Abstract
Introduction:
Seals, protected wild marine mammals, are widely found in waters around the world. However, rising concerns about their increasing numbers in some areas have led to potential worries regarding microbiological contamination of coastal areas by their feces, which could impact bathing and shellfish-harvesting activities. To the best of our knowledge, no study has been conducted on the bacterial and RNA viral communities present in the feces of both grey and harbor seals, which are the two main seal species observed in mainland France and overseas.
Methods:
Fecal bacterial (n = 132) and RNA viral (n = 40) communities of seals were analyzed using 16S rRNA gene amplicon high-throughput sequencing and viral RNA sequencing methods, respectively. In addition, to identify the specific characteristics of seal fecal microbial communities compared to other animal fecal microbial communities that may also contaminate coastal areas, the bacterial communities of seals were compared to those of wild waterbirds and breeding animals (i.e., cattle and pigs) which could be present in upstream catchments of coastal areas. Finally, ANCOM was used to identify unique and seal-associated Amplicon Sequence Variants (ASVs), aiming to develop a Microbial Source Tracking (MST) bacterial qPCR marker associated with seals.
Results and discussion:
The bacterial communities of grey and harbor seals were not found to be significantly different and were characterized by a predominance of Firmicutes, including the genera Clostridium sensu stricto 1 and Peptoclostridium, followed by Fusobacteriota with the genus Fusobacterium, and Bacteroidota with the genus Bacteroides. However, variations in bacterial communities between sites and individuals were observed. Similar observations were made for the RNA viral communities being characterized by a predominance of Picobirnaviridae (44% of total reads) and Astroviridae (15%). This study successfully developed a sensitive (89.8%) and specific (97.1%) MST qPCR marker targeting grey seal-associated bacteria belonging to the Bifidobacteriaceae family. This marker can be used to identify potential fecal contamination of coastal areas by seals and complements the MST toolboxes of markers already developed for humans, wild birds and livestock.
1 Introduction
Marine mammals such as pinnipeds, diving marine predators, play crucial ecological roles in the oceans, but little is known about their microbiotas (Bik et al., 2016; Numberger et al., 2016; Pacheco-Sandoval et al., 2019). The present study focused on the two most frequent seal species in France along the English Channel coast and in Saint-Pierre et Miquelon overseas (Figure 1): i.e. grey seals (Halichoerus grypus) and harbor seals, also named common seals (Phoca vitulina) (den Heyer et al., 2021; Poncet et al., 2021; Savouré-Soubelet et al., 2016).
Figure 1
Location of seal feces sampling sites.
Grey and harbor seals in mainland France are at the southern limit of the species’ ranges in the North-eastern Atlantic (Vincent et al., 2017). In Saint-Pierre et Miquelon, both species occur in smaller numbers that in the adjacent Canadian waters, although recent and frequent abundance estimates are only available for the grey seal (den Heyer et al., 2021). Most recent counts on haulout sites gave an annual maximum of 2,635 grey seals and 1,319 harbor seals in mainland France, and 218 grey seals and 1,652 harbor seals in Saint-Pierre et Miquelon, in 2021 (Poncet et al., 2021; Poncet et al., 2023). Both these seal species are protected at the international and European level (Appendix III of the Bern Convention and Annex II of the Bonn Convention EEC, 1992).
Grey and harbor seals are considered opportunistic predators, top-tier consumers that feed on benthic or/and pelagic species, mainly fish and cephalopods (Pacheco-Sandoval et al., 2019; Planque et al., 2021). They are sympatric in the North Atlantic and can share the same haul-out sites as well as potentially similar foraging grounds (Planque et al., 2021). Furthermore, they move and feed within short distances of the coast, i.e., few tens of kilometers from the shore for harbor seals and few hundreds of kilometers for some grey seal individuals (Cunningham et al., 2009; Härkönen et al., 1999; Harvey et al., 2008; Thompson et al., 1996; Vincent et al., 2017). The diet of seals could be different according to species and location. Seal feces analyzed in this study came from four different locations: i.e. Baie de Somme (BDS), Walde (WAL), Molène archipelago (MOL; Ile de Morgol), and Saint-Pierre et Miquelon (SPM; Le grand barachois, Miquelon).
In the Baie de Somme and Walde, grey seals feed predominantly on flatfish (61%), including Pleuronectidae (plaice and flounder) and Soleidae (mainly common sole and sand sole), and, to a lesser extent, on herring (18%) and Gadidae, such as whiting and poulting (Planque et al., 2018). Their diet also includes cephalopods, such as squid, and Chondrichtyes (rays or ground sharks, found in 3.2% of the samples). Harbor seals mainly feed on flatfish (86%): common sole, sand sole, yellow sole, thickback sole, plaice and flounder, and, to a lesser extent, on common dragonet (6%) (Planque et al., 2018). In the Iroise Sea (i.e., Molene archipelago), grey seals prefer ballan wrasse and the other round fish such as conger, pollack, sea bass or pout (Ridoux et al., 2007). Finally, the diet of grey and harbor seals in Saint-Pierre et Miquelon is currently being studied. Preliminary results suggest grey seal mainly feed on sandeels (Ammodytidae) and cod, but may also consume crustaceans, while harbor seals mainly feed on sticklebacks (Gasterosteidae), although this diet estimate may be biased seasonally (Vincent et al., 2022).
Seals, mainly piscivorous, have a diet high in proteins and polyunsaturated fatty acids, different from diet of other animals, such as terrestrial carnivorous mammals (Ley et al., 2008; Nelson et al., 2013). When they compared the fecal bacterial community of a variety of terrestrial mammals, Ley et al. (2008) identified that diet in addition to host phylogeny and, to a lesser extent, gut morphology influenced the composition of the gut bacterial community.
Due to their high trophic level diet but also to their long life, wide distribution, coastal habitat and possible infection with bacteria, viruses, and parasites that could also be pathogens for humans, marine mammals such as seals can serve as sentinel species for coastal ecosystem health (Hughes et al., 2013; Baily et al., 2015; Simeone et al., 2015). Indeed, the presence of bacterial, viral, and fungal agents of marine mammal zoonoses has been reported in seals: among the bacteria, e.g., Brucella spp., Clostridium spp., Erysipelotrix spp., Mycobacterium spp., Mycoplasma spp., Photobacterium spp., Vibrio spp. (reviewed in Waltzek et al., 2012; Hughes et al., 2013; Siebert et al., 2017; Pacheco-Sandoval et al., 2024). In addition, pathogenic bacteria, that can cause food poisoning in humans, such as Campylobacter spp., Salmonella spp. and Clostridium perfringens, have also been isolated from seal feces (Nelson et al., 2013; Greig et al., 2014; Lisle et al., 2004; Baily et al., 2015; Stoddard et al., 2007). In addition, fecal indicator bacteria (FIB) such as Escherichia coli which are used to highlight a fecal contamination in bathing and shellfish-harvesting areas, have been found to be present in seal feces, as they are in the feces of other marine and terrestrial mammals (Calambokidis and McLaughlin, 1987).
More globally, the microbial communities present in seal feces have been the subject of little research (Numberger et al., 2016; Pacheco-Sandoval et al., 2019; Switzer et al., 2023; Pacheco-Sandoval et al., 2024). They have been shown to differ from those of terrestrial mammals and humans, with, e.g., a significantly higher average relative abundance of a phylum such as Fusobacteriodota (Nelson et al., 2013; Switzer et al., 2023).
Questions may also be raised about viruses shed by these animals. The marine environment is known for its huge diversity of viruses (Sunagawa et al., 2020) and the potential emergence of human pathogenic viruses has been raised mainly for RNA viruses belonging to the Picornaviridae or Caliciviridae families (Bergh et al., 1989). Recently, few studies have been conducted in fur seals to describe the virome, either in feces (Kluge et al., 2016; Martínez-Puchol et al., 2022), or from spleen and lung samples (Canova et al., 2021). DNA viruses, such as circovirus and adenovirus, have also been described in fur seals feces (Chiappetta et al., 2017). In addition to human contamination, such as wastewater treatment plant discharges and livestock effluents, wildlife can potentially introduce fecal contamination into coastal waters (Boukerb et al., 2021; Rince et al., 2018). Indeed, a high diversity of viral sequences was detected in shellfish samples in a wildlife-contaminated area (Bonny et al., 2021). One possible consequence is the negative impact on human health through shellfish consumption or water ingestion, and the increased risk of zoonotic transmission through contaminated shellfish (Zakhour et al., 2010). The ability to describe the different communities may help to prevent zoonotic events in a One Health approach (Santiago-Rodriguez and Hollister, 2023).
The microbiological quality of coastal waters can be affected by fecal material originated from anthropogenic sources (e.g., wastewater treatment plant effluent, agricultural run-off) and from wildlife sources (e.g., wild bird droppings and the feces of marine mammals such as seals). In fact, seal haul-out sites can result in large amounts of feces being deposited in coastal waters, such as recreational areas, which can lead to poor water quality and beach closures (Paar et al., 2024). Microbial Source Tracking (MST) is a discipline which uses host-associated (e.g., bacterial, viral or chemical) markers to discriminate between human and animal excreta and to distinguish animal hosts as an approach to identify, and ultimately remediate sources of fecal contamination (Harwood et al., 2014). Tested and validated for source specificity and sensitivity, MST markers can be used as an MST toolbox for routine water quality monitoring in coastal environments (Gourmelon et al., 2010; Jarde et al., 2018; Sinigalliano et al., 2021). The MST markers were essentially based on qPCR bacterial markers associated with different hosts, but other markers developed using, for example, RNA viral sequencing could also be useful tools for tracing sources of fecal contamination (Sinigalliano et al., 2021; Santiago-Rodriguez and Hollister, 2023). Few microbial MST markers associated with wildlife have been developed and a seal-associated microbial marker to assess potential seal contamination of coastal waters is still lacking (Paar et al., 2024).
The main objectives of this study were the identification of microbial (i.e., bacterial and viral) communities in the feces of the two most frequent seal species (grey seal: Halicheorus grypus and harbor seal: Phoca vitulina) in France (Saint-Pierre et Miquelon island and mainland France) using 16S rRNA gene amplicon high-throughput sequencing and viral RNA sequencing and the investigation of MST microbial markers associated with seal fecal contamination.
2 Materials and methods
2.1 Fecal sample collection and location
Fresh fecal samples from seals [96 grey seals, 29 harbor seals and seven unidentified seals for which species was not certain, n = 132] were collected from 2015 to 2020 in mainland France as well as in Saint-Pierre et Miquelon (SPM) (Table 1). Individual feces were collected with a sterile spatula, retaining the inner part and ensuring no cross-contamination from the environment. The samples were either frozen at −20°C on site [e.g. SPM and Bay de Somme (BDS) samples] or sent on dry ice within 2 days to the laboratory where they were then frozen at −80°C until use. All samples were aseptically homogenized.
Table 1
| Mainland France | Saint-Pierre et Miquelon | |||
|---|---|---|---|---|
| Sites | BDS | WAL | MOL | SPM |
| Grey seals | 32 | 11 | 20 (20)* | 33 (8)* |
| Harbor seals | 15 | 0 | 0 | 14 (12)* |
| Not specified species seals | 0 | 0 | 0 | 7 ++ (2)* |
| Total number of seal feces | 47 | 11 | 20 | 54 |
| Date of collection (years) | 2017–2019 | 2017 | 2020 | 2015–2020 |
Description of seal fecal samples (n = 132).
*() number of seal fecal samples analyzed for viral communities. ++Seal fecal samples not analyzed for bacterial communities.BDS, Baie de Somme; WAL, Walde; MOL, Molène archipelago; SPM, Saint-Pierre et Miquelon; Le grand barachois, Miquelon.
In addition, fecal samples collected from other sources [wild waterbirds, livestock (i.e., pigs and cattle); n = 322] from mainland France and described in a previous study (Boukerb et al., 2021) were used to compare bacterial communities in the seal samples described above to those of other sources.
2.2 Fecal sample treatments for RNA viral analysis
Fecal samples were thawed and mixed, with 1 g (wet weight) of the sample diluted in 9 mL of PBS. The mixture was vortexed at high speed for 30 s and then mixed vigorously for 20 s, repeated three times using a Fast-Prep apparatus. Sodium pyrophosphate was added to a final concentration of 10 mM, and the mixture was incubated for 40 min with gentle agitation at room temperature. This was followed by 3 cycles of sonication for 1 min each at maximum intensity using a cup-horn adaptor (Bandelin, HD 2200), with 1 min of cooling on ice in between. After centrifugation at 8,000 g for 20 min, the supernatants were collected, the pH was adjusted to 7 using HCl, and then 5 mL of 12% polyethylene glycol (PEG 60000) (Sigma-Aldrich, St-Quentin, France) was added. Following overnight incubation at 4°C with gentle agitation, the mixture was centrifuged at 13,000 g for 20 min at 4°C. The resulting pellet was resuspended in 2 mL of 0.05 M glycine buffer (pH 9) (Strubbia et al., 2019).
One negative control was prepared using 10 mL of the PBS used for fecal dilution, with the same processing steps applied as to the fecal samples. A positive control was made by inoculating 1 g of seal feces with an RNA virus, specifically norovirus GII.17 [P17], at a final concentration of 107 genome copies/g. This positive control underwent all extraction and library preparation steps in the same manner as the other samples.
