- 1Department of Food Science and Technology, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia
- 2Department of Biotechnology, Jožef Stefan Institute, Ljubljana, Slovenia
The pathogenic bacterium Campylobacter jejuni is a major food safety concern as it can form biofilms that increase its survival and infective potential. Biofilms consist of microbial cells and extracellular matrix (ECM), which is made of water and extracellular polymeric substances (EPS), which are critical for structural integrity and pathogenicity. The aim of this study was to optimize a protocol for the isolation of C. jejuni ECM. We employed eight physical and chemical isolation methods to extract and purify ECM, followed by different qualitative and quantitative analyses using gel electrophoresis and spectroscopy. This comprehensive approach enabled the evaluation of ECM composition in terms of polysaccharides, proteins, and extracellular DNA. The isolation methods resulted in different yields and purities of the extracted ECM components. Centrifugation in combination with chemical treatments proved to be most effective, isolating higher concentrations of polysaccharides and proteins. Additionally, extraction with ether solution facilitated the recovery of high-molecular-weight extracellular DNA. Overall, we provide a refined methodology for ECM extraction from C. jejuni. As polysaccharides and proteins participate in biofilm stability and microbial communication, and extracellular DNA participates in genetic exchange and virulence, our study contributes towards a better understanding of the persistence of this pathogen in the food industry.
1 Introduction
The Gram-negative bacterium Campylobacter jejuni is found in the intestines of many wild and domestic animals, making them potential asymptomatic carriers or zoonotic transmission (Blaser, 1997; Snelling et al., 2005; Burnham and Hendrixson, 2018). It is one of the main causes of bacterial foodborne gastroenteritis worldwide and the most common cause of foodborne zoonotic infections (EFSA and ECDC, 2023). Infections in humans usually occur through the ingestion of contaminated food of animal origin or untreated water or direct contact with infected animals, particularly pets. However, most human cases are associated with the consumption of contaminated poultry (Blaser, 1997; Sheppard and Maiden, 2015). C. jejuni has an optimal growth temperature of around 42°C, which facilitates its colonization in chicken intestine and makes poultry an important vector for its transmission into the human food chain (Snelling et al., 2005; Levin, 2007; Sheppard and Maiden, 2015; Burnham and Hendrixson, 2018).
Contrary to previous assumptions that C. jejuni cannot survive outside hosts in aerobic natural environments or in the food chain (Solomon and Hoover, 1999; Klančnik et al., 2013, 2014; Giaouris et al., 2015), it now shows a wide distribution in the environment and has been detected in food, water, and microbial biofilms on microplastics from seawater (Good et al., 2019; Tram et al., 2020; Kolenc et al., 2024). Recent studies have further elucidated its pathogenesis, persistence, and resilience and have linked these properties to genomic polymorphism, limited catabolic capacity, abnormalities in gene regulation, and a protective biofilm matrix that shields it from environmental stressors (Klančnik et al., 2021; Sabotič et al., 2023).
The formation of biofilms is an important survival strategy for C. jejuni, providing protection against environmental stress and increasing its infectivity. These biofilms comprise dynamic microbial communities that form on both abiotic and biotic surfaces and are driven by multiple cellular interactions and complex adhesion mechanisms. Biofilms can rapidly (within 48 h) develop into dense structures with strong adhesion and structural complexity and are thus difficult to treat (Sabotič et al., 2023; Joshua et al., 2006; Teh et al., 2010; Reuter et al., 2010; Püning et al., 2021; Ramić et al., 2023; Ma et al., 2022; Carpentier and Cerf, 1993; Sulaeman et al., 2012; Kemper and Hensel, 2023).
Biofilms are complex assemblies of microorganisms embedded in an extracellular matrix (ECM) of water and extracellular polymeric substances (EPS; Aguilera et al., 2008). EPS are crucial for the formation, architecture, and functionality of biofilms and account for 50–90% of biofilm mass. They include polysaccharides, proteins, lipids, and extracellular DNA (eDNA) at different concentrations, depending on environmental conditions and nutrient availability (Donlan, 2002; Vu et al., 2009; Flemming et al., 2016; Karygianni et al., 2020; Flemming et al., 2023).
ECM plays a crucial role in improving the resistance of C. jejuni biofilms to environmental stress. It forms a protective barrier around cells that not only protects against physical disturbances, antimicrobial agents, bacteriophages, and biocides but also contributes to the mechanical strength and stability of the biofilm. This barrier increases the antimicrobial resistance of the biofilm by hindering the diffusion of antibiotics and complicating the treatment of associated infections by promoting the intercellular exchange of resistance genes (Donlan, 2002; Karygianni et al., 2020; Flemming et al., 2016). The bacterial ECM also significantly affects the heterogeneity of biofilms by influencing porosity, density, and water content, thereby improving the adaptability of biofilms to different environmental conditions. In addition, bacterial ECM promotes unique microenvironments by enriching biofilms with nutrients and supporting vital functions, such as resource acquisition, hydration, and external digestion, all of which are important for cell survival, metabolic activity, and intercellular interactions. The bacterial ECM contains environmental materials, such as dissolved nutrients, humic substances, and exopolymer particles, which are crucial for cellular metabolism and the structural integrity of the biofilm. Furthermore, most bacterial ECM consist of neutral or polyanionic polysaccharides, which are crucial for the attraction of divalent cations such as calcium and magnesium. These cations bind to bacterial ECM polymers and form hydrogen bonds that stabilize biofilm structure, provide structural cohesion, and protect the biofilm from desiccation (Donlan, 2002). Environmental factors, such as temperature, pH, nutrient availability, and stress, influence bacterial regulatory pathways and lead to increased bacterial ECM production and modifications (Moorhead and Griffiths, 2011; Lu et al., 2012; Feng et al., 2016). Components of ECM, such as eDNA, can be degraded by enzymes or external factors, leading to the dissolution of biofilms and potential bacterial proliferation (Brown et al., 2015; Flemming et al., 2016; Karygianni et al., 2020). This degradation underlines the dynamic nature of biofilms and their susceptibility to environmental influences.
ECM is also central to the formation and maintenance of C. jejuni biofilms, thereby increasing biofilm resistance and facilitating C. jejuni spread. In C. jejuni, ECM plays a complex role in biofilms and are thus important for microbial ecology, pathogenesis, and treatment resistance (Table 1). Different methods have been used to study ECM characteristics, composition and function (Table 2). Crystal violet staining was usually used to quantify biofilms, which is essential for the estimation of ECM content (Feng et al., 2016; Oh et al., 2018). Fluorescence microscopy was used for ECM visualization, with methods such as high-content screening with TAMRA and SytoX fluorescent markers providing quantitative insights into the integrity and composition of ECM (Oh et al., 2018; Whelan et al., 2021).
