- 1School of Life Science and Technology, Wuhan Polytechnic University, Wuhan, China
- 2Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 3Eco-Environmental Monitoring and Research Center, Pearl River Valley and South China Sea Ecology and Environment Administration, Ministry of Ecology and Environment, Guangzhou, China
- 4Ecological Environment Science and Technology Center, Wuhan, China
Introduction: Anaerobic–anoxic–oxic (A2/O) and deoxidation ditch (DOD) processes are being increasingly preferred owing to their effectiveness in treating various wastes in wastewater treatment plants (WWTPs). Considering seasonal variations is crucial in optimizing treatment processes, ensuring compliance with regulations, and maintaining the overall efficiency and effectiveness of WWTPs. This study aimed to determine the influence of seasonality on nitrogen removing microbes and functional genes within A2/O and DOD processes in the humid Wuhan and semi-arid Xi’an cities, China.
Methods: The physicochemical parameters of water quality were determined, and molecular and bioinformatic analyses of the bacterial community and nitrogen metabolism functional genes in the two different treatment processes of two WWTPs were performed over four seasons.
Results and discussion: Our analyses revealed a significant difference in all physicochemical parameters across all experimental groups (p < 0.05). At the genus level, the abundance of Dokdonella, one unidentified genus of Nitrospiraceae, Terrimonas, and one unidentified genus of Chloroflexi was the highest in all groups. Generally, warmer seasons exhibited higher biodiversity indices. The A2/O system exhibited higher values in terms of most nitrogen metabolism functional genes than those of the DOD sewage treatment system. In both WWTPs, the abundance of most genes in spring and summer were higher than that of autumn and winter seasons. Taken together, changes in temperature, caused by seasonal changes, may contribute to changes in abundance of nitrogen metabolic functional genes.
1 Introduction
Several emerging techniques are applied in wastewater treatment plants (WWTPs) to efficiently treat different wastes. Anaerobic–anoxic–oxic (A2/O) and deoxidation ditch (DOD) processes have increasingly been preferred because of their efficiency in treating various wastewater within WWTPs (Das and Singh, 2022). A2/O, a wastewater treatment process, involves a sequence of treatment zones designed to eliminate organic matter and nutrients of nitrogen and phosphorus from wastewater (Moharram et al., 2017). The A2/O process is a cyclic operation, with the wastewater flowing sequentially through anaerobic, anoxic, and oxic zones. This configuration allows the efficient removal of organic matter, nitrogen, and phosphorus, making it an effective treatment method for wastewater before its discharge into receiving bodies of water or reuse in various applications (Gizińska-Górna and Wasąg, 2022; Moharram et al., 2017). Therefore, it is commonly used in biological nutrient removal processes to effectively reduce nitrogen and phosphorus concentrations in wastewater. DOD, another wastewater treatment method, represents a specific activated sludge (AS) process to remove organic matter and nutrients from wastewater (Shammas and Wang, 2009). It is commonly employed in municipal and industrial WWTPs. In the DOD process, wastewater flows into a circular or rectangular basin, known as the ditch, equipped with mechanical or diffused aerators. The wastewater is subjected to a combination of aerobic and anoxic conditions to facilitate the treatment process (Shammas and Wang, 2009; Stamou, 1997). A2/O and DOD processes affect microbial ecology due to variations in their operating conditions, treatment mechanisms, and the types of microorganisms involved (Feng et al., 2022; Park et al., 2021). Both processes have demonstrated efficiency in nitrogen removal, but their performance can vary significantly due to seasonal and geographic factors, which influence microbial activity and gene expression (Gizińska-Górna and Wasąg, 2022; Park et al., 2021; Shammas and Wang, 2009).
Seasonality can have a significant impact on WWTP performance. Moreover, seasonality can influence WWTP performance by affecting flow rates, organic load, nutrient balance, temperature conditions, algal blooms, and sludge handling (Thobejane et al., 2023). Understanding and adapting to these seasonal variations are crucial for optimizing treatment processes, ensuring compliance with regulations, and maintaining the overall efficiency and effectiveness of WWTP operations (Makuwa et al., 2022). Cold temperatures can significantly impact WWTP efficiency and performance. The performance and microbial conditions of a WWTP during autumn or winter can equally be influenced by various factors, including the characteristics of the influent wastewater, weather conditions, and seasonal variations in pollutant loadings (Dai et al., 2023; Guerrero et al., 2011). The performance and microbial conditions of WWTPs depend on various factors, including specific characteristics of the influent wastewater, the design and capacity of the treatment plant, and any seasonal variations in pollutant loadings (Bhatt et al., 2020; Bwapwa, 2022). These variations can be driven by regional and local factors, including climate, geographic location, and specific seasonal patterns. Monitoring and adapting treatment processes based on influent characteristics, flow rates, and other seasonal variations are essential to maintain optimal performance and meet regulatory standards (Guerrero et al., 2011; Liu et al., 2016).
Recent studies have emphasized the importance of nitrogen metabolism functional genes, including amoA (ammonia-oxidizing bacteria), nirS and nirK (denitrification genes), and nosZ (nitrous oxide reductase), in the biological nitrogen removal process (Arumugham et al., 2024; Galloway et al., 2008; Zheng et al., 2024). These genes regulate the nitrification and denitrification stages, which are sensitive to environmental factors like temperature and dissolved oxygen (DO) levels (Li et al., 2018; Potgieter et al., 2020; Zhang et al., 2019). Seasonal variations in nitrogen functional genes and nitrogen metabolism in WWTPs can occur due to changes in influent characteristics, temperature, hydraulic loading, and microbial community dynamics (Liu et al., 2016). These variations can be noticeable in nitrification and denitrification genes, nitrogen metabolism, temperature effects, organic loadings, and nutrient levels (Li et al., 2015). Specific dynamics of nitrogen functional genes and nitrogen metabolism depend on the design and operation of WWTPs, regional climate, and the specific characteristics of the influent wastewater. Regular monitoring of nitrogen-related parameters, including gene expression, nutrient concentrations, and treatment performance, can provide insights into seasonal variations and guide operational adjustments to optimize nitrogen removal efficiency throughout the year (Li et al., 2015; Zhang et al., 2019).
Despite the known influence of seasonality, few studies have conducted comprehensive comparisons of microbial dynamics and functional gene abundance across different WWTP processes in distinct climatic regions. Previous research has primarily focused on how operational parameters such as hydraulic retention time and sludge age affect nitrogen removal, but the response of A2/O and DOD systems to geographical and seasonal variations remains less explored (Song et al., 2021; Thobejane et al., 2023). Moreover, understanding how these factors impact the abundance of specific nitrogen metabolism functional genes can provide deeper insights into microbial activity and system performance under varying conditions.
Therefore, this study aimed to evaluate the differences in the abundance of nitrogen removal functional microbes based on high-throughput experimental results between different sewage treatment processes within A2/O and DOD processes in the humid Wuhan and semiarid Xi’an cities, China. The study focused on analyzing the process function differences and comparing and analyzing the functional genes of nitrogen removal microorganisms in different seasons of the two sewage treatment processes through quantitative polymerase chain reaction (qPCR) technology. The overall goal was to compare the effects of the treatment process and environmental conditions such as geographic and seasonal variations on the nitrogen removal performance of sewage treatment plants and provide the theoretical basis and implications for optimizing nitrogen removal-related processes. It was hypothesized that there is no variation in the levels of functional genes and nitrogen metabolism between treatment systems and across different seasons.
2 Materials and methods
2.1 Experimental design and sampling
This study was undertaken in two WWTPs marked as B and T. WWTP B in Xi’an City primarily accepts and treats production wastewater from industrial enterprises and domestic sewage from residential areas in the surrounding areas of the plant. It has a wastewater mixing ratio of ~3:7, a service area of 85 km2, a service population of 1.2 million people, and daily treatment volume of ~150,000 tons. The daily treatment capacity of DOD is ~100,000 tons and that of modified A2/O is ~50,000 tons; the sewage treatment follows the national level-A effluent standard. The main pollutants in the influent water of this WWTP are biological oxygen demand (BOD), suspended solids, chemical oxygen demand (COD), ammonia–nitrogen (NH4+–N), and other pollutants. The main industrial wastewater discharges in the basin are primarily electronics, pharmaceuticals, leather, coking, paper, etc. WWTP T in Wuhan primarily accepts and treats domestic wastewater and possible partial production wastewater from residents in the surrounding areas of the plant, with a service area of 43 km2, a service population of 330,000 people, and a small number of biopharmaceutical enterprises and electronic product processing plants in the service area. The plant’s current daily treatment capacity is 100,000 tons. DOD treats 40,000 tons and A2/O treats 60,000 tons daily; the wastewater treatment follows the national level-A effluent standard. The main operating parameters of the two WWTPs are shown in Supplementary Table S1. The experimental design had 16 samples categories: BA1 (WWTP B with A2/O process sampled in spring), BA2 (WWTP B with A2/O process sampled in summer), BA3 (WWTP B with A2/O process sampled in autumn), BA4 (WWTP B with A2/O process sampled in winter), BD1 (WWTP B with DOD process sampled in spring), BD2 (WWTP B with DOD process sampled in summer), BD3 (WWTP B with DOD process sampled in autumn), BD4 (WWTP B with DOD process sampled in winter), TA1 (WWTP T with A2/O process sampled in spring), TA2 (WWTP T with A2/O process sampled in summer), TA3 (WWTP T with A2/O process sampled in autumn), TA4 (WWTP T with A2/O process sampled in winter), TD1 (WWTP T with DOD process sampled in spring), TD2 (WWTP T with DOD process sampled in summer), TD3 (WWTP T with DOD process sampled in autumn), and TD4 (WWTP T with DOD process sampled in winter).
Based on the knowledge that the sample of the aeration tank in the biochemical section can wholly represent the entire sewage treatment plant, the slurry mixed with water at the outlet of the aeration tank was selected as the representative sample. To achieve that, a mixture of 500 mL of AS was uniformly collected in triplicate in a sterile sampling bottle. The influent sample from each sampling point was also collected similarly for water quality determination.
2.2 Measurement of physicochemical parameters
The collected AS and sewage samples were immediately transferred to the laboratory in an ice box and temporarily stored at 4°C. AS samples were centrifuged at 10,000 × g at 4°C for 15 min and frozen at −80°C for DNA extraction. The water quality indexes of the influent water samples were determined by national standards: total nitrogen (TN) was determined by alkaline potassium persulfate digestion ultraviolet spectrophotometry (HJ 636–2012), NH4+–N was determined by Knotler reagent spectrophotometry (HJ 535–2009), total phosphorus (TP) was determined by ammonium molybdate spectrophotometry (GB 11893–89), BOD5 was determined by dilution and inoculation (GB 7488–87), and COD was determined by rapid digestion spectrophotometry (HJ/T 399–2007).
2.3 Sample DNA extraction
For DNA extraction, 0.9 g of the collected AS sample was weighed, DNA was extracted using a DNA Isolation Kit (E.Z.N.A., Omega, Norcross, GA, United States) and DNA concentration and purity were determined. The entire procedure was performed according to the manufacturer’s instructions. The obtained DNA solution was analyzed by 1% agarose electrophoresis, and its concentration and purity were verified by a Nanodrop microspectrophotometer.
