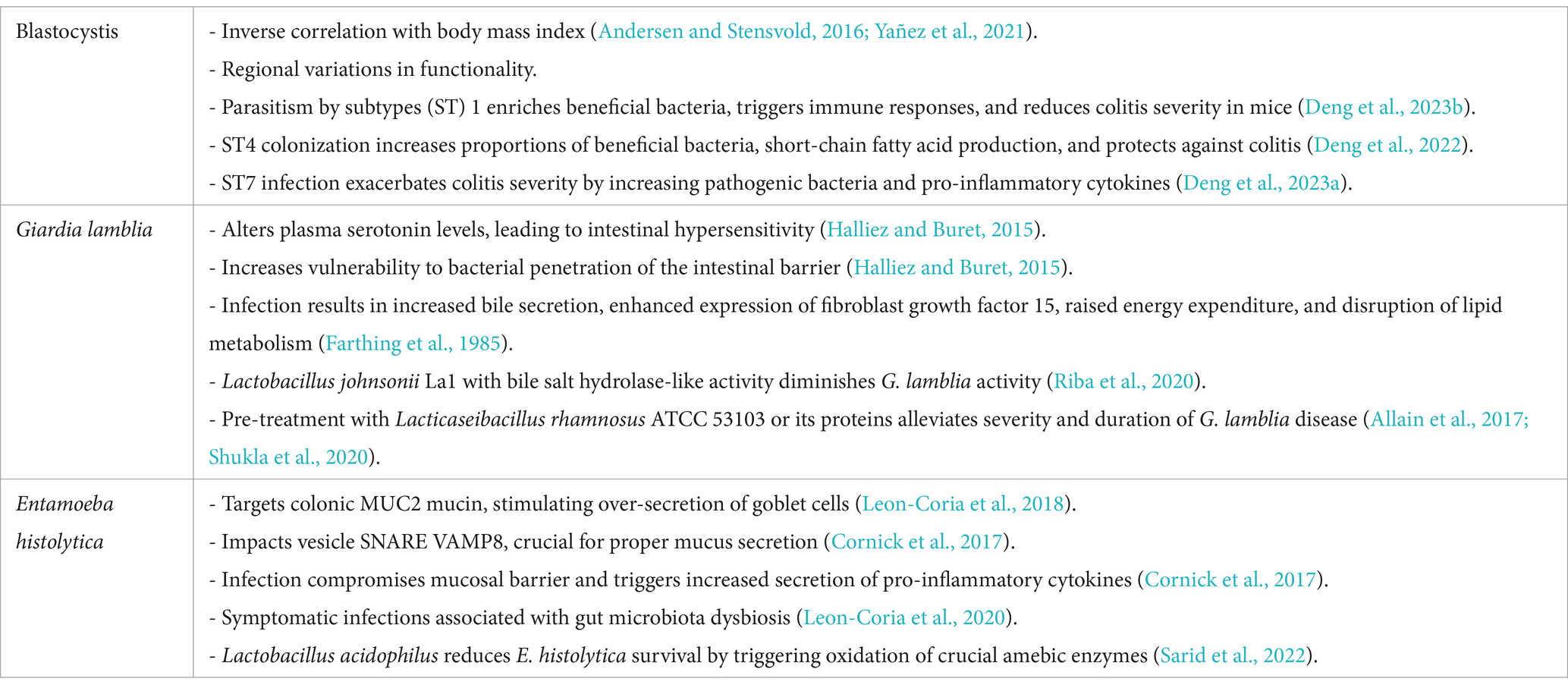
The most prevalent protozoans in the human gut are Blastocystis, Giardia lamblia, and Entamoeba histolytica (Chabé et al., 2017; Dagne and Alelign, 2021; von Huth et al., 2021). Therefore, we will explore the potential relationships between these protozoans and NAFLD in the following sections.
Blastocystis
Obesity and insulin resistance are closely linked to inflammation and oxidative stress originating from white adipose tissue (Caudet et al., 2022). These factors can lead to metabolic complications. Interestingly, the presence of Blastocystis, a common gut protozoan, has been inversely correlated with body mass index (BMI) (Andersen and Stensvold, 2016; Yañez et al., 2021).
The impact of Blastocystis on gut health appears to vary by subtype and region. For instance, studies in China found positive associations between various Blastocystis subtypes (STs) and beneficial bacteria like Akkermansia (Huang et al., 2024), while research in Belgium reported negative correlations between Blastocystis ST3 and ST4 and Akkermansia (Tito et al., 2019). In Singapore, infection with Blastocystis ST1 enriched beneficial bacteria such as Alloprevotella and Akkermansia, induced immune responses involving Th2 and Treg cells, and reduced the severity of DSS-induced colitis in mice (Deng et al., 2023b). Fecal microbiota transplantation from these mice conferred protection by inducing Treg cells and increasing SCFA production (Deng et al., 2023b). Similarly, ST4 colonization increased beneficial bacteria like Clostridia vadinBB60 and Lachnospiraceae NK4A136. It enhanced SCFA production and upregulated the proportions of Foxp3+ and IL-10-producing CD4+ T cells. These changes contribute to anti-inflammatory responses and provide protection against DSS-induced colitis through Th2 responses and increased production of anti-inflammatory cytokine IL-10 (Deng et al., 2022). In contrast, infection with ST7 worsened colitis by increasing pathogenic bacteria and pro-inflammatory cytokines IL-17A and TNF-α in CD4+ T cells (Deng et al., 2023a). ST7 also reduced levels of Bifidobacterium longum and lactobacilli, which are important for maintaining gut barrier function (Yason et al., 2019). Additionally, it disrupts intestinal barrier integrity by degrading tight junction proteins such as occludin and ZO-1. This degradation allows more pathogens and toxins to cross the intestinal barrier, further intensifying the inflammatory response. Moreover, Blastocystis ST7 infection can activate Th17 and Th1 cells, leading to an increased release of pro-inflammatory cytokines, including IL-17A and TNF-α (Yason et al., 2019; Deng et al., 2023a).
Blastocystis subtypes exhibit distinct genomic characteristics, with significant differences in amino acid sequences, particularly in genes related to protein kinases and proteases, which may account for variations in their pathogenic potential (Gentekaki et al., 2017). Interestingly, the same Blastocystis subtype may interact differently with the gut microbiota in different regions, possibly due to adaptation through horizontal gene transfer from prokaryotic and eukaryotic sources. This gene acquisition allows Blastocystis to adapt to the gut environment by acquiring new functional genes involved in crucial metabolic pathways. These include carbohydrate utilization, anaerobic amino acid and nitrogen metabolism, oxidative stress resistance, and pH regulation (Eme et al., 2017). These acquired genes may influence the composition of the gut microbiota and its inflammatory status, highlighting the parasite’s adaptive nature and its complex interplay with the gut environment. Given these effects on gut microbiota composition, immune responses, and inflammation, Blastocystis may play a role in modulating metabolic disorders like NAFLD. However, further research is needed to elucidate these connections.
Giardia lamblia
Giardia lamblia infection affects host health by altering plasma serotonin levels, leading to intestinal hypersensitivity and increased susceptibility to bacterial translocation across the intestinal barrier (Halliez and Buret, 2015). It also induces changes in the expression of genes related to immune responses, cell cycle regulation, and apoptosis in intestinal epithelial cells, further contributing to cellular damage. Specifically, immune-related genes such as CXCL1, CXCL2, CXCL3, CCL2, and CCL20 are upregulated, promoting inflammation. Cell cycle disruption involves G1/S phase arrest due to DNA damage and oxidative stress. Apoptosis-related genes like Bax are upregulated, Bcl-2 is downregulated, and caspases (Caspase-9 and Caspase-3) are activated, leading to mitochondrial apoptosis (Cotton et al., 2015).
