- 1Engineering Research Center of Health Medicine Biotechnology of Institution of Higher Education of Guizhou Province, School of Biology and Engineering (School of Modern Industry for Health and Medicine), Guizhou Medical University, Guiyang, China
- 2Guiyang Maternal and Child Health Hospital, Guiyang, China
- 3Shanghai Institute of Phage, Shanghai Public Health Clinical Center, Fudan University, Shanghai, China
Phages and phage-encoded lytic enzymes are promising antimicrobial agents. In this study, we report the isolation and identification of bacteriophage KP2025 from Klebsiella pneumoniae. Bioinformatics analysis of KP2025 revealed a putative endolysin, LysKP213, containing a T4-like_lys domain. Purified LysKP213 was found to be highly thermostable, retaining approximately 44.4% of its lytic activity after 20 h of incubation at 95°C, and approximately 57.5% residual activity after 30 min at 121°C. Furthermore, when administered in combination with polymyxin B or fused at the N-terminus with the antimicrobial peptide cecropin A (CecA), LysKP213 exhibited increased antibacterial activity against Gram-negative pathogens, including K. pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, and Escherichia coli, both in vitro and in vivo. These results indicated that LysKP213 is a highly thermostable endolysin that, when combined with or fused with an outer membrane permeabilizer, has enhanced antibacterial activity and is a candidate agent for the control of infections by Gram-negative pathogens.
Introduction
The emergence of multidrug-resistant (MDR) microorganisms has led to a serious health challenge worldwide due to the limitations of currently available antibiotics (Hu et al., 2021; Bharadwaj et al., 2022; Kocsis, 2023; Zhu et al., 2023). The World Health Organization (WHO) has published a list of “priority pathogens,” which includes Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Staphylococcus aureus, and Enterobacter spp. (Tacconelli et al., 2018). Polymyxins (polymyxin B and colistin [polymyxin E]) are the main class of antibiotics currently used against MDR Gram-negative bacteria in the clinic (Roberts et al., 2015; Nang et al., 2021). However, the utility of polymyxins as “last-line antibiotics” is limited due to high rates of toxicity (Nang et al., 2021). Consequently, alternative antibacterial agents for the treatment of infections by MDR microorganisms are urgently required.
Endolysins are lytic enzymes produced by bacteriophages during the last step of their lifecycle to digest the peptidoglycan layer of the bacterial cell wall (Gondil et al., 2020; Abdelrahman et al., 2021). Endolysin has a low probability of developing resistance. Therefore, endolysins are considered promising alternatives to counter drug-resistant infections (Danis-Wlodarczyk et al., 2021; Khan et al., 2021; Liu B. et al., 2023; Liu et al., 2023a). In addition, in recent years, increasingly animal models have shown efficacy of endolysins against bacterial infections in vivo (Fursov et al., 2020; Schmelcher and Loessner, 2021). However, most endolysins are limited in their ability to degrade the peptidoglycan layer owing to the presence of an outer membrane (OM) in Gram-negative bacteria (Lai et al., 2020; Shah et al., 2023; Wang et al., 2024). Thus, numerous studies have focused on combining endolysins with an OM permeabilizer, such as EDTA (a chelating agent), organic acids, and colistin (Thummeepak et al., 2016; Blasco et al., 2020; Hong et al., 2022; Sitthisak et al., 2023). Despite its usefulness as an OM permeabilizer, EDTA inhibits blood clotting at low concentrations; thus, the potential applications of an endolysin-EDTA combination are limited to the topical treatment of localized bacterial infections. Combinations of different endolysins with colistin have been shown to have marked antibacterial activity against A. baumannii infection, resulting in lower doses of colistin and, consequently, reduced toxicity (Thummeepak et al., 2016; Blasco et al., 2020; Hong et al., 2022; Sitthisak et al., 2023). Additionally, several studies have recently highlighted the substantial antimicrobial activity of the combination of phage endolysins with antimicrobial peptides (AMPs) capable of permeabilizing the OM of Gram-negative bacteria (Abdelkader et al., 2022; Hong et al., 2022; Lim et al., 2022; Zhang et al., 2022; Xuan et al., 2023).
In this study, we report the isolation and identification of an endolysin, LysKP213, from the K. pneumoniae bacteriophage KP2025. LysKP213 displayed antimicrobial activity against a relatively broad spectrum of Gram-negative bacteria after EDTA treatment. In addition, when combined with polymyxin B or fused with the AMP cecropin A (CecA), LysKP213 exhibited enhanced antibacterial activity against the tested strains both in vitro and in vivo relative to that seen with polymyxin B or CecA alone. These results highlight the potential of endolysin LysKP213 as a candidate antibacterial agent for the control of infections with Gram-negative pathogens.
Materials and methods
Bacterial strains and growth conditions
The tested bacteria were obtained from clinical samples (Supplementary Tables S1, S4). The bacteria were mixed with Luria-Bertani (LB) broth, cultured at 37°C with shaking and stored at −80°C in 50% glycerol (Wei et al., 2023).
Bacteriophage isolation
Hospital sewage was centrifuged at 8,000 rpm for 10 min, filtered through a 0.22-μm filter, and stored at 4°C for later use. Bacteriophage isolation was performed as previously described (Hua et al., 2017; Luo et al., 2018), with minor modifications. K. pneumoniae strain 12092025 served as the host. Briefly, the filtrate was mixed with an equal volume of 3× LB broth and, to enrich K. pneumoniae phages, 1 mL of K. pneumoniae (OD600 = 0.5) was added to the mixture, followed by incubation for 8 h at 37°C with shaking. The mixture was collected by centrifugation after 8 h and passed through a 0.22-μm filter. A double-layer agar technique was used to detect and purify phages. 50 μL of filtrate and 200 μL of K. pneumoniae strain 12092025 were mixed with Semi-solid LB agar medium and mixtures were poured onto LB agar plates, incubating the plates overnight at 37°C. A single plaque was picked and resuspended in 500 μL of K. pneumoniae strain 12092025 and incubated on 200 rpm at 37°C for 2 h. Then, bacterium-phage mixture was mixed with 10 mL of the Semi-solid LB agar and poured onto LB agar plates and incubated overnight at 37°C. This process was repeated at least three times to obtain pure phage, which was named KP2025.
Electron microscopy
A carbon copper membrane was coated with 20 μL of a pure phage sample for transmission electron microscopy (TEM). The membranes were negatively stained with 2% (w/v) phosphotungstic acid for 20 min and were then allowed to dry at room temperature. The morphology of the phage was then observed by TEM (JEM-1200EX, Japan) at an accelerating voltage of 100 kV (Mendes et al., 2014).
Host range determination
The host range was determined using different bacterial strains kept in our laboratory as described previously (Chang et al., 2005), with the following modifications: 50 μL of a phage suspension and 200 μL of the cells from an overnight culture of bacterial were mixed with 10 mL of the Semi-solid LB agar and poured on the surface of LB agar plates, incubating the plates overnight and observe the plaques to confirm host range. 10 mL Semi-solid LB agar was mixed with 100 μL of the cells, which was then overlaid on the surface of the solidified LB agar. 10 μL (ca. 1.0 × 109 PFU/mL) of a phage suspension was spotted onto the plate, which was then incubated overnight, and host range was established by bacterial lysis at the spot where the phage was deposited. Each experiment was repeated three times.
Multiplicity of infection and one-step growth curve
The number of cells of the host bacterium K. pneumoniae strain 12092025 was adjusted to 1 × 108 colony-forming units (CFU)/mL in a turbidimeter. According to the MOIs, the ratios between KP2025 and the host bacteria were 0.001, 0.01, 0.1, 1, 10 and 100. The phage-bacteria mixture was incubated at 37°C for 4 h and centrifuged at 8,000 rpm for 3 min. To calculate the phage titer, the supernatant was serially diluted. The MOI resulting in the release of the highest number of progeny phages was considered the ideal MOI (Wei et al., 2023).
The one-step growth curve was determined as described previously (Yuan et al., 2019; Cong et al., 2021). Briefly, K. pneumoniae strain 12092025 was cultured to the mid-exponential phase, collected by centrifugation at 8,000 rpm for 3 min, and resuspended in LB broth to a density of 1 × 108 CFU/mL. Phage KP2025 was added at the ideal MOI of 0.01, followed by incubation at 37°C, with shaking. Measurements were taken for a total of 3.5 h. Samples were taken every 10 min for the first 40 min, then every 20 min until 120 min, and subsequently every 30 min. Phage titers were determined using the double-layer agar plate method.
Bacteriophage DNA extraction and genome sequencing
Phenol and chloroform-isoamyl alcohol (24:1) was used to extract phage genomic DNA. A total of 200 μL of phage was 100 μL phenol, 100 μL chloroform-isoamyl alcohol mixed, vortexed for 1 min, and centrifuged at 12,000 rpm for 5 min. The above steps were repeated once. Whole-genome sequencing was performed on the Illumina NovaSeq platform.
Whole-genome bioinformatic analysis
Raw data quality was assessed using FastQC (v.0.11.5, default settings) (Bolger et al., 2014). The sequences were assembled and spliced with SPAdes (v.3.15.3, default settings) (Bankevich et al., 2012). Open reading frames encoded in the phage genome were predicted by RAST1 (Aziz et al., 2008). Similarities between phage KP2025 and other phage genomes were compared using BLASTn (O'Leary et al., 2016). BLASTp2 (Johnson et al., 2008) was used to determine protein function and similarity. The circular map of the genome was plotted using Proksee.3 The gene encoding tRNA was predicted by tRNAscan-SE v.2.04 (Lowe and Eddy, 1997). The online prediction platforms Virulence Finder5 and ResFinder6 (Kleinheinz et al., 2014) were employed to predict whether the genome of phage KP2025 encodes virulence and drug resistance genes. The conserved domains of LysKP213 were analyzed using the NCBI conserved domain database. Multiple sequence alignments were performed by the Clustal Omega7 and rendered with Espript.8 Three-dimensional protein models were visualized by PyMOL and WinCoot. Each online prediction tool was then run with default settings (RAST, BLASTn, BLASTp, Proksee, tRNAscan-SE, Virulence Finder, ResFinder, NCBI conserved domain database, Clustal Omega, and Espript).
The production and purification of recombinant LysKP213 and CecA-LysKP213
The genomic DNA of phage KP2025 was used as a template for the PCR amplification of the LysKP213-encoding gene. PCR was performed using the gene-specific primer pair 5′-CGGGATCCATGG CTAGAGTAGTAGAT-3′ (forward) and 5′-CCCAAGCTTGTTGTA AGCATCAAGT-3′ (reverse). BamH I and Hind III restriction sites, present, respectively, at the 5′ ends of the forward and reverse primers (underlined), were used for cloning the amplified sequences into the pET-28a vector. The PCR conditions were as follows: pre-denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 45 s, and extension at 72°C for 90 s, with a final extension step at 72°C for 10 min, 30 cycles in total. The PCR products were detected using 1% agarose gel electrophoresis and the LysKP213-encoding gene size was found to contain 501 bp. Escherichia coli BL21 (DE3) carrying pET-28a-LysKP213 was used for the expression of recombinant LysKP213 (Wang et al., 2022). CecA-LysKP213 was constructed by fusing CecA (NCBI PRF 0708214A) to the 5′ end of LysKP213 with the (GGGGS)3 linker (Hong et al., 2022). Specifically, cells expressing the recombinant protein were cultured in LB broth containing 50 μg/mL kanamycin at 37°C with shaking. At an OD600 of 0.5, isopropyl-β-D-thiogalactopyranoside (IPTG) was added at the final concentration of 0.5 mM, and the bacteria were incubated at 28°C (LysKP213) or 22°C (CecA-LysKP213) for 8 h with shaking. Following centrifugation at 12,000 rpm for 10 min, the bacterial pellet was resuspended in 10 mL of 5 mM imidazole containing NaH2PO4 and NaCl supplemented with 10 μL of protease inhibitor. Ultrasonication was performed with an ultrasonic crusher (135 W, 70% power, 3 s on and 3 s off, for a total of 30 min). Recombinant LysKP213 and CecA-LysKP213 were Ni-affinity purified using a His Trap HP column. The proteins were collected by eluting with a gradient of 5–250 mM imidazole and dialyzed against 20 mM Tris–HCl and 10% glycerol (pH 7.4). Recombinant LysKP213 and CecA-LysKP213 were validated using 12% SDS–PAGE.
Analysis of the antibacterial activity of LysKP213 and CecA-LysKP213
As described previously, the antibacterial activity of LysKP213 was performed by Lu et al. (2021). Briefly, the bacteria were cultured at 37°C with shaking to an OD600 of 0.6–0.8 and centrifuged at 12,000 rpm for 1 min. The bacterial pellet was washed once with 20 mM Tris–HCl and treated with 0.5 M EDTA for 5 min, washed again with Tris–HCl, centrifuged at 12,000 rpm for 1 min, and finally resuspended in Tris–HCl. After the addition of LysKP213, the bacteria were incubated at 37°C. The standard turbidity measurement method was used and the OD600 was recorded using a plate reader. Additionally, serially diluted bacteria were spotted on LB agar, and the number of CFUs was calculated. The bacteria inhibitory activity of LysKP213 was calculated as [OD600 (bacteria only) − OD600 sample (endolysin added)]/[OD600 (bacteria only)] (Plotka et al., 2015).
The antibacterial activity of CecA-LysKP213 was determined using K. pneumoniae strain 12092025, P. aeruginosa strain PAO1, A. baumannii strain 12091082, and E. coli strain B5 as described previously (Lim et al., 2022). Briefly, after culturing to the exponential phase (OD600 = 0.6–0.8), the bacteria were collected by centrifugation at 12,000 rpm for 1 min and adjusted to 1 × 108 CFU/mL in 20 mM Tris–HCl after washing. The bacteria were then mixed with CecA-LysKP213 (21 μg/mL), incubated at 37°C for 2 h, serially diluted, and spotted on LB agar to calculate the number of bacteria.
