- 1Department of Sustainable Crop Production (DI.PRO.VE.S.), Università Cattolica del Sacro Cuore, Piacenza, Italy
- 2Research Center on Plant Health Modelling (PHeM), Università Cattolica del Sacro Cuore, Piacenza, Italy
Sour rot (SR) is a grapevine disease complex that is not completely understood in its etiology and epidemiology. Recently, SR has received special attention due to its increasing economic importance due to crop losses and reduced wine quality. In this study, the fungal and bacterial microbiota of healthy (i.e., without rot symptoms) and rotten (i.e., exhibiting visual and olfactory SR symptoms) ripe bunches were characterized across 47 epidemics (39 vineyards in six Italian grape-growing areas) over three years. The 16S rRNA gene, ITS high-throughput amplicon sequencing, and quantitative PCR were used to assess the relative abundance and dynamic changes of microorganisms associated with SR. The estimators of genera richness of fungal communities within samples indicated a significantly different diversity between healthy and rotten bunches. For bacterial communities, the healthy and rotten bunches significantly differed in the total number of species, but not in abundance distribution across species. The bunch status (i.e., healthy and rotten) was a significant source of diversity (p < 0.01) when the community composition between samples was evaluated, indicating that microbiome composition varied between healthy and rotten bunches. In particular, healthy and rotten bunches shared 43.1 and 54.8% of fungal and bacterial genera, respectively; 31.3% (fungal) and 26.2% (bacterial) genera were associated with rotten bunches only. The yeast genera Zygosaccharomyces, Zygoascus, Saccharomycopsis, Issatchenkia, and Pichia and the bacterial genera Orbus, Gluconobacter, Komagataeibacter, Gluconacetobacter, and Wolbachia were strongly associated with bunches showing SR symptoms based on a linear discriminant analysis. These microorganisms have been associated with Drosophila insects in literature. The relationships between the microflora associated with SR-affected bunches and the roles of Drosophila in SR development need further investigation, which may open perspectives for more effective disease control.
1 Introduction
Grapes are affected by some diseases, collectively known as bunch rots, which affect bunches during ripening (Pearson and Goheen, 1988). Botrytis bunch rot (or gray mold) is caused by Botryotinia fuckeliana and is undoubtedly the most widespread rot. The non-Botrytis rots are sour rot (SR) (caused by a complex of microorganisms), ripe rot (caused by Colletotrichum spp.), bitter rot (caused by the fungus Greeneria uvicola), and Botryosphaeria rot (caused by Botryosphaeria spp.), which is commonly associated with trunk disease. Minor rots are caused by Aspergillus spp. (mainly black Aspergilla), Cladosporium spp., Penicillium spp., and Rhizopus spp. These bunch rots impact wine composition and quality through the production of compounds potentially responsible for off-flavors and aromas in wine or the production of mycotoxins, such as ochratoxin A and patulin (Battilani and Pietri, 2002; Barata et al., 2008a; Steel et al., 2013; Rousseaux et al., 2014). SR has recently become increasingly relevant (Brischetto et al., 2024).
Grape berries affected by SR show oxidation of the grape skin, which turns brown in red and white varieties and then becomes extremely fragile and cracks. The softening of the berry follows the disaggregation of the internal berry tissue (Gravot et al., 2001; Hall et al., 2018b). Rotten berries are characterized by a strong and pungent smell as the result of the production of several chemical compounds, such as acetic acid, glycerol, ethyl acetate, ethanol, galacturonic acid, acetaldehyde, and gluconic acid (Marchetti et al., 1984; Zoecklein et al., 2000). The etiology of SR is complex. Multiple microorganisms, including yeasts, bacteria, and filamentous fungi (Barata, 2011; Hewstone et al., 2007; Steel et al., 2013), have been isolated from affected berries, with high variability among the studies conducted in different years, regions, and viticultural contexts (Brischetto et al., 2024). SR depends on infestation by Drosophila spp. flies (Diptera: Drosophilidae) (Hall et al., 2018a). Adults of D. melanogaster, the vinegar fly, and other Drosophila spp. (Vercesi and Bisiach, 1987) deposit eggs onto exposed fruit pulp and larvae preventing the healing of wounds through their movements and favoring the penetration of SR-related microorganisms in the presence of lesions on the berry skin (Barata et al., 2012a; Fermaud et al., 2002). In contrast, D. suzukii, the spotted wing fly, can lay eggs in unwounded grapes (Atallah et al., 2014; Rombaut et al., 2017).
A recent systematic literature review (Brischetto et al., 2024) showed that there is still uncertainty about the microorganisms primarily involved in SR etiology and whether the microorganisms involved differ by region or vary in abundance between symptomatic and asymptomatic grape berries. Indeed, only 10 papers have focused on the differences between microbial communities associated with healthy and rotten grapes. Most of these papers were based on classical cultural techniques, which led to the misestimation of microbial communities (Lleixà et al., 2018). More recent molecular methods provide a better picture of microbial populations (Nocker et al., 2007) associated with plant diseases (Huang et al., 2017; Shen et al., 2018; Brady et al., 2017). Hall et al. (2019) used high-throughput sequencing to characterize the microbiome of SR-affected grapes in New York, US, and Tasmania, AUS. Acetobacter spp. were more abundant in rotten berries than in healthy ones. The yeast genera Candida, Hanseniaspora, Pichia, and Saccharomyces were abundant in healthy and rotten berries. However, SR-associated organisms were grouped primarily by location, not by presence/absence of symptoms or grapes. Gao et al. (2020) conducted a metagenomic analysis to determine the diversity and abundance of bacteria and fungi in spoiled table grapes collected in eastern coastal China. The dominant bacteria genera in SR-affected grapes were Acetobacter, Gluconobacter, Bacillus, and Lactococcus. Issatchenkia terricola, Colletotrichum viniferum, Hanseniaspora vineae, Saprochaete gigas, and Candida diversa were dominant among fungi. Finding robust relationships between grape microflora and SR is relevant for extending the research to other grape-growing areas.
This study aimed to (i) determine the fungal and bacterial microbiota of healthy and rotten (i.e., exhibiting visual and olfactory SR symptoms) ripe bunches from different grape-growing areas of Italy over three years, (ii) characterize the diversity and composition of these microbiomes, and (iii) identify the microorganisms significantly associated with SR.
2 Materials and methods
2.1 Sample collection
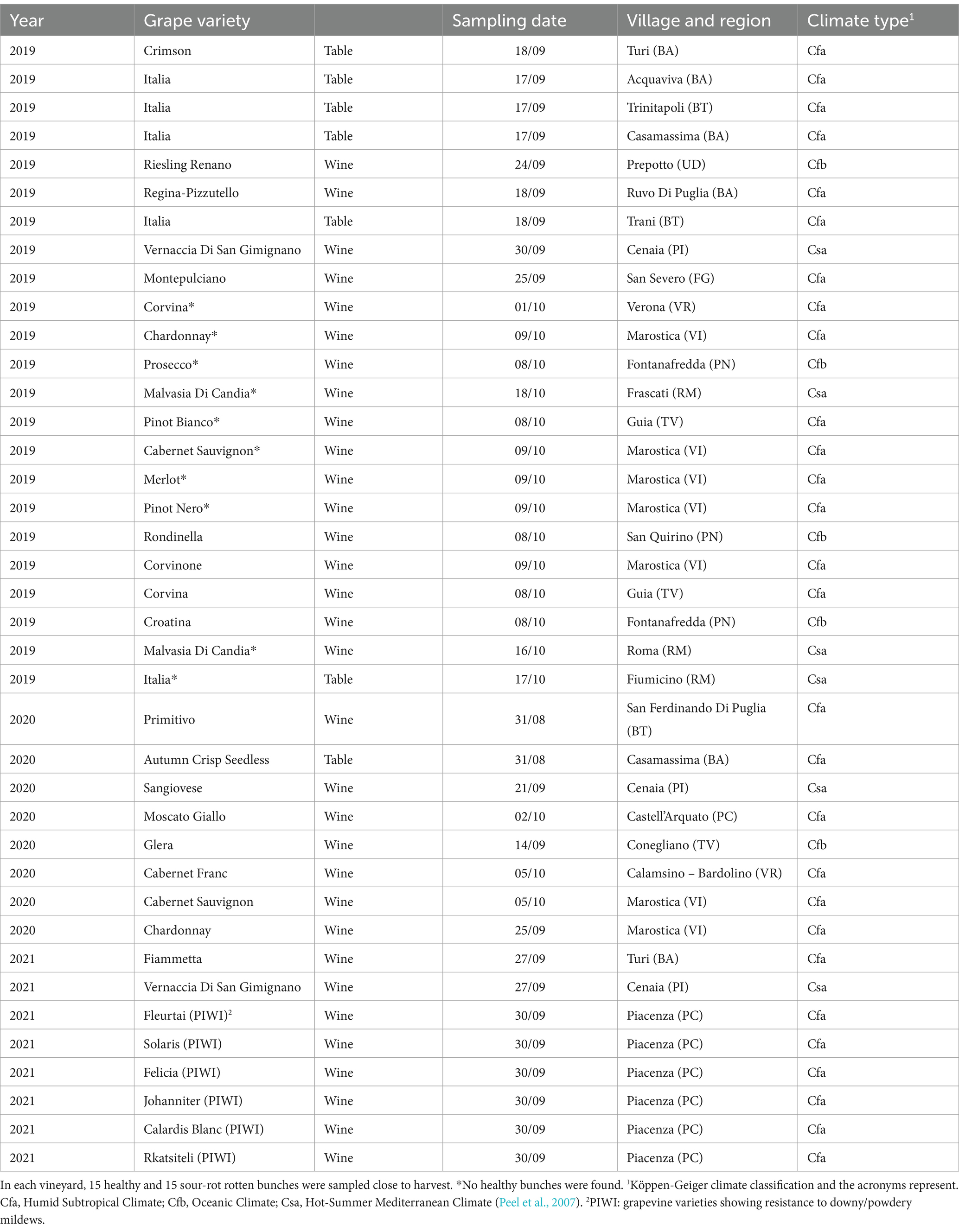
Grape samples were collected from 39 vineyards in six Italian grape-growing regions (i.e., Veneto, Friuli Venezia Giulia, Emilia Romagna, Toscana, Lazio, and Puglia) in 2019, 2020, and 2021 (Table 1). In each vineyard, ripe bunches (BBCH 89; Lorenz et al., 1995) were harvested and divided into two categories: healthy (i.e., without any rot symptoms) and rotten (i.e., exhibiting visual and olfactory SR symptoms). Fifteen random bunches were collected for each category.
For each sample, 100 berries were randomly removed from the bunches with sterilized scissors, placed in a plastic bag, and manually pressed. Then, 100 mL of the obtained must (i.e., a blend of pulp and juice obtained from the crushing of the berries) was extracted and placed into two 50 mL Falcon tubes. The samples were stored at −20°C until molecular analysis was performed.
2.2 DNA extraction, amplification, and sequencing
Must samples were sent to WineSeq laboratories1, 2 for total DNA extraction and high-throughput sequencing. Samples were processed using the Qiagen PowerSoil® DNA isolation Kit and analyzed for the 16 s rRNA V4 region and the ITS by amplification of the ITS1 region using WineSeq® custom primers (Patent WO2017096385). After quality control by electrophoresis gel, each library (16S and ITS) was pooled in equimolar amounts and subsequently sequenced on an Illumina MiSeq instrument (Illumina, San Diego, CA, USA) using 2×301 paired-end reads and according to the Biome-Makers implemented protocol. All the data produced and collected were subsequently analyzed using a QIIME-based custom bioinformatics pipeline (Patent WO2017096385, Biome Makers). The first quality control was used to remove adapters and chimeras. Later, the readings were trimmed, and sequence variant (SV) clusters were performed using 97% identity. SV clusters were compared with the WineSeq® taxonomy database (Patent WO2017096385) to identify the entire microbial population (bacteria, yeasts, and filamentous fungi) (Belda et al., 2017).
2.3 Data analysis
The fungal and bacterial SVs shared among bunch status were obtained using a Venn diagram analysis using the software available at: http://bioinformatics.psb.ugent.be/, accessed in June 2023.
