- Department of Bioscience and Technology for Food, Agriculture and Environment, University of Teramo, Teramo, Italy
Introduction: The cheese microbiota is very complex and is made up of technologically-relevant, spoilage, opportunistic and pathogenic microorganisms. Among them lactic acid bacteria and yeasts are the main ones. One of the most interesting dairy yeasts is Kluyveromyces marxianus because of its technological properties including the ability to produce aroma compounds.
Methods: This study investigated the contribution of Kluyveromyces marxianus to the gross composition and aroma profile of cow cheeses. Experimental cheeses were prepared by inoculating a co-culture of K. marxianus FM09 and a commercial strain of Lacticaseibacillus casei and compared with cheeses obtained with only L. casei. The gross composition was determined by a FoodScan™ 2 Dairy Analyser, and free amino acids were evaluated at 507 nm after reaction with Cd-ninhydrin. The volatile organic compounds were extracted by head-space solid phase micro-extraction and analyzed by gas chromatography–mass spectrometry coupled with odor activity values. qRT-PCR was applied to determine the expression of genes involved in esters synthesis and degradation.
Results: The inoculation of K. marxianus induced an increase of pH and a reduction of protein content of cheeses, in agreement with the stronger proteolysis detected in these cheeses. K. marxianus influenced the content of aroma compounds both quantitatively and qualitatively. In particular, an increase of higher alcohols, esters and organic acids was observed. Moreover, 12 compounds were detected only in cheeses obtained with the co-culture. These differences were in agreement with the odor activity values (OAV). In fact, only 11 compounds showed OAV > 1 in cheeses obtained with the commercial strain, and 24 in those obtained with the co-culture. The qPCR analysis revealed an over expression of ATF1, EAT1, and IAH1 genes.
Conclusion: Kluyveromyces marxianus could act as an important auxiliary starter for cheese production through the development and diversification of compounds related to flavor in short-aged cow cheeses.
1 Introduction
The global cheese production is consistently growing, with the global cheese market expected to reach $287.12 billion by 2032, increasing from $191.94 billion in 2024, at a compound annual growth rate (CAGR) of 4.61% during the forecast period. The dairy industry has to meet the growing consumer demand for dairy products with unique flavors. Cheese flavor is one of the most relevant attributes influencing consumers’ acceptance and preference (Arora et al., 1995). The taste and aroma of cheese are determined by a complex combination of volatile and non-volatile components. Each compound contributes to the overall flavor, but cannot fully represent it on its own (Padilla et al., 2014).
The characteristics of cheese are influenced by several factors, including the type of milk used, the diet of the animals, the ripening conditions, the length of ripening, and the microbiota (Mayo et al., 2021). Microorganisms play a crucial role in the fermentation process and in the biochemical reactions that take place during the manufacturing and ripening processes. The cheese microbiota consists of a diverse group of microorganisms, including prokaryotes, eukaryotes, and viruses. Among them, lactic acid bacteria (LAB) are the most abundant and play a significant role in the production and aging of cheese. However, yeasts represent an important component of the microbiota of several cheese varieties (Bintsis, 2021). The occurrence of yeasts in these products is related to their ability to develop at low temperatures, assimilate/ferment lactose, and assimilate organic acids, besides proteolytic and lipolytic activities and resistance against high salt concentrations (Bintsis, 2021; Padilla et al., 2014). Yeasts are involved in the increase in pH on the cheese surface, which is caused by the metabolization of lactate into CO2 and H2O; and they use amino acids as energy sources after lactate exhaustion, producing considerable amounts of ammonia (Padilla et al., 2014). Nevertheless, yeasts can also induce cheese deterioration through early gas production, creating numerous small holes. Yeasts may also cause pink discoloration, brown patches, and deacidification of the cheese surface when they catabolize amino acids to produce NH3 (Bintsis, 2021; Zheng et al., 2021).
One of the most interesting dairy yeasts is Kluyveromyces marxianus. This yeast is thermotolerant, since it has been shown to grow at temperatures as high as 42°C. However, it is also able to develop at refrigerated temperatures (Fonseca et al., 2008; Tofalo et al., 2014). Kluyveromyces marxianus strains affect the ripening of cheese by their proteolytic and lipolytic activities, as well as the formation of volatile aroma compounds (Binetti et al., 2013; Padilla et al., 2014). This yeast species metabolizes lactose as carbon source due to the expression of the LAC12 and LAC4 genes, which code for a lactose permease and a β-galactosidase, respectively (Varela et al., 2017). The strains metabolize the lactose to generate ethanol and carboxylic acids, which are then converted into esters (resulting in a fruity flavor) and acetaldehyde. K. marxianus has been linked to the development of acidic, cidery, alcoholic, fermented, and fruity flavors (Zheng et al., 2021). This species has also acquired the QPS (Qualified Presumption of Safety) and GRAS (Generally Recognized as Safe) status from the European Food Safety Authority (EFSA) (EFSA Panel on Additives and Products or Substances used in Animal Feed [FEEDAP], 2004; Wang et al., 2018; Perpetuini et al., 2020).
As stated above, yeasts have a beneficial effect on the development of cheese aroma, which has generated commercial interest in utilizing specific strains as starter cultures together with LAB (Fröhlich-Wyder et al., 2019). In this study the effect of K. marxianus on cow cheese aroma profile was investigated. K. marxianus FM09 was used to make experimental cheese, with cheese made using a LAB commercial starter being used as the control group. The gross composition and the volatile profile of the cheeses were determined. Moreover, the expression of genes involved in esters synthesis and degradation in K. marxianus was evaluated.
2 Materials and methods
2.1 Strains used in this study
A commercial LAB strain of Lacticaseibacillus casei (CC) and the K. marxianus FM09 strain were used. K. marxianus FM09 was isolated from fermented milk (Fasoli et al., 2016). FM09 and CC strains were routinely grown in YPD or MRS, respectively. Strains were stored at −80°C in YPD or MRS broth supplemented with glycerol (20% v/v final concentration). Both strains belong to the Culture Collection of the Microbial Biotechnology Laboratory (University of Teramo, Teramo, Italy).
2.2 Cheesemaking procedure
Cheeses were manufactured on a laboratory scale under aseptic conditions. The cow pasteurized milk (72°C for 15 s) was distributed in 5 L containers. Three cheesemaking trials were carried out in 3 different days using the milk collected that day. In each trial, two cheese batches were simultaneously produced using two different starter combinations: the first inoculating only the CC and the second with a co-culture of CC and K. marxianus FM09. Therefore, a total of 6 trials were obtained, 3 inoculated with CC and 3 with CC + FM09. To prepare the inoculum for cheesemaking, the strains were inoculated in pasteurized cow milk incubated at 30°C for 48 h and inoculated at a final 7 Log CFU/mL concentration. A commercial bovine rennet was added according to the manufacturer’s instructions. After 15 min at 37°C the curd was cut and the whey was eliminated. The containers were stored at 15°C for 30 days.
2.3 Gross composition of cheeses
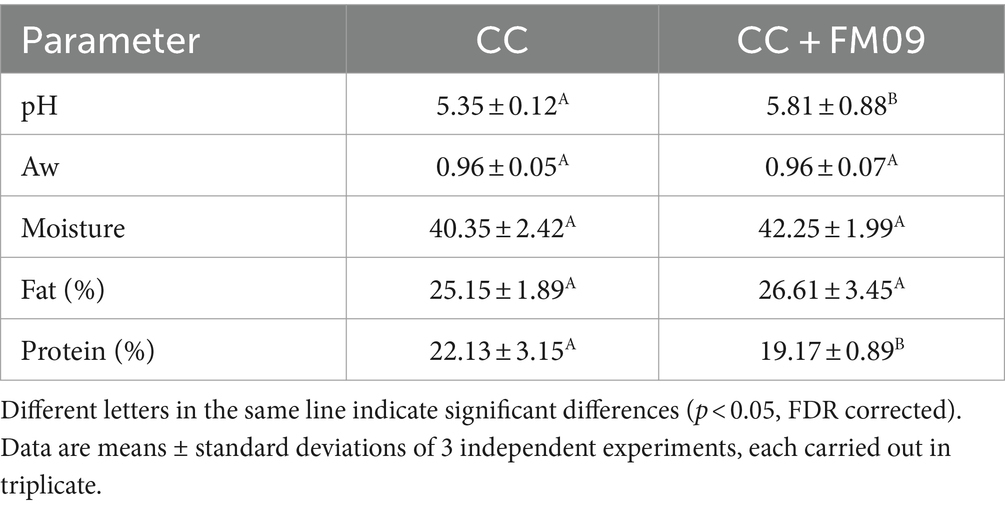
The main chemical characteristics (moisture, fat, and protein) were determined by a FoodScan™ 2 Dairy Analyser (Foss, Padova, Italy). The pH was determined by the pHmeter (Mettler Toledo, Milan, Italy) and the Aw was determined by Water Lab (Steroglass, Perugia, Italy) in agreement with the manufacturer’s instructions. Free amino acids (FAAs, expressed as mg leucine/g) were evaluated at 507 nm after reaction with Cd-ninhydrin according to Folkertsma and Fox (1992). Three technical replicates were performed on each sample.