Analysis of the effects of pH, NaCl concentration, human serum, and temperature on LysKP213 activity
To study the effect of temperature on endolysin activity, the endolysin was preincubated at various temperatures (4–100°C) for 30 min. K. pneumoniae strain 12092025 was washed once with 20 mM Tris–HCl (pH 7.4) and treated with 0.5 M EDTA for 5 min, washed again with Tris–HCl, centrifuged at 12,000 rpm for 1 min, and finally resuspended in Tris–HCl. All groups were then added to the cell preparations for 30 min at 37°C. After incubation, the OD600 value was determined (Yu et al., 2019).
To investigate the effect of pH on the activity of LysKP213, 0.5 M EDTA-treated K. pneumoniae strain 12092025 was resuspended in buffer solutions of different pH (20 mM citrate buffer pH 4–5, 20 mM imidazole-HCl buffer pH 6, and 20 mM tris–HCl buffer pH 7–12) and incubated with the endolysin for 30 min at 37°C. After incubation, the OD600 value was determined (Yu et al., 2019).
The activity of LysKP213 at NaCl concentrations from 0 to 500 mM was measured at optimal pH conditions (Kim et al., 2020). Some modifications were made to the endolysin antibacterial activity method in different human serum described by Chen et al. (2021).
To test the endolysin antibacterial activity in human serum, K. pneumoniae strain 12092025 was washed once with 20 mM Tris–HCl (pH 7.4) and treated with 0.5 M EDTA for 5 min, washed again with Tris–HCl, centrifuged at 12,000 rpm for 1 min, and finally resuspended in Tris–HCl. Then, cells were treated with 21 μg/mL LysKP213 in the presence of 1–10% human serum (Sigma-Aldrich, Shanghai, China) at 37°C for 30 min. Endolysin treatment in the absence of human serum served as the control. After incubation, the OD600 value was determined. The antibacterial activity of LysKP213 was calculated as [OD600 (bacteria only) − OD600 sample (endolysin added)]/[OD600 (bacteria only)]. Each experiment was repeated three times (Chen et al., 2021).
Assessment of thermal stability
Some modifications were made to the endolysin thermal stability method described by Plotka et al. (2014). Briefly, aliquots of endolysin LysKP213and hen egg white lysozyme (HEWL; 21 μg/mL) were heated at 95°C for increasing durations in a Super Gradient Thermal Cycler (LongGene) or autoclaved at 121°C for 30 min. Then, 0.5 M EDTA-treated K. pneumoniae strain 12092025 incubated with the endolysin for 30 min at 37°C. After incubation, the OD600 value was determined.
Determination of the minimum inhibitory concentration (MIC)
The CLSI-recommended broth microdilution method was used to determine the MIC (the lowest concentration at which an antibiotic inhibits bacterial growth) of the antibiotic polymyxin B in Mueller Hinton broth (MHB). The obtained MIC value was used for the subsequent combination experiment.
The combination of LysKP213 with Polymyxin B
Some modifications were made to the endolysin LysKP213 combined with polymyxin B method described by Sitthisak et al. (2023). Klebsiella pneumoniae strain 12092025, A. baumannii strain 12091082, P. aeruginosa strain PAO1, and E. coli strain B5 were used as bacterial substrates. The bacteria were cultured in LB broth and the turbidity of the bacterial suspension was measured using a densitometer. The turbidity was corrected to an equivalent 0.5 McFarland standard or approximately 1 × 108 CFU/mL, and the cell suspension was diluted 50 times with 2× MHB. Aliquots of 100 μL were transferred to 96-well plates, and 50 μL of LysKP213 at different concentrations (0–85 μg/mL) and 50 μL of different concentrations of polymyxin B were added to the wells. The bacteria were then cultured in an incubator at 37°C protected from light for 16 h, following which the OD600 was determined using a spectrophotometer. Each experiment was repeated three times.
Galleria mellonella larval infection model
The Galleria mellonella larval model study was conducted following the procedures by Kim et al. with some minor modifications (Kim et al., 2018) and referring to other Galleria mellonella larval studies (Chen et al., 2021). Briefly, Galleria mellonella was used as the experimental animal. G. mellonella larvae with a length of 20–25 mm and weighing 250–350 mg are stored in a dark environment at 4°C for 24 h before infection. The larvae were injected through the last right proleg with 10 μL of a bacterial solution containing 1 × 107 CFU/mL of K. pneumoniae strain 12092025 or 1 × 108 CFU/mL of A. baumannii strain 12091082, P. aeruginosa strain PAO1, or E. coli strain B5. These doses were previously determined to yield 80% mortality after 96 h. The infected larvae were divided into the following six groups (10 larvae per group): (i) a control group, which received phosphate-buffered saline (PBS); (ii) a polymyxin B treatment group, which received either 14 μg/mL polymyxin B (for K. pneumoniae strain 12092025-infected larvae), 12 μg/mL polymyxin B (for P. aeruginosa strain PAO1-infected larvae), 10 μg/mL polymyxin B (for A. baumannii strain 12091082-infected larvae), or 8 μg/mL polymyxin B (for E. coli strain B5-infected larvae); (iii) a LysKP213 treatment group, which received 21 μg/mL LysKP213; (iv) a combination treatment group, which received polymyxin B plus LysKP213 treatment; (v) a CecA (GL Biochem (Shanghai) Ltd., China) treatment group, which received 3 μg/mL CecA; and (vi) a CecA-LysKP213 treatment group, which received 21 μg/mL CecA-LysKP213. All treatments were administered via injection (10 μL) into the last left proleg 30 min following infection. After treatment, the larvae of each group were placed in Petri dishes and cultured in darkness at 37°C for 96 h. Mortality was assessed every 12 h. All the experiments were independently performed three times.
Statistical analysis
GraphPad Prism v.8.0 was used for data analysis. Differences between datasets were evaluated using two-tailed Student’s t-tests; for survival experiments, a log-rank (Mantel-Cox) test was employed. Data are presented as means ± SD, with p-values <0.05 being considered significant.
Results
Biological characteristics and host range of phage KP2025
The isolated phage KP2025 was initially screened against K. pneumoniae strain 12092025 through a spot test. KP2025 produced clear plaques of similar morphology (Figure 1A). Transmission electron micrograph analysis showed that phage KP2025 has an icosahedral head and slender tail, characteristic of Myoviridae (Figure 1B).
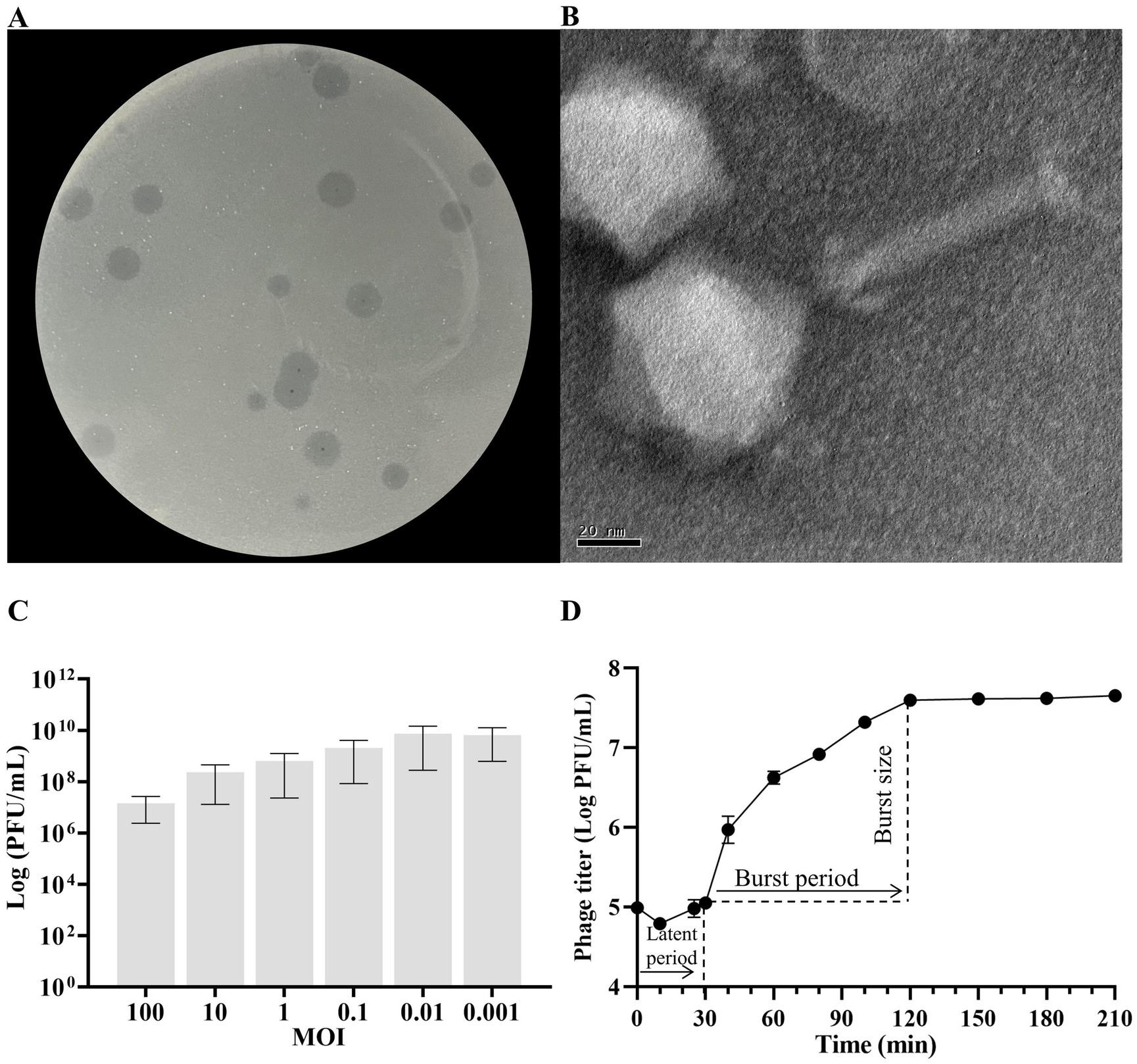
Figure 1. Biological characterization of Klebsiella pneumoniae phage KP2025. (A) Clear plaques of phage KP2025 on double-layer agar. (B) Transmission electron micrograph of phage KP2025. Scale bar = 20 nm. (C) Titers of phage KP2025 produced at different multiplicities of infection (MOIs). Values represent means ± SD (n = 3). (D) One-step growth curve for phage KP2025. Values represent means ± SD (n = 3).
The MOI that yielded the highest phage titer among the assessed MOIs was 0.01 (Figure 1C). A one-step growth curve reflects the important parameters of phage growth, including the latent period, burst size, and release period. As illustrated in Figure 1D, the latent and rising periods of KP2025 lasted for 30 and 90 min, respectively, and the average burst size (the number of phage particles released by each infected host cell) was 100 ± 10 PFU/mL. KP2025 exhibited good lytic properties against the clinical K. pneumoniae and E. coli strains, as evidenced by the results of the host range assay (Supplementary Table S1).
Identification of LysKP213 from the genome of phage KP2025
The genome of phage KP2025 contains 149,523 bp and has a GC content of 40.4% (GenBank accession no. PP919962). The genome contains 275 predicted open reading frames (ORFs) and 19 tRNAs (Supplementary Table S2). BLASTp analysis revealed that ORF213 (length: 501 bp) encodes a putative endolysin, which we named LysKP213 (Figure 2A). This endolysin contains a conserved T4-like_lys domain (NCBI domain architecture ID: 10091399; conserved domain accession: cd00735) and belongs to the Lyz-like superfamily (Figure 2B).
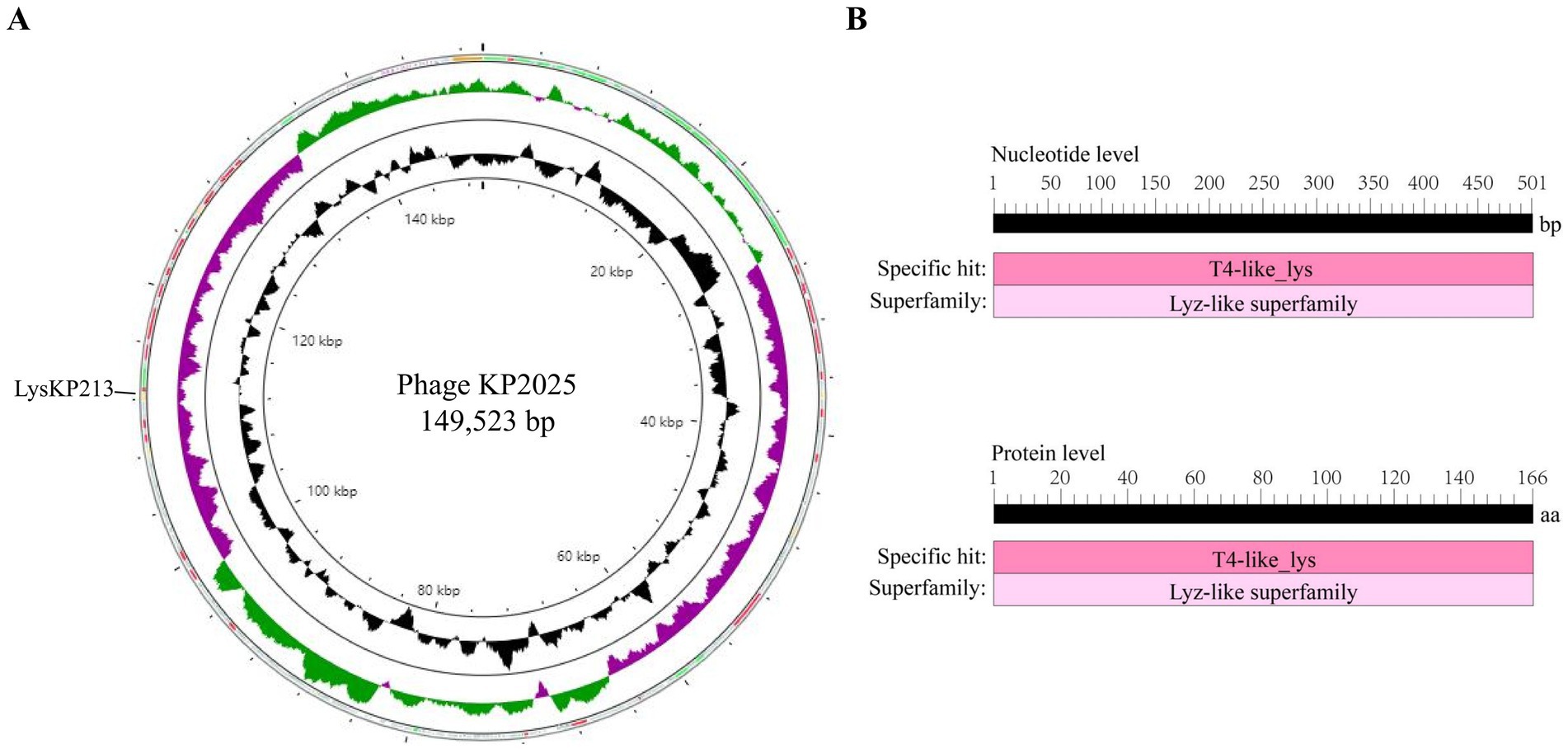
Figure 2. Whole-genome sequencing of phage KP2025 and identification of the endolysin LysKP213. (A) The genomic map of phage KP2025. The inner to outer circles were as: genome size (with an interval of 4 kb); GC content; GC skew [(G − C)/(G + C)]; and open reading frames (ORFs). (B) Conserved domain analysis of LysKP213 at the nucleic acid and protein levels.
