- 1School of Public Health, Shantou University, Shantou, China
- 2Key Laboratory of Bovine Disease Control in Northeast China, Ministry of Agriculture and Rural Affairs, College of Animal Science and Veterinary Medicine, Heilongjiang Bayi Agricultural University, Daqing, China
- 3State Key Laboratory for Diagnosis and Treatment of Severe Zoonotic Infectious Diseases, Key Laboratory for Zoonosis Research of the Ministry of Education, Institute of Zoonosis, and College of Veterinary Medicine, Jilin University, Changchun, China
- 4State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
Tabanids, commonly known as horseflies and belonging to the family Tabanidae, are blood-feeding arthropods (BFA) found worldwide. They are known for their ability to mechanically and biologically transmit various animal pathogens. Tabanids are potential vectors for diseases such as Francisella tularensis, Anaplasma marginale, Theileria spp., and contributors to lumpy skin diseases. Despite their involvement in common BFA studies, tabanids have not been extensively explored in microbiome research. In this study, the microbiota structure and composition in various organs of four distinct genera of tabanids: Atylotus, Haematopota, Tabanus, and Hybomitra were examined. High-throughput sequencing of the bacterial 16S rRNA gene was performed to gain insights into the microbial communities associated with the different tabanid species. Result display that microbiota composition and diversity, including Firmicutes, Proteobacteria, and Bacteroidetes, varied significantly among the different organs, with the ovaries exhibiting significantly higher diversity. Apart from the Haematopota genus, Tenericutes were enriched in the midgut of other tabanid species, whereas the Malpighian tubules exhibited a higher abundance of Bacteroides. Notably, the ovarian microbiota structure was conserved among the four tabanid species, indicating its potential association with reproductive development. Evaluation of the potential pathogen risk revealed putative pathogens in over 100 genera associated with these tabanid commensal organisms. Twenty genera were annotated as zoonotic agents with a high abundance of Citrobacter and Brucella, highlighting the presence of this important group of zoonotic pathogens. Functional predictions of vector-microbiota interactions indicate that microbiota significantly affects vector biological traits and can influence pathogen transmission via direct interactions or by regulating host immunity and nutrition. For the first time, the distribution characteristics and functions of four genera of horsefly microbiota were analyzed, revealing the presence of multiple potential pathogenic microorganisms. These findings provide valuable insights for future research and the development of symbiotic-based strategies to control insect-borne diseases among tabanids.
1 Introduction
Tabanids, commonly known as horseflies, belong to the family Tabanidae and are widespread blood-feeding arthropods (BFA) within the order Diptera, encompassing four subfamilies and 144 genera (Sevidzem et al., 2021). They are crucial in transmitting pathogens through mechanical and biological means, affecting wild animals, domestic livestock, and humans (Maity et al., 2017; Whyte et al., 2020). Members of the family Tabanidae are vectors implicated in transmitting more than 80 viral, bacterial, and protozoan agents (Krinsky, 1976), rendering them of notable medical and veterinary significance because of the substantial risks they pose to livestock and wildlife.
Numerous metagenomic and 16S-rRNA amplicon sequencing studies have extensively explored the symbiotic microbiota relationships in various arthropods (Shi et al., 2022; Wang et al., 2023; Song et al., 2022; Ahmad et al., 2022; Murakami et al., 2018). The microbiota functionally complements the host biology and helps synthesize essential vitamins and cofactors (Song et al., 2022). The focus has predominantly been on well-known species such as bees, mosquitoes, and ticks (Feng et al., 2022; Liberti et al., 2022; Kurokawa et al., 2020). These species tend to receive more attention because they transmit diseases such as Plasmodium spp. and other deadly pathogens, making them economically and medically important (Wang et al., 2023). In contrast, hematophagous arthropods such as horseflies, deerflies, and blackflies have received comparatively less attention. Despite being less harmful than well-known vector insects, these arthropods remain crucial for pathogen transmission, particularly in endemic regions with inadequate medical and sanitary conditions (Rodrigues et al., 2022; Olkeba et al., 2022; Sitarz et al., 2022). The application of high-throughput sequencing technology provides a promising avenue for investigating the microbial diversity of arthropods (Gómez et al., 2022; Swe et al., 2019; Estrada-Peña et al., 2018). Gaining deeper insight into the intricate relationship between arthropods and their microbiota will contribute to a broader understanding of the ecology and evolution of these organisms.
Tabanids exhibit remarkable diversity, comprising over 4,500 species and subspecies, and are widely distributed worldwide. Tabanus has a global distribution and is particularly abundant in subtropical and tropical regions (Mullens et al., 2022; Keita et al., 2020; Votýpka et al., 2019). Haematopota spp. and Chrysops spp. are the most prevalent in Europe (Dörge et al., 2020) whereas in Northeast China, the dominant species is Hybomitra sp., especially in the Palearctic region, followed by Tabanus, Haematopota turkestanica, Haematopota and Atylotus. Tabanids are 6–30 mm long and possess wings and large compound eyes. They are primarily active around animal colonies during the daytime in the summer. Many female tabanids require blood for oogenesis. Upon biting, the saliva containing various anticoagulants and allergens are injected into the host (Veraldi and Esposito, 2017; Maity et al., 2017), leading to allergic reactions and pain. Consequently, they pose significant challenges to grazing areas, causing weight loss, discomfort, and agitation. Moreover, tabanids have medical significance as vectors of numerous viruses, bacteria, parasites, and pathogens (Keita et al., 2020; Baldacchino et al., 2014) contributing to the spread of diseases in humans and animals through biological and mechanical transmission. Studies have highlighted their role in transmitting Francisella tullarensis, Anaplasma marginale Theileria spp., and causing lumpy skin disease (Janse et al., 2017; Hornok, 2008; Sohier et al., 2019).
The microbiota plays a crucial role in the development of BFA and can significantly affect their vectorial capacity and host immune system signaling (Husnik, 2018; Sonenshine and Stewart, 2021). They colonize various tissues within insects. For instance, in mosquitoes, the salivary glands exhibit the highest microbiota diversity (Sharma et al., 2014), a feature closely linked to their life history traits. The microbiota present in the midgut or salivary glands can interact directly with pathogens, thereby affecting the vectorial capacity of BFA (Gómez et al., 2022; Aguilar-Díaz et al., 2021). Consequently, manipulating the bacterial populations inhabiting these BFAs has emerged as a promising strategy for developing novel strategies to control the transmission of vector-borne diseases.
Despite the numerous microbiome studies conducted on common BFA, similar investigations in tabanids remain undocumented. Given the potential effect of bacteria on vector capacity, understanding the composition of the microbiota within tabanids could aid in assessing whether certain tabanid populations are more prone to transmitting pathogens than others (Zhu et al., 2024). In this study, we assessed the microbiota structure of four tabanid species by analyzing their presence in various organs across the four tabanid genera. These findings contribute to advancing our understanding of the bacterial communities associated with tabanids, thereby establishing a foundation for future research and implementing control measures targeting tabanids.
2 Methodology
2.1 Tabanids sample collection and cleaning
Tabanids were captured near horse and cattle farms in Daqing City, Heilongjiang Province, Northeastern China, using the net method from July to September 2021. Collectors held the end of the net handle and waved it in an “X” pattern, allowing for easy maneuvering while walking. Tabanids of the same genus were placed in specialized cages to shield them from direct sunlight and ensure their activity. Species identification was based on morphological characteristics, including the shape of the basal scapula of the head, as well as the length, width, and degree of dorsal protrusion of the antennae. We also examined the colour of the back plate on the abdomen and wing pattern (China fauna editorial committee of the Chinese Academy of Sciences, 1998). The cleaning procedures were as follows: (1) The tabanids were placed into a sterile centrifuge tube, followed by the addition of 75% ethanol solution. The tube was then gently shaken for 1 min; (2) the tabanid samples were transferred to a new centrifuge tube, and 0.1% sodium hypochlorite solution was added. The tube was again shaken gently for 1 min; (3) the samples were transferred once more to a new centrifuge tube and cleaned twice with distilled water; (4) distilled water was added again, and after gentle agitation, the liquid was poured out for preservation; and (5) and the cleaned tabanids were fixed onto wax plates and dissected under sterile conditions, and tissue samples were collected from the Malpighian tubules, midgut, and ovary. The tissue samples were stored at −80°C for subsequent use. A total of 432 tabanids were collected. Among them, 135 tabanids were used in the study, including 40 T. griseinus, 45 Atylotus sp., 25.
H. turkestanica and 25 Hybomitra sp. After dissecting the tabanids five samples was combined into one pool and submitted to sequencing. All tabanids were adults, and the detailed sample information is shown in Table 1.
2.2 DNA extraction and 16S rRNA sequencing
DNA was extracted from the Malpighian tubules, midgut, and ovary tissues using the DNA Stool Mini Kit, following the manufacturer’s protocol. Subsequently, the extracted DNA was utilized for Illumina sequencing using primers 338F: 5′- ACT CCT ACG GGA GGC AGC AG-3′ and 806R: 5′- GGA CTA CCA GGG TATC-TAA TCC-3′, targeting the 16S (V3–V4) region. PCR products were subjected to 250 bp paired-end sequencing on an Illumina NovaSeq 6,000 platform.
