- RNA Biology Laboratory, Department of Chemistry, Indian Institute of Technology Delhi, New Delhi, India
The efficacy of the tuberculosis treatment is restricted by innate drug resistance of Mycobacterial tuberculosis and its ability to acquire resistance to all anti-tuberculosis drugs in clinical use. A profound understanding of bacterial ploys that decrease the effectiveness of drugs would identify new mechanisms for drug resistance, which would subsequently lead to the development of more potent TB therapies. In the current study, we identified a virulence-associated small RNA (sRNA) MTS1338-driven drug efflux mechanism in M. tuberculosis. The treatment of a frontline antitubercular drug rifampicin upregulated MTS1338 by >4-fold. Higher intrabacterial abundance of MTS1338 increased the growth rate of cells in rifampicin-treated conditions. This fact was attributed by the upregulation of an efflux protein CydC by MTS1338. Gel-shift assay identified a stable interaction of MTS1338 with the coding region of cydC mRNA thereby potentially stabilizing it at the posttranscriptional level. The drug efflux measurement assays revealed that cells with higher MTS1338 abundance accumulate less drug in the cells. This study identified a new regulatory mechanism of drug efflux controlled by an infection-induced sRNA in M. tuberculosis.
1 Introduction
Persistence of Mycobacterial tuberculosis, the aetiological agent of tuberculosis (TB), in the hostile environment of host macrophages relies on fast and precise reprogramming of transcriptional profile in response to external stimuli (Miotto et al., 2022; Arnvig and Young, 2012; Dutta and Srivastava, 2018; Taneja and Dutta, 2019). An array of complex regulatory networks controlled by transcriptional modulators, two-component systems, sigma factors, riboswitches, and small RNAs (sRNAs) fine-tune the mycobacterial gene expression for adaptation to external challenges in real-time (Miotto et al., 2022; Taneja and Dutta, 2019; Smith et al., 2013; Petchiappan and Chatterji, 2017), which render them notoriously difficult to eliminate from infected patients. Consequently, prolonged treatment with frequent administration of multiple antibiotics is required to treat TB (Ginsberg and Spigelman, 2007; Dhar and McKinney, 2010; van den Boogaard et al., 2009). M. tuberculosis exploits a plethora of intrinsic mechanisms to combat anti-TB drug therapy (Remm et al., 2022). Its complex hydrophobic cell envelope is impermeable to many drugs and is considered as one of the key features of defense against drug influx (Sarathy et al., 2012). M. tuberculosis possesses additional machinery for drug tolerance, which includes multiple efflux pumps that deport antimycobacterial compounds out of the cells, mycobacterial enzymes capable of chemical modification of antibiotics, and alterations in gene expression to adapt to antibiotic-exposed conditions (Briffotaux et al., 2019; Nasiri et al., 2017). Antibiotics also lead to the development of acquired M. tuberculosis resistance against it through target mutations (Takiff and Feo, 2015), which is associated with the reduced fitness of the resistant mutants (Andersson and Hughes, 2010). Because of the above-mentioned capabilities of M. tuberculosis, illegitimate drug usage by patients frequently causes a high degree of treatment failure leading to disease relapse and emergent drug resistance (Dhar and McKinney, 2010; van den Boogaard et al., 2009) triggering onward transmission and amplification of drug-resistant strain (Barilar and Battaglia, 2024).
Traditional antimycobacterial compounds interfere with the biosynthetic processes, e.g., biogenesis of cell wall, transcription, and protein synthesis, which are necessitated for cell growth (Dhar and McKinney, 2010). A few of these antibiotics like isoniazid, ethambutol, etc. for inhibition of cell wall formation, rifampicin for interference with transcription, and streptomycin for the inhibition of protein synthesis of M. tuberculosis are regarded as the frontline anti-TB drugs (Briffotaux et al., 2019). M. tuberculosis cells although highly susceptible to death in the presence of these antibiotics in vitro, are less affected by the same antimycobacterial agents while grown in vivo in mammalian hosts (Dhar and McKinney, 2010; Young et al., 2008; McKinney, 2000). Therefore, chronic TB is more difficult to control through a single antibiotic therapy and calls for second-line regimens. Patient nonadherence to such an extended therapeutical procedure, insufficient health infrastructure, and lack of antibiotics help promote the development of resistant strains of exceedingly slow-growing M. tuberculosis (Briffotaux et al., 2019). Hence, multidrug-resistant TB (MDR-TB) and extremely drug-resistant TB (XDR-TB) strains have evolved (Almeida Da Silva and Palomino, 2011; Udwadia et al., 2012) which accentuates the requirement for new strategies to develop novel anti-TB drugs. For that reason, it is essential to comprehensively understand the regulatory networks promoting resistance in M. tuberculosis against antibiotics.
Although sRNAs have been identified to be a ubiquitous class of regulators contributing to antibacterial resistance in pathogenic bacteria (Mediati et al., 2021), sRNA-mediated regulation of antibiotic resistance in M. tuberculosis has not been investigated yet. Only a handful of sRNAs among a few dozen discovered in M. tuberculosis until now have been identified with associated functions given the difficulty of understanding sRNA function in vivo in M. tuberculosis (Dutta and Srivastava, 2018; Arnvig et al., 2011; Singh et al., 2021). MTS1338 (or ncRv11733) is one among those sRNAs, which exists only in the pathogenic mycobacterial genome (Dutta and Srivastava, 2018; Arnvig et al., 2011; Moores et al., 2017), and is highly accumulated during M. tuberculosis infection (Arnvig et al., 2011). Consequently, MTS1338 is highly upregulated in response to stresses associated with infection (Singh et al., 2021; Salina et al., 2019) and configures a stress-resistance signature in M. tuberculosis (Martini et al., 2023). Substantial alteration of the transcriptomic profile of M. tuberculosis overexpressing MTS1338 suggests its potential role in the global gene regulatory network, which promotes the survival of M. tuberculosis in host macrophages through an adaptive mechanism (Martini et al., 2023). Participation of MTS1338 in the regulation of numerous physiological processes firmly indicated that MTS1338 might play an important role in antimycobacterial resistance.
In the current study, we identified that MTS1338 is highly induced while the cells are exposed to antibiotic like rifampicin. Cells with higher intracellular MTS1338 accumulation were ascertained to grow at a higher rate than the cells with basal MTS1338 level. Investigations on how MTS1338 in its higher abundance promotes M. tuberculosis growth affirmed that MTS1338 increases the abundance of the mRNA of an efflux protein CydC by binding to its coding region potentially protecting it at the posttranscriptional stage from cellular endoribonucleases. Inhibition of efflux proteins of the cells undergone a prior treatment with rifampicin resulted in a better mycobacterial growth rate for the cells overexpressing MTS1338. A higher abundance of intracellular MTS1338 resulted in a lower accumulation of antibiotics in the cell suggesting a potential role of this sRNA in dealing with mycobacterial adaptation to antibiotics.
2 Materials and methods
2.1 Materials
Middlebrook 7H11 agar, Middlebrook 7H9 broth, and OADC (oleic acid-albumin-dextrose-catalase) were purchased from Difco Laboratories (United States). Tween 80, lysozyme, and antibiotics (rifampicin, tetracycline, kanamycin) were procured from Sigma-Aldrich. RNeasy Mini Kit and UniPro GelEx/clean-up kit were obtained from Qiagen (Germany). Genomic DNA extraction kit, GoTaq green master mix, and nuclease-free water were from Promega. Oligonucleotides were synthesized by Sigma Chemical Co. (United States). SYBR green master mix and iScript™ cDNA synthesis kit were procured from Bio-Rad (Bio-Rad). Restriction enzymes, e.g., BamHI and HindIII were obtained from New England Biolabs. MEGAshortscript™ T7 transcription kit and Max™ Prehyb/hyb buffer solution were purchased from Invitrogen. Hybond-N+ nylon membrane was purchased from GE Healthcare. carbonyl cyanide m-chlorophenylhydrazone (CCCP) was purchased from Sigma Chemical Co. (United States). Plasmid pST-Ki was a generous gift from Dr. V. K. Nandicoori of the National Institute of Immunology, India. All other chemicals were of molecular biology grade.
2.2 Bacterial strain and growth conditions
The M. tuberculosis avirulent strain H37Ra was used as a model strain. The frozen glycerol stock was streaked on a Middlebrook 7H11 solid agar plate and stored at 37°C for 4–5 weeks. In a culture tube, one colony from the growing plate was inoculated with Middlebrook 7H9 broth (Difco Laboratories) supplemented with 10% OADC (oleic acid-albumin-dextrose-catalase; Difco Laboratories) and 0.05% (v/v) polysorbate 80 (Sigma-Aldrich). Subsequently, the cells were incubated at 37°C under shaking at 200 rpm. Cell growth while attaining OD600 0.6–0.9, was regarded as the exponential growth phase of M. tuberculosis.