Data analyses were performed using MicrobiomeAnalyst (Dhariwal et al., 2017; Chong et al., 2020). Data were filtered before the analysis based on the following criteria to remove low-quality or uninformative features: (i) SVs with less than four reads in a minimum of 20% of samples, and (ii) SVs with less than 10% inter-quantile range were excluded, because very small counts in very few samples are likely due to sequencing errors or low-level contaminations, and those that are close to constant throughout the experiment conditions are unlikely to be associated with the conditions under study.
Alpha diversity was calculated using Shannon and Chao1 indices in the Phyloseq package, and beta diversity was estimated using a principal coordinates analysis (PCoA) based on Bray–Curtis metrics (Vázquez-Baeza et al., 2013) with MicrobiomeAnalyst. PERMANOVA analysis was used to evaluate which SVs significantly differed in abundance among the experimental factors.
The linear discriminant analysis effect size (LEfSe) algorithm was used to identify taxa at the genus level that differed in relative abundance between bunch status (healthy and rotten) (Segata et al., 2011). MicrobiomeAnalyst LEfSe implementation was used. The threshold for the logarithmic linear discriminant analysis (LDA) score was set at 2.0, and the FDR-adjusted p-value cutoff was set at 0.1.
A correlation network analysis was performed using MicrobiomeAnalyst based on the SparCC algorithm (Friedman and Alm, 2012). The permutation was settled at 100, with a p-value threshold of 0.01 and a correlation threshold of 0.5 at the genus taxonomic level.
3 Results
A total of 313 fungal SVs were annotated in our study for a total of 8,260,221 reads; 41.2% of these SVs were present in both healthy and rotten bunches, and 31.3% were associated with rotten bunches only (Figure 1A). A total of 405 bacterial SVs were annotated for 3,468,723 reads; 26.2 and 19.1% of bacterial SVs were associated with rotten and healthy bunches, respectively, while the majority of bacterial SVs (54.7%) were in common (Figure 1B).
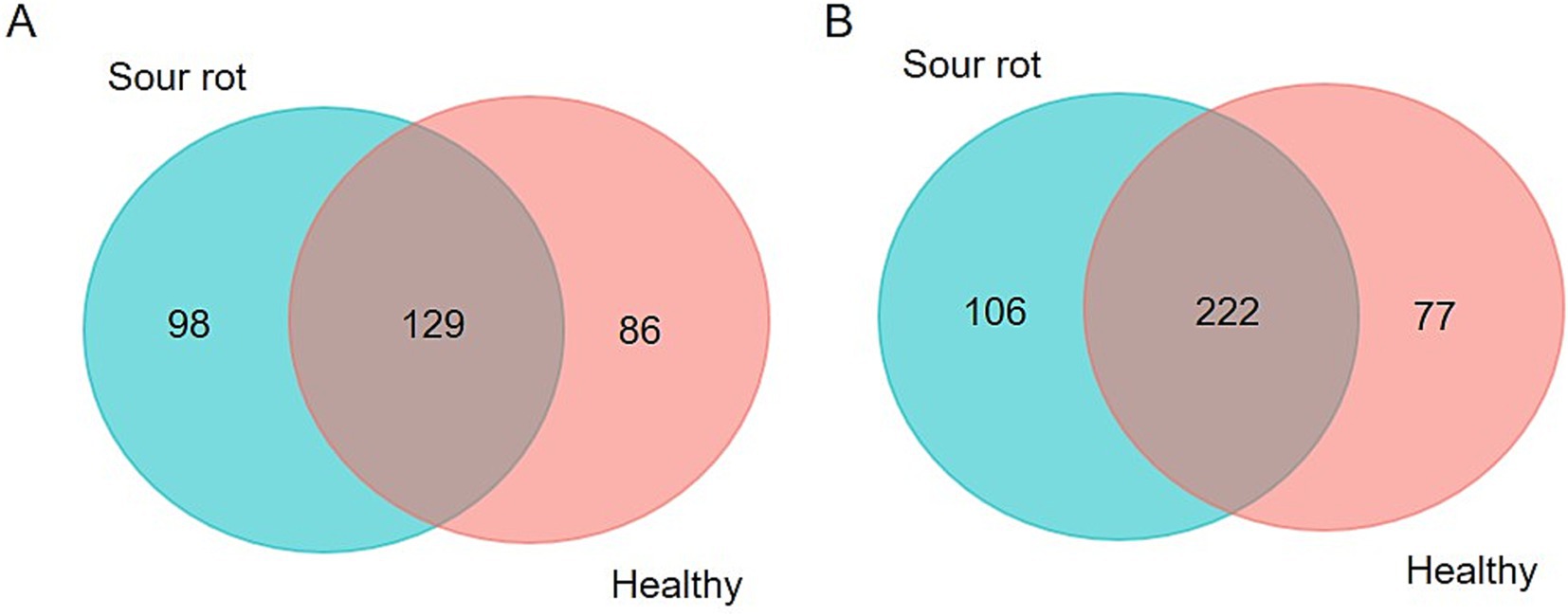
Figure 1. Venn diagram illustrating the overlap of the number of SVs identified in the fungal (A) and bacterial (B) microbiota between grapevine bunches showing sour rot symptoms or not (referred to as healthy).
For robust statistical analysis, 146 fungal SVs were eliminated for low abundance and four for low variance. Therefore, the analysis was conducted on 27 SVs. For bacterial SVs, 219 were eliminated for low abundance and seven for low variance, so 59 SVs (2,852,372 reads) were used. The numbers of reads for all SVs are shown in the Supplementary Table S1.
Figure 2 shows the relative abundance of fungal and bacterial genera detected in healthy and rotten bunches. The most abundant genera in healthy bunches were Hanseniaspora (13.4%), Candida (12.6%), Starmerella (9.9%), and Alternaria (9%) (Figure 2A). In rotten bunches, Candida was the most abundant genus (24.6%), followed by Starmerella (21.8%), Botryotinia (13.2%), Hanseniaspora (10.8%), and Botrytis (8.9%) (Figure 2A). Sphingomonas (10.6%), Gluconobacter (10.3%), and Bacillus (8.65%) were the most abundant genera in the healthy bunches (Figure 2B). The abundance of Gluconobacter spp. increased in rotten bunches (18.8%), while that of Sphingomonas spp. and Bacillus spp. decreased (3.6 and 2%, respectively). Komagataeibacter spp. (27.4%), Orbus spp. (11.8%), and Gluconacetobacter spp. (7.7%) were also abundant in rotten bunches. Pantoea spp. were present in healthy and rotten bunches with lower abundance (5.9 and 1.5%, respectively) (Figure 2B).
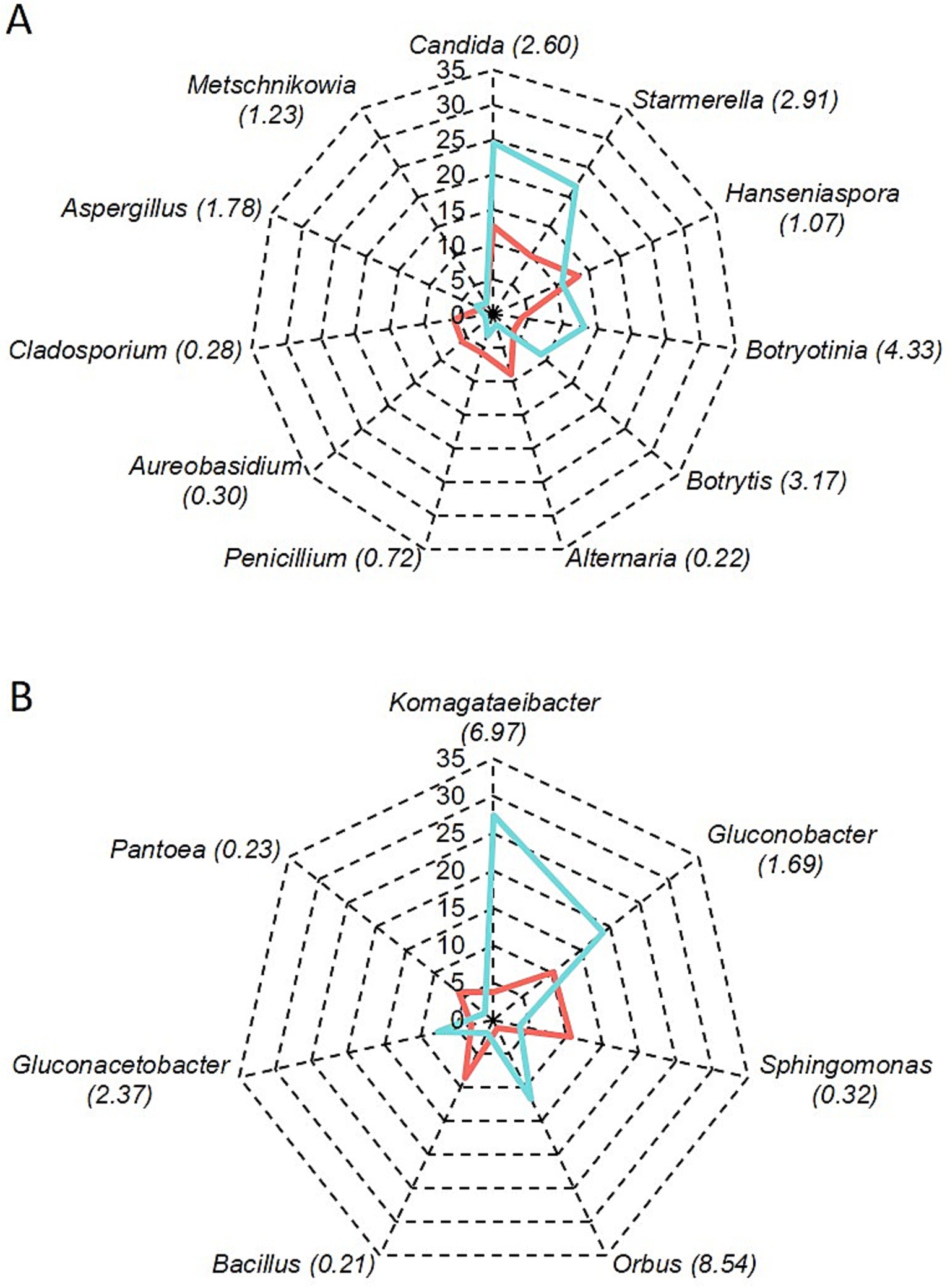
Figure 2. Relative abundance of fungal (A) and bacterial (B) genera in grapevine bunches showing sour rot symptoms (light blue line) or not (red line); the number between brackets shows the ratio of the number of reads in rotten vs. healthy bunches.
The alpha diversity of fungal communities, which reflects the distribution of SV abundances in a single sample, differed among the samples. Chao1, which is based on the abundance of SVs belonging to a genus in a sample, and Shannon, which accounts for both the number of SVs and their relative abundance, both indicated that fungal diversity in healthy bunches was significantly different from that in rotten bunches, with p = 0.008 and 0.047, respectively (Figures 3A,B). The alpha diversity of bacterial communities was significantly different in healthy and rotten bunches only for the Shannon estimator (Chao1: p = 0.460; Shannon: p = 0.030) (Figures 3C,D). These results indicated that healthy and rotten bunches differed in the total number of species, but not in abundance distribution across species.
