- 1Third Hospital of Shanxi Medical University, Shanxi Bethune Hospital, Shanxi Academy of Medical Sciences, Tongji Shanxi Hospital, Taiyuan, China
- 2School of Pharmacy, Shanxi Medical University, Taiyuan, China
- 3Shanxi Academy of Advanced Research and Innovation (SAARI), Taiyuan, China
Objective: The purpose of this study was to explore the potential mechanism of Helicobacter pylori (Hp) eradication by probiotic therapy through 16S rRNA gene sequencing technology and untargeted metabolomics.
Methods: Twenty four Hp-infected children were recruited from the Shanxi Bethune Hospital, and 24 healthy children were recruited as a blank control group. Group A: fecal samples from 24 healthy children. Group B: fecal samples of 24 children with Hp infection. Group B1 (n = 15): fecal samples of group B treated with probiotic therapy for 2 weeks. Group B2 (n = 19): fecal samples of group B treated with probiotic therapy for 4 weeks. The above fecal samples were analyzed by 16S rRNA gene sequencing technology and untargeted metabolomics.
Results: There was no significant difference in alpha diversity and beta diversity among the four groups, but many bacteria with statistical difference were found in each group at the bacterial genus level and phylum level. LEfSe results showed that in group B, Porphyromonadaceae, Shigella and other microorganisms related to intestinal microecological dysbiosis were enriched. And in group B2, abundant characteristic microorganisms were found, namely Bacillales and Prevotella. KEGG metabolic pathway enrichment analysis showed that groups B1 and B2 were involved in 10 metabolic pathways potentially related to probiotic treatment: purine metabolism, nitrogen metabolism, arginine biosynthesis, alanine, aspartic acid and glutamate metabolism, glyoxylic acid and dicarboxylic acid metabolism, unsaturated fatty acid biosynthesis, fatty acid extension, fatty acid degradation, pyrimidine metabolism, fatty acid biosynthesis.
Conclusion: Probiotic therapy can inhibit Hp to some extent and can relieve gastrointestinal symptoms, making it a preferred therapy for children with Hp infection and functional abdominal pain. Hp infection can reduce the diversity of intestinal microbes, resulting in the disturbance of intestinal microbiota and changes in the relative abundance of microbiota in children, while probiotic therapy can restore the diversity of intestinal microbes and intestinal microecological balance.
1 Introduction
Helicobacter pylori (Hp) is a gram-negative bacterium that is mainly found in the gastric mucosa. Hp is one of the common chronic infections in humans. Hp infection usually occurs in childhood and lasts until adulthood, affecting adult health. Once infected, it is difficult to be cleared naturally (Jiang, 2023). Globally, infection rates in developed and developing countries fluctuated between 8.9 and 72.8% (Segal et al., 2008). According to the epidemiological survey, the Hp infection rate of children in Europe, Asia, South America, and South Africa was 7.0–33.0%, 37.5–66.0%, 48.0–78.0%, and 87.0%, respectively (Goh et al., 2011). According to a study in 2021, the rate of Hp infection among children and adolescents in China was 20.55% (Zhou et al., 2023). The guidelines of the American Society of Hematology state that routine Hp testing and eradication treatment are not recommended for asymptomatic children infected with Hp (Neunert et al., 2011). However, Hp detection is recommended for children with gastrointestinal symptoms such as repeated abdominal discomfort, nausea, vomiting, belching, etc. And Hp eradication therapy can be considered for those with positive test results (Subspecialty Group of Gastroenterology, the Society of Pediatrics, Chinese Medical Association, National Children’s Medical Center Digestive Specialist Alliance, Editorial Board, Chinese Journal of Pediatrics, 2023). For Hp-positive children with functional gastrointestinal disorders, the eradication of Hp could lead to long-term relief of symptoms in some patients, and could be considered as a preferred treatment (Sugano et al., 2015). Therefore, Hp eradication therapy is recommended for children with Hp infection and functional abdominal pain.
Currently recognized first-line and second-line regimens to eradicate Hp infection in children require at least two antibiotics. These antibiotic therapy has many disadvantages (Seo et al., 2020) in the eradication of Hp infection in children, such as the limited choice of antibiotics in the eradication therapy, the increasing resistance rate of bacteria to antibiotics, and the low tolerance of children to adverse drug reactions (such as abdominal pain, diarrhea, nausea, vomiting, etc.). In recent years, the eradication of probiotics for Hp infection has gradually become a focal point in the research field. Probiotics have been proven to inhibit Hp infection, indicating their potential significance in combating this issue (Lv et al., 2015; Subspecialty Group of Gastroenterology, the Society of Pediatrics, Chinese Medical Association, National Children’s Medical Center Digestive Specialist Alliance, Editorial Board, Chinese Journal of Pediatrics, 2023). Compared to antibiotic therapy, probiotics rarely cause adverse reactions and do not increase strain resistance. Additionally, children have a higher tolerance and better compliance with probiotics. Therefore, the development of probiotic therapy for treating Hp infection in children is of great significance in academic research.
Studies have shown that probiotics can secrete antimicrobial substances, mainly including lactic acid, short-chain fatty acids, hydrogen peroxide, bactericin and so on. Lactic acid and short-chain fatty acids can reduce cytoplasmic pH and prevent the colonization of Hp, while hydrogen peroxide is associated with the destruction of pathogenic proteins, membrane lipids, and DNA of bacterial cells (Ji and Yang, 2020). In addition, probiotics can affect immune function and indirectly inhibit Hp infection (Sanders et al., 2019). Clostridium butyricum produces butyrate, which may exhibit bactericidal effects on Hp. In vitro studies utilizing the supernatant of butyrate culture and butyrate-producing bacteria have revealed their ability inhibit the growth of Hp, which is attributed to the destruction of the envelope of Hp cells (Yonezawa et al., 2012). And previous studies have proved that there is antagonistic interaction between Clostridium butyricum and Hp in vitro and in vivo, and the supernatant of Clostridium butyricum culture can inhibit the growth of Hp before and after pH adjustment (Takahashi et al., 2000), so it has a promising prospect in the treatment of Hp infection. In addition, supplementation of Bacillus coagulans has a positive effect on the function of the human immune system (Fu et al., 2021). It has been reported that the supplementation of Bacillus coagulans in the treatment of Hp infection with triple therapy can significantly improve the eradication rate and effectively reduce the incidence of adverse reactions (Wang, 2013). Currently, researches have demonstrated that the combination of Clostridium butyricum and Bacillus coagulans is capable of effectively eradicating Hp, with a higher eradication rate compared to either Clostridium butyricum or Bacillus coagulans alone (Zhang et al., 2020; Zhu, 2023). While there is compelling evidence suggesting that probiotic strains may not exert their effectiveness through colonization in the gastrointestinal tract, but rather by sharing genes and metabolites and directly influencing the intestinal barrier and immune cells (Wieërs et al., 2019), the precise mechanism of action of probiotics remains unclear. In particular, the mechanism of eradication of Hp by Clostridium butyricum and Bacillus coagulans is rarely reported. Based on this, this study intends to explore the potential impact of probiotic therapy on Hp infection in children through 16S rRNA gene sequencing technology and untargeted metabolomics.
2 Materials and methods
2.1 Study design
In this study, 24 children with Hp infection and functional abdominal pain were recruited from the pediatric clinic of Shanxi Bethune Hospital, Shanxi Province, China. Besides, 24 healthy children were recruited as a control group. The inclusion criteria were as follows: (1) Aged between 5 and 14 years old; (2) Hp infection confirmed by 13C urea breath test; (3) Functional abdominal pain meeting the diagnostic criteria of Rome IV, with no other organic diseases; (4) No previous Hp eradication therapy. The exclusion criteria included: (1) Children who do not follow medical advice resulting in invalid results; (2) Patients with a history of allergy to microecological preparations; (3) Individuals with a history of Hp eradication; (4) Those who have taken antibacterial drugs within the past month.
The administration regimen for children infected with Hp was as follows: Clostridium butyricum Powder, Live (Qingdao Eastsea Pharmaceutical Co., Ltd., lot No: B202301038), 1 bag (1.5 × 107 CFU) per dose, 3 times a day; Bacillus coagulans Tablets, Live (Qingdao Eastsea Pharmaceutical Co., Ltd., lot No: S202302029), 3 tablets (5.25 × 107 CFU) per dose, 3 times a day. Additionally, to alleviate functional abdominal pain in the included population as soon as possible, Lamb’s Tripe Extract and Vitamin B12 Granules (Xinjiang Biochemical Pharmaceutical Co., Ltd., lot No: 202210035, 1 bag per dose, 3 times a day) was added. The indications of this drug are upper abdominal discomfort, abdominal distention, loss of appetite caused by chronic gastritis, which is a protective agent for gastric mucosa with high safety and has a strong gastric acid buffer effect. The above three drugs were combined to treat Hp infected children for 4 weeks. And healthy children in the control group did not receive any interventions.
The gender, age, and weight data of each child were collected. After 4 weeks of probiotic medication, the subjects underwent the 13C urea breath test again at the gastroenterology department of Shanxi Bethune Hospital. We collected the Delta Over Baseline (DOB) value from two breath tests before and after medication, and determined whether Hp has been eradicated (DOB < 4.0 represents normal; DOB ≥ 4.0 represents Hp infection). In addition, gastrointestinal symptom rating scale (GSRS) scores were obtained before and 4 weeks after treatment. These measures were used to evaluate the antibacterial effect of probiotic therapy on Hp and the improvement of functional abdominal pain in the children, respectively. In addition, 10 mL sterile fecal sampling tubes were utilized to collect stool samples from healthy children and Hp infection children. For Hp infection children, the first stool samples were collected prior to treatment, the second after 2 weeks of treatment, and the third after 4 weeks of treatment. Following each collection, the samples were promptly stored in a refrigerator at −80°C for subsequent 16S rRNA gene sequencing analysis and untargeted metabolomic analysis.
This study was approved by the Ethics Review Committee of Shanxi Bethune Hospital (approval number: YXLL-2022-064), and all participants provided signed informed consent. Furthermore, this study has been registered in the Chinese Clinical Trial1 with the registration number ChiCTR2200062024.
2.2 16S rRNA gene sequencing analysis
The specific process for 16S rRNA gene sequencing analysis of children’s stool samples was as follows. Total genomic DNA was extracted from fecal samples using the M5635-02 OMEGA Soil DNA Kit (Omega Bio-Tek, Norcross, GA, United States). NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis were then used to measure the quantity and quality of the extracted DNAs. Then, forward primer 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used in PCR amplification of the bacterial 16S rRNA gene V3-V4 region. The PCR amplification products were purified by VAHTSTM DNA Clean Beads (Nazyme, Nanjing, China) and quantified by the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, amplicons were pooled in equal amounts, and pair-end 2 × 250 bp sequencing was performed using the Illlumina NovaSeq platform with NovaSeq 6,000 SP Reagent Kit (500 cycles) at Shanghai Metabo-Profile Biotechnology Co., Ltd. (Shanghai, China). Sequences were quality filtered, denoised, merged and chimera removed using the DADA2 plugin, then the effective sequences of each sample were obtained. USEARCH (version 10.0) was used to assign qualified sequences with similarity threshold exceeding 97% to an operation classification unit (OTU) for subsequent analysis.