2.3 Antibiotic treatment
M. tuberculosis H37Ra cells were grown at 37°C in an orbital shaker incubator at 200 rpm until OD600 of the culture reached 0.9. The cells were then harvested at room temperature. After that, the cells were again suspended in 7H9 media and treated with rifampicin (2 μg/mL) for 30 min. Cells cultured under identical conditions without the addition of antibiotics were taken as control.
2.4 RNA isolation and RT-qPCR
Cell pellets with or without prior antibiotic treatment were re-suspended in 4 mL of PBS containing lysozyme (4 mg/mL). Lysozyme treatment was continued for 2 h at 37°C in a water bath with circulation. After that, cells were sonicated for 90 s (with alternate 10 s on or off pulse) using a Branson sonicator at 4°C. Total RNA was extracted using RNeasy Mini Kit (Qiagen). RNA concentration was measured using a JENWAY Genova Nanodrop spectrophotometer and RNA quality was assessed in a 1% agarose gel.
Total RNA (1 μg) from treated/untreated cells was used to synthesize cDNA using iScriptTM cDNA synthesis kit (Bio-Rad), along with random hexamer (Bio-Rad C1000, United States) as a primer. Complementary DNAs were diluted 100 times with nuclease-free water and 1 μL of diluted cDNA was employed in a quantitative real-time PCR reaction using SYBR green utilizing a CFX96 Touch™ Real-Time PCR detection system (Bio-Rad). A representative qPCR reaction of 20 μL contains 20 ng of cDNA, 1 μM of forward and reverse primers (P1 & P2 for MTS1338, P5 & P6 for 5S RNA, P7 & P8 for cydC mRNA in Supplementary Table S1), 10 μL of SYBR green master mix, and 8 μL of nuclease-free water mix. 5S RNA gene of M. tuberculosis was taken as an internal control. The 2-ΔΔCt technique was used to calculate relative gene expression. Every experiment was carried out in compliance with the manufacturer’s guidelines.
2.5 Northern blot
Total RNA (15 μg), isolated from untreated or treated M. tuberculosis cells, were separated on 2% agarose-formaldehyde gel using 1X MOPS [3-(N-morpholino) propane sulfonic acid] buffer, which was then washed with deionized water. RNA was transferred from agarose gel to Hybond-N+ nylon membrane (GE Healthcare) by downward capillary transfer in 10X SSC buffer at room temperature overnight and UV crosslinked to the membrane by UV crosslinker (Herolab, Germany). A 5′-Cy5 labeled DNA probe (P11 for MTS1338 and P12 for 5S RNA in Supplementary Table S1) was used to detect MTS1338. Hybridization (Shake ‘n’ Stack™ Hybridization Chamber, Thermo Fisher Scientific, United States) of Cy5-labeled probe to crosslinked RNA on the membrane was promoted by incubating them in Northern Max™ Prehyb/hyb buffer solution (Invitrogen) overnight. The membrane was subsequently washed with pre-warmed SSC buffer three times and the bands were visualized by high-resolution gel imaging for the fluorescence system (G:Box Chemi-XX9, SYNGENE). Bands were quantitated by ImageJ.
2.6 Cloning and overexpression of MTS1338
The genomic DNA of M. tuberculosis H37Ra cells was extracted using the Genomic DNA Extraction Kit (Promega Co., United States). The gene encoding MTS1338 was amplified from genomic DNA in a PCR reaction using KOD hot start polymerase (Invitrogen) with primers P3 and P4 listed in Supplementary Table S1. The amplicon was purified by gel extraction (Qiagen). Both the PCR products and the shuttle vector pST-Ki were digested with BamHI and HindIII for 2 h at 37°C. Digested products were purified through phenol: chloroform: isoamyl alcohol (25, 24:1) extraction and were ligated by T4 DNA ligase (New Eng Biolab). The resulting recombinant plasmids (pMTS1338) were transformed into Escherichia coli DH5α cells, which were grown on an LB (Kan+) plate overnight at 37°C. Cloning of gene encoding MTS1338 under the control of tetracycline promoter was confirmed by DNA sequencing. Recombinant pMTS1338 from E. coli cells was electroporated into M. tuberculosis H37Ra cells and subsequent MTS1338 overexpression was monitored upon the addition of tetracycline (1 μg/mL) for 72 h.
2.7 Growth analysis
The 7H9 medium supplemented with 10% OADC and 0.05% Tween 80 was used to grow the mycobacterial bacilli. Carbonyl cyanide 3-chlorophenylhydrazone (4 μg/mL) was added to the cells 5 days after the cells (OD600-0.9) were treated with rifampicin (2 μg/mL). The cells were continued to grow for 16 days under shaking (200 rpm) at 37°C. Growth was monitored by measuring the optical density of the culture at 600 nm every 24 h with a UV spectrophotometer (JASCO).
2.8 RNA decay
Overnight cultures were diluted 1:100 into fresh 7H9 medium supplemented with 10% OADC and 0.05% Tween 80 and grown to exponential phase (OD600-0.9) followed by treatment with 500 μg/mL rifampicin (Chen et al., 2019). Equal volumes of cells were removed for the zero-time sample after 1 min of rifampicin addition to allow complete inhibition of new transcription and immediately mixed with ice-cold stop solution (95% ethanol, 5% phenol). Additional samples were removed at indicated time points in the figure legends. Changes in cydC mRNA accumulation were determined by RT-qPCR analysis.
2.9 In vitro transcription
Genomic DNA of M. tuberculosis H37Ra cells was used as a template for the synthesis of genes of CydC and MTS1338 by PCR amplification using primers (P8 & P14 for cydC, P2 & P13 for MTS1338 and P10 & P11 for katG mRNA) listed in Supplementary Table S1. The forward primer was fused with a promoter sequence of T7 RNA polymerase. Templates were gel extracted using a QIAquick® gel extraction kit. Each template of 100 nM was added in an in vitro transcription reaction using a MEGAshortscript™ T7 transcription kit. Transcripts were extracted with phenol: chloroform: isoamyl alcohol (25: 24: 1) and precipitation overnight with ethanol at-20°C. The concentration of RNA was estimated spectrophotometrically by NanoDrop® spectrophotometer (Maestrogen, Taiwan) and RNA quality was checked on 6% polyacrylamide, 7.5 M urea gel.
2.10 Electrophoretic mobility shift assay
Transcripts of MTS1338 and cydC mRNA fragment (916–1,030 residues) were denatured at 80°C for 10 min and then cooled on ice. Binding reactions (10 μL) were conducted by incubating cydC mRNA (10 nM) with increasing amounts of MTS1338 (0–8 μM) at 37°C for 30 min in 1X binding buffer [100 mM Tris acetate pH 7.6, 500 mM NaOAc, and 25 mM Mg (OAc)2]. Aliquots of each binding reaction were separated in an agarose gel (2%) using 1X MOPS buffer at 4°C. Gel was stained with 1X SYBR™ Gold Nucleic Acid Gel Stain (Invitrogen) and bands were visualized by high-resolution gel imaging for the fluorescence system (G: Box Chemi-XX9, SYNGENE).
2.11 EtBr accumulation assay
H37Ra cells harboring empty pST-Ki or recombinant pMTS1338 plasmid were grown in 7H9 medium supplemented with 10% OADC until the mid-exponential phase (OD600-0.9). Cells were then treated with tetracycline (1 μg/mL) for 72 h for the extrachromosomal induction of MTS1338 followed by the addition of EtBr (1 μg/mL). After 10 min of EtBr exposure at 37°C, cells were treated with an efflux pump inhibitor CCCP with a final concentration of 6.25 μM. Cells were then pelleted down by centrifugation at 3500 g for 5 min and then resuspended in 1 mL of Phosphate buffer saline with/without CCCP. Intracellular accumulation of EtBr was measured in situ by fluorimetry with excitation at 520 nm and emission at 590 ± 20 nm. Fluorescence was monitored every 20 min, and the measurements were continued for 60 min.
2.12 Statistical analysis
Data obtained from experimental studies were presented as mean ± standard deviation. One-way analysis of variance was conducted, and a difference of p < 0.05 was considered statistically significant.
