- 1School of Traditional Chinese Medicine, Hunan University of Chinese Medicine, Changsha, Hunan, China
- 2Department of Endocrinology, The First Hospital of Hunan University of Chinese Medicine, Changsha, Hunan, China
Objective: The relationship between gut microbiome and trimethylamine oxide (TMAO) has not been fully elucidated. We aimed to assess the causal effects of different gut microbes on TMAO using Mendelian randomization (MR).
Methods: Gut microbiome and TMAO datasets were acquired from genome-wide association studies and screened for single nucleotide polymorphisms according to the basic assumptions of MR. Inverse variance weighted was used as the main method in MR analysis to assess the causal relationship between the gut microbiome and TMAO. Finally, the MR-Egger intercept, Cochran's Q test, and leave-one-out sensitivity analysis were used to assess the horizontal pleiotropy, heterogeneity, and robustness of the results, respectively.
Results: MR analysis revealed that the species Bacteroides finegoldii (odds ratio [OR] 1.064, 95% confidence interval [CI] 1.003 to 1.128, p = 0.039), family Sutterellaceae (OR 1.188, 95% CI 1.003 to 1.407, p = 0.047), and phylum Pseudomonadota (OR 1.205, 95% CI 1.036 to 1.401, p = 0.016), as well as the species Bacteroides uniformis (OR 1.263, 95% CI 1.039 to 1.535, p = 0.019), were positively associated with increased genetic susceptibility to TMAO. In contrast, the species Bacteroides thetaiotaomicron (OR 0.813, 95% CI 0.696 to 0.950, p = 0.009) and Bilophila wadsworthia (OR 0.828, 95% CI 0.690 to 0.995, p = 0.044) were associated with reduced genetic susceptibility to TMAO. Additionally, the MR-Egger intercept indicated no horizontal pleiotropy (p ≥ 0.05), and Cochran's Q test and sensitivity analysis demonstrated that the results were not heterogeneous (p ≥ 0.05) and were robust.
Conclusion: Our findings revealed the role of the phylum Pseudomonadota, family Sutterellaceae, species Bacteroides finegoldii, and Bacteroides uniformis in increasing TMAO, as well as the species Bacteroides thetaiotaomicron and Bilophila wadsworthia in decreasing TMAO. This study provides new insights into the relationship between the gut microbiome and TMAO levels.
1 Introduction
Gut microbiome is the microbial community in the gut composed of bacteria, fungi, archaea, prokaryotes, eukaryotes, and viruses (Hillman et al., 2017). It is associated with the health of its host organism and plays a role in the digestion, absorption, metabolism, and synthesis of vitamins as well as the regulation of immune function (Tzeng and Lee, 2024; Dong et al., 2024). The gut microbiome is predominantly composed of bacteria, with the most common phyla being Firmicutes, Bacteroidetes, Pseudomonadota, and Fusobacteria (Hillman et al., 2017). Among them, the phyla Firmicutes and Bacteroidetes account for more than 90% of the total number of gut microbe, and their ratio serves as a biological marker for several diseases (Yañez et al., 2021). Previous studies have shown that gut microbes are associated with many diseases such as cardiovascular diseases, neurodegenerative diseases, diabetes, and osteoporosis (Xie et al., 2024; Andreu-Sánchez et al., 2024). Subsequent studies have found that the mechanisms by which gut microbes influence diseases may be related to the regulation of metabolites such as trimethylamine N-oxide (TMAO) (Li et al., 2019, 2022). The metabolites represented by TMAO may be essential mediators of gut microbes that influence the health of organisms.
TMAO is a metabolite of gut microbes, whose synthesis is influenced by factors such as diet, intestinal microbiota, and the activity of flavin-containing monooxygenase 3 (FMO3) (Zhang et al., 2023). Trimethylamine (TMA), a precursor of TMAO, is generated from choline, phosphatidylcholine, and carnitine through metabolism by intestinal microbes (Badaoui et al., 2024). Subsequently, TMA enters blood circulation and is oxidized in the liver by FMO3 to generate TMAO (Zhang et al., 2023). As research has progressed, TMAO is associated with diseases such as cardiovascular disease, kidney disease, diabetes, cancer, and Alzheimer's disease and is increasingly recognized as the link between gut microbes and diseases (Subramaniam and Fletcher, 2018; Wang et al., 2023). Subsequent studies have pointed out that TMAO induces disease through the activation of NF-κB-mediated inflammatory responses and is considered an important metabolite associated with disease (Zhen et al., 2023; Constantino-Jonapa et al., 2023). However, < 1% of the vast number of gut microbes possess genes required to produce TMAO (Rath et al., 2017). Although previous studies have reported that several intestinal microbiota are involved in its synthesis (Zhang and Jian, 2023), the relationship between different gut microbes and TMAO has not been comprehensively assessed. Additionally, limited by the nature of the study, the causal attributes of different gut microbes and TMAO require further clarification. A novel and effective method is needed to comprehensively assess the causal effects of the gut microbiome on TMAO.
Mendelian randomization (MR) is a cutting-edge epidemiological research method based on genetic epidemiology (Smith and Ebrahim, 2003). It uses genetic variants as instrumental variables to assess the causal effects of exposure on outcomes, effectively overcoming the limitations of traditional observational studies (Birney, 2022). As MR follows the rule of random assignment, it is less susceptible to confounding variables and reverse causality than traditional cross-sectional and observational studies (Birney, 2022). Additionally, because of its vast dataset, MR can provide a more comprehensive analysis of causal effects. In this study, we assessed the relationship between different gut microbes and TMAO using two-sample MR.
2 Materials and methods
2.1 Study design
MR stands out as a robust approach for evaluating causal relationships. It serves as a valuable analytical technique when experimental data are limited and is particularly effective in addressing confounding and reverse causality concerns within observational data. Its strength lies in its ability to simulate the control of randomization over the experimental design, thus reducing possible confounding variables and biases as well as improving the accuracy of causal inferences. When experimental evidence is sufficient and the results are not reverse causal or confounding, MR analysis is no longer of sufficient value.
The causal effects of different gut microbiomes on TMAO have not been fully elucidated; therefore, we employed MR analysis to explore the gut microbiomes that have a significant effect on TMAO. The principle of MR is to analyze the causal effect of exposure on outcomes through single nucleotide polymorphisms (SNPs), which are common variations in the genome. This requires the MR design to meet three basic assumptions (Davies et al., 2018) (Figure 1): assumption 1 (association assumption) is that SNPs are strongly associated with radiation exposure, assumption 2 (independence assumption) is that SNPs are independent of confounding variables, and assumption 3 (exclusivity assumption) is that SNPs cannot affect outcomes through pathways other than exposure. Based on these assumptions, the MR analysis identified SNPs that were significantly associated with each gut microbiota. We then assessed the independent impact of these SNPs on TMAO to uncover certain microbiota within the gut microbiome dataset that had a notable influence on TMAO.
