- 1Department of Clinical Laboratory Medicine, Guangdong Provincial Key Laboratory of Major Obstetric Diseases, Guangdong Center for Provincial Clinical Research Obstetrics and Gynecology, The Third Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
- 2School of Pharmacy, Guangzhou Xinhua University, Guangzhou, China
- 3Department of Immunology and Microbiology, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, China
- 4Advanced Medical Technology Center, The First Affiliated Hospital, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, China
- 5Key Laboratory of Tropical Diseases Control (Sun Yat-sen University), Ministry of Education, Guangzhou, China
Colistin (CT) is the last-resort of antibiotic against multidrug-resistance (MDR) Acinetobacter baumannii (A. baumannii) infection. However, colistin resistance is increasingly reported in A. baumannii isolates partially due to the global emergence and dissemination of plasmid-borne mobile colistin resistance (mcr) gene and is a threat to human health. Thus, available treatment strategies urgently required in the fight against colistin-resistant A. baumannii. Here, we showed that mcr confers damaged outer membrane (OM) permeability in A. baumannii, which could compromise the viability of A. baumannii. Consistently, A. baumannii with colistin resistance exhibits increased susceptibility to macromolecular antibiotics such as rifampicin (RIF) and erythromycin (ERY). Moreover, the combination therapy of colistin and rifampicin demonstrates efficacy against colistin-resistant A. baumannii, regardless of the presence of mcr. Altogether, our data suggest that the synergy of colistin in combination with macromolecular hydrophobic antibiotics poses a promising therapeutic alternative for colistin-resistant A. baumannii.
1 Introduction
Acinetobacter baumannii is identified as a clinically significant opportunistic pathogen causing extensively nosocomial infections, especially in intensive care unit (ICU) patients, patients with prolonged hospitalization and patients undergoing central vascular catheterization and tracheostomy (Morris et al., 2019; Ayoub Moubareck and Hammoudi Halat, 2020). A. baumannii is intrinsically resistant to penicillins and cephalosporins, which were previously considered the first-line treatments for A. baumannii infections (Dijkshoorn et al., 2007). Except that, A. baumannii are resistant to almost all available antibiotics (Gordon and Wareham, 2010). Hence, colistin (also known as polymyxin E), which target the negatively charged phosphate groups of lipid A, is considered one of the final therapeutic options for managing MDR A. baumannii infections (Novović and Jovčić, 2023).
The recent surge in colistin usage in clinical practice has led to the rapid spread of resistance in A. baumannii (Novović and Jovčić, 2023). There are two primary mechanisms of colistin resistance in A. baumannii: complete loss or modifications of the target Lipopolysaccharide (LPS), resulting in the elimination or reduction of its negative charge (Olaitan et al., 2014). Among these, plasmid-mediated colistin resistance encoded by mcr genes has been identified as a significant factor driving rapid dissemination through horizontal gene transfer in A. baumannii. Since the first mobilized colistin resistance gene, mcr-1, was founded in Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) in China in 2016 (Liu et al., 2016), mcr-positive A. baumannii has been isolated from various sources: human feces, human clinical samples (Martins-Sorenson et al., 2020), livestock, food origin, aquaculture products, retail meat etc. (Hameed et al., 2019; Kareem, 2020). The mcr-1 and mcr-4.3 are most commonly detected in A. baumannii (Novović and Jovčić, 2023). The rapid spread of colistin resistance in A. baumannii has made it essential to thoroughly investigate new potential last-resort therapeutic options.
Recently, promising results have been demonstrated in vitro for cefiderocol (Stefano Stracquadanio et al., 2022), intravenous fosfomycin (Marino et al., 2022), and combination therapy with sulbactam–durlobactam (Karlowsky et al., 2022) in the treatment of infections caused by colistin-resistance A. baumannii. Macromolecular-hydrophobic antibiotics such as rifampicin and erythromycin have shown significant synergy with colistin against mcr-1-positive Enterobacteriaceae, like E. coli and K. pneumoniae (MacNair et al., 2018). MacNair et al. propose that the mechanism behind this antibiotic potentiation is due to MCR-1 providing minimal protection against outer membrane perturbation. Indeed, concurrently conferring colistin resistance to the bacteria, MCR-1 disrupts lipid homeostasis, affecting outer membrane permeability, and subsequently leading to compromised viability of E. coli and K. pneumoniae (Feng et al., 2022). Similar to the impact of MCR, colistin resistance mediated by mutation of lpxA results in the complete loss of LPS/Lipooligosaccharide (LOS), leading to a reduction in membrane integrity of A. baumannii (Moffatt et al., 2010). However, it remains unclear whether MCR influences the viability of A. baumannii and subsequently affects the effectiveness of synergy antibiotic treatment.
Here, we show that the increased OM permeability affected the viability of mcr-1.1 and mcr-4.3 positive A. baumannii strains in the stationary phase. Moreover, the damage to the OM leads to an increased sensitivity of mcr-positive A. baumannii to a variety of macromolecular antibiotics such as rifampicin and erythromycin. Consistently, colistin potentiates rifampicin and erythromycin in colistin-resistance A. baumannii with or without mcr gene. Our study would help to develop therapeutic strategies to improve effectiveness of treatment for A. baumannii infections.
2 Materials and methods
2.1 Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. The primers used in this study are listed in Supplementary Table S2. Unless otherwise stated, all A. baumannii strains were grown in Luria–Bertani (LB) broth or on LB agar plates at 37°C. Antibiotics and other chemicals were used at the following final concentrations: gentamicin, 10 μg/mL; colistin, 4 μg/mL; kanamycin, 50 μg/mL.
2.2 Strain construction
To generate pMMB67EH-NP-mcr-1.1 and pMMB67EH-NP-mcr-4.3, the mcr-1.1/mcr-4.3 gene and their native promoter were amplified from an mcr-1-carrying IncX4 (Feng et al., 2018) plasmid and mcr-4.3 carrying plasmid (Ma et al., 2019) and then cloned into pMMB67EH-spy, a low-copy number plasmid (containing the RSF1010 origin of replication). The map of plasmids and recombinant plasmids used for constructing mcr-positive A. baumannii strains are listed into Supplementary Figure S1.
2.3 Growth kinetics
The A. baumannii strains were grown to mid-log phase (OD600 = 0.6–0.8) in LB medium. The cultures were pelleted, washed once with PBS, adjusted to an OD600 of 0.5 with PBS, and diluted 1: 10. Then, 10 μL of the diluted culture was added into 90ul of LB. The OD600 of the culture within 24 h was determined using a microplate reader (BioTek). Three replicates were analyzed for each strain.
2.4 Antimicrobial susceptibility testing
The MIC of the antimicrobial agents against A. baumannii with or without MCR were determined using the standard broth microdilution method, according to the CLSI 2020 guidelines. In brief, all antibiotics were 2-fold diluted in Mueller-Hinton broth (MHB), and 90 μL of this solution was mixed with 10 μL of diluted culture, containing approximately 1.5 × 106 CFU/mL in a 96-well microtiter plate. After 16–20 h of incubation at 37°C, the MIC values were defined as the lowest concentration of antibiotics with no visible bacterial growth. Experiments were performed with 3 biological replicates.