In addition, we performed a multiple sequence alignment between LysKP213 and four representative members of the Lyz-like superfamily, including the peptidoglycan hydrolases from the A. baumannii 10 (PHAb10), the peptidoglycan hydrolases from the A. baumannii 8 (PHAb8), the E. coli O157:H7 phage FAHEc1 endolysin (LysF1), and the well-studied T4 lysozyme (T4L) (Weaver and Matthews, 1987; Love et al., 2021; Zhang et al., 2024). The result showed that they possessed the conserved Glu-Asp-Thr catalytically active sites and are mainly composed of α-helices and β-folds, characteristic of the Lyz-like superfamily (Figure 3A; Supplementary Table S3). Further three-dimensional structure analysis indicated that LysKP213, PHAb10, PHAb8, and LysF1 superimposed well with T4L, especially in the conserved Glu-Asp-Thr catalytically active sites (Figure 3B). Thus, these results indicated that LysKP213, PHAb10, PHAb8, and LysF1 were highly similar to T4L and might share similar catalytic mechanisms of action.
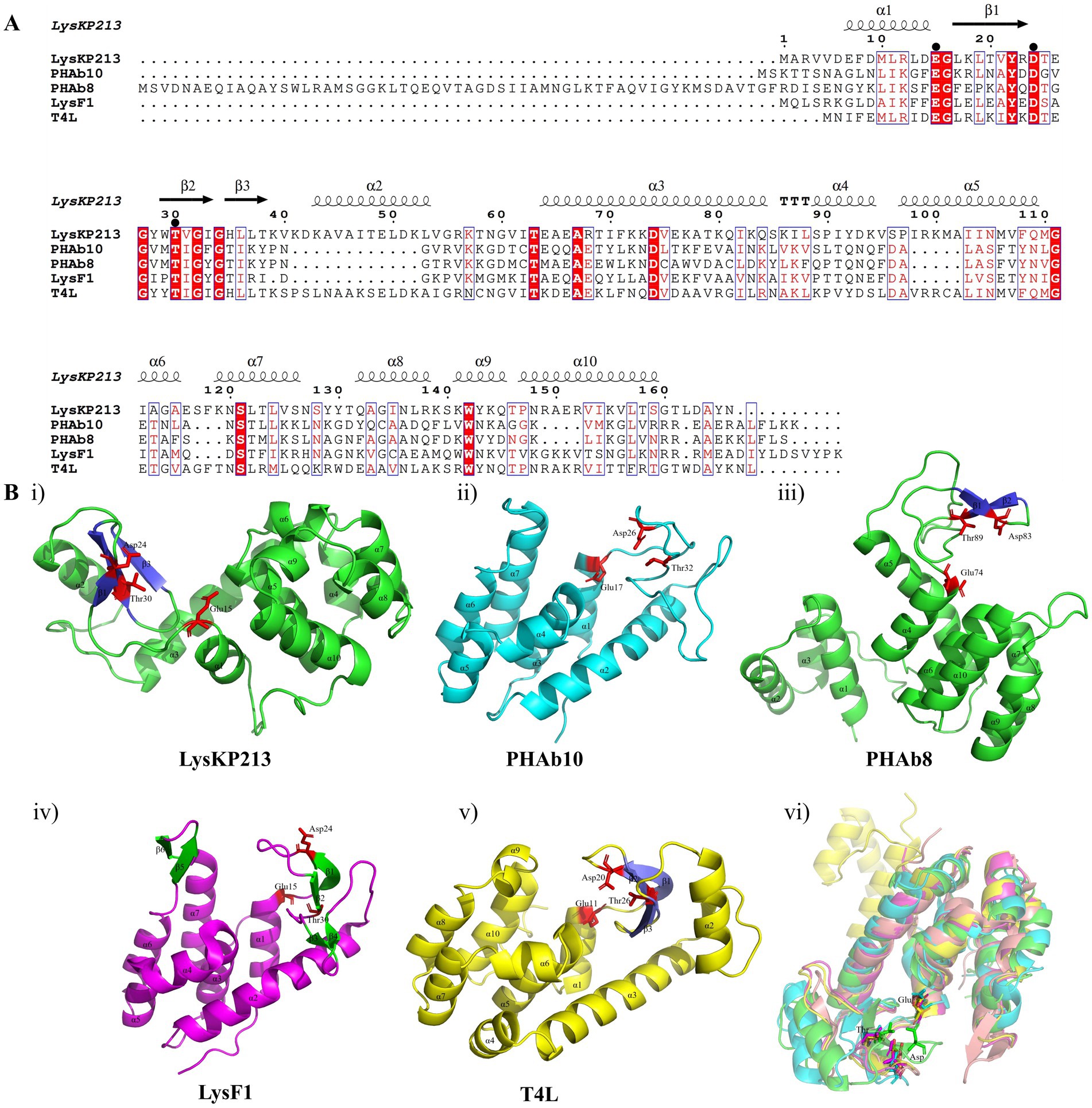
Figure 3. Multiple sequence alignment and structure analysis of the endolysin LysKP213. (A) Multiple sequence comparison between LysKP213 and four representative members of the Lyz-like superfamily; black circles indicate key catalytic sites of the enzyme. (B) Three-dimensional structure of LysKP213 (i), PHAb10 (ii), PHAb8 (iii), LysF1 (iv), and T4L (v). (vi) Structure superposition of LysKP213 and four classical T4L-like lysozymes. LysKP213 is shown in cyan, PHAb10 in magenta, PHAb8 in yellow, T4L in green, and LysF1 in pink. Residues in the conservative catalytic triad are shown as sticks.
Molecular cloning, expression, purification, and activity validation of recombinant LysKP213
The LysKP213-encoding gene segment was amplified from the genome of phage KP2025 by PCR (Figure 4A). The IPTG-induced expression of recombinant endolysin LysKP213 was carried out in genetically engineered E. coli BL21 (DE3). Subsequently, a HisTrap affinity column was used to obtain purified LysKP213. The band size was further confirmed by 12% SDS–PAGE (Figure 4B).
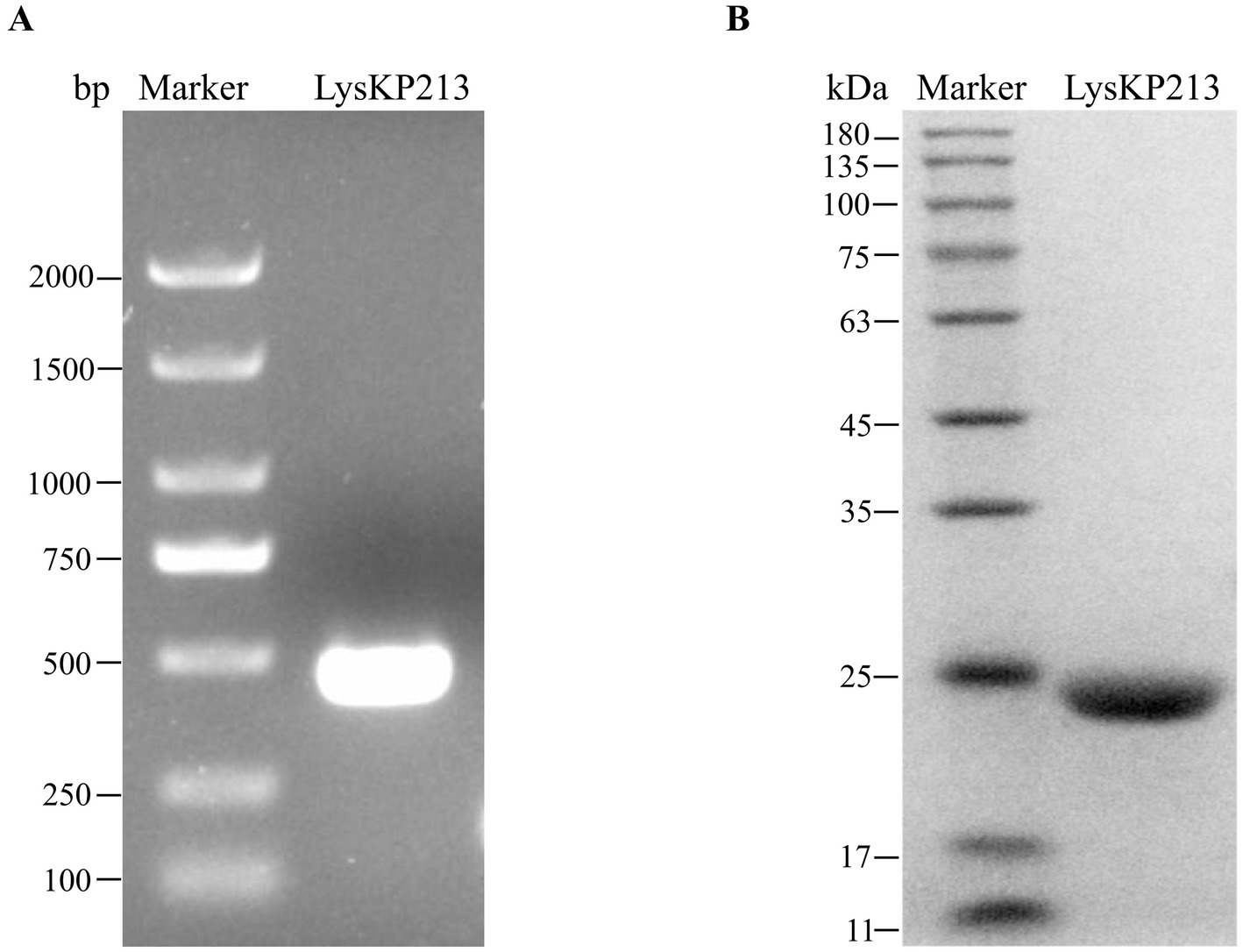
Figure 4. Gene cloning, expression, and purification of the endolysin LysKP213. (A) The genome of bacteriophage KP2025 was used as a template for the PCR amplification of the LysKP213-encoding gene. (B) IPTG (final concentration 0.5 mM) was used to induce the overexpression of recombinant LysKP213; the final purified endolysin LysKP213 was analyzed using 12% SDS–PAGE.
Next, the antimicrobial activity of the purified LysKP213 was assessed using the turbidity reduction assay. K. pneumoniae strain 12092025 served as the host strain and phage KP2025 treated with 0.5 M EDTA served as the test strain. The results showed that the antimicrobial activity of LysKP213 increased in a concentration-dependent manner; however, this activity decreased at concentrations over 21 μg/mL. In addition, the highest antimicrobial activity of LysKP213 was observed at the concentration of 21 μg/mL after 30 min of incubation (Figure 5).
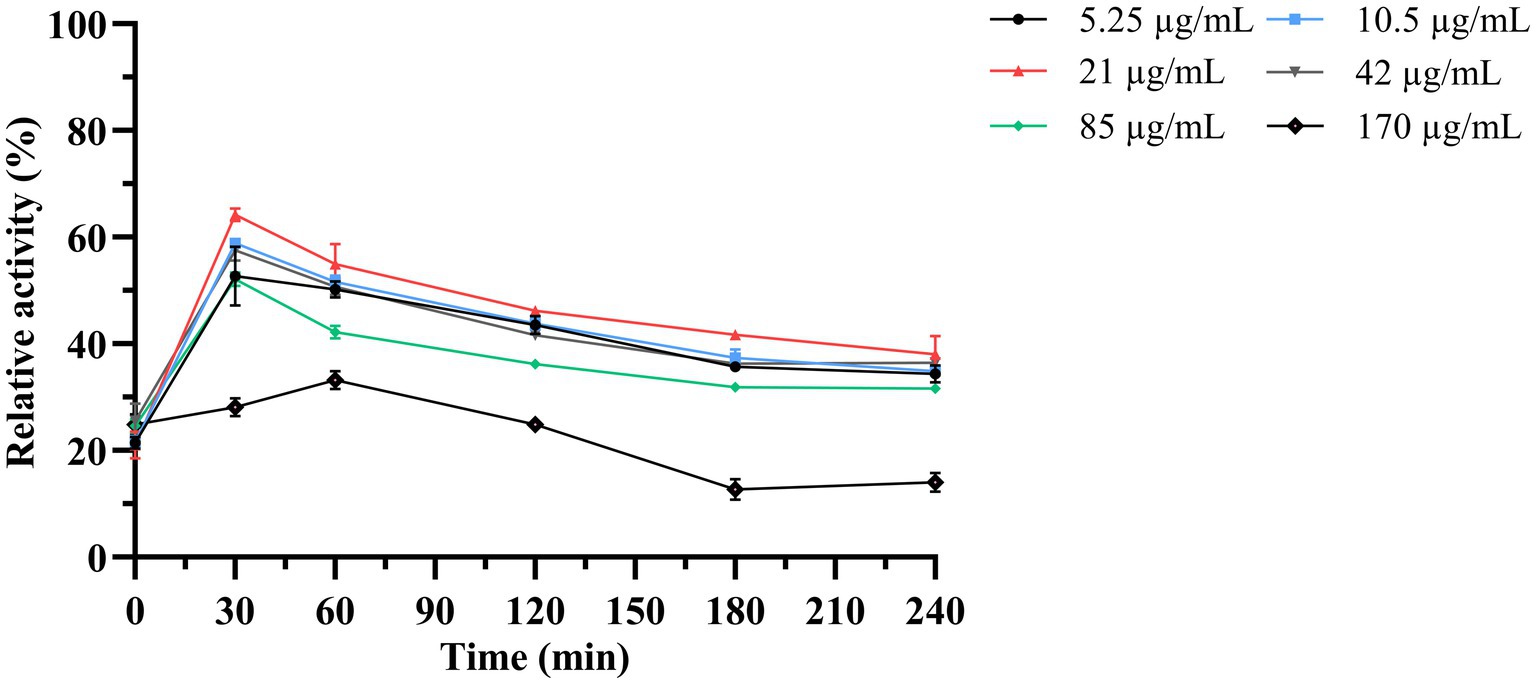
Figure 5. The antimicrobial activity of different concentrations of endolysin LysKP213 against K. pneumoniae strain 12092025. Each test were repeated three times. The values are the means and standard deviations from triplicate assays.
