- 1Department of Quality Assurance and Laboratory Management, School of Biomedical and Laboratory Science, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia
- 2Department of Medical Microbiology, Amhara National Regional State Public Health Institute, Bahir Dar, Ethiopia
- 3Department of Molecular Laboratory, Trachoma Elimination Program, The Carter Center, Bahir Dar, Ethiopia
- 4Microbiology Laboratory, University of Gondar Comprehensive Specialized Hospital, Gondar, Ethiopia
Background: Neonatal sepsis is a major cause of morbidity and mortality worldwide. Understanding the bacterial profiles and antibiotic susceptibility patterns causing neonatal sepsis is crucial for guiding appropriate treatment, improving patient outcomes, and combating the emergence of antibiotic resistance. Despite its importance, data regarding neonatal sepsis in the study area is limited. Therefore, this study aimed to characterize the bacterial pathogens and identify associated factors among neonates with suspected sepsis at the University of Gondar Comprehensive Specialized Hospital, Ethiopia.
Methods: A cross-sectional study was conducted by reviewing laboratory records of neonates admitted for suspected sepsis from January 2019 to December 2021. Data were checked for completeness and encoded in a spreadsheet program. Then, data were exported to STATA version 17 for analysis. Descriptive statistics such as frequency and percentage were computed. The association between neonatal sepsis and potential risk factors was assessed using Pearson’s chi-square test. A p-value of < 0.05, was considered statistically significant.
Results: A total of 1,236 neonates were included. Of these, 96.2% (1,190/1,236) had a fever before admission. The prevalence of culture-confirmed sepsis was 25.4% (314/1,236). Bacterial pathogens accounted for 23% (284/1,236) of these isolates, with Gram-negative bacteria being more prevalent at 75.3% (214/284) than Gram-positive bacteria at 24.7% (70/284). The most frequently isolated bacterial pathogens were K. pneumoniae 38.7% (110/284) and S. aureus 13% (37/284). The isolates demonstrated a high resistance level to commonly used antibiotics, with 61.6% exhibiting multidrug resistance. K. pneumoniae showed the highest rate of multidrug resistance (90.9%). Neonatal sepsis was associated with several factors, including fever before and after admission, hypothermia, increased respiration, suspected pneumonia, and suspected meningitis.
Conclusion: This study identified a high prevalence of culture-confirmed sepsis in neonates at UoGCSH, with Gram-negative bacteria, especially K. pneumoniae, dominating the isolated pathogens. The isolated bacteria exhibited alarming resistance to commonly used antibiotics, with a high proportion demonstrating multidrug resistance. Implementing effective antibiotic stewardship programs is crucial to optimize antibiotic use, reduce unnecessary prescriptions, and curb the spread of resistant strains.
Background
Neonatal sepsis is a significant global health challenge, particularly in developing countries, where it is the leading cause of morbidity and mortality. It accounts for an estimated 3 million cases annually with a high mortality rate of 11–19% (Glaser et al., 2021; Fleischmann-Struzek et al., 2018). Immature immune systems and barriers heighten neonatal susceptibility to infections (Georges Pius et al., 2022). Sub-Saharan Africa bears a substantial burden, with neonatal sepsis leading to an estimated annual loss of 5.3–8.7 million disability-adjusted life years and an economic impact ranging from $10 billion to $469 billion (Ranjeva et al., 2018; Molloy et al., 2020). This issue is especially pronounced in East Africa, where prevalence reaches 29.7% (Abate et al., 2020).
In Ethiopia, the prevalence of neonatal sepsis is alarmingly high, with rates ranging from 45.8 to 78.3% in different regions (Bayih et al., 2021; Belachew and Tewabe, 2020; Mustefa et al., 2020; Bekele et al., 2022; Roble et al., 2022). This burden is further exacerbated by the increasing antimicrobial resistance (Chaurasia et al., 2019). Multiple risk factors are associated with neonatal infection, including low birth weight (Bayih et al., 2021; Belachew and Tewabe, 2020; Mustefa et al., 2020; Li et al., 2023), maternal history of urinary tract infection (Abate et al., 2020; Bayih et al., 2021; Belachew and Tewabe, 2020; Mustefa et al., 2020), formula feeding, cesarean section (Li et al., 2023), preterm birth (Abate et al., 2020; Bayih et al., 2021; Belachew and Tewabe, 2020; Mustefa et al., 2020; Li et al., 2023; Satar and Özlü, 2012), home delivery, prolonged labor, and premature rupture of membranes (Abate et al., 2020), antenatal urinary tract infection, and intrapartum fever (Bayih et al., 2021; Belachew and Tewabe, 2020; Mustefa et al., 2020). Despite advancements in neonatal care, diagnosing neonatal sepsis remains challenging, underscoring the importance of prompt antibiotic treatment (Ershad et al., 2019).
Regional variations exist in the spectrum of bacterial pathogens causing neonatal sepsis (Poyekar, 2022; Moni et al., 2020). While the predominant bacteria vary geographically, Gram-negative bacteria like Klebsiella spp. (Pokhrel et al., 2018; Acheampong et al., 2022; Bhat et al., 2011; Almohammady et al., 2020; Fenta et al., 2022; Pataskar et al., 2023; Bai et al., 2021; Siddiqui et al., 2023; Weldu et al., 2020), E. coli (Pokhrel et al., 2018; Acheampong et al., 2022; Bhat et al., 2011; Almohammady et al., 2020; Fenta et al., 2022; Pataskar et al., 2023; Bai et al., 2021), and E. cloacae complex (Pataskar et al., 2023; Bai et al., 2021) are commonly isolated. Particularly in developing countries, Gram-negative bacteria stand as the leading cause of morbidity and mortality (Kamalakannan, 2018; Bethou and Bhat, 2022). Among Gram-positive isolates, S. aureus (Pokhrel et al., 2018; Acheampong et al., 2022; Almohammady et al., 2020; Tessema et al., 2021; Yadav et al., 2018; Negussie et al., 2015) and CoNS (Pokhrel et al., 2018; Acheampong et al., 2022; Almohammady et al., 2020; Tessema et al., 2021; Yadav et al., 2018; Negussie et al., 2015; Aku et al., 2018) are frequently identified. Antibiotic resistance is a significant concern (Negussie et al., 2015), with alarmingly high levels of multidrug-resistant strains negatively impacting treatment outcomes (Uwe et al., 2022). This emphasizes the need for strict antibiotic use guidelines. Isolated bacteria often show high resistance to conventional antibiotics like ampicillin, cephalosporins, and piperacillin-tazobactam (Uwe et al., 2022; Verma et al., 2015). Studies from India report high rates of multidrug resistance in Acinetobacter spp. (82%), Klebsiella spp. (54%), and E. coli (38%) isolates (Chaurasia et al., 2016). Another Indian study found bacterial isolates resistant to aminoglycosides (74%), third/fourth-generation cephalosporins (95%), and carbapenems (56%) (Shah et al., 2022). Others indicate that 54% of isolated bacteria were resistant to at least one antibiotic (Sands et al., 2021). In Iran, K. pneumoniae showed the highest resistance to Cefixime (80.6%), while E. coli exhibited significant resistance to Ampicillin (61.8%) (Moftian et al., 2023). Even Gram-negative bacteria harbor multiple cephalosporin and carbapenem resistance genes, highlighting widespread antimicrobial resistance (Sands et al., 2021).
Local data is crucial for informing treatment strategies due to regional variations in bacterial spectrum and antimicrobial sensitivity patterns (Poyekar, 2022; Moni et al., 2020; Uwe et al., 2022; Verma et al., 2015). The burden of neonatal sepsis is worsened by the scarcity of accurate information on its causes and consequences in developing countries, including Ethiopia. Additionally, most studies are limited by small sample sizes (Sands et al., 2021).
Therefore, this study aimed to address the data gap by determining the most common bacterial etiologies of neonatal sepsis in Gondar Comprehensive Specialized Hospital (UoGCSH), a critical healthcare hub in northwestern Ethiopia, and assessing the antibiotic resistance patterns of these key pathogens. The findings will inform targeted treatment strategies to improve outcomes for critically ill neonates in the local hospital’s care and contribute valuable regional data to national efforts to combat the public health challenge of neonatal sepsis.
Materials and methods
Study design and setting
A cross-sectional study was conducted on neonates (aged ≤28 days) suspected of bloodstream infections who were admitted to the UoGCSH from January 2019 to December 2021. The hospital is located in the center of Gondar town, approximately 747 kilometers northwest of Addis Ababa, the capital city of Ethiopia. The hospital is a leading healthcare institution in the region and serves as a referral center for over 7 million people, catering to a diverse population from both urban and rural areas (Gobezie et al., 2023). The hospital is equipped with specialized facilities and a dedicated neonatal unit, which is essential for managing cases of neonatal sepsis. It offers various specialized diagnostic services to neonates and children. The neonatal unit is equipped with advanced medical facilities and staffed by skilled healthcare professionals, including pediatricians and nurses, who are dedicated to managing complex cases of neonatal sepsis. The hospital has several laboratory departments, including medical microbiology, clinical chemistry, hematology, serology, medical parasitology, and one main laboratory room. The microbiology laboratory plays a vital role in enabling the collection and analysis of blood samples to identify bacterial pathogens and assess their antibiotic susceptibility profiles.
