- 1Maximum Containment Facility, ICMR-National Institute of Virology, Pune, India
- 2Animal House Division, ICMR-National Institute of Virology, Pune, India
- 3Field Unit, ICMR-National Institute of Virology, Dibrugarh, India
- 4National Institute of One Health, Nagpur, India
Introduction: India has experienced seven outbreaks of the Nipah virus (NiV) since 2001, primarily occurring in the southern and eastern regions of the country. The southern region has been the main site for these outbreaks. In contrast, the eastern region, which borders Bangladesh, has not reported any outbreaks since 2007. However, Bangladesh continues to experience nearly annual outbreaks, indicating a significant lack of surveillance in that area. To improve the country’s preparedness and to gather support for enhancing public health surveillance in eastern and northeastern states near the area affected by the NiV, a cross-sectional survey was conducted to determine the prevalence of NiV in the bat species Pteropus medius in Bihar, West Bengal, Assam, and Meghalaya states in India, which are adjacent to Bangladesh.
Methods: Throat and rectal swabs, blood samples, and organ samples were collected. Real-time quantitative reverse transcription PCR (qRT-PCR) was utilized for the detection of Nipah viral RNA, and sequencing was conducted for further confirmation. Bat IgG enzyme-linked immunosorbent assay (ELISA) was employed for antibody detection.
Results: Throat and rectal swab samples of 212 P. medius tested for NiV using qRT- PCR were found negative, whereas organ samples of two (one each from West Bengal and Bihar) out of the 10 bats collected tested positive. The retrieved NiV genome (~91%) showed close homology to the NiV-Bangladesh genotype indicating the circulation of two geographically distinct NiV strains in India. The seroprevalence estimated by ELISA ranged from 23 to 65% in the studied states.
Discussion: The serological and virological evidence obtained from the study indicates that a broader geographical area is under threat of spillover in India. It’s crucial to implement a One Health approach connecting bat surveillance studies with human surveillance and risk factor studies in the region.
1 Introduction
Nipah virus (NiV) is a paramyxovirus that causes a highly fatal viral infection characterized by severe respiratory and neurological signs in humans. The virus is considered a priority pathogen by the World Health Organization (WHO, n.d.). The NiV is a negative sense single-stranded RNA virus with the genome coding for six structural and 3 non-structural proteins. The nucleoprotein (N), phosphoprotein (P), glycoprotein (G), matrix (M), fusion (F), and RNA polymerase (L) constitute the structural proteins. The P gene encodes for three non-structural proteins, i.e., V, W, and C. The NiV strains fall into separate clades like Malaysia, Bangladesh, and India. The Bangladesh and Indian clades differ by about 9% in nucleotide substitutions from Malaysian strains (Whitmer et al., 2021).
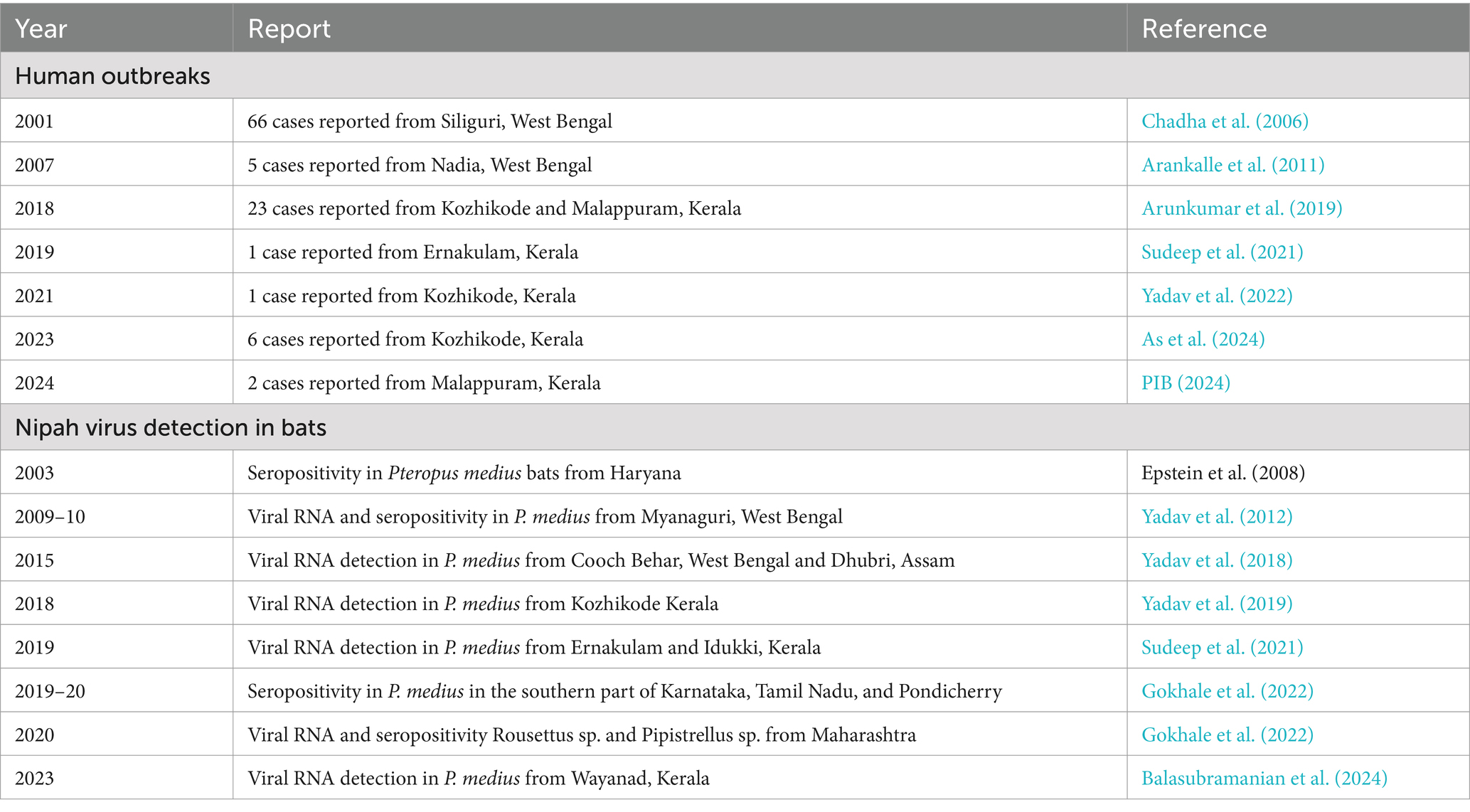
Outbreaks of the NiV disease have been reported in several countries, including Malaysia, Singapore, Bangladesh, India, and the Philippines (Chua et al., 1999; Paton et al., 1999; Chadha et al., 2006; Rahman and Chakraborty, 2012; Ching et al., 2015). The disease was first identified in Malaysia in the late 1990s, where it spread from fruit bats to pig populations and then to humans (Chua et al., 1999). In Singapore, the disease was reported among abattoir workers handling pigs imported from Malaysia during the same period (Paton et al., 1999). In the Philippines, henipavirus infection was reported among people who came in contact with sick horses (Ching et al., 2015). Unlike Malaysia, Singapore and Philippines, multiple outbreaks of NiV disease have been reported in Bangladesh and India. Since 2001, Bangladesh has been facing annual outbreaks in the northwestern districts, known as the “Nipah belt, “and Kerala state in India has been experiencing frequent outbreaks since 2018 (Arunkumar et al., 2019; Sudeep et al., 2021; Yadav et al., 2022; As et al., 2024; PIB, 2024; Nipah virus infection, n.d.).
There have been seven known outbreaks of the NiV disease in India. Two of them occurred in the Siliguri and Nadia districts of West Bengal, which are located near the border with Bangladesh, in the years 2001 and 2007, respectively (Chadha et al., 2006; Arankalle et al., 2011). The other five outbreaks in India occurred in the Kozhikode, Ernakulam, and Malappuram districts of the southern Indian state of Kerala after 2018 (Figure 1, Table 1) (Arunkumar et al., 2019; Sudeep et al., 2021; Yadav et al., 2022; As et al., 2024). Surveys conducted in various species of bats in India have detected the presence of NiV RNA in Pteropus medius, Rousettus sp., and Pipistrellus sp. (Table 1 and Figure 1).
