
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 September 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1466990
Introduction: Enterococci are commensals of the gastrointestinal tract of humans and animals that evolved into opportunistic pathogens with high antimicrobial resistance and virulence. Multidrug-resistant Enterococcus is a major cause of hospital-acquired infections worldwide. For this reason, the characterization of non-clinical reservoirs of Enterococci and their epidemiological link to resistant hospital isolates is crucial for controlling their spread.
Methods: A total of 295 samples collected from livestock (pigs and cows, n = 135) and environment (public buses, passengers hands, and urban environments, n = 160) were screened for Enterococcus spp. E. faecium antimicrobial resistance profiles, virulence potential, and clonal population were further characterized.
Results: Enterococci were detected in 90.5% (n = 267) of the samples, with a higher prevalence in livestock (100%) than the environment (82.5%, p < 0.0001), but none of the isolates exhibited vancomycin resistance. E. faecalis was the most prevalent species (51.7%), predominantly found in livestock (62.2%), while E. faecium was more common in the environment. Of the 59 E. faecium isolates, 78% showed resistance to ≥3 antibiotic classes and contained associated resistance genes, namely tetracyclines (tetM and tetL), beta-lactams (mutations in pbp5), and high-level resistance to aminoglycosides (ant(6)-Ia and aac(6′)-aph(2″)). A wide array of virulence factors was detected among E. faecium, associated with adherence, biofilm formation, and adaptation to host response, while hospital-associated virulence markers, such as IS16, were less frequent, probably due to the non-clinical nature of the isolates. Clonal population analysis revealed a diverse E. faecium population. Although no direct epidemiological link could be traced between our isolates and specific clinical isolates, infection-associated genetic backgrounds were identified in non-clinical isolates: one isolate from pigs belonged to CC17 (ST32), while four isolates belonged to CC94, including one recovered from pigs (ST296), one from cows (ST2206), one from the urban environment (ST1205), and other from buses (ST800).
Discussion: This study underscores a high prevalence of clinically relevant Enterococcus species among healthy livestock and the environment. Despite the absence of vancomycin resistance and limited hospital infection-associated clonal lineages, the presence of E. faecium with significant virulence potential and resistance to critical antibiotics in human and veterinary medicine highlights the need for continuing surveillance of non-clinical reservoirs.
Enterococci are Gram-positive acid lactic bacteria ubiquitously found in nature in a wide range of habitats, including the gastrointestinal tract (GIT) of humans, vertebrate and invertebrate animals as part of its microbiome, as well as in the natural environment in plants, soil, fresh, and marine waters and in food (Murray, 1990; Byappanahalli et al., 2012; Lebreton et al., 2014; Cattoir, 2022). While the Enterococcus genus comprises >50 species, its distribution in nature is highly variable and associated with host species factors such as age and diet or environmental stress conditions (Lebreton et al., 2014; Cattoir, 2022). Enterococcus faecalis, Enterococcus faecium, Enterococcus hirae, and Enterococcus durans are the species most commonly found in the gut of mammals, with E. faecium and E. faecalis representing up to 1% of human adult microbiota (Torres et al., 2018; Cattoir, 2022). Some of these Enterococcus species, namely E. faecalis and E. faecium, evolved as human opportunistic pathogens being responsible for a broad range of infections from the urinary tract, intra-abdominal infections or wounds, to more severe life-threatening conditions such as bacteremia and endocarditis usually associated with high antibiotic resistance and virulence (Cetinkaya et al., 2000; Arias and Murray, 2012; Zhong et al., 2019; Cattoir, 2022; Wei et al., 2024). Although E. faecalis accounts for the majority of enterococcal infections in humans, E. faecium has been increasingly reported in hospitals usually associated with severe conditions (Gao et al., 2017; ECDC, 2023; Wei et al., 2024). In fact, a major hallmark of enterococci that allowed the success of nosocomial pathogens is its capacity to acquire antimicrobial resistance and adaptation to hostile environments. Enterococci are intrinsically resistant to a variety of antimicrobials and have remarkable genome plasticity that enables the accumulation of mutations and acquisition of external antimicrobial resistance determinants (Cetinkaya et al., 2000; Arias and Murray, 2012; Guzman Prieto et al., 2016).
Of major concern was the emergence of vancomycin-resistant isolates, which through the acquisition of the van operon were able to produce altered cell wall peptidoglycan precursors not recognized by glycopeptides (Uttley et al., 1988; Cetinkaya et al., 2000). Vancomycin-resistant E. faecium (VRE) become endemic in US hospitals as a consequence of the increasing usage of vancomycin in difficult-to-treat infections, while in European hospitals, VRE infections have been increasing since the COVID-19 pandemics, reaching a mean rate of 17.6% in 2022 (ECDC, 2023). Antibiotic-resistant E. faecium is among the top ten bacterial pathogens with the highest antimicrobial resistance-associated mortality, with treatment often restricted to last-resort antibiotics such as linezolid, tigecycline, and daptomycin. However, sporadic-resistant isolates to these antibiotics have been already reported (Bender et al., 2018a), leading VRE to be considered as a high-priority pathogen for drug development by WHO (2024).
Colonized individuals or animals are potential reservoirs of multidrug-resistant Enterococcus spp., representing a significant source for pathogen transmission (Garcia-Solache and Rice, 2019; Cattoir, 2022; Wei et al., 2024). As VRE have a high capacity to persist in environmental surfaces and be maintained as commensals among humans and animals in the gut until they achieve optimal conditions to develop infection (Chavers et al., 2003; Zaheer et al., 2020; Wada et al., 2021), the screening of community and environment reservoirs for multidrug-resistant E. faecium emerged as an urgent tool to support nosocomial infection control strategies.
In Portugal, VRE infections have been increasingly detected in hospitals paralleling the European scenario (ECDC, 2023). Moreover, healthy individuals without hospital and/or antibiotic exposure were identified as asymptomatic carriers of antibiotic-resistant Enterococci (Novais et al., 2006; Correia et al., 2014; Freitas et al., 2018). While VRE was sporadically detected in food-producing animals and pets (Zaheer et al., 2020; Cattoir, 2022; Gião et al., 2022), a comprehensive screening of environmental reservoirs in the country remains scarce.
Hence, in this study we applied a One Health approach to unveil the major VRE and antibiotic-resistant Enterococci reservoirs among healthy livestock and urban environment in Portugal. We will assess the significance of these reservoirs on the maintenance of hospital-associated multidrug-resistant Enterococcus, namely E. faecium, clonal lineages, and/or antibiotic resistance and virulence determinants in natural ecosystems, responding to the need for valuable epidemiological information to guide the design of targeted and effective infection control measures.
Samples from livestock and environmental sources, previously recovered in the scope of MRSA surveillance studies in Portugal (Conceição et al., 2013, 2017a,b; Lopes et al., 2019), were screened for the presence of vancomycin-resistant enterococci.
A total of 294 samples included in this study were selected in order to include the highest variability and representativeness of collection dates, source, and positivity for other bacterial carriage or contamination (namely MRSA):
A total of 80 nasal swabs from healthy swine were collected in two independent Portuguese farms producing animals for human consumption, in two screening periods in 2016 and 2018 (Conceição et al., 2017a; Lopes et al., 2019). These farms used amoxicillin (0.5%), colistin (0.5%), and zinc oxide (0.15%) in the feed regimen of all animals until 2016, but since then, one farm banned colistin usage, while the other farm completely eliminated antibiotics, keeping zinc oxide (0.15%) only to prevent gastrointestinal diseases.
A collection of 55 samples (36 swabs from the nasal cavity and 19 from the inguinal region) were recovered from calves and bovines in a beef production herd in the south of Portugal in January 2016 (Conceição et al., 2017b). At the time of sampling, none of the animals presented apparent clinical symptoms of infection and only two cows had been treated with antibiotics (oxytetracycline).
Eighty-nine samples recovered from highly touched surfaces in public buses in Lisbon, between May 2011 and May 2012, and eighteen samples of passengers’ hands previously identified as positives for MRSA in May 2012 and in January 2013 were selected for VRE screening (Conceição et al., 2013). Bus sampling was performed before any cleaning procedure in the vehicles allocated to different travel routes, some of them serving hospitals, while the hands of individuals were screened immediately after leaving the buses (Conceição et al., 2013).
All 52 environmental samples, recovered in June 2013 in the outdoor areas around major hospitals in Lisbon, were included. Samples were previously recovered by swabbing the surfaces of parking meters, stairwells, automated teller machines, bus stops, public phones, and treadmill stops with sterile cotton gauzes humidified with sterile water and further placed into tryptic soy broth (TSB; Becton, Dickinson & Co, New Jersey, United States) tubes and transported to the laboratory. Two samples were found to be contaminated with MRSA.
These non-clinical settings were shown to be reservoirs of opportunistic pathogens such as S. aureus/MRSA (Conceição et al., 2013, 2017a,b; Lopes et al., 2019); the samples included in the study were considered as convenience samples, available in the laboratory for the screening of additional bacterial pathogens.
All samples were previously stored at −80°C at the Laboratory of Molecular Genetics culture collection at ITQB-NOVA. An aliquot (100 μL) of each sample was added to brain heart infusion (BHI; Becton, Dickinson & Co, New Jersey, United States) tubes supplemented with 6 μg/mL of vancomycin and incubated at 37°C with overnight agitation, following the Clinical and Laboratory Standards Institute (CLSI, 2021) guidelines for the detection of vancomycin-resistant enterococcus, except for the usage of broth media to enable a higher inoculum volume to be used directly from the stored samples. Samples were then plated in parallel in non-selective tryptic soy agar (TSA; Becton, Dickinson & Co, New Jersey, United States) plates and chromogenic selective media for Enterococcus spp. (Compass Enterococcus agar; BIOKAR Diagnosis, Beauvais, France) and for vancomycin-resistant enterococci (CHROMagar VRE; CHROMagar, Paris, France) and incubated at 35 ± 2°C for 24 h. After incubation, plates were visually inspected and presumptive enterococci colonies with distinct morphologies (different sizes, shapes, or colors) were streaked on BHI agar (OXOID, Thermo Fisher Scientific, Maryland, USA) until purity.
Enterococcus species were identified by PCR amplification of internal fragments of the species-specific ddl gene, for E. faecium and E. faecalis, and the sodA gene, for E. casseliflavus, E. gallinarum, E. raffinosus, E. hirae, and E. durans, as previously described (Dutka-Malen et al., 1995; Jackson et al., 2004).
The presence of van genes (vanA, vanB, vanC1, and vanC2/3) was detected by multiplex PCR (Dutka-Malen et al., 1995; Poulsen et al., 1999).
Given the clinical significance of E. faecium carriage and infection in both humans and animals, we performed additional characterization of isolates, resistance mechanisms, and clonal population structure of the E. faecium recovered from the different collection samples.
