- 1Department of Food, Environmental and Nutritional Sciences (DeFENS), University of Milan, Milan, Italy
- 2Department of Environmental and Resource Engineering, Technical University of Denmark, Kongens Lyngby, Denmark
- 3CNR – IRSA Water Research Institute, Molecular Ecology Group (MEG), Verbania, Italy
- 4National Biodiversity Future Center, Palermo, Italy
- 5Department of Biological and Chemical Engineering, Center for Water Technology, Aarhus University, Aarhus, Denmark
Introduction: Horizontal gene transfer (HGT) of antibiotic resistance genes (ARGs) is one of the primary routes of antimicrobial resistance (AMR) dissemination. In the One Health context, tracking the spread of mobile genetic elements (MGEs) carrying ARGs in agri-food ecosystems is pivotal in understanding AMR diffusion and estimating potential risks for human health. So far, little attention has been devoted to plant niches; hence, this study aimed to evaluate the conjugal transfer of ARGs to the bacterial community associated with the plant rhizosphere, a hotspot for microbial abundance and activity in the soil. We simulated a source of AMR determinants that could enter the food chain via plants through irrigation.
Methods: Among the bacterial strains isolated from treated wastewater, the strain Klebsiella variicola EEF15 was selected as an ARG donor because of the relevance of Enterobacteriaceae in the AMR context and the One Health framework. The strain ability to recolonize lettuce, chosen as a model for vegetables that were consumed raw, was assessed by a rifampicin resistant mutant. K. variicola EEF15 was genetically manipulated to track the conjugal transfer of the broad host range plasmid pKJK5 containing a fluorescent marker gene to the natural rhizosphere microbiome obtained from lettuce plants. Transconjugants were sorted by fluorescent protein expression and identified through 16S rRNA gene amplicon sequencing.
Results and discussion: K. variicola EEF15 was able to colonize the lettuce rhizosphere and inhabit its leaf endosphere 7 days past bacterial administration. Fluorescence stereomicroscopy revealed plasmid transfer at a frequency of 10−3; cell sorting allowed the selection of the transconjugants. The conjugation rates and the strain’s ability to colonize the plant rhizosphere and leaf endosphere make strain EEF15::lacIq-pLpp-mCherry-gmR with pKJK5::Plac::gfp an interesting candidate to study ARG spread in the agri-food ecosystem. Future studies taking advantage of additional environmental donor strains could provide a comprehensive snapshot of AMR spread in the One Health context.
1 Introduction
Horizontal gene transfer (HGT) is an evolutionary process that confers new phenotypes and metabolic activities to bacteria, allowing their rapid adaptation to new environmental selective pressures such as antibiotic presence (Aminov, 2011; Saak et al., 2020). Through various HGT mechanisms, mobile genetic elements (MGEs) harboring antibiotic-resistant genes (ARGs) can spread among microorganisms (Smalla et al., 2018). The diffusion of antimicrobial resistance (AMR) has been recognized as one of the serious global challenges of this century; as a consequence, treatments against bacterial infections are gradually losing efficacy, representing important threats to human health (Pinilla-Redondo et al., 2018; Larsson and Flach, 2022). Thus, it is crucial to track how AMR determinants can move—individually and collectively—among environmental bacterial communities and to understand how their transfer is influenced by the complexity of the ecosystems (Saak et al., 2020).
In the One Health perspective, studies on the spread of AMR determinants should include the agri-food ecosystems. The presence of AMR determinants has been documented in specific hotspots of the agri-food and related ecosystems, such as managed fields, rhizosphere, and wastewater treatment plants (WWTPs) (Larsson and Flach, 2022; Riva V. et al., 2020). Treated wastewater, in particular, can convey and promote the maintenance of an antibiotic resistome in freshwater habitats (Corno et al., 2019; Di Cesare et al., 2015), and, through the reuse of treated wastewater for irrigation, ARGs and antibiotic-resistant bacteria (ARB) might enter the food chain via plants (Riva F. et al., 2020; Riva et al., 2022a; Musiyiwa et al., 2024). ARB and ARGs have indeed been found in fresh products and farm environments, underlining the risks to which humans are exposed (Perera et al., 2020; Araújo et al., 2017; Yang et al., 2020; Nüesch-Inderbinen et al., 2015; Schierstaedt et al., 2019; Cerqueira et al., 2019).
Among HGT mechanisms, conjugation usually takes place in environments with high cell density and high metabolic activity (Ulrich et al., 2015), i.e., where high density of cells can increase chance for contacts (Riva V. et al., 2020), such as rhizosphere, mycelia, sewage sludge and host-associated microbial communities (Van Elsas et al., 2003; Qiu et al., 2018; Berthold et al., 2016; Virolle et al., 2020). Conjugation is influenced by different factors, e.g., temperature, nutrient ability, and cell-to-cell contact (Zarei-Baygi and Smith, 2020), as well as the presence of emerging pollutants, which could enhance the spread of ARGs (Feng et al., 2021; Yu et al., 2021). The transfer can occur between bacteria from the same species or between taxonomically distant ones (Zarei-Baygi and Smith, 2020; Klümper et al., 2015); for instance, the taxonomic relatedness between donors belonging to Escherichia coli species and recipient bacteria has been recently investigated in a meta-analysis and found to influence conjugal rate in liquid conditions, but not on solid surfaces (Alderliesten et al., 2020). The genetic background of E. coli donor strains has also been shown to affect the conjugal transfer of the broad host range IncP-1 plasmid pKJK10 when transferred to a certain E. coli recipient strain in genome-wide association studies, which encompassed deletions, insertions, recombinations, as well as the presence or absence of genes (Van Wonterghem et al., 2022). Focusing on the identification of genes involved in conjugation, authors have, indeed, found several chromosomally encoded genes of E. coli that positively impact transfer, i.e., genes fliF and fliK involved in flagella assembly, ucpA expressing an oxidoreductase, and kefB encoding a potassium transporter (Van Wonterghem et al., 2022).
Several approaches have been developed to track conjugation events in specific environmental niches, such as soil (Klümper et al., 2015; Fan et al., 2019) or sewage treatment plant communities (Li et al., 2020). The rhizosphere is the portion of soil strictly attached to the plant roots and influenced by root exudates. Because of its high content of nutrients, as well as its bacterial abundance and mobility, it has been indicated as a hotspot of HGT, especially via conjugation (Van Elsas et al., 2003). However, ARG transfer in the rhizosphere and other plant compartments has been poorly investigated so far (Sánchez-Salazar et al., 2023; Xu et al., 2021; Musiyiwa et al., 2024), but this knowledge is necessary to understand the magnitude of AMR diffusion, also considering how microbiomes respond to changing environmental conditions (Smalla et al., 2018; Pinilla-Redondo et al., 2018; Larsson et al., 2018; Saak et al., 2020; Abs et al., 2023). Thus, this study aimed to evaluate the conjugal transfer of plasmids within the rhizosphere microbiome, taking advantage of a bacterial donor strain of environmental origin. We focused our attention on the Enterobacteriaceae family because of its relevance in the One Health context: this family is known to include both pathogenic and commensal bacteria and can colonize several environments linked to the agri-food system, e.g., soil, plant niches, freshwater bodies, and human and animal gut (Touchon et al., 2020; Riva F. et al., 2020; Leimbach et al., 2013; Nüesch-Inderbinen et al., 2015; Wang et al., 2020; Schikora et al., 2012; Zhang et al., 2020). Specifically, we selected an Enterobacteriaceae strain isolated from the effluent from a municipal WWTP to mimic a real scenario of ARB released by WWTP and entering the agri-food system. The selected strain was genetically modified and used as an environmental donor strain to study ARG spread to the rhizosphere bacterial community associated with lettuce, chosen as a model of vegetables eaten raw. The results of conjugation experiments were then compared to the ones obtained with a strain of laboratory origin belonging to the Enterobacteriaceae family (i.e., E. coli MG1655).
2 Materials and methods
2.1 Bacteria isolation and identification
A sample of 1.1L of water was collected from the effluent from the largest municipal WWTP of Milan municipality (Milano-Nosedo, Northern Italy) and filtered through a 0.22-μm membrane filter GSWP04700 by vacuum pump. Bacteria on the filter were resuspended in saline solution (0.9% NaCl), serially diluted, plated on Violet Red Bile Lactose Agar (Thermo Fisher Scientific) supplemented with 100 μg/mL cycloheximide and incubated at 30°C for 48 h. Single colonies were randomly picked and spread 3 times on the same medium to obtain pure cultures. Pure cultures were stored at −80°C in 20% glycerol stocks. Genomic DNA was extracted from strains by boiling cell lysis (Ferjani et al., 2015) and the bacteria were identified by 16S rRNA gene amplification and partial sequencing followed by basic local alignment search tool (BLAST) alignment on the NCBI database (Callegari et al., 2020).
2.2 Antibiotic sensitivity
The bacterial collection was screened for strains sensitivity to kanamycin (100 μg/mL) and rifampicin (RIF) (100 μg/mL). Briefly, isolates’ growth was checked on Luria Bertani agar plates supplemented with the aforementioned antibiotics, after an incubation of 72 h at 30°C. Klebsiella variicola EEF15, selected as the donor strain, was also screened for sensitivity against gentamycin (100 μg/mL), as reported by Klümper et al. (2015).
2.3 Lettuce plants and agricultural soils
Experiments were conducted with two different setups. Colonization experiments in the lettuce rhizosphere were carried out with Lactuca sativa seedlings transplanted in agricultural soil collected in Triuggio (Monza and Brianza, Italy). In contrast, filter mating conjugation experiments between the donor strain and the lettuce rhizosphere community were carried out with L. sativa seedlings transplanted in agricultural soil collected from CRUCIAL (closing the rural–urban nutrient cycle) agricultural field site (Taastrup, Denmark), from a plot fertilized with NPK (Fernández-Calviño et al., 2021; Bollmann et al., 2017; Fernández-Calviño et al., 2017). Experiments were performed at least 1 week after the transplantation of lettuce plants from pot soil to agricultural soil. Lettuce plants were grown as described by Riva F. et al. (2020).
