- 1National Engineering Research Center for Biotechnology, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing, China
- 2State Key Laboratory of Materials-Oriented Chemical Engineering, College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing, China
- 3Soochow University, Suzhou, China
lysine is an essential amino acid with significant importance, widely used in the food, feed, and pharmaceutical industries. To meet the increasing demand, microbial fermentation has emerged as an effective and sustainable method for L-lysine production. Escherichia coli has become one of the primary microorganisms for industrial L-lysine production due to its rapid growth, ease of genetic manipulation, and high production efficiency. This paper reviews the recent advances in E. coli strain engineering and fermentation process optimization for L-lysine production. Additionally, it discusses potential technological breakthroughs and challenges in E. coli-based L-lysine production, offering directions for future research to support industrial-scale production.
1 Introduction
L-lysine is an essential amino acid that plays a crucial role in the growth and development of both animals and humans, particularly in protein synthesis, bone development, immune function, and hormone production (Matthews, 2020). Since animals cannot synthesize lysine on their own and must obtain it through diet or supplements, L-lysine is especially important as a feed additive in the livestock industry (Bampidis et al., 2022). Additionally, lysine is widely used in the food processing industry and the pharmaceutical sector, such as in the production of health products, medicines, and nutritional supplements (Cheng et al., 2018; Pandey et al., 2020). In 2021, the global lysine market was valued at $6.96 billion, and it is projected to reach $12.38 billion by 2030, with a compound annual growth rate of 6.5% (Leinonen et al., 2019). The continuous growth in global demand for L-lysine is mainly driven by the increasing consumption of meat and the rising demand for high-protein foods (Wang L. et al., 2021). According to market research, the growth of the lysine market is primarily concentrated in the Asia-Pacific, North America, and Europe regions, with particularly strong demand in areas with advanced livestock industries (Parisi et al., 2020). In recent years, lysine production has gradually shifted from traditional chemical synthesis methods to biological fermentation methods to meet market demands for environmental protection and sustainable development (He et al., 2024; Félix et al., 2019).
In industrial production, L-lysine is primarily produced through microbial fermentation (Ikeda, 2017). Microbial fermentation for L-lysine production offers multiple advantages: first, this method utilizes microbial metabolic pathways to efficiently synthesize L-lysine through the fermentation process, resulting in high production efficiency; second, the fermentation method is relatively environmentally friendly, reducing potential pollutants generated during chemical synthesis; third, microbial fermentation can use renewable resources as raw materials, thereby lowering production costs; and finally, through genetic engineering techniques, microbial strains can be further optimized to enhance the yield and purity of L-lysine (Anaya-Reza and Lopez-Arenas, 2017; D'Este et al., 2018). These advantages make microbial fermentation the preferred method for L-lysine production, bringing significant economic benefits while meeting modern industrial requirements for environmental protection and sustainable development.
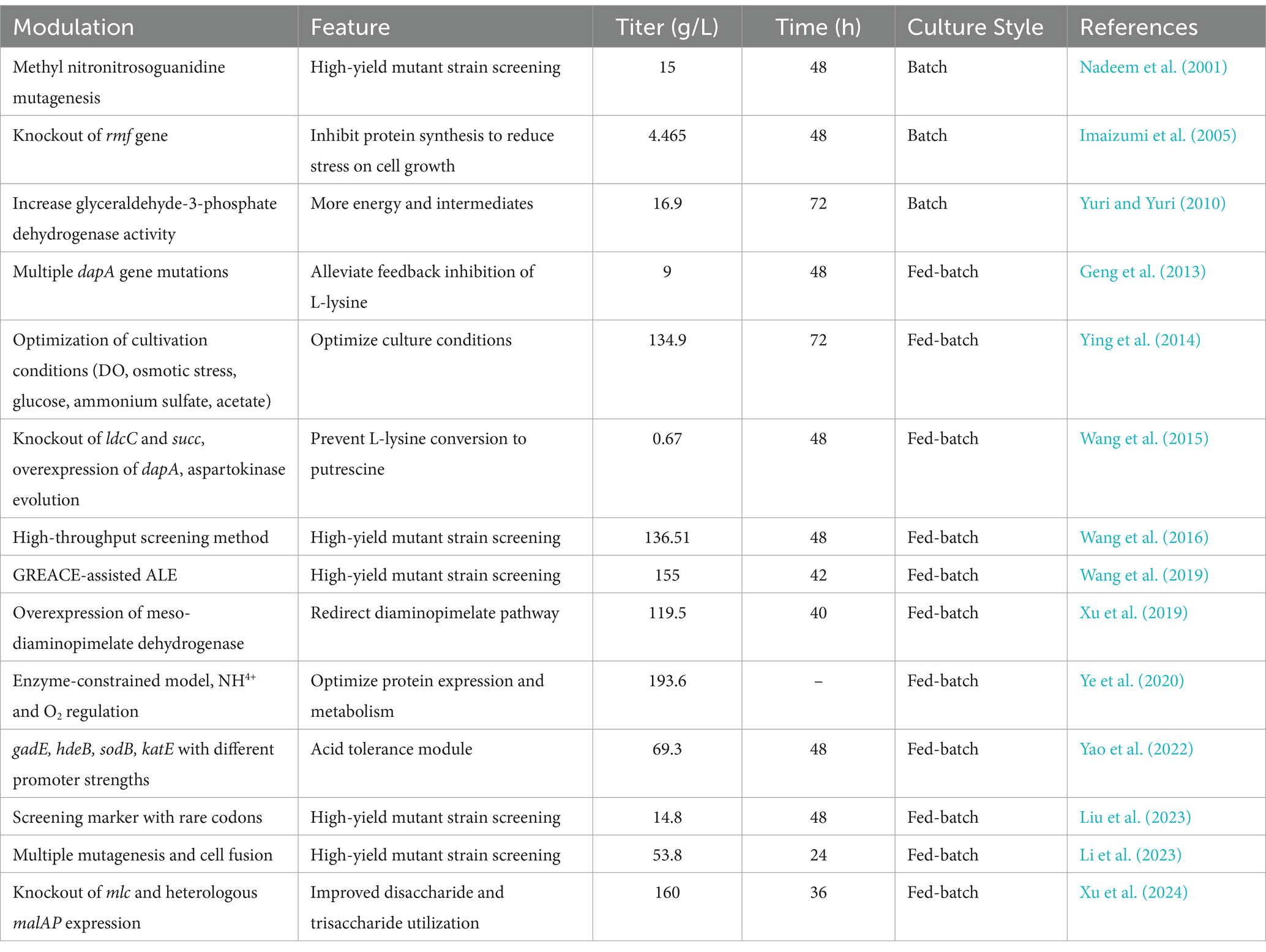
The main strains used in microbial fermentation for L-lysine production include Escherichia coli, Corynebacterium glutamicum, and Brevibacterium flavum. Among these, Escherichia coli has become the dominant strain for L-lysine production due to its well-characterized genetic background, extensive gene editing tools, and short growth cycle (Mir et al., 2024; Chen et al., 2013). The different methods for producing L-lysine in Escherichia coli are shown in Table 1.
In recent years, significant progress has been made in the mutagenesis breeding of L-lysine-producing E. coli (Tadepally, 2019). Through physical, chemical, and biological mutagenesis methods, researchers have been able to induce genetic mutations in E. coli and screen for mutants with high lysine production. UV irradiation and chemical mutagens such as nitrosoguanidine (NTG) are commonly used in mutagenesis breeding (Wang J. et al., 2021; Xu Z. et al., 2016). High-throughput screening can rapidly identify mutant strains with high L-lysine production (Zeng et al., 2020; Kaczmarek and Prather, 2021). With the development of metabolomics, transcriptomics, and other technologies, the efficiency of mutagenesis breeding has continuously improved, allowing researchers to more accurately analyze and screen for mutants with desirable traits. However, due to the randomness of mutagenesis, mutagenesis breeding is often combined with genetic engineering to further optimize strain performance (Chen et al., 2023).
Genetic engineering breeding plays an increasingly important role in the improvement of L-lysine-producing E. coli (Xu et al., 2015). Through gene editing, metabolic engineering, and synthetic biology techniques, researchers can precisely modify the metabolic pathways of E. coli to enhance its lysine production capacity (Liu et al., 2022). Common strategies include the overexpression of key enzyme genes, the knockout of competing metabolic pathways, and the optimization of transcriptional regulatory networks (Dasgupta et al., 2020). By utilizing gene editing technologies such as CRISPR-Cas9, scientists can simultaneously modify multiple genes to achieve global optimization of metabolic networks (Wu et al., 2020). Genetic engineering breeding has already been applied in industrial production, significantly improving lysine production efficiency. In the future, by integrating intelligent methods such as machine learning and metabolic modeling, genetic engineering breeding is expected to further enhance the efficiency and automation of lysine production (Xu et al., 2020).
Optimizing the fermentation process of E. coli for lysine production is a crucial step in improving production efficiency. Current research focuses primarily on the composition of the culture medium, fermentation parameters, and process control (Du Y. H. et al., 2022). By optimizing the carbon-to-nitrogen ratio, trace elements, and vitamins in the culture medium, the growth rate of E. coli and lysine production can be significantly increased (Hao et al., 2020). Additionally, controlling temperature, pH, and dissolved oxygen levels during fermentation can effectively maintain the metabolic activity of E. coli and reduce the generation of by-products (Wang et al., 2018). Furthermore, the application of novel fermentation processes, such as continuous fermentation and fed-batch fermentation, has achieved positive results in industrial production, allowing for extended fermentation times and increased lysine accumulation. In recent years, intelligent fermentation systems have gradually emerged, utilizing sensors and automated control technologies to achieve real-time monitoring and dynamic adjustment of the fermentation process, further enhancing production efficiency and process stability (Egbune et al., 2024; Chen T. et al., 2019). These advancements have laid a significant foundation for the large-scale industrial production of lysine in Escherichia coli.
This paper reviews the research progress on enhancing L-lysine production in Escherichia coli through strain engineering and fermentation optimization. It also highlights newly developed strategies for L-lysine production in E. coli, as well as various value-added chemicals derived from L-lysine. The review provides future directions for improving L-lysine yields through innovations in systems biology and bioprocess engineering.
2 Strain modification
2.1 Traditional mutation breeding
Mutagenesis breeding of L-lysine-producing Escherichia coli began in the mid-20th century when scientists started using physical and chemical mutagens, such as ultraviolet (UV) radiation, X-rays, and N-methyl-N′-nitro-N-nitrosoguanidine (NTG), to induce genetic mutations in E. coli to screen for strains with enhanced L-lysine production (Nakamori, 2017). Early applications of this method successfully developed several industrially valuable mutants, significantly improving L-lysine production capacity. For instance, by mutagenizing E. coli with NTG, researchers were able to screen for L-lysine auxotrophic strains (Lys-), followed by penicillin enrichment to eliminate wild-type strains (Choi et al., 2016). Additionally, UV and ethyl methanesulfonate (EMS) mutagenesis, both individually and in combination, were employed to screen for L-lysine auxotrophic mutants, optimizing mutagenesis conditions (Liu and Jiang, 2015). As technology advanced, the efficiency of mutagenesis breeding continued to improve, especially with the integration of modern high-throughput screening techniques, allowing researchers to more rapidly identify superior mutants. This has enabled the continued application of this traditional breeding method in L-lysine production (Wang et al., 2012).
Currently, research on the mutagenesis breeding of L-lysine-producing E. coli is ongoing, with a focus on enhancing mutagenesis efficiency, screening for superior high-yield mutants, and further optimizing the metabolic pathways of mutants through metabolic flux analysis (Hirasawa and Shimizu, 2016). Although mutagenesis breeding involves randomness and unpredictability, its simplicity, ease of implementation, and low cost make it an important tool in industrial microbial breeding (Li et al., 2022; Sanghavi et al., 2020). In the future, with the advancement of genome editing technologies and systems biology, mutagenesis breeding is expected to combine with precise genetic engineering methods, such as CRISPR-Cas9, to achieve more efficient targeted mutagenesis and further increase L-lysine production. Additionally, the GREACE method (Genomic Replication Engineering Assisted Continuous Evolution) is an evolutionary engineering approach used to enhance microbial tolerance and production capabilities (Lee and Kim, 2020). The core principle of this method involves the introduction of proofreading-deficient mutants of the DNA polymerase complex, which promotes continuous mutations in vivo, thereby accelerating the evolutionary process. In practical applications, the GREACE method employs a “mutation combined with selection” strategy to rapidly screen for strains with superior phenotypes under stress conditions (Zhao et al., 2022; Luan et al., 2013). For instance, in studies aimed at improving L-lysine production, an E. coli strain RS3 modified using the GREACE method achieved a final yield of 155.0 g/L, with a glucose conversion rate of 0.59 g/g and a total output of 605.6 g in a 5-liter fermenter (Wang et al., 2019). This breeding strategy, which integrates traditional and modern technologies, will provide new directions for the industrial application of E. coli in L-lysine production (Nakamori, 2017).
