- 1Institute of Veterinary Medicine and Immunology, Sichuan Agricultural University, Chengdu, China
- 2Sichuan Agricultural University, Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
- 3Sichuan Agricultural University, International Joint Research Center for Animal Disease Prevention and Control of Sichuan Province, Chengdu, China
- 4Engineering Research Center of Southwest Animal Disease Prevention and Control Technology, Ministry of Education of the People's Republic of China, Chengdu, China
- 5Research Center of Avian Disease, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 6Sinopharm Yangzhou VAC Biological Engineering Co., Ltd., Yangzhou, China
Herpesviruses are a family of double-stranded DNA viruses with a tegument structure and a genome composed of a single sequence and terminal repeat (TR) sequences. The herpesvirus UL14 gene encodes the protein UL14 (pUL14), which has various subcellular localizations and plays a vital role in regulating immediate–early (IE) gene transcription and expression, influences the intracellular localization patterns of several proteins belonging to the capsid and the DNA packaging machinery, participates in secondary envelopment, and influences viral particle release. Additionally, pUL14 has roles in maintaining cellular homeostasis and preventing apoptosis. This review discusses how pUL14 engages in the life cycle of herpesviruses and provides new ideas for further research on pUL14’s function in viral infection.
1 Introduction
The Herpesviridae is a family of large double-stranded DNA-enveloped viruses (Connolly et al., 2021) that includes many important human and animal pathogens. At present, herpes simplex virus 1 (HSV-1; Xu et al., 2023), herpes simplex virus 2 (HSV-2; Truffot et al., 2023), varicella-zoster virus (VZV; Bloch et al., 2023), Epstein–Barr virus (EBV; Li et al., 2023), human cytomegalovirus (HCMV; Nuévalos et al., 2023), and human herpesvirus 6, 7, and 8 (HHV-6, HHV-7, and HHV-8; Hamada et al., 2023; Naipauer and Mesri, 2023) are associated with human diseases. Animal herpesviruses include bovine herpes virus type 1 (BoHV-1; Jones, 2019), pseudorabies virus (PRV; Pomeranz et al., 2005), Marek’s disease virus (MDV; Teng et al., 2023) and duck plague virus (DPV), which primarily cause diseases in animals (Shen et al., 2023). On the basis of differences in genomic homology, host range of infection, replication efficiency, latent cell species, and pathogenicity, the herpesvirus family is divided into the Alphaherpesvirinae (α), Betaherpesvirinae (β), and Gammaherpesvirinae (γ) subfamilies by the International Commission on Taxonomy of Viruses (ICTV; Whitley et al., 1998; Gatherer et al., 2021). HSV-1, HSV-2, VZV, BoHV-1, PRV, MDV, and DPV are members of the alphaherpesvirus subfamily. HCMV, HHV-6, and HHV-7 belong to the betaherpesvirus subfamily, and HHV-8 and EBV belong to the gammaherpesvirus subfamily. The virion is comprised, from inside to outside, of four layers: the double-stranded DNA genome, capsid, tegument, and envelope (Boehmer and Lehman, 1997; Hulo et al., 2011; Figure 1). The tegument is a unique structure of herpesviruses that plays critical roles in the life cycle of the virus (Compans et al., 2008), such as regulating kinase activity, modulating innate immunity, and modulating viral gene transcription and expression (Burgess and Mohr, 2018; Van Gent et al., 2022; Sanders et al., 2023). In addition, the tegument protein, as a structural protein of the virion, connects the capsid with the envelope and plays a key role in the process of virus packaging, maturation, and release (Thomas et al., 2022; Uddin et al., 2023; Wu et al., 2023). At present, studies on the tegument have focused mainly on viral gene transcriptional regulatory proteins with specific molecular biological significance and functional proteins with specific effects on cells, such as VP22 (Pheasant et al., 2023) and VP16 (Ma et al., 2023). In contrast, studies on small tegument proteins, such as pUL14, are relatively rare. However, this protein also plays an important role in regulating immediate–early (IE) gene transcription and expression, influencing the intracellular localization patterns of a number of proteins belonging to the capsid and the DNA packaging machinery, participating in secondary envelopment, and influencing viral particle release. In this review, we summarize the general structural features and functions of pUL14 and how pUL14 participates in the life cycle of herpesviruses and provide new ideas for further research on pUL14’s function in viral infection.
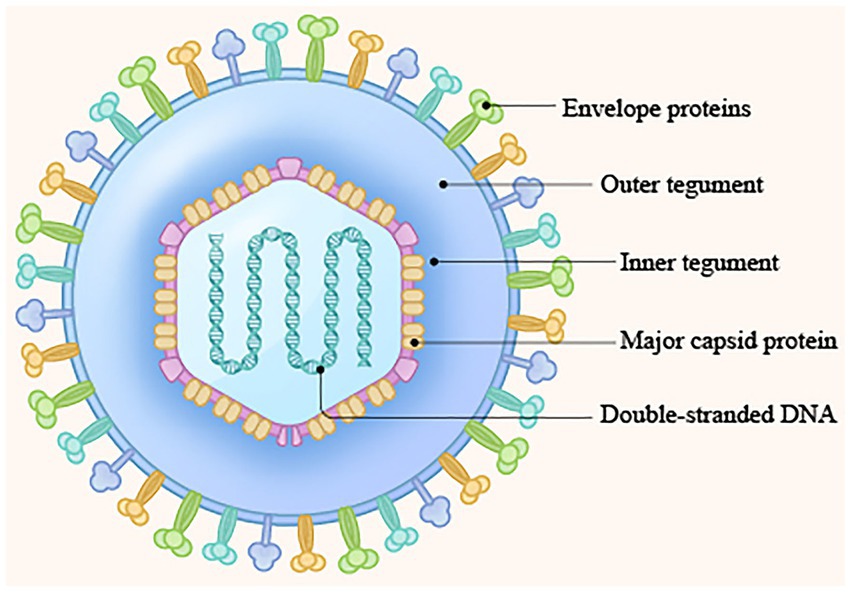
Figure 1. Structure of the herpesvirus genome. (1) Double-stranded DNA genome; (2) Capsid protein; (3) Tegument protein; (4) Envelope protein (Boehmer and Lehman, 1997; Hulo et al., 2011).
2 Characteristics of the herpesvirus UL14 gene
2.1 The herpesvirus UL14 gene is located on the viral genome
The arrangement of reiterated sequences in the herpesvirus genome, whether as direct or inverted repeats at the genome termini or internally, leads to the formation of various classes of genome architecture. Some of these classes include orientation isomers resulting from recombination between terminal and internal inverted repeats. The type A genome, represented by HHV-6/7 (Berneman et al., 1992; Braun et al., 1997; Dominguez et al., 1999), has a unique sequence flanked by terminal repeat (TR) sequences on both sides, and the HHV-8 genome (Russo et al., 1996) has a similar arrangement. Type C genomes, represented by the EBV (Baer et al., 1984) genome, have a sequence of the unique long (UL) component and a sequence of the unique short (US) component, separated by a middle internal repeat (MIR). In the D-type genomes, as represented by VZV (Scott and Davison, 1986), a TR sequence undergoes an internal inversion, and the US surrounded by the inverted repeat sequence can invert relative to the UL. The E-type viral genomes, represented by the HSV-1/2 (McGeoch et al., 1988; Dolan et al., 1998) and HCMV genomes (Akrigg et al., 1985; Dunn et al., 2003; Rigoutsos et al., 2003), are composed of UL, US, TR, and internal repeat (IR) sequences (Figure 2A).
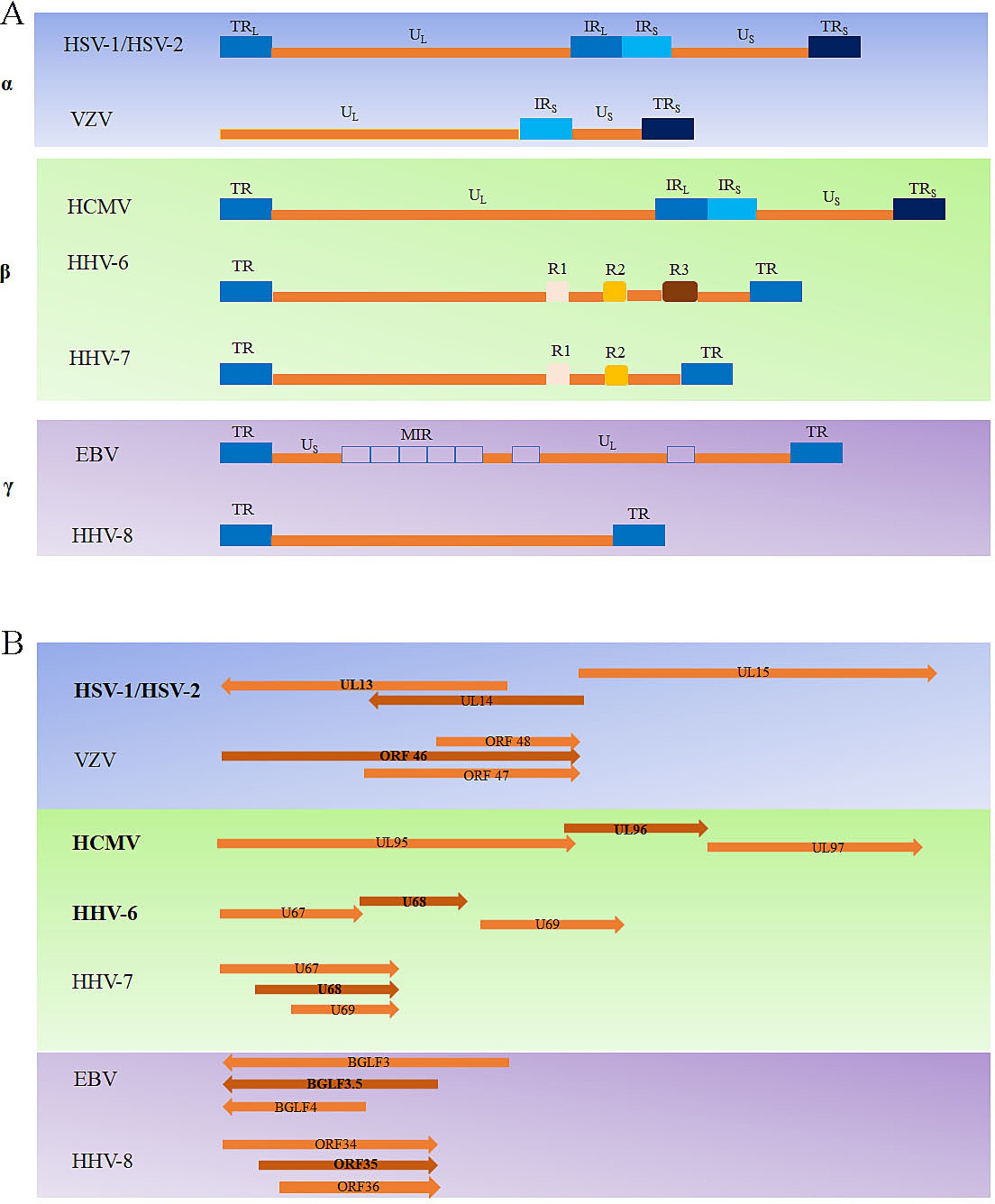
Figure 2. Overview of the types of herpesvirus genomes (A) and the herpesvirus UL14 genes located in the genome (B).