2.5 In vitro competition assays
In vitro competition experiments were used to measure the competitiveness of the ATCC 17978 (mcr-1.1/pMMB67EH), ATCC 17978 (mcr-4.3/pMMB67EH), and ATCC 17978 (pMMB67EH only). The mcr-positive strains (both resistant to gentamicin and colistin) were competed against ATCC 17978 (pMMB67EH only, only resistant to gentamicin) and plating assays on LB agar plates with colistin/gentamicin or gentamicin was used to measure changes in the CFU of the two strains during competition. All competitions were carried out in M9 medium with three replicates per strain. The bacteria were cultured overnight in LB supplemented with the appropriate antibiotics. After the cell density was normalized (OD600 = 0.5), the bacteria solution was diluted 1: 100 in M9 broth and mixed at 1: 1 ratio. Cultures were incubated at 37°C with shaking (220 rpm). After 12, 24, 48, 72 h, aliquots were serially diluted 10-fold in PBS, and 10 μL of each dilution was spotted onto LB agar with colistin/gentamicin (CFU mcr ) or gentamicin (CFU total). The plates were incubated at 37°C and enumerate the rate by CFU count after 16 h: rate (%) = CFU mcr /CFU total * 100%.
2.6 SDS sensitivity assay
Bacterial cells were grown overnight in LB broth at 37°C. After the cell density was normalized (OD600 = 0.5), the resulting cultures were serially diluted, and 10 μL of each dilution was spotted onto LB agar or LB agar supplemented with the indicated concentrations of sodium dodecyl sulfate (SDS) and ethylenediaminetetraacetic acid (EDTA) (Figure 1A: 0.1% SDS + 0.2 mM EDTA or 0.1% SDS + 1.0 mM EDTA; Figure 1B: 0.1% SDS + 0.4 mM EDTA or 0.5% SDS + 0.4 mM EDTA). The plates were incubated at 37°C and photographed after ∼24 h (Feng et al., 2022). Experiments were performed with 3 biological replicates.
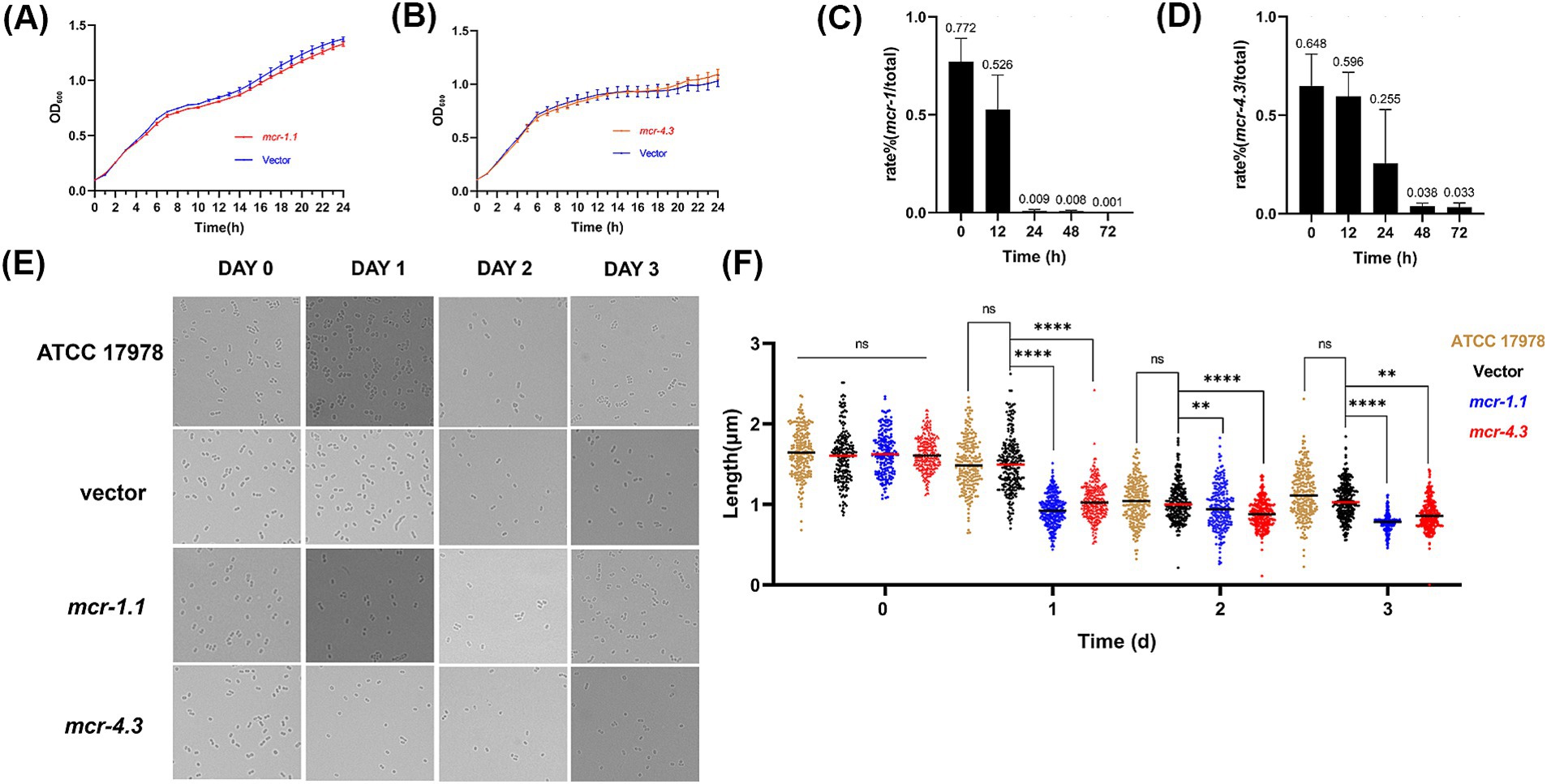
Figure 1. MCR affect the morphology and growth of A. baumannii in the stationary phase. (A,B) Growth curve of A. baumannii ATCC17978 carrying empty vector or expressing MCR were shown. (A) ATCC17978 carrying empty vector or expressing MCR-4.3; (B) ATCC17978 carrying empty vector or expressing MCR-1.1. The Y-axes showed optical densities at OD600 of broth cultures; X-axes showed time of growth (hours). (C,D) In vitro competition experiments were also performed to determine the effect of mcr-1.1 and mcr-4.3 expression on bacterial growth. Rate (%) = CFU mcr/ CFU total * 100%. CFU total is all A. baumannii (culture on LB agar supplemented with gentamicin) and CFU mcr is from MCR-expressing A. baumannii (culture on LB agar supplemented with colistin/gentamicin). (E) Images of ATCC 17978 and ATCC 17978 carrying empty vector or mcr showing morphology variant were induced during the stationary phase where bacteria are subjected to increased survival pressures and the MCR-expressing A. baumannii was too weak to grow better (>200 cells for each group). (F) Scatterplots of cell length in ATCC 17978 (gold), Vector cells (black), MCR-1.1-expressing cells (blue) and MCR-4.3-expressing cells (red) at the indicated time points. Middle lines represented median values.
