- 1Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Neston, United Kingdom
- 2Centre for Genomic Research, University of Liverpool, Liverpool, United Kingdom
Introduction: Raw-meat diets (RMD) for dogs, comprising unprocessed or non-heat-treated animal material, are increasingly popular. However, RMDs have been demonstrated to be contaminated with antimicrobial resistant (AMR) bacteria, and there is concern that such diets may pose a zoonotic disease risk. Additionally, dogs fed RMD may shed more AMR- fecal bacteria compared to those fed conventional cooked diets. Data from the UK remain limited; the present study investigated the presence of AMR-Escherichia coli in the feces of RMD and non-RMD (NRMD)-fed dogs in the UK, the E. coli AMR gene complement, and the lifestyle risk factors associated with AMR- E. coli carriage.
Methods: Fecal samples from UK-owned dogs (N = 193 RMD, N = 239 NRMD) and questionnaires discussing lifestyle factors, were obtained between October 2020-August 2021. Samples underwent culture and antimicrobial susceptibility testing to determine the presence of AMR-E. coli. Whole genome sequencing determined AMR gene carriage. Risk factors for the presence of AMR-E. coli were determined by multivariable modeling.
Results: RMD dogs carried significantly more fecal AMR E. coli (p < 0.001), including third-generation cephalosporin resistant, extended-spectrum beta-lactamase (ESBL) producing, and multidrug resistant isolates and multivariable modeling confirmed raw-meat diets to be a significant risk factor. The blaCTX–M–15 gene was the most frequently identified blaESBL gene. The blaCTX–M–55 and blaSHV–66 genes were also prevalent and were only found in RMD dogs. The mobile colistin resistance gene, mcr-4 was identified in one ESBL-producing E. coli isolate from a NRMD-fed dog.
Conclusion: This study has shown that dogs fed RMD in the UK are significantly more likely to shed E. coli which is resistant to highest priority critically important antibiotics, and multidrug resistant E. coli, than dogs fed NRMD. Additionally, AMR-E. coli isolates from RMD-fed dogs harbor multiple, diverse, and novel AMR genes. Therefore, provision of RMD to dogs could pose an important potential threat to human and animal health, especially given the close nature of the relationship many owners share with their pets. Awareness of these findings should be shared with pet owners, veterinary and medical professionals, pet food manufacturers and public health to mitigate potential risks.
Introduction
Raw meat diets (RMD) for pets remain a popular alternative diet choice, and while conventional cooked kibble-based diets continue to be a staple for the majority of dogs, RMD is increasingly fed as at least a constituent part of the diet for many (Dodd et al., 2020; Morgan et al., 2022; PDSA, 2022). The 2022 PDSA PAW report estimated that 7% of UK dogs were fed RMD, equating to 790,000 dogs (PDSA, 2022). RMDs are comprised of muscle, bone, skin, cartilage, tendon and organs from livestock and wild animals, which have not undergone heat treatment or cooking during the food production process (Freeman et al., 2013; Davies et al., 2019), and may be provided in a commercial pre-prepared food, or home-prepared. RMDs for dogs and cats have been demonstrated to harbor pathogenic and zoonotic organisms, including E. coli O157:H7, Salmonella spp., Listeria monocytogenes, Campylobacter spp., amongst others (Davies et al., 2019; Kaindama et al., 2020) and such bacteria have been found to be shed by dogs and cats fed RMD globally (Morley et al., 2006; Finley et al., 2007; Lefebvre et al., 2008; Leonard et al., 2015; Baede et al., 2017; Runesvärd et al., 2020; Viegas et al., 2020; Groat et al., 2022).
Furthermore, antimicrobial-resistant (AMR) bacteria have been isolated from samples of pre-prepared RMD for pets in mainland Europe (Nilsson, 2015; Baede et al., 2017; Nüesch-Inderbinen et al., 2019) and the UK (Morgan et al., 2024) and shedding of AMR bacteria by companion animals fed RMD is of increasing concern. Provision of RMD has been identified as a risk factor for canine fecal carriage of AMR E. coli in mainland Europe (van den Bunt et al., 2020), with greater proportions of dogs fed RMD shedding extended-spectrum beta-lactamase producing (ESBL)-E. coli demonstrating resistance to highest priority critically important antibiotics (HPCIAs), including third-generation cephalosporins and quinolones than those fed non-raw diets (NRMD) (Runesvärd et al., 2020).
In the UK, provision of RMD has previously been identified as a risk factor for fecal carriage of AMR E. coli in both healthy, non-veterinary visiting dogs (Schmidt et al., 2015) and those visiting veterinary practices (Wedley et al., 2017). Additionally, feeding RMD was identified as a risk factor for carriage of third-generation cephalosporin resistant (3GCR) E. coli in rural-living dogs (Sealey et al., 2022). A small study indicated that AMR, 3GCR and multidrug resistant (MDR) E. coli were significantly more likely to be shed by dogs fed RMD, compared to those fed NRMD, with 31% of dogs fed RMD shedding 3GCR-E. coli, compared to 4% of dogs fed NRMD (Groat et al., 2022).
Genes encoding 3GCR (such as blaESBL genes including those of blaCTX–M group 1 and blaCMY) and resistance to quinolones (such as qnr genes), may also be co-harbored and plasmid encoded, thus mobile, increasing the potential for transmission of MDR. Plasmid-mediated AMR genes, have been identified in E. coli isolated from companion animals, and have been associated with those fed RMD (Groat et al., 2022; Mounsey et al., 2022; Sealey et al., 2022).
Dogs and their owners share close and frequent contact, therefore the risk posed by RMD with regards to zoonotic disease and AMR is a potential public health concern. Despite the popularity and interest surrounding RMD, data from larger scale studies surrounding the potential AMR risks associated with their provision as a diet for dogs, particularly in the UK, remain limited, and associated AMR-E. coli genome sequencing data is sparse.
Aims
The aims of this study were to determine the presence of AMR E. coli in the feces of dogs fed either RMD or NRMD in the UK, with focus on 3GCR- E. coli, ESBL-producing E. coli and MDR-E. coli, alongside investigation of the AMR genes harbored by E. coli isolates via whole genome sequencing (WGS). Additionally, this study aimed to determine the dog and owner lifestyle risk factors for the carriage of such AMR E. coli in canine feces.
Materials and methods
This study was cross-sectional in design. Data were collected between October 2020–August 2021. Participant recruitment was via email contact of dog owners who had previously participated in related studies (Morgan et al., 2022) and had agreed to be contacted further, and additionally, through social media. Following recruitment, participating households were sent a questionnaire and a fecal sample collection kit via post.
Dog owners were requested to collect one sample from a freshly evacuated stool at one time point from their dog. For multi-dog households, owners were requested to select one dog at random to participate in the study. Completed questionnaires and collected canine fecal samples were received by the laboratory by prepaid first class return post. Samples were stored at 4°C and tested within 48 h of sample delivery. Participant details were anonymised, and each sample and corresponding questionnaire was assigned a unique identification number.
The questionnaire asked about dog lifestyle and clinical factors including diet, recent antibiotic treatment, and veterinary visits, recent diarrhea, and treatment, contact with other animals and access to communal areas such as dog kennels, dog shows and public parks. It also collected data on owner factors including age, location in the country, receipt of antibiotics and place of work. Questions were multiple choice, with additional free text boxes included for owners to expand on their answers where appropriate.
Based on prior research from the UK (Groat et al., 2022), we hypothesized that the percentage of RMD-fed dogs carrying ESBL-producing E. coli was 30%, compared to 5% (or less) in dogs fed NRMD. A sample size of at least 32 in each group would provide 80% power to detect this difference in ESBL-producing E. coli with 95% confidence.
Microbiological methods
A 1 g sample of each fecal sample was homogenized in 4 ml buffered peptone water (BPW) at room temperature and incubated aerobically at 35°C ± 1 for 18–20 h. Following incubation, a 5 μl loopful of the homogenate was inoculated onto plain chromogenic Harlequin E. coli/Coliform Agar (HECA) (Neogen, UK) and HECA infused with 1 μg/ml cefotaxime, a third-generation cephalosporin (HECA+Cx); all plates were incubated at 35°C ± 1 for 18–20 h. Following incubation, plates were analyzed for the presence of typical E. coli colonies (dark blue-violet colonies, 0.1–2 mm diameter). To explore wider AMR diversity without selection, four typical colonies, where present, were randomly picked from each HECA plate, and two colonies were picked from each HECA+Cx plate to select for cephalosporin resistant E. coli specifically, then individually plated onto nutrient agar (NA) (Neogen, UK) plates and incubated at 35°C ± 1 for 18–20 h.
E. coli isolates from plain HECA plates underwent antimicrobial susceptibility testing (AST) via the disk diffusion method. Antibiotic disks were chosen representing antimicrobials used in dogs and humans, and susceptibility tested in compliance with European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations (EUCAST, 2022). Isolates were inoculated into sterile saline to 0.5 McFarland then a 5 μl loopful was spread onto Mueller-Hinton agar (Neogen, UK) and antibiotic disks applied. Plates were incubated aerobically at 35°C ± 1 for 18–20 h. Antimicrobials tested were ampicillin 10 μg, amoxicillin-clavulanate 20 μg/10 μg, ciprofloxacin 5 μg, tigecycline 15 μg, trimethoprim-sulphamethoxazole 1.25 μg/23.75 μg, amikacin 30 μg and meropenem 10 μg (MAST Group Ltd, Liverpool UK). A susceptible control strain of E. coli (ATCC 25922) was also tested.
Following incubation, zones of inhibition (ZOI) for each antibiotic disk were measured to the nearest millimeter. Human clinical breakpoints used for interpretation were as recommended by EUCAST (EUCAST, 2022) for all antibiotics other than amoxycillin-clavulanate, where the breakpoint used for interpretation was as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2020). Multidrug resistance (MDR) was defined as demonstrated phenotypic resistance to three or more classes of antibiotics (Magiorakos et al., 2012).
E. coli isolates from HECA+Cx plates initially underwent the extended-spectrum beta-lactamase (ESBL) double-disk test using cefotaxime 5 μg, cefotaxime 5 μg +clavulanic acid 10 μg, ceftazidime 10 μg and ceftazidime 10 μg +clavulanic acid 10 μg disks (EUCAST ESBL detection set, MAST Group Ltd, Liverpool UK). Plates were incubated at 35°C ± 1 for 18–20 h. For isolates where the ZOI surrounding the cephalosporin +clavulanic acid disk was a minimum of 5 mm larger than the ZOI for the corresponding cephalosporin disk alone for ≥ 1 antibiotic pair, an ESBL-producing phenotype was confirmed, and isolates underwent full AST as described above. Non-ESBL producing third-generation cephalosporin-resistant (3GCR) E. coli isolates which did not demonstrate a typical positive result for ESBL production on the double disk test, but which demonstrated a pattern suggestive of AmpC production whereby there was no, or minimal, ZOI present surrounding the clavulanic acid disk(s), were also subject to full AST.