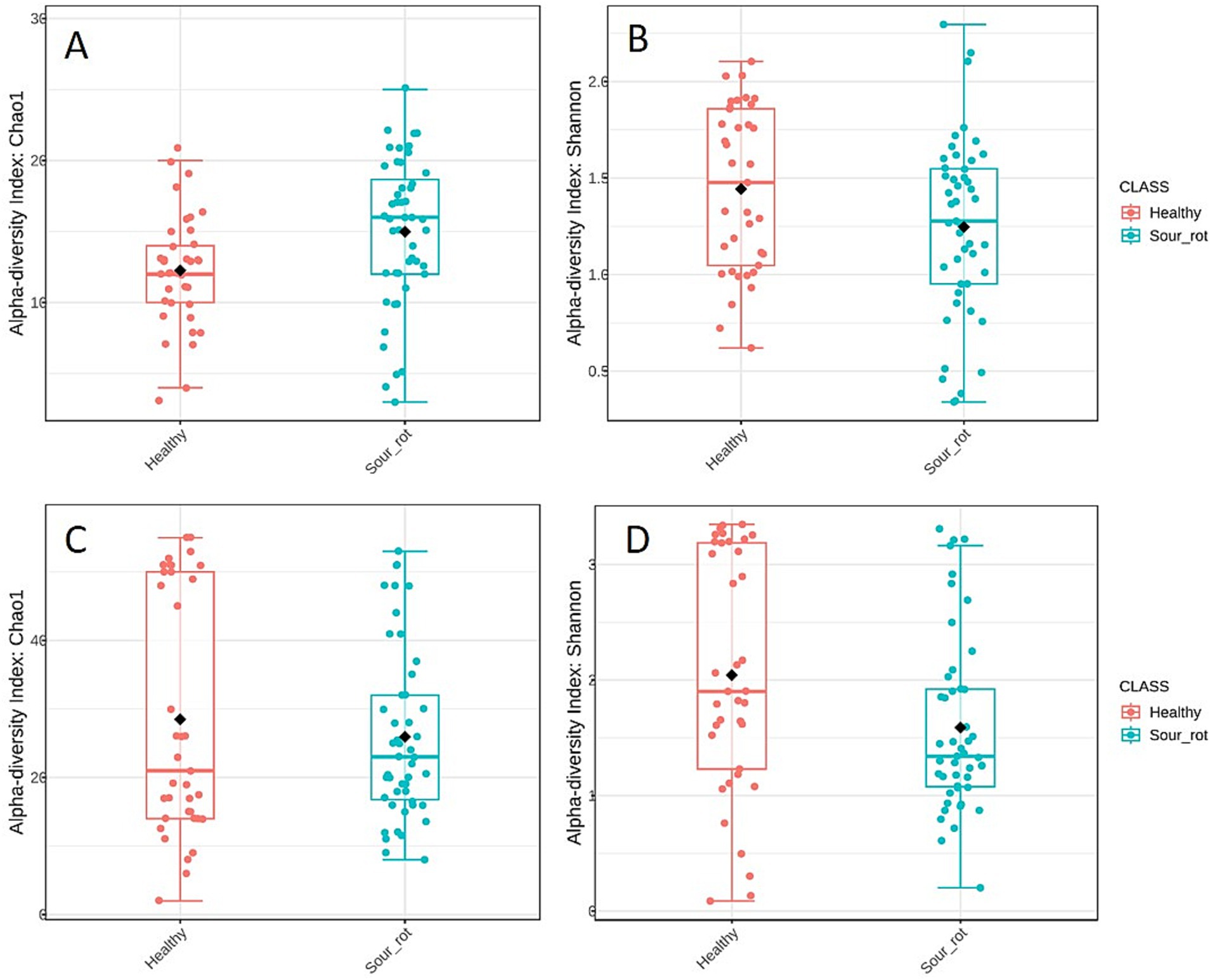
Figure 3. Boxplot illustrating the differences in the fungal (A,B) and bacterial (C,D) communities in healthy (red) and rotten (light blue) bunches based on Chao1 (A,C) and Shannon (B,D) diversity indicators. The box extends from the 25th to the 75th quartile of the data distribution, the line crossing the box represents the median, and the black diamond indicates the average; whiskers extend to the maximum and minimum.
The beta diversity of fungal and bacterial communities, which focuses on SV dissimilarities between samples, measured with the Bray–Curtis dissimilarity, is presented using the PCoA in Figure 4. The bunch status (healthy or rotten) was a significant source of beta diversity for both fungal (R2 = 0.074, p = 0.002; Figure 4A) and bacterial (R2 = 0.012, p = 0.001; Figure 4B) communities. This result indicated that the microbiome composition differed in healthy and rotten bunches.
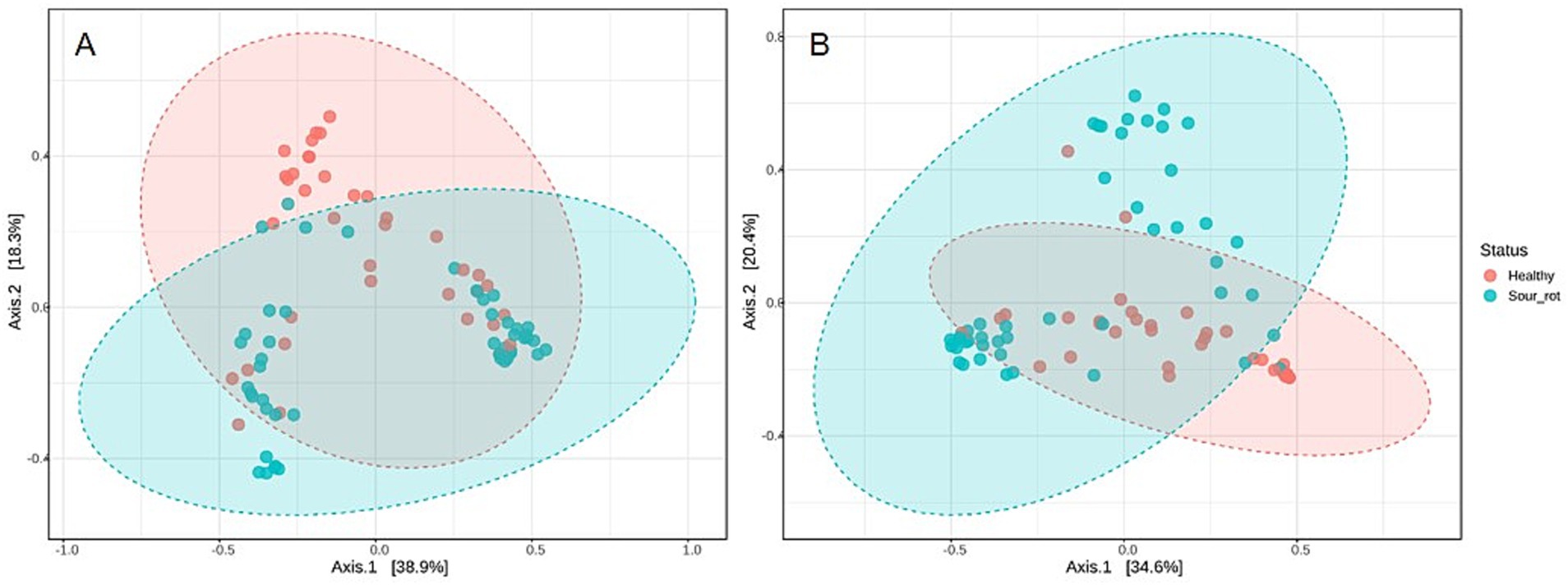
Figure 4. Principal coordinate analysis (PCoA) based on Bray–Curtis dissimilarity metrics, showing the distance in the fungal (A) and bacterial (B) communities present in healthy (red dots) and rotten (light blue dots) bunches. Areas show distinct clustering of healthy (red) and rotten (light blue) bunches.
LEfSe detected 10 fungal genera as the main determinants of the dissimilarities between healthy and rotten bunches (Figure 5A). Zygosaccharomyces (p ≤ 0.001), Zygoascus (p < 0.001), Saccharomycopsis (p = 0.004), Issatchenkia (p = 0.004), and Pichia (p = 0.002) were the most important fungal genera that distinguished rotten bunches, with LDA scores between −3.33 and − 4.85 (Figure 5A), even though they accounted for approximately 3.5% of the total reads in rotten bunches. Fifteen bacterial genera differed between healthy and rotten bunches, which specifically were Orbus (p < 0.001), Gluconobacter (p < 0.001), and Wolbachia (p = 0.004), and with lower significance, Komagataeibacter (p = 0.030) and Gluconacetobacter (p = 0.011), showing an LDA score < −6. Therefore, the rotten bunches had statistically consistent differences (Figure 5B).
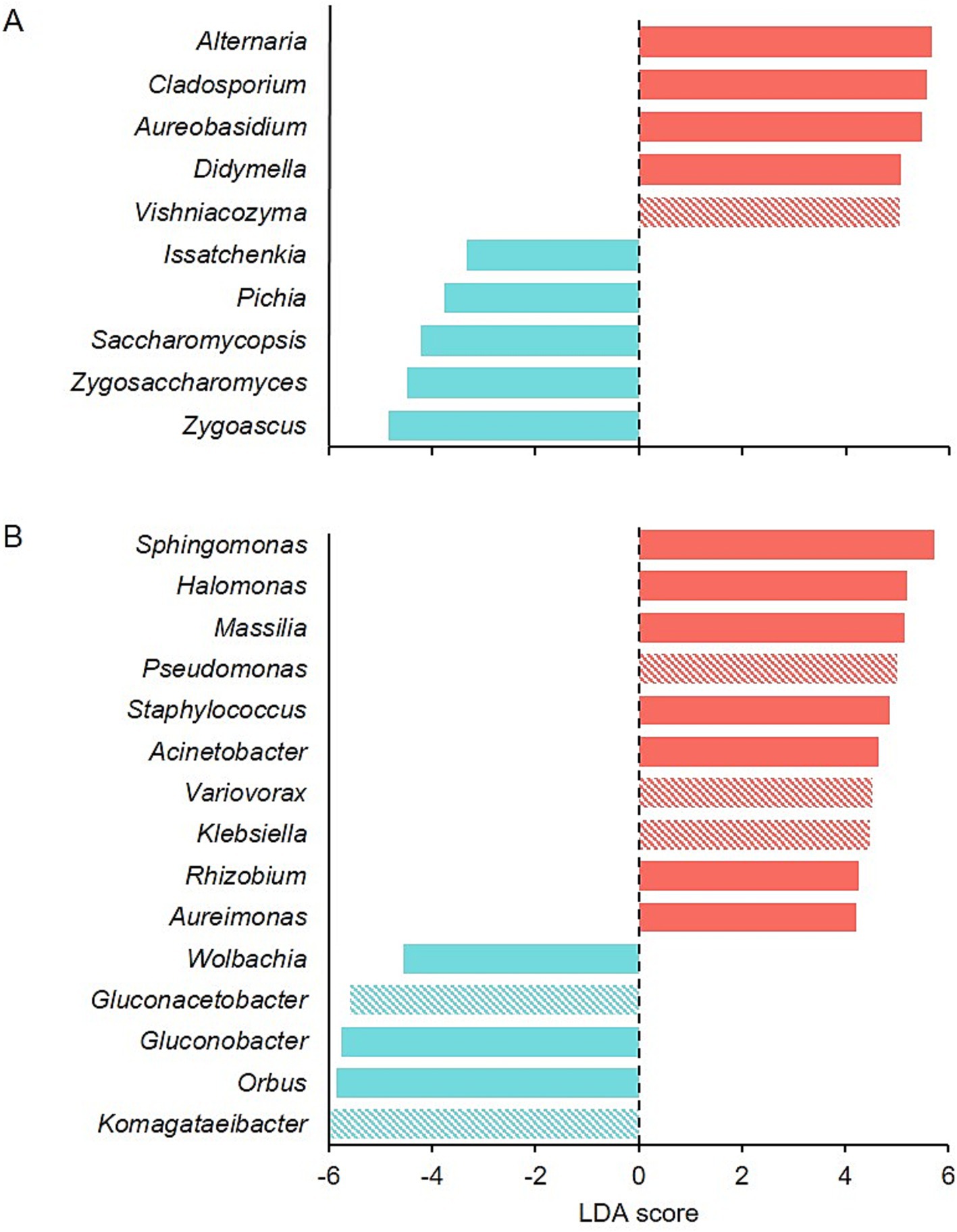
Figure 5. Graphical summary of LEfSe analysis for fungal (A) and bacterial (B) communities in healthy (red) and rotten bunches (light blue). The LDA score represents the extent to which the genera differ among the groups: the higher the positive score, the higher the increase in the relative abundance of the genus concerning rotten bunches, and the lower the negative score, the higher the increase in the relative abundance of the genus in rotten concerning healthy bunches. Full and diagonally striped bar colors mean p-values <0.01 and <0.05, respectively.
In rotten bunches, 96 and 383 significant edges and connections were observed through the correlation network analysis between the fungal (Figure 6) and bacterial (Figure 7) genera, respectively. Positive correlations indicated that genera are likely to coexist, while negatively related genera competitively exclude each other. In particular, the most important fungal genera that characterized rotten bunches, such as Issatchenkia and Pichia, correlated positively among them (0.761) and Candida (0.869 and 0.787, respectively), which was the most abundant genus in rotten bunches. Concerning the bacteria, the genera strongly associated with bunches showing SR symptoms, such as Gluconacetobacter, Orbus, and Wolbachia, were positively correlated with each other, with the strongest correlation between Gluconacetobacter and Orbus (0.956). However, the latter was negatively correlated with Komagataeibacter (−0.514), one of the most abundant genera characterizing rotten bunches.
