
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 30 October 2024
Sec. Microbiotechnology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1473396
This article is part of the Research Topic Recent Advances in Cellulosomes and Their Application in Bioenergy Production View all articles
 Christine M. Minor1,2†
Christine M. Minor1,2† Allen Takayesu1,2†
Allen Takayesu1,2† Sung Min Ha3
Sung Min Ha3 Lukasz Salwinski4
Lukasz Salwinski4 Michael R. Sawaya1
Michael R. Sawaya1 Matteo Pellegrini4
Matteo Pellegrini4 Robert T. Clubb1,2,5*
Robert T. Clubb1,2,5*Introduction: Several species of cellulolytic bacteria display cellulosomes, massive multi-cellulase containing complexes that degrade lignocellulosic plant biomass (LCB). A greater understanding of cellulosome structure and enzyme content could facilitate the development of new microbial-based methods to produce renewable chemicals and materials.
Methods: To identify novel cellulosome-displaying microbes we searched 305,693 sequenced bacterial genomes for genes encoding cellulosome proteins; dockerin-fused glycohydrolases (DocGHs) and cohesin domain containing scaffoldins.
Results and discussion: This analysis identified 33 bacterial species with the genomic capacity to produce cellulosomes, including 10 species not previously reported to produce these complexes, such as Acetivibrio mesophilus. Cellulosome-producing bacteria primarily originate from the Acetivibrio, Ruminococcus, Ruminiclostridium, and Clostridium genera. A rigorous analysis of their enzyme, scaffoldin, dockerin, and cohesin content reveals phylogenetically conserved features. Based on the presence of a high number of genes encoding both scaffoldins and dockerin-fused GHs, the cellulosomes in Acetivibrio and Ruminococcus bacteria possess complex architectures that are populated with a large number of distinct LCB degrading GH enzymes. Their complex cellulosomes are distinguishable by their mechanism of attachment to the cell wall, the structures of their primary scaffoldins, and by how they are transcriptionally regulated. In contrast, bacteria in the Ruminiclostridium and Clostridium genera produce ‘simple’ cellulosomes that are constructed from only a few types of scaffoldins that based on their distinct complement of GH enzymes are predicted to exhibit high and low cellulolytic activity, respectively. Collectively, the results of this study reveal conserved and divergent architectural features in bacterial cellulosomes that could be useful in guiding ongoing efforts to harness their cellulolytic activities for bio-based chemical and materials production.
Lignocellulosic plant biomass (LCB) is the largest source of carbon in the biosphere and a promising feedstock for the production of renewable materials, biofuels, and chemicals (Bar-On et al., 2018). LCB’s utility is limited by its recalcitrance to hydrolysis which makes it costly to degrade at an industrial scale (Chundawat et al., 2011; Liu et al., 2021). LCB consists of crystalline cellulose (32–47% dry weight) and hemicellulose (19–27%) encased within a web of cross-linked monolignols that forms lignin (5–24%; Shukla et al., 2023). Several species of highly cellulolytic anaerobic bacteria have garnered significant interest as potential tools to efficiently deconstruct LCB into its component sugars for use in bio-commodity production (Wang et al., 2024). These microbes display massive multi-cellulase containing complexes called cellulosomes that degrade LCB’s cellulose and hemicellulose components (Dassa et al., 2017; Artzi et al., 2017; Gilbert, 2007; Bayer et al., 2004; Smith and Bayer, 2013; Doi et al., 2003). A deeper understanding of the structural diversity of these complexes in bacteria could facilitate their usage in industrial applications.
Cellulosomes were first discovered in Acetivibrio thermocellus (formerly known as Clostridium thermocellum and Hungateiclostridium thermocellum) as a “cellulose-binding factor,” and subsequently have been found in a range of anaerobic eubacteria (Artzi et al., 2017; Lamed et al., 1983). These multi-cellulase complexes are constructed from scaffolding proteins (called scaffoldins) that coordinate the binding of an array of glycoside hydrolases (GH) (Dassa et al., 2017; Artzi et al., 2017; Gilbert, 2007; Bayer et al., 2004; Smith and Bayer, 2013). Each scaffoldin contains one or more cohesin domains that bind noncovalently to dockerin domains that are genetically fused to the GHs (called DocGH enzymes). In addition, scaffoldins can harbor carbohydrate binding module (CBM) domains, cell wall (e.g., S-layer homology (SLH) domains) binding modules, and dockerin domains that interact with other scaffoldins on the cell surface. For example, the ScaA scaffoldin in A. thermocellus contains multiple cohesin domains that bind DocGH enzymes, an internal CBM type-3 domain (CBM3) that binds cellulose, and a C-terminal dockerin domain that enables it to associate with a series of cell wall associated scaffoldins (ScaB, ScaC, ScaD, and ScaF) (Hong et al., 2014). Three major types of GHs function synergistically to degrade cellulose: endoglucanases, exoglucanases, and β-glucosidases (Bhardwaj et al., 2021; Sharma et al., 2016). Endoglucanases hydrolyze internal β-(1,4)-glycosidic bonds in cellulose (e.g., GH7, GH12), creating reducing and non-reducing ends that are further hydrolyzed by exoglucanases (e.g., GH5, GH9). The resulting cellodextrin carbohydrate oligomers are then degraded into glucose by β-glucosidases. The carbohydrate substrate specificities of cellulosomal DocGH enzymes vary, but members of the GH5, GH10, GH11, GH43, and GH48 families are frequently present in cellulosome producing bacteria (Artzi et al., 2017). Other types of carbohydrate active enzymes (CAZymes) are also fused to dockerin domains enabling their incorporation into cellulosomes, including polysaccharide lyases (PLs), and carbohydrate esterases (CEs). Collectively, the diversity of DocGH enzymes, CAZymes, and CBM modules within the cellulosome enable bacteria to degrade LCB more efficiently than microbes that simply secrete GHs, because enzyme colocalization by the cellulosome promotes enzyme–enzyme synergy, enzyme-proximity enhancement, and cellulose-enzyme-microbe interactions (Lu et al., 2006; Barba-Cedillo and Montanier, 2023; Smith et al., 2017).
Several species of mesophilic and thermophilic anaerobic bacteria display cellulosomes that vary in their complexity and composition. These include complex, simple, and cell-free cellulosomes that differ in both the number and types of cohesin-containing scaffoldins they possess (Dassa et al., 2017; Artzi et al., 2017; Bule et al., 2018; Bae et al., 2013). Three types of cohesin (Coh1, Coh2, and Coh3) and dockerin (Doc1, Doc2, and Doc3) domains have been identified based on their primary sequences (Bayer et al., 2004). Biochemical experiments have shown that these domains typically interact with one another in a species- and type-specific manner (e.g., Doc1 binds to Coh1 domains, but not with Coh2 or Coh3 domains within the same species; Leibovitz and Béguin, 1996; Pagès et al., 1997), however, there are several exceptions (Hamberg et al., 2014; Phitsuwan et al., 2019; Artzi et al., 2014). It has also been noted that there are two distinct conformations Doc1 modules can bind to Coh1 modules, further expanding the possible cellulosome architectures (Carvalho et al., 2007; Cameron et al., 2015). Complex cellulosomes, typified by the one present in Acetivibrio cellulolyticus, contain a primary scaffoldin that harbors several Coh1 modules for DocGH binding and a C-terminal Doc2 that enables it to interact with Coh2 modules presented in cell wall associated anchoring scaffoldins (Figure 1, right; Brás et al., 2016). In many cases, microbes containing this type of primary scaffoldin also possess adaptor scaffoldins that harbor both cohesin and dockerin domains that are believed to expand both the number and types of DocGH proteins that are incorporated into the cellulosome [e.g., Acetivibrio clariflavus (Artzi et al., 2014), Pseudobacteroides cellulosolvens (Zhivin et al., 2017), A. cellulolyticus (Dassa et al., 2012), and Acetivibrio alkalicellulosi (Phitsuwan et al., 2019)]. Other bacterial species typified by Clostridium acetobutylicum produce simple cellulosomes that contain a singular multi-cohesin primary scaffoldin that houses an N-terminal CBM3 and interspersed X2 domains (e.g., Clostridium cellulovorans, Ruminiclostridium cellulolyticum, Clostridium josui; Figure 1, left; Dassa et al., 2017). The mechanism of cell surface attachment by simple cellulosomes is poorly understood but is likely mediated by the N-terminal CBM3 present on their primary scaffoldins as seen for R. cellulolyticum (Tao et al., 2022). Lastly, many microbes possess cell-free cellulosomes composed of multi-cohesin scaffoldins bound with DocGH enzymes which are secreted into the environment to degrade LCB (e.g., A. clariflavus) (Artzi et al., 2014; Raman et al., 2009).
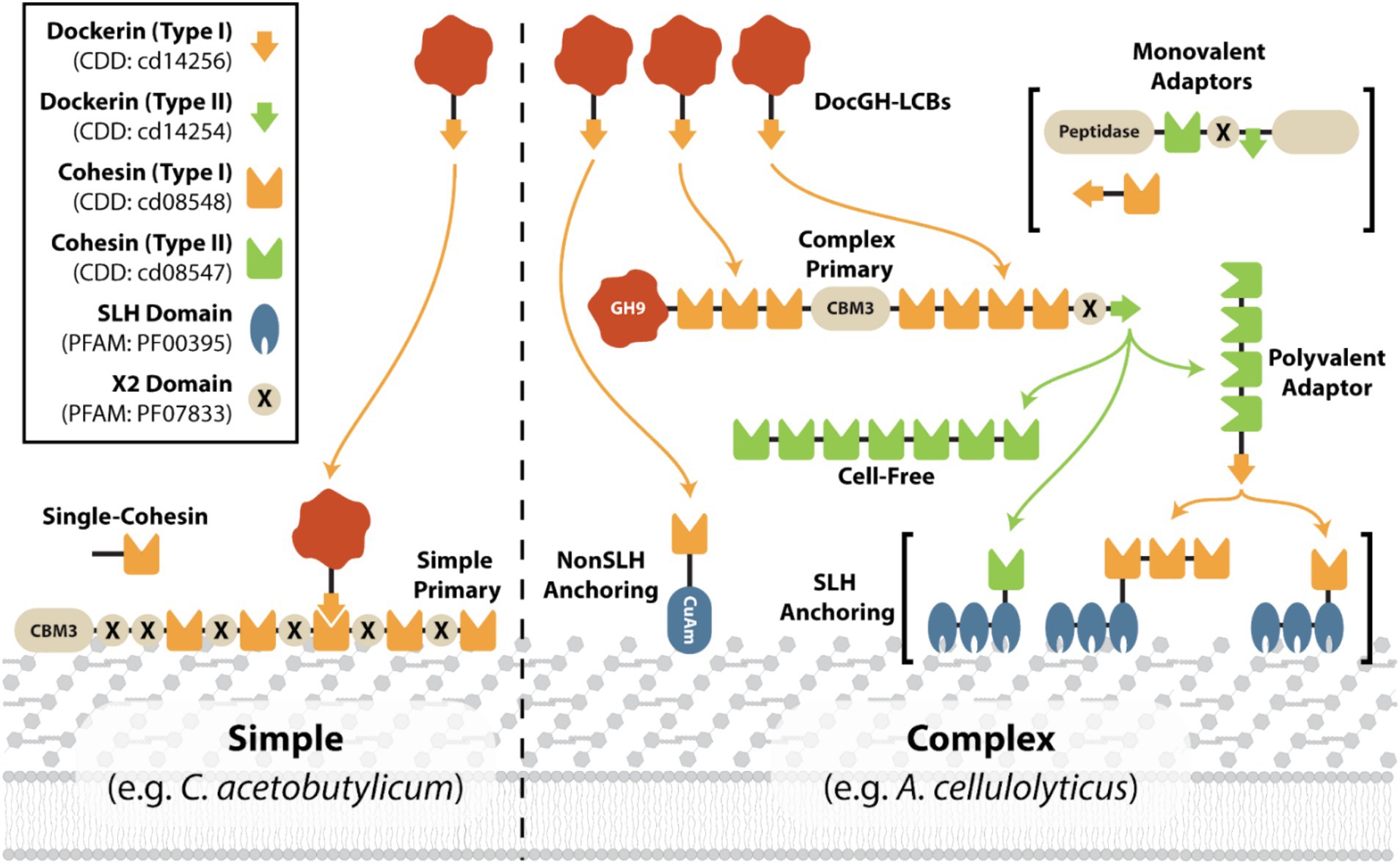
Figure 1. Representative scaffoldin compositions in simple and complex cellulosomes. Cartoon showing the different types of scaffoldins that are found in either simple (e.g., C. acetobutylicum; Nölling et al., 2001) or complex (e.g., A. cellulolyticus; Hamberg et al., 2014; Dassa et al., 2012) cellulosomes. Simple cellulosomes typically contain a simple primary scaffoldin and a small number of accessory scaffoldins. Complex cellulosomes contain a multi-cohesin complex primary scaffoldin with an associated anchoring scaffoldin, either a SLH or nonSLH-anchoring scaffoldin. Many complex cellulosomes also contain adaptor scaffoldins (polyvalent or monovalent) and cell-free scaffoldins that are presumably secreted. An example of a cohesin:DocGH interaction is shown on the left.
While a number of reviews have been written describing cellulosomes (Dassa et al., 2017; Artzi et al., 2017; Bae et al., 2013; Fontes and Gilbert, 2010), to the best of our knowledge, a systematic analysis of sequenced genomes to identify bacteria that are capable of producing these structures has not been performed. To gain a comprehensive understanding of cellulosome displaying bacteria that could have applications in LCB degradation, we analyzed >305 k complete and draft microbial genomes for genes encoding cellulosomal proteins. This analysis revealed a total of 33 bacterial species have the capacity to produce cellulosomes, including 10 species not previously reported in the literature. These microbes produce simple or complex cellulosomes that are populated with either small or large numbers of DocGH enzymes that are known to degrade LCB. The majority of cellulosome-producing bacteria are members of the Acetivibrio, Clostridium, Ruminiclostridium, and Ruminococcus genera and exhibit phylogenetically conserved properties when their scaffoldins, enzymes, and mechanisms of cellulosomal gene regulation are compared (Figure 2).