All isolates were confirmed as E. coli by PCR of the uspA gene on cell lysates (Anastasi et al., 2010). Primers used were CCGATACGCTGCCAATCAGT (forward) and ACGCAGACCGTAGGCCAGAT (reverse), with an amplicon size of 884 base pairs, and 5× HOT FIREPol® Ready To Load Master Mix (Solis Biodyne, Estonia).
Whole genome sequencing
DNA extraction was performed on ESBL-producing E. coli isolates using the QIAamp® DNA mini kit (Qiagen, Crawley, UK).
Genomic DNA samples were submitted to the Centre for Genomic Research, University of Liverpool for Illumina NEBNext Ultra II FS DNA Library Prep, completed following the manufacturer’s protocol. Each library was quantified using Qubit and the size distribution assessed using the fragment analyser. These final libraries were pooled in equimolar amounts using the Qubit and fragment analyser data. The quantity and quality of the pool was assessed by Bioanalyzer and subsequently by qPCR using the Illumina Library Quantification Kit from Kapa (KK4854) on a Roche Light Cycler LC480II according to manufacturer’s instructions.
Following calculation of the molarity using qPCR data, template DNA was diluted to 300pM and denatured for 8 min at room temperature using freshly diluted 0.2 N sodium hydroxide (NaOH) and the reaction was subsequently terminated by the addition of 400 mM TrisCl pH = 8. To improve sequencing quality control 1% PhiX was spiked in. The libraries were sequenced on the Illumina® NovaSeq 6000 platform (Illumina®, San Diego, USA) following the standard workflow over 1 lane of an S4 flow cell, generating 2 × 150 bp paired end reads.
Quality- and adapter-trimmed reads were assembled using SPAdes v3.13.1 (Bankevich et al., 2012). Contigs shorter than 200 bp were removed and assemblies were included in the analyses if they had (1) assembly size +/− 50% of 4.5Mb, (2) genome completeness > 90% and duplication < 10% using BUSCO v4.0.4 (Simão et al., 2015) with the gammaproteobacteria database and (3) < 10% sample read assignment to non-Enterobacteriaceae taxa using MetaPhlAn v2.8.1 (Segata et al., 2012). MLST profiles and allele sequences were obtained from pubmlst.org. Allele sequences were aligned to assemblies using Bowtie2 version 2.3.5.1 (Langmead and Salzberg, 2012) in sensitive mode, and best aligned alleles for each locus were selected and used to determine sequence type. Sequence types were used to infer eBURST groups, using goeBURST,1 where group members shared at least 2 ST locus alleles. Genes were predicted using PROKKA version 1.14.0 (Seemann, 2014) and used to reconstruct the core genome across samples, using Panaroo v1.2.4 (Tonkin-Hill et al., 2020) in “moderate” mode. Samples were filtered to only include those with fewer than 30% of core bases missing with subsequent recalculation. Phylogenetic estimation was carried out using the Panaroo core gene alignment, with IQ-TREE v2.0 (Nguyen et al., 2015), with 1,000 bootstrap replicates using the GTR model. Tree visualization was carried out at microreact.org.
AMR genes were identified by interrogating genome assemblies with RGI version 5.1.0 (Alcock et al., 2020). Plasmids were identified using PlasmidFinder v2.1.6 (Carattoli et al., 2014) and the Enterobacteriaceae plasmid marker database.
Data analysis
Data analysis was undertaken in SPSS 27 [IBM Corp. (released 2020). IBM SPSS Statistics for Windows, Version 27.0. Armonk, NY: IBM Corp.]. Descriptive analysis was undertaken to determine the frequency and percentage (with 95% confidence intervals) of antimicrobial-resistant E. coli present at sample and isolate level for dogs fed RMD or NRMD. Comparisons between dogs fed RMD and NRMD were undertaken using the chi-square test (Fisher’s exact test for groups of N < 5), and significance was set at p < 0.05.
Descriptive analysis of categorical questionnaire response data (frequency, percentage) was undertaken. Based on the accompanying laboratory results, three outcomes were analyzed; “presence of ESBL- producing E. coli,” “presence of 3GCR-E. coli” and “presence of MDR-E. coli.” Odds ratios and 95% confidence intervals were generated by univariable logistic regression to identify explanatory variables associated with the three outcomes. Variables with a liberal p-value of < 0.3 were selected for inclusion into each multivariable model. Correlations between each variable were assessed, and where a high correlation coefficient (> 0.7) was identified, only the variable deemed most suitable was selected for inclusion into the model. Multivariable logistic regression models were built using a backward elimination method to sequentially remove variables with a p-value of > 0.05 until all remaining variables were significant at p < 0.05. Variables which had been eliminated were individually reinserted back into the model and checked to ensure that any confounding or significant variables had not been omitted. Plausible interactions between variables were also tested in the model to ensure no significant interactions had been missed and then “goodness of fit” of the final model was tested using the Hosmer-Lemeshow test.
Results
A total of 432 (193 RMD-fed, 239 NRMD-fed) canine fecal samples were received. E. coli was isolated from 92.6% (400/432; 191 RMD, 209 NRMD) of samples and E. coli which demonstrated resistance to at least one class of antibiotics was isolated from 39.4% (76/193) of RMD-fed and 13.8% (N = 33/239) of NRMD-fed dogs (p < 0.001) (Table 1). Dogs which were fed RMD carried significantly more 3GCR-E. coli (p < 0.001), ESBL-producing E. coli (p < 0.001), multidrug-resistant (MDR) ESBL-producing E. coli (p < 0.001) and fluoroquinolone-resistant (FQR) ESBL-producing E. coli (p < 0.001) than dogs fed NRMD (Table 1).
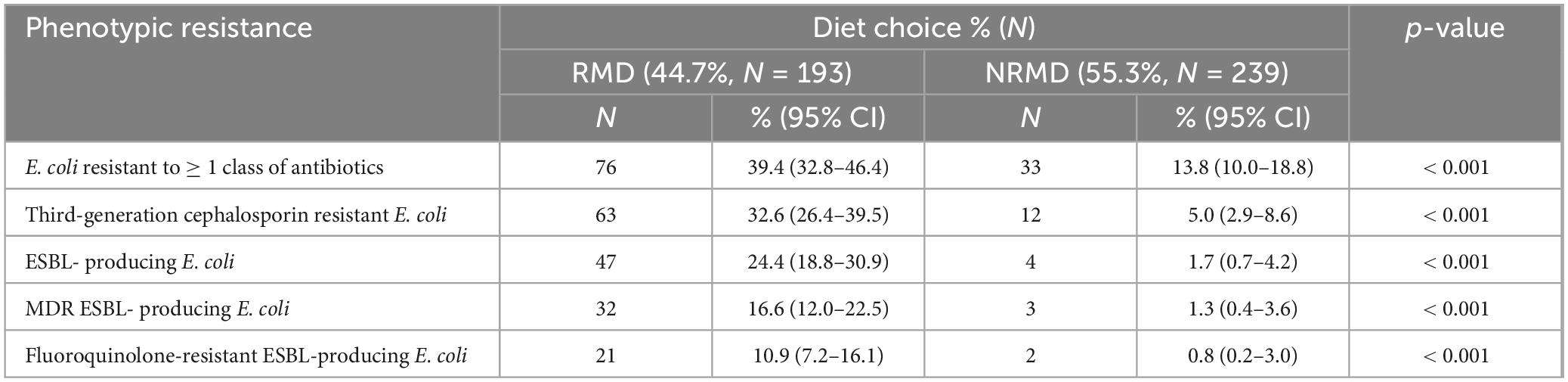
Table 1. Sample level data [Number (N) and percentage (%)] describing the overall phenotypic antimicrobial resistance demonstrated by E. coli isolated from the feces of dogs fed either a raw (RMD, N = 193) or non-raw (NRMD, N = 239) diet.
Of the dogs fed RMD, approximately one third shed 3GCR-E. coli in their feces and a quarter shed ESBL-producing E. coli. Additionally, 17% of RMD-fed dogs shed MDR ESBL-producing E. coli, compared to 1% of those fed NRMD.
Eighty-seven (N = 75 RMD, N = 12 NRMD) 3GCR E. coli isolates demonstrated unique resistance profiles on AST within a sample. Twenty separate resistance profiles were identified from RMD isolates, and just seven from NRMD isolates. The most frequently observed resistance profiles are presented in Table 2. The most frequently observed profile in both RMD (14.7%, N = 11) and NRMD (N = 25.0%, N = 3) isolates was resistance to ampicillin, amoxycillin-clavulanate, cefotaxime and ceftazidime.
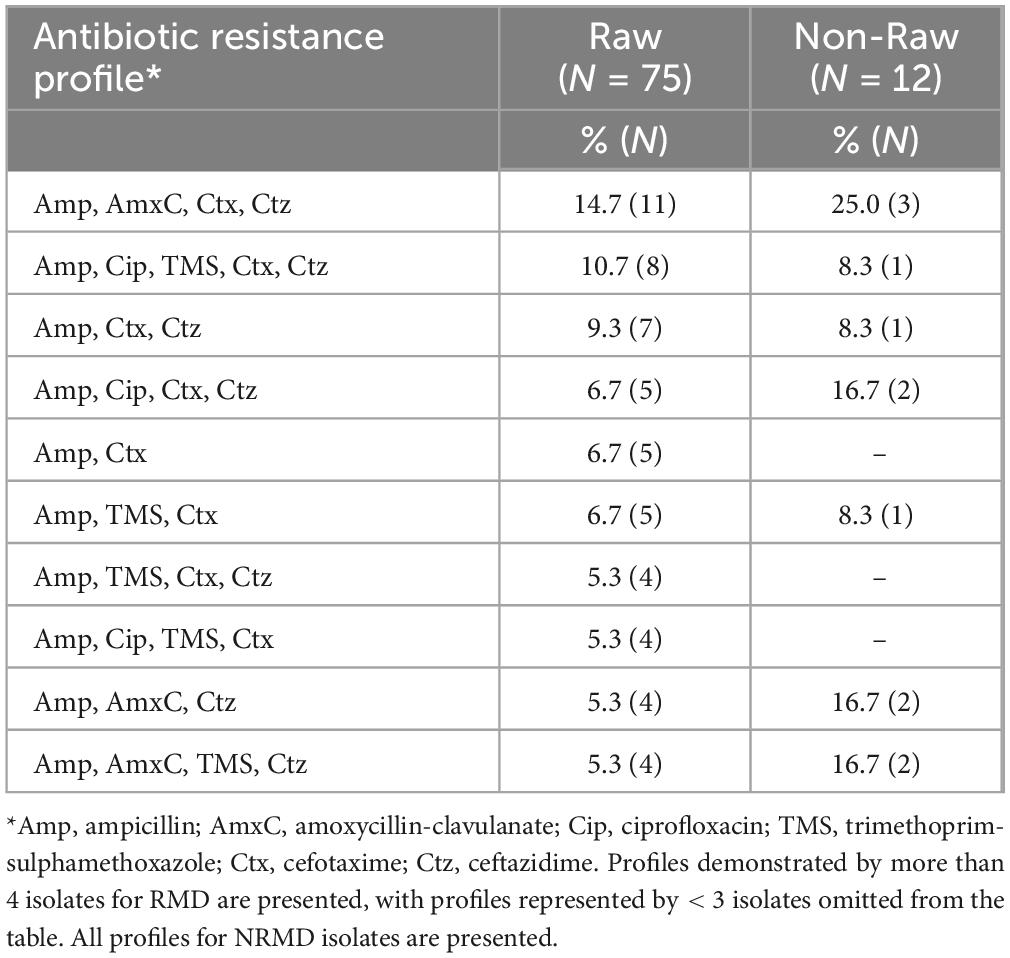
Table 2. Resistance profiles of isolates submitted for whole genome sequencing (N = 87; RMD = 75, NRMD = 12).