2.3 Sequence data processing and generation of amplicon sequence variants table
Raw reads were initially filtered using Trimmomatic (v0.33) (Bolger et al., 2014). Subsequently, the primer sequences were identified and eliminated using Cutadapt (v1.9.1) (Martin, 2011), resulting in the generation of high-quality reads without primer sequences. The clean reads were then spliced and clustered on the QIIME2 platform (Bolyen et al., 2019) using the DADA2 algorithm (Callahan et al., 2016) to obtain amplicon sequence variants (ASVs). Taxonomic annotation of the feature sequences was processed using a Bayes classifier with SILVA (release 138.1) as the reference database (Quast et al., 2012). The Shannon diversity index and Bray-Curtis distance of the samples were calculated using the “diversity” and “vegdist” functions in the R package: vegan. Permutational multivariate analysis of variance (PERMANOVA) was performed using the adonis function in vegan. The ggplot2 package was used to visualize data such as species composition and principal component analysis. Statistics on the composition of each sample were calculated at various taxonomic levels, including phylum, class, order, family, genus, and species. The abundance of each species in the samples and distribution histograms at each taxonomic level were obtained using QIIME2.
2.4 Statistical analysis
All analyses were conducted using R version 4.2.1. Rarefaction curves based on the number of feature sequences were generated using “estimate R” function in the package vegan v2.5–7 (Dixon, 2003). The profile was transformed into a table of relative abundances prior to statistical analyses. Wilcoxon rank sum test was employed to assess differences in species between groups, and a p-value of <0.05 was considered statistically significant. The Shannon index was used to evaluate microbiota diversity. Principal coordinate analysis (PCoA) was conducted based on the Bray-Curtis distance and permutational multivariate analysis of variance (PERMANOVA) with a permutation of 999. Linear discriminant analysis of effect size (LEfSe) was performed to identify the key species that most likely explained the differences between the groups. Multiple bacterial pathogen detection (MBPD), which can detect a broad range of animal, plant, and zoonotic pathogens based on 16S rRNA gene sequencing (Yang et al., 2023), was employed to explore putative pathogens in horsefly microbial communities.
3 Results
3.1 Morphological characteristics
Phenotypic sex determination relies on secondary sex characteristics, particularly eye shape. Morphological identification was conducted individually for each collected specimen, using morphological descriptions to ascertain the genus or species level. The captured tabanids were classified into four genera: Atylotus: Usually small sized fly, frons with spotted calli or without it. Colour of eyes in living specimen green or yellow. Basal plate of flagellum broad and obtuse dorsal angle. Basicosta pale to brown setulose. Haematopota: Generally small and slender flies of brownish to blackish grey in colour; eyes with several wavy bands in live condition; frons with velvety black spot on each side above the frontal callus and often a mid-frontal spot above these; picture wing pattern, i.e., dark wing with pattern of pale spots; mid tibiae and hind tibiae often with pale rings. Tabanus: Frons with prominent callus. Hybomitra: Medium sized fly; vertex with ocellar tubercle; eyes with dense pubescence and 3 green or purple band in live condition; basal and median callus usually broad; body blackish to dark greyish often with orangish side markings in at least 2 to 3 anterior abdominal segments. Two of these were identified at the genus level (Atylotus sp. and Hybomitra sp.), while the other two were identified at the species level (Haematopota turkestanica and Tabanus griseinus). The morphological characteristics of the cells are depicted in Figure 1A.
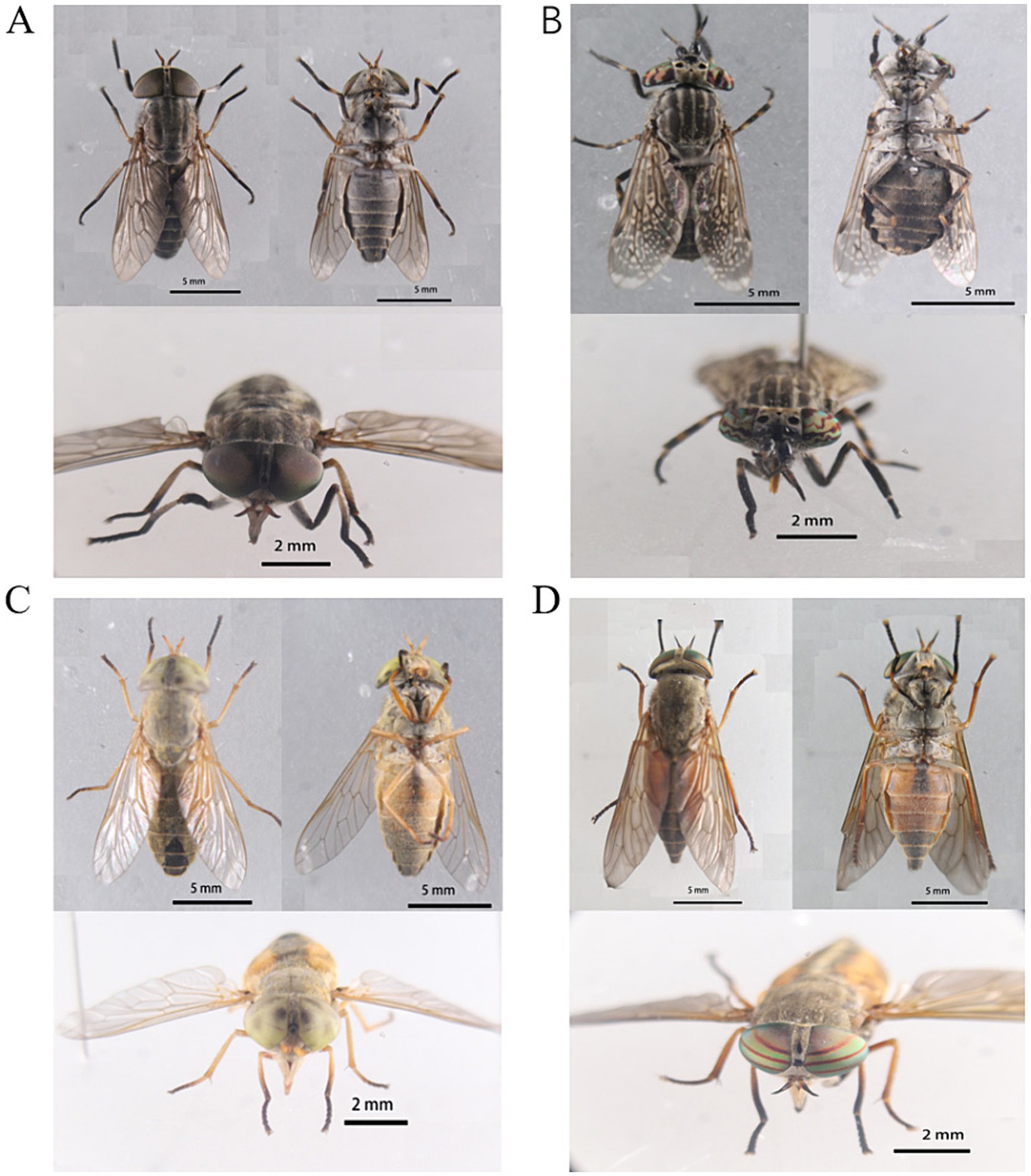
Figure 1. Morphological characteristics of tabanids. (A) Tabanus griseinus. (B) Haematopota turkestanica. (C) Atylotus sp. (D) Hybomitra sp.
3.2 Summary of 16S-rRNA sequencing
After dissecting the tabanids We characterized different organs bacterial community composition by sequencing the V3–V4 region of the 16S rRNA (16S) gene. In total, 7,516,786 pairs of raw reads were obtained. After quality control, 7,486,165 clean reads were obtained, with an average of 77,981 clean reads per sample. A total of 1,338 ASVs were obtained following the splicing and clustering of clean reads. Notably, more than 1,000 ASVs were shared among the different organs of the four tabanid species, indicating the presence of a relatively stable core community within each organ. Taxonomic classification revealed that nearly all ASVs could be classified above the genus level, indicating that the majority of the tabanid microbiota comprised known bacteria (Figures 1B–D).
3.3 Diversity of microbiota in Tabanidae
To investigate the diversity of the microbiota in Tabanidae, the microbiota present in the midgut, Malpighian tubules, and ovary of four genera of Tabanidae were analyzed. The rarefaction analysis indicated that the sampling number curve for each organ tended to saturate at 20 (Figure 2A). Moreover, the number of species observed in the ovaries exceeded that in the other two organs with the same sample size, indicating higher species richness in the ovaries.