Following Giardia lamblia infection, there is a significant decrease in the activities of sucrase, maltase, and lactase—the enzymes responsible for breaking down carbohydrates to release monosaccharides (Solaymani-Mohammadi and Singer, 2011). This impairment in carbohydrate digestion leads to malabsorption and nutrient deficiencies. Additionally, infected children often exhibit widespread amino acid deficiencies and elevated levels of specific phenolic acids, which are byproducts of bacterial amino acid metabolism (Halliez and Buret, 2015). Furthermore, G. lamblia utilizes bile salts for growth, and its infection can increase bile secretion. This alteration changes the gut microbiota composition, increases unconjugated bile acids, enhances the expression of fibroblast growth factor 15, raises host energy expenditure, disrupts lipid metabolism, and ultimately reduces adipose tissue (Farthing et al., 1985). Collectively, these effects demonstrate that G. lamblia infection significantly impairs nutrient absorption and metabolism, contributing to the host’s nutritional deficiencies and metabolic disturbances.
Certain probiotics can mitigate the effects of G. lamblia infection. Lactobacillus johnsonii La1, which possesses bile salt hydrolase activity regulated by the bsh47 and bsh56 genes, can reduce G. lamblia activity (Riba et al., 2020). Moreover, pre-treatment with inactivated Lacticaseibacillus rhamnosus ATCC 53103 or its proteins has shown potential in reducing the severity and duration of G. lamblia infection by modulating the gut microbiota and mucosal immunity. Supplementation with live bacteria is even more effective, as observed in mouse studies (Allain et al., 2017; Shukla et al., 2020). These findings suggest that G. lamblia infection can impact host metabolism and gut barrier function, which may have implications for metabolic disorders such as NAFLD.
Entamoeba histolytica
Intestinal mucus secretion is essential for maintaining the mucosal barrier and protecting epithelial cells from pathogens. Entamoeba histolytica targets colonic MUC2 mucin, stimulating over-secretion from goblet cells (Leon-Coria et al., 2018). The vesicle-associated membrane protein 8 (VAMP8), present on mucin granules in goblet cells, regulates mucus secretion during defense against E. histolytica. Activation of VAMP8 is crucial for proper mucus production; its absence impairs mucus secretion. In VAMP8-deficient animals, E. histolytica infection compromises the mucosal barrier and increases the secretion of pro-inflammatory cytokines IL-1α, IL-1β, and TNF-α (Cornick et al., 2017).
E. histolytica trophozoites typically exist as commensal organisms in the colons of most infected individuals, with 90% remaining asymptomatic (Nagaraja and Ankri, 2019). In germ-free animals, amoeba infection does not cause disease, but pathogenicity is restored in the presence of gut microbiota, indicating that E. histolytica’s pathogenicity is closely related to the gut microbiota (Chadee et al., 1987). Symptomatic infections are often associated with gut microbiota dysbiosis, which reduces CXCR2 expression on neutrophils, hindering their migration to the intestine and worsening amoebic colitis (Watanabe et al., 2017; Leon-Coria et al., 2020).
Certain bacteria and their metabolites can reduce the pathogenicity of E. histolytica. Supplementation with Clostridium scindens enhances granulocyte-monocyte progenitor cells (GMPs) in the gut, increasing neutrophil populations and strengthening the immune response against E. histolytica (Watanabe et al., 2017). Transplantation of bone marrow from mice colonized with C. scindens into uninfected mice also increases intestinal neutrophils and provides protection against amoebic colitis (Burgess et al., 2020). Deoxycholate, a metabolite produced by C. scindens through bile acid metabolism, similarly increases GMPs and offers immune protection (Burgess et al., 2020). While Enterobacteriaceae induce E. histolytica genes associated with oxidative stress survival, this effect is not observed when amoebas are co-cultured with probiotics (Leon-Coria et al., 2020). Lactobacillus acidophilus, a probiotic in the human gut, reduces E. histolytica survival by 50% after a 2-h co-culture by promoting the oxidation of essential amoebic enzymes like pyruvate oxidoreductase, Gal/GalNAc lectin, and cysteine proteases (Sarid et al., 2022). The 2-h co-culture period demonstrates that significant enzyme oxidation and a reduction in parasite viability can be observed within this timeframe. However, while these results underscore the potential efficacy of L. acidophilus in rapidly impacting E. histolytica survival, further studies with extended co-culture durations and more complex, gut-like conditions are needed to confirm its long-term effects and clinical relevance. These interactions suggest that alterations in gut microbiota can influence the pathogenicity of E. histolytica, which may have implications for intestinal health and metabolic disorders such as NAFLD (Table 4).
Future perspectives and research gaps
Despite significant advancements in elucidating the role of the gut microbiome in the pathogenesis of NAFLD, several research gaps persist that warrant further investigation. Firstly, while numerous studies have highlighted the impact of bacterial dysbiosis on NAFLD, the contributions of other microbial communities—including viruses (especially bacteriophages), fungi, and parasites—remain underexplored. Comprehensive multi-omics approaches are necessary to understand the complex interactions among these microorganisms and their collective influence on hepatic metabolism and inflammation.
Secondly, the causal relationships between specific microbial alterations and NAFLD progression are not yet firmly established. Most existing studies are cross-sectional, making it challenging to discern whether microbiota changes are a cause or consequence of NAFLD. Longitudinal cohort studies and well-designed interventional trials are essential to determine causality and to identify potential microbial biomarkers for early diagnosis and prognosis.
Thirdly, the mechanisms by which the gut microbiota modulate host metabolic pathways and immune responses in NAFLD are not fully elucidated. While pathways involving LPS-mediated activation of TLR4 and subsequent NF-κB signaling are recognized, other pathways—such as those involving SCFAs, bile acids, and microbial metabolites affecting regulatory T cells—require deeper exploration. Unraveling these mechanisms could reveal novel therapeutic targets and inform the development of microbiome-based interventions.
Moreover, individual variability in microbiota composition, influenced by genetics, diet, environment, and lifestyle factors, poses a challenge for translating findings into clinical practice. Personalized medicine approaches that account for these variables are needed to design effective prevention and treatment strategies.
Lastly, while therapeutic interventions like fecal microbiota transplantation (FMT), probiotics, and prebiotics show promise, their long-term efficacy and safety in NAFLD patients are not well-established. Rigorous clinical trials with standardized protocols are required to assess their therapeutic potential and to optimize dosing regimens.
In conclusion, addressing these research gaps through interdisciplinary and translational research will enhance our understanding of the gut-liver axis in NAFLD. Such efforts are crucial for the development of innovative microbiome-targeted therapies that could improve patient outcomes and reduce the global burden of NAFLD.
Summary
This review assimilates extensive research focusing on the intricate associations between gut microbiota and NAFLD. It highlights the potential therapeutic impact of certain microorganisms on NAFLD management. The dynamic interrelationships among bacteria, viruses, fungi, parasites, and the host, including the modulation of microbiota pathogenicity and virulence, are crucial factors to consider in devising comprehensive strategies to combat NAFLD. Understanding and harnessing the complex interactions within the gut microbiome will be instrumental in advancing our approach to address the challenges posed by NAFLD.