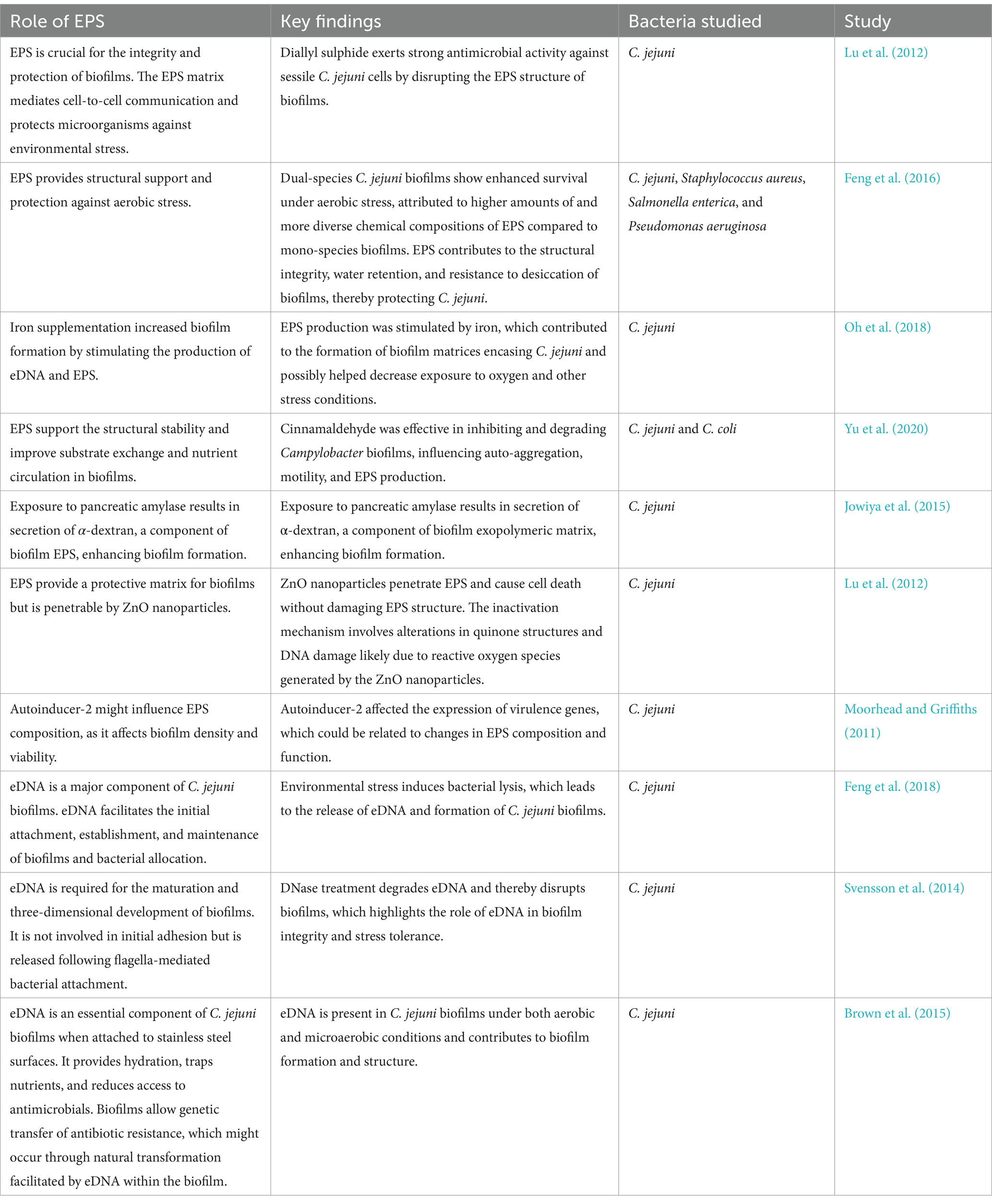
Table 1. The roles of extracellular polymeric substances (EPS) in Campylobacter jejuni biofilms. eDNA: extracellular DNA.
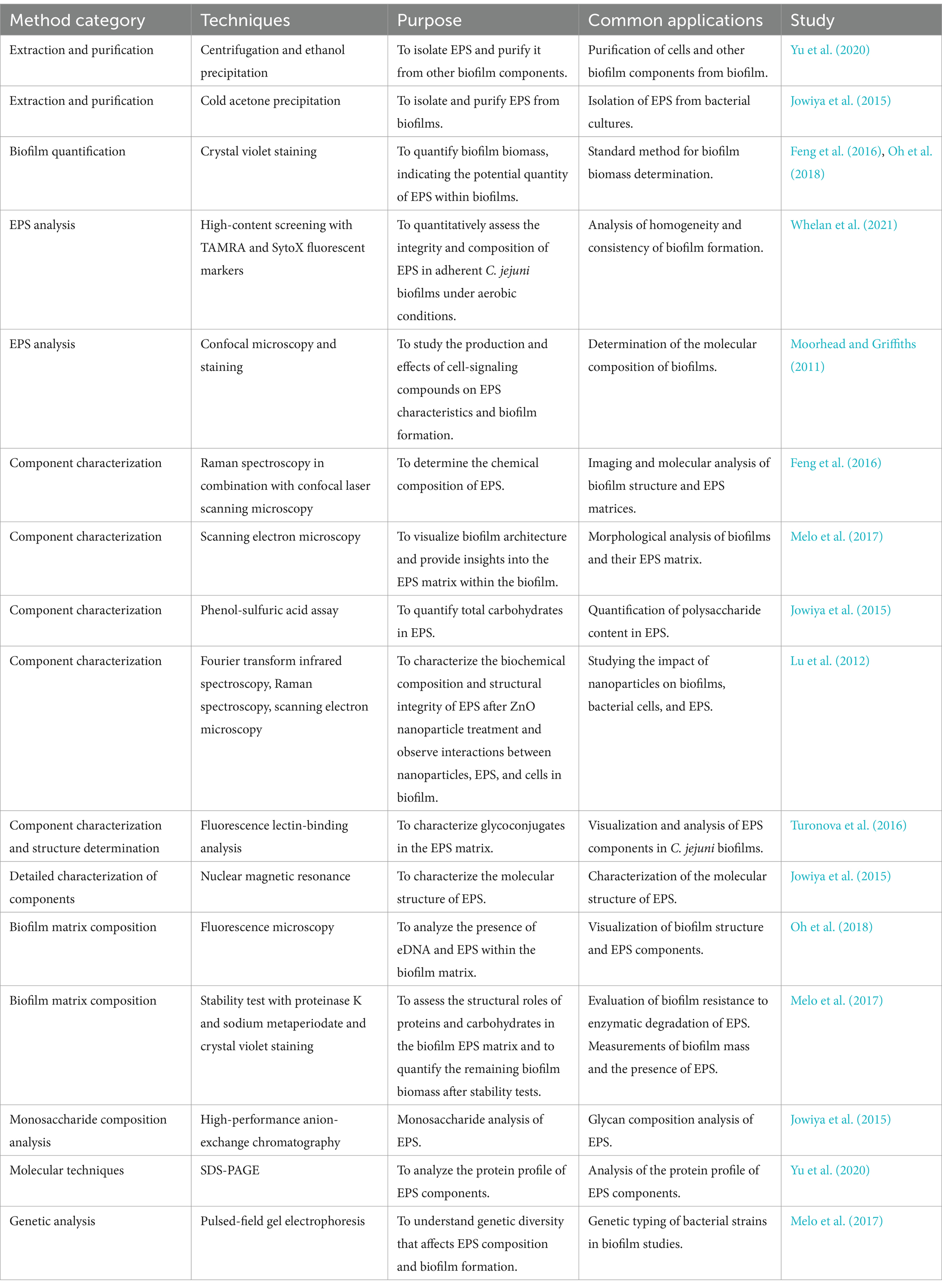
Table 2. The methods used in studies of Campylobacter jejuni extracellular polymeric substances (EPS).
The techniques used to characterize ECM components range from microscopic methods such as confocal microscopy and scanning electron microscopy to spectroscopic methods such as Raman spectroscopy and Fourier transform infrared spectroscopy, which help determine the biochemical composition of ECM in detail (Moorhead and Griffiths, 2011; Feng et al., 2016; Melo et al., 2017). In addition, high-performance anion exchange chromatography and nuclear magnetic resonance provide more precise details about the molecular structure of ECM components (Jowiya et al., 2015). Moreover, molecular techniques and genetic analyses link ECM components to their functional roles (Melo et al., 2017; Yu et al., 2020). Quantitative PCR with SYBR Green I and specific primers is used to investigate specific components (e.g., eDNA), also in combination with confocal microscopy (Feng et al., 2018). DNase-I is used to investigate the roles of flagella-mediated adhesion and eDNA in biofilm formation and maturation (Svensson et al., 2014; Feng et al., 2018; Brown et al., 2015). Fluorescence lectin binding analysis is used to characterize glycoconjugates in ECM, providing insights into the complex interactions within biofilms (Turonova et al., 2016).
The aim of this study was to optimize a protocol that isolates the ECM of C. jejuni and to evaluate its composition. We investigated different physical and chemical methods for isolating essential ECM components, such as polysaccharides, proteins, and eDNA, to determine the most effective techniques to extract these molecules at high concentrations.
2 Materials and methods
2.1 Growth conditions
Cultures of C. jejuni ATCC 11168 were stored at −80°Cin 20% glycerol (Kemika, Croatia) and 80% Mueller Hinton broth (MHB, Oxoid, UK). C. jejuni was incubated on Karmali agar (Oxoid, UK) supplemented with Campylobacter-selective Karmali supplement (Oxoid, UK) for 24 h under microaerobic conditions (5% O2, 10% CO2, and 85% N2; Thermo Scientific Oxoid CampyGen atmosphere, USA) in anaerobic jar (3.5 L, Oxoid, UK) in incubator (Kambič, Slovenia) at 42°C. The pure culture was transferred to Mueller Hinton agar (MHA, Oxoid, UK) and incubated under the same conditions overnight.
