- 1Bioplastics Innovation Hub, Food Futures Institute, Murdoch University, Murdoch, WA, Australia
- 2School of Medical, Molecular, and Forensic Sciences, Murdoch University, Murdoch, WA, Australia
- 3Department of Biology, University of Waterloo, Waterloo, ON, Canada
- 4SoilsWest, Centre for Sustainable Farming Systems, Food Futures Institute, Murdoch University, Murdoch, WA, Australia
- 5CSIRO Environment, Black Mountain Science and Innovation Park, Canberra, ACT, Australia
This review examines the role of Pseudomonas spp. bacteria as biocontrol agents against crop diseases, focusing on their mechanisms of action, efficacy, and potential applications in sustainable agriculture. Pseudomonas spp., ubiquitous in soil ecosystems and root microbiomes, have attracted attention for their ability to suppress phytopathogens and enhance plant health through various mechanisms. These include direct competition for nutrients, production of antimicrobial compounds and volatile organic compounds, competition using type VI secretion systems, and indirect induction of systemic resistance. Our review shows that Pseudomonas strains effectively control a wide range of diseases across diverse plant species, with some strains demonstrating efficacy comparable to chemical fungicides. However, the review also highlights challenges in achieving consistent performance when using Pseudomonas inoculants under field conditions due to various biotic and abiotic factors. Strategies to optimize biocontrol potential, such as formulation techniques, application methods, and integration with other management practices, are discussed. The advantages of Pseudomonas-based biocontrol for sustainable agriculture include reduced reliance on chemical pesticides, enhanced crop productivity, and improved environmental sustainability. Future research directions should focus on understanding the complex interactions within the plant microbiome, optimizing delivery systems, and addressing regulatory hurdles for commercial deployment. This review underscores the significant potential of Pseudomonas spp. in sustainable crop protection while acknowledging the need for further research to fully harness their capabilities in agricultural systems.
1 Introduction
The UN’s Sustainable Development Goals, released in 2015, outline 17 urgent objectives to be achieved through global collaboration. Among these, Goal 2 aims to end hunger, achieve worldwide food security, improve nutrition, and promote sustainable agriculture. As the world’s population increases, the demand for food production also rises. This increases the pressure on agricultural systems globally (United Nations, 2015). One important development that will help address these issues is the growing popularity in some countries of plant-based diets as an alternative to traditional meat and dairy consumption.
Chemical fertilizers play a crucial role in meeting the increasing demand for plant-based food by maximizing crop yields (Bhatti et al., 2017; Hera, 1995; Pahalvi et al., 2021). The three primary nutrients in commercial fertilizers, nitrogen (N), phosphate (P), and potassium (K) are used extensively in modern agriculture (McGuire, 2015). However, these fertilizers can also disrupt natural soil processes, leading to reduced water retention, imbalanced soil fertility, declined the agricultural soil quality with the reduction in soil organic matter (Dar et al., 2016; Dinesh et al., 2010; Ongley et al., 2010).
Similarly, the widespread use of pesticides to combat crop diseases poses significant environmental and health risks. Many pesticides are toxic to humans, animals, and non-target organisms, including essential pollinators (Brevik and Burgess, 2012; Geiger et al., 2010; Lee and Choi, 2020; Sponsler et al., 2019). The ecological impacts of many pesticides extend to soil and water systems. These chemicals disrupt microbial communities and reduce soil fertility (Law et al., 2017; Pimentel et al., 1993; Viaene et al., 2016). The loss of beneficial microbial species in the agricultural soil can exacerbate pathogen invasions and compromise ecosystem resilience (Jacobsen and Hjelmsø, 2014; Meena et al., 2020). Since microbial communities are the primary drivers of soil nutrient cycling, due to their various metabolic activities, their relevance in moderating ecosystem function cannot be understated (Balser et al., 2002).
In response to these challenges, there is a growing imperative to explore sustainable alternatives to conventional agricultural practices (Vasilescu et al., 2023). In this regard, microbiological tools, such as biofertilizers and biocontrol agents, have emerged as promising solutions. Biofertilizers, containing beneficial microorganisms, enhance soil fertility and promote plant growth through natural processes (Verma et al., 2019). Biocontrol agents offer non-chemical methods for managing plant diseases by leveraging the antagonistic properties of microorganisms against pathogens (Bhardwaj et al., 2014; Bonaterra et al., 2022; Parani and Saha, 2012).
The bacterial genus Pseudomonas, characterized by its metabolic diversity and abundance in various environments, has garnered particular attention for its potential applications in agriculture (Palleroni, 2015). Members of this genus show considerable metabolic and genetic diversity (Peix et al., 2009); its various secondary metabolites are known to form virulence factors, pigments, and biofilms (Drenkard and Ausubel, 2002; Moissenet and Khedher, 2011; Raio and Puopolo, 2021; Stover et al., 2000). A large number of Pseudomonas strains are known for plant growth-promoting potential by producing various substances such as siderophores, 1-aminocyclopropane-1-carboxylate (ACC) deaminase and lipopeptides (Leontidou et al., 2020; Pršić and Ongena, 2020). Using microbes such as Pseudomonas offers a promising approach to sustainable farming, offering solutions that support plant health while minimizing environmental impacts (Hamid et al., 2021; Khatoon et al., 2020; Nikel et al., 2014; Sharma and Archana, 2016).
Here, we provide a comprehensive overview of Pseudomonas spp. as microbiological tools for sustainable agriculture, examining their status and potential applications in crop production, particularly in cereal crops. We discuss the mechanisms by which Pseudomonas spp. act as biocontrol agents, including direct inhibition of phytopathogens and induction of systemic resistance in plants. Additionally, we explore approaches for optimizing the efficacy of Pseudomonas-based biocontrol strategies, such as improved delivery methods and genetic engineering. Finally, we address the challenges and future directions in harnessing Pseudomonas for sustainable crop protection, emphasizing the importance of ongoing research in advancing environmentally friendly agricultural practices.
2 Diversity and plant interactions of Pseudomonas species
Key interactions between plants and microbes occur in the soil surrounding plant roots (Berg et al., 2014; Gaiero et al., 2013; Jain et al., 2020). The rhizosphere, the immediate soil layer influenced by root exudates, is a hotspot for microbial colonization, fostering diverse communities of bacteria, fungi, and other microorganisms (Berg et al., 2014; Gaiero et al., 2013; Jain et al., 2020). Pseudomonas spp. are often major components of the rhizosphere microbiome and engage in multifaceted interactions with plants, exerting significant influence on plant health, nutrient cycling, and ecosystem functioning (Botelho and Mendonça-Hagler, 2006; Chaudhary et al., 2021; Raio and Puopolo, 2021).
Pseudomonas spp. display remarkable adaptability to diverse environmental conditions, thriving in various soil types and agricultural settings (Palleroni, 2015). Their metabolic versatility and genetic plasticity enable them to colonize plant roots and establish intricate symbiotic relationships with their host plants (Palleroni, 2015; Peix et al., 2009). Many studies have demonstrated the enrichment of Pseudomonas spp. in the roots and rhizosphere of diverse plants such as Arabidopsis thaliana, potatoes, rice, wheat, and barley (Andreote et al., 2009; Buddrus-Schiemann et al., 2010; Jain et al., 2020; Lawongsa et al., 2008; Persello-Cartieaux et al., 2001; Raio and Puopolo, 2021; Wang et al., 2012). The adaptability of Pseudomonas spp. to different plant environments might have evolved due to selective pressures, leading to the development of metabolic pathways conducive to nutrient acquisition from plant-derived compounds (Imperato et al., 2019; Rainey, 1999).
At the molecular level, plant-Pseudomonas interactions are orchestrated through intricate signaling pathways and molecular dialogs (Girard et al., 2020; Preston, 2004). Root exudates, comprising a diverse array of largely low molecular weight, organic compounds serve as chemoattractants, guiding Pseudomonas migration toward the rhizosphere (Drigo et al., 2009; Jain et al., 2020; Marilley et al., 1999; Waldon et al., 1989; Wang et al., 2017). Upon encountering plant roots, Pseudomonas spp. employ chemotaxis and quorum sensing systems to modulate their behavior and adapt to changing environmental cues (Loh et al., 2002; Schikora et al., 2016).
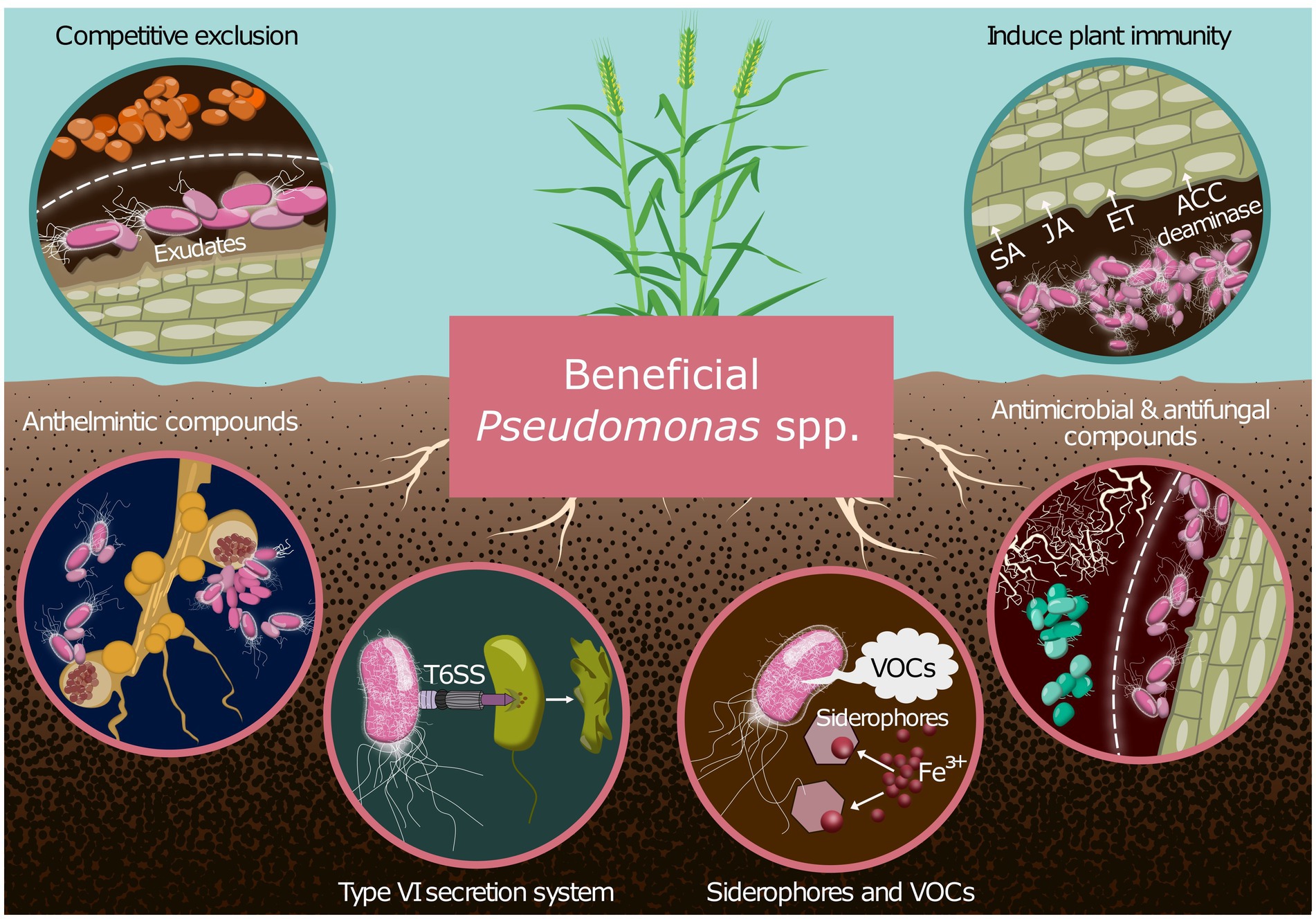
Pseudomonas species actively participate in nutrient cycling processes, facilitating the uptake and assimilation of essential nutrients by plants (Botelho and Mendonça-Hagler, 2006; Chaudhary et al., 2021; Raio and Puopolo, 2021). Through the production of plant growth-promoting substances, such as phytohormones and siderophores (Leontidou et al., 2020), Pseudomonas species stimulate root growth, enhance nutrient acquisition, and confer resistance to environmental stresses (Raio and Puopolo, 2021; Sun et al., 2022). Moreover, Pseudomonas spp. produce a wide range of antimicrobial secondary metabolites. This diversity enables them to outcompete other microorganisms for niche space and carbon resources provided by plants (Figure 1) (Raio and Puopolo, 2021; Validov et al., 2005).
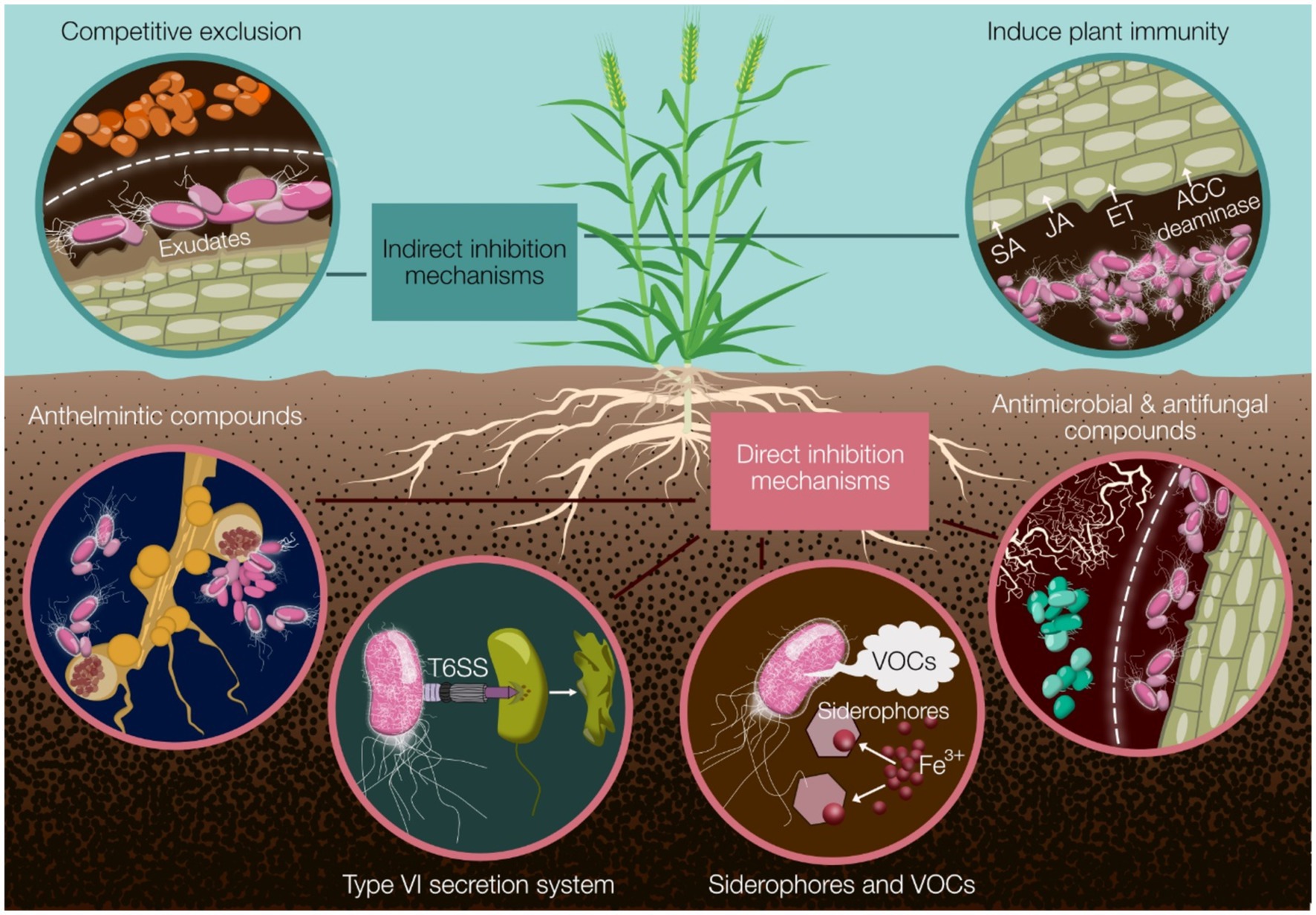
Figure 1. A summary of the major mechanisms by which Pseudomonas biocontrol strains can protect cereal crops from disease. Indirect mechanisms include competitive exclusion and induction of plant immunity through phytohormone modulation (SA, JA, ET) and ACC deaminase activity. Direct mechanisms encompass the production of anthelmintic, antimicrobial, and antifungal compounds; deployment of the type VI secretion system (T6SS); and secretion of siderophores and volatile organic compounds (VOCs). These diverse strategies collectively contribute to effective disease suppression and improved crop health. SA, salicylic acid; JA, jasmonic acid; ET, ethylene; ACC, 1-aminocyclopropane-1-carboxylate; T6SS, type VI secretion system; VOCs, volatile organic compounds.
In addition to promoting plant growth and nutrient acquisition, Pseudomonas spp. play a pivotal role in suppressing plant pathogens and mitigating disease incidence (Hernández-León et al., 2015; Lahlali et al., 2022; Validov et al., 2005; Vivekananthan et al., 2004). Through the production of antimicrobial compounds, lytic enzymes, and volatile organic compounds, Pseudomonas spp. inhibit the proliferation of phytopathogens and confer protection to their host plants (Hernández-León et al., 2015; Mehmood et al., 2023; Raio and Puopolo, 2021; Validov et al., 2005; Vivekananthan et al., 2004). Furthermore, Pseudomonas-mediated induction of systemic resistance within host plants primes the plants for enhanced defense responses, bolstering their resilience against pathogen attacks (Haney et al., 2018; Leeman et al., 1995).
These attributes position Pseudomonas spp. as promising candidates for biocontrol agents in agriculture, as they not only contribute to plant protection but also promote plant growth under various environmental conditions, including high salinity and drought (Cheng et al., 2007; Forni et al., 2017; Orozco-Mosqueda et al., 2019; Rangarajan et al., 2003). However, it is essential to acknowledge that while many Pseudomonas strains act as beneficial or passive colonizers of the plant microbiome, certain species may exhibit phytopathogenic traits (Leontidou et al., 2020; Montes-Osuna et al., 2022; Pršić and Ongena, 2020; Young, 1991). One well-documented example is Pseudomonas syringae, known for causing various plant diseases by producing virulence factors such as coronafacic acid, ice nucleation protein, and the type III secretion system (T3SS) and its associated type III secreted effector (T3SE) proteins (Alattas et al., 2023; Bundalovic-Torma et al., 2022; Büttner, 2016; Weiler et al., 1994; Xin et al., 2018).
3 Pseudomonas as a plant pathogen
P. syringae is a highly diverse bacterial species complex known for its ability to cause disease in a wide range of plant hosts. The complex is currently divided into at least 13 phylogenetic groups or phylogroups, with 7 considered primary phylogroups that contain most of the recognized pathogenic strains (Dillon et al., 2021). Although strains are often classified into pathovars based on the host they were isolated from, there is not always a strong correlation between phylogeny and host specificity. Many closely related strains can infect different hosts, while strains isolated from the same host may be phylogenetically diverse (Baltrus et al., 2017; Morris et al., 2019).
Key factors in P. syringae pathogenicity are the T3SS and T3SE proteins. The T3SS protein allows the bacteria to inject effector proteins directly into plant cells, where they can manipulate host processes to promote infection (Büttner, 2016). P. syringae strains typically possess 20–30 different T3SE proteins, with over 70 distinct T3SE families identified across the species complex (Dillon et al., 2019). However, only a small number of core effectors are conserved across most strains. The diversity and composition of effector repertoires plays a major role in determining host range and virulence capabilities (Laflamme et al., 2020).