These 87 isolates underwent whole genome sequencing (WGS) and included ESBL-producing isolates, and those which were potential pAmpC based on their cephalosporin resistance profile but were not confirmed as ESBL-producing.
Multiple and varied AMR genes were identified on WGS and full results are presented in Supplementary Appendix Table 1 with a summary presented in Table 3. The E. coli isolates detected from dogs fed RMD had a wider variety of AMR genes than dogs fed NRMD (Table 3). The predominant ESBL-genes in both RMD and NRMD isolates were blaCTX–M, with blaCTX–M–15 being the most frequently found (19%, 14/75 RMD; 25% 3/12 NRMD). A wide range of blaCTX–M genes was present in RMD isolates, with 12 different genes being identified, compared to two different blaCTX–M genes identified in NRMD isolates (blaCTX–M–15 and blaCTX–M–1). Within the RMD isolates, blaCTX–M–55 was the second-most frequently isolated ESBL-gene (12%, 9/75), however, this gene was not present in NRMD-originating isolates. Multiple blaTEM genes were identified in the isolates, with blaTEM–1 being the most frequently isolated (Supplementary Appendix Table 1), however, in terms of ESBL-producing blaTEM genes, blaTEM–52 and blaTEM–60 were isolated in RMD E. coli isolates only. Additionally, two inhibitor-resistant blaTEM genes were identified, blaTEM–78 (N = 3 RMD, N = 1 NRMD) and blaTEM–185 (N = 3 RMD only). The ESBL-producing blaSHV–66 gene was only identified in RMD isolates. The ESBL blaOXA–45 gene was infrequently observed and was identified in one isolate each from RMD and NRMD-fed dogs.
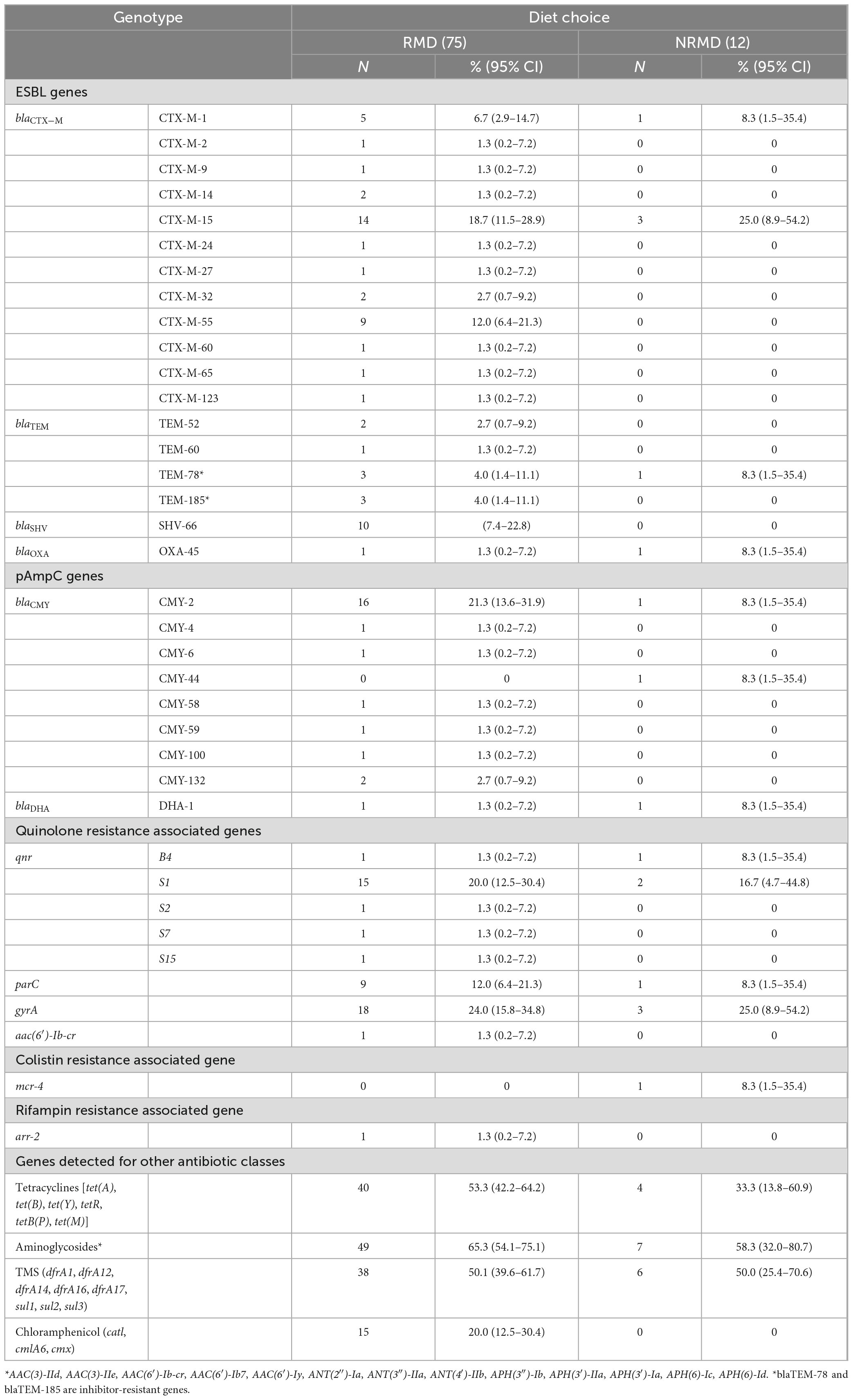
Table 3. Summary table of ESBL and pAmpC genes identified in E. coli isolates from RMD-fed (N = 75 isolates) and NRMD-fed (N = 12 isolates) via whole genome sequencing, demonstrating percentage (%) and number (N) of genes present within the isolates submitted for sequencing.
Additionally, pAmpC genes were mainly observed in RMD isolates. By far the most frequently observed pAmpC gene was blaCMY–2, present in 21% (16/75) of RMD E. coli isolates, whereas this gene was only identified in one NRMD isolate. With regards to genes associated with plasmid-mediated quinolone resistance, five separate qnr genes were observed in RMD isolates, and only two in NRMD isolates. The qnrS1 gene was most frequently isolated, Both RMD and NRMD isolates demonstrated the presence of variants of the parC and gyrA genes which mediate quinolone resistance. Concerningly, one RMD isolate carried the aac(6′)-Ib-cr gene, which can simultaneously result in resistance to fluoroquinolones with a piperazinyl group (e.g., ciprofloxacin) and aminoglycoside resistance. The mobile colistin-resistance encoding mcr-4 gene was identified in one NRMD-originating isolate.
The ESBL, pAmpC and plasmid-mediated quinolone-resistance associated qnr genes present for each isolate, alongside the phenotypic AST results and the associated sequence type (ST) and clonal complex (CC) identified are presented in Figure 1. All isolates demonstrated resistance to ampicillin, and phenotypic 3GCR was indicated in all but three isolates. Ciprofloxacin resistance was demonstrated by 31% (27/87) of isolates (N = 24 RMD, N = 3 NRMD) and resistance to TMS was observed in 41% (36/87) of isolates (N = 32 RMD, N = 4 NRMD). No phenotypic resistance to tigecycline, amikacin or meropenem was identified. MDR phenotypes were present in 79% (69/87) of isolates (N = 59 RMD, N = 10 NRMD). STs, their associated bla genes and their phylogenetic relationships are presented in Figure 2, with an interactive phylogenetic tree available at https://microreact.org/project/jD612SJW4MUv28tQ1BmMpX-genever-v2. Fifty-two distinct E. coli sequence types (STs) were identified (N = 42 STs RMD only, N = 5 STs NRMD only, N = 5 STs both RMD and NRMD), including STs of concern such as ST10, ST38, ST58, ST69, ST155 and ST410. Two novel STs were identified across three RMD isolates. The most frequently observed STs from RMD dogs were ST38 (N = 5), ST117 (N = 4), ST602 (N = 4) and ST752 (N = 5), whereas the most common STs in isolates from NRMD dogs were ST75 (N = 2) and ST88 (N = 2).

Figure 1. ESBL, pAmpC and quinolone resistance associated qnr genes associated with each isolate which underwent whole genome sequencing, alongside the sequence type (ST) and clonal complex (CC) identified and phenotypic resistance demonstrated via disk diffusion. For the “raw” column, a yellow box denotes a raw-fed dog isolate, whereas a blue box denotes a non-raw fed dog isolate. For the genes, a colored box indicates presence of a gene. For the phenotypic resistance, a black box denotes a resistance, and a gray box denotes susceptible. Although amikacin, tigecycline and meropenem were all tested via disk diffusion, no resistance was observed, and they have been omitted from this figure. *Amp, ampicillin; AmxC, amoxycillin-clavulanate; Cip, ciprofloxacin; TMS, trimethoprim-sulphamethoxazole; Ctx, cefotaxime; Ctz, ceftazidime; MDR, multidrug resistance. ^inhibitor-resistant genes.