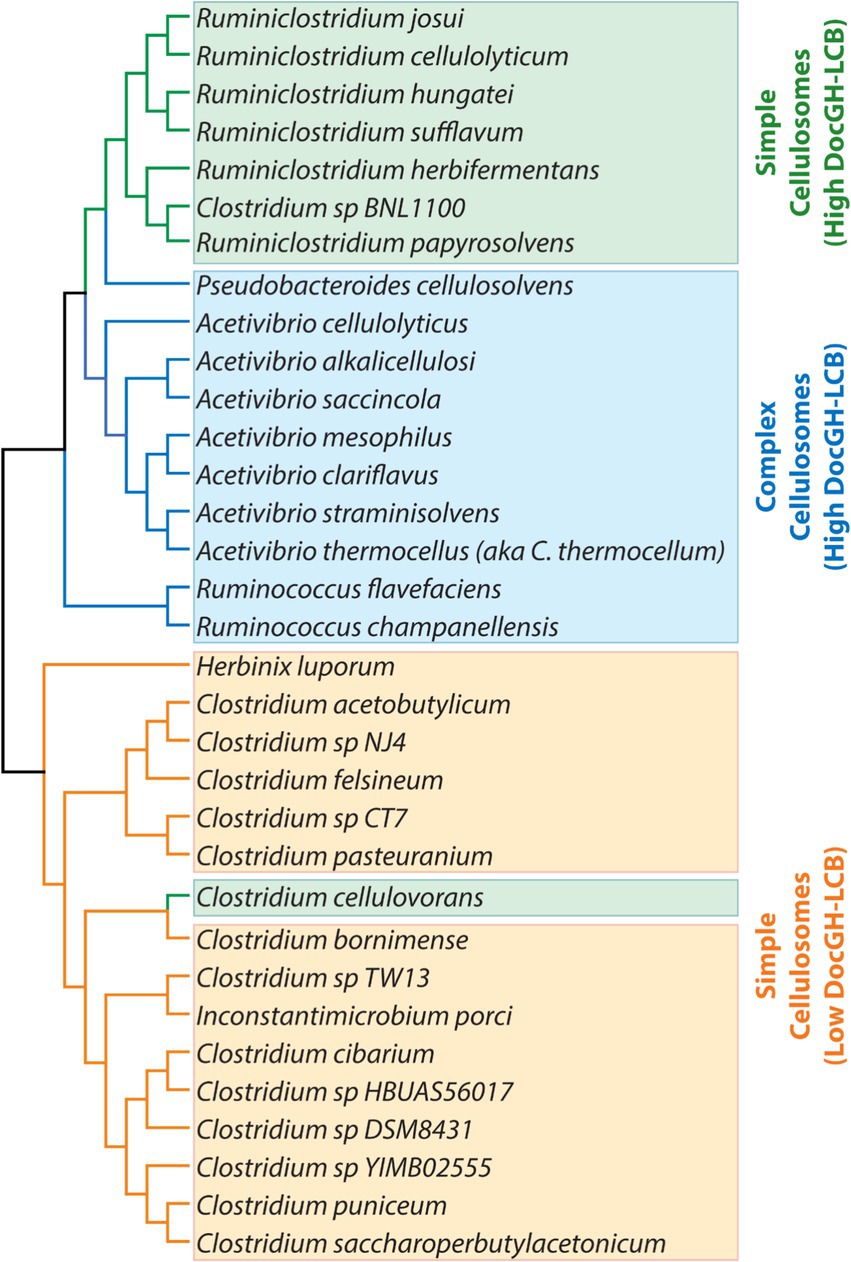
Figure 2. Taxonomic diversity of cellulosome producing bacteria. Phylogenetic tree of the bacteria listed in Tables 1, 2 (excluding the “scaffolding-containing” organisms). Bacteria are colored according to their predicted cellulosome type and number of genes encoding DocGH-LCB enzymes: Complex cellulosomes with high numbers of DocGH-LCB genes (blue); Simple cellulosome producing bacteria with high DocGH-LCB gene counts (green); Simple cellulosome producing bacteria with a low number of DocGH-LCB genes (orange). 16S rRNA sequences were aligned with ClustalW and the tree was constructed using MEGA11.
To identify bacteria that display cellulosomes we searched a total of 305,693 prokaryotic genome assemblies within the NCBI Reference Sequence Database (RefSeq), which contains both complete and draft genomes (Figure 3; Li et al., 2021). Initially, the program HMMER (Eddy, 2009) was used to search the RefSeq-annotated protein-encoding genes for dockerin, cohesin, and GH domains using hidden Markov models (HMMs). A total of 255 genomes were carried forward for further analysis as they harbored at least one gene encoding a multi-cohesin containing scaffoldin (≥ 2 cohesin domains) and one gene encoding a DocGH protein. For cases where multiple genome sequences were available for the same bacterial species, only the representative genome within the ProGenomes database that contained the fewest number of contigs was analyzed (Mende et al., 2017). An exception was made for the three sequenced genomes from A. thermocellus (ATCC 27405, DSM 1313, and AD2), as it is a prototypical cellulosome-producing organism. After eliminating redundancies, a total of 139 distinct microbial genomes were retained for a more extensive and computationally demanding analysis using the program InterProScan (v. 5.59-91.0) (Quevillon et al., 2005). The analysis by InterProScan revealed a total of 37 bacterial species containing genes encoding putative cellulosomes (their genomes have at least one multi-cohesin and one DocGH encoding gene). Of these, 33 species likely produce conventional cellulosomes that are related to those in C. acetobutylicum and A. cellulolyticus (Figure 1), whereas 4 species may produce non-conventional cellulosomes (described below).
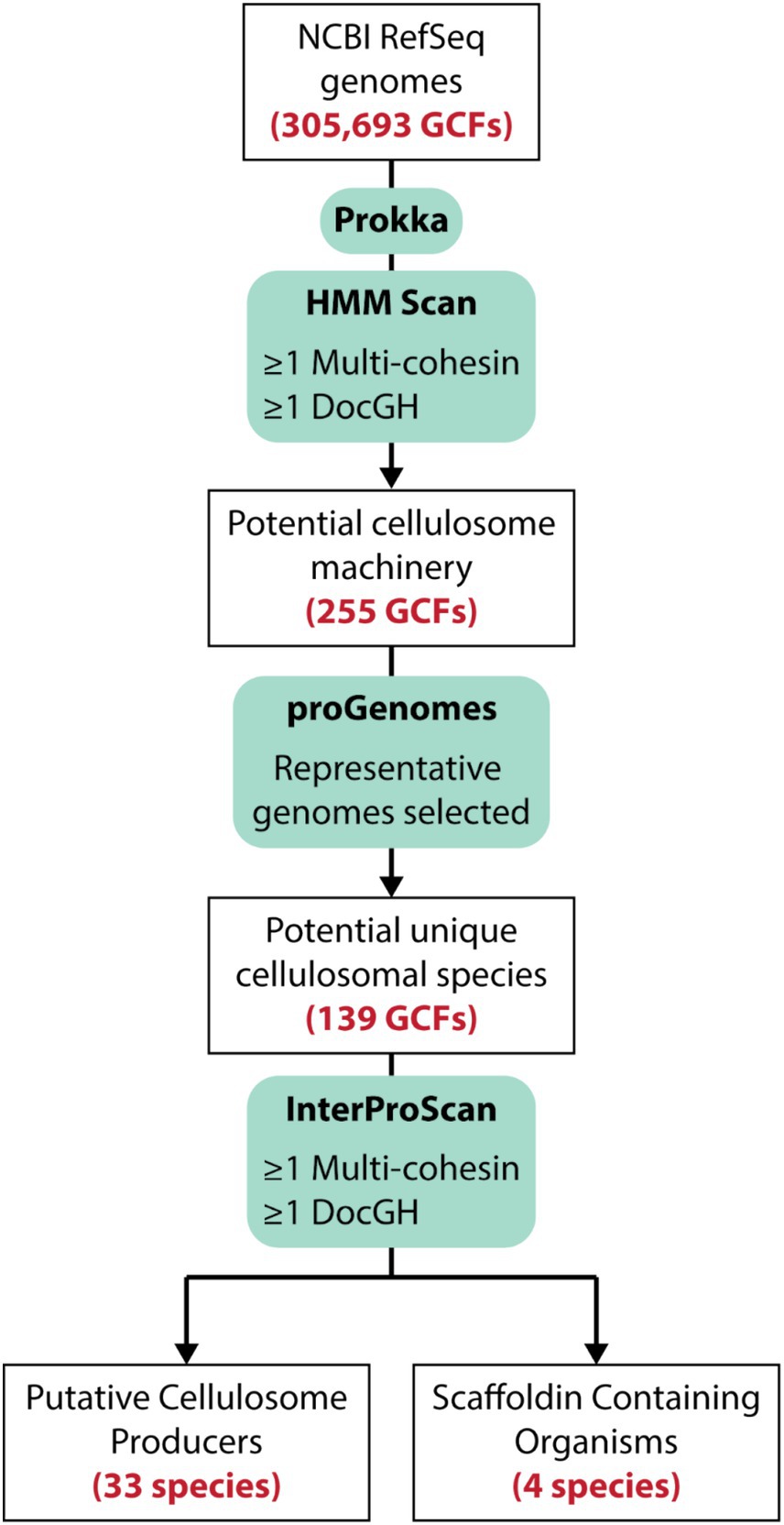
Figure 3. Strategy used to identify cellulosome producing microbes. More than 305 k sequenced bacterial genomes within the NCBI Reference Sequence database (RefSeq). Translated protein-encoding genes were then initially searched using HMM profiles to identify genomes that contained cohesin and DocGH enzymes. A subset of those genomes were then submitted for de novo annotation via Prokka (v. 1.14.16; Seemann, 2014) followed by a more stringent analysis of these protein sequences using InterProScan (v. 5.59-91.0; Quevillon et al., 2005) which identified 33 species that likely produce conventional cellulosomes based on the types of cohesin-containing proteins they possess. A total of 4 species contain multi-cohesin scaffoldins that are unlikely to assemble into conventional cellulosomes (Scaffoldin Containing Organisms).
To gain insight into the structure and composition of each bacterium’s cellulosome, we systematically classified their scaffoldins into eight categories based on their domain content (Figure 4). The eight scaffoldin types and their functions in cellulosome assembly are demonstrated for C. acetobutylicum and A. cellulolyticus, which produce simple and complex cellulosomes, respectively (Figure 1; Dassa et al., 2012; Sabathé et al., 2002). These types include: (1) “simple primary” scaffoldins that are produced by C. acetobutylicum and other mesophiles which contain an N-terminal CBM3 and multiple cohesin domains (either Coh1 or Coh2) that are often interspersed with X2 domains (Artzi et al., 2017), (2) “single-cohesin” containing proteins of unknown function, (3) “complex primary” scaffoldins found in A. cellulolyticus and related species that harbor multiple Coh1 modules, an internal CBM3, and a single C-terminal Doc2 domain that enables it to bind to cell wall anchoring scaffoldins (Dassa et al., 2012), (4) “SLH-anchoring” scaffoldins that contain at least one cohesin module paired with a SLH-domain, (5) “nonSLH-anchoring” scaffoldins that contain at least one cohesin module and a known cell wall interacting domain/motif (e.g., LPxTG sorting signal, Lysin motif, C-terminal TM-helix, or Cu-Amine Oxidase-like domains), (6) “Monovalent adaptor” scaffoldins that contain a dockerin and a single cohesin module which have been proposed to facilitate type-switching between different types of cohesins and dockerin-fused enzymes (Artzi et al., 2017), (7) “Polyvalent adaptor” scaffoldins that contain a dockerin and several cohesins that suggest that more elaborate cellulosome architectures can be constructed by increasing the number of binding sites for DocGH enzyme proteins and/or scaffoldins (Artzi et al., 2017), and (8) “cell-free” scaffoldins that are presumably secreted to degrade LCB as they contain several cohesin domains that are capable of binding to DocGH enzymes (Raman et al., 2009). A list of the domain and motif identifiers used to discover cellulosome components within the cellulosome is provided in Supplementary Tables S1, S5.
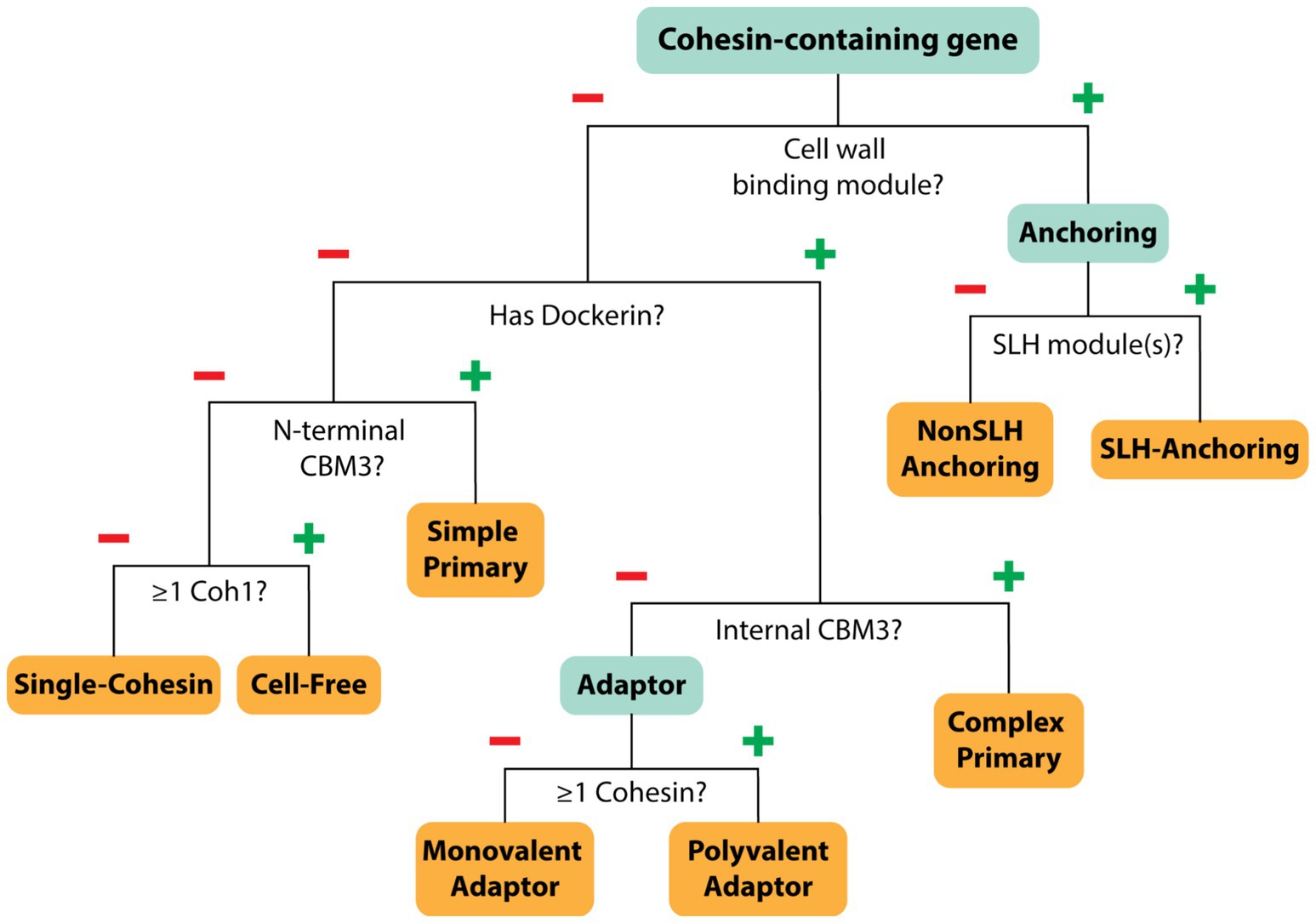
Figure 4. Scaffoldin categorization. Decision tree showing how each cohesin-containing protein was classified. Each cohesin-containing protein was classified into one of eight categories based on its type and abundance (Coh1, Coh2, Coh3), as well as the presence of additional domains such as dockerin, CBM3, and cell wall binding domains.
