- 1Department of Food Science and Biotechnology, Carbohydrate Bioproduct Research Center, College of Life Science, Sejong University, Seoul, Republic of Korea
- 2Department of Food Biotechnology, Dong-A University, Busan, Republic of Korea
- 3Department of Food and Animal Biotechnology, Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul, Republic of Korea
- 4Department of Agricultural Biotechnology, Seoul National University, Seoul, Republic of Korea
- 5Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul, Republic of Korea
- 6Center for Food and Bioconvergence, Seoul National University, Seoul, Republic of Korea
Introduction: Cronobacter sakazakii is a notorious foodborne pathogen, frequently contaminating powdered infant formula and causing life-threatening diseases in infants. The escalating emergence of antibiotics-resistant mutants has led to increased interest in using bacteriophage as an alternative antimicrobial agent.
Methods: Two phages, CR8 and S13, were isolated from feces and soil samples and their morphology, physiology, and genomics were characterized. Phage receptor was determined using deletion mutants lacking flgK, rfaC, fhuA, btuB, lamb, or ompC genes, followed by complementation. Phage-resistant mutants were analyzed for phenotypic changes and fitness trade-offs using motility assays and Caco-2 cell invasion models.
Results: CR8 and S13 were identified as members of Caudoviricetes. Phage CR8 and phage S13 utilize flagella and LPS, respectively, to adhere to host cells. Bacterial challenge assay demonstrated delayed emergence of the resistant mutant as well as stronger lytic activity of a phage cocktail consisting of CR8 and S13 than the single phage treatment. Phenotypic analysis of the phage cocktail resistant strain, designated as CSR strain, revealed that the resistance resulted from the impaired receptor proteins for phage, such as defects in motility and alteration in LPS structure. CSR strain exhibited significant attenuation in invading human intestinal epithelial Caco-2 cells compared to WT cells.
Conclusion: This study demonstrates that the development of the phage cocktail targeting distinct host receptors can serve as a promising antimicrobial strategy to effectively control C. sakazakii.
1 Introduction
Cronobacter sakazakii, a gram-negative food-borne pathogen, has been classified by International Commission on Microbiological Specification for Foods as a “Severe hazard for restricted populations, causing life threatening or substantial chronic sequelae or long duration” (Tompkin, 2002). C. sakazakii is particularly notorious to neonates due to its ability to resist desiccation, which allows it to survive in dry conditions such as powdered infant formula. Infection caused by C. sakazakii can lead to necrotizing enterocolitis, septicemia, and meningitis in infants and immunocompromised individuals with a mortality rate up to 55% and even survivors often suffer from significant neurological sequelae (Muytjens et al., 1983; Harris and Oriel, 1989; Lucas and Cole, 1990; Van Acker et al., 2001; Dumen, 2010). Moreover, C. sakazakii is capable of forming biofilms, commonly found in food processing areas, heightening the risk of infection among vulnerable groups (Iversen et al., 2004; Lehner et al., 2005; Hurrell et al., 2009).
Unfortunately, antibiotic therapies for controlling C. sakazakii are limited due to the emergence of resistant strains to many conventional antibiotics (Pitout et al., 1997; Dennison and Morris, 2002; Kilonzo-Nthenge et al., 2012). These strains exhibit resistance to a range of commonly used antibiotics, including ampicillin, cefotaxime, cephalothin, and ceftriaxone. Such resistance not only complicates treatment options but also elevates the risk of potentially severe public health outbreaks (Kim et al., 2007; Zuber et al., 2008). Given these challenges, there are urgent needs to explore new approaches for controlling C. sakazakii.
Bacteriophage therapy presents a promising alternative to antibiotics. Specially, phages with lytic life cycle are of significant interest because they target and kill specific bacterial strains without combining their DNA with the host genetic materials (Hagens and Loessner, 2007; Strauch et al., 2007). Several studies have demonstrated the efficacy of phage therapy against C. sakazakii infection. For example, the application of C. sakazakii-infecting phage CR5 to artificially contaminated infant formula resulted in complete inhibition of bacterial growth at multiplicity of infection (MOI) of 105 (Lee et al., 2016). Additionally, Wang et al. reported that the new C. sakazakii phage JK004 significantly reduced bacterial counts in an infant formula milk at 37°C (Wang et al., 2022). However, such phage treatments that involve only a single type of phage may often lead to the development of phage resistance. This issue can be addressed by introducing a cocktail of multiple phages instead of a single phage. Multiple studies have indicated that phage therapy using phage cocktails can be more effective than individual phages alone, especially in cases where the cocktail includes phages that target distinct receptors on the bacterial surface (Bai et al., 2019; Yang et al., 2020). A phage cocktail has been shown to improve bacterial eradication and delay or prevent the development of phage resistance (Tanji et al., 2004; Tanji et al., 2005; Regeimbal et al., 2016). However, only a few studies have specifically examined phage cocktails targeting C. sakazakii.
In this study, we isolated two novel C. sakazakii-specific phages, CR8 and S13, from environmental samples. We analyzed their biological and genomic characteristics and developed a phage cocktail containing the two phages. The efficacy of this phage cocktail in controlling C. sakazakii was evaluated. Additionally, we characterized a phage-resistant C. sakazakii isolate, which exhibited the loss of motility as well as altered LPS structures. The phage-resistant isolate demonstrated reduced fitness in invasion assay, suggesting a trade-off between the development of resistance and bacterial fitness.
2 Materials and methods
2.1 Bacterial strains and growth condition
Bacterial strains used in this study are listed in Table 1. Cronobacter sakazakii ATCC 29544 (FARMER et al., 1980) was used as a host strain for bacteriophage isolation and propagation. All the bacterial strains were cultured in tryptic soy broth (TSB, BD Bacto, Franklin Lakes, NJ) or tryptic soy agar (TSA, BD Difco, Franklin Lakes, NJ) at 37°C for 12 h with an agitation speed of 220 rpm.
2.2 Isolation and propagation of Cronobacter sakazakii-specific bacteriophages
Feces and soil samples were collected from University animal farm of Seoul National University (Suwon, South Korea) for isolation of C. sakazakii-targeting bacteriophages. The 25 g samples were homogenized in 225 mL sterile Butterfield’s phosphate-buffered dilution water (0.25 M KH2PO4 adjusted to pH 7.2 with NaOH) using a blender (BacMixer 400, Interscience Laboratory Inc., France). The 25 mL of each homogenized sample was mixed with 25 mL of 2X TSB and incubated at 37°C for 8 h with 220 rpm agitation. After incubation, the culture was centrifuged (9,000 × g, 10 min, 4°C) and the supernatant was filtered using 0.22-μm-pore-size filters (Millipore, Billerica, MA). The 10 mL filtrate was mixed with 50 mL of TSB containing 5 mM CaCl2 and 500 μL overnight culture of C. sakazakii ATCC 29544 and incubated at 37°C for 8 h with 220 rpm agitation, followed by centrifugation and filtration as described above. This supernatant was used for plaque assay. Tenfold serial dilutions of the last filtrate were dotted on the bacterial lawn (5 mL of 0.4% soft TSA containing 100 μL of overnight culture of C. sakazakii ATCC 29544 and being poured into TSA plate) and the plate was incubated overnight at 37°C to confirm the presence of bacteriophages via formation of plaques. Each single plaque on dotted or overlaid plate was picked with a sterile tip and dissolved in sterilized sodium chloride-magnesium sulfate (SM) buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10 mM MgSO4·7H2O). This process was repeated five times to isolate a single bacteriophage.
To propagate and concentrate the isolated phages, a culture of C. sakazakii ATCC 29544 with an optical density at 600 nm = 0.5 was incubated with SM buffer containing a single phage plaque and the incubation was carried out at 37°C until the culture became clear. After centrifugation of the cleared culture (15,000 × g, 10 min, 4°C), the supernatant was filtered with 0.22-μm-pore-size filter to remove cell debris and subsequently precipitated by the addition of polyethylene glycol (PEG) 6,000 (Junsei chemical, Japan) dissolved in 1 M NaCl solution at 4°C for 12 h. Centrifuged and precipitated phages were dissolved in SM buffer and the phage particles were separated by cesium chloride (CsCl) density gradient ultracentrifugation (25,000 rpm, 2 h, 4°C), followed by dialysis using SM buffer and storage at 4°C.
2.3 Thermal and pH stability analysis
To determine the stability of CR8 and S13 under various temperature and pH conditions, each phage (~1 × 108 PFU/mL) was incubated overnight at varying temperatures (4–75°C) and pH values (2 to 12). Thermal stability was estimated by incubating 1 mL of phage lysate (~108 PFU/mL) across a temperature range from 4 to 80°C for 1 h. The stability of phages across different pH levels was assessed by exposing the phage lysate to SM buffers with varied pH values for 1 h. The viability of both phages was determined by measuring the concentration of the remaining activated phages using a spotting assay.