Antimicrobial susceptibility testing was performed for all 59 E. faecium isolates through the disk diffusion method for 16 antibiotics: ampicillin (10 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), erythromycin (15 μg), gentamicin (30 μg), levofloxacin (5 μg), linezolid (10 μg), linezolid (30 μg), nitrofurantoin (100 μg), quinupristin–dalfopristin (15 μg), teicoplanin (30 μg), tetracycline (15 μg), tigecycline (30 μg), trimethoprim (5 μg), trimethoprim-sulfamethoxazole (25 μg), and vancomycin (30 μg), according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2022) guidelines or Clinical and Laboratory Standards Institute (CLSI, 2021) guidelines, whenever EUCAST breakpoints were not available.
Vancomycin and teicoplanin minimal inhibitory concentrations (MIC) were determined by E-test according to manufacturer instructions (bioMérieux, Marcy-l’Étoile, France). Linezolid MIC was assessed by E-test in isolates phenotypically resistant by the disk diffusion method only. Detection of linezolid resistance genes was also performed in a multiplex PCR including primers for cfr, optrA, and poxtA genes (Bender et al., 2019).
Additionally, high-level aminoglycoside resistance (HLAR) was detected by agar dilution method, according to the CLSI guidelines, on BHI agar plates supplemented with gentamicin (500 μg/mL) or streptomycin (2000 μg/mL).
A total of 23 E. faecium representative isolates were selected for whole genome sequencing (WGS) based on their phenotypic antimicrobial resistance profiles, in order to include the highest variability of resistant patterns concerning the different sources of the isolates in the study (Supplementary Table S1). Total DNA was extracted by the DNeasy Blood and Tissue Kit according to the manufacturer’s protocol (Qiagen, Hilden, Germany). Genomic libraries were created using the Nextera XT DNA Sample Preparation Kit (Illumina, Little Chesterford, United Kingdom) followed by a 150-bp paired-end sequencing with an estimated coverage of 100X. Raw data were assembled de novo using the INNUca v3.1 pipeline.1 MLST 22 was used to predict in situ multilocus sequence type (MLST). Novel identified gene allele sequences or MLST profiles were submitted to the PubMLST website3 for new allele or sequence types (ST) assignments, respectively.
Genomic data analysis of antimicrobial resistance, virulence, and plasmids was performed in a batch mode using ABRicate4 and through the available resources at the Center for Genomic Epidemiology.5 To identify antimicrobial resistance genes or mutations responsible for resistant phenotypes, we used ResFinder (Zankari et al., 2012) and Linezolid Resistant Enterococci Finder (LRE-finder) 1.0,6 the Comprehensive Antibiotic Resistance Database (CARD) platform (Jia et al., 2017), and the National Center for Biotechnology Information (NCBI AMRFinderPlus).7 Virulence determinants were identified through VirulenceFinder 2.08 using the Virulence Factor Database (VFDB) for Enterococcus and the E. faecium and Enterococcus lactis database recently published (Roer et al., 2024). Detection of bacteriocin determinants was performed through the MyDbFinder 2.0 tool using the enterococcus bacteriocin database described by Tedim et al. (2023). The assessment of rep genes for identification of families of plasmids was performed using PlasmidFinder.9 All screenings were carried out with default parameters for a minimum identity of 90% and minimum coverage of 60%, except for PlasmidFinder which had a defined default ID threshold of 95%.
Clonal population structure and prevalent clonal lineages were determined based on MLST and on the core genome MLST (cgMLST). cgMLST allele calling was performed using the chewBBACA v2.5 pipeline.10 The visualization of relationships between isolates was performed on a minimum spanning tree constructed on PHYLOViZ 2.0.11 Isolates with ≤20 cgMLST allele differences were considered clonally related and belonging to the same clonal type (CT) (de Been et al., 2015). Additionally, a distance matrix-based phylogeny was constructed on FastME 2.0 Web server12 and the tree was further visualized on Microreact.13 A collection of 132 genomes of E. faecium infection isolates recovered in Portuguese hospitals between 2019 and 2021 (Conceição et al., 2022) available at the Laboratory of Molecular Genetics (ITQB-NOVA) and BioProject PRJNA1146999 were included for additional analysis and comparison to the livestock and environmental E. faecium isolates identified in this study.
Nominal categorical variables such as species prevalence, resistance patterns, and sample collections were compared using Fisher’s exact test, with p-values <0.05 being considered statistically significant. All statistical analyses were performed using GraphPad Prism software version 9.4 (GraphPad Software, La Jolla California, United States).
An oral informed consent was obtained at the time of screening of the passengers’ hands for each human participant. Moreover, samples from livestock and environmental sources included in this study were previously recovered in the scope of MRSA surveillance studies in Portugal, which were approved by ethical committees and published. Ethical approval is detailed in several published papers (Conceição et al., 2013, 2017a,b; Lopes et al., 2019). Regarding infection isolates from Portuguese hospitals included in the clonal population analysis, the isolates were recovered as part of the normal clinical routine of each hospital, without impacting the patient’s clinical status. The protocol was approved by the ethics committee of the participating institutions under the approval reference number CA1622.19-3. All ethics statements presented comply with publisher ethics guidelines and policies.
A total of 295 samples [135 samples from livestock (pigs, n = 80; cows n = 55) and 160 samples from the environment (public buses, n = 90; passenger hands, n = 18; urban environment, n = 52)] were screened for the presence of enterococci. The overwhelming majority (90.5%, n = 267) of the samples contained enterococci, but none of them showed vancomycin resistance. Enterococci were more prevalent among livestock samples (100%) compared to the environment (82.5%) (p < 0.0001), namely due to the lower prevalence detected among bus passengers’ hands and urban environment (61% prevalence each). Considering the different species, E. faecalis was the most prevalent species detected in half of the enterococcus-positive samples (51.7%), followed by E. casseliflavus (35.3%), E. gallinarum (24%), E. faecium (21.7%), and E. hirae (9.4%); E. durans was detected in two samples only. While E. faecalis seems to be more associated with livestock samples (62.2% versus 40.9%, p = 0.0006), being equally detected in pigs (66.3%) and cows (56.4%), E. faecium was most prevalent in environment associated samples (31.8% versus 11.9%, p < 0.0001) (Figure 1). Moreover, E. faecalis and E. faecium were the unique species detected in all collections, although at different frequencies (Table 1). E. casseliflavus and E. gallinarum were not identified among the hands of bus passengers, whereas the two unique E. durans isolates were recovered from a bus and a passenger’s hands. E. raffinosus was not identified in the study, and other enterococci that could not be classified at the species level (n = 84) due to the limited range of species-specific primers available for PCR assays were detected in all collections (Table 1). Buses and the urban environment showed the highest diversity of enterococci, with six different species identified in each collection. Pigs were mostly colonized by E. faecalis (66.3%) followed by E. gallinarum (43.8%) while the overwhelming majority of cows carried E. casseliflavus (70.9%) and E. faecalis (56.4%). E. faecium was prevalent in the urban environment (37.5%), while the remaining environmental samples were mostly contaminated by E. faecalis (Table 1).
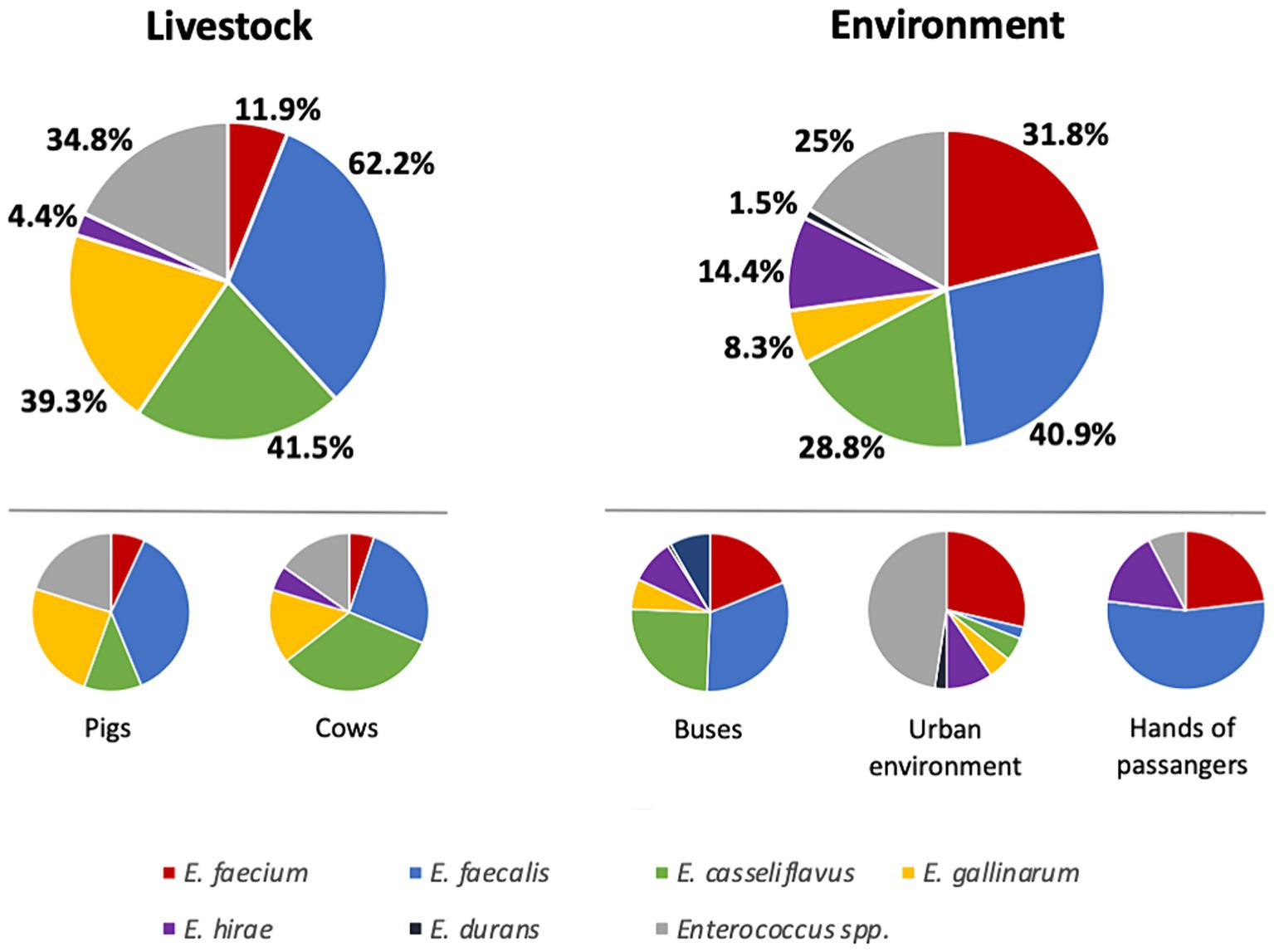
Figure 1. Distribution of enterococcus species by sample collection individually and regarding livestock and environment. Percentages referred to the total of positive samples in each collection.