3 Results
3.1 MTS1338 is upregulated following rifampicin treatment
H37Ra cells were used as a model strain for our experiments as both strains of M. tuberculosis, H37Ra, and H37Rv exhibit equivalent minimum inhibitory concentrations to most antituberculosis drugs (Heinrichs et al., 2018). Given the induction of MTS1338 in response to multiple infection-associated stresses and its regulation to remodel the global gene expression of M. tuberculosis, it insinuated that MTS1338 might play some role in mycobacterial adaptation to antibiotic treatment. To address that hypothesis M. tuberculosis H37Ra cells were grown in Middlebrook 7H9 broth supplemented with 10% OADC and 0.05% (v/v) polysorbate 80, until OD600 reached 0.9 and then treated with rifampicin (2 mg/mL), a frontline antimycobacterial drug, for 30 min at 37°C under shaking condition. The concentration of rifampicin was used in a similar experiment as described (Lake et al., 2023). Another M. tuberculosis culture identically treated except for rifampicin was considered as a control. MTS1338 abundance in the total RNA isolated from rifampicin-treated and untreated cells was measured by RT-qPCR analysis. Figure 1A clearly shows that cells accumulate as much as ~5-fold more MTS1338 in 30 min following rifampicin treatment. In addition, northern blotting for MTS1338 in the total RNA isolated from rifampicin-treated or control cells revealed a 4-fold more accumulation of MTS1338 in rifampicin-treated cells (Figures 1B,C) than in the control untreated cells. These observations lead to the conclusion that MTS1338 is a rifampicin-induced sRNA that is potentially involved in adaptation to rifampicin-treated conditions.
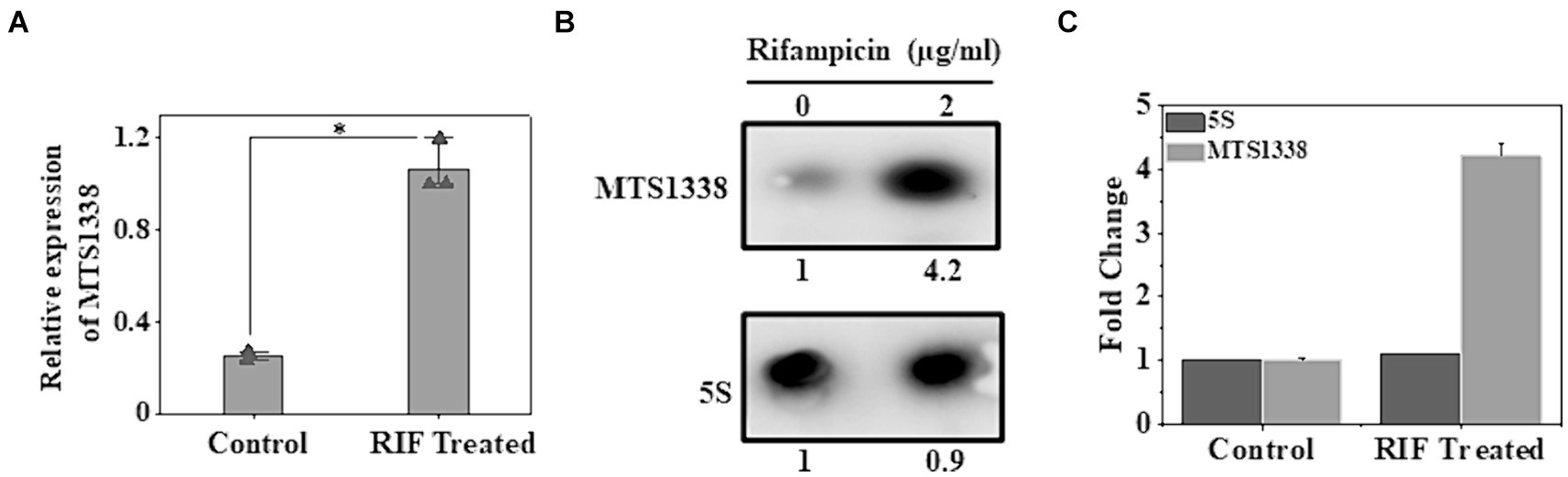
Figure 1. Effect of rifampicin (A) treatment on the intracellular abundance of MTS1338 in Mycobacterium tuberculosis cells. Mid-exponentially grown cells (OD600–0.9) were exposed to rifampicin (2 μg/mL) and then were harvested. Total RNA was isolated and MTS1338 abundance was estimated by RT-qPCR analysis. 5S gene was taken as an internal control. The relative amounts of the transcripts are presented as the average ± standard deviation (SD) from three separate experiments and normalized to the level of the untreated sample. One-way ANOVA was conducted and a difference of *p < 0.05 was considered statistically significant. (B) Total RNA (15 μg) isolated from the cells treated with/without rifampicin (2 μg/mL) were separated in a 2% agarose formaldehyde gel and analyzed by northern blot. The number under each lane represents the relative amount of MTS1338 when compared to untreated sample, which was set as 1. 5S rRNA gene was taken as a loading control. Northern blots were carried out three times and one representative blot is presented. (C) Northern blot experiments were carried out three times with 5′-Cy5 labeled DNA oligonucleotide probes for RNAs. Bands were quantitated by ImageJ. The amount of MTS1338 and 5S RNA in the untreated cell was set at 1.
3.2 Enhanced intracellular MTS1338 abundance increases the growth rate in the presence of rifampicin
As MTS1338 accumulates to a higher degree following antibiotic treatment on M. tuberculosis cells, it was of immense interest to investigate whether overexpression of MTS1338 ameliorates the growth rate of the cells. To address that question, the gene encoding MTS1338 was cloned in the pST-Ki shuttle vector under the control of Ptet promoter, and the recombinant plasmid thus obtained (pMTS1338) was electroporated in M. tuberculosis H37Ra cells. Overexpression of MTS1338 was monitored by adding tetracycline (1 μg/mL) to the culture media for 72 h. Estimation of intracellular MTS1338 abundance upon addition of tetracycline resulted in ~3.5-fold more accumulation of MTS1338 when compared to the control cells analyzed by RT-qPCR (Figure 2A). Northern blot analysis (Figure 2B) of MTS1338 in the total RNA isolated from the cells with/without MTS1338 overexpression also substantiated ~4-fold more accumulation of MTS1338 in the cells harboring pMTS1338 plasmid (Figure 2C).
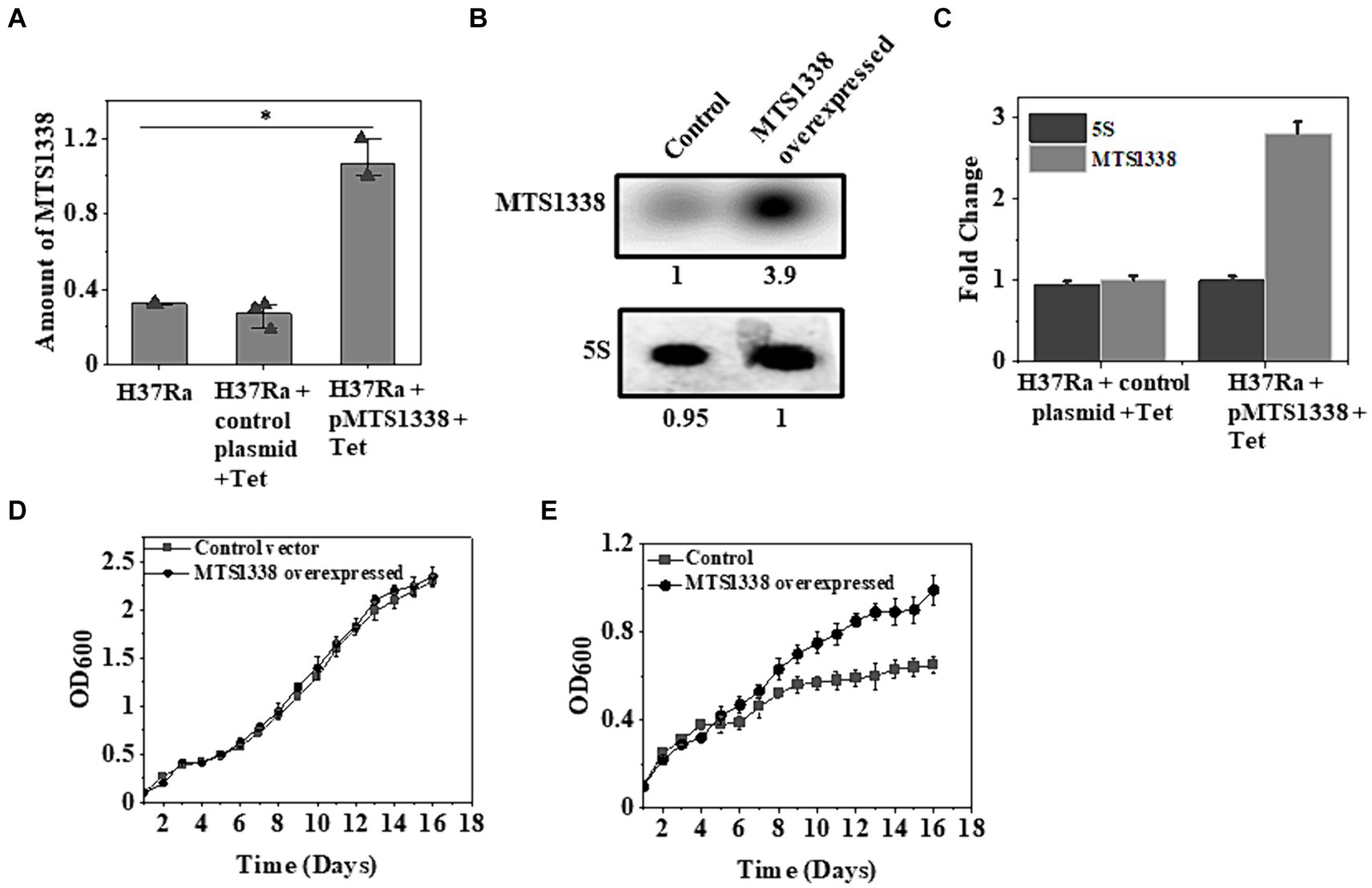