The UL14 gene is conserved in the alphaherpesviruses, and the coding region overlaps that of UL13, which encodes a protein kinase (Cunningham et al., 2000). The homologous genes in other herpesviruses include VZV ORF46, HCMV UL96, HHV-6 U68, HHV-7 U68, Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF35, and EBV BGLF3.5. The position of the UL14 gene in the genome varies among the different human herpesviruses: HHV-6/7 U68 and KSHV ORF35 are unique sequences in the middle of the genome, and the rest are located in the UL region. HSV-1/2 UL14, HCMV UL96, and HHV-6 U68 partially overlap with the neighboring genes UL13, UL95, and U67 on one side. However, VZV ORF46, HHV-7 U68, EBV BGLF3.5, and KSHV ORF35 partially overlap with neighboring genes (Figure 2B). Overall, owing to the large differences in genome arrangement among members of the α-herpesvirus, β-herpesvirus, and γ-herpesvirus subfamilies, there are also differences in the arrangement of the genes homologous to the UL14 gene in these three subfamilies.
2.2 The types of herpesviruses UL14 genes
Herpesvirus genes are transcribed by the cell’s DNA-dependent RNA transcriptase. The genes fall into three main transcription regulation classes: IE, early genes (E), and late genes (L), of which late genes can be divided into early late genes (partly dependent on the synthesis of viral DNA) and late genes (highly dependent on the synthesis of viral DNA; Subak-Sharpe and Dargan, 1998). The IE gene is the first to be transcribed and the first to be expressed. It initiates and guides the transcription and expression of early and late genes. The early genes are transcribed before viral DNA replication after IE protein activation and reach the highest transcription levels after replication begins. Finally, the late genes, which encode mainly structural proteins of the virus, are expressed (Honess and Roizman, 1974). HSV-1/2 pUL14 is expressed late in infection and is considered a late gene (Cunningham et al., 2000; Yamauchi et al., 2001). However, HCMV UL96 is an early gene (Tandon and Mocarski, 2011). These findings suggest that the UL14 gene in various herpesvirus subfamilies exhibits different gene transcription patterns, implying that its functions might also vary.
3 Characteristics of the protein encoded by the herpesvirus UL14 gene
3.1 The molecular weight of the protein encoded by the UL14 gene
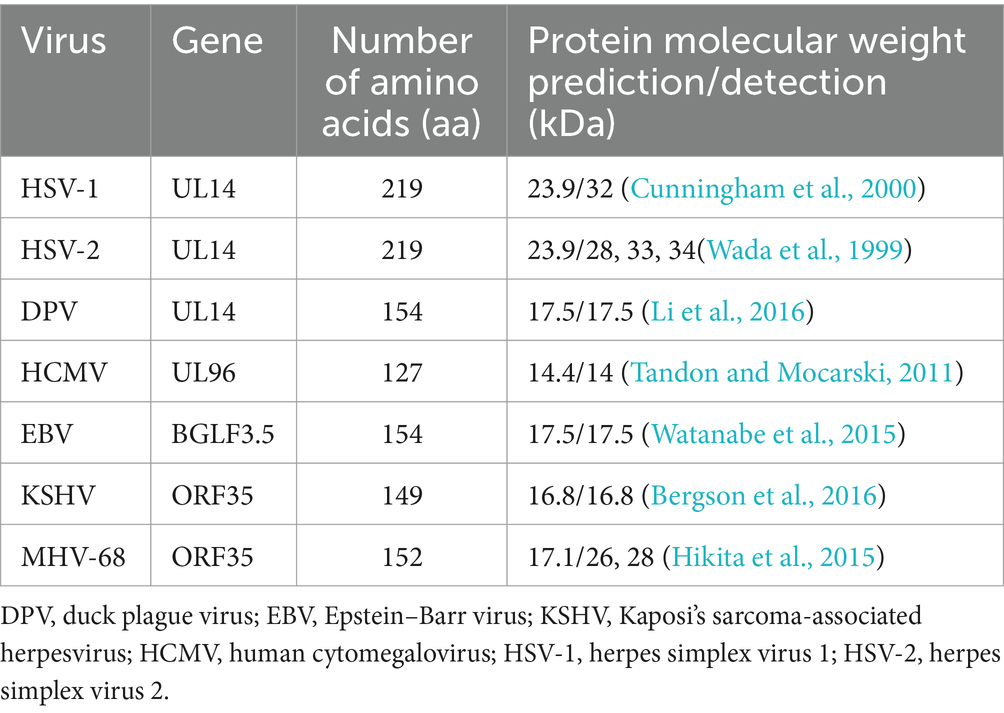
The herpesvirus UL14 gene encodes a relatively small protein (Table 1), and phosphorylation results in different molecular masses of pUL14. HSV-1/2 pUL14 is predicted to be a 219 amino acid protein with a molecular mass of 23 kDa.
However, in HSV-1-infected rabbit skin fibroblast (RS) cells, the UL14 gene product was identified as a 32 kDa protein, whereas in HSV-2-infected Vero cell lysates, 34, 33, and 28 kDa proteins were detected. Cunningham C et al. speculated that these differences might result from phosphorylation based on their phosphatase treatment and immunoblotting experiments (Wada et al., 1999; Cunningham et al., 2000). Additionally, in HEK293T cells transfected with MHV-68 ORF35 (the gene homologous to HSV-1 UL14), 26 and 28 kDa proteins were detected, which was attributed to phosphorylation at S124, S126, and S129 (Hikita et al., 2015). In summary, the size of pUL14 varies among herpesviruses.
3.2 Localization of the protein encoded by the UL14 gene
3.2.1 Subcellular localization of pUL14
The most common subcellular localizations of proteins are whole-cell localization, cytoplasmic localization, nuclear localization, and nucleolar localization. The subcellular localization of a protein is crucial for understanding its function (Nair, 2003). For viruses, the varying subcellular localizations of their proteins indicate distinct functions. After synthesis in the cytoplasm, viral proteins are transported to specific locations where they can effectively carry out their functions (Xu and Massagué, 2004; Terry et al., 2007; Liu et al., 2016; Wubben et al., 2020; Li J. et al., 2021; Wing et al., 2022). The herpesvirus viral DNA genome replicates in the nucleus and assembles with the capsid protein to form a nucleocapsid (Johnson and Baines, 2011; DeRussy et al., 2016). The nucleocapsid moves to the inner nuclear membrane for primary envelopment (Mettenleiter et al., 2013; Takeshima et al., 2019). The nucleocapsid escapes from the nucleus, enters the cytoplasm, and undergoes secondary envelopment (Owen et al., 2015). Therefore, their localization can preliminarily predict the possible functions of viral proteins in the cell.
pUL14 shows different intracellular distribution patterns when expressed alone. Indirect immunofluorescence assay (IFA) results revealed that the HCMV UL96 (Tandon and Mocarski, 2011), MHV-68 ORF35 (Hikita et al., 2015), and KSHV ORF35 (Masa et al., 2008) gene products were localized only in the cytoplasm and were not affected by other viral proteins. However, the subcellular localization of alphaherpesvirus pUL14 varies and includes whole-cell, cytoplasmic, and nuclear localization. In transfected cells, this diversity in localization was observed simultaneously at the same time. For example, HSV-2 pUL14, when expressed alone, exhibited a varied intracellular distribution 24 h after transfection: it localized to the cytoplasm in 70% of the expressing cells, was found in both the cytoplasm and nucleus in 15% of the expressing cells, and was exclusively nuclear in 15% of the expressing cells (Yamauchi et al., 2001). Additionally, the distribution of pUL14 in the cells is dynamic: in DPV-infected duck embryo fibroblasts (DEFs), pUL14 was mainly distributed in the cytoplasm in the early stages of viral infection but migrated to the nucleus in the late stage of infection (Li et al., 2016). Currently, there is no research indicating that HSV-1/2 pUL14 has a nuclear localization signal (NLS); however, a region that is highly conserved between HSV-1 and HSV-2, especially 60RLKSRARLE68, is similar to an α-helix contained within the substrate-binding region of Hsp70. One study showed that pUL14 translocates to the nucleus and nucleolus upon stress, such as heat shock and osmotic shock (Yamauchi et al., 2002a, 2002b, 2003). In addition, when pUL14 was cotransfected with other viral proteins, such as VP16, VP26, and UL17, it not only facilitated the nuclear localization of these other viral proteins but also enhanced their nuclear localization ability; furthermore, these proteins colocalized within the nucleus. Notably, a bimolecular fluorescence complementation (BiFC) assay was used to confirm a direct interaction between DPV pUL14 and VP16 (Yamauchi et al., 2001; Ohta et al., 2011). In summary, pULl4 displays a variety of modes of subcellular localization through the nuclear membrane and is regulated by various factors, which also suggests that pUL14 may have multiple functions.