2.4 Transmission electron microscopy (TEM)
Phage CR8 and S13 stocks were diluted (about 11010 pfu/ml) with SM buffer and placed on carbon-coated copper grids. Following the removal of the excess phage suspension, negative staining was performed by adding 2% aqueous uranyl acetate (pH 4.0). The grids were examined by TEM (LEO 912 AB; Carl Zeiss, Germany) at 80 kV at the National Academy of Agricultural Science (Suwon, South Korea). Based on the examined morphologies, each phage was classified according to the guidelines of the International Committee on Taxonomy of Viruses (Fauquet et al., 2005).
2.5 Genome sequencing and bioinformatics analysis
Bacteriophage genomic DNA was extracted from the phage lysate according to Lambda DNA extraction protocol using the Phase lock gel (PLG; 5 PRIME, Hamburg, Germany). Whole-genome sequencing of the extracted phage DNA was carried out with a Genome Sequencer FLX (GS-FLX) titanium sequencer (Roche, Mannheim, Germany) and assembled with Newbler v2.3 (Roche) at Macrogen Inc., South Korea. Open reading frames (ORFs) were predicted using GeneMarkS (Besemer et al., 2001), Glimmer v3.02 (Delcher et al., 2007), and FgenesB softwares (Softberry, Inc. Mount Kisco, NY, USA), and ribosomal binding sites were identified using RBSfinder (J. Craig Venter Institute, Rockville, MD, USA). BLASTP (Altschul et al., 1990) and InterProScan (Zdobnov and Apweiler, 2001) programs are employed to annotate predicted ORFs, and the sequencing and annotation data were processed by Artemis14 (Carver et al., 2008). The CGView server program was used to generate a circular genome map of the phages (Stothard and Wishart, 2005). Sequence manipulations and genomic analysis were performed using CLC Genomics work-bench version 23 on a workstation at the Biopolymer Research Center for Advanced Material, Sejong University.
Phylogenetic analysis of amino acid sequences of the large subunit of the phage terminase was conducted. Sequence alignment was performed using MUSCLE, and the evolutionary distances among phages were calculated (Edgar, 2004). The phylogenetic tree was constructed using the neighbor-joining method in MEGA 11.0.13, with 1,000 bootstrap replicates (Saitou and Nei, 1987; Tamura et al., 2021).
For genomic comparison between phage CR8 and S13, their genomes were aligned using BLASTn and the results were visualized by Easyfig (Sullivan et al., 2011).
2.6 Construction of candidates for phage receptor deletion mutants and complementation
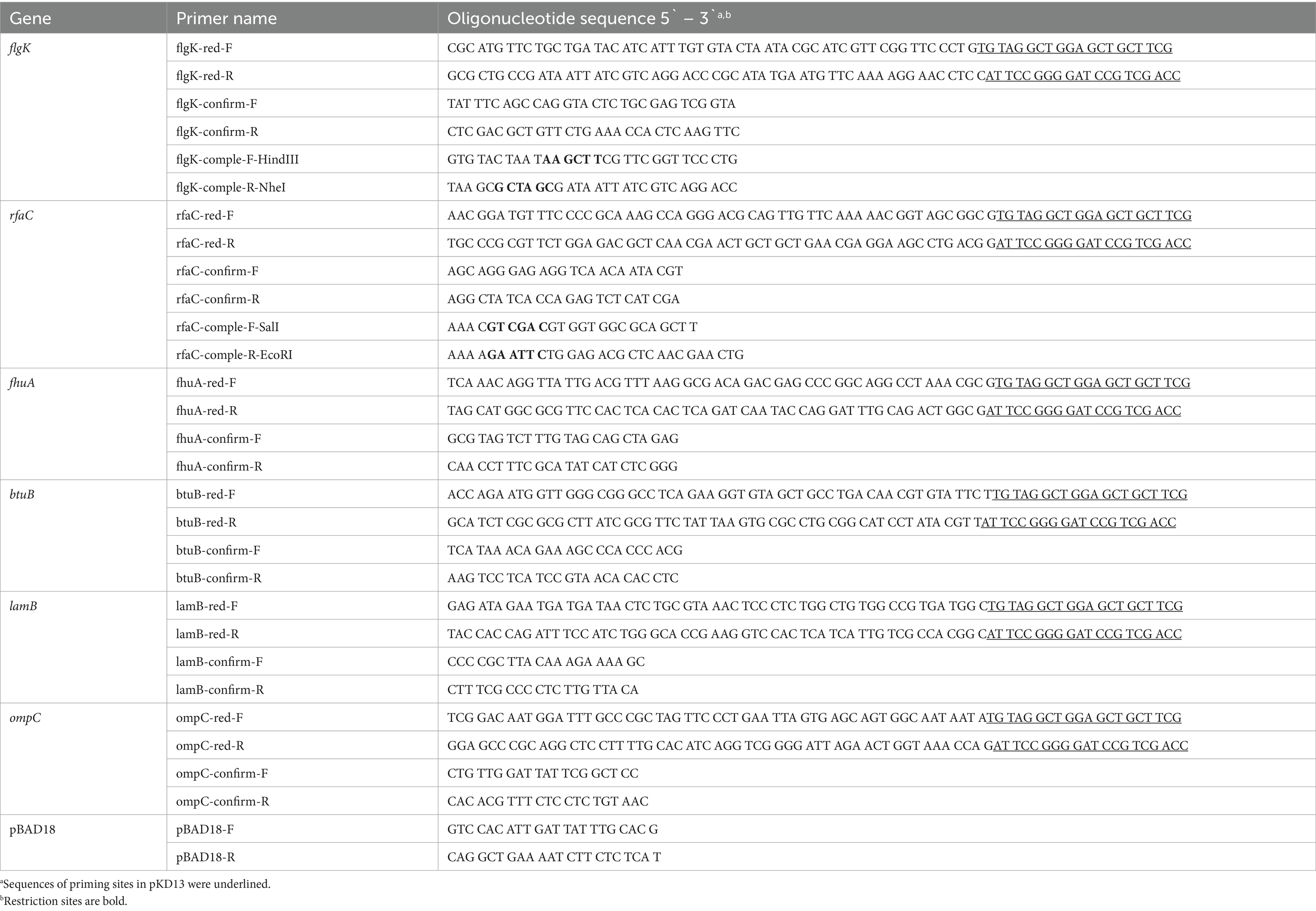
Cronobacter sakazakii ATCC 29544 derivatives with deletion of flgK, rfaC, fhuA, btuB, lamb, or ompC genes were constructed using the one-step gene inactivation method as described previously (Datsenko and Wanner, 2000). Briefly, the kanamycin resistance (Kmr) cassette from plasmid pKD13 was amplified by PCR using primers specific for each gene. Sequences of all primers for these constructs are listed in Table 2. The resulting PCR products were used to transform into C. sakazakii ATCC 29544 strain containing pKD46, and and integrated into the flgK, rfaC, fhuA, btuB, lamb, or ompC genes. Subsequently, the Kmr cassette was removed by introducing plasmid pCP20. For complementation of the deletion mutations, each gene of C. sakazakii ATCC 29544 was amplified by PCR using the primers containing restriction site (HindIII and NheI). The PCR products were digested with HindIII and NheI and ligated into HindIII/NheI-digested pBAD18 vector (Guzman et al., 1995). Plasmid constructs were confirmed by sequencing, and transformed into each deletion mutant strains. The protein synthesis was induced by the addition of 0.2% arabinose (final concentration) and phage infectivity was examined by spotting assay.
2.7 Adsorption assay
Cronobacter sakazakii ATCC 29544 wild-type or deletion mutant strains were grown exponentially until OD600 reached to 1.0 to 1.5 and 900 μL of the culture was aliquoted into 6 microtubes (900 μL culture per 1.75-ml tube). Each culture was infected by phage (MOI = 0.01) with 5 min intervals and incubated until the first infected tube was incubated for 20 min. After incubation, all tubes were centrifuged (16,000 rpm, 1 min, 4°C) and the supernatant was filtrated (0.22-μm-pore-size filter) to remove bacterial cells. Overlay assay was carried out as previously described (Kim and Ryu, 2011), to determine the number of unadsorbed phage particles. For control, phage was added to fresh TSB without host bacteria.
2.8 Host range analysis
An overnight culture of each tested bacteria (100 μL) was inoculated into 5 mL of 0.4% soft TSA and mixed by gentle vortexing. The mixture was poured into the 1.5% TSA plate (with 50 μg/mL ampicillin or kanamycin, if necessary) and placed at room temperature (RT) for 20 min to allow the agar to solidify. Then, 10 μL of tenfold serial dilutions of phage stock was dotted on the bacterial lawn. The plates were dried at RT for 20 min, followed by overnight incubation at 37°C. The susceptibility of the tested bacteria to phages was examined on the following day.
2.9 Growth inhibition assay
An overnight culture of C. sakazakii ATCC 29544 was 1% subinoculated into 50 mL of fresh TSB medium and incubated at 37°C with agitation for 1.5 h (OD at 600 nm = 0.4 ~ 0.6). Phages were added to the culture at an MOI of 1. Bacterial growth was monitored by measuring the OD600 every hour after phage infection. An uninfected culture was used as a negative control.