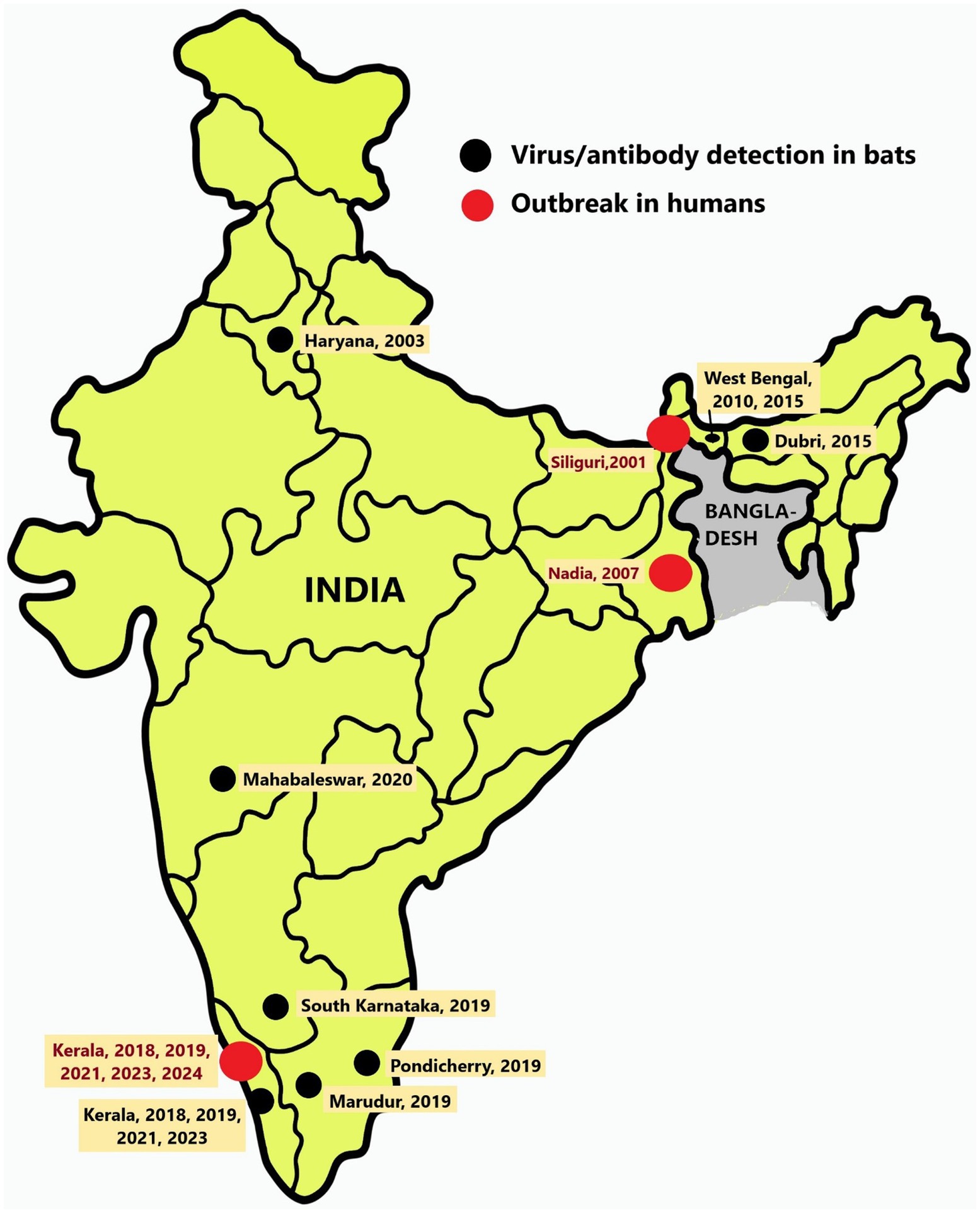
Figure 1. Location of Nipah virus outbreaks and detection in bats: Map of India showing the locations of Nipah virus outbreaks in humans (red dots) and the regions from where bats were found harboring Nipah virus (black dots).
Fruit bats belonging to the Pteropus species act as reservoir hosts for the NiV and play a crucial role in disease maintenance and transmission (Epstein et al., 2020). The ecological and economic importance of fruit bats has been widely documented. They contribute to pollination, seed dispersal, pest control, predation, and nutrient distribution across ecosystems, serving as an important ecological indicator (Kasso and Balakrishnan, 2013). The serological or molecular evidence of henipavirus infection has been detected in Pteropid bats in countries that have experienced outbreaks and in other countries, including Cambodia, China, East Timor, Indonesia, Madagascar, Papua New Guinea, Thailand, and Vietnam (Reynes et al., 2005; Wacharapluesadee et al., 2005; Iehlé et al., 2007; Li et al., 2008; Breed et al., 2010; Sendow et al., 2010; Hasebe et al., 2012).
Bangladesh shares 75% of its international border with the eastern and northeastern states of India, and the region known as the Nipah belt is near India. The surveys conducted on P. medius bats in eight districts of Bangladesh between 2006 and 2012 revealed virus detection throughout the country (Epstein et al., 2016, 2020). This included bat colonies from Thakurgaon [~30 km from the India (West Bengal) border], Kushtia [~30 km from the India (West Bengal) border], Cumilla [~2 km from India (Tripura) border], Sylhet [~30 km from India (Meghalaya) border], Chattogram [~70 km from India (Tripura/Mizoram) border], and Khulna [~70 km from India (West Bengal) border] (Epstein et al., 2020). Even though Bangladesh reports outbreaks annually, there are no reports of NiV human outbreaks after 2007 from the state of West Bengal or other neighboring states of India bordering Bangladesh (Nipah virus infection, n.d.).
The Nipah sentinel surveillance system established in 2006 in Bangladesh has helped detect Nipah virus cases and contain outbreaks in the country (Satter et al., 2023). However, India lacks a similar coordinated surveillance system. Syndromic surveillance has been strengthened in the state of Kerala following the highly fatal outbreak in 2018, leading to an effective containment response in the subsequent years (Yadav et al., 2022; As et al., 2024). Here we present our findings of the cross-sectional survey in P. medius taken up in the states (West Bengal, Meghalaya, and Assam) sharing the borders with Bangladesh and an adjoining state, Bihar, between 2022 and 2023. These crucial efforts were focused on advocating for the enhancement of public health surveillance activities in the states neighboring the Nipah belt.
2 Materials and methods
2.1 Sample collection
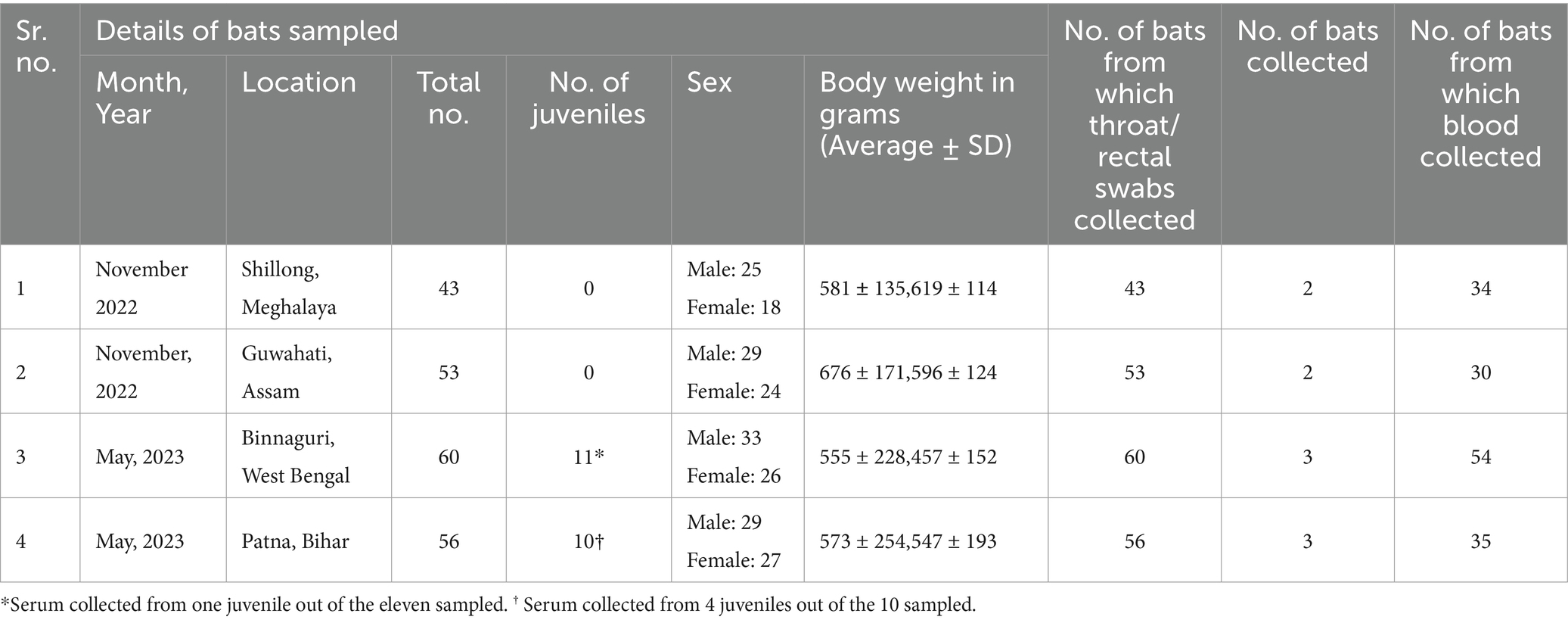
The Institute Animal Ethics Committee of ICMR-National Institute of Virology, Pune, and the Principal Chief Conservator of Forests of Meghalaya, Assam, West Bengal, and Bihar approved the study. The national guidelines of the Committee for Control and Supervision of Experiments on Animals, Government of India followed for the care and use of animals in the study. The bat samples were collected from Patna, Bihar (~400 km from Naogaon, the nearest location in Bangladesh from where NiV cases were reported in 2023 and ~ 350 km to Siliguri, West Bengal from where NiV outbreaks reported in India), Binnaguri, West Bengal (~200 km from Naogaon and ~ 60 km from Siliguri, West Bengal); from Guwahati, Assam (~260 km from Narsingdi, nearest location in Bangladesh from where NiV cases are reported in 2023) and Shillong, Meghalaya (~200 km from Narsingdi) (Figure 2).
