- 1Institute of Human Nutrition and Food Science, Division of Food Science, Faculty of Agricultural and Nutritional Sciences, University of Kiel, Kiel, Germany
- 2Instituto de Lactología Industrial (CONICET-UNL), Faculty of Chemical Engineering, National University of Litoral, Santa Fe, Argentina
The filamentous fungus Aspergillus oryzae has a long tradition in East Asian food processing. It is therefore not surprising that in recent years fermentation products of A. oryzae have attracted attention in the emerging field of postbiotics. This review aims to provide a comprehensive summary of the potential postbiotic effects of fermentation products from A. oryzae, by discussing possible mechanisms of action against the background of the molecular composition determined so far. In particular, cell wall constituents, enzymes, extracellular polymeric substances, and various metabolites found in A. oryzae fermentation preparations are described in detail. With reference to the generally assumed key targets of postbiotics, their putative beneficial bioactivities in modulating the microbiota, improving epithelial barrier function, influencing immune responses, metabolic reactions and signaling through the nervous system are assessed. Drawing on existing literature and case studies, we highlight A. oryzae as a promising source of postbiotics, particularly in the context of animal health and nutrition. Challenges and opportunities in quality control are also addressed, with a focus on the necessity for standardized methods to fully harness the potential of fungal-based postbiotics. Overall, this article sheds light on the emerging field of A. oryzae-derived postbiotics and emphasizes the need for further research to fully realize their therapeutic potential.
1 Introduction
Fungi, with their exceptional diversity and versatility, play pivotal roles in ecological (Coleine et al., 2022; Bahram and Netherway, 2022), industrial (Sharma and Rath, 2021; Kumar et al., 2023; Arnau et al., 2020), and biomedical domains (Bachheti et al., 2021; Gupta et al., 2020; Hashem et al., 2023). As natural decomposers, they are fundamental to nutrient cycling within ecosystems, by breaking down complex organic materials into simpler monomeric forms (Kong et al., 2022; Lustenhouwer et al., 2020; Fukasawa and Matsukura, 2021). This enzymatic process, facilitated by a robust arsenal of different hydrolases not only supports ecosystem functioning but also enables the production of commercially valuable substances like ethanol (Nogueira et al., 2020; Tse et al., 2021) and organic acids (Dusengemungu et al., 2021; Andrino et al., 2021). Additionally, fungi enhance various processes in the food industry such as fermentation which is vital for the production of bread (Dahiya et al., 2020), cheese and meat (Ropars and Giraud, 2022), and beverages (Takeshita and Oda, 2023). In the medical field, fungi provide potent pharmaceuticals like antibiotics and are investigated for their ability to produce diverse secondary metabolites with therapeutic properties, including statins and other bioactive compounds (Devi et al., 2020; Adeleke and Babalola, 2021; Ancheeva et al., 2020). While historically recognized for their nutritional and medicinal value (Nagaoka and Hara, 2019; Pandey et al., 2019; Fleming, 1929; Wasser, 2002), fungi also produce mycotoxins such as aflatoxins, ochratoxin A, and fumonisins, which pose significant health risk (Perrone and Gallo, 2017).
Aspergillus oryzae, commonly known as “Koji mold,” plays a crucial role in the fermentation of various food products in East Asian cuisine (Liu et al., 2024; Yamashita, 2021). The A. oryzae-based preparation of koji, which has a long tradition of more than a 1000 years (Kusumoto et al., 2021), is used in the production of sake (rice wine), shoyu (soy sauce), amazake (rice koji beverage), osu (rice vinegar), kurosu (black rice vinegar), shochu (distilled alcoholic beverage fermented with koji), and miso (soybean paste) (Kusumoto et al., 2021; Ichishima, 2022; Murooka, 2020; Takeshita and Oda, 2023; Gall and Benkeblia, 2023). To start the fermentation process, the fungus is added to a substrate such as rice or soybeans, where it grows and produces enzymes that break down the starches, proteins, and fats of the substrate into simpler compounds including sugars, amino acids, and fatty acids. These compounds are then further fermented by other microorganisms, such as yeast and lactic acid bacteria, to generate the final food products (Liu et al., 2024). The beneficial involvement of koji molds in Japanese culture has propelled research and advancements in areas such as academia, industry, medicine, and agriculture (Yamashita, 2021; Daba et al., 2021; Kitagaki, 2021).
A. oryzae belongs to a genus of filamentous fungi (its taxonomic classification is depicted Figure 1) that includes a variety of species, some of which are beneficial for human use, while others are pathogenic or produce toxic compounds. Aspergillus molds often reproduce through both sexual (Asgarivessal et al., 2023) and asexual methods (Ojeda-López et al., 2018), although some species, including A. oryzae, are primarily recognized for reproducing exclusively asexually (Wada et al., 2012). The structure of the conidiophore (see the morphology of Aspergillus in Figure 1), which carries the asexual spores, is a crucial characteristic used for taxonomic classification in Aspergillus (Suleiman, 2023). Among the approximately 400 Aspergillus species (Ibrahim et al., 2023), the most widely recognized members are A. flavus and A. parasiticus, which are mycotoxin producers (Ting et al., 2020; Priesterjahn et al., 2020). Their infestation of crops like maize, peanuts, and beans, coupled with the production of highly carcinogenic and mutagenic mycotoxins, notably aflatoxin B1, poses significant health threats to both humans and animals (Fouad et al., 2019). A. fumigatus is another significant species, predominantly known as a pathogen that can cause invasive aspergillosis, particularly in immunocompromised individuals (Latgé and Chamilos, 2019). The beneficial Aspergillus species A. oryzae and the closely related A. sojae are regarded by most taxonomists as the domesticated forms of A. flavus and are widely used in food industry (Frisvad et al., 2018; Daba et al., 2021; Frisvad et al., 2019). Typically, A. oryzae thrives best at temperatures between 32 and 36°C and is unable to grow beyond 44°C. It prefers a pH range of 5.0 to 6.0 for growth, but can germinate in conditions with pH levels from 2.0 to 8.0 (Daba et al., 2021).
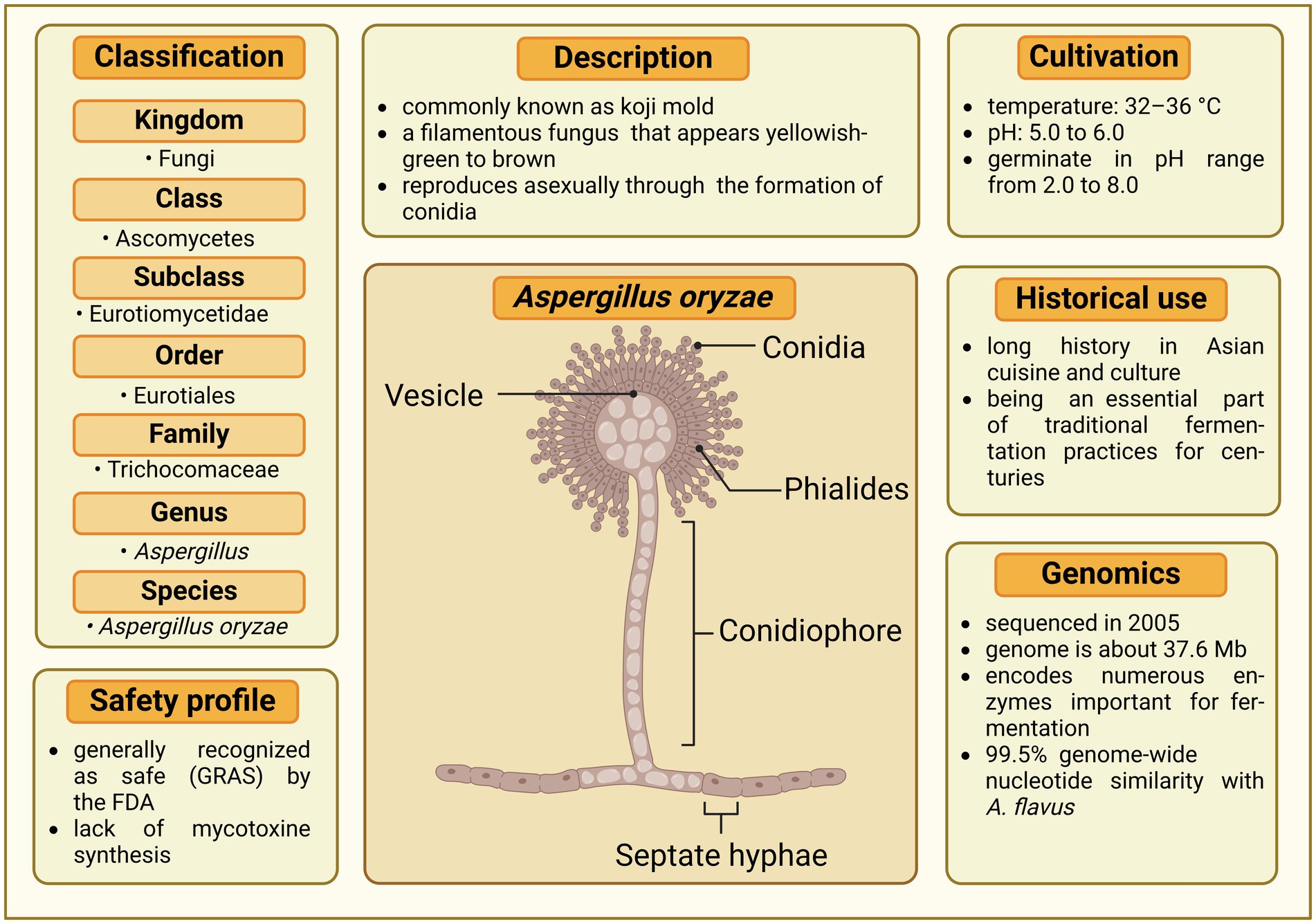
Figure 1. Comprehensive profile of Aspergillus oryzae. The central illustration depicts the morphological structures of A. oryzae, including vesicle, conidia, phialides, conidiophore, and septate hyphae. Surrounding the illustration are key aspects of the fungus, covering its classification, safety profile, description, cultivation requirements, historical use, and genomic characteristics. Created in BioRender. Seidler, Y. (2024) BioRender.com/i28s583.
The genome of A. oryzae has a size of 37.6 Mb (Machida et al., 2005), which is 20–30% larger than the genomes of other Aspergillus species such as A. nidulans and A. fumigatus (Nierman et al., 2005; Galagan et al., 2005). This difference in genome size is attributed to the presence of a higher number of transposable elements and gene duplications in A. oryzae. Remarkably, despite the larger genome of A. oryzae, there are almost no genotypic differences between A. oryzae and A. flavus, with the two species sharing 99.5% genome-wide nucleotide similarity (Han et al., 2024). The A. oryzae genome contains approximately 12,000 genes distributed on eight chromosomes that encode a wide variety of enzymes, including amylases, proteases, and lipases, which contribute to the fungus’s ability to degrade complex organic materials and are essential for its use in fermentation processes (Machida et al., 2005). While the genetic structure of numerous A. oryzae strains has been decoded (Machida et al., 2005; Zhong et al., 2018; Deng et al., 2018; Umemura et al., 2012; Zhao et al., 2012) functional genomics is still in its nascent stage in terms of developing strains for industrial purposes. This is evident from the fact that so far only around 200 genes have been functionally verified, making up a minimal of 1.7% of the whole genome (He et al., 2019).
A. oryzae is commonly classified as a fungus that is not pathogenic (Kitagaki, 2021). Moreover, it has not been associated with any carcinogenic compounds, including aflatoxins (Barbesgaard et al., 1992). This has been confirmed for different koji-mold strains of A. oryzae, which were all tested negative for aflatoxins (Tanaka et al., 2006). While there are Aspergillus strains that can produce certain types of mycotoxins (Kato et al., 2011), the fermentation industry consistently verifies that the levels of mycotoxins in their products are within the limits set by health authorities. Interestingly, A. oryzae has also been shown to degrade aflatoxins. A product called “D-Tox,” developed from A. oryzae, can reduce aflatoxin B1 (AFB1) by up to 90%, offering a promising approach to improving food safety (Choi et al., 2024).
Although A. oryzae and A. flavus are genotypically very similar (Han et al., 2024), A. oryzae is used in food production and listed as “Generally Recognized as Safe” (GRAS) status by the Food and Drug Administration (FDA) and has been approved as a safe microorganism by the World Health Organisation (WHO) (He et al., 2018a), whereas A. flavus is harmful for human, plants, and animals (Fouad et al., 2019; Amaike and Keller, 2011). It is suspected that there are disabling mutations in the gene cluster, which result in A. oryzae not producing aflatoxins, whereas A. flavus does (Tominaga et al., 2006; Tao and Chung, 2014). Subsequent studies, particularly at the molecular level, reinforced these findings. The aflatoxin production mechanism in A. flavus involves a series of over 25 genes. Through polymerase chain reaction (PCR) evaluations, it was observed that 15 out of 39 A. oryzae strains had missing sequences in five genes that align with the A. flavus aflatoxin gene cluster. In the other strains analyzed, the genes in the corresponding cluster were found to be non-functional (Kusumoto et al., 2000). Chang (2019) compared 13 A. flavus and 11 A. oryzae genome sequences based on genome-wide total single nucleotide polymorphism (total SNPs). The study proved a new technique to distinguish between A. flavus and A. oryzae.
No cases of invasive growth or systemic infections caused by A. oryzae have been reported in healthy individuals. However, there have been rare instances where strains identified as A. oryzae were isolated from individuals with compromised health, suggesting that while A. oryzae possesses a low potential for pathogenicity, it can grow in human tissue under extraordinary conditions, similar to many other detrimental microorganisms (Stelmaska et al., 1974; Liao et al., 1988; Tomizawa, 1981). There have been a limited number of reported cases of allergic reactions primarily attributed to A. oryzae, but these cases likely involved individuals already prone to allergic reactions and who were exposed to a significant amount of conidia via inhalation (Kino et al., 1982; Barbesgaard et al., 1992). Taken together, the demonstrated safety of A. oryzae qualifies it as a favored progenitor organism for not only the fermentation of foods but also the synthesis of a wide array of enzymes and chemicals with prospective medicinal benefits that could be harnessed for forthcoming medical procedures (Yu et al., 2004).
In the realm of microbiology and food science, A. oryzae represents a subject of study, bridging ancient culinary practices with modern biotechnological applications. Esteemed for its pivotal role in the fermentation of a vast array of East Asian foods (Daba et al., 2021), A. oryzae is not merely a workhorse of traditional fermentation but also of interest in the burgeoning field of postbiotics. For example, recent investigations into its postbiotic potential reveal promising health benefits in animal models (Kaufman et al., 2021; Ríus et al., 2022), suggesting a broader applicability of A. oryzae-derived products in promoting well-being.
In this review, we explore the various constituents, molecules, and cell wall components of A. oryzae, including enzymes and both primary and secondary metabolites. Additionally, we introduce the concept of postbiotics and link the metabolites, cell wall components, and enzymes of A. oryzae with the five proposed modes of action of postbiotics (Salminen et al., 2021). These modes of action are crucial in understanding how compounds from A. oryzae can potentially confer benefits to the host, which is a defining feature of postbiotics. Each metabolite’s role is examined in the context of these bioactive mechanisms, helping to clarify their potential health benefits. Thus, this work not only compiles existing research on A. oryzae but also identifies knowledge gaps and suggests directions for future studies. Finally, we focus on the emerging challenges and opportunities in the quality control of postbiotics derived from A. oryzae, a novel scientific topic that has not been extensively discussed in the literature. This review aims to set a foundation for advancing the understanding and development of A. oryzae as a postbiotic.
2 Methodology
The content synthesized in this narrative review is derived from an exhaustive literature search employing various scholarly databases and scientific websites, including Scopus, Web of Science, PubMed, Google Scholar, and Science Direct. This search utilized a comprehensive set of keywords to capture the broad spectrum of research concerning A. oryzae including the term “Aspergillus oryzae” in combination with the term’s “enzymes,” “compounds,” “fermentation,” “primary compounds,” “secondary compounds,” “secondary metabolism,” “cell wall,” “biological activity,” “prebiotic,” “probiotic,” “postbiotic,” “heat inactivated,” “inactivated,” “killed,” “dried,” “inanimated,” and “non-viable,” respectively.
Additionally, the selection process prioritized peer-reviewed articles, reviews, and significant research reports, while conference papers and abstracts were excluded unless they provided novel insights or data unavailable elsewhere. The search strategy included scanning titles, abstracts, and full-texts to ensure relevance to A. oryzae’s diverse roles and applications in enzymatic processes, fermentation, and bioactivity.
Each keyword was carefully selected to ensure that all relevant aspects of A. oryzae, including its primary and secondary metabolites, and its use as a biotic agent, were thoroughly explored. Furthermore, we ensured that studies addressing safety, industrial applications, and advances in biotic and postbiotic research were also covered, thereby providing a holistic overview of the literature.
3 Fermentation of Aspergillus oryzae
3.1 Solid state and submerged fermentation
In general, fermentation is defined as a biochemical process in which microorganisms such as bacteria and fungi break down complex compounds into simpler substances and generate energy under anaerobic conditions (Nout, 2005). Yet, fermentation in a biotechnical context can be aerobe (culturing) and anaerobe as for instance in soy sauce production (Ito and Matsuyama, 2021). During this process, microorganisms produce primary and secondary metabolites. These bioactive compounds, which include antibiotics, peptides, enzymes, and growth factors, have significant industrial and economic value (Robinson et al., 2001; Balakrishnan, 1996). As the demand for these compounds has grown, techniques have been scaled up from laboratory settings to industrial levels. This scaling presents challenges in maintaining controlled environments for microbial growth, as deviations can lead to undesired (by-)products (Subramaniyam and Vimala, 2012). Depending on the nature of the substrate, solid state fermentation (SSF) is distinguished from submerged fermentation (SmF). The former is characterized as a fermentation procedure where microorganisms grow on solid substrates in the absence of free-flowing liquid (Wang J. et al., 2023). One of the primary advantages of utilizing solid substrates is the opportunity to efficiently recycle nutrient-rich waste (El Sheikha and Ray, 2023; Paz-Arteaga et al., 2023). In this approach, substrates undergo a slow and consistent consumption, facilitating prolonged fermentation periods and a controlled release of nutrients. SSF is especially advantageous for fungi and certain microorganisms that prefer environments with reduced moisture content. On the other hand, SmF is a sophisticated fermentation technique utilizing liquid media and substrates, including molasses and various broths. This method facilitates the secretion of bioactive compounds directly into the fermentation broth (Xv et al., 2024). SmF offers precise control over essential parameters, such as temperature, aeration, and agitation, all pivotal for maximizing product yield. The meticulous regulation of these factors, especially temperature, impacts the growth rate, oxygen dynamics, and overall product synthesis of the microorganism (Esmeralda-Guzmán et al., 2024). The choice between SSF and SmF often depends on the specific bioactive compound being produced and the substrate used in the fermentation process (Premalatha et al., 2023; Sankar et al., 2023).
As far as A. oryzae is concerned, the production of various foods through SSF (Rousta et al., 2021) on diverse substrates such as rice, wheat bran, and soybeans has a long tradition. Moreover, SmF with A. oryzae is frequently employed for the production of enzymes (Masuda et al., 2009; Shah et al., 2014) and organic acids (Singh et al., 2022; Badar et al., 2021). Both fermentation techniques have been developed for large-scale production.
3.2 Stress response and physiological adaptation during fermentation
During fermentation, the microorganism used has to adapt to various abiotic factors including temperature, pH, oxidative, and osmotic conditions, which can sometimes cause stress (Hagiwara et al., 2015). In the context of soy sauce fermentation, for instance, A. oryzae contends with elevated salt concentrations (17–18%) and an acidic pH (He et al., 2018b). The high salt concentrations induces stress due to the disruption of osmotic potential (Taymaz-Nikerel et al., 2016). Research by He et al. (2018b) highlighted that salt stress led to changes both at the transcriptome and metabolome levels in A. oryzae with upregulated expression of genes related to arginine accumulation and oleic acid synthesis. Additionally, variations in lipid metabolism in response to salt stress were observed, notably leading to an increase in intracellular linoleic acid. Recent studies further show that reducing salt during soy sauce production enhances A. oryzae’s carbon and protein metabolism, which not only improves substrate efficiency but also supports Lactobacillus growth and balances the microecology. This adaptation also boosts the production of smoky, nutty, and malty aromas in soy sauce while reducing rancid odors by suppressing fatty aldehyde production through enhanced β-oxidation (Liu et al., 2024).
During rice wine fermentation, A. oryzae undergoes ethanol-induced stress. Ma et al. (2019) elucidated that ethanol absorption led to cellular perturbations and a rise in fatty acid unsaturation. This was evidenced by the conversion of stearic acid to linoleic acid and the heightened expression of related fatty acid desaturases. Temperature fluctuations present another challenge for A. oryzae during fermentation. Both high and low temperatures can hinder the growth and conidial formation. On a molecular level, temperature changes were reported to influence the expression of genes associated with sugar and lipid metabolism. Specifically, low temperatures stimulated genes related to trehalose synthesis and starch metabolism, while high temperatures suppressed genes involved in the metabolism of fructose, galactose, and glucose (Jiang et al., 2022). Furthermore, in the context of glucosamine (GlcN) production from A. oryzae NCH-42, environmental factors such as nitrogen sources, temperature, and pH play a pivotal role in determining the cell wall composition. Li W. et al., 2021 found that acidic stress, particularly at a pH of 2.5, significantly enhanced GlcN production, with the fungal biomass yielding up to 0.31 g/g of GlcN. This was corroborated by scanning electron microscopy (SEM) examinations that revealed a robust mycelial structure under these conditions (Li J. S. et al., 2021).
The research on A. oryzae demonstrates that a variety of abiotic factors, along with the type of fermentation and the substrate used, influence the chemical composition of A. oryzae fermentation end products. Consequently, it can be inferred that depending on the actual fermentation conditions end products from the same species may vary considerably in their effectiveness to cause beneficial effects and may not necessarily operate through the same mechanisms.
4 Postbiotics – an emerging concept in the field of “biotics”
4.1 From prebiotics and probiotics to postbiotics
Prebiotics were originally defined as “a non-digestible food ingredient that beneficially affects the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria already resident in the colon” (Gibson and Roberfroid, 1995). This concept has its roots in early research, such as the study by Rettger and Cheplin (1921) who observed the enrichment of human microbiota with lactobacilli after carbohydrate consumption. Over time, the definition of prebiotics has evolved. The most recent version, provided by the International Scientific Association for Probiotics and Prebiotics (ISAPP), defines a prebiotic as “a substrate that is selectively utilized by host microorganisms conferring a health benefit” (Dubos et al., 1965; Savage, 1977). Common examples of prebiotics include inulin, fructooligosaccharides (FOS), and galactooligosaccharides (GOS), which are preferentially metabolized by bifidobacterial (Gibson et al., 2017).
In contrast, probiotics—first introduced by Lilly and Stillwell (1965)—refer to live microorganisms. In, 2001, the WHO and FAO defined probiotics as “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” (FAO/WHO, 2001). Synbiotics refer to a combination of probiotics and prebiotics that either function independently or synergistically to provide health benefits (Markowiak and Śliżewska, 2018; Swanson et al., 2020).
As research has advanced, additional terms such as paraprobiotics (non-active probiotic cells) and postbiotics have been introduced, emphasizing that non-living cells, whether whole or fragmented, can also positively affect human health. The ISAPP defines postbiotics as “a preparation of inanimate microorganisms and/or their components that confers a health benefit on the host” (Salminen et al., 2021). The core of the definition are inanimate microbes, metabolites can be present, or not, in the final product. Metabolites alone, in the absence of inanimate cells, are not regarded as postbiotics according to the definition. Common misunderstanding around the definition of postbiotics was recently clarified (Vinderola et al., 2024).
Besides inanimate microorganisms, a postbiotic product may include metabolites generated by microorganisms, such as short chain fatty acids (SCFA), exopolysaccharides, cell wall fragments, enzymes/proteins, cell-free supernatant, and other metabolites (Żółkiewicz et al., 2020). In a 2021 publication by ISAPP, various proposed mechanisms of postbiotics were outlined. Accordingly, postbiotics are suggested to potentially modulate gut microbiota dynamics, booster intestinal barrier integrity, exhibit immunomodulatory effects, regulate systemic metabolism, and/or engage in signaling through the nervous system. A comprehensive description of these mechanisms can be found in the publication by Salminen et al. (2021). The differences between prebiotics, probiotics, and postbiotics are illustrated in Figure 2.
