- 1Lab of Microbiological Engineering (Infection and Immunity), School of Life and Health Sciences, Hainan Province Key Laboratory of One Health, Collaborative Innovation Center of One Health, Hainan University, Haikou, China
- 2Hainan International One Health Institute, Hainan University, Haikou, China
- 3School of Chemical Engineering and Technology, Hainan University, Haikou, China
Background: Sheep are a rarely raised livestock in Hainan Island, China, because of the unfavorable tropical marine climate. Here, this article reports a severe pneumonia in the sheep breeding and domestication facility caused acute mortality during the winter 2021–2022.
Methods: Six sheep were clinically dissected and histopathologically observed. The bacteria were isolated and cultured by traditional methods and identified by 16S rRNA sequencing. The genotypes, serotypes, virulence genes and antimicrobial resistance genes were analyzed by PCR and whole genome sequencing. The pubMLST website was used for phylogenetic analysis of related strains. Kirby-Bauer disk diffusion method was used for antimicrobial susceptibility test. The antimicrobial susceptibility test standard was referred to the Clinical and Laboratory Standards Institute (CLSI). The virulence of bacteria was detected by mouse infection model.
Results: Etiology and histopathology examination of the pneumonia reveled pulmonary abscess and alveolar neutrophilia and pulmonary fibrinous exudates. Escherichia coli was the only bacterial species isolated, primarily from the lungs and blood of the six dead or moribund sheep, a total of 29 E. coli strains were isolated. Antimicrobial resistance profiling shows that all the isolates were resistant to six agents (penicillin, ampicillin, cephalothin, neomycin, erythromycin, and vancomycin) belonging to five classes of antibiotics, classifying them as multi drug resistant (MDR). Furthermore, genotyping analysis revealed all strains were common with 11–17 virulence factors indicating high pathogenicity. The lab mice infection model shows that all strains severely affect the health status particularly weight loss, lethargy, pneumonia and shortly lead to death. The molecular epidemiological analysis indicated most strains share the same genotype as previously reported strains in humans and other farmed animals this suggests a high possibility of cross-species transmission (CST) of virulent and MDR isolates. This CST could be from sheep to humans and other farmed animals or from humans and other farmed animals to sheep.
Conclusion: Therefore, this study indicates that E. coli is an emerging threat that causes sheep pneumonia in Hainan, and the quarantine of contacts is important to control the spread of virulent E. coli and the transmission of acquired resistance genes between humans and farmed animals such as sheep.
Background
Escherichia coli, a gram negative, genetically diverse bacterium includes both beneficial strains colonizing the gut and highly pathogenic strains which is responsible for intestinal and extraintestinal diseases. The emergence of intestinal pathogenic E. coli (IPEC) and extraintestinal pathogenic E. coli (ExPEC) presents ongoing health concerns (Poolman and Wacker, 2016). These pathogenic strains cause a wide range of host infective syndromes, ranging from simple diarrhea and urinary tract infections (UTIs) to life-threatening septicemia or bacteremia. Furthermore, the appearance of multidrug-resistant (MDR) strains poses a significant challenge to the current treatments, fueling the global spread of antimicrobial resistance (Russo and Johnson, 2003; Mellata, 2013).
Certain epidemiological surveillance findings highlight the zoonotic risks posed by the highly virulent and MDR strains (Desvars-Larrive et al., 2024). These measures include using multi-locus sequence typing (MLST), pulsed field gel electrophoresis (PFGE), and virulence gene profiles (Maluta et al., 2014; Rodriguez-Siek et al., 2005) to compare the genetic relatedness between human and avian isolates acquired from poultry products, as well as between avian and human isolates from urine or blood sources. Previous studies have reported cross-species transmission from human to poultry and from poultry to mice (Tivendale et al., 2010; Zhao et al., 2009). Sheep, a widely distributed livestock species in the mainland China are rarely raised in the southernmost areas, such as Hainan Island due to the hot and humid climate (Gencay, 2014). To meet the demand of the growing tourist industry and provide for the nutritional needs of locals, sheep farming has been introduced to the area. However, this harsh and new habitat can adversely affect the farmed sheep, making them more susceptible to various infections including bacterial, viral and fungal diseases (Zhao J. Y. et al., 2021; Zhao X. et al., 2021; Kuraz Abebe, 2022). This can lead to a decline in livestock productivity and profitability, consequently damaging the economic health of the industry. Previous studies reported that the most common diseases among sheep are respiratory diseases such as pneumonia (Scott, 2011; Mekibib et al., 2019). Bacterial bronchopneumonia tends to occur most frequently in sheep that have experienced recent stressors, such as transportation, dietary changes, or mixing with herds from unrelated farms (Aiello and Moses, 2016). The most prevalent bacterial pathogens associated with the primary or secondary bronchopneumonia in sheep are including Mannheimia haemolytica, Mycoplasma ovipneumoniae, Pasteurella multocida, Bibersteinia trehalosi, Chlamydia pneumoniae, and Salmonella spp. (Besser et al., 2013; Alley et al., 1999). In rare cases, however, E. coli species have also been isolated from lung or heart blood samples of sheep with pneumonia and septicemia (Singh et al., 2019).
Here, we provide evidence that E. coli was the sole predisposing agent responsible for severe respiratory infections in a newly established sheep-breeding enterprise in Wenchang, Hainan (E110.78°, N19.96°), during winter 2021 to 2022. Additionally, the risks of cross-species transmission were investigated by ascertaining the antimicrobial resistance, virulence, and genetic characteristics of the E. coli isolates.
Materials and methods
Animal collection and gross examination
Sporadic deaths were reported in a newly established breeding farm in Wenchang, Hainan during the winter 2021–2022 following the introduction of new breeds. During this period, a total of six dead sheep including five adults and a lamb were collected for necropsy. According to a previous study (Rahsan et al., 2018), clinical signs including wounds, ulcers, skin lesions, excrements, and secretions from nose, eyes and mouth of each carcass were recorded before autopsy. Following dissection, bronchial exudates and lesions of the internal organs were examined visually and multiple systematic incisions.
Histopathological examination
Following collection, tissue samples from lung, liver and other organs were fixed with 10% neutral buffered formalin. Subsequently, the samples were sent to Wuhan Saville Biotechnology Co., Ltd.1 for paraffin embedding, sectioning and staining. The stained slides were examined under a light microscope at 10× and 100× for histopathological evaluation (Rehman et al., 2021; Mekibib et al., 2019).
Bacterial examination
For mycoplasma detection, a piece of lung tissue (0.2–0.5 g) was homogenized and centrifuged at 8, 000 rpm for 10 min followed by DNA extraction from 200 μL supernatant using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China). Specific primers were designed (Supplementary Table S1) and used in a polymerase chain reaction (PCR) (Young et al., 2010). In this paper, double-distilled water (ddH₂O) is employed as the negative control in all PCR. Furthermore, for bacterial isolation samples from various internal organs, blood, pleural effusions and bronchial exudates were inoculated onto brain heart infusion (BHI) agar plates following the method described by Rehman et al. (2021). Subsequently, the colonies were then primarily identified based on the conventional biochemical methods (Jia et al., 2022). For molecular identification, DNA was extracted from the samples using DNA isolation mini kit (Novazan, Nanjing, China) and 16S rRNA gene was amplified with the universal primers listed in Supplementary Table S1 followed by sequencing (Nanshan Biotech). Finally, the species level of each isolate was identified as matching the reference strain based on the new canonical clustering threshold of >99% similarity of full-length 16S rRNA gene (Edgar, 2018).
Extraintestinal pathogenic Escherichia coli identification
E. coli isolates were tested by PCR with primer sets (Supplementary Table S1) of the following five virulence marker genes associated with ExPEC: papA/C (P fimbriae; counted as 1), sfa/foc (S and F1C fimbriae), afa/dra (A fimbrial and Dr-binding adhesion), kpsM II (group 2 capsule), and iutA (aerobactin system). Isolates positive for two or more of these markers were classified as ExPEC (Johnson et al., 2003).
Phylogrouping and MLST analysis
The E. coli isolates were characterized according to phylogrouping and multilocus sequence typing (MLST) analysis. Phylogroups were determined using the Clermont PCR method (Clermont et al., 2013) (primers listed in Supplementary Table S1). In addition, MLST was performed using two schemes: the Achtman scheme targeting seven house-keeping genes (adk, fumC, gyrB, icd, mdh, recA and purA) (Manges et al., 2015), and the Pasteur scheme targetting eight housekeeping genes (dinB, icdA, pabB, polB, putP, trpA, trpB and uidA) (Primers in Supplementary Table S1) (Clermont et al., 2015).