2.2 Genetically engineered breeding
Genetic engineering of strains is a comprehensive process involving the knockout of specific genes, targeted mutagenesis, overexpression of key enzyme genes, and the modification of metabolic pathways. Through precise modifications and optimizations, the lysine production capacity of Escherichia coli can be significantly enhanced, providing highly efficient and stable strain resources for industrial production (Leong et al., 2020).
2.2.1 Biosynthesis pathway of lysine in Escherichia coli
The genomic modification strategies for lysine production in Escherichia coli are primarily designed based on the lysine biosynthesis pathway in E. coli (Figure 1). The synthesis of lysine begins with L-aspartate, which is catalyzed by aspartate aminotransferase (AST) from oxaloacetate. Oxaloacetate is a key intermediate in the tricarboxylic acid (TCA) cycle. Under the catalysis of aspartate kinase (AK), L-aspartate first reacts with ATP to form L-4-asparty phosphate, a crucial initial step in the aspartate pathway. Subsequently, L-4-asparty phosphate undergoes a series of enzymatic reactions to convert into L-aspartate-4-semialdehyde, which is then catalyzed by dihydrodipicolinate synthase (DHDPS) to produce L-2,3-dihydrodipicolinate, a key intermediate in lysine biosynthesis. L-2,3-dihydrodipicolinate is then reduced by dihydrodipicolinate reductase (DHDPR) to tetrahydrodipicolinate. Through a series of enzymatic reactions, tetrahydrodipicolinate is ultimately converted into meso-2,6-diaminopimelate (DAP). This process involves several enzymes, including DAP aminotransferase and DAP decarboxylase. Finally, DAP decarboxylase catalyzes the conversion of meso-2,6-diaminopimelate into L-lysine.
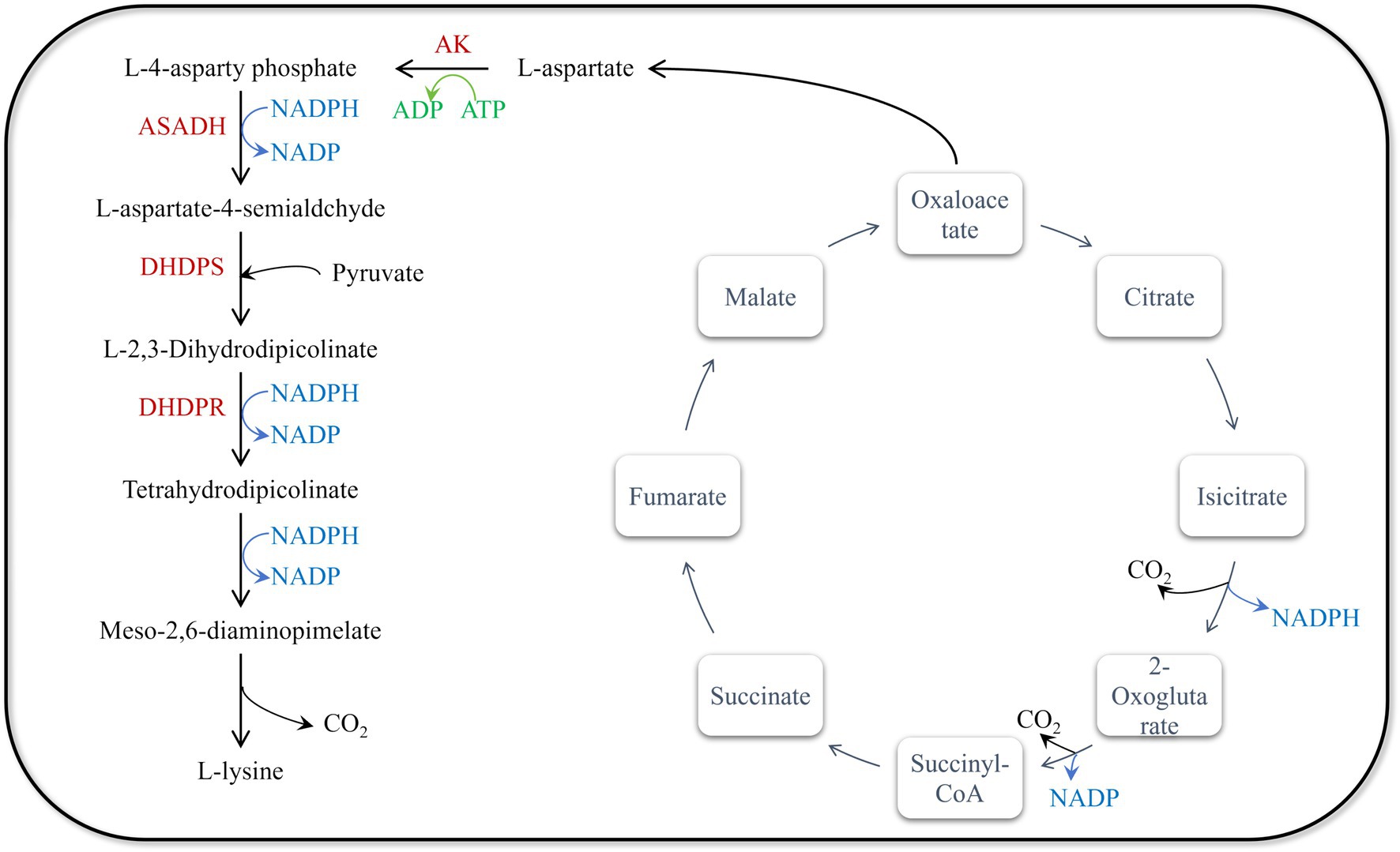
Figure 1. Synthesis pathway of L-lysine in Escherichia coli. There are six enzymes related to L-lysine biosynthesis in E. coli: ASADH (aspartic semialdehyde dehydrogenase, encoded by asd), AK (aspartic kinase, encoded by lysC), DHDPR (dihydropyridine dicarboxylic acid reductase, encoded by dapB), DHDPS (dihydropyridine dicarboxylic acid synthase, encoded by dapA), DAPDC (diaminopimelate decarboxylase, encoded by lysA) and DAPDH (diaminopimelate dehydrogenase, encoded by ddh).
2.2.2 Gene knockout
Gene knockout breeding is a method of increasing the lysine production of Escherichia coli through genetic engineering (Theisen and Liao, 2017). This process typically involves targeted knockout or mutation of certain genes in the Escherichia coli genome to optimize metabolic pathways, reduce byproduct production, and increase lysine production. By knocking out metabolic pathway genes that compete with lysine, such as tyrosine and tryptophan synthesis genes, as well as key enzyme genes involved in lysine degradation pathways, the production of by-products can be reduced, lysine consumption can be reduced, and more carbon can be directed toward lysine synthesis and accumulation can be increased (Ohno et al., 2014). By knocking out or mutating feedback inhibition genes (such as dapA) and enhancing key enzymes in the lysine synthesis pathway (such as LysA), the efficiency of lysine synthesis can be further improved. By reducing the generation of by-products such as acetic acid and lactic acid, optimizing the metabolic network, and introducing new metabolic pathways, lysine production can be further optimized (Liu et al., 2023).
Gene editing techniques, including CRISPR/Cas9 and λ—Red homologous recombination, have been applied to modify the genome of Escherichia coli (Zhao et al., 2016). Composite gene editing methods have also been explored, such as combining lambda Red recombination with homologous endonucleases I-SceI or CRISPR/Cas9. Although CRISPR-Cas9 is commonly used for gene knockout, altering multiple genes remains challenging (Su et al., 2020; Feng et al., 2018). Researchers combined CRISPR-Cas9 and MetClo assembly to design a standardized iterative genome editing system for Escherichia coli. This method simplifies the process of introducing multiple gene modifications, making it more efficient (Dong et al., 2021). These techniques have enabled researchers to study gene function and optimize metabolic pathways to produce high-value products.
2.2.3 Targeted mutagenesis and promoter replacement
Targeted mutagenesis refers to the precise mutation of specific genes in the Escherichia coli genome using gene-editing technologies to enhance lysine biosynthesis or reduce lysine degradation pathways (Simon et al., 2018). By mutating key enzyme genes in the pathway, it is possible to decrease by-product formation or increase the activity of critical enzymes. For example, mutating the gene encoding phosphoenolpyruvate carboxylase (PPC) can reduce pyruvate consumption, thereby increasing the supply of lysine precursors (Shimizu and Matsuoka, 2022; Yin et al., 2024). Alternatively, mutating genes encoding regulatory factors can alleviate feedback inhibition in lysine biosynthesis. For instance, in the L-lysine biosynthesis pathway, targeted mutagenesis of the lysC gene, which encodes aspartokinase, and the dapA gene, which encodes dihydrodipicolinate synthase, can relieve feedback inhibition by L-lysine on its biosynthetic pathway (Li et al., 2023).
Promoter replacement is a method used to regulate the expression of target genes by replacing the promoters of genes in Escherichia coli (Marschall et al., 2017). By replacing the original weak promoter with a strong promoter, the transcription level of the target gene can be increased, thereby enhancing lysine synthesis (Jin et al., 2019). For example, replacing the promoter of the dapA gene with the constitutive tac promoter can achieve stable and efficient gene expression (Ying et al., 2014). Through promoter engineering, promoters that are sensitive to environmental conditions (such as temperature, pH, and nutrients) can be designed, enabling dynamic regulation of lysine synthesis (Wang et al., 2020).
2.2.4 Overexpression of key enzyme genes
Overexpressing key enzyme genes in the lysine biosynthesis pathway can enhance the activity of these enzymes, thereby increasing lysine production (Li et al., 2017). Aspartokinase is a crucial regulatory enzyme in this pathway, whose activity is subject to feedback inhibition by lysine (Lo et al., 2009). By constructing a feedback-resistant mutant of lysC and overexpressing this gene in Escherichia coli, this feedback inhibition can be alleviated, leading to increased lysine production. Dihydrodipicolinate synthase is the rate-limiting enzyme in lysine biosynthesis, and its overexpression can significantly accelerate lysine synthesis. In addition to lysC and dapA, overexpression of genes such as dapB, dapD, and lysA can also enhance the efficiency of lysine synthesis (Zhang et al., 2023). For instance, Xu J. et al. (2016) studied key enzymes in the L-lysine ester synthesis pathway of a high-yield L-lysine strain, including dihydrodipicolinate synthase (DHDPS) and aspartokinase III (AK III). They modified these enzymes to protect them from feedback inhibition and enhance their activity.
2.2.5 Optimization of metabolic pathways
In Escherichia coli, lysine metabolism involves the tricarboxylic acid (TCA) cycle, glycolytic pathway, diaminopimelate pathway, and pentose phosphate pathway (Figure 2). By modulating the carbon flux ratio between the glycolytic and pentose phosphate pathways, the production of NADPH can be increased, which favors lysine biosynthesis (Tsuge and Kondo, 2017; Wendisch, 2017). Additionally, the introduction of a heterologous multifunctional enzyme, which replaces multiple original steps of enzyme-catalyzed reactions, can simplify the lysine biosynthetic pathway, thereby enhancing production efficiency. Furthermore, increasing the concentration of lysine synthesis precursors within the metabolic pathway can effectively boost lysine yield (Banno et al., 2018).