2.4 Microbiological analysis
For the microbiological analysis, serial dilutions in sodium citrate (2% w/v) were prepared starting from 10 g of cheese. In both trials yeasts and LAB were enumerated. Yeasts were enumerated on Yeast Peptone Dextrose Agar (YPD; 1% w/v yeast extract, 2% w/v peptone, 2% w/v glucose and 2%w/v agar; Oxoid, Milan, Italy) supplemented with chloramphenicol (150 mg/L) at 28°C for 2 days, while LAB on DeMan-Rogosa-Sharp Agar (MRS) (Oxoid), acidified to pH 5.4 with acetic acid, and supplemented with 100 ppm cycloheximide (Merk Life Science S.r.l., Milan, Italy). Plates were incubated at 30°C for 2 days in anaerobic conditions using the Gas-Pack anaerobic system (AnaeroGen; Oxoid). Cell counts were performed in triplicate on each sample.
2.5 Determination of aroma compounds
Solid-phase microextraction (SPME) was used to extract volatile organic compounds (VOCs) according to Li et al. (2022). The gas chromatography–mass spectrometry (GC–MS) analysis was carried out using a gas chromatograph (Clarus 580; Perkin Elmer, Waltham, MA, United States) coupled with a mass spectrometer (SQ8S; Perkin Elmer). The gas chromatograph was equipped with an Elite-5MS column (length × internal diameter: 30 × 0.25 mm; film thickness: 0.25 μm; Perkin Elmer). An 85 μm fiber coated with carboxen-polydimethylsiloxane (Sigma-Aldrich, Milan, Italy) was used. Aroma components were identified by comparing the retention times of pure reference standards tested under the same conditions. The following standards were selected according to Sandoval-Copado et al. (2018) and Padilla et al. (2014): diacetyl, 2,3-pentanedione, 2-butanone, 2-nonanone, 2-hexanone, acetic acid, lactic acid, butyric acid, isovaleric acid, valeric acid, acetoin, propionic acid, decanoic acid, hexanoic acid, furfural, acetaldehyde, decanal, butyraldehyde, δ-decalactone, γ-dodecalactone, δ-dodecalactone, 1-octanol, 2,3-butanediol, 2-phenylethanol, isoamyl alcohol, 1-hexanol, ethyl hexanoate, phenylethyl acetate, isoamyl acetate, ethyl octanoate, diethyl succinate, ethyl acetate, ethyl butanoate. The standard solutions were purchased from Merk Life Science S.r.l (Mila, Italy) and were of the highest purity available. 2-Methyl-hexanol was used as internal standard. A minimum similarity criterion of 85% was used to compare mass spectra with MS fragmentation patterns in the National Institute for Standards and Technology database (NIST version 2005) in order to identify the compounds provisionally. The volatile compounds were quantified using the calibration curve of standards. The analyses were performed in triplicate. The odor activity values (OAV) were used to describe the intensity of odor compounds in the sample. The OAV is the ratio of a single compound’s concentration to that compound’s odor threshold. When a compound has OAV > 1, it is considered to contribute to the overall aroma of the sample. The analyses were performed in triplicate.
2.6 qRT-PCR analysis
RNA was extracted using RNeasy PowerSoil Total RNA Kit (Qiagen, Mila, Italy) according to the manufacturer’s instructions in order to determine the expression of the following genes: ATF1, EAT1, and IAH1. RNA concentration was determined spectrophotometrically with a Nanodrop spectrophotometer ND-1000 and 1 μg total RNA was retrotranscribed into cDNA, using the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s instructions. qRT-PCR was performed according to Muñoz-Miranda et al. (2024). Briefly, the following primer pairs were used:
IAH1_F 5’ CAGCGGAAAGGGTTACGAAG 3′ and IAH1_R 5’ AGGGGTCAAGAGTAGAGCCA 3′, ATF1_F 5’ AAAATCCCAAG AAACGAACCCA 3′ and ATF1_R 5’ TACGGCATCTCTGTCCCCG 3′, EAT1_F 5’ GCCCTTGTGAAATCTTGCGTT 3′ and EAT1_R 5’ CCGAGGCTTTGTGTCTCCATAG 3′. The qRT-PCR mixture (25 μL) contained: 12.5 μL SYBR™ Green PCR Master Mix (Thermo Fisher Scientific, Milan, Italy), 1 μL of template (100 ng/μL), 0.25 μL of each primer (10 μM) and diethylpyrocarbonate-treated water (DEPC-water). The thermal cycling conditions were as follows: 95°C for 10 min, 40 cycles of 30 s at 95°C, and 60 s at 60°C. ALG9 (coding for alpha-1,2-mannosyltransferase, ALG9_F 5’ CCATCTCAGGATCCC TCTTC 3′, ALG9_R GCATTCCAGCGAATAGTTGA 3′) and ACT1 (coding for actin, ACT1_F 5’ GGCTGAACGTGGTTACTCCT 3′ and ACT1_R 5’ AGAAGCGGTTTGCATTTCTT 3′) were used as reference genes (Perpetuini et al., 2019). qRT-PCR was performed using an iCycler IQ realtime PCR Detection System (Bio-Rad, Milan, Italy). Melting temperature analysis of the PCR products was performed to determine the specificity of the PCR reactions. Calculation of relative transcript levels (RTLs) was carried out using the comparative Ct method [2–ΔCt gene – ΔCt reference gene], as described by Livak and Schmittgen (2001). All analyses were performed in triplicate.
2.7 Statistical analysis
Prism 7.0 program (GraphPad Software Inc., La Jolla, CA, United States) was used to analyze data and prepare graph. Results were expressed as mean value ± standard deviation. t-test was used to determine significance (p < 0.05) concerning the gross composition and volatile compounds. In order to avoid increased chance of false positive for the comparison between CC and FM09 + CC, multiple comparison correction, accounting for the main volatile compounds, was performed through the False Discovery Rate (FDR) algorithm (Genovese et al., 2002).
3 Results
3.1 Gross composition and microbial count of cheeses
The physico-chemical composition of cheeses is reported in Table 1. The main differences were detected for the pH and the protein content. In particular, K. marxianus FM09 induced an increase of pH, probably because of the lactic acid utilization by this yeast, and a reduction of protein content in agreement with the stronger proteolysis detected in these cheeses (Figure 1). Figure 1 showed the evolution of FAAs during the ripening period. The FAAs content increased significantly during ripening reaching a concentration of 36.98 mg leucine/g and 51.87 mg leucine/g in cheeses obtained with CC and CC + FM09, respectively.

Figure 1. Evolution of free amino acids throughout ripening in cheeses produced with the commercial starter (CC) and CC + K. marxianus FM09. Data are means ± standard deviations of 3 independent experiments, each carried out in triplicate. * p<0.05.
The addition of K. marxianus FM09 did not influence the other parameters which did not show significant differences.
In both cheeses the number of LAB increased during the first 15 days and then decreased. In particular, the number of LAB reached a value of 8.8 Log CFU/g and 9.15 Log CFU/g, in cheeses obtained with CC and CC + FM09, respectively. After 30 days of ripening the number of LAB was about a Log higher in the cheeses obtained with the co-culture (8.71 Log CFU/g) than in the others (7.92 Log CFU/g) (Figure 2). The number of yeasts reached the highest concentration after 15 days (8.2 Log UFC/g), and decreased after 30 days of ripening (7.45 Log CFU/g) (Figure 2). The yeasts were absent in cheeses obtained with CC.