3.2.2 Location of the UL14 protein in the virus structure
To determine whether pUL14 is a component of virion particles and what its location is in virion particles, mass spectrometry and Western blotting were used, which revealed that HSV-1 and HSV-2 pUL14 are minor components of the virion tegument (Cunningham et al., 2000; Yamauchi et al., 2001). Kerstin Radtke further identified HSV-1 pUL14 as an inner tegument protein (Radtke et al., 2010). The localization of pUL14 in the structure of other herpesviruses has not yet been reported. Overall, the results indicate that HSV-1/2 pUL14 is a tegument protein.
4 Function of herpesvirus pUL14
4.1 Role of pUL14 in virus replication
Herpesvirus proliferation mainly includes entry, replication, virion assembly, primary and secondary envelopment, and release of mature virions (Demangone et al., 1987; Boehmer and Lehman, 1997; Figure 3). The herpes simplex virus enters cells through the action of four glycoproteins, gB, gD, gH, and gL, and the viral nucleocapsid is then released into the cytoplasm (steps 1 and 2; Gerber et al., 1995; Herold et al., 1996; Cheshenko and Herold, 2002; Atanasiu et al., 2007; Gianni et al., 2009; Connolly et al., 2011; Karasneh and Shukla, 2011). After entering the cell, most viral tegument proteins are dissociated by the actions of pUS3 and pUL13, and the remaining few tegument proteins associate with the nucleocapsid, participating in microtubule-dependent cellular molecular movement, specifically the transport of the nucleocapsid by the cytoplasmic dynein/dynactin complex (step 3; Granzow et al., 2005; Maurer et al., 2008), which transports the nucleocapsid to the nuclear pore and releases viral DNA into the nucleus with the assistance of various viral proteins (step 4; Batterson et al., 1983; Ojala et al., 2000; Copeland et al., 2009). The cyclization of linear double-stranded DNA molecules into the nucleus activates the transcription and translation of viral IE genes under the action of transcriptional activators (such as VP16; step 5; Batterson and Roizman, 1983), and the translated IE proteins can transactivate the transcription and translation of early genes. The translated early proteins (mainly enzymes related to viral DNA replication), in turn, activate the replication, transcription, and translation of the late genes, and the translated late proteins (mainly the structural proteins of the virus, including cortical proteins, membrane proteins, and capsid proteins; steps 6–8; Kelly et al., 2009; Melchjorsen et al., 2009). In the nucleus, the replicated viral DNA is wrapped by capsid protein to form the nuclear capsid (step 9), and newly formed nucleocapsids bud at the inner nuclear membrane, creating primary enveloped virions within the perinuclear space. These primary envelopes then fuse with the outer nuclear membrane, allowing the nucleocapsids to be translocated into the cytoplasm (steps 10–12). Subsequently, the nucleocapsids acquire a set of tegument proteins in the cytoplasm and bud into vesicles originating from host organelles to gain the viral envelope (steps 13 and 14). These vesicles are then transported to the plasma membrane, where they fuse and release mature virions into the extracellular space (Mettenleiter, 2002; Lv et al., 2019; steps 15–16).
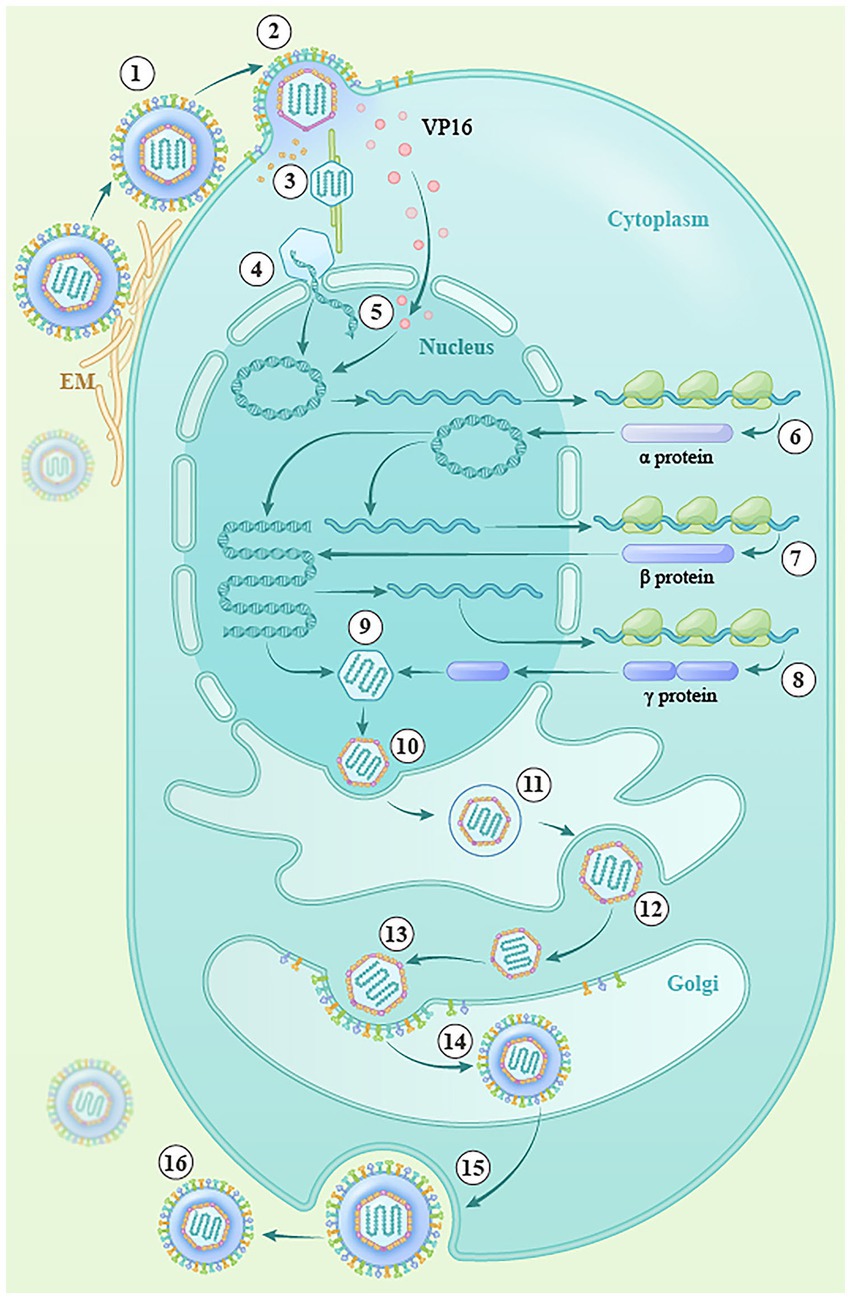
Figure 3. The lytic replication cycle of herpesviruses. (1) Entry into the cell (steps ①, ②), (2) transfer of the capsid to the nucleus (steps③–⑤), (3) viral DNA replication and transcription (steps ⑥–⑧), (4) capsid assembly (step ⑨), (5) egress from the nucleus (steps ⑩–⑫), (6) maturation and envelopment of viral particles in the cytoplasm (steps ⑬, ⑭), and (7) release of mature virions (steps ⑮, ⑯).
The UL14 gene and its homologs are essential for efficient viral growth. Studies have shown that a UL14 deletion virus forms very small plaques, produces fewer viruses, and exhibits a slightly delayed replication kinetics at a low multiplicity of infection in infected cells (Wada et al., 1999; Cunningham et al., 2000; Tandon and Mocarski, 2011; Brechtel et al., 2014). The impact of the UL14 gene-encoded protein on the viral replication cycle is specifically reflected in its regulation of IE gene transcription and expression, modulation of the nuclear localization of capsid assembly and genome packaging proteins, involvement in secondary envelopment, and influence on virion release.
4.1.1 Regulation of the expression of the viral IE genes ICP0 and ICP4
During a productive virus infection, the tegument protein pUL48, a component of the HSV-1 virion that is a gene alpha transinducing factor (α-TIF, also known as VP16), interacts with the host cell transcription factor Oct-1 to initiate host cell RNA polymerases. Host RNA polymerase II (RNAP II) is recruited to the IE gene promoter to mediate IE viral gene transcription and produce IE proteins (Lu and Misra, 2000). Research has shown that HSV-1/DPV pUL14 enhances the nuclear localization of VP16, that HSV-1 pUL14 indirectly regulates the expression of the IE genes ICP4, and that DPV pUL14 significantly promotes the transcriptional activation of VP16 (Lu and Misra, 2000; Li et al., 2016; Zhou et al., 2021). These results suggest that pUL14 may indirectly regulate the transcription and expression of the IE genes ICP0 and ICP4 by enhancing the nuclear localization of VP16.