2.4 Root and leaf endosphere colonization by Klebsiella variicola EEF15 RIF-R
A rifampicin-resistant (RIF-R) mutant of the strain K. variicola EEF15, isolated from the effluent of the WWTP, was obtained as described in Riva F. et al. (2020) and used to test its ability to colonize the lettuce rhizosphere. Bacterial suspensions were grown overnight growth, and the following day, they were prepared to bacterize plant roots with a final concentration of 1 × 108 and 1 × 109 cells of EEF15 RIF-R per gram of soil (indicatively 300 g of soil were added per pot). Plant bacterization was performed by applying the bacterial suspensions to the soil surrounding the collar of potted lettuce plants (n = 3). After 1 week, the rhizosphere was collected (Marasco et al., 2018), and EEF15 RIF-R colonization was assessed by plating different serial dilutions of the rhizosphere samples on Luria broth (LB) medium supplemented with 100 μg/mL RIF in triplicate (Riva F. et al., 2020). The identity of randomly selected RIF-R colonies (n = 18) was confirmed by an intergenic transcribed spacer (ITS)-polymerase chain reaction (PCR) fingerprinting, comparing ITS-PCR profiles with the one of the EEF15 RIF-R strain (Mapelli et al., 2013).
K. variicola EEF15 RIF-R to colonize leaf endosphere, lettuce plants (n = 3) were bacterized with 1 × 109 cells of EEF15 RIF-R per gram of commercial soil. After 1 week, the leaf endosphere microbiome was extracted as per Klerks et al. (2007). Approximately 10 g of lettuce leaves per plant (sampling both grown and newly formed leaves) were washed twice for 3 min in sterile distilled water, followed by a wash for 30 s with a solution of ethanol (70%), and finally washed for 3 times for 3 min in sterile distilled water. After the washing procedures, lettuce leaves were smashed in 10 mL of distilled water. The extract from the leaves was subsequently diluted and plated on LB and added with RIF (100 μg/mL). The distilled water used during the last washing procedure was plated on LB Petri dishes with RIF (100 μg/mL) to verify the sterilization of lettuce leaves. As per the rhizosphere, the identity of randomly selected RIF-R colonies (n = 12) was confirmed by ITS-PCR fingerprinting (Mapelli et al., 2013).
2.5 Donor strain construction
The strain K. variicola EEF15 was chromosomally tagged with a cassette containing a mCherry gene, a constitutively expressed lacIq gene, and a gentamycin resistance gene (gmR); the broad host range plasmid pKJK5::Plac::gfp, carrying a green fluorescent protein gene under the control of laclq gene repressible promoter and a kanamycin resistance gene (kmR), was then introduced into the strain. Insertion of the gene cassette on the chromosome and the consequent loss of the plasmid used to insert the gene cassette was carried out as previously described (McKenzie and Craig, 2006; Fournet-Fayard et al., 1995) and verified by fluorescence microscopy and PCR analysis (Lambertsen et al., 2004). Briefly, the suicide plasmid pGRG36 containing the lacIq-pLpp-mCherry-gmR cassette was inserted by electroporation at 1800 V (Fournet-Fayard et al., 1995) and transformed cells were selected on LB agar plates with gentamicin (100 μg/mL). As explained by Mckenzie and Craig (2006), to ensure the complete loss of the plasmid, red fluorescent colonies grown on the selective media were streaked twice and incubated at 42°C. Primers used to verify the loss of the plasmid were (i) Backbone pGRG36Fw 1 5′-TAG AGC GTC GCT ATT GGC AG-3′; Backbone pGRG36Rv 1 5′-TGC TCA ACG AGT TCG CTT CT-3′ and (ii) Backbone pGRG36Fw 8 5′-TGC TAG AGG CAT TAC GCT CG-3′; Backbone pGRG36Rv 8 5′-GCA AAG CGG GCA AAT ACC AA-3′. Primers were designed using the Primer designing tool—National Center for Biotechnology Information (NCBI).1 Insertion of the plasmid pKJK5::Plac::gfp in electrocompetent cells was carried out by electroporation at 1800 V (Fournet-Fayard et al., 1995); transformed cells were selected on LB agar plates with kanamycin (100 μg/mL). The acquisition of the plasmid carrying the gfp was verified by fluorescence microscopy (gfp expression), adding 0.1 μM IPTG in the growth media. The strain EEF15::lacIq-pLpp-mCherry-gmR with plasmid pKJK5::Plac::gfp was then used in conjugation experiments.
2.6 Conjugal transfer to the selected recipient strain Pseudomonas putida KT2440
Conjugation assays were performed using filter mating assays between the donor strain K. variicola EEF15::lacIq-pLpp-mCherry-gmR with pKJK5::Plac::gfp and the recipient P. putida KT2440 (Schlechter and Remus-Emsermann, 2019). Bacteria were grown separately in LB liquid media, and 109 cells/ml of each strain (calculated by using a Thoma cell-counting chamber) were mixed and inoculated on a nitrocellulose membrane filter (GSWP, 25-mm diameter, 0.22 μm pore size, Millipore, Burlington, MA, USA) placed on LB agar plates. The filters were incubated for 24 h at 30°C. Transconjugants were selected by plating the conjugation mix on cetrimide agar (Millipore) added with kanamycin (100 μg/mL): donor growth was inhibited on cetrimide agar. In contrast, the growth of P. putida KT2440 without the conjugative plasmid was inhibited by the presence of kanamycin. Transfer of the plasmid in recipient cells was verified by fluorescent microscopy (monitoring the gfp expression) and PCR amplification (of the gfp gene; Riva et al., 2022a), and by carrying out ITS-PCR amplification to confirm strain identification (Mapelli et al., 2013).
2.7 Conjugal transfer to the bacterial community of lettuce rhizosphere
Rhizosphere was collected from lettuce plants as described by Marasco et al. (2018). Briefly, lettuce roots were collected in a 50-ml tube, immersed in physiological water, and vortexed to remove the attached rhizosphere; roots were then separated from the rhizosphere and the rhizosphere was used for conjugation assays. Conjugation between strain EEF15::lacIq-pLpp-mCherry-gmR tagged with plasmid pKJK5::Plac::gfp and the lettuce rhizosphere community was conducted as already performed by Klümper et al. (2014), modifying two steps of the protocol. In particular, (i) the microbiome of the rhizosphere was extracted as described by using the Nycodenz density gradient (Fan et al., 2019); and (ii) a cell density of 3.38 × 108 cells/ml, instead of 3.38 × 107 cells/ml, for the donor strains and the pool of recipient bacterial cells were used. Cell concentrations were estimated using a Thoma cell-counting chamber. To estimate the conjugation transfer rate of plasmid pKJK5::Plac::gfp from strain EEF15::lacIq-pLpp-mCherry-gmR, filter mating experiments were performed also using E. coli MG1655::lacIq-pLpp-mCherry-kmR with plasmid pKJK5::Plac::gfp, as donor strain (Klümper et al., 2015; Supplementary Table 1). Filters placed on plates containing 10% soil extract medium were incubated at 25°C for a period of 48 h. The ability of the donor strains to transfer pKJK5::Plac::gfp plasmid to the rhizosphere microbiome was evaluated by fluorescence stereomicroscopy and calculated as conjugal transfer rate, as reported in the following formula: [Transconjugants per pictures × filter area (µm2)]/[picture area (µm2) × recipients introduced originally]. The number of transconjugants was estimated by counting the green microcolonies using fluorescence stereomicroscopy.
2.8 Sorting and identification of transconjugants
Transconjugants were sorted and identified as per Klümper et al. (2014). Briefly, bacteria on the filter were detached and resuspended in PBS solution, added with 30% glycerol, and stored at −80°C. After 4 weeks, the samples were thawed and sorted on a BD FACSAria. Transconjugants were selected by gfp expression, excluding non-fluorescent soil bacteria and all the background particles. DNA from the sorted samples was extracted using InstaGene™ Matrix (Bio-Rad); hypervariable regions V3-V4 were amplified, and the Illumina library was prepared as described in Li et al. (2021). Amplicons were further sequenced through the Illumina MiSeq platform using Reagents Kit v3 (2 × 300 cycles) in the paired-end mode at Copenhagen University.
2.9 Metataxonomic sequencing analyses
Reads were filtered, merged, and grouped in amplicon sequence variants (ASVs) with the DADA2 pipeline (Callahan et al., 2016; Callahan et al., 2017) as suggested in the online tutorial, except for filtering that was done with maxEE “2” for R1 and “10” for R2. Taxonomy was assigned using the Silva Database version 138 (Yilmaz et al., 2014), while the microbiome data were analyzed by phyloseq R package (McMurdie and Holmes, 2013). β-Diversity was calculated as weighted UniFrac Distance and plotted as principal coordinate analysis (PCoA). α-Diversity was calculated as Richness (number of ASVs), Shannon, and Simpson indices. To determine differences between the communities that were treated with the two donors, analysis of compositions of microbiomes with bias correction (ANCOMBC) using the ANCOMBC package in R was conducted for both the ASV table and genera table, and the results were inspected for differentially distributed taxa (Lin and Peddada, 2020). All analyses were performed using RStudio.2 Sequences affiliated with the Klebsiella and Escherichia genera (6.5% of all reads) were removed from the amplicon metagenomics sequencing dataset to eliminate any possible contamination due to their presence in sorted transconjugants. These genera were also removed from the other samples and the control to not create artificial differences between the treatments, even though this can cause the loss of some transconjugants from the results.