Next, we closely examined the enzyme composition of each bacterial species. The Carbohydrate-Active Enzyme database (CAZy) is a well-curated resource that classifies glycoside hydrolases (GHs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), and glycosyl transferases (GTs) into families based on experimentally published data and publicly available sequences (Cantarel et al., 2009). GHs, PLs, and CEs are of particular interest in cellulosome-producing bacteria since their activities are directly linked to polymer breakdown. These enzymes are listed in Supplementary Table S1 and collectively referred to as CAZymes. Many of these CAZymes are often fused to a dockerin domain to facilitate their incorporation into cellulosomes, while enzymes not fused to dockerins are presumably secreted by the bacterium. Using the criteria outlined in Supplementary Table S1 we identified dockerin proteins fused GHs with activity against cellulose (DocGH-Cell), hemicellulose (DocGH-Hemi), and oligosaccharides (DocGH-Oligo), as well their corresponding non-dockerin-fused GHs (referred to as FreeGH-Cell, FreeGH-Hemi, and FreeGH-Oligo, respectively) (Supplementary Table S4). DocGH-Cell and DocGH-Hemi enzymes are of particular interest as they contribute significantly to cellulolytic activity against LCB and are distinguished as DocGH-LCB enzymes (DocGH-Cell plus DocGH-Hemi). A similar category was defined for non-dockerin-fused GHs with LCB activity (FreeGH-LCB). Based on their DocGH-LCB count there are two broad types of organisms that display cellulosomes, “high DocGH-LCB” microbes that contain genes encoding a large number of these enzymes (22 to 70) and “low DocGH-LCB” microbes that have fewer DocGH-LCBs (1 to 10) (Figure 5).
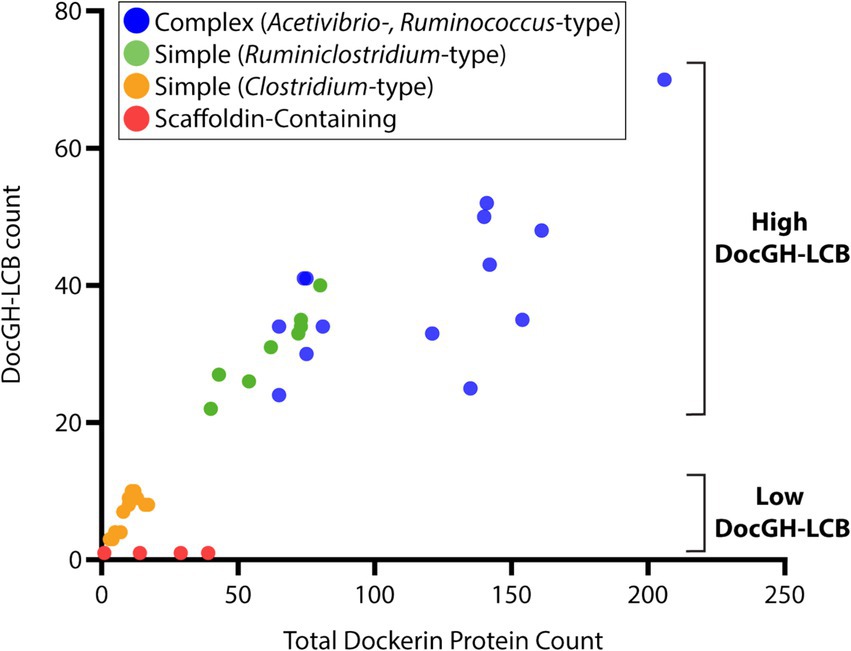
Figure 5. Dockerin-protein and DocGH-LCB distribution. Correlation between total dockerin protein count and DocGH-LCB protein count (R2 = 0.81, p < 0.05). Reported here are the number of genes encoding dockerin-fusion and DocGH-LCB proteins for all microbes listed in Tables 1, 2. Classes of microbes that are shown are indicated in the key and correspond to: Complex Acetivibrio and Ruminococcus cellulosomes (blue), Simple Ruminiclostridium cellulosomes (green), Simple Clostridium cellulosomes (orange), and bacteria containing scaffoldin encoding genes that are unlikely to produce conventional cellulosomes (red).
Our genomic screen detected all multi-cohesin cellulosome bacteria previously documented to produce cellulosomes, as well as several new species (Tables 1, 2). However, we detected fewer cohesin domain-containing proteins than previously reported in the literature for three genomes from two bacterial species, R. champanellensis (18P13) and R. flavefaciens (strains 17, 007c) (Ben David et al., 2015; Dassa et al., 2014). The genome for R. flavefaciens strain FD-1 previously described in the literature as containing a cellulosome was not analyzed by us because its sequenced genome is suppressed in the RefSeq database and because this species is already represented by several sequenced genomes (Dassa et al., 2014). For the remaining three genomes, there are two main reasons why their cohesin modules were undercounted. First, each genome contains a large number of contigs that lead to sequencing truncations in their cohesin-containing scaffoldin genes (Supplementary Table S2 lists the sequencing statistics for the genomes analyzed in this study). These truncations led to nine abbreviated cohesin-containing genes in R. flavefaciens strain 17, two in R. flavefaciens strain 007c, and four in R. champanellensis, lowering the number of detectable cohesins. For example, sequencing truncations in R. flavefaciens strain 17 occur in several of its scaffoldin genes (ScaB, ScaE, ScaG, ScaI, and orf02408) that previously were identified by sequencing a single contig from this microbe (Ding et al., 2001; Rincon et al., 2003; Rincón et al., 2004).
A second reason for the cohesin undercount is that the primary sequences of these modules in Ruminococcus species are highly divergent and thus not detectable using the HMM profiles employed by InterProScan (Coh1: cd08548, Coh2: cd08547, Coh3: cd08759, Coh: PF00963). For example, the protein encoded by the scaA gene in R. flavefaciens strain 007c has been reported to contain four cohesins and a C-terminal dockerin domain when its primary sequence was searched using BLAST and an unknown query sequence (Dassa et al., 2014). Instead, three of the modules are annotated as members of the CBM2/3 superfamily (SSF49384) and the fourth is not assigned to a protein family (Supplementary Table S3). Furthermore, in other Ruminococcus scaffoldins, previously reported cohesins are identified by InterProScan as G3DSA:2.60.40.680 superfamily members (CATH-3D) and not cohesins (Dawson et al., 2017). To investigate this issue we used AlphaFold2 to predict the atomic structures of scaffoldins within the R. champanellensis and R. flavefaciens genomes that contained cohesins that could not be identified by InterProScan. The predicted structures of these ‘undetected’ domains were compared to experimentally determined cohesin structures [Coh1 (PDB:1OHZ), Coh2 (PDB:2BM3) and Coh3 (PDB:2ZF9) cohesins] and their similarity determined by calculating a template modeling (TM) score (Dong et al., 2018). In all organisms, AlphaFold2-based predictions identified additional cohesin domains within scaffoldins that could not be detected by InterProScan (Supplementary Table S3). Given that we undercounted the cohesin domains in the three Ruminococcus species above, we extended the structural analysis to three additional R. flavefaciens strains (AE3010, SAb67, YL228) that were identified through our analysis. There is no literature reported cohesin domains for these strains. In R. flavefaciens strain AE3010, only three cohesin containing proteins (7 cohesin domains) were detected using InterProScan but based on AlphaFold2 a total of 18 domains (across 11 proteins) with cohesin folds are detected (summarized in Supplementary Table S3 and Supplementary Figure S2). Collectively, these results suggest that more complete genomic sequencing of Ruminococcus genomes is needed to fully define their scaffoldin complement and it highlights the utility of employing structure-based approaches to identify their cohesins.
Harnessing the potent cellulolytic activity of cellulosome producing microbes could lead to improved methods to convert abundant LCB into renewable chemicals and materials. To gain insight into their structures and distribution in nature, we searched 305,693 prokaryotic genomes for genes that encode cellulosome components—at least one multi-cohesin containing protein and one DocGH enzyme. This analysis identified 33 bacterial species that likely produce conventional cellulosomes that resemble those present in C. acetobutylicum and A. cellulolyticus, as well as 4 species that produce scaffoldin-containing structures that could bind DocGH enzymes and other dockerin-fusion proteins. The cellulosomes within the 33 species can be classified as having either complex or simple structures following the convention established by Bayer and colleagues (Figure 1; Tables 1 and 2) (Artzi et al., 2017). With only two exceptions, these microbes originate from four genera of gram-positive bacteria: Acetivibrio, Ruminococcus, Ruminiclostridium, and Clostridium (Figure 2). Based on their predicted complement of scaffoldin proteins, 10 species produce complex cellulosomes that are related to the one in A. cellulolyticus, while the remaining 23 species may produce less complex (simple) structures that resemble C. acetobutylicum’s cellulosome (Figure 1). We expect the cellulosomes in these bacteria to exhibit varying levels of cellulolytic activity based on their DocGH-LCB profiles (Figure 5; Supplementary Table S4). As documented in Table 1, all bacteria with the genetic capacity to produce complex cellulosomes also contain a large number of genes that encode DocGH-LCB proteins (called “high DocGH-LCB” microbes) and cellulose-binding CBM modules suggesting that they are cellulolytic. In contrast, the genomes of bacteria that display simple cellulosomes can either have high or low numbers of DocGH-LCB encoding genes, implying differences in their cellulolytic activities (Table 2). To the best of our knowledge, over a quarter of the cellulosome producing species discovered in this search (10 total) have not been previously described in the literature. Notably, although R. cellobioparum (subsp. termitidis; Dassa et al., 2017) and R. bromii (Ze et al., 2015) have previously been reported to produce cellulosomes, the sequenced genomes for these microbes lack genes for a multi-cohesin containing protein and therefore were not classified by us as containing a bona fide cellulosome. Below we summarize these findings.
A number of bacterial species have been shown to display multi-scaffoldin containing complexes that have been referred to as “complex”/ “highly-structured” cellulosomes (Dassa et al., 2017; Artzi et al., 2017). Here we broadly define a complex cellulosome as containing at least two scaffoldins, a cell wall associated anchoring scaffoldin that contains a cohesin domain that could potentially bind to a dockerin domain located within a second multi-cohesin containing scaffoldin (Figure 1). Using this definition, complex cellulosomes contain either a complex primary scaffoldin (multi-cohesin, dockerin domain, and internal CBM3 domain containing) or a polyvalent adaptor (multi-cohesin and dockerin domain containing) that has the potential to bind via cohesin-dockerin interactions to the cell surface by interacting with an anchoring scaffoldin (either a SHL- or nonSLH-anchoring scaffoldin) (Figure 1). Based on this definition several Acetivibrio and Ruminococcus bacterial species produce complex cellulosomes (Table 1). Acetivibrio bacteria produce “classical” complex cellulosomes related the prototypical cellulosome from A. thermocellus in which DocGH enzymes bind to a complex primary scaffoldin that is tethered to the cell-surface via cohesin-dockerin interactions with a SLH-anchoring scaffoldin (Figure 1), whereas in Ruminococcus bacteria the enzymes bind to a polyvalent adaptor scaffoldin that form dockerin-cohesin interactions with a nonSLH-anchoring protein.