Author contributions
LL: Writing – original draft, Writing – review & editing. FC: Writing – original draft, Writing – review & editing. CG: Data curation, Writing – review & editing. ZL: Writing – review & editing. JQ: Data curation, Writing – review & editing. JH: Conceptualization, Methodology, Project administration, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abenavoli, L., Maurizi, V., Boccuto, L., Di Berardino, A., Giostra, N., Santori, P., et al. (2022). Nutritional support in acute liver failure. Diseases 10:108. doi: 10.3390/diseases10040108
Allain, T., Chaouch, S., Thomas, M., Vallée, I., Buret, A. G., Langella, P., et al. (2017). Bile-salt-hydrolases from the probiotic strain Lactobacillus johnsonii La1 mediate anti-giardial activity in vitro and in vivo. Front. Microbiol. 8:2707. doi: 10.3389/fmicb.2017.02707
Alrafaie, A. M., and Stafford, G. P. (2023). Enterococcal bacteriophage: a survey of the tail associated lysin landscape. Virus Res. 327:199073. doi: 10.1016/j.virusres.2023.199073
Andersen, L. O., and Stensvold, C. R. (2016). Blastocystis in health and disease: are we moving from a clinical to a public health perspective? J. Clin. Microbiol. 54, 524–528. doi: 10.1128/JCM.02520-15
Appleby, R. N., Moghul, I., Khan, S., Yee, M., Manousou, P., Neal, T. D., et al. (2019). Non-alcoholic fatty liver disease is associated with dysregulated bile acid synthesis and diarrhea: a prospective observational study. PLoS One 14:e0211348. doi: 10.1371/journal.pone.0211348
Aron-Wisnewsky, J., Vigliotti, C., Witjes, J., Le, P., Holleboom, A. G., Verheij, J., et al. (2020). Gut microbiota and human NAFLD: disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 17, 279–297. doi: 10.1038/s41575-020-0269-9
Bach, J.-F. (2018). The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat. Rev. Immunol. 18, 105–120. doi: 10.1038/nri.2017.111
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Bahmani, P., Maleki, A., Sadeghi, S., Shahmoradi, B., and Ghahremani, E. (2017). Prevalence of intestinal Protozoa infections and associated risk factors among schoolchildren in Sanandaj City, Iran. J Parasitol 12, 108–116
Baron, S. ed. (1996). Medical Microbiology., 4th ed. Galveston, TX: University of Texas Medical Branch at Galveston. Available at: http://www.ncbi.nlm.nih.gov/books/NBK7627/ (Accessed November 9, 2024).
Basaranoglu, M., Basaranoglu, G., and Bugianesi, E. (2015). Carbohydrate intake and nonalcoholic fatty liver disease: fructose as a weapon of mass destruction. Hepatobil. Surg. Nutr. 4, 109–116. doi: 10.3978/j.issn.2304-3881.2014.11.05
Belzer, C., and de Vos, W. M. (2012). Microbes inside--from diversity to function: the case of Akkermansia. ISME J. 6, 1449–1458. doi: 10.1038/ismej.2012.6
Boulangé, C. L., Neves, A. L., Chilloux, J., Nicholson, J. K., and Dumas, M.-E. (2016). Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 8:42. doi: 10.1186/s13073-016-0303-2
Boursier, J., Mueller, O., Barret, M., Machado, M., Fizanne, L., Araujo-Perez, F., et al. (2016). The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 63, 764–775. doi: 10.1002/hep.28356
Breuninger, T. A., Wawro, N., Breuninger, J., Reitmeier, S., Clavel, T., Six-Merker, J., et al. (2021). Associations between habitual diet, metabolic disease, and the gut microbiota using latent Dirichlet allocation. Microbiome 9:61. doi: 10.1186/s40168-020-00969-9
Brown, G. D., and Gordon, S. (2001). Immune recognition. A new receptor for beta-glucans. Nature 413, 36–37. doi: 10.1038/35092620
Burgess, S. L., Leslie, J. L., Uddin, J., Oakland, D. N., Gilchrist, C., Moreau, G. B., et al. (2020). Gut microbiome communication with bone marrow regulates susceptibility to amebiasis. J. Clin. Invest. 130, 4019–4024. doi: 10.1172/JCI133605
Cai, D., Yuan, M., Frantz, D. F., Melendez, P. A., Hansen, L., Lee, J., et al. (2005). Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat. Med. 11, 183–190. doi: 10.1038/nm1166
Cao, Z., Sugimura, N., Burgermeister, E., Ebert, M. P., Zuo, T., and Lan, P. (2022). The gut virome: a new microbiome component in health and disease. EBioMedicine 81:104113. doi: 10.1016/j.ebiom.2022.104113
Carey, J. N., Mettert, E. L., Fishman-Engel, D. R., Roggiani, M., Kiley, P. J., and Goulian, M. (2019). Phage integration alters the respiratory strategy of its host. eLife 8:e49081. doi: 10.7554/eLife.49081
Castro-Rodríguez, D. C., Juárez-Pilares, G., Cano-Cano, L., Pérez-Sánchez, M., Ibáñez, C. A., Reyes-Castro, L. A., et al. (2020). Impact of Leuconostoc SD23 intake in obese pregnant rats: benefits for maternal metabolism. J. Dev. Orig. Health Dis. 11, 533–539. doi: 10.1017/S2040174420000367
Caudet, J., Trelis, M., Cifre, S., Tapia, G., Soriano, J. M., Rodrigo, R., et al. (2022). Do intestinal unicellular parasites have a role in the inflammatory and redox status among the severely obese? Antioxidants 11:2090. doi: 10.3390/antiox11112090
Chabé, M., Lokmer, A., and Ségurel, L. (2017). Gut Protozoa: friends or foes of the human gut microbiota? Trends Parasitol. 33, 925–934. doi: 10.1016/j.pt.2017.08.005
Chadee, K., Petri, W. A., Innes, D. J., and Ravdin, J. I. (1987). Rat and human colonic mucins bind to and inhibit adherence lectin of Entamoeba histolytica. J. Clin. Invest. 80, 1245–1254. doi: 10.1172/JCI113199
Chang, P. V., Hao, L., Offermanns, S., and Medzhitov, R. (2014). The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 111, 2247–2252. doi: 10.1073/pnas.1322269111
Collins, S. L., Stine, J. G., Bisanz, J. E., Okafor, C. D., and Patterson, A. D. (2023). Bile acids and the gut microbiota: metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 21, 236–247. doi: 10.1038/s41579-022-00805-x
Cornick, S., Moreau, F., Gaisano, H. Y., and Chadee, K. (2017). Entamoeba histolytica-induced mucin exocytosis is mediated by VAMP8 and is critical in mucosal innate host defense. mBio 8:e01323-17. doi: 10.1128/mBio.01323-17
Cotton, J. A., Amat, C. B., and Buret, A. G. (2015). Disruptions of host immunity and inflammation by Giardia Duodenalis: potential consequences for co-infections in the gastro-intestinal tract. Pathogens 4, 764–792. doi: 10.3390/pathogens4040764