A phage cocktail consisting of phage CR8 and S13 was evaluate for their ability to inhibit the growth of host bacteria. The phages CR8 and S13 were added to the bacterial culture prepared as above at an MOI of 1 each. To count the number of viable cells, samples taken from the culture were centrifuged (15,000 rpm, 1 min) to remove free phage and the cell pellet was resuspended in same volume of phosphate-buffered saline (PBS, GeneDEPOT, Barker, TX, USA) (pH 7.4). Then, it was 10-fold diluted with PBS, plated onto TSA plate, and followed incubation at 37°C for 12 h.
2.10 Isolation and characterization of phage cocktail-resistant mutants
To isolate phage cocktail-resistant mutants, the high-titer phage cocktail overlay assay was conducted as described previously (Capparelli et al., 2010). Briefly, 100 μL of each phage (about 1010 PFU/mL, for each) and the same volume of overnight bacterial culture were added to molten 5-ml soft TSA (0.4% agar). The mixture was poured onto TSA plate and incubated at 37°C until resistant colonies appeared. Three colonies were picked and streak-purified 20 times on fresh TSA plate in the absence of phages. One of them was selected for further study and the maintenance of phage resistance was certified by spotting assay.
2.11 Motility assay
One microliter of the overnight culture was injected onto soft-agar motility plates (TSA medium containing 0.3% agar) to allow bacteria to swim in the plates. The plates were dried for 30 min at room temperature and incubated for 8 h at 37°C. As a positive and negative control, C. sakazakii ATCC 29544 wild type strain and flagella (ΔflgK) defective mutant strain were used, respectively.
2.12 LPS extraction
LPS was extracted from bacteria grown overnight using hot phenol-water extraction as previously described (Kim and Ryu, 2012; Kim et al., 2018). The extracted LPS was subjected to deoxycholate polyacrylamide gel electrophoresis (DOC-PAGE) followed by fluorescent staining using the Pro-Q® Emerald 300 Lipopolysaccharide Gel Stain Kit (Molecular Probes, Cat. No. P20495; Eugene, Oregon, USA) according to manufacturer’s instructions. The gel was visualized under 300 nm UV by the Gel doc™ EZ System (Bio-rad). As a positive and negative control, C. sakazakii ATCC 29544 wild type strain and LPS (ΔrfaC) defective mutant strain were used, respectively.
2.13 Autoaggregation assay
Autoaggregation assay was conducted as previously described (Kim et al., 2018). Briefly, bacteria were cultured overnight, subcultured in fresh TSB medium at 1%, and then incubated at 37°C with constant shaking. Mid-exponentially grown bacteria were diluted with fresh TSB broth to achieve an OD600 of 1.5 in glass tubes and left at room temperature.
2.14 Invasion assay
Bacterial invasion ability into mammalian Caco-2 cells was assessed using a modified gentamicin protection assay (Kim et al., 2010). Bacteria were cultured overnight, then transferred to fresh LB medium at a 1% concentration and incubated at 37°C for 3 h with 220 rpm agitation. The OD600-adjusted bacterial cells were pelleted, resuspended in 400 μL of EMEM for infection and used to infect monolayered cells. The infection was carried out for 1.5 h in the presence of 5% CO2 and washed 3X with PBS. Gentamicin was treated to eliminate any bacteria that were adhering to cells for 1.5 h and washed 3X with PBS. Cells were then lysed with 1% Triton X-100 (500 μL/ well) for 15 min, cell lysates were serially diluted and plated onto LB plates to enumerate CFUs. Relative invasion capability was calculated as described previously (Kim et al., 2010) using following formula:
2.14.1 Data processing and statistical analysis
Statistical analysis was performed using Student’s unpaired t-test in Prism (version 10.3.0), with significance represented as ****p < 0.0001 for invasion into host cells.
3 Results
3.1 Isolation of phages and their host range determination
Out of five C. sakazakii-infecting phages isolated from environmental samples, the phages CR8 and S13 were selected for this study due to their superior lytic activity against C. sakazakii and their ability to target different host receptors, ensuring a more effective and robust phage cocktail. The two phages formed clear plaques without halos on the lawn of C. sakazakii ATCC29544. The stability of CR8 and S13 under various temperature and pH conditions were evaluated. Both phages maintained high thermal stability from 4°C to 45°C, but a significant decrease was evident at 55°C, with a notable drop in titer for both CR8 and S13. In a wide pH range (2 to 12), the phages remained highly stable, indicating that both CR8 and S13 phages have robust pH tolerance, which may be advantageous for various application where pH fluctuations occur (Supplementary Figure S1). Their host range studies determined by spotting assay on strains of Cronobacter, Escherichia coli, Salmonella Typhimurium, Listeria monocytogenes, and Bacillus cereus showed that these phages formed plaques only on Cronobacter genus (Table 1).
3.2 Morphological analysis of phage CR8 and S13 by TEM
Morphological analysis revealed that both phages have a characteristic of the family Caudoviricetes with a contractile tail (Figure 1). Phage CR8 has an isometric head with the mean diameter of 78.6 ± 1.7 nm, and phage S13 has longish head with the major axis of 103.7 ± 4.1 nm and the minor axis of 74.7 ± 6.0 nm. Their tail length is similar to each other – non-contracted tail and contracted tail length of phage CR8 are 122.1 ± 2.3 nm and 58.1 nm, respectively, and those of phage S13 are 112.0 ± 2.1 nm and 55.0 ± 2.1 nm (Figure 1). According to the International Committee on Taxonomy of Viruses (ICTV, https://ictv.global/, accessed on 20 August 2024), CR8 is predicted to belong to the Certrevirus genus and S13 shares genetic homologies with phages in the Straboviridae family, genus Slopekvirus.
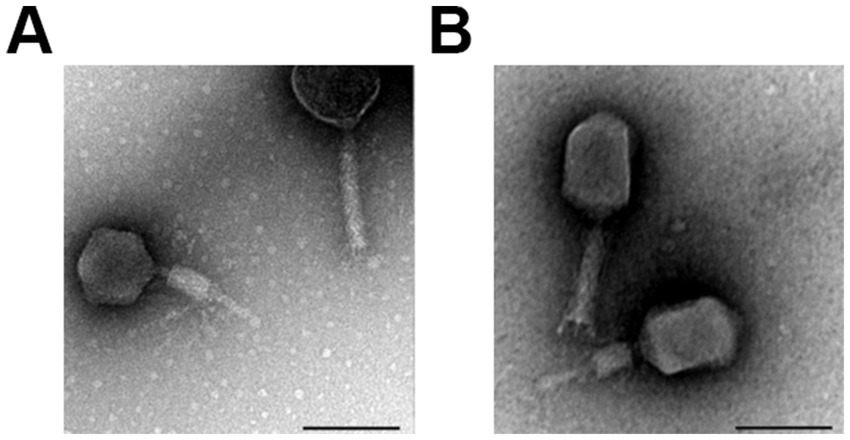
Figure 1. Morphology of phage CR8 and S13. TEM images of phage CR8 (A), S13 (B) with contractile tail. Scale bar, 100 nm.
3.3 Bacteriophage genome analysis
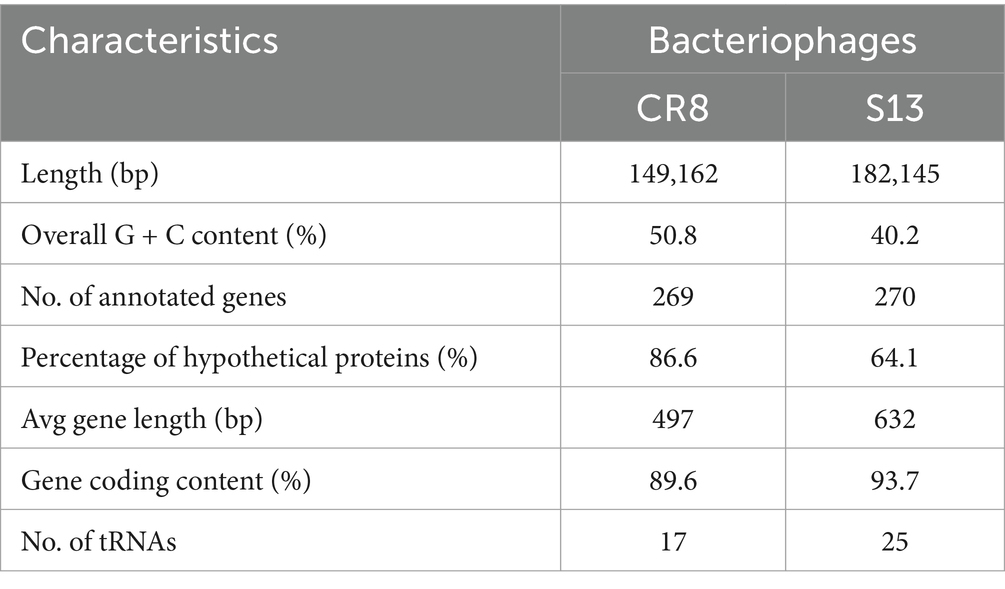
Whole genome analysis revealed that the genome sizes of CR8 and S13 are 149,162-bp and 182,145-bp, respectively and the GC contents are 50.8 and 40.2%, respectively. Phages CR8 and S13 contain 269 ORFs with 17 tRNAs and 270 ORFs with 25 tRNAs, respectively (Table 3; Figure 2). No genes related to virulence and antibiotic resistance were identified in the genome of both phages, demonstrating their safety in application. The majority of predicted genes encoded hypothetical proteins (86.6% for CR8 and 64.1% for S13). Additional details on each ORF are provided in the Supplementary Table S1. Meanwhile, phage CR8 genome shares 97% nucleotide identity with that of phage CR3, a C. sakazakii infecting phage (Shin et al., 2012), but they did not have similarity with other phages genomes publicized in GenBank database. Phage S13 genome showed little sequence homology with well-known T4 phage. Additionally, no sequence similarity between CR8 and S13 was observed at the nucleotide level (Supplementary Figure S2).