Related complex cellulosomes are produced by 7 species of Acetivibrio bacteria [A. alkalicellulosi (Phitsuwan et al., 2019), A. cellulolyticus (Hamberg et al., 2014), A. clariflavus (Artzi et al., 2015), A. mesophilus (Rettenmaier et al., 2019), A. saccincola (Aikawa et al., 2018), A. straminisolvens (Kato et al., 2004), A. thermocellus (Lamed et al., 1983), and P. cellulosolvens (Zhivin et al., 2017)]. In the Acetivibrio bacteria, their complex primary scaffoldins contain multiple Coh1 domains that bind to DocGH enzymes through Doc1-Coh1 interactions and to the cell-surface through Doc2-Coh2 interactions with an SLH anchoring scaffoldin (Xu et al., 2004). P. cellulosolvens is the single exception, as it originates from the Pseudobacteroides genus and the roles of the cohesin-dockerin interactions are reversed (i.e., it uses Doc2/Coh2 and Doc1/Coh1 interactions to mediate DocGH and scaffoldin-scaffoldin binding, respectively) (Xu et al., 2004). In all of these bacteria the primary complex scaffoldin follow a similar domain arrangement, it contains multiple cohesins, a C-terminal dockerin module, and an internal CBM3 module that presumably enables each microbe to adhere to cellulose. These bacteria also possess Coh2-containing SLH-anchoring scaffoldins that bind to the bacterium’s peptidoglycan to coordinate the binding of the complex primary scaffoldin via its Doc2 domain. The lone exception is A. straminisolvens, which lacks a primary complex scaffoldin, but nevertheless contains a large number of other types of scaffoldins, including two SLH-anchoring scaffoldins. All of these bacteria are “high DocGH-LCB” producers and are characterized by the presence of a large number of accessory scaffoldins that increase the number of enzymes that can be incorporated into the cellulosome (Table 1). These accessory scaffoldins include monovalent adaptors, polyvalent adaptors, SLH anchoring, nonSLH anchoring, and cell-free scaffoldins (Figure 1). Both types of adaptor scaffoldins act to increase the size and complexity of the cellulosomes by extending the existing structure and allowing for type-switching within type-specific cohesin-dockerin interactions (Artzi et al., 2017). Aside from traditional SLH anchoring proteins, these bacteria have cohesin-containing nonSLH-anchoring scaffoldins that contain Cu-Amine Oxidase-like domains associated with secondary cell wall polymers (Dassa et al., 2012). The nonSLH-anchoring scaffoldins contain either Coh1 or Coh2 modules, suggesting they, respectively, facilitate either individual DocGH enzyme or complex primary scaffoldin binding to the cell surface. While not scaffoldins per se, Acetivibrio complex cellulosome producers also contain a large number of genes encoding CBM3-SLH fusion proteins that may function to tether the microbe to cellulose (Wang et al., 2022). The genomes of these microbes also contain a large number of genes encoding LCB active GHs that are fused to cellulose-binding CBM modules (Table 1, CBM(cell)-GH-LCB) as compared to other types of cellulosome producers. The cellulosomes in Acetivibrio bacteria are also unique because unlike other microbes, they uniformly produce multi-cohesin-containing cell-free scaffoldins that presumably form higher-order structures containing DocGH-LCB enzymes that are secreted into the environment to degrade LCB (Artzi et al., 2014; Chow and Wu, 2017).
P. cellulosolvens produces a complex cellulosome that is most closely related to those found in Acetivibrio species, as it contains SLH-anchoring scaffoldins that coordinate the binding of a complex primary scaffoldin bearing a C-terminal Doc1. Moreover, as compared to Acetivibrio and Ruminococcus bacteria, it is phylogenetically more closely related to Acetivibrio bacteria based on its 16S rRNA sequence (Figure 2). It stands out as producing the most complex cellulosome, since its genome encodes genes for an astounding 79 cohesin domains that are distributed between 33 scaffoldins: three primary, 10 monovalent adaptor, three polyvalent adaptor, eight SLH-anchoring, three nonSLH-anchoring, three cell-free scaffoldins, and three single cohesin domain containing proteins. P. cellulosolvens is also unique because it has genes encoding a complex primary scaffoldin and two primary scaffoldins that are typically found in simple cellulosomes within Ruminiclostridium and Clostridium bacteria (previously referred to as ScaM1 and ScaM2 in P. cellulosolvens; Zhivin et al., 2017). As noted previously, the usage of the cohesin and dockerin domains in P. cellulosolvens is reversed as compared to other bacteria in the Acetivibrio category.
Another conserved feature in Acetivibrio bacteria and P. cellulosolvens is the manner in which they control the expression of the DocGH enzymes and scaffoldin proteins to construct their cellulosome. Microbes have been shown to alter the complement of their DocGH enzymes when different types of LCB substrates are encountered (Artzi et al., 2015; Blouzard et al., 2010) by regulating gene expression using either two-component systems (Celik et al., 2013; Kampik et al., 2020), selective RNA transcript stabilization (Bhaskar et al., 2021; Xu et al., 2015), or transmembrane biomass-sensing RsgI-type anti-σ factors that regulate σI-factors (Nataf et al., 2010; Muñoz-Gutiérrez et al., 2016; Kahel-Raifer et al., 2010). Our analysis suggests that in P. cellulosolvens and all Acetivibrio complex cellulosome producing bacteria, RsgI-type anti-σ factors are used to dictate cellulosomal gene expression, as each species contains genes encoding 7 to 14 of these factors (Table 1).
Previous studies have shown that Ruminococcus flavefaciens and Ruminococcus champanellensis produce cellulosomes, which like their Acetivibrio counterparts contain an array of primary, cell wall anchoring, and adaptor scaffoldins (Artzi et al., 2017). Prior studies highlighted several unique features in Ruminococcus cellulosomes. First, cell surface attachment in Ruminococcus cellulosomes is often mediated by an anchoring scaffoldin that is covalently linked to the cell wall by a sortase enzyme instead of by a SLH-anchoring scaffoldin as observed in Acetivibrio species (Dassa et al., 2014; Rincon et al., 2007). Second, the primary scaffoldins in Ruminococcus species lacks a characteristic internal CBM3 that can mediate direct attachment to cellulosic substrates. Third, unlike Acetivibrio bacteria, these species contain a unique monovalent adaptor scaffoldin called ScaC which is often used as the genomic signature to identify Ruminococcus cellulosomes (Rincón et al., 2004; Jindou et al., 2008). Lastly, it has been noted in the literature that the scaffoldins in the Ruminococcus cellulosomes contain dockerin and cohesin modules with divergent primary sequences (frequently Doc3-and Coh3-types; Ding et al., 2001; Salama-Alber et al., 2012).
The scaffoldin proteins were identified in R. champanellensis 18P13 and R. flavefaciens (strains 17 and 007c) before their genomes were sequenced (Dassa et al., 2014). Surprisingly, we identified fewer cohesin domains and scaffoldin proteins encoded in these genomes than previously reported. In many cases this occurred because their sequenced genomes contain a large number of sequence contigs that caused scaffoldin gene truncations (Supplementary Table S2). In other instances, even if an intact, non-truncated scaffoldin encoding gene was present, it was not possible to detect the full complement of cohesins within the translated protein product using the sequence profiles employed by InterProScan (Coh1: cd08548, Coh2: cd08547, Coh3: cd08759, Coh: PF00963). Indeed, only when AlphaFold2 was used to predict the atomic structures of these proteins was the full complement of previously reported cohesin modules identified. For example, an InterProScan analysis of translated genes in R. flavefaciens (strain 007c) identified a single scaffoldin, whereas 10 scaffoldins have been reported in the literature (Dassa et al., 2014). However, when AlphaFold2 was employed, 9 of these 10 scaffoldins were detected. Similar results were obtained when AlphaFold2 was applied to other ruminococcal genomes documented to contain genes for cellulosomes (summarized in Supplementary Table S3). Interestingly, even though Coh3 domains are a signature feature of ruminococcal cellulosomes, InterProScan did not identify these domains in R. champanellensis 18P13 or R. flavefaciens (using the cd14255 profile for a Coh3 domain). We conclude that the sequence signatures employed by InterProScan are not sufficiently robust to identify Coh3 cohesins, consistent with the findings reported by Flint and colleagues who have subdivided R. flavefaciens’ cohesins into 6 different groups based on sequence homology (Ding et al., 2001). Our results also demonstrate the utility of using AlphaFold2 structure predictions to identify cohesins with divergent primary sequences.
Our analysis identified a previously unrecognized bacterium as a “classical” complex cellulosome producer, Acetivibrio mesophilus N2K1 (formerly known as Hungateiclostridium mesophilum) (Tindall, 2019). This gram-positive anaerobic bacterium was first isolated from a mesophile consortium in a biogas fermenter fed with maize silage (Rettenmaier et al., 2019). It has an optimal growth temperature of 45°C and expresses two hemicellulases that have been biochemically characterized, but the presence of a cellulosome has not been reported to the best of our knowledge (Liu et al., 2021). Its cellulosome is likely cellulolytic because its genome contains 40 genes encoding DocGH-LCBs. Based on our analysis, A. mesophilus’ cellulosome is strikingly similar to the archetypal cellulosome produced by A. thermocellus (Figure 6). Specifically, both species produce a “classical” complex primary scaffoldin (ScaA-like: WP_128706406.1) that contains nine Coh1 modules, an internal CBM3, and a C-terminal Doc2 module for cell surface attachment via interactions with a SLH-anchoring scaffoldin (ScaF-like: WP_128705811.1). A. mesophilus has an additional three genes for SLH-anchoring scaffoldins (ScaB-like: WP_235832675.1, ScaC-like: PROKKA_02165, and ScaD-like: WP_069196093.1) that are related to A. thermocellus’ ScaB, ScaC, and ScaD scaffoldins, as well as one nonSLH-anchoring scaffoldin (ScaG-like: WP_235832552.1) that is similar to ScaG. A. mesophilus also produces an additional scaffoldin that closely resembles scaffoldins found in C. alkalicellulosi (ScaO2; Phitsuwan et al., 2019) and A. cellulolyticus (ScaO; Dassa et al., 2012). This scaffoldin (WP_128706311.1) contains a C-terminal Coh1-Doc1 bi-domain unit, three Fibronectin-type III (FN3) repeats, as well as S8-peptidase-like and galactose oxidase-like domains of unknown function. Given the presence of a C-terminal Doc1 domain in this scaffoldin, it is tempting to speculate that it binds to the primary scaffoldin and/or the ScaD-like and ScaG-like scaffoldins that contain complementary Coh1 modules.
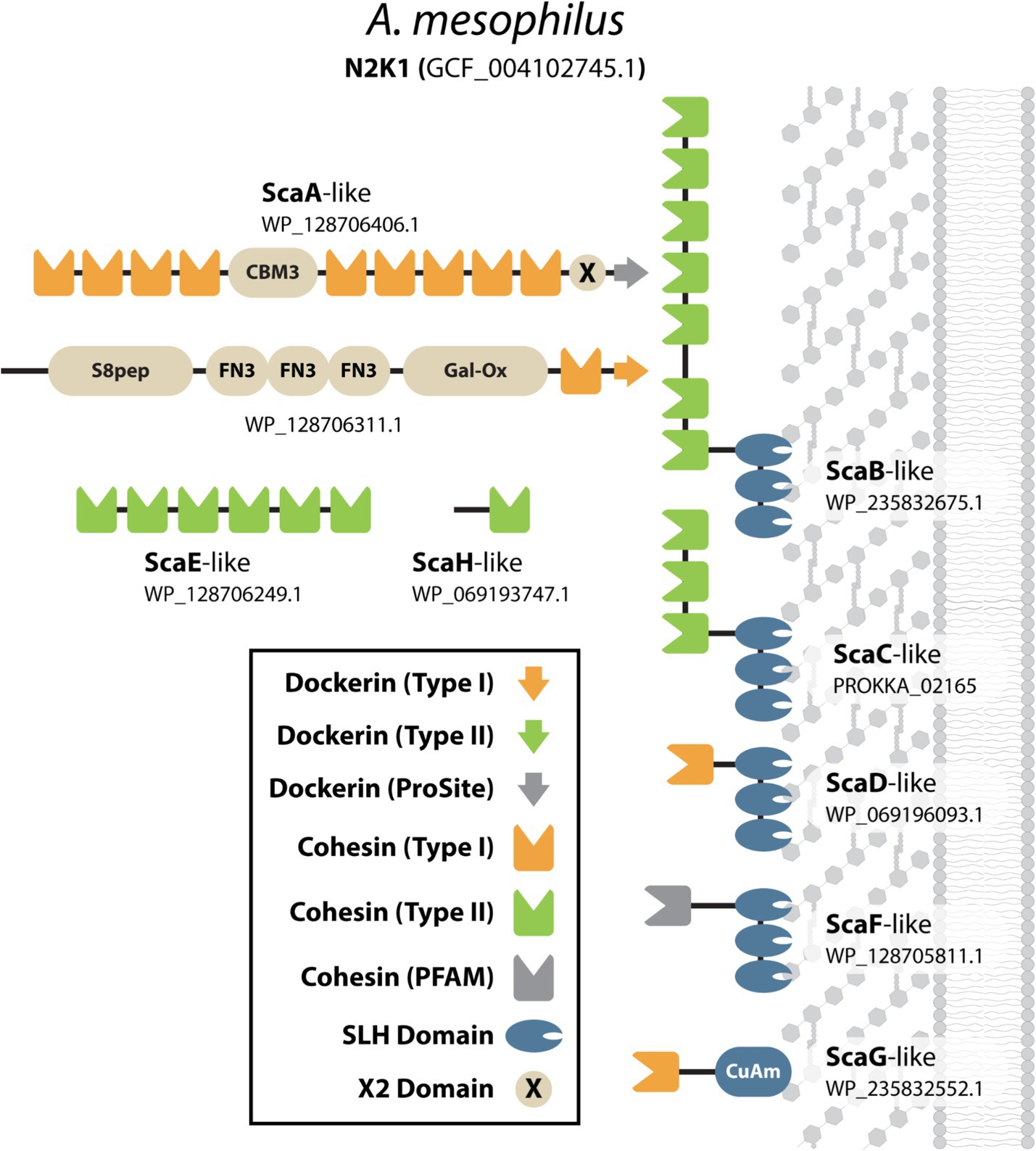
Figure 6. Scaffoldin proteins in A. mesophilus (N2K1). Cartoon representation of all of the cohesin-containing proteins in A. mesophilus (N2K1). The domains they possess are defined in the key and also include; Fibronectin Type-III like (FN3), CBM type-3 domain (CBM3), Copper Amine Oxidase-Like (CuAm), S8-subtilase/peptidase (S8pep), and Galactose Oxidase-like (Gal-Ox) domains. Proteins were named based on their sequence homology to scaffoldins present in A. thermocellus.
We identified 23 species of anaerobic mesophilic bacteria that display less complex cellulosomes and fewer types of scaffoldins. The cellulosomes in these microbes always contain a simple primary scaffoldin that houses several Coh1 domains and an N-terminal CBM3 module (Figure 1). In all cases the gene encoding the primary scaffoldin protein (cipA) is located within a cipA operon that also contains genes for DocGH enzymes. The vast majority of DocGHs in these microbes contain complementary Doc1 modules for direct interaction with the simple primary scaffoldin. The lone exception is Clostridium sp. HBUAS56017, whose simple primary scaffoldin lacks a CBM.