T3SE proteins have a range of virulence functions in plants, including suppressing immune responses, altering hormone signaling, disrupting cellular processes, and creating favorable conditions for bacterial growth (Khan et al., 2018). Some effectors, like HopZ1a, demonstrate remarkable functional diversity, targeting multiple unrelated plant proteins to suppress immunity, alter phytohormone signaling, and disrupt microtubule integrity (Jiang et al., 2013; Lee et al., 2012; Rufián et al., 2021). However, plants have evolved immune receptors capable of recognizing many effectors, triggering strong defense responses called effector-triggered immunity (ETI) (Jones and Dangl, 2006). This creates an evolutionary arms race, with bacteria evolving new or modified effectors to evade detection, and plants evolving new receptors.
Interestingly, recent research has found that a relatively small number of plant immune receptors are capable of recognizing effectors from a wide range of P. syringae strains, providing broad-spectrum resistance. For example, in Arabidopsis thaliana, the ZAR1 and CAR1 receptors can detect effectors from 95% of P. syringae strains (Laflamme et al., 2020). The ZAR1 receptor in particular shows a remarkably broad recognition profile, capable of detecting at least six unrelated T3SE families through association with multiple kinase proteins (Martel et al., 2020).
The evolution and diversity of P. syringae effector repertoires is shaped by several key processes (Dillon et al., 2019). Horizontal gene transfer allows effectors to move between strains, potentially expanding host range. Gene loss or pseudogenization can occur when effectors are recognized by plant defenses. Mutational changes in effector sequences may alter their function or allow evasion of host recognition (Dillon et al., 2019). Some effectors show functional redundancy, allowing loss of one effector to be compensated for by others (Kvitko et al., 2009). Additionally, some effectors can suppress the immune responses triggered by other effectors, a phenomenon known as meta effector interactions (Wei et al., 2018).
The importance of environmental factors in P. syringae infections should not be overlooked. Temperature, humidity, and leaf wetness play crucial roles in disease development. For example, optimal conditions for infection include temperatures around 15–25°C and periods of high humidity or leaf wetness (Xin et al., 2018). These environmental factors influence both bacterial growth and the plant’s immune responses. P. syringae is not limited to agricultural settings, it has also been isolated from various environmental sources including aquatic habitats, rain, and wild plants (Morris et al., 2013). This environmental ubiquity may contribute to the emergence of new plant diseases as strains adapt to new hosts or environmental conditions change.
Despite the existence of pathogenic strains, the vast majority of isolated and characterized Pseudomonas strains are beneficial or benign (Palleroni, 2015; Peix et al., 2009). Only a fraction of Pseudomonas strains, out of the numerous species identified, are pathogenic (Palleroni, 2015), highlighting the potential for screening diverse strains to identify those suitable for biocontrol applications. Below, we examine the diverse mechanisms through which Pseudomonas spp. contribute to the suppression of cereal crop diseases. Additionally, we explore the practical applications of these strains in agriculture and discuss the factors influencing their competitiveness and efficacy as biocontrol agents.
4 Control of plant diseases in cereal crops
Pseudomonas spp. offer significant potential for managing plant diseases in cereal crops. For instance, when rice seeds undergo treatment with Pseudomonas fluorescens PF1 before sowing, at the time of sowing, and again at 30 days after sowing, the resulting seedlings display enhanced resistance to Xanthomonas oryzae pv. pryzae (Vidhyasekaran et al., 2001). This treatment led to a significant decrease in disease incidence from 6.8 to 1.2%. In both greenhouse and field conditions, P. fluorescens strains PF1 and FP7 exhibit the ability to inhibit the mycelial growth of the sheath blight fungus Rhizoctonia solani. This inhibition contributes to improved seedling vigor and increased yield in rice plants (Nandakumar et al., 2001). Moreover, the treatment of rice cv. IR50 with the same Pseudomonas strains induces systematic resistance against R. solani, accompanied by an increase in chitinase and peroxidase activity (Nandakumar et al., 2001).
Field trials conducted with rice across different seasons evaluate the efficacy of P. fluorescens strain Pf-1 in controlling Hirschmanniella gracilis, root endoparasitic nematodes that are common in aquatic environments. The seed treatment with this biocontrol strain leads to a high level of bacterial colonization, suppression of nematodes, and a 13% increase in rice yield (Ramakrishnan et al., 1998).
In the case of wheat (Triticum aestivum L.), studies have shown that two Pseudomonas strains, GRP3 and PRS9, play a role in promoting wheat growth in terms of root-shoot length and plant weight (Sharma et al., 2011). P. fluorescens strain PSR21, when applied as a wheat seed treatment and later as a foliar spray during the spring, results in a significant decrease in the average degree of culm damage (where the culm is the above-ground stem of a grass or sedge) (Kita et al., 2004). Further research has highlighted the efficacy of P. fluorescens in reducing disease incidence, such as that caused by the fungal pathogen Helminthosporium sativum in wheat. Interestingly, following treatment, the bacterial population tends to decrease toward the root tip in wheat plants (Srivastava et al., 1999; Wang et al., 1999). Moreover, a recent study has investigated the effectiveness of eight Pseudomonas strains as biocontrol agents (Clough et al., 2022). Among these, Pseudomonas protegens CHA0 was identified as the most potent strain, significantly inhibiting the growth of Ralstonia solanacearum, a pathogen causing bacterial wilt in plants. This inhibitory activity was linked to the production of key secondary metabolites, including orfamides, pyoluteorin, and 2,4-diacetylphloroglucinol (DAPG) (Clough et al., 2022).
Pseudomonas spp. hold immense potential for combating plant diseases in cereal crops, offering multifaceted strategies for disease management. From rice to wheat, the application of various Pseudomonas strains has shown remarkable results, including reduced disease incidence, enhanced seedling vigor, and increased crop yield. These findings underscore the importance of developing microbial-based solutions in agriculture to mitigate the impact of plant pathogens on crop production.
5 Direct inhibition of phytopathogens
5.1 Antimicrobial and antifungal compounds
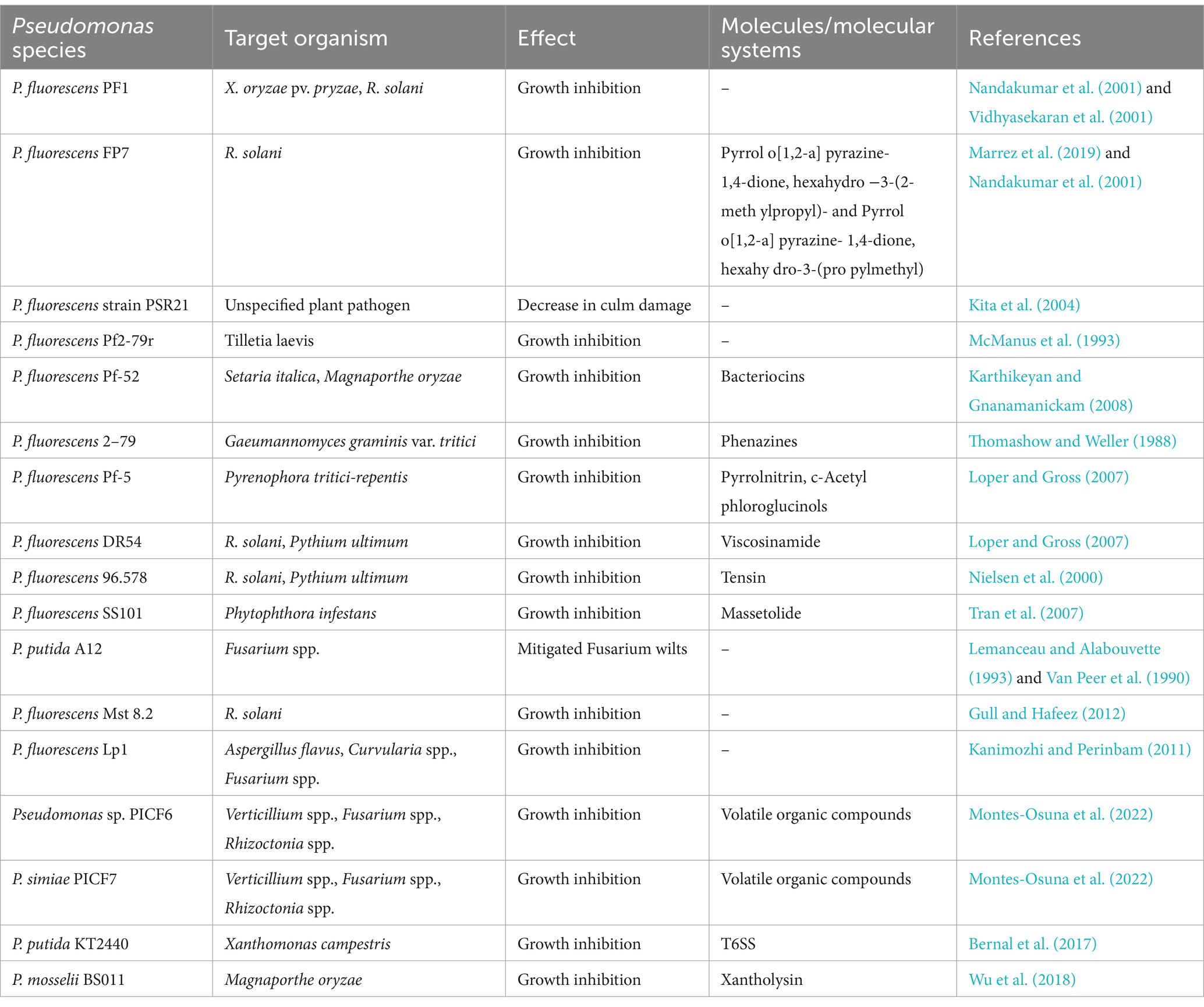
In the pursuit of novel biocontrol agents, Pseudomonas spp. have garnered considerable attention for their potential to inhibit some of the most damaging cereal crop pathogens. Efforts to isolate Pseudomonas strains from diverse environments have revealed promising candidates capable of combating phytopathogens (Chaudhary et al., 2021; Lv et al., 2023). Several studies have identified Pseudomonas species with the ability to inhibit numerous cereal crop pathogens, including Magnaporthe oryzae (causing rice blast), Gaeumannomyces graminis var. tritici (a fungus responsible for wheat take-all disease), Fusarium spp. (associated with head blight, root rot, wilt, and grain contamination), Pyrenophora tritici-repentis (a fungal pathogen that is the cause of tan spot disease), and Rhizoctania solani (a wide-host-range soil-borne pathogen) (Garbeva et al., 2004; Karthikeyan and Gnanamanickam, 2008; Lemanceau and Alabouvette, 1993; Rangarajan et al., 2003; Thomashow and Weller, 1988) (Table 1). These findings mark the initial steps in the quest to identify effective biocontrol agents, demonstrating the ability of isolates to confer plant protection in vivo, through greenhouse experiments and field trials.
Several greenhouse and growth chamber studies have explored the use of bioactive compounds purified from cultures of Pseudomonas species (Table 1) (Ligon et al., 2000; Wu et al., 2018). For instance, metabolites from Pseudomonas mosselii BS011 exhibited broad-spectrum inhibition against phytopathogenic fungi including M. oryzae (Wu et al., 2018). Importantly, Pseudomonas strains have demonstrated equal inhibition of pathogens compared to standard chemical fungicides (Ligon et al., 2000). For example, culture filtrates of P. fluorescens strain BL915 exhibited equal efficacy against R. solani compared to quintozene, a conventional chemical fungicide (Ligon et al., 2000).
Pseudomonas spp. Metabolites multiple compounds which contribute to their antagonistic properties. For example, the antagonistic properties of P. protegens FD6 were examined against pathogenic fungi such as Botrytis cinerea and Monilinia fructicola (Zhang et al., 2020). Genomic analysis identified 12 gene clusters responsible for the production of key secondary metabolites, including 2,4-diacetylphloroglucinol (2,4-DAPG), pyoluteorin (PLT), and pyrrolnitrin (PRN), all essential for its antifungal activities. Mutant analysis revealed that the pltD mutant, lacking PLT production, exhibited only 30% inhibition of grey mold disease, while the phlC mutant, deficient in 2,4-DAPG production but with a marked increase in PLT, demonstrated 65% inhibition. In contrast, the wild-type FD6 strain showed complete suppression of the pathogen, with no visible disease lesions on tomato fruits after 5 days of treatment. These findings underscore the critical roles of PLT and 2,4-DAPG, with PLT being particularly crucial for the high-level inhibition of fungal pathogens. The inverse relationship between the production of these two metabolites highlights the complexity of the biocontrol mechanisms in P. protegens FD6 (Zhang et al., 2020).
For wheat take-all disease, Pseudomonas spp. have been used as potential alternatives to conventional chemical treatments. Pseudomonas species offer resilience under unfavorable conditions and exhibit promising colonization abilities with cereal crop roots (Al Zadjali et al., 2023; Capper and Higgins, 1993; Dowling and O'Gara, 1994; Thomashow and Weller, 1988; Xu et al., 2021). Thus, the exploration of Pseudomonas-based biocontrol agents holds significant promise in augmenting sustainable agricultural practices, heralding a new era in crop protection strategies.
5.2 Siderophores
Siderophores constitute a group of secondary metabolites synthesized by various bacteria, fungi, yeast, and specific plants in response to iron scarcity (Dwivedi and Johri, 2003). These low molecular weight molecules, typically ranging from 500–1,500 Daltons, exhibit a strong binding affinity for iron (III) (Hider and Kong, 2010). The term “siderophore” is derived from the Greek, meaning “iron carrier” (Neilands, 1995); the first siderophore was isolated during 1949–1952 (Hider and Kong, 2010). The first crystalline form of siderophore was isolated by Neilands (Neilands, 1952), sparking research into these iron-chelating compounds (Chincholkar et al., 2000). Kloepper et al. (1980) provided initial evidence of siderophores from plant growth-promoting bacteria (PGPB) acting as biocontrol agents. The ability of an organism to produce siderophores is closely related to cyanide production, and their absence can impact the biocontrol activity of the microbes and their ability to restrict iron access to target pathogens (Ho et al., 2021; Jha et al., 2011). Iron plays an important role in the life of nearly all living organisms (Krewulak and Vogel, 2008).
Plants rely heavily on iron for various vital processes such as photosynthesis, oxygen metabolism, DNA and RNA synthesis, and more (Aznar et al., 2015; Rout and Sahoo, 2015). However, in many natural environments with aerobic conditions and neutral pH levels, iron exists mainly in its oxidized ferric (Fe3+) state, which is largely insoluble and limits its availability to plants (Colombo et al., 2014). The siderophores act as high-affinity chelating agents to solubilize ferric ions and transport them to the plant or bacterial cell where it is converted to Fe2+ (Kramer et al., 2020). Moreover, restricting the supply of iron to pathogens acts as a strategy to hinder their growth, owing to the insolubility of ferric iron forms (Kramer et al., 2020; Lau et al., 2016).
Siderophores can take up the iron available in the surrounding environment, and subsequently, iron is acquired by the organisms possessing receptors for that siderophore-iron complex, making it unavailable to their competitors (Hakim et al., 2021). Besides being used as iron carriers, bacterial siderophore-iron complexes can also be utilized by certain plants aiding in their better growth (Pathak et al., 2017). Among the Gram-negative bacteria where over 90% of the bacterial population produces siderophores, the most dominant genera are Enterobacter and Pseudomonas (Tian et al., 2009). Among Pseudomonas species, pyoverdine is the predominant siderophore observed (Cornelis and Matthijs, 2002; Ghssein and Ezzeddine, 2022).
Siderophores primarily contribute to the survival and fitness of microbial populations, directly and indirectly enhancing the activity of biocontrol species (Gull and Hafeez, 2012; Kanimozhi and Perinbam, 2011). For example, siderophores from P. fluorescens strain Mst 8.2 were shown to be a potent inhibitor against R. solani, with 70% disease reduction in wheat (Gull and Hafeez, 2012). Similarly, siderophores from P. fluorescens Lp1 have been used to inhibit plant fungal pathogens such as Aspergillus flavus, Curvularia spp., and Fusarium spp. (Kanimozhi and Perinbam, 2011).
5.3 Volatile organic compounds
Pseudomonas species are known to produce a wide array of volatile organic compounds (VOCs) with various functions and applications in agricultural settings (Hernández-León et al., 2015). These VOCs are small molecules characterized by low molecular weights and high vapor pressures, allowing them to diffuse effectively through soil pores and influence microbial communities and plant health (Wheatley, 2002).
Pseudomonas strains can generate complex mixtures of VOCs, although the full extent of their functional diversity is still being elucidated (Schmidt et al., 2015). Some VOCs produced by Pseudomonas spp. exhibit antimicrobial properties against phytopathogens, making them potential candidates for biocontrol applications. For instance, studies have identified Pseudomonas-derived VOCs with antifungal activity against a range of plant pathogens, including Verticillium spp., Fusarium spp., and Rhizoctonia spp. (Cordero et al., 2014; Elkahoui et al., 2015; Montes-Osuna et al., 2022). Pseudomonas sp. PICF6 produces 20 VOCs, that include compounds with reported antifungal (e.g., 1-undecene, (methyldisulfanyl) methane and 1-decene) or plant growth promoting (e.g., tridecane, 1-decene) activities (Montes-Osuna et al., 2022). These findings suggest that Pseudomonas-derived VOCs could serve as biofumigants to mitigate the proliferation of pathogenic species and promote plant health.
Further studies have investigated the antifungal activity of VOCs produced by P. fluorescens ZX against postharvest fungal pathogens (Wang et al., 2021; Yue et al., 2023). The research demonstrates that P. fluorescens ZX VOCs effectively inhibit the growth of both Penicillium italicum and Botrytis cinerea, causative agents of blue mold in citrus and gray mold in various fruits, respectively (Wang et al., 2021; Yue et al., 2023). The VOCs significantly suppressed mycelial growth, conidial germination, and sporulation of these pathogens in vitro and reduced disease incidence and lesion size on infected fruits in vivo. Mechanistically, the VOCs primarily act by damaging the pathogens’ cell membrane integrity and permeability, leading to cellular content leakage, decreased ergosterol biosynthesis, and increased malondialdehyde content (Wang et al., 2021; Yue et al., 2023). Additionally, the VOCs interfere with pathogen respiration by inhibiting key enzymes such as ATPase, malate dehydrogenase, and succinate dehydrogenase, causing energy metabolism disruption and reactive oxygen species accumulation. Transcriptomic analysis revealed significant changes in gene expression related to membrane components and amino acid metabolism pathways in treated pathogens (Wang et al., 2021; Yue et al., 2023).
However, to fully harness the potential of Pseudomonas VOCs for practical applications, further investigations are necessary to fully characterize their production within the plant root system and assess their efficacy under natural environmental conditions. By exploring the roles of Pseudomonas-derived VOCs in plant-microbe interactions, researchers can unlock valuable insights into sustainable strategies for crop protection and soil management.