Serotype identification
Whole genome sequencing (WGS) was performed using Illumina HiSeq™ platform (PE150) at Sangon Bioengineering Co., Ltd. (Shanghai, China) on representative isolate of each E. coli sequence type. Serotypes were determined for these isolates using online SerotypeFinder2.0 services (Joensen et al., 2015). PCR based serotyping with primers designed for the representative isolates (Supplementary Table S1) was subsequently used to identify serotypes of the remaining isolates.
Antimicrobial susceptibility assay
For antimicrobial susceptibility assay, Kirby-Bauer disc diffusion method was used for 19 drugs following the CLSI guidelines (Humphries et al., 2021). However, for two drugs polymyxin B and tigecycline (Solarbio Life Sciences Co, Beijing, China), the minimal inhibitory concentration (MIC) was determined according to the 2017 EUCAST methods.2 The antibiotics employed altogether fall into 11 major categories. Firstly, β-lactams include ampicillin, amoxicillin, and penicillin. Secondly, aminoglycosides consist of streptomycin, neomycin, and gentamicin. Thirdly, chloramphenicols are represented by chloramphenicol and florfenicol. Fourthly, fluoroquinolones comprise norfloxacin, ofloxacin, ciprofloxacin, and nalidixic acid. Fifthly, tetracyclines consist of tetracycline and tigecycline. Sixthly, cephalosporins are exemplified by cephalothin. Seventhly, sulfonamides include sulfisoxazole and trimethoprim. Eighthly, polypeptides are represented by polymyxin B. Ninthly, carbapenems are embodied by imipenem. Tenthly, macrolides are exemplified by erythromycin. Lastly, glycopeptides are represented by vancomycin. While E. coli ATCC 25,922 was subjected as quality control. Furthermore, the resistance gene pattern of each isolate was screened by PCR with the primer sets of 19 acquired antimicrobial resistance genes (Supplementary Table S1).
Virulence gene profiling
Based on the WGS of each representative E. coli isolate, the virulence factors were further obtained by VFDB tool3 following the method of Rehman et al. (2023).
Core genome multilocus phylogenetic analysis
Phylogenetic relationship between the isolated strains in this study and previously observed E. coli strains was determined according to a previous study with slight modifications (Rehman et al., 2023). The GrapeTree tool was used from pubMLST4 to analyze core genome multilocus phylogenetic analysis (cgMLPA) data for the isolated strains and other E. coli strains (Supplementary Table S2). iTOL v.45 program was used for visualization and incorporation of available metadata into a phylogenetic tree.
Experimental animal and ethics statement
Specific pathogen free (SPF) KM mice were employed from the Laboratory Animal Center, School of Pharmaceutical Sciences, Hainan University (SYXK0230021). The experimental protocol was approved by the Animal Management and Ethics Committee of Hainan University (HNUAUCC-2023-00020), and adhered to the recommendations of the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
Experimental design and infection model in mice
Mice were randomly allocated into six groups (five challenge groups and one control group) with six mice per group. The weight of each mouse was documented. Intraperitoneal injections were administered following the method of Long et al. (2022). Challenge groups were injected with 0.2 mL of E. coli solution (approximately 2 × 106 CFU/mL), while control group received 0.2 mL PBS solution. Mice mortality rate and weight fluctuations were monitored every 6 h for 72 h. Pathological changes in the vital organs (heart, lung, liver, kidney and spleen) were recorded, and lungs were sent to Wuhan Saville Biotechnology Co., Ltd. (see text footnote 1) for further histopathological analysis.
Results
Necropsy and histopathological examination
Bronchopneumonia was diagnosed in six autopsied sheep collected at different time intervals. The common signs of the disease were sticky mucus in nostrils (Figure 1a) and trachea (Figure 1b), as well as irregular red consolidation and large-scale abscesses on the surfaces of extremely enlarged lungs (Figure 1c). These abscesses likely resulted from the internal accumulation of abundant pale-yellow mucoid exudate. (Figure 1d). Histopathological sectioning of lungs further revealed the features of lobular pneumonia of alveoli obliteration with the infiltration of neutrophils and capillary occlusion with intra-lumen fibrin exudates (Figure 1e,f).
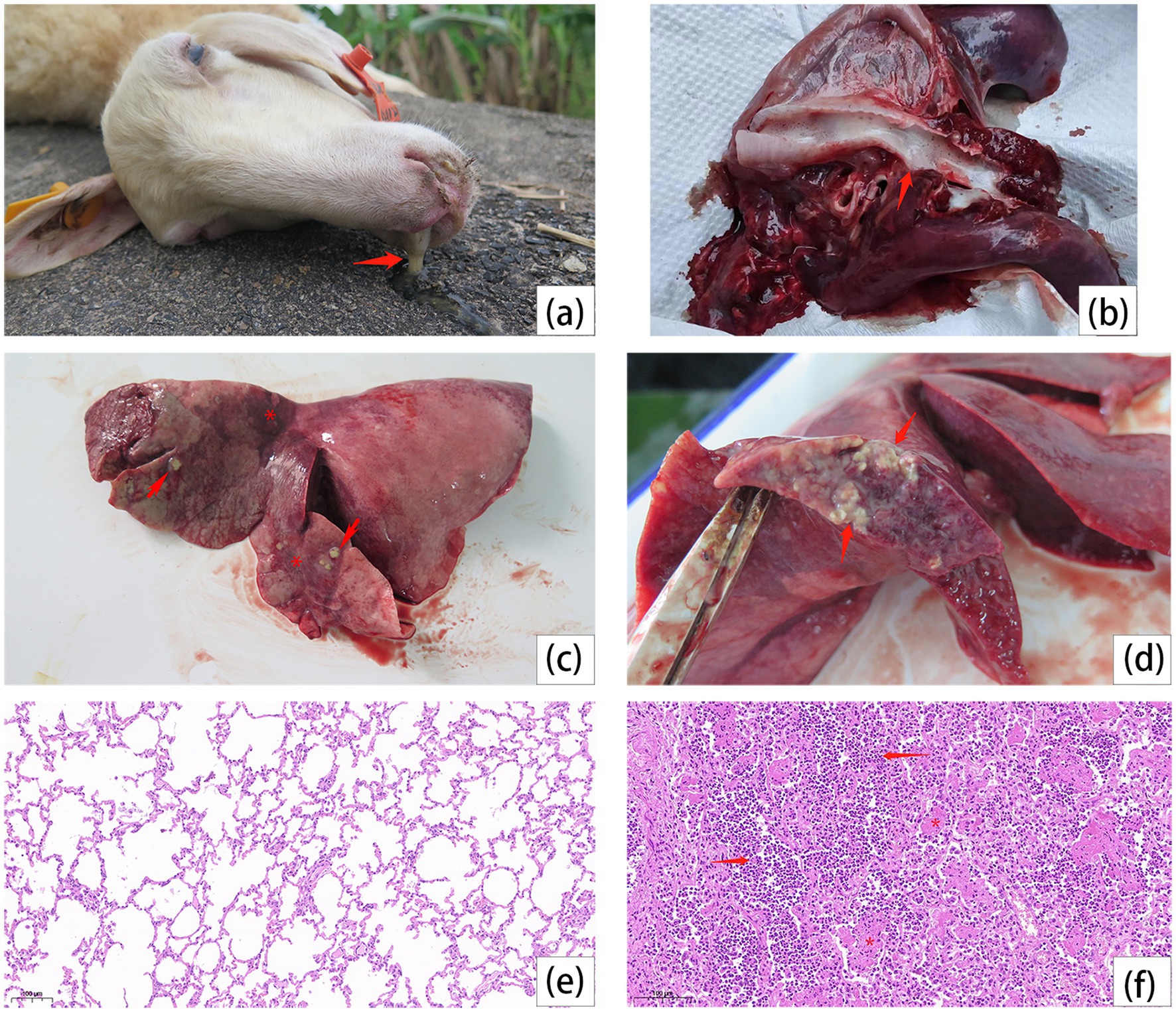
Figure 1. Dead sheep autopsy, lesions on trachea and lungs, and lung histopathology. (a) Secretions from the nasal cavity of the dead sheep (arrow). (b) White secretions in the trachea (arrow). (c) Lobar pneumonia in the left cranial and medial lobes. Irregular red consolidation in the affected lobe (stars), with varying sizes of abscesses (arrows). (d) The abscess infiltrates the lung (arrows). (e) The histopathological observation of lung in healthy sheep. (f) The alveoli are filled with dense neutrophils (arrows) and some alveoli have fibrin exudate in the lumen (stars).
Bacterial identification
Gram-negative, rod-shape bacteria were isolated from various tissues, especially lungs, liver, and blood. Further, PCR detection with specific primers for M. ovipneumoniae and M. mycoides was negative. Based on traditional 16S rRNA sequencing, all the isolates were identified as E. coli (Table 1).