3 Results
3.1 Donor strain isolation, selection, and construction
The selective medium Violet Red Bile Lactose Agar was used to obtain a collection of Enterobacteriaceae strains from the effluent of a municipal WWTP. A total number of 14 isolates, including 9 Klebsiella, 2 Enterobacter, 1 Citrobacter, 1 Kluyvera, and 1 Raoultella, were purified and identified by 16S rRNA gene sequencing (Table 1). All the isolates showed sensitivity against kanamycin and RIF (Table 1). Specifically, RIF sensitivity was investigated to further select a RIF-sensitive strain to obtain a RIF-resistant (RIF-R) mutant to be used for plant colonization experiments; sensitivity against kanamycin was instead tested since needed for the construction of the donor strain (the plasmid pKJK5::Plac::gfp carries a kmR gene). Among RIF- and kanamycin-sensitive isolates, the strain K. variicola EEF15 was selected as a donor for conjugation experiments because the Klebsiella genus resulted in the most representative one among the obtained isolates. The strain EEF15 was genetically modified on the chromosome by adding a cassette carrying a red fluorescent protein (mCherry), a lacIq repressor gene, and a gentamycin resistance gene (gmR). Finally, the plasmid pKJK5::Plac::gfp carrying a gfp gene, the expression of which was under the control of lacIq gene carried on the chromosome, was introduced in the strain. The resulting mutant was gentamycin-resistant and expressed red fluorescence. The gene gfp, plasmid encoded, would be only expressed in a recipient strain of the rhizosphere microbiome missing lacIq gene. The strategy we used to evaluate HGT in the rhizosphere microbiome consisted of two steps: initially, we extracted the cells of the microbial community associated with the lettuce rhizosphere by using a density gradient, and we then exposed it to the donor strain obtained as previously described (Supplementary Figure 1).
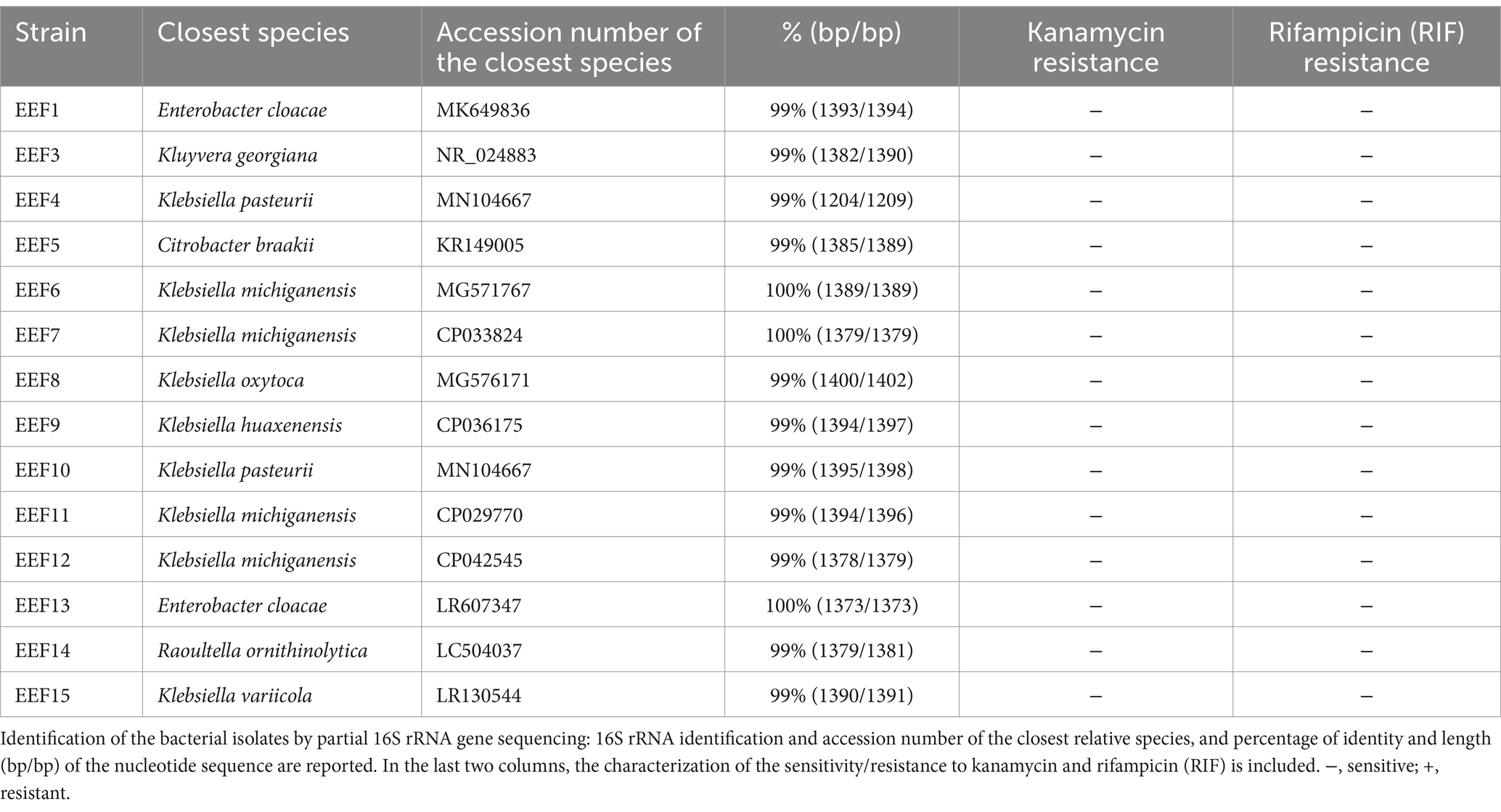
Table 1. List of strains isolated from the effluent of the wastewater treatment plant (WWTP) of Nosedo (Milan, Italy).
3.2 Rhizosphere and endosphere colonization by the donor strain
The ability of the EEF15 strain to colonize the lettuce rhizosphere and endosphere was verified using the RIF-R strain EEF15 RIF-R. This ability is indeed important to study the spread of AMR at the plant level, considering that an antibiotic-resistant strain could reach the plant due to the reuse of treated wastewater as an irrigation source. The results showed that 1 week after the bacterization of the lettuce rhizosphere, 2.62 × 107 ± 1.21 × 107 CFU/g of the soil of RIF-R bacteria were present in the rhizosphere when 1 × 108 EEF15 RIF-R cells/g of soil were applied (Figure 1; Supplementary Table 2). Similarly, when 1 × 109 EEF15 RIF-R cells/g of soil were used, 2.87 × 108 ± 1.53 × 107 CFU/g of soil of RIF-R bacteria were isolated from bacterized plant rhizosphere (Figure 1; Supplementary Table 2). No RIF-R CFUs were detected in the negative control (i.e., not bacterized lettuce plants). ITS-PCR fingerprinting obtained from rhizosphere colonization experiments confirmed their identity as EEF15 RIF-R (Supplementary Figure 2A). Seven days after the rhizosphere bacterization with 1 × 109 cells/ml, EEF15 RIF-R was isolated from the leaf endosphere, resulting in an average of 4.12 CFU/g of leaves (Figure 1; Supplementary Table 2). No RIF-R CFUs were found in the negative control (i.e., not bacterized lettuce plants) and in the distilled water used for washing procedures. ITS-PCR fingerprinting on selected colonies, obtained from endosphere colonization experiments, confirmed their identity as EEF15 RIF-R (Supplementary Figure 2B).
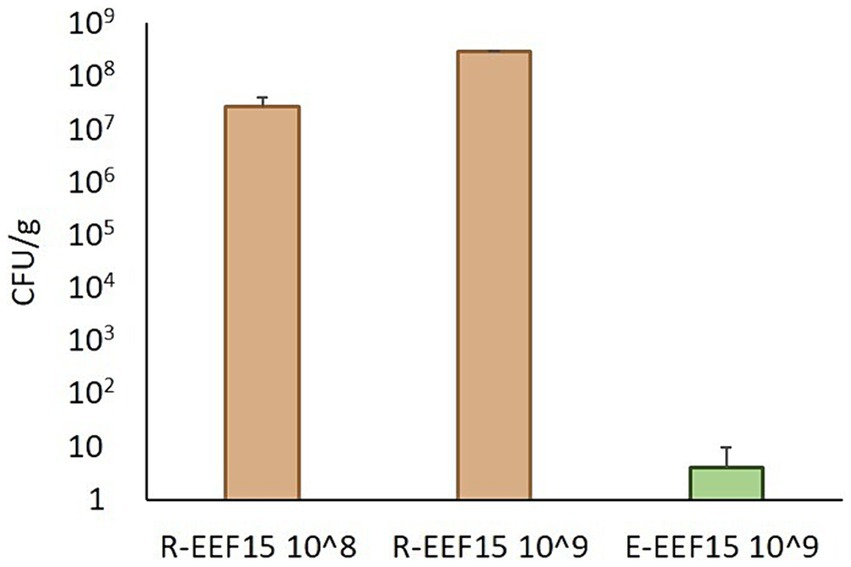
Figure 1. Lettuce colonization by Klebsiella variicola EEF15 RIF-R when lettuce rhizosphere was bacterized with 108 and 109 cells/g of soil (indicated as “108” and “109” in the sample names). EEF15 RIF-R abundance in the rhizosphere (“R” samples) and in the leaf endosphere (“E” sample) is indicated as CFU/g of soil or leaf tissue, respectively.
3.3 Transfer of plasmid pKJK5::Plac::gfp to a known bacterial recipient
We first assessed the transfer of plasmid pKJK5::Plac::gfp from the donor K. variicola EEF15::lacIq-pLpp-mCherry-gmR to the strain P. putida KT2440, used as the recipient in previous studies (Berthold et al., 2016). Filter mating experiments, based on Schlechter and Remus-Emserman, 2019, were performed using a ratio between the donor and the recipient strains equal to 1:1. The identity of random kanamycin-resistant transconjugants colonies was verified by ITS-PCR fingerprinting, showing to be the same of the recipient P. putida KT2440. Plasmid acquisition by transconjugants was verified using fluorescence microscopy, antibiotic selection, and gfp amplification through PCR (Supplementary Figure 3A).