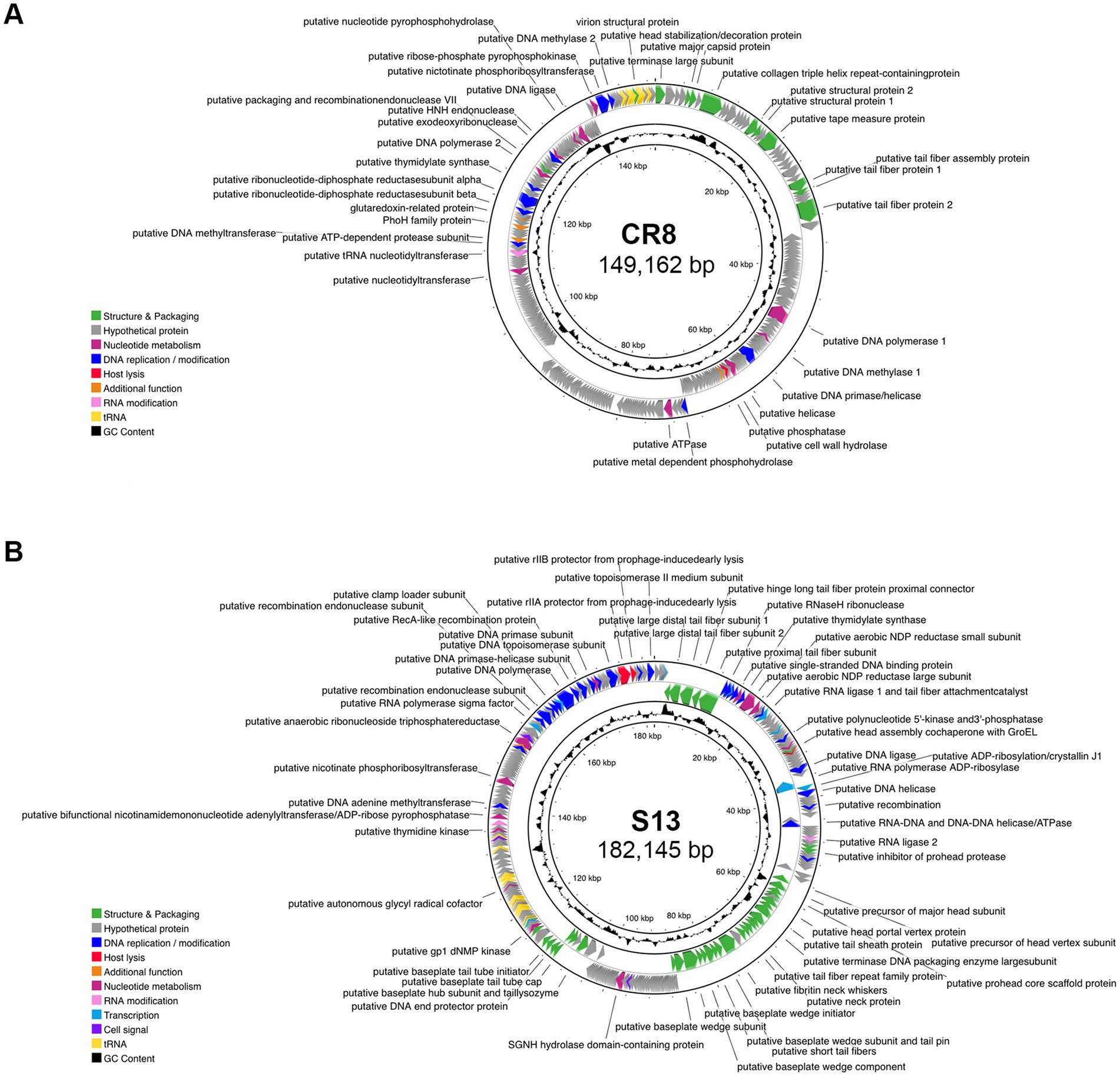
Figure 2. Genomic features of phage CR8 and S13. Genome maps of CR8 (A) and S13 (B). Predicted ORFs in the genome were annotated and colored by functional groups as follows: phage structure and packaging (green), nucleotide metabolism (magenta), DNA replication/modification (blue), host lysis (red), RNA modification (pink), transcription (teal), cell signal (purple) and additional function (orange).
The ORFs were categorized into eight functional groups; structure/packaging, DNA replication/modification, host lysis, nucleotide metabolism, RNA modification, transcription, cell signal and other groups. Genes related to lysogenic functions such as integrase, transposase, excisionase, repressor, and the genome attachment site (attP) were absent in the genome of CR8 and S13, suggesting that the two phages are likely lytic phages. Phage CR8 has two genes encoding tail fiber proteins, CR8_028 and CR8_033, which are likely receptor binding protein (RBP) targeting the flagella of C. sakazakii. Meanwhile, phage S13 possesses genes encoding for multiple tail fiber subunits and putative RBP (S13_008), which likely targets the LPS of C. sakazakii. Interestingly, S13 contains a gene encoding putative RNA polymerase sigma factor which is utilized in gene expression at the middle and late stages (Reznikoff et al., 1985), suggesting their potential role in preferential transcription of phages genes over host genes. Genes related to host lysis, such as holin and endolysin, were not detected in the phage CR8 genome, but it contains a cell wall hydrolase (CR8_090), which shows 99% amino acid sequence identity with an endolysin of phage CR3. A gene encoding a putative lysozyme (S13_190) was identified in the phage S13 genome, but its amino acid sequence showed no homology with any known lysozyme from Cronobacter phages. It exhibited 71 and 69% homology with the lysozyme from Citrobacter phage Ci1 and Shigella phage Esh27, respectively.
The phylogenetic analysis of the two phages was conducted based on their large subunit of terminase (Supplementary Figure S3). The resulting trees include Cronobacter phages, alongside other phages from different bacterial hosts. CR8 closely aligns with Pectobacterium phage vB_PcaM_P7_Pc, a member of the genus Cetrevirus. Phage vB_PcaM_P7_Pc exhibited myovirus-like morphology with lytic life cycle. Phage S13 is positioned near the Cronobacter phage LPCS28, which is also lytic phage. The findings highlight potential of phages CR8 and S13 for use as biocontrol or as therapeutic agents.
3.4 Identification of phage receptor
To identify bacterial components that are required for phage infection, we decided to test six receptors known in Gram-negative bacteria (Rakhuba et al., 2010). To this end, we constructed deletion mutants of C. sakazakii ATCC 29544, each lacking one of the followings: flagella (ΔflgK), LPS (ΔrfaC), outer membrane transporter for ferrichrome (ΔfhuA), vitamin B12 (ΔbtuB), maltose (ΔlamB), and outer membrane porin OmpC (ΔompC). These mutants were challenged to test susceptibility to specific phages. Phage CR8 was unable to form any plaques on the lawns of ΔflgK deletion mutant and phage S13 failed to form plaques on the lawns of ΔrfaC deletion mutant (Figures 3A,B). However, the infectivity of both phages to C. sakazakii ATCC 29544 was fully restored when flgK and rfaC genes were reintroduced into their respective mutants (Figures 3A,B). To determine whether these genes are required for the initial step of phage infection, phage adsorption assay was conducted with the WT and deletion mutants. The ability of phage CR8 to bind to its host receptor decreased in the absence of flgK (Figure 3C). Most of the phage S13 efficiently bound to the WT strain within 20 min, while its binding to the ΔrfaC deletion mutant was completely abolished (Figure 3D). These results indicate that flagella and LPS serve as receptors for the phage CR8 and S13, respectively.