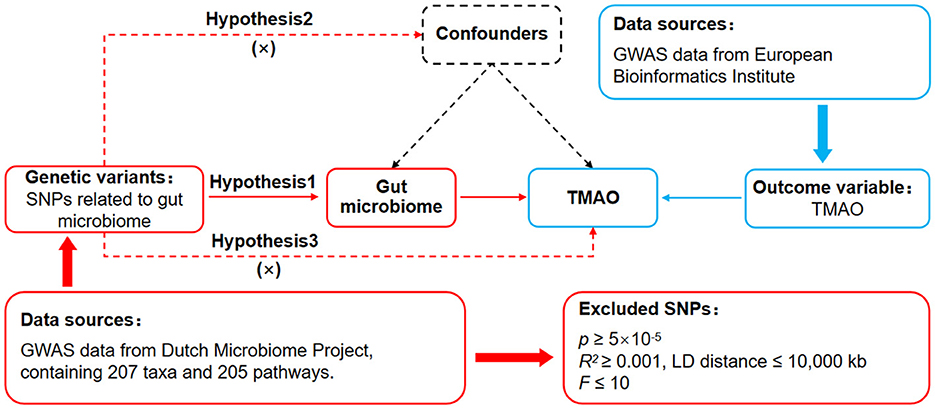
Figure 1. MR design to analyze the effects of the gut microbiome on genetic susceptibility to TMAO. MR, Mendelian randomization; TMAO, trimethylamine N-oxide.
2.2 Data sources
Gut microbiome data were obtained from the Dutch Microbiome Project (Lopera-Maya et al., 2022). Shotgun metagenomic sequencing was used to report genome-wide association study (GWAS) data for 207 taxa and 205 pathways by testing the feces of 7,738 Europeans. The data were derived from a secondary analysis of two longitudinal cohort data by Panyard et al. (2021) and were included in the European Bioinformatics Institute. They performed metabolomic analysis on 291 individuals of European ancestry by ultrahigh-performance liquid chromatography-tandem mass spectrometry and finalized 338 metabolites, including TMAO, referenced as ebi-a-GCST90026279.
2.3 Genetic tool variable selection
First, qualifying p < 5 × 10−5, SNPs that strongly correlated with the gut microbiome were searched to satisfy Assumption 1. Qualifying R2 < 0.001 and kb = 10,000, we then searched for independent SNPs to exclude interference from linkage disequilibrium. F > 10 was then limited to searching for strongly correlated SNPs to exclude interference from weakly correlated variables. F was calculated as F = [R2/(1-R2)] * [(N-K-1)/K], where K refers to the number of paired samples, N is the total number of samples, and R2 is the cumulative explained variance. Afterward, SNPs containing confounding variables were excluded using PhenoScanner to satisfy Assumption 2. Palindromic and ambiguous SNPs were then excluded based on the effect of allele frequency when adjusting the allele orientation for exposure and outcome. Finally, the MR-Pleiotropy RESidual Sum and Outlier were used to exclude aberrant SNPs (p < 1) to ensure the correctness of the causal inference.
2.4 Data analysis
This study followed the STROBE-MR guidelines (Skrivankova et al., 2021). The “TwoSampleMR (0.5.7)” program package for R 4.3.1 was used to conduct a two-sample MR analysis of the gut microbiome with TMAO. Inverse variance weighted (IVW) was set as the primary tool for the MR analysis, whereas the MR-Egger, weighted mode, weighted median, and simple mode were set as secondary tools. IVW is typically used as the primary tool for MR analysis when the assumptions of instrument strength and the absence of pleiotropy hold. Additionally, MR-Egger and weighted mode are worth referring to when the results have pleiotropy; the weighted median is beneficial when there is potential heterogeneity in genetic instruments affecting the analysis results, and the simple mode is a commonly used secondary tool that does not consider horizontal pleiotropy or other potential complexities.
Subsequently, the MR-Egger intercept was used to evaluate horizontal pleiotropy, with p ≥ 0.05 indicating that the results had no horizontal pleiotropy to satisfy Assumption 3. Cochran's Q was used to assess heterogeneity, with p ≥ 0.05 indicating no significant heterogeneity of the results. Finally, a leave-one-out sensitivity analysis was used to assess the robustness of the results. The lack of significant changes in the pooled effect size indicated that the results were robust.
3 Results
3.1 Genetic instrumental variables
After testing the assumptions of association, independence, and exclusivity, MR analysis revealed that six gut microbes were significantly associated with genetic susceptibility to TMAO. Among them, 11 SNPs for the phylum Pseudomonadota, nine SNPs for the family Sutterellaceae, seven for the species Bilophila wadsworthia, 18 for the species Bacteroides finegoldii, eight for the species Bacteroides uniformis, and eight SNPs for the species Bacteroides thetaiotaomicron were included in the MR analysis (Supplementary Table S1).
3.2 Two-sample MR analysis
MR analysis based on IVW revealed that the species Bacteroides finegoldii (odds ratio [OR] 1.064, 95% confidence interval [CI] 1.003 to 1.128, p = 0.039), family Sutterellaceae (OR 1.188, 95% CI 1.003 to 1.407, p = 0.047), and phylum Pseudomonadota (OR 1.205, 95% CI 1.036 to 1.401, p = 0.016), as well as the species Bacteroides uniformis (OR 1.263, 95% CI 1.039 to 1.535, p = 0.019), were positively associated with increased genetic susceptibility to TMAO. In contrast, the species Bacteroides thetaiotaomicron (OR 0.813, 95% CI 0.696 to 0.950, p = 0.009) and Bilophila wadsworthia (OR 0.828, 95% CI 0.690 to 0.995, p = 0.044) were associated with reduced genetic susceptibility to TMAO (Figure 2). Figure 3 summarizes the causal effects of the gut microbiome on TMAO obtained using the five methods, and Figure 4 shows the independent effects of each SNP. The MR-Egger intercept indicated no horizontal pleiotropy (p ≥ 0.05) (Supplementary Table S2).

Figure 2. Forest plot of MR analysis for the effects of the gut microbiome on genetic susceptibility to TMAO. MR, Mendelian randomization; TMAO, trimethylamine N-oxide.
