- 1Department of Plant Pathology, University of Minnesota, St. Paul, MN, United States
- 2Department of Ecology, Evolution and Behavior, University of Minnesota, St. Paul, MN, United States
- 3United States Department of Agriculture-Agricultural Research Service (USDA-ARS) Plant Science Research Unit, St. Paul, MN, United States
- 4Department of Microbiology, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
- 5Department of Microbiology and Plant Pathology, University of California, Riverside, Riverside, CA, United States
- 6Center for Biogeochemistry and Microbial Ecology (Soil BioME), University of New Hampshire, Durham, NC, United States
- 7Department of Natural Resources and the Environment, University of New Hampshire, Durham, NC, United States
Plants serve as critical links between above- and below-ground microbial communitites, both influencing and being influenced by microbes in these two realms. Below-ground microbial communities are expected to respond to soil resource environments, which are mediated by the roots of plants that can, in turn, be influenced by the above-ground community of foliar endophytes. For instance, diverse plant communities deposit more, and more diverse, nutrients into the soil, and this deposition is often increased when foliar pathogens are removed. Differences in soil resources can alter soil microbial composition and phenotypes, including inhibitory capacity, resource use, and antibiotic resistance. In this work, we consider plots differing in plant richness and application of foliar fungicide, evaluating consequences on soil resource levels and root-associated Streptomyces phenotypes. Soil carbon, nitrogen, phosphorus, potassium, and organic matter were greater in samples from polyculture than monoculture, yet this increase was surprisingly offset when foliar fungal communities were disrupted. We find that Streptomyces phenotypes varied more between richness plots—with the Streptomyces from polyculture showing lower inhibitory capacity, altered resource-use profiles, and greater antibiotic resistance—than between subplots with/without foliar fungicide. Where foliar fungicide affected phenotypes, it did so differently in polyculture than in monoculture, for instance decreasing niche width and overlap in monoculture while increasing them in polyculture. No differences in phenotype were correlated with soil nutrient levels, suggesting the need for further research looking more closely at soil resource diversity and particular compounds that were found to differ between treatments.
1 Introduction
While these two worlds can influence one another through myriad direct and indirect pathways, plants are critical links between above- and below-ground microbial ecosystems (Masters et al., 1993; Bardgett et al., 1998; Van der Putten et al., 2001; Bezemer and van Dam, 2005). In this work, we focus on the role of plants in facilitating biota in above-ground compartments to indirectly impact soil communities, by modifying plant productivity, growth, and morphology (Foley et al., 2021; Bagchi et al., 2017; Casas et al., 2011; Bardgett et al., 1998).
The phyllosphere represents a vast and functionally diverse microbial habitat, with complex effects on plant health and productivity. Plant-beneficial phyllosphere inhabitants can provide defense from pathogens or herbivores (Liu et al., 2020; Ritpitakphong et al., 2016), induce resistance (Bezemer and van Dam, 2005), produce plant hormones (Abadi et al., 2020; Lu et al., 2018), fix nitrogen (Madhaiyan et al., 2015), or confer resistance to abiotic stressors (Qu et al., 2020; Xu et al., 2022). However, foliar pathogens, especially fungi, often have strong negative effects on plant health and productivity and result in major crop losses worldwide (Teng et al., 1984).
Despite multiple studies focusing on herbivory by relatively large animals (insects, vertebrates), there are few data on linkages between foliar and soil microbial communities. Yet, impacts of foliar microbes on host plants are likewise expected to have consequences for below-ground communities. For example, fungicide treatments have been shown to increase above- and below-ground plant biomass (Seabloom et al., 2017). Luo et al. (2022) found that infection of Panax notoginseng with the foliar pathogen Alternaria panax enhanced root exudation of organic acids, sugars, and amino acids, as well as modified rhizosphere soil microbial community composition. Rudrappa et al. (2008) demonstrated that foliar infection of Arabidopsis thaliana by Pseudomonas syringae increased root exudation of D-malic acid and promoted root colonization by beneficial Bacillus subtilis. Over multiple growing seasons, changes in plant phenotypes or productivity because of plant-microbe interactions in the phyllosphere may modify soil resource concentrations (e.g., carbon, nitrogen, phosphorous, and potassium) in ways that can also impact soil microbial communities.
In contrast, plant host community richness has been repeatedly shown to exert broad influence on soil microbial communities through alteration of the above- and below-ground nutrient environments. Specifically, diverse plant communities are often substantially more productive (Tilman et al., 2001) and often experience altered pathogen effects (Mitchell et al., 2002; Seabloom et al., 2017) than monoculture. Moreover, plant richness has been hypothesized to be a crucial driver of soil bacterial community composition and function through the provision of greater abundances and diversities of plant-derived resources (Schlatter et al., 2015; Meier and Bowman, 2008; Bakker et al., 2013b; Ng et al., 2014). For example, greater abundances of carbon compounds support higher soil microbial densities, alter microbial community structure and diversity, and impact soil enzyme activities (Griffiths et al., 1998; Hernández and Hobbie, 2010). Further, greater diversity of carbon compounds in soil is hypothesized to increase the number of ecological niches available to soil microbes and has been found to increase microbial diversity and shift microbial carbon use capacities (Orwin et al., 2006; Essarioui et al., 2016; Kinkel et al., 2011).
In addition to ecological effects on soil microbes, resource environments likewise influence the co-evolutionary dynamics of microbial populations, including the structuring of inter-microbial interactions (Craig MacLean et al., 2005; Lawrence et al., 2012). For instance, Kinkel et al. (2011) have theorized that differing levels of disturbance and soil resource diversity can drive selection pressures for alternative evolutionary trajectories—favoring antagonism or niche differentiation (Pekkonen et al., 2013; Fiegna et al., 2015). Specifically, low-diversity resource environments, where microbes must compete for resource pools, are predicted to favor high frequencies of antagonistic microbes (e.g., antibiotic producers) and may generate a co-evolutionary arms-race (Bakker et al., 2013b; Kinkel et al., 2014, 2011). In contrast, a greater diversity of resources is hypothesized to promote adaptive radiation and niche differentiation among microbial taxa, as a means to minimize resource competition (Craig MacLean et al., 2005; Kinkel et al., 2014). Finally, greater resource abundance, by supporting higher microbial densities and encounter rates, may strengthen selection along either pathway, speeding the rate of co-evolution (Lopez Pascua et al., 2014).
Streptomyces are filamentous, gram-positive bacteria that are found ubiquitously in soils and are often closely associated with plant roots in the rhizosphere or endosphere (Viaene et al., 2016). As producers of antibiotic compounds, Streptomyces are important in clinical medicine and in suppressing soil-borne plant pathogens in natural and agricultural systems (Viaene et al., 2016; Barka et al., 2016; Schlatter and Kinkel, 2014; Jauri et al., 2016; Liu et al., 2019). In addition to their antibiotic-producing capacities, Streptomyces are a substantial natural reservoir of antibiotic resistance genes (D'Costa et al., 2007) and are highly diverse in their abilities to utilize resources for growth (Schlatter et al., 2013). Together, Streptomyces antibiotic inhibition, resistance, and resource use traits are all hypothesized to be critical for their interactions with other microbes, and in their capacities to respond to variation in resource inputs and impact plant health (Kinkel et al., 2014; Schlatter et al., 2009, 2013).
Variation in microbial interaction traits in soil are likely to reflect the abundance and diversity of soil resources, with consequences for microbial community diversity and function (Schlatter et al., 2009). Yet, despite the importance of plant-derived resources as a mechanistic link between plants and soil microbial communities, our understanding of the specific impacts of the abundance and diversity of plant-derived resources on traits of soil microbes in field settings remains limited. Here, we test how chronic disruption of foliar fungi (by fungicide applications) affects soil resources and Streptomyces interaction phenotypes (inhibition, resistance, and nutrient-use). We conduct this test in two communities that differ in plant richness (a monoculture and a polyculture with sixteen species). In agreement with previous research, we hypothesize that disruption of the foliar fungal community will increase below-ground carbon accumulation (Kohli et al., 2021; Zaret et al., 2022; Yang et al., 2009; Cong et al., 2014; Seabloom et al., 2017; but see Luo et al., 2022), and that these effects will be greater (in both absolute and relative terms) in diverse vs. monoculture plant communities (Seabloom et al., 2017). We expect these differences in soil nutrient levels to effect differences in Streptomyces community composition, as well as antibiotic inhibitory/resistance and resource use phenotypes, especially through an increase in inhibitory phenotypes in monoculture compared to more diverse plant communities (Bakker et al., 2013b). In accordance with the co-evolutionary theory put forward by Kinkel et al. (2011), we expect the disruption of the foliar fungal communities to favor escalation of antagonism, while an increase in plant (and therefore soil carbon) diversity will favor character displacement.
2 Methods
2.1 Experimental design and soil sampling
Soils for this study were collected in November 2013 from a long-term grassland biodiversity experiment at the University of Minnesota Cedar Creek Ecosystem Science Reserve (CCESR), part of the U.S Long Term Ecological Research (LTER) Network (45.4°N, 93.2°W). Soils at this site are sandy and nutrient poor (Grigal et al., 1974; Fay et al., 2015), and all topsoil was removed prior to the establishment of the biodiversity experiment (Tilman et al., 1997). The entire experimental field is burned each spring to remove any litter remaining from the previous year.
The two 9 × 9 m biodiversity plots from which our data were collected were established in 1994 (19 years prior to sampling); one was maintained as an Andropogon gerardii monoculture and the other was maintained as a 16 species native perennial polyculture containing Andropogon gerardii. In 2008, a foliar fungicide treatment [2 L m-2 Quilt (Syngenta Crop Protection, Inc.), a combination of Azoxystrobin (7.5 %) and Propiconazole (12.5 %)] applied biweekly) was nested into the plant diversity experimental plots to create 1.5 × 2 m fungicide treatment and control subplots within the 1 and 16 species whole plots. Care was taken in the application of the foliar fungicide to minimize contamination of the soil surface. In addition, Seabloom et al. (2017) found no direct effects of fungicide on plant biomass when grown in sterile soil. A more detailed description can be found in Borer et al. (2015), Seabloom et al. (2017). The current study sampled within the paired untreated control and fungicide treated subplots at each of the two levels of plant diversity.