Figure 2. A higher MTS1338 abundance increases the growth rate in the presence of rifampicin. (A) Tetracycline (1 μg/mL) was added to the exponentially grown M. tuberculosis cells (OD600 = 0.9) harboring empty or recombinant pMTS1338 plasmids. Cells continued to grow for 72 h and the total RNA was extracted from each sample. MTS1338 abundance was estimated by RT-qPCR analysis. 5S gene was taken as an internal control. The relative amounts of the transcripts are presented as the average ± SD from three separate experiments and normalized to the level of the untreated sample. One-way ANOVA was conducted and a difference of *p < 0.05 was considered statistically significant. (B) Northern blot was carried out using 15 μg of total RNA isolated from the cells with/without overexpressing MTS1338. 5S RNA gene was taken as loading control. Northern blot experiments were carried out three times and identical results were obtained. A representative of Northern blots was presented. The number under overexpressed lane represents the relative amount of RNA when compared with control lane, which was set 1.0. (C) Bands of three independent northern blots were quantitated by ImageJ. Data were presented as the average ± SD. The amount of MTS1338 and 5S RNA in the untreated cell was set at 1. (D) H37Ra cells harboring recombinant pMTS1338 plasmid or empty vector were grown at 37°C. OD600 was monitored in regular interval and was plotted against time. The data presented was the average ± S.D. of three independent experiments. Growth experiments were also carried out in presence of rifampicin (5 μg/mL) added at the day 1 (E).
To examine whether higher MTS1338 abundance affects cell growth or not, we monitored the growth of cells with/without extrachromosomal MTS1338 overexpression and found that overexpression of MTS1338 in H37Ra cells under normal conditions did not show any alteration of growth rate (Figure 2D). This fact indicated that a higher accumulation of MTS1338 is not detrimental to cell viability. However, growth analysis in the presence of rifampicin revealed a significant difference in the growth rate of the cells with or without MTS1338 overexpression (Figure 2E). H37Ra cells overexpressing MTS1338 appeared to grow at a higher rate with a generation time of 6 days than the cells containing empty vector (generation time 9 days). This firmly established that MTS1338 promotes cell growth in the presence of first-line antibiotic rifampicin.
3.3 Elevated MTS1338 induces the gene encoding efflux protein CydC
MTS1338-driven increased cellular robustness in opposition to rifampicin apprised us to investigate how MTS1338 acts to relieve the antagonistic effect created by rifampicin. To address that question, we aimed to examine the expression of genes encoding ABC transporters since it is one of the most predominant intrinsic mechanisms by which M. tuberculosis can ride out antibiotic treatment is drug efflux (Remm et al., 2022). The mycobacterial cydC gene, transcribed from the cydABDC gene cluster, encodes an efflux protein. The absence of CydC in M. tuberculosis alleviates its ability to survive the transition from acute to chronic infection. Expression of cydABDC has also been shown to be induced by hypoxia and NO stress (Shi et al., 2005). We selected cydC gene for our investigation of whether MTS1338 exerts any effect on cydDC expression as in many instances cydDC transporters are recognized to help persist M. tuberculosis against drugs like isoniazid (8). To settle that hypothesis, we compared the expression of cydC gene in H37Ra cells harboring either pMTS1338 or empty pST-Ki vectors and found >3-fold induction of cydC in MTS1338 overexpressing cells (Figure 3A). Northern blot analysis (Figure 3B) for the determination of cydC accumulation revealed that the cells with higher MTS1338 abundance upregulated cydC transcripts by >3-fold (Figure 3C), which reinforced RT-qPCR data. These experiments pointed to the involvement of MTS1338 in inducing the cydC gene.
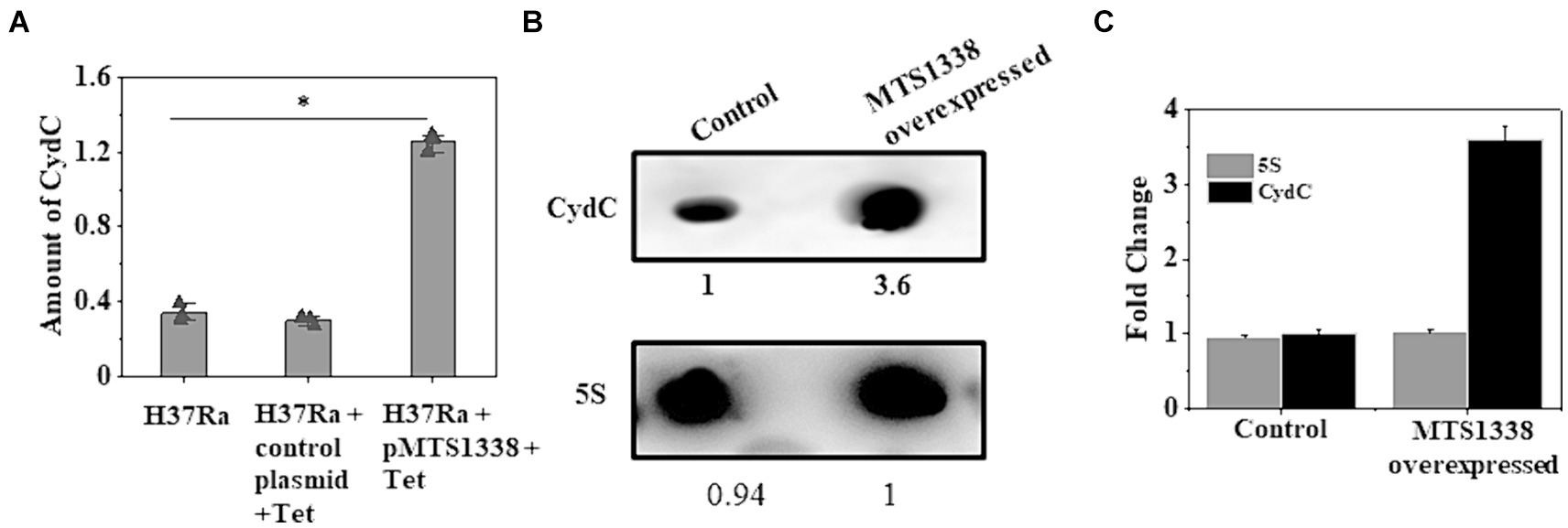
Figure 3. (A) Relative expression of cydC mRNA was estimated by RT-qPCR analysis in the exponentially grown M. tuberculosis cells (OD600 = 0.9) harboring control or recombinant pMTS1338 vectors. 5S gene was taken as an internal control. The relative amounts of cydC mRNA are presented as the average ± standard deviation from three separate experiments and normalized to the level of the untreated sample. One-way ANOVA was conducted and a difference of *p < 0.05 was considered statistically significant. (B) Northern blot was carried out using 5’-Cy5-labeled DNA probe to detect cydC mRNA present in the total RNA isolated from the cells with/without overexpressing MTS1338. 5S was taken as a loading control. A representative of three independent northern blots with identical results was presented. The number under each lane represents the relative amount of cydC when compared with control. (C) Bands of three independent northern blots were quantitated by ImageJ. Data were presented as the average ± SD. The amount of cydC and 5S RNA in the control cell was set at 1.
