- 1Centre for Soil and Environmental Research, Lincoln University, Christchurch, New Zealand
- 2Key Laboratory of Humid Subtropical Eco-geographical Process of Ministry of Education, School of Geographical Sciences/School of Carbon Neutrality Future Technology, Fujian Normal University, Fuzhou, China
- 3Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing, China
The contrasting response of AOA, AOB, and comammox Nitrospira amoA transcript abundance to temperature, moisture, and nitrogen was investigated using soil microcosms. The moisture, temperature, and nitrogen treatments were selected to represent conditions typically found in a New Zealand (NZ) dairy farm. AOB dominated all synthetic urine treated soils. Peak AOB amoA transcript abundance was positively correlated with estimated soil ammonia availability. While AOB gDNA abundance and nitrification rate trends were similar. AOA were strongly influenced by soil temperature. At 20°C, AOA amoA peak transcript abundance averaged over 1 order of magnitude higher than at 8°C. Within the AOA community a member of the Nitrosocosmicus clade was positively correlated with ammonium and estimated ammonia concentrations. The presence and relative increase of an AOA community member in a high nitrogen environment poses an interesting contrast to current scientific opinion in NZ. Comammox Nitrospira abundance showed no correlation with soil moisture. This suggests that previously found associations are more complex than originally thought. Further research is required to determine the drivers of comammox Nitrospira abundance in a high moisture environment. Overall, these results indicate that AOB are the main drivers of nitrification in New Zealand dairy farm soils.
Introduction
Comammox Nitrospira are a newly discovered group of bacteria that are able to undertake both steps of the nitrification process, the oxidation of ammonia (NH3) via nitrite () to nitrate () (Van Kessel et al., 2015; Daims et al., 2015).
Originally, it was thought that comammox Nitrospira preferred substrate-limited conditions, such as ammonia-depleted biofilms (Van Kessel et al., 2015; Daims et al., 2015). However, subsequent studies have identified that certain members of the comammox Nitrospira community may favor a copiotrophic environment (Li et al., 2019; Chisholm et al., 2023). The environmental preference of comammox Nitrospira may be clade-specific, as several studies have suggested that comammox Nitrospira clade A and clade B respond differently to certain conditions, such as soil pH and ammonia availability (Wang et al., 2021; Li et al., 2022).
While some clade A members have demonstrated a positive response to nitrogen amendment (Orellana et al., 2018; Li et al., 2019; Takahashi et al., 2020), the response of clade B is less understood. Preliminary genomic studies suggest that clade B prefers a low nitrogen environment as their genome contains MEP-type ammonia transporters, which have a high affinity but low uptake capacity (Walter et al., 2008; Palomo et al., 2018; Li et al., 2022; Chisholm et al., 2024; Shah et al., 2024).
In terrestrial ecosystems, studies have shown that typically, clade A are more abundant than clade B (Li et al., 2022). However, Chisholm et al. (2023) found that comammox Nitrospira clade B was the most abundant ammonia oxidiser in a high-rainfall, high-fertility, pasture-based dairy farm soil and shared a strong positive correlation with soil moisture. This suggests that under certain environmental conditions, comammox Nitrospira clade B may be the dominant nitrifier.
There is a wealth of available information on ammonia oxidizing archaea (AOA) and ammonia oxidizing bacteria (AOB) in soil ecosystems (e.g. Prosser and Nicol, 2008; Di et al., 2009; Prosser and Nicol, 2012; Di et al., 2014; Taylor et al., 2017). While both are typically present in soil, they seemingly occupy different ecological niches. Historically, AOA are thought to prefer oligotrophic/extreme environments such as low ammonia availability, low pH, and high temperatures (Könneke et al., 2005; Francis et al., 2007; Di et al., 2009, 2010; Taylor et al., 2017). While AOB are functionally dominant in a nitrogen rich environment (Rütting et al., 2021). Within the context of New Zealand soils, AOB are typically more abundant than AOA (Di et al., 2009, 2010; Chisholm et al., 2023).
In order to investigate this further, an incubation experiment was conducted using a New Zealand dairy farm soil. The focus of the experiment was to compare comammox Nitrospira abundance, activity, and community composition to AOB and AOA under three different moisture levels: High (46.5% θg); Medium (37.5% θg); and Low (28.5% θg). Each moisture level was also run at two different temperatures to simulate the lowest (8°C) and highest (20°C) monthly average soil temperature in Canterbury, New Zealand. Two nitrogen levels were also employed to mimic common NZ dairy farm N inputs. These were urea-N fertilizer applied at 50 kg N ha−1 and synthetic urine-N, a concentrated localized application of N equivalent to 700 kg N ha−1 to simulate a dairy cow urine deposition during outdoor grazing (Cameron et al., 2013). Canonical ammonia oxidiser abundance, activity and active community composition was also analyzed to provide a useful comparison as the response of AOB and AOA to nitrogen amendment in various environments is well documented, and are generally contrasting (Di et al., 2009, 2010).
It is expected that comammox Nitrospira will share a positive and negative correlation with soil moisture and temperature, respectively. AOA will prefer the high temperature environment and AOB will dominate the synthetic urine treatments.
Methods
A Templeton silt loam [typic immature pallic (Hewitt, 2010)] was used in this study. Soil from the top 10 cm was collected in May 2022 from the Lincoln University Research Dairy Farm (LURDF; 43°38′24.3″S 172°27′20.1″E; Lincoln, Canterbury New Zealand). The soil was sieved through a 6 mm sieve. A subsample was taken for soil physiochemical analysis (Supplementary Table 1).
Physicochemical analysis of soil samples
Soil pH was determined by mixing soil and water at a ratio of 1:2.5. Samples were then shaken for 1 h and left to settle overnight. Soil pH was measured using a pH meter (Mettler-Toledo, Switzerland) the following morning. Olsen P was determined using the methodology explained in Olsen (1954). Soil cations (K, Ca, Mg, and Na) were quantified using a soil solution ratio of 1:20 1M ammonium acetate at pH 7 followed by atomic adsorption spectroscopy (Rayment and Higginson, 1992). Total N was analyzed by combustion method using an Elementor Vario Max Cube Analyser. The methods used to determine Organic C and CEC are explained in Blakemore (1987) and Brown (1943), respectively. Soil gravimetric water content was calculated by drying 10 g of soil at 105°C overnight. Soil ammonium, nitrate and nitrite were extracted using 2 M KCl. Briefly, 5 g of soil were mixed with 25 ml of 2 M KCl and shaken for 1 h. After centrifugation for 10 min, the supernatant was filtered through Whatman No. 41 filter paper. The solution was analyzed for and concentrations using a Flow Injection Analyser (FIA; FOSS FIA star 5,000 triple channel analyser). Soil nitrite concentration was not measured in this study. Ammonia availability was estimated using the Visual MinTeq modeling software (Gustafsson, 2011). The average nitrification rate was estimated by dividing the change in nitrate concentration by the number of days. The average nitrification rate was estimated by dividing the change in nitrate concentration by the number of days.
Incubation experiment
Five-hundred grams (dry weight basis) was weighed into the plastic incubation containers (1 L volume, 10.0 cm diameter). Two holes were made in the lid of the containers to allow for gas exchange. The soil moisture was adjusted to 28.5% (L), 37.5% (M), and 46.5% (H) gravimetric water content [θ g; equivalent to 45%, 60%, and 75% water filled pore space (WFPS), respectively] and placed into an incubator set at either 8°C or 20°C to represent the Low and High temperature treatments, respectively. The soil was left in the incubators for 6 weeks for acclimatization before the nitrogen (N) treatments were applied. The following N-treatments were then applied: control, N0; 50 kg urea-N ha−1 equivalent, N50; and 700 kg synthetic urine-N ha−1, N700 (synthetic urine formula Clough et al., 1998). Each treatment had 4 replicates, totalling 72 experimental units. Samples were taken on days 1, 7, 14, 29, 57, 92, and 120. Soil pH, moisture, ammonia, nitrate, and AOB, AOA, and comammox amoA genomic DNA (gDNA) abundance were measured at each sampling date. AOB, AOA, and comammox amoA transcript abundance was measured from samples taken at days 7, 14, 29, and 57. Day 14 samples were selected for active AOB, AOA, and comammox community analysis. Soil samples used for molecular analysis were stored at −80°C for DNA and RNA extraction. The soil moisture content was maintained twice a week by monitoring the weight of the incubation vessels and adding deionised water to achieve the desired weight.
The genomic DNA (gDNA) was extracted from each sample using a NucleoSpin® Soil Kit (Macherey-Nagel, Düren, Germany) following the manufacturer's instructions.
The extracted DNA was diluted 20-fold using deionised water in a CAS-1200 Robotic liquid handling system (Corbett Life Science, Australia), and stored at −80°C until ready for analysis.
