
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 09 January 2025
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1483633
This article is part of the Research TopicHorizontal Transfer of Antibiotic Resistance Genes in the Environment: Dynamic, Contributing Factors, and ControlView all 8 articles
 Hongmei Liu1,2,3†
Hongmei Liu1,2,3† Na Xia1†
Na Xia1† Fanan Suksawat3
Fanan Suksawat3 Bundit Tengjaroenkul3
Bundit Tengjaroenkul3 Yue Hu1
Yue Hu1 Xiaofeng Zhou1
Xiaofeng Zhou1 Xiaojiang Li1
Xiaojiang Li1 Cuiqin Huang2
Cuiqin Huang2 Yinli Bao2
Yinli Bao2 Qiong Wu2
Qiong Wu2 Chunrong Zhang2
Chunrong Zhang2 Sunpetch Angkititrakul3*
Sunpetch Angkititrakul3* Bin Xiang1*
Bin Xiang1* Xin Wu1*
Xin Wu1*Background: Proteus mirabilis is a conditionally pathogenic bacterium that is inherently resistant to polymyxin and tigecycline, largely due to antibiotic resistance genes (ARGs). These ARGs can be horizontally transferred to other bacteria, raising concerns about the Inc plasmid-mediated ARG transmission from Proteus mirabilis, which poses a serious public health threat. This study aims to investigate the presence of Inc plasmid types in pig-derived Proteus mirabilis in Kunming, Yunnan, China.
Methods: Fecal samples were collected from pig farms across six districts of Kunming (Luquan, Jinning, Yiliang, Anning, Songming, and Xundian) from 2022 to 2023. Proteus mirabilis isolates were identified using IDS and 16S rRNA gene sequencing. Then, positive strains underwent antimicrobial susceptibility testing and incompatibility plasmid typing. Multi-drug-resistant isolates with positive incompatibility plasmid genes were selected for whole-genome sequencing. Resistance and Inc group data were then isolated and compared with 126 complete genome sequences from public databases. Whole-genome multi-locus sequence typing, resistance group analysis, genomic island prediction, and plasmid structural gene analysis were performed.
Results: A total of 30 isolates were obtained from 230 samples, yielding a prevalence of 13.04%. All isolates exhibited multi-drug resistance, with 100% resistance to cotrimoxazole, erythromycin, penicillin G, chloramphenicol, ampicillin, and streptomycin. Among these, 15 isolates tested positive for the IncQ1α plasmid repC gene. The two most multi-drug-resistant and repC-positive strains, NO. 15 and 21, were sequenced to compare genomic features on Inc groups and ARGs with public data. Genome analysis revealed that the repC gene was primarily associated with IncQ1α, with structural genes from other F-type plasmids (TraV, TraU, TraN, TraL, TraK, TraI, TraH, TraG, TraF, TraE/GumN, and TraA) also present. Strain NO. 15 carried 33 ARGs, and strain NO. 21 carried 38 ARGs, conferring resistance to tetracyclines, fluoroquinolones, aminoglycosides, sulfonamides, peptides, chloramphenicol, cephalosporins, lincomycins, macrolides, and 2-aminopyrimidines.
Conclusion: The repC gene is primarily associated with IncQ1α, with structural genes from other F-type plasmids. A comparison with 126 public genome datasets confirmed this association.
The issue of antibiotic resistance (AMR) in Enterobacteriaceae bacteria is a serious and growing global public health problem. AMR increases mortality and prolongs disease progression. A meta-analysis reported that the mortality relative risk of carbapenem-resistant Enterobacteriaceae to carbapenem-susceptible ones is 2.14 (95%CI 1.85 to 2.48; I2 = 80.0%) (Zhou et al., 2021). Carbapenems are often used as “last-line agents” to defend against multidrug-resistant Gram-negative organisms. Tigecycline and polymyxin have been used to treat serious infections caused by carbapenemase-producing Enterobacteriaceae.
However, one member of the Enterobacteriaceae family, Proteus mirabilis (P. mirabilis), has been largely overlooked. P. mirabilis was reported as inherently resistant to polymyxin and tigecycline (Alqurashi et al., 2022). However, only a few articles have reported the risks associated with this inherent resistance, which can horizontally transferred to other Enterobacteriaceae.
P. mirabilis is a conditional pathogen primarily found in the intestines of animals, which was first identified and named by Hauser in 1885 (Drzewiecka, 2016). It belongs to the Enterobacteriaceae family within the genus Proteus, along with P. penneri, P. vulgaris, P. myxofaciens, P. hauseris, and three unnamed genomospecies (Proteus genomospecies 4, 5, and 6) (O’Hara et al., 2000). It is mainly found in the gastrointestinal tracts of humans and animals, and humans (Sanches et al., 2021), pigs (Qu et al., 2022), dogs (Kyung et al., 2024), chickens (Ramatla et al., 2024), fish (Anifowose et al., 2024), and other animals (Kang et al., 2021). If hosts become infected, it can lead to gastroenteritis (Ravindran et al., 2023), urinary tract infections (Chakkour et al., 2024), meningitis (Costa Filho et al., 2023), and other diseases. P. mirabilis has been used as an indicator of food and fecal contamination, which has led to an underestimation of its pathogenicity (Yu et al., 2021). In recent years, there have been increasing reports of P. mirabilis causing diseases in animals, which has had significant adverse effects on the livestock and poultry industry (Li et al., 2023). Due to the overuse of antibiotics, the losses caused by multidrug-resistant P. mirabilis are increasing. A prevalence of AMR P. mirabilis isolated from meat products in southern Brazil reported the high prevalence of multi-drug resistance (MDR) isolates in chicken (76.5%), which threatens the breeding industry seriously (Sanches et al., 2023).
The critical issue of antibiotic resistance extends beyond its direct impact to include the horizontal transfer of antibiotic resistance genes (ARGs) (Elhoshi et al., 2023). ARGs carried on plasmids can confer acquired antimicrobial resistance to recipient bacteria through mechanisms such as conjugation, transformation, and transduction (Mei et al., 2024). This HGT allows ARGs to spread rapidly among different bacterial species, exacerbating the problem of MDR (Darby et al., 2023). This spread of ARGs through plasmid-mediated HGT can rapidly transform sensitive bacteria into MDR. Typically, plasmids exhibiting MDR can be classified according to their incompatibility (Inc) because of the feature that plasmids cannot stably coexist with other plasmids within the same bacterial strains (Meinersmann, 2019). According to the Inc classification, plasmids could be divided into several groups (IncA, IncB, IncC, IncD, IncF, IncH, IncI, IncJ, IncK, IncL, IncM, IncN, IncO, IncQ, and IncP), with each group having distinct characteristics (Foley et al., 2021). The Inc plasmid typing method has become the most common approach for plasmid typing. Research on the correlation of incompatibility groups from different regions is a crucial tool for studying the genetic characteristics of plasmids.