3.4 Conjugation with the bacterial community of lettuce rhizosphere
Klebsiella variicola EEF15::lacIq-pLpp-mCherry-gmR harboring the plasmid pKJK5::Plac::gfp was incubated with the natural bacterial community obtained from lettuce rhizosphere. Likely, also the strain E. coli MG1655::lacIq-pLpp-mCherry-kmR harboring the plasmid pKJK5::Plac::gfp was used as a donor in conjugal experiments with the bacterial community obtained from the lettuce rhizosphere (Supplementary Table 1). Mating filters were analyzed using fluorescence stereomicroscopy, showing the presence of red- and green-tagged colonies, hence confirming both the presence of donors as well as the transfer of plasmid in bacterial recipients, respectively (Supplementary Figure 3B). Transfer frequencies from Klebsiella or Escherichia donor strains to the recipient community are listed in Table 2, while Supplementary Table 3 reports the number of transconjugants detected in each of the 20–21 analyzed portions for each filter. The conjugal transfer rate from K. variicola EEF15 donor strain, even if resulted similar and with the same order of magnitude (i.e., 2.41 × 10−3 ± 5.71 × 10−4 for K. variicola EEF15 and 3.50 × 10−3 ± 1.60 × 10−4 for E. coli MG1655), was significantly lower than the one from E. coli MG1655 (analysis of variance [ANOVA], p < 0.05). No green events (presence of transconjugants) were detected when only the donors or the recipient cells were placed separately on the filters.
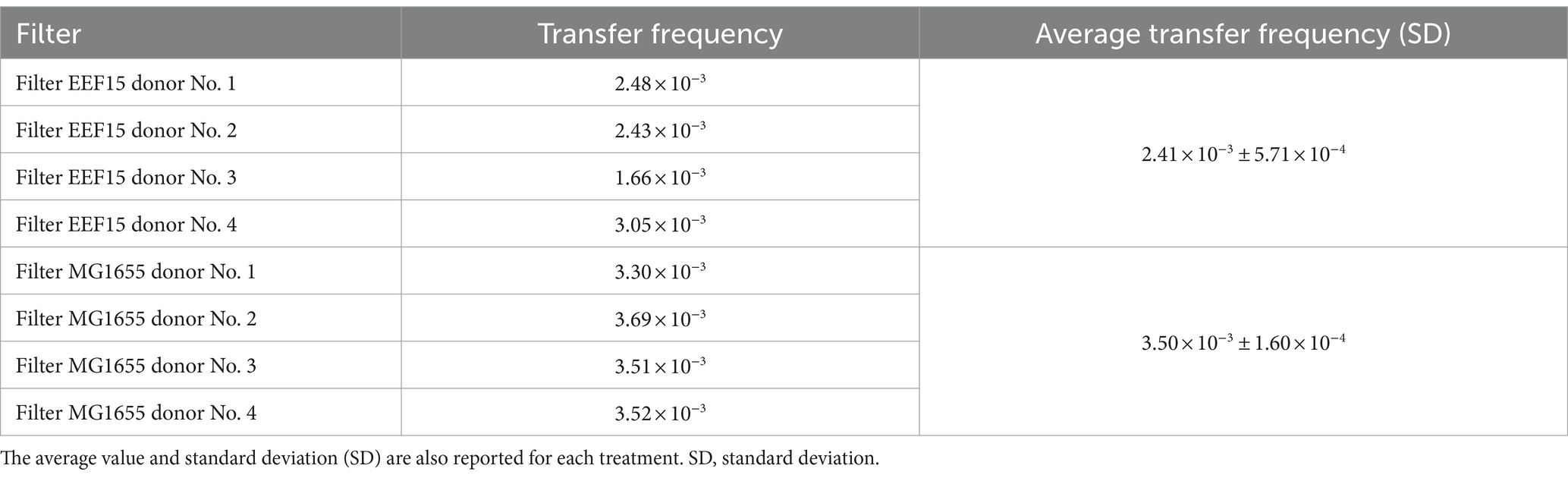
Table 2. Transfer frequency of plasmid pKJK5::Plac::gfp between Klebsiella variicola EEF15::lacIq-pLpp-mCherry-gmR or Escherichia coli MG1655::lacIq-pLpp-mCherry-kmR, and the bacterial community of lettuce rhizosphere, calculated per each filter.
Following cell sorting and subsequent DNA extraction from sorted cells, the identity of transconjugants was assessed through 16S rRNA gene amplicon sequencing, retrieving a total number of 112,748 reads, corresponding to 1,280 ASVs (Supplementary Table 4). All the samples showed sufficient sequencing depth to capture taxa diversity (Supplementary Figure 4). According to ANOVA analysis, the α-diversity indices (Richness, Shannon, and Simpson) of the transconjugant bacterial populations did not show statistical differences depending on the bacterial donors used in the experiments (ANOVA, p > 0.05; Supplementary Table 5). β-Diversity was checked at ASV level: Multidimensional scaling (MDS)/PCoA on weighted UniFrac Distance is shown in Figure 2, showing a distinct clustering of the transconjugants compared with the original lettuce bacterial community. Among the transconjugants, a secondary grouping by the donor (E. coli/K. variicola) was revealed. ANCOMBC analysis showed that none of the ASVs or genera were significantly differently distributed between the communities derived from the two donors (data deposited in GitHub as indicated in the “Data Availability Statement” section). Figure 3 shows the average relative abundance of ASVs at the family level considering donor strains or original rhizosphere community (Supplementary Table 4), whereas Supplementary Figure 5 depicts the relative ASV abundance considering each sample. Eighty-two ASVs with a relative abundance lower than 0.05% for at least one treatment were classified as “Others.”
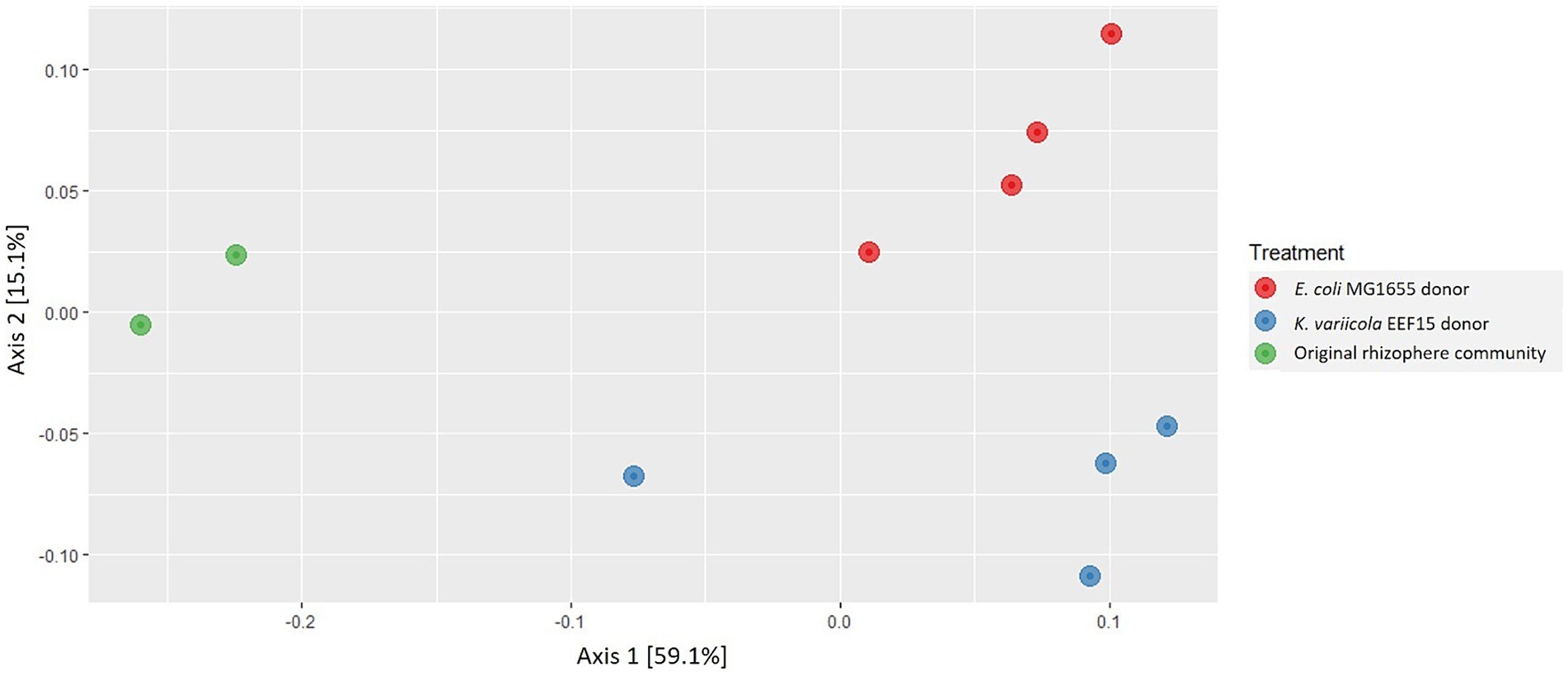
Figure 2. Multidimensional scaling (MDS)/principal coordinate analysis (PCoA) on weighted UniFrac Distance of the sample replicates of each treatment, i.e., transconjugants obtained using Escherichia coli MG1655::lacIq-pLpp-mCherry-kmR donor strain, transconjugants obtained using Klebsiella variicola EEF15::lacIq-pLpp-mCherry-gmR donor strain and the original bacterial community of lettuce rhizosphere.