2.4 High-throughput sequencing and data processing
This experiment utilized the Ion Torrent IonS5™ XL high-throughput sequencing technology. The bacterial 16S rDNA V4 region of DNA samples which passed quality control was amplified using barcoded specific primers 515F/806R and Phusion® High-Fidelity PCR Master Mix with GC Buffer from New England Biolabs (Li et al., 2024). Sequencing data underwent initial quality control using Cutadapt version 1.9.1 to obtain raw reads, followed by chimera removal using Vsearch to yield clean reads (Rognes et al., 2016). All sequencing data for microbial communities of municipal WWTPs were submitted to the National Center of Biotechnology Information (NCBI) Sequence Read Archive under accession numbers PRJNA605862 and PRJNA606090.
During data processing, clean data were clustered into Operational Taxonomic Units (OTUs) with 97% identity using Uparse version 7.0 (Haas et al., 2011). OTU sequences were annotated using Mothur version 1.40 and the SILVA132 SSU rRNA database with a threshold of 0.8 ~ 1 to obtain taxonomic information and to analyze community composition at various taxonomic levels (Edgar, 2013). Multiple sequence alignment of all OTU sequences was performed using MUSCLE version 3.8 to establish their phylogenetic relationships (Quast et al., 2012).
2.5 Primer selection for denitrification-related genes
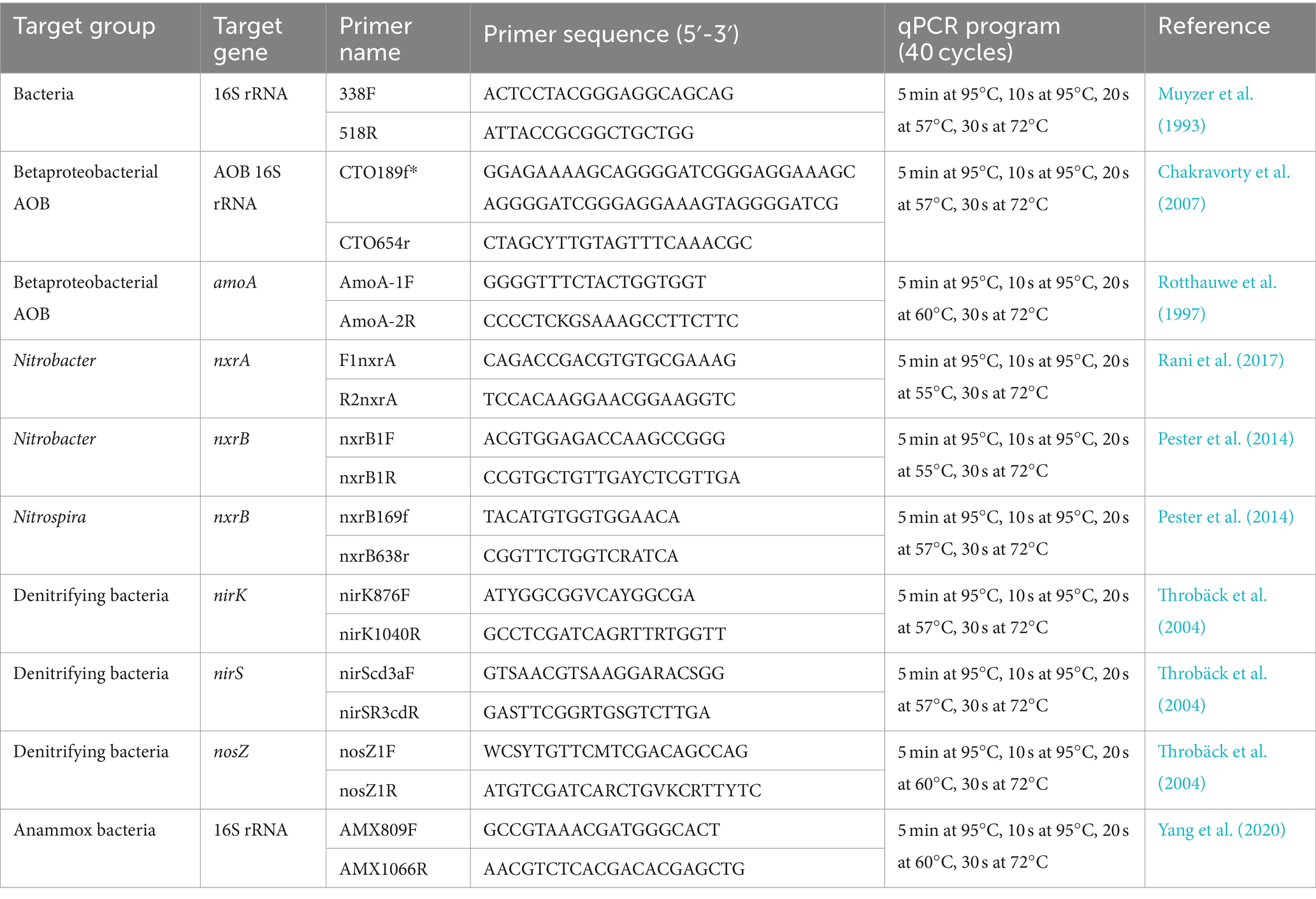
To study the difference in nitrogen removal performance between different samples, qPCR was performed for the relevant functional genes in the main nitrogen removal process. Ammonia-oxidizing bacteria (AOB) 16S rRNA genes, which cover a wider area, and amoA, which provides more functionally relevant information about nitrifying bacterial populations, were selected as primers for AOB studies. Owing to the sequence diversity of nitrite-oxidizing bacteria (NOB) and to fully reflect the abundance of the taxa in the sample, multiple primer pairs were used to analyze the characteristics of the taxa. These included nxrA and nxrB for Nitrobacter and nxrB for Nitrospira to distinguish specific nitrite-oxidizing microbes. For the comammox study, although relevant or combination primers were used to study the ecological abundance, their effect in pre-experiment was stable due to the source of primers or samples, so this part of the experiment was omitted. For denitrifying bacteria, nirS and nirK genes, with the same function but within a different niche, were selected to detect the abundance of nitrite-reducing bacteria. Primers targeting the nosZ gene were selected to measure the ability to perform complete nitrification and denitrification. The anammox 16S rRNA gene was selected as a primer to determine the abundance of anaerobic AOB. Information and references related to primers are detailed in Table 1.
2.6 Gene cloning and plasmid construction and extraction
Using primers described in Table 1, the DNA sample in Supplementary Table S1 was selected as the template, and PCR amplification of functional genes was performed using PCR 2× Es Taq Master Mix (Kangwei CW0690L, China). The PCR procedure was described previously (Braker et al., 1998; Geets et al., 2007). The PCR products were detected by 1% agarose electrophoresis, fragment size was compared to those existing in the literature, and the gel recovered after detection was confirmed to be accurate. The Agarose Gel DNA Recovery Kit (Kangwei CW2302S, China) was used according to the manufacturer’s instructions.
For the plasmid construction experiment, the pMD® 18-T Vector Cloning Kit (TaKaRa, Dalian, China) was used for T-carrier ligation and transferred into DH5α-competent cells with a plasmid size of 2,692 bp. The experimental steps included T vector ligation, heat shock conversion, culture, and identification. After incubating the correct clone in LA liquid bares overnight, the plasmid was extracted using the Fast Plasmid DNA Small Quantity Kit (Simgen, Hangzhou, China) according to the manufacturer’s instructions. The plasmids used in the experiment are listed in Supplementary Table S2.
2.7 qPCR of samples and establishment of their standard curves
For the qPCR of samples and establishment of their standard curves, SYBR Green I dye with Hieff® qPCR SYBR® Green Master Mix (No Rox; Yisheng, Shanghai) was used as the reagent. The Light Cycler 480 instrument (Roche, Switzerland) was used for this process (Yang et al., 2020).
The standard plasmid copy number was calculated by applying the plasmid concentration value to Equations 1, 2.
The obtained plasmid stock solution was diluted to 10−8 by a 10-fold concentration gradient, the reaction system was prepared using each concentration gradient as a template, and the corresponding annealing temperature was set for qPCR. The Ct value corresponding to each measured concentration was recorded, the Ct value was denoted on the X-axis and the log (copies) value was denoted on the Y-axis to establish a standard curve.
The extracted DNA was used as the template for qPCR, with three parallel experiments per sample, and the reaction system was the same as mentioned in Table 1. The Ct value measured by each sample was marked into the corresponding standard curve to calculate the copy number and corresponding copy number concentration value.
2.8 Statistical analysis
For data analysis and visualization, Qiime version 1.9 software (Caporaso et al., 2010) was used to calculate diversity indices such as Observed-otus, Chao1, Shannon, Simpson, Ace, and Goods-coverage. α- and β-Diversity indices and group differences were analyzed using R version 2.15 software. Qiime was used to calculate Unifrac distances and construct an unweighted pair group method with arithmetic mean sample cluster trees. R version 22.15, along with its vegan, weighted gene co-expression network analysis, stat, FactoMineR, and ggplot2 packages, was used to plot rarefaction curves, rank abundance curves, species accumulation curves, principal component analysis (PCA), principal coordinates analysis (PCoA), and non-metric multidimensional scaling plots, and conduct canonical correspondence analysis, redundancy analysis, and environmental factor correlation analysis (Gupta et al., 2018).
Independent- and paired-sample t-tests were performed to analyze the process (A and D) and sewage treatment plant (B and T), respectively, and to compare the differences in functional genes between different processes and sewage treatment plants. One-way analysis of variance (ANOVA) was used for differences between seasons; two-way ANOVA was used for interaction between processes, WWTPs, and seasons; and the Student’s–Newman–Keuls’ method was used for multiple comparisons. The above analysis was done using SPSS version 23.0 (IBM, United States). The relationship between the physical and chemical indexes of influent water and the abundance of different denitrification functional genes was analyzed by CANOCO 5.0 for RDA. PAST version 4.0 was used for the PCA of individual gene abundance differences between samples.
3 Results
3.1 WWTP operation and performances
The physicochemical parameters across different WWTPs during different seasons are provided in Supplementary Table S1 and Supplementary Figure S1. Statistics results showed that there were significant differences (p < 0.05) in BOD5, TP, and SV30 between T and B, but there was no difference in temperature, pH, COD, TN, and NH4+–N, and no significant difference in all parameters in different seasons. On average, temperature and pH values were higher in WWTP T than those in WWTP B. However, COD, BOD5, TP, TN, NH4+–N, and SV30 values were lower in WWTP T than those in WWTP B. SV30 values were generally lower in the A2/O system than those in the DOD sewer system. The concentration of each pollutant in the influent of WWTP B was significantly higher than that of WWTP T, which may be due to the presence of industrial effluent in the influent.
While ensuring the operation and normal effluent quality of WWTPs, SV30 values, which reflect sludge settling performance, compared across various processes of the same WWTP revealed that the sludge settling performance between the two processes was partially different, possibly because of the maintenance of different sludge concentrations and sludge return ratios. Furthermore, the SV30 value of WWTP B was relatively high, especially in winter, indicating that the sludge settling performance of WWTP B was poor during the winter, with sludge bulking even occurring.
3.2 Sequencing statistics and microbial diversity
3.2.1 Sequencing data
The quality control procedure, obtained 584,822 sequences and 130,189 OTUs were obtained via high-throughput sequencing. The rarefaction curve (Supplementary Figure S2) demonstrates that the curve levels off when the sequencing depth reaches 40,000 tags, indicating that the present sequencing data is sufficient and reasonable (Lundberg et al., 2013).