2.3 Total DNA extraction
Aliquots of approximately 0.25 g (wet weight) of seal feces were used for direct extraction of total genomic DNA using the FASTDNA™ Spin Kit for Soil (MP Biomedicals, Illkirch, France) as recommended by the manufacturer’s protocol and prepared under the conditions detailed by Boukerb et al. (2021). DNA was eluted in 100 μL sterile DNA/RNA free water. DNA quality and concentrations were assessed using a spectrophotometer (Epoch, BioTek) and a Qubit 4 fluorometer (Thermo Fisher Scientific). All DNA extracts were preserved at −80°C until the 16S rRNA gene amplicon library preparation.
2.4 RNA viral purification steps and nucleic acid extraction
The resuspended pellets were filtered through a series of filters with pore sizes of 5 μm, 1.2 μm, and 0.45 μm (Minisart NML 17594, NML17593, PES16533). The filtrates containing viral particles were recovered and free DNA and RNA were degrading by adding 2,000 units of OmniCleave Endonuclease (Lucigen, Wisconsin, United States) and the mixture was incubated for 1 h at 37°C. Nucleic acids (NA) were then extracted by adding 10 mL of lysis buffer (bioMérieux, Lyon, France) and 50 μL of paramagnetic beads (NucliSens kit, bioMérieux) (Strubbia et al., 2020). After performing washing steps using the eGENE-UP® apparatus (bioMérieux) according to the manufacturer’s instructions, the extracted NA were treated with 25 U TURBO™ DNase (Ambion, Thermo Fisher Scientific, France) for 30 min at 37°C. An additional RNA purification step was carried out using the RNA Clean & Concentrator™ -5 kit (Zymo Research, Irvine, USA) to remove DNase and PCR inhibitors.
2.5 16S rRNA gene library generation and MiSeq sequencing
The libraries were prepared (343F/784R, targeting V3-V4 hypervariable region of the 16S rRNA gene; Andersson et al., 2008; Liu et al., 2008) as detailed in Boukerb et al. (2021) and sequenced in three runs using the 2 × 250 paired-end method on an Illumina MiSeq instrument: (i) by the GeT-PlaGe platform (Toulouse, France) for the seal feces samples FPh1 to FPh20 and for the fecal samples from other sources [pig, bovine, poultry, and wild birds; described in Boukerb et al. (2021); Supplementary Tables S1A,B; May and November 2017], with a MiSeq Reagent Kit V3 chemistry (Illumina) and (ii) by the Bacterial Communication and Anti-infectious Strategies laboratory facility (Rouen, France) for samples FPh21 to FPh129 (Supplementary Table S1A; run performed 20 May 2021) with a MiSeq Reagent Kit V2 chemistry (Illumina).
2.6 Library and sequencing for viral analysis
All RNA extracts were converted to cDNA in triplicate using Superscript II (Invitrogen, Saint-Aubin, France) and random hexamers (New England Biolabs (NEB), United States) (Bonny et al., 2021). Negative and positive controls were transcribed only once. To minimize bias during library preparation, samples were processed in batches rather than individually. After double-strand DNA synthesis with the second strand reaction buffer from NEBNext Ultra RNA Library Prep, DNA fragmentation was carried out in 68 tubes using sonication (Ultrasonicator Covaris M220, duty factor: 5%, peak power: 75, cycles per burst: 200 for 195 s, Woburn, MA). Libraries were prepared using the KAPA Hyper Prep kit (Roche), following the manufacturer’s protocol. Each run included a negative control with sterile RNase-free water. The 128 libraries were sequenced in two separate runs using Illumina NextSeq 500 technology to generate 2×150 bp reads (iGenSeq, ICM, Paris, France).
2.7 Data availability
The 16S rRNA gene and RNA viral datasets generated for this study can be found in the European Nucleotide Archive (ENA) database (Study: ERP149173; Samples from ERS15965056 to ERS15970887 and from ERS16078207 to ERS16078302; details in the Supplementary Table S1C).
2.8 Bioinformatic analysis
2.8.1 Bacterial communities
Raw data were analyzed using the SAMBA v4 workflow1 a Standardized and Automatized MetaBarcoding Analysis workflow using DADA2 (Callahan et al., 2016) and QIIME 2 (Bolyen et al., 2019) with default parameters unless otherwise indicated. Two SAMBA runs were performed, one including all seal fecal samples (n = 132) in addition to eight DNA extraction controls and one qPCR control (used at the microDecon step McKnight et al., 2019) and a second one including a selection of seal samples (i.e., only mainland France seal samples) and fecal samples from other sources also collected in mainland France (described in Boukerb et al., 2021).
This pipeline, developed by SeBiMER (Ifremer’s Bioinformatics Core Facility), is an open-source, modular NextFlow workflow (v22.10.4) (Di Tommaso et al., 2017) designed to process eDNA metabarcoding data. SAMBA is structured around three main parts: data integrity checking, bioinformatics processing, and statistical analysis. Firstly, the SAMBA verification step checked the integrity of the raw data. Then, sequencing primers were removed from the reads using Cutadapt (Martin, 2011) via QIIME 2 (v2022.11) (Bolyen et al., 2019), and reads without detected primers were discarded. Subsequently, the DADA2 package (v3.6.1) (Callahan et al., 2016) was used to filter out low-quality reads, correct sequencing errors, merge paired reads, infer Amplicon Sequence Variants (ASVs), and eliminate chimeras. Given the known issue of DADA2’s tendency to overestimate diversity, an additional ASV clustering step (OTU calling) was performed using the dbOTU3 algorithm (Olesen et al., 2017). Taxonomic classification was carried out using a naïve Bayesian classification against the SILVA database 138 (Glöckner et al., 2017; Quast et al., 2013), which was pre-filtered for the sequenced region (V3-V4).
Finally, SAMBA performed extensive analyses of the alpha- and beta-diversity using homemade R scripts (R Core Team, 2022) and ‘vegan’ package in R v4.0.2. Differences in alpha diversity indices among different fecal samples from different seal species and seal fecal samples from different sites were analyzed using Adonis statistical test and pairwise t-test.
ASV abundances for each sample were calculated at the phylum, family, and genus taxonomic levels. Alpha diversity within bacterial communities was characterized using the observed species richness and Shannon indices. Beta diversity was assessed through ordination analyses, specifically non-metric multidimensional scaling (NMDS) based on a weighted UniFrac distance matrix, with data normalized via Cumulative Sum Scaling (CSS) (Lozupone et al., 2011). To evaluate significant variance differences across indices by group or species, a permutational multivariate analysis of variance (PERMANOVA) was performed, followed by post-hoc pairwise comparisons.
To visualize the number of total, exclusive, and shared ASVs and genera among seal species, UpSet plots were created. Similarly, to Pacheco-Sandoval et al. (2024), to analyze the core group across species (i.e., grey seal and harbor seal), ASVs with a prevalence of 80% or higher in grey seal and harbor seal samples were considered, respectively, without setting a minimum relative abundance threshold.
2.8.2 Viral communities
Bioinformatic analysis was conducted using a Nextflow pipeline as outlined by Bonny et al. (2021). For each sample, reads from the three libraries were combined for subsequent analysis (Schaeffer et al., 2023). Briefly, Fastq files were trimmed with fastp using a quality threshold of 25. Clean reads were deduplicated (CD-hit) and mapped to remove RNA contaminants (Silva RNA database) and PCR duplicates. De novo assembly was performed using metaSPAdes with k-mer lengths of 21, 33, 55, 77, and 99 (Nurk et al., 2017). Contigs longer than 300 bp were filtered and identified using BLASTn and BLASTx with an E-value threshold of 10−5 and nr bank using diamond with an e-value of 10−3. When both methods (i.e., BLASTn and BLASTx) provided results, the BLASTn assignment is retained. To assess contig coverage, post-processed reads were mapped to the metaSPAdes contigs using Bowtie2 (v2.3.0) (Langmead and Salzberg, 2012; Nurk et al., 2017). Multi-mapped reads were excluded to avoid potential overestimation of abundance. Only merged reads were considered for each sample after this step.
Taxonomic identification was performed using the Entrez Direct tool, which enabled extraction of information at specified taxonomic levels. A heatmap was generated using R. Reads per million (rpm) were calculated by dividing the number of reads per family by the total number of trimmed and deduplicated reads.
2.9 Identification of potential seal bacterial MST markers
Differences in the mean abundance of microbial taxa in seal feces compared to other fecal sources (e.g., pigs, cattle, and wild waterbirds) (Boukerb et al., 2021) were identified using ANCOM (Analysis of Composition of Microbiomes). In ANCOM analysis, the W value represents the number of times a particular ASV is found to be significantly different in terms of abundance in pairwise log-ratio comparisons with all other ASVs (Mandal et al., 2015).
Among the 83 unique and seal-associated ASVs identified by ANCOM, the sequences of the 20 ASVs with the higher W value were retained and subsequently compared to the sequences of the NCBI nucleotide database,2 including, e.g., other marine mammals such as dolphin sequences (Bik et al., 2016) and other animals, to identify, for each ASV, the top 20 matches with the greatest host diversity (seals and other non-targeted animals). These sequences were then aligned using BioEdit software with ClustalW Multiple Alignment to identify variable regions (specific to the target sequence) and conserved regions (shared with non-target sequences).
When seal-specific regions were identified, primers were drawn manually or using Primer3,3 OligoCalc (v3.27) (Kibbe, 2007) and Multiple Primer Analyzer (ThermoFisher).
2.10 qPCR assays
Pairs of primers were designed to target the four ASVs that were most specific for seals out of the 20 ASVs selected. One of these four ASVs was from Fournierella genus (Firmicutes), one from Atopobium genus (Actinobacteriodota), one from Slackia genus (Actinobacteriodota), and the last from the family Bifidobacteriaceae (Actinobacteriodota). Details of the marker genes, primer sequences and qPCR reactions are listed in the Supplementary Table S2.
Quantitative PCR assays were conducted using the SybrGreen Mix kit (Invitrogen) on BioRad CX96 instrument under the following conditions: an initial step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by the completion of a melting curve from 55°C to 95°C in 0.5°C increments. Reactions were carried out in 25 μL volume, with forward and reverse primers at a final concentration of 200 nM (Eurogentec, France), and 2 μL of DNA template. To prevent inhibitors from affecting the PCRs, DNA samples were diluted 10- and 100-fold and the weaker dilution without inhibitors was retained.
Negative controls (without template DNA) were included in triplicate for each run. A gBlocks Gene fragment (IDT, Integrated DNA Technology) containing a 406 bp partial sequence of 16S rRNA gene belonging to the Bifidobacteriaceae family was used as standard. This synthetic oligonucleotide was used at 10-fold dilutions ranging from 106 to 1 copies per qPCR. PCR efficiency varied between 85.1 and 99.8% and coefficient of determination (R2) for all the standard curves were > 0.99.
Then, the sensitivity and specificity of the primers targeting the ASV belonging to the family Bifidobacteriaceae were assayed on target (n = 83) and non-target fecal (n = 54) samples using qPCR assays.
3 Results
3.1 Bacterial communities in seal feces
3.1.1 Analysis of raw data of metabarcoding 16S
Illumina sequencing of the V3-V4 hypervariable region of the 16S rRNA gene yielded a total of 5,820,125 reads for the 125 seal fecal samples. After verification of sequence quality, assembly and chimeras, a range of 3,340 to 80,009 reads per sample with an average of 26,325 ± 15,061 reads per sample were retained (56.5% of the raw data) for further analysis. After the dbOTU3, ASV clustering and microDecon steps, a total of 1,184 ASVs were obtained ranging from 21 to 187 per sample, where 98.8% of the ASVs were assigned taxonomically to the phylum level, 91.6% to the genus level, and 61.4% to the species level.
3.1.2 Taxonomic composition of seal fecal bacterial communities
Five phyla were found to be the most abundant in the feces of grey seals (n = 96): Firmicutes (67.3% ± 4.4%), Fusobacteriota (15.4% ± 24.6%), Bacteroidota (13.1% ± 19.8%), Proteobacteria (2.7% ± 7.9%) and Actinobacteriota (1.5% ± 3.4%; Figure 2A).
Figure 2
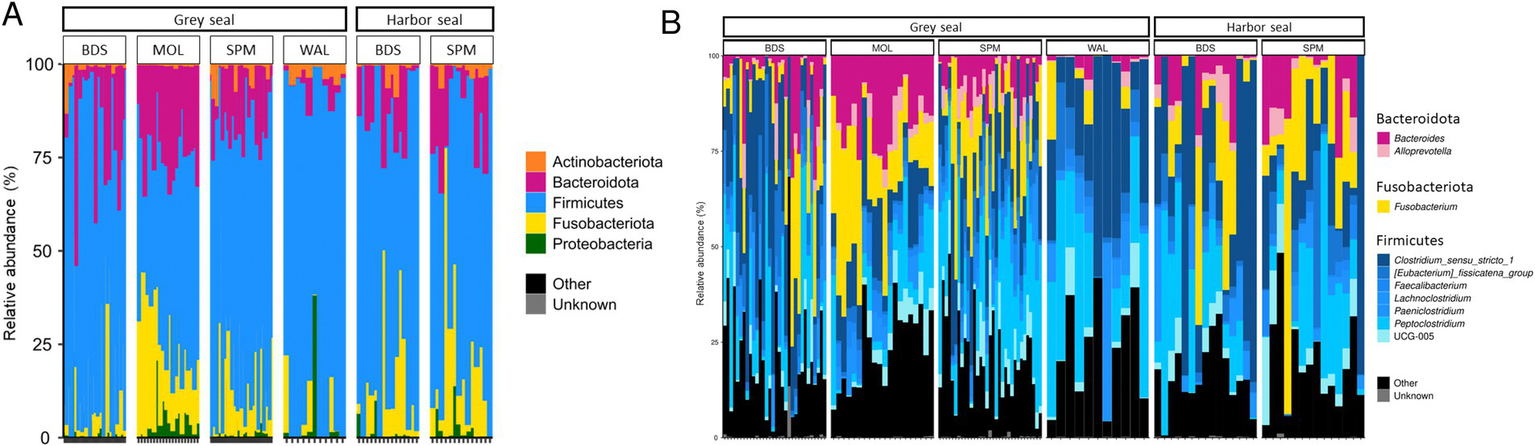
Phylum-level (A) and genus-level (B) bacterial community composition in two seal species: grey seal (n = 96) and harbor seal (n = 29). BDS (Baie de Somme); WAL (Walde), MOL (Molène archipelargo), Saint-Pierre et Miquelon (SPM, Le grand barachois, Miquelon).