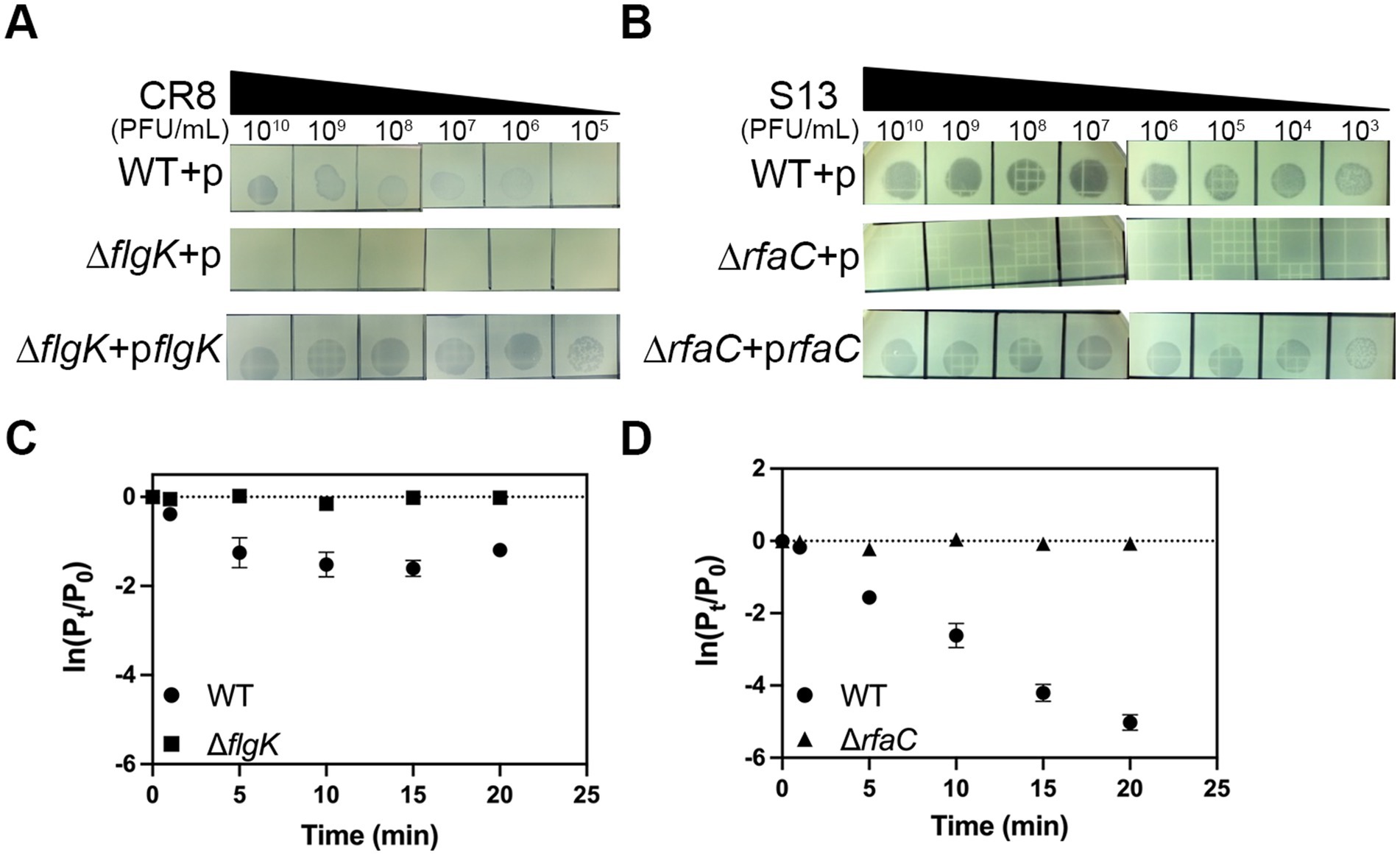
Figure 3. Flagella and LPS are receptors for CR3 and S13, respectively. The sensitivity of the bacteria to CR3 (A) and S13 (B) phages was determined by spotting assay. Complementation of flagella and LPS completely restored the phage susceptibility. Noted strains are WT + p, wild type harboring pBAD18; ΔflgK + p, ΔflgK harboring pBAD18; ΔflgK + pflgK, ΔflgK expressing flgK, ΔrfaC + p, ΔflgK harboring pBAD18; ΔrfaC + prfaC ΔrfaC expressing rfaC. (C,D) adsorption kinetics of phage CR8 (C) and S13 (D) against C. sakazakii ATCC 29544 (circle), ΔflgK (square) and ΔrfaC (triangle). All bacterial strains grown exponentially were challenged with each phage at an MOI of 0.01. Data displayed are mean ± SEM for three biological replicates. Pt, phage titer (PFU/ mL) at the indicated time; P0, initial phage titer.
3.5 Bacterial challenge assay of a phage cocktail targeting two different host receptors
The ability of a phage cocktail to inhibit the growth of host bacteria was evaluated using bacterial challenge assays. Phage CR8 and phage S13 showed similar ability to lyse host bacteria with rapid early cell lysis, and the phage-resistance appeared 5 h after infection. On the other hand, a phage cocktail consisting of phage CR8 and phage S13 showed the prolonged period of growth inhibition up to 11 h, resulting in the delayed emergence of the resistant mutant (Figure 4A) and there was a significant reduction in the number of viable cells compared to single-phage treatments (Figure 4B).
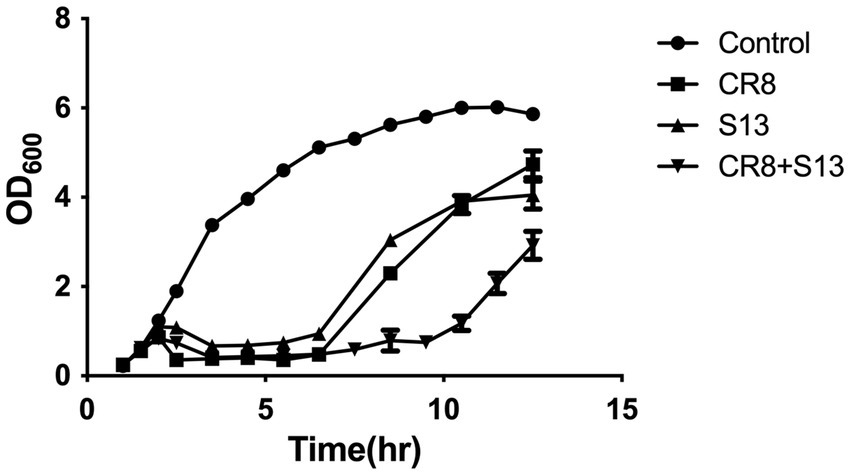
Figure 4. Bacterial challenge assay with phages CR8 and S13 against C. sakazakii ATCC 29544. Each phage or a phage cocktail was added at a MOI of 1 to the bacterial culture after 1.5 h incubation. Bacterial growth was determined by measuring absorbance at 600 nm hourly. Data shown and error bars are means ± SEM for three biological replicates at 1 h increments.
3.6 Isolation and characterization of phage cocktail-resistant mutants
Three phage cocktail-resistant C. sakazakii colonies were picked from high-titer overlay plates and streak-purified 20 times onto fresh TSA plates in the absence of phages to isolate a single mutant strain. At each streaking-purifying step, the newly formed colonies were subjected to spotting assay to confirm phage resistance and a single colony that exhibited consistent resistance across generations was selected for further study and named CSR strain. Given that CR8 and S13 target flagella and LPS as receptors, we hypothesized that the resistant mutant might lack flagella and LPS or exhibit loss of motility and altered LPS structure. To test these hypotheses, motility and flagella structure were examined. The CSR strain was defective in motility like ΔflgK deletion mutant, yet still displayed flagella on its surface (Figures 5A,B). LPS structure of CSR strain differed from that of WT and ΔrfaC deletion mutant; some parts of inner core and O-antigen regions of LPS were absent in CSR strain (Figure 5C). It should be noted that nothing was extracted in ΔrfaC deletion mutant, which produces a lipophilic moiety (lipid A-Kdo2; rough-type of LPS) (Figure 5C; Sirisena et al., 1992), because the hot phenol-water method is selective for smooth types of LPS over rough types (Henderson et al., 2013). Previous studies have demonstrated that the alteration in LPS leads to autoaggregation in C. sakazakii (Wang et al., 2012; Kim et al., 2018). Consistent with these findings, CSR cells precipitated more quickly than WT cells when left in broth culture at room temperature, an indicative of enhanced autoaggregation (Figure 5D).