P. mirabilis is widely distributed in the natural environment, with animal gastrointestinal tracts serving as essential vectors for the horizontal transmission of antibiotic resistance genes (ARGs). Due to its low pathogenicity, P. mirabilis has often been overlooked. However, it is inherently resistant to polymyxin tigecycline, raising concerns about its potential role in ARG transmission, which poses a significant public health threat.
This study aims to investigate the presence of Inc plasmid types in P. mirabilis isolated from pigs in Kunming, Yunnan. Specifically, 30 P. mirabilis strains were isolated from 230 pig fecal samples collected in Kunming between 2022 and 2023. These isolates were analyzed for AMR and the prevalence of Inc plasmid rep genes. Based on these results, two isolates with notable AMR profiles and mobile genetic elements were selected for whole-genome sequencing. Then, the complete genome data from this study were then compared with publicly available P. mirabilis data from NCBI. This study could provide a theoretical foundation for subsequent research on the transfer and dissemination of ARGs, as well as the mechanisms of drug resistance in P. mirabilis.
From 2022 to 2023, a total of 230 fecal swab samples were collected from swine farms across various districts of Kunming. Specifically, 34 samples were collected from the Luquan district of Kunming in March 2022, 22 samples from the Jinning district in June 2022, 52 samples from the Yiliang district in August 2022, 33 samples from the Anning district in March 2023, 39 samples from Songming in June 2023, and 42 samples from the Xundian district in August 2023. The samples were placed in sterilized centrifuge tubes containing Brain Heart Infusion (BHI) agar, refrigerated, and promptly transported to the Yunnan Joint International R&D Center of Veterinary Public Health for bacterial culture. Initial characterization of the samples began on the day they arrived at the laboratory. This formula, n = (z)2p(1–p)/d2, was applied to calculate the required sample size. In the formula, the ‘n’ represents the required sample size; ‘z’ the level of confidence according to a standard normal distribution (for a level of confidence of 95%, z = 1.96); the ‘p’ the expected prevalence, and the ‘d’ allowable error (here it was set to be 5%). The expected prevalence was approximately 18.03% for P. mirabilis, according to the report by Rui et al. (2016).
The fecal swabs were inoculated in buffered peptone water (BPW, Huankai) and incubated in a constant temperature shaker at 37°C, 140 rpm/min for 12 h. Subsequently, 100 μL of the culture was transferred onto Xylose Lysine Deoxycholate (XLD) agar plates and incubated under the same conditions for an additional 12 h. Suspected colonies were selected, re-inoculated onto fresh XLD agar, and incubated at 37°C for 18 h. Finally, a single colony from suspected isolates was then purified by culturing on lysogeny broth (LB) medium. To confirm purity, all isolates were triple-passaged to obtain fresh colonies, followed by Gram staining and biochemical tests, including TSI (Triple Sugar Iron), urea, indole, phenylalanine deaminase, ornithine decarboxylase, and lactose utilization.
Preliminary identification was conducted by amplifying the sequences based on the GenBank-registered ids gene cluster of P. mirabilis. Bacteria with positive ids gene clusters were further analyzed by blasting the 16S rRNA genes. Primers are shown in Table 1.
A total of 30 isolates were subjected to antimicrobial susceptibility testing using the disk diffusion method and minimum inhibitory concentration (MIC) assays. Both methods were conducted according to the guidelines of the American Clinical and Laboratory Standards Institute (CLSI). MIC results were interpreted according to the recommendations of the Clinical Laboratory Standard Institute guidelines (CLSI, 2018; Humphries et al., 2021). Isolates were classified as multidrug-resistant if they were resistant to three or more antimicrobial drugs in the panel (Dargatz et al., 2016). The 19 antibiotics from 8 categories included: aminoglycoside antibiotic have Neomycin (NEO, 30 μg) and Streptomycin (STR, 10 μg); Beta-lactam antibiotics have Cefaclor (CEC, 30 μg), Cefotaxime (CTX, 30 μg), Ceftriaxone (CRO, 30 μg), Cefepime (FEP, 30 μg), Penicillin (PEN,10 units) (for Staphylococcus spp.) or 1 unit (for Streptococcus spp.), Ampicillin (AMP, 10 μg), Imipenem (IPM, 10 μg); tetracycline class of antibiotics have Tigecycline (TGC, 15 μg) and Tetracycline (TC, 30 μg); Fluoroquinolones have Norfloxacin (NOR, 10 μg) and Ciprofloxacin (CIP, 5 μg); Amphenicols have Chloramphenicol (CHL, 30 μg); Macrolide antibiotics have Erythromycin (ERY, 15 μg); lincosamide antibiotics have Clindamycin (CLI, 2 μg); Sulfonamides and trimethoprim have Cotrimoxazole (SXT, 1.25/23.75 μg); While polymyxin antibiotics contain polymyxin E (colistin) and polymyxin B, they were tested using the MIC. The antimicrobial susceptibility results were evaluated based on the CLSI M100-S23 criteria.
The inc/rep PCR method was used to detect replicons on reference plasmids (Carattoli et al., 2005). Eighteen pairs of primers were designed for 18 Inc plasmid genes, including rep and par. Primers are shown in Table 2. After the amplification reaction, the results were observed using 1% agarose gel electrophoresis.
The characteristic gene-positive isolates from the 30 strains were used as donor strains in a conjugation test. The recipient strain was an engineered Escherichia coli DH5α. Donor and recipient strains were mixed in a 4:1 ratio and inoculated into LB medium, followed by incubation at 37°C for 4 h. Subsequently, 100 μL of the donor, recipient, and conjugated cultures were plated on MacConkey agar containing antibiotics to which all strains were resistant. The plates were then incubated at 37°C for 24 h.
Isolates exhibiting severe MDR and containing transfer elements were selected for whole-genome sequencing. DNA was extracted using SDS combined with a purification column. Then, the genome was quality-checked. Qualified DNA was randomly fragmented using Covaris. The ONT SQK-LSK109 and EXP-NBD104/114 kits (Oxford Nanopore Technologies1) were used for online library construction: the qualified DNA samples were purified using magnetic beads to select DNA fragments with an average size of 200–400 bp.
The 1 μL sample was taken for Qubit quantification, damage repair, end-repair, barcode labeling, pooling library preparation, and sequencing junction ligation. Their whole genomes were sequenced using Illumina MiSeq and Oxford Nanopore MinION platforms. The genomes of this study were assembled using the NECAT software, which used long-read data from the Nanopore. Subsequently, the final high-quality assembly was achieved using Pilon software (v.1.22) to correct the Illumina MiSeq sequencing data based on NGS reads. Multiple assembly versions were mirrored to create the final map.
This study employed the complete level whole genome data from both self-isolated and sequenced strains, as well as 126 publicly available strains. The public complete genome data for 126 strains of P. mirabilis were downloaded from the NCBI database on March 17, 2024. The whole genome data of the self-isolated strains were annotated using the GO, KEGG, and RAST2 databases.