Sequence data analysis were mainly performed using QIIME2 2019.4 (according to the official tutorials)2 and R packages (v3.2.0). Briefly, raw sequence data were demultiplexed using the demux plugin following by primers cutting with cutadapt plugin (Martin, 2011). Sequences were then quality filtered, denoised, merged and chimera removed using the DADA2 plugin (Callahan et al., 2016). Non-singleton amplicon sequence variants (ASVs) were aligned with mafft (Katoh et al., 2002) and used to construct a phylogeny with fasttree2 (Price et al., 2009). Alpha diversity metrics (Chao1, Shannon, Simpson), beta diversity metrics (weighted UniFrac, unweighted UniFrac) were estimated using the diversity plugin. Alpha diversity indices were visualized as box plots and betadiversity analysis was visualized via principal coordinate analysis (PCoA). Permutational multivariate analysis of variance (PERMANOVA) was used to compare bacterial abundance and diversity. Moreover, the online analysis platform Huttentower Lab Galaxy Huttentower was used for linear discriminant analysis (LEfSe).
2.3 Untargeted metabolomics analysis
The 20 mg freeze-dried fecal sample was transferred to a 1.5 mL centrifuge tube, followed by the addition of 1 mL of iced water for thorough mixing, and centrifugation for 15 min (13,000 rpm, 4°C) after ultrasonic processing for 20 min. The supernatant was then transferred. Then 1 mL of cold methanol (LC–MS grade, Thermo Fisher Scientific) was added to the remaining precipitate in the centrifuge tube and thoroughly mixed. After 20 min of ultrasonic processing, the mixture was centrifuged for 15 min (13,000 rpm, 4°C), and the supernatant was retained. The two kinds of supernatant obtained from the above two operations were, respectively, absorbed 500 μL and mixed, and then added 1 mL of cold acetonitrile (LC–MS grade, Thermo Fisher Scientific). After vortex mixing, the supernatant was retained after ultrasonic processing for 20 min and centrifugation for 15 min (13,000 rpm, 4°C). The supernatant was then evaporated to dryness using a high-speed vacuum concentrator. After drying, the product was redissolved in 100 μL 80% methanol solution and centrifuged for 15 min (13,000 rpm, 4°C) to obtain the supernatant for untargeted metabolomics analysis.
The LC–MS system for metabolomics analysis was composed of UltiMate 3,000 high performance liquid chromatography (Thermo Fisher Scientific, Waltham, MA, United States) and Thermo Scientific™ Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The liquid chromatography column used in the study was Waters Acquity UPLC HSS T3 column (1.8 μm, 2.1 mm × 100 mm) purchased from Waters, USA. The injection volume was 2 μL and the flow rate was 0.30 mL/min. The mobile phase consisted of 0.1% formic acid (LC–MS grade, Tokyo Chemical Industry) aqueous solution (A) and 0.1% formic acid-acetonitrile solution (B). The gradient elution conditions were as follows: 0 ~ 2 min, 98% A and 2% B; 3 ~ 5 min, 75% A and 25% B; 6 ~ 9 min, 50% A and 50% B; 10 ~ 12 min, 30% A and 70% B; 13 ~ 16 min, 15% A and 85% B; 17 ~ 18 min, 2% A and 98% B; 18.5 ~ 20 min, 98% A and 2% B. In this study, the ionization method of heated electrospray ionization (HESI) was utilized. The specific parameters were as follows: a spray voltage of 3,500 V (positive) /2500 V (negative); a capillary temperature of 320°C and a heater temperature of 300°C; the sheath gas flow rate was set at 35 arb and the auxiliary gas flow rate at 10 arb. The scanning mode was Full Scan/dd-MS2, with an m/z collection range of 100 ~ 1,500 in positive and negative ion switching collection mode. The resolution was MS Full Scan 35,000 FWHM and MS/MS 17500 FWHM. Additionally, the secondary fragmentation energies for MS were specified as 12.5 eV, 25 eV and 37.5 eV.
The original mass spectrometry data were processed using Compound Discover (version 3.3 SP2) software. Peak extraction, peak comparison, compound identification, and other data processing operations were conducted to obtain the peak table data of metabolites in the sample. Subsequently, the peak table data of metabolites were imported into SIMCA (version 14.1) software for further analysis. Principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were initially used to analyze the overall differences among the groups, among which OPLS-DA analysis also needs to use permutation test to verify the reliability of the model. Once the reliability of the OPLS-DA model was confirmed, group B-A, group B1-B, and group B2-B were analyzed separately by OPLS-DA to obtain variable importance in projection (VIP). By combining VIP values with Student-t test and fold change (FC) values, differential metabolites in group B, group B1, and group B2 were screened under specific conditions: VIP > 1, p < 0.05, FC > 1.5 or FC < 2/3. Finally, the differential metabolites were introduced into the online analysis platform of MetaboAnalyst 6.03 for enrichment analysis of KEGG metabolic pathway.
2.4 Data processing and analysis
The sex ratio was analyzed using the Chi-square test in SPSS software, while other demographic data was analyzed using the Student’s t test. A significance level of p < 0.05 was employed to determine whether there were statistically significant differences between the data.
3 Results
3.1 Baseline characteristics of children
Stool samples of healthy children were categorized as group A (n = 24), while stool samples of children with Hp infection and functional abdominal pain were classified as group B (n = 24). After 2 weeks of treatment with probiotics, 15 children in group B submitted stool samples, which were recorded as group B1, numbered B1_1 ~ B1_15. And after 4 weeks of treatment with probiotics, 19 children in group B submitted stool samples, which were recorded as group B2, numbered B2_1 ~ B2_19. The demographic data of children in groups A and B, along with the DOB value, Hp eradication rate, and GSRS scores of children in group B before and after probiotic therapy are presented in Table 1. A total of 12 children with Hp infection underwent Hp reexamination after the completion of the four-week probiotic therapy period. Statistical analysis revealed a decreasing trend in the DOB value, and the Hp eradication rate was 33.33%. The decrease in DOB value meant that probiotic therapy may reduce the bacterial load of Hp, but the difference was not significant (p = 0.065) probably due to insufficient sample size. In addition, the eradication rate also proved that probiotic therapy had an inhibitory effect on Hp. Following the four-week probiotic therapy period, there was a significant reduction in GSRS scores (p < 0.001), indicating that the child’s gastrointestinal symptoms were reduced after medication. Therefore, it proved that probiotic therapy can effectively alleviate symptoms associated with functional abdominal pain among infected children.
3.2 Effect of probiotic therapy on gut microbiota
3.2.1 Alpha diversity and beta diversity
Alpha diversity is an index that can reflect the species richness and species diversity of samples. In this study, three alpha diversity indices (Chao 1, Shannon, Simpson) were chosen to assess the microbial community diversity in the stool samples, and the results are shown in Figure 1. The Chao 1 index was utilized to characterize species richness (Figure 1A), while the Shannon and Simpson indices were used to characterize species diversity (Figures 1B,C). It was found that there were no significant differences in the three indices among the four groups in this study.
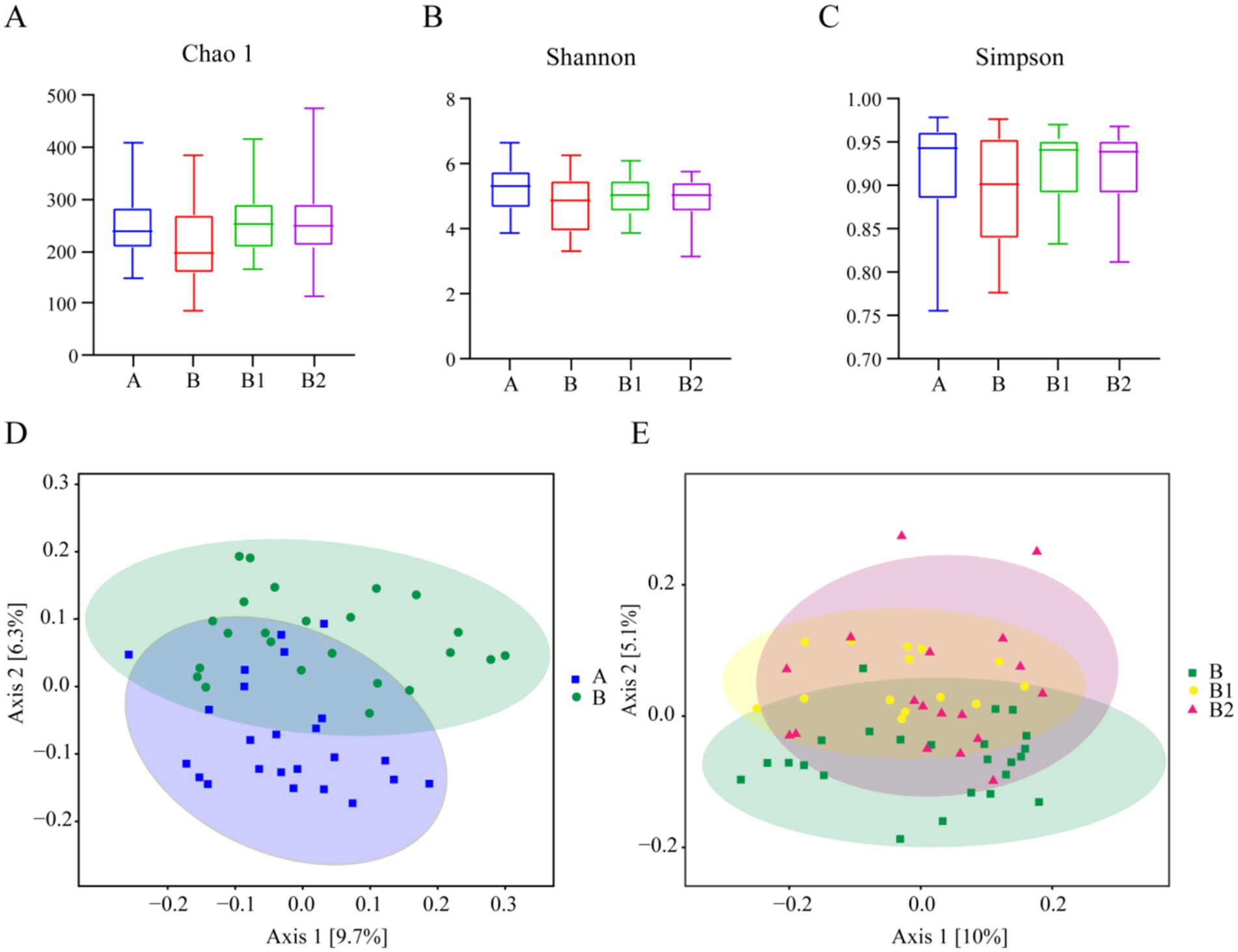
Figure 1. Alpha diversity and beta diversity. Alpha diversity of Group A, B, B1 and B2: (A) Chao1 index, (B) Shannon index, (C) Simpson index. Beta diversity of Group A and B: (D); beta diversity of Group B, B1 and B2: (E).