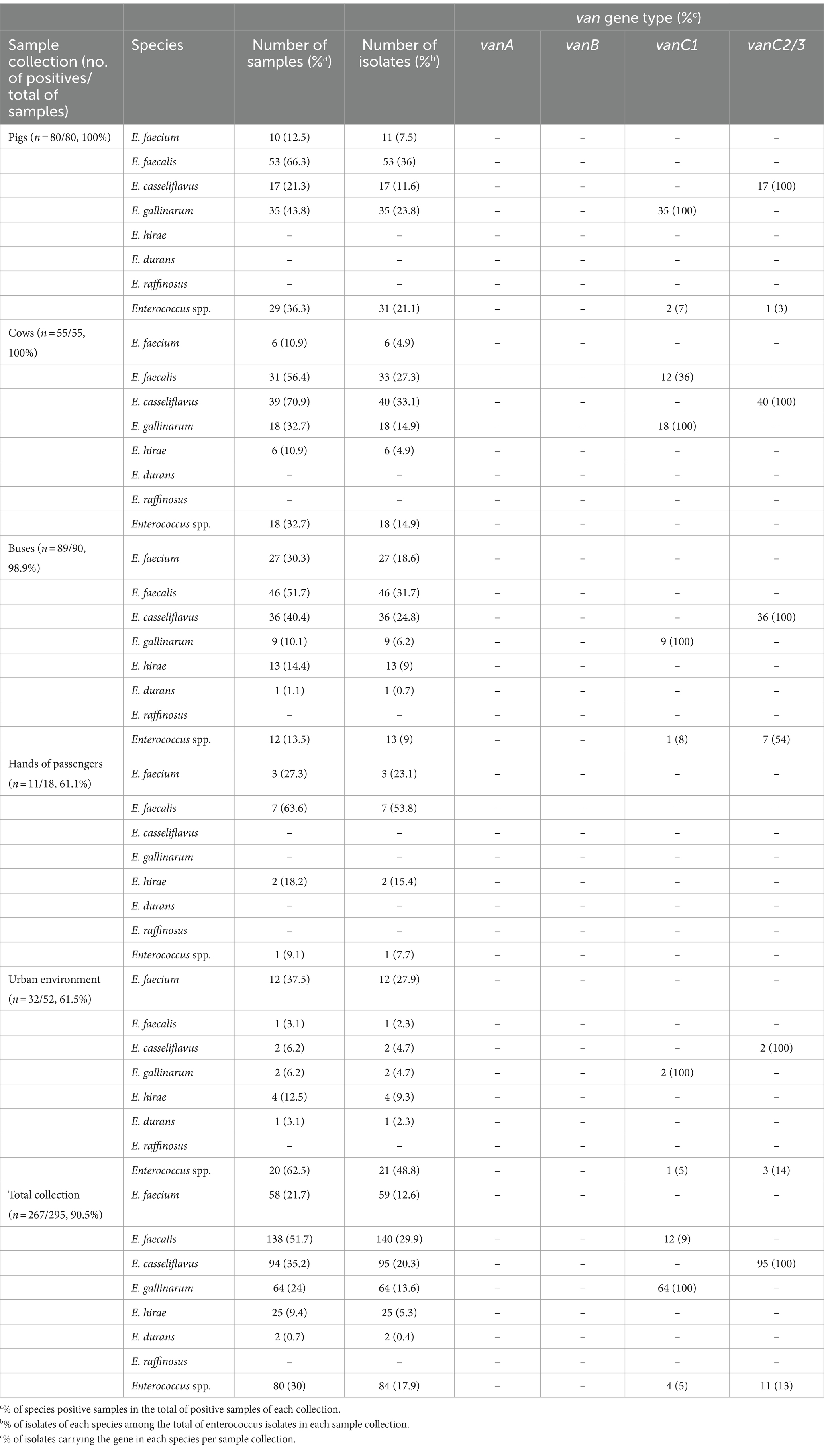
Table 1. Prevalence of the different Enterococcus species and distribution of van genes in each sample collection screened.
Considering vancomycin-resistant determinants, neither vanA nor vanB genes were detected. However, vanC genes were detected in 40% (186 out of 468) of the isolates, namely vanC1 detected in 80 (17%) isolates and vanC2/3 detected in 106 (23%) (Table 1). It is known that vanC genes could be intrinsically present in specific enterococcus species, which could be confirmed in our results by the detection of vanC1 genes in all E. gallinarum isolates while vanC2/3 was exclusively present in all E. casseliflavus. Moreover, vanC1 was additionally detected in 36% of the E. faecalis isolated from cows (Table 1), but none of them was identified in samples co-contaminated with vanC1 E. gallinarum.
A total of 59 E. faecium isolates (Table 1), recovered from pigs (n = 11), cows (n = 6), buses (n = 27), hands of bus passengers (n = 3), and urban environment (n = 12), were more thoroughly characterized.
The overwhelming majority (78%) of E. faecium isolates showed phenotypic resistance to ≥3 antibiotic classes and were considered multidrug-resistant (MDR) (Magiorakos et al., 2012). High resistance rates were related to nitrofurans (nitrofurantoin, 97%), sulfonamides (trimethoprim–sulfamethoxazole, 86%; trimethoprim, 81%), tetracyclines (tetracycline, 36%; tigecycline, 32%), macrolides (erythromycin, 34%), penicillin (ampicillin, 27%), and high-level resistance to aminoglycosides (namely streptomycin, 22%) (Figure 2). None of the isolates showed phenotypic resistance to glycopeptides, namely vancomycin (MICs 0.38–3 μg/mL) and teicoplanin (MICs 0.023–1 μg/mL), nor to linezolid (MIC 1.5–3 μg/mL).
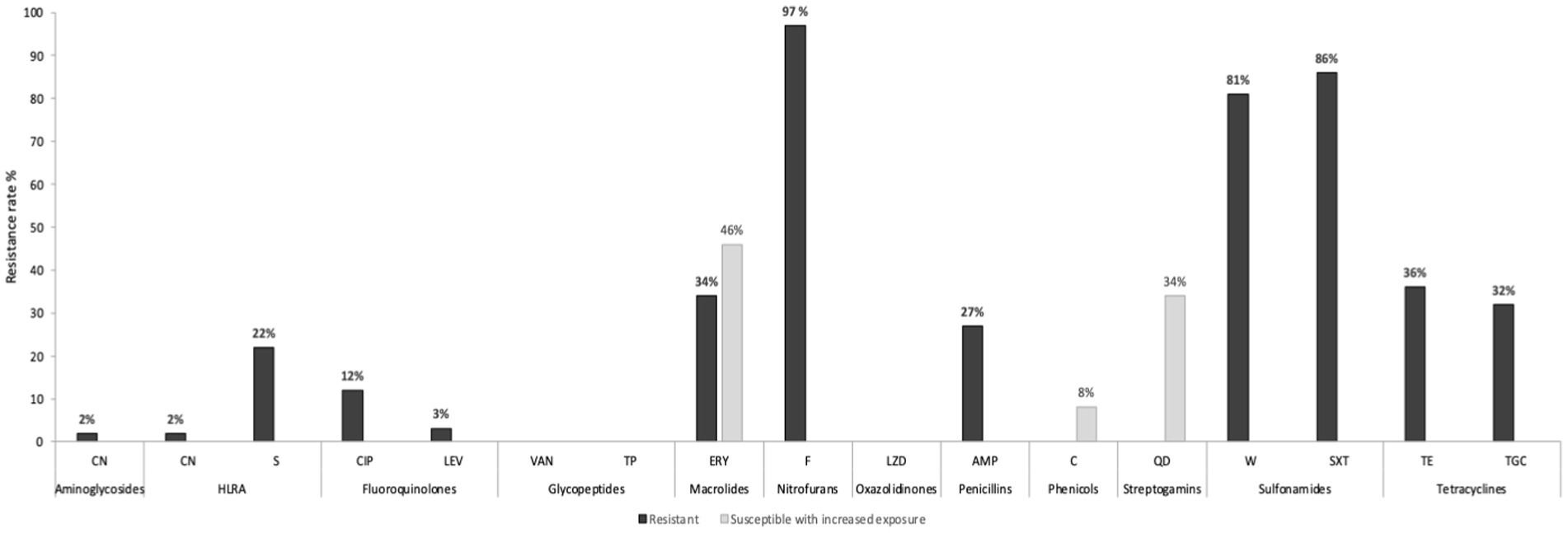
Figure 2. Phenotypic antibiotic resistance profiles of the 59 E. faecium isolates. CN, gentamicin; S, streptomycin; HLAR, high-level aminoglycosides resistance; CIP, ciprofloxacin; LEV, levofloxacin; VAN, vancomycin; TC, teicoplanin; ERY, erythromycin; F, nitrofurantoin; LNZ, linezolid; AMP, ampicillin; C, chloramphenicol; QD, quinupristin–dalfopristin; W, trimethoprim; SXT, trimethoprim–sulfamethoxazole; TE, tetracycline; TGC, tigecycline.
MDR phenotype was highly detected in livestock isolates (94%) compared to environment samples (71%) although this difference was not statistically significant (p = 0.0838). On the other hand, tetracycline and ampicillin resistances were significantly associated with livestock. Tetracycline was detected in 64.7% of livestock isolates versus 23.8% in the environment (p = 0.0059), while ampicillin was identified in 52.9% of animal isolates (pigs n = 7; cows n = 2) compared to 17% of the environmental E. faecium (p = 0.0085). Globally, high-level resistance (HLR) to gentamycin was detected in 2% of E. faecium, while 22% showed HLR to streptomycin, mainly associated with livestock isolates (90%) (p = 0.0002).
Genomic characterization of 23 selected E. faecium isolates allowed the identification of resistance genes associated with MDR phenotype in 87% (20/23) of the isolates (Figure 3). A single isolate, recovered from the hands of a bus passenger, contained resistance genes associated with five antimicrobial classes (aminoglycosides, amphenicols, β-lactam, macrolides, and tetracyclines).