Blood culture and bacterial identification
Standardized protocol-guided blood collection and bacterial identification were implemented. Two milliliters of blood were aseptically collected from each neonate and inoculated at a 1:10 ratio (blood: broth) into sterile Tryptone Soy Broth. Culture bottles incubated at 35-37°C for up to 7 days with daily monitoring for growth signs (hemolysis, turbidity, clot formation). Positive cultures underwent Gram staining and subculture onto various selective and differential media (blood agar, chocolate agar with 5% CO2, MacConkey agar, and mannitol salt agar) for further differentiation. These plates were incubated aerobically at 37°C for 18–24 h.
A two-step approach identified bacterial isolates. Initially, colony characteristics (color, size, shape, texture) were examined macroscopically. Subsequently, Gram-negative isolates underwent various conventional biochemical tests (indole, urease, lysine decarboxylase, triple sugar iron agar, citrate utilization, oxidase, and motility tests) for further differentiation. Gram-positive identification relied on Gram staining, catalase activity, coagulase testing, and hemolytic pattern analysis. This combined approach ensured comprehensive and reliable bacterial pathogen identification (Arega et al., 2018; Arega et al., 2017).
Antimicrobial susceptibility testing
The Kirby-Bauer disk diffusion method determined antimicrobial susceptibility patterns. Briefly, a standardized suspension of bacterial isolates was prepared in saline and adjusted to a 0.5 McFarland standard. This suspension was then inoculated onto Mueller-Hinton agar (non-fastidious bacteria) or Mueller-Hinton agar supplemented with 5% sheep blood (fastidious bacteria). Following inoculation, commercially available antibiotic disks (erythromycin, clindamycin, ampicillin, etc.) were applied, and plates were incubated at 37°C for 18–24 h. The diameters of inhibition zones surrounding each disk were measured, and susceptibility was categorized as sensitive, intermediate, or resistant according to the 2019 CLSI guidelines (Clinical and Laboratory Standards Institute, 2019).
Data extraction
The primary data source for this study was the records from the microbiology laboratory at UoGCSH. Six experienced laboratory professionals were involved in the data collection process guided by a standardized checklist. This checklist captured demographic information (age, gender), clinical setting (ICU admission), admission date, presenting complaints (fever, hypothermia), prior antibiotic use, culture and identification results, and susceptibility testing results for a broad spectrum of antibiotics. Antibiotic susceptibility results of intermediate susceptibility were categorized as “resistant” for analysis purposes.
Operational and case definitions
Antimicrobial susceptibility pattern
The response of specific bacterial isolates to various antibiotics, categorized as resistant, intermediate, or susceptible based on inhibition zone diameters. We categorized both “resistant” and “intermediate” patterns as resistant.
Multidrug resistance
The ability of a bacterial strain to resist three or more antimicrobial agents from different classes (Alam et al., 2011).
Data management and analysis
Data were checked for completeness and encoded in an Excel spreadsheet. Then, the data were exported to STATA version 17 for analysis. Descriptive statistics (frequency and percentage) were computed. Pearson’s chi-square test was used to assess the association between neonatal sepsis and potential risk factors. A p-value of less than 0.05 was considered statistically significant. Finally, the study results are presented in text, tables, and figures as appropriate.
Sample and data quality control
Standard operating procedures for microbiological techniques were followed throughout blood sample collection, transportation, culture media inoculation and incubation, and biochemical testing. Culture media sterility was ensured by random selection and incubation of 5% of prepared media. Media performance was regularly evaluated using known standard strains of E. coli (ATCC 25922), S. aureus (ATCC 25923), and P. aeruginosa (ATCC 27853). Microbiology experts monitored culture media inoculation, colony characterization, measurement, and interpretation of antibiotic susceptibility tests. The investigators developed a standardized data extraction form, and its accuracy, completeness, consistency, and reliability were assessed using a pilot study involving a random sample of 100 patient records.
Ethical consideration
Before commencing the research, the authors ensured adherence to ethical guidelines. They obtained ethical approval from the University of Gondar Institutional Review Board (IRB). Additionally, a letter of support from the College of Medicine and Health Sciences facilitated data collection. To ensure participant anonymity, patient personal information was omitted, and data were analyzed anonymously. Since the study was retrospective, the IRB waived the requirement for informed consent as obtaining consent from past participants would be impractical. Furthermore, to strengthen confidentiality, no personal identifiers were used, and only the investigator had access to the collected data. The research was conducted following the Declaration of Helsinki.
Results
Socio-demographic and clinical characteristics of study participants
A total of 1,236 study participants were enrolled in the study. The majority, 747 (60.4%), were male, and 772 (62.4%) were aged less than 7 days. The primary clinical setting was the NICU, which accounted for 1,215 (98.2%) of the participants. The distribution of cases by year of diagnosis showed that the highest number of cases, 680 (55.0%), occurred in 2021. A significant proportion of patients, 1,174 (95.2%), experienced fever, with 1,190 (96.2%) reporting fever onset before admission. Furthermore, 384 (31.0%) had received antibiotic therapy within 2 days before admission. Increased respiration was observed in 503 (40.7%) participants, and a similar proportion, 503 (40.7%), were suspected of having pneumonia (Table 1).
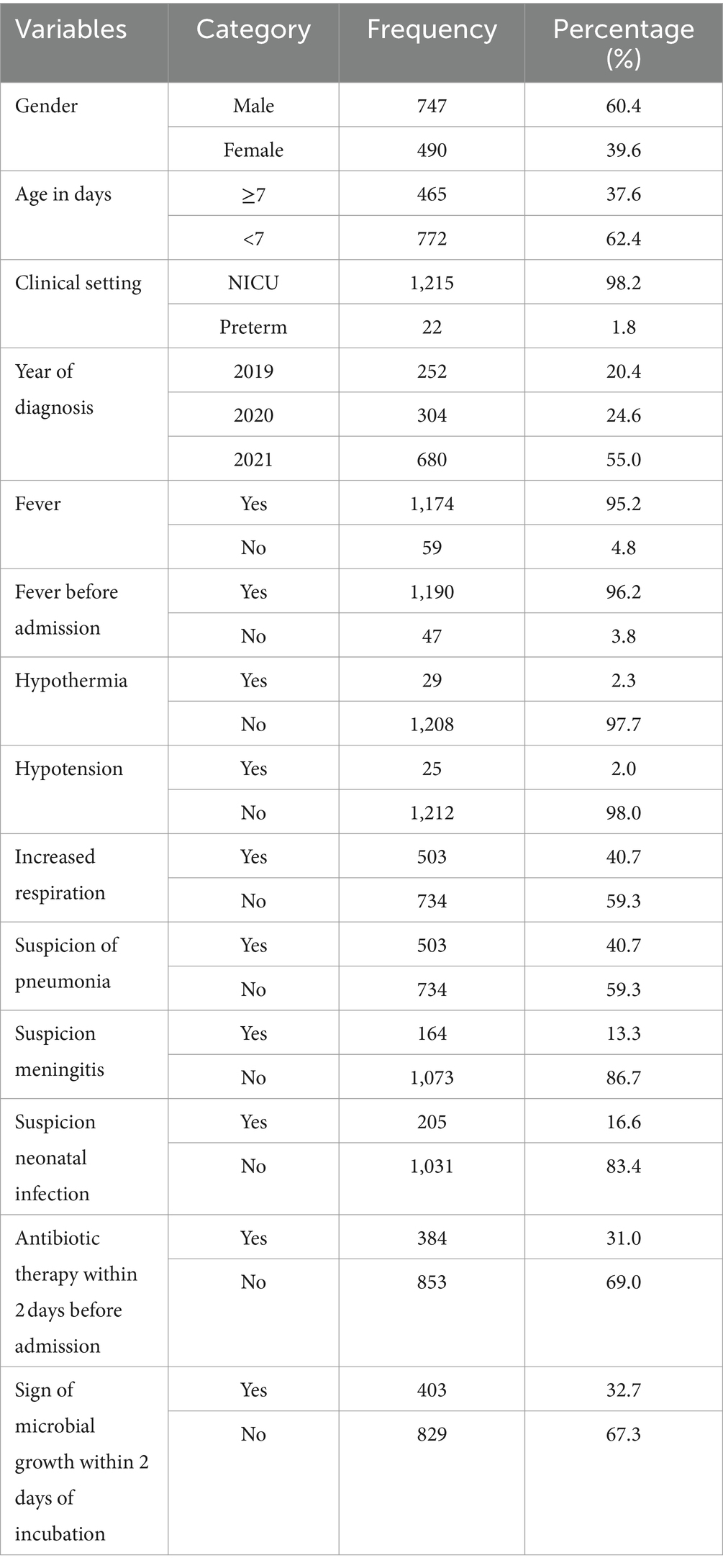
Table 1. Socio-demographic characteristics of bloodstream infections suspected study participants at UoGCSH, Ethiopia (n = 1,236).