In addition, beta diversity is utilized to assess the similarity of different samples in terms of species diversity. PCoA was employed in this study to analyze beta diversity among various groups, and the results are also presented in Figure 1. The closer the points on the coordinate diagram are, the higher the similarity between the samples is. The findings revealed no significant difference in beta diversity between healthy children and Hp-infected children, and probiotic therapy did not result in a significant alteration in the diversity of gut microbes among children after 2 and 4 weeks of treatment.
3.2.2 Gut microbiota composition
LEfSe analysis is commonly utilized to identify the enriched gut microbiota within each group. In this study, LDA > 2.0 was used as the criterion (Segata et al., 2011) to screen the enriched gut microbiota from phylum to genus level between group A and group B at first. The aim was to identify variations in gut microbiota between healthy children and those infected with Hp. The results are presented in Figure 2. The characteristic gut microbiota species enriched in group A and group B were 21 and 12, respectively. In Group A, the enriched gut microbiota included Erysipelotrichi, Erysipelotrichaceae, Lactobacillus, Lactococcus, Lactobacillaceae, etc. Meanwhile, the enriched gut microbiota in group B were Bacteroides, Eggerthella, Porphyromonadaceae, Parabacteroides, Pseudomonadales, etc. Besides, using LDA > 2.0 as the criterion, this study conducted LEfSe analysis to compare the differences in gut microbiota composition from phylum to genus level among groups B, B1 and B2, in order to identify characteristic microorganisms enriched in the gut of children infected with Hp before treatment, 2 weeks after probiotic treatment, and 4 weeks after probiotic treatment. The results are presented in Figure 2, showing that only two characteristic microorganisms, Bacillales and Prevotella, were found in group B2.
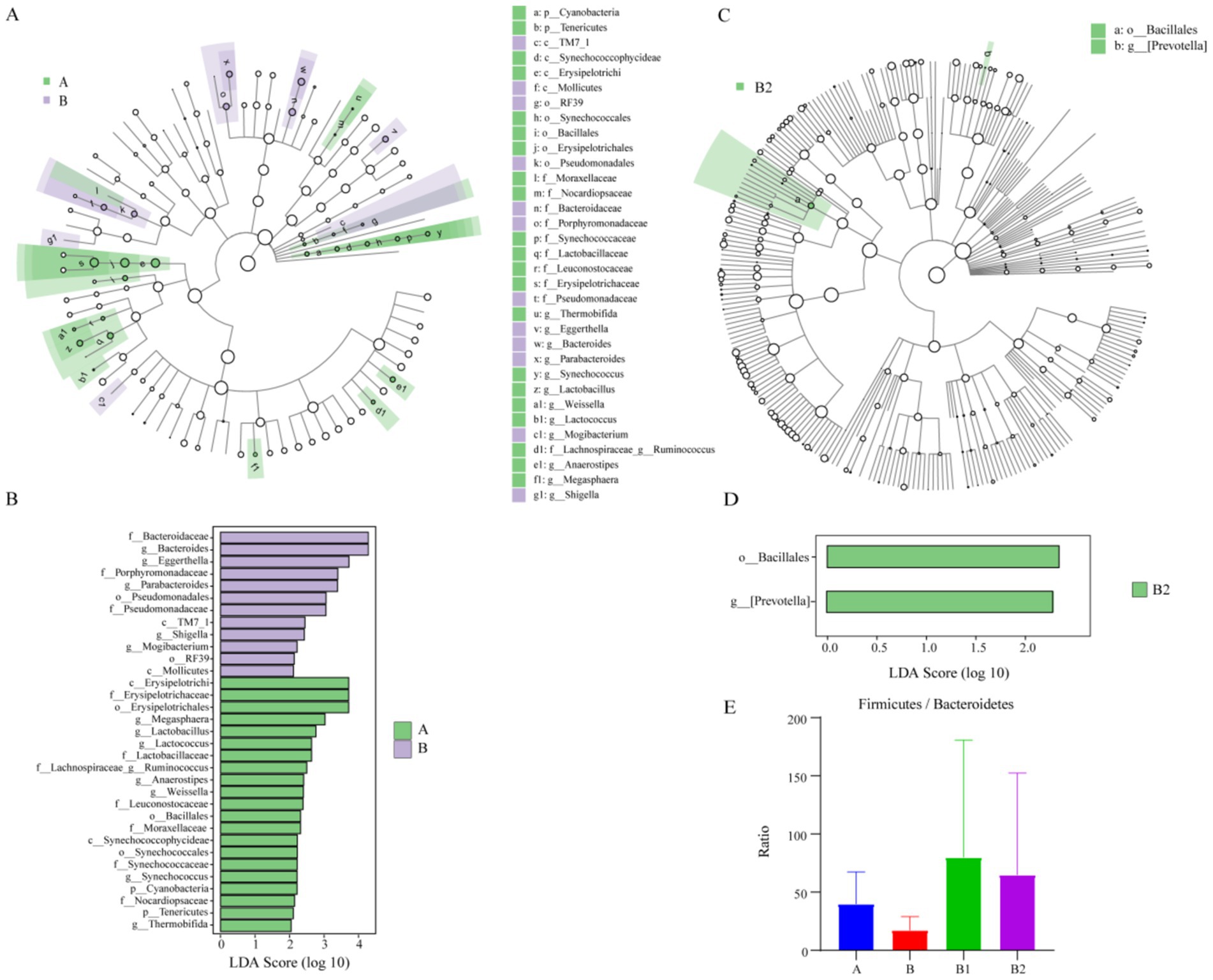
Figure 2. Results of LEfSe analysis and F/B value. The enriched gut microbiota in group A and group B: (A,B); the enriched gut microbiota in group B2: (C,D). Ratio of Firmicutes to Bacteroidetes among different groups: (E).
The ratio of the relative content of Firmicutes and Bacteroidetes (F/B value) is commonly utilized to assess the health of the intestinal microecological environment (Wei et al., 2021). Therefore, the F/B value of each group was analyzed in this study, and the results indicated that Hp infection was associated with a decrease in F/B value, while probiotic therapy was associated with an increase in F/B value, as depicted in Figure 2. However, possibly due to the small sample size, these differences did not show statistical significance.
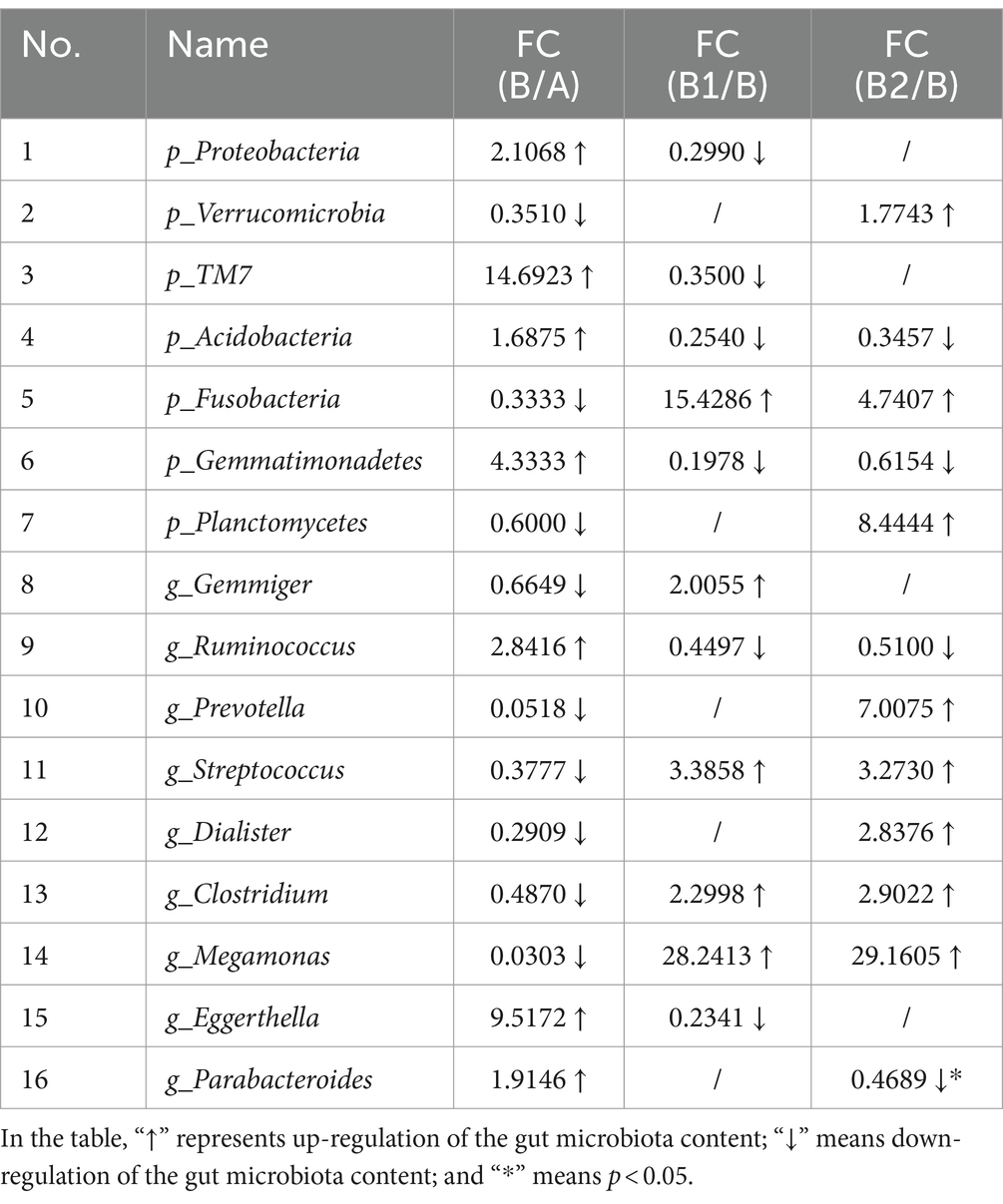
In order to comprehensively evaluate the effects of probiotic therapy on the microbiota composition of children infected with Hp, this study also conducted statistical analysis on the top 20 gut microbiota with relative abundance at phylum level and genus level, and the results are shown in Figure 3. At phylum level and genus level, with FC > 1.5 or FC < 2/3 as the criterion, the gut microbiota with recovery trends in relative content in group B1 and B2 were determined, respectively. As displayed in Table 2, at the phylum level, there were each 5 gut microbiota with recovery trends in group B1 and group B2. Meanwhile, at the genus level, 6 gut microbiota in group B1 and 7 gut microbiota in group B2 were found to have recovery trends, and the abundance of Parabacteroides decreased significantly in group B2 (p = 0.0465).