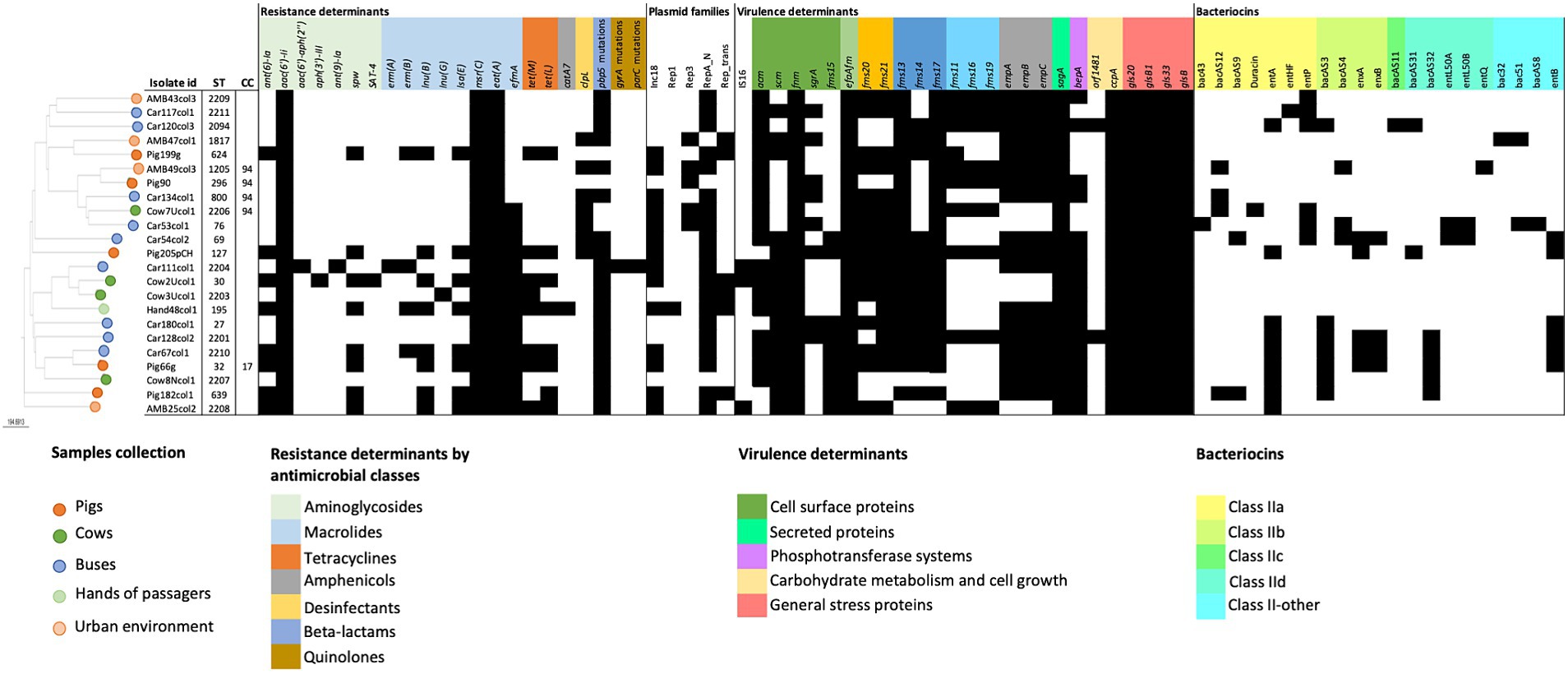
Figure 3. Genomic data analysis regarding cgMLST distance-based matrix phylogeny, antimicrobial resistance, virulence, and plasmids of the 23 E. faecium selected for WGS. Antimicrobial resistance genes and mutations (vanA, vanB, cfr, optrA, or poxtA and 23S rRNA mutations) or virulence determinants (esp., ptsD, hylfm, cylA, or gelE) not detected in any isolate were not included. Dark full squares indicate the presence of the gene or mutation; ST, sequence type; CC, clonal complex.
None of the E. faecium isolates contained vancomycin-resistant genes, supporting the previous PCR and phenotypic antimicrobial susceptibility results. Moreover, linezolid phenotypic susceptibility was confirmed by the non-detection of resistance genes (cfr, optrA, and poxtA) nor 23S rRNA mutations.
Low-level resistance to aminoglycosides could be attributed to the ubiquitous presence of the aminoglycoside 6′ acetyltransferase [AAC(6′)-Ii] in this species, which was confirmed by the detection of aac(6′)-Ii genetic determinant in all E. faecium isolates in this study. On the other hand, the genomic analysis showed that the phenotypically detected high-level aminoglycoside resistance was mostly associated with the presence of the ant(6)-la gene in the case of streptomycin and to aac(6′)-aph(2″) gene in the single high-level gentamicin-resistant isolate (Figure 3). Other aminoglycoside resistance-associated genes were also detected, but at a lower frequency as spw (35%), aph(3′)-IIIa, sat-4, and ant(9)-la with 4% each.
Regarding resistance determinants of macrolides and lincosamides, in addition to msr(C) that was intrinsically present in all E. faecium isolates, other genes, such as efmA, lnu(B), lnu(G), lsa(E), erm(B), and erm(A), were also detected in the sampled collections, although at different percentages (Figure 3).
Resistance to β-lactams, namely ampicillin, was mainly conferred by mutations in the pbp5 gene that were detected in 83% of the isolates (n = 19) (Figure 3), varying from 2 to 18 mutations per isolate. Seven isolates (from pigs, cows, urban environments, and buses) contained the clpL gene, commonly associated with decreased penicillin susceptibility and to thermoresistant phenotypes. In addition, 39% (8/23) of E. faecium harbored tetracycline resistance genes tet(M) and/or tet(L), including two isolates phenotypically resistant also to tigecycline. The genes tet(M) and tet(L) are commonly associated with tetracycline-resistant enterococcus from animals, which is in accordance with the high tetracycline resistance prevalence of 73% among pigs and 50% among cows observed in this study.
E. faecium isolated from buses showed a higher diversity of resistance genes (n = 15), while the single sequenced E. faecium from the hands of a passenger combined the higher number of resistance genes (n = 12), including the catA7 gene associated with phenotypic resistance to chloramphenicol.
A high prevalence of putative virulence markers was detected among E. faecium isolates from all collections studied, including 18 out of the 32 tested determinants identified in >70% of the isolates (Figure 3). Almost half of the isolates (43%) contained ≥20 virulence markers, mostly environmental isolates, 57% versus 33% of livestock, although the difference was not significant (p = 0.1068). Cell surface proteins associated with bacterial adherence including adhesins genes efaAfm, fnm (100% each), acm (83%), and pili-associated proteins fms13 and fms17 (91% each) were identified in more than 80% of the isolates. All isolates (100%) contained determinants involved in biofilm production and cell growth such as sagA, a peptidoglycan secreted hydrolase, and ccpA, a catabolite control protein A, as well as gls 20-glsB and gls 33-glsB1 general stress response regulators influencing colonization and adaptation to host response.
On the other hand, virulence determinants with proven impact in biofilm and endocarditis infection such as empA/empB/empC cluster (78%) or bepA (74%) were widely detected.
Three isolates, from the urban environment, a bus, and a cow sample, contained the insertion sequence 16 (IS16), a marker associated with hospital strains that contribute to E. faecium genomic plasticity and host adaptation. Other virulence determinants associated with clinical isolates or antibiotic-resistant strains as esp., ptsD, hylfm, cylA, or gelE were not detected in our collections (Figure 3).
Genes associated with bacteriocins were detected in 78% of the E. faecium isolates. Environmental isolates showed a high variability of bacteriocin genes (n = 20), and a high prevalence of bacteriocin E. faecium producers (n = 12.86%) compared to livestock isolates (n = 11 different bacteriocin genes and 67% (n = 6) bacteriocin producers). Differences were observed regarding the variability of bacteriocin gene content among the different collections, with bus isolates containing 18 different bacteriocin genes compared to urban environments (n = 6), pigs (n = 9), and cows (n = 7) (Figure 3). Moreover, bus isolates showed also the highest bacteriocin content per isolate, with Car54 and Car53 harboring, respectively, 8 and 7 bacteriocin genes. None of the tested bacteriocins could be detected among the isolate of hands. Globally, the most prevalent toxins detected in ≥25% of the isolates included enterocin A (entA, 39%), enterocin B (entB, 26%), enterocin P (entP, 26%), and bacAS3 (30%). Among these bacteriocins, entA was the unique gene detected in all studied collections, although at different frequencies. The gene entB was present in bus and isolates of the pig while entP was found in all except isolates of pigs. The gene bacAS3 was not detected among urban environmental isolates.
Globally, the in silico analysis identified a high diversity of plasmids among the E. faecium genomes. Plasmids from RepA_N and Inc18 rep families were the most prevalent, detected in 86.9 and 60.8% of the isolates, respectively, and were present in all five sample collections (Figure 3). No major associations were detected between rep genes and specific resistance or virulence determinants in our E. faecium isolates, except for the Rep_1 plasmid found in the unique isolate from the hands of a passenger. Rep_1 was probably associated with chloramphenicol resistance detected in this isolate, given the co-localization of rep and the catA7 gene.
Clonal population analysis showed a highly diverse E. faecium population structure. Each isolate was characterized by a different sequence type (ST), including eight new STs (ST2203, ST2204, ST2206, ST2207, ST2208, ST2209, ST2210, and ST2211) (Figure 3). Five STs could be assigned to two major E. faecium clonal complexes (CC): ST32 which belonged to CC17 and was identified in a single isolate from a pig; and ST296, ST2206, ST1205, and ST800, belonging to CC94 and recovered from different sources (one from pigs, one from cows, one from the urban environment, and other from buses). The remaining 18 isolates were considered as single STs. A more detailed cgMLST analysis confirmed the high variability in the population with all cgMLST profiles showing more than 20 allele differences between them and therefore were assigned to different clonal types (CT) (de Been et al., 2015).
Clonal population analysis, including genomic data of E. faecium isolated from infection in Portuguese hospitals, showed that none of the E. faecium isolates from this study and clinical isolates shared the same ST (Figure 4; Supplementary Tables S1, S2). However, a few related STs could be identified between both collections, namely related to CC94: ST800 identified in a bus isolate and ST1205 isolated in the environment were, respectively, single-locus and triple-locus variants of ST94; ST296 from a pig is a double-locus variant of the clinical ST2206. Two livestock isolates characterized by ST32 and ST2203, belonged to CC17 and were highly related to ST17 and ST494, the two most prevalent STs among clinical E. faecium in Portugal between 2019 and 2020. Moreover, isolate Car180col1 shared six out of the seven MLST alleles with a clinical ST2020 isolate.