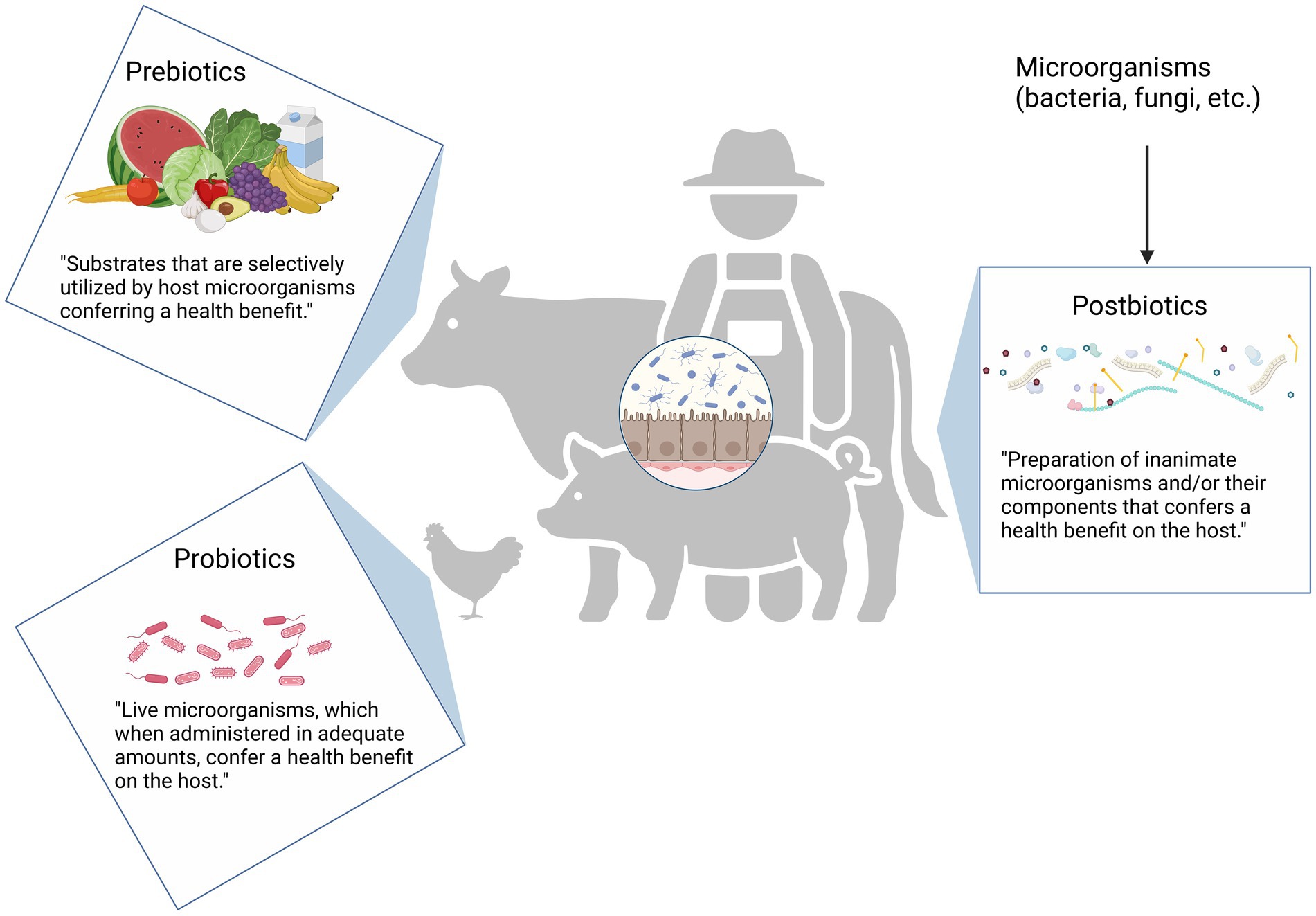
Figure 2. Definition of prebiotics, probiotics, and postbiotics. Prebiotics are specific dietary compounds that beneficially nurture the host’s existing microorganisms. They do not contain live organisms but fuel beneficial bacteria already present in the gut. Examples include soluble fibers such as inulin, fructooligosaccharides, and galactooligosaccharides. Probiotics refer to live microorganisms that, when consumed in adequate amounts, confer health benefits on the host. These are the “good” bacteria introduced into the system to enhance gut health and other functions. Only those strains with scientifically demonstrated health effects are termed as “probiotic-like.” Postbiotics, as the name suggests, are derived “after life.” They may include substances obtained from microbial activity once the microorganisms are no longer alive. This category can encompass a range of components, from cell wall fragments and enzymes to amino acids, organic acids, and various metabolites. Created in BioRender. Seidler, Y. (2024) BioRender.com/h54l897.
5 Possible bioactive compounds in Aspergillus oryzae fermentation end products
According to the definition proposed by Salminen et al. (2021), fermentation end products from A. oryzae fermentation can be categorized as postbiotics as long as they do not contain living cells. Given that postbiotics can include a combination of non-viable cells, cell fragments and metabolites of the progenitor microorganism, it is expected that their components bearing bioactivity can exhibit considerable diversity. This diversity will be examined in the subsequent section, with a primary emphasis on compounds derived from A. oryzae. However, the specific bioactivities and health benefits associated with these compounds will be addressed in detail in section 7, in order to avoid redundancy and ensure clarity.
5.1 Cell wall fragments of Aspergillus species
The cell walls of filamentous fungi are complex structures composed mainly of polysaccharides (90%) (Latgé, 2010), which differ significantly from the cellulose-based plant cell walls (Bowman and Free, 2006). The fungal cell wall has been simplistically separated into a fibrillar alkali-insoluble skeleton and an amorphous alkali-soluble cement (Latgé, 2010). At present, there is no detailed description of the cell wall of A. oryzae. For this reason, we have mainly reviewed evidence from A. fumigatus, which cell wall has been dissected in depth at the molecular level. However, it is important to note that while cell wall components of A. fumigatus are implicated in causing 50–90% of human aspergillosis cases (Latgé and Chamilos, 2019), A. oryzae is a non-pathogenic species considered as safe. Even though we generalize between the two Aspergillus species in this review due to their taxonomic proximity, the reader should be aware that biologically relevant differences in their cell wall composition and/or structure likely exist.
The main polysaccharides in the Aspergillus cell wall are α-glucans (α-1,3-glucan and α-1,4 glucan), β-glucans (β-1,6-branched β-1,3-glucan and linear β-1,3/1,4-glucan), galactosaminogalactan (GAG), galactomannan (GM), and chitin (linked via β-1,4 linkage to β-1,3-glucan) (Figure 3). These polysaccharides have been characterized in detail in A. fumigatus (Fontaine et al., 2000; Mouyna and Fontaine, 2008).
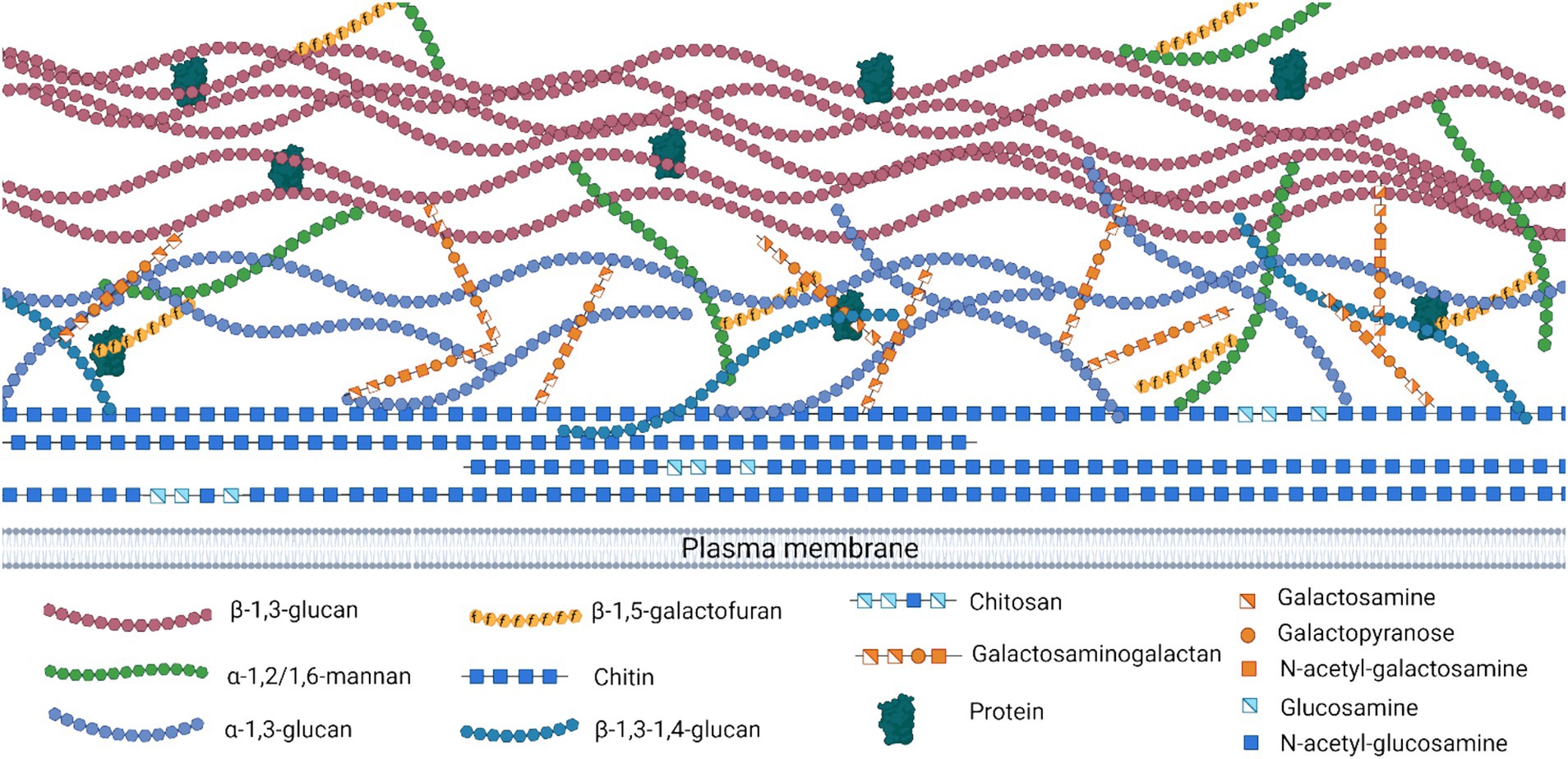
Figure 3. Aspergillus spp. cell wall organization and polysaccharides. Structural depiction of the cell wall polysaccharides showing linkages and monosaccharide components. Created with Biorender.com.
The polysaccharide α-1,3-glucan stands as a hallmark component of the outer cell wall in A. fumigatus (Lee and Sheppard 2016). Comprised of glucose units interconnected through α-1,3 linkages, its presence, while not unique to A. fumigatus, holds relevance in the context of bioactivity-bearing compounds. In the case of A. oryzae, the production of α-1,3-glucan is orchestrated by three synthases: Ags1, Ags2, and Ags3 (Miyazawa et al., 2016; Zhang et al., 2017). In studies focusing on A. oryzae, ags gene knockouts have led to enhanced recombinant enzyme production (Miyazawa et al., 2016), and it was observed that α-1,3-glucan acts as a potential inhibitory factor for enzymes such as α-amylase (Zhang et al., 2017). Additionally, the deletion of α-1,4-glucan in A. oryzae results in an increased accumulation of α-1,3 glucan (Koizumi et al., 2023).
β-1,3-glucan is a primary structural component of the A. fumigatus hyphal cell wall, consisting of glucose residues connected by β-1,3 linkages (Lee and Sheppard 2016). In a comparative study involving Saccharomyces cerevisiae, Xanthomonas campestris, and Bacillus natto, A. oryzae stood out by producing the highest mass of β-glucan during SmF conditions (Utama et al., 2021). Additionally, SSF with A. oryzae was shown to increase the β-glucan content of brown rice used as a substrate (Ji and Ra, 2021).
In A. fumigatus, the inner cell wall structural integrity is mainly attributed to the presence of chitin, a pivotal structural polysaccharide. Chitin, consisting of N-acetyl-glucosamine (GlcNAc) residues linked by β-1,4 bonds, is a fundamental component of the fungal cell wall carbohydrate structure, though its prevalence differs among fungal species (Lenardon et al., 2010; Munro and Gow, 2001; Latgé, 2007). For instance, while it constitutes a mere 1–2% of the cell wall in S. cerevisiae, in filamentous fungi, it can represent up to 10–20% of the mycelial dry weight (Bartnicki-Garcia, 1968).
In A. oryzae, chitin significantly influences the structural integrity of the cell wall. Recent research has advanced the understanding of chitin’s biosynthesis in A. oryzae through the characterization of chitin synthase genes. The novel gene chsZ suggests a unique role in chitin synthesis, highlighted by its classification into a newly proposed class VI of chitin synthases (Chigira et al., 2002). Moreover, studies on the disruption of chitin synthase genes like chsB and csmA have shown that these alterations affect fungal morphology and growth dynamics without impacting α-amylase productivity, which influences the rheological properties of the cultivation broth in industrial fermentations (Müller et al., 2002a; Müller et al., 2002b).
Adjacent to chitin is its derivative, chitosan, which also composes the cell walls of numerous fungi. This compound emerges from the deacetylation of chitin, a process mediated by chitin deacetylases (CDA), culminating in the formation of glucosamine residues. This was described for the first time by Kreger (1954). Since then, two putative CDA-encoding genes have been identified in A. fumigatus, but their precise roles and contributions remain enigmatic (Gastebois et al., 2009). Chitosan’s physicochemical properties, notably its solubility and rigidity, are influenced by its degree of deacetylation (Kurita, 2006; El Knidri et al., 2018; Dash et al., 2011), which is defined as the molar fraction of deacetylated units in the polymer chain (Zhang et al., 2005). This has propelled chitosan to the forefront of various industrial applications, with notable inroads in the medical sector, particularly for the prevention of biofilm formation on medical devices (No et al., 2007; Salamanca et al., 2006; Boateng et al., 2008). A study investigated chitosan production by novel A. oryzae isolates grown under SmF conditions. One isolate (A2) identified by a 99% genetic similarity to known A. oryzae genome sequences, yielded the highest level of chitosan at 352 mg/L and a biomass of 9.48 g/L. Structural validation revealed a 55.23% degree of deacetylation for the chitosan, which displayed antimicrobial activity against several pathogens, most effectively against Salmonella typhimurium (Jebur et al., 2019).
GM is an immunoreactive substance (Latgé et al., 1994) widely distributed among most Aspergillus species (Barreto-Berter et al., 1980; Bardalaye and Nordin, 1977; Bennett et al., 1985; Reiss and Lehmann, 1979). In A. fumigatus, its complex structure is made up of mannose and galactofuranose (Galf). A mannan chain serves as the backbone, with mannose residues connected via α-1,3 or α-1,6 linkages, while the side chains of the galactomannan consist of an average of 4 to 5 β-1,5-Galf units (Latgé et al., 1994). The exact role of GM in virulence is still a topic of debate. While the mannan components, which are vital for maintaining cell wall structure, are non-antigenic, the role of the Galf side chain in virulence remains not clearly defined (Lee and Sheppard, 2016; Lamarre et al., 2009; Schmalhorst et al., 2008). The absence of Galf in A. fumigatus resulted in attenuated virulence (Schmalhorst et al., 2008) and GM detection using a monoclonal antibody that specifically reacts with Galf-containing glycostructures serves as a diagnostic marker for Aspergillus infection, which emphasize the importance of understanding its synthesis and regulation (Heesemann et al., 2011; Marino et al., 2017). Nakajima and Ichishima (1994) isolated a GM-protein complex from A. oryzae’s hyphal walls, composed of 89% carbohydrates and 11% proteins, featuring a mannan backbone with predominantly β-Galf-capped mannose side chains. Structural changes induced by chemical treatments indicated a complex architecture involving both N- and O-linked carbohydrate chains. Another study explored the biosynthesis and functional role of D-Galf-containing glycans in A. oryzae. The deletion of the ugmA gene, encoding uridine diphosphate (UDP)-galactopyranose mutase, significantly impaired mycelial elongation, underscoring the importance of these glycans in maintaining cell wall integrity. Nuclear magnetic resonance analysis of the ΔugmA mutant confirmed the presence of core mannan backbones, despite the absence of Galf-containing sugar chains (Kadooka et al., 2023).
GAG is a complex sugar molecule present within the extracellular matrix as well as the inner and outer layers of the cell walls in A. fumigatus hyphae (Lee and Sheppard, 2016). This compound, made up of galactose and GalNAc connected through α-1,4 bonds, has a diverse structure due to the non-uniform positioning of its galactose and GalNAc units (Gravelat et al., 2013; Fontaine et al., 2011). The heterogeneity is unique to the GAG because most of the cell wall polysaccharides are homopolymers (chitin, glucans). Building upon the structural diversity of GAG in Aspergillus species, recent studies have elucidated its functional role in A. oryzae, particularly in mediating hyphal aggregation, which significantly impacts industrial fermentation processes. Research identified that GAG, alongside α-1,3-glucan, contributes to hyphal pellet formation. This was demonstrated by disrupting GAG biosynthesis in α-1,3-glucan-deficient mutants (AGΔ), resulting in the AGΔ-GAGΔ double mutant with fully dispersed hyphae in liquid culture, confirming GAG’s essential role in aggregation (Miyazawa et al., 2019). Further experimental addition of a partially purified GAG fraction to AGΔ-GAGΔ cultures induced mycelial pellet formation, highlighting GAG’s aggregation mechanism through acetylated galactosamine-mediated hydrogen bonding. This property not only affects hyphal structure but also enhances bioreactor production efficiency (Ichikawa et al., 2022). Additionally, A. oryzae NCH-42 was studied as a non-animal source of glucosamine (GlcN). Environmental factors like pH were examined, revealing that acidic conditions (pH 2.5) significantly boosted GlcN content (Li J. S. et al., 2021).
While sphingolipids are typically associated with the cell membrane rather than the structural framework of the cell wall, their inclusion in this context is warranted due to their relevance to A. oryzae. Investigations have revealed the presence and functional significance of glycosylceramides, shedding light on their potential health-promoting effects. For instance, Tani et al. (2014) identified β-glucosylceramide and β-galactosylceramide in A. oryzae. Similarly, Miyagawa et al. (2019) explored the cosmetic implications of glycosylceramides derived from various sources, including A. oryzae, on gene expression in human keratinocytes. These findings suggest that the abundance of glycosylceramides in A. oryzae may contribute to ceramide biosynthesis and tight junction formation in the skin, potentially elucidating the cosmetic benefits associated with koji. Additionally, Hamajima et al. (2016) investigated the prebiotic effects of koji glycosylceramide in murine models, unveiling its capacity to stimulate the proliferation of beneficial gut microbiota such as Blautia coccoides. The author suggest that the collective findings hint at a plausible nexus between Japanese dietary practices, gut microbiome modulation, and longevity via the consumption of koji-derived glycosylceramides.
5.2 Extracellular polymeric substances
Filamentous hyphae of Aspergillus grow embedded within an extracellular polymeric substance (EPS). In 1999, EPS were defined as all polymers outside the cell wall, which are not directly anchored to the outer membrane or murein-protein layer (Nielsen and Jahn, 1999). This extracellular matrix mediated adherence to inorganic substances and host cells and enhanced resistance to host defense and antifungal agents. The matrix is composed of heterogeneous macromolecules of extracellular DNA, proteins, lipids, and polyols, and exopolysaccharides including α-glucans, GM, and GAG, being carbohydrates and proteins usually the major components of EPS (Sheng et al., 2010). In A. fumigatus, the exopolysaccharides GAG, GM, α- and β-glucans are found, whereby GAG plays a critical role in the maintenance of the extracellular matrix of A. fumigatus (Gravelat et al., 2013; Lee et al., 2015). The bioactive properties of exopolysaccharides are known to depend on many factors, such as the monosaccharide components, molecular mass, conformation, and linkage type (Zhou and Chen, 2011; Elsehemy et al., 2020; Xu et al., 2006). Research on A. oryzae has shown that its EPS play a crucial role in supporting microalgal-fungal co-cultivation systems, particularly in wastewater treatment applications. The EPS, consisting of high molecular weight substances including proteins, polysaccharides, and enzymes like amylase, protease, and lipase, contribute significantly to the structural integrity and functionality of these systems (Nie et al., 2022). EPS components facilitate the formation and stabilization of microalgal-fungal aggregates and enhance nutrient removal and biomass production (Wu et al., 2023).
5.3 Enzyme production by Aspergillus oryzae fermentation
Enzymes are proteins produced by living organisms that serve as catalysts to facilitate specific biochemical reactions (Sundarram and Murthy, 2014; Gurung et al., 2013). Owing to their broad range of applications, the biotechnical production of enzymes through microbial fermentation is an important technology in diverse areas such as production of food, pharmaceuticals, and therapeutics (Cruz-Casas et al., 2021). Fungi are key sources of hydrolytic digestive enzymes, which also play a decisive role in fermentation processes. In the case of A. oryzae, several biocatalysts of potential commercial interest have been identified that can be produced via SSF and/or SmF (Table 1). These enzymes, their potential and, in some cases, their current applications are summarized in the following paragraphs.
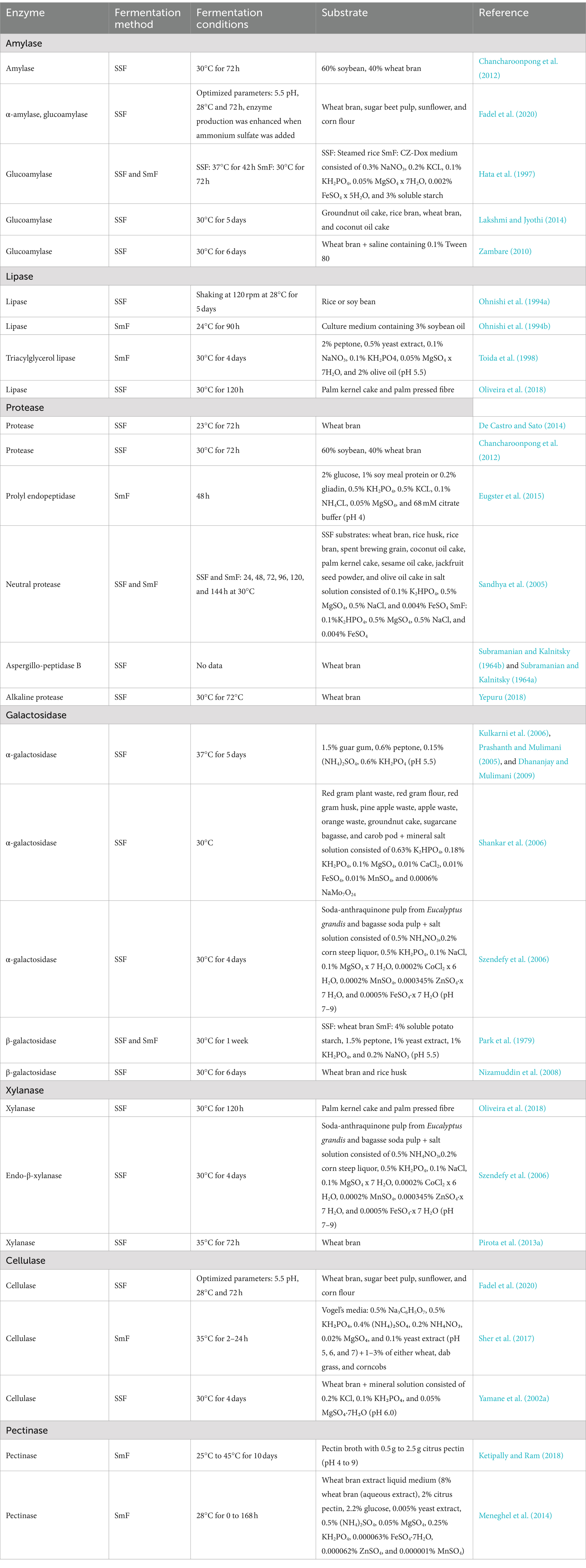
Table 1. Enzymes produced by Aspergillus oryzae by solid-state fermentation (SSF) or submerged-fermentation (SmF).