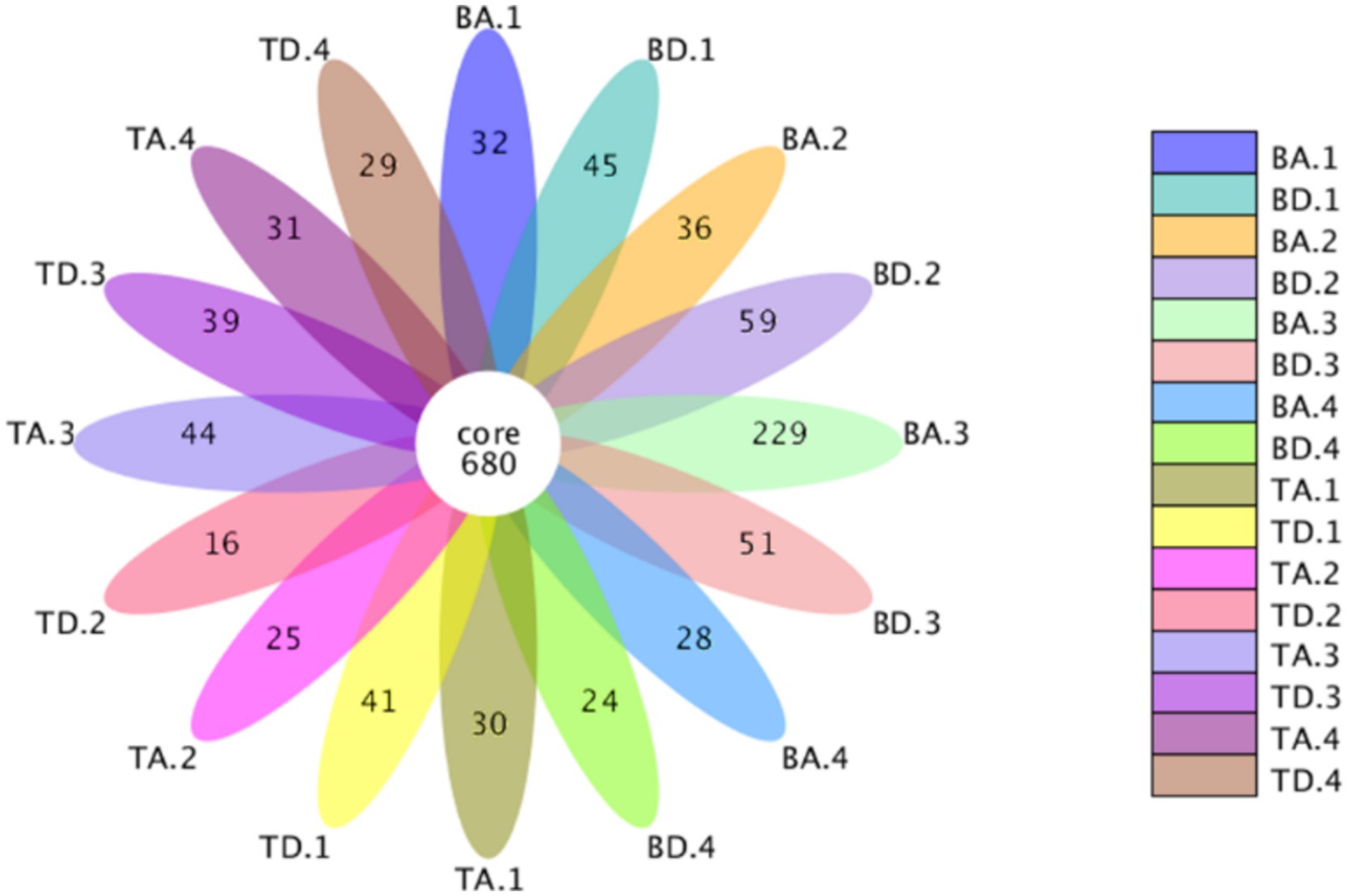
Operational Taxonomic Units analysis in the different groups. When samples were grouped according to different processes and seasons between the two WWTPs, 680 core OTUs were shared among all groups (Figure 1), 1,228 core OTUs were shared among groups at WWTP B (Supplementary Figure S3), and 835 core OTUs among groups at WWTP T (Supplementary Figure S3), indicating that WWTP B possessed a larger core microbial community. Under consistent sequencing depth conditions, OTU values at WWTP B were slightly higher than those at WWTP T (Supplementary Table S3), suggesting that WWTP B may have a higher community diversity than WWTP T.
3.2.2 α-diversity
The results on α-diversity across groups are presented in Supplementary Table S3, with the boxplot of the Shannon Index for different groups shown in Figure 2A. Comparing the different processes, the α-diversity of the A2/O system was partially higher and more stable than that of DOD processes across all seasons. Although there were some variations in Shannon Indices, these differences were not statistically significant, except for a significant difference observed in summer (Wilcoxon rank-sum test, p < 0.05), suggesting that the more complex habitat of the A2/O process supports a more diverse microbial community structure.
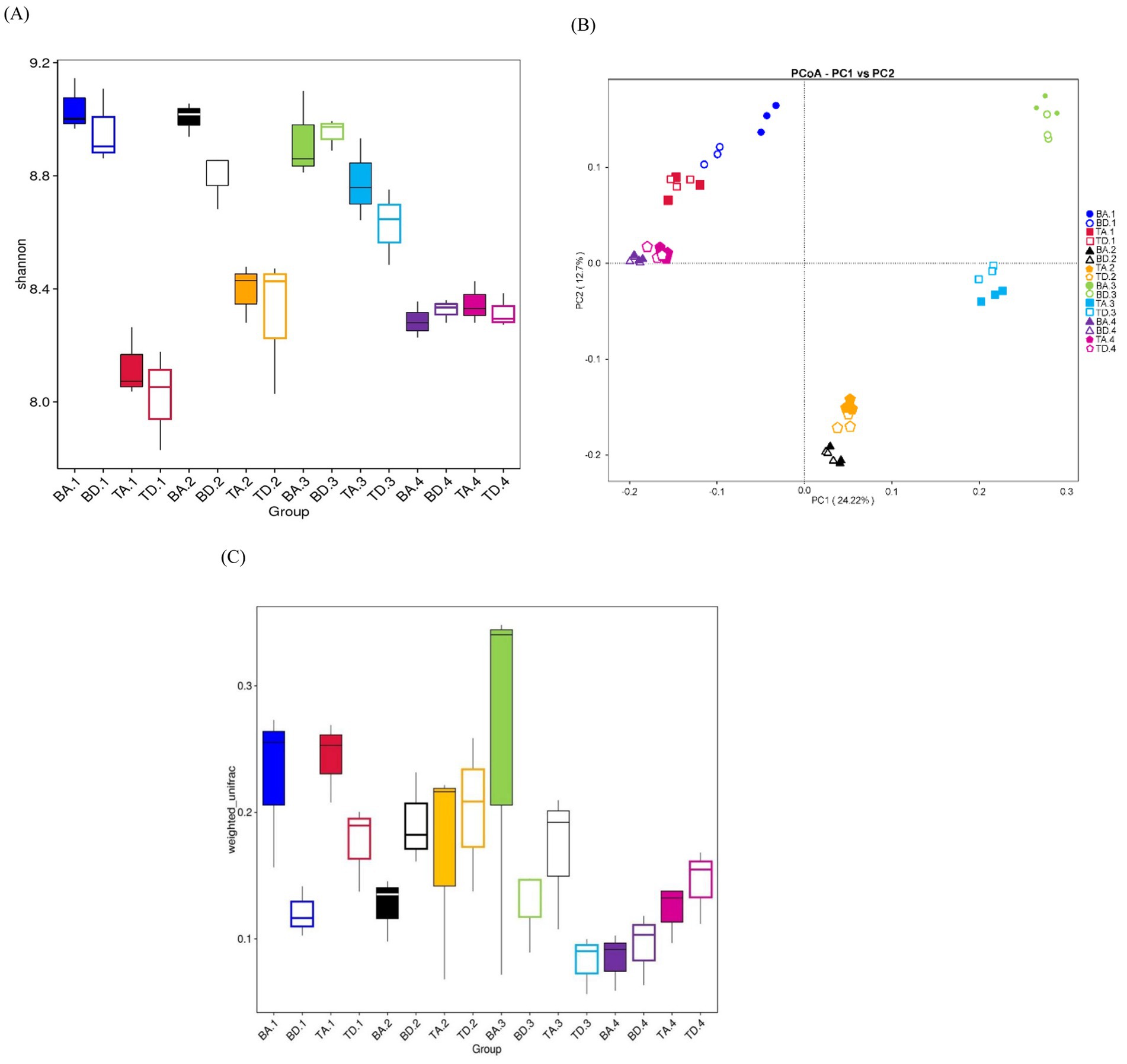
Figure 2. α- and β-diversity among all groups. (A) Boxplots for α-diversity of Shannon-Wilcox indices; same color indicates the same season and location, and the same color with a different shape indicates a different process. (B) PCoA plot based on unweighted_unifrac distances of each group. (C) Boxplots for β-diversity indices based on unweighted_unifrac distances of each group.
On comparing different sites, the α-diversity of WWTP B was significantly higher than WWTP T in all seasons except winter, in which the difference between the two WWTPs was insignificant (Wilcoxon rank-sum test, p < 0.05). This could be attributed to WWTP B receiving a certain amount of industrial wastewater with a higher pollutant load, leading to a more complex microbial community in AS.
3.2.3 β-diversity
The PCoA plot shows that samples from the same city and season consistently cluster together in proximity, indicating that the β-diversity difference between the A2/O and DOD processes is insignificant. However, the distance between samples from different seasons within the same city is greater than that between samples from different locations in the same season, suggesting a seasonal variation in β-diversity, with differences between seasons being more pronounced than those between processes and locations (Figure 2B).
The β-diversity of A2/O processes is generally higher than that of DOD processes in spring and autumn, but lower in summer and winter. This suggests that the differences in β-diversity between the A2/O and DOD processes are influenced by seasonal factors, possibly because of the presence of temperature-sensitive taxa in A2/O processes (Figure 2C).
3.2.4 Microbial community analysis
The taxonomic annotation results of OTUs at the phylum level revealed that the predominant phyla across all samples included Proteobacteria, Bacteroidetes, and Chloroflexi, collectively accounting for 68% of the total abundance. Other phyla, such as Nitrospirae, Actinobacteria, Planctomycetes, Firmicutes, and Acidobacteria, each accounted for >1% of the total abundance (Supplementary Figure S4). Within these major phyla, the predominant class level included Alphaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria within Proteobacteria, Bacteroidia within the Phylum Bacteroidetes; and Anaerolineae along with unidentified_Chloroflexi within the Chloroflexi. The composition of these phyla is consistent with previous reports on the microbial community structure of AS. The seasonal fluctuations in the abundance of these major microbial groups were more pronounced in WWTP B than in WWTP T. Among the dominant phyla with abundances >1%, Nitrospirae, Planctomycetes, and Firmicutes showed significant differences between the A2/O and DOD processes. Nitrospirae and Planctomycetes were more abundant in the DOD processes, whereas Firmicutes had higher abundances in the A2/O processes (Supplementary Figure S4).