PubMLST3 was used to analyze the genomic relationship of selected P. mirabilis and the published strains based on their location and sampling time. The 16 representative strains from various years and regions were selected. Gene loci were plotted using Entro-wgMLST v1.0. The phylogenetic trees were graphed using ggplot2 in R.
The resistome (the entire set of ARGs) of P. mirabilis was analyzed using the Comprehensive Antibiotic Resistance Database (CARD4).
The expression of resistance gene types and the phylogenetic analysis of ARGs based on sequence site information across the whole genome datasets were conducted.
IslandPath 1.0.6 software was employed to predict the genome islands and potential horizontal gene transfers using the sequence composition prediction method. The expression of genome island-related genes in the whole-genome-sequenced strains in this study was analyzed. SnapGene Viewer 6.1.2 software was used for the gene island visualization.
The annotation results of these genomes were examined to determine the expression of transfer-related genes. The proportion of Inc plasmid structure genes in P. mirabilis was determined. The presence or absence of repA and repC, along with their sequence site information, was analyzed for evolutionary relationships.
The Ids gene was amplified by PCR. The positive P. mirabilis product was 829 bp. The 16S rRNA gene PCR amplification produced a band at 1,300 bp, which was sequenced and compared using NCBI’s BLAST software.5 It revealed that all isolates had more than 98% nucleic acid sequence homology with P. mirabilis. Ultimately, 30 isolates were obtained from 230 samples, resulting in a prevalence of 13.04% (see Figure 1).
The results were evaluated according to the CLSI M100-S23 criteria by the diameter of the inhibitory zone. The summary results are presented in Figure 2. Overall, among all P. mirabilis isolates (n = 30), the highest frequency of resistance was observed against SXT, ERY, PEN, AMP, CHI, STR, TC, and TGC (100%), followed by CEC (97%), CLI (97%), and CIP (70%). All 30 isolates exhibited MDR. Specifically, 4 isolates were resistant to 16 drugs is strain 16, 21, 22, 25 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-NEO-CET-CRO-NOR-CIP-CHI), 3 isolates to 15 drugs is strain 13, 17 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-CET-CRO-NOR-CIP-CHI), and strain 15 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-NEO-CRO-NOR-CIP-CHI), 1 isolate to 14 drugs is strain 8 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-CET-FEP-CIP-CHI), 8 isolates to 13 drugs is strain 3, 23, 24 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CLI-NEO-CET-CIP-CHI); and strain 7, 9 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-FEP-CIP- CHI); and strain 7 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-NOR-CIP-CHI); and strain 10 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CET-CRO-CIP-CHI); and strain 26 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-NEO-CRO-CHI); 8 isolates to 12 drugs is strain 2, 4, 5, 19, 20 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-CIP-CHI); and strain 1 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CLI-NOR-CIP-CHI); and strain 14 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-CET-CHI); and strain 30 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-NOR-CHI), 6 isolates to 11 drugs is strain 11, 12, 18, 27, 28, 29 (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI-CHI).
For antimicrobial resistance, 14 resistance patterns were identified, with the most common being the 10-resistance pattern (AMP-ERY-PEN-SXT-STR-PMB-PMC-TC-CEC-CLI), accounting for 20%, followed by 12-resistance (17%), 15-resistance (14%). The MDR rate (resistant to over nine types of antimicrobials) reached 100%, as shown in Figure 2. According to Magiorakos et al. (2012), the isolated strains were classified into MDR, XDR, and PDR categories, which can be viewed in the attached table.
The results indicated that 15 isolates were positive for the IncQ1α plasmid repC gene, specifically isolates 4–9, 11, 15, 19–21, 23, 24, 26, and 27, with a detection rate of 50%. The repC gene amplification results for IncQ1α were shown in Figure 3, with amplified bands appearing at 802 bp, consistent with the expected results. The remaining isolates were negative for the Inc marker genes. According to the antimicrobial susceptibility results, two multi-drug-resistant and growth-dominant strains were selected for subsequent experiments. Considering the number of multi-drug-resistant in the repC-positive isolates, it can be inferred that NO. 15 and NO. 21 were suitable subjects for further research.
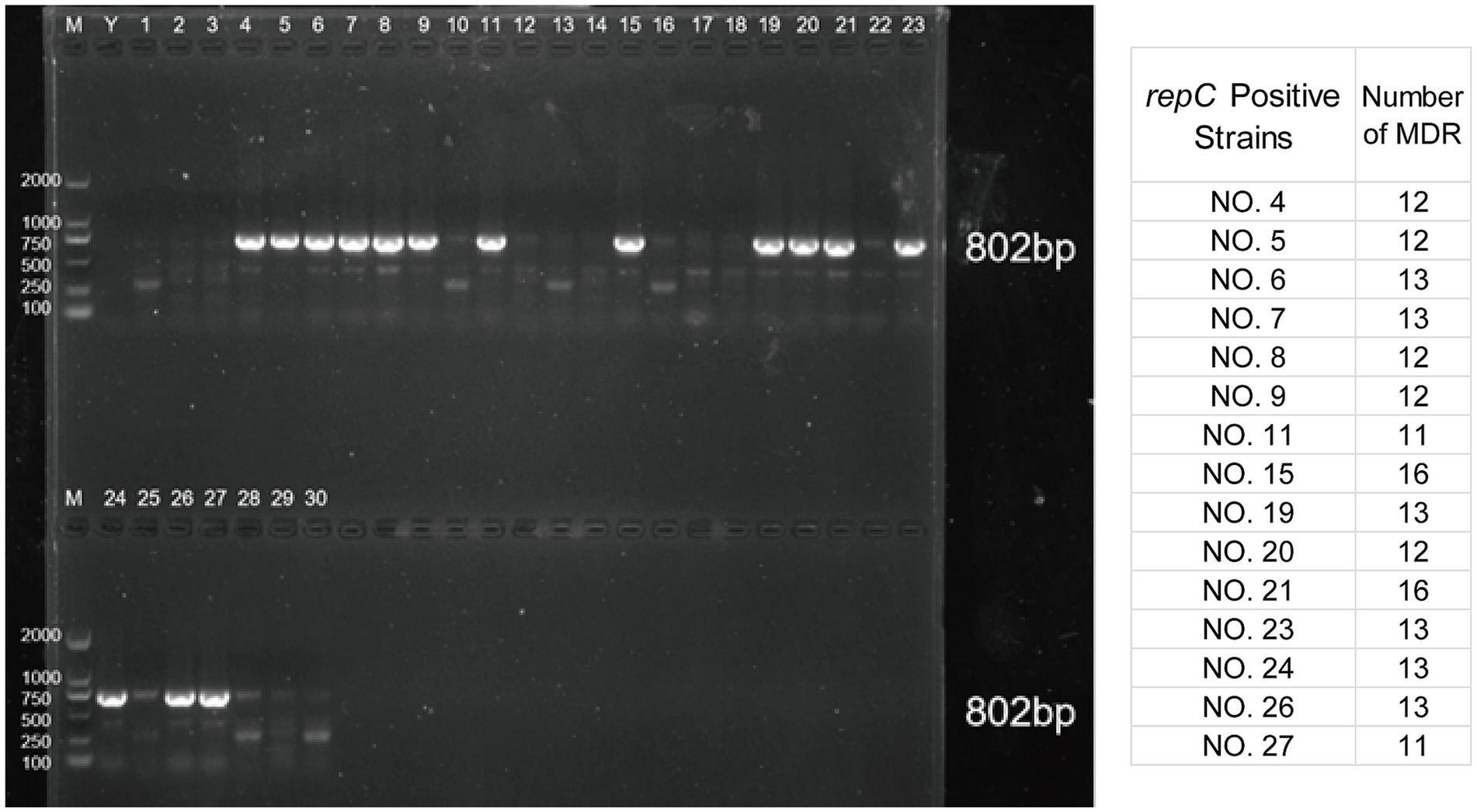
Figure 3. PCR amplification results of IncQ1𝛼 plasmid repC gene. M lane is DL2000 Maker, Y is a negative sample, and lanes 1–30 are samples.