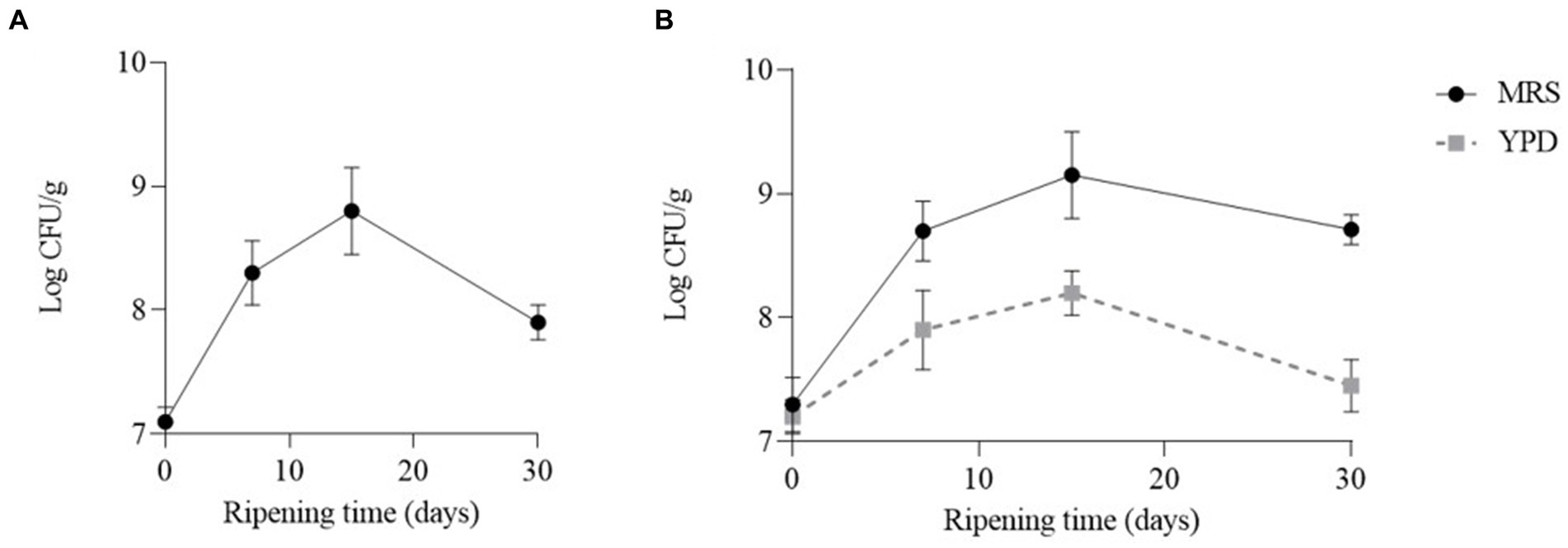
Figure 2. Evolution of lactic acid bacteria and yeasts in the cheeses obtained with CC (A) and CC + FM09 (B). Data are means ± standard deviations of 3 independent experiments, each carried out in triplicate.
3.2 Aroma compounds
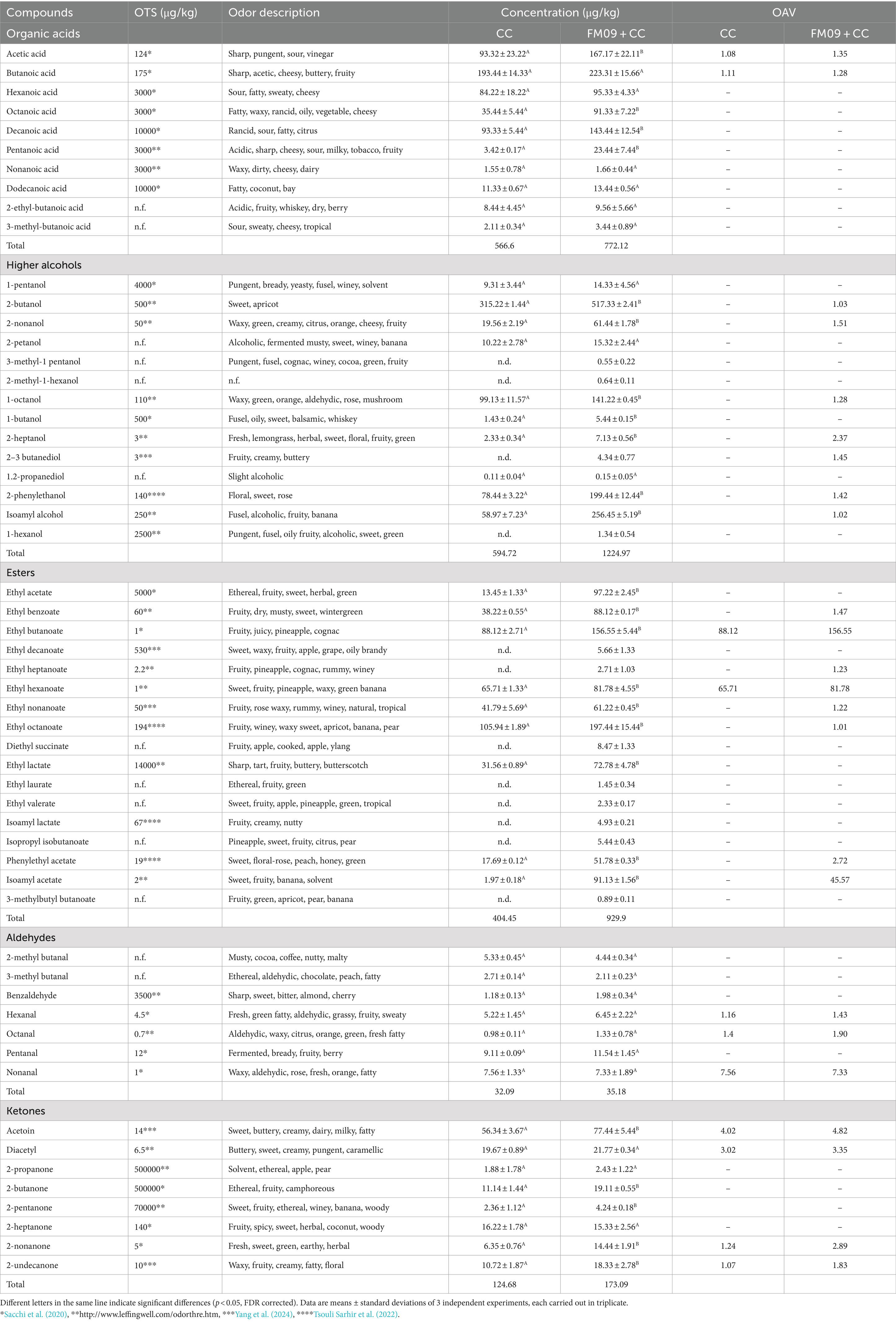
A total of 55 volatile compounds were detected in the cheeses, including 10 organic acids, 13 higher alcohols, 17 esters, 7 aldehydes, and 8 ketones (Table 2). The content of higher alcohols was higher in cheeses obtained with FM09 + CC (1,079 μg/kg) than in the others (660.19 μg/kg). The most abundant were isoamyl alcohol (fusel, alcoholic, whiskey, fruity, banana), 1-octanol (waxy, green, orange, aldehydic, rose, mushroom), 2-butanol (sweet, apricot), and 2-phenylethanol (floral, sweet, rose). Their concentrations were 58.97 ± 7.23 μg/kg and 56.45 ± 5.19 μg/kg, 99.13 ± 11.57 μg/kg and 141.22 ± 0.45 μg/kg, 315.22 ± 1.45 μg/kg and 517.33 ± 2.41 μg/kg, 78.44 ± 3.22 μg/kg and 199.44 ± 12.44 μg/kg, in cheeses obtained with CC and FM09 + CC, respectively.
The inoculation of K. marxianus induced an increase of esters’ content. In both cheeses ethyl esters were more abundant than acetate ones. These compounds were mainly present in cheeses obtained with CC + FM09. The main esters detected were ethyl butanoate (88.12 ± 2.71 μg/kg and 156.55 ± 5.44 μg/kg in cheeses obtained with CC and FM09 + CC, respectively), ethyl hexanoate (65.71 ± 1.33 μg/kg and 81.78 ± 4.55 μg/kg in cheeses obtained with CC and FM09 + CC, respectively), ethyl octanoate (105.94 ± 1.89 μg/kg and 197.44 ± 15.44 μg/kg in cheeses obtained with CC and FM09 + CC, respectively).
Acetic (93.32 ± 23.22 μg/kg and167.17 ± 22.11 μg/kg in cheeses obtained with CC and FM09 + CC, respectively) and butanoic acids (193.44 ± 14.33 μg/kg and 223.31 ± 15.66 μg/kg in cheeses obtained with CC and FM09 + CC, respectively) were the main acids detected, followed by hexanoic (84.22 ± 18.22 μg/kg and 95.33 ± 4.33 μg/kg in cheeses obtained with CC and FM09 + CC, respectively), and decanoic (93.33 ± 5.44 μg/kg and 143.44 ± 12.54 μg/kg in cheeses obtained with CC and FM09 + CC, respectively) acids.