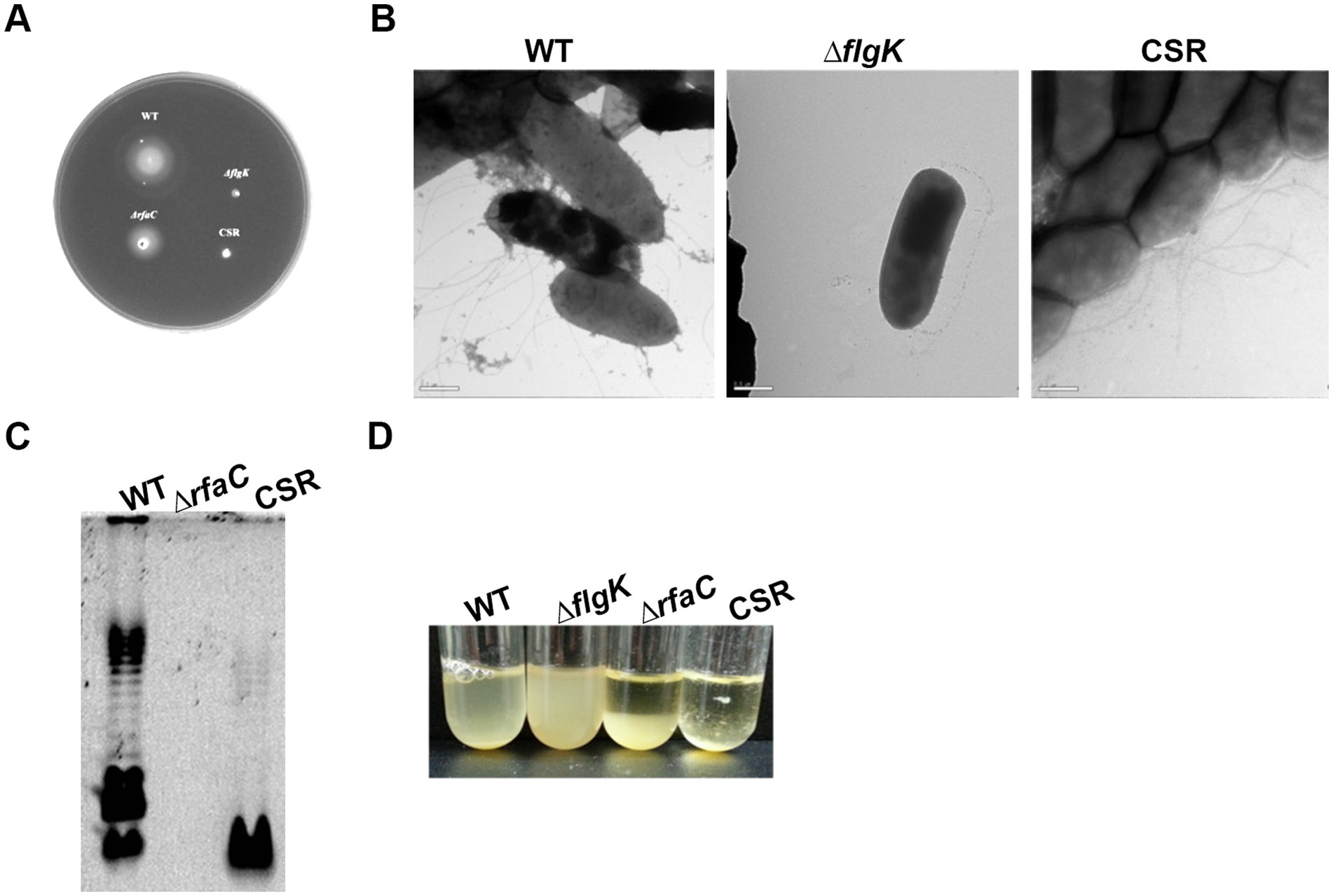
Figure 5. Characterization of CSR strain resistant to a phage cocktail infection. (A) Motility of C. sakazakii strains on semisolid agar plates: WT, ΔflgK, ΔrfaC and CSR, respectively. (B) TEM analysis. Scale bars, 500 nm. (C) LPS profiles. LPS fractions were extracted using hot phenol-water method, fractionated by DOC-PAGE and fluorescently stained. (D) Autoaggregation of the CSR strain. Bacterial cultures of WT, ΔflgK, ΔrfaC, and CSR strain were diluted at an OD600 of 1.5 using fresh TSB broth and incubated statically at room temperature. The data displayed are a representative of three biological replicates.
3.7 Decreased fitness in virulence resulting from acquiring phage-resistance
Considering that the development of phage resistance often incurs fitness costs, including reduced virulence and heightened vulnerability to antimicrobials (Chart et al., 1989; Chan et al., 2016; Gordillo Altamirano et al., 2021; Salazar et al., 2021; Wang et al., 2021) or immune clearance (Roach et al., 2017), we aimed to evaluate invasion capability of the CSR strain to mammalian intestinal epithelial Caco-2cells. Consistent with previous studies, invasiveness of the CSR strain was significantly reduced by 90% compared to WT cells (Figure 6).
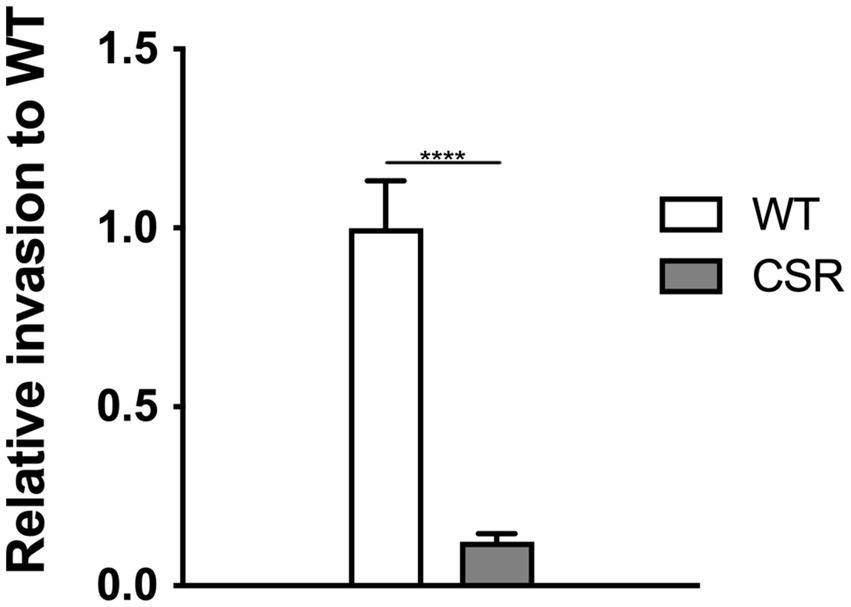
Figure 6. Attenuation in invasion resulted from the acquisition of phage resistance. Invasive ability into human epithelial cells, Caco-2. Confluent monolayers of Caco-2 epithelial cells were challenged with WT C. sakazakii or the CSR strain, incubated for 1.5 h, followed by gentamicin treatment at a concentration of 100 μg/mL for a further 1.5 h. The Caco-2 cells were lysed with 1% TritonX-100 to harvest the intracellular bacteria. The data shown and error bars are mean ± SEM for three biological replicates. Statistical significance was conducted using unpaired two- tailed Student’s t test and significance displayed as ****p < 0.0001.
4 Discussion
Cronobacter sakazakii has been detected in various foods and environmental sources (Kandhai et al., 2004), but it is difficult to destroy the pathogen without affecting the quality of foods (Dumen, 2010). When the foods were stored improperly, the pathogen can propagate to cause the disease in immunocompromised human. Outbreaks of disease associated with C. sakazakii have occurred sporadically and are particularly dangerous to infants, suggesting that new measures to protect infants from the pathogen are needed. In the food safety aspect, bacteriophages have been studied and evaluated with promising results due to their host specificity (Garcia et al., 2008). The specificity of phage is related to their adsorption on bacterial cell surface, especially receptors (Rakhuba et al., 2010). In the present study, we isolated novel phages which utilize different receptor each other and characterized them to construct effective phage cocktail.
Phage CR8 and phage S13 belong to the Caudoviricetes family, and interestingly, the head of phage S13 is not isometric; rather, it is slightly elongated (Figure 1). This prolate head is unusual because isometric heads are in a majority (85%) of the tailed phages (Calendar and Abedon, 2005). In our genome analysis, phage CR8 has a genome size of 149,162 bp, and phage S13 has 182,145 bp. The larger head size of S13 compared to CR8 corroborates previous findings that phage head size varies proportionally with genome size (Pariza and Iandolo, 1974). Interestingly, the majority of genes in the phage CR8 and S13 encode hypothetical proteins, suggesting the phages are quite distinct from those previously reported. Additionally, limited genetic information is available on C. sakazakii phages, indicating the need for further research in this area.
Phage cocktail containing phages that target different host receptors has been effective in phage therapy. Bai et al. reported that a phage cocktail containing three Salmonella phages, each targeting different host receptors such as flagella, O-antigen and BtuB, significantly reduced the development of bacterial resistance (Bai et al., 2019) Similarly, a phage cocktail comprising five Pseudomonas infecting phages, each targeting distinct receptors, was shown to effectively constrain the emergence of phage resistant Pseudomonas (Yang et al., 2020). Treatment of the phage cocktail containing phage CR8 and S13 delayed the development of resistant bacteria by several hours compared to the treatment with each single phage, demonstrating that a phage cocktail could be a powerful strategy to control C. sakazakii. Bacteria can evolve to gain resistance to phage through a range of mechanisms (Labrie et al., 2010), notable one of which is loss or modification of receptor, which prevents phage adsorption (Filippov et al., 2011). We observed the loss of motility and the auto-aggregation in CSR, leading us to initially assume that the resistant strains have no flagella and LPS. However, TEM analysis demonstrated that the CSR still possess flagella similar to wild-type strain. These findings suggest that the resistance may arise from events occurring during the phage replication process following adsorption, rather than from the absence of structural receptors such as flagella. It should be noted that the direction of flagellar rotation, in addition to retaining the flagellar structure itself, is required for the Salmonella phage iEPS5 to adsorb and successfully infect the host bacteria (Choi et al., 2013). DOC-PAGE analysis of LPS revealed that CSR has defective O-antigen and core region in their LPS (Figure 5C). Incomplete deletion of O-antigen suggest that LPS core domain of C. sakazakii contains branched oligosaccharide and that CSR was affected in some branch as reported in E. coli K1 strain (Jiménez et al., 2012). It remains unclear which gene in CSR was affected, thus genetic analyses such as whole genome sequencing are necessary for future study.