The RNA extraction was conducted using RNeasy® PowerSoil® Total RNA Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. In short, 2 g of fresh soil was extracted using phenol/chloroform/isoamyl alcohol mixture (SigmaAldrich, St. Louis, MO, United States) and final RNA pellets were resuspended in 100 μl of elution buffer. The extracted total RNA was then treated with RNase-free DNase I (ThermoFisher Scientific, Waltham, MA, United States) according to the manufacturer's instructions. First-strand complementary DNA (cDNA) was synthesized using SuperScript IV One-Step RT-PCR I (InvitrogenTM, Thermo Fisher Scientific). The cDNA was diluted two-fold and stored at −80°C for downstream analysis.
Real-time quantitative PCR (qPCR) analysis was carried out using a QuantStudio™ 5 Real-Time PCR system (Thermo Fisher Scientific, Christchurch, New Zealand). Each qPCR reaction had a final volume of 16 μl containing 8 μl PowerTrack SYBR Green Master Mix (Thermo Fisher Scientific, Christchurch, New Zealand), 3.2 μl Invitrogen UltraPure Distilled H2O (Life Technologies, New York USA), 0.65 μl of each primer and 3.5 μl gDNA or cDNA template. Standard curves were generated using 10-fold serial dilutions of plasmids containing correct inserts of the target genes. Controls (DNase treated RNA) were included to ensure all DNA was removed from the RNA. The primer sets comamoA F/comamoA R (Zhao et al., 2019), Arch—amoA F/Arch—amoA R (Francis et al., 2005), and amoA1F/amoA2R (Rotthauwe et al., 1997) were used to quantify comammox Nitrospira, AOA, and AOB amoA gene abundances, respectively. The amplification conditions are given in Supplementary Table 2. Melting curve analysis was performed at the end of each amplification cycle to confirm the reactions specificity. Real-time qPCR data analysis was carried out by using QuantStudio Design and Analyse Software (Thermo Fisher Scientific, Christchurch, New Zealand).
Day 14 cDNA was selected for Next Generation Sequencing (NGS). Before sequencing, the samples underwent two rounds of PCR. The first round used adapted ComamoA F/ComamoA R, Arch-amoA F/Arch-amoA R, and amoA 1F/amoA 2R—overhang primers for comammox, AOA and AOB, respectively. The second used Nextera XT Index Kit (Illumina, San Diego, CA, USA) to attach barcodes to the sequences from each sample. The resulting PCR products were purified using AMPure XP beads (Beckman Coulter, Brea, CA). MiSeq Reagent kit v3 was used for library construction. The libraries were sequenced via the MiSeq platform (Illumina, San Diego, CA, USA). Sequencing was undertaken by Auckland Genome Services (Auckland, New Zealand).
The sequencing reads were imported into QIIME2 (version 2021-11) (Bolyen et al., 2019). Low-quality sequences with a quality score < 20, ambiguous nucleotides, short and chimeric sequences were discarded using the DaDa2 pipeline (Callahan et al., 2016). Amplicon Sequence Variants (ASVs) with < 0.005% relative abundance were removed (Bokulich et al., 2013). The ASVs were exported into the Geneious Prime software (version 2022.0.2), where the nucleotide sequences were in silico translated into amino acid sequences. Protein ASV sequences along with reference protein sequences collected from the NCBI database were used to construct a Bayesian phylogenetic tree (Supplementary Figures 3–5) (Huelsenbeck and Ronquist, 2001). Produced ASVs are given in the Supplementary material.
Statistical analysis
Prior to statistical analysis, the transcript and gDNA amoA abundance values were log10 transformed. Repeated measures analysis of variance (ANOVA) were used to investigate the effect of temperature, moisture, and nitrogen on comammox Nitrospira, AOA and AOB transcript and gDNA amoA gene abundance over time.
A two-way ANOVA followed by an LSD test was used to determine if temperature, moisture, or nitrogen significantly affected the transcript and gDNA abundance of comammox, AOA, or AOB amoA at each sampling date (Genstat 22nd edition) (VSN International, 2022).
The Shannon alpha diversity indices were calculated in R with the “Vegan” package to summarize the diversity of active AOB, AOA, and comammox within samples (Dixon, 2003).
Weighted unifrac and Bray-Curtis analysis were conducted using the Qiime2 “qiime diversity core-metrics-phylogenetic” plugin (Bolyen et al., 2019). Heatmaps were produced in Qiime2 using the “qiime feature-table heatmap” plugin (Hunter, 2007). For AOA and AOB, two heatmaps were produced. One using the common (present in all experimental units) and the other using the clades obtained from the phylogenetic tree. Relative abundance was log10 transformed.
Using the “Hmisc” package in R, the Spearman correlation was employed to investigate any correlations between ASV relative abundance, environmental parameters, and the various nitrogen species concentrations (Harrell Jr and Harrell Jr, 2019). Only ASVs that were present in more than one-third of the samples and had more than 0.1% relative abundance were included.
The Spearman correlation was also employed to investigate the correlations between Shannons entropy and environmental parameters.
Results
amoA gene transcript abundance
AOB
Throughout the trial, AOB amoA transcript abundance was significantly influenced by nitrogen (P < 0.001, v.r. = 149.79), while temperature (P = 0.029, v.r. = 5.56) and moisture (P = 0.012, v.r. = 3.27) had a relatively small effect (Figure 1). While AOB amoA transcript abundance significantly increased in response to N700, there was no significant response in the N0 and N50 treatments (Figure 1).
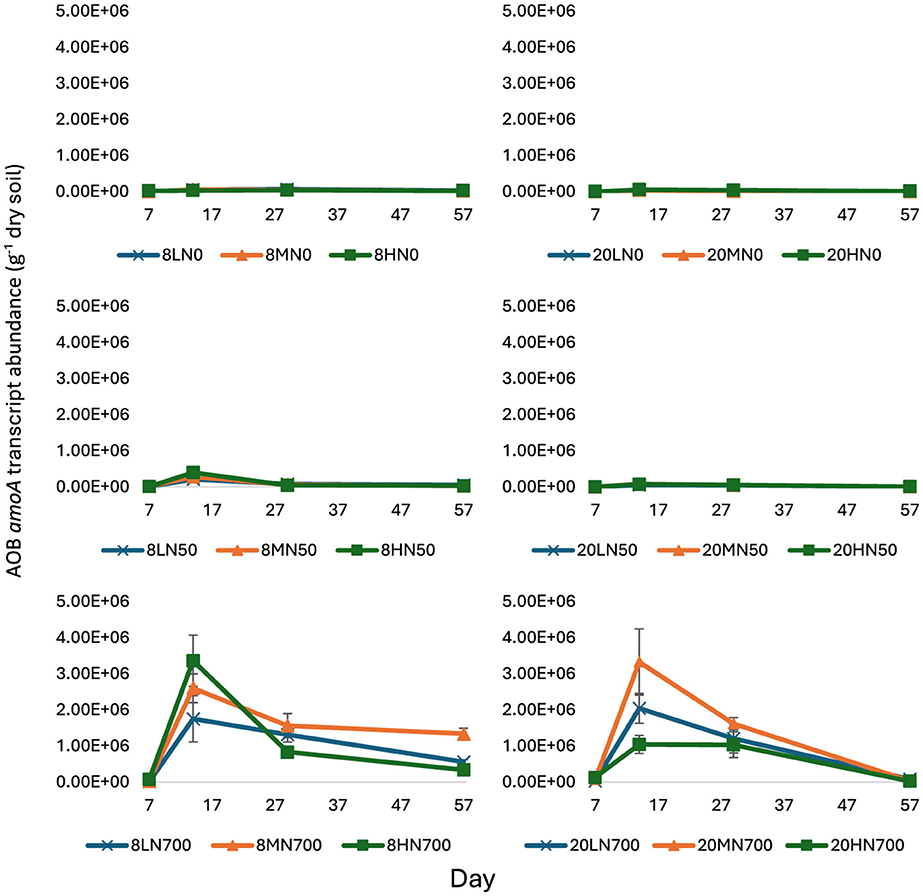
Figure 1. AOB amoA transcript abundance from day 7 to 57. Error bars represent standard error of the mean (n = 4). 8, 8°C; 20, 20°C; L, low moisture (28.5% θ g); M, medium moisture (37.5% θ g); H, high moisture (46.5% θ g); N0, no added nitrogen; N50, N applied at a rate of 50 kg N ha−1 as urea; N700, N applied at a rate of 700 kg N ha−1 as synthetic urine.
At day 7, moisture, nitrogen, and temperature did not significantly affect AOB amoA transcript abundance.
At day 14, AOB amoA transcript counts peaked at 3.36 × 106 g−1 dry soil (8HN700) and 3.33 × 106 g−1 dry soil (20MN700) for the 8 and 20°C treatments, respectively (Figure 1). Peak AOB amoA transcript abundance was one order of magnitude higher in the N700 treatments, relative to their respective N0 treatments. AOB amoA transcript abundance decreased after day 14.
At day 57, temperature (P < 0.001, v.r. = 146.64), moisture (P < 0.001, v.r. 22.35), and nitrogen (P < 0.001, v.r. = 149.51) had a significant effect on AOB amoA transcript abundance (Figure 1).