These five phyla were also the most abundant in the feces of harbor seals (n = 29) with similar values: Firmicutes (74.1% ± 65.9%), Fusobacteriota (12.9% ± 15%), Bacteroidota (10.2% ± 14.2%), Proteobacteria (1.6% ± 2.7%) and Actinobacteriota (1.2 ± 2.1; Figure 2A).
In the grey seal, the main genus found was the genus Clostridium sensu stricto 1 (17.8% ± 11.1%), followed by the genus Peptoclostridium (16.4% ± 11.3%), Fusobacterium (14.8% ± 13.0%) and, to a lesser extent, Bacteroides (9.1% ± 8.8%), [Eubacterium] Fissicatena group (5.6% ± 3.9%), Ruminococcaceae UCG-005 (4.1% ± 3.8%), Paeniclostridium (2.6 ± 3.0%), Faecalibacterium (3.3% ± 2.8%) and Alloprevotella (2.2 ± 2.2%; Figure 2B).
In the harbor seal, a similar distribution of genera was observed with the main genus found Clostridium sensu stricto 1 (28.7% ± 29.1%), followed by the genera Peptoclostridium (14.8% ± 11.5%), Fusobacterium (12.1% ± 8.1%) and, to a lesser extent, the genera Bacteroides (6.9% ± 6.31%), [Eubacterium] Fissicatena group (5.4% ± 5.5%), Ruminococcaceae UCG-005 (3.7% ± 2.9%), Paeniclostridium (3.7% ± 5.9%), Alloprevotella (2.3% ± 2.2%) and Faecalibacterium (2.9% ± 2.2%; Figure 2B).
Of the genera identified, 34.4% occurred exclusively in one seal species and one site, while 27.8% occurred in both two species and different sites (Figure 3B). For ASVs, the majority of ASVs are found exclusively in one species and one site (57.9%), while only 91 ASVs (7.2%) are shared between the two species and the four sites (Figure 3A).
Figure 3
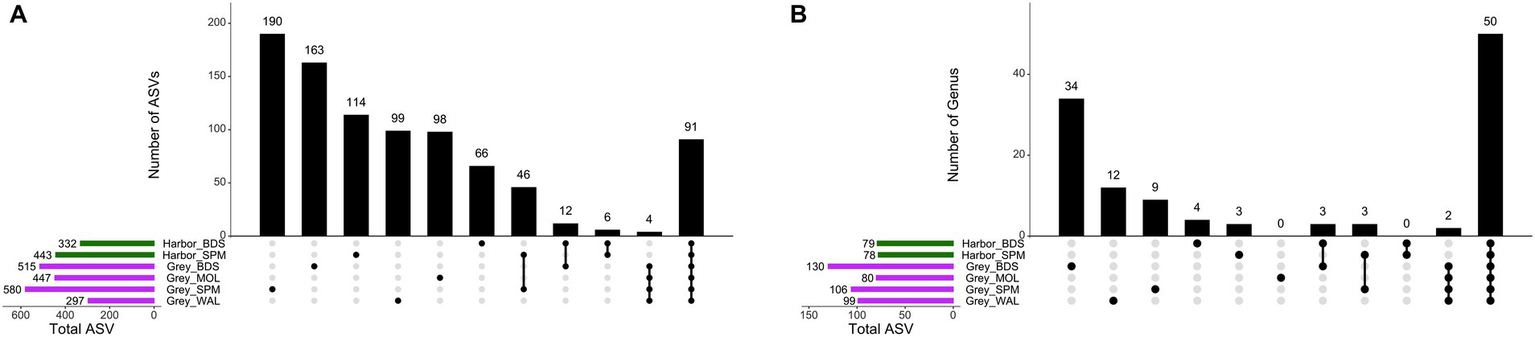
UpSetR plots showing the number of unique (single dot), shared (connected dots), and total (horizontal bars) ASVs (A) and genera (B) with an 80% prevalence in the fecal samples.
The core group of harbor seals consisted of nine ASVs belonging to the [Eubacterium] fissicatena group, Blautia, Clostridium sensu stricto 1 (two ASVs), Fusobacterium (two ASVs), Peptoclostridium, Ruminiclostridium 9, Ruminococcaceae UCG-005. The core group of grey seals consisted of these nine ASVs and a further five ASVs belonging to the genera Bacteroides, Faecalibacterium, Fournierella, Lachnoclostridium genera and to the family Erysipelotrichaceae.
3.1.3 Alpha diversity
Two indices were calculated to evaluate alpha diversity within the analyzed dataset: the observed species richness (quantitative species richness) and the Shannon index (non-parametric quantitative species richness) (Figure 4).
Figure 4
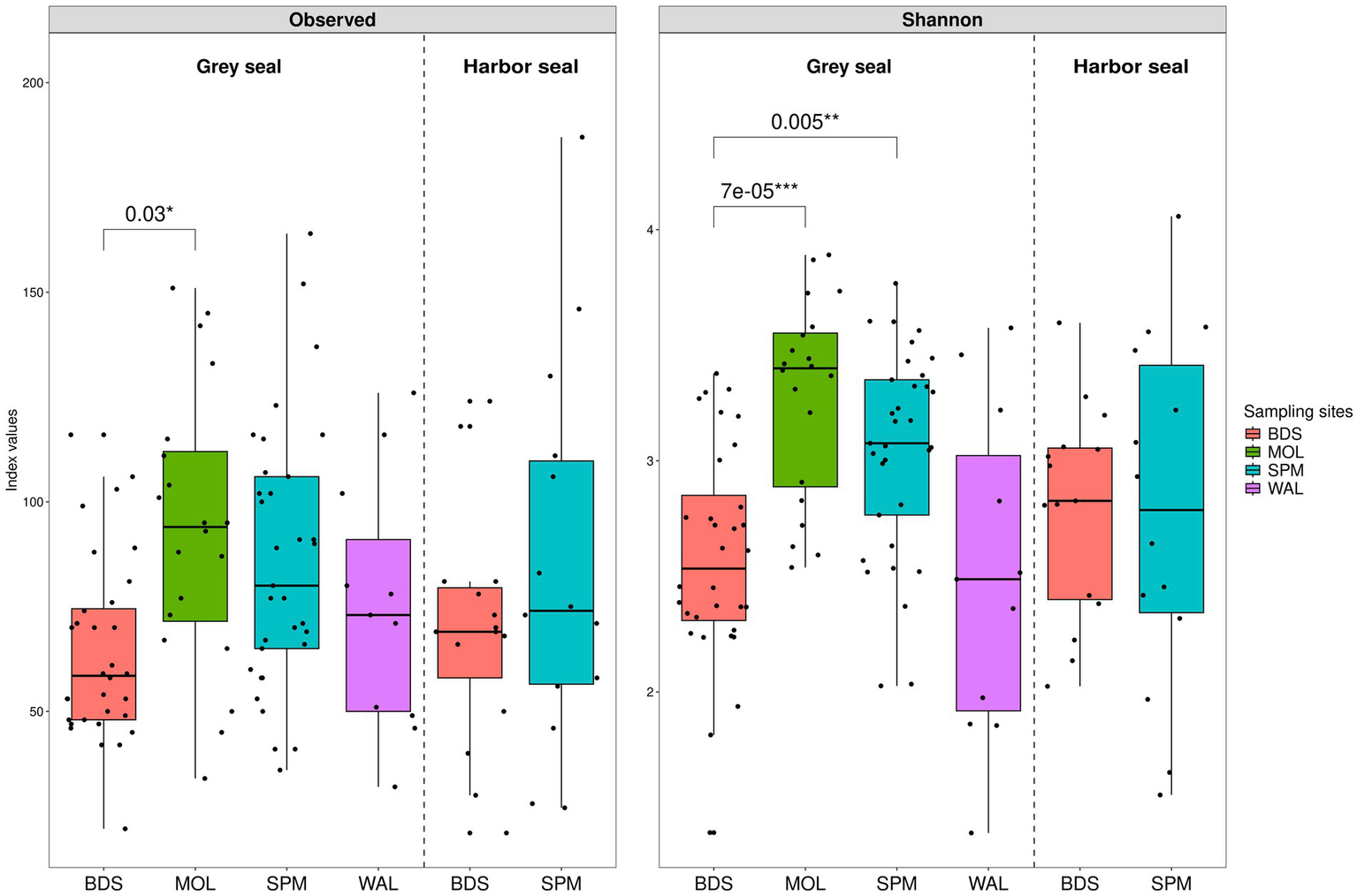
Boxplots illustrating alpha diversity in grey seal (n = 96) and harbor seal (n = 29) feces. The following indices were calculated: the observed species richness (A) and the Shannon diversity (B) index calculated from a contingency table of ASVs.
The observed species richness index shows a similar alpha diversity for grey seals, ranging from 22 to 164 (median value 73), and harbor seals, ranging from 21 to 187 (median value 71) (Figure 4A). The Adonis statistical test concludes that there is no significant difference between the two seal species (p-value 0.54074 > 0.05).
However, a slight significant difference was observed according to the sampling location: BDS, SPM, WAL, and MOL (Adonis p-value 0.01768). The pairwise t-test shows a difference between grey seals (GS) from MOL and grey seals from BDS (p-value 0.03) [GS-MOL from 34 to 151 (median 94) versus GS-BDS from 22 to 116 (median 58.5)].
Similarly, for the Shannon index, the Adonis statistical test shows that there is non-significant difference between the two seal species (p-value 0.37156; Figure 4B). The Shannon index varies for grey seals from 1.4 to 3.9 (median 2.9) and for harbor seals from 1.5 to 4.1 (median 2.8). However, a significant difference was observed according to the sampling location: BDS, SPM, WAL, and MOL (Adonis p-value 0.00697). The pairwise t-test shows a difference between grey seals from MOL and from BDS (p-value <0.001) [GS-MOL from 2.5 to 3.9 (median 3.4) versus GS-BDS from 1.4 to 3.4 (median 2.5)], and between grey seals from BDS and SPM (p-value 0.005) [GS-BDS from 1.4 to 3.4 (median 2.5) versus GS-SPM from 2.0 to 3.8 (median 3.1)].
3.1.4 Beta diversity
NMDS (weighted UniFrac index, on CSS normalized data) of the seal feces dataset was used to plot the beta diversity between samples (Figure 5). There was a slight significant difference in the bacterial community between groups (seal species and sites) (Adonis p-value: 0.015). Bacterial communities between the grey seals of MOL were significantly different from those of WAL (pairwise Adonis p.adjusted 0.015).
Figure 5
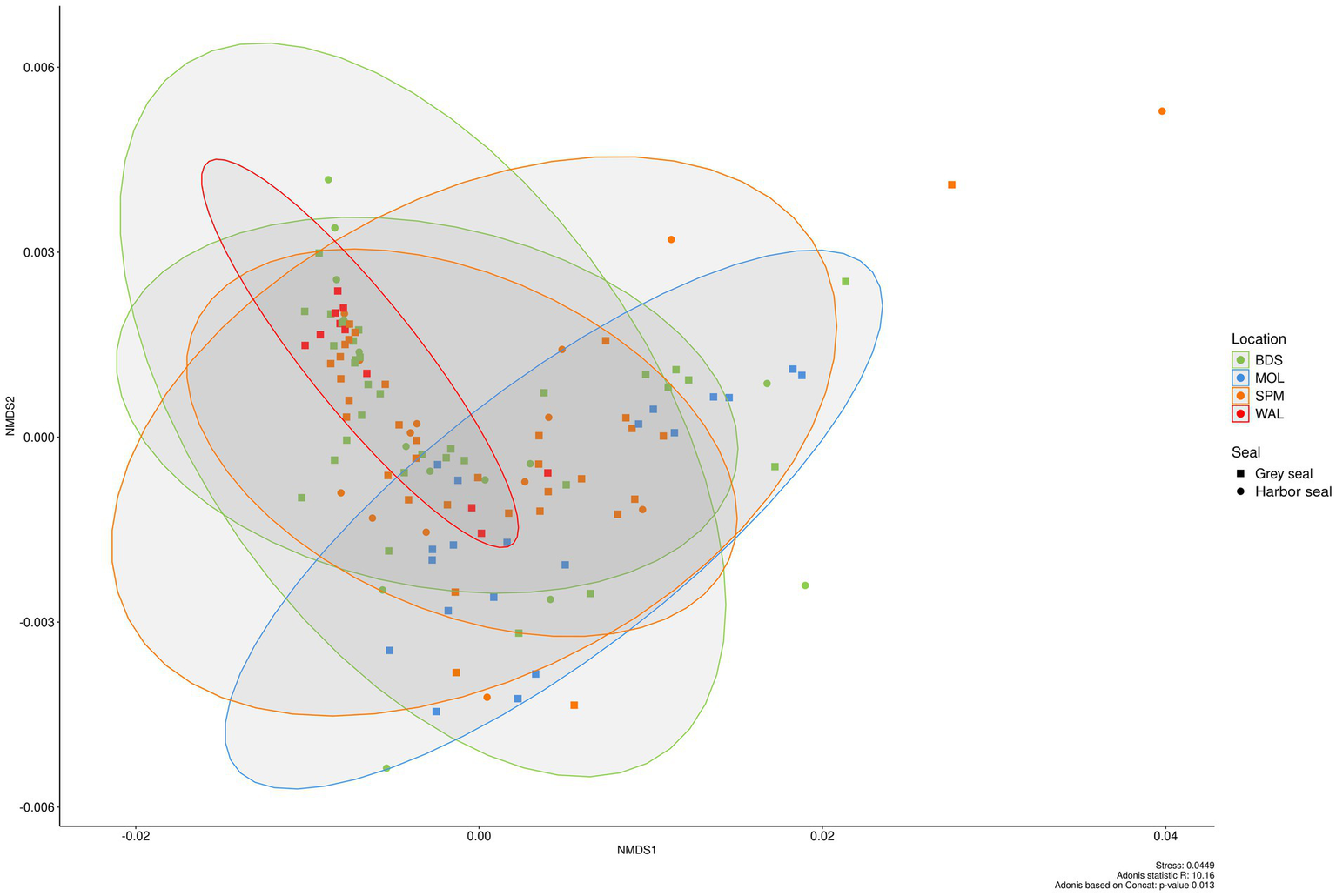
Beta diversity in the feces of two seal species (grey seal and harbor seal) collected in four sites (BDS, POL, WAL, and SPM) represented as NMDS obtained from a distance matrix calculated with the weighted UniFrac index. The data were previously normalized with CSS.