Critically, due to the timing of fungicide application and sample collection, we do not interpret application of fungicide to mean foliar fungi were eliminated from these plants. While prior literature has highlighted the reduction in pathogenic fungi following fungicide application in these experimental plots (with consequent beneficial effects on host plants; Borer et al., 2015; Seabloom et al., 2017; Kohli et al., 2021), there has been less investigation into the consequences for foliar fungal communities as a whole. In other experimental systems, fungicide application was not found to reduce overall fungal diversity, richness, and colonization rate (Lane et al., 2023; Chen et al., 2020). In the absence of definitive evidence of the impact of our fungicidal treatment on the foliar community abundance and composition, in this work we consider this treatment as a temporary disruption of the foliar fungal community, with possible consequences for plant health and resource allocation.
Triplicate (1 cm diameter, 10 cm depth) soil cores were collected from within 10 cm of the central axis of 6 individual Andropogon gerardii plants from each subplot. Soil cores from the same individual plant were bulked in the field (n = 6 plants × 2 diversity treatments × 2 fungicide treatments = 24 composite samples), transported in a cooler to the lab, sieved (using a 2 mm mesh), and stored at -20 °C until processing. All processing, isolation, and phenotype characterization experiments took place in 2013–14.
2.2 Streptomyces densities and inhibitor densities
Streptomyces densities (total and proportion demonstrating inhibitory phenotypes) were determined as described previously (Bakker et al., 2013b). Briefly, soil samples were dried under two layers of sterile cheesecloth in a fume hood overnight. Dry soil samples were finely ground and 5 g of each soil was added to 25 mL of sterile deionized (DI) water and placed on an orbital shaker at 175 rpm at 4 °C for 60 min. Soil suspensions were serially diluted in sterile DI water and 100 μL of each dilution was spread onto 1 % water agar. After plates were allowed to dry, 5 mL of 1 % starch casein agar (SCA; Küster and Williams, 1964) was pipetted to cover the entire plate. Prior to overlaying, SCA was allowed to cool to prevent the medium from killing microorganisms. SCA is a semi-selective medium for Streptomyces and allows filamentous microbes to grow through the medium while suppressing non-filamentous bacteria (Oskay et al., 2009). Plates were incubated for 3 d at 28 °C and Streptomyces densities were evaluated by counting the number of colonies exhibiting characteristic Streptomyces morphology. After determining densities, a modified Herr's assay (Wiggins and Kinkel, 2005a,b) was used to assess inhibitory Streptomyces densities. Briefly, plates were overlaid with 10 mL of 1 % SCA, dried until solid, and then overlaid with 150 μL of spore suspension of an indicator strain (Streptomyces strains LK1324.2 or DL87). After 3 d of growth at 28 °C, Streptomyces colonies inhibiting the indicator overlay were counted and inhibition zone sizes were measured twice from the edge of the colony to the edge of clear overlay inhibition at right angles to one another. Overlay DL87 is a Streptomyces scabies isolate, which causes common scab disease of potato (Liu et al., 1996), and LK1324.2 is a non-pathogenic isolate previously obtained from CCESR soil (Davelos et al., 2004). For each composite soil sample, densities (CFU/g) were averaged over three replicate plates for each overlay isolate (n = 6 plates per soil sample).
2.3 Streptomyces isolation
After processing, soils from two individual plants/treatment (eight of the original 24 total bulked samples; two from each subplot) were randomly selected for Streptomyces isolation. Soils were serially diluted in sterile DI and 10 μL was spread on SCA. After 5 d of growth, colonies exhibiting characteristic Streptomyces morphology were selected by dividing each plate into a grid and randomly picking grid cells from which to select colonies. Five colonies were collected from three separate plates for each soil sample. Streptomyces colonies were picked with a sterile toothpick and streaked onto oatmeal agar (OA; Kharel et al., 2010). After 5 d of growth, single colonies were swabbed with a cotton applicator and spread as a lawn on OA. After ≈7 d growth, spores were collected by gently swabbing Streptomyces lawns with 4 mL of a 20 % glycerol solution. Spore stocks were stored at -20 °C until further use. Eighty Streptomyces isolates (n = 10 isolates per plant) were randomly selected for further characterization.
2.4 Streptomyces resource use characterization
For each Streptomyces isolate, resource use phenotypes on 95 distinct carbon sources were determined using Biolog SF-P2 plates (Biolog, Inc. Hayward, CA) as described previously (Schlatter et al., 2013). Briefly, fresh spore suspensions of Streptomyces isolates were adjusted to an OD590 of 0.22 and 100 μL of a spore suspension was inoculated into each well of a Biolog SF-P2 plate. After 3 d of growth at 28 °C, the absorbance in each well at 590 nm (AU590) was measured using a BioTek Synergy H1 plate reader (BioTek Instruments, Inc. Winooski, VT, USA). The absorbance in the water control well was subtracted from all other wells prior to subsequent analyses. A carbon source was considered to be used by an isolate if the AU590 was >0.01 above the water control well. Niche width and growth efficiency were determined for each isolate, where the niche width of an isolate is the number of used resources and resource use efficiency is the mean absorbance value for used resources. Niche overlap, a measure of shared resource use among isolates, was calculated for each pair of isolates a and b as the average pairwise niche overlap: , where is the absorbance (i.e., growth) of isolate x on carbon source i. Notably, this metric is asymmetric, such that, in general, .
2.5 16S rRNA gene sequencing
Partial 16S rRNA gene sequences were obtained for n = 76 isolates using previously described protocols (Davelos et al., 2004). Briefly, genomic DNA was extracted from cultures of each isolate grown in Yeast-Dextrose broth for 3 d (28 °C, 175 rpm) using the Wizard Genomic DNA Purification kit (Promega Corp., Madison, WI). PCR reactions consisted of 12.5 μL HotStart Master Mix (Qiagen), 0.75 μL of pA (10 pM), 0.75 μL of pH (10 pM), 1.0 μL of Streptomyces DNA (25 ng), and 10.0 μL of PCR grade H2O. Thermocycling conditions consisted of an initial denaturation of 95 °C for 4 min, followed by 30 cycles of 95 °C for 30 s, 50 °C for 30 s, 72 °C for 90 s, and a final extension at 72 °C for 7 min. PCR products were checked for the expected product on a 1 % agarose gel, purified with the Qiaquick PCR cleanup kit (Qiagen, Valencia, CA), and sequenced using the forward primer (pA) at ACGT, Inc (Wheeling, IL). Sequences were edited manually, classified using the RDP Classifier (Wang et al., 2007), and aligned using MUSCLE (v3.8.1551) (Edgar, 2004). Gaps were removed using trimAl (v1.4.rev22) (Capella-Gutiérrez et al., 2009) and a maximum likelihood phylogenetic tree was constructed using RAxML (v8.2.12) (Stamatakis, 2014) using a GTR+γ+I substitution model, Embleya hyalina strain NBRC 13850 as an outgroup, and 1,000 bootstraps to assess confidence. Phylogenetic diversity of isolates from each treatment was quantified with Faith's Phylogenetic Diversity using the pd function in the picante (v. 1.8.2) R package (Faith, 1992).
2.6 Antibiotic resistance
Resistance profiles against nine clinical antibiotics [kanamycin (30 μg), streptomycin (10 μg), erythromycin (15 μg), vancomycin (30 μg), amoxicillin/clavulanic acid (30 μg), novobiocin (5 μg), chloramphenicol (30 μg), rifampin (5 μg), and tetracycline (30 μg) (BD BBL Sensi-Disc; Becton, Dickinson and Company, Sparks, MD)] were determined for each isolate using a disk-diffusion assay as detailed in Otto-Hanson et al. (2013). Five of these antibiotics are produced by Streptomyces; and their modes of action include inhibition of protein, RNA, DNA gyrase, and cell wall synthesis.
In brief, 100 μL of each isolate was spread on plates containing 15 mL of SCA. Four different antibiotic discs were then applied to each plate and plates were incubated at 27 °C for 3 d. Inhibition zones were calculated by averaging two right-angle radial measurements. Each isolate-antibiotic combination was replicated twice.
2.7 Soil nutrient characteristics
Total soil carbon (C; %) and nitrogen (N; %) were determined for each composite soil sample (≈20 g/sample) at the University of Nebraska-Lincoln Ecosystem Analysis Laboratory using a Costech ECS 4010 elemental analyzer (Costech Analytical Technologies, Inc.). Soil (≈20 g/sample) from each composite sample was analyzed for soil organic matter (%), Bray-1 extracted phosphorus (P; ppm), NH4OAc extracted potassium (K; ppm), and pH at the University of Minnesota Research Analytical Laboratory (https://ral.cfans.umn.edu) using standard protocols.
2.8 Statistical analysis
Variance in phylogenetic (unweighted unifrac) distance was partitioned across treatments (plant diversity and fungicide application) with a permutational multivariate analysis of variance using the adonis2 function in the vegan (v. 2.6-4) R package (Oksanen et al., 2022). Differences in Streptomyces populations (total densities, inhibitory densities, and proportions), resource use (niche width, mean efficiency, niche overlap), and soil characteristics with plant richness and fungicides were assessed with a nested ANOVA, where the fungicide factor was nested within plant richness. All statistical analyses were performed in R (v. 4.3.1) (R Core Team, 2023) unless otherwise indicated.
3 Results
In this work, we examine differences in phenotypes across 80 Streptomyces isolates collected from plots that vary in plant richness and disruption of the foliar fungal community via fungicide application. Specifically, the fungicide treatment is nested within the richness plots, leading to a hierarchical design. We consider phylogenetic relatedness, inhibitory resource-use, and antibiotic phenotypes, as well as their relationship to in situ soil nutrient levels. In these results, we generally present statistical differences using post-hoc test results, focusing on the nested interaction between fungicide and plant richness; however, in some cases, we instead focus on differences between richness treatments that do not appear clearly when fungicide is additionally taken into account. Importantly, Streptomyces community composition is expected to vary seasonally, and in response to the pulse perturbation of foliar fungicide. Thus, our results represent a snapshot of time within these wider dynamics. Also note that, though we have replicated plants within each plot, our experimental design contains only one plot for each level of plant richness. As such, care must be taken to not overinterpret these results and to consider them in the context of the wider literature on plant richness and fungicide effects on soil Streptomyces communities.