At all tested time points, AOB amoA transcript abundance exhibited a positive correlation with estimated ammonia concentration (expressed as mg L−1), amount of ammonium (expressed as kg N ha−1), and soil pH (except for day 29; Table 1). At day 14, the correlation between AOB amoA transcript abundance and soil NH4 and estimated NH3 concentrations was 0.85 and 0.74, respectively.
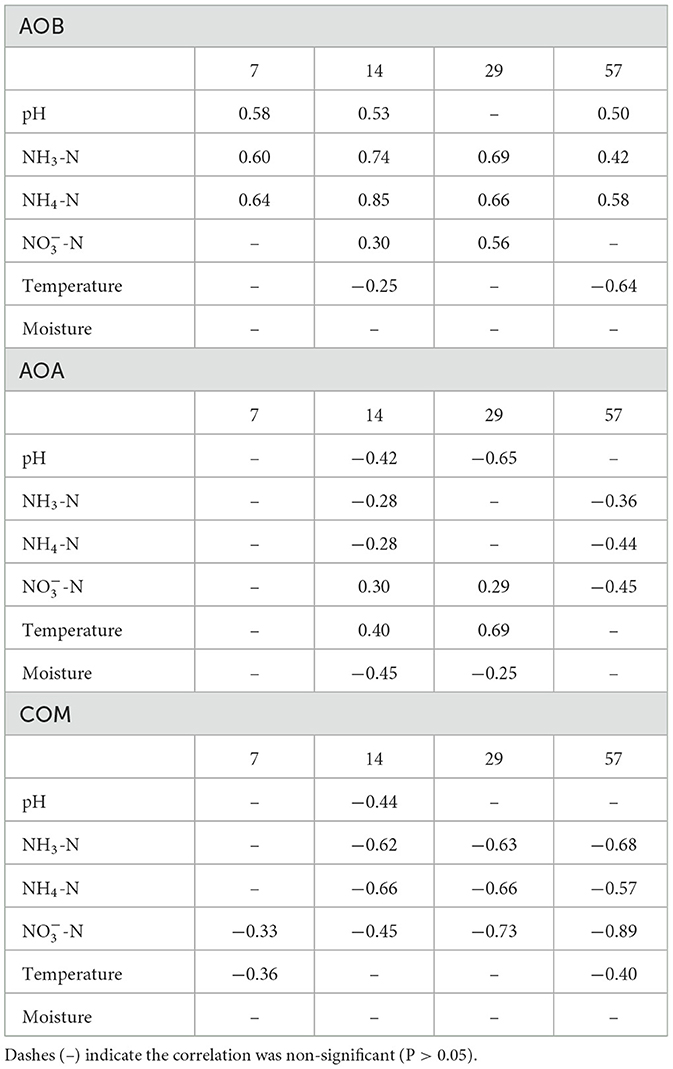
Table 1. Spearman correlation matrix for AOB, AOA, and Comammox Nitrospira (COM) amoA transcript abundance and select environmental parameters from day 7 to 57.
Within the 20HN700 treatment, AOB amoA gDNA growth began on day 1 and peaked on day 29 (Supplementary Figure 1). This coincided with the peak nitrification rate (day 1–29, 31.2 kg -N ha−1 day−1). Comparatively, AOB growth within the 8HN700 treatment began 14 days later and peaked at day 57. This also coincided with the peak nitrification rate of 19.2 kg -N ha−1 day−1 (from day 14–57). However, peak nitrification in the 8HN700 treatment was 38.6% less than the 20HN700 treatment (Supplementary Figure 2). This coincides with differences in AOB amoA gDNA accumulation rates over the same period (8HN700 44.8% less than the 20HN700 treatment). AOB amoA transcript abundance did not show this trend.
AOA
Throughout the trial, AOA amoA transcript abundance was strongly influenced by temperature (P < 0.001, v.r. = 71.81). A significant moisture effect (P = 0.003, v.r. = 6.52) was also observed, while the nitrogen effect was non-significant (P = 0.302; Figure 2).
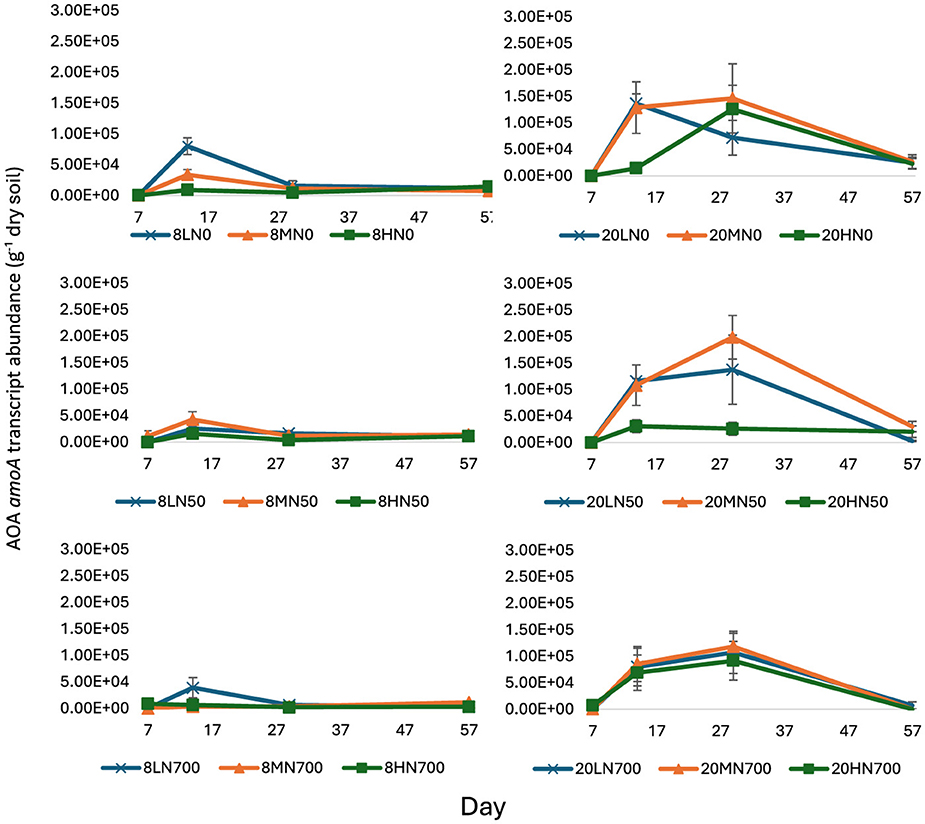
Figure 2. AOA amoA transcript abundance from day 7 to 57. Error bars represent the standard error of the mean (n = 4). 8, 8°C; 20, 20°C; L, low moisture (28.5% θ g); M, medium moisture (37.5% θg); H, high moisture (46.5% θ g); N0, no added nitrogen; N50, N applied at a rate of 50 kg N ha−1 as urea; N700, N applied at a rate of 700 kg N ha−1 as synthetic urine.
At day 7, AOA amoA transcript was almost undetected. From day 14 to 29, AOA amoA transcript abundance was significantly higher in the 20°C treatments (Figure 2).
In the 20°C treatments, AOA amoA transcript abundance peaked at day 29 (except for 20LN0 and 20HN50), where the highest counts were 1.46 × 105 g−1 dry soil (20MN0), 1.99 × 105 g−1 dry soil (20MN50), and 1.18 × 105 g−1 dry soil (20MN700) for 20N0, 20N50, and 20N700 treatments, respectively (Figure 2). In the 8°C treatments, AOA amoA transcript abundance peaked 15 days earlier, with 8.01 × 104 g−1 dry soil (8LN0), 3.90 × 104 g−1 dry soil (8MN50), and 4.24 × 104 g−1 dry soil (8LN700) for 8N0, 8N50, and 8N700 treatments, respectively. Interestingly, the peak abundance in the 20N50 and 20N700 treatments was almost one order of magnitude higher than their 8°C counterparts (Figure 2).
At day 57, AOA amoA transcript abundance was significantly lower in the N700 treatments relative to the N50 and N0 treatments (P < 0.001). At peak abundance, high nitrogen-induced inhibition was higher in the 8°C treatments (51.3 vs. 21.3%, 90 vs. 19.1%, and 28.5 vs. 27.3% for the low, medium, and high moisture treatments, respectively).
AOA amoA transcript abundance shared a negative correlation with soil pH at days 14 and 29 (−0.42 and −0.65, respectively).
COM
Throughout the trial, nitrogen (P < 0.001, v.r. = 71.66) had the most influence on comammox Nitrospira amoA transcript abundance. Temperature had a minor effect (P = 0.03. v.r. = 5.01). While the moisture effect (P = 0.935) was calculated to be non-significant (Figure 3).