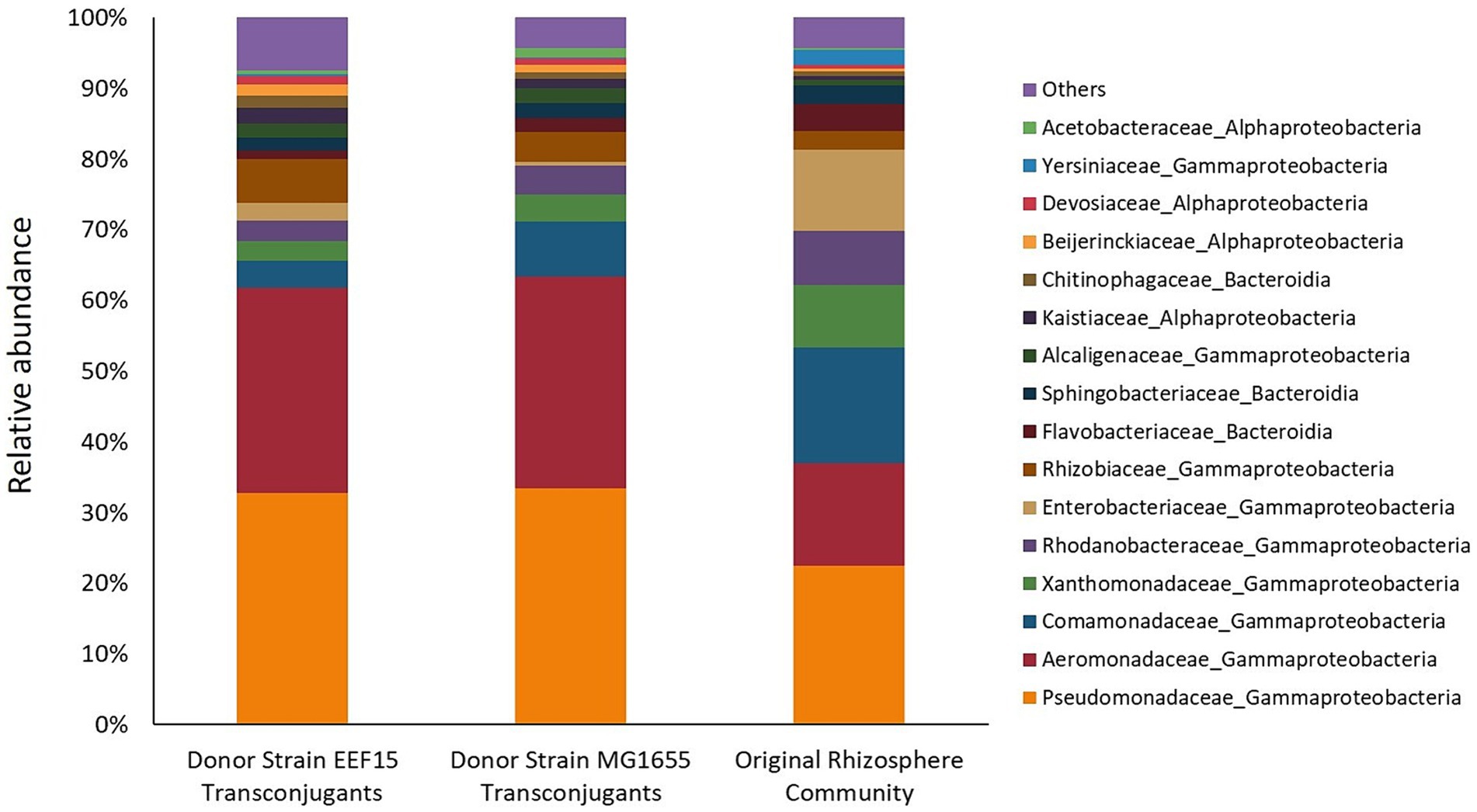
Figure 3. Relative abundance of ASVs related to filter mating assays and from the original rhizosphere bacterial community (higher than 0.05% for at least one treatment) at the family level. Eighty-two ASVs, whose relative abundance is lower than 0.05%, are classified as “Others.” From the left to the right, histograms refer to transconjugants obtained with Klebsiella variicola EEF15::lacIq-pLpp-mCherry-gmR and Escherichia coli MG1655::lacIq-pLpp-mCherry-kmR donor strains. The last histogram showed the relative abundance of bacterial families of the bacterial community originally associated with the lettuce rhizosphere.
The results show that transconjugants were affiliated with the major bacterial phyla found in the original rhizosphere bacterial community. The most represented bacterial phylum was Pseudomonadota, with the Gammaproteobacteria class comprising approximately 60% of the transconjugants. Families of Pseudomonadaceae and Aeromonadaceae were the most abundant ones, representing, respectively, approximately 30 and 29% of the transconjugants. Alphaproteobacteria class represented 12.9 and 8.2% of the total ASVs retrieved in experiments with EEF15 and MG1655 donor strains, respectively. Alphaproteobacteria class was mainly represented by the families of Rhizobiaceae, Kaistiaceae, and Beijerinckiaceae, belonging to the Rhizobiales order. Other bacterial phyla, i.e., Bacteroidota, Actinomycetota, Bacillota, Deinococcota, and Cyanobacteriota, constituted a total of 8.1% (with K. variicola EEF15 donor) and 7.2% (with E. coli MG1655 donor) of the transconjugants. The bacterial community of lettuce rhizosphere showed the presence of the same bacterial families identified in conjugative experiments but at different relative abundance. For instance, some taxa were less abundant in the original rhizosphere bacterial community compared to the transconjugant pools, e.g., families Pseudomonadaceae and Aeromonadaceae that were, respectively, the 22 and 14% of the total ASVs, suggesting that these families are more prone to the acquisition of the conjugal broad host range plasmid compared to the rest of the community.
4 Discussion
Considering the reuse of treated wastewater as an irrigation source in the One Health context, K. variicola was chosen as a donor strain since indicated as an emerging pathogen for humans (Rodríguez-Medina et al., 2019). K. variicola (belonging to the Klebsiella pneumoniae complex in the Enterobacteriaceae family) has been reported to be associated with different environments, such as plant niches (e.g., rhizosphere), freshwater bodies, humans, insects, and other animals, as suggested by its name (the term “variicola,” coming from Latin, indicates that the bacterium can be found in different places) (Rodríguez-Medina et al., 2019; Barrios-Camacho et al., 2019). In addition to its pathogenicity and wide distribution, it is intrinsically resistant to ampicillin (blaLEN), and reports on multidrug-resistant isolates of K. variicola, not only from clinical settings, are increasing (Barrios-Camacho et al., 2019; Rodríguez-Medina et al., 2019). For these reasons and considering the strain’s ability to colonize the lettuce rhizosphere (as shown in this study), where bacterial activity is enhanced if compared to bulk soil (Zhu et al., 2018), and to reach the endosphere, the strain K. variicola EEF15 has been selected as the perfect candidate to study conjugation events in complex plant-associated communities.
Gaining insight into plasmid dissemination when treated wastewater is reused as an irrigation source is of compelling interest. To this aim, the EEF15 donor strain of environmental origin (isolated from treated wastewater) was genetically manipulated to assess plasmid transfer to the complex natural bacterial community associated with the lettuce rhizosphere. Notably, plasmid conjugal transfer was specifically verified without the application of a selective agent (Klümper et al., 2015; Berthold et al., 2016; Fan et al., 2019; Xu et al., 2021). Our environmental donor strain was obtained following a strategy widely employed in previous studies (Klümper et al., 2015; Li et al., 2020): the selected strain was chromosomally tagged with a cassette carrying a red fluorescence protein (mCherry), a lacIq repressor gene, and transformed with a plasmid of interest encoding a gfpmut3 gene downstream of a LacI-repressible promoter (Musovic et al., 2010), enabling the tracking of the plasmid movement to the recipient bacteria by flow cytometry. The following sorting of GFP-tagged cells and the application of 16S rRNA gene amplicon sequencing on the sorted cells allowed the identification of transconjugants (Klümper et al., 2014). This approach, based on cultivation-independent techniques, allowed to avoid the use of cultivation-based methods which are biased by the very low bacterial culturability in laboratory conditions (Joseph et al., 2003): estimates have indicated that only 1.4–14.1% of the total cells in a soil sample are indeed cultivable (Janssen et al., 2002).
In our study, we showed that the transfer of pKJK5::Plac::gfp plasmid occurred from the donor strain K. variicola EEF15::lacIq-pLpp-mCherry-gmR to both a selected recipient (i.e., a P. putida strain) and the complex bacterial community of plant rhizosphere, as confirmed by PCR and fluorescence (stereo)microscopy. We observed conjugal transfer to the rhizosphere community with a frequency of 10−3 with both donors. Lower values of conjugation frequency (i.e., 10−4) have been reported in the recent publication by Macedo et al. (2022) in which authors described the conjugal transfer of the plasmid pKJK5 in soil microcosms with or without manure. However, considering the diversity of the experimental set-ups (microcosms vs. filter mating assays, as well as differences in cell density, temperature, contact period, and nutrient availability), comparisons among studies should be made with caution (He et al., 2024; Pallares-Vega et al., 2021). Moreover, it is noteworthy to mention that, even if we observed a statistically significantly lower conjugal transfer rate with the environmental K. variicola donor strain than with the laboratory E. coli one, EEF15- and MG1655-derived pools of ASV transconjugants shared the same taxonomy and taxonomic composition (Figures 2, 3). A lower frequency of plasmid transfer by an environmental isolate cannot, nevertheless, be considered a lower risk for ARG circulation. Finally, we need to consider that the use of laboratory strains could not provide an exhaustive estimation of the impact of conjugal transfer since they might have lost important functions or regulatory genes that might be essential in “real-world” situations, e.g., proteins related to the osmotic stress or genes involved in cell to cell aggregation and biofilm generation (Sim et al., 2014; Tchagang et al., 2018), in comparison to environmental strains, which are adapted to thrive in specific habitats (Fux et al., 2005). Since their first isolation, researchers have sub-cultured laboratory strains such as E. coli K12 (from which derives the laboratory donor strain MG1655, used in this study) or P. aeruginosa PAO1 for many years, likely differentiating new phenotypes (Fux et al., 2005). For instance, if we compare uropathogenic and enteropathogenic strains to the avirulent laboratory strain of E. coli K12 through genomics analysis, only 39% of their combined set of proteins are shared (Welch et al., 2002; Fux et al., 2005). Future studies should aim to obtain a better comprehension of the magnitude of the phenomenon, and this should not disregard the use of environmental donor strains and experimental settings closer to real environmental situations. The use of environmentally relevant strains, which could own some specific traits influencing HGT outside of laboratory conditions, should help to disentangle the significance and mechanisms of AMR diffusion in different ecosystems, providing a more reliable description of AMR spread (Bernardy et al., 2016; Pinilla-Redondo et al., 2018; Fan et al., 2019; Riva F. et al., 2020; Riva V. et al., 2020; Di Cesare et al., 2022).