Although significant variations were observed among subgroups at the genus level, the genera Dokdonella, unidentified_Nitrospiraceae, Terrimonas, and unidentified_Chloroflexi were present in all sample groups with abundances exceeding 0.5%. The abundances of Terrimonas, unidentified_Nitrospiraceae, and Rhodococcus, which were reported as dominant genera (Zhang et al., 2019), varied notably, as shown in Supplementary Figure S4. This indicates that while the abundance of microbial communities in AS significantly fluctuates in the genus level because of environmental influences, the core community structure remains relatively stable. Significant differences in abundance and composition were observed across groups from different locations and seasons (Supplementary Figure S4). In WWTP B, the microbial community structure remained relatively stable during spring, summer, and autumn, however, the abundance of unidentified_Nitrospiraceae and Dokdonella increased substantially in winter. In contrast, in WWTP T, Terrimonas, unidentified_Nitrospiraceae, and Dokdonella were more abundant in spring, with their proportions gradually decreasing through summer, autumn, and winter, whereas the abundance of unidentified_Flavobacteriales and Candidatus_Competibacter steadily increased across all four seasons. Among the top 30 dominant genera, Candidatus Competibacter was more abundant in DOD processes, whereas Haliangium, Lactobacillus, and Deinococcus were more prevalent in the A2/O process in WWTP B (Figure 3A). In WWTP T, Lactobacillus, unidentified Bacteria, Rhodobacter, and Iamia had higher abundance in the A2/O process, whereas Dechloromonas was more abundant in DOD processes (Figure 3B).
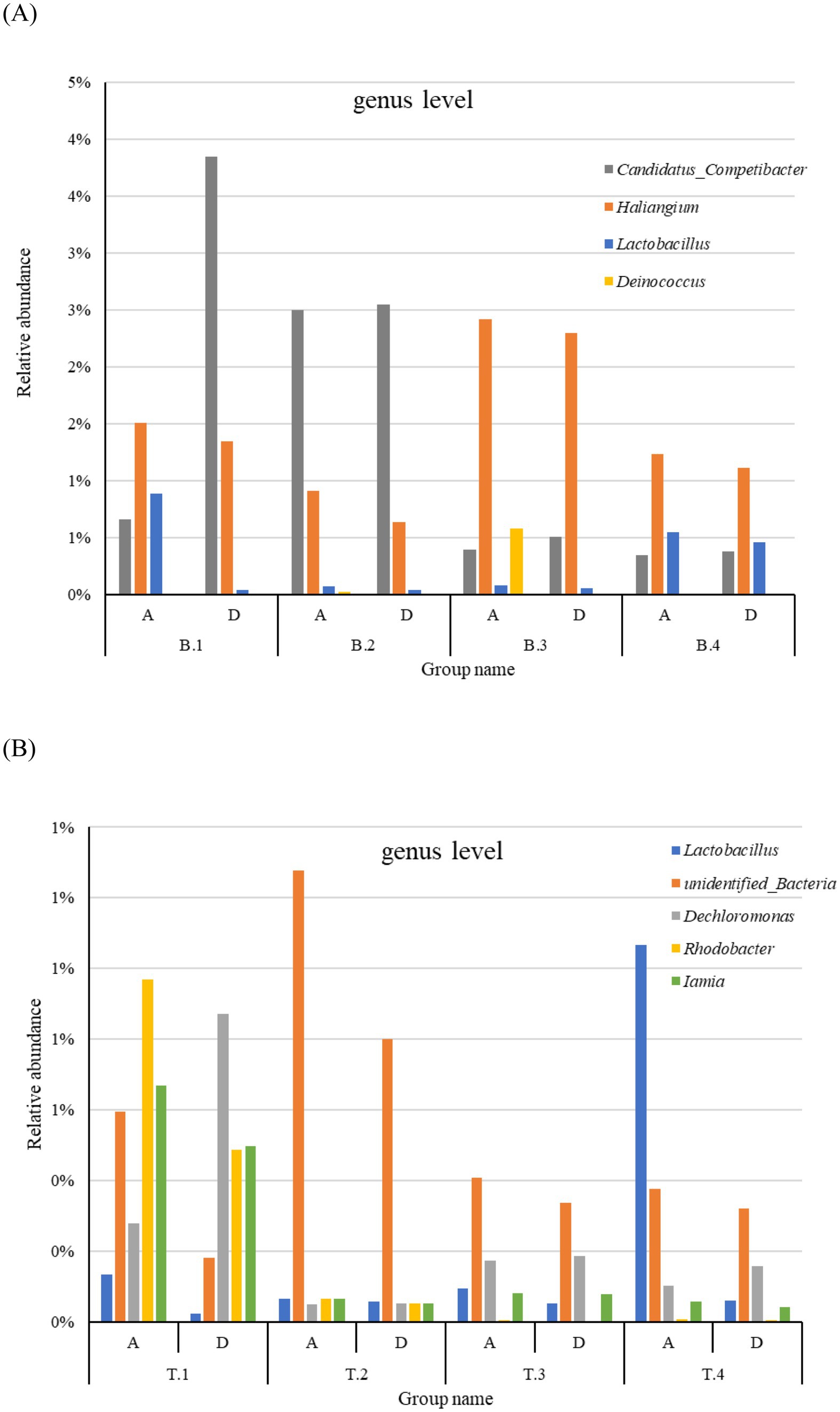
Figure 3. Relative abundance at the genus level showing differences between the A2/O and DOD processes in WWTP B (A) and WWTP T (B). T and B Representing Wuhan and Xi’an respectively; A and D represents A2/O and DOD processes respectively; 1, 2, 3, and 4, respectively, represent the four seasons of spring, summer, autumn and winter.
3.3 Abundance of total bacterial 16S rRNA genes and nitrogen metabolism functional genes
This study employed a comparative approach to investigate the differences in abundance of functional genes involved in nitrogen metabolism, including in 16S rRNA, AOB-16S rRNA, AOB-amoA, Nitrobacter nxrA, Nitrobacter nxrB, Nitrospira nxrB, nirS, nirK, nosZ, and anammox 16S rRNA. The results are presented in Supplementary Table S4, which lists the copy number differences between groups for these genes.
3.3.1 Total bacterial 16S rRNA gene abundance
The absolute abundance of the total bacterial 16S rRNA gene in WWTP T samples was 1.94 × 107 to 4.35 × 107 copies/ng, and the abundance of total bacterial 16S rRNA genes in WWTP B samples was 1.40 × 107 to 6.50 × 107 copies/ng, among which the fluctuation in WWTP B was more obvious, and the difference between the two processes was relatively large. OTUs results were similar, and the microbial diversity of WWTP B was slightly higher than that of WWTP T (Supplementary Figure S5).
3.3.2 AOB-associated genes
The absolute abundance of AOB amoA and 16S rRNA genes in the samples was detected in both WWTPs. The abundance of AOB amoA and 16S rRNA genes in WWTP B was 1.94 × 102 to 5.18 × 102 and 3.53 × 102, respectively, whereas the abundance of AOB amoA and 16S rRNA genes in WWTP T samples was 5.55 × 102 to 1.14 × 103 and 9.23 × 102 to 1.66 × 103, respectively (Figure 4A). Due to the coverage of different primers, the AOB 16S rRNA gene had more copies than AOB amoA (Abellan-Schneyder et al., 2021). Comparative analysis between Wuhan and Xi’an revealed significant differences in the absolute abundance of AOB amoA and 16S rRNA genes between WWTPs T and B (p < 0.05). Overall, the absolute abundance was higher in WWTP T than in WWTP B. When comparing the copy number of the two genes relative to the bacterial 16S rRNA gene, only AOB amoA showed a significant difference between the two locations. Although the NH4+–N concentration in the influent water of WWTP B was generally higher than that in WWTP T, the AOB amoA abundance of each group of samples from WWTP B was lower than that of WWTP T.
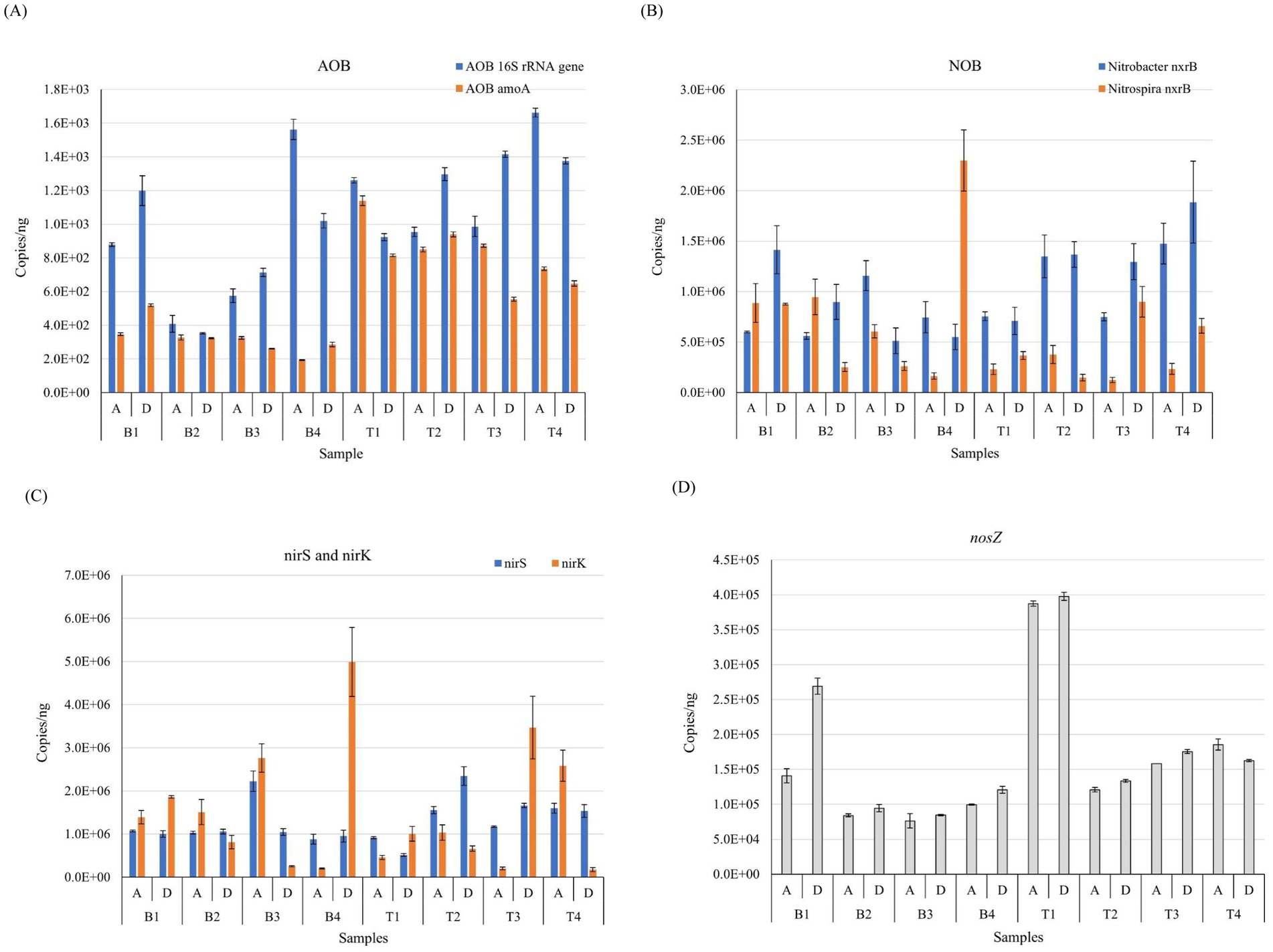
Figure 4. Comparison of the absolute abundance of nitrogen metabolism functional genes including AOB 16S rRNA gene and AOB amoA gene (A), Nitrobacter nxrB and Nitrospira nxrB gene (B), nirS and nirK gene (C), and nosZ gene (D) by qPCR (copies/ng DNA) in different seasons and processes of WWTPs B and T. T and B represent Wuhan and Xi’an respectively; A and D represent A2/O and DOD processes respectively; 1, 2, 3, and 4 represent the four seasons of spring, summer, autumn and winter, respectively.