5.4 Anthelmintic compounds
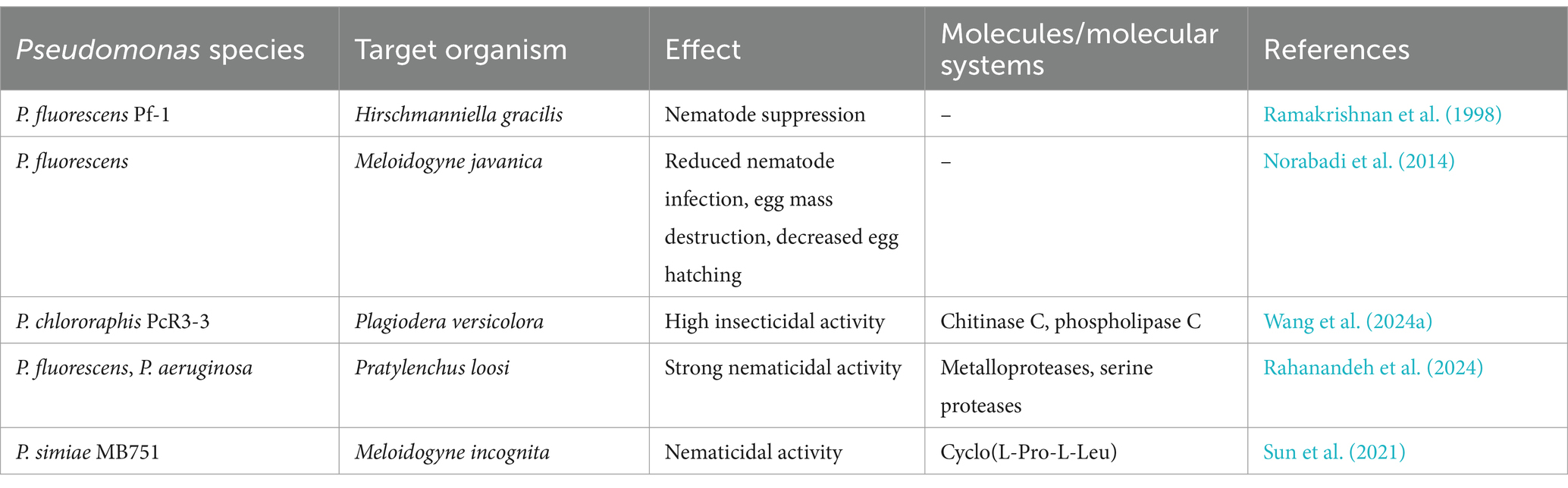
Pseudomonas spp., in addition to their antimicrobial activities, have also demonstrated the capability to produce potent anthelmintic compounds, they can destroy parasitic worms and nematodes (Devaraj et al., 2019; Lee C. H. et al., 2000). While not as extensively studied in Pseudomonas as in Streptomyces, the potential for nematode control exists within this bacterial genus (Table 2) (Devaraj et al., 2019; Lee C. H. et al., 2000; Rahanandeh et al., 2024; Spiegel et al., 1991; Wang et al., 2024a; Wang et al., 2024b).
Root-knot nematodes (Meloidogyne spp.) are among the most economically damaging plant-parasitic nematodes worldwide, causing significant yield losses in many crops (Sun et al., 2021). These microscopic roundworms infect plant roots, forming characteristic galls or “knots” that disrupt water and nutrient uptake. Meloidogyne incognita is one of the most widespread and studied species, with a broad host range and short generation time that makes it particularly difficult to control.
One study investigated the potential of a strain of P. fluorescens as a biocontrol agent against Meloidogyne javanica, another important root-knot nematode species (Norabadi et al., 2014). In greenhouse and laboratory experiments, P. fluorescens application significantly reduced nematode infection compared to the control group. The bacteria destroyed the nematode egg mass matrix and decreased egg-hatching levels. Additionally, P. fluorescens inoculation led to increased activities of peroxidase (POX) and phenylalanine ammonia lyase (PAL), key enzymes associated with plant defense mechanisms (Norabadi et al., 2014). These observations suggest that P. fluorescens effectively suppress nematode populations by disrupting egg structures and inducing systemic resistance in plants.
Recent research has expanded our understanding of Pseudomonas spp. potential for control of both nematodes and insect pests (Wang et al., 2024a; Wang et al., 2024b). For example, Pseudomonas chlororaphis PcR3-3, isolated from willow roots, exhibited high insecticidal activity against the coleopteran pest Plagiodera versicolora (Wang et al., 2024a). This leaf beetle is a significant pest of willow and poplar trees. While this study primarily focused on insecticidal activity, it highlights the diverse capabilities of Pseudomonas strains against different invertebrates. The genome of P. chlororaphis PcR3-3 was found to contain several genes potentially involved in insect pathogenicity, including those encoding chitinase C and phospholipase C (Wang et al., 2024b). These enzymes have been previously associated with the disruption of insect gut structures and could potentially play a role in nematode control as well.
Other studies have demonstrated the nematicidal potential of Pseudomonas spp. against the root lesion nematode Pratylenchus loosi, a significant pest of tea plants (Rahanandeh et al., 2024). It was found that P. fluorescens and Pseudomonas aeruginosa strains, which were isolated from tea rhizosphere, produced proteases with strong nematicidal activity. These proteases were capable of completely degrading Pratylenchus loosi nematodes within 8 h of exposure (Rahanandeh et al., 2024). Characterization of the proteases revealed them to be metalloproteases and serine proteases, classes of enzymes known to be involved in the degradation of nematode cuticles.
Although the research exploring Pseudomonas spp. as a source of anthelmintic compounds is relatively limited in Pseudomonas compared to other prokaryotes, these studies have indicated the potential of certain Pseudomonas strains to control populations of both plant-parasitic nematodes and insect pests. Given the diverse array of natural products synthesized by Pseudomonas strains and their interactions within soil ecosystems, it is likely that additional anthelmintic compounds may be discovered among Pseudomonas spp. Metabolites (Devaraj et al., 2019). This highlights the potential of Pseudomonas species as versatile biocontrol agents capable of addressing multiple agricultural pest problems.
5.5 Type VI secretion system
The Type VI Secretion System (T6SS) has emerged as a pivotal weapon in bacterial warfare, allowing bacteria to deliver toxic effectors directly into neighboring cells (Mougous et al., 2006; Pukatzki et al., 2006). T6SSs are present in more than 25% of gram-negative bacteria (Ho et al., 2014). The T6SS is a contact-dependent secretion system that plays a crucial role in interbacterial competition (Mougous et al., 2006; Pukatzki et al., 2006). It provides a selective advantage to producer strains by annihilating competitors either in an indiscriminate manner or in response to danger signals (Basler et al., 2013; Durán et al., 2021; Ho et al., 2014; Hood et al., 2010). The T6SS was originally described in Vibrio cholerae and P. aeruginosa as a proteinaceous nanomachine that translocates specific proteins directly into target cells (Mougous et al., 2006; Pukatzki et al., 2006). It was later observed and analyzed in many other bacterial pathogens (Burtnick et al., 2011; De Pace et al., 2010; Murdoch et al., 2011; Suarez et al., 2008). However, the analytical description of T6SS in non-pathogenic bacteria is underrepresented in the scientific literature (Bernal et al., 2017; Durán et al., 2021; Marchi et al., 2013).
The killing activity conferred by antibacterial T6SSs can be extremely potent during in vitro co-culture experiments, with T6SS-wielding cells often able to virtually eliminate similar numbers of susceptible competitor cells within a few hours (Basler et al., 2013; Durán et al., 2021; Ho et al., 2014). Antibacterial T6SSs are frequently found in both pathogenic and symbiotic or beneficial plant-associated bacteria (Bernal et al., 2018; Durán et al., 2021). This suggests that antibacterial T6SSs may be involved in establishing and protecting beneficial plant-associated communities from invasion of these communities by pathogens. For example, the T6SS-dependent antibacterial activity in the rhizosphere bacterium P. putida KT2440 contributes to reducing colonization and necrosis induced by the phytopathogen Xanthomonas campestris when both are co-infiltrated into Nicotiana benthamiana leaves (Bernal et al., 2017).
Further in planta studies show that T6SS in P. fluorescens MFE01 play crucial role in protecting potato tubers from Pectobacterium atrosepticum infection (Bourigault et al., 2023). Fluorescence microscopy revealed that MFE01 exhibits an aggressive T6SS behavior with continuous and intense firing activity, causing rounding and lysis of target P. atrosepticum cells. The study identified a putative T6SS-secreted amidase effector, Tae3Pf, as a major contributor to MFE01’s antibacterial activity. This effector was found to be toxic when produced in the periplasm of Escherichia coli, with its toxicity neutralized by the inner membrane immunity protein Tai3Pf. While Tae3Pf plays a significant role, the research suggests that other T6SS effectors likely contribute to MFE01’s overall antibacterial activity (Bourigault et al., 2023). These findings highlight the importance of the T6SS in P. fluorescens MFE01’s biocontrol capabilities, offering new perspectives on bacterial antagonism in the context of plant protection and potentially leading to the development of more effective biocontrol strategies in agriculture.
6 Indirect inhibition of phytopathogens
In addition to direct inhibition via the production of antagonistic compounds, Pseudomonas spp. demonstrate a remarkable capacity to indirectly inhibit plant pathogens (Haney et al., 2018; Leeman et al., 1995). Competitive exclusion is one of the primary mechanisms through which Pseudomonas strains exert their inhibitory effects (Archetti et al., 2011; Buddrus-Schiemann et al., 2010; Rainey, 1999). By occupying niche spaces and consuming available resources, these strains effectively thwart the colonization efforts of pathogens, preventing them from establishing robust populations and causing harm to plants (Archetti et al., 2011; Buddrus-Schiemann et al., 2010; Rainey, 1999). This strategy, however, does not operate in isolation; rather, it complements direct antagonism, wherein Pseudomonas spp. secretes antimicrobial compounds that further hinder pathogen growth and proliferation (Haney et al., 2018; Leeman et al., 1995). These antimicrobials may arise as byproducts of interference competition over resources provided via plant root exudates or organic matter in the soil, illustrating the multifaceted nature of Pseudomonas-mediated inhibition.
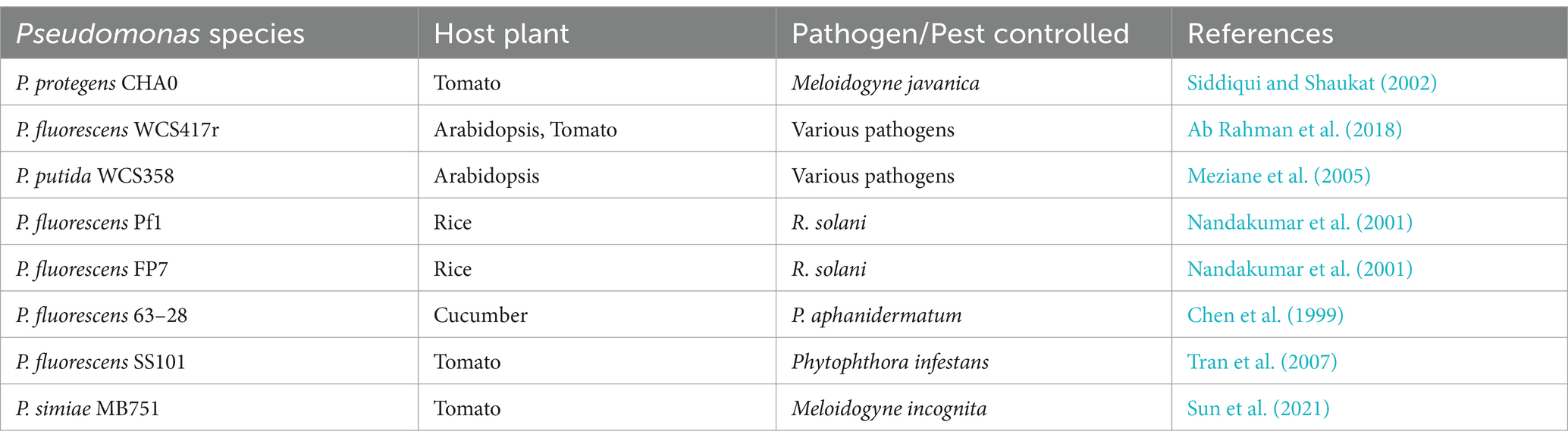
Pseudomonas spp. possess the ability to activate host resistance pathways, thereby inducing systemic resistance (ISR) in plants (Table 3) (Chen et al., 1999; Elsharkawy et al., 2022; Haney et al., 2018; Hoffland et al., 1996; Leeman et al., 1995; Meziane et al., 2005; Nandakumar et al., 2001). ISR is a state of enhanced defensive capacity developed by a plant reacting to specific biotic or chemical stimuli. Pseudomonas spp. mediate ISR through phytohormone defense signaling pathways such as jasmonic acid/ethylene (JA/ET) or salicylic acid (SA) (Chen et al., 1999; Pangesti et al., 2016). For instance, a study investigated the efficacy of Pseudomonas corrugata strain 13 and Pseudomonas aureofaciens strain 63–28 in inducing systemic resistance against Pythium aphanidermatum in cucumber roots (Chen et al., 1999). Both bacterial strains were found to produce SA in vivo and induced cucumber roots to accumulate endogenous SA within a day of inoculation (Chen et al., 1999). Notably, plants treated with a Pseudomonas strain showed significantly elevated SA levels compared to the control, persisting from 1 to 5 days post-bacterization (Chen et al., 1999). Interestingly, SA inhibited the mycelial growth of the pathogen P. aphanidermatum at higher SA concentrations (Chen et al., 1999).
In response to infections by various pathogens, such as fungi, bacteria, or nematodes, plants often produce stress ethylene (Abeles et al., 1992; Iqbal et al., 2017). The symptoms observed in an infected plant largely stem from the stress caused by the pathogen (Iqbal et al., 2017; Van Loon, 1984). A significant portion of the damage to infected plants results from the plant’s response to increased levels of stress ethylene (Stearns and Glick, 2003). Exogenous ethylene can exacerbate pathogen infections, while chemical inhibitors of ethylene synthesis can reduce their severity (Stearns and Glick, 2003; Yang et al., 2017). Additionally, pretreating plants with ACC deaminase-containing plant growth-promoting bacteria can provide significant protection against ethylene-induced damage from pathogen infections (Glick et al., 1995; Hao et al., 2011; Toklikishvili et al., 2010; Wang et al., 2000). Some bacteria that promote plant growth encode the enzyme ACC deaminase, which breaks down ACC exudates from plants, the direct precursor of ethylene production, and lowers the amount of ethylene that plants produce in response to different stressors (Glick et al., 1995; Hao et al., 2011). Many other studies showed that the application of ACC-deaminase-producing plant-growth-promoting bacteria was found to inhibit the growth of phytopathogens in crops (Toklikishvili et al., 2010; Wang et al., 2000).
The ability of Pseudomonas spp. to induce plant disease resistance holds significant promise for its application as a biocontrol agent (Haney et al., 2018; Leeman et al., 1995). Strains that exhibit such abilities may prove highly effective at protecting their plant hosts against pathogenic infections in situ, even if they demonstrate poor bioactivity against phytopathogens in vitro. This underscores the importance of screening for biocontrol strains based not only on their performance in traditional in vitro bioactivity assays but also on their ability to elicit host defenses and confer protection to host plant species in vivo. As research in this field progresses, better proxies, and methodologies for evaluating the efficacy of Pseudomonas strains as biocontrol agents should be developed, considering the intricate interplay between bacterial-induced plant defenses and pathogen susceptibility.
7 The potential use of Pseudomonas species as effective biocontrol agents
Many studies demonstrate promise in laboratory settings but exhibit inconsistent efficacy in agricultural settings (Arora et al., 2008; Garbeva et al., 2004; Lee H. S. et al., 2000; Rini and Sulochana, 2007). The inconsistent results of biocontrol treatments show we need to better understand what affects soil and root microbes. Abiotic factors such as soil type, climate, and farming practices, along with biotic factors like host crop species and root exude profiles, profoundly influence microbial assemblages and biocontrol success (Babin et al., 2019; Classen et al., 2015; Edwards et al., 2015; Glick and Gamalero, 2021; Rousk et al., 2010). Efforts such as the Microbiome Stress project aim to understand bacterial community responses to environmental stressors, aiding in the development of robust biocontrol strategies resilient to changing environmental conditions and advancing sustainable agriculture practices (Rocca et al., 2019).
7.1 Abiotic factors influencing biocontrol efficacy
Numerous studies have focused on enhancing the effectiveness and consistency of Pseudomonas spp. biocontrol agents through various agricultural practices and interventions (Zhang et al., 2015). This includes the identification of several minerals and carbon sources that have a differential influence on the production of the antibiotics 2,4-diacetylphloroglucinol, pyoluteorin and pyrrolnitrin and the salicylic acid and pyochelin by disease-suppressive strains of P. fluorescens (Duffy and Défago, 1999). For example, the addition of glycerol, zinc and ammonium molybdate to P. protegens CHA0 has shown promise in increasing its biocontrol efficacy against Meloidogyne javanica (Hamid et al., 2003; Siddiqui and Shaukat, 2002). Additionally, adding zinc to soil containing P. fluorescens PfAs1 has enhanced the expression of genes resistant to bacterial leaf blight in rice, while also significantly reducing bacterial leaf blight in rice caused by X. oryzae pv. oryzae (Sharma et al., 2020). Similarly, supplementing P. fluorescens NK2 with the zinc oxide nanoparticle (ZnO-NP) enhances the inhibition efficacy against the pathogenic bacterium Pseudomonas viridiflava NK2 in cucumber (Al-Karablieh et al., 2022).
Beyond strain inoculation, various agricultural practices significantly impact the composition and establishment of species within the plant root microbiome. Practices like irrigation, tillage, and cropping methods can play crucial roles (Babin et al., 2019; Dennert et al., 2018; Mavrodi et al., 2018). Agro-chemicals such as pesticides and fertilizers also shape the plant root and soil microbiome, offering protection against crop diseases (Ding et al., 2013; Shen et al., 2019). For instance, ammonia fumigation effectively suppresses Fusarium wilt disease in bananas (Musa acuminate Cavendish) while inducing shifts in the soil microbial community, notably reducing Fusarium species abundance (Shen et al., 2019). Studies suggest that combining organic fertilizers with biocontrol strains enhances disease suppression further. Thus, the addition of biocontrol strains to organic fertilizers before application creates a more conducive soil environment, fostering nutrient availability, root colonization, and biocontrol effectiveness, a strategy known as bio-organic fertilizer application, widely recognized for its efficacy in disease suppression (Ding et al., 2013; Ketabchi et al., 2016; Watanabe et al., 1987).
Conversely, certain chemical additives and carbon compounds have been found to impede the biocontrol efficacy of Pseudomonas spp. For example, the addition of glucose can hinder the efficacy of P. protegens CHA0 against Meloidogyne javanica (Siddiqui and Shaukat, 2002). However, some Pseudomonas strains exhibit a high tolerance to commonly used fungicidal compounds, paving the way for synergistic approaches where chemical and biological pest control methods can be combined to enhance efficacy while reducing the overall pesticide dose (Anand et al., 2010; Kataria et al., 2002).
While these findings suggest the potential for optimizing farming practices to maximize disease suppression, comprehensive research in this area remains limited. The complexity of agricultural ecosystems and the variability in pathogen dynamics necessitates further investigation to identify the most effective strategies tailored to specific pathogens, climatic conditions, and soil characteristics. Nonetheless, exploring the interplay between Pseudomonas spp. biocontrol agents, agricultural practices, and chemical additives hold promise for sustainable disease management and crop protection.
Optimizing biocontrol delivery systems involving Pseudomonas
Numerous methods exist for delivering biocontrol strains to crops, each potentially influencing the consistency of biocontrol strategies (Jambhulkar et al., 2016; Preininger et al., 2018). Foliar spraying may seem appealing, especially in developed countries with available spraying equipment, but microbial suspensions can damage or clog machinery due to settling out of solution, and stresses from spraying apparatus (e.g., heat stress, shearing forces) can reduce biocontrol strain viability (Figure 2) (Preininger et al., 2018). Moreover, foliar spray that is suitable for microbial inoculants targeting foliar diseases (Jambhulkar et al., 2016) is often less effective for controlling root diseases like wheat take-all. Soil inoculation, another recommended method, is often employed if biocontrol strains are susceptible to desiccation (Jambhulkar et al., 2016). As discussed earlier, methods such as bio-organic fertilizer application (Ding et al., 2013; Ketabchi et al., 2016; Watanabe et al., 1987) can enhance biocontrol strain success. However, these strategies may increase the expense and complexity of applying disease-suppressive measures, while also potentially altering soil chemistry and microbiome composition with unknown or conflicting effects (Babin et al., 2019; Shen et al., 2019).