Considering the identification of taxa of the bacterial community associated with the lettuce rhizosphere having the potential to acquire exogenous plasmids by a conjugative donor, the phylum Pseudomonadota with the class Gammaproteobacteria was the dominant one, as reported in other soil-related studies on HGT evaluation (Klümper et al., 2015; Fan et al., 2019). However, Klümper et al. (2015) studied the conjugal transfer between several donors and the bacterial community extracted from the CRUCIAL soil, which was indeed the same soil that we considered in our experiments but to which lettuce plants applied a selection, recruiting bacteria in the rhizosphere. As already reported, the broad host range plasmid pKJK5 (a plasmid belonging to the IncP-1ε subgroup) was transferred to recipients belonging to different bacterial phyla in addition to Pseudomonadota (Klümper et al., 2015), i.e., Actinomycetota and Bacteroidota, for some of which poor culturability has been reported (Joseph et al., 2003): conjugation events between the Gram-negative donors and Gram-positive recipient bacteria have been indeed detected in both transconjugant pools, confirming what already documented in other studies, about the plasmid transmissibility to a broad phylogenetic range (Klümper et al., 2015; Fan et al., 2019; Qiu et al., 2018). Contrary to Klümper et al. (2015), we did not detect any differences between the transconjugant pools obtained from the two donor strains in terms of ASV taxonomy and taxonomic composition, indicating that in our study, the broad host range plasmid pKJK5, independently from the donor strain considered, could be acquired by the same bacterial taxa even if with a different conjugal frequency. However, the reasons behind the plasmid ability to be transferred to the same taxa are unclear (Klümper et al., 2015): we hypothesized that EEF15 and MG1655 donor strains, even if belonging to different species within the Enterobacteriaceae family, shared some characteristics which could favor cell-to-cell contact at the same extent, e.g., mating pair stabilization (Hirt et al., 2002; Virolle et al., 2020; Low et al., 2022). Moreover, the method we applied cannot allow us to discriminate between the horizontal plasmid transfer and the subsequent vertical transmission due to the growth of transconjugants on filter mating plates incubated at 25°C for 48 h (Klümper et al., 2015): the relative abundance of transconjugant taxa could be thus influenced by their growth rates, especially when comparisons with the taxa relative abundance of native rhizosphere are made. Finally, as also indicated by Klümper et al. (2015), the use of a Nycodenz-based extraction protocol resulted in the selection of a part of the bacterial microbiome associated with the lettuce rhizosphere, which was further exposed to plasmid transfer experiments: although we were not able to recover all the rhizospheric bacteria, it is noteworthy to mention that the same bias is applied with both donors; this could also allow to speculate that conjugation rates might be even higher in natural settings.
Among transconjugants, several bacterial families known to be able to thrive in the agri-food ecosystem, e.g., in plant and water-related microbiomes, as well as putative pathogenic bacteria derived from anthropogenic environments, have been detected. The best example is given by the most abundant family represented by Pseudomonadaceae with the genus Pseudomonas, known to be part of the plant and soil microbial communities, to be capable of suppressing plant pathogens, but also including possible pathogenic species/strains (Lyng and Kovács, 2023; Vieira et al., 2024; Dsouza et al., 2024). We retrieved several families (e.g., Comamonadaceae, Xanthomonadaceae, Enterobacteriaceae, Rhizobiaceae, and Acetobacteraceae) which are commonly retrieved in the plant microbiome (Riva et al., 2022b) and are known to be involved in plant physiology and fitness, i.e., including plant growth promoting rhizobacteria, bacterial taxa able to limit the growth of microbial phytopathogens, and plant pathogens (Vicente et al., 2023; Kaur et al., 2023; Bhadrecha et al., 2023; Leach et al., 2017). Likely, among the most abundant transconjugants there were also several families primarily linked to AMR spread in aquatic environments, such as Aeromonadaceae and Legionellaceae (Suyamud et al., 2024; Falkinham, 2020), and Flavobacteriaceae and Enterobacteriaceae families known to be hosted by aquatic animal species and plants (Burbick et al., 2023). Finally, we observed the presence of families typically associated with humans, able to thrive in anthropized environments, such as Streptococcaceae and Staphylococcaceae (Ziarati et al., 2022; Lawhon et al., 2023). This highlights that the transfer of ARG-carrying plasmids can occur among bacteria relevant in the agri-food ecosystem, possibly contributing to the spread of ARGs to pathogens in human-related habitats.
5 Conclusion
In this study, we have evaluated the ability of an environmental donor strain, isolated from treated wastewater and genetically manipulated, to transfer a conjugal broad host range plasmid to the rhizospheric microbial community of lettuce plants grown in agricultural soil to assess ARGs spread. Plasmid transfer was demonstrated to occur in a wide phylogenetic group of the rhizosphere microbiome without any selective pressure. This, together with the donor strain’s ability to colonize the plant rhizosphere and to reach the leaf endosphere, highlights the importance of studying the magnitude of the diffusion of AMR determinants in the agri-food ecosystem, particularly related to vegetables eaten raw, which have the potential to become in direct contact with the human microbiome. Future studies taking advantage of environmental donor strains could allow us to obtain a better comprehension of AMR spread under the context of the One Health approach to appraise possible risks related to human health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
FR: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing, Data curation, Investigation, Visualization. AD: Formal analysis, Investigation, Writing – review & editing. EE: Data curation, Formal analysis, Writing – review & editing. VR: Data curation, Investigation, Writing – review & editing, Visualization. SB: Resources, Supervision, Writing – review & editing. FM: Funding acquisition, Writing – review & editing. BS: Resources, Supervision, Writing – review & editing. EC: Conceptualization, Formal analysis, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Italian Ministry of University and Research through the PRIN2020 No. 2020T8T5MM (“Eco-evolutionary approach to identification and isolation of freshwater bacteria harboring antibiotic resistance genes—IdeARG”) and supported by Cariplo Foundation through the project No. 2018-0995 (“Novel wastewater disinfection treatments to mitigate the spread of antibiotic resistance in agriculture—WARFARE”).
Acknowledgments
We acknowledge help from Zhiming He (DTU, Denmark) for the gene transfer experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1457854/full#supplementary-material
Footnotes
References
Abs, E., Chase, A. B., and Allison, S. D. (2023). How do soil microbes shape ecosystem biogeochemistry in the context of global change? Environ. Microbiol. 25, 780–785. doi: 10.1111/1462-2920.16331
Alderliesten, J. B., Duxbury, S. J., Zwart, M. P., de Visser, J. A. G., Stegeman, A., and Fischer, E. A. (2020). Effect of donor-recipient relatedness on the plasmid conjugation frequency: a meta-analysis. BMC Microbiol. 20, 135–110. doi: 10.1186/s12866-020-01825-4
Aminov, R. I. (2011). Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2:158. doi: 10.3389/fmicb.2011.00158
Araújo, S., Silva, A. T. I., Tacão, M., Patinha, C., Alves, A., and Henriques, I. (2017). Characterization of antibiotic resistant and pathogenic Escherichia coli in irrigation water and vegetables in household farms. Int. J. Food Microbiol. 257, 192–200. doi: 10.1016/j.ijfoodmicro.2017.06.020
Barrios-Camacho, H., Aguilar-Vera, A., Beltran-Rojel, M., Aguilar-Vera, E., Duran-Bedolla, J., Rodríguez-Medina,, et al. (2019). Molecular epidemiology of Klebsiella variicola obtained from different sources. Sci. Rep. 9:10610. doi: 10.1038/s41598-019-46998-9
Bernardy, E. E., Turnsek, M. A., Wilson, S. K., Tarr, C. L., and Hammer, B. K. (2016). Diversity of clinical and environmental isolates of Vibrio cholerae in natural transformation and contact-dependent bacterial killing indicative of type VI secretion system activity. App. Environ. Microbiol. 82, 2833–2842. doi: 10.1128/AEM.00351-16
Berthold, T., Centler, F., Hübschmann, T., Remer, R., Thullner, M., Harms, H., et al. (2016). Mycelia as a focal point for horizontal gene transfer among soil bacteria. Sci. Rep. 6:36390. doi: 10.1038/srep36390
Bhadrecha, P., Singh, S., and Dwibedi, V. (2023). ‘A plant’s major strength in rhizosphere’: the plant growth promoting rhizobacteria. Arch. Microbiol. 205:165. doi: 10.1007/s00203-023-03502-2
Bollmann, U. E., Fernández-Calviño, D., Brandt, K. K., Storgaard, M. S., Sanderson, H., and Bester, K. (2017). Biocide runoff from building facades: degradation kinetics in soil. Environment 51, 3694–3702. doi: 10.1021/acs.est.6b05512