Aldehydes were present in similar concentration in both cheeses (32.09 μg/kg in CC cheeses and 35.18 μg/kg in FM09 + CC cheeses). The main aldehydes detected were 2-methylbutanal (5.33 ± 0.45 μg/kg and 4.44 ± 0.34 μg/kg in cheeses obtained with CC and FM09 + CC, respectively), hexanal (5.22 ± 1.45 μg/kg and 6.45 ± 2.22 μg/kg in cheeses obtained with CC and FM09 + CC, respectively), and nonanal (7.56 ± 1.33 μg/kg and 7.33 ± 1.89Aμg/kg in cheeses obtained with CC and FM09 + CC, respectively).
The inoculation of K. marxianus FM09 influenced the content of ketones, in fact the cheeses made only with CC had a ketone content approximately 50 μg/kg lower than the others. The main ketones detected were acetoin (56.34 ± 3.67 μg/kg and 77.44 ± 5.44 μg/kg in cheeses obtained with CC and FM09 + CC, respectively) and diacetyl (19.67 ± 0.89 μg/kg and 21.77 ± 0.34 μg/kg in cheeses obtained with CC and FM09 + CC, respectively).
The Venn diagram combined with the calculation of OAV method can explore the unique volatile compounds of cheeses obtained with CC and CC + FM09. The Venn diagram showed that the cheeses shared 44 compounds, while 12 were found only in cheeses obtained with CC + FM09 (Figure 3). This data suggested a specific effect of the inoculated yeast in the definition of cheese aroma; and this effect was particularly evident for esters. In fact, 8 esters out of 17 were specifically associated to the presence of K. marxianus FM09.
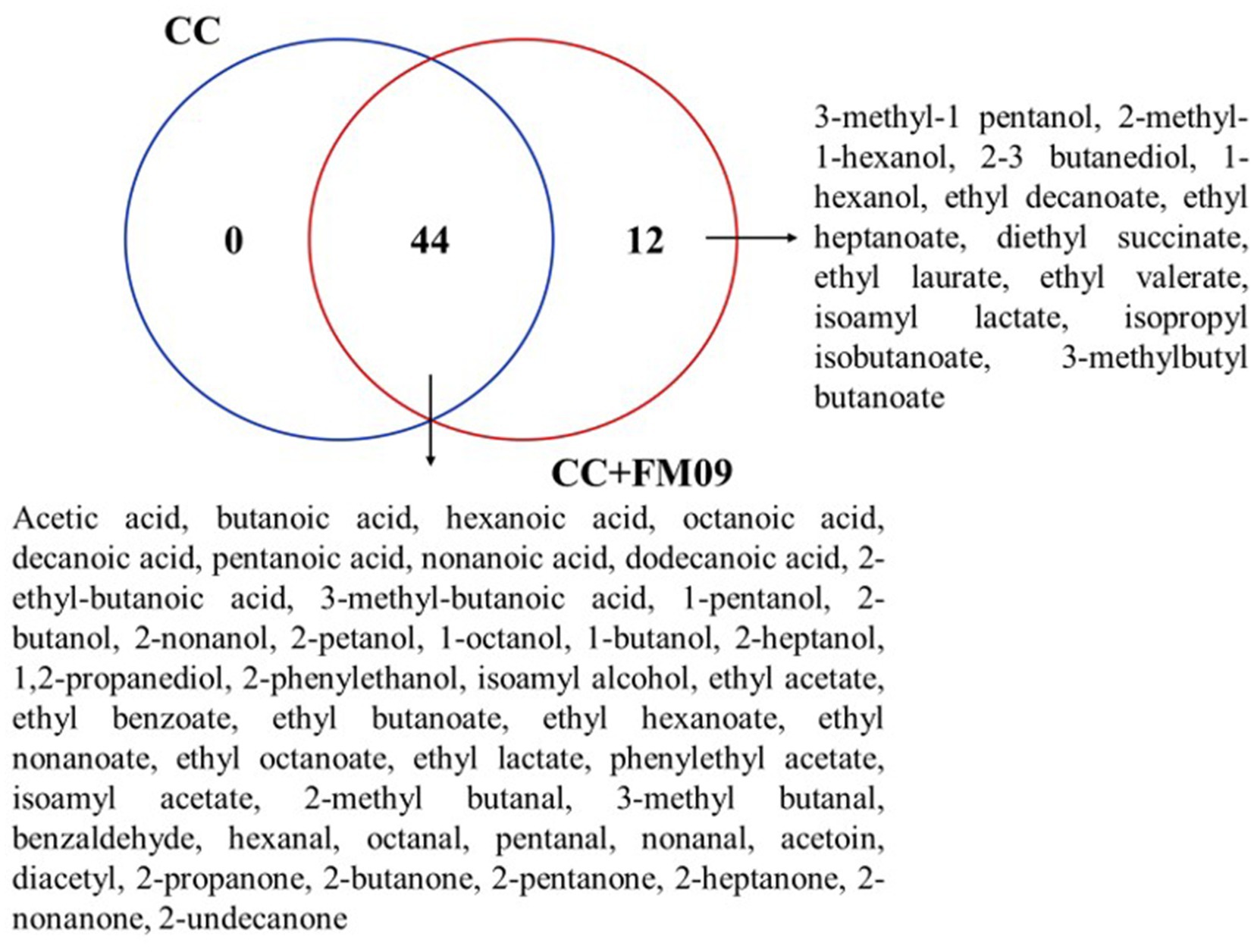
Figure 3. Venn diagram showing the similarities and differences of volatile compounds between cheese samples The Venn diagram was generated with the web tool provided by the Bioinformatics and Systems Biology of Gent.
The contribution of aroma is not only determined by the concentration but also by the odor threshold values. Therefore, the OAV were calculated to further analyze the contributions of aroma-active compounds (Table 2). Cheeses obtained with CC + FM09 showed a richer flavor than the control. In fact, only 11 compounds showed OAV > 1 in cheeses obtained with CC, and 24 in those obtained with CC + FM09. Moreover, OAV were higher in these last cheeses. This result suggested once again the effect of K. marxianus on cheese aroma profile. The main differences were detected for higher alcohols, and esters. In particular, 2-butanol, 2-nonanol, 1-octanol, 2-heptanol, 2,3-butanediol, 2-phenylethylalcohol and isoamyl alcohol showed OAV >1 only in cheeses produced with FM09 + CC. Two (ethyl butanoate and ethyl hexanoate) and 8 (ethyl benzoate, ethyl butanoate, ethyl heptanoate, ethyl hexanoate, ethyl nonanoate, ethyl octanoate, phenylethyl acetate, and isoamyl acetate) esters showed OAV > 1 in cheeses produced with CC and FM09 + CC, respectively. The majority of compounds with OAV > 1 detected in cheeses obtained with CC + FM09 showed fruity notes, and the main contributors were ethyl butanoate, ethyl hexanoate, and isoamyl acetate. The evaluation of aroma characteristics other than fruity, showed that cheeses produced with FM09 + CC showed higher buttery notes than the others, due to the contribution of acetoin, diacetyl and 2,3-butanediol. Moreover, these cheeses were also characterized by green and fresh aroma thanks to the OAV of 2-heptanol, hexanal, octanal, 1-octanol, 2-nonanol.
3.3 qPCR analysis
Kluyveromyces marxianus mainly influenced the content of esters, therefore the expression of some genes involved in their production was analyzed. Since the activity of ester-synthesizing enzymes and ester-hydrolyzing enzymes are related to the ester accumulation the following genes were tested: ATF1 and EAT1 genes encode for alcohol acetyl-transferases (AATase), and IAH1 gene for an esterase. ATF1 and EAT1 genes were upregulated and showed a fold change of 4.96 and 6.19, respectively (Figure 4). The gene IAH1 was upregulated, with a fold change of 3.34.
