- 1State Key Laboratory of Animal Nutrition and Feeding, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 2School of Agriculture and Food Science, University College Dublin, Dublin, Ireland
- 3Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences, Beijing, China
- 4National Center of Technology Innovation for Dairy, Inner Mongolia, China
Ruminants excrete both undigested and excess consumed nitrogen (N) through feces and urine, which results in the waste of feed protein and environmental pollution. This study aims to screen bacteria to identify strains with potential to improve nitrogen utilization in the rumen. In a tube screening test, when 30 mmol/L ammonium sulfate was added as the only nitrogen source in liquid medium, among 115 bacterial strains belonging to Bacillus and lactic-acid bacteria, 10 strains with the highest growth (OD600nm level) in comparison to the other evaluated strains were identified, and of these, Ligilactobacillus agilis W70 and Limosilactobacillus fermentum M50, had the highest ammonia nitrogen utilizing capabilities of ammonium sulfate (32.09 and 40.64%, respectively). A subsequent in vitro fermentation experiment was conducted with the diet consisting of 0.5 g TMR, 50 mL buffer solution, and 25 mL fresh rumen fluid per serum bottle which was incubated at 39°C for 24 h. The experiment consisted of 3 treatments, CTL, L. agilis W70 (1 × 109 cfu/mL), and L. fermentum M50 (1 × 109 cfu/mL), each treatment had 6 replicates, repeated across 3 batches. The addition of L. agilis W70 decreased NH3N (p < 0.01), the ratio of acetate to propionate (A:P) (p < 0.01), and increased the yields of microbial protein (MCP) (p < 0.01) and enzymatic activities of glutamate dehydrogenase (GDH) (p < 0.05) and glutamine synthetase (GS) (p < 0.05). Further genome analysis revealed that the L. agilis W70 harbors the glutamate dehydrogenase and glutamine synthetase-encoding genes gdhA and glnA that play key roles in ammonia utilization. This study identified L. agilis W70 as a strain which exhibits high NH3N utilization capability and enhances ammonia assimilation in vitro. The strain was further characterized to elucidate this ammonia assimilation potential. Further studies will be conducted to develop strain L. agilis W70 as a new feed additive to improve the nitrogen utilization efficiency in ruminant animals.
1 Introduction
Ruminants have low nitrogen utilization efficiency of around 25% due to their unique digestive tract structure (Bach et al., 2005; Lavery and Ferris, 2021), where rumen microorganisms play a role in nitrogen metabolism (Liu et al., 2023), with some fermented protein converted into ammonia (Eschenlauer et al., 2002). If unutilized this ammonia excreted in urine and feces leads to both a waste of protein resources and environmental pollution, with nitrogen in urine being especially harmful due to its high potential for volatilization (Qi et al., 2023).
Supplementation of direct fed microbials (DFMs) in feed is an environmentally friendly, cost-effective, and biologically safe practice to reduce ruminal ammonia emissions and to increase nitrogen utilization efficiency (Callaway et al., 2023). Moreover, DFMs have potential to stabilize ruminal microbiota, improve nitrogen flow to the lower digestive tract, and increase milk protein yield (Karimi, 2022). Bacillus species are non-toxic, spore-forming bacteria that can colonize the intestinal tract; they are aerobic and well-suited to maintain viability during industrial production and storage (Cappellozza et al., 2023). Similarly, lactic acid bacteria (LAB) are non-toxic, produce lactic acid to help regulate rumen pH, and are facultative anaerobes, making them highly compatible with production and storage requirements (Kesen and Aiyegoro, 2018). Among them, ammonia utilizing bacteria assimilate ammonia nitrogen as a nitrogen source to produce microbial protein (MCP) in the rumen, (Zurak et al., 2023), which can be transformed to amino acids in the small intestine and transferred to the mammary gland to produce milk protein (Griswold et al., 2003). In view of the importance of improving animal productivity, it is a priority to screen and identify ammonia utilizing bacteria to regulate rumen fermentation to reduce rumen ammonia nitrogen emissions and enhance MCP production.
Studies on the isolation and functional characterization of rumen-originated ammonia nitrogen utilizing bacteria so far have been scarce. The advantage of this study is that our previous research cultured many rumen-derived strains using the culturomics method. From these, we intend to screen ammonia utilizing strains in this study, ensuring they are well-adapted to the rumen environment (Li et al., 2022). Screening ammonia-utilizing bacteria with ammonium sulfate as the sole nitrogen source is effective because it ensures that only bacteria proficient in ammonia metabolism will grow (Liu et al., 2020). In vitro studies offer a controlled setting to assess the impact of DFMs on rumen fermentation, which is crucial for evaluating their efficacy before applying them in vivo (Mi et al., 2019). Additionally, genetic analysis can offer insights into the specific genes and metabolic pathways involved in ammonia assimilation (Kim et al., 2014). This study aimed to screen rumen bacteria for higher ammonia nitrogen utilizing capability, evaluate their potential to reduce ammonia nitrogen emission and improve MCP production in the rumen, and explore potential genomic mechanisms of the identified ammonia nitrogen utilizing bacteria.
2 Materials and methods
2.1 Screening and evaluation of rumen bacteria in medium
2.1.1 Primary screening test
One hundred and fifteen strains derived from rumen contents, belonging to Bacillus and lactic-acid bacteria (Ligilactobacillus, Limosilactobacillus, Lacticaseibacillus, Weissella, Pediococcus, and Enterococcus) identified previously (Duan et al., 2023) and conserved in the Ruminant Nutrition Laboratory in the Institute of Animal Science, Chinese Academy of Agricultural Sciences were selected for the primary screening test. Of these, 26 strains grown under facultative anaerobic gas conditions and 89 strains grown in strictly anaerobic gas conditions were inoculated at the ratio of 10% (v/v) into a 50 mL flask containing 15 mL liquid medium (modified MRS for lactic-acid bacteria and modified LB for Bacillus respectively). For facultative anaerobic gas conditions, the liquid medium was dispensed into screw-cap tubes sealed with butyl rubber stoppers, autoclaved, inoculated with bacteria, and then cultivated in a microbiological incubator under static conditions. For strictly anaerobic gas condition, the liquid medium was dispensed into screw-cap tubes sealed with butyl rubber stoppers, deoxygenated by gassing with 100% N2, autoclaved, inoculated with bacteria, and then cultivated in an anaerobic workstation under static conditions. The gas composition of the anaerobic workstation (Thermo 1,029, Waltham, USA) consisted of 85% N2, 10% H2, and 5% CO2. An oxygen indicator pack (Mitsubishi Gas Chemical Company, Tokyo, Japan) was used to monitor oxygen levels in the anaerobic workstation, displaying blue at oxygen levels above 0.5%, an intermediate pink-blue at 0.1–0.5%, and pink below 0.1%, indicating a stable anaerobic environment when consistently pink. MRS medium contains the following components: 10 g/L peptone, 8 g/L meat extract, 4 g/L yeast extract, 20 g/L C6H12O6, 2 g/L K2HPO4, 5 g/L sodium acetate, 2 g/L triammonium citrate, 0.2 g/L MgSO4 7H2O, 0.05 g/L MnSO4 4H2O, and 1 mL/L Tween 80. The modified MRS medium was prepared by replacing the nitrogen sources—peptone, meat extract, and yeast extract—with ammonium sulfate as the sole nitrogen source. The modified LB medium was prepared following the method of Liu et al. (2020), also using ammonium sulfate as the sole nitrogen source. Ammonium sulfate concentrations were set at 5, 10, 15, 20, 25, and 30 mmol/L. The strains were incubated at 39°C for 72 h, and the optical density (OD) values at 600 nm were determined.
2.1.2 Comparison of ammonia utilizing capability
Based on the screening results of the primary test, 10 strains with the highest OD600nm levels on 30 mmol/L ammonium sulfate were selected for the secondary screening test. The bacteria were inoculated and cultured as described above. The OD600nm was monitored at 0 h, 24 h, 48 h, and 72 h. In addition, the NH3N contents in the supernatant and MCP yields were determined using an automated phenol-hypochlorite method (Broderick and Kang, 1980) and Folin phenol method (Makkar et al., 1982) respectively.
2.1.3 Strain growth curve and microbial protein synthesis under various ammonium sulfate concentrations
Based on the results of the secondary screening test, Ligilactobacillus agilis W70 and Limosillactobacillus fermentum M50 were cultured in modified MRS medium containing 5, 10, 15, 20, 25, 30 mmol/L ammonium sulfate as the sole nitrogen source for 72 h. The OD600nm were determined at 0 h, 24 h, 48 h, and 72 h, and the NH3N contents and MCP yields were determined at 0 h and 72 h as described above.
2.1.4 Primary screening of cellulase, amylase, protease, and lipase activities
The W70 and M50 strains were cultured separately on carboxymethyl cellulose (CMC) agar, starch agar, skim milk agar, and Tween 80 agar media to assess their cellulolytic, amylolytic, proteolytic, and lipolytic activities, respectively, for 72 h at 39°C (Masi et al., 2023). The concentration and enzymatic activity produced were indicated by the diameter of the hydrolysis zone in each specific medium.
