- 1Integrative Science Center of Germplasm Creation in Western China (CHONGQING) Science City, Biological Science Research Center, Southwest University, Chongqing, China
- 2Chongqing Engineering and Technology Research Center for Novel Silk Materials, Southwest University, Chongqing, China
The Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway plays a crucial role in the viral immune processes of organisms, with STAT being a key transcription factor downstream in this pathway. The STAT gene of Bombyx mori has two splicing forms, named BmSTAT-S and BmSTAT-L. This study compared the effects of the two splicing forms on Bombyx mori nucleopolyhedrovirus (BmNPV) infection through cell-level interference and further explored whether BmSTAT participates in the immune response to BmNPV infection via transgenic intervention at the individual level. Our research results indicated that BmNPV upregulates the expression of BmSTAT-S and BmSTAT-L in Bombyx mori BmE cells and larvae. Furthermore, BmE cells with interference of BmSTAT-S or BmSTAT-L displayed significantly higher expression levels of the viral gene GP41 and increased viral fluorescence compared to the control group after 48 h of infection with BmNPV. Then, we constructed transgenic silkworms with genetic interference, and the results showed that both the transgenic silkworms with systemic interference and midgut-specific interference of the two splice forms of BmSTAT exhibited significantly reduced survival rates and increased viral replication numbers after infection with BmNPV. The above results indicated that the BmSTAT gene is involved in the immune response of Bombyx mori to BmNPV and these findings lay the foundation for further research on the mechanism of JAK/STAT signaling pathway involvement in BmNPV infection.
1 Introduction
Bombyx mori, a significant economic insect, holds considerable importance in sericulture and life science research (Goldsmith et al., 2005; Jiang et al., 2021b). In China, diseases in silkworms account for over 20% of the annual cocoon yield losses, with viral infections contributing to more than 80% of these losses, predominantly due to the Bombyx mori nucleopolyhedrovirus (BmNPV) induced hemorrhagic septicemia (Jiang and Xia, 2014; Jiang, 2021). BmNPV, a rod-shaped virus with circular double-stranded DNA, occurs across various silkworm-producing regions globally, particularly causing outbreaks during the summer and autumn seasons that lead to significant production losses. Although studies have indicated that multiple signaling pathways, including RNA interference (RNAi), the stimulator of interferon genes (STING) signaling pathway, and the NF-kB signaling pathway, are involved in the immune response of Bombyx mori to viral infections (Jiang, 2021), the immune mechanisms between Bombyx mori and BmNPV have not yet been effectively elucidated.
The Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway is a ubiquitously expressed intracellular signal transduction pathway, with related proteins and fundamental functions are relatively conserved across organisms. This pathway has been demonstrated to participate in numerous critical biological processes including cell proliferation, differentiation, apoptosis, and immune regulation (Hombría and Brown, 2002; Myllymäki and Rämet, 2014; Herrera and Bach, 2019). The human STAT protein family consists of seven members, and comparative studies on the sequences of different STAT gene products reveal their common origin and evolutionary process. However, research discovered that these seven STAT exhibited a wide range of biological functions. Specifically, mice with STAT1 or STAT2 genes knocked out exhibit impaired antiviral responses, while the knockout of STAT3 results in embryonic lethality. Furthermore, the deletion of STAT4 leads to impaired T-helper 1 (Th1) cells development, and the absence of STAT5 causes defects in mammary gland development, infertility, and the absence of natural killer (NK) cells. Additionally, the knockout of STAT6 impairs T-helper 2 (Th2) cells development (Ihle, 1996; Morris et al., 2018). In Anopheles gambiae, two STAT genes exist: AgSTAT-A and a second intronless STAT gene, AgSTAT-B, which is produced by replication of AgSTAT-A in vivo. AgSTAT-B acts upstream to regulate the expression of AgSTAT-A, jointly responding to bacterial and Plasmodium infections (Gupta et al., 2009). In Caenorhabditis elegans, STA-1 regulates TGF-β signaling, while STA-2 plays a crucial role in maintaining the integrity and barrier function of the epidermis (Wang and Levy, 2006; Zhang et al., 2015). Additionally, in the fall armyworm Spodoptera frugiperda, two splice variants of STAT are found in the Sf9 cell line derived from pupal ovarian tissues, but no studies have yet demonstrated functional differences between these splice variants (Yan et al., 1996; Yeh et al., 2008). In Drosophila, STAT is critically important for various developmental processes, including embryonic segmentation (Hou et al., 1996), formation of wings and eyes (Hou et al., 1996), development of the hindgut and midgut (Schindler and Darnell, 1995), proliferation and differentiation of blood cells (Myllymäki and Rämet, 2014), and maintenance of germline stem cells (Hombría and Brown, 2002).
In addition, the JAK/STAT pathway has also been proven to be widely involved in the viral immune response of various organisms. In mammals, the JAK/STAT pathway can induce the upregulation of multiple interferon-stimulated genes (ISGs), thereby rapidly eliminating viruses within infected cells (Iwasaki and Pillai, 2014; Schoggins et al., 2014). Moreover, STAT plays a pivotal role in the antiviral defense of Drosophila, studies found that the number of factors specifically binding to the optimal DNA binding site of STAT92E in whole fly nuclear extracts of Drosophila infected with Drosophila C virus (DCV) increased, indicated that the infection of Drosophila with DCV induced the DNA binding activity of STAT (Yan et al., 1996; Dostert et al., 2005; Kemp et al., 2013). The infection with invertebrate iridescent virus 6 (IIV-6) also triggers the JAK/STAT pathway, which is crucial for the host’s defense against IIV-6 infection (West and Silverman, 2018). The pathway has also been shown to contribute to the immune response between Aedes aegypti and dengue virus (DENV)(Souza-Neto et al., 2009), and Culex mosquitoes and West Nile virus (WNV) (Paradkar et al., 2012). Additionally, studies indicated that the JAK/STAT pathway can promote viral proliferation during viral infection responses. For instance, the cytokine receptor Domeless in Litopenaeus vannamei (LvDOME) may be hijacked by the white spot syndrome virus (WSSV), facilitating viral replication (Yan et al., 2015), the CqDOME in Cherax quadricarinatus plays a crucial role in enhancing WSSV infection (Liu et al., 2020), and WSSV utilizes the transcription factor PmSTAT in Penaeus monodon to augment the expression of viral genes within host cells (Liu et al., 2007). These observations suggest that the immunological functions and mechanisms of the JAK/STAT pathway vary among different species.
The fundamental components of the JAK/STAT signaling pathway in Bombyx mori have been identified, including key genes such as BmDOME, BmSTAT, and BmHOP. The BmSTAT gene in Bombyx mori exists in two splice forms, named BmSTAT-S and BmSTAT-L. Previous studies reported that the open reading frames (ORFs) of BmSTAT-S and BmSTAT-L comprise 2,202 and 2,313 nucleotides, encoding 733 and 770 amino acids, respectively. The last 44 amino acid residues of BmSTAT-L differ from the final 7 amino acid residues of BmSTAT-S, with BmSTAT-S being spliced from exons 1–19 and BmSTAT-L from exons 1–18 and exon 20. Amino acid homology analysis revealed that the BmSTAT gene shares similarities of 92, 43, 35, and 42% with Spodoptera frugiperda STAT (SfSTAT), Danio rerio STAT (DrSTAT), Drosophila melanogaster STAT (DmSTAT), and Homo sapiens STAT (HsSTAT), respectively (Zhang et al., 2016). Research indicated that JAK/STAT signaling pathway participates in cellular proliferation (Hu et al., 2015), and the development of wing primordia in Bombyx mori (Zhang et al., 2020). Moreover, it had been demonstrated to play a significant role in the immune response of Bombyx mori, BmNPV infection can activate the JAK/STAT pathway (Liu et al., 2015), and under Beauveria bassiana stress, this pathway may be involved in the synthesis and secretion of antifungal substances (Geng et al., 2016; Zhang et al., 2016). Inhibition of heat shock protein 90 (Hsp90) can suppress BmNPV proliferation in Bombyx mori and leads to the upregulation of STAT and downregulation of suppressor of cytokine signalling protein 2 (SOCS2) and SOCS6, suggesting a potential cooperative role of Hsp90 with the JAK/STAT pathway in viral resistance (Shang et al., 2020). However, whether the key downstream transcription factor BmSTAT in the JAK/STAT signaling pathway participates in the BmNPV infection and its mechanisms has not yet been reported. This study explored the relationship between BmSTAT and BmNPV replication from the cellular and individual levels, laying a foundation for further research on the mechanism of JAK/STAT pathway involvement in BmNPV infection and how to inhibit the replication of BmNPV in Bombyx mori.