4.1.2 Facilitation of the localization of crucial proteins involved in viral capsid assembly or DNA packaging
A-, B-and C-capsids were found in HSV-infected cells using sucrose density gradient ultracentrifugation. A-capsids are empty, lacking viral DNA and internal scaffolds (Gibson and Roizman, 1972; Sherman and Bachenheimer, 1988; Schrag et al., 1989; Booy et al., 1991). B-capsids contain some viral DNA and are considered intermediates in the assembly process. They do not directly transition into C-capsids but are crucial for the maturation of C-capsids, which are filled with viral DNA. Scaffold proteins, including VP22a, and considerably lower amounts of the UL26 gene products VP21 and VP24 play a role in the structural organization of B-capsids but are not merely precursors for C-capsids (Liu and Roizman, 1991, 1993; Newcomb and Brown, 1991); C-capsids contain the viral genome and are further assembled into infectious virions (Heming et al., 2017; Freeman et al., 2021). The B-type nucleocapsid was purified and analyzed using cryo-electron microscopy. The results revealed that the difference between the A-type nucleocapsid and B-type nucleocapsid particles was that the B-type capsid contained the ICP35 protein, a product of the UL26.5 gene (Newcomb et al., 1993). The capsid scaffold protein ICP35 is a capsid structural protein that temporarily operates within the capsid during viral assembly (Thomsen et al., 1995; Li X. et al., 2021). In infected Vero cells, a specific fluorescence signal of HSV-2 pUL14 was observed in the cytoplasm and nucleus at 6-h postinfection. UL14 was mainly observed in the nucleus as fine, discrete particles colocalized with the scaffolding protein ICP35. However, at 12 h postinfection, the ICP35 protein had aggregated in the perinuclear region of the cytoplasm and was located primarily in the nucleus for assembly, and colocalization with pUL14 was no longer observed. It is speculated that pUL14 may be involved in viral capsid assembly or egress (Wada et al., 1999).
DNA cleavage/packaging is mediated by proteins encoded by the UL6, UL15, UL17, UL25, UL28, UL32, and UL33 genes (Yu and Weller, 1998; Wills et al., 2006; Albright et al., 2015). Deleting any of these genes results in the aggregation of empty capsid and viral DNA duplicates in the nucleus, which cannot be assembled into the nucleocapsid. These genes have been shown to play important roles in viral assembly rather than capsid synthesis and DNA replication (Sherman and Bachenheimer, 1987; Stow, 2001; Yang et al., 2011; Borst et al., 2013). In the DNA assembly process of the nucleocapsid, the UL17 protein is a DNA assembly protein that plays a key role in the early stage of capsid assembly, participating in the entire process of cleavage and assembly. In the absence of the UL17 protein, DNA assembly cannot take place (Salmon et al., 1998; Thurlow et al., 2005). pUL33 acts as a terminal enzyme subunit, a component of the “molecular motor” involved in the cleavage/packaging of viral genomic DNA. The absence of pUL33 leads to the formation of only type B capsids and not type C capsids in the nucleus of infected cells, greatly reducing the infectivity of the virus (Al-Kobaisi et al., 1991). The UL33 protein of HSV-2 typically localizes to the cytoplasm when expressed alone and is translocated to the nucleus when co-expressed with the UL14 protein in the absence of viral infection (Yamauchi et al., 2001). These findings indicate that pUL14 can modulate the localization of pUL33; however, the exact mechanisms and effects, such as whether pUL14 is involved in DNA cleavage/packaging, should be verified.
Additionally, U2OS cells synchronously infected with UL14-deficient virus (14D) or rescued virus (14R) at an MOI of 40 PFU/cell in the presence of cycloheximide were fixed 2 h after being shifted to 37°C and permeabilized, followed by incubation with an anti-VP5 mAb for the indirect immunofluorescence analysis of the major capsid protein VP5. These 14R-infected cells showed a marked accumulation of capsids at the rim of the nucleus. In contrast, in 14D-infected cells, capsids were found at the nuclear rim to a much lesser extent. These results suggest that HSV-1 pUL14 is crucial for transporting the viral capsid protein VP5 to the nuclear rim. In addition, when HSV-2 UL14 was co-expressed with the capsid protein VP26, the cytoplasmic localization of VP26 changed to a nuclear localization (Yamauchi et al., 2001, 2008). Therefore, HSV-1/2 pUL14 can affect the localization of the capsid proteins VP5 and VP26.
4.1.3 Participation in secondary envelopment
After nuclear egress, the nucleocapsid must obtain the complete set of tegument proteins and acquire its final (secondary) envelope (Mettenleiter et al., 2009; Yanagi et al., 2019). Recent studies have confirmed that the Golgi apparatus, particularly the trans-Golgi network, plays a crucial role in the envelopment of HSV (Harley et al., 2001; McMillan and Johnson, 2001), and HSV-1 pUL51 closely colocalized with Golgi marker proteins, including the Golgi-58 k protein and GM130, in transfected cells expressing only UL51 during HSV infection; however, the UL51 protein only partially colocalized with the Golgi markers. pUL51 is incorporated into virions during envelopment and localizes primarily to the inside of cytoplasmic vesicles and/or the viral envelope (Nozawa et al., 2003). Research has shown that the HSV-1 pUL51 amino acid residues Leu-111, Ile-119, and Tyr-123 are required for interaction with pUL14 in HSV-1-infected cells. In addition, in cells infected with wild-type HSV-1, 9.3% of the virus particles were unenveloped or partially enveloped nucleocapsids in the cytoplasm. However, in cells infected with ΔUL14, ΔUL51, UL51LIY/AAA, or ΔUL51/ΔUL14 viruses, 38 to 44% of the virus particles were unenveloped or partially enveloped nucleocapsids in the cytoplasm, which was 4 to 5 times greater than that in cells infected with wild-type HSV-1. These results indicated that the mutations in UL14 and/or UL51 induced comparable levels of aberrant accumulation of unenveloped capsids and partially enveloped nucleocapsids in the cytoplasm (Oda et al., 2016).
4.1.4 Affects the production of virions released into cells
The nucleocapsid, after completing tegumentation and secondary envelopment in the cytoplasm, enters vesicles derived from the host membrane structure and releases virus particles from the cell (Crump, 2018). When RS cells were infected with a high MOI (5 PFU/cell) for 24 h to determine the proportion of total virus released to total virus production, the results revealed that the average release rates of an HSV-1 mutated virus lacking UL14 were 19.1 and 17.1% of the proportion of virus released into the medium by the parent and the UL14-reverted virus, respectively (Cunningham et al., 2000). In addition, Human foreskin-derived fibroblast (HF) cells were infected with HCMV UL96DDBAC (HCMV ΔUL96 BAC was also engineered to insert an FKBP-DD tag at the C-terminus) or parental virus at an MOI of 3.0 and incubated for 4 days in the presence of the encapsidation inhibitor BDCRB (20 μM) to synchronize infection. The BDCRB block was then removed, and the cells were incubated for 2 days. Analysis of cell-associated and released virus infectivity revealed a delay in UL96DDBAC virus replication compared to the parental virus (Tandon and Mocarski, 2011). These results indicate that HSV-1 UL14 or HCMV UL96 affect a virus maturation step involving efficient accumulation of cell-associated virus preceding release.
4.2 Maintaining cell homeostasis
pUL14 likely contributes to maintaining cellular homeostasis, such as cytoskeletal organization, after HSV-1 infection. To investigate whether pUL14 could alter the distribution of cytoskeletal proteins, immunofluorescence analysis with phalloidin and an anti-α-tubulin antibody was used to detect filamentous actin (F-actin) and microtubules in cells infected with an HSV-1 UL14-defective virus or UL14R. The results revealed that infection with the HSV-1 virus with the UL14 gene deletion led to cytoskeletal tissue destruction earlier than that observed in wild-type infected cells, suggesting that pUL14 plays a positive role in maintaining the cytoskeletal morphology of infected cells (Ohta et al., 2011).
4.3 pUL14 and neurovirulence
pUL14 is related to the neurotropic properties of herpesvirus. In mice intracranially injected with HSV-1 UL14-deficient viruses, the 50% lethal dose of the mutant virus was reduced by more than 30,000-fold. Additionally, recovery of the mutant virus from latently infected sacral ganglia in mice injected peripherally was significantly lower than that of the wild-type virus (Cunningham et al., 2000). In mice infected with the MHV-68 ORF35stop virus, lytic replication in the lung after intranasal infection and the frequency of ex vivo reactivation from latency after intraperitoneal infection were lower in ORF35stop virus-infected mice than in mice infected with the WT or marker-reverted virus (Hikita et al., 2015). These results indicate that UL14 plays an important role in establishing or reactivating latent infection.