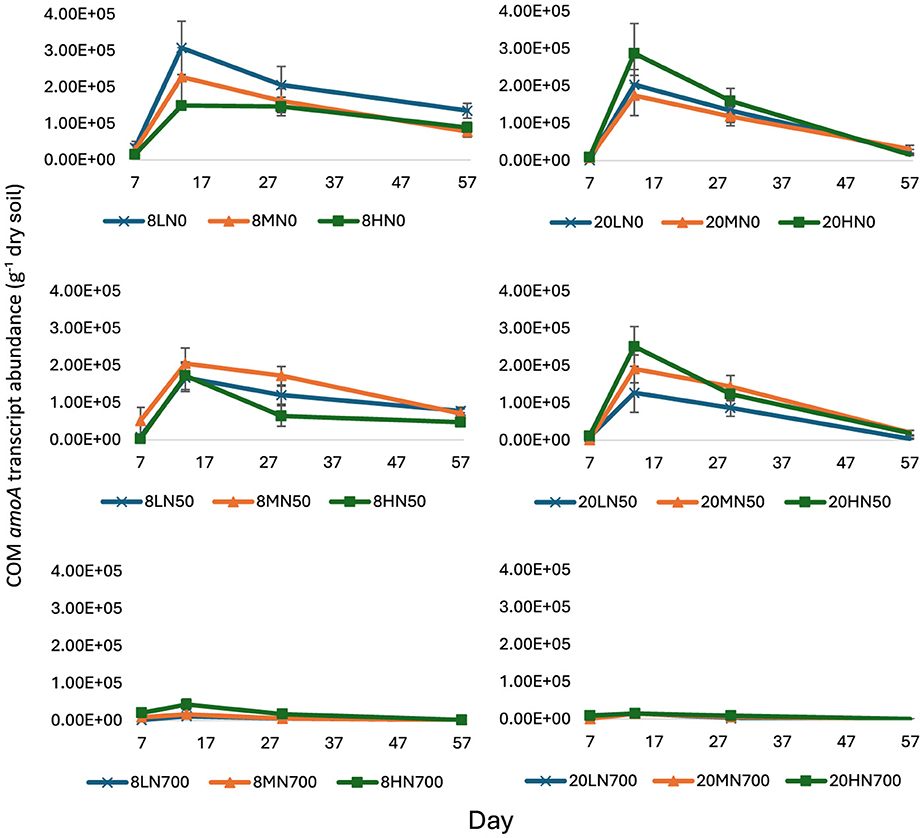
Figure 3. Comammox Nitrospira amoA transcript abundance from day 7 to 57. Error bars represent standard error of the mean (n = 4). 8, 8°C; 20, 20°C; L, low moisture (28.5% θ g); M, medium moisture (37.5% θ g); H, high moisture (46.5% θ g); N0, no added nitrogen; N50, N applied at a rate of 50 kg N ha−1 as urea; N700, N applied at a rate of 700 kg N ha−1 as synthetic urine.
From day 14 onwards, comammox Nitrospira amoA transcript abundance was significantly lower in the N700 treatments (P < 0.001).
Except for 8HN0, comammox Nitrospira amoA transcript abundance peaked at day 14, where counts were highest in the 8LN0 (3.08 × 105 g−1 dry soil), 20HN50 (2.52 × 105 g−1 dry soil), and 8HN700 (4.27 × 104 g−1 dry soil) for the respective N treatments (Figure 3). Peak comammox Nitrospira amoA transcript abundance in the N0 and N50 treatments was almost one order of magnitude higher than the N700 treatments.
Peak comammox Nitrospira amoA transcript abundance in the 8HN700 treatment was almost four times higher than the other 8N700 treatments (Figure 3).
Comammox Nitrospira amoA transcript abundance shared a strong negative correlation with soil ammonia and ammonium concentrations from day 14 to 57. At day 14, comammox Nitrospira amoA transcript abundance also shared a negative correlation with soil pH and soil nitrate concentrations (Table 1).
Diversity analysis
AOB
A total of 2,770,824 high-quality AOB amoA transcript sequences were obtained from 72 samples. The sequencing analysis revealed that the AOB community was made up of 58 ASVs, grouped into five clades; Nitrosospira 3a.1 (81.5%), Nitrosospira 10/11 (8.42%), Nitrosospira 3b (1.69%), Nitrosospira 2/4 (0.826%), and Nitrosomonas (7.9%). ASV003 made up 80.7% of the AOB community (Supplementary Figure 3).
Temperature shared contrasting correlations with Nitrosospira 3a.1 and Nitrosomonas, with an r2 of −0.46 and 0.47, respectively (P < 0.0001; Supplementary Table 3).
Nitrosomonas also shared a significant positive correlation with soil nitrate and estimated ammonia concentration (P < 0.0001, r2 = 0.50, P = 0.0051, r2 = 0.33, respectively; Supplementary Table 3). Comparatively, the relative abundance of all Nitrosospira clusters were not positively correlated with soil ammonia or ammonium concentrations (Supplementary Table 3).
AOA
A total of 2,840,949 high-quality AOA amoA transcript sequences were obtained from 72 samples. The sequencing analysis revealed that the AOA community was made up of 22 ASVs, grouped into two clades: Nitrososphaerales (86.2%), and Nitrosotaleales (13.8%). The most abundant ASV, ASV011 made up 63.5% of the total community (Supplementary Figure 4). ASV002 was the only AOA ASV that was positively correlated with estimated ammonia availability (Supplementary Table 4). ASV002 showed 100% similarity to Nitrosocomicus sp.
AOA community diversity was significantly higher in the 8°C treatments relative to the 20°C treatments (P < 0.001; Figure 4). No treatment effect was observed within each temperature block.
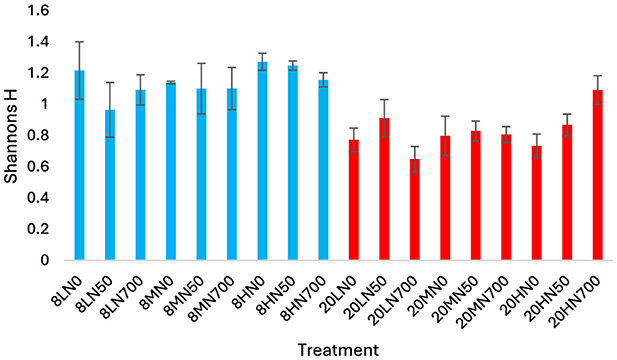
Figure 4. Shannons entropy of the AOA community in each treatment. Error bars represent standard error of the mean. 8, 8°C; 20, 20°C; L, low moisture (28.5% θ g); M, medium moisture (37.5% θ g); H, high moisture (46.5% θg); N0, no added nitrogen; N50, N applied at a rate of 50 kg N ha−1 as urea; N700, N applied at a rate of 700 kg N ha−1 as synthetic urine.
Figure 5 shows that at 8°C, less abundant ASVs (members of Nitrososphaerales) made up a larger proportion of the AOA community. At 20°C, the relative abundance of the Nitrosotaleales clade significantly increased, while the proportion of ASVs associated with Nitrosophaerales decreased. Within the 20°C treatments, the relative abundance of Nitrosotaleales was reduced by synthetic urine. The relative abundance of ASV009 and ASV011 were positively correlated with temperature, while all other significant ASVs were negatively correlated (Supplementary Table 4). Similarly, ASV009 was the only ASV that was negatively correlated with pH and ammonium (Supplementary Table 4).