Simple cellulosome producing bacteria originate from the Ruminiclostridium and Clostridium genera (Figure 2). The Ruminiclostridium bacteria produce simple cellulosomes that likely have high cellulolytic activity as their genomes contain a large number of DocGH-LCB genes [R. hungatei (Monserrate et al., 2001), R. papyrosolvens (Ren et al., 2019), R. josui (Kakiuchi et al., 1998), R. herbifermentans (Rettenmaier et al., 2019), R. cellulolyticum (Desvaux, 2005), and R. sufflavum (Nishiyama et al., 2009) (Figure 5)]. In contrast, nearly all of the clostridial species are low DocGH-LCB producers suggesting their simple cellulosomes have limited cellulolytic activity [C. felsineum (previously known as C. roseum; Collins et al., 1994), C. acetobutylicum (Sabathé et al., 2002; López-Contreras et al., 2003; López-Contreras et al., 2004), C. bornimense (Tomazetto et al., 2016), C. cibarium (Gilroy et al., 2021), C. puniceum (Dassa et al., 2017), C. saccharoperbutylacetonicum (Levi Hevroni et al., 2020), and several additional Clostridium spp. (CT7, HBUAS56017, DSM 8431, NJ4, TW13, YIM B02555)]. This notion is consistent with experimental data, as the simple cellulosomes produced by high number DocGH-LCB bacteria, R. cellulolyticum, R. papyrosolvens, and R. herbifermentans, are shown to have potent cellulolytic activity (Ren et al., 2019; Rettenmaier et al., 2019; You et al., 2023; Giallo et al., 1985), whereas the simple cellulosomes in C. acetobutylicum and C. saccharoperbutylacetonicum that have a low number of DocGH-LCB genes are less cellulolytic (Sabathé et al., 2002; López-Contreras et al., 2003; López-Contreras et al., 2004; Levi Hevroni et al., 2020). The only two exceptions to the idea that simple cellulosomes in clostridial bacteria are less cellulolytic are from C. cellulovorans (Usai et al., 2020) and Clostridium sp. BNL1100 (Li et al., 2012). These species are presumably cellulolytic as their genomes contain a high number of DocGH-LCB encoding genes. However, it is noteworthy that based on its 16S rRNA sequence, Clostridium sp. BNL1100 can be classified as a member of the Ruminiclostridium genus (Figure 2). Finally, there are two “non-clostridial” species with genomes encoding simple cellulosomes and a low number of DocGH-LCB enzymes, Herbinix luporum (Koeck et al., 2016) and Inconstantimicrobium porci (Wylensek et al., 2020). Based on their 16S rRNA sequence both species are most closely related to clostridial bacteria.
The mechanism(s) through which simple cellulosomes are attached to the cell surface remains incompletely understood. This is because their primary simple scaffoldins lack obvious cell wall binding modules or dockerin domains that could mediate their attachment to anchoring scaffoldins (either SLH-anchoring or nonSLH-anchoring scaffoldins; Figure 1). Cell wall attachment by simple primary scaffoldins may be mediated by their N-terminal CBM3 domain as supported by recent Western blot data demonstrating cell surface binding by a CBM3a module (Tao et al., 2022). In addition, all of the simple primary scaffoldins contain X2 domains that have been implicated in both cell wall attachment (Kosugi et al., 2004) and cellular interactions with cellulose (Tao et al., 2022; Tarraran et al., 2021). Notably, two bacterial species contain genes for more than one primary simple scaffoldin, R. herbifermentans and C. cellulovorans (Table 2). R. herbifermentans’ genome has genes encoding four simple primary scaffoldins that are located in a single cipA operon (Rettenmaier et al., 2019), with each scaffoldin possessing 5 to 14 Coh1 modules. In the case of C. cellulovorans, genes for 3 primary scaffoldin proteins are present (WP_010073402.1, WP_010073403.1, and WP_013291799.1). Of these, only one scaffoldin (WP_013291799.1, CbpA) is encoded by a gene located within a cipA gene cluster (Dassa et al., 2017).
Some species of simple cellulosome-producing bacteria harbor genes encoding unique accessory scaffoldins with large numbers of FN3 domains that have been proposed to disrupt crystalline polysaccharide structures or solubilize large protein complexes (Kataeva et al., 2002; Alahuhta et al., 2010). Simple cellulosome displaying bacteria containing FN3 scaffoldins include: R. cellulolyticum, R. herbifermentans, R. hungatei, C. josui, R. sufflavum, C. felsineum, C. pasteurianum, and Clostridium sp. CT7. C. pasteurianum and Clostridium sp. CT7 are notable as they each contain a scaffoldin with 30 FN3 repeats that is capped by a C-terminal Coh2-Doc2 module whose binding partners are unknown, as no other proteins in these microbes contain Doc2 or Coh2 modules. A limited number of complex cellulosome-displaying bacteria also contain scaffoldins with FN3 domains, but typically only 1–3 copies of this module are present. Finally, three simple cellulosome-producing bacteria produce cell-free scaffoldins that may be secreted as they lack dockerin domains and cell wall binding modules (R. sufflavum, Clostridium sp. HBUAS56017, and C. bornimense).
We identified 9 new bacterial species that based on their genome sequences produce simple cellulosomes: Clostridium cibarium, Clostridium pasteurianum, Clostridium sp. CT7, Clostridium sp. DSM 8431, Clostridium sp. HBUAS56017, Clostridium sp. NJ4, Clostridium sp. TW13, Clostridium sp. YIM B02555, and I. porci. Supplementary Figure S1 shows their predicted scaffoldins, while Supplementary Table S4 enumerates their dockerin, cohesin, and enzyme content. They are all low DocGH producers that phylogenetically cluster with clostridial bacteria (Figure 2). Each contains at least one simple primary scaffoldin that is a hallmark of simple cellulosome producers—defined as a scaffoldin that contains two or more cohesin domains and an N-terminal CBM3 (a lone exception is Clostridium sp. CT7 that lacks an N-terminal CBM3). As with other simple cellulosome producers, their primary scaffoldins contain X2 and Coh1 modules and each microbe almost exclusively produces DocGH enzymes containing Doc1 modules. Several of these microorganisms also produce a single scaffoldin that encodes 1–2 cohesins which are either Coh1-or Coh2-type (Clostridium sp. NJ4, Clostridium sp. YIM B02555, Clostridium sp. HBUAS56017, Clostridium sp. DSM 8431, Clostridium sp. CT7, Clostridium cibarium).
Our analysis predicts for the first time the scaffoldin and enzyme composition in 4 microbes previously noted to produce cellulosomes: R. hungatei, C. bornimense, C. felsineum, and H. luporum (Supplementary Table S4). All have the genomic capacity to produce simple primary scaffoldins that are the core of a simple cellulosome (Figure 1). R. hungatei DSM 14427 is of particular interest as it is the only one in this group whose genome contains a high number of DocGH-LCB encoding genes, as well as genes encoding 3 accessory scaffoldins that contain 7–9 FN3 modules and a C-terminal cohesin domain. Based on their primary sequences, 2 of these scaffoldins contain Coh2 domains whose binding partners are not known because R. hungatei’s genome lacks genes that encode for Doc2 containing proteins. Finally, the genomes of H. luporum and C. cibarium encode for proteins that may function as nonSLH-anchoring scaffoldins, as they contain a single Coh2 domain and a C-terminal transmembrane helix that may be embedded in the bilayer. However, the binding partners for these scaffoldins also remain unclear, since only in C. cibarium are genes encoding complementary Doc2 containing proteins identifiable.
We used broad search criteria to identify cellulosome producing bacteria—any genome that contained a gene for at least one multi-cohesin and one DocGH protein. Four microbial genomes barely satisfied these criteria and are unlikely to produce conventional cellulosomes because their largest scaffoldin contains only two cohesin modules. These include three species from the gram-positive Bacillota phylum whose members are known to display cellulosomes, Iocasia fonsfrigidae (Zhang et al., 2021), Lachnoclostridium sp. MSJ-17, and Paenibacillus guangzhouensis (Li et al., 2014), as well as Limihaloglobus sulfuriphilus (Pradel et al., 2020) which is a member of the rare Planctomycetes–Verrucomicrobia–Chlamydiae (PVC) superphylum. Lachnoclostridium sp. MSJ-17 encodes four cohesin-containing scaffoldins, three proteins that contain a single cohesin, and one larger scaffoldin that contains two cohesins and an N-terminal Doc1 domains. Two of the single cohesin-containing scaffoldins may be cell-associated as they contain C-terminal transmembrane helices (WP_216523161.1 and WP_216522914.1). Collectively, these scaffoldins could bind as many as 39 distinct dockerin-containing proteins, but the microbe is presumably non-cellulolytic as its genome encodes only a single DocGH-LCB. I. fonsfrigidae was isolated from deep sea sediment and has the potential to produce two large scaffoldins. One of them contains two cohesins, an FN3 module, and a CBM3 domain that could mediate cellulose binding. The second scaffoldin is also sizable (662 amino acids) but is predicted to contain only a single C-terminal cohesin domain. Interestingly, this microbe’s genome contains only a single gene encoding one DocGH-LCB enzyme. The gram-positive soil bacterium P. guangzhouensis is the most impressive of the scaffoldin-containing microbes as its genome encodes 35 cohesin-containing proteins. A total of 28 of these proteins are monovalent adaptors that contain a single dockerin-cohesin domain pair and in many instances, they possess an additional N-terminal GH enzyme that could degrade hemicellulose or oligosaccharide polymers (GH2, GH20, GH29, GH31, and GH43). There are also five SLH-anchoring scaffoldins to which these proteins could dock onto, potentially creating an enzyme rich surface that would be architecturally distinct from conventional cellulosomes (Phitsuwan et al., 2019). Fascinatingly, in this microbe, nearly all of its dockerin domains are located within its cohesin-containing scaffoldins (only one dockerin is located in a non-scaffoldin protein). This raises the possibility that the full-complement of its dockerin proteins were not detected by InterProScan and/or its scaffoldins act to opportunistically scavenge dockerin-fusion proteins that are produced by other microbes. Notably, P. guangzhouensis’ genome also contains a large number of genes that encode for CAZymes, but most of these are not of the family-type that is known to degrade LCB. The genome in the PVC superphylum bacterium L. sulfuriphilus contains genes encoding a single Coh2-Doc1-Coh1 scaffoldin-like protein and 13 dockerin-fusion proteins. It is presumably non-cellulolytic as it contains only a single DocGH-LCB and a limited number of CAZymes. Notably, the dockerin proteins in L. sulfuriphilus are fused to domains not commonly found in cellulosome producing bacteria, including FAD/NAD-binding, aspartic peptidase-like domains, and HdrA-like domains.
Across the studied bacterial species in this study, an examination of their dockerin-fusion proteins provides insight into both the numbers and types of proteins that are incorporated into cellulosomes (Figures 5, 7). In general, the genomes of Acetivibrio and Ruminococcus bacteria produce complex cellulosomes containing a high number of DocGH-LCB genes (>20) suggesting that these structures are cellulolytic (Figure 5). In contrast, Ruminiclostridium and Clostridium bacteria that have the genomic capacity to produce simple cellulosomes contain either high or low numbers of DocGH-LCB genes, respectively. Interestingly, a near linear relationship is observed between the total number of dockerin and DocGH-LCB encoding genes within an organism (R2 = 0.81), which is consistent with the primary function of cellulosomes being to degrade LCB (Figure 5). Thus, only bacteria whose genomes contain a large and diverse set of DocGH-LCBs (Ruminiclostridium, Acetivibrio, and Ruminococcus) also possess a high number of genes that encode for other types of dockerin-fusion proteins. For example, on average high DocGH-LCB producers contain ~40 genes encoding DocGH-LCBs and an impressive ~80 genes that encode other types of dockerin-fusion proteins (Figure 7). This number is much smaller in the low DocGH-LCB producers (Clostridium), as they only contain on average ~ 2 and ~ 3 genes that encode DocGH-LCBs and other types of dockerin-fusions, respectively. There is significant variability amongst the high DocGH-LCB producing microbes, as P. cellulosolvens (classified by us as an Acetivibrio-type) contains a total of 206 dockerin-fusion genes (of which 70 are DocGH-LCB genes), whereas R. hungatei (Ruminiclostridium-type) contains only 40 (20 DocGH-LCB genes). Interestingly, our analysis reveals that bacteria frequently supplement their DocGH-LCBs with a similar set of dockerin-fused accessory proteins that may facilitate LCB degradation. These include dockerin-fusion proteins containing: CBMs that bind carbohydrates (Doc-CBM, orange), carbohydrate active hydrolases such as pectin lyases and carbohydrate esterases (Doc-CAZymes, red), cohesin domains that are part of scaffoldins that construct the cellulosome (Doc-cohesin, yellow), and proteins with other functions (Doc-other, gray; Figure 7). This enrichment supports the idea that the primary function of the cellulosomes in Acetivibrio, Ruminococcus, and Ruminiclostridium bacteria is to degrade LCB or related carbohydrate polymers. Interestingly, perhaps to compensate for their deficiency in DocGH-LCBs and complementary proteins, the genomes of some clostridial bacteria contain larger numbers of genes encoding carbohydrate active hydrolases that are freely secreted (Figure 7).