Seasonal comparative analysis indicated that the absolute abundance of AOB amoA varied significantly across seasons (p < 0.05), although there was no significant difference in the AOB 16S rRNA gene. The abundance of the AOB 16S rRNA gene and amoA in summer and autumn was relatively consistent, but there were substantial differences between spring and winter, particularly in WWTP B. This variation may be attributed to the greater impact of seasonal temperature differences on AOB amoA. Comparative analysis of different processes showed that the process type did not significantly affect the abundance of AOB 16S rRNA and amoA genes. There was no significant correlation between the interactions of the two gene abundances across different processes, locations, and seasons.
3.3.3 NOB-associated genes
The absolute abundance of Nitrobacter nxrA and nxrB and Nitrospira nxrB in WWTP B samples were 8.62 × 101 to 2.99 × 102, 5.12 × 105 to 1.41 × 106, and 1.63 × 105 to 2.30 × 106, respectively. In contrast, the abundance of Nitrobacter nxrA and nxrB and Nitrospira nxrB in samples from WTTP T was 1.15 × 102 to 1.83 × 102, 7.10 × 105 to 1.89 × 106, and 1.23 × 105 to 8.99 × 105, respectively. Because the copy number of Nitrobacter nxrA differed from that of Nitrobacter nxrB and Nitrospira nxrB by nearly three orders of magnitude in samples (Figure 4B; Supplementary Figure S5). The absolute abundance of Nitrobacter nxrB was higher than that of Nitrospira nxrB in most groups, and the difference between them was smaller in WWTP B than in WWTP T. This may be due to differences in influent water quality or operational conditions, resulting in a higher abundance of the R-countermeasure Nitrobacter better adapted to the environment of high nitrite and high oxygen concentrations than the K-countermeasure Nitrospira which was better adapted to the environment of low nitrite and low oxygen concentrations (Blackburne et al., 2007; Dytczak et al., 2008; Kim and Kim, 2006). The absolute abundance of Nitrobacter nxrA did not differ significantly between processes, locations, seasons, and interactions with each other.
3.3.4 Denitrification-related genes
The absolute abundance of nirS, nirK, and nosZ genes in the samples revealed copy number ranges of 8.80 × 105 to 2.22 × 106, 2.03 × 105 to 4.99 × 106, and 7.63 × 104 to 2.69 × 105 in WWTB B and 5.18 × 105 to 2.35 × 106, 1.75 × 105 to 3.47 × 106, and 1.21 × 105 to 3.98 × 105, respectively, in WWTP T (Figures 4C,D). Results on the differences in copy number of nirS, nirK, and nosZ in different processes, locations, and seasons showed that the absolute abundance of nirS and nirK was insignificant between different seasons and locations, but there was a significant difference in the copy number of nosZ between seasons and locations (p < 0.05), and multiple comparisons showed that the difference between the first and other quarters was the most pronounced among seasonal differences. Among different processes in all other subgroups, except for the fourth quarter of WWTP T, the absolute abundance of nosZ in DOD was higher than in A2/O. The highest and average values of absolute nosZ abundance were significantly higher in WWTP T than those in WWTP B at different sites. Although nirS and nirK in different ecological niches had the same function, they were unrelated. In the denitrification process, WWTP T produced less N2O than WWTP B.
3.3.5 Anaerobic ammonia oxidation
Results on the absolute abundance for anammox 16S rRNA gene in the samples showed that WWTP B had a copy number of 4.66 × 104 to 5.12 × 105, whereas the WWTP T copy number was 6.46 × 104 to 4.43 × 105 (Supplementary Figure S5). Comparative analysis of the anammox 16S rRNA gene showed that differences were not significant across processes, locations, seasons, and interactions, and seasonal differences were random, possibly due to the sensitivity of functional taxa to dissolved oxygen (DO).
3.4 Effects of influent water quality on denitrification gene abundance
The results of the RDA analysis of the abundance of each gene and physicochemical properties of the influent water are shown in Figure 5. The abundance of the total bacterial 16S rRNA gene was positively correlated with BOD, NH4+–N, TP, and TN; that is, the total bacterial growth was promoted when there was nutrient-rich status. nirS had a strong positive correlation with pH during the denitrification process, and a positive correlation between nirK and nutrient sources was observed. In the final step of denitrification, the abundance of nosZ was only positively correlated with temperature, comparable to the significant difference between seasons. Results showed that the significant difference in nosZ between seasons was primarily affected by temperature changes. Pearson correlation analysis of the relationship between the main NOB and nitrite-reducing bacteria revealed a significant positive correlation between nirS and the copy number of the Nitrobacter nxrB gene (r = 0.621, p < 0.05). Consequently, there was a significant positive correlation between nirK and Nitrospira nxrB gene copy number (r = 0.793; p < 0.01). nirS and nirK in different ecological niches may have a certain correspondence between different taxa of nxrB, Nitrobacter nxrB, and Nitrospira nxrB, thus, portraying mutual support.
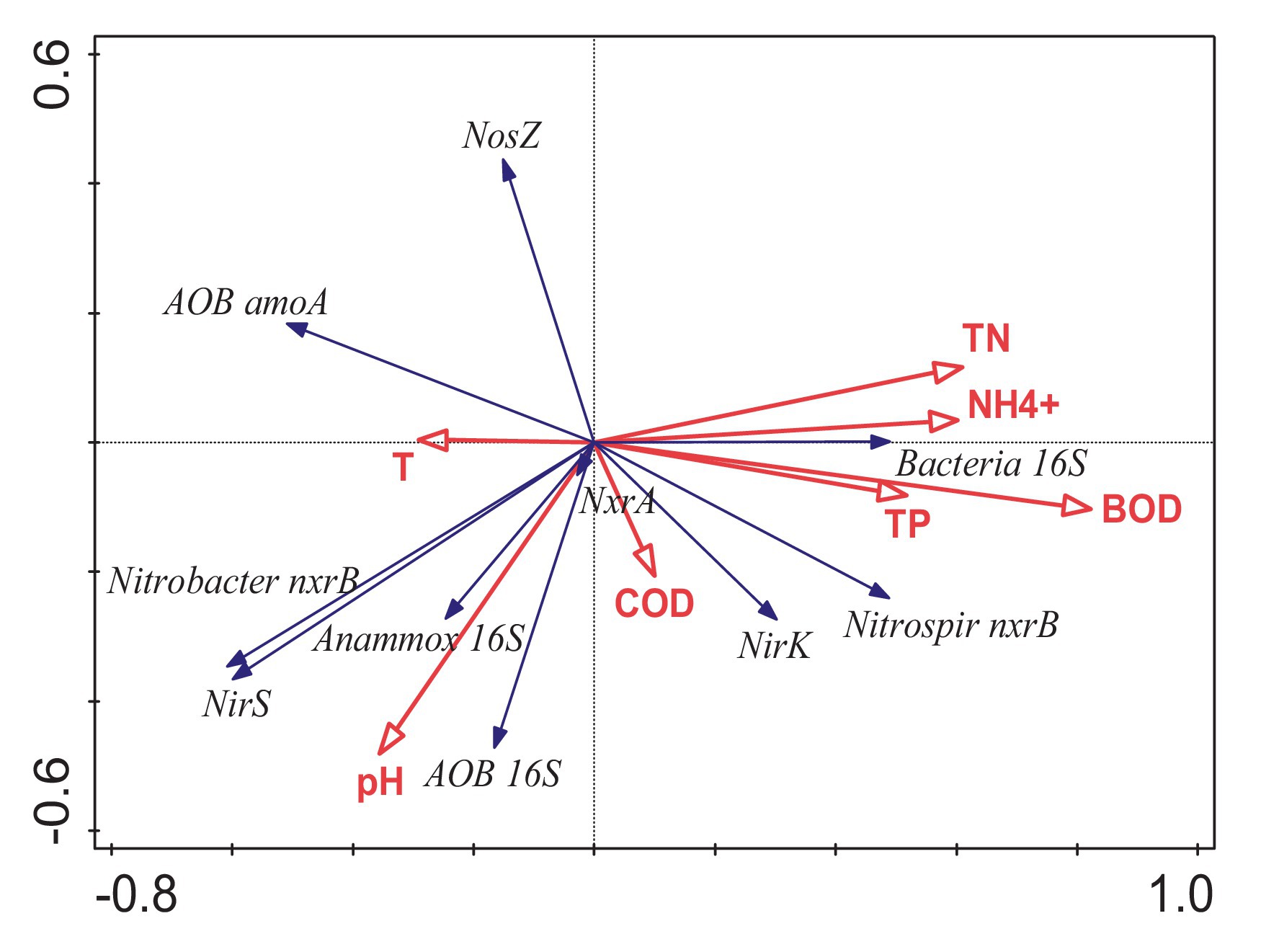
Figure 5. RDA of the relationship between the influent environmental variables and the absolute abundance of genes; arrow length represents the strength of the correlation between the environmental variables and the absolute abundance of genes; longer the arrow length, the stronger the correlation. The perpendicular distance between the influent environmental variables and absolute abundance of genes axes in the plot reflects their correlations. The smaller the distance, the stronger the correlation. BOD, biochemical oxygen demand for 5 days; pH, pH value; NH4+–N, ammonia nitrogen concentration; TN, total nitrogen concentration; COD, chemical oxygen demand concentration; DO, dissolved oxygen; T, temperature.
4 Discussion
Differences in microbial community composition and their variations are often key to understanding the mechanisms of microbial community assembly and ecosystem functioning (Wu et al., 2019; Zhou et al., 2013). Different processes maintain distinct operational parameters, creating differentiated habitats within the biochemical reaction tanks, leading to microbial community structure variations (Park et al., 2021). Herein, the findings indicated that a comparative analysis of microbial community structures across different processes using the same influent did not reveal significant consistent differences. However, some general distinctions were observed. For instance, among the phyla with abundances >1%, Nitrospirae and Planctomycetes were more abundant in the DOD processes, whereas Firmicutes showed higher abundance in the A2/O process. Reportedly, Nitrospirae is the primary nitrifying phylum in AS microbial community, while Planctomycetes is known for anaerobic ammonia oxidation (Zheng et al., 2020). DOD processes exhibit a stronger nitrogen removal capacity compared to A2/O processes (Zhou et al., 2012). Therefore, the difference in the abundance of Nitrospirae and Planctomycetes may be crucial in differing nitrogen removal efficiencies observed between A2/O and DOD processes.