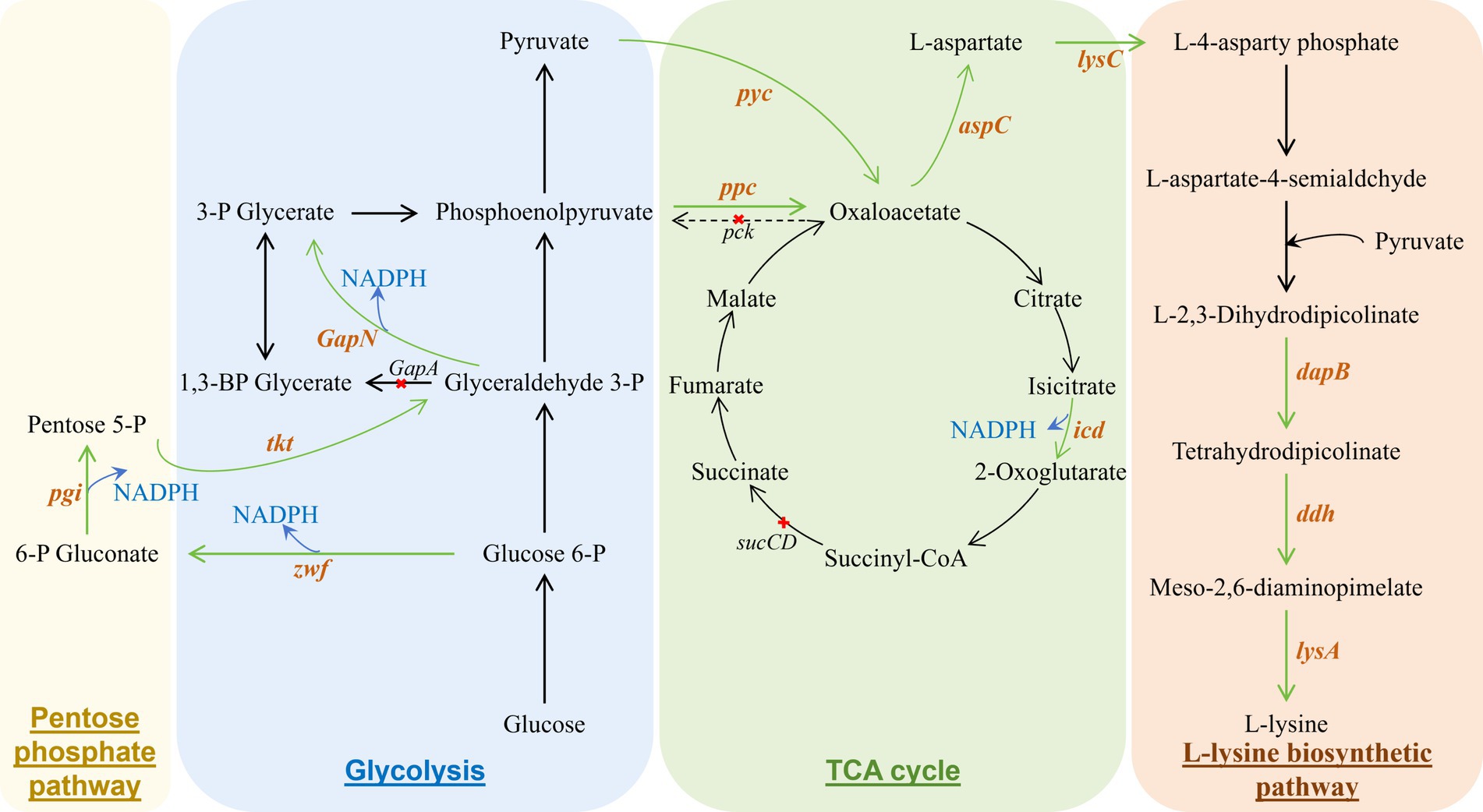
Figure 2. Metabolic engineering modification of Escherichia coli to produce L-lysine. The green arrows indicate the enhancement of the synthetic pathway by overexpressing the relevant enzyme gene; the dashed line with “X” indicates the inactivated response. These genes encode the following enzymes: pgi, glucose-6-phosphate isomerase; zwf, Glucose-6-phosphate 1-dehydrogenase; tkt, Transketolase; pck, Phosphoenolpyruvate carboxykinase; pyc, Pyruvate carboxylase; ppc, Phosphoenolpyruvate caroxylase; icd, Isocitrate dehydrogenase; sucCD, succinyl-CoA synthetase; aspC, Aspartate aminotransferase; ddh, Diaminopimelate dehydroganase; lysA, Diaminopimelate decarboxylase.
2.2.6 Coenzyme balance
In the lysine biosynthesis pathway, NADH and NADPH are crucial cofactors. Optimizing the balance of these cofactors can significantly enhance lysine production (Wang et al., 2017). By introducing or overexpressing NADH oxidase, the accumulation of NADH can be reduced, thereby increasing the availability of NADPH. Researchers can also optimize the NADPH/NADH ratio by modulating the cellular cofactor regeneration systems (Liu et al., 2018). Alternatively, by knocking out or downregulating metabolic pathways that competitively consume NADPH, more NADPH can be directed toward lysine biosynthesis. For example, knocking out certain genes involved in fatty acid synthesis in Escherichia coli can reduce NADPH consumption. Lysine synthesis requires a substantial amount of ATP. Enhancing ATP generation pathways can also improve lysine production. For instance, overexpressing ATP synthase can increase intracellular ATP levels (Gao et al., 2021). Moreover, through metabolic flux analysis (MFA), researchers can quantitatively analyze the intracellular flow of cofactors and strategically adjust metabolic pathways to optimize lysine production.
In summary, genome editing technologies hold great promise for boosting lysine production in E. coli, benefiting industries such as animal feed and biotechnology. Researchers continue to refine these methods to improve strain performance and yield (Chen et al., 2024a; Eggeling et al., 2015).
2.3 Omics analysis and computer simulation
Omics analysis and computational simulation techniques have played a pivotal role in the breeding research of L-lysine-producing Escherichia coli (St. John and Bomble, 2019). These technologies provide a novel perspective for understanding and optimizing the metabolic pathways of E. coli. Through multi-layered Omics analyses, including genomics, transcriptomics, proteomics, and metabolomics, researchers can comprehensively reveal the key genes, regulatory networks, and metabolic bottlenecks that influence L-lysine production in E. coli. This information is crucial for designing more efficient L-lysine-producing strains (Becker and Wittmann, 2018). For instance, metabolomics analysis allows researchers to identify significant intermediate metabolites in the L-lysine production process and their accumulation patterns, thereby enabling gene modifications to enhance production efficiency (Ahmed et al., 2024).
On the other hand, computational simulation techniques, particularly metabolic network modeling and dynamic simulations, have further enhanced the precision of breeding efforts. Using these tools, researchers can predict the potential impacts of metabolic engineering strategies, assess the effectiveness of different genetic modifications, and conduct virtual experiments to reduce blind spots and time costs in actual experiments (Wang L. et al., 2021). In recent years, machine learning-based computational simulation tools have also begun to be applied in optimizing L-lysine-producing strains. These tools analyze vast amounts of Omics data to predict the optimal gene modification targets (Costello and Martin, 2018).
Manipulating single or multiple key genes within a metabolic pathway to increase the production of desired metabolites requires several rounds of experimentation to identify enzymes that impact productivity or yield (Chen et al., 2018). The utility of mathematical tools can effectively accelerate this process. Ensemble modeling (EM) utilizes phenotypic data (such as the impact of enzyme overexpression or knockout on steady-state yield) to select plausible models that can describe the available data, thereby improving L-lysine-producing strains. Recently, a strategy was explored for forming a set of dynamic models that capture the cellular dynamics through phenotypic data screening, followed by consecutive enzyme overexpression, which demonstrated some predictive capacity. Referring to the resulting strain (overexpression of desensitized diaminopimelate synthase dapA), this set of models could predict the desensitization of aspartokinase lysC as a subsequent rate-controlling step in the L-lysine pathway. This work illustrates the usability, applicability, and broad application of an ensemble modeling framework for constructing production strains (Brockman and Prather, 2015). Recently, researchers have also developed an innovative computational strategy that combines flux balance models with statistical methods, proposing a method to redirect flux by analyzing correlations in flux within the metabolic network to optimize product yield (Lee and Kim, 2015).
2.4 Innovative technology
2.4.1 Biosensor
The application of biosensors in breeding lysine-producing Escherichia coli offers a novel, rapid, and sensitive method for screening and optimizing high-efficiency strains. Biosensors typically consist of a receptor element that specifically binds to the target product, such as lysine, and a reporter element that generates a measurable signal (Newman et al., 2011). By integrating these sensors into the genome of E. coli, researchers can monitor lysine concentrations in real time and select high-producing strains based on signal intensity. This approach significantly reduces the cumbersome quantitative detection steps in traditional high-throughput screening processes and greatly improves breeding efficiency. For example, Wang et al. (2016) utilized an L-lysine sensor in E. coli to screen mutant strain MU-1 from an atmospheric and room temperature plasma (ARTP) mutagenesis library, achieving a 9.05% increase in conversion rate and a 21% increase in yield compared to the original strain.
In recent years, significant progress has been made in the field of biosensor breeding for lysine-producing E. coli, driven by advancements in synthetic biology and gene editing technologies. Recent research has focused on developing more precise and multifunctional biosensors. For instance, one type of sensor is based on the transcriptional regulator LysG, which responds to high concentrations of L-lysine by promoting the expression of the lysE gene (Steffen et al., 2016). By fusing the lysE gene with a fluorescent protein gene and using fluorescence-activated cell sorting (FACS) to select cells with strong fluorescence signals, high L-lysine-producing strains can be obtained (Ye et al., 2020).
Another type of sensor is based on the interaction between L-lysine and the L-lysine aptamer riboswitch, which results in a regulatory effect (Shi et al., 2018; Wang et al., 2016). For instance, Yang et al. (2013) used an L-lysine riboswitch to regulate the expression of tetA. When the intracellular concentration of L-lysine is high, the expression of tetA is blocked, rendering the cells sensitive to tetracycline. TetA can transport Ni2+ into the cytoplasm, leading to cell death. If its expression is blocked, Ni2+ intracellular transport is prevented, allowing the cells to grow in a NiCl2 environment. By selecting an appropriate concentration of NiCl2 as a selection pressure, strains with high L-lysine production can be screened. They used this biosensor to optimize the promoter of phosphoenolpyruvate carboxylase (encoded by the ppc gene), which plays a key role in the distribution of metabolic flux in L-lysine production, and identified promoters from a constructed ppc promoter library that favored L-lysine ester production (Wang W. et al., 2022).
Moreover, researchers have developed multiplex biosensor systems capable of detecting multiple metabolites or intermediate products simultaneously, providing a more comprehensive assessment of metabolic pathway optimization (Chu et al., 2022). Another important advancement is the integration of machine learning with biosensors, allowing for the analysis of large datasets to predict and design more efficient sensor systems. The combination of these technologies has greatly enhanced the potential applications of biosensors in the breeding of lysine-producing strains, further advancing the production efficiency of lysine in E. coli.
2.4.2 High-throughput screening
In recent years, the combination of lysine biosensors and fluorescence-activated cell sorting (FACS) has established a classical high-throughput screening strategy (Della Corte et al., 2020). This system enables the screening of cells containing plasmid libraries as well as cells with genomic mutations. For instance, the study by Wang Yan et al. introduced a novel approach for evolving industrial lysine-producing strains using an L-lysine-inducible promoter. Based on intracellular enhanced green fluorescent protein (EGFP) levels controlled by the promoter, fluorescence-activated cell sorting was initially used to screen 186 strains from a library of 10 million mutants derived from a high-lysine-producing Escherichia coli strain. A multiparameter evaluation of lysine concentration, yield, and cell-specific productivity in these strains identified two mutants, MU-1 and MU-2, which achieved lysine yields of 136.51 ± 1.55 g/L and 133.29 ± 1.42 g/L, respectively, in a 5-liter fermenter. Compared to the original strain, the lysine concentration and yield of the MU-1 strain increased by 21.00 and 9.05%, respectively, while those of the MU-2 strain increased by 18.14 and 10.41%, respectively. The mutant selection and evaluation system developed in this study can be applied to continuously enhance lysine strains for industrial production (Fang et al., 2024; Schendzielorz et al., 2014).
Additionally, droplet microfluidics technology allows for high-throughput screening in tiny droplets. Using Mass-Activated Droplet Sorting (MADS), cell variants with high lysine production can be screened based on signal intensity measured by electrospray ionization mass spectrometry (ESI-MS) (Payne et al., 2023). Strategies based on tRNA modification can be used to effectively identify and screen strains with high amino acid yields (Blay et al., 2020). For example, a research group at Beijing Institute of Technology proposed a strategy called AMINO, which screens high-producing amino acid strains based on synthetic tRNA CUA. This strategy utilizes aminoacylation reactions to reduce the affinity of tRNA for aminoacyl-tRNA synthetase by modifying the anticodon of tRNA, thereby screening for strains with high yields of the target amino acid (Guo et al., 2023). Other studies have employed large-scale activated droplet sorting (MADS) technology to screen E. coli mutants growing in microdroplets in a high-throughput manner. This method selects droplets based on signal intensity measured by electrospray ionization mass spectrometry (ESI-MS), enabling screening at a rate of 0.5 samples per second, thus rapidly identifying high-lysine-producing mutants (Payne et al., 2023).
2.4.3 Directed evolution
Directed evolution simulates the natural selection pressures in vitro, allowing Escherichia coli to gradually adapt to high-lysine production environments, thereby selecting mutant strains capable of maintaining high growth and production levels under elevated lysine concentrations (Matsui and Asano, 2015). Recently, Xurong Yao and colleagues designed a synthetic acid-tolerance module to finely regulate the expression of multifactorial gene blocks. Through directed evolution, they developed an acid-responsive asr promoter library, from which four variants were selected for constructing the synthetic module. The laboratory strain Escherichia coli MG1655 was first selected for growth under mildly acidic conditions (pH 5–6), followed by an evaluation of the lysine production performance of the industrial lysine-producing E. coli strain SCEcL3. The results demonstrated that this synthetic biology strategy significantly enhanced the robustness and productivity of both strains in mildly acidic environments (Yao et al., 2022).
2.4.4 Microbial symbiont design
Significant progress has been made in breeding Escherichia coli for lysine production using microbial consortia design. Researchers have combined genetically engineered lysine-assimilating E. coli with glutamate-assimilating Corynebacterium glutamicum to form a synthetic symbiotic consortium. In this alliance, E. coli secretes fructose as a carbon source, which C. glutamicum utilizes for growth and metabolism. Meanwhile, C. glutamicum synthesizes and secretes L-lysine, which is then utilized by E. coli. This mutualistic relationship effectively converts sucrose into L-lysine (Jia et al., 2016). This approach not only increases L-lysine production and reduces costs but also provides environmental sustainability benefits. By leveraging the metabolic capabilities of each organism, the consortium optimizes resource utilization and enhances overall process efficiency. Future research may further refine these microbial communities to expand their applications in biotechnology and industrial processes (Figure 3).