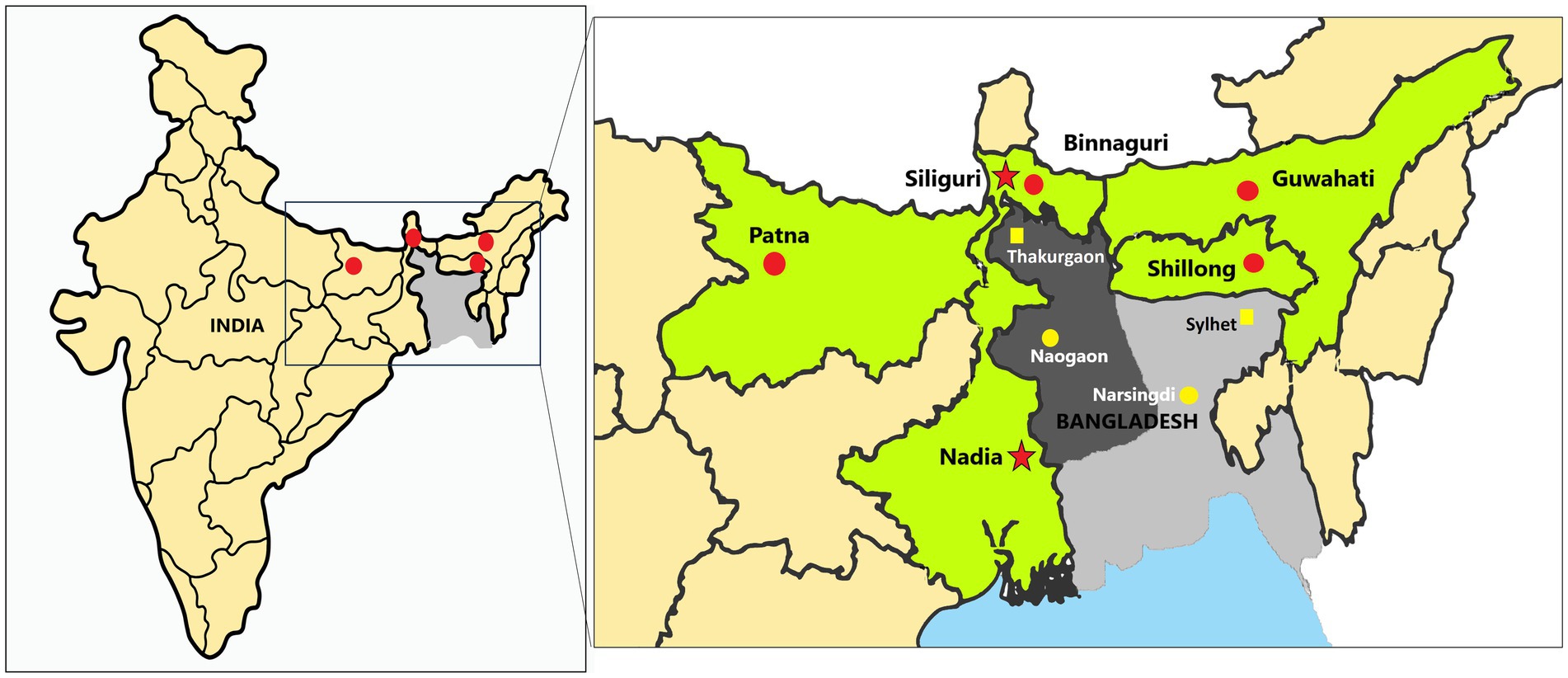
Figure 2. Sample collection sites of the study: The left map of India shows the sampled sites marked as red dots, while the right magnified image displays the sampled sites (red dots) in each state (marked as green). The red dots on the map indicate the locations of the sampling sites in the states of Bihar (Patna), West Bengal (Binnaguri), Assam (Guwahati), and Meghalaya (Shillong) in India. The red stars denote the previous Nipah virus outbreak locations in India, i.e., Siliguri and Nadia. The dark grey region in the central and northwest areas of Bangladesh is known as the Nipah belt, while the rest of the country is colored light grey. The yellow dots mark the cities of Naogaon and Narsingdi (used to measure the approximate distance to each sampling site). Thakurgaon and Sylhet in Bangladesh are marked as yellow squares places in Bangladesh that have reported NiV positivity in bats near sampling locations in India.
Mist nets were set up on poles near the bat roosts to trap the bats. The bats were identified by their morphological features and their body weight, sex, and reproductive status were recorded. The bats were classified into juveniles and adults based on their body weight and secondary sexual characteristics (Epstein et al., 2020). A total of 212 P. medius were trapped which included 43 adult bats from Shillong, Meghalaya, 53 adult bats from Guwahati, Assam, 60 bats (Adults =49, Juveniles =11) from Binnaguri, West Bengal, and 56 bats (Adults = 39; Juveniles = 10) from Patna, Bihar (Table 2).
The captured bats were anesthetized using isoflurane and swabbed (throat and rectal) for testing, and blood samples were also collected. The swabs were collected using a sterile swab applicator in a virus transport medium (Himedia, India). Blood samples (2 mL) were collected from the wing vein using a sterile 25G needle in a serum separation gel tube and serum was separated by centrifugation after 45 min. The bats were released to their natural habitat after recovery from anesthesia. Few bats (2 bats from Assam and Bihar each, 3 bats each from Meghalaya, and West Bengal) which were injured during trapping were euthanized and transported to the containment laboratory in liquid nitrogen for organ collection. The kidney, spleen, liver, intestine, and reproductive organs were collected for NiV RNA testing.
2.2 Real-time quantitative reverse transcription PCR (qRT-PCR)
The RNA was extracted from the samples using the magnetic bead-based method in the Kingfisher Flex machine (Thermo Fischer Scientific, USA) using the Magmax Viral RNA isolation kit (Thermo Scientific, USA) as per the manufacturer’s instructions. The real-time qRT-PCR was carried out with published NiV-specific primers and probes targeting the N gene (Guillaume et al., 2004).
2.3 Partial and whole genome sequencing
The qRT-PCR positive samples were further confirmed by Sanger sequencing. For sequencing, a nested RT-PCR was performed with the primers (1st PCR: forward: CGTGGTTATCTTGAACCTATGTACTTCAG and reverse: CGCAACTTTAATGTAATTGGTCCCTTAGTG, 2nd PCR: forward: CAGAGAAGCTAAATTTGCTGCAGGAGG and Reverse: TCACACATCAGCTCTGACAAAGTCAAG) specific for the NiV N-gene. The PCR products were electrophoresed on 1.5% agarose gel and bands (342 bp) were purified using the QIAquick Gel Extraction Kit (Qiagen, Germany). The Big Dye™ Terminator Cycle Sequencing Kit (Applied Biosystems, USA) was used for sequencing PCR and the DyeEx 2.0 kit (Qiagen, Germany) was used for the PCR product purification. The ABI 3130 XL genetic analyzer (Applied Biosystems, USA) was used for sequencing and the Sequencher 5.1 software (Accelrys Inc., USA) was used for analysis. The retrieved partial NiV N gene sequences were submitted to GenBank and the accession number was obtained.