2.2 Evaluation of bacteria using in vitro rumen fermentation
2.2.1 Preparation of rumen fluid solution and diets
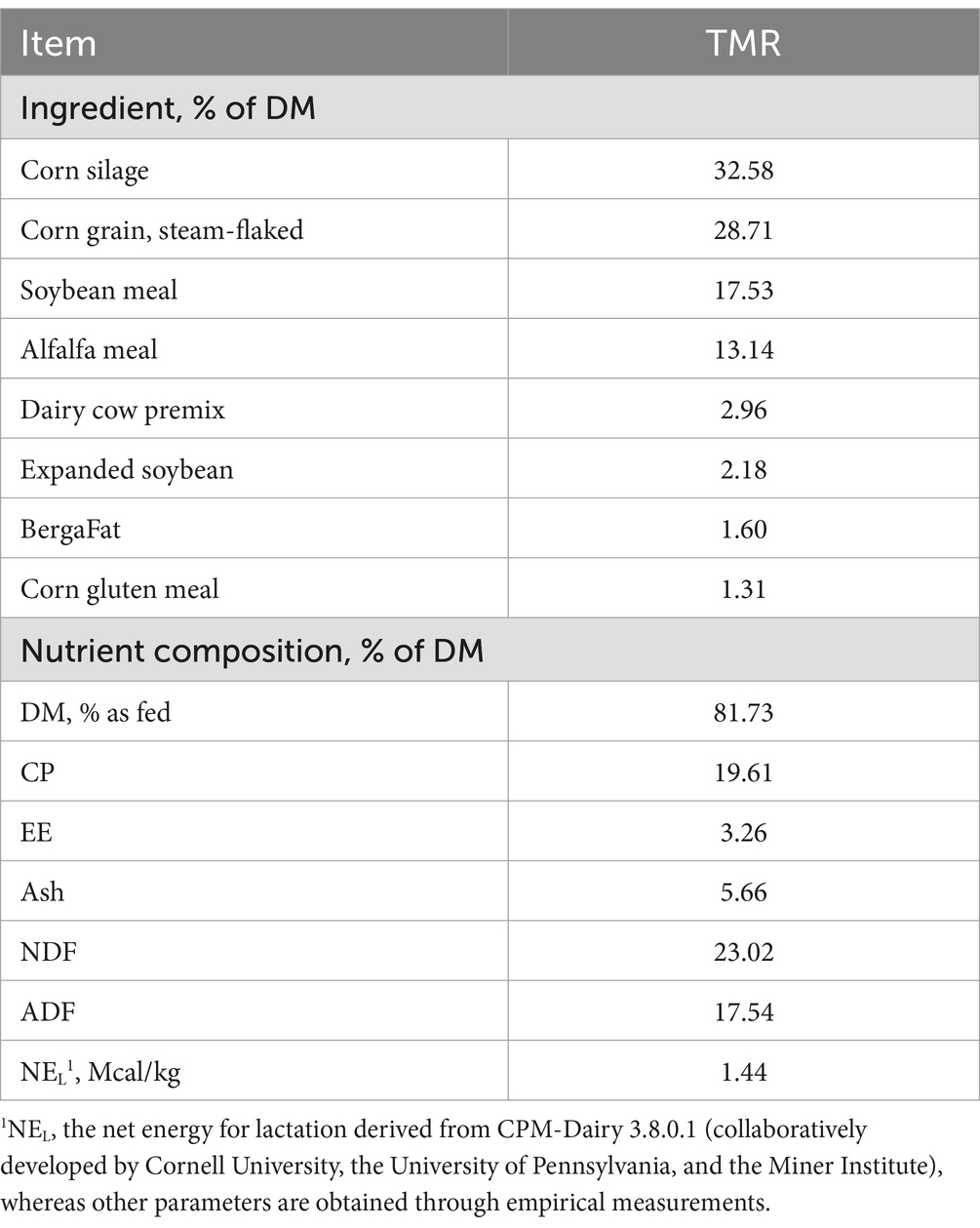
All animal procedures were approved by the Institutional Animal Care and Use Committee of the Institute of Animal Science, Chinese Academy of Agricultural Sciences and complied with the protocol IAS2022-91. Three lactating Holstein cows with the same parity, health condition, and fitted with a permanent rumen fistula were used as rumen fluid donors. The cows were fed total mixed ration (TMR) twice a day (7:00 and 19:00) and drank freely. The Total Mixed Ration (TMR) diet consisting of corn silage (57.61%), alfalfa meal (12.39%), steam-flaked corn grain (8.91%), grind corn grain (7.61%), soybean meal (6.74%), soybean hulls (2.61%), VitTM premix (1.41%), corn gluten meal (1.09%), molasses (0.65%), cottonseed (0.43%), sodium bicarbonate (0.33%), and Bergafat (0.22%). The nutrient composition of the diet is as follows: DM 43.1% as fed, CP 14.7% DM, Ash 5.5% DM, ADF 22.5% DM, NDF 37.2% DM, NEL 1.74 Mcal/kg. The rumen fluid was collected 2 h after morning feeding, mixed and put into an autoclaved thermos bottle, and brought back to the laboratory. Half a gram of TMR was put into each serum bottle as a fermentation substrate. The ingredient and chemical composition of the diets in vitro rumen fermentation are shown in Table 1. Under anaerobic and sterile conditions, 25 mL rumen fluid was filtered through four layers of cheesecloth, and then diluted with 50 mL of modified Menke and Steingass buffer solution (Menke, 1988).
2.2.2 In vitro rumen fermentation
Inocula of the W70 and M50 strains were prepared in the form of freeze-dried powder and added to serum bottles at a final concentration of 1 × 109 cfu/g and cultured at 39°C for 24 h. The experiment contained three treatment groups: CTL group (control), Group W70 (1 × 109 cfu/g W70), and Group M50 (1 × 109 cfu/g M50). The choice of 1 × 109 cfu for in vitro rumen fermentation was based on prior literature regarding the effects of direct fed microbials (DFM) on in vitro fermentation (Opsi et al., 2012; Cappellozza et al., 2023; Fitriyah et al., 2024). The in vitro fermentation was conducted using 6 replicates per treatment and the experiment was repeated 3 times giving 18 replicates per treatment in total.
2.2.3 Sample collection and analysis
After 24 h fermentation, the gas volume of each serum bottle was measured, and the pH values of fermentation mixtures were determined immediately after uncapping. The fermentation mixtures were then collected and filtered through nylon bags. For NH3N and volatile fatty acid (VFA) analyses, 25% meta-phosphoric acid was added to the fermentation supernatant at the ratio of 20% (v/v), centrifuged at 10,000 × g at 4°C for 10 min, and stored at −20°C. The VFA contents were determined by gas chromatography as described previously (Stewart and Duncan, 1985). The NH3N contents and MCP yields were determined as described above. Enzymatic activities of glutamate dehydrogenase (GDH) and glutamine synthetase (GS) were assessed colourimetrically using an assay kit (Beijing Boxbio Science & Technology Co. Ltd., Beijing, China) according to the manufacturer’s protocol. The residue DM (dry matter) was estimated after drying and compared with initial DM to calculate the dry matter digestibility (DMD) as described previously (Van Soest and Robertson, 1985).
2.3 Phenotype and genome analysis of Ligilactobacillus agilis W70
2.3.1 Phenotypic characterization and identification
Gram staining of the W70 strain was performed using the modified Hucker’s method (Hucker and Conn, 1923). Plate streaking of the W70 strain was performed on an MRS agar plate. The growth of the W70 strain under facultative anaerobic conditions at 39°C was monitored for 48 h. To confirm the identity of the W70 strain, genomic DNA was extracted using a Bacterial DNA kit (Solarbio, Beijing, China) according to the manufacturer’s instructions. The 16S rRNA gene fragment of the W70 strain was amplified from genomic DNA using the universal primers 27F (5’-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3′) (Frank et al., 2008). The PCR reaction was performed with a MyCycler Thermal Cycler (Bio-Rad, Hercules, CA, USA) in a volume of 20 μL containing 10 μL of 2 × TaqPCR StarMix (Genstar, Beijing, China), 8.2 μL of ddH2O, 1 μL of bacterial DNA template, and 0.4 μL of each primer. The cycling conditions were as follows: 2 min at 94°C and 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. The PCR products were sequenced using a Sanger sequencer (Applied Biosystems 3730XL, Foster City, CA, USA). The 16S rRNA sequence of the W70 strain was aligned with those of Ligilactobacillus type species using Clustal W, and a phylogenetic analysis was performed using the maximum likelihood method in MEGA 61 (Li et al., 2020). The ribosomal protein of the W70 strain was extracted in 1 mL of lysis buffer (70% formic acid) and 1 mL of matrix solution (saturated α-cyano acid-4-hydroxycinnamic in 50% acetonitrile and 2.5% trifluoroacetic acid) and identified by MALDI-TOF MS using the EXS3000 system (Zybio Inc., Chongqing, China). The spectrum was compared with those of known bacteria in the database. Isolates were accurately identified to the species and genus levels with the score values of greater than 1.9 and 1.7–1.9.
2.3.2 Genomic DNA extraction
A single colony of the W70 strain on the MRS agar plate was inoculated into 2 mL of MRS medium and grown at 39°C for approximately 15 h (maximum growth). The cell biomass was harvested by 10 min centrifugation at 12,000 × g. Genomic DNA was then extracted using the Bacterial/fungal DNA extraction kit (Majorbio, Shanghai, China) according to the manufacturer’s protocol. Genomic DNA was gel-purified and quantified for further study.
2.3.3 Library construction and genome sequencing
The whole genome sequencing and assembly were carried out by the Majorbio Bio-pharm Technology Co., Ltd., Shanghai, China. Genomic DNA was sequenced using the combination of PacBio Sequel IIe and Illumina sequencing platforms. For Illumina sequencing, genomic DNA was sheared into 400–500 bp fragments using a Covaris M220 Focused Acoustic Shearer following the manufacturer’s protocol. Illumina sequencing libraries were prepared from the sheared fragments using the NEXTFLEX Rapid DNA-Seq Kit. Briefly, the 5′ prime ends were first end-repaired and phosphorylated; the 3′ ends were then A-tailed and ligated to sequencing adapters; and the adapters-ligated products were enriched using PCR. The libraries were paired-end sequenced (2 × 150 bp) on Illumina Novaseq 6,000. For PacBio sequencing, genomic DNA was fragmented at ~10 kb. The fragments were then purified, end-repaired, and ligated with SMRT bell sequencing adapters following manufacturer’s recommendations (Pacific Biosciences, Menlo Park, CA, USA). The PacBio library was p sequenced on one SMRT cell using standard methods.
2.3.4 Genome assembly and annotation
The raw Illumina sequencing reads generated from the paired-end library were subjected to quality-filtering using fastp v0.23.0. High-fidelity (HiFi) sequencing reads were generated from the PacBio platform for further analysis. Clean short reads and HiFi reads were assembled to construct the complete genome using Unicycle v0.4.8 (Wick et al., 2017) and Pilon v1.22 to polish the assembly using short-read alignments, reducing the error rate. The coding sequences (CDs) of chromosome and plasmid were predicted using the Glimmer or Prodigal v2.6.3 (Hyatt et al., 2010) and GeneMarkS (Besemer and Borodovsky, 2005) respectively. tRNA-scan-SE (v2.0) (Chan and Lowe, 2019) and Barrnap v0.92 were used for tRNA and rRNA prediction, respectively. The CDs were annotated from NR, Swiss-Prot, Pfam, GO, COG, and KEGG database using sequence alignment tools such as BLAST, Diamond and HMMER. Briefly, each set of query proteins were aligned with the databases, and annotations of best-matched subjects (e-value <10−5) were obtained for gene annotation. Biosynthetic gene clusters (BGCs) of secondary metabolites were identified by the antiSMASH v5.1.2.