2.7 Whole-genome sequencing and SNP analysis
The genomic DNA of the F7-AB was extracted using the Qiagen Blood and Tissue kit (Qiagen, Hilden, Germany). DNA libraries were constructed with 350 bp paired-end fragments and sequenced using an Illumina HiSeq 2000 platform. The Illumina reads of the strain were mapped against the reference genome sequence of A. baumannii ATCC 19606 (GenBank accession: GCF_009035845.1) using the program MAQ. In our WGS analysis, we used a variety of programs for different steps. We started with FastQC (Babraham Bioinformatics, 2010) and Trimmomatic (Bolger et al., 2014) for quality control and preprocessing of raw sequencing data. Trimmed reads with length > 30 and Phred scores > 20 were retained for subsequent analyzes. Then, we employed BWA (Li and Durbin, 2009) for aligning the reads to a reference genome (A. baumannii ATCC 19606; GenBank accession: GCF_009035845.1). Variant calling was done using Genome Analysis Toolkit (GATK) (McKenna et al., 2010; DePristo et al., 2011). After obtaining the variant calls from the WGS data, we further filtered and annotated the SNPs using VCFtools (Danecek et al., 2011) and SnpEff (Cingolani et al., 2014) for SNP analysis. We excluded ambiguous calls as wells as indels and MNVs. We also excluded calls with low coverage using a minimum depth of either 10% of the mean coverage and calls with mapping quality or base quality scores < 30. Consistent with previous findings (Oikonomou et al., 2015; Nhu et al., 2016; Zhang et al., 2017; Nurtop et al., 2019; Jovcic et al., 2021; Kabic et al., 2023), four mutations [LpxD (E117K), PmrA (I13N), PmrB (A138T and A444V)] linked to colistin resistance were detected among the screened SNPs (Supplementary Table S6).
2.8 Colistin combination susceptibility testing
Synergy measurement was utilized to determine the antimicrobial effect of colistin and macromolecular hydrophobic antibiotics (rifampicin, erythromycin and cefoxitin) upon colistin-resistance A. baumannii (MCR-1.1-expressing cells, MCR-4.3-expressing cells, pig feces-derived MCR-4.3-expressing A. baumannii strain AB18PR065, clinical A. baumannii strain F7-AB). The checkerboard assay was set up in a 96-well plate. Briefly, columns 1 to 8 contained 2-fold serial dilutions of colistin, and rows A to H contained 2-fold serial dilutions of macromolecular hydrophobic antibiotics. Then the strains were cultured to exponential phase and adjusted to a density of OD600 = 0.5 by diluting with LB broth. After dilution at a ratio of 1: 10, 20 μL of diluted culture was added to 180 μL of LB broth containing colistin and another indicated antibiotics in a 96-well plate. A nanophotometer (NP80, IMPLEN) was utilized to measure the optical density at 600 nm (OD600) in each well before and after incubation at 37°C for 16 h. The fractional inhibitory concentration index (FICI) was calculated according to a previously published protocol (Pál et al., 2023). Briefly, FICI = (MIC of drug A in combination) / (MIC of drug A alone) + (MIC of drug B in combination) / (MIC of drug B alone), Synergy is defined as FICI ≤ 0.5, Indifference is defined as 0.5 < FICI ≤ 4, Antagonism is defined as FICI > 4 (Jenkins and Schuetz, 2012).
2.9 Synthesis of cDNA and quantitative real-time PCR
Exponentially growing bacterial cultures (optical density at OD600 = 0.4–0.6) of ATCC 17978 (mcr-1.1/pMMB67EH), ATCC 17978 (mcr-4.3/pMMB67EH) and ATCC 17978 (pMMB67EH only, as a control group) were pelleted. After the supernatant was removed, the cell pellet was resuspended in RNA-easy Isolation Reagent (Vazyme). The total RNA was precipitated by the addition of isopropanol and collected by centrifugation. The supernatant was discarded, and the RNA pellet was washed with 75% ethanol. After the pellet was air-dried, the mRNA was dissolved in RNase-free H2O. Any contaminating genomic DNA was digested with gDNA wiper Mix (Vazyme). The purified mRNA was reverse transcribed to cDNA with HiScript II qRT SuperMix (Vazyme). The cDNA levels of the target genes were then quantified with quantitative real-time PCR (qPCR) on CFX Opus 96 Real-Time PCR Instrument (Bio-Rad) using AceQ Universal SYBR qPCR Master Mix (Vazyme). All qPCR primers were determined to be >95% efficient, and the cDNA molecular masses were experimentally confirmed to be within the linear dynamic range of the assay. The signals were normalized to those of the housekeeping 16S rRNA transcript and quantified with the ΔΔCt method. The error bars are the 95% confidence intervals of three technical replicates.
2.10 Bright-field microscopy for measuring cell length
The A. baumannii strains were grown to mid-log phase (OD600 = 0.6–0.8) in LB medium. The cultures were pelleted, washed once with PBS, adjusted to an OD600 of 0.5 with PBS, and diluted 1: 10. Then, 200 μL of the diluted culture was added into 1800ul of LB. Cultures were incubated at 37°C with shaking (220 rpm). After 0, 1, 2, 3 day, bright-field imaging was performed on an inverted microscope (Olympus BX63). The ImageJ software was used to analyze the cell length.
2.11 Statistical analysis
Statistical analysis was performed using Prism (version 8.3.0, GraphPad Software). Data were analyzed using the paired Student’s t test, and for comparison of data from three or more conditions, analysis of variance (ANOVA) was used. p value of 0.05 or less was considered statistically significant.
3 Results
3.1 MCR affects the morphology and growth of Acinetobacter baumannii in the stationary phase
Although evidence of our previous study showed that expression of MCR-1 induces cell shrinkage and death in E. coli and K. pneumonia during the stationary phase (Feng et al., 2022), the impact of MCR on the viability of A. baumannii remains unclear. To study the impact of MCR on the physiology of A. baumannii, the plasmids pMMB67EH-NP-mcr-1.1 and pMMB67EH-NP-mcr-4.3 carrying mcr-1.1 and mcr-4.3 and their native promoters, respectively, were generated, and the low-copy number pMMB67EH-spy plasmid served as the empty vector control. We then constructed three strains, A. baumannii strain ATCC 17978 carrying pMMB67EH-NP-mcr-1.1 (MCR-1.1-expressing cells), strain ATCC 17978 carrying pMMB67EH-NP-mcr-4.3 (MCR-4.3-expressing cells) and ATCC 17978 carrying the empty vector pMMB67EH-spy (Vector cells). Minimum inhibitory concentration (MIC) of colistin against the three strains above were then determined (Table 1), and the growth of MCR-1.1-expressing cells, MCR-4.3-expressing cells and Vector cells in LB medium were monitored (Figures 2A,B). Like previously observed in the E. coli (Feng et al., 2022), the expression of MCR did not affect the growth of A. baumannii ATCC 17978 (Figures 2A,B). However, when we extended the incubation time, we found that the MCR-expressing cells have shorter length compared to vector cells in the stationary phase (Figure 2E). Meanwhile, in vitro competition experiments were also performed to determine the effect of mcr-1.1 expression on bacterial growth in the stationary phase. As shown in Figures 2C,D, after 24 h, the growth rates of A. baumannii strains expressing MCR showed significant decrease when compared to Vector cells (pMMB67EH-spy, without mcr). These results suggest that the viability and competitiveness of mcr-positive A. baumannii strains was impaired in the stationary phase.