Burbick, C. R., Munson, E., Lawhon, S. D., Zapp, A., Villaflor, M., and Thelen, E. (2023). An update on novel taxa and revised taxonomic status of bacteria (including members of the phylum Planctomycetota) isolated from aquatic host species described in 2018 to 2021. J. Clin. Microbiol. 61:e0142622. doi: 10.1128/jcm.01426-22
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. doi: 10.1038/ismej.2017.119
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nMeth.3869
Callegari, M., Jucker, C., Fusi, M., Leonardi, M. G., Daffonchio, D., Borin, S., et al. (2020). Hydrolytic profile of the culturable gut bacterial community associated with Hermetia illucens. Front. Microbiol. 11:1965. doi: 10.3389/fmicb.2020.01965
Cerqueira, F., Matamoros, V., Bayona, J. M., Berendonk, T. U., Elsinga, G., Hornstra, L. M., et al. (2019). Antibiotic resistance gene distribution in agricultural fields and crops. A soil-to-food analysis. Environ. Res. 177:108608. doi: 10.1016/j.envres.2019.108608
Corno, G., Yang, Y., Eckert, E. M., Fontaneto, D., Fiorentino, A., Galafassi, S., et al. (2019). Effluents of wastewater treatment plants promote the rapid stabilizationof the antibiotic resistome in receiving freshwater bodies. Water Res. 158, 72–81. doi: 10.1016/j.watres.2019.04.031
Di Cesare, A., Eckert, E. M., Teruggi, A., Fontaneto, D., Bertoni, R., Callieri, C., et al. (2015). Constitutive presence of antibiotic resistance genes within the bacterial community of a large subalpine lake. Mol. Ecol. 24, 3888–3900. doi: 10.1111/mec.13293
Di Cesare, A., Riva, F., Colinas, N., Borgomaneiro, G., Borin, S., Cabello-Yeves, P. J., et al. (2022). Zooplankton as a transitional host for Escherichia coli in freshwater. Appl. Environ. Microbiol. 88:e0252221. doi: 10.1128/aem.02522-21
Dsouza, F. P., Dinesh, S., and Sharma, S. (2024). Understanding the intricacies of microbial biofilm formation and its endurance in chronic infections: a key to advancing biofilm-targeted therapeutic strategies. Arch. Microbiol. 206:85. doi: 10.1007/s00203-023-03802-7
Falkinham, J. O. III (2020). Living with Legionella and other waterborne pathogens. Microorganisms 8:2026. doi: 10.3390/microorganisms8122026
Fan, X. T., Li, H., Chen, Q. L., Zhang, Y. S., Ye, J., Zhu, Y. G., et al. (2019). Fate of antibiotic resistant Pseudomonas putida and broad host range plasmid in natural soil microcosms. Front. Microbiol. 10:194. doi: 10.3389/fmicb.2019.00194
Feng, G., Huang, H., and Chen, Y. (2021). Effects of emerging pollutants on the occurrence and transfer of antibiotic resistance genes: a review. J. Hazard. Mater. 420:126602. doi: 10.1016/j.jhazmat.2021.126602
Ferjani, R., Marasco, R., Rolli, E., Cherif, H., Cherif, A., Gtari, M., et al. (2015). The date palm tree rhizosphere is a niche for plant growth promoting bacteria in the oasis ecosystem. Biomed. Res. Int. 2015:153851. doi: 10.1155/2015/153851
Fernández-Calviño, D., Rousk, J., Bååth, E., Bollmann, U. E., Bester, K., and Brandt, K. K. (2017). Ecotoxicological assessment of propiconazole using soil bacterial and fungal growth assays. Appl. Ecol. 115, 27–30. doi: 10.1016/j.apsoil.2017.03.009
Fernández-Calviño, D., Rousk, J., Bååth, E., Bollmann, U. E., Bester, K., and Brandt, K. K. (2021). Short-term toxicity assessment of a triazine herbicide (terbutryn) underestimates the sensitivity of soil microorganisms. Soil Biol. Biochem. 154:108130. doi: 10.1016/j.soilbio.2021.108130
Fournet-Fayard, S., Joly, B., and Forestier, C. (1995). Transformation of wild type Klebsiella pneumoniae with plasmid DNA by electroporation. J. Microbiol. Methods 24, 49–54. doi: 10.1016/0167-7012(95)00053-4
Fux, C. A., Shirtliff, M., Stoodley, P., and Costerton, J. W. (2005). Can laboratory reference strains mirror ‘real-world’ pathogenesis? Trends Microbiol. 13, 58–63. doi: 10.1016/j.tim.2004.11.001
He, Z., Smets, B. F., and Dechesne, A. (2024). Mating assay: plating below a cell density threshold is required for unbiased estimation of transfer frequency or transfer rate. Res. Square. doi: 10.21203/rs.3.rs-4373740/v1
Hirt, H., Schlievert, P. M., and Dunny, G. M. (2002). In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect. Immun. 70, 716–723. doi: 10.1128/iai.70.2.716-723.2002
Janssen, P. H., Yates, P. S., Grinton, B. E., Taylor, P. M., and Sait, M. (2002). Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68, 2391–2396. doi: 10.1128/aem.68.5.2391-2396.2002
Joseph, S. J., Hugenholtz, P., Sangwan, P., Osborne, C. A., and Janssen, P. H. (2003). Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69, 7210–7215. doi: 10.1128/AEM.69.12.7210-7215.2003
Kaur, T., Khanna, K., Sharma, S., and Manhas, R. K. (2023). Mechanistic insights into the role of actinobacteria as potential biocontrol candidates against fungal phytopathogens. J. Basic Microbiol. 63, 1196–1218. doi: 10.1002/jobm.202300027
Klerks, M. M., Franz, E., van Gent-Pelzer, M., Zijlstra, C., and Van Bruggen, A. H. (2007). Differential interaction of Salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 1, 620–631. doi: 10.1038/ismej.2007.82
Klümper, U., Dechesne, A., and Smets, B. F. (2014). “Protocol for evaluating the permissiveness of bacterial communities toward conjugal plasmids by quantification and isolation of transconjugants” in Hydrocarbon and lipid microbiology protocols. Eds. T. J. McGenity, K. N. Timmis, and B. Nogales (Berlin, Heidelberg: Springer), 275–288.
Klümper, U., Riber, L., Dechesne, A., Sannazzarro, A., Hansen, L. H., Sørensen, S. J., et al. (2015). Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 9, 934–945. doi: 10.1038/ismej.2014.191
Lambertsen, L., Sternberg, C., and Molin, S. (2004). Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6, 726–732. doi: 10.1111/j.1462-2920.2004.00605.x
Larsson, D. J., Andremont, A., Bengtsson-Palme, J., Brandt, K. K., de Roda Husman, A. M., Fagerstedt, P., et al. (2018). Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Intern. 117, 132–138. doi: 10.1016/j.envint.2018.04.041
Larsson, D. G., and Flach, C. F. (2022). Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269. doi: 10.1038/s41579-021-00649-x
Lawhon, S. D., Burbick, C. R., Munson, E., Thelen, E., Zapp, A., and Wilson, A. (2023). Update on novel validly published taxa of bacteria isolated from domestic animals described in 2022. J. Clin. Microbiol. 61:e0083923. doi: 10.1128/jcm.00839-23
Leach, J. E., Triplett, L. R., Argueso, C. T., and Trivedi, P. (2017). Communication in the phytobiome. Cell 169, 587–596. doi: 10.1016/j.cell.2017.04.025
Leimbach, A., Hacker, J., and Dobrindt, U. (2013). E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr. Top. Microbiol. Immunol. 358, 3–32. doi: 10.1007/82_2012_303
Li, L., Dechesne, A., Madsen, J. S., Nesme, J., Sørensen, S. J., and Smets, B. F. (2020). Plasmids persist in a microbial community by providing fitness benefit to multiple phylotypes. ISME J. 14, 1170–1181. doi: 10.1038/s41396-020-0596-4
Li, L., Nesme, J., Quintela-Baluja, M., Balboa, S., Hashsham, S., and Smets, B. F. (2021). Extended-spectrum β-lactamase and carbapenemase genes are substantially and sequentially reduced during conveyance and treatment of urban sewage. Environ. Sci. Technol. 55, 5939–5949. doi: 10.1021/acs.est.0c08548
Lin, H., and Peddada, S. D. (2020). Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11:3514. doi: 10.1038/s41467-020-17041-7
Low, W. W., Wong, J. L. C., Beltran, L. C., Seddon, C., David, S., Kwong, H.-S., et al. (2022). Mating pair stabilization mediates bacterial conjugation species specificity. Nat. Microbiol. 7, 1016–1027. doi: 10.1038/s41564-022-01146-4
Lyng, M., and Kovács, Á. T. (2023). Frenemies of the soil: Bacillus and Pseudomonas interspecies interactions. Trends Microbiol. 31, 845–857. doi: 10.1016/j.tim.2023.02.003
Macedo, G., Olesen, A. K., Maccario, L., Leal, L. H. V. D., Maas, P., Heederik, D., et al. (2022). Horizontal gene transfer of an IncP1 plasmid to soil bacterial community introduced by Escherichia coli through manure amendment in soil microcosms. Environm. Sci. Technol. 56, 11398–11408. doi: 10.1021/acs.est.2c02686
Mapelli, F., Marasco, R., Rolli, E., Barbato, M., Cherif, H., Guesmi, A., et al. (2013). Potential for plant growth promotion of rhizobacteria associated with Salicornia growing in Tunisian hypersaline soils. Biomed. Res. Int. 2013:248078. doi: 10.1155/2013/248078
Marasco, R., Rolli, E., Fusi, M., Michoud, G., and Daffonchio, D. (2018). Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 6, 3–17. doi: 10.1186/s40168-017-0391-2
McKenzie, G. J., and Craig, N. L. (2006). Fast, easy and efficient: site-specific insertion of transgenes into Enterobacterial chromosomes using Tn 7 without need for selection of the insertion event. BMC Microbiol. 6:39. doi: 10.1186/1471-2180-6-39
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Musiyiwa, K., Simbanegavi, T. T., Marumure, J., Makuvara, Z., Chaukura, N., and Gwenzi, W. (2024). The soil-microbe-plant resistome: a focus on the source-pathway-receptor continuum. Environ. Sci. Pollut. Res. 31, 12666–12682. doi: 10.1007/s11356-023-31788-8
Musovic, S., Dechesne, A., Sørensen, J., and Smets, B. F. (2010). Novel assay to assess permissiveness of a soil microbial community toward receipt of mobile genetic elements. Appl. Environ. Microbiol. 76, 4813–4818. doi: 10.1128/AEM.02713-09