In fact, higher abundances of Fusobacteriota (23.1% ± 28.5% vs. 4.7% ± 16.9%) and Bacteroidota (22.7% ± 20.3% vs. 2.6% ± 7.8%) were obtained in MOL seal feces whereas a higher abundance of Firmicutes (83.8% ± 31.6% vs. 50.1% ± 46.9%) was observed in WAL fecal samples. More precisely, higher abundances of the genera Fusobacterium (22.0% ± 19.4% vs. 3.5% ± 5.3%) and Bacteroides (16.8% ± 12.9% vs. 1.6% ± 1.6%) were obtained in MOL seal feces whereas higher abundances of the genera Clostridium sensu stricto 1 (28.7% ± 19.7% vs. 11.6% ± 8.5%), Peptoclostridium (23.6% ± 13.7% vs. 9.0% ± 11.9%), [Eubacterium] fissicatena group (5.8% ± 4.6% vs. 2.5% ± 2.8%) were obtained in WAL seal feces.
3.2 Analysis of viral communities
The two sequencing runs provided about the same number of reads (about 3.8 million of reads for run 1 and 3.1 for run 2). After cleaning and deduplication, reads obtained for each sample triplicates were merged, decreasing the variability of read numbers obtained for each library. After this step, number of reads varied from four to 10 million per sample. More than 50% of the samples (24 samples out of 42 samples) displayed number of reads comprised between five to 7 million. This is an important point as despite efforts during sample preparation, the read number identified as bacteria was still high and represented 64% of total read numbers. Only three samples provided less than 10 thousand viral reads, while 24 samples displayed more than 100 thousand reads (data not shown). Overall, viral reads represent around 6%, and were identified as belonging to different viral families, and except two samples (Fph102 and Fph106) all samples provided enough reads to build contigs and identified some viral families.
As frequently observed with virome analysis, many reads (26%) cannot be classified, raising the need to identify all these data (Figure 6A). Reads belonging to diverse families such as Discistroviridae, Nodaviridae, Marnaviridae, Totiviridae, Tymorviridae, Tombusviridae, Solemoviridae, represented 23% of total viral reads, being the only reads identified in some samples (Figure 6B; Figure 7). Within these families, some were identified in all samples as for the Nodaviridae or Picornaviridae families or, to a lesser extent, the Tymoviridae family. Many reads (44%) were identified as belonging to the Picobinaviridae family (Figure 6A). This percentage varied among the different samples but seems to be more frequently detected in samples collected in SPM. This viral family that includes small viruses with a segmented dsRNA genome, was first suggested to infect mammals but now has been described in a large number of invertebrates and bacteria. Recently, an emerging picobirnavirus genotype was identified in patient with an acute respiratory virus, with presumably zoonotic origin (Berg et al., 2021). Then most of the reads were identified as belonging to the Astroviridae reads (15%) or Caliciviridae reads (9%) (Figure 6A). Picornaviridae reads were identified mainly in grey seal from MOL, but most of contigs could not be further identified, while in samples collected in SPM site, some contigs belonging to different genera were identified (Figure 8). Caliviridae reads were predominantly identified in samples collected in SPM whatever the seal species but the observed diversity was comparable in the two sites with contigs belonging to different genera identified (Figure 8). Reads identified as belonging to the Astroviridae family were detected in a large range of samples (Figure 7). Within this family, most of the reads were unassigned, however mamastrovirus reads were identified mainly from MOL samples. This family represented 98% of the reads detected in one sample (Fph90), and almost 92% of the reads of another sample (Fph91) (Figure 6B).
Figure 6
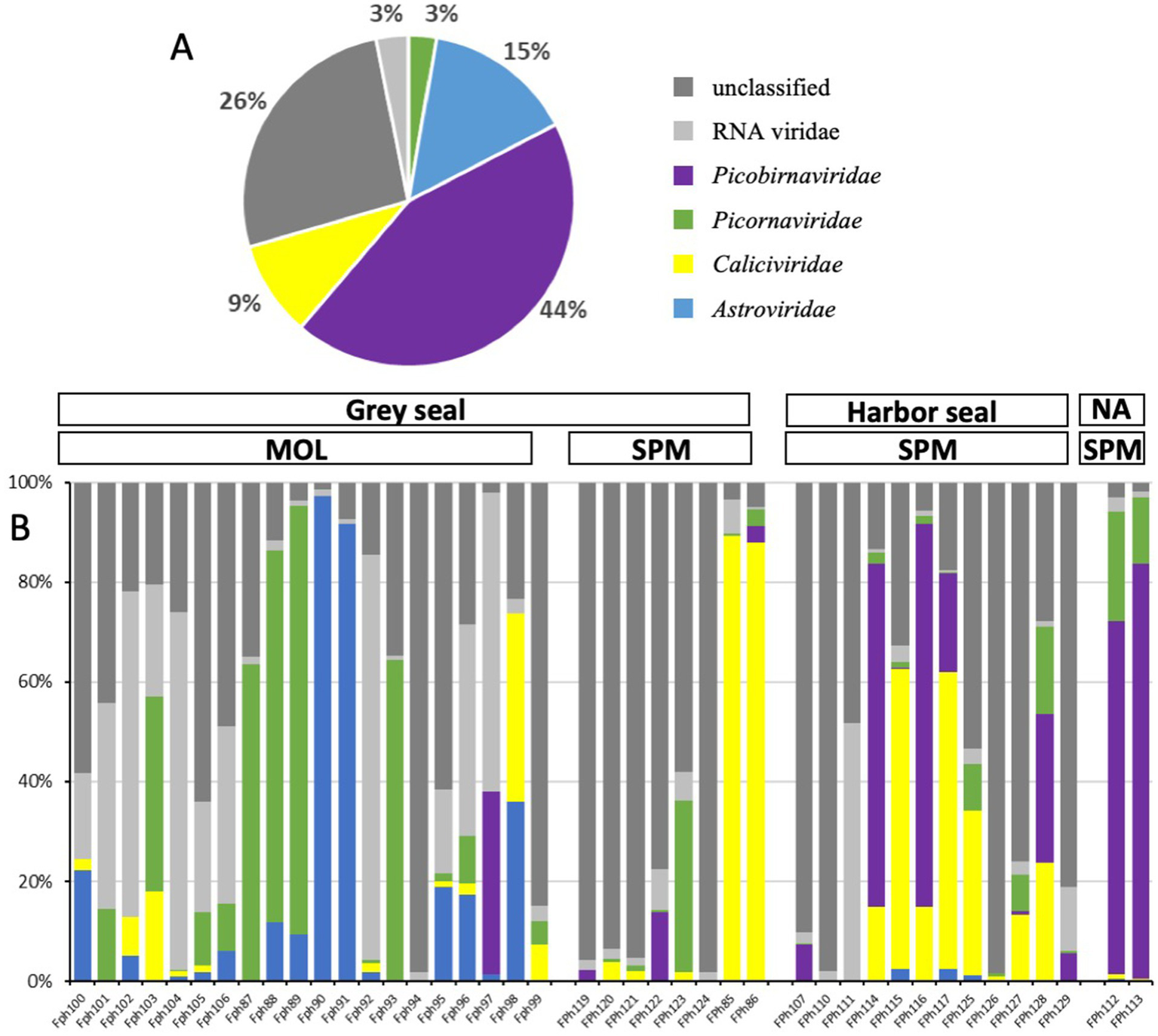
Viral reads distribution among viral families for the two seal species and the two sample sites. (A) Distribution of cumulative reads obtained for all samples expressed in % for the different families. (B) Relative abundance of the different families identified in each sample including unclassified or RNA viridae reads.
Figure 7
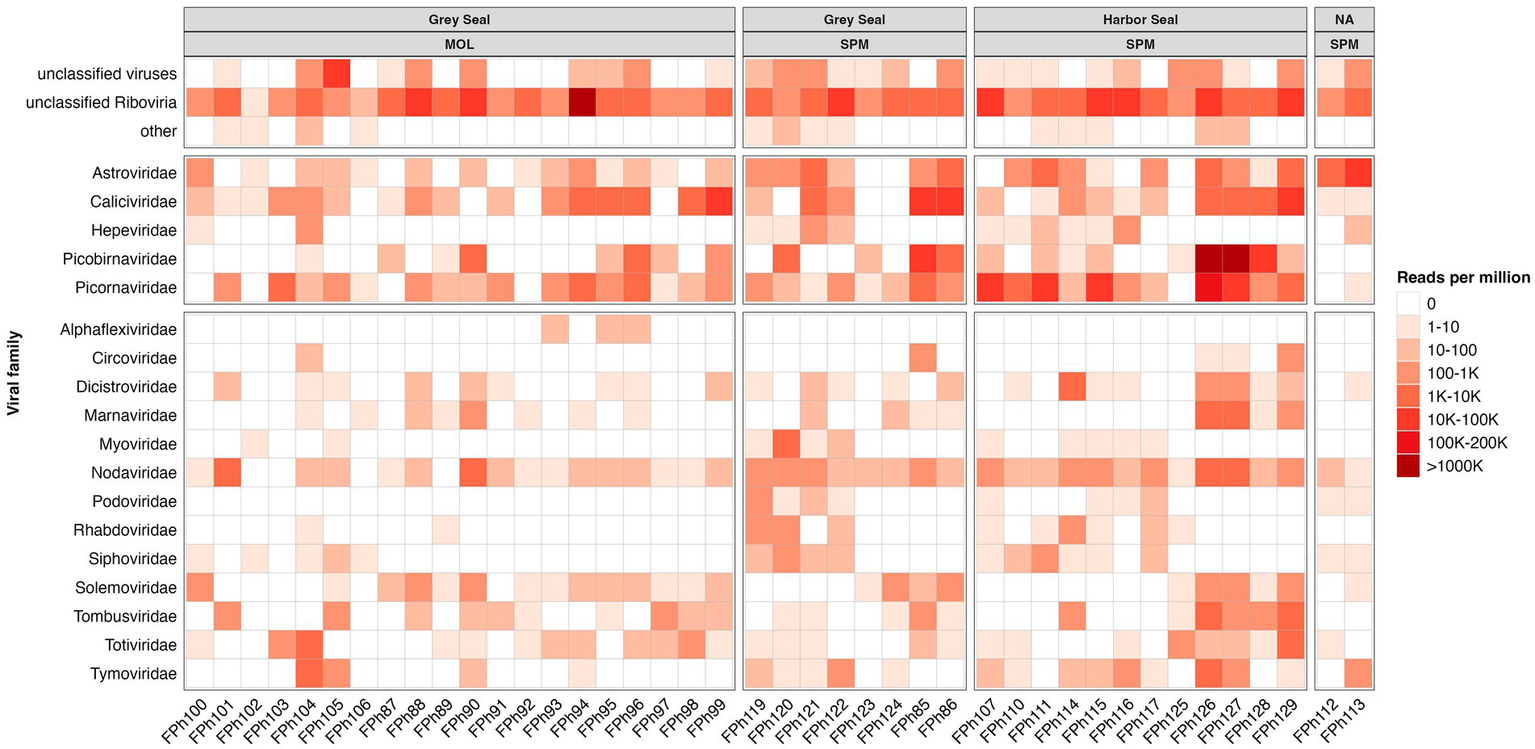
Heatmap representing the abundance of the 13 most abundant families and the other viral read abundance all included as ‘other families’, the scale bar expressed the number of reads par million (rpm).