ExPEC identification
A total of 29 isolates were recovered and considered for ExPEC identification. In term of pathogenic E. coli, ExPEC strains are responsible for the majority of human or animal extraintestinal infections, including pneumonia, urinary tract infections, and sepsis. However, according to the classic ExPEC virulence genes defined by Johnson et al. (2003), our results demonstrated that only three are categorized in ExPEC (Table 1). Interestingly, all 29 isolates originated from outside of the gastrointestinal tract. This finding suggests that the current method may not effectively differential between facultative ExPEC pathogens and commensal E. coli.
Phylogrouping and MLST analysis
A total of three phylogroups were predicted and among 29 isolates, five belonged to A, 23 belonged to B1, and only one belonged to E (Figure 2). According to the two MLST schemes, the isolates were further subdivided into nine sequence types (ST) (Figure 2). Three isolates (spleen, blood, and trachea) from the second sheep exhibited novel ST according to Achtman scheme. This new ST was closely related to ST1704, but it remained as unassigned as a new identifier. Interestingly, Pasteur scheme classified this same ST as the existing ST1107 (Figure 2). Moreover, most of the STs in this study were very consistent between the both MLST schemes, except ST2521 (Achtman), which was subdivided into ST1109 and ST1110 in the Pasteur MSLT scheme, and ST1107 of Pasteur scheme into ST1704 and ST14878 based on the Achtman scheme (Figure 2).
Classification of serotype
WGS and PCR data were combined to classify the 29 isolates. According to the analysis, eight serotypes were obtained including both lipopolysaccharide O antigen and flagella H antigen. One exception case was the O-antigen serotype of three isolates (Y2XX1 from blood and Y2P1 and Y2P3 from sheep 2 spleen) was completely unmatched in the database of the Center for Genomic Epidemiology.6 This could be due to sequencing gaps within O-antigen and H-antigen gene clusters in their assembled genomes (Figure 2). By comparing the correlations of serotypes and ST, the serotypes O147:H7 and O153:H12 subdivided the isolates of ST2521 (Achtman scheme) into ST1109 and ST1110 (Pasteur scheme). Similarly, the O26:H11 and O8:H7 serotypes distinguished isolates ST1107 (Pasteur scheme) into ST1704 and ST14878 (Achtman scheme) (Figure 2). These finding suggest that the serotyping may be more effective in resolving the genetic distance between isolates than sequence typing. However, the two MLST schemes (Achtman and Pasteur) can be applied for co-analysis.
Virulence factors detection
WGS data of 10 isolates representing unique serotypes were analyzed using the VFDB database. A total of 25 virulence factors categorized into seven classes were predicted. The most prevalent virulence factors included six adhesion-associated elements (CFA/I fimbriae, pilus, laminin-binding fimbriae, EaeH, hemorrhagic E. coli pilus, and type I fimbriae), two autotransporter associated elements (EhaA/B and UpaG adhesin), one invasion-associated element (invasion protein), two secretion system factors (T3SS translocator protein and Esp family protein), and one toxin factor (hemolysin/cytolysin A). All strains harbored at least 11 virulence factors. The highest and lowest virulence factors were predicted in isolates Y13F1 (17) and Y3X1 (11), respectively (Figure 2).
Mouse infection model
To assess the pathogenicity of isolated strains with different virulence factor combinations, a mouse infection model was established. Mortality rate and weight changes were analyzed after intraperitoneal injection of isolates. The Y13F1 isolate exhibited the most lethal effect, resulting in the death of all mice within 36 h. Conversely, Y2P1 isolate demonstrated the least lethal effect and did not cause any mortality (Figure 3A). By examining the average weight changes of mice post-intraperitoneal injection, a significant increase in weight was noticed for the control group. All mice within the challenge group experienced weight loss, with Y5YF5 resulting in the most significant weight loss and Y13F1 the least (Figure 3B). Gross pathology examination shows that the main organs of all experimental mice exhibited either localized or extensive hemorrhages except control mice. Lung hemorrhages were most severe in mice challenged with Y13F1 and Y3YF1 isolates compared to other experimental groups (Figure 3C). Additionally, the spleens of Y2P1 and Y3YF1 challenged mice were notably swollen, while the spleens of Y5YF3 and Y3YF1 challenged mice appeared more blackened (Figure 3C).
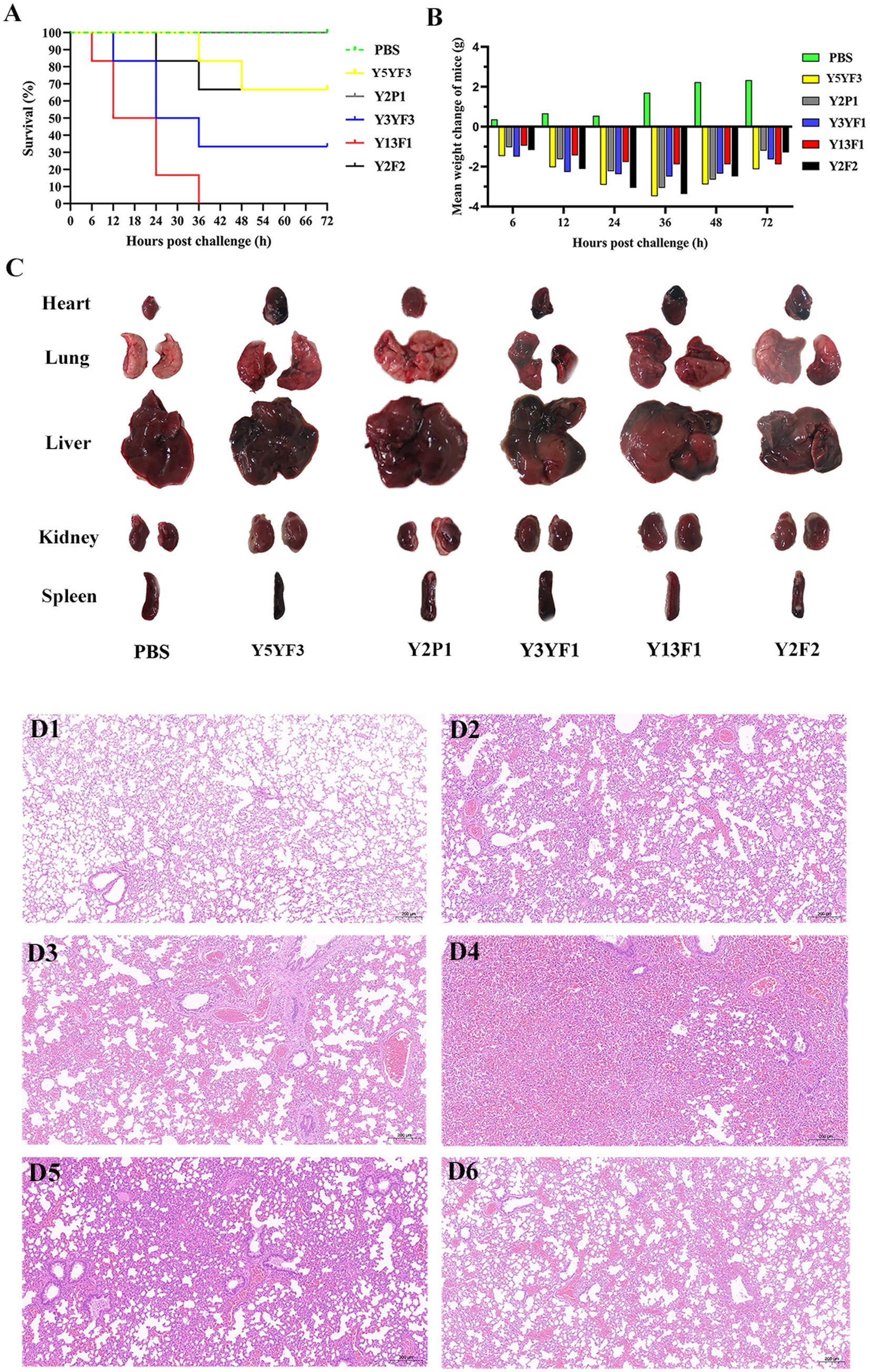
Figure 3. The results of pathogenicity test on mice. (A) Mouse survival rate curve. (B) Mean weight change of mice. (C) The main organs of the experimental mouse. (D) The histopathological observation of lung in healthy sheep (D1–D6 separately represent the group of PBS, Y5YF3, Y2P1, Y3YF1, Y13F1, Y2F2).