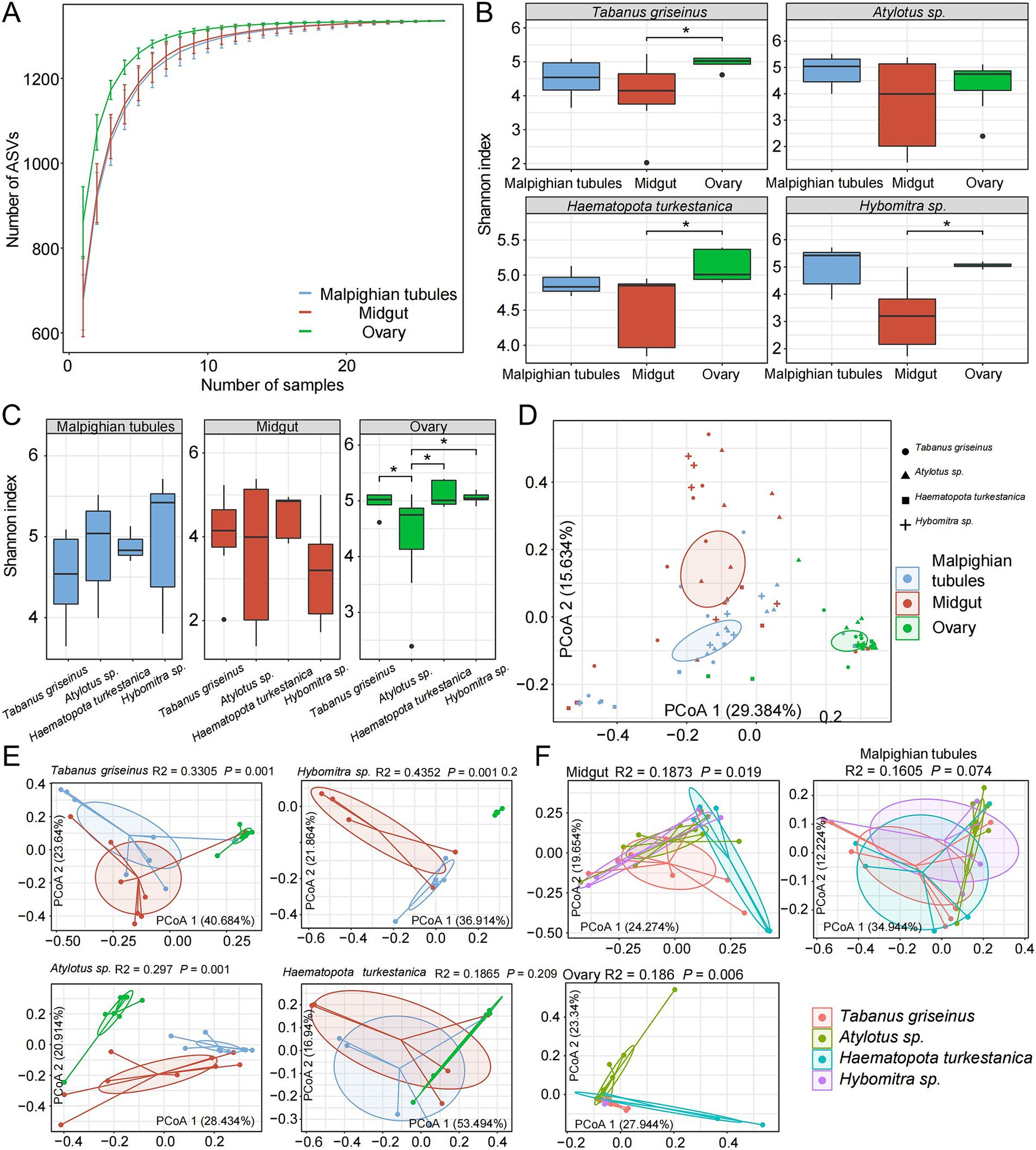
Figure 2. Symbiotic microbial diversity in tabanids. (A) The dilution curve shows that the number of observed feature sequences increases with the number of samples. (B,C) Show a comparison of the α diversity of microbes across different organs and species, respectively. (D) PCoA analysis based on Bray-Curtis distances revealed a distinct separation among samples, with four shapes representing different species and three colors used to distinguish various organ groups. (E,F) Show the separation between different organs and different species, respectively. R2 and p values represent the results of PERMANOVA.
Subsequently, the Shannon index was used to evaluate the microbial diversity in each organ. The midgut exhibited significantly lower microbial diversity than the other organs, whereas the ovaries displayed the highest microbial diversity (p < 0.05). The rank-sum test revealed that Tabanus griseinus, Hybomitra sp., and Haematopota turkestanica exhibited significantly higher microbiota diversity in their ovaries than in the other organs (p < 0.05; Figure 2B). Furthermore, microbiota diversity within the same organ among different tabanids was compared. The microbiota diversity of Atylotus sp. ovaries was low (p < 0.05, compared with other groups), whereas the differences among the other groups were not statistically significant (Figure 2C).
PCoA was conducted using the Bray-Curtis distance metric revealed significant effects of different organs and tabanids on the microbiota composition (Figure 2D). Notably, the organs exhibited stronger explanatory power for microbiota variation (R2 = 0.2264, p < 0.001) than the tabanids. Further analysis of the characteristics of microbial diversity among the different tabanids involved the calculation of the distance between samples at the species and organ levels. We observed significant differences in the β-diversity of the microbiota across different organs within the same species (Figure 2E). Among different species, the β-diversity of the Malpighian tubules microbiota is relatively similar, whereas the midgut and ovary microbiota exhibit substantial differences (Figure 2F).
3.4 Composition of microbiota in Tabanidae
A total of 1,338 ASVs were classified into 20 phyla, 225 families, and 506 genera. Firmicutes (37.6% ± 13.7%), Proteobacteria (32.7% ± 20.4%) and Bacteroidetes (11.6% ± 6.3%) were the dominant phyla. Other phyla, including Tenericutes (7.8% ± 17.4%), Epsilonbacteraeota (2.3% ± 3.5%), Cyanobacteria (2.3 ± 2.0%), Actinobacteria (1.9% ± 1.0%), and Acidobacteria (1.4% ± 1.0%) were also observed (Figure 3A). At the family level, Enterobacteriaceae (22.8% ± 20.0%), Lachnospiraceae (9.5% ± 8.8%), Ruminococcaceae (8.7% ± 4.2%), and Spiroplasmataceae (7.4% ± 17.5%) were predominant (Figure 3B).
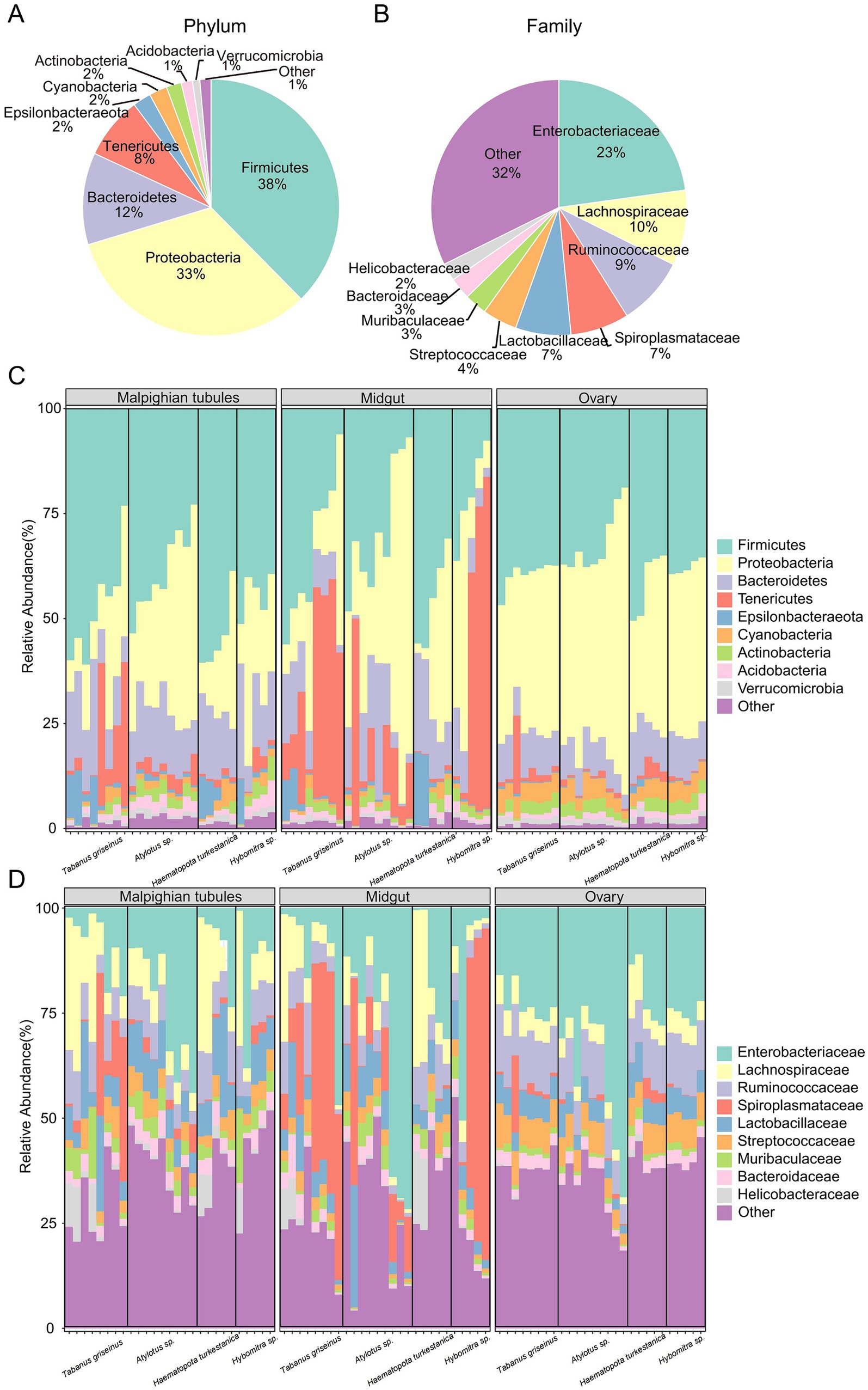
Figure 3. Microbiota composition in tabanids. (A,B) Show bacterial taxa in all samples, at phyla and family level, respectively; (C,D) Show bacterial taxa in difference group, at phyla and family level, respectively. Only the top ten phyla or families with the highest abundance are shown.