3.1 Streptomyces community composition
The composition of Streptomyces communities differed among treatments (Figure 1; p = 0.048). When comparing plant richness or fungicide treatments, communities were significantly phylogenetically clustered according to plant richness (p = 0.020), but not disruption of foliar communities within monoculture or polyculture (p = 0.458). If the one outlying isolate is removed, monoculture communities were found to be more phylogenetically diverse than communities from polyculture (p = 0.003), but the effect of fungicide more nuanced, with untreated polyculture communities being less diverse than monoculture communities, and those from fungicide treated polyculture falling in between.
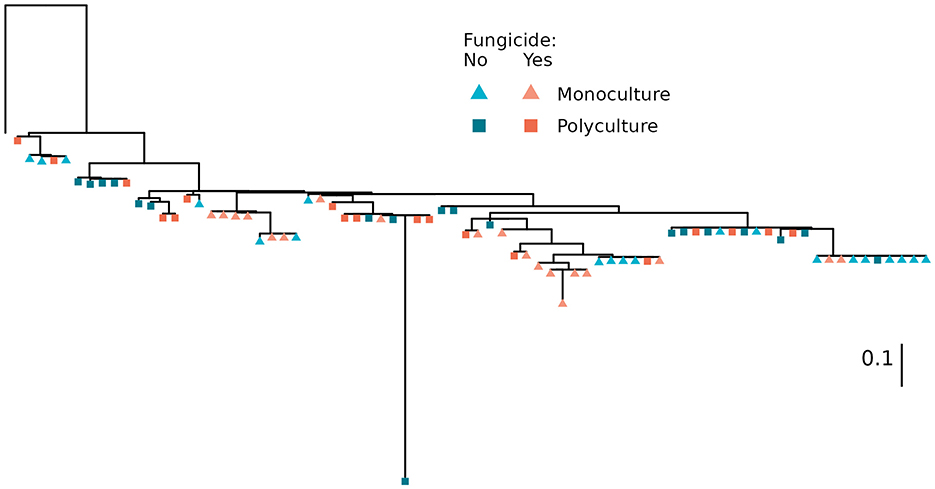
Figure 1. Phylogenetic relationships between isolates. Isolates are colored according to the plant richness of the sample plot and shaped according to foliar-community disruption via application of a foliar fungicide. Tree was generated with RAxML (v8.2.12) (Stamatakis, 2014) using a GTR+γ+I substitution model, Embleya hyalina strain NBRC 13850 as an outgroup (leftmost, unlabeled tip), and 1,000 bootstraps to assess confidence. The scale bar indicates evolutionary distance (substitution/site). Branch support values and isolate identity can be found in Supplementary Figure S1.
3.2 Soil nutrient characteristics
Soil resources [total carbon (C; %), total nitrogen (N; %), and organic matter (%)], as well as soil pH in the rooting zone of Andropogon gerardii varied significantly with foliar community disruption, but only in polyculture (Figure 2; Supplementary Table S1). Surprisingly, in polyculture, the untreated subplot tended to have greater C, N, and organic matter than the treated subplot, and than both treated and untreated monoculture. Soil in the monoculture plot was more acidic than polyculture soil. Moreover, in polyculture, the fungicide-treated subplot had lower pH (that is, more similar to monoculture) than untreated ones. Potassium (K; ppm) was higher in polyculture than monoculture, but with no differences between foliar fungicide treatments, and phosphorus (P; ppm) was unchanged across all treatments. In summary, resource abundance was found to vary significantly across treatments (Supplementary Table S2): polyculture had higher resource abundances than monoculture, and foliar fungal disruption was more influential in polyculture, where untreated polyculture samples had the highest resource abundances and treated polyculture samples were usually indistinguishable from monoculture samples.
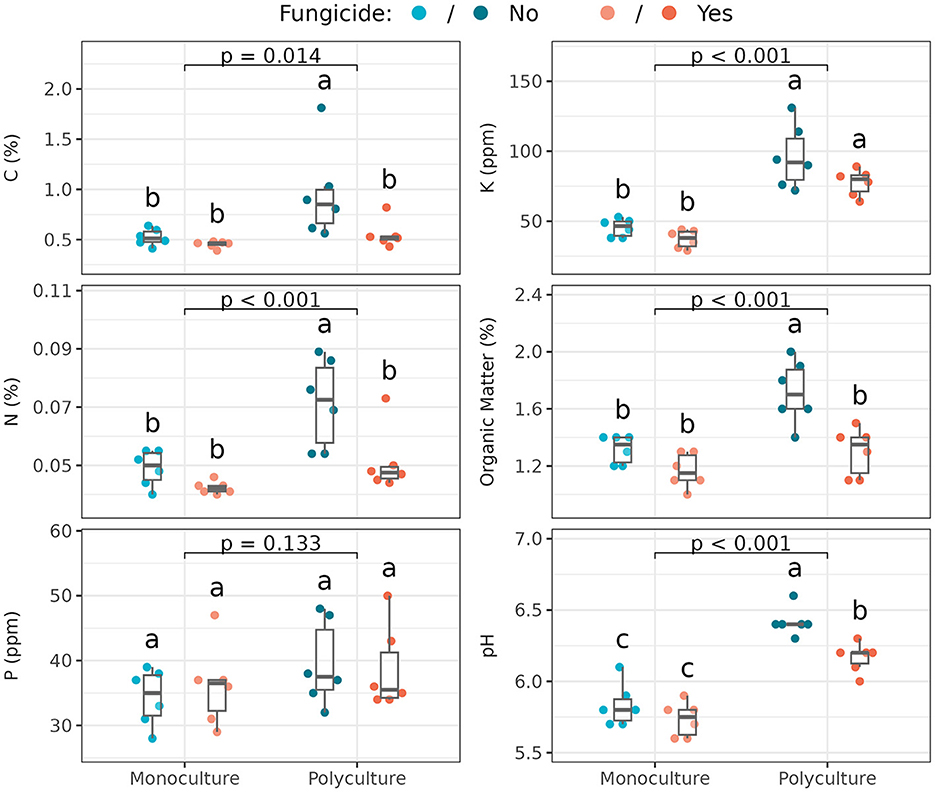
Figure 2. Soil nutrient characteristics among plant richness and fungicide treatments (n = 6 samples per treatment, i.e,. subplot). Box plots indicate 25th, 50th, and 75th percentiles of the data, with whiskers extending to the largest (smallest) value no further than 1.5 times the interquartile range beyond the 75th (25th) percentile. The reported p-value is for the plant richness effect, while different letters above boxplots indicate statistically significant differences between all four subplots using Tukey's HSD post-hoc test (p < 0.05). Means and standard deviations for each measure, as well as full ANOVA results are reported in Supplementary Tables S1, S2.
3.3 Streptomyces densities and inhibitory activities and their relationships with soil nutrient characteristics
Total Streptomyces densities and densities of inhibitory Streptomyces differed significantly among plant richness and fungicide treatments (Figure 3). Streptomyces densities were lowest in monoculture whose foliar fungal community was disrupted and higher in treated polyculture and untreated monoculture (Figure 3, Top). Considering only Streptomyces exhibiting inhibitory phenotypes, the monoculture plot supported significantly higher densities of inhibitors than did the polyculture plot, though there was no significant effect of foliar fungicide treatment on inhibitor densities in either plant richness treatment (Figure 3, Middle). Proportions of Streptomyces exhibiting inhibitory phenotypes varied significantly with both plant richness and disruption of foliar fungal communities (Figure 3, Bottom). Inhibitory Streptomyces comprised ≈ 10–20% more of the total Streptomyces community in monoculture vs. polyculture. Disruption of foliar fungal communities further increased the relative frequency of inhibitory Streptomyces in monoculture, but not in polyculture. Together, these data suggest that antibiotic-producing Streptomyces are selectively enriched in monoculture vs. polyculture settings, and that disruption of foliar community structure increases the selection for antibiotic inhibition, but only in monoculture (Essarioui et al., 2017). In contrast with densities and proportions of inhibitory Streptomyces, mean inhibition zone sizes, or the intensity of Streptomyces inhibitory phenotypes, did not vary significantly among treatments (data not shown).
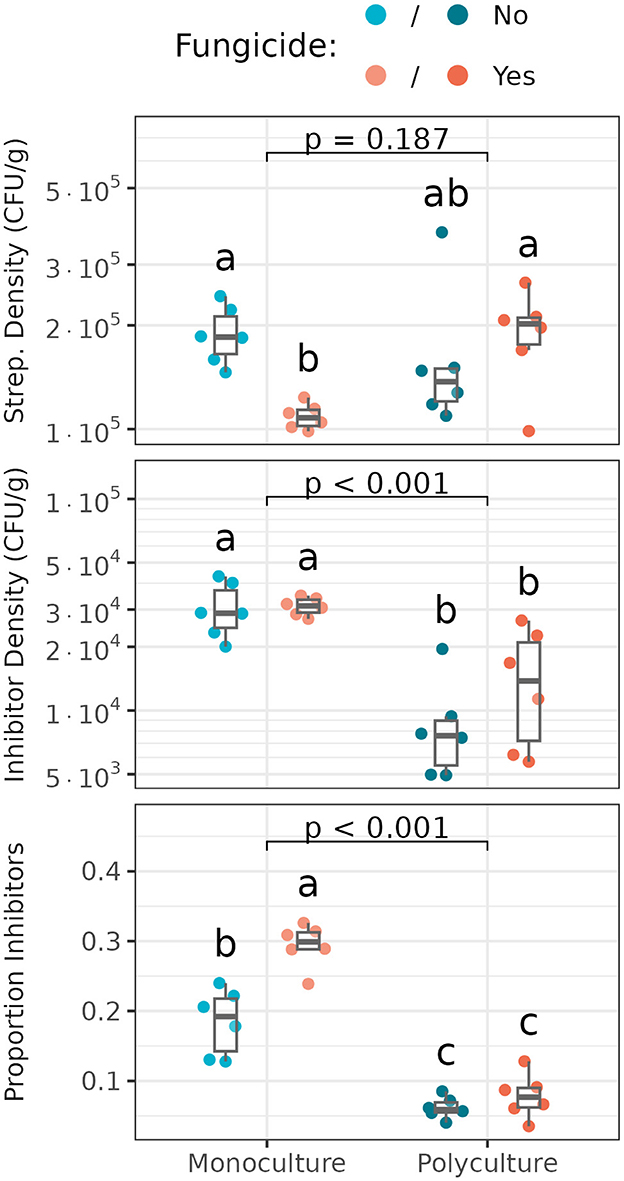
Figure 3. Densities of Streptomyces (Top), inhibitory Streptomyces (Middle), and proportions of Streptomyces that are inhibitory (Bottom) in untreated (blue) and fungicide-treated (orange) monoculture and polyculture. Box plots indicate 25th, 50th and 75th percentiles of the data, with whiskers extending to the largest (smallest) value no further than 1.5 times the interquartile range beyond the 75th (25th) percentile. The reported p-value is for the plant richness effect, while different letters above boxplots indicate statistically significant differences between all four subplots using Tukey's HSD post-hoc test (p < 0.05). Means and standard deviations for each measure, as well as full ANOVA results are reported in Supplementary Tables S7, S8.