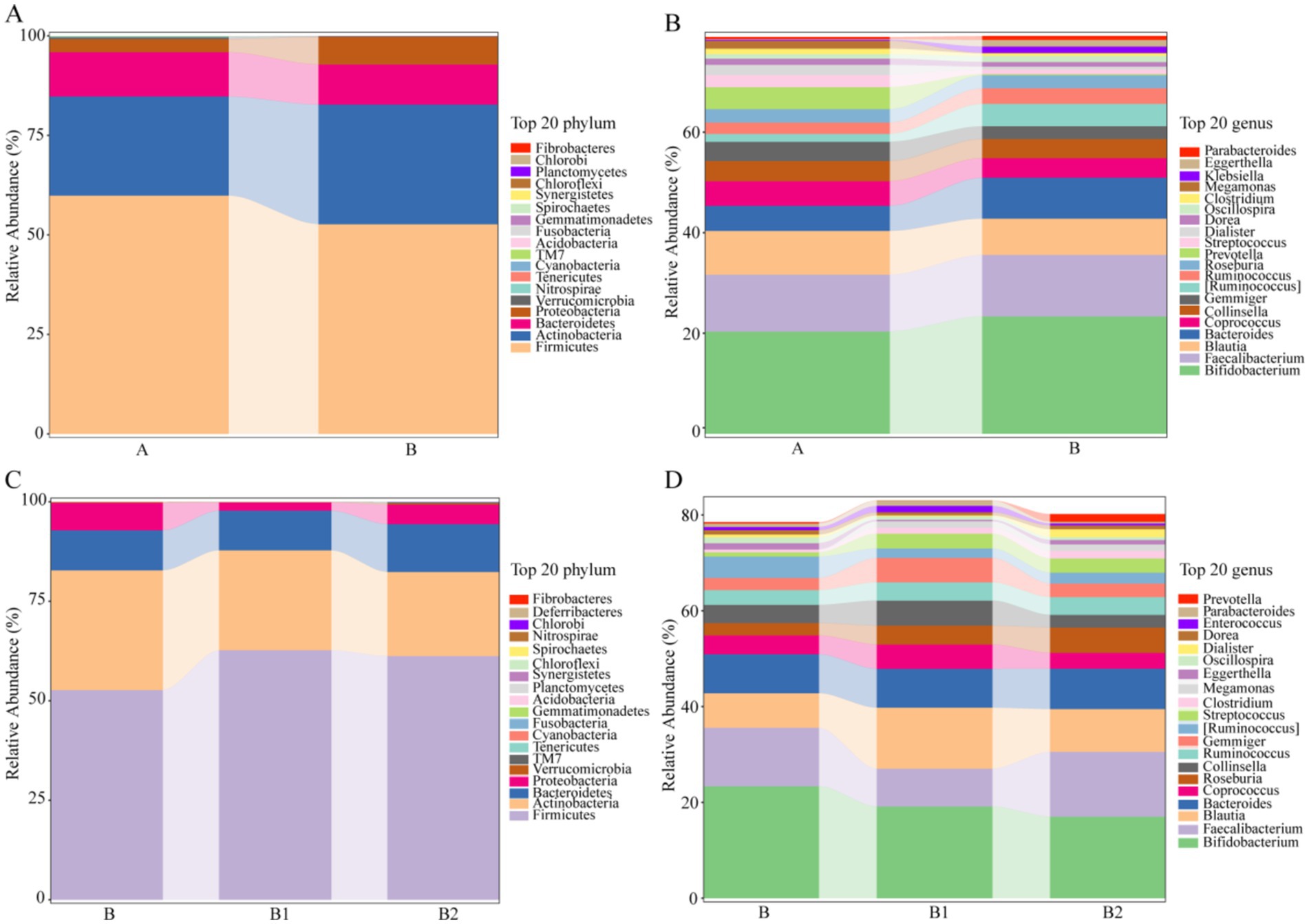
Figure 3. The relative abundance of the top 20 gut microbiota in different groups at the phylum/genus level. (A,B) The relative abundance of the top 20 gut microbiota in group A and group B at the phylum/genus level. (C,D) The relative abundance of the top 20 gut microbiota in group B, group B1 and group B2 at the phylum/genus level.
3.3 Effect of probiotic therapy on metabolites
3.3.1 Variety of fecal metabolic profiles
Unsupervised PCA analysis can effectively capture the overall differences between various sample groups and the degree of variation between samples within the group. In this study, PCA analysis was performed on group A and group B, as well as on group B, group B1 and group B2, in order to examine the disparities in metabolic profiles between Hp infected children and healthy children, as well as the differences among Hp infected children before medication and at 2 and 4 weeks post-medication. The 3D score charts of PCA are depicted in Figure 4, demonstrating discernible separation trends between group A and group B, as well as among group B, group B1, and group B2. These observation suggested that there were differences in the metabolite profiles between Hp infected children and healthy children, and Hp infected children exhibit changes in their metabolite profiles following probiotic therapy. Additionally, the tightly clustered QC samples indicated that the analysis method is stable and exhibits good repeatability.
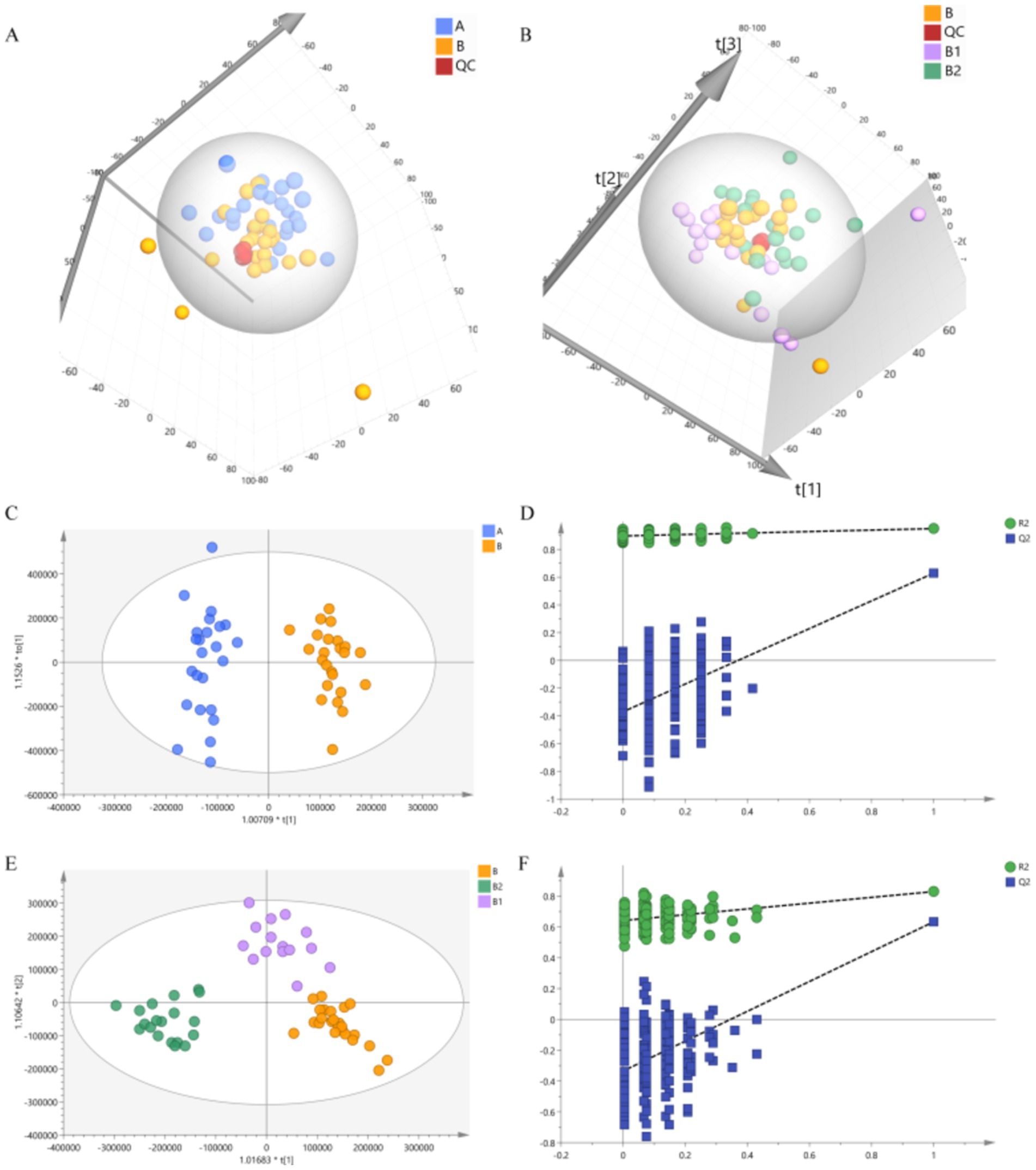
Figure 4. Results of PCA and OPLS-DA. (A) PCA result of group A and group B. (B) PCA result of group B, group B1 and group B2. (C,D) Results of OPLS-DA and permutations test of group A and group B. (E,F) Results of OPLS-DA and permutations test of group B, group B1 and group B2.
To further analyze the differences between the groups, this study utilized a supervised OPLS-DA analysis method. In comparison to PCA analysis, OPLS-DA analysis is more sensitive to inter-group differences and can achieve better separation results (Thévenot et al., 2015). The OPLS-DA analysis results are presented in Figure 4, clear separation were observed among the groups, which indicate significant differences in the metabolite profiles between each group. Additionally, the permutations test was conducted to verify the reliability of the OPLS-DA model. The verification results depicted in Figure 4 showed that Q2 intersected with the vertical axis on the negative half axis, confirming the reliability of the OPLS-DA model.