Histopathological changes in the lungs show inflammatory responses compared to the control group. The lung inflammatory response in the Y3YF1 group was particularly intense, characterized by inflammatory cell infiltration, fibrinoid exudation, and bronchial epithelial cell shedding (Figure 3D). This study demonstrates that isolates with different virulence factor combinations exhibit variation in lethality, target organ tropism, and their capacity to cause pneumonia. However, all strains displayed a degree of pathogenicity, highlighting the importance of further investigation into these isolates.
cgMLPA analysis
Genotypes of E. coli isolated in this study were compared to the pubMLST database. We discovered that E. coli with the same genotype as Y2F2 has only been reported in cows from the USA, UK, and Kenya, and in birds from Denmark (Figure 4A). However, E. coli with the same genotype as Y11S2 in this study exhibited a broader range of sources, including healthy individuals’ feces in China and France; clinical patients in the USA; cows from the USA, UK, Japan, and France, and chickens from Kenya and Peru (Figure 4B). E. coli with the same genotypes as Y5YF3 and Y2S3 have been found in clinical patients in the United States; feces from healthy people in France; cows in the USA, UK, and France; and pets, pigs, rabbits, and other animals in various countries (Figure 4C). E. coli with the Y2P1 genotype has only been reported in cows from the UK while the Y3X1 genotype was found only in individuals from China and Kenya (Supplementary Table S2). No reports of E. coli with the same genotypes as Y2XX1 and Y2P2 were found (Supplementary Table S2). The aforementioned E. coli genotypes were initially discovered and reported in sheep. Similarly, E. coli with the Y13F1 genotype in this study was primarily found in sheep, with a few individual cases detected in pigs and healthy people in China (Figure 4D).
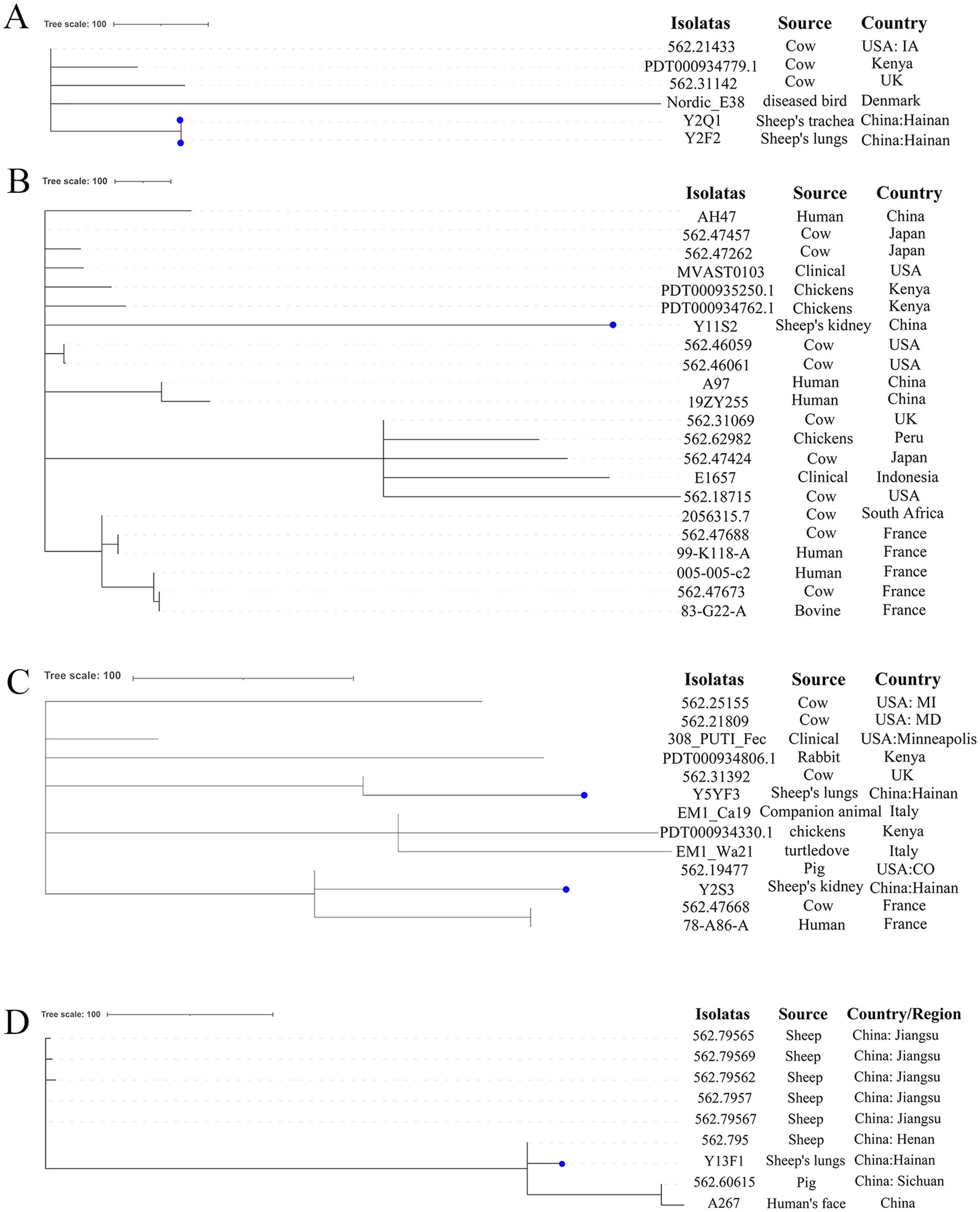
Figure 4. The phylogenetic analysis results of E. coli in this study and E. coli with the same genotypes in the pubMLST database based on cgMLST. (A–D) Separately represent the strains of ST1146, ST1308, ST2521 and ST971 strains in pubMLST database, the dots represent the strains in this study.
Antimicrobial susceptibility assay
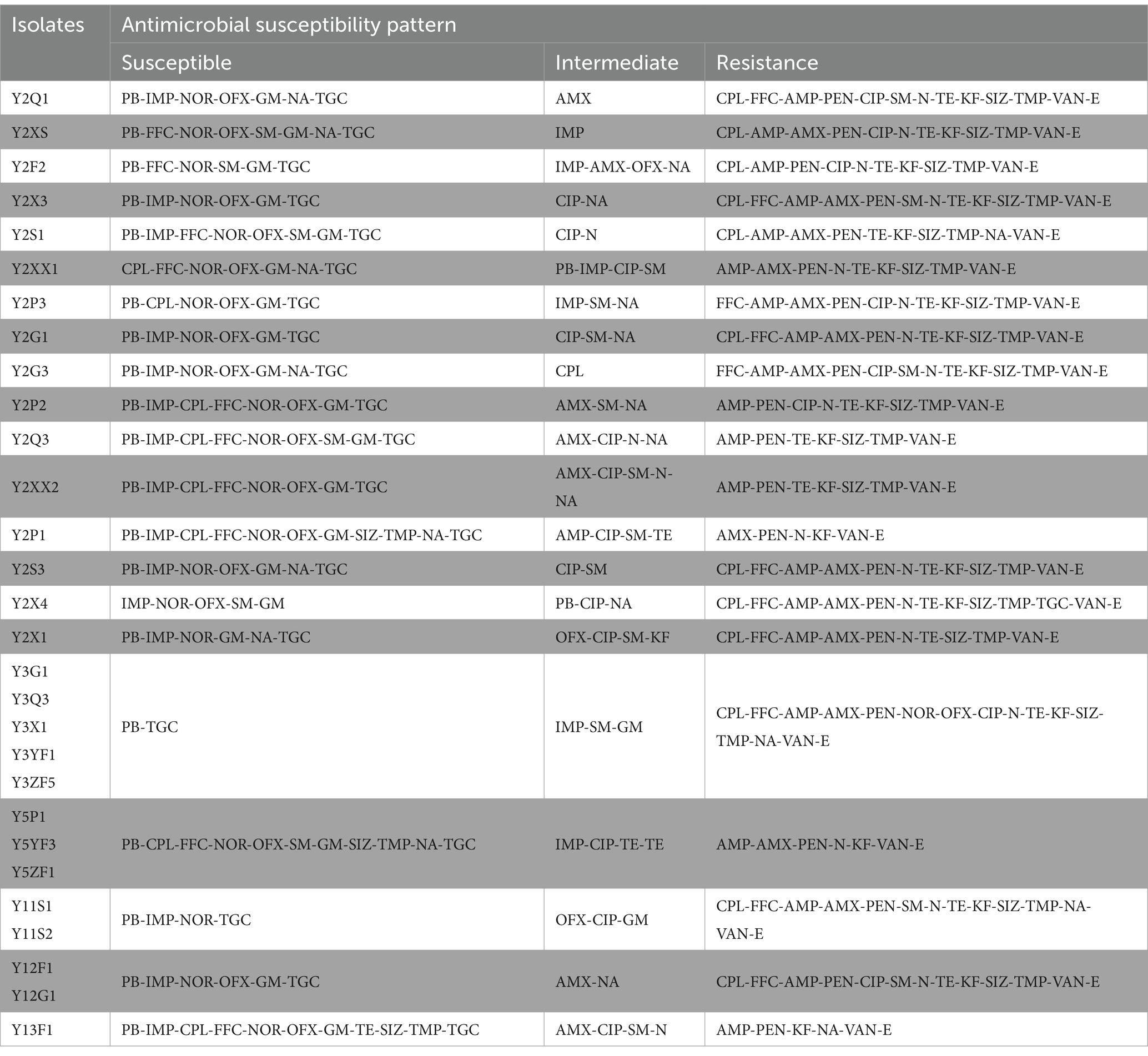
Twenty-one antibiotics were used in susceptibility tests, with isolates showing resistance to at least six and up to sixteen antibiotics (Figure 5 and Table 2). The total resistance rates were 80% to tetracycline and sulfonamides (sulfamethoxazole and trimethoprim), 72% to amoxicillin, 66% to chloramphenicol, 59% to florfenicol, 45% to ciprofloxacin, 31% to nalidixic acid, 24% to streptomycin, and 17% to norfloxacin and ofloxacin. High or intermediate susceptibility was observed for four drugs (polymyxin B, imipenem, tigecycline, and gentamicin) by all isolates. On the other hand, all isolates were resistant to six agents (penicillin, ampicillin, cephalothin, neomycin, erythromycin, and vancomycin) (Figure 5).