2 Materials and methods
2.1 Cell, silkworm strain, and viruses
The silkworm embryonic cell line BmE, the silkworm strain Dazao (DZ), wild BmNPV, and BmNPV-GFP expressing green fluorescent protein were maintained at Biological Science Research Center (Southwest University, Chongqing, China). BmE cells were cultured at 27°C in Grace medium. Silkworms were fed with mulberry leaves under conditions of 25°C. The silkworm larvae were orally inoculated with wild BmNPV and BmE cells were infected with BmNPV-GFP.
2.2 Subcellular localization
The cDNA of silkworm gonad was used as the template for polymerase chain reaction (PCR), and the cell expression vectors pSL1180 [Hr3-A4-RED-BmSTAT-S-SV40] and pSL1180 [Hr3-A4-RED-BmSTAT-L-SV40] were constructed. The related sequences are shown in Supplementary data and the basic vector pSL1180 [Hr3-A4-RED-SV40] is conserved in our laboratory. Cell transfection was conducted using X-tremeGENE HP DNA transfection reagent (Roche, Mannheim, Germany) according to the manufacturer’s instructions, and incubated for 8 h with serum-free media without antibiotics. Then, cells were cultured for another 48 h in fresh completed medium before other treatments. After 48 h of transfection, the cells were fixed for 10 min with 4% paraformaldehyde at room temperature and washed three times with phosphate buffered saline (PBS). Then cells were incubated with 4′-6-diamidino-2-phenylindole (DAPI) for 10 min, and observed under a fluorescence microscope after washing three times with PBS (Kang et al., 2021).
2.3 Viral infection
Monolayer cultures of BmE cells were seeded in 12-well plates for 12 h. BmNPV-GFP was added to the cells at a multiplicity of infection (MOI) of 1 (200 μl/well in 12-well plates) and the supernatant was removed after 1 h of absorption. Then, fresh complete medium was added, and culturing continued at 27°C (Wang et al., 2017). Cell RNA was extracted at 0, 3, 6, 12, and 24 h after BmNPV infection, and quantitative polymerase chain reaction (qPCR) was performed to detect the expression levels of BmSTAT-S and BmSTAT-L. For transfected cells, infected with BmNPV-GFP at 48 h after transfection, the operation method is the same as above. Total DNA was extracted for qPCR analysis of BmNPV gene GP41 and control gene glyceraldehyde3-phosphate dehydrogenase (BmGAPDH) (Guo et al., 2016) at 24 and 48 h post infection (hpi). The viral fluorescence was observed at 72 hpi.
The transgenic and wild silkworms were raised to fourth instar, 300 healthy larvae were randomly selected from each transgenic strain and non-transgenic strain. The experimental group was infected with BmNPV, and the control group was not infected with the virus. Each treatment contained three replicates, and each replicate contained 50 larvae. Orally infected with wild BmNPV using 2 × 106 occlusion bodies (OB)/larva. Total DNA was extracted from six larvae of transgenic and non-transgenic silkworms at 48 h after infection with BmNPV, which was used for qPCR analysis of the BmNPV gene GP41 and the control gene BmGAPDH. The mortality of larvae was recorded daily until day 10 post infection (Guo et al., 2019).
2.4 Quantitative polymerase chain reaction analysis
The total RNA was extracted using Trizol (Invitrogen, USA). 1 μg of total RNA was used for cDNA synthesis following the manufacturer’s protocol of the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time; TaKaRa, Japan). RNA samples were reverse transcribed into cDNA and used as templates. Genomic DNA samples were diluted with ddH2O to 200 ng/μl and used as templates. qPCR was performed with a NovoStart SYBR qPCR SuperMix Plus (Novoprotein, China) and a 7500 Fast Real-time PCR System (Applied Biosystems, USA). The silkworm BmGAPDH gene (Guo et al., 2019) was used as the internal control. All primer sequences are shown in Supplementary Table S1. All experiments were independently performed with three biological replicates, and each detection was performed three times. Finally, relative gene expression levels were analyzed using the 2−△△Ct method, and statistical analyses were performed using GraphPad Prism version 8 (Tang et al., 2020).
2.5 RNA interference
For RNA interference (RNAi) of BmSTAT, about 500 bp fragment was amplified by PCR from cDNA using a pair of primers with T7 RNA polymerase-binding site attached to the 5′-end of each primer for BmSTAT (related primers are shown in Supplementary Table S1). First, the DNA fragment was amplified using PCR, and the double-stranded RNAs (dsRNAs) were synthesized in vitro at 37°C by T7 RNA polymerase (Promega, Madison, WI, USA) for 12 h using the DNA fragment as a template (Kang et al., 2021). The Red gene dsRNA (dsRed) was used as a control. Specific sequence for BmSTAT-S and BmSTAT-L were selected, and small interfering RNAs (siRNAs) were designed and synthesized by Sangon Biotech (Shanghai, China). Related sequences are shown in Supplementary Table S2. A 2 μg sample of dsRNA or siRNA was transfected into BmE cells. After 48 h, RNA was extracted for qPCR analysis.
2.6 Establishment and detection of transgenic interference silkworm strains
Transgenic interference vectors were constructed using dsRNA fragments. The synthesis of pMV [BamHI-dsBmSTAT + A3intron-EcoR I] and pMV [EcoR I-FXHBdsBmSTAT-Not I] was carried out at Sangon Biotech (Shanghai, China). dsBmSTAT and FXHBdsBmSTAT are reverse complementary sequences. Then, the two target fragments were connected to vectors for whole-body overexpress, pSL1180 [Hr3-A4-SV40], and midgut specific overexpression, pSL1180 [P3P + 5UI-SV40], producing two expression vectors, pSL1180 [Hr3-A4-dsBmSTAT-SV40] and pSL1180 [P3P + 5UI-dsBmSTAT-SV40]. Finally, the fragments were connected to the piggyBac [3 × P3-EGFP] vector, generating the piggyBac [3 × P3-EGFP, A4-dsBmSTAT] systemic transgenic interference vector and the piggyBac [3 × P3-EGFP, P3P + 5UI-dsBmSTAT] midgut-specific transgenic interference vector. The A3 intron was cloned from the genomic DNA of DZ silkworm and used as a spacer between antisense and sense fragments. The related sequences are shown in the Supplementary data and the basic vectors are conserved in our laboratory.