Proteases are enzymes that catalyze the hydrolysis of peptide bonds in proteins, resulting in the formation of smaller peptides or individual amino acids. Their significance in the food industry is multifaceted and has been extensively studied. According to Machida et al. (2005), A. oryzae has the largest expansion of hydrolytic genes with 135 proteinase genes including both endo- and exoproteases. Throughout the fermentation process, endoproteases primarily create numerous free termini, facilitating the action of exoproteases (Eugster et al., 2015). In broad bean paste fermentation, the use of a strain of A. oryzae with higher protease activity compared to commonly used strains significantly enhances flavor profiles by increasing umami amino acids and volatile flavors, thus improving the sensory characteristics of the final product (Niu et al., 2023). During A. oryzae mediated soy sauce fermentation, diverse enzymes are leveraged by the fungus to break down proteins and carbohydrates under high-salt conditions. Early-stage enzymes such as peptidases break down proteins into peptides, while later-stage enzymes like metallopeptidases and extracellular proteinases continue this proteolysis (Zhao et al., 2018). Specifically, leucine aminopeptidase II from A. oryzae plays a notable role, accounting for almost 80% of the glutamic acid release from soybean proteins, which essential for the umami flavor of soy sauce (Zhao et al., 2018).
Lipases function as versatile biocatalysts, facilitating a range of reactions such as esterification, hydrolysis, alcoholysis, transesterification, aminolysis, and acidolysis (Zhang et al., 2020). These enzymes have found extensive applications in sectors like the chemical, food, pharmaceutical, and detergent industries (Verma et al., 2012). During the miso fermentation process, lipase from A. oryzae facilitates the hydrolysis of glycerides in soybeans. This not only results in the development of distinct flavors and aromas but also leads to notable changes in lipid composition, contributing to the formation of characteristic taste profiles (Ohnishi, 1982).
Esterases are enzymes that break down esters into alcohols and acids. While they share similarities with lipases in breaking down glycerides, esterases specifically target short-chain glycerides (Okpara, 2022). In the beverage industry, feruloyl esterase plays a role in producing ferulic acid. This acid then serves as a starting material for creating the aromatic compound, vanillin, a primary component of vanilla that enhances beverage flavors (Gallage et al., 2014). In brewing sake, A. oryzae produces feruloyl esterase, which facilitates the release of ferulic acid from the rice endosperm cell walls. A study examining the effects of this enzyme, specifically the FaeA variant produced by A. oryzae, highlighted its significant role in flavor formation (Todokoro et al., 2022).
The enzyme group of amylases, responsible for breaking down starches, consists of two primary categories: amylases and glucoamylase (Lakshmi and Jyothi, 2014). α-amylase acts on starch, converting it into maltose, glucose, and maltotriose by targeting the α-1,4-D-glucosidic bonds between glucose molecules in the straight amylase chain. On the other hand, glucoamylase releases individual glucose molecules from the non-reducing ends of both amylose and amylopectin, resulting in the exclusive production of glucose from starches and related compounds (Zong et al., 2022). In a recent study, twenty-five filamentous fungal isolates were tested as potential α-amylase sources under SSF conditions using wheat bran as substrate. Among those, an A. oryzae isolate (F-923) was the most promising candidate for the production of the target enzyme. Ammonium sulfate supplementation and the addition of soluble starch were found to enhance the α-amylase yield (Fadel et al., 2020).
Galactosidases are a group of enzymes that catalyze the hydrolysis of α- and β-galactosidic bonds in certain carbohydrates or glycosides, respectively. These enzymes play a pivotal role in the digestion and assimilation of carbohydrates in various organisms. Among the microorganisms known to produce these enzymes, A. oryzae stands out due to its widespread use in industrial applications (Nath et al., 2014). α-galactosidases are specialized in cleaving α-galactosidic bonds, particularly those found in oligosaccharides like raffinose, stachyose, and verbascose. These sugars are commonly found in legumes and certain vegetables and are not digestible by human, which can cause digestive discomfort in some people (Ibrahim et al., 2010). Accordingly, fungal α-galactosidases have the capacity to remove the flatulence-inducing sugars of soymilk and soybean. The results are interesting for their potential use in the food industry (Patil and Mulimani, 2008; Guo YuHan et al., 2018). β-galactosidases target the β-galactosidic bonds, which are present in certain disaccharides like lactose (Saqib et al., 2017). This enzyme is therefore crucial for the digestion of lactose by converting it into glucose and galactose (Swallow, 2003). Lactose intolerance in humans arises from a deficiency of this enzyme. β-galactosidases are frequently used in the dairy industry, especially in the manufacture of lactose-free products (Saqib et al., 2017). The A. oryzae β-galactosidase is employed in particular for the production of lactose-free and no-sugar-added yoghurt (Miao et al., 2024). In soy sauce fermentation, β-glucosidase and β-xylanase (see below) are essential for glucose metabolism throughout fermentation, contributing to the development of flavors like alcohols, acids, and esters (Nakadai et al., 1972).
Xylanases are a class of enzymes that degrade the β-1,4-glycosidic linkage between xylose residues of linear polysaccharides (Pirota et al., 2013). As such, they play a crucial role in the decomposition of hemicellulose, one of the major components of plant cell walls (Dodd and Cann, 2009). Given their ability to break down complex carbohydrates, xylanases have garnered significant attention in various industries, particularly in the paper and pulp industry for bleaching paper (Walia et al., 2017) and in the feed industry to improve the digestibility of animal feeds (Ho, 2017; Ojha et al., 2019). Given these applications, A. oryzae is used for the production of xylanases due to its efficiency in enzyme synthesis (Pirota et al., 2013; Kimura et al., 2000; Chipeta et al., 2008). Within A. oryzae, several specialized xylanases have been identified, including ethanol-tolerant xylanase (Sato et al., 2010), thermo-acid/alkali stable xylanase (Bhardwaj et al., 2019), thermo-alkali-stable xylanase (Bhardwaj et al., 2020), thermostable xylanase (He et al., 2015), and low-molecular-weight xylanase (Duarte et al., 2012). These specialized enzymes highlight the versatility and adaptability of A. oryzae in producing xylanases that meet specific industrial needs.
Cellulases are complex enzyme systems responsible for the breakdown of cellulose, the primary structural component of plant cell walls. Cellulose is made up of glucose molecules linked together by β-1,4-glycosidic bonds (Sher et al., 2017). Comprising multiple enzymes, cellulases catalyze the hydrolysis of cellulose into glucose, which can then be utilized by microorganisms or converted into other valuable products. The industrial relevance of cellulases is vast. It plays a pivotal role in several sectors, including the biofuel industry, textile industry, paper and pulp industry, and food industry (Ejaz et al., 2021; Saranraj et al., 2012). In a SSF with A. oryzae, four cellulose-degrading enzymes were identified. Notably, one of these identified cellulases significantly enhanced material utilization and alcohol yield during sake mash fermentation (Yamane et al., 2002b).
Pectinases are enzymes that break down pectin and they are categorized based on their action mechanism as polygalacturonases, lyases, and pectin methylesterases (Sudeep et al., 2020). Pectin, a key component in the cell walls of higher plants, consists of high molecular weight acidic heteropolysaccharides primarily made of α-1,4-linked D-galacturonic acid residues (Kavuthodi and Sebastian, 2018). Pectinases, primarily produced through microbes, play a pivotal role in the food and beverage industry, enhancing juice extraction, improving coffee, cocoa, and tea quality, and increasing oil yield in refineries (Sudeep et al., 2020; Tapre and Jain, 2014; Anand et al., 2017). Additionally, they enhance the digestibility of pectin-rich animal feeds (Azzaz et al., 2021; Tahir et al., 2006). A. oryzae is often used as the chosen microorganism for pectinase production (Ketipally and Ram, 2018; Meneghel et al., 2014; Chen et al., 2014; Hoa and Hung, 2013; Jaramillo et al., 2015; Hoa and Hung, 2013).
5.4 Primary and secondary metabolites produced by Aspergillus oryzae fermentation
Numerous analytical studies including targeted and untargeted approaches revealed that A. oryzae generate a wide variety of primary and secondary metabolites (Table 2). Typically, primary metabolites are directly associated with an organism’s growth and development and are crucial for its regular physiological activities. Secondary metabolites are not directly linked to normal growth and development, but rather play important roles in virulence, host defense, and environmental survival (Vining, 1990; Pagare et al., 2015; Bennett and Wallsgrove, 1994).
Table 2. Metabolites found in Aspergillus oryzae fermentation products and their described bioactivity.
A. oryzae has a long history in fermentative metabolite production. A standout feature of A. oryzae is the production of kojic acid, a versatile secondary metabolite (Rosfarizan et al., 2010; Yabuta, 1924; Yan et al., 2014) first isolated in 1907 from koji-culture (Bentley, 2006). Kojic acid finds applications as antibiotic (Morton et al., 1945; Rodrigues et al., 2022), food preservative (Wang et al., 2021), and antioxidant (Ermis et al., 2023; Yi and Kim, 1982). Its role as a tyrosinase inhibitor (Saruno et al., 1979) has also propelled its use in cosmetics for skin-lightening (Phasha et al., 2022) and in medicine for chloasma treatments (Monteiro et al., 2013). The biosynthesis pathway of kojic acid in A. oryzae is well known (Marui et al., 2011; Terabayashi et al., 2010; Chib et al., 2023).
Aspergilli are very suitable for the industrial production of organic acids. Notably, they play a role in the synthesis of the 1,4-dicarboxylic acids like succinic, malic, and fumaric acids. These acids, integral to the tricarboxylic acid (TCA) cycle in all living organisms, have been highlighted by the U.S. Department of Energy as some of the top high-value chemicals derived from biomass (Werpy and Petersen, 2004). While Aspergillus species are adept at producing fumaric and malic acids, especially under stress (Shigeo et al., 1962), there are variances in organic acid production capabilities across different strains (Goldberg et al., 2006; Yang et al., 2016). A. oryzae is rarely used for the production of succinic acid and fumaric acids, but it has a remarkable potential to produce significant quantities of the former under specific fermentation conditions (Brown et al., 2013; Geyer et al., 2018; Knuf et al., 2013; Knuf et al., 2014; Kövilein et al., 2021; Shigeo et al., 1962). The primary application of malic acid is in the food and beverage sector, where it enhances flavor in products like candies and soft drinks (Kövilein et al., 2020). Its unique taste profile also aids in masking the aftertaste of artificial sweeteners (Aldrich et al., 1979). Beyond this, it finds use in cleaning products (Zhang et al., 2016) and animal feed additive (Stallcup, 1979).
Secondary metabolite production by Aspergillus species varies based on fermentation type and conditions (Son et al., 2018). A. oryzae produces various secondary metabolites, including terpenoids, coumarins, and oxylipins (summarized in Table 2).
5.5 Aspergillus oryzae derived postbiotics
In the previous sections we have described various compounds derived from the fermentation with A. oryzae, the bioactivity of which will next be discussed against the context of their potential use as postbiotics (Figure 4) to confer health benefits on humans or animals. Additionally, it is important to note that the composition and the existing molecules in a potential A. oryzae postbiotic are influenced by the inactivation process. Different drying methods can alter the composition of primary and secondary metabolite profile (Managa et al., 2020; Lu et al., 2024; Zagórska et al., 2022), enzymatic activity (Valadez-Carmona et al., 2017; Wang et al., 2018), and the morphology, conformation, and molecular weight of polysaccharides (Kong et al., 2015; Li W. et al., 2021).
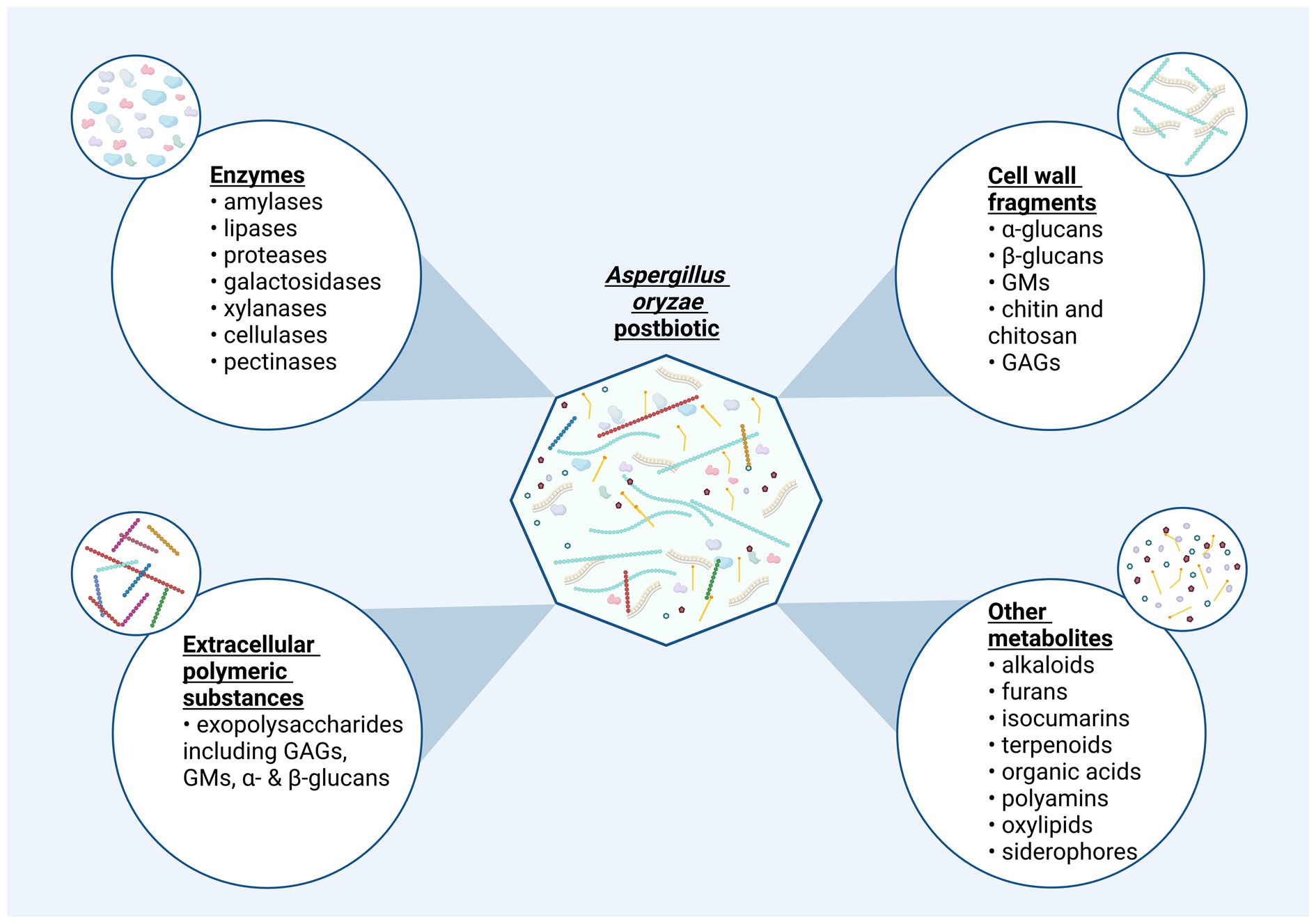
Figure 4. The potential composition of a postbiotic product derived from Aspergillus oryzae. Postbiotics produced via fermentation with A. oryzae may display a diverse molecular composition. Bioactive compounds in this inanimated fermentation preparations can be organized in four categories: “Non-viable cells or cell-fragments,” “enzymes,” “extracellular polymeric substances,” and “other metabolites”. The latter category is reserved for primary and secondary metabolites not fitting the prior classifications. This categorization provides a structured insight into potential ingredients present in postbiotics derived from A. oryzae. Created in BioRender. Seidler, Y. (2024) BioRender.com/f31t998
6 Case studies with Aspergillus oryzae postbiotics
The exploration of A. oryzae for its health-promoting potential has predominantly been centered around its probiotic (Konishi et al., 2021; Dawood et al., 2020a; Lee et al., 2006; Iwashita et al., 2015) or prebiotic capabilities (Hamajima et al., 2016; Kim et al., 2014; Podversich et al., 2023), which are well-documented in the scientific literature. However, the concept of utilizing A. oryzae as a progenitor microorganism for the production of a postbiotic product (or preparation) is a relatively new area of research, as evidenced by the paucity of literature on the postbiotic potential of A. oryzae. Certainly, only three published studies have been found (Kaufman et al., 2021; Ríus et al., 2022), which represent pioneering efforts to characterize and harness the postbiotic properties of A. oryzae. By employing the fruit fly model Drosophila melanogaster, Kaufman et al. (2021) showed that the supplementation of fly diet with an A. oryzae postbiotic improved heat stress tolerance. Remarkably, the same A. oryzae postbiotic increased milk production and reduced inflammatory markers when fed to dairy cows that were exposed to elevated ambient temperatures. In a follow-up study, Ríus et al. (2022) found that the feeding of A. oryzae postbiotic to dairy calves not only mitigated heat-induced reductions in the efficiency of energy use for growth but also improved intestinal roles like barrier function and water absorption, although it did not significantly decrease markers of systemic inflammation. More recently, Junior et al., 2024 examined the effects of A. oryzae postbiotics during gestation and lactation in sows, showing a reduction in body weight loss among sows but no significant effect on litter or nursery performance. These findings suggest A. oryzae postbiotics could be a valuable tool in improving heat tolerance, physiological functions in various animal species, and potentially reducing weight loss in sows during critical periods.
The term “postbiotic” itself is relatively new to the scientific community, which partly explains the scarcity of studies explicitly addressing A. oryzae in this context. Nevertheless, by expanding the search terms to include “inanimate,” “heat killed,” “non-viable,” “inactivated,” “killed,” and “dried,” three additional papers were uncovered (Hymes-Fecht and Casper, 2021; Gomez-Alarcon et al., 1990; Nomura et al., 2022). Although these studies do not label A. oryzae explicitly as a postbiotic, they provide valuable insights into its application and potential health benefits in a state that aligns with the broader definition of postbiotics. Investigating the impact on livestock digestive efficiency, Hymes-Fecht and Casper (2021) found that a dried A. oryzae fermentation product enhanced the degradation of neutral detergent fiber in selected forages. This led to improvements in nutrient absorption and feed efficiency, suggesting beneficial applications in ruminant diets. In a related study, the effectiveness of dried A. oryzae cultures on nutrient utilization was assessed in mature Holstein cows. Their trials showed that dried A. oryzae cultures increase the rumen and total tract digestibility of fiber fractions, facilitating better energy extraction from feed and contributing to enhance animal health and productivity (Gomez-Alarcon et al., 1990). Further emphasizing A. oryzae’s gastrointestinal benefits, Nomura et al. (2022) demonstrated that heat-killed A. oryzae spores significantly increase the population of Bifidobacterium pseudolongum in mice. This beneficial anti-inflammatory gut microbe and the alleviation of colitis symptoms underscore A. oryzae’s potential to enhance gut health and mitigate inflammatory responses. Overall, these studies (Table 3) can be seen as proof of principle that A. oryzae fermentation products can act as postbiotics and help, for instance, to promote digestive health, improve nutrient utilization and increase resistance to inflammation in animals.
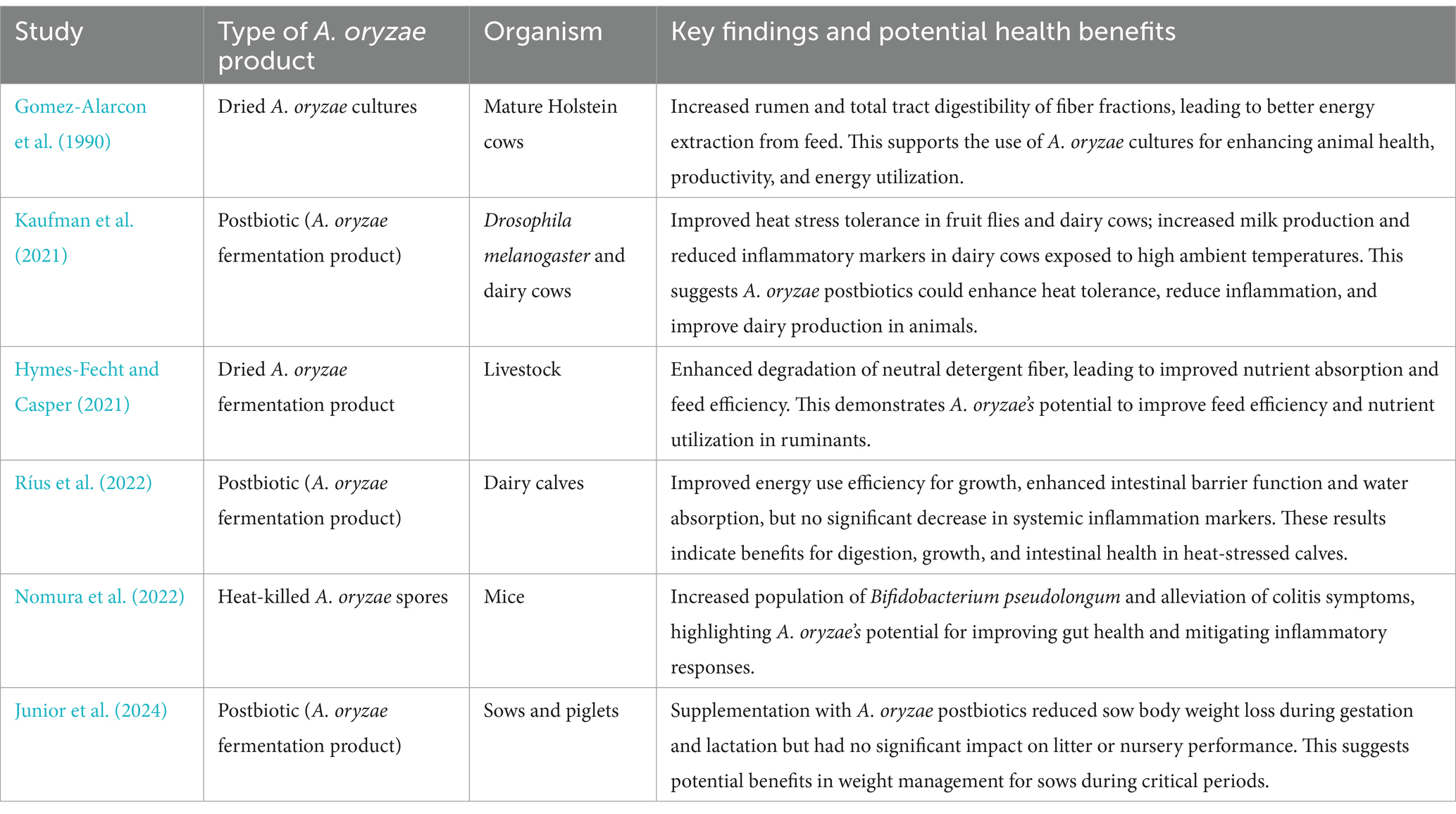
Table 3. Summary of studies investigating the health and performance benefits of Aspergillus oryzae products.
7 Potential mode of action of Aspergillus oryzae postbiotics
The following sections delve into the five key mechanistic components proposed by Salminen et al. (2021) to underpin the beneficial effects of postbiotics. These include modulation of the resident gut microbiota, enhancement of the epithelial barrier function, modulation of local and systemic immune responses, modulation of systemic metabolic responses, and systemic signaling via the nervous system (Salminen et al., 2021). Each mechanism is introduced and explained using examples from organisms other than A. oryzae. Subsequently, we proposed associations between these general mechanisms and the specific constituents or molecules found in A. oryzae. It is important to note that while we attempted to delineate specific and independent effects, there are considerable overlaps between some mechanistic components that are not easily separable. For example, an influence on the gut microbiota composition can alter the endogenous production of downstream effectors like secondary bile acids (Ridlon et al., 2014) and SCFAs (Silva et al., 2020).