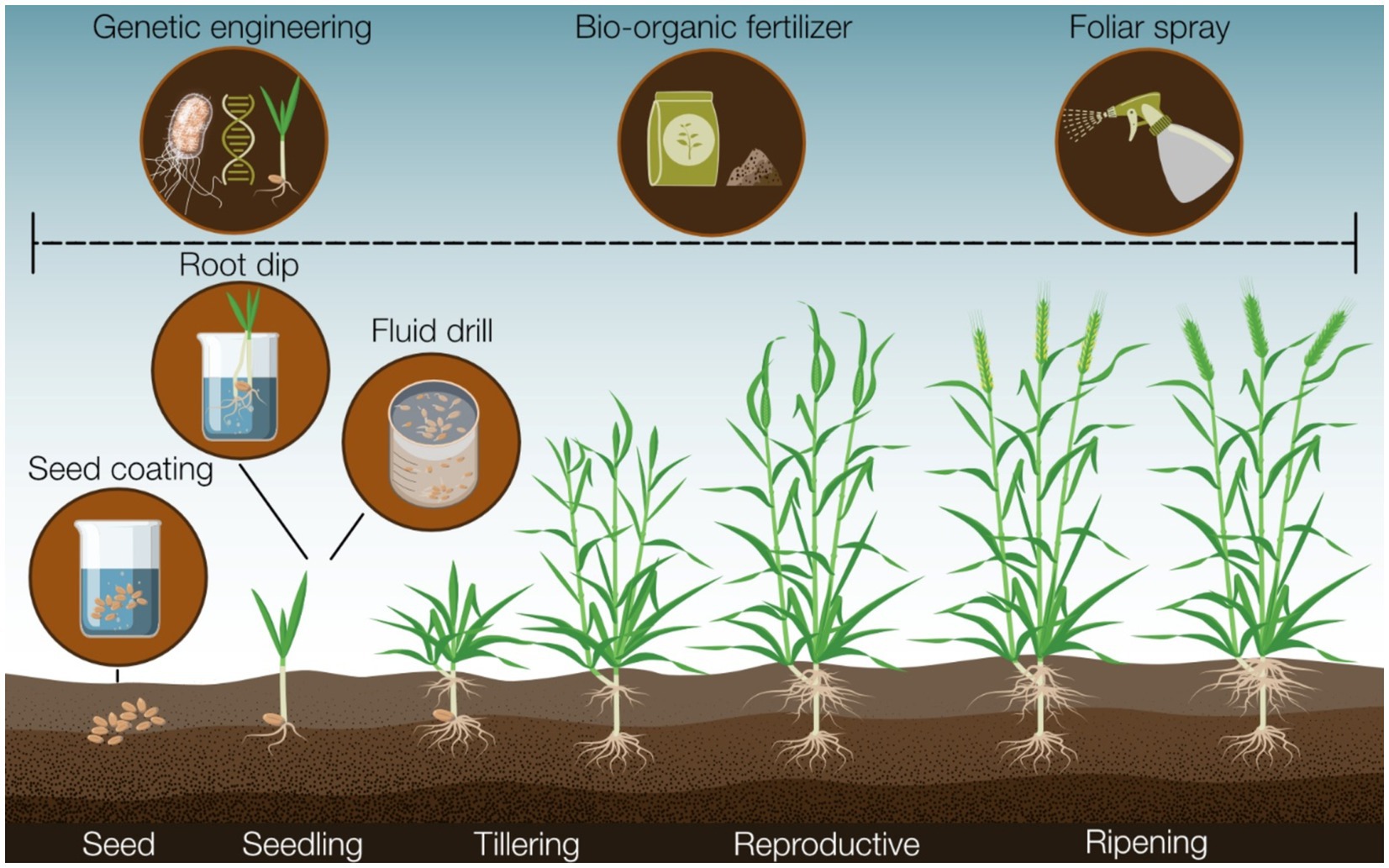
Figure 2. A summary of the major tools and methods available that could facilitate the application of biocontrol strains that can influence biocontrol efficacy in the field.
Direct inoculation of biocontrol strains onto plant roots offers a strategy to bypass challenges associated with soil-survivability as the strain bypasses exposure to an environmental medium before colonizing roots (Jambhulkar et al., 2016; Pill, 1991). Techniques like fluid drill inoculation and root transplant dip exemplify this approach, allowing biocontrol agents to colonize roots under controlled conditions (Jambhulkar et al., 2016; Pill, 1991). In root dip, seedling roots are immersed in a liquid cell suspension before field transfer, while in fluid drill methods, seeds pre-germinate within a gel containing the biocontrol strain (Jambhulkar et al., 2016; Pill, 1991). Root dip has demonstrated enhanced root colonization by Pseudomonas compared to soil inoculation in some cases (Pathak et al., 2004), effectively protecting crops against R. solani. Other studies showed that root dip was effective for P. fluorescens SBW25 to colonize the root of wheat (Guan et al., 2024). However, pre-germinating plants and manually inoculating roots are labor-intensive processes compared to purchasing pre-coated seeds (O’Callaghan, 2016). Similarly, fluid drill methods have shown increased root colonization by inoculated bacterial strains, in limited studies, indicating efficient disease suppression (Clarkson et al., 2002; Hardaker and Hardwick, 1978). Nevertheless, research exploring the ecological impact of fluid drill gel application remains sparse.
As mentioned, plants can also undergo colonization by coating the seed with a formulation of biocontrol strain spores or cells, employing various methods to adhere biocontrol strains to the seed surface. For instance, seeds may be submerged in a microbial suspension and subsequently dried before seed germination (known as bio-priming) (Callan et al., 1990), or a liquid cell suspension or adhesive may be utilized to coat the seed in bacterial cells (referred to as film coating) (O’Callaghan, 2016). Seed coating technologies effectively deliver biocontrol strains directly to the soil surrounding a germinating seed and the rhizosphere (Jambhulkar et al., 2016; O’Callaghan, 2016), with numerous instances demonstrating the efficacy of seed coating approaches in disease suppression in both field and laboratory settings (Ardakani et al., 2010; Callan et al., 1990; El-Mougy and Abdel-Kader, 2008; Ma et al., 2019; Prathuangwong et al., 2013). This includes several studies showcasing the effectiveness of seed coatings as delivery methods for Pseudomonas spp. biocontrol strains (Ardakani et al., 2010; Prathuangwong et al., 2013). In addition, scientists have recently reported developing a unique procedure to introduce endophytic bacteria into plant seeds so that it was no longer necessary to treat the seeds before planting (Glick and Gamalero, 2021).
While the focus of this review is on Pseudomonas as biocontrol agents, it is worth noting an emerging trend in the use of nanoparticle-based Pseudomonas biofertilizers. Although primarily applied for plant growth promotion rather than disease control, this innovative approach may have implications for biocontrol strategies. Nanoparticle-based biocontrol formulations have shown significant potential in overcoming delivery challenges (El-Ghamry et al., 2018). In these formulations, plant growth-promoting bacteria and nutrients are encapsulated within nanoscale polymers (nanoencapsulation) (Golbashy et al., 2017). This method enhances soil microbial activity, improves aeration, and supports natural fertilization (Itelima et al., 2018). For instance, selenium nanoparticles combined with Pseudomonas spp. have been used to improve crop nutrition and soil quality (Sonali et al., 2024) However, not all nanoparticles interact synergistically with Pseudomonas spp. (Dimkpa et al., 2012a; Dimkpa et al., 2012b). For example, zinc nanoparticles were found to enhance siderophore production more effectively than copper nanoparticles, which inhibited the expression of genes related to siderophore transport (Dimkpa et al., 2012a). These findings suggest that while nanoparticle-based biocontrol formulations hold significant promise in enhancing soil health and crop productivity, their interactions with specific biocontrol strains, such as Pseudomonas spp., are complex and not always synergistic.
The successful delivery of biocontrol strains remains a key challenge in modern agriculture, with methods ranging from foliar sprays to seed coatings and root inoculation. While each approach offers distinct advantages, the complexity of balancing cost, labor, and ecological impact must be carefully considered. Recent advancements, such as nanoparticle-based biocontrol formulations, hold great potential for improving efficacy and sustainability. However, the variability in interactions between nanoparticles and biocontrol strains like Pseudomonas spp. underscores the need for further research to optimize these technologies for widespread agricultural use.
7.3 Exploiting plant recruitment mechanisms to improve biocontrol agent outcomes
In addition to increasing the competitiveness of bacterial strains when applied to seeds and soil, leveraging the mechanisms by which plants selectively recruit microbial species from the soil could enhance the efficacy of biocontrol strains (Ryan et al., 2009; Zhang et al., 2015). Plants release approximately 20–40% of photosynthetically fixed carbon through their roots into the soil, comprising various compounds like ions, amino acids, sugars, phenolics, mucilage, polysaccharides, and proteins (Badri et al., 2013; Bais et al., 2006; el Zahar Haichar et al., 2016; Zhalnina et al., 2018). This root exudation triggers a surge in microbial abundance and activity in the soil surrounding the roots, known as the “rhizosphere effect,” as many microbes are attracted to the carbon-rich nutrients exuded from the roots (Bais et al., 2006); soil microbes come to plant rhizospheres to feed. Moreover, exudates potentially act as a filtering mechanism, enabling plants to selectively enrich for specific microbial species with certain metabolic capabilities (Bais et al., 2006). Experiments profiling the root exudates of Arabidopsis thaliana have shown that specific exudate compounds correlate with the abundances of specific bacterial taxa, indicating their role in microbial recruitment (Badri et al., 2013; Chaparro et al., 2013; Chaparro et al., 2014). Stable isotope probing experiments have further revealed that different microbial taxa metabolize plant root exudates based on differences in exudate composition (el Zahar Haichar et al., 2016; Haichar et al., 2008; Haichar et al., 2012). Additionally, root exudation can be influenced by abiotic and biotic factors, including responses to pathogens, which alter the microbial community composition (Jousset et al., 2011; Lanoue et al., 2010).
Beyond changes in their relative abundance, root exudates may alter the functionality of the root microbiome by modifying microbial gene expression (Chaparro et al., 2014). For instance, benzoxazinoid-related compounds, exuded by maize roots, correlate with an increased number of microbial transcripts related to beneficial P. putida, which resulted in significantly higher numbers of P. putida cells colonizing the root (Neal et al., 2012).
Understanding the relationships between root exudate composition, microbial community structure, and microbiome functionality offers promising avenues for enhancing crop productivity and health. Engineering plants to produce specific root exudates could improve the colonization potential and efficacy of beneficial species and biocontrol agents, such as Pseudomonas spp. (Ali and Glick, 2024). Arabidopsis mutant lines engineered with altered root exudation profiles have shown increased recruitment of beneficial rhizobacteria, suggesting similar approaches could be applied to cereal crops through breeding or genetic modification (Badri et al., 2009; Bressan et al., 2009; Huang et al., 2019). However, there remains a substantial knowledge gap regarding the compounds acting as signals and nutrients for bacteria of interest. While cues are well-defined for certain plant-microbe symbioses, many other systems lack a detailed understanding. Advanced tools like stable isotope probing, metabolomics, dual RNA sequencing, and imaging mass spectrometry are beginning to elucidate some of these interactions, paving the way for a deeper understanding of plant-microbe dynamics in the future (Berry et al., 2013; Camilios-Neto et al., 2014; el Zahar Haichar et al., 2016; Haichar et al., 2012; Zhalnina et al., 2018).
7.4 Engineered Pseudomonas for enhanced efficacy
It is generally assumed that suppression of plant pathogens by Pseudomonas spp. is based on two primary features: (1) production of natural products (direct inhibition), and (2) stimulation of ISR, which activates the plant defense system against harmful microbes and viruses. The expression of genes and natural products of Pseudomonas is influenced by growth and environmental conditions (Chin-A-Woeng et al., 2003; Hamid et al., 2003). These conditions can negatively affect the efficacy of Pseudomonas spp. as biocontrol strains (Nadeem et al., 2016). For instance, biocontrol Pseudomonas strains may perform well in one location or field season but not the next, owing to a wide range of biotic and abiotic factors in the soil environment that can adversely impact the Pseudomonas strain’s root colonization, expression of genes involved in biocontrol and/or activity of biocontrol metabolites (Babin et al., 2019; Classen et al., 2015; Edwards et al., 2015; Rousk et al., 2010).
Based on the increasing knowledge of the molecular mechanisms that underline natural product synthesis, the use of improved genetically modified biocontrol agents in agricultural seems feasible. However, the intentional release of genetically modified organisms (GMOs) requires extensive analysis of their potential ecological impact (Glick and Skof, 1986; Kolseth et al., 2015; Then et al., 2020). Moreover, the issue of whether GMOs could affect non-target organisms needs to be addressed. Laboratory experiments must be conducted before the release of GMOs into the natural environment.
Many studies have engineered Pseudomonas strains to enhance their biocontrol efficacy (Bakker et al., 2002; Barahona et al., 2011; Glandorf et al., 2001; Leeflang et al., 2002; Niemann et al., 1997; Shaukat and Siddiqui, 2003; Viebahn et al., 2003; Xiao-Jing et al., 2005). For instance, P. putida WCS358 was genetically modified to overproduce the antifungal compound 2,4-diacetylphloroglucinol (Phl) (Bakker et al., 2002; Glandorf et al., 2001; Leeflang et al., 2002), this genetically modified strain displays enhanced antifungal activity (Glandorf et al., 2001). Modified derivatives of strain P. putida WCS358 caused transient shifts in the composition of bacterial and fungal communities in the rhizosphere of wheat. However, they had no effect on soil metabolic activities (Bakker et al., 2002; Glandorf et al., 2001; Leeflang et al., 2002). In addition, a hypermotile mutant of P. fluorescens F113 was superior to the wild-type strain in colonizing the plant rhizosphere and controlling Fusarium oxysporum and Phytophthora cactorum pathogenesis (Barahona et al., 2011).
By harnessing the power of genetic modification, researchers may be able to tailor Pseudomonas strains to target specific pathogens while minimizing unintended ecological impacts (Barahona et al., 2011; Glandorf et al., 2001; Leeflang et al., 2002; Viebahn et al., 2003). These advancements pave the way for sustainable and environmentally friendly strategies to combat crop diseases, contributing to the overall resilience and productivity of agricultural systems. Continued research and innovation in this area holds promise for further enhancing the biocontrol potential of genetically modified Pseudomonas strains, ultimately benefiting farmers and ecosystems alike.
7.5 The biosafety of Pseudomonas-based biocontrol agents
Beyond these logistical hurdles, the safety concerns regarding clinical toxicity and environmental persistence pose additional obstacles to the commercialization of Pseudomonas spp. as biocontrol agents, necessitating extensive screening processes that impede progress (Fravel, 2005). Many candidate biocontrol agents are not well characterized and lack a thorough assessment of their non-target effects, leaving gaps in our understanding of their ecological impacts (Viaene et al., 2016; Winding et al., 2004).
An underlying concern with using antagonistic bacteria like Pseudomonas spp. as biocontrol agents is the diverse array of secondary metabolites they produce, which could potentially exert unintended non-target effects, including those detrimental to human health (Deising et al., 2017). While these metabolites effectively target pathogens, some may also pose risks to human cells by interacting with essential human metabolites, such as cholesterol (Zotchev, 2003). The actual production and concentration of antimicrobials by biocontrol agents in the soil remains inadequately explored (Deising et al., 2017; Winding et al., 2004). Some evidence suggests that the role of antimicrobials in inhibiting pathogens within the plant root niche might be minor compared to other traits like resource competition and immune system activation (Hennessy et al., 2017; Koch et al., 2018). The tight regulation of gene clusters encoding secondary metabolites raises the possibility that their activation depends on the direct interaction with specific pathogenic species, potentially minimizing non-target effects compared to the widespread use of purified antimicrobial molecules (Bertrand et al., 2014; Van der Meij et al., 2017). Nonetheless, deeper insights into the in vivo behavior and concentration of secondary metabolites produced by biocontrol agents are essential to develop strategies that mitigate non-target effects.
Notwithstanding several studies highlighting the influence of Pseudomonas spp. on individual plant symbionts, broader investigations into the impacts of these bacteria on the soil ecosystem and root microbiome are relatively scant (Balser et al., 2002; Glick and Gamalero, 2021; Khatoon et al., 2020; Lv et al., 2023). The introduction of biocontrol strains at low densities, such as through seed coatings, may have limited effects on the indigenous microbial community, especially in highly diverse soils (Koch et al., 2018). However, larger inoculations could modify existing soil communities, with some studies demonstrating short-term alterations in microbial composition following biocontrol strain application (Schmidt et al., 2014; Winding et al., 2004). Understanding the long-term implications of biocontrol strain application on pathogen populations, soil microbiome functionality, and environmental processes requires meticulously designed experiments, incorporating indicators like mycorrhizal abundance and functional gene expression related to plant-beneficial processes (Schmidt et al., 2014; Sisaphaithong et al., 2012; Winding et al., 2004). Rigorous controls are imperative to delineate the specific impacts of biocontrol Pseudomonas strains on soil microbiomes, paving the way for informed decision-making in sustainable agriculture (Winding et al., 2004).
Genetically modified biocontrol agents offer several potential advantages over conventional chemical pesticides. They can be engineered to have increased efficacy, broader host range, improved persistence in the environment, and enhanced production of antimicrobial compounds (Glandorf, 2019; Scheepmaker et al., 2016; Tonui et al., 2022). For example, Pseudomonas strains have been modified to overproduce antifungal metabolites, leading to better suppression of plant pathogens (Glandorf, 2019; Scheepmaker et al., 2016; Tonui et al., 2022). Genetically modified biocontrol agents could provide more targeted pest control while reducing reliance on chemical pesticides, aligning with goals for integrated pest management and sustainable agriculture (Scheepmaker et al., 2016).
However, the development and use of genetically modified biocontrol agents also raise important biosafety concerns that require careful consideration. One primary concern is the potential for unintended ecological impacts. While these biocontrol agents are designed to target specific pests, their release into complex ecosystems could have unforeseen consequences on non-target organisms or broader ecological processes. For instance, there are questions about whether genetically modified biocontrol agents could outcompete native microbial populations or disrupt existing ecological balances (Glandorf, 2019; Scheepmaker et al., 2016; Tonui et al., 2022).
Another significant concern is the potential for horizontal gene transfer. Genetically modified biocontrol agents carry modified genetic material, and there is a possibility that these genes could be transferred to other microorganisms in the environment. This could potentially lead to the spread of engineered traits beyond the intended biocontrol agent, with unknown consequences for microbial ecology and potentially human health (Glandorf, 2019; Scheepmaker et al., 2016; Tonui et al., 2022). The likelihood and potential impacts of such gene transfer events need to be thoroughly assessed as part of the risk evaluation process for genetically modified biocontrol agents.
8 Registration and commercialization of Pseudomonas spp. as biocontrol strains
In contrast to the established practice of biocontrol for insects, biocontrol methods targeting plant diseases represent a relatively recent development. This notwithstanding, the bacterium, Agrobacterium radiobacter strain K 84 was registered by the United States Environmental Protection Agency (EPA) as a means of controlling crown gall disease in 1979. A decade later, the first fungus, Trichoderma harzianum ATCC 20476, received EPA approval and registration for combating plant diseases.