In the context of Red Queen dynamics, the emergence of bacterial resistance to phages is inevitable. As anticipated, we observed the emergence of mutants resistant to a phage cocktail. Among these, we isolated and characterized a mutant, designated CSR, which exhibited stable resistance to phage infections (Figures 5, 6). Notably, CSR exhibited a loss of motility despite retaining flagellar structure (Figure 5) and displayed alteration in its LPS structures compared to WT strain (Figure 5). It is noteworthy that the emergence of phage-resistant bacteria is considered clinically significant, as they often become susceptible to antibiotics or hypovirulent (Niu et al., 2020; Gordillo Altamirano et al., 2021; Majkowska-Skrobek et al., 2021; Song et al., 2021). Consistent with the fact that phage selective pressure on hypervirulent bacteria reduces virulence by altering bacterial surface appendages, including LPS and capsule (Tang et al., 2023), the CSR isolate showed highly impaired invasion ability into intestinal epithelial cells. This finding supports the potential use of the phage cocktail in this study as a novel antibacterial agent.
5 Conclusion
We evaluated the efficacy of a phage cocktail consisting of two distinct phages, CR3 and S13, which target different host receptors, for potential use as a biocontrol agent. Morphological and genomic analyses confirmed that both phages belong to members of the Caudoviricetes class with contractile tails. Phage CR8 targets flagella, while phage S13 targets LPS as their respective host receptors. The combination of these two phages significantly delayed the emergence of the resistant mutant by prolonging period of growth inhibition and demonstrated stronger lytic activity compared to individual phage treatments. Notably, we isolated a phage-resistant strain with phenotypic defects in motility and altered LPS structure. The resistant strain exhibited severe impaired invasion ability to human epithelial cells, likely due to a fitness tradeoff between acquisition of the resistance and virulence. Overall, the phage cocktail presents a promising and sustainable alternative to antibiotics for controlling Cronobacter.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/genbank, accession numbers KC954774 and KC954775.
Author contributions
SK: Data curation, Visualization, Writing – original draft, Writing – review & editing. BS: Data curation, Visualization, Writing – original draft, Writing – review & editing. YK: Data curation, Methodology, Writing – review & editing. HK: Data curation, Methodology, Writing – review & editing. GN: Formal analysis, Writing- review & editing. HS: Conceptualization, Supervision, Writing – original draft, Writing – review & editing. SR: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by a grant from the Ministry of Science and ICT (MSIT) through the National Research Foundation (NRF) of Korea (NRF-RS-2023-0021200) (BS), and by Biomaterials Specialized Graduate Program through the Korea Environmental Industry & Technology Institute (KEITI) funded by the Ministry of Environment (MOE) (BS). This research was also supported by Korea Basic Science Institute (National research Facilities and Equipment Center) grant funded by the Ministry of Education (2023R1A6C101A045), and NRF funded by the Ministry of Education (2022R1A6A1A03055869).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1468686/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bai, J., Jeon, B., and Ryu, S. (2019). Effective inhibition of Salmonella Typhimurium in fresh produce by a phage cocktail targeting multiple host receptors. Food Microbiol. 77, 52–60. doi: 10.1016/j.fm.2018.08.011
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Capparelli, R., Nocerino, N., Lanzetta, R., Silipo, A., Amoresano, A., Giangrande, C., et al. (2010). Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS One 5:e11720. doi: 10.1371/journal.pone.0011720
Carver, T., Berriman, M., Tivey, A., Patel, C., Böhme, U., Barrell, B. G., et al. (2008). Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–2676. doi: 10.1093/bioinformatics/btn529
Chan, B. K., Sistrom, M., Wertz, J. E., Kortright, K. E., Narayan, D., and Turner, P. E. (2016). Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 6:26717. doi: 10.1038/srep26717
Chart, H., Row, B., Threlfall, E. J., and Ward, L. R. (1989). Conversion of Salmonella enteritidis phage type 4 to phage type 7 involves loss of lipopolysaccharide with concomitant loss of virulence. FEMS Microbiol. Lett. 60, 37–40. doi: 10.1016/0378-1097(89)90073-6
Choi, Y., Shin, H., Lee, J. H., and Ryu, S. (2013). Identification and characterization of a novel flagellum-dependent Salmonella-infecting bacteriophage, iEPS5. Appl. Environ. Microbiol. 79, 4829–4837. doi: 10.1128/AEM.00706-13
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645. doi: 10.1073/pnas.120163297
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Dennison, S. K., and Morris, J. (2002). Multiresistant Enterobacter sakazakii wound infection in an adult. Inf. Med. 19, 533–535.
Dumen, E. (2010). Coronabacter sakazakii (Enterobacter sakazakii) only an infant problem. Kafkas Univ Vet Fak Derg 16, S171–S178. doi: 10.9775/kvfd.2010.1949
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Farmer, J. J., Asbury, M. A., Hickman, F. W., and Brenner, D. J. (1980). Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int. J. Syst. Bacteriol. 30, 569–584. doi: 10.1099/00207713-30-3-569
Fauquet, C. M., Mayo, M., Maniloff, J., Desselberger, U., and Ball, L. A. (2005). Virus taxonomy: VIIIth report of the international committee on taxonomy of viruses. Cambridge: Academic Press.
Filippov, A. A., Sergueev, K. V., He, Y., Huang, X.-Z., Gnade, B. T., Mueller, A. J., et al. (2011). Bacteriophage-resistant mutants in Yersinia pestis: identification of phage receptors and attenuation for mice. PLoS One 6:e25486. doi: 10.1371/journal.pone.0025486
Garcia, P., Martinez, B., Obeso, J., and Rodríguez, A. (2008). Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47, 479–485. doi: 10.1111/j.1472-765X.2008.02458.x
Gordillo Altamirano, F., Forsyth, J. H., Patwa, R., Kostoulias, X., Trim, M., Subedi, D., et al. (2021). Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat. Microbiol. 6, 157–161. doi: 10.1038/s41564-020-00830-7
Guzman, L.-M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
Hagens, S., and Loessner, M. J. (2007). Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 76, 513–519. doi: 10.1007/s00253-007-1031-8
Harris, L. S., and Oriel, P. J. (1989). “Heteropolysaccharide produced by Enterobacter sakazakii ” U.S. Patent number 4:636–806.