Upregulation of cydC in response to higher intracellular MTS1338 abundance and enhanced accumulation of MTS1338 following antibiotic treatment instigated us to investigate whether cydC is also upregulated in response to antibiotic treatment. To examine that objective, we compared the intracellular cydC levels in the cells treated with/without rifampicin by RT-qPCR and northern blot analysis. It is conspicuous from Figure 4A that cydC accumulation analyzed through RT-qPCR was induced by ~4-fold following rifampicin treatment. An analogous result of rifampicin-driven cydC induction was obtained through northern blot analysis (Figures 4B,C). Investigation of the expression of cydD, which encodes a different efflux protein, revealed that rifampicin treatment did not show any significant effect on its intracellular abundance in M. tuberculosis (Singh and Dutta, unpublished observation). These experiments firmly established that MTS1338 has a profound effect in maintaining intracellular cydC levels against antibiotic treatment.
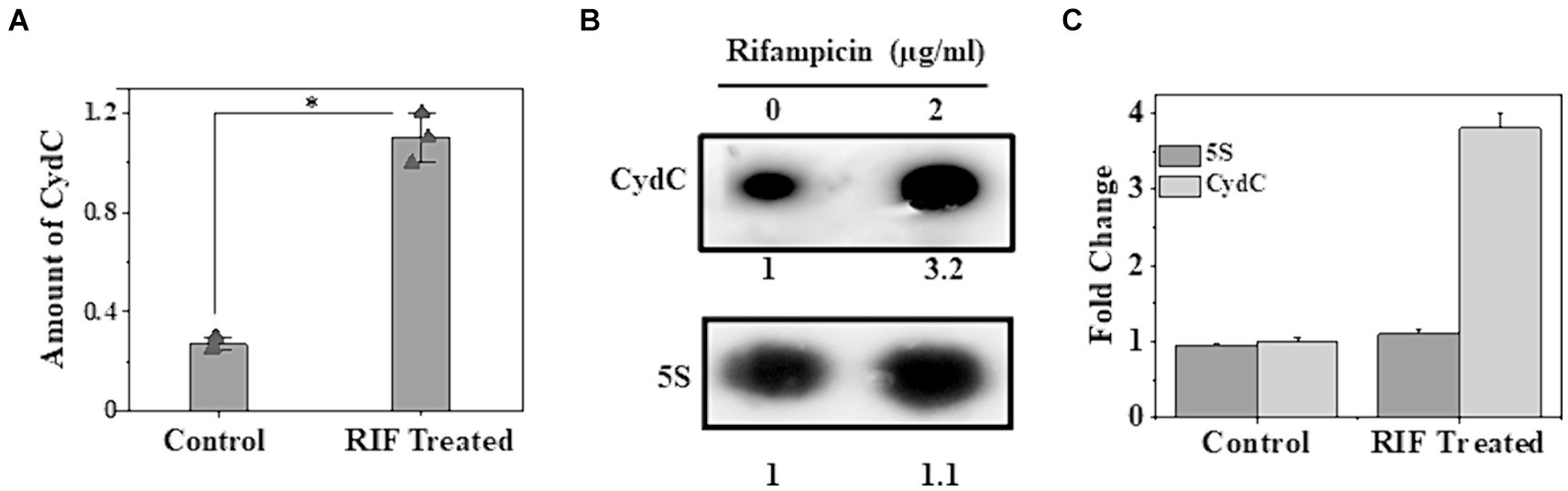
Figure 4. Accumulation of cydC in M. tuberculosis while treated with rifampicin (2 μg/mL). (A) Mid-log phase H37Ra cells (OD600 = 0.9) were exposed to antibiotics for 30 min at 37°C. Relative amount of cydC in the total RNA was measured by RT-qPCR analysis. 5S gene was taken as an internal control. The relative amounts of cydC mRNA are presented as the average ± standard deviation from three separate experiments and normalized to the level of the untreated sample. One-way ANOVA was conducted and a difference of *p < 0.05 was considered statistically significant. (B) Total RNA (15 μg) from rifampicin treated and untreated samples were separated in in a 2% agarose formaldehyde gel and analyzed by northern blot. The number under each lane represents the relative amount of cydC when compared to untreated sample, which was set as 1. 5S RNA gene was taken as a loading control. Northern blots were carried out three times and one representative blot is presented. (C) Northern blot experiments were carried out three times with 5′-Cy5 labeled DNA oligonucleotide probes for RNAs. Bands were quantitated by ImageJ. The amount of MTS1338 and 5S RNA in the untreated cell was set at 1.
3.4 MTS1338 directly interacts with the coding region of cydC mRNA
MTS1338-mediated upregulation of cydC mRNA brought up the question of whether MTS1338 regulates cydC abundance by directly interacting with it. To address that question, both cydC mRNA and MTS1338 sequences were used in the IntaRNA (Mann et al., 2017) to identify if any complementary region(s) exists between them. A region of 15-nt on cydC mRNA (966–980 residues) was found to be completely complementary to an intermediate portion (56–70 residues) of MTS1338 (Figure 5A). To confirm that hypothesis, both MTS1338 RNA and cydC mRNA fragments surrounding residues 916 to 1,030 that possesses region complementary to MTS1338 were transcribed and purified in vitro. Incubation of MTS1338 with cydC fragment resulted in the formation of cydC-MTS1338 binary complex, which was observed as a high molecular weight band generated in the gel-shift assay (Figure 5B). Addition of increasing amounts of MTS1338 to a constant amount of cydC mRNA gave rise to a concomitant increase in the population of cydC-MTS1338 binary complex. This experiment confirmed that MTS1338 directly interacts with the coding region of cydC mRNA potentially stabilizing it at the posttranscriptional level. A control experiment with katG mRNA did not show any interaction with MTS1338 (Figure 5C).
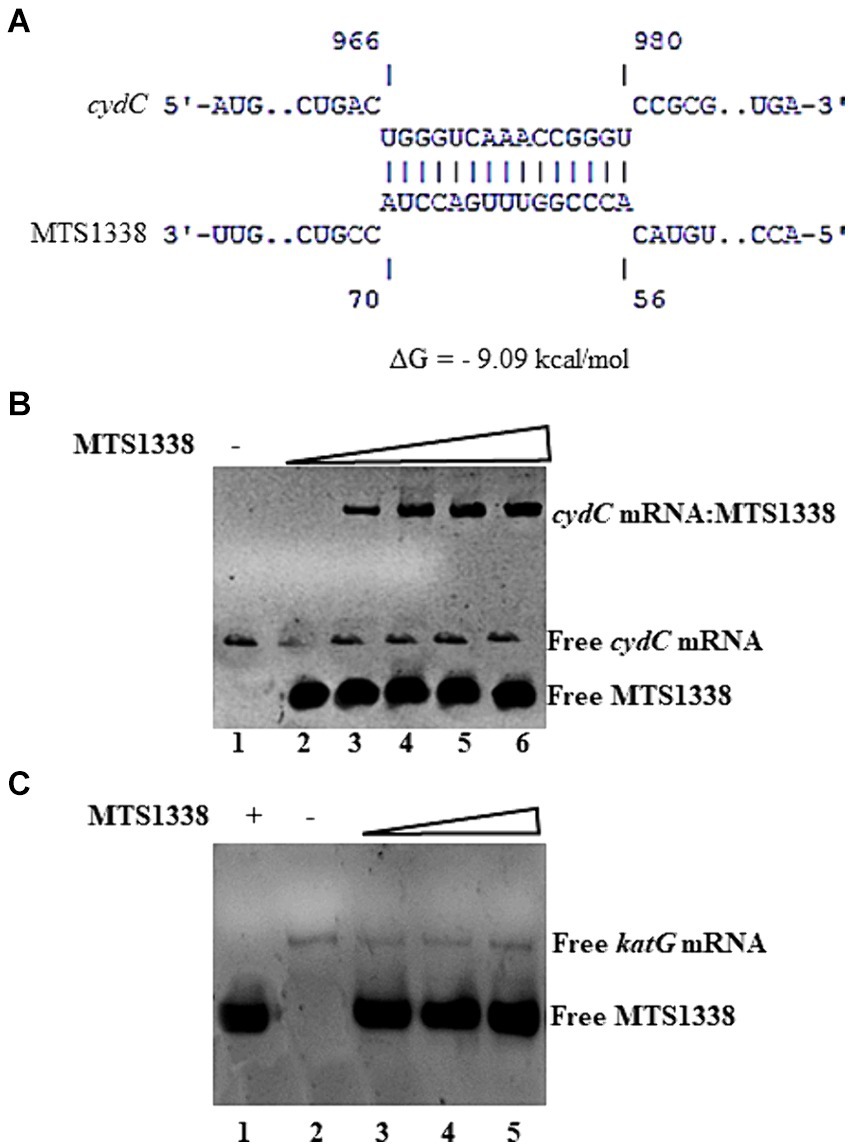
Figure 5. (A) Base pairing region between MTS1338 and cydC mRNA fragment as predicted by IntaRNA program. The numbering of nucleotides on MTS1338 and cydC mRNA is relative to the transcription start site of each RNA. (B,C) Interaction between MTS1338 and cydC mRNA (B) and katG (C). Fragment of cydC and katG mRNA (10 pmol) was incubated with 10, 20 and 30 pmol of MTS1338 (lanes 4, 5, and 6 respectively) in a 20 μL reaction. Numbers at the bottom of the gels represent the lanes (C).