To date, biocontrol agents represent <5% of the entire crop protection industry (Devaraj et al., 2019), despite their well-documented efficacy. The limited availability of licensed products for plant disease biocontrol is largely attributed to challenges in technology transfer, particularly in developing countries, where the economic potential of these agents is not completely recognized. Moreover, the mass production of promising microbial candidates for phytopathogen biocontrol often faces obstacles due to a lack of organism-specific research methods and the high costs associated with in vivo production and licensing, making them less competitive compared to existing chemical agents (Gehlot and Singh, 2018; Sundh and Goettel, 2013).
Ensuring effective deployment of biocontrol agents presents logistical challenges, including timing and density of application, as well as maintaining their presence in the target environment. Ecological concerns also arise from the introduction of non-native organisms, which may become invasive and disrupt local ecosystems. Despite promising results demonstrated in laboratory settings, many biocontrol agents have shown limited efficacy under real-world conditions (Backer et al., 2018; Capper and Higgins, 1993; Deacon, 1991; Mark et al., 2006), making them much less attractive to end users compared to chemical pesticides.
The release of GMOs, including engineered Pseudomonas strains, adds another layer of complexity to the registration and commercialization process. Regulatory frameworks for GMOs are typically more stringent than those for non-modified organisms, reflecting concerns about potential ecological impacts and gene transfer to wild populations. In the United States, for instance, the EPA regulates genetically engineered microbial pesticides under the Federal Insecticide, Fungicide, and Rodenticide Act (FIFRA) and the Toxic Substances Control Act (TSCA). These regulations require extensive risk assessments, including evaluations of genetic stability, potential for horizontal gene transfer, and impacts on non-target organisms (Angelo, 2023; Jones et al., 2024). The European Union has even more restrictive policies on GMOs, which mandate a case-by-case environmental risk assessment and post-market (Mizoguchi et al., 2024; Zimny, 2023).
The assessment of genetically modified biocontrol agents under existing regulations such as EC 1107/2009 (concerning the placing of plant protection products on the market) and Directive 2001/18/EC (on the deliberate release of GMOs into the environment) provides a foundation for evaluating their safety. However, these frameworks may need to be adapted or expanded to fully address the unique characteristics of living, replicating microbial agents (Glandorf, 2019; Scheepmaker et al., 2016; Tonui et al., 2022). This is particularly true for novel approaches like gene drive systems, which could potentially lead to the spread of engineered traits through wild populations more rapidly than traditional genetic modifications. These regulatory hurdles, while necessary for ensuring safety, can significantly delay the commercialization of genetically engineered Pseudomonas spp. biocontrol agents and increase development costs.
Public perception of GMOs significantly influences research trajectories in genetically modified biocontrol agent development. The reluctance to invest in technologies facing public skepticism has created a feedback loop, potentially impeding progress in sustainable pest management strategies (Lucht, 2015; Tonui et al., 2024). This phenomenon extends beyond funding constraints, affecting the entire research ecosystem.
The hesitancy to pursue genetically modified biocontrol research has broader implications for agricultural biotechnology. The overlap in expertise between genetically modified biocontrol development and other agricultural biotechnology applications suggests that reduced focus on gene modification research could lead to a general decline in capacity across the field (Qaim, 2020). This potential loss of human capital is particularly concerning in the context of early-career researchers, who may be dissuaded from specializing in gene modification-related fields due to perceived limited prospects (Merkley, 2020; Vanloqueren and Baret, 2017).
However, recent meta-analyses indicate a gradual shift in public attitudes toward GMOs in certain regions. It was reported an increasing trend of support for genetically modified foods in the United States over two decades, particularly among younger demographics and individuals with higher scientific literacy (Cui and Shoemaker, 2018). Similarly in Europe, an upward trend was observed in positive attitudes toward genetically modified crops between 2002 and 2019 (Woźniak et al., 2021). Multiple factors contribute to this attitudinal shift, including increased familiarity with GMOs, improved science communication, and growing recognition of biotechnology’s potential in addressing food security and environmental challenges (Fernbach et al., 2019).
Despite evolving public sentiment and scientific advancements, regulatory frameworks governing GMOs have remained largely static, creating a misalignment between current knowledge and regulatory approaches. Many jurisdictions maintain approval processes for GMOs established during periods of greater public skepticism (Eriksson et al., 2019). Additionally, current regulations often fail to account for advancements in genetic engineering techniques, such as CRISPR-Cas9, which allow for more precise genetic modifications. The regulatory focus on the process of genetic modification rather than the properties of the resulting organism may lead to over-regulation of genetically modified biocontrol agents that pose no greater risk than traditionally bred alternatives (Lassoued et al., 2019).
Current regulation and registration procedures, largely derived from chemical pesticide frameworks, do not adequately address the unique considerations of microbial biocontrol agents, contributing to their slow implementation in the field (Sundh and Goettel, 2013). As research on genetically modified biocontrol agents progresses, it will be crucial to conduct comprehensive, long-term studies on their ecological impacts. This should include assessments of their effects on soil microbial communities, potential impacts on beneficial insects and other non-target organisms, and their persistence in various environmental conditions. Such research will be essential not only for refining risk assessment procedures but also for building public trust in the technology (Glandorf, 2019; Scheepmaker et al., 2016; Tonui et al., 2022).
9 Conclusions and future perspectives
In the realm of sustainable agriculture, the use of microorganisms to combat plant diseases and enhance crop yield represents a promising alternative to chemical interventions. Pseudomonas spp., among others, have garnered attention for their potential as biocontrol agents due to their intricate interactions with plants and the environment; they are typically equipped with an arsenal of secondary metabolites and enzymes adept at engaging with host organisms and outcompeting adversaries. These bioactive molecules offer substantial benefits to crop plants by fostering plant growth and curbing disease incidence. Despite their resilience to environmental pressures, the efficacy of Pseudomonas-based biocontrol strategies hinges on a multifaceted interplay of factors, as elucidated in this review. While strides have been made to optimize delivery methods, mitigate stressors, and enhance soil, and root colonization, gaps persist in understanding the broader spectrum of influences, including agricultural practices, which shape plant microbiome dynamics and biocontrol efficacy.
To realize the full potential of biocontrol agents, comprehensive research efforts are imperative, with a focus on elucidating pre-existing signaling pathways between plants and microbes to bolster the colonization potential of desirable microbial strains. Moreover, investigations into the impact of candidate biocontrol strains on native microbial communities and ecosystem functions are paramount for deploying biocontrol agents with minimal non-target effects on a wider scale. The development of more consistent biocontrol strategies necessitates a holistic, combinatorial approach encompassing diverse aspects including delivery mechanisms, formulation enhancements, agricultural methodologies, and the intricate differences of plant-microbe symbioses. By embracing this integrated framework, it should be possible to realize effective, sustainable biocontrol solutions tailored to the complexities of modern agricultural landscapes.
Author contributions
HA: Investigation, Writing – original draft, Writing – review & editing. BG: Writing – review & editing. DM: Supervision, Writing – review & editing. CS: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. HA was supported by a Murdoch International Postgraduate Scholarship and top-up scholarship through the CSIRO-Murdoch University-Industry Bioplastics Innovation Hub.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ab Rahman, S. F. S., Singh, E., Pieterse, C. M., and Schenk, P. M. (2018). Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 267, 102–111. doi: 10.1016/j.plantsci.2017.11.012
Abeles, F. B., Morgan, P. W., and Saltveit, M. E. Jr. (1992). Ethylene in plant biology. New York, NY: Academic Press.
Al Zadjali, M., Rabiey, M., McMillan, V., Shaw, L. J., Hammond-Kosack, K., Malone, J. G., et al. (2023). Characterising the influence of first-year wheat cultivar on pseudomonas selection and function in a take-all infected field. Crops 3, 195–208. doi: 10.3390/crops3030019
Alattas, H., Ardley, J., Swift, R., Jackson, S., Biddulph, B., Bekuma, A., et al. (2023). Complete genome sequence of the ice-nucleation-active Pseudomonas syringae strain MUP20, isolated from frost-damaged wheat (Triticum aestivum cv Scepter) in Western Australia. Microbiol. Res. Announcements 12, e01275–e01222. doi: 10.1128/mra.01275-22
Ali, S., and Glick, B. R. (2024). Root exudate metabolites Alter food crops microbiomes, impacting plant biocontrol and growth. Crops 4, 43–54. doi: 10.3390/crops4010004
Al-Karablieh, N., Al-Shomali, I., Al-Elaumi, L., and Hasan, K. (2022). Pseudomonas fluorescens NK4 siderophore promotes plant growth and biocontrol in cucumber. J. Appl. Microbiol. 133, 1414–1421. doi: 10.1111/jam.15645
Anand, T., Chandrasekaran, A., Kuttalam, S., Senthilraja, G., and Samiyappan, R. (2010). Integrated control of fruit rot and powdery mildew of chilli using the biocontrol agent Pseudomonas fluorescens and a chemical fungicide. Biol. Control 52, 1–7. doi: 10.1016/j.biocontrol.2009.09.010
Andreote, F. D., de Araújo, W. L., de Azevedo, J. L., van Elsas, J. D., da Rocha, U. N., and van Overbeek, L. S. (2009). Endophytic colonization of potato (Solanum tuberosum L.) by a novel competent bacterial endophyte, Pseudomonas putida strain P9, and its effect on associated bacterial communities. Appl. Environ. Microbiol. 75, 3396–3406. doi: 10.1128/AEM.00491-09
Angelo, M. J. (2023). XII. 14 pesticide regulation in the US. Chem. Risk Gov. 12:290. doi: 10.4337/9781785369520.XII.14
Archetti, M., Úbeda, F., Fudenberg, D., Green, J., Pierce, N. E., and Yu, D. W. (2011). Let the right one in: a microeconomic approach to partner choice in mutualisms. Am. Nat. 177, 75–85. doi: 10.1086/657622
Ardakani, S. S., Heydari, A., Khorasani, N., and Arjmandi, R. (2010). Development of new bioformulations of Pseudomonas fluorescens and evaluation of these products against damping-off of cotton seedlings. J. Plant Pathol. 1, 83–88.
Arora, N. K., Khare, E., Oh, J. H., Kang, S. C., and Maheshwari, D. K. (2008). Diverse mechanisms adopted by fluorescent Pseudomonas PGC2 during the inhibition of Rhizoctonia solani and Phytophthora capsici. World J. Microbiol. Biotechnol. 24, 581–585. doi: 10.1007/s11274-007-9505-5
Aznar, A., Chen, N. W., Thomine, S., and Dellagi, A. (2015). Immunity to plant pathogens and iron homeostasis. Plant Sci. 240, 90–97. doi: 10.1016/j.plantsci.2015.08.022
Babin, D., Deubel, A., Jacquiod, S., Sørensen, S. J., Geistlinger, J., Grosch, R., et al. (2019). Impact of long-term agricultural management practices on soil prokaryotic communities. Soil Biol. Biochem. 129, 17–28. doi: 10.1016/j.soilbio.2018.11.002
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Badri, D. V., Chaparro, J. M., Zhang, R., Shen, Q., and Vivanco, J. M. (2013). Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288, 4502–4512. doi: 10.1074/jbc.M112.433300
Badri, D. V., Quintana, N., El Kassis, E. G., Kim, H. K., Choi, Y. H., Sugiyama, A., et al. (2009). An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 151, 2006–2017. doi: 10.1104/pp.109.147462
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Bakker, P. A., Glandorf, D. C., Viebahn, M., Ouwens, T. W., Smit, E., Leeflang, P., et al. (2002). Effects of Pseudomonas putida modified to produce phenazine-1-carboxylic acid and 2, 4-diacetylphloroglucinol on the microflora of field grown wheat. Antonie Van Leeuwenhoek 81, 617–624. doi: 10.1023/A:1020526126283
Balser, T. C., Kinzig, A. P., and Firestone, M. K. (2002). Linking soil microbial communities and ecosystem functioning. Funct. Consequences Biodiv. Emp. Progr. Theor. Ext. 2022, 265–293. doi: 10.1515/9781400847303.265
Baltrus, D. A., McCann, H. C., and Guttman, D. S. (2017). Evolution, genomics and epidemiology of Pseudomonas syringae: challenges in bacterial molecular plant pathology. Mol. Plant Pathol. 18, 152–168. doi: 10.1111/mpp.12506
Barahona, E., Navazo, A., Martínez-Granero, F., Zea-Bonilla, T., Pérez-Jiménez, R. M., Martín, M., et al. (2011). Pseudomonas fluorescens F113 mutant with enhanced competitive colonization ability and improved biocontrol activity against fungal root pathogens. Appl. Environ. Microbiol. 77, 5412–5419. doi: 10.1128/AEM.00320-11
Basler, M., Ho, B. T., and Mekalanos, J. J. (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894. doi: 10.1016/j.cell.2013.01.042
Berg, G., Grube, M., Schloter, M., and Smalla, K. (2014). Unraveling the plant microbiome: looking back and future perspectives. Front. Microbiol. 5:148. doi: 10.3389/fmicb.2014.00148
Bernal, P., Allsopp, L. P., Filloux, A., and Llamas, M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987. doi: 10.1038/ismej.2016.169
Bernal, P., Llamas, M. A., and Filloux, A. (2018). Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 20, 1–15. doi: 10.1111/1462-2920.13956
Berry, D., Stecher, B., Schintlmeister, A., Reichert, J., Brugiroux, S., Wild, B., et al. (2013). Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc. Natl. Acad. Sci. 110, 4720–4725. doi: 10.1073/pnas.1219247110
Bertrand, S., Bohni, N., Schnee, S., Schumpp, O., Gindro, K., and Wolfender, J.-L. (2014). Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 32, 1180–1204. doi: 10.1016/j.biotechadv.2014.03.001
Bhardwaj, D., Ansari, M. W., Sahoo, R. K., and Tuteja, N. (2014). Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb. Cell Factories 13, 1–10. doi: 10.1186/1475-2859-13-66
Bhatti, A. A., Haq, S., and Bhat, R. A. (2017). Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 111, 458–467. doi: 10.1016/j.micpath.2017.09.036
Bonaterra, A., Badosa, E., Daranas, N., Francés, J., Roselló, G., and Montesinos, E. (2022). Bacteria as biological control agents of plant diseases. Microorganisms 10:1759. doi: 10.3390/microorganisms10091759
Botelho, G. R., and Mendonça-Hagler, L. C. (2006). Fluorescent pseudomonads associated with the rhizosphere of crops: an overview. Braz. J. Microbiol. 37, 401–416. doi: 10.1590/S1517-83822006000400001
Bourigault, Y., Dupont, C. A., Desjardins, J. B., Doan, T., Bouteiller, M., Le Guenno, H., et al. (2023). Pseudomonas fluorescens MFE01 delivers a putative type VI secretion amidase that confers biocontrol against the soft-rot pathogen Pectobacterium atrosepticum. Environ. Microbiol. 25, 2564–2579. doi: 10.1111/1462-2920.16492
Bressan, M., Roncato, M.-A., Bellvert, F., Comte, G., Haichar, F., Achouak, W., et al. (2009). Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 3, 1243–1257. doi: 10.1038/ismej.2009.68
Buddrus-Schiemann, K., Schmid, M., Schreiner, K., Welzl, G., and Hartmann, A. (2010). Root colonization by Pseudomonas sp. DSMZ 13134 and impact on the indigenous rhizosphere bacterial community of barley. Microb. Ecol. 60, 381–393. doi: 10.1007/s00248-010-9720-8
Bundalovic-Torma, C., Lonjon, F., Desveaux, D., and Guttman, D. S. (2022). Diversity, evolution, and function of Pseudomonas syringae effectoromes. Annu. Rev. Phytopathol. 60, 211–236. doi: 10.1146/annurev-phyto-021621-121935
Burtnick, M. N., Brett, P. J., Harding, S. V., Ngugi, S. A., Ribot, W. J., Chantratita, N., et al. (2011). The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79, 1512–1525. doi: 10.1128/IAI.01218-10
Büttner, D. (2016). Behind the lines–actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40, 894–937. doi: 10.1093/femsre/fuw026
Callan, N. W., Mathre, D., and Miller, J. B. (1990). Bio-priming seed treatment for biological control of Pythium ultimum preemergence damping-off in sh-2 sweet corn. Plant Dis. 74, 368–372. doi: 10.1094/PD-74-0368
Camilios-Neto, D., Bonato, P., Wassem, R., Tadra-Sfeir, M. Z., Brusamarello-Santos, L. C., Valdameri, G., et al. (2014). Dual RNA-seq transcriptional analysis of wheat roots colonized by Azospirillum brasilense reveals up-regulation of nutrient acquisition and cell cycle genes. BMC Genomics 15, 1–13. doi: 10.1186/1471-2164-15-378
Capper, A., and Higgins, K. (1993). Application of Pseudomonas fluorescens isolates to wheat as potential biological control agents against take-all. Plant Pathol. 42, 560–567. doi: 10.1111/j.1365-3059.1993.tb01536.x
Chaparro, J. M., Badri, D. V., Bakker, M. G., Sugiyama, A., Manter, D. K., and Vivanco, J. M. (2013). Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 8:e55731. doi: 10.1371/journal.pone.0055731
Chaparro, J. M., Badri, D. V., and Vivanco, J. M. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. doi: 10.1038/ismej.2013.196
Chaudhary, P., Khati, P., Chaudhary, A., Maithani, D., Kumar, G., and Sharma, A. (2021). Cultivable and metagenomic approach to study the combined impact of nanogypsum and Pseudomonas taiwanensis on maize plant health and its rhizospheric microbiome. PLoS One 16:e0250574. doi: 10.1371/journal.pone.0250574
Chen, C., Bélanger, R. R., Benhamou, N., and Paulitz, T. C. (1999). Role of salicylic acid in systemic resistance induced by Pseudomonas spp. against Pythium aphanidermatum in cucumber roots. Eur. J. Plant Pathol. 105, 477–486. doi: 10.1023/A:1008743502784
Cheng, Z., Park, E., and Glick, B. R. (2007). 1-Aminocyclopropane-1-carboxylate deaminase from Pseudomonas putida UW4 facilitates the growth of canola in the presence of salt. Can. J. Microbiol. 53, 912–918. doi: 10.1139/w07-050
Chin-A-Woeng, T. F., Bloemberg, G. V., and Lugtenberg, B. J. (2003). Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol. 157, 503–523. doi: 10.1046/j.1469-8137.2003.00686.x
Chincholkar, S., Chaudhari, B., Talegaonkar, S., and Kothari, R. (2000). Microbial iron chelators: A sustainable tool for the biocontrol of plant diseases. Biocontrol Potential Expl. Sust. Agric. Crop Dis. Weeds Nematodes 1, 49–70. doi: 10.1007/978-1-4615-4209-4_5
Clarkson, J. P., Payne, T., Mead, A., and Whipps, J. (2002). Selection of fungal biological control agents of Sclerotium cepivorum for control of white rot by sclerotial degradation in a UK soil. Plant Pathol. 51, 735–745. doi: 10.1046/j.1365-3059.2002.00787.x
Classen, A. T., Sundqvist, M. K., Henning, J. A., Newman, G. S., Moore, J. A., Cregger, M. A., et al. (2015). Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: what lies ahead? Ecosphere 6, 1–21. doi: 10.1890/ES15-00217.1
Clough, S. E., Jousset, A., Elphinstone, J. G., and Friman, V. P. (2022). Combining in vitro and in vivo screening to identify efficient Pseudomonas biocontrol strains against the phytopathogenic bacterium Ralstonia solanacearum. Microbiology 11:e1283. doi: 10.1002/mbo3.1283
Colombo, C., Palumbo, G., He, J.-Z., Pinton, R., and Cesco, S. (2014). Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J. Soils Sediments 14, 538–548. doi: 10.1007/s11368-013-0814-z
Cordero, P., Príncipe, A., Jofré, E., Mori, G., and Fischer, S. (2014). Inhibition of the phytopathogenic fungus Fusarium proliferatum by volatile compounds produced by Pseudomonas. Arch. Microbiol. 196, 803–809. doi: 10.1007/s00203-014-1019-6
Cornelis, P., and Matthijs, S. (2002). Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4, 787–798. doi: 10.1046/j.1462-2920.2002.00369.x
Cui, K., and Shoemaker, S. P. (2018). Public perception of genetically-modified (GM) food: a nationwide Chinese consumer study. NPJ Sci. Food 2:10. doi: 10.1038/s41538-018-0018-4
Dar, G., Kamili, A., Chishti, M., Dar, S., Tantry, T., and Ahmad, F. (2016). Characterization of Aeromonas sobria isolated from fish Rohu (Labeo rohita) collected from polluted pond. J. Bacteriol. Parasitol. 7, 1–5. doi: 10.4172/2155-9597.1000273
De Pace, F., Nakazato, G., Pacheco, A., Boldrin de Paiva, J., Sperandio, V., and Dias da Silveira, W. (2010). The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect. Immun. 78, 4990–4998. doi: 10.1128/IAI.00531-10
Deacon, J. (1991). Significance of ecology in the development of biocontrol agents against soil-borne plant pathogens. Biocontrol Sci. Tech. 1, 5–20. doi: 10.1080/09583159109355181
Deising, H. B., Gase, I., and Kubo, Y. (2017). The unpredictable risk imposed by microbial secondary metabolites: how safe is biological control of plant diseases? J. Plant Dis. Prot. 124, 413–419. doi: 10.1007/s41348-017-0109-5
Dennert, F., Imperiali, N., Staub, C., Schneider, J., Laessle, T., Zhang, T., et al. (2018). Conservation tillage and organic farming induce minor variations in Pseudomonas abundance, their antimicrobial function and soil disease resistance. FEMS Microbiol. Ecol. 94:fiy075. doi: 10.1093/femsec/fiy075
Devaraj, S., Amirthalingam, M., Sabapathy, P. C., Govindan, S., Palanisamy, S., and Kathirvel, P. (2019). Anthelmintic efficacy of glycolipid biosurfactant produced by Pseudomonas plecoglossicida: an insight from mutant and transgenic forms of Caenorhabditis elegans. Biodegradation 30, 203–214. doi: 10.1007/s10532-018-9831-3
Dillon, M. M., Almeida, R. N., Laflamme, B., Martel, A., Weir, B. S., Desveaux, D., et al. (2019). Molecular evolution of Pseudomonas syringae type III secreted effector proteins. Front. Plant Sci. 10:418. doi: 10.3389/fpls.2019.00418
Dillon, M. M., Ruiz-Bedoya, T., Bundalovic-Torma, C., Guttman, K. M., Kwak, H., Middleton, M. A., et al. (2021). Comparative genomic insights into the epidemiology and virulence of plant pathogenic pseudomonads from Turkey. Microbial. Genom. 7:000585. doi: 10.1099/mgen.0.000585
Dimkpa, C. O., Mclean, J. E., Britt, D. W., and Anderson, A. J. (2012a). CuO and ZnO nanoparticles differently affect the secretion of fluorescent siderophores in the beneficial root colonizer, Pseudomonas chlororaphis O6. Nanotoxicology 6, 635–642. doi: 10.3109/17435390.2011.598246
Dimkpa, C. O., Zeng, J., McLean, J. E., Britt, D. W., Zhan, J., and Anderson, A. J. (2012b). Production of indole-3-acetic acid via the indole-3-acetamide pathway in the plant-beneficial bacterium Pseudomonas chlororaphis O6 is inhibited by ZnO nanoparticles but enhanced by CuO nanoparticles. Appl. Environ. Microbiol. 78, 1404–1410. doi: 10.1128/AEM.07424-11
Dinesh, R., Srinivasan, V., Hamza, S., and Manjusha, A. (2010). Short-term incorporation of organic manures and biofertilizers influences biochemical and microbial characteristics of soils under an annual crop [turmeric (Curcuma longa L.)]. Bioresour. Technol. 101, 4697–4702. doi: 10.1016/j.biortech.2010.01.108
Ding, C., Shen, Q., Zhang, R., and Chen, W. (2013). Evaluation of rhizosphere bacteria and derived bio-organic fertilizers as potential biocontrol agents against bacterial wilt (Ralstonia solanacearum) of potato. Plant Soil 366, 453–466. doi: 10.1007/s11104-012-1425-y
Dowling, D. N., and O'Gara, F. (1994). Metabolites of Pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol. 12, 133–141. doi: 10.1016/0167-7799(94)90091-4
Drenkard, E., and Ausubel, F. M. (2002). Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416, 740–743. doi: 10.1038/416740a
Drigo, B., Van Veen, J. A., and Kowalchuk, G. A. (2009). Specific rhizosphere bacterial and fungal groups respond differently to elevated atmospheric CO2. ISME J. 3, 1204–1217. doi: 10.1038/ismej.2009.65
Duffy, B. K., and Défago, G. V. (1999). Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65, 2429–2438. doi: 10.1128/AEM.65.6.2429-2438.1999
Durán, D., Bernal, P., Vazquez-Arias, D., Blanco-Romero, E., Garrido-Sanz, D., Redondo-Nieto, M., et al. (2021). Pseudomonas fluorescens F113 type VI secretion systems mediate bacterial killing and adaption to the rhizosphere microbiome. Sci. Rep. 11:5772. doi: 10.1038/s41598-021-85218-1
Dwivedi, D., and Johri, B. (2003). Antifungals from fluorescent pseudomonads: biosynthesis and regulation. Curr. Sci. 25, 1693–1703.