Non-diapause DZ embryos were used for microinjection with the transgenic vector, this generation was called G0. The G1 embryos were screened for GFP-positive expression using a fluorescent microscope (Olympus, Japan). Two transgenic lines were obtained and named as A4-dsBmSTAT and Mg-dsBmSTAT. RNA from the midgut was extracted from A4-dsBmSTAT, Mg-dsBmSTAT and non-transgenic silkworm on the third day of fifth instar for qPCR analysis of BmSTAT-S, BmSTAT-L and the control BmGAPDH. Each detection was performed three times. All the primers are shown in Supplementary Table S1.
2.7 Statistical analysis
Statistical data were presented as means ± standard deviation (SD). Three independent experiments were performed to ensure validity, with at least three samples per test taken for statistical analysis. Statistical analyses were conducted using GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA). Significance (p value) was determined using the Student’s t-test and denoted as follows: *p < 0.05, **p < 0.01, and ***p < 0.001.
3 Results
3.1 Expression and localization of BmSTAT-S and BmSTAT-L in Bombyx mori
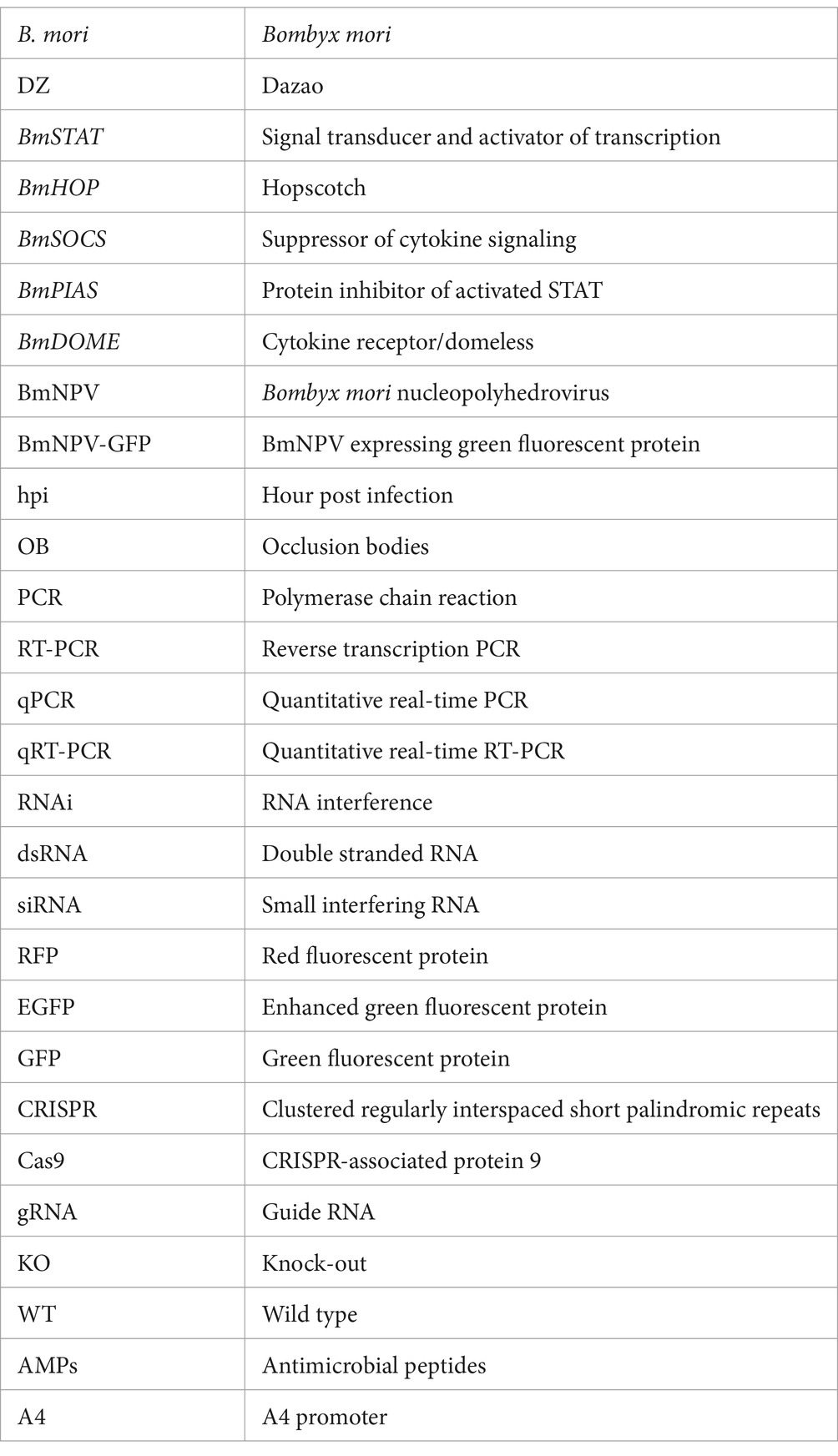
The midgut is an important organ of innate immunity in silkworms, and to resist the infection of pathogenic microorganisms, the silkworm midgut has developed a complete immune response system. In our earlier study, we found that the expression level of BmSTAT in the midgut was relatively high (Wang et al., 2024), suggesting that it might be involved in the immune response to pathogens. To explore the temporal expression patterns of BmSTAT in the midgut of Bombyx mori, quantitative reverse transcription PCR (qRT-PCR) was employed to monitor the midgut from day-3 of the fourth instar to day-5 of the pupal stage. The findings revealed that both splice forms of BmSTAT were expressed at all stages examined, with BmSTAT-L consistently showing higher expression levels than BmSTAT-S. The expression levels of both splice forms in the midgut began to decrease from day-3 of the fourth instar, followed by a gradual increase by day-3 of the fifth instar (Figure 1A).
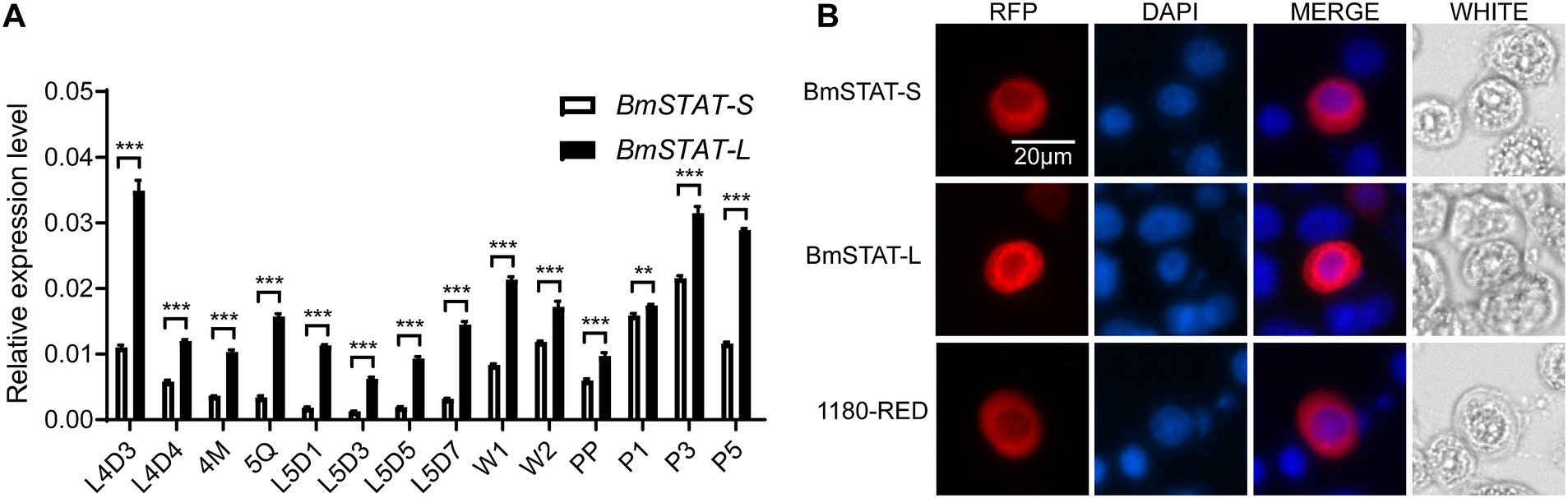
Figure 1. Expression and localization of BmSTAT-S and BmSTAT-L in B. mori. (A) Expression profile analysis of BmSTAT in midgut. L4D3 and L4D4 represent day 3 and day 4 of the fourth instar; 4 M: the fourth molting; 5Q: fifth instar larva; L5D1, L5D3, L5D5, and L5D7 correspond to day 1, day 3, day 5, and day 7 of the fifth instar, respectively; W1 and W2 represent day 1 and day 2 at the wandering stage; PP: prepupa; P1, P3, P5 represent day 1, day 3, day 5 of the pupal stage, respectively. (B) Subcellular localization analysis of BmSTAT. RFP: Red fluorescence protein fusion expression vector; DAPI: Stained nucleus (blue); 1,180-RED is the control group. Data are presented as means ± SD (n = 3). Significance levels: *p < 0.05, **p < 0.01, and ***p < 0.001; ns indicates no significance.