The characteristics of the microbial composition in different organs of the four Tabanidae species were examined to analyze the variation in microbiota in tabanids. Firmicutes, Proteobacteria, and Bacteroidetes were the most dominant bacterial phyla in the various organs (Figure 3C). Notably, a high abundance of Tenericutes (20.9% ± 24.9%) was observed in the midguts of most tabanids. However, Haematopota turkestanica exhibited a lower abundance of Tenericutes (1.2% ± 0.7%) in its midgut than other tabanids. In Malpighian tubules, Tabanus griseinus and Haematopota turkestanica displayed higher abundances of Epsilonbacteraeota(5.0 ± 5.6 and 5.6% ± 7.5%). Furthermore, Tabanus griseinus exhibited a higher abundance of Tenericutes (11.3% ± 14.8%) and a lower abundance of Proteobacteria (16.8% ± 10.1%) in the Malpighian tubule. However, the overall structure of ovarian microbiota across the four tabanid species remained relatively consistent.
At the family level, Enterobacteriaceae (19.8% ± 22.9%), Lachnospiraceae (10.9% ± 11.1%), and Ruminococcaceae (9.7% ± 4.3%) emerged as the dominant families across the organs (Figure 3D). Notably, the abundance of Spiroplasmataceae (11.1% ± 14.8%) in the Malpighian tubules of Tabanus griseinus surpassed that in other tabanids. Additionally, higher levels of Enterobacteriaceae (23.7% ± 14.3%) and lower levels of Lachnospiraceae (7.1% ± 1.9) were observed in the Malpighian tubules of Atylotus sp. Spiroplasmataceae (29.1 ± 21.1 and 41.3% ± 38.3%) was notably abundant in the midgut of Tabanus griseinus and Hybomitra sp. Interestingly, the midgut of Haematopota turkestanica contains more Lachnospiraceae (6.7% ± 4.1%) but almost no Spiroplasmataceae (0.6% ± 0.6). It is noteworthy that compared to other tabanids, Atylotus sp. exhibited a higher enrichment of Enterobacteriaceae (33.2 ± 26.8 and 39.0% ± 15.9%) in both the midgut and ovary. The structures of the ovary microbiota of Tabanus griseinus, Haematopota turkestanica, and Hybomitra sp. were consistent, suggesting that the composition of the ovary microbiota may not be significantly influenced by genetic background.
The main changes observed at the family level extended downstream to the hierarchical groups; therefore, we further examined the characteristics at the genus level. The relative abundances of dominant bacterial genera were not evenly distributed among each group (Supplementary Table S1), therefore, LEfSe analysis was used to detect characteristic genera in each group. This study confirmed consistent enrichment of Lachnospiraceae NK4A136 group and Helicobacter (LDA > 3, p < 0.05) in the Malpighian tubules of Tabanus griseinus, Atylotus sp., and Hybomitra sp. (Figures 4A,B,D). Notably, there was minimal difference in the microbiota of each organ in Haematopota turkestanica, with only three enriched genera (Figure 4C) found in the Malpighian tubules after lowering the threshold of the LDA score (LDA > 2, p < 0.05). A few bacteria were enriched in the midgut; however, Photobacterium was uniformly enriched in Atylotus spp., Haematopota turkestanica, and Hybomitra spp. In addition, only Photobacterium was significantly enriched in the midguts of Hybomitra spp. (Figure 4D).
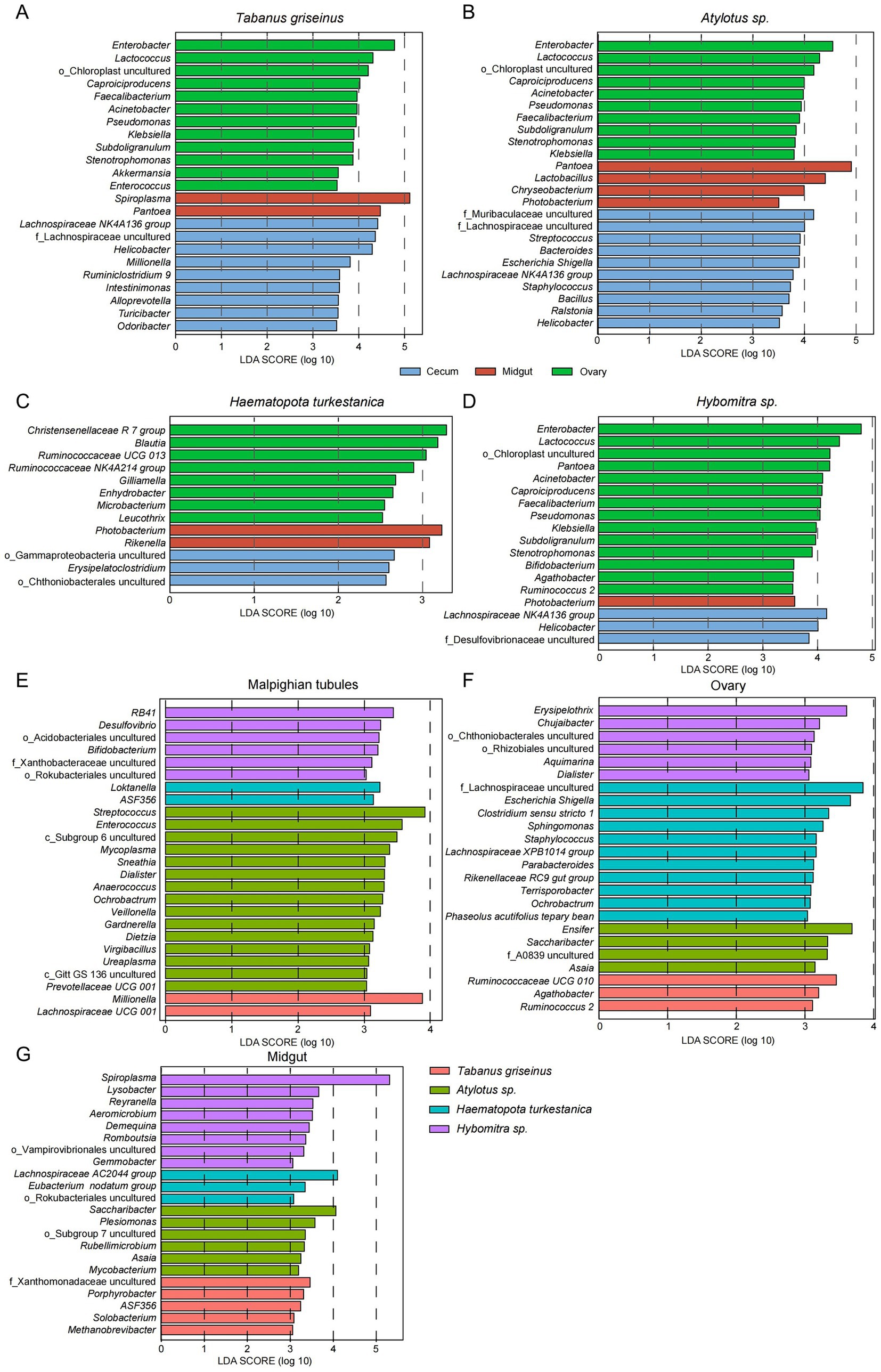
Figure 4. LEfSe analysis revealed differences in microbiota characteristics of the tabanids. The characteristic bacteria (Genus level) of organs of Tabanus griseinus (A), Atylotus sp. (B), Haematopota turkestanica (C), and Hybomitra sp. (D) were analyzed using LEfSe. Then the bacteria characteristics (genus level) of different Tabanus were compared in Malpighian tubules (E), ovary (F), and midgut (G) respectively.