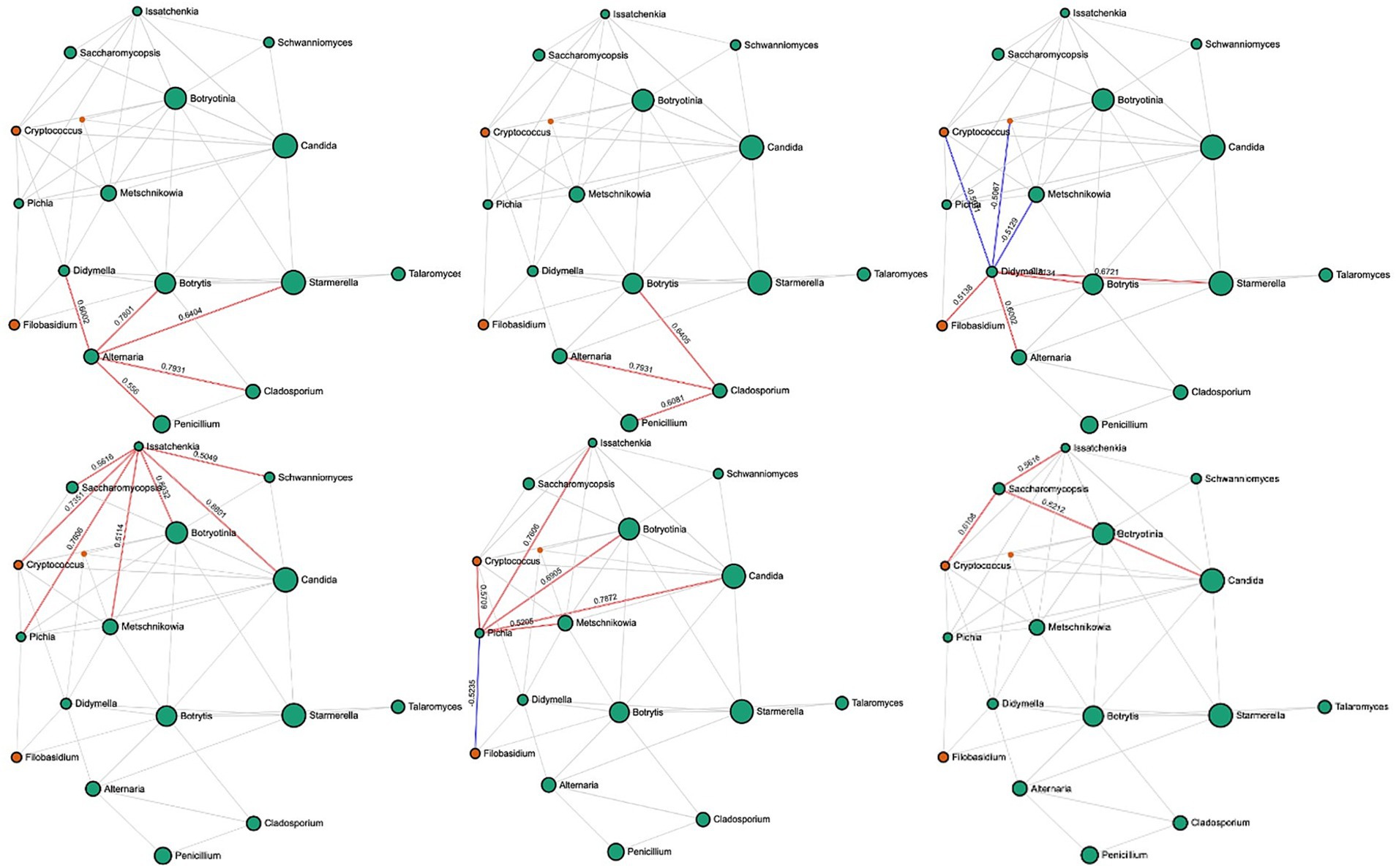
Figure 6. SparCC correlation analysis for fungal communities in rotten bunches. Nodes represent taxa at the genus level. Node size is based on the number of connections to each taxon. Edges represent correlations between pairs: red and blue edges represent positive and negative correlations, respectively; the value is the correlation coefficient between taxa. The nodes are colored based on phyla: green for Ascomycota and orange for Basidiomycota.
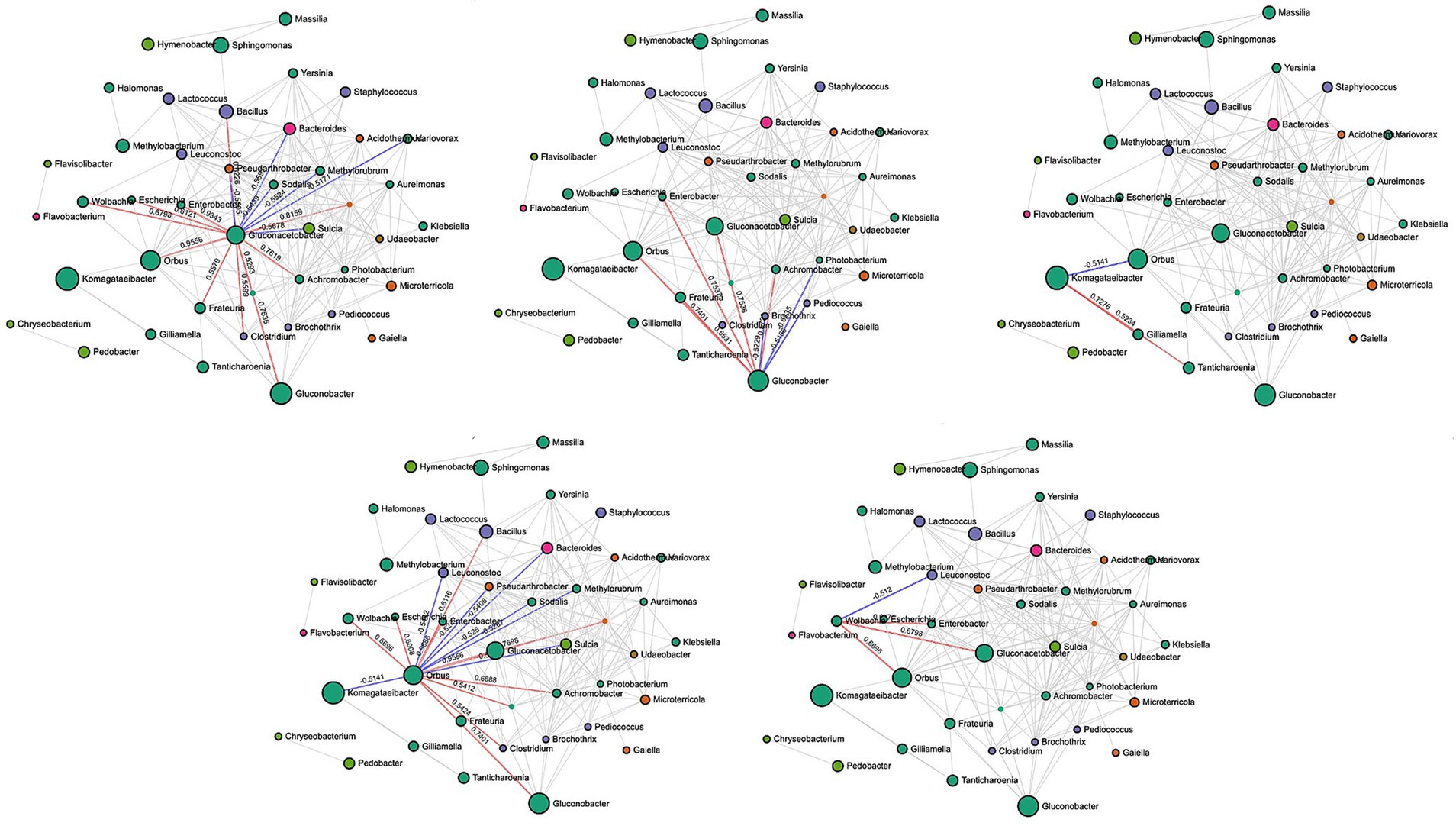
Figure 7. SparCC correlation analysis for bacterial communities in rotten bunches. Nodes represent taxa at the genus level. Node size is based on the number of connections to each taxon. Edges representing correlations between pairs: red and blue edges represent positive and negative correlations, respectively; the value is the correlation coefficient between taxa. The nodes are colored based on phyla: green for Proteobacteria, orange for Actinobacteria, purple for Firmicutes, pink for Bacteroidetes, green for Gemmatimonadota, yellow for Bacteroidota, and brown for Verrucomicrobiota.
4 Discussion
Our study analyzed the fungal and bacterial microbiomes of grape bunches affected by SR to determine the main differences from healthy bunches. Information about the microorganisms more likely to be associated with the disease was also inferred.
Grape berries host a complex microbial community comprising bacteria, yeasts, and filamentous fungi (Barata et al., 2012b; Rousseaux et al., 2014; Fleet, 2003; Ribéreau-Gayon et al., 2006), which play a prominent role in the winemaking process and wine quality (Barbe et al., 2001; Nisiotou et al., 2011; Verginer et al., 2010; Pretorius, 2000). Some yeasts can benefit winemaking, while others can negatively affect wine quality (Martins et al., 2014). Other microorganisms are considered spoilage agents, such as filamentous fungi, which may influence the safety or sensory quality of wines (Barata et al., 2011; Rousseaux et al., 2014; Steel et al., 2013; Barbe et al., 2001; Nisiotou et al., 2011; Verginer et al., 2010). Similarly, some bacteria participate in wine fermentation, such as lactic acid bacteria (LAB), which conduct malolactic fermentation, improving wine flavor and stability (Lonvaud-Funel, 1999). Other bacteria, such as Acetobacter spp., are detrimental to wine quality because of acetic acid production (Barata et al., 2012b).
Our analysis showed that the number of fungal and bacterial SVs was higher in SR-affected than in healthy bunches. This result agrees with the literature review by Brischetto et al. (2024), which revealed that 69 and 128 microorganisms were isolated from unaffected and affected berries, respectively. Thirty different genera were found in the previous literature on SR (Brischetto et al., 2024), and 28 were found in our samples, even though there was no complete agreement about whether they were associated with healthy or rotten bunches (Supplementary Figure S1). For example, Botrytis spp. were previously found occasionally in rotten bunches only. Still, this genus and its teleomorph, Botryotinia, accounted for 22.7% of reads in rotten bunches and 7.8% in healthy ones (Supplementary Figure S1A). Of the 30 bacterial genera mentioned in previous literature on SR (Brischetto et al., 2024), 25 (86.2%) were also found in our samples, and their presence in healthy/rotten bunches was not always in agreement with the literature (Supplementary Figure S1B). For instance, Bacillus spp. was reported to be prevalent in rotten bunches (Brischetto et al., 2024), but it was more abundant in healthy (8.7% of the total reads) than in rotten (2%) bunches in our samples.
Our microbiome analysis revealed that the richness and evenness of both species (as shown by significant alpha diversity estimators) were influenced by bunch status, and the microbiome composition significantly varied between healthy and rotten bunches (as demonstrated by a significant beta diversity indicator). Overall, the microbial profile of rotten bunches was characterized by the yeast genera Zygosaccharomyces, Zygoascus, Saccharomycopsis, Issatchenkia, and Pichia and by the bacterial genera Orbus, Gluconobacter, Wolbachia, Komagataeibacter, and Gluconacetobacter, which frequently coexisted, being closely correlated with each other. The contemporary presence of yeasts and bacteria, especially the so-called acetic acid bacteria (AAB), in grape berries showing typical SR symptoms has been previously documented (see Brischetto et al., 2024). Hall et al. (2018a) postulated a succession of these microorganisms during disease development, with yeasts producing ethanol from sugars and AAB using ethanol to produce acetic acid.
Our analysis showed that Zygosaccharomyces, Zygoascus, Saccharomycopsis, Issatchenkia, and Pichia were characteristic of SR-affected bunches, even though they accounted for less than 3.5% of the total reads. These yeast genera belong to the order Saccharomycetales, families Saccharomycetaceae, Trichomonascaceae, Saccharomycopsidaceae, and Pichiaceae, respectively, which globally accounted for 58.3% of the total reads in our SR-affected samples.