Coyte, K. Z., and Rakoff-Nahoum, S. (2019). Understanding competition and cooperation within the mammalian gut microbiome. Curr. Biol. 29, R538–R544. doi: 10.1016/j.cub.2019.04.017
d’Enfert, C., Kaune, A.-K., Alaban, L.-R., Chakraborty, S., Cole, N., Delavy, M., et al. (2021). The impact of the fungus-host-microbiota interplay upon Candida albicans infections: current knowledge and new perspectives. FEMS Microbiol. Rev. 45:fuaa060. doi: 10.1093/femsre/fuaa060
Dabke, K., Hendrick, G., and Devkota, S. (2019). The gut microbiome and metabolic syndrome. J. Clin. Invest. 129, 4050–4057. doi: 10.1172/JCI129194
Dagne, N., and Alelign, A. (2021). Prevalence of intestinal protozoan parasites and associated risk factors among school children in Merhabete District, Central Ethiopia. J. Parasitol. Res. 2021:9916456. doi: 10.1155/2021/9916456
Dai, X., Hou, H., Zhang, W., Liu, T., Li, Y., Wang, S., et al. (2020). Microbial metabolites: critical regulators in NAFLD. Front. Microbiol. 11:567654. doi: 10.3389/fmicb.2020.567654
De Filippo, C., Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 107, 14691–14696. doi: 10.1073/pnas.1005963107
Deleu, S., Machiels, K., Raes, J., Verbeke, K., and Vermeire, S. (2021). Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? EBioMedicine 66:103293. doi: 10.1016/j.ebiom.2021.103293
Demir, M., Lang, S., Hartmann, P., Duan, Y., Martin, A., Miyamoto, Y., et al. (2022). The fecal mycobiome in non-alcoholic fatty liver disease. J. Hepatol. 76, 788–799. doi: 10.1016/j.jhep.2021.11.029
Deng, L., Wojciech, L., Png, C. W., Kioh, D. Y. Q., Gu, Y., Aung, T. T., et al. (2023a). Colonization with two different Blastocystis subtypes in DSS-induced colitis mice is associated with strikingly different microbiome and pathological features. Theranostics 13, 1165–1179. doi: 10.7150/thno.81583
Deng, L., Wojciech, L., Png, C. W., Kioh, Y. Q. D., Ng, G. C., Chan, E. C. Y., et al. (2023b). Colonization with ubiquitous protist Blastocystis ST1 ameliorates DSS-induced colitis and promotes beneficial microbiota and immune outcomes. NPJ Biof. Microb. 9:22. doi: 10.1038/s41522-023-00389-1
Deng, L., Wojciech, L., Png, C. W., Koh, E. Y., Aung, T. T., Kioh, D. Y. Q., et al. (2022). Experimental colonization with Blastocystis ST4 is associated with protective immune responses and modulation of gut microbiome in a DSS-induced colitis mouse model. Cell. Mol. Life Sci. 79:245. doi: 10.1007/s00018-022-04271-9
Dillon, S., Agrawal, S., Banerjee, K., Letterio, J., Denning, T. L., Oswald-Richter, K., et al. (2006). Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J. Clin. Invest. 116, 916–928. doi: 10.1172/JCI27203
Dubik, M., Pilecki, B., and Moeller, J. B. (2022). Commensal intestinal Protozoa-underestimated members of the gut microbial community. Biology 11:1742. doi: 10.3390/biology11121742
Dunstan, R. A., Bamert, R. S., Belousoff, M. J., Short, F. L., Barlow, C. K., Pickard, D. J., et al. (2021). Mechanistic insights into the capsule-targeting Depolymerase from a Klebsiella pneumoniae bacteriophage. Microbiol. Spectr. 9:e0102321. doi: 10.1128/Spectrum.01023-21
Elsayed, A., Ismaiel, A., Procopio, A. C., Luzza, F., Abenavoli, L., and Dumitrascu, D. L. (2022). Noninvasive biochemical markers and surrogate scores in evaluating nonalcoholic steatohepatitis. Minerva Med. 113, 864–874. doi: 10.23736/S0026-4806.22.08185-X
Eme, L., Gentekaki, E., Curtis, B., Archibald, J. M., and Roger, A. J. (2017). Lateral gene transfer in the adaptation of the anaerobic parasite Blastocystis to the gut. Curr. Biol. 27, 807–820. doi: 10.1016/j.cub.2017.02.003
Eslam, M., Sanyal, A. J., and George, J.International Consensus Panel (2020). MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology 158, 1999–2014.e1. doi: 10.1053/j.gastro.2019.11.312
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 110, 9066–9071. doi: 10.1073/pnas.1219451110
Fagundes, R. R., Belt, S. C., Bakker, B. M., Dijkstra, G., Harmsen, H. J. M., and Faber, K. N. (2024). Beyond butyrate: microbial fiber metabolism supporting colonic epithelial homeostasis. Trends Microbiol. 32, 178–189. doi: 10.1016/j.tim.2023.07.014
Fang, J., Yu, C.-H., Li, X.-J., Yao, J.-M., Fang, Z.-Y., Yoon, S.-H., et al. (2022). Gut dysbiosis in nonalcoholic fatty liver disease: pathogenesis, diagnosis, and therapeutic implications. Front. Cell. Infect. Microbiol. 12:997018. doi: 10.3389/fcimb.2022.997018
Farthing, M. J., Keusch, G. T., and Carey, M. C. (1985). Effects of bile and bile salts on growth and membrane lipid uptake by Giardia lamblia. Possible implications for pathogenesis of intestinal disease. J. Clin. Invest. 76, 1727–1732. doi: 10.1172/JCI112162
Fay, K. T., Ford, M. L., and Coopersmith, C. M. (2017). The intestinal microenvironment in sepsis. Biochim. Biophys. Acta Mol. basis Dis. 1863, 2574–2583. doi: 10.1016/j.bbadis.2017.03.005
Feingold, K. R., Hardardottir, I., Memon, R., Krul, E. J., Moser, A. H., Taylor, J. M., et al. (1993). Effect of endotoxin on cholesterol biosynthesis and distribution in serum lipoproteins in Syrian hamsters. J. Lipid Res. 34, 2147–2158. doi: 10.1016/S0022-2275(20)35355-4
Feingold, K. R., Memon, R. A., Moser, A. H., Shigenaga, J. K., and Grunfeld, C. (1999). Endotoxin and interleukin-1 decrease hepatic lipase mRNA levels. Atherosclerosis 142, 379–387. doi: 10.1016/s0021-9150(98)00265-2
Filik, K., Szermer-Olearnik, B., Oleksy, S., Brykała, J., and Brzozowska, E. (2022). Bacteriophage tail proteins as a tool for bacterial pathogen recognition-a literature review. Antibiotics 11:555. doi: 10.3390/antibiotics11050555
Fischetti, V. A. (2008). Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11, 393–400. doi: 10.1016/j.mib.2008.09.012
Forlano, R., Mullish, B. H., Roberts, L. A., Thursz, M. R., and Manousou, P. (2022). The intestinal barrier and its dysfunction in patients with metabolic diseases and non-alcoholic fatty liver disease. Int. J. Mol. Sci. 23:662. doi: 10.3390/ijms23020662
Gantner, B. N., Simmons, R. M., and Underhill, D. M. (2005). Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24, 1277–1286. doi: 10.1038/sj.emboj.7600594
Gentekaki, E., Curtis, B. A., Stairs, C. W., Klimeš, V., Eliáš, M., Salas-Leiva, D. E., et al. (2017). Extreme genome diversity in the hyper-prevalent parasitic eukaryote Blastocystis. PLoS Biol. 15:e2003769. doi: 10.1371/journal.pbio.2003769