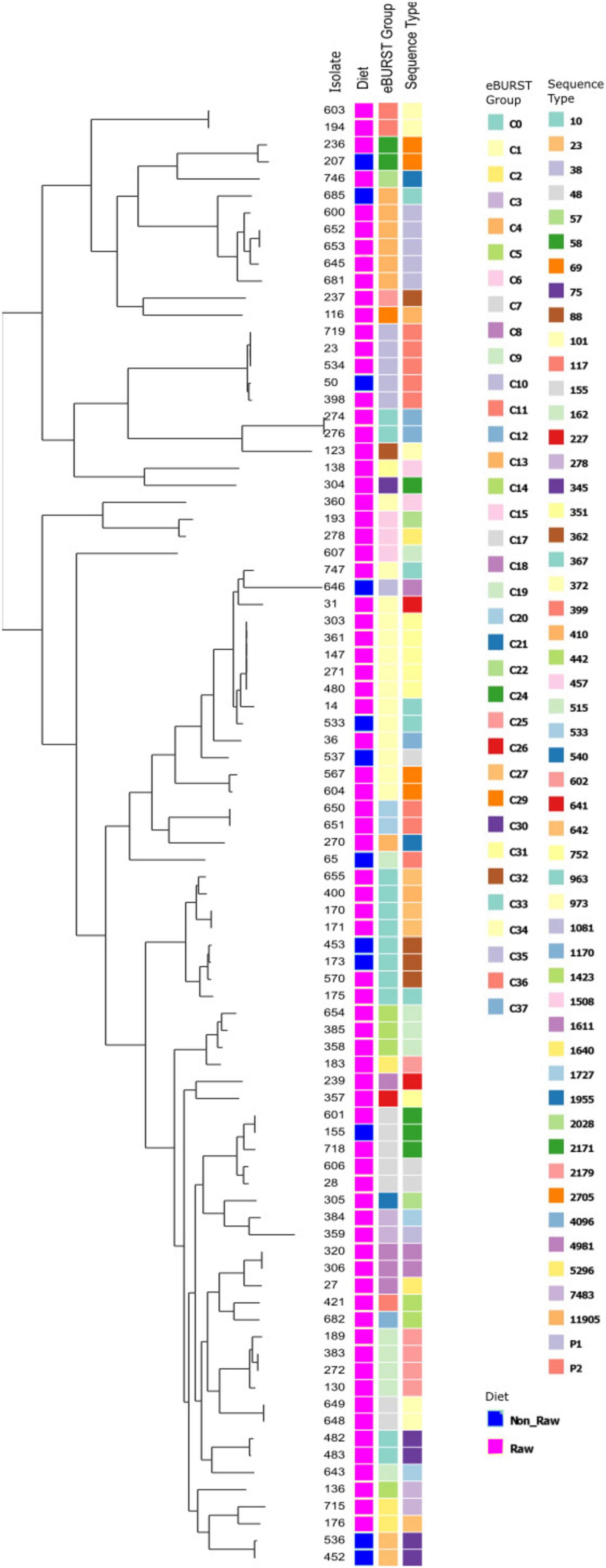
Figure 2. Phylogenetic tree demonstrating the eBURST groups and sequence types (ST) present, and their relationships, for E. coli isolates which underwent whole genome sequencing. For the “group” column, a pink box denotes raw-fed, blue denotes non-raw-fed isolate.
Multiple plasmid-mediated AMR genes were often observed concurrently, particularly within RMD-isolates, however, presence of the AMR-genes was not always associated with phenotypic resistance. The presence of the blaCTX–M–15 gene was frequently associated with the presence of qnrS1 across a range of STs. This was the case for 9 isolates (N = 7 RMD, N = 2 NRMD), and of these, 8 isolates demonstrated MDR on AST. One isolate (ST533), from a single RMD-fed dog harbored both qnrS1 and qnrS15, was MDR, and demonstrated FQR and 3GCR. It was, however, not associated with the presence of blaCTX–M genes, but blaSHV–66 and blaCMY–2 were both present. A further isolate of interest from an RMD-fed dog (ST351) carried blaCTX–M–27, blaCTX–M–123, blaTEM–185 and qnrS1. and was phenotypically MDR, with FQR and 3GCR. Both isolates which carried the blaDHA–1 gene (N = 1 RMD, N = 1 NRMD) also concurrently carried qnrB4, and were the only isolates associated with the carriage of this qnr gene. Both isolates demonstrated phenotypic resistance to amoxycillin-clavulanate, but only one demonstrated phenotypic FQR (RMD-fed). All ST101 and ST752 isolates harbored the blaCTX–M–55 gene. All but one of the isolates which carried blaCTX–M–55 demonstrated phenotypic MDR to combinations of ampicillin, ciprofloxacin, TMS, cefotaxime and ceftazidime. All isolates which harbored the blaTEM–78 gene demonstrated phenotypic amoxycillin-clavulanate resistance, alongside being MDR.
Multiple combinations of plasmid groups were associated with blaESBL gene carriage in the present study (Table 4), including IncF, IncB/O/K/Z, IncI, IncH and IncX groups. There were some associations of plasmids with specific STs; plasmid IncFII was identified in all ST752 isolates.
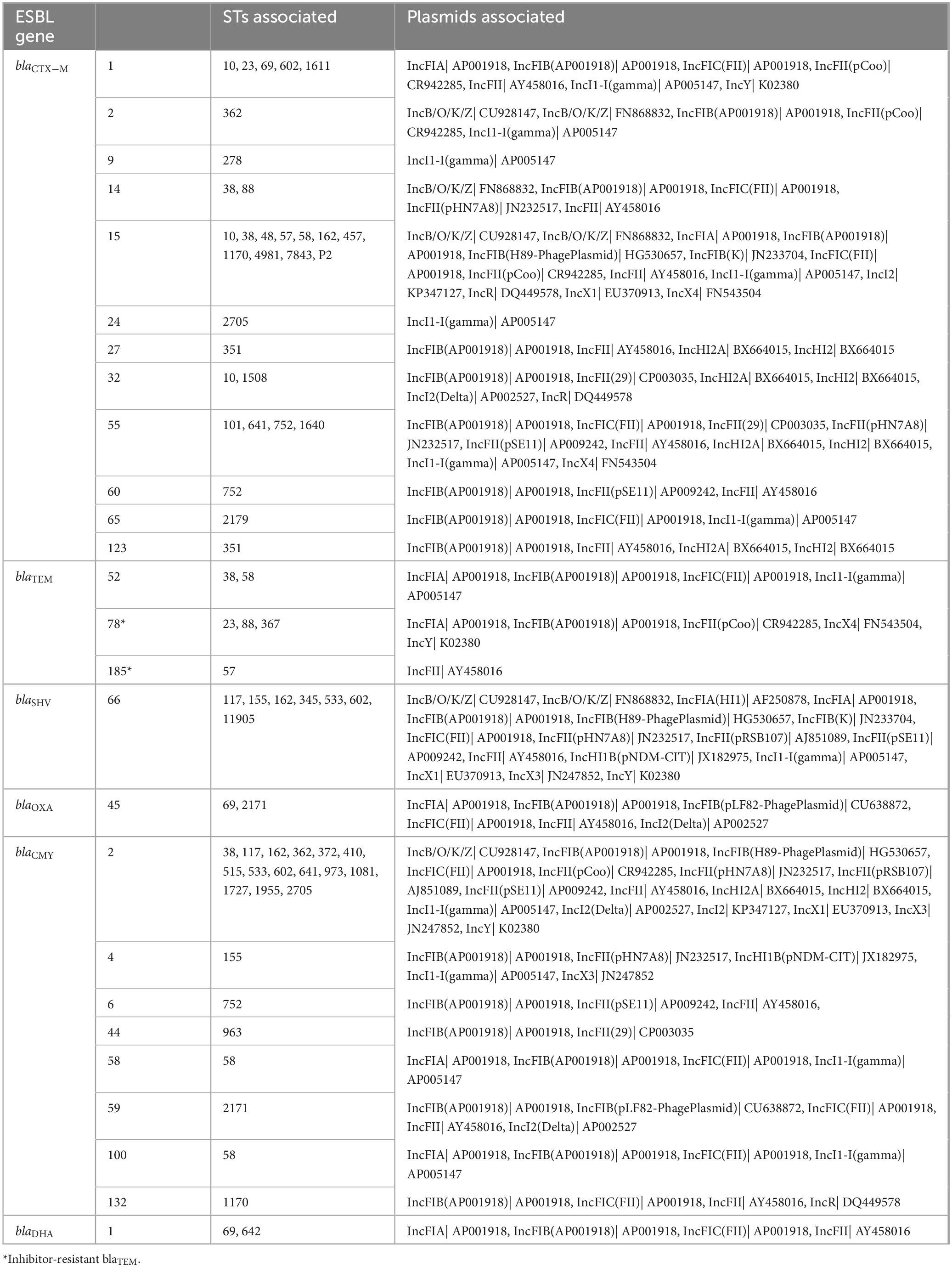
Table 4. Inc group plasmids associated with sequence types (STs) and ESBL genes of interest from ESBL-producing E. coli isolated from RMD-fed (N = 75 isolates) and NRMD-fed (N = 12 isolates) dog feces in the present study.
Survey data analysis
A total of 432 surveys were received. Participant demographics and univariable logistic regression results are presented in Supplementary Appendix Tables 2–4. Multivariable analysis demonstrated several dog and owner lifestyle risk factors for carriage of 3GCR-E. coli, ESBL-producing E. coli and MDR E. coli by dogs (Table 5).
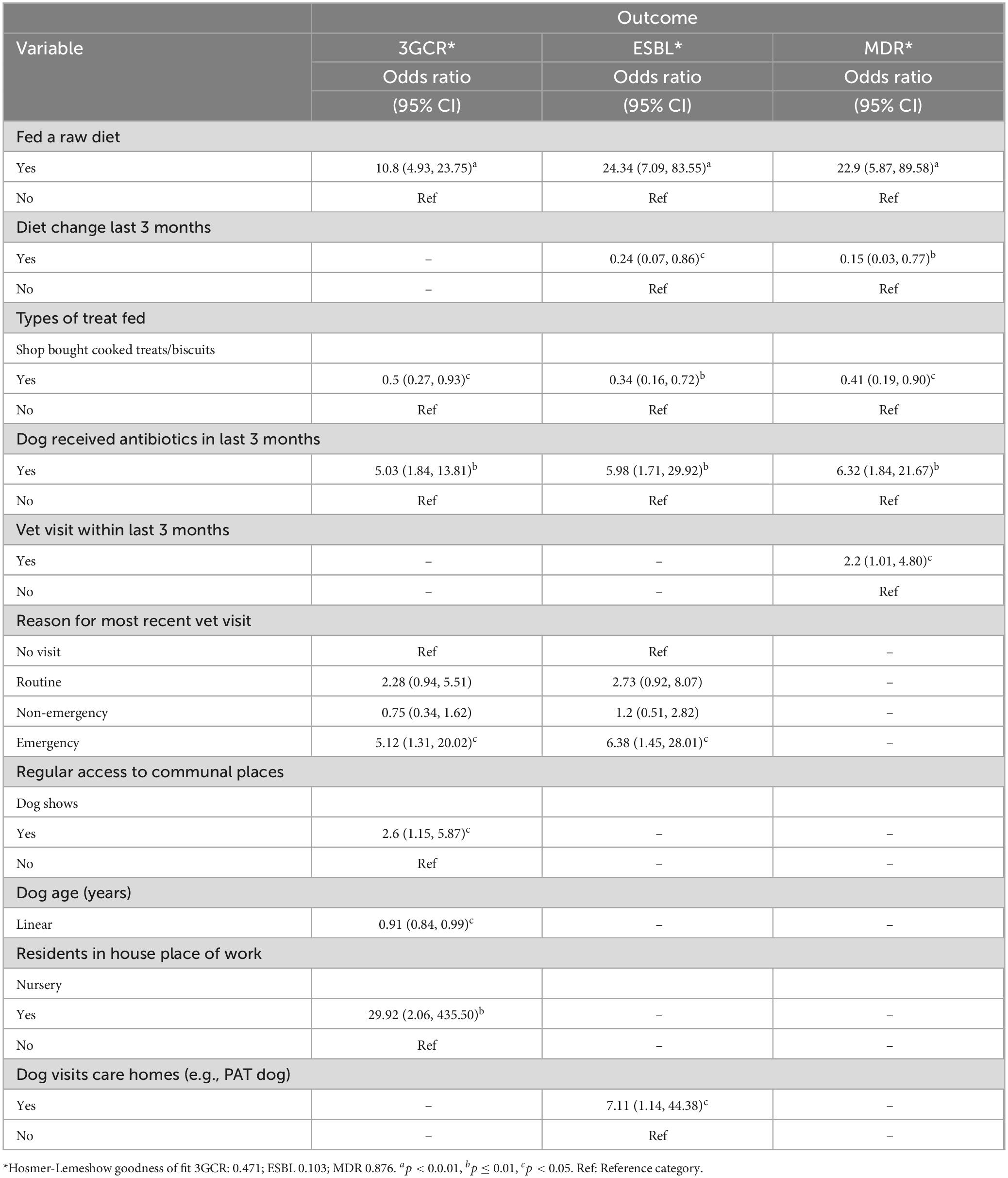
Table 5. Final multivariable regression models describing explanatory variables significantly associated with dog (N = 432) fecal carriage of 3GCR-, ESBL-producing and MDR-E. coli in the present study.