To investigate the subcellular localization of BmSTAT, the BmSTAT gene was transfected into Bombyx mori embryonic cells (BmE). The results revealed that both splice variants of BmSTAT predominantly localize in the cytoplasm of BmE cells, with minor distribution also observed in the nucleus (Figure 1B).
3.2 The expression levels of BmSTAT-S and BmSTAT-L are upregulated by BmNPV induction in Bombyx mori
To determine whether BmSTAT responds to BmNPV infection, BmE cells were infected with BmNPV-GFP (MOI = 1), and fourth-instar larvae were infected with wild-type BmNPV (2 × 106 OB/larva). The transcriptional levels of BmSTAT-S and BmSTAT-L were assessed using qRT-PCR. The results showed that in BmE cells, the expression levels of BmSTAT-S and BmSTAT-L significantly increased at 6, 12, and 24 h post-infection (Figures 2A,B). In Bombyx mori larvae, the expression levels of BmSTAT-S and BmSTAT-L were significantly elevated 48 h post-infection, reaching 1.18 and 1.41 times the control levels, respectively (Figures 2C,D). These experimental results indicate that BmNPV can activate the JAK/STAT pathway and induce an increase in the expression of BmSTAT-S and BmSTAT-L.
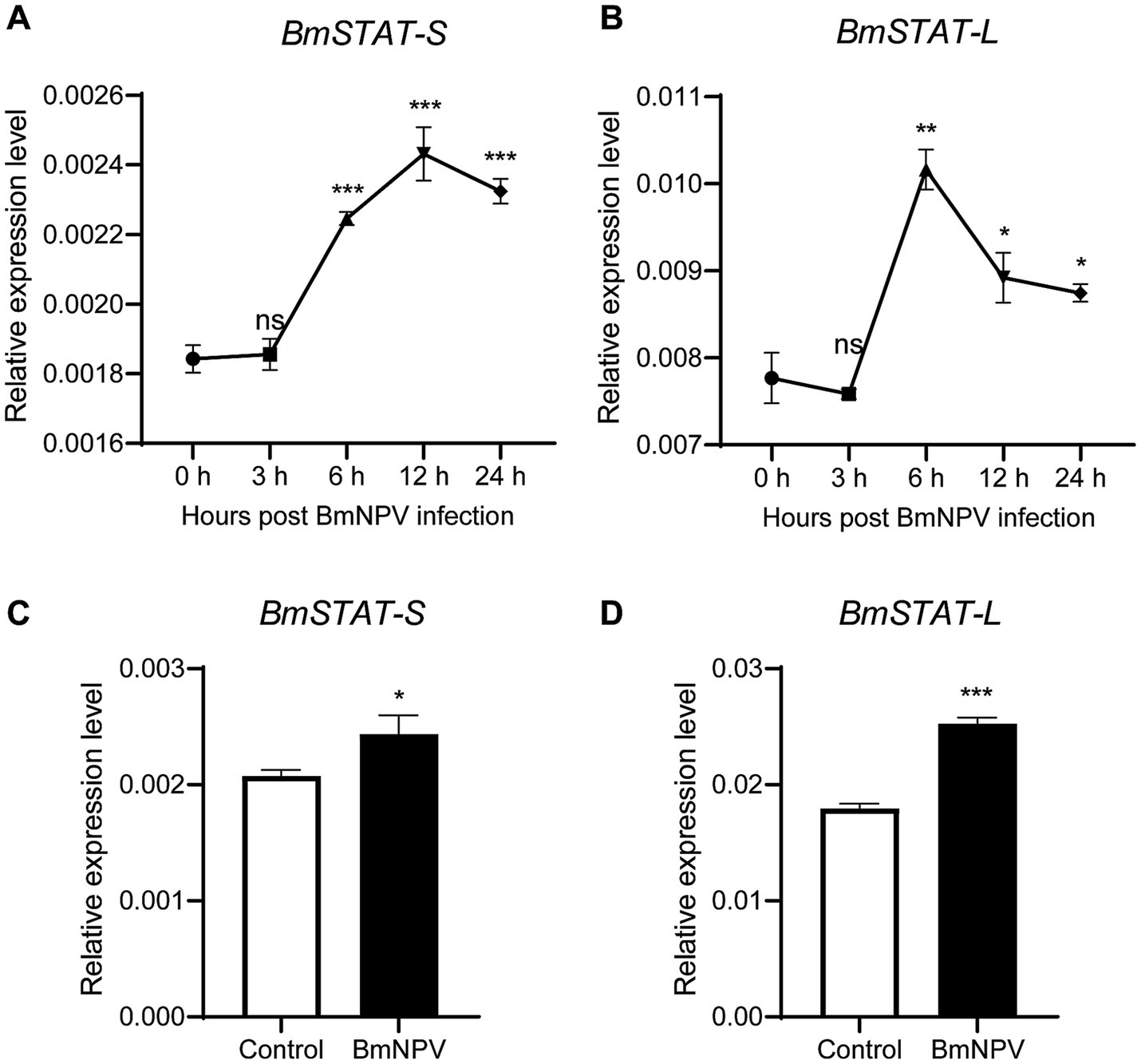
Figure 2. qRT-PCR analysis of BmSTAT-S and BmSTAT-L after BmNPV infection. (A,B) Detection of BmSTAT-S and BmSTAT-L in cells post-BmNPV infection. BmE cells were infected with BmNPV-GFP, and RNA was extracted at 0, 3, 6, 12, and 24 hpi to detect the expression of BmSTAT-S and BmSTAT-L. (C,D) Detection of BmSTAT-S and BmSTAT-L in silkworm post-BmNPV infection. Fourth-instar,1-day-old silkworm larvae were orally infected with wild-type BmNPV, and RNA was extracted at 48 hpi to detect BmSTAT-S and BmSTAT-L expression. Control, non-infected silkworms; BmNPV, infected silkworms. Data are presented as means ± SD (n = 3). Significance levels: *p < 0.05, **p < 0.01, and ***p < 0.001; ns indicates no significance.