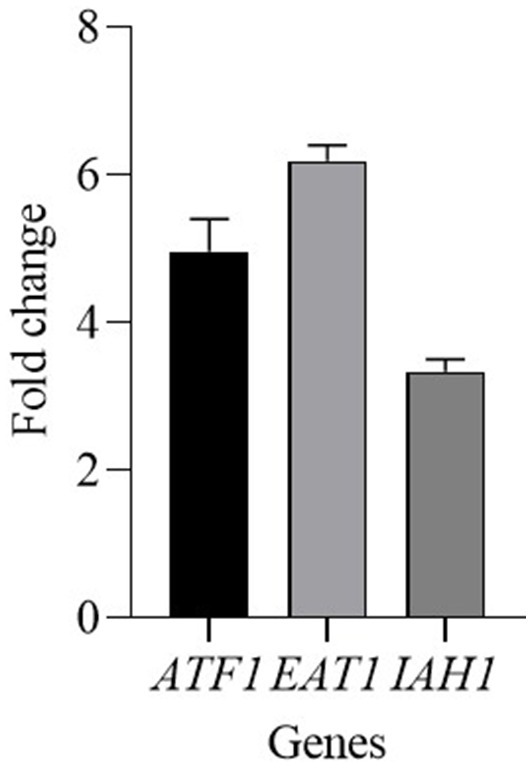
Figure 4. Relative transcript levels of genes involved in ester content regulation in K. marxianus after 30 days of ripening of cheeses made with CC + FM09. Transcript levels are expressed as x-fold increase in comparison with the expression at T0. Three biological replicates were performed, with 3 technical repeats.
4 Discussion
This study investigated the effect of K. marxianus FM09 on cow cheese characteristics, and the metabolic potential of this strain to impact cheese flavor. The inoculation of FM09 induced an increase of pH, probably because of the lactic acid utilization by this yeast, and favored the proteolysis. A similar result concerning pH was obtained by Pisano et al. (2020) when K. lactis was used as adjunct culture for Caciotta cheese. K. marxianus participates in deacidification of the curd through lactate/lactose consumption with a notable increase in pH that enables the acid-sensitive bacteria to develop at the cheese surface (Zheng et al., 2021). The higher proteolysis detected in cheeses made with FM09 + CC is in agreement with Xiao et al. (2020) who used K. marxianus A2 as adjunct culture for the production of Kazak cheeses. The authors highlighted that this yeast contributed to the formation of FAAs, in fact the cheeses showed significant higher amounts of amino acids, except for valine and leucine, than the other cheeses obtained with Pichia fermentans A19 and P. kudriavzevii A11. The proteolytic activity of K. marxianus, due to peptidases like aminopeptidases and carboxypeptidases, is well known and could result in great amounts of volatile compounds, contributing, thus, to the development of aroma and flavor of cheese (Tofalo et al., 2014; Xiao et al., 2020). Moreover, it should be postulated a synergistic effect between K. marxianus and LAB resulting in stronger proteolysis. A similar effect has been observed between D. hansenii and LAB (Ferreira and Viljoen, 2003). The highest proteolysis detected in these cheeses can also explain the increase of pH. In fact, the acids can be neutralized by some alkaline substances from protein decomposition, which will cause the increase of pH (Gori et al., 2007). The other parameters were similar to those obtained for mountain Caciotta cheese (Cardin et al., 2024).
The microbiological analyses revealed that L. casei reached a higher cell concentration in presence of K. marxianus FM09. Similar results were obtained by Huang et al. (2020) who noticed that the population of total LAB was significantly higher in multi-starters fermentation system (Streptococcus thermophilus + K. marxianus and Lactobacillus delbrueckii spp. bulgaricus + K. marxianus) (log 9.65 CFU/mL) than single culture (log 9.45 CFU/mL). Probably the metabolites produced by yeast decomposition of the substrate such as amino acids and vitamins are used by LAB to further promote growth (Zhang et al., 2017). Meanwhile, some LAB metabolites also provide an energy source for yeast growth (Zhang et al., 2017). In this study, the combination of this species/strains was useful for the production cow cheeses since non-competitive or inhibitory effects between these 2 strains were detected.
The inoculation of K. marxianus influenced the volatile profile of cheeses. In fact, the cheeses obtained with FM09 + CC showed qualitative and quantitative differences if compared to cheeses obtained only with CC. The content of organic acids was increased of 37%, and the main acids interested were acetic, butanoic, octanoic, decanoic and pentanoic ones (Table 2). Carboxylic acids can be originated from three main biochemical pathways: (1) lipolysis (hydrolysis of triglycerides into free fatty acids, such as butanoic, pentanoic, hexanoic, heptanoic, octanoic, nonanoic, decanoic and undecanoic acids); (2) proteolysis (breakage of caseins into peptides and amino acids, such as 2-methylpropanoic and 3-methylbutanoic acids); and (3) lactate metabolism (acetic and propanoic acids) (Delgado et al., 2011).
Kluyveromyces marxianus is known to produce a variety of organic acids. These include acetic acid, propionic acid, butanoic acid, octanoic acid, among others (Pentjuss et al., 2017; Fasoli et al., 2015; Cheon et al., 2014). The highest increase was recorded for octanoic acid (160%) and acetic acid (79%). Acetic and butanoic acids contributes to the typical flavor of most types of cheese, giving rise to vinegar-like, fruity, and sharp odors (Zheng et al., 2017). In particular, butanoic acid has a rancid cheese-like odor and plays an important role in the flavor of many cheese types such as Camembert, Cheddar (aged, regular and low fat), Grana Padano, Gruyere, Pecorino, Ragusano and Roncal cheese (Curioni and Bosset, 2002). Hexanoic and decanoic acids have been detected in Grana Padano and Roncal cheeses (Curioni and Bosset, 2002). The increase detected in this study is in agreement with previous observations. Acetic acid is the main organic acid produced by K. marxianus, in fact its accumulation during exponential growth phase is typical for K. marxianus fermentations (Pentjuss et al., 2017; Löser et al., 2013; Signori et al., 2014). The production of octanoic acid has been already reported by other authors (Zheng et al., 2017; Fasoli et al., 2015). In particular, Fasoli et al. (2015) detected an increase of octanoic acid concentration when K. marxianus was grown in raw cheese whey (the aqueous part of milk that remains after the separation of the curd during cheese making) containing 4.8% lactose, 1.2% proteins and 0.75% fats.
The presence of K. marxianus induced an increase of alcohol content of 63%. All detected alcohols were more present in cheeses obtained with CC + FM09; and this increase was particularly evident for 2-phenylethanol (109%). Kluyveromyces marxianus is able to produce various alcohols. For instance, it has been utilized for ethanol production from crude whey, or furfuryl alcohol from furfural (Gethins et al., 2015). Moreover, alcohol dehydrogenases from K. marxianus have been investigated, showing the yeast’s capability to produce different alcohols with various chain lengths (Liang et al., 2014). The production of 2-phenylethanol has been largely demonstrated in K. marxianus (Fasoli et al., 2015; Perpetuini et al., 2022; Gethins et al., 2015; Alonso-Vargas et al., 2022). This compound is produced from the catabolism of L-phenylalanine via the Ehrlich pathway or by de novo synthesis from sugars, through the Shikimate pathway (Wittmann et al., 2002). Some studies demonstrated that cheese whey represented a good carbon source for 2-phenylethanol production by K. marxianus (Alonso-Vargas et al., 2022; Fasoli et al., 2015; Han et al., 2024). In particular, Han et al. (2024) demonstrated that co-culture of Pediococcus lactis and K. marxianus improved the metabolic capacity for producing 2-PE. This may be because P. lactis effectively activated the Ehrlich pathway of K. marxianus and enhanced the robustness of K. marxianus through improving the fluidity and stability of cell membranes.