Figure 8
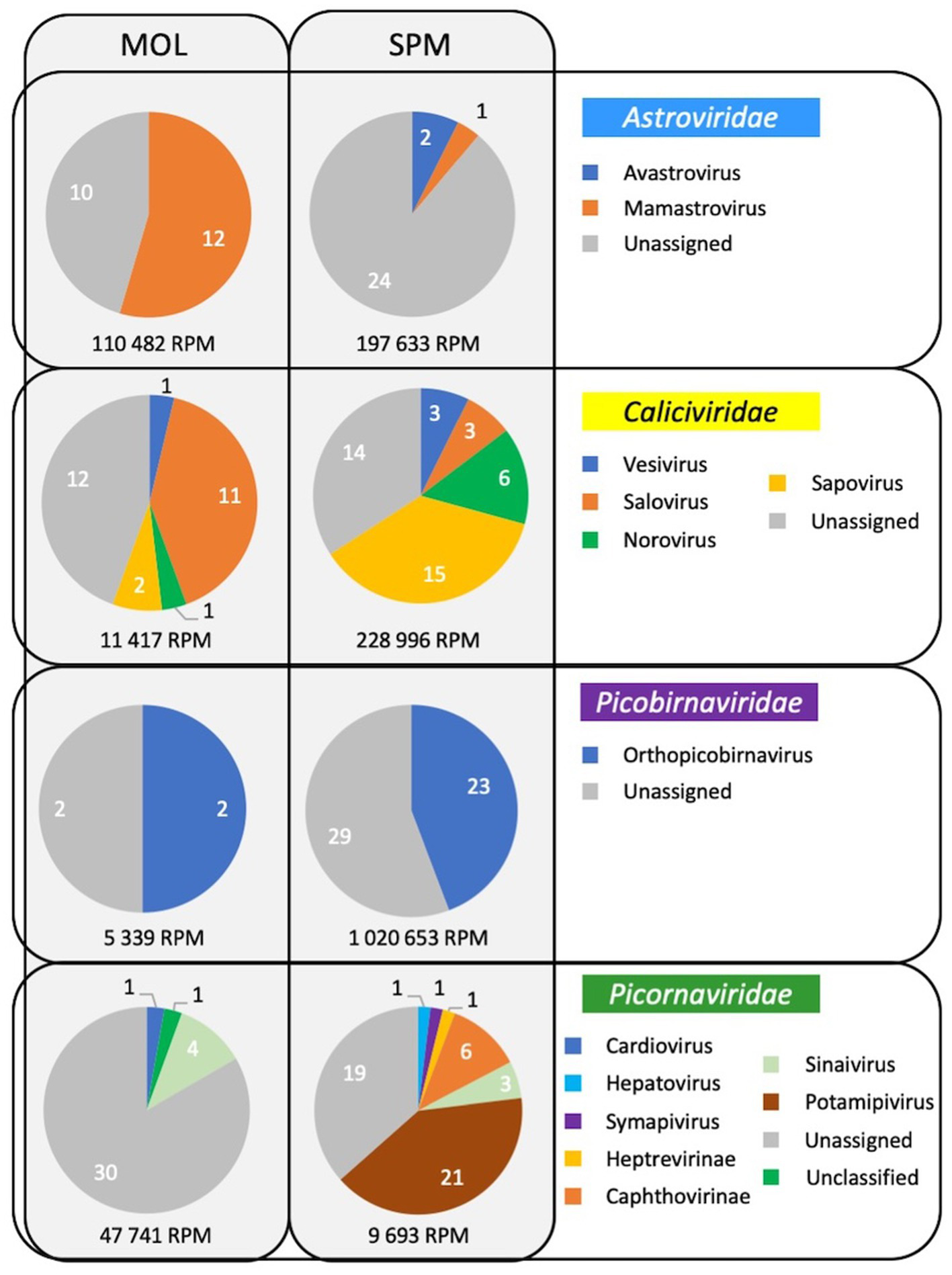
Viral diversity identified in samples collected in the two sites for four families infecting mammals.
3.3 Animal microbial communities and MST marker
3.3.1 Comparison of bacterial communities of seal fecal samples to other animal fecal sources in metropolitan France
In order to identify the specificities of seal fecal microbial communities and develop thus a seal-associated MST marker, the bacterial communities of seals were first compared to those of wild waterbirds and livestock (i.e., cattle and pigs).
The observed species richness and Shannon indexes show low alpha diversity in the grey and harbor seal feces similar to those observed previously for wild waterbird fecal samples and significantly different from the high ones observed in the cattle and pig samples (Adonis statistical test; p < 0.0001; Figure 9A). For example, the Shannon index of the cattle fecal samples ranged from 4.8 to 5.4 (median value 5.3) whereas in the grey seal feces samples the Shannon index ranged from 1.4 to 3.9 (median value 2.8).
Figure 9
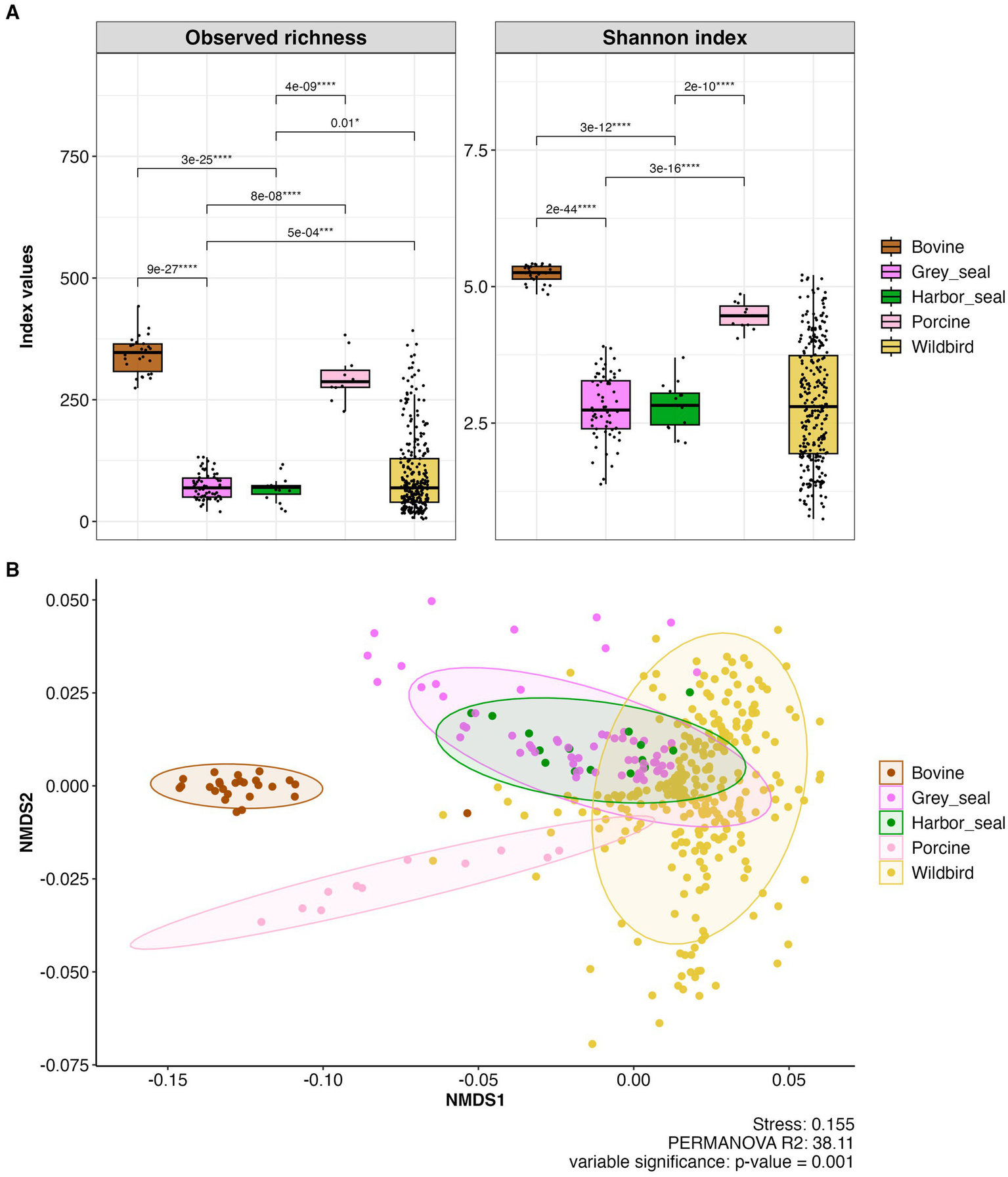
(A) Boxplots showing alpha diversity (observed species richness and Shannon indices) in fecal samples of grey and harbor seal fecal samples (n = 78), wild waterbirds (wildbird; n = 275), cattle (bovine; n = 26), pigs (porcine; n = 11) (total number of samples n = 390; metropolitan France). (B) Beta diversity in the feces of seals, wild waterbirds, cattle and pigs represented as NMDS obtained from a distance matrix calculated with the weighted UniFrac index. The data were previously normalized with CSS.
Concerning beta diversity, NMDS analysis of the entire dataset using the weighted UniFrac distance metric demonstrated that individuals from the same group (i.e., grey seals, harbor seals, wild waterbirds, cattle, or pigs) clustered together. However, there was notably greater diversity among the wild waterbird samples (stress value 0.157; Permanova; R2 = 52.21; p = 0.001; Figure 9B).
The bacterial communities observed in grey seal and harbor seal feces are significantly different from those present in the fecal samples of all the other animals (i.e., cattle, pig, and wild waterbirds) (pairwise- Adonis p-value: 0.001 and p.adjusted 0.01).
The ANCOM (Analysis of Composition of Microbiomes) analysis conducted on the five groups (i.e., grey seals, harbor seals, wild waterbirds, pigs, and cattle) revealed that the seal groups are distinguished from the others primarily by a significantly higher presence of sequences from the following genera: the [Clostridium] inoculum group, [Eubacterium] Fissicatena group, Lachnoclostridium, Peptoclostridium, Tuzzerella, belonging to Firmicutes; Anaerobiospirillum and Edwardsiella, belonging to Proteobacteria; Fusobacterium, belonging to Fusobacteroidota; and Atopobium, belonging to Actinobacteriodota (Supplementary Figures S1A,B).
Furthermore, we can note that the fecal samples closer to seal samples belong to wild waterbirds and more precisely shorebirds such as knots, oystercatchers, and curlews (Supplementary Table S1).
In addition, ANCOM identified 83 ASVs that were present only in seal feces and/or at higher levels in seal feces than in other animal sources. The 20 most specific ASVs for seals were selected and pairs of primers were designed targeting seal-associated members of the genera Fournierella, Atopobium, Slackia and the family Bifidobacteriaceae (i.e., four of these ASVs) (Supplementary Table S2).
3.3.2 Seal-associated bacterial MST qPCR marker selected
The seal-associated marker targeting the Bifidobacteriaceae family (Seal_Bifido) was found to be the most efficient. This MST marker presented a sensitivity of 89.8% on grey seal feces (n = 49) [i.e. 82.6% on seal feces from BDS (n = 23); 95% on seal feces from MOL (n = 20), and 100% on seal feces from SPM (n = 6)] while it presented a sensitivity of 100% on harbor seal feces (n = 14) from SPM and no harbor seal feces (n = 15) from BDS was found positive for this MST marker (only one detected). A specificity of 97.1% was obtained when tested on 69 non-target fecal samples [human feces (n = 9), WWTP effluent (n = 4), cattle fecal samples (n = 9), pig fecal samples (n = 10), horse feces (n = 9), poultry (n = 4; chicken and guinea fowl), pigeon (n = 1), wild waterbirds (n = 23; mallard, n = 3; knot and dunlin, n = 3; Brent goose, n = 3; seagull, n = 3; oystercatcher, n = 5; common shelduck, n = 3; great cormorant, n = 3)]. Only two cormorant feces (n = 3; FO199 and FO211) were found positive with the seal-associated marker. Interestingly, the melt curve for these non-target samples showed a single peak at a slightly different melting temperature (Tm) to that obtained for the seal feces samples (i.e., 85.5°C vs. 84.5°C).
3.3.3 Viral MST marker
Unlike the bacterial communities, the analysis of the RNA virus communities obtained in this study did not allow us to identify viral sequences specifically associated with seals that could be used to develop a viral qRT-PCR MST marker. This is due to the fact that viral communities were not characterized in other animals in this study and the percentage of unassigned reads obtained was high.
4 Discussion
This study describes the microbial communities (both bacterial and viral) present in the two most common seal species in France (i.e., grey seal and harbor seal) and across several contrasting sites. It included 132 fecal samples (from 96 grey seals, 29 harbor seals, and seven unspecified seal species) collected from various locations, including SPM and three French mainland sites. The inclusion of a large number of fecal samples from wild free-ranging seals and the analysis of both bacteria diversity and virus diversity set this study apart from previous research, which often had a smaller sample size, and frequently from dead or sick individuals. In our study, samples were collected randomly without any a priori of the status health of the seals.
4.1 Bacterial communities in seals compared to other sources
The fecal bacterial communities found in seals were significantly different from those of terrestrial domesticated mammals such as pigs and cattle (omnivorous and herbivores, respectively) and wild waterbirds in France (Boukerb et al., 2021). This difference in bacterial communities between marine and terrestrial mammals was also previously described and can be attributed to a significantly higher relative abundance of members of the Fusobacteroidota phylum in seals compared to terrestrial mammals (Nelson et al., 2013; this study). Such results were observed in previous studies on harbor seals (Switzer et al., 2023), Australian fur seals (Glad et al., 2010; Nelson et al., 2013; Smith et al., 2013) and sea lions (Bik et al., 2016).
Members of the Fusobacteroidota phylum are gram-negative bacteria that range from facultative anaerobes to obligate anaerobes. They ferment carbohydrates or amino acids to produce various organic acids, including acetic, formic, and butyric acid (Bennett and Eley, 1993; Olsen, 2014). These bacteria can be found in various habitats, such as sediments and the gut microbiomes of strict carnivores adapted to diets rich in proteins, purines, and polyunsaturated fatty acids.