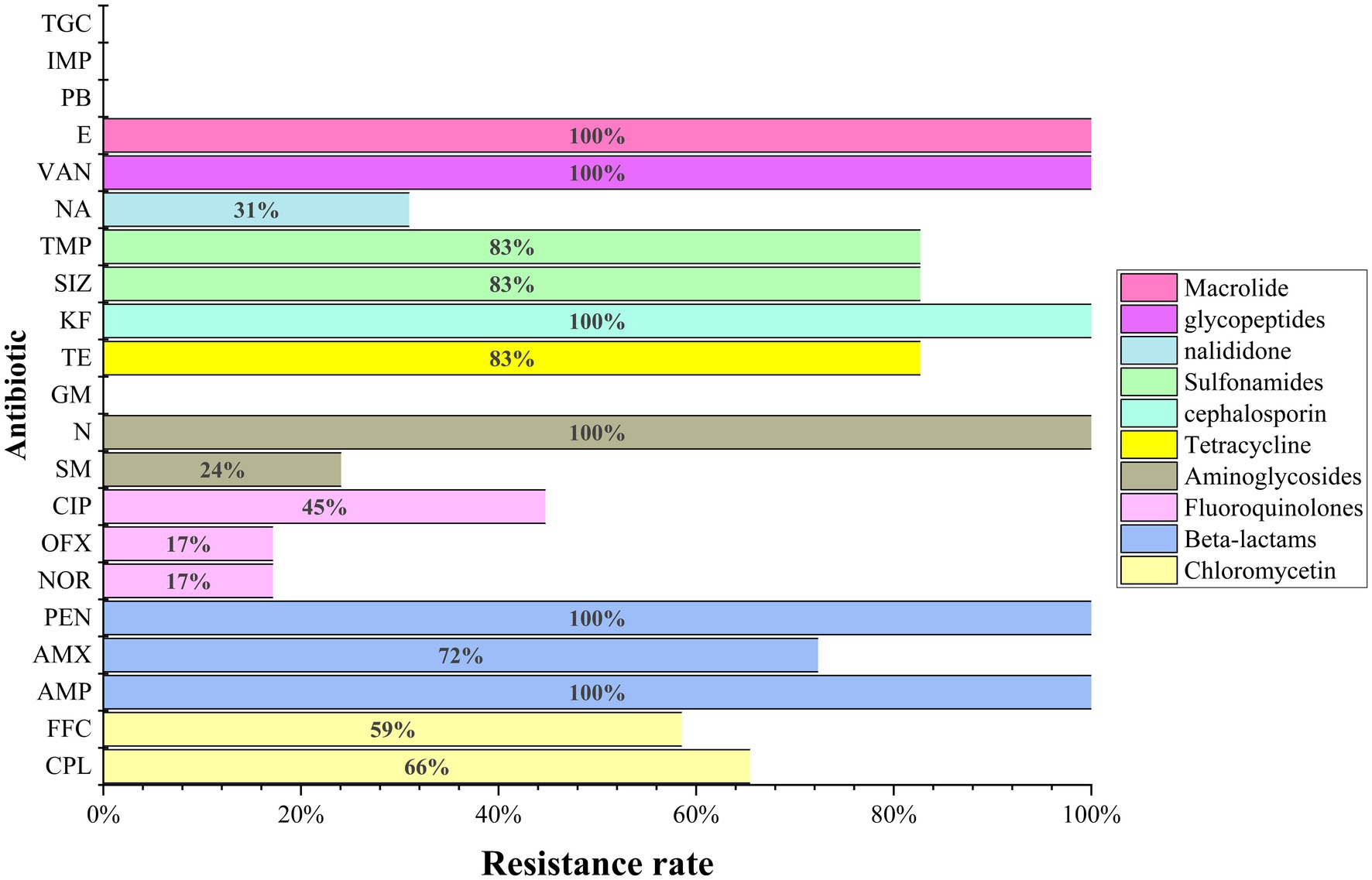
Figure 5. The results of antimicrobial susceptibility tests of the 29 E. coli in this study. AMX (amoxicillin), PEN (penicillin), IMP (imipenem), GM (gentamicin), TGC (tigecycline), VAN (vancomycin).
Detection of antimicrobial resistance genes
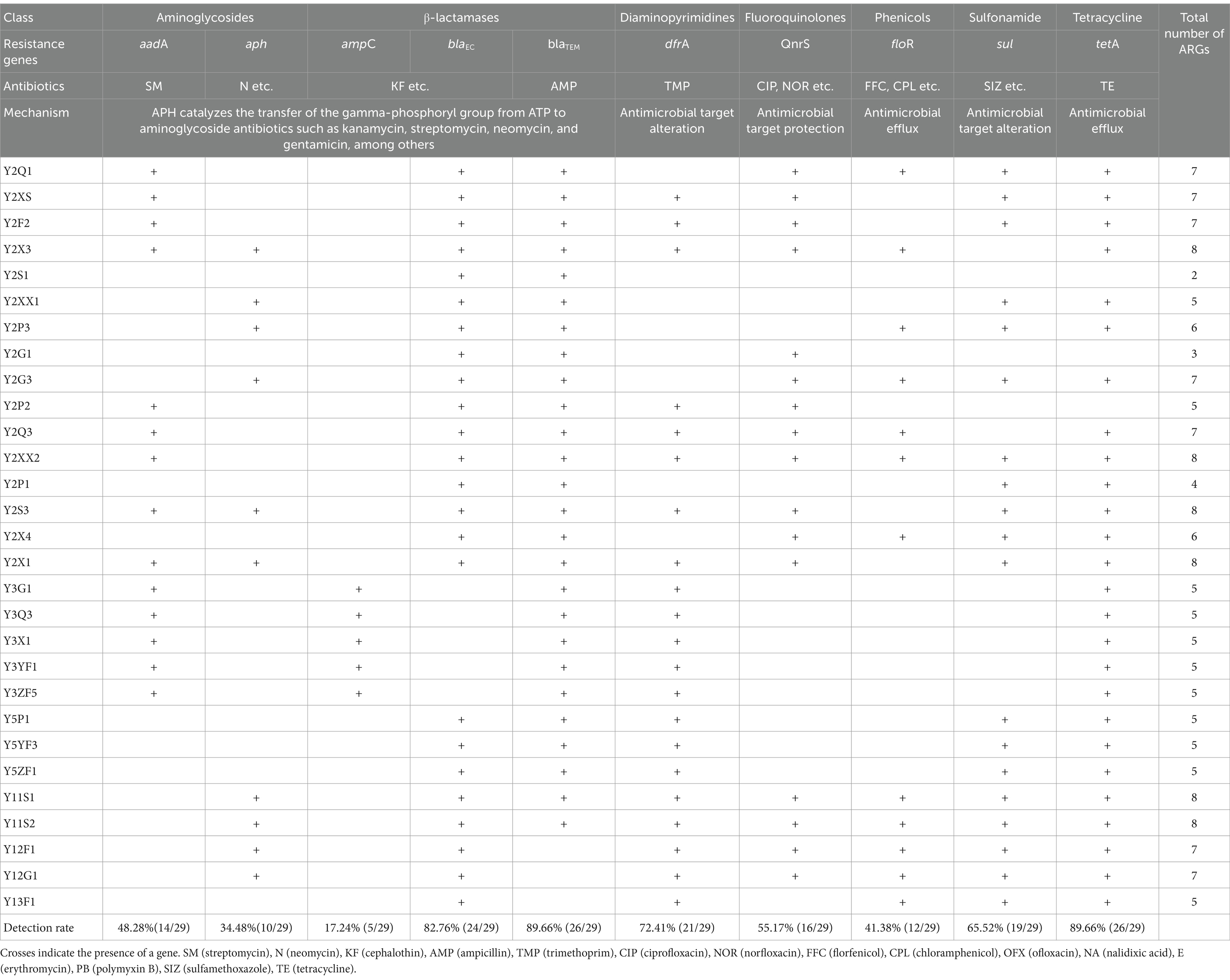
A total of ten antimicrobial resistance genes (ARGs) or their variants (such as aadA1, aadA5, and aadA8 of aadAs) were detected by PCR among the 21 isolates of E. coli (Table 3). The number of ARGs harbored by each isolate varied, ranging from 2 (observed in 1 isolate from sheep Y2) to 8 (observed in 6 isolates: 4 from sheep Y2 and 2 from sheep Y11). However, no single ARG was consistently present in all isolates. The frequencies, from high to low, were 89.7% (26/29) for blaTEM and tetA, 82.8% (24/29) for blaEC, 72.4% (21/29) for dfrA, 65.5% (19/29) for sul, 55.2% (16/29) for qnrS, 48.3% (14/29) for aadA, 41.4% (12/29) for floR, 34.5% (10/29) for aph, and 17.2% (5/29) for ampC (blaCMY). Furthermore, at least one of the three ARGs targeting β-lactamases was found in all 29 isolates.