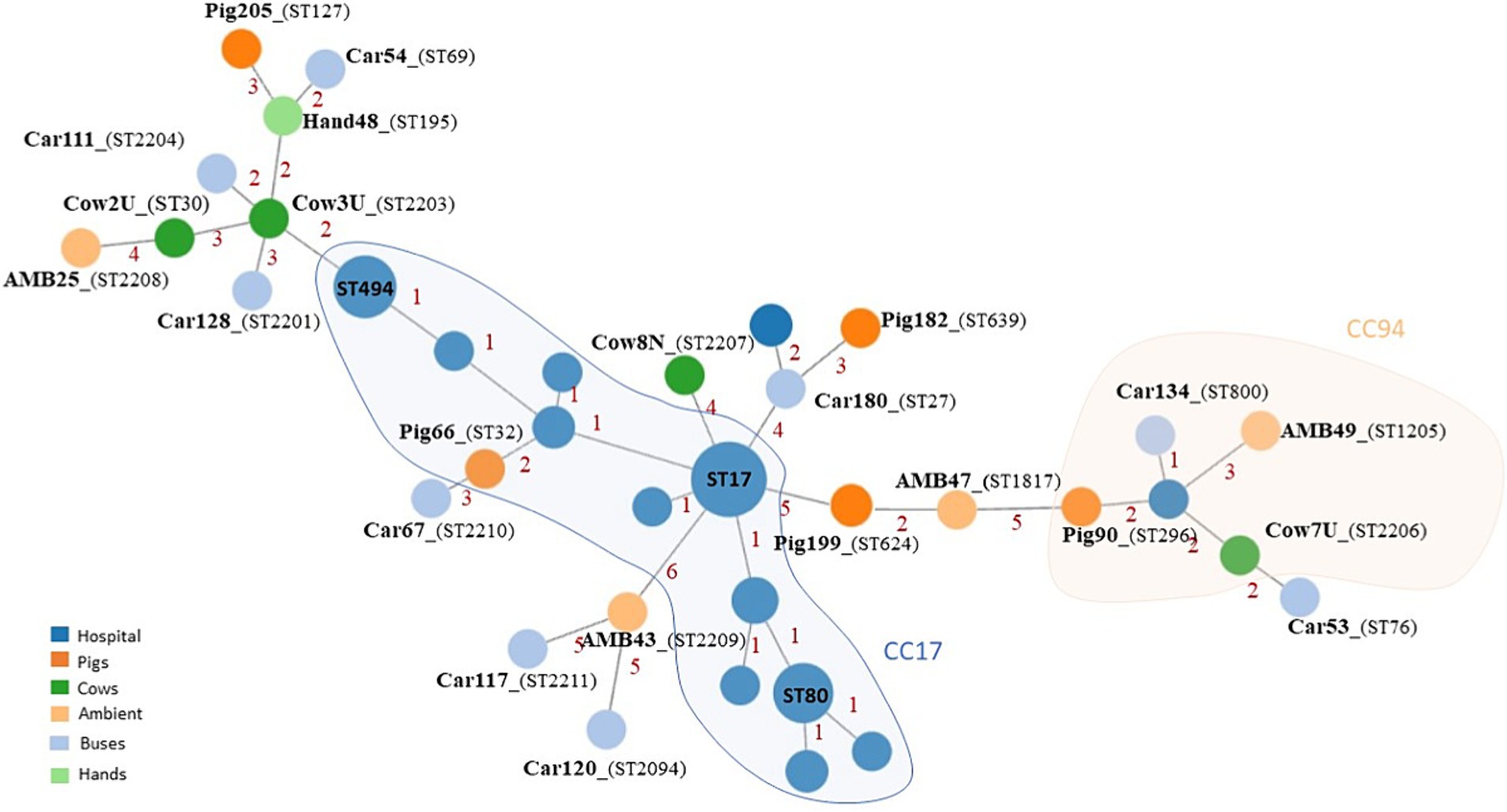
Figure 4. Minimum spanning tree based on MLST analysis of 23 E. faecium isolates from livestock and environment and E. faecium isolated from infections in Portuguese hospitals between 2019 and 2020. AMB, Urban environment; representative isolates of E. faecium derived from this study are labeled in boldface.
On the other hand, based on the 1,159 loci common to all E. faecium isolates, cgMLST analysis confirmed a high population diversity between isolates identified in this study and the hospital-associated population. None of the isolates from both collections belonged to common CTs, differing between 377 and 1,082 allelic differences. Therefore, no direct epidemiological link between E. faecium derived from this study and the E. faecium recovered from infection in Portuguese hospitals could be drawn, when considering a more discriminatory method as cgMLST.
This study aimed at the identification of enterococci reservoirs outside hospitals, namely healthy livestock or environmental surfaces, providing valuable knowledge on the dissemination and resistance mechanisms of this major human pathogen. To the best of our knowledge, our study represents the first surveillance of non-clinical settings as the hands of passengers and outdoor urban surfaces near hospitals as possible reservoirs for VRE. Still, this is the first study in Portugal to explore livestock nares and udder regions as potential VRE colonization reservoirs.
In this study, E. faecalis and E. faecium were the unique species identified in all collections, although at different percentages depending on the sample source. Our results are in line with a recent One Health continuum surveillance of Enterococcus spp. that reveals E. faecium and E. faecalis as the predominant species related to human-associated environments (Zaheer et al., 2020). In fact, buses and hands of passengers were highly contaminated with E. faecalis and E. faecium, confirming that community settings could be reservoirs of major human pathogens, as previously shown for methicillin-resistant Staphylococcus aureus (Conceição et al., 2013). A similar study in Turkey supported also the role of public transports as reservoirs of Enterococcus spp.; however, isolates were not identified at the species level (Birteksöz Tan and Erdogdu, 2017). Moreover, we identified clinically relevant Enterococcus spp. in other urban environmental public surfaces highly touched by the hands of individuals, as previously identified in shared urban bicycles in China (Gu et al., 2020). As we screened urban environmental surfaces close to hospitals, and it is known that individuals with direct contact with the nosocomial environment have a high probability of having hands contaminated by VRE and transmitting these pathogens to the hospitals’ outside environment (Hayden et al., 2008), our study highlights the importance of hands’ hygiene measures to prevent human pathogens dissemination.
In our livestock samples, E. faecalis was the most prevalent species detected, as recently reported in pigs and to a lesser extent in cows cecal swabs collected at slaughterhouses in Portugal (Gião et al., 2022). Moreover, contrary to other studies that showed E. hirae as a prevalent species colonizing the gut of cattle and further detected in the food production chain (Zaheer et al., 2020; Gião et al., 2022), in our collection E. hirae was randomly detected comparing to the predominance of E. casseliflavus. We swabbed the nares and the udder region of cattle animals, which could justify this difference among the enterococcus species identified.
Pigs were highly nasally colonized by Enterococcus spp., namely E. faecalis has recently been reported in four pig farms in Spain (Abdullahi et al., 2023), which suggest that although the nares are not the common habitat for Enterococcus species, nasal sampling might be valuable for enterococci monitoring in animals.
Vancomycin resistance was not detected in our samples. Although isolates were able to initially grow in media supplemented with 6 μg/mL of vancomycin, none of them grew in VRE-selective media. The presence of other bacteria in our initial multibacterial samples might produce factors that protect enterococci from vancomycin or alter the local environment in a way that reduces the effective concentration of the antibiotic to concentrations that allow the growth of susceptible isolates, or the low-level resistance to vancomycin phenotype intrinsic of some enterococcus species could support this observation. VRE has already been identified in animals in Portugal (Freitas et al., 2009; Ramos et al., 2012; Braga et al., 2013; Novais et al., 2013), but there are no data regarding VRE in non-clinical environmental settings. Our study contributes to fulfilling this gap, identifying multidrug-resistant E. faecium isolates in buses, passengers, and urban environments, in addition to vancomycin resistance.
The vanC genes are considered to be intrinsic to E. gallinarum (vanC1), E. casseliflavus (vanC2), and E. flavescens (vanC3) (Ahmed and Baptiste, 2018; Torres et al., 2018). However, vanC1 was detected in 36% of our E. faecalis isolates from cows, which is in line with the increasing reports of vanC1 in E. faecalis from animal-associated environment samples (Schwaiger et al., 2012; de Garnica et al., 2013; Nishiyama et al., 2015). On the other hand, none of the E. faecium isolated in our study harbor van genes, although vanC1 was already reported in E. faecium from human infection (Sun et al., 2014), evidencing that vanC1 could be spread to other clinically relevant Enterococcus species. Although a possible bias toward the selection of intrinsic vancomycin-resistant enterococcus could be suggested by the usage of 6 μg/mL of vancomycin in the first screening of samples, no other van genes or vancomycin non-susceptibility could be detected in the majority of the Enterococcus isolates in addition to naturally intrinsic resistant E. casseliflavus and E. gallinarum and 9% of the total E. faecalis isolates.
Given the clinical relevance of E. faecium in human infection, namely VRE that is considered a high-priority pathogen by the WHO (2024), a comprehensive characterization of enterococci isolated in the present study was focused on E. faecium isolates.
The rationale to further study the E. faecium isolates was supported by a significant increase of E. faecium in nosocomial infections worldwide, namely in Europe, associated not only with vancomycin resistance but also with other antibiotics used in human clinical practice and in veterinary medicine as ampicillin, aminoglycosides (high-level resistance), or last-resort antibiotics like linezolid and tigecycline. In the most recent Antimicrobial Surveillance Report 2022, the ECDC reported a significantly increasing trend of antimicrobial-resistant E. faecium in some individual countries in Europe and highlighted the urgent need for close monitoring to better understand the epidemiology, clonal diversity, and risk factors associated with vancomycin-resistant E. faecium infections (ECDC, 2023). In Portugal, the country where the study was carried out, VRE has been increasingly detected in hospitals compared to a decreasing trend observed in E. faecalis resistant to aminoglycosides since 2018, which heightened concern among hospitals about these resistant E. faecium infections and the need for monitoring reservoirs and the development of effective infection control guidelines (ECDC, 2023). Additionally, E. faecium reservoirs have been identified in a One Health continuum (Zaheer et al., 2020), suggesting that this relevant species for human health, may be prevalent in non-clinical environments and could potentially be transmitted to humans.
We identified high rates of antimicrobial resistance among the isolates independently of the source. Resistance of ampicillin and tetracycline was significantly associated with livestock samples, which could be a consequence of the usage of a β-lactam (amoxicillin) as prophylaxis in the feed regimen of pigs at the time of sampling, or with the global and widespread use of tetracycline for prophylactic purposes in livestock (Roberts and Schwarz, 2016). The gene tet(M) is considered the most frequent mechanism of tetracycline resistance in enterococci from humans, animals, food, and the environment (Cho et al., 2020), even though in this study tet(M) and tet(L) were equally detected. In addition, 32% of E. faecium from non-clinical settings presented phenotypic resistance to tigecycline, but only two isolates harbored tet(L) and tet(M), suggesting that other resistance mechanisms could be involved that deserve further investigation.