Several species in the yeast genus Zygosaccharomyces are well-known spoilage microorganisms for their high sugar, ethanol, and acetic acid tolerance (Palma et al., 2018; James and Stratford, 2003). These microorganisms are considered rare contaminants of grapes but are among the most dangerous wine spoilers (Fleet et al., 2002; Barata et al., 2008a). Fermentative species belonging to Zygoascus have been characterized as producers of biogenic amines in wine (Tristezza et al., 2013). In particular, Z. hellenicus (teleomorph of Candida steatolytica) has been described as a contaminant often associated with damaged grapes (Barata et al., 2008b). Some Saccharomycopsis (specifically, S. vini and S. crataegensis) have been previously associated with SR (Barata et al., 2008a, 2008b; Bisiach et al., 2021; Guerzoni and Marchetti, 1982; Marchetti et al., 1984), with S. crataegensis being characterized by lipolytic activity and reproducing the disease symptoms when inoculated in combination with other microorganisms, such as Issatchenkia occidentalis and Kloeckera apiculata (Guerzoni and Marchetti, 1987).
Pichia spp. are yeasts that consume glucose without ethanol formation (Varela and Varela, 2019; Vicente et al., 2021), and Issatchenkia spp. can ferment glucose to ethanol in acidic media (Hisamatsu et al., 2006). These genera are closely related, and some species of Issatchenkia have been proposed to be classified within Pichia (Kurtzman et al., 2008). All of these yeasts have been previously isolated from SR-affected bunches (Guerzoni and Marchetti, 1987; Fleet et al., 2002; Nisiotou and Nychas, 2007; Barata et al., 2008a, 2008b, 2012a), and the yeasts Zygoascus hellenicus and Issatchenkia spp. have been proposed as biomarkers for SR (Barata et al., 2012b).
The yeasts are also part of the D. melanogaster microbiome (Broderick and Lemaitre, 2012), with Hanseniaspora, Candida, Zygoascus, Saccharomycopsis, and Pichia being commonly associated with natural Drosophila spp. populations (Chandler et al., 2012; Stamps et al., 2012; Hamby et al., 2012; Scheidler et al., 2015; Begon, 1982; Ganter, 2006; Barata et al., 2012a). The interactions between Drosophila spp. and yeasts appear mutualistic, as yeasts affect several aspects of insect physiology, behavior, and immunity (Hoang et al., 2015). Ingestion by the larvae of some yeasts speeds up larval development time and increases adult body weight (Anagnostou et al., 2010). Yeasts can survive digestion by D. melanogaster, so flies serve as yeast vectors under natural conditions (Reuter et al., 2007; Coluccio et al., 2008).
In our samples, bacteria of the family Acetobacteraceae accounted for 56.5% of total bacterial reads in SR-affected bunches, with the genera Komagataeibacter, Gluconacetobacter, and Gluconobacter representing 53.8% of these reads. Komagataeibacter was the most abundant genus found in our samples (27.4% of the total reads in rotten bunches). This genus was recently defined to include several species previously classified as Gluconacetabacter (Yamada et al., 2012; Mateo et al., 2014). These bacteria have been isolated from rotten grape bunches (Mateo et al., 2014; Gopu and Govindan, 2018; Gao et al., 2020; Srivastava and Mathur, 2022; Ryngajłło et al., 2020) and can produce gluconic acid from glucose and other sugars, and oxidate ethanol to acetic acid (Gomes et al., 2021). Komagataeibacter spp. is also an efficient bacterial cellulose producer from various carbon and nitrogen sources (Islam et al., 2017), including grape pomace (Gorgieva et al., 2023). Gluconacetobacter spp. and Gluconobacter spp. were among the most prevalent bacteria in the affected berries in previous studies (Brischetto et al., 2024). Together with Acetobacter spp., they have been associated with grape and wine spoilage (Joyeux et al., 1984; Navarro et al., 2013; Hall et al., 2019). Species of Acetobacter are often isolated from wine due to their ethanol tolerance, whereas Gluconobacter spp. prefer sugar-rich environments with low amounts of alcohol (Campaniello and Sinigaglia, 2017). Gluconobacter spp. oxidize grape sugars primarily using gluconic acid (Batt and Tortorello, 2014). Some Gluconacetobacter spp. produce thick leathery pellicles in the air liquid during winemaking, which is considered a contaminant (Rani et al., 2011). Unlike previous studies (Hall et al., 2019), Acetobacter spp. had a low abundance in our samples (1.76% of total fungal reads).
In addition to yeasts, Acetobacteraceae has been frequently associated with Drosophila spp. in nature (Staubach et al., 2013). Gluconobacter spp. was the most prevalent bacteria in wild-caught flies in some studies (Staubach et al., 2013; Corby-Harris et al., 2007; Ryu et al., 2008), and present, but not prevalent in others (Chandler et al., 2011). Komagataeibacter spp. have also been found in the Drosophila spp. gut as a valuable microbiota member in overcoming environmental stress (Beribaka et al., 2021). The relationship between AAB, Drosophila spp., and SR has been demonstrated by Barata et al. (2012a). These authors did not observe SR when bunches inoculated with AAB were physically separated from insects, even when berries were artificially injured, because wounds in berry skin healed in the absence of Drosophila spp., thus preventing SR development. The authors then concluded that, in the vineyard, the induction of SR depends on the contamination of wounded berries by a microbial consortium transported by Drosophila spp. that act as vectors for microorganisms associated with grape SR. Hall et al. (2018a) postulated that the role of Drosophila spp. go beyond vectoring because, in artificial inoculation studies, SR symptoms developed only in the presence of D. melanogaster, either wild type or axenic. Softening of the berry pulp by the enzymes released by larvae to facilitate consumption (Gregg et al., 1990) may be an aspect to be considered. However, the AAB–Drosophila spp.–SR relationship seems even more complex.
AAB are considered ubiquitous symbionts of Drosophila spp. (Rosenberg et al., 2007; Douglas, 2018). AAB and other microorganisms are part of the microbial community within the intestine of D. melanogaster (Ryu et al., 2008). AAB and LAB metabolize ethanol and acetic acid within the gut of D. melanogaster, and the secondary metabolites produced are beneficial for the growth and development of the insect, both directly (Douglas, 2017; Fischer et al., 2017) and indirectly (Keebaugh et al., 2018). AAB and other bacteria are also transported in bristled areas or tarsal segments on the fly surface (Barata et al., 2012a; Hong et al., 2022), forming biofilms (Ren et al., 2007) and promoting the dispersal and establishment of these bacteria in fruit (Barata et al., 2012a). The fly surface microbiota is complex, and the bacterial richness of surface microbiomes is much higher than that of gut microbiota. Such microbiota may defend insects against fungal and parasitic infections (e.g., Beauveria brassiana and Metarhizium robertsii), inhibiting spore germination (Hong et al., 2022).
The presence of AAB in the substrate has beneficial effects on D. melanogaster larval growth and development time (Shin et al., 2011). Indeed, these bacteria are ingested by insects and become part of their gut microbiota (Wong et al., 2011), playing a significant functional role in the life of the host, including innate immunity (Ryu et al., 2008), lifespan (Clark et al., 2015; Lee et al., 2019), nutrition (Beribaka et al., 2021), reproduction (Leitão-Gonçalves et al., 2017), and behavior (Silva et al., 2021). Rosenberg et al. (2007) speculated that the microbial community associated with Drosophila spp. can be seen as an external organ of the fly holobiont. It is also known that bacteria are attractive to Diptera because of the production of a range of volatile compounds, including ammonia (Bateman and Morton, 1981; Robacker et al., 1998; MacCollum, 1992; Lauzon et al., 1998; Robacker and Lauzon, 2002). Gluconobacter spp. and Komagataeibacter spp. rapidly produced acetic acid and ethanol, which are attractive to Drosophila spp. (West, 1961; Landolt et al., 2012; Mazzetto et al., 2016). The volatile compounds produced by mutualistic microorganisms living inside host insects with a symbiotic relationship with plants trigger their trophic interaction (Frago et al., 2012) and strengthen the insect–bacteria relationship.
The genus Orbus represented 11.8% of the reads in SR-affected bunches. This genus was not previously reported as being associated with SR-affected berries (Brischetto et al., 2024). It was initially classified as Enterobacteriaceae, a large family that includes many animal- and plant-associated bacteria. It was reclassified into the family Orbaceae (order Orbales) within γ-Proteobacteria (Kwong and Moran, 2013). These bacteria were found to be free-living associates of many insects, including the gut of D. melanogaster (Cox and Gilmore, 2007), with several lineages being endosymbiotic and required for insect nutrition, defense from parasites, and tolerance of heat stress (Douglas, 1998; Montllor et al., 2002; Moran et al., 2005). Chandler et al. (2011) designated this entire lineage as “Enterobacteriaceae Group Orbus” and found it abundantly in Drosophila spp. samples, representing over 21% of all bacteria in natural Drosophila spp. populations. Other Enterobacteriaceae, such as Enterobacter/Pantoea and Klebsiella (present in our SR-affected samples with 1.9% abundance), participate in nitrogen cycling within the dipteran gut and serve as important contributors to insect survival in nature (Lauzon et al., 2000).
The genus Wolbachia represented 0.6% of the reads in SR-affected bunches. Wolbachia spp. bacteria are α-Proteobacteria and obligate endosymbionts that are extremely widespread in approximately 40% of all insect species and cause various types of reproductive phenotypes that favor vertical transmission and spread in populations (Werren et al., 2008; Fast et al., 2011). In Drosophila spp., Wolbachia spp. can have both deleterious and beneficial effects on different fitness components, such as fecundity, lifespan, and stress tolerance (Serga et al., 2021). Beneficial effects include improved reproduction related to a shortened lifespan and lower stress resistance (Serga et al., 2021). The presence of Wolbachia spp. changes the composition of the bacterial communities in the Drosophila spp. fly gut, decreasing its biodiversity, particularly by reducing the abundance of Acetobacter spp. (Yixin et al., 2017). It may be speculated that the presence of Orbus and Wolbachia genera in SR-affected berries and other Enterobacteriaceae of the Drosophila spp. gut microbiota is related to the presence of Drosophila spp. in those berries.
In conclusion, our analysis revealed that even if approximately 570 microorganisms were found in grape bunches affected by SR collected in 39 vineyards in six Italian grape-growing regions characterized by different pedoclimatic conditions over three years, few bacteria and yeast were closely related to the presence of the disease. These microorganisms were also found in previous studies, so we can consider our results sufficiently robust. All these microorganisms have been previously associated with wild Drosophila spp. in the literature, with a complex relationship that can be depicted in Figure 8.