3.3 Interference of BmSTAT-S, BmSTAT-L promotes BmNPV proliferation in BmE cells
To confirm the impact of BmSTAT on BmNPV, double-stranded RNA targeting the common sequence of BmSTAT-S and BmSTAT-L was synthesized. The results demonstrated that, 48 h post-transfection, the transcription level of BmSTAT was successfully reduced by 45% (Figure 3A). Cells with successful BmSTAT interference were infected with BmNPV (MOI = 1). Compared to the control group transfected with dsRED double-stranded RNA, the experimental group transfected with dsBmSTAT showed an increase in viral copy gene GP41 to 1.13 times and 1.26 times that of the control at 24 and 48 h, respectively (Figure 3B). At 72 h, the number of virus-infected cells exhibiting green fluorescence was significantly higher than that in the control group (Figure 3C).
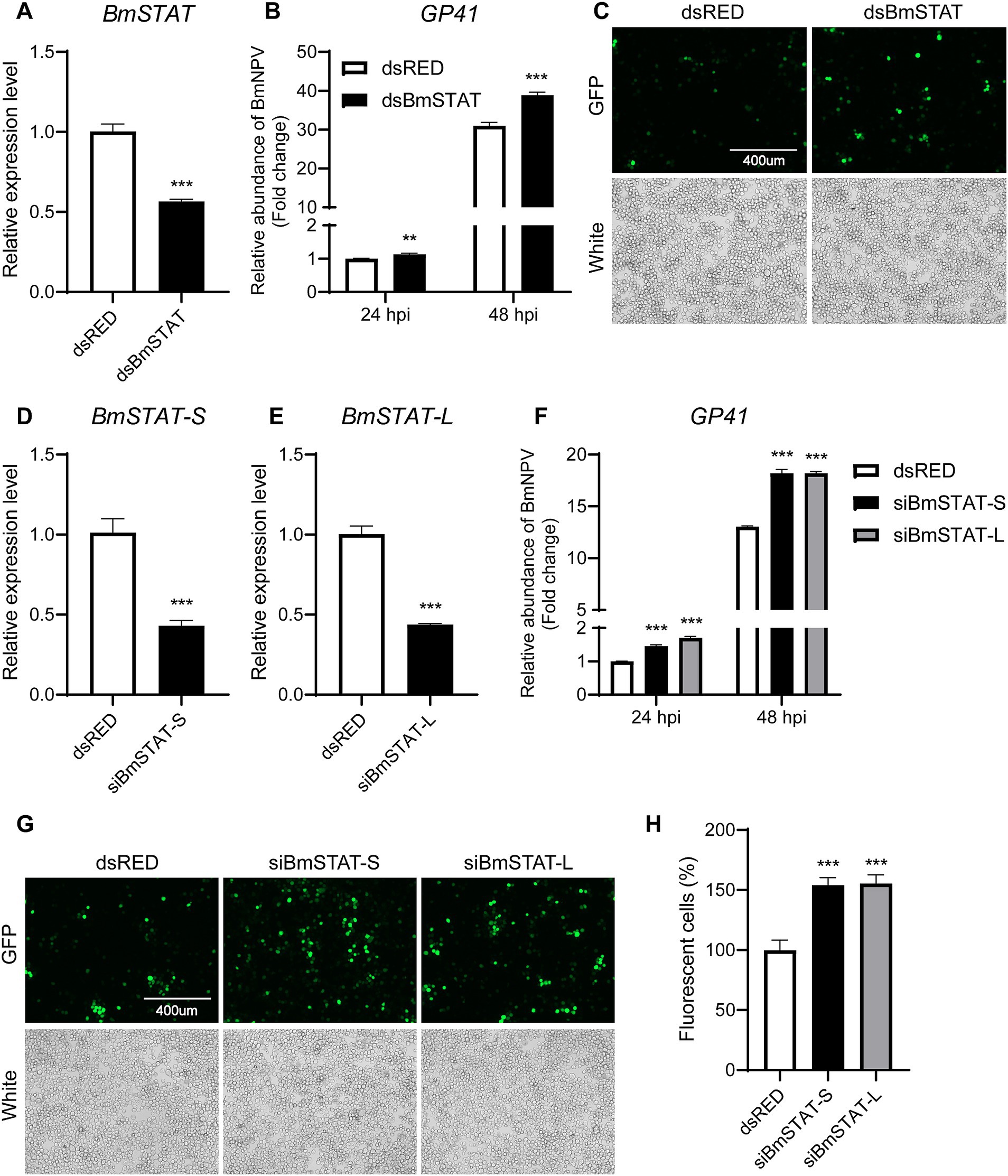
Figure 3. The downregulation of BmSTAT-S or BmSTAT-L promoted BmNPV multiplication. (A) qRT-PCR analysis of BmSTAT expression in BmE cells after transfection with dsBmSTAT and dsRED. (B) Analysis of viral DNA content. The cells were infected with BmNPV-GFP (MOI = 1) 48 h after transfection with dsRed and dsBmSTAT. Total DNA was extracted from infected cells at 24 and 48 hpi. The cumulative viral DNA content was assessed by qPCR analysis of GP41. (C) Observation of viral fluorescence. The green fluorescence of the virus was observed at 72 h after infection. (D) Analysis of BmSTAT-S expression by qRT-PCR after interference with BmSTAT-S in BmE cells. (E) Analysis of BmSTAT-L expression by qPCR after interference with BmSTAT-L in BmE cells. (F) Analysis of viral DNA content. After interference with BmSTAT-S and BmSTAT-L, the cells were infected with BmNPV-GFP (MOI = 1), and total DNA was extracted at 24 and 48 h post-infection for qPCR to detect the virus gene GP41 to determine the proliferation of the virus. (G) The green fluorescence of the virus was observed at 72 h after infection. (H) Quantification of fluorescent cells. The numbers of infected BmE cells in images shown in (G) were counted. The average level of dsRed was set at 100%. The data presented are the means ± SD (n = 3). For the significance analysis: **p < 0.01, ***p < 0.001.
To investigate the differential effects of the two splice forms of BmSTAT on BmNPV, siRNA was employed to specifically target BmSTAT-S and BmSTAT-L in Bombyx mori cells, followed by infection with BmNPV to assess viral proliferation. The siRNAs synthesized reduced the transcription levels of BmSTAT-S by 55% and BmSTAT-L by 56% (Figures 3D,E). Cells with successful interference of BmSTAT-S or BmSTAT-L were infected with BmNPV at an MOI of 1. Compared to the control group transfected with dsRED double-stranded RNA, the group with BmSTAT-S interference exhibited 1.45 times and 1.40 times the viral copy number at 24 and 48 h, respectively (Figure 3F). For the BmSTAT-L interference group, the viral copy numbers were 1.70 times and 1.40 times that of the control at 24 and 48 h, respectively (Figure 3F). Additionally, the number of cells displaying viral green fluorescence significantly increased by 72 h in both the BmSTAT-S and BmSTAT-L interference groups compared to the control (Figures 3G,H). The experimental results indicated that the interference with either BmSTAT-S or BmSTAT-L individually, or with both splice forms of BmSTAT simultaneously, leads to the enhancement of BmNPV replication in BmE cells.
3.4 Transgenic silkworm with BmSTAT interference has lower resistance to BmNPV
To determine the impact of interfering with BmSTAT on BmNPV in individuals, double-stranded RNA was designed to target a common sequence fragment of BmSTAT-S and BmSTAT-L, and transgenic interference vectors were constructed to obtain transgenic silkworms with gene interference. Initially, we acquired a systemic interference vector, piggyBac [3 × P3-EGFP, A4-dsBmSTAT], and a midgut-specific interference vector, piggyBac [3 × P3-EGFP, P3P + 5UI-dsBmSTAT] (Figure 4A). Subsequently, both plasmids were microinjected to produce two transgenic interference silkworms, named A4-dsBmSTAT (systemic transgenic interference silkworm) and Mg-dsBmSTAT (midgut-specific transgenic interference silkworm; Figure 4B). On day three of the fifth instar, midguts were sampled from three randomly selected transgenic positive individuals and wild-type silkworms to extract RNA for qRT-PCR analysis. The expression levels of BmSTAT-S and BmSTAT-L were found to be significantly reduced in both transgenic lines (Figure 4C).