Ghabrial, S. A., Castón, J. R., Jiang, D., Nibert, M. L., and Suzuki, N. (2015). 50-plus years of fungal viruses. Virology 479-480, 356–368. doi: 10.1016/j.virol.2015.02.034
Gottlieb, A., and Canbay, A. (2019). Why bile acids are So important in non-alcoholic fatty liver disease (NAFLD) progression. Cells 8:1358. doi: 10.3390/cells8111358
Hagbom, M., De Faria, F. M., Winberg, M. E., Westerberg, S., Nordgren, J., Sharma, S., et al. (2020). Neurotrophic factors protect the intestinal barrier from rotavirus insult in mice. mBio 11:e02834-19. doi: 10.1128/mBio.02834-19
Halliez, M. C. M., and Buret, A. G. (2015). Gastrointestinal parasites and the neural control of gut functions. Front. Cell. Neurosci. 9:452. doi: 10.3389/fncel.2015.00452
He, C., Cheng, D., Peng, C., Li, Y., Zhu, Y., and Lu, N. (2018). High-fat diet induces Dysbiosis of gastric microbiota prior to gut microbiota in association with metabolic disorders in mice. Front. Microbiol. 9:639. doi: 10.3389/fmicb.2018.00639
Howard-Varona, C., Hargreaves, K. R., Abedon, S. T., and Sullivan, M. B. (2017). Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11, 1511–1520. doi: 10.1038/ismej.2017.16
Hrncir, T., Hrncirova, L., Kverka, M., Hromadka, R., Machova, V., Trckova, E., et al. (2021). Gut microbiota and NAFLD: Pathogenetic mechanisms, microbiota signatures, and therapeutic interventions. Microorganisms 9:957. doi: 10.3390/microorganisms9050957
Hsu, C. L., Duan, Y., Fouts, D. E., and Schnabl, B. (2021). Intestinal virome and therapeutic potential of bacteriophages in liver disease. J. Hepatol. 75, 1465–1475. doi: 10.1016/j.jhep.2021.08.003
Huang, L.-S., Yeh, Y.-M., Chiu, S.-F., Huang, P.-J., Chu, L. J., Huang, C.-Y., et al. (2024). Intestinal microbiota analysis of different Blastocystis subtypes and Blastocystis-negative individuals in Taiwan. Biom. J. 47:100661. doi: 10.1016/j.bj.2023.100661
Ianiro, G., Iorio, A., Porcari, S., Masucci, L., Sanguinetti, M., Perno, C. F., et al. (2022). How the gut parasitome affects human health. Ther. Adv. Gastroenterol. 15:17562848221091524. doi: 10.1177/17562848221091524
Iliev, I. D., and Cadwell, K. (2021). Effects of intestinal Fungi and viruses on immune responses and inflammatory bowel diseases. Gastroenterology 160, 1050–1066. doi: 10.1053/j.gastro.2020.06.100
Jasirwan, C. O. M., Muradi, A., Hasan, I., Simadibrata, M., and Rinaldi, I. (2021). Correlation of gut Firmicutes/Bacteroidetes ratio with fibrosis and steatosis stratified by body mass index in patients with non-alcoholic fatty liver disease. Biosci. Microb. Food Health 40, 50–58. doi: 10.12938/bmfh.2020-046
Khan, A., Ding, Z., Ishaq, M., Bacha, A. S., Khan, I., Hanif, A., et al. (2021). Understanding the effects of gut microbiota dysbiosis on nonalcoholic fatty liver disease and the possible probiotics role: recent updates. Int. J. Biol. Sci. 17, 818–833. doi: 10.7150/ijbs.56214
Kim, C. H. (2021). Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell. Mol. Immunol. 18, 1161–1171. doi: 10.1038/s41423-020-00625-0
Kimura, I., Inoue, D., Hirano, K., and Tsujimoto, G. (2014). The SCFA receptor GPR43 and energy metabolism. Front. Endocrinol. 5:85. doi: 10.3389/fendo.2014.00085
Kneeman, J. M., Misdraji, J., and Corey, K. E. (2012). Secondary causes of nonalcoholic fatty liver disease. Ther. Adv. Gastroenterol. 5, 199–207. doi: 10.1177/1756283X11430859
Krüger, W., Vielreicher, S., Kapitan, M., Jacobsen, I. D., and Niemiec, M. J. (2019). Fungal-bacterial interactions in health and disease. Pathogens 8:70. doi: 10.3390/pathogens8020070
Kwon, H.-K., Lee, C.-G., So, J.-S., Chae, C.-S., Hwang, J.-S., Sahoo, A., et al. (2010). Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. Sci. USA 107, 2159–2164. doi: 10.1073/pnas.0904055107
Lang, S., Demir, M., Martin, A., Jiang, L., Zhang, X., Duan, Y., et al. (2020). Intestinal Virome signature associated with severity of nonalcoholic fatty liver disease. Gastroenterology 159, 1839–1852. doi: 10.1053/j.gastro.2020.07.005
Lee, M.-F., Wang, N. M., Chu, Y.-W., Wu, C.-S., and Lin, W.-W. (2024). The anti-inflammatory effect of Lactococcus lactis-Ling-Zhi 8 on ameliorating atherosclerosis and nonalcoholic fatty liver in high-fat diet rabbits. Int. J. Mol. Sci. 25:11278. doi: 10.3390/ijms252011278
Leon-Coria, A., Kumar, M., and Chadee, K. (2020). The delicate balance between Entamoeba histolytica, mucus and microbiota. Gut Microbes 11, 118–125. doi: 10.1080/19490976.2019.1614363
Leon-Coria, A., Kumar, M., Moreau, F., and Chadee, K. (2018). Defining cooperative roles for colonic microbiota and Muc2 mucin in mediating innate host defense against Entamoeba histolytica. PLoS Pathog. 14:e1007466. doi: 10.1371/journal.ppat.1007466
Lian, C.-Y., Zhai, Z.-Z., Li, Z.-F., and Wang, L. (2020). High fat diet-triggered non-alcoholic fatty liver disease: a review of proposed mechanisms. Chem. Biol. Interact. 330:109199. doi: 10.1016/j.cbi.2020.109199
Liang, G., and Bushman, F. D. (2021). The human virome: assembly, composition and host interactions. Nat. Rev. Microbiol. 19, 514–527. doi: 10.1038/s41579-021-00536-5
Lin, D. M., and Lin, H. C. (2019). A theoretical model of temperate phages as mediators of gut microbiome dysbiosis. F1000Res 8:F1000 Faculty Rev-997. doi: 10.12688/f1000research.18480.1
Liu, L., Fu, Q., Li, T., Shao, K., Zhu, X., Cong, Y., et al. (2022). Gut microbiota and butyrate contribute to nonalcoholic fatty liver disease in premenopause due to estrogen deficiency. PLoS One 17:e0262855. doi: 10.1371/journal.pone.0262855
Liu, L., Yin, M., Gao, J., Yu, C., Lin, J., Wu, A., et al. (2023). Intestinal barrier function in the pathogenesis of nonalcoholic fatty liver disease. J. Clin. Transl. Hepatol. 000–458. doi: 10.14218/JCTH.2022.00089
Loke, P., and Harris, N. L. (2023). Networking between helminths, microbes, and mammals. Cell Host Microbe 31, 464–471. doi: 10.1016/j.chom.2023.02.008
Macias-Paz, I. U., Pérez-Hernández, S., Tavera-Tapia, A., Luna-Arias, J. P., Guerra-Cárdenas, J. E., and Reyna-Beltrán, E. (2023). Candida albicans the main opportunistic pathogenic fungus in humans. Rev. Argent. Microbiol. 55, 189–198. doi: 10.1016/j.ram.2022.08.003