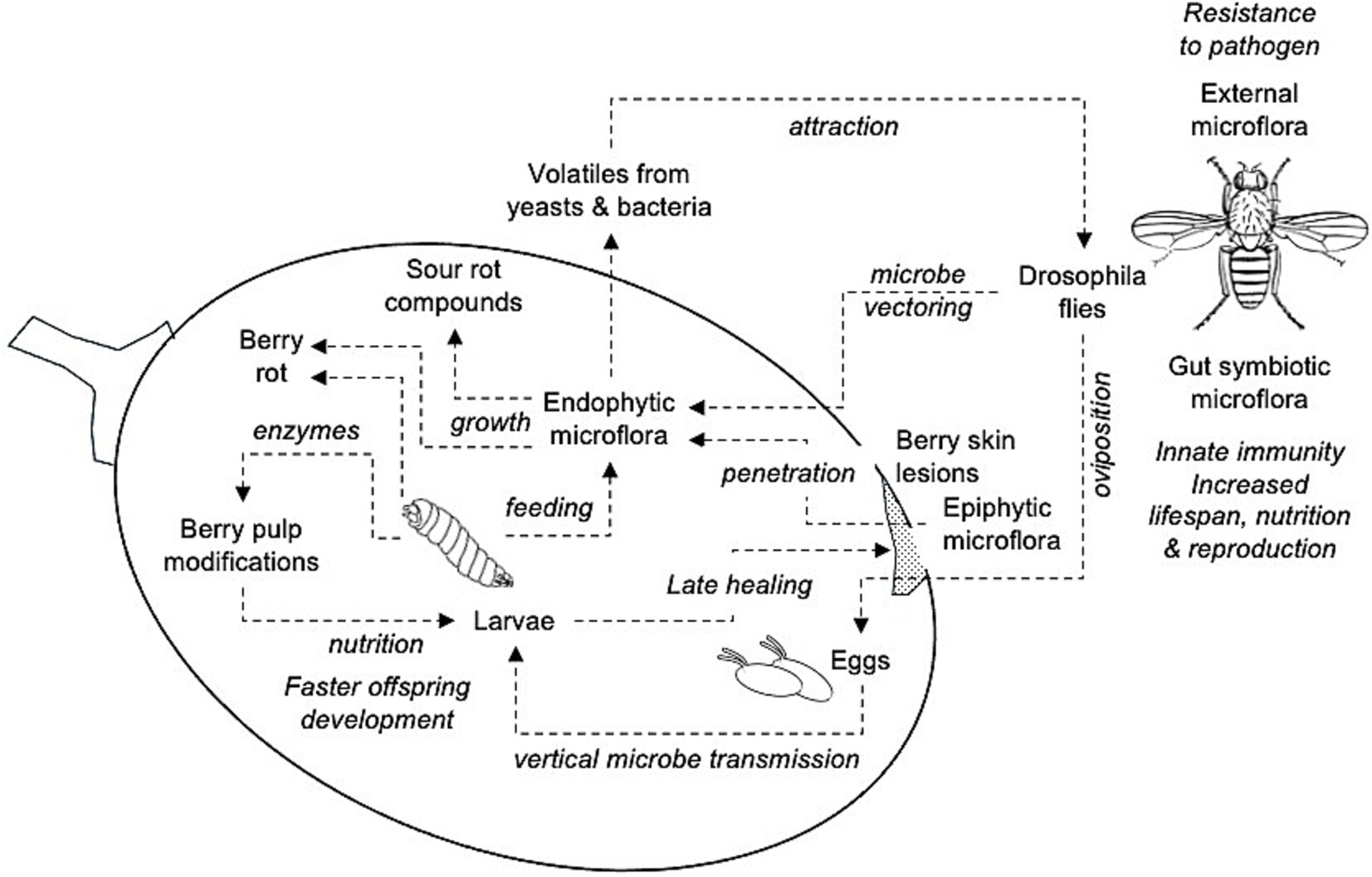
Figure 8. Schematic relationship of the interactions between yeasts and bacteria associated with grape sour rot and Drosophila spp., as from literature. These relationships agree with the microbial composition of the SR-affected bunches in this study. Flying adults carry microorganisms on both external parts of their bodies and in the gut as symbionts, which provide multiple benefits to insects. Adults deposit eggs in the berry pulp through berry skin lesions (e.g., D. melanogaster) or directly (e.g., D. suzukii) so that the epiphytic yeasts and bacteria can penetrate the pulp or enter the pulp through vectoring on fly body parts or vertical transmission (adults to eggs). Larvae develop into the berry feeding the endophytic microorganisms and berry pulp components through modifications induced by insect-released enzymes; this results in faster offspring development. Larvae also delay wound healing through movement. Endophytic microorganisms grow and produce berry rot and the compounds associated with sour rot (ethanol, acetic acid, gluconic acid, etc.). Some of these compounds are volatiles that attract flies.
Further studies on the association between SR microorganisms and Drosophila spp. could contribute to explaining the differences in the microflora composition and abundance between different studies. Indeed, the gut microbiota composition of Drosophila species varies in association with diet, genotype, laboratory, and age (Broderick and Lemaitre, 2012; Chandler et al., 2011; Staubach et al., 2013; Clark and Walker, 2018; Marra et al., 2021). For instance, the microbial diversity in the guts of D. suzukii differs from that of D. melanogaster because of the adaptation of the former to its high-sugar ecological niche (Lin et al., 2021). A better understanding of the SR–Drosophila spp. relationships could also open up perspectives for disease control. The literature review by Brischetto et al. (2024) showed that disease control based on using fungicides, natural products, and biocontrol microorganisms, either alone or in an integrated pest management strategy, provided inconsistent, often poor, control. An indirect SR control targeted at flies has been proposed by Bisiach et al. (2021). In some viticultural areas of the US, fly control was achieved by using various insecticides (Weigle et al., 2020), primarily pyrethroid zeta-cypermethrin, which however, has been associated with increased fly resistance to insecticides (Sun et al., 2019; Mertz et al., 2022, 2023; Hubhachen et al., 2022). Developments in the control of D. suzukii to limit the damage caused by this insect (Tait et al., 2021) could be considered in further studies for less insecticide-dependent control of the flies related to grape SR; these include biocontrol (using predators, parasitoids, or entomopathogens), mass trapping, attract, and kill, repellents, and oviposition repellents.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
CB: Formal analysis, Methodology, Writing – original draft. VR: Conceptualization, Formal analysis, Methodology, Writing – review & editing. GF: Conceptualization, Formal analysis, Methodology, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
CB conducted this study within the Doctoral School on the Agro-Food System (AgriSystem) at the Università Cattolica del Sacro Cuore, Piacenza, Italy.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1450443/full#supplementary-material
Footnotes
References
Anagnostou, C., Dorsch, M., and Rohlfs, M. (2010). Influence of dietary yeasts on Drosophila melanogaster life-history traits. Entomol. Exp. Appl. 136, 1–11. doi: 10.1111/j.1570-7458.2010.00997.x
Atallah, J., Teixeira, L., Salazar, R., Zaragoza, G., and Kopp, A. (2014). The making of a pest: the evolution of a fruit-penetrating ovipositor in Drosophila suzukii and related species. Proc. Biol. Sci. 281:20132840. doi: 10.1098/rspb.2013.2840
Barata, A. (2011). Microbial ecology of sour rotten grapes and their influence onchemical and sensorial wine quality. Tese apresentada Para obtenc¸ ão do grau deDoutor em Engenharia Alimentar. Portugal: Instituto Superior de Agronomía, UniversidadTécnica de Lisboa, 6.
Barata, A., Campo, E., Malfeito-Ferreira, M., Loureiro, V., Cacho, J., and Ferreira, V. (2011). Analytical and sensorial characterization of the aroma of wines produced with sour rotten grapes using GC-O and GC-MS: identification of key aroma compounds. J. Agric. Food Chem. 59, 2543–2553. doi: 10.1021/jf104141f
Barata, A., González, S., Malfeito-Ferreira, M., Querol, A., and Loureiro, V. (2008a). Sour rot-damaged grapes are sources of wine spoilage yeasts. FEMS Yeast Res. 8, 1008–1017. doi: 10.1111/j.1567-1364.2008.00399.x
Barata, A., Malfeito-Ferreira, M., and Loureiro, V. (2012b). The microbial ecology of wine grape berries. Int. J. Food Microbiol. 153, 243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025
Barata, A., Santos, S. C., Malfeito-Ferreira, M., and Loureiro, V. (2012a). New insights into the ecological interaction between grape berry microorganisms and Drosophila flies during the development of sour rot. Microb. Ecol. 64, 416–430. doi: 10.1007/s00248-012-0041-y
Barata, A., Seborro, F., Belloch, C., Malfeito-Ferreira, M., and Loureiro, V. (2008b). Ascomycetous yeast species recovered from grapes damaged by honeydew and sour rot. J. Appl. Microbiol. 104, 1182–1191. doi: 10.1111/j.1365-2672.2007.03631.x
Barbe, J. C., De Revel, G., Joyeux, A., Bertrand, A., and Lonvaud-Funel, A. (2001). Role of botrytized grape microorganisms in SO2 binding phenomena. J. Appl. Microbiol. 90, 34–42. doi: 10.1046/j.1365-2672.2001.01200.x
Bateman, M. A., and Morton, T. C. (1981). The importance of ammonia in proteinaceous attractants for fruit flies (family: Tephritidae). Aust. J. Agric. Res. 32, 883–903. doi: 10.1071/AR9810883
Batt, C. A., and Tortorello, M. L. (2014). Encyclopedia of food microbiology. 2nd Edn. Academic Press: Massachusetts.
Battilani, P., and Pietri, A. (2002). Ochratoxin A in grapes and wine. Mycotoxins in plant disease: under the aegis of COST action 835 “agriculturally important toxigenic Fungi 1998-2003”, EU project (QLK 1-CT-1998-01380), and ISPP Fusarium committee, 639–643
Begon, M. (1982). “Yeasts and drosophila” in The genetics and biology of Drosophila. eds. M. Ashburner, H. Carson, and J. Thompson Jr., 345–384.
Belda, I., Palacios, A., Fresno, J., Ortega, H., and Acedo, A. (2017). “WineSeq®: a new tool for the study of the functional biodiversity of soils, and its use as a biomarker and guide for vitiviniculture practices” in BIO web of conferences, vol. 9 (Les Ulis: EDP Sciences), 1012.
Beribaka, M., Jelić, M., Tanasković, M., Lazić, C., and Stamenković-Radak, M. (2021). Life history traits in two Drosophila species differently affected by microbiota diversity under lead exposure. Insects 12:1122. doi: 10.3390/insects12121122
Bisiach, M., Minervini, G., and Zerbetto, F. (2021). “Possible integrated control of grapevine sour-rot” in Integrated pest control in viticulture (Boca Raton, FL: CRC Press), 265–275.
Brady, C., Arnold, D., McDonald, J., and Denman, S. (2017). Taxonomy and identification of bacteria associated with acute oak decline. World J. Microbiol. Biotechnol. 33:143. doi: 10.1007/s11274-017-2296-4
Brischetto, C., Rossi, V., and Fedele, G. (2024). Knowledge gaps on grape sour rot inferred from a systematic literature review. Front. Plant Sci. 15:1415379. doi: 10.3389/fpls.2024.1415379
Broderick, N. A., and Lemaitre, B. (2012). Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3, 307–321. doi: 10.4161/gmic.19896
Campaniello, D., and Sinigaglia, M. (2017). “Wine spoiling phenomena” in Microbiological quality of food (Sawston: Woodhead Publishing), 237–255.
Chandler, J. A., Eisen, J. A., and Kopp, A. (2012). Yeast communities of diverse Drosophila species: comparison of two symbiont groups in the same hosts. Appl. Environ. Microbiol. 78, 7327–7336. doi: 10.1128/AEM.01741-12
Chandler, J. A., Lang, J. M., Bhatnagar, S., Eisen, J. A., and Kopp, A. (2011). Bacterial communities of diverse Drosophila species: ecological context of a host–microbe model system. PLoS Genet. 7:e1002272. doi: 10.1371/journal.pgen.1002272
Chong, J., Liu, P., Zhou, G., and Xia, J. (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15, 799–821. doi: 10.1038/s41596-019-0264-1
Clark, R. I., Salazar, A., Yamada, R., Fitz-Gibbon, S., Morselli, M., Alcaraz, J., et al. (2015). Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Rep. 12, 1656–1667. doi: 10.1016/j.celrep.2015.08.004
Clark, R. I., and Walker, D. W. (2018). Role of gut microbiota in aging-related health decline: insights from invertebrate models. Cell. Mol. Life Sci. 75, 93–101. doi: 10.1007/s00018-017-2671-1
Coluccio, A. E., Rodriguez, R. K., Kernan, M. J., and Neiman, A. M. (2008). The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS One 3:e2873. doi: 10.1371/journal.pone.0002873
Corby-Harris, V., Pontaroli, A. C., Shimkets, L. J., Bennetzen, J. L., Habel, K. E., and Promislow, D. E. L. (2007). Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl. Environ. Microbiol. 73, 3470–3479. doi: 10.1128/AEM.02120-06
Cox, C. R., and Gilmore, M. S. (2007). Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun. 75, 1565–1576. doi: 10.1128/IAI.01496-06
Dhariwal, A., Chong, J., Habib, S., King, I. L., Agellon, L. B., and Xia, J. (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45, W180–W188. doi: 10.1093/nar/gkx295
Douglas, A. E. (1998). Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43, 17–37. doi: 10.1146/annurev.ento.43.1.17
Douglas, A. E. (2017). The B vitamin nutrition of insects: the contributions of diet, microbiome and horizontally acquired genes. Curr. Opin. Insect Sci. 23, 65–69. doi: 10.1016/j.cois.2017.07.012
Douglas, A. E. (2018). The Drosophila model for microbiome research. Lab. Anim. 47, 157–164. doi: 10.1038/s41684-018-0065-0
Fast, E. M., Toomey, M. E., Panaram, K., Desjardins, D., Kolaczyk, E. D., and Frydman, H. M. (2011). Wolbachia enhance Drosophila stem cell proliferation and target the germline stem cell niche. Science 334, 990–992.