Prevalence of bacterial isolates
The overall prevalence of microbial isolates was 25.4% (314/1236). Among this, bacterial pathogens and yeast cells accounted for 90.5% (284/314) and 9.6% (30/314), respectively. The prevalence of bacterial pathogens from the overall septicemia suspected patients was 23% (284/1236). Among the bacterial isolates, Gram-negative bacteria were predominant, comprising 75.3% (214/284) of the identified pathogens, while Gram-positive bacteria accounted for 24.7% (70/284). Additionally, 4 Bacillus spp. isolates were identified as contaminants (Figure 1). The most frequently isolated bacterial pathogens were K. pneumoniae at 38.7% (110/284), followed by S. aureus at 13% (37/284), and Acinetobacter spp. at 8.1% (30/284). Other significant findings included E. coli at 6.3% (18/284), and Non-fermenting Gram-negative rods (NFGNR) at 6% (17/284) (Table 2).
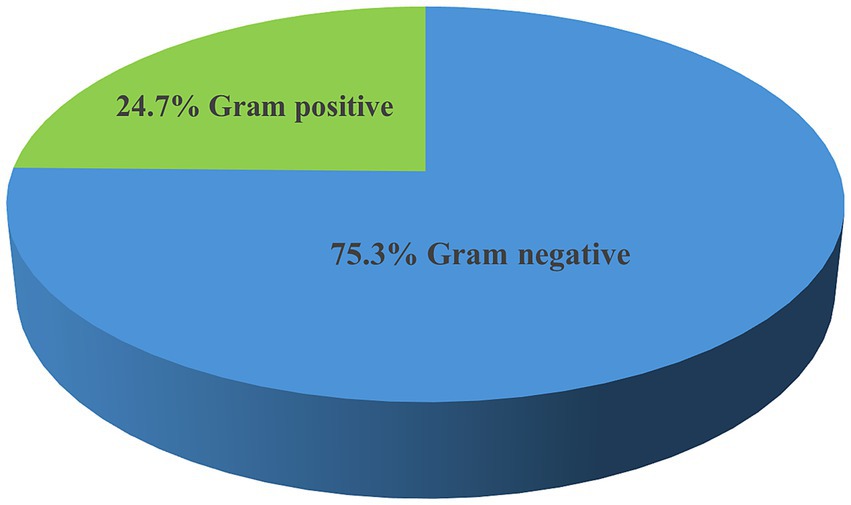
Figure 1. Frequency of bacterial pathogens isolated from neonates suspected of bloodstream infections at UoGCSH, 2024.
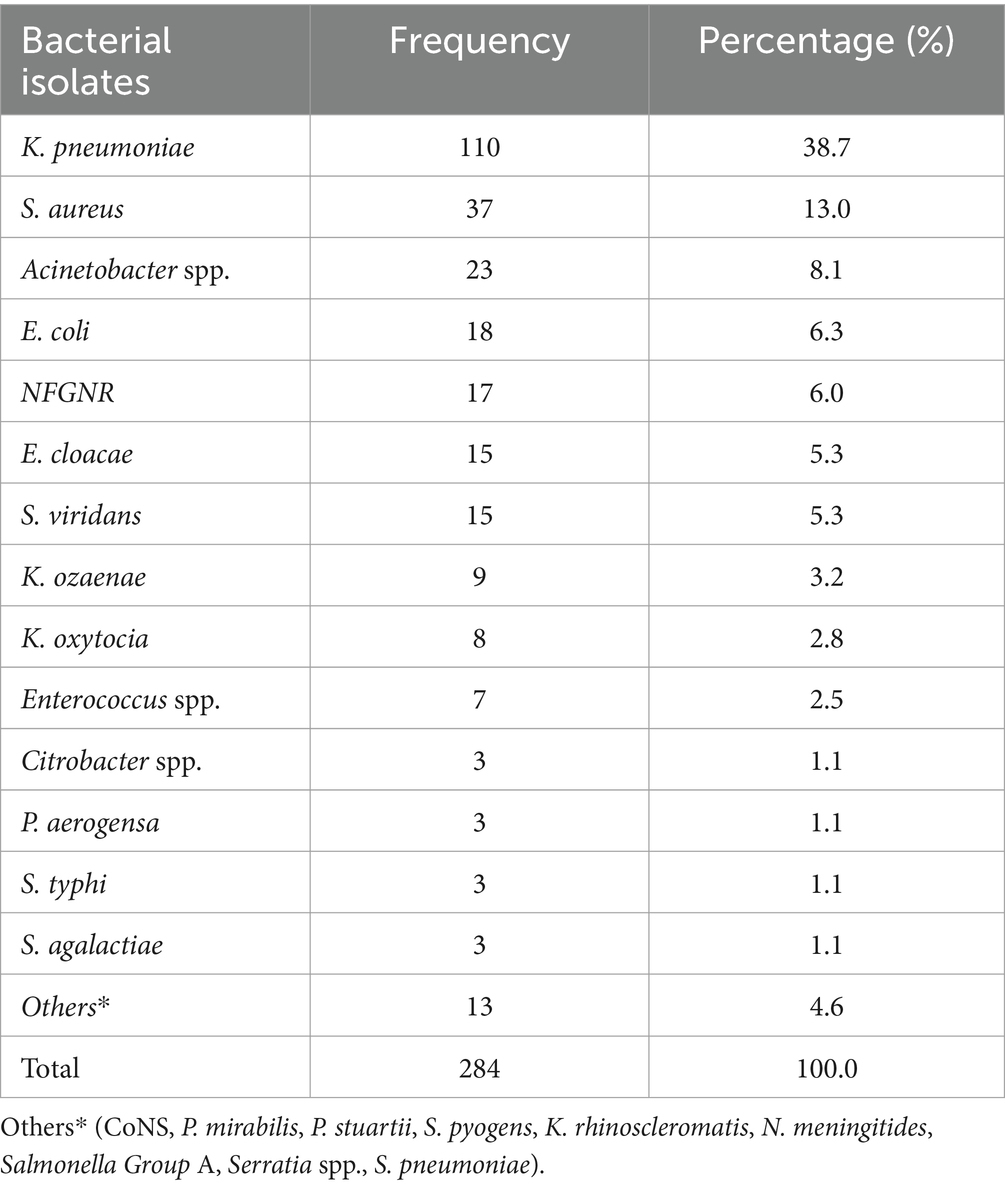
Table 2. Frequency of bacterial pathogens isolated from patients suspected of septicemia at the UoGCSH, Ethiopia, 2024 (n = 284 isolates).
Antimicrobial susceptibility patterns of bacterial isolates
Among Gram-positive isolates, CoNS exhibited 100% (2/2) resistance to penicillin, oxacillin, and gentamicin. S. aureus showed a high resistance rate of 94.6% (35/37) of isolates to penicillin, 64.9% to oxacillin, and 40.5% to gentamicin. Similarly, for S. viridans 73.3% (11/15) of isolates were resistant to ampicillin and 20% (3/15) were resistant to vancomycin. The single isolate of S. pneumoniae showed no resistance to the tested antibiotics. Enterococcus species demonstrated 85.7% (6/7) resistance to penicillin, 57% (4/7) to vancomycin, and 71.4% (5/7) to chloramphenicol. S. agalactiae exhibited no resistance to ampicillin and vancomycin antimicrobial agents. S. pyogenes showed 50% (1/2) resistance to vancomycin. This study highlights significant resistance patterns, especially in CoNS, S. aureus, and Enterococcus spp., necessitating careful consideration of antibiotic selection for effective treatment (Table 3).

Table 3. Antimicrobial sensitivity test result for Gram-positive bacterial pathogens isolated from suspected bloodstream infections at UoGCSH, Ethiopia, 2024.
A significant burden of antimicrobial resistance among Gram-negative bacterial isolates recovered among the study participants. Among the isolated bacterial pathogens, K. pneumoniae was the most concerning resistance profile for meropenem (88.1%), ceftazidime (83.6%), ceftriaxone (83.6%), and amoxicillin-clavulanate (69%). Conversely, E. coli showed relatively lower resistance rates, with the highest resistance observed for ampicillin (66.7%), gentamicin (61.1%), and amoxicillin-clavulanate (55.6%). Acinetobacter spp. exhibited moderate resistance to ceftriaxone (52.2%), ceftazidime (47.8%), and amoxicillin-clavulanate (47.8%), but remained largely susceptible to meropenem (13%) and ciprofloxacin (21.7%). E. cloacae isolate showed high resistance to ampicillin (93.3%) and amoxicillin-clavulanate (86.7%), with moderate resistance to other antibiotics tested. The NFGNR group displayed variable resistance patterns, with the highest rates observed for ceftazidime (64.7%) and ceftriaxone (64.7%). Among bacterial isolates, Bacillus spp. were more likely contaminants and has not been tested for antimicrobial agents. Furthermore, K. ozaenae isolates exhibited particularly high resistance to ceftazidime (88.9%), ceftriaxone (88.9%), and trimethoprim-sulfamethoxazole (88.9%) (Table 4).