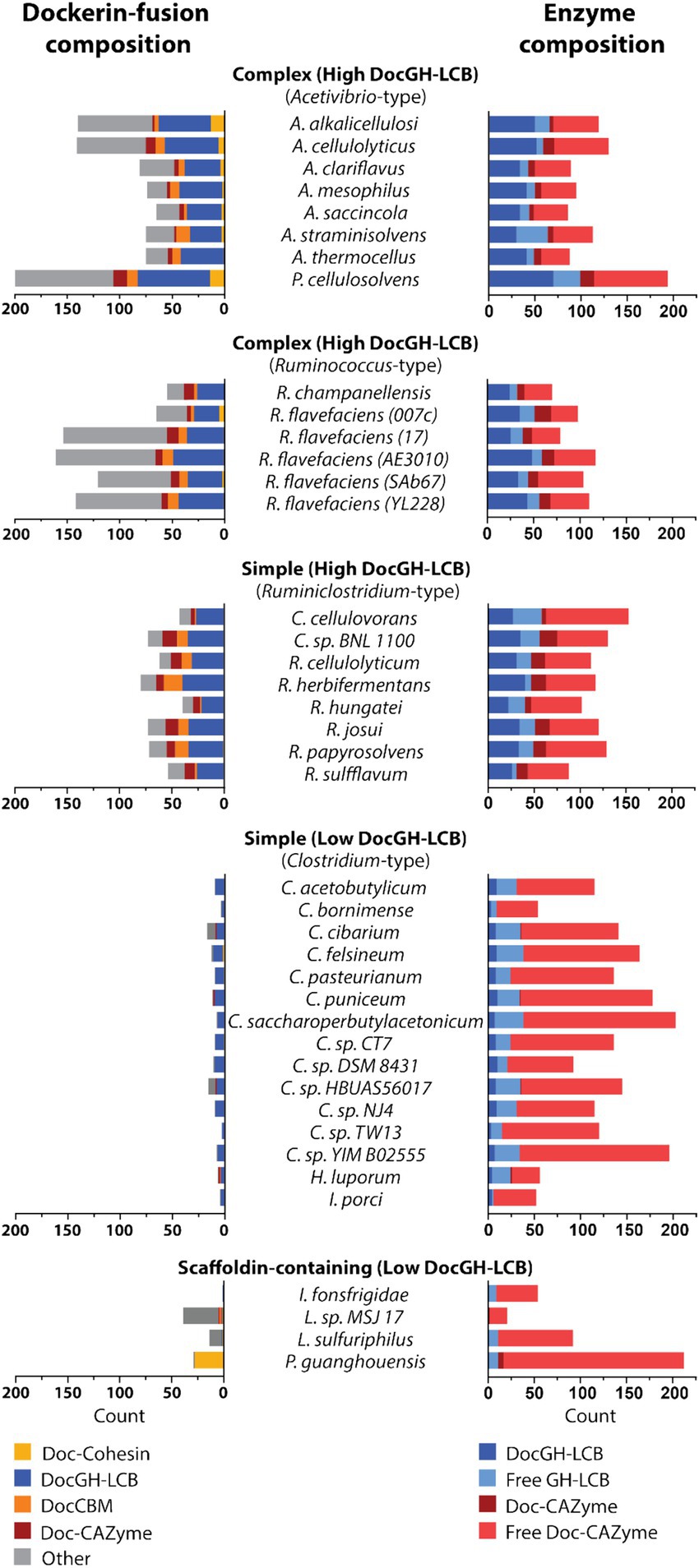
Figure 7. Domain and enzyme composition in dockerin-fused proteins. Plots show the types of domains that are fused to dockerin proteins in microbes that produce Acetivirbio-type complex cellulosomes, Ruminococcus-type complex cellulosomes, Ruminiclostridium-type simple cellulosomes, Clostridium-type simple cellulosomes, and other bacteria with scaffoldin-containing proteins that are unlikely to form conventional cellulosomes (see Tables 1, 2 for a complete list). (Left) Stacked bar plot representation of the number of dockerin-fusion proteins based on the types of domains they contain. Co-occurring domain type is color coded according to the following: Yellow: Cohesin, Dark Blue: Glycoside Hydrolase with LCB activity, Orange: Carbohydrate Binding Module, Red: Carbohydrate Active Enzymes (CAZyme), Grey: other. (Right) Stacked bar plot representation of cellulosomal enzyme composition for each cellulosome-producing microbe, including; Dark Blue: DocGH-LCB, Light Blue: Free GH-LCB, Red: Dockerin-fused CAZyme, and Light Red: Free CAZyme.
Our results indicate that with only a few exceptions, bacteria with the genetic capacity to produce cellulosomes originate from four genera: Acetivibrio-, Ruminococcus-, Ruminiclostridium- and Clostridium-type cellulosomes. Acetivibrio-type cellulosomes, including the one found in P. cellulosolvens, are complex and can be populated with a high number distinct LCB active enzymes. They are characterized by the presence of a conserved dockerin-containing primary scaffoldin, SLH-anchoring scaffoldins that tether the cellulosome to the cell surface, cell-free scaffoldins that are presumably secreted to form multi-enzyme complexes, and CBM3-SLH proteins that may enable microbial tethering to LCB. Dockerin-fused enzymes bind to the scaffoldins via Doc1-Coh1 interactions, whereas Doc2-Coh2 interactions mediate scaffoldin-scaffoldin interactions (the exception is P. cellulosolvens in which these interactions are reversed). Only microbes that harbor Acetivibrio-type cellulosomes use a dedicated suite of polysaccharide-sensing RsgI transmembrane receptors to regulate its composition. Ruminococcus bacteria (R. champanellensis or R. flavefaciens) can also produce complex cellulosomes that contain a high number of distinct DocGH-LCB enzymes. However, they are unique because their anchoring scaffoldins are attached to the cell wall via sortase enzymes instead of SLH domains, and many of their dockerin-fusion proteins contain Doc3 modules. It was challenging to define the components of these structures from genomic sequence data as their cohesins frequently have divergent primary sequences that could not be detected using InterProScan. Indeed, we detected no Coh3-type modules based on their primary sequence, and only when AlphaFold2 was employed to predict their structures were several cohesins identified. Ruminiclostridium and Clostridium bacteria produce ‘simple’ cellulosomes that contain only a limited number of scaffoldins. They are further distinguished by the presence of a primary scaffoldin that contains an N-terminal CBM3, X2, and multiple cohesin domains. Their genomes encode only a limited number of scaffoldins and their primary scaffoldins adhere to the microbial surface through a poorly understand mechanism as they lack obvious domains that are capable of binding to the cell wall. These simple cellulosomes can be subdivided further by the number of distinct DocGH-LCB enzymes they house, with Ruminiclostridium and Clostridium genomes typically encoding for high and low numbers of DocGH-LCB enzymes, respectively. Finally, several species of simple cellulosome displaying bacteria are unique as they contain scaffoldins harboring up to 30 FN3 repeats that may disrupt crystalline polysaccharide structures and/or solubilize large protein complexes. Mapping the precise architectures of these cellulosomes requires additional experimental studies to define the specific set of cohesin-dockerin interactions that form the “glue” that hold these structures together, since at present it is not always possible to reliably predict the specificity of these interactions using only primary sequence data.
This comparative genomic analysis identified 33 bacterial species with the capacity to produce cellulosomes, including 10 previously unreported species. The actual number of distinct cellulosome producing species in nature is likely much larger, as up to 1.6 million operational taxonomic units (a proxy for bacterial species) are estimated to exist (Louca et al., 2019), of which only ~2.1% have had their genomes completely sequenced (Zhang et al., 2020). This undersampling is evident from metagenomics data, which reveals the presence of cellulosome displaying bacteria with incompletely sequenced genomes. Here we focused our efforts only on bacteria with completely sequenced genomes, as significant genome gaps exist in metagenomic data that make it extremely difficult to identify the complete set of cellulosome encoding genes in these microbes (Nam et al., 2023). Collectively, the results of this study provide insight into the structural diversity of bacterial cellulosomes, and they reveal conserved architectural features that may be useful in guiding ongoing engineering efforts to produce bio-based chemicals and materials from plant biomass.
The retrieval of cellulosome-displaying bacteria consists of two components: (i) a pre-scan phase to select for genomes that potentially contain cellulosomes, and (ii) an in-depth scan of each of these filtered genomes. While metagenomic sequencing has strengths in identifying novel microbes, the aim of this study is to categorize and take inventory of the diversity of cellulosomes displayed by fully sequenced microbes. For this analysis we therefore did not include metagenome-based sequences because genomes resolved by this method are often incomplete and constructed from many contigs (Nam et al., 2023). These two shortcomings result in significant gaps in an organism’s genome making it difficult to resolve all protein coding genes. Therefore, for the pre-scan phase, only the RefSeq database (O'Leary et al., 2016; July 10, 2023 release) was used, which contained 138,491 organisms with 371,291,248 records. The genome of Acetivibrio alkalicellulosi is not deposited in the RefSeq database, because it has been suppressed due to contamination (Supplementary Table S2). However, it has been reported to produce a cellulosome and therefore was manually included in our analysis (Phitsuwan et al., 2019). From the RefSeq database we retrieved only prokaryotic genomes (305,693) and performed a hidden Markov model (HMM) search (Potter et al., 2018) using dockerin (cd14256, cd14254, cd14255), cohesin (cd08548, cd08547, cd08759), and GHs (PF00150, PF03537, PF01670, PF12891, PF02015, PF02011, PF01270, PF00759) domain profiles obtained from the Conserved Domains Database (CDD) and Pfam database (Wang et al., 2023; Mistry et al., 2021). Genomes were targeted for the next in-depth phase only if they met the following criteria: i) they must have at least one dockerin-fused GH protein, and ii) they must have at least one protein with two or more cohesins.
In the in-depth scanning phase, the 139 genomes from the prescan phase were re-annotated using Prokka (v. 1.14.6; Seemann, 2014), and all the locus tags from NCBI’s annotations were mapped using BLAST (v. 2.13.0+; Camacho et al., 2009) to maintain consistency with the existing gene naming convention. After re-annotation, InterProScan (v. 5.59-91.0; Jones et al., 2014) was performed, which includes the following member databases: Phobius (v. 1.01; Käll et al., 2004), SUPERFAMILY (v. 1.75; Gough et al., 2001), ProSiteProfiles (v. 2022_01; Sigrist et al., 2013), SMART (v. 7.1; Letunic et al., 2021), CDD (v. 3.18; Wang et al., 2023), PRINTS (v. 42.0; Attwood et al., 2012), and Pfam (v. 35.0; Mistry et al., 2021). Additionally, three databases were added to capture signal peptide regions (SignalP v. 6.0; Teufel et al., 2022), transmembrane regions (DeepTMHMM v. 1.0.19; Hallgren et al., 2022), and subcellular localization information (PSORTb v. 3.0; Yu et al., 2010).
After the in-depth genome analysis, we applied the same criteria as before (i) ≥1 DocGH fusion protein and (ii) at least one protein with ≥2 cohesin domains) and identified 37 genomes that met our criteria. 132 genomes did not pass our criteria using the InterProScan data because the initial HMMER search was not conducted with stringent threshold values, which inflated the initial number of domains each genome had. However, this genome-based analysis procedure did not identify several bacteria which have previously been shown to display cellulosomes (e.g., Ruminococcus flavefaciens 007c, FD-1, 007c, and Ruminococcus champanellensis18P13), presumably because RefSeq uses stringent criteria for inclusion of genomes in their database and these genomes were of insufficient quality. These species genomes were manually added into our analysis for completeness’ sake. A custom python (v. 3.11.5) script was used to analyze the domain composition in the scaffoldins and dockerin-fused proteins in the 37 bacterial species that produce cellulosomes. The domain identifiers used for this analysis are listed in Supplementary Table S5.
We observed that 34 protein sequences (from 3 organisms: R. flavefaciens 17, R. flavefaciens 007c, and R. champanellensis) are reported in the literature to possess sequence homology to cohesin domains but we could not identify them as such using the HMM profiles provided by InterProScan (Dassa et al., 2014). Similarly, our analysis showed significantly lower numbers of scaffoldin proteins for additional R. flavefaciens strains (AE3010, SAb67, YL228) that we suspected also encode cohesin domains that are too divergent to be identified by the HMM profiles. Recognizing that structure is better conserved than sequence, we used AlphaFold2 (Jumper et al., 2021) to build atomic models for any protein across these six organisms that contained at least one of the following domains: Coh1 (cd08548), Coh2 (cd08547), Coh3 (cd08759), general Coh (PF00963), CBM2/3 Superfamily (IPR008965), CATH3D entry (G3DSA:2.60.40.680). This resulted in the predictions of 90 putative cohesin-containing proteins. We requested three models to be built for each sequence and used only the model with highest pLDDT score for further structural analysis. For multi-domain proteins, we divided the model into individual domains using the program UniDoc (Zhu et al., 2023), producing 311 domains. The structure of each domain was compared to four reference models: Coh1 (PDB:1OHZ), Coh2 (PDB:1TYJ), Coh3 (PDB:2FZ9), and CBM3 (PDB:6UFW). Structural comparisons were performed with the program TMalign (Dong et al., 2018). TM-scores for all comparisons were normalized to 140 residues to simplify comparison. Query domains with TM-scores greater than 0.50 were considered as matches to the reference model. If a domain scored higher than 0.50 for multiple reference models, then we attribute the domain identity to match the highest scoring reference model.
Information for existing publicly accessible datasets is contained within the article. The genome accession numbers presented in this study can be found in supplementary table 4 and online through the RefSeq or GenBank repositories. The original InterProScan analaysis will be provided upon request. Any further inquiries can be directed to the corresponding author(s).
CM: Writing – original draft, Writing – review & editing, Data curation, Formal analysis, Investigation, Methodology. AT: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. SH: Data curation, Formal analysis, Writing – original draft, Writing – review & editing, Software. LS: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. MS: Data curation, Formal analysis, Writing – original draft, Writing – review & editing, Investigation. MP: Writing – original draft, Writing – review & editing, Conceptualization, Supervision. RC: Conceptualization, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition, Project administration, Resources.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This material is based upon work supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research program under Award Number DE-FC02-02ER63421. We also acknowledge support from NIH grant R01-AI052217 (R.T.C), a Cellular and Molecular Biology training grant T32GM145388 (CM), and the UCLA Quantitative & Computational Biosciences Collaboratory for assistance.