3.5 MTS1338 overabundance increases the stability of cydC mRNA
Direct interaction of MTS1338 to the coding region of cydC mRNA raised the question of whether MTS1338 binding to cydC mRNA increased its stability in vivo. To help answer this question, we first overexpressed MTS1338 in exponentially growing cells (OD600–0.9) and subsequently rifampicin (500 μg/mL) was added to it. The use of this high concentration of rifampicin completely inhibits new transcription. The amount of cydC mRNA in the cells with/without overexpressing MTS1338 was measured at different time points after rifampicin addition by RT-qPCR analysis. As can be seen in Figure 6, overexpression of MTS1338 led to dramatic stabilization of the cydC message, increasing its half-life from ~4 min to ~15 min. This firmly indicated that the binding of MTS1338 to cydC mRNA protected from intracellular degradation possibly by ribonucleases.
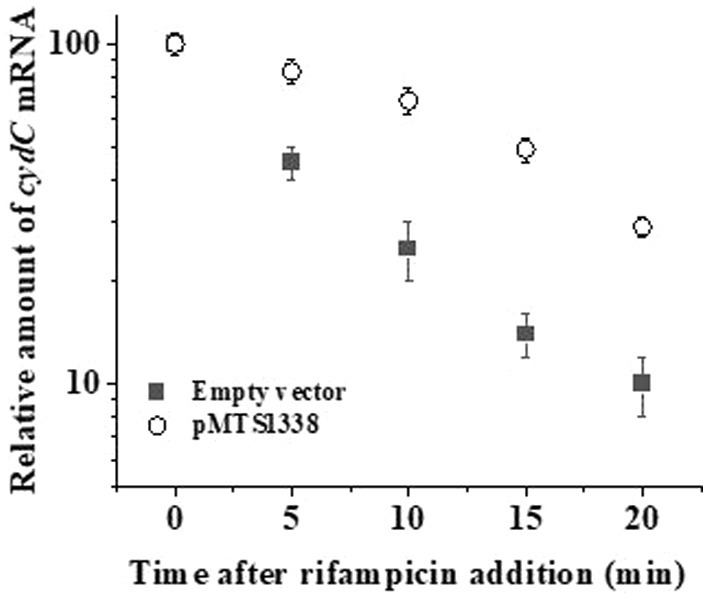
Figure 6. Stability of cydC mRNA with/without MTS1338 overexpression. Cells harboring either empty or pMTS1338 plasmids were grown to exponential phase (OD600–0.9) and then tetracycline (1 μg/mL) was added to cells. Cells were further grown for 72 h and were treated with 500 mg/liter rifampicin. Samples were withdrawn at the indicated time points and the amount of cydC mRNA was determined by RT-qPCR analysis.
3.6 A higher accumulation of MTS1338 increases rifampicin efflux from axenically grown Mycobacterium tuberculosis
We measured intrabacterial ethidium bromide (EtBr) accumulation as a substitute for rifampicin efflux as EtBr has been considered as an artificial efflux pump substrate (Lake et al., 2023). The fluorescence of EtBr increases by ~20-fold when intercalated with DNA (Jernaes and Steen, 1994). Thus, occluding EtBr efflux from M. tuberculosis cells by efflux pump inhibitor renders it more fluorescent, which can be measured by spectrofluorimetry. To examine the effect of a higher accumulation of MTS1338 on rifampicin efflux, M. tuberculosis cells with/without overexpressing MTS1338 were exposed to EtBr for 10 min at 37°C and the efflux pump inhibitor CCCP was subsequently added to the cells. Cells were then centrifuged and resuspended in PBS and CCCP was added to the resuspended cells. Figure 7A shows the accumulation of EtBr in M. tuberculosis with/without overexpressing MTS1338 treated with CCCP. It is evident in Figure 7A that cells with a higher intracellular MTS1338 abundance accumulated as much as 50% less EtBr than what was obtained in the cells with a low abundance of MTS1338. This experiment firmly suggested that a higher abundance of MTS1338 promoted a lower accumulation of EtBr potentially deporting it from the cells.
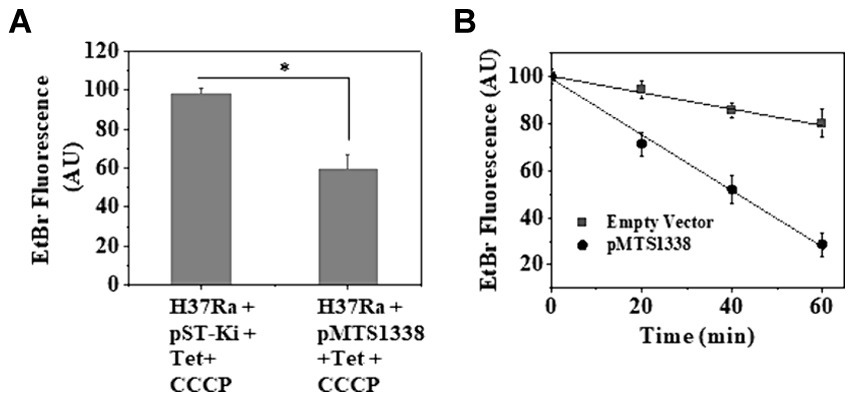
Figure 7. (A) Accumulation of EtBr in M. tuberculosis cells with/without MTS1338 overexpression. Cells (OD600 = 0.9) containing empty or recombinant pMTS1338 plasmids were allowed to grow in presence of tetracycline (1 μg/mL) for 72 h followed by the addition of EtBr (1 μg/mL). After 10 min CCCP (6.25 μM) was added to the culture. Cells were then pelleted down and resuspended in PBS containing CCCP. Intracellular EtBr fluorescence was measured by fluorimetry. Data was presented as the average ± SD from three separate experiments. One-way ANOVA was conducted and a difference of *p < 0.05 was considered statistically significant. (B) After the treatment with EtBr and CCCP, cells were resuspended in PBS and intracellular accumulation of EtBr was measured by fluorimetry at indicated time points. Data was presented as the average ± SD from three separate experiments.
In a similar line, to investigate the rate of EtBr efflux in cells with empty/recombinant pMTS1338 plasmids, EtBr-containing cells were treated with CCCP and then resuspended in PBS to substantially diminish the inhibitory effect of CCCP on efflux proteins. After that intracellular EtBr accumulation was measured by spectrofluorimetry every 20 min up to 60 min. Figure 7B shows that the decrease in the inhibitory effect of CCCP resulted in the outflow of EtBr from the cells, which was reflected by the concomitant decrease in EtBr fluorescence with increasing time. Interestingly, a nearly 75% decrease in EtBr fluorescence was observed for the cells with higher MTS1338 abundance, whereas for the cells with low intracellular MTS1338 population, only a ~ 20% decrease in EtBr fluorescence was measured after 60 min. This experiment established that MTS1338 facilitates the deportation of EtBr, a surrogate of rifampicin, from the cells.
4 Discussion
Although M. tuberculosis possesses an array of eccentric mechanisms to withstand antibiotic action, sRNA-mediated drug tolerance of mycobacteria has not been reported earlier. In the current study, we systematically analyzed the remodeling of an intracellular regulatory circuit of M. tuberculosis in response to antibiotic treatment. MTS1338, a distinctive sRNA exists only in pathogenic mycobacterial strains (Arnvig et al., 2011) and can confer pathogenic properties to nonpathogenic mycobacteria while expressed in a heterologous manner (Bychenko et al., 2021). Treatment of first-line antitubercular drug rifampicin on M. tuberculosis upregulated MTS1338, which in turn was found to ameliorate its survival in the presence of the drug. The treatment of a second-line drug kanamycin on M. tuberculosis also showed an identical effect on MTS1338. Examination of how MTS1338 promotes M. tuberculosis survival under rifampicin-exposed conditions revealed that a higher MTS1338 abundance induced an efflux protein CydC that minimizes drug concentration in the cell.