Furthermore, this study enabled the comparison of bacterial communities of wild marine mammals and wild waterbirds from coastal areas, which had not been done to the best of our knowledge. The bacterial communities in seals were found to be more similar to those of wild waterbirds, especially shorebirds (as shown in the NMDS Figure 9), than to those of terrestrial mammals (i.e., pigs and cattle). Both seals and wild waterbirds exhibited a high average relative abundance of members from the phyla Firmicutes and Fusobacteriota, as well as the following genera: Clostridium sensu stricto 1, Peptoclostridium, Bacteroides, Fusobacterium, and Cetobacterium. However, wild waterbirds had a greater relative abundance of members from the phyla Proteobacteria and Actinobacteriota compared to seals.
4.2 Bacterial communities in seals
This study described the bacterial communities of grey seals and harbor seals. To our knowledge, only two studies reported bacterial data on grey seals (based a single seal, not using metabarcoding 16S, and focused on seal pups and yearlings, respectively) (Glad et al., 2010; Watkins et al., 2022), whereas the gut microbiota of harbor seals has been described in several studies (Numberger et al., 2016; Pacheco-Sandoval et al., 2019; Pacheco-Sandoval et al., 2022; Switzer et al., 2023).
Five main phyla (i.e., Firmicutes, Fusobacteriodota, Bacteroidota; Proteobacteria, and Actinobacteria) were observed in both grey seals and harbor seals in this study, consistent with previous studies on different seals species (Glad et al., 2010; Tian et al., 2022). However, the relative abundance of these main phyla differed in the feces of harbor seals in France compared to those of harbor captive seals in Germany (Baltic sea) or harbor wild seals in Mexico (Baja California) (Numberger et al., 2016; Pacheco-Sandoval et al., 2019). Specifically, the abundance of Firmicutes was nearly twice as high in the feces of French harbor seals (SPM; 65.4% ± 23.3%; n = 14 and BDS; 74.1% ± 24.0%; n = 15) compared to German (32.2%; n = 5) and Mexican (21–57%; five different seal colonies; n = 20) harbor seals. On the other hand, the relative abundance of the phylum Bacteroidota (SPM: 12.3% ± 11.6% and BDS: 9.8% ± 9.6%), was approximately half that of German and Mexico seals (27.7% and 19–36%, respectively). The relative abundance of Fusobacteriota (SPM: 18.2% ± 20.0% and BDS: 13.2% ± 15.7%) was lower than the one of German seals (27.3%) whereas the relative abundance of Fusobacteriota in Mexico seals was variable according to the seal colonies (4–42%).
Furthermore, the high Firmicutes to Bacteroidota ratio observed both in grey seals and harbor seals has been previously reported in grey, harbor, and spotted seals (Glad et al., 2010; Numberger et al., 2016; Pacheco-Sandoval et al., 2019; Tian et al., 2022). This high ratio observed may be related to their fat reserves (blubber).
There was a significant inter-individual variability in bacterial communities among the seal feces collected in this study, consistent with previous studies (Pacheco-Sandoval et al., 2019). The difference in relative abundance can be partly explained by the diet, which is known to be one of the main factors impacting the composition of the gut microbiota. In fact, diet of seals varies geographically, seasonally, inter-annually, but also inter-individually (Brown and Pierce, 1998; Gosch et al., 2019; Walton and Pomeroy, 2003; Wilson and Hammond, 2019). Seals are opportunistic hunters and adapt their carnivorous diet to local conditions, specializing in different hunting techniques, such as shredding fishing nets or traps containing shellfish. In addition to diet, other factors can contribute to the inter-individual differences in bacterial communities. These include variations in seal populations and species, the behavior and habitat of seals, such as wild free-ranging behavior and the type of habitat [e.g., sand (SPM, BDS and WAL) or rocks (MOL)]. Furthermore, individual characteristics such as age, sex, and health status can impact microbial composition. For example, Tian et al. (2020) and Pacheco-Sandoval et al. (2022) identified age-related differences in the composition of the gut microbiota in spotted seals (Phoca largha) and harbor seals in Mexico, respectively and Switzer et al. (2023) sex- and age-related differences in the composition of rectal swabs in harbor seals (Phoca vitulina richardii) in California.
Unfortunately, despite collecting freshly collected feces from the environment throughout the study, host characteristics such as age, behavior and health status could not be obtained. Among the available parameters, a significant difference in bacterial communities was observed based on site (i.e., grey seals from WAL and MOL), but no significant differences were found between the two seal species.
The fecal microbial communities can also be used to study the presence of bacteria responsible for zoonoses in marine mammals and human pathogenic bacteria in these seals, which could be sentinels for marine and human health in coastal areas.
Genera already isolated from seals such as Bisgaardia spp., Campylobacter spp., Clostridium sensus stricto 1 including Clostridium perfringens, Edwardsiella spp., Mycobacterium spp., Mycoplasma spp., Photobacterium damnselae, streptococci, and Vibrio spp. as well as genera of the family Erisipelotrichaceae, all known to contain marine mammal or human pathogenic species were found in the present study (Waltzek et al., 2012; Hughes et al., 2013; Greig et al., 2014; Lisle et al., 2004; Nelson et al., 2013; Stoddard et al., 2007; Siebert et al., 2017; Pacheco-Sandoval et al., 2024).
However, other genera also responsible for zoonotic diseases such as Brucella spp., Klebsiella pneumoniae, Leptospira spp., Pleisomonas shigelloides, Salmonella spp., Staphylococcus aureus (Waltzek et al., 2012; Greig et al., 2014; Siebert et al., 2017) were not found in these seal bacterial communities. One explanation could be that these feces were from healthy animals and the bacteria belonging to these genera were absent or present at very low concentrations in the feces analyzed in this study.
Fecal indicator bacteria (FIB) such as Escherichia coli (E. coli) are present in seal feces, as they are in the feces of other marine and terrestrial mammals (Fulham et al., 2020). Concentrations of E. coli ranging from 2.5 106 ± 4.9 106 CFU/g of feces were found in the feces of SPM seals (n = 28) in 2020 (data not shown), consistent with those ranging from 4.0 106 to 9.2 108 MPN per g of feces (n = 10) found in harbor seals in the United States (Calambokidis and McLaughlin, 1987). In addition, the genus Escherichia/Shigella was reported in the seal bacterial communities in this study, as previously reported in seals in Germany, Antarctica, Scotland, and the United States (Nelson et al., 2013; Greig et al., 2014; Numberger et al., 2016; Switzer et al., 2023).
The ubiquitous presence of E. coli in seals, as well as in other fecal sources highlights the need to develop host-associated MST bacterial markers to identify potential sources of E. coli contamination in coastal bathing areas and shellfish-harvesting areas.
4.3 Viral communities in seals
As mentioned above, there is a paucity of data on the seal virome in the literature. Despite methodological improvements in recent years, virome identification is still challenging. Indeed, most viruses are small particles with short genomes and the amount of unknown or unclassified sequences is very important, as highlighted by the Tara Oceans expeditions (Alberti et al., 2017). One possible approach is to enrich for viral sequences during the library preparation using probes targeting a large diversity of viral sequences (Briese et al., 2015). Such an approach was found to be valuable in our hands to identify sequences related to human viruses in bivalve mollusks exposed to wildlife (Bonny et al., 2021), and was used in seal samples (serum and feces) collected in an area with a high human density (Martínez-Puchol et al., 2022). As our study aimed to assess the diversity of viral sequences in an area with a very low human population density such as Saint-Pierre et Miquelon, we did not perform any enrichment step during the library preparation. This may explain why a large number of reads were identified as fish viruses or phages. Such a preponderance of phages or unknown riboviruses was previously described in subantartic and South American fur seals, where up to 89% of reads were identified as phages (Kluge et al., 2016). Our results are more comparable to their findings in Arctocephalus tropicalis than to the virus distribution obtained on samples collected from Arctocephalus australis (Kluge et al., 2016). Nevertheless, our data showed a high percentage of reads identified as belonging to four viral families, that we selected because they can infect mammals but also humans, such as Picobirnaviridae, Picornaviridae, Caliciviridae, and Astroviridae. These four families accounted for approximately 3 to 8% of the reads from fecal samples collected from subantarctic and South American fur seals (Kluge et al., 2016). As with bacteria, some differences between the two collection sites can be highlighted, with Astroviridae and Picornaviridae reads being more frequently identified at one site. However, the relatively small number of samples collected prevents any further analysis of such distributions between sites or seal species. The detection of picornaviruses in marine mammals has been described previously (Kapoor et al., 2008). More recently, some strains have been characterized such as an aquamavirus A, with the description of two novel strains, such as a harbor seal picornavirus and a ribbon seal picornavirus (Rodrigues et al., 2020). In our study, some sequences related to potamipivirus, a genus related to the aquamivirus were detected in the SPM site, but a more precise identification cannot be obtained. Some other RNA viruses such as the phocine distemper virus which belongs to the morbilivirus (Paramyxoviridae family), have been responsible for large outbreaks in different seal populations, but as enveloped viruses they may be less resistant in stool samples (VanWormer et al., 2019). As mentioned above, no data were collected on the health status of the seals, so the sequences detected may be associated with healthy seals and very little is known about viruses that cause disease in seals. In this study, we focused on RNA viruses as they are more likely to mutate than DNA viruses and can jump from one species to another. Recent works have shown that seals are infected and die from some avian influenza viruses or the human pandemic H1N1 virus (Liang et al., 2023; Mirolo et al., 2023; Plancarte et al., 2023). If the route of transmission from humans to seals is not clear, the reverse route (seals to humans) can be hypothesized through the consumption of foods such as shellfish, justifying the four viral families selected for this study, for which the enteric route is the main route of transmission (Santiago-Rodriguez and Hollister, 2023). For example, it might be interesting to find some mamastrovirus or norovirus contigs, as these viruses are also frequently detected in humans. In any case, our study therefore confirms the need to further investigate the degree of homology of these viral sequences with other known viruses that may be detected in coastal environments. For example, it has been hypothesized that some of the vesiviruses (Caliciviridae family) may have originated from marine reservoirs, with the detection of antibodies against the San Miguel sea lion viruses (a marine calicivirus) raising the possibility of the emergence of new strains (Smith et al., 1998).
4.4 Microbial source tracking seal marker
The difference between seals, wild waterbirds and terrestrial mammals allowed for the identification of specific bacteria to serve as MST markers.
Microbial Source Tracking involves identifying and applying markers associated with different hosts to the environment. Previous studies have developed markers for pigs, cattle, humans, and birds, and the seal marker developed in this study complements the existing panel of markers (Mieszkin et al., 2009, 2010). This study developed the first seal bacterial marker, to our knowledge, and it proved to be sensitive and specific to grey seals. However, the marker was found to be less prevalent in the samples of harbor seals at the BDS site.
In the future, this marker could be applied to areas with high seal numbers, such as in the Wadden Sea (Brasseur, 2017; Brasseur et al., 2015), and could be used in combination with mitochondrial DNA seal markers that can even identify the species of seals (Arnason et al., 1993), providing a complementary approach to identify a source of seal contamination.
This study did not identify any seal-specific viral sequences that could be used to develop a viral MST marker. However, it provided useful results for future comparison with viral communities from other animal sources. By providing a better identification of unclassified riboviruses or other viral families, the virome could also be useful for marker development (Santiago-Rodriguez and Hollister, 2023). More data are needed to complement such an approach.
5 Conclusion
The bacterial communities of grey and harbor seals were not significantly different, which is consistent with the partial overlap in the ecological niches (and more specifically their diet, where they co-occur) as already shown in the Eastern Channel (Planque et al., 2021) and presumed in Saint-Pierre et Miquelon (Vincent et al., 2022). They were characterized by a majority abundance of Firmicutes including the genera Clostridium sensu stricto 1 and Peptoclostridium followed by Fusobacteriota with the genus Fusobacterium, and Bacteroiota with the genus Bacteroides. However, variations in bacterial communities between sites and between individuals were observed. A similar observation was also done for the virome, raising questions about sources of contamination for the seals (food, water quality…), but also some relationship between bacteria and viruses, other than phages.
In this study, a sensitive and specific MST qPCR bacterial marker belonging to the Bifidobacteriaceae family was developed, which could be used to identify potential fecal contamination of coastal areas by seals.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/ena, ERP149173.
Author contributions
AGS: Formal analysis, Investigation, Visualization, Writing – review & editing. JoS: Formal analysis, Investigation, Writing – review & editing. CM: Formal analysis, Investigation, Writing – review & editing. CN: Formal analysis, Visualization, Writing – review & editing. JuS: Formal analysis, Investigation, Writing – review & editing. HG: Funding acquisition, Investigation, Resources, Writing – review & editing. CV: Funding acquisition, Investigation, Resources, Writing – original draft. TV: Formal analysis, Investigation, Writing – review & editing. FG: Conceptualization, Funding acquisition, Supervision, Writing – original draft. MG: Conceptualization, Formal analysis, Funding acquisition, Investigation, Supervision, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work has been supported by various projects. The “PHOBI” project was found by the scientific direction of IFREMER (Inter-center project 2019–2021). The “COPEMAM” project in SPM was funded by the OFB (French Biodiversity Agency) and the DTAM-SPM, while the “Eco-Phoques” project was funded by the Regional Council of “Hauts-de-France” and the OFB.Finally, the work was also funded by the VEO European project H2020 SC1-2019-874735.