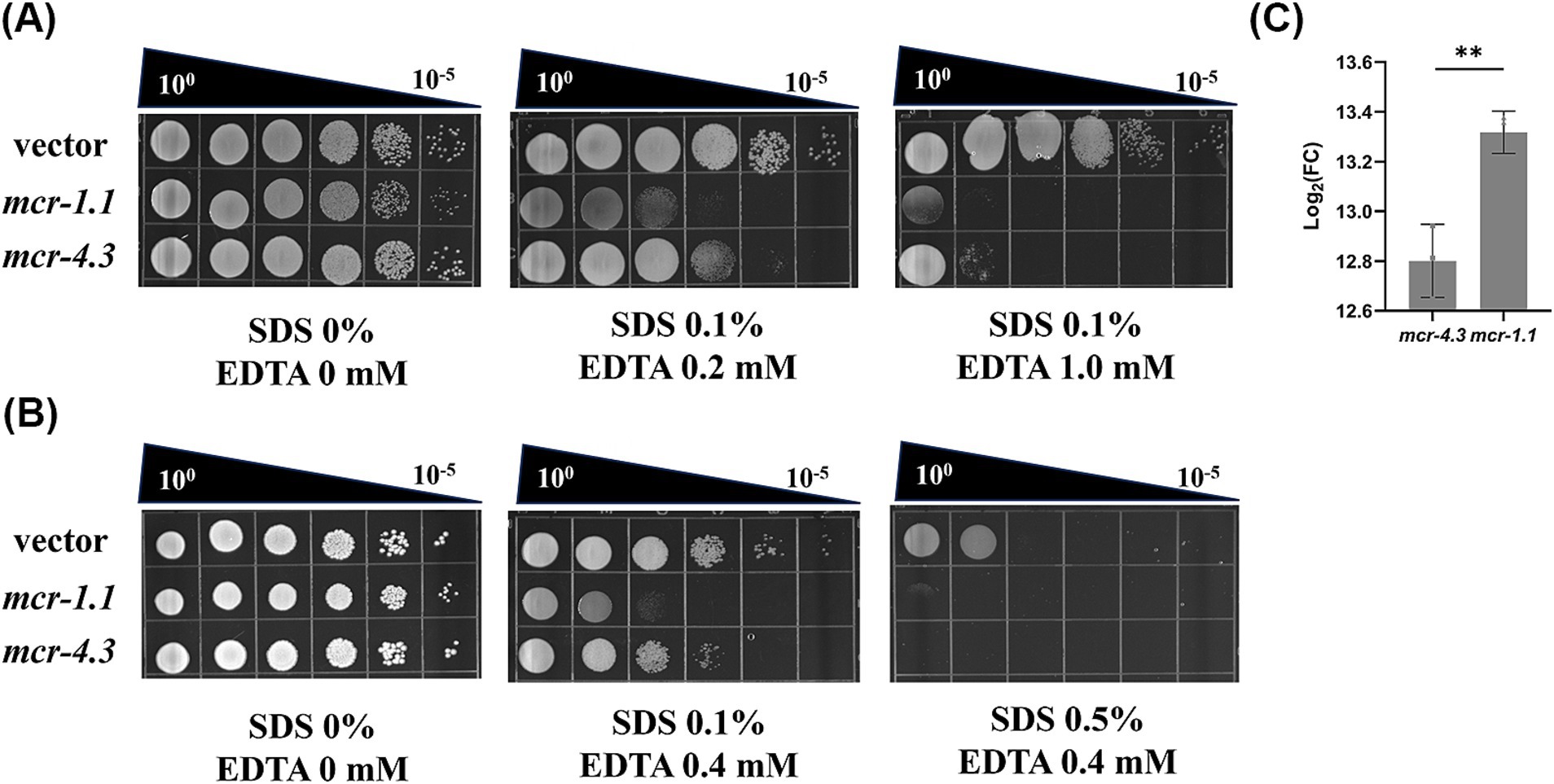
Figure 2. MCR confer an outer membrane permeability defect to A. baumannii. (A) Efficiency of plating assays on LB agar plates containing 0.1% SDS and 0.2 mM or 1.0 mM EDTA were shown on 10-fold dilutions of cultures. Cells expressing mcr-1.1 and mcr-4.3 were more sensitive to SDS/EDTA, indicating OM permeability impair. (B) Efficiency of plating assays on LB agar plates containing 0.1% or 0.5% SDS and 0.4 mM EDTA were shown on 10-fold dilutions of cultures. Each assay was repeated 3 times with similar results. (C) Levels of mcr mRNA in MCR-1.1-expressing or MCR-4.3-expressing A. baumannii ATCC 17978.
3.2 MCR impairs OM permeability in Acinetobacter baumannii
To understand how MCR damage the viability of A. baumannii strains, we investigated the mechanism underlying this phenomenon. Overexpression of mcr-1 results in significant degradation in cell membrane and cytoplasmic structures (Yang et al., 2017). Therefore, we wonder whether the bacterial death caused by MCR in A. baumannii strains results from increased OM permeability. To test this, we employed SDS/EDTA sensitivity assays to measure the OM permeability of MCR-1.1-expressing cells, MCR-4.3-expressing cells, vector cells (Figures 3A,B). As expected, all the MCR-expressing cells displayed increased sensitivity to detergents SDS/EDTA compared with the vector cells, indicating that MCR-1.1 and MCR-4.3 both increased the permeability of OM in A. baumannii. Moreover, MCR-1.1-expressing cells were more sensitive to SDS/EDTA than MCR-4.3-expressing cells, which may be associated with the different expression level of MCR protein (Figure 3C). These data demonstrated that the increased OM permeability affected the viability of mcr-positive A. baumannii strains in the stationary phase.