The influence of temperature, pH, human serum, and NaCl on the lytic activity of LysKP213
Subsequently, we used the host strain of bacteriophage KP2025, K. pneumoniae strain 12092025, as the effector substrate to measure the effects of temperature, pH, human serum, and salinity (NaCl concentrations) on LysKP213 activity. We found that the thermostability of LysKP213 was very broad, ranging from 4 to 100°C (Figure 6A). The pH values tested ranged from 4 to 12 and LysKP213 had relatively high lytic activity in the 7–11 pH range (Figure 6B). The concentration of human serum significantly affected the activity of LysKP213; nevertheless, relatively high lytic activity was observed at concentrations between 1 and 3% (Figure 6C). The most suitable pH identified in the pH test was used to assess the effect of salinity (NaCl concentration) on the activity of LysKP213. We noted that LysKP213 retained 60% relative activity against K. pneumoniae strain 12092025 at NaCl concentrations of up to 500 mM (Figure 6D).
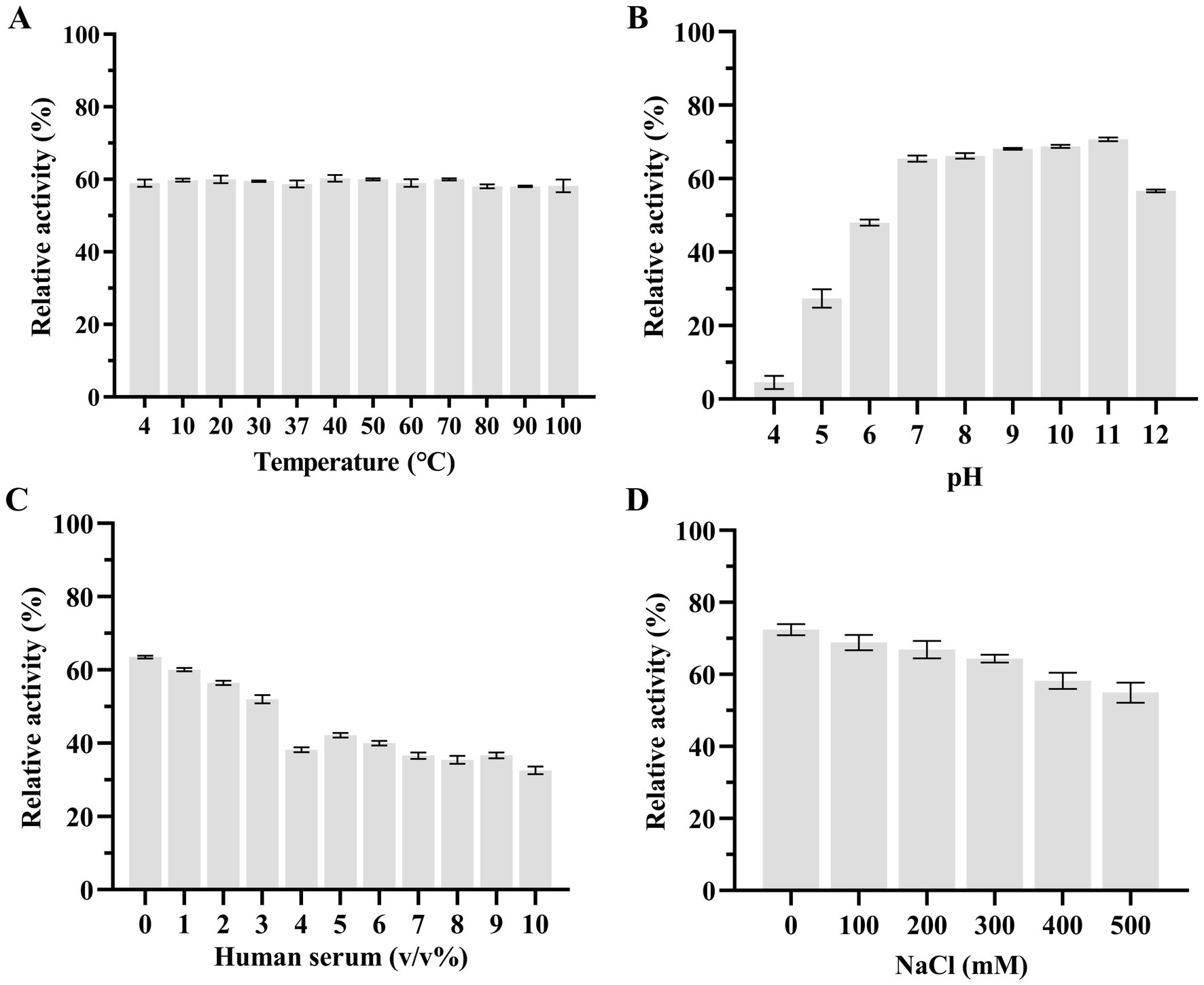
Figure 6. The effects of temperature (A), pH (B), human serum (C), and NaCl (D) on the lytic activity of the endolysin LysKP213. Klebsiella pneumoniae strain 12092025, the host strain for bacteriophage KP2025, was used as an effector substrate. In addition, the maximum activity of LysKP213 was set as 100% under various test conditions to facilitate comparisons. Each experiment was performed in triplicate. The error bars represent the SD.
We further estimated the thermostability of endolysin LysKP213. For this, LysKP213 was incubated for increasing durations (up to 24 h) at 95°C or autoclaved at 121°C for 30 min, followed by an activity assay (Figure 7). The results indicated that LysKP213 is highly thermostable. After incubation at 95°C for 20 h, LysKP213 retained 44.4% of its lytic activity. Furthermore, LysKP213 retained 15.8% residual activity after 24 h at 95°C, and 57.5% residual activity after 30 min at 121°C. Meanwhile, HEWL, used as a control, was completely inactivated after 6 h at 95°C.
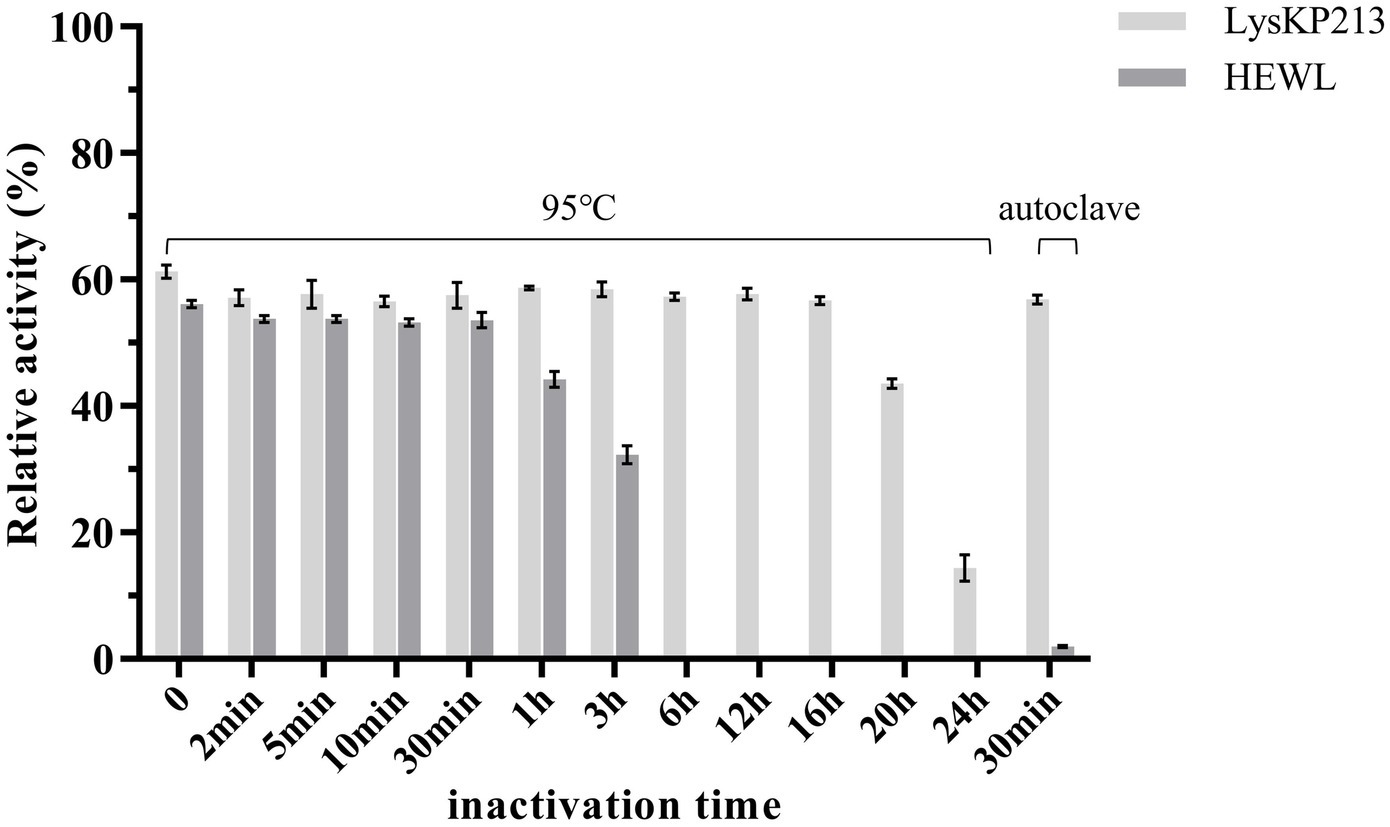
Figure 7. Examination of the thermal stability of the endolysin LysKP213. LysKP213 (21 μg/mL) and hen egg white lysozyme (HEWL) (21 μg/mL) were incubated at 95°C for 0–24 h or autoclaved for 30 min at 121°C, following which their activity against Klebsiella pneumoniae strain 12092025 cells was measured. The activities of LysKP213 and HEWL are indicated as percentages relative to untreated samples (0 min). Each experiment was performed in triplicate. The error bars indicate the standard deviation.
Analysis of the lytic activity spectrum of LysKP213
Like other phage-encoded endolysins, LysKP213 could not directly lyse Gram-negative bacteria owing to the protection afforded by the OM. Thus, K. pneumoniae, A. baumannii, P. aeruginosa, and E. coli were treated with EDTA to remove the OM before the addition of LysKP213. We observed that LysKP213 could lyse most of the Gram-negative bacterial strains tested as determined by the measurement of turbidity (Figure 8; Supplementary Table S4). Moreover, LysKP213 exhibited substantial lytic activity against K. pneumoniae strain 12092025, A. baumannii strain 19606, P. aeruginosa strain PAO1, and E. coli strain B5; however, LysKP213 had no lytic activity against the Gram-positive bacterium Staphylococcus aureus (Figure 8; Supplementary Table S4).