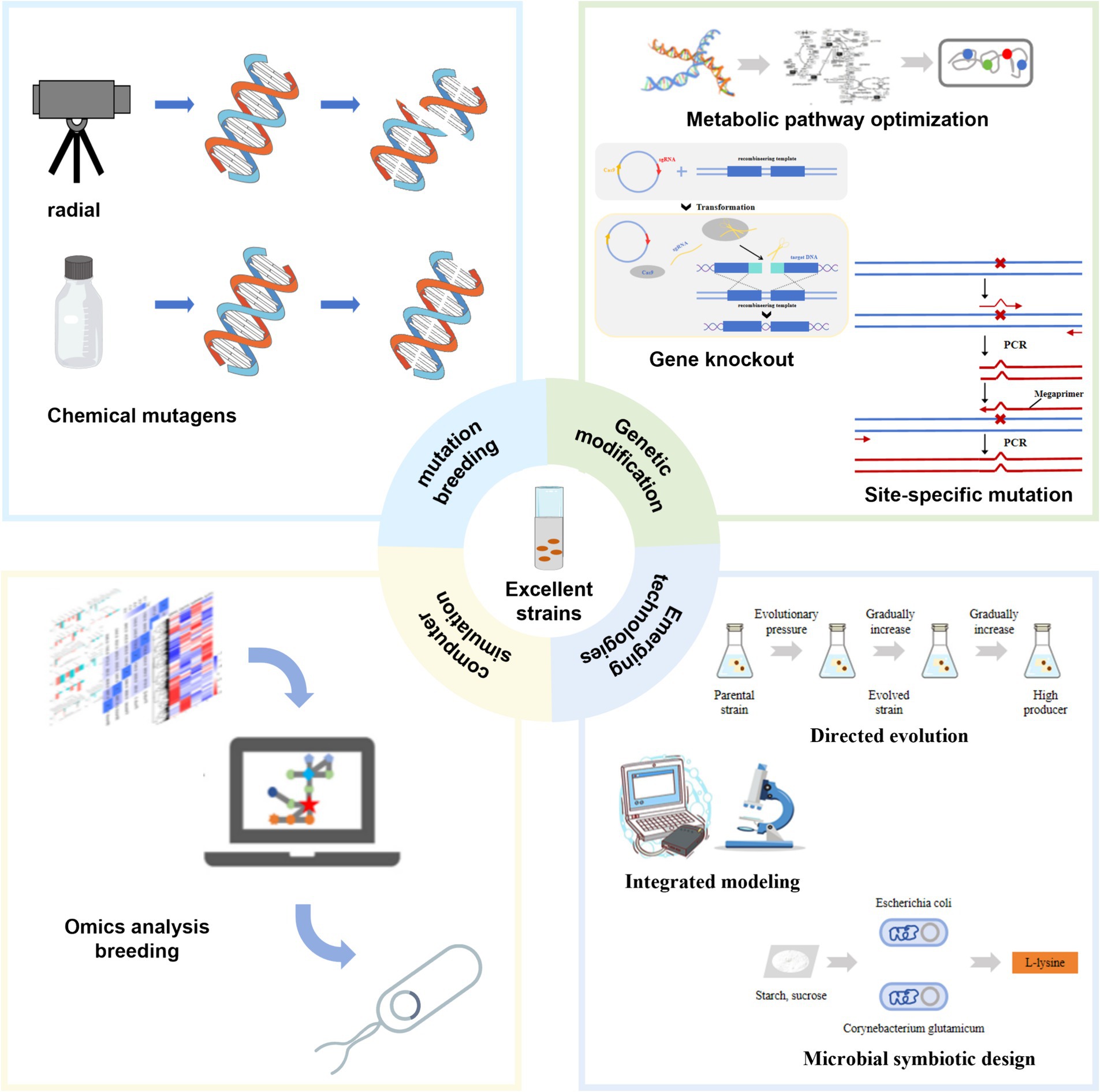
Figure 3. Strain transformation strategy. This figure illustrates various techniques for modifying Escherichia coli strains to increase L-lysine production.
3 Optimization of fermentation process
Constructing or breeding superior L-lysine-producing strains is only the initial stage of industrial production. To fully realize the production potential of these strains, it is necessary to optimize the fermentation process to achieve higher production efficiency and product concentration.
3.1 Cultivation methods
The fermentation methods for L-lysine production by Escherichia coli mainly include continuous fermentation and fed-batch fermentation. In continuous fermentation, the fermentation broth is continuously replenished and removed, maintaining microbial growth in a stable state, thereby extending the production cycle and increasing L-lysine accumulation. This method is suitable for long-term efficient production but requires a high level of system stability, necessitating precise control of fermentation conditions. Researchers have developed intelligent control systems that enable automated process control by real-time monitoring of fermentation parameters such as pH, dissolved oxygen, and glucose concentration, further enhancing production stability and L-lysine yield. Additionally, improvements in the design of novel bioreactors, such as the application of membrane bioreactors, have significantly increased the efficiency of continuous fermentation (Chen et al., 2024b).
Fed-batch fermentation, on the other hand, involves the gradual addition of nutrients during the fermentation process to avoid substrate inhibition effects and the accumulation of by-products. Due to its flexibility and ease of control, fed-batch fermentation is widely used in industrial production. By finely tuning feeding strategies, fed-batch fermentation can significantly improve L-lysine yield and production efficiency (Rajpurohit and Eiteman, 2022). Researchers have explored various feeding strategies, such as tricalcium phosphate, sulfuric acid, and activated carbon pretreatment methods (TPSA), to enhance sugar consumption rates and L-lysine production. Moreover, multi-mode feeding strategies, such as staged feeding and multi-substrate feeding, have been developed to maximize yield and production efficiency (He et al., 2015).
3.2 Fermentation conditions
3.2.1 Fermentation temperature
Fermentation temperature is one of the critical factors affecting lysine production efficiency in Escherichia coli. Temperature directly influences the metabolic activity, enzyme functionality, and growth rate of the cells. During lysine fermentation, a temperature of approximately 37°C is typically chosen, as it is the optimal growth temperature for E. coli, maximizing biomass accumulation and lysine yield. However, excessively high temperatures can intensify cellular stress responses and reduce enzyme activity, leading to a decrease in lysine production. Conversely, lower temperatures may slow metabolic rates but can help reduce by-product formation and extend the fermentation period. Therefore, precise temperature control is crucial for optimizing lysine production (Ma et al., 2021).
Recent advancements have shown that dynamic regulation of fermentation temperature can further enhance lysine production efficiency. For instance, maintaining a higher temperature during the initial phase of fermentation can promote rapid E. coli growth and early product accumulation, while lowering the temperature in the later stages can reduce by-product formation and prolong cell viability, thereby improving the final lysine yield. Additionally, some studies have explored the application of temperature-sensitive regulatory elements, enabling E. coli to automatically adjust its metabolic pathways in response to temperature changes, maximizing lysine synthesis. By utilizing computer simulations and systems biology approaches, researchers can predict cellular metabolic behavior under various temperature conditions, optimizing the fermentation process. The application of these dynamic temperature control strategies and intelligent regulatory methods holds significant promise for advancing the industrial-scale production of lysine.
3.2.2 Dissolved oxygen in fermentation
Dissolved oxygen (DO) plays a crucial role in the fermentation process of L-lysine production by Escherichia coli, influencing cell growth, metabolic activities, and the final yield of L-lysine. E. coli is an aerobic microorganism, and during fermentation, an adequate oxygen supply can promote efficient carbon source metabolism, thereby increasing cell growth rates and product accumulation. However, excessively high DO levels may induce oxidative stress, inhibiting L-lysine synthesis, while insufficient DO can lead to the accumulation of metabolic byproducts and reduced fermentation efficiency. Therefore, maintaining an optimal DO level is essential for the efficient production of L-lysine by E. coli.
The intelligent monitoring and dynamic control of DO during fermentation have become focal points of research. Advanced sensor technology is utilized to monitor DO levels in real time, and in combination with computer control systems, enables dynamic adjustment of DO to meet the changing oxygen demands at different stages of the fermentation process (Zhou et al., 2011). Additionally, research has explored methods such as optimizing agitation speed, aeration rates, and employing microbubble oxygenation techniques to improve oxygen transfer efficiency, ensuring that E. coli obtains sufficient oxygen supply under high-density culture conditions.
3.2.3 Fermentation pH
pH directly influences the physiological state of cells, enzyme activity, and the balance of metabolic pathways. During the initial stages of fermentation, a lower pH favors rapid growth of Escherichia coli; however, as fermentation progresses, the accumulation of L-lysine can lead to acidification of the culture medium. If not controlled, the resulting pH drop may inhibit cellular activity and reduce L-lysine yield. Therefore, maintaining an optimal pH is crucial for achieving efficient L-lysine production. Buffer systems can be employed to stabilize the pH during fermentation and minimize its impact on L-lysine production. For example, adding pH buffers like phosphate or adjusting the initial pH of the medium can provide high buffering capacity and consistent pH levels (Ferreira et al., 2015). The addition of weak bases such as calcium carbonate to neutralize fermentation acids can also help regulate pH. Additionally, the use of ammonia for neutralization is also another popular approach.
3.2.4 Culture medium formula
The composition of the fermentation medium is crucial for enhancing lysine production in Escherichia coli. Traditional fermentation media typically consist of carbon sources, nitrogen sources, inorganic salts, trace elements, and vitamins. Common carbon sources include glucose, sucrose, and corn steep liquor, where the choice and concentration of the carbon source directly affect the growth rate of the microbial cells and lysine accumulation. Nitrogen sources usually involve yeast extract, soybean meal hydrolysate, or inorganic nitrogen (such as ammonium salts) to provide essential amino acids and other nitrogenous compounds necessary for cell growth. Additionally, inorganic salts and trace elements like phosphates, sulfates, calcium, magnesium, and zinc play vital roles in metabolic activities, while vitamins such as biotin and niacin are important for maintaining enzyme activity and promoting metabolism (Wang et al., 2023). Through optimization methods like response surface analysis, the optimal conditions for the medium composition can be systematically determined. In recent years, attention has also been given to non-traditional carbon sources, such as glycerol and cellulosic hydrolysates, as potential medium alternatives. These novel carbon sources not only enhance lysine yield but also offer advantages like reducing production costs and minimizing waste discharge. Furthermore, the development of E. coli specific media that can adapt to high-density fermentation, as well as multifunctional media formulations that integrate pH adjustment and foam control, has become a current research focus.
3.3 Extraction and purification of lysine
A critical factor in production is the separation and purification of lysine. Developing appropriate separation and extraction methods is essential for enhancing product yield and reducing production costs.
3.3.1 Traditional separation methods
3.3.1.1 Extraction methods: reverse micelle extraction and solvent extraction
Reverse micelle extraction and solvent extraction are among the most commonly used methods. Reverse micelle extraction is straightforward, allows for continuous operation, and produces products with high catalytic activity. However, its main limitation is that effective separation requires a very low ion concentration in the solution (Chaurasiya and Hebbar, 2017). Additionally, electrostatic interactions between surfactants and amino acid ions can affect extraction efficiency. In recent years, researchers have made progress in improving the stability and selectivity of reverse micelle systems. For instance, by developing novel gemini surfactants and multifunctional additives, it is possible to enhance the stability of reverse micelles and increase the extraction efficiency of lysine. Moreover, integrating reverse micelle extraction with membrane separation techniques is being explored to improve extraction efficiency and reduce energy consumption.
Solvent extraction leverages the solubility differences of lysine in various solvents, using organic solvents to extract lysine from the fermentation broth. Although solvent extraction is simple and requires relatively basic equipment, organic solvents pose health risks due to their volatility, and the separation efficiency may not be optimal (Sánchez-Velázquez et al., 2023). Recently, the development of environmentally friendly solvents has become a research hotspot in solvent extraction. The use of green solvents, such as ionic liquids and deep eutectic solvents, can reduce environmental pollution and improve lysine extraction efficiency. Additionally, researchers are exploring the combination of solvent extraction with other separation techniques, such as ultrasound-assisted extraction or microwave-assisted extraction, to further enhance extraction efficiency and yield.
3.3.1.2 Acid precipitation
Acid precipitation involves adding acids (such as hydrochloric acid, sulfuric acid, or other organic acids) to the fermentation broth, lowering the pH, and causing lysine to precipitate out of the solution. The isoelectric point of lysine is typically around pH 9.5, and in an acidic environment, lysine molecules become positively charged, forming lysine salts with reduced solubility, leading to precipitation. Acid precipitation is simple to operate and effectively separates lysine in a short time. By studying the effects of different acids (organic and inorganic) and their concentrations on lysine precipitation, researchers have found that selective acid use and pH optimization can improve lysine precipitation efficiency and purity. For example, some studies suggest that using milder organic acids, such as citric acid, can reduce co-precipitation of impurities and increase lysine recovery. Combining acid precipitation with membrane separation can further purify lysine precipitates through membrane retention, thereby enhancing the purity of the final product (Chen et al., 2020). The use of more environmentally friendly acids or acidic solution systems can reduce environmental impact. Additionally, some studies have explored the possibility of recycling and reusing waste acids during the precipitation process to lower production costs and reduce waste liquid discharge. Real-time optimization of lysine precipitation processes can be achieved through online monitoring and feedback control of pH and precipitation conditions, ensuring consistent product quality.