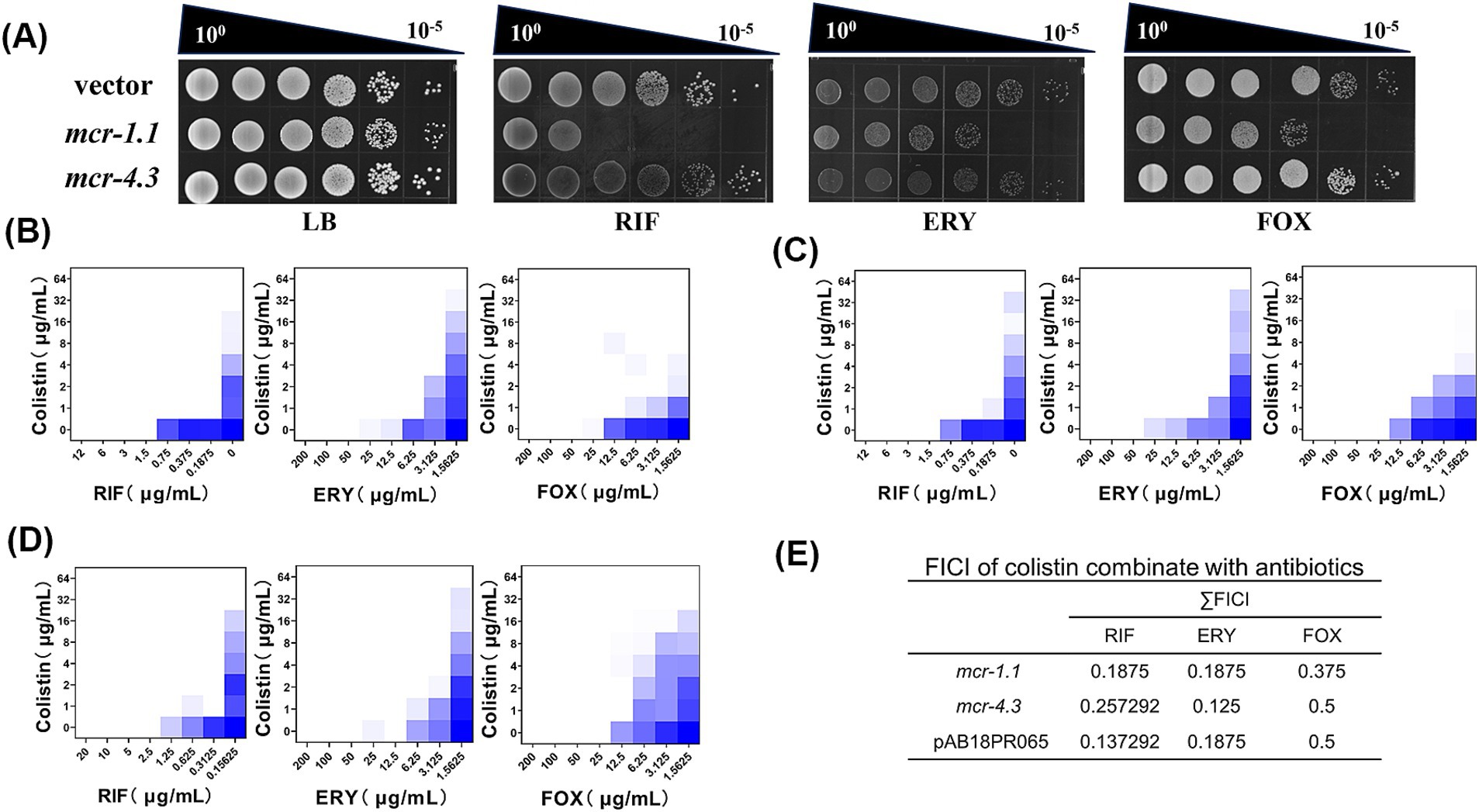
Figure 3. Colistin potentiates macromolecular hydrophobic antibiotics in mcr-positive A. baumannii. (A) Efficiency of plating assays on LB agar plates with 0.75 μg/mL RIF, 10 μg/mL ERY, 15 μg/mL FOX. Ten-fold dilutions of cultures are indicated above the left plate. MCR-expressing cells exhibited increased sensitivity to macromolecular antibiotics. (B–D) Checkerboard broth microdilution assays showing dose-dependent potentiation of rifampicin, erythromycin and cefoxitin by colistin against A. baumannii expressing (B) MCR-1.1; (C) MCR-4.3; and (D) a pig feces-derived MCR-4.3-expressing A. baumannii strain (AB18PR065). The calculated values of FICI are listed in (E). (E) The FICI is the sum of the two FICIs, with an FICI index ≤0.5 considered synergistic. Dark blue regions represent higher cell density. Data in (B–D) represent the mean OD600 of two biological replicates. RIF, rifampicin; ERY, erythromycin; FOX, cefoxitin.
3.3 Colistin potentiates antibiotics in mcr-positive Acinetobacter baumannii
Acinetobacter is intrinsically resistant toward many antibiotics because of the low permeability of its outer membrane (Zahn et al., 2016), especially resistant to OM-impermeable large molecule antimicrobials due to the absence of large-channel general porins such as OmpF and OmpC (Sugawara and Nikaido, 2012). Therefore, we speculated that mcr-positive A. baumannii might be more sensitive to certain antimicrobials. To do it, we screened nearly 10 antibiotics (Supplementary Table S3), covering most of macromolecular hydrophobic antibiotics for change in MIC with or without the presence of MCR protein. As we had expected, we found that MCR-expressing cells were more sensitive to rifampicin (RIF), erythromycin (ERY), and MCR-1.1-expressing cells were more sensitive to cefoxitin (FOX), in contrast to MCR-negative cells (Figure 1A). Next, we aimed to investigate whether antibiotics could potentiate this effect. In the presence of colistin, several antibiotics were highly potentiated. Checkerboard broth microdilution assays showed potentiation of RIF, ERY, and FOX by colistin against MCR-expressing A. baumannii (Figures 1B–D). By calculating their Fractional Inhibitory Concentration Index (FICI), in which ≤0.5 indicating synergistic, we found that there was potent synergy of colistin combination with rifampicin, erythromycin and cefoxitin versus MCR-expressing A. baumannii (Figures 1B–D).
To investigate whether colistin potentiation is conserved beyond laboratory-generated MCR-expressing A. baumannii, we tested the FICI of a MCR-4.3-expressing A. baumannii strain (AB18PR065) derived from pig feces (Ma et al., 2019) in colistin combination with rifampicin, erythromycin and cefoxitin. As expected, the FICI was less than or equal to 0.5 for all three drugs (Figures 1D,E), indicating that MCR-4.3 also rendered the potentiation of large molecule antimicrobials in clinical A. baumannii expressing MCR.
3.4 Colistin potentiates RIF in clinical colistin-resistance Acinetobacter baumannii
Given the therapeutic potential of combination therapy in mcr-positive A. baumannii, we wondered whether this potentiation could be applicated to clinical chromosomally mediated colistin resistance A. baumannii, since the complete loss or modifications of the target LPS are two main mechanisms of chromosomally mediated colistin resistance A. baumannii (Novović and Jovčić, 2023), which is similar to the acquired mechanism of mcr encoding a phosphoethanolamine (pEtN) transferase that adds pEtN to the lipid A component of LPS. To test this, we fist the FICI of a clinical A. baumannii strain F7-AB which is intrinsically resisted to colistin [LpxD (E117K), PmrA (I13N), PmrB (A138T and A444V)] (Oikonomou et al., 2015; Nhu et al., 2016; Zhang et al., 2017; Nurtop et al., 2019; Jovcic et al., 2021; Kabic et al., 2023). Encouragingly, the result of the checkerboard assay was similar with that of mcr-positive A. baumannii except erythromycin and cefoxitin (Figure 4). These observations indicate that the combination of colistin with rifampicin is not only able to overcome mcr mediated acquired colistin resistance, but also able to treat chromosomal colistin resistance.