Mandrekar, P., and Szabo, G. (2009). Signalling pathways in alcohol-induced liver inflammation. J. Hepatol. 50, 1258–1266. doi: 10.1016/j.jhep.2009.03.007
Manrique, P., Dills, M., and Young, M. J. (2017). The human gut phage community and its implications for health and disease. Viruses 9:141. doi: 10.3390/v9060141
Mbaye, B., Borentain, P., Magdy Wasfy, R., Alou, M. T., Armstrong, N., Mottola, G., et al. (2022). Endogenous ethanol and triglyceride production by gut Pichia kudriavzevii, Candida albicans and Candida glabrata yeasts in non-alcoholic steatohepatitis. Cells 11:3390. doi: 10.3390/cells11213390
Memaj, P., and Jornayvaz, F. R. (2022). Non-alcoholic fatty liver disease in type 1 diabetes: prevalence and pathophysiology. Front. Endocrinol. 13:1031633. doi: 10.3389/fendo.2022.1031633
Mendes, K. L., Lelis, D. F., de Freitas, D. F., da Silveira, L. H., de Paula, A. M. B., Guimarães, A. L. S., et al. (2021). Acute oral treatment with resveratrol and Lactococcus lactis Subsp. Lactis decrease body weight and improve liver proinflammatory markers in C57BL/6 mice. Mol. Biol. Rep. 48, 1725–1734. doi: 10.1007/s11033-021-06190-7
Mims, T. S., Abdallah, Q. A., Stewart, J. D., Watts, S. P., White, C. T., Rousselle, T. V., et al. (2021). The gut mycobiome of healthy mice is shaped by the environment and correlates with metabolic outcomes in response to diet. Commun. Biol. 4:281. doi: 10.1038/s42003-021-01820-z
Miquel, S., Martín, R., Rossi, O., Bermúdez-Humarán, L. G., Chatel, J. M., Sokol, H., et al. (2013). Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 16, 255–261. doi: 10.1016/j.mib.2013.06.003
Mirmiran, P., Amirhamidi, Z., Ejtahed, H.-S., Bahadoran, Z., and Azizi, F. (2017). Relationship between diet and non-alcoholic fatty liver disease: a review article. Iran. J. Public Health 46, 1007–1017
Mouzaki, M., Wang, A. Y., Bandsma, R., Comelli, E. M., Arendt, B. M., Zhang, L., et al. (2016). Bile acids and Dysbiosis in non-alcoholic fatty liver disease. PLoS One 11:e0151829. doi: 10.1371/journal.pone.0151829
Munukka, E., Pekkala, S., Wiklund, P., Rasool, O., Borra, R., Kong, L., et al. (2014). Gut-adipose tissue axis in hepatic fat accumulation in humans. J. Hepatol. 61, 132–138. doi: 10.1016/j.jhep.2014.02.020
Nagaraja, S., and Ankri, S. (2019). Target identification and intervention strategies against amebiasis. Drug Resist. Updat. 44, 1–14. doi: 10.1016/j.drup.2019.04.003
Naka, S., Nomura, R., Takashima, Y., Okawa, R., Ooshima, T., and Nakano, K. (2014). A specific Streptococcus mutans strain aggravates non-alcoholic fatty liver disease. Oral Dis. 20, 700–706. doi: 10.1111/odi.12191
Nakajima, H., Watanabe, T., Miyazaki, T., Takeuchi, M., Honda, Y., Shimada, N., et al. (2012). Acute liver dysfunction in the course of norovirus gastroenteritis. Case Rep. Gastroenterol. 6, 69–73. doi: 10.1159/000336202
Naudin, C. R., Maner-Smith, K., Owens, J. A., Wynn, G. M., Robinson, B. S., Matthews, J. D., et al. (2020). Lactococcus lactis subspecies cremoris elicits protection against metabolic changes induced by a Western-style diet. Gastroenterology 159, 639–651.e5. doi: 10.1053/j.gastro.2020.03.010
Neish, A. S. (2009). Microbes in gastrointestinal health and disease. Gastroenterology 136, 65–80. doi: 10.1053/j.gastro.2008.10.080
Netea, M. G., Gow, N. A. R., Munro, C. A., Bates, S., Collins, C., Ferwerda, G., et al. (2006). Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and toll-like receptors. J. Clin. Invest. 116, 1642–1650. doi: 10.1172/JCI27114
Netea, M. G., Van Der Graaf, C. A. A., Vonk, A. G., Verschueren, I., Van Der Meer, J. W. M., and Kullberg, B. J. (2002). The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J. Infect. Dis. 185, 1483–1489. doi: 10.1086/340511
Niu, C., Tu, Y., Jin, Q., Chen, Z., Yuan, K., Wang, M., et al. (2023). Mapping the human oral and gut fungal microbiota in patients with metabolic dysfunction-associated fatty liver disease. Front. Cell. Infect. Microbiol. 13:1157368. doi: 10.3389/fcimb.2023.1157368
Pane, J. A., Webster, N. L., and Coulson, B. S. (2014a). Rotavirus activates lymphocytes from non-obese diabetic mice by triggering toll-like receptor 7 signaling and interferon production in plasmacytoid dendritic cells. PLoS Pathog. 10:e1003998. doi: 10.1371/journal.ppat.1003998
Pane, J. A., Webster, N. L., Zufferey, C., and Coulson, B. S. (2014b). Rotavirus acceleration of murine type 1 diabetes is associated with increased MHC class I-restricted antigen presentation by B cells and elevated proinflammatory cytokine expression by T cells. Virus Res. 179, 73–84. doi: 10.1016/j.virusres.2013.11.009
Parada Venegas, D., De la Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/fimmu.2019.00277
Pasechnek, A., Rabinovich, L., Stadnyuk, O., Azulay, G., Mioduser, J., Argov, T., et al. (2020). Active Lysogeny in Listeria monocytogenes is a Bacteria-phage adaptive response in the mammalian environment. Cell Rep. 32:107956. doi: 10.1016/j.celrep.2020.107956
Pearson, J. A., Tai, N., Ekanayake-Alper, D. K., Peng, J., Hu, Y., Hager, K., et al. (2019). Norovirus changes susceptibility to type 1 diabetes by altering intestinal microbiota and immune cell functions. Front. Immunol. 10:2654. doi: 10.3389/fimmu.2019.02654
Peleg, A. Y., Hogan, D. A., and Mylonakis, E. (2010). Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 8, 340–349. doi: 10.1038/nrmicro2313
Pérez, J. C. (2021). Fungi of the human gut microbiota: roles and significance. Int. J. Med. Microbiol. 311:151490. doi: 10.1016/j.ijmm.2021.151490
Peroumal, D., Sahu, S. R., Kumari, P., Utkalaja, B. G., and Acharya, N. (2022). Commensal fungus Candida albicans maintains a long-term mutualistic relationship with the host to modulate gut microbiota and metabolism. Microbiol. Spectr. 10:e0246222. doi: 10.1128/spectrum.02462-22
Pickert, G., Neufert, C., Leppkes, M., Zheng, Y., Wittkopf, N., Warntjen, M., et al. (2009). STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206, 1465–1472. doi: 10.1084/jem.20082683
Pierce, E. C., Morin, M., Little, J. C., Liu, R. B., Tannous, J., Keller, N. P., et al. (2021). Bacterial-fungal interactions revealed by genome-wide analysis of bacterial mutant fitness. Nat. Microbiol. 6, 87–102. doi: 10.1038/s41564-020-00800-z