Correlations between Streptomyces/inhibitor densities and soil nutrient characteristics differed between polyculture and monoculture plots. In monoculture, soil C, N, K, and organic matter were all positively correlated with Streptomyces densities (Pearson's R2 ≥0.36, p < 0.041; Supplementary Table S3). In contrast, the proportion of Streptomyces that exhibited inhibitory phenotypes were significantly negatively correlated with N and organic matter in monoculture (R2 ≥0.42, p < 0.022; Supplementary Table S3). In polyculture, however, there was only one significant correlation between Streptomyces density metrics and soil nutrients: a negative relationship between the density of inhibitors and soil P (R2 = 0.53, p = 0.017; Supplementary Table S4). This suggests that Streptomyces densities and inhibitory phenotypes in polyculture are less responsive to soil resource levels than those in monoculture.
Interestingly, the proportions of inhibitory Streptomyces were negatively correlated with overall Streptomyces densities in monoculture (R2 = 0.81, p < 0.001), but not polyculture (R2 = 0.05, p = 0.490), while absolute densities of inhibitors showed the opposite trend, significantly increasing with Streptomyces density in polyculture (R2 = 0.64, p = 0.002), but not in monoculture (R2 = 0.02, p = 0.691). Taken together, these suggest that as Streptomyces abundance increases in monoculture, non-inhibitory Streptomyces increase faster than their inhibitory counterparts, resulting in non-inhibitory Streptomyces representing a larger proportion of the total Streptomyces community than they did at lower abundances. However, in polyculture, the relative frequency of inhibitory Streptomyces remains largely constant, with both inhibitory and non-inhibitory Streptomyces increasing in tandem.
3.4 Nutrient use and niche overlap among Streptomyces isolates
Niche width, or the number of resources that individual Streptomyces could grow on, varied significantly with disruption of the foliar fungal community, but not with plant richness. However, the effect of this disruption on Streptomyces niche widths differed between monoculture vs. polyculture (Figure 4). Specifically, in the monoculture subplot where foliar fungicides were applied, Streptomyces had reduced niche widths compared to the subplot with undisturbed foliar fungal communities, while in polyculture, niche widths were increased where foliar fungicide was applied.
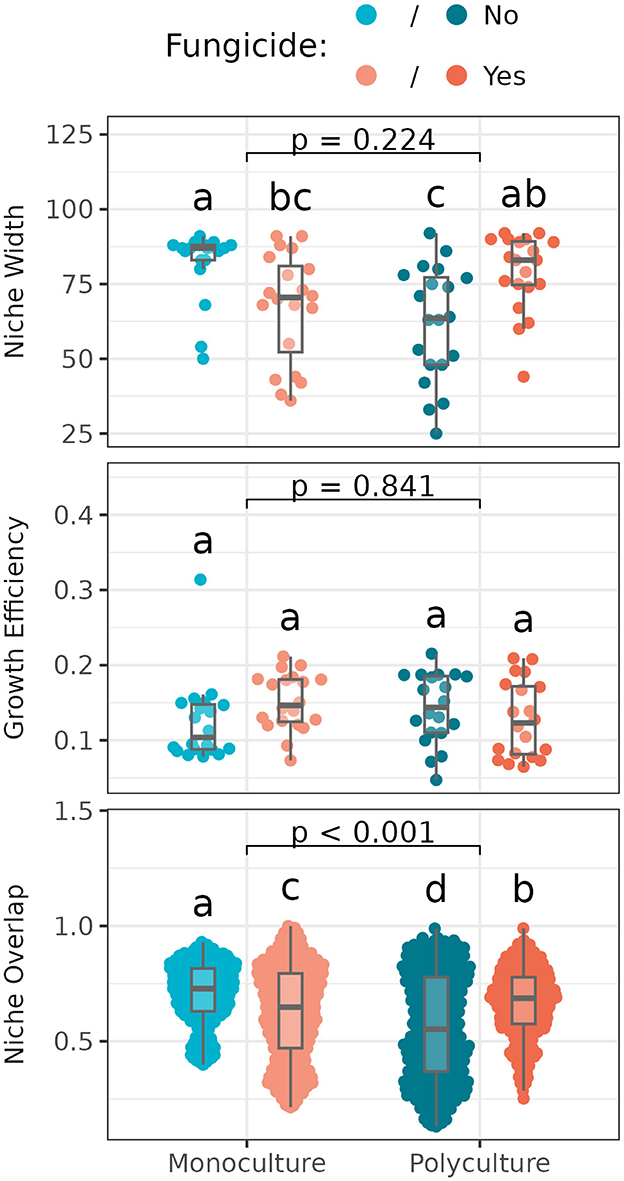
Figure 4. Niche width (number of carbon sources utilized; Top), growth efficiency (average optical density across utilized carbon sources; Middle), and niche overlap (shared use of carbon sources between paired isolates; Bottom) among Streptomyces from different plant richness and fungicide treatments. Box plots indicate 25th, 50th, and 75th percentiles of the data, with whiskers extending to the largest (smallest) value no further than 1.5 times the interquartile range beyond the 75th (25th) percentile. The reported p-value is for the plant richness effect, while different letters above boxplots indicate statistically significant differences between all four subplots using Tukey's HSD post-hoc test (p < 0.05). Means and standard deviations for each measure, as well as full ANOVA results are reported in Supplementary Tables S9, S10.
Growth efficiency, or the average growth of individual Streptomyces isolates across used resources, did not vary significantly with plant richness or fungicide treatment (Figure 4). Niche overlap, or the degree to which isolates sampled from the same plant grow on the same resources, varied significantly across both foliar fungicide and plant richness treatments (Figure 4), following a similar pattern as noted for niche width (with which niche overlap is strongly correlated; R2 = 0.79, p < 0.001).
3.5 Antibiotic resistance
For six of the nine antibiotics tested, we found no significant difference in inhibition zone size across treatments (Supplementary Tables S5, S6). For amoxicillin, erythromycin, and chloramphenicol, however, we found Streptomyces from monoculture were significantly more susceptible (i.e., had larger inhibition zones in response to antibiotic disks) than those from polyculture (N.b. that the difference in chloramphenicol was significant in an analysis of variance despite no significant pairwise differences found in the post-hoc analysis; Figure 5; Supplementary Table S6).
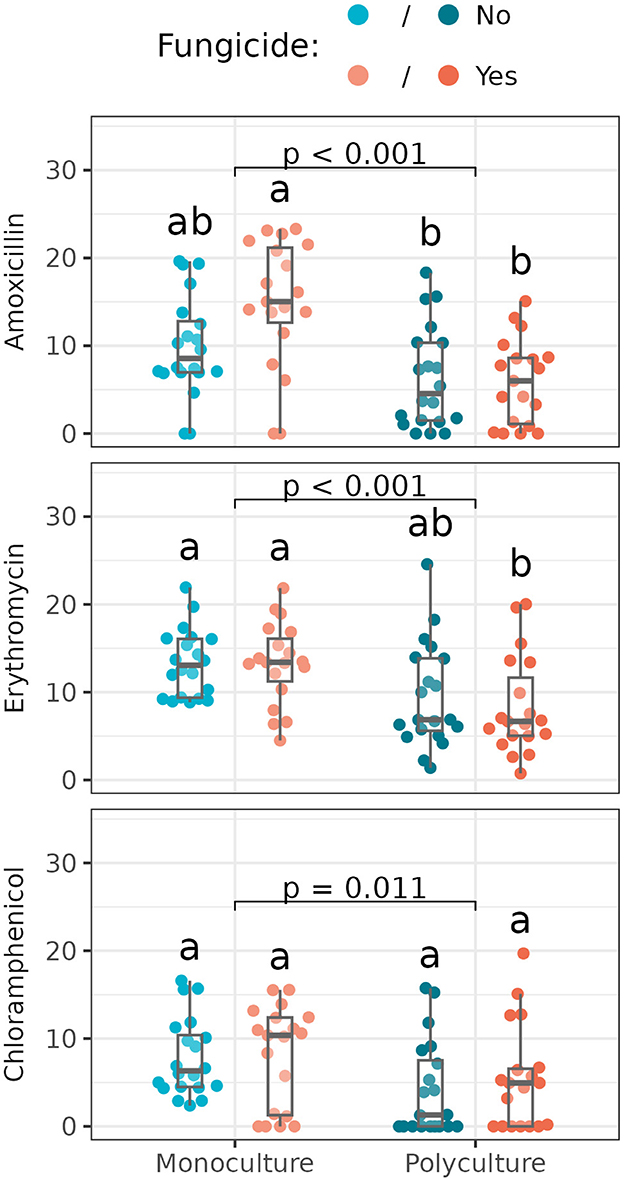
Figure 5. Resistance zone sizes of Streptomyces isolates from different plant richness and fungicide treatments (n = 18–20 isolates/treatment). Box plots indicate 25th, 50th, and 75th percentiles of the data, with whiskers extending to the largest (smallest) value no further than 1.5 times the interquartile range beyond the 75th (25th) percentile. The reported p-value is for the plant richness effect, while different letters above boxplots indicate statistically significant differences between all four subplots using Tukey's HSD post-hoc test (p < 0.05). Antibiotics without significant differences across treatments (and all means and standard deviations) are shown in Supplementary Table S5. An analysis of variance reports a significant difference between Streptomyces from monoculture vs. polyculture for all three antibiotics listed here, with more resistance (i.e., smaller resistance zone sizes) in Streptomyces from polyculture (Supplementary Table S6).