The whole genome sequencing for the Nipah virus qRT-PCR positive samples was performed using the Virus Surveillance Panel (VSP) Illumina RNA Prep with Enrich A kit (Illumina, USA). A total of 8.5 microliters of extracted RNA was denatured and combined with random hexamers in a thermal cycler at 65°C for 5 min. Following this, the hexamer-primed RNA fragments underwent reverse transcription by adding the reverse transcriptase and first strand synthesis master mix in the thermal cycler. The conditions for this step were as follows: 25°C for 10 min, 42°C for 15 min, and 70°C for 15 min. In the subsequent step, the RNA template was removed, and a replacement strand was synthesized to generate blunt-ended double-stranded cDNA fragments. The cDNA was tagmented using enrichment bead-linked transposomes after purification. The tagmented cDNA were purified and amplified to add adapter sequences for dual indexing, and P7 and P5 sequences for clustering. Four index sets (A, B, C, and D), each containing 96 unique, single-use Illumina® DNA/RNA UD indexes, were used. Libraries were quantified using the Invitrogen Qubit dsDNA HS Assay Kit (Thermo Fischer Scientific, USA). Subsequently, libraries (200 ng library or 600 ng pool) were hybridized using oligos from the VSP panel, followed by bead-based capture of hybridized probes, amplification, clean-up, and quantification. Normalized libraries, diluted to an equimolar concentration of 4 pM, were then pooled. Using a 150 bp paired-end sequencing approach and 300 cycles, the libraries were sequenced on the Miniseq sequencer (Illumina, USA) utilizing a high output kit (Illumina, USA). The fastq (raw read files) were mapped with the Nipah virus (NC_002728.1) reference strain using CLC genomics 24.02 software. The coverage above 10X and mutation frequency above 50% were considered for generating variant calling files. Clustal Omega was used to visualize and represent the nucleotide changes.
2.4 Phylogenetic analysis
The phylogenetic analysis of the sequence retrieved in the study was performed with the reference sequences of Nipah virus (NC_002728.1) along with a subset of NiV whole genome sequences from the NCBI database. The analysis included 7 full genome sequences (6 human and 1 bat) from India, 17 (16 human and 1 bat) from Bangladesh, 1 (bat) from Cambodia, and 7 (3 human and 4 swine) from Malaysia. For alignment and visualization, we utilized the MAFFT tool and MEGAX (Katoh et al., 2019). We used IQTREE with the Tamura-Nei + F model and 1,000 ultrafast bootstrap replicates to generate the maximum likelihood tree (Minh et al., 2020). The phylogenetic tree was visualized using the Interactive Tree of Life (iTOL) (Letunic and Bork, 2021).
2.5 Virus isolation
Vero cells (ATCC® CCL-81™, ATCC, USA) and infant CD1 (ICR) laboratory mice (3–4 days old) were used for virus isolation. The CD1 mouse is an outbred mouse strain procured from the CCSEA-licensed laboratory animal house of ICMR-National Institute of Virology, Pune. The samples were inoculated to 24-well plates with Vero cells and were incubated for 1 h. After washing, tissue culture media with 2% fetal bovine serum were added to each well, and plates were incubated in a CO2 incubator for 5 days. The samples were blindly passaged three times. Simultaneously, samples were also intracranially inoculated in the mouse model and the animals were observed for sickness. Brain samples were collected 7 days after inoculation from mice and were homogenized in sterile media. The cell culture supernatants and the mice brain suspension were tested for NiV by real-time qRT-PCR after each passage.
2.6 Anti-Nipah bat IgG ELISA
P. medius bat sera samples (n = 153) were tested by an in-house enzyme-linked immunosorbent assay as described earlier (Gokhale et al., 2022). Heat-inactivated bat sera samples were added to gamma-irradiated whole virion NiV antigen-coated plates and were incubated at 37°C for 1 h. After washing, anti-bat IgG HRP conjugate was added to the plate and incubated for 1 h. After washing, 3,3′,5,5′-Tetramethylbenzidine substrate was added and plates were kept for incubation at 37°C for half an hour. Using 1 N H2SO4, the color reaction was stopped. The plate was read at 450 nm. The serum samples with an optical density (OD) value of ≥0.3 were considered positive for anti-Nipah IgG antibodies.
2.7 Serum neutralization test
A micro-neutralization test (MNT) was conducted on a subset of bat serum samples found positive (n = 19) and negative (n = 6) by IgG ELISA. The assay was performed in Vero (ATCC® CCL − 81™) cell monolayers in 96-well cell culture plates in the containment facility. The bat serum samples were treated by heat inactivation at 56°C for 1 h. Dilutions (two-fold) of the bat sera were prepared by using a sterile tissue culture media. The virus-serum mixture prepared in 1:1 dilution with 100 median tissue culture infectious dose of Nipah virus (GenBank accession ID: MH523642) was then incubated at 37°C for 1 h in a CO2 incubator and was then added to the cell monolayer in 96-well plates in duplicate. The assay included positive (anti-Nipah IgG positive mice serum), negative (anti-Nipah IgG negative mice serum), and cell controls. After an incubation period of 5 days, the cells were observed for cytopathic changes, and the neutralization titer was calculated.
3 Results
3.1 Viral RNA detection in P. medius by qRT-PCR
The throat and rectal swab samples of all the bats collected during the study (n = 212) were negative for viral RNA whereas viral RNA could be detected in the organ samples of two bats out of the 10 bats from which organs were collected. Kidney (Viral RNA copy number = 1.26 × 104/ml) and spleen (Viral RNA copy number = 9.5 × 104/ml) samples of one bat collected from Binnaguri, West Bengal, and spleen (Viral RNA copy number = 2.7 × 105/ml) sample of one bat collected from Patna, Bihar were found positive by qRT-PCR.
3.2 Partial and whole genome sequencing
By Sanger’s sequencing, a partial N gene (GenBank Accession no. OR765959) sequence was retrieved from one spleen sample (from Patna, Bihar) out of the three found positive by real-time qRT-PCR. By whole genome sequencing, 91.9% of the NiV genome (GenBank Accession no. PP554504) could be retrieved from the above sample (Supplementary Figure S1, Supplementary Table S1).
3.3 Phylogenetic analysis
The phylogenetic analysis showed close homology of the sequences retrieved to the NiV Bangladesh genotype (Figure 3, Supplementary Figure S2). Percent identity ranging from 98.8 to 99.4% was observed with the NiV-Bangladesh-2 clade which comprises of sequences from Bangladesh as well as one sequence obtained from an outbreak in West Bengal, India (Table 3, Figure 3). A percent identity of about ~98% with NiV-Bangladesh clade 1, ~97% with NiV-Indian clade, and ~ 92% with NiV-Malaysian clade was observed (Table 3, Figure 3). The analysis of amino acid differences showed the reported substitutions present in the Bangladesh genotype sequences along with a few additional substitutions in P/V/W (Q247H, S335F, A383V), F (P298H, R300S, F301Y, N302K), and L (Q2087R) protein (Supplementary Table S2).