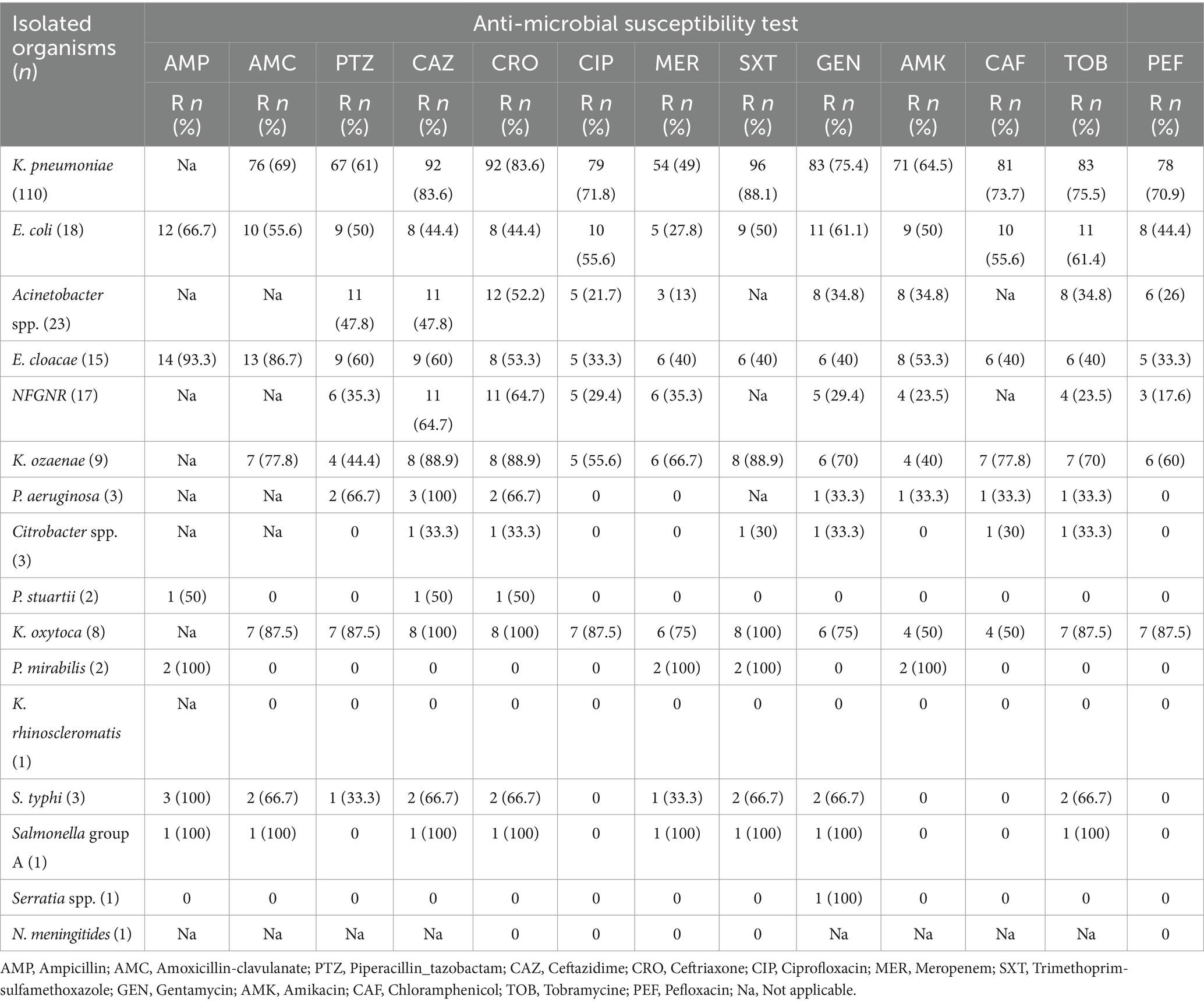
Table 4. Antimicrobial sensitivity test results for Gram-negative bacterial isolates from bloodstream infections suspected neonates at UoGCSH, Ethiopia.
Multidrug resistance pattern of bacterial isolates
The isolates exhibited a concerning level of multidrug resistance with 61.6% (175/284) of the isolates being resistant to three or more antibiotic classes. The data shows that K. pneumoniae had the highest rate of multidrug resistance, with 90.9% (100/284) of the isolates being resistant to three or more antibiotic classes. Notably, K. ozaenae, K. oxytocia, and P. mirabilis exhibited 100% (2/2) multidrug resistance. Other bacteria such as S. aureus, Acinetobacter spp., E. coli, and NFGNR also demonstrated high MDR rates at 40.5% (15/284), 43.5% (10/284), 61.1% (11/284), and 41.2% (7/284), respectively. On the other hand, Enterococcus spp. and S. viridans S. agalactiae, CoNS, P. stuartii, and S. pyogens did not show any multidrug resistance. The data indicates a concerning level of multidrug resistance among the bacterial isolates, with 61.6% of the total isolates being resistant to three or more antibiotic classes (Table 5).
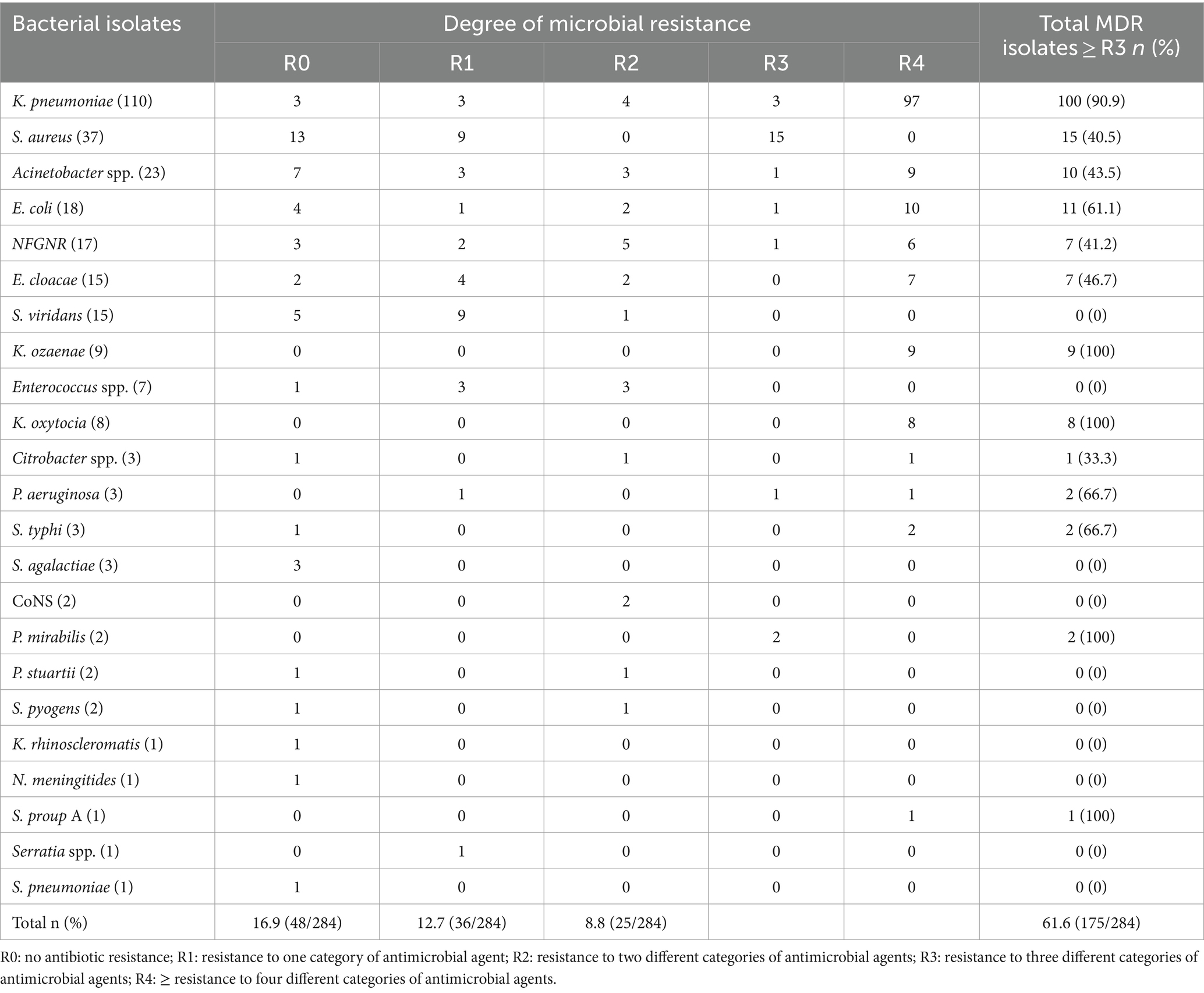
Table 5. Multidrug resistance patterns of bacterial isolates from neonates suspected of bloodstream infections at UoGCSH, 2024.
Factors associated with bloodstream infections
This analysis investigated factors associated with bloodstream infections using Pearson’s chi-square test. While no significant association was found between positive blood cultures and gender, fever before admission, fever after admission, and hypothermia, several other factors emerged as important. Patients with increased respiration and suspected pneumonia were more likely to have positive cultures (p = 0.004). Likewise, suspected meningitis also showed a significant association (p = 0.009). Interestingly, prior antibiotic use did not statistically influence the results. Notably, the year of admission was the only demographic factor with a significant association. Patients admitted in 2021 had a higher proportion of positive cultures compared to those admitted in 2019 or 2020 (p < 0.001) (Table 6).