The ampicillin resistance rate of 56% among livestock in our study surpasses the 30% resistance reported in poultry E. faecium in Portugal (Poeta et al., 2006), but parallels the increasing rates of vancomycin-susceptible ampicillin-resistant E. faecium causing infections in European hospitals (Garcia-Solache and Rice, 2019). On the other hand, the significant use of aminoglycosides in veterinary medicine reflects the rates of high-level aminoglycosides resistance in E. faecium from livestock and human isolates (McGlinchey et al., 2008; Li et al., 2015; Diab et al., 2019). We detected approximately 24% HLAR in E. faecium isolates from non-clinical settings, associated with the presence of the ant(6)-la gene, as previously reported in animal E. faecium isolates (Han et al., 2011; El-Mahdy et al., 2018; Cagnoli et al., 2022) and suggesting a livestock reservoir for this acetyltransferase gene. In 2022, 34.9% of E. faecium isolated from invasive infections in Europe showed aminopenicillins and gentamicin high-level resistance (ECDC, 2023). We identified non-clinical environments and healthy livestock as reservoirs of ampicillin and high-level aminoglycosides-resistant E. faecium in Portugal. This finding is of major concern as the spread of these resistant isolates compromises the clinical utility of the commonly used synergistic protocol for aminoglycosides with a cell wall-active antibiotic such as ampicillin in the treatment of severe enterococcal infections (Miller et al., 2014; Echeverria-Esnal et al., 2023).
Moreover, in our study, almost all isolates presented phenotypic resistance to nitrofurantoin, a synthetic antibiotic used as the first-line therapy of uncomplicated lower urinary tract infection caused by HLAR Enterococcus and VRE (Mahdizade Ari et al., 2023). High nitrofurantoin resistance rates (56–84%) have been reported in clinical E. faecium isolates (Wang et al., 2013), but the identification of such a high prevalence of resistance in non-clinical isolates in our study is of major concern.
In our collections, E. faecium harbored an arsenal of virulence determinants associated with bacterial adherence and biofilm production such as efaAfm, fnm, acm, and pili-associated proteins, which provide them an advantage on environmental persistence. Moreover, adhesion is usually the first step required for infection, suggesting that these environmental surfaces and healthy livestock represent reservoirs of potentially high pathogenic E. faecium isolates. This assumption is even supported by the high prevalence of virulence determinants with proven impact in biofilm and endocarditis infection as empA/empB/empC cluster, bepA or efaAfm, widespread in E. faecium isolates, regardless of the isolation source. Putative virulence markers for E. faecium hospital-associated strains, such as insertion sequence 16 (IS16) or acm and scm adhesins (Werner et al., 2011), were also widely detected supporting the pathogenic potential of these environmental and livestock-associated E. faecium and showed a different scenario previously reported across a One Health continuum in Canada where livestock-associated Enterococcus spp. lacked virulence genes associated with clinical isolates (Zaheer et al., 2020). In addition, three genes (sgrA, fms15, and orf1481) exclusively associated with E. faecium hospital variants (Roer et al., 2024) were detected among E. faecium in this study, namely in bus isolates, suggesting a possible hospital-associated origin and dissemination to the community.
Bacteriocin production ensures niche competition of enterococci in the gut, namely against particular pathogens or adaptation to different environmental conditions (Tedim et al., 2023). In fact, in our collection, environmental isolates harbored a higher bacteriocin genetic determinants content, belonging exclusively to the heat-stable, non-lantibiotic class II bacteriocins. entA, a pediocin-like bacteriocin (class IIa) with strong antilisterial effects, and bacAS3 (subclass Iib) were the most prevalent bacteriocin determinants identified in our study (39 and 30%, respectively) as previously reported in a large and diverse collection of E. faecium isolates (Tedim et al., 2023). Enterocin gene entA was already identified not only in food-associated E. faecium but also in clinical outbreaks of VRE- and vancomycin-susceptible E. faecium in different European hospitals (Freitas et al., 2016). Moreover, 22% of our isolates, mainly isolates from the bus, contained >5 different bac genes, which is much higher than the previously bac mean content (1 to 5) among E. faecium (Tedim et al., 2023), suggesting a high adaptive potential of our environmental isolates.
Population analysis of the 23 E. faecium representative isolates showed a highly polyclonal bacterial collection, regardless of the isolation source. This variability in clonal population structure was not surprising given the non-clinical nature of these E. faecium isolates. Moreover, the identification of new STs, namely in cows (ST2203, ST2206, and ST2207), buses (ST2204, ST2210, and ST2211), and urban environmental surfaces (ST2208 and ST2209) suggests the acquisition of genetic variability to improve adaptation and maintenance in adverse environmental conditions or hosts.
Although cgMLST analysis could not trace a direct transmission link between E. faecium from invasive infections in Portuguese hospitals and our non-clinical E. faecium isolates, the genetic background of several isolates from pigs (ST32-CC17; ST624; ST127; ST296), cows (ST30; ST2206-CC94), buses (ST27, ST2201; ST800-CC94), and urban environment (ST1205-CC94) were already identified in human clinical infections and carriage (see text footnote 3) (Werner et al., 2012; Bender et al., 2018b; Zhou et al., 2019; Peng et al., 2022). In line with our observation, recent studies suggested that instead of an evident gene flow from livestock E. faecium to hospital E. faecium, it is supported by distinctive niche adaptations for the distribution of antimicrobial resistance genes, host adaptation, and virulence determinants among E. faecium population (Gouliouris et al., 2018; Arredondo-Alonso et al., 2020).
The identification in our study of E. faecium isolates belonging to genetic backgrounds prevalent in infection and associated with antimicrobial resistance to last-resort antibiotics is a relevant finding. Therefore, ST32 belonged to the CC17, the major hospital-associated clonal lineage responsible for human infections worldwide. While ST32 was detected in a pig nasal swab, it was already identified from carriage and infection in other livestock and humans as well as from environmental wastewaters (see text footnote 3) (Werner et al., 2012). Recent findings from Portugal reported linezolid-resistant ST32 E. faecium in a healthy human rectal swab (Freitas et al., 2020), highlighting its significant adaptive and pathogenic potential.
On the other hand, ST30 identified in our study in a cow sample was recently reported in German hospitals containing the optrA linezolid resistance gene (Bender et al., 2018b). In Portugal, ST30 E. faecium isolates were reported in a pig, dust, and food of pigs in the same farm (Novais et al., 2013). Our detection of ST30 in a cow sample underscores its capacity for host diversification.
CC94 E. faecium lineage is mostly associated with human community and commensal isolates that can cause sporadic infections in animals and humans (Peng et al., 2022) In our study, single isolates from livestock (a pigs ST296 isolate and a cow ST2206), a bus (ST800), and urban environment (ST1205) exhibited loci variations of the ST94, indicating their affiliation with the widespread CC94 lineage.
E. faecium isolated in this study and the E. faecium recovered from infection in Portuguese hospitals differed in the isolation period, which could be a limitation in the genomic comparison analysis. In fact, a more global approach as MLST identified isolates in this study sharing the same genetic background as infection isolates from hospitals, but the high discriminatory power of cgMLST could not identify a direct link between these isolates, which could be due to evolutionary pressure that allows the accumulation of small genetic events along time, promoting adaptation to selective pressure of each environment. An example to support this assumption was the identification in this study of an E. faecium bus isolate sampled in 2012 and characterized by ST27 that belonged to the same genetic background (sharing five MLST alleles) of an ST2020 isolate causing infection in hospitals in the same geographic area approximately 8 years later.
Another possible limitation of the study was the inclusion of livestock nasal swabs, which is not the primary niche of Enterococcus colonization in humans and animals. However, it has been shown that Enterococcus can be found in colonization of the anterior nares and co-habit with other pathogens such as S. aureus. Therefore, the previous identification of these samples as reservoirs of S. aureus and MRSA (Conceição et al., 2013, 2017a,b; Lopes et al., 2019) as well as the close living conditions of the animals (namely pigs, that live approximately 32 to 33 animals in 8–12 m2 stockyards) made the samples to be considered potential reservoirs for multidrug-resistant VRE and therefore selected for this study.
Major achievements of this study included the unveiling of a global high prevalence of Enterococcus spp. in non-clinical settings such as livestock- and human-associated environments, including clinically relevant species such as E. faecium and E. faecalis. Although no vancomycin resistance could be detected, livestock and environmental surfaces showed to be reservoirs of E. faecium isolates with antimicrobial resistance to clinically relevant antibiotics, namely aminoglycosides (high-level resistance), ampicillin and tetracyclines including tigecycline. Moreover, these E. faecium isolates harbor a wide arsenal of virulence determinants associated with increased adaptation to hostile environments and different hosts, evasion of the immune system, and capacity of infection development, associated with genetic backgrounds already identified in infection isolates. Our results evidenced that surveillance of Enterococcus spp., with a special focus on E. faecium outside nosocomial settings is of paramount importance for effective infection control strategies and reduction of antimicrobial resistance spread, given its proven role as reservoirs of multidrug resistance and virulent strains.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The studies involving humans were approved by the Ethics Committee of ESSCVP as part of prior research study for bus passengers hands screening, as detailed in the manuscript. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin because a verbal informed consent was obtained for each participant (all aged over 18) at the time of hand screenings, which was registered in the respective record sheet at time of sampling as part of prior research study, as detailed in the manuscript. Ethical approval was not required for the studies involving animals in accordance with the local legislation and institutional requirements because the animal samples used in this study were obtained as part of prior research studies, as detailed in the manuscript. These samples comprised exclusively of nasal or udder swabs. Collection of the samples was conducted by the veterinarians responsible for each farm, ensuring that no stress or injuries were inflicted to the livestock. The research was executed in direct collaboration with the veterinarians from each respective farm. Written informed consent was obtained from the owners for the participation of their animals in this study.