We found additional effects of foliar fungicide application on isolate susceptibility to amoxicillin: isolates from subplots in which the foliar fungal communities were disrupted were more susceptible to amoxicillin, compounding the difference seen between plant richnesses (Figure 5). Yet, in all three cases, more variance was explained by the plot-level richness treatment than disruption of the foliar fungal community (Supplementary Figure S2).
3.6 Differentially used nutrients among Streptomyces
Streptomyces growth rates differed between fungicide treatments for twenty-three carbon sources in monoculture and nine in polyculture (Figure 6; Supplementary Figure S3); however only seven of the significant relationships in monoculture (and none in polyculture) remained significant following correction for multiple hypothesis testing by controlling the false discovery rate (Benjamini and Hochberg, 1995; Benjamini and Yekutieli, 2001). Five of those seven showed a reduction in Streptomyces growth on the carbon source when foliar fungal communities were disrupted. Likewise, Fifteen comparisons were significant across plant richness treatment (Supplementary Figure S4), with none significant after correction. Applying a nested analysis of variance structure as used elsewhere in this work, we found nine carbon sources for which richness was significant, and sixteen for which disruption of the foliar fungal community (nested within richness) was significant (Supplementary Figures S5, S6).
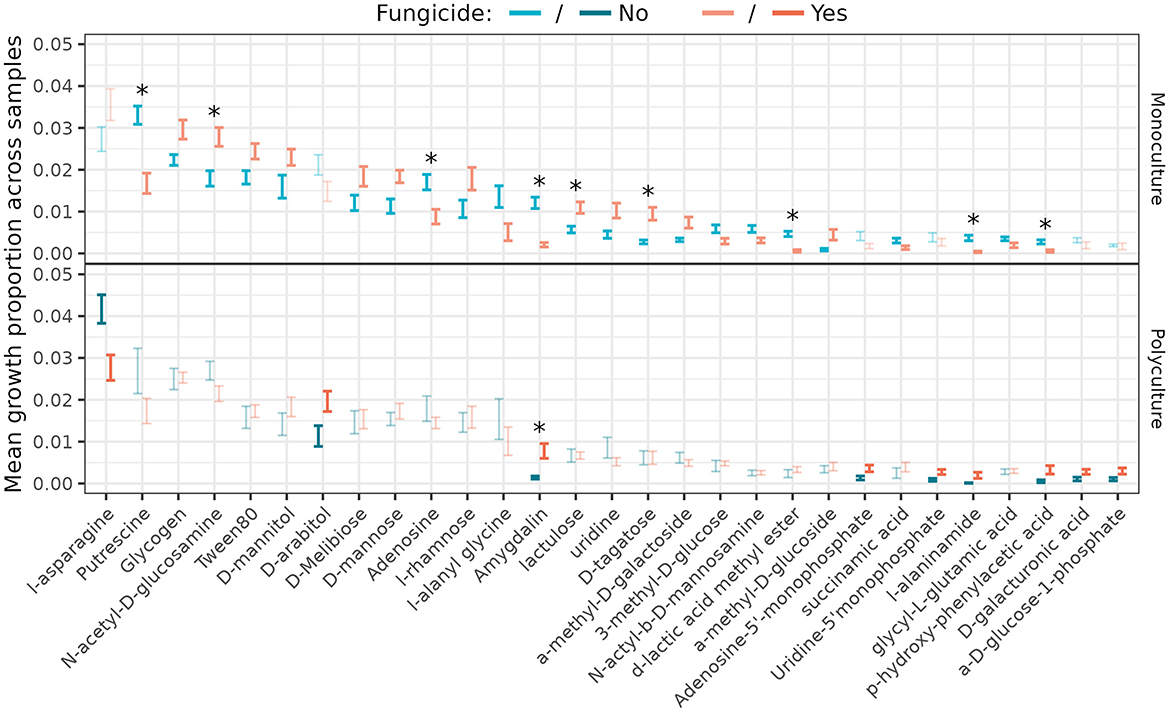
Figure 6. Proportion of total resource use by Streptomyces isolates from different plant richness (monoculture—top, polyculture—bottom) and fungicide (yes—orange, no—blue) treatments (n = 20 isolates/treatment). Error bars represent standard error around the mean. Significant (p < 0.05) pairwise differences are darker/thicker than non-significant comparisons, and those comparisons that remained significant following correction for multiple comparisons are marked with an asterisk. Only carbon sources for which the difference across fungicide treatments was significant for at least one level of plant richness were included. See Supplementary Figure S3 for all 95 carbon sources.
4 Discussion
The role of plant hosts and microbial species interactions in mediating selection for microbial phenotypes in soil remains poorly understood. Plant species richness is associated with both greater above-ground plant productivity and increased soil nutrient abundance and diversity (Tilman et al., 2001; Ng et al., 2014; Steinauer et al., 2016; El Moujahid et al., 2017; Yang et al., 2019; Furey and Tilman, 2021), in turn producing microbial communities with greater niche differentiation and increased inhibitory phenotypes in monoculture compared to polyculture plots (Orwin et al., 2006; Essarioui et al., 2016; Bakker et al., 2013b). These data suggest a key role for species interactions, modulated by plant species richness, in determining a microbial community's co-evolutionary trajectory (Kinkel et al., 2011).
Here we leverage a long-term experiment in which plant productivity in both monoculture and polyculture was enhanced through regular disruption of foliar fungal communities to test the hypothesis that enhanced plant productivity heightens selection for antagonistic vs. niche-differentiated Streptomyces populations. We hypothesized that plant productivity-driven increases in nutrient fluxes to soil microbes drive niche differentiation as an alternative to antagonistic phenotypes, which are posited to provide a competitive advantage in low-nutrient soils. Consistent with theoretical expectations, we found greater niche differentiation (Kinkel et al., 2011) and reduced inhibitory phenotypes (Bakker et al., 2013b) among Streptomyces from higher plant richness. However, despite enhanced above-ground productivity associated with foliar fungal community disruption, effects on Streptomyces populations were nuanced. Disruption of foliar fungal communities in monoculture increased inhibitory phenotypes and reduced niche width and niche overlap, while in polyculture, this treatment had no effect on inhibition and increased niche widths and niche overlap. Moreover, contrary to our expectations, none of the observed differences in Streptomyces inhibitory, resource use, and resistance phenotypes could be correlated to any soil resource measures.
Critically, our experimental design contains only one plot for each level of plant richness. While replicate samples were taken within each subplot, this design constrains our inference as to the importance of plant richness as a driver of microbial phenotypes, rather than, for example, a plot effect. Nevertheless, there is substantial evidence that plant richness is a key factor differentiating Streptomyces phenotypes in these plots (Steinauer et al., 2016; El Moujahid et al., 2017; Schlatter et al., 2015; Bakker et al., 2013b; Essarioui et al., 2017, 2016; Bakker et al., 2013a, 2010), and that it has consequences for soil nutrient diversity (Steinauer et al., 2016; El Moujahid et al., 2017) and abundance (Yang et al., 2019; Furey and Tilman, 2021). Moreover, we see consistent differences in Streptomyces phenotypes across fungicide treatments within the polyculture plot, even though the treated subplot has resource levels largely indistinguishable from either of the monoculture subplots. Thus, while we cannot exclude the possibility that plot effects contribute to the observed resource and phenotypic differences among Streptomyces, we focus here on changes in the context of differing plant richness.
4.1 Soil nutrients vary with plant richness and foliar fungicide
As expected, we found higher resource levels in soil from the Andropogon gerardii rhizosphere in polyculture compared to monoculture (Anacker et al., 2021; Lange et al., 2015; Furey and Tilman, 2021), in particular in measures of carbon, nitrogen, organic matter, and potassium. Work in these same experimental plots has shown that polycultures produce more diverse carbon compounds in the soil (Supplementary Figure S7), in agreement with prior literature (Steinauer et al., 2016; El Moujahid et al., 2017). Yet, we saw either no appreciable change in (in monoculture) or a decrease in total nutrient abundance (and pH) following fungicide application (in polyculture), in contrast to prior literature (Seabloom et al., 2017). Importantly, the soil carbon metrics used in this work may fail to capture highly-labile carbon substrates that are rapidly metabolized by soil microbes, or the diversity of carbon substrates, either one of which can differ with plant species richness. A more detailed look at the soil chemistry, perhaps focused on compounds for which we saw differences in Streptomyces consumption across treatments, in the future could confirm the inferences put forth here.
For carbon, nitrogen, and organic matter, however, the disruption of the foliar fungal communities had the opposite effect to expectation in polyculture: offsetting the relative increase from monoculture and producing nutrient levels similar to those observed in the monoculture. Considering pH, soil from polyculture was significantly less acidic than monoculture, with foliar community disruption reducing this difference (making soil more acidic), but not as acidic as monoculture. This could suggest an overall positive effect of foliar fungi on soil nutrients in polyculture [as has, for instance, been noted for endophytic and rhizospheric fungi (Van Der Heijden et al., 2016; Baron and Rigobelo, 2022; Chetia et al., 2019) and even, in some cases, pathogens (Luo et al., 2022)], one which is mitigated by the application of foliar fungicide. We did not find an effect of foliar fungal community disruption on any measure of soil nutrients in monoculture. However, in light of extensive previous literature demonstrating the effects of plant richness (Steinauer et al., 2016; Furey and Tilman, 2021; Anacker et al., 2021; Lange et al., 2015; El Moujahid et al., 2017) and foliar fungicide application (Luo et al., 2022; Zaret et al., 2023) on soil resource levels, it is possible we do not see it here in part due to spatio-temporal heterogeneity.