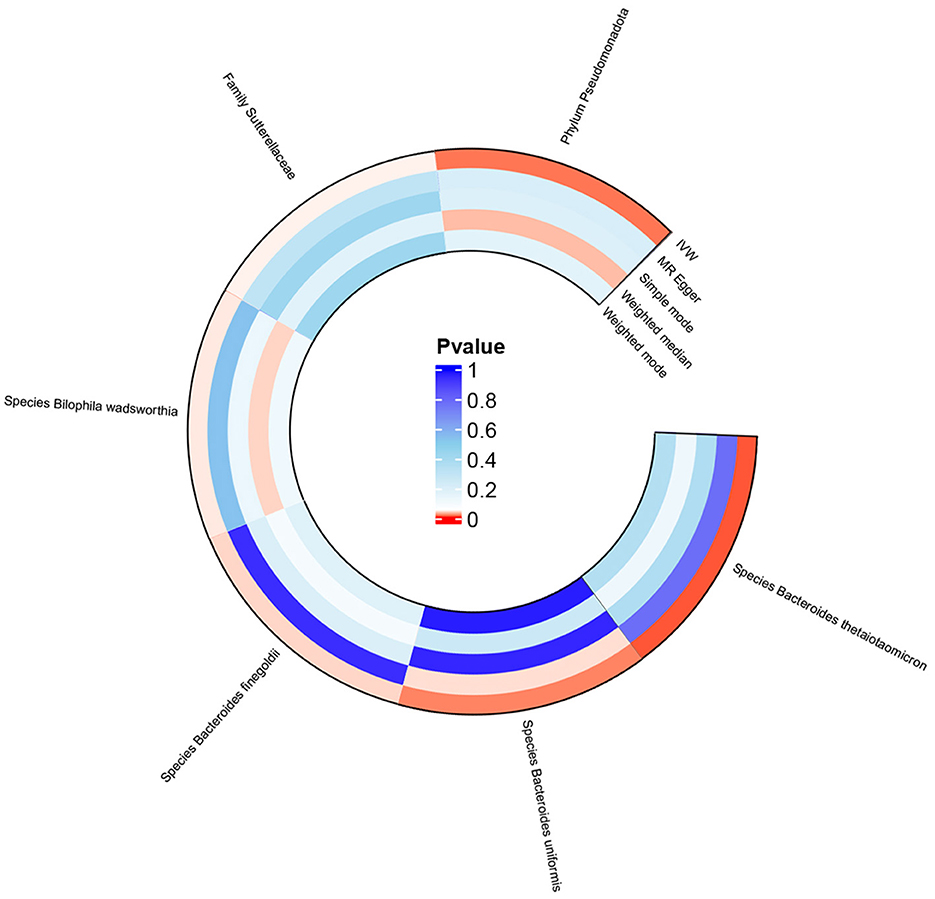
Figure 3. Circle plot of MR analysis for the effects of the gut microbiome on genetic susceptibility to TMAO. MR, Mendelian randomization; TMAO, trimethylamine N-oxide.
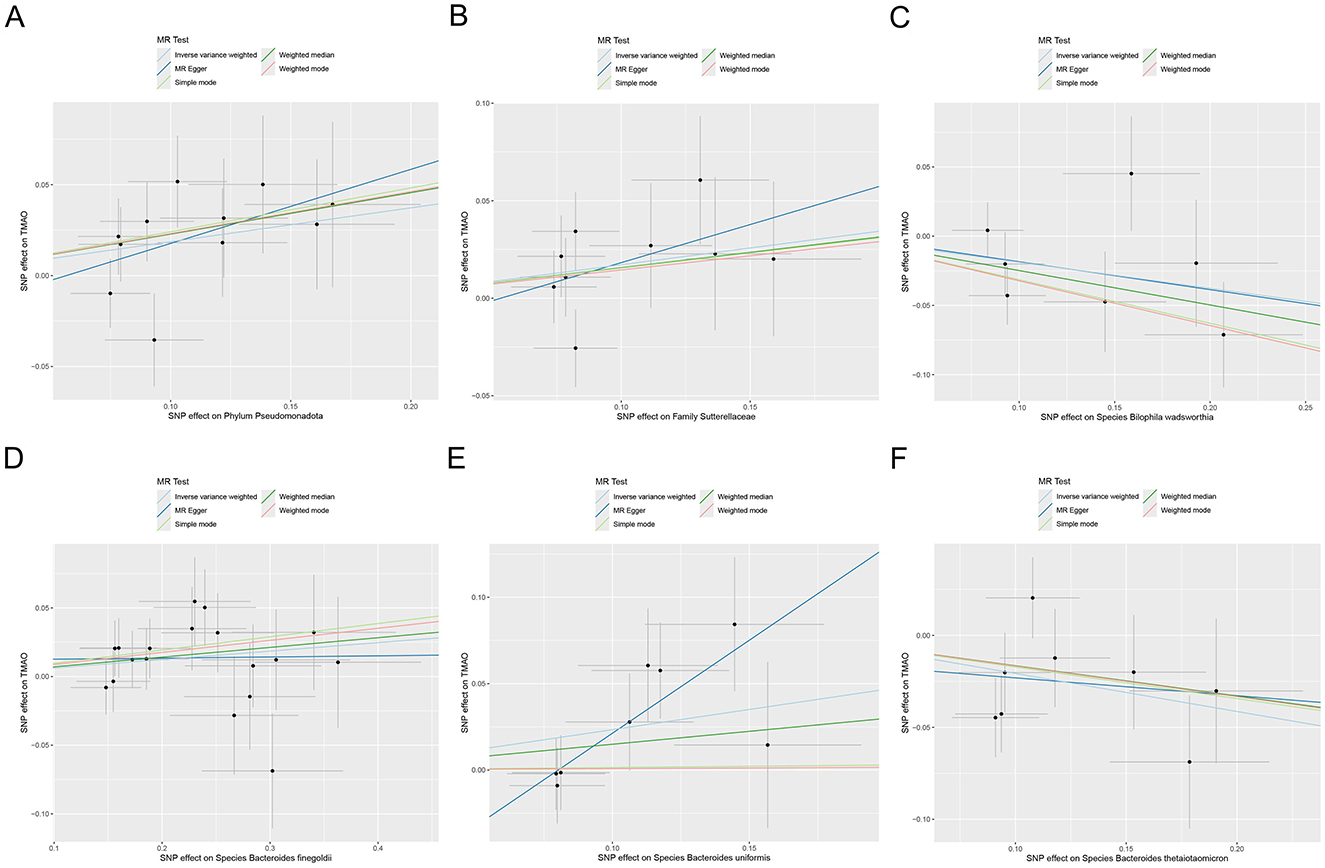
Figure 4. Scatter plot of MR analysis for the effects of the gut microbiome on genetic susceptibility to TMAO. (A) Phylum Pseudomonadota; (B) family Sutterellaceae; (C) species Bilophila wadsworthia; (D) species Bacteroides finegoldii; (E) species Bacteroides uniformis; (F) species Bacteroides thetaiotaomicron. MR, Mendelian randomization; TMAO, trimethylamine N-oxide.
3.3 Heterogeneity and sensitivity analysis
Cochran's Q demonstrated no significant heterogeneity (p ≥ 0.05) in the results (Supplementary Table S3, Figure 5). The leave-one-out sensitivity analysis revealed that the MR results were robust (Figure 6).