2.3.5 The homology models of deduced protein of gdhA and glnA
The deduced protein sequences of gdhA and glnA were blasted against the databases of Swiss-prot (Uniprot)3 and PDB4. Homology-modeled using the Swiss Model5 with the PDB templates 5GUD for gdhA and 7TDP for glnA, respectively. Alignment with protein structure-resolved homologs of >50% identities using ClustalW and identification of catalytic domains, catalytic residues, and substrate binding sites using ESPript6.
2.3.6 Gene expression analysis of gdhA and glnA by quantitative real-time PCR
To evalute the gene expression of gdhA and glnA between the different nitrogen concentrations, quantitative real-time PCR (RT-qPCR) analyses were performed. The total RNA was extracted from W70 grown in 5 mmol/L and 30 mmol/L ammonium sulfate as the sole nitrogen source modified MRS medium by SteadyPure Universal RNA Extraction Kit (Accurate Biotechnology Co. Ltd., Changsha, Hunan, China) and reverse transcribed into cDNA by using cDNA Synthesis Kit (Beijing Solarbio Science & Technology Co. Ltd., Beijing, China). Quantitative PCR was performed using SYBR Green (Thermo Fisher Scientific, Waltham, MA, USA) by using a Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) in a 10 μL mixture containing cDNA template 1 μL, forward and reverse primer 0.5 μL each, SYBR Green qPCR Mix 5 μL, and ddH2O 3.5 μL. The PCR amplification was performed under the following conditions: 95°C for 2 min, followed by 40 cycles at 94°C for 15 s, and 60°C for 60 s of annealing extension, finally at 95°C for 15 s, 60°C for 60 s and 95°C for 15 s, for dissociation curve analysis. Specific qPCR primers were designed by Primer-BLAST7 based on the gdhA and glnA sequences of W70 and synthesized by BGI, Shenzhen, China (Supplementary Table S1). The relative expression ratio of gdhA and glnA genes versus the 16S rDNA gene was calculated using the 2-ΔΔCT method, and all data were given in terms of relative mRNA expression (Liu et al., 2020).
2.4 Data analysis and availability
The OD600nm, MCP, NH3N and RT-qPCR data of 10 strains were analyzed by one-way ANOVA in SAS 9.4 (SAS Institute, Cary, NC, USA). The results are presented as histograms made using Origin (Version 8.0.0). Least square means were reported, and statistical significance was set at p < 0.05.
The data were analyzed using the PROC GLM procedure of SAS 9.4 (SAS Institute, Cary, NC, USA) to compare the means of gas, pH, total VFA, acetate, propionate, isobutyrate, butyrate, isovalerate, valerate, the ratio of acetate and propionate (A:P), DMD, NH3N, MCP, GDH, and GS in vitro rumen fermentation using the W70 and M50 strains. Treatment was set as the fixed effect and batch was set as a random effect in the model. Differences were tested by the tukey method. Statistical significance was declared at p < 0.05.
3 Results
3.1 Screening for ammonia nitrogen utilizing strains
A total of 115 rumen bacteria were initially screened for their ability to utilize ammonia nitrogen with ammonium sulfate as the sole nitrogen source. In facultative anaerobic gas conditions, Bacillus safenisis B55, Ligilactobacillus agilis W70, Bacillus safenisis B52, Bacillus safenisis F79, and Weissella confuse W26 were the strains with the highest growth (OD600nm level) in 30 mmol/L ammonium sulfate (Supplementary Figure S1). In strictly anaerobic gas conditions, Enterococcus faecium F42, Limosilactobacillus fermentum M50, Enterococcus lactis M21, Enterococcus faecium B2, and Lactobacillus salivarius X36 showed the highest growth (OD600nm level) in 30 mmol/L ammonia nitrogen (Supplementary Figure S2).
To further analyze these strain’s ammonia nitrogen utilizing capacity, a secondary screening test, using 30 mmol/L ammonium sulfate as the sole nitrogen source was conducted. L. agilis W70 and L. fermentum M50 outperformed the other four tested strains following 72 h incubation under facultative anaerobic and strictly anaerobic gas conditions, respectively, (Figures 1A,B), and showed greater capabilities to reduce ammonia nitrogen and produce MCP (Figures 1C,D).
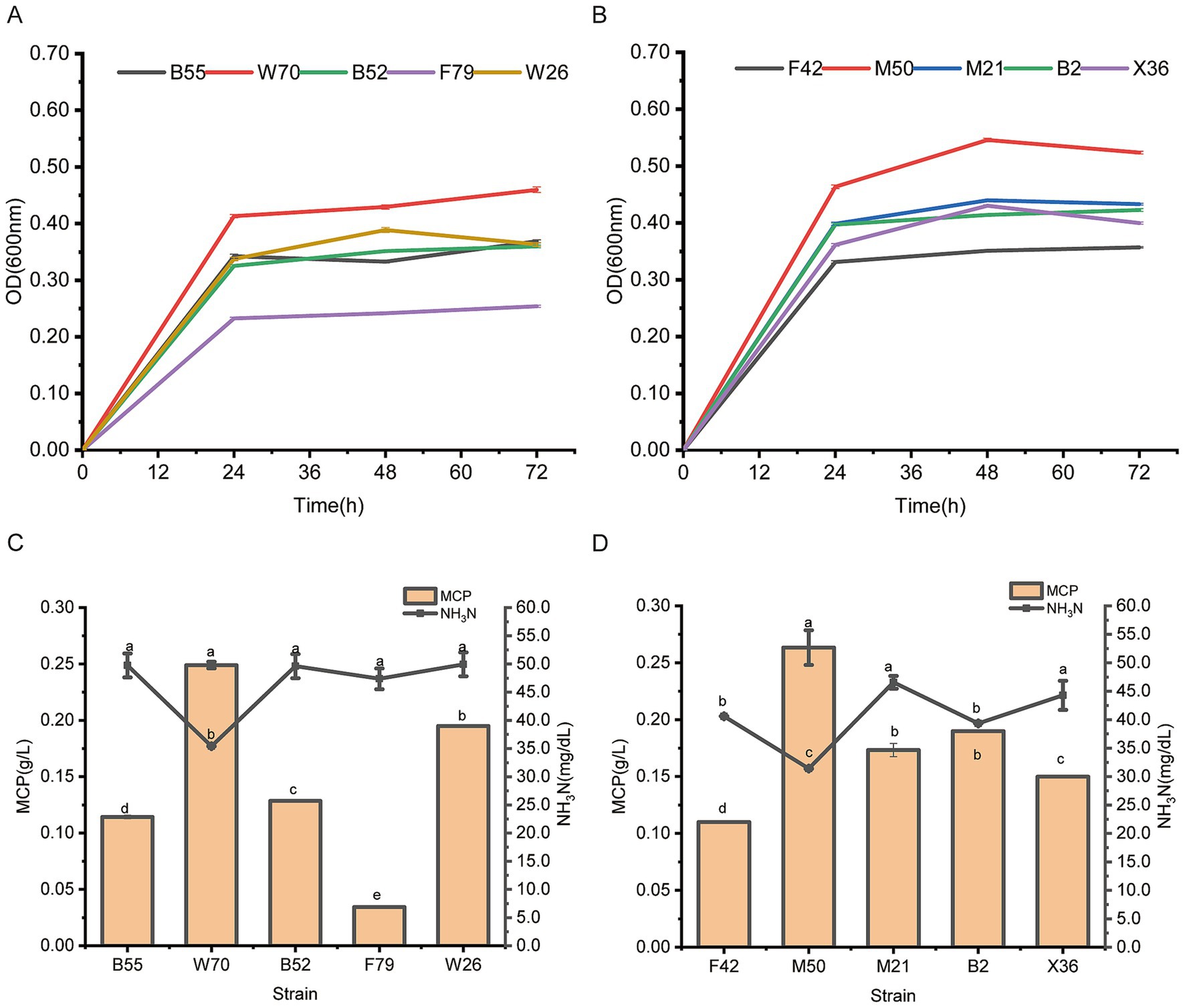
Figure 1. Growth curve and ammonia nitrogen utilizing capacities of Bacillus safenisis B55, Lactobacillus agilis W70, Bacillus safenisis B52, Bacillus safenisis F79, Weissella confuse W26, Enterococcus faecium F42, Lactobacillus fermentum M50, Enterococcus lactis M21, Enterococcus faecium B2, and Lactobacillus salivarius X36 with 30 mmol/L ammonium sulfate as the sole nitrogen source. (A) Growth curves of five strains under facultative anaerobic conditions. (B) Growth curves of five strains under strictly anaerobic conditions. (C) Ammonia nitrogen utilizing capacities of five strains under facultative anaerobic conditions. (D) Ammonia nitrogen utilizing capacities of five strains under strictly anaerobic conditions. Data are shown as mean ± standard deviation of three experiments. Different letters refer to statistically significant differences at p < 0.05.
Testing different ammonium sulfate concentrations helps assess the strains’ stability and adaptability under varying nitrogen source, ensuring that the selected strains exhibit consistent and reliable performance in vitro and in vivo. When ammonium sulfate was provided as the sole nitrogen sources at concentrations of 5, 10, 15, 20, 25, and 30 mmol/L, the W70 and M50 strains exhibited higher growth (OD600nm level), indicating their tolerance to high levels of ammonium sulfate, which is a necessary prerequisite (Figures 2A,B). With the increase of ammonium sulfate concentration, the W70 and M50 strains displayed a trend of increased MCP. The ammonia nitrogen concentrations of 25 mmol/L and 30 mmol/L were optimal for the MCP production of the W70 and M50 strains, respectively, suggesting that the strain requires a certain nitrogen concentration to support effective utilization of ammonia nitrogen (Figures 2C,D). When 30 mmol/L of ammonium sulfate was used as the sole nitrogen source, the W70 strain showed the highest ammonia nitrogen utilization, with ammonium sulfate levels decreasing from 52.13 mg/dL at 0 h to 35.4 mg/dL at 72 h, a 32.09% reduction (Figure 2E). Similarly, strain M50 demonstrated a strong capacity for ammonia nitrogen utilization at 30 mmol/L of ammonium sulfate, with concentrations dropping from 52.88 mg/dL to 31.39 mg/dL over 72 h, resulting in a 40.64% reduction (Figure 2F). The ammonia consumption rates of W70 and M50 were 23.24 mg/L/h and 29.85 mg/L/h, respectively, with 30 mmol/L ammonium sulfate as the sole nitrogen source. Correspondingly, the bacterial growth rates were 0.10 h−1 and 0.10 h−1 under the same conditions.