4.2 Soil nutrients do not explain most differences in Streptomyces phenotypes
In line with theoretical expectations suggesting that microbes are less likely to invest in direct competition when resources are plentiful (Kinkel et al., 2011; Granato et al., 2019), we see lower levels of inhibition and niche overlap in polyculture (Bakker et al., 2013b). Interestingly, however, we also saw significant changes in soil microbial phenotypes that were independent of changes in soil resource abundance. For instance, while most measures of soil chemistry did not differ under monoculture between fungicide treatments, we nevertheless observed differences in Streptomyces densities, proportion of inhibitory Streptomyces, niche width, and niche overlap. For Streptomyces densities and the proportion of Streptomyces that were inhibitory, the disruption of foliar fungal communities in monoculture appeared to preferentially reduce the densities of non-inhibitory Streptomyces: leaving the total number of inhibitors unchanged, but substantially increasing their relative abundance. In contrast, the polyculture did not exhibit different proportions of inhibitory Streptomyces, but did show differences in nutrient abundance, with higher resource abundance (untreated subplot) corresponding to narrower niches, in contrast with prior work (Lin et al., 2021; Dundore-Arias et al., 2019). Importantly, our assessment of inhibitory capacity was limited to two indicator strains, and the addition of additional strains might alter our conclusions.
Despite no difference in Streptomyces abundance between fungicide treated/untreated subplots in monoculture, Streptomyces abundance was nevertheless positively correlated with greater soil nutrients, while the proportion of inhibitory Streptomyces decreased with greater soil resources (there were few significant correlations in polyculture). Consistent with previous work, these data suggest the significance of constitutive antibiotic production to Streptomyces fitness is lower in high resource environments (Bakker et al., 2013b); however, plant richness has been shown to increase both resource abundance and diversity (Steinauer et al., 2016; Yang et al., 2019; Furey and Tilman, 2021; El Moujahid et al., 2017), somewhat confounding our analyses.
4.3 Streptomyces phenotypes varied more across plant richness than foliar fungicide
Rather than foliar fungicide application or soil nutrient abundance, most variation in Streptomyces inhibition, nutrient-use, and resistance in our experiment was between the two plots differing in the richness of their host plant communities. In particular, we found that Streptomyces clustered phylogenetically according to source plot/plant richness and that the monoculture plot had both more a more diverse community (Schlatter et al., 2015) and consistently had higher rates of inhibition (Bakker et al., 2013b), higher levels of pairwise niche-overlap, and less resistance to specific antibiotics than did Streptomyces from polyculture (Bakker et al., 2010; Vetsigian et al., 2011).
This discordance between inhibitory and resistance phenotypes builds on current understanding about the relationship between diversity and microbial antagonistic phenotypes. One explanation could be the relative fitness costs associated with maintaining resistance vs. inhibitory capacity (Gerardin et al., 2016; Czárán et al., 2002). If the former is more costly in the face of the diversity of antibiotics one's neighbors could produce, it might be selectively lost in favor of strong offensive capabilities in a resource-rich environment (Vetsigian et al., 2011). Alternatively, taken together with changes in niche width/overlap in monoculture, this could indicate a community-level trade-off between maintaining broad niches and inhibitory (but not resistance) phenotypes (Schlatter and Kinkel, 2015). However, the absence of this relationship in Streptomyces from polyculture argues against this interpretation. Finally, at least some of these resistance phenotypes could be evolutionary spandrels, serving primarily some other purpose in Streptomyces, but having the convenient side effect of detoxification (González-Candelas et al., 2011; Fajardo et al., 2008). Further research could explore this more comprehensively by, for instance, seeing whether these results are consistent when resistance is evaluated with respect to antibiotics produced by sympatric Streptomyces, rather than standard clinical antibiotics.
If this dominating effect of plot is driven by plant richness, it would be both interesting and in agreement with results in the study of disease-suppressive soils and, in particular, monoculture decline (Papavizas and Lumsden, 1980), where sustained low-diversity plant communities dramatically impact microbial interaction phenotypes and the functioning of soil communities (Weller et al., 2002). Suppressive soils develop in agricultural systems after long-term monoculture where high densities and/or frequencies of antagonistic microbes accumulate and limit plant disease development (typically species of Streptomyces, Pseudomonas, Bacillus, or Trichoderma; Schlatter et al., 2017). In contrast, introducing plant richness (temporally) with crop rotation can disrupt the development of disease-suppressive soil communities (Schlatter et al., 2017). Similarly, in experimental prairie plant richness manipulations, the soils of long-term monocultures contain higher frequencies of pathogen-antagonists than more diverse plant communities. This reduced soil antagonist community with increasing plant richness may feedback to contribute to plant fitness in polyculture (Bakker et al., 2013b; Essarioui et al., 2016). Though the mechanisms of general pathogen suppression are poorly understood, both ecological and evolutionary dynamics are likely to be crucial to the generation of disease-suppressive microbial communities.
4.4 Conclusion
Streptomyces phenotypes of inhibition, antibiotic resistance, and nutrient-use differed across our treatments. We found that soil from polyculture tended to have higher resource abundances and diversities than monoculture, resulting in Streptomyces populations with lower proportions of inhibitors and less competition for resources (i.e., less niche overlap). Yet, many phenotypic differences appeared to be unrelated to our measures of soil resource abundance. Disrupting the foliar fungal community did not result in the expected increase in measured soil resources in monoculture, and led to reduced soil resources in polyculture. Nevertheless, we saw differences in soil Streptomyces inhibition and resource use phenotypes across fungicide treatments in monoculture, suggesting that disruption of the above-ground, foliar fungal community has ramifications for below-ground microbes beyond the resource pathways we focused on here. Moreover, the effect of foliar fungal disruption on Streptomyces resource use in monoculture (reduced niche width and overlap) was opposite for Streptomyces from polyculture (increased niche width and overlap). The relatively larger and more consistent phenotypic effects across plant richness as compared to fungicide treatment highlights the importance of plants in linking above- and below-ground ecosystems, as well as suggesting a potentially greater role for soil resource diversity over soil resource abundance in phenotypic selection.
Data availability statement
The data presented in the study are deposited in the Environmental Data Initiative (https://portal.edirepository.org/nis/mapbrowse?scope=knb-lter-cdr&identifier=736), doi: 10.6073/pasta/f921a2a9ad3eba4789c9ec7fe8bc1ab7.
Author contributions
MM-S: Formal analysis, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. DS: Conceptualization, Methodology, Supervision, Writing – review & editing. NP: Conceptualization, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. SC: Resources, Visualization, Writing – review & editing. AG: Writing – review & editing. EB: Funding acquisition, Project administration, Writing – review & editing. ES: Funding acquisition, Project administration, Writing – review & editing. LK: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by funding from projects 2018-67013-27403 and 2018-67103-28061 from the U.S. Department of Agriculture's National Institute of Food and Agriculture, and by the Minnesota Agricultural Experiment Station. This work was further supported by grants from the US National Science Foundation Long-Term Ecological Research Program (LTER) including DEB-0620652, DEB-1234162, and DEB-1831944 and from the U.S. National Science Foundation grant MSB-1241895. Further support was provided by the Cedar Creek Ecosystem Science Reserve and the University of Minnesota. This research project was supported by The Office of the Permanent Secretary of the Ministry of Higher Education, Science: Research and Innovation Fund (RGNS65-005) to Nuttapon Pombubpa.