2.2 ECM isolation
Eight different methods were used to isolate the ECM, all of which were preceded by the same step. C. jejuni biomass was scraped off four plates using a sterile disposable cotton swab and added to 1.5 ml microcentrifuge tubes containing 1 ml of phosphate-buffered saline (PBS; 10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.4). Suspension was centrifuged at 12,000 × g for 3 min at 4°C. The supernatant with weakly bound ECM components (named as method SV) was then removed using an automated pipette, filtered through a 0.2 μm pore size membrane (Whatman, UK), and stored at −20°C until further use. The obtained biomass pellet was used for the eight isolation methods (named A–H) described below. The isolated ECM was stored at −20°C until further use.
2.2.1 Isolation with weakly bound ECM (named as method SV)
The supernatant containing weakly bound ECM components was obtained after centrifugation of the resuspended C. jeuni biomass at 12,000 × g for 3 min at 4°C. It was removed using an automated pipette, filtered through a 0.2 μm pore size membrane (Whatman, UK), and stored at −20°C until further use.
2.2.2 Isolation with NaCl (named as method A)
The pellet was resuspended in 1 ml of 1.5 M NaCl (KEFO 7647-14-5) solution by vortexing. This suspension was centrifuged at 5,000 × g for 10 min at 25°C, and the supernatant, containing the isolated ECM, was filtered through a 0.2 μm pore size membrane (Whatman, UK; Chiba et al., 2015).
2.2.3 Isolation by centrifugation (named as method B)
The pellet was resuspended in 1 ml of PBS (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.4) by vortexing. This suspension was centrifuged at 20,000 × g for 20 min at 4°C, and the supernatant, containing the isolated ECM, was filtered through a 0.2 μm pore size membrane (Whatman, UK; Liu and Fang, 2002).
2.2.4 Isolation by heating in Na2CO3 (named as method C)
The pellet was resuspended in 1 ml of PBS by vortexing, and this suspension was transferred into a new 1.5 ml microcentrifuge tube with 5 mg of Na2CO3 (Honeywell Fluka 31432). The solution was then incubated for 35 min in a ThermoShaker thermoblock at 80°C with simultaneous stirring at 400 rpm and then cooled at room temperature and centrifuged at 12,000 × g for 20 min at 4°C (Felz et al., 2016). The supernatant, containing the isolated ECM, was filtered through a 0.2 μm pore size membrane (Whatman, UK).
2.2.5 Isolation with ethylenediaminetetraacetic acid (EDTA; named as method D)
The pellet was resuspended in 1 ml of PBS by vortexing, and the suspension was divided into two 2 ml microcentrifuge tubes for further steps, to each of which 500 μl of 2% EDTA (Serva 11280.02) was added to give a final EDTA concentration of 1%. This was followed by incubation with agitation on an orbital shaker for 3 h at 4°C and then centrifugation at 12,000 × g for 20 min at 4°C (Jachlewski et al., 2015). The supernatant, containing the isolated ECM, was filtered through a 0.2 μm pore size membrane (Whatman, UK).
2.2.6 Isolation with NaOH (named as method E)
The pellet was resuspended in 1 ml of PBS by vortexing, and 0.4 g of NaOH (Fisher 1310-73-2) was added. This was followed by incubation with agitation on an orbital shaker for 3 h at 4°C and centrifugation at 20,000 × g for 20 min at 4°C (Jachlewski et al., 2015). The supernatant, containing the isolated ECM, was filtered through a 0.2 μm pore size membrane (Whatman, UK).
2.2.7 Isolation with formaldehyde and NaOH (named as method F)
The pellet was resuspended in 1 ml of PBS by vortexing, and 6 μl of 37% formaldehyde (Sigma-Aldrich 1.04003.1000, Merck, Germany) was added. This was followed by incubation with agitation on an orbital shaker for 1 h at 4°C. Next, 0.4 ml of 1 M NaOH was added, followed by incubation with agitation on an orbital shaker for 3 h at 4°C and centrifugation at 20,000 × g for 20 min at 4°C (Liu and Fang, 2002). The supernatant, containing the isolated ECM, was filtered through a 0.2 μm pore size membrane (Whatman, UK).
2.2.8 Isolation with a Dowex cation exchange resin (named as method G)
First, the extraction buffer and Dowex cation exchange resin were prepared. The extraction buffer contained 17.8 mg of Na2HPO4 · 2H2O (Serva 30200.01), 27.5 mg of NaH2PO4 · H2O (Serva 30186), 26 mg of NaCl (KEFO 7647-14-5), 3.7 mg of KCl (Serva 26868.02), and 50 ml of distilled water. The Dowex cation exchanger was prepared by adding 1 g of Dowex cation exchange resin (Supelco 44514, Merck, Germany) to 10 ml of extraction buffer, mixing well with an automatic pipette, and incubating for 15 min. The extraction buffer was then removed using an automated pipette, and the washing procedure repeated. The extraction buffer was then removed again using an automated pipette, and 10 ml of extraction buffer was added. The prepared cation exchanger was stored at 4°C until use.
The pellet was resuspended in 1 ml of PBS by vortexing, and the suspension was divided into two 2 ml microcentrifuge tubes, to each of which 1 ml of Dowex cation exchanger was added. This was followed by incubation with agitation on an orbital shaker for 3 h at 4°C and centrifugation at 12,000 × g for 10 min at 4°C (Frølund et al., 1996). The supernatant, containing the isolated ECM, was filtered through a 0.2 μm pore size membrane (Whatman, UK).
2.2.9 Isolation with ether solution (named as method H)
First, 10 ml of 30 mM ether solution was prepared from 112 mg of dicyclohexano-18-crown-6 (Sigma-Aldrich 158402, Merck, Germany) and 50 mM Tris–HCl, pH 8 (Tris, Serva 37180.04; HCl, VWR Chemicals BDH 20252.290).
The pellet was resuspended in 1 ml of PBS by vortexing, and the suspension was divided into two 2 ml microcentrifuge tubes, to each of which 500 μl of 30 mM ether solution was added. This was followed by incubation with agitation on an orbital shaker for 3 h at 4°C and then centrifugation at 16,000 × g for 20 min at 4°C (Jachlewski et al., 2015). The supernatant, containing the isolated ECM, was filtered through a 0.2 μm pore size membrane.
2.3 Preparation of total cell lysate (named as method CL)
Before cell lysate preparation, empty 15 ml centrifuge tubes were weighed. Next, C. jejuni biomass was scraped off eight plates with a sterile disposable cotton swab and added to tubes containing 4 ml of cell lysate buffer (2 mM EDTA and 1% Triton X-100 (9036-19-5, Merck) in PBS). This was followed by centrifugation at 4,400 × g for 20 min at 4°C. The supernatant was then removed, and the remaining pellet was weighed to calculate the amount of biomass scraped from the plates. The pellet was then resuspended in 4 ml of cell lysate buffer. This was followed by sonication with the Hielscher UP200St (Ultrasound Technology, Germany) sonicator (cycle, 90%; amplitude, 90; power 200 W) for four rounds of 5 min each. This was followed by centrifugation at 18,000 × g for 5 min at 4°C. Cell lysates were filtered through a 0.2 μm pore size membrane and stored at −20°C until further use.
2.4 Analysis of isolated ECM
The isolated ECM samples were analyzed for polysaccharides, proteins and eDNA. The polysaccharides were analyzed using the phenol-sulfuric acid method. In addition, SDS-PAGE and periodic acid-Schiff staining were used to detect glycoproteins and polysaccharides and SDS-PAGE and Alcian blue staining were used to detect acidic polysaccharides and polysaccharides with sulfate groups. The total protein content of the samples was determined using the commercial colorimetric DC protein assay (Bio-Rad, USA). The proteins in the sample were also analyzed by SDS-PAGE and Coomassie staining. The eDNA content was determined by agarose gel electrophoresis.