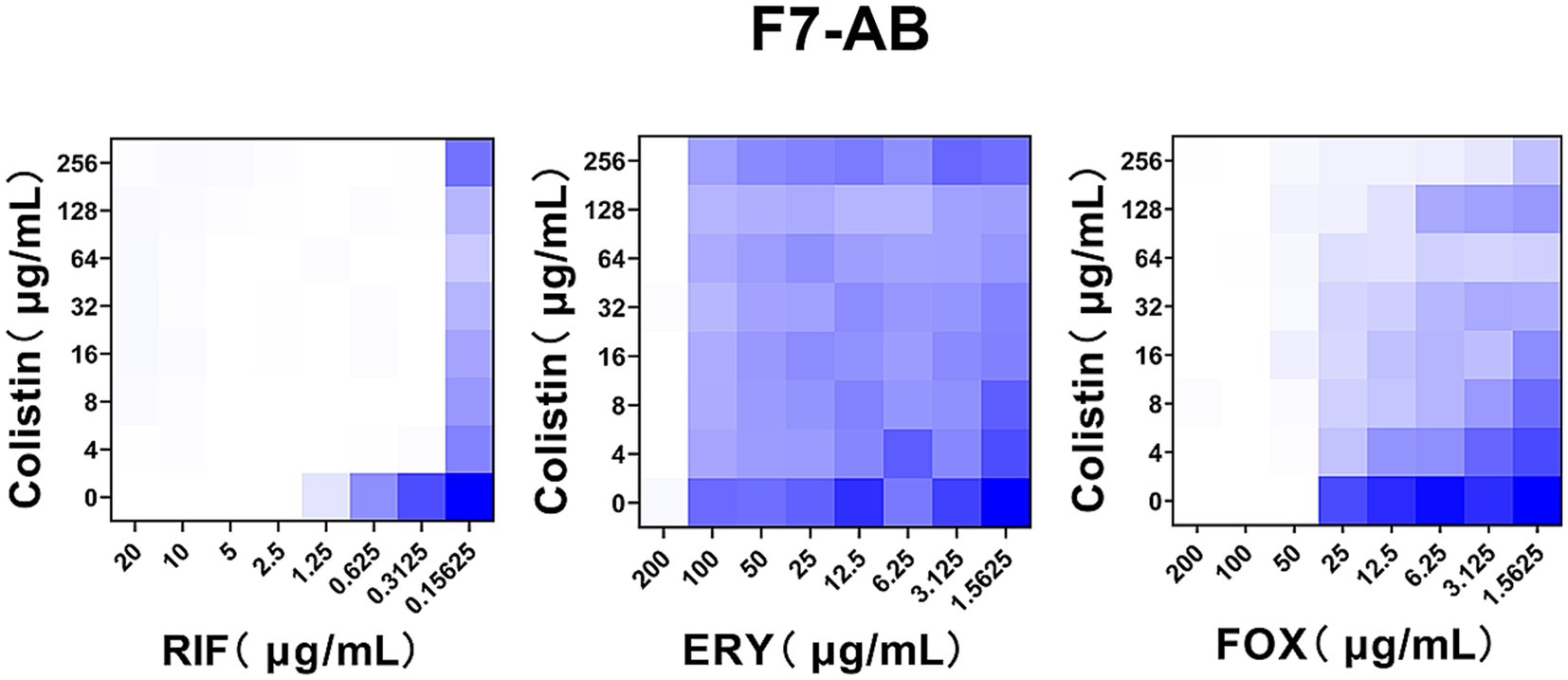
Figure 4. Colistin potentiates RIF in clinical colistin-resistance A. baumannii. Checkerboard broth microdilution assays showed dose-dependent potentiation of rifampicin but not erythromycin and cefoxitin by colistin against clinical A. baumannii strain F7-AB which is intrinsically resistant to colistin. Dark blue regions represent higher cell density. Data represent the mean OD600 of two biological replicates. RIF, rifampicin; ERY, erythromycin; FOX, cefoxitin.
4 Discussion
In this study, we were aimed at better understanding impact of MCR on mcr-positive A. baumannii in the stationary phase and bridging the gap between widespread colistin resistance and the development of new treatments. Our data founded that MCR shortened the cell lengths and decreased the competitiveness of A. baumannii. Then using SDS sensitivity assay, we found that MCR provides serious disruption to the OM of A. baumannii, which was thought to be resistant to many large molecule antibiotics due to its lower membrane permeability (Zahn et al., 2016). Therefore, we used antimicrobial susceptibility testing (AST) and checkerboard assay to find that although mcr-1.1/mcr-4.3 confers no change in susceptibility to all selected antibiotics, colistin and rifampicin, erythromycin and cefoxitin combination therapy shows efficacy against colistin-resistance A. baumannii.
A recent study reported that MCR-1 damages the OM permeability barrier by disrupting lipid homeostasis in E. coli and K. pneumonia, resulting in cell shrinkage and death during stationary phase (Feng et al., 2022). Furthermore, overexpression of MCR results in impaired competitive ability and drastic degradation in Gram-negative cell membrane (Yang et al., 2017). The same phenomena were observed in our study on the outer membrane of mcr-positive A. baumannii. Given the significance of A. baumannii infection and its prevalence in hospital setting (Magill et al., 2014; Lob et al., 2016), we wonder whether the strains expressing MCR will be more vulnerable than vector cells so that the colistin-resistance cells could be eliminated by some large antibiotics capable of penetrating the cell membrane. Unexpectedly, the susceptibility of intracellular antibiotics does not change when cells expressed MCR-1.1 or MCR-4.3. This observation is contrary to previous reports that the large OM-impermeable antimicrobials such as vancomycin, rifampicin and erythromycin would be more lethal to gram-negative bacteria with severe damaged outer membrane (Muheim et al., 2017; Mecsas et al., 2021). Indeed, more and more evidences support that OM permeability impaired strains need more stress conditions to display decreased growth rate, cell viability, competitive ability and more sensitive to large-scaffold antibiotics (Heesterbeek et al., 2019; Mecsas et al., 2021; Feng et al., 2022). And the stressful environment can be osmotic stresses (Ohnishi et al., 2004; Virolle et al., 2018), oxidative stress responses (Sun et al., 2014; Mancini and Imlay, 2015) and starvation in stationary phase. Accordingly, we show that MCR did not damage the growth of A. baumannii during the first day of culture, but impair the cells viability in the stationary phase.