JL: Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. HL: Writing – review & editing, Validation, Supervision, Project administration, Funding acquisition, Conceptualization. TC: Writing – review & editing, Writing – original draft, Validation, Supervision, Project administration, Methodology, Investigation, Formal analysis, Conceptualization.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Samples included in this study were previously collected under the scope of published surveillance studies financially supported by: Projects PTDC/DTP-EPI/0842/2014 from FCT - Fundação para a Ciência e a Tecnologia, Portugal (PI: Marta Aires-de-Sousa); Project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER funds through COMPETE2020 - Programa Operacional Competitividade e Internacionalização (POCI) and ONEIDA project (LISBOA-01-0145-FEDER-016417) co-funded by FEEI - “Fundos Europeus Estruturais e de Investimento” from “Programa Operacional Regional Lisboa 2020” and by national funds from FCT (Coordinated by Raquel Sá-Leão and Mónica Serrano).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1466990/full#supplementary-material
1. ^https://github.com/B-UMMI/INNUca
2. ^https://github.com/tseemann/mlst
4. ^https://github.com/tseemann/abricate
5. ^http://www.genomicepidemiology.org
6. ^https://cge.food.dtu.dk/services/LRE-finder/
7. ^https://github.com/ncbi/amr/wiki/Running-AMRFinderPlus
8. ^https://cge.food.dtu.dk/services/VirulenceFinder/
9. ^https://cge.food.dtu.dk/services/PlasmidFinder/
10. ^https://github.com/B-UMMI/chewBBACA
11. ^https://online2.phyloviz.net/index
Abdullahi, I. N., Lozano, C., Juárez-Fernández, G., Höfle, U., Simón, C., Rueda, S., et al. (2023). Nasotracheal enterococcal carriage and resistomes: detection of optrA-, poxtA- and cfrD-carrying strains in migratory birds, livestock, pets, and in-contact humans in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 42, 569–581. doi: 10.1007/s10096-023-04579-9
Ahmed, M. O., and Baptiste, K. E. (2018). Vancomycin-resistant enterococci: a review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microb. Drug Resist. 24, 590–606. doi: 10.1089/mdr.2017.0147
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Arredondo-Alonso, S., Top, J., McNally, A., Puranen, S., Pesonen, M., Pensar, J., et al. (2020). Plasmids shaped the recent emergence of the major nosocomial pathogen Enterococcus faecium. MBio 11, 1–17. doi: 10.1128/mbio.03284-19
Bender, J. K., Cattoir, V., Hegstad, K., Sadowy, E., Coque, T. M., Westh, H., et al. (2018a). Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug Resist. Up 40, 25–39. doi: 10.1016/j.drup.2018.10.002
Bender, J. K., Fleige, C., Klare, I., and Werner, G. (2019). Development of a multiplex-PCR to simultaneously detect acquired linezolid resistance genes cfr, optrA and in enterococci of clinical origin. J. Microbiol. Methods 160, 101–103. doi: 10.1016/j.mimet.2019.03.025
Bender, J. K., Fleige, C., Lange, D., Klare, I., and Werner, G. (2018b). Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int. J. Antimicrob. Agents 52, 819–827. doi: 10.1016/j.ijantimicag.2018.09.009
Birteksöz Tan, A., and Erdogdu, G. (2017). Microbiological burden of public transport vehicles. Istanbul. J. Phar. 47, 52–56. doi: 10.5152/IstanbulJPharm.2017.008
Braga, T. M., Pomba, C., and Lopes, M. F. S. (2013). High-level vancomycin resistant Enterococcus faecium related to humans and pigs found in dust from pig breeding facilities. Vet. Microbiol. 161, 344–349. doi: 10.1016/j.vetmic.2012.07.034
Byappanahalli, M. N., Nevers, M. B., Korajkic, A., Staley, Z. R., and Harwood, V. J. (2012). Enterococci in the environment. Microbiol. Mol. Biol. Rev. 76, 685–706. doi: 10.1128/MMBR.00023-12
Cagnoli, G., Bertelloni, F., Interrante, P., Ceccherelli, R., Marzoni, M., and Ebani, V. V. (2022). Antimicrobial-resistant Enterococcus spp. in wild avifauna from Central Italy. Antibiotics 11:852. doi: 10.3390/antibiotics11070852
Cattoir, V. (2022). The multifaceted lifestyle of enterococci: genetic diversity, ecology and risks for public health. Curr. Opin. Microbiol. 65, 73–80. doi: 10.1016/j.mib.2021.10.013
Cetinkaya, Y., Falk, P., and Mayhall, C. G. (2000). Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13, 686–707. doi: 10.1128/CMR.13.4.686
Chavers, L. S., Moser, S. A., Benjamin, W. H., Banks, S. E., Steinhauer, J. R., Smith, A. M., et al. (2003). Vancomycin-resistant enterococci: 15 years and counting. J. Hosp. Infect. 53, 159–171. doi: 10.1053/jhin.2002.1375
Cho, S., Barrett, J. B., Frye, J. G., and Jackson, C. R. (2020). Antimicrobial resistance gene detection and plasmid typing among multidrug resistant enterococci isolated from freshwater environment. Microorganisms 8:1338. doi: 10.3390/microorganisms8091338
CLSI (2021). Performance standards for antimicrobial susceptibility testing. 31st ed. CLSI Supplement M100 (ISBN 978-1-68440-105-5 [electronic]). Clinical and Laboratory Standards Institute, USA.
Conceição, T., de Lencastre, H., and Aires-de-Sousa, M. (2017a). Frequent isolation of methicillin resistant Staphylococcus aureus (MRSA) ST398 among healthy pigs in Portugal. PLoS One 12:e0175340. doi: 10.1371/journal.pone.0175340
Conceição, T., de Lencastre, H., and Aires-de-Sousa, M. (2017b). Healthy bovines as reservoirs of major pathogenic lineages of Staphylococcus aureus in Portugal. Microb. Drug Resist. 23, 845–851. doi: 10.1089/mdr.2017.0074
Conceição, T., Diamantino, F., Coelho, C., de Lencastre, H., and Aires-de-Sousa, M. (2013). Contamination of public buses with MRSA in Lisbon, Portugal: a possible transmission route of major MRSA clones within the community. PLoS One 8:e77812. doi: 10.1371/journal.pone.0077812
Conceição, T., Ribeiro, M., Carvalho, L., Matos, S., Pinto, M., Almeida, A. M., et al. (2022). Drivers of Enterococcus faecium infection in Portuguese hospitals: spread of major clonal lineages. 32nd European congress of Clinical Microbiology & Infectious Diseases (ECCMID), Lisbon, Portugal, P0474.
Correia, S., Ponce, P., Jones-Dias, D., Caniça, M., Igrejas, G., and Poeta, P. (2014). Vancomycin-resistant enterococci among haemodialysis patients in Portugal: prevalence and molecular characterization of resistance, virulence and clonality. Enf. Inf. Microbiol. Clin. 32, 174–176. doi: 10.1016/j.eimc.2013.09.001
de Been, M., Pinholt, M., Top, J., Bletz, S., Mellmann, A., van Schaik, W., et al. (2015). Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J. Clin. Microbiol. 53, 3788–3797. doi: 10.1128/JCM.01946-15
de Garnica, M.-L., Valdezate, S., Gonzalo, C., and Saez-Nieto, J. A. (2013). Presence of the vanC1 gene in a vancomycin-resistant Enterococcus faecalis strain isolated from ewe bulk tank milk. J. Med. Microbiol. 62, 494–495. doi: 10.1099/jmm.0.052274-0
Diab, M., Salem, D., El-Shenawy, A., El-Far, A., Abdelghany, A., Awad, A. R., et al. (2019). Detection of high level aminoglycoside resistance genes among clinical isolates of Enterococcus species. Egyptian J. Med. Human Gen. 20:28. doi: 10.1186/s43042-019-0032-3
Dutka-Malen, S., Evers, S., and Courvalin, P. (1995). Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33, 24–27. doi: 10.1128/jcm.33.1.24-27.1995
ECDC (2023). Antimicrobial resistance in the EU/EEA (EARS-net) – annual epidemiological report 2022. Stockholm, Sweden, November 2023.
Echeverria-Esnal, D., Sorli, L., Navarrete-Rouco, M. E., Prim, N., Barcelo-Vidal, J., Conde-Estévez, D., et al. (2023). Ampicillin-resistant and vancomycin-susceptible Enterococcus faecium bacteremia: a clinical narrative review. Expert Rev. Anti-Infect. Ther. 21, 759–775. doi: 10.1080/14787210.2023.2223977
El-Mahdy, R., Mostafa, A., and El-Kannishy, G. (2018). High level aminoglycoside resistant enterococci in hospital-acquired urinary tract infections in Mansoura, Egypt. Germs 8, 186–190. doi: 10.18683/germs.2018.1145
EUCAST (2022). Breakpoint tables for interpretation of MICs and zone diameters. Version 12.0, 2022. Available at: http://www.eucast.org
Freitas, A. R., Novais, C., Duarte, B., Pereira, A. P., Coque, T. M., and Peixe, L. (2018). High rates of colonisation by ampicillin-resistant enterococci in residents of long-term care facilities in Porto, Portugal. Int. J. Antimicrob. Agents 51, 503–507. doi: 10.1016/j.ijantimicag.2017.11.007
Freitas, A. R., Novais, C., Ruiz-Garbajosa, P., Coque, T. M., and Peixe, L. (2009). Dispersion of multidrug-resistant Enterococcus faecium isolates belonging to major clonal complexes in different Portuguese settings. Appl. Environ. Microbiol. 75, 4904–4908. doi: 10.1128/AEM.02945-08
Freitas, A. R., Tedim, A. P., Duarte, B., Elghaieb, H., Abbassi, M. S., Hassen, A., et al. (2020). Linezolid-resistant (Tn6246::fexB-poxtA) Enterococcus faecium strains colonizing humans and bovines on different continents: similarity without epidemiological link. J. Antimicrob. Chemother. 75, 2416–2423. doi: 10.1093/jac/dkaa227
Freitas, A. R., Tedim, A. P., Francia, M. V., Jensen, L. B., Novais, C., Peixe, L., et al. (2016). Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986–2012). J. Antimicrob. Chemother. 71, 3351–3366. doi: 10.1093/jac/dkw312
Gao, W., Howden, B. P., and Stinear, T. P. (2017). Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 41, 76–82. doi: 10.1016/j.mib.2017.11.030
Garcia-Solache, M., and Rice, L. B. (2019). The Enterococcus: a model of adaptability to its environment. Clin. Microbiol. Rev. 32, 1–28. doi: 10.1128/CMR.00058-18
Gião, J., Leão, C., Albuquerque, T., Clemente, L., and Amaro, A. (2022). Antimicrobial susceptibility of Enterococcus isolates from cattle and pigs in Portugal: linezolid resistance genes optrA and poxtA. Antibiotics 11:615. doi: 10.3390/antibiotics11050615
Gouliouris, T., Raven, K. E., Ludden, C., Blane, B., Corander, J., Horner, C. S., et al. (2018). Genomic surveillance of Enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. MBio 9, 1–15. doi: 10.1128/mbio.01780-18
Gu, J., Xie, X.-J., Liu, J.-X., Shui, J.-R., Zhang, H.-Y., Feng, G.-Y., et al. (2020). Prevalence and transmission of antimicrobial-resistant staphylococci and enterococci from shared bicycles in Chengdu, China. Sci. Total Environ. 738:139735. doi: 10.1016/j.scitotenv.2020.139735
Guzman Prieto, A. M., van Schaik, W., Rogers, M. R., Coque, T. M., Baquero, F., Corander, J., et al. (2016). Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones? Front. Microbiol. 7:788. doi: 10.3389/fmicb.2016.00788
Han, D., Unno, T., Jang, J., Lim, K., Lee, S.-N., Ko, G., et al. (2011). The occurrence of virulence traits among high-level aminoglycosides resistant Enterococcus isolates obtained from feces of humans, animals, and birds in South Korea. Int. J. Food Microbiol. 144, 387–392. doi: 10.1016/j.ijfoodmicro.2010.10.024
Hayden, M. K., Blom, D. W., Lyle, E. A., Moore, C. G., and Weinstein, R. A. (2008). Risk of hand or glove contamination after contact with patients colonized with vancomycin-resistant Enterococcus or the colonized patients’ environment. Inf. Control Hosp. Epidemiol. 29, 149–154. doi: 10.1086/524331
Jackson, C. R., Fedorka-Cray, P. J., and Barrett, J. B. (2004). Use of a genus- and species-specific multiplex PCR for identification of enterococci. J. Clin. Microbiol. 42, 3558–3565. doi: 10.1128/JCM.42.8.3558-3565.2004
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Lebreton, F., Willems, R. J. L., and Gilmore, M. S. (2014). “Enterococcus diversity, origins in nature, and gut colonization” in Enterococci: from commensals to leading causes of drug resistant infection. eds. M. S. Gilmore, D. B. Clewell, and Y. Ike, et al. (Boston: Massachusetts Eye and Ear Infirmary).