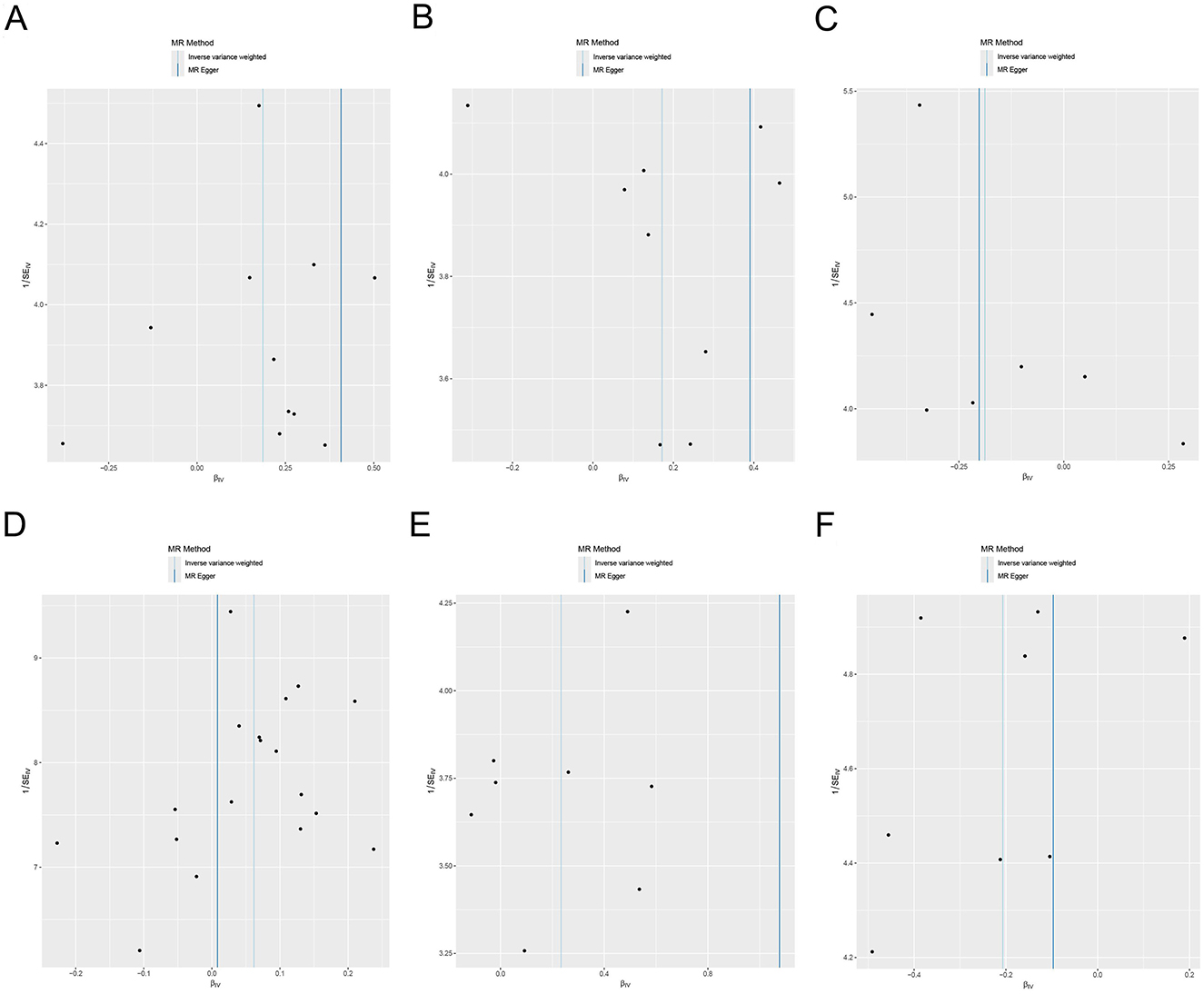
Figure 5. Funnel plot of heterogeneity analysis for the effects of the gut microbiome on genetic susceptibility to TMAO. (A) Phylum Pseudomonadota; (B) family Sutterellaceae; (C) species Bilophila wadsworthia; (D) species Bacteroides finegoldii; (E) species Bacteroides uniformis; (F) species Bacteroides thetaiotaomicron. TMAO, trimethylamine N-oxide.
Figure 6. Leave-one-out sensitivity analysis for the effects of the gut microbiome on genetic susceptibility to TMAO. (A) Phylum Pseudomonadota; (B) family Sutterellaceae; (C) species Bilophila wadsworthia; (D) species Bacteroides finegoldii; (E) species Bacteroides uniformis; (F) species Bacteroides thetaiotaomicron. TMAO, trimethylamine N-oxide.
4 Discussion
4.1 Research background and results
TMAO, a significant metabolite produced by gut microbes, has critical implications in the development of many diseases (Spasova et al., 2024). Although previous studies have provided preliminary insights into the relationship between gut microbes and TMAO (Romano et al., 2015), they are insufficient to adequately explain causal effects (Jin et al., 2024). To the best of our knowledge, this is the first MR analysis to assess the causal relationship between the gut microbiome and TMAO using GWAS. Our findings revealed the role of Bacteroides finegoldii, Bacteroides uniformis, Bacteroides thetaiotaomicron, and Bilophila wadsworthia at the species level, Sutterellaceae at the family level, and Pseudomonadota (formerly Proteobacteria) at the phylum level in modulating TMAO levels (Figure 7). As these gut microbiota belong to the phyla Pseudomonadota and Bacteroidetes, MR analysis revealed the potential effects of these phyla on TMAO levels in Europeans. A previous genetic study reported contrasting results to ours (Andreu-Sánchez et al., 2024), in which they found that the phyla Pseudomonadota and Bacteroidetes were not associated with TMAO levels. We speculate that this discrepancy may stem from different datasets, as the dataset used for this MR analysis was from the Dutch Microbiome Project, whereas the dataset used for the previous genetic study was from the Rotterdam Study III-2 and LifeLines-DEEP (Andreu-Sánchez et al., 2024). Additionally, although previous studies support that the phyla Pseudomonadota and Bacteroidetes affect the prognosis of cardiovascular disease by increasing TMAO levels (Martins et al., 2024; Melnychuk and Lizogub, 2022), the MR analysis by Wang et al. (2023) did not find a significant association between these phyla and cardiovascular disease. We speculate that this may be because this previous study concentrated solely on species with consistent positive findings across multiple databases, leading to a focus on the role of genus RuminococcusUCG010 in cardiovascular disease.