Acknowledgments
We gratefully acknowledge technical and field support from Lindsey Otto-Hanson and Jon Anderson. Mention of any trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1452534/full#supplementary-material
References
Abadi, V. A. J. M., Sepehri, M., Rahmani, H. A., Zarei, M., Ronaghi, A., Taghavi, S. M., et al. (2020). Role of dominant phyllosphere bacteria with plant growth-promoting characteristics on growth and nutrition of maize (Zea mays L.). J. Soil Sci. Plant Nutr. 20, 2348–2363. doi: 10.1007/s42729-020-00302-1
Anacker, B., Seastedt, T., Halward, T., and Lezberg, A. (2021). Soil carbon and plant richness relationships differ among grassland types, disturbance history and plant functional groups. Oecologia 196, 1153–1166. doi: 10.1007/s00442-021-04992-x
Bagchi, S., Roy, S., Maitra, A., and Sran, R. S. (2017). Herbivores suppress soil microbes to influence carbon sequestration in the grazing ecosystem of the trans-himalaya. Agric. Ecosyst. Environ. 239, 199–206. doi: 10.1016/j.agee.2017.01.033
Bakker, M. G., Bradeen, J. M., and Kinkel, L. L. (2013a). Effects of plant host species and plant community richness on streptomycete community structure. FEMS Microbiol. Ecol. 83, 596–606. doi: 10.1111/1574-6941.12017
Bakker, M. G., Glover, J. D., Mai, J. G., and Kinkel, L. L. (2010). Plant community effects on the diversity and pathogen suppressive activity of soil streptomycetes. Appl. Soil Ecol. 46, 35–42. doi: 10.1016/j.apsoil.2010.06.003
Bakker, M. G., Otto-Hanson, L., Lange, A., Bradeen, J. M., and Kinkel, L. L. (2013b). Plant monocultures produce more antagonistic soil Streptomyces communities than high-diversity plant communities. Soil Biol. Biochem. 65, 304–312. doi: 10.1016/j.soilbio.2013.06.007
Bardgett, R. D., Wardle, D. A., and Yeates, G. W. (1998). Linking above-ground and below-ground interactions: how plant responses to foliar herbivory influence soil organisms. Soil Biol. Biochem. 30, 1867–1878. doi: 10.1016/S0038-0717(98)00069-8
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Klenk, H.-P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Baron, N. C., and Rigobelo, E. C. (2022). Endophytic fungi: a tool for plant growth promotion and sustainable agriculture. Mycology 13, 39–55. doi: 10.1080/21501203.2021.1945699
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Benjamini, Y., and Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188. doi: 10.1214/aos/1013699998
Bezemer, T. M., and van Dam, N. M. (2005). Linking aboveground and belowground interactions via induced plant defenses. Trends Ecol. Evol. 20, 617–624. doi: 10.1016/j.tree.2005.08.006
Borer, E. T., Lind, E. M., Ogdahl, E. J., Seabloom, E. W., Tilman, D., Montgomery, R. A., et al. (2015). Food-web composition and plant diversity control foliar nutrient content and stoichiometry. J. Ecol. 103, 1432–1441. doi: 10.1111/1365-2745.12461
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Casas, C., Omacini, M., Montecchia, M. S., and Correa, O. S. (2011). Soil microbial community responses to the fungal endophyte neotyphodium in italian ryegrass. Plant Soil 340, 347–355. doi: 10.1007/s11104-010-0607-8
Chen, J., Akutse, K. S., Saqib, H. S. A., Wu, X., Yang, F., Xia, X., et al. (2020). Fungal endophyte communities of crucifer crops are seasonally dynamic and structured by plant identity, plant tissue and environmental factors. Front. Microbiol. 11:1519. doi: 10.3389/fmicb.2020.01519
Chetia, H., Kabiraj, D., Bharali, B., Ojha, S., Barkataki, M. P., Saikia, D., et al. (2019). Exploring the benefits of endophytic fungi via omics. Adv. Endophyt. Fung. Res. 51–81. doi: 10.1007/978-3-030-03589-1_4
Cong, W.-F., van Ruijven, J., Mommer, L., De Deyn, G. B., Berendse, F., and Hoffland, E. (2014). Plant species richness promotes soil carbon and nitrogen stocks in grasslands without legumes. J. Ecol. 102, 1163–1170. doi: 10.1111/1365-2745.12280
Craig MacLean, R., Dickson, A., and Bell, G. (2005). Resource competition and adaptive radiation in a microbial microcosm. Ecol. Lett. 8, 38–46. doi: 10.1111/j.1461-0248.2004.00689.x
Czárán, T. L., Hoekstra, R. F., and Pagie, L. (2002). Chemical warfare between microbes promotes biodiversity. Proc. Nat. Acad. Sci. U. S. A. 99, 786–790. doi: 10.1073/pnas.012399899
Davelos, A. L., Kinkel, L. L., and Samac, D. A. (2004). Spatial variation in frequency and intensity of antibiotic interactions among streptomycetes from prairie soil. Appl. Environ. Microbiol. 70, 1051–1058. doi: 10.1128/AEM.70.2.1051-1058.2004
D'Costa, V. M., Griffiths, E., and Wright, G. D. (2007). Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10, 481–489. doi: 10.1016/j.mib.2007.08.009
Dundore-Arias, J. P., Felice, L., Dill-Macky, R., and Kinkel, L. L. (2019). Carbon amendments induce shifts in nutrient use, inhibitory, and resistance phenotypes among soilborne Streptomyces. Front. Microbiol. 10:420796. doi: 10.3389/fmicb.2019.00498
Edgar, R. C. (2004). Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
El Moujahid, L., Le Roux, X., Michalet, S., Bellvert, F., Weigelt, A., and Poly, F. (2017). Effect of plant diversity on the diversity of soil organic compounds. PLoS ONE 12:e0170494. doi: 10.1371/journal.pone.0170494
Essarioui, A., Kistler, H. C., and Kinkel, L. L. (2016). Nutrient use preferences among soil Streptomyces suggest greater resource competition in monoculture than polyculture plant communities. Plant Soil 409, 329–343. doi: 10.1007/s11104-016-2968-0
Essarioui, A., LeBlanc, N., Kistler, H. C., and Kinkel, L. L. (2017). Plant community richness mediates inhibitory interactions and resource competition between Streptomyces and Fusarium populations in the rhizosphere. Microb. Ecol. 74, 157–167. doi: 10.1007/s00248-016-0907-5
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Fajardo, A., Martínez-Martín, N., Mercadillo, M., Galán, J. C., Ghysels, B., Matthijs, S., et al. (2008). The neglected intrinsic resistome of bacterial pathogens. PLoS ONE 3:e1619. doi: 10.1371/journal.pone.0001619
Fay, P. A., Prober, S. M., Harpole, W. S., Knops, J. M., Bakker, J. D., Borer, E. T., et al. (2015). Grassland productivity limited by multiple nutrients. Nat. Plants 1:15080. doi: 10.1038/nplants.2015.80
Fiegna, F., Moreno-Letelier, A., Bell, T., and Barraclough, T. G. (2015). Evolution of species interactions determines microbial community productivity in new environments. ISME J. 9, 1235–1245. doi: 10.1038/ismej.2014.215
Foley, K. M., Beard, K. H., Atwood, T. B., and Waring, B. G. (2021). Herbivory changes soil microbial communities and greenhouse gas fluxes in a high-latitude wetland. Microb. Ecol. 83, 127–136. doi: 10.1007/s00248-021-01733-8
Furey, G. N., and Tilman, D. (2021). Plant biodiversity and the regeneration of soil fertility. Proc. Nat. Acad. Sci. U. S. A. 118:e2111321118. doi: 10.1073/pnas.2111321118
Gerardin, Y., Springer, M., and Kishony, R. (2016). A competitive trade-off limits the selective advantage of increased antibiotic production. Nat. Microbiol. 1:16175. doi: 10.1038/nmicrobiol.2016.175
González-Candelas, F., Comas, I., Martínez, J. L., Galán, J. C., and Baquero, F. (2011). “The evolution of antibiotic resistance,” in Genetics and Evolution of Infectious Disease, ed. M. Tibayrenc (San Diego, CA: Elsevier), 305–337.
Granato, E. T., Meiller-Legrand, T. A., and Foster, K. R. (2019). The evolution and ecology of bacterial warfare. Curr. Biol. 29, R521–R537. doi: 10.1016/j.cub.2019.04.024
Griffiths, B., Ritz, K., Ebblewhite, N., and Dobson, G. (1998). Soil microbial community structure: effects of substrate loading rates. Soil Biol. Biochem. 31, 145–153. doi: 10.1016/S0038-0717(98)00117-5
Grigal, D., Chamberlain, L., Finney, H., Wroblewski, D., and Gross, E. (1974). Soils of the Cedar Creek Natural History Area. Miscellaneous report 123 of the Agricultural Experiment Station, University of Minnesota.
Hernández, D. L., and Hobbie, S. E. (2010). The effects of substrate composition, quantity, and diversity on microbial activity. Plant Soil 335, 397–411. doi: 10.1007/s11104-010-0428-9
Jauri, P. V., Altier, N., and Kinkel, L. L. (2016). Streptomyces for sustainability. Microb. Models 1, 251–276. doi: 10.1007/978-981-10-2555-6_12
Kharel, M. K., Shepherd, M. D., Nybo, S. E., Smith, M. L., Bosserman, M. A., and Rohr, J. (2010). Isolation of Streptomyces species from soil. Curr. Protoc. Microbiol. 19, 10E-4. doi: 10.1002/9780471729259.mc10e04s19
Kinkel, L. L., Bakker, M. G., and Schlatter, D. C. (2011). A coevolutionary framework for managing disease-suppressive soils. Annu. Rev. Phytopathol. 49, 47–67. doi: 10.1146/annurev-phyto-072910-095232
Kinkel, L. L., Schlatter, D. C., Xiao, K., and Baines, A. D. (2014). Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among streptomycetes. ISME J. 8, 249–256. doi: 10.1038/ismej.2013.175
Kohli, M., Henning, J. A., Borer, E. T., Kinkel, L. L., and Seabloom, E. W. (2021). Foliar fungi and plant diversity drive ecosystem carbon fluxes in experimental prairies. Ecol. Lett. 24, 487–497. doi: 10.1111/ele.13663
Küster, E., and Williams, S. (1964). Selection of media for isolation of streptomycetes. Nature 202, 928–929. doi: 10.1038/202928a0
Lane, B. R., Kendig, A. E., Wojan, C. M., Adhikari, A., Jusino, M. A., Kortessis, N., et al. (2023). Fungicide-mediated shifts in the foliar fungal community of an invasive grass. Phytob. J. 7, 198–207. doi: 10.1094/PBIOMES-03-22-0018-R
Lange, M., Eisenhauer, N., Sierra, C. A., Bessler, H., Engels, C., Griffiths, R. I., et al. (2015). Plant diversity increases soil microbial activity and soil carbon storage. Nat. Commun. 6:6707. doi: 10.1038/ncomms7707
Lawrence, D., Fiegna, F., Behrends, V., Bundy, J. G., Phillimore, A. B., Bell, T., et al. (2012). Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 10:e1001330. doi: 10.1371/journal.pbio.1001330
Lin, Q., Li, L., Adams, J. M., Heděnec, P., Tu, B., Li, C., et al. (2021). Nutrient resource availability mediates niche differentiation and temporal co-occurrence of soil bacterial communities. Appl. Soil Ecol. 163:103965. doi: 10.1016/j.apsoil.2021.103965
Liu, D., Anderson, N. A., and Kinkel, L. L. (1996). Selection and characterization of strains of Streptomyces suppressive to the potato scab pathogen. Can. J. Microbiol. 42, 487–502. doi: 10.1139/m96-066
Liu, D., Kinkel, L. L., Eckwall, E. C., Anderson, N. A., and Schottel, J. L. (2019). “Biological control of plant disease using antagonistic Streptomyces,” in Ecological Interactions and Biological Control, eds. D. A. Andow, D. W. Ragsdale, and R. F. Nyvall (Boca Raton, FL: CRC Press), 224–239.