3.3.1.3 Ion exchange resin
Ion exchange resins are porous polymers with functional ionic groups that can separate charged lysine molecules from solutions through reversible ion exchange reactions. Cation exchange resins are typically used in lysine extraction because lysine behaves as a positively charged cation under most fermentation broth pH conditions. Ion exchange resins have high selectivity for lysine and operate under mild conditions. In recent years, researchers have developed resins with larger specific surface areas and higher ion exchange capacities, significantly improving lysine adsorption efficiency. Additionally, resins with special functional groups have been introduced to enhance lysine selective adsorption, reducing co-adsorption of impurities (Chen X. et al., 2019). The application of immobilized resins and multifunctional resins is becoming more common to improve overall process efficiency and stability. Recent studies have explored efficient resin regeneration methods, such as optimizing the composition of regeneration solutions and using auxiliary regeneration techniques (e.g., ultrasound or microwave assistance), to extend the lifespan of resins while maintaining their high efficiency (Zhang et al., 2011; Ecker et al., 2012; Li et al., 2012; Yokoyama et al., 2013).
3.3.1.4 Membrane separation
Membrane separation technology uses membranes with specific pore sizes to separate different components in a solution. Depending on pore size and membrane characteristics, membrane separation technology is primarily divided into microfiltration (MF), ultrafiltration (UF), nanofiltration (NF), and reverse osmosis (RO) (Wang K. et al., 2022). Ultrafiltration and nanofiltration are commonly used in lysine extraction. Membrane fouling is a major bottleneck limiting the development of membrane separation technology. Recently, various anti-fouling membrane technologies have been developed, such as surface coating technology, dynamic membrane technology, and surface functionalization technology. These methods involve introducing hydrophobic, antibacterial, or superwetting coatings on the membrane surface, significantly reducing membrane fouling and improving the stability and efficiency of the separation process. Membrane bioreactors, which combine membrane separation technology with bioreactors, enable integrated operation during fermentation and separation extraction (Du Y. et al., 2022).
3.3.1.5 Electrodialysis
Electrodialysis uses membranes with selective ion exchange functions (cation and anion membranes) to separate charged molecules like lysine from fermentation broth under the influence of a direct current electric field (Doble, 2016). Lysine exists as a positively charged ion in solution and moves through cation exchange membranes under an electric field, while other impurities pass through anion exchange membranes or are retained, thereby concentrating and purifying lysine. Electrodialysis is energy-efficient but requires precise control of operating conditions, such as current density and flow rate. Additionally, electrodialysis membranes are susceptible to fouling and scaling, which can reduce separation efficiency.
3.3.2 Emerging separation technology
3.3.2.1 Ultrasonic-assisted extraction
Ultrasonic-assisted extraction technology utilizes the cavitation effect generated by ultrasound waves propagating in liquid. This effect creates localized high temperatures and pressures, which disrupt the cell wall and increase the permeability of intracellular substances, thereby improving extraction efficiency (Shen et al., 2023). This method, known for its efficiency, energy-saving, and environmental-friendly characteristics, shows great potential in the extraction and separation of lysine produced by Escherichia coli.
3.3.2.2 Enzymatic hydrolysis
Enzymatic hydrolysis refers to the selective degradation of the cell wall or membrane by enzymes, allowing the release of intracellular lysine for extraction and separation (Yi et al., 2023). This method offers high specificity and mild reaction conditions, avoiding the degradation of lysine or the formation of by-products that can occur during chemical extraction.
3.3.2.3 Reverse-phase high-performance liquid chromatography
Reverse-phase high-performance liquid chromatography (RP-HPLC) is a widely used method for the separation, purification, and analysis of biomolecules, including lysine. RP-HPLC separates lysine based on differences in the partition coefficients between the non-polar stationary phase and the polar mobile phase (Zhang et al., 2024). This method is renowned for its high resolution, excellent selectivity, and efficiency, enabling precise separation and quantitative analysis of lysine (Figure 4).
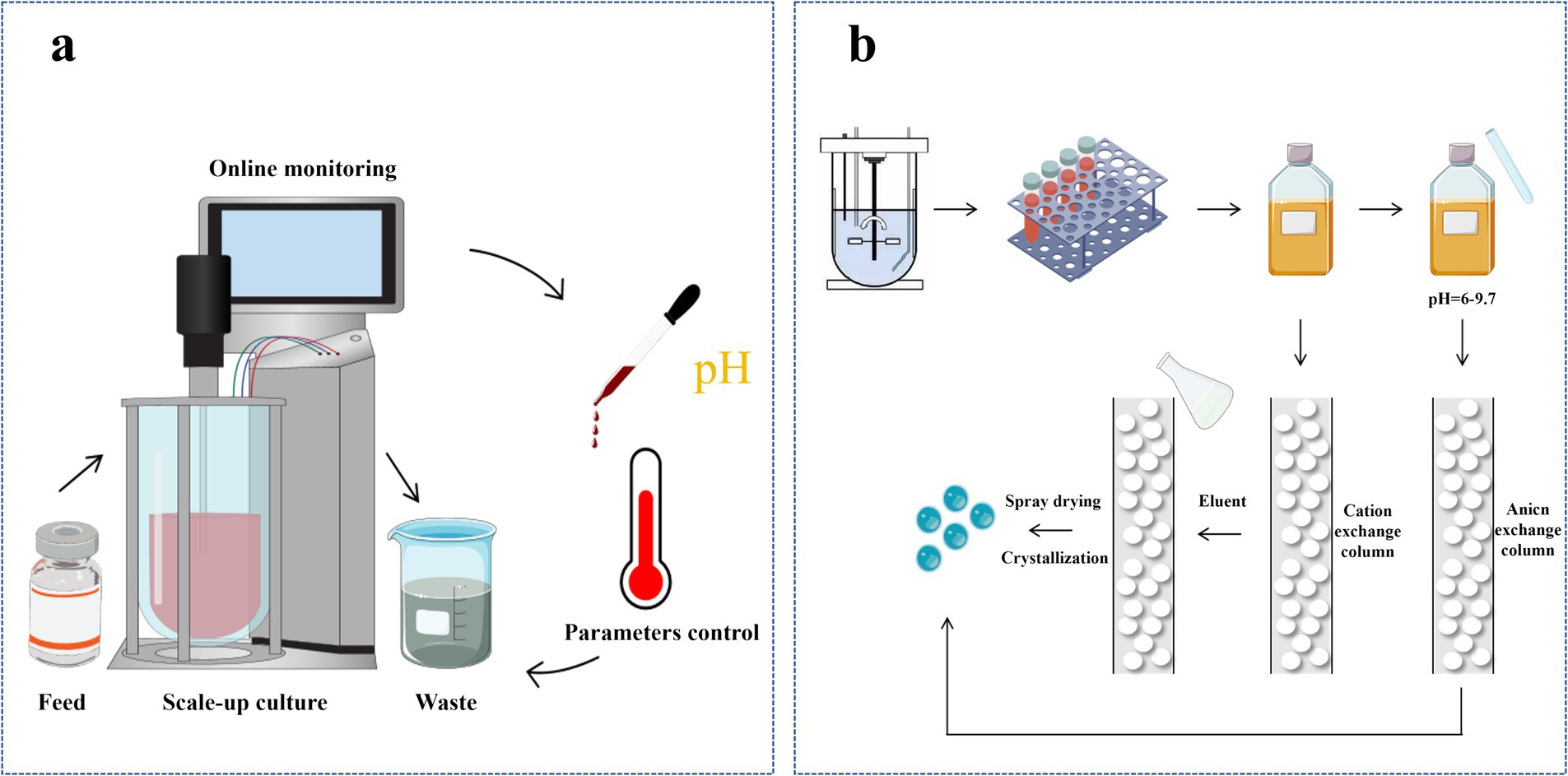
Figure 4. Production of L-lysine. (a) Continuous process of fermentation for L-lysine production; (b) Isolation and refinement of L-lysine fermentation broth.
4 Lysine derivatives
Lysine is widely used as a nutritional supplement and chemical intermediate in the beverage, feed, and food industries. Additionally, lysine holds promise as a precursor for the production of high-value chemicals used in pharmaceuticals, active pharmaceutical ingredients, and materials.
4.1 Lysine derivatives for bio-based materials
Putrescine (1,5-diaminopentane) has multiple uses in both industrial and agricultural fields. Bio-based putrescine is an important platform chemical for bio-based polyamides such as PA 54, PA 510, and PA 512 (Kim et al., 2017). The production of putrescine can be achieved through whole-cell biocatalysis or microbial fermentation, involving recombinant Escherichia coli and Corynebacterium glutamicum (Gevrekci, 2017). Engineered strains have demonstrated increased putrescine yield, with E. coli achieving a maximum yield of 9.61 g/L (Li et al., 2020). Additionally, whole-cell biotransformation is a feasible method for efficiently synthesizing putrescine from L-lysine.
5-Aminovaleric acid (5AVA) is a precursor for the production of nylon 5, nylon 6,5, and other five-carbon derivatives such as valerolactam, glutaric acid, 1,5-pentanediol, and 5-hydroxyvaleric acid. It is a promising platform compound (Liu et al., 2014). These compounds have widespread applications in plastics, solar cells, and artificial antigens. Various biosynthetic pathways have been explored for the production of 5AVA. One approach involves the natural pathway of Pseudomonas putida; another method uses a biosynthesis route mediated by δ-aminovaleramidase (DavA) and Lys 2-monooxygenase (DavB), starting from L-lysine, which is first converted to 5-aminovaleramide and then hydrolyzed to 5AVA. This strategy has been implemented in engineered E. coli, achieving a final yield of up to 57.52 g/L (Jorge et al., 2017). Additionally, a novel pathway involves using Trichoderma reesei L-lysine α-oxidase (LysOx) to oxidize the α-carbon of L-lysine esters to 6-amino-2-ketohexanoic acid, which is then decarboxylated to produce 5AVA without the need for catalase.
δ-Valerolactam is an organic compound typically obtained by cyclization and dehydration of 5-aminovaleric acid (5AVA) under vacuum conditions (Park et al., 2014). These compounds are used as precursors for nylon 5 and nylon 6,5. Recombinant E. coli strains overexpressing Streptomyces acyl-CoA ligase ORF26 produce δ-valerolactam and ε-caprolactam (Zhang et al., 2017). Additionally, an efficient metabolic pathway for synthesizing ε-caprolactam and δ-valerolactam from ω-amino acids was constructed by activating β-alanine CoA transferase (Act) from Clostridium propionicum (Chae et al., 2017). Another innovative L-PA-based approach involves converting L-PA to δ-valerolactam under the catalysis of oxidative decarboxylase.
ε-Caprolactam is the starting material for the synthesis of nylon 6,5 and is also a primary chemical used in the synthesis of nylon 6. Researchers have developed a route for the chemical conversion of biomass-derived lysine into ε-caprolactam (Kumar et al., 2019). This process involves cyclization and subsequent transformation steps to produce α-amino-ε-caprolactam, which can be further utilized in the preparation of nylon 6. Additionally, ε-caprolactam can be produced by cyclization of purified biosynthetic intermediates, a method that holds promise for replacing the conventional process of producing ε-caprolactam from petroleum.
4.2 Lysine-based compounds for pharmaceuticals
L-Pipecolic acid (L-PA) is a crucial non-proteinogenic amino acid widely used as a key intermediate in the synthesis of various pharmaceuticals and biochemical compounds. Currently, L-PA synthesis is primarily achieved through two methods: biosynthesis and chemical synthesis. Due to the cumbersome procedures and low yields associated with chemical synthesis, biosynthesis has garnered significant attention from global chemical companies for its efficiency and environmental benefits. There are four known biosynthetic pathways for L-PA, including the production of L-PA through α-nitrogen loss and ε-nitrogen condensation from lysine, as well as through ε-nitrogen loss with the incorporation of lysine and α-nitrogen. These methods offer economically viable routes for the large-scale production of chiral L-PA.
6-Aminocaproic acid (6ACA) is a widely used non-natural amino acid known for its ability to inhibit the activity of enzymes such as plasmin, pepsin, and elastase, thereby effectively treating certain hemorrhagic conditions (Grottke et al., 2013). Additionally, 6ACA is a crucial component in the production of Nylon 6. Producing 6ACA from biological sources, particularly from lysine, has the potential to significantly reduce refinery emissions and enhance biosecurity. Although various biosynthetic pathways have been explored, further experimental validation of these engineered approaches is necessary.
5 Conclusion
In this review, we have explored various strategies for optimizing Escherichia coli strains and fermentation processes to enhance L-lysine production. Key approaches include genetic engineering of metabolic pathways, modification of regulatory mechanisms, and optimization of fermentation conditions. By targeting bottlenecks in the L-lysine biosynthetic pathway, significant improvements in yield and productivity have been achieved. Additionally, advances in synthetic biology and genome editing technologies, such as CRISPR/Cas9, have opened new avenues for more precise and efficient strain engineering (Pu et al., 2023).