3.3.2 Screening of differential metabolites
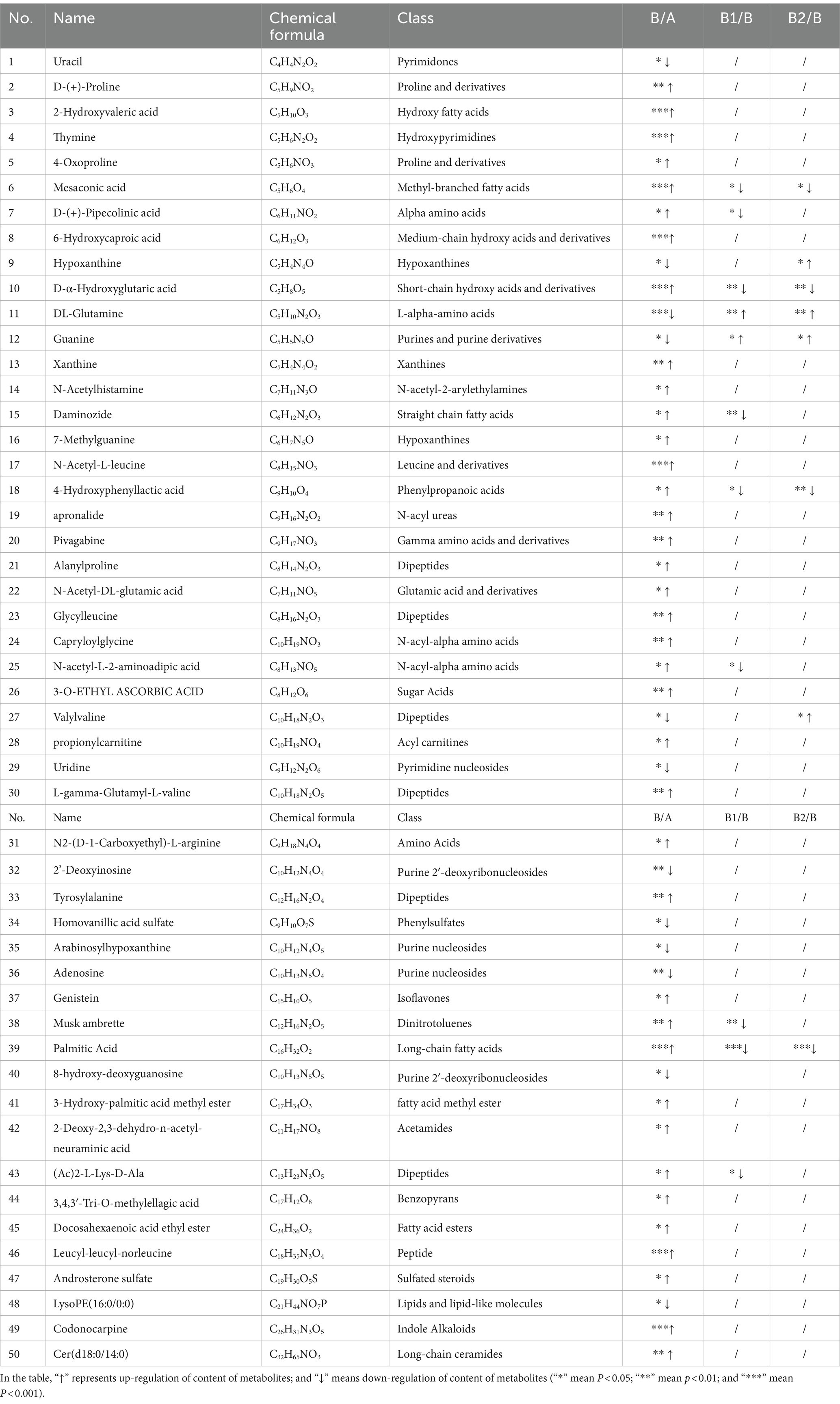
OPLS-DA was employed to analyze group B and group A, and VIP values were obtained. In this study, these values were combined with the Student’s t-test and FC value to screen for differential metabolites between the groups. The screening conditions were set as VIP > 1, p < 0.05, FC > 1.5 or FC < 2/3, and the resulting differential metabolites are presented in Table 3. Group B exhibited 50 differential metabolites, indicating that Hp infection may lead to significant changes in these metabolites.
Subsequently, further analyses were conducted on the metabolites of group B1 and group B, as well as group B2 and group B. Similarly, using VIP > 1, p < 0.05, FC > 1.5 or FC < 2/3 as criteria, metabolites with opposite adjustment trends compared to those in group B were screened for group B1 and group B2. In the end, there were 11 metabolites exhibiting opposite adjustment trends in the B1 group and 8 metabolites showing opposite adjustment trends in the B2 group. These metabolites were closely associated with the eradication of Hp through probiotic therapy.
3.3.3 Analysis of metabolic pathway
Firstly, the differential metabolites of group B were introduced into the online analysis platform, MetaboAnalyst 6.0, for enrichment analysis of KEGG metabolic pathway, and the results are shown in Figure 5. Hp infection in children was closely related to 14 metabolic pathways including purine metabolism, pyrimidine metabolism, arginine biosynthesis, nitrogen metabolism, pantothenate and CoA biosynthesis, beta-alanine metabolism, alanine, aspartate and glutamate metabolism, glutathione metabolism, glyoxylate and dicarboxylate metabolism, arginine and proline metabolism, biosynthesis of unsaturated fatty acids, fatty acid elongation, fatty acid degradation, and fatty acid biosynthesis.
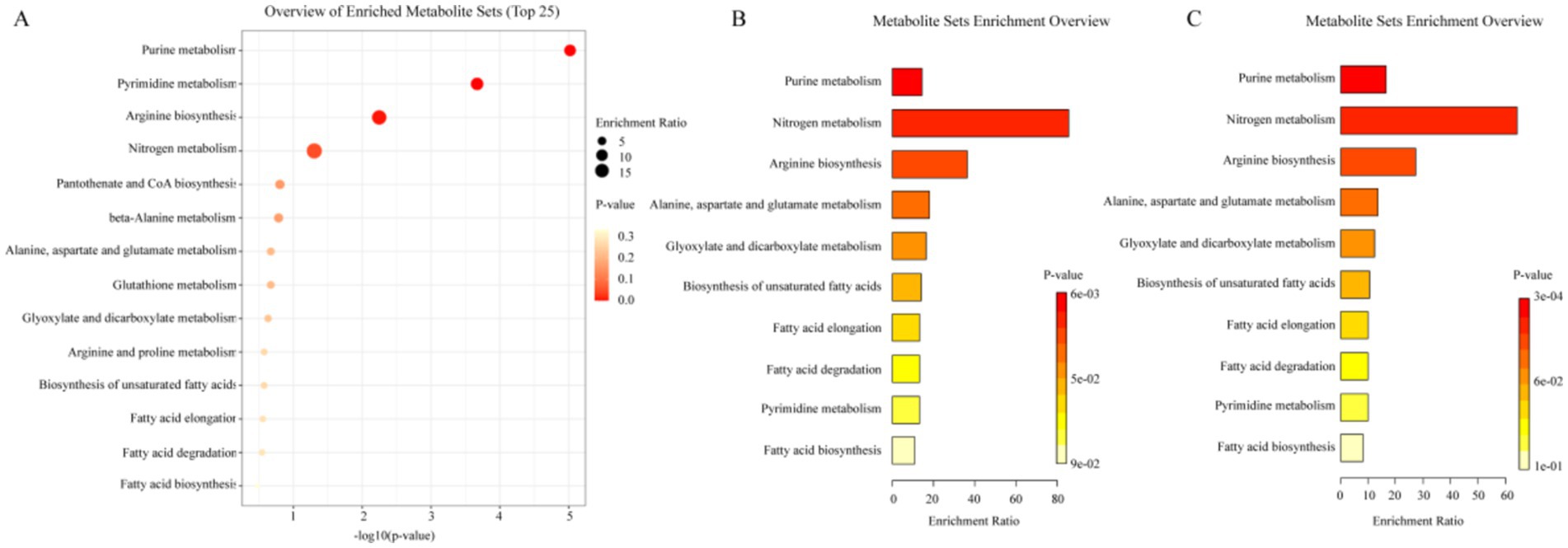
Figure 5. Analysis results of metabolic pathway. (A) Metabolic pathways of differential metabolites of group B. (B) Metabolic pathways of differential metabolites of group B1. (C) Metabolic pathways of differential metabolites of group B2.
Subsequently, the metabolites in group B1 and group B2 with regulatory trends opposite to those in group B were also incorporated into the analysis platform for KEGG metabolic pathway enrichment analysis. The results are presented in Figure 5. These metabolites in both B1 and B2 group encompassed 10 metabolic pathways, namely purine metabolism, nitrogen metabolism, arginine biosynthesis, alanine, aspartate and glutamate metabolism, glyoxylate and dicarboxylate metabolism, biosynthesis of unsaturated fatty acids, fatty acid elongation, fatty acid degradation, pyrimidine metabolism, fatty acid biosynthesis. This suggested that these 10 metabolic pathways might be the key pathways for the efficacy of probiotic therapy, and the prolonged duration of medication (2 weeks/4 weeks) did not produce additional changes in metabolic pathways.
4 Discussion
4.1 Impact of probiotic therapy on gut bacteria composition
After 4 weeks of probiotic therapy, the DOB value of Hp infected children showed a decreasing trend, and the Hp eradication rate was 33.33%, suggesting that probiotic therapy could inhibit Hp to a certain extent. In addition, the GSRS scores were significantly reduced after 4 weeks of probiotic therapy, indicating that probiotic therapy can significantly alleviate functional abdominal pain symptoms in children with Hp infection.
Although Hp infection did not significantly alter alpha diversity of the gut microbiota in children, the alpha diversity exhibited a decrease tendency, which was in line with previous studies reporting an inverse relationship between microbial diversity and Hp abundance (Das et al., 2017). There was no significant difference in beta diversity between healthy children and those infected with Hp, and probiotic therapy failed to significantly alter the beta diversity of the gut microbiota in children after 2 and 4 weeks of treatment. Whether probiotics can significantly alter gut microbiota diversity is still a controversial issue (Kristensen et al., 2016). Most randomized controlled trials indicated that there were no significant differences in alpha diversity and beta diversity between the probiotic group and the placebo group (Chen et al., 2021). But some studies have also reported contrasting findings, indicating that certain probiotics can significantly influence alpha and beta diversity during intervention periods of 90 days (Rahayu et al., 2021) and 4 weeks (Ferrario et al., 2014), respectively. Interindividual variations, duration of treatment, smaller sample sizes, diverse evaluation tools, and various types of probiotics may account for the inconsistent results observed. The F/B ratio, a commonly employed metric to reflect the status of gut microbial ecosystem health, showed a downward trend in children with Hp infection in this study. Furthermore, the intestinal tracts of children infected with Hp demonstrated a notable enrichment of microorganisms implicated in gut dysbiosis, such as Porphyromonadaceae and Shigella (Laffin et al., 2019). And probiotic therapy was related to the recovery of F/B values. The enriched Bacillales in the probiotic treatment group was also one of the enriched species in the gut of healthy children. In Hp-infected children, an increasing trend in the abundance of Proteobacteria was observed, whereas a corresponding decrease in the abundance of Streptococcus was noted. These outcomes were congruent with the findings reported in other relevant studies (Klymiuk et al., 2017; Schulz et al., 2018; He et al., 2019). Following probiotic therapy, the abundances of both microbiota were modulated toward their original levels. Previous studies have reported that probiotic supplementation can enhance the abundance of Prevotella (Rahayu et al., 2021). The enrichment of Prevotella in the probiotic-treated group observed in this study further validated this notion. Moreover, probiotic therapy significantly reduced the abundance of Parabacteroides, downregulated the levels of Ruminococcus, Eggerthella and TM7, while upregulating the abundances of Planctomycetes, Gemmiger, Dialister, and others. The aforementioned results suggested that Hp infection could lead to an imbalance in gut microbiota, causing alterations in microbial community structure. Conversely, probiotic therapy could modulate the microbial community structure, restore intestinal microecological balance and exert an inhibitory effect on Hp.