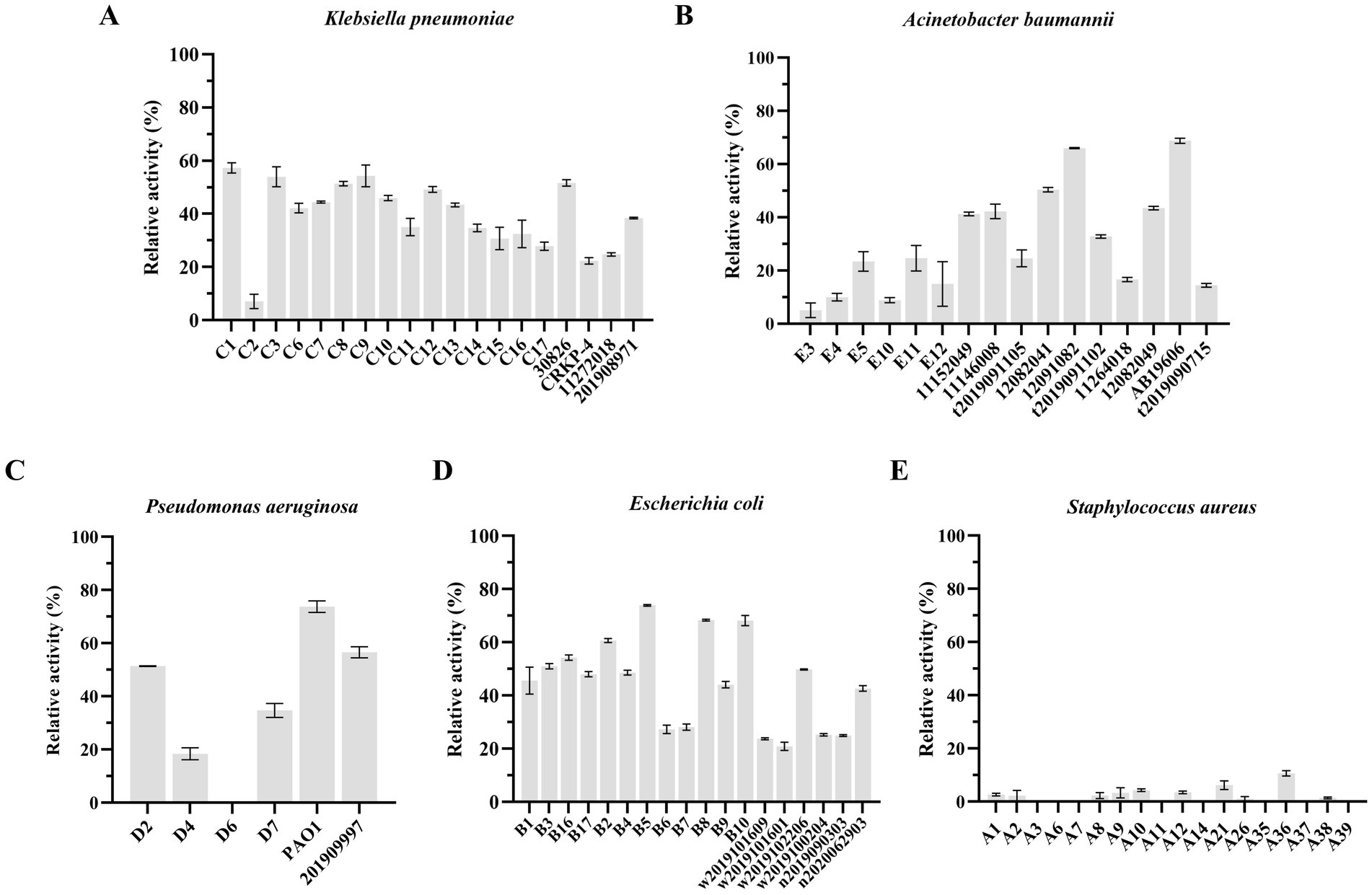
Figure 8. The lytic activity spectrum of endolysin LysKP213 against Klebsiella pneumoniae (A), Acinetobacter baumannii (B), Pseudomonas aeruginosa (C), Escherichia coli (D), and Staphylococcus aureus (E) as determined using the standard turbidity measurement method. Each experiment was performed in triplicate.
We further investigated the lytic activity of LysKP213 against K. pneumoniae strain 12092025, P. aeruginosa strain PAO1, A. baumannii strain 12091082, E. coli strain B5, and S. aureus strain A36 using CFU reduction analysis. Compared to the control group, the Gram-negative bacterial cells showed a significant reduction in the number of CFUs after treatment with LysKP213 (Figure 9). These findings were consistent with those observed using the turbidity measurement method described above and indicated that, when the OM is removed, LysKP213 has relatively broad lytic activity against most Gram-negative bacteria, including K. pneumoniae, A. baumannii, P. aeruginosa, and E. coli.
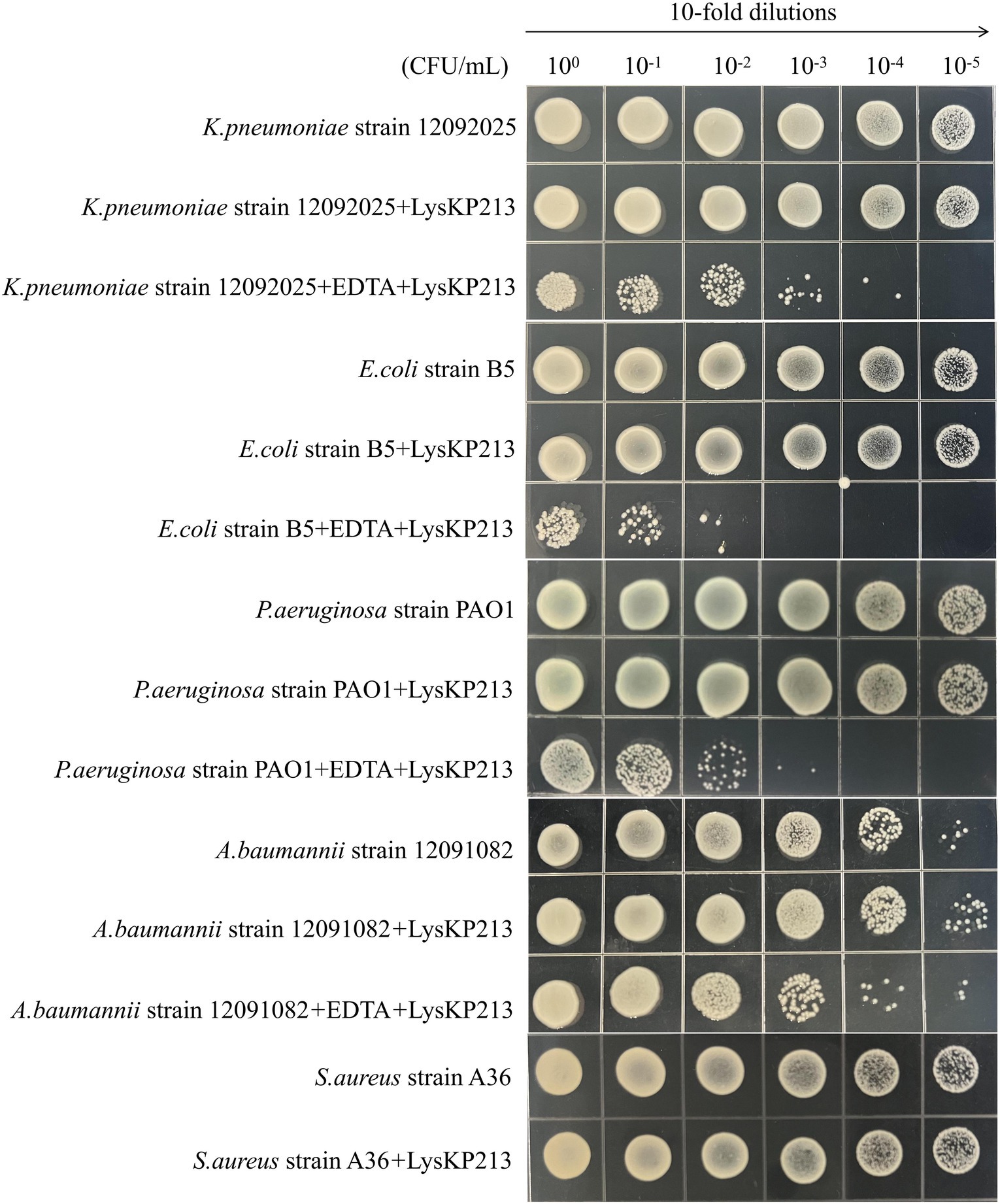
Figure 9. Representative images depicting bacterial cell viability before and after LysKP213 treatment. To measure viability, 10-fold dilutions of log-phase bacterial cells (Klebsiella pneumoniae strain 12092025, Pseudomonas aeruginosa strain PAO1, Acinetobacter baumannii strain 12091082, and Escherichia coli strain B5) that were untreated, treated with EDTA, or treated with EDTA plus LysKP213 were spotted onto agar plates. Additionally, 10-fold dilutions of log-phase Staphylococcus aureus strain A36 cells, untreated or treated with LysKP213, were also spotted on agar plates. LysKP213 in combination with EDTA reduced the numbers of viable K. pneumoniae, P. aeruginosa, A. baumannii, and E. coli cells by 1–3 log units; however, LysKP213 treatment did not affect S. aureus strain A36. The numbers above the colony represent the dilution ratio.
The synergistic effects of endolysin LysKP213 in combination with polymyxin B in vitro
The antibiotic colistin disrupts the OM of Gram-negative bacteria. Recently, it was shown that colistin has synergistic effects against Gram-negative bacteria when administered in combination with phage lysins (Thummeepak et al., 2016). Accordingly, we next evaluated the putative synergistic effect of the endolysin LysKP213 in combination with polymyxin B in vitro using the microdilution checkboard assay. First, we measured the MICs of polymyxin B alone against K. pneumoniae strain 12092025, E. coli strain B5, P. aeruginosa strain PAO1, and A. baumannii strain 12091082 (Supplementary Table S5). Next, we measured the effects of LysKP213 in combination with polymyxin B and found that this combination inhibited the viability of the four bacterial strains at the concentrations of 0.5 and 21 μg/mL, 0.5 and 10 μg/mL, 0.25 and 5 μg/mL, and 0.25 and 85 μg/mL, respectively (Figure 10). The addition of LysKP213 yielded a two-fold reduction in the MICs of polymyxin B against K. pneumoniae strain 12092025, E. coli strain B5, and P. aeruginosa strain PAO1, and an eight-fold reduction in the polymyxin B MIC against A. baumannii strain 12091082 (Figure 10).
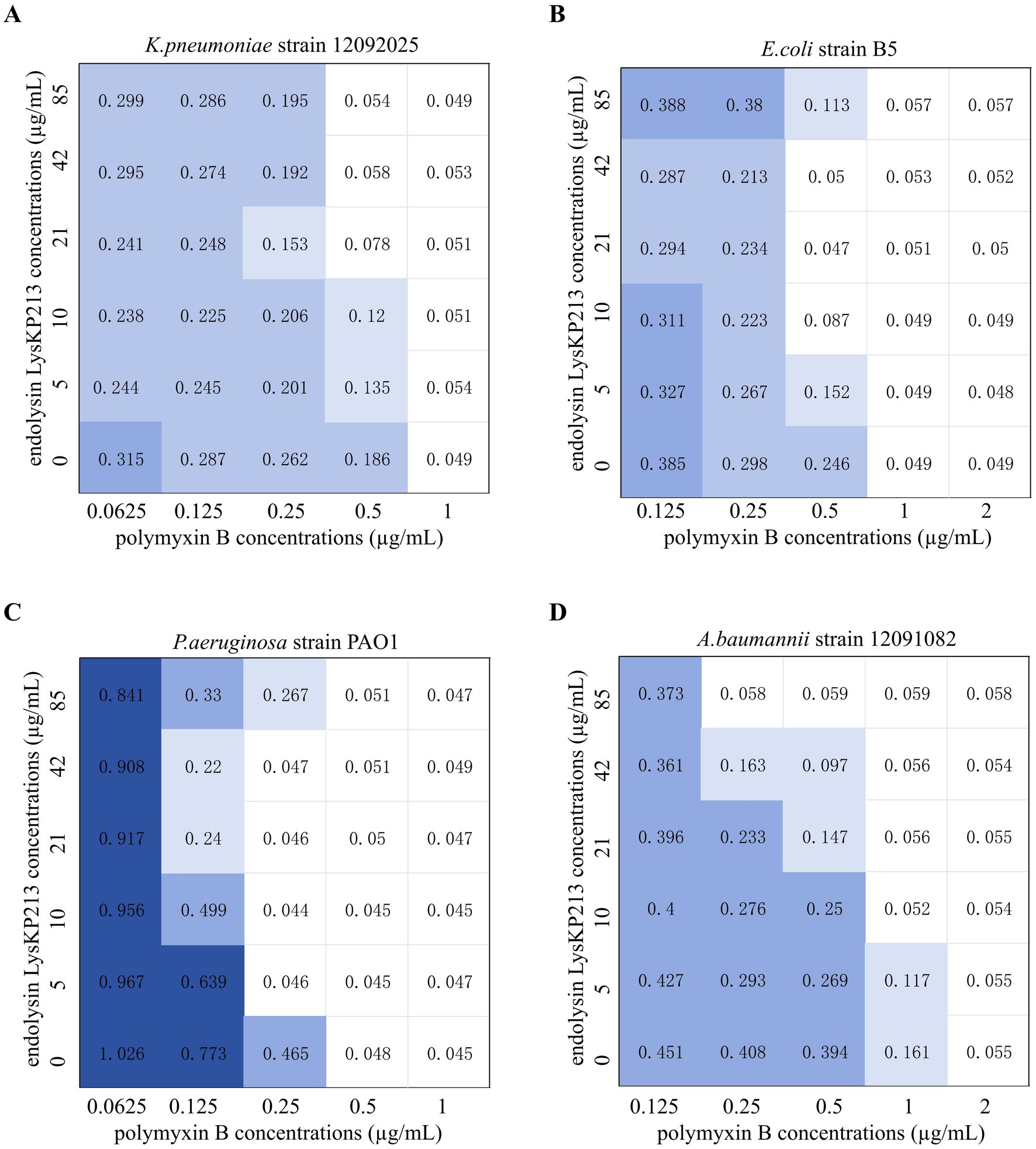
Figure 10. The microdilution checkboard assay for the synergistic effects of LysKP213 in combination with polymyxin B on Klebsiella pneumoniae strain 12092025 (A), Escherichia coli strain B5 (B), Pseudomonas aeruginosa strain PAO1 (C), and Acinetobacter baumannii strain 12091082 (D). The results are representative of at least three independent experiments.
The antibacterial activity of endolysin LysKP213 in combination with polymyxin B in vivo
Next, we evaluated the antibacterial activity of the endolysin LysKP213 in combination with polymyxin B in vivo using a G. mellonella larvae infection model with K. pneumoniae strain 12092025, P. aeruginosa strain PAO1, A. baumannii strain 12091082, and E. coli strain B5. The survival rate of G. mellonella larvae infected with any of the four tested strains was significantly higher after combination treatment than after treatment with polymyxin B or LysKP213 alone (**p < 0.01 and ***p < 0.001, respectively) (Figure 11).