Acknowledgments
We thank Jonathan Porter (National Laboratory Service, Environment Agency, Starcross, Devon, United Kingdom) for critical reading of the manuscript. We are also grateful for the contribution of Amine Boukerb and the Normandy network of microbial metagenomics RNAmb supported by Région Normandie and European Union (FEDER) for sequencing a part of seal fecal samples. We thank Yann Planque and Jerome Spitz (University of La Rochelle), Sarah Monnet (Picardie Nature), Frank Utizbérea, Daniel Koelsch and Gianni Boissel (DTAM) and Bruno Letournel (OFB) from SPM and Cécile Gicquel from PMI for sampling seal fecal samples from BDS and WAL, from SPM, and from MOL, respectively and Vicky Detcheverry (DTAM) for performing bacterial analyses (E. coli) on a part of SPM seal fecal samples. Finally, we are grateful to Pauline Auffret and Elisabeth Hellec from SeBiMER, Ifremer, for preparing and submitting the datasets to ENA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1484094/full#supplementary-material
Footnotes
1.^ https://gitlab.ifremer.fr/bioinfo/workflows/samba
2.^ https://blast.ncbi.nlm.nih.gov/Blast.cgi; NCBI nt; retrieved in May 2021
3.^ v4.1.0; https://primer3.ut.ee/
References
1
Alberti A. Poulain J. Engelen S. Labadie K. Romac S. Ferrera F. et al . (2017). Viral to metazoan marine plankton nucleotide sequences from the Tara oceans expedition. Sci. Data4:170093. doi: 10.1038/sdata.2017.93
2
Andersson A. F. Lindberg M. Jakobsson H. Bäckhed F. Nyrén P. Engstrand L. (2008). Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One3:e2836. doi: 10.1371/journal.pone.0002836
3
Arnason U. Gullberg A. Johnsson E. Ledje C. (1993). The nucleotide sequence of the mitochondrial DNA molecule of the grey seal, Halichoerus grypus, and a comparison with mitochondrial sequences of other true seals. J. Mol. Evol.37, 323–330. doi: 10.1007/BF00178862
4
Baily J. L. Méric G. Bayliss S. Foster G. Moss S. E. Watson E. et al . (2015). Evidence of land-sea transfer of the zoonotic pathogen Campylobacter to a wildlife marine sentinel species. Mol. Ecol.24, 208–221. doi: 10.1111/mec.13001
5
Bennett K. W. Eley A. (1993). Fusobacteria: new taxonomy and related diseases. J. Med. Microbiol.39, 246–254. doi: 10.1099/00222615-39-4-246
6
Berg M. G. Forberg K. Perez L. J. Luk K.-C. Meyer T. V. Cloherty G. A. (2021). Emergence of a distinct Picobirnavirus genotype circulating in patients hospitalized with acute respiratory illness. Viruses13:2534. doi: 10.3390/v13122534
7
Bergh Ø. BØrsheim K. Y. Bratbak G. Heldal M. (1989). High abundance of viruses found in aquatic environments. Nature340, 467–468. doi: 10.1038/340467a0
8
Bik E. M. Costello E. K. Switzer A. D. Callahan B. J. Holmes S. P. Wells R. S. et al . (2016). Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat. Commun.7:10516. doi: 10.1038/ncomms10516
9
Bolyen E. Rideout J. R. Dillon M. R. Bokulich N. A. Abnet C. C. Al-Ghalith G. A. et al . (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol.37, 852–857. doi: 10.1038/s41587-019-0209-9
10
Bonny P. Schaeffer J. Besnard A. Desdouits M. Ngang J. J. E. Le Guyader F. S. (2021). Human and animal RNA virus diversity detected by metagenomics in cameroonian clams. Front. Microbiol.12:385. doi: 10.3389/fmicb.2021.770385
11
Boukerb A. M. Noël C. Quenot E. Cadiou B. Chevé J. Quintric L. et al . (2021). Comparative analysis of fecal microbiomes from wild waterbirds to poultry, cattle, pigs, and wastewater treatment plants for a microbial source tracking approach. Front. Microbiol.12:553. doi: 10.3389/fmicb.2021.697553
12
Brasseur S. M. J. (2017). Seals in motion: How movements drive population development of harbour seals and grey seals in the North Sea. phD thesis. Wageningen, The Netherlands: Wageningen University, p. 176.
13
Brasseur S. M. J. M. van Polanen Petel T. D. Gerrodette T. Meesters E. H. W. G. Reijnders P. J. H. Aarts G. (2015). Rapid recovery of Dutch gray seal colonies fuelled by immigration. Mar. Mamm. Sci.31, 405–426. doi: 10.1111/mms.12160
14
Briese T. Kapoor A. Mishra N. Jain K. Kumar A. Jabado O. J. et al . (2015). Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. MBio6, e01491–e01415. doi: 10.1128/mBio.01491-15
15
Brown E. Pierce G. (1998). Monthly variation in the diet of harbour seals in inshore waters along the southeast Shetland (UK) coastline. Mar. Ecol. Prog. Ser.167, 275–289. doi: 10.3354/meps167275
16
Calambokidis J. McLaughlin B. (1987). Harbor seal populations and their contributions to fecal coliform pollution in Quilcene Bay, Washington. Final report for contract from Jefferson County and the Washington Department of Ecology, Olympia, Washington, p. 29.
17
Callahan B. J. McMurdie P. J. Rosen M. J. Han A. W. Johnson A. J. A. Holmes S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–583. doi: 10.1038/nmeth.3869
18
Canova R. Budaszewski R. F. Weber M. N. da Silva M. S. Puhl D. E. Battisti L. O. et al . (2021). Spleen and lung virome analysis of south American fur seals (Arctocephalus australis) collected on the southern Brazilian coast. Infect. Genet. Evol.92:104862. doi: 10.1016/j.meegid.2021.104862
19
Chiappetta C. M. Cibulski S. P. Lima F. E. S. Varela A. P. M. Amorim D. B. Tavares M. et al . (2017). Molecular detection of circovirus and adenovirus in feces of fur seals (Arctocephalus spp.). EcoHealth14, 69–77. doi: 10.1007/s10393-016-1195-8
20
Cunningham L. Baxter J. M. Boyd I. L. Duck C. D. Lonergan M. Moss S. E. et al . (2009). Harbour seal movements and haul-out patterns: implications for monitoring and management. Aquat. Conserv. Mar. Freshw19, 398–407. doi: 10.1002/aqc.983
21
den Heyer C. E. Bowen W. D. Dale J. Gosselin J.-F. Hammill M. O. Johnston D. W. et al . (2021). Contrasting trends in gray seal (Halichoerus grypus) pup production throughout the increasing Northwest Atlantic metapopulation. Mar. Mamm. Sci.37, 611–630. doi: 10.1111/mms.12773
22
Di Tommaso P. Chatzou M. Floden E. W. Barja P. P. Palumbo E. Notredame C. (2017). Nextflow enables reproducible computational workflows. Nat. Biotechnol.35, 316–319. doi: 10.1038/nbt.3820
23
EEC . Council Directive 92/43/EEC of 21 May 1992 on the conservation of natural habitats and of wild fauna and flora. No. 206. pp. 7–50. (1992).
24
Fulham M. Power M. Gray R. (2020). Diversity and distribution of Escherichia coli in three species of free-ranging Australian pinniped pups. Front. Mar. Sci.7:1171. doi: 10.3389/fmars.2020.571171
25
Glad T. Kristiansen V. F. Nielsen K. M. Brusetti L. Wright A.-D. G. Sundset M. A. (2010). Ecological characterisation of the colonic microbiota in arctic and sub-arctic seals. Microb. Ecol.60, 320–330. doi: 10.1007/s00248-010-9690-x
26
Glöckner F. O. Yilmaz P. Quast C. Gerken J. Beccati A. Ciuprina A. et al . (2017). 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol.261, 169–176. doi: 10.1016/j.jbiotec.2017.06.1198
27
Gourmelon M. Caprais M. P. Mieszkin S. Marti R. Wery N. Jarde E. et al . (2010). Development of microbial and chemical MST tools to identify the origin of the faecal pollution in bathing and shellfish harvesting waters in France. Water Res.44, 4812–4824. doi: 10.1016/j.watres.2010.07.061
28
Gosch M. Cronin M. Rogan E. Hunt W. Luck C. Jessopp M. (2019). Spatial variation in a top marine predator’s diet at two regionally distinct sites. PLoS One14:e0209032. doi: 10.1371/journal.pone.0209032
29
Greig D. J. Gulland F. M. D. Smith W. A. Conrad P. A. Field C. L. Fleetwood M. et al . (2014). Surveillance for zoonotic and selected pathogens in harbor seals Phoca vitulina from Central California. Dis. Aquat. Org.111, 93–106. doi: 10.3354/dao02762
30
Härkönen T. Hårding K. C. Lunneryd S. G. (1999). Age- and sex-specific behaviour in harbour seals Phoca vitulina leads to biased estimates of vital population parameters. J. Appl. Ecol.36, 825–841. doi: 10.1046/j.1365-2664.1999.00434.x
31
Harvey V. Côté S. D. Hammill M. O. (2008). The ecology of 3-D space use in a sexually dimorphic mammal. Ecography31, 371–380. doi: 10.1111/j.0906-7590.2008.05218.x
32
Harwood V. J. Staley C. Badgley B. D. Borges K. Korajkic A. (2014). Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol. Rev.38, 1–40. doi: 10.1111/1574-6976.12031
33
Hughes S. N. Greig D. J. Miller W. A. Byrne B. A. Gulland F. M. D. Harvey J. T. (2013). Dynamics of Vibrio with virulence genes detected in Pacific harbor seals (Phoca vitulina richardii) off California: implications for marine mammal health. Microb. Ecol.65, 982–994. doi: 10.1007/s00248-013-0188-1
34
Jarde E. Jeanneau L. Harrault L. Quenot E. Solecki O. Petitjean P. et al . (2018). Application of a microbial source tracking based on bacterial and chemical markers in headwater and coastal catchments. Sci. Total Environ.610-611, 55–63. doi: 10.1016/j.scitotenv.2017.07.235
35
Kapoor A. Victoria J. Simmonds P. Wang C. Shafer R. W. Nims R. et al . (2008). A highly divergent picornavirus in a marine mammal. J. Virol.82, 311–320. doi: 10.1128/JVI.01240-07
36
Kibbe W. A. (2007). OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res.35, W43–W46. doi: 10.1093/nar/gkm234
37
Kluge M. Campos F. S. Tavares M. de Amorim D. B. Valdez F. P. Giongo A. et al . (2016). Metagenomic survey of viral diversity obtained from feces of subantarctic and south American fur seals. PLoS One11:e0151921. doi: 10.1371/journal.pone.0151921
38
Langmead B. Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods9, 357–359. doi: 10.1038/nmeth.1923
39
Ley R. E. Hamady M. Lozupone C. Turnbaugh P. J. Ramey R. R. Bircher J. S. et al . (2008). Evolution of mammals and their gut microbes. Science320, 1647–1651. doi: 10.1126/science.1155725
40
Liang Y. Hjulsager C. K. Jensen T. K. Hammer A. S. V. Ovesen M. T. Larsen L. E. (2023). Characterization of high pathogenicity avian influenza H5Nx viruses from a wild harbor seal and red foxes in Denmark, 2021 and 2022. Influenza Other Respir. Viruses17:e13208. doi: 10.1111/irv.13208
41
Lisle J. T. Smith J. J. Edwards D. D. McFeters G. A. (2004). Occurrence of microbial indicators and Clostridium perfringens in wastewater, water column samples, sediments, drinking water, and Weddell seal feces collected at McMurdo Station, Antarctica. Appl. Environ. Microbiol.70, 7269–7276. doi: 10.1128/AEM.70.12.7269-7276.2004
42
Liu Z. DeSantis T. Z. Andersen G. L. Knight R. (2008). Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res.36:e120. doi: 10.1093/nar/gkn491
43
Lozupone C. Lladser M. E. Knights D. Stombaugh J. Knight R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J.5, 169–172. doi: 10.1038/ismej.2010.133
44
Mandal S. Van Treuren W. White R. A. Eggesbø M. Knight R. Peddada S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis.26:27663. doi: 10.3402/mehd.v26.27663
45
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J.17, 10–12. doi: 10.14806/ej.17.1.200
46
Martínez-Puchol S. Cardona L. Drago M. Gazo M. Bofill-Mas S. (2022). Viral metagenomics reveals persistent as well as dietary acquired viruses in Antarctic fur seals. Sci. Rep.12:18207. doi: 10.1038/s41598-022-23114-y
47
McKnight D. T. Huerlimann R. Bower D. S. Schwarzkopf L. Alford R. A. Zenger K. R. (2019). microDecon: a highly accurate read-subtraction tool for the post-sequencing removal of contamination in metabarcoding studies. Environ. DNA1, 14–25. doi: 10.1002/edn3.11
48
Mieszkin S. Furet J.-P. Corthier G. Gourmelon M. (2009). Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific Bacteroidales 16S rRNA genetic markers. Appl. Environ. Microbiol.75, 3045–3054. doi: 10.1128/AEM.02343-08
49
Mieszkin S. Yala J.-F. Joubrel R. Gourmelon M. (2010). Phylogenetic analysis of Bacteroidales 16S rRNA gene sequences from human and animal effluents and assessment of ruminant fecal pollution by real-time PCR. J. Appl. Microbiol.108, 974–984. doi: 10.1111/j.1365-2672.2009.04499.x
50
Mirolo M. Pohlmann A. Ahrens A. K. Kühl B. Rubio-Garcìa A. Kramer K. et al . (2023). Highly pathogenic avian influenza a virus (HPAIV) H5N1 infection in two European grey seals (Halichoerus grypus) with encephalitis. Emerg. Microbes Infect.12:e2257810. doi: 10.1080/22221751.2023.2257810
51
Nelson T. M. Rogers T. L. Brown M. V. (2013). The gut bacterial community of mammals from marine and terrestrial habitats. PLoS One8:e83655. doi: 10.1371/journal.pone.0083655
52
Numberger D. Herlemann D. P. R. Jürgens K. Dehnhardt G. Schulz-Vogt H. (2016). Comparative analysis of the fecal bacterial community of five harbor seals (Phoca vitulina). MicrobiologyOpen5, 782–792. doi: 10.1002/mbo3.369
53
Nurk S. Meleshko D. Korobeynikov A. Pevzner P. A. (2017). metaSPAdes: a new versatile metagenomic assembler. Genome Res.27, 824–834. doi: 10.1101/gr.213959.116
54
Olesen S. W. Duvallet C. Alm E. J. (2017). dbOTU3: a new implementation of distribution-based OTU calling. PLoS One12:e0176335. doi: 10.1371/journal.pone.0176335
55
Olsen I. (2014). “The family Fusobacteriaceae” in The prokaryotes: Firmicutes and Tenericutes. eds. RosenbergE.DeLongE. F.LoryS.StackebrandtE.ThompsonF. (Berlin, Heidelberg: Springer), 109–132.