7.1 Modulation of the resident gut microbiota
Postbiotics can influence the composition and functionality of the human intestinal microbiome through both direct and indirect means (Ozma et al., 2022). Substances contained in postbiotics, like SCFAs, directly affect the microbiome (Rad et al., 2021). Additionally, EPS from organisms like Bifidobacterium interact with the gut flora (Salazar et al., 2008), and certain metabolites like bacteriocins (Huang et al., 2021) and organic acids (Dittoe et al., 2018) can suppress pathogenic activity within the gut.
Additionally, it’s important to note that postbiotic products often include indigestible fibers that modulate the microbiome. This modulation represents a prebiotic-like effect, indicating that the use of postbiotics does not exclude prebiotic benefits. The presence of these indigestible polysaccharides in postbiotics demonstrates their dual functionality, contributing to microbiota modulation through mechanisms typically associated with prebiotics. A. oryzae is rich in various polysaccharides, primarily known as indigestible fibers. These compounds, including β-glucans (Lam and Chi-Keung Cheung, 2013; Russo et al., 2012), GM (Zartl et al., 2018; Wang H. et al., 2023), chitin and chitosan (Liu et al., 2018b; Guan and Feng, 2022) can act as prebiotics, fostering an environment conducive to the growth of beneficial gut microbiota. Recent research has utilized β-galactosidase from A. oryzae to produce novel galactooligosaccharides from lactose. Purified and analyzed oligosaccharides mainly consist of galactose and glucose. Prebiotic activity tests have shown that these oligosaccharides significantly enhance the growth of Bifidobacterium infantis, especially at higher concentrations, indicating their potential as prebiotics (Kim et al., 2014). Furthermore, koji contains glycosylceramide, which serves as a prebiotic for B. coccoides. Ingestion of purified koji glycosylceramide by mice has led to increased B. coccoides abundance in the intestinal microbiota (Hamajima et al., 2016).
7.2 Enhancement of epithelial barrier function
The integrity of the gut epithelium is crucial for maintaining overall health, serving as a critical barrier against pathogenic invaders and harmful substances (Celebi Sözener et al., 2020). Strategies to influence the epithelial barrier function include promoting the secretion of proteins such as HM0539 (Gao et al., 2019), reducing inflammation (Schiavi et al., 2016), supporting the functioning of tight junctions (Engevik et al., 2019), and providing protection against LPS-induced disruption (Feng et al., 2018). These effects are well-documented for postbiotics, including preparations from Lactobacillus plantarum (Izuddin et al., 2019), Lactobacillus rhamnosus GG (Gao et al., 2019), and Bifidobacterium longum (Martorell et al., 2021). The use of such postbiotics enhance the integrity and functionality of the epithelial barrier, which is crucial for maintaining gut health and preventing the entry of pathogens.
Enzymes produced during A. oryzae fermentation may play a role in this context, potentially influencing the digestibility and bioavailability of nutrients that support epithelial health (Li et al., 2018; He et al., 2020). Xylanase supplementation has been found to improve the intestinal health of broiler chickens, particularly in alleviating barrier impairments caused by Clostridium perfringens infections, according to research by Liu et al. (2012). In a separate study, Petry et al. (2020) demonstrated that xylanase also increases gut barrier integrity in growing pigs. The enhancement of the gut barrier can lead to better nutrient absorption and overall improved health outcomes for the pigs, showcasing the broad applicability of xylanase across different species. Further, research has highlighted the role of phytase in the expression of intestinal tight junction and nutrient transporter genes in pigs (Lu et al., 2020). Lastly, a study by Moita et al. (2021) showed that phytase supplementation enhances intestinal health in broiler chickens by potentially modulating the gut microbiota. It promotes the growth of beneficial bacteria and reduces harmful bacteria, which in turn improves intestinal morphology. These changes are associated with increased nutrient digestibility and improved bone parameters, indicating a direct link between enzyme supplementation and enhanced physiological development in poultry. Additionally, potential constituents of postbiotics from A. oryzae, such as certain furans and alkaloids, may possess the ability to impact directly tight junction proteins (Guo et al., 2020). Furthermore, the expected prebiotic-like effect of preparations from A. oryzae could also influence epithelial barrier function through alterations in the gut microbiome (Rose et al., 2021; Camilleri, 2021).
7.3 Modulation of local and systemic immune responses
Postbiotics are primarily associated with immunoregulatory functions, as they activate both the adaptive and innate immune responses, maintain the integrity of the intestinal mucosal barrier, and counteract microorganisms through the production of antibiotic substances (Ozma et al., 2022). Building upon the comprehensive understanding of the Aspergillus species’ cell wall intricacies, it is imperative to explore the broader biological implications of these molecular constituents. The fungal cell wall, abundant with diverse polysaccharides and biomolecules, serves as the primary interface between the fungus and its host, orchestrating a myriad of immunological interactions. These components, recognized as pathogen-associated molecular patterns (PAMPs), are adeptly discerned by the host’s immune surveillance mechanisms. This recognition is facilitated by a sophisticated network of immune cell receptors, notably the Toll-like receptors (TLRs) and C-type lectin receptors (CLRs). Tailored to detect specific fungal PAMPs, these receptors initiate a series of immune responses aimed at countering the fungal intrusion (see Figure 5).
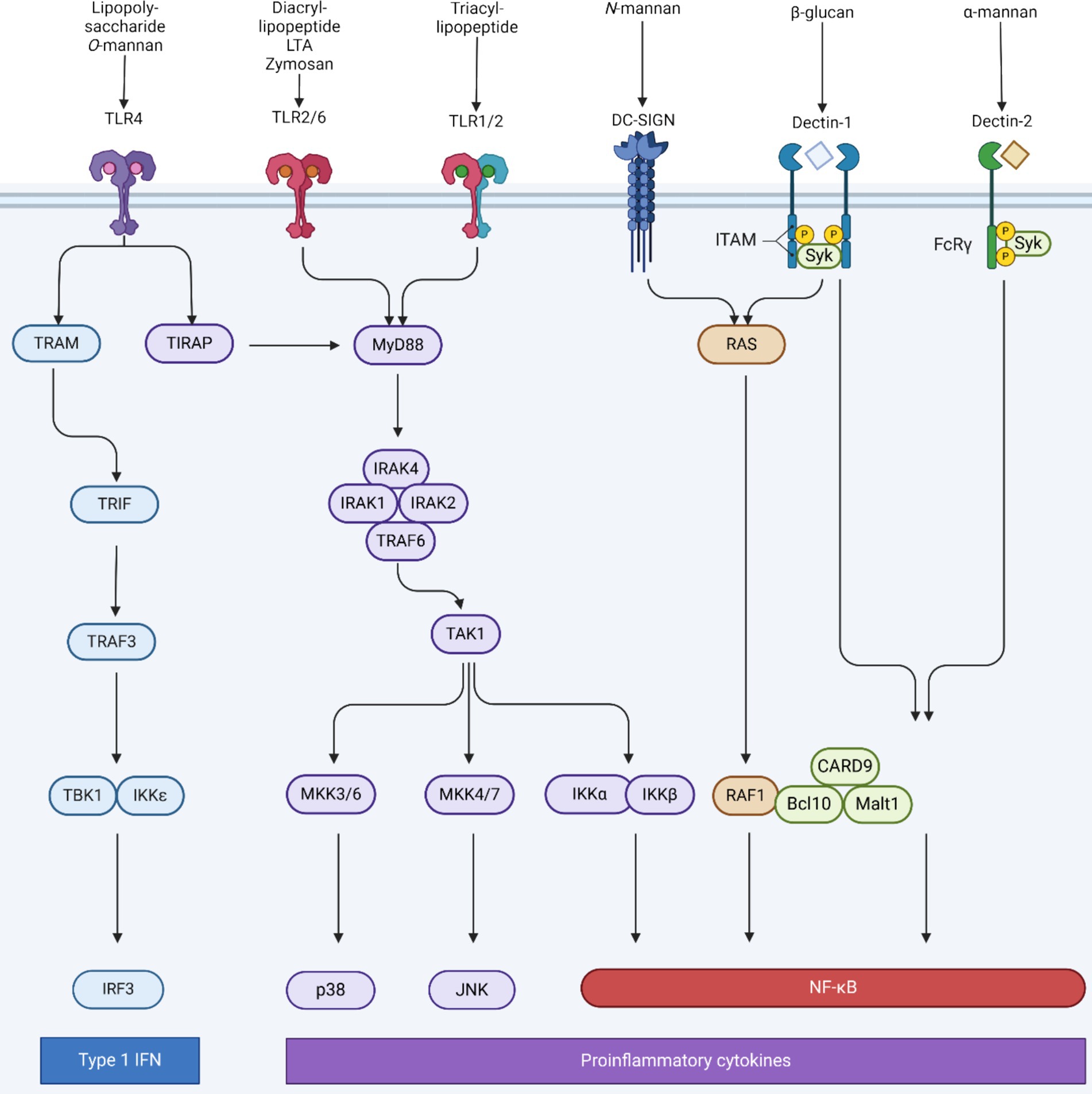
Figure 5. Receptors involved in antifungal immunity, their intracellular signaling pathways and their corresponding fungal PAMPs. Recognition of fungi cell wall components of the cell membrane is mainly mediated by toll-like receptors (TLRs) and C-type lectin receptor (CLRs). TLR2 and TLR4 both signal to interleukin-1 receptor-associated kinase (IRAKs) through myeloid differentiation primary response 88 (MyD88). An IRAK4/IRAK1/IRAK2/tumor necrosis factor receptor-associated factor 6 (TRAF6) complex activates transforming growth factor-beta activated kinase 1 (TAK1) which leads to the activation of the IkappaB kinase (IKK) complex and the mitogen activated protein (MAP) kinase cascade (mitogen activated protein kinase kinase (MKK) 3/6 and MKK4/7), which finally leads to the nuclear translocation of pro-inflammatory transcription factor nuclear factor-kappa B (NF-κB), activator protein (AP-1), and interferons regulatory factor 3 (IRF3). Each transcription factor is responsible for the transcription of specific genes that encodes different set or proteins such as pro-inflammatory cytokines. TLR4 activation also triggers the secretion of interferon α (IFN-α) and IFN-β promoted by Toll/IL-1R domain-containing adapter-inducing factor (TRIF) -mediated interferons regulatory factor 3 (IRF3). Dectin-1 and dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) also activate NF-κB signaling via the rat sarcoma-rapidly accelerated fibrosarcoma 1 (RAS–RAF-1) pathway. Moreover, dectin-1 and dectin-2 recruit the spleen tyrosine kinase (Syk) to form a caspase recruitment domain containing protein 9 (CARD9)/ B-cell lymphoma 10 (BCL-10) / mucosa-associated lymphoid tissue lymphoma translocation protein 1. (MALT1) complex, which activates NF-κB. FcγR, Fcγ receptor; IL, interleukin; JNK, Jun N-terminal kinase; ITAM, immunoreceptor tyrosine-based activation motif. Created with Biorender.com.
Central to the recognition of fungi by the immune system are pattern recognition receptors (PRRs) that identify fungal PAMPs (Amarante-Mendes et al., 2018). There are four major sub-families of PRRs, namely (i) the TLRs, (ii) the nucleotide-binding oligomerization domain (NOD)- like receptors (NLR), (iii) the retinoic acid-inducible gene (RIG)-like receptors, and (iv) the CLRs (Walsh et al., 2013). Among the PRRs, TLRs play a pivotal role in fungal recognition. For instance, TLR2 and TLR4 recognize fungal cell wall components like phospholipomannan and O-linked mannan, respectively (Netea et al., 2008). However, the recognition of fungi is not limited to TLRs. CLRs such as Dectin-1 and Dectin-2 receptor are paramount in sensing β-glucans and the macrophage mannose receptor (MR) recognized N-linked mannans in the fungal cell wall (Netea et al., 2008). NLRs and RIG-like receptors are another layer to the immune surveillance against bacteria. So far, no studies have documented the involvement of NLRs and RIG-I-like receptors in the recognition of fungi (Netea et al., 2008). It is known that the addition of A. oryzae to primary human corneal epithelial cells (HCEC) leads to the upregulation of TLR2 and TLR4 (Ballal et al., 2017). Similarly, in male broiler chicks, the mRNA expression of immune mediators in the intestine was studied, showing that TLR4 expression in the A. oryzae and antibiotic groups was higher than in the control group (Takahashi, 2012).
In many biological scenarios, the activation of cellular receptors is not confined to a singular entity; rather, multiple receptors can be concurrently activated. It is important to acknowledge that PRRs can exhibit inhibitory interactions, particularly when faced with a variety of pathogens. Ferwerda et al. (2008) showed that Dectin-1 receptor has potent synergistic effects with both TLR2 and TLR4 in human peripheral blood mononuclear cells (PBMCs) and macrophages. Loures et al. (2015) indicated that TLRs and CLRs are both involved in the induction of lymphocyte proliferation and Th17/Tc17 differentiation mediated by Paracoccidiodes brasiliensis activated dendritic cells (DCs), but a synergist action was restricted to Dectin-1 receptor, TLR-4, and MR. Other research showed that pure TLR2 and TLR4 ligands generate macrophages with a diminished ability to produce inflammatory cytokines. In contrast, mouse hematopoietic stem and progenitor cells (HSPCs) activation in response to Candida albicans leads to the generation of macrophages that are better prepared to deal with the infection, as they produce higher amounts of inflammatory cytokines and have higher fungicidal capacity than control macrophages (Megías et al., 2016). It was reported that simultaneous stimulation with TLR2 and TLR4 ligands results in the production of tumor necrosis factor alpha (TNF-α) at levels much greater than that observed for each of the ligands alone (Sato et al., 2000). Research has also identified other combinations of TLR ligands in DCs that can boost the production of IL-12 and IL-23, leading to DCs with enhanced and sustained T helper type 1–polarizing capacity (Napolitani et al., 2005). This combined TLR stimulation has been used to create an adjuvant to improve T cell responses to vaccines. For instance, stimulating with three TLRs (TLR2/6, TLR3, and TLR9) can not only increase the number of T cells but also change the quality of the immune response, promoting the growth of regulatory T cells (Zhu et al., 2010). In summary, the combined stimulation of TLRs and CLRs, especially Dectin-1 receptor, significantly influences immune responses. The distinct reactions of macrophages and DCs highlight their unique roles in immunity. The synergistic effects of specific combinations, particularly in vaccine adjuvants, open promising therapeutic avenues, emphasizing the potential of targeted immune modulation in addressing infections.
In Nile tilapia, various studies have demonstrated that the use of A. oryzae as probiotic positively impacts the immune system, oxidative stress response, growth, and disease resistance (Dawood et al., 2020b; Dawood et al., 2020a; Iwashita et al., 2015). In red sea breams fed with A. oryzae fermented rapeseed meal, a significantly enhanced immunological response was observed (Dossou et al., 2018). In a study in which mice underwent acute inflammation caused by ear edema, the feeding of a diet containing 10% rice bran with A. oryzae reduced inflammation severity (Umeyama et al., 2021). Similarly, it has been shown in cattle that supplementation with an A. oryzae postbiotic reduced inflammation in response to heat stress (Kaufman et al., 2021). In a recent in vitro study, A. oryzae fermentation extract inhibited Mycoplasma pneumoniae growth and invasion into A549 lung epithelial cells and reduced the production of TNF-α and IL-6 in murine MH-S alveolar macrophages. Subsequently, in an in vivo mouse model of pneumonia, the extract decreased neutrophil infiltration and lung inflammation, demonstrating its potential as a therapeutic agent (Lee et al., 2023).
In summary, the comprehensive understanding of Aspergillus species’ cell wall components and their interactions with the host’s immune system highlights the potential for A. oryzae-derived postbiotics to exert significant immunomodulatory effects. These postbiotics may have the capability to modulate both local and systemic immune responses, thereby potentially enhancing host defense mechanisms against pathogens.
7.4 Modulation of systemic metabolic responses
The influence on systemic metabolic responses could stem directly from the metabolites or enzymes harbored within inactive microorganisms commonly found in postbiotics (Salminen et al., 2021). For instance, a recent study conducted by Travers et al. (2016) showcased that the supernatant of Lactobacillus johnsonii, regarded as a postbiotic, harbors active elements proficient in transforming benign bile constituents into markedly toxic compounds for Giardia duodenalis. This exemplifies how postbiotics can influence bile acid metabolism, underscoring their potential in addressing parasitic infections. Furthermore, A. oryzae fermentation end products have been implicated in influencing bile acids. Feeding obese mice koji glycosylceramide led to a significant reduction in liver cholesterol levels, likely due to cholesterol conversion into bile acids. This indicates that koji glycosylceramide impacts both bile acid and cholesterol metabolism in obese mice (Hamajima et al., 2019). The secondary metabolites produced by A. oryzae have been shown to possess various biological properties, including antioxidative, antimicrobial, and antitumor effects (see Table 2). These properties suggest a potential role for A. oryzae postbiotics in modulating systemic metabolic responses, although the specific mechanisms through which these effects are mediated remain to be fully elucidated. The concentration and bioavailability of these metabolites in the final product are likely crucial factors dictating their biological efficacy.
7.5 Systemic signaling via the nervous system
By signaling through the nervous system and ultimately altering the delivery of neuroactive compounds, microorganisms can influence host behavior and cognitive function. This interplay, enriched by microbiome biotransformation, exposes the host to bioactive products affecting gastrointestinal-central nervous system communication. These compounds modulate central physiological and pathological processes via receptor binding, vagus nerve stimulation, neurotransmission, and neuroinflammation. Therefore, understanding the impact of SCFA, bile acids, neurotransmitters, and other microbial products in the gut-brain axis is crucial (Caspani and Swann, 2019).
While direct evidence linking A. oryzae postbiotics to systemic signaling via the nervous system is limited, the potential for β-glucans (Singh and Bhardwaj, 2023), glucosylceramides (Hamajima et al., 2019), and other polysaccharides (Nomura et al., 2022) to impact the gut-brain axis is an area of great interest. These components could modulate neurological health and behavior through their effects on the gut microbiota, bile acid metabolism, and immune function, highlighting an exciting frontier for future research.
8 Challenges and opportunities in quality control of fungal postbiotics
Within the realm of postbiotic research, the advent of fungal-derived postbiotics represents a step toward exploring a previously untapped source of beneficial compounds for human and animal health. The process of inactivating microbial cells to produce postbiotics poses no greater challenge for fungi than it does for bacteria.
Techniques such as spray drying (Stephan and Zimmermann, 1998), high-pressure processing (Pinto et al., 2020), heat inactivation (Silva, 2020), thermosonication (Silva, 2020), and the creation of lysates are equally applicable to fungal cells, providing a robust framework for the development of fungal postbiotics. However, the rigorous quality control and quantification of these fungal postbiotics emerge as a paramount challenge, underscored by the unique biological characteristics of fungi and the complex biochemical composition of their derivatives.
In their publication, Salminen et al. (2021) recognized the necessity of providing clear guidance on the technical aspects of postbiotic characterization and quantification. For bacteria, metrics such as colony-forming units (CFU) and cell sorting techniques offer straightforward quantification methods. In contrast, the fungal domain presents a more intricate scenario. Fungal cells, with their varied sizes, multicellular structures, and resilient spore forms, necessitate the development of bespoke quantification strategies that go beyond the bacterial paradigms. It is noteworthy that in most studies focusing on koji production, the quantification of GlcNAc has been utilized as an index for the mycelial weight of A. oryzae (Ferdouse et al., 2019). In the study by Ferdouse et al. (2019), alongside the quantification of GlcNAc, the content of glycosylceramide was measured.
Current methodologies, including flow cytometry, can adeptly measure inanimate intact microbes, distinguishing between live, dead, and damaged cells (Robertson et al., 2021; Duquenoy et al., 2020; Baymiev et al., 2020). However, analyzing fungi is, often requiring indirect quantification like biomass estimation, which may not fully capture the postbiotic potential. Flow cytometry is underutilized for filamentous fungi due to the size of hyphae, which cannot pass through the system, limiting analysis to early-stage spores (Mathis et al., 2020). Despite this, it shows promise for spore sorting and viability testing. For fungi like Trichoderma, conidia can be effectively analyzed using various dyes and markers like GFP, with protocols for preparing and staining cells (Steiger, 2021).
Moreover, the identification and quantification of metabolites, which is crucial for understanding the bioactive profile of fungal postbiotics, may demand the use of sophisticated analytical techniques such as high-performance liquid chromatography (HPLC) and mass spectrometry. Yet, the heterogeneity of fungal metabolites and the need for specific markers for bioactivity assessment pose significant hurdles in standardizing these measures.
9 Conclusion and future perspectives
Although A. oryzae has long been used in the food sector, its application as a postbiotic is still in its infancy. An important basis for assessing its postbiotic potential is the systematic analysis of its components, which has only recently been initiated. As summarized here, numerous interesting molecules have already been identified, including secondary metabolites, cell wall components and enzymes, whose different bioactivities could potentially cover all five of the postbiotic targets described by Salminen et al. (2021), suggesting a broad spectrum of health benefits. Nevertheless, these analyses have not yet been completed. For example, a detailed understanding of the structure of the cell wall is still lacking. It can also be assumed that further bioactive substances produced by A. oryzae will be discovered in the future. Most of all, however, there is currently a lack of controlled studies investigating the postbiotic potential of A. oryzae in vivo. The limited published research primarily focused on the impacts of A. oryzae postbiotics on animal health and nutrition. Findings from those studies suggest that A. oryzae fermentation end products, whether in dried or otherwise inactivated form, may confer beneficial effects by improving gut health, promoting immune responses, and enhancing nutrient absorption. Hence, we conclude that the exploration of A. oryzae as a source of postbiotics represents a promising avenue in the field of biotics. However, to unlock the full therapeutic potential of A. oryzae-derived postbiotics and deepen our understanding of their role in promoting health and well-being, future research should aim to elucidate the specific mechanisms of action of A. oryzae postbiotics and investigate their therapeutic applications in different health and disease contexts.
As postbiotics from fungi such as A. oryzae is a relatively new research field, it also presents challenges in manufacturing and quality control. Standardized fermentation protocols are required to guarantee reproducible compositions of postbiotic products. Moreover, while the methods for inactivating fungi are broadly similar to those for the much better established bacterial postbiotics, a major challenge is the development of rigorous quality assurance and quantification methods of these novel fungal products. Meeting this demand will require a concerted effort by researchers utilizing state-of-the-art technologies and interdisciplinary approaches to create robust standards for the development of fungal postbiotics. Indeed, commitment in this area will be crucial to realize the full potential of fungal postbiotics for supporting human and animal health.
Author contributions
YS: Writing – review & editing, Writing – original draft. GR: Writing – review & editing. KL: Writing – review & editing. GV: Writing – review & editing. II: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors acknowledge the financial support by Land Schleswig-Holstein within the funding program “Open Access Publikationsfonds.”
Conflict of interest
The authors declare that the Institute of Human Nutrition and Food Science (including YS, GR, II, and KL) received funding from BioZyme Incorporated. YS and II received consulting fees from BioZyme Incorporated. GV is a board member of the ISAPP.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adeleke, B. S., and Babalola, O. O. (2021). Pharmacological potential of fungal endophytes associated with medicinal plants: a review. J. Fungi 7:147. doi: 10.3390/jof7020147
Akasaka, N., Kato, S., Kato, S., Hidese, R., Wagu, Y., Sakoda, H., et al. (2018). Agmatine production by Aspergillus oryzae is elevated by low pH during solid-state cultivation. Appl. Environ. Microbiol. 84:18. doi: 10.1128/AEM.00722-18
Aldrich, D., Vink, W., Deptula, R. W., Muskus, D. J., Fronczkowski, P. R., and Chrusch, M. (1979). Sugarless candies. Google patents.