Edwards, J., Johnson, C., Santos-Medellín, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. 112, E911–E920. doi: 10.1073/pnas.1414592112
el Zahar Haichar, F., Heulin, T., Guyonnet, J. P., and Achouak, W. (2016). Stable isotope probing of carbon flow in the plant holobiont. Curr. Opin. Biotechnol. 41, 9–13. doi: 10.1016/j.copbio.2016.02.023
El-Ghamry, A., Mosa, A. A., Alshaal, T., and El-Ramady, H. (2018). Nanofertilizers vs. biofertilizers: new insights. Environ. Biodiv. Soil Secur. 2, 51–65. doi: 10.21608/jenvbs.2019.6775.1043
Elkahoui, S., Djébali, N., Yaich, N., Azaiez, S., Hammami, M., Essid, R., et al. (2015). Antifungal activity of volatile compounds-producing Pseudomonas P2 strain against Rhizoctonia solani. World J. Microbiol. Biotechnol. 31, 175–185. doi: 10.1007/s11274-014-1772-3
El-Mougy, N., and Abdel-Kader, M. (2008). Long-term activity of bio-priming seed treatment for biological control of faba bean root rot pathogens. Australas. Plant Pathol. 37, 464–471. doi: 10.1071/AP08043
Elsharkawy, M. M., Sakran, R. M., Ahmad, A. A., Behiry, S. I., Abdelkhalek, A., Hassan, M. M., et al. (2022). Induction of systemic resistance against sheath blight in rice by different Pseudomonas isolates. Life 12:349. doi: 10.3390/life12030349
Eriksson, D., Kershen, D., Nepomuceno, A., Pogson, B. J., Prieto, H., Purnhagen, K., et al. (2019). A comparison of the EU regulatory approach to directed mutagenesis with that of other jurisdictions, consequences for international trade and potential steps forward. New Phytol. 222, 1673–1684. doi: 10.1111/nph.15627
Fernbach, P. M., Light, N., Scott, S. E., Inbar, Y., and Rozin, P. (2019). Extreme opponents of genetically modified foods know the least but think they know the most. Nat. Hum. Behav. 3, 251–256. doi: 10.1038/s41562-018-0520-3
Forni, C., Duca, D., and Glick, B. R. (2017). Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 410, 335–356. doi: 10.1007/s11104-016-3007-x
Fravel, D. (2005). Commercialization and implementation of biocontrol. Annu. Rev. Phytopathol. 43, 337–359. doi: 10.1146/annurev.phyto.43.032904.092924
Gaiero, J. R., McCall, C. A., Thompson, K. A., Day, N. J., Best, A. S., and Dunfield, K. E. (2013). Inside the root microbiome: bacterial root endophytes and plant growth promotion. Am. J. Bot. 100, 1738–1750. doi: 10.3732/ajb.1200572
Garbeva, P., Van Veen, J. A., and Van Elsas, J. D. (2004). Assessment of the diversity, and antagonism towards Rhizoctonia solani AG3, of Pseudomonas species in soil from different agricultural regimes. FEMS Microbiol. Ecol. 47, 51–64. doi: 10.1016/S0168-6496(03)00234-4
Gehlot, P., and Singh, J. (2018). Fungi and their role in sustainable development: Current perspectives. Cham: Springer.
Geiger, F., Bengtsson, J., Berendse, F., Weisser, W. W., Emmerson, M., Morales, M. B., et al. (2010). Persistent negative effects of pesticides on biodiversity and biological control potential on European farmland. Basic Appl. Ecol. 11, 97–105. doi: 10.1016/j.baae.2009.12.001
Ghssein, G., and Ezzeddine, Z. (2022). A review of Pseudomonas aeruginosa metallophores: pyoverdine, pyochelin and pseudopaline. Biology 11:1711. doi: 10.3390/biology11121711
Girard, L., Höfte, M., and Mot, R. D. (2020). Lipopeptide families at the interface between pathogenic and beneficial Pseudomonas-plant interactions. Crit. Rev. Microbiol. 46, 397–419. doi: 10.1080/1040841X.2020.1794790
Glandorf, D. C. M. (2019). Re-evaluation of biosafety questions on genetically modified biocontrol bacteria. Eur. J. Plant Pathol. 154, 43–51. doi: 10.1007/s10658-018-1598-1
Glandorf, D. C., Verheggen, P., Jansen, T., Jorritsma, J.-W., Smit, E., Leeflang, P., et al. (2001). Effect of genetically modified Pseudomonas putida WCS358r on the fungal rhizosphere microflora of field-grown wheat. Appl. Environ. Microbiol. 67, 3371–3378. doi: 10.1128/AEM.67.8.3371-3378.2001
Glick, B. R., and Gamalero, E. (2021). Recent developments in the study of plant microbiomes. Microorganisms 9:1533. doi: 10.3390/microorganisms9071533
Glick, B. R., Karaturovíc, D. M., and Newell, P. C. (1995). A novel procedure for rapid isolation of plant growth promoting pseudomonads. Can. J. Microbiol. 41, 533–536. doi: 10.1139/m95-070
Glick, B. R., and Skof, Y. C. (1986). Environmental implications of recombinant DNA technology. Biotechnol. Adv. 4, 261–277. doi: 10.1016/0734-9750(86)90312-5
Golbashy, M., Sabahi, H., Allahdadi, I., Nazokdast, H., and Hosseini, M. (2017). Synthesis of highly intercalated urea-clay nanocomposite via domestic montmorillonite as eco-friendly slow-release fertilizer. Arch. Agron. Soil Sci. 63, 84–95. doi: 10.1080/03650340.2016.1177175
Guan, Y., Bak, F., Hennessy, R. C., Horn Herms, C., Elberg, C. L., Dresbøll, D. B., et al. (2024). The potential of Pseudomonas fluorescens SBW25 to produce viscosin enhances wheat root colonization and shapes root-associated microbial communities in a plant genotype-dependent manner in soil systems. Msphere 9, e00294–e00224. doi: 10.1128/msphere.00294-24
Gull, M., and Hafeez, F. Y. (2012). Characterization of siderophore producing bacterial strain Pseudomonas fluorescens Mst 8.2 as plant growth promoting and biocontrol agent in wheat. Afr. J. Microbiol. Res. 6, 6308–6318. doi: 10.5897/AJMR12.1285
Haichar, F., Marol, C., Berge, O., Rangel-Castro, J. I., Prosser, J. I., Balesdent, J., et al. (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2, 1221–1230. doi: 10.1038/ismej.2008.80
Haichar, F., Roncato, M. A., and Achouak, W. (2012). Stable isotope probing of bacterial community structure and gene expression in the rhizosphere of Arabidopsis thaliana. FEMS Microbiol. Ecol. 81, 291–302. doi: 10.1111/j.1574-6941.2012.01345.x
Hakim, S., Naqqash, T., Nawaz, M. S., Laraib, I., Siddique, M. J., Zia, R., et al. (2021). Rhizosphere engineering with plant growth-promoting microorganisms for agriculture and ecological sustainability. Front. Sust. Food Syst. 5:617157. doi: 10.3389/fsufs.2021.617157
Hamid, M., Siddiqui, I., and Shahid Shaukat, S. (2003). Improvement of Pseudomonas fluorescens CHA0 biocontrol activity against root-knot nematode by the addition of ammonium molybdate. Lett. Appl. Microbiol. 36, 239–244. doi: 10.1046/j.1472-765X.2003.01299.x
Hamid, B., Zaman, M., Farooq, S., Fatima, S., Sayyed, R. Z., Baba, Z. A., et al. (2021). Bacterial plant biostimulants: a sustainable way towards improving growth, productivity, and health of crops. Sustain. For. 13:2856. doi: 10.3390/su13052856
Haney, C. H., Wiesmann, C. L., Shapiro, L. R., Melnyk, R. A., O'Sullivan, L. R., Khorasani, S., et al. (2018). Rhizosphere-associated Pseudomonas induce systemic resistance to herbivores at the cost of susceptibility to bacterial pathogens. Mol. Ecol. 27, 1833–1847. doi: 10.1111/mec.14400
Hao, Y., Charles, T. C., and Glick, B. R. (2011). ACC deaminase activity in avirulent Agrobacterium tumefaciens D3. Can. J. Microbiol. 57, 278–286. doi: 10.1139/w11-006
Hardaker, J., and Hardwick, R. (1978). A note on Rhizobium inoculation of beans (Phaseolus vulgaris) using the fluid drill technique. Exp. Agric. 14, 17–21. doi: 10.1017/S0014479700008280
Hennessy, R. C., Glaring, M. A., Olsson, S., and Stougaard, P. (2017). Transcriptomic profiling of microbe–microbe interactions reveals the specific response of the biocontrol strain P. fluorescens In5 to the phytopathogen Rhizoctonia solani. BMC. Res. Notes 10, 1–9. doi: 10.1186/s13104-017-2704-8
Hera, C. (1995). The role of inorganic fertilizers and their management practices. Fertilizer Res. 43, 63–81. doi: 10.1007/BF00747684
Hernández-León, R., Rojas-Solís, D., Contreras-Pérez, M., del Carmen Orozco-Mosqueda, M., Macías-Rodríguez, L. I., Reyes-de la Cruz, H., et al. (2015). Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol. Control 81, 83–92. doi: 10.1016/j.biocontrol.2014.11.011
Hider, R. C., and Kong, X. (2010). Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657. doi: 10.1039/b906679a
Ho, B. T., Dong, T. G., and Mekalanos, J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21. doi: 10.1016/j.chom.2013.11.008
Ho, Y.-N., Hoo, S. Y., Wang, B.-W., Hsieh, C.-T., Lin, C.-C., Sun, C.-H., et al. (2021). Specific inactivation of an antifungal bacterial siderophore by a fungal plant pathogen. ISME J. 15, 1858–1861. doi: 10.1038/s41396-020-00871-0
Hoffland, E., Hakulinen, J., and Van Pelt, J. A. (1996). Comparison of systemic resistance induced by avirulent and nonpathogenic Pseudomonas species. Phytopathology 86, 757–762. doi: 10.1094/Phyto-86-757
Hood, R. D., Singh, P., Hsu, F., Güvener, T., Carl, M. A., Trinidad, R. R., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Huang, A. C., Jiang, T., Liu, Y.-X., Bai, Y.-C., Reed, J., Qu, B., et al. (2019). A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364:eaau6389. doi: 10.1126/science.aau6389
Imperato, V., Portillo-Estrada, M., McAmmond, B. M., Douwen, Y., Van Hamme, J. D., Gawronski, S. W., et al. (2019). Genomic diversity of two hydrocarbon-degrading and plant growth-promoting Pseudomonas species isolated from the oil field of Bóbrka (Poland). Genes 10:443. doi: 10.3390/genes10060443
Iqbal, N., Khan, N. A., Ferrante, A., Trivellini, A., Francini, A., and Khan, M. (2017). Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Front. Plant Sci. 8:475. doi: 10.3389/fpls.2017.00475
Itelima, J., Bang, W., Onyimba, I., Sila, M., and Egbere, O. (2018). Bio-fertilizers as key player in enhancing soil fertility and crop productivity: A review. Cham: Springer.
Jacobsen, C. S., and Hjelmsø, M. H. (2014). Agricultural soils, pesticides and microbial diversity. Curr. Opin. Biotechnol. 27, 15–20. doi: 10.1016/j.copbio.2013.09.003
Jain, A., Chakraborty, J., and Das, S. (2020). Underlying mechanism of plant–microbe crosstalk in shaping microbial ecology of the rhizosphere. Acta Physiol. Plant. 42:8. doi: 10.1007/s11738-019-3000-0
Jambhulkar, P. P., Sharma, P., and Yadav, R. (2016). “Delivery systems for introduction of microbial inoculants in the field” in Microbial inoculants in sustainable agricultural productivity: Vol. 2: Functional applications. eds. D. P. Singh, H. B. Singh, and R. Prabha (New Delhi: Springer), 199–218.
Jha, C. K., Aeron, A., Patel, B. V., Maheshwari, D. K., and Saraf, M. (2011). Enterobacter: role in plant growth promotion. Bac. Agrobiol. Plant Growth Resp. 2011, 159–182. doi: 10.1007/978-3-642-20332-9_8
Jiang, S., Yao, J., Ma, K.-W., Zhou, H., Song, J., He, S. Y., et al. (2013). Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 9:e1003715. doi: 10.1371/journal.ppat.1003715
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, M. G. K., Iqbal, S., and Fosu-Nyarko, J. (2024). “Chapter 13 - regulation of CRISPR-edited plants in Australia and New Zealand” in Global regulatory outlook for CRISPRized plants. eds. K. A. Abd-Elsalam and A. Ahmad (New York, NY: Academic Press), 281–292.
Jousset, A., Rochat, L., Lanoue, A., Bonkowski, M., Keel, C., and Scheu, S. (2011). Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol. Plant-Microbe Interact. 24, 352–358. doi: 10.1094/MPMI-09-10-0208
Kanimozhi, S., and Perinbam, K. (2011). Siderophore production by Pseudomonas fluorescens Lp1 and its application as biocontrol agent. J. Pharm. Res. 4, 3175–3179.
Karthikeyan, V., and Gnanamanickam, S. S. (2008). Biological control of Setaria blast (Magnaporthe grisea) with bacterial strains. Crop Prot. 27, 263–267. doi: 10.1016/j.cropro.2007.05.013
Kataria, H., Wilmsmeier, B., and Buchenauer, H. (2002). Efficacy of Pseudomonas fluorescens strains and some modern fungicides for control of Rhizoctonia solani AG-4 in bean and cucumber/Wirksamkeit von Pseudomonas fluorescens-Stāmmen und einigen modernen Fungiziden bei der Bekämpfung von Rhizoctonia solani AG-4 an Bohnen und Gurken. J. Plant Dis. Prot. 1, 384–400.
Ketabchi, S., Charehgani, H., and Majzoob, S. (2016). Impact of rhizosphere antagonistic Bacteria and urea fertilizer on root knot nematode (Meloidogyneincognita) under Green house condition. JAPS 26, 1780–1786.
Khan, M., Seto, D., Subramaniam, R., and Desveaux, D. (2018). Oh, the places they'll go! A survey of phytopathogen effectors and their host targets. Plant J. 93, 651–663. doi: 10.1111/tpj.13780
Khatoon, Z., Huang, S., Rafique, M., Fakhar, A., Kamran, M. A., and Santoyo, G. (2020). Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J. Environ. Manag. 273:111118. doi: 10.1016/j.jenvman.2020.111118
Kita, W., Pietr, S., Nowak, W., and Sowiński, J. (2004). The effect of biological and conventional method of plant protection on the yield and healthiness of two wheat cultivars. Lublin: Annales Universitatis Mariae Curie-Sklodowska.