The ovaries were enriched with the most characteristic bacteria, and similar to the Malpighian tubules, Tabanus griseinus, Atylotus spp., and Hybomitra spp. showed significant consistency (Figures 4A,B,D). Enterobacter, Lactococcus, Caproiciproducens, Faecalibacterium, Acinetobacter, Pseudomonas, Klebsiella, Subdoligranulum, and Stenotrophomonas were enriched in the ovaries (LDA > 3, p < 0.05). Interestingly, the ovarian microbiota of Haematopota turkestanica differed from that of the other tabanids, with bacteria such as Christensenellaceae R 7 Group, Blautia, and Ruminococcaceae UCG 013 being significantly enriched in the ovaries (Figure 4C).
LEfSe was employed to analyze how the microbiota varied across the organs of different species, revealing bacterial composition differences in the Malpighian tubules, midgut, and ovaries of each tabanid species. In the Malpighian tubules, Atylotus sp. was the most prominent genus (Figure 4E), with Streptococcus displaying the highest LDA value and was identified as the most significantly enriched genus (LDA > 3, p < 0.05). Furthermore, the genera most significantly enriched in Tabanus griseinus, Haematopota turkestanica, and Hybomitra sp. were Millionella, Loktanella, and RB41 (LDA >3, p < 0.05). In the midgut samples collected from the Hybomitra sp., yet in the context of an expected microbiome diversity, we observed Spiroplasma being the most significantly enriched (Figure 4G). The uncultured F_Xanthomonadaceae, Saccharibacter, and Lachnospiraceae AC2044 group were the most significantly enriched genera in Tabanus griseinus, Atylotus sp., and Haematopota turkestanica, respectively.
Consistent with the organ-level analysis, Haematopota turkestanica exhibited the most specialized ovarian microbiota, thus having the highest number of marked genera (Figure 4F). F_Lachnospiraceae uncultured and Escherichia/Shigella were the most abundant bacteria in the ovaries. The most significantly enriched genera in the ovaries of Tabanus griseinus, Atylotus spp. and Hybomitra spp. were Ruminococcaceae UCG 010, Ensifer, and Erysipelothrix, respectively.
3.5 Putative pathogens in the tabanids
Putative pathogens within the microbiota of tabanids were identified using a pathogen detection pipeline to assess the potential public health risks associated with tabanids. The MBPD is known for its ability to detect a broad spectrum of animal, plant, and zoonotic pathogens. The results revealed over 100 genera of putative animal pathogens were the most abundant (Figure 5). Among these, 20 genera were annotated as zoonotic agents, with Citrobacter exhibiting the highest abundance among the zoonotic pathogens. Interestingly, Brucella species had varying abundance distributions among the four fly species, highlighting the presence of this important group of zoonotic pathogens (Figure 5).
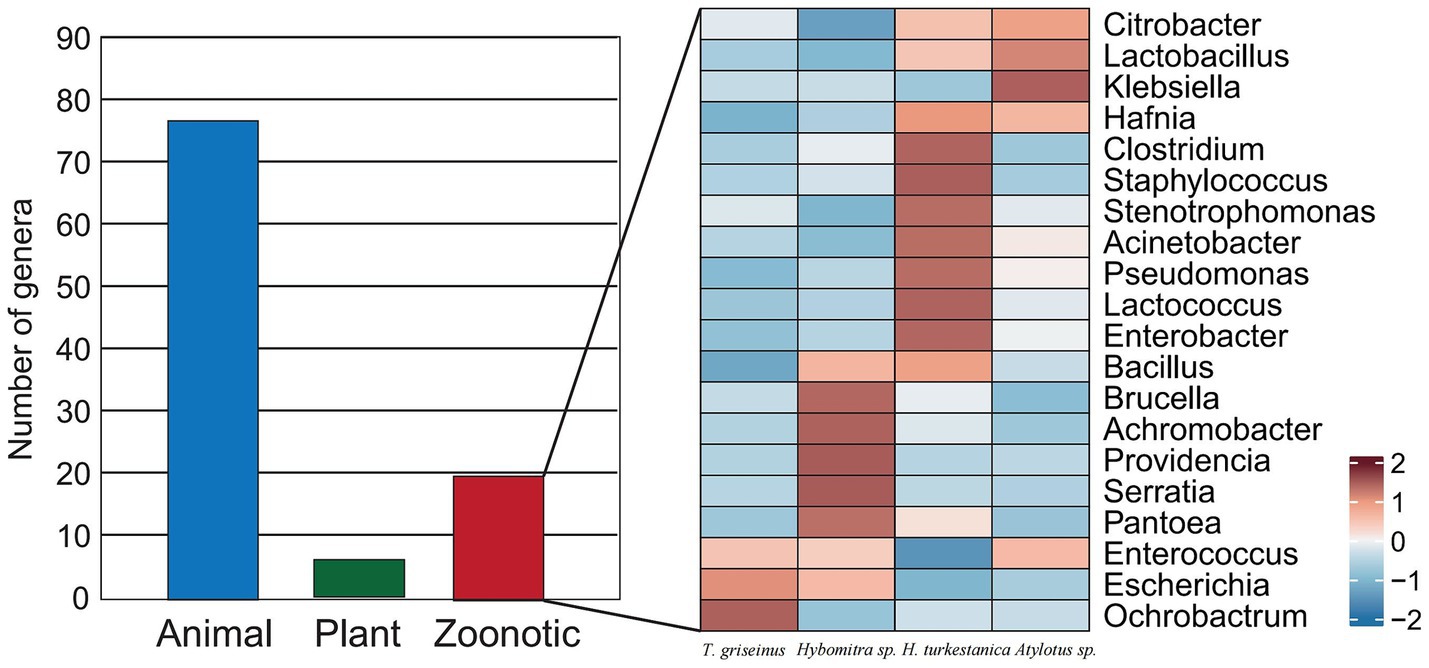
Figure 5. The composition of putative pathogens and the microbial index of pathogenic bacteria in the tabanid microbiota.
3.6 Microbiota impacting on vectors
Research on vector-microbiota interactions has revealed the significant impact that microbiota can have on various biological traits in disease-transmitting vectors. This change in the composition of the vector microbiota can potentially influence pathogenic infections through diverse mechanisms. Microbiota may directly interact with vector pathogens or modulate pathogen infection by regulating host immune defenses and nutritional status. The influence of the microbiota on other aspects of vector physiology and pathogen transmission, as observed in the microbiome of tabanids, is summarized in Table 2.
4 Discussion
Interest in microbial communities associated with disease vectors has grown in recent years. This interest stems from understanding that interactions between bacteria can influence the survival and transmission of pathogens (Song et al., 2022; Laukaitis and Macaluso, 2021; Wu et al., 2019). Despite this increased focus, many arthropod-associated human diseases remain undiagnosed, and knowledge of the prevalence, diversity, and pathogenicity of novel arthropod-borne pathogens is limited. The necessity for ongoing microbiological surveillance of vectors is evident. Differences in microbiota composition have been extensively documented in other vector arthropods, including variations in vector species, sex, organs, and developmental stages (Strand, 2018; Muturi et al., 2017; Choubdar et al., 2021). This study focused on the differences in the microbiota among tabanids, considering two factors: organs and species. The research observed that diversity of the microbiota in the ovaries was greater than that in other organs across all tabanids. However, differences in microbiota diversity among different tabanid species were only evident in the ovarian samples of Atylotus spp., suggesting that microbiota diversity in different organs may be independent.
Interestingly, previous studies have found that the microbiota diversity in the ovary is higher than that in the digestive tract (Malpighian tubules and midgut), which contrasts with observations in hard ticks (Guizzo et al., 2020). This reduction in diversity may be associated with blood feeding (Qiu et al., 2022). Studies have also revealed that the gut bacterial diversity of Aedes aegypti decreases after feeding on blood (Muturi et al., 2019), although it returns to its original pre-blood meal levels as the blood meal is digested. Considering that wild tabanids typically feed on various hosts and conducting artificial feeding experiments under laboratory conditions presents challenges, it is important to acknowledge the potential influence of blood meal sources on microbiota dynamics. Hence, additional research is necessary to clarify the effect of blood meals on the microbiota of tabanids. The midgut is the primary site for bacterial infection, colonization, and subsequent spread among BFA, with the relative abundance of these bacteria varying from one host to another. The microbiota found in the tabanids exhibited a core composition of Firmicutes and Proteobacteria at the phylum level. Tenericutes were the dominant phyla in Tabanus griseinus and Hybomitra sp., whereas Proteobacteria were the dominant phyla in Atylotus sp. and Haematopota turkestanica. Tenericutes are frequently found in insects such as spiders and bees (Magagnoli et al., 2022). Most Tenericute species in the midgut of Tabanus griseinus and Hybomitra spp. belong to Spiroplasmataceae, which were recently reclassified under the phylum Mycoplasmatota (Oren and Garrity, 2021). This bacterium is widespread in various arthropods and has been extensively described in ticks (Binetruy et al., 2019). Spiroplasmataceae are transmitted between arthropods through maternal inheritance and horizontal transfer (Bell-Sakyi et al., 2015). Enterobacteriaceae, a representative family of Proteobacteria in Atylotus sp. and Haematopota turkestanica, has been reported in A. aegypti mosquitoes (Macleod et al., 2021; David et al., 2016) and is considered a potential pathogen. Members of this family are closely associated with viral infection and transmission (Wu et al., 2019; Kozlova et al., 2021). The representative family of Firmicutes is Lachnospiraceae, which has also been reported in A. aegypti (Arévalo-Cortés et al., 2022) and is associated with mucin degradation in the gut (Vacca et al., 2020).