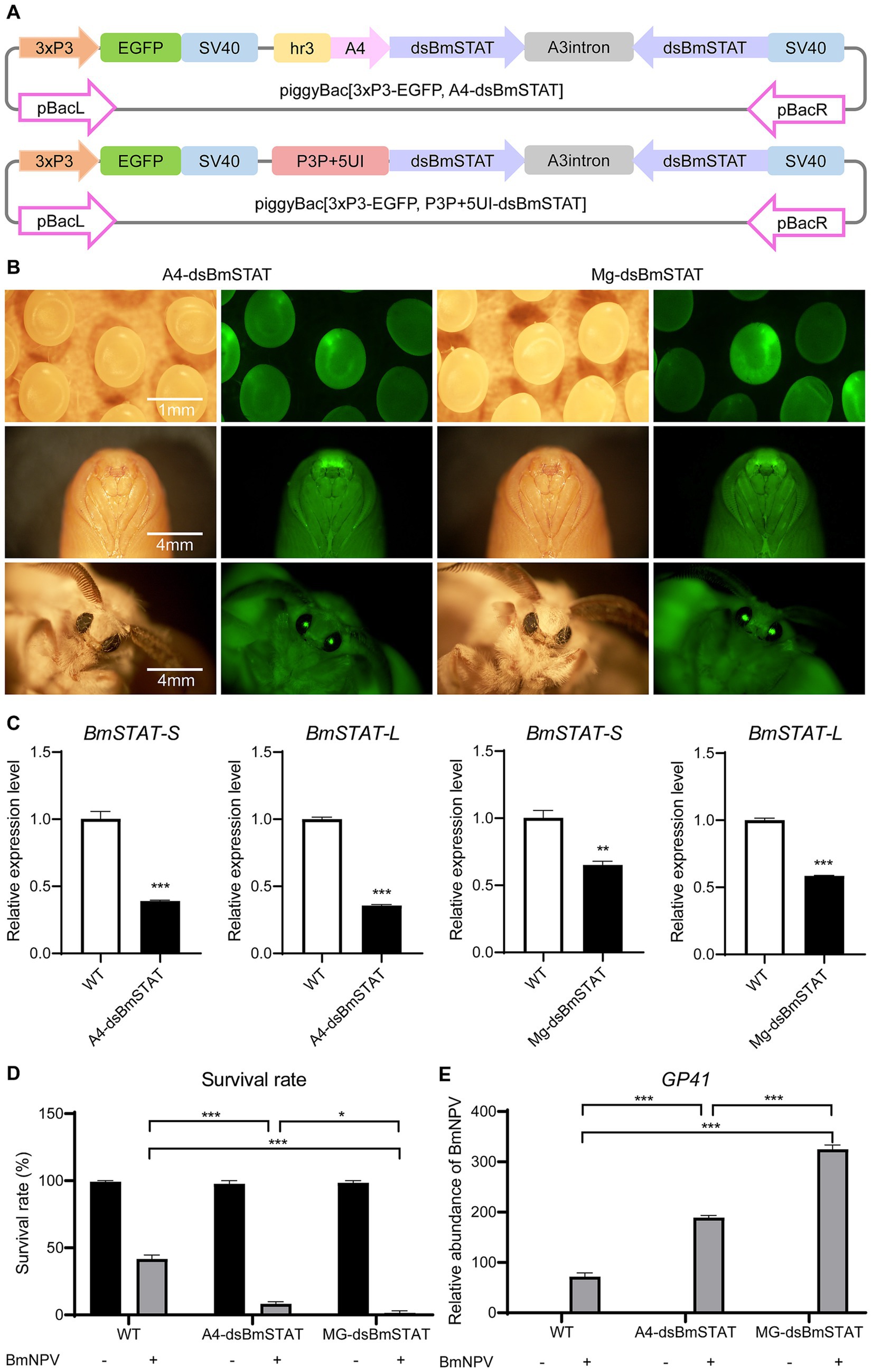
Figure 4. Transgenic silkworms with BmSTAT interference had lower resistance to BmNPV. (A) Schematic of the piggyBac[3xP3-EGFP, A4-dsBmSTAT] and piggyBac [3xP3-EGFP, P3P + 5UI-dsBmSTAT] vectors. Hr3, enhancer; A4, systemic promoter; dsBmSTAT, double-stranded RNA of BmSTAT; A3 intron, a spacer; SV40, polyadenylation signal as terminator; P3P + 5UI, midgut-specific promoter. (B) The transgenic silkworm A4-dsBmSTAT and Mg-dsBmSTAT were observed by fluorescence microscope and photographed. (C) RNA was extracted from the midgut of A4-dsBmSTAT and Mg-dsBmSTAT on the 3rd day of 5th instar, and qRT-PCR was used to detect the expression of BmSTAT-S and BmSTAT-L. (D) The survival rates of A4-dsBmSTAT and Mg-dsBmSTAT infected with BmNPV. (E) Detection of viral replication. After A4-dsBmSTAT and Mg-dsBmSTAT were infected with BmNPV, the number of viral replications was detected at 48 h. The data presented are the means ± SD (n = 3). For the significance analysis: **p < 0.01, ***p < 0.001.
Fourth instar, day-one larvae of A4-dsBmSTAT and Mg-dsBmSTAT were fed a BmNPV-infected diet containing 2 × 106 occlusion bodies (OB) per larva. The results demonstrated that both A4-dsBmSTAT and Mg-dsBmSTAT exhibited reduced survival rates compared to the control group. The survival rate of the control group was 40%, while it was 8% for A4-dsBmSTAT and 1% for Mg-dsBmSTAT (Figure 4D). Further examination of viral replication 48 h post-infection revealed that the viral copy numbers in both types of transgenic interfered silkworms were significantly higher than those in the control group, being 2.6 and 4.4 times higher, respectively (Figure 4E). These findings indicated that transgenic interference of BmSTAT in Bombyx mori results in decreased resistance to BmNPV.
4 Discussion
The JAK/STAT signaling pathway is one of the most crucial cytokine-mediated signal transduction pathways in organisms and has been demonstrated to participate in numerous key biological processes. Our findings indicate that BmNPV can activate the JAK/STAT signaling pathway and induce an increase in the expression levels of BmSTAT-S and BmSTAT-L. Moreover, reducing the expression level of BmSTAT promotes the proliferation of BmNPV and leads to a decrease in the resistance of silkworms to BmNPV. This demonstrats that BmSTAT plays a crucial role in the defense mechanism of silkworms against BmNPV infection and virus replication.