The ethyl esters were the main esters detected in this study in both cheeses. They are considered to be major contributors to overall flavor and were the most abundant esters in Grana Padano cheese (Moio and Addeo, 1998), in Brie cheese (Karahadian et al.,1985), in bovine Mozzarella cheese (Moio et al., 1993), and in Parmesan cheese (Nursten, 1997), and could contribute to the aroma of cheese by minimizing the sharpness and bitterness derived from carboxylic acids (Curioni and Bosset, 2002). The use of K. marxianus induced an increase of esters content of 174%. All esters showed a higher concentration in cheeses obtained with CC + FM09 than in the others. This result is in agreement with the recognized ability of K. marxianus to produce esters (Morrissey et al., 2015; Reyes-Sánchez et al., 2019). The most common esters produced by K. marxianus are 2-phenylethyl acetate, isoamyl acetate, and ethyl acetate. These esters are synthesized from 2-phenylethanol, isoamyl alcohol, and ethanol, respectively (Morrissey et al., 2015). Their content showed the highest percentage increase in cheeses obtained with K. marxianus. This data is in agreement with the content of higher alcohols detected. In fact, according to Morrissey et al. (2015) the conditions that give rise to elevated levels of higher alcohols thus also provide the circumstances in which elevated levels of acetate esters are possible. The qPCR analysis was applied to better demonstrate the influence of K. marxianus FM09 on esters content. The increased fold change detected for ATF1 and EAT1 genes is in agreement with previous observations which showed that their deletion induced a reduction of ethyl acetate, isoamyl acetate, and 2-phenylethyl acetate (Löbs et al., 2018). The highest expression of EAT1 gene is in agreement with Löbs et al. (2018) who revealed the key role played by this gene in K. marxianus esters production. In fact, its deletion caused a reduction of esters content of more than 80%. Concerning the over expression of IAH1 gene, it should be noted that the role of IAH1 gene in ester metabolisms is not clear. In fact, some authors supposed that IAH1 could have an ester synthase activity (Vidal et al., 2013; Löser et al., 2014; Löbs et al., 2017; Muñoz-Miranda et al., 2024; Dank et al., 2018). In particular, Dank et al. (2018) hypothesize that IAH1 gene may have promiscuous activity on ethyl esters, resulting in a redirection of intracellular fluxes of amino acid catabolism intermediates to ethyl esters. Moreover, it should be noted that the accumulation of esters relies on the balance between esters synthesis and degradation (van Rijswijck et al., 2019). Dank et al. (2018) showed that the deletion of the IAH1 and TIP1 genes in S. cerevisiae results in an intracellular metabolite imbalance, triggering a decrease in ester substrates, a reduction in ester synthase activity to compensate for the decreased hydrolyzing activity, or an upregulation of promiscuous esterases.
The inoculation of K. marxianus favored the accumulation of ketones in the cheese. Ketones are considered key component of the flavor of several cheeses (Nogueira et al., 2005) and are generally produced by the enzymatic oxidative decarboxylation of fatty acids by LAB. The main ketones detected were acetoin and diacetyl. Acetoin can be derived from diacetyl metabolism (Rehman et al., 2000) or from pyruvate metabolism during the conversion of lactose to lactic acid (Nogueira et al., 2005). It is characterized by buttery notes, and has been found in several cheeses above the aroma threshold (Sablé and Cottenceau, 1999). Diacetyl is obtained from pyruvate stemming from lactose and citrate metabolism. It is appreciated for its buttery and nut-like notes and was identified as a key aroma component of Camembert, Cheddar and Emmental (Curioni and Bosset et al., 2002). K. marxianus ability to produce ketones has been shown also by Risner et al. (2019). These authors found differences in ketones production by K. marxianus between acid and sweet whey distillates. Sweet whey distillate contained significantly greater concentration of ketones than the others, and 2-Heptanone, 2-pentadecanone, 2-tridecanone, and 2-undecanone were identified exclusively in these distillates. Perpetuini et al. (2022) reported that the production of ketones was influenced by K. marxianus aggregation state, and they were mainly produced by sessile cells and the main ketones detected were 2-nonanone, and 8-hydroxyoctan-2-one.
Aldehydes are transitory compounds in cheese because they are rapidly reduced to primary alcohols or even oxidized to the corresponding acids (Curioni and Bosset, 2002). However, they can have an important impact on cheese aroma since they have low threshold values. It should be noted that their contribution to cheese aroma depends not only on the concentration of single compound, since a synergistic effect can be also present. For example, 2-methylbutanal and 3-methylbutanal, both found in this study, can have a synergistic effect. 2-methylbutanal produced malty, cacao, and apple-like aromas, while 3-methylbutanal produced malty, coffee, and cacao aromas. Therefore, the mixture of these compounds emitted a pleasant nutty and malty odor with a threshold of 29.29% of the single compound thresholds (Chen et al., 2020). Both cheeses were characterized by the presence of pentanal, hexanal, and nonanal. These aldehydes are commonly found in cheeses (Grana Padano, Mozzarella, among others) and confer green grass and herbaceous aromas (Curioni and Bosset, 2002). Such compounds may also result from β-oxidation of unsaturated fatty acids or even light-induced reactions, although such reactions are not likely to occur in cheese (Borle et al., 2002). K. marxianus can produce these compounds as part of its metabolic processes for xylitol, ethanol, lactate, and aroma compounds production (Baptista and Domingues, 2022). Additionally, the YGL157w gene product of Kluyveromyces marxianus DMB1 strain has been characterized as a broad specificity NADPH-dependent aldehyde reductase, indicating its involvement in aldehyde metabolism (Akita et al., 2015). The obtained results suggested a synergistic effect between K. marxianus and L. casei in terms of aroma compounds production. A synergistic effect between K. marxianus and LAB (L. helveticus and L. delbrueckii subsp. bulgaricus) concerning the increase of lactic acid production has been already reported especially (Plessas et al., 2008; Lee, 2005). This positive effect between yeast and LAB could be due to the fact that yeasts provide LAB with growth factors such as vitamins that promote growth and consequently leading to increased lactic acid production, while the yeasts uses the bacterial end-products of LAB as energy sources (Plessas et al., 2008).
This study established the effect of K. marxianus FM09 in flavor development and gross composition of short-aged cow cheese. The inoculation of this yeasts induced an increase of esters, higher alcohols, organic acids, and ketones, as well as the production of key volatile compounds was observed. These results demonstrated the potential of K. marxianus FM09 as co-adjunct culture even if further studies are necessary to better investigate the interactions of the strains used and the cheese microbiota. The metabolic pathways of K. marxianus involved in the definition of cheese characteristics should be highlighted using multi -omics approaches.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
GP: Writing – review & editing, Data curation, Writing – original draft. ARo: Writing – review & editing, Formal analysis, Investigation, Methodology. ARa: Investigation, Methodology, Formal analysis, Writing – review & editing. RT: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The publishing fees were supported by University of Teramo.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akita, H., Watanabe, M., Suzuki, T., Nakashima, N., and Hoshino, T. (2015). Characterization of the Kluyveromyces marxianus strain DMB1 YGL157w gene product as a broad specificity NADPH-dependent aldehyde reductase. AMB Expr. 5:17. doi: 10.1186/s13568-015-0104-9
Alonso-Vargas, M., Téllez-Jurado, A., Gómez-Aldapa, C. A., Ramírez-Vargas, M., Conde-Báez, L., Castro-Rosas, J., et al. (2022). Optimization of 2-Phenylethanol production from sweet whey fermentation using Kluyveromyces marxianus. Fermentation 8:39. doi: 10.3390/fermentation8020039
Arora, G., Cormier, F., and Lee, B. (1995). Analysis of odor-active volatiles in Cheddar cheese headspace by multidimensional GC/MS/sniffing. J. Agric. Food Chem. 43, 748–752. doi: 10.1021/jf00051a035
Baptista, M., and Domingues, L. (2022). Kluyveromyces marxianus as a microbial cell factory for lignocellulosic biomass valorisation. Biotechnol. Adv. 60:108027. doi: 10.1016/j.biotechadv.2022.108027
Binetti, A., Carrasco, M., Reinheimer, J., and Suárez, V. (2013). Yeasts from autochthonal cheese starters: technological and functional properties. J. Appl. Microbiol. 115, 434–444. doi: 10.1111/jam.12228
Bintsis, T. (2021). Yeasts in different types of cheese. AIMS Microbiol. 7, 447–470. doi: 10.3934/microbiol.2021027
Borle, F., Bosset, J. O., and Sieber, R. (2002). Photo-oxidation and photoprotection of foods, especially of dairy products. An update of a review article (1996–2000). Sci. Alim. 21, 579–598. doi: 10.3166/sda.21.571-590
Cardin, M., Cardazzo, B., Coton, M., Carraro, L., Lucchini, R., Novelli, E., et al. (2024). Ecological diversity and associated volatilome of typical mountain Caciotta cheese from Italy. Int. J. Food Microbiol. 411:110523. doi: 10.1016/j.ijfoodmicro.2023.110523
Chen, C., Zhou, W., Yu, H., Yuan, J., and Tian, H. (2020). Evaluation of the perceptual interactions among aldehydes in a Cheddar cheese matrix according to odor threshold and aroma intensity. Molecules 25:4308. doi: 10.3390/molecules25184308
Cheon, Y., Kim, J. S., Park, J. B., Heo, P., Lim, J. H., Jung, G. Y., et al. (2014). A biosynthetic pathway for hexanoic acid production in Kluyveromyces marxianus. J. Biotechnol. 182, 30–36. doi: 10.1016/j.jbiotec.2014.04.010
Curioni, P. M. G., and Bosset, J. O. (2002). Key odorants in various cheese types as determined by gas chromatography-olfactometry. Int. Dairy J. 12, 959–984. doi: 10.1016/S0958-6946(02)00124-3
Dank, A., Smid, E. J., and Notebaart, R. A. (2018). CRISPR-Cas genome engineering of esterase activity in Saccharomyces cerevisiae steers aroma formation. BMC. Res. Notes 11, 4–9. doi: 10.1186/s13104-018-3788-5
Delgado, F. J., González-Crespo, J., Cava, R., and Ramírez, R. (2011). Formation of the aroma of a raw goat milk cheese during maturation analysed by SPME–GC–MS. Food Chem. 129, 1156–1163. doi: 10.1016/j.foodchem.2011.05.096
EFSA Panel on Additives and Products or Substances used in Animal Feed [FEEDAP] (2004). Scientific panel on additives and products or substances used in animal feed on the safety of the microorganism product Turval BO399R for use as feed additive for weaned piglets. Question N° EFSA2003-051. EFSA J. 28, 1–6.