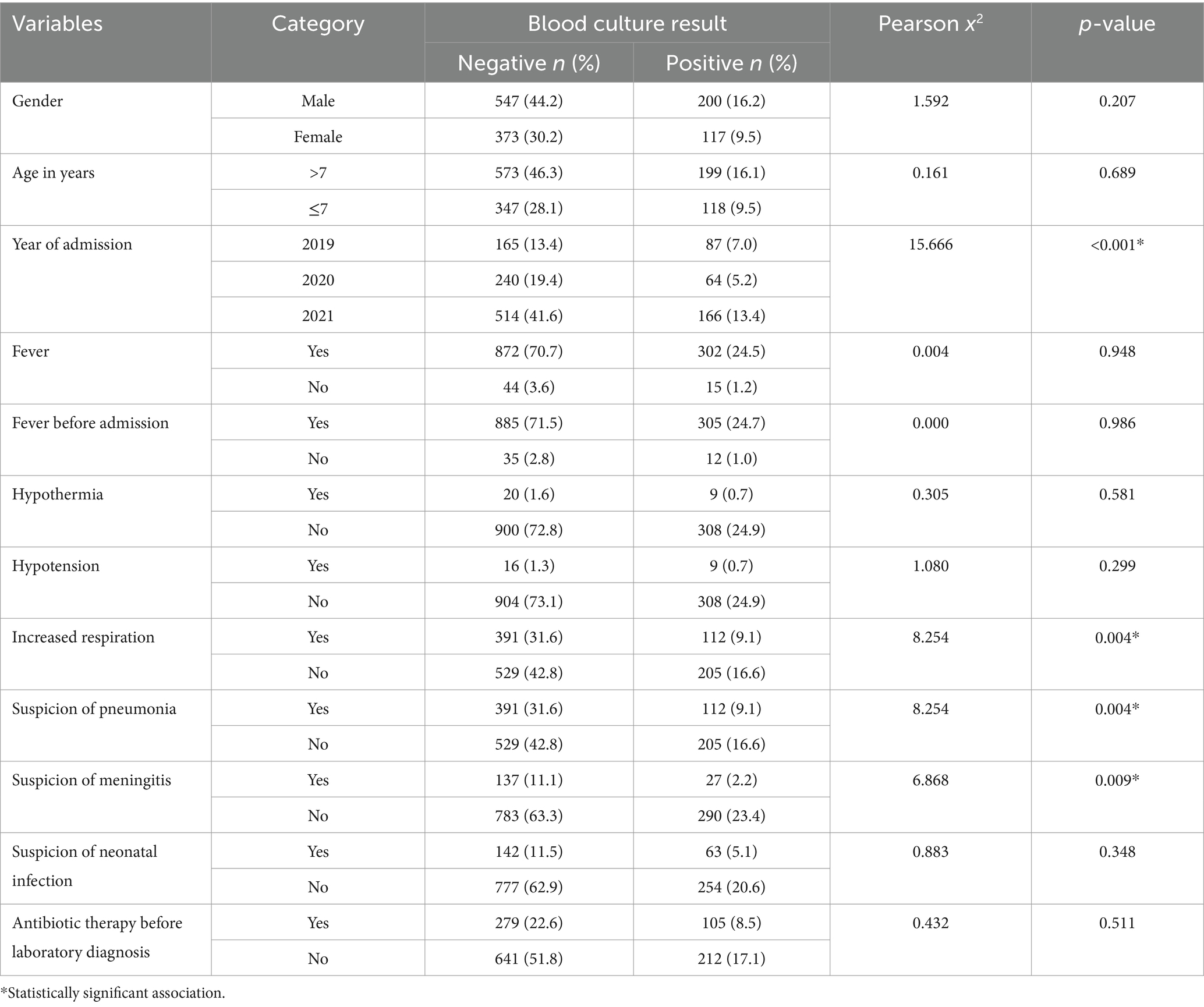
Table 6. Factors associated with positive bacterial blood cultures among neonates with suspected bloodstream infection at UoGCSH (2019–2021).
Discussion
Neonatal sepsis is a significant threat to newborn health, particularly in developing countries like Ethiopia, where it is a leading cause of mortality and morbidity (Weldu et al., 2020; Wen et al., 2021; Klingenberg et al., 2018; Sherif et al., 2023). Accurately identifying bacterial pathogens causing neonatal sepsis and their antibiotic susceptibility patterns is crucial for effective patient management. This information helps guide treatment strategies and improves patient outcomes. Therefore, this study aimed to characterize the bacterial profiles and antibiotic susceptibility at UoGCSH in Ethiopia.
The current study found that nearly a quarter of neonates suspected of sepsis had confirmed bacterial infections (23%). This prevalence is comparable to studies conducted in China (23%) (Fang et al., 2023), Tanzania (24%) (Mhada et al., 2012), Ghana (21.0%) (Acheampong et al., 2022), Nepal (20.5%) (Pokhrel et al., 2018), and Ethiopia (21%) (Sherif et al., 2023). However, the finding is higher than results from Bhutan (14%) (Jatsho et al., 2020), Nepal (10.8%) (Thapa and Sapkota, 2019), Uganda (12.8%) (Tumuhamye et al., 2020), Iran (15.98%) (Akbarian-Rad et al., 2020), South Africa (11.0%) (Reddy et al., 2021), Pakistan (8.9%) (Atif et al., 2021), and Ghana (17.3%) (Aku et al., 2018). Conversely, the current finding is much lower than those reported in various settings in Ethiopia (36.5–46.6%) (Weldu et al., 2020; Worku et al., 2022; Mezgebu et al., 2023; Assemie et al., 2020; Geyesus et al., 2017), Zambia (38%) (Egbe et al., 2023), Nigeria (49.6%) (Peterside et al., 2015), Tanzania (72%) (Majigo et al., 2023), and Uganda (59.0%) (Zamarano et al., 2021). The discrepancy in prevalence rates across different geographical regions could be attributed to factors such as differences in hygiene practices, antibiotic use patterns, variations in study design, and broader epidemiological factors. Additionally, improvements in diagnostic techniques, changes in hospital practices, and seasonal trends could potentially influence the incidence and prevalence of neonatal sepsis.
Among the neonates suspected of septicemia, only 25.4% tested positive for cultures. Several factors could explain the relatively low rate of culture positivity. These include the use of antibiotic treatment before admission, which could suppress bacterial growth, the limitations of conventional culture methods, and the possibility of non-bacterial infections presenting with similar clinical signs and symptoms. Nevertheless, in cases where classical sepsis symptoms were present, but no microbial isolates were obtained, management primarily involved empirical antibiotic therapy based on clinical judgment and local guidelines.
The study revealed that Gram-negative bacteria were predominant, accounting for 75.3% of the isolates, compared to Gram-positive bacteria. This result aligns with previous research from Ethiopia, South Africa, Germany, Tanzania, Uganda, and Nepal, where Gram-negative bacteria were also the majority (Pokhrel et al., 2018; Tessema et al., 2021; Sherif et al., 2023; Majigo et al., 2023; Zamarano et al., 2021; Thomas et al., 2024). However, this percentage is significantly higher than the 58.1% Gram-negative bacteria predominance reported in a systematic review from Iran (Moftian et al., 2023). Among the bacterial pathogens isolated, K. pneumoniae was the most frequently isolated followed by S. aureus and Acinetobacter spp. This finding is supported by a systematic review and a review in Sub-Saharan Africa, Uganda, and Pakistan (Tumuhamye et al., 2020; Atif et al., 2021; Okomo et al., 2019). The high frequency of K. pneumoniae isolates in the current study is particularly concerning, as this pathogen is known to be a common cause of healthcare-associated infections and is often associated with multidrug resistance. The prevalence of S. aureus and Acinetobacter spp. is also significant, as these bacteria can be challenging to treat due to their ability to develop resistance to various antimicrobial agents.
The antimicrobial susceptibility data revealed alarming levels of resistance among both gram-positive and gram-negative bacterial isolates. This concerning trend underscores the critical need for alternative therapeutic strategies and judicious antibiotic use to combat the growing challenge of antimicrobial resistance. Among the gram-positive bacteria, CoNS exhibited 100% resistance to penicillin, oxacillin, and gentamicin, indicating that these common antimicrobial agents are no longer effective against CoNS infections. Similarly, S. aureus showed high resistance rates to penicillin (94.6%), oxacillin (64.9%), and gentamicin (40.5%), suggesting that empiric treatment with these drugs may be increasingly ineffective. S. viridans also demonstrated significant resistance to ampicillin (73.3%) and vancomycin (20%), which are commonly used therapeutic options for streptococcal infections. These findings highlight the urgent need for new antimicrobial agents and strategies to address the rising threat of antimicrobial resistance.
Gram-negative bacterial isolates also exhibited substantial resistance patterns. K. pneumoniae showed the most alarming resistance to highly active antimicrobial agents, including meropenem (88.1%), ceftazidime (83.6%), ceftriaxone (83.6%), and amoxicillin-clavulanate (69%). This indicates that clinicians may have limited treatment options for K. pneumoniae infections, as these drugs are often considered last-line or “reserve” antimicrobials. Although E. coli showed relatively lower resistance rates compared to K. pneumoniae, it still presented significant resistance to ampicillin (66.7%), gentamicin (61.1%), and amoxicillin-clavulanate (55.6%). These findings are consistent with the previous study from the Tigray region, Ethiopia, in which K. pneumoniae and E. coli were resistant to common antimicrobial agents (Weldu et al., 2020). These observed patterns of resistance to commonly used antimicrobial drugs, such as ampicillin, ceftazidime, ceftriaxone, gentamicin, and amoxicillin-clavulanic acid, have been reported in other studies as well (Bai et al., 2021; Weldu et al., 2020; Sherif et al., 2023), suggesting that these resistance trends are widespread and not limited to the specific setting of this study.