There were also some common risk factors across all three outcomes, dogs fed a raw diet and dogs which had received antibiotics in the last 3 months were significantly more likely to shed 3GCR, ESBL-producing and MDR E. coli. Dog owners were asked to report the type of antibiotic prescribed (if known), these descriptive results are shown in Supplementary Appendix Table 5. The most frequently prescribed antibiotic was amoxycillin-clavulanate (N = 19 dogs), followed by metronidazole (N = 7 dogs). Veterinary visits in the last 3 months were also common to all outcomes, dogs which had visited for an emergency appointment were more likely to shed 3GCR and ESBL-producing E. coli, whereas dogs which had attended a veterinary clinic in general were more likely to shed MDR E. coli. Dogs which were fed shop bought cooked treats/biscuits were less likely to shed 3GCR, ESBL-producing or MDR E. coli.
There were some risk factors which were unique for 3GCR and ESBL-producing E. coli carriage. Dogs which attended dog shows or whose owner worked in a nursery were more likely to carry 3GCR-E. coli, however, dogs were less likely to carry 3GCR-E. coli with increasing age. Dogs that visited care homes, for example “Pets as Therapy” (PAT) dogs were more likely to carry ESBL-producing E. coli in their feces.
Discussion
This study specifically aimed to investigate the effect of diet on fecal AMR E. coli carriage by dogs and to determine the dog and owner lifestyle risk factors associated with canine carriage of AMR E. coli. It has provided strong evidence that provision of RMD to dogs in the UK is a significant risk factor, and that dogs fed RMD are significantly more likely to shed MDR E. coli and E. coli demonstrating resistance to HPCIAs in their feces than those fed a cooked diet.
In the present study, a quarter of dogs fed RMD carrying ESBL-producing E. coli, and approximately one third of dogs fed RMD carrying 3GCR-E. coli. The provision of raw meat to dogs has been identified as a risk factor for AMR E. coli carriage globally (Lefebvre et al., 2008; Baede et al., 2015; Leonard et al., 2015; Runesvärd et al., 2020; van den Bunt et al., 2020) as well as in previous smaller studies in the UK (Wedley et al., 2017; Sealey et al., 2022). Additionally, small studies in Sweden and the UK have also observed significantly greater 3GCR- and ESBL-producing E. coli carriage in dogs fed RMD than those fed NRMD (Runesvärd et al., 2020; Groat et al., 2022). The findings of the present study support those of previous studies, albeit on a larger scale, highlighting the concerning prevalence of E. coli demonstrating resistance to HPCIAs in the feces of dogs fed RMD in the UK, and providing further detail surrounding the phenotypic AMR profiles of E. coli carried by RMD-fed dogs, and the associated genotypes.
There was a significantly greater prevalence of phenotypic resistance to ampicillin, amoxycillin-clavulanate, TMS and ciprofloxacin in the AMR-E. coli isolated from dogs fed RMD than NRMD. High levels of phenotypic resistance to ampicillin, amoxycillin-clavulanate and/or TMS have been reported in dogs fed RMD previously in the UK and Sweden (Schmidt et al., 2015; Runesvärd et al., 2020; Groat et al., 2022), and a UK study of 16-week-old puppies identified that provision of a raw diet was the most substantial risk factor for FQR E. coli carriage (Mounsey et al., 2022). The findings of the present study are interesting as two previous studies demonstrated no (Groat et al., 2022) or uncommon (Schmidt et al., 2015) phenotypic fluoroquinolone resistance in E. coli isolated from healthy adult dogs in the UK. However, in the present study approximately 11% of RMD-fed dogs carried ciprofloxacin-resistant ESBL-producing E. coli (compared to < 1% of NRMD-fed dogs), and this was frequently associated with MDR. No carbapenem resistance was demonstrated in dogs fed either diet in the present study, a finding which echoes that of Runesvärd et al. (2020).
In addition, isolates from RMD-fed dogs demonstrated more varied STs and a great diversity of ESBL genes. Furthermore, STs observed in E. coli isolated from RMD-fed dogs were similar those present in samples of UK RMDs (Morgan et al., 2024), and included globally disseminated uropathogenic STs 58 and 69; ST155, which has major importance in the plasmid mediated spread of ESBL-genes from animals to humans (Matamoros et al., 2017); and ST602, which is commonly isolated from UK livestock (Ludden et al., 2019). The most frequently observed genes in E. coli isolates from RMD-fed dogs in the present study were blaCTX–M–15, blaCTX–M–55 and blaSHV–66. While blaCTX–M–15 was identified, albeit far less frequently, in dogs fed NRMD, no blaCTX–M–55 or blaSHV–66 was present in isolates from NRMD-fed dogs. The presence of blaCTX–M–15 was frequently associated with concurrent qnrS1 carriage, as well as MDR, and was present across a range of STs. There are few studies which have specifically investigated the resistance genes present in E. coli isolated from dogs fed raw diets. However, blaCTX–M–15 has been identified as the most prevalent blaESBL gene in samples of UK raw pet food (Morgan et al., 2024), and has been isolated from poultry and pigs at slaughter in the UK (Randall et al., 2011; Veterinary Medicines Directorate, 2021, 2022) While previous studies have demonstrated a predominance of blaCTX–M–1 in the UK healthy dog population (Wedley et al., 2017; Mounsey et al., 2022), this gene was only observed in 5 isolates from RMD-fed and one isolate from NRMD-fed dogs in the present study. The blaCTX–M–15 gene is the most frequently isolated blaCTX–M gene in canine E. coli in other countries including the USA (Lv et al., 2013), Canada (Cormier et al., 2019) and Portugal (Carvalho et al., 2021). It is also the most commonly identified blaESBL gene associated with human E. coli infections in the UK (Woodford, 2008; Woodford et al., 2011). The dominance of blaCTX–M–15 across a range of STs in the present study is interesting and, along with the WGS findings from other studies (Timofte et al., 2016; Singleton et al., 2021; Sealey et al., 2022), may demonstrate an increase in this particular gene within the canine population in the UK in general, alongside a decrease in blaCTX–M–1 carriage, as well as potentially an increased risk of blaCTX–M–15 carriage in RMD-fed dogs. A recent study of canine fecal E. coli from dogs in the South West of England demonstrated a predominance of the blaCTX–M–15 gene in urban dogs, but not rural dogs, however, excretion of E. coli with blaCTX–M genes was significantly associated with RMD-feeding in both urban and rural dogs (Sealey et al., 2022).
The blaCTX–M–55 gene is derived from blaCTX–M–15 (He et al., 2015) and is frequently identified in humans, as well as being reported in food-producing animals and pets, in China (Sun et al., 2010; Lv et al., 2013; Zhang et al., 2014). However, blaCTX–M–55 is infrequently identified in dogs in other countries, and has been reported in low numbers previously in studies from Korea (Tamang et al., 2012), Canada (Cormier et al., 2019), Portugal (Carvalho et al., 2021), France (Lupo et al., 2018), Switzerland (Zogg et al., 2018) and the Netherlands (Baede et al., 2015). Interestingly, a study of dogs fed either RMD or a conventional cooked diet in Brazil found that ESBL-producing E. coli was only shed by RMD-fed dogs, and the most commonly identified blaESBL gene was blaCTX–M–55 (Ramos et al., 2022). The high prevalence of blaCTX–M–55 in the present study (12% of RMD isolates) is a particularly intriguing finding; it has only been reported once before in dogs in the UK, in E. coli isolates from clinical samples (Bortolami et al., 2019). All isolates which carried blaCTX–M–55 in this study except one demonstrated MDR. blaCTX–M–55 has been identified in healthy pigs at slaughter in the UK (Veterinary Medicines Directorate, 2022), and was the most frequently identified blaESBL gene in healthy broilers (Veterinary Medicines Directorate, 2021). It has also been identified in pre-prepared RMD containing duck in the UK (Morgan et al., 2024). Therefore blaCTX–M–55 could be an emerging blaESBL gene of interest within Europe, as well as within the UK dog population and may be associated with provision of raw meat, particularly poultry.
The identification of blaSHV–66 in E. coli isolated from dogs fed RMD, which was not present in E. coli isolated from NRMD-fed dogs, is also of relevance. blaSHV–66 is usually more frequently associated with Klebsiella spp (Shibu et al., 2021), although has been reported in equine clinical E. coli isolates from one study in the UK (Isgren, 2020). Other ESBL-producing blaSHV genes, in particular blaSHV–12, have been associated with E. coli isolated from dogs in Spain, Switzerland and France (Alonso et al., 2017; Zogg et al., 2018; Dupouy et al., 2019). Two studies in the UK have identified blaSHV–12 carriage in canine E. coli from single dogs (Singleton et al., 2021; Sealey et al., 2022), however, other UK studies did not isolate any blaSHV genes from canine fecal E. coli (Wedley et al., 2017; Schmidt et al., 2018; Groat et al., 2022; Mounsey et al., 2022). This is the first report of blaSHV–66 presence in ESBL-producing E. coli isolated from dogs which may suggest that blaSHV–66 is an emerging blaESBL gene of concern.