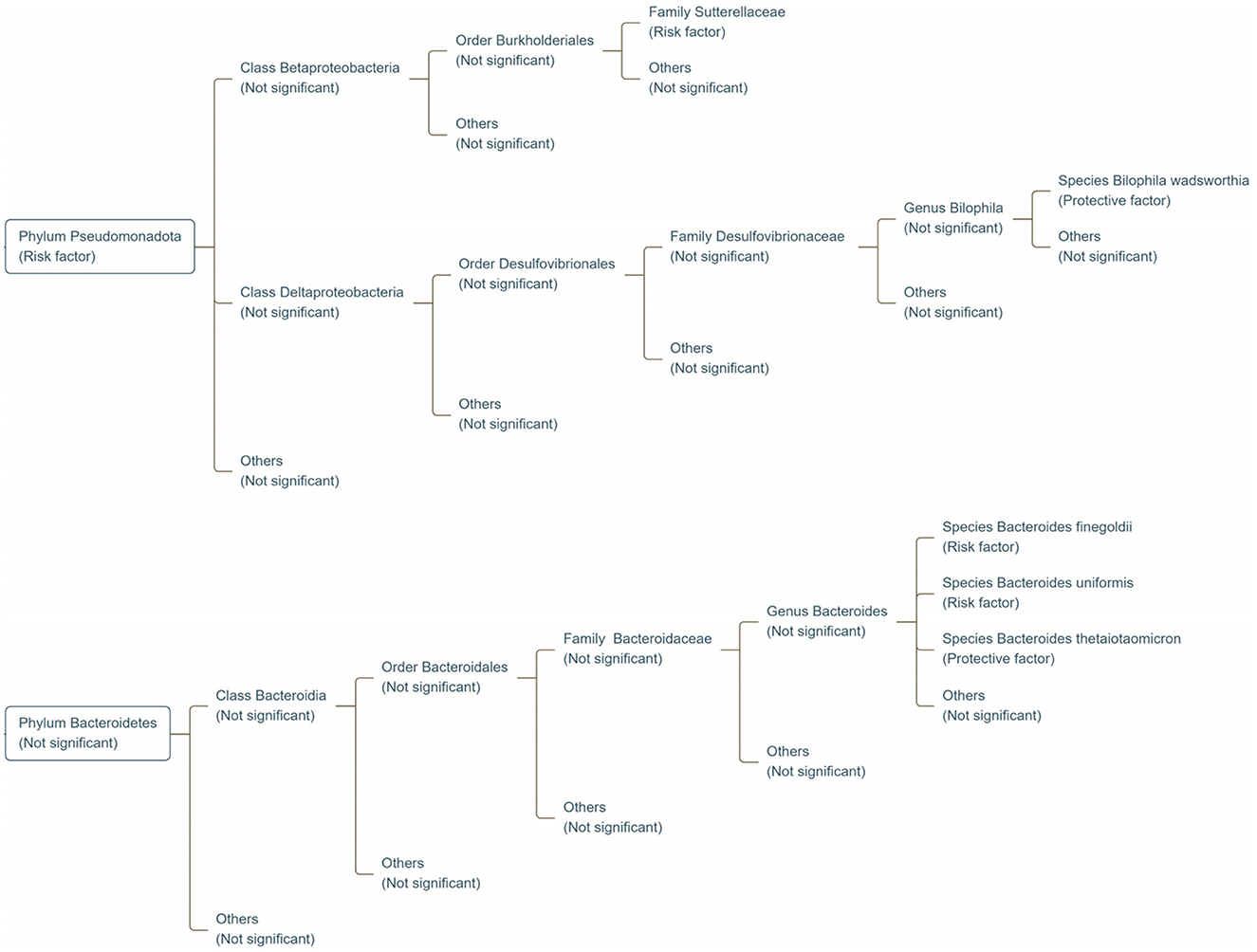
Figure 7. Classification map of gut microbiota significantly associated with genetic susceptibility to TMAO. Risk factor refers to the factors associated with increased genetic susceptibility to TMAO; protective factor refers to those associated with reduced genetic susceptibility to TMAO; not significant refers to no significant relationship with TMAO.
4.2 Effects of phylum Pseudomonadota on TMAO
Pseudomonadota is one of the most abundant and diverse phyla in the bacterial community, containing several common beneficial and pathogenic bacteria such as the species Escherichia coli, Serratia marcescens, and Helicobacter pylori (Rizzatti et al., 2017). A clinical study conducted in China revealed that the abundance of phylum Pseudomonadota was significantly higher in patients with preeclampsia (4.51%) than in healthy controls (2.56%) (Wang et al., 2019). Similarly, the plasma concentration of TMAO was higher in preeclampsia patients (2.17 ± 1.35 μmol/L) compared to controls (1.36 ± 0.72 μmol/L) (Wang et al., 2019). A meta-analysis involving 67 studies also found a potential association between the phylum Pseudomonadota and serum TMAO, which demonstrated that cardiovascular patients had significantly higher levels of both the phylum Pseudomonadota (standardized mean difference [SMD]: 0.33, 95% CI: 0.18–0.49) and plasma TMAO than healthy controls (SMD: 0.42, 95% CI: 0.17–0.66) (Martins et al., 2024). Another study in China found that a daily calorie-restricted diet increased the relative abundance of the phylum Pseudomonadota by 3.39 times and dramatically increased circulating TMAO in mice compared to normal feeding (Zhang et al., 2022). This evidence suggests that the abundance of the phylum Pseudomonadota may be positively correlated with the serum TMAO concentration. In addition, subsequent studies have indicated that the phylum Pseudomonadota may increase the synthesis of TMAO by regulating CutC (Huang et al., 2024). CutC is a choline TMA lyase commonly found in choline-fermenting bacteria that converts choline into the TMAO precursor TMA (Martínez-del Campo et al., 2015). A Chinese animal study indicated that a methionine-restricted diet significantly inhibited the growth of TMA-producing bacteria in the phylum Pseudomonadota by downregulating CutC expression, which in turn reduced TMAO levels in mice fed a high-fat diet (Lu et al., 2023). In summary, the phylum Pseudomonadota may synthesize TMA to increase TMAO levels, and its synthesis is regulated by CutC. This suggests that the abundance of the phylum Pseudomonadota may be positively correlated with serum TMAO concentrations.
Additionally, this MR study revealed that the family Sutterellaceae within phylum Pseudomonadota is associated with increased TMAO levels. Family Sutterellaceae is a family of Gram-negative bacteria that includes genera such as Sutterella, Mesosutterella, Parasutterella, and Turicimonas. A clinical study in Korea found that patients with polycystic ovary syndrome showed a significant increase in the abundance of the intestinal family Sutterellaceae, which led to elevated circulating TMAO levels and promoted the progression of cardiovascular disease, diabetes mellitus, and non-alcoholic fatty liver disease (Singh et al., 2024). Another animal experiment in China indicated that the relative abundance of genus Sutterella and circulating TMAO were significantly elevated in diabetic rats with erectile dysfunction compared to healthy rats (Li et al., 2017). These findings support a positive correlation between TMAO and the abundance of family Sutterellaceae. In addition, we speculate that the family Sutterellaceae may elevate TMAO levels by promoting TMA synthesis.
Our findings also indicate that the species Bilophila wadsworthia, which belongs to the phylum Pseudomonadota, is associated with decreased TMAO levels. Species Bilophila wadsworthia is a species belonging to the phylum Pseudomonadota family Desulfovibrionaceae genus Bilophila, and is the most studied and well-documented model species of genus Bilophila. Unfortunately, there is currently no literature documenting a direct relationship between the species Bilophila wadsworthia and TMAO, though the available literature suggests that the Bilophila genus is associated with decreased TMAO. A study in the United States suggested that the genus Bilophila reduced TMAO synthesis by converting TMA to dimethylamine (DMA) (Kivenson and Giovannoni, 2020). It has also been found that the genus Bilophila reduces cardiovascular disease by lowering circulating TMAO levels (Kivenson and Giovannoni, 2020), supporting TMAO as a link between gut microbes and disease. Another study in Sweden reported that the abundance of genus Bilophila was associated with reduced blood lipid levels and cardiovascular risk markers (Marungruang et al., 2018); this effect may have been achieved by modulating TMAO. These findings indicate that the genus Bilophila inhibits the synthesis of TMAO by converting TMA to DMA, suggesting that the species Bilophila wadsworthia may also be associated with reduced TMAO.