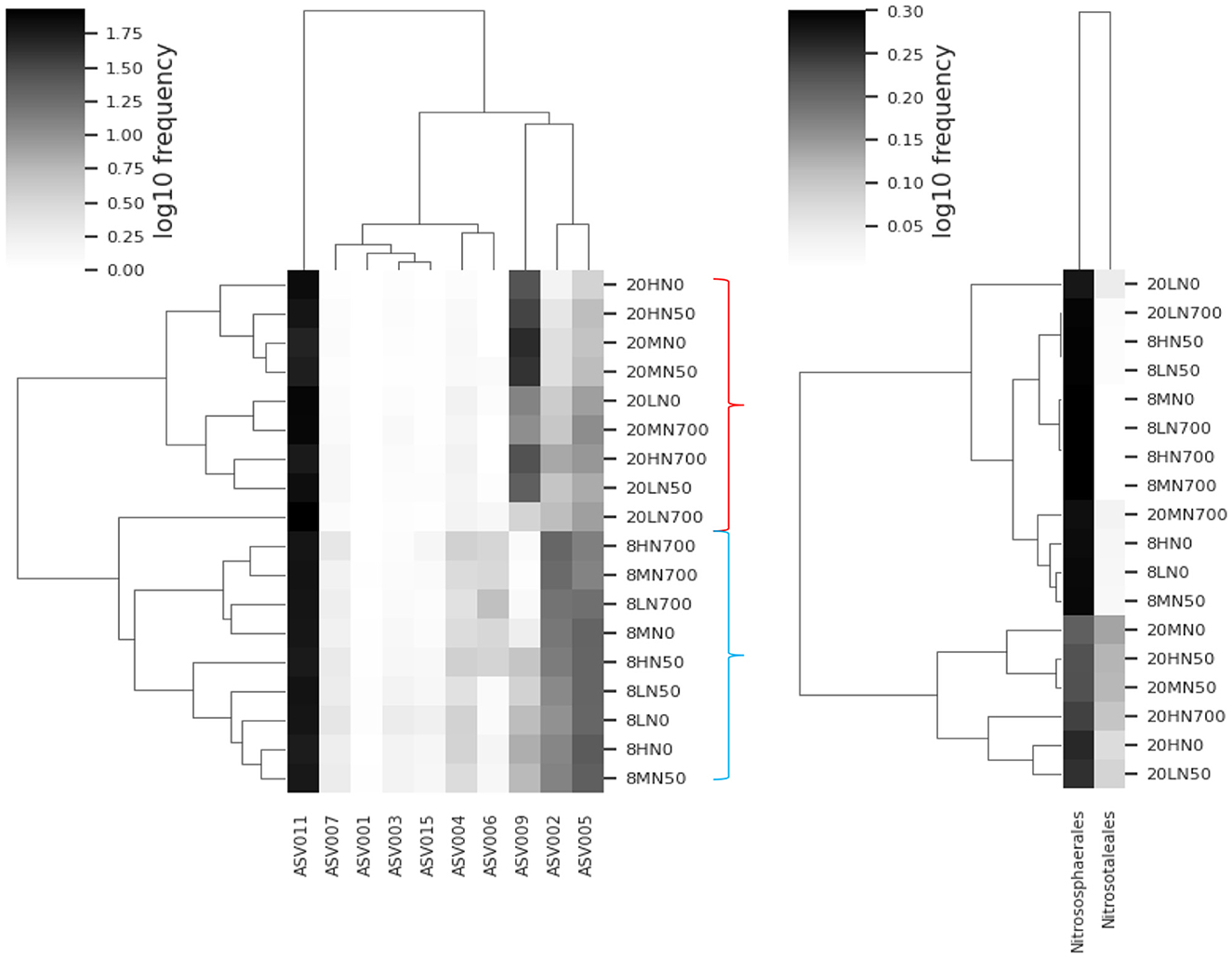
Figure 5. Heatmap of the common AOA ASVs (left) and the AOA clades within (right) each treatment. Relative abundance of clades is based on the associated ASVs shown in the phylogenetic tree (Supplementary Figure 4). Red bracket indicates the 20°C treatments. Blue bracket indicates the 8°C treatments. 8, 8°C; 20, 20°C; L, low moisture (28.5% θ g); M, medium moisture (37.5% θ g); H, high moisture (46.5% θg); N0, no added nitrogen; N50, N applied at a rate of 50 kg N ha−1 as urea; N700, N applied at a rate of 700 kg N ha−1 as synthetic urine.
The Bray-Curtis analysis shows groupings of AOA beta diversity based on the two temperatures used (Supplementary Figure 13). Within the 8°C treatment, the application of synthetic urine seemed to also cause a community shift.
COM
The analysis of the 72 samples yielded 2,757,309 high-quality comammox Nitrospira amoA sequences. The comammox Nitrospira community was composed of 46 ASVs, divided into 3 clades. Clade B.2 was the most abundant, accounting for 78.6% of the population, followed by clade B.1 and clade A.2, which made up 21.3% and 0.021% of the population, respectively. Only 1 ASV, ASV005 was associated with comammox Nitrospira clade A.2 (Supplementary Figure 5).
Comammox Nitrospira alpha diversity was significantly affected by Temperature (P < 0.001), moisture (P = 0.002), and nitrogen (P < 0.001; Supplementary Figure 11). Moreover, comammox Nitrospira community diversity had a contrasting response to synthetic urine at 8 and 20°C. At 8°C, alpha diversity increased, while at 20°C, it decreased (Supplementary Figure 11).
The most abundant ASV, ASV002 (a member of clade B.2) was reduced under the 8N700 treatments. In comparison, the relative abundance of both ASV009 and ASV016 (members of clade B.1) increased under the 8N700 treatments (Supplementary Figure 12).
Discussion
The contrasting response of AOA, AOB, and comammox Nitrospira amoA transcript abundance to temperature, moisture, and nitrogen investigated in this study gives novel insight into their respective roles in New Zealand's fertile dairy farm soils.
Similar to previous studies, AOB positively responded to the application of synthetic urine (Nicol et al., 2008; Di et al., 2009, 2014; Chisholm et al., 2024). Synthetic urine increased AOB amoA cDNA abundance to 1–3 orders of magnitude higher than AOA and comammox Nitrospira amoA transcript abundance. The relative response of AOB, plus the strong positive correlation between NH3-N, NH4-N, and AOB amoA transcript abundance, suggests that they are the main contributors to nitrification in a nitrogen-rich environment.
Peak AOB amoA gDNA abundance in the 8 and 20°C N700 treatments was similar (Supplementary Figure 1). However, at 20°C, the peak occurred 30 days earlier. Within the N700 treatments, AOB growth (AOB gDNA amoA abundance) followed a similar trend to accumulation/nitrification rate (Supplementary Figures 1, 2). The AOB amoA transcript abundance did not follow the nitrate accumulation trend. This suggests that while the soil ammonia/ammonium concentration dictates the AOB amoA transcript response, ultimately, the AOB amoA gDNA community abundance is indicative of the soil nitrification rate.
The relative abundance of Nitrosomonas members was higher in the N700 treatments. This was more noticeable at 20°C (Supplementary Figure 9). All of the Nitrosomonas ASVs detected in this study were closely associated with Nitrosomonas europaea. This was expected as they: (i) possess a high ammonia saturation constant (Suzuki et al., 1974; Jiang and Bakken, 1999), (ii) are more tolerant to free nitrous acid than other AOB species (Liu et al., 2023), (iii) are more responsive to high ammonium availability after a period of starvation (Bollmann et al., 2005). However, it does raise the question: “why are Nitrosospira the dominant member of the AOB community?”. The answer is likely multifactorial. A possible partial explanation is that they are more tolerant to low pH, and are more suited to an environment with fluctuating ammonia availability (Groeneweg et al., 1994; Jiang and Bakken, 1999; Burton and Prosser, 2001; Terada et al., 2013). While Nitrosomonas europaea are more responsive to sudden and significant increases in ammonia availability, they are also less tolerant to low ammonia environments.
As stated previously, AOB amoA gDNA abundance was highest in the high moisture treatments. Di et al. (2014) also concluded that AOB amoA gDNA abundance responded positively to soil moisture. This was attributed to the presence of nirK in some AOB genomes. The nirK gene facilitates the production of nitrite reductase enzyme, which allows AOB to undertake a process known as nitrifier denitrification in oxygen limited environments (Wrage et al., 2001; Wrage-Mönnig et al., 2018). However, there was no significant variation in nitrate accumulation within the N700 treatments, despite the varying rate at which the concentration of ammonium (and estimated ammonia availability) decreased (Supplementary Figures 6, 7). This trend was particularly noticeable in the 20N700 treatments, most likely due to the relatively lower soil pH (Wrage-Mönnig et al., 2018). A possible explanation is reciprocal feeding within the AOB community, whereby is reduced to NO and oxidized back to with fluctuating O2 availability (Caranto and Lancaster, 2017).
Within the N0 and N50 treatments, the concentration of was lower at high moisture contents, suggesting that traditional nitrifier denitrification rather than the proposed reciprocal feeding occurred.
As far as the authors are aware, this is the first study to investigate the effect of temperature on AOA activity and abundance in New Zealand soils.
Both AOA amoA transcript and gDNA abundance were significantly higher in the 20°C treatments (Figure 2, Supplementary Figure 8). Comparatively, temperature did not alter AOB peak transcript or gDNA amoA abundance. Taylor et al. (2017) showed that AOA and AOB were active across different temperature ranges. However, the two temperatures used in this study were within the range dominated by AOB activity/abundance (Taylor et al., 2017). Previous studies have suggested that for nitrification to be dominated by AOA, temperatures must exceed 30°C (Tourna et al., 2011; Lehtovirta-Morley et al., 2016b; Taylor et al., 2017). This exceeds all monthly average soil temperatures in New Zealand. It is also worth noting that the nitrification potential outlined in Taylor et al. (2017) was without nitrogen fertilizer application. Many AOA possess a high substrate affinity for ammonia (Offre et al., 2014), which allows them to thrive in an oligotrophic/nutrient deficient environment (Lehtovirta-Morley et al., 2016b; Herbold et al., 2017; Ouyang et al., 2017). Because of this, their relative growth potential is limited under nitrogen-amended soils. Furthermore, AOA amoA transcript and gDNA growth was inhibited in the N700 treatments.
AOA may have significantly contributed to ammonia oxidation in the 50 kg ha−1 urea amended soils at 20°C. However, AOA amoA transcriptional response occurred 14 days after peak AOB amoA transcript abundance. This was also after much of the soil ammonium was consumed (Supplementary Figure 6). This indicates that AOA only responded after the soil ammonium concentration became non-limiting.