A single food source, such as blood, causes the arthropod host to be deficient in essential metabolites, such as thiamine, pyridoxine, folic acid, and other B vitamins (Rio et al., 2016). These metabolites are crucial for arthropods, and ectoparasites rely on their microbiota to compensate for the missing nutrients. Unlike non-blood-feeding insects, certain gut bacteria in blood-sucking arthropods benefit their hosts by upregulating numerous genes that encode essential nutrients (Sonenshine and Stewart, 2021). Different blood-sucking arthropods harbor diverse composition of their microbial communities that provide essential nutrients to the host during the long-term adaptation process to compensate for genome loss (Duron and Gottlieb, 2020). Three phyla (Firmicutes, Actinomycetes, and Proteobacteria) and eight bacterial families (Bacteriaceae, Rickettsiaceae, Anaplasmodiaceae, Enterobacteriaceae, Sphingomonas, Moraxellaceae, Pseudomonas, and Staphylococcaceae) were identified in most blood-sucking arthropods (Clow et al., 2018). Ruminococcaceae are abundant in the cecum and midgut of tabanids, and members of this family have been reported in both hard ticks (Xu et al., 2023) and black soldier flies (Wu et al., 2022). This family comprises an important class of organolytic bacteria that aid in the digestion of host and metabolism nutrients (Alvarado et al., 2021).
Our study revealed a relatively consistent microbiome in the ovaries of various tabanid species, suggesting the conservation of vertically transmitted bacteria across the tabanid family. The predominant bacterial families identified in ovaries were Rumenococcaceae, Enterobacteriaceae, Trichomyceaceae, Lactobacillaceae, and Streptococcaceae. The microbiota present in the ovaries may play a role in facilitating viral infection of arthropods, as observed in mosquitoes (Díaz et al., 2021; Caragata et al., 2021). Further studies examining viral carriage in horsefly ovaries are necessary to elucidate the relationship between these bacteria and pathogens.
The midgut and Malpighian tubules of BFA serve as primary sites of viral invasion (Wu et al., 2019). The intricate community of symbiotic microbiota in these organs can regulate host defense against viral invasion or entry into the intestinal epithelium. For instance, Wolbachia, a symbiotic bacterium in mosquitoes, interferes with mosquito-borne viral replication, likely through alterations in immune or physiological responses (Shi et al., 2023; Hajdušek et al., 2013). Similarly, Serratia marcescens, a bacterium known to promote dengue virus infection in mosquitoes (Wu et al., 2019) was identified in the midgut samples of tabanids. Therefore, targeting midgut microbiota is a promising approach for interventions to control and prevent insect-borne diseases.
Spiroplasma is a bacterium widely distributed in various tissues such as the hemolymph, adipose bodies, and salivary glands of ticks, and can be transmitted vertically through the ovaries (Ballinger and Perlman, 2017). Previous studies have demonstrated that Spiroplasma can confer protection against parasitic wasps and nematodes (Wang et al., 2023). However, the role of Spiroplasma in susceptibility to arbovirus vectors remains poorly understood. Our findings indicate that Spiroplasma is highly abundant in the ovaries of tabanids, suggesting that tabanids may harbor a vertically transmitted protective microbiota similar to that observed in other arthropods.
The presence of pathogenic bacteria in blood-sucking arthropods poses a significant public health risk, and is a key area of current research (Van Treuren et al., 2015). However, tabanids have received relatively little attention compared to other hematophagous Diptera species. The MBPD results highlight the potential risks associated with tabanids. For instance, Brucella is an important pathogen that affects both humans and animals, and can cause abortion in animals, leading to economic losses and impacting animal health (Suárez-Esquivel et al., 2020). Although cases transmitted by horseflies are rarely reported, the possibility that these flies serve as neglected vectors for transmission cannot be disregarded, whereas other detected pathogens rarely infect healthy populations. However, infections caused by opportunistic pathogens can complicate diagnosis and treatment and pose a serious threat to immunocompromised patients owing to their multidrug resistance (Sheppard, 2022). Therefore, despite conducting an in-depth analysis of potentially pathogenic bacteria in the tabanid microbiota, further research is warranted to obtain a comprehensive understanding of the public health risks associated with pathogenic microorganisms carried by the microbiota of tabanids.
The microbiota of BFA can be influenced by a variety of biological and abiotic factors at both the macro (population) and micro (organ/tissue) levels. Geography is a significant factor that influences variation in the bacterial communities of BFA (Thapa et al., 2019; Yu et al., 2022). It is influenced by factors such as host activity, temperature, humidity, and sunlight exposure. This suggests the potential for tracing the origin of quarantined insects using microbial community characteristics. The sampling site for this study was Daqing City, Heilongjiang Province, China, known for its abundant wetland resources supporting various tabanid species. However, the limited scope of our sampling site may constrain the generalizability of our findings, highlighting the need for a broader sample collection to comprehensively understand the symbiotic microbiota of tabanids.
5 Conclusion
This study comprehensively analyzed the microbiota composition of four common tabanid species. Significant variations in microbial community characteristics were observed across various organs, with notably high diversity detected in the ovaries. The similarities in bacterial taxa among the ovaries of the four tabanid species suggest potential conservation of vertically transmitted bacteria in these BFA. Moreover, distinct microbiota characteristics in the digestive tracts of the four tabanid species may be closely linked to their specific ecological niches and behaviors. Future studies should explore the intricate relationships among tabanid microbiota, habits, and habitats, as well as their potential roles as vectors of pathogens. These insights could aid in the geographical tracking of parasite species and the development of symbiotic-based strategies for controlling insect-borne diseases.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1119251/.
Author contributions
H-YQ: Writing – original draft. Q-BL: Writing – original draft. C-RW: Writing – review & editing. HJ: Writing – original draft. C-FL: Writing – review & editing. S-SL: Writing – review & editing. M-HN: Writing – review & editing. Q-CC: Writing – review & editing. J-FJ: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Key Research and Development Program of China (2019YFC1200501), National Natural Science Foundation of China (32072885) and Natural Science Foundation of Heilongjiang Province (ZD2022C006).
Acknowledgments
We would like to acknowledge the staff and workers in the filed who helped in the collection of horseflies.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1467875/full#supplementary-material
References
Aguilar-Díaz, H., Quiroz-Castañeda, R. E., Cobaxin-Cárdenas, M., Salinas-Estrella, E., and Amaro-Estrada, I. (2021). Advances in the study of the tick cattle microbiota and the influence on vectorial capacity. Front. Vet. Sci. 8:710352. doi: 10.3389/fvets.2021.710352
Ahmad, F., Yang, G., Zhu, Y., Poulsen, M., Li, W., Yu, T., et al. (2022). Tripartite symbiotic digestion of lignocellulose in the digestive system of a fungus-growing termite. Microbiol. Spectr. 10, e01234–e01222. doi: 10.1128/spectrum.01234-22
Alvarado, W. A., Agudelo, S. O., Velez, I. D., and Vivero, R. J. (2021). Description of the ovarian microbiota of Aedes aegypti (L) Rockefeller strain. Acta Trop. 214:105765. doi: 10.1016/j.actatropica.2020.105765
Arévalo-Cortés, A., Damania, A., Granada, Y., Zuluaga, S., Mejia, R., and Triana-Chavez, O. (2022). Association of Midgut bacteria and their metabolic pathways with Zika infection and insecticide resistance in Colombian Aedes aegypti populations. Viruses 14:2197. doi: 10.3390/v14102197
Baldacchino, F., Desquesnes, M., Mihok, S., Foil, L. D., Duvallet, G., and Jittapalapong, S. (2014). Tabanids: neglected subjects of research, but important vectors of disease agents! Infect. Genet. Evol. 28, 596–615. doi: 10.1016/j.meegid.2014.03.029
Ballinger, M. J., and Perlman, S. J. (2017). Generality of toxins in defensive symbiosis: ribosome-inactivating proteins and defense against parasitic wasps in Drosophila. PLoS Pathog. 13:e1006431. doi: 10.1371/journal.ppat.1006431
Bell-Sakyi, L., Palomar, A. M., and Kazimirova, M. (2015). Isolation and propagation of a Spiroplasma sp. from Slovakian Ixodes ricinus ticks in Ixodes spp. cell lines. Ticks Tick Borne Dis. 6, 601–606. doi: 10.1016/j.ttbdis.2015.05.002
Binetruy, F., Bailly, X., Chevillon, C., Martin, O. Y., Bernasconi, M. V., and Duron, O. (2019). Phylogenetics of the Spiroplasma ixodetis endosymbiont reveals past transfers between ticks and other arthropods. Ticks Tick Borne Dis. 10, 575–584. doi: 10.1016/j.ttbdis.2019.02.001
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caragata, E. P., Dutra, H. L., Sucupira, P. H., Ferreira, A. G., and Moreira, L. A. (2021). Wolbachia as translational science: controlling mosquito-borne pathogens. Trends Parasitol. 37, 1050–1067. doi: 10.1016/j.pt.2021.06.007
China fauna editorial committee of the Chinese Academy of Sciences (1998). Fauna of China. Beijing: Science Press.