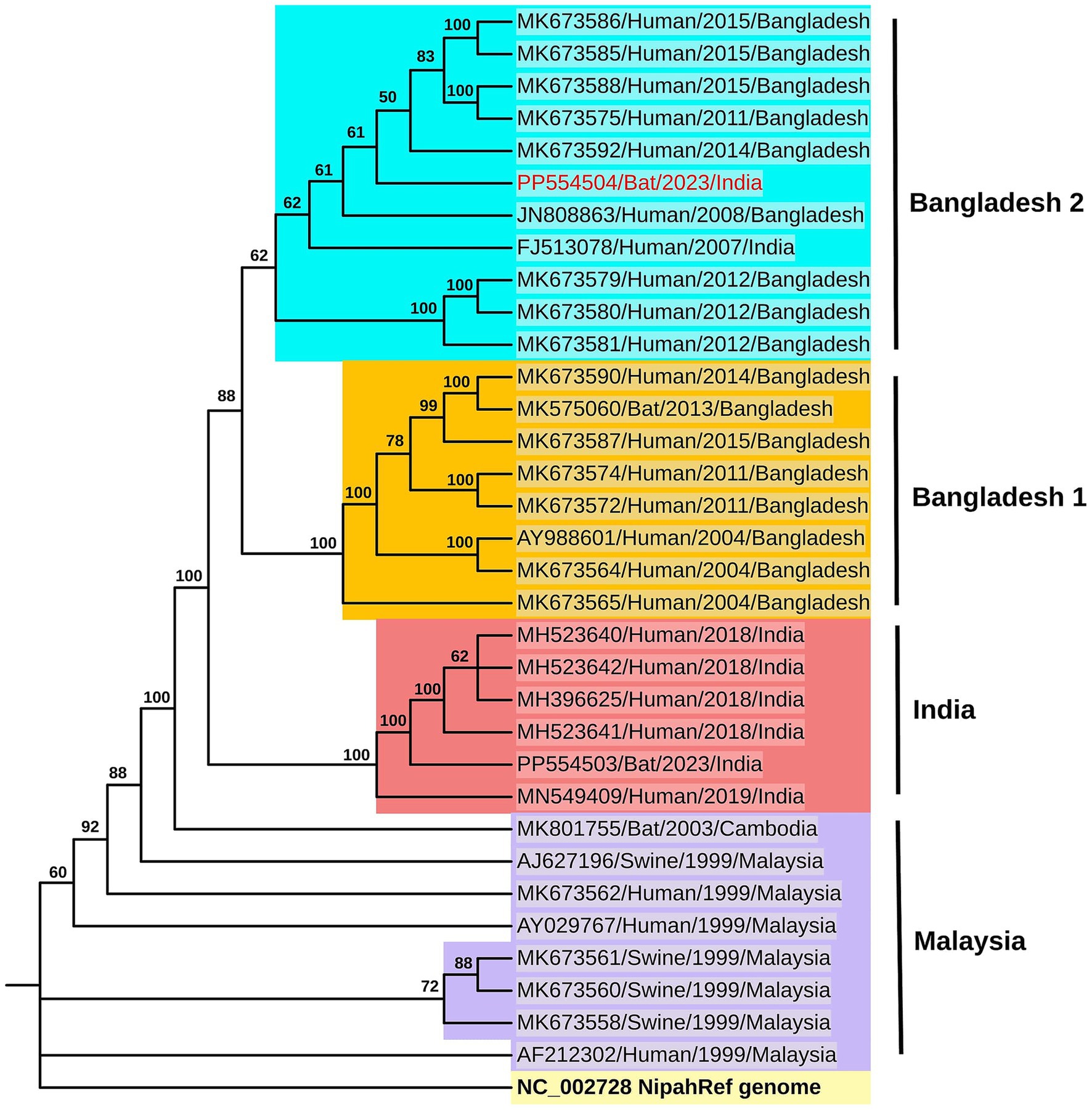
Figure 3. Phylogenetic analysis: The maximum likelihood phylogenetic tree generated using the TN + F model in IQTREE for the genome sequence of the Nipah virus (Accession ID: PP554504) retrieved from the P. medius (colored in red) along with the NiV reference sequence (light yellow background) as well as sequences from the NiV-Bangladesh clade-2 (cyan blue background) and clade-1 (dark yellow background), India (red background) and Malaysia (lavender background) clades. The sequences are labeled with GenBank accession number, host, year, and country, and the bootstrap values are marked along the branches.
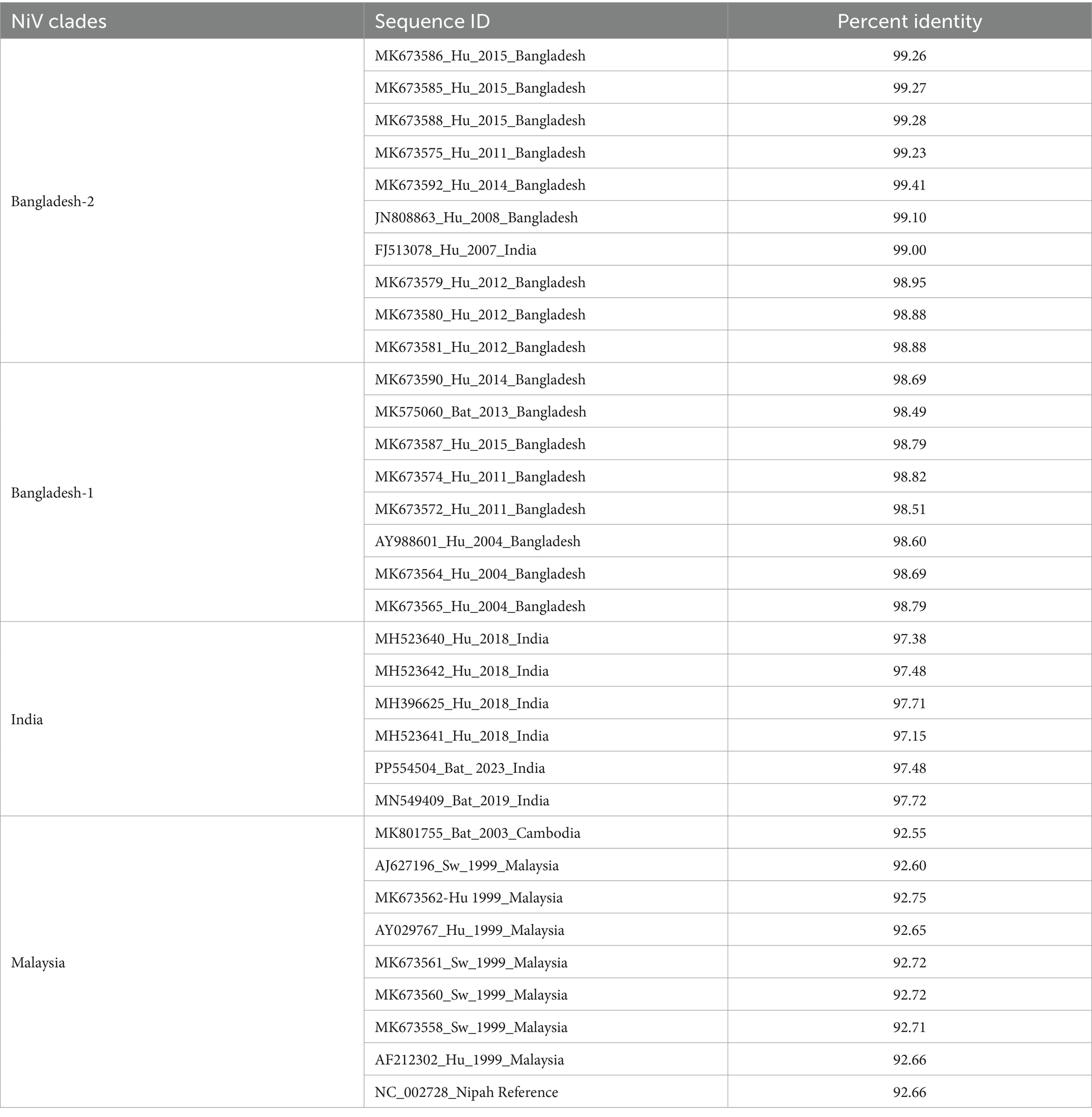
Table 3. The percent identity of the NiV genome retrieved in the study (Accession ID: PP554504) with the Nipah virus reference strain (NC_002728) and a subset of full genome sequences available in the database.
3.4 Virus isolation
The virus isolation attempts were unsuccessful in both cell culture and animal models from Nipah virus PCR-positive samples.
3.5 Anti-Nipah IgG antibody detection by ELISA and neutralization test
The NiV antibody positivity estimated by ELISA in each state was 23.5% (8/34) in Shillong Meghalaya, 33.3% (10/30) in Guwahati, Assam, 62.9% (34/54) in Binnaguri, West Bengal and 65.7% (23/35) in Patna, Bihar (Figure 4A). The seroprevalence was higher in May, at 64% (57/89), compared to November, which had a rate of 28.13% (18/64) (Figure 4B). Out of the 34 seropositive bats from Binnaguri, four were juvenile and out of the 23 positives from Bihar, one was from a juvenile bat. A subset of positive samples (n = 19) and negative samples (n = 6) was confirmed for neutralizing antibodies by neutralization assay. All the IgG-positive samples tested for neutralizing antibodies were found to have neutralization antibody titer ≥40 (Figure 4C, Supplementary Table S3).