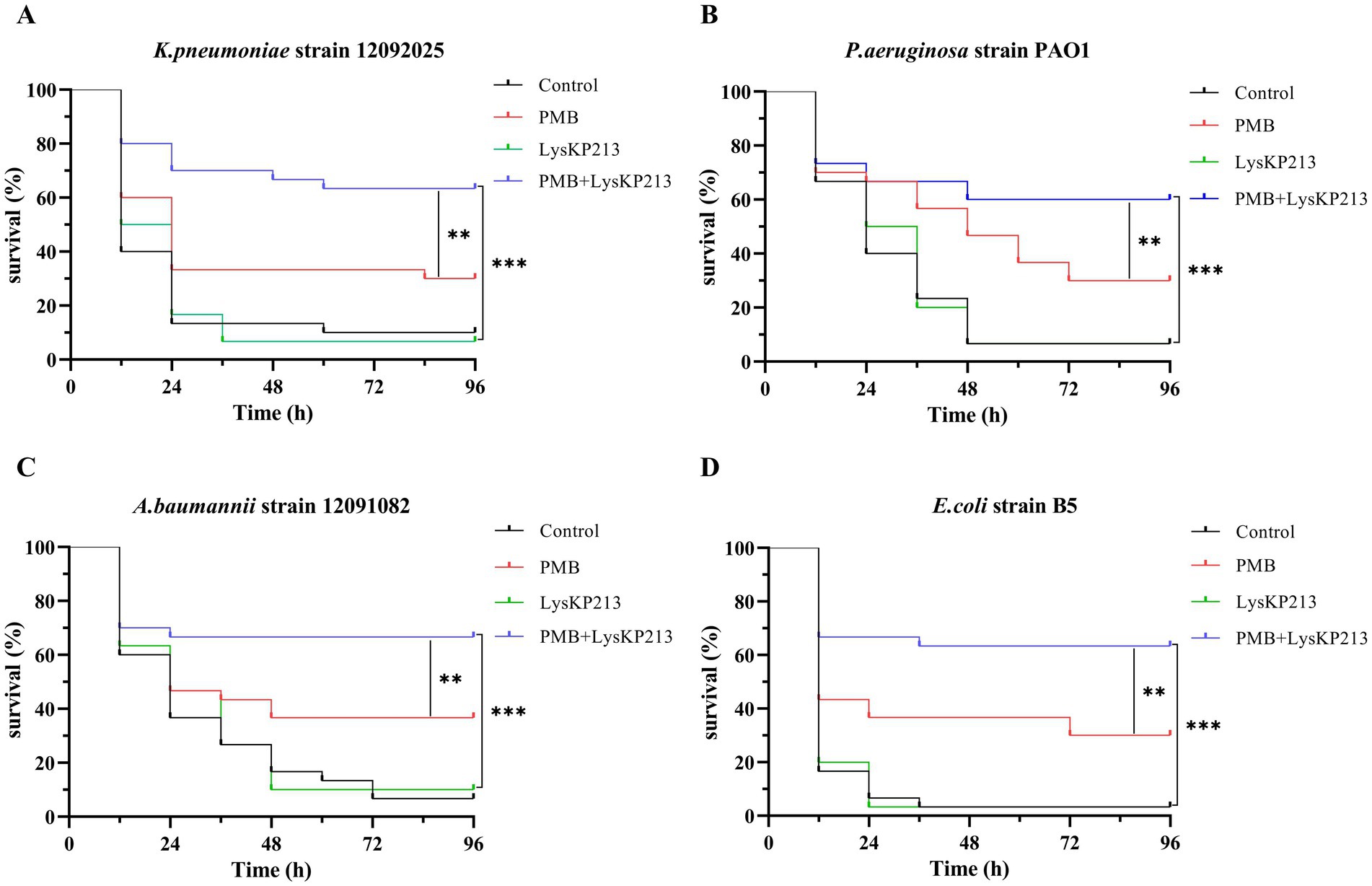
Figure 11. The antibacterial activity of endolysin LysKP213 in combination with polymyxin B in vivo. Survival curves for Galleria mellonella larvae infected with Klebsiella pneumoniae strain 12092025 (A), Pseudomonas aeruginosa strain PAO1 (B), Acinetobacter baumannii strain 12091082 (C), and Escherichia coli strain B5 (D). G. mellonella larvae were separately infected with each of the four bacterial strains and then treated with PBS (control), polymyxin B (14 μg/mL), LysKP213 (21 μg/mL), or polymyxin B (14 μg/mL) in combination with LysKP213 (21 μg/mL). **p < 0.01, ***p < 0.001.
The synergistic effects of the engineered fusion protein CecA-LysKP213 in vitro
Phage endolysin has difficulty reaching the peptidoglycan layer of Gram-negative bacteria owing to the presence of an OM. Here, we tested whether the inclusion of the membrane-destabilizing peptide, CecA, would increase membrane permeability and thus enhance the antibacterial activity of LysKP213. For this, we fused CecA to the N-terminus of LysKP213 (CecA-LysKP213) (Supplementary Figure S1) and found that CecA-LysKP213 reduced the numbers of K. pneumoniae strain12092025, P. aeruginosa strain PAO1, A. baumannii strain 12091082, and E. coli strain B5 cells by 1–2 log units (Figure 12). This demonstrated the antibacterial activity of LysKP213 against Gram-negative bacteria in vitro was improved when it was fused to CecA.
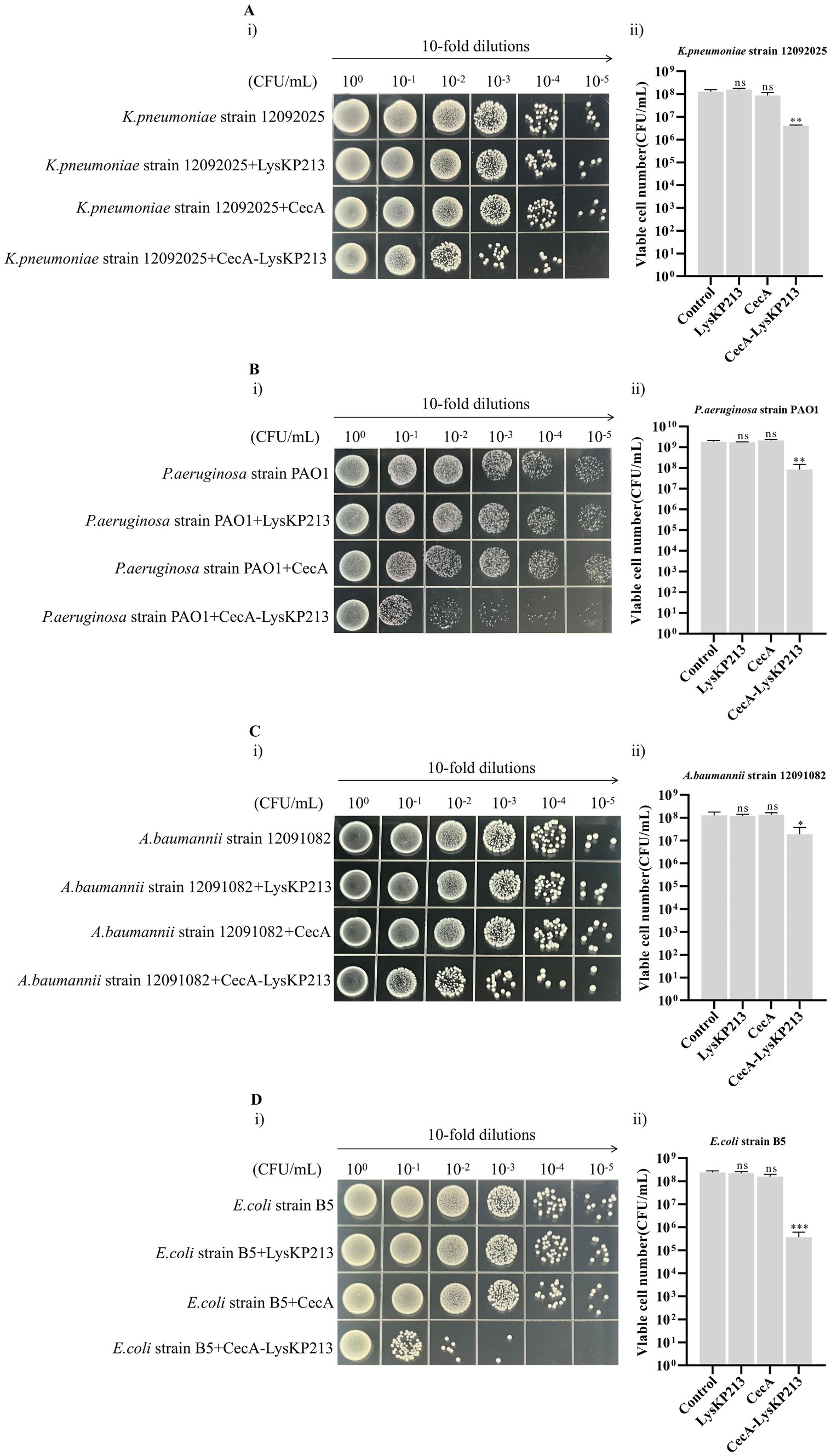
Figure 12. The antibacterial activity of CecA-LysKP213 against Gram-negative bacteria in vitro. The antibacterial activity of CecA-LysKP213 was tested against Klebsiella pneumoniae strain 12092025 (A), Pseudomonas aeruginosa strain PAO1 (B), Acinetobacter baumannii strain 12091082 (C), and Escherichia coli strain B5 (D) using a colony forming unit (CFU) reduction assay. The bacteria were treated with PBS (control), CecA-LysKP213 (21 μg/mL), CecA (3 μg/mL), or LysKP213 (21 μg/mL). The experiments were repeated three times and the data are presented as means ± SD. *p < 0.05, **p < 0.01, ***p < 0.001; n.s., not significant.
The antibacterial activity of the engineered fusion protein CecA-LysKP213 in vivo
We further evaluated the antibacterial activity of CecA-LysKP213 in vivo using a G. mellonella larvae infection model with K. pneumoniae strain 12092025, P. aeruginosa strain PAO1, A. baumannii strain 12091082, and E. coli strain B5 (Figure 13). Only 10% of G. mellonella larvae infected with K. pneumoniae survived without CecA-LysKP213 treatment, whereas approximately 40% of the infected larvae survived after CecA-LysKP213 treatment (Figure 13A). Only 2% of G. mellonella larvae infected with P. aeruginosa strain PAO1 survived without CecA-LysKP213 treatment; however, approximately 30% of the infected larvae survived after CecA-LysKP213 treatment (Figure 13B). Only 10% of infected G. mellonella larvae infected with A. baumannii strain 12091082 survived without CecA-LysKP213 treatment; in contrast, approximately 16% of the infected larvae survived following CecA-LysKP213 treatment (Figure 13C). Finally, only 1% of G. mellonella larvae infected with E. coli strain B5 survived without CecA-LysKP213 treatment, while approximately 30% of the infected worms survived when after CecA-LysKP213 treatment (Figure 13D). Together, these results revealed that the in vivo antibacterial activity of endolysin LysKP213 against Gram-negative bacteria was improved with the addition of CecA.
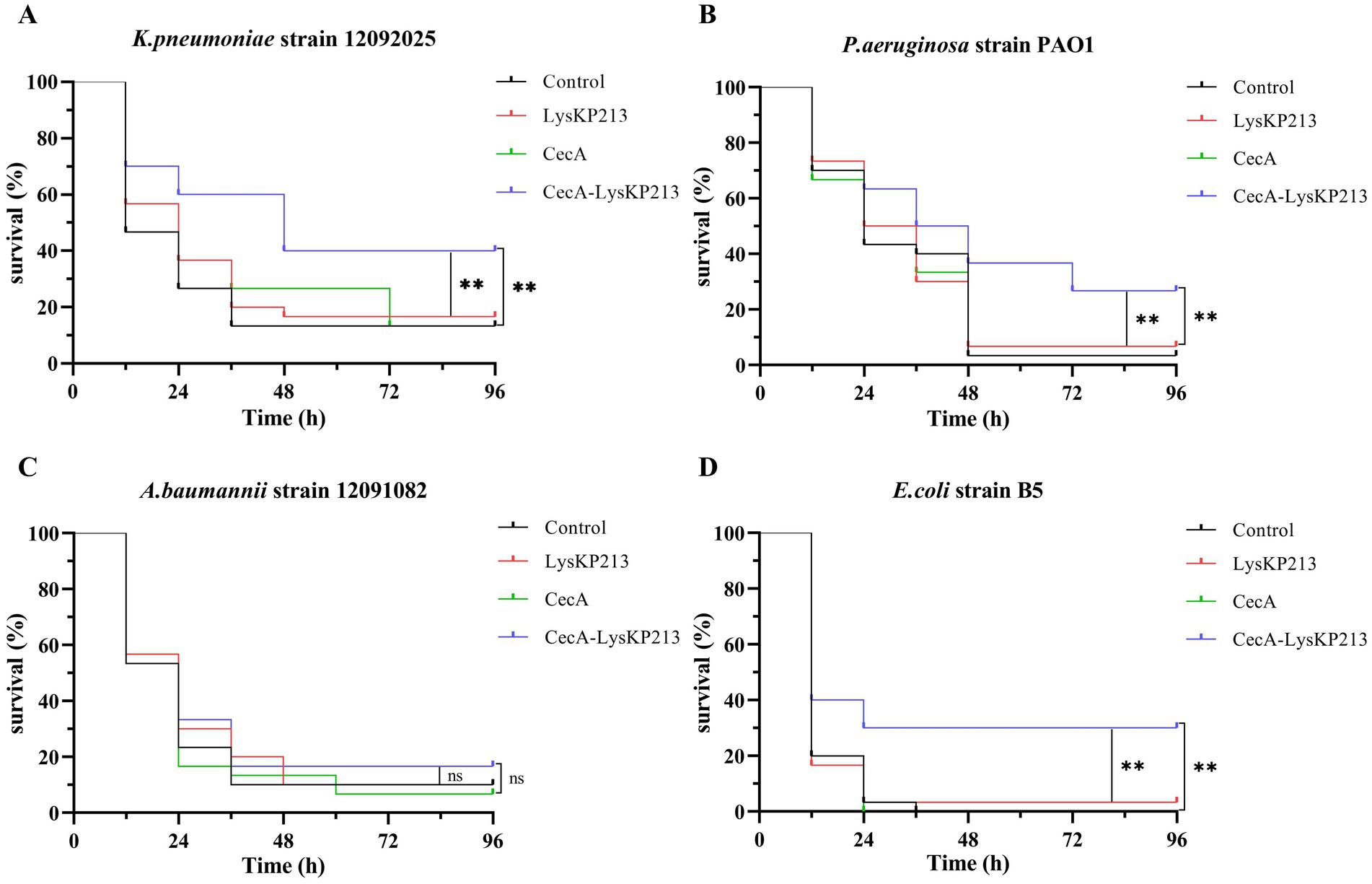
Figure 13. In vivo antibacterial activity of the engineered fusion protein CecA-LysKP213. Survival curves for Galleria mellonella larvae infected with Klebsiella pneumoniae strain 12092025 (A), Pseudomonas aeruginosa strain PAO1 (B), Acinetobacter baumannii strain 12091082 (C), and Escherichia coli strain B5 (D). G. mellonella larvae were separately infected with each of the four bacterial strains and then treated with PBS (control), CecA-LysKP213 (21 μg/mL), CecA (3 μg/mL), or LysKP213 (21 μg/mL). Larval survival was monitored after 96 h of incubation at 37°C (n = 10 per group). The experiment was repeated three times. **p < 0.01, n.s., not significant.