The global emergence of pathogens resistant to antimicrobial agents accounts for the increasing health crisis of people. Multifaceted approaches for the mitigation of disease prevalence by promoting antibiotic stewardship are required to control antimicrobial resistance and tolerance. Understanding how pathogens remodel their gene regulatory pathways against the existing antibiotic arsenal is a critical step for the development of new antimicrobial drugs or improving the self-life of current antibiotics (Mediati et al., 2021). Although multiple mechanisms have been elucidated for mycobacterial resistance against antibiotics, a small RNA regulator contributing to antimycobacterial resistance has never been reported before this study. However, the instances of sRNAs exist in pathogens other than mycobacteria acting as regulatory hubs for antibacterial resistance. A highly conserved sRNA SprX in Staphylococcus aureus causing bacteraemia, infective endocarditis, and osteomyelitis in humans alters antibiotic sensitivity by negatively regulating transcription factor SpoVG (Mediati et al., 2021; Eyraud et al., 2014; Liu et al., 2016). Two sRNAs, Sr0161 and ErsA, in multidrug-resistant Pseudomonas aeruginosa negatively regulate a major outer-membrane porin OprD, which is responsible for the uptake of carbapenem antibiotic. Thus, sRNA-mediated downregulation of OprD leads to bacterial resistance against carbapenem (Ochs et al., 1999; Zhang et al., 2017). Numerous sRNA in E. coli, e.g., MicF, GcvB, RyhB, etc. have been shown to modulate antibiotic sensitivity leading to resistance (Dersch et al., 2017). Many other pathogens like Salmonella sp. (Acuña et al., 2016), Vibrio sp. (Peschek et al., 2020), etc. exploit sRNAs to develop resistance against antibiotics. MTS1338-mediated growth advantage of M. tuberculosis under antibiotic-treated conditions confirms the sRNA orchestration of cydC upregulation for mycobacterial adaptation to antibiotic treatment.
Efflux of drugs through membrane transporters or efflux pump proteins is one of the paramount strategies of M. tuberculosis for acquiring antibiotic resistance (10). Efflux pumps are membrane-spanning proteins primarily controlled by regulatory systems that have evolved to counter antibiotics (Smith et al., 2013). In the current context, MTS1338 after being induced by rifampicin acts as a pivot that upregulates a higher intracellular abundance of efflux proteins CydC thereby promoting the deportation of drugs leading to the improvement of cellular fitness (Andersson and Hughes, 2010). Thus, CydC activity in M. tuberculosis is coupled to MTS1338 regulation and is conceivable to be involved during mycobacterial infection owing to MTS1338’s role under that condition. M. tuberculosis possesses at least 18 different transporters related to antibiotic susceptibility (Viveiros et al., 2012). Thus, it would not be surprising if MTS1338 controls multiple transporters in response to antibiotic treatment. An example lies in M. tuberculosis, as a transcription regulator Lsr2 in this bacterium being upregulated by isoniazid and ethambutol negatively controlled the expression of two transporters IniBAC and EpfA, which led to obtaining resistance against antibiotics (Colangeli et al., 2007). CydC also can be controlled by multiple regulators as it happens in E. coli where three regulatory systems for antibiotics controlled a single transporter protein AcrB, a major determinant for multidrug resistance (Alekshun and Levy, 1997). Additional studies are required to unfold the MTS1338-driven regulatory processes how MTS1338 facilitates the outflow of antibiotics through CydC.
Given the multidimensional role of MTS1338, it would not be surprising if MTS1338 is accountable for turning on other pathways leading to antimycobacterial resistance. As MTS1338 is induced in response to both front-line and second-line drugs, it is conceivable that MTS1338 might promote antibacterial resistance against a broad range of antibiotics. Several sigma factors and WhiB protein transcription factors have been identified to be responsible for antibiotic susceptibility (Miotto et al., 2022). The key among the WhiB proteins that promote antibacterial drug resistance is WhiB7 (Burian et al., 2013; Reeves et al. 2013; Pisu et al., 2017). WhiB7 is upregulated by several 100-folds upon exposure to front-line drugs like rifampicin and is barely controlled by MTS1338 as increased intracellular abundance does not significantly alter the intracellular accumulation of whiB7 transcript (Singh and Dutta, unpublished observation). Likewise, SigE in M. tuberculosis was reported to be one of the leading among 13 Sigma factors gaining resistance against antibiotics (Pisu et al., 2017). M. tuberculosis also exploits many other transcription factors like GntR, XRE, etc. to obtain resistance (Miotto et al., 2022). Additional work is required to establish whether MTS1338 is involved in all the abovementioned mechanisms.
The current study depicted a novel antimycobacterial resistance mechanism regulated by a distinctive sRNA MTS1338 in M. tuberculosis. This adds substantially to our knowledge of how M. tuberculosis triggers a regulatory cascade for acquiring resistance against antimycobacterial drugs controlled by a sRNA, which was never unveiled before.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SS: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. TD: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Indian Council for Medical Research, Govt. of India for funding research grants (no-52/18/2022-BMS and 52/07/2019-BMS) to TD. TD also thanks IIT Delhi for funding the FIRP grant (MI02367G) to initiate this research work.
Acknowledgments
SS acknowledges the Indian Council for Medical Research, Govt. of India for providing her Research Associateship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1469280/full#supplementary-material
References
Acuña, L. G., Barros, M. J., Peñaloza, D., Rodas, P. I., Paredes-Sabja, D., Fuentes, J. A., et al. (2016). A feed-forward loop between SroC and MgrR small RNAs modulates the expression of eptB and susceptibility of polymyxin B in Salmonella Typhimurium. Microbiology 162, 1996–2004. doi: 10.1099/mic.0.000365
Alekshun, M. N., and Levy, S. B. (1997). Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41, 2067–2075. doi: 10.1128/AAC.41.10.2067
Almeida Da Silva, P. E., and Palomino, J. C. (2011). Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J. Antimicrob. Chemother. 66, 1417–1430. doi: 10.1093/jac/dkr173
Andersson, D. I., and Hughes, D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271. doi: 10.1038/nrmicro2319
Arnvig, K. B., Comas, I., Thomson, N. R., Houghton, J., Boshoff, H. I., Croucher, N. J., et al. (2011). Sequence-based analysis uncovers an abundance of noncoding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 7:e1002342. doi: 10.1371/journal.ppat.1002342
Arnvig, K. B., and Young, D. B. (2012). Noncoding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol. 9, 427–436. doi: 10.4161/rna.20105
Barilar, I., and Battaglia, S. (2024). Quantitative measurement of antibiotic resistance in Mycobacterium tuberculosis reveals genetic determinants of resistance and susceptibility in a target gene approach. Nat. Commun. 15:488. doi: 10.1038/s41467-023-03651-4
Briffotaux, J., Liu, S., and Gicquel, B. (2019). Genome-wide transcriptional responses of Mycobacterium to antibiotics. Front. Microbiol. 10:249. doi: 10.3389/fmicb.2019.00249
Burian, J., Yim, G., Hsing, M., Axerio-Cilies, P., Cherkasov, A., Spiegelman, G. B., et al. (2013). The mycobacterial antibiotic resistance determinant WhiB7 acts as a transcriptional activator by binding the primary sigma factor SigA (RpoV). Nucleic Acids Res. 41, 10062–10076. doi: 10.1093/nar/gkt751
Bychenko, O., Skvortsova, Y., Ziganshin, R., Grigorov, A., Aseev, L., Ostrik, A., et al. (2021). Mycobacterium tuberculosis small RNA MTS1338 confers pathogenic properties to nonpathogenic Mycobacterium smegmatis. Microorganisms 9:414. doi: 10.3390/microorganisms9020414