The assembly of microbiomes in WWTPs is closely related to absolute latitude, mean annual temperature, and influent COD and BOD concentrations (Song et al., 2021; Wu et al., 2019). WWTPs B and T are located in Wuhan and Xi’an, respectively, in different climatic regions of China, with distinct northern and southern climates. Variations in climate and temperature due to differing latitudes, along with differences in local dietary habits, can lead to differences in the composition of major pollutants in domestic wastewater and changes in the AS state. These factors may be the primary reasons for seasonal differences in the microbial community structure observed between the two locations. Dokdonella is considered a dominant genus composition in WWTPs for the antibiotic pharmaceutical industry. The elevated concentration of pharmaceutical wastewater in the influent of WWTP B during winter may explain the higher abundance of Dokdonella in that season. Candidatus Competibacter and Flavobacterium exhibit strong capabilities in removing COD, NH4+–N, and TN, and the NH4+–N concentration in the influent of WWTP T in the fall and winter was nearly twice as much as that in the spring and summer (Supplementary Table S1), which may be the primary reason for the increased abundance of denitrifying bacteria such as Candidatus Competibacter and Flavobacterium in autumn and winter.
Seasonality can exert a significant influence on the performance and dynamics of nitrogen functional genes within A2/O and DOD processes, which are commonly used in WWTPs and designed to remove nitrogen compounds through microbial processes (Li et al., 2018; Zhang et al., 2016). Nitrogen functional genes are responsible for various transformations of nitrogen within these systems (Myrold, 2021). The key genes involved in nitrogen removal include AOB genes (such as amoA), NOB genes (such as nxrA), and denitrifying bacteria genes (such as nirS and nirK) (Galloway et al., 2008; Liu et al., 2022). The activities and abundance of these genes are greatly influenced by environmental factors, including temperature, DO levels, and organic loading rates, which normally exhibit seasonal variations. Therefore, the influence of seasonality on nitrogen functional genes within A2/O and DOD systems depends on the specific system design, operational parameters, and geographical location. Furthermore, advancements in process control and optimization techniques, such as real-time monitoring and adaptive control strategies, can help mitigate the effects of seasonality and enhance the overall performance of these systems throughout the year (Holmes et al., 2019; Hu et al., 2023; Kuypers et al., 2018; Potgieter et al., 2020; Zhao et al., 2023). This study covered the contribution of seasonality on nitrogen functional genes within A2/O and DOD processes in Wuhan and Xi’an cities, China.
The diversity and population of AOB abundance in AS are the main factors affecting the nitrification efficiency of AS systems, and the increase in AOB diversity and population density can enhance the intensity of nitrification (Awolusi et al., 2018), indicating that WWTP B with a higher AS concentration had a higher NH4+–N removal capacity than WWTP T. AOB amoA could be more effective than AOB 16S rRNA or bacterial 16S rRNA genes, reflecting the differences in bacterial composition in different AS samples (Wang et al., 2021). Hence, AOB amoA is highly targeted in analyzing of ammonia oxidation functional microbes and can more accurately reflect AOB and ammonia oxidation intensity in AS. Studies have reported that NOB found in AS are primarily Nitrobacter and Nitrospira, with Nitrospira being relatively more dominant (Daims et al., 2015; Gonzalez-Martinez et al., 2016; Johnston et al., 2019). Nitrobacter nxrB was relatively dominant in most grouped samples, and the difference between the two WWTP B samples was smaller than that in the WWTP T sample. This could have been due to the difference in influent water quality of the sewage treatment plant or the operating conditions, resulting in a relatively high nitrite concentration in the biochemical pool, making it more suitable for high nitrite and oxygen concentrations. Indeed, the abundance of R-strategist Nitrobacter was higher than that of Nitrospira, adapted to low nitrite and low oxygen concentration of K-strategist (Han et al., 2017).
Nitrite oxidation is an essential step in fixed nitrogen transformation. The physiology of NOB implies that nitrite oxidation rates should be controlled by the concentration of their substrate, nitrite, and oxygen (terminal electron acceptor). Consequently, potential nitrite oxidation (PNO) is significantly positively correlated with Nitrobacter nxrA copy number in soil, and changes in PNO may mediate changes in the abundance ratio of Nitrospira nxrB/Nitrobacter nxrA (Attard et al., 2010; Cebron and Garnier, 2005; Han et al., 2017). Therefore, the abundance ratio can better predict the change in PNO (Florido et al., 2015). In this study, unlike the soil environment, the ratio trend of Nitrospira nxrB to Nitrobacter nxrA in the sample was almost consistent with that of Nitrospira nxrB, which made it difficult to comprehensively reflect the relative abundance of these two taxa of NOB, making it necessary to find suitable characterization indicators of AS microorganisms.
The changes in influent water quality often significantly affect AS microbial communities (Florido et al., 2015; Yang et al., 2013). During nitrification, the abundance of the AOB 16S rRNA gene was positively correlated with pH, consistent with other reports (Abellan-Schneyder et al., 2021; Deissová et al., 2023). However, AOB amoA had a strong positive correlation with temperature, and this phenomenon was corroborated by the significant difference in AOB amoA copy number between seasons, indicating that the microbes encoding AOB amoA could be more sensitive to temperature changes, as reported previously (Lu et al., 2015; Song and Niu, 2022). Although AOB amoA and 16S rRNA genes reflected the abundance of AOB, the target genes of different primers reflected different biota characteristics of the same function, that is some AOB were sensitive to pH, whereas some AOB were more sensitive to temperature.
Among NOB-related genes, Nitrobacter nxrA and nxrB and Nitrospira nxrB, pH and COD were the most influential on Nitrobacter nxrA, and all indicated a positive correlation. Nitrobacter nxrB had a stronger positive correlation with temperature and pH than Nitrospira nxrB, whereas Nitrospira nxrB was not sensitive to pH. Because no specific functional marker gene can target the broad functional population of NOB, the specific role of pH in influencing NOB growth remains unclear (Msimbira and Smith, 2020). Studies showed that Nitrobacter nxrA may positively correlate with pH and organic matter concentration (Bartosch et al., 2002; Li et al., 2019). Therefore, similar conclusions can be made in this study, although Nitrobacter nxrA was slightly affected by environmental factors. Nitrospira nxrB was more associated with NH4+–N concentration in influent water than other functional genes, indicating that Nitrospira plays an important role in nitrite oxidation in AS (Mehrani et al., 2020). Indeed, the absolute abundance of the anammox 16S rRNA gene during anaerobic ammonia oxidation had a strong positive correlation with pH and temperature, indicating stable anaerobic conditions and that environmental parameters are important factors affecting anammox bacterial communities that are rarely affected by nutrient source restriction (Chen et al., 2023).
As reported previously, changes in the abundance of denitrification genes nxrB or nirS were accompanied by changes in the abundance of microbes, such as Flavobacterium or Candidatus Competibacter (Braker et al., 2000; Wei et al., 2015; Yang et al., 2022). Flavobacterium has strong nitrogen and phosphorus removal ability (Enisoglu-Atalay et al., 2018), and Candidatus Competibacter plays a key role in denitrification processes (Rubio-Rincón et al., 2017). Genomic analysis of existing Candidatus Competibacter-related genera, which is the main nitrogen removal functional microbes in AS, through the NCBI database found a nirS reductase gene, indicating an association between the relative abundance of nitrogen removal microbes and the nitrogen removal functional gene. The unidentified genus of Flavobacteriales and Candidatus Competibacter may be related to the nitrogen metabolism functional genes nirS and Nitrobacter nxrB, and the main nitrogen removal functional microbes, the unidentified genus of Nitrospiraceae, may be related to the nitrogen metabolism functional genes nirK and Nitrospira nxrB, and these two groups could play their own roles in different ecological niches.
This study showed that the A2/O system exhibited higher levels of most nitrogen metabolism functional genes than those of the DOD system in WWTPs of the two locations. Spring and summer recorded higher values of most genes across all WWTPs than during autumn and winter. The finding concurred with studies that compared the A2/O and DOD systems in terms of nitrogen removal efficiency and abundance of nitrogen metabolism functional genes. Studies have reported higher values of key functional genes involved in nitrification (such as amoA for AOB) and denitrification (such as nirS and nirK for denitrifying bacteria) in the A2/O system compared to the DOD system (Kim and Nakhla, 2010; Xu et al., 2014). Other studies investigated the performance and microbial community composition of full-scale WWTPs employing the A2/O and DOD processes by analyzing the abundance of nitrogen metabolism functional genes and found that the A2/O system exhibited higher values for key genes involved in nitrification and denitrification compared to the DOD process (Ding et al., 2006; Wang et al., 2019; Xu et al., 2014). Other studies compared A2/O system to the sequencing batch reactor (SBR) process in a full-scale WWTP. It evaluated the nitrogen removal performance and analyzed the microbial community, including the abundance of nitrogen metabolism functional genes. Results indicated that the A2/O system had higher values for genes associated with nitrification and denitrification than the SBR process (Baeza et al., 2002; Chen ZhaoBo et al., 2011). Thus, understanding the composition and activity of nitrogen metabolism functional genes in different WWTP types is important for optimizing WWTP processes and improving nitrogen removal efficiency. It can help identify the key microbial populations involved in nitrogen transformation and guide the design and operation of WWTPs to achieve effective nitrogen removal and minimize environmental impacts (Holmes et al., 2019; Xu et al., 2022).