Furthermore, the majority (61.6%) of the bacterial isolates were MDR, although this rate is lower than those reported in some other studies where the proportion of MDR was as high as 84% (Sherif et al., 2023; Zenebe et al., 2021). Among specific bacterial pathogens, K. pneumoniae had the highest rate of MDR, with 90.9% of the isolates being resistant to three or more antibiotic classes. Other bacterial pathogens, including K. ozaenae, K. oxytoca, and P. mirabilis, also exhibited 100% MDR. Additionally, S. aureus, Acinetobacter spp., E. coli, and non-fermenting Gram-negative rods demonstrated high MDR rates of 40.5, 43.5, 61.1, and 41.2%, respectively. The high levels of MDR observed in this study are concerning and call for stringent antibiotic stewardship programs to mitigate the spread of resistant strains. Multidrug resistance is particularly alarming as it severely limits the available treatment options and increases the risk of treatment failures. These resistance patterns highlight the urgent need for continuous surveillance and the development of new antimicrobial agents to effectively address the growing threat of antimicrobial resistance.
The study identified several key factors significantly associated with neonatal sepsis. One notable finding was the correlation between the year of admission and the incidence of neonatal sepsis. This temporal trend could be influenced by various factors, including changes in hospital practices, hygiene protocols, and seasonal trends. Additionally, variations in antibiotic resistance patterns, broader epidemiological factors, and improvements in diagnostic techniques could also contribute to these yearly fluctuations. Another important risk factor identified was rapid breathing, which can serve as an early clinical sign of sepsis, often indicating an underlying infection that requires immediate medical attention. The study found that neonates presenting with increased respiration were more likely to develop sepsis. Furthermore, the association between suspected pneumonia and meningitis with neonatal sepsis highlights the potential for overlapping clinical presentations. These symptoms had a higher likelihood of developing into sepsis, underscoring the importance of vigilant clinical assessment and timely intervention in neonatal care.
Limitations
This study has several important limitations that should be considered when interpreting the results. Since the research was conducted at a single healthcare facility, it may limit the generalizability of the findings to other geographical regions or healthcare settings. The patient population and antimicrobial resistance patterns observed at this one site may not be representative of broader regional or national trends. Furthermore, the study did not include molecular typing of bacterial isolates, which could have provided more detailed insights into the genetic mechanisms underlying antimicrobial resistance and the epidemiology of the infections. The absence of this molecular data limits the depth of understanding regarding the specific strains and resistance patterns present in the bacterial population studied.
Conclusion
Newborn sepsis caused by highly resistant bacteria presents a significant challenge at the UoGCSH. This study identified a high prevalence of culture-confirmed sepsis, with Gram-negative bacteria, especially K. pneumoniae, dominating the isolated pathogens. These bacteria exhibited alarming resistance to commonly used antibiotics, with a very high proportion demonstrating multidrug resistance. K. pneumoniae displayed the most concerning resistance rates. Additionally, the study linked specific factors like year of admission, rapid breathing, suspected pneumonia, and suspected meningitis to an increased risk of neonatal sepsis.
Recommendation
• There should be continuous surveillance of bacterial pathogens causing neonatal sepsis, monitoring their evolving antibiotic susceptibility patterns, which are crucial to inform effective treatment guidelines.
• The hospital should improve diagnostic techniques for the early and accurate identification of bacterial pathogens causing neonatal sepsis.
• The hospital should tailor antibiotic regimens based on the specific bacterial profiles and resistance patterns identified in the local hospital setting to improve the effectiveness of treatments.
• The Federal Ministry of Health and the regional health bureau should develop and implement robust antibiotic stewardship programs to optimize the use of antibiotics, reduce unnecessary prescriptions, and curb the spread of resistant strains.
• The Federal Ministry of Health should develop training programs and provide healthcare providers with the latest guidelines for managing neonatal sepsis.
• Further study should be conducted using molecular techniques to improve the detection of causative pathogens, particularly for those culture-negative patients.
• Researchers should invest significant effort to discover new antimicrobial agents that can effectively combat the resistant strains and provide effective treatment options for neonatal sepsis.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by University of Gondar College of Medicine and Health Science Institution Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin because this research was conducted retrospectively using medical record data; it analyzes data collected in the past. Due to this retrospective nature, obtaining informed consent from participants was not considered practical. Informed consent typically involves explaining the research to potential participants or guardians and obtaining their permission to be involved. In a retrospective study, we are looking back at existing data, and contacting past participants to get their consent now would be difficult or impossible. Therefore, with the approval of the IRB, the requirement for informed consent was waived.
Author contributions
TD: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. GB: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – review & editing. GA: Investigation, Methodology, Resources, Visualization, Writing – review & editing. WF: Investigation, Methodology, Resources, Visualization, Writing – review & editing. MW: Investigation, Methodology, Resources, Visualization, Writing – review & editing. TF: Investigation, Methodology, Resources, Visualization, Writing – review & editing. MM: Investigation, Methodology, Resources, Visualization, Writing – review & editing. SB: Investigation, Methodology, Resources, Visualization, Writing – review & editing. MG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to express their sincere gratitude to the participants from the UoGCSH for their valuable contributions to this study. We would also like to thank the dedicated data collectors for their diligent efforts in gathering the required data that made this research possible.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AMR, Antimicrobial resistance; BSI, Blood stream infection; CLSI, Clinical Laboratory Standards Institute; CoNS, Coagulase negative staphylococcus; ERC, Ethical review committee; NFGNR, Non-fermenting gram-negative rods; NICU, Neonatal intensive care unit; MDR, Multi-drug resistance; MRSA, Methicillin resistant Staphylococcus aureus; NFGNR, Non-fermenter gram negative rods; UoGCSH, University of Gondar Comprehensive Specialized Hospital.
References
Abate, B. B., Kasie, A. M., Reta, M. A., and Kassaw, M. W. (2020). Neonatal sepsis and its associated factors in East Africa: a systematic review and meta-analysis. Int. J. Public Health 65, 1623–1633. doi: 10.1007/s00038-020-01489-x
Acheampong, E. N., Tsiase, J. A., Afriyie, D. K., and Amponsah, S. K. (2022). Neonatal Sepsis in a resource-limited setting: causative microorganisms and antimicrobial susceptibility profile. Interdiscip. Perspect. Infect. Dis. 2022, 1–7. doi: 10.1155/2022/7905727
Akbarian-Rad, Z., Riahi, S. M., Abdollahi, A., Sabbagh, P., Ebrahimpour, S., Javanian, M., et al. (2020). Neonatal sepsis in Iran: a systematic review and meta-analysis on national prevalence and causative pathogens. PLoS One 15:e0227570. doi: 10.1371/journal.pone.0227570
Aku, F. Y., Akweongo, P., Nyarko, K., Sackey, S., Wurapa, F., Afari, E. A., et al. (2018). Bacteriological profile and antibiotic susceptibility pattern of common isolates of neonatal sepsis, ho municipality, Ghana-2016. Mat. Health Neonatol. Perinatol. 4:2. doi: 10.1186/s40748-017-0071-z