Amaike, S., and Keller, N. P. (2011). Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133. doi: 10.1146/annurev-phyto-072910-095221
Amarante-Mendes, G. P., Adjemian, S., Branco, L. M., Zanetti, L. C., Weinlich, R., and Bortoluci, K. R. (2018). Pattern recognition receptors and the host cell death molecular machinery. Front. Immunol. 9:2379. doi: 10.3389/fimmu.2018.02379
Anand, G., Yadav, S., and Yadav, D. (2017). Production, purification and biochemical characterization of an exo-polygalacturonase from Aspergillus niger MTCC 478 suitable for clarification of orange juice. 3 Biotech 7:122. doi: 10.1007/s13205-017-0760-3
Ancheeva, E., Daletos, G., and Proksch, P. (2020). Bioactive secondary metabolites from endophytic fungi. Curr. Med. Chem. 27, 1836–1854. doi: 10.2174/0929867326666190916144709
Andrino, A., Guggenberger, G., Kernchen, S., Mikutta, R., Sauheitl, L., and Boy, J. (2021). Production of organic acids by arbuscular mycorrhizal fungi and their contribution in the mobilization of phosphorus bound to iron oxides. Front. Plant Sci. 12:842. doi: 10.3389/fpls.2021.661842
Antipova, T. V., Zaitsev, K. V., Oprunenko, Y. F., Zherebker, A. Y., Rystsov, G. K., Zemskova, M. Y., et al. (2019). Austalides V and W, new meroterpenoids from the fungus Aspergillus ustus and their antitumor activities. Bioorg. Med. Chem. Lett. 29:126708. doi: 10.1016/j.bmcl.2019.126708
Arnau, J., Yaver, D., and Hjort, C. M. (2020). “Strategies and challenges for the development of industrial enzymes using fungal cell factories” in Grand Challenges in fungal biotechnology. ed. H. Nevalainen (Berlin: Springer Nature).
Asgarivessal, M., Mirzaei, S., and Soltani, J. (2023). Heterothallism and sexual reproduction in the Iranian isolates of Aspergillus flavus. Mycol. Iran. 10, 33–44. doi: 10.22043/MI.2023.362488.1260
Azzaz, H. H., Kholif, A. E., Murad, H. A., El-Bordeny, N. E., Ebeid, H. M., Hassaan, N. A., et al. (2021). A new pectinase produced from Aspergillus terreus compared with a commercial pectinase enhanced feed digestion, milk production and milk fatty acid profile of Damascus goats fed pectin-rich diet. Ann. Anim. Sci. 21, 639–656. doi: 10.2478/aoas-2020-0083
Bachheti, R. K., Abate, L., Bachheti, A., Madhusudhan, A., and Husen, A. (2021). “Algae-, fungi-, and yeast-mediated biological synthesis of nanoparticles and their various biomedical applications” in Handbook of greener synthesis of nanomaterials and compounds. eds. B. I. Kharisov and O. Kharissova (Amsterdam, The Netherlands: Elsevier), 701–734.
Badar, R., Yaqoob, S., Ahmed, A., and Shaoor, Q. U. (2021). Screening and optimization of submerged fermentation of Aspergillus species for kojic acid production. Res. Square 2021:3340. doi: 10.21203/rs.3.rs-334019/v1
Bahram, M., and Netherway, T. (2022). Fungi as mediators linking organisms and ecosystems. FEMS Microbiol. Rev. 46:58. doi: 10.1093/femsre/fuab058
Balakrishnan, K. (1996). Production of biologically active secondary metabolites in solid state fermentation. J. Sci. Ind. Res. 55, 365–372.
Ballal, S., Belur, S., Laha, P., Roy, S., Swamy, B. M., and Inamdar, S. R. (2017). Mitogenic lectins from Cephalosporium curvulum (CSL) and Aspergillus oryzae (AOL) mediate host–pathogen interactions leading to mycotic keratitis. Mol. Cell. Biochem. 434, 209–219. doi: 10.1007/s11010-017-3050-9
Barbesgaard, P., Heldt-Hansen, H. P., and Diderichsen, B. (1992). On the safety of Aspergillus oryzae. A review. Appl. Microbiol. Biotechnol. 36:230. doi: 10.1007/BF00183230
Bardalaye, P. C., and Nordin, J. H. (1977). Chemical structure of the galactomannan from the cell wall of Aspergillus niger. J. Biol. Chem. 252. doi: 10.1016/S0021-9258(17)40498-4
Barreto-Berter, E. M., Travassos, L. R., and Gorin, P. A. J. (1980). Chemical structure of the D-galacto-D-mannan component from hyphae of Aspergillus niger and other Aspergillus spp. Carbohydr. Res. 86. doi: 10.1016/S0008-6215(00)85904-2
Bartnicki-Garcia, S. (1968). Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu. Rev. Microbiol. 22. doi: 10.1146/annurev.mi.22.100168.000511
Baymiev, A. K., Baymiev, A. K., Kuluev, B. R., Shvets, K. Y., Yamidanov, R. S., Matniyazov, R. T., et al. (2020). Modern approaches to differentiation of live and dead bacteria using selective amplification of nucleic acids. Microbiology 89, 13–27. doi: 10.1134/S0026261720010038
Bennett, J. E., Bhattacharjee, A. K., and Glaudemans, C. P. J. (1985). Galactofuranosyl groups are immunodominant in Aspergillus fumigatus galactomannan. Mol. Immunol. 22. doi: 10.1016/0161-5890(85)90158-0
Bennett, R. N., and Wallsgrove, R. M. (1994). Secondary metabolites in plant defence mechanisms. New Phytol. 127, 617–633. doi: 10.1111/j.1469-8137.1994.tb02968.x
Bentley, R. (2006). From miso, sake and shoyu to cosmetics. A century of science for kojic acid. Nat. Prod. Rep. 23, 1046–1062. doi: 10.1039/b603758p
Bhardwaj, N., Kumar, B., Agarwal, K., Chaturvedi, V., and Verma, P. (2019). Purification and characterization of a thermo-acid/alkali stable xylanases from Aspergillus oryzae LC1 and its application in Xylo-oligosaccharides production from lignocellulosic agricultural wastes. Int. J. Biol. Macromol. 122, 1191–1202. doi: 10.1016/j.ijbiomac.2018.09.070
Bhardwaj, N., Verma, V. K., Chaturvedi, V., and Verma, P. (2020). Cloning, expression and characterization of a thermo-alkali-stable xylanase from Aspergillus oryzae LC1 in Escherichia coli BL21 (DE3). Protein Expr. Purif. 168:105551. doi: 10.1016/j.pep.2019.105551
Boateng, J. S., Matthews, K. H., Stevens, H. N. E., and Eccleston, G. M. (2008). Wound healing dressings and drug delivery systems: A review. J. Pharm. Sci. 97. doi: 10.1002/jps.21210
Bowman, S. M., and Free, S. J. (2006). The structure and synthesis of the fungal cell wall. BioEssays 28, 799–808. doi: 10.1002/bies.20441
Brown, S. H., Bashkirova, L., Berka, R., Chandler, T., Doty, T., McCall, K., et al. (2013). Metabolic engineering of Aspergillus oryzae NRRL 3488 for increased production of L-malic acid. Appl. Microbiol. Biotechnol. 97, 8903–8912. doi: 10.1007/s00253-013-5132-2
Camilleri, M. (2021). Human intestinal barrier: effects of stressors, diet, prebiotics, and probiotics. Clin. Transl. Gastroenterol. 12:e00308. doi: 10.14309/ctg.0000000000000308
Caspani, G., and Swann, J. (2019). Small talk: microbial metabolites involved in the signaling from microbiota to brain. Curr. Opin. Pharmacol. 48, 99–106. doi: 10.1016/j.coph.2019.08.001
Celebi Sözener, Z., Cevhertas, L., Nadeau, K., Akdis, M., and Akdis, C. A. (2020). Environmental factors in epithelial barrier dysfunction. J. Allergy Clin. Immunol. 145, 1517–1528. doi: 10.1016/j.jaci.2020.04.024
Chancharoonpong, C., Hsieh, P.-C., and Sheu, S.-C. (2012). Enzyme production and growth of Aspergillus oryzae S. On soybean koji fermentation. APCBEE Proc. 2, 57–61. doi: 10.1016/j.apcbee.2012.06.011
Chang, P.-K. (2019). Genome-wide nucleotide variation distinguishes Aspergillus flavus from Aspergillus oryzae and helps to reveal origins of atoxigenic A. flavus biocontrol strains. J. Appl. Microbiol. 127, 1511–1520. doi: 10.1111/jam.14419
Cheah, I. K., and Halliwell, B. (2012). Ergothioneine; antioxidant potential, physiological function and role in disease. Biochim. Biophys. Acta 1822, 784–793. doi: 10.1016/j.bbadis.2011.09.017
Chen, J., Yang, R., Chen, M., Wang, S., Li, P., Xia, Y., et al. (2014). Production optimization and expression of pectin releasing enzyme from Aspergillus oryzae PO. Carbohydr. Polym. 101, 89–95. doi: 10.1016/j.carbpol.2013.09.011
Chen, T., Yang, W., Li, T., Yin, Y., Liu, Y., Wang, B., et al. (2022). Hemiacetalmeroterpenoids A-C and Astellolide Q with antimicrobial activity from the marine-derived fungus Penicillium sp. N-5. Mar. Drugs 20:514. doi: 10.3390/md20080514
Chib, S., Jamwal, V. L., Kumar, V., Gandhi, S. G., and Saran, S. (2023). Fungal production of kojic acid and its industrial applications. Appl. Microbiol. Biotechnol. 107, 2111–2130. doi: 10.1007/s00253-023-12451-1
Chigira, Y., Abe, K., Gomi, K., and Nakajima, T. (2002). chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr. Genet. 41, 261–267. doi: 10.1007/s00294-002-0305-z
Chipeta, Z. A., Du Preez, J. C., and Christopher, L. (2008). Effect of cultivation pH and agitation rate on growth and xylanase production by Aspergillus oryzae in spent sulphite liquor. J. Ind. Microbiol. Biotechnol. 35, 587–594. doi: 10.1007/s10295-008-0320-2
Choi, D., Alshannaq, A. F., and Yu, J.-H. (2024). Safe and effective degradation of aflatoxins by food-grade culture broth of Aspergillus oryzae. PNAS Nexus 3:271. doi: 10.1093/pnasnexus/pgae271
Coleine, C., Stajich, J. E., and Selbmann, L. (2022). Fungi are key players in extreme ecosystems. Trends Ecol. Evol. 37, 517–528. doi: 10.1016/j.tree.2022.02.002
Courteix, C., Privat, A.-M., Pélissier, T., Hernandez, A., Eschalier, A., and Fialip, J. (2007). Agmatine induces Antihyperalgesic effects in diabetic rats and a Superadditive interaction with R(−)-3-(2-Carboxypiperazine-4-yl)-propyl-1-phosphonic acid, a N-methyl-d-aspartate-receptor antagonist. J. Pharmacol. Exp. Ther. 322, 1237–1245. doi: 10.1124/jpet.107.123018
Cruz-Casas, D. E., Aguilar, C. N., Ascacio-Valdés, J. A., Rodríguez-Herrera, R., Chávez-González, M. L., and Flores-Gallegos, A. C. (2021). Enzymatic hydrolysis and microbial fermentation: the most favorable biotechnological methods for the release of bioactive peptides. Food Chem. 3:100047. doi: 10.1016/j.fochms.2021.100047
Daba, G. M., Mostafa, F. A., and Elkhateeb, W. A. (2021). The ancient koji mold (Aspergillus oryzae) as a modern biotechnological tool. Bioresour. Bioprocess. 8:52. doi: 10.1186/s40643-021-00408-z
Daengrot, C., Rukachaisirikul, V., Tansakul, C., Thongpanchang, T., Phongpaichit, S., Bowornwiriyapan, K., et al. (2015). Eremophilane Sesquiterpenes and diphenyl Thioethers from the soil fungus Penicillium copticola PSU-RSPG138. J. Nat. Prod. 78, 615–622. doi: 10.1021/np5005328
Dahiya, S., Bajaj, B. K., Kumar, A., Tiwari, S. K., and Singh, B. (2020). A review on biotechnological potential of multifarious enzymes in bread making. Process Biochem. 99, 290–306. doi: 10.1016/j.procbio.2020.09.002
Dash, M., Chiellini, F., Ottenbrite, R. M., and Chiellini, E. (2011). Chitosan—A versatile semi-synthetic polymer in biomedical applications. Prog. Polym. Sci. 36. doi: 10.1016/j.progpolymsci.2011.02.001
Dawood, M. A. O., Eweedah, N. M., Moustafa, E. M., and Farahat, E. M. (2020a). Probiotic effects of Aspergillus oryzae on the oxidative status, heat shock protein, and immune related gene expression of Nile tilapia (Oreochromis niloticus) under hypoxia challenge. Aquaculture 520:734669. doi: 10.1016/j.aquaculture.2019.734669
Dawood, M. A. O., Eweedah, N. M., Moustafa, E. M., and Shahin, M. G. (2020b). Synbiotic effects of Aspergillus oryzae and β-glucan on growth and oxidative and immune responses of nile Tilapia, Oreochromis niloticus. Probiotics Antimicrob. Proteins 12, 172–183. doi: 10.1007/s12602-018-9513-9
De Castro, R. J. S., and Sato, H. H. (2014). Protease from Aspergillus oryzae. Biochemical characterization and application as a potential biocatalyst for production of protein hydrolysates with antioxidant activities. J. Food Proc. 2014:372352. doi: 10.1155/2014/372352
Deng, S., Pomraning, K. R., Bohutskyi, P., and Magnuson, J. K. (2018). Draft genome sequence of Aspergillus oryzae ATCC 12892. Genome Announc. 6:18. doi: 10.1128/genomeA.00251-18
Devi, R., Kaur, T., Guleria, G., Rana, K. L., Kour, D., Yadav, N., et al. (2020). “Fungal secondary metabolites and their biotechnological applications for human health” in New and future developments in microbial biotechnology and bioengineering. ed. V. G. Gupta (Amsterdam, The Netherlands: Elsevier), 147–161.
Dhananjay, S. K., and Mulimani, V. H. (2009). Three-phase partitioning of α-galactosidase from fermented media of Aspergillus oryzae and comparison with conventional purification techniques. J. Ind. Microbiol. Biotechnol. 36, 123–128. doi: 10.1007/s10295-008-0479-6
Dittoe, D. K., Ricke, S. C., and Kiess, A. S. (2018). Organic acids and potential for modifying the avian gastrointestinal tract and reducing pathogens and disease. Front. Vet. Sci. 5:216. doi: 10.3389/fvets.2018.00216
Dodd, D., and Cann, I. K. O. (2009). Enzymatic deconstruction of xylan for biofuel production. GCB Bioenergy 1, 2–17. doi: 10.1111/j.1757-1707.2009.01004.x
Dossou, S., Koshio, S., Ishikawa, M., Yokoyama, S., Dawood, M. A., El Basuini, M. F., et al. (2018). Growth performance, blood health, antioxidant status and immune response in red sea bream (Pagrus major) fed Aspergillus oryzae fermented rapeseed meal (RM-Koji). Fish Shellfish Immunol. 75, 253–262. doi: 10.1016/j.fsi.2018.01.032
Duarte, G. C., Moreira, S., Gómez-Mendoza, D. P., Siqueira, F. G., Batista, L. R., Amaral, L. I. V., et al. (2012). Use of residual biomass from the textile industry as carbon source for production of a low-molecular-weight xylanase from Aspergillus oryzae. Appl. Sci. 2, 754–772. doi: 10.3390/app2040754
Dubos, R., Schaedler, R. W., Costello, R., and Hoet, P. (1965). Indigenous, normal, and autochthonous flora of the gastrointestinal tract. J. Exp. Med. 122, 67–76. doi: 10.1084/jem.122.1.67
Duquenoy, A., Bellais, S., Gasc, C., Schwintner, C., Dore, J., and Thomas, V. (2020). Assessment of gram-and viability-staining methods for quantifying bacterial community dynamics using flow cytometry. Front. Microbiol. 11:1469. doi: 10.3389/fmicb.2020.01469
Dusengemungu, L., Kasali, G., Gwanama, C., and Mubemba, B. (2021). Overview of fungal bioleaching of metals. Environ. Adv. 5:100083. doi: 10.1016/j.envadv.2021.100083
Ejaz, U., Sohail, M., and Ghanemi, A. (2021). Cellulases. From bioactivity to a variety of industrial applications. Biomimetics 6:44. doi: 10.3390/biomimetics6030044
El Knidri, H., Belaabed, R., Addaou, A., Laajeb, A., and Lahsini, A. (2018). Extraction, chemical modification and characterization of chitin and chitosan. Int. J. Biol. Macromol. 120, 1181–1189.
El Sheikha, A. F., and Ray, R. C. (2023). Bioprocessing of horticultural wastes by solid-state fermentation into value-added/innovative bioproducts: a review. Food Rev. Intl. 39, 3009–3065. doi: 10.1080/87559129.2021.2004161
Elsehemy, I. A., Noor El Deen, A. M., Awad, H. M., Kalaba, M. H., Moghannem, S. A., Tolba, I. H., et al. (2020). Structural, physical characteristics and biological activities assessment of scleroglucan from a local strain Athelia rolfsii TEMG. Int. J. Biol. Macromol. 163, 1196–1207. doi: 10.1016/j.ijbiomac.2020.06.272
Endo, A., Hasum, K., Hasumi, K., Sakai, K., and Kanbe, T. (1985). Specific inhibition of glyceraldehyde-3-phosphate dehydrogenase by koningic acid (heptelidic acid). J. Antibiot. 38, 920–925. doi: 10.7164/antibiotics.38.920
Engevik, M. A., Luk, B., Chang-Graham, A. L., Hall, A., Herrmann, B., Ruan, W., et al. (2019). Bifidobacterium dentium fortifies the intestinal mucus layer via autophagy and calcium signaling pathways. MBio 10:01087-19. doi: 10.1128/mBio.01087-19
Ermis, N., Zare, N., Darabi, R., Alizadeh, M., Karimi, F., Singh, J., et al. (2023). Recent advantage in electrochemical monitoring of gallic acid and kojic acid. A new perspective in food science. J. Food Meas. Charact. 1, 1–10. doi: 10.1007/s11694-023-01881-0
Esmeralda-Guzmán, M., Juan, J. B. F., Aguilar, C. N., Juan, A. A. V., and Sepúlveda, L. (2024). Parameters that influence the fermentation submerged process: bioprocess development in biotechnology. In: L. S. Torre, J. C. Contreras-Esquivel, A. R. Abraham, and A. K. Haghi Bioresources and Bioprocess in Biotechnology for a Sustainable Future. Boca Raton, FL: CRC Press, pp. 179–200.
Eugster, P. J., Salamin, K., Grouzmann, E., and Monod, M. (2015). Production and characterization of two major Aspergillus oryzae secreted prolyl endopeptidases able to efficiently digest proline-rich peptides of gliadin. Microbiology 161, 2277–2288. doi: 10.1099/mic.0.000198
Fadel, M., AbdEl-Halim, S., Sharada, H., Yehia, A., and Ammar, M. (2020). Production of glucoamylase, α-amylase and cellulase by Aspergillus oryzae F-923 cultivated on wheat bran under solid state fermentation. J. Adv. Biol. Biotechnol. 23, 8–22. doi: 10.9734/jabb/2020/v23i430149
FAO/WHO (2001). Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria: Joint FAO/WHO expert consultation. Cordoba, Argentina: FAO/WHO.
Feng, Y., Wang, Y., Wang, P., Huang, Y., and Wang, F. (2018). Short-chain fatty acids manifest stimulative and protective effects on intestinal barrier function through the inhibition of NLRP3 inflammasome and autophagy. Cell. Physiol. Biochem. 49, 190–205. doi: 10.1159/000492853
Ferdouse, J., Miyagawa, M., Hirano, M., Kitajima, Y., Inaba, S., and Kitagaki, H. (2019). A new method for determining the mycelial weight of the koji-mold Aspergillus oryzae by measuring its glycosylceramide content. J. Gen. Appl. Microbiol. 65, 34–38. doi: 10.2323/jgam.2018.04.003
Ferwerda, G., Meyer-Wentrup, F., Kullberg, B.-J., Netea, M. G., and Adema, G. J. (2008). Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell. Microbiol. 10, 2058–2066. doi: 10.1111/j.1462-5822.2008.01188.x
Fleming, A. (1929). On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. Influenzae. Br. J. Exp. Pathol. 10:226.
Fontaine, T., Delangle, A., Simenel, C., Coddeville, B., van Vliet, S. J., van Kooyk, Y., et al. (2011). Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus. PLOS Pathogens 7. doi: 10.1371/journal.ppat.1002372
Fontaine, T., Simenel, C., Dubreucq, G., Adam, O., Delepierre, M., Lemoine, J., et al. (2000). Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 275, 27594–27607. doi: 10.1074/jbc.M909975199
Fouad, A. M., Ruan, D., El-Senousey, H. K., Chen, W., Jiang, S., and Zheng, C. (2019). Harmful effects and control strategies of aflatoxin b1 produced by Aspergillus flavus and Aspergillus parasiticus strains on poultry. Toxins 11:176. doi: 10.3390/toxins11030176
Frisvad, J. C., Hubka, V., Ezekiel, C. N., Hong, S.-B., Novßkovß, A., Chen, A. J., et al. (2019). Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 93, 1–63. doi: 10.1016/j.simyco.2018.06.001
Frisvad, J. C., Møller, L. L. H., Larsen, T. O., Kumar, R., and Arnau, J. (2018). Safety of the fungal workhorses of industrial biotechnology. Update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Appl. Microbiol. Biotechnol. 102, 9481–9515. doi: 10.1007/s00253-018-9354-1
Fukasawa, Y., and Matsukura, K. (2021). Decay stages of wood and associated fungal communities characterise diversity–decomposition relationships. Sci. Rep. 11:8972. doi: 10.1038/s41598-021-88580-2
Galagan, J. E., Calvo, S. E., Cuomo, C., Ma, L.-J., Wortman, J. R., Batzoglou, S., et al. (2005). Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438, 1105–1115. doi: 10.1038/nature04341
Gall, E., and Benkeblia, N. (2023). Mycoagroecology: Integrating fungi into agroecosystems. First Edn. Boca Raton, FL: CRC Press.
Gallage, N. J., Hansen, E. H., Kannangara, R., Olsen, C. E., Motawia, M. S., Jørgensen, K., et al. (2014). Vanillin formation from ferulic acid in Vanilla planifolia is catalysed by a single enzyme. Nat. Commun. 5:5037. doi: 10.1038/ncomms5037
Gao, J., Li, Y., Wan, Y., Hu, T., Liu, L., Yang, S., et al. (2019). A novel Postbiotic from Lactobacillus rhamnosus GG with a beneficial effect on intestinal barrier function. Front. Microbiol. 10:477. doi: 10.3389/fmicb.2019.00477
Gastebois, A., Clavaud, C., Aimanianda, V., and Latgé, J.-P. (2009). Aspergillus fumigatus. Cell wall polysaccharides, their biosynthesis and organization. Future Microbiol. 4. doi: 10.2217/fmb.09.29
Geyer, M., Onyancha, F. M., Nicol, W., and Brink, H. G. (2018). Malic acid production by Aspergillus oryzae: The role of CaCO3. Ital. Assoc. Chem. Eng. 70:301. doi: 10.3303/CET1870301
Gibson, G. R., Hutkins, R., Sanders, M. E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). Expert consensus document. The international scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14:75. doi: 10.1038/nrgastro.2017.75
Gibson, G. R., and Roberfroid, M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125, 1401–1412. doi: 10.1093/jn/125.6.1401
Goldberg, I., Rokem, J. S., and Pines, O. (2006). Organic acids. Old metabolites, new themes. J. Chem. Technol. Biotechnol. 81, 1601–1611. doi: 10.1002/jctb.1590
Gomez-Alarcon, R. A., Dudas, C., and Huber, J. T. (1990). Influence of cultures of Aspergillus oryzae on rumen and total tract digestibility of dietary components. J. Dairy Sci. 73, 703–710. doi: 10.3168/jds.S0022-0302(90)78723-1
Gravelat, F. N., Beauvais, A., Liu, H., Lee, M. J., Snarr, B. D., Chen, D., et al. (2013). Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal β-glucan from the immune system. PLoS Pathog. 9:3575. doi: 10.1371/journal.ppat.1003575
Guan, Z., and Feng, Q. (2022). Chitosan and Chitooligosaccharide: the promising non-plant-derived prebiotics with multiple biological activities. Int. J. Mol. Sci. 23:761. doi: 10.3390/ijms23126761
Guo, G., Shi, F., Zhu, J., Shao, Y., Gong, W., Zhou, G., et al. (2020). Piperine, a functional food alkaloid, exhibits inhibitory potential against TNBS-induced colitis via the inhibition of IκB-α/NF-κB and induces tight junction protein (claudin-1, occludin, and ZO-1) signaling pathway in experimental mice. Hum. Exp. Toxicol. 39, 477–491. doi: 10.1177/0960327119892042
Guo YuHan, G. Y., Yang YongZhi, Y. Y., Guo HeNan, G. H., Wang Jian, W. J., and Cao YunHe, C. Y. (2018). Expression of α-galactosidase gene gal a from Aspergillus oryzae in Pichia pastoris and its enzymolysis effect on soybean oligosaccharides in soymilk. Chin. J. Anim. Nutr. 11, 4569–4579.