Discussion
In recent years, an increasing number of studies have reported E. coli infections from non-intestinal organs of sick and dead sheep, raising concerns about animal health risks. Supporting these concerns, Kjelstrup et al. (2013) isolated E. coli from vital organs of lambs that died from septicemia and Singh et al. (2019) found it in the hearts and lungs of sheep and goats suffering from pneumonia or septicemia. Our study, similar to previous findings, isolated E. coli from various organs (liver, heart, kidneys, and lungs) of sheep with pneumonia. Strikingly, we found a rare case where E. coli was identified in multiple vital organs of a single sheep (sheep 1). We postulate that the systemic infection is likely to be related to the high virulency of the pathogens and low immunity of the sheep 1. Furthermore, not only sheep 1 but also other sheep in this study exhibit the phenomenon of mixed infection with multiple strains. This could be related to the fact that the sheep on this farm are introduced from inland China to Hainan Province. As they are not acclimatized to the tropical climate of Hainan Province and are highly vulnerable to attacks by local pathogens. As described in previous studies, pathogens have a very strong ability to spread across regions (Cheng et al., 2015). Coupled with the inherent pathogens carried by the sheep themselves upon introduction, it is very likely to lead to the risk of mixed infection with multiple strains for these sheep. In the field of veterinary science, understanding such factors is crucial for formulating effective prevention and control strategies.
It is noteworthy that as time goes on, the number of E. coli isolated at different times decreases in turn (Table 1). This is hypothesized to be related to two factors. On one hand, the sheep introduced from mainland China to Hainan Province become more adapted to the local climate and show increased resistance to pathogens over time. On the other hand, at the initial stage of the epidemic outbreak, the farm employed antibiotic treatment, which eliminated most of the pathogens sensitive to antibiotics.
Subsequent identification shows that three isolates belonged to ExPEC, which were isolated from the spleen (one strain) and lungs (two strains) of sheep 1 (Table 1). Genotyping and serotyping are both essential methods within current bacterial typing systems. However, the correlation between the two for bacterial classification is infrequently reported. In our study, we employed the two prevailing E. coli MLST typing methods (the Achtman scheme and Pasteur scheme) to investigate the STs. We discovered a certain correlation between STs and serotypes. However, the STs and serotypes could not be matched one by one using a single MLST scheme. Strains belonging to the same serotype could exhibit different STs. This is because strains within the same serotype group might have variations in the specific genes analyzed by the chosen MLST scheme. Since they target different conserved genes, combining both schemes can circumvent this issue (Figure 2). Further optimization of this combined MLST scheme holds promise a more suitable typing method for E. coli that would be advantageous for the prevention and management of pathogenic E. coli.
This study identified a concerning number of virulence factors (25) and various combinations (8) within the isolates (Figure 2). A number of previous studies have suggested that the diversity and combination of virulence factors may intensify the challenges of infection control (Liu et al., 2015). In this study, strains with varying combinations of virulence factors demonstrated distinct lethality and pneumonia (Figure 3), with Y13F1 the most lethal strain (Figure 3A). This may be attributed to its possession of the highest number of virulence factors, including two related to iron uptake. Studies have shown that strains carrying such virulence factors possess greater virulence and environmental adaptability (Lemos and Balado, 2020), as well as a higher propensity to cause urinary tract infections and even septicemia (Frick-Cheng et al., 2022). Interestingly, isolate Y3X1, despite carrying the lowest number of virulence factors, caused the most severe lung inflammatory response in the mouse infection model (Figure 3D). This phenomenon might be explained by its possession of specific virulence factors, Tia and Hek, which are related to invasins activity. Maciel et al. (2017) demonstrated that this virulence factor plays a crucial role in avian septicemia caused by E. coli. ExPEC is a primary pathogen of hospital-acquired infections, frequently causing septicemia and urinary tract infections and exhibiting high pathogenicity (Shulman et al., 2018). Consistent with previous findings, the ExPEC (Y5YF3) isolate also exhibited strong pathogenicity, and causing weight loss, lung inflammation, and severe melanization in the spleen (Figures 3B,C).
While the presence of drug-resistance genes does not guarantee the development of antimicrobial resistance, it can slightly increase the chances of bacteria becoming resistant. This phenomenon adds to the complexity of treatment and prevention. All isolates, with the exception of “Y2S1” and “Y2G1,” carried at least four types of ARGs (Table 3). This number is significantly higher than previous studies. Furlan et al. (2019) isolated E. coli from sheep feces primarily carried one or two types of ARGs. Moreover, Gencay (2014) isolated 24 strains of E. coli O157 from sheep having only 3 strains with 3 types of ARGs. In addition, the prevalence of certain ARGs found in our study was much higher than previously reported in sheep. For instance, the blaTEM gene, one of the most prevalent ESBLs genes in E. coli, can lead to bacterial resistance to third-generation cephalosporin antibiotics (Gundran et al., 2019). Among the 17 E. coli strains isolated from sheep by Gupta et al. (2022), the prevalence of blaTEM was 35%, which was significantly lower than our study (89.66%). TetA and tetB are the most important ARGs for tetracycline resistance in E. coli isolated from different animals (Bryan et al., 2004). In the 19 strains of E. coli isolated from sheep reported by Furlan et al. (2019), the detection rate of tetA or tetB was 36.84%, which is also much lower than our study (89.66%).
All isolates were categorized as MDR bacteria showing resistance to at least 6 out of 21 antibiotics tested, and while 75.86% (22 isolates) were resistant to over 10 antibiotics. Of note, 5 E. coli strains isolated from various organs of “sheep 2” were entirely resistant to 15 of the 21 antibiotics, intermediately resistant to 4, and only fully susceptible to polymyxin B and tetracycline (Table 2). Furthermore, all isolates were completely resistant to 6 of the 21 antibiotics, and over half of the isolates were completely resistant to 6 of the remaining 15 antibiotics (Figure 5). This finding highlights a significant increase in antimicrobial resistance compared to previous studies. In a study by Gencay (2014), only four out of 24 E. coli isolates from sheep showed intermediate resistance to cephalosporin antibiotics, with none exhibiting complete resistance to the remaining. The rate of complete resistance to antibiotics in E. coli isolated from sheep in Bangladesh by Gupta et al. (2022) did not exceed 50% for 10 tested drugs. Antimicrobial resistance occurs primarily due to misuse and overuse. Consequently, farms should be counseled to employ rational antibiotic usage to preclude abuse and misuse. Concurrently, relevant departments can provide training on “rational antibiotic use.”
Multiple previous studies have demonstrated that E. coli isolated from poultry and birds (particularly ExPEC) possess zoonotic potential (Hu et al., 2022; Xia et al., 2022; Skarżyńska et al., 2021). Based on the virulence overlap between E. coli isolated from sheep and enteropathogenic E. coli, Zhao J. Y. et al. (2021) and Zhao X. et al. (2021) suggested that E. coli derived from sheep may also have zoonotic potential. According to the Achtman scheme, all isolates were categorized into eight sequence types. Further analysis of strain information from pubMLST databases and cgMLST, excluding Y13F1, which is a common genotype in sheep, showed the strains were first reported in sheep. In previous studies, these strains were mainly found in human feces, patients in hospitals, cows (Figure 4 and Supplementary Table S2), and other animals such as (Zhang et al., 2021), pigs (Hsueh et al., 2020), and horses (Elias et al., 2019). A multi-host source of strains of the same genotype indicating interspecies transmission of virulent and multidrug-resistant isolates from sheep to humans and other farmed animals or from humans and other farmed animals to sheep is highly possible. Most importantly, sheep are a vital source of meat globally, including China. Isolating such highly virulent E. coli strains from sheep poses a serious risk to human health.