IncQ plasmid-positive isolates formed colorless and transparent colonies on MacConkey’s medium supplemented with 10 μg/mL ampicillin, 30 μg/mL tetracycline, 5 μg/mL ciprofloxacin, and 15 μg/mL erythromycin. The control strain, E. coli DH5α, did not grow under these conditions. The control strain, E. coli DH5α, did not grow under these conditions. No transfer events were observed in any of the conjugation experiments.
Isolates NO. 15 and NO. 21 were both 16-drug-resistant strains and tested positive for the IncQα1 plasmid repC gene.
The assembled genome sequences and functional annotation were mapped using the BLAST Ring Image Generator (BRIG 0.95). The whole genome landscapes of strains 15 and 21 are shown in Figure 4.
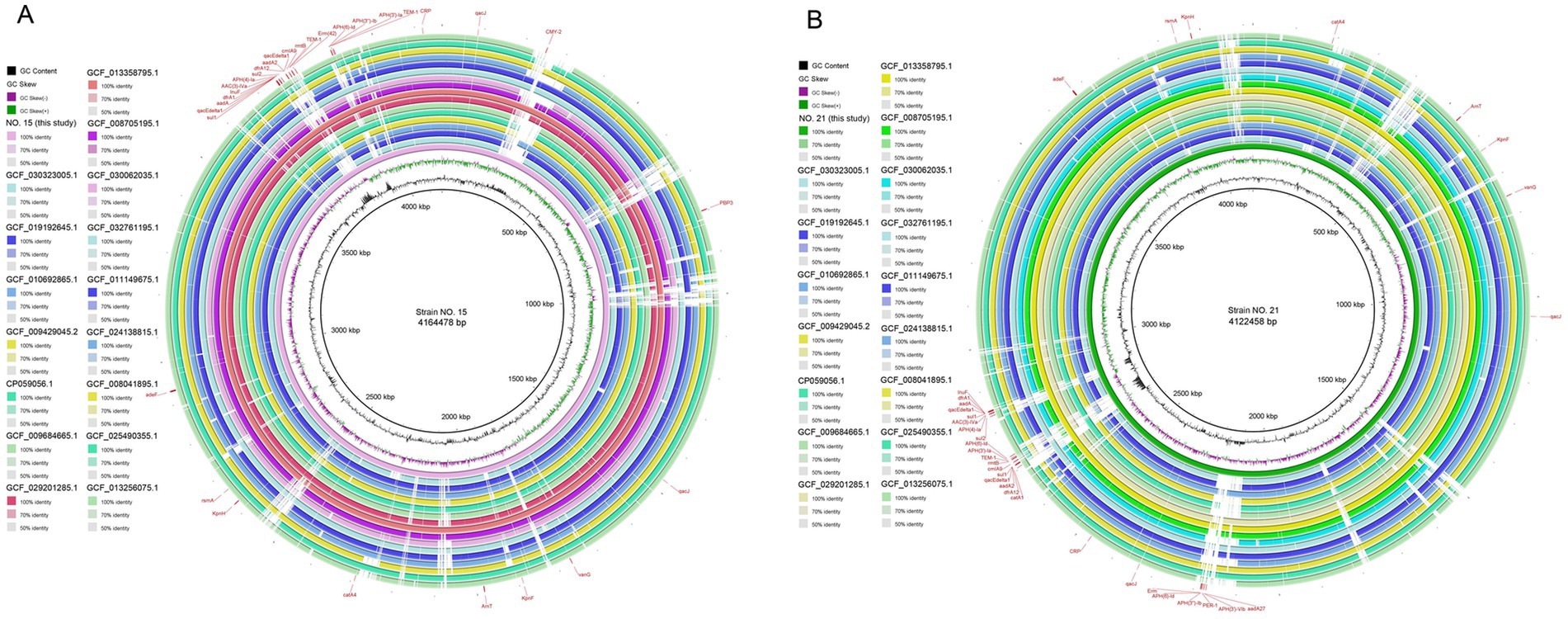
Figure 4. Parts A and B are a whole genome mapping of strains 15 and 21. From the outer to the inner circle: Target strain genome location coordinates; positive chain restriction-modification enzyme coding genes (color corresponding to COG classification); negative chain restriction-modification enzyme coding genes (color corresponding to COG classification); rRNA and tRNA distribution; Genome GC skew value (the specific algorithm is G−C/G + C). The inward purple part indicates that the content of G is lower than that of C in this region, and the outward pink part is the opposite: genomic GC content (the outward green part indicates that the GC content of this region is higher than the average GC content of the whole genome, and the inward blue part is the opposite, and the higher the peak value, the greater the difference from the average GC content).
This study analyzed two strains of P. mirabilis and 16 strains from diverse global sources using whole-genome phylogenetic tree analysis. The results revealed three major strain groups. Interestingly, no correlations were observed among P. mirabilis strains from human, food, or animal origins. This indicates a close relationship between human-derived P. mirabilis and those found in food, animals, and pets. The two strains studied closely resembled porcine-derived P. mirabilis, reported in 2019 in Henan, China (NCBI genome assembly number GCA_013358795.1) (see Figure 5).
The sequences of strains NO. 15 and 21 were compared with the CARD databases using the Resistance Gene Identifier (RGI). Resistance gene annotation and statistical analysis were conducted for sequences with an identity greater than 95%. The antimicrobial resistance gene information for strains 15 and 21 is presented in Table 3.