4.3 Effects of phylum Bacteroidetes on TMAO
Phylum Bacteroidetes consists of Gram-negative bacteria that are non-endospore, anaerobic, aerobic, or rod-shaped, and is the most abundant Gram-negative phylum residing in the gastrointestinal tract (Gibiino et al., 2018; Pan et al., 2023). A clinical study conducted in China revealed that, compared to healthy controls, patients with acute myocardial infarction exhibited significantly elevated levels of phylum Bacteroidetes in their gut as well as increased TMAO levels (Qian et al., 2022). Another study in Ukraine demonstrated that the intestinal abundance of phylum Bacteroidetes and plasma TMAO levels were significantly increased in atherosclerotic patients with arrhythmia compared to those without (Melnychuk and Lizogub, 2022). Subsequently, an animal experiment in China revealed that compared with normal controls, mice in the diarrhea model group had a significantly increased abundance of phylum Bacteroidetes and significantly higher levels of CutC activity as well as TMAO in the cecum of mice (Guo et al., 2024). CutC is a choline TMA lyase whose main function is to convert choline to TMA. It indicates that the phylum Bacteroidetes may increase TMA levels by enhancing CutC activity, thereby providing raw materials for TMAO synthesis.
This MR also revealed that the species Bacteroides finegoldii and Bacteroides uniformis, belonging to the phylum Bacteroidetes, were associated with increased levels of TMAO. Species Bacteroides finegoldii and Bacteroides uniformis are Gram-negative, rod-shaped, obligate anaerobic bacteria. Previous studies have suggested that the species Bacteroides uniformis plays an important role in lipid metabolism. A clinical study in China demonstrated that the species Bacteroides uniformis reduced the risk of hepatic steatosis and hyperlipidemia in mice, and its abundance was negatively correlated with low density lipoprotein cholesterol (LDL-C) levels (Yan et al., 2022). Another clinical study in China noted that carnitine (a TMAO precursor) levels were negatively correlated with LDL-C levels (Xiong et al., 2022). This implies that the species Bacteroides uniformis may increase TMAO levels by promoting carnitine synthesis, which, in turn, reduces LDL-C levels. Subsequently, a clinical study in the Netherlands noted that the species Bacteroides uniformis was positively correlated with levels of betaine, another TMAO precursor (R = 0.59, p = 0.04) (Hartstra et al., 2020). This suggests that the species Bacteroides uniformis may increase TMAO levels by promoting the synthesis of carnitine and betaine. However, currently no studies report the relationship between the species Bacteroides finegoldii and TMAO; this relationship requires future validation. We did not find any other Bacteroidetes species that were associated with increased TMAO levels. From this, we speculate that the effect of phylum Bacteroides on TMAO levels may be mediated specifically by the species Bacteroides finegoldii and Bacteroides uniformis; however, further studies are needed to explore and elucidate the effects of different phylum Bacteroides species on TMAO levels.
Species Bacteroides thetaiotaomicron is a polysaccharide-degrading bacterium also belonging to the phylum Bacteroides. Our MR demonstrated that the species Bacteroides finegoldii and Bacteroides uniformis were associated with increased TMAO. In contrast, the species Bacteroides thetaiotaomicron was associated with reduced TMAO. A longitudinal clinical study in the United States revealed that a healthy plant-based diet is positively associated with the relative abundance of species Bacteroides thetaiotaomicron and negatively associated with TMAO (Shen et al., 2024). Although this study did not directly confirm that reduced TMAO was caused by the species Bacteroides thetaiotaomicron, it suggests a potential relationship between the two. We speculate that a healthy plant-based diet may reduce TMAO by decreasing the intake of TMA precursor substances and increasing their consumption by bacteria. However, the causal effects of the phylum Bacteroides and its subspecies on TMAO need to be further explored.
4.4 Implications for clinical applications
The intricate interplay between specific gut microbes and TMAO susceptibility revealed in this study highlights potential interventions to modulate TMAO-related health outcomes and offers significant implications for clinical applications and therapeutic approaches. Phylum Pseudomonadota, family Sutterellaceae, and species Bacteroides finegoldii were associated with increased susceptibility to TMAO. Based on the results of previous studies, we hypothesize that these gut microbes increase the risk of cardiovascular disease by increasing TMAO levels, and may serve as key targets for personalized therapy aimed at reducing the cardiovascular risks associated with elevated TMAO levels. Conversely, microbial species such as Bacteroides thetaiotaomicron and Bilophila wadsworthia, which reduce genetic susceptibility to TMAO, suggest promising avenues for therapeutic interventions that may help lower TMAO levels and potentially improve overall health outcomes. In light of these findings, future research and clinical endeavors should explore the development of tailored probiotic or prebiotic interventions to effectively modulate gut microbiome composition and target TMAO-related pathways with precision, offering novel therapeutic strategies for managing TMAO-associated conditions. This study elucidated the practical implications of these microbial relationships and their implications for future interventions, setting the stage for innovative approaches to address TMAO-related health challenges in clinical settings.
4.5 Limitations and prospects
Although this MR study provides genetic evidence of a causal relationship between the gut microbiome and TMAO, it has some limitations. Both the gut microbiome and TMAO datasets used in this study were from Europe, thus potentially limiting the generalizability of our findings to other ethnicities. Additionally, although this study revealed the significant causal effects of six gut microbes on TMAO, the biological mechanisms by which they modulate it remain unclear. There may also be unrecognized confounding variables between the gut microbiome and TMAO, which increases the potential risk of bias. Given these limitations, future studies should expand the datasets of the gut microbiome and TMAO to other races, aiming for multiracial MR analysis and equitable health outcomes. We expect more detailed and comprehensive cohort studies in the future to validate the relationship between these six gut microbes and TMAO. Finally, we anticipate that further animal experiments will explore the mechanisms through which these gut microbes modulate TMAO, thereby offering objective biological insights.
5 Conclusion
Our findings suggest that the phylum Pseudomonadota, family Sutterellaceae, and species Bacteroides finegoldii, as well as Bacteroides uniformis are associated with increased TMAO. In contrast, the species Bilophila wadsworthia and Bacteroides thetaiotaomicron are associated with reduced TMAO. This study provides novel insights into the relationship between the gut microbiome and TMAO levels. However, owing to the current scarcity of evidence, further biological studies are necessary to explore the mechanisms by which these gut microbes regulate TMAO.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YYu: Conceptualization, Supervision, Writing – original draft. YYi: Formal analysis, Writing – original draft. JD: Methodology, Writing – original draft. XY: Data curation, Writing – original draft. SB: Writing – original draft. RY: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (U21A20411) and Hunan University of Chinese Medicine Disciplinary Construction' Revealing the List and Appointing Leaders' Project (22JBZ002).