Previously, we discovered that both BmSTAT-S and BmSTAT-L are highly expressed in the midgut, a crucial immune organ in Bombyx mori (Wang et al., 2024). During BmNPV infection, the midgut serves as the initial site of infection and the first line of defense, implying that the BmSTAT gene may play a significant role in immune function. Subcellular localization studies showed that BmSTAT-S and BmSTAT-L were distributed in both the cytoplasm and the nucleus (Figure 1B), consistent with the function of BmSTAT as a transcription factor that is activated in the cytoplasm and then enters the nucleus to regulate the expression of downstream target genes. Previous studies have shown that the expression levels of BmSTAT in Bombyx mori hemolymph significantly increase following infection with both BmNPV and BmBDV (Liu et al., 2015). Post-infection with BmNPV, immunofluorescence of hemocytes showed an increased expression of BmSTAT-S (Zhang et al., 2016). We found that after BmNPV infected BmE cells, the expression levels of BmSTAT-S and BmSTAT-L increased at 6, 12, and 24 h. After stage 4 larvae infected with BmNPV, the expression levels of BmSTAT-S and BmSTAT-L significantly rose at 48 h post-infection (Figure 2). Understanding the impact of BmNPV on BmSTAT facilitates further exploration of the functional relationship between BmSTAT and BmNPV. Our research findings also indicated that the simultaneous interference of the two splicing forms of BmSTAT in BmE cells promotes the replication of BmNPV. It was further discovered that separate interference of BmSTAT-S or BmSTAT-L also enhances the proliferation of BmNPV (Figure 3). Therefore, both simultaneous and separate interference yield similar experimental results, suggesting that BmSTAT is involved in the in vivo response after BmNPV infection and plays a crucial role in resisting BmNPV infection.
Previous studies have shown that the silencing of genes associated with the JAK/STAT signaling pathway leads to a significant increase in viral titers and mortality following viral infections. The knockout of STAT1 increases susceptibility to mouse hepatitis virus (MHV) and results in a faster mortality rate following vesicular stomatitis virus (VSV) infection compared to controls (Durbin et al., 1996; Park et al., 2000). Knocking out STAT92E in Drosophila significantly increased lethality after infection with the IIV-6 (West and Silverman, 2018). In addition, studies have shown that the JAK/STAT signaling pathway plays a role in promoting viral proliferation in response to viral infection. The promoter of the shrimp WSSV ie1 gene contains a STAT binding site, and overexpression of LvDOME can increase the activity of the ie1 gene promoter by 20.2 times (Yan et al., 2015). Furthermore, WSSV uses PmSTAT as a transcription factor to enhance the expression of viral genes in host cells (Liu et al., 2007). Based on results obtained from the silkworm cellular levels, we chosen to concurrently interfere with both splice forms of STAT at the individual level for further experimentation. Our results indicated that reducing BmSTAT expression at the individual level significantly decreases survival rates following BmNPV infection and results in copy numbers significantly higher than those in the control group (Figure 4). This suggests that BmSTAT may play a role in resisting viral infection during the process of BmNPV infection, similar to that in mammals and Drosophila. Additionally, our research found that Bombyx mori with midgut-specific interference of the BmSTAT gene exhibited a higher mortality rate post-BmNPV infection compared to those with systemic interference. The midgut serves as the initial target organ for BmNPV invasion, where intact occlusion-derived viruses (ODVs) traverse the peritrophic membrane, and nucleocapsids enter the midgut epithelial cells through envelope-mediated membrane fusion, causing primary infection (Jiang et al., 2021a). It is hypothesized that midgut-specific interference of the BmSTAT gene facilitates viral invasion, leading to a rapid increase in viral load within the body and consequently higher mortality rates.
In summary, we found that BmNPV upregulates the expression of BmSTAT-S and BmSTAT-L in both BmE cells and larvae. Interfering with either BmSTAT-S or BmSTAT-L in BmE cells enhances the proliferation of BmNPV, and systemic and midgut-specific interference of the two splice forms of BmSTAT in transgenic Bombyx mori results in reduced resistance to BmNPV. Consistent experimental outcomes were obtained at both the cellular and individual levels, indicating that the BmSTAT gene is involved in the immune response of Bombyx mori to BmNPV and plays a significant role during the immune process. However, the interaction mechanism between the JAK/STAT signaling pathway and BmNPV still requires further study.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found in the article/Supplementary material.
Author contributions
WL: Data curation, Investigation, Writing – original draft. LZ: Formal analysis, Methodology, Writing – original draft. ZD: Validation, Visualization, Writing – original draft. SW: Data curation, Writing – original draft. PZ: Resources, Writing – review & editing. QX: Funding acquisition, Supervision, Writing – review & editing. ZL: Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (grant no. 2022YFD1201600), the National Science Foundation of China (32030103 and 32172798). Thanks for the support of the Academy for Advanced Interdisciplinary Studies Device Sharing Service Platform (Equipment No. 18A13430), Southwest University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1485951/full#supplementary-material
References
Dostert, C., Jouanguy, E., Irving, P., Troxler, L., Galiana-Arnoux, D., Hetru, C., et al. (2005). The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat. Immunol. 6, 946–953. doi: 10.1038/ni1237
Durbin, J. E., Hackenmiller, R., Simon, M. C., and Levy, D. E. (1996). Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450. doi: 10.1016/S0092-8674(00)81289-1
Geng, T., Lv, D. D., Huang, Y. X., Hou, C. X., Qin, G. X., and Guo, X. J. (2016). JAK/STAT signaling pathway-mediated immune response in silkworm (Bombyx mori) challenged by Beauveria bassiana. Gene 595, 69–76. doi: 10.1016/j.gene.2016.09.043
Goldsmith, M. R., Shimada, T., and Abe, H. (2005). The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 50, 71–100. doi: 10.1146/annurev.ento.50.071803.130456
Guo, H., Jiang, L., and Xia, Q. (2016). Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol. Gen. Genomics. 291, 999–1004. doi: 10.1007/s00438-015-1125-4
Guo, H., Sun, Q., Wang, B., Wang, Y., Xie, E., Xia, Q., et al. (2019). Spry is downregulated by multiple viruses to elevate ERK signaling and ensure viral reproduction in silkworm. Dev. Comp. Immunol. 98, 1–5. doi: 10.1016/j.dci.2019.04.001
Gupta, L., Molina-Cruz, A., Kumar, S., Rodrigues, J., Dixit, R., Zamora, R. E., et al. (2009). The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe 5, 498–507. doi: 10.1016/j.chom.2009.04.003
Herrera, S. C., and Bach, E. A. (2019). JAK/STAT signaling in stem cells and regeneration: from Drosophila to vertebrates. Development 146:7643. doi: 10.1242/dev.167643