4.4 pUL14 and apoptosis
Apoptosis, also known as programmed cell death, is one of the innate defense strategies of host cells against the invasion of foreign microorganisms such as bacteria or viruses. The replication and transmission of the virus depend on the host cell system; therefore, the host cell inhibits the proliferation and transmission of the virus through apoptosis. The virus has also evolved corresponding strategies to regulate the signaling pathway of host cell apoptosis to maintain the survival of virus-infected cells, thus facilitating viral proliferation and spread. Herpesviruses can inhibit cell apoptosis at multiple stages of viral infection to prevent premature cell death and ensure viral proliferation (Goodkin et al., 2004; Nguyen and Blaho, 2006). BHV-1, HSV-1, and HSV-2 pUL14 contain an amino acid sequence homologous to a portion of the peptide-binding domain conserved in the Hsp70 family. Various stress conditions, such as increased temperature, ATP depletion, and hyperosmotic shock, trigger the accumulation of UL14 protein in the nucleus. BHV-1 UL14 can inhibit sorbitol-induced apoptosis. Cell lines expressing HSV-2 and HSV-1 pUL14 have a greater ability to inhibit apoptosis than parental cell lines under osmotic shock and treatment with certain drugs (Yamauchi et al., 2002a, 2002b, 2003; De Martino et al., 2007). These findings indicate that pUL14 is closely related to apoptosis and that the mechanism by which pUL14 inhibits apoptosis may be related to its structural similarity to heat shock proteins.
5 Interaction with other host proteins
A high-throughput yeast two-hybrid system was used to identify the cellular factors that interact with UL14, which identified the potential interacting protein TSC22D3. The IFA results revealed that pUL14 and TSC22D3 were colocalized in the nucleus, whereas TSC22D3, when expressed alone, was present throughout the cell. The Co-IP results reveal that pUL14 can interact with TSC22D3 (D’Adamio et al., 1997; Wu et al., 2011). TSC22D3, also known as glucocorticoid-induced leucine zipper (GILZ), is a protein of the β-transforming growth factor-stimulated protein 22 (TSC-22) family (D’Adamio et al., 1997). Structurally, the GILZ gene consists of three domains: an N-terminal domain that includes the TSC box, a leucine zipper, and a C-terminal region that is rich in proline and glutamic acid. As a multifunctional protein, TSC22D3 is involved in interactions with many signaling molecules, including Raf, Ras, AP-1, and NF-KB, thereby influencing downstream signaling pathways (Mittelstadt and Ashwell, 2001; Marco et al., 2006). When stimulated by glucocorticoids (GCs), the glucocorticoid receptor (GR) in the cytoplasm is activated and subsequently translocated to the nucleus, where it binds to the glucocorticoid response element (GRE) on DNA, inducing the synthesis of TSC22D3 and regulating other signaling pathways (Ayroldi and Riccardi, 2009). Previous results indicate that HSV-1 can remain latent in the trigeminal ganglia and reactivate upon stimulation. During this process, glucocorticoid levels are modulated, inducing the expression of TSC22D3, which plays a regulatory role (Bener et al., 2007). These results suggest that TSC22D3 is involved in various signaling pathways and may be influenced by glucocorticoids, but the molecular mechanism of the interaction between TSC22D3 and pUL14 needs further study.
6 Future prospects
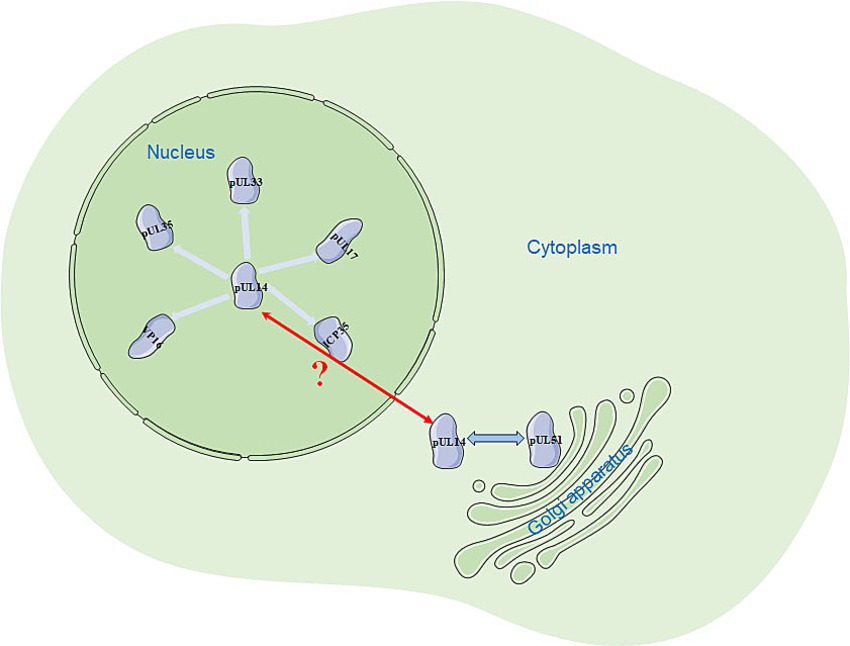
Currently, studies on the herpesvirus UL14 gene have focused mainly on human herpesviruses. Studying the UL14 gene in various herpesviruses is beneficial for obtaining a more comprehensive understanding and elucidation of the functions of the UL14 gene. The UL14 gene and its homologs can present diverse subcellular localizations. In the nucleus, they can colocalize with pUL17, pUL33, pUL35, VP16, and ICP35; in the cytoplasm, they can also interact with pUL51 (Figure 4). The predicted molecular structure of this protein suggests that it may possess a potential NLS, which is due to the presence of several arginine (R)-rich regions in its N-terminus, as this basic amino acid-rich region is likely to mediate nuclear input (Lee et al., 2023). pUL14 may also have a nuclear export signal (NES; Zhou et al., 2023) that mediates protein nuclear export. Further research is needed to understand how the NLS and NES mediate the nuclear import and export of pUL14, whether other nuclear import and export receptors (such as importin α/β) are involved, and whether this process depends on Ran-GTP. The differences in the subcellular localization of pUL14 observed under transfection and viral infection conditions may be due to the influence of other viral proteins on the localization of pUL14 after viral infection. Therefore, investigating the causes behind the various localizations of pUL14 and its impact on viral processes such as budding, packaging, envelopment, and release, which are related to viral replication, can provide valuable insights into the viral replication cycle. Additionally, this research will aid in understanding the pathogenic mechanisms of herpesviruses.
Author contributions
JW: Writing – original draft, Writing – review & editing, Formal Analysis. MW: Supervision, Writing – review & editing. AC: Funding acquisition, Writing – review & editing. WZ: Conceptualization, Writing – review & editing. QY: Data curation, Writing – review & editing. BT: Investigation, Writing – review & editing. XO: Formal Analysis, Writing – review & editing. DS: Data curation, Writing – review & editing. YH: Formal Analysis, Writing – review & editing. XZ: Investigation, Writing – review & editing. YW: Investigation, Writing – review & editing. SZ: Investigation, Writing – review & editing. JH: Investigation, Writing – review & editing. ZW: Investigation, Writing – review & editing. YY: Investigation, Writing – review & editing. LZ: Investigation, Writing – review & editing. DZ: Investigation, Writing – review & editing. ML: Investigation, Writing – review & editing. SC: Investigation, Writing – review & editing. RJ: Investigation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the China Agricultural Research System (CARS-42-17), the Sichuan Veterinary Medicine, and the Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
Conflict of interest
WZ is employed by Sinopharm Yangzhou Vac Biological Engineering Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akrigg, A., Wilkinson, G. W., and Oram, J. D. (1985). The structure of the major immediate early gene of human cytomegalovirus strain AD169. Virus Res. 2, 107–121. doi: 10.1016/0168-1702(85)90242-4
Albright, B. S., Kosinski, A., Szczepaniak, R., Cook, E. A., Stow, N. D., Conway, J. F., et al. (2015). The putative herpes simplex virus 1 chaperone protein UL32 modulates disulfide bond formation during infection. J. Virol. 89, 443–453. doi: 10.1128/JVI.01913-14
Al-Kobaisi, M. F., Rixon, F. J., McDougall, I., and Preston, V. G. (1991). The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180, 380–388. doi: 10.1016/0042-6822(91)90043-B
Atanasiu, D., Whitbeck, J. C., Cairns, T. M., Reilly, B., Cohen, G. H., and Eisenberg, R. J. (2007). Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. 104, 18718–18723. doi: 10.1073/pnas.0707452104
Ayroldi, E., and Riccardi, C. (2009). Glucocorticoid-induced leucine zipper (GILZ): a new important mediator of glucocorticoid action. FASEB J. 23, 3649–3658. doi: 10.1096/fj.09-134684
Baer, R., Bankier, A. T., Biggin, M. D., Deininger, P. L., Farrell, P. J., Gibson, T. J., et al. (1984). DNA sequence and expression of the B95-8 Epstein—Barr virus genome. Nature 310, 207–211. doi: 10.1038/310207a0
Batterson, W., Furlong, D., and Roizman, B. (1983). Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45, 397–407. doi: 10.1128/jvi.45.1.397-407.1983
Batterson, W., and Roizman, B. (1983). Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46, 371–377. doi: 10.1128/jvi.46.2.371-377.1983
Bener, D., Wohlman, A., Itzik, A., Yirmiya, R., Ben-Hur, T., and Weidenfeld, J. (2007). Glucocorticoid resistance following herpes Simplex-1 infection: role of hippocampal glucocorticoid receptors. Neuroendocrinology 85, 207–215. doi: 10.1159/000102976