Gupta, S., Chaturvedi, P., Kulkarni, M. G., and van Staden, J. (2020). A critical review on exploiting the pharmaceutical potential of plant endophytic fungi. Biotechnol. Adv. 39:107462. doi: 10.1016/j.biotechadv.2019.107462
Gurung, N., Ray, S., Bose, S., and Rai, V. (2013). A broader view: microbial enzymes and their relevance in industries, medicine, and beyond. Biomed. Res. Int. 2013, 1–18. doi: 10.1155/2013/329121
Hagiwara, D., Yoshimi, A., Sakamoto, K., Gomi, K., and Abe, K. (2015). “Response and adaptation to cell wall stress and osmotic stress in Aspergillus species” in Stress biology of yeasts and fungi: applications for industrial brewing and fermentation (Berlin: Springer).
Hamajima, H., Matsunaga, H., Fujikawa, A., Sato, T., Mitsutake, S., Yanagita, T., et al. (2016). Japanese traditional dietary fungus koji Aspergillus oryzae functions as a prebiotic for Blautia coccoides through glycosylceramide. Japanese dietary fungus koji is a new prebiotic. Springerplus 5:1321. doi: 10.1186/s40064-016-2950-6
Hamajima, H., Tanaka, M., Miyagawa, M., Sakamoto, M., Nakamura, T., Yanagita, T., et al. (2019). Koji glycosylceramide commonly contained in Japanese traditional fermented foods alters cholesterol metabolism in obese mice. Biosci. Biotechnol. Biochem. 83, 1514–1522. doi: 10.1080/09168451.2018.1562877
Han, D. M., Baek, J. H., Choi, D. G., Jeon, M.-S., Eyun, S., and Jeon, C. O. (2024). Comparative pangenome analysis of Aspergillus flavus and Aspergillus oryzae reveals their phylogenetic, genomic, and metabolic homogeneity. Food Microbiol. 119:104435. doi: 10.1016/j.fm.2023.104435
Hashem, A. H., Attia, M. S., Kandil, E. K., Fawzi, M. M., Abdelrahman, A. S., Khader, M. S., et al. (2023). Bioactive compounds and biomedical applications of endophytic fungi: a recent review. Microb. Cell Factories 22:107. doi: 10.1186/s12934-023-02118-x
Hata, Y., Ishida, H., Kojima, Y., Ichikawa, E., Kawato, A., Suginami, K., et al. (1997). Comparison of two glucoamylases produced by Aspergillus oryzae in solid-state culture (koji) and in submerged culture. J. Ferment. Bioeng. 84, 532–537. doi: 10.1016/S0922-338X(97)81907-1
He, B., Hu, Z., Ma, L., Li, H., Ai, M., Han, J., et al. (2018a). Transcriptome analysis of different growth stages of Aspergillus oryzae reveals dynamic changes of distinct classes of genes during growth. BMC Microbiol. 18:12. doi: 10.1186/s12866-018-1158-z
He, B., Ma, L., Hu, Z., Li, H., Ai, M., Long, C., et al. (2018b). Deep sequencing analysis of transcriptomes in Aspergillus oryzae in response to salinity stress. Appl. Microbiol. Biotechnol. 102, 897–906. doi: 10.1007/s00253-017-8603-z
He, H., Qin, Y., Li, N., Chen, G., and Liang, Z. (2015). Purification and characterization of a thermostable hypothetical xylanase from Aspergillus oryzae HML366. Appl. Biochem. Biotechnol. 175, 3148–3161. doi: 10.1007/s12010-014-1352-x
He, B., Tu, Y., Jiang, C., Zhang, Z., Li, Y., and Zeng, B. (2019). Functional genomics of Aspergillus oryzae. Strategies and Progress. Microorganisms 7:103. doi: 10.3390/microorganisms7040103
He, X., Yu, B., He, J., Huang, Z., Mao, X., Zheng, P., et al. (2020). Effects of xylanase on growth performance, nutrients digestibility and intestinal health in weaned piglets. Livest. Sci. 233:103940. doi: 10.1016/j.livsci.2020.103940
Heesemann, L., Kotz, A., Echtenacher, B., Broniszewska, M., Routier, F., Hoffmann, P., et al. (2011). Studies on galactofuranose-containing glycostructures of the pathogenic mold Aspergillus fumigatus. Int. J. Med. Microbiol. 301, 523–530.
Hoa, B. T., and Hung, P. V. (2013). Optimization of nutritional composition and fermentation conditions for cellulase and pectinase production by Aspergillus oryzae using response surface methodology. Int. Food Res. J. 20:3269.
Horie, Y., Goto, A., Imamura, R., Itoh, M., Ikegawa, S., Ogawa, S., et al. (2020). Quantification of ergothioneine in Aspergillus oryzae-fermented rice bran by a newly-developed LC/ESI-MS/MS method. LWT 118:108812. doi: 10.1016/j.lwt.2019.108812
Huang, F., Teng, K., Liu, Y., Cao, Y., Wang, T., Ma, C., et al. (2021). Bacteriocins: potential for human health. Oxidative Med. Cell. Longev. 2021, 1–17. doi: 10.1155/2021/5518825
Hymes-Fecht, U. C., and Casper, D. P. (2021). Adaptation and withdrawal of feeding dried Aspergillus oryzae fermentation product to dairy cattle and goats on in vitro NDF digestibility of selected forage sources. Transl. Anim. Sci. 5:51. doi: 10.1093/tas/txab051
Ibrahim, S. A., Alazzeh, A. Y., Awaisheh, S. S., Song, D., Shahbazi, A., and AbuGhazaleh, A. A. (2010). Enhancement of α- and β-galactosidase activity in Lactobacillus reuteri by different metal ions. Biol. Trace Elem. Res. 136, 106–116. doi: 10.1007/s12011-009-8519-2
Ibrahim, S. R. M., Mohamed, S. G. A., Alsaadi, B. H., Althubyani, M. M., Awari, Z. I., Hussein, H. G. A., et al. (2023). Secondary metabolites, biological activities, and industrial and biotechnological importance of Aspergillus sydowii. Mar. Drugs 21:441. doi: 10.3390/md21080441
Ichikawa, H., Miyazawa, K., Komeiji, K., Susukida, S., Zhang, S., Muto, K., et al. (2022). Improved recombinant protein production in Aspergillus oryzae lacking both α-1,3-glucan and galactosaminogalactan in batch culture with a lab-scale bioreactor. J. Biosci. Bioeng. 133, 39–45. doi: 10.1016/j.jbiosc.2021.09.010
Ichishima, E. (2022). Summarized enzymology of Aspergillus oryzae: the national microorganism of Japan. Curr. Top. Chem. Biochem. 2, 20–44. doi: 10.9734/bpi/ctcb/v2/15725D
Ito, K., and Matsuyama, A. (2021). Koji molds for Japanese soy sauce brewing: characteristics and key enzymes. J. Fungi 7:658. doi: 10.3390/jof7080658
Iwashita, M. K. P., Nakandakare, I. B., Terhune, J. S., Wood, T., and Ranzani-Paiva, M. J. T. (2015). Dietary supplementation with Bacillus subtilis, Saccharomyces cerevisiae and Aspergillus oryzae enhance immunity and disease resistance against Aeromonas hydrophila and Streptococcus iniae infection in juvenile tilapia Oreochromis niloticus. Fish Shellfish Immunol. 43, 60–66. doi: 10.1016/j.fsi.2014.12.008
Izuddin, W. I., Loh, T. C., Foo, H. L., Samsudin, A. A., and Humam, A. M. (2019). Postbiotic L. plantarum RG14 improves ruminal epithelium growth, immune status and upregulates the intestinal barrier function in post-weaning lambs. Sci. Rep. 9:9938. doi: 10.1038/s41598-019-46076-0
Jaramillo, P. M. D., Andreaus, J., Neto, G. P., Castro, C. F. d. S., Filho, E. X. F., Souza, D., et al. (2015). The characterization of a pectin-degrading enzyme from Aspergillus oryzae grown on passion fruit peel as the carbon source and the evaluation of its potential for industrial applications. Biocatal. Biotransformation 33, 310–322. doi: 10.3109/10242422.2016.1168817
Jebur, H. A., Abdulateef, A. A., and Thbit, Z. A. (2019). Chitosan production from Aspergillus oryzae SU-B2 by submerged fermentation and studying some of its physiochemical and antibacterial characteristics. J. Pharm. Sci. Res. 11, 609–613.
Ji, S. B., and Ra, C. H. (2021). Coproduction of enzymes and Beta-glucan by Aspergillus oryzae using solid-state fermentation of Brown Rice. J. Microbiol. Biotechnol. 31, 1028–1034. doi: 10.4014/jmb.2105.05005
Jiang, C., Ge, J., He, B., Zhang, Z., Hu, Z., Li, Y., et al. (2022). Transcriptomic analysis reveals Aspergillus oryzae responds to temperature stress by regulating sugar metabolism and lipid metabolism. PLoS One 17:e0274394. doi: 10.1371/journal.pone.0274394
Junior, S. R. S., Johnston, L. J., Manu, H., Gomez, A., Ocazio, C., Ipharraguerre, I. R., et al. (2024). 281 effects of dietary Aspergillus oryzae postbiotic during gestation and lactation on sow and progeny performance. J. Anim. Sci. 102, 175–176. doi: 10.1093/jas/skae102.194
Kadooka, C., Tanaka, Y., Hira, D., Maruyama, J., Goto, M., and Oka, T. (2023). Identification of galactofuranose antigens such as galactomannoproteins and fungal-type galactomannan from the yellow koji fungus (Aspergillus oryzae). Front. Microbiol. 14:1110996. doi: 10.3389/fmicb.2023.1110996
Kato, N., Tokuoka, M., Shinohara, Y., Kawatani, M., Uramoto, M., Seshime, Y., et al. (2011). Genetic safeguard against mycotoxin cyclopiazonic acid production in Aspergillus oryzae. Chembiochem 12, 1376–1382. doi: 10.1002/cbic.201000672
Kaufman, J. D., Seidler, Y., Bailey, H. R., Whitacre, L., Bargo, F., Lüersen, K., et al. (2021). A postbiotic from Aspergillus oryzae attenuates the impact of heat stress in ectothermic and endothermic organisms. Sci. Rep. 11:6407. doi: 10.1038/s41598-021-85707-3
Kavuthodi, B., and Sebastian, D. (2018). Review on bacterial production of alkaline pectinase with special emphasis on Bacillus species. Biosci. Biotechnol. Res. Commun. 11, 18–30. doi: 10.21786/bbrc/11.1/4
Ketipally, R., and Ram, M. R. (2018). Optimization of pectinase production by Aspergillus oryzae RR 103. Curr. Agric. Res. J. 6, 37–44. doi: 10.12944/CARJ.6.1.05
Kim, S. Y., Jeong, H.-S., Ahn, S.-W., and Shin, K.-S. (2014). Prebiotic effects of structurally identified galacto-oligosaccharides produced by β-galactosidase from Aspergillus oryzae. Food Sci. Biotechnol. 23, 823–830. doi: 10.1007/s10068-014-0111-7
Kimura, T., Suzuki, H., Furuhashi, H., Aburatani, T., Morimoto, K., Karita, S., et al. (2000). Molecular cloning, overexpression, and purification of a major xylanase from Aspergillus oryzae. Biosci. Biotechnol. Biochem. 64, 2734–2738. doi: 10.1271/bbb.64.2734
Kino, T., Chihara, J., Mitsuyasu, K., Kitaichi, M., Honda, K., Kado, M., et al. (1982). A case of allergic bronchopulmonary aspergillosis caused by Aspergillus oryzae which is used for brewing bean paste (miso) and soy sauce (shoyu). Jpn. J. Thorac. Dis. 20, 467–475
Kitagaki, H. (2021). Medical application of substances derived from non-pathogenic fungi Aspergillus oryzae and A. luchuensis-containing Koji. J. Fungi 7:243. doi: 10.3390/jof7040243
Knuf, C., Nookaew, I., Brown, S. H., McCulloch, M., Berry, A., and Nielsen, J. (2013). Investigation of malic acid production in Aspergillus oryzae under nitrogen starvation conditions. Appl. Environ. Microbiol. 79, 6050–6058. doi: 10.1128/AEM.01445-13
Knuf, C., Nookaew, I., Remmers, I., Khoomrung, S., Brown, S., Berry, A., et al. (2014). Physiological characterization of the high malic acid-producing Aspergillus oryzae strain 2103a-68. Appl. Microbiol. Biotechnol. 98, 3517–3527. doi: 10.1007/s00253-013-5465-x
Koizumi, A., Miyazawa, K., Ogata, M., Takahashi, Y., Yano, S., Yoshimi, A., et al. (2023). Cleavage of α-1, 4-glycosidic linkages by the glycosylphosphatidylinositol-anchored α-amylase AgtA decreases the molecular weight of cell wall α-1, 3-glucan in Aspergillus oryzae. Front. Fungal Biol. 3:1061841. doi: 10.3389/ffunb.2022.1061841
Kong, Z., Wang, M., Shi, X., Wang, X., Zhang, X., Chai, L., et al. (2022). The functions of potential intermediates and fungal communities involved in the humus formation of different materials at the thermophilic phase. Bioresour. Technol. 354:127216. doi: 10.1016/j.biortech.2022.127216
Kong, L., Yu, L., Feng, T., Yin, X., Liu, T., and Dong, L. (2015). Physicochemical characterization of the polysaccharide from Bletilla striata: effect of drying method. Carbohydr. Polym. 125, 1–8. doi: 10.1016/j.carbpol.2015.02.042
Konishi, H., Isozaki, S., Kashima, S., Moriichi, K., Ichikawa, S., Yamamoto, K., et al. (2021). Probiotic Aspergillus oryzae produces anti-tumor mediator and exerts anti-tumor effects in pancreatic cancer through the p38 MAPK signaling pathway. Sci. Rep. 11:11070. doi: 10.1038/s41598-021-90707-4
Kövilein, A., Kubisch, C., Cai, L., and Ochsenreither, K. (2020). Malic acid production from renewables. A review. J. Chem. Technol. Biotechnol. 95, 513–526. doi: 10.1002/jctb.6269
Kövilein, A., Umpfenbach, J., and Ochsenreither, K. (2021). Acetate as substrate for l-malic acid production with Aspergillus oryzae DSM 1863. Biotechnol. Biofuels 14, 1–15. doi: 10.1186/s13068-021-01901-5
Kreger, D. R. (1954). Observations on cell walls of yeasts and some other fungi by X-ray diffraction and solubility tests. Biochim. Biophys. Acta 13. doi: 10.1016/0006-3002(54)90264-4
Kulkarni, D. S., Kapanoor, S. S., Girigouda, K., Kote, N. V., and Mulimani, V. H. (2006). Reduction of flatus-inducing factors in soymilk by immobilized α-galactosidase. Biotechnol. Appl. Biochem. 45, 51–57. doi: 10.1042/BA20060027
Kumar, A., Verma, V., Dubey, V. K., Srivastava, A., Garg, S. K., Singh, V. P., et al. (2023). Industrial applications of fungal lipases: a review. Front. Microbiol. 14:1142536. doi: 10.3389/fmicb.2023.1142536
Kurita, K. (2006). Chitin and chitosan. Functional biopolymers from marine crustaceans. Mar. Biotechnol. 8, 203–226.
Kusumoto, K., Nogata, Y., and Ohta, H. (2000). Directed deletions in the aflatoxin biosynthesis gene homolog cluster of Aspergillus oryzae. Curr. Genet. 37, 104–111. doi: 10.1007/s002940050016
Kusumoto, K.-I., Yamagata, Y., Tazawa, R., Kitagawa, M., Kato, T., Isobe, K., et al. (2021). Japanese traditional miso and Koji making. J. Fungi 7:579. doi: 10.3390/jof7070579
Lakshmi, M., and Jyothi, P. (2014). Production and optimization of glucoamylase from Aspergillus oryzae NCIM 1212 using wheat bran, varying chemical parameters under solid state fermentation. Int. J. Curr. Microbiol. App. Sci. 3, 70–76.
Lam, K.-L., and Chi-Keung Cheung, P. (2013). Non-digestible long chain beta-glucans as novel prebiotics. Bioact. Carbohydr. Diet. Fibre 2, 45–64. doi: 10.1016/j.bcdf.2013.09.001
Lamarre, C., Beau, R., Balloy, V., Fontaine, T., Hoi, J. W. S., Guadagnini, S., et al. (2009). Galactofuranose attenuates cellular adhesion of Aspergillus fumigatus. Cell. Microbiol. 11, 1612–1623.
Latgé, J.-P. (2007). The cell wall. A carbohydrate armour for the fungal cell. Mol. Microbiol. 66. doi: 10.1111/j.1365-2958.2007.05872.x
Latgé, J.-P. (2010). Tasting the fungal cell wall. Cell. Microbiol. 12, 863–872. doi: 10.1111/j.1462-5822.2010.01474.x
Latgé, J.-P., and Chamilos, G. (2019). Aspergillus fumigatus and aspergillosis in 2019. Clin. Microbiol. Rev. 33:18. doi: 10.1128/CMR.00140-18
Latgé, J.-P., Kobayashi, H., Debeaupuis, J.-P., Diaquin, M., Sarfati, J., Wieruszeski, J.-M., et al. (1994). Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect. Immun. 62, 5424–5433.
Lavinsky, D., Arteni, N. S., and Netto, C. A. (2003). Agmatine induces anxiolysis in the elevated plus maze task in adult rats. Behav. Brain Res. 141, 19–24. doi: 10.1016/S0166-4328(02)00326-1
Lee, H.-Y., Chen, C.-C., Pi, C.-C., and Chen, C.-J. (2023). Aspergillus oryzae fermentation extract alleviates inflammation in Mycoplasma pneumoniae pneumonia. Molecules (Basel, Switzerland) 28:1127. doi: 10.3390/molecules28031127
Lee, K., Lee, S. K., and Lee, B. D. (2006). Aspergillus oryzae as probiotic in poultry-a review. Int. J. Poult. Sci. 5, 1–3.
Lee, M. J., Liu, H., Barker, B. M., Snarr, B. D., Gravelat, F. N., Al Abdallah, Q., et al. (2015). The fungal exopolysaccharide galactosaminogalactan mediates virulence by enhancing resistance to neutrophil extracellular traps. PLoS Pathog. 11:e1005187. doi: 10.1371/journal.ppat.1005187
Lee, M. J., and Sheppard, D. C. (2016). Recent advances in the understanding of the Aspergillus fumigatus cell wall. J. Microbiol. (Seoul, Korea) 54. doi: 10.1007/s12275-016-6045-4
Lenardon, M. D., Munro, C. A., and Gow, N. A. R. (2010). Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol. 13. doi: 10.1016/j.mib.2010.05.002
Li, J.-S., Chew, Y.-M., Lin, M.-C., Lau, Y.-Q., and Chen, C.-S. (2021). Enhanced glucosamine production from Aspergillus oryzae NCH-42 via acidic stress under submerged fermentation. CyTA 19, 614–624. doi: 10.1080/19476337.2021.1946158
Li, Q., Gabler, N. K., Loving, C. L., Gould, S. A., and Patience, J. F. (2018). A dietary carbohydrase blend improved intestinal barrier function and growth rate in weaned pigs fed higher fiber diets1. J. Anim. Sci. 96, 5233–5243. doi: 10.1093/jas/sky383
Li, W., Wu, D.-T., Li, F., Gan, R.-Y., Hu, Y.-C., and Zou, L. (2021). Structural and biological properties of water soluble polysaccharides from lotus leaves: effects of drying techniques. Molecules (Basel, Switzerland) 26:4395. doi: 10.3390/molecules26154395
Liao, W., Shao, J., Li, S., Wan, G., Da, Z., Sun, Y., et al. (1988). Mycological identification of pulmonary aspergilloma caused by Aspergillus oryzae with proliferating heads. Chin. Med. J. 101, 601–604
Lilly, D. M., and Stillwell, R. H. (1965). Growth promoting factors produced by probiotics. Science 147, 747–748. doi: 10.1126/science.147.3659.747
Liu, L., Bao, L., Wang, L., Ma, K., Han, J., Yang, Y., et al. (2018a). Asperorydines A–M: Prenylated tryptophan-derived alkaloids with neurotrophic effects from Aspergillus oryzae. J. Org. Chem. 83, 812–822. doi: 10.1021/acs.joc.7b02802
Liu, D., Guo, S., and Guo, Y. (2012). Xylanase supplementation to a wheat-based diet alleviated the intestinal mucosal barrier impairment of broiler chickens challenged by Clostridium perfringens. Avian Pathol. 41, 291–298. doi: 10.1080/03079457.2012.684089
Liu, Y., Sun, G., Li, J., Cheng, P., Song, Q., Lv, W., et al. (2024). Starter molds and multi-enzyme catalysis in koji fermentation of soy sauce brewing: a review. Food Res. Int. 184:114273. doi: 10.1016/j.foodres.2024.114273
Liu, L., Wang, Y., Kong, M., and Li, X. (2018b). Prebiotic-like effects of water soluble chitosan on the intestinal microflora in mice. Int. J. Food Eng. 14:20180089. doi: 10.1515/ijfe-2018-0089
Loures, F. V., Araújo, E. F., Feriotti, C., Bazan, S. B., and Calich, V. L. G. (2015). TLR-4 cooperates with Dectin-1 and mannose receptor to expand Th17 and Tc17 cells induced by Paracoccidioides brasiliensis stimulated dendritic cells. Front. Microbiol. 6:261. doi: 10.3389/fmicb.2015.00261
Lu, H., Peng, S., Xu, N., Shang, X., Liu, J., Xu, Z., et al. (2024). Exploring the effects of different drying methods on related differential metabolites of Pleurotus citrinopileatus singer based on untargeted metabolomics. Plan. Theory 13:1594. doi: 10.3390/plants13121594
Lu, H., Shin, S., Kuehn, I., Bedford, M., Rodehutscord, M., Adeola, O., et al. (2020). Effect of phytase on nutrient digestibility and expression of intestinal tight junction and nutrient transporter genes in pigs. J. Anim. Sci. 98:skaa206. doi: 10.1093/jas/skaa206
Lustenhouwer, N., Maynard, D. S., Bradford, M. A., Lindner, D. L., Oberle, B., Zanne, A. E., et al. (2020). A trait-based understanding of wood decomposition by fungi. Proc. Natl. Acad. Sci. 117, 11551–11558. doi: 10.1073/pnas.1909166117
Ma, L., Fu, L., Hu, Z., Li, Y., Zheng, X., Zhang, Z., et al. (2019). Modulation of fatty acid composition of Aspergillus oryzae in response to ethanol stress. Microorganisms 7:158. doi: 10.3390/microorganisms7060158
Machida, M., Asai, K., Sano, M., Tanaka, T., Kumagai, T., Terai, G., et al. (2005). Genome sequencing and analysis of Aspergillus oryzae. Nature 438, 1157–1161. doi: 10.1038/nature04300
Managa, M. G., Sultanbawa, Y., and Sivakumar, D. (2020). Effects of different drying methods on untargeted phenolic metabolites, and antioxidant activity in Chinese cabbage (Brassica rapa L. subsp. chinensis) and nightshade (Solanum retroflexum dun.). Molecules (Basel, Switzerland) 25:1326. doi: 10.3390/molecules25061326
Marino, C., Rinflerch, A., and de Lederkremer, R. M. (2017). Galactofuranose antigens, a target for diagnosis of fungal infections in humans. Future Science OA 3, FSO199.