Mouse infection models have a crucial role in assessing the public health safety risks posed by highly virulent pathogens to mammals (Sarkar and Heise, 2019). Mouse infection model results reveal that strains Y13F1 and Y3YF3 cause the most severe pneumonia and highest mortality in mice. The relationship between the ability to cause pneumonia or death and the phenotypes/genotypes of these strains merits further study.
As an international free-trade port, Hainan Island’s frequent trade exchanges make it highly vulnerable to biosecurity threats from foreign pathogens. The multidrug-resistant and potentially zoonotic E. coli strains identified in this study underscore the importance of the One Health concept (Atlas, 2013). Collaborative efforts among different departments are essential to strengthen quarantine measures. This will prevent the introduction of highly virulent, multidrug-resistant, and zoonotic pathogens to Hainan Island, along with the spread of invasive species.
Conclusion
Sheep are a vital source of meat for human consumption. The occurrence of pneumonia not only jeopardizes the health of sheep flocks but also threatens the economic stability of farms. Our findings confirmed the significant role of E. coli in causing suppurative pneumonia in sheep. Diseased sheep have become a novel reservoir of highly virulent and multidrug-resistant E. coli strains with the potential to transmit across species and pose a zoonotic threat. We also characterized the diversity of sequence types, virulence factor combinations, and antimicrobial resistance profiles of E. coli within sheep breeding populations affected by pneumonia in Hainan Province. These results have significant implications for developing effective control measures to prevent the spread of this pathogen within sheep farms and potentially to humans.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1142750.
Ethics statement
The animal studies were approved by Animal Management and Ethics Committee of Hainan University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
MW: Formal analysis, Investigation, Writing – original draft. XL: Funding acquisition, Writing – review & editing. GG: Software, Writing – review & editing. MR: Conceptualization, Investigation, Writing – review & editing. XG: Data curation, Validation, Writing – review & editing. LF: Project administration, Software, Validation, Writing – review & editing. NY: Software, Visualization, Writing – review & editing. JZe: Funding acquisition, Resources, Writing – review & editing. JZh: Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (32360879 to XL, 32060131 to JZh, and 32060788 to JZe), and the Key Research and Development Program of Hainan Province (ZDYF2024SHFZ142 to JZe). Hainan University High-level Talent Scientific Research Start-up Fund (RZ2300002761 to XL), after the Key Research and Development Program of Hainan Province (ZDYF2024SHFZ142 to JZh).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1479759/full#supplementary-material
Footnotes
1. ^https://www.servicebio.cn/
2. ^http://www.eucast.org/clinical_breakpoints/
References
Aiello, S. E., and Moses, M. A. (2016). The Merck veterinary manual. 11th Edn. New Jersey, USA: Wiley.
Alley, M. R., Ionas, G., and Clarke, J. K. (1999). Chronic non-progressive pneumonia of sheep in New Zealand—a review of the role of Mycoplasma ovipneumoniae. N. Z. Vet. J. 47, 155–160. doi: 10.1080/00480169.1999.36135
Atlas, R. M. (2013). One Health: its origins and future. Curr. Top. Microbiol. Immunol. 365, 1–13. doi: 10.1007/82_2012_223
Besser, T. E., Frances Cassirer, E., Highland, M. A., Wolff, P., Justice-Allen, A., Mansfield, K., et al. (2013). Bighorn sheep pneumonia: sorting out the cause of a polymicrobial disease. Prev. Vet. Med. 108, 85–93. doi: 10.1016/j.prevetmed.2012.11.018
Bryan, A., Shapir, N., and Sadowsky, M. J. (2004). Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70, 2503–2507. doi: 10.1128/AEM.70.4.2503-2507.2004
Cheng, C., Jun, Q., Qingling, M., Zhengxiang, H., Yu, M., Xuepeng, C., et al. (2015). Serological and molecular survey of sheep infected with Mycoplasma ovipneumoniae in Xinjiang, China. Trop. Anim. Health Prod. 47, 1641–1647. doi: 10.1007/s11250-015-0908-2
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Clermont, O., Gordon, D., and Denamur, E. (2015). Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 161, 980–988. doi: 10.1099/mic.0.000063
Desvars-Larrive, A., Vogl, A. E., Puspitarani, G. A., Yang, L., Joachim, A., and Käsbohrer, A. (2024). A One Health framework for exploring zoonotic interactions demonstrated through a case study. Nat. Commun. 15:5650. doi: 10.1038/s41467-024-49967-7
Edgar, R. C. (2018). Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34, 2371–2375. doi: 10.1093/bioinformatics/bty113
Elias, L., Gillis, D. C., Gurrola-Rodriguez, T., Jeon, J. H., Lee, J. H., Kim, T. Y., et al. (2019). The occurrence and characterization of extended-spectrum-beta-lactamase-producing Escherichia coli isolated from clinical diagnostic specimens of equine origin. Animals 10:28. doi: 10.3390/ani10010028
Frick-Cheng, A. E., Sintsova, A., Smith, S. N., Pirani, A., Snitkin, E. S., and Mobley, H. L. T. (2022). Ferric citrate uptake is a virulence factor in uropathogenic Escherichia coli. mBio 13:e0103522. doi: 10.1128/mbio.01035-22
Furlan, J. P. R., Gallo, I. F. L., de Campos, A. C. L. P., Navarro, A., Kobayashi, R. K. T., Nakazato, G., et al. (2019). Characterization of non-O157 Shiga toxin-producing Escherichia coli (STEC) obtained from feces of sheep in Brazil. World J. Microbiol. Biotechnol. 35:134. doi: 10.1007/s11274-019-2712-z
Gencay, Y. E. (2014). Sheep as an important source of E. coli O157/O157:H7 in Turkey. Vet. Microbiol. 172, 590–595. doi: 10.1016/j.vetmic.2014.06.014
Gundran, R. S., Cardenio, P. A., Villanueva, M. A., Sison, F. B., Benigno, C. C., Kreausukon, K., et al. (2019). Prevalence and distribution of Bla CTX-M, Bla SHV, bla TEM genes in extended-spectrum β-lactamase-producing E. coli isolates from broiler farms in the Philippines. BMC Vet. Res. 15:227. doi: 10.1186/s12917-019-1975-9
Gupta, M. D., Sen, A., Shaha, M., Dutta, A., and Das, A. (2022). Occurrence of Shiga toxin-producing Escherichia coli carrying antimicrobial resistance genes in sheep on smallholdings in Bangladesh. Vet. Med. Sci. 8, 2616–2622. doi: 10.1002/vms3.935
Hsueh, S. C., Lai, C. C., Huang, Y. T., Liao, C. H., Chiou, M. T., Lin, C. N., et al. (2020). Molecular evidence for intra-and inter-farm spread of porcine mcr-1-carrying Escherichia coli in Taiwan. Front. Microbiol. 11:1967. doi: 10.3389/fmicb.2020.01967
Hu, J., Afayibo, D. J. A., Zhang, B., Zhu, H., Yao, L., Guo, W., et al. (2022). Characteristics, pathogenic mechanism, zoonotic potential, drug resistance, and prevention of avian pathogenic Escherichia coli (APEC). Front. Microbiol. 13:1049391. doi: 10.3389/fmicb.2022.1049391
Humphries, R., Bobenchik, A. M., Hindler, J. A., and Schuetz, A. N. (2021). Overview of changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st edition. J. Clin. Microbiol. 59:e0021321. doi: 10.1128/JCM.00213-21
Jia, Y., Mao, W., Liu, B., Zhang, S., Cao, J., and Xu, X. (2022). Study on the drug resistance and pathogenicity of Escherichia coli isolated from calf diarrhea and the distribution of virulence genes and antimicrobial resistance genes. Front. Microbiol. 13:992111. doi: 10.3389/fmicb.2022.992111
Joensen, K. G., Tetzschner, A. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Johnson, J. R., Murray, A. C., Gajewski, A., Sullivan, M., Snippes, P., Kuskowski, M. A., et al. (2003). Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47, 2161–2168. doi: 10.1128/AAC.47.7.2161-2168.2003