The abundance of Verrucomicrobia is closely associated with intestinal health, as it has the capability to produce short chain fatty acids (SCFAs) such as propionic acid and butyric acid. These SCFAs play a crucial role in maintaining intestinal health and regulating the immune system (Schlesner, 2006). Furthermore, like Verrucomicrobia, Clostridium, Fusobacteria, and Megamonas were also identified as producers of SCFAs like butyrate (Brennan and Garrett, 2019; Martín-Núñez et al., 2019; Cao et al., 2022), but their abundance in HP-infected children were lower than in healthy children. Hp can overcome the epithelial barrier and further destroy the tight mucosal defense barrier (Marques et al., 2021), eventually triggering chronic inflammation. Butyrate, which is the primary SCFA produced through bacterial fermentation of dietary fiber, can enhance mucosal homeostasis by exerting beneficial effects on innate and adaptive immune cells and epithelial barrier function (Cushing et al., 2015). Previous studies have demonstrated that butyrate may exhibit a bactericidal effect on Hp. Researchers utilized butyrate and supernatant of butyrate-producing bacteria for in vitro research, and the results showed that both of them had inhibitory effects on the growth of Hp, and potentially destructive abilities on Hp biofilm (Yonezawa et al., 2012). Compared with healthy children, the abundance of these butyrate-producing enterobacteria decreased in Hp-infected children, but increased after probiotic treatment, suggesting that probiotic therapy can adjust the abundance of these butyrate-producing enterobacteria to inhibit Hp.
N-nitroso compounds are often considered as carcinogenic factors, which are related to chronic bacterial infection. The risk of Hp-related gastric cancer was related to the increase of nitrite in the stomach and the formation of N-nitroso compounds (Leach et al., 1987). Recent studies showed that nitrate and nitrite were biological activity reservoirs of nitric oxide (NO), and NO might play an important regulatory role in human body, but this biological activation required the existence of symbiotic bacteria, because mammals lacked specific reductase (Lundberg et al., 2008). Moreover, some studies showed that acidified nitrite had bactericidal effect on Hp (Dykhuizen et al., 1998). Therefore, nitrate-nitrite-NO pathway might play an important regulatory mechanism in people infected with Hp (Chen et al., 2018). Gemmatimonadetes might be involved in the nitrogen cycle, which could use a variety of organic and inorganic substances as carbon and energy sources, including nitrate (Oshiki et al., 2022). Additionally, Acidobacteria could also play a role in nitrogen cycle, as previous studies have demonstrated its ability to utilize nitrite as a nitrogen source (Kielak et al., 2016). In this study, the abundance of the above two kinds of gut microbiota related to nitrate-nitrite-NO pathway were up-regulated in children infected with Hp, but down-regulated in children treated with probiotics, suggesting that probiotic therapy may inhibit Hp by regulating such intestinal bacteria.
4.2 Impact of probiotic therapy on metabolic profile
Purine and pyrimidine are typically utilized for the synthesis of nucleotides, which serve as precursors of nucleic acids and significant participants in diverse biological processes (Marais et al., 1999). According to studies, numerous microorganisms, including infectious pathogens, lacked the pathways for de novo purine biosynthesis. Consequently, these microorganisms could only retrieve free purine and purine nucleotides from the environment to fulfill their purine requirements, a phenomenon known as the salvage pathways (Ullman and Carter, 1995; El Kouni, 2003). In recent years, it has been discovered that Hp also falls into this category of microorganisms lacking the pathways for de novo purine biosynthesis, yet it possessed potent salvage pathways of purine (Liechti and Goldberg, 2012). In vitro studies have demonstrated that substances targeting the purine uptake and processing mechanism of Hp have potential antibacterial effects (Gollapalli et al., 2010; Hedstrom et al., 2011). Furthermore, a previous study which using bismuth to inhibit Hp showed significant adjustments in the metabolic pathways of purine and pyrimidine in Hp positive patients after treatment (Yao et al., 2021), which was consistent with the findings of this study. The results of KEGG metabolic pathway analysis of differential metabolites in children infected with Hp indicated that Hp infection could impact purine and pyrimidine metabolism in children. Additionally, the KEGG metabolic pathway analysis of significantly reversed metabolites in the probiotic treatment group also implicated purine and pyrimidine metabolism, suggesting that probiotic therapy may influence nucleotide metabolism of Hp by regulating these pathways to inhibit its growth.
Ammonia serves as the primary nitrogen source for Hp, and it has demonstrated the existence of a nitrogen-or ammonia-dependent regulatory system in Hp metabolism (Mobley et al., 2001). Hp facilitates the generation of reactive nitrogen species (RNS) in the human stomach, which have been implicated in Hp-associated inflammation as well as DNA damage (Shimizu et al., 2017). Macrophages can utilize L-arginine as a substrate to produce NO via the induction of inducible nitric oxide synthase (Tsuji et al., 1996). Furthermore, in vitro experiments have demonstrated the bactericidal effect of NO against Hp (Kuwahara et al., 2000). Relativistically, Hp can also inhibit the production of NO by inducing macrophage apoptosis and synthesizing arginase, among other mechanisms (Gobert et al., 2001; Bussière et al., 2005). In this study, nitrogen metabolism pathway was enriched in Hp-infected children and was also one of the enriched pathways in children after probiotic treatment. This phenomenon may be attributed to the aforementioned interplay between NO and Hp, suggesting that probiotic therapy potentially inhibit Hp by modulating nitrogen metabolism in the human body.
Studies have demonstrated that amino acids not only serve as energy and nitrogen sources for Hp, but also play a pivotal role in protein synthesis and bacterial colonization, and they can determine the virulence and stress resistance of Hp strains (Leduc et al., 2010). Substances with bactericidal effects against Hp strains, such as bismuth agents, can lead to a decrease in amino acid abundance within the Hp growth environment, thereby inhibiting the growth of Hp strains (Han et al., 2018). Furthermore, it has been reported in other study that following the use of Hp eradication medications, the downregulated amino acid metabolic pathways in Hp include the biosynthesis of phenylalanine, tyrosine, tryptophan, and lysine, as well as the metabolism of alanine, aspartic acid, glutamic acid, arginine, and proline (Yao et al., 2021). In the present study, probiotic therapy primarily modulated the arginine biosynthesis pathway and the metabolism of alanine, aspartic acid, and glutamic acid, suggesting that these two pathways may represent potential mechanisms underlying the antibacterial effects of probiotics.
The probiotic therapy utilized in this study included Clostridium butyricum, which can increase the abundance of SCFAs in the body (Okamoto et al., 2000). SCFAs, including acetate, propionate, butyrate and valeric acid, play a crucial role in maintaining the normal biological activity of the host (Agus et al., 2021). Besides, SCFAs serve as one of the primary antibacterial substances secreted by probiotics and have various functions such as preserving intestinal barrier integrity (Dalile et al., 2019), promoting the induction and expansion of intestinal regulatory T cells and macrophages (Kim, 2021; Traxinger et al., 2022), combating cancer and oxidation (Liu et al., 2021), inhibiting inflammation induced by pathogens (He et al., 2020), among others. However, it was noteworthy that SCFAs was not prominently featured among the differential metabolites identified by LC–MS in this study due to its strong volatility (He et al., 2020). Additionally, it has been reported that palmitic acid, a long-chain fatty acid, can impact the virulence of Hp strains and regulate gene expression (Valdez-Salazar et al., 2021), thereby enhancing the motility and adhesion of Hp. The results of this study showed a significant increase in palmitic acid levels in children infected with Hp (p < 0.001), but a significant decrease in the probiotic treatment group (p < 0.001). This suggested that probiotic therapy may influence the motility and adhesion of Hp by regulating the content of palmitic acid. Some studies have suggested that certain fatty acids, particularly polyunsaturated fatty acids, exhibited a bactericidal effect on Hp, and the efficacy of their bactericidal activity would increase with the degree of unsaturation (Sun et al., 2003). In addition, it has been found that certain fatty acids could inhibit the growth of Hp in a dose-dependent manner and induce the transformation of bacteria from bacillus to coccus, leading to a decrease in bacterial activity (Correia et al., 2012). In summary, fatty acids play a significant role in inhibiting Hp, and probiotic therapy in this study may exert an antibacterial effect by regulating metabolic pathways related to fatty acids in vivo.
Glyoxylic acid and dicarboxylic acid are crucial metabolites in organisms, playing a role in energy metabolism. The tricarboxylic acid cycle is the primary source of cell energy (Duggleby and Jackson, 2002), and the metabolism of glyoxylic acid and dicarboxylic acid can regulate this cycle (Kang et al., 2021). This study suggested that Hp infection may impact the metabolism of glyoxylic acid and dicarboxylic acid in children, potentially affecting the energy metabolism pathway. Furthermore, an experiment involving bismuth eradication of Hp indicated that bismuth can affect various metabolic pathways, including glyoxylic acid or dicarboxylic acid metabolism, suggesting a relationship between this metabolic pathway and bismuth’s inhibition of Hp growth (Yao et al., 2021). In conclusion, regulating the metabolic pathways of glyoxylic acid and dicarboxylic acid may be a potential mechanism for probiotic therapy to inhibit Hp.