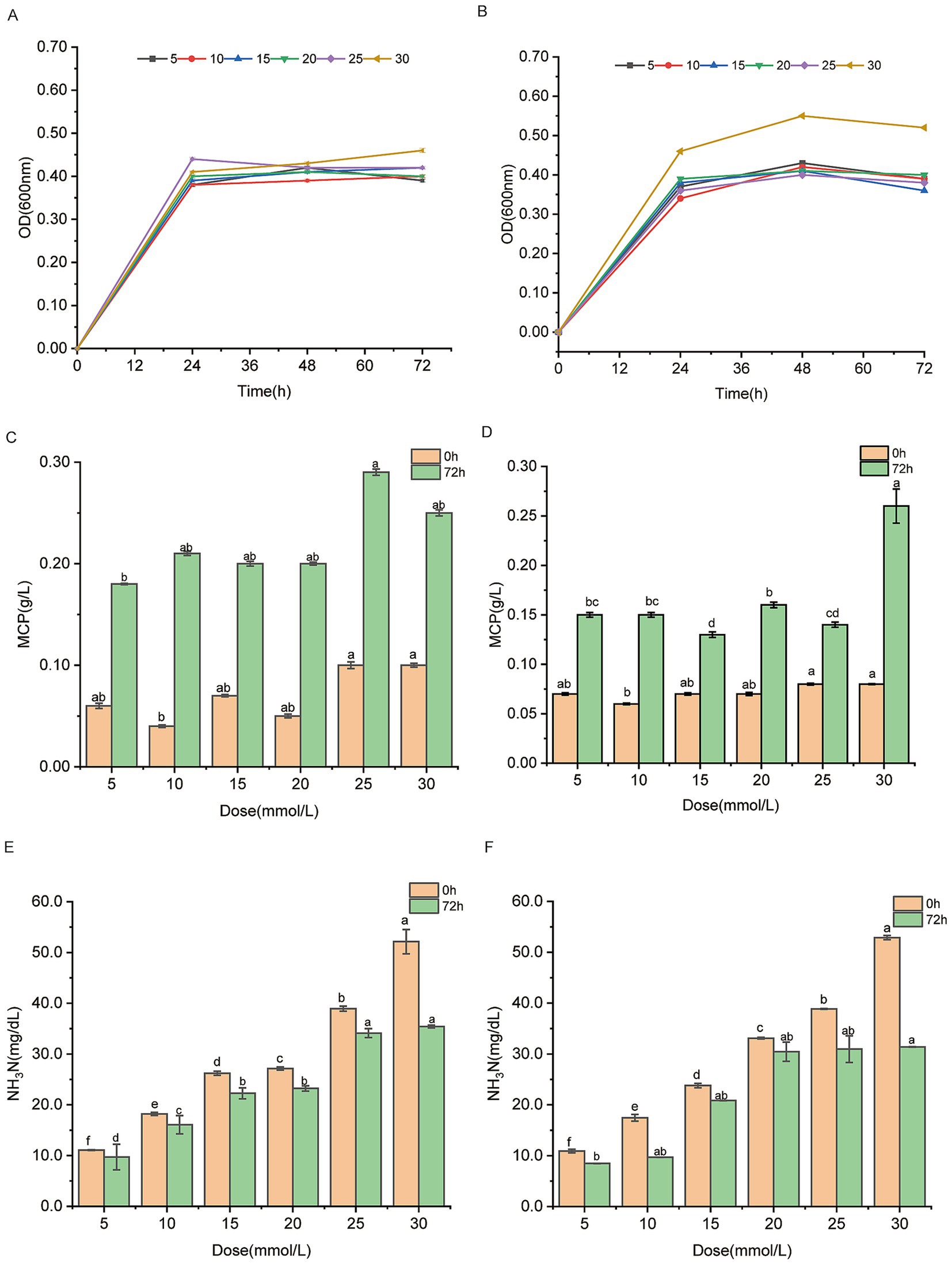
Figure 2. Ammonia nitrogen utilizing capacities of Lactobacillus agilis W70 and Lactobacillus fermentum M50 with 5, 10, 15, 20, 25, 30 mmol/L ammonium sulfate as the sole nitrogen source. (A,B) Growth curves over 72 h. (C,D) MCP yields at 0 and 72 h. (E,F) Ammonia nitrogen utilizing capacities at 0 and 72 h. Data are shown as mean ± standard deviation of three experiments. Different letters refer to statistically significant differences at p < 0.05.
The ability of the identified strains to degrade cellulose, starch, protein, and lipid was tested using cellulase, amylase, protease, and lipase activity plates. The results reveal that neither the W70 nor the W50 strain produced enzymes to degrade these substrates.
3.2 In vitro rumen fermentation
Facultative anaerobic L. agilis W70 and strictly anaerobic L. fermentum M50 were selected for the ruminal fermentation experiment. Supplementation of 1 × 109 cfu/g of W70 and M50 had no effects on the pH, gas volume, and DMD (Table 2). In comparison with the CTL group, supplementation with the W70 strain significantly decreased the NH3N concentration (p < 0.01) and A:P ratio (p < 0.01), and increased the yields of total VFA (p < 0.05), acetate (p < 0.05), propionate (p < 0.05), isobutyrate (p < 0.01), butyrate (p < 0.01), isovalerate (p < 0.01), valerate (p < 0.01), MCP (p < 0.01), GDH (p < 0.05), and GS (p < 0.05). Supplementation with M50 strain increased acetate and MCP yields compared to the CTL group, without affecting NH3N levels. In combination with the screening and in vitro fermentation results, the W70 strain has more potential for NH3N utilization, total VFA/MCP production, and enzyme GDH/GS production. Therefore, this strain was characterized in further detail.
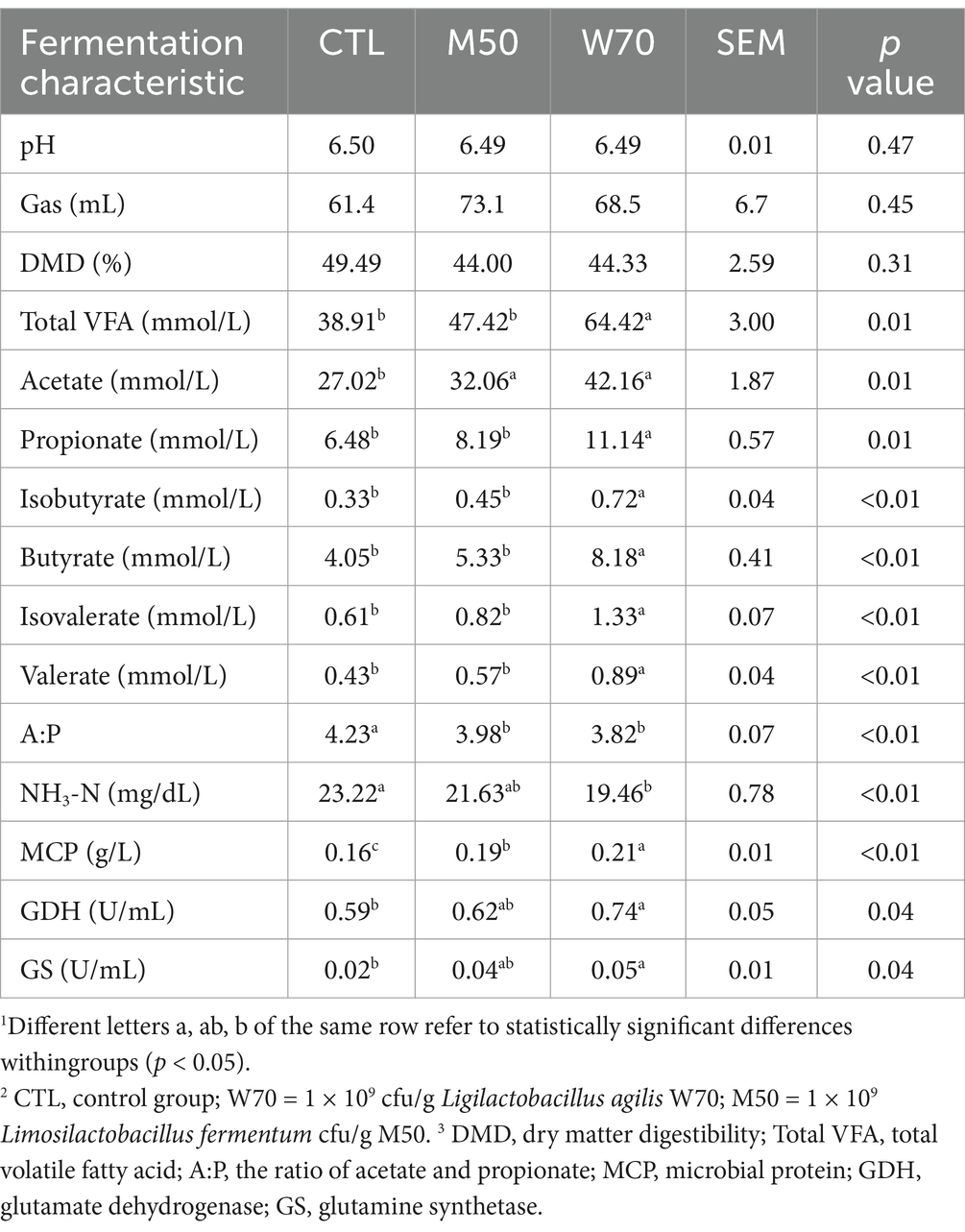
Table 2. Effect of L. agilis W70 and L. fermentum M50 on rumen fermentation in vitro1.
3.3 Phenotypic and genetic analysis of Ligilactobacillus agilis W70
The W70 strain, identified as a gram-positive bacterium following gram staining, has rod-shaped cells measuring 0.14–0.57 μm × 1.03–1.90 μm (width × length) (Figures 3A,B). The cells were found singly or in pairs as shown by gram staining (Figure 3A). In the MRS liquid medium, the W70 strain cultured under facultative anaerobic condition at 39°C for 48 h showed an OD600 nm maximum of 1.10 ± 0.01 after incubation for 15 h (Figure 3C). Based on the partial sequences of 16S rRNA gene and spectra of ribosomal proteins, the W70 strain was phylogenetically related to the genus Ligilactobacillus (Figures 3D,E).