2.4.1 Phenol-sulfuric acid method
The phenol-sulphuric acid method is a quantitative spectrophotometric method for determining the concentration of carbohydrates in a sample. A calibration curve was established with glucose standard solutions prepared from a stock concentration of 1 mg/ml. A glucose solution of the indicated concentration was prepared in sterile glass tubes to a final volume of 100 μl in two technical replicates of each concentration. Samples were prepared in two technical replicates by adding 50 μl of dH2O to 50 μl of the sample. 50 μl of 80% phenol (Sigma-Aldrich P9346) was then added to all tubes and the contents shaken. Then 2 ml of sulfuric acid (Carlo Erba Reagents 410301, Italy) was added to each tube and incubated for 10 min at room temperature. After 10 min, 1 ml of each solution was transferred to cuvettes and the absorbance was measured at 490 nm using a Lambda-25 spectrophotometer (PerkinElmer, USA). The concentrations of polysaccharides in each ECM sample were then determined using the calibration curve. For the statistical analysis of the results, an ordinary one-way ANOVA with Dunnett’s comparison tests was performed in GraphPad Prism (GraphPad Software, San Diego, United States).
2.4.2 DC protein assay kit analysis
The commercial DC Protein Assay Kit (Bio-Rad, USA) was used to determine the protein concentration in ECM samples. A calibration curve for bovine serum albumin was established to determine the protein concentration. The calibration curve for bovine serum albumin (Sigma-Aldrich 9048-46-8) was generated with a stock concentration of 10 mg/ml. 5 μl of the standard solution of bovine serum albumin or 5 μl of each ECM sample was applied to a microtiter plate. To ensure comparability with the calibration curve, 2- and 5-fold dilutions were also prepared for the samples. Subsequently, 25 μl of reagent A’ (Bio-Rad, USA) and 200 μl of reagent B (Bio-Rad, USA) were added to all wells of the microtiter plate and incubated on an orbital shaker for 5 s while shaking. This was followed by a 15-min incubation at room temperature. After 15 min, the absorbance was measured at 750 nm using a Tecan Infinite M1000 spectrophotometer (Tecan, Switzerland). The protein concentration of each sample was determined using the calibration curve. For the statistical analysis of the results, an ordinary one-way ANOVA with Dunnett’s comparison tests was performed in GraphPad Prism (GraphPad Software, San Diego, United States).
2.4.3 SDS-PAGE and different staining methods
The protein content of the ECM samples was analyzed by SDS-PAGE. 30 μl of ECM was mixed with 5 μl of 6× loading buffer and loaded onto a 1.5 mm 10% polyacrylamide geL. The Amersham Low Molecular Weight Calibration Kit for SDS electrophoresis (Cytiva 17-0446-01, GE HealthCare, USA) was used as a size marker. Electrophoresis was performed in 1X SDS buffer in a SDS-PAGE device (Mini Protean II, Bio-Rad, USA). A constant current of 35 mA/gel was used for protein separation.
2.4.3.1 Glycoprotein and polysaccharide detection
Periodic acid staining and Schiff’s reagent (Sigma-Aldrich, USA) were used to detect glycoproteins and polysaccharides following manufacturer’s instructions. The SDS-PAGE gel was first fixed in 50% methanol (Carlo Erba Reagents 412721) for 30 min. The gel was then washed twice with 3% glacial acetic acid (J.T.Baker 64–19-7) solution, each wash lasting 20 min. This was followed by incubation with oxidation reagent (to prepare 100 ml of oxidation reagent, 1 g of periodic acid (Sigma-Aldrich P7875) was weighed, 3 ml of 100% glacial acetic acid and up to 100 ml of dH2O were added) for 15 min with constant stirring. The gel was then washed three times with 3% glacial acetic acid solution, each wash lasting 15 min. The gel was then incubated with Schiff’s reagent (Sigma-Aldrich S5133) for 35 min and then with reducing reagent (0.5 g Na₂S₂O₅ (Sigma-Aldrich 71932, Merck) was used to prepare 100 ml reducing reagent and added up to 100 ml dH2O) for 5 min. It was washed with 3% glacial acetic acid solution for 5 min and finally with ultrapure water for 5 min. The gel was photographed with the camera of a cell phone.
2.4.3.2 Polysaccharide detection
To detect acidic polysaccharides and polysaccharides with sulphate groups, staining with the dye Alcian blue was performed. The SDS-PAGE gel was first fixed in EAW (Ethanol, Acetic acid and distilled Water) solution for 4 h (to prepare 100 ml of EAW, 40 ml of absolute ethanol, 5 ml of 100% glacial acetic acid and up to 100 ml of dH2O were added). The gel was then incubated in Alcian blue solution for 30 min (to prepare 100 ml of Alcian blue solution, 0.5 g of Alcian blue dye (Merck 1.05234.0010) was weighed out, 2 ml of 100% glacial acetic acid and up to 100 ml of dH2O were added). After incubation, the gel was washed twice with 2% glacial acetic acid solution, each wash lasting 15 min, and then incubated overnight. The gel was photographed the next day using a ChemiDoc gel documenter (Bio-Rad, USA) and a cell phone camera.
2.4.3.3 Protein detection
The proteins on the SDS-PAGE gels were detected using Coomassie Brilliant Blue staining. After electrophoresis, the gel was transferred to a Petri dish containing Coomassie dye (ThermoScientific, USA). The gel was incubated in the solution for 1 h on an orbital shaker at room temperature. After 1 h of incubation, destaining was performed with 30% destaining solution (300 ml ethanol (Carlo Erba Reagents 4146072), 100 ml glacial acetic acid, and 600 ml dH2O were used to prepare 1 L solution), which was changed three times every 15 min. A 10% destaining solution (140 ml ethanol, 50 ml glacial acetic acid and 810 ml dH2O) was then added and the gel was incubated overnight. The gel was photographed the next day using a ChemiDoc gel documentation device (Bio-Rad, USA).
2.4.4 Agarose gel electrophoresis
Agarose gel electrophoresis was used to analyze the DNA content of the ECM. A 1% agarose gel was prepared. The gel was prepared by weighing 0.5 g of agarose (Sigma-Aldrich A9539) into a flask and dissolving it in 50 ml of TAE (40 mM Tris-acetate, pH 8, 1 mM EDTA) electrophoresis buffer. The melted agarose was cooled to 60°C and 5 μl of the intercalating dye SYBR Safe (Invitrogen S33102) was added. The solution was poured into an electrophoresis beaker (Bio-Rad, USA). After the gel had set, 1X TAE buffer was added and 20 μl of the sample was applied to the pockets to which 4 μl of 6-fold loading buffer had already been added. The DNA size marker 1 kb GeneRuler (Thermo Scientific, USA) for the determination of fragment size above 1 kbp (6 μl) was also applied to the gel to which 1 μl of 6-fold loading buffer had been added. Agarose gel electrophoresis was performed at a constant voltage of 80 V for approximately 40 min. After completion of electrophoresis, the gel was transferred from the beaker to a UV beam gel imager (UVItec, UK) or an Image Lab Touch gel imager (Bio-Rad, USA).
3 Results and discussion
After extraction, we determined the eDNA content in each sample, the protein content and the polysaccharide content to evaluate the yield of these macromolecules for each protocol. We used eight different protocols that utilized NaCl, centrifugation, heating in Na2CO3, EDTA, NaOH, formaldehyde and NaOH, the cation exchanger Dowex, and an ether solution. Different protocols for isolating the ECM of C. jejuni yielded variable amounts of polysaccharides, proteins, and eDNA. Interestingly, Flemming and Wingender (2010) and Aguilera et al. (2008) came to similar conclusions for ECM of other types of microbial biofilms. Numerous studies have endeavoured to develop a protocol for the optimal isolation of ECM components (Liang et al., 1992; Tabouret et al., 1992; Tapia et al., 2009; Wu and Xi, 2009), however, a universal method for ECM isolation remains elusive and challenging in terms of cost-effectiveness, simplicity, and applicability to different types of components and bacteria (Aguilera et al., 2008; Chiba et al., 2015).
To evaluate the isolated ECM samples, we assessed which isolation methods are most suitable for (1) isolating the ECM of C. jejuni, (2) isolating individual major components of the ECM, and (3) yielding the highest concentrations of individual ECM components. Table 3 summarizes the main components of the ECM in samples prepared by different isolation methods in our study.