Li, W., Li, J., Wei, Q., Hu, Q., Lin, X., Chen, M., et al. (2015). Characterization of aminoglycoside resistance and virulence genes among Enterococcus spp. isolated from a hospital in China. Int. J. Environ. Res. Public Health 12, 3014–3025. doi: 10.3390/ijerph120303014
Lopes, E., Conceição, T., Poirel, L., de Lencastre, H., and Aires-de-Sousa, M. (2019). Epidemiology and antimicrobial resistance of methicillin-resistant Staphylococcus aureus isolates colonizing pigs with different exposure to antibiotics. PLoS One 14:e0225497. doi: 10.1371/journal.pone.0225497
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Inf. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mahdizade Ari, M., Dashtbin, S., Ghasemi, F., Shahroodian, S., Kiani, P., Bafandeh, E., et al. (2023). Nitrofurantoin: properties and potential in treatment of urinary tract infection: a narrative review. Front. Cell. Infect. Microbiol. 13, 1–13. doi: 10.3389/fcimb.2023.1148603
McGlinchey, T. A., Rafter, P. A., Regan, F., and McMahon, G. P. (2008). A review of analytical methods for the determination of aminoglycoside and macrolide residues in food matrices. Anal. Chim. Acta 624, 1–15. doi: 10.1016/j.aca.2008.05.054
Miller, W. R., Munita, J. M., and Arias, C. A. (2014). Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther. 12, 1221–1236. doi: 10.1586/14787210.2014.956092
Murray, B. E. (1990). The life and times of the Enterococcus. Clin. Microbiol. Rev. 3, 46–65. doi: 10.1128/CMR.3.1.46
Nishiyama, M., Iguchi, A., and Suzuki, Y. (2015). Identification of Enterococcus faecium and Enterococcus faecalis as vanC -type vancomycin-resistant enterococci (VRE) from sewage and river water in the provincial city of Miyazaki, Japan. J. Environ. Sci. Health, Part A 50, 16–25. doi: 10.1080/10934529.2015.964599
Novais, C., Coque, T. M., Sousa, J. C., and Peixe, L. V. (2006). Antimicrobial resistance among faecal enterococci from healthy individuals in Portugal. Clin. Microbiol. Inf. 12, 1131–1134. doi: 10.1111/j.1469-0691.2006.01542.x
Novais, C., Freitas, A. R., Silveira, E., Antunes, P., Silva, R., Coque, T. M., et al. (2013). Spread of multidrug-resistant Enterococcus to animals and humans: an underestimated role for the pig farm environment. J. Antimicrob. Chemother. 68, 2746–2754. doi: 10.1093/jac/dkt289
Peng, Z., Yan, L., Yang, S., and Yang, D. (2022). Antimicrobial-resistant evolution and global spread of Enterococcus faecium clonal complex (CC) 17: progressive change from gut colonization to hospital-adapted pathogen. China CDC Weekly 4, 17–21. doi: 10.46234/ccdcw2021.277
Poeta, P., Costa, D., Rodrigues, J., and Torres, C. (2006). Antimicrobial resistance and the mechanisms implicated in faecal enterococci from healthy humans, poultry and pets in Portugal. Int. J. Antimicrob. Agents 27, 131–137. doi: 10.1016/j.ijantimicag.2005.09.018
Poulsen, R. L., Pallesen, L. V., Frimodt-Møller, N., and Espersen, F. (1999). Detection of clinical vancomycin-resistant enterococci in Denmark by multiplex PCR and sandwich hybridization. APMIS 107, 404–412. doi: 10.1111/j.1699-0463.1999.tb01573.x
Ramos, S., Igrejas, G., Rodrigues, J., Capelo-Martinez, J.-L., and Poeta, P. (2012). Genetic characterisation of antibiotic resistance and virulence factors in vanA-containing enterococci from cattle, sheep and pigs subsequent to the discontinuation of the use of avoparcin. Vet. J. 193, 301–303. doi: 10.1016/j.tvjl.2011.12.007
Roberts, M. C., and Schwarz, S. (2016). Tetracycline and phenicol resistance genes and mechanisms: importance for agriculture, the environment, and humans. J. Environ. Qual. 45, 576–592. doi: 10.2134/jeq2015.04.0207
Roer, L., Kaya, H., Tedim, A. P., Novais, C., Coque, T. M., Aarestrup, F. M., et al. (2024). VirulenceFinder for Enterococcus faecium and Enterococcus lactis: an enhanced database for detection of putative virulence markers by using whole-genome sequencing data. Microbiol. Spect. 12, e03724–e03723. doi: 10.1128/spectrum.03724-23
Schwaiger, K., Bauer, J., Hörmansdorfer, S., Mölle, G., Preikschat, P., Kämpf, P., et al. (2012). Presence of the resistance genes vanC1 and pbp5 in phenotypically vancomycin and ampicillin susceptible Enterococcus faecalis. Microb. Drug Resist. 18, 434–439. doi: 10.1089/mdr.2011.0227
Sun, M., Wang, Y., Chen, Z., Zhu, X., Tian, L., and Sun, Z. (2014). The first report of the vanC1 gene in Enterococcus faecium isolated from a human clinical specimen. Mem. Inst. Oswaldo Cruz 109, 712–715. doi: 10.1590/0074-0276140019
Tedim, A. P., Almeida-Santos, A. C., Lanza, V. F., Novais, C., Coque, T. M., Freitas, A. R., et al. (2023). Bacteriocin distribution patterns in Enterococcus faecium and Enterococcus lactis: bioinformatic analysis using a tailored genomics framework. bioRxiv :2023.11.13.566347. doi: 10.1101/2023.11.13.566347
Torres, C., Alonso, C. A., Ruiz-Ripa, L., León-Sampedro, R., Campo, R., and Coque, T. (2018). Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol. Spect. 6, 1–41. doi: 10.1128/microbiolspec.ARBA-0032-2018
Uttley, A. H., Collins, C. H., Naidoo, J., and George, R. C. (1988). Vancomycin-resistant enterococci. Lancet 1, 57–58. doi: 10.1016/S0140-6736(88)91037-9
Wada, Y., Irekeola, A. A., E A R, E. N. S., Yusof, W., Lih Huey, L., Ladan Muhammad, S., et al. (2021). Prevalence of vancomycin-resistant Enterococcus (VRE) in companion animals: the first meta-analysis and systematic review. Antibiotics 10:138. doi: 10.3390/antibiotics10020138
Wang, J.-T., Chang, S.-C., Wang, H.-Y., Chen, P.-C., Shiau, Y.-R., and Lauderdale, T.-L. (2013). High rates of multidrug resistance in Enterococcus faecalis and E. faecium isolated from inpatients and outpatients in Taiwan. Diag. Microbiol. Infect. Dis. 75, 406–411. doi: 10.1016/j.diagmicrobio.2013.01.004
Wei, Y., Palacios Araya, D., and Palmer, K. L. (2024). Enterococcus faecium: evolution, adaptation, pathogenesis and emerging therapeutics. Nat. Rev. Microbiol. 1–17. doi: 10.1038/s41579-024-01058-6
Werner, G., Fleige, C., Feßler, A. T., Timke, M., Kostrzewa, M., Zischka, M., et al. (2012). Improved identification including MALDI-TOF mass spectrometry analysis of group D streptococci from bovine mastitis and subsequent molecular characterization of corresponding Enterococcus faecalis and Enterococcus faecium isolates. Vet. Microbiol. 160, 162–169. doi: 10.1016/j.vetmic.2012.05.019
Werner, G., Fleige, C., Geringer, U., van Schaik, W., Klare, I., and Witte, W. (2011). IS element IS16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect. Dis. 11:80. doi: 10.1186/1471-2334-11-80
WHO (2024). WHO Bacterial Priority Pathogens List, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Geneva: World Health Organization; 2024. Licence: CC BY-NC-SA 3.0 IGO.
Zaheer, R., Cook, S. R., Barbieri, R., Goji, N., Cameron, A., Petkau, A., et al. (2020). Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a one-health continuum. Sci. Rep. 10:3937. doi: 10.1038/s41598-020-61002-5
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhong, Z., Kwok, L.-Y., Hou, Q., Sun, Y., Li, W., Zhang, H., et al. (2019). Comparative genomic analysis revealed great plasticity and environmental adaptation of the genomes of Enterococcus faecium. BMC Gen 20:602. doi: 10.1186/s12864-019-5975-8
Keywords: Enterococcus spp., livestock, environment, Enterococcus faecium , antimicrobial resistance, non-clinical enterococcus reservoirs, aminoglycosides high-level resistance, ampicillin resistant E. faecium
Citation: Lopes J, de Lencastre H and Conceição T (2024) Genomic analysis of Enterococcus faecium from non-clinical settings: antimicrobial resistance, virulence, and clonal population in livestock and the urban environment. Front. Microbiol. 15:1466990. doi: 10.3389/fmicb.2024.1466990
Received: 18 July 2024; Accepted: 20 August 2024;
Published: 11 September 2024.
Edited by:
Miklos Fuzi, Independent Researcher, Seattle, WA, United StatesReviewed by:
Salome N. Seiffert, Zentrum für Labormedizin (ZLM), SwitzerlandCopyright © 2024 Lopes, de Lencastre and Conceição. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teresa Conceição, dGVyZXNhZ2NAaXRxYi51bmwucHQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.