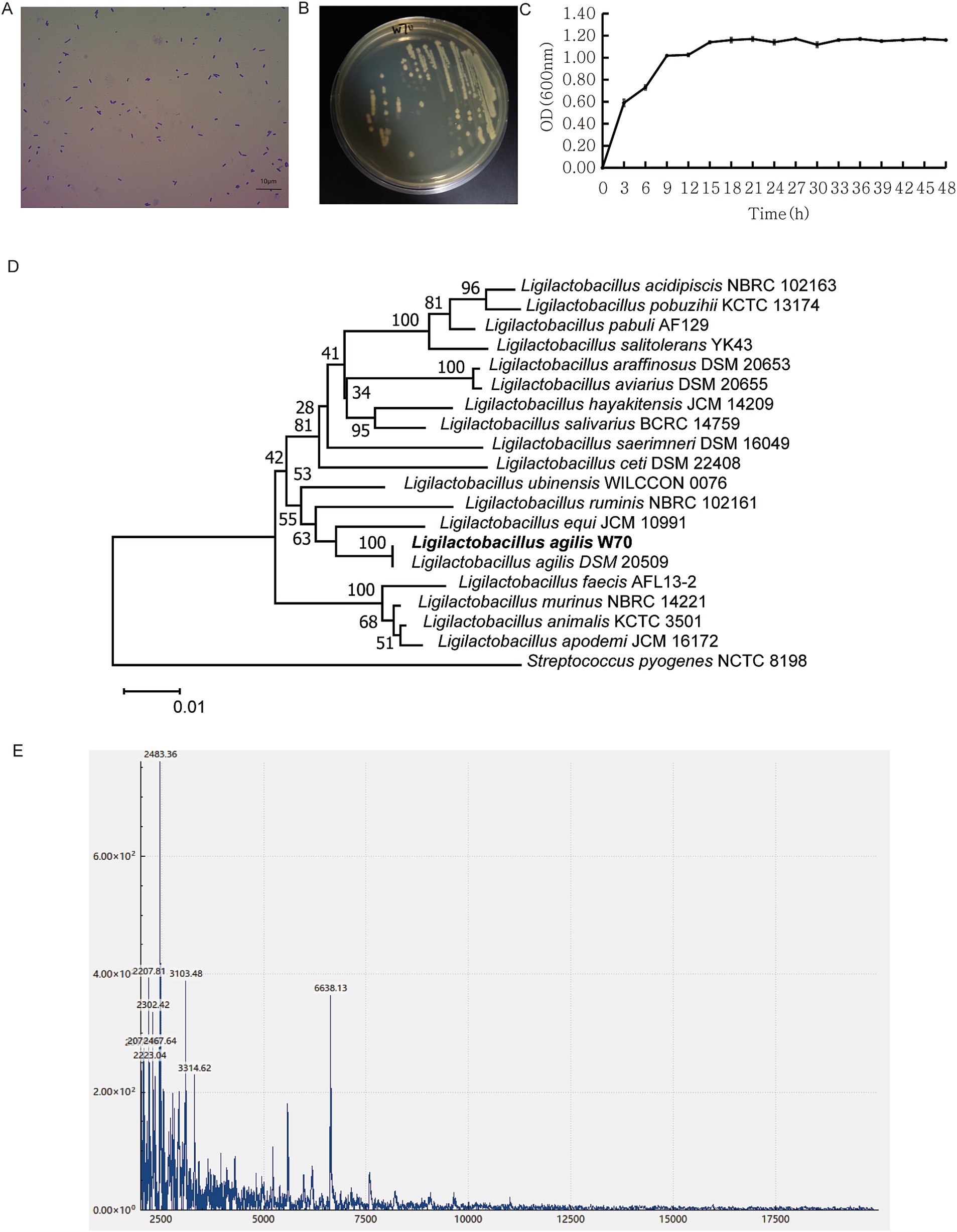
Figure 3. Morphological and physiochemical characters of Ligilactobacillus agilis W70. (A) Gram staining. (B) Colonies on MRS plate at 39°C. (C) Growth curve over 48 h in MRS broth at 39°C. Data are shown as mean ± standard deviation of triplicate. (D) Phylogenetic tree of the genus Ligilactobacillus constructed using the neighbor joining method based on the partial sequence of 16S rRNA gene. (E) MALDI-TOF MS profile of the ribosomal protein.
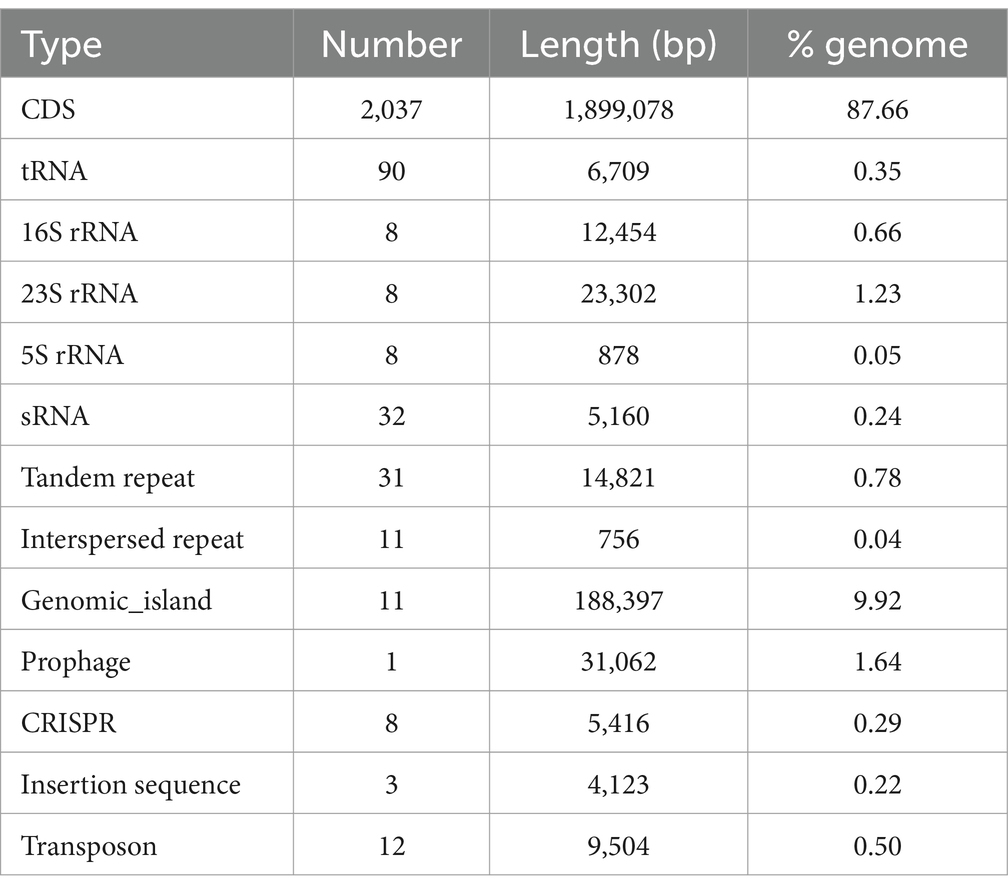
The genome of the W70 strain is 2.17 Mb in size, with a G + C content of 41.7% (Figure 4A), and contains 2037 coding sequences, 90 tRNAs, 24 rRNA genes (8 copies of each 5S rRNA, 16S rRNA, and 23S rRNA), 32 sRNAs, 31 tandem repeats, 11 interspersed repeats, 11 gene islands, 1 prophage, 8 CRISPR repeats, 3 insertion sequences, and 12 transposons (Table 3). KEGG analysis indicated that most of the annotated genes are involved in carbohydrate metabolism (138 genes), membrane transport (109 genes), amino acid metabolism (89 genes), and metabolism of other amino acids (29 genes) (Figure 4B). GO analysis identified 834 genes related to molecular function (MF), 616 genes related to biological processes (BP), and 557 genes related to cellular components (CC) (Supplementary Figure S3A). COG analysis classified the putative proteins in three main groups, translation, ribosomal structure and biogenesis (193), carbohydrate transport and metabolism (172 genes), and amino acid transport and metabolism (144) (Supplementary Figure S3B).