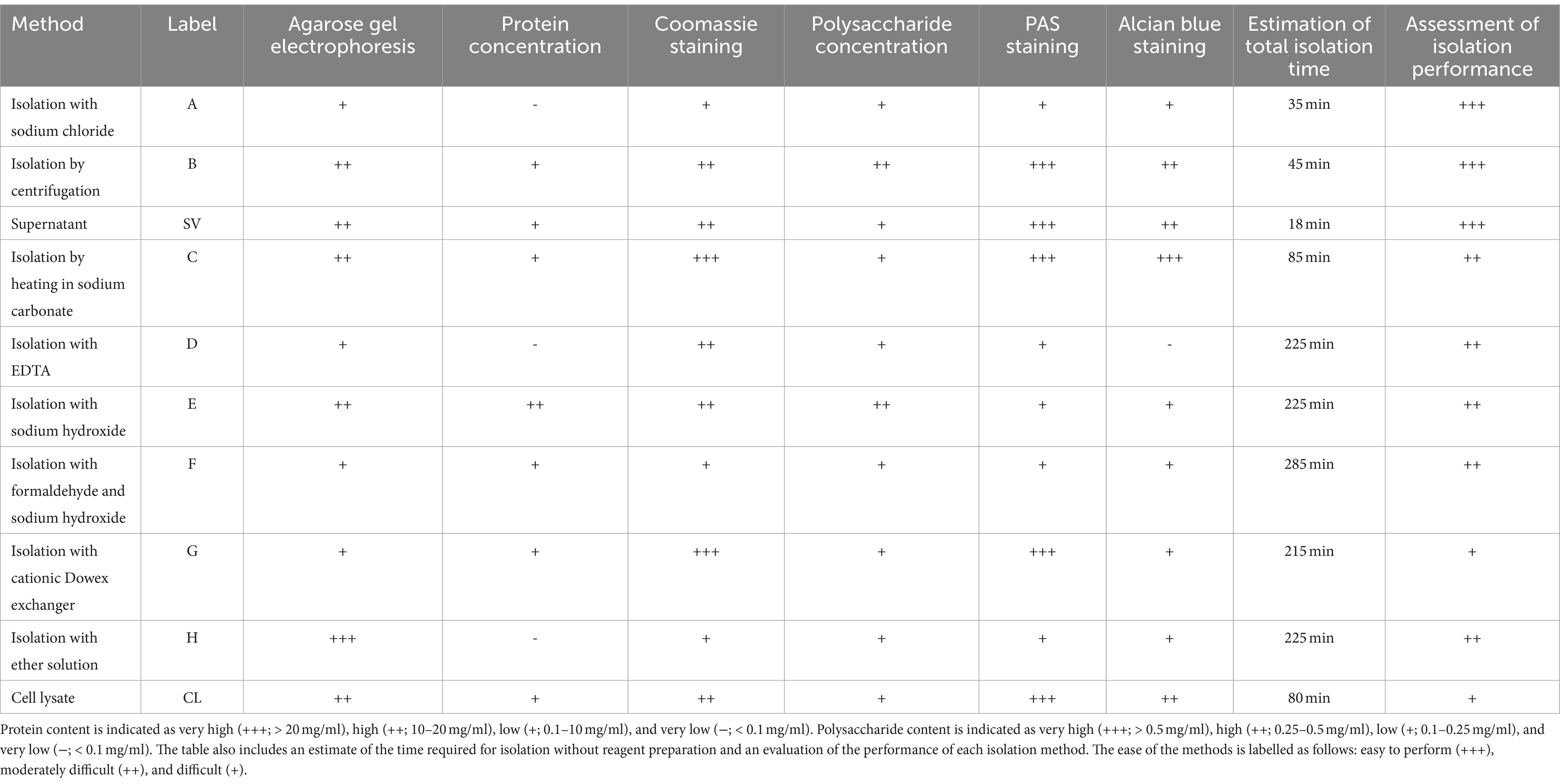
Table 3. Analysis results for determining the presence of major extracellular matrix components in samples prepared using different methods for isolating the extracellular matrix of C. jejuni, including the estimation of total isolation time and assessment of isolation performance.
3.1 Polysaccharides
The polysaccharide content in samples obtained according to different extraction protocols was analyzed using the phenol-sulfuric acid method and SDS-PAGE followed by periodic acid-Schiff or Alcian blue staining. First, polysaccharide concentrations were determined using the phenol-sulfuric acid method (Figure 1A). The highest polysaccharide concentrations were found in isolates obtained with both centrifugation methods (B and SV) and with NaOH (E). In other ECM isolates and in the cell lysate (CL), lower but comparable polysaccharide concentrations were found. The lowest polysaccharide concentrations were found in isolates obtained with NaCl (A), EDTA (D), and by heating in Na2CO3 (C). Despite these differences, all the methods yielded polysaccharide concentrations of less than 0.5 mg/ml. Our results indicate that the methods that most effectively isolate high polysaccharide concentrations from the ECM of C. jejuni employ either NaOH (E) or centrifugation only (B and SV).
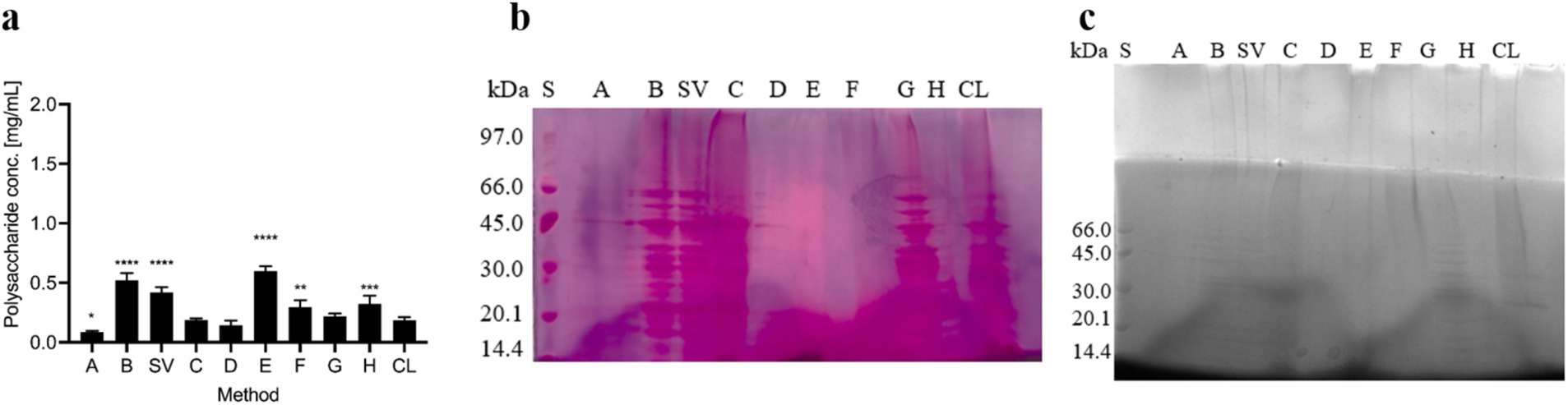
Figure 1. Polysaccharides in the ECM isolates of Campylobacter jejuni. (A) Polysaccharide content as revealed by the phenol-sulphuric acid method. The mean values and standard deviations of two measurements are shown. Dunnett’s multiple comparisons with the control cell lysate tested at significance levels of * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. The ANOVA results were significant [F (9, 3) = 60.98, p < 0.0001]. (B) SDS-PAGE analysis with periodic acid-Schiff staining. (C) SDS-PAGE analysis with Alcian blue staining. Samples were obtained by sodium chloride isolation (A), centrifugation (B), a procedure that yielded a supernatant of weakly bound extracellular matrix components (SV), heating in sodium carbonate (C), EDTA (D), sodium hydroxide (E), formaldehyde and sodium hydroxide (F), Dowex cation exchanger (G) and ether solution (H). Total cell lysate (CL) was also tested.
SDS-PAGE and periodic acid-Schiff staining (Figure 1B) showed that many individual bands were present in the isolates obtained by centrifugation (B and SV), heating in Na2CO3 (C), and the cation exchanger Dowex (G), and in the cell lysate. The patterns of these bands resemble those of SDS-PAGE and Coomassie blue staining (Figure 2B), suggestive of glycoproteins. In other isolates, no glycoproteins or polysaccharides were isolated, or the stains were less distinct.