Acknowledgments
We would like to express our gratitude to Dr. Xinran Wei from Hunan University of Chinese Medicine for reviewing the statistical analysis of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1465455/full#supplementary-material
Abbreviations
CI, confidence interval; FMO3, flavin-containing monooxygenase 3; GWAS, genome-wide association studies; OR, odds ratio; IVW, inverse variance weighted; MR, Mendelian randomization; SNPs, single nucleotide polymorphisms; TMA, trimethylamine; TMAO, trimethylamine N-oxide.
References
Andreu-Sánchez, S., Ahmad, S., Kurilshikov, A., Beekman, M., Ghanbari, M., van Faassen, M., et al. (2024). Unraveling interindividual variation of trimethylamine N-oxide and its precursors at the population level. iMeta 3:e183. doi: 10.1002/imt2.183
Badaoui, W., Marhuenda-Egea, F. C., Valero-Rodriguez, J. M., Sanchez-Jerez, P., Arechavala-Lopez, P., Toledo-Guedes, K., et al. (2024). Metabolomic and lipidomic tools for tracing fish escapes from aquaculture facilities. ACS Food Sci. Technol. 4, 871–879. doi: 10.1021/acsfoodscitech.3c00589
Birney, E. (2022). Mendelian randomization. Cold Spring Harb. Perspect. Med. 12:a041302. doi: 10.1101/cshperspect.a041302
Constantino-Jonapa, L. A., Espinoza-Palacios, Y., Escalona-Montaño, A. R., Hernández-Ruiz, P., Amezcua-Guerra, L. M., Amedei, A., et al. (2023). Contribution of trimethylamine N-oxide (TMAO) to chronic inflammatory and degenerative diseases. Biomedicines 11:431. doi: 10.3390/biomedicines11020431
Davies, N. M., Holmes, M. V., and Davey Smith, G. (2018). Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362:k601. doi: 10.1136/bmj.k601
Dong, X., Zhang, J., Li, W., Li, Y., Jia, L., Liu, Z., et al. (2024). Yi-Shen-Hua-Shi regulates intestinal microbiota dysbiosis and protects against proteinuria in patients with chronic kidney disease: a randomized controlled study. Pharm. Biol. 62, 356–366. doi: 10.1080/13880209.2024.2345080
Gibiino, G., Lopetuso, L. R., Scaldaferri, F., Rizzatti, G., Binda, C., Gasbarrini, A., et al. (2018). Exploring Bacteroidetes: Metabolic key points and immunological tricks of our gut commensals. Dig. Liver Dis. 50, 635–639. doi: 10.1016/j.dld.2018.03.016
Guo, M., Fang, L., Chen, M., Shen, J., Tan, Z., He, W., et al. (2024). Dysfunction of cecal microbiota and CutC activity in mice mediating diarrhea with kidney-yang deficiency syndrome. Front. Microbiol. 15:1354823. doi: 10.3389/fmicb.2024.1354823
Hartstra, A. V., Schüppel, V., Imangaliyev, S., Schrantee, A., Prodan, A., Collard, D., et al. (2020). Infusion of donor feces affects the gut–brain axis in humans with metabolic syndrome. Molec. Metab. 42:101076. doi: 10.1016/j.molmet.2020.101076
Hillman, E. T., Lu, H., Yao, T., and Nakatsu, C. H. (2017). Microbial ecology along the gastrointestinal tract. Microbes Environ. 32, 300–313. doi: 10.1264/jsme2.ME17017
Huang, Y., Zhang, J., Zhang, Y., Wang, W., Li, M., Chen, B., et al. (2024). Red meat intake, faecal microbiome, serum trimethylamine N-oxide and hepatic steatosis among Chinese adults. Liver Int. 44, 1142–1153. doi: 10.1111/liv.15860
Jin, Q., Zhang, C., Chen, R., Jiang, L., Li, H., Wu, P., et al. (2024). Quinic acid regulated TMA/TMAO-related lipid metabolism and vascular endothelial function through gut microbiota to inhibit atherosclerotic. J. Transl. Med. 22:352. doi: 10.1186/s12967-024-05120-y
Kivenson, V., and Giovannoni, S. J. (2020). An expanded genetic code enables trimethylamine metabolism in human gut bacteria. mSystems 5, e00413–e00420. doi: 10.1128/mSystems.00413-20
Li, D., Lu, Y., Yuan, S., Cai, X., He, Y., Chen, J., et al. (2022). Gut microbiota-derived metabolite trimethylamine-N-oxide and multiple health outcomes: an umbrella review and updated meta-analysis. Am. J. Clin. Nutr. 116, 230–243. doi: 10.1093/ajcn/nqac074
Li, H., Qi, T., Huang, Z., Ying, Y., Zhang, Y., Wang, B., et al. (2017). Relationship between gut microbiota and type 2 diabetic erectile dysfunction in Sprague-Dawley rats. J. Huazhong Univ. Sci. Technol. 37, 523–530. doi: 10.1007/s11596-017-1767-z
Li, L., Chen, B., Zhu, R., Li, R., Tian, Y., Liu, C., et al. (2019). Fructus Ligustri Lucidi preserves bone quality through the regulation of gut microbiota diversity, oxidative stress, TMAO and Sirt6 levels in aging mice. Aging (Albany NY) 11, 9348–9368. doi: 10.18632/aging.102376
Lopera-Maya, E. A., Kurilshikov, A., van der Graaf, A., Hu, S., Andreu-Sánchez, S., Chen, L., et al. (2022). Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch Microbiome Project. Nat. Genet. 54, 143–151. doi: 10.1038/s41588-021-00992-y
Lu, M., Yang, Y., Xu, Y., Wang, X., Li, B., Le, G., et al. (2023). Dietary methionine restriction alleviates choline-induced tri-methylamine-N-oxide (TMAO) elevation by manipulating gut microbiota in mice. Nutrients 15:206. doi: 10.3390/nu15010206
Martínez-del Campo, A., Bodea, S., Hamer, H. A., Marks, J. A., Haiser, H. J., Turnbaugh, P. J., et al. (2015). Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. MBio 6, e00042–e00015. doi: 10.1128/mBio.00042-15
Martins, D., Silva, C., Ferreira, A. C., Dourado, S., Albuquerque, A., Saraiva, F., et al. (2024). Unravelling the gut microbiome role in cardiovascular disease: a systematic review and a meta-analysis. Biomolecules 14:731. doi: 10.3390/biom14060731
Marungruang, N., Tovar, J., Björck, I., and Hållenius, F. F. (2018). Improvement in cardiometabolic risk markers following a multifunctional diet is associated with gut microbial taxa in healthy overweight and obese subjects. Eur. J. Nutr. 57, 2927–2936. doi: 10.1007/s00394-017-1563-3
Melnychuk, I., and Lizogub, V. G. (2022). Gut microbiota composition and its metabolites changes in patients with atherosclerosis and atrial fibrillation. Wiad. Lek. 75, 2994–2999. doi: 10.36740/WLek202212117