Despite these advancements, there remain several challenges that need to be addressed. One major hurdle is the balance between growth and production, as maximizing L-lysine output often compromises the overall fitness of the microbial cells (Liu and Wang, 2021). Moreover, the complex interplay between metabolic fluxes and regulatory networks in engineered strains can lead to unexpected outcomes, requiring further fine-tuning and iterative testing (Pang et al., 2022). In large-scale industrial production, rising production costs, increasing environmental pollution, and high substrate consumption pose significant challenges. Current separation and purification processes require substantial amounts of acid–base reagents and water, leading to escalating costs and environmental burdens, which result in resource waste and hinder the development of lysine production. Membrane separation technology, with its advantages of energy savings, no phase transition, simplicity of use, scalability, and elimination of chemical reagents, can significantly enhance the separation and purification of L-lysine, thereby facilitating green production for enterprises (Zhang et al., 2019).
Looking forward, future research should focus on integrating systems biology approaches with machine learning and artificial intelligence to predict and design more efficient production strains. Additionally, exploring co-culture systems and dynamic control strategies could provide new opportunities for overcoming limitations associated with single-strain fermentations (Van Lent et al., 2023). With the continuous advancement of metabolic engineering technologies, we can anticipate more innovative approaches to further improve the yield and quality of L-lysine.
Author contributions
ZW: Conceptualization, Investigation, Writing – original draft. TC: Conceptualization, Supervision, Writing – review & editing. WS: Conceptualization, Supervision, Writing – review & editing, Funding acquisition. YC: Conceptualization, Funding acquisition, Writing – review & editing. HY: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key R&D Program of China (2022YFC2105400); the Key R&D project of Ningxia Hui Autonomous Region (2023BEE01011); the National Natural Science Foundation of China (22178176; 22208157); Jiangsu National Synergetic Innovation Center for Advanced Materials (SICAM-XTA2201); the Youth Fund of Natural Science Foundation of Jiangsu Province (BK20220334); the State Key Laboratory of Materials-Oriented Chemical Engineering (SKL-MCE-22A04); Nanjing Carbon Peak and Carbon Neutrality Technology Innovation Plan (202211019).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, Z., Wan, S., Zhang, F., and Zhong, W. (2024). Artificial intelligence for omics data analysis. BMC Methods 1:4. doi: 10.1186/s44330-024-00004-5
Anaya-Reza, O., and Lopez-Arenas, T. (2017). Comprehensive assessment of the L-lysine production process from fermentation of sugarcane molasses. Bioprocess Biosyst. Eng. 40, 1033–1048. doi: 10.1007/s00449-017-1766-2
Bampidis, V., Azimonti, G., de Lourdes Bastos, M., Christensen, H., Dusemund, B., Durjava, M. F., et al. (2022). Safety and efficacy of a feed additive consisting of concentrated liquid l-lysine, l-lysine monohydrochloride and concentrated liquid L-lysine monohydrochloride produced by Escherichia coli NITE BP-02917 for all animal species (Metex NoovistaGo). EFSA J. 20:e07612. doi: 10.2903/j.efsa.2022.7612
Banno, S., Nishida, K., Arazoe, T., Mitsunobu, H., and Kondo, A. (2018). Deaminase-mediated multiplex genome editing in Escherichia coli. Nat. Microbiol. 3, 423–429. doi: 10.1038/s41564-017-0102-6
Becker, J., and Wittmann, C. (2018). From systems biology to metabolically engineered cells—an omics perspective on the development of industrial microbes. Curr. Opin. Microbiol. 45, 180–188. doi: 10.1016/j.mib.2018.06.001
Blay, V., Tolani, B., Ho, S. P., and Arkin, M. R. (2020). High-throughput screening: today’s biochemical and cell-based approaches. Drug Discov. Today 25, 1807–1821. doi: 10.1016/j.drudis.2020.07.024
Brockman, I. M., and Prather, K. L. (2015). Dynamic metabolic engineering: new strategies for developing responsive cell factories. Biotechnol. J. 10, 1360–1369. doi: 10.1002/biot.201400422
Chae, T. U., Ko, Y. S., Hwang, K. S., and Lee, S. Y. (2017). Metabolic engineering of Escherichia coli for the production of four-, five-and six-carbon lactams. Metab. Eng. 41, 82–91. doi: 10.1016/j.ymben.2017.04.001
Chaurasiya, R. S., and Hebbar, H. U. (2017). Reverse micelles for nanoparticle synthesis and biomolecule separation. Nanosci. Food Agricult. 24, 181–211. doi: 10.1007/978-3-319-53112-0_5
Chen, X., Diao, W., Ma, Y., and Mao, Z. (2020). Extraction and purification of ε-poly-l-lysine from fermentation broth using an ethanol/ammonium sulfate aqueous two-phase system combined with ultrafiltration. RSC Adv. 10, 29587–29593. doi: 10.1039/D0RA04245E
Chen, L., Duan, L., Sun, M., Yang, Z., Li, H., Hu, K., et al. (2023). Current trends and insights on EMS mutagenesis application to studies on plant abiotic stress tolerance and development. Front. Plant Sci. 13:1052569. doi: 10.3389/fpls.2022.1052569
Chen, X., Gao, C., Guo, L., Hu, G., Luo, Q., Liu, J., et al. (2018). DCEO biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals. Chem. Rev. 118, 4–72. doi: 10.1021/acs.chemrev.6b00804
Chen, Y., Gustafsson, J., Tafur Rangel, A., Anton, M., Domenzain, I., Kittikunapong, C., et al. (2024a). Reconstruction, simulation and analysis of enzyme-constrained metabolic models using GECKO toolbox 3.0. Nat. Protoc. 19, 629–667. doi: 10.1038/s41596-023-00931-7
Chen, X., Li, Q., He, H., Zhang, J., and Mao, Z. (2019). Effect of ion form of the ion-exchange resin on ε-poly-l-lysine purification from microbial fermentation broth. RSC Adv. 9, 12174–12181. doi: 10.1039/C9RA00493A
Chen, T., Liu, N., Ren, P., Xi, X., Yang, L., Sun, W., et al. (2019). Efficient biofilm-based fermentation strategies for L-threonine production by Escherichia coli. Front. Microbiol. 10:1773. doi: 10.3389/fmicb.2019.01773
Chen, Y., Song, W., Wang, G., Wang, Y., Dong, S., Wu, Y., et al. (2024b). Metabolic engineering of high L-lysine-producing Escherichia coli for de novo production of L-lysine-derived compounds. ACS Synth. Biol. 13, 2948–2959. doi: 10.1021/acssynbio.4c00356
Chen, X., Zhou, L., Tian, K., Kumar, A., Singh, S., Prior, B. A., et al. (2013). Metabolic engineering of Escherichia coli: a sustainable industrial platform for bio-based chemical production. Biotechnol. Adv. 31, 1200–1223. doi: 10.1016/j.biotechadv.2013.02.009
Cheng, J., Chen, P., Song, A., Wang, D., and Wang, Q. (2018). Expanding lysine industry: industrial biomanufacturing of lysine and its derivatives. J. Ind. Microbiol. Biotechnol. 45, 719–734. doi: 10.1007/s10295-018-2030-8
Choi, K. R., Shin, J. H., Cho, J. S., Yang, D., and Lee, S. Y. (2016). Systems metabolic engineering of Escherichia coli. EcoSal Plus 7, 10–1128. doi: 10.1128/ecosalplus.esp-0010-2015
Chu, S. S., Nguyen, H. A., Zhang, J., Tabassum, S., and Cao, H. (2022). Towards multiplexed and multimodal biosensor platforms in real-time monitoring of metabolic disorders. Sensors 22:5200. doi: 10.3390/s22145200
Costello, Z., and Martin, H. G. (2018). A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data. NPJ Syst. Biol. Appl. 4, 19–14. doi: 10.1038/s41540-018-0054-3
Dasgupta, A., Chowdhury, N., and De, R. K. (2020). Metabolic pathway engineering: perspectives and applications. Comput. Methods Prog. Biomed. 192:105436. doi: 10.1016/j.cmpb.2020.105436
Della Corte, D., van Beek, H. L., Syberg, F., Schallmey, M., Tobola, F., Cormann, K. U., et al. (2020). Engineering and application of a biosensor with focused ligand specificity. Nat. Commun. 11:4851. doi: 10.1038/s41467-020-18400-0
D'Este, M., Alvarado-Morales, M., and Angelidaki, I. (2018). Amino acids production focusing on fermentation technologies–a review. Biotechnol. Adv. 36, 14–25. doi: 10.1016/j.biotechadv.2017.09.001
Doble, M. (Ed.) (2016). Principles of downstream techniques in biological and chemical processes : Apple Academic Press.
Dong, H., Cui, Y., and Zhang, D. (2021). CRISPR/Cas technologies and their applications in Escherichia coli. Front. Bioeng. Biotechnol. 9:762676. doi: 10.3389/fbioe.2021.762676
Du, Y., Pramanik, B. K., Zhang, Y., Dumée, L., and Jegatheesan, V. (2022). Recent advances in the theory and application of nanofiltration: a review. Curr. Pollut. Rep. 8, 51–80. doi: 10.1007/s40726-021-00208-1
Du, Y. H., Wang, M. Y., Yang, L. H., Tong, L. L., Guo, D. S., and Ji, X. J. (2022). Optimization and scale-up of fermentation processes driven by models. Bioengineering 9:473. doi: 10.3390/bioengineering9090473
Ecker, J. R., Bickmore, W. A., Barroso, I., Pritchard, J. K., Gilad, Y., and Segal, E. (2012). ENCODE explained. Nature. 489, 52–54. doi: 10.1038/489052a
Egbune, E. O., Ezedom, T., Odeghe, O. B., Orororo, O. C., Egbune, O. U., Ehwarieme, A. D., et al. (2024). Solid-state fermentation production of L-lysine by Corynebacterium glutamicum (ATCC 13032) using agricultural by-products as substrate. World J. Microbiol. Biotechnol. 40:20. doi: 10.1007/s11274-023-03822-x
Eggeling, L., Bott, M., and Marienhagen, J. (2015). Novel screening methods—biosensors. Curr. Opin. Biotechnol. 35, 30–36. doi: 10.1016/j.copbio.2014.12.021
Fang, H., Zhao, J., Zhao, X., Dong, N., Zhao, Y., and Zhang, D. (2024). Standardized iterative genome editing method for Escherichia coli based on CRISPR-Cas9. ACS Synth. Biol. 13, 613–623. doi: 10.1021/acssynbio.3c00585
Félix, F. K. D. C., Letti, L. A. J., de Melo, V., Pereira, G., Bonfim, P. G. B., Soccol, V. T., et al. (2019). L-lysine production improvement: a review of the state of the art and patent landscape focusing on strain development and fermentation technologies. Crit. Rev. Biotechnol. 39, 1031–1055. doi: 10.1080/07388551.2019.1663149
Feng, X., Zhao, D., Zhang, X., Ding, X., and Bi, C. (2018). CRISPR/Cas9 assisted multiplex genome editing technique in Escherichia coli. Biotechnol. J. 13:e1700604. doi: 10.1002/biot.201700604
Ferreira, C. M., Pinto, I. S., Soares, E. V., and Soares, H. M. (2015). (un) suitability of the use of pH buffers in biological, biochemical and environmental studies and their interaction with metal ions–a review. RSC Adv. 5, 30989–31003. doi: 10.1039/C4RA15453C
Gao, H., Tuyishime, P., Zhang, X., Yang, T., Xu, M., and Rao, Z. (2021). Engineering of microbial cells for L-valine production: challenges and opportunities. Microb. Cell Factories 20, 172–116. doi: 10.1186/s12934-021-01665-5
Geng, F., Chen, Z., Zheng, P., Sun, J., and Zeng, A. P. (2013). Exploring the allosteric mechanism of dihydrodipicolinate synthase by reverse engineering of the allosteric inhibitor binding sites and its application for lysine production. Appl. Microbiol. Biotechnol. 97, 1963–1971. doi: 10.1007/s00253-012-4062-8