However, the relative abundance of AOA ASV002 increased in the N700 treatments. ASV002 shared 100% identity with Candidatus Nitrosocosmicus franklandus. Ca. N. franklandus has been characterized as having a comparable ammonia affinity to soil borne AOB and responsive to high urea concentration (Lehtovirta-Morley et al., 2016a). Lehtovirta-Morley et al. (2016a) showed that Ca. N. franklandus had an optimum temperature of 40°C, and did not grow at 25°C. ASV002 shared a negative correlation with soil temperature, which suggests that in soil borne Nitrosocosmicus may be tolerant to a wider range of temperatures. The presence and relative increase of Nitrosocosmicus AOA in a high nitrogen environment poses an interesting contrast from conclusions derived from Di et al. (2009). However, as the AOA amoA transcript and gDNA abundance was significantly inhibited in the N700 treatments, it is unlikely that the overall AOA community significantly contribute to nitrification in New Zealand dairy farm soils.
Temperature was the sole treatment to affect soil pH (Supplementary Table 5). At a higher temperature, soil pH was significantly lower. Because of this, the observed temperature effect may be confounded by a soil pH effect (Lu et al., 2018). Both of these influence AOA activity and abundance (Herbold et al., 2017; Gubry-Rangin et al., 2017; Taylor et al., 2017).
Gubry-Rangin et al. (2017) observed that the AOA temperature response was pH dependant. In a low soil pH environment, optimum temperature was at 20°C, while in neutral soils, it was 30°C. It was concluded that two abundant acidic clusters (Nitrososphaera and Nitrosotalea) may prefer lower temperatures, relative to neutrophilic thaumarchaeotal clusters.
The relative abundance of AOA amoA ASV009 increased in all 20°C treatments, while the relative abundance of all other ASVs decreased (ASV009 is a member of Nitrosotaleales).
Previous studies have reported that Nitrosotaleales can thrive at a low soil pH. Their optimal growth temperature is one of the lowest recorded at 25°C (Lehtovirta-Morley et al., 2011). Comparatively, the optimum temperature for Nitrososphaeales (Nitrososphaeales viennensis) was 35°C (Tourna et al., 2011). This provides a possible explanation for the relative increase in Nitrosotaleales, while the relative abundance of other AOA community members decreased or remained unchanged. It is likely that the positive relationship between AOA activity and temperature is due to the relative increase of the Nitrosotaleales clade, while the Nitrososphaeales clade had a smaller effect.
The relative abundance of Nitrosotaleales decreased under the synthetic urine treatments at 20°C. A possible explanation is inhibition by free nitrous acid. Lehtovirta-Morley et al. (2011) demonstrated that Nitrosotalea devanterra was inhibited by 2.53 μM free nitrous acid (40 μM nitrite at pH 4.5).
Comammox Nitrospira amoA transcript and gDNA abundance did not respond to soil moisture. This suggests that the moisture-comammox Nitrospira association found in Chisholm et al. (2023) is more complex than previously thought. It is possible that the high moisture found in Chisholm et al. (2023) facilitated an interaction between comammox Nitrospira and another microorganism, such as anammox (Vilardi et al., 2023; Guo et al., 2024). Or the high moisture facilitated a change in environment that was undetected. Chisholm et al. (2023) sampled a diverse range of terrestrial ecosystems. These sites contained a wide variety of plant species and soil types which could be indirectly linked to soil moisture. Wang et al. (2021) indicated that comammox Nitrospira community composition was significantly influenced by plant type. Comammox Nitrospira clade B have the ability to oxidize formate and other metabolites suggestive of a mixotrophic nature (Palomo et al., 2018; Palatinszky et al., 2024). Formate is produced from the decomposition of organic matter, plant root exudation or excreted as a microbial metabolic byproduct (Adeleke et al., 2017). Therefore, future studies should carefully consider biotic interactions to explain the relative distribution of comammox Nitrospira clade B.
Despite the presence of genes suggestive of a microaerophilic preference in the comammox Nitrospira genome (Palomo et al., 2019), the results presented in Masta et al. (2022) indicate that comammox Nitrospira amoA abundance remain unchanged with changing oxygen availability, further indicating that this relationship is more complex than previously thought. Clearly, additional research is required to understand this phenomenon.
Similarly to AOB, comammox Nitrospira did not respond to temperature. This may be because comammox Nitrospira obtained the ability to oxidize ammonia through a horizontal gene transfer event with β-AOB, indicating that their ammonia monooxygenase enzymes are physiologically similar (Palomo et al., 2018). However, Feng et al. (2022) concluded that the comammox Nitrospira abundance shared a negative correlation with soil temperature. This was attributed to either a low optimum temperature for clade A.2 or the decrease in soil pH caused by a temperature-induced higher nitrification rate. Based on enriched and isolated cultures, the optimum temperature for comammox Nitrospira is 37 and 23°C for N. Inopinata and N. Nitrosa, respectively (Van Kessel et al., 2015; Kits et al., 2017). However, all current isolated and enriched cultures were obtained from an aquatic environment and are members of clade A.1. Whereas the community in this study was dominated by clade B and derived from soil.
It is well established that the relative abundance of comammox Nitrospira clades is determined by key environmental parameters (Wang et al., 2021; Gao et al., 2023). Physiochemical properties like soil pH are considered niche separating (Wang et al., 2021).
It is also possible that comammox Nitrospira was limited by other environmental conditions within this study. Therefore, their response to desirable conditions was limited. A possible example of this could be soil ammonia availability.
Comammox Nitrospira transcript amoA abundance was negatively correlated with estimated ammonia, ammonium, and nitrate concentrations (Table 1). Isolated cultures suggest that comammox Nitrospira prefer an oligotrophic environment (Kits et al., 2017) as genomic analysis revealed that they possess MEP-type ammonia transporters (Palomo et al., 2018). These ammonia transporters have a high affinity (micromolar range) and low uptake capacity (Walter et al., 2008). The genomic analysis also revealed an extra non-operon amoC gene, which is responsible for adaption to low nitrosative stress and ammonia starvation conditions (Palomo et al., 2018; Koch et al., 2019; Xu et al., 2020). The lack of response to changes in soil moisture suggest that there is an additional abiotic or biotic parameter that has not been considered.
The disparity between comammox Nitrospira amoA transcript and amoA gDNA abundance trend indicate that comammox may have gained energy from a source other than ammonia oxidation. Kits et al. (2017) showed that comammox Nitrospira inopinata have a lower nitrite affinity than canonical Nitrospira (canonical NitrospiraKm: 6–27 μM vs. Comammox Km(app): 449.2 μM ). Therefore, it is possible that comammox Nitrospira preferentially undertake the second step of nitrification in a high nitrogen environment, while acting similar to canonical ammonia oxidisers in a low nitrogen environment.
As far as the authors are aware, this is the first paper to find comammox Nitrospira clade A.2 in New Zealand soils. Previous papers found that the community solely consisted of clade B (Hsu et al., 2022; Chisholm et al., 2023, 2024; Shah et al., 2024). The discovery of clade A.2 rules out geographical isolation as a potential explanation for their absence in previous studies. Therefore, a different explanation must be explored. Previous papers suggests that soil pH is a possible driver of clade A.2 abundance and comammox Nitrospira niche specificity (Wang et al., 2021). However, no other studies have found the almost complete dominance of clade B. These results indicate that there is another, previously unidentified environmental parameter (either biotic or abiotic) that heavily influences the abundance of comammox Nitrospira clade B in terrestrial ecosystems. However, based on our current understanding, it is fair to assume that comammox Nitrospira clade B do not significantly contribute to nitrification in New Zealand soils.
Overall, the results presented in this paper support the current paradigm that AOB are the major contributor to nitrification in a nitrogen-rich soil. While both the AOA and comammox Nitrospira communities are inhibited by high ammonia availability. It is also possible that, while capable of ammonia oxidation, both AOA and comammox Nitrospira are capable of a wide range of metabolic pathways that allow them to thrive in a low nitrogen environment. However, the relative abundance of Nitrosocosmicus AOA did increase in response to the application of synthetic urine. However, they were not a major member of the AOA community. Their presence and relative increase pose an interesting question: “are there nutrient rich environments in which they are the dominant ammonia oxidiser?”.
Conclusion
Ammonia oxidizing bacteria dominated soil treated with synthetic urine at low, medium, and high soil moisture, and at high and low temperature. AOB amoA gDNA abundance was highest in the high moisture treatments. This was attributed to the presence of nirK in some AOB genomes. The nirK gene facilitates the production of nitrite reductase enzyme, which allows AOB to undertake a process known as nitrifier denitrification. However, soil ammonium and nitrate concentrations suggest that the AOB present in the synthetic urine treatments participated in reciprocal feeding, whereby is reduced to NO and oxidized back to with fluctuating O2 availability.
Ammonia oxidizing archaea amoA abundance and community composition was strongly influenced by soil temperature. At 20°C, AOA amoA peak transcript abundance averaged over 1 order of magnitude higher. ASV002 (a member of the Nitrosocosmicus clade) was positively correlated to ammonium and estimated soil ammonia concentrations. The presence and relative increase of Nitrosocosmicus AOA in a high nitrogen environment poses an interesting contrast from current scientific opinion.