Pan, X., Raaijmakers, J. M., and Carrión, V. J. (2023). Importance of Bacteroidetes in host–microbe interactions and ecosystem functioning. Trends Microbiol. 31, 959–971. doi: 10.1016/j.tim.2023.03.018
Panyard, D. J., Kim, K. M., Darst, B. F., Deming, Y. K., Zhong, X., Wu, Y., et al. (2021). Cerebrospinal fluid metabolomics identifies 19 brain-related phenotype associations. Commun. Biol. 4:63. doi: 10.1038/s42003-020-01583-z
Qian, X., Liu, A., Liang, C., He, L., Xu, Z., Tang, S., et al. (2022). Analysis of gut microbiota in patients with acute myocardial infarction by 16S rRNA sequencing. Ann. Transl. Med. 10:1340. doi: 10.21037/atm-22-5671
Rath, S., Heidrich, B., Pieper, D. H., and Vital, M. (2017). Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome 5:54. doi: 10.1186/s40168-017-0271-9
Rizzatti, G., Lopetuso, L. R., Gibiino, G., Binda, C., and Gasbarrini, A. (2017). Proteobacteria: a common factor in human diseases. Biomed. Res. Int. 2017:9351507. doi: 10.1155/2017/9351507
Romano, K. A., Vivas, E. I., Amador-Noguez, D., and Rey, F. E. (2015). Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio 6:e02481. doi: 10.1128/mBio.02481-14
Shen, X., Tilves, C., Kim, H., Tanaka, T., Spira, A. P., Chia, C. W., et al. (2024). Plant-based diets and the gut microbiome: findings from the Baltimore longitudinal study of aging. Am. J. Clin. Nutr. 119, 628–638. doi: 10.1016/j.ajcnut.2024.01.006
Singh, V., Mahra, K., Jung, D., and Shin, J.-. H. (2024). Gut microbes in polycystic ovary syndrome and associated comorbidities; type 2 diabetes, non-alcoholic fatty liver disease (NAFLD), cardiovascular disease (CVD), and the potential of microbial therapeutics. Probiotics Antimicrob. Proteins 16, 1744–1761. doi: 10.1007/s12602-024-10262-y
Skrivankova, V. W., Richmond, R. C., Woolf, B. A. R., Yarmolinsky, J., Davies, N. M., Swanson, S. A., et al. (2021). Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA 326, 1614–1621. doi: 10.1001/jama.2021.18236
Smith, G. D., and Ebrahim, S. (2003). “Mendelian randomization”: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32, 1–22. doi: 10.1093/ije/dyg070
Spasova, N., Somleva, D., Krastev, B., Ilieva, R., Borizanova, A., Svinarov, D., et al. (2024). Association of the trimethylamine N-oxide with cardiovascular risk and vascular alterations in middle-aged patients with risk factors for cardiovascular diseases. Biosci. Rep. 45:BSR20232090. doi: 10.1042/BSR20232090
Subramaniam, S., and Fletcher, C. (2018). Trimethylamine N-oxide: breathe new life. Br. J. Pharmacol. 175, 1344–1353. doi: 10.1111/bph.13959
Tzeng, H. T., and Lee, W, C. (2024). Impact of transgenerational nutrition on nonalcoholic fatty liver disease development: interplay between gut microbiota, epigenetics and immunity. Nutrients 16:1388. doi: 10.3390/nu16091388
Wang, D., Chen, X., Li, Z., and Luo, Y. (2023). Association of the gut microbiota with coronary artery disease and myocardial infarction: a mendelian randomization study. Front. Genet. 14:1158293. doi: 10.3389/fgene.2023.1158293
Wang, J., Gu, X., Yang, J., Wei, Y., and Zhao, Y. (2019). Gut microbiota dysbiosis and increased plasma LPS and TMAO levels in patients with preeclampsia. Front. Cell. Infect. Microbiol. 9:409. doi: 10.3389/fcimb.2019.00409
Xie, L., Pan, L., Liu, B., Cheng, H., and Mao, X. (2024). Research progress on the association between trimethylamine/trimethylamine-N-oxide and neurological disorders. Postgrad. Med. J. 100, 283–288. doi: 10.1093/postmj/qgad133
Xiong, X., Zhou, J., Fu, Q., Xu, X., Wei, S., Yang, S., et al. (2022). The associations between TMAO-related metabolites and blood lipids and the potential impact of rosuvastatin therapy. Lipids Health Dis. 21:60. doi: 10.1186/s12944-022-01673-3
Yan, H., Qin, Q., Chen, J., Yan, S., Li, T., Gao, X., et al. (2022). Gut microbiome alterations in patients with visceral obesity based on quantitative computed tomography. Front. Cell. Infect. Microbiol. 11:823262. doi: 10.3389/fcimb.2021.823262
Yañez, C. M., Hernández, A. M., Sandoval, A. M., Domínguez, M. A. M., Muñiz, S. A. Z., Gómez, J. O. G., et al. (2021). Prevalence of Blastocystis and its association with Firmicutes/Bacteroidetes ratio in clinically healthy and metabolically ill subjects. BMC Microbiol. 21:339. doi: 10.1186/s12866-021-02402-z
Zhang, J., Zhu, P., Li, S., Gao, Y., and Xing, Y. (2023). From heart failure and kidney dysfunction to cardiorenal syndrome: TMAO may be a bridge. Front. Pharmacol. 14:1291922. doi: 10.3389/fphar.2023.1291922
Zhang, X., Shi, L., Li, Q., Song, C., Han, N., Yan, T., et al. (2022). Caloric restriction, friend or foe: effects on metabolic status in association with the intestinal microbiome and metabolome. J. Agric. Food Chem. 70, 14061–14072. doi: 10.1021/acs.jafc.2c06162
Zhang, Y., and Jian, W. (2023). Signal pathways and intestinal flora through trimethylamine N-oxide in alzheimer's disease. Curr. Protein Pept. Sci. 24, 721–736. doi: 10.2174/1389203724666230717125406
Keywords: gut microbiome, trimethylamine N-oxide, relationship, genome-wide association studies, Mendelian randomization
Citation: Yu Y, Yin Y, Deng J, Yang X, Bai S and Yu R (2024) Unveiling the causal effects of gut microbiome on trimethylamine N-oxide: evidence from Mendelian randomization. Front. Microbiol. 15:1465455. doi: 10.3389/fmicb.2024.1465455
Received: 16 July 2024; Accepted: 07 October 2024;
Published: 25 October 2024.
Edited by:
Astghik Zaveni Pepoyan, Armenian National Agrarian University, ArmeniaReviewed by:
Natalya Harutyunyan, Armenian National Agrarian University, ArmeniaMudasir Rashid, Howard University Hospital, United States
Copyright © 2024 Yu, Yin, Deng, Yang, Bai and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Yu, eXVyb25nMTk2OTA1QDE2My5jb20=
 Yunfeng Yu
Yunfeng Yu Yuman Yin1
Yuman Yin1 Juan Deng
Juan Deng Xinyu Yang
Xinyu Yang Siyang Bai
Siyang Bai Rong Yu
Rong Yu