56
Paar J. Willis J. R. Sette L. Wood S. A. Bogomolni A. Dulac M. et al . (2024). Occurrence of recreational water quality monitoring general fecal indicator bacteria and fecal source identification genetic markers in gray seal scat. Sci. Total Environ.934:173220. doi: 10.1016/j.scitotenv.2024.173220
57
Pacheco-Sandoval A. Lago-Lestón A. Abadía-Cardoso A. Solana-Arellano E. Schramm Y. (2022). Age as a primary driver of the gut microbial composition and function in wild harbor seals. Sci Rep12:14641. doi: 10.1038/s41598-022-18565-2
58
Pacheco-Sandoval A. Schramm Y. Heckel G. Brassea-Pérez E. Martínez-Porchas M. Lago-Lestón A. (2019). The Pacific harbor seal gut microbiota in Mexico: its relationship with diet and functional inferences. PLoS One14:e0221770. doi: 10.1371/journal.pone.0221770
59
Pacheco-Sandoval A. Schramm Y. Heckel G. Giffard-Mena I. Lago-Lestón A. , (2024). Unraveling the gut microbiota of Mexican pinnipeds: the dominance of life histories over phylogeny. Appl. Environ. Microbiol.90:e02030–23. doi: 10.1128/aem.02030-23
60
Plancarte M. Kovalenko G. Baldassano J. Ramírez A. L. Carrillo S. Duignan P. J. et al . (2023). Human influenza a virus H1N1 in marine mammals in California, 2019. PLoS One18:e0283049. doi: 10.1371/journal.pone.0283049
61
Planque Y. Huon M. Caurant F. Picard M. H. Beaudet M. Besson K. A. et al . (2018). Ecologie alimentaire des phoques. In: Eco-Phoques - Bases scientifiques pour une meilleure connaissance des phoques et de leurs interactions avec les activités humaines en Manche Nord-Est. Rapport final pour la région Hauts-de-France et l’AFB. pp. 41–80.
62
Planque Y. Spitz J. Authier M. Guillou G. Vincent C. Caurant F. (2021). Trophic niche overlap between sympatric harbour seals (Phoca vitulina) and grey seals (Halichoerus grypus) at the southern limit of their European range (eastern English Channel). Ecol. Evol.11, 10004–10025. doi: 10.1002/ece3.7739
63
Poncet S. Mercereau I. Couvrat C. Baron M. Francou M. Hemon A. et al . (2023). Monitoring seals in France – 2020-2021: Extended summary of the collective report of the National Seal Network.
64
Poncet S. Sicard M. Baron M. Francou M. Hemon A. Fremau M. H. et al . (2021). Recensement des colonies et reposoirs de phoques en France en 2019.
65
Quast C. Pruesse E. Yilmaz P. Gerken J. Schweer T. Yarza P. et al . (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res.41, D590–D596. doi: 10.1093/nar/gks1219
66
R Core Team . (2022). R: A Language and Environment for Statistical Computing BibSonomy. Available at: https://www.bibsonomy.org/bibtex/7469ffee3b07f9167cf47e7555041ee7 (Accessed August 25, 2023).
67
Ridoux V. Spitz J. Vincent C. Walton M. (2007). Grey seal diet at the southern limit of its European distribution: combining dietary analyses and fatty acid profiles. J. Mar. Biol. Assoc.87, 255–264. doi: 10.1017/S002531540705463X
68
Rince A. Baliere C. Hervio-Heath D. Cozien J. Lozach S. Parnaudeau S. et al . (2018). Occurrence of bacterial pathogens and human noroviruses in shellfish-harvesting areas and their catchments in France. Front. Microbiol.9:2443. doi: 10.3389/fmicb.2018.02443
69
Rodrigues T. C. S. Nielsen O. Burek-Huntington K. A. Popov V. L. Raverty S. Lambourn D. M. et al . (2020). Genomic characterization of picornaviruses isolated from ribbon (Histriophoca fasciata) and harbor (Phoca vitulina) seals. Front. Vet. Sci.7:716. doi: 10.3389/fvets.2020.554716
70
Santiago-Rodriguez T. M. Hollister E. B. (2023). Viral metagenomics as a tool to track sources of fecal contamination: a one health approach. Viruses15:236. doi: 10.3390/v15010236
71
Savouré-Soubelet A. Aulagnier S. Haffner P. Moutou F. Van Canneyt O. Charrassin J. B. et al . (2016). Atlas des mammifères sauvages de France, volume 1: Mammifères marins. 1st ed, Patrimoines naturels. Muséum national d’Histoire naturelle.
72
Schaeffer J. Desdouits M. Besnard A. Le Guyader F. S. (2023). Looking into sewage: how far can metagenomics help to detect human enteric viruses?Front. Microbiol.14:1161674. doi: 10.3389/fmicb.2023.1161674
73
Siebert U. Rademaker M. Ulrich S. A. Wohlsein P. Ronnenberg K. Prenger-Berninghoff E. (2017). Bacterial microbiota in harbor seals (Phoca vitulina) from the North Sea of Schleswig-Holstein, Germany, around the time of morbillivirus and influenza epidemics. J. Wildl. Dis.53, 201–214. doi: 10.7589/2015-11-320
74
Simeone C. A. Gulland F. M. D. Norris T. Rowles T. K. (2015). A systematic review of changes in marine mammal health in North America, 1972-2012: the need for a novel integrated approach. PLoS One10:e0142105. doi: 10.1371/journal.pone.0142105
75
Sinigalliano C. Kim K. Gidley M. Yuknavage K. Knee K. Palacios D. et al . (2021). Microbial sourcet of fecal indicating bacteria in coral reef waters, recreational waters, and groundwater of Saipan by real-time quantitative PCR. Front. Microbiol.11:650. doi: 10.3389/fmicb.2020.596650
76
Smith S. C. Chalker A. Dewar M. L. Arnould J. P. Y. (2013). Age-related differences revealed in Australian fur seal Arctocephalus pusillus doriferus gut microbiota. FEMS Microbiol. Ecol.86, 246–255. doi: 10.1111/1574-6941.12157
77
Smith A. W. Skilling D. E. Cherry N. Mead J. H. Matson D. O. (1998). Calicivirus emergence from ocean reservoirs: zoonotic and interspecies movements. Emerg. Infect. Dis.4, 13–20. doi: 10.3201/eid0401.980103
78
Stoddard R. A. Miller W. G. Foley J. E. Lawrence J. Gulland F. M. D. Conrad P. A. et al . (2007). Campylobacter insulaenigrae isolates from northern elephant seals (Mirounga angustirostris) in California. Appl. Environ. Microbiol.73, 1729–1735. doi: 10.1128/AEM.01816-06
79
Strubbia S. Schaeffer J. Besnard A. Wacrenier C. Le Mennec C. Garry P. et al . (2020). Metagenomic to evaluate norovirus genomic diversity in oysters: impact on hexamer selection and targeted capture-based enrichment. Int. J. Food Microbiol.323:108588. doi: 10.1016/j.ijfoodmicro.2020.108588
80
Strubbia S. Schaeffer J. Oude Munnink B. B. Besnard A. Phan M. V. T. Nieuwenhuijse D. F. et al . (2019). Metavirome sequencing to evaluate norovirus diversity in sewage and related bioaccumulated oysters. Front. Microbiol.10:2394. doi: 10.3389/fmicb.2019.02394
81
Sunagawa S. Acinas S. G. Bork P. Bowler C. Coordinators T. O. Eveillard D. et al . (2020). Tara oceans: towards global ocean ecosystems biology. Nat. Rev. Microbiol.18, 428–445. doi: 10.1038/s41579-020-0364-5
82
Switzer A. D. Callahan B. J. Costello E. K. Bik E. M. Fontaine C. Gulland F. M. D. et al . (2023). Rookery through rehabilitation: microbial community assembly in newborn harbour seals after maternal separation. Environ. Microbiol.25, 2182–2202. doi: 10.1111/1462-2920.16444
83
Thompson P. M. Mcconnell B. J. Tollit D. J. Mackay A. Hunter C. Racey P. A. (1996). Comparative distribution, movements and diet of harbour and grey seals from Moray firth, N. E. Scotland. J. Appl. Ecol.33:1572. doi: 10.2307/2404795
84
Tian J. Du J. Han J. Wang Z. Fu Z. Lu Z. (2020). Gut microbiota variations in a rescued spotted seal Phoca largha pup. Aquat. Biol.29, 105–109. doi: 10.3354/ab00729
85
Tian J. Sanganyado E. Wang Z. Kong Z. Han J. Lu Z. et al . (2022). Spotted seals (Phoca largha) harbor unique gut microbiota shaped by their host habitat. Sci. Total Environ.832:155015. doi: 10.1016/j.scitotenv.2022.155015
86
VanWormer E. Mazet J. A. K. Hall A. Gill V. A. Boveng P. L. London J. M. et al . (2019). Viral emergence in marine mammals in the North Pacific may be linked to Arctic Sea ice reduction. Sci. Rep.9:15569. doi: 10.1038/s41598-019-51699-4
87
Vincent C. Huon M. Caurant F. Dabin W. Deniau A. Dixneuf S. et al . (2017). “Grey and harbour seals in France: distribution at sea, connectivity and trends in abundance at haulout sites” in Deep Sea Research Part II: Topical Studies in Oceanography, Abundance, distribution and habitats of Atlantic and Mediterranean marine megafauna. ed. VincentC., vol. 141 (Amsterdam, Netherlands: Elsevier), 294–305.
88
Vincent C. Kiszka J. Garnier T. Wynn-Simmonds S. Planque Y. Caputo M. et al . (2022). COPEMAM - Mieux comprendre les conflits entre pêches et mammifères marins dans l’archipel de Saint Pierre et Miquelon.
89
Walton M. Pomeroy P. (2003). Use of blubber fatty acid profiles to detect inter-annual variations in the diet of grey seals Halichoerus grypus. Mar. Ecol. Prog. Ser.248, 257–266. doi: 10.3354/meps248257
90
Waltzek T. B. Cortés-Hinojosa G. Wellehan J. F. X. Gray G. C. (2012). Marine mammal zoonoses: a review of disease manifestations. Zoonoses Public Health59, 521–535. doi: 10.1111/j.1863-2378.2012.01492.x
91
Watkins C. A. Gaines T. Strathdee F. Baily J. L. Watson E. Hall A. J. et al . (2022). A comparative study of the fecal microbiota of gray seal pups and yearlings - a marine mammal sentinel species. MicrobiologyOpen11:e1281. doi: 10.1002/mbo3.1281
92
Wilson L. J. Hammond P. S. (2019). The diet of harbour and grey seals around Britain: examining the role of prey as a potential cause of harbour seal declines. Aquat. Conserv.29, 71–85. doi: 10.1002/aqc.3131
93
Zakhour M. Maalouf H. Di Bartolo I. Haugarreau L. Le Guyader F. S. Ruvoën-Clouet N. et al . (2010). Bovine norovirus: carbohydrate ligand, environmental contamination, and potential cross-species transmission via oysters. Appl. Environ. Microbiol.76, 6404–6411. doi: 10.1128/AEM.00671-10
Summary
Keywords
Grey seal, harbor seal, bacterial communities, fecal contamination, microbial source tracking, RNA virome
Citation
Godino Sanchez A, Serghine J, Le Mennec C, Noël C, Schaeffer J, Goraguer H, Vincent C, Vitré T, Le Guyader FS and Gourmelon M (2024) Grey and harbor seals in France (mainland and Saint-Pierre et Miquelon): microbial communities and identification of a microbial source tracking seal marker. Front. Microbiol. 15:1484094. doi: 10.3389/fmicb.2024.1484094
Received
21 August 2024
Accepted
07 November 2024
Published
04 December 2024
Volume
15 - 2024
Edited by
Chih-Horng Kuo, Academia Sinica, Taiwan
Reviewed by
Po-Yu Liu, National Sun Yat-sen University, Taiwan
Asunción Lago-Lestón, Center for Scientific Research and Higher Education in Ensenada, Mexico
Updates
Copyright
© 2024 Godino Sanchez, Serghine, Le Mennec, Noël, Schaeffer, Goraguer, Vincent, Vitré, Le Guyader and Gourmelon.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michèle Gourmelon, Michele.Gourmelon@ifremer.fr
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.