5 Conclusion
The effects of different processes, locations, and seasons on the nitrogen removal microbes and functional genes in WWTPs were investigated. Seasonal variations or differences in influent water quality had a greater effect on the assembly of microbiomes in WWTPs than location differences, and location differences had a greater effect than process differences. The absolute abundance of AOB amoA varied significantly over seasons due to temperature changes. Among NOB, no significant differences were found in Nitrobacter nxrA in different processes, geographic locations, and seasons. Interestingly, the absolute abundance of Nitrobacter nxrB in AS was higher than that of Nitrospira, which could be attributed to differences in influent water quality or operating conditions in sewage treatment plants. The difference in absolute abundance between nirS and nirK in different locations and seasons was insignificant. The absolute abundance of nosZ in DOD was generally higher than that in A2/O, and the absolute abundance of nosZ in the WWTP of Wuhan (T) was higher than that of the WWTP of Xi’an (B). There was variation in the absolute abundance of anammox 16S rRNA gene between different processes, locations, and seasons, which could be attributed to the sensitivity of functional microbes to DO. Bacteria in terms of the AOB 16S rRNA gene were sensitive to pH, whereas the communities corresponding to AOB amoA were more sensitive to temperature proving that the absolute abundance of the anammox 16S rRNA gene in anaerobic AOB may be affected by temperature changes and was hardly affected by seasonal differences. Comprehensive analysis of nitrogen metabolism functional microbes and genes revealed that nitrogen metabolism functional genes nirS and Nitrobacter nxrB in AS could be related to the unidentified genus of Flavobacteriales and Candidatus Competibacter as the main nitrogen removal functional microbes, whereas nitrogen metabolism functional genes nirK and Nitrospira nxrB could be associated with the unidentified genus of Nitrospiraceae as the major denitrification functional microbes, which may play significant ecological roles in different niches. This study provides critical insights into the seasonal and geographic factors affecting nitrogen removal processes in WWTPs. By understanding these influences, treatment plants can implement targeted strategies to enhance nitrogen removal efficiency and meet regulatory standards, minimizing the environmental impact of wastewater discharge.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
SL: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Writing – original draft, Writing – review & editing. YL: Data curation, Formal analysis, Writing – original draft. LY: Data curation, Methodology, Writing – original draft. EX: Data curation, Investigation, Validation, Writing – original draft. DX: Conceptualization, Resources, Validation, Visualization, Writing – original draft. HC: Writing – review & editing. JD: Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft, Writing – review & editing. DQ: Conceptualization, Data curation, Investigation, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (no. 32102769), Natural Science Foundation of Hubei Province of China (no. 2022CFB403), Scientific Research Items Foundation of Hubei Educational Committee (no. Q20221614), Guangzhou Science and Technology Programme (2023A04J1029) and Research and Innovation Initiatives of WHPU (2023Y14).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1498681/full#supplementary-material
References
Abellan-Schneyder, I., Matchado, M. S., Reitmeier, S., Sommer, A., Sewald, Z., Baumbach, J., et al. (2021). Primer, pipelines, parameters: issues in 16S rRNA gene sequencing. Msphere 6, 01202–01220. doi: 10.1128/mSphere.01202-20
Arumugham, T., Khudzari, J., Abdullah, N., Yuzir, A., Iwamoto, K., and Homma, K. (2024). Research trends and future directions on nitrification and denitrification processes in biological nitrogen removal. J. Environ. Chem. Eng. 2:111897. doi: 10.1016/j.jece.2024.111897
Attard, E., Poly, F., Commeaux, C., Laurent, F., Terada, A., Smets, B. F., et al. (2010). Shifts between Nitrospira-and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ. Microbiol. 12, 315–326. doi: 10.1111/j.1462-2920.2009.02070.x
Awolusi, O. O., Kumari, S., and Bux, F. (2018). Evaluation of ammonia oxidizing bacterial community structure of a municipal activated sludge plant by 454 high-throughput pyrosequencing. Environ. Process. 5, 43–57. doi: 10.1007/s40710-018-0319-2
Baeza, J., Gabriel, D., and Lafuente, J. (2002). Improving the nitrogen removal efficiency of an A2/O based WWTP by using an on-line knowledge based expert system. Water Res. 36, 2109–2123. doi: 10.1016/S0043-1354(01)00402-X
Bartosch, S., Hartwig, C., Spieck, E., and Bock, E. (2002). Immunological detection of -like bacteria in various soils. Microb. Ecol. 43, 26–33. doi: 10.1007/s00248-001-0037-5
Bhatt, P., Mathur, N., Singh, A., Pareek, H., and Bhatnagar, P. (2020). Evaluation of factors influencing the environmental spread of pathogens by wastewater treatment plants. Water Air Soil Pollut. 231:440. doi: 10.1007/s11270-020-04807-4
Blackburne, R., Vadivelu, V. M., Yuan, Z., and Keller, J. (2007). Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter. Water Res. 41, 3033–3042. doi: 10.1016/j.watres.2007.01.043
Braker, G., Fesefeldt, A., and Witzel, K.-P. (1998). Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64, 3769–3775. doi: 10.1128/AEM.64.10.3769-3775.1998
Braker, G., Zhou, J., Wu, L., Devol, A. H., and Tiedje, J. M. (2000). Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific northwest marine sediment communities. Appl. Environ. Microbiol. 66, 2096–2104. doi: 10.1128/AEM.66.5.2096-2104.2000
Bwapwa, J. (2022). “Factors affecting the bioremediation of industrial and domestic wastewaters” in Microbial biodegradation and bioremediation. eds. S. Das and H. R. Dash (Amsterdam, Netherlands: Elsevier), 461–472.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cebron, A., and Garnier, J. (2005). Nitrobacter and Nitrospira genera as representatives of nitrite-oxidizing bacteria: detection, quantification and growth along the lower Seine River (France). Water Res. 39, 4979–4992. doi: 10.1016/j.watres.2005.10.006
Chakravorty, S., Helb, D., Burday, M., Connell, N., and Alland, D. (2007). A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 69, 330–9. doi: 10.1016/j.mimet.2007.02.005
Chen, X., Liu, L., Bi, Y., Meng, F., Wang, D., Qiu, C., et al. (2023). A review of anammox metabolic response to environmental factors: characteristics and mechanisms. Environ. Res. 223:115464. doi: 10.1016/j.envres.2023.115464
Chen ZhaoBo, C. Z., Nie ShuKai, N. S., Ren NanQi, R. N., Chen ZhiQiang, C. Z., Wang HongCheng, W. H., and Cui MinHua, C. M. (2011). Improving the efficiencies of simultaneous organic substance and nitrogen removal in a multi-stage loop membrane bioreactor-based PWWTP using an on-line knowledge-based expert system. Water Res. 45, 5266–5278. doi: 10.1016/j.watres.2011.07.032
Dai, H., Zhao, J., Wang, Z., Chen, C., Liu, X., Guo, Z., et al. (2023). Optimal control of sewage treatment process using a dynamic multi-objective particle swarm optimization based on crowding distance. J. Environ. Chem. Eng. 11:109484. doi: 10.1016/j.jece.2023.109484
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Das, P., and Singh, K. (2022). “Wastewater remediation: Emerging technologies and future prospects” in Environmental degradation: challenges and strategies for mitigation, eds. V. S. Singh, S. Yadav, and P. Singh (Cham, Switzerland: Springer Nature). 227–250.
Deissová, T., Zapletalová, M., Kunovský, L., Kroupa, R., Grolich, T., Kala, Z., et al. (2023). 16S rRNA gene primer choice impacts off-target amplification in human gastrointestinal tract biopsies and microbiome profiling. Sci. Rep. 13:12577. doi: 10.1038/s41598-023-39575-8
Ding, Y.-W., Lin, W., Wang, B.-Z., and Zheng, W. (2006). Removal of nitrogen and phosphorus in a combined A2/O-BAF system with a short aerobic SRT. J. Environ. Sci. 18, 1082–1087. doi: 10.1016/s1001-0742(06)60043-0
Dytczak, M. A., Londry, K. L., and Oleszkiewicz, J. A. (2008). Activated sludge operational regime has significant impact on the type of nitrifying community and its nitrification rates. Water Res. 42, 2320–2328. doi: 10.1016/j.watres.2007.12.018
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Enisoglu-Atalay, V., Atasever-Arslan, B., Yaman, B., Cebecioglu, R., Kul, A., Ozilhan, S., et al. (2018). Chemical and molecular characterization of metabolites from Flavobacterium sp. PLoS One 13:e0205817. doi: 10.1371/journal.pone.0205817
Feng, Z., Li, T., Lin, Y., and Wu, G. (2022). Microbial communities and interactions in full-scale A2/O and MBR wastewater treatment plants. J. Water Proc. Engineer. 46:102660. doi: 10.1016/j.jwpe.2022.102660
Florido, E., Zurita, A., Pérez, I., Pérez-Montero, E., Coelho, P., and Gadotti, D. (2015). Central enhancement of the nitrogen-to-oxygen abundance ratio in barred galaxies. Astron. Astrophy. 584:A88. doi: 10.1051/0004-6361/201526191
Galloway, J. N., Townsend, A. R., Erisman, J. W., Bekunda, M., Cai, Z., Freney, J. R., et al. (2008). Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892. doi: 10.1126/science.1136674
Geets, J., De Cooman, M., Wittebolle, L., Heylen, K., Vanparys, B., De Vos, P., et al. (2007). Real-time PCR assay for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Appl. Microbiol. Biotechnol. 75, 211–221. doi: 10.1007/s00253-006-0805-8
Gizińska-Górna, M., and Wasąg, Z. (2022). Reliability and efficiency of the removal of pollutants in the mechanical–biological wastewater treatment plant A2/O. Desalin. Water Treat. 246, 120–138. doi: 10.5004/dwt.2022.28006
Gonzalez-Martinez, A., Rodriguez-Sanchez, A., Van Loosdrecht, M., Gonzalez-Lopez, J., and Vahala, R. (2016). Detection of comammox bacteria in full-scale wastewater treatment bioreactors using tag-454-pyrosequencing. Environ. Sci. Pollut. Res. 23, 25501–25511. doi: 10.1007/s11356-016-7914-4
Guerrero, J., Guisasola, A., Vilanova, R., and Baeza, J. A. (2011). Improving the performance of a WWTP control system by model-based setpoint optimisation. Environ. Model Softw. 26, 492–497. doi: 10.1016/j.envsoft.2010.10.012
Gupta, A., Dutta, A., Sarkar, J., Panigrahi, M. K., and Sar, P. (2018). Low-abundance members of the Firmicutes facilitate bioremediation of soil impacted by highly acidic mine drainage from the Malanjkhand copper project, India. Front. Microbiol. 9:2882. doi: 10.3389/fmicb.2018.02882
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Han, S., Luo, X., Liao, H., Nie, H., Chen, W., and Huang, Q. (2017). Nitrospira are more sensitive than Nitrobacter to land management in acid, fertilized soils of a rapeseed-rice rotation field trial. Sci. Total Environ. 599, 135–144. doi: 10.1016/j.scitotenv.2017.04.086
Holmes, D. E., Dang, Y., and Smith, J. A. (2019). Nitrogen cycling during wastewater treatment. Adv. Appl. Microbiol. 106, 113–192. doi: 10.1016/bs.aambs.2018.10.003
Hu, P., Qian, Y., Liu, J., Gao, L., Li, Y., Xu, Y., et al. (2023). Delineation of the complex microbial nitrogen-transformation network in an anammox-driven full-scale wastewater treatment plant. Water Res. 235:119799. doi: 10.1016/j.watres.2023.119799
Johnston, J., LaPara, T., and Behrens, S. (2019). Composition and dynamics of the activated sludge microbiome during seasonal nitrification failure. Sci. Rep. 9:4565. doi: 10.1038/s41598-019-40872-4
Kim, D.-J., and Kim, S.-H. (2006). Effect of nitrite concentration on the distribution and competition of nitrite-oxidizing bacteria in nitratation reactor systems and their kinetic characteristics. Water Res. 40, 887–894. doi: 10.1016/j.watres.2005.12.023
Kim, M., and Nakhla, G. (2010). Comparative performance of A2/O and a novel membrane-bioreactor-based process for biological nitrogen and phosphorus removal. Water Environ. Res. 82, 69–76. doi: 10.2175/106143009X447966
Kuypers, M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Li, X., Gong, Q., and Li, Z. (2024). Response characteristics of soil microorganisms under strong disturbance conditions in the riparian zone of the three gorges reservoir area. Sci. Rep. 14:18394. doi: 10.1038/s41598-024-69533-x
Li, Y., Xi, R., Wang, W., and Yao, H. (2019). The relative contribution of nitrifiers to autotrophic nitrification across a pH-gradient in a vegetable cropped soil. J. Soils Sediments 19, 1416–1426. doi: 10.1007/s11368-018-2109-x
Li, X., Zhang, M., Liu, F., Chen, L., Li, Y., Li, Y., et al. (2018). Seasonality distribution of the abundance and activity of nitrification and denitrification microorganisms in sediments of surface flow constructed wetlands planted with Myriophyllum elatinoides during swine wastewater treatment. Bioresour. Technol. 248, 89–97. doi: 10.1016/j.biortech.2017.06.102
Li, X., Zhang, M., Liu, F., Li, Y., He, Y., Zhang, S., et al. (2015). Abundance and distribution of microorganisms involved in denitrification in sediments of a Myriophyllum elatinoides purification system for treating swine wastewater. Environ. Sci. Pollut. Res. 22, 17906–17916. doi: 10.1007/s11356-015-5041-2
Liu, Y., Chi, Q., Cheng, H., Ding, H., Wen, T., Zhao, J., et al. (2022). Comparative microbial nitrogen functional gene abundances in the topsoil vs. subsoil of three grassland habitats in northern China. Front. Plant Sci. 12:792002. doi: 10.3389/fpls.2021.792002
Liu, T., Liu, S., Zheng, M., Chen, Q., and Ni, J. (2016). Performance assessment of full-scale wastewater treatment plants based on seasonal variability of microbial communities via high-throughput sequencing. PLoS One 11:e0152998. doi: 10.1371/journal.pone.0152998
Lu, S., Liao, M., Xie, C., He, X., Li, D., He, L., et al. (2015). Seasonal dynamics of ammonia-oxidizing microorganisms in freshwater aquaculture ponds. Ann. Microbiol. 65, 651–657. doi: 10.1007/s13213-014-0903-2
Lundberg, D. S., Yourstone, S., Mieczkowski, P., Jones, C. D., and Dangl, J. L. (2013). Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002. doi: 10.1038/nmeth.2634
Makuwa, S., Tlou, M., Fosso-Kankeu, E., and Green, E. (2022). The effects of dry versus wet season on the performance of a wastewater treatment plant in north West Province, South Africa. Water SA 48, 40–49. doi: 10.17159/wsa/2022.v48.i1.3897
Mehrani, M.-J., Sobotka, D., Kowal, P., Ciesielski, S., and Makinia, J. (2020). The occurrence and role of Nitrospira in nitrogen removal systems. Bioresour. Technol. 303:122936. doi: 10.1016/j.biortech.2020.122936
Moharram, M., Abdelhalim, H., and Rozaik, E. (2017). Performance appraisal of the A2/O process in domestic wastewater treatment replacing the anaerobic unit with UASB. HBRC J. 13, 98–105. doi: 10.1016/j.hbrcj.2015.01.004
Msimbira, L. A., and Smith, D. L. (2020). The roles of plant growth promoting microbes in enhancing plant tolerance to acidity and alkalinity stresses. Front. Sustain. Food Syst. 4:106. doi: 10.3389/fsufs.2020.00106
Muyzer, G., de Waal, E. C., and Uitterlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 59, 695–700. doi: 10.1128/aem.59.3.695-700.1993
Myrold, D. D. (2021). “Transformations of nitrogen” in Principles and applications of soil microbiology. D. M. Sylvia, J. J. Fuhrmann, P. G. Hartel, and D. A. Zuberer (Amsterdam, Netherlands: Elsevier), 385–421.