Markowiak, P., and Śliżewska, K. (2018). The role of probiotics, prebiotics and synbiotics in animal nutrition. Gut Pathog. 10, 1–20. doi: 10.1186/s13099-018-0250-0
Martorell, P., Alvarez, B., Llopis, S., Navarro, V., Ortiz, P., Gonzalez, N., et al. (2021). Heat-treated Bifidobacterium longum CECT-7347: a whole-cell postbiotic with antioxidant, anti-inflammatory, and gut-barrier protection properties. Antioxidants 10:536. doi: 10.3390/antiox10040536
Marui, J., Yamane, N., Ohashi-Kunihiro, S., Ando, T., Terabayashi, Y., Sano, M., et al. (2011). Kojic acid biosynthesis in Aspergillus oryzae is regulated by a Zn (II) 2Cys6 transcriptional activator and induced by kojic acid at the transcriptional level. J. Biosci. Bioeng. 112, 40–43. doi: 10.1016/j.jbiosc.2011.03.010
Masuda, S., Kikuchi, K., Matsumoto, Y., Sugimoto, T., Shoji, H., and Tanabe, M. (2009). Analysis of enzyme production by submerged culture of Aspergillus oryzae using whole barley. Biosci. Biotechnol. Biochem. 73, 2190–2195. doi: 10.1271/bbb.90270
Mathis, H., Margeot, A., and Bouix, M. (2020). Optimization of flow cytometry parameters for high-throughput screening of spores of the filamentous fungus Trichoderma reesei. J. Biotechnol. 321, 78–86. doi: 10.1016/j.jbiotec.2020.05.015
Megías, J., Martínez, A., Yáñez, A., Goodridge, H. S., Gozalbo, D., and Gil, M. L. (2016). TLR2, TLR4 and Dectin-1 signalling in hematopoietic stem and progenitor cells determines the antifungal phenotype of the macrophages they produce. Microbes Infect. 18, 354–363. doi: 10.1016/j.micinf.2016.01.005
Meneghel, L., Reis, G. P., Reginatto, C., Malvessi, E., and da Silveira, M. M. (2014). Assessment of pectinase production by Aspergillus oryzae in growth-limiting liquid medium under limited and non-limited oxygen supply. Process Biochem. 49, 1800–1807. doi: 10.1016/j.procbio.2014.07.021
Miao, M., Li, S., Yang, S., Yan, Q., Xiang, Z., and Jiang, Z. (2024). Engineering the β-galactosidase from Aspergillus oryzae for making lactose-free and no-sugar-added yogurt. J. Dairy Sci. 107, 6602–6613. doi: 10.3168/jds.2023-24310
Miyagawa, M., Fujikawa, A., Nagadome, M., Kohama, K., Ogami, T., Kitamura, S., et al. (2019). Glycosylceramides purified from the Japanese traditional non-pathogenic fungus Aspergillus and koji increase the expression of genes involved in tight junctions and ceramide delivery in normal human epidermal keratinocytes. Fermentation 5:43. doi: 10.3390/fermentation5020043
Miyazawa, K., Yoshimi, A., Sano, M., Tabata, F., Sugahara, A., Kasahara, S., et al. (2019). Both galactosaminogalactan and α-1, 3-glucan contribute to aggregation of Aspergillus oryzae hyphae in liquid culture. Front. Microbiol. 10:2090. doi: 10.3389/fmicb.2019.02090
Miyazawa, K., Yoshimi, A., Zhang, S., Sano, M., Nakayama, M., Gomi, K., et al. (2016). Increased enzyme production under liquid culture conditions in the industrial fungus Aspergillus oryzae by disruption of the genes encoding cell wall α-1,3-glucan synthase. Biosci. Biotechnol. Biochem. 80, 1853–1863. doi: 10.1080/09168451.2016.1209968
Moita, V. H. C., Duarte, M. E., and Kim, S. W. (2021). Supplemental effects of phytase on modulation of mucosa-associated microbiota in the jejunum and the impacts on nutrient digestibility, intestinal morphology, and bone parameters in broiler chickens. Animals 11:3351. doi: 10.3390/ani11123351
Monteiro, R. C., Kishore, B. N., Bhat, R. M., Sukumar, D., Martis, J., and Ganesh, H. K. (2013). A comparative study of the efficacy of 4% hydroquinone vs 0.75% kojic acid cream in the treatment of facial melasma. Indian J. Dermatol. 58:157. doi: 10.4103/0019-5154.108070
Monti, F., Ripamonti, F., Hawser, S. P., and Islam, K. (1999). Aspirochlorine. A highly selective and potent inhibitor of fungal protein synthesis. J. Antibiot. 52, 311–318. doi: 10.7164/antibiotics.52.311
Morton, H. E., Kocholaty, W., Junowicz-Kocholaty, R., and Kelner, A. (1945). Toxicity and antibiotic activity of kojic acid produced by Aspergillus luteo-virescens. J. Bacteriol. 50, 579–584. doi: 10.1128/jb.50.5.579-584.1945
Mouyna, I., and Fontaine, T. (2008). “Cell wall of Aspergillus fumigatus. A dynamic structure” in Aspergillus fumigatus and Aspergillosis. eds. J. P. Latgé and W. J. Steinbach (Washington, DC: ASM Press), 169–183.
Müller, C., Hjort, C. M., Hansen, K., and Nielsen, J. (2002a). Altering the expression of two chitin synthase genes differentially affects the growth and morphology of Aspergillus oryzae. Microbiology 148, 4025–4033. doi: 10.1099/00221287-148-12-4025
Müller, C., McIntyre, M., Hansen, K., and Nielsen, J. (2002b). Metabolic engineering of the morphology of Aspergillus oryzae by altering chitin synthesis. Appl. Environ. Microbiol. 68, 1827–1836. doi: 10.1128/AEM.68.4.1827-1836.2002
Munro, C. A., and Gow, N. A. (2001). Chitin synthesis in human pathogenic fungi. Med. Mycol. 39, 41–53.
Murooka, Y. (2020). Japanese food for health and longevity: The science behind a great culinary tradition. Newcastle upon Tyne: Cambridge Scholars Publishing.
Nagaoka, S., and Hara, Y. (2019). Compactin. The discovery of statin, the “penicillin” for cholesterol. In: S. Nagaoka Drug Discovery in Japan: Investigating the Sources of Innovation. Berlin: Springer Nature, pp. 15–34.
Nakadai, T., Nasuno, S., and Iguchi, N. (1972). The action of peptidases from Aspergillus oryzae in digestion of soybean proteins. Agric. Biol. Chem. 36, 261–268. doi: 10.1080/00021369.1972.10860243
Nakajima, T., and Ichishima, E. (1994). Chemical structure of the galactomannan moiety in the cell wall glycoproteins of Aspergillus oryzae. J. Ferment. Bioeng. 78, 472–475. doi: 10.1016/0922-338X(94)90050-7
Napolitani, G., Rinaldi, A., Bertoni, F., Sallusto, F., and Lanzavecchia, A. (2005). Selected toll-like receptor agonist combinations synergistically trigger a T helper type 1–polarizing program in dendritic cells. Nat. Immunol. 6, 769–776. doi: 10.1038/ni1223
Nath, A., Mondal, S., Chakraborty, S., Bhattacharjee, C., and Chowdhury, R. (2014). Production, purification, characterization, immobilization, and application of β-galactosidase: a review. Asia Pac. J. Chem. Eng. 9, 330–348. doi: 10.1002/apj.1801
Netea, M. G., Brown, G. D., Kullberg, B. J., and Gow, N. A. R. (2008). An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 6, 67–78. doi: 10.1038/nrmicro1815
Nie, Y., Wang, Z., Wang, W., Zhou, Z., Kong, Y., and Ma, J. (2022). Bio-flocculation of Microcystis aeruginosa by using fungal pellets of Aspergillus oryzae: performance and mechanism. J. Hazard. Mater. 439:129606. doi: 10.1016/j.jhazmat.2022.129606
Nielsen, P. H., and Jahn, A. (1999). “Extraction of EPS” in Microbial extracellular polymeric substances. eds. J. Wingender, T. R. Neu, and H. C. Flemming (Berlin: Springer Science and Business Media).
Nierman, W. C., Pain, A., Anderson, M. J., Wortman, J. R., Kim, H. S., Arroyo, J., et al. (2005). Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438, 1151–1156. doi: 10.1038/nature04332
Niu, C., Xing, X., Yang, X., Zheng, F., Liu, C., Wang, J., et al. (2023). Isolation, identification and application of Aspergillus oryzae BL18 with high protease activity as starter culture in doubanjiang (broad bean paste) fermentation. Food Biosci. 51:102225. doi: 10.1016/j.fbio.2022.102225
Nizamuddin, S., Sridevi, A., and Narasimha, G. (2008). Production of [beta]-galactosidase by Aspergillus oryzae in solid-state fermentation. Afr. J. Biotechnol. 7:1096.
No, H. K., Meyers, S. P., Prinyawiwatkul, W., and Xu, Z. (2007). Applications of chitosan for improvement of quality and shelf life of foods. J. Food Sci. 72, R87–R100.
Nogueira, K. M. V., Mendes, V., Carraro, C. B., Taveira, I. C., Oshiquiri, L. H., Gupta, V. K., et al. (2020). Sugar transporters from industrial fungi: key to improving second-generation ethanol production. Renew. Sust. Energ. Rev. 131:109991. doi: 10.1016/j.rser.2020.109991
Nomura, R., Tsuzuki, S., Kojima, T., Nagasawa, M., Sato, Y., Uefune, M., et al. (2022). Administration of Aspergillus oryzae suppresses DSS-induced colitis. Food Chem. Mol. Sci. 4:100063. doi: 10.1016/j.fochms.2021.100063
Nout, M. R. (2005). “Food fermentation: an introduction” in Food fermentation (Wageningen, Netherlands: Wageningen Academic Publishers), 13–18.
Ohnishi, K. (1982). Effect of lipase activity on aging and quality of miso. Nippon Shokuhin Kogyo Gakkaishi 29, 85–92. doi: 10.3136/nskkk1962.29.2_85
Ohnishi, K., Yoshida, Y., and Sekiguchi, J. (1994a). Lipase production of Aspergillus oryzae. J. Ferment. Bioeng. 77, 490–495. doi: 10.1016/0922-338X(94)90116-3
Ohnishi, K., Yoshida, Y., Toita, J., and Sekiguchi, J. (1994b). Purification and characterization of a novel lipolytic enzyme from Aspergillus oryzae. J. Ferment. Bioeng. 78, 413–419. doi: 10.1016/0922-338X(94)90039-6
Ojeda-López, M., Chen, W., Eagle, C. E., Gutiérrez, G., Jia, W. L., Swilaiman, S. S., et al. (2018). Evolution of asexual and sexual reproduction in the aspergilli. Stud. Mycol. 91, 37–59. doi: 10.1016/j.simyco.2018.10.002
Ojha, B. K., Singh, P. K., and Shrivastava, N. (2019). Enzymes in the animal feed industry: Enzymes in food biotechnology. Amsterdam, The Netherlands: Elsevier, 93–109.
Okpara, M. O. (2022). Microbial enzymes and their applications in food industry. A mini-review. Adv. Enzyme Res. 10, 23–47. doi: 10.4236/aer.2022.101002
Oliveira, A. C., Amorim, G. M., Azevêdo, J. A. G., Godoy, M. G., and Freire, D. M. G. (2018). Solid-state fermentation of co-products from palm oil processing. Production of lipase and xylanase and effects on chemical composition. Biocatal. Biotransformation 36, 381–388. doi: 10.1080/10242422.2018.1425400
Ozma, M. A., Abbasi, A., Akrami, S., Lahouty, M., Shahbazi, N., Ganbarov, K., et al. (2022). Postbiotics as the key mediators of the gut microbiota-host interactions. Infez. Med. 30:3002. doi: 10.53854/liim-3002-3
Pagare, S., Bhatia, M., Tripathi, N., Pagare, S., and Bansal, Y. K. (2015). Secondary metabolites of plants and their role: Overview. Curr. Trends Biotechnol. Pharm. 9, 293–304.
Pandey, V. V., Varshney, V. K., and Pandey, A. (2019). Lovastatin. A journey from ascomycetes to basidiomycetes fungi. J. Biol. Act. Prod. Nat. 9: 162–178. doi: 10.1080/22311866.2019.1622452
Park, Y. K., De Santi, M. S., and Pastore, G. M. (1979). Production and characterization of β-galactosidase from Aspergillus oryzae. J. Food Sci. 44, 100–103. doi: 10.1111/j.1365-2621.1979.tb10016.x
Patil, A. G. G., and Mulimani, V. H. (2008). Removal of flatulence-inducing sugars by using free and polyvinyl alcohol immobilized α-galactosidase from Aspergillus oryzae. Biotechnol. Bioprocess Eng. 13:3. doi: 10.1007/s12257-008-0024-5
Paz-Arteaga, S. L., Ascacio-Valdés, J. A., Aguilar, C. N., Cadena-Chamorro, E., Serna-Cock, L., Aguilar-González, M. A., et al. (2023). Bioprocessing of pineapple waste for sustainable production of bioactive compounds using solid-state fermentation. Innovative Food Sci. Emerg. Technol. 85:103313. doi: 10.1016/j.ifset.2023.103313
Peng, J., Zhang, X., Wang, W., Zhu, T., Gu, Q., and Li, D. (2016). Austalides SU, new meroterpenoids from the sponge-derived fungus Aspergillus aureolatus HDN14-107. Mar. Drugs 14:131. doi: 10.3390/md14070131
Perrone, G., and Gallo, A. (2017). Aspergillus species and their associated mycotoxins. Methods Mol. Biol. 154, 33–49. doi: 10.1007/978-1-4939-6707-0_3
Petry, A. L., Huntley, N. F., Bedford, M. R., and Patience, J. F. (2020). Xylanase increased the energetic contribution of fiber and improved the oxidative status, gut barrier integrity, and growth performance of growing pigs fed insoluble corn-based fiber. J. Anim. Sci. 98:skaa233. doi: 10.1093/jas/skaa233
Pfefferle, W., Anke, H., Bross, M., Steffan, B., Vianden, R., and Steglich, W. (1990). Asperfuran, a novel antifungal metabolite from Aspergillus oryzae. J. Antibiot. 43, 648–654. doi: 10.7164/antibiotics.43.648
Phasha, V., Senabe, J., Ndzotoyi, P., Okole, B., Fouche, G., and Chuturgoon, A. (2022). Review on the use of kojic acid—a skin-lightening ingredient. Cosmetics 9:64. doi: 10.3390/cosmetics9030064
Pinto, C. A., Moreira, S. A., Fidalgo, L. G., Inácio, R. S., Barba, F. J., and Saraiva, J. A. (2020). Effects of high-pressure processing on fungi spores: factors affecting spore germination and inactivation and impact on ultrastructure. Compr. Rev. Food Sci. Food Saf. 19, 553–573. doi: 10.1111/1541-4337.12534
Pirota, R. D. P., Tonelotto, M., da Silva Delabona, P., Fonseca, R. F., Paixão, D. A. A., Baleeiro, F. C. F., et al. (2013). Enhancing xylanases production by a new Amazon Forest strain of Aspergillus oryzae using solid-state fermentation under controlled operation conditions. Ind. Crop. Prod. 45, 465–471. doi: 10.1016/j.indcrop.2013.01.010
Podversich, F., Tarnonsky, F., Bollatti, J. M., Silva, G. M., Schulmeister, T. M., Martinez, J. J. V., et al. (2023). Effects of Aspergillus oryzae prebiotic on animal performance, nutrients digestibility, and feeding behavior of backgrounding beef heifers fed with either a sorghum silage-or a byproducts-based diet. J. Anim. Sci. 101:skac312. doi: 10.1093/jas/skac312
Prashanth, S. J., and Mulimani, V. H. (2005). Soymilk oligosaccharide hydrolysis by Aspergillus oryzae α-galactosidase immobilized in calcium alginate. Process Biochem. 40, 1199–1205. doi: 10.1016/j.procbio.2004.04.011
Premalatha, A., Vijayalakshmi, K., Shanmugavel, M., and Rajakumar, G. S. (2023). Optimization of culture conditions for enhanced production of extracellular α-amylase using solid-state and submerged fermentation from Aspergillus tamarii MTCC5152. Biotechnol. Appl. Biochem. 70, 835–845. doi: 10.1002/bab.2403
Priesterjahn, E.-M., Geisen, R., and Schmidt-Heydt, M. (2020). Influence of light and water activity on growth and mycotoxin formation of selected isolates of Aspergillus flavus and Aspergillus parasiticus. Microorganisms 8:2000. doi: 10.3390/microorganisms8122000
Rad, A. H., Aghebati-Maleki, L., Kafil, H. S., and Abbasi, A. (2021). Molecular mechanisms of postbiotics in colorectal cancer prevention and treatment. Crit. Rev. Food Sci. Nutr. 61, 1787–1803. doi: 10.1080/10408398.2020.1765310
Reiss, E., and Lehmann, P. F. (1979). Galactomannan antigenemia in invasive aspergillosis. Infect. Immun. 25, 357–365.
Ren, R., Chen, C.-J., Hu, S.-S., Ge, H.-M., Zhu, W.-Y., Tan, R.-X., et al. (2015). Drimane Sesquiterpenoids from the Aspergillus oryzae QXPC-4. Chem. Biodivers. 12, 371–379. doi: 10.1002/cbdv.201400119
Rettger, L. F., and Cheplin, H. A. (1921). A treatise on the transformation of the intestinal flora: With special reference to the implantation of Bacillus acidophilus. New Haven, Connecticut: Yale University Press.
Ridlon, J. M., Kang, D. J., Hylemon, P. B., and Bajaj, J. S. (2014). Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30, 332–338. doi: 10.1097/MOG.0000000000000057
Ríus, A. G., Kaufman, J. D., Li, M. M., Hanigan, M. D., and Ipharraguerre, I. R. (2022). Physiological responses of Holstein calves to heat stress and dietary supplementation with a postbiotic from Aspergillus oryzae. Sci. Rep. 12:1587. doi: 10.1038/s41598-022-05505-3
Robertson, J., McGoverin, C., White, J. R., Vanholsbeeck, F., and Swift, S. (2021). Rapid detection of Escherichia coli antibiotic susceptibility using live/dead spectrometry for lytic agents. Microorganisms 9:924. doi: 10.3390/microorganisms9050924
Robinson, T., Singh, D., and Nigam, P. (2001). Solid-state fermentation. A promising microbial technology for secondary metabolite production. Appl. Microbiol. Biotechnol. 55, 284–289. doi: 10.1007/s002530000565
Rodrigues, J. C., Lima da Silva, W., Ribeiro da Silva, D., Maia, C. R., Santos Goiabeira, C. V., Figueiredo Chagas, H. D., et al. (2022, 2022). Antimicrobial activity of Aspergillus sp. from the Amazon biome. Isolation of kojic acid. Int. J. Microbiol. :4010018. doi: 10.1155/2022/4010018
Ropars, J., and Giraud, T. (2022). Convergence in domesticated fungi used for cheese and dry-cured meat maturation: beneficial traits, genomic mechanisms, and degeneration. Curr. Opin. Microbiol. 70:102236. doi: 10.1016/j.mib.2022.102236
Rose, E. C., Odle, J., Blikslager, A. T., and Ziegler, A. L. (2021). Probiotics, prebiotics and epithelial tight junctions: a promising approach to modulate intestinal barrier function. Int. J. Mol. Sci. 22:6729. doi: 10.3390/ijms22136729
Rosfarizan, M., Mohd, S. M., Nurashikin, S., Madihah, M. S., and Arbakariya, B. A. (2010). Kojic acid. Applications and development of fermentation process for production. Biotechnol. Mol. Biol. Rev. 5, 24–37.
Rousta, N., Hellwig, C., Wainaina, S., Lukitawesa, L., Agnihotri, S., Rousta, K., et al. (2021). Filamentous fungus Aspergillus oryzae for food: from submerged cultivation to fungal burgers and their sensory evaluation—a pilot study. Foods (Basel, Switzerland) 10:2774. doi: 10.3390/foods10112774
Russo, P., López, P., Capozzi, V., De Palencia, P. F., Dueñas, M. T., Spano, G., et al. (2012). Beta-glucans improve growth, viability and colonization of probiotic microorganisms. Int. J. Mol. Sci. 13, 6026–6039. doi: 10.3390/ijms13056026
Sakata, K., Kuwatsuka, T., Sakurai, A., Takahashi, N., and Tamura, G. (1983). Isolation of aspirochlorine (= antibiotic A30641) as a true antimicrobial constituent of the antibiotic, oryzachlorin, from Aspergillus oryzae. Agric. Biol. Chem. 47, 2673–2674. doi: 10.1271/bbb1961.47.2673
Salamanca, A. E. de, Diebold, Y., Calonge, M., García-Vazquez, C., Callejo, S., Vila, A., et al. (2006). Chitosan nanoparticles as a potential drug delivery system for the ocular surface. Toxicity, uptake mechanism and in vivo tolerance. Invest. Ophthalmol. Visual Sci. 47, 1416–1425.
Salazar, N., Gueimonde, M., Hernández-Barranco, A. M., Ruas-Madiedo, P., and de los Reyes-Gavilán, C. G. (2008). Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal bacteria. Am. Soc. Microbiol. 74, 4737–4745. doi: 10.1128/AEM.00325-08
Salminen, S., Collado, M. C., Endo, A., Hill, C., Lebeer, S., Quigley, E. M. M., et al. (2021). The international scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667. doi: 10.1038/s41575-021-00440-6
Sandhya, C., Sumantha, A., Szakacs, G., and Pandey, A. (2005). Comparative evaluation of neutral protease production by Aspergillus oryzae in submerged and solid-state fermentation. Process Biochem. 40, 2689–2694. doi: 10.1016/j.procbio.2004.12.001
Sankar, M., Mathew, R. M., Puthiyamadam, A., Sreeja-Raju, A., Christopher, M., Gokhale, D. V., et al. (2023). Comparison of the solid-state and submerged fermentation derived secretomes of hyper-cellulolytic Penicillium janthinellum NCIM 1366 reveals the changes responsible for differences in hydrolytic performance. Bioresour. Technol. 371:128602. doi: 10.1016/j.biortech.2023.128602
Saqib, S., Akram, A., Halim, S. A., and Tassaduq, R. (2017). Sources of β-galactosidase and its applications in food industry. 3 Biotech 7:79. doi: 10.1007/s13205-017-0645-5
Saranraj, P., Stella, D., and Reetha, D. (2012). Microbial cellulases and its applictions. Int. J. Biochem. Biotechnol. Sci. 1, 1–12.
Saruno, R., Kato, F., and Ikeno, T. (1979). Kojic acid, a tyrosinase inhibitor from Aspergillus albus. Agric. Biol. Chem. 43, 1337–1338. doi: 10.1271/bbb1961.43.1337
Sato, Y., Fukuda, H., Zhou, Y., and Mikami, S. (2010). Contribution of ethanol-tolerant xylanase G2 from Aspergillus oryzae on Japanese sake brewing. J. Biosci. Bioeng. 110, 679–683. doi: 10.1016/j.jbiosc.2010.07.015
Sato, S., Nomura, F., Kawai, T., Takeuchi, O., Mühlradt, P. F., Takeda, K., et al. (2000). Synergy and cross-tolerance between toll-like receptor (TLR) 2-and TLR4-mediated signaling pathways. J. Immunol. 165, 7096–7101. doi: 10.4049/jimmunol.165.12.7096
Savage, D. C. (1977). Microbial ecology of the gastrointestinal tract. Ann. Rev. Microbiol. 31, 107–133. doi: 10.1146/annurev.mi.31.100177.000543
Schiavi, E., Gleinser, M., Molloy, E., Groeger, D., Frei, R., Ferstl, R., et al. (2016). The surface-associated exopolysaccharide of Bifidobacterium longum 35624 plays an essential role in dampening host proinflammatory responses and repressing local TH17 responses. Appl. Environ. Microbiol. 82, 7185–7196. doi: 10.1128/AEM.02238-16
Schmalhorst, P. S., Krappmann, S., Vervecken, W., Rohde, M., Müller, M., Braus, G. H., et al. (2008). Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryot. Cell 7, 1268–1277.
Servillo, L., D'Onofrio, N., and Balestrieri, M. L. (2017). Ergothioneine antioxidant function. From chemistry to cardiovascular therapeutic potential. J. Cardiovasc. Pharmacol. 69, 183–191. doi: 10.1097/FJC.0000000000000464
Shah, I. J., Gami, P. N., Shukla, R. M., and Acharya, D. K. (2014). Optimization for α-amylase production by Aspergillus oryzae using submerged fermentation technology. Basic Res. J. Microbiol. 1, 1–10.