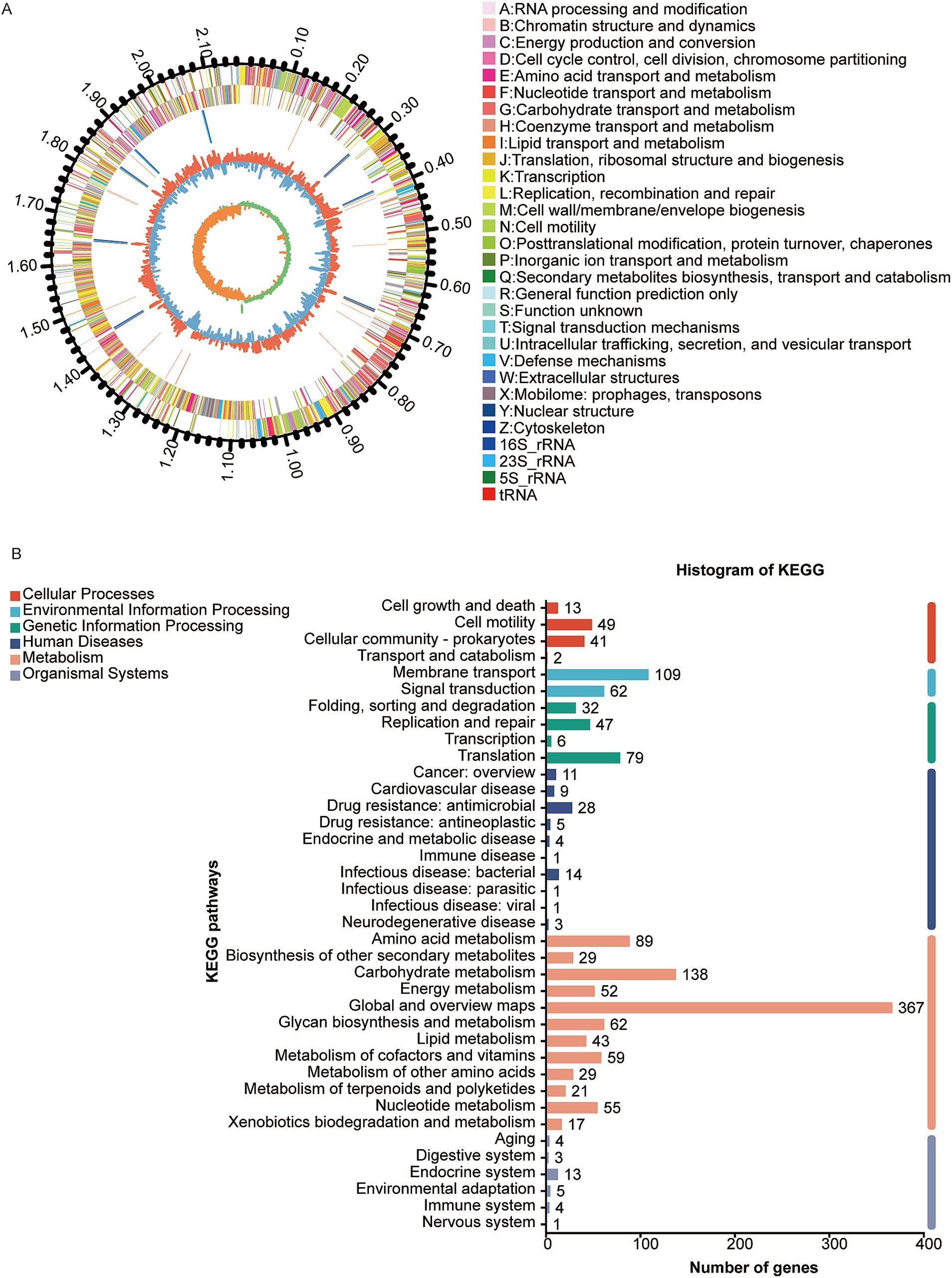
Figure 4. Genome analysis of Ligilactobacillus agilis W70. (A) Circle diagram of the nuclear genome. From the outside to the inside are the genome size, coding genes (sense), coding genes (antisense), tRNAs (red) and rRNAs (5S rRNA,16S rRNA and 23S rRNA,), prophage (purple), gene islands (yellow), insertion sequence (black), GC ratio and GC-skew. (B) KEGG classification of putative proteins.
The pathways of nitrogen metabolism in the W70 strain were predicted and presented in Supplementary Figure S4. The genes involved in ammonia assimilation, such as glnA and gdhA, were found to play key roles in the glutamate metabolism (Supplementary Table S2). The gdhA gene is 1,335 bp in length, encoding a deduced protein of 444 amino acids. Its similarity to sequences in the Swiss-Prot database is <62.67%, and < 62.90% in the PDB database. The homology model, based on the PDB template 5GUD, is presented in Table 4. As shown in Supplementary Figure S5 alignment with protein structure-resolved homologs of >50% identities, similar to other prokaryotic GDHs (5GUD and 5IJZ from Corynebacterium glutamicum, 4FCC and 3SBO from Escherichia coli, 3R3J from Plasmodium falciparum 3D7, and 1BGV, 1 K89, and 1AUP from Clostridium symbiosum), the gdhA-encoded protein consists of domain I (catalytic domain) that covers Met1-Ala204 and Asn369-Ile405, and domain II (nucleotide-binding domain) that covers Arg205-Ser368. Of them, Lys136, Ser 260, and Arg287 are predicted to be catalytic residues, and Arg94, Asp166, and Asn344 are putative binding sites to NADP+, respectively. The glnA gene is 1,344 bp in length, encoding a deduced protein of 447 amino acids. Its similarity to sequences in the Swiss-Prot database is <71.36%, and < 71.14% in the PDB database. The homology model, based on the PDB template 7TDP, is shown in Table 4. As shown in Supplementary Figure S6 alignment with protein structure-resolved homologs of >50% identities, similar to other prokaryotic GSs (7TDP from Paenibacillus polymyxa, 7TF6 from Staphylococcus aureus, 7TEN from Listeria monocytogenes, 4S0R from Bacillus subtilis, 8OOW from Methermicoccus shengliensis DSM 18856, 8UFJ from Methanosarcina mazei Go1, and 8OOL from Methanothermococcus thermolithotrophicus), the glnA-encoded protein is conserved in the catalytic domain and substrate binding site n. The expression of gdhA and glnA was significantly increased in modified MRS medium containing 30 mmol/L ammonium sulfate as the sole nitrogen source compared to 5 mmol/L ammonium sulfate (Supplementary Figure S7). These findings suggest that gdhA and glnA play a regulatory role in ammonia assimilation in W70.
The pathways of biosynthesis of amino acids in the W70 strain were predicted and presented in Supplementary Figure S8. Genes involved in the biosynthesis of amino acids, such as proline, arginine, lysine, aspartate, asparagine, isoleucine, leucine, valine, methionine, serine, and histidine were also identified in the genome (Supplementary Table S3). The pathways of propionate metabolism in the W70 strain were predicted and presented in Supplementary Figure S9. The genes involved in propionate metabolism, such as ldh, pflD, pta, and ackA, were found to play key roles in the propionate production (Supplementary Table S4).
4 Discussion
Nitrogen is an essential macronutrient for microbial growth and basic metabolic processes (Chen et al., 2023). The degradation and utilization rate of nitrogen sources varies depending on the species and strains of ruminal bacteria (Cantalapiedra-Hijar et al., 2023). Some ruminal bacteria contribute to excess ammonia accumulation, resulting in a low yield of MCP (Qi et al., 2024). Therefore, identifying novel ammonia nitrogen utilizing bacteria and understanding their role in promoting nitrogen conversion in the rumen will provide knowledge to manipulate the functional microbes for potential application in the ruminant industry (Pfau et al., 2023).
4.1 Ligilactobacillus agilis W70 and Limosilactobacillus fermentum M50 as ammonia nitrogen utilizing strains
In the present study, 115 ruminal bacteria isolated from rumen fluid samples were tested in a medium with ammonium sulfate as the sole nitrogen source. The OD600nm values were employed as indicators to identify the strain’s ammonia nitrogen utilizing capacity. Only the strains capable of utilizing ammonium sulfate were able to grow; those that could not utilize this nitrogen source did not survive. A previous study on ruminal ammonia nitrogen utilizing bacteria indicated that Ruminococcus albus 8 exhibits a preference for ammonia due to its higher growth rate (Kim et al., 2014). Among the tested strains, 10 strains which included Bacillus safenisis B55, Lactobacillus agilis W70, Bacillus safenisis B52, Bacillus safenisis F79, Weissella confuse W26, Enterococcus faecium F42, Lactobacillus fermentum M50, Enterococcus lactis M21, Enterococcus faecium B2, and Lactobacillus salivarius X36 had the highest OD600nm levels in the presence of 30 mmol/L ammonium sulfate compared to others. Similarly, previous research has shown that certain probiotics, including Bacillus and lactic acid bacteria, can reduce NH3 emissions in rumen gas fermentation and animal studies (Sharma et al., 2017; Park et al., 2016; Mi et al., 2019) which agrees with the findings of the current study.
In this study, ammonium sulfate at 30 mmol/L was used for the secondary screening test of ammonia nitrogen utilization capacity. This concentration was selected based on findings by Liu et al. (2020), who reported that Bacillus velezensis LG37, isolated from an aquaculture pond, exhibited increased growth with rising ammonium sulfate concentrations (5, 10, 15, 20, 25, and 30 mmol/L), reaching maximum growth at 30 mmol/L. This screening allowed us to conduct an accurate evaluation of not only bacterial growth but also the strains’ ability to utilize ammonia nitrogen and produce metabolic products. As a result, L. agilis W70 and L. fermentum M50 decreased ammonia nitrogen and increased MCP to a greater extent than the four other tested strains. The optimal ammonia nitrogen concentrations for MCP production were identified as 25 mmol/L for L. agilis W70 and 30 mmol/L for L. fermentum M50 in this study, with excessive nitrogen concentrations not promoting further MCP production, consistent with findings by Zurak et al. (2023). This limitation may be due to metabolic overload or interference with other physiological functions of the strains (He et al., 2023). Previous research has shown that Lactobacillus plantarum, when added to pig manure, reduced NH3 emissions by 36% (Huang and Li, 2006), supporting our findings on the ammonia utilizing capacity of Lactobacillus in lowering ammonia levels, even in different contexts. In this study, L. agilis W70 and L. fermentum M50, which exhibited the highest ammonia nitrogen utilization capacity at 30 mmol/L ammonium sulfate, reduced ammonia nitrogen by 32.09 and 40.64%, respectively.
4.2 Ligilactobacillus agilis W70 reduced ammonia concentration and increased MCP production in vitro rumen fermentation
In order to make a direct comparison of ammonia nitrogen utilizing ability between the W70 and M50 strains, in vitro rumen fermentation was carried out. The in vitro fermentation replicates the rumen environment to provide a realistic assessment of the strain’s performance under conditions similar to those in the actual rumen, while also evaluating whether they have any negative impact on rumen function in a more complex and representative fermentation setting. The pH in this study was within the optimal rumen range of 6.0–7.0, as indicated by Wang et al. (2023). pH influences the activity of rumen microorganisms and the efficiency of rumen fermentation, as strain functionality is influenced by the complex rumen environment (Sanjorjo et al., 2023). Therefore, the lack of influence on pH is a positive finding in this case. In addition supplementation of 1 × 109 cfu/mL of the W70 strain had no effect on the gas volume and DMD, emphasizing no negative influence on rumen fermentation. The W70 strain accelerated the utilization of NH3N in the rumen through MCP synthesis, thereby maximizing MCP production without wasting nitrogen in this study. MCP is synthesized by rumen microorganisms from NH3N, peptides, and amino acids in the rumen, which subsequently provides 50–90% rumen bypass protein for dairy cows (Li et al., 2023). Maintaining a proper NH3N balance is essential, as insufficient NH3N limits MCP production, while excess NH3N can result in nitrogen loss and waste (Zurak et al., 2023). The reduction in ammonia and increase in MCP is potentially associated with the higher expression levels of enzymes GDH and GS of strain W70 in the rumen. Although this mechanism needs to be further clarified as the enzymes were measured in the rumen fluid not the strains. VFAs are the primary energy pools in rumen and hindgut in ruminants for physiological activities, including growth, reproduction, and lactation (Sanz-Fernandez et al., 2020). The decrease in the A:P ratio of the W70 strain in the TMR diet indicated a shift toward more propionate production relative to acetate, which is beneficial for ruminants due to the difference in energy yield, improved feed efficiency, and reduced methane emissions (Russell, 2002). The production of propionate may be linked to the W70 strain itself, as its genome includes a KEGG carbohydrate metabolism pathway related to propionate synthesis from propionyl-CoA. The lower NH3N concentrations and A:P ratio and higher MCP indicated that the W70 strain contributed to nitrogen transformation and propionate production, without impairing effects in vitro rumen fermentation.