Park, J.-G., Lee, B., Heo, T.-Y., Cheon, A.-I., and Jun, H.-B. (2021). Metagenomics approach and canonical correspondence analysis of novel nitrifiers and ammonia-oxidizing archaea in full scale anaerobic-anoxic-oxic (A2/O) and oxidation ditch processes. Bioresour. Technol. 319:124205. doi: 10.1016/j.biortech.2020.124205
Pester, M., Maixner, F., Berry, D., Rattei, T., Koch, H., Lücker, S., et al. (2014). NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol. 16, 3055–71. doi: 10.1111/1462-2920.12300
Potgieter, S. C., Dai, Z., Venter, S. N., Sigudu, M., and Pinto, A. J. (2020). Microbial nitrogen metabolism in chloraminated drinking water reservoirs. Msphere 5:00274-20. doi: 10.1128/mSphere.00274-20
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rani, S., Koh, H. W., Rhee, S. K., Fujitani, H., and Park, S. J. (2017). Detection and diversity of the nitrite oxidoreductase alpha subunit (nxrA) gene of nitrospina in marine sediments. Microb Ecol. 73, 111–122. doi: 10.1007/s00248-016-0897-3
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 63, 4704–12. doi: 10.1128/aem.63.12.4704-4712.1997
Rubio-Rincón, F., Lopez-Vazquez, C., Welles, L., Van Loosdrecht, M., and Brdjanovic, D. (2017). Cooperation between Candidatus Competibacter and Candidatus Accumulibacter clade I, in denitrification and phosphate removal processes. Water Res. 120, 156–164. doi: 10.1016/j.watres.2017.05.001
Shammas, N. K., and Wang, L. K. (2009). Oxidation ditch. In Biological treatment processes, eds. L. K. Wang, N. C. Pereira, and Y.-T. HungL. (Cham: Springer Nature Press) 513–538.
Song, Y., Mhuantong, W., Liu, S.-Y., Pisutpaisal, N., Wongwilaiwalin, S., Kanokratana, P., et al. (2021). Tropical and temperate wastewater treatment plants assemble different and diverse microbiomes. Appl. Microbiol. Biotechnol. 105, 853–867. doi: 10.1007/s00253-020-11082-0
Song, L., and Niu, S. (2022). Increased soil microbial AOB amoA and narG abundances sustain long-term positive responses of nitrification and denitrification to N deposition. Soil Biol. Biochem. 166:108539. doi: 10.1016/j.soilbio.2021.108539
Stamou, A. I. (1997). Modelling of oxidation ditches using an open channel flow 1-D advection-dispersion equation and ASM1 process description. Water Sci. Technol. 36, 269–276. doi: 10.2166/wst.1997.0214
Thobejane, M. P., van Blerk, N., and Welz, P. J. (2023). Influence of seasonality, wastewater treatment plant process, geographical location and environmental parameters on bacterial community selection in activated sludge wastewater treatment plants treating municipal sewage in South Africa. Environ. Res. 222:115394. doi: 10.1016/j.envres.2023.115394
Throbäck, I. N., Enwall, K., Jarvis, A., and Hallin, S. (2004). Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol. 49, 401–17. doi: 10.1016/j.femsec.2004.04.011
Wang, H., Bagnoud, A., Ponce-Toledo, R. I., Kerou, M., Weil, M., Schleper, C., et al. (2021). Linking 16S rRNA gene classification to amoA gene taxonomy reveals environmental distribution of ammonia-oxidizing archaeal clades in peatland soils. Msystems 6:00546-21. doi: 10.1128/msystems.00546-21
Wang, C., Liu, Y., Lv, W., Xia, S., Han, J., Wang, Z., et al. (2019). Enhancement of nitrogen removal by supplementing fluidized-carriers into the aerobic tank in a full-scale A2/O system. Sci. Total Environ. 660, 817–825. doi: 10.1016/j.scitotenv.2019.01.046
Wei, W., Isobe, K., Nishizawa, T., Zhu, L., Shiratori, Y., Ohte, N., et al. (2015). Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. 9, 1954–1965. doi: 10.1038/ismej.2015.9
Wu, L., Ning, D., Zhang, B., Li, Y., Zhang, P., Shan, X., et al. (2019). Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiol. 4, 1183–1195. doi: 10.1038/s41564-019-0426-5
Xu, S., Bernards, M., and Hu, Z. (2014). Evaluation of anaerobic/anoxic/oxic (A2/O) and reverse A2/O processes in biological nutrient removal. Water Environ. Res. 86, 2186–2193. doi: 10.2175/106143014X14062131178394
Xu, S.-Q., Qian, X.-X., Jiang, Y.-H., Qin, Y.-L., Zhang, F.-Y., Zhang, K.-Y., et al. (2022). Genetic foundations of direct ammonia oxidation (Dirammox) to N2 and MocR-like transcriptional regulator DnfR in Alcaligenes faecalis strain JQ135. Appl. Environ. Microbiol. 88, e02261–e02221. doi: 10.1128/aem.02261-21
Yang, Y., Li, M., Li, H., Li, X.-Y., Lin, J.-G., Denecke, M., et al. (2020). Specific and effective detection of anammox bacteria using PCR primers targeting the 16S rRNA gene and functional genes. Sci. Total Environ. 734:139387. doi: 10.1016/j.scitotenv.2020.139387
Yang, Y., Liu, H., and Lv, J. (2022). Response of N2O emission and denitrification genes to different inorganic and organic amendments. Sci. Rep. 12:3940. doi: 10.1038/s41598-022-07753-9
Yang, L., Mei, K., Liu, X., Wu, L., Zhang, M., Xu, J., et al. (2013). Spatial distribution and source apportionment of water pollution in different administrative zones of Wen-Rui-tang (WRT) river watershed, China. Environ. Sci. Pollut. Res. 20, 5341–5352. doi: 10.1007/s11356-013-1536-x
Zhang, Q., Yang, W., Ngo, H., Guo, W., Jin, P., Dzakpasu, M., et al. (2016). Current status of urban wastewater treatment plants in China. Environ. Int. 92, 11–22. doi: 10.1016/j.envint.2016.03.024
Zhang, X., Zha, L., Jiang, P., Wang, X., Lu, K., He, S., et al. (2019). Comparative study on nitrogen removal and functional genes response between surface flow constructed wetland and floating treatment wetland planted with Iris pseudacorus. Environ. Sci. Pollut. Res. 26, 23696–23706. doi: 10.1007/s11356-019-05580-6
Zhao, W., Bi, X., Bai, M., and Wang, Y. (2023). Research advances of ammonia oxidation microorganisms in wastewater: metabolic characteristics, microbial community, influencing factors and process applications. Bioprocess Biosyst. Eng. 46, 621–633. doi: 10.1007/s00449-023-02866-5
Zheng, C., He, T., Wang, C., Zhang, M., Yang, L., and Yang, L. (2024). Key enzymes, functional genes, and metabolic pathways of the nitrogen removal-related microorganisms. Crit. Rev. Environ. Sci. Technol. 54, 1672–1691. doi: 10.1080/10643389.2024.2350317
Zheng, M., Li, S., Ni, G., Xia, J., Hu, S., Yuan, Z., et al. (2020). Critical factors facilitating candidatus nitrotoga to be prevalent nitrite-oxidizing bacteria in activated sludge. Environ. Sci. Technol. 54, 15414–15423. doi: 10.1021/acs.est.0c04192
Zhou, X., Guo, X., Han, Y., Liu, J., Ren, J., Wang, Y., et al. (2012). Enhancing nitrogen removal in an Orbal oxidation ditch by optimization of oxygen supply: practice in a full-scale municipal wastewater treatment plant. Bioprocess Biosyst. Eng. 35, 1097–1105. doi: 10.1007/s00449-012-0694-4
Keywords: anaerobic–anoxic–oxic system, deoxidation ditch process, nitrogen metabolism genes, seasonal variation, wastewater treatment
Citation: Liu S, Liu Y, Ye L, Xiao E, Xu D, Chao H, Dai J and Qiu D (2024) Comparative analyses on nitrogen removal microbes and functional genes within anaerobic–anoxic–oxic and deoxidation ditch sewage-treating processes in Wuhan and Xi’an cities, China. Front. Microbiol. 15:1498681. doi: 10.3389/fmicb.2024.1498681
Edited by:
Nan Wu, Tianjin Agricultural University, ChinaReviewed by:
Zhe Lu, South China Botanical Garden, ChinaQingwei Lin, Henan Normal University, China
Copyright © 2024 Liu, Liu, Ye, Xiao, Xu, Chao, Dai and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingcheng Dai, ZGFpamluZ2NoZW5nMTIzQDE2My5jb20=; Dongru Qiu, cWl1QGloYi5hYy5jbg==
 Shuangyuan Liu
Shuangyuan Liu Yaqi Liu2
Yaqi Liu2 Jingcheng Dai
Jingcheng Dai Dongru Qiu
Dongru Qiu