Nüesch-Inderbinen, M., Zurfluh, K., Peterhans, S., Hächler, H., and Stephan, R. (2015). Assessment of the prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in ready-to-eat salads, fresh-cut fruit, and sprouts from the Swiss market. J. Food Prot. 78, 1178–1181. doi: 10.4315/0362-028X.JFP-15-018
Pallares-Vega, R., Macedo, G., Brouwer, M. S., Hernandez Leal, L., van der Maas, P., van Loosdrecht, M. C., et al. (2021). Temperature and nutrient limitations decrease transfer of conjugative IncP-1 plasmid pKJK5 to wild Escherichia coli strains. Front. Microbiol. 12:656250. doi: 10.3389/fmicb.2021.656250
Perera, L. N., Mafiz, A. I., Amarasekara, N. R., Chang, E., Rao, V. B. K., and Zhang, Y. (2020). Antimicrobial-resistant E. coli and Enterococcus spp. recovered from urban community gardens. Food Control 108:106857. doi: 10.1016/j.foodcont.2019.106857
Pinilla-Redondo, R., Riber, L., and Sørensen, S. J. (2018). Fluorescence recovery allows the implementation of a fluorescence reporter gene platform applicable for the detection and quantification of horizontal gene transfer in anoxic environments. Appl. Environ. Microbiol. 84:e02507-17. doi: 10.1128/AEM.02507-17
Qiu, Y., Zhang, J., Li, B., Wen, X., Liang, P., and Huang, X. (2018). A novel microfluidic system enables visualization and analysis of antibiotic resistance gene transfer to activated sludge bacteria in biofilm. Sci. Total Environ. 642, 582–590. doi: 10.1016/j.scitotenv.2018.06.012
Riva, V., Mapelli, F., Bagnasco, A., Mengoni, A., and Borin, S. (2022b). A meta-analysis approach to defining the culturable core of plant endophytic bacterial communities. Appl. Environ. Microbiol. 88:e0253721. doi: 10.1128/aem.02537-21
Riva, V., Patania, G., Riva, F., Vergani, L., Crotti, E., and Mapelli, F. (2022a). Acinetobacter baylyi strain BD413 can acquire an antibiotic resistance gene by natural transformation on lettuce phylloplane and enter the endosphere. Antibiotics 11:1231. doi: 10.3390/antibiotics11091231
Riva, F., Riva, V., Eckert, E. M., Colinas, N., Di Cesare, A., Borin, S., et al. (2020). An environmental Escherichia coli strain is naturally competent to acquire exogenous DNA. Front. Microbiol. 11:574301. doi: 10.3389/fmicb.2020.574301
Riva, V., Riva, F., Vergani, L., Crotti, E., Borin, S., and Mapelli, F. (2020). Microbial assisted phytodepuration for water reclamation: environmental benefits and threats. Chemosphere 241:124843. doi: 10.1016/j.chemosphere.2019.124843
Rodríguez-Medina, N., Barrios-Camacho, H., Duran-Bedolla, J., and Garza-Ramos, U. (2019). Klebsiella variicola: an emerging pathogen in humans. Emerg. Microbes Infect. 8, 973–988. doi: 10.1080/22221751.2019.1634981
Saak, C. C., Dinh, C. B., and Dutton, R. J. (2020). Experimental approaches to tracking mobile genetic elements in microbial communities. FEMS Microbiol. Rev. 44, 606–630. doi: 10.1093/femsre/fuaa025
Sánchez-Salazar, A. M., Taparia, T., Olesen, A. K., Acuña, J. J., Sørensen, S. J., and Jorquera, M. A. (2023). An overview of plasmid transfer in the plant microbiome. Plasmid 127:102695. doi: 10.1016/j.plasmid.2023.102695
Schierstaedt, J., Grosch, R., and Schikora, A. (2019). Agricultural production systems can serve as reservoir for human pathogens. FEMS Microbiol. Lett. 366:fnaa016. doi: 10.1093/femsle/fnaa016
Schikora, A., Garcia, A. V., and Hirt, H. (2012). Plants as alternative hosts for Salmonella. Trends Plant Sci. 17, 245–249. doi: 10.1016/j.tplants.2012.03.007
Schlechter, R. O., and Remus-Emsermann, M. N. (2019). Delivering "chromatic Bacteria" fluorescent protein tags to Proteobacteria using conjugation. Bio Protoc. 9:e3199. doi: 10.21769/BioProtoc.3199
Sim, M., Lim, B., Sim, S. H., Kim, D., Jung, E., Lee, Y., et al. (2014). Two tandem RNase III cleavage sites determine betT mRNA stability in response to osmotic stress in Escherichia coli. PLoS One 9:e100520. doi: 10.1371/journal.pone.0100520
Smalla, K., Cook, K., Djordjevic, S. P., Klümper, U., and Gillings, M. (2018). Environmental dimensions of antibiotic resistance: assessment of basic science gaps. FEMS Microbiol. Ecol. 94:fiy195. doi: 10.1093/femsec/fiy195
Suyamud, B., Chen, Y., Quyen, D. T. T., Dong, Z., Zhao, C., and Hu, J. (2024). Antimicrobial resistance in aquaculture: occurrence and strategies in Southeast Asia. Sci. Total Environ. 907:167942. doi: 10.1016/j.scitotenv.2023.167942
Tchagang, C. F., Xu, R., Doumbou, C. L., and Tambong, J. T. (2018). Genome analysis of two novel Pseudomonas strains exhibiting differential hypersensitivity reactions on tobacco seedlings reveals differences in nonflagellar T3 SS organization and predicted effector proteins. Microbiol. Open 7:e00553. doi: 10.1002/mbo3.553
Touchon, M., Perrin, A., de Sousa, J. A. M., Vangchhia, B., Burn, S., O'Brien, C. L., et al. (2020). Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet. 16:e1008866. doi: 10.1371/journal.pgen.1008866
Ulrich, A., Becker, R., Ulrich, K., and Ewald, D. (2015). Conjugative transfer of a derivative of the IncP-1α plasmid RP4 and establishment of transconjugants in the indigenous bacterial community of poplar plants. FEMS Microbiol. Lett. 362:fnv201. doi: 10.1093/femsle/fnv201
Van Elsas, J. D., Turner, S., and Bailey, M. J. (2003). Horizontal gene transfer in the phytosphere. New Phytol. 157, 525–537. doi: 10.1046/j.1469-8137.2003.00697.x
Van Wonterghem, L., De Chiara, M., Liti, G., Warringer, J., Farewell, A., Verstraeten, N., et al. (2022). Genome-wide association study reveals host factors affecting conjugation in Escherichia coli. Microorganisms 10:608. doi: 10.3390/microorganisms10030608
Vicente, T. F. L., Félix, C., Félix, R., Valentão, P., and Lemos, M. F. L. (2023). Seaweed as a natural source against phytopathogenic bacteria. Mar. Drugs 21:23. doi: 10.3390/md21010023
Vieira, M. E. O., Nunes, V. V., Calazans, C. C., and Silva-Mann, R. (2024). Unlocking plant defenses: harnessing the power of beneficial microorganisms for induced systemic resistance in vegetables – a systematic review. Biol. Control 188:105428. doi: 10.1016/j.biocontrol.2023.105428
Virolle, C., Goldlust, K., Djermoun, S., Bigot, S., and Lesterlin, C. (2020). Plasmid transfer by conjugation in gram-negative bacteria: from the cellular to the community level. Genes 11:1239. doi: 10.3390/genes11111239
Wang, C. H., Hsieh, Y. H., Powers, Z. M., and Kao, C. Y. (2020). Defeating antibiotic-resistant bacteria: exploring alternative therapies for a post-antibiotic era. Int. J. Mol. Sci. 21:1061. doi: 10.3390/ijms21031061
Welch, R. A., Burland, V., Plunkett, G. III, Redford, P., Roesch, P., Rasko, D., et al. (2002). Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99, 17020–17024. doi: 10.1073/pnas.252529799
Xu, H., Chen, Z., Huang, R., Cui, Y., Li, Q., Zhao, Y., et al. (2021). Antibiotic resistance gene-carrying plasmid spreads into the plant endophytic bacteria using soil bacteria as carriers. Environ. Sci. Technol. 55, 10462–10470. doi: 10.1021/acs.est.1c01615
Yang, X., Wu, Q., Huang, J., Wu, S., Zhang, J., Chen, L., et al. (2020). Prevalence and characterization of Salmonella isolated from raw vegetables in China. Food Control 109:106915. doi: 10.1016/j.foodcont.2019.106915
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Yu, Z., Wang, Y., Lu, J., Bond, P. L., and Guo, J. (2021). Nonnutritive sweeteners can promote the dissemination of antibiotic resistance through conjugative gene transfer. ISME J. 15, 2117–2130. doi: 10.1038/s41396-021-00909-x
Zarei-Baygi, A., and Smith, A. L. (2020). Intracellular versus extracellular antibiotic resistance genes in the environment: prevalence, horizontal transfer, and mitigation strategies. Bioresour. Technol. 319:124181. doi: 10.1016/j.biortech.2020.124181
Zhang, H., Zhang, Q., Chen, S., Zhang, Z., Song, J., Long, Z., et al. (2020). Enterobacteriaceae predominate in the endophytic microbiome and contribute to the resistome of strawberry. Sci. Tot. Environ. 727:138708. doi: 10.1016/j.scitotenv.2020.138708
Zhu, H., Zhang, L., Li, S., Wang, Y., Sun, S., Chen,, et al. (2018). The rhizosphere and root exudates of maize seedlings drive plasmid mobilization in soil. Appl. Soil Ecol. 124, 194–202. doi: 10.1016/j.apsoil.2017.10.039
Keywords: horizontal gene transfer, conjugation, rhizosphere, endosphere, agri-food system, ready-to-eat vegetables, microbial community
Citation: Riva F, Dechesne A, Eckert EM, Riva V, Borin S, Mapelli F, Smets BF and Crotti E (2024) Conjugal plasmid transfer in the plant rhizosphere in the One Health context. Front. Microbiol. 15:1457854. doi: 10.3389/fmicb.2024.1457854
Edited by:
Anna Maria Puglia, University of Palermo, ItalyReviewed by:
Aabid Hussain, Cleveland Clinic, United StatesRachele Isticato, University of Naples Federico II, Italy
Copyright © 2024 Riva, Dechesne, Eckert, Riva, Borin, Mapelli, Smets and Crotti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elena Crotti, ZWxlbmEuY3JvdHRpQHVuaW1pLml0
†Present address: Arnaud Dechesne, Department of Biotechnology and Biomedicine, Technical University of Denmark, Kongens Lyngby, Denmark
 Francesco Riva
Francesco Riva Arnaud Dechesne
Arnaud Dechesne Ester M. Eckert
Ester M. Eckert Valentina Riva
Valentina Riva Sara Borin
Sara Borin Francesca Mapelli
Francesca Mapelli Barth F. Smets2,5
Barth F. Smets2,5 Elena Crotti
Elena Crotti