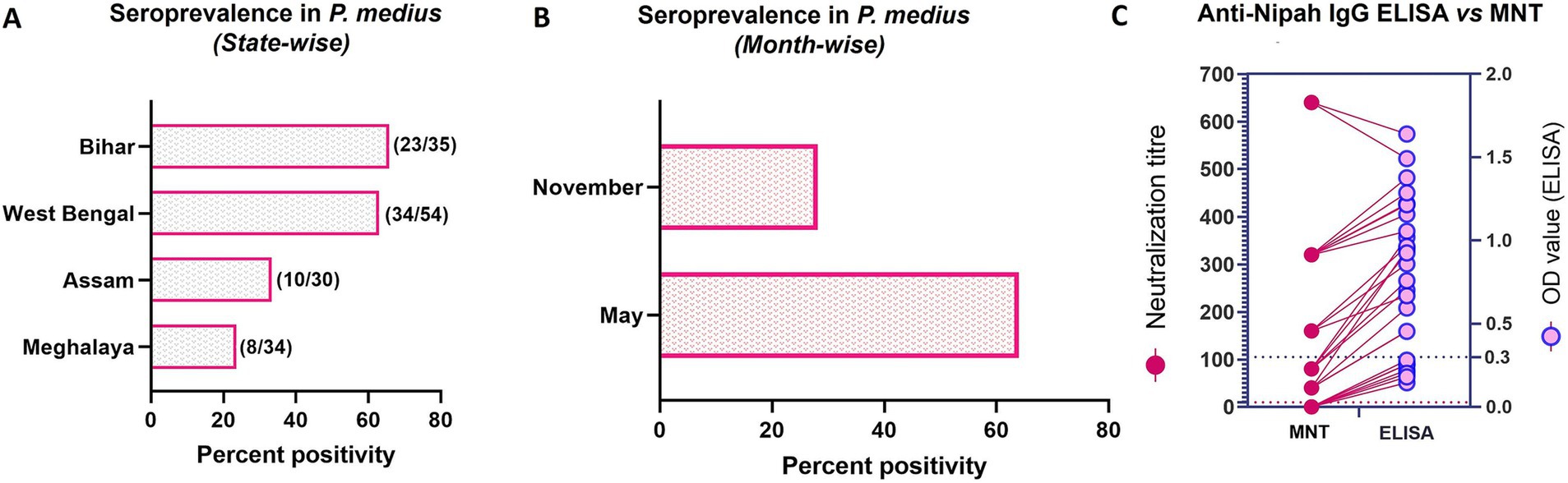
Figure 4. Nipah virus seroprevalence in P. medius. (A) The horizontal bar graph shows the percent positivity of bats with anti-Nipah IgG antibodies estimated by ELISA in Bihar, West Bengal, Assam, and Meghalaya states (the total number of IgG-positive samples out of the total serum samples tested is marked on the right of each bar). (B) The horizontal bar graph shows the percent positivity of bats with anti-Nipah IgG antibodies estimated by ELISA during the November and May months. (C) The graph shows the comparison of the neutralization titer (on the left y-axis) with the OD value of ELISA (on the right y-axis) of a subset of bat serum (19 ELISA positive and 6 ELISA negative samples). The cut-off of the microneutralization test (MNT) is marked as dotted lines (dark pink) at 10 on the left y-axis and ELISA as blue dotted lines at 0.3 on the right y-axis. The lines connect the values of the same sample by MNT and ELISA.
4 Discussion
The states of Bihar, West Bengal, Meghalaya, and Assam in India, which are adjacent to Bangladesh, showed a high seroprevalence in P. medius during the study. Among these states, West Bengal, Assam, and Meghalaya share a border with Bangladesh. During the sampling period of the study, fourteen cases of NiV infection were reported in several districts of Bangladesh, including Naogaon, Natore, Pabna, Rajshahi, Narsingdi, Rajbari, and Shariatpur. The case fatality rate was 71.4% (Nipah virus infection, n.d.; Gavi, n.d.). While Naogaon and Rajshahi districts share borders with the Indian state of West Bengal, the other districts where cases were reported are also situated closer to India.
The prevalence of NiV in bats in Bihar and Meghalaya states of India has not been studied previously. This study reports the first instance of viral RNA positivity in bats from Bihar state. Additionally, the Meghalaya state, which shares a border with the non-Nipah belt of Bangladesh, showed seroprevalence. A previous study demonstrated the presence of viral RNA and seroprevalence in P. medius in districts outside the Nipah belt of Bangladesh (Epstein et al., 2020). The sampling site in Meghalaya is only about 75 km far from Sylhet in Bangladesh which has reported NiV bat positivity (Anderson et al., 2019). Previous studies has also reported NiV detection in bats from the North Bengal region of India in Myanaguri in 2010 and Cooch Behar in 2015 (Yadav et al., 2012, 2018). These areas are located close to the site that was sampled for the present study in West Bengal, where NiV RNA was also detected. In 2015, the Dhubri district, located in the western part of Assam near the Nipah belt of Bangladesh, was surveyed, and viral RNA was detected in P. medius (Yadav et al., 2018). For the current study, a new urban location in central Assam, far from the previous surveyed site was chosen. This site is about 140 km from Sylhet, Bangladesh, and approximately 170 km from Dhubri, India where NiV positivity in bats was reported earlier. It was found that bats in this region have seropositivity.
Higher seroprevalence was observed in the West Bengal and Bihar sites, which were sampled during May, compared to the sites in Assam and Meghalaya, which were sampled in November. The higher prevalence of antibodies against NiV in bats, including juveniles, and the virus detection indicate that the virus is actively circulating in this area. Fluctuating seroprevalence (30 to 80%) and low NiV prevalence in P. medius have been reported in a longitudinal study performed at Faridpur, Bangladesh. There was also no evidence of seasonal fluctuations in seroprevalence or virus detection in this longitudinal study (Epstein et al., 2020). A three-year longitudinal study conducted on P. hypomelanus bats in Malaysia also showed a non-seasonal fluctuating seroprevalence ranging from 1 to 20% (Rahman et al., 2013). Our earlier studies in the northeast region of India in 2009–10 (May to October in Myanaguri) and 2015 (March–May in Jalpaiguri, Cooch Behar, and Dhubri; November in Dhubri) have shown low seropositivity (Yadav et al., 2012, 2018). Studies in Kerala state during the outbreak and non-outbreak periods have also shown seropositivity ranging between 9 to 21% in bats (Sudeep et al., 2021; Gokhale et al., 2022; Yadav et al., 2022; Balasubramanian et al., 2024).
The current study confirms active Nipah virus circulation among bats in eastern and northeastern India, with all surveyed sites showing seropositivity and viral RNA detected in Bihar and West Bengal. The absence of active human surveillance in India might have been one of the reasons for the absence of Nipah virus case detection from these studied regions. The hospital-based national surveillance system which includes active and passive surveillance strategies established since 2006 has played a key role in the detection and response of the outbreaks in Bangladesh (Satter et al., 2023). With the repeated outbreaks in Kerala state of India, syndromic surveillance was initiated in the state and this has resulted in early detection of cases and containment (Yadav et al., 2022). All these points towards the necessity of strengthening public health surveillance in the region. Recently, ICMR has included the Nipah virus in the list of pathogens for respiratory and acute encephalitis surveillance through its virus disease research network laboratories and associated tertiary care hospitals in West Bengal and Kerala (Unpublished data). This approach should be expanded to other regions of the country.
The throat and rectal swab samples collected during the present study tested negative for NiV, suggesting the absence of viral shedding through the gastrointestinal tract. The absence of swab positivity in organ-positive bats suggests that viral excretion in bats is episodic. Organ viral positivity in Pteropus bats has been reported in field and experimental studies (Middleton et al., 2007; Yadav et al., 2018). During experimental infection in P. poliocephalus bats, a transient presence of NiV was found in the kidney and uterus (Middleton et al., 2007). Multiple field studies have reported NiV RNA detection in spleen and kidney samples of P. medius (Epstein et al., 2016; Yadav et al., 2018; Balasubramanian et al., 2024). Urine and urogenital swabs, which are important samples to understand virus shedding, were not collected in the study. Efforts to isolate the virus were unsuccessful. Based on previous reports and our observations, virus isolation is challenging, and successful virus isolation does not appear to correlate with virus copy numbers (Anderson et al., 2019; Yadav et al., 2019; Balasubramanian et al., 2024).