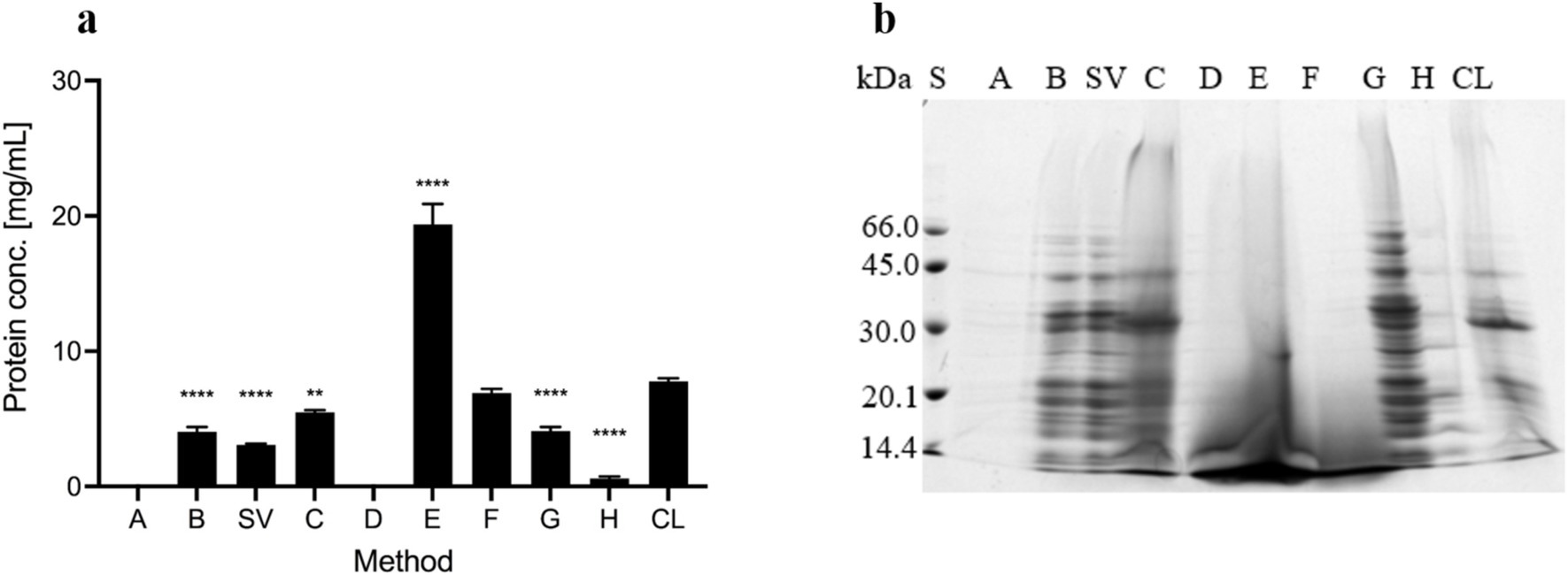
Figure 2. Proteins in the ECM isolates of Campylobacter jejuni. (A) Protein content as revealed by the DC Protein Assay. The mean values and standard deviations of three measurements are shown. Dunnett’s multiple comparisons with the control cell lysate tested at significance levels of p** < 0.01 and ****p < 0.0001. The ANOVA results were significant [F (7, 16) = 286.1, p < 0.0001]. (B) SDS-PAGE analysis with Coomassie Brilliant Blue staining. Samples were obtained by sodium chloride isolation (A), centrifugation (B), a procedure that yielded a supernatant of weakly bound extracellular matrix components (SV), heating in sodium carbonate (C), EDTA (D), sodium hydroxide (E), formaldehyde and sodium hydroxide (F), Dowex cation exchanger (G) and ether solution (H). Total cell lysate (CL) was also tested.
SDS-PAGE and Alcian blue staining (Figure 1C) revealed similar patterns of acidic polysaccharides or polysaccharides with sulfate groups as SDS-PAGE and Coomassie blue staining (Figure 2B), indicating the presence of glycoproteins. No or very few glycoproteins were isolated with NaCl (A), EDTA (D), and ether solution (H). The presence of high-molecular-weight acidic polysaccharides or polysaccharides with sulfate groups was also detected by the methods using centrifugation (B and SV), heating in Na2CO3 (C), formaldehyde and NaOH (F), and the cation exchanger (G).
3.2 Proteins
The protein concentration was measured with the DC Protein Assay kit (Figure 2A). No protein was found in the isolates obtained with NaCl (A) and EDTA (D). The highest protein concentration was detected in the isolate obtained with NaOH (E), which is probably due to protein precipitation by denaturation with NaOH. Lower and comparable protein concentrations were detected in all other isolates and the cell lysate (CL). Protein concentrations were below 10 mg/ml in all samples except the sample obtained with NaOH, in which the protein concentration was 19 mg/ml ± 1.5 mg/ml. As such, this method is the most effective for isolating the highest protein concentrations from the ECM of C. jejuni, however, these proteins are most likely denatured and have lost their functionality.
SDS-PAGE and Coomassie staining (Figure 2B) detected proteins in all the isolates. Based on the intensity of the stains, a significant amount of proteins was detected in the cell lysates and all isolates, except isolates obtained with NaCl (A) and ether solution (H), in which fewer proteins were present. The same protein patterns were determined in isolates obtained with both centrifugation methods (B and SV), the cation exchanger Dowex (G), NaCl (A), and ether solution (H). The protein patterns of these samples differed from those of the cell lysate (CL). The protein pattern of the isolate obtained by heating in Na2CO3 (C) was similar to the protein pattern of the cell lysate. In other isolates (D, E, and F), predominantly low-molecular-weight, possibly denatured proteins (< 30 kDa) were detected (Figure 2B).
3.3 eDNA
Agarose gel analysis (Figure 3) revealed the presence of high-molecular-weight eDNA in isolates obtained with NaCl (A), both centrifugation methods (B and SV), EDTA (D), and ether solution (H), the latter showing the strongest signal. The isolate obtained by heating in Na2CO3 (C) contained fragments of >10 kbp and < 2 kbp. The isolate obtained with NaOH (E) contained fragments of 1–10 kbp. The isolate obtained with formaldehyde and NaOH (F) contained fragments of 0.5–2 kbp. The isolate obtained with the cation exchanger Dowex (G) contained a weak band of a fragment of >6 kbp. Cell lysates contained fragments of <1 kbp.
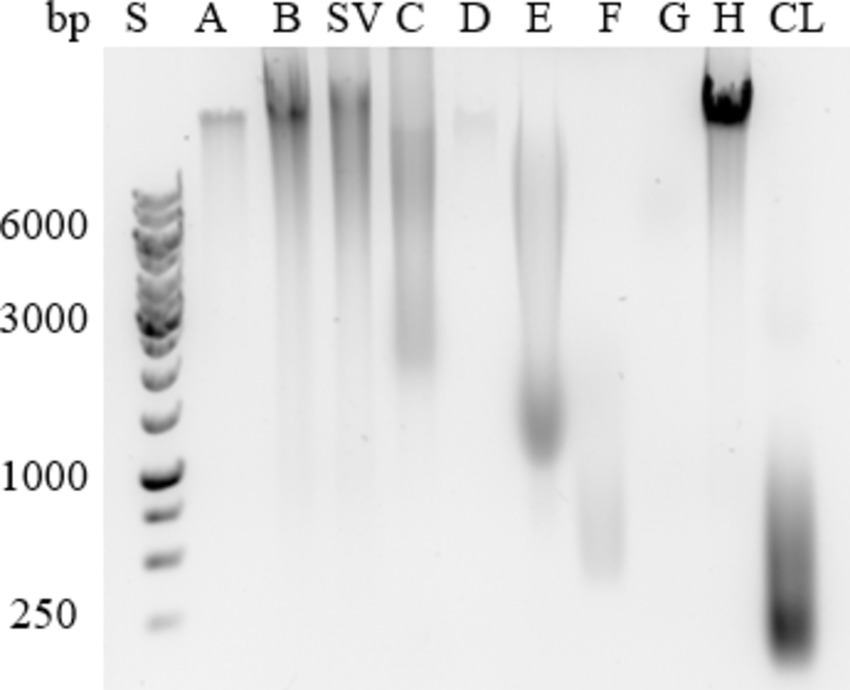
Figure 3. Extracellular DNA in the ECM isolates of Campylobacter jejuni. A representative example of agarose gel electrophoresis. Samples were obtained by sodium chloride isolation (A), centrifugation (B), a procedure that yielded a supernatant of weakly bound extracellular matrix components (SV), heating in sodium carbonate (C), EDTA (D), sodium hydroxide (E), formaldehyde and sodium hydroxide (F), Dowex cation exchanger (G) and ether solution (H). Total cell lysate (CL) was also tested.