It is unsurprising that the most prevalent pAmpC gene in this study was blaCMY–2, present across a range of STs, as this is the most frequently isolated pAmpC gene from E. coli of animal and human origin (Denisuik et al., 2013; Hansen et al., 2016). Additionally, blaCMY–2 has been demonstrated in E. coli isolated from raw pet food in the UK and mainland Europe (Nilsson, 2015; Baede et al., 2017; Morgan et al., 2024). Dogs have been frequently shown to carry E. coli which harbors blaCMY–2 in previous studies from South Korea, the Netherlands, Denmark, Costa Rica, France and the UK (Tamang et al., 2012; Baede et al., 2015; Hansen et al., 2016; Rodríguez-González et al., 2020; Haenni et al., 2022; Sealey et al., 2022). However, although it was isolated from E. coli from one NRMD-fed dog in the present study, far more E. coli isolates from RMD-fed dogs were demonstrated to carry this gene, suggesting that provision of RMD could be a risk for blaCMY–2 carriage. This finding is also supported by the multivariable model results demonstrating provision of RMD to be a risk factor for phenotypic 3GCR-E. coli carriage by dogs.
Of concern was the identification of the mcr-4 (ST4981, isolated from a NRMD-fed dog) gene in this study, which confers plasmid-mediated resistance to colistin. This isolate was also phenotypically MDR. The mcr-4 gene has previously been reported in K. pneumoniae isolated from canine feces in China (Wang et al., 2021), however, to the author’s knowledge, this is the first report of isolation of this gene from canine E. coli.
Although the greatest odds for AMR E. coli shedding by dogs was associated with RMD provision, there were additional risk factors identified across the three models tested (ESBL-producing E. coli, 3GCR-E. coli and MDR-E. coli). The provision of antibiotics in the last 3 months was a significant risk factor, and has been identified as a risk factor for carriage of AMR E. coli by dogs over this timeframe previously (Gandolfi-Decristophoris et al., 2013; Wedley et al., 2017). Treatment with specific antibiotics has been linked with AMR E. coli carriage in dogs; the provision of oral cephalexin has been associated with selection of blaCMY–2 producing E. coli (Damborg et al., 2011), and carriage of MDR E. coli has been attributed to the use of fluoroquinolones (Gibson et al., 2011; Leite-Martins et al., 2014), amoxycillin-clavulanate and cefovecin (Schmidt et al., 2018). Fluoroquinolone use was not widely reported in the present study, with amoxycillin-clavulanate being the most frequently prescribed antibiotic reported.
Visiting a veterinary practice in the last 3 months was a further risk factor for AMR E. coli carriage, with an emergency visit specifically being significant for ESBL-producing and 3GCR E. coli. Previous studies have identified veterinary hospitals as sources of ESBL-producing E. coli (Timofte et al., 2016; Schmitt et al., 2021), with carriage by staff (Royden et al., 2019) and patients being reported. A further study identified frequent carriage of AMR E. coli by vet-visiting dogs, with resistance to ampicillin, tetracycline and trimethoprim most commonly detected (Wedley et al., 2017). As opposed to previous studies where hospitalization and length of stay was a significant risk factor for MDR E. coli (Gibson et al., 2011; Tuerena et al., 2016; Haenni et al., 2022), hospitalization was not significant for any of the AMR outcomes in the present study.
Research is required to investigate the potential for transmission and co-carriage of AMR E. coli between dogs, in-contact people, and the environment. Dogs and their owners frequently share close contact, especially within the home where behaviors such as sharing of soft furnishings and beds, dogs sitting on the owners lap, and dogs licking owners hands and faces occur (Westgarth et al., 2008), as well as owners kissing their pets (do Vale et al., 2021). It is this close relationship, and the behaviors associated with it, which may pose a particularly high risk for transmission of AMR-bacteria between pets and their owners. In particular, risky behaviors around food such as sharing plates, utensils and allowing pets to eat from bare hands is reported, despite owners potentially being aware of the zoonotic disease potential (Dickson et al., 2019). Dogs and humans in close contact, either within the home or within another close-contact environment such as a shelter or veterinary hospital environment, have been demonstrated to share AMR E. coli with similar resistance genes and resistance patterns (Sidjabat et al., 2006; Toombs-Ruane et al., 2020; Cozma et al., 2022; Naziri et al., 2022), and AMR E. coli of the same sequence type (Johnson et al., 2016; Grönthal et al., 2018). ESBL and AmpC-producing E. coli of the same strain has been identified between human patients with urinary tract infections and pet dogs in the same household, suggesting within-household transmission does occur (Johnson et al., 2016; Toombs-Ruane et al., 2020).
The findings of the present study highlight that such close contact should be of particular concern with dogs fed a raw meat diet, where the potential for contact with foodborne zoonotic pathogens is greater. Few studies have investigated the risks of transmission and co-carriage of AMR E. coli within a pet-owning household in relation to provision of a raw diet specifically. A study from the UK identified a common E. coli lineage (ST744) carried by a raw fed puppy and isolated from a human urinary tract infection within a local area (Mounsey et al., 2022). A previous study from The Netherlands identified co-carriage of ESBL-producing E. coli between dogs and their owners in a small number of households, and observed that provision of RMD was a risk factor for ESBL-producing E. coli carriage in dogs (van den Bunt et al., 2020). Carriage of AMR E. coli of STs which are known to be of clinical importance in human medicine has been identified in the present study to a greater degree in dogs fed RMD, associated with mobile transmissible genetic elements. Therefore, it stands to reason that dogs fed RMD could pose an increased public health risk for transmission of AMR E. coli, however, further research is required to investigate this risk.
This study relied on direct contact using email of dog owners who had previously volunteered to take part in related studies, and via social media, thus there may have been an element of bias; certain owner demographics (such as those without access to social media), and populations of dogs where raw feeding may be regularly utilized (such as hunt kennels) may be underrepresented. Owner responses within the survey could be subject to recall bias, however, this is unlikely to affected recall around the overall food type.
The HECA media used for bacterial isolation allowed easy recognition of E. coli colonies. However, it is possible some colonies could be missed if there was a slight deviation from the expected color for any reason. This could lead to underestimation of E. coli presence at sample level. A set number of E. coli picks were taken from each agar plate. This method aims to obtain a representative sample by sampling multiple colonies at random, however, does mean that there could be an over- or underrepresentation of the level of AMR present, depending on the colonies picked. Finally, the presence of the AMR-genes identified by WGS in this study was not always associated with phenotypic resistance; interpretation of the AMR genes must be undertaken with caution as their presence does not necessarily indicate that resistance will be demonstrated. Further research is needed to determine the transmissibility of genes, however, the identification of these plasmid-mediated genes should be cause for concern surrounding the potential for spread of genes capable of mediating resistance to HPCIAs to other bacteria.
Conclusion
This study has contributed to the growing body of evidence to suggest that provision of RMD to dogs is a public health concern. Dogs fed RMD were demonstrated to shed significantly greater proportions of E. coli resistant to HPCIAs than dogs fed NRMD. STs and ESBL genes were identified which are linked to those identified in livestock and humans, and associated with clinical disease in both humans and animals, and novel AMR genes not previously identified in healthy dogs were detected. This constitutes a potential One Health concern, as well as a concern for animal health and welfare. Further research is required to investigate the risks of co-carriage and transmission of AMR E. coli with respect to dogs, their owners and their environment, nevertheless, provision of RMD as a pet food choice should be considered with caution and efforts should be made to continue to engage with pet owners, pet food retailers, veterinary and medical professional with regards to the AMR bacteria risks associated with RMD feeding.
Data availability statement
The study questionnaire is available on request. The whole genome sequencing data presented in the study are deposited in the ENA repository, accession number PRJEB77569.
Ethics statement
All participation was anonymous and ethical approval was granted by the University of Liverpool Veterinary Ethics Committee (approval number VREC935).
Author contributions
GM: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. GP: Conceptualization, Formal analysis, Funding acquisition, Supervision, Writing – review & editing. SH: Data curation, Methodology, Software, Visualization, Writing – review & editing. VS: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. NW: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This study was joint funded by the University of Liverpool and the Veterinary Medicines Directorate (VMD) as part of a larger Ph.D. project.
Acknowledgments
We thanks to the Leahurst laboratory technicians (Jenny Llewelyn, Grace Higham, Cathy Glover) at the University of Liverpool for preparing the media required for this study, to the Centre for Genomic Research at the University of Liverpool for their assistance with whole genome sequencing and bioinformatics, and to the Veterinary Medicines Directorate (VMD) for their review and feedback on this manuscript. We thanks to the University of Liverpool and VMD for funding this work. Additionally, we extend our great thanks to all the dog owners who participated in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1460143/full#supplementary-material
Footnotes
References
Alcock, B., Raphenya, A., Lau, T., Tsang, K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Alonso, C., Michael, G., Li, J., Somalo, S., Simón, C., Wang, Y., et al. (2017). Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J. Antimicrob. Chemother. 72, 1589–1596. doi: 10.1093/jac/dkx024
Anastasi, E., Matthews, B., Gundogdu, A., Vollmerhausen, T., Ramos, N., Stratton, H., et al. (2010). Prevalence and persistence of Escherichia coli strains with uropathogenic virulence characteristics in sewage treatment plants. Appl. Environ. Microbiol. 76, 5882–5886. doi: 10.1128/AEM.00141-10
Baede, V., Broens, E., Spaninks, M., Timmerman, A., Graveland, H., Wagenaar, J., et al. (2017). Raw pet food as a risk factor for shedding of extended-spectrum beta-lactamase-producing Enterobacteriaceae in household cats. PLoS One 12:e0187239. doi: 10.1371/journal.pone.0187239
Baede, V., Wagenaar, J., Broens, E., Duim, B., Dohmen, W., Nijsse, R., et al. (2015). Longitudinal study of extended-spectrum-β-lactamase- and AmpC-producing Enterobacteriaceae in household dogs. Antimicrob. Agents Chemother. 59, 3117–3124. doi: 10.1128/AAC.04576-14
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A., Dvorkin, M., Kulikov, A., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bortolami, A., Zendri, F., Maciuca, E., Wattret, A., Ellis, C., Schmidt, V., et al. (2019). Diversity, virulence, and clinical significance of extended-spectrum β-lactamase- and pAmpC-producing Escherichia coli from companion animals. Front. Microbiol. 10:1260. doi: 10.3389/fmicb.2019.01260
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carvalho, I., Cunha, R., Martins, C., Martínez-Álvarez, S., Safia Chenouf, N., Pimenta, P., et al. (2021). Antimicrobial resistance genes and diversity of clones among faecal ESBL-producing Escherichia coli isolated from healthy and sick dogs living in portugal. Antibiotics (Basel) 10:1013. doi: 10.3390/antibiotics10081013
CLSI (2020). CLSI M100-ED29: 2021 Performance standards for antimicrobial susceptibility testing, 30th Edn. Wayne, PA: CLSI.