Strain NO. 15 carried 33 ARGs, with qacJ present in two subtypes. Ten were efflux pump genes, 14 genes in synthesizing antibiotic-inactivating enzymes, five genes in altering therapeutic targets, and three genes in substituting therapeutic targets. One gene was involved in protecting the therapeutic target. These genes confer resistance to a wide range of antibiotics, such as tetracyclines, fluoroquinolones, aminoglycosides, sulfonamides, polypeptides, chloramphenicols, cephalosporins, lincosamides, macrolides, and 2-aminopyrimidine.
Strain NO. 21 carried 38 ARGs, including two subtypes of qacJ, sul1, catA1, and APH (6)-Id genes. Among the ARGs, 11 were involved in efflux pumps, 16 in synthesizing antibiotic-inactivating enzymes, 7 in target alteration, 3 in target substitution, and 1 in target protection. These genes mediate resistance to tetracyclines, fluoroquinolones, aminoglycosides, sulfonamides, polypeptides, chloramphenicols, cephalosporins, lincosamides, macrolides, and 2-aminopyrimidines.
To identify the most prevalent resistance genes in P. mirabilis, a meta-resistome analysis based on 128 strains (the 126 publicly available data and two strains of this study) of P. mirabilis was performed. According to the annotation results, 105 antimicrobial resistance genes were identified. These genes resisted various antibiotics, including aminoglycosides, beta-lactams, cephalosporins, chloramphenicols, chloromycetin, diaminopyrimidines, elfamycin, glycopeptides, oxazolidinone, phenicol, pleuromutilin, macrolide, lincosamide, streptogramin, monobactam, carbapenem, cephalosporin, penam, nucleoside, phosphonic acid, polypeptides, quinolone, rifamycins, and tetracyclines. The MFS, SMR, and RND efflux pumps were also discovered. There were 12 genes expressed in 90% of the strains. They were cephalosporins ARG PBP3; chloramphenicols ARG catA4; elfamycin ARG EF-Tu; glycopeptides ARG vanG; polypeptides ARG ArnT; quinolone ARG gyrB; MFS efflux pump-related genes KpnE, KpnF; the RND efflux pump-related genes adeF, rsmA, and CRP, the SMR efflux pump-related gene qacJ. Notably, the polypeptide gene ArnT was present in 99.2% of P. mirabilis strains. Alternatively, it might elucidate the principal cause of its intrinsic resistance to polymyxin. Details are shown in Figure 6. Supplementary material presents the phylogenetic relationships of the primary resistance genes (vanG, rsmA, PBP3, KpnH, KpnF, gyrB, EF-TU, CRP, catA4, ArnT, and adeF), including those from 2 strains in this experiment and 16 representative various region strains from public data.
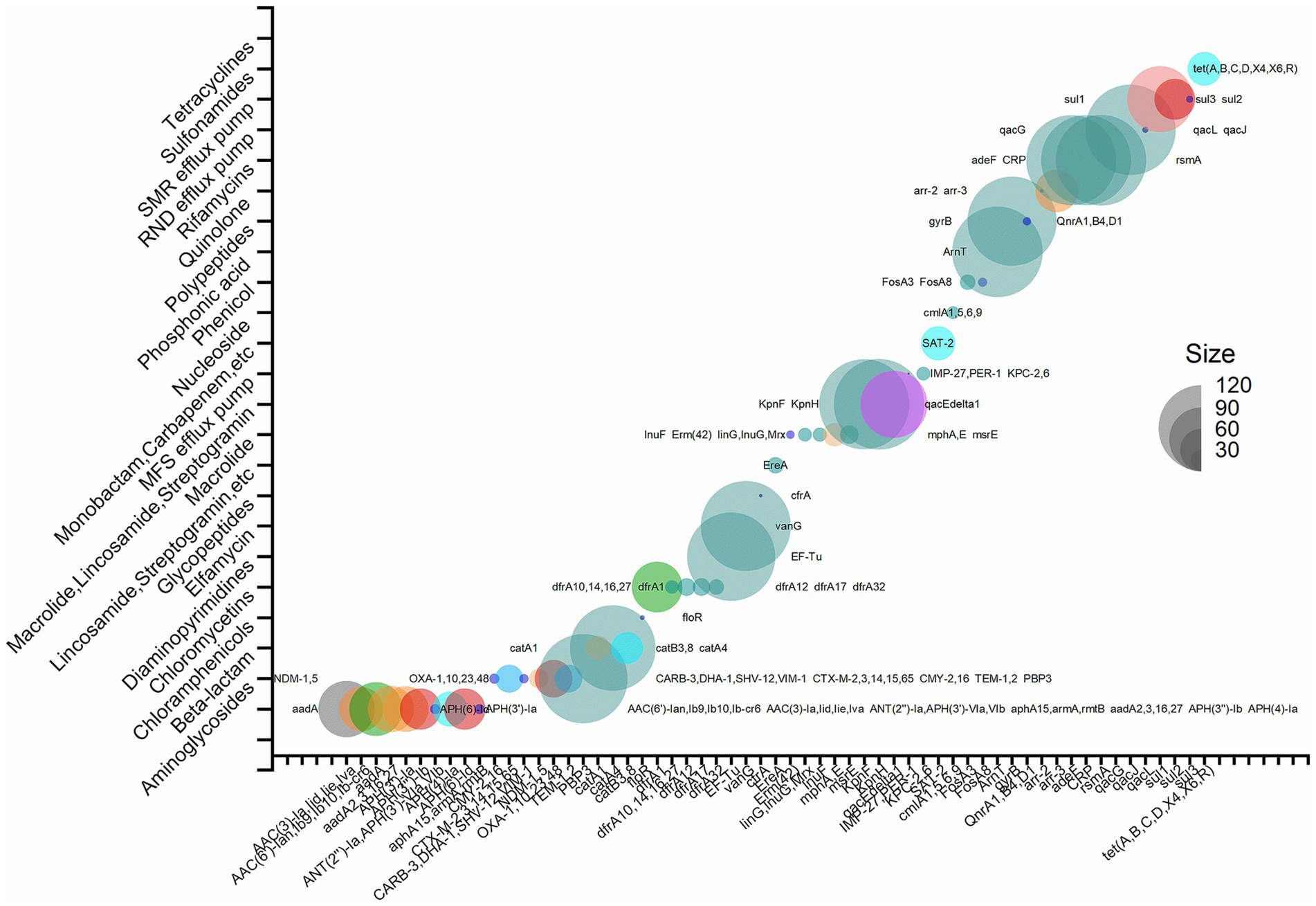
Figure 6. Resistome bubble plots of 128 strains of P. mirabilis (126 strains with publicly available data and two strains in this study).
Strain No. 15 contained nine genome islands, totaling 350,110 bp and an average length of 38,901 bp. Strain No. 21 contained eight genome islands, with a total length of 343,701 bp and an average length of 42,962 bp, as shown in Table 4. Strain NO. 15 contains 20 ARGs across nine genome islands, representing 60.61% (20/33) of its total ARGs. Similarly, NO. 21 has 27 ARGs distributed over eight genome islands, accounting for 71.05% (27/38) of its total ARGs.