Alam, M., Pillai, P., Kapur, P., and Pillai, K. (2011). Resistant patterns of bacteria isolated from bloodstream infections at a university hospital in Delhi. J. Pharm. Bioallied Sci. 3, 525–530. doi: 10.4103/0975-7406.90106
Almohammady, M. N., Eltahlawy, E. M., and Reda, N. M. (2020). Pattern of bacterial profile and antibiotic susceptibility among neonatal sepsis cases at Cairo University children hospital. J. Taibah Univ. Med. Sci. 15, 39–47. doi: 10.1016/j.jtumed.2019.12.005
Arega, B., Wolde-Amanuel, Y., Adane, K., Belay, E., Abubeker, A., and Asrat, D. (2017). Rare bacterial isolates causing bloodstream infections in Ethiopian patients with cancer. Infect. Agents Cancer 12, 1–6. doi: 10.1186/s13027-017-0150-9
Arega, B., Woldeamanuel, Y., Adane, K., Sherif, A. A., and Asrat, D. (2018). Microbial spectrum and drug-resistance profile of isolates causing bloodstream infections in febrile cancer patients at a referral hospital in Addis Ababa, Ethiopia. Infect. Drug Resist. 11, 1511–1519. doi: 10.2147/IDR.S168867
Assemie, M. A., Alene, M., Yismaw, L., Ketema, D. B., Lamore, Y., Petrucka, P., et al. (2020). Prevalence of neonatal Sepsis in Ethiopia: a systematic review and Meta-analysis. Int. J. Pediatr. 2020, 1–9. doi: 10.1155/2020/6468492
Atif, M., Zia, R., Malik, I., Ahmad, N., and Sarwar, S. (2021). Treatment outcomes, antibiotic use and its resistance pattern among neonatal sepsis patients attending Bahawal Victoria Hospital, Pakistan. PLoS One 16:e0244866. doi: 10.1371/journal.pone.0244866
Bai, X., Wei, Q., Duan, T., Yi, Y., Peng, H., and Hu, L. (2021). Predominance of gram-negative infections a cause of neonatal sepsis among low birth weight preterm infants. J. Lab. Med. 45, 7–12. doi: 10.1515/labmed-2020-0022
Bayih, W. A., Ayalew, M. Y., Chanie, E. S., Abate, B. B., Alemayehu, S. A., Belay, D. M., et al. (2021). The burden of neonatal sepsis and its association with antenatal urinary tract infection and intra-partum fever among admitted neonates in Ethiopia: a systematic review and meta-analysis. Heliyon 7:e06121. doi: 10.1016/j.heliyon.2021.e06121
Bekele, K., Bekele, F., Edosa, D., Mekonnen, M., and Benayew, M. (2022). Magnitude and associated factors of neonatal sepsis among neonates admitted to neonatal intensive care unit of northern Oromia hospitals, Ethiopia: a multicenter cross-sectional study. Ann. Med. Surg. 78:103782. doi: 10.1016/j.amsu.2022.103782
Belachew, A., and Tewabe, T. (2020). Neonatal sepsis and its association with birth weight and gestational age among admitted neonates in Ethiopia: systematic review and meta-analysis. BMC Pediatr. 20, 1–7. doi: 10.1186/s12887-020-1949-x
Bethou, A., and Bhat, B. V. (2022). Neonatal sepsis—newer insights. Indian J. Pediatr. 89, 267–273. doi: 10.1007/s12098-021-03852-z
Bhat, Y. R., Lewis, L. E. S., and Ke, V. (2011). Bacterial isolates of early-onset neonatal sepsis and their antibiotic susceptibility pattern between 1998 and 2004: an audit from a center in India. Ital. J. Pediatr. 37:32. doi: 10.1186/1824-7288-37-32
Chaurasia, S., Jeeva Sankar, M., Agarwal, R., Yadav, C. P., Arya, S., Kapil, A., et al. (2016). Characterisation and antimicrobial resistance of sepsis pathogens in neonates born in tertiary care centres in Delhi, India: a cohort study. Lancet Glob. Health 4, e752–e760. doi: 10.1016/S2214-109X(16)30148-6
Chaurasia, S., Sivanandan, S., Agarwal, R., Ellis, S., Sharland, M., and Sankar, M. J. (2019). Neonatal sepsis in South Asia: huge burden and spiralling antimicrobial resistance. BMJ 364:k5314. doi: 10.1136/bmj.k5314
Clinical and Laboratory Standards Institute. (2019). Performance Standards for Antimicrobial Susceptibility Testing, Twenty-ninth CLSI supplement, M100. Wayne, Pennsylvania: Clinical and Laboratory Standards Institute.
Egbe, F. N., Cowden, C., Mwananyanda, L., Pierre, C., Mwansa, J., Lukwesa Musyani, C., et al. (2023). Etiology of bacterial Sepsis and isolate resistance patterns in hospitalized neonates in Zambia. Pediatr. Infect. Dis. J. 42, 921–926. doi: 10.1097/INF.0000000000004008
Ershad, M., Mostafa, A., Dela Cruz, M., and Vearrier, D. (2019). Neonatal sepsis. Curr. Emerg. Hosp. Med. Rep. 7, 83–90. doi: 10.1007/s40138-019-00188-z
Fang, P., Gao, K., Yang, J., Li, T., Gong, W., Sun, Q., et al. (2023). Prevalence of multidrug-resistant pathogens causing neonatal early and late onset Sepsis, a retrospective study from the tertiary referral Children's hospital. Infect. Drug Resist. 16, 4213–4225. doi: 10.2147/IDR.S416020
Fenta, G. M., Woldemariam, H. K., Metaferia, Y., Seid, A., and Gebretsadik, D. (2022). Admission outcome and antimicrobial resistance pattern of bacterial isolates among neonates with suspected Sepsis in neonatal intensive care unit at Dessie comprehensive specialized hospital, Dessie, northeastern Ethiopia. Interdiscip. Perspect. Infect. Dis. 2022, 1–13. doi: 10.1155/2022/1318295
Fleischmann-Struzek, C., Goldfarb, D. M., Schlattmann, P., Schlapbach, L. J., Reinhart, K., and Kissoon, N. (2018). The global burden of paediatric and neonatal sepsis: a systematic review. Lancet Respir. Med. 6, 223–230. doi: 10.1016/S2213-2600(18)30063-8
Georges Pius, K., Aurore Albane, E., Marie-Paul, B., Komba, D., Ngando, V., and Eteme, A. (2022). Neonatal sepsis: highlights and controversies. J. Pediatr. Neonatal. 4, 1–5. doi: 10.33425/2689-1085.1035
Geyesus, T., Moges, F., Eshetie, S., Yeshitela, B., and Abate, E. (2017). Bacterial etiologic agents causing neonatal sepsis and associated risk factors in Gondar, Northwest Ethiopia. BMC Pediatr. 17:137. doi: 10.1186/s12887-017-0892-y
Glaser, M. A., Hughes, L. M., Jnah, A., and Newberry, D. (2021). Neonatal sepsis: a review of pathophysiology and current management strategies. Adv. Neonatal Care 21, 49–60. doi: 10.1097/ANC.0000000000000769
Gobezie, N. Z., Endalew, N. S., Tawuye, H. Y., and Aytolign, H. A. (2023). Prevalence and associated factors of postoperative orthostatic intolerance at University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia, 2022: cross sectional study. BMC Surg. 23:108. doi: 10.1186/s12893-023-02015-5
Jatsho, J., Nishizawa, Y., Pelzom, D., and Sharma, R. (2020). Clinical and bacteriological profile of neonatal Sepsis: a prospective hospital-based study. Int. J. Pediatr. 2020, 1–9. doi: 10.1155/2020/1835945
Kamalakannan, S. K. (2018). Neonatal sepsis past to present. Biomed. J. Sci. Tech. Res. 3, 3309–3314. doi: 10.26717/BJSTR.2018.03.000909
Klingenberg, C., Kornelisse, R. F., Buonocore, G., Maier, R. F., and Stocker, M. (2018). Culture-negative early-onset neonatal Sepsis — at the crossroad between efficient Sepsis care and antimicrobial stewardship. Front. Pediatr. 6:285. doi: 10.3389/fped.2018.00285
Li, J., Xiang, L., Chen, X., Li, S., Sun, Q., Cheng, X., et al. (2023). Global, regional, and national burden of neonatal sepsis and other neonatal infections, 1990–2019: findings from the global burden of disease study 2019. Eur. J. Pediatr. 182, 2335–2343. doi: 10.1007/s00431-023-04911-7
Majigo, M., Makupa, J., Mwazyunga, Z., Luoga, A., Kisinga, J., Mwamkoa, B., et al. (2023). Bacterial Aetiology of neonatal Sepsis and antimicrobial resistance pattern at the regional referral hospital, Dar es Salam, Tanzania; a call to strengthening antibiotic stewardship program. Antibiotics (Basel) 12:767. doi: 10.3390/antibiotics12040767
Mezgebu, T., Ossabo, G., Zekiwos, A., Mohammed, H., and Demisse, Z. (2023). Neonatal sepsis and its associated factors among neonates admitted to the neonatal intensive care unit in Wachemo university comprehensive specialized hospital, southern Ethiopia, 2022. Front. Pediatr. 11:1184205. doi: 10.3389/fped.2023.1184205
Mhada, T. V., Fredrick, F., Matee, M. I., and Massawe, A. (2012). Neonatal sepsis at Muhimbili National Hospital, Dar Es Salaam, Tanzania; aetiology, antimicrobial sensitivity pattern and clinical outcome. BMC Public Health 12:904. doi: 10.1186/1471-2458-12-904
Moftian, N., Rezaei-hachesu, P., Arab-Zozani, M., Samad-soltani, T., Esfandiari, A., Tabib, M. S., et al. (2023). Prevalence of gram-negative bacteria and their antibiotic resistance in neonatal sepsis in Iran: a systematic review and meta-analysis. BMC Infect. Dis. 23:534. doi: 10.1186/s12879-023-08508-1