Colistin acts by associating with the anionic LPS component of the Gram-negative outer membrane, causing membrane destabilization that leads to cell envelope permeability, leakage of cellular contents, and ultimately lytic cell death (Schindler and Osborn, 1979). When in combination with colistin, RIF or ERY can overcome mcr-1 mediated colistin resistance in Enterobacteriaceae. Likewise, we found that as Gram-negative bacteria, mcr-positive A. baumannii is more sensitive to the combination of colistin and RIF or ERY. In this case, we guess that colistin just like a stress which aggravate the OM damage in mcr-positive A. baumannii, then large antibiotics like RIF can enter the cells and function more efficiently. Similar to the mechanism of resistance mediated by mcr, colistin resistance cause by chromosomal mutations include complete loss of LPS by inactivation of the biosynthetic pathway or modifications of target LPS driven by the addition of pEtN moieties to lipid A (Adams et al., 2009; Moffatt et al., 2010; Olaitan et al., 2014). The protective function of outer membrane that serves as a barrier is dependent on LPS and the OM permeability will increase when LPS was changed (Sherman et al., 2018). On this account, our data suggested that the combination of colistin with rifampicin also have the ability to overcome chromosomal colistin resistance in clinical A. baumannii strains F7-AB.
In conclusion, our findings suggest that mcr-1.1 and mcr-4.3 can induce OM defect in A. baumannii then leading to growth inhibition. Due to the increased permeability, synergy with colistin potentiates rifampicin in colistin-resistance A. baumannii. These findings would help develop strategies to improve the therapeutic effects of colistin-resistance A. baumannii infection.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1138508.
Author contributions
ML: Data curation, Writing – original draft, Writing – review & editing. FM: Data curation, Funding acquisition, Writing – original draft. HZ: Data curation, Writing – original draft. DZ: Methodology, Writing – original draft. LL: Methodology, Writing – original draft. RL: Methodology, Writing – original draft. JL: Methodology, Writing – original draft. YW: Methodology, Writing – original draft. LX: Methodology, Writing – original draft. CL: Methodology, Writing – original draft. G-BT: Methodology, Writing – original draft. SF: Conceptualization, Funding acquisition, Writing – review & editing, Writing – original draft. YX: Conceptualization, Funding acquisition, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant number 82072319 to YX, grant numbers 82002173, 82272378 to SF), Medical Scientific Research Foundation of Guangdong Province of China (grant number B2023250 to FM).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1468682/full#supplementary-material
References
Adams, M. D., Nickel, G. C., Bajaksouzian, S., Lavender, H., Murthy, A. R., Jacobs, M. R., et al. (2009). Resistance to Colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53, 3628–3634. doi: 10.1128/aac.00284-09
Ayoub Moubareck, C., and Hammoudi Halat, D. (2020). Insights into Acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 9:119. doi: 10.3390/antibiotics9030119
Babraham Bioinformatics (2010). FastQC a quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed December 2, 2023).
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2014). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6, 80–92. doi: 10.4161/fly.19695
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
DePristo, M. A., Banks, E., Poplin, R., Garimella, K. V., Maguire, J. R., Hartl, C., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. doi: 10.1038/ng.806
Dijkshoorn, L., Nemec, A., and Seifert, H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5, 939–951. doi: 10.1038/nrmicro1789
Feng, S., Liang, W., Li, J., Chen, Y., Zhou, D., Liang, L., et al. (2022). MCR-1-dependent lipid remodelling compromises the viability of gram-negative bacteria. Emerg. Microb. Infect. 11, 1236–1249. doi: 10.1080/22221751.2022.2065934
Feng, S., Shen, C., Chen, H., Zheng, X., Xia, Y., Zhong, L.-L., et al. (2018). Co-production of MCR-1 and NDM-5 in Escherichia coli isolated from a colonization case of inpatient. Infect. Drug Resist. 11, 1157–1161. doi: 10.2147/idr.S171164
Gordon, N. C., and Wareham, D. W. (2010). Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Agents 35, 219–226. doi: 10.1016/j.ijantimicag.2009.10.024
Hameed, F., Khan, M. A., Muhammad, H., Sarwar, T., Bilal, H., and Rehman, T. U. (2019). Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: first report from Pakistan. Rev. Soc. Bras. Med. Trop. 52:e20190237. doi: 10.1590/0037-8682-0237-2019
Heesterbeek, D. A. C., Martin, N. I., Velthuizen, A., Duijst, M., Ruyken, M., Wubbolts, R., et al. (2019). Complement-dependent outer membrane perturbation sensitizes gram-negative bacteria to gram-positive specific antibiotics. Sci. Rep. 9:3074. doi: 10.1038/s41598-019-38577-9
Jenkins, S. G., and Schuetz, A. N. (2012). Current concepts in laboratory testing to guide antimicrobial therapy. Mayo Clin. Proc. 87, 290–308. doi: 10.1016/j.mayocp.2012.01.007
Jovcic, B., Novovic, K., Dekic, S., and Hrenovic, J. (2021). Colistin resistance in environmental isolates of Acinetobacter baumannii. Microb. Drug Resist. 27, 328–336. doi: 10.1089/mdr.2020.0188
Kabic, J., Novovic, K., Kekic, D., Trudic, A., Opavski, N., Dimkic, I., et al. (2023). Comparative genomics and molecular epidemiology of colistin-resistant Acinetobacter baumannii. Comput. Struct. Biotechnol. J. 21, 574–585. doi: 10.1016/j.csbj.2022.12.045
Kareem, S. M. (2020). Emergence of mcr-and fosA3-mediated colistin and fosfomycin resistance among carbapenem-resistant Acinetobacter baumannii in Iraq. Meta Gene 25:100708. doi: 10.1016/j.mgene.2020.100708
Karlowsky, J. A., Hackel, M. A., McLeod, S. M., and Millerc, A. A. (2022). In vitro activity of Sulbactam-Durlobactam against global isolates of Acinetobacter baumannii-calcoaceticus complex collected from 2016 to 2021. Antimicrob. Agents Chemother. 66:e0078122. doi: 10.1128/aac.00781-22
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Lob, S. H., Hoban, D. J., Sahm, D. F., and Badal, R. E. (2016). Regional differences and trends in antimicrobial susceptibility of Acinetobacter baumannii. Int. J. Antimicrob. Agents 47, 317–323. doi: 10.1016/j.ijantimicag.2016.01.015