The NiV strains found in Bangladesh have a nucleotide difference of around 9% from the Malaysian genotype and 3% from the virus sequences found in South India. The genome obtained from Bihar in the present study is over 99% similar to the NiV sequence of the 2007 outbreak from West Bengal in India, suggesting that a persistent virus strain is circulating in the region. The evolutionary rate estimated for NiV is 2.18 × 10−4 substitutions/site/year, which is relatively slow compared to similar viruses (Whitmer et al., 2021). It has been reported that there is a high level of sequence identity among NiV isolates found in bats in Bangladesh (Epstein et al., 2016; Anderson et al., 2019). The amino acid differences in the sequence retrieved in the present study were almost similar to those reported earlier for NiV-B sequences from Bangladesh and India (Lo et al., 2012; Mohandas et al., 2023). However, the significance of many of these changes is yet to be explored.
Fruit bats play a critical role in the ecosystem. The foraging range of Pteropus bats ranges from a few kilometers to 20–30 km and occasional reports of long-distance movement have also been reported. Migrant bats typically have a metapopulation structure with distinct roosts connected by migration, potentially exposing resident and migrant populations to viruses (Fleming, 2010). Migrations associated with breeding season, food scarcity, different seasons, and environmental disturbances have been reported (Epstein et al., 2020). Many studies have correlated spillover events with changing bat ecology associated with land use change, habitat loss, and periodic food resource shortages (McKee et al., 2021; Eby et al., 2023). Deforestation and urbanization have decreased forest cover and natural foraging resources for bats (McKee et al., 2021; Roy et al., 2024). As a result, bats are being forced to move and search for food in cultivated areas such as farms and home gardens. This could potentially increase the risk of disease spillover and such opportunistic feeding behavior has been documented in Malaysia and Bangladesh during previous outbreaks (Chua et al., 1999; McKee et al., 2021). Hence, it is crucial to prioritize the conservation of bat roosting trees, tree cover, and foraging sites to reduce human-bat interaction events. P. medius are seen throughout the Indian subcontinent (Saikia, 2018). The studies on bat ecology, behavior, foraging resources, migration, and land-use changes in the region could enhance the understanding of spillover threats. Increasing community awareness of potential risk factors and strategies to minimize exposure is essential.
The findings of this study indicate that a larger geographical area in India is at risk for Nipah virus spillover. These findings highlight the necessity of enhancing human surveillance for Nipah virus infection and investigating the disease risk factors in these regions of India. The implementation of a One Health approach is critical in preventing virus spillovers.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by Institutional Animal Ethics Committee, ICMR-National Institute of Virology, Pune. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SM: Formal analysis, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. DiP: Investigation, Methodology, Supervision, Writing – review & editing. BM: Investigation, Methodology, Supervision, Writing – review & editing. VR: Investigation, Methodology, Writing – review & editing. AS: Formal analysis, Methodology, Supervision, Writing – review & editing. SB: Investigation, Methodology, Writing – review & editing. AK: Methodology, Writing – review & editing. RS: Methodology, Writing – review & editing. DeP: Methodology, Writing – review & editing. PY: Conceptualization, Data curation, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study is supported by the extramural grant of the Indian Council of Medical Research for the project 'Countrywide survey of Nipah virus in Pteropus bats.
Acknowledgments
The authors acknowledge the Director, ICMR-National Institute of Virology, Pune for the support. We would also like to acknowledge the support of the office of the Principal Chief Conservator of Forests (Wildlife) of Bihar, West Bengal, Assam and Meghalaya states, Divisional Forest Officers of Shillong, Guwahati, Jalpaiguri and Patna, Office of the Guwahati Municipal Corporation, Office of the Shillong Cantonment Board, Station Commander, Binnaguri Military station and Office of the Chief Justice, High Court of Judicature at Patna for the sampling from the respective locations. We would also like to thank Sanjay Gopale, Ganesh Chopade, Manjunath Holeppanavar., Vishal Gaikwad, Vitthal P. Sasane, Annasaheb Suryawanshi, Deepak Mali, and Machhindra Karanjawane for the sample collection activities in the field. We would also like to acknowledge the support received from Rajlaxmi Jain for the logistic support as well as laboratory sample testing, Juhi Khurana, Prasad Sarkale, Triparna Majumdar, Savita Patil, Abhimanyu Kumar, Kundan Wakchaure, Pranita Gawande, Priya Wadhwania, Jyoti Yemul, Vaishnavi and Yash Joshi for laboratory sample processing and testing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1493428/full#supplementary-material
References
Anderson, D. E., Islam, A., Crameri, G., Todd, S., Islam, A., Khan, S. U., et al. (2019). Isolation and full-genome characterization of Nipah viruses from bats, Bangladesh. Emerg. Infect. Dis. 25, 166–170. doi: 10.3201/eid2501.180267
Arankalle, V. A., Bandyopadhyay, B. T., Ramdasi, A. Y., Jadi, R., Patil, D. R., Rahman, M., et al. (2011). Genomic characterization of Nipah virus, West Bengal, India. Emerg. Infect. Dis. 17, 907–909. doi: 10.3201/eid1705.100968
Arunkumar, G., Chandni, R., Mourya, D. T., Singh, S. K., Sadanandan, R., Sudan, P., et al. (2019). Outbreak investigation of Nipah virus disease in Kerala, India, 2018. J. Infect. Dis. 219, 1867–1878. doi: 10.1093/infdis/jiy612
As, A. K., et al. (2024). Clinico-epidemiological presentations and management of Nipah virus infection during the outbreak in Kozhikode district, Kerala state, India 2023. J. Med. Virol. 96:e29559. doi: 10.1002/jmv.29559
Balasubramanian, R., Mohandas, S., Thankappan, U. P., Shete, A., Patil, D., Sabarinath, K., et al. (2024). Surveillance of Nipah virus in Pteropus medius of Kerala state, India, 2023. Front. Microbiol. 15:1342170. doi: 10.3389/fmicb.2024.1342170
Breed, A. C., Yu, M., Barr, J. A., Crameri, G., Thalmann, C. M., and Wang, L. F. (2010). Prevalence of Henipavirus and Rubulavirus antibodies in Pteropid bats, Papua New Guinea. Emerg. Infect. Dis. 16, 1997–1999. doi: 10.3201/eid1612.100879
Chadha, M. S., Comer, J. A., Lowe, L., Rota, P. A., Rollin, P. E., Bellini, W. J., et al. (2006). Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 12, 235–240. doi: 10.3201/eid1202.051247
Ching, P. K. G., de los Reyes, V. C., Sucaldito, M. N., Tayag, E., Columna-Vingno, A. B., Malbas, F. F. Jr., et al. (2015). Outbreak of Henipavirus infection, Philippines, 2014. Emerg. Infect. Dis. 21, 328–331. doi: 10.3201/eid2102.141433
Chua, K. B., Goh, K. J., Wong, K. T., Kamarulzaman, A., Tan, P. S. K., Ksiazek, T. G., et al. (1999). Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354, 1257–1259. doi: 10.1016/S0140-6736(99)04299-3
Eby, P., Peel, A. J., Hoegh, A., Madden, W., Giles, J. R., Hudson, P. J., et al. (2023). Pathogen spillover driven by rapid changes in bat ecology. Nature 613, 340–344. doi: 10.1038/s41586-022-05506-2
Epstein, J. H., Anthony, S. J., Islam, A., Kilpatrick, A. M., Ali Khan, S., Balkey, M. D., et al. (2020). Nipah virus dynamics in bats and implications for spillover to humans. Proc. Natl. Acad. Sci. 117, 29190–29201. doi: 10.1073/pnas.2000429117
Epstein, J. H., Anthony, S. J., Islam, A., Kilpatrick, A. M., Khan, S. A., Ross, N., et al. (2016). Nipah virus ecology and infection dynamics in its bat reservoir, Pteropus medius, in Bangladesh. Int. J. Infect. Dis. 53, 20–21. doi: 10.1016/j.ijid.2016.11.056
Fleming, T. H. (2010). Bat migration. Encycl. Anim. Behav., 145–149. doi: 10.1016/B978-0-08-045337-8.00074-7
Gavi Nipah virus keeps the health system on its toes. Available at: https://www.gavi.org/vaccineswork/bangladesh-nipah-virus-keeps-health-system-its-toes (Accessed May 10, 2024).