Plovier, H., Everard, A., Druart, C., Depommier, C., Van Hul, M., Geurts, L., et al. (2017). A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113. doi: 10.1038/nm.4236
Portincasa, P., Bonfrate, L., Vacca, M., De Angelis, M., Farella, I., Lanza, E., et al. (2022). Gut microbiota and Short chain fatty acids: implications in glucose homeostasis. Int. J. Mol. Sci. 23:1105. doi: 10.3390/ijms23031105
Quesada-Vázquez, S., Bone, C., Saha, S., Triguero, I., Colom-Pellicer, M., Aragonès, G., et al. (2022). Microbiota Dysbiosis and gut barrier dysfunction associated with non-alcoholic fatty liver disease are modulated by a specific metabolic cofactors’ combination. Int. J. Mol. Sci. 23:13675. doi: 10.3390/ijms232213675
Raeisi, H., Noori, M., Azimirad, M., Mohebbi, S. R., Asadzadeh Aghdaei, H., Yadegar, A., et al. (2023). Emerging applications of phage therapy and fecal virome transplantation for treatment of Clostridioides difficile infection: challenges and perspectives. Gut. Pathog. 15:21. doi: 10.1186/s13099-023-00550-3
Rasmussen, T. S., Mao, X., Forster, S., Larsen, S. B., Von Münchow, A., Tranæs, K. D., et al. (2024). Overcoming donor variability and risks associated with fecal microbiota transplants through bacteriophage-mediated treatments. Microbiome 12:119. doi: 10.1186/s40168-024-01820-1
Rasmussen, T. S., Mentzel, C. M. J., Kot, W., Castro-Mejía, J. L., Zuffa, S., Swann, J. R., et al. (2020). Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut 69, 2122–2130. doi: 10.1136/gutjnl-2019-320005
Rau, M., Rehman, A., Dittrich, M., Groen, A. K., Hermanns, H. M., Seyfried, F., et al. (2018). Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol J 6, 1496–1507. doi: 10.1177/2050640618804444
Riazi, K., Azhari, H., Charette, J. H., Underwood, F. E., King, J. A., Afshar, E. E., et al. (2022). The prevalence and incidence of NAFLD worldwide: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 7, 851–861. doi: 10.1016/S2468-1253(22)00165-0
Riba, A., Hassani, K., Walker, A., van Best, N., von Zezschwitz, D., Anslinger, T., et al. (2020). Disturbed gut microbiota and bile homeostasis in Giardia-infected mice contributes to metabolic dysregulation and growth impairment. Sci. Transl. Med. 12:eaay7019. doi: 10.1126/scitranslmed.aay7019
Rothschild-Rodriguez, D., Hedges, M., Kaplan, M., Karav, S., and Nobrega, F. L. (2022). Phage-encoded carbohydrate-interacting proteins in the human gut. Front. Microbiol. 13:1083208. doi: 10.3389/fmicb.2022.1083208
Round, J. L., and Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323. doi: 10.1038/nri2515
Sarid, L., Zanditenas, E., Ye, J., Trebicz-Geffen, M., and Ankri, S. (2022). Insights into the mechanisms of Lactobacillus acidophilus activity against Entamoeba histolytica by using thiol redox proteomics. Antioxidants 11:814. doi: 10.3390/antiox11050814
Sarkar, A., Mitra, P., Lahiri, A., Das, T., Sarkar, J., Paul, S., et al. (2023). Butyrate limits inflammatory macrophage niche in NASH. Cell Death Dis. 14:332. doi: 10.1038/s41419-023-05853-6
Sebaa, S., Behnke, J. M., Baroudi, D., Hakem, A., and Abu-Madi, M. A. (2021). Prevalence and risk factors of intestinal protozoan infection among symptomatic and asymptomatic populations in rural and urban areas of southern Algeria. BMC Infect. Dis. 21:888. doi: 10.1186/s12879-021-06615-5
Seelbinder, B., Chen, J., Brunke, S., Vazquez-Uribe, R., Santhaman, R., Meyer, A.-C., et al. (2020). Antibiotics create a shift from mutualism to competition in human gut communities with a longer-lasting impact on fungi than bacteria. Microbiome 8:133. doi: 10.1186/s40168-020-00899-6
Shi, H., Kokoeva, M. V., Inouye, K., Tzameli, I., Yin, H., and Flier, J. S. (2006). TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Invest. 116, 3015–3025. doi: 10.1172/JCI28898
Shiha, G., Korenjak, M., Eskridge, W., Casanovas, T., Velez-Moller, P., Högström, S., et al. (2021). Redefining fatty liver disease: an international patient perspective. Lancet Gastroenterol. Hepatol. 6, 73–79. doi: 10.1016/S2468-1253(20)30294-6
Shukla, G., Kamboj, S., and Sharma, B. (2020). Comparative analysis of Antigiardial potential of heat inactivated and probiotic protein of probiotic Lactobacillus rhamnosus GG in murine giardiasis. Probiot. Antimicrob. Proteins 12, 271–279. doi: 10.1007/s12602-018-9506-8
Silveira, C. B., and Rohwer, F. L. (2016). Piggyback-the-winner in host-associated microbial communities. NPJ Biof. Microb. 2:16010. doi: 10.1038/npjbiofilms.2016.10
Solaymani-Mohammadi, S., and Singer, S. M. (2011). Host immunity and pathogen strain contribute to intestinal disaccharidase impairment following gut infection. J. Immunol. 187, 3769–3775. doi: 10.4049/jimmunol.1100606
Spencer, L., Olawuni, B., and Singh, P. (2022). Gut Virome: role and distribution in health and gastrointestinal diseases. Front. Cell. Infect. Microbiol. 12:836706. doi: 10.3389/fcimb.2022.836706
Su, Z., and Tao, X. (2021). Current understanding of IL-37 in human health and disease. Front. Immunol. 12:696605. doi: 10.3389/fimmu.2021.696605
Sun, M., Wang, Q., Zhang, M., Zhang, G., Wu, T., Liu, R., et al. (2020). Leuconostoc pseudomesenteroides improves microbiota dysbiosis and liver metabolism imbalance and ameliorates the correlation between dihydroceramide and strains of Firmicutes and Proteobacteria in high fat diet obese mice. Food Funct. 11, 6855–6865. doi: 10.1039/d0fo01009j
Sutcliffe, S. G., Reyes, A., and Maurice, C. F. (2023). Bacteriophages playing nice: lysogenic bacteriophage replication stable in the human gut microbiota. iScience 26:106007. doi: 10.1016/j.isci.2023.106007
Sutton, T. D. S., and Hill, C. (2019). Gut bacteriophage: current understanding and challenges. Front. Endocrinol. 10:784. doi: 10.3389/fendo.2019.00784
Szóstak, N., Figlerowicz, M., and Philips, A. (2023). The emerging role of the gut mycobiome in liver diseases. Gut Microbes 15:2211922. doi: 10.1080/19490976.2023.2211922
Taslem Mourosi, J., Awe, A., Guo, W., Batra, H., Ganesh, H., Wu, X., et al. (2022). Understanding bacteriophage tail Fiber interaction with host surface receptor: the key “blueprint” for reprogramming phage host range. Int. J. Mol. Sci. 23:12146. doi: 10.3390/ijms232012146