Mostly high-molecular-weight eDNA was present in the ECM of C. jejuni, and only cell lysates contained low-molecular-weight eDNA. This is expected because the cell lysates were sonicated during preparation, which fragmented the DNA. Our findings indicate that the most suitable method for isolating high concentrations of high-molecular-weight DNA is extraction with ether solution (H; Figure 3).
3.4 ECM
An overview of the methods previously used for investigating ECM, divided into extraction and purification techniques, biofilm quantification and ECM analysis (Table 2) shows the variety of methods used for ECM studies. For accurate analysis of the ECM in C. jejuni biofilms, the ECM must first be extracted and purified, which was usually achieved by ethanol or acetone precipitation in combination with centrifugation (Jowiya et al., 2015; Yu et al., 2020), which favours the isolation of polysaccharides and proteins. Each method can illuminate a part of the ECM mosaic from a different angle, so that one can only interpret the results correctly if one knows the method well. In ex situ research, sample isolation and extraction is the first step, and it is extremely important to know and understand which part of the mosaic we can investigate with it. Here we have provided an overview of how the different isolation or extraction protocols can lead to different ratios and qualities of ECM components. The selection of the extraction method is an important step that can influence the outcome of the study. However, when selecting the method, not only the quantity of isolated macromolecules should be considered, but also their quality, as some extraction methods that yield the largest quantity are also destructive (e.g., NaOH for proteins). Therefore, when selecting the method for ECM extraction, the intended downstream analysis should be considered, e.g., preference of proteins for proteomics and polysaccharides for glycomics, but also the impact of the chemicals used in the purification as they may affect the quality of samples. Finally, it is recommended to use more than one method for ECM isolation in a study to compensate for any bias in ECM composition due to the isolation method.
4 Conclusion
Different isolation and extraction protocols for the extracellular matrix enriched different molecular components, resulting in very different ECM samples. For the isolation of C. jejuni ECM and its major components, the centrifugation method, the method in which the supernatant is obtained from weakly bound components, and heating in Na2CO3 were found to be the most suitable methods. These methods isolated all biopolymers and are simple, reliable and fast. The isolation methods using NaCl and EDTA were less suitable because they isolated lower amounts of eDNA and polysaccharides and failed to isolate proteins. All other isolation methods were considered suitable. The isolation method using NaOH isolated proteins and polysaccharides at the highest concentrations most effectively, but they were degraded. The ether dissolution method was best suited for the isolation of eDNA because it isolates high-molecular-weight DNA. Depending on the intended subsequent use or analytical method for the ECM sample, the isolation protocol should be carefully selected, ideally using more than one protocol to obtain more meaningful conclusions, as different protocols result in different compositions of the main ECM components.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
NP: Writing – original draft, Writing – review & editing, Formal analysis, Investigation. AK: Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition, Project administration. JS: Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Slovenian Research and Innovation Agency by projects numbers J4-4555, J4-4550, J7-4420, J4-3088, J4-4548, J2-50064, P4-0432, and P4-0116.
Acknowledgments
We thank Eva Lasic for editing and reviewing a draft of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aguilera, A., Souza-Egipsy, V., San Martín-Úriz, P., and Amils, R. (2008). Extraction of extracellular polymeric substances from extreme acidic microbial biofilms. Appl. Microbiol. Biotechnol. 78, 1079–1088. doi: 10.1007/s00253-008-1390-9
Blaser, M. J. (1997). Epidemiologic and clinical features Ofcampylobacter jejuniinfections. J. Infect. Dis. 176, S103–S105. doi: 10.1086/513780
Brown, H. L., Hanman, K., Reuter, M., Betts, R. P., and van Vliet, A. H. M. (2015). Campylobacter jejuni biofilms contain extracellular DNA and are sensitive to DNase I treatment. Front. Microbiol. 6:699. doi: 10.3389/fmicb.2015.00699
Burnham, P. M., and Hendrixson, D. R. (2018). Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol. 16, 551–565. doi: 10.1038/s41579-018-0037-9
Carpentier, B., and Cerf, O. (1993). Biofilms and their consequences, with particular reference to hygiene in the food industry. J. Appl. Bacteriol. 75, 499–511. doi: 10.1111/j.1365-2672.1993.tb01587.x
Chiba, A., Sugimoto, S., Sato, F., Hori, S., and Mizunoe, Y. (2015). A refined technique for extraction of extracellular matrices from bacterial biofilms and its applicability. Microb. Biotechnol. 8, 392–403. doi: 10.1111/1751-7915.12155
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063
EFSA and ECDC (2023). The European Union one health 2022 Zoonoses report. EFSA J. 21:e8442. doi: 10.2903/j.efsa.2023.8442
Felz, S., Al-Zuhairy, S., Aarstad, O. A., van Loosdrecht, M. C., and Lin, Y. M. (2016). Extraction of Structural Extracellular Polymeric Substances from Aerobic Granular Sludge. J. Vis. Exp. e54534. doi: 10.3791/54534
Feng, J., Lamour, G., Xue, R., Mirvakliki, M. N., Hatzikiriakos, S. G., Xu, J., et al. (2016). Chemical, physical and morphological properties of bacterial biofilms affect survival of encased Campylobacter jejuni F38011 under aerobic stress. Int. J. Food Microbiol. 238, 172–182. doi: 10.1016/j.ijfoodmicro.2016.09.008
Feng, J., Ma, L., Nie, J., Konkel, M. E., and Lu, X. (2018). Environmental stress-induced bacterial lysis and extracellular DNA release contribute to Campylobacter jejuni biofilm formation. Appl. Environ. Microbiol. 84, e02068–e02017. doi: 10.1128/aem.02068-17
Flemming, H.-C., van Hullebusch, E. D., Neu, T. R., Nielsen, P. H., Seviour, T., Stoodley, P., et al. (2023). The biofilm matrix: multitasking in a shared space. Nat. Rev. Microbiol. 21, 70–86. doi: 10.1038/s41579-022-00791-0
Flemming, H.-C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Flemming, H.-C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Frølund, B., Palmgren, R., Keiding, K., and Nielsen, P. H. (1996). Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res. 30, 1749–1758. doi: 10.1016/0043-1354(95)00323-1
Giaouris, E., Heir, E., Desvaux, M., Hébraud, M., Møretrø, T., Langsrud, S., et al. (2015). Intra- and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front. Microbiol. 6:841. doi: 10.3389/fmicb.2015.00841
Good, L., Miller, W. G., Niedermeyer, J., Osborne, J., Siletzky, R. M., Carver, D., et al. (2019). Strain-specific differences in survival ofCampylobacterspp. In naturally contaminated Turkey feces and water. Appl. Environ. Microbiol. 85, e01579–e01519. doi: 10.1128/aem.01579-19
Jachlewski, S., Jachlewski, W. D., Linne, U., Bräsen, C., Wingender, J., and Siebers, B. (2015). Isolation of extracellular polymeric substances from biofilms of the thermoacidophilic archaeon Sulfolobus acidocaldarius. Front. Bioeng. Biotechnol. 3:123. doi: 10.3389/fbioe.2015.00123
Joshua, G. W. P., Guthrie-Irons, C., Karlyshev, A. V., and Wren, B. W. (2006). Biofilm formation in Campylobacter jejuni. Microbiology 152, 387–396. doi: 10.1099/mic.0.28358-0