Fasoli, G., Barrio, E., Tofalo, R., Suzzi, G., and Belloch, C. (2016). Multilocus analysis reveals large genetic diversity in Kluyveromyces marxianus strains isolated from Parmigiano Reggiano and pecorino di Farindola cheeses. Int. J. Food Microbiol. 233, 1–10. doi: 10.1016/j.ijfoodmicro.2016.05.028
Fasoli, G., Tofalo, R., Lanciotti, R., Schirone, M., Patrignani, F., Perpetuini, G., et al. (2015). Chromosome arrangement, differentiation of growth kinetics and volatile molecule profiles in Kluyveromyces marxianus strains from Italian cheeses. Int. J. Food Microbiol. 214, 151–158. doi: 10.1016/j.ijfoodmicro.2015.08.001
Ferreira, A. D., and Viljoen, B. C. (2003). Yeasts as adjunct starters in matured Cheddar cheese. Int. J. Food Microbiol. 86, 131–140. doi: 10.1016/S0168-1605(03)00252-6
Folkertsma, B., and Fox, P. F. (1992). Use of the cd-ninhydrin reagent to assess proteolysis in cheese during ripening. J. Dairy Res. 59, 217–224. doi: 10.1017/S0022029900030466
Fonseca, G. G., Heinzle, E., Wittmann, C., and Gombert, A. K. (2008). The yeast Kluyveromyces marxianus and its biotechnological potential. Appl. Microbiol. Biotechnol. 79, 339–354. doi: 10.1007/s00253-008-1458-6
Fröhlich-Wyder, M. T., Arias-Roth, E., and Jakob, E. (2019). Cheese yeasts. Yeast 36, 129–141. doi: 10.1002/yea.3368
Genovese, C. R., Lazar, N. A., and Nichols, T. (2002). Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage 15, 870–878. doi: 10.1006/nimg.2001.1037
Gethins, L., Guneser, O., Demirkol, A., Rea, M. C., Stanton, C., Ross, R. P., et al. (2015). Influence of carbon and nitrogen source on production of volatile fragrance and flavour metabolites by the yeast Kluyveromyces marxianus. Yeast 32, 67–76. doi: 10.1002/yea.3047
Gori, K., Mortensen, H. D., Arneborg, N., and Jespersen, L. (2007). Ammonia production and its possible role as a mediator of communication for Debaryomyces hansenii and other cheese-relevant yeast species. J. Dairy Sci. 90, 5032–5041. doi: 10.3168/jds.2006-750
Han, Y., Meng, J., Zhu, L., Zhang, M., Zhang, L., Shi, J., et al. (2024). Multi-mode enhancement and co-fermentation mechanism of 2-phenylethanol production by Kluyveromyces marxianus. Food Biosci. 59:103916. doi: 10.1016/j.fbio.2024.103916
Huang, Z., Huang, L., Xing, G., Xu, X., Tu, C., and Dong, M. (2020). Effect of co-fermentation with lactic acid bacteria and K. Marxianus on physicochemical and sensory properties of goat milk. Food Secur. 9:299. doi: 10.3390/foods9030299
Karahadian, C., Josephson, D. B., and Lindsay, R. C. (1985). Contribution of Penicillium sp. to the flavors of brie and camembert cheese. J. Dairy Sci. 68, 1865–1877. doi: 10.3168/jds.S0022-0302(85)81043-2
Lee, K. (2005). Comparison of fermentative capacities of lactobacilli in single and mixed culture in industrial media. Process Biochem. 40, 1559–1564. doi: 10.1016/j.procbio.2004.04.017
Li, Y., Wang, T., Li, S., Yin, P., Sheng, H., Wang, T., et al. (2022). Influence of GABA- producing yeasts on cheese quality, GABA content, and the volatilome. LWT Food Sci. Technol. 154:112766. doi: 10.1016/j.lwt.2021.112766
Liang, J. J., Zhang, M. L., Ding, M., Mai, Z. M., Wu, S. X., Du, Y., et al. (2014). Alcohol dehydrogenases from Kluyveromyces marxianus: heterologous expression in Escherichia coli and biochemical characterization. BMC Biotechnol. 14:45. doi: 10.1186/1472-6750-14-45
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Löbs, A.-K., Engel, R., Schwartz, C., Flores, A., and Wheeldon, I. (2017). CRISPR-Cas9-enabled genetic disruptions for understanding ethanol and ethyl acetate biosynthesis in Kluyveromyces marxianus. Biotechnol. Biofuels 10, 1–14. doi: 10.1186/s13068-017-0854-5
Löbs, A. K., Schwartz, C., Thorwall, S., and Wheeldon, I. (2018). Highly multiplexed CRISPRi repression of respiratory functions enhances mitochondrial localized ethyl acetate biosynthesis in Kluyveromyces marxianus. ACS Synth. Biol. 7, 2647–2655. doi: 10.1021/acssynbio.8b00331
Löser, C., Urit, T., and Bley, T. (2014). Perspectives for the biotechnological production of ethyl acetate by yeasts. Appl. Microbiol. Biotechnol. 98, 5397–5415. doi: 10.1007/s00253-014-5765-9
Löser, C., Urit, T., Stukert, A., and Bley, T. (2013). Formation of ethyl acetate from whey by Kluyveromyces marxianus on a pilot scale. J. Biotechnol. 163, 17–23. doi: 10.1016/j.jbiotec.2012.10.009
Mayo, B., Rodríguez, J., Vázquez, L., and Flórez, A. B. (2021). Microbial interactions within the cheese ecosystem and their application to improve quality and safety. Food Secur. 10:602. doi: 10.3390/foods10030602
Moio, L., and Addeo, F. (1998). Grana Padano cheese aroma. J. Dairy Res. 65, 317–333. doi: 10.1017/S0022029997002768
Moio, L., Langlois, D., Etievant, P. X., and Addeo, F. (1993). Powerful odorants in water buffalo and bovine mozzarella cheese by use of extract dilution sniffing analysis. Italian J. Food Sci. 3, 227–237.