Cormier, A., Zhang, P., Chalmers, G., Weese, J., Deckert, A., Mulvey, M., et al. (2019). Diversity of CTX-M-positive Escherichia coli recovered from animals in Canada. Vet. Microbiol. 231, 71–75. doi: 10.1016/j.vetmic.2019.02.031
Cozma, A., Rimbu, C., Zendri, F., Maciuca, I., and Timofte, D. (2022). Clonal dissemination of extended-spectrum cephalosporin-resistant enterobacterales between dogs and humans in households and animal shelters of Romania. Antibiotics (Basel) 11:1242. doi: 10.3390/antibiotics11091242
Damborg, P., Gaustad, I., Olsen, J., and Guardabassi, L. (2011). Selection of CMY-2 producing Escherichia coli in the faecal flora of dogs treated with cephalexin. Vet. Microbiol. 151, 404–408. doi: 10.1016/j.vetmic.2011.03.015
Davies, R., Lawes, J., and Wales, A. (2019). Raw diets for dogs and cats: A review, with particular reference to microbiological hazards. J. Small Anim. Pract. 60, 329–339. doi: 10.1111/jsap.13000
Denisuik, A., Lagacé-Wiens, P., Pitout, J., Mulvey, M., Simner, P., Tailor, F., et al. (2013). Molecular epidemiology of extended-spectrum β- lactamase-, AmpC β-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007-11. J. Antimicrob. Chemother. 68, i57–i65. doi: 10.1093/jac/dkt027
Dickson, A., Smith, M., Smith, F., Park, J., King, C., Currie, K., et al. (2019). Understanding the relationship between pet owners and their companion animals as a key context for antimicrobial resistance-related behaviours: An interpretative phenomenological analysis. Health Psychol. Behav. Med. 7, 45–61. doi: 10.1080/21642850.2019.1577738
do Vale, B., Lopes, A., Fontes, M., Silvestre, M., Cardoso, L., and Coelho, A. C. (2021). A cross-sectional study of knowledge on ownership, zoonoses and practices among pet owners in Northern Portugal. Animals (Basel) 11:3543. doi: 10.3390/ani11123543
Dodd, S., Cave, N., Abood, S., Shoveller, A., Adolphe, J., and Verbrugghe, A. (2020). An observational study of pet feeding practices and how these have changed between 2008 and 2018. Vet. Rec. 186, 643. doi: 10.1136/vr.105828
Dupouy, V., Abdelli, M., Moyano, G., Arpaillange, N., Bibbal, D., Cadiergues, M., et al. (2019). Prevalence of beta-lactam and quinolone/fluoroquinolone resistance in Enterobacteriaceae from dogs in france and spain-characterization of ESBL/PAMPC isolates, genes, and conjugative plasmids. Front. Vet. Sci. 6:279. doi: 10.3389/fvets.2019.00279
EUCAST (2022). Breakpoint tables for interpretation of MICs and zone diameters. Version 12.0, 2022. Växjö: The European Committee on Antimicrobial Susceptibility Testing.
Finley, R., Ribble, C., Aramini, J., Vandermeer, M., Popa, M., Litman, M., et al. (2007). The risk of Salmonellae shedding by dogs fed Salmonella-contaminated commercial raw food diets. Can. Vet. J. 48, 69–75.
Freeman, L., Chandler, M., Hamper, B., and Weeth, L. (2013). Current knowledge about the risks and benefits of raw meat-based diets for dogs and cats. J. Am. Vet. Med. Assoc. 243, 1549–1558. doi: 10.2460/javma.243.11.1549
Gandolfi-Decristophoris, P., Petrini, O., Ruggeri-Bernardi, N., and Schelling, E. (2013). Extended-spectrum β-lactamase-producing Enterobacteriaceae in healthy companion animals living in nursing homes and in the community. Am. J. Infect. Control 41, 831–835. doi: 10.1016/j.ajic.2012.11.013
Gibson, J., Morton, J., Cobbold, R., Filippich, L., and Trott, D. (2011). Risk factors for multidrug-resistant Escherichia coli rectal colonization of dogs on admission to a veterinary hospital. Epidemiol. Infect. 139, 197–205. doi: 10.1017/S0950268810000798
Groat, E., Williams, N., Pinchbeck, G., Warner, B., Simpson, A., and Schmidt, V. M. (2022). UK dogs eating raw meat diets have higher risk of Salmonella and antimicrobial-resistant Escherichia coli faecal carriage. J. Small Anim. Pract. 63, 435–441. doi: 10.1111/jsap.13488
Grönthal, T., Österblad, M., Eklund, M., Jalava, J., Nykäsenoja, S., Pekkanen, K., et al. (2018). Sharing more than friendship - transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill. 23:1700497. doi: 10.2807/1560-7917.ES.2018.23.27.1700497
Haenni, M., Boulouis, H., Lagrée, A., Drapeau, A., Va, F., Billet, M., et al. (2022). Enterobacterales high-risk clones and plasmids spreading blaESBL/AmpC and blaOXA-48 genes within and between hospitalized dogs and their environment. J. Antimicrob. Chemother. 77, 2754–2762. doi: 10.1093/jac/dkac268
Hansen, K., Bortolaia, V., Nielsen, C., Nielsen, J., Schønning, K., Agersø, Y., et al. (2016). Host-specific patterns of genetic diversity among IncI1-Iγ and IncK plasmids encoding CMY-2 β-lactamase in Escherichia coli isolates from humans, poultry meat, poultry, and dogs in Denmark. Appl. Environ. Microbiol. 82, 4705–4714. doi: 10.1128/AEM.00495-16
He, D., Chiou, J., Zeng, Z., Liu, L., Chen, X., Zeng, L., et al. (2015). Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid β-lactamases derived from CTX-M-14 and CTX-M-15. Antimicrob. Agents Chemother. 59, 5976–5983. doi: 10.1128/AAC.04920-14
Isgren, C. M. (2020). The emerging problem of antimicrobial resistance in horses: Investigating faecal carriage and environmental contamination with resistant Escherichia coli in equine hospitals and clinical infections with multidrug resistant bacteria, [Ph.D. thesis]. Liverpool: University of Liverpool.
Johnson, J., Davis, G., Clabots, C., Johnston, B., Porter, S., DebRoy, C., et al. (2016). Household clustering of Escherichia coli sequence type 131 clinical and fecal isolates according to whole genome sequence analysis. Open Forum Infect. Dis. 3:ofw129. doi: 10.1093/ofid/ofw129
Kaindama, L., Jenkins, C., Aird, H., Jorgensen, F., Stoker, K., and Byrne, L. (2020). A cluster of Shiga Toxin-producing Escherichia coli O157:H7 highlights raw pet food as an emerging potential source of infection in humans. Epidemiol. Infect. 149:e124. doi: 10.1017/S0950268821001072
Langmead, B., and Salzberg, S. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lefebvre, S., Reid-Smith, R., Boerlin, P., and Weese, J. (2008). Evaluation of the risks of shedding Salmonellae and other potential pathogens by therapy dogs fed raw diets in Ontario and Alberta. Zoonoses Public Health 55, 470–480. doi: 10.1111/j.1863-2378.2008.01145.x
Leite-Martins, L., Mahú, M., Costa, A., Mendes, A., Lopes, E., Mendonça, D., et al. (2014). Prevalence of antimicrobial resistance in enteric Escherichia coli from domestic pets and assessment of associated risk markers using a generalized linear mixed model. Prev. Vet. Med. 117, 28–39. doi: 10.1016/j.prevetmed.2014.09.008
Leonard, E., Pearl, D., Janecko, N., Finley, R., Reid-Smith, R., Weese, J., et al. (2015). Risk factors for carriage of antimicrobial-resistant Salmonella spp and Escherichia coli in pet dogs from volunteer households in Ontario, Canada, in 2005 and 2006. Am. J. Vet. Res. 76, 959–968. doi: 10.2460/ajvr.76.11.959
Ludden, C., Raven, K., Jamrozy, D., Gouliouris, T., Blane, B., Coll, F., et al. (2019). One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 10:e002693–18. doi: 10.1128/mBio.02693-18
Lupo, A., Saras, E., Madec, J., and Haenni, M. (2018). Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimicrob. Chemother. 73, 867–872. doi: 10.1093/jac/dkx489
Lv, L., Partridge, S., He, L., Zeng, Z., He, D., Ye, J., et al. (2013). ‘Genetic characterization of inci2 plasmids carrying blaCTX-M-55 spreading in both pets and food animals in China Luchao’. Antimicrob. Agents Chemother. 57, 2824–2827. doi: 10.1128/AAC.02155-12
Magiorakos, A., Srinivasan, A., Carey, R., Carmeli, Y., Falagas, M., Giske, C., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Matamoros, S., van Hattem, J., Arcilla, M., Willemse, N., Melles, D., Penders, J., et al. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. doi: 10.1038/s41598-017-15539-7
Morgan, G., Pinchbeck, G., Taymaz, E., Chattaway, M., Schmidt, V., and Williams, N. (2024). An investigation of the presence and antimicrobial susceptibility of Enterobacteriaceae in raw and cooked kibble diets for dogs in the United Kingdom. Front. Microbiol. 14:1301841. doi: 10.3389/fmicb.2023.1301841
Morgan, G., Williams, N., Schmidt, V., Cookson, D., Symington, C., and Pinchbeck, G. A. (2022). Dog’s Dinner: Factors affecting food choice and feeding practices for UK dog owners feeding raw meat-based or conventional cooked diets. Prev. Vet. Med. 208:105741. doi: 10.1016/j.prevetmed.2022.105741
Morley, P., Strohmeyer, R., Tankson, J., Hyatt, D., Dargatz, D., and Fedorka-Cray, P. (2006). Evaluation of the association between feeding raw meat and Salmonella enterica infections at a Greyhound breeding facility. J. Am. Vet. Med. Assoc. 228, 1524–1532. doi: 10.2460/javma.228.10.1524
Mounsey, O., Wareham, K., Hammond, A., Findlay, J., Gould, V., Morley, K., et al. (2022). Evidence that faecal carriage of resistant Escherichia coli by 16-week-old dogs in the United Kingdom is associated with raw feeding. One Health 14:100370. doi: 10.1016/j.onehlt.2022.100370
Naziri, Z., Poormaleknia, M., and Ghaedi Oliyaei, A. (2022). Risk of sharing resistant bacteria and/or resistance elements between dogs and their owners. BMC Vet. Res. 18:203. doi: 10.1186/s12917-022-03298-1
Nguyen, L., Schmidt, H., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nilsson, O. (2015). Hygiene quality and presence of ESBL-producing Escherichia coli in raw food diets for dogs. Infect. Ecol. Epidemiol. 5:28758. doi: 10.3402/iee.v5.28758
Nüesch-Inderbinen, M., Treier, A., Zurfluh, K., and Stephan, R. (2019). Raw meat-based diets for companion animals: A potential source of transmission of pathogenic and antimicrobial-resistant Enterobacteriaceae. R. Soc. Open Sci. 6:191170. doi: 10.1098/rsos.191170
PDSA (2022). PDSA animal wellbeing PAW report 2022: The essential insight into the wellbeing of UK pets, PDSA animal wellbeing report. Birmingham: PDSA.