Kloepper, J. W., Leong, J., Teintze, M., and Schroth, M. N. (1980). Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286, 885–886. doi: 10.1038/286885a0
Koch, E., Ole Becker, J., Berg, G., Hauschild, R., Jehle, J., Köhl, J., et al. (2018). Biocontrol of plant diseases is not an unsafe technology! J. Plant Dis. Prot. 125, 121–125. doi: 10.1007/s41348-018-0158-4
Kolseth, A.-K., D’Hertefeldt, T., Emmerich, M., Forabosco, F., Marklund, S., Cheeke, T. E., et al. (2015). Influence of genetically modified organisms on agro-ecosystem processes. Agric. Ecosyst. Environ. 214, 96–106. doi: 10.1016/j.agee.2015.08.021
Kramer, J., Özkaya, Ö., and Kümmerli, R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163. doi: 10.1038/s41579-019-0284-4
Krewulak, K. D., and Vogel, H. J. (2008). Structural biology of bacterial iron uptake. Biochimica et Biophysica Acta (BBA). Biomembranes 1778, 1781–1804. doi: 10.1016/j.bbamem.2007.07.026
Kvitko, B. H., Park, D. H., Velásquez, A. C., Wei, C.-F., Russell, A. B., Martin, G. B., et al. (2009). Deletions in the repertoire of Pseudomonas syringae pv. Tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5:e1000388. doi: 10.1371/journal.ppat.1000388
Laflamme, B., Dillon, M. M., Martel, A., Almeida, R. N., Desveaux, D., and Guttman, D. S. (2020). The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science 367, 763–768. doi: 10.1126/science.aax4079
Lahlali, R., Ezrari, S., Radouane, N., Kenfaoui, J., Esmaeel, Q., El Hamss, H., et al. (2022). Biological control of plant pathogens: a global perspective. Microorganisms 10:596. doi: 10.3390/microorganisms10030596
Lanoue, A., Burlat, V., Henkes, G. J., Koch, I., Schurr, U., and Röse, U. S. (2010). De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New Phytol. 185, 577–588. doi: 10.1111/j.1469-8137.2009.03066.x
Lassoued, R., Macall, D. M., Hesseln, H., Phillips, P. W., and Smyth, S. J. (2019). Benefits of genome-edited crops: expert opinion. Transgenic Res. 28, 247–256. doi: 10.1007/s11248-019-00118-5
Lau, C. K., Krewulak, K. D., and Vogel, H. J. (2016). Bacterial ferrous iron transport: the Feo system. FEMS Microbiol. Rev. 40, 273–298. doi: 10.1093/femsre/fuv049
Law, J. W.-F., Ser, H.-L., Khan, T. M., Chuah, L.-H., Pusparajah, P., Chan, K.-G., et al. (2017). The potential of Streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Front. Microbiol. 8:3. doi: 10.3389/fmicb.2017.00003
Lawongsa, P., Boonkerd, N., Wongkaew, S., O’Gara, F., and Teaumroong, N. (2008). Molecular and phenotypic characterization of potential plant growth-promoting Pseudomonas from rice and maize rhizospheres. World J. Microbiol. Biotechnol. 24, 1877–1884. doi: 10.1007/s11274-008-9685-7
Lee, G.-H., and Choi, K.-C. (2020). Adverse effects of pesticides on the functions of immune system. Comp. Biochem. Physiol. Toxicol. Pharmacol. 235:108789. doi: 10.1016/j.cbpc.2020.108789
Lee, H. S., Han, D. S., Choi, S. J., Choi, S. W., Kim, D. S., Bai, D. H., et al. (2000). Purification, characterization, and primary structure of a chitinase from Pseudomonas sp. YHS-A2. Appl. Microbiol. Biotechnol. 54, 397–405. doi: 10.1007/s002530000408
Lee, A. H.-Y., Hurley, B., Felsensteiner, C., Yea, C., Ckurshumova, W., Bartetzko, V., et al. (2012). A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 8:e1002523. doi: 10.1371/journal.ppat.1002523
Lee, C.-H., Kempf, H.-J., Lim, Y.-H., and Cho, Y.-H. (2000). Biocontrol activity of Pseudomonas cepacia AF2001 and anthelmintic activity of its novel metabolite, cepacidine A. J. Microbiol. Biotechnol. 10, 568–571.
Leeflang, P., Smit, E., Glandorf, D., Van Hannen, E., and Wernars, K. (2002). Effects of Pseudomonas putida WCS358r and its genetically modified phenazine producing derivative on the Fusarium population in a field experiment, as determined by 18S rDNA analysis. Soil Biol. Biochem. 34, 1021–1025. doi: 10.1016/S0038-0717(02)00035-4
Leeman, M., Van Pelt, J., Den Ouden, F., Heinsbroek, M., Bakker, P., and Schippers, B. (1995). Induction of systemic resistance by Pseudomonas fluorescens in radish cultivars differing in susceptibility to fusarium wilt, using a novel bioassay. Eur. J. Plant Pathol. 101, 655–664. doi: 10.1007/BF01874869
Lemanceau, P., and Alabouvette, C. (1993). Suppression of Fusarium wilts by fluorescent pseudomonads: mechanisms and applications. Biocontrol Sci. Tech. 3, 219–234. doi: 10.1080/09583159309355278
Leontidou, K., Genitsaris, S., Papadopoulou, A., Kamou, N., Bosmali, I., Matsi, T., et al. (2020). Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 10:14857. doi: 10.1038/s41598-020-71652-0
Ligon, J. M., Hill, D. S., Hammer, P. E., Torkewitz, N. R., Hofmann, D., Kempf, H.-J., et al. (2000). Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag. Sci. 56, 688–695. doi: 10.1002/1526-4998(200008)56:8<688::AID-PS186>3.0.CO;2-V
Loh, J., Pierson, E. A., Pierson, L. S. III, Stacey, G., and Chatterjee, A. (2002). Quorum sensing in plant-associated bacteria. Curr. Opin. Plant Biol. 5, 285–290. doi: 10.1016/S1369-5266(02)00274-1
Loper, J. E., and Gross, H. (2007). Genomic analysis of antifungal metabolite production by Pseudomonas fluorescens Pf-5. New Persp. Appr. Plant Growth-Promoting Rhizobacteria Res. 119, 265–278. doi: 10.1007/978-1-4020-6776-1_4
Lucht, J. M. (2015). Public acceptance of plant biotechnology and GM crops. Viruses 7, 4254–4281. doi: 10.3390/v7082819
Lv, N., Tao, C., Ou, Y., Wang, J., Deng, X., Liu, H., et al. (2023). Root-associated antagonistic Pseudomonas spp. contribute to soil Suppressiveness against Banana Fusarium wilt disease of Banana. Microbiol. Spectr. 11:e0352522. doi: 10.1128/spectrum.03525-22
Ma, Y., Látr, A., Rocha, I., Freitas, H., Vosátka, M., and Oliveira, R. S. (2019). Delivery of inoculum of Rhizophagus irregularis via seed coating in combination with Pseudomonas libanensis for cowpea production. Agronomy 9:33. doi: 10.3390/agronomy9010033
Marchi, M., Boutin, M., Gazengel, K., Rispe, C., Gauthier, J. P., Guillerm-Erckelboudt, A. Y., et al. (2013). Genomic analysis of the biocontrol strain P seudomonas fluorescens pf 29 Arp with evidence of T 3 SS and T 6 SS gene expression on plant roots. Environ. Microbiol. Rep. 5, 393–403. doi: 10.1111/1758-2229.12048
Marilley, L., Hartwig, U., and Aragno, M. (1999). Influence of an elevated atmospheric CO2 content on soil and rhizosphere bacterial communities beneath Lolium perenne and Trifolium repens under field conditions. Microb. Ecol. 38, 39–49. doi: 10.1007/s002489900155
Mark, G. L., Morrissey, J. P., Higgins, P., and O'Gara, F. (2006). Molecular-based strategies to exploit Pseudomonas biocontrol strains for environmental biotechnology applications. FEMS Microbiol. Ecol. 56, 167–177. doi: 10.1111/j.1574-6941.2006.00056.x
Marrez, D. A., Abdel-Rahman, G. N., and Salem, S. H. (2019). Evaluation of Pseudomonas fluorescens extracts as biocontrol agents against some foodborne microorganisms. Jordan J. Biol. Sci. 12, 535–541.
Martel, A., Laflamme, B., Seto, D., Bastedo, D. P., Dillon, M. M., Almeida, R. N., et al. (2020). Immunodiversity of the Arabidopsis ZAR1 NLR is conveyed by receptor-like cytoplasmic kinase sensors. Front. Plant Sci. 11:1290. doi: 10.3389/fpls.2020.01290
Mavrodi, D. V., Mavrodi, O. V., Elbourne, L. D., Tetu, S., Bonsall, R. F., Parejko, J., et al. (2018). Long-term irrigation affects the dynamics and activity of the wheat rhizosphere microbiome. Front. Plant Sci. 9:345. doi: 10.3389/fpls.2018.00345
McGuire, S. (2015). FAO, IFAD, and WFP. The state of food insecurity in the world 2015: meeting the 2015 international hunger targets: taking stock of uneven progress. Adv. Nutr. 6, 623–624
McManus, P., Ravenscroft, A., and Fulbright, D. (1993). Inhibition of Tilletia laevis teliospore germination and suppression of common bunt of wheat by Pseudomonas fluorescens. Plant Dis. 77:1012. doi: 10.1094/PD-77-1012
Meena, R. S., Kumar, S., Datta, R., Lal, R., Vijayakumar, V., Brtnicky, M., et al. (2020). Impact of agrochemicals on soil microbiota and management: a review. Land 9:34. doi: 10.3390/land9020034
Mehmood, N., Saeed, M., Zafarullah, S., Hyder, S., Rizvi, Z. F., Gondal, A. S., et al. (2023). Multifaceted impacts of plant-beneficial pseudomonas spp. in managing various plant diseases and crop yield improvement. ACS Omega 8, 22296–22315. doi: 10.1021/acsomega.3c00870
Merkley, E. (2020). Anti-intellectualism, populism, and motivated resistance to expert consensus. Public Opin. Q. 84, 24–48. doi: 10.1093/poq/nfz053
Meziane, H., Van Der Sluis, I., Van Loon, L. C., Höfte, M., and Bakker, P. A. (2005). Determinants of Pseudomonas putida WCS358 involved in inducing systemic resistance in plants. Mol. Plant Pathol. 6, 177–185. doi: 10.1111/j.1364-3703.2005.00276.x
Mizoguchi, H., Fleischmann, T., Komuro, M., Hirai, T., Ikeda, A., Saito, K., et al. (2024). Comparative analysis of GMO regulatory requirements for AAV vectors in the EU and Japan focusing on the shedding data and containment measures. Exp. Opin. Biol. Ther. 24, 529–542. doi: 10.1080/14712598.2024.2371042
Moissenet, D., and Khedher, M. (2011). Virulence factors in Pseudomonas aeruginosa: mechanisms and modes of regulation. Ann. Biol. Clin. 69, 393–403. doi: 10.1684/abc.2011.0589
Montes-Osuna, N., Cernava, T., Gómez-Lama Cabanás, C., Berg, G., and Mercado-Blanco, J. (2022). Identification of volatile organic compounds emitted by two beneficial endophytic pseudomonas strains from olive roots. Plan. Theory 11:318. doi: 10.3390/plants11030318
Morris, C. E., Lamichhane, J. R., Nikolić, I., Stanković, S., and Moury, B. (2019). The overlapping continuum of host range among strains in the Pseudomonas syringae complex. Phytopathol. Res. 1, 1–16. doi: 10.1186/s42483-018-0010-6
Morris, C. E., Monteil, C. L., and Berge, O. (2013). The life history of Pseudomonas syringae: linking agriculture to earth system processes. Annu. Rev. Phytopathol. 51, 85–104. doi: 10.1146/annurev-phyto-082712-102402
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Murdoch, S. L., Trunk, K., English, G., Fritsch, M. J., Pourkarimi, E., and Coulthurst, S. J. (2011). The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 193, 6057–6069. doi: 10.1128/JB.05671-11
Nadeem, S. M., Naveed, M., Ayyub, M., Khan, M. Y., Ahmad, M., and Zahir, Z. A. (2016). Potential, limitations and future prospects of Pseudomonas spp. for sustainable agriculture and environment: a review. Soil and Environ. 35, 106–145.
Nandakumar, R., Babu, S., Viswanathan, R., Raguchander, T., and Samiyappan, R. (2001). Induction of systemic resistance in rice against sheath blight disease by Pseudomonas fluorescens. Soil Biol. Biochem. 33, 603–612. doi: 10.1016/S0038-0717(00)00202-9
Neal, A. L., Ahmad, S., Gordon-Weeks, R., and Ton, J. (2012). Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One 7:e35498. doi: 10.1371/journal.pone.0035498
Neilands, J. (1952). A crystalline organo-iron pigment from a rust fungus (Ustilago sphaerogena). J. Am. Chem. Soc. 74, 4846–4847. doi: 10.1021/ja01139a033
Neilands, J. (1995). Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726. doi: 10.1074/jbc.270.45.26723
Nielsen, T., Thrane, C., Christophersen, C., Anthoni, U., and Sørensen, J. (2000). Structure, production characteristics and fungal antagonism of tensin–a new antifungal cyclic lipopeptide from Pseudomonas fluorescens strain 96.578. J. Appl. Microbiol. 89, 992–1001. doi: 10.1046/j.1365-2672.2000.01201.x
Niemann, S., Keel, C., Pühler, A., and Selbitschka, W. (1997). Biocontrol strain Pseudomonas fluorescens CHA0 and its genetically modified derivative with enhanced biocontrol capability exert comparable effects on the structure of a Sinorhizobium meliloti population in gnotobiotic systems. Biol. Fertil. Soils 25, 240–244. doi: 10.1007/s003740050309
Nikel, P. I., Martínez-García, E., and De Lorenzo, V. (2014). Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 12, 368–379. doi: 10.1038/nrmicro3253
Norabadi, M. T., Sahebani, N., and Etebarian, H. R. (2014). Biological control of root-knot nematode (Meloidogyne javanica) disease by Pseudomonas fluorescens (Chao). Arch. Phytopathol. Plant Protect. 47, 615–621. doi: 10.1080/03235408.2013.816102
O’Callaghan, M. (2016). Microbial inoculation of seed for improved crop performance: issues and opportunities. Appl. Microbiol. Biotechnol. 100, 5729–5746. doi: 10.1007/s00253-016-7590-9
Ongley, E. D., Xiaolan, Z., and Tao, Y. (2010). Current status of agricultural and rural non-point source pollution assessment in China. Environ. Pollut. 158, 1159–1168. doi: 10.1016/j.envpol.2009.10.047
Orozco-Mosqueda, M. D. C., Duan, J., DiBernardo, M., Zetter, E., Campos-García, J., Glick, B. R., et al. (2019). The production of ACC deaminase and Trehalose by the plant growth promoting bacterium Pseudomonas sp. UW4 synergistically protect tomato plants against salt stress. Front. Microbiol. 10:1392. doi: 10.3389/fmicb.2019.01392
Pahalvi, H. N., Rafiya, L., Rashid, S., Nisar, B., and Kamili, A. N. (2021). “Chemical fertilizers and their impact on soil health” in Microbiota and biofertilizers, Vol 2: Ecofriendly tools for reclamation of degraded soil environs. eds. G. H. Dar, R. A. Bhat, M. A. Mehmood, and K. R. Hakeem (Cham: Springer International Publishing), 1–20. doi: 10.1007/978-3-030-61010-4_1
Palleroni, N. J. (2015). “Pseudomonas” in Bergey's manual of systematics of Archaea and Bacteria. ed. W. B. Whitman (Hoboken, NJ: Wiley).
Pangesti, N., Reichelt, M., van de Mortel, J. E., Kapsomenou, E., Gershenzon, J., van Loon, J. J., et al. (2016). Jasmonic acid and ethylene signaling pathways regulate glucosinolate levels in plants during rhizobacteria-induced systemic resistance against a leaf-chewing herbivore. J. Chem. Ecol. 42, 1212–1225. doi: 10.1007/s10886-016-0787-7
Parani, K., and Saha, B. (2012). Prospects of using phosphate solubilizing Pseudomonas as bio fertilizer. Eur. J. Biol. Sci. 4, 40–44. doi: 10.5829/idosi.ejbs.2012.4.2.63117
Pathak, A., Sharma, A., Johri, B., and Sharma, A. (2004). Pseudomonas strain GRP3 induces systemic resistance to sheath blight in rice. Los Baños: International Rice Research Notes.
Pathak, R., Shrestha, A., Lamichhane, J., and Gauchan, D. P. (2017). PGPR in biocontrol: mechanisms and roles in disease suppression. Int. J. Agron. Agri. R 11, 69–80.
Peix, A., Ramírez-Bahena, M.-H., and Velázquez, E. (2009). Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 9, 1132–1147. doi: 10.1016/j.meegid.2009.08.001
Persello-Cartieaux, F., David, P., Sarrobert, C., Thibaud, M.-C., Achouak, W., Robaglia, C., et al. (2001). Utilization of mutants to analyze the interaction between Arabidopsis thaliana and its naturally root-associated Pseudomonas. Planta 212, 190–198. doi: 10.1007/s004250000384
Pill, W. G. (1991). Advances in fluid drilling. HortTechnology 1, 59–65. doi: 10.21273/HORTTECH.1.1.59
Pimentel, D., McLaughlin, L., Zepp, A., Lakitan, B., Kraus, T., Kleinman, P., et al. (1993). Environmental and economic effects of reducing pesticide use in agriculture. Agric. Ecosyst. Environ. 46, 273–288. doi: 10.1016/0167-8809(93)90030-S
Prathuangwong, S., Athinuwat, D., Chuaboon, W., Chatnaparat, T., and Buensanteai, N. (2013). Bioformulation Pseudomonas fluorescens SP007s against dirty panicle disease of rice. Afr. J. Microbiol. Res. 7, 5274–5283. doi: 10.5897/AJMR2013.2503
Preininger, C., Sauer, U., Bejarano, A., and Berninger, T. (2018). Concepts and applications of foliar spray for microbial inoculants. Appl. Microbiol. Biotechnol. 102, 7265–7282. doi: 10.1007/s00253-018-9173-4
Preston, G. M. (2004). Plant perceptions of plant growth-promoting Pseudomonas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 907–918. doi: 10.1098/rstb.2003.1384
Pršić, J., and Ongena, M. (2020). Elicitors of plant immunity triggered by beneficial bacteria. Front. Plant Sci. 11:594530. doi: 10.3389/fpls.2020.594530
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Qaim, M. (2020). Role of new plant breeding technologies for food security and sustainable agricultural development. Appl. Econ. Perspect. Policy 42, 129–150. doi: 10.1002/aepp.13044
Rahanandeh, H., Safari Motlagh, M. R., and Hamzeh, A. (2024). Protease enzymes from Pseudomonas fluorescens and Pseudomonas aeruginosa as potential bioagents against Pratylenchus loosi. Can. J. Plant Pathol. 46, 465–477. doi: 10.1080/07060661.2024.2361628
Rainey, P. B. (1999). Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1, 243–257. doi: 10.1046/j.1462-2920.1999.00040.x
Raio, A., and Puopolo, G. (2021). Pseudomonas chlororaphis metabolites as biocontrol promoters of plant health and improved crop yield. World J. Microbiol. Biotechnol. 37:99. doi: 10.1007/s11274-021-03063-w
Ramakrishnan, S., Sivakumar, C., and Poornima, K. (1998). Management of rice root nematode, Hirschmanniella gracilis (de man) Luc and Goodey with Pseudomonas fluorescens Migula. J. Biol. Control. 22, 135–141.