In this study, healthy children and Hp infected children were recruited, and probiotic therapy was provided for Hp infected children for 4 weeks. Fecal samples were collected from healthy children, as well as from Hp infected children before and after probiotic therapy. Subsequently, 16S rRNA gene sequencing analysis and untargeted metabonomics analysis were conducted on the aforementioned samples to investigate the potential mechanism of probiotic therapy in eradicating Hp. Although the eradication rate of probiotic therapy currently lags behind that of antibiotics, the emergence of this therapy offers hope to individuals who are unable to tolerate conventional treatment. This study thoroughly analyzes the potential mechanism of probiotic therapy in eradicating Hp, providing a valuable reference for identifying potential targets and developing new drugs to combat Hp. This study also has several limitations. Firstly, due to the cost limitation, the sample size in this study is not large, and it needs to be expanded for further research in the future. Secondly, this study found that probiotic therapy can affect the abundance of some intestinal microorganisms that can produce SCFAs, but SCFAs was not found in the results of untargeted metabonomics based on LC–MS, which was caused by the volatile characteristics of SCFAs, and it needs to be verified by targeted metabonomics based on GC–MS in the future.
5 Conclusion
In conclusion, this study demonstrated that probiotic therapy can help maintain intestinal microecological balance, regulate the abundance of butyrate-producing bacteria and intestinal microbiota related to the nitrate-nitrite-NO pathway. Furthermore, probiotic therapy was found to regulate pyrimidine and purine metabolic pathways, nitrogen metabolic pathways, amino acid-related metabolic pathways, fatty acid-related metabolic pathways, as well as glyoxylic acid and dicarboxylic acid metabolic pathways in children. The aforementioned approaches might be potential mechanisms of probiotic therapy for Hp infection.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.chictr.org.cn, ChiCTR2200062024.
Ethics statement
The studies involving humans were approved by the Ethics Review Committee of Shanxi Bethune Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the minor(s)’ legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
Author contributions
YY: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. LD: Data curation, Investigation, Methodology, Software, Writing – original draft. JX: Funding acquisition, Resources, Writing – review & editing. ZZ: Formal analysis, Writing – review & editing. PJ: Formal analysis, Writing – review & editing. JZ: Investigation, Writing – review & editing. WC: Project administration, Resources, Writing – review & editing. WG: Data curation, Methodology, Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Fundamental Research Program of Shanxi Province (202203021212109), and WU JIEPING Medical Foundation (320.6750.2021-08-12).
Acknowledgments
We would like to express our sincere appreciation to all those who have contributed to the completion of the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Agus, A., Clément, K., and Sokol, H. (2021). Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 70, 1174–1182. doi: 10.1136/gutjnl-2020-323071
Brennan, C. A., and Garrett, W. S. (2019). Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166. doi: 10.1038/s41579-018-0129-6
Bussière, F. I., Chaturvedi, R., Cheng, Y., Gobert, A. P., Asim, M., Blumberg, D. R., et al. (2005). Spermine causes loss of innate immune response to Helicobacter pylori by inhibition of inducible nitric-oxide synthase translation. J. Biol. Chem. 280, 2409–2412. doi: 10.1074/jbc.C400498200
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., Holmes, S., et al. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cao, W., Zheng, C., Xu, X., Jin, R., Huang, F., Shi, M., et al. (2022). Clostridium butyricum potentially improves inflammation and immunity through alteration of the microbiota and metabolism of gastric cancer patients after gastrectomy. Front. Immunol. 13:1076245. doi: 10.3389/fimmu.2022.1076245
Chen, M. J., Chen, C. C., Huang, Y. C., Tseng, C. C., Hsu, J. T., Lin, Y. F., et al. (2021). The efficacy of Lactobacillus acidophilus and rhamnosus in the reduction of bacterial load of Helicobacter pylori and modification of gut microbiota-a double-blind, placebo-controlled, randomized trial. Helicobacter 26:e12857. doi: 10.1111/hel.12857
Chen, L., Xu, W., Lee, A., He, J., Huang, B., Zheng, W., et al. (2018). The impact of Helicobacter pylori infection, eradication therapy and probiotic supplementation on gut microenvironment homeostasis: an open-label, randomized clinical trial. EBioMedicine 35, 87–96. doi: 10.1016/j.ebiom.2018.08.028
Correia, M., Michel, V., Matos, A. A., Carvalho, P., Oliveira, M. J., Ferreira, R. M., et al. (2012). Docosahexaenoic acid inhibits Helicobacter pylori growth in vitro and mice gastric mucosa colonization. PLoS One 7:e35072. doi: 10.1371/journal.pone.0035072
Cushing, K., Alvarado, D. M., and Ciorba, M. A. (2015). Butyrate and mucosal inflammation: new scientific evidence supports clinical observation. Clin. Transl. Gastroenterol. 6:e108. doi: 10.1038/ctg.2015.34
Dalile, B., Oudenhove, L. V., Vervliet, B., and Verbeke, K. (2019). The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 16, 461–478. doi: 10.1038/s41575-019-0157-3
Das, A., Pereira, V., Saxena, S., Ghosh, T. S., Anbumani, D., Bag, S., et al. (2017). Gastric microbiome of Indian patients with Helicobacter pylori infection, and their interaction networks. Sci. Rep. 7:15438. doi: 10.1038/s41598-017-15510-6
Duggleby, S. L., and Jackson, A. A. (2002). Protein, amino acid and nitrogen metabolism during pregnancy: how might the mother meet the needs of her fetus? Curr. Opin. Clin. Nutr. Metab. Care 5, 503–509. doi: 10.1097/00075197-200209000-00008
Dykhuizen, R. S., Fraser, A., McKenzie, H., Golden, M., Leifert, C., and Benjamin, N. (1998). Helicobacter pylori is killed by nitrite under acidic conditions. Gut 42, 334–337. doi: 10.1136/gut.42.3.334
El Kouni, M. H. (2003). Potential chemotherapeutic targets in the purine metabolism of parasites. Pharmacol. Ther. 99, 283–309. doi: 10.1016/S0163-7258(03)00071-8
Ferrario, C., Taverniti, V., Milani, C., Fiore, W., Laureati, M., Noni, I. D., et al. (2014). Modulation of fecal Clostridiales bacteria and butyrate by probiotic intervention with Lactobacillus paracasei DG varies among healthy adults. J. Nutr. 144, 1787–1796. doi: 10.3945/jn.114.197723
Fu, R., Liang, C., Chen, D., Yan, H., Tian, G., Zheng, P., et al. (2021). Effects of dietary Bacillus coagulans and yeast hydrolysate supplementation on growth performance, immune response and intestinal barrier function in weaned piglets. J. Anim. Physiol. Anim. Nutr. 105, 898–907. doi: 10.1111/jpn.13529
Gobert, A. P., McGee, D. J., Akhtar, M., Mendz, G. L., Newton, J. C., Cheng, Y., et al. (2001). Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 98, 13844–13849. doi: 10.1073/pnas.241443798
Goh, K. L., Chan, W. K., Shiota, S., and Yamaoka, Y. (2011). Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter 16, 1–9. doi: 10.1111/j.1523-5378.2011.00874.x
Gollapalli, D. R., Macpherson, I. S., Liechti, G., Gorla, S. K., Goldberg, J. B., and Hedstrom, L. (2010). Structural determinants of inhibitor selectivity in prokaryotic IMP dehydrogenases. Chem. Biol. 17, 1084–1091. doi: 10.1016/j.chembiol.2010.07.014
Han, B., Zhang, Z., Xie, Y., Hu, X., Wang, H., Xia, W., et al. (2018). Multi-omics and temporal dynamics profiling reveal disruption of central metabolism in Helicobacter pylori on bismuth treatment. Chem. Sci. 9, 7488–7497. doi: 10.1039/C8SC01668B
He, C., Peng, C., Wang, H., Ouyang, Y., Zhu, Z., Shu, X., et al. (2019). The eradication of Helicobacter pylori restores rather than disturbs the gastrointestinal microbiota in asymptomatic young adults. Helicobacter 24:e12590. doi: 10.1111/hel.12590
He, J., Zhang, P., Shen, L., Niu, L., Tan, Y., Chen, L., et al. (2020). Short-chain fatty acids and their association with Signalling pathways in inflammation, glucose and lipid metabolism. Int. J. Mol. Sci. 21:6356. doi: 10.3390/ijms21176356
Hedstrom, L., Liechti, G., Goldberg, J. B., and Gollapalli, D. R. (2011). The antibiotic potential of prokaryotic IMP dehydrogenase inhibitors. Curr. Med. Chem. 18, 1909–1918. doi: 10.2174/092986711795590129
Ji, J., and Yang, H. (2020). Using probiotics as supplementation for Helicobacter pylori antibiotic therapy. Int. J. Mol. Sci. 21:1136. doi: 10.3390/ijms21031136
Jiang, M. Z. (2023). Further standardize the diagnosis and treatment of Helicobacter pylori infection in children. Zhonghua Er Ke Za Zhi 61, 577–579. doi: 10.3760/cma.j.cn112140-20230510-00326
Kang, W., Suzuki, M., Saito, T., and Miyado, K. (2021). Emerging role of TCA cycle-related enzymes in human diseases. Int. J. Mol. Sci. 22:13057. doi: 10.3390/ijms222313057
Katoh, K., Misawa, K., Kuma, K. I., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., Veen, J. A. V., and Kuramae, E. E. (2016). The ecology of acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7:744. doi: 10.3389/fmicb.2016.00744
Kim, C. H. (2021). Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell. Mol. Immunol. 18, 1161–1171. doi: 10.1038/s41423-020-00625-0
Klymiuk, I., Bilgilier, C., Stadlmann, A., Thannesberger, J., Kastner, M. T., Högenauer, C., et al. (2017). The human gastric microbiome is predicated upon infection with Helicobacter pylori. Front. Microbiol. 8:2508. doi: 10.3389/fmicb.2017.02508
Kristensen, N. B., Bilgilier, C., Stadlmann, A., Thannesberger, J., Kastner, M. T., Högenauer, C., et al. (2016). Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 8:52. doi: 10.1186/s13073-016-0300-5
Kuwahara, H., Miyamoto, Y., Akaike, T., Kubota, T., Sawa, T., Okamoto, S., et al. (2000). Helicobacter pylori urease suppresses bactericidal activity of peroxynitrite via carbon dioxide production. Infect. Immun. 68, 4378–4383. doi: 10.1128/IAI.68.8.4378-4383.2000
Laffin, M., Fedorak, R., Zalasky, A., Park, H., Gill, A., Agrawal, A., et al. (2019). A high-sugar diet rapidly enhances susceptibility to colitis via depletion of luminal short-chain fatty acids in mice. Sci. Rep. 9:12294. doi: 10.1038/s41598-019-48749-2
Leach, S. A., Thompson, M., and Hill, M. (1987). Bacterially catalysed N-nitrosation reactions and their relative importance in the human stomach. Carcinogenesis 8, 1907–1912. doi: 10.1093/carcin/8.12.1907
Leduc, D., Gallaud, J., Stingl, K., and Reuse, H. (2010). Coupled amino acid deamidase-transport systems essential for Helicobacter pylori colonization. Infect. Immun. 78, 2782–2792. doi: 10.1128/IAI.00149-10
Liechti, G., and Goldberg, J. B. (2012). Helicobacter pylori relies primarily on the purine salvage pathway for purine nucleotide biosynthesis. J. Bacteriol. 194, 839–854. doi: 10.1128/JB.05757-11
Liu, P., Wang, Y., Yang, G., Zhang, Q., Meng, L., Xin, Y., et al. (2021). The role of short-chain fatty acids in intestinal barrier function, inflammation, oxidative stress, and colonic carcinogenesis. Pharmacol. Res. 165:105420. doi: 10.1016/j.phrs.2021.105420
Lundberg, J. O., Weitzberg, E., and Gladwin, M. T. (2008). The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 7, 156–167. doi: 10.1038/nrd2466
Lv, Z., Wang, Y., Yang, G., Zhang, Q., Meng, L., Xin, Y., et al. (2015). Efficacy and safety of probiotics as adjuvant agents for Helicobacter pylori infection: a meta-analysis. Exp. Ther. Med. 9, 707–716. doi: 10.3892/etm.2015.2174
Marais, A., Marais, A., Mendz, G. L., Hazell, S. L., and Mégraud, F. (1999). Metabolism and genetics of Helicobacter pylori: the genome era. Microbiol. Mol. Biol. Rev. 63, 642–674. doi: 10.1128/MMBR.63.3.642-674.1999
Marques, M. S., Costa, A. C., Osório, H., Pinto, M., Relvas, S., Dinis-Ribeiro, M., et al. (2021). Helicobacter pylori Pqq E is a new virulence factor that cleaves junctional adhesion molecule a and disrupts gastric epithelial integrity. Gut Microbes 13, 1–21. doi: 10.1080/19490976.2021.1921928
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200
Martín-Núñez, G. M., Cornejo-Pareja, I., Coin-Aragüez, L., Roca-Rodríguez, D. M., Araceli Muñoz-Garach, A., Clemente-Postigo, M., et al. (2019). H. pylori eradication with antibiotic treatment causes changes in glucose homeostasis related to modifications in the gut microbiota. PLoS One 14:e0213548. doi: 10.1371/journal.pone.0213548
Mobley, H. L. T., and Hazell, S. L. (Eds.) (2001). Helicobacter pylori: physiology and genetics. Washington, DC: ASM Press.