Jowiya, W., Brunner, K. T., Abouelhadid, S., Hussain, H., Nair, S. P., Sadiq, S., et al. (2015). Pancreatic amylase is an environmental signal for regulation of biofilm formation and host interaction in Campylobacter jejuni. Infect. Immun. 83, 4884–4895. doi: 10.1128/iai.01064-15
Karygianni, L., Ren, Z., Koo, H., and Thurnheer, T. (2020). Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 28, 668–681. doi: 10.1016/j.tim.2020.03.016
Kemper, L., and Hensel, A. (2023). Campylobacter jejuni: targeting host cells, adhesion, invasion, and survival. Appl. Microbiol. Biotechnol. 107, 2725–2754. doi: 10.1007/s00253-023-12456-w
Klančnik, A., Šimunović, K., Sterniša, M., Ramić, D., Smole Možina, S., and Bucar, F. (2021). Anti-adhesion activity of phytochemicals to prevent Campylobacter jejuni biofilm formation on abiotic surfaces. Phytochem. Rev. 20, 55–84. doi: 10.1007/s11101-020-09669-6
Klančnik, A., Vučković, D., Jamnik, P., Abram, M., and Smole Možina, S. (2014). Stress response and virulence of heat-stressed Campylobacter jejuni. Microbes Environ. 29, 338–345. doi: 10.1264/jsme2.me14020
Klančnik, A., Vučković, D., Plankl, M., Abram, M., and Smole Možina, S. (2013). In vivo modulation of Campylobacter jejuni virulence in response to environmental stress. Foodborne Pathog. Dis. 10, 566–572. doi: 10.1089/fpd.2012.1298
Kolenc, Ž., Kovač Viršek, M., Klančnik, A., and Janecko, N. (2024). Microbial communities on microplastics from seawater and mussels: insights from the northern Adriatic Sea. Sci. Total Environ. 949:175130. doi: 10.1016/j.scitotenv.2024.175130
Levin, R. E. (2007). Campylobacter jejuni: a review of its characteristics, pathogenicity, ecology, distribution, subspecies characterization and molecular methods of detection. Food Biotechnol. 21, 271–347. doi: 10.1080/08905430701536565
Liang, O. D., Ascencio, F., Fransson, L. A., and Wadström, T. (1992). Binding of heparan sulfate to Staphylococcus aureus. Infect. Immun. 60, 899–906. doi: 10.1128/iai.60.3.899-906.1992
Liu, H., and Fang, H. H. P. (2002). Extraction of extracellular polymeric substances (EPS) of sludges. J. Biotechnol. 95, 249–256. doi: 10.1016/s0168-1656(02)00025-1
Lu, X., Weakley, A. T., Aston, D. E., Rasco, B., Wang, S., and Konkel, M. E. (2012). Examination of nanoparticle inactivation of Campylobacter jejuni biofilms using infrared and Raman spectroscopies. J. Appl. Microbiol. 113, 952–963. doi: 10.1111/j.1365-2672.2012.05373.x
Ma, L., Feng, J., Zhang, J., and Lu, X. (2022). Campylobacter biofilms. Microbiol. Res. 264:127149. doi: 10.1016/j.micres.2022.127149
Melo, R. T., Mendonça, E. P., Monteiro, G. P., Siqueira, M. N., Pereira, C., Peres, P. A. B. M., et al. (2017). Intrinsic and extrinsic aspects on Campylobacter jejuni biofilms. Front. Microbiol. 8:1332. doi: 10.3389/fmicb.2017.01332
Moorhead, S. M., and Griffiths, M. W. (2011). Expression and characterization of cell-signalling molecules in Campylobacter jejuni. J. Appl. Microbiol. 110, 786–800. doi: 10.1111/j.1365-2672.2010.04934.x
Oh, E., Andrews, K. J., and Jeon, B. (2018). Enhanced biofilm formation by ferrous and ferric iron through oxidative stress in Campylobacter jejuni. Front. Microbiol. 9:1204. doi: 10.3389/fmicb.2018.01204
Püning, C., Su, Y., Lu, X., and Gölz, G. (2021). Molecular mechanisms of Campylobacter biofilm formation and quorum sensing. Curr. Top. Microbiol. Immunol. 431, 293–319. doi: 10.1007/978-3-030-65481-8_11
Ramić, D., Jug, B., Šimunović, K., Tušek Žnidarič, M., Kunej, U., Toplak, N., et al. (2023). The role ofluxSin Campylobacter jejuni beyond intercellular signaling. Microbiol. Spectrum 11:e0257222. doi: 10.1128/spectrum.02572-22
Reuter, M., Mallett, A., Pearson, B. M., and van Vliet, A. H. M. (2010). Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl. Environ. Microbiol. 76, 2122–2128. doi: 10.1128/aem.01878-09
Sabotič, J., Janež, N., Volk, M., and Klančnik, A. (2023). Molecular structures mediating adhesion of Campylobacter jejuni to abiotic and biotic surfaces. Vet. Microbiol. 287:109918. doi: 10.1016/j.vetmic.2023.109918
Sheppard, S. K., and Maiden, M. C. J. (2015). The evolution of Campylobacter jejuni and Campylobacter coli. Cold Spring Harb. Perspect. Biol. 7:a018119. doi: 10.1101/cshperspect.a018119
Snelling, W. J., Matsuda, M., Moore, J. E., and Dooley, J. S. G. (2005). Campylobacter jejuni. Lett. Appl. Microbiol. 41, 297–302. doi: 10.1111/j.1472-765x.2005.01788.x
Solomon, E. B., and Hoover, D. G. (1999). Campylobacter jejuni: a bacterial paradox. J. Food Saf. 19, 121–136. doi: 10.1111/j.1745-4565.1999.tb00239.x
Sulaeman, S., Hernould, M., Schaumann, A., Coquet, L., Bolla, J.-M., Dé, E., et al. (2012). Enhanced adhesion of Campylobacter jejuni to abiotic surfaces is mediated by membrane proteins in oxygen-enriched conditions. PLoS One 7:e46402. doi: 10.1371/journal.pone.0046402
Svensson, S. L., Pryjma, M., and Gaynor, E. C. (2014). Flagella-mediated adhesion and extracellular DNA release contribute to biofilm formation and stress tolerance of Campylobacter jejuni. PLoS One 9:e106063. doi: 10.1371/journal.pone.0106063
Tabouret, M., De Rycke, J., and Dubray, G. (1992). Analysis of surface proteins of Listeria in relation to species, serovar and pathogenicity. J. Gen. Microbiol. 138, 743–753. doi: 10.1099/00221287-138-4-743
Tapia, J., Muñoz, J. A., González, F., Blázquez, M. L., Malki, M., and Ballester, A. (2009). Extraction of extracellular polymeric substances from the acidophilic bacterium Acidiphilium 3.2Sup(5). Water Sci. Technol. 59, 1959–1967. doi: 10.2166/wst.2009.192
Teh, K. H., Flint, S., and French, N. (2010). Biofilm formation by Campylobacter jejuni in controlled mixed-microbial populations. Int. J. Food Microbiol. 143, 118–124. doi: 10.1016/j.ijfoodmicro.2010.07.037
Tram, G., Day, C. J., and Korolik, V. (2020). Bridging the gap: a role for Campylobacter jejuni biofilms. Microorganisms 8:452. doi: 10.3390/microorganisms8030452
Turonova, H., Neu, T. R., Ulbrich, P., Pazlarova, J., and Tresse, O. (2016). The biofilm matrix of Campylobacter jejuni determined by fluorescence lectin-binding analysis. Biofouling 32, 597–608. doi: 10.1080/08927014.2016.1169402
Vu, B., Chen, M., Crawford, R., and Ivanova, E. (2009). Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554. doi: 10.3390/molecules14072535
Whelan, M. V. X., Simpson, J. C., and Ó Cróinín, T. (2021). A novel high-content screening approach for the elucidation of C. jejuni biofilm composition and integrity. BMC Microbiol. 21:2. doi: 10.1186/s12866-020-02062-5
Wu, J., and Xi, C. (2009). Evaluation of different methods for extracting extracellular DNA from the biofilm matrix. Appl. Environ. Microbiol. 75, 5390–5395. doi: 10.1128/aem.00400-09
Keywords: Campylobacter jejuni , biofilm, extracellular polymeric substances, isolation methods, extraction and purification
Citation: Pavlinjek N, Klančnik A and Sabotič J (2024) Evaluation of physical and chemical isolation methods to extract and purify Campylobacter jejuni extracellular polymeric substances. Front. Microbiol. 15:1488114. doi: 10.3389/fmicb.2024.1488114
Edited by:
Ozan Gundogdu, University of London, United KingdomReviewed by:
Andreas Erich Zautner, University Hospital Magdeburg, GermanyBassam A. Elgamoudi, Griffith University, Australia
Copyright © 2024 Pavlinjek, Klančnik and Sabotič. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anja Klančnik, YW5qYS5rbGFuY25pa0BiZi51bmktbGouc2k=
 Natalija Pavlinjek
Natalija Pavlinjek Anja Klančnik
Anja Klančnik Jerica Sabotič
Jerica Sabotič