Bergson, S., Itzhak, I., Wasserman, T., Gelgor, A., Kalt, I., and Sarid, R. (2016). The Kaposi’s-sarcoma-associated herpesvirus orf35 gene product is required for efficient lytic virus reactivation. Virology 499, 91–98. doi: 10.1016/j.virol.2016.09.008
Berneman, Z. N., Ablashi, D. V., Li, G., Eger-Fletcher, M., Reitz, M. S., Hung, C. L., et al. (1992). Human herpesvirus 7 is a T-lymphotropic virus and is related to, but significantly different from, human herpesvirus 6 and human cytomegalovirus. Proc. Natl. Acad. Sci. 89, 10552–10556. doi: 10.1073/pnas.89.21.10552
Bloch, K. C., Glaser, C., Gaston, D., and Venkatesan, A. (2023). State of the art: acute encephalitis. Clin. Infect. Dis. 77, e14–e33. doi: 10.1093/cid/ciad306
Boehmer, P. E., and Lehman, I. R. (1997). Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66, 347–384. doi: 10.1146/annurev.biochem.66.1.347
Booy, F. P., Newcomb, W. W., Trus, B. L., Brown, J. C., Baker, T. S., and Steven, A. C. (1991). Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64, 1007–1015. doi: 10.1016/0092-8674(91)90324-R
Borst, E. M., Kleine-Albers, J., Gabaev, I., Babić, M., Wagner, K., Binz, A., et al. (2013). The human cytomegalovirus UL51 protein is essential for viral genome cleavage-packaging and interacts with the terminase subunits pUL56 and pUL89. J. Virol. 87, 1720–1732. doi: 10.1128/JVI.01955-12
Braun, D. K., Dominguez, G., and Pellett, P. E. (1997). Human herpesvirus 6. Clin. Microbiol. Rev. 10, 521–567. doi: 10.1128/CMR.10.3.521
Brechtel, T. M., Mocarski, E. S., and Tandon, R. (2014). Highly acidic C-terminal region of cytomegalovirus pUL96 determines its functions during virus maturation independently of a direct pp 150 interaction. J. Virol. 88, 4493–4503. doi: 10.1128/JVI.03784-13
Burgess, H. M., and Mohr, I. (2018). Defining the role of stress granules in innate immune suppression by the herpes simplex virus 1 endoribonuclease VHS. J. Virol. 92, e00829–e00818. doi: 10.1128/JVI.00829-18
Cheshenko, N., and Herold, B. C. (2002). Glycoprotein B plays a predominant role in mediating herpes simplex virus type 2 attachment and is required for entry and cell-to-cell spread. J. Gen. Virol. 83, 2247–2255. doi: 10.1099/0022-1317-83-9-2247
Compans, R. W., Cooper, M. D., Honjo, T., Koprowski, H., Melchers, F., Oldstone, M. B. A., et al. (2008). Structure and formation of the cytomegalovirus virion. Curr. Top. Microbiol. Immunol. 325, 187–204. doi: 10.1007/978-3-540-77349-8_11
Connolly, S. A., Jackson, J. O., Jardetzky, T. S., and Longnecker, R. (2011). Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9, 369–381. doi: 10.1038/nrmicro2548
Connolly, S. A., Jardetzky, T. S., and Longnecker, R. (2021). The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 19, 110–121. doi: 10.1038/s41579-020-00448-w
Copeland, A. M., Newcomb, W. W., and Brown, J. C. (2009). Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83, 1660–1668. doi: 10.1128/JVI.01139-08
Crump, C. (2018). Virus assembly and egress of HSV. Adv. Exp. Med. Biol. 1045, 23–44. doi: 10.1007/978-981-10-7230-7_2
Cunningham, C., Davison, A. J., MacLean, A. R., Taus, N. S., and Baines, J. D. (2000). Herpes simplex virus type 1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J. Virol. 74, 33–41. doi: 10.1128/JVI.74.1.33-41.2000
D’Adamio, F., Zollo, O., Moraca, R., Ayroldi, E., Bruscoli, S., Bartoli, A., et al. (1997). A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity 7, 803–812. doi: 10.1016/S1074-7613(00)80398-2
De Martino, L., Marfe, G., Consalvo, M. I., Di Stefano, C., Pagnini, U., and Sinibaldi-Salimei, P. (2007). Antiapoptotic activity of bovine herpesvirus type-1 (BHV-1) UL14 protein. Vet. Microbiol. 123, 210–216. doi: 10.1016/j.vetmic.2007.02.026
Demangone, M., Hill, J. M., and Kwon, B. S. (1987). Effects of acyclovir therapy during simultaneous reactivation of latent HSV-1 in rabbits. Antivir. Res. 7, 237–243. doi: 10.1016/0166-3542(87)90032-5
DeRussy, B. M., Boland, M. T., and Tandon, R. (2016). Human cytomegalovirus pUL93 links nucleocapsid maturation and nuclear egress. J. Virol. 90, 7109–7117. doi: 10.1128/JVI.00728-16
Dolan, A., Jamieson, F. E., Cunningham, C., Barnett, B. C., and McGeoch, D. J. (1998). The genome sequence of herpes simplex virus type 2. J. Virol. 72, 2010–2021. doi: 10.1128/JVI.72.3.2010-2021.1998
Dominguez, G., Dambaugh, T. R., Stamey, F. R., Dewhurst, S., Inoue, N., and Pellett, P. E. (1999). Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73, 8040–8052. doi: 10.1128/JVI.73.10.8040-8052.1999
Dunn, W., Chou, C., Li, H., Hai, R., Patterson, D., Stolc, V., et al. (2003). Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100, 14223–14228. doi: 10.1073/pnas.2334032100
Freeman, K. G., Huffman, J. B., Homa, F. L., and Evilevitch, A. (2021). UL25 capsid binding facilitates mechanical maturation of the herpesvirus capsid and allows retention of pressurized DNA. J. Virol. 95:e0075521. doi: 10.1128/JVI.00755-21
Gatherer, D., Depledge, D. P., Hartley, C. A., Szpara, M. L., Vaz, P. K., Benkő, M., et al. (2021). ICTV virus taxonomy profile: Herpesviridae 2021. J. Gen. Virol. 102:001673. doi: 10.1099/jgv.0.001673
Gerber, S. I., Belval, B. J., and Herold, B. C. (1995). Differences in the role of glycoprotein C of HSV-1 and HSV-2 in viral binding may contribute to serotype differences in cell tropism. Virology 214, 29–39. doi: 10.1006/viro.1995.9957
Gianni, T., Amasio, M., and Campadelli-Fiume, G. (2009). Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL in part through the C-terminal profusion domain. J. Biol. Chem. 284, 17370–17382. doi: 10.1074/jbc.M109.005728
Gibson, W., and Roizman, B. (1972). Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10, 1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972
Goodkin, M. L., Morton, E. R., and Blaho, J. A. (2004). Herpes simplex virus infection and apoptosis. Int. Rev. Immunol. 23, 141–172. doi: 10.1080/08830180490265574
Granzow, H., Klupp, B. G., and Mettenleiter, T. C. (2005). Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79, 3200–3205. doi: 10.1128/JVI.79.5.3200-3205.2005
Hamada, N., Shigeishi, H., Oka, I., Sasaki, M., Kitasaki, H., Nakamura, M., et al. (2023). Associations between oral human herpesvirus-6 and-7 and periodontal conditions in older adults. Life 13:324. doi: 10.3390/life13020324
Harley, C. A., Dasgupta, A., and Wilson, D. W. (2001). Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75, 1236–1251. doi: 10.1128/JVI.75.3.1236-1251.2001
Heming, J. D., Conway, J. F., and Homa, F. L. (2017). Herpesvirus capsid assembly and DNA packaging. Adv. Anat. Embryol. Cell Biol. 223, 119–142. doi: 10.1007/978-3-319-53168-7_6
Herold, B. C., Gerber, S. I., Belval, B. J., Siston, A. M., and Shulman, N. (1996). Differences in the susceptibility of herpes simplex virus types 1 and 2 to modified heparin compounds suggest serotype differences in viral entry. J. Virol. 70, 3461–3469. doi: 10.1128/jvi.70.6.3461-3469.1996
Hikita, S., Yanagi, Y., and Ohno, S. (2015). Murine gammaherpesvirus 68 ORF35 is required for efficient lytic replication and latency. J. Gen. Virol. 96, 3624–3634. doi: 10.1099/jgv.0.000310
Honess, R. W., and Roizman, B. (1974). Regulation of herpesvirus macromolecular synthesis I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14, 8–19. doi: 10.1128/jvi.14.1.8-19.1974
Hulo, C., De Castro, E., Masson, P., Bougueleret, L., Bairoch, A., Xenarios, I., et al. (2011). ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 39, D576–D582. doi: 10.1093/nar/gkq901
Johnson, D. C., and Baines, J. D. (2011). Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9, 382–394. doi: 10.1038/nrmicro2559
Jones, C. (2019). Bovine herpesvirus 1 counteracts immune responses and immune-surveillance to enhance pathogenesis and virus transmission. Front. Immunol. 10:1008. doi: 10.3389/fimmu.2019.01008
Karasneh, G. A., and Shukla, D. (2011). Herpes simplex virus infects most cell types in vitro: clues to its success. Virol. J. 8:481. doi: 10.1186/1743-422X-8-481
Kelly, B. J., Fraefel, C., Cunningham, A. L., and Diefenbach, R. J. (2009). Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145, 173–186. doi: 10.1016/j.virusres.2009.07.007