4.3 Genomic information of nitrogen metabolism of Ligilactobacillus agilis W70
Ammonia nitrogen utilizing bacteria typically use ammonia assimilation pathways to convert ammonia into glutamate and glutamine (Zhu et al., 2023), which are then involved in protein synthesis in the rumen. To examine the functionality of the W70 strain, a genome sequence analysis was conducted. KEGG analysis revealed that annotated genes of the W70 strain are involved in various metabolic processes which include: carbohydrate metabolism (138 genes), membrane transport (109 genes), amino acid metabolism (89 genes), and metabolism of other amino acids (29 genes). The W70 strain contains the genes gdhA and glnA, which are crucial for glutamate metabolism. Specifically, gdhA encodes GDH, responsible for utilizing ammonia to synthesize glutamate (De Mendonça Lopes et al., 2023), while glnA encodes GS, which converts glutamate to glutamine (Zhao et al., 2023). GDH and GS are well documented in ammonia assimilation processes (Wang et al., 2015), and are probably key to the enhanced ammonia utilization observed in the W70 strain. The assimilation of ruminal ammonia is typically mediated by GDH or GS (Nguyen et al., 2023). GDH is particularly effective when ammonia concentrations are high (Shilpha et al., 2023). Furthermore, gdhA and glnA have been identified as key genes of ammonia emission reduction in Enterococcus faecium strain C2 and Bacillus coagulans strain B1, reducing ammonia emission rates by 53.60 and 31.38%, respectively (Xiao et al., 2021). The high ammonia concentration used in this study (up to 30 mmol/L) might activate the GDH pathway in the W70 strain. As reported by Tomita et al. (2017), the GDH from Corynebacterium glutamicum, CgGDH (PDB: 5GUD), showed the highest catalytic efficiency against NADPH. Thus the gdhA encoded protein with high identity to CgGDH (62.90%) is inferred to own similar high catalytic efficiency and make great contribution to the ammonia utilization of strain W70. And as reported by Travis et al. (2022), the deduced protein of glnA forms a ligand with Met-Sox-P in the presence of ATP. In the complexes, the Glu307-Asp55 interaction would shield the Met-Sox-P from attack by bulk solvent as well as facilitate proton abstraction of the ammonium to form glutamine. These findings imply that glnA may encode highly efficient enzymes for ammonia utilization in strain W70.
In addition to gdhA and glnA, W70’s genome also includes genes involved in the biosynthesis of various amino acids such as proline, arginine, lysine, aspartate, asparagine, isoleucine, leucine, valine, methionine, serine, and histidine. Genes related to nitrogen metabolism are advantageous for further applications in the ruminant industry to enhance nitrogen utilization efficiency (Zhang et al., 2023). Similarly, genes involved in ammonia assimilation and the biosynthesis of alanine, aspartate, glutamate, and lysine were found in Succinivibrio dextrinosolvens strain Z6, which showed maximum growth in ammonia containing medium (Hailemaria et al., 2020). Moreover, supplementing dairy cows with 15 g/day of Bacillus megaterium BM1259 (1 × 108 cfu/g) improved nitrogen utilization and milk production, which was closely related to the nitrate assimilation-related genes and pathways suggesting this strain may be advantageous when fed in vivo (Deng et al., 2021). Therefore, the gdhA and glnA genes in the W70 strain, along with genes involved in amino acid biosynthesis, contribute to its effective ammonia nitrogen utilization capability in vitro and suggest potential applications for improving nitrogen utilization efficiency in vivo.
5 Conclusion
In summary, Lactobacillus agilis W70 and Lactobacillus fermentum M50 isolated from rumen fluid were found to have greater ammonia nitrogen utilizing capabilities than other tested strains. Among them, the W70 strain showed higher efficiency in NH3N utilization and MCP synthesis in vitro rumen fermentation. Genome analysis revealed that gdhA and glnA coding for putative glutamate dehydrogenase and glutamine synthetase, respectively, are potentially key genes of the W70 strain in regulating nitrogen metabolism, as demonstrated in tube tests and in vitro experiments. Further studies on application of W70 strain in feed will be conducted to improve the nitrogen utilization efficiency in the ruminant industry.
Data availability statement
The datasets presented in this article are not readily available because the relevant strains are patented. Requests to access the datasets should be directed to the corresponding author.
Ethics statement
The animal study was approved by the Institutional Animal Care and Use Committee of the Institute of Animal Science, Chinese Academy of Agricultural Sciences and complied with the protocol IAS2022-91. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XD: Data curation, Formal analysis, Methodology, Writing – original draft. SV: Supervision, Validation, Writing – review & editing. RM: Methodology, Software, Visualization, Writing – review & editing. LM: Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing. JG: Supervision, Writing – review & editing, Resources, Software. DB: Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The National Center of Technology Innovation for Dairy (No.2024-KFKT-025), the Key Research and Development Program of the Ningxia Hui Autonomous Region (2022BBF02016), and the Agriculture Science and Technology Innovation Program (ASTIP-IAS-17).
Acknowledgments
We sincerely thank the China Scholarship Council’s State Scholarship Fund for XD’s study in University College Dublin as a Visiting Ph. D Student (No. 202303250001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1498109/full#supplementary-material
Footnotes
1. ^http://www.megasoftware.net/
2. ^https://github.com/tseemann/barrnap
3. ^https://www.uniprot.org/blast
5. ^https://swissmodel.expasy.org/interactive
References
Bach, A., Calsamiglia, S., and Stern, M. D. (2005). Nitrogen metabolism in the rumen. J. Dairy Sci. 88, E9–E21. doi: 10.3168/jds.S0022-0302(05)73133-7
Besemer, J., and Borodovsky, M. (2005). GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33, W451–W454. doi: 10.1093/nar/gki487
Broderick, G., and Kang, J. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63, 64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
Callaway, T. R., Koyun, O., Corcionivoschi, N., Baloyi, J., Ateba, C., Stef, L., et al. (2023). “Practical applications of probiotics in beef cattle production” in Direct-fed Microbials and prebiotics for animals: Science and mechanisms of action. eds. T. R. Callaway and S. C. Ricke (Cham: Springer International Publishing), 301–322.
Cantalapiedra-Hijar, G., Martinez-fernandez, G., Forano, E., Denman, S. E., Morgavi, D., and Mcsweeney, C. S. (2023). The extent of nitrogen isotopic fractionation in rumen bacteria is associated with changes in rumen nitrogen metabolism. PLoS One 18:e0291243. doi: 10.1371/journal.pone.0291243
Cappellozza, B. I., Joergensen, J. N., Copani, G., Bryan, K. A., Fantinati, P., Bodin, J. C., et al. (2023). Evaluation of a Bacillus-based direct-fed microbial probiotic on in vitro rumen gas production and nutrient digestibility of different feedstuffs and total mixed rations. Transl. Anim. Sci. 7:044. doi: 10.1093/tas/txad044
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chen, H., Liu, K., Yang, E., Chen, J., Gu, Y., Wu, S., et al. (2023). A critical review on microbial ecology in the novel biological nitrogen removal process: dynamic balance of complex functional microbes for nitrogen removal. Sci. Total Environ. 857:159462. doi: 10.1016/j.scitotenv.2022.159462
De Mendonça Lopes, A. S., De Oliveira, J. S., De Lima Cruz, G. F., De Sousa Vieira, D., De Sousa Santos, F. N., Lemos, M. L. P., et al. (2023). Effects of non-protein nitrogen on buffel grass fiber and ruminal bacterial composition in sheep. Livest. Sci. 272:105237. doi: 10.1016/j.livsci.2023.105237
Deng, B., Wang, L., Ma, Q., Yu, T., Liu, D., Dai, Y., et al. (2021). Genomics analysis of Bacillus megaterium 1259 as a probiotic and its effects on performance in lactating dairy cows. Animals 11:397. doi: 10.3390/ani11020397
Duan, X., Ma, R., Vigors, S., Ma, L., Gu, J., and Bu, D. (2023). Insight into the ruminal microbiota of dairy cows using the culturomics and amplicon-based sequencing approaches. J. Dairy Sci. 106, 292–293.
Eschenlauer, S., Mckain, N., Walker, N., Mcewan, N., Newbold, C., and Wallace, R. (2002). Ammonia production by ruminal microorganisms and enumeration, isolation, and characterization of bacteria capable of growth on peptides and amino acids from the sheep rumen. Appl. Environ. Microbiol. 68, 4925–4931. doi: 10.1128/AEM.68.10.4925-4931.2002
Fitriyah, N. S., Oluodo, L. A., Hnokaew, P., Umsook, S., Thirawong, P., Khamlor, T., et al. (2024). Optimum level of Lactobacillus plantarum supplementation as probiotic on in vitro digestibility and rumen fermentation products in thai native cattle. Vet. Integr. Sci. 22, 489–500. doi: 10.12982/VIS.2024.034
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Griswold, K., Apgar, G., Bouton, J., and Firkins, J. (2003). Effects of urea infusion and ruminal degradable protein concentration on microbial growth, digestibility, and fermentation in continuous culture. J. Anim. Sci. 81, 329–336. doi: 10.2527/2003.811329x
Hailemaria, M. S., Zhao, S., and Wang, J. (2020). Complete genome sequencing and transcriptome analysis of nitrogen metabolism of Succinivibrio dextrinosolvens strain Z6 isolated from dairy cow rumen. Front. Microbiol. 11:1826. doi: 10.3389/fmicb.2020.01826
He, H., Li, Y., Zhang, L., Ding, Z., and Shi, G. (2023). Understanding and application of Bacillus nitrogen regulation: a synthetic biology perspective. J. Adv. Res. 49, 1–14. doi: 10.1016/j.jare.2022.09.003
Huang, C., and Li, J. (2006). Effect of adding Lactobacillus plantarum and soluble carbohydrates to swine manure on odorous compounds, chemical composition and indigenous flora. J. Environ. Sci. 18, 201–206.