Fermaud, M., Gravot, E., and Blancard, D. (2002). La pourriture acide dans le vignoble bordelais [France]. 2. Vection par les drosophiles des micro-organismes pathogènes. Phytoma. La Défense des Végétaux (France) 547, 41–44.
Fischer, C. N., Trautman, E. P., Crawford, J. M., Stabb, E. V., Handelsman, J., and Broderick, N. A. (2017). Metabolite exchange between microbiome members produces compounds that influence Drosophila behavior. eLife 6:e18855. doi: 10.7554/eLife.18855
Fleet, G. H. (2003). Yeast interactions and wine flavour. Int. J. Food Microbiol. 86, 11–22. doi: 10.1016/S0168-1605(03)00245-9
Fleet, G. H., Prakitchaiwattana, C., Beh, A. I., and Heard, G. (2002). “The yeast ecology of wine grapes” in Biodiversity and Biotechnology of Wine Yeasts. ed. M. Ciani (Kerala, India: Res Signpost), 1–17.
Frago, E., Dicke, M., and Godfray, H. C. J. (2012). Insect symbionts as hidden players in insect–plant interactions. Trends Ecol. Evol. 27, 705–711. doi: 10.1016/j.tree.2012.08.013
Friedman, J., and Alm, E. J. (2012). Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8. doi: 10.1371/journal.pcbi.1002687
Ganter, P. F. (2006). “Yeast and invertebrate associations” in Biodiversity and ecophysiology of yeasts (Berlin, Heidelberg: Springer), 303–370.
Gao, H., Yin, X., Jiang, X., Shi, H., Yang, Y., Wang, C., et al. (2020). Diversity and spoilage potential of microbial communities associated with grape sour rot in eastern coastal areas of China. PeerJ 8:e9376. doi: 10.7717/peerj.9376
Gomes, R. J., de Sousa Faria-Tischer, P. C., Tischer, C. A., Constantino, L. V., de Freitas Rosa, M., Chideroli, R. T., et al. (2021). Komagataeibacter intermedius V-05: an acetic acid bacterium isolated from vinegar industry, with high capacity for bacterial cellulose production in soybean molasses medium. Food Technol. Biotechnol. 59, 432–442. doi: 10.17113/ftb.59.04.21.7148
Gopu, G., and Govindan, S. (2018). Production of bacterial cellulose from Komagataeibacter saccharivorans strain BC1 isolated from rotten green grapes. Prep. Biochem. Biotechnol. 48, 842–852. doi: 10.1080/10826068.2018.1513032
Gorgieva, S., Jančič, U., Cepec, E., and Trček, J. (2023). Production efficiency and properties of bacterial cellulose membranes in a novel grape pomace hydrolysate by Komagataeibacter melomenusus AV436T and Komagataeibacter xylinus LMG 1518. Int. J. Biol. Macromol. 244:125368. doi: 10.1016/j.ijbiomac.2023.125368
Gravot, E., Blancard, D., Fermaud, M., Lonvaud, A., and Joyeux, A. (2001). Sour rot. I: etiology. Research into the causes of this form of rot of grapes in Bordeaux vineyards. Phytoma 543, 36–39.
Gregg, T. G., McCrate, A., Reveal, G., Hall, S., and Rypstra, A. L. (1990). Insectivory and social digestion in Drosophila. Biochem. Genet. 28, 197–207. doi: 10.1007/BF00561337
Guerzoni, M. E., and Marchetti, R. (1982). Microflora associata al marciume acido della vite e modificazioni indotte dalla malattia sulla composizione di uve e mosti. Difesa Piante 4, 231–246.
Guerzoni, E., and Marchetti, R. (1987). Analysis of yeast flora associated with grape sour rot and of the chemical disease markers. Appl. Environ. Microbiol. 53, 571–576. doi: 10.1128/aem.53.3.571-576.1987
Hall, M. E., Loeb, G. M., Cadle-Davidson, L., Evans, K. J., and Wilcox, W. F. (2018a). Grape sour rot: a four-way interaction involving the host, yeast, acetic acid bacteria, and insects. Phytopathology 108, 1429–1442. doi: 10.1094/PHYTO-03-18-0098-R
Hall, M. E., Loeb, G. M., and Wilcox, W. F. (2018b). Control of sour rot using chemical and canopy management techniques. Am. J. Enol. Vitic. 69, 342–350. doi: 10.5344/ajev.2018.17091
Hall, M. E., O’Bryon, I., Wilcox, W. F., Osier, M. V., and Cadle-Davidson, L. (2019). The epiphytic microbiota of sour rot-affected grapes differs minimally from that of healthy grapes, indicating causal organisms are already present on healthy berries. PLoS One 14:e0211378. doi: 10.1371/journal.pone.0211378
Hamby, K. A., Hernández, A., Boundy-Mills, K., and Zalom, F. G. (2012). Associations of yeasts with spotted-wing drosophila (Drosophila suzukii; Diptera: Drosophilidae) in cherries and raspberries. Appl. Environ. Microbiol. 78, 4869–4873. doi: 10.1128/AEM.00841-12
Hewstone, N., Valenzuela, J., and Muñoz, C. (2007). Nueva variedad de uva de mesa. Agric. Cult. Tec. 67, 201–204. doi: 10.4067/S0365-28072007000200011
Hisamatsu, M., Furubayashi, T., Karita, S., Mishima, T., and Isono, N. (2006). Isolation and identification of a novel yeast fermenting ethanol under acidic conditions. J. Appl. Glycosci. 53, 111–113. doi: 10.5458/jag.53.111
Hoang, D., Kopp, A., and Chandler, J. A. (2015). Interactions between Drosophila and its natural yeast symbionts—is Saccharomyces cerevisiae a good model for studying the fly-yeast relationship? PeerJ 3:e1116. doi: 10.7717/peerj.1116
Hong, S., Sun, Y., Sun, D., and Wang, C. (2022). Microbiome assembly on Drosophila body surfaces benefits the flies to combat fungal infections. Iscience 25:104408. doi: 10.1016/j.isci.2022.104408
Huang, N., Wang, W., Yao, Y., Zhu, F., Wang, W., and Chang, X. (2017). The influence of different concentrations of bio-organic fertilizer on cucumber Fusarium wilt and soil microflora alterations. PLoS One 12:e0171490. doi: 10.1371/journal.pone.0171490
Hubhachen, Z., Pointon, H., Perkins, J. A., Van Timmeren, S., Pittendrigh, B., and Isaacs, R. (2022). Resistance to multiple insecticide classes in the vinegar fly Drosophila melanogaster (Diptera: Drosophilidae) in Michigan vineyards. J. Econ. Entomol. 115, 2020–2028. doi: 10.1093/jee/toac155
Islam, M. U., Ullah, M. W., Khan, S., Shah, N., and Park, J. K. (2017). Strategies for cost-effective and enhanced production of bacterial cellulose. Int. J. Biol. Macromol. 102, 1166–1173. doi: 10.1016/j.ijbiomac.2017.04.110
James, S. A., and Stratford, M. (2003). “Spoilage yeasts with emphasis on the genus Zygosaccharomyces” in Yeasts in Food. eds. T. Boekhout and V. Robert (Amsterdam: Elsevier), 171–196.
Joyeux, A., Lafon-Lafourcade, S., and Ribéreau-Gayon, P. (1984). Metabolism of acetic acid bacteria in grape must. Consequences on alcoholic and malolactic fermentation. Sci. Aliments 4, 247–255.
Keebaugh, E. S., Yamada, R., Obadia, B., Ludington, W. B., and Ja, W. W. (2018). Microbial quantity impacts Drosophila nutrition, development, and lifespan. Iscience 4, 247–259. doi: 10.1016/j.isci.2018.06.004
Kurtzman, C. P., Robnett, C. J., and Basehoar-Powers, E. (2008). Phylogenetic relationships among species of pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. Nov., Lindnera gen. Nov. and Wickerhamomyces gen. Nov. FEMS Yeast Res. 8, 939–954. doi: 10.1111/j.1567-1364.2008.00419.x
Kwong, W. K., and Moran, N. A. (2013). Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. Nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. Nov., sp. nov., a member of Orbaceae fam. Nov., Orbales Ord. Nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 63, 2008–2018. doi: 10.1099/ijs.0.044875-0
Landolt, P. J., Adams, T., and Rogg, H. (2012). Trapping spotted wing drosophila, Drosophila suzukii (Matsumura) (Diptera: Drosophilidae), with combinations of vinegar and wine, and acetic acid and ethanol. J. Appl. Entomol. 136, 148–154. doi: 10.1111/j.1439-0418.2011.01646.x
Lauzon, C. R., Sjogren, R. E., and Prokopy, R. J. (2000). Enzymatic capabilities of bacteria associated with apple maggot flies: a postulated role in attraction. J. Chem. Ecol. 26, 953–967. doi: 10.1023/A:1005460225664
Lauzon, C. R., Sjogren, R. E., Wright, S. E., and Prokopy, R. J. (1998). Attraction of Rhagoletis pomonella (Diptera: Tephritidae) flies to odor of bacteria: apparent confinement to specialized members of Enterobacteriaceae. Environ. Entomol. 27, 853–857. doi: 10.1093/ee/27.4.853
Lee, H. Y., Lee, S. H., Lee, J. H., Lee, W. J., and Min, K. J. (2019). The role of commensal microbes in the lifespan of Drosophila melanogaster. Aging (Albany NY) 11, 4611–4640. doi: 10.18632/aging.102073
Leitão-Gonçalves, R., Carvalho-Santos, Z., Francisco, A. P., Fioreze, G. T., Anjos, M., Baltazar, C., et al. (2017). Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol. 15:e2000862. doi: 10.1371/journal.pbio.2000862
Lin, Q., Zhai, Y., Chen, H., Qin, D., Zheng, L., and Gao, H. (2021). Analyses of the gut bacteriomes of four important Drosophila pests. Can. Entomol. 153, 757–773. doi: 10.4039/tce.2021.45
Lleixà, J., Kioroglou, D., Mas, A., and Portillo, M. D. C. (2018). Microbiome dynamics during spontaneous fermentations of sound grapes in comparison with sour rot and Botrytis infected grapes. Int. J. Food Microbiol. 281, 36–46. doi: 10.1016/j.ijfoodmicro.2018.05.016
Lonvaud-Funel, A. (1999). “Lactic acid bacteria in the quality improvement and depreciation of wine” in Lactic acid Bacteria: Genetics, metabolism and applications. Proceedings of the sixth symposium on lactic acid Bacteria: Genetics, metabolism and applications, 19–23 September 1999 (Veldhoven: Springer Netherlands), 317–331.
Lorenz, D. H., Eichhorn, K. W., Bleiholder, H., Klose, R., Meier, U., and Weber, E. (1995). Phenological growth stages of the grapevine (Vitis vinifera L. ssp. vinifera)- codes and descriptions according to the extended BBCH scale. Aust. J. Grape Wine Res. 1, 100–103. doi: 10.1111/j.1755-0238.1995.tb00085.x
MacCollom, G. B., Lauzon, C. R., Weires, Jr, R. W., and Rutkowski, A. A. (1992). Attraction of adult apple maggot (Diptera: Tephritidae) to microbial isolates. J. Econ. Entomol. 85, 83–87.
Marchetti, R., Guerzoni, M. E., and Gentile, M. (1984). Research on the etiology of a new disease of grapes: sour rot. Vitis 23, 55–65.