Ramos, C., Kamei, C., Viegas, F., de Melo Barbieri, J., Cunha, J., Hounmanou, Y., et al. (2022). Fecal shedding of multidrug resistant Escherichia coli isolates in dogs fed with raw meat-based diets in Brazil. Antibiotics (Basel) 11:534. doi: 10.3390/antibiotics11040534
Randall, L., Clouting, C., Horton, R., Coldham, N., Wu, G., Clifton-Hadley, F., et al. (2011). Prevalence of Escherichia coli carrying extended-spectrum β-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J. Antimicrob. Chemother. 66, 86–95. doi: 10.1093/jac/dkq396
Rodríguez-González, M., Jiménez-Pearson, M., Duarte, F., Poklepovich, T., Campos, J., Araya-Sánchez, L., et al. (2020). Multidrug-resistant CTX-M and CMY-2 producing Escherichia coli isolated from healthy household dogs from the great Metropolitan Area, Costa Rica. Microb. Drug Resist. 26, 1421–1428. doi: 10.1089/mdr.2020.0146
Royden, A., Ormandy, E., Pinchbeck, G., Pascoe, B., Hitchings, M., Sheppard, S., et al. (2019). Prevalence of faecal carriage of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli in veterinary hospital staff and students. Vet. Rec. Open 6:e000307. doi: 10.1136/vetreco-2018-000307
Runesvärd, E., Wikström, C., Fernström, L., and Hansson, I. (2020). Presence of pathogenic bacteria in faeces from dogs fed raw meat-based diets or dry kibble. Vet. Rec. 187:e71. doi: 10.1136/vr.105644
Schmidt, V., Pinchbeck, G., McIntyre, K., Nuttall, T., McEwan, N., Dawson, S., et al. (2018). Routine antibiotic therapy in dogs increases the detection of antimicrobial-resistant faecal Escherichia coli. J. Antimicrob. Chemother. 73, 3305–3316. doi: 10.1093/jac/dky352
Schmidt, V., Pinchbeck, G., Nuttall, T., McEwan, N., Dawson, S., and Williams, N. (2015). Antimicrobial resistance risk factors and characterisation of faecal E. coli isolated from healthy Labrador retrievers in the United Kingdom. Prev. Vet. Med. 119, 31–40. doi: 10.1016/j.prevetmed.2015.01.013
Schmitt, K., Kuster, S., Zurfluh, K., Jud, R., Sykes, J., Stephan, R., et al. (2021). Transmission chains of extended-spectrum beta-lactamase-producing Enterobacteriaceae at the companion animal veterinary clinic-household interface. Antibiotics (Basel) 10:171. doi: 10.3390/antibiotics10020171
Sealey, J., Hammond, A., Mounsey, O., Gould, V., Reyher, K., and Avison, M. (2022). Molecular ecology and risk factors for third-generation cephalosporin-resistant Escherichia coli carriage by dogs living in urban and nearby rural settings. J. Antimicrob. Chemother. 77, 2399–2405. doi: 10.1093/jac/dkac208
Seemann, T. (2014). Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Segata, N., Waldron, L., Ballarini, A., Narasimhan, V., Jousson, O., and Huttenhower, C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814. doi: 10.1038/nmeth.2066
Shibu, P., McCuaig, F., McCartney, A., Kujawska, M., Hall, L., and Hoyles, L. (2021). Improved molecular characterization of the Klebsiella oxytoca complex reveals the prevalence of the kleboxymycin biosynthetic gene cluster. Microb. Genom. 7:000592. doi: 10.1099/mgen.0.000592
Sidjabat, H., Townsend, K., Lorentzen, M., Gobius, K., Fegan, N., Chin, J., et al. (2006). Emergence and spread of two distinct clonal groups of multidrug-resistant Escherichia coli in a veterinary teaching hospital in Australia. J. Med. Microbiol. 55, 1125–1134. doi: 10.1099/jmm.0.46598-0
Simão, F., Waterhouse, R., Ioannidis, P., Kriventseva, E., and Zdobnov, E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Singleton, D., Pongchaikul, P., Smith, S., Bengtsson, R., Baker, K., Timofte, D., et al. (2021). Temporal, spatial, and genomic analyses of Enterobacteriaceae clinical antimicrobial resistance in companion animals reveals phenotypes and genotypes of one health concern. Front. Microbiol. 12:700698. doi: 10.3389/fmicb.2021.700698
Sun, Y., Zeng, Z., Chen, S., Ma, J., He, L., Liu, Y., et al. (2010). High prevalence of bla(CTX-M) extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 16, 1475–1481. doi: 10.1111/j.1469-0691.2010.03127.x
Tamang, M., Nam, H., Jang, G., Kim, S., Chae, M., Jung, S., et al. (2012). Molecular characterization of extended-spectrum-β-lactamase-producing and plasmid-mediated AmpC β-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob. Agents Chemother. 56, 2705–2712. doi: 10.1128/AAC.05598-11
Timofte, D., Maciuca, I., Williams, N., Wattret, A., and Schmidt, V. (2016). Veterinary hospital dissemination of CTX-M-15 extended-spectrum beta-lactamase-producing Escherichia coli ST410 in the United Kingdom. Microb. Drug Resist. 22, 609–615. doi: 10.1089/mdr.2016.0036
Tonkin-Hill, G., MacAlasdair, N., Ruis, C., Weimann, A., Horesh, G., Lees, J., et al. (2020). Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 21:180. doi: 10.1186/s13059-020-02090-4
Toombs-Ruane, L., Benschop, J., French, N., Biggs, P., Midwinter, A., Marshall, J., et al. (2020). Carriage of extended-spectrum-beta-lactamase- and AmpC beta-lactamase-producing Escherichia coli strains from humans and pets in the same households. Appl. Environ. Microbiol. 86, e1613–e1620. doi: 10.1128/AEM.01613-20
Tuerena, I., Williams, N., Nuttall, T., and Pinchbeck, G. (2016). Antimicrobial-resistant Escherichia coli in hospitalised companion animals and their hospital environment. J. Small Anim. Pract. 57, 339–347. doi: 10.1111/jsap.12525
van den Bunt, G., Fluit, A., Spaninks, M., Timmerman, A., Geurts, Y., Kant, A., et al. (2020). Faecal carriage, risk factors, acquisition and persistence of ESBL-producing Enterobacteriaceae in dogs and cats and co-carriage with humans belonging to the same household. J. Antimicrob. Chemother. 75, 342–350. doi: 10.1093/jac/dkz462
Veterinary Medicines Directorate (2021). Supplementary material (UK-VARSS 2020). Veterinary antibiotic resistance and sales surveillance report (UK-VARSS 2020). Addlestone: Veterinary Medicines Directorate.
Veterinary Medicines Directorate (2022). UK - veterinary antibiotic resistance and sales surveillance report: 2021. Supplementary material 3 – resistance data’, Veterinary antibiotic resistance and sales surveillance report (UK-VARSS 2021). Birmingham: Veterinary Medicines Directorate.
Viegas, F., Ramos, C., Xavier, R., Lopes, E., Júnior, C., Bagno, R., et al. (2020). Fecal shedding of Salmonella spp., Clostridium perfringens, and Clostridioides difficile in dogs fed raw meat-based diets in Brazil and their owners’ motivation. PLoS One 15:e0231275. doi: 10.1371/journal.pone.0231275
Wang, G., Liu, H., Feng, Y., Zhang, Z., Hu, H., Liu, J., et al. (2021). Colistin-resistance mcr genes in Klebsiella pneumoniae from companion animals. J. Glob. Antimicrob. Resist. 25, 35–36. doi: 10.1016/j.jgar.2021.02.023
Wedley, A., Dawson, S., Maddox, T., Coyne, K., Pinchbeck, G., Clegg, P., et al. (2017). Carriage of antimicrobial resistant Escherichia coli in dogs: Prevalence, associated risk factors and molecular characteristics. Vet. Microbiol. 199, 23–30. doi: 10.1016/j.vetmic.2016.11.017
Westgarth, C., Pinchbeck, G., Bradshaw, J., Dawson, S., Gaskell, R., and Christley, R. (2008). Dog-human and dog-dog interactions of 260 dog-owning households in a community in Cheshire. Vet. Rec. 162, 436–442. doi: 10.1136/vr.162.14.436
Woodford, N. (2008). Successful, multiresistant bacterial clones. J. Antimicrob. Chemother. 61, 233–234. doi: 10.1093/jac/dkm474
Woodford, N., Turton, J., and Livermore, D. (2011). Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35, 736–755. doi: 10.1111/j.1574-6976.2011.00268.x
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659. doi: 10.1186/s12879-014-0659-0
Zogg, A., Simmen, S., Zurfluh, K., Stephan, R., Schmitt, S., and Nüesch-Inderbinen, M. (2018). High prevalence of extended-spectrum β-lactamase producing Enterobacteriaceae among clinical isolates from cats and dogs admitted to a veterinary hospital in Switzerland. Front. Vet. Sci. 5:62. doi: 10.3389/fvets.2018.00062
Keywords: raw meat diet, dog, AMR, E. coli, carriage, One Health
Citation: Morgan G, Pinchbeck G, Haldenby S, Schmidt V and Williams N (2024) Raw meat diets are a major risk factor for carriage of third-generation cephalosporin-resistant and multidrug-resistant E. coli by dogs in the UK. Front. Microbiol. 15:1460143. doi: 10.3389/fmicb.2024.1460143
Received: 05 July 2024; Accepted: 15 August 2024;
Published: 09 September 2024.
Edited by:
Fohad Mabood Husain, King Saud University, Saudi ArabiaReviewed by:
Tales Fernando Da Silva, Federal University of Minas Gerais, BrazilSalome N. Seiffert, Zentrum für Labormedizin (ZLM), Switzerland
Copyright © 2024 Morgan, Pinchbeck, Haldenby, Schmidt and Williams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Genever Morgan, Z21vcmdhbkBsaXZlcnBvb2wuYWMudWs=
 Genever Morgan
Genever Morgan Gina Pinchbeck
Gina Pinchbeck Sam Haldenby2
Sam Haldenby2 Vanessa Schmidt
Vanessa Schmidt Nicola Williams
Nicola Williams