Ma, F., Shen, C., Zheng, X., Liu, Y., Chen, H., Zhong, L., et al. (2019). Identification of a novel plasmid carrying mcr-4.3 in an Acinetobacter baumannii strain in China. Antimicrob. Agents Chemother. 63:e00133-19. doi: 10.1128/AAC
MacNair, C. R., Stokes, J. M., Carfrae, L. A., Fiebig-Comyn, A. A., Coombes, B. K., Mulvey, M. R., et al. (2018). Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat. Commun. 9:458. doi: 10.1038/s41467-018-02875-z
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., et al. (2014). Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med. 370, 1198–1208. doi: 10.1056/NEJMoa1306801
Mancini, S., and Imlay, J. A. (2015). The induction of two biosynthetic enzymes helps Escherichia coli sustain heme synthesis and activate catalase during hydrogen peroxide stress. Mol. Microbiol. 96, 744–763. doi: 10.1111/mmi.12967
Marino, A., Stracquadanio, S., Campanella, E., Munafò, A., Gussio, M., Ceccarelli, M., et al. (2022). Intravenous Fosfomycin: a potential good partner for Cefiderocol. Clinical experience and considerations. Antibiotics 12:49. doi: 10.3390/antibiotics12010049
Martins-Sorenson, N., Snesrud, E., Xavier, D. E., Cacci, L. C., Iavarone, A. T., McGann, P., et al. (2020). A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J. Antimicrob. Chemother. 75, 60–64. doi: 10.1093/jac/dkz413
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mecsas, J., Jung, H.-J., Sorbara, M. T., and Pamer, E. G. (2021). TAM mediates adaptation of carbapenem-resistant Klebsiella pneumoniae to antimicrobial stress during host colonization and infection. PLoS Pathog. 17:e1009309. doi: 10.1371/journal.ppat.1009309
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D. F., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977. doi: 10.1128/aac.00834-10
Morris, F. C., Dexter, C., Kostoulias, X., Uddin, M. I., and Peleg, A. Y. (2019). The mechanisms of disease caused by Acinetobacter baumannii. Front. Microbiol. 10:1601. doi: 10.3389/fmicb.2019.01601
Muheim, C., Götzke, H., Eriksson, A. U., Lindberg, S., Lauritsen, I., Nørholm, M. H. H., et al. (2017). Increasing the permeability of Escherichia coli using MAC13243. Sci. Rep. 7:17629. doi: 10.1038/s41598-017-17772-6
Nhu, T. K., Riordan, D. W., Do Hoang Nhu, T., Thanh, D. P., Thwaites, G., Huong Lan, N. P., et al. (2016). The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 6:28291. doi: 10.1038/srep28291
Novović, K., and Jovčić, B. (2023). Colistin resistance in Acinetobacter baumannii: molecular mechanisms and epidemiology. Antibiotics 12:516. doi: 10.3390/antibiotics12030516
Nurtop, E., Bayindir Bilman, F., Menekse, S., Kurt Azap, O., Gonen, M., Ergonul, O., et al. (2019). Promoters of Colistin resistance in Acinetobacter baumannii infections. Microb. Drug Resist. 25, 997–1002. doi: 10.1089/mdr.2018.0396
Ohnishi, H., Mizunoe, Y., Takade, A., Tanaka, Y., Miyamoto, H., Harada, M., et al. (2004). Legionella dumoffiiDjlA, a member of the DnaJ family, is required for intracellular growth. Infect. Immun. 72, 3592–3603. doi: 10.1128/iai.72.6.3592-3603.2004
Oikonomou, O., Sarrou, S., Papagiannitsis, C. C., Georgiadou, S., Mantzarlis, K., Zakynthinos, E., et al. (2015). Rapid dissemination of colistin and carbapenem resistant Acinetobacter baumannii in Central Greece: mechanisms of resistance, molecular identification and epidemiological data. BMC Infect. Dis. 15:559. doi: 10.1186/s12879-015-1297-x
Olaitan, A. O., Morand, S., and Rolain, J.-M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Pál, C., Liang, L., Zhong, L.-L., Wang, L., Zhou, D., Li, Y., et al. (2023). A new variant of the colistin resistance gene MCR-1 with co-resistance to β-lactam antibiotics reveals a potential novel antimicrobial peptide. PLoS Biol. 21:e3002433. doi: 10.1371/journal.pbio.3002433
Schindler, M., and Osborn, M. J. (1979). Interaction of divalent cations and Polymyxin B with lipopolysaccharide. Biochemistry 18, 4425–4430. doi: 10.1021/bi00587a024
Sherman, D. J., Xie, R., Taylor, R. J., George, A. H., Okuda, S., Foster, P. J., et al. (2018). Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359, 798–801. doi: 10.1126/science.aar1886
Stracquadanio, S., Bonomo, C., Marino, A., Bongiorno, D., Privitera, G. F., Bivona, D. A., et al. (2022). Acinetobacter baumannii and Cefiderocol, between Cidality and adaptability. Microbiol. Spectr. 10:e0234722. doi: 10.1128/spectrum.02347-22
Sugawara, E., and Nikaido, H. (2012). OmpA is the principal nonspecific slow Porin of Acinetobacter baumannii. J. Bacteriol. 194, 4089–4096. doi: 10.1128/jb.00435-12
Sun, W.-S. W., Syu, W. Jr., Ho, W.-L., Lin, C.-N., Tsai, S.-F., and Wang, S.-H. (2014). SitA contributes to the virulence of Klebsiella pneumoniae in a mouse infection model. Microbes Infect. 16, 161–170. doi: 10.1016/j.micinf.2013.10.019
Virolle, M.-J., Kang, Y., and Hwang, I. (2018). Glutamate uptake is important for osmoregulation and survival in the rice pathogen Burkholderia glumae. PLoS One 13:e0190431. doi: 10.1371/journal.pone.0190431
Yang, Q., Li, M., Spiller, O. B., Andrey, D. O., Hinchliffe, P., Li, H., et al. (2017). Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat. Commun. 8:2054. doi: 10.1038/s41467-017-02149-0
Zahn, M., Bhamidimarri, S. P., Baslé, A., Winterhalter, M., and van den Berg, B. (2016). Structural insights into outer membrane permeability of Acinetobacter baumannii. Structure 24, 221–231. doi: 10.1016/j.str.2015.12.009
Keywords: Acinetobacter baumannii , colistin, rifampicin, MCR, outer membrane permeability
Citation: Li M, Ma F, Zhao H, Zhou D, Liang L, Lv R, Li J, Wang Y, Xu L, Liu C, Tian G-B, Feng S and Xia Y (2024) Outer membrane permeability of mcr-positive bacteria reveals potent synergy of colistin and macromolecular antibiotics against colistin-resistant Acinetobacter baumannii. Front. Microbiol. 15:1468682. doi: 10.3389/fmicb.2024.1468682
Edited by:
Nabil Karah, Umeå University, SwedenReviewed by:
Nalumon Thadtapong, National Institute of Health of Thailand, ThailandFirdoos Ahmad Gogry, Washington State University, United States
Bingbing Sun, Center for Excellence in Molecular Plant Sciences (CAS), China
Copyright © 2024 Li, Ma, Zhao, Zhou, Liang, Lv, Li, Wang, Xu, Liu, Tian, Feng and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siyuan Feng, ZnN5MDU5M0AxNjMuY29t; Yong Xia, MjAwODY5MTA3NUBnemhtdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Meisong Li
Meisong Li Furong Ma1†
Furong Ma1† Hui Zhao
Hui Zhao Dianrong Zhou
Dianrong Zhou Lujie Liang
Lujie Liang Jiachen Li
Jiachen Li Guo-Bao Tian
Guo-Bao Tian