Tito, R. Y., Chaffron, S., Caenepeel, C., Lima-Mendez, G., Wang, J., Vieira-Silva, S., et al. (2019). Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut 68, 1180–1189. doi: 10.1136/gutjnl-2018-316106
Tokuhara, D. (2021). Role of the gut microbiota in regulating non-alcoholic fatty liver disease in children and adolescents. Front. Nutr. 8:700058. doi: 10.3389/fnut.2021.700058
van Tilburg Bernardes, E., Pettersen, V. K., Gutierrez, M. W., Laforest-Lapointe, I., Jendzjowsky, N. G., Cavin, J.-B., et al. (2020). Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat. Commun. 11:2577. doi: 10.1038/s41467-020-16431-1
Visekruna, A., and Luu, M. (2021). The role of Short-chain fatty acids and bile acids in intestinal and liver function, inflammation, and carcinogenesis. Front. Cell Dev. Biol. 9:703218. doi: 10.3389/fcell.2021.703218
von Huth, S., Thingholm, L. B., Kofoed, P.-E., Bang, C., Rühlemann, M. C., Franke, A., et al. (2021). Intestinal protozoan infections shape fecal bacterial microbiota in children from Guinea-Bissau. PLoS Negl. Trop. Dis. 15:e0009232. doi: 10.1371/journal.pntd.0009232
Wang, Z., Li, Y., Liao, W., Huang, J., Liu, Y., Li, Z., et al. (2022). Gut microbiota remodeling: a promising therapeutic strategy to confront hyperuricemia and gout. Front. Cell. Infect. Microbiol. 12:935723. doi: 10.3389/fcimb.2022.935723
Wang, T., Liu, J., Luo, Y., Yu, B., Kong, X., Zheng, P., et al. (2023). Combined effects of host genetics and diet on porcine intestinal fungi and their pathogenic genes. Front. Microbiol. 14:1192288. doi: 10.3389/fmicb.2023.1192288
Wang, Q., Zhang, G., An, C., Hambly, B. D., and Bao, S. (2024). The role of IL-37 in gastrointestinal diseases. Front. Immunol. 15:1431495. doi: 10.3389/fimmu.2024.1431495
Wang, L., Zhang, K., Zeng, Y., Luo, Y., Peng, J., Zhang, J., et al. (2023). Gut mycobiome and metabolic diseases: the known, the unknown, and the future. Pharmacol. Res. 193:106807. doi: 10.1016/j.phrs.2023.106807
Watanabe, K., Gilchrist, C. A., Uddin, M. J., Burgess, S. L., Abhyankar, M. M., Moonah, S. N., et al. (2017). Microbiome-mediated neutrophil recruitment via CXCR2 and protection from amebic colitis. PLoS Pathog. 13:e1006513. doi: 10.1371/journal.ppat.1006513
Wu, J.-W., Wang, J.-T., Lin, T.-L., Liu, Y.-Z., Wu, L.-T., and Pan, Y.-J. (2023). Identification of three capsule depolymerases in a bacteriophage infecting Klebsiella pneumoniae capsular types K7, K20, and K27 and therapeutic application. J. Biomed. Sci. 30:31. doi: 10.1186/s12929-023-00928-0
Wu, X., Xia, Y., He, F., Zhu, C., and Ren, W. (2021). Intestinal mycobiota in health and diseases: from a disrupted equilibrium to clinical opportunities. Microbiome 9:60. doi: 10.1186/s40168-021-01024-x
Xie, C., and Halegoua-DeMarzio, D. (2019). Role of probiotics in non-alcoholic fatty liver disease: does gut microbiota matter? Nutrients 11:2837. doi: 10.3390/nu11112837
Xiong, J., Chen, X., Zhao, Z., Liao, Y., Zhou, T., and Xiang, Q. (2022). A potential link between plasma short-chain fatty acids, TNF-α level and disease progression in non-alcoholic fatty liver disease: a retrospective study. Exp. Ther. Med. 24:598. doi: 10.3892/etm.2022.11536
Yañez, C. M., Hernández, A. M., Sandoval, A. M., Domínguez, M. A. M., Muñiz, S. A. Z., and Gómez, J. O. G. (2021). Prevalence of Blastocystis and its association with Firmicutes/Bacteroidetes ratio in clinically healthy and metabolically ill subjects. BMC Microbiol. 21:339. doi: 10.1186/s12866-021-02402-z
Yao, Y., Cai, X., Fei, W., Ye, Y., Zhao, M., and Zheng, C. (2022). The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit. Rev. Food Sci. Nutr. 62, 1–12. doi: 10.1080/10408398.2020.1854675
Yason, J. A., Liang, Y. R., Png, C. W., Zhang, Y., and Tan, K. S. W. (2019). Interactions between a pathogenic Blastocystis subtype and gut microbiota: in vitro and in vivo studies. Microbiome 7:30. doi: 10.1186/s40168-019-0644-3
Yasutake, K., Kohjima, M., Kotoh, K., Nakashima, M., Nakamuta, M., and Enjoji, M. (2014). Dietary habits and behaviors associated with nonalcoholic fatty liver disease. World J. Gastroenterol. 20, 1756–1767. doi: 10.3748/wjg.v20.i7.1756
You, N., Xu, J., Wang, L., Zhuo, L., Zhou, J., Song, Y., et al. (2021). Fecal Fungi Dysbiosis in nonalcoholic fatty liver disease. Obesity 29, 350–358. doi: 10.1002/oby.23073
Young, R. (2014). Phage lysis: three steps, three choices, one outcome. J. Microbiol. 52, 243–258. doi: 10.1007/s12275-014-4087-z
Zhang, F., Aschenbrenner, D., Yoo, J. Y., and Zuo, T. (2022). The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe 3, e969–e983. doi: 10.1016/S2666-5247(22)00203-8
Zhang, Z., Zhang, H., Chen, T., Shi, L., Wang, D., and Tang, D. (2022). Regulatory role of short-chain fatty acids in inflammatory bowel disease. Cell Commun. Signal 20:64. doi: 10.1186/s12964-022-00869-5
Zheng, Y., Valdez, P. A., Danilenko, D. M., Hu, Y., Sa, S. M., Gong, Q., et al. (2008). Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289. doi: 10.1038/nm1720
Zhu, L., Baker, S. S., Gill, C., Liu, W., Alkhouri, R., Baker, R. D., et al. (2013). Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57, 601–609. doi: 10.1002/hep.26093
Zuppi, M., Hendrickson, H. L., O’Sullivan, J. M., and Vatanen, T. (2021). Phages in the gut ecosystem. Front. Cell. Infect. Microbiol. 11:822562. doi: 10.3389/fcimb.2021.822562
Keywords: NAFLD, gut microbiome, microbial therapeutics, gut virome, gut fungi, parasites
Citation: Li L, Cai F, Guo C, Liu Z, Qin J and Huang J (2024) Gut microbiome and NAFLD: impact and therapeutic potential. Front. Microbiol. 15:1500453. doi: 10.3389/fmicb.2024.1500453
Edited by:
Hyun-Seob Song, University of Nebraska-Lincoln, United StatesReviewed by:
Jeehwan Oh, University of Wisconsin-Madison, United StatesBhagavathi Sundaram Sivamaruthi, Chiang Mai University, Thailand
Anna Caterina Procopio, Magna Græcia University, Italy
Copyright © 2024 Li, Cai, Guo, Liu, Qin and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiean Huang, aGphZ3htdUAxNjMuY29t
 Liwei Li
Liwei Li Fuqing Cai
Fuqing Cai Jiean Huang
Jiean Huang