Gevrekci, A. Ö. (2017). The roles of polyamines in microorganisms. World J. Microbiol. Biotechnol. 33:204. doi: 10.1007/s11274-017-2370-y
Grottke, O., Frietsch, T., Maas, M., Lier, H., and Rossaint, R. (2013). Umgang mit Massivblutungen und assoziierten perioperativen Gerinnungsstörungen. Anaesthesist 62, 213–224. doi: 10.1007/s00101-012-2136-8
Guo, H., Wang, N., Ding, T., Zheng, B., Guo, L., Huang, C., et al. (2023). A tRNA modification-based strategy for identifying amiNo acid overproducers (AMINO). Metab. Eng. 78, 11–25. doi: 10.1016/j.ymben.2023.04.012
Hao, Y., Ma, Q., Liu, X., Fan, X., Men, J., Wu, H., et al. (2020). High-yield production of L-valine in engineered Escherichia coli by a novel two-stage fermentation. Metab. Eng. 62, 198–206. doi: 10.1016/j.ymben.2020.09.007
He, X., Chen, K., Li, Y., Wang, Z., Zhang, H., Qian, J., et al. (2015). Enhanced L-lysine production from pretreated beet molasses by engineered Escherichia coli in fed-batch fermentation. Bioprocess Biosyst. Eng. 38, 1615–1622. doi: 10.1007/s00449-015-1403-x
He, J., Tang, M., Zhong, F., Deng, J., Li, W., Zhang, L., et al. (2024). Current trends and possibilities of typical microbial protein production approaches: a review. Crit. Rev. Biotechnol. 1-18, 1–18. doi: 10.1080/07388551.2024.2332927
Hirasawa, T., and Shimizu, H. (2016). Recent advances in amino acid production by microbial cells. Curr. Opin. Biotechnol. 42, 133–146. doi: 10.1016/j.copbio.2016.04.017
Ikeda, M. (2017). Lysine fermentation: history and genome breeding. Adv. Biochem. Eng. Biotechnol. 159, 73–102. doi: 10.1007/10_2016_27
Imaizumi, A., Takikawa, R., Koseki, C., Usuda, Y., Yasueda, H., Kojima, H., et al. (2005). Improved production of L-lysine by disruption of stationary phase-specific rmf gene in Escherichia coli. J. Biotechnol. 117, 111–118. doi: 10.1016/j.jbiotec.2004.12.014
Jia, X., Liu, C., Song, H., Ding, M., Du, J., Ma, Q., et al. (2016). Design, analysis and application of synthetic microbial consortia. Synth. Syst. Biotechnol. 1, 109–117. doi: 10.1016/j.synbio.2016.02.001
Jin, L. Q., Jin, W. R., Ma, Z. C., Shen, Q., Cai, X., Liu, Z. Q., et al. (2019). Promoter engineering strategies for the overproduction of valuable metabolites in microbes. Appl. Microbiol. Biotechnol. 103, 8725–8736. doi: 10.1007/s00253-019-10172-y
Jorge, J. M., Pérez-García, F., and Wendisch, V. F. (2017). A new metabolic route for the fermentative production of 5-aminovalerate from glucose and alternative carbon sources. Bioresour. Technol. 245, 1701–1709. doi: 10.1016/j.biortech.2017.04.108
Kaczmarek, J. A., and Prather, K. L. (2021). Effective use of biosensors for high-throughput library screening for metabolite production. J. Ind. Microbiol. Biotechnol. 48:kuab049. doi: 10.1093/jimb/kuab049
Kim, J. H., Seo, H. M., Sathiyanarayanan, G., Bhatia, S. K., Song, H. S., Kim, J., et al. (2017). Development of a continuous L-lysine bioconversion system for cadaverine production. J. Ind. Eng. Chem. 46, 44–48. doi: 10.1016/j.jiec.2016.09.038
Kumar, R., Shah, S., Paramita Das, P., Bhagavanbhai, G. G. K., Al Fatesh, A., and Chowdhury, B. (2019). An overview of caprolactam synthesis. Catal. Rev. 61, 516–594. doi: 10.1080/01614940.2019.1650876
Lee, S. Y., and Kim, H. U. (2015). Systems strategies for developing industrial microbial strains. Nat. Biotechnol. 33, 1061–1072. doi: 10.1038/nbt.3365
Lee, S., and Kim, P. (2020). Current status and applications of adaptive laboratory evolution in industrial microorganisms. J. Microbiol. Biotechnol. 30, 793–803. doi: 10.4014/jmb.2003.03072
Leinonen, I., Iannetta, P. P., Rees, R. M., Russell, W., Watson, C., and Barnes, A. P. (2019). Lysine supply is a critical factor in achieving sustainable global protein economy. Front. Sustain. Food Syst. 3:27. doi: 10.3389/fsufs.2019.00027
Leong, Y. K., Chen, C. H., Huang, S. F., Lin, H. Y., Li, S. F., Ng, I. S., et al. (2020). High-level l-lysine bioconversion into cadaverine with enhanced productivity using engineered Escherichia coli whole-cell biocatalyst. Biochem. Eng. J. 157:107547. doi: 10.1016/j.bej.2020.107547
Li, G., Huang, D., Sui, X., Li, S., Huang, B., Zhang, X., et al. (2020). Advances in microbial production of medium-chain dicarboxylic acids for nylon materials. React. Chem. Eng. 5, 221–238. doi: 10.1039/C9RE00338J
Li, N., Lu, J., Wang, Z., Du, P., Li, P., Su, J., et al. (2023). Improving the regeneration rate of deep lethal mutant protoplasts by fusion to promote efficient L-lysine fermentation. BMC Biotechnol. 23:22. doi: 10.1186/s12896-023-00792-8
Li, S., Mao, Y., Zhang, L., Wang, M., Meng, J., Liu, X., et al. (2022). Recent advances in microbial ε-poly-L-lysine fermentation and its diverse applications. Biotechnol. Biofuels Bioprod. 15:65. doi: 10.1186/s13068-022-02166-2
Li, Y., Wei, H., Wang, T., Xu, Q., Zhang, C., Fan, X., et al. (2017). Current status on metabolic engineering for the production of l-aspartate family amino acids and derivatives. Bioresour. Technol. 245, 1588–1602. doi: 10.1016/j.biortech.2017.05.145
Li, J., Zhang, Y., Shen, F., and Yang, Y. (2012). Comparison of magnetic carboxymethyl chitosan nanoparticles and cation exchange resin for the efficient purification of lysine-tagged small ubiquitin-like modifier protease. J. Chromatogr. B. 907, 159–162. doi: 10.1016/j.jchromb.2012.08.029
Liu, W., and Jiang, R. (2015). Combinatorial and high-throughput screening approaches for strain engineering. Appl. Microbiol. Biotechnol. 99, 2093–2104. doi: 10.1007/s00253-015-6400-0
Liu, J., Li, H., Zhao, G., Caiyin, Q., and Qiao, J. (2018). Redox cofactor engineering in industrial microorganisms: strategies, recent applications and future directions. J. Ind. Microbiol. Biotechnol. 45, 313–327. doi: 10.1007/s10295-018-2031-7
Liu, Z., and Wang, Y. (2021). An evolving and flourishing metabolic engineering. Sheng wu gong Cheng xue bao=. Chin. J. Biotechnol. 37, 1494–1509. doi: 10.13345/j.cjb.200729
Liu, Y., Wang, K., Pan, L., and Chen, X. (2022). Improved production of ε-poly-L-lysine in Streptomyces albulus using genome shuffling and its high-yield mechanism analysis. Front. Microbiol. 13:923526. doi: 10.3389/fmicb.2022.923526
Liu, H., Yang, C., Yang, L., Wang, R., Li, P., Du, B., et al. (2023). Screening l-lysine-overproducing Escherichia coli using artificial rare codons and a rare codon-rich marker. Fermentation 9:899. doi: 10.3390/fermentation9100899
Liu, P., Zhang, H., Lv, M., Hu, M., Li, Z., Gao, C., et al. (2014). Enzymatic production of 5-aminovalerate from L-lysine using L-lysine monooxygenase and 5-aminovaleramide amidohydrolase. Sci. Rep. 4:5657. doi: 10.1038/srep05657
Lo, C. C., Bonner, C. A., Xie, G., D'Souza, M., and Jensen, R. A. (2009). Cohesion group approach for evolutionary analysis of aspartokinase, an enzyme that feeds a branched network of many biochemical pathways. Microbiol. Mol. Biol. Rev. 73, 594–651. doi: 10.1128/mmbr.00024-09
Luan, G., Cai, Z., Li, Y., and Ma, Y. (2013). Genome replication engineering assisted continuous evolution (GREACE) to improve microbial tolerance for biofuels production. Biotechnol. Biofuels 6, 1–11. doi: 10.1186/1754-6834-6-137
Ma, Q., Xia, L., Tan, M., Sun, Q., Yang, M., Zhang, Y., et al. (2021). Advances and prospects in metabolic engineering for the production of amino acids. Sheng wu gong Cheng xue bao=. Chin. J. Biotechnol. 37, 1677–1696. doi: 10.13345/j.cjb.200588
Marschall, L., Sagmeister, P., and Herwig, C. (2017). Tunable recombinant protein expression in E. coli: promoter systems and genetic constraints. Appl. Microbiol. Biotechnol. 101, 501–512. doi: 10.1007/s00253-016-8045-z
Matsui, D., and Asano, Y. (2015). Heterologous production of L-lysine ε-oxidase by directed evolution using a fusion reporter method. Biosci. Biotechnol. Biochem. 79, 1473–1480. doi: 10.1080/09168451.2015.1034654
Matthews, D. E. (2020). Review of lysine metabolism with a focus on humans. J. Nutr. 150, 2548S–2555S. doi: 10.1093/jn/nxaa224
Mir, S., Faheem, M., Sial, M. A., Ullah, G., and Leghari, K. A. (2024). “Mutagenesis: exploring genetic diversity of industrial crop plants” in Industrial crop plants (Singapore: springer Nature Singapore), 73–100. doi: 10.1007/978-981-97-1003-4_3
Nadeem, S., Ikram, A., Rana, S. M., Yaqoob, N., Qureshi, M. J., and Shakoori, A. R. (2001). Enhanced l-lysine production by an Escherichia coli mutant warn 30522 after MNNG treatment. Int. J. Agric. Biol. 48:96.
Nakamori, S. (2017). Early history of the breeding of amino acid-producing strains. Adv. Biochem. Eng. Biotechnol. 159, 35–53. doi: 10.1007/10_2016_25
Newman, R. H., Fosbrink, M. D., and Zhang, J. (2011). Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 111, 3614–3666. doi: 10.1021/cr100002u
Ohno, S., Shimizu, H., and Furusawa, C. (2014). FastPros: screening of reaction knockout strategies for metabolic engineering. Bioinformatics 30, 981–987. doi: 10.1093/bioinformatics/btt672
Pandey, A. K., Pandey, K., and Singh, L. K. (2020). “Microbial production and applications of L-lysine” in Innovations in Food Technology: Current Perspectives and Future Goals, vol. 211-229.
Pang, Q., Han, H., Qi, Q., and Wang, Q. (2022). Application and population control strategy of microbial modular co-culture engineering. Sheng wu gong Cheng xue bao=. Chin. J. Biotechnol. 38, 1421–1431. doi: 10.13345/j.cjb.210555
Parisi, G., Tulli, F., Fortina, R., Marino, R., Bani, P., Dalle Zotte, A., et al. (2020). Protein hunger of the feed sector: the alternatives offered by the plant world. Ital. J. Anim. Sci. 19, 1204–1225. doi: 10.1080/1828051X.2020.1827993
Park, S. J., Oh, Y. H., Noh, W., Kim, H. Y., Shin, J. H., Lee, E. G., et al. (2014). High-level conversion of L-lysine into 5-aminovalerate that can be used for nylon 6, 5 synthesis. Biotechnol. J. 9, 1322–1328. doi: 10.1002/biot.201400156
Payne, E. M., Murray, B. E., Penabad, L. I., Abbate, E., and Kennedy, R. T. (2023). Mass-activated droplet sorting for the selection of lysine-producing Escherichia coli. Anal. Chem. 95, 15716–15724. doi: 10.1021/acs.analchem.3c03080
Pu, W., Chen, J., Wang, Y., Zheng, P., and Sun, J. (2023). Advances of development and application amino acid biosensors. Sheng wu gong Cheng xue bao=. Chin. J. Biotechnol. 39, 2485–2501. doi: 10.13345/j.cjb.221003
Rajpurohit, H., and Eiteman, M. A. (2022). Nutrient-limited operational strategies for the microbial production of biochemicals. Microorganisms 10:2226. doi: 10.3390/microorganisms10112226
Sánchez-Velázquez, O. A., Manzanilla-Valdez, M. L., Wang, Y., Mondor, M., and Hernández-Álvarez, A. J. (2023). “Micellar precipitation and reverse micelle extraction of plant proteins” in Green protein processing technologies from plants: Novel extraction and purification methods for product development (Cham: springer International Publishing), 237–263. doi: 10.1007/978-3-031-16968-7_10
Sanghavi, G., Gupta, P., Rajput, M., Oza, T., Trivedi, U., and Singh, N. K. (2020). “Microbial strain engineering” in Engineering of Microbial Biosynthetic Pathways, 11–32.