Gokhale, M., Sudeep, A. B., Mathapati, B., Balasubramanian, R., Ullas, P. T., Mohandas, S., et al. (2022). Serosurvey for Nipah virus in bat population of southern part of India. Comp. Immunol. Microbiol. Infect. Dis. 85:101800. doi: 10.1016/j.cimid.2022.101800
Guillaume, V., Lefeuvre, A., Faure, C., Marianneau, P., Buckland, R., Lam, S. K., et al. (2004). Specific detection of Nipah virus using real-time RT-PCR (TaqMan). J. Virol. Methods 120, 229–237. doi: 10.1016/j.jviromet.2004.05.018
Hasebe, F., Thuy, N. T. T., Inoue, S., Yu, F., Kaku, Y., Watanabe, S., et al. (2012). Serologic evidence of nipah virus infection in bats, Vietnam. Emerg. Infect. Dis. 18, 536–537. doi: 10.3201/eid1803.111121
Iehlé, C., Razafitrimo, G., Razainirina, J., Andriaholinirina, N., Goodman, S. M., Faure, C., et al. (2007). Henipavirus and Tioman virus antibodies in Pteropodid bats, Madagascar. Emerg. Infect. Dis. 13, 159–161. doi: 10.3201/eid1301.060791
Kasso, M., and Balakrishnan, M. (2013). Ecological and Economic Importance of Bats (Order Chiroptera). International Scholarly Research Notices 2013:187415, 1–9. doi: 10.1155/2013/187415
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, Y., Wang, J., Hickey, A. C., Zhang, Y., Li, Y., Wu, Y., et al. (2008). Antibodies to Nipah or Nipah-like viruses in bats, China. Emerg. Infect. Dis. 14, 1974–1976. doi: 10.3201/eid1412.080359
Lo, M. K., Lowe, L., Hummel, K. B., Sazzad, H. M. S., Gurley, E. S., Hossain, M. J., et al. (2012). Characterization of Nipah virus from outbreaks in Bangladesh, 2008-2010. Emerg. Infect. Dis. 18, 248–255. doi: 10.3201/eid1802.111492
McKee, C. D., Islam, A., Luby, S. P., Salje, H., Hudson, P. J., Plowright, R. K., et al. (2021). The ecology of Nipah virus in Bangladesh: a Nexus of land-use change and opportunistic feeding behavior in bats. Viruses 13:169. doi: 10.3390/v13020169
Middleton, D. J., Morrissy, C. J., van der Heide, B. M., Russell, G. M., Braun, M. A., Westbury, H. A., et al. (2007). Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J. Comp. Pathol. 136, 266–272. doi: 10.1016/j.jcpa.2007.03.002
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mohandas, S., Shete, A., Sarkale, P., Kumar, A., Mote, C., and Yadav, P. (2023). Genomic characterization, transcriptome analysis, and pathogenicity of the Nipah virus (Indian isolate). Virulence 14:2224642. doi: 10.1080/21505594.2023.2224642
Nipah virus infection Nipah virus infection – Bangladesh. Available at: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON442 (Accessed November 14, 2023).
Paton, N. I., Leo, Y. S., Zaki, S. R., Auchus, A. P., Lee, K. E., Ling, A. E., et al. (1999). Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet 354, 1253–1256. doi: 10.1016/S0140-6736(99)04379-2
PIB A Nipah virus patient in Mallapuram district of Kerala, confirmed by NIV, Pune succumbs to the disease (2024). Available at: https://pib.gov.in/pib.gov.in/Pressreleaseshare.aspx?PRID=2034771 (Accessed October 22, 2024).
Rahman, M., and Chakraborty, A. (2012). Nipah virus outbreaks in Bangladesh: a deadly infectious disease. WHO South-East Asia Journal of Public Health 1, 208–212. doi: 10.4103/2224-3151.206933
Rahman, S. A., Hassan, L., Epstein, J. H., Mamat, Z. C., Yatim, A. M., Hassan, S. S., et al. (2013). Risk factors for Nipah virus infection among Pteropid bats, peninsular Malaysia. Emerg. Infect. Dis. 19, 51–60. doi: 10.3201/eid1901.120221
Reynes, J.-M., Counor, D., Ong, S., Faure, C., Seng, V., Molia, S., et al. (2005). Nipah virus in Lyle’s flying foxes, Cambodia. Emerg. Infect. Dis. 11, 1042–1047. doi: 10.3201/eid1107.041350
Roy, K., Saha, G. K., and Mazumdar, S. (2024). How free-ranging Indian flying fox (Pteropus medius) forage in urban areas? A study from Kolkata, India. J. Urban Ecol. 10:juae007. doi: 10.1093/jue/juae007
Saikia, U. (2018). A review of Chiropterological studies and a distributional list of the bat Fauna of India. Rec. Zool. Surv. India. 118, 242–280. doi: 10.26515/rzsi/v118/i3/2018/121056
Satter, S. M., Aquib, W. R., Sultana, S., Sharif, A. R., Nazneen, A., Alam, M. R., et al. (2023). Tackling a global epidemic threat: Nipah surveillance in Bangladesh, 2006–2021. PLoS Negl. Trop. Dis. 17:e0011617. doi: 10.1371/journal.pntd.0011617
Sendow, I., Field, H. E., Adjid, A., Ratnawati, A., Breed, A. C., Morrissy, C., et al. (2010). Screening for Nipah virus infection in West Kalimantan province, Indonesia. Zoonoses Public Health 57, 499–503. doi: 10.1111/j.1863-2378.2009.01252.x
Sudeep, A. B., Yadav, P. D., Gokhale, M. D., Balasubramanian, R., Gupta, N., Shete, A., et al. (2021). Detection of Nipah virus in Pteropus medius in 2019 outbreak from Ernakulam district, Kerala, India. BMC Infect. Dis. 21:162. doi: 10.1186/s12879-021-05865-7
Wacharapluesadee, S., Lumlertdacha, B., Boongird, K., Wanghongsa, S., Chanhome, L., Rollin, P., et al. (2005). Bat Nipah Virus, Thailand. Emerg. Infect. Dis. 11, 1949–1951. doi: 10.3201/eid1112.050613
Whitmer, S. L. M., Lo, M. K., Sazzad, H. M. S., Zufan, S., Gurley, E. S., Sultana, S., et al. (2021). Inference of Nipah virus evolution, 1999–2015. Virus Evol. 7:veaa 062. doi: 10.1093/ve/veaa062
WHO Nipah baseline situation analysis. Available at: https://www.who.int/publications/m/item/nipah-baseline-situation-analysis (Accessed November 14, 2023).
Yadav, P. D., Raut, C. G., Shete, A. M., Mishra, A. C., Towner, J. S., Nichol, S. T., et al. (2012). Detection of Nipah virus RNA in fruit bat (Pteropus giganteus) from India. Am. J. Trop. Med. Hygiene 87, 576–578. doi: 10.4269/ajtmh.2012.11-0416
Yadav, P. D., Sahay, R. R., Balakrishnan, A., Mohandas, S., Radhakrishnan, C., Gokhale, M. D., et al. (2022). Nipah virus outbreak in Kerala state, India amidst of COVID-19 pandemic. Front. Public Health 10:818545. doi: 10.3389/fpubh.2022.818545
Yadav, P. D., Shete, A. M., Kumar, G. A., Sarkale, P., Sahay, R. R., Radhakrishnan, C., et al. (2019). Nipah virus sequences from humans and bats during Nipah outbreak, Kerala, India, 2018. Emerg. Infect. Dis. 25, 1003–1006. doi: 10.3201/eid2505.181076
Keywords: Nipah virus, eastern India, Pteropus medius , Bihar, West Bengal, Assam, Meghalaya
Citation: Mohandas S, Patil D, Mathapati B, Rai V, Shete A, Belani S, Kumar A, Sahay R, Patil D and Yadav PD (2024) Nipah virus survey in Pteropus medius of eastern and northeastern region of India, 2022–2023. Front. Microbiol. 15:1493428. doi: 10.3389/fmicb.2024.1493428
Edited by:
Junki Maruyama, University of Texas Medical Branch at Galveston, United StatesReviewed by:
Stephanie Salyer, Centers for Disease Control and Prevention (CDC), United StatesHongzhao Li, Canadian Food Inspection Agency (CFIA), Canada
Copyright © 2024 Mohandas, Patil, Mathapati, Rai, Shete, Belani, Kumar, Sahay, Patil and Yadav. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pragya D. Yadav, aGVsbG9wcmFneWEyMkBnbWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
 Sreelekshmy Mohandas
Sreelekshmy Mohandas Dilip Patil
Dilip Patil Basavaraj Mathapati
Basavaraj Mathapati Vishal Rai
Vishal Rai Anita Shete
Anita Shete Sujeet Belani4
Sujeet Belani4 Abhinendra Kumar
Abhinendra Kumar Rima Sahay
Rima Sahay Deepak Patil
Deepak Patil Pragya D. Yadav
Pragya D. Yadav