Molloy, E. J., Wynn, J. L., Bliss, J., Koenig, J. M., Keij, F. M., McGovern, M., et al. (2020). Neonatal sepsis: need for consensus definition, collaboration and core outcomes. Pediatr. Res. 88, 2–4. doi: 10.1038/s41390-020-0850-5
Moni, S. C., Mollah, A. H., Banerjee, M., Khan, T. H., Sejuti, A., and Morshed, S. S. (2020). Neonatal Sepsis: clinical characteristics, epidemiology and antibiotic sensitivity pattern of the bacterial pathogens in neonatal intensive care unit of a tertiary care hospital. Mymensingh Med. J. 29, 366–375
Mustefa, A., Abera, A., Aseffa, A., Abathun, T., Degefa, N., and Tadesse, H. (2020). Prevalence of neonatal sepsis and associated factors amongst neonates admitted in arbaminch general hospital, arbaminch, southern Ethiopia, 2019. J. Pediatr. Neonatal Care 10, 1–7. doi: 10.15406/jpnc.2020.10.00404
Negussie, A., Mulugeta, G., Bedru, A., Ali, I., Shimeles, D., Lema, T., et al. (2015). Bacteriological profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia suspected children in selected hospitals Addis Ababa, Ethiopia. Int. J. Biol. Med. Res. 6, 4709–4717
Okomo, U., Akpalu, E. N. K., Le Doare, K., Roca, A., Cousens, S., Jarde, A., et al. (2019). Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: a systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect. Dis. 19, 1219–1234. doi: 10.1016/S1473-3099(19)30414-1
Pataskar, A., Chandel, A., Chauhan, V., and Jain, M. (2023). Gram-negative late onset neonatal Sepsis in a tertiary care center from Central India: a retrospective analysis. Clin. Med. Insights Pediatr. 17:11795565231189595. doi: 10.1177/11795565231189595
Peterside, O., Pondei, K., and Akinbami, F. O. (2015). Bacteriological profile and antibiotic susceptibility pattern of neonatal Sepsis at a teaching Hospital in Bayelsa State, Nigeria. Trop. Med. Health 43, 183–190. doi: 10.2149/tmh.2015-03
Pokhrel, B., Koirala, T., Shah, G., Joshi, S., and Baral, P. (2018). Bacteriological profile and antibiotic susceptibility of neonatal sepsis in neonatal intensive care unit of a tertiary hospital in Nepal. BMC Pediatr. 18:208. doi: 10.1186/s12887-018-1176-x
Poyekar, S. S. (2022). Neonatal sepsis – microbiological spectrum, antimicrobial sensitivity, and risk factors for mortality in newborn unit of rural teaching hospital: a retrospective study. Med. J. Dr DY Patil Vidyapeeth 15, 331–334. doi: 10.4103/mjdrdypu.mjdrdypu_558_20
Ranjeva, S. L., Warf, B. C., and Schiff, S. J. (2018). Economic burden of neonatal sepsis in sub-Saharan Africa. BMJ Glob. Health 3:e000347. doi: 10.1136/bmjgh-2017-000347
Reddy, K., Bekker, A., Whitelaw, A. C., Esterhuizen, T. M., and Dramowski, A. (2021). A retrospective analysis of pathogen profile, antimicrobial resistance and mortality in neonatal hospital-acquired bloodstream infections from 2009-2018 at Tygerberg hospital, South Africa. PLoS One 16:e0245089. doi: 10.1371/journal.pone.0245089
Roble, A. K., Ayehubizu, L. M., and Olad, H. M. (2022). Neonatal sepsis and associated factors among neonates admitted to neonatal intensive care unit in general hospitals, eastern Ethiopia 2020. Clin. Med. Insights 16:117955652210983. doi: 10.1177/11795565221098346
Sands, K., Carvalho, M. J., Portal, E., Thomson, K., Dyer, C., Akpulu, C., et al. (2021). Characterization of antimicrobial-resistant gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat. Microbiol. 6, 512–523. doi: 10.1038/s41564-021-00870-7
Satar, M., and Özlü, F. (2012). Neonatal sepsis: a continuing disease burden. Turk. J. Pediatr. 54, 449–457
Shah, M. H., McAleese, S., Kadam, S., Parikh, T., Vaidya, U., Sanghavi, S., et al. (2022). Emerging antibiotic resistance patterns in a neonatal intensive care unit in Pune, India: a 2-year retrospective study. Front. Pediatr. 10:10. doi: 10.3389/fped.2022.864115
Sherif, M., Abera, D., and Desta, K. (2023). Prevalence and antibiotic resistance pattern of bacteria from sepsis suspected neonates at St. Paul’s hospital millennium medical college, Addis Ababa, Ethiopia. BMC Pediatr. 23:575. doi: 10.1186/s12887-023-04399-y
Siddiqui, T., Dubey, A., Kar, M., Patel, S. S., Sahu, C., and Ghoshal, U. (2023). Bacteriological profiles and antibiotic susceptibility of neonatal sepsis in a university hospital of northern India. J. Family Med. Prim. Care 12, 493–498. doi: 10.4103/jfmpc.jfmpc_1535_22
Tessema, B., Lippmann, N., Knüpfer, M., Sack, U., and König, B. (2021). Antibiotic resistance patterns of bacterial isolates from neonatal Sepsis patients at University Hospital of Leipzig, Germany. Antibiotics 10:323. doi: 10.3390/antibiotics10030323
Thapa, S., and Sapkota, L. B. (2019). Changing trend of neonatal septicemia and antibiotic susceptibility pattern of isolates in Nepal. Int. J. Pediatr. 2019, 1–7. doi: 10.1155/2019/3784529
Thomas, R., Ondongo-Ezhet, C., Motsoaledi, N., Sharland, M., Clements, M., and Velaphi, S. (2024). Incidence, pathogens and antimicrobial resistance of blood and cerebrospinal fluid isolates from a tertiary neonatal unit in South Africa: a 10 year retrospective review. PLoS One 19:e0297371. doi: 10.1371/journal.pone.0297371
Tumuhamye, J., Sommerfelt, H., Bwanga, F., Ndeezi, G., Mukunya, D., Napyo, A., et al. (2020). Neonatal sepsis at Mulago national referral hospital in Uganda: etiology, antimicrobial resistance, associated factors and case fatality risk. PLoS One 15:e0237085. doi: 10.1371/journal.pone.0237085
Uwe, N. O., Ezenwa, B. N., Fajolu, I. B., Oshun, P., Chukwuma, S. T., and Ezeaka, V. C. (2022). Antimicrobial susceptibility and neonatal sepsis in a tertiary care facility in Nigeria: a changing trend? JAC Antimicrob. Resist. 4:4. doi: 10.1093/jacamr/dlac100
Verma, P. K., Berwal, P. K., Nagaraj, N., Swami, S. S. S., Jivaji, P., and Narayan, S. (2015). Neonatal sepsis: epidemiology, clinical spectrum, recent antimicrobial agents and their antibiotic susceptibility pattern. Int. J. Contemp. Pediatr. 2, 176–180. doi: 10.18203/2349-3291.ijcp20150523
Weldu, Y., Naizgi, M., Hadgu, A., Desta, A. A., Kahsay, A., Negash, L., et al. (2020). Neonatal septicemia at intensive care unit, Ayder comprehensive specialized hospital, Tigray, North Ethiopia: bacteriological profile, drug susceptibility pattern, and associated factors. PLoS One 15:e0235391. doi: 10.1371/journal.pone.0235391
Wen, S. C. H., Ezure, Y., Rolley, L., Spurling, G., Lau, C. L., Riaz, S., et al. (2021). Gram-negative neonatal sepsis in low- and lower-middle-income countries and WHO empirical antibiotic recommendations: a systematic review and meta-analysis. PLoS Med. 18:e1003787. doi: 10.1371/journal.pmed.1003787
Worku, E., Fenta, D. A., and Ali, M. M. (2022). Bacterial etiology and risk factors among newborns suspected of sepsis at Hawassa, Ethiopia. Sci. Rep. 12:20187. doi: 10.1038/s41598-022-24572-0
Yadav, N. S., Sharma, S., Chaudhary, D. K., Panthi, P., Pokhrel, P., Shrestha, A., et al. (2018). Bacteriological profile of neonatal sepsis and antibiotic susceptibility pattern of isolates admitted at Kanti Children’s hospital, Kathmandu, Nepal. BMC. Res. Notes 11:301. doi: 10.1186/s13104-018-3394-6
Zamarano, H., Musinguzi, B., Kabajulizi, I., Manirakiza, G., Guti, W., Muhwezi, I., et al. (2021). Bacteriological profile, antibiotic susceptibility and factors associated with neonatal Septicaemia at Kilembe mines hospital, Kasese District Western Uganda. BMC Microbiol. 21:303. doi: 10.1186/s12866-021-02367-z
Keywords: neonate sepsis, bacterial profiles, antibiotic susceptibility, University of Gondar Comprehensive Specialized Hospital, Ethiopia
Citation: Deress T, Belay G, Ayenew G, Ferede W, Worku M, Feleke T, Mulu M, Belay S and Getie M (2024) Bacterial profiles and their antibiotic susceptibility patterns in neonatal sepsis at the University of Gondar Comprehensive Specialized Hospital, Ethiopia. Front. Microbiol. 15:1461689. doi: 10.3389/fmicb.2024.1461689
Edited by:
Valério Monteiro-Neto, Federal University of Maranhão, BrazilReviewed by:
Ashutosh Pathak, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGI), IndiaOkon Okwong Kenneth, Federal Medical Center Makurdi, Nigeria
Copyright © 2024 Deress, Belay, Ayenew, Ferede, Worku, Feleke, Mulu, Belay and Getie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teshiwal Deress, dGVzaGl3YWxkZXJlc3NAZ21haWwuY29t
†ORCID: Teshiwal Deress, http://orcid.org/0000-0002-1678-604X
Michael Getie, https://orcid.org/0000-0001-7461-859X
 Teshiwal Deress
Teshiwal Deress Gizeaddis Belay
Gizeaddis Belay Getahun Ayenew
Getahun Ayenew Worku Ferede4
Worku Ferede4