Marra, A., Hanson, M. A., Kondo, S., Erkosar, B., and Lemaitre, B. (2021). Drosophila antimicrobial peptides and lysozymes regulate gut microbiota composition and abundance. MBio 12:e0082421. doi: 10.1128/mBio.00824-21
Martins, G., Vallance, J., Mercier, A., Albertin, W., Stamatopoulos, P., Rey, P., et al. (2014). Influence of the farming system on the epiphytic yeasts and yeast-like fungi colonizing grape berries during the ripening process. Int. J. Food Microbiol. 177, 21–28. doi: 10.1016/j.ijfoodmicro.2014.02.002
Mateo, E., Torija, M. J., Mas, A., and Bartowsky, E. J. (2014). Acetic acid bacteria isolated from grapes of south Australian vineyards. Int. J. Food Microbiol. 178, 98–106. doi: 10.1016/j.ijfoodmicro.2014.03.010
Mazzetto, F., Gonella, E., Crotti, E., Vacchini, V., Syrpas, M., Pontini, M., et al. (2016). Olfactory attraction of Drosophila suzukii by symbiotic acetic acid bacteria. J. Pest. Sci. 89, 783–792. doi: 10.1007/s10340-016-0754-7
Mertz, R. W., DeLorenzo, S., Sun, H., Loeb, G., and Scott, J. G. (2023). Selection for, and characterization of, malathion and zeta-cypermethrin resistance in vineyard-collected drosophila melanogaster. Pest Manag. Sci. 79, 1623–1627. doi: 10.1002/ps.7335
Mertz, R. W., Hesler, S., Pfannenstiel, L. J., Norris, R. H., Loeb, G., and Scott, J. G. (2022). Insecticide resistance in Drosophila melanogaster in vineyards and evaluation of alternative insecticides. Pest Manag. Sci. 78, 1272–1278. doi: 10.1002/ps.6745
Montllor, C. B., Maxmen, A., and Purcell, A. H. (2002). Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27, 189–195. doi: 10.1046/j.1365-2311.2002.00393.x
Moran, N. A., Russell, J. A., Koga, R., and Fukatsu, T. (2005). Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71, 3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005
Navarro, D., Mateo, E., Torija, M., and Mas, A. (2013). Acetic acid bacteria in grape must. Acetic Acid Bacteria 2:e4. doi: 10.4081/aab.2013.s1.e4
Nisiotou, A. A., and Nychas, G. J. E. (2007). Yeast populations residing on healthy or Botrytis-infected grapes from a vineyard in Attica, Greece. Appl. Environ. Microbiol. 73, 2765–2768. doi: 10.1128/AEM.01864-06
Nisiotou, A. A., Rantsiou, K., Iliopoulos, V., Cocolin, L., and Nychas, G. J. E. (2011). Bacterial species associated with sound and Botrytis-infected grapes from a Greek vineyard. Int. J. Food Microbiol. 145, 432–436. doi: 10.1016/j.ijfoodmicro.2011.01.017
Nocker, A., Burr, M., and Camper, A. K. (2007). Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 54, 276–289. doi: 10.1007/s00248-006-9199-5
Palma, M., Guerreiro, J. F., and Sá-Correia, I. (2018). Adaptive response and tolerance to acetic acid in Saccharomyces cerevisiae and Zygosaccharomyces bailii: a physiological genomics perspective. Front. Microbiol. 9:274. doi: 10.3389/fmicb.2018.00274
Peel, M. C., Finlayson, B. L., and McMahon, T. A. (2007). Updated world map of the Köppen-Geiger climate classification. Hydrol. Earth Syst. Sci. 11, 1633–1644. doi: 10.5194/hess-11-1633-2007
Pretorius, I. S. (2000). Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16, 675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B
Rani, M. U., Udayasankar, K., and Appaiah, K. A. A. (2011). Properties of bacterial cellulose produced in grape medium by native isolate Gluconacetobacter sp. J. Appl. Polym. Sci. 120, 2835–2841. doi: 10.1002/app.33307
Ren, C., Webster, P., Finkel, S. E., and Tower, J. (2007). Increased internal and external bacterial load during Drosophila aging without lifespan trade-off. Cell Metab. 6, 144–152. doi: 10.1016/j.cmet.2007.06.006
Reuter, M., Bell, G., and Greig, D. (2007). Increased outbreeding in yeast in response to dispersal by an insect vector. Curr. Biol. 17, R81–R83. doi: 10.1016/j.cub.2006.11.059
Ribéreau-Gayon, P., Dubourdieu, D., Donèche, B., and Lonvaud, A. (2006). “The Microbiology of wine and vinifications” in Handbook of Enology, (England: John Wiley and Sons,West Sussex), 1, 1–51.
Robacker, D. C., and Lauzon, C. R. (2002). Purine metabolizing capability of Enterobacter agglomerans affects volatiles production and attractiveness to Mexican fruit fly. J. Chem. Ecol. 28, 1549–1563. doi: 10.1023/A:1019920328062
Robacker, D. C., Martinez, A. J., Garcia, J. A., and Bartelt, R. J. (1998). Volatiles attractive to the Mexican fruit fly (Diptera: Tephritidae) from eleven bacteria taxa. Fla. Entomol. 81, 497–508. doi: 10.2307/3495948
Rombaut, A., Guilhot, R., Xuéreb, A., Benoit, L., Chapuis, M. P., Gibert, P., et al. (2017). Invasive Drosophila suzukii facilitates Drosophila melanogaster infestation and sour rot outbreaks in the vineyards. R. Soc. Open Sci. 4:170117. doi: 10.1098/rsos.170117
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Rousseaux, S., Diguta, C. F., Radoï-Matei, F., Alexandre, H., and Guilloux-Bénatier, M. (2014). Non-Botrytis grape-rotting fungi responsible for earthy and moldy off-flavors and mycotoxins. Food Microbiol. 38, 104–121. doi: 10.1016/j.fm.2013.08.013
Ryngajłło, M., Jędrzejczak-Krzepkowska, M., Kubiak, K., Ludwicka, K., and Bielecki, S. (2020). Towards control of cellulose biosynthesis by Komagataeibacter using systems-level and strain engineering strategies: current progress and perspectives. Appl. Microbiol. Biotechnol. 104, 6565–6585. doi: 10.1007/s00253-020-10671-3
Ryu, J. H., Kim, S. H., Lee, H. Y., Bai, J. Y., Nam, Y. D., Bae, J. W., et al. (2008). Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. doi: 10.1126/science.1149357
Scheidler, N. H., Liu, C., Hamby, K. A., Zalom, F. G., and Syed, Z. (2015). Volatile codes: correlation of olfactory signals and reception in Drosophila-yeast chemical communication. Sci. Rep. 5:14059. doi: 10.1038/srep14059
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Serga, S. V., Maistrenko, O. M., Matiytsiv, N. P., Vaiserman, A. M., and Kozeretska, I. A. (2021). Effects of Wolbachia infection on fitness-related traits in Drosophila melanogaster. Symbiosis 83, 163–172. doi: 10.1007/s13199-020-00743-3
Shen, Z., Penton, C. R., Lv, N., Xue, C., Yuan, X., Ruan, Y., et al. (2018). Banana Fusarium wilt disease incidence is influenced by shifts of soil microbial communities under different monoculture spans. Microb. Ecol. 75, 739–750. doi: 10.1007/s00248-017-1052-5
Shin, S. C., Kim, S. H., You, H., Kim, B., Kim, A. C., Lee, K. A., et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. doi: 10.1126/science.1212782
Silva, V., Palacios-Muñoz, A., Okray, Z., Adair, K. L., Waddell, S., Douglas, A. E., et al. (2021). The impact of the gut microbiome on memory and sleep in Drosophila. J. Exp. Biol. 224:jeb233619. doi: 10.1242/jeb.233619
Srivastava, S., and Mathur, G. (2022). Komagataeibacter saccharivorans strain BC-G1: an alternative strain for production of bacterial cellulose. Biologia 77, 3657–3668. doi: 10.1007/s11756-022-01222-4
Stamps, J. A., Yang, L. H., Morales, V. M., and Boundy-Mills, K. L. (2012). Drosophila regulate yeast density and increase yeast community similarity in a natural substrate. PLoS One 7:e42238. doi: 10.1371/journal.pone.0042238
Staubach, F., Baines, J. F., Künzel, S., Bik, E. M., and Petrov, D. A. (2013). Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. PLoS One 8:e70749. doi: 10.1371/journal.pone.0070749
Steel, C. C., Blackman, J. W., and Schmidtke, L. M. (2013). Grapevine bunch rots: impacts on wine composition, quality, and potential procedures for the removal of wine faults. J. Agric. Food Chem. 61, 5189–5206. doi: 10.1021/jf400641r
Sun, H., Loeb, G., Walter-Peterson, H., Martinson, T., and Scott, J. G. (2019). Insecticide resistance in Drosophila melanogaster (Diptera: Drosophilidae) is associated with field control failure of sour rot disease in a New York vineyard. J. Econ. Entomol. 112, 1498–1501. doi: 10.1093/jee/toz039
Tait, G., Mermer, S., Stockton, D., Lee, J., Avosani, S., Abrieux, A., et al. (2021). Drosophila suzukii (Diptera: Drosophilidae): a decade of research towards a sustainable integrated pest management program. J. Econ. Entomol. 114, 1950–1974. doi: 10.1093/jee/toab158
Tristezza, M., Vetrano, C., Bleve, G., Spano, G., Capozzi, V., Logrieco, A., et al. (2013). Biodiversity and safety aspects of yeast strains characterized from vineyards and spontaneous fermentations in the Apulia region, Italy. Food Microbiol. 36, 335–342. doi: 10.1016/j.fm.2013.07.001
Varela, J., and Varela, C. (2019). Microbiological strategies to produce beer and wine with reduced ethanol concentration. Curr. Opin. Biotechnol. 56, 88–96. doi: 10.1016/j.copbio.2018.10.003
Vázquez-Baeza, Y., Pirrung, M., Gonzalez, A., and Knight, R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. doi: 10.1186/2047-217X-2-16
Vercesi, A., and Bisiach, M. (1987). Studies on the correlation between the presence of some Drosophila species and the appearance of grape-vine sour-rot in Lombardy (Italy) GC Lozzia and a Cantoni Istituto di Entomologia Agraria dell’Università di Milano, Italy. Integr. Pest Control. Vitic. 10104:173.
Verginer, M., Leitner, E., and Berg, G. (2010). Production of volatile metabolites by grape-associated microorganisms. J. Agric. Food Chem. 58, 8344–8350. doi: 10.1021/jf100393w
Vicente, J., Calderón, F., Santos, A., Marquina, D., and Benito, S. (2021). High potential of Pichia kluyveri and other Pichia species in wine technology. Int. J. Mol. Sci. 22:1196. doi: 10.3390/ijms22031196
Weigle, T. H., Muza, A. J., Brown, B., Hed, B. E., Helms, M., Landers, A. J., et al. (2020). New York and Pennsylvania pest management guidelines for grapes. Ithaca, NY: Cornell Univ.
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nature Reviews Microbiology 6, 741–751.
West, A. S. (1961). Chemical attractants for adult Drosophila species. J. Econ. Entomol. 54, 677–681. doi: 10.1093/jee/54.4.677
Wong, C. N. A., Ng, P., and Douglas, A. E. (2011). Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13, 1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x
Yamada, Y., Yukphan, P., Lan Vu, H. T., Muramatsu, Y., Ochaikul, D., Tanasupawat, S., et al. (2012). Description of Komagataeibacter gen. Nov., with proposals of new combinations (Acetobacteraceae). J. Gen. Appl. Microbiol. 58, 397–404. doi: 10.2323/jgam.58.397
Ye, Y. H., Seleznev, A., Flores, H. A., Woolfit, M., and McGraw, E. A. (2017). Gut microbiota in Drosophila melanogaster interacts with Wolbachia but does not contribute to Wolbachia-mediated antiviral protection. J. Invertebr. Pathol. 143, 18–25. doi: 10.1016/j.jip.2016.11.011
Yixin, H. Y., Seleznev, A., Flores, H. A., Woolfit, M., and McGraw, E. A. (2017). Gut microbiota in Drosophila melanogaster interacts with Wolbachia but does not contribute to Wolbachia-mediated antiviral protection. Journal of Invertebrate Pathology 143, 18–25.
Keywords: Vitis vinifera, minor grape rots, bunch microflora, high-throughput sequencing, acetic acid bacteria, Enterobacteriaceae, non-Saccharomyces yeasts
Citation: Brischetto C, Rossi V and Fedele G (2024) The microbiome analysis of ripen grape berries supports the complex etiology of sour rot. Front. Microbiol. 15:1450443. doi: 10.3389/fmicb.2024.1450443
Edited by:
Anna Maria Pirttilä, University of Oulu, FinlandReviewed by:
Rajib Majumder, Macquarie University, AustraliaRajesh Ramdas Waghunde, Navsari Agricultural University, India
Copyright © 2024 Brischetto, Rossi and Fedele. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giorgia Fedele, Z2lvcmdpYS5mZWRlbGVAdW5pY2F0dC5pdA==
 Chiara Brischetto
Chiara Brischetto Vittorio Rossi
Vittorio Rossi Giorgia Fedele
Giorgia Fedele