Discussion
Bacteriophages and bacteriophage-encoded endolysins display marked antibacterial activity. Moreover, bacteriophage-encoded endolysins specifically degrade the peptidoglycans in the bacterial cell wall and have low resistance potential. Nevertheless, most bacteriophage-encoded endolysins cannot easily reach the cell wall of Gram-negative bacteria owing to the presence of an OM. Consequently, different strategies have been employed aiming to solve this problem, including disrupting the OM with OM permeabilizers, such as EDTA (a chelating agent), organic acids, and colistin, or engineering endolysins such as Artilysin and CecA::ST01 (Gerstmans et al., 2016; Lim et al., 2022; Carratala et al., 2023; Zheng and Zhang, 2024). Thus, although bacteriophage-encoded endolysin therapy currently has some technology, regulatory, and logistical hurdles, the future looks promising with the emergence of engineered endolysins.
In this study, we isolated and characterized endolysin LysKP213 from K. pneumoniae bacteriophage KP2025. LysKP213 was found to be a lysozyme-like protein harboring a T4-like_lys domain. Recently, P9ly, which also contains a conserved T4-like_lys domain, was reported to exert antibacterial activity against different strains of bacteria, including MDR Gram-negative Shigella dysenteriae and Gram-positive S. aureus (Wang et al., 2022). Here, we evaluated the antibacterial activity spectrum of LysKP213 after EDTA treatment and observed that it possesses antibacterial activity against Gram-negative pathogens, including K. pneumoniae, P. aeruginosa, A. baumannii, and E. coli, but not against S. aureus, a Gram-positive bacterium.
We further found that LysKP213 retains approximately 44.4% of its lytic activity after 20 h of incubation at 95°C and approximately 57.5% residual activity after 30 min at 121°C. This indicates that LysKP213 is a highly thermostable phage-encoded endolysin. The lytic activity for most phage endolysin is within 4–40°C, which limits their application, including vaccine design, in food processing, and biotechnological production (Liu et al., 2023b). By contrast, endolysins have good (thermo)stability properties and can have a broader range of applications in human, plant/veterinary, and environmental health (Żebrowska et al., 2022; Choi and Kong, 2023; Liu et al., 2023b; Shin et al., 2023; Golosova et al., 2024). Thus, LysKP213, as a highly thermostable phage-encoded endolysin, could become useful to combat Gram-negative pathogens in the agricultural, food, and medical industries. In addition, an open question remains: what are the mechanisms responsible for the high thermal stability of LysKP213? PHAb10, a hyper-thermostable lysozyme, retains almost 100% bactericidal activity after treatment at 100°C for 1 h (Zhang et al., 2024). Here, we found that LysKP213 was similar to PHAb10 and might share similar catalytic mechanisms of action. Thus, we speculated that LysKP213 and PHAb10 might share a similar mechanism of action responsible for the high thermal stability. However, the thermal stability of phage endolysin seems to be a complex phenomenon that can be affected by many factors. Further studies are necessary to verify the mechanisms responsible for the high thermal stability of LysKP213.
Colistin (polymyxin E) and polymyxin B, the only polymyxins available commercially, have similar mechanisms of action, antimicrobial spectra, clinical uses, and toxicity. However, they also differ in several aspects, including chemical structure, formulation, potency, dosage, and pharmacokinetic properties (Kwa et al., 2007). These two medications disrupt the OM of Gram-negative bacteria via electrostatic interactions with lipopolysaccharides and phospholipids present in the OM (Kwa et al., 2007). Recently, several studies revealed that colistin in combination with phage endolysins improved antimicrobial activity against Gram-negative pathogens relative to either treatment alone (Thummeepak et al., 2016; Blasco et al., 2020; Sitthisak et al., 2023). Furthermore, one study indicated that the combined effect of polymyxin B in combination with phage endolysin, despite the absence of a synergistic effect on the inhibitory concentration, exerted a synergistic antibacterial effect against Gram-negative pathogens, including K. pneumoniae and Salmonella enterica serovar Typhimurium (Gontijo et al., 2024).
Accordingly, in this study, we further used the cationic antibiotic polymyxin B in combination with LysKP213 to overcome the barrier represented by the OM of Gram-negative bacteria. Similar to that reported for the cationic antibiotic colistin in combination with phage endolysin, the combination of LysKP213 with polymyxin B enhanced the antibacterial activity of the former against Gram-negative pathogens, including K. pneumoniae, P. aeruginosa, A. baumannii, and E. coli, both in vitro and in vivo.
AMPs with membrane-disrupting ability are found in all kingdoms of life and show promise as agents for the control of antimicrobial resistance (Zhang et al., 2021). CecA was among the first cecropins discovered in Hyalophora cecropia and exhibits marked antibacterial activity against Gram-negative pathogens (Moore et al., 1996; Brady et al., 2019). Several studies have reported that fusing CecA, an OM permeabilizing peptide, to the N-terminus of phage endolysins increases the antibacterial activity of the latter (Abdelkader et al., 2022; Hong et al., 2022; Lim et al., 2022). However, it was unknown whether the fusion of CecA to a phage endolysin harboring a T4-like_lys domain exerted similarly enhanced antibacterial activity. Here, we found that an engineered CecA-LysKP213 fusion protein improved the antibacterial activity of this endolysin against K. pneumoniae, P. aeruginosa, A. baumannii, and E. coli in vitro as well as in vivo.
Over the years, murine models have been regarded as the gold standard for studying microbial infections (Gunatilake, 2018; Hugenholtz and de Vos, 2018). However, because they have animal protection acts, ethical concerns and other drawbacks include the expensive feed costs and the special biosafety conditions. In recent years, G. mellonella larvae have been an alternative model to study microbial infections, including the use of G. mellonella larvae to evaluate the antimicrobial activity of phage and phage lysins in vivo (Ramarao et al., 2012; Tsai et al., 2016; Kim et al., 2018; Feng et al., 2023). The main advantage of the G. mellonella larval is that although it lacks an adaptive immune system, its innate immune system is remarkably similar to that of the murine innate immune system in function and structure. Secondly, G. mellonella larvae can survive at 37°C, equivalent to the temperature in murine. Thirdly, G. mellonella larvae have no ethical constraints, are cheaper to feed and breed, are easy and safe to manipulate, and have a short life cycle. Thus, the G. mellonella larvae in vivo models have been used to examine the antimicrobial activity of LysKP213 and CecA-LysKP213 and were compared with the turbidity reduction assay and CFU reduction analysis in vitro.
In summary, in this study, we isolated and identified endolysin LysKP213 from the K. pneumoniae bacteriophage KP2025. When combined with polymyxin B or fused with CecA, LysKP213 displayed enhanced antibacterial activity against K. pneumoniae, P. aeruginosa, A. baumannii, and E. coli compared to treatment with either polymyxin B alone or CecA alone, both in vitro and in vivo. These results highlight the potential of endolysin LysKP213 as a candidate antibacterial agent for the control of infections with Gram-negative pathogens.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PP919962.
Author contributions
DC: Investigation, Writing – original draft, Data curation, Formal analysis, Methodology, Project administration, Software, Visualization. JL: Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Writing – original draft. LuL: Formal analysis, Methodology, Software, Resources, Writing – review & editing. KX: Formal analysis, Resources, Software, Data curation, Writing – review & editing. LiL: Data curation, Formal analysis, Resources, Software, Writing – review & editing. LY: Formal analysis, Resources, Software, Methodology, Writing – review & editing. HL: Funding acquisition, Investigation, Writing – review & editing. TZ: Funding acquisition, Investigation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32200125) and the special funds from the central finance to support the development of local universities (Qian Jiao Ji No. [2023]036).
Acknowledgments
We thank Lu Liang and Kaide Xia for providing experimental strains.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1454618/full#supplementary-material
Supplementary Figure S1 | Purification of the fusion protein CecA-LysKP213. IPTG (final concentration 0.5 mM) was used to induce the overexpression of the fusion protein CecA-LysKP213; the final purified fusion protein CecA-LysKP213 was analyzed using 12% SDS-PAGE.
Footnotes
2. ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
4. ^ http://lowelab.ucsc.edu/cgi-bin/tRNAscan-SE2.cgi
5. ^ https://cge.cbs.dtu.dk/services/VirulenceFinder/
6. ^ https://cge.cbs.dtu.dk/services/ResFinder/
References
Abdelkader, K., Gutierrez, D., Tames-Caunedo, H., Ruas-Madiedo, P., Safaan, A., Khairalla, A. S., et al. (2022). Engineering a Lysin with intrinsic antibacterial activity (LysMK34) by cecropin A fusion enhances its antibacterial properties against Acinetobacter baumannii. Appl. Environ. Microbiol. 88:e0151521. doi: 10.1128/AEM.01515-21
Abdelrahman, F., Easwaran, M., Daramola, O. I., Ragab, S., Lynch, S., Oduselu, T. J., et al. (2021). Phage-encoded endolysins. Antibiotics (Basel) 10:124. doi: 10.3390/antibiotics10020124
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bharadwaj, A., Rastogi, A., Pandey, S., Gupta, S., and Sohal, J. S. (2022). Multidrug-resistant bacteria: their mechanism of action and prophylaxis. Biomed. Res. Int. 2022, 5419874–5419817. doi: 10.1155/2022/5419874
Blasco, L., Ambroa, A., Trastoy, R., Bleriot, I., Moscoso, M., Fernandez-Garcia, L., et al. (2020). In vitro and in vivo efficacy of combinations of colistin and different endolysins against clinical strains of multi-drug resistant pathogens. Sci. Rep. 10:7163. doi: 10.1038/s41598-020-64145-7
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brady, D., Grapputo, A., Romoli, O., and Sandrelli, F. (2019). Insect cecropins, antimicrobial peptides with potential therapeutic applications. Int. J. Mol. Sci. 20:5862. doi: 10.3390/ijms20235862
Carratala, J. V., Aris, A., Garcia-Fruitos, E., and Ferrer-Miralles, N. (2023). Design strategies for positively charged endolysins: insights into Artilysin development. Biotechnol. Adv. 69:108250. doi: 10.1016/j.biotechadv.2023.108250
Chang, H. C., Chen, C. R., Lin, J. W., Shen, G. H., Chang, K. M., Tseng, Y. H., et al. (2005). Isolation and characterization of novel giant Stenotrophomonas maltophilia phage phiSMA5. Appl. Environ. Microbiol. 71, 1387–1393. doi: 10.1128/AEM.71.3.1387-1393.2005
Chen, X., Liu, M., Zhang, P., Leung, S. S. Y., and Xia, J. (2021). Membrane-permeable antibacterial enzyme against multidrug-resistant Acinetobacter baumannii. ACS Infect. Dis. 7, 2192–2204. doi: 10.1021/acsinfecdis.1c00222
Choi, D., and Kong, M. (2023). LysGR1, a novel thermostable endolysin from Geobacillus stearothermophilus bacteriophage GR1. Front. Microbiol. 14:1178748. doi: 10.3389/fmicb.2023.1178748
Cong, C., Wei, B., Cui, H., Li, X., Yuan, Y., Wang, L., et al. (2021). Isolation, characterization and comparison of lytic Epseptimavirus phages targeting Salmonella. Food Res. Int. 147:110480. doi: 10.1016/j.foodres.2021.110480
Danis-Wlodarczyk, K. M., Wozniak, D. J., and Abedon, S. T. (2021). Treating bacterial infections with bacteriophage-based enzybiotics: in vitro, in vivo and clinical application. Antibiotics (Basel) 10, 10:1497. doi: 10.3390/antibiotics10121497
Feng, J., Li, F., Sun, L., Dong, L., Gao, L., Wang, H., et al. (2023). Characterization and genome analysis of phage vB_KpnS_SXFY507 against Klebsiella pneumoniae and efficacy assessment in galleria mellonella larvae. Front. Microbiol. 14:1081715. doi: 10.3389/fmicb.2023.1081715
Fursov, M. V., Abdrakhmanova, R. O., Antonova, N. P., Vasina, D. V., Kolchanova, A. D., Bashkina, O. A., et al. (2020). Antibiofilm activity of a broad-range recombinant endolysin LysECD7: in vitro and in vivo study. Viruses 12:545. doi: 10.3390/v12050545