Morrissey, J. P., Etschmann, M. M., Schrader, J., and de Billerbeck, G. M. (2015). Cell factory applications of the yeast Kluyveromyces marxianus for the biotechnological production of natural flavour and fragrance molecules. Yeast 32, 3–16. doi: 10.1002/yea.3054
Muñoz-Miranda, L. A., Zepeda-Peña, A. C., Casas-Godoy, L., Pereira-Santana, A., Méndez-Zamora, A., Barrera-Martínez, I., et al. (2024). CRISPRi-induced transcriptional regulation of IAH1 gene and its influence on volatile compounds profile in Kluyveromyces marxianus DU3. World J. Microbiol. Biotechnol. 40:121. doi: 10.1007/s11274-023-03811-0
Nogueira, M. C. L., Lubachevsky, G., and Rankin, S. A. (2005). A study of the volatile composition of Minas cheese. LWT Food Sci. Technol. 38, 555–563. doi: 10.1016/j.lwt.2004.07.019
Nursten, H. E. (1997). The flavour of Milk and dairy products: I. Milk of different kinds, milk powder, butter and cream. Int. J. Dairy Technol. 50, 48–56. doi: 10.1111/j.1471-0307.1997.tb01735.x
Padilla, B., Belloch, C., López-Díez, J. J., Flores, M., and Manzanares, P. (2014). Potential impact of dairy yeasts on the typical flavour of traditional ewes' and goats' cheeses. Int. Dairy J. 35, 122–129. doi: 10.1016/j.idairyj.2013.11.002
Pentjuss, A., Stalidzans, E., Liepins, J., Kokina, A., Martynova, J., Zikmanis, P., et al. (2017). Model-based biotechnological potential analysis of Kluyveromyces marxianus central metabolism. J. Ind. Microbiol. Biotechnol. 44, 1177–1190. doi: 10.1007/s10295-017-1946-8
Perpetuini, G., Tittarelli, F., Battistelli, N., Suzzi, G., and Tofalo, R. (2020). γ-Aminobutyric acid production by Kluyveromyces marxianus strains. J. Appl. Microbiol. 129, 1609–1619. doi: 10.1111/jam.14736
Perpetuini, G., Tittarelli, F., Perla, C., and Tofalo, R. (2022). Influence of different aggregation states on volatile organic compounds released by dairy Kluyveromyces marxianus strains. Food Secur. 11:2910. doi: 10.3390/foods11182910
Perpetuini, G., Tittarelli, F., Suzzi, G., and Tofalo, R. (2019). Cell wall surface properties of Kluyveromyces marxianus strains from dairy-products. Front. Microbiol. 10:79. doi: 10.3389/fmicb.2019.00079
Pisano, M. B., Rosa, A., Putzu, D., Marincola, F. C., Mossa, V., Viale, S., et al. (2020). Influence of autochthonous putative probiotic cultures on microbiota, lipid components and metabolome of Caciotta cheese. Front. Microbiol. 11:583745. doi: 10.3389/fmicb.2020.583745
Plessas, S., Bosnea, L., Psarianos, C., Koutinas, A. A., Marchant, R., and Banat, I. M. (2008). Lactic acid production by mixed cultures of Kluyveromyces marxianus, Lactobacillus delbrueckii ssp. bulgaricus and Lactobacillus helveticus. Bioresour. Technol. 99, 5951–5955. doi: 10.1016/j.biortech.2007.10.039
Rehman, S., McSweeney, P. L. H., Banks, J. M., Brechany, E. Y., Muir, D. D., and Fox, P. F. (2000). Ripening of Cheddar cheese made from blends of raw and pasteurized milk. Int. Dairy J. 10, 33–44. doi: 10.1016/S0958-6946(00)00024-8
Reyes-Sánchez, F. J., Páez-Lerma, J. B., Rojas-Contreras, J. A., López-Miranda, J., Soto-Cruz, N. Ó., and Reinhart-Kirchmayr, M. (2019). Study of the enzymatic capacity of Kluyveromyces marxianus for the synthesis of esters. J. Mol. Microbiol. Biotechnol. 29, 1–9. doi: 10.1159/000507551
Risner, D., Tomasino, E., Hughes, P., and Meunier-Goddik, L. (2019). Volatile aroma composition of distillates produced from fermented sweet and acid whey. J. Dairy Sci. 102, 202–210. doi: 10.3168/jds.2018-14737
Sablé, S., and Cottenceau, G. (1999). Current knowledge of soft cheeses flavor and related compounds. J. Agric. Food Chem. 47, 4825–4836. doi: 10.1021/jf990414f
Sacchi, R., Marrazzo, A., Masucci, F., Di Francia, A., Serrapica, F., and Genovese, A. (2020). Effects of inclusion of fresh forage in the diet for lactating buffaloes on volatile organic compounds of milk and mozzarella cheese. Molecules 25:1332. doi: 10.3390/molecules25061332
Sandoval-Copado, J. R., Orozco-Villafuerte, J., García-Fabila, M. M., and Colín-Cruz, M. A. (2018). Optimization of headspace solid-phase microextraction and gas chromatography coupled with mass spectrometry technique for the quantification of volatile compounds in a fresh cheese (Oaxaca). J Chromatogr Sep Tech. 9, 1–7. doi: 10.4172/2157-7064.1000415
Signori, L., Passolunghi, S., Ruohonen, L., Porro, D., and Branduardi, P. (2014). Effect of oxygenation and temperature on glucose-xylose fermentation in Kluyveromyces marxianus CBS712 strain. Microb. Cell Factories 13:51. doi: 10.1186/1475-2859-13-51
Tofalo, R., Fasoli, G., Schirone, M., Perpetuini, G., Pepe, A., Corsetti, A., et al. (2014). The predominance, biodiversity and biotechnological properties of Kluyveromyces marxianus in the production of pecorino di Farindola cheese. Int. J. Food Microbiol. 187, 41–49. doi: 10.1016/j.ijfoodmicro.2014.06.029
Tsouli Sarhir, S., Amanpour, A., Bouseta, A., and Selli, S. (2022). Potent odorants and sensory characteristics of the soft white cheese “Jben”: effect of salt content. Flavour Frag. J. 37, 243–253. doi: 10.1002/ffj.3696
van Rijswijck, I. M. H., Kruis, A. J., Wolkers-Rooijackers, J. C. M., Abee, T., and Smid, E. J. (2019). Acetate-ester hydrolase activity for screening of the variation in acetate ester yield of Cyberlindnera fabianii, Pichia kudriavzevii and Saccharomyces cerevisiae. LWT Food Sci. Technol. 104, 8–15. doi: 10.1016/j.lwt.2019.01.019
Varela, J. A., Montini, N., Scully, D., Van der Ploeg, R., Oreb, M., Boles, E., et al. (2017). Polymorphisms in the LAC12 gene explain lactose utilisation variability in Kluyveromyces marxianus strains. FEMS Yeast Res. 17:fox021. doi: 10.1093/femsyr/fox021
Vidal, E. E., de Billerbeck, G. M., Simões, D. A., Schuler, A., François, J. M., and de Morais, M. A. (2013). Influence of nitrogen supply on the production of higher alcohols/esters and expression of flavour-related genes in cachaça fermentation. Food Chem. 138, 701–708. doi: 10.1016/j.foodchem.2012.10.147
Wang, W., Li, Z., Gan, L., Fan, H., and Guo, Y. (2018). Dietary supplemental Kluyveromyces marxianus alters the serum metabolite profile in broiler chickens. Food Funct. 17, 3776–3787. doi: 10.1039/C8FO00268A
Wittmann, C., Hans, M., and Bluemke, W. (2002). Metabolic physiology of aroma-producing Kluyveromyces marxianus. Yeast 19, 1351–1363. doi: 10.1002/yea.920
Xiao, J., Chen, Y., Li, J., Shi, X., Deng, L., and Wang, B. (2020). Evaluation of the effect of auxiliary starter yeasts with enzyme activities on Kazak cheese quality and flavor. Front. Microbiol. 11:614208. doi: 10.3389/fmicb.2020.614208
Yang, Y., Xia, Y., Li, C., Wang, G., Xiong, Z., Song, X., et al. (2024). Metabolites, flavor profiles and ripening characteristics of Monascus-ripened cheese enhanced by Ligilactobacillus salivarius AR809 as adjunct culture. Food Chem. 436:137759. doi: 10.1016/j.foodchem.2023.137759
Zhang, D. D., Liu, J. L., Jiang, T. M., Li, L., Fang, G. Z., Liu, Y. P., et al. (2017). Influence of Kluyveromyces marxianus on proteins, peptides, and amino acids in lactobacillus-fermented milk. Food Sci. Biotechnol. 26, 739–748. doi: 10.1007/s10068-017-0094-2
Zheng, X., Li, K., Shi, X., Ni, Y., Li, B., and Zhuge, B. (2017). Potential characterization of yeasts isolated from Kazak artisanal cheese to produce flavoring compounds. Microbiol. Open 7:e00533. doi: 10.1002/mbo3.533
Keywords: Kluyveromyces marxianus , aroma compounds, OAV, qPCR, cheese
Citation: Perpetuini G, Rossetti AP, Rapagnetta A and Tofalo R (2024) Unlocking the potential of Kluyveromyces marxianus in the definition of aroma composition of cheeses. Front. Microbiol. 15:1464953. doi: 10.3389/fmicb.2024.1464953
Edited by:
Marta Laranjo, University of Évora, PortugalReviewed by:
Giacomo Zara, University of Sassari, ItalyHosam Elhalis, Agency for Science, Technology and Research (A*STAR), Singapore
Copyright © 2024 Perpetuini, Rossetti, Rapagnetta and Tofalo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosanna Tofalo, cnRvZmFsb0B1bml0ZS5pdA==
 Giorgia Perpetuini
Giorgia Perpetuini Alessio Pio Rossetti
Alessio Pio Rossetti Arianna Rapagnetta
Arianna Rapagnetta Rosanna Tofalo
Rosanna Tofalo