Rangarajan, S., Saleena, L. M., Vasudevan, P., and Nair, S. (2003). Biological suppression of rice diseases by Pseudomonas spp. under saline soil conditions. Plant Soil 251, 73–82. doi: 10.1023/A:1022950811520
Rini, C., and Sulochana, K. (2007). Usefulness of Trichoderma and Pseudomonas against Rhizoctonia solani and Fusarium oxysporum infecting tomato. J. Trop. Agric. 45, 21–28.
Rocca, J. D., Simonin, M., Blaszczak, J. R., Ernakovich, J. G., Gibbons, S. M., Midani, F. S., et al. (2019). The microbiome stress project: toward a global meta-analysis of environmental stressors and their effects on microbial communities. Front. Microbiol. 9:3272. doi: 10.3389/fmicb.2018.03272
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Rout, G. R., and Sahoo, S. (2015). Role of iron in plant growth and metabolism. Rev. Agric. Sci. 3, 1–24. doi: 10.7831/ras.3.1
Rufián, J. S., Rueda-Blanco, J., López-Márquez, D., Macho, A. P., Beuzón, C. R., and Ruiz-Albert, J. (2021). The bacterial effector HopZ1a acetylates MKK7 to suppress plant immunity. New Phytol. 231, 1138–1156. doi: 10.1111/nph.17442
Ryan, P. R., Dessaux, Y., Thomashow, L. S., and Weller, D. M. (2009). Rhizosphere engineering and management for sustainable agriculture. Plant Soil 321, 363–383. doi: 10.1007/s11104-009-0001-6
Scheepmaker, J. W. A., Hogervorst, P. A., and Glandorf, D. C. M. (2016). Future introductions of genetically modified microbial biocontrol agents in the EU: are current EU legislation and risk assessment fit for purpose? RIVM Letter Rep. 2016-0057, P20–P56.
Schikora, A., Schenk, S. T., and Hartmann, A. (2016). Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl homoserine lactone group. Plant Mol. Biol. 90, 605–612. doi: 10.1007/s11103-016-0457-8
Schmidt, R., Cordovez, V., De Boer, W., Raaijmakers, J., and Garbeva, P. (2015). Volatile affairs in microbial interactions. ISME J. 9, 2329–2335. doi: 10.1038/ismej.2015.42
Schmidt, R., Köberl, M., Mostafa, A., Ramadan, E. M., Monschein, M., Jensen, K. B., et al. (2014). Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 5:64. doi: 10.3389/fmicb.2014.00064
Sharma, R. K., and Archana, G. (2016). Cadmium minimization in food crops by cadmium resistant plant growth promoting rhizobacteria. Appl. Soil Ecol. 107, 66–78. doi: 10.1016/j.apsoil.2016.05.009
Sharma, P., Bora, L. C., Nath, P. D., Acharjee, S., Bora, P., and Vasantrao, J. M. (2020). Zinc enriched Pseudomonas fluorescens triggered defense response in rice against bacterial leaf blight. Indian J. Agric. Sci. 90, 593–596. doi: 10.56093/ijas.v90i3.101491
Sharma, S. K., Johri, B. N., Ramesh, A., Joshi, O. P., and Sai Prasad, S. (2011). Selection of plant growth-promoting Pseudomonas spp. that enhanced productivity of soybean-wheat cropping system in Central India. J. Microbiol. Biotechnol. 21, 1127–1142. doi: 10.4014/jmb.1012.12018
Shaukat, S., and Siddiqui, I. (2003). Impact of biocontrol agents Pseudomonas fluorescens CHA0 and its genetically modified derivatives on the diversity of culturable fungi in the rhizosphere of mungbean. J. Appl. Microbiol. 95, 1039–1048. doi: 10.1046/j.1365-2672.2003.02074.x
Shen, Z., Xue, C., Penton, C. R., Thomashow, L. S., Zhang, N., Wang, B., et al. (2019). Suppression of banana Panama disease induced by soil microbiome reconstruction through an integrated agricultural strategy. Soil Biol. Biochem. 128, 164–174. doi: 10.1016/j.soilbio.2018.10.016
Siddiqui, I., and Shaukat, S. (2002). Zinc and glycerol enhance the production of nematicidal compounds in vitro and improve the biocontrol of Meloidogyne javanica in tomato by fluorescent pseudomonads. Lett. Appl. Microbiol. 35, 212–217. doi: 10.1046/j.1472-765X.2002.01162.x
Sisaphaithong, T., Kondo, D., Matsunaga, H., Kobae, Y., and Hata, S. (2012). Expression of plant genes for arbuscular mycorrhiza-inducible phosphate transporters and fungal vesicle formation in sorghum, barley, and wheat roots. Biosci. Biotechnol. Biochem. 76, 2364–2367. doi: 10.1271/bbb.120782
Sonali, J. M. I., Gayathri, K. V., Rangasamy, G., Kumar, P. S., and Rajagopal, R. (2024). Efficacy of biogenic selenium nanoparticles from Pseudomonas libanesis towards growth enhancement of okra. Waste Biomass Valor. 15, 1793–1806. doi: 10.1007/s12649-023-02233-1
Spiegel, Y., Cohn, E., Galper, S., Sharon, E., and Chet, I. (1991). Evaluation of a newly isolated bacterium, Pseudomonas chitinolytica sp. nov., for controlling the root-knot nematode Meloidogynejavanica. Biocontrol Sci. Tech. 1, 115–125. doi: 10.1080/09583159109355191
Sponsler, D. B., Grozinger, C. M., Hitaj, C., Rundlöf, M., Botías, C., Code, A., et al. (2019). Pesticides and pollinators: A socioecological synthesis. Sci. Total Environ. 662, 1012–1027. doi: 10.1016/j.scitotenv.2019.01.016
Srivastava, R., Johri, B., and Sharma, A. (1999). Colonization of wheat (Triticum aestivum L.) root by fluorescent pseudomonads (GRP3 and PRS9). Ind. J. Microbiol. 39, 205–210.
Stearns, J. C., and Glick, B. R. (2003). Transgenic plants with altered ethylene biosynthesis or perception. Biotechnol. Adv. 21, 193–210. doi: 10.1016/S0734-9750(03)00024-7
Stover, C., Pham, X., Erwin, A., Mizoguchi, S., Warrener, P., Hickey, M., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. doi: 10.1038/35023079
Suarez, G., Sierra, J. C., Sha, J., Wang, S., Erova, T. E., Fadl, A. A., et al. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44, 344–361. doi: 10.1016/j.micpath.2007.10.005
Sun, X., Xu, Z., Xie, J., Hesselberg-Thomsen, V., Tan, T., Zheng, D., et al. (2022). Bacillus velezensis stimulates resident rhizosphere Pseudomonas stutzeri for plant health through metabolic interactions. ISME J. 16, 774–787. doi: 10.1038/s41396-021-01125-3
Sun, X., Zhang, R., Ding, M., Liu, Y., and Li, L. (2021). Biocontrol of the root-knot nematode Meloidogyne incognita by a nematicidal bacterium Pseudomonas simiae MB751 with cyclic dipeptide. Pest Manag. Sci. 77, 4365–4374. doi: 10.1002/ps.6470
Sundh, I., and Goettel, M. S. (2013). Regulating biocontrol agents: a historical perspective and a critical examination comparing microbial and macrobial agents. BioControl 58, 575–593. doi: 10.1007/s10526-012-9498-3
Then, C., Kawall, K., and Valenzuela, N. (2020). Spatiotemporal controllability and environmental risk assessment of genetically engineered gene drive organisms from the perspective of European Union genetically modified organism regulation. Integr. Environ. Assess. Manag. 16, 555–568. doi: 10.1002/ieam.4278
Thomashow, L. S., and Weller, D. M. (1988). Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170, 3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988
Tian, F., Ding, Y., Zhu, H., Yao, L., and Du, B. (2009). Genetic diversity of siderophore-producing bacteria of tobacco rhizosphere. Braz. J. Microbiol. 40, 276–284. doi: 10.1590/S1517-83822009000200013
Toklikishvili, N., Dandurishvili, N., Vainstein, A., Tediashvili, M., Giorgobiani, N., Lurie, S., et al. (2010). Inhibitory effect of ACC deaminase-producing bacteria on crown gall formation in tomato plants infected by Agrobacterium tumefaciens or A. vitis. Plant Pathol. 59, 1023–1030. doi: 10.1111/j.1365-3059.2010.02326.x
Tonui, W. K., Ahuja, V., Beech, C. J., Connolly, J. B., Dass, B., Glandorf, D. C. M., et al. (2022). Points to consider in seeking biosafety approval for research, testing, and environmental release of experimental genetically modified biocontrol products during research and development. Transgenic Res. 31, 607–623. doi: 10.1007/s11248-022-00311-z
Tonui, W. K., Ogoyi, D., Thuo, C., Tareh, C., Alukhaba, C. L., Ouedraogo, A., et al. (2024). Communicating the uncertainties associated with genetic biocontrol approaches: insights from communicators, science journalists and scientists in Africa. J. Sci. Commun. 23:N02. doi: 10.22323/2.23060802
Tran, H., Ficke, A., Asiimwe, T., Höfte, M., and Raaijmakers, J. M. (2007). Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol. 175, 731–742. doi: 10.1111/j.1469-8137.2007.02138.x
United Nations (2015). Transforming our world: the 2030 agenda for sustainable development. New York, NY: United Nations, Department of Economic and Social Affairs, 1–41.
Validov, S., Mavrodi, O., la Fuente, L., Boronin, A., Weller, D., Thomashow, L., et al. (2005). Antagonistic activity among 2, 4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. FEMS Microbiol. Lett. 242, 249–256. doi: 10.1016/j.femsle.2004.11.013
Van der Meij, A., Worsley, S. F., Hutchings, M. I., and van Wezel, G. P. (2017). Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 41, 392–416. doi: 10.1093/femsre/fux0005
Van Loon, L. (1984). “Regulation of pathogenesis and symptom expression in diseased plants by ethylene,” in Ethylene: Biochemical, Physiological and Applied Aspects, An International Symposium, Oiryat Anavim, Israel held January 9–12.
Van Peer, R., Van Kuik, A., Rattink, H., and Schippers, B. (1990). Control of Fusarium wilt in carnation grown on rockwool by Pseudomonas sp. strain WCS417r and by Fe-EDDHA. Neth. J. Plant Pathol. 96, 119–132. doi: 10.1007/BF01974251
Vanloqueren, G., and Baret, P. V. (2017). “How agricultural research systems shape a technological regime that develops genetic engineering but locks out agroecological innovations 1” in Food sovereignty, agroecology and biocultural diversity. ed. M. Pimbert (London: Routledge), 57–92.
Vasilescu, A. G., Pleşoianu, A.-I., and Pătru-Stupariu, I. (2023). Aspects of traditional agricultural landscapes: potential alternative development paths for sustainable agriculture—A review. Biodivers. Conserv. 32, 3703–3730. doi: 10.1007/s10531-023-02658-z
Verma, M., Mishra, J., and Arora, N. K. (2019). Plant growth-promoting rhizobacteria: diversity and applications. Environ. Biotechnol. Sust. Future 6, 129–173. doi: 10.1007/s10531-023-02658-z
Viaene, T., Langendries, S., Beirinckx, S., Maes, M., and Goormachtig, S. (2016). Streptomyces as a plant's best friend? FEMS Microbiol. Ecol. 92:fiw119. doi: 10.1093/femsec/fiw119
Vidhyasekaran, P., Kamala, N., Ramanathan, A., Rajappan, K., Paranidharan, V., and Velazhahan, R. (2001). Induction of systemic resistance by Pseudomonas fluorescens Pf1 against Xanthomonas oryzae pv. Oryzae in rice leaves. Phytoparasitica 29, 155–166. doi: 10.1007/BF02983959
Viebahn, M., Glandorf, D., Ouwens, T., Smit, E., Leeflang, P., Wernars, K., et al. (2003). Repeated introduction of genetically modified Pseudomonas putida WCS358r without intensified effects on the indigenous microflora of field-grown wheat. Appl. Environ. Microbiol. 69, 3110–3118. doi: 10.1128/AEM.69.6.3110-3118.2003
Vivekananthan, R., Ravi, M., Ramanathan, A., and Samiyappan, R. (2004). Lytic enzymes induced by Pseudomonas fluorescens and other biocontrol organisms mediate defence against the anthracnose pathogen in mango. World J. Microbiol. Biotechnol. 20, 235–244. doi: 10.1023/B:WIBI.0000023826.30426.f5
Waldon, H. B., Jenkins, M. B., Virginia, R. A., and Harding, E. E. (1989). Characteristics of woodland rhizobial populations from surface-and deep-soil environments of the Sonoran Desert. Appl. Environ. Microbiol. 55, 3058–3064. doi: 10.1128/aem.55.12.3058-3064.1989
Wang, P., Feng, X., and Wang, G. (1999). Screening and identification of PGPR strains isolated from the rhizosphere of winter wheat (Triticum aestivum L.). J. Huazhong Agric. Univ. 18, 352–356.
Wang, Y.-N., He, W.-H., He, H., Du, X., Jia, B., Zeng, Z.-P., et al. (2012). Pseudomonas nitritireducens sp. nov., a nitrite reduction bacterium isolated from wheat soil. Arch. Microbiol. 194, 809–813. doi: 10.1007/s00203-012-0838-6
Wang, C., Knill, E., Glick, B. R., and Défago, G. (2000). Effect of transferring 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase genes into Pseudomonas fluorescens strain CHA0 and its gac A derivative CHA96 on their growth-promoting and disease-suppressive capacities. Can. J. Microbiol. 46, 898–907. doi: 10.1139/w00-071
Wang, P., Marsh, E. L., Ainsworth, E. A., Leakey, A. D., Sheflin, A. M., and Schachtman, D. P. (2017). Shifts in microbial communities in soil, rhizosphere and roots of two major crop systems under elevated CO2 and O3. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-14936-2
Wang, H., Zhang, Y., Dai, D., Fu, J., Sung Kim, D., Li, S., et al. (2024b). Genomic insight into the insecticidal potential of a new Pseudomonas chlororaphis isolate. J. Econ. Entomol. 117, 82–92. doi: 10.1093/jee/toad232
Wang, H., Zhang, F., Zhang, Y., Wang, M., Zhang, Y., and Zhang, J. (2024a). Enrichment of novel entomopathogenic Pseudomonas species enhances willow resistance to leaf beetles. Microbiome 12:169. doi: 10.1186/s40168-024-01884-z
Wang, Z., Zhong, T., Chen, K., Du, M., Chen, G., Chen, X., et al. (2021). Antifungal activity of volatile organic compounds produced by Pseudomonas fluorescens ZX and potential biocontrol of blue mold decay on postharvest citrus. Food Control 120:107499. doi: 10.1016/j.foodcont.2020.107499
Watanabe, N., Lewis, J. A., and Papavizas, G. C. (1987). Influence of nitrogen fertilizers on growth, spore production and germination, and biocontrol potential of Trichoderma and Gliocladium. J. Phytopathol. 120, 337–346. doi: 10.1111/j.1439-0434.1987.tb00497.x
Wei, H.-L., Zhang, W., and Collmer, A. (2018). Modular study of the type III effector repertoire in Pseudomonas syringae pv. Tomato DC3000 reveals a matrix of effector interplay in pathogenesis. Cell Rep. 23, 1630–1638. doi: 10.1016/j.celrep.2018.04.037
Weiler, E. W., Kutchan, T. M., Gorba, T., Brodschelm, W., Niesel, U., and Bublitz, F. (1994). The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett. 345, 9–13. doi: 10.1016/0014-5793(94)00411-0
Wheatley, R. (2002). The consequences of volatile organic compound mediated bacterial and fungal interactions. Antonie Van Leeuwenhoek 81, 357–364. doi: 10.1023/A:1020592802234
Winding, A., Binnerup, S. J., and Pritchard, H. (2004). Non-target effects of bacterial biological control agents suppressing root pathogenic fungi. FEMS Microbiol. Ecol. 47, 129–141. doi: 10.1016/S0168-6496(03)00261-7
Woźniak, E., Tyczewska, A., and Twardowski, T. (2021). Bioeconomy development factors in the European Union and Poland. New Biotechnol. 60, 2–8. doi: 10.1016/j.nbt.2020.07.004
Wu, L., Xiao, W., Chen, G., Song, D., Khaskheli, M. A., Li, P., et al. (2018). Identification of Pseudomonas mosselii BS011 gene clusters required for suppression of rice blast fungus Magnaporthe oryzae. J. Biotechnol. 282, 1–9. doi: 10.1016/j.jbiotec.2018.04.016
Xiao-Jing, X., Li-Qun, Z., You-Yong, Z., and Wen-Hua, T. (2005). Improving biocontrol effect of Pseudomonas fluorescens P5 on plant diseases by genetic modification with chitinase gene. Chin. J. Agric. Biotechnol. 2, 23–27. doi: 10.1079/CJB200553
Xin, X.-F., Kvitko, B., and He, S. Y. (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328. doi: 10.1038/nrmicro.2018.17
Xu, W., Xu, L., Deng, X., Goodwin, P. H., Xia, M., Zhang, J., et al. (2021). Biological control of take-all and growth promotion in wheat by Pseudomonas chlororaphis YB-10. Pathogens 10:903. doi: 10.3390/pathogens10070903
Yang, C., Li, W., Cao, J., Meng, F., Yu, Y., Huang, J., et al. (2017). Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 89, 338–353. doi: 10.1111/tpj.13388
Young, J. (1991). Pathogenicity and identification of the lilac pathogen, Pseudomonas syringae pv. Syringae van hall 1902. Ann. Appl. Biol. 118, 283–298. doi: 10.1111/j.1744-7348.1991.tb05629.x
Yue, Y., Wang, Z., Zhong, T., Guo, M., Huang, L., Yang, L., et al. (2023). Antifungal mechanisms of volatile organic compounds produced by Pseudomonas fluorescens ZX as biological fumigants against Botrytis cinerea. Microbiol. Res. 267:127253. doi: 10.1016/j.micres.2022.127253
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., Da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, Q., Kong, X., Li, S., Chen, X. J., and Chen, X. J. (2020). Antibiotics of Pseudomonas protegens FD6 are essential for biocontrol activity. Australas. Plant Pathol. 49, 307–317. doi: 10.1007/s13313-020-00696-7
Zhang, Y., Ruyter-Spira, C., and Bouwmeester, H. J. (2015). Engineering the plant rhizosphere. Curr. Opin. Biotechnol. 32, 136–142. doi: 10.1016/j.copbio.2014.12.006
Zimny, T. (2023). Regulation of GMO field trials in the EU and new genomic techniques: will the planned reform facilitate experimenting with gene-edited plants? Biotechnologia 104, 75–83. doi: 10.5114/bta.2023.125086
Keywords: biocontrol, sustainable agriculture, plant-microbe interactions, rhizosphere, crop protection, phytopathogen suppression
Citation: Alattas H, Glick BR, Murphy DV and Scott C (2024) Harnessing Pseudomonas spp. for sustainable plant crop protection. Front. Microbiol. 15:1485197. doi: 10.3389/fmicb.2024.1485197
Edited by:
Febri Doni, Padjadjaran University, IndonesiaReviewed by:
Nurul Shamsinah Mohd Suhaimi, University of Malaya, MalaysiaDedat Prismantoro, Padjadjaran University, Indonesia
Copyright © 2024 Alattas, Glick, Murphy and Scott. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hussain Alattas, aHVzc2Fpbi5hbGF0dGFzQG11cmRvY2guZWR1LmF1
 Hussain Alattas
Hussain Alattas Bernard R. Glick
Bernard R. Glick Daniel V. Murphy
Daniel V. Murphy Colin Scott
Colin Scott