Liu, H., Brettell, L. E., and Singh, B. (2020). Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 25, 841–844. doi: 10.1016/j.tplants.2020.06.003
Lopez Pascua, L., Hall, A. R., Best, A., Morgan, A. D., Boots, M., and Buckling, A. (2014). Higher resources decrease fluctuating selection during host-parasite coevolution. Ecol. Lett. 17, 1380–1388. doi: 10.1111/ele.12337
Lu, T., Ke, M., Lavoie, M., Jin, Y., Fan, X., Zhang, Z., et al. (2018). Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome 6, 1–12. doi: 10.1186/s40168-018-0615-0
Luo, L., Zhang, J., Ye, C., Li, S., Duan, S., Wang, Z., et al. (2022). Foliar pathogen infection manipulates soil health through root exudate-modified rhizosphere microbiome. Microbiol. Spectr. 10, e02418–e02422. doi: 10.1128/spectrum.02418-22
Madhaiyan, M., Alex, T. H. H., Ngoh, S. T., Prithiviraj, B., and Ji, L. (2015). Leaf-residing methylobacterium species fix nitrogen and promote biomass and seed production in jatropha curcas. Biotechnol. Biofuels 8, 1–14. doi: 10.1186/s13068-015-0404-y
Masters, G., Brown, V., and Gange, A. (1993). Plant mediated interactions between above-and below-ground insect herbivores. Oikos 66, 148–151. doi: 10.2307/3545209
Meier, C. L., and Bowman, W. D. (2008). Phenolic-rich leaf carbon fractions differentially influence microbial respiration and plant growth. Oecologia 158, 95–107. doi: 10.1007/s00442-008-1124-9
Mitchell, C. E., Tilman, D., and Groth, J. V. (2002). Effects of grassland plant species diversity, abundance, and composition on foliar fungal disease. Ecology 83, 1713–1726. doi: 10.1890/0012-9658(2002)083[1713:EOGPSD]2.0.CO;2
Ng, E. L., Patti, A. F., Rose, M. T., Schefe, C. R., Wilkinson, K., and Cavagnaro, T. R. (2014). Functional stoichiometry of soil microbial communities after amendment with stabilised organic matter. Soil Biol. Biochem. 76, 170–178. doi: 10.1016/j.soilbio.2014.05.016
Oksanen, J., Simpson, G. L., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., et al. (2022). vegan: Community Ecology Package. R package version 2, 6–4.
Orwin, K. H., Wardle, D. A., and Greenfield, L. G. (2006). Ecological consequences of carbon substrate identity and diversity in a laboratory study. Ecology 87, 580–593. doi: 10.1890/05-0383
Oskay, M. (2009). Comparison of Streptomyces diversity between agricultural and non-agricultural soils by using various culture media. Sci. Res. Essays 4, 997–1005.
Otto-Hanson, L., Grabau, Z., Rosen, C., Salomon, C., and Kinkel, L. (2013). Pathogen variation and urea influence selection and success of Streptomyces mixtures in biological control. Phytopathology 103, 34–42. doi: 10.1094/PHYTO-06-12-0129-R
Papavizas, G., and Lumsden, R. (1980). Biological control of soilborne fungal propagules. Annu. Rev. Phytopathol. 18, 389–413. doi: 10.1146/annurev.py.18.090180.002133
Pekkonen, M., Ketola, T., and Laakso, J. T. (2013). Resource availability and competition shape the evolution of survival and growth ability in a bacterial community. PLoS ONE 8:e76471. doi: 10.1371/journal.pone.0076471
Qu, Q., Zhang, Z., Peijnenburg, W., Liu, W., Lu, T., Hu, B., et al. (2020). Rhizosphere microbiome assembly and its impact on plant growth. J. Agric. Food Chem. 68, 5024–5038. doi: 10.1021/acs.jafc.0c00073
R Core Team (2023). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ritpitakphong, U., Falquet, L., Vimoltust, A., Berger, A., Métraux, J.-P., and L'Haridon, F. (2016). The microbiome of the leaf surface of arabidopsis protects against a fungal pathogen. New Phytol. 210, 1033–1043. doi: 10.1111/nph.13808
Rudrappa, T., Czymmek, K. J., Paré, P. W., and Bais, H. P. (2008). Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. doi: 10.1104/pp.108.127613
Schlatter, D. C., Bakker, M. G., Bradeen, J. M., and Kinkel, L. L. (2015). Plant community richness and microbial interactions structure bacterial communities in soil. Ecology 96, 134–142. doi: 10.1890/13-1648.1
Schlatter, D. C., DavelosBaines, A. L., Xiao, K., and Kinkel, L. L. (2013). Resource use of soilborne Streptomyces varies with location, phylogeny, and nitrogen amendment. Microb. Ecol. 66, 961–971. doi: 10.1007/s00248-013-0280-6
Schlatter, D. C., Fubuh, A., Xiao, K., Hernandez, D., Hobbie, S., and Kinkel, L. (2009). Resource amendments influence density and competitive phenotypes of Streptomyces in soil. Microb. Ecol. 57, 413–420. doi: 10.1007/s00248-008-9433-4
Schlatter, D. C., and Kinkel, L. L. (2014). Global biogeography of Streptomyces antibiotic inhibition, resistance, and resource use. FEMS Microbiol. Ecol. 88, 386–397. doi: 10.1111/1574-6941.12307
Schlatter, D. C., and Kinkel, L. L. (2015). Do tradeoffs structure antibiotic inhibition, resistance, and resource use among soil-borne Streptomyces? BMC Evol. Biol. 15, 1–11. doi: 10.1186/s12862-015-0470-6
Schlatter, D. C., Kinkel, L. L., Thomashow, L., Weller, D., and Paulitz, T. (2017). Disease suppressive soils: new insights from the soil microbiome. Phytopathology 107, 1284–1297. doi: 10.1094/PHYTO-03-17-0111-RVW
Seabloom, E. W., Kinkel, L., Borer, E. T., Hautier, Y., Montgomery, R. A., and Tilman, D. (2017). Food webs obscure the strength of plant diversity effects on primary productivity. Ecol. Lett. 20, 505–512. doi: 10.1111/ele.12754
Stamatakis, A. (2014). Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steinauer, K., Chatzinotas, A., and Eisenhauer, N. (2016). Root exudate cocktails: the link between plant diversity and soil microorganisms? Ecol. Evol. 6, 7387–7396. doi: 10.1002/ece3.2454
Teng, P., Shane, W., and MacKenzie, D. R. (1984). Crop losses due to plant pathogens. CRC Crit. Rev. Plant Sci. 2, 21–47. doi: 10.1080/07352688409382187
Tilman, D., Knops, J., Wedin, D., Reich, P., Ritchie, M., and Siemann, E. (1997). The influence of functional diversity and composition on ecosystem processes. Science 277, 1300–1302. doi: 10.1126/science.277.5330.1300
Tilman, D., Reich, P. B., Knops, J., Wedin, D., Mielke, T., and Lehman, C. (2001). Diversity and productivity in a long-term grassland experiment. Science 294, 843–845. doi: 10.1126/science.1060391
Van Der Heijden, M. G., De Bruin, S., Luckerhoff, L., Van Logtestijn, R. S., and Schlaeppi, K. (2016). A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J. 10, 389–399. doi: 10.1038/ismej.2015.120
Van der Putten, W. H., Vet, L. E., Harvey, J. A., and Wäckers, F. L. (2001). Linking above-and belowground multitrophic interactions of plants, herbivores, pathogens, and their antagonists. Trends Ecol. Evol. 16, 547–554. doi: 10.1016/S0169-5347(01)02265-0
Vetsigian, K., Jajoo, R., and Kishony, R. (2011). Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 9:e1001184. doi: 10.1371/annotation/1d584443-c6b8-423b-8027-5f9034d4599f
Viaene, T., Langendries, S., Beirinckx, S., Maes, M., and Goormachtig, S. (2016). Streptomyces as a plant's best friend? FEMS Microbiol. Ecol. 92:fiw119. doi: 10.1093/femsec/fiw119
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Weller, D. M., Raaijmakers, J. M., Gardener, B. B. M., and Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.030402.110010
Wiggins, B., and Kinkel, L. (2005a). Green manures and crop sequences influence potato diseases and pathogen inhibitory activity of indigenous streptomycetes. Phytopathology 95, 178–185. doi: 10.1094/PHYTO-95-0178
Wiggins, B. E., and Kinkel, L. L. (2005b). Green manures and crop sequences influence alfalfa root rot and pathogen inhibitory activity among soil-borne streptomycetes. Plant Soil 268, 271–283. doi: 10.1007/s11104-004-0300-x
Xu, N., Zhao, Q., Zhang, Z., Zhang, Q., Wang, Y., Qin, G., et al. (2022). Phyllosphere microorganisms: sources, drivers, and their interactions with plant hosts. J. Agric. Food Chem. 70, 4860–4870. doi: 10.1021/acs.jafc.2c01113
Yang, Y., Fang, J., Ji, C., and Han, W. (2009). Above-and belowground biomass allocation in tibetan grasslands. J. Veg. Sci. 20, 177–184. doi: 10.1111/j.1654-1103.2009.05566.x
Yang, Y., Tilman, D., Furey, G., and Lehman, C. (2019). Soil carbon sequestration accelerated by restoration of grassland biodiversity. Nat. Commun. 10:718. doi: 10.1038/s41467-019-08636-w
Zaret, M., Kinkel, L., Borer, E. T., and Seabloom, E. W. (2023). Soil nutrients cause threefold increase in pathogen and herbivore impacts on grassland plant biomass. J. Ecol. 111, 1629–1640. doi: 10.1111/1365-2745.14111
Keywords: soil nutrients, microbe-microbe interactions, phenotypes, co-evolution, phylogeny, plant diversity, soil ecology
Citation: Michalska-Smith M, Schlatter DC, Pombubpa N, Castle SC, Grandy AS, Borer ET, Seabloom EW and Kinkel LL (2024) Plant community richness and foliar fungicides impact soil Streptomyces inhibition, resistance, and resource use phenotypes. Front. Microbiol. 15:1452534. doi: 10.3389/fmicb.2024.1452534
Received: 21 June 2024; Accepted: 27 August 2024;
Published: 07 October 2024.
Edited by:
Claudia Knief, University of Bonn, GermanyReviewed by:
Cyril Bontemps, Université de Lorraine, FranceLouise Thatcher, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia
Mousumi Das, Atmiya University, India
Copyright © 2024 Michalska-Smith, Schlatter, Pombubpa, Castle, Grandy, Borer, Seabloom and Kinkel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew Michalska-Smith, bWljaGFsc2thLXNtaXRoQHBtLm1l
 Matthew Michalska-Smith
Matthew Michalska-Smith Daniel C. Schlatter
Daniel C. Schlatter Nuttapon Pombubpa
Nuttapon Pombubpa Sarah C. Castle
Sarah C. Castle A. Stuart Grandy
A. Stuart Grandy Elizabeth T. Borer
Elizabeth T. Borer Eric W. Seabloom
Eric W. Seabloom Linda L. Kinkel1
Linda L. Kinkel1