Hayashi, K., Morooka, N., Yamamoto, Y., Fujita, K., Isono, K., Choi, S., et al. (2006). Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2:0007. doi: 10.1038/msb4100049
Henderson, J. C., O’Brien, J. P., Brodbelt, J. S., and Trent, M. S. (2013). Isolation and chemical characterization of lipid a from gram-negative bacteria. J. Vis. Exp. 79:e50623. doi: 10.3791/50623
Hurrell, E., Kucerova, E., Loughlin, M., Caubilla-Barron, J., and Forsythe, S. J. (2009). Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, Salmonella serovars and other Enterobacteriaceae. Int. J. Food Microbiol. 136, 227–231. doi: 10.1016/j.ijfoodmicro.2009.08.007
Iversen, C., Lane, M., and Forsythe, S. J. (2004). The growth profile, thermotolerance and biofilm formation of Enterobacter sakazakii grown in infant formula milk. Lett. Appl. Microbiol. 38, 378–382. doi: 10.1111/j.1472-765X.2004.01507.x
Jiménez, N., Senchenkova, S. N., Knirel, Y. A., Pieretti, G., Corsaro, M. M., Aquilini, E., et al. (2012). Effect of LPS biosynthesis mutants on K1 polysaccharide association with Escherichia coli cell surface. J. Bacteriol. 194, 3356–3367. doi: 10.1128/jb.00329-12
Kandhai, M. C., Reij, M. W., Gorris, L. G. M., Guillaume-Gentil, O., and van Schothorst, M. (2004). Occurrence of Enterobacter sakazakii in food production environments and households. Lancet 363, 39–40. doi: 10.1016/S0140-6736(03)15169-0
Kilonzo-Nthenge, A., Rotich, E., Godwin, S., Nahashon, S., and Chen, F. (2012). Prevalence and antimicrobial resistance of Cronobacter sakazakii isolated from domestic kitchens in middle Tennessee, United States. J. Food Prot. 75, 1512–1517. doi: 10.4315/0362-028X.JFP-11-442
Kim, K., Kim, K.-P., Choi, J., Lim, J.-A., Lee, J., Hwang, S., et al. (2010). Outer membrane proteins a (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 76, 5188–5198. doi: 10.1128/AEM.02498-09
Kim, K.-P., Klumpp, J., and Loessner, M. J. (2007). Enterobacter sakazakii bacteriophages can prevent bacterial growth in reconstituted infant formula. Int. J. Food Microbiol. 115, 195–203. doi: 10.1016/j.ijfoodmicro.2006.10.029
Kim, M., and Ryu, S. (2011). Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar typhimurium and Escherichia coli. Appl. Environ. Microbiol. 77, 2042–2050. doi: 10.1128/AEM.02504-10
Kim, M., and Ryu, S. (2012). Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar typhimurium. Mol. Microbiol. 86, 411–425. doi: 10.1111/j.1365-2958.2012.08202.x
Kim, S., Yoon, H., and Ryu, S. (2018). New virulence factor CSK29544_02616 as LpxA binding partner in Cronobacter sakazakii. Sci. Rep. 8:835. doi: 10.1038/s41598-018-19306-0
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Lee, J.-H., Bai, J., Shin, H., Kim, Y., Park, B., Heu, S., et al. (2016). A novel bacteriophage targeting Cronobacter sakazakii is a potential biocontrol agent in foods. Appl. Environ. Microbiol. 82, 192–201. doi: 10.1128/AEM.01827-15
Lee, J. H., Choi, Y., Shin, H., Lee, J., and Ryu, S. (2012). Complete genome sequence of Cronobacter sakazakii temperate bacteriophage phiES15. J. Virol. 86, 7713–7714. doi: 10.1128/jvi.01042-12
Lehner, A., Riedel, K., Eberl, L., Breeuwer, P., Diep, B., and Stephan, R. (2005). Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various strains:: aspects promoting environmental persistence. J. Food Prot. 68, 2287–2294. doi: 10.4315/0362-028x-68.11.2287
Lucas, A., and Cole, T. (1990). Breast milk and neonatal necrotising enterocolitis. Lancet 336, 1519–1523. doi: 10.1016/0140-6736(90)93304-8
Majkowska-Skrobek, G., Markwitz, P., Sosnowska, E., Lood, C., Lavigne, R., and Drulis-Kawa, Z. (2021). The evolutionary trade-offs in phage-resistant Klebsiella pneumoniae entail cross-phage sensitization and loss of multidrug resistance. Environ. Microbiol. 23, 7723–7740. doi: 10.1111/1462-2920.15476
Muytjens, H., Zanen, H., Sonderkamp, H., Kollee, L., Wachsmuth, I. K., and Farmer, J. (1983). Analysis of eight cases of neonatal meningitis and sepsis due to Enterobacter sakazakii. J. Clin. Microbiol. 18, 115–120. doi: 10.1128/jcm.18.1.115-120.1983
Niu, T., Guo, L., Luo, Q., Zhou, K., Yu, W., Chen, Y., et al. (2020). Wza gene knockout decreases Acinetobacter baumannii virulence and affects Wzy-dependent capsular polysaccharide synthesis. Virulence 11, 1–13. doi: 10.1080/21505594.2019.1700659
Pariza, M. W., and Iandolo, J. J. (1974). Determination of genome size of selected typing bacteriophages of Staphylococcus aureus. Appl. Environ. Microbiol. 28, 510–512. doi: 10.1128/am.28.3.510-512.1974
Pitout, J., Moland, E., Sanders, C., Thomson, K., and Fitzsimmons, S. (1997). Beta-lactamases and detection of beta-lactam resistance in Enterobacter spp. Antimicrob Agents Chemother. 41, 35–39. doi: 10.1128/AAC.41.1.35
Rakhuba, D., Kolomiets, E., Szwajcer Dey, E., and Novik, G. (2010). Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 59, 145–155. doi: 10.33073/pjm-2010-023
Regeimbal, J. M., Jacobs, A. C., Corey, B. W., Henry, M. S., Thompson, M. G., Pavlicek, R. L., et al. (2016). Personalized therapeutic cocktail of wild environmental phages rescues mice from Acinetobacter baumannii wound infections. Antimicrob. Agents Chemother. 60, 5806–5816. doi: 10.1128/AAC.02877-15
Reznikoff, W. S., Siegele, D. A., Cowing, D. W., and Gross, C. A. (1985). The regulation of transcription initiation in bacteria. Annu. Rev. Genet. 19, 355–387. doi: 10.1146/annurev.ge.19.120185.002035
Roach, D. R., Leung, C. Y., Henry, M., Morello, E., Singh, D., Di Santo, J. P., et al. (2017). Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 22:38-+. doi: 10.1016/j.chom.2017.06.018
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Salazar, K. C., Ma, L., Green, S. I., Zulk, J. J., Trautner, B. W., Ramig, R. F., et al. (2021). Antiviral resistance and phage counter adaptation to antibiotic-resistant Extraintestinal pathogenic Escherichia coli. MBio 12:211. doi: 10.1128/mBio.00211-21
Shin, H., Lee, J.-H., Kim, Y., and Ryu, S. (2012). Complete genome sequence of Cronobacter sakazakii bacteriophage CR3. J. Virol. 86, 6367–6368. doi: 10.1128/jvi.00636-12
Sirisena, D. M., Brozek, K. A., MacLachlan, P. R., Sanderson, K. E., and Raetz, C. R. (1992). The rfaC gene of Salmonella typhimurium. Cloning, sequencing, and enzymatic function in heptose transfer to lipopolysaccharide. J. Biol. Chem. 267, 18874–18884. doi: 10.1016/S0021-9258(19)37042-5
Song, L., Yang, X., Huang, J., Zhu, X., Han, G., Wan, Y., et al. (2021). Phage selective pressure reduces virulence of Hypervirulent Klebsiella pneumoniae through mutation of the wzc gene. Front. Microbiol. 12:739319. doi: 10.3389/fmicb.2021.739319
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Strauch, E., Hammerl, J., and Hertwig, S. (2007). Bacteriophages: new tools for safer food? J. Verbr. Lebensm. 2, 138–143. doi: 10.1007/s00003-007-0188-5
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tang, M., Huang, Z., Zhang, X., Kong, J., Zhou, B., Han, Y., et al. (2023). Phage resistance formation and fitness costs of hypervirulent Klebsiella pneumoniae mediated by K2 capsule-specific phage and the corresponding mechanisms. Front. Microbiol. 14:1156292. doi: 10.3389/fmicb.2023.1156292
Tanji, Y., Shimada, T., Fukudomi, H., Miyanaga, K., Nakai, Y., and Unno, H. (2005). Therapeutic use of phage cocktail for controlling Escherichia coli O157:H7 in gastrointestinal tract of mice. J. Biosci. Bioeng. 100, 280–287. doi: 10.1263/jbb.100.280
Tanji, Y., Shimada, T., Yoichi, M., Miyanaga, K., Hori, K., and Unno, H. (2004). Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotechnol. 64, 270–274. doi: 10.1007/s00253-003-1438-9
Tompkin, R. B. (2002). Microorganisms in foods 7: Microbiological testing in food safety management. New york: Kluwer Academic/Plenum publishiers.
Van Acker, J., De Smet, F., Muyldermans, G., Bougatef, A., Naessens, A., and Lauwers, S. (2001). Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J. Clin. Microbiol. 39, 293–297. doi: 10.1128/JCM.39.1.293-297.2001
Wang, L., Hu, X., Tao, G., and Wang, X. (2012). Outer membrane defect and stronger biofilm formation caused by inactivation of a gene encoding for heptosyltransferase I in Cronobacter sakazakii ATCC BAA-894. J. Appl. Microbiol. 112, 985–997. doi: 10.1111/j.1365-2672.2012.05263.x
Wang, X., Loh, B., Gordillo Altamirano, F., Yu, Y., Hua, X., and Leptihn, S. (2021). Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg. Microbes Infect. 10, 2205–2219. doi: 10.1080/22221751.2021.2002671
Wang, L., Pang, X., Zhao, J., Jin, H., Yang, X., Fu, S., et al. (2022). Isolation and characteristics of new phage JK004 and application to control Cronobacter sakazakii on material surfaces and powdered infant formula. LWT 153:112571. doi: 10.1016/j.lwt.2021.112571
Yang, Y., Shen, W., Zhong, Q., Chen, Q., He, X., Baker, J. L., et al. (2020). Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 11:327. doi: 10.3389/fmicb.2020.00327
Zdobnov, E. M., and Apweiler, R. (2001). InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848. doi: 10.1093/bioinformatics/17.9.847
Keywords: Cronobacter sakazakii, phage cocktail, flagella, lipopolysaccharides, bacteriophage therapy, phage resistance, fitness trade-off
Citation: Kim S, Son B, Kim Y, Kim H, Nam G, Shin H and Ryu S (2024) Targeted dual-receptor phage cocktail against Cronobacter sakazakii: insights into phage-host interactions and resistance mechanisms. Front. Microbiol. 15:1468686. doi: 10.3389/fmicb.2024.1468686
Edited by:
Aneta Skaradzinska, Wroclaw University of Environmental and Life Sciences, PolandReviewed by:
Qingli Dong, University of Shanghai for Science and Technology, ChinaSylwia Bloch, University of Gdansk, Poland
Copyright © 2024 Kim, Son, Kim, Kim, Nam, Shin and Ryu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hakdong Shin, aHNoaW5Ac2Vqb25nLmFjLmty; Sangryeol Ryu, c2FuZ3J5dUBzbnUuYWMua3I=
†These authors have contributed equally to this work and share first authorship
 Seongok Kim
Seongok Kim Bokyung Son
Bokyung Son Yeran Kim3,4
Yeran Kim3,4 Hakdong Shin
Hakdong Shin Sangryeol Ryu
Sangryeol Ryu