Contrasting to a previous study, the abundance of comammox Nitrospira was not positively correlated with soil moisture. This suggests that the association is more complex than previously thought. Further research is required to determine the drivers of comammox Nitrospira abundance in a high moisture environment. The majority of the comammox Nitrospira community was associated with clade B, with a small clade A.2 component. As far as the authors are aware, this is the first New Zealand paper to identify sequences associated with clade A.2. Overall, these results indicate that AOB are the main drivers of nitrification in New Zealand dairy farm soils.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
CC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. HD: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. KC: Conceptualization, Funding acquisition, Resources, Supervision, Validation, Visualization, Writing – review & editing. AP: Conceptualization, Investigation, Methodology, Supervision, Validation, Visualization, Writing – review & editing. JS: Validation, Visualization, Writing – review & editing. LZ: Validation, Visualization, Writing – review & editing. KS: Investigation, Validation, Writing – review & editing. XC: Data curation, Methodology, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank the New Zealand Ministry of Business, Innovation and Employment (MBIE), and the New Zealand Agricultural Greenhouse Gas Research Centre (NZAGRC) for funding some of the operational costs, Lincoln University for a Doctoral Scholarship for Chris Chisholm, and Jie Lei, Steve Moore, Trevor Hendry, and Rosemary Dorsey for technical support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1466991/full#supplementary-material
References
Adeleke, R., Nwangburuka, C., and Oboirien, B. (2017). Origins, roles and fate of organic acids in soils: a review. S. Afr. J. Bot. 108, 393–406. doi: 10.1016/j.sajb.2016.09.002
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Bollmann, A., Schmidt, I., Saunders, A. M., and Nicolaisen, M. H. (2005). Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 71, 1276–1282. doi: 10.1128/AEM.71.3.1276-1282.2005
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Brown, I. C. (1943). A rapid method of determining exchangeable hydrogen and total exchangeable bases of soils. Soil Sci. 56, 353–358. doi: 10.1097/00010694-194311000-00004
Burton, S. A., and Prosser, J. I. (2001). Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl. Environ. Microbiol. 67, 2952–2957. doi: 10.1128/AEM.67.7.2952-2957.2001
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., Holmes, S. P., et al. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cameron, K. C., Di, H. J., and Moir, J. L. (2013). Nitrogen losses from the soil/plant system: a review. Ann. Appl. Biol. 162, 145–173. doi: 10.1111/aab.12014
Caranto, J. D., and Lancaster, K. M. (2017). Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc. Nat. Acad. Sci. 114, 8217–8222. doi: 10.1073/pnas.1704504114
Chisholm, C., Di, H., Cameron, K., Podolyan, A., Shen, J., Zhang, L., et al. (2024). Contrasting response of comammox Nitrospira, ammonia oxidising bacteria, and archaea to soil pH and nitrogen inputs. Sci. Total Environ. 924:171627. doi: 10.1016/j.scitotenv.2024.171627
Chisholm, C., Di, H. J., Cameron, K., Podolyan, A., Shah, A., Hsu, L., et al. (2023). Soil moisture is a primary driver of comammox Nitrospira abundance in New Zealand soils. Sci. Total Environ. 858:159961. doi: 10.1016/j.scitotenv.2022.159961
Clough, T. J., Ledgard, S. F., Sprosen, M. S., and Kear, M. J. (1998). Fate of 15 N labelled urine on four soil types. Plant Soil 199, 195–203. doi: 10.1023/A:1004361009708
Daims, H., Lebedeva, E. V., Pjevac, P., Han, P., Herbold, C., Albertsen, M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Di, H. J., Cameron, K. C., Podolyan, A., and Robinson, A. (2014). Effect of soil moisture status and a nitrification inhibitor, dicyandiamide, on ammonia oxidizer and denitrifier growth and nitrous oxide emissions in a grassland soil. Soil Biol. Biochem. 73, 59–68. doi: 10.1016/j.soilbio.2014.02.011
Di, H. J., Cameron, K. C., Shen, J.-P., Winefield, C. S., O'Callaghan, M., Bowatte, S., et al. (2010). Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol. Ecol. 72, 386–394. doi: 10.1111/j.1574-6941.2010.00861.x
Di, H. J., Cameron, K. C., Shen, J. P., Winefield, C. S., O'Callaghan, M., Bowatte, S., et al. (2009). Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat. Geosci. 2, 621–624. doi: 10.1038/ngeo613
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Feng, M., He, Z.-Y., Fan, J., Ge, A.-H., Jin, S., Lin, Y., et al. (2022). Temperature has a strong impact on the abundance and community structure of comammox Nitrospira in an Ultisol. J. Soils Sediments 22, 2593–2603. doi: 10.1007/s11368-022-03261-5
Francis, C. A., Beman, J. M., and Kuypers, M. M. M. (2007). New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1, 19–27. doi: 10.1038/ismej.2007.8
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Nat. Acad. Sci. 102, 14683–14688. doi: 10.1073/pnas.0506625102
Gao, F., Li, Y., Fan, H., Xue, J., and Yao, H. (2023). Main environmental drivers of abundance, diversity and community structure of comammox Nitrospira in paddy soils. Pedosphere 33, 808–818. doi: 10.1016/j.pedsph.2022.06.061
Groeneweg, J., Sellner, B., and Tappe, W. (1994). Ammonia oxidation in nitrosomonas at NH3 concentrations near km: effects of pH and temperature. Water Res. 28, 2561–2566. doi: 10.1016/0043-1354(94)90074-4
Gubry-Rangin, C., Novotnik, B., Mandič-Mulec, I., Nicol, G. W., and Prosser, J. I. (2017). Temperature responses of soil ammonia-oxidising archaea depend on pH. Soil Biol. Biochem. 106, 61–68. doi: 10.1016/j.soilbio.2016.12.007
Guo, Z., Ma, X. S., and Ni, S.-Q. (2024). Journey of the swift nitrogen transformation: unveiling comammox from discovery to deep understanding. Chemosphere 358:142093. doi: 10.1016/j.chemosphere.2024.142093
Gustafsson, J. P. (2011). Visual MINTEQ 3.0 User Guide. Stockholm: KTH, Department of Land and Water Recources, 550.
Herbold, C. W., Lehtovirta-Morley, L. E., Jung, M.-Y., Jehmlich, N., Hausmann, B., Han, P., et al. (2017). Ammonia-oxidising archaea living at low pH: insights from comparative genomics. Environ. Microbiol. 19, 4939–4952. doi: 10.1111/1462-2920.13971
Hsu, P.-C., Di, H., Cameron, K., Podolyan, A., Chau, H., Luo, J., et al. (2022). Comammox Nitrospira Clade B is the most abundant complete ammonia oxidizer in a dairy pasture soil and inhibited by dicyandiamide and high ammonium concentrations. Front. Microbiol. 13:1048735. doi: 10.3389/fmicb.2022.1048735
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Hunter, J. D. (2007). Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95. doi: 10.1109/MCSE.2007.55
Jiang, Q. Q., and Bakken, L. R. (1999). Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 30, 171–186. doi: 10.1111/j.1574-6941.1999.tb00646.x
Kits, K. D., Sedlacek, C. J., Lebedeva, E. V., Han, P., Bulaev, A., Pjevac, P., et al. (2017). Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 549, 269–272. doi: 10.1038/nature23679
Koch, H., van Kessel, M. A., and Lücker, S. (2019). Complete nitrification: insights into the ecophysiology of comammox Nitrospira. Appl. Microbiol. Biotechnol. 103, 177–189. doi: 10.1007/s00253-018-9486-3
Könneke, M., Bernhard, A. E., José, R., Walker, C. B., Waterbury, J. B., Stahl, D. A., et al. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Lehtovirta-Morley, L. E., Ross, J., Hink, L., Weber, E. B., Gubry-Rangin, C., Thion, C., et al. (2016a). Isolation of ‘Candidatus Nitrosocosmicus franklandus', a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol. Ecol. 92:fiw057. doi: 10.1093/femsec/fiw057
Lehtovirta-Morley, L. E., Sayavedra-Soto, L. A., Gallois, N., Schouten, S., Stein, L. Y., Prosser, J. I., et al. (2016b). Identifying potential mechanisms enabling acidophily in the ammonia-oxidizing archaeon “Candidatus Nitrosotalea devanaterra.” Appl. Environ. Microbiol. 82, 2608–2619. doi: 10.1128/AEM.04031-15
Lehtovirta-Morley, L. E., Stoecker, K., Vilcinskas, A., Prosser, J. I., and Nicol, G. W. (2011). Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Nat. Acad. Sci. 108, 15892–15897. doi: 10.1073/pnas.1107196108
Li, C., He, Z.-Y., Hu, H.-W., and He, J.-Z. (2022). Niche specialization of comammox Nitrospira in terrestrial ecosystems: Oligotrophic or copiotrophic? Crit. Rev. Environ. Sci. Technol. 53, 161–176. doi: 10.1080/10643389.2022.2049578
Li, C., Hu, H.-W., Chen, Q.-L., Chen, D., and He, J.-Z. (2019). Comammox Nitrospira play an active role in nitrification of agricultural soils amended with nitrogen fertilizers. Soil Biol. Biochem. 138:107609. doi: 10.1016/j.soilbio.2019.107609
Liu, Y., Zhu, Y., Wu, D., Wang, Z., Wang, Y., Wang, G., et al. (2023). Effect of free nitrous acid on nitritation process: microbial community, inhibitory kinetics, and functional biomarker. Bioresour. Technol. 371:128595. doi: 10.1016/j.biortech.2023.128595
Lu, X., Nicol, G. W., and Neufeld, J. D. (2018). Differential responses of soil ammonia-oxidizing archaea and bacteria to temperature and depth under two different land uses. Soil Biol. Biochem. 120, 272–282. doi: 10.1016/j.soilbio.2018.02.017
Masta, M., Espenberg, M., Gadegaonkar, S. S., Pärn, J., Sepp, H., Kirsimäe, K., et al. (2022). Integrated isotope and microbiome analysis indicates dominance of denitrification in N2O production after rewetting of drained fen peat. Biogeochemistry 161, 119–136. doi: 10.1007/s10533-022-00971-3
Nicol, G. W., Leininger, S., Schleper, C., and Prosser, J. I. (2008). The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10, 2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x
Offre, P., Kerou, M., Spang, A., and Schleper, C. (2014). Variability of the transporter gene complement in ammonia-oxidizing archaea. Trends Microbiol. 22, 665–675. doi: 10.1016/j.tim.2014.07.007
Olsen, S. R. (1954). Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate. Washington, DC: US Department of Agriculture.