This work used computational and storage services associated with the Hoffman2 Cluster which is operated by the UCLA Office of Advanced Research Computing’s Research Technology Group.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1473396/full#supplementary-material
Aikawa, S., Baramee, S., Sermsathanaswadi, J., Thianheng, P., Tachaapaikoon, C., Shikata, A., et al. (2018). Characterization and high-quality draft genome sequence of Herbivorax saccincola A7, an anaerobic, alkaliphilic, thermophilic, cellulolytic, and xylanolytic bacterium. Syst. Appl. Microbiol. 41, 261–269. doi: 10.1016/j.syapm.2018.01.010
Alahuhta, M., Xu, Q., Brunecky, R., Adney, W. S., Ding, S. Y., Himmel, M. E., et al. (2010). Structure of a fibronectin type III-like module from Clostridium thermocellum. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 66, 878–880. doi: 10.1107/S1744309110022529
Artzi, L., Bayer, E. A., and Moraïs, S. (2017). Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 15, 83–95. doi: 10.1038/nrmicro.2016.164
Artzi, L., Dassa, B., Borovok, I., Shamshoum, M., Lamed, R., and Bayer, E. A. (2014). Cellulosomics of the cellulolytic thermophile Clostridium clariflavum. Biotechnol. Biofuels 7:100. doi: 10.1186/1754-6834-7-100
Artzi, L., Morag, E., Barak, Y., Lamed, R., and Bayer, E. A. (2015). Clostridium clariflavum: key cellulosome players are revealed by proteomic analysis. MBio 6, e00411–e00415. doi: 10.1128/mBio.00411-15
Attwood, T. K., Coletta, A., Muirhead, G., Pavlopoulou, A., Philippou, P. B., Popov, I., et al. (2012). The PRINTS database: a fine-grained protein sequence annotation and analysis resource--its status in 2012. Database (Oxford) 2012:bas019. doi: 10.1093/database/bas019
Bae, J., Morisaka, H., Kuroda, K., and Ueda, M. (2013). Cellulosome complexes: natural biocatalysts as arming microcompartments of enzymes. J. Mol. Microbiol. Biotechnol. 23, 370–378. doi: 10.1159/000351358
Barba-Cedillo, V., and Montanier, C. Y. (2023). Effect of multimodularity and spatial organization of glycoside hydrolases on catalysis. Essays Biochem. 67, 629–638. doi: 10.1042/EBC20220167
Bar-On, Y. M., Phillips, R., and Milo, R. (2018). The biomass distribution on earth. Proc. Natl. Acad. Sci. USA 115, 6506–6511. doi: 10.1073/pnas.1711842115
Bayer, E. A., Belaich, J.-P., Shoham, Y., and Lamed, R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Ann. Rev. Microbiol. 58, 521–554. doi: 10.1146/annurev.micro.57.030502.091022
Ben David, Y., Dassa, B., Borovok, I., Lamed, R., Koropatkin, N. M., Martens, E. C., et al. (2015). Ruminococcal cellulosome systems from rumen to human. Environ. Microbiol. 17, 3407–3426. doi: 10.1111/1462-2920.12868
Bhardwaj, N., Kumar, B., Agrawal, K., and Verma, P. (2021). Current perspective on production and applications of microbial cellulases: a review. Bioresour. Bioprocess. 8:95. doi: 10.1186/s40643-021-00447-6
Bhaskar, Y., Su, X., Xu, C., and Xu, J. (2021). Predicting selective RNA processing and stabilization operons in Clostridium spp. Front. Microbiol. 12:673349. doi: 10.3389/fmicb.2021.673349
Blouzard, J.-C., Coutinho, P. M., Fierobe, H. P., Henrissat, B., Lignon, S., Tardif, C., et al. (2010). Modulation of cellulosome composition in Clostridium cellulolyticum: adaptation to the polysaccharide environment revealed by proteomic and carbohydrate-active enzyme analyses. Proteomics 10, 541–554. doi: 10.1002/pmic.200900311
Brás, J. L. A., Pinheiro, B. A., Cameron, K., Cuskin, F., Viegas, A., Najmudin, S., et al. (2016). Diverse specificity of cellulosome attachment to the bacterial cell surface. Sci. Rep. 6:38292. doi: 10.1038/srep38292
Bule, P., Pires, V. M., Fontes, C. M., and Alves, V. D. (2018). Cellulosome assembly: paradigms are meant to be broken! Curr. Opin. Struct. Biol. 49, 154–161. doi: 10.1016/j.sbi.2018.03.012
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Cameron, K., Najmudin, S., Alves, V. D., Bayer, E. A., Smith, S. P., Bule, P., et al. (2015). Cell-surface attachment of bacterial multienzyme complexes involves highly dynamic protein-protein anchors. J. Biol. Chem. 290, 13578–13590. doi: 10.1074/jbc.M114.633339
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Carvalho, A. L., Dias, F. M. V., Nagy, T., Prates, J. A. M., Proctor, M. R., Smith, N., et al. (2007). Evidence for a dual binding mode of dockerin modules to cohesins. Proc. Natl. Acad. Sci. USA 104, 3089–3094. doi: 10.1073/pnas.0611173104
Celik, H., Blouzard, J. C., Voigt, B., Becher, D., Trotter, V., Fierobe, H. P., et al. (2013). A two-component system (XydS/R) controls the expression of genes encoding CBM6-containing proteins in response to straw in Clostridium cellulolyticum. PLoS One 8:e56063. doi: 10.1371/journal.pone.0056063
Chow, N., and Wu, J. H. D. (2017). “The cellulosome” in Biotechnology of microbial enzymes. ed. G. Brahmachari (Elsevier: Biotechnology of Microbial Enzymes, Academic Press), 243–266. doi: 10.1016/B978-0-12-803725-6.00010-8
Chundawat, S. P. S., Beckham, G. T., Himmel, M. E., and Dale, B. E. (2011). Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu. Rev. Chem. Biomol. Eng. 2, 121–145. doi: 10.1146/annurev-chembioeng-061010-114205
Collins, M. D., Lawson, P. A., Willems, A., Cordoba, J. J., Fernandez-Garayzabal, J., Garcia, P., et al. (1994). The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44, 812–826. doi: 10.1099/00207713-44-4-812
Dassa, B., Borovok, I., Lamed, R., Henrissat, B., Coutinho, P., Hemme, C. L., et al. (2012). Genome-wide analysis of Acetivibrio cellulolyticus provides a blueprint of an elaborate cellulosome system. BMC Genomics 13:210. doi: 10.1186/1471-2164-13-210
Dassa, B., Borovok, I., Lombard, V., Henrissat, B., Lamed, R., Bayer, E. A., et al. (2017). Pan-Cellulosomics of mesophilic Clostridia: variations on a theme. Microorganisms 5:74. doi: 10.3390/microorganisms5040074
Dassa, B., Borovok, I., Ruimy-Israeli, V., Lamed, R., Flint, H. J., Duncan, S. H., et al. (2014). Rumen cellulosomics: divergent fiber-degrading strategies revealed by comparative genome-wide analysis of six ruminococcal strains. PLoS One 9:e99221. doi: 10.1371/journal.pone.0099221
Dawson, N. L., Lewis, T. E., das, S., Lees, J. G., Lee, D., Ashford, P., et al. (2017). CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 45, D289–D295. doi: 10.1093/nar/gkw1098
Desvaux, M. (2005). Clostridium cellulolyticum: model organism of mesophilic cellulolytic clostridia. FEMS Microbiol. Rev. 29, 741–764. doi: 10.1016/j.femsre.2004.11.003
Ding, S. Y., Rincon, M. T., Lamed, R., Martin, J. C., McCrae, S. I., Aurilia, V., et al. (2001). Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183, 1945–1953. doi: 10.1128/JB.183.6.1945-1953.2001
Doi, R. H., Kosugi, A., Murashima, K., Tamaru, Y., and Han, S. O. (2003). Cellulosomes from mesophilic bacteria. J. Bacteriol. 185, 5907–5914. doi: 10.1128/JB.185.20.5907-5914.2003
Dong, R., Pan, S., Peng, Z., Zhang, Y., and Yang, J. (2018). mTM-align: a server for fast protein structure database search and multiple protein structure alignment. Nucleic Acids Res. 46, W380–W386. doi: 10.1093/nar/gky430
Eddy, S. R. (2009). A new generation of homology search tools based on probabilistic inference. Genome Inform. Int. Conf. Genome Inform. 23, 205–211.
Fontes, C. M. G. A., and Gilbert, H. J. (2010). Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79, 655–681. doi: 10.1146/annurev-biochem-091208-085603
Giallo, J., Gaudin, C., and Belaich, J. P. (1985). Metabolism and solubilization of cellulose by Clostridium cellulolyticum H10. Appl. Environ. Microbiol. 49, 1216–1221. doi: 10.1128/aem.49.5.1216-1221.1985
Gilbert, H. J. (2007). Cellulosomes: microbial nanomachines that display plasticity in quaternary structure. Mol. Microbiol. 63, 1568–1576. doi: 10.1111/j.1365-2958.2007.05640.x
Gilroy, R., Ravi, A., Getino, M., Pursley, I., Horton, D. L., Alikhan, N. F., et al. (2021). Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture. PeerJ 9:e10941. doi: 10.7717/peerj.10941
Gough, J., Karplus, K., Hughey, R., and Chothia, C. (2001). Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313, 903–919. doi: 10.1006/jmbi.2001.5080
Hallgren, J., Tsirigos, K. D., Pedersen, M. D., Armenteros, J. J. A., Marcatili, P., Nielsen, H., et al. (2022). DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv. doi: 10.1101/2022.04.08.487609
Hamberg, Y., Ruimy-Israeli, V., Dassa, B., Barak, Y., Lamed, R., Cameron, K., et al. (2014). Elaborate cellulosome architecture of Acetivibrio cellulolyticus revealed by selective screening of cohesin-dockerin interactions. PeerJ 2:e636. doi: 10.7717/peerj.636
Hong, W., Zhang, J., Feng, Y., Mohr, G., Lambowitz, A. M., Cui, G. Z., et al. (2014). The contribution of cellulosomal scaffoldins to cellulose hydrolysis by Clostridium thermocellum analyzed by using thermotargetrons. Biotechnol. Biofuels 7:80. doi: 10.1186/1754-6834-7-80
Jindou, S., Brulc, J. M., Levy-Assaraf, M., Rincon, M. T., Flint, H. J., Berg, M. E., et al. (2008). Cellulosome gene cluster analysis for gauging the diversity of the ruminal cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol. Lett. 285, 188–194. doi: 10.1111/j.1574-6968.2008.01234.x
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinforma. Oxf. Engl. 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kahel-Raifer, H., Jindou, S., Bahari, L., Nataf, Y., Shoham, Y., Bayer, E. A., et al. (2010). The unique set of putative membrane-associated anti-sigma factors in Clostridium thermocellum suggests a novel extracellular carbohydrate-sensing mechanism involved in gene regulation. FEMS Microbiol. Lett. 308, 84–93. doi: 10.1111/j.1574-6968.2010.01997.x
Kakiuchi, M., Isui, A., Suzuki, K., Fujino, T., Fujino, E., Kimura, T., et al. (1998). Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180, 4303–4308. doi: 10.1128/JB.180.16.4303-4308.1998
Käll, L., Krogh, A., and Sonnhammer, E. L. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036. doi: 10.1016/j.jmb.2004.03.016
Kampik, C., Denis, Y., Pagès, S., Perret, S., Tardif, C., Fierobe, H.-P., et al. (2020). A novel two-component system, XygS/XygR, positively regulates xyloglucan degradation, import, and catabolism in Ruminiclostridium cellulolyticum. Appl. Environ. Microbiol. 86, e01357–e01320. doi: 10.1128/AEM.01357-20
Kataeva, I. A., Seidel, R. D. III, Shah, A., West, L. T., Li, X. L., and Ljungdahl, L. G. (2002). The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68, 4292–4300. doi: 10.1128/AEM.68.9.4292-4300.2002
Kato, S., Haruta, S., Cui, Z. J., Ishii, M., Yokota, A., and Igarashi, Y. (2004). Clostridium straminisolvens sp. nov., a moderately thermophilic, aerotolerant and cellulolytic bacterium isolated from a cellulose-degrading bacterial community. Int. J. Syst. Evol. Microbiol. 54, 2043–2047. doi: 10.1099/ijs.0.63148-0
Koeck, D. E., Hahnke, S., and Zverlov, V. V. (2016). Herbinix luporum sp. nov., a thermophilic cellulose-degrading bacterium isolated from a thermophilic biogas reactor. Int. J. Syst. Evol. Microbiol. 66, 4132–4137. doi: 10.1099/ijsem.0.001324
Kosugi, A., Amano, Y., Murashima, K., and Doi, R. H. (2004). Hydrophilic domains of scaffolding protein CbpA promote glycosyl hydrolase activity and localization of cellulosomes to the cell surface of Clostridium cellulovorans. J. Bacteriol. 186, 6351–6359. doi: 10.1128/JB.186.19.6351-6359.2004
Lamed, R., Setter, E., and Bayer, E. A. (1983). Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156, 828–836. doi: 10.1128/jb.156.2.828-836.1983
Leibovitz, E., and Béguin, P. (1996). A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178, 3077–3084. doi: 10.1128/jb.178.11.3077-3084.1996
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Levi Hevroni, B., Moraïs, S., Ben-David, Y., Morag, E., and Bayer, E. A. (2020). Minimalistic cellulosome of the butanologenic bacterium Clostridium saccharoperbutylacetonicum. MBio 11, e00443–e00420. doi: 10.1128/mBio.00443-20
Li, J., Lu, Q., Liu, T., Zhou, S., Yang, G., and Zhao, Y. (2014). Paenibacillus guangzhouensis sp. nov., an Fe(III)- and humus-reducing bacterium from a forest soil. Int. J. Syst. Evol. Microbiol. 64, 3891–3896. doi: 10.1099/ijs.0.067173-0
Li, W., O’Neill, K. R., Haft, D. H., DiCuccio, M., Chetvernin, V., Badretdin, A., et al. (2021). RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 49, D1020–D1028. doi: 10.1093/nar/gkaa1105
Li, L.-L., Taghavi, S., Izquierdo, J. A., and van der Lelie, D. (2012). Complete genome sequence of Clostridium sp. strain BNL1100, a cellulolytic mesophile isolated from corn Stover. J. Bacteriol. 194, 6982–6983. doi: 10.1128/JB.01908-12
Liu, Y., Vanderhaeghen, S., Feiler, W., Angelov, A., Baudrexl, M., Zverlov, V., et al. (2021). Characterization of two α-l-Arabinofuranosidases from Acetivibrio mesophilus and their synergistic effect in degradation of arabinose-containing substrates. Microorganisms 9:1467. doi: 10.3390/microorganisms9071467