Gerstmans, H., Rodriguez-Rubio, L., Lavigne, R., and Briers, Y. (2016). From endolysins to Artilysin(R)s: novel enzyme-based approaches to kill drug-resistant bacteria. Biochem. Soc. Trans. 44, 123–128. doi: 10.1042/BST20150192
Golosova, N. N., Khlusevich, Y. A., Morozova, V. V., Matveev, A. L., Kozlova, Y. N., Tikunov, A. Y., et al. (2024). Characterization of a thermostable endolysin of the Aeribacillus phage AeriP45 as a potential Staphylococcus biofilm-removing agent. Viruses 16:93. doi: 10.3390/v16010093
Gondil, V. S., Harjai, K., and Chhibber, S. (2020). Endolysins as emerging alternative therapeutic agents to counter drug-resistant infections. Int. J. Antimicrob. Agents 55:105844. doi: 10.1016/j.ijantimicag.2019.11.001
Gontijo, M., Pereira Teles, M., Martins Correia, H., Pérez Jorge, G., Rodrigues Santos Goes, I. C., Fasabi Flores, A. J., et al. (2024). Combined effect of SAR-endolysin LysKpV475 with polymyxin B and Salmonella bacteriophage phSE-5. Microbiology (Reading) 170:001462. doi: 10.1099/mic.0.001462
Gunatilake, M. (2018). History and development of laboratory animal science in Sri Lanka. Animal Model Exp. Med. 1, 3–6. doi: 10.1002/ame2.12003
Hong, H. W., Kim, Y. D., Jang, J., Kim, M. S., Song, M., and Myung, H. (2022). Combination effect of engineered endolysin EC340 with antibiotics. Front. Microbiol. 13:821936. doi: 10.3389/fmicb.2022.821936
Hu, Y., Anes, J., Devineau, S., and Fanning, S. (2021). Klebsiella pneumoniae: prevalence, reservoirs, antimicrobial resistance, pathogenicity, and infection: a hitherto unrecognized zoonotic bacterium. Foodborne Pathog. Dis. 18, 63–84. doi: 10.1089/fpd.2020.2847
Hua, Y., Luo, T., Yang, Y., Dong, D., Wang, R., Wang, Y., et al. (2017). Phage therapy as a promising new treatment for lung infection caused by Carbapenem-resistant Acinetobacter baumannii in mice. Front. Microbiol. 8:2659. doi: 10.3389/fmicb.2017.02659
Hugenholtz, F., and De Vos, W. M. (2018). Mouse models for human intestinal microbiota research: a critical evaluation. Cell. Mol. Life Sci. 75, 149–160. doi: 10.1007/s00018-017-2693-8
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Khan, F. M., Gondil, V. S., Li, C., Jiang, M., Li, J., Yu, J., et al. (2021). A novel Acinetobacter baumannii bacteriophage endolysin LysAB54 with high antibacterial activity against multiple gram-negative microbes. Front. Cell. Infect. Microbiol. 11:637313. doi: 10.3389/fcimb.2021.637313
Kim, H., Lee, H. G., Kwon, I., and Seo, J. (2020). Characterization of endolysin LyJH307 with antimicrobial activity against Streptococcus Bovis. Animals (Basel) 10:963. doi: 10.3390/ani10060963
Kim, N. H., Park, W. B., Cho, J. E., Choi, Y. J., Choi, S. J., Jun, S. Y., et al. (2018). Effects of phage endolysin SAL200 combined with antibiotics on Staphylococcus aureus infection. Antimicrob. Agents Chemother. 62, e00731–e00718. doi: 10.1128/AAC.00731-18
Kleinheinz, K. A., Joensen, K. G., and Larsen, M. V. (2014). Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4:e27943. doi: 10.4161/bact.27943
Kocsis, B. (2023). Hypervirulent Klebsiella pneumoniae: an update on epidemiology, detection and antibiotic resistance. Acta Microbiol. Immunol. Hung. 70, 278–287. doi: 10.1556/030.2023.02186
Kwa, A., Kasiakou, S. K., Tam, V. H., and Falagas, M. E. (2007). Polymyxin B: similarities to and differences from colistin (polymyxin E). Expert Rev. Anti-Infect. Ther. 5, 811–821. doi: 10.1586/14787210.5.5.811
Lai, W. C. B., Chen, X., Ho, M. K. Y., Xia, J., and Leung, S. S. Y. (2020). Bacteriophage-derived endolysins to target gram-negative bacteria. Int. J. Pharm. 589:119833. doi: 10.1016/j.ijpharm.2020.119833
Lim, J., Hong, J., Jung, Y., Ha, J., Kim, H., Myung, H., et al. (2022). Bactericidal effect of cecropin A fused endolysin on drug-resistant gram-negative pathogens. J. Microbiol. Biotechnol. 32, 816–823. doi: 10.4014/jmb.2205.05009
Liu, B., Guo, Q., Li, Z., Guo, X., and Liu, X. (2023). Bacteriophage endolysin: a powerful weapon to control bacterial biofilms. Protein J. 42, 463–476. doi: 10.1007/s10930-023-10139-z
Liu, H., Hu, Z., Li, M., Yang, Y., Lu, S., and Rao, X. (2023a). Therapeutic potential of bacteriophage endolysins for infections caused by gram-positive bacteria. J. Biomed. Sci. 30:29. doi: 10.1186/s12929-023-00919-1
Liu, H., Kheirvari, M., and Tumban, E. (2023b). Potential applications of thermophilic bacteriophages in one health. Int. J. Mol. Sci. 24:8222. doi: 10.3390/ijms24098222
Love, M. J., Coombes, D., Manners, S. H., Abeysekera, G. S., Billington, C., and Dobson, R. C. J. (2021). The molecular basis for Escherichia coli O157:H7 phage FAHEc1 endolysin function and protein engineering to increase thermal stability. Viruses 13:1101. doi: 10.3390/v13061101
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Lu, Y., Wang, Y., Wang, J., Zhao, Y., Zhong, Q., Li, G., et al. (2021). Phage endolysin LysP108 showed promising antibacterial potential against methicillin-resistant Staphylococcus aureus. Front. Cell. Infect. Microbiol. 11:668430. doi: 10.3389/fcimb.2021.668430
Luo, J., Jiang, M., Xiong, J., Li, J., Zhang, X., Wei, H., et al. (2018). Exploring a phage-based real-time PCR assay for diagnosing Acinetobacter baumannii bloodstream infections with high sensitivity. Anal. Chim. Acta 1044, 147–153. doi: 10.1016/j.aca.2018.09.038
Mendes, J. J., Leandro, C., Mottola, C., Barbosa, R., Silva, F. A., Oliveira, M., et al. (2014). In vitro design of a novel lytic bacteriophage cocktail with therapeutic potential against organisms causing diabetic foot infections. J. Med. Microbiol. 63, 1055–1065. doi: 10.1099/jmm.0.071753-0
Moore, A. J., Beazley, W. D., Bibby, M. C., and Devine, D. A. (1996). Antimicrobial activity of cecropins. J. Antimicrob. Chemother. 37, 1077–1089. doi: 10.1093/jac/37.6.1077
Nang, S. C., Azad, M. A. K., Velkov, T., Zhou, Q. T., and Li, J. (2021). Rescuing the last-line polymyxins: achievements and challenges. Pharmacol. Rev. 73, 679–728. doi: 10.1124/pharmrev.120.000020
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Plotka, M., Kaczorowska, A. K., Morzywolek, A., Makowska, J., Kozlowski, L. P., Thorisdottir, A., et al. (2015). Biochemical characterization and validation of a catalytic site of a highly thermostable Ts2631 endolysin from the Thermus scotoductus phage vB_Tsc2631. PLoS One 10:e0137374. doi: 10.1371/journal.pone.0137374
Plotka, M., Kaczorowska, A. K., Stefanska, A., Morzywolek, A., Fridjonsson, O. H., Dunin-Horkawicz, S., et al. (2014). Novel highly thermostable endolysin from Thermus scotoductus MAT2119 bacteriophage Ph2119 with amino acid sequence similarity to eukaryotic peptidoglycan recognition proteins. Appl. Environ. Microbiol. 80, 886–895. doi: 10.1128/AEM.03074-13
Ramarao, N., Nielsen-Leroux, C., and Lereclus, D. (2012). The insect galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J. Vis. Exp. 70:e4392. doi: 10.3791/4392
Roberts, K. D., Azad, M. A., Wang, J., Horne, A. S., Thompson, P. E., Nation, R. L., et al. (2015). Antimicrobial activity and toxicity of the major lipopeptide components of polymyxin B and colistin: last-line antibiotics against multidrug-resistant gram-negative Bacteria. ACS Infect. Dis. 1, 568–575. doi: 10.1021/acsinfecdis.5b00085
Schmelcher, M., and Loessner, M. J. (2021). Bacteriophage endolysins – extending their application to tissues and the bloodstream. Curr. Opin. Biotechnol. 68, 51–59. doi: 10.1016/j.copbio.2020.09.012
Shah, S., Das, R., Chavan, B., Bajpai, U., Hanif, S., and Ahmed, S. (2023). Beyond antibiotics: phage-encoded lysins against gram-negative pathogens. Front. Microbiol. 14:1170418. doi: 10.3389/fmicb.2023.1170418
Shin, D., Ha, E., Kong, M., and Ryu, S. (2023). Characterization of thermostable bacteriophage CPD2 and its endolysin LysCPD2 as biocontrol agents against Clostridium perfringens. Food Sci. Biotechnol. 32, 2069–2077. doi: 10.1007/s10068-023-01314-0
Sitthisak, S., Manrueang, S., Khongfak, S., Leungtongkam, U., Thummeepak, R., Thanwisai, A., et al. (2023). Antibacterial activity of vB_AbaM_PhT2 phage hydrophobic amino acid fusion endolysin, combined with colistin against Acinetobacter baumannii. Sci. Rep. 13:7470. doi: 10.1038/s41598-023-33822-8
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Thummeepak, R., Kitti, T., Kunthalert, D., and Sitthisak, S. (2016). Enhanced antibacterial activity of Acinetobacter baumannii bacteriophage OABP-01 endolysin (LysABP-01) in combination with colistin. Front. Microbiol. 7:1402. doi: 10.3389/fmicb.2016.01402
Tsai, C. J., Loh, J. M., and Proft, T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7, 214–229. doi: 10.1080/21505594.2015.1135289
Wang, Y., Wang, X., Liu, X., and Lin, B. (2024). Research progress on strategies for improving the enzyme properties of bacteriophage endolysins. J. Microbiol. Biotechnol. 34, 1189–1196. doi: 10.4014/jmb.2312.12050
Wang, F., Xiao, Y., Lu, Y., Deng, Z. Y., Deng, X. Y., and Lin, L. B. (2022). Bacteriophage lytic enzyme P9ly as an alternative antibacterial agent against antibiotic-resistant Shigella dysenteriae and Staphylococcus aureus. Front. Microbiol. 13:821989. doi: 10.3389/fmicb.2022.821989
Weaver, L. H., and Matthews, B. W. (1987). Structure of bacteriophage T4 lysozyme refined at 1.7 Å resolution. J. Mol. Biol. 193, 189–199. doi: 10.1016/0022-2836(87)90636-x
Wei, B., Cong, C., Zheng, L., Chen, L., and Yan, X. (2023). Isolation, characterization and whole genome analysis of the novel genus Lederbergvirus, phage vB_EcoP_E212 infecting enterotoxigenic Escherichia coli K88. Virus Res. 331:199125. doi: 10.1016/j.virusres.2023.199125
Xuan, J., Feng, W., Wang, J., Wang, R., Zhang, B., Bo, L., et al. (2023). Antimicrobial peptides for combating drug-resistant bacterial infections. Drug Resist. Updat. 68:100954. doi: 10.1016/j.drup.2023.100954
Yu, J. H., Lim, J. A., Chang, H. J., and Park, J. H. (2019). Characteristics and lytic activity of phage-derived peptidoglycan hydrolase, LysSAP8, as a potent alternative biocontrol agent for Staphylococcus aureus. J. Microbiol. Biotechnol. 29, 1916–1924. doi: 10.4014/jmb.1908.08021
Yuan, Y., Zhao, F., Wang, L., Tan, D., Cong, C., Li, X., et al. (2019). Complete genome analysis of the novel Enterococcus faecalis phage vB_EfaS_AL3. Arch. Virol. 164, 2599–2603. doi: 10.1007/s00705-019-04341-7
Żebrowska, J., Żołnierkiewicz, O., Ponikowska, M., Puchalski, M., Krawczun, N., Makowska, J., et al. (2022). Cloning and characterization of a thermostable endolysin of bacteriophage TP-84 as a potential disinfectant and biofilm-removing biological agent. Int. J. Mol. Sci. 23:7612. doi: 10.3390/ijms23147612
Zhang, L., Hu, F., Zhao, Z. R., Li, X. F., Zhong, M. Y., He, J. J., et al. (2024). Dimer-monomer transition defines a novel hyper-thermostable peptidoglycan hydrolase mined from bacterial proteome. bioRxiv. doi: 10.1101/2024.04.26.591248
Zhang, R., Xu, L., and Dong, C. (2022). Antimicrobial peptides: an overview of their structure, function and mechanism of action. Protein Pept. Lett. 29, 641–650. doi: 10.2174/0929866529666220613102145
Zhang, Q. Y., Yan, Z. B., Meng, Y. M., Hong, X. Y., Shao, G., Ma, J. J., et al. (2021). Antimicrobial peptides: mechanism of action, activity and clinical potential. Mil. Med. Res. 8:48. doi: 10.1186/s40779-021-00343-2
Zheng, T., and Zhang, C. (2024). Engineering strategies and challenges of endolysin as an antibacterial agent against gram-negative bacteria. Microb. Biotechnol. 17:e14465. doi: 10.1111/1751-7915.14465
Keywords: phage, phage endolysin, Gram-negative bacteria, polymyxin B, cecropin A, synergy
Citation: Chu D, Lan J, Liang L, Xia K, Li L, Yang L, Liu H and Zhang T (2024) The antibacterial activity of a novel highly thermostable endolysin, LysKP213, against Gram-negative pathogens is enhanced when combined with outer membrane permeabilizing agents. Front. Microbiol. 15:1454618. doi: 10.3389/fmicb.2024.1454618
Edited by:
Tamas Feher, HUN-REN Biological Research Centre, HungaryReviewed by:
Swapnil Ganesh Sanmukh, Université Clermont Auvergne, FranceVijay Singh Gondil, University of Rochester Medical Center, United States
Copyright © 2024 Chu, Lan, Liang, Xia, Li, Yang, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tingting Zhang, enR0LWdkQDE2My5jb20=; Hongmei Liu, aG1saXVAZ21jLmVkdS5jbg==
†These authors have contributed equally to this work
 Dingjian Chu1†
Dingjian Chu1† Kaide Xia
Kaide Xia Tingting Zhang
Tingting Zhang