Lee, J., Kwon, Y.-E., Edwards, S. D., Guim, H., and Jae Jeong, K. (2023). Improved biocompatibility of dendrimer-based gene delivery by histidine-modified nuclear localization signals. Int. J. Pharm. 644:123299. doi: 10.1016/j.ijpharm.2023.123299
Li, J. S. Z., Abbasi, A., Kim, D. H., Lippman, S. M., Alexandrov, L. B., and Cleveland, D. W. (2023). Chromosomal fragile site breakage by EBV-encoded EBNA1 at clustered repeats. Nature 616, 504–509. doi: 10.1038/s41586-023-05923-x
Li, J., Guo, Y., Deng, Y., Hu, L., Li, B., Deng, S., et al. (2021). Subcellular localization of Epstein-Barr virus BLLF2 and its underlying mechanisms. Front. Microbiol. 12:672192. doi: 10.3389/fmicb.2021.672192
Li, X., Wang, J., Mou, T., Gao, Y., Wang, L., Fan, S., et al. (2021). Immunological identification and characterization of the capsid scaffold protein encoded by UL26.5 of herpes simplex virus type 2. Front. Cell. Infect. Microbiol. 11:649722. doi: 10.3389/fcimb.2021.649722
Li, F., Zhang, Y., Chen, S., Wang, M., Jia, R., Zhu, D., et al. (2016). Identification of the nuclear localization signal region of duck enteritis virus UL14 and its interaction with VP16. Intervirology 59, 187–196. doi: 10.1159/000452711
Liu, C., Cheng, A., Wang, M., Chen, S., Jia, R., Zhu, D., et al. (2016). Characterization of nucleocytoplasmic shuttling and intracellular localization signals in duck enteritis virus UL54. Biochimie 127, 86–94. doi: 10.1016/j.biochi.2016.05.003
Liu, F. Y., and Roizman, B. (1991). The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 65, 5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991
Liu, F., and Roizman, B. (1993). Characterization of the protease and other products of amino-terminus-proximal cleavage of the herpes simplex virus 1 UL26 protein. J. Virol. 67, 1300–1309. doi: 10.1128/jvi.67.3.1300-1309.1993
Lu, R., and Misra, V. (2000). Potential role for luman, the cellular homologue of herpes simplex virus VP16 (alpha gene trans-inducing factor), in herpesvirus latency. J. Virol. 74, 934–943. doi: 10.1128/JVI.74.2.934-943.2000
Lv, Y., Zhou, S., Gao, S., and Deng, H. (2019). Remodeling of host membranes during herpesvirus assembly and egress. Protein Cell 10, 315–326. doi: 10.1007/s13238-018-0577-9
Ma, Y., Deng, X., Zhou, L., Dong, H., and Xu, P. (2023). HSV-1 selectively packs the transcription factor Oct-1 into EVs to facilitate its infection. Front. Microbiol. 14:1205906. doi: 10.3389/fmicb.2023.1205906
Marco, B. D., Massetti, M., Bruscoli, S., Macchiarulo, A., Virgilio, R. D., Velardi, E., et al. (2006). Glucocorticoid-induced leucine zipper (GILZ)/NF-B interaction: role of GILZ homo-dimerization and C-terminal domain. Nucleic Acids Res. 35, 517–528. doi: 10.1093/nar/gkl1080
Masa, S.-R., Lando, R., and Sarid, R. (2008). Transcriptional regulation of the open reading frame 35 encoded by Kaposi’s sarcoma-associated herpesvirus. Virology 371, 14–31. doi: 10.1016/j.virol.2007.08.023
Maurer, U. E., Sodeik, B., and Grünewald, K. (2008). Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc. Natl. Acad. Sci. 105, 10559–10564. doi: 10.1073/pnas.0801674105
McGeoch, D. J., Dalrymple, M. A., Davison, A. J., Dolan, A., Frame, M. C., McNab, D., et al. (1988). The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69, 1531–1574. doi: 10.1099/0022-1317-69-7-1531
McMillan, T. N., and Johnson, D. C. (2001). Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75, 1928–1940. doi: 10.1128/JVI.75.4.1928-1940.2001
Melchjorsen, J., Matikainen, S., and Paludan, S. R. (2009). Activation and evasion of innate antiviral immunity by herpes simplex virus. Viruses 1, 737–759. doi: 10.3390/v1030737
Mettenleiter, T. C. (2002). Herpesvirus assembly and egress. J. Virol. 76, 1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002
Mettenleiter, T. C., Klupp, B. G., and Granzow, H. (2009). Herpesvirus assembly: an update. Virus Res. 143, 222–234. doi: 10.1016/j.virusres.2009.03.018
Mettenleiter, T. C., Müller, F., Granzow, H., and Klupp, B. G. (2013). The way out: what we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 15, 170–178. doi: 10.1111/cmi.12044
Mittelstadt, P. R., and Ashwell, J. D. (2001). Inhibition of AP-1 by the glucocorticoid-inducible protein GILZ. J. Biol. Chem. 276, 29603–29610. doi: 10.1074/jbc.M101522200
Naipauer, J., and Mesri, E. A. (2023). The Kaposi’s sarcoma progenitor enigma: KSHV-induced MEndT-EndMT axis. Trends Mol. Med. 29, 188–200. doi: 10.1016/j.molmed.2022.12.003
Nair, R. (2003). LOC3D: annotate sub-cellular localization for protein structures. Nucleic Acids Res. 31, 3337–3340. doi: 10.1093/nar/gkg514
Newcomb, W. W., and Brown, J. C. (1991). Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 65, 613–620. doi: 10.1128/jvi.65.2.613-620.1991
Newcomb, W. W., Trus, B. L., Booy, F. P., Steven, A. C., Wall, J. S., and Brown, J. C. (1993). Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232, 499–511. doi: 10.1006/jmbi.1993.1406
Nguyen, M. L., and Blaho, J. A. (2006). “Apoptosis during herpes simplex virus infection” in Advances in virus research (New York, NY: Elsevier), 67–97.
Nozawa, N., Daikoku, T., Koshizuka, T., Yamauchi, Y., Yoshikawa, T., and Nishiyama, Y. (2003). Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J. Virol. 77, 3204–3216. doi: 10.1128/JVI.77.5.3204-3216.2003
Nuévalos, M., García-Ríos, E., Mancebo, F. J., Martín-Martín, C., and Pérez-Romero, P. (2023). Novel monoclonal antibody-based therapies: implications for the treatment and prevention of HCMV disease. Trends Microbiol. 31, 480–497. doi: 10.1016/j.tim.2022.12.003
Oda, S., Arii, J., Koyanagi, N., Kato, A., and Kawaguchi, Y. (2016). The interaction between herpes simplex virus 1 tegument proteins UL51 and UL14 and its role in Virion morphogenesis. J. Virol. 90, 8754–8767. doi: 10.1128/JVI.01258-16
Ohta, A., Yamauchi, Y., Muto, Y., Kimura, H., and Nishiyama, Y. (2011). Herpes simplex virus type 1 UL14 tegument protein regulates intracellular compartmentalization of major tegument protein VP16. Virol. J. 8:365. doi: 10.1186/1743-422X-8-365
Ojala, P. M., Sodeik, B., Ebersold, M. W., Kutay, U., and Helenius, A. (2000). Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20, 4922–4931. doi: 10.1128/MCB.20.13.4922-4931.2000
Owen, D., Crump, C., and Graham, S. (2015). Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses 7, 5084–5114. doi: 10.3390/v7092861
Pheasant, K., Perry, D., Wise, E. L., Cheng, V., and Elliott, G. (2023). Dysregulated metabolism of the late herpes simplex virus 1 transcriptome through the vhs-VP22 axis uncouples virus cytopathic effect and virus production. PLoS Pathog. 19:e1010966. doi: 10.1371/journal.ppat.1010966
Pomeranz, L. E., Reynolds, A. E., and Hengartner, C. J. (2005). Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69, 462–500. doi: 10.1128/MMBR.69.3.462-500.2005
Radtke, K., Kieneke, D., Wolfstein, A., Michael, K., Steffen, W., Scholz, T., et al. (2010). Plus-and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991. doi: 10.1371/journal.ppat.1000991
Rigoutsos, I., Novotny, J., Huynh, T., Chin-Bow, S. T., Parida, L., Platt, D., et al. (2003). In silico pattern-based analysis of the human cytomegalovirus genome. J. Virol. 77, 4326–4344. doi: 10.1128/JVI.77.7.4326-4344.2003
Russo, J. J., Bohenzky, R. A., Chien, M.-C., Chen, J., Yan, M., Maddalena, D., et al. (1996). Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. 93, 14862–14867. doi: 10.1073/pnas.93.25.14862
Salmon, B., Cunningham, C., Davison, A. J., Harris, W. J., and Baines, J. D. (1998). The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72, 3779–3788. doi: 10.1128/JVI.72.5.3779-3788.1998
Sanders, L. S., Comar, C. E., Srinivas, K. P., Lalli, J., Salnikov, M., Lengyel, J., et al. (2023). Herpes simplex Virus-1 ICP27 nuclear export signal mutants exhibit cell type-dependent deficits in replication and ICP4 expression. J. Virol. 97:e0195722. doi: 10.1128/jvi.01957-22
Schrag, J. D., Prasad, B. V. V., Rixon, F. J., and Chiu, W. (1989). Three-dimensional structure of the HSV1 nucleocapsid. Cell 56, 651–660. doi: 10.1016/0092-8674(89)90587-4
Scott, J. E., and Davison, A. J. (1986). The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67, 1759–1816. doi: 10.1099/0022-1317-67-9-1759