Chen, H., Previero, A., and Deutscher, M. P. (2019). A novel mechanism of ribonuclease regulation: GcvB and Hfq stabilize the mRNA that encodes RNase BN/Z during exponential phase. J. Biol. Chem. 294, 19997–20008. doi: 10.1074/jbc.RA119.011367
Colangeli, R., Helb, D., Vilchèze, C., Hazbón, M. H., Lee, C. G., Safi, H., et al. (2007). Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 3:e87. doi: 10.1371/journal.ppat.0030087
Dersch, P., Khan, M. A., Mühlen, S., and Görke, B. (2017). Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 8:803. doi: 10.3389/fmicb.2017.00803
Dhar, N., and McKinney, J. D. (2010). Mycobacterium tuberculosis persistence mutants identified by screening in isoniazid-treated mice. Proc. Natl. Acad. Sci. USA 107, 12275–12280. doi: 10.1073/pnas.1003219107
Dutta, T., and Srivastava, S. (2018). Small RNA mediated regulation in bacteria: a growing palette of diverse mechanisms. Gene 658, 105–112. doi: 10.1016/j.gene.2018.03.021
Eyraud, A., Tattevin, P., Chabelskaya, S., and Felden, B. (2014). A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res. 42, 4892–4905. doi: 10.1093/nar/gku149
Ginsberg, A. M., and Spigelman, M. (2007). Challenges in tuberculosis drug research and development. Nat. Med. 13, 290–294. doi: 10.1038/nm0307-290
Heinrichs, M. T., May, R. J., Heider, F., Reimers, T., Peloquin, C. A., and Derendorf, H. (2018). Mycobacterium tuberculosis strains H37Ra and H37Rv have equivalent minimum inhibitory concentrations to most antituberculosis drugs. Int. J. Mycobacteriol. 7, 156–161. doi: 10.4103/ijmy.ijmy_33_18
Jernaes, M. W., and Steen, H. B. (1994). Staining of Escherichia coli for flow cytometry: influx and efflux of ethidium bromide. Cytometry 17, 302–309. doi: 10.1002/cyto.990170405
Lake, M. A., Adams, K. N., Nie, F., Fowler, E., Verma, A. K., Dei, S., et al. (2023). The human proton pump inhibitors inhibit Mycobacterium tuberculosis rifampicin efflux and macrophage-induced rifampicin tolerance. Proc. Natl. Acad. Sci. USA 120, 1–10. doi: 10.1073/pnas.2215512120
Liu, X., Zhang, S., and Sun, B. (2016). SpoVG regulates cell wall metabolism and oxacillin resistance in methicillin-resistant Staphylococcus aureus strain N315. Antimicrob. Agents Chemother. 60, 3455–3461. doi: 10.1128/AAC.00026-16
Mann, M., Wright, P. R., and Backofen, R. (2017). IntaRNA2.0: enhanced and customizable prediction of RNA–RNA interactions. Nucleic Acids Res. 45, W435–W439. doi: 10.1093/nar/gkx279
Martini, B. A., Grigorov, A. S., Skvortsova, Y. V., Bychenko, O. S., Salina, E. G., and Azhikina, T. L. (2023). Small RNA MTS1338 configures a stress resistance signature in Mycobacterium tuberculosis. Int. J. Mol. Sci. 24:7928. doi: 10.3390/ijms24097928
McKinney, J. D. (2000). In vivo veritas: the search for TB drug targets goes live. Nat. Med. 6, 1330–1333. doi: 10.1038/82142
Mediati, D., Wu, S., and Wu, W. (2021). Networks of resistance: small RNA control of antibiotic resistance. Trends Genet. 37, 35–45. doi: 10.1016/j.tig.2020.08.016
Miotto, P., Sorrentino, R., de Giorgi, S., Provvedi, R., Cirillo, D. M., and Manganelli, R. (2022). Transcriptional regulation and drug resistance in Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 12:990312. doi: 10.3389/fcimb.2022.990312
Moores, A., Riesco, A. B., Schwenk, S., and Arnvig, K. B. (2017). Expression, maturation and turnover of DrrS, an unusually stable, DosR regulated small RNA in Mycobacterium tuberculosis. PLoS One 12:e0174079. doi: 10.1371/journal.pone.0174079
Nasiri, M. J., Haeili, M., Ghazi, M., Goudarzi, H., Pormohammad, A., Imani Fooladi, A. A., et al. (2017). New insights into the intrinsic and acquired drug resistance mechanisms in mycobacteria. Front. Microbiol. 8:681. doi: 10.3389/fmicb.2017.00681
Ochs, M. M., McCusker, M. P., Bains, M., and Hancock, R. E. W. (1999). Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob. Agents Chemother. 43, 1085–1090. doi: 10.1128/AAC.43.5.1085
Peschek, N., Herzog, R., Singh, P. K., Sprenger, M., Meyer, F., Fröhlich, K. S., et al. (2020). RNA-mediated control of cell shape modulates antibiotic resistance in Vibrio cholerae. Nat. Commun. 11:6067. doi: 10.1038/s41467-020-19890-8
Petchiappan, A., and Chatterji, D. (2017). Antibiotic resistance: current perspectives. ACS Omega 2, 7400–7409. doi: 10.1021/acsomega.7b01368
Pisu, D., Provvedi, R., and Espinosa, D. M. (2017). The alternative sigma factors SigE and SigB are involved in tolerance and persistence to antitubercular drugs. Antimicrob. Agents Chemother. 61, e01596–e01516. doi: 10.1128/AAC.01596-16
Reeves, A. Z., Campbell, P. J., Sultana, R., Malik, S., Murray, M., Plikaytis, B. B., et al. (2013). Aminoglycoside cross-resistance in Mycobacterium tuberculosis due to mutations in the 5′ untranslated region of whiB7. Antimicrob. Agents Chemother. 57, 1857–1865. doi: 10.1128/AAC.02191-12
Remm, S., Earp, J. C., Dick, T., Dartois, V., and Seeger, M. A. (2022). Critical discussion on drug efflux in Mycobacterium tuberculosis. FEMS Microbiol. Rev. 46, 1–15. doi: 10.1093/femsre/fuab042
Salina, E. G., Grigorov, A., Skvortsova, Y., Majorov, K., Bychenko, O., Ostrik, A., et al. (2019). MTS1338, a small Mycobacterium tuberculosis RNA, regulates transcriptional shifts consistent with bacterial adaptation for entering into dormancy and survival within host macrophages. Front. Cell. Infect. Microbiol. 9:405. doi: 10.3389/fcimb.2019.00405
Sarathy, J. P., Dartois, V., and Lee, E. J. D. (2012). The role of transport mechanisms in Mycobacterium tuberculosis drug resistance and tolerance. Pharmaceuticals 5, 1210–1235. doi: 10.3390/ph5111210
Shi, L., Sohaskey, C. D., Kana, B. D., Dawes, S., North, R. J., Mizrahi, V., et al. (2005). Change in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. USA 102, 15629–15634. doi: 10.1073/pnas.0507850102
Singh, S., Nirban, R., and Dutta, T. (2021). MTS1338 in Mycobacterium tuberculosis promotes detoxification of reactive oxygen species under oxidative stress. Tuberculosis 131:102142. doi: 10.1016/j.tube.2021.102142
Smith, T., Wolff, K. A., and Nguyen, L. (2013). Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr. Top. Microbiol. Immunol. 358, 53–80. doi: 10.1007/82_2012_303
Takiff, H. E., and Feo, O. (2015). Clinical value of whole-genome sequencing of Mycobacterium tuberculosis. Lancet Infect. Dis. 15, 1077–1090. doi: 10.1016/S1473-3099(15)00071-7
Taneja, S., and Dutta, T. (2019). On a stake out: mycobacterial small RNA identification and regulation. Non-coding RNA Res. 4, 86–95. doi: 10.1016/j.ncrna.2019.05.001
Udwadia, Z. F., Amale, R. A., Ajbani, K. K., and Rodrigues, C. (2012). Totally drug resistant tuberculosis in India. Clin. Infect. Dis. 54, 579–581. doi: 10.1093/cid/cir889
van den Boogaard, J., Lyimo, R., Irongo, C. F., Boeree, M. J., Schaalma, H., Aarnoutse, R. E., et al. (2009). Community vs. facility-based directly observed treatment for tuberculosis in Tanzania’s Kilimanjaro region. Int. J. Tuberc. Lung Dis. 13, 1524–1529. doi: 10.5588/ijtld.09.0032
Viveiros, M., Martins, M., Rodrigues, L., Machado, D., Couto, I., Ainsa, J., et al. (2012). Inhibitors of mycobacterial efflux pumps as potential boosters for TB drugs. Expert Rev. Anti-Infect. Ther. 10, 983–998. doi: 10.1586/eri.12.89
Young, D. B., Perkins, M. D., Duncan, K., and Barry, C. E. III. (2008). Confronting the scientific obstacles to global control of tuberculosis. J. Clin. Invest. 118, 1255–1265. doi: 10.1172/JCI34614
Keywords: small RNAs, gene regulation, antimicrobial resistance, efflux protein, drug efflux
Citation: Singh S and Dutta T (2024) A virulence-associated small RNA MTS1338 activates an ABC transporter CydC for rifampicin efflux in Mycobacterium tuberculosis. Front. Microbiol. 15:1469280. doi: 10.3389/fmicb.2024.1469280
Edited by:
Shicheng Chen, Northern Illinois University, United StatesReviewed by:
Runhua Han, University of Manitoba, CanadaDexi Li, Henan Agricultural University, China
Copyright © 2024 Singh and Dutta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:Tanmay Dutta, ZHRhbm1heUBjaGVtaXN0cnkuaWl0ZC5hYy5pbg==
 Saumya Singh
Saumya Singh Tanmay Dutta
Tanmay Dutta