In these two P. mirabilis strains, several typical F-type plasmid conjugation transfer region structure genes of the T4SS secretion system, such as traF, traH, traG, traN, traU, traW, traC, traV, traB, traA, traE, traD, and traI, were present in Island 1 of strain No. 15 and Island 4 of strain NO. 21. These structural genes were predominantly clustered together. Both plasmids contain transposons such as the integrator int., the flhCD promoters, and tnpA (see Figure 7).
In the islands containing the repC gene of IncQ1α, both NO. 15 island 8 and NO.21 island 6 harbor the repC and repA genes. The related ARGs included the mercury resistance genes, bla-TEM, strA, strB, sul genes, and various transposons such as tnp (see Figure 8).
The annotation results of 128 genomes were analyzed to determine the proportion and composition of each structural gene. The proportions were as follows: TraV (50.78%), TraU (51.56%), TraN (62.50%), TraL (50.78%), TraK (50.00%), TraI (43.75%), TraH (61.72%), TraG (84.38%), TraF (15.63%), TraE/GumN (100%), TraA (50.00%), and TraW (1.56%). Additionally, icmH was 92.19%, mobA was 99.22%, mobB was 99.22%, mobP1 was 10.94%, mobH was 44.53%, and moBI was 50.78%. Other genes had proportions less than 5%. Classification of their expression patterns revealed that they all contained type-F plasmid structural genes; the patterns included 48 distinct types among the 128 bacterial strains, indicating no correlation between them (see Figure 9).
From 128 whole genome datasets, 26 repC genes were identified. Among these, the repC genes of GCF013357505 and GCF011383025 were incomplete, resulting in 24 complete datasets. In this study, 15 repC gene sequences were obtained through sequencing. Notably, all 41 repC genes belong to the IncQ1 group. Combining these 15 sequences with the 24 publicly available ones, an evolutionary tree was constructed using the maximum likelihood method using MEGA 5.0 software. These repC genes were sorted into three groups. The overall mean genetic distance of the repC genes (15 sequences in this study and the 24 publicly available ones) was 0.006 ± 0.001.
A total of 14 repA genes were annotated across 126 public data strains. Among these, the repA of GCF002197405.1 was identified as IncN and GCF0230895.1 as IncFII. The remaining 11 strains (GCF_014843115.1, GCF_015169015.1, GCF_018972025.2, GCF_025490355.1, GCF_026016045.1, GCF_026016105.1, GCF_026016125.1, GCF_026016145.1, GCF_033170445.1, GCF_033215415.1, and GCF_033439945.1) contained incomplete repA genes with consistent sequence expression. These genes are closely related to sequences such as the Vibrio alginolyticus strain C1579 plasmid pC1579 and Shewanella aestuarii strain PN3F2 plasmid pPN3F2_1, among others. Additionally, the repA strain GCF002180235.1 is closely related to the Escherichia coli strain BK31611 plasmid pBK31611. The two repA strains analyzed in this study show a close plasmid relationship with RSF1010 (IncQ-1α plasmid), pO26-CRL-125 (IncK2 plasmid), and p0716-KPC (IncFII plasmid) (see Figure 10).
Proteus mirabilis is a conditionally pathogenic bacterium that is commonly found in the natural environment and animals (Girlich et al., 2020). It was reported to be inherently resistant to polymyxin and tigecycline. Horizontal transfer of resistance genes can transform drug-sensitive bacteria into multidrug-resistant strains rapidly. The inherent resistance of P. mirabilis poses significant risks if such a transfer occurs. However, this topic has been largely overlooked due to the severe neglect of its conditional pathogenicity. This study aims to investigate the presence of Inc plasmid types in pig-derived P. mirabilis isolated from Kunming, Yunnan. The 30 strain of P. mirabilis were successfully isolated from 230 pig fecal samples in Kunming, with a prevalence of 13.04% (30/230). This was consistent with that of Chinnam et al. (2021), who found 23 and 26 strains in 160 pork samples and 163 rectal swabs of normal pigs in Andhra Pradesh, India, with prevalences of 14.38 and 15.95%, respectively.
The wgMLST results included 16 representative strains from various years and regions, along with two strains from this study. They exhibited no relationship among P. mirabilis strains from human, food, or animal origins, meaning a close genomic relationship between human-derived P. mirabilis and the strains found in food, animals, and pets. The two strains studied closely resembled porcine-derived P. mirabilis, which was reported in 2019 in Henan, China (NCBI genome assembly number GCA_013358795.1).
The antimicrobial susceptibility results exhibited a serious AMR in the pig-derived P. mirabilis in Kunming, Yunnan. All the strains were MDR, with 100% resistance to cotrimoxazole, erythromycin, penicillin G, chloramphenicol, ampicillin, and streptomycin. P. mirabilis was naturally resistant to tetracycline and polymyxin with a resistance rate of 100% (Alqurashi et al., 2022), which is consistent with the current experimental results. The MDR rate in this study reached 100%, which was higher than the 76.7% reported in Northeast China (Sun et al., 2020) and the 78.13% reported in Brazil (Sanches et al., 2019) of the P. mirabilis isolates from chickens, which fully reflects the MDR of P. mirabilis. These results highlighted a significant drug resistance issue in Kunming pig-derived P. mirabilis, warranting attention. It may serve as a reservoir of resistance genes within the gut microbiota.
The resistome results of the two whole-genome-sequenced strains revealed that strain NO. 15 harbored a total of 33 ARGs, while strain NO. 21 carried 38 ARGs. These genes confer resistance to a wide range of antibiotics, including tetracyclines, fluoroquinolones, aminoglycosides, sulfonamides, polypeptides, chloramphenicols, cephalosporins, lincosamides, macrolides, and 2-aminopyrimidine. The resistome results from the genome of 126 strains of public data and two strains in this study revealed that 12 genes were expressed in 90% of the strains, including Cephalosporins ARG PBP3, Chloramphenicols ARG catA4, Elfamycin ARG EF-Tu, Glycopeptides ARG vanG, Polypeptides ARG ArnT, Quinolone ARG gyrB, MFS efflux pump-related genes KpnE and KpnF, RND efflux pump-related genes adeF, rsmA, and CRP, and the SMR efflux pump-related gene qacJ. Notably, the polypeptide gene ArnT was present in 99.2% of P. mirabilis strains, potentially explaining its intrinsic resistance to polymyxin (Petrou et al., 2016).
Among the 30 P. mirabilis strains, 15 contained the Inc plasmid characteristic gene, repC of IncQ1α. Analysis of Inc plasmid characteristic genes in 126 public databases of P. mirabilis revealed the presence of repC genes. A combined analysis of all repC genes from these 15 strains and public data showed high similarity, with an overall mean genetic distance of 0.006 ± 0.001 across the 15 sequences in this study and the 24 publicly available ones.