Choubdar, N., Karimian, F., Koosha, M., and Oshaghi, M. A. (2021). An integrated overview of the bacterial flora composition of Hyalomma anatolicum, the main vector of CCHF. PLoS Negl. Trop. Dis. 15:e0009480. doi: 10.1371/journal.pntd.0009480
Clow, K. M., Weese, J. S., Rousseau, J., and Jardine, C. M. (2018). Microbiota of field-collected Ixodes scapularis and Dermacentor variabilis from eastern and southern Ontario, Canada. Ticks Tick Borne Dis. 9, 235–244. doi: 10.1016/j.ttbdis.2017.09.009
David, M. R., Santos, L. M., Vicente, A. C., and Maciel-de-Freitas, R. (2016). Effects of environment, dietary regime and ageing on the dengue vector microbiota: evidence of a core microbiota throughout Aedes aegypti lifespan. Mem. Inst. Oswaldo Cruz 111, 577–587. doi: 10.1590/0074-02760160238
Díaz, S., Camargo, C., and Avila, F. W. (2021). Characterization of the reproductive tract bacterial microbiota of virgin, mated, and blood-fed Aedes aegypti and Aedes albopictus females. Parasit. Vectors 14, 592–512. doi: 10.1186/s13071-021-05093-7
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Dörge, D. D., Cunze, S., and Klimpel, S. (2020). Incompletely observed: niche estimation for six frequent European horsefly species (Diptera, Tabanoidea, Tabanidae). Parasit. Vectors 13:461. doi: 10.1186/s13071-020-04316-7
Duron, O., and Gottlieb, Y. (2020). Convergence of nutritional symbioses in obligate blood feeders. Trends Parasitol. 36, 816–825. doi: 10.1016/j.pt.2020.07.007
Estrada-Peña, A., Cabezas-Cruz, A., Pollet, T., Vayssier-Taussat, M., and Cosson, J. F. (2018). High throughput sequencing and network analysis disentangle the microbial communities of ticks and hosts within and between ecosystems. Front. Cell. Infect. Microbiol. 8:236. doi: 10.3389/fcimb.2018.00236
Feng, Y., Peng, Y., Song, X., Wen, H., An, Y., Tang, H., et al. (2022). Anopheline mosquitoes are protected against parasite infection by tryptophan catabolism in gut microbiota. Nat. Microbiol. 7, 707–715. doi: 10.1038/s41564-022-01099-8
Gómez, M., Martinez, D., Muñoz, M., and Ramírez, J. D. (2022). Aedes aegypti and ae. Albopictus microbiome/virome: new strategies for controlling arboviral transmission? Parasit. Vectors 15:287. doi: 10.1186/s13071-022-05401-9
Guizzo, M. G., Neupane, S., Kucera, M., Perner, J., Frantová, H., da Silva Vaz, I., et al. (2020). Poor unstable midgut microbiome of hard ticks contrasts with abundant and stable monospecific microbiome in ovaries. Front. Cell. Infect. Microbiol. 10:211. doi: 10.3389/fcimb.2020.00211
Hajdušek, O., Šíma, R., Ayllón, N., Jalovecká, M., Perner, J., De La Fuente, J., et al. (2013). Interaction of the tick immune system with transmitted pathogens. Front. Cell. Infect. Microbiol. 3:26. doi: 10.3389/fcimb.2013.00026
Hornok, S. A. (2008). Molecular identification of Anaplasma marginale and rickettsial endosymbionts in blood-sucking flies (Diptera: Tabanidae, Muscidae) and hard ticks (Acari: Ixodidae). Vet. Parasitol. 154, 354–359. doi: 10.1016/j.vetpar.2008.03.019
Husnik, F. (2018). Host--symbiont--pathogen interactions in blood-feeding parasites: nutrition, immune cross-talk and gene exchange. Parasitology 145, 1294–1303. doi: 10.1017/S0031182018000574
Janse, I., Maas, M., Rijks, J. M., Koene, M., van der Plaats, R. Q. J., Engelsma, M., et al. (2017). Environmental surveillance during an outbreak of tularaemia in hares, the Netherlands, 2015. Eur. Secur. 22:30607. doi: 10.2807/1560-7917.ES.2017.22.35.30607
Keita, M. L., Medkour, H., Sambou, M., Dahmana, H., and Mediannikov, O. (2020). Tabanids as possible pathogen vectors in Senegal (West Africa). Parasit. Vectors 13, 500–515. doi: 10.1186/s13071-020-04375-w
Kozlova, E. V., Hegde, S., Roundy, C. M., Golovko, G., Saldaña, M. A., Hart, C. E., et al. (2021). Microbial interactions in the mosquito gut determine Serratia colonization and blood-feeding propensity. ISME J. 15, 93–108. doi: 10.1038/s41396-020-00763-3
Krinsky, W. L. (1976). Animal disease agents transmitted by horse flies and deer flies (Diptera: Tabanidae). J. Med. Entomol. 13, 225–275. doi: 10.1093/jmedent/13.3.225
Kurokawa, C., Lynn, G. E., Pedra, J. H., Pal, U., Narasimhan, S., and Fikrig, E. (2020). Interactions between Borrelia burgdorferi and ticks. Nat. Rev. Microbiol. 18, 587–600. doi: 10.1038/s41579-020-0400-5
Laukaitis, H. J., and Macaluso, K. R. (2021). Unpacking the intricacies of Rickettsia-vector interactions. Trends Parasitol. 37, 734–746. doi: 10.1016/j.pt.2021.05.008
Liberti, J., Kay, T., Quinn, A., Kesner, L., Frank, E. T., and Cabirol, A. E. (2022). The gut microbiota affects the social network of honeybees. Nat. Ecol. Evol. 6, 1471–1479. doi: 10.1038/s41559-022-01840-w
Macleod, H. J., Dimopoulos, G., and Short, S. M. (2021). Larval diet abundance influences size and composition of the midgut microbiota of Aedes aegypti mosquitoes. Front. Microbiol. 12:645362. doi: 10.3389/fmicb.2021.645362
Magagnoli, S., Alberoni, D., Baffoni, L., Martini, A., Marini, F., Di Gioia, D., et al. (2022). The ground beetle Pseudoophonus rufipes gut microbiome is influenced by the farm management system. Sci. Rep. 12:22638. doi: 10.1038/s41598-022-25408-7
Maity, A., Naskar, A., Hazra, S., Sengupta, J., Parui, P., Homechaudhuri, S., et al. (2017). Taxonomic accounts of horse flies (Diptera: Tabanidae) from arid zone, part of Chotanagpur plateau, West Bengal. Munis Entomol. Zool. 12, 419–429.