Shankar, S. K., Girigouda, K., and Mulimani, V. H. (2006). Production of alpha-galactosidase by Aspergillus oryzae using solid state fermentation. Bioresour. Technol. 98, 958–961. doi: 10.1016/j.biortech.2006.03.013
Sharma, P., and Rath, S. K. (2021). “Potential applications of fungi in the remediation of toxic effluents from pulp and paper industries” in Fungi Bio-Prospects in Sustainable Agriculture, Environment and Nano-Technology. ed. P. Sharma (Amsterdam, The Netherlands: Elsevier), 193–211.
Sheng, G.-P., Yu, H.-Q., and Li, X.-Y. (2010). Extracellular polymeric substances (EPS) of microbial aggregates in biological wastewater treatment systems. A review. Biotechnol. Adv. 28, 882–894. doi: 10.1016/j.biotechadv.2010.08.001
Sher, H., Faheem, M., Ghani, A., Mehmood, R., Rehman, H., and Bokhari, S. A. I. (2017). Optimization of cellulase enzyme production from Aspergillus oryzae for industrial applications. World J. Biol. Biotechnol. 2, 155–158. doi: 10.33865/wjb.002.02.0088
Shigeo, A., Akira, F., and Ichiro, T. K. (1962). Method of producing L-malic acid by fermentation. Google patents.
Silva, F. V. M. (2020). Ultrasound assisted thermal inactivation of spores in foods: pathogenic and spoilage bacteria, molds and yeasts. Trends Food Sci. Technol. 105, 402–415. doi: 10.1016/j.tifs.2020.09.020
Silva, Y. P., Bernardi, A., and Frozza, R. L. (2020). The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 11:508738. doi: 10.3389/fendo.2020.00025
Singh, R. P., and Bhardwaj, A. (2023). β-Glucans: a potential source for maintaining gut microbiota and the immune system. Front. Nutr. 10:1143682. doi: 10.3389/fnut.2023.1143682
Singh, S., Srivastava, P., Deep, S., Ashish, A., Priya, S., Huria, R., et al. (2022). Fermentation strategies for organic acid production. Ind. Microbiol. Biotechnol., 379–425. doi: 10.1007/978-981-16-5214-1_14
Son, S. Y., Lee, S., Singh, D., Lee, N.-R., Lee, D.-Y., and Lee, C. H. (2018). Comprehensive secondary metabolite profiling toward delineating the solid and submerged-state fermentation of Aspergillus oryzae KCCM 12698. Front. Microbiol. 9:1076. doi: 10.3389/fmicb.2018.01076
Steiger, M. G. (2021). Flow cytometry for filamentous Fungi. Methods Mol. Biol. 2234, 147–155. doi: 10.1007/978-1-0716-1048-0_13
Stelmaska, E., Pietkiewicz, D., and Dabrowski, W. (1974). Aspergillosis of the external ear. Pol Tyg Lek 29, 2039–2040.
Stephan, D., and Zimmermann, G. (1998). Development of a spray-drying technique for submerged spores of entomopathogenic fungi. Biocontrol Sci. Tech. 8, 3–11. doi: 10.1080/09583159830388
Subramanian, A. R., and Kalnitsky, G. (1964a). The major alkaline proteinase of Aspergillus oryzae, aspergillopeptidase B. II. Partial specific volume, molecular weight, and amino acid composition. Biochemistry 3, 1868–1874. doi: 10.1021/bi00900a013
Subramanian, A. R., and Kalnitsky, G. (1964b). The major alkaline proteinase of Aspergillus oryzae, Aspergillopeptidase B.* I. Isolation in homogeneous form. Biochemistry 3, 1861–1867. doi: 10.1021/bi00900a012
Subramaniyam, R., and Vimala, R. (2012). Solid state and submerged fermentation for the production of bioactive substances. A comparative study. Int J Sci Nat 3, 480–486.
Sudeep, K. C., Upadhyaya, J., Joshi, D. R., Lekhak, B., Kumar Chaudhary, D., Raj Pant, B., et al. (2020). Production, characterization, and industrial application of pectinase enzyme isolated from fungal strains. Fermentation 6:59. doi: 10.3390/fermentation6020059
Suleiman, W. B. (2023). A multi-aspect analysis of two analogous Aspergillus spp. belonging to section Flavi: Aspergillus flavus and Aspergillus oryzae. BMC Microbiol. 23:71. doi: 10.1186/s12866-023-02813-0
Sundarram, A., and Murthy, T. P. K. (2014). α-Amylase production and applications. A review. J. Appl. Environ. Microbiol. 2, 166–175. doi: 10.12691/jaem-2-4-10
Swallow, D. M. (2003). Genetics of lactase persistence and lactose intolerance. Annu. Rev. Genet. 37, 197–219. doi: 10.1146/annurev.genet.37.110801.143820
Swanson, K. S., Gibson, G. R., Hutkins, R., Reimer, R. A., Reid, G., Verbeke, K., et al. (2020). The international scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 17, 687–701. doi: 10.1038/s41575-020-0344-2
Szendefy, J., Szakacs, G., and Christopher, L. (2006). Potential of solid-state fermentation enzymes of Aspergillus oryzae in biobleaching of paper pulp. Enzym. Microb. Technol. 39, 1354–1360. doi: 10.1016/j.enzmictec.2006.06.016
Tahir, M., Saleh, F., Ohtsuka, A., and Hayashi, K. (2006). Pectinase plays an important role in stimulating digestibility of a corn-soybean meal diet in broilers. J. Poult. Sci. 43, 323–329. doi: 10.2141/jpsa.43.323
Takahashi, K. (2012). Effect of a cultures of Aspergillus oryzae on inflammatory response and mRNA expression in intestinal immune-related mediators of male broiler chicks. J. Poult. Sci. 49, 94–100. doi: 10.2141/jpsa.011079
Takeshita, N., and Oda, K. (2023). Japanese traditional fermentation uses solid-state fungal cultivation to produce sake. Microb. Ferm. Nat. Des. Proc. 1, 255–260. doi: 10.1002/9781119850007.ch9
Tamogami, S., Katayama, M., Marumo, S., and Isobe, M. (1996). Synthesis of the 5-Demethyl-6-deoxy analogue of Sporogen AO-1, a Sporogenic substance of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 60, 1372–1374. doi: 10.1271/bbb.60.1372
Tanaka, K., Kushiro, M., and Manabe, M. (2006). A review of studies and measures to improve the mycotoxicological safety of traditional Japanese mold-fermented foods. Mycotoxin Res. 22, 153–158. doi: 10.1007/BF02959268
Tanaka, Y., Shiomi, K., Kamei, K., Sugoh-Hagino, M., Enomoto, Y., Fang, F., et al. (1998). Antimalarial activity of radicicol, heptelidic acid and other fungal metabolites. J. Antibiot. 51, 153–160. doi: 10.7164/antibiotics.51.153
Tanaka, S., Wada, K., Katayama, M., and Marumo, S. (1984). Isolation of sporogen-AO1, a sporogenic substance, from Aspergillus oryzae. Agric. Biol. Chem. 48, 3189–3191. doi: 10.1271/bbb1961.48.3189
Tani, Y., Amaishi, Y., Funatsu, T., Ito, M., Itonori, S., Hata, Y., et al. (2014). Structural analysis of cerebrosides from Aspergillus fungi: the existence of galactosylceramide in A. oryzae. Biotechnol. Lett. 36, 2507–2513. doi: 10.1007/s10529-014-1631-1
Tao, L., and Chung, S. H. (2014). Non-aflatoxigenicity of commercial Aspergillus oryzae strains due to genetic defects compared to aflatoxigenic Aspergillus flavus. J. Microbiol. Biotechnol. 24, 1081–1087. doi: 10.4014/jmb.1311.11011
Tapre, A. R., and Jain, R. K. (2014). Pectinases. Enzymes for fruit processing industry. Int. Food Res. J. 21:447.
Taymaz-Nikerel, H., Cankorur-Cetinkaya, A., and Kirdar, B. (2016). Genome-wide transcriptional response of Saccharomyces cerevisiae to stress-induced perturbations. Front. Bioeng. Biotechnol. 4:17. doi: 10.3389/fbioe.2016.00017
Terabayashi, Y., Sano, M., Yamane, N., Marui, J., Tamano, K., Sagara, J., et al. (2010). Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 47, 953–961. doi: 10.1016/j.fgb.2010.08.014
Ting, W. E., Chang, C.-H., Szonyi, B., and Gizachew, D. (2020). Growth and aflatoxin B1, B2, G1, and G2 production by Aspergillus flavus and Aspergillus parasiticus on ground flax seeds (Linum usitatissimum). J. Food Prot. 83, 975–983. doi: 10.4315/JFP-19-539
Todokoro, T., Fukuda, K., Matsumura, K., Irie, M., and Hata, Y. (2016). Production of the natural iron chelator deferriferrichrysin from Aspergillus oryzae and evaluation as a novel food-grade antioxidant. J. Sci. Food Agric. 96, 2998–3006. doi: 10.1002/jsfa.7469
Todokoro, T., Negoro, H., Kotaka, A., Hata, Y., and Ishida, H. (2022). Aspergillus oryzae FaeA is responsible for the release of ferulic acid, a precursor of off-odor 4-vinylguaiacol in sake brewing. J. Biosci. Bioeng. 133, 140–145. doi: 10.1016/j.jbiosc.2021.11.001
Toida, J., Arikawa, Y., Kondou, K., Fukuzawa, M., and Sekiguchi, J. (1998). Purification and characterization of triacylglycerol lipase from Aspergillus oryzae. Biosci. Biotechnol. Biochem. 62, 759–763. doi: 10.1271/bbb.62.759
Tominaga, M., Lee, Y.-H., Hayashi, R., Suzuki, Y., Yamada, O., Sakamoto, K., et al. (2006). Molecular analysis of an inactive aflatoxin biosynthesis gene cluster in Aspergillus oryzae RIB strains. Appl. Environ. Microbiol. 72, 484–490. doi: 10.1128/AEM.72.1.484-490.2006
Travers, M.-A., Sow, C., Zirah, S., Deregnaucourt, C., Chaouch, S., Queiroz, R. M. L., et al. (2016). Deconjugated bile salts produced by extracellular bile-salt hydrolase-like activities from the probiotic Lactobacillus johnsonii La1 inhibit Giardia duodenalis in vitro growth. Front. Microbiol. 7:1453. doi: 10.3389/fmicb.2016.01453
Tse, T. J., Wiens, D. J., and Reaney, M. J. T. (2021). Production of bioethanol—a review of factors affecting ethanol yield. Fermentation 7:268. doi: 10.3390/fermentation7040268
Umemura, M., Koike, H., Yamane, N., Koyama, Y., Satou, Y., Kikuzato, I., et al. (2012). Comparative genome analysis between Aspergillus oryzae strains reveals close relationship between sites of mutation localization and regions of highly divergent genes among Aspergillus species. DNA Res. 19, 375–382. doi: 10.1093/dnares/dss019
Umeyama, L., Kasahara, S., Sugawara, M., Yokoyama, S., Saiki, I., and Hayakawa, Y. (2021). Anti-inflammatory effect of fermented brown rice and rice bran with Aspergillus oryzae on mice. Tradit. Kampo Med. 8, 60–65. doi: 10.1002/tkm2.1270
Utama, G. L., Dio, C., Sulistiyo, J., Yee Chye, F., Lembong, E., Cahyana, Y., et al. (2021). Evaluating comparative β-glucan production aptitude of Saccharomyces cerevisiae, Aspergillus oryzae, Xanthomonas campestris, and Bacillus natto. Saudi J. Biol. Sci. 28, 6765–6773. doi: 10.1016/j.sjbs.2021.07.051
Valadez-Carmona, L., Plazola-Jacinto, C. P., Hernández-Ortega, M., Hernández-Navarro, M. D., Villarreal, F., Necoechea-Mondragón, H., et al. (2017). Effects of microwaves, hot air and freeze-drying on the phenolic compounds, antioxidant capacity, enzyme activity and microstructure of cacao pod husks (Theobroma cacao L.). Innovative Food Sci. Emerg. Technol. 41, 378–386. doi: 10.1016/j.ifset.2017.04.012
Verma, N., Thakur, S., and Bhatt, A. K. (2012). Microbial lipases. Industrial applications and properties (a review). Int. Res. J. Biol. Sci 1, 88–92.
Vinderola, G., Sanders, M. E., Cunningham, M., and Hill, C. (2024). Frequently asked questions about the ISAPP postbiotic definition. Front. Microbiol. 14:1324565. doi: 10.3389/fmicb.2023.1324565
Vining, L. C. (1990). Functions of secondary metabolites. Ann. Rev. Microbiol. 44, 395–427. doi: 10.1146/annurev.mi.44.100190.002143
Wada, R., Maruyama, J., Yamaguchi, H., Yamamoto, N., Wagu, Y., Paoletti, M., et al. (2012). Presence and functionality of mating type genes in the supposedly asexual filamentous fungus Aspergillus oryzae. Appl. Environ. Microbiol. 78, 2819–2829. doi: 10.1128/AEM.07034-11
Walia, A., Guleria, S., Mehta, P., Chauhan, A., and Parkash, J. (2017). Microbial xylanases and their industrial application in pulp and paper biobleaching. A review. 3 Biotech 7, 1–12. doi: 10.1007/s13205-016-0584-6
Walsh, D., McCarthy, J., O’Driscoll, C., and Melgar, S. (2013). Pattern recognition receptors—molecular orchestrators of inflammation in inflammatory bowel disease. Cytokine Growth Factor Rev. 24, 91–104. doi: 10.1016/j.cytogfr.2012.09.003
Wang, R., Hu, X., Agyekumwaa, A. K., Li, X., Xiao, X., and Yu, Y. (2021). Synergistic effect of kojic acid and tea polyphenols on bacterial inhibition and quality maintenance of refrigerated sea bass (Lateolabrax japonicus) fillets. LWT 137:110452. doi: 10.1016/j.lwt.2020.110452
Wang, J., Huang, Z., Jiang, Q., Roubík, H., Xu, Q., Cai, M., et al. (2023). Fungal solid-state fermentation of crops and their by-products to obtain protein resources: The next frontier of food industry. Trends Food Sci. Technol. 138, 628–644. doi: 10.1016/j.tifs.2023.06.020
Wang, W., Jung, J., McGorrin, R. J., Traber, M. G., Leonard, S. W., Cherian, G., et al. (2018). Investigation of drying conditions on bioactive compounds, lipid oxidation, and enzyme activity of Oregon hazelnuts (Corylus avellana L.). LWT 90, 526–534. doi: 10.1016/j.lwt.2018.01.002
Wang, H., Lai, C., Tao, Y., Zhou, M., Tang, R., and Yong, Q. (2023). Evaluation of the enzymatic production and prebiotic activity of galactomannan oligosaccharides derived from Gleditsia microphylla. Fermentation 9:632. doi: 10.3390/fermentation9070632
Wasser, S. P. (2002). Medicinal mushrooms as a source of antitumor and immunomodulating polysaccharides. Appl. Microbiol. Biotechnol. 60, 258–274. doi: 10.1007/s00253-002-1076-7
Werpy, T., and Petersen, G. (2004). Top value added chemicals from biomass: Volume I--results of screening for potential candidates from sugars and synthesis gas.
Wu, Q., Li, S., Wang, H., Wang, W., Gao, X., Guan, X., et al. (2023). Construction of an efficient microalgal-fungal co-cultivation system for swine wastewater treatment: nutrients removal and extracellular polymeric substances (EPS)-mediated aggregated structure formation. Chem. Eng. J. 476:146690. doi: 10.1016/j.cej.2023.146690
Xu, C. P., Kim, S. W., Hwang, H. J., and Yun, J. W. (2006). Production of exopolysaccharides by submerged culture of an enthomopathogenic fungus, Paecilomyces tenuipes C240 in stirred-tank and airlift reactors. Bioresour. Technol. 97, 770–777. doi: 10.1016/j.biortech.2005.01.042
Xv, W., Zheng, Q., Ye, Z.-W., Wei, T., Guo, L.-Q., Lin, J.-F., et al. (2024). Submerged culture of edible and medicinal mushroom mycelia and their applications in food products: a review. Int. J. Med. Mushrooms 26, 1–13. doi: 10.1615/IntJMedMushrooms.2023052039
Yabuta, T. (1924). The constitution of Koji acid, a γ-Pyrone derivative formed by Aspergillus oryzae from carbohydrates. Bull. Agric. Chem. Soc. Jpn 1, 1–3. doi: 10.1271/bbb1924.1.1
Yamane, Y.-I., Fujita, J., Izuwa, S., Fukuchi, K., Shimizu, R.-I., Hiyoshi, A., et al. (2002a). Properties of cellulose-degrading enzymes from Aspergillus oryzae and their contribution to material utilization and alcohol yield in sake mash fermentation. J. Biosci. Bioeng. 93, 479–484. doi: 10.1016/S1389-1723(02)80095-0
Yamane, Y.-I., Fujita, J., Shimizu, R.-I., Hiyoshi, A., Fukuda, H., Kizaki, Y., et al. (2002b). Production of cellulose- and xylan-degrading enzymes by a koji mold, Aspergillus oryzae, and their contribution to the maceration of rice endosperm cell wall. J. Biosci. Bioeng. 93, 9–14. doi: 10.1016/S1389-1723(02)80046-9
Yan, S., Tang, H., Wang, S., Xu, L., Liu, H., Guo, Y., et al. (2014). Improvement of kojic acid production in Aspergillus oryzae B008 mutant strain and its uses in fermentation of concentrated corn stalk hydrolysate. Bioprocess Biosyst. Eng. 37, 1095–1103. doi: 10.1007/s00449-013-1081-5
Yang, L., Lübeck, M., Ahring, B. K., and Lübeck, P. S. (2016). Enhanced succinic acid production in Aspergillus saccharolyticus by heterologous expression of fumarate reductase from Trypanosoma brucei. Appl. Microbiol. Biotechnol. 100, 1799–1809. doi: 10.1007/s00253-015-7086-z
Yepuru, S. K. (2018). Production of alkaline protease from Aspergillus oryzae isolated from seashore of bay of Bengal. J. Appl. Nat. Sci. 10, 1210–1215. doi: 10.31018/jans.v10i4.1905
Yi, B.-H., and Kim, D.-H. (1982). Antioxidant activity of maltol, kojic acid, levulinic acid, furfural, 5-hydroxymethyl furfural, and pyrazine. Korean J. Food Sci. Technol. 14, 265–270.
Yu, J., Proctor, R. H., Brown, D. W., Abe, K., Gomi, K., Machida, M., et al. (2004). Genomics of economically significant Aspergillus and Fusarium species. Appl. Mycol. Biotechnol. 4, 249–283. doi: 10.1016/S1874-5334(04)80013-3
Zagórska, J., Czernicka-Boś, L., Kukula-Koch, W., Szalak, R., and Koch, W. (2022). Impact of thermal processing on the composition of secondary metabolites of ginger rhizome—a review. Foods (Basel, Switzerland) 11:3484. doi: 10.3390/foods11213484
Zambare, V. (2010). Solid state fermentation of Aspergillus oryzae for glucoamylase production on agro residues. Int. J. Life Sci 4, 16–25. doi: 10.3126/ijls.v4i0.2892
Zartl, B., Silberbauer, K., Loeppert, R., Viernstein, H., Praznik, W., and Mueller, M. (2018). Fermentation of non-digestible raffinose family oligosaccharides and galactomannans by probiotics. Food Funct. 9, 1638–1646. doi: 10.1039/C7FO01887H
Zhang, M., Li, Q., Lan, X., Li, X., Zhang, Y., Wang, Z., et al. (2020). Directed evolution of Aspergillus oryzae lipase for the efficient resolution of (R,S)-ethyl-2-(4-hydroxyphenoxy) propanoate. Bioprocess Biosyst. Eng. 43:393. doi: 10.1007/s00449-020-02393-7
Zhang, S., Sato, H., Ichinose, S., Tanaka, M., Miyazawa, K., Yoshimi, A., et al. (2017). Cell wall α-1,3-glucan prevents α-amylase adsorption onto fungal cell in submerged culture of Aspergillus oryzae. J. Biosci. Bioeng. 124, 47–53. doi: 10.1016/j.jbiosc.2017.02.013
Zhang, Z., Wang, B., Zhou, P., Guo, D., Kang, R., and Zhang, B. (2016). A novel approach of chemical mechanical polishing using environment-friendly slurry for mercury cadmium telluride semiconductors. Sci. Rep. 6:22466. doi: 10.1038/srep22466
Zhang, Y., Xue, C., Xue, Y., Gao, R., and Zhang, X. (2005). Determination of the degree of deacetylation of chitin and chitosan by X-ray powder diffraction. Carbohydr. Res. 340, 1914–1917.
Zhao, G., Ding, L.-L., Yao, Y., Cao, Y., Pan, Z.-H., and Kong, D.-H. (2018). Extracellular proteome analysis and flavor formation during soy sauce fermentation. Front. Microbiol. 9:1872. doi: 10.3389/fmicb.2018.01872
Zhao, G., Yao, Y., Qi, W., Wang, C., Hou, L., Zeng, B., et al. (2012). Draft genome sequence of Aspergillus oryzae strain 3.042. Am. Soc. Microbiol. 11:1178. doi: 10.1128/EC.00160-12
Zhong, Y., Lu, X., Xing, L., Ho, S. W. A., and Kwan, H. S. (2018). Genomic and transcriptomic comparison of Aspergillus oryzae strains. A case study in soy sauce koji fermentation. J. Ind. Microbiol. Biotechnol. 45, 839–853. doi: 10.1007/s10295-018-2059-8
Zhou, L.-B., and Chen, B. (2011). Bioactivities of water-soluble polysaccharides from Jisongrong mushroom. Anti-breast carcinoma cell and antioxidant potential. Int. J. Biol. Macromol. 48, 1–4. doi: 10.1016/j.ijbiomac.2010.09.004
Zhou, Y., Debbab, A., Wray, V., Lin, W., Schulz, B., Trepos, R., et al. (2014). Marine bacterial inhibitors from the sponge-derived fungus Aspergillus sp. Tetrahedron Lett. 55, 2789–2792. doi: 10.1016/j.tetlet.2014.02.062
Zhou, M., Zhou, K., He, P., Wang, K.-M., Zhu, R.-Z., Wang, Y.-D., et al. (2016). Antiviral and cytotoxic isocoumarin derivatives from an endophytic fungus Aspergillus oryzae. Planta Med. 82, 414–417. doi: 10.1055/s-0035-1558331
Zhu, Q., Egelston, C., Gagnon, S., Sui, Y., Belyakov, I. M., Klinman, D. M., et al. (2010). Using 3 TLR ligands as a combination adjuvant induces qualitative changes in T cell responses needed for antiviral protection in mice. J. Clin. Invest. 120, 607–616. doi: 10.1172/JCI39293
Żółkiewicz, J., Marzec, A., Ruszczyński, M., and Feleszko, W. (2020). Postbiotics-a step beyond pre- and probiotics. Nutrients 12:2189. doi: 10.3390/nu12082189
Zomkowski, A. D. E., Hammes, L., Lin, J., Calixto, J. B., Santos, A. R. S., and Rodrigues, A. L. S. (2002). Agmatine produces antidepressant-like effects in two models of depression in mice. Neuroreport 13, 387–391. doi: 10.1097/00001756-200203250-00005
Keywords: Aspergillus oryzae , postbiotic, immune modulation, fungal metabolites, gut microbiota
Citation: Seidler Y, Rimbach G, Lüersen K, Vinderola G and Ipharraguerre IR (2024) The postbiotic potential of Aspergillus oryzae – a narrative review. Front. Microbiol. 15:1452725. doi: 10.3389/fmicb.2024.1452725
Edited by:
Arun K. B., Christ University, IndiaCopyright © 2024 Seidler, Rimbach, Lüersen, Vinderola and Ipharraguerre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yvonne Seidler, c2VpZGxlckBmb29kc2NpLnVuaS1raWVsLmRl; Ignacio R. Ipharraguerre, aXBoYXJyYWd1ZXJyZUBmb29kc2NpLnVuaS1raWVsLmRl
 Yvonne Seidler
Yvonne Seidler Gerald Rimbach
Gerald Rimbach Kai Lüersen
Kai Lüersen Gabriel Vinderola
Gabriel Vinderola Ignacio R. Ipharraguerre
Ignacio R. Ipharraguerre