Shen, B., Ruan, P., Cheng, A., Wang, M., Zhang, W., Wu, Y., et al. (2023). Characterization of a unique novel LORF3 protein of duck plague virus and its potential pathogenesis. J. Virol. 97:e0157722. doi: 10.1128/jvi.01577-22
Sherman, G., and Bachenheimer, S. L. (1987). DNA processing in temperature-sensitive morphogenic mutants of HSV-1. Virology 158, 427–430. doi: 10.1016/0042-6822(87)90214-5
Sherman, G., and Bachenheimer, S. L. (1988). Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology 163, 471–480. doi: 10.1016/0042-6822(88)90288-7
Stow, N. D. (2001). Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75, 10755–10765. doi: 10.1128/JVI.75.22.10755-10765.2001
Subak-Sharpe, J. H., and Dargan, D. J. (1998). HSV molecular biology: general aspects of herpes simplex virus molecular biology. Virus Genes 16, 239–251. doi: 10.1023/A:1008068902673
Takeshima, K., Arii, J., Maruzuru, Y., Koyanagi, N., Kato, A., and Kawaguchi, Y. (2019). Identification of the capsid binding site in the herpes simplex virus 1 nuclear egress complex and its role in viral primary envelopment and replication. J. Virol. 93, e01290–e01219. doi: 10.1128/JVI.01290-19
Tandon, R., and Mocarski, E. S. (2011). Cytomegalovirus pUL96 is critical for the stability of pp150-associated Nucleocapsids. J. Virol. 85, 7129–7141. doi: 10.1128/JVI.02549-10
Teng, M., Zhu, Z.-J., Yao, Y., Nair, V., Zhang, G.-P., and Luo, J. (2023). Critical roles of non-coding RNAs in lifecycle and biology of Marek’s disease herpesvirus. Sci. China Life Sci. 66, 251–268. doi: 10.1007/s11427-022-2258-4
Terry, L. J., Shows, E. B., and Wente, S. R. (2007). Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416. doi: 10.1126/science.1142204
Thomas, E. C. M., Bossert, M., and Banfield, B. W. (2022). The herpes simplex virus tegument protein pUL21 is required for viral genome retention within capsids. PLoS Pathog. 18:e1010969. doi: 10.1371/journal.ppat.1010969
Thomsen, D. R., Newcomb, W. W., Brown, J. C., and Homa, F. L. (1995). Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J. Virol. 69, 3690–3703. doi: 10.1128/jvi.69.6.3690-3703.1995
Thurlow, J. K., Rixon, F. J., Murphy, M., Targett-Adams, P., Hughes, M., and Preston, V. G. (2005). The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79, 150–158. doi: 10.1128/JVI.79.1.150-158.2005
Truffot, A., Noble, J., Dartevel, A., Chevalier, E., Dard, C., Giovannini, D., et al. (2023). Fatal HSV-2 primary infection most likely acquired by kidney transplantation: a case report. Int. J. Antimicrob. Agents 61:106769. doi: 10.1016/j.ijantimicag.2023.106769
Uddin, M. K., Watanabe, T., Arata, M., Sato, Y., Kimura, H., and Murata, T. (2023). Epstein-Barr virus BBLF1 mediates secretory vesicle transport to facilitate mature Virion release. J. Virol. 97:e0043723. doi: 10.1128/jvi.00437-23
Van Gent, M., Chiang, J. J., Muppala, S., Chiang, C., Azab, W., Kattenhorn, L., et al. (2022). The US3 kinase of herpes simplex virus phosphorylates the RNA sensor RIG-I to suppress innate immunity. J. Virol. 96, e01510–e01521. doi: 10.1128/JVI.01510-21
Wada, K., Goshima, F., Takakuwa, H., Yamada, H., Daikoku, T., and Nishiyama, Y. (1999). Identification and characterization of the UL14 gene product of herpes simplex virus type 2. J. Gen. Virol. 80, 2423–2431. doi: 10.1099/0022-1317-80-9-2423
Watanabe, T., Fuse, K., Takano, T., Narita, Y., Goshima, F., Kimura, H., et al. (2015). Roles of Epstein-Barr virus BGLF3.5 gene and two upstream open reading frames in lytic viral replication in HEK293 cells. Virology 483, 44–53. doi: 10.1016/j.virol.2015.04.007
Whitley, R. J., Kimberlin, D. W., and Roizman, B. (1998). Herpes simplex viruses. Clin. Infect. Dis. 26, 541–553. doi: 10.1086/514600
Wills, E., Scholtes, L., and Baines, J. D. (2006). Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid. J. Virol. 80, 10894–10899. doi: 10.1128/JVI.01364-06
Wing, C. E., Fung, H. Y. J., and Chook, Y. M. (2022). Karyopherin-mediated nucleocytoplasmic transport. Nat. Rev. Mol. Cell Biol. 23, 307–328. doi: 10.1038/s41580-021-00446-7
Wu, L., Wang, M., Cheng, A., Tian, B., Huang, J., Wu, Y., et al. (2023). Duck plague virus tegument protein vp22 plays a key role in the secondary envelopment and cell-to-cell spread. Vet. Res. 54:60. doi: 10.1186/s13567-023-01191-9
Wu, F., Xing, J., Wang, S., Li, M., and Zheng, C. (2011). Screening and identification of host factors interacting with UL14 of herpes simplex virus 1. Med. Microbiol. Immunol. (Berl.) 200, 203–208. doi: 10.1007/s00430-011-0196-z
Wubben, J. M., Atkinson, S. C., and Borg, N. A. (2020). The role of protein disorder in nuclear transport and in its subversion by viruses. Cells 9:2654. doi: 10.3390/cells9122654
Xu, L., and Massagué, J. (2004). Nucleocytoplasmic shuttling of signal transducers. Nat. Rev. Mol. Cell Biol. 5, 209–219. doi: 10.1038/nrm1331
Xu, X.-Q., Xu, T., Ji, W., Wang, C., Ren, Y., Xiong, X., et al. (2023). Herpes simplex virus 1-induced Ferroptosis contributes to viral encephalitis. MBio 14:e0237022. doi: 10.1128/mbio.02370-22
Yamauchi, Y., Daikoku, T., Goshima, F., and Nishiyama, Y. (2003). Herpes simplex virus UL14 protein blocks apoptosis. Microbiol. Immunol. 47, 685–689. doi: 10.1111/j.1348-0421.2003.tb03432.x
Yamauchi, Y., Goshima, F., Yoshikawa, T., Nozawa, N., Koshizuka, T., and Nishiyama, Y. (2002a). Intercellular trafficking of herpes simplex virus type 2 UL14 deletion mutant proteins. Biochem. Biophys. Res. Commun. 298, 357–363. doi: 10.1016/S0006-291X(02)02452-X
Yamauchi, Y., Kiriyama, K., Kubota, N., Kimura, H., Usukura, J., and Nishiyama, Y. (2008). The UL14 tegument protein of herpes simplex virus type 1 is required for efficient nuclear transport of the alpha transinducing factor VP16 and viral capsids. J. Virol. 82, 1094–1106. doi: 10.1128/JVI.01226-07
Yamauchi, Y., Wada, K., Goshima, F., Daikoku, T., Ohtsuka, K., and Nishiyama, Y. (2002b). Herpes simplex virus type 2 UL14 gene product has heat shock protein(HSP)-like functions. J. Cell Sci. 115, 2517–2527. doi: 10.1242/jcs.115.12.2517
Yamauchi, Y., Wada, K., Goshima, F., Takakuwa, H., Daikoku, T., Yamada, M., et al. (2001). The UL14 protein of herpes simplex virus type 2 translocates the minor capsid protein VP26 and the DNA cleavage and packaging UL33 protein into the nucleus of coexpressing cells. J. Gen. Virol. 82, 321–330. doi: 10.1099/0022-1317-82-2-321
Yanagi, Y., Masud, H. M. A. A., Watanabe, T., Sato, Y., Goshima, F., Kimura, H., et al. (2019). Initial characterization of the Epstein⁻Barr virus BSRF1 gene product. Viruses 11:285. doi: 10.3390/v11030285
Yang, K., Wills, E. G., and Baines, J. D. (2011). A mutation in UL15 of herpes simplex virus 1 that reduces packaging of cleaved genomes. J. Virol. 85, 11972–11980. doi: 10.1128/JVI.00857-11
Yu, D., and Weller, S. K. (1998). Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72, 7428–7439. doi: 10.1128/JVI.72.9.7428-7439.1998
Zhou, T., Fan, D., Wang, M., Cheng, A., Wu, Y., Yang, Q., et al. (2021). Duck plague virus pUL48 protein activates the immediate-early gene to initiate the transcription of the virus gene. Front. Microbiol. 12:795730. doi: 10.3389/fmicb.2021.795730
Keywords: herpesvirus, pUL14, tegument proteins, viral replication, viral infection
Citation: Wan J, Wang M, Cheng A, Zhang W, Yang Q, Tian B, Ou X, Sun D, He Y, Zhao X, Wu Y, Zhang S, Huang J, Wu Z, Yu Y, Zhang L, Zhu D, Liu M, Chen S and Jia R (2024) Multiple functions of the herpesvirus UL14 gene product in viral infection. Front. Microbiol. 15:1483022. doi: 10.3389/fmicb.2024.1483022
Edited by:
Tejabhiram Yadavalli, University of Illinois Chicago, United StatesReviewed by:
Madavaraju Krishnaraju, Northwestern University, United StatesRahul K. Suryawanshi, Gladstone Institutes, United States
Copyright © 2024 Wan, Wang, Cheng, Zhang, Yang, Tian, Ou, Sun, He, Zhao, Wu, Zhang, Huang, Wu, Yu, Zhang, Zhu, Liu, Chen and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
 Jieyu Wan
Jieyu Wan Mingshu Wang
Mingshu Wang Anchun Cheng
Anchun Cheng Wei Zhang6
Wei Zhang6 Qiao Yang
Qiao Yang Bin Tian
Bin Tian Xumin Ou
Xumin Ou Di Sun
Di Sun Shaqiu Zhang
Shaqiu Zhang Juan Huang
Juan Huang Dekang Zhu
Dekang Zhu Mafeng Liu
Mafeng Liu Shun Chen
Shun Chen