Schendzielorz, G., Dippong, M., Grunberger, A., Kohlheyer, D., Yoshida, A., Binder, S., et al. (2014). Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS Synth. Biol. 3, 21–29. doi: 10.1021/sb400059y
Shen, L., Pang, S., Zhong, M., Sun, Y., Qayum, A., Liu, Y., et al. (2023). A comprehensive review of ultrasonic assisted extraction (UAE) for bioactive components: principles, advantages, equipment, and combined technologies. Ultrason. Sonochem. 101:106646. doi: 10.1016/j.ultsonch.2023.106646
Shi, S., Ang, E. L., and Zhao, H. (2018). In vivo biosensors: mechanisms, development, and applications. J. Ind. Microbiol. Biotechnol. 45, 491–516. doi: 10.1007/s10295-018-2004-x
Shimizu, K., and Matsuoka, Y. (2022). Feedback regulation and coordination of the main metabolism for bacterial growth and metabolic engineering for amino acid fermentation. Biotechnol. Adv. 55:107887. doi: 10.1016/j.biotechadv.2021.107887
Simon, A. J., Morrow, B. R., and Ellington, A. D. (2018). Retroelement-based genome editing and evolution. ACS Synth. Biol. 7, 2600–2611. doi: 10.1021/acssynbio.8b00273
St. John, P. C., and Bomble, Y. J. (2019). Approaches to computational strain design in the multiomics era. Front. Microbiol. 10:597. doi: 10.3389/fmicb.2019.00597
Steffen, V., Otten, J., Engelmann, S., Radek, A., Limberg, M., Koenig, B. W., et al. (2016). A toolbox of genetically encoded FRET-based biosensors for rapid l-lysine analysis. Sensors 16:1604. doi: 10.3390/s16101604
Su, B., Song, D., and Zhu, H. (2020). Homology-dependent recombination of large synthetic pathways into E. coli genome via λ-red and CRISPR/Cas9 dependent selection methodology. Microb. Cell Factories 19, 108–111. doi: 10.1186/s12934-020-01360-x
Tadepally, H. D. (2019). “Recent advances in the industrial production of l-lysine by Bacteria” in Recent Developments in Applied Microbiology and Biochemistry, 97–106.
Theisen, M., and Liao, J. C. (2017). Industrial biotechnology: Escherichia coli as a host. Indust. Biotechnol. 1, 149–181. doi: 10.1002/9783527807796.ch5
Tsuge, Y., and Kondo, A. (2017). “Production of amino acids (L-glutamic acid and L-lysine) from biomass” in Production of Platform Chemicals from Sustainable Resources, 437–455.
Van Lent, P., Schmitz, J., and Abeel, T. (2023). Simulated design–build–test–learn cycles for consistent comparison of machine learning methods in metabolic engineering. ACS Synth. Biol. 12, 2588–2599. doi: 10.1021/acssynbio.3c00186
Wang, M., Chen, B., Fang, Y., and Tan, T. (2017). Cofactor engineering for more efficient production of chemicals and biofuels. Biotechnol. Adv. 35, 1032–1039. doi: 10.1016/j.biotechadv.2017.09.008
Wang, J., Gao, C., Chen, X., and Liu, L. (2021). Expanding the lysine industry: biotechnological production of L-lysine and its derivatives. Adv. Appl. Microbiol. 115, 1–33. doi: 10.1016/bs.aambs.2021.02.001
Wang, J., Gao, D., Yu, X., Li, W., and Qi, Q. (2015). Evolution of a chimeric aspartate kinase for L-lysine production using a synthetic RNA device. Appl. Microbiol. Biotechnol. 99, 8527–8536. doi: 10.1007/s00253-015-6615-0
Wang, T., Jia, S., Tan, Z., Dai, Y., Song, S., and Wang, G. (2012). Mutagenesis and selective breeding of a high producing ɛ-poly-L-lysine strain. Front. Chem. Sci. Eng. 6, 179–183. doi: 10.1007/s11705-012-1273-6
Wang, X., Li, Q., Sun, C., Cai, Z., Zheng, X., Guo, X., et al. (2019). GREACE-assisted adaptive laboratory evolution in endpoint fermentation broth enhances lysine production by Escherichia coli. Microb. Cell Factories 18, 106–113. doi: 10.1186/s12934-019-1153-6
Wang, Y., Li, Q., Zheng, P., Guo, Y., Wang, L., Zhang, T., et al. (2016). Evolving the L-lysine high-producing strain of Escherichia coli using a newly developed high-throughput screening method. J. Ind. Microbiol. Biotechnol. 43, 1227–1235. doi: 10.1007/s10295-016-1803-1
Wang, J., Lu, X., Ying, H., Ma, W., Xu, S., Wang, X., et al. (2018). A novel process for cadaverine bio-production using a consortium of two engineered Escherichia coli. Front. Microbiol. 9:1312. doi: 10.3389/fmicb.2018.01312
Wang, K., Wang, X., Januszewski, B., Liu, Y., Li, D., Fu, R., et al. (2022). Tailored design of nanofiltration membranes for water treatment based on synthesis–property–performance relationships. Chem. Soc. Rev. 51, 672–719. doi: 10.1039/D0CS01599G
Wang, Y., Wang, H., Wei, L., Li, S., Liu, L., and Wang, X. (2020). Synthetic promoter design in Escherichia coli based on a deep generative network. Nucleic Acids Res. 48, 6403–6412. doi: 10.1093/nar/gkaa325
Wang, J., Wang, S., Zhao, S., Sun, P., Zhang, Z., and Xu, Q. (2023). Productivity enhancement in L-lysine fermentation using oxygen-enhanced bioreactor and oxygen vector. Front. Bioeng. Biotechnol. 11:1181963. doi: 10.3389/fbioe.2023.1181963
Wang, W., Zhang, J., Tao, H., Lv, X., Deng, Y., and Li, X. (2022). E. coli biosensor based on modular GFP and luxI/luxR cyclic amplification circuit for sensitive detection of lysine. Anal. Bioanal. Chem. 414, 8299–8307. doi: 10.1007/s00216-022-04364-1
Wang, L., Zhang, C., Zhang, J., Rao, Z., Xu, X., Mao, Z., et al. (2021). Epsilon-poly-L-lysine: recent advances in biomanufacturing and applications. Front. Bioeng. Biotechnol. 9:748976. doi: 10.3389/fbioe.2021.748976
Wu, Y., Liu, Y., Lv, X., Li, J., Du, G., and Liu, L. (2020). Applications of CRISPR in a microbial cell factory: from genome reconstruction to metabolic network reprogramming. ACS Synth. Biol. 9, 2228–2238. doi: 10.1021/acssynbio.0c00349
Xu, J., Han, M., Ren, X., and Zhang, W. (2016). Modification of aspartokinase III and dihydrodipicolinate synthetase increases the production of L-lysine in Escherichia coli. Biochem. Eng. J. 114, 79–86. doi: 10.1016/j.bej.2016.06.025
Xu, N., Liu, Y., Jiang, H., Liu, J., and Ma, Y. (2020). Combining protein and metabolic engineering to construct efficient microbial cell factories. Curr. Opin. Biotechnol. 66, 27–35. doi: 10.1016/j.copbio.2020.06.001
Xu, J. Z., Ruan, H. Z., Liu, L. M., Wang, L. P., and Zhang, W. G. (2019). Overexpression of thermostable meso-diaminopimelate dehydrogenase to redirect diaminopimelate pathway for increasing L-lysine production in Escherichia coli. Sci. Rep. 9:2423. doi: 10.1038/s41598-018-37974-w
Xu, X., Wang, H., Chen, X., Wu, J., Gao, C., Song, W., et al. (2024). Metabolic engineering of the substrate utilization pathway in Escherichia coli increases L-lysine production. Sheng Wu Gong Cheng Xue Bao 40, 2513–2527. doi: 10.13345/j.cjb.240049
Xu, Z., Xu, Z., Feng, X., Xu, D., Liang, J., and Xu, H. (2016). Recent advances in the biotechnological production of microbial poly (ɛ-L-lysine) and understanding of its biosynthetic mechanism. Appl. Microbiol. Biotechnol. 100, 6619–6630. doi: 10.1007/s00253-016-7677-3
Xu, J., Zhang, J., Guo, Y., and Zhang, W. (2015). Genetically modifying aspartate aminotransferase and aspartate ammonia-lyase affects metabolite accumulation in l-lysine producing strain derived from Corynebacterium glutamicum ATCC13032. J. Mol. Catal. B Enzym. 113, 82–89. doi: 10.1016/j.molcatb.2014.12.015
Yang, J., Seo, S. W., Jang, S., Shin, S. I., Lim, C. H., Roh, T. Y., et al. (2013). Synthetic RNA devices to expedite the evolution of metabolite-producing microbes. Nat. Commun. 4:1413. doi: 10.1038/ncomms2404
Yao, X., Liu, P., Chen, B., Wang, X., Tao, F., Lin, Z., et al. (2022). Synthetic acid stress-tolerance modules improve growth robustness and lysine productivity of industrial Escherichia coli in fermentation at low pH. Microb. Cell Factories 21:68. doi: 10.1186/s12934-022-01795-4
Ye, C., Luo, Q., Guo, L., Gao, C., Xu, N., Zhang, L., et al. (2020). Improving lysine production through construction of an Escherichia coli enzyme-constrained model. Biotechnol. Bioeng. 117, 3533–3544. doi: 10.1002/bit.27485
Yi, Z. H. U., Mingqiang, X. U., Yanna, R. E. N., and Menghao, C. A. I. (2023). Recombinant expression, refolding, and purification of Lysobacter enzymogenes Lys-C in Escherichia coli. J. East China Univ. Sci. Technol. 49, 702–711. doi: 10.14135/j.cnki.1006-3080.20220607001
Yin, L., Zhou, Y., Ding, N., and Fang, Y. (2024). Recent advances in metabolic engineering for the biosynthesis of Phosphoenol pyruvate–oxaloacetate–pyruvate-derived amino acids. Molecules 29:2893. doi: 10.3390/molecules29122893
Ying, H., He, X., Li, Y., Chen, K., and Ouyang, P. (2014). Optimization of culture conditions for enhanced lysine production using engineered Escherichia coli. Appl. Biochem. Biotechnol. 172, 3835–3843. doi: 10.1007/s12010-014-0820-7
Yokoyama, Y., Yokokawa, A., Noguchi, K., and Tanabe, T. (2013). Polyfunctional low-capacity cation-exchange packing material for the separation of underivatized amino acids. Talanta. 103, 245–251. doi: 10.1016/j.talanta.2012.10.039
Zeng, W., Guo, L., Xu, S., Chen, J., and Zhou, J. (2020). High-throughput screening technology in industrial biotechnology. Trends Biotechnol. 38, 888–906. doi: 10.1016/j.tibtech.2020.01.001
Zhang, J., Barajas, J. F., Burdu, M., Wang, G., Baidoo, E. E., and Keasling, J. D. (2017). Application of an acyl-CoA ligase from Streptomyces aizunensis for lactam biosynthesis. ACS Synth. Biol. 6, 884–890. doi: 10.1021/acssynbio.6b00372
Zhang, Y., Chen, Y., Yue, M., and Ji, W. (2011). Recovery of L-lysine from L-lysine monohydrochloride by ion substitution using ion-exchange membrane. Desalination. 271, 163–168. doi: 10.1016/j.desal.2010.12.016
Zhang, Z., Chu, R., Wei, W., Song, W., Ye, C., Chen, X., et al. (2024). Systems engineering of Escherichia coli for high-level glutarate production from glucose. Nat. Commun. 15:1032. doi: 10.1038/s41467-024-45448-z
Zhang, J., Luo, J., Ren, Z., Zhang, H., Chen, X., and Wan, Y. (2019). Advanced applications of membrane technology in biological detection. Sheng wu gong Cheng xue bao=. Chin. J. Biotechnol. 35, 2257–2268. doi: 10.13345/j.cjb.190229
Zhang, C. Y., Yang, H., Zhu, D. J., Liu, T. Y., Wang, L., Zhang, H. J., et al. (2023). Effect of overexpression of key genes in L-lysine biosynthesis pathway and ε-poly-L-lysine synthase gene on ε-poly-L-lysine production. Food Ferment. Indust. 49, 1–9. doi: 10.13995/j.cnki.11-1802/ts.031337
Zhao, M., Gao, M., Xiong, L., Liu, Y., Tao, X., Gao, B., et al. (2022). CRISPR-Cas assisted shotgun mutagenesis method for evolutionary genome engineering. ACS Synth. Biol. 11, 1958–1970. doi: 10.1021/acssynbio.2c00112
Zhao, D., Yuan, S., Xiong, B., Sun, H., Ye, L., Li, J., et al. (2016). Development of a fast and easy method for Escherichia coli genome editing with CRISPR/Cas9. Microb. Cell Factories 15, 205–209. doi: 10.1186/s12934-016-0605-5
Keywords: L-lysine, Escherichia coli , genome modification, fermentation optimization, separation and purification
Citation: Wu Z, Chen T, Sun W, Chen Y and Ying H (2024) Optimizing Escherichia coli strains and fermentation processes for enhanced L-lysine production: a review. Front. Microbiol. 15:1485624. doi: 10.3389/fmicb.2024.1485624
Edited by:
Mariusz Cycoń, Medical University of Silesia, PolandReviewed by:
Aamir Rasool, University of Balochistan, PakistanMeijie Li, Qingdao Agricultural University, China
Copyright © 2024 Wu, Chen, Sun, Chen and Ying. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjun Sun, c3Vud2VuanVuQG5qdGVjaC5lZHUuY24=; Yong Chen, Y2hlbnlvbmcxOTgyQG5qdGVjaC5lZHUuY24=
 Zijuan Wu1,2
Zijuan Wu1,2 Tianpeng Chen
Tianpeng Chen Yong Chen
Yong Chen Hanjie Ying
Hanjie Ying