Kjelstrup, C. K., Arnesen, L. P., Granquist, E. G., and L'Abée-Lund, T. M. (2013). Characterization of Escherichia coli O78 from an outbreak of septicemia in lambs in Norway. Vet. Microbiol. 166, 276–280. doi: 10.1016/j.vetmic.2013.05.004
Kuraz Abebe, B. (2022). A review of the potential and constraints for crossbreeding as a basis for goat production by smallholder farmers in Ethiopia. Bull. Natl. Res. Cent. 46:80. doi: 10.1186/s42269-022-00763-7
Lemos, M. L., and Balado, M. (2020). Iron uptake mechanisms as key virulence factors in bacterial fish pathogens. J. Appl. Microbiol. 129, 104–115. doi: 10.1111/jam.14595
Liu, X., Thungrat, K., and Boothe, D. M. (2015). Multilocus sequence typing and virulence profiles in uropathogenic Escherichia coli isolated from cats in the United States. PLoS One 10:e0143335. doi: 10.1371/journal.pone.0143335
Long, N., Deng, J., Qiu, M., Zhang, Y., Wang, Y., Guo, W., et al. (2022). Inflammatory and pathological changes in Escherichia coli infected mice. Heliyon 8:e12533. doi: 10.1016/j.heliyon.2022.e12533
Maciel, J. F., Matter, L. B., Trindade, M. M., Camillo, G., Lovato, M., de Ávila, B. S., et al. (2017). Virulence factors and antimicrobial susceptibility profile of extraintestinal Escherichia coli isolated from an avian colisepticemia outbreak. Microb. Pathog. 103, 119–122. doi: 10.1016/j.micpath.2016.12.020
Maluta, R. P., Logue, C. M., Casas, M. R., Meng, T., Guastalli, E. A., Rojas, T. C., et al. (2014). Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS One 9:e105016. doi: 10.1371/journal.pone.0105016
Manges, A. R., Harel, J., Masson, L., Edens, T. J., Portt, A., Reid-Smith, R. J., et al. (2015). Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog. Dis. 12, 302–310. doi: 10.1089/fpd.2014.1860
Mekibib, B., Mikir, T., Fekadu, A., and Abebe, R. (2019). Prevalence of pneumonia in sheep and goats slaughtered at Elfora Bishoftu export abattoir, Ethiopia: a pathological investigation. J. Vet. Med. 2019, 5169040–5169010. doi: 10.1155/2019/5169040
Mellata, M. (2013). Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 10, 916–932. doi: 10.1089/fpd.2013.1533
Poolman, J. T., and Wacker, M. (2016). Extraintestinal pathogenic Escherichia coli, a common human pathogen: challenges for vaccine development and Progress in the field. J. Infect. Dis. 213, 6–13. doi: 10.1093/infdis/jiv429
Rahsan, Y., Nihat, Y., Bestami, Y., Adnan, A., and Nuran, A. (2018). Histopathological, immunohistochemical, and parasitological studies on pathogenesis of Coenurus cerebralis in sheep. J. Vet. Res. 62, 35–41. doi: 10.2478/jvetres-2018-0005
Rehman, M. N. U., Dawar, F. U., Zeng, J., Fan, L., Feng, W., Wang, M., et al. (2023). Complete genome sequence analysis of Edwardsiella tarda SC002 from hatchlings of Siamese crocodile. Front. Vet. Sci. 10:1140655. doi: 10.3389/fvets.2023.1140655
Rehman, M. N. U., Yu, W., Pan, J., Han, Y., Yang, N., Wang, X., et al. (2021). Histological and molecular characterization of Edwardsiella tarda infection in Siamese crocodile (Crocodylus siamensis) hatchlings. Aquaculture 535:736367. doi: 10.1016/j.aquaculture.2021.736367
Rodriguez-Siek, K. E., Giddings, C. W., Doetkott, C., Johnson, T. J., Fakhr, M. K., and Nolan, L. K. (2005). Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151, 2097–2110. doi: 10.1099/mic.0.27499-0
Russo, T. A., and Johnson, J. R. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5, 449–456. doi: 10.1016/s1286-4579(03)00049-2
Sarkar, S., and Heise, M. T. (2019). Mouse models as resources for studying infectious diseases. Clin. Ther. 41, 1912–1922. doi: 10.1016/j.clinthera.2019.08.010
Scott, P. R. (2011). Treatment and control of respiratory disease in sheep. Vet. Clin. North Am. Food Anim. Pract. 27, 175–186. doi: 10.1016/j.cvfa.2010.10.016
Shulman, A., Yair, Y., Biran, D., Sura, T., Otto, A., Gophna, U., et al. (2018). The Escherichia coli type III secretion system 2 has a global effect on cell surface. mBio 9, e01070–e01018. doi: 10.1128/mBio.01070-18
Singh, F., Sonawane, G. G., Kumar, J., Dixit, S. K., Meena, R. K., and Tripathi, B. N. (2019). Antimicrobial resistance and phenotypic and molecular detection of extended-spectrum ß-lactamases among extraintestinal Escherichia coli isolated from pneumonic and septicemic sheep and goats in Rajasthan, India. Turk. J. Vet. Anim. Sci. 43, 754–760. doi: 10.3906/vet-1905-1
Skarżyńska, M., Zaja, C. M., Bomba, A., Bocian, Ł., Kozdruń, W., Polak, M., et al. (2021). Antimicrobial resistance glides in the sky-free-living birds as a reservoir of resistant Escherichia coli with zoonotic potential. Front. Microbiol. 12:656223. doi: 10.3389/fmicb.2021.656223
Tivendale, K. A., Logue, C. M., Kariyawasam, S., Jordan, D., Hussein, A., Li, G., et al. (2010). Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect. Immun. 78, 3412–3419. doi: 10.1128/IAI.00347-10
Xia, F., Jiang, M., Wen, Z., Wang, Z., Wang, M., Xu, Y., et al. (2022). Complete genomic analysis of ST117 lineage extraintestinal pathogenic Escherichia coli (ExPEC) to reveal multiple genetic determinants to drive its global transmission: ST117 E. coli as an emerging multidrug-resistant foodborne ExPEC with zoonotic potential. Transbound. Emerg. Dis. 69, 3256–3273. doi: 10.1111/tbed.14678
Young, L., Sung, J., Stacey, G., and Masters, J. R. (2010). Detection of mycoplasma in cell cultures. Nat. Protoc. 5, 929–934. doi: 10.1038/nprot.2010.43
Zhang, Y., Kuang, X., Liu, J., Sun, R. Y., Li, X. P., Sun, J., et al. (2021). Identification of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli isolates from migratory birds in Guangdong, China. Front Microbiol. 12:755233. doi: 10.3389/fmicb.2021.755233
Zhao, J. Y., Du, Y. Z., Song, Y. P., Zhou, P., Chu, Y. F., and Wu, J. Y. (2021). Investigation of the prevalence of Mycoplasma Ovipneumoniae in Southern Xinjiang, China. J. Vet. Res. 65, 155–160. doi: 10.2478/jvetres-2021-0021
Zhao, L., Gao, S., Huan, H., Xu, X., Zhu, X., Yang, W., et al. (2009). Comparison of virulence factors and expression of specific genes between uropathogenic Escherichia coli and avian pathogenic E. coli in a murine urinary tract infection model and a chicken challenge model. Microbiology 155, 1634–1644. doi: 10.1099/mic.0.024869-0
Zhao, X., Lv, Y., Adam, F. E. A., Xie, Q., Wang, B., Bai, X., et al. (2021). Comparison of antimicrobial resistance, virulence genes, phylogroups, and biofilm formation of Escherichia coli isolated from intensive farming and free-range sheep. Front. Microbiol. 12:699927. doi: 10.3389/fmicb.2021.699927
Keywords: Escherichia coli , pneumonia, multidrug-resistance, highly virulent, crosshost transmission, zoonotic potential
Citation: Wang M, Li X, Guo G, Rehman MNU, Gao X, Fan L, Yang N, Zeng J and Zheng J (2024) Emergence of highly virulent and multidrug-resistant Escherichia coli in breeding sheep with pneumonia, Hainan Province, China. Front. Microbiol. 15:1479759. doi: 10.3389/fmicb.2024.1479759
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Kálmán Imre, Banat University of Agricultural Sciences and Veterinary Medicine, RomaniaRosario Morales-Espinosa, National Autonomous University of Mexico, Mexico
Copyright © 2024 Wang, Li, Guo, Rehman, Gao, Fan, Yang, Zeng and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiping Zheng, amlwaW5nLnpoZW5nQGhhaW5hbnUuZWR1LmNu
†These authors have contributed equally to this work
 Mengqi Wang1,2†
Mengqi Wang1,2† Xuesong Li
Xuesong Li Muhammad Nafees Ur Rehman
Muhammad Nafees Ur Rehman Jiping Zheng
Jiping Zheng