Hombría, J. C., and Brown, S. (2002). The fertile field of Drosophila Jak/STAT signalling. Curr. Biol. 12, R569–R575. doi: 10.1016/S0960-9822(02)01057-6
Hou, X. S., Melnick, M. B., and Perrimon, N. (1996). Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell 84, 411–419. doi: 10.1016/S0092-8674(00)81286-6
Hu, X., Zhang, X., Wang, J., Huang, M., Xue, R., Cao, G., et al. (2015). Transcriptome analysis of BmN cells following over-expression of BmSTAT. Mol. Gen. Genomics. 290, 2137–2146. doi: 10.1007/s00438-015-1065-z
Ihle, J. N. (1996). STATs: signal transducers and activators of transcription. Cell 84, 331–334. doi: 10.1016/s0092-8674(00)81277-5
Iwasaki, A., and Pillai, P. S. (2014). Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14, 315–328. doi: 10.1038/nri3665
Jiang, L. (2021). Insights into the antiviral pathways of the silkworm Bombyx mori. Front. Immunol. 12:639092. doi: 10.3389/fimmu.2021.639092
Jiang, L., Goldsmith, M. R., and Xia, Q. (2021a). Advances in the arms race between silkworm and Baculovirus. Front. Immunol. 12:628151. doi: 10.3389/fimmu.2021.628151
Jiang, L., and Xia, Q. (2014). The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 48, 1–7. doi: 10.1016/j.ibmb.2014.02.003
Jiang, L., Xie, E., Guo, H., Sun, Q., Liuli, H., Wang, Y., et al. (2021b). Heat shock protein 19.9 (Hsp19.9) from Bombyx mori is involved in host protection against viral infection. Dev. Comp. Immunol. 114:103790. doi: 10.1016/j.dci.2020.103790
Kang, X., Wang, Y., Liang, W., Tang, X., Zhang, Y., Wang, L., et al. (2021). Bombyx mori nucleopolyhedrovirus downregulates transcription factor BmFoxO to elevate virus infection. Dev. Comp. Immunol. 116:103904. doi: 10.1016/j.dci.2020.103904
Kemp, C., Mueller, S., Goto, A., Barbier, V., Paro, S., Bonnay, F., et al. (2013). Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 190, 650–658. doi: 10.4049/jimmunol.1102486
Liu, W. J., Chang, Y. S., Wang, A. H., Kou, G. H., and Lo, C. F. (2007). White spot syndrome virus annexes a shrimp STAT to enhance expression of the immediate-early gene ie1. J. Virol. 81, 1461–1471. doi: 10.1128/JVI.01880-06
Liu, L. K., Chen, X. X., Gao, R. L., Wang, K. J., Zheng, W. Y., and Liu, H. P. (2020). A cytokine receptor domeless promotes white spot syndrome virus infection via JAK/STAT signaling pathway in red claw crayfish Cherax quadricarinatus. Dev. Comp. Immunol. 111:103749. doi: 10.1016/j.dci.2020.103749
Liu, W., Liu, J., Lu, Y., Gong, Y., Zhu, M., Chen, F., et al. (2015). Immune signaling pathways activated in response to different pathogenic micro-organisms in Bombyx mori. Mol. Immunol. 65, 391–397. doi: 10.1016/j.molimm.2015.02.018
Morris, R., Kershaw, N. J., and Babon, J. J. (2018). The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 27, 1984–2009. doi: 10.1002/pro.3519
Myllymäki, H., and Rämet, M. (2014). JAK/STAT pathway in Drosophila immunity. Scand. J. Immunol. 79, 377–385. doi: 10.1111/sji.12170
Paradkar, P. N., Trinidad, L., Voysey, R., Duchemin, J. B., and Walker, P. J. (2012). Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. USA 109, 18915–18920. doi: 10.1073/pnas.1205231109
Park, C., Li, S., Cha, E., and Schindler, C. (2000). Immune response in Stat2 knockout mice. Immunity 13, 795–804. doi: 10.1016/S1074-7613(00)00077-7
Schindler, C., and Darnell, J. E. Jr. (1995). Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64, 621–652. doi: 10.1146/annurev.bi.64.070195.003201
Schoggins, J. W., MacDuff, D. A., Imanaka, N., Gainey, M. D., Shrestha, B., Eitson, J. L., et al. (2014). Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695. doi: 10.1038/nature12862
Shang, Q., Wu, P., Huang, H. L., Zhang, S. L., Tang, X. D., and Guo, X. J. (2020). Inhibition of heat shock protein 90 suppresses Bombyx mori nucleopolyhedrovirus replication in B. mori. Insect Mol. Biol. 29, 205–213. doi: 10.1111/imb.12625
Souza-Neto, J. A., Sim, S., and Dimopoulos, G. (2009). An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. USA 106, 17841–17846. doi: 10.1073/pnas.0905006106
Tang, X., Liu, H., Shi, Z., Chen, Q., Kang, X., Wang, Y., et al. (2020). Enhanced silk yield in transgenic silkworm (Bombyx mori) via ectopic expression of BmGT1-L in the posterior silk gland. Insect Mol. Biol. 29, 452–465. doi: 10.1111/imb.12655
Wang, Y., and Levy, D. E. (2006). C. elegans STAT cooperates with DAF-7/TGF-beta signaling to repress dauer formation. Curr. Biol. 16, 89–94. doi: 10.1016/j.cub.2005.11.061
Wang, L., Xiao, Q., Zhou, X. L., Zhu, Y., Dong, Z. Q., Chen, P., et al. (2017). Bombyx mori nuclear Polyhedrosis virus (BmNPV) induces host cell autophagy to benefit infection. Viruses 10:14. doi: 10.3390/v10010014
Wang, S., Zhou, L., Liang, W., Li, S., Zou, Y., Xuan, Q., et al. (2024). STAT transcription factor is indispensable for oogenesis in silkworm. Int. J. Biol. Macromol. 278:133864. doi: 10.1016/j.ijbiomac.2024.134032
West, C., and Silverman, N. (2018). p38b and JAK-STAT signaling protect against invertebrate iridescent virus 6 infection in Drosophila. PLoS Pathog. 14:e1007020. doi: 10.1371/journal.ppat.1007020
Yan, M., Li, C., Su, Z., Liang, Q., Li, H., Liang, S., et al. (2015). Identification of a JAK/STAT pathway receptor domeless from Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 44, 26–32. doi: 10.1016/j.fsi.2015.01.023
Yan, R., Small, S., Desplan, C., Dearolf, C. R., and Darnell, J. E. Jr. (1996). Identification of a Stat gene that functions in Drosophila development. Cell 84, 421–430. doi: 10.1016/S0092-8674(00)81287-8
Yeh, M. S., Cheng, C. H., Chou, C. M., Hsu, Y. L., Chu, C. Y., Chen, G. D., et al. (2008). Expression and characterization of two STAT isoforms from Sf9 cells. Dev. Comp. Immunol. 32, 814–824. doi: 10.1016/j.dci.2007.12.001
Zhang, X., Guo, R., Kumar, D., Ma, H., Liu, J., Hu, X., et al. (2016). Identification, gene expression and immune function of the novel Bm-STAT gene in virus-infected Bombyx mori. Gene 577, 82–88. doi: 10.1016/j.gene.2015.11.027
Zhang, Y., Li, W., Li, L., Li, Y., Fu, R., Zhu, Y., et al. (2015). Structural damage in the C. elegans epidermis causes release of STA-2 and induction of an innate immune response. Immunity 42, 309–320. doi: 10.1016/j.immuni.2015.01.014
Zhang, X. J., Li, D. D., Xu, G. F., Chen, Y. Q., and Zheng, S. C. (2020). Signal transducer and activator of transcription is involved in the expression regulation of ecdysteroid-induced insulin-like growth factor-like peptide in the pupal wing disc of silkworm, Bombyx mori. Insect Sci. 27, 1186–1197. doi: 10.1111/1744-7917.12736
Glossary
Keywords: BmSTAT, BmNPV, transgene, Bombyx mori, immune response
Citation: Liang W, Zhou L, Dang Z, Wang S, Zhao P, Xia Q and Lu Z (2024) Downregulation of BmSTAT transcription factor promoted nucleopolyhedrovirus replication in Bombyx mori. Front. Microbiol. 15:1485951. doi: 10.3389/fmicb.2024.1485951
Edited by:
Tejabhiram Yadavalli, University of Illinois Chicago, United StatesReviewed by:
Guan-Jun Yang, Ningbo University, ChinaQiuning Liu, Yancheng Teachers University, China
Shudi Zhao, Zhejiang University, China
Copyright © 2024 Liang, Zhou, Dang, Wang, Zhao, Xia and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongyan Lu, bHV6aG9uZ3lAc3d1LmVkdS5jbg==
 Wenjuan Liang
Wenjuan Liang Li Zhou1,2
Li Zhou1,2 Ping Zhao
Ping Zhao Qingyou Xia
Qingyou Xia Zhongyan Lu
Zhongyan Lu