Hucker, G. J., and Conn, H. J. (1923). Methods of gram staining. New York: Agricultural Experiment Station.
Hyatt, D., Chen, G. L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Karimi, A. (2022). Effect of probiotics (direct-fed microbials) in poultry production: a comprehensive review. Rev. Electron. Vet. 23, 72–82.
Kesen, M. A., and Aiyegoro, O. A. (2018). Beneficial characteristics and evaluation criteria of probiotics. Int. J. Food Biosci. 1, 19–26.
Kim, J. N., Henriksen, E. D., Cann, I. K., and Mackie, R. I. (2014). Nitrogen utilization and metabolism in Ruminococcus albus 8. Appl. Enviro. Microbiol. 80, 3095–3102. doi: 10.1128/AEM.00029-14
Lavery, A., and Ferris, C. P. (2021). Proxy measures and novel strategies for estimating nitrogen utilisation efficiency in dairy cattle. Animals 11:343. doi: 10.3390/ani11020343
Li, Y., Hu, J., Xiang, Y., Zhang, Y., Chen, D., and Liu, F. (2020). Identification and comparative expression profiles of chemosensory genes in major chemoreception organs of a notorious pests, Laodelphax striatellus. Comp. Biochem. Physiol. Part D Genomics Proteomics 33:100646. doi: 10.1016/j.cbd.2019.100646
Li, B., Jia, G., Wen, D., Zhao, X., Zhang, J., Xu, Q., et al. (2022). Rumen microbiota of indigenous and introduced ruminants and their adaptation to the Qinghai–Tibetan plateau. Front. Microbiol. 13:1027138. doi: 10.3389/fmicb.2022.1027138
Li, S., Zeng, H., Wang, C., and Han, Z. (2023). Effect of methionine hydroxy analog on hu sheep digestibility, rumen fermentation, and rumen microbial community in vitro. Meta 13:169. doi: 10.3390/metabo13020169
Liu, Y., Hu, J., Li, M. M., and Zhao, G. (2023). Effects of taurine on rumen fermentation, nutrient digestion, rumen bacterial community and metabolomics and nitrogen metabolism in beef steers. J. Sci. Food Agric. 103, 3414–3426. doi: 10.1002/jsfa.12474
Liu, G., Vijayaraman, S. B., Dong, Y., Li, X., Andongmaa, B. T., Zhao, L., et al. (2020). Bacillus velezensis LG37: transcriptome profiling and functional verification of GlnK and MnrA in ammonia assimilation. BMC Genomics 21:215. doi: 10.1186/s12864-020-6621-1
Makkar, H., Sharma, O., Dawra, R., and Negi, S. (1982). Simple determination of microbial protein in rumen liquor. J. Dairy Sci. 65, 2170–2173. doi: 10.3168/jds.S0022-0302(82)82477-6
Masi, C., Tebiso, A., and Kumar, K. S. (2023). Isolation and characterization of potential multiple extracellular enzyme-producing bacteria from waste dumping area in Addis Ababa. Heliyon 9:e12645. doi: 10.1016/j.heliyon.2022.e12645
Menke, K. H. (1988). Estimation of the energetic feed value obtained from chemical analysis and in vitro gas production using rumen fluid. Anim. Res. Dev. 28, 7–55.
Mi, J., Chen, X., and Liao, X. (2019). Screening of single or combined administration of 9 probiotics to reduce ammonia emissions from laying hens. Poultry Sci. 98, 3977–3988. doi: 10.3382/ps/pez138
Nguyen, T. T. M., Badhan, A. K., Reid, I. D., Ribeiro, G., Gruninger, R., Tsang, A., et al. (2023). Comparative analysis of functional diversity of rumen microbiome in bison and beef heifers. Appl. Environ. Microbiol. 89, e01320–e01323. doi: 10.1128/aem.01320-23
Opsi, F., Fortina, R., Tassone, S., Bodas, R., and López, S. (2012). Effects of inactivated and live cells of Saccharomyces cerevisiae on in vitro ruminal fermentation of diets with different forage: concentrate ratio. J. Agric. Sci. 150, 271–283. doi: 10.1017/S0021859611000578
Park, J. W., Jeong, J. S., Lee, S. I., and Kim, I. H. (2016). Effect of dietary supplementation with a probiotic (Enterococcus faecium) on production performance, excreta microflora, ammonia emission, and nutrient utilization in ISA brown laying hens. Poultry Sci. 95, 2829–2835. doi: 10.3382/ps/pew241
Pfau, A. P., Shepherd, E. A., Martin, M. G., Ascolese, S., Mason, K. M., Egertmclean, A. M., et al. (2023). Beta-adrenergic agonists, dietary protein, and rumen bacterial community interactions in beef cattle: a review. Vet. Sci. 10:579. doi: 10.3390/vetsci10090579
Qi, W., Xue, M. Y., Jia, M. H., Zhang, S., Yan, Q., and Sun, H. Z. (2024). Understanding the functionality of the rumen microbiota: searching for better opportunities for rumen microbial manipulation. Anim. Biosci. 37, 370–384. doi: 10.5713/ab.23.0308
Qi, J., Yang, H., Wang, X., Zhu, H., Wang, Z., Zhao, C., et al. (2023). State of the art on animal manure pollution control and resource utilization. J. Environ. Chem. Eng. 11:110462. doi: 10.1016/j.jece.2023.110462
Russell, J. (2002). Rumen microbiology and its role in ruminant nutrition. Ithaca: Cornell University Press.
Sanjorjo, R. A., Tseten, T., Kang, M. K., Kwon, M., and Kim, S. W. (2023). In pursuit of understanding the rumen microbiome. Fermentation 9:114. doi: 10.3390/fermentation9020114
Sanz-Fernandez, M. V., Daniel, J. B., Seymour, D. J., Kvidera, S. K., Bester, Z., Doelman, J., et al. (2020). Targeting the hindgut to improve health and performance in cattle. Animals 10:1817. doi: 10.3390/ani10101817
Sharma, N. K., Choct, M., Wu, S., and Swick, R. A. (2017). Nutritional effects on odour emissions in broiler production. Worlds Poult. Sci. J. 73, 257–280. doi: 10.1017/S0043933917000046
Shilpha, J., Song, J., and Jeong, B. R. (2023). Ammonium phytotoxicity and tolerance: an insight into ammonium nutrition to improve crop productivity. Agronomy 13:1487. doi: 10.3390/agronomy13061487
Stewart, C. S., and Duncan, S. H. (1985). The effect of avoparcin on cellulolytic bacteria of the ovine rumen. Microbiology 131, 427–435. doi: 10.1099/00221287-131-3-427
Tomita, T., Yin, L., Nakamura, S., Kosono, S., Kuzuyama, T., and Nishiyama, M. (2017). Crystal structure of the 2-iminoglutarate-bound complex of glutamate dehydrogenase from Corynebacterium glutamicum. FEBS Lett. 591, 1611–1622. doi: 10.1002/1873-3468.12667
Travis, B. A., Peck, J. V., Salinas, R., Dopkins, B., Lent, N., Nguyen, V. D., et al. (2022). Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in gram-positive bacteria. Nat. Commun. 13:3793. doi: 10.1038/s41467-022-31573-0
Van Soest, P. J., and Robertson, J. (1985). Analysis of forages and fibrous foods. Ithaca: Cornell University Press.
Wang, R., He, S., Huang, D., Wu, D., Peng, H., He, S., et al. (2023). The response of rumen pH, fermentation parameters and rumen bacteria to feeds of different concentrate to roughage ratios in buffalos. Front. Microbiomes 1:1053794. doi: 10.3389/frmbi.2022.1053794
Wang, P., Tan, Z., Guan, L., Tang, S., Zhou, C., Han, X., et al. (2015). Ammonia and amino acids modulates enzymes associated with ammonia assimilation pathway by ruminal microbiota in vitro. Livest. Sci. 178, 130–139. doi: 10.1016/j.livsci.2015.05.033
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Xiao, S., Zhang, H., Zhu, R., Liao, X., Wu, Y., Mi, J., et al. (2021). Ammonia reduction by the gdhA and glnA genes from bacteria in laying hens. Ecotoxicol. Environ. Saf. 222:112486. doi: 10.1016/j.ecoenv.2021.112486
Zhang, X. M., Chen, W. X., Yan, Q. X., Wang, C., Lin, B., Yi, S. Y., et al. (2023). Low-protein diet promotes nitrogen retention efficiency via enhanced renal urea reabsorption and microbial hydrogen incorporation in the rumen of goats. Anim. Feed Sci. Technol. 305:115762. doi: 10.1016/j.anifeedsci.2023.115762
Zhao, X., Song, Y., Wang, T., Hua, C., Hu, R., Shang, Y., et al. (2023). Glutamine synthetase and GlnR regulate nitrogen metabolism in Paenibacillus polymyxa WLY78. Appl. Environ. Microbiol. 89:e0013923. doi: 10.1128/aem.00139-23
Zhu, L., Huang, C., Li, W., Wu, W., Tang, Z., Tian, Y., et al. (2023). Ammonia assimilation is key for the preservation of nitrogen during industrial-scale composting of chicken manure. Waste Manag. 170, 50–61. doi: 10.1016/j.wasman.2023.07.028
Keywords: Ligilactobacillus agilis , NH3N, rumen fermentation, screening, functional, physicochemical characterization
Citation: Duan X, Vigors S, Ma R, Ma L, Gu J and Bu D (2025) Ligilactobacillus agilis W70: a probiotic with capacity to enhance ammonia assimilation in the rumen. Front. Microbiol. 15:1498109. doi: 10.3389/fmicb.2024.1498109
Edited by:
Oleksandr S. Yushchuk, University of Insubria, ItalyReviewed by:
Ping Li, Guizhou University, ChinaJavier Carrillo-Campos, Autonomous University of Chihuahua, Mexico
Copyright © 2025 Duan, Vigors, Ma, Ma, Gu and Bu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dengpan Bu, YnVkZW5ncGFuQDEyNi5jb20=
 Xiaowei Duan1,2
Xiaowei Duan1,2 Stafford Vigors
Stafford Vigors Rui Ma
Rui Ma Lu Ma
Lu Ma Dengpan Bu
Dengpan Bu