Analysis of whole genome data from 2 strains and 126 public datasets revealed that both strains isolated in this experiment contain structural genes of type-F plasmids, such as TraV, TraU, TraN, TraL, TraK, TraI, TraH, TraG, TraF, TraE/GumN, and TraA. Among all 128 datasets, the proportion of structural genes in type-F plasmids is relatively high, with the majority of structural genes ranging over 40%. The other Inc structural genes that mediated the conjugation transfer, such as MobC, trbB, trbD, and so on, mainly account for less than 5%. Analysis of type-F plasmid structural genes in 128 strains showed that all contained these genes, but their compositions varied significantly. The 128 strains exhibited 48 patterns, with no correlation found.
An equilibrium in the transmission process may favor the persistence of P. mirabilis. The repC gene was primarily associated with IncQ1α plasmids, along with structural genes of other F-type plasmids. Evidence includes the pattern indicating no regularity. Genes such as TraV, TraU, TraN, TraL, TraK, TraI, TraH, TraG, TraF, TraE/GumN, and TraA have higher expression levels. The repC expresses only one type. The lack of regularity suggested the multi-source origins, while the high expression levels of F-type plasmid structural genes indicated a significant prevalence. The repC gene has a relatively close genetic distance. More extensive epidemiological studies to understand its severity were needed.
The P. mirabilis isolates derived from pigs in Kunming were predominantly positive for the repC gene associated with the IncQ1α plasmid and also carried structural genes of the F-type plasmid. This trend was similarly observed in the 126 publicly available genome datasets used for comparison. P. mirabilis may maintain this plasmid composition to achieve a balance in its propagation and survival.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
HL: Writing – original draft, Writing – review & editing. NX: Investigation, Writing – review & editing. FS: Formal analysis, Writing – review & editing. BT: Visualization, Writing – review & editing. YH: Data curation, Methodology, Writing – review & editing. XZ: Methodology, Writing – review & editing. XL: Software, Writing – review & editing. CH: Writing – review & editing, Validation. YB: Writing – review & editing, Project administration. QW: Methodology, Writing – review & editing. CZ: Writing – review & editing, Methodology. SA: Supervision, Writing – review & editing. BX: Conceptualization, Resources, Writing – review & editing. XW: Funding acquisition, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Yunnan Provincial Innovation Team of Key Technologies for Prevention and Control of Important Livestock and Poultry Diseases [Grant No. 202405AS350004], Yunnan Province Science and Technology Department, the Agricultural Joint Special Project [grant number 202301BD070001-113], Yunnan Province Science and Technology Department, the Basic Research Special Project [grant number 202201AU070181], the Yunnan Joint International R&D Center of Veterinary Public Health [grant number 2023BI60619], and Open Fund Project from Engineering Research Center for the Prevention and Control of Animal Original Zoonosis of Fujian Province University, College of Life Science, Longyan University [Grant No. 2022K001].
The authors thank the Ruili Penghe Agricultural Food Development Co., Ltd, Knorigene TechnologiesLtd. and Wuhan Generead Biotechnologies Co. Ltd. for their kindly helps in lab work. Thanks also go to Doubao of ByteDance for grammar revision.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1483633/full#supplementary-material
Alqurashi, E., Elbanna, K., Ahmad, I., and Abulreesh, H. H. (2022). Antibiotic resistance in Proteus mirabilis: mechanism, status, and public health significance. J. Pure Appl. Microbiol. 16, 1550–1561. doi: 10.22207/JPAM.16.3.59
Anifowose, O. R., Oladosu, G. A., and Omotosho, O. O. (2024). Occurrence and characterization of Proteus mirabilis from infected farmed African catfish in Ogun state, Nigeria. Mol. Biol. Rep. 51:446. doi: 10.1007/s11033-023-08973-6
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Chakkour, M., Hammoud, Z., Farhat, S., El Roz, A., Ezzeddine, Z., and Ghssein, G. (2024). Overview of Proteus mirabilis pathogenicity and virulence. Insights into the role of metals. Front. Microbiol. 15:1383618. doi: 10.3389/fmicb.2024.1383618
Chinnam, B. K., Nelapati, S., Tumati, S. R., Bobbadi, S., Chaitanya Peddada, V., and Bodempudi, B. (2021). Detection of β-lactamase-producing Proteus mirabilis strains of animal origin in Andhra Pradesh, India and their genetic diversity. J. Food Prot. 84, 1374–1379. doi: 10.4315/JFP-20-399
Costa Filho, F. F., Furlan, A., and Avner, B. S. (2023). Spontaneous Proteus mirabilis meningitis in adults requiring an extended antibiotic course: case report and literature review. Cureus 15:e39225. doi: 10.7759/cureus.39225
Darby, E. M., Trampari, E., Siasat, P., Gaya, M. S., Alav, I., Webber, M. A., et al. (2023). Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 21, 280–295. doi: 10.1038/s41579-022-00820-y
Dargatz, D. A., Kopral, C. A., Erdman, M. M., and Fedorka-Cray, P. J. (2016). Prevalence and antimicrobial resistance of salmonella isolated from cattle feces in United States Feedlots in 2011. FBD 13, 483–489. doi: 10.1089/fpd.2016.2128
Drzewiecka, D. (2016). Significance and roles of Proteus spp. Bacteria in natural environments. Microb. Ecol. 72, 741–758. doi: 10.1007/s00248-015-0720-6
Elhoshi, M., El-Sherbiny, E., Elsheredy, A., and Aboulela, A. G. (2023). A correlation study between virulence factors and multidrug resistance among clinical isolates of Proteus mirabilis. Braz. J. Microbiol. 54, 1387–1397. doi: 10.1007/s42770-023-01080-5
Foley, S. L., Kaldhone, P. R., Ricke, S. C., and Han, J. (2021). Incompatibility group I1 (IncI1) plasmids: their genetics, biology, and public health relevance. Microbiol. Mol. Biol. Rev. 85. doi: 10.1128/MMBR.00031-20
Girlich, D., Bonnin, R. A., Dortet, L., and Naas, T. (2020). Genetics of acquired antibiotic resistance genes in Proteus spp. Front. Microbiol. 11:256. doi: 10.3389/fmicb.2020.00256