Liu, Y., Tang, Y., Gao, H., Zhang, W., Jiang, Y., Xin, F., et al. (2021). Challenges and future perspectives of promising biotechnologies for lignocellulosic biorefinery. Mol. Basel Switz. 26:5411. doi: 10.3390/molecules26175411
López-Contreras, A. M., Gabor, K., Martens, A. A., Renckens, B. A., Claassen, P. A., van der Oost, J., et al. (2004). Substrate-induced production and secretion of cellulases by Clostridium acetobutylicum. Appl. Environ. Microbiol. 70, 5238–5243. doi: 10.1128/AEM.70.9.5238-5243.2004
López-Contreras, A. M., Martens, A. A., Szijarto, N., Mooibroek, H., Claassen, P. A. M., van der Oost, J., et al. (2003). Production by Clostridium acetobutylicum ATCC 824 of CelG, a cellulosomal glycoside hydrolase belonging to family 9. Appl. Environ. Microbiol. 69, 869–877. doi: 10.1128/AEM.69.2.869-877.2003
Louca, S., Mazel, F., Doebeli, M., and Parfrey, L. W. (2019). A census-based estimate of Earth’s bacterial and archaeal diversity. PLoS Biol. 17:e3000106. doi: 10.1371/journal.pbio.3000106
Lu, Y., Zhang, Y.-H. P., and Lynd, L. R. (2006). Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc. Natl. Acad. Sci. USA 103, 16165–16169. doi: 10.1073/pnas.0605381103
Mende, D. R., Letunic, I., Huerta-Cepas, J., Li, S. S., Forslund, K., Sunagawa, S., et al. (2017). proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res. 45, D529–D534. doi: 10.1093/nar/gkw989
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Monserrate, E., Leschine, S. B., and Canale-Parola, E. (2001). Clostridium hungatei sp. nov., a mesophilic, N2-fixing cellulolytic bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 51, 123–132. doi: 10.1099/00207713-51-1-123
Muñoz-Gutiérrez, I., Ortiz de Ora, L., Rozman Grinberg, I., Garty, Y., Bayer, E. A., Shoham, Y., et al. (2016). Decoding biomass-sensing regulons of Clostridium thermocellum alternative sigma-I factors in a heterologous Bacillus subtilis host system. PLoS One 11:e0146316. doi: 10.1371/journal.pone.0146316
Nam, N. N., Do, H. D. K., Loan Trinh, K. T., and Lee, N. Y. (2023). Metagenomics: an effective approach for exploring microbial diversity and functions. Foods Basel Switz. 12:2140. doi: 10.3390/foods12112140
Nataf, Y., Bahari, L., Kahel-Raifer, H., Borovok, I., Lamed, R., Bayer, E. A., et al. (2010). Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proc. Natl. Acad. Sci. USA 107, 18646–18651. doi: 10.1073/pnas.1012175107
Nishiyama, T., Ueki, A., Kaku, N., and Ueki, K. (2009). Clostridium sufflavum sp. nov., isolated from a methanogenic reactor treating cattle waste. Int. J. Syst. Evol. Microbiol. 59, 981–986. doi: 10.1099/ijs.0.001719-0
Nölling, J.̈., Breton, G., Omelchenko, M. V., Makarova, K. S., Zeng, Q., Gibson, R., et al. (2001). Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183, 4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Pagès, S., Bélaïch, A., Bélaïch, J. P., Morag, E., Lamed, R., Shoham, Y., et al. (1997). Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29, 517–527. doi: 10.1002/(SICI)1097-0134(199712)29:4<517::AID-PROT11>3.0.CO;2-P
Phitsuwan, P., Moraïs, S., Dassa, B., Henrissat, B., and Bayer, E. A. (2019). The Cellulosome paradigm in an extreme alkaline environment. Microorganisms 7:347. doi: 10.3390/microorganisms7090347
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Pradel, N., Fardeau, M.-L., Tindall, B. J., and Spring, S. (2020). Anaerohalosphaera lusitana gen. nov., sp. nov., and Limihaloglobus sulfuriphilus gen. nov., sp. nov., isolated from solar saltern sediments, and proposal of Anaerohalosphaeraceae fam. nov. within the order Sedimentisphaerales. Int. J. Syst. Evol. Microbiol. 70, 1321–1330. doi: 10.1099/ijsem.0.003919
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. doi: 10.1093/nar/gki442
Raman, B., Pan, C., Hurst, G. B., Rodriguez, M., McKeown, C. K., Lankford, P. K., et al. (2009). Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One 4:e5271. doi: 10.1371/journal.pone.0005271
Ren, Z., You, W., Wu, S., Poetsch, A., and Xu, C. (2019). Secretomic analyses of Ruminiclostridium papyrosolvens reveal its enzymatic basis for lignocellulose degradation. Biotechnol. Biofuels 12:183. doi: 10.1186/s13068-019-1522-8
Rettenmaier, R., Gerbaulet, M., Liebl, W., and Zverlov, V. V. (2019). Hungateiclostridium mesophilum sp. nov., a mesophilic, cellulolytic and spore-forming bacterium isolated from a biogas fermenter fed with maize silage. Int. J. Syst. Evol. Microbiol. 69, 3567–3573. doi: 10.1099/ijsem.0.003663
Rettenmaier, R., Kowollik, M.-L., Klingl, A., Liebl, W., and Zverlov, V. (2019). Ruminiclostridium herbifermentans sp. nov., a mesophilic and moderately thermophilic cellulolytic and xylanolytic bacterium isolated from a lab-scale biogas fermenter fed with maize silage. Int. J. Syst. Evol. Microbiol. 71:4692. doi: 10.1099/ijsem.0.004692
Rincon, M. T., Cepeljnik, T., Martin, J. C., Barak, Y., Lamed, R., Bayer, E. A., et al. (2007). A novel cell surface-anchored cellulose-binding protein encoded by the sca gene cluster of Ruminococcus flavefaciens. J. Bacteriol. 189, 4774–4783. doi: 10.1128/JB.00143-07
Rincon, M. T., Ding, S. Y., McCrae, S. I., Martin, J. C., Aurilia, V., Lamed, R., et al. (2003). Novel organization and divergent dockerin specificities in the cellulosome system of Ruminococcus flavefaciens. J. Bacteriol. 185, 703–713. doi: 10.1128/JB.185.3.703-713.2003
Rincón, M. T., Martin, J. C., Aurilia, V., McCrae, S. I., Rucklidge, G. J., Reid, M. D., et al. (2004). ScaC, an adaptor protein carrying a novel cohesin that expands the dockerin-binding repertoire of the Ruminococcus flavefaciens 17 cellulosome. J. Bacteriol. 186, 2576–2585. doi: 10.1128/JB.186.9.2576-2585.2004
Sabathé, F., Bélaïch, A., and Soucaille, P. (2002). Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 217, 15–22. doi: 10.1111/j.1574-6968.2002.tb11450.x
Salama-Alber, O., Gat, Y., Lamed, R., Shimon, L. J. W., Bayer, E. A., and Frolow, F. (2012). Crystallization and preliminary X-ray characterization of a type III cohesin-dockerin complex from the cellulosome system of Ruminococcus flavefaciens. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 68, 1116–1119. doi: 10.1107/S1744309112033088
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinforma. Oxf. Engl. 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sharma, A., Tewari, R., Rana, S. S., Soni, R., and Soni, S. K. (2016). Cellulases: classification, methods of determination and industrial applications. Appl. Biochem. Biotechnol. 179, 1346–1380. doi: 10.1007/s12010-016-2070-3
Shukla, A., Kumar, D., Girdhar, M., Kumar, A., Goyal, A., Malik, T., et al. (2023). Strategies of pretreatment of feedstocks for optimized bioethanol production: distinct and integrated approaches. Biotechnol. Biofuels Bioprod. 16:44. doi: 10.1186/s13068-023-02295-2
Sigrist, C. J. A., de Castro, E., Cerutti, L., Cuche, B. A., Hulo, N., Bridge, A., et al. (2013). New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347. doi: 10.1093/nar/gks1067
Smith, S. P., and Bayer, E. A. (2013). Insights into cellulosome assembly and dynamics: from dissection to reconstruction of the supramolecular enzyme complex. Curr. Opin. Struct. Biol. 23, 686–694. doi: 10.1016/j.sbi.2013.09.002
Smith, S. P., Bayer, E. A., and Czjzek, M. (2017). Continually emerging mechanistic complexity of the multi-enzyme cellulosome complex. Curr. Opin. Struct. Biol. 44, 151–160. doi: 10.1016/j.sbi.2017.03.009
Tao, X., Liu, J., Kempher, M. L., Xu, T., and Zhou, J. (2022). In vivo functional characterization of hydrophilic X2 modules in the cellulosomal scaffolding protein. Front. Microbiol. 13:861549. doi: 10.3389/fmicb.2022.861549
Tarraran, L., Gandini, C., Luganini, A., and Mazzoli, R. (2021). Cell-surface binding domains from Clostridium cellulovorans can be used for surface display of cellulosomal scaffoldins in Lactococcus lactis. Biotechnol. J. 16:e2100064. doi: 10.1002/biot.202100064
Teufel, F., Almagro Armenteros, J. J., Johansen, A. R., Gíslason, M. H., Pihl, S. I., Tsirigos, K. D., et al. (2022). SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025. doi: 10.1038/s41587-021-01156-3
Tindall, B. J. (2019). Replacement of the illegitimate genus name Hungateiclostridium Zhang et al. 2018 in Hungateiclostridium mesophilum Rettenmaier et al. 2019 by Acetivibrio Patel et al. 1980, creating Acetivibrio mesophilus (Rettenmaier et al. 2019). Int. J. Syst. Evol. Microbiol. 69, 3967–3968. doi: 10.1099/ijsem.0.003810
Tomazetto, G., Hahnke, S., Koeck, D. E., Wibberg, D., Maus, I., Pühler, A., et al. (2016). Complete genome analysis of Clostridium bornimense strain M2/40(T): a new acidogenic Clostridium species isolated from a mesophilic two-phase laboratory-scale biogas reactor. J. Biotechnol. 232, 38–49. doi: 10.1016/j.jbiotec.2015.08.001
Usai, G., Cirrincione, S., Re, A., Manfredi, M., Pagnani, A., Pessione, E., et al. (2020). Clostridium cellulovorans metabolism of cellulose as studied by comparative proteomic approach. J. Proteome 216:103667. doi: 10.1016/j.jprot.2020.103667
Wang, J., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, S., et al. (2023). The conserved domain database in 2023. Nucleic Acids Res. 51, D384–D388. doi: 10.1093/nar/gkac1096
Wang, Y., Li, L., Xia, Y., and Zhang, T. (2022). Reliable and scalable identification and prioritization of putative cellulolytic anaerobes with large genome data. Front. Bioinform. 2:813771. doi: 10.3389/fbinf.2022.813771
Wang, Y., Zhang, Y., Cui, Q., Feng, Y., and Xuan, J. (2024). Composition of lignocellulose hydrolysate in different biorefinery strategies: nutrients and inhibitors. Mol. Basel Switz. 29:2275. doi: 10.3390/molecules29102275
Wylensek, D., Hitch, T. C. A., Riedel, T., Afrizal, A., Kumar, N., Wortmann, E., et al. (2020). A collection of bacterial isolates from the pig intestine reveals functional and taxonomic diversity. Nat. Commun. 11:6389. doi: 10.1038/s41467-020-19929-w
Xu, Q., Barak, Y., Kenig, R., Shoham, Y., Bayer, E. A., and Lamed, R. (2004). A novel Acetivibrio cellulolyticus anchoring scaffoldin that bears divergent cohesins. J. Bacteriol. 186, 5782–5789. doi: 10.1128/JB.186.17.5782-5789.2004
Xu, Q., Bayer, E. A., Goldman, M., Kenig, R., Shoham, Y., and Lamed, R. (2004). Architecture of the Bacteroides cellulosolvens cellulosome: description of a cell surface-anchoring scaffoldin and a family 48 cellulase. J. Bacteriol. 186, 968–977. doi: 10.1128/JB.186.4.968-977.2004
Xu, C., Huang, R., Teng, L., Jing, X., Hu, J., Cui, G., et al. (2015). Cellulosome stoichiometry in Clostridium cellulolyticum is regulated by selective RNA processing and stabilization. Nat. Commun. 6:6900. doi: 10.1038/ncomms7900
You, M., Zhao, Q., Liu, Y., Zhang, W., Shen, Z., Ren, Z., et al. (2023). Insights into lignocellulose degradation: comparative genomics of anaerobic and cellulolytic Ruminiclostridium-type species. Front. Microbiol. 14:1288286. doi: 10.3389/fmicb.2023.1288286
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinforma. Oxf. Engl. 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Ze, X., Ben David, Y., Laverde-Gomez, J. A., Dassa, B., Sheridan, P. O., Duncan, S. H., et al. (2015). Unique Organization of Extracellular amylases into amylosomes in the resistant starch-utilizing human colonic firmicutes bacterium Ruminococcus bromii. MBio 6, e01058–e01015. doi: 10.1128/mBio.01058-15
Zhang, Z., Wang, J., Wang, J., Wang, J., and Li, Y. (2020). Estimate of the sequenced proportion of the global prokaryotic genome. Microbiome 8:134. doi: 10.1186/s40168-020-00903-z
Zhang, J., Zhang, Y., Liu, R., Cai, R., Liu, F., and Sun, C. (2021). Iocasia fonsfrigidae NS-1 gen. nov., sp. nov., a novel deep-sea bacterium possessing diverse carbohydrate metabolic pathways. Front. Microbiol. 12:725159. doi: 10.3389/fmicb.2021.725159
Zhivin, O., Dassa, B., Moraïs, S., Utturkar, S. M., Brown, S. D., Henrissat, B., et al. (2017). Unique organization and unprecedented diversity of the Bacteroides (Pseudobacteroides) cellulosolvens cellulosome system. Biotechnol. Biofuels 10:211. doi: 10.1186/s13068-017-0898-6
Keywords: Cellulosome, biomass, lignocellulose, comparative genomics, cohesin, dockerin, AlphaFold2
Citation: Minor CM, Takayesu A, Ha SM, Salwinski L, Sawaya MR, Pellegrini M and Clubb RT (2024) A genomic analysis reveals the diversity of cellulosome displaying bacteria. Front. Microbiol. 15:1473396. doi: 10.3389/fmicb.2024.1473396
Received: 30 July 2024; Accepted: 04 October 2024;
Published: 30 October 2024.
Edited by:
Anandharaj Marimuthu, Berkeley Lab (DOE), United StatesReviewed by:
Eliane Ferreira Noronha, University of Brasilia, BrazilCopyright © 2024 Minor, Takayesu, Ha, Salwinski, Sawaya, Pellegrini and Clubb. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert T. Clubb, cmNsdWJiQG1iaS51Y2xhLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.