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mullens, B. A., Trout Fryxell, R., Masonick, P. K., Yanega, D. A., and Davis, T. M. (2022). Hiding in plain sight: an abundant and widespread north American horse fly (Diptera: Tabanidae) in the Tabanus sulcifrons group, Tabanus variegatus Fabricius, redescribed. J. Med. Entomol. 59, 1217–1235. doi: 10.1093/jme/tjac057
Murakami, T., Segawa, T., Takeuchi, N., Barcaza Sepúlveda, G., Labarca, P., Kohshima, S., et al. (2018). Metagenomic analyses highlight the symbiotic association between the glacier stonefly Andiperla willinki and its bacterial gut community. Environ. Microbiol. 20, 4170–4183. doi: 10.1111/1462-2920.14420
Muturi, E. J., Dunlap, C., Ramirez, J. L., Rooney, A. P., and Kim, C. H. (2019). Host blood-meal source has a strong impact on gut microbiota of Aedes aegypti. FEMS Microbiol. Ecol. 95:fiy213. doi: 10.1093/femsec/fiy213
Muturi, E. J., Ramirez, J. L., Rooney, A. P., and Kim, C. H. (2017). Comparative analysis of gut microbiota of mosquito communities in Central Illinois. PLoS Negl. Trop. Dis. 11:e0005377. doi: 10.1371/journal.pntd.0005377
Olkeba, B. K., Mereta, S. T., Goethals, P. L., Yewhalaw, D., Debesa, G., Ambelu, A., et al. (2022). Habitat preference of blackflies in Omo gibe river basin (Southwest Ethiopia): implications for onchocerciasis elimination and control. PLoS One 17:e0264750. doi: 10.1371/journal.pone.0264750
Oren, A., and Garrity, G. M. (2021). Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 71:005056. doi: 10.1099/ijsem.0.005056
Qiu, H., Lv, Q., Chang, Q., Ju, H., Wu, T., Liu, S., et al. (2022). Microbiota community structure and interaction networks within Dermacentor silvarum, Ixodes persulcatus, and Haemaphysalis concinna. Animals (Basel) 12:3237. doi: 10.3390/ani12233237
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rio, R. V., Attardo, G. M., and Weiss, B. L. (2016). Grandeur alliances: symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol. 32, 739–749. doi: 10.1016/j.pt.2016.05.002
Rodrigues, G. D., Lucas, M., Ortiz, H. G., Dos Santos Gonçalves, L., Blodorn, E., Domingues, W. B., et al. (2022). Molecular of Anaplasma marginale Theiler (Rickettsiales: Anaplasmataceae) in horseflies (Diptera: Tabanidae) in Uruguay. Sci. Rep. 12:22460. doi: 10.1038/s41598-022-27067-0
Sevidzem, S. L., Koumba, A. A., Yao-Acapovi, G. L., and Mavoungou, J. F. (2021). A nationwide survey of the tabanid fauna of Cameroon. Parasit. Vectors 14:392. doi: 10.1186/s13071-021-04894-0
Sharma, P., Sharma, S., Maurya, R. K., de, T. D., Thomas, T., Lata, S., et al. (2014). Salivary glands harbor more diverse microbial communities than gut in Anopheles culicifacies. Parasit. Vectors 7, 1–7. doi: 10.1186/1756-3305-7-235
Sheppard, S. K. (2022). Strain wars and the evolution of opportunistic pathogens. Curr. Opin. Microbiol. 67:102138. doi: 10.1016/j.mib.2022.01.009
Shi, C., Beller, L., Wang, L., Rosales Rosas, A., De Coninck, L., Héry, L., et al. (2022). Bidirectional interactions between arboviruses and the bacterial and viral microbiota in Aedes aegypti and Culex quinquefasciatus. MBio 13:e0102122. doi: 10.1128/mbio.01021-22
Shi, H., Yu, X., and Cheng, G. (2023). Impact of the microbiome on mosquito-borne diseases. Protein Cell 14, 743–761. doi: 10.1093/procel/pwad021
Sitarz, M., Buczek, A. M., Buczek, W., Buczek, A., and Bartosik, K. (2022). Risk of attacks by blackflies (Diptera: Simuliidae) and occurrence of severe skin symptoms in bitten patients along the eastern border of the European Union. Int. J. Environ. Res. Public Health 19:7610. doi: 10.3390/ijerph19137610
Sohier, C., Haegeman, A., Mostin, L., De Leeuw, I., Campe, W. V., de Vleeschauwer, A., et al. (2019). Experimental evidence of mechanical lumpy skin disease virus transmission by Stomoxys calcitrans biting flies and Haematopota spp. horseflies. Sci. Rep. 9:20076. doi: 10.1038/s41598-019-56605-6
Sonenshine, D. E., and Stewart, P. E. (2021). Microbiomes of blood-feeding arthropods: genes coding for essential nutrients and relation to vector fitness and pathogenic infections. A review. Microorganisms 9:2433. doi: 10.3390/microorganisms9122433
Song, X., Zhong, Z., Gao, L., Weiss, B. L., and Wang, J. (2022). Metabolic interactions between disease-transmitting vectors and their microbiota. Trends Parasitol. 38, 697–708. doi: 10.1016/j.pt.2022.05.002
Strand, M. R. (2018). Composition and functional roles of the gut microbiota in mosquitoes. Curr. Opin. Insect Sci. 28, 59–65. doi: 10.1016/j.cois.2018.05.008
Suárez-Esquivel, M., Chaves-Olarte, E., Moreno, E., and Guzmán-Verri, C. (2020). Brucella genomics: macro and Micro evolution. Int. J. Mol. Sci. 21:7749. doi: 10.3390/ijms21207749
Swe, P. M., Zakrzewski, M., Waddell, R., Sriprakash, K. S., and Fischer, K. (2019). High-throughput metagenome analysis of the Sarcoptes scabiei internal microbiota and in-situ identification of intestinal Streptomyces sp. Sci. Rep. 9:11744. doi: 10.1038/s41598-019-47892-0
Thapa, S., Zhang, Y., and Allen, M. S. (2019). Bacterial microbiomes of Ixodes scapularis ticks collected from Massachusetts and Texas, USA. BMC Microbiol. 19:138. doi: 10.1186/s12866-019-1514-7
Vacca, M., Celano, G., Calabrese, F. M., Portincasa, P., Gobbetti, M., and De Angelis, M. (2020). The controversial role of human gut lachnospiraceae. Microorganisms 8:573. doi: 10.3390/microorganisms8040573
Van Treuren, W., Ponnusamy, L., Brinkerhoff, R. J., Gonzalez, A., Parobek, C. M., Juliano, J. J., et al. (2015). Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl. Environ. Microbiol. 81, 6200–6209. doi: 10.1128/aem.01562-15
Veraldi, S., and Esposito, L. (2017). Skin lesions caused by Tabanus bovinus bites. J. Travel Med. 24:tax049. doi: 10.1093/jtm/tax049
Votýpka, J., Brzoňová, J., Ježek, J., and Modrý, D. (2019). Horse flies (Diptera: Tabanidae) of three west African countries: a faunistic update, barcoding analysis and trypanosome occurrence. Acta Trop. 197:105069. doi: 10.1016/j.actatropica.2019.105069
Wang, J., Gao, L., and Aksoy, S. (2023). Microbiota in disease-transmitting vectors. Nat. Rev. Microbiol. 21, 604–618. doi: 10.1038/s41579-023-00901-6
Whyte, A. F., Popescu, F. D., and Carlson, J. (2020). Tabanidae insect (horsefly and deerfly) allergy in humans: a review of the literature. Clin. Exp. Allergy 50, 886–893. doi: 10.1111/cea.13677
Wu, T., Han, B., Wang, X., Tong, Y., Liu, F., Diao, Q., et al. (2022). Chlorothalonil alters the gut microbiota and reduces the survival of immature honey bees reared in vitro. Pest Manag. Sci. 78, 1976–1981. doi: 10.1002/ps.6816
Wu, P., Sun, P., Nie, K., Zhu, Y., Shi, M., Xiao, C., et al. (2019). A gut commensal bacterium promotes mosquito permissiveness to arboviruses. Cell Host Microbe 25, 101–112.e5. doi: 10.1016/j.chom.2018.11.004
Xu, Z., Wu, X., Zhang, J., Cheng, P., Xu, Z., Sun, W., et al. (2023). Microplastics existence intensified bloom of antibiotic resistance in livestock feces transformed by black soldier fly. Environ. Pollut. 317:120845. doi: 10.1016/j.envpol.2022.120845
Yang, X., Jiang, G., Zhang, Y., Wang, N., Zhang, Y., Wang, X., et al. (2023). MBPD: a multiple bacterial pathogen detection pipeline for one health practices. iMeta 2:e82. doi: 10.1002/imt2.82
Yu, Y., Wang, Q., Zhou, P., Lv, N., Li, W., Zhao, F., et al. (2022). First glimpse of gut microbiota of quarantine insects in China. Genomics Proteomics Bioinformatics 20, 394–404. doi: 10.1016/j.gpb.2022.04.005
Keywords: arthropods, Malpighian tubules, microbiota, midgut, ovaries, tabanids
Citation: Qiu H-Y, Lv Q-B, Wang C-R, Ju H, Luo C-F, Liu S-S, Na M-H, Chang Q-C and Jiang J-F (2024) Microbiota profile in organs of the horseflies (Diptera: Tabanidae) in Northeastern China. Front. Microbiol. 15:1467875. doi: 10.3389/fmicb.2024.1467875
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Naima Bel Mokhtar, University of Patras, GreeceNikolaos Remmas, Democritus University of Thrace, Greece
Marcos Rogério André, São Paulo State University, Brazil
Copyright © 2024 Qiu, Lv, Wang, Ju, Luo, Liu, Na, Chang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiao-Cheng Chang, Y2hhbmdxaWFvY2hlbmcyMDAxQDE2My5jb20=; Jia-Fu Jiang, amlhbmdqZjIwMDhAMTM5LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Hong-Yu Qiu
Hong-Yu Qiu Qing-Bo Lv
Qing-Bo Lv Chun-Ren Wang
Chun-Ren Wang Hao Ju2
Hao Ju2 Chun-Feng Luo
Chun-Feng Luo Shun-Shuai Liu
Shun-Shuai Liu Qiao-Cheng Chang
Qiao-Cheng Chang Jia-Fu Jiang
Jia-Fu Jiang