Humphries, R., Bobenchik, A. M., Hindler, J. A., and Schuetz, A. N. (2021). Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st Edition. J. Clin. Microbiol. 59:e0021321. doi: 10.1128/JCM.00213-21
Kang, Q., Wang, X., Zhao, J., Liu, Z., Ji, F., Chang, H., et al. (2021). Multidrug-resistant Proteus mirabilis isolates carrying Bla(OXA-1) and Bla(NDM-1) from wildlife in China: increasing public health risk. Integr. Zool. 16, 798–809. doi: 10.1111/1749-4877.12510
Kyung, S. M., Lee, J. H., Lee, E.-S., Xiang, X.-R., and Yoo, H. S. (2024). Emergence and genomic chion of Proteus mirabilis harboring Bla(NDM-1) in Korean companion dogs. Vet. Res. 55:50. doi: 10.1186/s13567-024-01306-w
Li, Y., Yin, M., Fang, C., Fu, Y., Dai, X., Zeng, W., et al. (2023). Genetic analysis of resistance and virulence characteristics of clinical multidrug-resistant Proteus mirabilis isolates. Front. Cell. Infect. Microbiol. 13:1229194. doi: 10.3389/fcimb.2023.1229194
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mei, L., Song, Y., Liu, X., Li, K., Guo, X., Liu, L., et al. (2024). Characterization and implications of IncP-2A plasmid pMAS152 harboring multidrug resistance genes in extensively drug-resistant Pseudomonas aeruginosa. Microorganisms 12. doi: 10.3390/microorganisms12030562
Meinersmann, R. J. (2019). The biology of IncI2 plasmids shown by whole-plasmid multi-locus sequence typing. Plasmid 106:102444. doi: 10.1016/j.plasmid.2019.102444
O’Hara, C. M., Brenner, F. W., and Miller, J. M. (2000). Classification, identification, and clinical significance of Proteus, Providencia, and Morganella. Clin. Microbiol. Rev. 13, 534–546. doi: 10.1128/CMR.13.4.534
Petrou, V. I., Herrera, C. M., Schultz, K. M., Clarke, O. B., Vendome, J., Tomasek, D., et al. (2016). Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid a glycosylation. Science 351, 608–612. doi: 10.1126/science.aad1172
Qu, X., Zhou, J., Huang, H., Wang, W., Xiao, Y., Tang, B., et al. (2022). Genomic investigation of Proteus mirabilis isolates recovered from pig farms in Zhejiang Province, China. Front. Microbiol. 13:952982. doi: 10.3389/fmicb.2022.952982
Ramatla, T., Ramaili, T., Lekota, K., Mileng, K., Ndou, R., Mphuthi, M., et al. (2024). Antibiotic resistance and virulence profiles of Proteus mirabilis isolated from broiler chickens at abattoir in South Africa. Vet. Med. Sci. 10:e1371. doi: 10.1002/vms3.1371
Ravindran, D. R., Kannan, S., Jeyakumar, D., and Marudhamuthu, M. (2023). Characterization of phenyl propiolic acid from Proteus mirabilis DMTMMR-11 and evaluation of its mode of action against Yersinia enterocolitica (MTCC-840) an in-vitro and in-vivo based approach. Microb. Pathog. 182:106258. doi: 10.1016/j.micpath.2023.106258
Rui, Y., Tingting, W., Xiaoyou, W., Hua, Z., and Haozhong, L. (2016). Investigation on the infection status of Proteus mirabilis in piglets with diarrhea in Chongqing. Animals Breeding and Feed 19, 1–23. doi: 10.13300/j.cnki.cn42-1648/s.2019.12.010
Sanches, M. S., Baptista, A. A. S., de Souza, M., Menck-Costa, M. F., Koga, V. L., Kobayashi, R. K. T., et al. (2019). Genotypic and phenotypic profiles of virulence factors and antimicrobial resistance of Proteus mirabilis isolated from chicken carcasses: potential zoonotic risk. Braz. J. Microbiol. 50, 685–694. doi: 10.1007/s42770-019-00086-2
Sanches, M. S., Rodrigues da Silva, C., Silva, L. C., Montini, V. H., Lopes Barboza, M. G., Migliorini Guidone, G. H., et al. (2021). Proteus mirabilis from community-acquired urinary tract infections (UTI-CA) shares genetic similarity and virulence factors with isolates from chicken, beef and pork meat. Microb. Pathog. 158:105098. doi: 10.1016/j.micpath.2021.105098
Sanches, M. S., Silva, L. C., Da Silva, C. R., Montini, V. H., de Oliva, B. H. D., Guidone, G. H. M., et al. (2023). Prevalence of antimicrobial resistance and clonal relationship in ESBL/AmpC-producing Proteus mirabilis isolated from meat products and community-acquired urinary tract infection (UTI-CA) in southern Brazil. Antibiotics (Basel, Switzerland) 12. doi: 10.3390/antibiotics12020370
Sun, Y., Wen, S., Zhao, L., Xia, Q., Pan, Y., Liu, H., et al. (2020). Association among biofilm formation, virulence gene expression, and antibiotic resistance in Proteus mirabilis isolates from diarrhetic animals in Northeast China. BMC Vet. Res. 16:176. doi: 10.1186/s12917-020-02372-w
Tingyin Zhou, Y. N. (2009). “Standardized clinical microbiology testing,” in Standardized clinical microbiology testing. Shanghai Science & Technology Press.
Yu, Z., Joossens, M., Van den Abeele, A.-M., Kerkhof, P.-J., and Houf, K. (2021). Isolation, characterization and antibiotic resistance of Proteus mirabilis from Belgian broiler carcasses at retail and human stool. Food Microbiol. 96:103724. doi: 10.1016/j.fm.2020.103724
Zhou, R., Fang, X., Zhang, J., Zheng, X., Shangguan, S., Chen, S., et al. (2021). Impact of carbapenem resistance on mortality in patients infected with Enterobacteriaceae: a systematic review and meta-analysis. BMJ Open 11:e054971. doi: 10.1136/bmjopen-2021-054971
Keywords: Proteus mirabilis , antimicrobial resistance, Inc plasmid, repC gene, whole genome sequence
Citation: Liu H, Xia N, Suksawat F, Tengjaroenkul B, Hu Y, Zhou X, Li X, Huang C, Bao Y, Wu Q, Zhang C, Angkititrakul S, Xiang B and Wu X (2025) Prevalence and characterization of IncQ1α-mediated multi-drug resistance in Proteus mirabilis Isolated from pigs in Kunming, Yunnan, China. Front. Microbiol. 15:1483633. doi: 10.3389/fmicb.2024.1483633
Received: 20 August 2024; Accepted: 07 November 2024;
Published: 09 January 2025.
Edited by:
Umer Zeeshan Ijaz, University of Glasgow, United KingdomReviewed by:
Razak Hussain, University of Illinois at Urbana-Champaign, United StatesCopyright © 2025 Liu, Xia, Suksawat, Tengjaroenkul, Hu, Zhou, Li, Huang, Bao, Wu, Zhang, Angkititrakul, Xiang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Wu, d3V4aW5kZGxAaG90bWFpbC5jb20=; Bin Xiang, eGlhbmdiaW4yMDE4QDEyNi5jb20=; Sunpetch Angkititrakul, c3VucGV0Y2hAa2t1LmFjLnRo
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.