Orellana, L. H., Chee-Sanford, J. C., Sanford, R. A., Löffler, F. E., and Konstantinidis, K. T. (2018). Year-round shotgun metagenomes reveal stable microbial communities in agricultural soils and novel ammonia oxidizers responding to fertilization. Appl. Environ. Microbiol. 84:e01646-17. doi: 10.1128/AEM.01646-17
Ouyang, Y., Norton, J. M., and Stark, J. M. (2017). Ammonium availability and temperature control contributions of ammonia oxidizing bacteria and archaea to nitrification in an agricultural soil. Soil Biol. Biochem. 113, 161–172. doi: 10.1016/j.soilbio.2017.06.010
Palatinszky, M., Herbold, C. W., Sedlacek, C. J., Pühringer, D., Kitzinger, K., Giguere, A. T., et al. (2024). Growth of complete ammonia oxidizers on guanidine. Nature 633, 646–653. doi: 10.1038/s41586-024-07832-z
Palomo, A., Dechesne, A., and Smets, B. F. (2019). Genomic profiling of Nitrospira species reveals ecological success of comammox Nitrospira. bioRxiv 612226. doi: 10.1101/612226
Palomo, A., Pedersen, A. G., Fowler, S. J., Dechesne, A., Sicheritz-Pontén, T., Smets, B. F., et al. (2018). Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 12, 1779–1793. doi: 10.1038/s41396-018-0083-3
Prosser, J. I., and Nicol, G. W. (2008). Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 10, 2931–2941. doi: 10.1111/j.1462-2920.2008.01775.x
Prosser, J. I., and Nicol, G. W. (2012). Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 20, 523–531. doi: 10.1016/j.tim.2012.08.001
Rayment, G., and Higginson, F. R. (1992). Australian Laboratory Handbook of Soil and Water Chemical Methods. Sydney, NSW: Inkata Press.
Rotthauwe, J.-H., Witzel, K.-P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997
Rütting, T., Schleusner, P., Hink, L., and Prosser, J. I. (2021). The contribution of ammonia-oxidizing archaea and bacteria to gross nitrification under different substrate availability. Soil Biol. Biochem. 160:108353. doi: 10.1016/j.soilbio.2021.108353
Shah, A. S., Hsu, P.-C. L., Chisholm, C., Podolyan, A., Cameron, K., Luo, J., et al. (2024). Nitrification inhibitor chlorate and nitrogen substrates differentially affect comammox Nitrospira in a grassland soil. Front. Microbiol. 15:1392090. doi: 10.3389/fmicb.2024.1392090
Suzuki, I., Dular, U., and Kwok, S. (1974). Ammonia or ammonium ion as substrate for oxidation by Nitrosomonas europaea cells and extracts. J. Bacteriol. 120, 556–558. doi: 10.1128/jb.120.1.556-558.1974
Takahashi, Y., Fujitani, H., Hirono, Y., Tago, K., Wang, Y., Hayatsu, M., et al. (2020). Enrichment of comammox and nitrite-oxidizing Nitrospira from acidic soils. Front. Microbiol. 11:1737. doi: 10.3389/fmicb.2020.01737
Taylor, A. E., Giguere, A. T., Zoebelein, C. M., Myrold, D. D., and Bottomley, P. J. (2017). Modeling of soil nitrification responses to temperature reveals thermodynamic differences between ammonia-oxidizing activity of archaea and bacteria. ISME J. 11, 896–908. doi: 10.1038/ismej.2016.179
Terada, A., Sugawara, S., Yamamoto, T., Zhou, S., Koba, K., Hosomi, M., et al. (2013). Physiological characteristics of predominant ammonia-oxidizing bacteria enriched from bioreactors with different influent supply regimes. Biochem. Eng. J. 79, 153–161. doi: 10.1016/j.bej.2013.07.012
Tourna, M., Stieglmeier, M., Spang, A., Könneke, M., Schintlmeister, A., Urich, T., et al. (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Nat. Acad. Sci. 108, 8420–8425. doi: 10.1073/pnas.1013488108
Van Kessel, M. A., Speth, D. R., Albertsen, M., Nielsen, P. H., Op den Camp, H. J., Kartal, B., et al. (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. doi: 10.1038/nature16459
Vilardi, K., Cotto, I., Bachmann, M., Parsons, M., Klaus, S., Wilson, C., et al. (2023). Co-occurrence and cooperation between comammox and anammox bacteria in a full-scale attached growth municipal wastewater treatment process. Environ. Sci. Technol. 57, 5013–5023. doi: 10.1021/acs.est.2c09223
VSN International (2022). Genstat for Windows 22nd Edition [Internet]. Genstat.co.uk. Available at: https://vsni.co.uk/software/genstat/
Walter, B., Küspert, M., Ansorge, D., Krämer, R., and Burkovski, A. (2008). Dissection of ammonium uptake systems in Corynebacterium glutamicum: mechanism of action and energetics of AmtA and AmtB. J. Bacteriol. 190, 2611–2614. doi: 10.1128/JB.01896-07
Wang, D.-Q., Zhou, C.-H., Nie, M., Gu, J.-D., and Quan, Z.-X. (2021). Abundance and niche specificity of different types of complete ammonia oxidizers (comammox) in salt marshes covered by different plants. Sci. Total Environ. 768:144993. doi: 10.1016/j.scitotenv.2021.144993
Wrage, N., Velthof, G. L., van Beusichem, M. L., and Oenema, O. (2001). Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol. Biochem. 33, 1723–1732. doi: 10.1016/S0038-0717(01)00096-7
Wrage-Mönnig, N., Horn, M. A., Well, R., Müller, C., Velthof, G., Oenema, O., et al. (2018). The role of nitrifier denitrification in the production of nitrous oxide revisited. Soil Biol. Biochem. 123, A3–A16. doi: 10.1016/j.soilbio.2018.03.020
Xu, S., Wang, B., Li, Y., Jiang, D., Zhou, Y., Ding, A., et al. (2020). Ubiquity, diversity, and activity of comammox Nitrospira in agricultural soils. Sci. Total Environ. 706:135684. doi: 10.1016/j.scitotenv.2019.135684
Keywords: Comammox Nitrospira, ammonia oxidizing bacteria, AOB, ammonia oxidizing archaea, AOA, nitrification
Citation: Chisholm C, Di H, Cameron K, Podolyan A, Shen J, Zhang L, Sirisena K and Che X (2025) Transcriptional activity of ammonia oxidisers in response to soil temperature, moisture and nitrogen amendment. Front. Microbiol. 15:1466991. doi: 10.3389/fmicb.2024.1466991
Received: 19 July 2024; Accepted: 23 December 2024;
Published: 15 January 2025.
Edited by:
Tiehang Wu, Georgia Southern University, United StatesReviewed by:
Samik Bagchi, Digested Organics, United StatesChris Sedlacek, University of Southern Indiana, United States
Copyright © 2025 Chisholm, Di, Cameron, Podolyan, Shen, Zhang, Sirisena and Che. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Di, aG9uZy5kaUBsaW5jb2xuLmFjLm56
 Chris Chisholm
Chris Chisholm Hong Di
Hong Di Keith Cameron1
Keith Cameron1 Andriy Podolyan
Andriy Podolyan Jupei Shen
Jupei Shen Limei Zhang
Limei Zhang