Neunert, C., Lim, W., Crowther, M., Cohen, A., Solberg, L. Jr., Crowther, M. A., et al. (2011). The American Society of Hematology 2011 evidence-based practice guideline for immune thrombocytopenia. Blood 117, 4190–4207. doi: 10.1182/blood-2010-08-302984
Okamoto, T., Sasaki, M., Tsujikawa, T., Fujiyama, Y., Bamba, T., and Kusunoki, M. (2000). Preventive efficacy of butyrate enemas and oral administration of Clostridium butyricum M588 in dextran sodium sulfate-induced colitis in rats. J. Gastroenterol. 35, 341–346. doi: 10.1007/s005350050358
Oshiki, M., Yuka Toyama, Y., Suenaga, T., Terada, A., Kasahara, Y., and Takashi Yamaguchi, T. (2022). N(2) O reduction by Gemmatimonas aurantiaca and potential involvement of Gemmatimonadetes Bacteria in N(2)O reduction in agricultural soils. Microbes Environ. 37:n/a. doi: 10.1264/jsme2.ME21090
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). Fast tree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Rahayu, E. S., Mariyatun, M., Putri Manurung, N. E., Hasan, P. N., Therdtatha, P., Mishima, R., et al. (2021). Effect of probiotic Lactobacillus plantarum Dad-13 powder consumption on the gut microbiota and intestinal health of overweight adults. World J. Gastroenterol. 27, 107–128. doi: 10.3748/wjg.v27.i1.107
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R., and Rastall, R. A. (2019). Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616. doi: 10.1038/s41575-019-0173-3
Schlesner, H. (2006). The phylum verrucomicrobia: a phylogenetically heterogeneous bacterial group. New York: Springer.
Schulz, C., Schütte, K., Koch, N., Vilchez-Vargas, R., Wos-Oxley, M. L., Oxley, A. P. A., et al. (2018). The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut 67, 216–225. doi: 10.1136/gutjnl-2016-312904
Segal, I., Otley, A., Issenman, R., Armstrong, D., Espinosa, V., Cawdron, R., et al. (2008). Low prevalence of Helicobacter pylori infection in Canadian children: a cross-sectional analysis. Can. J. Gastroenterol. 22, 485–489. doi: 10.1155/2008/410176
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Seo, J. H., Bortolin, K., and Jones, N. L. (2020). Review: Helicobacter pylori infection in children. Helicobacter 25:e12742. doi: 10.1111/hel.12742
Shimizu, T., Chiba, T., and Marusawa, H. (2017). Helicobacter pylori-mediated genetic instability and gastric carcinogenesis. Curr. Top. Microbiol. Immunol. 400, 305–323.
Subspecialty Group of Gastroenterology, the Society of Pediatrics, Chinese Medical Association, National Children’s Medical Center Digestive Specialist Alliance, Editorial Board, Chinese Journal of Pediatrics (2023). Expert consensus on the diagnosis and management of Helicobacter pylori infection in Chinese children (2022). Zhonghua Er Ke Za Zhi 61, 580–587. doi: 10.3760/cma.j.cn112140-20220929-00849
Sugano, K., Tack, J., Kuipers, E. J., Graham, D. Y., El-Omar, E. M., Miura, S., et al. (2015). Kyoto global consensus report on Helicobacter pylori gastritis. Gut 64, 1353–1367. doi: 10.1136/gutjnl-2015-309252
Sun, C. Q., O'Connor, C. J., and Roberton, A. M. (2003). Antibacterial actions of fatty acids and monoglycerides against Helicobacter pylori. FEMS Immunol. Med. Microbiol. 36, 9–17. doi: 10.1016/S0928-8244(03)00008-7
Takahashi, M., Taguchi, H., Yamaguchi, H., Osaki, T., and Kamiya, S. (2000). Studies of the effect of Clostridium butyricum on Helicobacter pylori in several test models including gnotobiotic mice. J. Med. Microbiol. 49, 635–642. doi: 10.1099/0022-1317-49-7-635
Thévenot, E. A., Roux, A., Xu, Y., Ezan, E., and Junot, C. (2015). Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 14, 3322–3335. doi: 10.1021/acs.jproteome.5b00354
Traxinger, B. R., Richert-Spuhler, L. E., and Lund, J. M. (2022). Mucosal tissue regulatory T cells are integral in balancing immunity and tolerance at portals of antigen entry. Mucosal Immunol. 15, 398–407. doi: 10.1038/s41385-021-00471-x
Tsuji, S., Kawano, S., Tsujii, M., Takei, Y., Tanaka, M., Sawaoka, H., et al. (1996). Helicobacter pylori extract stimulates inflammatory nitric oxide production. Cancer Lett. 108, 195–200. doi: 10.1016/S0304-3835(96)04410-2
Ullman, B., and Carter, D. (1995). Hypoxanthine-guanine phosphoribosyltransferase as a therapeutic target in protozoal infections. Infect. Agents Dis. 4, 29–40
Valdez-Salazar, H. A., Ares, M. A., Fernández, F. J., Ibarra, J. A., Torres, J., Bustamante, V. H., et al. (2021). Long-chain fatty acids alter transcription of Helicobacter pylori virulence and regulatory genes. Peer J 9:e12270. doi: 10.7717/peerj.12270
Wang, Q. (2013). The clinical trial on the eradication of Helicobacter pylori using Bacillus coagulans tablets and triple therapy. Chin. J. Microecol. 25, 418–421.
Wei, S. D., Bahl, M. L., Baunwall, S. M. D., Hvas, C. L., and Licht, T. R. (2021). Determining gut microbial Dysbiosis: a review of applied indexes for assessment of intestinal microbiota imbalances. Appl. Environ. Microbiol. 87:e00395-21. doi: 10.1128/AEM.00395-21
Wieërs, G., Belkhir, L., Enaud, R., Leclercq, S., Philippart de Foy, J. M., Dequenne, I., et al. (2019). How probiotics affect the microbiota. Front. Cell. Infect. Microbiol. 9:454. doi: 10.3389/fcimb.2019.00454
Yao, X., Xiao, S., and Zhou, L. (2021). Integrative proteomic and metabolomic analyses reveal the mechanism by which bismuth enables Helicobacter pylori eradication. Helicobacter 26:e12846. doi: 10.1111/hel.12846
Yonezawa, H., Osaki, T., Hanawa, T., Kurata, S., Zaman, C., Woo, T. D. H., et al. (2012). Destructive effects of butyrate on the cell envelope of Helicobacter pylori. J. Med. Microbiol. 61, 582–589. doi: 10.1099/jmm.0.039040-0
Zhang, J., Guo, J., Li, D., Chen, M., Liu, J., Feng, C., et al. (2020). The efficacy and safety of Clostridium butyricum and Bacillus coagulans in Helicobacter pylori eradication treatment an open-label, single-arm pilot study. Medicine 99:e22976. doi: 10.1097/MD.0000000000022976
Zhou, X. Z., Lyu, N. H., Zhu, H. Y., Cai, Q. C., Kong, X. Y., Xie, P., et al. (2023). Large-scale, national, family-based epidemiological study on Helicobacter pylori infection in China: the time to change practice for related disease prevention. Gut 72, 855–869. doi: 10.1136/gutjnl-2022-328965
Keywords: Helicobacter pylori, probiotic therapy, mechanism, 16S rRNA gene sequencing, untargeted metabolomics
Citation: Yan Y, Dong L, Xu J, Zhang Z, Jia P, Zhang J, Chen W and Gao W (2024) Preliminary study on the potential impact of probiotic combination therapy on Helicobacter pylori infection in children using 16S gene sequencing and untargeted metabolomics approach. Front. Microbiol. 15:1487978. doi: 10.3389/fmicb.2024.1487978
Edited by:
Juan Xicohtencatl-Cortes, Hospital Infantil de México Federico Gómez, MexicoReviewed by:
Norma Velazquez-Guadarrama, Federico Gómez Children’s Hospital, MexicoOpeyemi U. Lawal, University of Guelph, Canada
Copyright © 2024 Yan, Dong, Xu, Zhang, Jia, Zhang, Chen and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weihong Chen, d2hoY2hlbkAxMjYuY29t; Weiqi Gao, Z2FveWFuZzA1MThAMTYzLmNvbQ==
†These authors share first authorship
 Ya Yan
Ya Yan Lingjun Dong
Lingjun Dong Juan Xu
Juan Xu Zhijiao Zhang
Zhijiao Zhang Pengyan Jia
Pengyan Jia Jingmin Zhang
Jingmin Zhang Weihong Chen
Weihong Chen Weiqi Gao
Weiqi Gao